Abstract
Barley Yellow Dwarf Virus (BYDV) is a positive strand RNA virus that lacks the canonical 5′ 7-methylguanosine cap and a 3′ poly-A tail. Instead, BYDV utilizes a cruciform cap independent translation element (CITE) in its 3′UTR RNA (BYDV-like CITE or BTE) that binds eukaryotic translation initiation factor (eIF) 4F and recruits 40S ribosomal subunits in the presence of active helicase factors (eIF4A, eIF4B, eIF4F and ATP). A long-range, 5-nucleotide, base-pairing kissing loop interaction between the 3′BTE and a 5′UTR stem-loop is necessary for translation to initiate. The 40S–eIF complex does not bind to the BYDV 5′UTR, suggesting the involvement of additional factors. We identified eIF3 as a component of the 3′BTE recruited complex using affinity-tagged 3′BTE RNA pull-down assays. Fluorescence anisotropy binding and gel shift assays showed that the 3′BTE and 5′UTR RNAs can simultaneously and non-competitively bind eIF3 in the presence of active helicase factors forming a single, macromolecular complex. Further, quantitative studies showed eIF3 increased recruitment of the 40S subunit by more than 25-fold. We propose a new role for eIF3, where eIF3 bridges BYDV’s UTRs, stabilizes the long-range 5′-3′ interaction, and facilitates recruitment of the 40S–eIF complex to the 5′UTR, leading to translation initiation.
INTRODUCTION
Plant RNA viruses pose a worldwide threat to the food supply. These viruses produce mRNAs that must outcompete host cell mRNAs for the protein synthesis machinery and have therefore evolved a myriad of non-canonical translation initiation mechanisms to circumvent canonical translation pathways (1–3). Many of these viruses are positive strand RNA viruses that lack the canonical 5′ 7-methylguanosine cap and 3′ poly-A tails in their mRNAs. Instead, they utilize RNA structures in their 3′ untranslated regions (UTR), known as 3′ cap-independent translation elements (CITEs) (2) to drive non-canonical translation initiation. Barley Yellow Dwarf Virus (BYDV, Genus Luteovirus), a widespread, devastating pest of small grain cereals (4) is a positive strand RNA virus which employs a non-canonical, cap-independent mechanism utilizing RNA structures in both its 5′ and 3′ mRNA UTRs for mRNA translation initiation and regulation (5–7) (Figure 1). The 3′UTR of BYDV mRNA harbors a cap independent translation element (CITE), commonly referred to as the 3′BTE (BYDV like CITE) which is essential for the translation at the proximal 5′AUG. The 3′BTE is one of the best characterized CITEs and is present in all Luteovirus members (family Luteoviridae) and some members of family Tombusviridae (2,8,9). All known 3′BTEs contain a highly conserved 17-nucleotide sequence GGAUCCUGGGAAACAGG, containing a six-base tract (bold, underlined) which is complementary to a stretch of bases in 18S rRNA (3,6). The BYDV 3′BTE is 105-nt long and has a cruciform secondary structure with three stem loops SL-I, SL-II, SL-III and a base stem (SL-IV) (Figure 1). The BYDV 5′UTR consists of four stem loops (SL-A, B, C and D) (Figure 1). Several reports have shown that the 5′UTR of BYDV is necessary for translation only when the 3′BTE is located at the 3′ end of the message, not when the 3′BTE is present in the 5′ context (6,7). Both these UTRs communicate via a long-distance RNA-RNA kissing-loop interaction (Watson-Crick base pairing) involving five complementary bases in the 3′BTE SL-III and a 3′BTE-complementary loop (BCL/SL-D) in the 5′UTR (Figure 1) (10,11). The long-distance kissing loop interaction helps bring the 5′-3′UTRs together, a step necessary for facilitating the transfer of 3′ recruited ribosomes/initiation factors to the 5′UTR for translation initiation (10,11).
Figure 1.

RNA structural elements involved in 3′BTE-mediated translation initiation (BYDV 5′UTR and 3′BTE). The 5′UTR consists of four stem loops (SL-A, B, C and D). The 3′BTE has a cruciform secondary structure with three stem loops (SL-I, II, III) and a base loop, SL-IV. Positions of the five complementary bases involved in long distance kissing loop interaction are shown in green (on 3′BTE SL-III and 5′UTR SL-D). The six-base tract complementary to 18S rRNA in the 3′BTE is shown in red.
The 3′BTE requires only a subset of the host's translation machinery to efficiently recruit the 40S ribosomal subunit. Previous reports have shown that eukaryotic initiation factor 4F (eIF4F) binds specifically to the 3′BTE via the eIF4G subunit and is crucial for 3′BTE-mediated translation (12,13). Although, the other eIF4F subunit, eIF4E does not bind directly to the 3′BTE, it promotes the binding of eIF4G to the 3′BTE via protein-protein interactions (14). Recent work with eIF4G deletion mutants has shown that only the core domain (eIF4G601–1196), comprising MIF4G (eIF4A, eIF3 and RNA binding sites) and a region immediately N-terminal of MIF4G, is necessary for 3′BTE binding and translation initiation (14). Translation efficiencies of the 3′BTE mutants are correlated with their different eIF4F binding affinities (12), which highlights eIF4F’s direct and central role in recruiting translation machinery. Recently, it was shown that eIF4F, along with eIF4A, eIF4B and ATP, can recruit the 40S ribosomal subunit to the 3′BTE (Kd = 120 ± 10 nM) (15). This 3′BTE-recruited complex must transfer to the 5′UTR for translation to initiate. This transfer is a prerequisite to AUG-recognition and elongation, yet no report has shown direct binding of the 40S ribosomal subunit to the BYDV 5′UTR. The exact mechanism for transfer of the 3′BTE recruited 40S–eIF complex to the 5′UTR, and involvement of other factors in the transfer remains unknown. In this report, we employed affinity pull down methods to identify eIF3 as a component of the BTE-recruited ribosomal complex. We propose that eIF3 is a component of the 3′BTE-mediated translation initiation complex that serves as a molecular bridge that brings the 5′- and 3′UTRs together, stabilizes the kissing loop interaction, and facilitates transfer of the 40S–eIF complex from the 3′BTE to the 5′UTR for translation to initiate. Our findings support a model (3,15,16) where eIF3 recruits the 40S ribosomal subunit (possibly as 43S pre-initiation complex, PIC) at the 3′BTE. The 40S–eIF complex at the 3′BTE is subsequently delivered to (or simultaneously interacts with) the 5′UTR via long distance RNA-RNA pairing and direct binding of eIF3 to the 5′UTR. We show here that ribosomes can bind directly and specifically to the 5′UTR in the presence of eIF3 and helicase unwinding factors (eIF4A, eIF4B, and eIF4F and ATP), which are reported to be involved in the recruitment of the 40S ribosomal subunit to the 3′BTE (15). Collectively the data presented in this report suggest that the 3′BTE, the 5′UTR, eIF3 and the 40S ribosomal subunit are components of a single macromolecular complex.
MATERIALS AND METHODS
Synthesis, purification, and fluorescein labeling of RNAs
A chimeric BYDV 3′BTE-streptotag construct was designed for the assembly and subsequent purification of the translation initiation complexes from wheat germ extracts (WGE). This designed construct contained the 105 nucleotide BTE RNA followed by a 3′ intrinsic streptomycin binding RNA aptamer tag, called streptotag. Streptotag is a 46nt RNA tag (5′-GGAUCGCAUUUGGACUUCUGCCCGCAAGGGCACCACGGUCGGAUCC-3′) that binds with micromolar affinity to streptomycin (17,18). DNA oligonucleotides used as templates for in vitro transcription of the chimeric RNA (BTE-Streptotag) were custom synthesized from IDT (Integrated DNA Technologies Inc.). The secondary structure of this construct was predicted using MFOLD (19,20) to make sure the BTE structure was not disrupted by the 3′streptotag (Supplementary Figure S1). Wild type 3′BTE RNA and 5′UTR RNA were also generated from synthetic DNA oligonucleotides obtained from IDT (Integrated DNA Technologies Inc.). A 100 nucleotide long polyUC RNA, an I-shaped CITE RNA (ISS, ∼100 nucleotide long) from Maize necrotic streak virus (MNeSV) and a 45 nucleotide long RNA corresponding to wheat 18S rRNA (18S rRNA, bases 1040–1084) were used as negative controls. The RNAs were in vitro transcribed using HiScribe™ T7 Quick High Yield RNA Synthesis Kit (NEB) as per manufacturer's instructions and were purified using RNA clean and concentrator™ kit (Zymo Research). The quality and quantity of purified RNAs were measured using a Thermo Scientific NanoDrop 1000 spectrophotometer. For anisotropy binding assays, in vitro transcribed RNAs were fluorescein labeled at the 5′-end using the 5′-end tag labeling kit (Vector labs) as per manufacturer's instructions and were purified with the RNA clean and concentrator™ kit. The purified RNAs were >99% pure with 15–20% labeling efficiency.
Assembly and pull down of the 3′BTE-mediated initiation complex
Raw wheat germ (Bob's Red Mill, Natural Foods, Inc., Milwaukie, OR, USA) was used to prepare wheat germ extract (WGE) as reported previously (21). The WGE was supplemented with 80 mM potassium acetate, 3 mM magnesium acetate, 120 μg/ml wheat germ tRNA (Sigma), 0.6mM spermidine, 20 mM creatine phosphate, 50 μg/ml creatine phosphokinase and RNase inhibitor (20 units) immediately before use. Dihydrostreptomycin was coupled to epoxy-activated Sepharose (GE Healthcare) as reported elsewhere (18,22) and the slurry was stored at 4°C in the dark. For assembly and purification of 48S initiation complexes, 1 ml of supplemented WGE was incubated with 1 ml of binding buffer (50 mM HEPES pH 7.3, 10 mM MgCl2, 120 mM KCl, 8% sucrose and 1 proteinase inhibitor tablet (EDTA free)/50 ml buffer) for 10 min at 37°C. Puromycin was added to a final concentration of 1 mM and the samples were incubated for 10 min on ice and then 10 min at 37°C. Finally, tagged mRNA (1 μM) was added along with 5 mM ATP (final concentration), and the reaction tubes were incubated at 37°C for 10 min. In the negative control sample, no exogenous RNA was added. Samples were loaded on a 1 ml dihydroxystreptomycin Sepharose column, which was pre-equilibrated with 1× binding buffer, and the column was washed with 10 ml 1× binding buffer. The initiation complexes/RNA binding proteins were eluted using 10 ml of 1× binding buffer supplemented with 100 μM streptomycin. The eluted fractions, along with the washes were analyzed on 10% SDS-PAGE gels and 1% native agarose gels. The eluted samples were layered onto a 10–30% sucrose density gradient in buffer I (50 mM Tris–HCl, 600 mM KCl, 1 mM Mg(OAc)2, 0.1 mM EDTA, 6 mM 2-mercaptoethanol and 5% sucrose) and centrifuged at 25 000 rpm in a Beckman SW28 rotor for 16 h at 4°C. Gradients were fractionated using a Brandel gradient density fractionator and were collected as 750 μl fractions (0.75 ml/minute). All the fractions were analyzed on a 1% agarose gel. The putative BTE-40S initiation complex fractions were pooled, and dialyzed against 20 mM Tris Acetate pH 7.5, 4 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol and 8% glycerol. The dialyzed samples were concentrated using 50 ml Centricons (10 kDa, Millipore), and were stored at –80°C. The samples were analyzed on 1% agarose gels. The sample corresponding to the putative 3′BTE recruited-40S initiation complex was sent for liquid chromatography–mass spectrometric (LC–MS) analysis (Proteomics Resource Center, NYU).
Purification of wheat initiation factors and 40S ribosomal subunits
Recombinant wheat eIF4A and eIF4B clones were generous gifts from Prof. D.R. Gallie (University of California, Riverside, CA, USA). The recombinant proteins were His-tagged and were purified from bacterial cultures using His trap HP columns (GE Healthcare Life Sciences). A dicistronic eIF4F clone containing both eIF4G and eIF4E genes for expression of the wheat eIF4F complex was a generous gift from Prof. K.S. Browning (University of Texas at Austin, Austin, TX, USA). The recombinant eIF4F protein was purified as described earlier (12,23). The 40S ribosomal subunits and native eIF3 protein were purified from wheat germ extract, using published protocols (24–26) with a few modifications. Briefly, eIF3 was purified from 0 to 40% ammonium sulfate fractions of wheat germ lysate postribosomal supernatants. The 0–40% ammonium sulfate pellets were suspended in buffer B-80 (20 mM HEPES pH 7.6, 0.1 M EDTA, 1 mM DTT, 10% glycerol and KCl as indicated) and dialyzed against buffer B-80 overnight. The dialyzed samples were clarified by centrifugation at 10 000 rpm for 10 min (at 4°C) in a SS-34 rotor using a Sorvall RC 5C plus centrifuge. The clarified samples were applied to a 50 ml DEAE-Sepharose column pre-equillibrated with buffer B-80. The column was washed with buffer B-80 and developed using a linear KCl gradient (150–300 mM) in buffer B. eIF3 eluted in the end fractions (250–300 mM KCl). The purified fractions were analyzed on 10% SDS-PAGE gels, pooled and subjected to a second ammonium sulfate purification at 50% saturation. The precipitated proteins were suspended in buffer B-100 and dialyzed overnight against the same buffer. The dialyzed samples were clarified by centrifugation at 10 000 rpm for 10 min in the cold, and were applied to a 10ml phosphocellulose column equilibrated in buffer B-100. The column was washed with buffers B-100 and B-150, and eIF3 was eluted using buffer B-300. The fractions were analyzed on 10% SDS-PAGE gels, pooled and the buffer was exchanged to buffer B-100 using 10kDa Centricons (Millipore). The KCl concentration of these samples was adjusted to 50 mM, right before loading on a 5 ml CM-Sephadex column (pre-equilibrated with B-50). The column was washed with buffer B-50 and eIF3 was eluted using buffer B-150. The purity of the isolated eIF3 protein was checked on a 10% SDS-PAGE (Figure 3A) and using total protein LC–MS (Proteomics Resource Center, NYU) (supplementary excel sheet 1). The presence of any residual copurified eIF4F was assessed by a western blot probed using anti-eIF4G antibodies (a generous gift from Prof. K.S. Browning, University of Texas at Austin, Austin, TX, USA) (Figure 3B).
Figure 3.
(A) A 10% SDS-PAGE gel showing the purified eIF3. The subunits are labeled according to their approximate mobilities. (B) A western blot probed with anti-eIF4G antibodies showing the absence of copurified eIF4G in the purified eIF3 preparation. Crude wheat germ lysate was used as a control. (C and D) show fluorescence anisotropy for the binding of 5′ fluorescein labeled BYDV UTRs to native wheat eIF3 (excitation, 495 nM and emission, 520 nM). The titrations were performed at 25°C for 30 min and 100 data points were collected. (C) The binding of the 3′BTE to eIF3 was non-specific in the absence of the helicase complex, but increased significantly in the presence of the helicase complex (Kd = 29.6 ± 1). The 3′BTE showed tight binding to eIF3 even in the absence of eIF4F (Kd = 19 ± 0.9). The negative control 18S rRNA, polyUC RNA and MNeSV ISS RNA showed no binding to eIF3, even in the presence of the helicase complex. (D) The binding of the 5′UTR to eIF3 was not significant in the absence of the helicase complex but increased significantly in presence of the helicase complex (Kd = 39.4 ± 2.9). The binding was abolished if no ATP was added to the helicase complex or if it was replaced by AMPPNP.
Fluorescence anisotropy binding experiments
Fluorescence anisotropy experiments were performed to assess binding of the 3′BTE and 5′UTR RNAs to eIF3 and 40S ribosomal subunits, in the presence and absence of the helicase complex (eIF4A, eIF4B, eIF4F and ATP) reported to be involved in 3′BTE-mediated translation initiation (15). Binding of eIF3 to the control RNAs (polyUC, MNeSV ISS and 18S rRNA derived oligonucleotide) was assessed in the presence of the helicase complex. Fluorescence anisotropy experiments use plane-polarized light to measure the rotational diffusion of a labeled molecule. Rotational diffusion decreases when an unlabeled factor binds the labeled molecule, increasing overall anisotropy. The increase in anisotropy represents a higher fraction of molecules bound and is used to measure the binding affinities of labeled RNAs to the titrated proteins. Changes in the fluorescence anisotropy of fluorescein labeled 3′BTE and 5′UTR (excitation wavelength, 495 nm) were measured using a stopped-flow model SF-300X (KinTek Corporation) set up with a titration module and equipped with two photomultiplier tubes each fitted with a polarizing filter and fluorescein filter (515 nm blocking edge BrightLine® long-pass filter, Semrock Inc.), arranged in a T-format configuration. For the binding assays, 5′-end fluorescein labeled RNAs were used at a final concentration of 50 nM in 200 μl of assay buffer (20 mM HEPES pH 7.6, 5 mM MgCl2, 200 mM KCl). Using an automated injection module, 20 μl of purified native eIF3 and/or 40S ribosomal subunits (2μM) were slowly injected in the cuvette containing the labeled RNA samples (with or without additional eIFs). The assays were performed at 25°C for 30 min, and 100 data points were collected. For assays with the helicase complex, the fluorescein labeled RNAs (50 nM) were pre-incubated with 300 nM each of eIF4A, eIF4B, eIF4F and 5 mM ATP, for 20 min at 25°C, and then titrated with eIF3 and/or 40S ribosomal subunits (2 μM). Binding affinities of the 3′BTE and 5′UTR RNA to 40S ribosomal subunits were measured in the presence of eIF3, and the helicase complex. Data from five independent binding assays were averaged and used for calculating the dissociation equilibrium constants (Kd) using Kaleidagraph software (Synergy software) as described in previous reports (12,15). The Kds were determined by fitting the anisotropy data to the equation:
 |
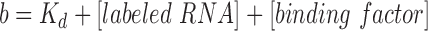 |
r obs is the observed anisotropy for any point in the titration curve, rmin is the minimum observed anisotropy in the absence of protein/40S ribosomal subunits, and rmax is the maximum anisotropy at saturation.
Gel shift assays
Gel shift assays were performed to confirm simultaneous binding of the 3′BTE and 5′UTRs to eIF3. Binding reactions for the individual RNAs (final concentration 250 nM) to eIF3 (final concentration 350 nM) were done in THEMK buffer (34 mM Tris–HCl pH 7.8, 66 mM HEPES, 0.1 mM EDTA, 2.5 mM MgCl2, 75 mM KCl) (27). Reactions were performed at 25°C for 20 min in the presence of the helicase unwinding complex (500 nM each of eIF4A, eIF4B, eIF4F and 5 mM ATP). For simultaneous binding, both the RNAs (250 nM of the 3′BTE, with 3-fold molar excess of the 5′ UTR RNA, 750 nM) were added together. The RNA–eIF3 complexes were resolved in native 1% agarose gel in cold TBE buffer at 4°C at 50 V for 1 h and were visualized using ethidium bromide staining. Similarly, gel shift assays were also performed using the control RNAs (polyUC, MNeSV ISS and 18S rRNA oligonucleotide).
RESULTS
Twelve eIF3 subunits were detected in the pull-down complex using BYDV 3′BTE RNA as bait
To identify any other factor(s) that may be involved in 3′BTE-mediated translation initiation, we utilized total proteome LC–MS characterization of the ribosomal complex pulled out from wheat germ extract using the BYDV 3′BTE RNA as bait. A chimeric BYDV 3′BTE-streptotag construct was designed for the assembly and subsequent purification of the translation initiation complexes. The affinity-pulled out complexes were fractionated by sucrose gradient and the components were analyzed using LC–MS (Figure 2). Apart from the ribosomal proteins, 12 eIF3 subunits were detected in this complex along with eIF4G, eIF4A and eIF2 subunit 3 (Supplementary excel sheet 1). The Uniprot wheat database was used for the searches, so proteins from bread wheat (Triticum aestivum) and related wild wheat varieties (Aegilops tauschii, Triticum urartu and Aegilops speltoides) were detected (Supplementary excel sheet 1).
Figure 2.
Schematics showing the experiments performed to isolate and identify protein/ribosomal complexes bound to the 3′BTE. The 3′BTE bound proteins/complexes were purified using a streptotagged 3′BTE RNA and were fractionated using sucrose gradients. The fractionated complexes were pooled and analyzed on 1% native agarose gel (lane 1: purified 40S and 60S ribosomal subunits for reference, lane 2: 3′BTE-ribosome complex, lane 3: the tagged 3′BTE RNA used for affinity purification) and the protein components were identified using LC–MS.
The 3′BTE RNA showed specific binding to purified native wheat eIF3
To quantitate the 3′BTE-eIF3 interaction and validate the pull-down results, we employed previously established fluorescence anisotropy binding assays (12,14,15). In the presence of the helicase complex (eIF4A, eIF4B, eIF4F and ATP), which has already been reported to enhance 40S ribosomal subunit recruitment to the 3′BTE (15); the binding of the 3′BTE RNA to native eIF3 protein (purified from WGE) was very tight (Figure 3C, Table 1) (Kd = 29.6 ± 1 nM). Since eIF3 is known to bind primarily to the 40S ribosomal subunit proteins and not to the ribosomal RNA (28–33) we used a 45 nucleotide RNA corresponding to wheat 18S rRNA (bases 1040–1084) as a negative control. A 100 nucleotide polyUC synthetic unstructured RNA and a 100 nucleotide MNeSV I-shaped CITE (ISS) were also used as controls. The ISS RNA forms an extended stem loop structure with 6 single stranded apical nucleotides which are complementary to an upstream 5′UTR loop (9). These complementary nucleotides may potentially be involved in a kissing loop interaction (9). The ISS RNA was included as a control to assess if an unrelated CITE element with potential kissing loop interaction binds to eIF3. The control RNA oligonucleotides showed no binding to eIF3, even in the presence of the helicase complex (Figure 3C). To test the possibility that eIF3 may be interacting indirectly with the 3′BTE, via binding of eIF3 to eIF4G (which binds very tightly and specifically to the 3′BTE) (13,14), we performed 3′BTE-eIF3 anisotropy binding assays in the absence of eIF4F (in the presence of only eIF4A–eIF4B–ATP). The 3′BTE showed very tight binding to eIF3, even in the absence of eIF4F, indicating that the 3′BTE-eIF3 binding is direct and not solely mediated through eIF4F (Figure 3C, Table 1, Kd = 19 ± 0.9 nM).
Table 1.
Equillibrium dissociation constants (Kd) for binding of the 3′BTE and 5′UTR RNA oligonucleotides to native wheat eIF3 protein
| eIF3 | |||
|---|---|---|---|
| RNA-eIFs | K d ± S.D. (nM) | ΔAnisotropy (rmax– rmin) | Goodness of fit (R2) |
| 3′BTE + helicase complex | 29.6 ± 1 | 0.063 | 0.996 |
| 3′BTE + helicase complex (*no eIF4F) | 19 ± 0.9 | 0.029 | 0.993 |
| 3′BTE + helicase complex + 3X unlabeled 5′UTR | 62 ± 3 | 0.047 | 0.997 |
| 5′UTR + helicase complex | 39.4 ± 2.9 | 0.042 | 0.993 |
| 5′UTR + helicase complex + 3X unlabeled 3′BTE | 86.8 ± 4.7 | 0.052 | 0.995 |
BYDV 5′UTR also showed specific binding to eIF3
To assess if eIF3 binds to the 5′UTR of BYDV, thus possibly bringing the two UTRs together, 5′ fluorescein labeled 5′UTR was titrated with purified, native eIF3. The BYDV 5′UTR showed very tight binding to eIF3 in the presence of an active helicase complex (Figure 3D, Table 1, Kd = 39.4 ± 2.9 nM). To assess the role of helicase complex assisted unwinding of the 5′UTR, in binding to eIF3, the anisotropy binding assays were done in the absence of ATP and in the presence of AMPPNP (a non-hydrolysable analogue of ATP) (Figure 3D). In the absence of ATP (when only eIF4A, eIF4B and eIF4F were added), or in the presence of the helicase complex and AMPPNP; eIF3–5′UTR binding was completely abolished, showing that an active helicase complex is necessary for binding of the 5′UTR RNA to eIF3.
Both BYDV UTRs can bind to eIF3 simultaneously
Gel shift assays were performed to confirm if the 5′UTR and 3′BTE RNAs bind to eIF3 simultaneously, possibly without competing (Figure 4A). The UTRs either individually, or together (with one UTR in 3-fold excess) were incubated with the helicase complex and eIF3. The complexes were resolved in 1% native agarose gels. The 5′UTR and the 3′BTE, both individually and together, showed clear shifts when bound to eIF3. The combined 3′ and 5′UTR shift displays a strong, simultaneous interaction of both the UTRs with eIF3 (Figure 4A). None of the negative control RNAs showed significant shifts in the presence of eIF3 (Figure 4A). These results further indicate that eIF3 may be involved in stabilizing the long-distance interaction between the two UTRs of BYDV, and may also help recruit/transfer 40S ribosomal subunits from the 3′BTE to the 5′UTR. In anisotropy binding assays, there was a slight change in the 5′UTR binding affinity to eIF3 in the presence of unlabeled 3-fold molar excess of the 3′BTE in the reaction mixture (Table 1, Kd = 86.8 ± 4.7 nM), with a slight increase in the overall anisotropy change, indicating that the 5′UTR and 3′BTE RNAs could bind to eIF3 simultaneously. A double reciprocal plot showed that a 3-fold molar excess of the 3′BTE did not compete for 5′UTR binding to eIF3 (Figure 4B). These data taken together with the gel shift data indicate that both the UTRs and eIF3 form a single ribonucleoprotein complex.
Figure 4.

(A) Gel shift assay showing simultaneous binding of BYDV UTRs to eIF3. Both the RNAs were pre-incubated with the helicase complex and then eIF3 was added. The MNeSV ISS RNA, polyUC RNA and 18S rRNA derived oligonucleotide were also assessed for binding to eIF3. Additional details are described in Materials and Methods. (B) For competition binding assays, 50 nM of labeled 5′UTR was pre-incubated with the helicase complex either with or without added 150 nM unlabeled 3′BTE for 20 min at 25°C. After the incubations, the sample was titrated with eIF3. The assays were performed in 20 mM HEPES pH 7.6, 5 mM MgCl2, 200 mM KCl.
eIF3 mediates 40S recruitment to both UTRs, increasing 40S-3′UTR interaction by >20-fold and is absolutely required for 40S-5′UTR binding
In the current model for 3′BTE-mediated translation, 40S ribosomal subunits bind to the 3′BTE (in the presence of the helicase complex, Kd = 120 ± 10 nM) (15), and are subsequently (or simultaneously) transferred to the 5′UTR for translation to initiate. To assess the effect of eIF3 in this recruitment, we performed fluorescence anisotropy assays in the presence of eIF3 and helicase complex. The 3′BTE-40S binding affinity increased significantly (Figure 5, Kd = 4.9 ± 0.4 nM). 5′UTR RNA showed no binding to 40S ribosomal subunits in the presence of only the helicase complex but showed a very tight and specific binding to 40S ribosomal subunits (Figure 5, Kd = 4.2 ± 0.9 nM) when eIF3 was added along with the helicase complex. There was no significant difference in the 5′UTR-40S binding affinity; even when 3-fold molar excess of the unlabeled 3′BTE was added during the titration (Figure 5, Kd = 2.7 ± 0.6 nM). These results confirmed that in the presence of eIF3, both the UTRs could bind to 40S ribosomal subunits simultaneously, with nanomolar affinities within the physiological range known for these interactions (34).
Figure 5.

Fluorescence anisotropy binding of BYDV UTRs to wheat 40S ribosomal subunit. The fluorescein labeled RNAs (50 nM) were pre-incubated with the helicase complex, eIF3 (150 nM) and ATP (5 mM) at 25°C for 20 min, and then titrated with 40S ribosomal subunits (2 μM). Both the UTRs showed very tight binding to 40S subunits in the presence of the helicase complex and eIF3 (BTE-40S, Kd = 4.9 ± 0.4 nM, and 5′UTR-40S, Kd = 4.2 ± 0.9 nM). The 5′UTR showed no binding to 40S subunits in the absence of eIF3, but in the presence of eIF3, the 5′UTR showed very tight binding to 40S ribosomal subunits even in the presence of a 3-fold molar excess of unlabeled 3′BTE RNA (Kd = 2.7 ± 0.6 nM).
DISCUSSION
BYDV utilizes an unconventional pathway for translation initiation by recruiting ribosomes at the 3′BTE, with the help of a subset of the host eIFs: namely eIF4F, eIF4A and eIF4B (15). However, for mRNA translation to begin, this recruited ribosomal complex must transfer to the 5′UTR and, at some point, must interact with both UTRs simultaneously. The only mechanism known to aid this transfer is the long distance, RNA–RNA kissing loop interaction between five bases in the 3′BTE and 5′UTR (10,11). It seems unlikely that the five Watson-Crick pairs alone could maintain a stable interaction long enough for the transfer of this large, ∼2 MDa complex. Moreover, in the presence of the eIFs reported to recruit 40S ribosomal subunits at the 3′BTE, the 5′UTR did not show binding to 40S ribosomal subunits suggesting involvement of additional factor(s) in the transfer/recruitment at the 5′UTR. To better understand the 5′UTR–3′UTR interaction and the transfer of the 40S–eIF complex to the 5′UTR, we identified additional proteins bound to the ribosomal complex recruited at the 3′BTE. To do this, we applied a pull-down based strategy using the 3′BTE as bait. As expected, eIF4G and multiple 40S ribosomal proteins were detected, along with eIF4A, eIF2 subunit 3 (eIF2 subunit gamma) and twelve subunits of eIF3 (subunits a, b, c, d, e, f, g, h, i, k, l, m) (supplementary excel sheet 1). Using fluorescence anisotropy binding measurements, we showed that purified native wheat germ eIF3 binds specifically to both the 3′BTE and the 5′UTR RNAs, in the presence of an active helicase unwinding complex. This is the first report showing direct, specific binding of the 40S–eIF complex to the BYDV 5′UTR. It was evident from the anisotropy binding and gel shift assays that both the UTRs could bind to eIF3 and the 40S–eIF complex simultaneously. In canonical translation initiation, eIF3 interacts with several initiation factors, the 40S ribosomal subunit and thus plays a central role in assembly of pre-initiation complexes (35–37). A few recent reports have shown that mammalian eIF3 binds specifically to defined structural elements in a set of mRNAs involved in cell growth control and regulates their expression (29,38,39). However, for some of the 5′UTRs that showed immunoprecipitation with eIF3 in cell lysates, it has been proposed that additional factors are required for binding of eIF3 to the target stem loops (29). BYDV UTRs are structured and it would be interesting to understand the sequence/structure motifs, and the mechanism involved in 3′BTE-5′UTR-eIF3 interactions. Our data indicate that eIF3′s binding to the 3′BTE is not solely indirect through interaction with the eIF4G subunit of eIF4F, because eIF3 could bind to the 3′BTE even when eIF4F was not added (Figure 3C). The additional roles of eIF3 in 3′BTE-mediated translation, apart from bridging the two UTRs and helping in the transfer of translation ribosomal machinery from the 3′ to 5′UTR during translation initiation, are currently not known.
Figure 6 summarizes the binding affinities of various eIFs to the 3′BTE and 5′UTR RNAs that have been reported so far (12,15), including the ones from this study. eIF4F plays a central role in BTE-mediated translation as it binds to the 3′BTE (via the eIF4G subunit), and determines the efficiency of mRNA translation (12–14). In a recent report, the core domain of eIF4G (MIF4G) along with a small region immediately upstream of MIF4G (eIF4G601–1196) (14,40) was shown to be crucial for efficient binding to the 3′BTE and for 3′BTE-mediated translation of a reporter gene in wheat germ lysates. This eIF4G601–1196 includes the eIF3 binding site, along with eIF4A/4B and RNA binding sites (14), suggesting possible roles of these eIFs in 3′BTE-mediated translation. eIF4F binds specifically and very tightly to the 3′BTE (29 ± 3 nM, Figure 6), and the helicase unwinding factors, eIF4A and eIF4B bind only moderately. All of these eIFs combined as a helicase unwinding complex (eIF4A, eIF4B, eIF4F and ATP) have been shown to stimulate binding of 40S ribosomal subunits to the 3′BTE (15). The 3′BTE can facilitate protein translation even when it is present in the 5′UTR context and in that case the 5′UTR of BYDV is dispensable (6,7,41). None of these eIFs have been shown to interact directly with the 5′UTR (13), making the 3′BTE a suitable candidate for initial recruitment of these eIFs and the 40S ribosomal subunit. We showed a direct and specific binding of 40S ribosomal subunits to the 5′UTR in the presence of the helicase complex and eIF3. We believe the helicase complex is necessary for eIF3 binding to partially unwind the complex 5′UTR structure and provide a ‘landing’ for eIF3 and that eIF3 must still recognize structural and likely sequence elements of the 5′ UTR. Collectively, our results expand the earlier proposed model (15) providing substantial evidence that 40S ribosomal subunits are initially recruited to the 3′BTE, identifying an essential role for eIF3 and providing insight into the mechanism of 40S PIC transfer from the 3′ BTE to the 5′ UTR. According to our new model (Figure 7), the first step in 3′BTE-mediated translation is binding of eIF4F (via the eIF4G subunit) to the 3′BTE. No helicase unwinding activity is required for this interaction, but recently it was shown that the helicase complex enhances binding of eIF4G to the 3′BTE (14). The conventional synergistic initiation factor interactions including helicase unwinding complex eIF4A (via binding to the eIF4G subunit of eIF4F), eIF4B and ATP stimulates and recruits the 40S ribosomal subunit possibly as the preformed 43S pre-initiation complex which includes eIF3, at the 3′BTE (3,14). This 43S PIC recruitment may also involve the complementary base pairing of the now unwound six base tract (GAUCCU) in the 17 nt conserved 3′BTE sequence, to the corresponding sequence in 18S rRNA acting as a lynchpin for proper alignment of the BTE and 40S subunit (3,6,15), although this interaction has not been proved experimentally. The kissing loop interaction is relatively weak and may serve to correctly align the 3′BTE and the 5′UTR. The presence of eIF3 serves to stabilize this interaction. The kissing loop interaction and simultaneous binding of both the UTRs to eIF3 ensure that the long-range RNA interactions enabling efficient 40S or PIC transfer and subsequent translation. The recruitment of 43S PIC at the 3′BTE has the added advantage of keeping the translation and transcription processes separate on the viral RNA templates (42,43).
Figure 6.
Summary of the binding affinities of BYDV UTRs for different eIFs (eIF4A, eIF4B, eIF4F) and 40S including eIF3. In the presence of eIF3, both the UTRs showed very tight binding to 40S subunits within the physiological range reported for these complexes.
Figure 7.
Model for recruitment of the pre-initiation complex to the 5′UTR via 3′BTE. The eIF4F complex binds directly to SLI of the 3′BTE, and recruits eIF4A and eIF4B via the conventional factor-factor interactions. In the presence of ATP, eIF3 (possibly as a part of the 43S pre-initiation complex) binds to the 3′BTE (via direct binding and through interaction of eIF3 with eIF4G) and the 40S/PIC is recruited. Subsequently (or simultaneously), the kissing loop interaction between SLIII (of the 3′BTE) and SL-D (of the 5′UTR, BCL) brings the UTRs together in appropriate alignment enabling eIF3 to stabilize the interaction through direct binding to the 5′ UTR. This stable complex, with the assistance of the helicase complex is able to transfer the 43S PIC to the 5′UTR for translation to begin. Additionally, eIF4A, eIF4B and ATP unwinding of the sequence GAUCCU in the 3′BTE, which is complementary to a stretch of bases in 18S rRNA may also act as a lynchpin to recruit the complex at the 3′BTE.
This model resembles the model proposed for Hepatitis C Virus internal ribosome entry site (HCV IRES) mediated translation initiation. For start codon recognition and translation initiation, HCV IRES only requires eIF2 and eIF3 (44,45). The simultaneous binding of the BYDV 3′BTE and 5′UTR to eIF3 and the 40S subunit resembles the HCV IRES and the 3′UTR interaction with eIF3 and the 40S ribosomal subunit (15,33,46–48). HCV IRES recruits the 40S ribosomal subunit at the 5′UTR while the HCV 3′UTR has been proposed to stimulate IRES-dependent translation by capturing and redelivering the 40S ribosomal subunit (and eIF3) to the IRES for subsequent rounds of translation (48). However, in 3′BTE-mediated translation, the 3′BTE plays a more direct role in recruiting necessary translational machinery. The binding site for HCV-like IRESs on the 40S ribosomal subunit has been shown to overlap with the binding site for eIF3, thus requiring rearrangements and displacement of eIF3 upon binding of the IRES to the 40S ribosomal subunit (32,33). Although displaced from the 40S ribosomal surface, eIF3 still remains attached to the 40S-IRES complex via interaction with the IRES. Structural studies are required to understand if the 3′BTE and/or 5′UTR employ similar strategies in BTE-mediated translation to capture and deliver the translation machinery from the 3′BTE to the 5′UTR.
We detected some ribosomal proteins in our mass spectrometry analysis (supplementary excel sheet 1) that have been shown to interact directly with the HCV IRES and the HCV 3′UTR (48). These included RPS3a (ribosomal protein S3a), which has been reported to interact with both the IRES and 3′UTR of HCV RNA. Ten unique peptides were detected for RPS4, which has been reported to interact with the 3′UTR of HCV (48) and has been proposed to be possibly involved in anchoring and positioning of HCV IRES 3′X region to make weak contacts with RPS8 and RPL22 in the context of the 80S ribosome. Many of the other detected 40S ribosomal proteins have been implicated in various cap-independent translation initiation mechanisms by different RNA viruses. RPS6 was detected with 8 unique peptides. RPS6 is located in the small head region of the 40S subunit and has been shown to be a critical host translational component involved in viral infections of cap-independent viruses (49). RPS6 silenced plants have been shown to support Tobacco mosaic virus accumulation (capped RNA) in the inoculated leaves but not Turnip mosaic virus and Tomato bushy stunt virus (both utilize cap-independent mechanisms) (49). RPS6 is also required for Drosophila C virus and Poliovirus, which both utilize cap-independent translation that is initiated by IRESs in their 5′UTRs (50). Collectively, these data indicate that the unique translation initiation mechanism utilized by BYDV which involves a CITE in its 3′UTR has similarities with a variety of other, non-canonical viral translation pathways involving IRESs. A better understanding of the BYDV mechanism may add to our understanding of the translation initiation mechanisms employed by these pathogens and could help us in devising universal methods to control them.
In conclusion, based on these observations, we propose here that in BYDV 3′BTE-mediated translation, eIF3 binds to both the UTRs, stabilizes the 3′UTR-5′UTR interaction and helps in the transfer of the translation machinery from the 3′BTE to the 5′UTR. Although we observed that 40S ribosomal subunit could directly bind to the 5′UTR in the presence of the helicase complex and eIF3 in the in vitro assays, the role of the 3′BTE in vivo seems to be more than just recruiting the 43S PIC. Upon 3′BTE binding, crucial conformational changes and structural rearrangements in the translation initiation complex may lead to locking the recruited complex in an active, scanning compatible form. Structural and kinetic studies of this 3′BTE recruited complex could prove invaluable in understanding this mechanism better.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Solomon Haizel, Dr Somdeb Mitra and Prof. Ruben Gonzalez (Columbia University, New York City, NY) for helpful discussions. We thank Prof. D.R. Gallie (University of California, Riverside, CA) and Prof. K.S. Browning (University of Texas at Austin, Austin, TX) for the recombinant eIF clones and wheat eIF4G antibodies used in this study. We acknowledge the proteomics support provided by Prof. Beatrix Ueberheide and Joshua Andrade (The Proteomics Resource Center, NYU Langone School of medicine). The Mass Spectrometric experiments were in part supported by NYU Langone School of Medicine.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [MCB 1513737 to D.J.G.]; Hunter College President's Fund for Faculty Advancement (to D.J.G.). P.P. was supported by the National Science Foundation [IGERT 0965983 to Hunter College]. Funding for open access charge: NSF grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Firth A.E., Brierley I.. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012; 93:1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon A.E., Miller W.A.. 3′ cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 2013; 67:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miras M., Miller W.A., Truniger V., Aranda M.A.. Non-canonical translation in plant RNA viruses. Front Plant Sci. 2017; 8:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller W.A., Rasochova L.. Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 1997; 35:167–190. [DOI] [PubMed] [Google Scholar]
- 5. Wang S., Miller W.A.. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J. Biol. Chem. 1995; 270:13446–13452. [DOI] [PubMed] [Google Scholar]
- 6. Wang S., Browning K.S., Miller W.A.. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997; 16:4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo L., Allen E., Miller W.A.. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000; 6:1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z., Kraft J.J., Hui A.Y., Miller W.A.. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology. 2010; 402:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Truniger V., Miras M., Aranda M.A.. Structural and functional diversity of plant virus 3′-Cap-Independent translation enhancers (3′-CITEs). Front. Plant Sci. 2017; 8:2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo L., Allen E.M., Miller W.A.. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001; 7:1103–1109. [DOI] [PubMed] [Google Scholar]
- 11. Rakotondrafara A.M., Polacek C., Harris E., Miller W.A.. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006; 12:1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banerjee B., Goss D.J.. Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3′-untranslated region correlates with translation efficiency. J. Biol. Chem. 2014; 289:4286–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Treder K., Kneller E.L., Allen E.M., Wang Z., Browning K.S., Miller W.A.. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008; 14:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao P., Liu Q., Miller W.A., Goss D.J.. Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3′ cap-independent translation element (BTE). J. Biol. Chem. 2017; 292:5921–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharma S.D., Kraft J.J., Miller W.A., Goss D.J.. Recruitment of the 40S ribosome subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J. Biol. Chem. 2015; 290:11268–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller W.A., Jackson J., Feng Y.. Cis- and trans-regulation of luteovirus gene expression by the 3′ end of the viral genome. Virus Res. 2015; 206:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bachler M., Schroeder R., von Ahsen U.. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA. 1999; 5:1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Locker N., Lukavsky P.J.. A practical approach to isolate 48S complexes: affinity purification and analyses. Methods Enzymol. 2007; 429:83–104. [DOI] [PubMed] [Google Scholar]
- 19. Zuker M., Jacobson A.B.. Using reliability information to annotate RNA secondary structures. RNA. 1998; 4:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morch M.D., Drugeon G., Zagorski W., Haenni A.L.. The synthesis of High-Molecular-Weight proteins in the Wheat-Germ translation system. Method Enzymol. 1986; 118:154–164. [Google Scholar]
- 22. Windbichler N., Schroeder R.. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat. Protoc. 2006; 1:637–640. [DOI] [PubMed] [Google Scholar]
- 23. Mayberry L.K., Dennis M.D., Leah Allen M., Ruud Nitka K., Murphy P.A., Campbell L., Browning K.S.. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol. 2007; 430:397–408. [DOI] [PubMed] [Google Scholar]
- 24. Montesano L., Glitz D.G.. Wheat germ cytoplasmic ribosomes. Localization of 7-methylguanosine and 6-methyladenosine by electron microscopy of immune complexes. J. Biol. Chem. 1988; 263:4939–4944. [PubMed] [Google Scholar]
- 25. Goss D.J., Rounds D.J.. A kinetic light-scattering study of the binding of wheat germ protein synthesis initiation factor 3 to 40S ribosomal subunits and 80S ribosomes. Biochemistry. 1988; 27:3610–3613. [DOI] [PubMed] [Google Scholar]
- 26. Lax S.R., Lauer S.J., Browning K.S., Ravel J.M.. Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol. 1986; 118:109–128. [DOI] [PubMed] [Google Scholar]
- 27. Sun C., Todorovic A., Querol-Audi J., Bai Y., Villa N., Snyder M., Ashchyan J., Lewis C.S., Hartland A., Gradia S. et al.. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20473–20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pisarev A.V., Kolupaeva V.G., Yusupov M.M., Hellen C.U., Pestova T.V.. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008; 27:1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee A.S., Kranzusch P.J., Cate J.H.. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015; 522:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. des Georges A., Dhote V., Kuhn L., Hellen C.U., Pestova T.V., Hashem Y.. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015; 525:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Erzberger J.P., Stengel F., Pellarin R., Zhang S., Schaefer T., Aylett C.H.S., Cimermancic P., Boehringer D., Sali A., Aebersold R. et al.. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014; 159:1227–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U., Pestova T.V., Frank J.. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013; 153:1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Pestova T.V., Hellen C.U., Frank J.. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013; 503:539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maag D., Fekete C.A., Gryczynski Z., Lorsch J.R.. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005; 17:265–275. [DOI] [PubMed] [Google Scholar]
- 35. Burks E.A., Bezerra P.P., Le H., Gallie D.R., Browning K.S.. Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J. Biol. Chem. 2001; 276:2122–2131. [DOI] [PubMed] [Google Scholar]
- 36. Hinnebusch A.G. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006; 31:553–562. [DOI] [PubMed] [Google Scholar]
- 37. Aitken C.E., Beznoskova P., Vlckova V., Chiu W.L., Zhou F., Valasek L.S., Hinnebusch A.G., Lorsch J.R.. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. Elife. 2016; 5:e20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee A.S., Kranzusch P.J., Doudna J.A., Cate J.H.. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016; 536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cate J.H. Human eIF3: from ‘blobology' to biological insight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017; 372:doi: 10.1098/rstb.2016.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kraft J.J., Treder K., Peterson M.S., Miller W.A.. Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res. 2013; 41:3398–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S., Guo L., Allen E., Miller W.A.. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999; 5:728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barry J.K., Miller W.A.. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller W.A., White K.A.. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006; 44:447–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kieft J.S., Zhou K., Jubin R., Doudna J.A.. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001; 7:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U.. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998; 12:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai Q., Todorovic A., Andaya A., Gao J., Leary J.A., Cate J.H.. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J. Mol. Biol. 2010; 403:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun C., Querol-Audi J., Mortimer S.A., Arias-Palomo E., Doudna J.A., Nogales E., Cate J.H.. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013; 41:7512–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bai Y., Zhou K., Doudna J.A.. Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 2013; 41:7861–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang C., Zhang C., Dittman J.D., Whitham S.A.. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology. 2009; 390:163–173. [DOI] [PubMed] [Google Scholar]
- 50. Cherry S., Doukas T., Armknecht S., Whelan S., Wang H., Sarnow P., Perrimon N.. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005; 19:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






