Self-perpetuating protein aggregates (prions) cause diseases in mammals. Yeast prions are heritable in cell divisions. Howie et al. demonstrate that the cellular apparatus responsible for the asymmetry of cell division controls destabilization of a yeast prion...
Keywords: Hsp104, [PSI+], Ssb, Sir2, stress
Abstract
Self-perpetuating transmissible protein aggregates, termed prions, are implicated in mammalian diseases and control phenotypically detectable traits in Saccharomyces cerevisiae. Yeast stress-inducible chaperone proteins, including Hsp104 and Hsp70-Ssa that counteract cytotoxic protein aggregation, also control prion propagation. Stress-damaged proteins that are not disaggregated by chaperones are cleared from daughter cells via mother-specific asymmetric segregation in cell divisions following heat shock. Short-term mild heat stress destabilizes [PSI+], a prion isoform of the yeast translation termination factor Sup35. This destabilization is linked to the induction of the Hsp104 chaperone. Here, we show that the region of Hsp104 known to be required for curing by artificially overproduced Hsp104 is also required for heat-shock-mediated [PSI+] destabilization. Moreover, deletion of the SIR2 gene, coding for a deacetylase crucial for asymmetric segregation of heat-damaged proteins, also counteracts heat-shock-mediated destabilization of [PSI+], and Sup35 aggregates are colocalized with aggregates of heat-damaged proteins marked by Hsp104-GFP. These results support the role of asymmetric segregation in prion destabilization. Finally, we show that depletion of the heat-shock noninducible ribosome-associated chaperone Hsp70-Ssb decreases heat-shock-mediated destabilization of [PSI+], while disruption of a cochaperone complex mediating the binding of Hsp70-Ssb to the ribosome increases prion loss. Our data indicate that Hsp70-Ssb relocates from the ribosome to the cytosol during heat stress. Cytosolic Hsp70-Ssb has been shown to antagonize the function of Hsp70-Ssa in prion propagation, which explains the Hsp70-Ssb effect on prion destabilization by heat shock. This result uncovers the stress-related role of a stress noninducible chaperone.
PROTEIN misfolding diseases, such as Alzheimer’s disease (AD), associated with fibrillar self-perpetuating protein aggregates (amyloids), are a rising cause of morbidity and mortality in developed countries with increasingly aged populations (Weuve et al. 2014). Recent studies indicate that AD is the third leading cause of death in the USA (James et al. 2014). Infectious amyloids (prions) cause transmissible spongiform encephalopathies such as “mad cow” disease in humans and other mammals [for review, see Prusiner (2013)]. In the budding yeast Saccharomyces cerevisiae, endogenous self-perpetuating amyloids (termed yeast prions) control heritable phenotypically detectable traits, making yeast a good model for studying general mechanisms of amyloid formation and propagation (Liebman and Chernoff 2012; Chernova et al. 2017a). In addition to causing pathogenic effects, prions, as carriers of protein-based inheritance, are proposed to play an important role in yeast biology [for review, see Byers and Jarosz (2014); Wickner et al. (2018)]. [PSI+], a prion form of the yeast translation termination factor Sup35, is one of the best-studied yeast prions to date. Formation of the Sup35 prion leads to a defect in translation termination that is easily detectable in specially designed strains. Moreover, Sup35 protein of the same sequence can form heritable variants of an amyloid (“prion strains”) of different characteristics, such as different phenotypic stringencies, that apparently reflect different structural parameters of respective amyloid fibrils [for review, see Liebman and Chernoff (2012)].
Like other organisms, yeast have a cellular response to proteotoxic stress, which is mediated by stress-induced chaperone proteins (Verghese et al. 2012). The protein complex, composed of the chaperone Hsp104, the major cytosolic member of the Hsp70 chaperone family (Ssa), and the cochaperone Hsp40, plays a major role in disaggregation and solubilization of stress-damaged proteins in yeast (Glover and Lindquist 1998). Notably, the same chaperone complex is crucial for the propagation of most known yeast prions, including [PSI+] [for review, see Liebman and Chernoff (2012); Chernova et al. (2017a)]. The current model of prion propagation in the yeast cell states that the balanced action of the Hsp104 and Hsp70-Ssa/Hsp40 proteins breaks prion fibrils into oligomers, resulting in prion proliferation (Reidy and Masison 2011; Chernova et al. 2014). When Hsp104 is artificially overproduced in excess of Hsp70-Ssa, it antagonizes propagation of [PSI+] and some (but not all) other prions (Chernoff et al. 1995; Chernova et al. 2017a; Matveenko et al. 2018). Existing data suggest that this antagonism occurs due to the ability of excess Hsp104 to bind prion fibrils independently of Hsp70-Ssa (Winkler et al. 2012). As Hsp104 is unable to break fibrils into oligomers on its own, this “nonproductive” binding results in prion loss. Prion malpartitioning in cell divisions is implicated as a cause of prion loss in the presence of excess Hsp104 (Ness et al. 2017), although potential contribution of polymer “trimming” by Hsp104 has also been proposed (Park et al. 2014; Greene et al. 2018). Various members of the Hsp40 family modulate effects of excess Hsp104 on [PSI+] in a prion-variant-specific fashion (Astor et al. 2018), although it is not known whether these effects occur due to direct interactions between Hsp104 and Hsp40, or via modulation of Hsp70-Ssa by Hsp40, which in turn shifts the balance between “productive” (that is, together with Hsp70-Ssa) or “nonproductive” binding of Hsp104 to prion polymers. Another member of the Hsp70 family, Ssb, which is normally associated with the ribosome and acts in folding newly synthesized polypeptides, promotes [PSI+] elimination by excess Hsp104 when accumulated in the cytosol instead of the ribosomal fraction (Chernoff et al. 1999; Kiktev et al. 2015; Chernoff and Kiktev 2016). Our data show that this effect of cytosolic Hsp70-Ssb is due to its ability to antagonize binding of Hsp70-Ssa to prion aggregates (Kiktev et al. 2015; Chernoff and Kiktev 2016).
Saccharomyces cerevisiae practices asymmetric cell division, so that mother and daughter (bud) cells are morphologically distinguishable from each other. After stress, asymmetric distribution of cytoplasm serves as a “last line” of defense, as stress-damaged proteins, which are not disaggregated by chaperones, are preferentially retained in the mother cell and cleared from the daughter cell, restoring its proliferation ability (Aguilaniu et al. 2003). Preferential recovery of daughter cells is an adaptive feature, as daughter cells are at the start of their replicative life span and will undergo more cell divisions than aging mothers. In yeast, asymmetric segregation of damaged proteins in cell divisions depends on several cellular components, including Hsp104, the actin cytoskeleton, and the NAD+-dependent deacetylase Sir2 (Aguilaniu et al. 2003; Erjavec et al. 2007; Tessarz et al. 2009; Liu et al. 2010). It has been proposed that Sir2 promotes asymmetric segregation via deacetylating the chaperonin complex which in turn modulates folding of actin (Liu et al. 2010).
The yeast prion [PSI+] is resistant to prolonged growth at high temperature when Hsp levels are proportionally increased; however, it is destabilized (most profoundly in weak [PSI+] strains) by short-term heat shock followed by a return to normal growth temperature (Tuite et al. 1981; Newnam et al. 2011; Klaips et al. 2014). Hsp104 exhibits lower background levels compared to Hsp70-Ssa; however, it is accumulated faster during heat shock, so that prion destabilization coincides with the period of a maximal imbalance between Hsp104 and Hsp70-Ssa (Newnam et al. 2011). Heat-shock-induced prion loss is facilitated in the absence of some cytoskeleton-associated proteins (Chernova et al. 2011; Ali et al. 2014), occurs primarily in cell divisions following heat shock, and is asymmetric, which may be explained either by asymmetric segregation of prion aggregates (Newnam et al. 2011; Ali et al. 2014) or by asymmetric accumulation of Hsp104 (Klaips et al. 2014). Thus, the behavior of prion aggregates during heat shock shows a resemblance to the behavior of stress-damaged aggregated proteins.
Here, we explore the cellular control of heat-shock-promoted destabilization of [PSI+]. Our data show that the same region of Hsp104 is involved in [PSI+] destabilization by artificial overproduction of Hsp104 and by heat shock, implicate the Sir2-dependent apparatus of cell asymmetry in heat-shock-dependent prion destabilization, and uncover a role for the heat-shock non-inducible chaperone Hsp70-Ssb in prion destabilization by stress.
Materials and Methods
Strains
S. cerevisiae strains used in this work were isogenic haploid derivatives of strain 1-1-74-694D (Derkatch et al. 1996, collection number OT55), which has the genotype MATa ade1-14 his3-Δ200 leu2-3,112 trp1-289 ura3-52, and contains a Rnq1 prion ([PIN+]) and a weak variant of the Sup35 prion ([PSI+]). Strains 2B-P2393 (MATa his5) and 78A-P2393 (MATα his5) originated from the collection of S.G. Inge-Vechtomov and were used as mating type testers. Deletion derivatives, with the exception of the strains containing ssb2Δ, zuo1Δ, or the hsp104-∆N allele, were constructed in the strain OT55 via the PCR-mediated direct transplacement approach (Longtine et al. 1998). Primers were designed containing the gene flanking sequences at the 5′ ends and sequences homologous to a replacement marker at the 3′ end. The replacement marker gene was PCR-amplified from a respective plasmid by using these primers, and transformed into the yeast cells via the lithium acetate protocol (Gietz et al. 1992). Potentials with phenotypes corresponding to a replacement marker were checked by PCR to confirm successful replacement. Strains containing ssb2Δ or zuo1Δ were constructed by designing primers ∼100 bp upstream and downstream of the gene in question, PCR amplifying the truncated version of the respective gene from strains GT127 (ssb2, Chernoff et al. 1999) and YGR285c (zuo1Δ, gift from K. Lobachev), and then transforming the fragment into the OT55 background and checking potentials by PCR as previously stated. This latter method was also used for construction of strains with Hsp104 tagged on the C-terminus with GFP. Primers were designed that are located at 250 bp upstream and downstream of the stop codon of the HSP104 gene. These primers were used to amplify the C-terminal region of HSP104 (fused to the GFP coding sequence), and the Schizosaccharomyces pombe HIS5 gene (homologous to the S. cerevisiae HIS3 gene), located immediately downstream, from the yeast strain YBD401 (Liu et al. 2010, gift from T. Nyström). The resulting PCR-amplified fragment was transformed into the strain OT55. His+ transformants were selected, and integration into the chromosome at the correct position was confirmed by both PCR and fluorescence microscopy (to ensure GFP expression).
The HSP104 allele with deletion of the N-terminal region encompassing codons 2–147, hsp104-∆N, was constructed by using the derivative of the integrative plasmid pFL34, pFL34-nAI, which contains a modified version of the URA3 gene with a substitution C378T, destroying the ApaI site but not changing the amino acid sequence of the Ura3 protein. The BamHI fragment of the plasmid pGCH16 (Hung and Masison 2006) containing the HSP104 gene with the deletion, eliminating codons 2–147, was ligated into the BamHI-cut plasmid pFL34-nAI, producing a plasmid that contains both HSP104 and URA3 ORFs in the same orientation. This plasmid, linearized by ApaI, was integrated into the HSP104 locus of the yeast strain OT55 by transformation, followed by the pop-out events, selected for on medium with 5-fluoroorotic acid (5-FOA) counterselecting against the URA3 marker. Derivatives that had lost the full-length HSP104 gene and maintained the shortened hsp104-ΔN version were identified by PCR. The resulting [PSI+] strain with the hsp104-ΔN (2–147) allele was named GT1819.
Plasmids
Plasmids pFA6a-His3MX6 (collection number 247) and pFA6a-KanMX6 (collection number 658) were used to construct gene deletions as described above (Longtine et al. 1998). Plasmid pGAL104-URA3 (collection number 46, gift of S. Lindquist), which contains the HSP104 gene under the galactose-inducible (PGAL) promoter, and control plasmid pRS316-GAL (collection number 3, gift of A. Bretscher) were used to check strains for curing by overexpression of HSP104. Plasmid pYCL-Sp-SUP35NMSc-DsRed (collection number 582, constructed by R. Wegrzyn, Chernoff Lab) containing the sequence coding for the S. cerevisiae Sup35NM, placed under the PSUP35 promoter and tagged with DsRed protein, was used to monitor aggregation of Sup35 by fluorescence microscopy.
Yeast media and phenotypic assays
Standard yeast media and protocols were used (Sherman 2002). Rich organic medium (YPD) contained 1% yeast extract, 2% peptone, and 2% dextrose (glucose). Synthetic medium (standard Difco medium, SD) contained 0.17% yeast nitrogen base without amino acids or ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, and 13 nutrition supplements (adenine, arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, tyrosine, uracil, and valine) unless some were specifically dropped as indicated (e.g., −Ade for synthetic medium lacking adenine). Solid media contained 1.5% agar (US Biologicals). Media used to induce the PGAL promoter contained 2% galactose (for solid media) or 2% galactose and 2% raffinose (for liquid media) instead of glucose. Presence of the [PSI+] prion in colonies growing on solid medium was assayed by suppression of the ade1-14 (UGA) reporter allele, leading to growth on –Ade medium and light pink (as opposed to red) color on YPD medium [for review, see Liebman and Chernoff (2012)]. In cases where temperature is not specifically mentioned, yeast cells were incubated at 30°, except for cultures grown before and after heat shock that were incubated at 25° to avoid uncontrolled partial induction of some Hsps.
Mating assay
To assay mating ability of the yeast strains, cultures were grown on YPD medium at 30° overnight and streaked across a fresh YPD plate (strains to be tested were streaked as horizontal lines, while the mating type tester strains of a or α mating types were streaked perpendicular to them, crossing the first streaks). Plates were incubated at 30° overnight, then velveteened to SD plates selective for diploids, which were incubated at 30° for 2 days and checked for growth every 24 hr. Nicotinamide (NAM) was added where shown at 10 mM concentration to both precultures and YPD and SD plates where mating was performed and detected.
Heat-shock assays
Mild heat shock was performed as previously described (Newnam et al. 2011). Fresh colonies grown on YPD plates at 25° were used to inoculate a preculture, grown in 10 ml of YPD medium overnight at 25° at 180 rpm, then diluted in 50 ml of fresh YPD medium to OD600 = 0.1, followed by growth at 25° at 180 rpm. After 2 hr of growth, cultures were moved to a 39° (or 42° where specifically mentioned) water bath with shaking at 180 rpm. Samples were taken before heat shock (T = 0), and after specified periods of heat shock, from 30 min to 4 hr. Each sample was immediately streaked out onto solid YPD medium to obtain individual colonies. Serial dilution spottings onto YPD plates were also performed in some experiments to check for cell viability. Plates were incubated at 25° for 2–6 days for viability assays, or for 7–10 days (to allow full color development) for prion destabilization assays. [PSI+] (pink), [psi−] (red), and mosaic [PSI+]/[psi−] (pink/red) colonies were detected by visual inspection and counted.
For heat-shock experiments with nicotinamide (NAM), yeast cells were incubated in YPD medium and on YPD plates containing 10 mM NAM prior to, during, and after heat shock.
[PSI+] curing by inactivation or overproduction of Hsp104
To determine whether deletion strains remained curable of [PSI+] by the Hsp104 inhibitor guanidine hydrochloride (GuHCl), yeast cells were incubated on YPD plates containing 5 mM (wild-type or sir2Δ strains) or 2 mM (zuo1Δ, ssz1Δ, ssb1/2Δ strains, which are more sensitive to GuHCl) GuHCl for three passages (∼20–40 generations) at 30° and then streaked out on YPD medium without GuHCl. Loss of the Ade+ phenotype was detected by color (red or mosaic vs. pink) and confirmed by growth after velveteen replica plating onto −Ade medium if necessary.
For curing yeast cells of the [PSI+] prion by transient overproduction of the Hsp104 chaperone, yeast cultures were transformed with a plasmid bearing either the HSP104 gene under the galactose-inducible (PGAL) promoter or a control empty vector plasmid. To quantify curing, cells bearing either the PGAL-HSP104 plasmid or control plasmid were pregrown overnight at 200 rpm, 30° in 10 ml of liquid synthetic glucose medium, selective for the plasmid. Preculture cells were spun down at 3500 rpm and washed with water, and a total of 2 × 105 cells were used to inoculate 50 ml of plasmid-selective synthetic media possessing 2% galactose and 2% raffinose instead of glucose to induce HSP104. Cultures were grown at 200 rpm, 30°. Aliquots were taken at the start and prior to dilution in induction media (T = 0) and after specified periods of growth; cells were pelleted, washed in water, and plated onto synthetic glucose medium selective for plasmid. Plates were grown at 30° for 3 days, then velveteen replica plated to YPD and –Ade (selective for the plasmid) media, and visually examined after 7 days of growth to identify and count the [PSI+], [psi−], and mosaic colonies by color and growth.
It should be noted that while heat shock or artificial overproduction of Hsp104 cause loss of prion, we have not detected any observable change in the variant-specific phenotypic patterns in the colonies retaining the [PSI+] prion after treatment (compared to the respective culture before treatment) in any strain used in our work.
Protein analysis
For protein isolation and analysis of Hsp levels, yeast cells from non-heat-shocked or heat-shocked cultures were precipitated by centrifugation at 3500 rpm, washed with and resuspended in 200 μl of lysis buffer [25 mM Tris-HCl, pH 7.5; 0.1 M NaCl; 10 mM EDTA; 4 mM PMSF; 200 μg/ml cycloheximide; 2 mM benzamidine; 20 μg/ml leupeptin; 4 μg/ml pepstatin A; 1 mM N-ethylmaleimide (NEM); and 1× complete mini protease inhibitor cocktail from Roche], and disrupted by agitation with one-third volume of glass beads for 8 min in a cold room at 4°. Cell debris was spun down at 4000 g for 2 min, and the supernatants were normalized with additional lysis buffer so that protein content was approximately equal by Bradford assay (BioRad). Extracts were then used for denaturing SDS-PAGE electrophoresis. For this purpose, from 15 to 21 μl of each protein sample was incubated with one-third of the volume of 4× loading buffer (240 mM Tris-HCl pH 6.8, 8% SDS, 40% glycerol, 12% 2-mercaptoethanol and 0.002% bromophenol blue) at room temperature for 10 min, boiled in a water bath for 10 min, and run in a 10% SDS-polyacrylamide gel with 4% stacking gel in Tris–Glycine–SDS running buffer (25 mM tris, 192 mM glycine, 0.1% SDS, pH 8.3) for 30 min at 85 V then for 1.5 hr at 100 V, followed by electrotransfer to a Hybond-ECL nitrocellulose membrane (GE Healthcare Life Sciences), preblocking with 5% nonfat milk, and probing with the appropriate antibody. Visualization utilized the appropriate secondary HRP (horseradish peroxidase) conjugate antibody and ECL chemiluminescent reagent (GE Healthcare). Amount of total protein loaded per lane varied from 4 to 13 g between experiments (the same amount of protein was loaded into each lane in each given gel). All protein isolation experiments were repeated with at least three (and in most cases, more) independent cultures. Densitometry was performed by using the program ImageJ, downloaded from https://imagej.nih.gov. Reaction to the Ade2-specific antibodies (in most cases) or Coomassie staining (for the same amounts of protein run on a separate gel) were employed as a loading control.
“Boiled gel” analysis (Kushnirov et al. 2006) was performed in the same manner as the SDS-PAGE, except that protein samples were not boiled prior to loading into the gel. Gels were run for an initial 30 min at 85 V then another 60 min at 100 V to allow monomeric protein enough time to travel for a sufficient distance. Electrophoresis was then paused, wells were filled with a new portion of polyacrylamide to trap aggregated protein that remained there, and the whole gel was placed into a boiling water bath for 10–15 min. After boiling, the gel was cooled for 5 min and placed back into the electrophoresis setup, so that electrophoresis could be resumed. As boiling solubilized polymers, protein from the aggregated fraction was now capable of moving in the gel. After ∼2 additional hr of electrophoresis, proteins were transferred to the membrane as described, followed by blocking and reaction to antibodies. The experiment was repeated three times with independent cultures and produced similar results. The amount of total protein loaded per lane varied from 4 to 16 µg between experiments (the same amount of protein was loaded into each lane in each given gel).
For fractionation of prion polymers by semidenaturing agarose gel electrophoresis, SDD-AGE (Kushnirov et al. 2006), yeast extracts were incubated with loading buffer as described above at room temperature for 10 min and run in 1.8% Tris-Acetate EDTA (TAE)-based agarose gel containing 0.1% SDS, followed by transfer to a nitrocellulose Protran membrane (Whatman) by capillary blotting. Membranes were reacted to appropriate antibodies after preblocking in 5% milk. The experiment was repeated four times with independent cultures and produced similar results. Amount of total protein loaded per lane varied from 25 to 88 µg between experiments (again, the same amount of protein was loaded into each lane in each given gel).
For sucrose density gradient centrifugation (Kiktev et al. 2015), cells were collected and disrupted as described above, except for using a gradient buffer (20 mM HEPES pH 7.5, 1 mM EGTA, 5 mM MgCl2, 10 mM KCl, 10% glycerol, 1× complete mini protease inhibitor cocktail from Roche, 3 mM phenylmethanesulfonylfluoride and 100 μg/ml of cycloheximide). One hundred microliters of a protein extract containing 30–160 μg of total protein as measured by Bradford assay was loaded on a sucrose cushion composed of 100 μl of 20%, 200 μl of 30%, and 200 μl of 40% sucrose in the gradient buffer. (The same amount of protein was always loaded into each sample in the given experiment.) Samples were centrifuged in 1.4 ml (11 × 34 mm) thick-wall polycarbonate tubes, using TLS-55 rotor on an Optima TLX-120 ultracentrifuge (Beckman, Brea, CA) at 259,000 g for 80 min. Fractions of 200 μl each were collected beginning from the top of the gradient; the solid pellet was resuspended in 200 μl of gradient buffer. Equal volumes of each fraction were loaded, resolved on SDS-polyacrylamide gel, and analyzed by Western immunoblotting as described above.
Antibodies
Rabbit polyclonal antibodies to Sup35C were a gift from Dr. D. Bedwell. Rabbit polyclonal antibody to Hsp104 was a gift from Dr. S. Lindquist. Rabbit polyclonal antibody to Hsp70-Ssa was a gift from Dr. E. Craig. Rabbit polyclonal antibody to Ade2 protein was kindly provided by Dr. V. Alenin. Mouse polyclonal antibody to Rpl3 (ribosomal protein marker) was a gift from Dr. J. Warner. Secondary anti-rabbit and anti-mouse HRP antibodies, used in immunoblotting, were purchased from Sigma-Aldrich.
Fluorescence microscopy
To visualize protein aggregates tagged by Hsp104-GFP, respective cultures were grown according to the procedures used in heat-shock experiments, heat shocked in a shaking water bath at 39 or 42° as specified, and moved back to 25° for recovery. Five-hundred-microliter samples were taken before and after heat shock, and after specified recovery time periods, and spun down at 3000 rpm for 2 min, after which the supernatant was removed and the cells were resuspended in 30–50 μl of water. Ten microliters of each sample was then placed onto a microscope slide and sealed with clear nail polish to prevent drying. Fluorescence was detected under a BX41 microscope (Olympus) at 100× (oil immersion) with the endow GFP bandpass emission (green) filter. For analysis of Sup35NM-DsRed aggregation and colocalization with Hsp104-GFP, cultures transformed with Sup35NM-DsRed were prepared and analyzed in the same way as above using a tetramethyl rhodamine isocyanate (rhodamine/Dil; red) filter for imaging red fluorescence prior to imaging for GFP. Images were taken using an Olympus DP-71 camera, and were overlaid using the program DP manager (Olympus) to determine colocalization.
Statistical analysis
Experimental means are depicted in graphs, with error bars typically representing SDs (unless stated otherwise), which were calculated according to a standard formula (McDonald 2009). These numbers are shown in the Supplemental Tables. In case of the fluorescence microscopy data, error bars represent standardized errors (SEs), calculated according to binomial or polynomial distribution formula (SEp = sqrt [p(1−p)/n], where p is the frequency of the given class, and n is the total number of cells in the sample). Statistical significance of differences was determined by Student’s t-test (for each time point in case of multiple time points) unless otherwise noted. Differences with P ≤ 0.05 were considered significant.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8171195.
Results
The N-terminal region of Hsp104 is required for [PSI+] destabilization by heat shock
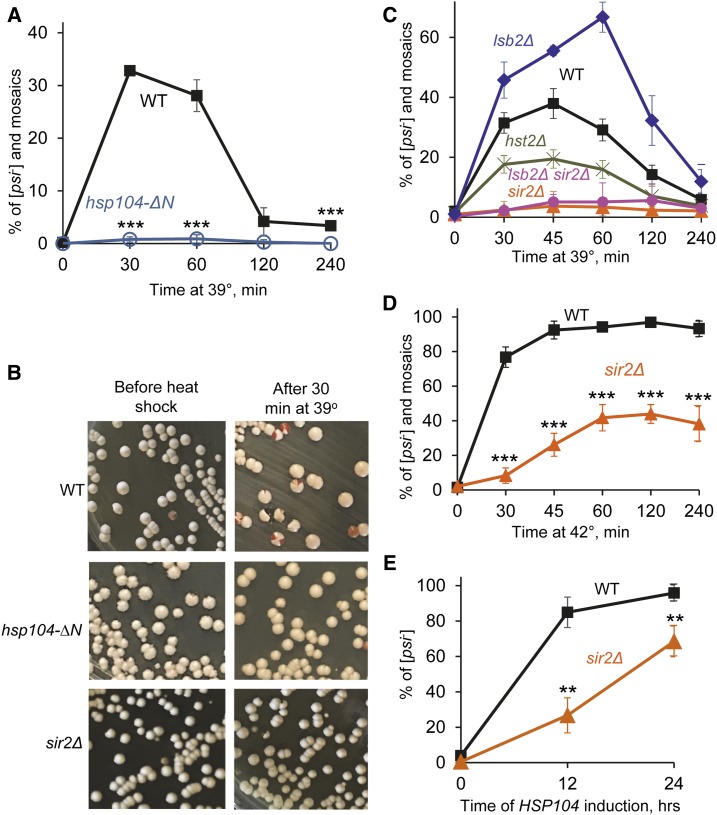
The N-terminal region, encompassing the first 147 amino acid residues of the Hsp104 protein, is not required for Hsp104 function in prion fragmentation and propagation, but is needed for [PSI+] curing by artificial overexpression of Hsp104 (Hung and Masison 2006). We checked whether this region is also required for curing by [PSI+] destabilization by short-term mild heat shock. For this purpose, a weak [PSI+] strain was constructed in which the N-terminal region of the HSP104 gene is deleted. The weak variant of the [PSI+] prion was chosen because it is destabilized by mild heat shock much more efficiently than the strong [PSI+] variant (Newnam et al. 2011). Effects of mild (39°) heat shock on the isogenic hsp104-∆N and wild-type strains were compared to each other. Neither wild-type nor the hsp104-∆N strain exhibited any observable decrease in viability during incubation at 39° (Supplemental Material, Figure S1 and Table S1). The wild-type strain produced 30% or more red ([psi−]) and mosaic (sectored [PSI+]/[psi−]) colonies after 30–60 min treatment at 39°; however, prion destabilization was ameliorated after prolonged incubation at 39° (Figure 1, A and B and Table S2). This confirms our previous observations (Newnam et al. 2011). Mosaic colonies prevailed among the colonies showing [PSI+] destabilization (Figure 1B and Table S2). This agrees with our previous findings showing that [PSI+] loss preferentially occurs in cell divisions following heat shock (Newnam et al. 2011; Ali et al. 2014). In contrast, the hsp104-∆N strain produced almost no red or mosaic colonies, indicating that deletion of the N-terminal region of Hsp104 impairs [PSI+] destabilization by heat shock. This result confirms that the same region of Hsp104 is crucial for both [PSI+] curing by Hsp104 overexpression and [PSI+] destabilization by heat shock.
Figure 1.
Effects of the deletions of HSP104N or SIR2 on [PSI+] destabilization. (A) Deletion of HSP104N (hsp104-∆N) prevents [PSI+] destabilization by short-term mild heat shock at 39°, compared to the isogenic wild-type (WT) strain. Numbers of colonies are shown in Table S2. (B) Examples of plates showing [PSI+] destabilization by mild heat shock in the WT strain (as detected by the appearance of red and mosaic pink-red colonies) and lack of destabilization in the hsp104-∆N and sir2∆ strains. (C) Quantitation of [PSI+] destabilization after mild heat shock at 39° in the WT, sir2∆, hst2∆, lsb2∆, and double lsb2∆ sir2∆ strains. Numbers of colonies are shown in Table S3. (D) Deletion of SIR2 (sir2∆) decreases [PSI+] destabilization by heat shock at 42°, compared to isogenic WT strain. Numbers of colonies are shown in Table S5. (E) Sir2∆ delays [PSI+] curing by Hsp104 overexpression, compared to isogenic WT strain. The PGAL-HSP104 construct was induced in galactose medium. Cells were sampled after indicated periods of time and plated onto the synthetic medium selected for the plasmids. The [psi−] colonies were identified after velveteen replica plating to the medium lacking adenine. Numbers of colonies are shown in Table S6. Error bars on A and C–E indicate SD. In general, the differences were statistically significant in all cases where deviations do not overlap. The P values are indicated by asterisks in parts A, D, and E: * P < 0.05, ** P < 0.01, *** P < 0.001. The P values for part C are shown in Table S4.
[PSI+] destabilization by mild heat shock is Sir2-dependent
Along with Hsp104, the NAD+-dependent protein deacetylase Sir2 (sirtuin) has been shown to control asymmetric segregation of aggregated oxidatively damaged proteins in cell divisions following heat shock (Aguilaniu et al. 2003; Tessarz et al. 2009; Liu et al. 2010). To determine whether Sir2 also plays a role in heat-shock-induced destabilization of [PSI+], we constructed a weak [PSI+] strain bearing the sir2∆ deletion and compared effects of mild heat shock at 39° on this strain and the isogenic wild-type strain containing the same variant of prion. Indeed, we observed that deletion of SIR2 dramatically reduced destabilization of [PSI+] by mild heat shock (Figure 1, B and C and Tables S3 and S4). Notably, deletion of HST2, another sirtuin not shown to be involved in asymmetric segregation of heat-damaged proteins (Liu et al. 2010), exhibited a significantly milder effect (Figure 1C and Tables S3 and S4). Neither sir2Δ (Figure S1 and Table S1) nor hst2Δ (Figure S1) exhibited any observable effect on the viability of yeast cells at 39°.
It has previously been demonstrated that Sir2 deficiency leads to actin cytoskeletal deficiencies (Erjavec et al. 2007; Liu et al. 2010), suggesting that association of aggregates with the actin cytoskeleton and actin cables prevents transmission of these aggregates into the daughter cell during asymmetric segregation (Tessarz et al. 2009; Liu et al. 2010, 2011). Indeed, deletion of the gene coding for the yeast actin assembly protein Lsb2 has been shown to increase [PSI+] destabilization by heat shock (Chernova et al. 2011). We have now shown that [PSI+] destabilization in the lsb2Δ strain is Sir2-dependent, as the lsb2Δ sir2Δ culture was essentially indistinguishable from the sir2Δ culture in regard to low frequencies of [PSI+] loss after mild heat shock (Figure 1C and Tables S3 and S4). This shows that the effect of sir2∆ on [PSI+] is epistatic to lsb2∆, in agreement with the notion that Sir2 might act on [PSI+] via modulation of the actin cytoskeleton. Notably, there was no difference in viability between the lsb2Δ and lsb2Δ sir2Δ strains, indicating that the effect of sir2Δ on prion destabilization is not due to any effect on cell viability.
As several previous studies of the effect of sir2∆ on aggregate localization used heat shock at 42° (Liu et al. 2010), we also checked [PSI+] destabilization by heat shock at that temperature. As we have reported previously and reproduced now, the weak [PSI+] prion is destabilized by heat shock at 42° in the wild-type strain, although in contrast to 39°, this destabilization is not ameliorated by prolonged incubation at high temperature (Figure 1D). The proportion of completely cured (red) colonies vs. mosaic colonies was higher after 42° treatment, compared to 39° (Table S5). No differences in viabilities at 42° were detected between the wild-type and sir2Δ cultures (Figure S1). We found that [PSI+] curing by 42° heat shock was significantly decreased although not entirely inhibited in the absence of Sir2, compared to the isogenic wild-type strain (Figure 1D and Table S1). This shows that the Sir2 protein is involved in prion destabilization at both 39 and 42°.
We then asked if sir2Δ acts on [PSI+] curing by artificial Hsp104 overexpression in the same direction as it does in the case of heat shock. For this purpose, we transformed wild-type and sir2Δ strains containing the weak [PSI+] variant with a plasmid containing the HSP104 gene under the control of a galactose-inducible (PGAL) promoter. The cultures were incubated in galactose medium to turn on the overexpression of Hsp104, then aliquots were plated onto glucose medium where overexpression is turned off, and the proportions of plasmid-containing cells that have either retained or lost [PSI+] were determined (Figure 1E and Table S6). We found that [PSI+] curing by artificial overexpression of Hsp104 was delayed, although not abolished, in sir2Δ cells as indicated by the increased proportion of colonies that grew on –Ade medium and retained lighter color on YPD, compared to the wild-type strain. Notably, the [PSI+] prion was as efficiently cured in the sir2Δ strain as in the wild-type strain by 5 mM guanidine hydrochloride (GuHCl), an agent inactivating Hsp104 (Figure S2). This indicates that Sir2 is involved in [PSI+] curing by Hsp104 overproduction but not by Hsp104 inactivation.
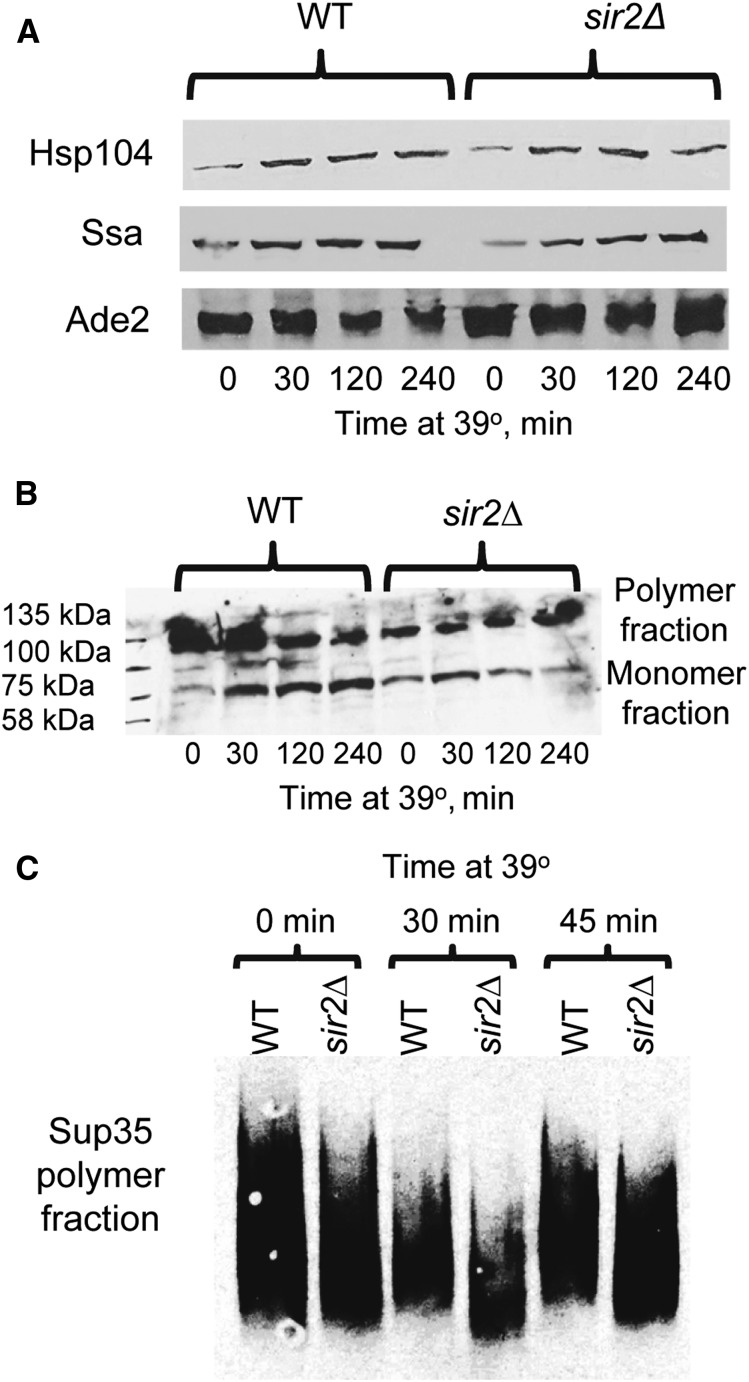
To determine if sir2Δ influences levels of heat-shock proteins (Hsp104 and Hsp70-Ssa) involved in [PSI+] propagation, and/or alters patterns of Sup35 aggregation, we compared levels of Hsps and aggregation patterns of Sup35 in the isogenic sir2Δ and wild-type strains before and during heat shock. Both Hsp104 and Hsp70-Ssa are induced by heat shock, with Hsp104 showing somewhat faster induction kinetics (see Newnam et al. 2011; also confirmed here, Figure S3). Western analysis indicated that Hsp104 and Hsp70-Ssa are produced at comparable levels in the sir2Δ and wild-type cultures before heat shock, and show nearly identical dynamics of heat-shock-induced accumulation in both cultures (Figure 2A and Table S7). The “Boiled gel” procedure (see Materials and Methods) showed that sir2Δ somewhat increases the proportion of monomeric vs. polymeric Sup35 protein in yeast cells prior to heat shock; however, further accumulation of monomeric protein during heat shock is not increased (and at a longer exposure to heat shock, the proportion of the monomeric Sup35 protein is decreased) in the sir2Δ cells, compared to wild-type cells (Figure 2B). Analysis of aggregate distribution by sizes, using semidenaturing gel electrophoresis (SDD-AGE), has demonstrated that heat-shocked sir2Δ cells show increased abundance of the smaller Sup35 polymers, compared to heat-shocked wild-type cells (Figure 2C). As smaller aggregates are more efficiently transmitted to daughter cells (see Derdowski et al. 2010) and therefore serve as more productive “seeds” for prion propagation, these data are in agreement with our observation that transmission of [PSI+] prion in post-heat-shock cell divisions is more efficient in the sir2∆ cells compared to wild-type cells.
Figure 2.
Hsp levels and Sup35 aggregation during heat shock. (A) Isogenic [PSI+] wild-type (WT) and sir2∆ cultures show similar background levels of Hsp104 or Ssa proteins, and similar dynamics of induction of these proteins during heat shock, as confirmed by SDS-PAGE and Western blotting, followed by the reaction to Hsp104-specific or Ssa-specific antibodies. Levels of Ade2 protein are shown as a loading control. Kinetics of Hsp104 and Ssa induction in the wild-type strain, based on densitometry measurements, are shown on Figure S3. Densitometry-based comparisons of the levels of Hsp104 and Ssa proteins in the wild-type and sir2Δ strains for each time point are shown in Table S7. (B) Sir2∆ increases proportion of monomeric vs. polymeric Sup35 protein in the [PSI+] culture prior to heat shock, but decreases accumulation of the monomeric fraction during heat shock, compared to the isogenic WT strain. Proteins were isolated from the cultures before heat shock and after heat shock for specified periods of time, and analyzed by the “boiled gel” assay (see Materials and Methods), followed by reaction to the Sup35C-specific antibody. Positions of molecular weight markers loaded at the beginning of electrophoresis are shown on the left. (C) During heat shock, Sup35 prion polymers of smaller size become less abundant in the WT culture but more abundant in the isogenic sir2∆ culture, as shown by SDD-AGE (see Materials and Methods), followed by reaction to the Sup35C-specific antibody.
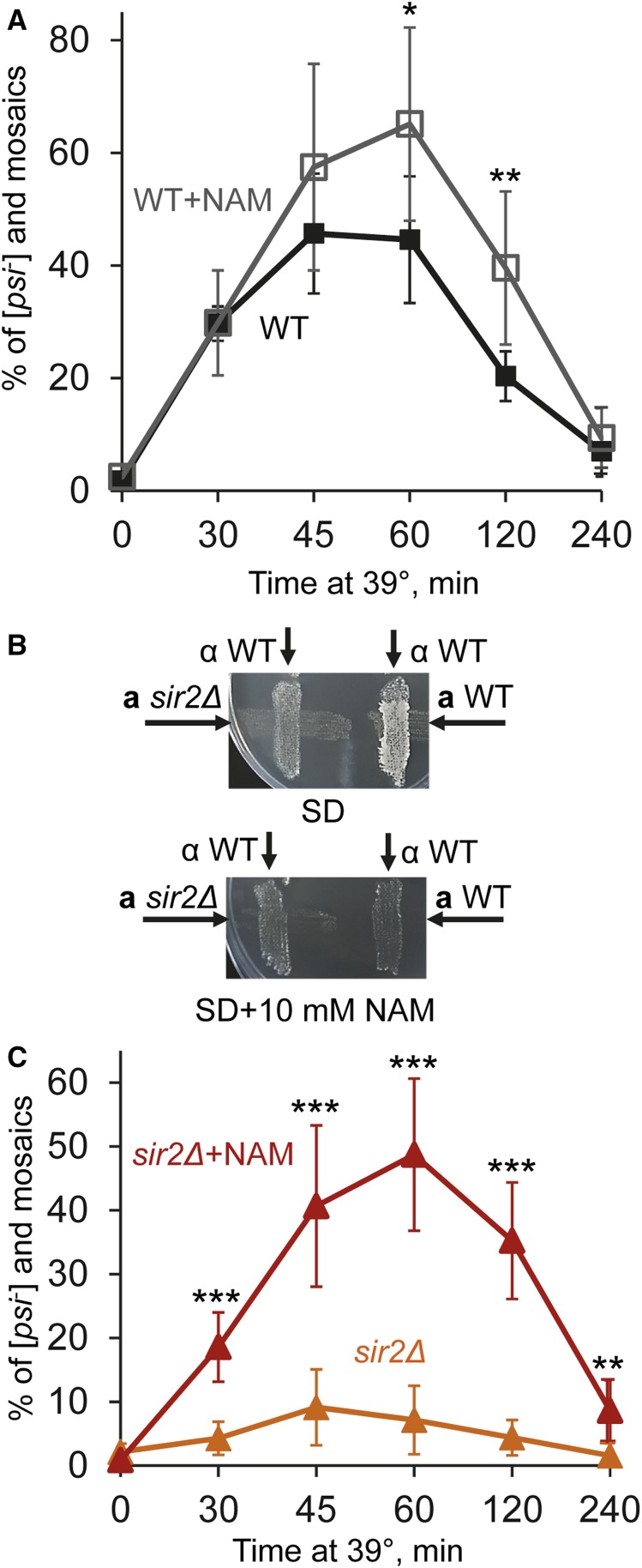
Addition of nicotinamide does not replicate effect of sir2Δ during heat shock
Nicotinamide (NAM), an agent known to inhibit Sir2, has been shown to inhibit Sir2-dependent gene silencing in yeast (Bitterman et al. 2002) and to antagonize asymmetric distribution of aggregated heat-damaged proteins in cell divisions (Liu et al. 2010). We checked whether or not addition of NAM is able to alter [PSI+] destabilization by heat shock to an extent similar to that of sir2∆. Surprisingly, addition of 10 mM NAM to yeast cultures during heat shock did not impair, and at least at some timepoints of the experiment, even somewhat increased [PSI+] curing in the wild-type strain (Figure 3A and Table S8). Notably, this concentration of NAM was sufficient to abolish the mating activity of yeast cells (Figure 3B), an effect that depends on inhibition of Sir2 and the resulting activation of normally silent mating type cassettes (Bitterman et al. 2002). This confirms that Sir2 was indeed inhibited by NAM. Potential explanation for the lack of impairment of [PSI+] curing by NAM is provided by our observation that NAM promotes [PSI+] curing by heat shock even in sir2∆ cells (Figure 3C and Table S9). Notably, both wild-type and sir2Δ cultures heat-shocked in the presence of NAM produced a somewhat increased proportion of completely cured (red) colonies vs. mosaic colonies, compared to cultures heat-shocked at 39° in the absence of NAM (Tables S8 and S9). Taken together, these data suggest that in addition to inhibiting Sir2, NAM facilitates heat-shock-mediated [PSI+] curing via a Sir2-independent mechanism.
Figure 3.
Effects of nicotinamide on heat-shock-mediated [PSI+] destabilization and mating. (A) Addition of nicotinamide (NAM) at the concentration of 10 mM does not decrease [PSI+] destabilization by mild heat shock in the wild-type (WT) strain. Colony numbers are shown in Table S8. (B) The sir2Δ strain is not capable of mating both in the absence of and in the presence of NAM, while the isogenic WT strain is capable of mating in the absence but not in the presence of NAM. Mating was detected in a combination with the tester WT strain of the opposite mating type (as indicated), by growth on synthetic medium selective for diploids (SD). Neither strain was capable of mating with the tester strain of the same mating type (data not shown). (C) Addition of NAM increases [PSI+] destabilization by mild heat shock in the sir2Δ strain. Colony numbers are shown in Table S9. Error bars in A and C indicate SD. Statistically significant differences in A and C are indicated by asterisks. Numbers of asterisks correspond to P values, as described in Figure 1 legend.
Mother–daughter distribution and colocalization of Hsp104-GFP and Sup35NM-DsRed foci in wild-type and sir2Δ cells
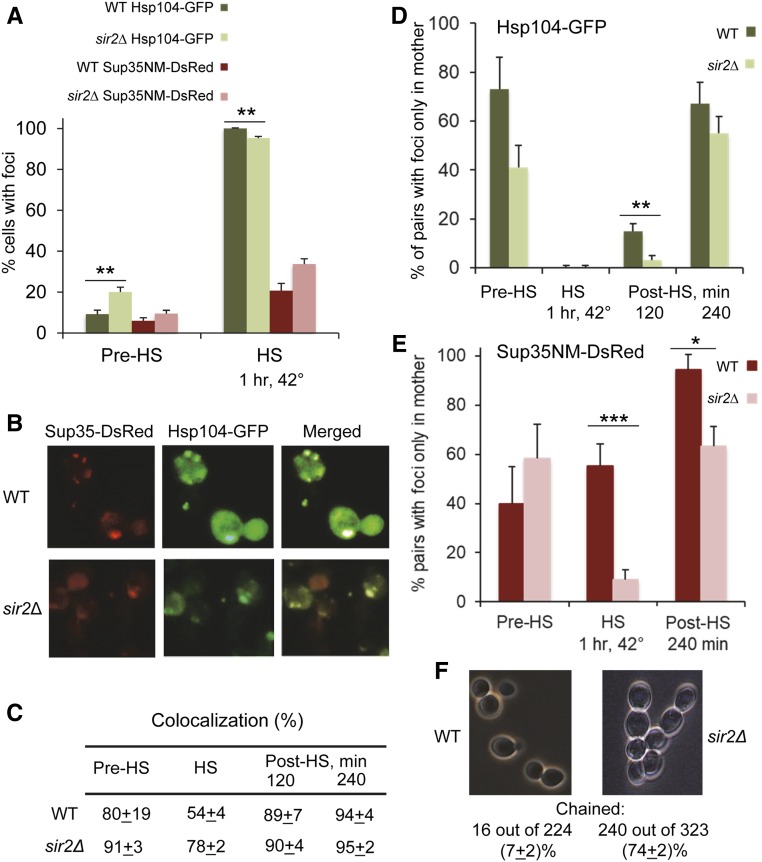
Our previous data indicate that [PSI+] loss in the first cell division after heat shock preferentially occurs in daughter cells (Newnam et al. 2011; Ali et al. 2014). Likewise, aggregates of oxidatively damaged proteins, associated with the Hsp104 chaperone, are known to be preferentially accumulated in mother cells (Erjavec et al. 2007). By using a yeast strain with an integrated cassette coding for GFP-tagged Hsp104, we confirmed that the Hsp104-GFP marked foci indeed appear during mild heat shock at 42° (Figure 4A and Table S10) as previously reported (Erjavec et al. 2007; Liu et al. 2010), or at 39° (Figure S4). To check the behavior of Sup35 prion aggregates in the same conditions, we then transformed a centromeric plasmid producing the chimeric Sup35NM-DsRed protein from the endogenous PSUP35 promoter into the same strains and subjected them to mild heat shock (42°, as in previously published experiments with Hsp104-GFP; Erjavec et al. 2007; Liu et al. 2010) followed by recovery at 25°. Dynamics of Sup35NM-DsRed aggregation were similar to those of Hsp104-GFP; however, aggregates were not as numerous and were formed in a smaller proportion of cells (Figure 4, A and B). Essentially all cells that possessed both Hsp104-GFP and Sup35NM-DsRed aggregates did show a high proportion of colocalization between them in both wild-type and sir2Δ strains (Figure 4, B and C), although some cells contained only Hsp104-GFP foci and did not show detectable Sup35NM-DsRed foci.
Figure 4.
Dynamics and localization of the Hsp104 and Sup35 aggregates during and after heat shock in the wild-type and sir2Δ cells. (A) Frequencies of cells with the Hsp104-GFP and Sup35NM-DsRed foci before (Pre-HS) and during (HS) heat shock for 1 hr at 42° in the wild-type (WT) and sir2Δ cells, as detected by fluorescence microscopy. Percentages of cells containing foci among all cells (Hsp104-GFP) or among all cells showing the red fluorescence (Sup35NM-DsRed) are shown. Foci were detected in one and the same culture, containing both the HSP104-GFP cassette on the chromosome and the SUP35NM-DsRed cassette on the plasmid. Numbers of cells are shown in Table S10. (B) Sup35NM-DsRed foci colocalize with Hsp104-GFP foci in both WT and sir2Δ cells. Heat shock was performed at 42° for 1 hr. (C) Quantitation of Hsp104-GFP and Sup35NM-DsRed colocalization. Percentages of colocalized dots or clumps out of all Hsp104-GFP and Sup35NM-DsRed dots or clumps detected in the cells containing both (green and red) types of aggregates are shown. In addition to before heat shock (Pre-HS) and 1 hr at 42° heat-shock (HS) data, (C) also shows data for the yeast cultures, recovering after heat shock (Post-HS) in YPD medium at 25° for specified periods of time. (D and E) Distribution of the Hsp104-GFP aggregates (D) and Sup35NM-DsRed aggregates (E), detectable by fluorescence microscopy between the mother and daughter cells before, during, and after heat shock. Numbers show percentages of cell pairs (cells with buds) containing foci only in the mother cell, out of all cell pairs where one or both cells have foci. In case of “chained” cells (see below), the mother cells producing buds at the end of the chain were considered. Designations are the same as in A and C. Cell counts are shown in Tables S11 (D) and S12 (E). (F) Sir2Δ leads to the “chained cells” phenotype, suggestive of delayed separation of cells. Numbers of chained vs. unchained cells were counted in the cultures prior to heat shock. Data are pooled from two experiments. Cells that are included in chains of three or more were considered “chained” cells. Only cells that were clearly either chained or not chained were counted. Differences between wild-type and sir2Δ cultures were statistically significant with P < 0.001. Standard deviations are shown in C. Standardized errors, calculated as described in Materials and Methods, are shown in A and D–F. Statistically significant differences in A, D, and E, as determined by Fischer’s exact test, are indicated by asterisks. Numbers of asterisks correspond to P values, as described in Figure 1 legend.
Importantly, the Hsp104-GFP foci did appear in both single and budded cells during heat shock, and in the case of budded cells, they were found in both mother and daughter (bud) cells. However, foci were preferentially retained in mother cells rather than in buds during recovery of wild-type culture after heat shock (Figure 4D, Figure S5, and Table S11), in agreement with previous observations (Erjavec et al. 2007). This mother-specific accumulation of the Hsp104-GFP foci was less pronounced in the sir2Δ strain (Figure 4D, Figure S5, and Table S11), as described previously (Liu et al. 2010). In the case of Sup35NM-DsRed, heat-shocked wild-type culture did show a high proportion of mother–daughter pairs with Sup35NM-DsRed foci appearing only in mother cells, compared to the sir2Δ culture (Figure 4, B and E and Table S12). This suggests that formation of some heat-shock-induced foci by Sup35NM-DsRed might be promoted by preexisting “nuclei,” more frequently present in mother cells, for example by protein deposits previously found in aged cells (Saarikangas and Barral 2015). Dynamics of the distribution of the Sup35NM-DsRed foci between the mother and daughter cells during recovery after heat shock generally followed that of the Hsp104-GFP foci, demonstrating increased accumulation of foci in mother cells (Figure 4E and Table S12). Once again, this asymmetry was impaired in part by sir2∆. Notably, the sir2Δ strain frequently formed “chains” with three or more cells, suggestive of a delay in cell separation after the cell division (Figure 4F). This delay in cell separation might provide an additional explanation for the impaired asymmetry of aggregate distribution in sir2∆ cells, as aggregates in sir2Δ cells have a longer time (compared to wild-type cells) for the transmission to daughters.
The heat-shock noninducible chaperone Hsp70-Ssb and its cochaperones influence [PSI+] destabilization by heat shock
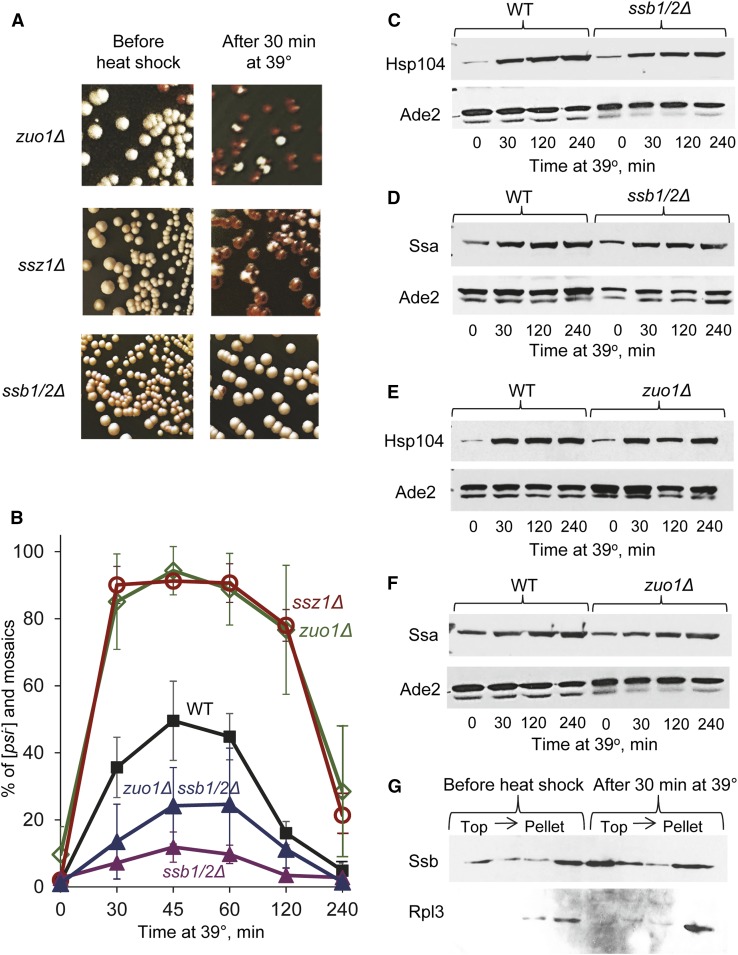
The ribosome-associated chaperone Hsp70-Ssb promotes [PSI+] curing by artificially overproduced Hsp104 (Chernoff et al. 1999), and release of Ssb from the ribosome to the cytosol in strains lacking one or both components of the ribosome-associated chaperone complex (RAC), Hsp40-Zuo1 and/or Hsp70-Ssz1, also leads to increased [PSI+] curing by excess Hsp104 and destabilization of some weak variants of [PSI+] (Kiktev et al. 2015). We have shown that Ssb release is accompanied by a decrease in the proportion of Ssa bound to the Sup35 aggregates, suggesting that cytosolic Ssb impairs the function of the Hsp104/Hsp70-Ssa/Hsp40 chaperone complex in prion propagation by antagonizing Ssa (Kiktev et al. 2015; Chernoff and Kiktev 2016). As the effect of heat shock on [PSI+] depends on the Hsp104/Ssa balance, we checked whether Ssb plays a role in [PSI+] destabilization by heat shock. For this purpose, we deleted either one or both SSB genes (SSB1 and/or SSB2) in the weak [PSI+] strain and subjected it to mild heat shock at 39°. Indeed, deletion of either one (ssb1∆ or ssb2∆, see Figure S6 and Tables S13 and S14) or both (ssb1/2∆, see Figure 5, A and B and Tables S13 and S14) SSB genes dramatically impaired [PSI+] destabilization by mild heat shock. In contrast, deletion of either ZUO1 or SSZ1, shown previously to release Ssb to the cytosol (Kiktev et al. 2015), dramatically increased heat-shock-mediated [PSI+] destabilization (Figure 5, A and B) and greatly increased the proportion of completely cured (red) colonies, relative to mosaic colonies (Table S13). Neither ssb1/2Δ nor zuo1Δ influenced cell viability at 39° (Figure S1 and Table S1) nor Hsp104 (Figure 5, C and D and Table S7) and Ssa (Figure 5, E and F and Table S7) levels before or during heat shock. While Ssb is not induced by heat shock (Iwahashi et al. 1995; Lopez et al. 1999, confirmed for our strains, see example in Figure S7), centrifugation of cell extracts in sucrose gradients demonstrated that a significant fraction of Ssb protein moves from the pellet fraction (corresponding to the ribosomal marker Rpl3) to the top (cytosolic) fractions in the extracts of heat-shocked cells, compared to the nonshocked culture (Figure 5G). These data demonstrate that heat shock induces a change in the intracellular localization of Ssb, and agree with our previous results showing that the release of Ssb from the ribosome to the cytosol antagonizes Ssa and impairs [PSI+] propagation (Kiktev et al. 2015; Chernoff and Kiktev 2016).
Figure 5.
Effects of Ssb and RAC proteins on [PSI+] destabilization by mild heat shock. (A and B) Deletion of both SSB coding genes (ssb1/2∆) decreases, while deletion of ZUO1 (zuo1Δ) or SSZ1 (ssz1Δ) increases [PSI+] destabilization by mild heat shock at 39°, as seen on plates (A) and confirmed by quantitation (B). Triple deletion ssb1/2Δ zuo1Δ greatly decreases [PSI+] destabilization as compared to zuo1Δ and wild-type strains, but shows somewhat increased [PSI+] destabilization as compared to the ssb1/2Δ strain (B). Bars indicate SD. Differences were statistically significant in all cases where deviations do not overlap. Numbers of colonies are shown in Table S13, while P values are shown in Table S14. (C and D) Isogenic [PSI+] wild-type (WT) and ssb1/2∆ cultures show similar background levels of the Hsp104 (C) and Ssa (D) proteins before and during heat shock, as confirmed by SDS-PAGE and Western blotting, followed by reaction to antibodies, specific to Hsp104 (C) or Ssa (D). (E and F) Isogenic [PSI+] WT and zuo1∆ cultures show similar levels of the Hsp104 (C) and Ssa (D) proteins at each time point before and during heat shock, as confirmed by SDS-PAGE and Western blotting, followed by reaction to antibodies, specific to Hsp104 (E) or Ssa (F). In C, D or F, levels of Ade2 protein in the same samples are shown as a loading control (second band seen on the Ade2 images represents a proteolytic product, detected in most experiments.) Densitometry-based comparisons of the levels of Hsp104 and Ssa proteins for each time point in the experiments are shown in Table S7. (G) Ssb moves to the cytosol during heat shock in wild-type cells. Protein extracts were isolated from yeast cultures either prior to heat shock or after heat shock for 30 min at 39°, and fractionated by sucrose gradient centrifugation, followed by SDS-PAGE and Western blotting with antibodies specific to Ssb or to ribosomal protein Rpl3. During heat shock, increased accumulation of Ssb in the top fraction of the gradient (corresponding to the cytosol fraction) is detected, while Rpl3 remains associated primarily with the pellet, containing ribosomes.
Increased [PSI+] destabilization in the zuo1∆ or ssz1∆ strains also agrees with previous observations showing that these deletions cause a release of all or most Ssb from the ribosome to the cytosol (Willmund et al. 2013; Kiktev et al. 2015). Indeed, the triple deletion ssb1/2Δ zuo1Δ showed a significantly decreased destabilization of [PSI+] by 39° heat shock, compared to the single deletion zuo1Δ (Figure 5B and Tables S13 and S14). However, the frequency of heat-shock-induced [PSI+] loss in the ssb1/2Δ zuo1Δ strain remains somewhat higher than that in the ssb1/2Δ strain, mostly due to mosaic colonies. No impact of triple ssb1/2Δ zuo1Δ deletion on cell viability at 39° was detected (Figure S1). Taken together, these data confirm that most of the increase in [PSI+] destabilization in the zuo1Δ deletion strain is mediated by Ssb; however, a minor Ssb-independent component of the effect of zuo1Δ on [PSI+] also exists.
Discussion
Hsp104 and prion destabilization by heat shock
While Hsp104 is capable of both promoting fragmentation/propagation of the Sup35 prion when working together with Hsp70-Ssa/Hsp40, and curing the Sup35 prion when artificially overexpressed in excess of Hsp70-Ssa, different regions of the Hsp104 protein are involved in these processes. Specifically, the N-terminal domain of Hsp104 was shown to be required for [PSI+] curing by Hsp104 overproduction, but not for the Hsp104 function in [PSI+] propagation (Hung and Masison 2006). Our data (Figure 1, A and B) show that heat-shock-mediated destabilization of [PSI+] also requires the N-terminal domain of Hsp104, further supporting the role of excess Hsp104 in this process. While it was suggested that excess Hsp104 solubilizes Sup35 prion polymers by “chopping” monomers from their ends (Park et al. 2014; Zhao et al. 2017; Greene et al. 2018), recent evidence rather indicates that [PSI+] loss in the presence of overproduced Hsp104 is due to prion malpartition in cell divisions, implying that excess Hsp104 impairs segregation of prion from mother into the daughter cell (Ness et al. 2017). Requirement of the same region of Hsp104 for both [PSI+] curing by artificial Hsp104 overproduction and [PSI+] destabilization by heat shock agrees with our previous data pointing to the role of excess Hsp104 in [PSI+] destabilization by heat shock (Newnam et al. 2011), while asymmetric loss of [PSI+] in the first cell division following heat shock (Newnam et al. 2011; Ali et al. 2014) supports the role of asymmetric segregation in heat-shock-induced prion destabilization.
Role of the cell asymmetry apparatus in prion destabilization by heat shock
In strong support of the role of asymmetric protein segregation in heat-shock-mediated destabilization of [PSI+], our data show that the deletion of SIR2, coding for a NAD+-dependent protein deacetylase (sirtuin), previously implicated in the control of cell asymmetry, decreases destabilization of the [PSI+] prion by heat shock (Figure 1, B–D). Asymmetric segregation of oxidatively damaged proteins has previously been shown to be dependent on both Sir2 and Hsp104 (Aguilaniu et al. 2003; Erjavec et al. 2007; Tessarz et al. 2009; Liu et al. 2010). It has been proposed that the effect of sir2Δ is due to increased acetylation of the chaperonin complex, which affects actin folding and thus influences actin-dependent retention and/or retrograde transport of heat-damaged proteins (Liu et al. 2010). Involvement of the actin cytoskeleton in [PSI+] destabilization by heat shock agrees with our previous results showing that the deletion of the actin assembly protein Lsb2 further increases [PSI+] loss at high temperature (Chernova et al. 2011; Ali et al. 2014). Moreover, we now show that sir2∆ is epistatic to Lsb2 in regard to heat-shock-mediated [PSI+] destabilization, indicating that both Sir2 and Lsb2 proteins are likely to act on [PSI+] via one and the same pathway (Figure 1C). However, our data also point to a delay of cell separation during cytokinesis in the sir2∆ strains (Figure 4F). This may also impair asymmetry of cytoplasm distribution due to a longer period of time available for the movement of cytoplasmic components from the mother to daughter cell, in accordance with the diffusion model of asymmetric segregation (Zhou et al. 2011). It should be noted that the “active” (via actin cytoskeleton) and “passive” (via diffusion) models of cell asymmetry are not mutually exclusive, as both processes may contribute to asymmetric distribution of protein aggregates. However, our observation that heat-shocked sir2∆ cells contain a large proportion of prion polymers of smaller size, compared to wild-type cells (Figure 2C), suggests that the role of Sir2 is achieved not only through a passive mechanism. It is possible that the increase in aggregate size during stress occurs due to inclusion of prion polymers into larger protein complexes. Assembly of these complexes may be promoted by Hsp104 and the actin cytoskeleton. Large complexes tend to segregate asymmetrically in cell divisions after stress, leading to prion loss. Assembly of these large complexes could be impaired in the sir2∆ cells. Decreased assembly of large complexes may increase accessibility of prion polymers for the newly immobilized protein and polymer fragmentation machinery, which further decreases the proportion of monomeric protein (Figure 2B) and polymer size (Figure 2C) in the heat-shocked sir2∆ cells.
In either case, the effects of sir2∆ on prion aggregates appear to parallel its effects on the aggregates of heat-damaged proteins. Two differences between these experimental models could, however, be detected. First, deletion of the gene HST2, coding for another sirtuin and not specifically implicated in chaperonin deacetylation, is not known to have any impact on asymmetric segregation of the aggregates of heat-damaged proteins (Liu et al. 2010). However, hst2∆ did somewhat decrease [PSI+] destabilization by heat shock in our hands (Figure 1C). Still, the effect of sir2∆ was much stronger than that of hst2∆. Second, treatment with NAM, an inhibitor of the deacetylase activity of Sir2, impaired asymmetric retention of heat-damaged proteins (Liu et al. 2010) but did not decrease [PSI+] destabilization in our assay (Figure 3A). This is due to the fact that NAM itself increases heat-shock promoted [PSI+] destabilization via a yet unknown Sir2-independent mechanism (Figure 3C).
The colocalization of Sup35NM with Hsp104 aggregates, observed in this work, indicates that Sup35 is segregated along with Hsp104-tagged aggregates into mother cells after heat stress. This asymmetric segregation may promote curing of [PSI+] and is impaired in sir2Δ cells. Two scenarios may explain the asymmetry of prion loss after heat shock: (1) asymmetric segregation of prion aggregates per se, leading to their accumulation in mother cells and resulting in their clearance from daughter cells, which therefore accumulate newly synthesized Sup35 protein in the nonprion form, as proposed by us (Newnam et al. 2011) and is in agreement with studies of the effects of artificial Hsp104 overexpression (Ness et al. 2017); and (2) asymmetric mother-specific accumulation of Hsp104, that in turn leads to a defect in prion maintenance, as proposed by Klaips et al. (2014). Experimental evidence could be provided in favor of both models. Indeed, fluorescently tagged Hsp104 is associated with aggregates of stress-damaged proteins and accumulates together with them in mother cells, apparently playing an important role in this process (Liu et al. 2010). Our data confirm both mother-specific accumulation of Hsp104 and Hsp104-marked protein aggregates in cell divisions following heat shock, and alterations of this asymmetry in sir2∆ cells (Figure 4, A and D). On the other hand, our data also show that aggregated clumps of Sup35 prion polymers, which are increased in size after stress, colocalize with the Hsp104-marked aggregates of heat-damaged proteins (Figure 4, B and C), and exhibit a tendency for Sir2-dependent mother-specific accumulation in cell divisions following stress (Figure 4E).
We believe that the two models described above are not mutually exclusive, especially taking into account recent data indicating that excess Hsp104 cures [PSI+] via a malpartition mechanism, rather than through monomerization (Ness et al. 2017). Apparently, Hsp104 may protect cells from toxic effects of protein aggregation in two ways: (a) via direct disaggregation and refolding of aggregated proteins when working in complex with Hsp70-Ssa/Hsp40; and (b) by promoting aggregate retention in the mother cell when acting in combination with the cytoskeletal machinery. The latter process represents a second line of defense that becomes crucial when levels of aggregate accumulation are too high and Hsp104/Hsp70-Ssa/Hsp40-promoted disaggregation is insufficient for survival. Aggregate retention is facilitated when Hsp104 is present in excess of Hsp70-Ssa, a situation that occurs after a short-term stress due to the fact that Ssa levels are not increased during stress as rapidly as are levels of Hsp104. As previously proposed by us (Liebman and Chernoff 2012), Hsp104 acts on prion aggregates in a manner similar to its effect on heat-damaged protein aggregates. Thus, in cell divisions following stress, both prion aggregates and aggregates of heat-damaged proteins, assembled into large structures via interactions with each other and Hsp104, are preferentially retained in mother cells. This results in preferential prion clearance from the daughters in the first poststress divisions as we previously reported. However, the mother cells retain prion aggregates as a part of larger protein complexes, having a lower ability of transmission into the daughters (see Derdowski et al. 2010), and contain Hsp104 at increased levels, promoting aggregate malpartitioning. This results in the unstable inheritance of prion aggregates in subsequent divisions of mother cells, as reported by Klaips et al. (2014). Sir2∆ partly restores symmetric distribution of both protein aggregates and Hsp104, thus antagonizing stress-induced prion loss. Our observation that sir2∆ also decreases [PSI+] curing by excess Hsp104 points to similarity between the effects of stress-induced and artificial Hsp104 overproduction. However, artificial Hsp104 overproduction is not accompanied by massive aggregation of other proteins, thus making the dependence of prion stabilization on Sir2 less evident.
Role of ribosome-associated chaperones during heat shock
Our work has also uncovered a new role for the Hsp70 chaperone Ssb during heat shock. Ssb is normally found in association with the RAC complex, composed of the proteins Hsp40-Zuo1 and Hsp70-Ssz, and is implicated in the prevention of misfolding of nascent polypeptides exiting the ribosome (Bukau et al. 2000; Koplin et al. 2010; Willmund et al. 2013). Ssb is also involved in a variety of other functions, such as ribosome biogenesis (Mudholkar et al. 2017), glucose repression (von Plehwe et al. 2009), and targeting some misfolded proteins for degradation (Ohba 1997; Kimura et al. 2016). Disruption of the RAC complex via deletion of the gene coding either for Zuo1 or for Ssz1 (or both) releases Ssb from the ribosome into cytosol (Gautschi et al. 2002; Willmund et al. 2013). Our previous data show that cytosolic Ssb antagonizes the binding of Ssa to the prion polymer, resulting effectively in a shift of the balance between Hsp104 and prion-bound Ssa, and therefore in a defect in prion fragmentation and propagation (Kiktev et al. 2015). Moreover, release of Ssb from the ribosome to cytosol is also observed in certain unfavorable growth conditions where translational rates go down and number of active polysomes is decreased, for example during growth in poor synthetic medium. Here we show that the release of Ssb from the ribosome to cytosol is also induced by heat shock (Figure 5G). Apparently, this release has functional consequences, as [PSI+] destabilization by mild heat shock is decreased in cells lacking one or both SSB genes (Figure 5, A and B and Figure S6), which release less or no Ssb, but this destabilization is increased in zuo1Δ cells (Figure 5, A and B), where binding of Ssb to the ribosome is known to be impaired (Willmund et al. 2013; Kiktev et al. 2015). Moreover, the increase of heat-shock-induced [PSI+] loss in zuo1Δ cells is to a significant extent dependent on Ssb (Figure 5B). Taken together, our findings support the concept that relocalization of Ssb into the cytosol mediates some of the effects of stress (specifically, heat shock) on aggregated proteins. Therefore, Ssb acts as an antiprion agent during stress, impairing proliferation of prion aggregates in poststress divisions. This effect is probably achieved via further altering the balance between the polymer-related Ssa and Hsp104 in favor of Hsp104. Heat shock induces massive protein aggregation (Wallace et al. 2015) and is shown to promote formation of some prions (Tyedmers et al. 2008; Chernova et al. 2017b,c). As at least some yeast prions are toxic to cells (Wickner et al. 2018), the antiprion effect of Ssb during stress may have an adaptive role. Notably, transcription of SSB genes is not induced by heat shock (Lopez et al. 1999), therefore our data show that in addition to changes in expression, stress-induced alterations in intracellular localization of some chaperone proteins may also play a functional, possibly adaptive, role. This parallels previous observations showing that relocalization of the Hsp40 protein Sis1 (a cochaperone of Ssa) between the cytosol and nucleus, mediated by the stress-inducible sorting factor Cur1, also has functional consequences, including the effect on heat-shock-dependent [PSI+] destabilization (Barbitoff et al. 2017; Matveenko et al. 2018).
The model for heat-shock-mediated destabilization of [PSI+] that emerges from our data explains this process via the shift in the balance of Hsp104 and Ssa proteins binding to prion polymers. This shift occurs not only because of rapid induction of Hsp104 during heat shock, but also due to release of Ssb from the ribosome to the cytosol in these conditions, as cytosolic Ssb antagonizes binding of Ssa to prions (Kiktev et al. 2015). Binding of Hsp104 in the absence of Ssa promotes Sir2-dependent asymmetric distribution of prion aggregates (coalescing with deposits of heat-damaged aggregated proteins) between the mother and daughter cells in poststress divisions, leading to malpartition and segregational loss of the prion. Such a mechanism links the effects of stress on heritable protein aggregation with the general cellular response to accumulation of aggregated proteins in stressed cells.
Acknowledgments
We thank V. Alenin, D. Bedwell, E. Craig, S. Lindquist, and J. Warner for antibodies, S.G. Inge-Vechtomov, K. Lobachev, and T. Nyström for strains, and G. Newnam for assistance in some experiments. This work was supported by grants MCB 1516872 and MCB 1817976 from National Science Foundation to Y.O.C. In addition, Y.O.C. was partially supported by project 15.61.2218.2013 from St. Petersburg State University, and Q.L.F. was partially supported by Presidential Undergraduate Research Award (PURA) from Georgia Institute of Technology.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8171195.
Communicating editor: O. Cohen-Fix
Literature Cited
- Aguilaniu H., Gustafsson L., Rigoulet M., Nyström T., 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299: 1751–1753. 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- Ali M., Chernova T. A., Newnam G. P., Yin L., Shanks J., et al. , 2014. Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J. Biol. Chem. 289: 27625–27639. 10.1074/jbc.M114.582429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astor M. T., Kamiya E., Sporn Z. A., Berger S. E., Hines J. K., 2018. Variant-specific and reciprocal Hsp40 functions in Hsp104-mediated prion elimination. Mol. Microbiol. 109: 41–62. 10.1111/mmi.13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbitoff Y. A., Matveenko A. G., Moskalenko S. E., Zemlyanko O. M., Newnam G. P., et al. , 2017. To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 105: 242–257. 10.1111/mmi.13697 [DOI] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A., 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 277: 45099–45107. 10.1074/jbc.M205670200 [DOI] [PubMed] [Google Scholar]
- Bukau B., Deuerling E., Pfund C., Craig E. A., 2000. Getting newly synthesized proteins into shape. Cell 101: 119–122. 10.1016/S0092-8674(00)80806-5 [DOI] [PubMed] [Google Scholar]
- Byers J. S., Jarosz D. F., 2014. Pernicious pathogens or expedient elements of inheritance: the significance of yeast prions. PLoS Pathog. 10: e1003992 10.1371/journal.ppat.1003992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Kiktev D. A., 2016. Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 62: 677–685. 10.1007/s00294-016-0586-2 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 268: 880–884. 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Newnam G. P., Kumar J., Allen K., Zink A. D., 1999. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI+] prion. Mol. Cell. Biol. 19: 8103–8112. 10.1128/MCB.19.12.8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Romanyuk A. V., Karpova T. S., Shanks J. R., Ali M., et al. , 2011. Prion induction by the short-lived, stress-induced protein lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell 43: 242–252. 10.1016/j.molcel.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Wilkinson K. D., Chernoff Y. O., 2014. Physiological and environmental control of yeast prions. FEMS Microbiol. Rev. 38: 326–344. 10.1111/1574-6976.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Wilkinson K. D., Chernoff Y. O., 2017aPrions, chaperones, and proteostasis in yeast. Cold Spring Harb. Perspect. Biol. pii: a023663. 10.1101/cshperspect.a023663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Kiktev D. A., Romanyuk A. V., Shanks J. R., Laur O., et al. , 2017b. Yeast short-lived actin-associated protein forms a metastable prion in response to thermal stress. Cell Rep. 18: 751–761. 10.1016/j.celrep.2016.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T. A., Chernoff Y. O., Wilkinson K. D., 2017c. Prion-based memory of heat stress in yeast. Prion 11: 151–161. 10.1080/19336896.2017.1328342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdowski A., Sindi S. S., Klaips C. L., DiSalvo S., Serio T. R., 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330: 680–683. 10.1126/science.1197785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W., 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144: 1375–1386. https://www.genetics.org/content/144/4/1375.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., Nyström T., 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21: 2410–2421. 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M., Mun A., Ross S., Rospert S., 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99: 4209–4214. 10.1073/pnas.062048599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H., 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425 10.1093/nar/20.6.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. R., Lindquist S., 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- Greene L. E., Zhao X., Eisenberg E., 2018. Curing of [PSI+] by Hsp104 overexpression: clues to solving the puzzle. Prion 12: 9–15. 10.1080/19336896.2017.1412911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung G. C., Masison D. C., 2006. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173: 611–620. 10.1534/genetics.106.056820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi H., Wu Y., Tanguay R. M., 1995. Detection and expression of the 70 kDa heat shock protein Ssb1p at different temperatures in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 213: 484–489. 10.1006/bbrc.1995.2157 [DOI] [PubMed] [Google Scholar]
- James B. D., Leurgans S. E., Hebert L. E., Scherr P. A., Yaffe K., et al. , 2014. Contribution of Alzheimer’s disease to mortality in the United States. Neurology 82: 1045–1050. 10.1212/WNL.0000000000000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev D. A., Melomed M. M., Lu C. D., Newnam G. P., Chernoff Y. O., 2015. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 96: 621–632. 10.1111/mmi.12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Kurata Y., Nakabayashi J., Kagawa H., Hirano H., 2016. N-Myristoylation of the Rpt2 subunit of the yeast 26S proteasome is implicated in the subcellular compartment-specific protein quality control system. J. Proteomics 130: 33–41. 10.1016/j.jprot.2015.08.021 [DOI] [PubMed] [Google Scholar]
- Klaips C. L., Hochstrasser M. L., Langlois C. R., Serio T. R., 2014. Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing. eLife 3: e04288 10.7554/eLife.04288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A., Preissler S., Ilina Y., Koch M., Scior A., et al. , 2010. A dual function for chaperones SSB–RAC and the NAC nascent polypeptide–associated complex on ribosomes. J. Cell Biol. 189: 57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V., Alexandrov I. M., Mitkevich O. V., Shkundina I. S., Ter-Avanesyan M. D., 2006. Purification and analysis of prion and amyloid aggregates. Methods 39: 50–55. 10.1016/j.ymeth.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Chernoff Y. O., 2012. Prions in yeast. Genetics 191: 1041–1072. 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Larsson L., Caballero A., Hao X., Oling D., et al. , 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140: 257–267. 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Liu B., Larsson L., Franssens V., Hao X., Hill S. M., et al. , 2011. Segregation of protein aggregates involves actin and the polarity machinery. Cell 147: 959–961. 10.1016/j.cell.2011.11.018 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez N., Halladay J., Walter W., Craig E. A., 1999. SSB, encoding a ribosome-associated chaperone, is coordinately regulated with ribosomal protein genes. J. Bacteriol. 181: 3136–3143. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC93769/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveenko A. G., Barbitoff Y. A., Jay-Garcia L. M., Chernoff Y. O., Zhouravleva G. A., 2018. Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 64: 317–325. 10.1007/s00294-017-0750-3 [DOI] [PubMed] [Google Scholar]
- McDonald J. H., 2009. Handbook of Biological Statistics. Sparky House Publishing, Baltimore. [Google Scholar]
- Mudholkar K., Fitzke E., Prinz C., Mayer M. P., Rospert S., 2017. The Hsp70 homolog Ssb affects ribosome biogenesis via the TORC1-Sch9 signaling pathway. Nat. Commun. 8: 937 10.1038/s41467-017-00635-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F., Cox B. S., Wongwigkarn J., Naeimi W. R., Tuite M. F., 2017. Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartition of [PSI+] propagons. Mol. Microbiol. 104: 125–143. 10.1111/mmi.13617 [DOI] [PubMed] [Google Scholar]
- Newnam G. P., Birchmore J. L., Chernoff Y. O., 2011Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 408: 432–448. 10.1016/j.jmb.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M., 1997. Modulation of intracellular protein degradation by SSB1–SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett. 409: 307–311. 10.1016/S0014-5793(97)00535-8 [DOI] [PubMed] [Google Scholar]
- Park Y. N., Zhao X., Yim Y. I., Todor H., Ellerbrock R., et al. , 2014. Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot. Cell 13: 635–647. 10.1128/EC.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B., 2013. Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 47: 601–623. 10.1146/annurev-genet-110711-155524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M., Masison D. C., 2011. Modulation and elimination of yeast prions by protein chaperones and co-chaperones. Prion 5: 245–249. 10.4161/pri.17749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., Barral Y., 2015. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife 4: e06197 10.7554/eLife.06197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 2002. Getting started with yeast. Methods Enzymol. 350: 3–41. 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- Tessarz P., Schwarz M., Mogk A., Bukau B., 2009. The yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol. Cell. Biol. 29: 3738–3745. 10.1128/MCB.00201-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Mundy C. R., Cox B. S., 1981. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 98: 691–711. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1214469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J., Madariaga M. L., Lindquist S., 2008. Prion switching in response to environmental stress. PLoS Biol. 6: e294 10.1371/journal.pbio.0060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J., Abrams J., Wang Y., Morano K. A., 2012. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 76: 115–158. 10.1128/MMBR.05018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Plehwe U., Berndt U., Conz C., Chiabudini M., Fitzke E., et al. , 2009. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 23: 2102–2115. 10.1101/gad.529409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E. W., Kear-Scott J. L., Pilipenko E. V., Schwartz M. H., Laskowski P. R., et al. , 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weuve J., Hebert L. E., Scherr P. A., Evans D. A., 2014. Deaths in the United States among persons with Alzheimer’s disease (2010–2050). Alzheimers Dement. 10: e40–e46. 10.1016/j.jalz.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Edskes H. K., Son M., Bezsonov E. E., DeWilde M., et al. , 2018. Yeast prions compared to functional prions and amyloids. J. Mol. Biol. 430: 3707–3719. 10.1016/j.jmb.2018.04.022 [DOI] [PubMed] [Google Scholar]
- Willmund F., del Alamo M., Pechmann S., Chen T., Albanèse V., et al. , 2013. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152: 196–209. 10.1016/j.cell.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Tyedmers J., Bukau B., Mogk A., 2012. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 198: 387–404. 10.1083/jcb.201201074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Rodriguez R., Silberman R. E., Ahearn J. M., Saidha S., et al. , 2017. Heat shock protein 104 (Hsp104)-mediated curing of [PSI+] yeast prions depends on both [PSI+] conformation and the properties of the Hsp104 homologs. J. Biol. Chem. 292: 8630–8641. 10.1074/jbc.M116.770719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Slaughter B. D., Unruh J. R., Eldakak A., Rubinstein B., et al. , 2011. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147: 1186–1196. 10.1016/j.cell.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8171195.