Abstract
Expression QTL (eQTL) detection has emerged as an important tool for unraveling the relationship between genetic risk factors and disease or clinical phenotypes. Most studies are predicated on the assumption that only a single causal variant explains the association signal in each interval. This greatly simplifies the statistical modeling, but is liable to biases in scenarios where multiple local causal-variants are responsible. Here, our primary goal was to address the prevalence of secondary cis-eQTL signals regulating peripheral blood gene expression locally, utilizing two large human cohort studies, each >2500 samples with accompanying whole genome genotypes. The CAGE (Consortium for the Architecture of Gene Expression) dataset is a compendium of Illumina microarray studies, and the Framingham Heart Study is a two-generation Affymetrix dataset. We also describe Bayesian colocalization analysis of the extent of sharing of cis-eQTL detected in both studies as well as with the BIOS RNAseq dataset. Stepwise conditional modeling demonstrates that multiple eQTL signals are present for ∼40% of over 3500 eGenes in both microarray datasets, and that the number of loci with additional signals reduces by approximately two-thirds with each conditioning step. Although <20% of the peak signals across platforms fine map to the same credible interval, the colocalization analysis finds that as many as 50–60% of the primary eQTL are actually shared. Subsequently, colocalization of eQTL signals with GWAS hits detected 1349 genes whose expression in peripheral blood is associated with 591 human phenotype traits or diseases, including enrichment for genes with regulatory functions. At least 10%, and possibly as many as 40%, of eQTL-trait colocalized signals are due to nonprimary cis-eQTL peaks, but just one-quarter of these colocalization signals replicated across the gene expression datasets. Our results are provided as a web-based resource for visualization of multi-site regulation of gene expression and its association with human complex traits and disease states.
Keywords: fine mapping, linkage disequilibrium, conditional association, colocalization, PolyQTL, gene regulation
SINCE the first genome-wide association study (GWAS) results were published in 2005 (Klein et al. 2005), thousands of genetic regions in human chromosomes have been found to be associated with human phenotypes, including disease states (Visscher et al. 2017). Since it is now assumed that the majority of single nucleotide polymorphism (SNP)-trait associations identified by GWAS can be attributed to effects on gene expression, precise estimation of the location and effect sizes of regulatory polymorphisms has become important for understanding the relationship between genetic and phenotypic variation (Maurano et al. 2012; Farh et al. 2015). The minimal expectation is that expression quantitative trait loci (eQTL) analysis can identify the gene within a locus that accounts for a GWAS signal, although it has become clear that even this goal is far from trivial (Chung et al. 2014; Pickrell 2014). Many investigators make the stronger assumption that colocalization of regulatory variants (eSNPs) and GWAS signals to a tight linkage disequilibrium interval implies the ability to define, if not the causal variant, then at least a credible set of SNPs that include the causal site (Trynka et al. 2013; Gaulton et al. 2015; Kichaev and Pasaniuc 2015; Liu et al. 2015).
However, high resolution fine mapping of eQTL results aligned with GWAS studies for diverse phenotypes has, as yet, provided only dozens of instances with unambiguous evidence that a specific variant affects a human complex trait or disease through its effect on gene expression. Several recent studies have begun to question the presumed identity of eQTL and GWAS hits: even though there is a highly significant overlap at the level of the locus, it is not so clear that the precise variants are the same. For example, Farh et al. (2015) estimated that only ∼10% of the GWAS hits take function as eQTL despite the vast majority of those hits mapping to noncoding DNA. Similarly, two recent studies of autoimmune disease have also argued that only around one-quarter of examined GWAS loci may act as eQTL in the profiled immune cells (Chun et al. 2017; Huang et al. 2017). Furthermore, work based on GTEx gene expression profiling aiming to integrate GWAS and eQTL results found that only a minority of GWAS loci match precisely to eQTL, while the diversity of regulatory effects across tissues can complicate interpretation (Hormozdiari et al. 2016; Gamazon et al. 2018). These results raise the question of why there are so many instances of discordant fine localization: are we simply limited by the low statistical power to detect association signals (Udler et al. 2010), is there mis-estimation of signal strength and location in the case of multiple eQTL per transcript (Zeng et al. 2017), or are regulatory effects so cell-type and context-specific that true colocalization is often missed? In this study, we will focus on the first two issues by addressing the concordance of signals in two large eQTL datasets where the expectation was that, despite technical differences between the platforms, shared cis-eQTL signals at the gene level would map to the same credible intervals.
The detection of eQTL is dependent on the accuracy of the technologies designed to estimate transcript abundance (gene expression) and to genotype or impute genetic variants. Genotype calling, whether based on gene chip platforms or whole-genome sequencing, is thought to be highly accurate and robust (1000 Genomes Project Consortium 2015), and methods for imputation of missing genotypes are now generally accepted to be valid for minor allele frequencies of 0.01 or higher. Constraints on gene expression measurement are more problematic, being subject both to the properties of the detection method and of the algorithms use to statistically analyze the data. Microarrays, principally Illumina- and Affymetrix-based for human studies, have been used widely to measure gene expression, and have supported the development of eQTL analyses. By far the largest published study is the 12,000 sample Blood eQTL compendium (Westra et al. 2013), now approaching 30,000. However, the nature of microarray probes provides incomplete coverage of the exons within genes, and there are analytical limitations due to the dynamic range of quantitative detection of expression, with the result that estimates of transcript abundance are strongly platform-specific. eQTL artifacts are also known to arise due to linkage disequilibrium between regulatory variants and SNPs located within transcript probes. Nevertheless, well-powered studies have detected primary eQTL for over half of all expressed genes in blood, providing ample opportunity to compare the fine-mapping of these signals (Lloyd-Jones et al. 2017).
A small number of studies have argued for high replicability of eQTL detected on the same platform. The Genotype-Tissue Expression (GTEx) project discovered eQTLs from postmortem analysis of over 40 tissues, finding extensive sharing of promoter-proximal signals for around half the loci (Ardlie et al. 2015). Zhernakova et al. (2017) found that 84% of previous cis-eQTL genes detected on an Illumina platform replicated in an RNAseq data set, the vast majority showing the same direction of allelic effect. Multiple Illumina-based peripheral blood studies carried out on different cohorts by different groups have also reported in excess of 70% shared signals for eQTL detected at 5% false discovery rates (FDRs; Zeller et al. 2010; Lloyd-Jones et al. 2017). However, differences between platforms seem to be much larger than expected; for example, only between one-quarter and one-third of eQTL association signals in the MRCE Illumina-based study replicated in a companion MRCE Affymetrix study (Liang et al. 2013). The differences may in part be due to the differential effects of alternative splicing on transcript abundance detected with probes that cover one or a few exons (Illumina) or more of the extent of each gene (Affymetrix), or to the effects of the normalization and other statistical procedures that are used to associate genotypes with transcript abundance estimates. It is also important to recognize that what is described as a shared signal where a genotype associates with gene expression in two studies may often simply reflect linkage disequilibrium between two independent signals.
Consequently, methods have been developed to evaluate and fine-map colocalization signals, whether across gene expression platforms, or between eQTL and GWAS signals. Most of the current methods seek to distinguish true colocalization from “shared” signal due to linkage disequilibrium (LD). COLOC was one of the first Bayesian methods which evaluates the relative statistical support of each eQTL-GWAS colocalization hypothesis contingent on LD (Giambartolomei et al. 2014). However, COLOC assumes the default model that a single-causal eQTL exists, which implies a strong prior that variants taking function as eQTL (or associated with a trait), also affect the trait (or expression), potentially leading to false positive colocalization. SMR, or Summary Mendelian Randomization, jointly evaluates the strength of eQTL and GWAS signals using a procedure known as HEIDI to filter heterogeneity of GWAS and eQTL signals in the presence of LD (Zhu et al. 2016). SMR is strongly dependent on the accuracy of LD inference from a reference panel, and the HEIDI test has been reported to be conservative. Another Bayesian method, eCAVIAR, calculates a posterior probability of eQTL-GWAS colocalization while allowing for multiple signals in the interval (Hormozdiari et al. 2016). The dependencies of all these methods on sample size have not been well characterized, and we found only ∼50% agreement between them in evaluation of causal variants in a Crohn’s disease study (Marigorta et al. 2017). Furthermore, lack of control for population structure or relatedness requires further modification when applied to data sets with large sample size.
In this study, we collected cis-eQTL results from three large data sets following a strategy summarized in Supplemental Material, Figure S1, and developed a statistical pipeline to achieve the following goals: (a) to evaluate the prevalence of multiple cis-eQTL regulation in human peripheral blood; (b) to estimate the extent of eQTL signal sharing across three expression platforms; and (c) to detect colocalization of eQTL signals with GWAS hits contingent on the LD at each locus, revealing the possible biological regulatory mechanisms linking genetic variants to complex human phenotypes.
Materials and Methods
Datasets
We analyzed three different peripheral blood eQTL data sets. The Consortium for the Architecture of Gene Expression (CAGE) dataset (Lloyd-Jones et al. 2017) consists of Illumina HT12 v3 microarray-based gene expression profiles, as well as whole genome genotype information, from five research studies: the Brisbane Systems Genetics Study (BSGS, N = 926) (Powell et al. 2012), Atlanta-based Centre for Health Discovery and Well-Being (CHDWB, N = 439) (Wingo and Gibson 2015) and Emory Cardiology Genebank (N = 147, Kim et al. 2014), Estonian Genome Centre—University of Tartu (EGCUT) study (N = 1065, Schramm et al. 2014), and the Morocco Lifestyle study (N = 188, Idaghdour et al. 2010), for a total of 2765 individuals. Institutional review board (IRB) approval was obtained for the combination of data into a mega-analysis both by the University of Queensland and for each participating site.
The second dataset from the Framingham Heart Study (FHS) (Huan et al. 2015) contains two-generation data generated on the Affymetrix Human Exon Array ST 1.0 for gene expression, and the Affymetrix 500K mapping array and Affymetrix 50K gene-focused MIP array for genotyping. A total of 5075 participants with both genotype and gene expression information from the offspring (N = 2119, eighth examination) and third-generation (N = 2956, second examination) cohorts were included in this study. Raw genotype and gene expression data were downloaded from dbGAP (phs000007.v25.p9) with IRB approval.
The BIOS RNAseq summary data were derived from a meta-analysis of results for a total of 2100 participants from four cohorts (Zhernakova et al. 2017): the Cohort on Diabetes and Atherosclerosis Maastricht (CODAM, 184 individuals included); LifeLines-DEEP (LLD, 626 individuals); the Leiden Longevity Study (LLS, 654 individuals); and the Rotterdam Study (RS, 652 individuals). We downloaded only the summary results of cis-eQTL signals from https://genenetwork.nl/biosqtlbrowser/, so were unable to perform the sequential stepwise regression analyses to detect secondary signals.
Genotype imputation
Genotype imputation for the CAGE cohort was performed jointly for the five contributing studies to ensure uniformity of assignment of strand identities of SNPs, and is described in detail in Lloyd-Jones et al. (2017) and at https://github.com/CNSGenomics/impute-pipe. Briefly, the pipeline involved preimputation quality control, and data-consistency checks, imputation to the 1000G reference panel with Impute2 (Howie et al. 2012), postimputation quality control (filtering on various data features), and merging of the datasets on common SNPs.
For the Framingham Heart Study data, there were a total of 6950 individuals before imputation, from which 29 individuals with genotype missing rate ≥5% were removed. Subsequently, any SNPs with genotype missing rate ≥5% were also removed along with SNPs with Hardy-Weinberg test P ≤ 10−6. Prior to imputation, the genotypes were prephased using Shapeit2 (Delaneau et al. 2013) using the “duohmm” parameter to account for pedigree information. Each chromosome was divided into 5 Mb chunks, incorporating the centromere-adjacent region (acen region) into the neighboring chunk, and similarly joining any chunk with <200 SNPs into a neighboring chunk. Imputation was performed with Impute2 (Howie et al. 2012), using qctool to convert gprobs to gen file format, and only SNPs with info value >0.3 were retained for subsequent analyses. The gen file was converted to plink file format, and SNPs with multiple alleles as well as InDel variants were filtered out. The remaining variants were further reduced to ∼6 million SNPs with a >95% call rate across all 5075 individuals represented by both genotype and gene expression data.
To be sure that imputation accuracy did not bias our results, we asked whether there is any relationship between imputation info score and peak eQTL signal detection. In CAGE, 72% of peak signals had info score >0.9, compared with 66% of all remaining SNPs—a slight but nonsignificant excess. Similarly, in FHS there was no relationship between info score and peak detection or number of peak. However, it should be recognized that missing variants not called by the imputation algorithm could account for ∼10% of all eQTL peaks (Zeng et al. 2017).
Probe reannotation
Since SNP imputation for the CAGE cohort was based on hg19/GRCH37, whereas the Illumina probe annotation was based on hg18/GRCH36, we reannotated the probe information by mapping the probe sequences to hg19/GRCH37 with BWA (Li and Durbin 2009), retaining only the uniquely mapped probes. All probe sequences were secondarily mapped to the reference genome with BLAT (Kent 2002), and only probe sequences uniquely mapped with both methods were determined to be high confidence and subsequently used for eQTL detection. Of a total of 45,931 probes mapped to the reference genome, 7349 probe sequences mapped to multiple regions or remained unmapped, leaving 38,582 probes taken forward for the eQTL analyses. See Table S1 for summary statistics. Since it is well known (Walter et al. 2007) that SNPs in a probe influence microarray hybridization, we also discarded 3856 Illumina probes containing SNPs with minor allele frequencies (maf) >1% in the 1000 Genomes European sample (Lappalainen et al. 2013). Similarly, SNPs in the Affymetrix probesets were also converted to positions in the hg19 assembly by applying liftOver (UCSC Genome Browser) to the GPL5188 annotation file downloaded from dbGAP, and annotated to the 1000 Genomes dataset. Any SNP with a maf >1% in the 1000 Genome European population, and located within a probeset, was deemed to be potentially unreliable, and was included as a covariate during the eQTL estimation steps. Among the 280,000 core probesets, 35,000 (12.5%) have such SNPs, and 15,368 transcripts contain at least one SNP in a probe.
Gene expression normalization
The gene expression normalization strategy for CAGE required aggressive procedures to account for study-specific biases, as described in detail in Lloyd-Jones et al. (2017). It consisted of five steps: (1) Variance stabilization using the vsn package (Lin et al. 2008); (2) Quantile normalization forcing the intensity distribution across all probes to have the same shape for all samples; (3) Batch effect correction via linear regression to account for known technical effects, such as RNA extraction date, and physical batch; (4) Batch effect correction [via principal component (PC) analysis, removing the first 10 PC to account for unknown confounding procedural, or population-based influences]; and (5) Rank normal transformation, namely a final transformation of each probe to a normal distribution with mean 0 and variance 1.
For the FHS data, raw gene expression processed by Affymetrix APT software (version 1.12.0) was downloaded from dbGAP, log2 transformed, and surrogate variable analysis (SVA) (Leek et al. 2012) was used to remove confounding factors, fitting a total of 62 surrogate variables by a linear regression model. Note that the published FHS gene expression study (Huan et al. 2015) reported results of a different normalization that included fitting blood cell counts, which we chose to avoid since a similar procedure was not applied to the CAGE data, and because the blood counts were abrogated by the SVA fitting.
Multi-site eQTL detection
For this study, local SNPs were stringently defined as SNPs located within 200 kb upstream or downstream of the gene (defined as the first TSS and last TES listed in the hg19 annotation) containing the probe. Sequential conditional analyses were performed for each probe, and the genes with significant eSNPs were called eGenes. Since both the CAGE and FHS cohorts contain family-based data [the former for a quarter of the samples, from the BSGS twin study (Powell et al. 2012); the latter for all participants], a mixed linear model was used for eQTL detection in GEMMA (Zhou and Stephens 2012), which fits a genetic relatedness matrix (GRM) as a covariate alongside fixed genotype effects. The multiTrans tool (Joo et al. 2016), which accounts for family structure, was used to specify a study-wise FDR of 5% for genes with multiple independent eSNPs, which was empirically observed to be approximately P < 10−5. After first scanning for evidence of at least one local eSNP at this threshold, the residuals after fitting the sentinel SNP were used in a sequential conditional scan for an independent secondary eSNP. This process was iterated until no more signals were observed below P = 10−5. SNPs in high LD with each previously detected signal (r2 ≥ 0.9) were also filtered out of each sequential analyses. The effect sizes of each discovered SNP were recorded as the sequential conditional estimates. Subsequently, for the multi-site effect size estimates, all discovered independent peak SNPs were fit with the GRM in one mixed model. However, since the GEMMA software does not report the effect sizes of all fixed effects simultaneously, we fit the multi-site models with one SNP specified as the target effect, including the other significant SNPs as fixed effects, as well as the GRM, as covariates. This estimation procedure was repeated for each included SNP, controlling for relatedness, recording the effect size of the target SNP as the multi-site effect. Note that the total amount of variance explained is the same for all such models fit for each gene. To control the influence of SNPs located in probes in the FHS data, we incorporated in-probe SNPs with an LD r2 cutoff 0.75 as covariates during the multi-site modeling step. This cutoff was chosen as a compromise between losing too many sites and controlling for LD between the eSNP and SNP-in-probe.
Fine-mapping with polyQTL
Fine-mapping to localize causal variants influencing gene expression was performed using PolyQTL (Zeng and Gibson 2018), a modification of DAP (Wen et al. 2016) which we developed to account for population structure and ancestry during Bayesian localization in the presence of multiple linked cis-acting variants. We incorporated an option for first performing sequential stepwise regression, using the mixed linear regression component of GEMMA (Zhou and Stephens 2012) as above to identify independent QTL regions for each transcript. PolyQTL also offers the option to estimate posterior probabilities for all eQTL at a locus simultaneously, but this was not performed here owing to the computational burden.
For each independent eQTL, we subsequently evaluated the importance of each variant in the LD region, defined as SNPs with r2 ≥ 0.3 with the peak variant. PolyQTL assumes that there is a single causal variant associated with each independent QTL, and evaluates the posterior probability, given the LD structure at the locus, that each variant in the interval is causal, such that the sum of the posterior probabilities for each independent QTL is between 0 and 1. Genes were modeled as being under partial control of local genotypes as well as the polygenetic background, expressed as y = Xiβi + G +ε, where y is a vector of transcript abundance phenotypes, G represents the influence of the polygenic background, Xi and βi are the genotype and effect of the explored variant, and ε is a random environmental factor also normally distributed N(0, ). PolyQTL uses REML to estimate genetic and environmental variances, and given the estimated GRM, K (Yang et al. 2010). To remove the influence of population structure, we transform the phenotype (y) and genotype (Xi) with the square root of the covariance of the phenotype, , where is the identity matrix, as this results in independent multivariate normal distributions. We then compute a posterior inclusion probability (PIP) for each variant, leading to a ranking of candidate causal variants (Zeng and Gibson 2018).
eQTL sharing across expression platforms
Despite the expectation that expression platform influences eQTL detection, we reasoned that cis-eQTL results can complement one another leading to enhanced detection of shared signals by overcoming false negative results from single studies. To this end, we performed joint analysis of the cis-eQTL signals obtained on all three platforms, namely Illumina, Affymetrix, and RNAseq. We devised a new method based on the eCAVIAR strategy (Hormozdiari et al. 2016), named DPolyQTL, which explores the signal sharing for two phenotypes (either molecular traits or phenotype traits) even where the collected samples are family-based or from diverse ethnicities. DPolyQTL calculates a posterior probability that the causal variants are shared for two phenotype traits, such as expression of a gene measured on two platforms, by multiplying the two posterior probabilities together to generate a colocalization posterior probability (CLPP: Hormozdiari et al. 2016). We validated the performance of DPolyQTL by performing 200 simulations assuming pairs of normally distributed gene expression traits in the absence of relatedness, but with mild population structure (Fst = 0.1), where an eQTL explains 4% of the variance in one study and from 2 to 10% of the variance in the second one. Table S2 shows the proportion of CLPP values at thresholds from 0.001 to 1 for N = 1000 or N = 1600 samples in each study. Power increases with effect size and sample size as expected, and is over 98% for CLPP 0.001, ∼80% for CLPP 0.01, dropping to between 25 and 35% for CLPP 0.5 and <8% variance explained for N = 1000.
Since interpretation of the calculated posterior probability as a measure of sharing of causal variants is confounded by the complex LD structure in the human genome, we conducted permutations to obtain the null distribution of the CLPP given that a true eQTL is detected in one of the datasets, the discovery dataset, and is replicated in the other one, the replication dataset. To do so, the phenotype was permuted in the replication dataset, and the posterior probability was recalculated. On this basis, the colocalization signal was determined to be true if the CLPP ≥ 0.001 and permutation P-value ≤ 0.05, similar to Hormozdiari et al. (2016).
eQTL and GWAS colocalization analysis
A curated summary of GWAS results for 1263 phenotype traits or disease was generated by eQTLgen Consortium members (UV, AC and LF) with citations listed in Table S3. For each trait or disease, we defined candidate independent candidate regions as all variants within 100 kb of a peak association signal at P ≤ 5 × 10−8, though we recognize that a minority of associations may be driven by more distant variants. To reduce the computational burden, we also excluded all variants in the interval with P ≥ 0.05.
Colocalization of the eQTL and GWAS signals was then assessed for all genes located within 1 Mb of the peak GWAS signal. Similar to the analysis of eQTL sharing between expression platforms, we used DPolyQTL on the GWAS and eQTL summary statistics to identify likely regulatory influences of gene expression on complex phenotypes or disease.
Data availability
Gene expression data for the CAGE studies is available from GEO under study accession numbers GSE61672 (CHDWB), GSE49925 (CAD), GSE17065 (Morocco), GSE53195 (BSGS), and GSE48348 (EGCUT), while the FHS data can be accessed from the dbGaP as phs000007.v25.p9. We have made all eQTL data publically available through two shiny apps, http://cnsgenomics.com/shiny/CAGE/, and for the conditional analysis of both CAGE and FHS at http://bloodqtlshiny.biosci.gatech.edu or in Excel format at https://ggibsongt.wixsite.com/gibsongatech/supplementary-data. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7175219.
Results
Multiple eQTL regulation is ubiquitous in human blood
Our first objective was to estimate the proportion of transcripts that are regulated by multiple independent eQTL in the two large cohort studies, CAGE and FHS. Since both datasets include siblings, we used GEMMA (Zhou and Stephens 2012) to perform sequential conditional eQTL analysis deploying a genetic relationship matrix based on all measured and imputed genotypes to model family structure and population structure. Applying sequential conditional analysis to CAGE, we detected 5974 eGenes (37.8% of 15,812 tested genes) with at least one significant eSNP at P < 10−5, corresponding to a FDR of ∼5%. Of these eGenes, 2187 (36.6%) contain probes influenced by more than one eSNP, and, hence, appear to be regulated by multiple regulatory elements. (Note that, in the case of genes with multiple probes on the Illumina platform, we only required that at least one probe was associated with an eQTL, and for multi-SNP regulation included only the probe with the largest number of significant independent eSNPs). Similarly, in the FHS data, we detected 5597 (35.3% of 15,853 tested genes), 2114 (37.8%) of which were regulated by multiple eQTLs. In CAGE, the average variance explained by detected eSNPs was 6.1%, the same as in FHS, 6.1%, and in both cases these estimates account for more than half of the previously estimated heritability attributed to the cis region (Lloyd-Jones et al. 2017). For those genes with multiple eQTL regulation in CAGE, which have a mean explained variance of 7.2%, the newly detected secondary eSNPs typically explained 20% more variance than the peak SNP alone, namely ∼1.2% of the phenotypic variance (6.0% vs. 7.2%), also in line with estimates from Lloyd-Jones et al. (2017) and Powell et al. (2013). For eGenes with multiple eQTLs in FHS, the mean explained variance is 6.3%, and the secondary signals increase the explained variance from 6.5 to 8.3%, an increase of ∼28%.
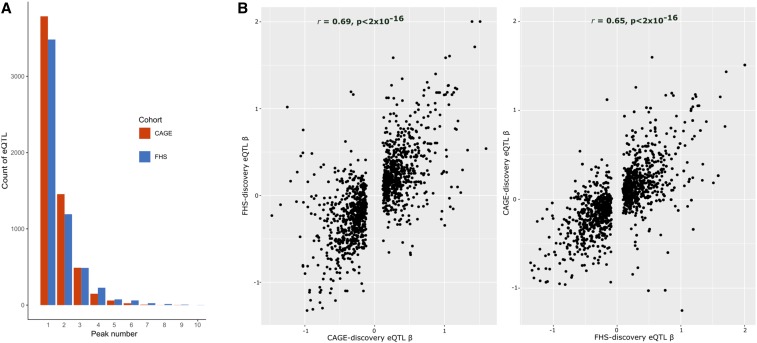
Figure 1 shows frequency histograms for the number of detected eQTL per gene after each sequential step in both studies: the number of loci with additional independent sites reduces by approximately two-thirds with each additional SNP in both CAGE and FHS, up to half a dozen variants, and a few loci have 10 sites. This reduction likely reflects the true prevalence of multi-site effects as well as reduced power to detect SNPs that explain less of the variance than the primary signal. A detailed example of multi-site association is shown for the HBZ locus in CAGE (Figure S2), where from left to right, and top to bottom are the results of stepwise conditional analysis yielding nine independent eQTL signals. The total explained variance is 39.8%, one-third more than that explained by the highest single peak (28.4%). An example from the FHS is ABHD2, where we detect five independent eQTLs explaining 9.3% of the variance, compared with 5.6% for the peak eSNP (note that the Affymetrix probeset contains a common variant, rs2283435, that is in linkage equilibrium with each of the five regulatory signals). All of our multiple eQTL results can be downloaded both in tabulated format and as locuszoom plots from our BloodQTL Shiny app at http://bloodqtlshiny.biosci.gatech.edu
Figure 1.
Discovery of eQTL in CAGE and FHS. (A) Counts of independent cis-eQTL detected in the CAGE and FHS cohorts. Red (CAGE) and blue (FHS) bars indicate the number of primary, secondary, tertiary, etc., eQTL detected conditional on the prior peaks at P < 10−5 in each sequential step. (B) Directional consistency of primary eQTL effects is indicated by plotting the magnitude and sign of eQTL effects (βs) in the validation study (y-axis) against discovery dataset (x-axis) for CAGE (left) or FHS (right).
We also computed the difference between the estimates fitting all discovered variants jointly and by summation of the conditional single-site estimates following eSNP sequential conditional discovery. The average change in estimated beta was just 0.04 ± 0.06 sdu, but with a long tail of large deviations.
Directional consistency of effects was evaluated by estimating the correlation between effect size estimates between the two platforms. Figure 1B shows that the primary eQTL discovered in CAGE replicate the sign of effect in 90% of cases with overall correlation of effect sizes 0.69 in FHS; and conversely those discovered in FHS have 92% replication of the sign, and overall correlation 0.65. These rates are similar to those observed across dozens of studies reported by the Blood eQTL Consortium (Võsa et al. 2019).
To directly compare signal replication from cohorts based on the same platform, we independently conducted eQTL detection for probes on chromosome 1 in the CHDWB and EGCUT cohorts in the CAGE study. Controlling FDR ≤ 0.01, we detected 665 significant e-signals (SNP to probe) in EGCUT, and 364 in CHDWB, among which 315 primary eQTL were in common (86.5% of the CHDWB eQTL), while 74.6% of the peak eSNPs were located in the same credible interval (genotype r2 ≥ 0.8). This high degree of concordance supports our mega-analysis strategy of combining cis-eQTL results from the different study cohorts, and provides a baseline for comparisons across platforms.
Limited overlap of cis-eQTL credible intervals between CAGE and FHS
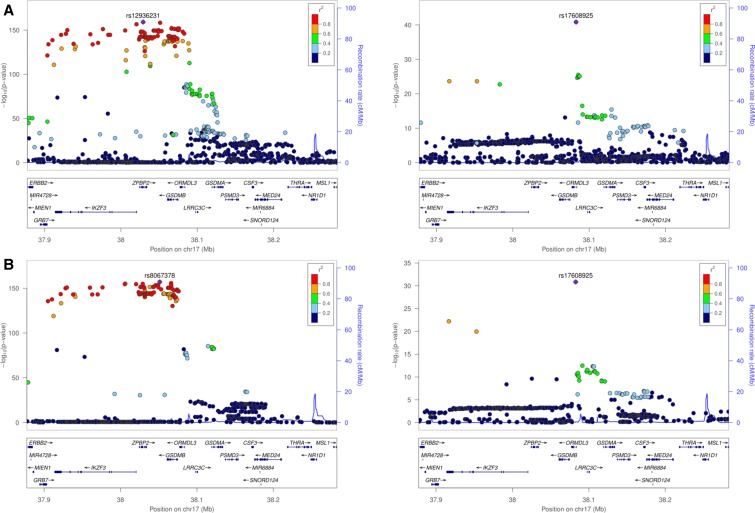
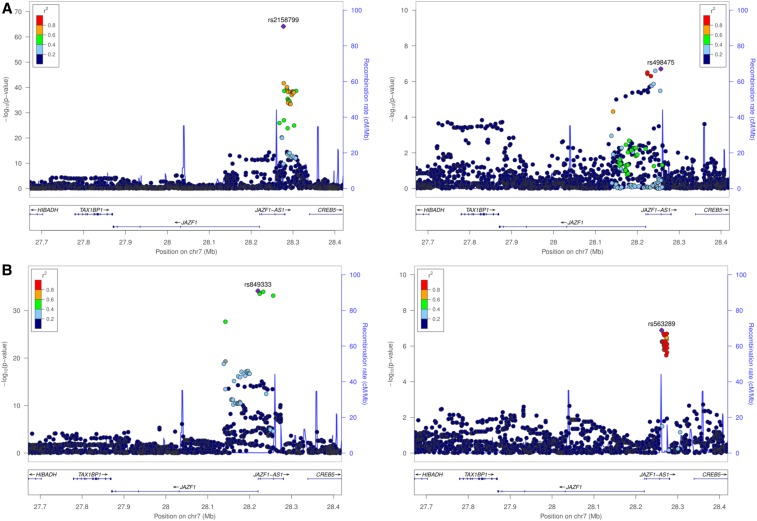
Despite the similar overall rates of eQTL detection, direct comparison of the results from the CAGE and FHS analyses suggested a disappointingly low level of replication. Primary peaks in CAGE were detected for 53.0% of the eGenes represented in the FHS, and reciprocally 56.5% of the FHS eGenes had primary signals in CAGE, very similar to the proportions reported for eSNPs at P < 10−8 across four peripheral blood studies (Zeller et al. 2010) that also had a variety of technical differences. However, the overall overlap between CAGE and FHS for eSNPs within credible intervals defined by LD r2 > 0.8 was just 29.1%. Furthermore, only 41.5% of the primary signals in FHS are in LD (r2 > 0.8) with the primary eSNP in CAGE, suggesting that different largest-effect regulatory variants are tagged in the two datasets. This overall eSNP replication rate was slightly higher (47.2%) when mapping to 1314 probesets that map directly to the same exon and have an eQTL signal on both platforms. Figure 2 and Figure 3 illustrate examples of loci each with two independent cis-eQTL signals associated with the same credible intervals. Whereas at ORMDL3 the relative magnitudes of the effects are the same, at JAZF1, the primary and secondary effects are rank-changed.
Figure 2.
Example of shared cis-eQTL signals, at the ORMDL3 locus in CAGE (A) and FHS (B). In both studies, two independent cis-eQTL were detected. rs12936231 and rs8067378 are the respective peak eSNPs for a credible interval of ∼50 SNPs, and are in complete LD (r2 = 1), whereas the peak conditional secondary association is at rs17608925 in both studies.
Figure 3.
Example of rank-changed cis-eQTL signals, at the JAZF1 locus in CAGE (A) and FHS (B). For CAGE, rs2158799 and rs498475 are the peak eSNPs in two independent credible intervals, the second of which is captured by rs849333 as the primary peak in the FHS. However, rs563289 is the secondary peak in FHS and appears to be a novel association, despite lying in the same physical region as the primary peak in CAGE.
The replication rate of secondary, tertiary, and quaternary signals in FHS irrespective of LD was just 19.0, 11.3, and 11.0%, indicating successive decay, likely due to reduced power for weaker signals. The reasons for the discrepancies between studies may have to do with collapsing of probe-level data down to gene-level signals losing information on splice isoforms, the different normalization strategies [which alone can double discovery rates (Qin et al. 2012)], and cross-study biological heterogeneity. Comparison of the percent variance explained by discovered SNPs on the two platforms in Figure S3 shows that effect sizes for many genes are disproportionately tagged by eQTL in the two studies, implying platform effects. Among 139 genes with >20% of the variance explained by cis-eSNPs, the replication rates are 64.7% for the primary signal, 34.8% for the secondary, 14.4% for the tertiary, and 8.5% for the quaternary. The subsequent panels in Figure S3 confirm that all of these replication rates are proportional to the percent variance explained overall, confirming that statistical power is a major source of low replication.
For the HT12 v3 Illumina probes, 10% of the uniquely mapping probes contain at least one SNP with MAF >1% in 1000 Genome European population samples. The prevalence of eQTL was twice as great for these probes (59% vs. 30% of 23,681 “clean” probes), so we just removed the Illumina probes containing SNPs in order to control the FDR. However, since most of the Affymetrix probesets contain at least one SNP, this was not practical for the FHS dataset, and instead we employed a conditional analysis strategy incorporating SNPs in probes as covariates. For ∼15,000 detected eSNPs, one-third of the association signals were abrogated by conditioning on the SNPs in probes, and the number of eGenes correspondingly reduced by 25%. Table 1 contrasts the eQTL results from both platforms before and after controlling for the SNP-in-probe effects. The first three data columns show the cumulative number of eGenes with at least 1, 2, 3, 4, or more detected eSNPs before SNP-in-probe removal, and the next three show the cumulative numbers after. The proportion of overlapping signals is not greatly affected. The last column shows that the number of eGenes where at least one detected signal is likely capturing the same variant is ∼44% (689/1562), and that the number where all of the multiply detected signals are within r2 > 0.8 is very small. There are 214 genes with at least two signals in high LD with one another.
Table 1. Cross-platform comparison of eSNP detection after adjustment for probe SNPs.
| With SNPs-in-probes | Without SNPs-in-probes | ||||||
|---|---|---|---|---|---|---|---|
| No. | CAGE | FHS | Both_anya | CAGE | FHS | Both_any | Both_highLDb |
| ≤1 | 3987 | 3881 | 1402 | 3787 | 3483 | 1182 | 474 |
| ≤2 | 5570 | 5983 | 1790 | 5242 | 4675 | 1433 | 616 (34) |
| ≤3 | 6115 | 6925 | 1902 | 5732 | 5163 | 1514 | 669 (7) |
| ≤4 | 6282 | 7327 | 1928 | 5881 | 5391 | 1534 | 686 (1) |
| >0 | 6383 | 7713 | 1970 | 5974 | 5605 | 1562 | 689 (0) |
Number of eGenes detected in both studies with at least the indicated number of independent associations, irrespective of whether they colocalise to the same fine-mapped interval.
Cumulative number of eGenes with at least one eSNP localized within r2 > 0.8 in both CAGE and FHS, namely eQTL that do fine-map to the same interval. Numbers in brackets indicates cases with 2, 3, or 4 eSNPs all in high LD between datasets.
DPolyQTL increases the proportion of cis-eQTL sharing across different expression platforms
Next, we used DPolyQTL to enhance the power to detect shared cis-eQTL credible intervals in the CAGE, FHS, and BIOS datasets. We extracted variants located in high LD with the reported peak variants in each eQTL locus for each pair of studies, and computed a posterior probability to evaluate the likelihood that each variant influences the trait, controlling for LD at the locus. Since the available BIOS dataset consisted only of summary results, it was used solely as a discovery dataset. Where genes in CAGE and FHS contained multiple probes or probesets, replication is reported where at least one probe in each dataset contained a signal.
Approximately one-third of all eQTL were detected by all three of the CAGE, FHS, and BIOS datasets. Shared signals are indicated for 62.6% of the detected cis-eQTL of CAGE in FHS, and for 53.3% of the FHS eGenes in CAGE. For the detected cis-eQTL in BIOS, we found a similar replication rate, namely 53.6 and 54.7%. However, considering that for some of the eGenes reported by BIOS, no expression information was available in CAGE and FHS, the replication rate considering all genes is 44.7% in CAGE, and 54.5% in FHS. Since DPolyQTL allows multiple eQTL signals to be explored simultaneously, we were able to estimate that whereas 43%–49% of primary eSNPs showed evidence for replication, the replication rate was considerably lower, only ∼10%, for secondary eSNPs.
Taken together, these results indicate >50% pairwise cross-platform replication of eGenes, with evidence that the majority of primary eQTL detected on one platform are also eQTL on another. However, the primary regulatory variant maps to a different credible interval in more than a third of cases, and replication of secondary variants is strongly reduced by low statistical power in the presence of multisite regulation.
Biological annotation of detected multiple eQTLs
Since chromatin marks are often used to enhance fine-mapping, on the basis that peak eSNPs are enriched in the vicinity of ENCODE features such as DNAse hypersensitivity, methylation, and histone modification, we asked whether there is a difference in functional attributes of primary and secondary eQTL. CADD (Kircher et al. 2014) and deltaSVM (Lee et al. 2015) are two commonly used scores that summarize multiple types of functional evidence. For CADD, we created a list of background SNPs with similar allele frequencies in the neighboring regions of peak eSNPs, and compared the distributions of the background and peak scores. Although the distribution of CADD scores was significantly higher for the reported-peak variants, suggesting elevated likelihood that they are pathogenic (Figure 4A for CAGE, and Figure 4B for FHS), the magnitude of the effect is small relative to the variance in CADD scores.
Figure 4.
Biological annotation of detected cis-eQTL signals. (A and B) Comparison of CADD score distributions for Primary and Secondary eSNPs and neighboring background SNPs at similar minor allele frequencies in the CAGE (A) and FHS (B) studies. (C and D) Relationship between observed beta and predicted deltaSVM score for significant peak eSNPs in the respective studies. P-values for comparisons are indicated.
Correspondingly, the positive predictive value for each SNP is low and functional discrimination of primary and secondary signals by this measure is poor (see also Liu et al. 2019). Potential causal variants defined by fine-mapping also have only a slightly elevated probability of locating within regulatory enhancers in the human genome as defined by the deltaSVM score. Setting any variant with a posterior probability ≥0.8 as a causal variant, we found that there is a weak, but significant, positive correlation between the reported beta value and deltaSVM (P ≤ 10−6 in both CAGE and FHS, Figure 4, C and D respectively). There is no difference between CAGE and FHS in the location of primary eQTL peaks relative to the transcription start site (TSS), although there is a slight increase in the dispersion of secondary relative to primary peak locations.
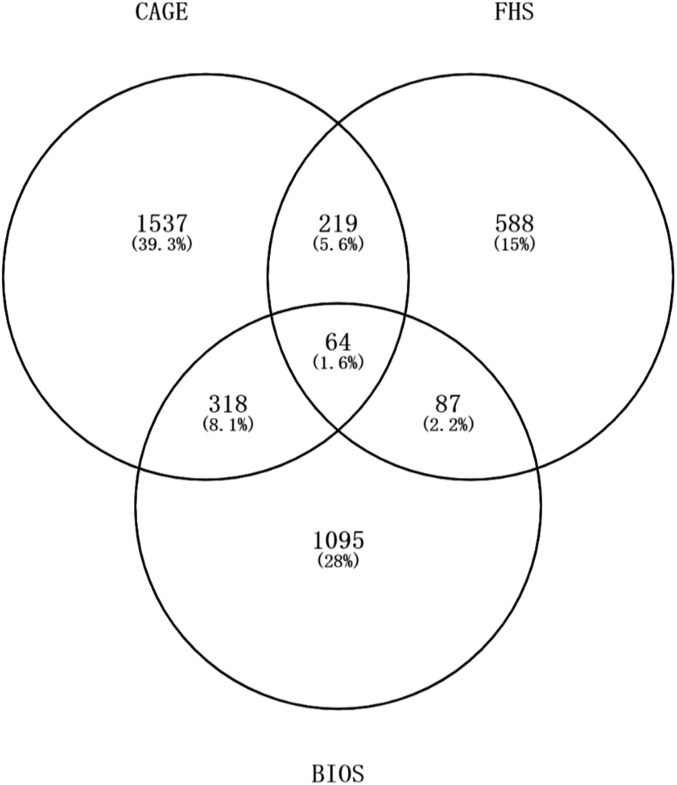
In order to evaluate whether eGenes associated with phenotypes are enriched for certain molecular functions, we next combined the full summary statistics of 1263 GWAS results with the eQTL signals from CAGE, FHS, and BIOS. Statistical power was maximized by performing colocalization analysis with DPolyQTL based on cis-eQTL detected on all three platforms. This colocalization analysis resulted in 1349 genes associated with 591 human complex phenotype traits or diseases, with a colocalization posterior probability (CLPP) > 0.001 (49. 8% of those explored). Simulations suggest that this CLPP cutoff captures almost all shared signals but with a FDR of ∼10%, whereas at the less permissive cutoff of 0.01, just 80% of signals are captured with FDR <5%. The highest single platform discovery rate was for the CAGE data on the Illumina platform, and the replication rate across platforms ranged from 24 to 30% as shown in Figure 5.
Figure 5.
Extent of replication of eQTL-GWAS colocalization with different expression platforms. The Venn diagram shows number of eQTL-GWAS joint associations (CLPP >0.001) in the three studies, and the percentage of all of the 3908 total associations in each sector.
PANTHER pathway analysis (Mi et al. 2017) revealed over-representation of colocalized genes annotated to protein kinase activity or to DNA binding activity. Among the most strongly implicated pathways are insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade (4.2-fold enrichment, 8.5 × 10−4), VEGF signaling pathway (3.2 fold enrichment, 4.4 × 10−4), interleukin signaling pathway (3.1 enrichment, 8.6 × 10−5), Ras Pathway (2.7-fold enrichment, 2.4 × 10−3), PDGF signaling pathway (2.3-fold enrichment, 6.0 × 10−4), gonadotropin-releasing hormone receptor pathway (2.0 fold enrichment, 7.2 × 10−4), and inflammation mediated by chemokine and cytokine signaling pathway (1.92-fold enrichment, 1.1 × 10−3). Furthermore, these 1349 detected genes were enriched for association with several diseases, notably with 327 causing Mendelian diseases (1.4-fold enrichment to background, P = 8.6 × 10−7), providing further evidence that genes defined by highly penetrant mutations also harbor quantitative regulatory variants that influence disease.
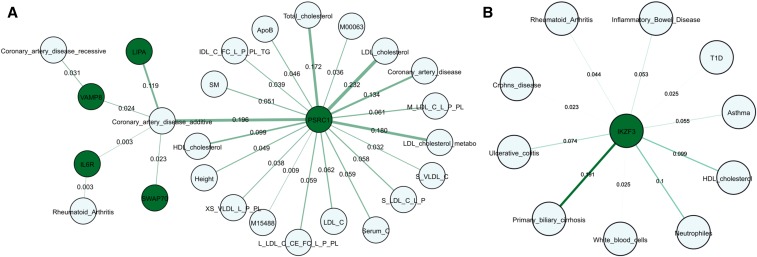
The colocalization results highlight a number of gene sets that interact together to influence disease susceptibility. For example, we found five genes associated with coronary artery disease, PSRC1, IL6R, LIPA, SWAP70, and VAMP8 in the CAGE dataset (Figure 6A). Four of these genes have previously been reported to be associated with coronary artery disease. PSRC1 encodes a cysteine protease that has been associated with HDL and LDL levels (Kathiresan et al. 2008), and its expression in mouse liver is significantly associated with plasma LDL cholesterol level (Schadt et al. 2008). LIPA encodes lipase A, which catalyzes the hydrolysis of cholesteryl esters and triglycerides, and is associated with CAD, where the lead CAD risk allele also associates with increased expression of LIPA mRNA in monocytes (Zeller et al. 2010) and liver [Coronary Artery Disease (C4D) Genetics Consortium 2011] SWAP70 encodes a signaling molecule involved in the regulation of filamentous-actin networks in cell migration and adhesion, and an intronic SNP has been reported to be a cis-eQTL in naïve and challenged monocytes (Nikpay et al. 2015). Notably, we found colocalization of eQTL at the IL6R gene with GWAS signals for both rheumatoid arthritis and coronary artery disease, hinting at a mechanistic basis for genetic correlation between these two conditions. Colocalization signals between the two different expression platforms complemented one another, since, for FHS gene expression, 11 genes also associated with CAD (ADAMTS7, CARF, CDKN2A, GGCX, HECTD4, IL6R, LIPA, PCSK9, PSRC1, USP39, VAMP8), including four of the CAGE genes (IL6R, LIPA, VAMP8, and PSRC1). Manual review of the literature finds that five of the remaining seven have previously been reported to be associated with CAD.
Figure 6.
Two examples of eQTL-GWAS colocalization. (A) Five loci with joint CAGE-eQTL and GWAS associations with CAD are joined in an extended network with seven additional loci in FHS. (B) IKZF3 was previously reported to be associated with three autoimmnune diseases; our analysis finds extended associations with other autoimmune diseases and blood traits. Genes are represented as solid dark green circles, and phenotypes or diseases as light blue circles. The thickness of edge line represents the strength of colocalization signal given by the indicated CLPP.
An example of colocalization linking a gene to multiple phenotypes is provided by IKZF3 (Figure 6B), which is known to associate with the autoimmune diseases Crohn’s disease, ulcerative colitis, and rheumatoid arthritis (Mancuso et al. 2017). These findings are replicated in our data, and enhanced further by associations with the additional autoimmune diseases, type 1 diabetes, and primary biliary cirrhosis as well as with asthma. Expression of IKZF3 is also associated with neutrophil cell and white blood cell counts. Most of these colocalization signals are replicated in either the FHS or BIOS data.
Gene expression regulated by nonprimary cis-eQTL mediates phenotype associations
Recent colocalization studies have reported that only a limited proportion of GWAS variants clearly function as eQTL (Farh et al. 2015; Chun et al. 2017; Huang et al. 2017). Since those studies focused on primary cis-eQTL, we asked whether conditional eQTL might increase the rate of discovery of colocalized eQTL-GWAS signals. For each expression-phenotype pair with at least nominally significant colocalized signals, we first identified variants where the credible interval maps to the primary cis-eQTL. If no such variants were found, we then evaluated joint signals at secondary cis-eQTL, tertiary signals, and so forth until no cis-eQTL remained to be evaluated.
For the 2138 colocalized signals in CAGE, we observed that 82.0% of the colocalized signals are with the primary eQTL, 11.0% from secondary signals, and 3.9% from tertiary signals. Similarly, in FHS, of 958 colocalized eQTL-GWAS signals, 69.5% are attributable to primary signals, 15.1% to secondary signals, and 11.9% to tertiary signals. For instance, in Figure 3, the secondary cis-eQTL for JAZF1 in CAGE, which is the primary cis-eQTL in FHS, locates in high LD with a type 2 diabetes GWAS hit. These colocalization results indicate a meaningful contribution of nonprimary cis-eQTL to trait variation.
Discussion
In summary, we find extensive evidence for secondary and tertiary cis-eQTL associations explaining gene expression variation in peripheral blood. At least a third of all highly expressed genes display such effects, consistent with recent evidence from very large-scale GWAS that at least one-quarter of all loci harbor multiple associations within a 1-Mb interval. However, the fine mapping of eQTL across platforms is considerably lower than expected, and, accordingly, replication of colocalization with visible phenotypes and disease risk is also modest, despite the large sample size of our two cohorts. Since secondary and tertiary effect sizes are generally smaller than primary ones, statistical power remains a major detriment to the joint fine mapping of regulatory variants to GWAS credible intervals.
Nevertheless, resolution of GWAS associations to single causal variants is a major current objective of human genetics. Four general strategies are being deployed: very large GWAS and eQTL studies, including cross-population analyses, intended to narrow peaks; sophisticated colocalization approaches; filtering on functional attributes associated with SNPs; and high-throughput experimental validation. The first objective is to define credible intervals that are highly likely to contain the causal variant or variants within a linkage disequilibrium block. However, several recent studies have reported that as few as one-third of disease associations map to the same credible interval as the lead eQTL, even in cases such an autoimmune Crohn’s disease, where the eQTL mapping is carried out in the, presumably relevant, peripheral blood tissue consisting of immune cells (Chun et al. 2017; Huang et al. 2017).
Two classes of explanation may account for this discrepancy between expectation and observation: biological and technical. The obvious biological explanation is that the causal variant detected by GWAS for some phenotypic trait does not directly regulate gene expression. It may, for example, influence chromatin structure, preparing the locus for induction under conditions not sampled in the transcriptomic study (Alasoo et al. 2018), and indeed there is some evidence for greater overlap of methylation QTL than expression QTL with Crohn’s disease associations (Huang et al. 2017). A corollary would be that the influence on gene expression would be seen only if the appropriate tissue (or the most important cell type within a mixture of cell types, such as peripheral blood) is sampled, or under more appropriate conditions of stimulation either ex vivo or in vivo (such as inflamed tissue-resident immune cells). The prevalence of response-eQTL provides good evidence in support of this claim (Fairfax and Knight 2014).
Technical explanations relate to statistical methodology and power, as well as platform effects. It is remarkable in our study that both the Illumina and Affymetrix datasets yielded very similar proportions of eGenes, as well as distributions of secondary and tertiary signals. Yet the overlap between these signals was only approximately one-half for the primary eSNPs, and <20% for conditional associations. Implementation of DPolyQTL provided evidence that statistical power is a major source of failure to replicate, both by enhancing the detection of shared primary signals between the datasets and showing that detection rates drop as effect sizes of secondary, tertiary, and quaternary associations reduce. Nevertheless, it is also clear that platform effects result in major differences in blood cis-eQTL detection. These are only partially ameliorated by focusing on probes that capture the same exon within a transcript, implying that detection of alternate splicing and isoform usage is just one aspect of the platform effect.
Irrespective of the causes of differential localization of primary eSNPs, an important practical implication of our findings is cautioning against the common use of summary eQTL statistics as evidence that a GWAS hit acts as an eQTL. Given the extensive linkage disequilibrium typically observed over long stretches of regulatory DNA, it is not uncommon for the GWAS variant to be included in a list of eQTL highly significant summary statistics listed on browsers such as the Blood eQTL browser. Visual inspection of the profile of association across the locus will often be sufficient to illustrate that the eQTL and GWAS peaks are not actually the same, as seen in Figure 3. Formal tests of the hypothesis of equivalence are provided by software tools such as COLOC (Giambartolomei et al. 2014), but these are designed for supervised analysis locus-by-locus and may be biased by the assumption that a single causal variant is responsible for each eQTL effect. The HEIDI test in SMR attempts to adjust the inference that an eQTL mediates the phenotypic association for local LD, providing genome-wide estimation of cases of heterogeneity of effects (Zhu et al. 2016). Alternatively, the Bayesian eCaviar approach (Hormozdiari et al. 2016), implemented here in DPolyeQTL to adjust for population structure and familial relatedness (Zeng and Gibson 2018), more directly adjusts for LD in the derivation of posterior probabilities of joint association. We recommend using a combination of these approaches to explore the likelihood that eQTL explain GWAS effects, and to this end have developed a web browser, which, for the first time, allows users to explore the profile of primary and secondary signals in peripheral blood.
Contrary to the expectation that mega-analysis of large eQTL studies would improve the resolution of eQTL signals, we instead find levels of complexity that complicate the ability to reduce genetic associations to single causal variants. Most clearly, it is apparent that multiple regulatory variants affect the expression of the majority of transcripts expressed in peripheral blood. Similarly, meta-analysis of GWAS including hundreds of thousands of individuals increasingly find secondary associations at individual loci (Wood et al. 2014; Yengo et al. 2018). We have previously shown by simulation that the presence of multiple variants in LD blocks typical of human genes biases both the localization of eSNPs and the estimation of their effect sizes, with as many as 20% of effects potentially located outside detected credible intervals (Zeng et al. 2017). While functional data collected by the ENCODE project and measures of evolutionary conservation are often used to filter or adjust eQTL estimation, our analyses only confirm a modest enrichment of such marks at eQTL peaks. Elsewhere, we show that this is in large part due to the high correlation of functional scores within credible intervals (Liu et al. 2019). Consequently, functional assays (Tewhey et al. 2016; Gasperini et al. 2017) will continue to provide the gold standard for demonstration that specific SNPs associate with traits function through gene regulation, though these too have yet to be shown to have the capacity to distinguish causal from background variants.
Acknowledgments
This research was primarily supported by the National Institutes of Health of the United States of America, R01 1-HG-008146 awarded to G.G. B.Z. was partially supported by the Chinese Scholarship Council. We are also grateful to the Australian National Health and Medical Research Council (NHMRC) and Estonian Research Council for earlier support in the development of the Consortium for the Architecture of Gene Expression (CAGE).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7175219.
Communicating editor: P. Scheet
Literature Cited
- 1000 Genomes Project Consortium. Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasoo K., Rodrigues J., Mukhopadhyay S., Knights A. J., Mann A. L., et al. , 2018. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat. Genet. 50: 424–431. 10.1038/s41588-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K. G., Deluca D. S., Segrè A. V., Sullivan T. J., Young T. R., et al. , 2015. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S., Casparino A., Patsopoulos N. A., Croteau-Chonka D. C., Raby B. A., et al. , 2017. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 49: 600–605. 10.1038/ng.3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D., Yang C., Li C., Gelernter J., Zhao H., 2014. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 10: e1004787 10.1371/journal.pgen.1004787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronary Artery Disease (C4D) Genetics Consortium , 2011. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43: 339–344. 10.1038/ng.782 [DOI] [PubMed] [Google Scholar]
- Delaneau O., Zagury J. F., Marchini J., 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10: 5–6. 10.1038/nmeth.2307 [DOI] [PubMed] [Google Scholar]
- Fairfax B. P., Knight J. C., 2014. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 30: 63–71. 10.1016/j.coi.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K. K., Marson A., Zhu J., Kleinewietfeld M., Housley W. J., et al. , 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518: 337–343. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon E. R., Segrè A. V., van de Bunt M., Wen X., Xi H. S., et al. , 2018. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 50: 956–967. 10.1038/s41588-018-0154-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M., Findlay G. M., McKenna A., Milbank J. H., Lee C., et al. , 2017. CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 101: 192–205. 10.1016/j.ajhg.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton K. J., Ferreira T., Lee Y., Raimondo A., Magi R., et al. , 2015. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 47: 1415–1425. 10.1038/ng.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C., Vukcevic D., Schadt E. E., Franke L., Hingorani A. D., et al. , 2014. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10: e1004383 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., van de Bunt M., Segrè A. V., Li X., Joo W. J., et al. , 2016. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 99: 1245–1260. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G. R., 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44: 955–959. 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T., Liu C., Joehanes R., Zhang X., Chen B. H., et al. , 2015. A systematic heritability analysis of the human whole blood transcriptome. Hum. Genet. 134: 343–358. 10.1007/s00439-014-1524-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Fang M., Jostins L., Umićević Mirkov M., Boucher G., et al. , 2017. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547: 173–178. 10.1038/nature22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idaghdour Y., Czika W., Shianna K. V., Lee S. H., Visscher P. M., et al. , 2010. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat. Genet. 42: 62–67. 10.1038/ng.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J. W., Hormozdiari F., Han B., Eskin E., 2016. Multiple testing correction in linear mixed models. Genome Biol. 17: 62 10.1186/s13059-016-0903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., et al. , 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197 (erratum: Nat. Genet. 40: 1384) 10.1038/ng.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Pasaniuc B., 2015. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97: 260–271 (erratum: Am. J. Hum. Genet. 97: 353) 10.1016/j.ajhg.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Ghasemzadeh N., Eapen D. J., Chung N. C., Storey J. D., et al. , 2014. Gene expression profiles associated with acute myocardial infarction and risk of cardiovascular death. Genome Med. 6: 40 10.1186/gm560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., et al. , 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. J., Zeiss C., Chew E. Y., Tsai J. Y., Sackler R. S., et al. , 2005. Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389. 10.1126/science.1109557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T., Sammeth M., Friedlander M. R., ’t Hoen P. A., Monlong J., et al. , 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Gorkin D. U., Baker M., Strober B. J., Asoni A. L., et al. , 2015. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 47: 955–961. 10.1038/ng.3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Morar N., Dixon A. L., Lathrop G. M., Abecasis G. R., et al. , 2013. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 23: 716–726. 10.1101/gr.142521.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. M., Du P., Huber W., Kibbe W. A., 2008. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 36: e11 10.1093/nar/gkm1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Z. S., van Sommeren H., Huang S. C., Ng R., Alberts R., et al. , 2015. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47: 979–986. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Sanderford M., Patel R., Chandrashekar P., Gibson G., et al. , 2019. Biological relevance of computationally predicted pathogenicity of noncoding variants. Nat. Commun. 10: 330 10.1038/s41467-018-08270-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones L. R., Holloway A., McRae A., Yang J., Small K., et al. , 2017. The Genetic architecture of gene expression in peripheral blood. Am. J. Hum. Genet. 100: 228–237 (erratum: Am. J. Hum. Genet. 100: 371) 10.1016/j.ajhg.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., et al. , 2017. Integrating Gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 100: 473–487. 10.1016/j.ajhg.2017.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta U. M., Denson L. A., Hyams J. S., Mondal K., Prince J., et al. , 2017. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 49: 1517–1521. 10.1038/ng.3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., et al. , 2017. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpay M., Goel A., Won H. H., Hall L. M., Willenborg C., et al. , 2015. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47: 1121–1130. 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., 2014. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94: 559–573 (erratum: Am. J. Hum. Genet. 95: 126) 10.1016/j.ajhg.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Henders A. K., McRae A. F., Caracella A., Smith S., et al. , 2012. The Brisbane systems genetics study: genetical genomics meets complex trait genetics. PLoS One 7: e35430 10.1371/journal.pone.0035430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Henders A. K., McRae A. F., Kim J., Hemani G., et al. , 2013. Congruence of additive and non-additive effects on gene expression estimated from pedigree and SNP data. PLoS Genet. 9: e1003502 10.1371/journal.pgen.1003502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Kim J., Arafat D., Gibson G., 2012. Effect of normalization on statistical and biological interpretation of gene expression profiles. Front. Genet. 3: 160 10.3389/fgene.2012.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E., Molony C., Chudin E., Hao K., Yang X., et al. , 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6: e107 10.1371/journal.pbio.0060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm K., Marzi C., Schurmann C., Carstensen M., Reinmaa E., et al. , 2014. Mapping the genetic architecture of gene regulation in whole blood. PLoS One 9: e93844 10.1371/journal.pone.0093844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R., Kotliar D., Park D. S., Liu B., Winnicki S., et al. , 2016. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165: 1519–1529 [corrigenda: Cell 172: 1132–1134 (2018)] 10.1016/j.cell.2016.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G., Sandor C., Han B., Xu H., Stranger B. E., et al. , 2013. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45: 124–130. 10.1038/ng.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler M. S., Tyrer J., Easton D. F., 2010. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genet. Epidemiol. 34: 463–468. 10.1002/gepi.20504 [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I., et al. , 2017. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Võsa U., Claringbould A., Westra H.-J., Bonder M. J., Deelen P., et al. , 2019. Unraveling the polygenic architecture of complex traits using blood eQTL meta-analysis. BioRxiv 10.1101/447367 [DOI] [Google Scholar]
- Walter N. A., McWeeney S. K., Peters S. T., Belknap J. K., Hitzemann R., et al. , 2007. SNPs matter: impact on detection of differential expression. Nat. Methods 4: 679–680. 10.1038/nmeth0907-679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Lee Y., Luca F., Pique-Regi R., 2016. Efficient integrative multi-SNP association analysis via deterministic approximation of posteriors. Am. J. Hum. Genet. 98: 1114–1129. 10.1016/j.ajhg.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H. J., Peters M. J., Esko T., Yaghootkar H., Schurmann C., et al. , 2013. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45: 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo A. P., Gibson G., 2015. Blood gene expression profiles suggest altered immune function associated with symptoms of generalized anxiety disorder. Brain Behav. Immun. 43: 184–191. 10.1016/j.bbi.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H., et al. , 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L., Sidorenko J., Kemper K. E., Zheng Z., Wood A. R., et al. , 2018. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27: 3641–3649. 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., et al. , 2010. Genetics and beyond – the transcriptome of human monocytes and disease susceptibility. PLoS One 5: e10693 10.1371/journal.pone.0010693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Gibson G., 2018. PolyQTL: Bayesian multiple eQTL detection with control for population structure and sample relatedness. Bioinformatics 35: 1061–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Lloyd-Jones L. R., Holloway A., Marigorta U. M., Metspalu A., et al. , 2017. Constraints on eQTL fine mapping in the presence of multisite local regulation of gene expression. G3 (Bethesda) 7: 2533–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova D. V., Deelen P., Vermaat M., van Iterson M., van Galen M., et al. , 2017. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 49: 139–145. 10.1038/ng.3737 [DOI] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M. R., et al. , 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48: 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Gene expression data for the CAGE studies is available from GEO under study accession numbers GSE61672 (CHDWB), GSE49925 (CAD), GSE17065 (Morocco), GSE53195 (BSGS), and GSE48348 (EGCUT), while the FHS data can be accessed from the dbGaP as phs000007.v25.p9. We have made all eQTL data publically available through two shiny apps, http://cnsgenomics.com/shiny/CAGE/, and for the conditional analysis of both CAGE and FHS at http://bloodqtlshiny.biosci.gatech.edu or in Excel format at https://ggibsongt.wixsite.com/gibsongatech/supplementary-data. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7175219.