Abstract
Neurospora crassa is an established reference organism to investigate carotene biosynthesis and light regulation. However, there is little evidence of its capacity to produce secondary metabolites. Here, we report the role of the fungal-specific regulatory velvet complexes in development and secondary metabolism (SM) in N. crassa. Three velvet proteins VE-1, VE-2, VOS-1, and a putative methyltransferase LAE-1 show light-independent nucleocytoplasmic localization. Two distinct velvet complexes, a heterotrimeric VE-1/VE-2/LAE-1 and a heterodimeric VE-2/VOS-1 are found in vivo. The heterotrimer-complex, which positively regulates sexual development and represses asexual sporulation, suppresses siderophore coprogen production under iron starvation conditions. The VE-1/VE-2 heterodimer controls carotene production. VE-1 regulates the expression of >15% of the whole genome, comprising mainly regulatory and developmental features. We also studied intergenera functions of the velvet complex through complementation of Aspergillus nidulans veA, velB, laeA, vosA mutants with their N. crassa orthologs ve-1, ve-2, lae-1, and vos-1, respectively. Expression of VE-1 and VE-2 in A. nidulans successfully substitutes the developmental and SM functions of VeA and VelB by forming two functional chimeric velvet complexes in vivo, VelB/VE-1/LaeA and VE-2/VeA/LaeA, respectively. Reciprocally, expression of veA restores the phenotypes of the N. crassa ve-1 mutant. All N. crassa velvet proteins heterologously expressed in A. nidulans are localized to the nuclear fraction independent of light. These data highlight the conservation of the complex formation in N. crassa and A. nidulans. However, they also underline the intergenera similarities and differences of velvet roles according to different life styles, niches and ontogenetic processes.
Keywords: velvet complex, Aspergillus nidulans, light control, Neurospora crassa, secondary metabolism
REGULATION of growth and development in response to environmental stimuli is an essential process in biology. Associated with morphogenetic programs, cells synchronize growth and metabolism in order to keep up with fluctuating environmental conditions. These stimuli are transduced to the nucleus to alter gene expression by signal transduction complexes composed of receptors, membrane-associated protein kinases, mitogen activated protein kinases (MAPK) cascades, and downstream regulatory protein complexes (Yu and Keller 2005; Bayram and Braus 2012; Frawley et al. 2018). A central complex in fungi is the heterotrimeric velvet complex that alters gene expression in response to environmental signals such as light, which results in different morphogenetic programs and production of secondary metabolites (Sarikaya-Bayram et al. 2015).
Fungi produce small bioactive compounds also named secondary metabolites (SMs) that have wide-ranging influences on cellular physiology such as antibiotics, mycotoxins, siderophores, antiviral, and cytotoxic molecules (Keller et al. 2005; Brakhage 2013). Each fungus can potentially produce up to 50–100 SMs depending on the genus. SM genes are often clustered and production of SMs is coordinately controlled by regulatory protein complexes in response to environmental stimuli such as light, carbon source, starvation, and pH (Keller et al. 2005; Brakhage 2013).
Neurospora crassa and Aspergillus nidulans are two model filamentous fungi with different characteristics and lifestyles, which have been used to understand fundamental questions for eukaryotic molecular genetics (Galagan et al. 2003, 2005; Borkovich et al. 2004; Park and Yu 2012; Fuller et al. 2016). However, it has been intriguing whether the regulatory protein complexes controlling developmental programs or SM production have been structurally and functionally conserved.
N. crassa has been used as a model system to study genetics, biochemistry, enzymology, chromatin biology, cell–cell fusion, light responses and circadian rhythms, and development (Springer 1993; Fleissner et al. 2008; Rountree and Selker 2010; Baker et al. 2012; Aramayo and Selker 2013; Hurley et al. 2015; Dunlap and Loros 2016). N. crassa can undergo three different sporulation pathways: two different asexual conidiation pathways produce macroconidia and microconidia, while a sexual sporulation pathway leads to formation of meiotic ascospores. Macroconidiation involves the formation of hyphal constrictions at the aerial hyphal tip, initially by a budding process, while microconidia, on the contrary, are produced from specialized hyphae in a process that involves the emergence of the microconidial bud and its liberation after breaking the cell wall (Springer 1993). N. crassa uses a heterothallic (self-sterile) system, which requires the fusion of two opposite mating types, a and α (Pöggeler et al. 2006). In brief, sexual development in N. crassa is initiated by the formation of protoperithecia (semiopen fruiting body, female organ), followed by the fertilization of the protoperithecia by microconidia from the opposite mating type (male hyphae). Fusion of two opposite mating type nuclei within the perithecia results in the formation of the zygote, which undergoes meiosis to generate sexually formed ascospores. N. crassa has a set of light receptors for blue, red, and green light but it uses mainly blue light as a signal to adjust cellular activities, sporulation and circadian rhythm, and the WC complex as the main photoreceptor (Baker et al. 2012; Fischer et al. 2016). The N. crassa genome is relatively poor in SM gene clusters (8–10 clusters) in comparison to Aspergilli (>50 clusters) (Kjærbølling et al. 2018) and only a few SMs from N. crassa have been identified: the antioxidant histidine-derived ergothioneine, the nonribosomal peptide coprogen, and the polyketide oxoalkylresorcylic acid (ORAS) (Huschka et al. 1985; Funa et al. 2007; Bello et al. 2012). Coprogen is a siderophore required for chelating iron ions from the environment, and is historically the oldest metabolite identified from N. crassa (Huschka et al. 1985). Microbes use siderophores for the utilization of environmental iron sources. Microbial pathogens sequester iron from high affinity iron-binding molecules such as ferritin, lactoferrin, and hemoglobin in the blood of mammals (Haas 2003, 2014; Haas et al. 2008). Furthermore, siderophores have significant potential to be used for treatment of various diseases, for drug delivery, for treatment of heavy metal pollution in the environment, and for the production of functional foods (Pócsi et al. 2008).
The velvet family of proteins is restricted to fungi and controls fundamental processes such as development and SM. The heterotrimeric velvet complex is formed by two velvet transcription factors, VeA and VelB, and the methyltransferase LaeA, and is a key element in the regulation of light-dependent fungal development and SM production in A. nidulans (Bayram et al. 2008a). Velvet proteins play essential roles in sporulation, pathogenicity, and SM production in different fungi including human pathogens, endophytic fungi, and plant pathogenic fungi (Calvo 2008; Sarikaya-Bayram et al. 2015). A. nidulans vegetative hypha differentiates upon reception of environmental signals. In the light, asexual sporulation (conidiation) is promoted, whereas in the dark, the sexual developmental program (formation of closed fruiting bodies named cleistothecia) is activated (Etxebeste et al. 2010; Rodriguez-Romero et al. 2010; Dyer and O’Gorman 2012). In contrast to N. crassa, A. nidulans has evolved a homothallic (self-fertile) system where the hyphae do not need an opposite mating partner (Galagan et al. 2003, 2005; Paoletti et al. 2007; Dyer and O’Gorman 2012). In A. nidulans under dark conditions, VeA and VelB form a heterodimer that enters the nucleus in a process mediated by an α-importin, then the VeA/VelB heterodimer forms a heterotrimeric velvet complex by interacting with the methyltransferase LaeA, which presumably regulates the transcription of genes for sexual development and SM production including polyketide mycotoxin sterigmatocystin. Although, LaeA was initially identified as a regulator of secondary metabolism, it also regulates fungal development (Bok and Keller 2004; Sarikaya Bayram et al. 2010). LaeA was shown to be a bona fide methyltransferase since it methylates itself on a methionine residue at position 207 (Patananan et al. 2013). However, other target substrates of LaeA are currently not known. VelB forms an additional heterodimeric complex with the velvet protein VosA, which is required for spore viability and stress tolerance (Ni and Yu 2007; Sarikaya Bayram et al. 2010). The VelB/VosA heterodimer activates the transcription of trehalose biosynthetic genes tpsA and orlA, as well as wetA, a gene essential for spore viability and conidiophore morphology (Park et al. 2012b). The structure of the velvet domain in the VelB-VosA heterodimer is similar to the DNA-binding motif of the rel homology domain of NF-kB transcription factors that activate inflammation pathways in mammals (Ahmed et al. 2013). The role of a fourth velvet protein, VelC, is less clear as the mutant shows slightly increased sexual development (Park et al. 2014). In addition to LaeA, VeA participates in the integration of other signals by interacting with other regulatory proteins. VeA interacts physically and functionally with the putative methyltransferase LlmF, the methyltransferase VipC/VapB heterodimer, the Far1-like DNA binding protein VipA, MAPK MpkB, and the red-light receptor phytochrome FphA (Purschwitz et al. 2008; Bayram et al. 2012; Palmer et al. 2013; Sarikaya-Bayram et al. 2014; Röhrig et al. 2017).
Mechanistic insights into the molecular functions of the velvet complex are mainly limited to Aspergilli. N. crassa and A. nidulans are distantly related, with an estimated divergence time of 394 MY (Kumar et al. 2017). N. crassa belongs to Sordariomycetes, which comprise many plant and human pathogenic fungi. In agreement with the phylogenetic distance, there are significant biological differences in response to light, sporulation pathways, and mode of reproduction between N. crassa and A. nidulans. Initial studies of the veA ortholog in N. crassa, ve-1, revealed that it was required for repression of asexual conidiation and the regulation of photocarotenogenesis (Bayram et al. 2008b; Olmedo et al. 2010; Gil-Sánchez et al., personal communication). However, the molecular functions of ve-1 and the other velvet complex members in N. crassa remain largely unknown. Here, we characterize the complete set of velvet proteins in N. crassa, and show that they form a regulatory velvet complex with a key role in the regulation of development and SM. The functional complementation of some of the velvet proteins between N. crassa and A. nidulans highlights the structural conservation of the velvet complex and its general role in fungal development and the regulation of SM.
Materials and Methods
Strains, media, and growth conditions
Strains used in this study are listed in Supplemental Material, Table S1. General genetic procedures and media used in the handling of N. crassa are available through the Fungal Genetics Stock Center (www.fgsc.net) (McCluskey 2003). N. crassa strains were mainly cultured in Vogel’s minimal medium (VMM, 2% sucrose w/v) and A. nidulans strains were cultured in glucose (1% w/v) minimal medium (GMM) supplemented with vitamins and trace elements. For iron (Fe) starvation experiments, trace elements were prepared without iron. The flasks were treated with HCl and EDTA as described in detail (Kragl et al. 2007; Schrettl et al. 2008).
For light induction of N. crassa, a total of 106 conidia were inoculated on 25 ml Vogel’s liquid medium in 90-mm plates, which were incubated in complete darkness at 22° for 48 hr. The cultures were then exposed to light for 30, 60, 120, and 300 min. For carotenoid induction, plates were exposed to light for 2 min, and then incubated at 8° for 24 hr prior to collection. Light exposure with different intensities was obtained with a quartz halogen lamp installed in a slide projector passed through a filter holder with two heat filters and neutral-density filters, as required to obtain the desired light intensity. A control treatment was always kept in the dark. Mycelia were collected, dried on paper, frozen in liquid nitrogen, and stored at −80°. Protein extraction, quantification, and western blotting were performed as described below. Three independent experiments were performed.
Plasmid construction and fungal expression of tagged proteins
Plasmid constructions, plasmids, oligonucleotides are shown in File S15.
Protein methods
Total protein isolation:
Protein extraction was performed either by bead beating or by grinding in liquid nitrogen. For bead beating, mycelia samples were placed in screw-cap microtubes containing 500 µl protein extraction buffer [50 mM HEPES pH 7.4, 10% (v/v) glycerol, 137 mM NaCl, 5 mM EDTA, 1 µM leupeptin, 1 µM pepstatin, 50 µM phenylmethylsulfonyl fluoride] and the same volume of 0.5 mm silica/zirconium beads. Cell lysis was carried out in a cell homogenizer (FastPrep-24; MP Biomedicals) by giving the samples three 30-sec pulses with 5 min incubations on ice in between pulses. Lysed samples were then centrifuged at 20,000 × g and 4° for 15 min. The supernatant was collected and either immediately used or stored at −80°. Unless otherwise stated, protein quantification was performed with the Bradford assay (Bradford 1976).
Preparation of nuclear and cytoplasmic protein extracts:
A total of 107 conidia were inoculated in 200 ml of VMM (liquid) and allowed to develop for 48 hr at 30° under agitation. Three different treatments were prepared: (1) continuous light; (2) continuous darkness; and (3) continuous darkness for 47.5 hr followed by a 30-min light exposure. Mycelia were fixed by adding 540 µl of formaldehyde for 15 min and then the reaction was stopped by adding glycine (125 mM). Mycelia were then collected by filtration and frozen in liquid nitrogen. Nuclear fraction extractions were performed as developed by Baum and Giles (1985) with minor modifications by (Schwerdtfeger and Linden 2000; Froehlich et al. 2002). Further details were given in File S15.
Antibodies used in this study:
Antibodies were used in the following indicated dilutions: Primary antibodies; monoclonal α-FLAG (1:10,000, F3165; Sigma), polyclonal α-histone 3 (H3) (ab1791, 1:5000; Abcam), monoclonal α-GFP (B2, Sc-9996, 1:2000; Santa Cruz), polyclonal α-human influenza hemagglutinin (HA) (1:4000, ab9110; Abcam), monoclonal α-tubulin (Sc-32293, 1:200; Santa Cruz). Secondary antibodies; HRP conjugated goat α-mouse IgG (#1721011, 1:10,000; Bio-Rad) was used for α-FLAG and α-tubulin detections. Goat α-mouse IgG (#1706516, 1:2000; Bio-Rad) was used for α-GFP and goat α-rabbit IgG (#1706515, 1:2000; Bio-Rad) was used for α-SkpA, α-HA, and α-H3 detections.
Co-immunoprecipitations of the velvet complexes in N. crassa:
The strains coexpressing VE-1FLAG VE-2HA, VE-1FLAG LAE-1HA, and VE-2FLAG LAE-1HA were grown for 48 hr as described for the cellular fractionation experiments, and the same three treatments were carried out: continuous light, complete darkness, and complete darkness followed by a 30-min light exposure. Mycelia were also fixed with formaldehyde and collected as described. Mycelial pads (8–10 g wet weight) were harvested by filtration, frozen in liquid nitrogen, and ground to fine powders, which were mixed in a 50 ml tube with 20 ml of IPB buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 0.1% (v/v) NP-40] with protease inhibitors (Roche complete protease inhibitor cocktail) and dissolved by vortexing on ice. After mixing, crude extracts were centrifuged at 14,300 × g, at 4° for 20 min. Supernatants were collected and transferred to ultracentrifuge tubes. Centrifugation was performed at 169,000 × g at 4° for 45 min. Then, a 1-ml fraction of each supernatant was transferred to a new tube, labeled “INPUT,” frozen in liquid nitrogen, and stored at −80° until use. The remaining supernatant was collected in 15-ml screw-cap tubes and kept at 4°. During the 45-min centrifugation, 300 μl of M2-FLAG agarose beads (Sigma) were equilibrated by washing them three times with 1 ml of IPB buffer followed by centrifugation at 1000 × g at 4°. After equilibration, 150 µl as added to the supernatant in 15 ml tubes, and the mix was incubated for 3 hr at 4° in a rotating mixer. Beads were then precipitated by centrifuging at 1500 × g for 10 min at 4°. A 1-ml sample of each supernatant were transferred to a new tube, labeled “flowthrough” (FT), frozen in liquid nitrogen, and stored at −80°. The beads were washed twice with 10 ml of IPB buffer by gentle rotation for 4 min at 4° and finally suspended with 500 μl of IPB buffer and transferred to new 2 ml tubes. Beads were washed twice with 1 ml IPB buffer by centrifugation at 1000 × g at 4°. At the end, 120 μl of NuPAGE buffer (Invitrogen) was added to the beads, vortexed, and incubated for 10 min at 72° with moderate shaking. The supernatant was collected by centrifuging at 1000 × g at 4° and transferred to new tubes labeled “IP.” Samples were treated with DTT (50 mM) and incubated again for 10 min at 72°. Protein concentrations from the INPUT samples were measured in a Nanodrop instrument (Absorbance set to 280 nm). INPUT, FT, and IP samples were used for western blot as described previously. Three independent experiments were performed.
GFP-TRAP and LC-MS/MS protein identifications:
Immunoprecipitation (IP) of VE-1GFP, VE-2GFP, VOS-1GFP, and LAE-1GFP were mainly performed as described (Bayram et al. 2012) by using GFP-TRAP magnetic beads (Chromotek). Further details are given in File S15.
Microscopy:
Spinning disc confocal microscopy of N. crassa and A. nidulans cells were performed as described earlier (Bayram et al. 2012; Dettmann et al. 2012). Details are given in File S15.
Secondary metabolite analysis
Thin layer chromatography for sterigmatocystin:
Extraction and running of mycotoxin sterigmatocystin (ST) from A. nidulans on silica thin layer chromatography plates (Macherey-Nagel) were carried out as described (Bayram et al. 2009).
RP-HPLC analysis:
RP-HPLC analysis was carried out using an Agilent Series 1200 HPLC System with a diode array (DAD) and separation across a water: acetonitrile gradient with 0.1% (v/v) TFA. Gradient conditions of 5–70% over 30 min at 2 ml/min were used for coprogen analysis with 100 μl injection. Gradient conditions of 5–100% acetonitrile over 30 min at 2 ml/min were used for organic extract analysis with 40 μl injection. Separation was carried out on a C18 column (Agilent Zorbax Eclipse XDB-C18 semi-preparative; 5 μm particle size; 9.4 × 250 mm) with DAD detection at 254 and 440 nm. Ferri-coprogen detection was carried out at 440 nm. The peak associated with coprogen was identified in previous analysis by LC-MS/MS detection of a fraction-collected peak. For quantification of coprogen, integration areas detected by HPLC were normalized to biomass (mAUs/g mycelia).
LC-MS/MS analysis:
For LC-MS/MS analysis of peaks from RP-HPLC analysis, peaks were collected by fractionation. Fractions were evaporated to dryness in a Speedivac and resuspended in water formic acid (0.1%). Samples were diluted 1 in 50 in water formic acid (0.1%) and spin filtered (0.2 μm) before LC-MS/MS with 1 μl injection. LC-MS/MS analysis was carried out on a nanoflow Agilent 1200 LC system and subjected to tandem mass spectrometry using an Agilent 6340 Ion Trap LC-MS System (Agilent Technologies). Samples were applied to a Zorbax SB-C18 HPLC-Chip with a 40 nl enrichment column and a 75 μm × 43 mm (5 μm particle and 300 Å pore size) analytical column. Identification of ferri-coprogen was confirmed via detection of a single charged ion in the MS spectrum (M: 821.3, [M+H]1+: observed m/z 822.1; expected m/z 822.3).
Carotenoid analysis:
Carotenoid analysis was performed as described earlier (Castrillo et al. 2018). Details were given in File S15.
RNA isolation andlibrary preparation:
RNA isolation, poly-A selection and library preparation was done as described in Klocko et al. (2016). The sequencing data were uploaded to the Galaxy web platform, and the public server at usegalaxy.org was used to analyze the data (Afgan et al. 2016). Specifically, the read quality was checked using FastQC, trimmed the adaptors using Trim Galore, aligned the reads and obtain read count using RNA STAR with intron length set to max 1000 bp. Differential gene expression analysis was done by DESeq2 (Love et al. 2014) to examine pairwise differences between WT and Δve-1 after 48 hr growth under standard or iron-free (Fe-Free) conditions. Genes with log2 ≥ 1.0 or ≤ −1.0 changed expression and adjusted P values ≤ 0.05 were used to create Venn diagrams using Biovenn (Hulsen et al. 2008). Genes were functionally annotated with Pfam domains and Gene Ontology terms using InterProScan (Jones et al. 2014).
Statistical analysis:
Most of the experiments were performed as three independent biological replicates and numerical data are expressed as the mean ± SD and SE. The mean data were compared for significant differences via the student t-test by using the software Graphpad Prism Version 6.
Data availability
Strains and plasmids are available upon request. RNA-seq data were deposited in the Gene Expression Omnibus public database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE123783. Supplemental files are available at FigShare. Table S1 Fungal (Aspergillus nidulans and Neurospora crassa) strains used in this study. Table S2 Oligonucleotides used in this study. Table S3 Plasmids used and constructed in this study. File S1 Interaction partners of VE-1GFP fusion in veA∆ strain of A. nidulans. File S2 Interaction partners of VE-2 GFP fusion in velB∆ strain of A. nidulans. File S3 Interaction partners of VOS-1GFP fusion in vosA∆ strain of A. nidulans. File S4 Interaction partners of LAE-1GFP fusion in laeA∆ strain of A. nidulans. File S5 Interaction partners of VE-1GFP fusion in N. crassa. File S6 Interaction partners of VE-2GFP fusion in N. crassa. File S7 Interaction partners of VOS-1GFP fusion in N. crassa. File S8 Interaction partners of LAE-1GFP fusion in N. crassa. File S9 List of predicted secondary metabolite gene clusters in N. crassa. File S10 List of up- and downregulated genes in ve-1 mutant under standard and iron starvation conditions. File S11 List of GO terms for upregulated genes in ve-1 mutant in both conditions. File S12 List of GO terms for downregulated genes in ve-1 mutant in both conditions. File S13 List of differentially expressed putative secondary metabolite genes in ve-1 mutant in both conditions. File S14 Top 20 up and downregulated genes under iron starvation and standard conditions in ve-1 mutant. File S15 Supplemental materials and methods. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8091182.
Results
N. crassa VE-1 and VE-2 are functional orthologs of A. nidulans VeA and VelB
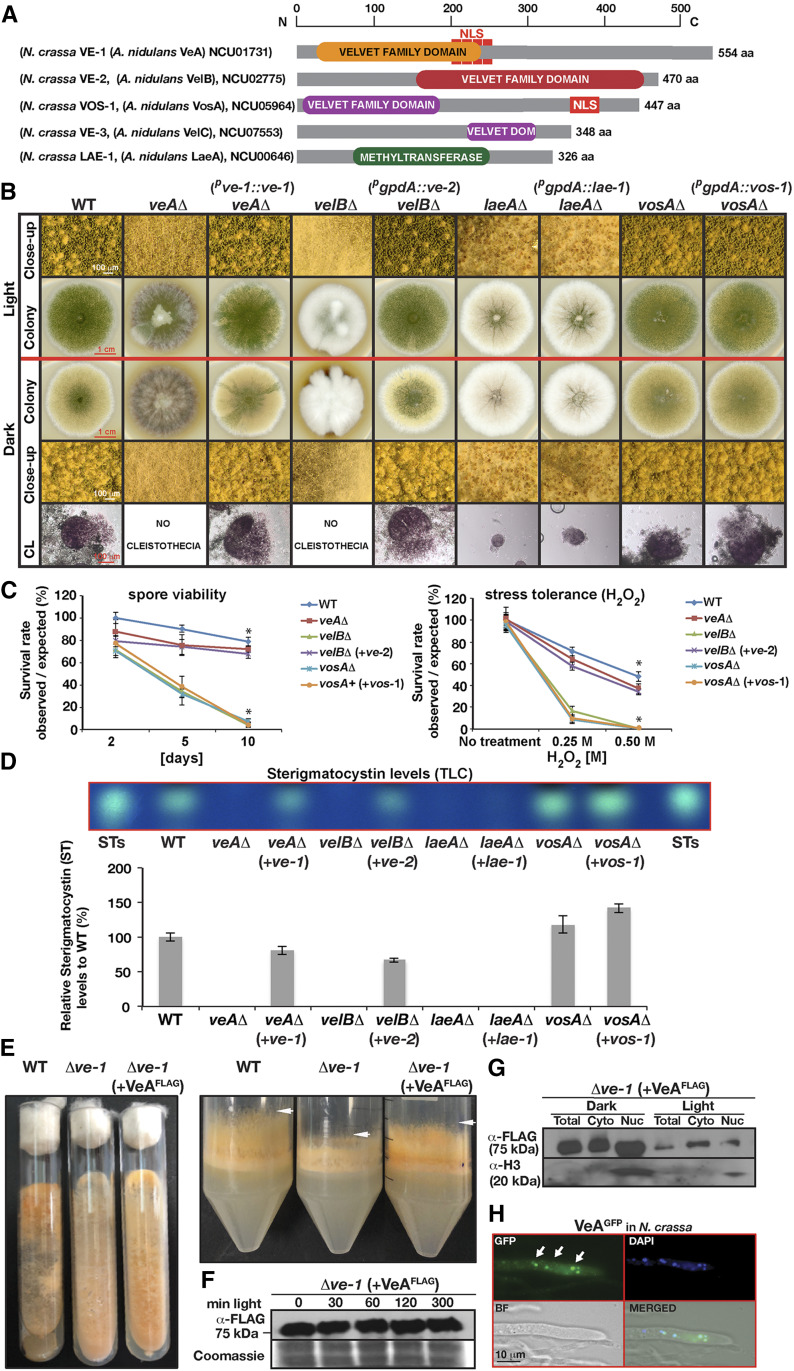
In addition to the characterized N. crassa VE-1 protein [NCU01731; 554 aa, 52% identical to A. nidulans VeA (Bayram et al. 2008b)], which contains a velvet superfamily domain in its N-terminus with a bipartite nuclear localization signal (NLS), the N. crassa genome encodes three additional velvet domain proteins as well as an ortholog of A. nidulans LaeA (Figure 1A and Ojeda-López et al. 2018). VE-2 (NCU02775; 470 aa; 47% identical to A. nidulans VelB) bears a velvet superfamily domain at its C-terminus, and, like VelB in A. nidulans, lacks an obvious NLS. In contrast, VOS-1 (NCU05964; 447 aa; 44% identical to A. nidulans VosA) possesses both an NLS sequence at its C-terminus and a velvet superfamily domain at its N-terminus. The domain structure of VE-3 (NCU07553) is similar to (40% identical) VelC but it appears to have a truncated velvet domain (Figure 1A and Ojeda-López et al. 2018). Sequence analysis of LAE-1 (NCU00646; 326 aa; 42% identical to A. nidulans LaeA) revealed an S-adenosyl methionine (SAM) binding site and a methyltransferase domain at the center of the protein.
Figure 1.
Interspecies functionality of velvet proteins in Aspergillus nidulans and Neurospora crassa for light-dependent development and secondary metabolite production. (A) Four velvet family proteins are shown encoded by N. crassa genome. Top scale indicates their length in amino acids (aa). VE-1 and VOS-1 contains a nuclear localization signal (NLS). (B) Growth and developmental responses of A. nidulans velvet complex mutants, veAΔ, velBΔ, laeAΔ, vosAΔ, and their complementation by N. crassa velvet orthologs (ve-1, ve-2, vos-1, lae-1) expressed under the control of the native (ve-1) promoter or the constitutive gpdA promoter (ve-2, vos-1, lae-1) in comparison to wild type (WT) grown under light and dark conditions. 5 × 103 spores were point-inoculated on GMM plates and grown at 37° for 5 days. (C) Spore viability and stress tolerance test veAΔ, velBΔ, laeAΔ, vosAΔ strains, and for strains expressing corresponding heterologous proteins. Spore viability tests were performed with 103 spores at 37°. (+) Represents complementations. Asterisks denote the significant difference (P < 0.05). (D) Examination of mycotoxin production in the velvet complex mutants and their respective complemented strains by thin layer chromatography (TLC). Strains were grown on GMM plates with oatmeal for 3 days in the dark. Quantification of the ST from two independent biological replicates of TLC plates. STs: Sterigmatocystin standard. (E) Complementation of Δve-1 phenotypes (reduced carotenogenesis, stunted hyphae) by A. nidulans VeA3XFLAG fusion expressed under N. crassa ve-1 promoter. (F) Expression of VeA3XFLAG in N. crassa under constant illumination. 30 µg total protein was loaded per lane. (G) Presence of VeA3XFLAG in total crude extract, cytoplasmic and nuclear fractions (50 µg in each lane) in the light and dark. Strain was grown in liquid VMM for 48 hr at 30° and then exposed to white light for 30 min or kept in the dark as a control. (H) Cellular localization of VeAGFP expressed under ccg-1 promoter in N. crassa grown in VMM on a coverslip overnight at 30°. DAPI stains DNA in the nuclei blue and arrows indicate the nuclear localization of VeAGFP.
In order to determine if the four N. crassa velvet and the lae-1 genes are functional orthologs of the A. nidulans genes, the ve-2, vos-1, and lae-1 genes were heterologously expressed under the control of the constitutive gpdA promoter in the A. nidulans mutants ΔvelB, ΔvosA, and ΔlaeA. In addition, ve-1 was expressed under control of its own native promoter in the A. nidulans ΔveA strain (Figure 1). The N. crassa ve-1 and ve-2 genes complemented the developmental defects of the A. nidulans strains that were characterized by loss of fruiting bodies (cleistothecia), brownish pigment secreted into media and reduced asexual sporulation (Figure 1B). Furthermore, ve-2 complemented the spore inviability (Figure 1C) and the defects of the ΔvelB mutant to oxidative stress (H2O2). However, the N. crassa vos-1 and lae-1 genes did not restore the mutant phenotypes of the A. nidulans ΔvosA and ΔlaeA strains, i.e., reduced spore viability and small cleistothecia formation defects, respectively. Moreover, we examined if the N. crassa velvet genes and lae-1 complemented the defect in the production of the mycotoxin sterigmatocystin (ST) observed in the A. nidulans mutants (Figure 1D). Expression of ve-1 and ve-2 in A. nidulans restored the lack of ST production in veA and velB mutants to the levels observed in the wild-type, as previously observed for the developmental defects. In contrast, expression of vos-1 and lae-1 did not lead to any significant changes in the chemical phenotypes of the ΔvosA and ΔlaeA strains.
As a further confirmation of the conserved nature of the velvet proteins, we complemented the N. crassa Δve-1 mutant with the A. nidulans veA gene. The two most evident phenotypes of Δve-1—reduced carotenoid accumulation and reduced growth of aerial hyphae (Bayram et al. 2008b)—were restored by the heterologous expression of the A. nidulans veA gene fused to the FLAG tag in N. crassa (Figure 1E). Furthermore, expression and localization of VeA in N. crassa were examined in response to light. The accumulation of VeA did not change under prolonged exposure to light (Figure 1F). VeA protein levels were mainly increased in the dark, and exhibited nuclear localization both under light and dark cultures (only dark is shown) (Figure 1, G and H).
In summary, our results indicate that the N. crassa ve-1 and ve-2 genes are functional orthologs of veA and velB in A. nidulans, while vos-1 and lae-1, seem not to be conserved. This stresses the similarities and differences between these two distantly related fungal species, even in proteins that are known to form complexes in order to function.
N. crassa VE-1, VE-2, VOS-1, and LAE-1 accumulate in A. nidulans nuclei, but only VE-1 and VE-2 interact with components of the heterotrimeric velvet complex in A. nidulans during vegetative growth
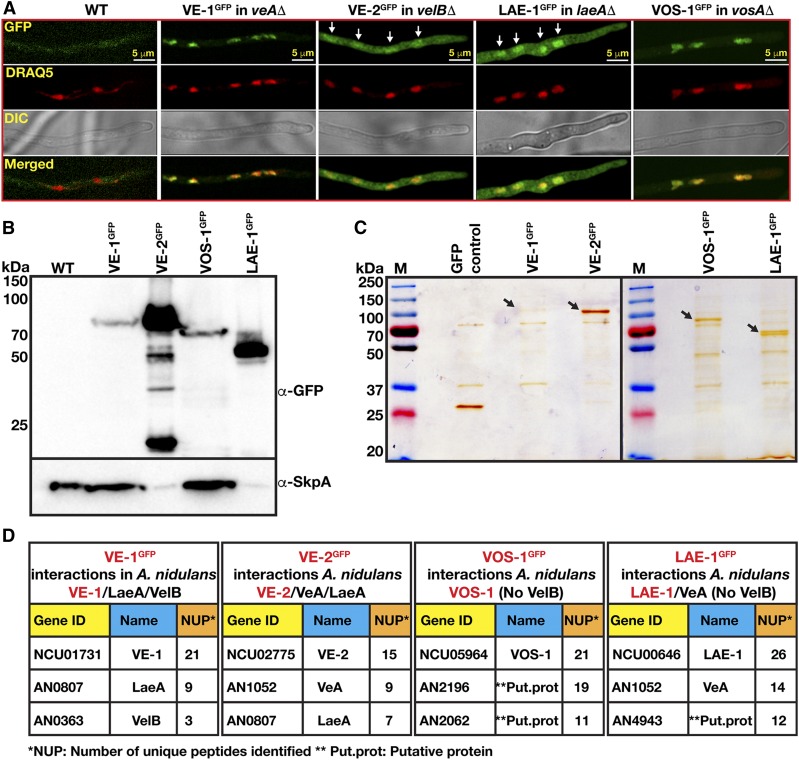
In A. nidulans, VeA displays light-dependent nuclear localization (Stinnett et al. 2007; Bayram et al. 2008a). In order to determine the subcellular localization of N. crassa VE-1 and VE-2 in A. nidulans, we heterologously expressed the two velvet proteins fused to synthetic green fluorescent proteins (sGFP) under the control of the gpdA promoter, and examined their localization in cultures of complemented A. nidulans mutants grown under light and dark conditions. We did not observe any effect of light on the localization of the fusion proteins, and, therefore, only dark grown cultures are shown (Figure 2, A and B). N. crassa VE-1GFP was localized in A. nidulans nuclei while the VE-2GFP was distributed evenly between nuclei and cytoplasm, resembling the subcellular localization of the corresponding A. nidulans proteins (Bayram et al. 2008a). Interestingly, VOS-1GFP had the strongest nuclear localization, whereas LAE-1GFP was found both in the nucleus and in the cytoplasm as shown after colocalization with colabeled red nuclei. Expression of all GFP fusions of VE-1, VE-2, VOS-1 were clearly detected in total protein extracts, confirming that all velvet family proteins and LAE-1 were properly expressed (Figure 2B).
Figure 2.
Localization and complex formation of Neurospora crassa velvet proteins in Aspergillus nidulans. (A) Subcellular localization of VE-1, VE-2, LAE-1, and VOS-1GFP fusion proteins in the respective A. nidulans velvet mutants (veAΔ, velBΔ, laeAΔ, vosAΔ). Chimeric fungi (103 spores) expressing the fusion proteins were grown at 30° for 16 hr and observed by confocal microscopy. DRAQ5 stains the nuclei red. (B) Protein expression of VE-1GFP, VE-2GFP, LAE-1GFP, VOS-1GFP fusion proteins assayed by a western blot analysis. In general, 80 µg total protein of crude cell extracts was loaded per lane, but, for the highly expressed VE-2 and LAE-1 samples, 40 µg was loaded. α-GFP detects GFP fusions and α-SkpA detects constitutively expressed SkpA protein, which serves as a loading control (therefore less SkpA is seen in the VE-2 and LAE-1 lanes. (C) Silver stained (10%) SDS gels of the GFP-TRAP pulldowns of fusion proteins. Arrows indicate the positions of strongly staining full-length precipitated fusion proteins. (D) Interaction partners of VE-1, VE-2, VOS-1, and LAE-1 in A. nidulans during vegetative growth. The results of GFP TRAP pulldowns digested with trypsin and identified by LC-MS. Gene ID (locus number), name and number of unique peptides found are given in the tables (see Files S1–S4). Chimeric velvet complexes found in each purification are given at the top of the table.
The A. nidulans VeA/VelB/LaeA complex controls sexual development and SM production. In addition, VelB forms a heterodimer with VosA that is required for spore viability and trehalose biogenesis. In order to determine if the N. crassa velvet proteins and LAE-1 have the capacity to interact with other members of the velvet proteins in A. nidulans, VE-1GFP, VE-2GFP, VOS-1GFP, and LAE-1GFP were immunoprecipitated, and interacting proteins were identified using liquid chromatography-mass spectrometry (LC-MS) (Figure 2, C and D and Files S1–S4). We detected the presence of VE-1 or VE-2 in chimeric heterotrimeric complexes (VE-1/VelB/LaeA and VeA/VE-2/LaeA) in vegetatively grown cultures of A. nidulans. However, we did not detect a chimeric VE-2/VosA complex and none of the velvet complex members were identified after immunoprecipitation of VOS-1GFP from vegetative cultures. Consistently, VosA was not identified by LC-MS in the N. crassa velvet protein precipitates. We detected the interaction between LAE-1GFP and VeA, but not VelB (File S4). In summary, these results suggest that only VE-1 and VE-2 from N. crassa form functional chimeric heterotrimeric velvet complexes with their counterparts in A. nidulans.
The N. crassa velvet proteins play different roles in asexual and sexual reproduction
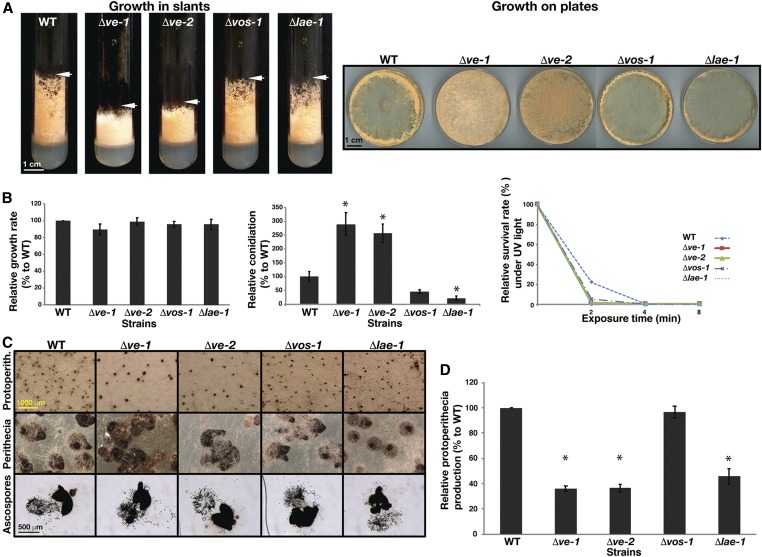
In order to understand the molecular and developmental functions of the velvet proteins and the putative methyltransferase LAE-1 in the biology of N. crassa, strains with deletions in each of these genes were subjected to development and growth tests (Figure 3). Δve-1, Δve-2, Δvos-1, or Δlae-1 deletion strains did not show any significant changes in growth rate (Figure 3, A and B). However, Δve-1 and Δve-2 exhibited reduced aerial hyphae in slants when compared to wild-type or to Δvos-1 and Δlae-1 (Figure 3A). Conidiation increased twofold in Δve-1 and Δve-2 compared to wild-type despite their reduced aerial growth, while Δvos-1 and Δlae-1 displayed slight or moderate reductions in conidiation (Figure 3B). The viability of conidia from the mutants did not change significantly. Sexual development was altered in all velvet and lae-1 mutants, and protoperithecia production was reduced to ∼50% of wild type. Fertilization of the remaining protoperithecia yielded viable ascospores, indicating a functional sexual cycle (Figure 3, C and D). In summary, the developmental alterations in the mutants revealed that VE-1 and VE-2 are required for the promotion of aerial hyphal growth and the repression of asexual spore formation. Moreover, VE-1, VE-2, and LAE-1 are important for the production of protoperithecia during sexual development. VOS-1 and LAE-1 play a minor role in the regulation of conidiation.
Figure 3.
Developmental functions of the velvet and Lae-1 orthologs of N. crassa (A) Slant growth phenotypes and hyphae formation of the wild type along with Δve-1, Δve-2, Δvos-1, and Δlae-1 strains on VMM after 4 days at RT (left panel) and on plates (right panel) under continuous illumination. (B) Quantification of relative growth rates, asexual conidiation and resistance to UV light. Growth rate was measured on VMM at RT for 24 hr. Conidiation was measured from plates grown for 4 days. For the UV test, 50 fresh spores spread on sorbose medium plates were exposed to UV for 0, 2, 4, or 8 min, incubated on plates at RT for 2–3 days, and colony forming units were counted. Asterisks denote the significant difference (P < 0.05) (C and D) Formation of protoperithecia, perithecia and ascospores of the WT and velvet mutants along with lae-1 mutant. Equal amounts of fungal spores were grown at the center of corn meal agar (CMA) plates for 7 days for protoperithecia formation, and 14 days at RT for perithecia maturation and ascospore analysis. Perithecia were squeezed open for photomicroscopy of ascospores.
Velvet proteins control production of secondary metabolites and carotenoid accumulation
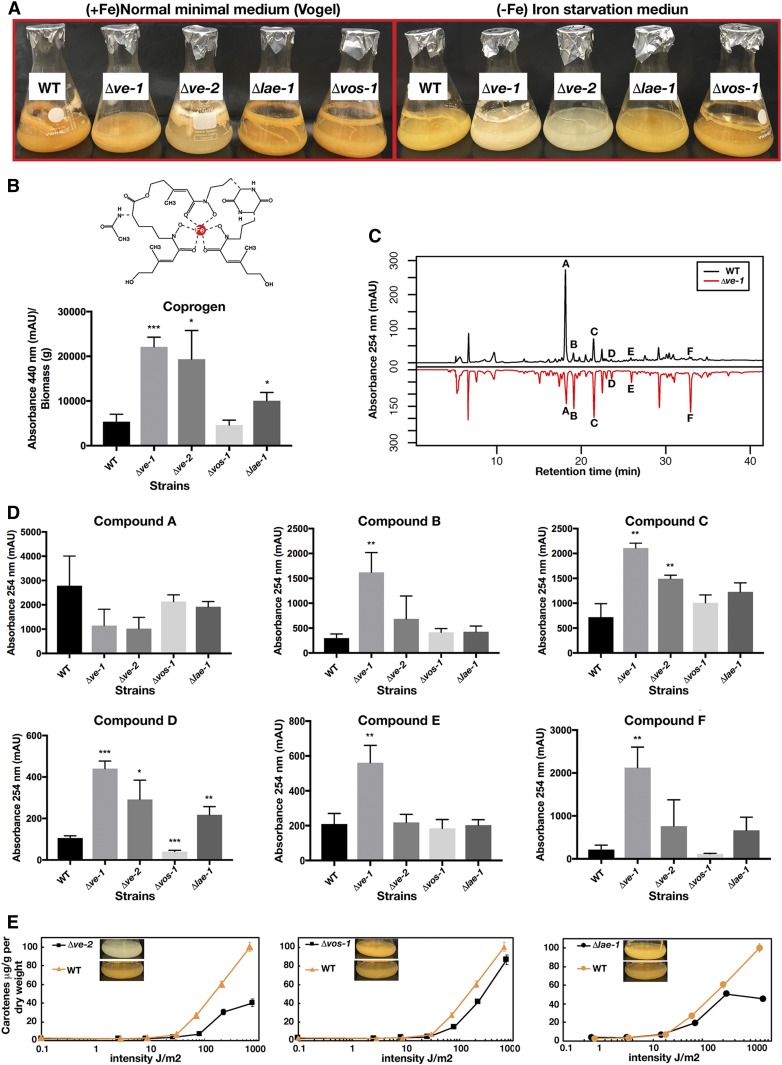
The N. crassa genome contains 10 putative SM gene clusters (File S9), and N. crassa produces several SMs including coprogen and ergothioneine (Tóth et al. 2009; Bello et al. 2012, 2014). In order to understand the role of the velvet proteins and of LAE-1 on the production of SMs, wild-type and deletion strains were grown in Vogel’s MM medium or in medium lacking iron to promote the production of nonribosomal peptide (NRPs) siderophores. Fe-free medium revealed a difference in SM production in the mutants, which was evident by the lighter colors of Δve-1 and Δve-2 liquid cultures (Figure 4A). Growth in Fe-free media resulted in at least fourfold to fivefold elevated levels of coprogen in Δve-1 and Δve-2 in comparison to wild type, suggesting that ve-1 and ve-2 are negative regulators of coprogen production (Figure 4B). Δlae-1 showed a slight increase in coprogen production, whereas the Δvos-1 mutant did not show any significant changes in coprogen production.
Figure 4.
Regulatory functions of the velvet orthologs in production of coprogen, carotenes and various other compounds. (A) A comparison of WT, ve-1, ve-2, vos-3, and lae-1 mutants of N. crassa grown in liquid VMM (left panel) with iron (Fe) and without (iron starvation) (right panel) at 37° for 7 days. (B) Production of the iron chelating and growth promoting compound coprogen was normalized to biomass (P < 0.05). (C) Mirror image comparison of the WT and ve-1 mutant’s organic extraction chromatogram by RP-HPLC (254 nm). (D) Production of five compounds significantly changed mainly in ve-1 or ve-2 mutants. Chromatograms were compared with WT and any compounds of interest were quantified by integrating areas associated with each peak. Quantifications were obtained from the supernatants of the three biological replicates grown in liquid iron-starvation Vogel’s media (FVMM) at 37° for 7 days. (E) Quantification of total carotene produced by ve-2, vos-1, and lae-1 mutants in comparison to WT under a gradient of illumination.
Comparison of organic extracts from 1-week-old cultures of wild-type and mutant strains showed at least six unique compounds that significantly changed their accumulation in the mutants when compared to wild type (Figure 4C; only the Δve-1 chromatogram is shown): A (RT ∼18 min), B (RT ∼19 min), C (RT ∼21 min), D (RT ∼23 min), E (RT ∼25 min), and F (RT ∼32 min). Deletion of ve-1 resulted in a major change in the accumulation of SMs, as it had a significant increase in the abundance of compounds B (P < 0.01), C (P < 0.01), D (P < 0.001), E (P < 0.01), and F (P < 0.01), and a decrease in the abundance of compound A. Deletion of the other genes led to minor changes in the accumulation of a smaller number of SMs. In the Δve-2 mutant, accumulation of compounds C and D increased, while accumulation of compound A decreased; Δlae-1 showed increased accumulation of compounds D and F, and in Δvos-1 decreased accumulation of compound D was observed.
The Δve-1 mutant accumulated less carotenoids than wild type, and it affected the light regulation of carotenoid biosynthesis (Bayram et al. 2008b; Gil-Sánchez et al., personal communication) (Figure 4A). We observed a light-orange color also in Δve-2, suggesting a role of VE-2 in carotenoid biosynthesis. Therefore, we measured accumulation of total carotenoids from all mutants under different light intensities, with the exception of Δve-1 (Figure 4E) (Gil-Sánchez et al., personal communication). The accumulation of carotenoids after light exposure was altered in all mutants, but the effect was more pronounced in Δve-2 and Δlae-1. Both mutants showed a reduction in the accumulation of carotenoids with high intensities of light (>100 J/m2). Our results suggest that VE-1, VE-2, and, partially, LAE-1 are involved in production of several unknown SMs, the siderophore coprogen, and carotenoids.
VE-1, VE-2, VOS-1, and LAE-1 are found in both nuclei and cytoplasm in a light-independent manner
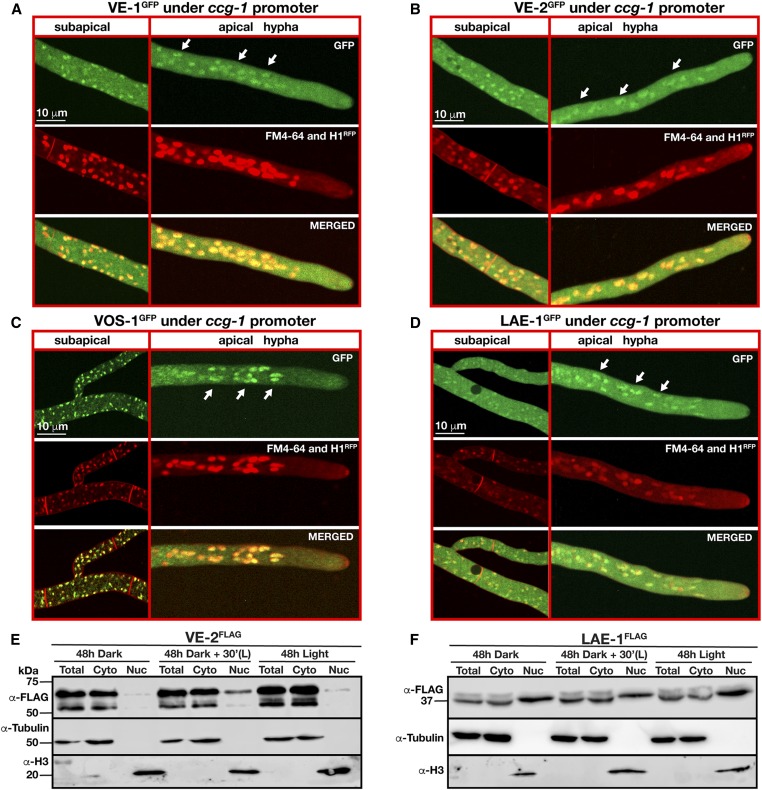
In order to identify the subcellular localization of the velvet proteins and of LAE-1 in N. crassa, GFP-fused versions of each protein were expressed under the control of the ccg-1 promoter at the his-3 locus. VE-1GFP displayed a nucleocytoplasmic localization in both apical and subapical N. crassa cells (Figure 5A). The VE-2GFP fusion was also found homogenously distributed in both nuclear and cytoplasmic fractions in apical and subapical cells. However, VE-2GFP showed a higher abundance in the cytoplasm (Figure 5, B and E). LAE-1GFP had a similar nucleocytoplasmic pattern of distribution (Figure 5, D and F). However, VOS-1GFP exhibited a more pronounced nuclear localization than the other two velvet proteins (Figure 5C), which is comparable to VosA localization in A. nidulans (Ni and Yu 2007). Since VE-2 and LAE-1 exhibited weak nuclear localization, we created strains with fusions of VE-2 or LAE-1 with a triple FLAG tag, and expressed the fused genes under their native promoters. Accumulation of the VE-2FLAG or LAE-1FLAG was examined in cytoplasmic and nuclear fractions of cultures grown in dark and light conditions (Figure 5, E and F). We detected both proteins in nuclear and cytoplasmic fractions, but VE-2FLAG was less abundant in the nucleus when compared to LAE-1FLAG. The nuclear subpopulation of LAE-1FLAG showed a higher molecular weight than cytoplasmic LAE-1FLAG, but the nature of the presumptive post-translational modification was not explored further. The exposure to light did not change the fractionation patterns of VE-2FLAG or LAE-1FLAG, confirming our localization experiments with GFP fusions of these proteins. Thus, the patterns of the velvet proteins in N. crassa showed that they are in both nuclei and cytoplasm, but their localization in the N. crassa nuclei suggests that these proteins might have important regulatory functions controlling gene expression.
Figure 5.
Expression and subcellular localization of N. crassa velvet orthologs. (A) Subcellular localizations of VE-1 GFP, (B) VE-2 GFP, (C) LAE-1 GFP, and (D) VOS-1GFP fusion proteins expressed under the control of the clock-controlled gene-1 (ccg-1) promoter at the his-3 locus of N. crassa. White arrows indicate the position of nuclei with GFP signals. Plasma membranes and septa were stained red by a membrane staining FM4-64 dye and nuclei were visualized using Histone 1 (1H)-RFP fusion. (E and F) Nuclear enrichment of VE-2 and LAE-1 triple FLAG fusion proteins expressed under the control of their native promoters extracted from cultures grown under light and dark conditions. Cultures were either grown in the dark for 48 hr, and then exposed to light for 30 min, or they were grown under continuous white light for 48 hr. Total, Cyto, and Nuc represent total, cytoplasmic, and nuclear extracts, respectively. Each lane was loaded with 70 µg protein. α-Tubulin and α-Histone 3 (H3) were used as loading controls for cytoplasmic and nuclear proteins, respectively.
Trimeric VE-1/VE-2/LAE-1 and dimeric VE-2/VOS-1 complexes coexist in N. crassa
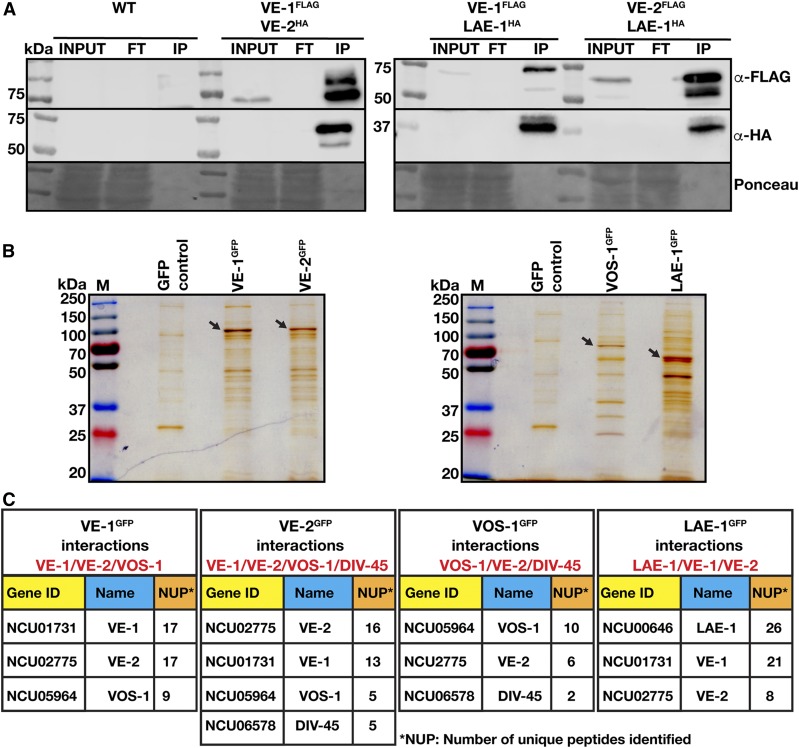
Next, we investigated whether the four N. crassa velvet proteins and LAE-1 interact to form regulatory protein complexes. Co-immunoprecipitation experiments (CoIPs) of VE-1, VE-2, and LAE-1 were performed using 3XFLAG- and HA-tagged fusion proteins expressed in VMM during vegetative growth under native conditions (Figure 6A). VE-1FLAG coprecipitated both VE-2HA and LAE-1HA. Furthermore, VE-2FLAG also coprecipitated LAE-1HA, consistent with formation of a heterotrimeric velvet complex in vivo, as shown in A. nidulans (Bayram et al. 2008a).
Figure 6.
In vivo interactions of the velvet protein complexes in N. crassa during vegetative growth (A) Co-immunoprecipitations (CoIPs) of the velvet complex using triple (3×) FLAG and HA-tagged versions of VE-1, VE-2, and LAE-1 expressed under their native promoters during vegetative growth. Triple HA-tagged VE-2 and LAE-1 were precipitated with VE-13xFLAG fusion. HA-tagged LAE-1 was also precipitated with VE-23XFLAG fusion. (B) Silver-stained SDS gels (10%) of the GFP-TRAP pulldowns of VE-1, VE-2, VOS-1, and LAE-1GFP fusion proteins from N. crassa grown for 24 hr at 30° in liquid Vogel’s MM medium. Arrows indicate the positions of precipitated full-length fusion proteins stained by Silver reagent. (C) Interaction partners of VE-1, VE-2, VOS-1, and LAE-1 after GFP-TRAP-MS in N. crassa during vegetative growth. Gene IDs are given as locus number (NCU). NUP; the number of unique peptides identified in MS. LC-MS identifications of interaction partners of respective proteins (see Files S5–S8).
To further support these results, and to identify additional protein interactions of the velvet proteins and of LAE-1, the GFP fusions of the velvet and LAE-1 proteins were used to identify by LC-MS their interacting proteins in vivo during vegetative growth (Figure 6, B and C). Protein extracts of the vegetative cultures were subjected to GFP-TRAP (pull-down). Silver-stained SDS-PAGE analysis of the GFP-Trap experiment confirmed the molecular sizes of the fusion proteins (Figure 6B). Copurifying proteins were identified by LC-MS, which suggested that VE-1 interacted with VE-2, but also with VOS-1, presumably via VE-2, which is similar to A. nidulans velvet complex (Bayram et al. 2008a) (Figure 6C and File S5). Interestingly, VE-2 reciprocally copurified VE-1 and also VOS-1. As a confirmation, VOS-1 reciprocally interacted with VE-2 but not with VE-1, indicating the presence of VE-2/VOS-1 heterodimers. Although LAE-1 was not found in purifications of VE-1 and VE-2, LAE-2GFP purifications led to identification of both VE-1 and VE-2, confirming the CoIP results (Figure 6C and Files S5–S8). In addition, we detected the interaction between the importin DIV-45—a homolog of the yeast karyopherin KAP114p involved in the nuclear import of specific proteins (Morehouse et al. 1999)—and VE-2 and VOS-1.
These results confirmed that the N. crassa velvet proteins form at least two distinct protein complexes, a heterotrimeric VE-1/VE-2/LAE-1 complex containing the putative LAE-1 methyltransferase (analogous to VeA/VelB/LaeA in A. nidulans), and a heterodimeric VE-2/VOS-1 (analogous to VelB/VosA) complex. The association of VE-2 and VOS-1 with DIV-45 suggest a role for this importin in the transport of the heterodimer and/or the velvet complex into the nucleus.
VE-1 controls expression of genes required for conidiation, secondary metabolite production, and carotenoid biosynthesis
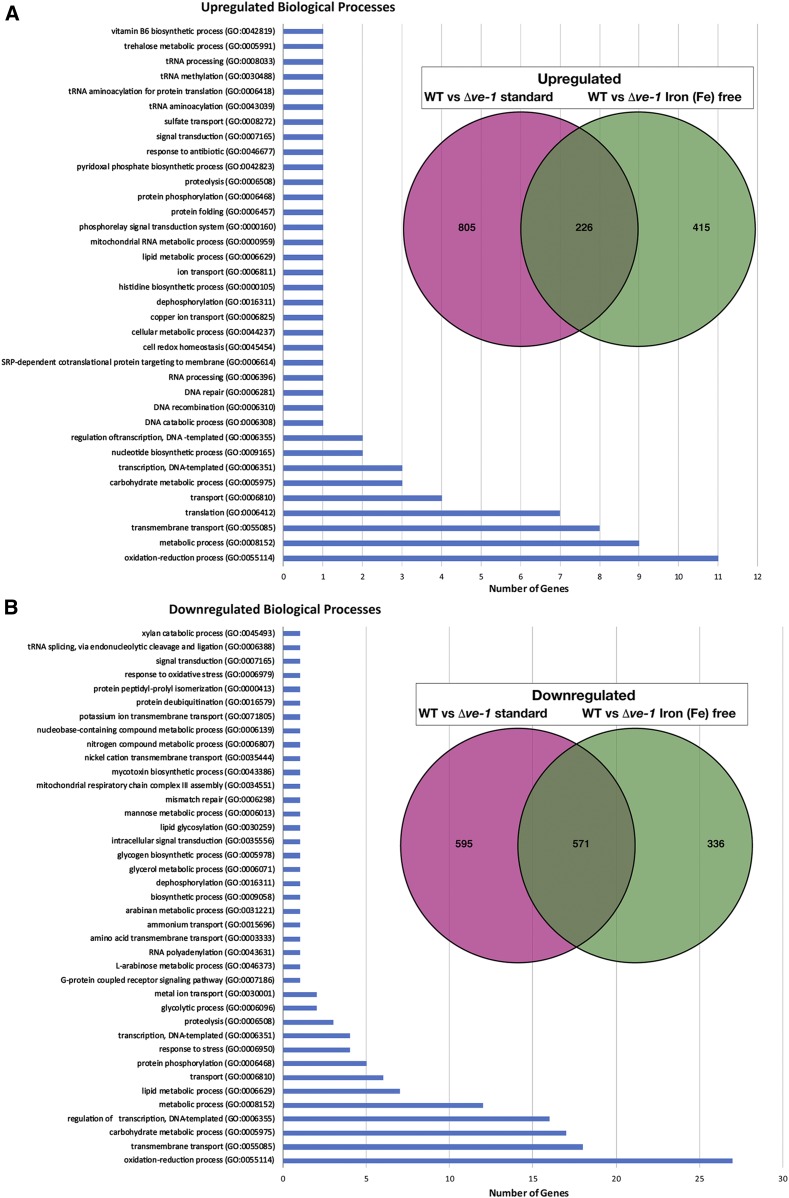
Our results suggest that VE-1 and VE-2 are the important members of the velvet protein family in N. crassa. They form a complex with LAE-1, and corresponding mutants show similar phenotypes in conidiation, sexual fruiting body formation, and SM production. In order to identify genes that are under their transcriptional control during different developmental and metabolic processes, we characterized the transcriptome of Δve-1 by RNA-seq experiments. We cultured wild-type and Δve-1 for 48 hr in the presence or absence of iron since the fungus activates SM gene expression in cultures older than 24 hr. The genes showing changes in mRNA accumulation at log2 > 1 were considered as upregulated or downregulated. A total of 2927 genes showed changes in mRNA accumulation under either of the two conditions (wild type vs. Δve-1 Vogel’s and Fe-free Vogel’s medium) (Figure 7 and Files S10–S14), which corresponds to ∼30% of N. crassa genes. Under standard growth conditions (with iron), expression of 1166 genes was downregulated and 1031 genes was upregulated (21% of genome), whereas, under Fe-free conditions, expression of 907 genes was downregulated and 641 genes was upregulated (15% of genome) in ve-1 compared to wild type.
Figure 7.
Control of gene expression by ve-1 gene. (A) Upregulated genes and biological processes in the ve-1 mutant grown under standard VMM and Fe-free conditions during 48 hr submerged culture. The Venn diagram represents the 805 upregulated genes (red) in the ve-1 mutant under standard conditions (iron containing) and 415 upregulated genes (green) in the ve-1 mutant specific to iron starvation. The overlapping 226 genes represent the genes upregulated under both conditions. Bar charts show the number of GO biological process terms upregulated (226 genes) in both conditions (see Files S10–S14). (B) Downregulated genes and biological processes in the ve-1 mutant under standard and Fe-free conditions during the 48 hr submerged culture; 595 and 336 genes were downregulated in the ve-1 mutant in standard and iron starvation conditions, respectively. Bar charts show the number of GO biological process terms downregulated (571 genes) in both conditions.
When we focused on the set of common genes that were upregulated (226) and downregulated (571) in the presence or absence of iron in Δve-1, we detected enrichment for the following biological processes: oxidation-reduction (11 genes up; 27 genes down), metabolic (9 up), transmembrane (8 up; 18 down), translation (7), transport (4), carbohydrate metabolism (3 up; 17 down), transcription (3 up; 16 down), nucleotide biosynthesis (2 up; 12 down) and other (27 up) were among the upregulated genes under both standard and iron-free conditions in Δve-1.
The A. nidulans genome encodes more than 10 LaeA-like methyltransferases (llmA to llmJ), which either control development or SM production (Palmer et al. 2013). Three methyltransferases LlmF and VipC-VapB heterodimers physically interfere with nuclear accumulation of VeA and therefore control coordination of development with SM production (Palmer et al. 2013; Sarikaya-Bayram et al. 2014). Two N. crassa orthologs of the A. nidulans LaeA-like methyltransferase, encoding NCU10101(llmG) and NCU10761 (llmA), were upregulated strongly under iron starvation conditions in Δve-1 (Table 1). In addition to these two methyltransferases, two sugar transporters, a trehalase enzyme, cytochrome c and an endonuclease were among the top 10 highly expressed genes (see also the top 20 list in File S14). Under standard growth conditions, mainly oxidation-reduction enzymes (luciferase-like monooxygenase) were at the top of the upregulated list, along with major facilitator superfamily genes (Table 2).
Table 1. Top 10 upregulated genes under iron starvation in ve-1 mutant.
| Gene ID | Log2Fold change | Biological process | Molecular function | PFAM domain(s) |
|---|---|---|---|---|
| NCU10101 (llmG) | 8.577 | — | — | Methyltransferase domain |
| NCU04537 | 7.338 | Transmembrane transport | Transmembrane transporter activity | Sugar (and other) transporter |
| NCU10761 (llmA) | 5.332 | — | — | Methyltransferase domain |
| NCU03016 | 4.990 | — | — | — |
| NCU00292 | 4.669 | — | — | Carboxylesterase family |
| NCU00943 | 4.617 | Trehalose metabolic process | Alpha, alpha-trehalase activity | Trehalase |
| NCU08648 | 4.595 | DNA catabolic process | Endonuclease activity nucleic acid binding | S1/P1 Nuclease |
| NCU09685 | 4.133 | — | — | Domain of unknown function (DUF1772) |
| NCU01808 | 4.047 | — | Heme binding electron transfer activity | Cytochrome c |
| NCU05627 | 4.0174 | Transmembrane transport | Transmembrane transporter activity | Sugar (and other) transporter |
Table 2. Top 10 upregulated genes under standard conditions in ve-1 mutant.
| Gene ID | Log2Fold change | Biological process | Molecular function | PFAM domain(s) |
|---|---|---|---|---|
| NCU10016 | 10.056 | Oxidation-reduction | FAD binding | Acyl CoA dehydrogenase |
| NCU07819 | 9.653 | Oxidation-reduction | Oxidoreductase activity | Taurine catabolism |
| NCU05883 | 9.128 | Oxidation-reduction | Oxidoreductase activity | Luciferase-like monooxygenase |
| NCU05888 | 9.016 | Oxidation-reduction | Oxidoreductase activity | Luciferase-li monooxygenase |
| NCU07610 | 8.904 | Oxidation-reduction | — | Taurine catabolism |
| NCU07820 | 8.608 | Transmembrane transport | — | Major facilitator superfamily |
| NCU05887 | 8.266 | — | — | — |
| NCU01095 | 7.972 | Transmembrane transport | — | Major facilitator superfamily |
| NCU09678 | 7.586 | Transmembrane transport | — | Major facilitator superfamily |
| NCU05884 | 7.488 | Transmembrane transport | — | Major facilitator superfamily |
Oxidation-reduction, proteolysis, bacterial cell-wall-degrading enzyme domain encoding (LysM domain) genes were the most drastically downregulated genes under iron starvation conditions in the Δve-1 mutant (Table 3). Interestingly, vos-1 was highly, and ve-2 was moderately, downregulated under normal conditions in the Δve-1 mutant, suggesting that VE-1 is required for proper expression of vos-1 and ve-2 (Table 4). In addition to the llmG and llmA homologs, which were upregulated in the Δve-1 mutant, the genes for many other methyltransferases, including NCU05841(llmB (vipC), two llmB-like methyltransferase NCU05501 and NCU05832, an O-methyltransferase domain encoding gene NCU05855, and two Lae-like genes NCU00304 and NCU00451, were downregulated under both standard and iron starvation conditions (File S14).
Table 3. Top 10 downregulated genes under iron starvation conditions in ve-1 mutant.
| Gene ID | Log2Fold change | Biological process | Molecular function | PFAM domain(s) |
|---|---|---|---|---|
| NCU05126 | −9.913 | — | Transferase activity | UbiA prenyltransferase |
| NCU00732 | −9.709 | Oxidation-reduction | Heme binding iron ion binding | Cytochrome P450 |
| NCU10865 | −8.695 | Metabolic process | Oxidoreductase activity | Central domain Tyrosinase |
| NCU04205 | −8.256 | Proteolysis | Aspartic type endopeptidase | Peptidase A4 family |
| NCU07033 | −8.160 | — | — | LysM domain |
| NCU04931 | −8.066 | — | — | — |
| NCU07034 | −7.793 | — | — | LysM domain |
| NCU08223 | −7.716 | — | — | — |
| NCU02919 | −7.522 | — | — | Cupin domain |
| NCU02369 | −7.321 | Carbohydrate metabolic process | Hydrolase activity | Glycosyl hydrolases Family 16 |
Table 4. Top 10 downregulated genes under standard conditions in ve-1 mutant.
| Gene ID | Log2Fold change | Biological process | Molecular function | PFAM domain(s) |
|---|---|---|---|---|
| NCU05126 | −9.789 | — | Transferase activity | UbiA prenyltransferase |
| NCU05567 | −9.636 | — | — | — |
| NCU09775 | −7.350 | Arabinan metabolic process | Alpha l arabinofuranosidase | Alpha l arabinofuranosidase B |
| NCU06912 | −7.305 | — | — | — |
| NCU00175 | −7.200 | — | — | — |
| NCU05964 vos-1 | −7.090 | — | — | Velvet domain protein |
| NCU06327 | −6.970 | Oxidation reduction | Heme binding, iron ion binding | Cytochrome P450 |
| NCU09702 | −6.845 | — | — | O-Glycosyl hydrolase family |
| NCU01092 | −6.830 | — | — | Enoyl (acyl carrier protein) reductase |
| NCU04482 | −6.734 | — | — | Putative necrosis inducing factor |
The Δve-1 mutant shows increased conidiation and reduced carotenoid production. In agreement with these phenotypes, conidiation genes con-6, and con-10 were moderately upregulated even in submerged media, which precludes conidiation of N. crassa. The carotenoid biosynthetic gene al-2 (phytoene synthase) as well as cao-1 (carotenoid oxygenase) were moderately downregulated under standard conditions in the ve-1 mutant. Expression of the of N. crassa clock components (wc-1, wc-2, frq, vvd) was unaltered in the absence of VE-1 (File S10).
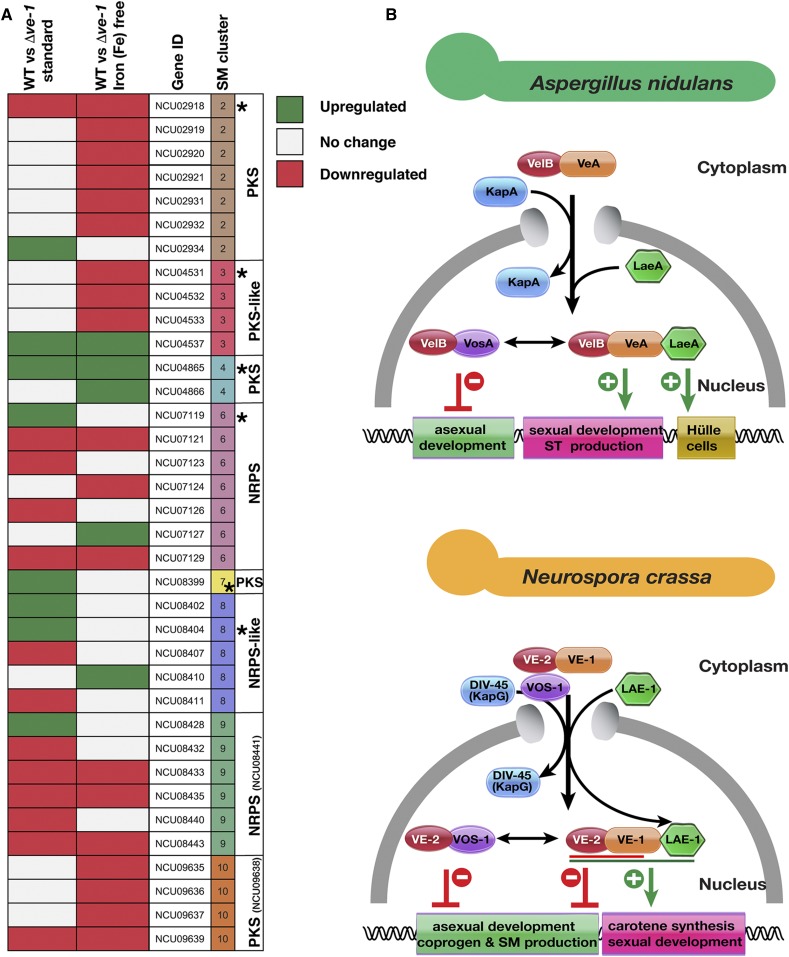
There are 10 predicted SM gene clusters with 94 genes in the N. crassa genome. Of these 94 genes, 36 showed changes in the accumulation of the corresponding mRNAs (either up or downregulation) in either of two conditions (Figure 8A). Expression of SM genes were mainly downregulated in Δve-1 mutant irrespective of iron starvation. Clusters 2, 9, and 10 were mostly downregulated, whereas clusters 4 and 8 were upregulated. Expression of the backbone enzymes of cluster 4 (PKS-like), 6 (NRPS), 7 (PKS) were particularly increased. Cluster 6 is responsible for production of coprogen with backbone enzyme NCU07119. Two genes belonging to coprogen production were upregulated in Δve-1 strain after 48 hr of growth.
Figure 8.
Expression of secondary metabolite genes and comparative model of the velvet complexes in A. nidulans and N. crassa. (A) Expression of eight putative secondary metabolite gene clusters of Neurospora in liquid 48 hr cultures under standard or iron starvation conditions in an Δve-1 strain in comparison to WT. PKS: Polyketide synthase, NRPS: Nonribosomal peptide synthase. SM cluster numbers, and the “backbone” genes (with NCU numbers indicated) in the clusters are identified. Backbone genes were indicated with an asterisk. Expression of some backbone genes did not change (those without the asterisks) (see Table S13). (B) Velvet complex has been studied mostly in A. nidulans (upper panel). The VelB-VeA heterodimer, formed by the two velvet family proteins (represented by red and orange spherical shapes), enters into the nucleus with help of importin α (KapA, Blue). In the nucleus VelB-VeA form a heterodimer with the methyltransferase LaeA. This complex has several functions; the VeA-VelB part of the complex is mainly responsible for sexual development and SM production (shown as green lines). In addition, LaeA has a particular function on formation of protective hülle cells required for protection and nursing of growing fruit bodies. In addition to its role in the velvet complex, VelB is also a part of heterodimeric complex formed with VosA, which activates spore viability, trehalose accumulation (green lines) and also represses asexual conidiation (red lines). In N. crassa, the situation is similar. The VE-1/VE-2 or VE-2/VOS-1 heterodimer enters into nucleus, presumably with help of importin DIV-45 (KAP114p ortholog) since DIV-45 was found in purifications of both VE-2 and VOS-1. The VE-2/VE-1 heterodimer is a major player to repress asexual conidiation and SM production including coprogen (red line) and to activate carotene biosynthesis (green line). The VE-1/VE-2/LAE-1 complex is required for maximum protoperithecia formation (sexual development). The VE-2/VOS-1 heterodimer presumably represses conidiation and hyphal growth.
These results reveal that VE-1 regulates >20% of the genome under different growth conditions. Furthermore, a great proportion of small methyltransferases, velvet genes, conidiation and carotenoid biosynthetic genes and SM genes require VE-1 for proper expression.
Discussion
Eukaryotes have developed different strategies to coordinate development and cellular physiology in response to environmental conditions. The velvet complex is one of the regulatory protein complexes that fungi use for the coordination of growth, development, and SM production. In this study, we have examined the cross-genus functions of the velvet complex by comparing its function in two model fungi from two different genera at the biochemical, cellular, and genetical levels.
The two N. crassa velvet genes ve-1 and ve-2 successfully complemented the deficiencies of the A. nidulans veA and velB mutants, indicating that VE-1 and VE-2 are functional orthologs of VeA and VelB. Intriguingly, VE-1 and VE-2 are functionally incorporated and form heterotrimeric chimeric complexes with either A. nidulans VelB/LaeA or VeA/LaeA, respectively. However, neither N. crassa VOS-1 or LAE-1 could complement the functions of vosA and laeA in A. nidulans. This might be due to the abolished/reduced ability of VOS-1 and LAE-1 to integrate into the A. nidulans complex. A residual interaction of N. crassa LAE-1 was observed with A. nidulans VeA but not VelB, while we did not observe complexes formed with VOS-1 in the vegetative growth stage. We did not test for interaction partners of VOS-1 in A. nidulans during asexual sporulation. Therefore, it is still possible that N. crassa VOS-1 might recruit A. nidulans VelB during asexual development. Nevertheless, N. crassa VOS-1 and LAE-1 were expressed and localized correctly within the A. nidulans nuclei. It is intriguing that N. crassa LAE-1 could interact with VeA in A. nidulans, but did not complement the functions of LaeA, presumably because the LAE-1/VeA heterodimer was not able to recruit VelB. In A. nidulans, VeA acts as a bridge between VelB and LaeA. Furthermore, in the absence of LaeA, VeA recruits a high level of VelB (Sarikaya Bayram et al. 2010). Presumably, the interaction of LAE-1 with VeA interferes with the VeA/VelB interaction in A. nidulans, consistent with the possibility that LAE-1 binds and masks the domain on VeA where VelB binds or changes the conformation of VeA, preventing VelB binding.
Our results allowed us to compare the current model for the mechanism of action of the velvet complexes in A. nidulans and N. crassa (Figure 8B). The A. nidulans velvet complex is formed in the nucleus after the entry of VeA/VelB heterodimers via the importin KapA. In N. crassa, however, another importin, DIV-45, which is an ortholog of the yeast karyopherine KAP114p, copurified with both VE-2 and VOS-1 and thus may promote nuclear translocation of the velvet proteins. Since VE-2 interacts with VE-1, this heterodimer is likely to use the DIV-45 importin for nuclear import as well. In A. nidulans, the VelB/VeA heterodimer recruits the LaeA methyltransferase upon nuclear entry, and the final heterotrimeric velvet complex participates in the regulation of sexual development, ST production, and hülle cell formation to protect growing fruiting bodies. VelB, in addition, forms a heterodimer with VosA that is essential for final spore maturation, spore viability, and trehalose accumulation.
In N. crassa, VE-1, VE-2, and LAE-1 form the heterotrimeric velvet complex. However, the VE-1/VE-2 heterodimer has more prominent roles in development than LAE-1, which is only required for full production of protoperithecia. Both VE-1 and VE-2 are equally important for the repression of conidiation and for the activation of carotenoid biosynthesis by light. This phenotypic observation was further confirmed by our transcriptomic results that showed the downregulation of two carotenoid biosynthetic genes. VE-2 forms a heterodimer with VOS-1, which presumably represses asexual conidiation. The slightly reduced conidiation in the Δvos-1 mutant and the minor alterations in SM and carotenoid accumulation suggest a minor role for VOS-1 in the regulation or activities of the velvet complexes.
Few cross-species functional studies with the velvet complex proteins have been reported. In the human pathogenic fungus A. fumigatus, ectopic introduction of the A. nidulans VeA-TAP fusion resulted in formation of the functional AnVeA–AfVelB–AfLaeA velvet complex (Park et al. 2012a). veA is required for production of gliotoxin (GT) and nitrogen source dependent sporulation in A. fumigatus (Krappmann et al. 2005; Dhingra et al. 2012). In an intergenera complementation experiment, the VeA ortholog of the dimorphic fungus Histoplasma capsulatum, Vea1, was able to restore cleistothecia formation, and partially complemented ST production in A. nidulans (Laskowski-Peak et al. 2012). Silencing of VEA1 facilitated the dimorphic switch from the yeast phase to the mycelial phase in H. capsulatum, and virulence of this strain is attenuated in murine and macrophage models.
The VosA/VelB heterodimer of A. nidulans has a DNA-binding domain that is similar to that of the Rel family of regulatory proteins in the mammalian immune system, and binds to DNA to regulate the transcription of several genes, including sporulation and trehalose genes (Ahmed et al. 2013). VeA was shown to bind to its own promoter (Rauscher et al. 2016). It is likely that VeA acts as a DNA binding domain protein since the velvet domain seems to be important for DNA binding (Sarikaya-Bayram et al. 2015). The H. capsulatum velvet family proteins Ryp2 (VelB) and Ryp3 (VosA) are important for the temperature-dependent dimorphic switch and also bind to a large set of H. capsulatum genes important for pathogenicity (Beyhan et al. 2013). Not only in H. capsulatum, but also in A. fumigatus, the veA and laeA members of the velvet complex control temperature-dependent processes like SM production (Lind et al. 2016), underlining that the velvet complex uses not only light but also temperature to regulate developmental and metabolic processes in at least two pathogenic fungi.
It is highly likely that the heterodimeric VE-1/VE-2 complex of N. crassa binds DNA, given the drastic changes in gene expression that we observed after deleting ve-1, since the velvet domains were shown to bind to DNA in several other fungi (Ahmed et al. 2013; Beyhan et al. 2013; Becker et al. 2016; Rauscher et al. 2016). A 21% change in the transcriptome was observed under standard growth conditions, and 15% under iron starvation conditions when the Δve-1 and the wild-type strains were compared (Figure 7). In contrast, loss of A. fumigatus veA changes expression of 10% of the genome including more than one-half of the genes in SM gene clusters (Lind et al. 2016). Interestingly, more than one-third of the SM gene clusters whose expression is differentially regulated (14 out of 37) are downregulated in the absence of veA in A. fumigatus. In the plant pathogenic fungus, A. flavus, expression of 5% of all genes were differentially expressed in a veA mutant (Cary et al. 2015). Out of 56 gene clusters, 28 contain at least one gene that is either up- or downregulated in veAΔ strain in A. flavus. Similarly, in N. crassa, expression of almost one-third of SM genes (36 out of 94) is either up- or downregulated in either normal or iron starvation media. Coprogen production increases threefold when ve-1 is missing. However, this metabolic change was not reflected in gene expression, because only two genes belonging to the coprogen cluster (cluster 6) were upregulated. This discrepancy between gene expression and coprogen production may be due to the different time points used for the assay of SM accumulation (7 days) and RNA extraction (48 hr) since it is extremely difficult to extract intact RNA from 7-day-old cultures in comparison to SMs, which can be easily extracted from old cultures.
In summary, our results suggest a strong conservation in the activities and interactions of the velvet family in the fungal kingdom given the biological activity of chimeric versions of the velvet complexes in A. nidulans. In N. crassa, however, our results show how the velvet complex has further evolved and specialized compared to the A. nidulans counterpart since the VE-1/VE-2 heterodimer plays a major regulatory role when compared to the trimeric VE-1/VE-2/LAE-1 complex. The key role of VE-1 in N. crassa biology, as shown by the developmental and metabolic phenotypes of the Δve-1 mutant, further indicates specialization when compared with the phenotype of the veA mutant in A. nidulans and other Aspergilli, and suggests that in-depth understanding of the role of the velvet complex in fungal biology will require detailed characterization in selected model fungi like N. crassa. It will be interesting to know whether, or where, the velvet proteins, VE-1, VE-2, and VOS-1 bind to in N. crassa genome. Molecular activities of A. nidulans LaeA are currently unknown except for automethylation activity. It will be also intriguing to learn whether N. crassa LAE-1 has similar or different biochemical activities than A. nidulans LaeA.
Fungi are prominent in human activities as they are sources of chemicals used as pharmaceuticals and drugs, and include pathogens of plants and animal, including humans. The key role of the velvet proteins in fungal development and SM production suggests that a full understanding of their complex dynamics in fungal biology and metabolism will improve our capacity to use fungi for food and drug production and may mitigate their detrimental effects on humans and plant hosts.
Acknowledgments
We thank Caroline Batchelor for LC-MS of the protein samples and Dean Frawley for proofreading the manuscript. This publication has emanated from research supported by a research grant from Science Foundation Ireland (SFI) under Grant Number 13/CDA/2142 to Özgür Bayram, 12/IP/1695 to Sean Doyle, the grants SE1054/6-2, SE1054/7-2 and SE1054/9-1 of the Deutsche Forschungsgemeinschaft to Stephan Seiler, National Institutes of Health (NIH) grants GM093061 and GM127142 to Eric Selker, Spanish Ministry of Science, Innovation and Universities and European funds (European Regional Development Fund, ERDF) (BIO2015-67148-R) to Luis M. Corrochano and David Cánovas. Özlem Sarikaya Bayram is supported by the Irish Research Council (IRC) Postdoctoral Fellowship (GOIPD/2014/178). Jamie McGowan is supported by an IRC Postgraduate Fellowship (GOIPG/2016/1112). Guilherme T. P. Brancini was supported by a Bolsa Estágio de Pesquisa no Exterior (BEPE) short-term fellowship from São Paulo Research Foundation (FAPESP) (2018/00355-7), Brazil. The mass spectrometry (MS) facility in Maynooth University was funded by SFI [12/RI/2346(3)] to S.D.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8091182.
Communicating editor: A. Mitchell
Literature Cited
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., et al. , 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44: W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Y. L., Gerke J., Park H. S., Bayram O., Neumann P., et al. , 2013. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-kappaB. PLoS Biol. 11: e1001750 [corrigenda: PLoS Biol. 12: e1001849 (2014)]. 10.1371/journal.pbio.1001750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Selker E. U., 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 5: a017921 10.1101/cshperspect.a017921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Loros J. J., Dunlap J. C., 2012. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 36: 95–110. 10.1111/j.1574-6976.2011.00288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J. A., Giles N. H., 1985. Genetic control of chromatin structure 5′ to the qa-x and qa-2 genes of Neurospora. J. Mol. Biol. 182: 79–89. 10.1016/0022-2836(85)90029-4 [DOI] [PubMed] [Google Scholar]
- Bayram O., Braus G. H., 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36: 1–24. 10.1111/j.1574-6976.2011.00285.x [DOI] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J., Helmstaedt K., et al. , 2008a. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506. 10.1126/science.1155888 [DOI] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Seiler S., Vogt N., Braus G. H., 2008b. Neurospora crassa ve-1 affects asexual conidiation. Fungal Genet. Biol. 45: 127–138. 10.1016/j.fgb.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Bayram O., Sari F., Braus G. H., Irniger S., 2009. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 71: 1278–1295. 10.1111/j.1365-2958.2009.06606.x [DOI] [PubMed] [Google Scholar]
- Bayram O., Bayram O. S., Ahmed Y. L., Maruyama J., Valerius O., et al. , 2012. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 8: e1002816 10.1371/journal.pgen.1002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Ziemons S., Lentz K., Freitag M., Kuck U., 2016. Genome-wide chromatin immunoprecipitation sequencing analysis of the Penicillium chrysogenum velvet protein PcVelA identifies methyltransferase PcLlmA as a novel downstream regulator of fungal development. MSphere 1: e00149-16 10.1128/mSphere.00149-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello M. H., Barrera-Perez V., Morin D., Epstein L., 2012. The Neurospora crassa mutant Nc Delta Egt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 49: 160–172. 10.1016/j.fgb.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Bello M. H., Mogannam J. C., Morin D., Epstein L., 2014. Endogenous ergothioneine is required for wild type levels of conidiogenesis and conidial survival but does not protect against 254 nm UV-induced mutagenesis or kill. Fungal Genet. Biol. 73: 120–127. 10.1016/j.fgb.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Beyhan S., Gutierrez M., Voorhies M., Sil A., 2013. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 11: e1001614 10.1371/journal.pbio.1001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J. W., Keller N. P., 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3: 527–535. 10.1128/EC.3.2.527-535.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., et al. , 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108. 10.1128/MMBR.68.1.1-108.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M., 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brakhage A. A., 2013. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11: 21–32. 10.1038/nrmicro2916 [DOI] [PubMed] [Google Scholar]
- Calvo A. M., 2008. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45: 1053–1061. 10.1016/j.fgb.2008.03.014 [DOI] [PubMed] [Google Scholar]
- Cary J. W., Han Z., Yin Y., Lohmar J. M., Shantappa S., et al. , 2015. Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 14: 983–997. 10.1128/EC.00092-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo M., Luque E. M., Pardo-Medina J., Limon M. C., Corrochano L. M., et al. , 2018. Transcriptional basis of enhanced photoinduction of carotenoid biosynthesis at low temperature in the fungus Neurospora crassa. Res. Microbiol. 169: 78–89. 10.1016/j.resmic.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Dettmann A., Illgen J., Marz S., Schurg T., Fleissner A., et al. , 2012. The NDR kinase scaffold HYM1/MO25 is essential for MAK2 map kinase signaling in Neurospora crassa. PLoS Genet. 8: e1002950 10.1371/journal.pgen.1002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra S., Andes D., Calvo A. M., 2012. VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 11: 1531–1543. 10.1128/EC.00222-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J. C., Loros J. J., 2016. Yes, circadian rhythms actually do affect almost everything. Cell Res. 26: 759–760. 10.1038/cr.2016.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P. S., O’Gorman C. M., 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36: 165–192. 10.1111/j.1574-6976.2011.00308.x [DOI] [PubMed] [Google Scholar]
- Etxebeste O., Garzia A., Espeso E. A., Ugalde U., 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18: 569–576. 10.1016/j.tim.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Fischer R., Aguirre J., Herrera-Estrella A., Corrochano L. M., 2016. The complexity of fungal vision. Microbiol. Spectr. 4 10.1128/microbiolspec.FUNK-0020-2016 [DOI] [PubMed] [Google Scholar]
- Fleissner A., Simonin A. R., Glass N. L., 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475: 21–38. 10.1007/978-1-59745-250-2_2 [DOI] [PubMed] [Google Scholar]
- Frawley D., Karahoda B., Sarikaya Bayram O., Bayram O., 2018. The HamE scaffold positively regulates MpkB phosphorylation to promote development and secondary metabolism in Aspergillus nidulans. Sci. Rep. 8: 16588 10.1038/s41598-018-34895-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A. C., Liu Y., Loros J. J., Dunlap J. C., 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819. 10.1126/science.1073681 [DOI] [PubMed] [Google Scholar]
- Fuller K. K., Cramer R. A., Zegans M. E., Dunlap J. C., Loros J. J., 2016. Aspergillus fumigatus photobiology illuminates the marked heterogeneity between isolates. MBio 7: e01517-16 10.1128/mBio.01517-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funa N., Awakawa T., Horinouchi S., 2007. Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 282: 14476–14481. 10.1074/jbc.M701239200 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., et al. , 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. 10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115. 10.1038/nature04341 [DOI] [PubMed] [Google Scholar]
- Haas H., 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62: 316–330. 10.1007/s00253-003-1335-2 [DOI] [PubMed] [Google Scholar]
- Haas H., 2014. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 31: 1266–1276. 10.1039/C4NP00071D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Eisendle M., Turgeon B. G., 2008. Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46: 149–187. 10.1146/annurev.phyto.45.062806.094338 [DOI] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J., Alkema W., 2008. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9: 488 10.1186/1471-2164-9-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J., Loros J. J., Dunlap J. C., 2015. Dissecting the mechanisms of the clock in Neurospora. Methods Enzymol. 551: 29–52. 10.1016/bs.mie.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huschka H., Naegeli H. U., Leuenberger-Ryf H., Keller-Schierlein W., Winkelmann G., 1985. Evidence for a common siderophore transport system but different siderophore receptors in Neurospora crassa. J. Bacteriol. 162: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H. Y., Fraser M., Li W., et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P., Turner G., Bennett J. W., 2005. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 3: 937–947. 10.1038/nrmicro1286 [DOI] [PubMed] [Google Scholar]
- Kjærbølling I., Vesth T. C., Frisvad J. C., Nybo J. L., Theobald S., et al. , 2018. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. USA 115: E753–E761. 10.1073/pnas.1715954115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko A. D., Ormsby T., Galazka J. M., Leggett N. A., Uesaka M., et al. , 2016. Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa. Proc. Natl. Acad. Sci. USA 113: 15048–15053. 10.1073/pnas.1615546113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl C., Schrettl M., Abt B., Sarg B., Lindner H. H., et al. , 2007. EstB-mediated hydrolysis of the siderophore triacetylfusarinine C optimizes iron uptake of Aspergillus fumigatus. Eukaryot. Cell 6: 1278–1285. 10.1128/EC.00066-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S., Bayram O., Braus G. H., 2005. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot. Cell 4: 1298–1307. 10.1128/EC.4.7.1298-1307.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S. B., 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34: 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Laskowski-Peak M. C., Calvo A. M., Rohrssen J., Smulian A. G., 2012. VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum. Fungal Genet. Biol. 49: 838–846. 10.1016/j.fgb.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Lind A. L., Smith T. D., Saterlee T., Calvo A. M., Rokas A., 2016. Regulation of secondary metabolism by the velvet complex is temperature-responsive in Aspergillus. G3 (Bethesda) 6: 4023–4033. 10.1534/g3.116.033084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K., 2003. The fungal genetics stock center: from molds to molecules. Adv. Appl. Microbiol. 52: 245–262. 10.1016/S0065-2164(03)01010-4 [DOI] [PubMed] [Google Scholar]
- Morehouse H., Buratowski R. M., Silver P. A., Buratowski S., 1999. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA 96: 12542–12547. 10.1073/pnas.96.22.12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Yu J. H., 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2: e970 10.1371/journal.pone.0000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda-López M., Chen W., Eagle C. E., Gutiérrez G., Jia L., et al. , 2018. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 91: 7–59. 10.1016/j.simyco.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M., Ruger-Herreros C., Luque E. M., Corrochano L. M., 2010. A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet. Biol. 47: 352–363. 10.1016/j.fgb.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Palmer J. M., Theisen J. M., Duran R. M., Grayburn W. S., Calvo A. M., et al. , 2013. Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 9: e1003193 10.1371/journal.pgen.1003193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., Seymour F. A., Alcocer M. J., Kaur N., Calvo A. M., et al. , 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17: 1384–1389. 10.1016/j.cub.2007.07.012 [DOI] [PubMed] [Google Scholar]
- Park H. S., Yu J. H., 2012. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15: 669–677. 10.1016/j.mib.2012.09.006 [DOI] [PubMed] [Google Scholar]
- Park H. S., Bayram O., Braus G. H., Kim S. C., Yu J. H., 2012a. Characterization of the velvet regulators in Aspergillus fumigatus. Mol. Microbiol. 86: 937–953. 10.1111/mmi.12032 [DOI] [PubMed] [Google Scholar]
- Park H. S., Ni M., Jeong K. C., Kim Y. H., Yu J. H., 2012b. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One 7: e45935 10.1371/journal.pone.0045935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. S., Nam T. Y., Han K. H., Kim S. C., Yu J. H., 2014. VelC positively controls sexual development in Aspergillus nidulans. PLoS One 9: e89883 10.1371/journal.pone.0089883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patananan A. N., Palmer J. M., Garvey G. S., Keller N. P., Clarke S. G., 2013. A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J. Biol. Chem. 288: 14032–14045. 10.1074/jbc.M113.465765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pócsi I., Jeney V., Kertai P., Pocsi I., Emri T., et al. , 2008. Fungal siderophores function as protective agents of LDL oxidation and are promising anti-atherosclerotic metabolites in functional food. Mol. Nutr. Food Res. 52: 1434–1447. 10.1002/mnfr.200700467 [DOI] [PubMed] [Google Scholar]
- Pöggeler S., Nowrousian M., Kück U., 2006. Fruiting-body development in ascomycetes, pp. 325–355 in The Mycota I Growth, Differentiation and Sexuality, edited by Fischer K. Springer-Verlag, Heidelberg: 10.1007/3-540-28135-5_16 [DOI] [Google Scholar]
- Purschwitz J., Muller S., Kastner C., Schoser M., Haas H., et al. , 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18: 255–259. 10.1016/j.cub.2008.01.061 [DOI] [PubMed] [Google Scholar]
- Rauscher S., Pacher S., Hedtke M., Kniemeyer O., Fischer R., 2016. A phosphorylation code of the Aspergillus nidulans global regulator VelvetA (VeA) determines specific functions. Mol. Microbiol. 99: 909–924. 10.1111/mmi.13275 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Romero J., Hedtke M., Kastner C., M¸ller S., Fischer R., 2010. Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol. 64: 585–610. 10.1146/annurev.micro.112408.134000 [DOI] [PubMed] [Google Scholar]
- Röhrig J., Yu Z., Chae K. S., Kim J. H., Han K. H., et al. , 2017. The Aspergillus nidulans Velvet-interacting protein, VipA, is involved in light-stimulated heme biosynthesis. Mol. Microbiol. 105: 825–838. 10.1111/mmi.13739 [DOI] [PubMed] [Google Scholar]
- Rountree M. R., Selker E. U., 2010. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity 105: 38–44. 10.1038/hdy.2010.44 [DOI] [PubMed] [Google Scholar]
- Sarikaya Bayram O., Bayram O., Valerius O., Park H. S., Irniger S., et al. , 2010. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6: e1001226 10.1371/journal.pgen.1001226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya-Bayram O., Bayram O., Feussner K., Kim J. H., Kim H. S., et al. , 2014. Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev. Cell 29: 406–420. 10.1016/j.devcel.2014.03.020 [DOI] [PubMed] [Google Scholar]
- Sarikaya-Bayram O., Palmer J. M., Keller N., Braus G. H., Bayram O., 2015. One Juliet and four Romeos: VeA and its methyltransferases. Front. Microbiol. 6: 1 10.3389/fmicb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C., et al. , 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70: 27–43. 10.1111/j.1365-2958.2008.06376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdtfeger C., Linden H., 2000. Localization and light-dependent phosphorylation of white collar 1 and 2, the two central components of blue light signaling in Neurospora crassa. Eur. J. Biochem. 267: 414–422. 10.1046/j.1432-1327.2000.01016.x [DOI] [PubMed] [Google Scholar]
- Springer M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. BioEssays 15: 365–374. 10.1002/bies.950150602 [DOI] [PubMed] [Google Scholar]
- Stinnett S. M., Espeso E. A., Cobeno L., Araujo-Bazan L., Calvo A. M., 2007. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 63: 242–255. 10.1111/j.1365-2958.2006.05506.x [DOI] [PubMed] [Google Scholar]
- Tóth V., Antal K., Gyemant G., Miskei M., Pocsi I., et al. , 2009. Optimization of coprogen production in Neurospora crassa. Acta Biol. Hung. 60: 321–328. 10.1556/ABiol.60.2009.3.9 [DOI] [PubMed] [Google Scholar]
- Yu J. H., Keller N., 2005. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43: 437–458. 10.1146/annurev.phyto.43.040204.140214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. RNA-seq data were deposited in the Gene Expression Omnibus public database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE123783. Supplemental files are available at FigShare. Table S1 Fungal (Aspergillus nidulans and Neurospora crassa) strains used in this study. Table S2 Oligonucleotides used in this study. Table S3 Plasmids used and constructed in this study. File S1 Interaction partners of VE-1GFP fusion in veA∆ strain of A. nidulans. File S2 Interaction partners of VE-2 GFP fusion in velB∆ strain of A. nidulans. File S3 Interaction partners of VOS-1GFP fusion in vosA∆ strain of A. nidulans. File S4 Interaction partners of LAE-1GFP fusion in laeA∆ strain of A. nidulans. File S5 Interaction partners of VE-1GFP fusion in N. crassa. File S6 Interaction partners of VE-2GFP fusion in N. crassa. File S7 Interaction partners of VOS-1GFP fusion in N. crassa. File S8 Interaction partners of LAE-1GFP fusion in N. crassa. File S9 List of predicted secondary metabolite gene clusters in N. crassa. File S10 List of up- and downregulated genes in ve-1 mutant under standard and iron starvation conditions. File S11 List of GO terms for upregulated genes in ve-1 mutant in both conditions. File S12 List of GO terms for downregulated genes in ve-1 mutant in both conditions. File S13 List of differentially expressed putative secondary metabolite genes in ve-1 mutant in both conditions. File S14 Top 20 up and downregulated genes under iron starvation and standard conditions in ve-1 mutant. File S15 Supplemental materials and methods. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8091182.