There is extensive variation in males for sperm competitive abilities, and in females for the ability to distinguish among sperm from different males. But it is still not known how females distinguish males...
Keywords: sperm competition, Drosophila, female choice, female nervous system, RNAi
Abstract
In many species, sperm can remain viable in the reproductive tract of a female well beyond the typical interval to remating. This creates an opportunity for sperm from different males to compete for oocyte fertilization inside the female’s reproductive tract. In Drosophila melanogaster, sperm characteristics and seminal fluid content affect male success in sperm competition. On the other hand, although genome-wide association studies (GWAS) have demonstrated that female genotype plays a role in sperm competition outcome as well, the biochemical, sensory, and physiological processes by which females detect and selectively use sperm from different males remain elusive. Here, we functionally tested 26 candidate genes implicated via a GWAS for their contribution to the female’s role in sperm competition, measured as changes in the relative success of the first male to mate (P1). Of these 26 candidates, we identified eight genes that affect P1 when knocked down in females, and showed that five of them do so when knocked down in the female nervous system. In particular, Rim knockdown in sensory pickpocket (ppk)+ neurons lowered P1, confirming previously published results, and a novel candidate, caup, lowered P1 when knocked down in octopaminergic Tdc2+ neurons. These results demonstrate that specific neurons in the female’s nervous system play a functional role in sperm competition and expand our understanding of the genetic, neuronal, and mechanistic basis of female responses to multiple matings. We propose that these neurons in females are used to sense, and integrate, signals from courtship or ejaculates, to modulate sperm competition outcome accordingly.
NATURAL and sexual selection increase the frequencies of alleles that boost an organism’s reproductive success. Sexual selection acts on precopulatory traits, such as male courtship behavior and female mate choice, as well as on postcopulatory processes. Sperm competition is one of these postcopulatory processes. Across vertebrates and invertebrates, it can be beneficial for females to obtain multiple mates (Jennions and Petrie 2000). If multiple mating occurs at a high-enough frequency, and/or if sperm is stored long-term, ejaculates from rival males will compete for oocyte fertilization (Parker 1970). This type of male–male postcopulatory sexual selection mediates the evolution of adaptations in males to mitigate the risk of sperm competition. One form of adaptation is to lower the chances of female remating with other males through transferring mating plugs (e.g., Parker 1970; Orr and Rutowski 1991) or seminal fluid proteins (e.g., Chapman et al. 2003; Liu and Kubli 2003), since the last male to mate often sires most of a female’s progeny. If a female does remate, characteristics of sperm and seminal fluid proteins influence a male’s ability to compete with ejaculates from other males. Drosophila melanogaster has proven to be an especially informative model to study these male × male genotypic interactions. Generally, longer and slower sperm are better at withstanding displacement in D. melanogaster (Lüpold et al. 2012). Genome-wide association studies (GWAS) further uncovered the genetic basis of male competitive ability. Besides genes encoding sperm components (Yeh et al. 2012), genes encoding seminal fluid proteins were discovered to play a role in sperm competition (Clark et al. 1995; Fiumera et al. 2005, 2007; Greenspan and Clark 2011). These proteins have a variety of functions, such as inducing female refractoriness to remating, stimulating egg laying [e.g., sex peptide (SP); Chapman et al. 2003; Liu and Kubli 2003], and promoting sperm storage (e.g., Acp36DE; Neubaum and Wolfner 1999; Acp29AB; Wong et al. 2008; and Acp62F; Mueller et al. 2008). Interestingly, many seminal fluid proteins evolve rapidly [reviewed in Swanson and Vacquier (2002)], and some were found to be harmful to females (Civetta and Clark 2000; Wigby and Chapman 2005; Mueller et al. 2007), suggesting that their evolution is mediated by sexual conflict: what makes a male a better competitor might actually be disadvantageous to females (Wigby and Chapman 2005; Hollis et al. 2019).
Although most studies of sperm competition have focused on the role of the male, a number of studies have argued that females are not “passive vessels” in this process. Cryptic female choice, whereby a female selectively uses sperm from ejaculates she received from multiple males, has been proposed as a powerful mechanism for female contributions to sperm competition (Eberhard 1996). A classic example of such female contribution has been observed in junglefowl, in which females were seen to eject sperm from subdominant males after forced copulation (Pizzari and Birkhead 2000). Studies in D. melanogaster, with standard male genotypes and varying female genotypes, also illustrate that male success depends not only on his genotype and the genotype of his competitor, but also on the genotype of the female (Clark et al. 1999, 2000; Lawniczak and Begun 2005; Chow et al. 2010; Giardina et al. 2011; Lüpold et al. 2013; Zhang et al. 2013; Reinhart et al. 2015). These three-way interactions have been suggested to be important for maintaining polymorphisms in populations (Clark et al. 2000; Clark 2002). However, despite the observation that female genotype plays a role, it has been difficult to disentangle female control from female × male interactions and to identify the genetic loci involved. Recent studies in Drosophila have begun to provide a way to dissect the female’s role in sperm competition, and to determine the genes and mechanisms that contribute to differences in sperm competition outcome. First, D. melanogaster males carrying sperm protamines labeled with GFP or red fluorescent protein enabled direct observation of competing sperm inside the female reproductive tract (Manier et al. 2010), and measurements of heritable variation across female genotypes in sperm ejection, storage, and displacement (Lüpold et al. 2013). Second, initial studies have been done of the female’s genetic makeup underlying variation in her contribution to sperm competition. Chow et al. (2013) identified SNPs whose presence in the female was associated with sperm competition outcome by performing sperm competition assays using two standard tester males and females from 39 Drosophila Genetic Reference Panel (DGRP) lines, a panel of wild-derived inbred lines whose genome sequences are available (Mackay et al. 2012). They found variation in the proportion of first male offspring (P1) across DGRP females, and a GWAS revealed correlations between P1 and SNPs in or close to 33 genes (Chow et al. 2013). However, roles for the majority of these genes in sperm competition were not known. Intriguingly, 15 of the 33 candidate genes identified by Chow et al. (2013) have expression biased to the nervous system or have known neural functions, encoding proteins such as ion channels, transcription factors involved in proneural development, or proteins with roles in vesicle trafficking. Moreover, when Chow et al. (2013) knocked down 4 of the 33 candidate genes in female sensory pickpocket (ppk+) neurons, which are required for female postmating responses (PMRs) (Yapici et al. 2008; Häsemeyer et al. 2009; Yang et al. 2009; Rezával et al. 2012), they found that knockdown of three of these four candidates mediated changes in P1, demonstrating a direct role for the female nervous system in impacting the paternity share of each male (Chow et al. 2013). This result supported the hypothesis from Arthur et al. (1998) that the female nervous system might influence sperm competition, based on the observation that the female nervous system is required for proper sperm storage. The importance of the female nervous system in sperm competition is further supported by findings regarding sex peptide receptor (SPR) (Chow et al. 2010) and Neprilysin 2 (Sitnik et al. 2014), which are two additional genes known to affect female contributions to sperm competition. This was determined in experiments that knocked down SPR or Neprilysin 2 in females ubiquitously, but both genes are known to be expressed in the female nervous system.
Nevertheless, many questions remain to be answered regarding the female’s involvement in sperm competition. For example, the relative contributions of neuronal vs. nonneuronal tissues to a female’s influence on sperm competition remain to be elucidated. In addition, we do not know if other neurons besides ppk+ neurons are involved. For example, the neuromodulator octopamine is required for sperm release from storage (Avila et al. 2012; Sitnik et al. 2014), and both octopamine and octopaminergic Tdc2+ neurons are required for ovulation (Monastirioti et al. 1996; Monastirioti 2003; Cole et al. 2005; Rubinstein and Wolfner 2013; Rezával et al. 2014), and refractoriness to remating (Rezával et al. 2014), suggesting a potential role for octopamine and Tdc2+ neurons in sperm competition. Here, we aimed to determine whether other candidate genes put forward by Chow et al. (2013) influence sperm competition, and whether or not most of them do this by acting through the female nervous system. We individually knocked down candidate genes using RNA interference (RNAi) in females, either ubiquitously or in the nervous system. Knockdown and control females were mated consecutively to two distinct tester males and we assessed the effect of knockdown on paternity ratios. Of 26 genes tested, 8 genes were found to affect the ratio of offspring sired by each male, significantly expanding the number of genetic loci known in females to influence sperm competition. The majority of these genes (five out of eight genes) affected sperm competition outcome when knocked down in the female nervous system, and we identified a role for not only ppk+, but also Tdc2+ neurons in sperm competition. Our results provide functional evidence that further emphasizes the crucial role of the female nervous system in sperm competition. These results will allow detailed dissection of the mechanisms of cryptic female choice and sperm competition inside the female reproductive tract, and by extension effects of postmating prezygotic sexual selection and sexual conflict.
Materials and Methods
Fly stocks and husbandry
The upstream activation sequence (UAS)/GAL4 system (Brand and Perrimon 1993) was used to individually knock down candidate genes ubiquitously, pan-neuronally, or in subsets of neurons in the female nervous system. Driver lines used were: ubiquitous drivers Tubulin-GAL4/TM3, Sb and Tubulin-GAL80ts; Tubulin-GAL4/TM3, Sb; and nervous system-specific drivers nSyb-GAL4 (Hindle et al. 2013), ppk-GAL4, and Tdc2-GAL4 (Cole et al. 2005). UAS-RNAi lines were ordered from the Vienna Drosophila Research Center (VDRC) for each candidate gene identified in a GWAS (Chow et al. 2013) with the following exceptions: CG34027, CG10858, RFeSP, and sti (no VDRC lines were available for these genes), and CG13594 (the only available VDRC line has 94 predicted off-targets). VDRC identifiers for all VDRC lines are available in Supplemental Material, Table S1. Lines were used from both the KK (attP background) and GD (w1118 background) RNAi libraries. Males used for the sperm competition assay had the cn bw or bwD genotypes. Males, and virgin knockdown and control females, were aged 3–7 days in single-sex vials before the start of each experiment.
Fly stocks were maintained at room temperature on standard yeast/glucose media on a 12-hr light/dark cycle. When using Tubulin-GAL80ts; Tubulin-GAL4/TM3, Sb, crosses were set up at room temperature, and knockdown and control virgin females were aged at 29°, and maintained at 29° throughout the sperm competition assay.
Verification of knockdown level
To verify knockdown level, UAS-RNAi lines were crossed to Tubulin-GAL4/TM3, Sb to generate Tubulin-GAL4 > UAS-RNAi knockdown flies. Control flies were generated in one of two ways: (1) TM3, Sb; UAS-RNAi siblings obtained from the same crosses, or (2) Tubulin-GAL4 > w1118 or Tubulin-GAL4 > attP flies generated by crossing Tubulin-GAL4/TM3, Sb to w1118 or attP (for GD and KK lines, respectively). Age-matched TM3, Sb; UAS-RNAi siblings, or Tubulin-GAL4 > w1118 or Tubulin-GAL4 > attP flies were collected at the same time as knockdown flies and tested as controls. Five candidate genes did not yield viable Tubulin-GAL4 > UAS-RNAi F1 progeny, suggesting that ubiquitous knockdown of the target gene was lethal and that the RNAi-mediated knockdown was successful at perturbing gene expression. For crosses that yielded viable Tubulin-GAL4 > UAS-RNAi F1 progeny, RT-PCR was used to assess the knockdown level of each UAS-RNAi line [methods described in Ravi Ram et al. (2006); Table S1]. Briefly, total RNA was isolated from 10 to 20 knockdown and control females using TRIzol according to the manufacturer’s instructions. RNA was DNAse treated (Promega, Madison, WI) and cDNA was synthesized (Clontech). PCR was used to amplify genes of interest and a housekeeping gene (Actin5C or Rp49), and the results were analyzed on a 1–2% agarose gel using gel electrophoresis. Dilutions were made of cDNA from knockdown and control females to compare their relative levels of expression.
Sperm competition experiments
In each experiment, knockdown females were generated by crossing UAS-RNAi lines for each candidate gene to a GAL4 driver. To obtain control females with wild-type gene expression, flies from the appropriate background stock (attP or w1118) were crossed with flies from the same GAL4 driver. Control and knockdown females were mated to cn bw males in single-pair matings on day 0 in vial 1. Copulations were observed. Males were removed after copulation ended and mated females were retained in the individual vials. In the evening of day 1, two bwD males were added to each vial and left with the female overnight. Both bwD males were removed in the morning of day 2, and each female was transferred to vial 2. Each female was transferred again every 48 hr to vials 3, 4, and 5 (on days 4, 6, and 8, respectively). All females were discarded on day 10. Progeny from eggs laid in vials 1–5 were reared to adulthood and the paternity of F1 female progeny was scored based on eye color: female offspring of cn bw males had red eyes and female offspring of bwD males had brown eyes. Male progeny were not scored because they were w, making it impossible to use eye color to assess their paternity. On average, each experiment consisted of 71.8 ± 25.1 control females and 65.9 ± 24.3 knockdown females who had mated at least once (mean ± SD). Of these females, 51.9 ± 21.7 control females and 46.9 ± 21.0 knockdown females in each experiment had mated with both males. Sample sizes for each experiment can be found in Table S2.
Since each female was paired with two bwD males and left overnight for the second mating, there was a chance for multiple remating events to occur, which would affect sperm competition. Nonetheless, in a separate experiment, we found that 0 out of 275 mated females remated twice within a 16-hr period.
Statistical analysis of remating rate, fertility, and P1
Remating rate, fertility, and P1 of knockdown and control females were calculated based on the number of first- and second-male progeny. All statistical analyses were performed using base R (version 3.3.1; R Core Team 2016) and the packages lme4 (Cole et al. 2005; Bates et al. 2015), lmerTest (Kuznetsova et al. 2017), and emmeans (https://cran.r-project.org/web/packages/emmeans/index.html).
Remating rate was calculated as the proportion of doubly mated females among all females who mated with the first male. Differences between remating rates of knockdown and control females were compared using Fisher’s exact test.
Because we only scored eye color in female offspring, we used the total number of female progeny produced by each doubly-mated female (rather than all progeny) as a proxy for fertility. Females who had mated only once (with the first or the second male, but not both males) were excluded from the analysis because we found a significant study-wide difference between the fertility of singly and doubly mated females (Figure S1). We compared the fertility of control and knockdown females by fitting linear models, or linear mixed models for experiments with multiple replicates, and by performing an ANOVA.
Finally, P1 was calculated for each doubly-mated female as the ratio of the number of female offspring sired by the first male vs. the total number of female offspring sired by either the first or second males in vials 2–5. Vial 1 was excluded from the calculation of P1 because both matings occurred in vial 1, and with this experimental setup we were unable to determine how many offspring were sired before the second mating. However, the presence of first- and/or second-male progeny in all vials was used to determine whether a female had mated with both males. For the statistical analysis of P1, we arcsine square-root-transformed P1 values before applying linear models, or linear mixed models for experiments with multiple replicates, to the transformed values. Temporal dynamics of P1 between control and knockdown females were compared using linear mixed models, with genotype and vial as fixed effects, and individual females as a random effect.
Since we analyzed 26 candidate genes using multiple GAL4 drivers, we performed a study-wide Benjamini Hochberg (Benjamini and Hochberg 1997) correction for multiple testing. This correction was performed on all nominal P-values for the test of overall P1. For the temporal analysis, we obtained four P-values for each gene/driver combination tested (one for each vial). We performed a false discovery rate (FDR) analysis on P-values for each of these vials separately, across gene/driver combinations.
Data availability statement
All supplemental tables, figures, R scripts, and progeny count data are available on FigShare. Table S1 includes an overview of VDRC lines and primer sequences used for RT-PCR. Table S2 contains sample sizes, summary statistics, and (adjusted) P-values for tests of remating, fertility, and overall P1. Table S3 contains nominal and adjusted P-values for P1 tests by vial. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7476407.
Results
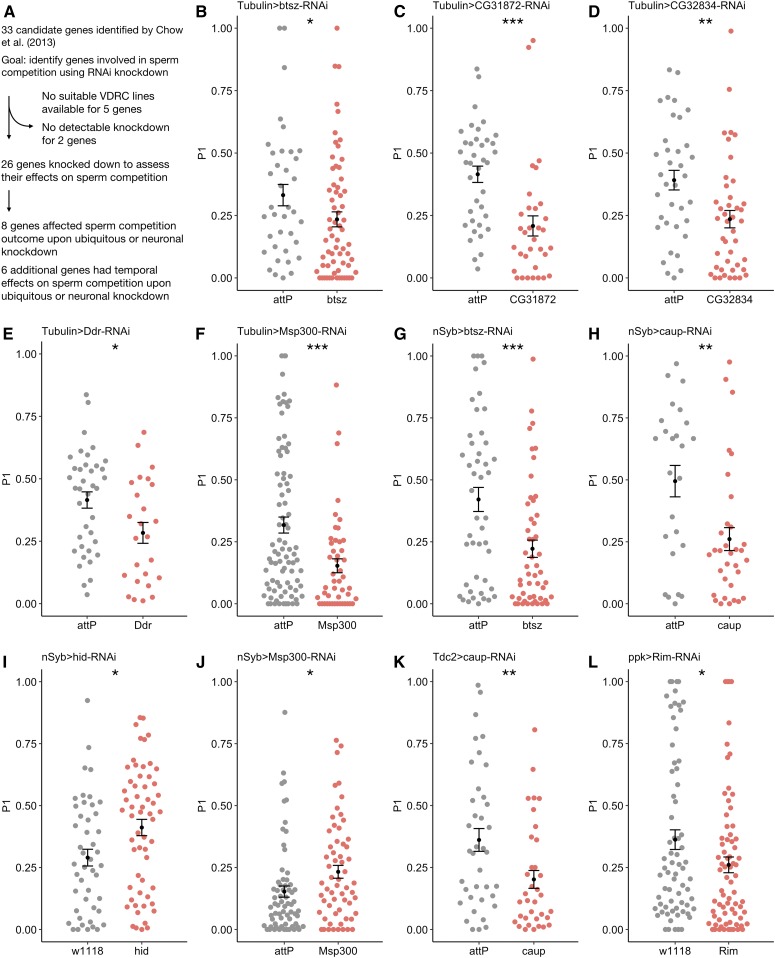
Chow et al. (2013) identified 33 top SNPs associated with sperm competition outcome in females. Not all of these SNPs were located within genes. Thus, to identify genes that directly affect sperm competition, we used RNAi to individually knock down genes that were put forward as candidates by Chow et al. (2013). We tested 26 of the 33 candidate genes for their roles in influencing the female’s contribution to sperm competition, which we scored as P1, the proportion of first-male progeny among total progeny after the second mating. Five of the genes that were identified by Chow et al. (2013) could not be tested because no suitable UAS-RNAi lines were available from the VDRC. RNAi lines for two additional genes, SK and CG33298, gave no detectable RNAi knockdown (Table S1), and thus, we were unable to assess these two genes’ role in sperm competition (Figure 1A).
Figure 1.
Out of 26 candidate genes tested, ubiquitous or tissue-specific knockdown of eight genes affects sperm competition. (A) Overview of the candidate genes from Chow et al. (2013) that were tested using RNAi knockdown. (B–L) Sperm competition was measured as the proportion of offspring sired by the first male (P1) over the course of 8 days. For all genes, controls were generated by crossing the appropriate background strain (attP or w1118) to the respective GAL4 driver line. Each dot represents overall P1 in vials 2–5 for an individual control or knockdown female. Significant differences in P1 between control and knockdown females were determined using linear models. Error bars represent the SEM. (B–F) Changes in P1 mediated by ubiquitous knockdown (Tubulin-GAL4/TM3, Sb). (G–J) Changes in P1 mediated by pan-neuronal knockdown (nSyb-GAL4). (K and L) Changes in P1 mediated by Tdc2+ neuron- or ppk+ neuron-specific knockdown. P1, proportion of offspring sired by the first male to mate; RNAi, RNA interference; VDRC, Vienna Drosophila Research Center. * P < 0.05, ** P < 0.01, *** P < 0.001.
Candidate gene knockdown caused changes in remating rate and fertility
How readily females remate after the first mating and how fertile they are can influence the risk and intensity of sperm competition. Therefore, we assessed the effect of knocking down each of the 26 genes on the remating rate and fertility. Of the 26 genes, ubiquitous knockdown of three genes (CG10962, CG33095 and Ddr) reduced female remating rate (Figure S2 and Table 1). The reduced remating rate observed upon ubiquitous knockdown needs to be interpreted with caution, since ubiquitous knockdowns could directly affect female receptivity to remating, or could have detrimental effects on overall female health or development, making females simply less inclined to mate. Hence, we also analyzed the effects of neuronal knockdown on remating rate. Remating rate was reduced by Tdc2+ neuron-specific knockdown of Rab2 and ppk+ neuron-specific knockdown of para (Figure S2 and Table 1). Finally, Tdc2+ and ppk+ neuron-specific knockdown of hid led to an increase in remating rate (Figure S2 and Table 1). Since hid expression stimulates apoptosis (Grether et al. 1995), differences in the numbers or innervation patterns of Tdc2+ and ppk+ neurons might be responsible for this effect.
Table 1. Remating rate for control and knockdown females.
| Genes | Drivers | Remating rate control | Remating rate KD | P-value |
|---|---|---|---|---|
| CG10962 | Tubulin | 0.88 | 0.62 | 0.000 |
| CG33095 | 0.81 | 0.63 | 0.039 | |
| Ddr | 0.64 | 0.37 | 0.004 | |
| Rab2 | Tdc2+ | 0.60 | 0.21 | 0.000 |
| para | ppk+ | 0.89 | 0.72 | 0.006 |
| hid | Tdc2+ | 0.53 | 0.86 | 0.000 |
| ppk+ | 0.66 | 0.91 | 0.000 |
Remating rate was calculated as the ratio of the number of females that mated with both males vs. the total number of females that mated with the first male. Differences between control and KD females were evaluated using a Fisher’s exact test. KD, knockdown.
Female fertility was affected by many candidate gene knockdowns. Ubiquitous knockdown of 18 of the 26 genes reduced female fertility (Figure S2 and Table 2). However, as mentioned above, these results could be either direct or indirect consequences of ubiquitous gene knockdown. Consistent with the latter hypothesis, we found that nervous system-specific knockdown of only five genes caused a decrease in female fertility (btsz, caup, Ddr, Rab2, Rim; Figure S2 and Table 2). Specifically, Tdc2+ neuron-specific knockdown of Rab2 mediated a substantial decrease in both fertility and remating rate, to the extent that very few doubly mated females were retrieved for sperm competition experiments (only 8 out of 50 females remated). Because the knockdown was tissue-specific, these results strongly suggest that Rab2 is essential for the proper functioning of Tdc2+ neurons, which are in turn known to be required for female remating and fecundity (Rezával et al. 2014). Interestingly, ubiquitous knockdown of Zasp66 and ppk+ neuron-specific knockdown of hid significantly increased fertility (Figure S2 and Table 2).
Table 2. Mean female fertility for control and knockdown females.
| Genes | Drivers | Mean fertility control (± SD) | Mean fertility KD (± SD) | P-value |
|---|---|---|---|---|
| Ddr | ppk+ | 180.83 (35.11) | 161.89 (34.81) | 0.004 |
| Tdc2+ | 113.07 (20.22) | 103.28 (22.98) | 0.042 | |
| nSyb | 144.53 (29.71) | 98.21 (32.63) | 0.000 | |
| Rim | Tdc2+ | 204.57 (47.96) | 181.50 (49.24) | 0.035 |
| Rab2 | Tdc2+ | 76.92 (21.09) | 14.25 (8.75) | 0.000 |
| caup | Tdc2+ | 133.95 (28.69) | 120.57 (22.99) | 0.012 |
| btsz | nSyb | 142.13 (31.65) | 117.89 (52.73) | 0.008 |
| Tubulin | 127.08 (21.89) | 93.77 (28.61) | 0.000 | |
| Zasp66 | Tubulin | 82.83 (17.18) | 103.48 (25.79) | 0.000 |
| hid | ppk+ | 177.24 (50.46) | 221.67 (41.59) | 0.000 |
| CG10962 | Tubulin | 91.28 (35.99) | 61.58 (30.12) | 0.000 |
| CG15800 | 113.92 (17.24) | 90.60 (21.05) | 0.000 | |
| CG31872 | 142.21 (22.79) | 117.15 (28.32) | 0.000 | |
| CG32532 | 144.13 (22.48) | 91.46 (40.95) | 0.000 | |
| CG32834 | 113.92 (17.24) | 100.37 (20.63) | 0.002 | |
| CG33095 | 128.31 (23.49) | 102.55 (28.22) | 0.000 | |
| 5-HT2B | 139.20 (38.85) | 72.72 (30.33) | 0.000 | |
| CG6163 | 128.31 (23.49) | 113.97 (27.49) | 0.007 | |
| sona | 142.16 (30.84) | 109.07 (31.19) | 0.000 | |
| Cyp313a2 | 135.10 (32.95) | 125.90 (26.78) | 0.014 | |
| Msp300 | 91.28 (35.99) | 56.27 (15.59) | 0.000 | |
| Rbp6 | 119.46 (19.49) | 94.39 (27.48) | 0.000 | |
| Shab | 139.20 (38.85) | 113.98 (24.63) | 0.000 | |
| sima | 139.20 (38.85) | 108.46 (26.81) | 0.000 | |
| spz5 | 135.10 (32.95) | 111.24 (35.96) | 0.000 | |
| uif | 142.16 (30.84) | 97.57 (22.37) | 0.000 |
Female fertility was calculated as the total number of female progeny produced by doubly mated females over the course of 10 days. Significant differences between control and KD females were evaluated using a linear model. KD, knockdown.
Although all 26 candidate genes were detected in a GWAS based on sperm competition outcomes, these results suggest that some of the genes also play roles in modulating other female reproductive traits. For the P1 measurements reported below, we found that candidate gene knockdown could affect P1 without affecting fertility (Figure S5, G and H), but we also observed cases in which both P1 and fertility differed between control and knockdown females. If fertility was reduced, we observed cases in which the reduction impacted only one of the two males (e.g., Figure S5F) and cases in which it impacted both males, but one male more than the other (e.g., Figure S5A). Both of these scenarios could lead to a change in P1.
Seven genes influence sperm competition outcome upon ubiquitous or pan-neuronal knockdown in females
Of the 26 candidate genes of interest, three (para, Rim, and Rab2) were reported to affect P1 when knocked down in ppk+ neurons by Chow et al. (2013). For the 23 remaining candidate genes, in an initial test we knocked down each candidate ubiquitously with Tubulin-GAL4. If constitutive ubiquitous knockdown was lethal, and/or if the gene of interest had a known neural function, Tubulin-GAL4; Tubulin-GAL80ts or the pan-neuronal driver nSyb-GAL4 were used instead of Tubulin-GAL4. In cases where ubiquitous knockdown produced a significant effect on overall P1, we proceeded to knock down the gene pan-neuronally, with the exception of CG31872 and CG32834. These two genes are not expressed in the nervous system, but are expressed in the female rectal pad and sperm storage organs, respectively (Leader et al. 2018). We hypothesize that the effects of knockdown on sperm competition outcome may be due to the importance of these genes’ expression in the female reproductive tract or other nonneural tissues.
Ubiquitous knockdown of five genes in females caused reduction of P1 (btsz, CG31872, CG32834, Ddr, and Msp300; Figure 1, B–F and Figure S2), attributable to fewer first-male progeny and more second-male progeny (CG31872 and CG32834, Figure S5, B and C), fewer first- and second-male progeny (btsz and Msp300, Figure S5, A and E), or fewer first-male progeny but similar numbers of second-male progeny relative to control females (Ddr, Figure S5D). The overall fertility of these knockdown females was also lower than that of control females for all five genes (Figure S2).
Additionally, we found four genes whose pan-neuronal knockdown caused an increase (hid and Msp300) or decrease (btsz and caup) in P1 (Figure 1, G–J and Figure S2). Pan-neuronal knockdown of btsz reduced the number of first-male progeny without affecting the number of second-male progeny, leading to an overall reduction in fertility (Figure S5F). Pan-neuronal knockdown of caup and hid affected the relative proportions of first- and second-male progeny without influencing overall fertility (Figure S5, G and H). Finally, Msp300 pan-neuronal knockdown females produced more first-male progeny but similar numbers of second-male progeny compared to control females, but the overall fertility difference between Msp300 knockdown and control females was not significant (Figure S5I). Intriguingly, ubiquitous knockdown of Msp300 lowered P1, while pan-neuronal knockdown increased P1 (Figure 1, F and J and Figure S2). This suggests that ubiquitous knockdown of Msp300 could be detrimental to females’ health, or that Msp300 expression in different tissues has distinct effects on sperm competition. Overall, we found seven genes that had effects on sperm competition when knocked down ubiquitously or pan-neuronally in females.
When analyzing P1 on a temporal, vial-by-vial basis, we found that at least two vials were significantly different between control and knockdown females for each of the genes that had an effect on overall P1 (Figure S4). Ddr, which affected overall P1 upon ubiquitous knockdown only, also showed significant effects on P1 in vials 2 and 3 with pan-neuronal knockdown (Figure S3F), suggesting some neuronal function for Ddr as well. Finally, five genes (sima, sona, spz5, CG33095, and Zasp66) did not change overall P1 when knocked down ubiquitously, but significantly affected P1 in at least one vial when ubiquitous knockdown was analyzed on a vial-by-vial basis (Figure S3, A–E). This result could have several explanations. The products of these five genes might influence processes that are important for sperm competition at specific times after the second mating. Alternatively, these gene products or the processes they mediate might have small roles, or are redundant players, in sperm competition.
Tdc2+ and ppk+ neurons play roles in sperm competition
Informed by the results of the initial test, we further asked in which of the female’s neurons the products of btsz, caup, hid, Msp300, and Ddr act to modulate sperm competition. In particular, we assessed the functions of these five genes in octopaminergic Tdc2+ neurons and proprioceptive ppk+ neurons, which have been implicated in female responses to mating (Cole et al. 2005; Yapici et al. 2008; Häsemeyer et al. 2009; Yang et al. 2009; Avila et al. 2012; Rezával et al. 2012, 2014; Rubinstein and Wolfner 2013). In addition to the five neural genes we identified from the initial test, three other genes had been reported to modulate sperm competition outcome through ppk+ neurons (para, Rab2, and Rim; Chow et al. 2013). Therefore, in the secondary test, we assessed the effect of knocking down each of these eight genes in Tdc2+ neurons and ppk+ neurons.
Of these eight genes, caup was the only gene that affected P1 when knocked down in Tdc2+ neurons (Figure 1K). Knockdown females produced much fewer first-male progeny and slightly more second-male progeny than control females over the course of the assay, resulting in an overall reduction in fertility and a significant decrease in P1 in vials 2–5 (Figure S4 and Figure S5J). Hid, one of the genes that affected P1 when knocked down pan-neuronally, had no overall effect on P1 when knocked down in Tdc2+ neurons. However, on a vial-by-vial basis, P1 in vials 2 and 3 was significantly higher, and in vial 5 significantly lower, in hid knockdown females relative to the P1 of controls (Figure S3G). This result suggests a weaker, but significant, role for hid in Tdc2+ neurons on sperm competition outcome. Similarly, Rab2 knockdown in Tdc2+ neurons mediated a significant increase in P1 only in vial 2 (Figure S3H).
We also corroborated earlier findings and showed that ppk+ neuron-specific knockdown of Rim caused females to have a lower P1 (Figure 1L), specifically by reducing the number of first-male progeny produced (Figure S5K). Temporal effects on P1 were observed for Ddr and hid knockdown in the ppk+ neurons, for which P1 was lower in knockdown females compared to controls in vial 5 only (Figure S3, I and J). None of the other genes affected P1 when knocked down in the ppk+ neurons. This included Rab2 and para, two genes that had been reported to affect P1 upon knockdown in ppk+ neurons by Chow et al. (2013). The previous study used a ppk-GAL4 driver generated in a different genetic background compared to the one used in our study, possibly explaining the discrepancy; alternatively, variable environmental factors could be the cause.
Multiple testing correction
We applied Benjamini Hochberg correction for multiple testing on (i) the P-values obtained for measurements of overall P1 and (ii) the P-values obtained for the analysis of temporal differences in P1. Out of 42 tests that were performed for overall P1, 11 tests had a nominal P-value < 0.05 (eight unique genes). Ten of these tests had an adjusted P-value < 0.1 (Table S2; this set still includes the eight unique genes). At an FDR of 0.1, we expect 9 out of these 10 tests to be true positives. For the temporal analysis, we treated each vial as a separate phenotype, and performed 42 tests on each of the four vials to analyze differences in P1 between control and knockdown females in each vial. Out of 168 tests, 44 had a nominal P-value < 0.05. Of these 44 tests, 31 had an adjusted P-value < 0.1 (Table S3). The only candidate gene that fell outside of the 0.1 FDR cutoff for the temporal analysis was spz5. Of the 31 tests with an adjusted P-value < 0.1, we expect ∼28 to be true positives.
Discussion
A number of approaches have suggested that females play an active role in sperm competition (Arthur et al. 1998; Chow et al. 2010, 2013; Sitnik et al. 2014), but the underlying mechanisms and genetics still remain poorly understood. Our group previously showed that the knockdown of three genes in the female nervous system changed the relative paternity success of two males (Chow et al. 2013). Here, we present eight genes that are important for the female’s involvement in sperm competition, including one gene reported in the previous study. We also show that the actions of five of these genes are required in the female nervous system, indicating a major role for the female nervous system in sperm competition. Further, we report that the ubiquitous or tissue-specific knockdown of six additional genes had time-specific effects on the outcome of sperm competition. Knockdown of 12 remaining candidate genes tested either had no detectable effect on sperm competition (perhaps another gene near the SNP is involved) or their role in sperm competition could not be identified given limitations of the RNAi method. Specifically, the SNPs were originally identified in DGRP lines, where their variation could have more subtle effects on the spatiotemporal dynamics of candidate gene expression. However, RNAi reduces gene expression continuously in all tissues where the GAL4 driver is active.
Understanding the functions of the genes we found to be involved in sperm competition can shed light on the mechanisms by which females contribute to this process. In this section, we discuss the potential biological significance of the genes we identified, in terms of the mechanism through which they could influence sperm competition and in terms of the evolutionary significance.
From a mechanistic point of view, most of the genes we found to affect P1 upon neuronal knockdown act during development or facilitate basic neuronal processes: btsz is a synaptotagmin-like protein involved in membrane trafficking (Serano and Rubin 2003), caup is involved in neuronal development (Gómez-Skarmeta and Modolell 1996), Msp300 has previously been found to play a role at the neuromuscular junction (Morel et al. 2014), the general function of Rim in the nervous system is to mediate efficient neurotransmitter secretion (Graf et al. 2012; Müller et al. 2012), and hid stimulates apoptosis (Grether et al. 1995). Rab2 has a temporal effect on sperm competition upon Tdc2+ neuron-specific knockdown and it encodes a GTPase involved in vesicle trafficking (Gaudet et al. 2011). We found that eight other genes affected sperm competition, overall or in specific vials, when knocked down ubiquitously. Of these eight genes, four have known functions in the nervous system or during development, and the other four are likely important in the female reproductive system. Ddr belongs to the family of receptor tyrosine kinases, but its exact function is unknown (Sopko and Perrimon 2013); spz5 is known for its function in the immune response, but is also involved in the development of the nervous system (Zhu et al. 2008); Zasp66 plays a role in muscle development (Katzemich et al. 2013); and sona encodes a metallopeptidase and is involved in Wg signaling (Kim et al. 2016). Based on their functions and the effects of knockdown in the female nervous system, it is likely that the genes themselves are not directly involved in sperm competition. Rather, knockdown of these genes can impair the function or connectivity of the female nervous system, which could then directly influence sperm competition. Potentially, knockdown of other genes that are required for neuronal development or signaling, or experimental manipulation of neuronal activity, can affect sperm competition in a similar way. For the same reason, neuronal or tissue-specific knockdowns are more likely to have direct effects on sperm competition, while ubiquitous knockdowns could lead to more widespread problems in female physiology.
Through which mechanisms can the female nervous system influence sperm competition? First, after mating, uterine conformational changes modulated by muscle contractions are needed to store sperm (Adams and Wolfner 2007; Mattei et al. 2015), and sperm in storage need to be maintained (Schnakenberg et al. 2012). At the same time, females are exposed to pheromones (Smith et al. 2017) and seminal fluid proteins like SP (Chapman et al. 2003; Liu and Kubli 2003). These molecules can affect her receptivity to remating, and directly impact the risk and intensity of sperm competition. The later she remates, the higher the success of the first male. Once a female is doubly-mated, the timing of sperm ejection after the second mating affects which sperm are stored and therefore contribute to the fertilization set (Manier et al. 2010; Lüpold et al. 2013). The diuretic hormone 44 (Dh44)+ neural circuit controls sperm ejection (Lee et al. 2015). However, we previously found that the pan-neuronal knockdown of Dh44 did not influence sperm competition (White 2017). Finally, once sperm from both males is stored in female sperm storage organs, there is an equal chance for each sperm to be used, regardless of the male of origin, according to the fair raffle hypothesis (Parker et al. 1990; Manier et al. 2010).
It is conceivable that our nervous system-specific gene knockdowns impact neuronal signaling and consequently female physiology, behavior, or muscle contractions, allowing for any of these female-mediated aspects of sperm competition to be affected. Since these aspects of sperm competition are important at different time points after both matings take place (e.g., sperm ejection and displacement occur early after the second mating, while sperm maintenance and use continue over the course of multiple days), we expect that the knockdown of some candidate genes would affect sperm competition outcome only in some vials. We performed a P1 by vial analysis to address this possibility, and we indeed identified candidates with such time-specific effects.
In line with the hypothesis that the female nervous system influences sperm competition, we identified a role for both sensory ppk+ neurons and octopaminergic Tdc2+ neurons in mediating sperm competition outcome. A population of sexually dimorphic Tdc2+ neurons located in the abdominal ganglion innervate the female reproductive tract extensively and regulate PMRs, including remating refractoriness and ovulation (Monastirioti et al. 1996; Rubinstein and Wolfner 2013; Rezával et al. 2014). Innervation of Tdc2+ neurons in the female sperm storage organs (seminal receptacle and paired spermathecae; Avila et al. 2012; Rezával et al. 2014) suggests that caup, hid, or Rab2 might affect the development or function of Tdc2+ neurons, which in turn could modulate sperm storage and sperm competition. In addition, sensory ppk+ neurons are also crucial for the female PMR (Häsemeyer et al. 2009; Yang et al. 2009). The male seminal fluid protein SP binds to the SPR expressed in female ppk+ neurons to silence these neurons and elicit the PMR (Yapici et al. 2008; Häsemeyer et al. 2009; Yang et al. 2009; Rezával et al. 2012; Lee et al. 2016). Both SP and SPR are known to influence sperm competition outcome (Chow et al. 2010; Castillo and Moyle 2014). Rim knockdown in the ppk+ neurons could affect these neurons’ signaling capabilities, thereby mediating a change in P1. SP and SPR silence the ppk+ neurons to induce increased egg production and lower remating rate. In this regard, it might be surprising that Rim knockdown does not mediate these PMRs. However, all females in our experiments are mated and thus exposed to SP, so the effect of Rim knockdown in a mated female might not have extra effects on PMR in addition to the ppk+ neuron-silencing effects that SP already has. Finally, although the female reproductive tract is extensively innervated, seminal fluid proteins can also enter the female’s hemolymph (Monsma et al. 1990; Lung and Wolfner 1999; Ram et al. 2005; Pilpel et al. 2008), and thus have the opportunity to directly interact with Tdc2+ or ppk+ neurons throughout the female body.
Besides a role for the female nervous system in sperm competition, other tissues are likely involved as well. Two of the eight genes that affected overall P1 were only tested with ubiquitous knockdown. CG32834, a predicted serine-type endopeptidase, is spermathecae-specific (Leader et al. 2018). The spermathecae are long-term sperm storage organs whose secretions affect sperm motility (Schnakenberg et al. 2011), so CG32834 has the potential to affect sperm storage, maintenance, or release from storage. In addition, a previous study found that CG32834 knockdown results in lower egg production and increased remating (Sirot et al. 2014a), in line with the results reported here. CG31872 is reported to be expressed in the female rectal pad (Leader et al. 2018), but it is not clear what its function is in female reproduction. In addition, two genes with temporal effects on P1 are also involved in female reproduction: sima plays a role in border cell migration in the ovary (Doronkin et al. 2010) and, CG33095, a gene with unknown function, is also expressed in the ovary (Leader et al. 2018). The roles of these genes in sperm competition can be further investigated in the future by testing tissue-specific knockdowns.
The question remains whether the genes and neurons we identified are required to assess and respond to male, and/or ejaculate, quality and effect cryptic female choice, or whether they influence sperm competition independently of male genotype and instead influence the paternity share based on male mating order. Across all tissues tested, six of the eight genes that affected sperm competition outcome when knocked down led to a decreased success for the first male. This suggests that in a wild-type situation, these genes, or the neurons in which they act, play a role in mediating a higher paternity success for the first male (P1) and a decreased success for the second male (P2). It is possible that, when these genes are knocked down, neuronal signaling in response to the first mating is impaired. This could lead to decreased storage of the first male’s sperm, increased loss or displacement of the first male’s sperm, or an incomplete switch from virgin to mated state. This could also explain the lower overall fertility that we often observed in knockdown females. A second mating, and a second exposure to mating signals, mechanical and/or molecular, might improve the response to mating, leading to a higher success for the second male.
From an evolutionary point of view, since the candidate genes tested here were identified based on natural variation across the DGRP, where females from some isofemale lines naturally have a lower P1 when doubly mated to standard tester males (Chow et al. 2013), it is possible that there is natural variation in how strongly females respond to mating due to variation in neural development or differences in neural gene expression. All SNPs in the 33 candidate genes identified by Chow et al. (2013) are in noncoding regions or are synonymous substitutions, suggesting that they can indeed affect gene expression. Durham et al. (2014) measured variation in fecundity across young and aged DGRP females, and identified associated candidate genes in a GWAS. Gene ontology categories overrepresented in those GWAS results included categories associated with neural development (Durham et al. 2014) and five of their candidate genes were also found in the GWAS from Chow et al. (2013) (Ddr, CG32834, sima, Rbp6, and CG15765). Genotype-specific differences in fecundity could exist because the optimal number and timing of egg production can be a source of sexual conflict: it is beneficial for males if a female produces many eggs shortly after mating (and before remating), while more reserved resource allocation can be beneficial for females (Sirot et al. 2014b; Wensing and Fricke 2018). In addition, there might be selection in females for a higher P1 and a lower P2. P2, or the paternity share of the second male, is usually high (> 50%) because of the mechanics of sperm displacement. It could be beneficial for females to counteract this process and to attempt to balance P1 and P2. Keeping a better balance in sperm from both males gives females the chance to maximize the genetic variation that can be passed on to her offspring.
Finally, an outstanding question in research on sexual conflict is concerned with the interplay of male signals that act on the female’s nervous system to influence her physiology and behavior, and the female’s processing of and response to male cues (Schnakenberg et al. 2012). Our findings regarding sensory ppk+ neurons and Tdc2+ neurons, which include neurons innervating the female reproductive tract, form an important step in understanding the mechanistic and molecular basis of that interplay in sperm competition.
Acknowledgments
We thank members of the Clark and Wolfner laboratories for helpful discussions and comments, and the editor and the anonymous reviewers for helpful comments and suggestions to improve the manuscript. We are grateful to the National Institutes of Health (grant R01 HD-059060 to A.G.C. and M.F.W.) for supporting this work. Several authors were supported for all or part of this study by fellowships: a State University of New York Diversity Fellowship, followed by a Howard Hughes Medical Institute Gilliam Fellowship (S.L.W.); a fellowship from the Belgian American Educational Foundation (S.Y.N.D.); and a summer internship from the National Science Foundation-funded Eco-Devo-Eco Network Research Collaboration Network (C. Extavour, Principal Investigator) for M.C.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7476407.
These authors contributed equally to this work.
Communicating editor: L. Moyle
Literature Cited
- Adams E. M., Wolfner M. F., 2007. Seminal proteins but not sperm induce morphological changes in the Drosophila melanogaster female reproductive tract during sperm storage. J. Insect Physiol. 53: 319–331. 10.1016/j.jinsphys.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur B., Hauschteck-Jungen E., Nöthiger R., Ward P. I., 1998. A female nervous system is necessary for normal sperm storage in Drosophila melanogaster: a masculinized nervous system is as good as none. Proc. R. Soc. Lond. B Biol. Sci. 265: 1749–1753. 10.1098/rspb.1998.0498 [DOI] [Google Scholar]
- Avila F. W., Bloch Qazi M. C., Rubinstein C. D., Wolfner M. F., 2012. A requirement for the neuromodulators octopamine and tyramine in Drosophila melanogaster female sperm storage. Proc. Natl. Acad. Sci. USA 109: 4562–4567. 10.1073/pnas.1117689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B., Walker S., 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Benjamini Y., Hochberg Y., 1997. Multiple hypotheses testing with weights. Scand. J. Stat. 24: 407–418. 10.1111/1467-9469.00072 [DOI] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Castillo D. M., Moyle L. C., 2014. Intraspecific sperm competition genes enforce post-mating species barriers in Drosophila. Proc. Biol. Sci. 281: 20142050 10.1098/rspb.2014.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T., Bangham J., Vinti G., Seifried B., Lung O., et al. , 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. 10.1073/pnas.1631635100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Wolfner M. F., Clark A. G., 2010. The genetic basis for male x female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics 186: 1355–1365. 10.1534/genetics.110.123174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y., Wolfner M. F., Clark A. G., 2013. Large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics 193: 177–185. 10.1534/genetics.112.146357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta A., Clark A. G., 2000. Correlated effects of sperm competition and postmating female mortality. Proc. Natl. Acad. Sci. USA 97: 13162–13165. 10.1073/pnas.230305397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., 2002. Sperm competition and the maintenance of polymorphism. Heredity 88: 148–153. 10.1038/sj.hdy.6800019 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Aguadé M., Prout T., Harshman L. G., Langley C. H., 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., Begun D. J., Prout T., 1999. Female x male interactions in Drosophila sperm competition. Science 283: 217–220. 10.1126/science.283.5399.217 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Dermitzakis E. T., Civetta A., 2000. Nontransitivity of sperm precedence in Drosophila. Evolution 54: 1030–1035. 10.1111/j.0014-3820.2000.tb00102.x [DOI] [PubMed] [Google Scholar]
- Cole S. H., Carney G. E., McClung C. A., Willard S. S., Taylor B. J., et al. , 2005. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 280: 14948–14955. 10.1074/jbc.M414197200 [DOI] [PubMed] [Google Scholar]
- Doronkin S., Djagaeva I., Nagle M. E., Reiter L. T., Seagroves T. N., 2010. Dose-dependent modulation of HIF-1alpha/sima controls the rate of cell migration and invasion in Drosophila ovary border cells. Oncogene 29: 1123–1134. 10.1038/onc.2009.407 [DOI] [PubMed] [Google Scholar]
- Durham M. F., Magwire M. M., Stone E. A., Leips J., 2014. Genome-wide analysis in Drosophila reveals age-specific effects of SNPs on fitness traits. Nat. Commun. 5: 4338 10.1038/ncomms5338 [DOI] [PubMed] [Google Scholar]
- Eberhard W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, NJ. [Google Scholar]
- Fiumera A. C., Dumont B. L., Clark A. G., 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169: 243–257. 10.1534/genetics.104.032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera A. C., Dumont B. L., Clark A. G., 2007. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics 176: 1245–1260. 10.1534/genetics.106.064915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P., Livstone M. S., Lewis S. E., Thomas P. D., 2011. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief. Bioinform. 12: 449–462. 10.1093/bib/bbr042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina T. J., Beavis A., Clark A. G., Fiumera A. C., 2011. Female influence on pre- and post-copulatory sexual selection and its genetic basis in Drosophila melanogaster. Mol. Ecol. 20: 4098–4108. 10.1111/j.1365-294X.2011.05253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta J. L., Modolell J., 1996. Araucan and caupolican provide a link between compartment subdivisions and patterning of sensory organs and veins in the Drosophila wing. Genes Dev. 10: 2935–2945. 10.1101/gad.10.22.2935 [DOI] [PubMed] [Google Scholar]
- Graf E. R., Valakh V., Wright C. M., Wu C., Liu Z., et al. , 2012. RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J. Neurosci. 32: 16586–16596. 10.1523/JNEUROSCI.0965-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan L., Clark A. G., 2011. Associations between variation in X chromosome male reproductive genes and sperm competitive ability in Drosophila melanogaster. Int. J. Evol. Biol. 2011: 214280 10.4061/2011/214280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M. E., Abrams J. M., Agapite J., White K., Steller H., 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9: 1694–1708. 10.1101/gad.9.14.1694 [DOI] [PubMed] [Google Scholar]
- Häsemeyer M., Yapici N., Heberlein U., Dickson B. J., 2009. Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron 61: 511–518. 10.1016/j.neuron.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Hindle S., Afsari F., Stark M., Middleton C. A., Evans G. J. O., et al. , 2013. Dopaminergic expression of the Parkinsonian gene LRRK2–G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Hum. Mol. Genet. 22: 2129–2140. 10.1093/hmg/ddt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis B., Koppik M., Wensing K. U., Ruhmann H., Genzoni E., et al. , 2019. Sexual conflict drives male manipulation of female postmating responses in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 116: 8437–8444. 10.1073/pnas.1821386116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennions M. D., Petrie M., 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. Camb. Philos. Soc. 75: 21–64. 10.1111/j.1469-185X.1999.tb00040.x [DOI] [PubMed] [Google Scholar]
- Katzemich A., Liao K.-A., Schoeck F., 2013. Zasp PDZ domain proteins cooperate in Z-disc formation and myofibril assembly. Biophys. J. 104: 447a 10.1016/j.bpj.2012.11.2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.-W., Won J.-H., Lee O.-K., Lee S.-S., Han J.-H., et al. , 2016. Sol narae (Sona) is a Drosophila ADAMTS involved in Wg signaling. Sci. Rep. 6: 31863 10.1038/srep31863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P. B., Christensen R. H. B., 2017. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82: 1–26. [Google Scholar]
- Lawniczak M. K. N., Begun D. J., 2005. A QTL analysis of female variation contributing to refractoriness and sperm competition in Drosophila melanogaster. Genet. Res. 86: 107–114. 10.1017/S0016672305007755 [DOI] [PubMed] [Google Scholar]
- Leader D. P., Krause S. A., Pandit A., Davies S. A., Dow J. A. T., 2018. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46: D809–D815. 10.1093/nar/gkx976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Choi H. W., Zhang C., Park Z.-Y., Kim Y.-J., 2016. A pair of oviduct-born pickpocket neurons important for egg-laying in Drosophila melanogaster. Mol. Cells 39: 573–579. 10.14348/molcells.2016.0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-M., Daubnerová I., Isaac R. E., Zhang C., Choi S., et al. , 2015. A neuronal pathway that controls sperm ejection and storage in female Drosophila. Curr. Biol. 25: 790–797. 10.1016/j.cub.2015.01.050 [DOI] [PubMed] [Google Scholar]
- Liu H., Kubli E., 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. 10.1073/pnas.1631700100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O., Wolfner M. F., 1999. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem. Mol. Biol. 29: 1043–1052. 10.1016/S0965-1748(99)00078-8 [DOI] [PubMed] [Google Scholar]
- Lüpold S., Manier M. K., Berben K. S., Smith K. J., Daley B. D., et al. , 2012. How multivariate ejaculate traits determine competitive fertilization success in Drosophila melanogaster. Curr. Biol. 22: 1667–1672. 10.1016/j.cub.2012.06.059 [DOI] [PubMed] [Google Scholar]
- Lüpold S., Pitnick S., Berben K. S., Blengini C. S., Belote J. M., et al. , 2013. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 110: 10693–10698. 10.1073/pnas.1300954110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manier M. K., Belote J. M., Berben K. S., Novikov D., Stuart W. T., et al. , 2010. Resolving mechanisms of competitive fertilization success in Drosophila melanogaster. Science 328: 354–357. 10.1126/science.1187096 [DOI] [PubMed] [Google Scholar]
- Mattei A. L., Riccio M. L., Avila F. W., Wolfner M. F., 2015. Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro-computed tomography scanning. Proc. Natl. Acad. Sci. USA 112: 8475–8480 [corrigenda: Proc. Natl. Acad. Sci. USA 114: E5485 (2017)]. 10.1073/pnas.1505797112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti M., 2003. Distinct octopamine cell population residing in the CNS abdominal ganglion controls ovulation in Drosophila melanogaster. Dev. Biol. 264: 38–49. 10.1016/j.ydbio.2003.07.019 [DOI] [PubMed] [Google Scholar]
- Monastirioti M., Linn C. E., Jr, White K., 1996. Characterization of Drosophila tyramine beta-hydroxylase gene and isolation of mutant flies lacking octopamine. J. Neurosci. 16: 3900–3911. 10.1523/JNEUROSCI.16-12-03900.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma S. A., Harada H. A., Wolfner M. F., 1990. Synthesis of two Drosophila male accessory gland proteins and their fate after transfer to the female during mating. Dev. Biol. 142: 465–475. 10.1016/0012-1606(90)90368-S [DOI] [PubMed] [Google Scholar]
- Morel V., Lepicard S., Rey A. N., Parmentier M.-L., Schaeffer L., 2014. Drosophila Nesprin-1 controls glutamate receptor density at neuromuscular junctions. Cell. Mol. Life Sci. 71: 3363–3379. 10.1007/s00018-014-1566-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J. L., Page J. L., Wolfner M. F., 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics 175: 777–783. 10.1534/genetics.106.065318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J. L., Linklater J. R., Ravi Ram K., Chapman T., Wolfner M. F., 2008. Targeted gene deletion and phenotypic analysis of the Drosophila melanogaster seminal fluid protease inhibitor Acp62F. Genetics 178: 1605–1614. 10.1534/genetics.107.083766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M., Liu K. S. Y., Sigrist S. J., Davis G. W., 2012. RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J. Neurosci. 32: 16574–16585. 10.1523/JNEUROSCI.0981-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum D. M., Wolfner M. F., 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A. G., Rutowski R. L., 1991. The function of the sphragis in Cressida cressida (Fab.) (Lepidoptera, Papilionidae): a visual deterrent to copulation attempts. J. Nat. Hist. 25: 703–710. 10.1080/00222939100770461 [DOI] [Google Scholar]
- Parker G. A., 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. Camb. Philos. Soc. 45: 525–567. 10.1111/j.1469-185X.1970.tb01176.x [DOI] [Google Scholar]
- Parker G. A., Simmons L. W., Kirk H., 1990. Analysing sperm competition data: simple models for predicting mechanisms. Behav. Ecol. Sociobiol. 27: 55–65. [Google Scholar]
- Pilpel N., Nezer I., Applebaum S. W., Heifetz Y., 2008. Mating-increases trypsin in female Drosophila hemolymph. Insect Biochem. Mol. Biol. 38: 320–330. 10.1016/j.ibmb.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Pizzari T., Birkhead T. R., 2000. Female feral fowl eject sperm of subdominant males. Nature 405: 787–789. 10.1038/35015558 [DOI] [PubMed] [Google Scholar]
- Ram K. R., Ji S., Wolfner M. F., 2005. Fates and targets of male accessory gland proteins in mated female Drosophila melanogaster. Insect Biochem. Mol. Biol. 35(9):1059-71. 10.1016/j.ibmb.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Ravi Ram K., Sirot L. K., Wolfner M. F., 2006. Predicted seminal astacin-like protease is required for processing of reproductive proteins in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 18674–18679. 10.1073/pnas.0606228103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2016. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Reinhart M., Carney T., Clark A. G., Fiumera A. C., 2015. Characterizing male-female interactions using natural genetic variation in Drosophila melanogaster. J. Hered. 106: 67–79. 10.1093/jhered/esu076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezával C., Pavlou H. J., Dornan A. J., Chan Y.-B., Kravitz E. A., et al. , 2012. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr. Biol. 22: 1155–1165. 10.1016/j.cub.2012.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezával C., Nojima T., Neville M. C., Lin A. C., Goodwin S. F., 2014. Sexually dimorphic octopaminergic neurons modulate female postmating behaviors in Drosophila. Curr. Biol. 24: 725–730. 10.1016/j.cub.2013.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein C. D., Wolfner M. F., 2013. Drosophila seminal protein ovulin mediates ovulation through female octopamine neuronal signaling. Proc. Natl. Acad. Sci. USA 110: 17420–17425. 10.1073/pnas.1220018110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnakenberg S. L., Matias W. R., Siegal M. L., 2011. Sperm-storage defects and live birth in Drosophila females lacking spermathecal secretory cells. PLoS Biol. 9: e1001192 10.1371/journal.pbio.1001192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnakenberg S. L., Siegal M. L., Bloch Qazi M. C., 2012. Oh, the places they’ll go: female sperm storage and sperm precedence in Drosophila melanogaster. Spermatogenesis 2: 224–235. 10.4161/spmg.21655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano J., Rubin G. M., 2003. The Drosophila synaptotagmin-like protein bitesize is required for growth and has mRNA localization sequences within its open reading frame. Proc. Natl. Acad. Sci. USA 100: 13368–13373. 10.1073/pnas.1835727100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot L. K., Findlay G. D., Sitnik J. L., Frasheri D., Avila F. W., et al. , 2014a Molecular characterization and evolution of a gene family encoding both female- and male-specific reproductive proteins in Drosophila. Mol. Biol. Evol. 31: 1554–1567. 10.1093/molbev/msu114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot L. K., Wong A., Chapman T., Wolfner M. F., 2014b Sexual conflict and seminal fluid proteins: a dynamic landscape of sexual interactions. Cold Spring Harb. Perspect. Biol. 7: a017533 10.1101/cshperspect.a017533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnik J. L., Francis C., Hens K., Huybrechts R., Wolfner M. F., et al. , 2014. Neprilysins: an evolutionarily conserved family of metalloproteases that play important roles in reproduction in Drosophila. Genetics 196: 781–797. 10.1534/genetics.113.160945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. T., Clarke N. V. E., Boone J. M., Fricke C., Chapman T., 2017. Sexual conflict over remating interval is modulated by the sex peptide pathway. Proc. Biol. Sci. 284: 20162394. 10.1098/rspb.2016.2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Perrimon N., 2013. Receptor tyrosine kinases in Drosophila development. Cold Spring Harb. Perspect. Biol. 5: a009050 10.1101/cshperspect.a009050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Vacquier V. D., 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. 10.1038/nrg733 [DOI] [PubMed] [Google Scholar]
- Wensing K. U., Fricke C., 2018. Divergence in sex peptide-mediated female post-mating responses in. Proc. Biol. Sci. 285: 20181563 10.1098/rspb.2018.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S. L., 2017 Investigating the female’s role in sperm competition in Drosophila melanogaster. Master Thesis, Cornell University, Ithaca, NY. [Google Scholar]
- Wigby S., Chapman T., 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15: 316–321. 10.1016/j.cub.2005.01.051 [DOI] [PubMed] [Google Scholar]
- Wong A., Albright S. N., Giebel J. D., Ram K. R., Ji S., et al. , 2008. A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics 180: 921–931. 10.1534/genetics.108.092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-H., Rumpf S., Xiang Y., Gordon M. D., Song W., et al. , 2009. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61: 519–526. 10.1016/j.neuron.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N., Kim Y.-J., Ribeiro C., Dickson B. J., 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451: 33–37. 10.1038/nature06483 [DOI] [PubMed] [Google Scholar]
- Yeh S.-D., Do T., Chan C., Cordova A., Carranza F., et al. , 2012. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc. Natl. Acad. Sci. USA 109: 2043–2048. 10.1073/pnas.1121327109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Clark A. G., Fiumera A. C., 2013. Natural genetic variation in male reproductive genes contributes to nontransitivity of sperm competitive ability in Drosophila melanogaster. Mol. Ecol. 22: 1400–1415. 10.1111/mec.12113 [DOI] [PubMed] [Google Scholar]
- Zhu B., Pennack J. A., McQuilton P., Forero M. G., Mizuguchi K., et al. , 2008. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 6: e284 10.1371/journal.pbio.0060284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supplemental tables, figures, R scripts, and progeny count data are available on FigShare. Table S1 includes an overview of VDRC lines and primer sequences used for RT-PCR. Table S2 contains sample sizes, summary statistics, and (adjusted) P-values for tests of remating, fertility, and overall P1. Table S3 contains nominal and adjusted P-values for P1 tests by vial. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7476407.