Abstract
The splicing factor Sam68 is upregulated in many human cancers, including prostate cancer (PCa) where it promotes cell proliferation and survival. Nevertheless, in spite of its frequent upregulation in cancer, the mechanism(s) underlying its expression are largely unknown. Herein, bioinformatics analyses identified the promoter region of the Sam68 gene (KHDRBS1) and the proto-oncogenic transcription factor c-MYC as a key regulator of Sam68 expression. Upregulation of Sam68 and c-MYC correlate in PCa patients. c-MYC directly binds to and activates the Sam68 promoter. Furthermore, c-MYC affects productive splicing of the nascent Sam68 transcript by modulating the transcriptional elongation rate within the gene. Importantly, c-MYC-dependent expression of Sam68 is under the tight control of external cues, such as androgens and/or mitogens. These findings uncover an unexpected coordination of transcription and splicing of Sam68 by c-MYC, which may represent a key step in PCa tumorigenesis.
INTRODUCTION
Prostate cancer (PCa) is among the most common tumors in adult men (1). PCa onset and progression rely on androgen receptor (AR) signaling and androgen deprivation therapy (ADT) represents the most effective cure (2). However, despite significant advances in treatments, PCa almost inevitably relapses with hormone-insensitive forms (CRPC) that are currently incurable (2). Among the factors contributing to PCa pathogenesis, aberrant splicing of cancer-specific genes contributes by promoting isoforms involved in growth, metastatic progression and hormone resistance of PCa cells (3,4). For example, a constitutively active splice variant of AR (AR-V7) is overexpressed in CRPC, where it promotes cell proliferation and migration by regulating a specific subset of genes even under ADT conditions (5). Likewise, the anti-apoptotic BCL-X long splice variant (BCL-XL) confers chemotherapy resistance (6), whereas the Cyclin D1b isoform promotes AR transcriptional activity (7). In support of their pro-oncogenic features, expression of these splice variants is linked to poor prognosis in PCa patients (7–9).
Precursor messenger RNA (pre-mRNA) splicing is operated by the spliceosome, a complex ribonucleoprotein machinery that removes introns and ligates exons to yield the mature mRNA (10). Although most exons are constitutively spliced in the mRNA, others are subjected to regulation through a process named alternative splicing (AS). Alternative recognition of exons is due to the presence of cis-regulatory RNA elements that recruit the spliceosome and sequence-specific trans-acting splicing factors (10). Moreover, since splicing is tightly coupled to transcription, mechanisms impacting on the RNA polymerase II (RNAPII) elongation rate, nucleosome positioning or histone modifications also contribute to exon recognition (10). These regulatory mechanisms are kept under tight control in the cell, whereas their dysregulation contributes to the onset and progression of human cancers (11,12), including PCa (3,4).
Splicing dysregulation is often determined by the altered expression of specific splicing factors (11,12). An example is provided by Sam68 (KHDRBS1), which is overexpressed in PCa and other human cancers (13). Sam68 promotes cell-cycle progression and survival to genotoxic stress of PCa cells (14), likely through induction of oncogenic splice variants like BCL-XL, cyclin D1b, CD44 variants and AR-V7 (3,4). Furthermore, Sam68 interacts with both AR and AR-V7, enhancing their transcriptional activity and androgen signaling (15). However, in spite of its well described upregulation in human cancers (13), no studies have directly addressed the mechanisms underlying its transcriptional regulation.
Herein, by carrying out a bioinformatics search for transcription factors that potentially regulate Sam68 expression we have identified the proto-oncogene c-MYC (MYC) as strong candidate. Our study documents the direct binding of c-MYC to the Sam68 promoter region as well as its effect on the transcriptional activation of the gene and productive splicing of the nascent transcript. Furthermore, we provide evidence that c-MYC-dependent expression of Sam68 is under the tight control of external cues and linked to favorable conditions, such as androgen and/or mitogen stimulation. These findings uncover an unexpected coordination of transcription and splicing of Sam68 by c-MYC, which may represent a vulnerable target for PCa treatment.
MATERIALS AND METHODS
Bioinformatics analyses
Sam68 promoter characterization was performed utilizing bioinformatics tools present in UCSC Genome Browser (https://genome.ucsc.edu; GRCh37/h19) and analyzing ChIP-seq data for RNAPII, H3K27Ac, H3K4Me1 and H3K4Me3 tracks from ENCODE project (https://www.encodeproject.org). Chromatin State Segmentation track displays a chromatin state segmentation from nine human cell types learned by computationally integrating ChIP-seq data for nine factors plus input using a Hidden Markov Model. In UCSC Genome Browser, in silico analysis of Sam68 promoter was performed using ‘HMR Conserved Transcription Factor Binding Site’ tool that use the Transfac Matrix Database (v.7.0). c-MYC ChIP-seq analyses was performed using ‘Transcription Factor ChIP-seq (161 factors) from ENCODE with Factorbook motifs’ tool.
Tumor Prostate Cancer datasets analysis was carried out utilizing Jenkins (GSE46691), Sawyers (GSE21034) and Sueltman (GSE29079) published datasets (16–18). Gene expression data for correlation analyses were downloaded from R2 genomics analysis and visualization platform (http://r2.amc.nl). Pearson’s correlation was used to evaluate the association between Sam68 and c-MYC expression. For gene expression analyses, the patients were divided into two groups according to the median of c-MYC gene expression. Then, Z-scores value of Sam68 was calculated in each sample and Mann–Whitney test was used to establish the significance level of Sam68 between the two groups (19).
Plasmid constructs and oligonucleotides
Intergenic region, human wild-type and deletion mutants of Sam68 and hnRNP A1 promoters were cloned in pGL3-basic vector (Promega). Human c-MYC and MAX expression plasmids were cloned in pCDNA3 vector (Invitrogen, Life technologies). Site-directed mutagenesis on pGL3-Sam68 promoter was performed using QuikChangeR II XL Site-Directed Mutagenesis Kit according to manufacturer's instruction (Stratagene). Sam68 minigene was cloned into the EcoRI/SalI restriction sites of pCI vector (Promega). All constructs were generated using genomic DNA or cDNA isolated from LNCaP cells as template. Inserts were amplified by the Phusion Hot Start High-Fidelity DNA polymerase (Thermo Fisher Scientific) and plasmids sequenced by Cycle Sequencing (Eurofins Genomics). Oligonucleotides used in this study are listed in the Supplementary Table S1.
Cell lines maintenance and transfection
Human prostate cancer cell lines LNCaP and 22Rv1 were cultured in RPMI 1640 medium (LONZA), while PC3 and DU145 PCa cell lines and human embryonic kidney cell line HEK293T were maintained in Dulbecco’s modified Eagle’s medium (LONZA). All media were supplemented with 10% Fetal Bovine Serum (FBS) (Gibco), penicillin (50 U/ml)/streptomycin (50 μg/ml) (Corning), 50 μg/ml gentamicin sulfate (Aurogene), 1% non-essential amino acids (Euroclone), 10 mM Hepes (Euroclone) and 1 mM sodium pyruvate (Aurogene). Cells were maintained in culture at 37°C under 5% CO2 in a humidified incubator, no longer than 3 months. Cell lines utilized were tested for mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (LONZA). LNCaP and HEK293T were transfected with the indicated reporter or expression vectors using Lipofectamine 2000 (Invitrogen) as previously described (20). For c-MYC (double transfection) and splicing factors (single transfection) RNAi experiments, PCa cells were transfected with 40 nM siRNAs using Lipofectamine RNAiMax (Invitrogen, Life technologies) according to manufacturer’s instruction. Control and c-MYC siRNAs were purchased from Dharmacon (On target plus human c-MYC L-003282-02 and On target plus non targeting pool D-001810-10) and Qiagen (Flexi Tube siRNA MYC SI03101847 and Negative control SI03650325). Sequences of control and splicing factors siRNAs were previously described (20).
Cell treatments and proliferation analysis
Growth arrest was induced by culturing PCa cells in 0% FBS and with 1 μM Enzalutamide (MDV3100; Sigma-Aldrich), or DMSO as vehicle control. For cell proliferation assays, cells were pulsed with 30 μM BrdU (Sigma-Aldrich) for 30 min. After PBS washes, cells were collected, fixed with 70% ethanol and stained with anti-BrdU antibody (347580, BD Biosciences) and propidium iodide (Sigma-Aldrich). Cell-cycle analysis was carried out using flow cytometry (FACSCalibur, BD Biosciences) according to manufacturer’s instructions.
Cell extracts and immunoblot analysis
Whole extract preparation was performed as previously described (21). Briefly, cells were resuspended in lysis buffer [50 mM Hepes, pH 7.4, 150 mM NaCl, 15 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 20 mM β-glycerophosphate, 0.5 mM NaVO4, protease inhibitor cocktail (Sigma-Aldrich), 0.5% Triton X-100]. After 10 min of ice incubation, cell suspension was centrifuged for 10 min at 12 000 ×g at 4°C and supernatant fractions were collected. Samples were denatured in Laemmli Sample buffer and 3–30 μg of total extract was separated on sodium dodecyl sulphate-polyacrylamide gelelectrophoresis (SDS-PAGE) and transferred to Polyvinylidene Fluoride (PVDF) membranes. Western blot analysis was carried using the following antibodies: anti-SAM68 (A302-110A, Bethyl Laboratories); anti-β-actin (A2066, Sigma-Aldrich); anti-β-tubulin (T4026, Sigma-Aldrich); anti-c-MYC (sc764, Santa Cruz Biotechnology); anti-hnRNP A1 (sc32301, Santa Cruz Biotechnology); anti-MAX (sc2011, Santa Cruz Biotechnology); anti-hnRNP I (sc16547 Santa Cruz Biotechnology); anti-hnRNP A2/B1 (sc393674, Santa Cruz Biotechnology); anti-GFP (sc9996, Santa Cruz Biotechnology); Anti-RNA polymerase II (sc899, Santa Cruz Biotechnology); anti-RNA polymerase II subunit B1, clone 3E10 (phospho CTD Ser-2, 04-1571, Millipore); anti-RNApolymerase II subunit B1, clone 3E8 (phospho-CTD Ser5; 04-1572, Millipore); anti-Flag (F3165, Millipore); anti-hnRNP H and anti-hnRNP F (kindly provided by Prof. B. Chabot, Université de Sherbrooke, Canada).
Luciferase-based report assay
Luciferase-based report assay was performed in HEK293T cells as previously described (22). Briefly, HEK293T cells were co-transfected with internal control (Renilla) and indicated (Firefly) luciferase reporters in presence or not of c-MYC, or MAX, expressing vectors. Luciferase assays were carried out at the indicated time points using the Dual Luciferase Reporter assay (Promega). Data were normalized for transfection efficiency by ratio between Firefly and Renilla luciferase activity, and represented as fold activation with respect to the control.
RNA extraction, gene expression and splicing assay analysis
RNA was extracted from cells using the Trizol reagent according to manufacturer’s instructions (Invitrogen, Life technologies) and treated with RNase-free DNase (Ambion). A total of 1 μg of RNA was retrotranscribed with M–MLV reverse transcriptase (Promega) and random primers (Roche). Gene expression and splicing patterns were evaluated by semi-quantitative (sqPCR) and quantitative (qPCR) PCR analyses using 10 ng of cDNA template. For splicing assay experiments, sqPCR analysis was performed using the following cycles: 95°C for 5 min (1 cycle) −95°C for 10 s, 56°C for 10 s, 72°C for 15 s (28 or 35 cycles for minigene-derived or endogenous isoforms, respectively)− and a final extension at 72°C for 5 min. The percentage Spliced-In Index (PSI/ψ) was calculated from densitometric analysis of PCR products as ψ = exon inclusion/ (exon inclusion + exon skipping) band intensities. qPCR analysis was carried out using PowerUp SYBR Green Master Mix (Applied Biosystems) and Applied Biosystems StepOnePlus Real-Time PCR system (Applied Biosystems) according to the manufacturer's instructions. Quantitative evaluation of gene and Sam68-ΔKH isoform expression was calculated relative to Histone 3 and Sam68-KH, respectively, by ΔΔCq method (21).
Analysis of RNA polymerase II (RNAPII) transcriptional rate on Sam68 pre-mRNA was performed as previously described (23). Briefly, LNCaP cells were grown overnight on 60-mm plates to 70–80% confluency and treated with 75 μM of 5,6-Dichlorobenzimidazole 1-β–D-ribofuranoside (DRB; Sigma-Aldrich) in culture medium for 6 h to reversibly halt transcription. After two washes in phosphate buffered saline (PBS), cells were incubated in fresh medium up to 30 min. Every 5 min cells were collected for RNA isolation. Fold change of Sam68 exon1–intron1 relative to exon8–intron8 expression was calculated by ΔΔCq method (21).
Chromatin immunoprecipitation
ChIP experiments were performed as previously described (22). Briefly, LNCaP cells were fixed by the addition of 1% (vol/vol) formaldehyde to the culture medium for 10 min at room temperature and then quenched in 125 mM glycine for 5 min. Cells were washed three times in PBS and lysed in nuclei extraction buffer (5 mM Pipes pH 8, 85 Mm KCl, 0.5% NP40) for 2 h, at 4°C under rotation. Lysate was centrifuged at 1200 × g for 5 min at 4°C. Nuclei pellet was resuspended in sonication buffer (10 mM ethylenediaminetetraacetic acid (EDTA) pH 8, 50 mM Tris–HCl pH 8, SDS 1%) and sonicated with Bioruptor (Dyagenode) to yield chromatin size of ∼400 bp and insoluble debris was removed by centrifugation. Cross-linked DNA was then quantified, diluted 1:10 with dilution buffer (0.01% SDS, 1.1% Triton X100, 1.2 mM EDTA, 16.7 mM Tris/HCl pH 8.0, 167 mM NaCl) and incubated with 5 μg of specific c-MYC antibody (sc-764X, Santa Cruz Biotechnologies), IgGs (Sigma-Aldrich) and no antibody, as a negative controls, under rotation at 4°C overnight. Dynabeads protein G (Invitrogen, Life technologies) were incubated with the mixture under rotation at 4°C for 2 h, then washed and heated at 65°C overnight to reverse formaldehyde cross-links. Immunoprecipitated DNA was recovered according to standard procedures and analyzed by qPCRs. DNA associated with c-MYC is represented as percentage of input, calculated by ΔCq method (21).
UV-crosslinked and RNA immunoprecipitation (CLIP) assays
UV-crosslinked and RNA immunoprecipitation (CLIP) assays were performed as previously described (24). Briefly, cells were washed once with PBS, UV-irradiated (400 mJ/cm2) and collected by scraping in lysis buffer [50 mM Tris pH 8, 100 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 1% NP40, 0.1% SDS, 0.5 mM Na3VO4, 1 mM dithiothreitol, protease inhibitor cocktail (Sigma-Aldrich), RNase inhibitor (Promega)]. After brief sonication, samples were incubated with DNase-RNase free (Ambion) for 3 min at 37°C and then centrifuged at 15000 × g for 3 min at 4°C. A total of 1 mg of supernatant (cell extract) was diluted to 1 ml with lysis buffer and immunoprecipitated with anti-hnRNP F (kindly provided by Prof. B. Chabot, Université de Sherbrooke, Canada) or IgGs (control) in the presence of Dynabeads protein G (Invitrogen, Life technologies) and 10 μl/ml of RNaseI 1:1000 (Ambion). Immunoprecipitates (IPs) were incubated for 2 h at 4°C under rotation. After two washes with high-salt buffer (50 mM Tris–HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% Igepal CA-630, 0.1% SDS, 0.5% sodium deoxycholate) and Proteinase K buffer (100 mM Tris–HCl, pH 7.4, 50 mM NaCl, 10 mM EDTA), the IPs were resuspended in Proteinase K buffer supplemented with 50 μg of Proteinase K and incubated 1 h at 55°C. RNA was isolated and retrotranscribed by standard procedures. About 10% of cell extract (0.1 mg) was treated with 50 μg of Proteinase K and RNA purified (input).
RESULTS
c-MYC binds the promoter region of Sam68
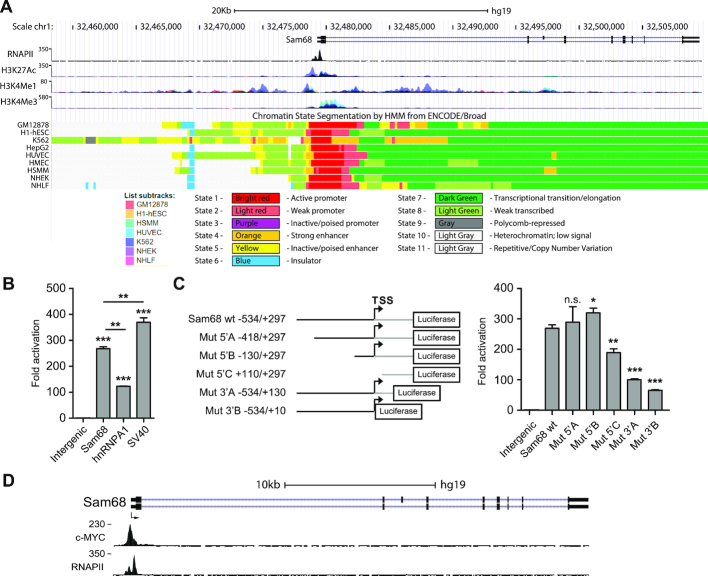
To identify the promoter region of the Sam68 gene (KHDRBS1), we queried the UCSC Genome Browser database (http://genome.ucsc.edu; GRCh37/h19) for RNA polymerase II (RNAPII) occupancy and chromatin features of active transcription within the gene and in the upstream intergenic region (∼20 kilobases). Chromatin immunoprecipitation (ChIP) sequencing datasets from seven human cell lines revealed that RNAPII peaks are present in the region spanning approximately −400 and +300 base pairs (bp) from the transcription start site (TSS) (Figure 1A and Supplementary Figure S1A). RNAPII peaks are flanked by deposition of histone marks typically enriched in active promoters (H3K4me3) and transcriptional regulatory elements (H4K3me1 and H3K27Ac) (Figure 1A). To evaluate the promoter activity of this region, we cloned the genomic sequence located between −534 and +297 bp from the TSS upstream of the luciferase reporter gene and transfected it into HEK293T cells. The putative promoter region of Sam68 displayed higher activity to that of the human HNRPA1 promoter, while being significantly weaker than the viral SV40 promoter (Figure 1B). No transcriptional activity was observed with an intergenic DNA region of the same length. Progressive deletion mutants indicated that the region between −130 bp and +297 bp from the TSS is required for the optimal activity of the Sam68 promoter (Figure 1C). These results suggest that a relatively small genomic region acts as promoter for Sam68 expression in human cells.
Figure 1.
Identification of the Sam68 promoter region. (A) UCSC Genome Browser snapshot of RNAPII, H3K27Ac, H3K4Me1 and H3K4Me3 ChIP-seq profiles and Chromatin State Segmentation of the Sam68 locus, including an ∼20 Kbp upstream intergenic region. Chromatin state segmentation (colored rectangles; state 1–11) and cell lines (colored squares; List subtracks) are indicated. (B and C) Bar graphs represent luciferase activity of Sam68, hnRNP A1 and SV40 promoters compared to an upstream intergenic region (intergenic; −17753 to −16920 bp from the TSS) used as negative control (B), and of Sam68 promoter deletion mutants (Mut5′A, Mut5′B, Mut5′C, Mut3′A, Mut3′B) compared to the wild-type (wt) and interegenic reporters (C). A schematic representation of wt and mutant reporters is also shown; the upstream (black line) and downstream (gray line) regions from the Sam68 TSS are indicated (C; left panel). All Luciferase assays were performed in HEK293T 48 h post-transfection. Data represent mean ± SD of three biological replicates. Statistical significance was calculated by Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant. (D) Representative ChIP-seq analysis of c-MYC and RNAPII binding to the Sam68 promoter region in NB4 cells with a schematic representation of the Sam68 gene structure showing predicted TSS (arrow), introns (horizontal lines) and exons (boxes).
To identified transcription factors that potentially bind cis-regulatory DNA elements in the Sam68 promoter, we queried the HMR Conserved Transcription Factor Binding Sites, the ENCODE Transcription Factor Binding Tracks datasets and ChIP-seq experiments deposited in the UCSC Genome Browser database. Among others, we found two potential binding sites for the oncogenic transcription factor c-MYC that are located at −64 and −45 bp from the Sam68 TSS (Supplementary Figure S1B), within the region required for optimal transcriptional activity (Figure 1C). Analysis of ChIP-seq experiments in multiple cell lines (Encode Project, https://www.encodeproject.org) confirmed the binding of c-MYC to the Sam68 promoter region (Figure 1D and Supplementary Figure S1C), suggesting a direct regulation of Sam68 expression by this transcription factor in cancer cells.
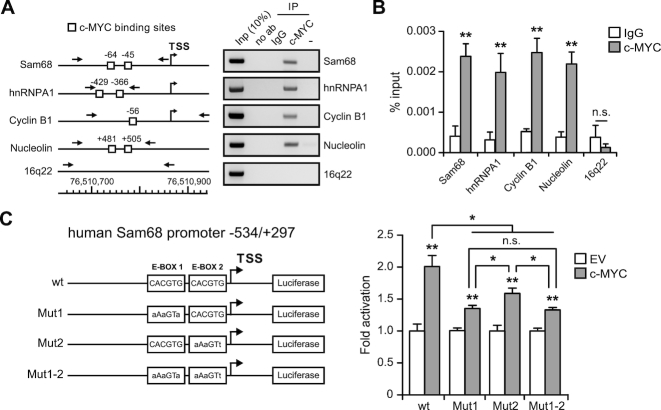
Sequence analysis of the Sam68 promoter highlighted two perfect c-MYC consensus E-box binding sites (CACGTG; Supplementary Figure S2A). To directly test whether c-MYC regulates Sam68 transcription, we performed ChIP experiments in LNCaP cells, an androgen-sensitive PCa cell line expressing high levels of Sam68 (14). Semiquantitative (sqPCR) and quantitative (qPCR) PCR analyses detected a significant enrichment of c-MYC in the promoter region of Sam68 (Figure 2A and B), which was comparable to that observed for other c-MYC targets, such as cyclin B1, nucleolin (25) and hnRNP A1 (26). By contrast, no binding was observed in the intergenic (16q22) control region of chromosome 16, indicating specificity of binding (Figure 2A and B).
Figure 2.
c-Myc binds to the Sam68 promoter region. (A) sqPCR and (B) qPCR analyses of ChIP experiments performed in LNCaP cells using c-MYC antibody and IgG (IgG), or no antibody (no ab), as negative control. Associated DNA is expressed as % of input (B). Schematic representation of the indicated gene promoters and of the 16q22 intergenic region is shown in (A) (left panel). c-MYC binding sites (solid box), TSS and primers position for PCR analyses (arrows) are indicated. (C) The bar graph represents the luciferase assay performed in HEK293T cells transfected with wt or mutated (Mut1, Mut2 and Mut1-2) Sam68 promoter reporters in combination, or not (empty vector, EV), with c-MYC-pCDNA3 vector (c-MYC). A schematic representation of wt and c-MYC binding site mutants (Mut1, Mut2 and Mut1-2) of the Sam68 promoter is also shown (left panel). (B and C) Data represent mean ± SD of three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant (Student's t-test).
Next, we tested whether c-MYC modulates Sam68 expression. Co-transfection of the Sam68 promoter reporter and c-MYC resulted in significant induction (∼2-fold) of the luciferase activity (Figure 2C), which was comparable to that observed with the hnRNPA1 promoter (Figure 2C and Supplementary Figure S2B). Individual (Mut1: CACGTG→AAAGTA; Mut2: CACGTG→AAAGTT) or double mutation (Mut1-2: CACGTG/CACGTG→AAAGTA/AAAGTT) of the putative E-box binding sites significantly impaired c-MYC-dependent activation of the Sam68 promoter (Figure 2C and Supplementary Figure S2C), with Mut1 causing the greatest influence. Interestingly, the double mutant (Mut1-2) did not show additive effect (Figure 2C), suggesting that c-MYC activity primarily relies on E-Box1 and that additional regions participate to regulation.
The c-MYC transcriptional activity often relies on its functional interaction with the co-regulator MAX (27). Thus, we also tested their possible cooperation on the regulation of Sam68 expression. Under sub-optimal conditions (24 h after transfection), MAX did not affect Sam68 expression when overexpressed alone, but it significantly enhanced the transcriptional activity of c-MYC on the Sam68 promoter in co-transfected cells (Supplementary Figure S2D). Collectively, these findings identify c-MYC as a direct regulator of Sam68 transcription.
c-MYC positively regulates Sam68 expression in prostate cancer
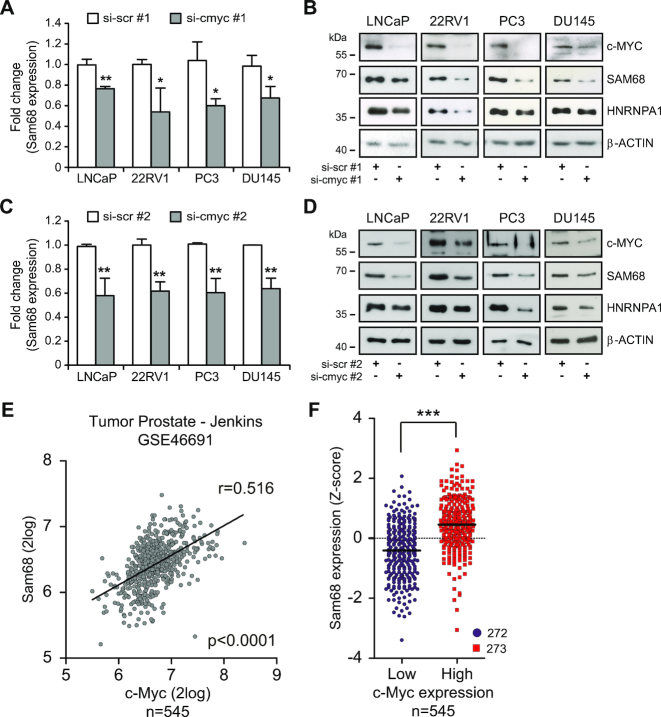
To test whether Sam68 expression is under the control of c-MYC in PCa cells, we knocked down its expression in four PCa cell lines (LNCaP, 22RV1, PC3 and DU145) using two pools of siRNAs (siRNA#1 and #2). c-MYC depletion caused a significant reduction in Sam68 expression at both mRNA and protein levels (Figure 3A–D), which was comparable to that exerted on hnRNP A1 (26) (Supplementary Figure S3A–D). These results indicate that c-MYC promotes Sam68 expression in PCa cells.
Figure 3.
c-MYC regulates Sam68 expression in PCa. (A–D) qPCR (A and C) and western blot (B and D) analyses of Sam68 expression in PCa cells lines transfected with two different pools of c-Myc (si-cmyc #1 and si-cmyc #2) or control (si-scr #1 and si-scr #2) siRNAs. (A and C) Fold change of Sam68 expression relative to Histone 3 expression was calculated by the ΔΔCq method. Data represent mean ± SD of three biological replicates. *P < 0.05; **P < 0.01 (Student's t-test). (E) Plot showing Sam68 and c-MYC expression in 545 PCa patients retrieved from the Jenkins (GSE46691) dataset. Pearson’s correlation coefficient (r) and P-value are reported. (F) Sam68 expression profile in PCa patients (Jenkins-GSE46691) classified according to Z-score normalization in c-MYClow (blue circles) and c-MYChigh (red squares) expressing group. The dot plot shows distribution and the median (horizontal line). Statistical significance was calculated by Mann–Whitney test, *** P < 0.001.
The pro-oncogenic role of c-MYC mainly relies on regulation of gene expression programs (28), including genes encoding core spliceosome components (29) and pro-oncogenic splicing factors (19,26). Since upregulation of both c-MYC (16,30) and Sam68 (14,15) have been reported in PCa, we asked whether their expression levels correlated in patients by analyzing public datasets (R2 genomics; http://r2.amc.nl). Pearson's correlation analysis using the Jenkins PCa dataset (GSE46691) deposited in R2 genomics (16) revealed a positive correlation (P < 0.0001) between c-MYC and Sam68 expression (Figure 3E). Similar positive correlation between c-MYC and Sam68 expression was found by analyzing other datasets of PCa patients (GSE21034 and GSE29079) (17,18) (Supplementary Figure S3E and F). Furthermore, Z-score classification of patients for low and high expression of c-MYC confirmed that Sam68 levels are significantly higher in the MYChigh compared to the MYClow group (Figure 3F; Supplementary Figure S3E and F). These findings suggest a direct link between upregulation of c-MYC and Sam68 transcription in PCa.
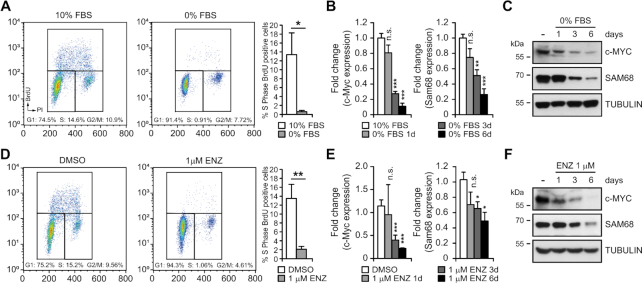
c-MYC downregulation impacts on Sam68 expression during cell growth arrest
Both c-MYC and Sam68 promote PCa cell proliferation (14,31), whereas cell growth arrest is linked to c-MYC downregulation (32). Thus, we asked whether Sam68 expression was perturbed under conditions that induce growth arrest. Serum deprivation of LNCaP cells for 6 days significantly impaired proliferation, as indicated by a reduction of BrdU positive cells (Figure 4A). Under these conditions, we also observed concomitant downregulation of both c-MYC and Sam68 mRNA and protein levels (Figure 4B and C). Serum deprivation and c-MYC silencing did not exert additive effects on Sam68 expression (Supplementary Figure A and B), suggesting that they function in the same pathway. Notably, a similar concomitant reduction of c-MYC and Sam68 expression was also observed in PC3 cells induced to growth arrest by serum deprivation (Supplementary Figure S4C–E).
Figure 4.
PCa cell growth arrest concomitantly induces c-MYC and Sam68 downregulation. (A–F) Representative dot plot profiles of cytometric analyses showing DNA content versus BrdU incorporation in LNCaP cells after 6 days of serum deprivation (A) or 1 μM Enzalutamide (D) conditions. Bar graphs (A and D) represents the percentage of S-phase BrdU positive cells after 6 days of treatment. qPCR (B and E) and western blot (C and F) time-course analyses of c-MYC and Sam68 expression (1d: 1 day; 3d: 3 days; 6d: 6 days). Fold change of Sam68 and c-MYC expression relative to Histone 3 expression was calculated by the ΔΔCq method. Data represent mean ± SD of three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant (Student's t-test).
Most PCas maintain dependency on androgens for cell proliferation and tumor growth. For this reason, inhibition of AR signaling represents the first-line therapy for this cancer. Thus, to impair PCa cell growth by an alternative approach, we treated the androgen-dependent LNCaP cells with Enzalutamide, a clinically approved AR inhibitor (2). Exposure of LNCaP cells to 1μM Enzalutamide for 6 days induced a significant reduction of proliferation (Figure 4D). Strikingly, also under this condition we observed a concomitant reduction in Sam68 and c-MYC levels (Figure 4E and F), indicating that the effect was linked to ADT-mediated growth arrest. As expected, proliferation of the androgen-insensitive PC3 cells was not affected by Enzalutamide treatment (Supplementary Figure S4F) and neither c-MYC nor Sam68 expression was significantly modulated (Supplementary Figure S4G and H). These results suggest that under adverse growth conditions, PCa cells concomitantly downregulate c-MYC and Sam68 expression to halt proliferation.
c-MYC affects alternative splicing of Sam68 through modulation of the RNAPII elongation rate
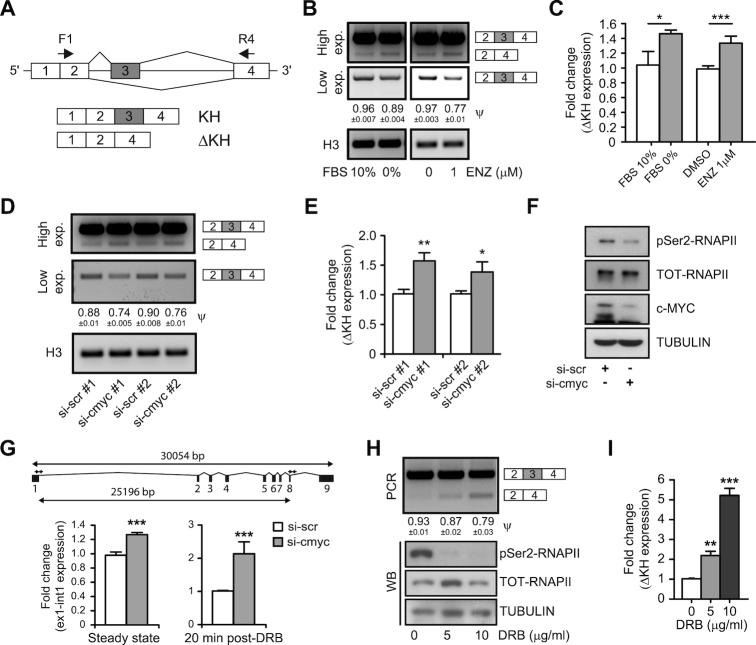
Regulation of exon 3 splicing in the Sam68 transcript has been previously related to growth conditions (33). Inclusion of exon 3 generates the full-length variant with an intact KH domain for RNA binding, whereas skipping of exon 3 generates an isoform that presents a 39-amino acid deletion in this domain (Sam68-ΔKH) (Figure 5A) and is unable to bind RNA (34). Although difficult to detect in cell lines under culture conditions, the Sam68-ΔKH splice variant was shown to be induced upon growth arrest in non-transformed fibroblasts (33). We observed an ∼30–50% increase in the levels of the Sam68-ΔKH variant in LNCaP cells exposed to either serum deprivation or Enzalutamide treatment, as determined by both sqPCR (Figure 5B) and qPCR (Figure 5C). These results indicate that growth arrest impacts on Sam68 AS in PCa cells. Since under these conditions expression of c-MYC is also halted (Figure 4), we asked whether c-MYC is involved in the Sam68 splicing switch. Strikingly, c-MYC depletion recapitulated the increase in exon 3 skipping observed under growth arrest in LNCaP cells (Figure 5D and E), indicating that c-MYC promotes both expression and productive splicing of the Sam68 full length variant.
Figure 5.
c-MYC modulates the AS of Sam68. (A) Diagram showing AS of Sam68. Exons (boxes), introns (horizontal lines) and position (arrows) of primers used for sqPCR are shown. (B–E) sqPCR (B and D) and qPCR (C and E) analyses of Sam68-ΔKH and KH splicing in LNCaP cells cultured under serum deprivation condition (0% FBS) or in presence of 1 μM Enzalutamide (Enz) (B and C), or transfected with control (si-scr #1 and #2) or c-MYC (si-myc #1 and #2) siRNAs (D and E). (F) Representative western blot analysis showing the expression level of c-MYC, total (TOT-RNAPII) and Serine2 (pSer2-RNAPII) RNAPII in LNCaP cells transfected with control (si-scr #1) or c-MYC (si-myc #1) siRNAs. Tubulin was used as loading control. (G) Bar graphs represent qPCR evaluation of Sam68 exon1-intron1 and exon8–intron8 ratio performed at steady state level (left) and 20min post-DRB release (right). LNCaP cells transfected with either control (si-scr #1) or c-MYC (si-cmyc #1) siRNAs were treated with 75 μM DRB for 6 h and RNA was extracted 20 min after DRB release. Data are reported as fold change of Sam68 exon1–intron1 relative to exon8–intron8 expression calculated by the ΔΔCq method. A scheme of Sam68 gene and primers position (arrows) used for qPCR is shown. (H and I) sqPCR (H) and qPCR (I) analyses showing Sam68-ΔKH and -KH ratio in LNCaP cells treated for 12 h with suboptimal DRB concentration. A representative western blot (WB) analysis (H) of the pSer2-RNAPII and TOT-RNAPII expression levels is also shown. Tubulin was used as loading control. (B, D and H) The Percent of Spliced-In Index (PSI; ψ) is reported below the gels. (C, E and I) Fold change of Sam68-ΔKH expression relative to Sam68-KH expression was calculated by the ΔΔCq method. (B–E, G–I) Mean ± SD of three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test).
Transcription and splicing are functionally coupled, and transcription-related processes influence splicing outcome (10). For instance, changes in the RNAPII elongation rate modulate splicing of hundreds of genes (35). Fast RNAPII elongation rate correlates with its phosphorylation in Serine 2 (p-Ser2) and we observed that c-MYC depletion reduced the levels of this post-translational modification in LNCaP cells (Figure 5F). Thus, to investigate whether the transcriptional activity of c-MYC influences Sam68 AS, we first tested the effect of c-MYC on the RNAPII elongation rate within the Sam68 transcription unit. To this end, we quantified RNAPII processivity as the ratio between promoter-distal and promoter–proximal regions of the Sam68 pre-mRNA at steady state (36). qRT-PCR analysis using primers located at the exon 1/intron 1 (430 bp from TSS) and exon 8/intron 8 boundaries (25707 bp from TSS) revealed a reduction in RNAPII processivity in c-MYC-depleted LNCaP cells (Figure 5G, steady state and Supplementary Figure S5A and B). This effect was even more evident in nascent pre-mRNA analyzed 20 min after release from the transcriptional block caused by incubation with the RNAPII inhibitor 5,6-dichloro-l-b-D-ribofuranosylbenzimidazole (DRB) (23). At this time point, when exon8/intron8 expression in the pre-mRNA reaches a plateau (Supplementary Figure S5C), depletion of c-MYC caused a two-fold reduction in RNAPII processivity within the Sam68 gene (Figure 5G, 20 min post-DRB).
These observations suggest that reduction of the RNAPII elongation rate may underlie the effect of c-MYC on Sam68 splicing. To test this possibility by an alternative approach, LNCaP cells were treated with suboptimal doses of DRB to reduce RNAPII p-Ser2 phosphorylation (Figure 5H) and its elongation rate without blocking transcription (35). Interestingly, we found that splicing of the Sam68-ΔKH isoform could be recapitulated in a dose-dependent manner in LNCaP cells treated with sub-optimal concentrations of DRB (Figure 5H and I; Supplementary Figure S5D). Similar results were obtained with two other RNAPII phosphorylation inhibitors, flavopiridol and LDC067 (Supplementary Figure S5E–H). Collectively these results suggest that the effect of c-MYC on Sam68 exon 3 splicing is mediated by a change in the RNAPII elongation rate.
A slow RNAPII elongation rate promotes recruitment of hnRNP F and skipping of Sam68 exon 3
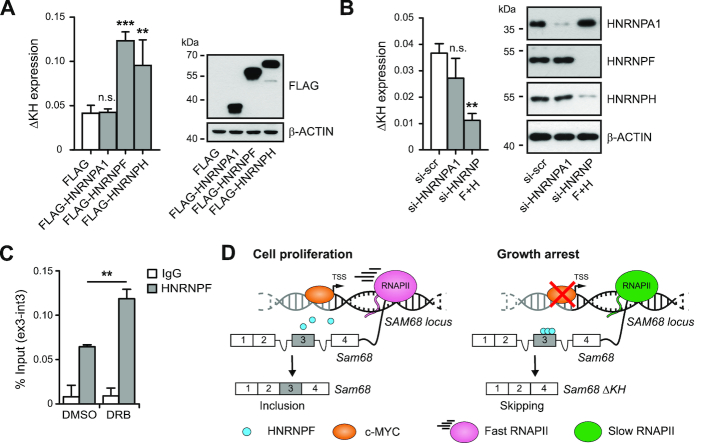
Changes in the RNAPII elongation rate may induce inclusion or skipping of alternative exons through kinetic or recruitment mechanisms (37), non mutually exclusive models that can also cooperate in splicing regulation (38). In the first case, a slow elongation offers a window of opportunity to the weaker splice sites before stronger splice sites of constitutive exons are transcribed. In the recruitment model, the slower RNAPII is able to recruit splicing regulators to modulate recognition of the alternative exon splice sites. Since phosphorylation of RNAPII mediates its interaction with splicing factors (SFs) during pre-mRNA transcription and processing (39), we hypothesized that reduction of p-Ser2 phosphorylation might affect the recruitment of specific SF(s) to the Sam68 pre-mRNA in addition to its effects on the elongation rate. To test this hypothesis, we searched for binding motifs of splicing factors within exon 3 and flanking intronic sequences using the SpliceAid 2 (http://193.206.120.249/splicing_tissue.html) (40) and RBP map (http://rbpmap.technion.ac.il/) (41) prediction tools (Supplementary Figure S6A). We found several potential binding sites for Sam68 itself, for c-MYC-regulated members of the hnRNP family (Supplementary Figure S6A), which generally act as splicing repressors, and for ETR-3, a splicing repressor that was recruited by slow RNAPII (38). To test whether these splicing factors could modulate exon 3 splicing, we constructed a minigene encompassing the whole alternatively spliced region of Sam68, from exon 2 to exon 4 including intervening introns (Supplementary Figure S6B). Splicing assays in HEK293T cells indicated that Sam68 does not regulate its own AS (Supplementary Figure S6B), nor this event was regulated by hnRNP A2, hnRNP I (PTBP1) and ETR-3 (Supplementary Figure S6C). By contrast, hnRNP A1, hnRNP F and hnRNP H promoted skipping of exon 3 (Supplementary Figure S6C). More importantly, qPCR analyses revealed that overexpression of the highly homologous hnRNP F and hnRNP H promoted exon 3 skipping also in the endogenous Sam68 transcript in LNCaP cells, whereas hnRNP A1 was ineffective (Figure 6A). Accordingly, hnRNP F/H depletion significantly reduced exon 3 skipping in the endogenous Sam68 transcript, whereas hnRNP A1 knockdown did not (Figure 6B). These results suggest that hnRNP F/H are strong candidates for regulation of Sam68 splicing in PCa cells.
Figure 6.
hnRNP F modulates the AS of Sam68 exon 3. (A and B) qPCR analyses of in vivo splicing assay performed in LNCaP cells transfected (A) or depleted (B) for the indicated splicing factors. Bar graphs (A and B; left panels) show the fold change of Sam68-ΔKH expression relative to Sam68-KH expression calculated by the ΔΔCq method. A representative western blot analysis (A and B; right panel) to asses the expression levels of the indicated splicing factors is shown. β-actin was used as loading control. (C) CLIP assays performed in LNCaP cells treated (DRB) or not (DMSO) with 5 μg/ml of DRB to detect the recruitment of hnRNP F on the endogenous Sam68 pre-mRNA. The immunoprecipitation was performed using hnRNP F antibody or IgGs, as negative control. RNA associated with hnRNP F was quantified by qPCR using primers located at the Sam68 exon 3–intron 3 boundary and represented as percentage (%) of input. (D) Schematic model for c-MYC-driven Sam68 gene expression and AS regulation. (A–C) Bars represent mean ± SD of three biological replicates. **P < 0.01; ***P < 0.001; n.s., not significant (Student's t-test).
c-MYC is known to regulate the expression of several splicing factors (19,42). Notably, we found that c-MYC depletion in LNCaP cells caused also mild downregulation of hnRNP F/H expression, in addition to that of its known targets (Supplementary Figure S6D). Since exon 3 skipping occurred in spite of the slightly reduced levels of hnRNP F/H, we asked whether modulation RNAPII elongation rate by c-MYC promoted recruitment of these factors on the Sam68 pre-mRNA. To test this possibility, we treated LNCaP cells with DRB, which mimicked the effect of c-MYC depletion on RNAPII phosphorylation and exon 3 splicing without significantly affecting expression of hnRNP F/H or other c-MYC-regulated splicing factors (Supplementary Figure S6E). CLIP experiments showed that recruitment of hnRNP F to the exon 3 region of the Sam68 pre-mRNA was significantly increased upon DRB treatment (Figure 6C).
DISCUSSION
Sam68 is upregulated and plays key roles in several human tumors (13), including PCa (14,15). However, despite its relevance in oncogenic processes, how Sam68 transcription is dysregulated in cancer cells remains unknown. Our study now demonstrates that the oncogenic transcription factor c-MYC promotes expression and proper splicing of Sam68 mRNA to yield high levels of this protein in PCa cells. Furthermore, we show that c-MYC and Sam68 expression are tightly linked to favorable environmental conditions, whereas downregulation of c-MYC under androgen and/or serum deprivation correlates with repression of Sam68 transcription and exon 3 splicing, yielding a variant unable to bind RNA (Figure 6D). Thus, our work uncovers the molecular mechanism underlying dysregulation of Sam68 expression in PCa, a finding that is probably extendible to other human cancers in which the c-MYC oncogene is upregulated.
To identify potential regulators of Sam68 transcription, we queried public datasets provided by the Encode project using the UCSC Genome Browser tool. This unbiased analysis identified a subset of transcription factors potentially involved in the regulation of Sam68 expression. Among them, we focused on c-MYC because of its well-established oncogenic function in human cancers (43). The activity of c-MYC profoundly alters the transcriptome of cancer cells (44). Furthermore, mounting evidence supports a key role for c-MYC also in the control of splicing, which further diversifyies the transcriptome of cancer cells. Indeed, spliceosome components (29,45) and splicing factors, including SRSF1, PTBP1, hnRNP A1 (19,26) are direct targets of c-MYC. Notably, SRSF1 expression promotes proliferation, cell survival and anchorage-independent growth in several tumors (19) and its phosphorylation by SRPK1 controls angiogenesis through the splicing regulation of VEGF (46), which is essential in highly vascularized cancers like PCa (47). On the other hand, the regulation of spliceosome components is required for c-MYC-driven malignant transformation (29,45), suggesting that safeguarding splicing fidelity is a key element for the pro-oncogenic function of c-MYC. Our study now identifies Sam68 as an additional splicing regulator under c-MYC control in cancer. We found a highly significant correlation between c-MYC and Sam68 expression in PCa patients, suggesting that this regulation is functional in vivo. Although our work was focused on PCa, it is likely that c-MYC more generally regulates Sam68 expression in cancer, as ChIP-seq peaks for this transcription factor within the Sam68 promoter region were found in cell lines of different origin.
Bioinformatics analyses revealed a relatively small region upstream of the first Sam68 coding exon that displays promoter features, such as histone marks of active transcription and RNAPII occupancy. Since the transcriptional activity of this region was confirmed in reporter assays, our study identifies a core promoter of human Sam68 that is active in multiple cell types. Our results also show that c-MYC mainly acts through binding to two E-boxes proximal to the TSS. Importantly, c-MYC expression is increased in PCa and correlates with disease severity (30,48). Since Sam68 and c-MYC upregulation is correlated in PCa patients, our findings suggest that Sam68 takes part to the oncogenic program activated by c-MYC in prostate cells.
c-MYC expression overrides growth arrest and reduces the capacity of cancer cells to differentiate (32). Sam68 appears to support the same functions, as its depletion impaired PCa cell proliferation (14) and induced neural stem cell differentiation (49). Thus, we tested the possibility that the expression of c-MYC and Sam68 responds to favorable growth conditions in PCa cells. As expected, we observed a concomitant reduction of c-MYC and Sam68 expression under conditions that cause proliferation arrest, such as serum deprivation and inhibition of AR signaling. These findings suggest the existence of a c-MYC-Sam68 functional axis in PCa and highlight a possible novel role for Sam68 as mediator of c-MYC function.
The KH domain is crucial for the RNA-binding activity of Sam68 (34). It was previously reported that a Sam68 splice variant lacking a functional KH domain (Sam68-ΔKH) is generated upon growth arrest (33) and this variant was proposed to participate to cell cycle arrest in fibroblasts. Nevertheless, whether the Sam68-ΔKH variant existed in cancer cells and the mechanisms underlying its expression had not been investigated. Sam68-ΔKH results from exon 3 skipping during pre-mRNA splicing. We observed an increase in exon 3 skipping upon growth arrest induced by either serum or androgen deprivation and this effect was recapitulated by knockdown of c-MYC. Thus, c-MYC likely controls both transcription and productive splicing of Sam68. This regulation may have widespread impact on the transcriptome of PCa cells. Expression of Sam68-ΔKH may not only impair splicing of the direct Sam68 targets, but also affect those of SRSF1 because Sam68 is required to inhibit non-sense mediated decay (NMD) of SRSF1 transcripts (50). These observations suggest that the c-MYC- mediated oncogenic program may also rely on a widespread splicing program empowered by the network of cross-regulation between its downstream effectors. Since c-MYC lacks molecular features that can be targeted by small molecule inhibitors, identification of downstream effectors may help develop treatments for MYC-driven cancers. For instance, antisense oligonucleotides (ASOs) targeting splicing of Sam68 exon 3 may weaken the c-MYC oncogenic program in PCa and enhance the efficacy of other anti-cancer treatments.
Mechanistically, we linked c-MYC action to modulation of RNAPII activity. Since c-MYC influences RNAPII occupancy across the transcribed unit of its target genes, as well as phosphorylation of RNAPII at Serine 2 (51), c-MYC might alter Sam68 splicing by affecting the RNAPII elongation rate within the locus. Indeed, we found that depletion of c-MYC decreased Ser-2 phosphorylation of RNAPII in LNCaP cells and reduced the RNAPII elongation rate in the Sam68 gene, which correlated with exon 3 skipping. In our experiments RNAPII processivity was calculated as ratio of regions in the same transcript, thus it should be independent of the total amount of transcript produced by the gene. Nevertheless, since transcription efficiency and RNAPII processivity are mechanistically linked (39), they could impact on each other and we cannot completely distinguish between these two aspects affected by c-MYC. However, the effect on Sam68 splicing could be recapitulated by treatment of cells with several CDK9 inhibitors. Thus, our results suggest that reducing the RNAPII elongation rate is sufficient for exon 3 splicing regulation and that c-MYC coordinates transcription and splicing of Sam68 by affecting the RNAPII elongation rate.
Changes in RNAPII dynamics can affect splicing by affecting recruitment of specific splicing factors to the regulated exons (37). Such model was recently demonstrated for the CFTR gene, where reduced RNAPII processivity allowed recruitment of the splicing repressor ETR-3 and promoted exon skipping (38). Herein, by performing bioinformatics analyses and splicing assays, we identified hnRNP F as a regulator of Sam68 exon 3 splicing, whose recruitment to the pre-mRNA is favored by a slow RNAPII in PCa cells. We focused on hnRNP F because its antibody functioned better than that of hnRNP H under CLIP conditions in LNCaP cells. However, given their high homology, this antibody also partially recognizes hnRNP H and it is likely that exon 3 skipping is due to binding of both hnRNPs to the Sam68 transcript.
In conclusion, our study reveals that c-MYC contributes to regulation of Sam68 transcription and productive splicing in PCa cells through modulation of RNAPII dynamics within the gene.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Ministry of Health ‘Ricerca Finalizata 2011’ [GR-2011-02348423]; Ricerca Finalizzata 2016 [RF-2016-02363460]; Associazione Italiana Ricerca sul Cancro (AIRC) [IG18790]. Funding for open access charge: Italian Ministry of Health “Ricerca Corrente".
Conflict of interest statement. None declared.
REFERENCES
- 1. Siegel R.L., Miller K.D., Jemal A.. Cancer Statistics, 2017. CA Cancer J. Clin. 2017; 67:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Pignot G., Maillet D., Gross E., Barthelemy P., Beauval J.B., Constans-Schlurmann F., Loriot Y., Ploussard G., Sargos P., Timsit M.O. et al.. Systemic treatments for high-risk localized prostate cancer. Nat. Rev. Urol. 2018; 15:498–510. [DOI] [PubMed] [Google Scholar]
- 3. Munkley J., Livermore K., Rajan P., Elliott D.J.. RNA splicing and splicing regulator changes in prostate cancer pathology. Hum. Genet. 2017; 136:1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sette C. Alternative splicing programs in prostate cancer. Int. J. Cell Biol. 2013; 2013:458727–458737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu R., Lu C., Mostaghel E.A., Yegnasubramanian S., Gurel M., Tannahill C., Edwards J., Isaacs W.B., Nelson P.S., Bluemn E. et al.. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012; 72:3457–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mercatante D.R., Mohler J.L., Kole R.. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem. 2002; 277:49374–49382. [DOI] [PubMed] [Google Scholar]
- 7. Burd C.J., Petre C.E., Morey L.M., Wang Y., Revelo M.P., Haiman C.A., Lu S., Fenoglio-Preiser C.M., Li J., Knudsen E.S. et al.. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2190–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castilla C., Congregado B., Chinchon D., Torrubia F.J., Japon M.A., Saez C.. Bcl-xL is overexpressed in hormone-resistant prostate cancer and promotes survival of LNCaP cells via interaction with proapoptotic Bak. Endocrinology. 2006; 147:4960–4967. [DOI] [PubMed] [Google Scholar]
- 9. Comstock C.E., Augello M.A., Benito R.P., Karch J., Tran T.H., Utama F.E., Tindall E.A., Wang Y., Burd C.J., Groh E.M. et al.. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin. Cancer Res. 2009; 15:5338–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Braunschweig U., Gueroussov S., Plocik A.M., Graveley B.R., Blencowe B.J.. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013; 152:1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anczukow O., Krainer A.R.. Splicing-factor alterations in cancers. RNA. 2016; 22:1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sveen A., Kilpinen S., Ruusulehto A., Lothe R.A., Skotheim R.I.. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016; 35:2413–2427. [DOI] [PubMed] [Google Scholar]
- 13. Bielli P., Busa R., Paronetto M.P., Sette C.. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer. 2011; 18:R91–R102. [DOI] [PubMed] [Google Scholar]
- 14. Busa R., Paronetto M.P., Farini D., Pierantozzi E., Botti F., Angelini D.F., Attisani F., Vespasiani G., Sette C.. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007; 26:4372–4382. [DOI] [PubMed] [Google Scholar]
- 15. Rajan P., Gaughan L., Dalgliesh C., El-Sherif A., Robson C.N., Leung H.Y., Elliott D.J.. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 2008; 215:67–77. [DOI] [PubMed] [Google Scholar]
- 16. Jenkins R.B., Qian J., Lieber M.M., Bostwick D.G.. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997; 57:524–531. [PubMed] [Google Scholar]
- 17. Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B. et al.. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010; 18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brase J.C., Johannes M., Mannsperger H., Falth M., Metzger J., Kacprzyk L.A., Andrasiuk T., Gade S., Meister M., Sirma H. et al.. TMPRSS2-ERG-specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011; 11:507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das S., Anczukow O., Akerman M., Krainer A.R.. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012; 1:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Annibalini G., Bielli P., De Santi M., Agostini D., Guescini M., Sisti D., Contarelli S., Brandi G., Villarini A., Stocchi V. et al.. MIR retroposon exonization promotes evolutionary variability and generates species-specific expression of IGF-1 splice variants. Biochim. Biophys. Acta. 2016; 1859:757–768. [DOI] [PubMed] [Google Scholar]
- 21. Bielli P., Busa R., Di Stasi S.M., Munoz M.J., Botti F., Kornblihtt A.R., Sette C.. The transcription factor FBI-1 inhibits SAM68-mediated BCL-X alternative splicing and apoptosis. EMBO Rep. 2014; 15:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paronetto M.P., Cappellari M., Busa R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K.E., Sette C.. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010; 70:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh J., Padgett R.A.. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009; 16:1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bielli P., Sette C.. Analysis of in vivo Interaction between RNA binding proteins and their RNA targets by UV Cross-linking and Immunoprecipitation (CLIP) Method. Bio Protoc. 2017; 7:e2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haggerty T.J., Zeller K.I., Osthus R.C., Wonsey D.R., Dang C.V.. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. David C.J., Chen M., Assanah M., Canoll P., Manley J.L.. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010; 463:364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amati B., Land H.. Myc-Max-Mad: a transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Genet. Dev. 1994; 4:102–108. [DOI] [PubMed] [Google Scholar]
- 28. Dang C.V., O’Donnell K.A., Zeller K.I., Nguyen T., Osthus R.C., Li F.. The c-Myc target gene network. Semin. Cancer Biol. 2006; 16:253–264. [DOI] [PubMed] [Google Scholar]
- 29. Koh C.M., Bezzi M., Low D.H., Ang W.X., Teo S.X., Gay F.P., Al-Haddawi M., Tan S.Y., Osato M., Sabo A. et al.. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015; 523:96–100. [DOI] [PubMed] [Google Scholar]
- 30. Gurel B., Iwata T., Koh C.M., Jenkins R.B., Lan F., Van Dang C., Hicks J.L., Morgan J., Cornish T.C., Sutcliffe S. et al.. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 2008; 21:1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rebello R.J., Pearson R.B., Hannan R.D., Furic L.. Therapeutic approaches targeting MYC-Driven prostate cancer. Genes (Basel). 2017; 8:71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sabo A., Kress T.R., Pelizzola M., de Pretis S., Gorski M.M., Tesi A., Morelli M.J., Bora P., Doni M., Verrecchia A. et al.. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014; 511:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barlat I., Maurier F., Duchesne M., Guitard E., Tocque B., Schweighoffer F.. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J. Biol. Chem. 1997; 272:3129–3132. [DOI] [PubMed] [Google Scholar]
- 34. Chen T., Damaj B.B., Herrera C., Lasko P., Richard S.. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol. Cell. Biol. 1997; 17:5707–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ip J.Y., Schmidt D., Pan Q., Ramani A.K., Fraser A.G., Odom D.T., Blencowe B.J.. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011; 21:390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Batsche E., Yaniv M., Muchardt C.. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006; 13:22–29. [DOI] [PubMed] [Google Scholar]
- 37. Naftelberg S., Schor I.E., Ast G., Kornblihtt A.R.. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015; 84:165–198. [DOI] [PubMed] [Google Scholar]
- 38. Dujardin G., Lafaille C., de la Mata M., Marasco L.E., Munoz M.J., Le Jossic-Corcos C., Corcos L., Kornblihtt A.R.. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell. 2014; 54:683–690. [DOI] [PubMed] [Google Scholar]
- 39. Hsin J.P., Manley J.L.. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012; 26:2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piva F., Giulietti M., Burini A.B., Principato G.. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 2012; 33:81–85. [DOI] [PubMed] [Google Scholar]
- 41. Paz I., Kosti I., Ares M. Jr., Cline M., Mandel-Gutfreund Y.. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014; 42:W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. David C.J., Manley J.L.. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010; 24:2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dang C.V. MYC on the path to cancer. Cell. 2012; 149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walz S., Lorenzin F., Morton J., Wiese K.E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M. et al.. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014; 511:483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S. et al.. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015; 525:384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amin E.M., Oltean S., Hua J., Gammons M.V., Hamdollah-Zadeh M., Welsh G.I., Cheung M.K., Ni L., Kase S., Rennel E.S. et al.. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011; 20:768–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mavrou A., Brakspear K., Hamdollah-Zadeh M., Damodaran G., Babaei-Jadidi R., Oxley J., Gillatt D.A., Ladomery M.R., Harper S.J., Bates D.O. et al.. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene. 2015; 34:4311–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hawksworth D., Ravindranath L., Chen Y., Furusato B., Sesterhenn I.A., McLeod D.G., Srivastava S., Petrovics G.. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis. 2010; 13:311–315. [DOI] [PubMed] [Google Scholar]
- 49. La Rosa P., Bielli P., Compagnucci C., Cesari E., Volpe E., Farioli Vecchioli S., Sette C.. Sam68 promotes self-renewal and glycolytic metabolism in mouse neural progenitor cells by modulating Aldh1a3 pre-mRNA 3′-end processing. Elife. 2016; 5:e20750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Valacca C., Bonomi S., Buratti E., Pedrotti S., Baralle F.E., Sette C., Ghigna C., Biamonti G.. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 2010; 191:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A.. c-Myc regulates transcriptional pause release. Cell. 2010; 141:432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.