Here, López-Cortegano and Caballero carried out a meta-analysis using records from a database of genome-wide studies in order to investigate the nature of missing heritability in humans...
Keywords: GWAS, missing heritability, prediction of complex traits, big data
Abstract
Thousands of genes responsible for many diseases and other common traits in humans have been detected by Genome Wide Association Studies (GWAS) in the last decade. However, candidate causal variants found so far usually explain only a small fraction of the heritability estimated by family data. The most common explanation for this observation is that the missing heritability corresponds to variants, either rare or common, with very small effect, which pass undetected due to a lack of statistical power. We carried out a meta-analysis using data from the NHGRI-EBI GWAS Catalog in order to explore the observed distribution of locus effects for a set of 42 complex traits and to quantify their contribution to narrow-sense heritability. With the data at hand, we were able to predict the expected distribution of locus effects for 16 traits and diseases, their expected contribution to heritability, and the missing number of loci yet to be discovered to fully explain the familial heritability estimates. Our results indicate that, for 6 out of the 16 traits, the additive contribution of a great number of loci is unable to explain the familial (broad-sense) heritability, suggesting that the gap between GWAS and familial estimates of heritability may not ever be closed for these traits. In contrast, for the other 10 traits, the additive contribution of hundreds or thousands of loci yet to be found could potentially explain the familial heritability estimates, if this were the case. Computer simulations are used to illustrate the possible contribution from nonadditive genetic effects to the gap between GWAS and familial estimates of heritability.
UNDERSTANDING the genetic architecture of complex traits has become a fundamental topic of study in human genetics (Gibson 2012; Timpson et al. 2018). In recent years, huge efforts have been made to investigate the genetic basis of human complex traits through Genome-Wide Association Studies (GWAS) or meta-analyses of their results (Paternoster et al. 2015; Gormley et al. 2016; Justice et al. 2017; Visscher et al. 2017). There has been a parallel increase in the number of big Consortiums able to carry out large GWAS with higher and higher numbers of individuals, and, therefore, with increasing statistical power (SIGMA Type 2 Diabetes Consortium et al. 2014; Yengo et al. 2018), as well as of genomic repositories and online resources, including databases specialized in published GWAS results (Sudlow et al. 2015; MacArthur et al. 2017; Canela-Xandri et al. 2018). To date, thousands of SNPs have been identified to be associated with hundreds of human diseases or other traits with genome-wide significance, according to data recorded by the NHGRI-EBI GWAS Catalog (MacArthur et al. 2017). However, SNP markers of known variants explain but a small percentage of the heritability measured by cohort studies for almost every studied trait, what has been referred to as “missing” heritability (Manolio et al. 2009; Nolte et al. 2017).
The most common assumption to explain the missing heritability is that many common variants of small effect pass unnoticed in most GWAS due to a lack of statistical power (Yang et al. 2010), and a number of loci on the order of hundreds to thousands are yet to be found (Visscher et al. 2017). In fact, the missing heritability gap of well-studied traits such as human height has been reduced as GWAS had been performed with increasingly larger sample sizes and statistic power (Wood et al. 2014; Yengo et al. 2018), although the newly found SNPs tend to have smaller effect sizes on average (Park et al. 2010), and the gap is reduced slowly (Nolte et al. 2017). In addition, common genotyped SNPs can capture up to 60% of familial heritability estimates (Yang et al. 2010; de los Campos et al. 2013; Nolte et al. 2017) or even higher proportions (Yang et al. 2015).
The narrow-sense heritability explained by SNPs found in GWA studies is compared with estimates obtained from family data, usually twin designs. These may involve nonadditive (dominance and epistasis) genetic components as well as other interaction terms including environmental effects (Zuk et al. 2012; Chen et al. 2015; Ni et al. 2018). Therefore, although it has been suggested that most genetic variation for human traits is of additive nature (Hill et al. 2008; Polderman et al. 2015; Zhu et al. 2015), some part of the gap between GWAS and familial heritability estimates may also be due to the bias involved in the familial estimates (Zuk et al. 2012; Hemani et al. 2013). One way to address this issue is to try getting the expected full contribution to narrow-sense heritability from loci detected by GWAS, and compare it with the familial estimates. In this work we attempt to do so by using information from the GWAS Catalog.
Our analysis consists of extracting information on effects and frequencies of variants for a number of human traits and diseases from the GWAS Catalog with the following objectives: (1) To investigate the nature of the distribution of locus effect sizes already discovered and their contribution to narrow-sense heritability, and (2) to predict the expected full distribution of effects and frequencies of loci in order to ascertain whether or not this could be able to explain the estimates of heritability obtained from family studies. Our results indicate that the familial heritability of 10 out of the 16 traits studied could be potentially explained by the contribution of the average effects of hundreds to thousands of loci yet to be found by GWAS. However, for the other six traits there is a substantial gap between the expected GWAS heritability and the familial heritability, suggesting that an additive contribution of single loci is unable to explain the familial heritability values.
Methods
In short, we began by processing the GWAS Catalog in two steps, in order to get a set of data with the most meaningful information associated to SNPs and GWA studies. First, by filtering incomplete or low informative data and, second, by clustering together traits with a highly overlapping genetic background. Additional processing was required for subsequent analyses involving locus effect sizes, frequencies, and contributions to heritability. Computer simulations were carried out to illustrate the possible impact of nonadditive genetic variation on familial estimates of heritability.
Processing of the GWAS catalog
All data manipulation, including statistical analysis, was carried out using the R language (R Core Team 2017). We worked with the NHGRI-EBI GWAS Catalog data (MacArthur et al. 2017), publicly available at https://www.ebi.ac.uk/gwas/, and accessed on December 5, 2017. We started by selecting a limited number of fields from the database for each scientific study PubMed ID (PMID), as the SNP ID itself, the mapped gene, the effect, reported as an odds ratio (OR) or beta-coefficient (BETA), the frequency of the risk allele, and the reported P-value. PMID-related variables were also gathered, as the name of the disease or trait examined in the study and the total population sample, computed from the information of the initial and replication samples used. The Catalog contains some ambiguity regarding the units of the effects registered. Doubtful cases were checked by looking at the corresponding publications, and, if their effect could not be assigned as BETA or, e.g., because it was measured in trait units rather than in standardized units, they were disregarded.
We checked for the occurrence of a list of necessary variables (effect, gene, P-value, SNP, and trait), and removed any row corresponding to a SNP without a complete information on these variables. We also limited our study to the most significant associations, eliminating SNPs with a significance level higher than the standard P-value = 5 × 10−8. A separate dataset without filtering for statistical significance was also considered for the final set of traits. For all purposes, only one SNP per associated Catalog gene (that with the lowest P-value) was considered. Thus, the gene or intergenic name was the unit considered in the analysis aimed at representing a potential causal locus. Thus, hereafter each different GWAS-Catalog gene represents a locus corresponding to the information of a single SNP. For example, for the trait Height, the Catalog version analyzed contained a total of 855 SNP entrances from 25 different PMIDs. Many of these SNPs were associated to the same Catalog gene or intergenic sequence. Considering only different gene names and selecting the SNP with the lowest P-value associated to each gene, only 370 different loci remained. Later, after filtering by type of effect (BETA), which implied removing one PMID with ambiguous type and another with OR type, the remaining final set of data for this trait contained 346 loci arising from 10 PMID.
Because we wanted to investigate the distribution of locus effects with robustness, we only considered traits with a wide and well-known genetic background composed by at least 30 unique genes detected. We initially differentiated as many traits as unique names were given to the mapped disease or trait field in the original GWAS Catalog. However, it often occurs that different researchers studying the same trait publish their results using different trait names (e.g., “LDL” and “LDL levels”). In order to avoid working with duplicated or redundant traits, we clustered some of them (see Supplemental Material, Table S1) on the basis of their common genetic background and carried out some additional processing steps, as explained in File S1. After this step, we restricted the traits analyzed to those represented by at least three different PMIDs.
Contribution of loci to heritability
From the filtered GWAS database, narrow-sense locus-specific heritability was estimated through the calculation of the contribution of each locus to the additive variance by using the classical formula (Falconer and Mackay 1996), , where is the risk allele frequency and is the average effect of the gene substitution for the locus (henceforth, the average effect or locus effect). For BETA traits, the additive variance equals the heritability , as the average effects are measured in phenotypic SD. For OR traits, we estimated the locus-specific heritability (i.e., variance in liability) following the method described by So et al. (2011), assuming additivity of SNP effects, and the prevalence values published in different epidemiology and genetic papers (Table S2). From the and frequency values, locus effects for OR traits were obtained in the same units of phenotypic SD as BETA traits. Finally, the contribution to heritability of all loci corresponding to a given trait were added together to obtain the GWAS heritability for each trait.
After all the above filtering steps, the dataset for locus effects and heritability analyses had a total of 7886 loci corresponding to 328 studies and 42 human traits. The estimated values are shown in Table S2 along with the reported values of familial heritability found in the literature.
In order to measure the proportional contribution of different classes of locus effects to global we defined three arbitrary, but well-defined, effect classes: low, medium, and high. These classes were assigned to each trait according to the mean and SD of their distribution of effect sizes. Low-effect sizes were defined as those with a value lower than e−1 SD below the mean effect size. Medium-effect sizes were those between e−1 SD below and above the mean, and high-effect sizes those with effects larger than e−1 SD above the mean. With these definitions, an average of ∼50% of the loci were in the low-effect class, ∼36% in the medium-effect class, and ∼14% in the high-effect class.
Analysis of the change in locus effect size, frequency, and explained heritability for increasing sample sizes
We assumed that locus effects and frequencies would be better estimated in studies with larger sample sizes (N), in agreement with previous studies (Auer and Lettre 2015; Visscher et al. 2017) as well as our own observation from the GWAS Catalog. Thus, estimates obtained in studies with larger N were reassigned to the corresponding gene identity, independently of the study. That is, if we consider two studies, PMID1 and PMID2, regarding the same human trait, with sample sizes , the SNP effects and frequencies associated to genes found in PMID1 that were also present in PMID2 were assigned the values of the corresponding genes in PMID2. Therefore, a locus found in different studies would have an associated effect and frequency corresponding to a single SNP from the study with the largest sample size, usually, but not always, the most recent one.
We tested three different regression models to measure the relationship between variables in the analyses of locus effects, frequencies (in terms of the minor allele frequency, MAF) or heritability. These regression models were: simple linear regression: ; two-parameter exponential regression: ; and four-parameter logistic regression: ; where the dependent variable Y may refer to the mean locus effect size, frequency, heritability, or any other related variable, such as the parameters of the distribution of locus effects, and the independent variable X is the number of loci found at a given stage in studies with increasing sample sizes.
When these models were performed using the accumulated number of loci as an independent variable, we only considered those traits that had at least three observations (i.e., three PMIDs) in which the cumulated number of loci was at least 30, so that every regression analysis had at least three points, each corresponding to an estimate obtained with at least 30 loci. This corresponds to a subset of 16 traits, 177 PMIDs, and 5692 loci. For model selection, the Akaike Information Criterion (AIC) was used (Akaike 1974). The final dataset with all unique loci described for each trait after all above processing steps, and after updating locus effect sizes and frequencies, is shown in Table S3. This is the dataset used for estimating heritabilities from the GWAS Catalog shown in Table S2.
Inferring the distribution of locus effects, the missing number of loci, and the expected value of heritability
Locus effects and MAFs were fitted into known probability distributions using the R package “fitdistrplus” (Delignette-Muller and Dutang 2015) using the maximum likelihood estimation method. In order to determine which distribution best fitted the observed locus effects, we considered the following possible continuous distributions: Beta, Exponential, Gamma, Gaussian, Logistic, and Log-normal. We then selected the best fit by using AIC (see Table S4).
Given that the change in the parameters of the distribution of locus effects and MAF as new loci are being discovered could also be predicted with the regression parameters described in the previous section, the expected distribution of locus effects and frequencies, including those yet unobserved, could be inferred. From these, we could obtain the number of loci necessary to explain the observed value of familial heritability , or the closest one. To do so, we assumed an increasing number of loci for each trait, and sampled that number from the predicted distribution of effect sizes and MAF. For each number of loci sampled, the distribution parameters were those predicted from the corresponding regression parameters (Table S5). This process was repeated 10,000 times for each set of loci that were added (up to 20,000 loci), providing a distribution of expected heritability values. From this distribution, the expected parameters and numbers of loci that could explain within 95% confidence intervals were chosen. If could not be explained by any expected distribution and any number of added loci, the median heritability estimate closest to was chosen. A detailed example of the prediction procedure is shown in File S2 and Figure S1 therein.
Cross-validation of predictions
We evaluated the accuracy of the predictions on a different set of data composed of new variants published in a more recent version of the GWAS Catalog accessed on August 27, 2018. For validation purposes, we only considered new gene-associated SNPs that belong to traits already present on our final dataset of 16 traits used for inferring the distribution of locus effects. This test set contained data of 153 SNPs mapping new gene names (loci) described in 11 different PMIDs corresponding to the following eight traits: Body mass index, Height, Prostate cancer, Psoriasis, Rheumatoid arthritis (including Rheumatoid arthritis ACPA-positive), Systemic lupus erythematosus, Type 2 diabetes, and Waist-to-hip ratio adjusted for BMI, i.e., Waist-to-hip-related traits (Table S6).
Computer simulations
Computer simulations were carried with an in-house C program to illustrate the possible biases inherent to estimates of heritability obtained from family data, particularly twin studies, when dominance and epistasis models are assumed. The expected distributions of locus effect sizes and frequencies for the trait “Digestive disease” were used for illustration. A population of size 2 × 106 randomly mated diploid individuals was considered where alleles for 660 biallelic loci have homozygous effects a, where a values are twice the allelic average effects sampled from the inferred log-normal distribution of average effects for the Digestive disease trait (mean a = 0.029). Heterozygous effects (ah) were assumed either additive (h = 0.5), partially recessive (h = 0.2), or fully recessive (h = 0). Allelic frequencies (q) were taken from the expected distribution of frequencies for the Digestive disease. Individual genotypes for the quantitative trait were the sum of the genotypic values for all loci involved, and phenotypic effects were obtained by adding an environmental deviation normally distributed with mean zero and variance VE adjusted such that the phenotypic variance is VP = 1.
The additive (VAg) and dominance (VDg) variances in the absence of epistasis (genic variances) were obtained from the sum of the variances of individual loci. Thus, VAg = Σ2α2pq and VDg = Σ(2dpq)2, where α = ah – 2dq, d = a(h – 1/2), the summation is over all loci, = VAg/VP is the narrow-sense genic heritability, and = VDg/VP the genic dominance contribution to phenotypic variance. The genotypic variance (VG) was calculated from the multilocus genotypic values of individuals, and the broad-sense heritability was obtained as H2 = VG/VP.
A twin design was carried out producing 106 families of two monozygotic and two dizygotic twins. The phenotypic correlations between monozygotic (tMZ) and dizygotic (tDZ) twins were obtained from ANOVA. Estimates of familial heritabilities were calculated as = 2(tMZ–tDZ). No shared environmental effects were assumed between twins. Thus, is expected to estimate + 1.5 in the absence of epistasis (Lynch and Walsh 1998, p. 538). Average locus effects (α) were estimated from the regression of the individual phenotypic values on the number of copies of the alleles for each locus, and the contribution of each locus to heritability was obtained as because VP = 1, where q is the allele frequency. The analogous to GWAS heritability was obtained as the sum of contributions from all loci.
A multilocus epistatic model was assumed where homozygous genotypes for the trait interact with one another. Thus, epistasis occurs only between homozygous loci such that their multilocus genotypic effects for the trait are doubled. Four scenarios were then considered combining within-locus additive or recessive gene action, and between-locus additive or epistatic gene action. Under dominance, the epistatic model assumed involves additive by additive (VAA), additive by dominance (VAD) and dominance by dominance (VDD) components. Allelic homozygous and dominance effects accounting for the epistatic effects imply an increase in the additive variance relative to the case of no epistasis (Cheverud and Routman 1995). The GWAS heritability is expected to estimate the narrow-sense heritability (h2) while the twins heritability is expected to estimate h2 + VD/VP + VAA/VP + VAD/VP + VDD/VP + higher order epistatic components (Lynch and Walsh 1998, p. 583). All simulation values and estimates were averaged over 20 replicates.
Data availability
The GWAS Catalog database is publicly accessible and downloadable from https://www.ebi.ac.uk/gwas/. The Supplemental Material contains two Files, five Figures, with Figure S1 included in File S2, and seven Tables. Relevant code has been made available in a public repository at Github (https://github.com/armando-caballero/Missing-heritability). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798580.
Results
The observed distribution of locus effect sizes
Locus effect sizes for most traits (90%) fitted better to a log-normal distribution than to any of the other distributions assessed (beta, exponential, gamma, Gaussian, and logistic), the remaining 10% fitting best to a beta distribution (Table S4).
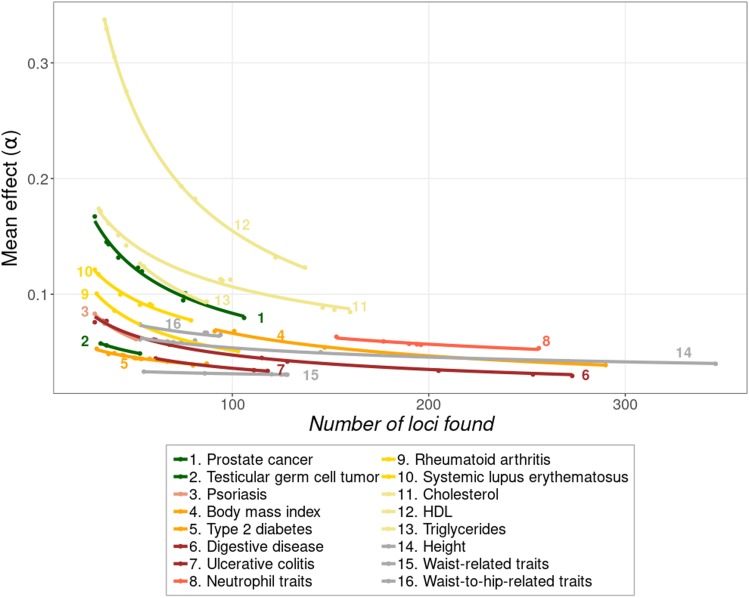
Figure 1 shows how the average locus effect (α) steadily declines as new loci are found with larger samples sizes. The total number of loci considered for each of the traits is available in Table S2. This decline is remarkably consistent across traits, with a two-parameter exponential model fitting best the observations (average R2 = 0.96). The trend observed is in accordance with the expectation that loci of large effect are likely to be found with low sample sizes, whereas decreasingly lower locus effects would only be found with larger and larger sample sizes. The rate of decline of α on number of accumulated loci was substantially lower for skeletal traits (b = −0.19 ± 0.05), i.e., Height- and Waist-related traits, than for the rest of traits (b = −0.48 ± 0.04) (see Figure 1 and Table S5A). Finally, higher average locus effects were associated with lower MAF for all 42 traits (see Table S4) with a linear regression of locus effects on MAF of b = −0.263 ± 0.033, averaged across traits.
Figure 1.
Decline of the average locus effect (α) with the number of loci found. The points represent the cumulated results of successive GWAS with increasing larger sample sizes. The first point at the left of the series is the mean effect of loci found in the GWAS with the lowest sample size (conditional on finding at least 30 loci), and the following points give the mean effect of loci as additional ones are found by studies with larger sample sizes (usually, but not always, by more recent studies). The lines are the fit of the observations to an exponential model (average = 0.96). Traits are colored depending on the functional domain they belong: cancer (green), dermatological (pink), endocrine (orange), gastrointestinal (brown), hematological (red), immunological (yellow), metabolic (beige), skeletal (gray). The final set of data corresponding to the last (right-hand side) points for each line are given in Table S3.
Loci contributions to heritability
Estimates of the heritability explained by the contribution of individual loci and of familial heritability estimates for 42 human traits are given in Table S2, with averages 0.13 ± 0.02 and 0.53 ± 0.03, respectively. The proportion of explained by was 25% on average, ranging widely from 1.6% (Migraine) to 100% (Basal cell carcinoma, and Red blood cell traits).
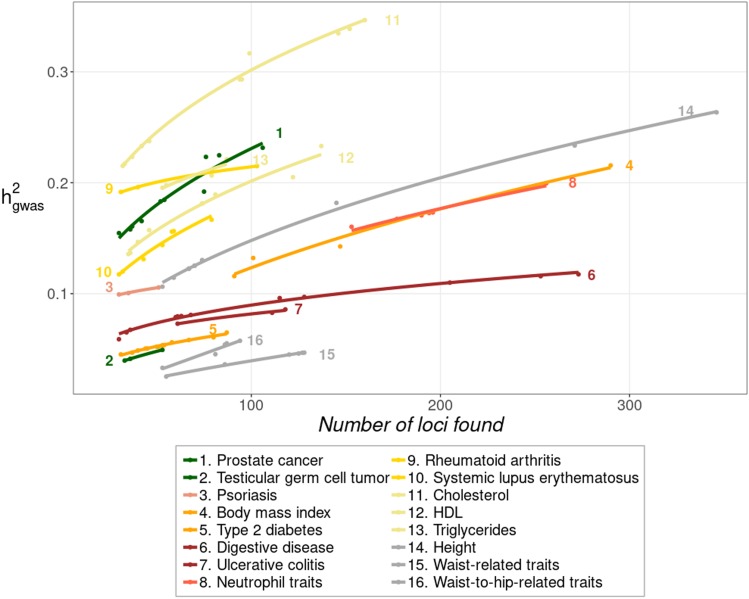
Figure 2 shows the increase in for each trait as more loci are found with higher sample sizes (as for Figure 1). A two-parameter exponential model gave the best fit to the data with average R2 = 0.97 (Table S5C). The figure shows that, for most traits, there is a substantial increase in the heritability explained as new loci have been found. However, for some traits (e.g., Digestive disease, number 6 in Figure 2) it looks like is approaching an asymptotic value. It can also be seen that in many cases the intercept is expected to be well above zero, suggesting that loci contributing most to the heritability were found in the studies with the lowest sample sizes, usually the earliest ones.
Figure 2.
Increase of heritability explained by loci found as the number of these increases. The points represent the observed values, while the lines are the fit to an exponential model (average = 0.97). Traits are colored depending on the functional domain they belong: cancer (green), dermatological (pink), endocrine (orange), gastrointestinal (brown), hematological (red), immunological (yellow), metabolic (beige), skeletal (gray). The final set of data corresponding to the last (right-hand side) points for each line are given in Table S3.
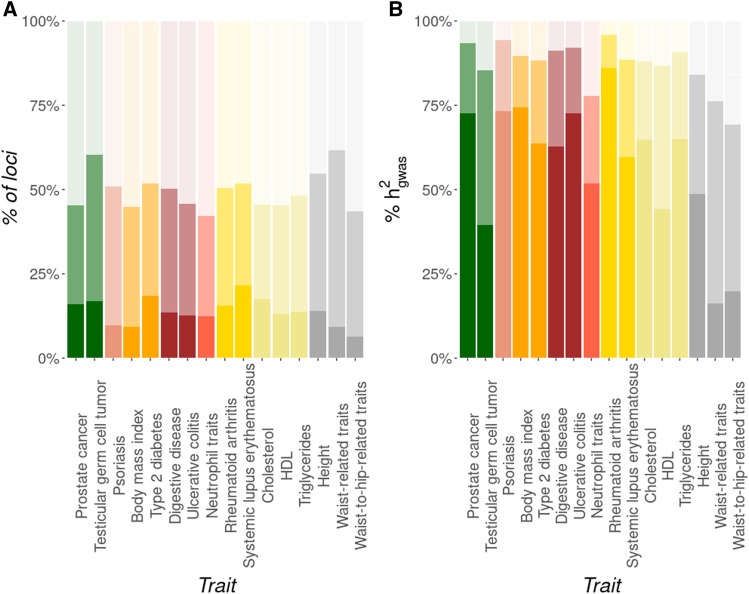
The proportional contribution of loci with different effect sizes to is shown in Figure 3. Three arbitrary classes of effect sizes were made such that ∼50% of loci were within the low-effect class, ∼36% within the medium-effect class, and ∼14% within the high-effect class (panel A). Most of the heritability, however, was explained by loci of large effect (57.2% ± 19.5; panel B), with those of medium and low effect explaining much lower proportions (29.8% ± 13.5 and 13.0% ± 7.2, respectively), even though there is a negative correlation between allele frequencies and locus effect sizes. Similar results are obtained when considering all 42 traits (data not shown).
Figure 3.
Percentage of loci for different classes of effect sizes and their contributions to heritability (in %). (A) Three arbitrary classes of locus effect sizes (high, medium, and low effects) are assumed such that ∼50% of loci are within the low-effect class (high transparency), ∼36% within the medium-effect class (low transparency), and ∼14% within the large-effect class (solid colors). (B) Contribution (in percentage) of the three classes to heritability. Traits are ordered and colored by functional domain: Cancer (green), dermatological (pink), endocrine (orange), gastrointestinal (brown), hematological (red), immunological (yellow), metabolic (beige), skeletal (gray).
Expected distribution of effects, and inference of the missing number of loci to explain the estimates of familial heritability
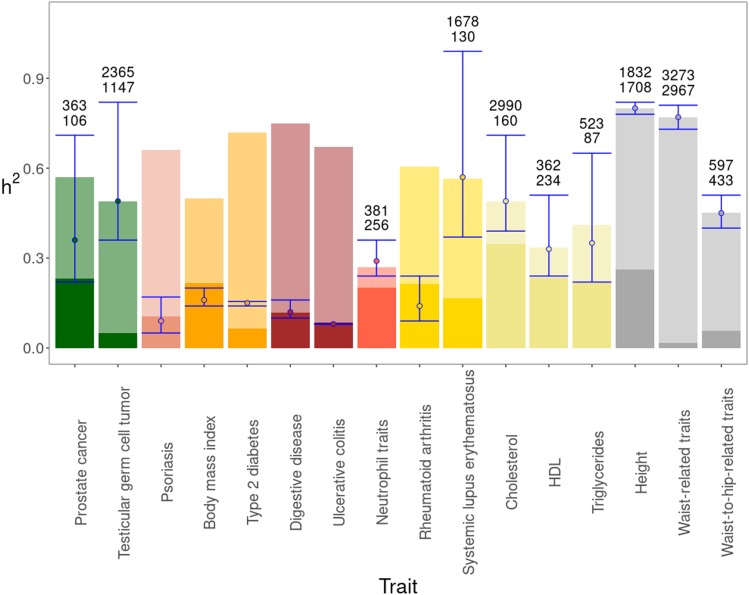
Because, as shown above, the observed distribution of locus effect sizes fitted well to a log-normal distribution, the MAFs to a normal distribution (Table S4), and the change of their distribution parameters with the number of loci found to an exponential regression model (Figure 1 and Table S5, D–G), we were able to predict their expected distributions for any given number of loci, and thus infer the expected heritability closest to . The results are summarized in Figure 4, which shows the current values of (dark bars) and (light bars). The heritability computed from the expected distribution of locus effects, which explains or approaches most to the familial heritability, is shown as a dot (median value) and a 95% confidence interval. The height of the error bar is highly related to the magnitude of the variance parameter (and therefore skewness) of the log-normal distribution. The expected number of loci necessary to explain the familial heritabilities (when reachable) are given over the bars. The expected distributions of effects obtained for all traits are shown in Figures S2 and S3, and the corresponding parameters in Table S7.
Figure 4.
Observed and expected values of heritability. The full length of bars indicate the mean familial heritability for the studied traits (average values are shown when there is a range of estimates from the Literature, Table S2). In solid color it is shown the heritability explained by the loci already found and available from the Catalog . The blue error bar gives the inferred value of heritability (the dot corresponds to the median value) that approaches most to the familial heritability with a 95% confidence interval, using data from the expected distribution of locus effects. The expected number of loci for each trait required to explain the familial heritabilities within the error bars assuming an additive contribution of single loci are given over the bars. Traits are colored depending on the functional domain they belong: Cancer (green), dermatological (pink), endocrine (orange), gastrointestinal (brown), hematological (red), immunological (yellow), metabolic (beige), skeletal (gray).
For 10 out of 16 traits, the expected distribution found would be able to predict the familial heritability accounting only for the contribution of average effects of single loci. Thus, if a number of loci (within those indicated over the bars) were found, and their contribution to heritability were added, the values of the familial heritability could be potentially reached, although this does not mean that this would actually be the case. In contrast, for the remaining six traits (Psoriasis, Body mass index, Type 2 diabetes, Digestive disease, Ulcerative colitis, and Rheumatoid arthritis), the familial heritability could not be reached when considering the average effects of single loci. Thus, even if an increasingly large number of loci from the expected distribution are considered, their additive contribution to would not be able to reach .
Cross-validation of predictions
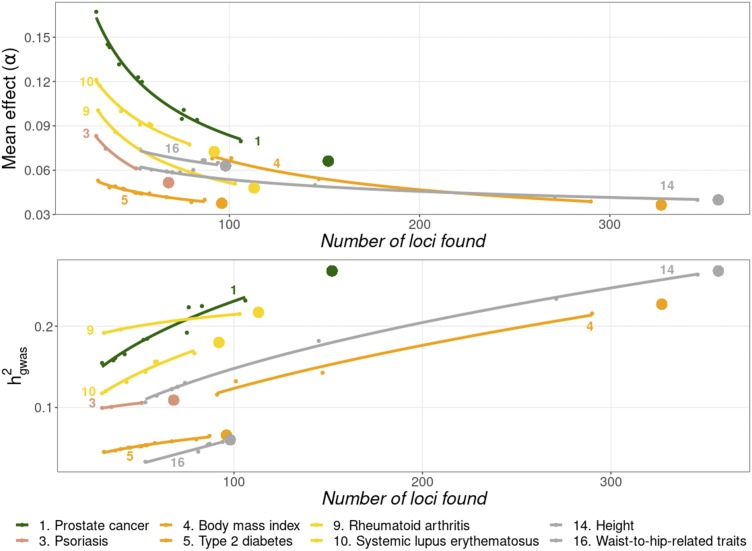
We tested the predictions on a set of new data from a more recent release of the Catalog, incorporating 11 new studies on eight of the 16 traits previously analyzed (Figure 5). The change in mean locus effect and was rather consistent with the previous results, as indicated by the approximate concordance between the large dots (new results) and the projections based on the previous data (lines). The inferred distribution parameters based on the cumulated number of loci showed a low bias when applied to the new data (Figure S4).
Figure 5.
Decline of the average locus effect (upper graph), and increase of the heritability explained (lower graph) as the number of loci found is increasing. The small points represent the observed values of the previous analyses (Figure 1 and Figure 2) and the large points those of a more recently collected set. Lines are the fit to an exponential model. Traits are colored depending on the functional domain they belong: Cancer (green), dermatological (pink), endocrine (orange), immunological (yellow), skeletal (gray).
Simulation results
Simulation results assuming the distribution of locus effects inferred for the Digestive disease trait under different models of gene action (additive or recessive within loci, and additive or epistatic between loci) are shown in Table 1. This shows the difference between the heritability estimated by a twin study and the GWAS heritability .
Table 1. Simulation results assuming the distribution of locus effects predicted for the Digestive disease trait.
| Within-locus | Additive | Additive | h = 0.2 | h = 0.2 | h = 0 |
|---|---|---|---|---|---|
| Between-locus | Additive | Epistatic | Additive | Epistatic | Epistatic |
| Parameters | |||||
| 0.103 | 0.102 | 0.068 | 0.068 | 0.050 | |
| 0.000 | 0.000 | 0.016 | 0.016 | 0.043 | |
| H2 | 0.103 | 0.332 | 0.084 | 0.347 | 0.370 |
| Estimations | |||||
| 0.104 | 0.282 | 0.069 | 0.234 | 0.196 | |
| tMZ | 0.104 | 0.332 | 0.084 | 0.347 | 0.370 |
| tDZ | 0.052 | 0.155 | 0.039 | 0.146 | 0.143 |
| = 2(tMZ – tDZ) | 0.103 | 0.355 | 0.091 | 0.403 | 0.455 |
| – | −0.001 | 0.073 | 0.022 | 0.169 | 0.259 |
| tMZ – 2tDZ | −0.000 | 0.022 | 0.007 | 0.055 | 0.085 |
genic narrow-sense heritability; genic contribution of dominance to phenotypic variance;H2, broad-sense heritability; GWAS estimate of heritability; tMZ, intraclass phenotypic correlation among monozygotic twins; tDZ, intraclass phenotypic correlation among dizygotic twins; estimate of heritability from twins. The phenotypic variance is one in all cases and no common environmental effects are assumed.
Under a full additive model, the estimates and are equal, as expected. In contrast, under a recessive-epistatic model, twin heritability can be substantially biased with respect to . Note, however, that the difference tMZ – 2tDZ is relatively small (< .1), which could suggest that there is no substantial deviation from a full additive model. Note that tMZ is expected to be equal to H2, as it is, and that the difference 4tDZ – tMZ is expected to be very close to the narrow-sense heritability, which agrees with the value of .
Discussion
By extracting the relevant data from the GWAS Catalog we have been able to infer the number and distribution of locus effects that could potentially explain the missing heritability assuming the cumulative contribution of average effects of single loci. Within the limitations of the data and the procedure followed, we found that, for 10 out of the 16 studied traits, this additive explanation appears, at least, feasible, whereas for the remainder is not.
Nature of the variation detected by GWAS
Our results show strong evidence indicating that the distribution of locus effects for different human traits fits better to a log-normal distribution than to other commonly used distributions, including the gamma distribution, widely assumed in population genetic studies (Pérez-Figueroa et al. 2009; Jiang et al. 2010; Schneider et al. 2011; Caballero et al. 2015; Keightley et al. 2016). In the field of genetics, the log-normal distribution has been previously suggested for Drosophila DNA polymorphism data (Loewe and Charlesworth 2006), and is usually assumed in models and natural processes arising not only in biology (Nei and Imaizumi 1966) but also in very different scientific disciplines (Limpert et al. 2001; Grönholm and Annila 2007).
We have also shown (Figure 1) that the average effect size tends to decrease for all traits studied as the number of discovered GWAS Catalog associated genes increases, the decrease fitting an exponential model. This supports the idea that higher-effect loci were discovered in the first GWAS (with lower sample sizes), while posterior analyses involving larger sample sizes allowed lower-effect loci to be discovered (Park et al. 2010; Simons et al. 2018).
We also observed a negative linear relationship (average across traits b = −0.263 ± 0.033) between the effect of loci and the minor allele frequency. This could be explained by a more likely detection by GWAS of loci of small effect when they are common than when they are rare. It could also (or in addition) be due to the action of purifying selection acting more strongly on large-effect than low-effect sizes. This is in agreement with previous evidence provided by Zeng et al. (2018), who detected signatures of negative (purifying) selection in multiple traits. It has been further described that nonsynonymous variants on core (coding) genes, as well as conserved regions, play an important role particularly for high effect mutations that segregate at lower frequencies (Gazal et al. 2018). If loci of large effect are maintained at low frequencies because of negative selection, they could contribute proportionately less to heritability than loci of small effect. Contrary to this, the results of Figure 3 show that there is a disproportionately larger contribution from loci of large effect to heritability, with those of small effect contributing generally little (Figure 3). This is in agreement with previous predictions (e.g., Caballero et al. 2015), and contradicts models suggesting that most of the heritability for complex traits in humans must be due to loci of small effect (Eyre-Walker 2010).
Expected distribution of locus effects and missing heritability
With the Catalog data, we could infer the expected distribution of locus effect sizes for a number of complex traits. This allowed us to investigate whether the cumulative contribution of the average effect of single loci would make it possible to explain familial heritability estimates, thus inferring the missing number of loci yet to be potentially found and their nature. In a pioneering work, Park et al. (2010) used information from the first round of GWA studies and an approach based on the power of detection of variants, to make predictions of the total number of loci needed to explain up to 20% of the genetic variance for height, Crohn’s disease and cancers (breast, prostate, and colorectal). With the limited information at that time it was not possible, however, to make predictions of whether genetic variance could ever be fully explained or not with additional findings. Currently, many more data are available, and we could base our approach on this cumulative data in a different way.
Our results concur with those of Park et al. (2010) in pointing to lower effect sizes discovered with higher sample sizes, and an increasingly lower contribution of these loci to heritability. We could additionally give evidence on the parametric nature of the distribution of effects (log-normal), and thus predict the expected number of loci needed to explain or not the estimates of familial heritability. Our results indicate that, for 10 out of 16 traits, the familial heritability could potentially be completely explained by the cumulative contribution of average effects of single loci found by GWAS (Figure 4). The number of loci required for this is < 1000 for Prostate cancer, Neutrophil traits, HDL, Triglycerides, and Waist-to-hip-related traits, or around a few thousand for Testicular germ cell tumor, Systemic lupus erythematosus, Cholesterol, Height, and Waist-related traits. Thus, according to our results, a few hundred or a few thousand loci would be able to explain the missing heritability for this set of traits, assuming an additive contribution of locus average effects to heritability, in line with previous predictions (Visscher et al. 2017). For example, for human height, we infer that ∼1800 loci would be necessary to explain the estimates of familial heritability (Figure 4). Yengo et al. (2018) have recently found 3290 SNPs for Height clustered to 712 genomic loci, which could account for ∼25% of the variation in Height. Our predictions thus suggest that a further 1000 loci would be necessary to explain the full familial heritability. This prediction should, however, be taken with reservations from a quantitative perspective, as the definitions of locus in both studies are different, the estimate of familial heritability for height could be overestimated (Yang et al. 2015), and because of the limitations of our approach (see below).
For 6 out of 16 traits, however, our results indicate that the additive contribution of effects of single loci, even in large numbers, cannot explain the familial heritability. For these traits, the expected heritability is close to, or slightly above, that already explain by GWAS (Figure 4). One anomalous result occurs with body mass index, for which the expected value of heritability was slightly below the currently observed value. The reason for this maladjustment is likely to be the bias generated when inferring the expected distribution for this trait. The observation that, for some traits, the expected heritability cannot reach the familial one, relies on the fact that, for these traits, the expected change in the shape of the distribution of locus effects predicts effect sizes too small to contribute significantly to heritability as the number of loci increases. In fact, the reason why some estimates of the missing number of loci to reach the familial heritability are rather high in Figure 4 (e.g., for waist-related-traits, requiring ∼3000 loci), is that the approach of the expected heritability to the familial one is rather slow as the number of loci found increases because these have lower and lower effect sizes.
Our inference that, for some traits, the familial heritability could not be retrieved by the accumulation of the contribution of average effects of single loci can also be deduced from Figure 2. If the increase in heritability with the accumulated number of loci is predicted from the figure for future numbers of loci to be found (regression parameters shown in Table S5C), it seems that, for some traits, such as Psoriasis, Type 2 diabetes, Digestive disease, Ulcerative colitis, and Rheumatoid arthritis, the heritability will reach an asymptotic value below even when up to thousands of loci are considered. It may be noted, however, that, for some traits, such as Psoriasis, the number of loci found so far is small, and predictions can be less accurate than for traits for which many data are available.
Our predictions should be taken with caution, and considered as mere approximations, given the assumptions on which are based and the possible sources of bias involved. We made a selection of the most informative SNPs available in the Catalog for each trait, i.e., those with P-value . The reason was to consider only those for which the evidence of association with the trait is strong. This means that the number of loci assumed to be found is generally lower than that provided by the GWAS Catalog. With this assumption, we would expect our predictions of number of loci and heritability from their effects to be underestimations. We made a reanalysis without filtering by P-value (Table S7). For a few traits was increased significantly, even >1, probably due to the presence of many false positives or overestimation because of linkage disequilibrium between loci (see below). However, the average predicted heritability across all traits was very similar when the restrictive filtered data were used (0.352) or not (0.358).
Another possible source of bias is that we took the SNP most significantly associated (with the lowest P-value) to a given GWAS-Catalog gene or intergenic sequence, and assumed that the estimated effect and frequency of that SNP is the same as for the corresponding associated gene (locus). Thus, we assumed that the selected SNPs were at complete LD with the associated locus. Therefore, the average effect size of the considered loci, and their contribution to heritability, would be expected to be overestimations. An additional source of overestimation of average effects could arise from the fact that, even though we considered different gene Catalog names as units of analysis (with a single associated SNP to each), different SNPs in high LD could be associated to different Catalog genes. These sources of overestimation can, in fact, be taking place in our analysis for some traits. For example, for Height, the 346 loci considered in our final set of analysis explain = 0.26, which is very close to that obtained by Yengo et al. (2018) ( = 0.25) but ascribed to 712 associated genomic loci (although the definitions of locus differ between both studies; see below). Nevertheless, the average obtained for the set of 16 traits analyzed is 0.16 on average (ranging from 0.05 to 0.35; Table S7), which is within the majority of estimates of observed for the analyzed traits (Speliotes et al. 2010; McAllister et al. 2011; Reiner et al. 2011; Jostins et al. 2012; Tsoi et al. 2012; Hara et al. 2014; Tada et al. 2014; Litchfield et al. 2015; Mancuso et al. 2015). In addition, the average from the loci considered in this study for all traits is, on average, 25% of familial heritability (Table S2), a proportion of the order of those found in the literature (Zuk et al. 2012).
In order to consider the possibility of an overestimation of loci effects because linkage disequilibrium can correlate effect sizes between close loci, we performed an additional filtering following the definition of locus by Wood et al. (2014) and Yengo et al. (2018), as one or multiple jointly associated SNPs located within 1-Mb window. This definition of locus does not coincide with that followed in our analysis, where a locus refers to a single gene or intergenic sequence referred to in the Catalog with attached estimates from the single most associated SNP. However, to apply the former definition, we removed from all our analyses all loci that were within 1 Mb distance of another one. With this approach, 24% of the loci were removed, and the final number of traits available for prediction was reduced from 16 to 11. The results of this analysis are given in Figure S5. As mentioned above, with the original analysis the average for the 16 traits was 0.16. After the 1-Mb pruning, the average was reduced down to 0.10 (Table S7). Our main predictions, however, did not change qualitatively from the previous ones for the 11 remaining traits, except for 1 trait. In the new analysis, the familial heritability for Prostate cancer could not be reached by adding more loci (Figure S5) whereas in the former analysis it could (Figure 4). For the other 10 traits, however, the same conclusion held regarding the possibility, or not, of explaining familial heritability, although there were substantial differences in the number of loci predicted to reach the familial heritability (e.g., >6000 missing loci for Height), always assuming an additive contribution of loci.
Our results could also be affected by Winner’s curse (Lohmueller et al. 2003), which causes estimates of genetic effects to be upwardly biased because only variants with highly significant evidence of association are considered. Xiao and Boehnke (2009) showed that the bias incurred by Winner’s curse in the estimation of average effects decreases with the power of the analysis, and that, for a fixed power, the bias is reduced as the cut-off significance level is more restrictive. In this respect, we used a rather restrictive genome-wide significance level in our analyses (5 × 10−8). It is, however, possible that the estimated effects in the earlier studies (with lower sample sizes and, therefore, lower power) were more upwardly biased by Winner’s curse than the estimates obtained in later studies (with higher statistical power). But, as explained in the methods section, we replaced the estimated effects of the loci in the earlier studies by the estimated effects in the later ones. In fact, 87% of the loci effects were obtained, or their effects were updated, in studies with the largest samples sizes. Therefore, we would expect a low impact of Winner’s curse on our results.
Another source of uncertainty could be the distribution of locus effects assumed. We fitted the locus effect sizes to a log-normal distribution, which was that fitting best to 90% of the traits studied (Table S4). We repeated the inferences of expected heritabilities assuming other distributions (beta, gamma and exponential; Table S7). These analyses result on inferences of that sometimes fit in appearance the expected . However, the apparent fit is likely to be the result of an upward bias due to overestimation of the average effects, as very often the estimates of heritability are well above 1. Thus, our inferences based on the log-normal distribution seem to be justified.
Finally, the estimates of familial heritability for the different traits vary between studies and populations. We used values available in the literature and averaged them when there was a range of values, but these are subject to some variation, and are lacking for some traits, so that values for analogous traits need to be used. In summary, the different possible sources of over and underestimations attached to the analysis, the scarcity of data available for some traits, the uncertainty of some estimates and the limitations of the data provided by the Catalog, require treating our results with caution.
Gap between the expected GWAS and familial heritabilities
Our results emphasize that, for 6 out of 16 traits, the expected decrease in effect size for new loci is such that it seems not feasible to explain the observed by , even if the contribution of thousands of loci were assumed. For 10 traits, however, it appears that the additive contribution of hundreds to thousands of further loci could potentially explain the familial heritabilities. This does not imply, however, that this will be the case. It is possible that the actual number of missing loci is lower than that predicted and, therefore, that the familial heritability will not ever be reached either. What we conclude here is that, according to the GWAS Catalog and its limitations, this appears to be possible.
The expected distributions of effects inferred in this study (Figure 4, Figures S2 and S3, and Table S7), show a main lack of loci of small effects to be found. This is in line with the observation that the mean effect size monotonically decreases as more loci are being discovered (Figure 1), in agreement with Park et al. (2010). Therefore, it is expected that the missing heritability gap will be reduced very slowly with higher sample sizes and statistic power (Kim et al. 2017). However, we find that not only loci of small effect are missing, and it is also expected to continue finding loci of moderate effect that have passed undetected so far, and that could still explain a substantial part of the missing heritability.
It has been suggested that our inability to find the remaining loci by GWAS may be explained on technical grounds (Manolio et al. 2009). Rare SNPs (say with MAF < 5%) have low coverage in current genotyping technology and are usually missing. Whole genome sequencing then could provide the clue to find the proportion of missing heritability attributable to moderate or high effect loci, but it is expected that SNPs with extremely low frequencies contribute little to heritability, which has been already reported for diseases as Type 2 diabetes (Fuchsberger et al. 2016). In fact, simulation studies (Thornton et al. 2013; Caballero et al. 2015) predict that full sequencing data accounting for SNP variation will not be able to increase substantially the estimates of heritability. However, it is possible that copy number variation, such as insertions and deletions that could be found by whole genome sequencing could make a substantial contribution to missing heritability (Locke et al. 2006; McCarroll 2008; Bassett et al. 2017). Furthermore, genome-wide markers may overcome other statistical limitations for SNPs of complex traits, as inconsistent estimations of the locus effects due to SNPs in LD with more than one QTL as well as imperfect LD (de los Campos et al. 2010, 2015).
For some traits, an asymptotic value of is expected to be substantially lower than , and other phenomena additional to the additive contribution of single average effect loci may be involved. In fact, most estimates of have been obtained with twin data designs, which are known to give estimates of broad-sense heritability that include contributions from dominance and epistasis. In a large meta-analysis of the heritability of human traits based on 50 years of twin studies including nearly 18,000 traits, Polderman et al. (2015) found that genetic variation for a majority of traits is inconsistent with a substantial influence from shared environment or nonadditive genetic sources. This conclusion was reached by testing the difference between the correlations of monozygotic twins (tMZ) and twice the correlation of dizygotic twins (tDZ). A positive value of this difference would imply a contribution from nonadditive (dominance and epistasis) variance whereas a negative difference would imply a substantial contribution of shared environment (Hill et al. 2008). Polderman et al. (2015) found that the difference was not significantly different from zero for ∼69% of the traits (they actually rather tested the ratio tMZ/tDZ = 2). Yet, in the remaining 31% there was a significant deviation, what would imply some contributions from nonadditive or environmental effects in twin heritability estimates. In addition, Zhu et al. (2015) analyzed the contribution of dominance to genetic variation for 79 human traits, concluding that the contribution of dominance variance is only about a fifth of the additive variance on average, suggesting a relatively low contribution from dominance to genetic variation, although for some traits this contribution could be very substantial. These theoretical studies and empirical analyses thus suggest that most variation for human traits is of additive nature. However, the contribution from nongenetic factors may be non-negligible for some traits.
Estimating the contribution of epistasis to genetic variation is elusive given the difficulties to evaluate it properly, and the empirical test carried out by Polderman et al. (2015) using the correlations between monozygotic and dizygotic twins may not fully consider the possibility that epistatic effects contribute substantially to variation. Therefore, it is possible that, for at least some traits, the difference between the additive contributions from average locus effects found from GWAS cannot reach the familial heritability estimates because these are broad-sense heritabilities inflated by nonadditive genetic components. In fact, our computing simulations assuming dominance and epistasis show that there may be a substantial gap (> 0.2) between the heritability obtained from GWAS and the estimate of heritability obtained from twin studies , even though the difference between the correlations of monozygotic twins and twice the correlation of dizygotic twins is < 0.1 (Table 1). The epistatic model assumed in our simulations, involving a doubling of the effect of homozygous loci is, of course, an arbitrary one, but allows for illustrating this issue.
There is increasing evidence that epistasis is a major determinant of additive variance (Bloom et al. 2013; Brookfield 2013; Mackay 2013; Monnahan and Kelly 2015; Huang and Mackay 2016; Csilléry et al. 2018). In fact, epistasis has already been described playing an important role in psoriasis through the interaction of the HLA–ERAP1 loci (Strange et al. 2010) and other immunity disorders (Cortes et al. 2015). Dominance could also take a place in biasing the familial estimates of heritability for some traits, including height and BMI (Chen et al. 2015; Zhu et al. 2015), and the contribution from dominance variance for life-history traits is of the same order as that from additive variance, according to the meta-analyses of Mousseau and Roff (1987) and Crnokrak and Roff (1995). In addition, estimates of familial heritability for some traits, such as human height, can also be overestimated if assortative mating is not properly modeled (Lynch and Walsh 1998; Yang et al. 2015). Finally, any source of genotype-covariate interaction is likely to have an effect on the estimates of SNP-heritability (Evans and Keller 2018; Ni et al. 2018). For example, genotype-environment interactions have also been proposed to explain part of the genetic variance of complex traits (Zheng et al. 2013; Robinson et al. 2017) and, thus, their heritability.
Concluding, GWA analyses are a powerful tool to discover variants associated to complex diseases, and the success in finding the missing heritability may depend, in many instances, on our ability to detect low variant effects with accuracy. For some traits, however, the contribution of single loci found by GWAS does not appear enough to explain the familial heritability, and other sources of genetic or environmental variation contributing to this may be involved.
Acknowledgments
We are grateful to Naomi Wray, S. Hong Lee, Humberto Quesada, and two anonymous referees for helpful comments, and to Aurora García-Dorado and Carlos López-Fanjul for helpful discussions. The analyses reported here were performed on the FinisTerrae machine provided by Centro de Supercomputación de Galicia (CESGA; Galicia Supercomputing Center). This work was funded by the Agencia Estatal de Investigación (AEI) (CGL2016-75904-C2-1-P), Xunta de Galicia (ED431C 2016-037) and Fondos Feder: “Unha maneira de facer Europa.”
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798580.
Communicating editor: N. Wray
Literature Cited
- Akaike H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19: 716–723. 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- Auer P. L., Lettre G., 2015. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 7: 16 10.1186/s13073-015-0138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. S., Lowther C., Merico D., Costain G., Chow E. W. C., et al. , 2017. Rare genome-wide copy number variation and expression of schizophrenia in 22q11.2 deletion syndrome. Am. J. Psychiatry 174: 1054–1063. 10.1176/appi.ajp.2017.16121417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W., Lite T. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. 10.1038/nature11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield J. F. Y., 2013. Quantitative genetics: heritability is not always missing. Curr. Biol. 23: R276–R278. 10.1016/j.cub.2013.02.040 [DOI] [PubMed] [Google Scholar]
- Caballero A., Tenesa A., Keightley P. D., 2015. The nature of genetic variation for complex traits revealed by GWAS and regional heritability mapping analyses. Genetics 201: 1601–1613. 10.1534/genetics.115.177220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela-Xandri O., Rawlik K., Tenesa A., 2018. An atlas of genetic associations in UK Biobank. Nat. Genet. 50: 1593–1599. 10.1038/s41588-018-0248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kuja-Halkola R., Rahman I., Aspegard J., Viktorin A., et al. , 2015. Dominant genetic variation and missing heritability for human complex traits: insights from twin vs. genome-wide common SNP models. Am. J. Hum. Genet. 97: 708–714. 10.1016/j.ajhg.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., Routman E. J., 1995. Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A., Pulit S. L., Leo P. J., Pointon J. J., Robinson P. C., et al. , 2015. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 6: 7146 10.1038/ncomms8146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnokrak P., Roff D. A., 1995. Dominance variance: associations with selection and fitness. Heredity 75: 530–540. 10.1038/hdy.1995.169 [DOI] [Google Scholar]
- Csilléry K., Rodríguez-Verdugo A., Rellstab C., Guillaume F., 2018. Detecting the genomic signal of polygenic adaptation and the role of epistasis in evolution. Mol. Ecol. 27: 606–612. 10.1111/mec.14499 [DOI] [PubMed] [Google Scholar]
- Delignette-Muller M. L., Dutang C., 2015. fitdistrplus. An R package for fitting distributions. J. Stat. Softw. 64: 1–34. 10.18637/jss.v064.i04 [DOI] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. 10.1038/nrg2898 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Vazquez A. I., Fernando R., Klimentidis Y. C., Sorensen D., 2013. Prediction of complex human traits using the genomic best linear unbiased predictor. PLoS Genet. 9: e1003608 10.1371/journal.pgen.1003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Sorensen D., Gianola D., 2015. Genomic heritability: what is it? PLoS Genet. 11: e1005048 10.1371/journal.pgen.1005048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. M., Keller M. C., 2018. Using partitioned heritability methods to explore genetic architecture. Nat. Rev. Genet. 19: 185 10.1038/nrg.2018.6 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2010. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl. Acad. Sci. USA 107: 1752–1756. 10.1073/pnas.0906182107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Longmans Green, Harlow, Essex. [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., et al. , 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. 10.1038/nature18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal S., Loh P., Finucane H., Ganna A., Schoech A., et al. , 2018. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 50: 1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G., 2012. Rare and common variants: twenty arguments. Nat. Rev. Genet. 13: 135–145. 10.1038/nrg3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley P., Anttila V., Winsvold B. S., Palta P., Esko T., et al. , 2016. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 48: 856–866 (erratum: Nat. Genet. 48: 1296). 10.1038/ng.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönholm T., Annila A., 2007. Natural distribution. Math. Biosci. 210: 659–667. 10.1016/j.mbs.2007.07.004 [DOI] [PubMed] [Google Scholar]
- Hara K., Shojima N., Hosoe J., Kadowaki T., 2014. Genetic architecture of type 2 diabetes. Biochem. Biophys. Res. Commun. 452: 213–220. 10.1016/j.bbrc.2014.08.012 [DOI] [PubMed] [Google Scholar]
- Hemani G., Knott S., Haley C., 2013. An evolutionary perspective on epistasis and the missing heritability. PLoS Genet. 9: e1003295 10.1371/journal.pgen.1003295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4: e1000008 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Mackay T. F. C., 2016. The genetic architecture of quantitative traits cannot be inferred from variance component analysis. PLoS Genet. 12: e1006421 10.1371/journal.pgen.1006421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Mu B., Huang Z., Zhang M., Wang X., et al. , 2010. Impacts of mutation effects and population size on mutation rate in asexual populations: a simulation study. BMC Evol. Biol. 10: 298 10.1186/1471-2148-10-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R. K., Duerr R. H., McGovern D. P., et al. , 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice A. E., Winkler T. W., Feitosa M. F., Graff M., Fisher V. A., et al. , 2017. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat. Commun. 8: 14977 10.1038/ncomms14977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Campos J. L., Booker T. R., Charlesworth B., 2016. Inferring the frequency spectrum of derived variants to quantify adaptive molecular evolution in protein-coding genes of Drosophila melanogaster. Genetics 203: 975–984. 10.1534/genetics.116.188102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Grueneberg A., Vazquez A. I., Hsu S., de Los Campos G., 2017. Will big data close the missing heritability gap? Genetics 207: 1135–1145. 10.1534/genetics.117.300271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert E., Stahel W. A., Abbt M., 2001. Log-normal distributions across the sciences: keys and clues. Bioscience 51: 341–352. 10.1641/0006-3568(2001)051[0341:LNDATS]2.0.CO;2 [DOI] [Google Scholar]
- Litchfield K., Thomsen H., Mitchell J. S., Sundquist J., Houlston R. S., et al. , 2015. Quantifying the heritability of testicular germ cell tumour using both population-based and genomic approaches. Sci. Rep. 5: 13889 10.1038/srep13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D. P., Sharp A. J., McCarroll S. A., McGrath S. D., Newman T. L., et al. , 2006. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79: 275–290. 10.1086/505653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Charlesworth B., 2006. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2: 426–430. 10.1098/rsbl.2006.0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K. E., Pearce C. L., Pike M., Lander E. S., Hirschhorn J. N., 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 33: 177–182. 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer, Sunderland, MA. [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., et al. , 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2013. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N., Rohland N., Rand K. A., Tandon A., Allen A., et al. , 2015. The contribution of rare variation to prostate cancer heritability. Nat. Genet. 48: 30–35. 10.1038/ng.3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister K., Eyre S., Orozco G., 2011. Genetics of rheumatoid arthritis: GWAS and beyond. Open Access Rheumatol. 3: 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S. A., 2008. Extending genome-wide association studies to copy-number variation. Hum. Mol. Genet. 17: R135–R142. 10.1093/hmg/ddn282 [DOI] [PubMed] [Google Scholar]
- Monnahan P. J., Kelly J. K., 2015. Epistasis is a major determinant of the additive genetic variance in Mimulus guttatus. PLoS Genet. 11: e1005201 10.1371/journal.pgen.1005201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau T. A., Roff D. A., 1987. Natural selection and the heritability of fitness components. Heredity 59: 181–197. 10.1038/hdy.1987.113 [DOI] [PubMed] [Google Scholar]
- Nei M., Imaizumi Y., 1966. Effects of restricted population size and increase in mutation rate on the genetic variation of quantitative characters. Genetics 54: 763–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni G., van der Werf J., Zhou X., Hypponen E., Wray N. R., et al. , 2018. Genotype-covariate correlation and interaction disentangled by a whole-genome multivariate reaction norm model. bioRxiv 10.1101/377796. [DOI] [PMC free article] [PubMed]
- Nolte I. M., van der Most P. J., Alizadeh B. Z., de Bakker P. I., Boezen H. M., et al. , 2017. Missing heritability: is the gap closing? An analysis of 32 complex traits in the Lifelines Cohort Study. Eur. J. Hum. Genet. 25: 877–885. 10.1038/ejhg.2017.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Wacholder S., Gail M. H., Peters U., Jacobs K. B., et al. , 2010. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42: 570–575. 10.1038/ng.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L., Standl M., Waage J., Baurecht H., Hotze M., et al. , 2015. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 47: 1449–1456. 10.1038/ng.3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Figueroa A., Caballero A., García-Dorado A., López-Fanjul C., 2009. The action of purifying selection, mutation and drift on fitness epistatic systems. Genetics 183: 299–313. 10.1534/genetics.109.104893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman T. J. C., Benyamin B., de Leeuw C. A., Sullivan P. F., van Bochoven A., et al. , 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47: 702–709. 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- R Core Team , 2017. R: A Language and Environment for Statistical Computing. R Found Stat Comput, Vienna: Available at: https://www.R-project.org/. [Google Scholar]
- Reiner A. P., Lettre G., Nalls M. A., Ganesh S. K., Mathias R., et al. , 2011. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT). PLoS Genet. 7: e1002108 10.1371/journal.pgen.1002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. R., English G., Moser G., Lloyd-Jones L. R., Triplett M. A., et al. , 2017. Genotype-covariate interaction effects and the heritability of adult body mass index. Nat. Genet. 49: 1174–1181. 10.1038/ng.3912 [DOI] [PubMed] [Google Scholar]
- Schneider A., Charlesworth B., Eyre-Walker A., Keightley P. D., 2011. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics 189: 1427–1437. 10.1534/genetics.111.131730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGMA Type 2 Diabetes Consortium, . A. L. Williams, Jacobs S. B., Moreno-Macías H., Huerta-Chagoya A., et al. , 2014. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506: 97–101. 10.1038/nature12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Y. B., Bullaughey K., Hudson R. R., Sella G., 2018. A population genetic interpretation of GWAS findings for human quantitative traits. PLoS Biol. 16: e2002985 10.1371/journal.pbio.2002985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So H., Gui A. H. S., Cherny S. S., Sham P. C., 2011. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 35: 310–317. 10.1002/gepi.20579 [DOI] [PubMed] [Google Scholar]
- Speliotes E. K., Willer C. J., Berndt S. I., Monda K. L., Thorleifsson G., et al. , 2010. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42: 937–948. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A., Capon F., Spencer C. C. A., Knight J., Michael E. W., et al. , 2010. Genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42: 985–990. 10.1038/ng.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C., Gallacher J., Allen N., Beral V., Burton P., et al. , 2015. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12: e1001779 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada H., Won H., Melander O., Yang J., Peloso G. M., et al. , 2014. Multiple associated variants increase the heritability explained for plasma lipids and coronary artery disease. Circ Cardiovasc Genet 7: 583–587. 10.1161/CIRCGENETICS.113.000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. R., Foran A. J., Long A. D., 2013. Properties and modeling of GWAS when complex disease risk is due to non-complementing, deleterious mutations in genes of large effect. PLoS Genet. 9: e1003258 10.1371/journal.pgen.1003258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson N. J., Greenwood C. M. T., Soranzo N., Lawson D. J., Richards J. B., 2018. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat. Rev. Genet. 19: 110–124. 10.1038/nrg.2017.101 [DOI] [PubMed] [Google Scholar]
- Tsoi L. C., Spain S. L., Knight J., Ellinghaus E., Stuart P. E., et al. , 2012. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44: 1341–1348. 10.1038/ng.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Wray N. R., Zhang Q., Sklar P., McCarthy M. I., et al. , 2017. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H., et al. , 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R., Boehnke M., 2009. Quantifying and correcting for the winner’s curse in genetic association studies. Genet. Epidemiol. 33: 453–462. 10.1002/gepi.20398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of heritability for human height. Nat. Genet. 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A. E. A., et al. , 2015. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 47: 1114–1120. 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L., Sidorenko J., Kemper K. E., Zheng Z., Wood A. R., et al. , 2018. Meta-analysis of genome-wide association studies for height and body mass index in ∼700,000 individuals of European ancestry. Hum. Mol. Genet. 27: 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., de Vlaming R., Wu Y., Robinson M. R., Lloyd-Jones L. R., et al. , 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 50: 746–753. 10.1038/s41588-018-0101-4 [DOI] [PubMed] [Google Scholar]
- Zheng J., Arnett D. K., Lee Y., Shen J., Parnell L. D., et al. , 2013. Genome-wide contribution of genotype by environment interaction to variation of diabetes-related traits. PLoS One 8: e77442 10.1371/journal.pone.0077442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Bakshi A., Vinkhuyzen A. A. E., Hemani G., Lee S. H., et al. , 2015. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am. J. Hum. Genet. 96: 377–385. 10.1016/j.ajhg.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R., Lander E. S., 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 109: 1193–1198. 10.1073/pnas.1119675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GWAS Catalog database is publicly accessible and downloadable from https://www.ebi.ac.uk/gwas/. The Supplemental Material contains two Files, five Figures, with Figure S1 included in File S2, and seven Tables. Relevant code has been made available in a public repository at Github (https://github.com/armando-caballero/Missing-heritability). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7798580.