Abstract
Suppressed recombination allows divergence between homologous sex chromosomes and the functionality of their genes. Here, we reveal patterns of the earliest stages of sex-chromosome evolution in the diploid dioecious herb Mercurialis annua on the basis of cytological analysis, de novo genome assembly and annotation, genetic mapping, exome resequencing of natural populations, and transcriptome analysis. The genome assembly contained 34,105 expressed genes, of which 10,076 were assigned to linkage groups. Genetic mapping and exome resequencing of individuals across the species range both identified the largest linkage group, LG1, as the sex chromosome. Although the sex chromosomes of M. annua are karyotypically homomorphic, we estimate that about one-third of the Y chromosome, containing 568 transcripts and spanning 22.3 cM in the corresponding female map, has ceased recombining. Nevertheless, we found limited evidence for Y-chromosome degeneration in terms of gene loss and pseudogenization, and most X- and Y-linked genes appear to have diverged in the period subsequent to speciation between M. annua and its sister species M. huetii, which shares the same sex-determining region. Taken together, our results suggest that the M. annua Y chromosome has at least two evolutionary strata: a small old stratum shared with M. huetii, and a more recent larger stratum that is probably unique to M. annua and that stopped recombining ∼1 MYA. Patterns of gene expression within the nonrecombining region are consistent with the idea that sexually antagonistic selection may have played a role in favoring suppressed recombination.
Keywords: sex chromosomes, whole genome sequencing, sex linkage, evolutionary strata, gene expression
THE evolution of dioecy (separate sexes) from hermaphroditism, which has occurred repeatedly in independent lineages of flowering plants (Renner 2014), is a prelude to the possible evolution of sex chromosomes. Early sex-chromosome evolution typically involves the accumulation of repetitive sequences in a nonrecombining region (J. Wang et al. 2012; Hobza et al. 2017), differences in codon use between homologs (Ono 1939; Qiu et al. 2010), different patterns of gene expression at sex-linked loci (Zemp et al. 2016), pseudogenization and gene loss (Papadopulos et al. 2015; Wu and Moore 2015), and, ultimately, divergence in chromosome length between homologs (Puterova et al. 2018). Extreme divergence is common in many animals, but it is also known in some plants in which the homologous chromosomes are heteromorphic and distinguishable by karyotype, e.g., in the plant species Silene latifolia (Ono 1939; Krasovec et al. 2018) and Rumex hastatulus (Smith 1955; Hough et al. 2014). In other plants, the sex chromosomes remain indistinguishable by karyotype, and gene function is only mildly compromised, e.g., Asparagus officinalis (Loeptien 1979; Telgmann-Rauber et al. 2007), Spinacia oleracea (Yamamoto et al. 2014), Diospyros lotus (Akagi et al. 2014), Fragaria chiloensis (Tennessen et al. 2016), Populus (Geraldes et al. 2015), Carica papaya (Horovitz and Jiménez 1967; Liu et al. 2004), and Salix (Pucholt et al. 2015). Because of this variation, and because dioecy in plants has often evolved recently, plants with young homomorphic sex chromosomes provide particularly good models for studying the very earliest stages in sex-chromosome divergence (Charlesworth 2016).
Two hypotheses have been proposed to explain the suppression of recombination in plants. First, if dioecy evolves through the spread of male- and female-sterility mutations, these mutations must become linked on opposite chromosomes to avoid the expression of either hermaphroditism, or both male and female sterility simultaneously (Charlesworth and Charlesworth 1978). The main experimental support for this two-locus model comes from classic genetic studies in S. latifolia (Westergaard 1958) that demonstrated the presence of two sex-determining factors on the Y-chromosome: the stamen-promoting factor (SPF) and gynoecium suppression factor (GSF). More recent work mapped the location of these genes on the S. latifolia Y-chromosome (Kazama et al. 2016), although the actual GSF and SPF genes have yet to be identified. Nevertheless, although there is some support for it, the two-locus model does not explain why nonrecombining regions on sex chromosomes often expand greatly (Bergero and Charlesworth 2009), well beyond the region harboring the original sex-determining genes.
A second hypothesis invokes selection favoring suppressed recombination between a sex-determining locus and loci elsewhere on the sex chromosome that have different allelic effects on the fitness of males and females, i.e., alleles with sexually antagonistic effects (Rice 1987; Charlesworth 1991; Gibson et al. 2002; Charlesworth et al. 2005; Bergero and Charlesworth 2009). The suppression of recombination is expected to extend consecutively to generate linkage between the sex-determining locus and more sexually antagonistic loci (Charlesworth 2015), and these extensions can be identified as discrete “strata,” with greater X/Y divergence in strata that ceased recombining earliest. Evidence for strata has been found both in animals (Lahn and Page 1999; Nam and Ellegren 2008) and plants (Bergero et al. 2007; K. Wang et al. 2012), but there is still little direct evidence for the role played by sexually antagonistic selection in bringing them about (Bergero et al. 2019). A recent study of guppy sex chromosomes claimed evidence for the evolution of strata consistent with the sexual-antagonism hypothesis (Wright et al. 2017), but subsequent work has shown that strata could actually not have been involved in the evolution of suppressed recombination (Bergero et al. 2019). Although our understanding of the implications of suppressed recombination is well developed, evidence for its driver therefore remains weak.
Sexually antagonistic selection may be resolved either through differential gene expression between males and females, or through sex linkage of the responsible loci, with the phenotypic expression of sexual dimorphism being the ultimate result. Sexual dimorphism is known in dioecious species for a wide range of morphological (Eckhart 1999), life-history (Delph 1999) and physiological traits (Dawson and Geber 1999), as well as for gene expression (Baker et al. 2007; Sharma et al. 2014; Mohanty et al. 2017; Sanderson et al. 2018). In some cases, the differentially expressed genes are enriched on the sex chromosomes (Parisi et al. 2003; Leder et al. 2010; Albritton et al. 2014; Pucholt et al. 2017), likely as a result of the degeneration of one of the diploid copies following the suppression of recombination, or due to interactions between sex and gene expression at the loci concerned (reviewed in Mank and Ellegren 2009; Parsch and Ellegren 2013). In the latter scenario, dosage compensation may subsequently evolve in response to selection to restore similar levels of expression in males and females. While dosage compensation is an important feature of gene expression in many animal lineages (Mank 2013), its extent in plants is understudied. The clearest analysis remains that for S. latifolia, in which Papadopulos et al. (2015) confirmed gene loss, or lost expression, for many Y-linked genes, with associated incomplete dosage compensation from X-linked homologs, including full compensation at some loci. Further study of sequence evolution and patterns of gene expression in other species would be valuable, particularly those with homomorphic sex chromosomes at the very earliest stages of their evolution, as well as for loci close to the sex-determining locus.
Here, we identify the homomorphic sex chromosomes of the wind-pollinated dioecious annual plant Mercurialis annua (Euphorbiacae) using genetic mapping, sequence analysis of the genome, and patterns of gene expression. Until recently, sex in dioecious M. annua was thought to be determined by allelic variation at three independent loci (Durand et al. 1987; Durand and Durand 1991), but it is now known to have a simple XY system (Khadka et al. 2005; Russell and Pannell 2015). YY males of M. annua, which lack an X chromosome, are viable but partially sterile (Kuhn 1939; Li et al. 2019), indicating that the Y chromosome is only mildly degenerate. Our present study confirms this suggestion, finding very limited gene loss, yet suppressed recombination over a large portion of the sex chromosomes and substantial differences in gene expression between males and females at sex-linked loci. It would thus appear that the sex chromosomes of the species may be at a particularly interesting and revealing stage in their evolution (Veltsos et al. 2018).
To assemble the genome of M. annua, we combined short-read sequencing on the Illumina platform with long-read technology developed by Pacific Biosciences. We also analyzed the karyotype of M. annua using current imaging technology in an attempt to identify heteromorphism at the cytological level, and we obtained SNPs from the transcriptomes of small families to construct a genetic map for M. annua, to identify nonrecombining (sex-linked) genes and scaffolds, and to compare them with non-sex-linked regions. We then sought evidence for the evolution of evolutionary strata and genetic degeneration on the Y chromosome, including the fixation of pseudogenising mutations in the inferred fully Y-linked sequences, and the proportion of genes deleted from the Y. Finally, we examined sex-biased gene expression, and assessed whether sex-biased genes might be enriched on the sex chromosomes, as expected by theory (Connallon and Clark 2010; Meisel et al. 2012). Our results suggest that the sex chromosomes of M. annua are ∼1.5 million years old. Early stages of Y-chromosome degeneration are clearly apparent, but there are also signs that patterns of gene expression might have been affected by the accumulation of sexually antagonistic mutations.
Materials and Methods
Hairy root culture, chromosome preparation, and cytogenetics
M. annua seeds were sterilized by incubation in 4% sodium hypochloride and subsequently washed in 50% ethanol and sterile water. Seeds were grown on MS medium, and leaf discs were taken from 2- to 3-week-old plants that had been sexed using a previously developed Sequence Characterized Amplified Region (SCAR) marker (Khadka et al. 2002). Agrobacterium rhizogenes strain ARqua1 (Quandt et al. 1993) was grown overnight at 28° in LB medium supplemented with 300 µM acetosyringone to OD600 = 0.6. The culture was resuspended in liquid 1/2 MS with 300 µM acetosyringone, and used for direct inoculation of M. annua leaf discs. Explants were cocultivated at 28° on MS medium with 300 µM acetosyringone for 2 days. After cocultivation, explants were moved to MS medium supplemented with 300 µg/liter Timentin. Media were changed every 2 weeks.
To synchronize the hairy roots of M. annua, the DNA polymerase inhibitor aphidicolin was added for 12 hr. Mitoses were then accumulated in protoplasts using oryzalin treatment, transferred to chromic-acid-washed slides by dropping, and stored at −20° until use. For fluorescence in situ hybridization (FISH) experiments, slides were denatured in 7:3 (v/v) formamide: 2xSSC for 2 min at 72°, immediately dehydrated through 50, 70, and 100% ethanol (−20°), and air dried. The hybridization mixture (30 µl per slide) consisted of 200 ng of labeled probe, 15 µl formamide, 6 µl 50% dextrane sulfate, and 3 µl of 20× SSC. The volume was brought to 30 µl by adding TE, pH 8. The probes were denatured at 70° for 10 min, and slides hybridized for 18 hr at 37° in a humid chamber. Slides were analyzed using an Olympus Provis AX70 microscope, and image analysis was performed using ISIS software (Metasystems). DNA was labeled with Fluorolink Cy3-dUTP (red labeling; Amersham Pharmacia Biotech) using a Nick Translation mix (Roche), or with SpectrumGreen direct-labeled dUTP (green labeling; Vysis) and a Nick Translation kit (Vysis).
Genome and transcriptome sequencing
DNA for genomic sequencing was taken from leaf tissue of a single male, M1. RNA samples were collected from leaves and flower buds of this individual, as well as from three unrelated females, G1, G2, and G3, and two additional unrelated males, M2 and M3, all sampled from north-western France and grown together in a glasshouse. Sixty-five F1 and F2 progeny (five families) were then produced by crossing G1xM1 and G2xM1 (F1) as well as three pairs of G1xM1 progeny (F2) (Supplemental Material, Table S1), which were also used for RNA extraction and transcriptome sequencing. DNA was extracted using a Qiagen Plant DNeasy kit (Qiagen, Hilden, Germany). Illumina paired-end and mate-pair sequencing was carried out by the Beijing Genomics Institute (BGI) using Illumina HiSeq 2000 technology (100 bp reads). Pacific Biosciences long-read sequencing was performed on the individual M1 by the Centre for Integrative Genomics hosted at the University of Lausanne. RNA was extracted from a mixture of flower buds and leaf tissues using the Qiagen plant RNAeasy kit (Qiagen), and individual libraries were prepared for all 65 individuals (Table S1), which were sequenced on three lanes of Illumina HiSeq 2000 at the Wellcome Trust Centre for Human Genetics, Oxford.
Transcriptome assembly and annotation
The SEX-DETector pipeline (http://lbbe.univ-lyon1.fr/-SEX-DETector-.html?lang=fr) was used to produce the reference transcriptome (henceforth “ORFs”), and to obtain allele-specific expression and SNPs for X and Y haplotypes. Sex linkage was inferred from genetic mapping (see below). To identify sex-linked haplotypes, we employed a probabilistic model based on maximum-likelihood inference, implemented in SEX-DETector (Muyle et al. 2016). SEX-DETector, which assumes that X-linked and Y-linked haplotypes are passed only from fathers to daughters or from fathers to sons, respectively, is embedded into a Galaxy workflow pipeline that includes extra-assembly, mapping and genotyping steps prior to sex-linkage inference, which has been shown to have greater sensitivity, without an increased false positive rate, than without these steps (Muyle et al. 2016). First, poly-A tails were removed from transcripts using PRINSEQ (Schmieder and Edwards 2011), with parameters -trim_tail_left 5 -trim_tail_right 5. rRNA-like sequences were removed using riboPicker version 0.4.3 (Schmieder and Edwards 2011) with parameters -i 90 -c 50 -l 50 and the following databases: SILVA Large subunit reference database, SILVA Small subunit reference database, the GreenGenes database, and the Rfam database. Transcripts were then further assembled within Trinity components using cap3 (Huang and Madan 1999), with parameter -p 90 and custom Perl scripts. Coding sequences were predicted using Trinity TransDecoder (Haas et al. 2013), including PFAM domain searches as ORF retention criteria; these sequences were taken as our reference transcriptome. The RNAseq reads from parents and progeny were mapped onto the reference transcriptome using BWA (Li and Durbin 2009). The alignments were analyzed using reads2snp, a genotyper for RNAseq data that gives better results than standard genotypers when X and Y transcripts have different expression levels (Tsagkogeorga et al. 2012).
The SEX-DETector transcripts were mapped onto the genome using GMAP v2017-06-20 (Wu and Watanabe 2005), disallowing chimeric genes, mapping 38,963 (99%) of them to 48,298 loci. We then collapsed the inferred ORFs into 34,105 gene models using gffread v0.9.9 (http://ccb.jhu.edu/software/stringtie/gff.shtml). In total, 26,702 (67.9%) of the ORFs could be annotated using fast blastx on the Swiss-Prot database and Blast2GO (Conesa et al. 2005) in Geneious v9 (Kearse et al. 2012). The resulting transcriptome annotation information, including GO terms, is presented in File S4. We characterized genes as candidate sex-determining genes by identifying those containing the following strings in their annotation: flower, auxin, cytokinin, pollen, kelch, ethylene, swi, retinol, calmodulin, short-root, jasmonic. This manual characterization allowed us to visualize the mapping locations, test for their enrichment in the nonrecombining region, and identify whether any were among the outliers in various transcript-associated metrics.
Genome size estimation, assembly, and annotation
The genome size was estimated from the distribution of k-mers (size 31) obtained from jellyfish v2.1.0 (Marçais and Kingsford 2011), and analyzed by the web version of GenomeScope v1 (Vurture et al. 2017), limiting max k-mer coverage to 1000.
Sliding-window trimming and adaptor removal were carried out using Trimmomatic v. 0.30, with default parameters (Bolger et al. 2014). Exact duplicate read pairs were collapsed using fastx-collapser from the Fastx-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Low-complexity masking was carried out using DUST (Morgulis et al. 2006), with the default parameters, and ambiguous reads were removed. Pacific Biosciences long reads were error-corrected using Bowtie2 version 2.1.0 (Langmead and Salzberg 2012) in combination with LSC version 0.3.1 (Au et al. 2012).
Filtered paired-end and mate-paired genomic reads were assembled using SOAPdenovo2 (Luo et al. 2012), with k-mer values between 35 and 55 (odd values only). The best assembly was chosen using REAPR (Hunt et al. 2013). GapCloser (Luo et al. 2012) was run on the best assembly to correct false joins and fill gaps. Error-corrected Pacific Biosciences reads were then used to extend scaffolds, to fill gaps, and to join scaffolds using PBJelly2 (English et al. 2012). Additional scaffolding was carried out using default parameters on SSPACE (Boetzer et al. 2011), which revisits gaps using existing paired-end and mate-paired sequences. This step is intended to correct for any ambiguity introduced by the low-coverage of PacBio reads. Finally, L_RNA_Scaffolder (Xue et al. 2013) was used to bridge genomic scaffolds using the transcript assembly.
Transposable elements (TEs) and tandem repeats were predicted using a combination of Tandem Repeats Finder (Benson 1999), RepeatModeler, and RepeatMasker (Smit et al. 2013). Repeat libraries from M. annua (from RepeatModeler), Euphorbiacae, and Vitis vinifera were used for masking the genome before further analyses.
Summary of SNP calling pipelines
We called SNPs using four different pipelines. (1) X- and Y-specific SNPs were called and haplotypes with premature stop codons, and allele-specific expression SNPs were inferred on the basis of RNAseq data from the two largest families by SEX-DETector. (2) SNPs were called from genome-capture data of multiple populations, for independent analysis of genomic regions of differing sex linkage, as inferred from the genetic map. (3) SNPs were called on all individuals from five families, to perform genetic mapping. (4) SNPs were called on transcripts from the three unrelated males (M1, M2, and M3) and the three unrelated females (G1, G2, and G3) for comparisons between ORFs of differing sex linkage and comparisons of pairwise diversity between species. The SNPs for which there was evidence of separate X and Y copies and which also had homologs in M. huetii and R. communis were analyzed in a phylogenetic framework to estimate the (tree-based) evolution rate of the Y sequences. Specific details of each pipeline are given in the corresponding sections below.
Genetic map construction
We called SNPs independently of the SEX-DETector pipeline, using all individuals available over the three generations of our family crosses. We first mapped RNAseq reads to the reference transcriptome using Bowtie2 v2.3.1 (Langmead and Salzberg 2012). The resulting sam files were converted to bam, sorted, and converted to mpileup format using samtools v 1.3 (Li et al. 2009). The resulting posterior file containing segregation information of transcripts in the five mapping families (>164,000 markers and their genotype likelihoods) was passed through the LepMap3 (LM3) pipeline (Rastas 2017). First, we calculated the relatedness between individuals using the IBD module (of LM3) using a random subset of 3000 markers. Three individuals were discarded because their relatedness to their putative father (M6) was <0.2, i.e., they were likely the result of a different cross through contamination. The parental genotypes were then called using ParentCall2, with parameter halfSibs = 1, to take into account the half-sib family structure, in addition to the genotypic information of offspring, parents, and grandparents. The 158,000 remaining markers were further filtered using the Filtering2 module, with parameter dataTolerance = 0.001, to remove markers with distorted segregating. We then combined markers of each transcript into pseudomarkers, as they were not sufficiently informative on their own to separate into linkage groups. This was done by running OrderMarkers2 separately for each transcript without recombination (parameter values: recombination1 = 0, recombination2 = 0), and then obtaining genotype likelihoods for each transcript with parameter outputPhasedData = 4. This resulted in 13,261 markers, each consisting of a single transcript, which we reformatted as input for LM3. We defined linkage groups by running SeparateChromosomes2 with default parameters (lodLimit = 10), adding additional markers to the resulting eight linkage groups using JoinSingles2 with lodLimit = 8 and lodDifference = 2. We were able to assign 10,076 transcripts to eight linkage groups, which were ordered using OrderMarkers2. The linkage maps were drawn with R/qtl v1.41 (Broman et al. 2003) running in R 3.4.2 (R Development Core Team 2007).
Sequence divergence in M. annua, and in comparison with closely related species
We aligned reads from six unrelated individuals from France (M1, M2, M3, G1, G2, G3) to the 39,302 ORFs from the reference transcriptome assembled via the SEX-DETector pipeline, based on the M1 male transcriptome and using the BWA-MEM algorithm of BWA v0.7.13 (Li and Durbin 2009). Picard Tools v2.2.1 (http://broadinstitute.github.io/picard/) was used to mark duplicate read pairs. Local realignment around insertions and deletions (indels) was performed with GATK v3.7 (DePristo et al. 2011), followed by SNP calling on each individual using the HaplotypeCaller module in GATK (McKenna et al. 2010). Joint SNP calling was performed using GATK’s GenotypeGVCFs module, overlapping SNPs and indels were filtered out with the VariantFiltration module, and SNPs and indels were separated and filtered to produce two high-quality variant sets with the following parameters in the VariantFiltration module: “QUAL <30” “DP < 30” “MQ0 >= 4 && ((MQ0/(1.0 * DP)) > 0.1)” “QD < 5.0”. The high-quality SNP set was used to perform variant quality score recalibration to filter the full SNP set. This set was used to calculate within-species nucleotide diversity (π) using vcftools v 0.1.13 (Danecek et al. 2011).
Pairwise dN and dS values from M. huetii and R. communis were obtained using yn00 from PAML 4.9 (Yang 2007). This analysis used 4761 and 2993 homologs from the M. huetii and R. communis (http://castorbean.jcvi.org/introduction.shtml) transcriptomes, based on a de novo assembly using default settings in Trinity (Haas et al. 2013; File S5). Sequences were aligned with LASTZ (Harris 2007). Effective codon usage was calculated using ENCprime (https://jnpopgen.org/software/).
To estimate the rate of Y-sequence evolution, we identified one-to-one orthologs between the M. annua sex linked genes, and the closely related species M. huetii and R. communis, through reciprocal best BLASTP, with an e-value of 1e−50, a culling limit of 1, and considering only transcripts that were not split across contigs. The 98 orthologs identified were aligned with LASTZ (Harris 2007) and analyzed with PAML 4.9 (Yang 2007) to obtain tree-based estimates of dN and dS values. We employed three models: the “null model” (M0), where all parts of the tree are assumed to have the same dN/dS values; the “branch-specific” model (M1), where each branch may have its own dN/dS value; and the “two-ratio” model (M2), where the branch of interest (Y) may have a dN/dS ratio that differs from the rest of the tree (Nielsen and Yang 1998). Preliminary analysis revealed similar results for the M1 and M2 models (results not shown); we thus chose M2 as the simpler model for further analysis. We tested whether the Y branch evolved differently from the rest of the tree, using a likelihood ratio test (LRT) between the M2 and M0 models, with one degree of freedom and LRT = 2 × abs(l2−l0), where l2 and l0 are the likelihoods for M2 and M0, respectively.
Expression analysis
Reads from the RNA libraries of all 30 females and 35 males were pseudoaligned to the reference transcriptome using Kallisto v0.43.1 (Bray et al. 2016). The resulting raw count data were compiled in a table, and differential expression analysis between male and female samples was conducted with edgeR v3.18.1 (Robinson et al. 2010), running in R v.3.4.0 (R Development Core Team 2007). Genes were filtered from the analysis if their average log2 count per million (as computed by edgeR’s aveLogCPM function) was negative. This retained genes with an average count of ≥24 per sample. Two individual libraries were removed that were obvious outliers in the MDS plots of the filtered data. Libraries were normalized with the default (TMM) normalization. Dispersion was measured with default parameters using a negative binomial model. Sex-biased transcripts were defined statistically, i.e., by a false discovery rate (FDR; Benjamini and Hochberg 1995) of 5%. A statistical definition for sex-biased genes is justified because our RNAseq analysis was based on many samples, and because the sex-biased categories (male-, female-, and unbiased) used in our analysis capture biological information in that they differed statistically for various measurements. Results for effects of sex bias should be conservative, because our approach diluted “real” sex-biased genes with unbiased genes. We note, however, that the lack of minimum log2FC in our definition of sex-bias means that we cannot differentiate between allometric differences in tissue composition or gene regulation differences between males and females (Montgomery and Mank 2016).
SEX-DETector calculates X- and Y-allele expression from each male. X and Y read numbers were summed for each contig and individual separately, divided by the number of X/Y SNPs of the contig, and adjusted for the library size of the respective individual. X and Y expression levels were then averaged among individuals, and the ratio of the means was computed. Gene ontology (GO) enrichment analysis was performed on the sex-biased genes, as well as separately for the male- and female-biased genes, using topGO with the weight01 algorithm, which accounts for GO topology (Alexa and Rahnenfuhrer 2010).
Comparison of expression between sex-linked genes and autosomal genes
We categorized transcripts based on their genomic location into fully sex-linked (SL; transcripts in the nonrecombining region of LG1 in males), pseudoautosomal (PAR; transcripts in the rest of LG1 that showed recombination in both sexes), and autosomal (Au; transcripts that mapped to the remaining seven linkage groups). We investigated the effects of sex linkage, sex-biased expression, and their interactions, through linear models on the following metrics, using R version 3.4.2: absolute male/female log2FC (fold-change, i.e., overall sex-bias intensity); nucleotide diversity (π); dN/dS from pairwise comparisons with M. huetii and R. communis; and Y/X allele-specific log2FC obtained from SEX-DETector. The data were not normally distributed, and were thus Box-Cox-transformed after adding 1e−11 to all values to allow inclusion of zero values and estimating the transformation parameter using the command boxcoxnc with the Anderson-Darling method in the R package AID v2.3 (Dag et al. 2014). Absolute male/female (log2FC) and π remained non-normal after transformation when unbiased genes were included in the analysis, because there were too many transcripts with ∼0 values. Exclusion of these transcripts did not qualitatively affect the results (data not shown). We also analyzed the sex-biased genes on their own within the same statistical framework. This allowed a more similar distribution between the sex-bias categories, and successful tests of normality after Box-Cox transformation. The summary tables from the models were generated using the R package sjPlot version 2.4 (Lüdecke 2017).
We also compared the proportions of sex-biased/unbiased genes and male-biased/female-biased genes between pairs of genomic regions of differing sex linkage using chi square tests (or Fishers’s exact tests when the gene numbers were small), employing the sjt.xtab function of the in R package sjPlot v 2.4 (Lüdecke 2017). The pairs of genomic regions compared were: PAR-SL, Au-LG1, and Au-SL. Finally, we looked for enrichment of candidate genes in the same genomic region comparisons using the same methodology.
Divergence between males and females from natural populations
To assess divergence between males and females, we examined FST and sex-biased heterozygosity from individuals sampled from across the species’ range. Seeds obtained from multiple M. annua populations (Table S2) were germinated and grown in a glasshouse at the University of Lausanne. Twenty individuals of each sex were selected for genomic analysis to achieve a balance between representation of many populations and sufficient common sequences of good quality. High-quality genomic DNA was extracted from ∼100 mg of frozen leaf material using the DNeasy Plant Mini Kit (Qiagen), following standard protocols. Based on the successful probes from a previous gene-capture experiment (see González-Martínez et al. 2017), ∼21 Mbp of the M. annua diploid genome (175,000 120-mer probes) were sequenced, with sequence capture using SureSelect DNA Capture technology (Agilent Technologies, Santa Clara, CA) followed by sequencing using Illumina HiSeq 2500, outsourced to Rapid Genomics (Gainesville, FL). Our gene-capture experiment included both genic and intergenic regions and avoided repetitive regions and organelle genomes. Raw data were provided by Rapid Genomics as FASTQ files.
Sequencing quality was assessed using FASTQC v.10.5 (www.bioinformatics.babraham.ac.uk/projects/fastqc). Adapter removal and trimming used Trimmomatic v.0.36 (Bolger et al. 2014), with options ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW: 4:15 MINLEN:36. After filtering, we retained an average of 13 million paired-end sequences per sample. We aligned sequences to the M. annua scaffolds using BWA-MEM v.0.7.12 (Li and Durbin 2009) and processed the alignments with Picard Tools v.1.141 (http://broadinstitute.github.io/picard/). We ran Samtools mpileup (Li et al. 2009) with the probabilistic alignment disabled, and called SNPs using Varscan (Koboldt et al. 2009), with a minimum variant allele frequency of 0.15, and a minimum threshold for homozygotes of 0.85. We required a minimum of 10 reads per site, and a Phred quality score >20.
The gene capture ORFs were aligned to the genomic scaffolds using BLAST v.2.2.31 (Altschul et al. 1990). We retained hits with a maximum e-value of e−20, and a minimum identity of 95%. In the case of multiple ORFs mapping to the same genomic location, we selected the longest alignment, and removed all overlapping ORFs. This resulted in 21,203 transcripts aligning to the genome, 8317 of which were also included in the linkage map. All scaffolds with a transcript were assigned a linkage group position in the map. A total of 31 scaffolds contained transcripts assigned to more than one linkage group; in these cases, we assigned the scaffold to the linkage group with >50% of the transcripts (in the case of equal assignment, we excluded the scaffold from the analysis). In total, we were able to assign 6458 scaffolds to linkage-group positions.
We used the BLAST output to construct a bed file that defined the coordinates of each transcript on a genomic scaffold. We then extracted genetic information for the transcripts specified in the bed file. Using this information, we calculated two complementary measures of differentiation between males and females for transcripts >100 bp. First, we computed FST for each transcript using VCFtools v.0.1.15 (Danecek et al. 2011). Similarly, we assessed sex-biased heterozygosity for each transcript (Toups et al. 2019), which we defined as the log10 of the male heterozygosity:female heterozygosity, where heterozygosity is measured as the fraction of sites that are heterozygous. We expect this ratio to be zero for autosomal transcripts, and elevated on young sex chromosomes due to excess heterozygosity in males. For transcripts on older sex chromosomes, the X and Y alleles may have diverged sufficiently to prevent Y alleles from mapping to the X allele reference; in this case, we should expect X-chromosome hemizygosity in males and no inflated female heterozygosity.
For both FST and sex-biased heterozygosity in the two sexes, we assessed differences between the autosomes, PAR, and the sex-linked region using Wilcoxon signed rank tests. We then computed moving averages in sliding windows of 20 transcripts on all chromosomes using the rollmean function, from package zoo v.1.8-0 in R v3.2.2. To identify regions of elevated FST and sex-biased heterozygosity, we computed 95% confidence intervals on the basis of an average of 30 consecutive transcripts on the autosomes calculated 1000 times. Finally, for both metrics, we assessed whether the top 5% of transcripts were overrepresented on LG1 using chi-squared tests.
Data availability
All raw DNA and RNA sequence data generated in this study have been submitted to NCBI under accession SRP098613 BioProject ID PRJNA369310. Scripts, supplemental files, and intermediate files are available at https://osf.io/a9wjb/. File S1 contains the genetic map data and other information on each mapped transcript. File S2 contains the Sanger sequence of clones containing the X and Y variants, where there is a premature stop codon on the Y, and the script used to make the phylogenetic tree. File S3 summarizes the GO results for sex-biased genes. File S4 contains transcriptome annotation information. File S5 contains the assembled transcriptome and X and Y haplotype predictions from SEX-DETector. File S6 contains the genome and predicted genes on it. File S7 is the M. annua repeat library.
Results
Male karyotype
The M. annua karyotype has eight linkage groups, with no difference between males and females, i.e., the sex chromosomes are homomorphic (Figure 1 and Figure S1). Idiogram data, chromosomal arm ratios, and relative length measurements from 50 images (obtained from five individuals; Table S3) indicate that all M. annua chromosomes can be discriminated from one another. A partial distinction between the one large, two medium, and five smaller chromosomes is possible on the basis of the two common ribosomal DNA probes (45S and 5S), with a secondary constriction often visible at the location of the 45S locus.
Figure 1.
Basic karyotype for a M. annua male individual. The top row shows DAPI counterstain. The middle row shows chromosomes counterstained with DAPI (blue) after bicolor FISH with 45S rDNA (red) and 5S rDNA (green). The bottom row shows an idiogram of the male chromosomes. The white bar in the middle row represents 5 µm.
M. annua genome assembly
Using k-mer distribution, we estimate the haploid genome size to be 325 Mb (Figure S2 and Table S4), which is similar to the previous estimate of 640 Mb for the diploid genome (Obbard et al. 2006). Using a combination of short-read Illumina and long-read Pacific Biosciences DNA sequencing, we generated ∼56.3 Gb of sequence data from the M. annua male M1, corresponding to a coverage of ∼86.6×. After read filtering, genome coverage dropped to ∼63.7× (Table S5). De novo assembly and scaffolding yielded a final assembly of 89% of the genome (78% without gaps), 65% of which was assembled in scaffolds >1 kb, with an N50 of 12,808 across 74,927 scaffolds (Table S6). Assembly statistics were consistent with those for other species of the Malpighiales (Table S7), as was our estimate of total genomic GC content (34.7%; Smarda et al. 2012). The M. annua assembly encompassed over 89% of the assembled transcriptome; most of the unassembled genome sequence data are therefore likely repetitive. Using BUSCO2 (Simão et al. 2015), we recovered 76.1% of the 1440 genes in the BUSCO embryophyte database (of which 3.9% were duplicated). A total of 10.3% of the genes were fragmented, and 13.6% were missing.
Repeat masking identified simple tandem repeats in >10% of the assembly. This proportion is likely to be an underestimate, given the difficulty in assembling microsatellites. We characterized 15% of the assembly on the basis of homology information and DNA transposon and retrotransposon masking, with an additional 33% that corresponds to 1472 predicted novel transposable elements. The most frequent transposable repeats were gypsy LTR, copia LTR, and L1 LINE retrotransposons (Table S8), similar to findings for other plant genomes (Chan et al. 2010; Sato et al. 2011; J. Wang et al. 2012; Rahman et al. 2013). Across all data, 58% of the ungapped M. annua assembly was repetitive (Table S8), corresponding to 44% of the genome. Given that our assembly covered 78% of the genome, and if we assume that the missing fraction comprises only repeats, up to 66% of the M. annua genome may be repetitive. This too would be consistent with what is known for other plants, which have a similarly high AT-rich repeat content and a similar number of unclassified repetitive elements (e.g., Chan et al. 2010). The M. annua genome appears to be ∼240 Mb larger than that of its relative R. communis (see Table S7), perhaps reflecting ongoing transposon activity.
Sex-linked transcripts and genetic map for diploid M. annua
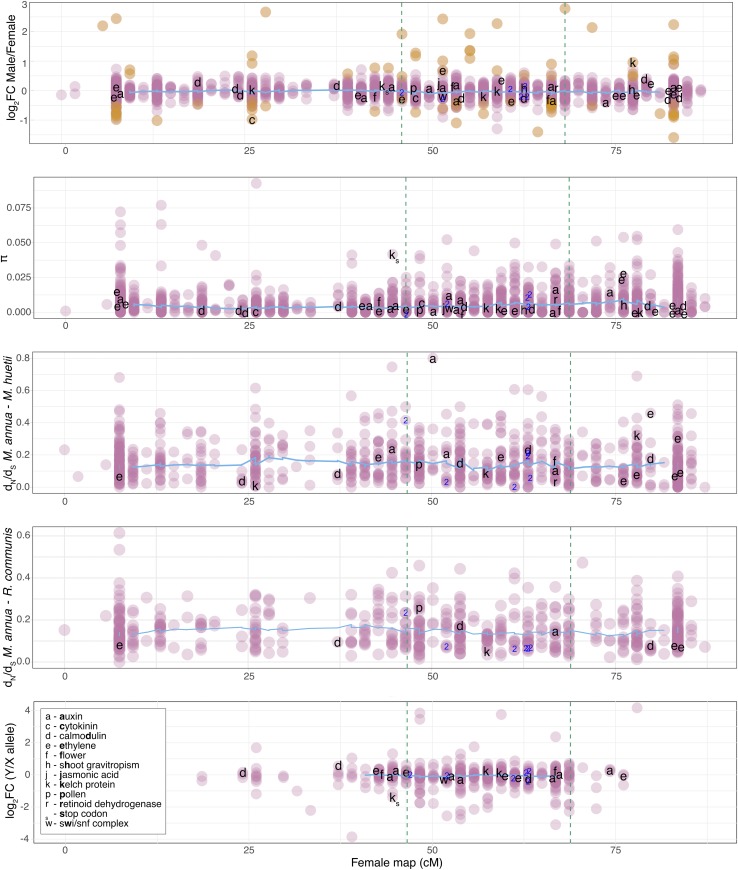
Genetic mapping recovered eight linkage groups (LG1 through LG8), corresponding to the expected chromosome number for the diploid M. annua karyotype (Durand 1963). LepMap3 (LM3) identified 568 sex-linked (SL) transcripts, of which 365 were also identified as sex-linked by SEX-DETector, and phased into X and Y haplotypes. The 568 SL transcripts mapped to LG1, which is thus the sex chromosome. An additional 1209 transcripts mapped to the two ends of LG1, representing two putative pseudoautosomal regions (PAR1 + PAR2; we refer to these as PAR and consider them together in all analyses); 8299 transcripts mapped to the other (autosomal) linkage groups. In total, 641 markers could be resolved from one another and were thus informative. We inferred the sex-linked region to comprise transcripts without recombination in males and in the same phase as that of their paternal grandfather. Assuming an equal transcript density across the genome, the sex-linked region represents 32.6% (about a third) of LG1, and 5.8% of the 320 Mb haploid genome (Obbard et al. 2006), or 18.6 Mb. This Y-linked region maps to LG1 at male position 53.85 cM and spans 22.23 cM on the female recombination map (from positions 46.44 to 68.67 cM; Figure 2). The total male and female recombination maps were 700.43 and 716.06 cM, respectively. All marker names and their associated positions and metrics are provided in File S1.
Figure 2.
Genetic map of the eight linkage groups (LG) for M. annua, with each LG represented by the male (left) and female (right) maps. Lines linking the two maps indicate the mapping position of groups of transcripts that segregated together (10,076 total transcripts). The sex-linked region is at 53.85 cM of chromosome 1 (LG1) on the male recombination map, and is highlighted by the gray vertical bar. The two pseudoautosomal regions (PAR) comprise the rest of LG1.
Characterization of contigs by sex linkage and ORF localization
We characterized the genome assembly by mapping all ORFs using GMAP and dividing genomic contigs into three groups: “sex-linked,” “autosomal,” and “expressed ORF” contigs (containing all contigs that mapped to a transcript, regardless of its inclusion on the genetic map; note that PAR contigs were included in the autosomal bin). There were 548 sex-linked contigs containing 1641 genes and a total of 8.3 Mb of sequence; 7579 autosomal contigs containing 23,502 genes, and 106 Mb sequence data; and 15,392 contigs with 34,105 expressed ORFs distributed across 176 Mb (Table 1).
Table 1. Summary statistics of contigs containing sex-linked, autosomal or all expressed ORFs.
| Whole genome | Expressed ORFs | Sex-linked | Autosomal | |
|---|---|---|---|---|
| Number of contigs | 720,537 | 15,392 | 538 | 7579 |
| Total base pairs | 546,375,413 | 176,423,112 | 8,271,542 | 105,835,810 |
| % GC | 34.7 | 34.5 | 34.5 | 34.3 |
| Average contig length | 15,330 (±20,028) | 11,462 (±17,207) | 15,374 (±21,615) | 13,964 (±19,678) |
| Total repeat density (%) | 51.5 | 18.7 | 18.4 | 18.3 |
| Final coding transcript number | 34,105 | 34,105 | 1641 | 23,502 |
| Average transcript length | 887 (±754) | 810 (±702) | 1265 (±878) | 1242 (±853) |
| Mean effective number of codons | 53.7 | 53.7 | 52.2 | 51.9 |
| Number of SNPs | 181,567 | 180,925 | 5274 | 102,881 |
| Number of ORFs transcripts with SNPs | 16,667 (49%) | 16,572 (49%) | 545 (33%) | 7714 (33%) |
| Median nucleotide diversity (π)/ORF | 0.0035 (±0.0113) | 0.0035 (±0.0113) | 0.006647 (±0.008079) | 0.007951 (±0.01016) |
Only ORFs supported by expression data were used for analysis of sex linkage (de novo predicted genes were excluded). Confidence intervals are 1 SD.
Sequence divergence, nucleotide diversity, and codon usage of X and Y haplotypes
We used the SNP and haplotype calls from SEX-DETector to compare nucleotide diversity and divergence between the X and Y ORFs within M. annua, as well as between X, Y, and autosomal ORFs of M. annua and closely related species. The mean pairwise dN/dS between X and Y haplotypes within M. annua was 0.182 (Table 2). For sex-linked transcripts, we excluded the few SNPs that did not show clear sex-linked segregation, but we could not apply such error correction for autosomes. Accordingly, π might be underestimated for sex-linked genes compared to autosomes (see Figure S3 and Table 1), so that metrics associated with sex-linked sequences should be regarded as conservative.
Table 2. Average pairwise dS and dN for ORFs of M. annua from X/Y haplotype comparison within M. annua or against M. huetii and R. communis sequences.
| dS | dN | dN/dS | |
|---|---|---|---|
| X/Y pairs | 0.011 | 0.002 | 0.182 |
| X/M. huetii | 0.112 | 0.016 | 0.143 |
| Y/M. huetii | 0.113 | 0.017 | 0.150 |
| X/R. communis | 0.548 | 0.070 | 0.128 |
| Y/R. communis | 0.555 | 0.070 | 0.126 |
| M. annua/M. huetii | 0.106 | 0.014 | 0.149 |
| M. annua/R. communis | 0.565 | 0.079 | 0.140 |
Autosomal ORF comparisons with the other species are also reported. Haplotypes with premature stop codons were excluded.
The pairwise dS and dN/dS values for all possible orthologs between M. annua and its relatives M. huetti and R. communis, and the X and Y haplotypes, are presented in Figure 3 and Table 2. For autosomal genes, dN/dS between M. annua and M. huetii and between M. annua and R. communis, respectively, was 0.149 and 0.140. Pairwise dS was lower between X/Y gene pairs within M. annua than between orthologous autosomal genes in M. annua and M. huetii or R. communis.
Figure 3.
Density plots of synonymous site divergence (dS) for: (A) all M. annua X/Y haplotypes, (B) autosomal M. annua/M. huetii homologs, (C) autosomal M. annua/R. communis homologs, (D) X-linked M. annua/M. huetii homologs, and (E) X-linked M. annua/R. communis homologs. The numbers on the bottom right indicate the sample size for each plot.
Codon usage in M. annua did not differ significantly between sex-linked, pseudoautosomal and autosomal (SL, PAR, and Au) ORFs (Nc = 54, Nc = 53, and Nc = 53, respectively; Figure S4).
We used PAML (Yang 2007) to analyze the 98 sex-linked genes with orthologs in M. annua, M. huetii, and R. communis, using the “two-ratio” M2 model (which accommodates differences in dN/dS between the Y branch and other branches of the tree; Nielsen and Yang 1998). Our analysis identified 84 sequences with 0.001 < dS < 2 on both X and Y branches. Variation for 74 of these sequences was consistent with a tree topology corresponding to the species tree and recent X-Y divergence for M. annua, i.e., trees with topology {[(M. annua-X, M. annua-Y), M. huetii], R. communis}; we focused further analysis on these genes. For 52 of these 74 genes (21 after FDR correction at 5% level; Benjamini and Hochberg 1995), the M2 model provided a better fit than the M0 model. Moreover, the Y lineage had more synonymous and more nonsynonymous mutations compared to the X lineage following divergence from M. huetii. Together, these results suggest that 28% of the Y-linked genes in our sample have been evolving faster than their corresponding X-linked orthologs, as inferred from the tree-based dN and dS values (Wilcoxon Rank Sum = 392 and 568, respectively; P < 0.001 for both tests; Figure 4), as well as under relaxed selection, as inferred from the tree-based dN/dS ratio (Wilcoxon Rank Sum = 267, P < 0.001; Figure 4). The seven sequences for which the Y variant clustered outside the clade (M. annua-X, M. huetii) were scattered across the genetic map of the nonrecombining region of the Y (Figure 5), i.e., they did not indicate a clear ancestral/common sex-determining region between M. annua and M. huetii. We did not investigate these genes further.
Figure 4.
Boxplots of tree-based estimates of dN/dS, dS, and dN for the X, Y, M. huetii, and R. communis branches of the 72 gene trees that correspond to tree topology {[(M. annua-X, M. annua-Y), M. huetii], R. communis}. A Y-sequence dN/dS outlier of 2.7 and an M. huetii-sequence dS outlier of 0.23 are not shown.
Figure 5.
Summary of transcript metrics calculated for sequences across the female recombination map of the putative sex chromosome LG1, based on analysis of X and Y sequences combined. The nonrecombining region in males is located between the vertical dotted lines. Orange points in the top panel indicate significant sex-bias (5% FDR). Potential candidate sex-determining genes are indicated by letters, with their putative function given in the inset legend. The only verified premature stop codon on the Y copy is indicated by ks beside the nonrecombining region (i.e., for a kelch protein gene). Transcripts for which the Y sequence diverged prior to the M. annua/M. huetii species split are labeled “2.”
Identification of a degenerated and duplicated sequence on the Y
The SEX-DETector pipeline detected putative premature stop codons in four Y and three X transcripts, although there was always a functional X haplotype for the same transcript. We confirmed one of the four Y-haplotype stop codons using PCR, through cloning and sequencing from five males and nine females sampled across the geographic range of diploid M. annua, which only showed the stop codons on sequences from males. Briefly, a phylogenetic tree was constructed for this gene (Figure S5) using RAxML v.8.2.10 using 1000 bootstraps (Stamatakis 2014) on sequences aligned with mafft v7.310 (Katoh and Standley 2013). Interestingly, two different Y-linked sequences could be obtained, sometimes from the same male, suggesting that the sequence has been duplicated in at least some males. All males and females analyzed also contained a functional copy that had 89% identity with an F-box/kelch repeat protein in R. communis. The transcript shows a lower Y/X expression ratio and a high π (marked “Ks” in Figure 5), suggesting that it represents a degenerated copy on the Y.
Identification and annotation of sex-biased genes
There were many more (1385) male-biased than female-biased genes (325), based on 5% FDR. This difference increased when bias was determined on the basis of a minimum log2 fold-change (log2FC) threshold of 1 (1141 vs. 140; Figure S6). Male-biased genes were enriched in biological functions such as anther-wall tapetum development, response to auxin, response to ethylene, cell-tip growth, pollen-tube growth, and floral-organ senescence, whereas female-biased genes were enriched for functions related to the maintenance of inflorescence meristem identity, jasmonic acid and ethylene-dependent systemic resistance, regulation of innate immune response, and seed maturation (File S3).
Identification of the ancestral evolutionary stratum on the Y
To identify a putative ancestral stratum in the nonrecombining region, we investigated variation in the following metrics across the female recombination map (Figure 5): the magnitude of sex-biased expression; the nucleotide diversity (π) of each transcript; and the pairwise dN/dS ratio based on comparison with homologous genes in the closely related dioecious sister species M. huetti, and its monoecious more distant relative R. communis (for which we could identify 4761 and 2993 homologs, respectively); and the expression ratio of X and Y haplotypes inferred from SEX-DETector (Muyle et al. 2016). We annotated Figure 5 with letters to indicate the position of candidate sex-determining genes, based on their blast hit descriptions. The sole obvious outlier for any of the metrics above was a transcript associated with auxin production, which had a high pairwise dN/dS value in the comparison with the close outgroup species, M. huetii.
Comparison of sex-linked, autosomal, and pseudoautosomal ORFs
We examined the effects of sex-bias (male-, female-, and unbiased genes) and genomic location (SL, PAR, or Au), as well as their interactions, on the five metrics plotted in Figure 5. We repeated the analyses after excluding unbiased transcripts for the four metrics with sufficient gene numbers (sex bias log2FC, π, and pairwise dN/dS using either M. huetii or R. communis) to directly compare male- and female-biased transcripts. The full models are presented in Tables S9–S11. Below, we summarize their significant results; Figure 6 shows the data distributions. The analyses revealed significantly higher differences in gene expression between the sexes (absolute log2FC) in the sex-linked region (P = 0.037; Table S9). These differences were largely due to significantly higher expression of male- than female-biased transcripts(male-bias*SL interaction: P = 0.036; Table S10).
Figure 6.
Graphical summary of the analysis of genes based on their sex-bias (blue: male-biased, red: female-biased, gray: unbiased) and sex linkage (SL: sex linked, PAR: remaining LG1, Au: LG 2-7). (A) absolute log2FC expression difference between the sexes, (B) nucleotide diversity (π), (C) pairwise dN/dS compared to M. huetii homologs, (D) pairwise dN/dS compared to R. communis homologs, (E) relative expression of Y over X alleles. The numbers to the right indicate the number of genes in each category. Data are plotted prior to Box-Cox transformation.
The increased magnitude of male-biased gene expression in fully sex-linked genes is consistent with a recent masculinization as sex-linked genes had slightly lower nucleotide diversity (π) than those in the PAR or autosomal genes (P = 0.039; Table S9), but male-biased genes were not affected as much (P = 0.017; Table S9). The pairwise dN/dS between M. annua and M. huetii was higher for sex-linked genes (P = 0.017; Table S9), as well as for female-biased genes localizing to the PAR (PAR*female-bias interaction: P = 0.005; Table S9). Sex-biased genes in the M. annua PAR had a lower π (PAR: P = 0.007; Table S10), and the dN/dS for sex-biased PAR genes between the M. annua and M. huetii was also lower (P = 0.018; Table S10). dN/dS with respect to the more distantly related R. communis was only higher for male-biased genes (P = 0.046; Table S9), though power was limited by the availability of fewer identified homologs (4761 in M. huetti compared to 2993 in R. communis). The Y-linked copy of female-biased genes had significantly lower expression than its X-linked counterpart (P = 0.035; Table S11).
Finally, we used chi-squared tests to compare the relative proportions of sex-biased vs. unbiased and male- vs. female-biased genes in different pairs of genomic regions (PAR-SL, Au-LG1, Au-SL; Table 3). The sex-linked region had significantly more sex-biased genes (compared to either PAR or Au regions). This enrichment was largely responsible for the significant sex-biased gene enrichment compared with the entire LG1 and the autosomes, and was largely due to more female-biased genes on LG1. The sex chromosomes thus appear to be feminized in terms of number of female-biased genes, in contrast to the genome as a whole, which was masculinized in terms of number of male-biased gene numbers, and, in contrast to absolute expression, which was higher for male-biased genes. We did not detect any enrichment for candidate genes with functions with obvious potential sexually antagonistic effects in the sex-linked region (Table 3).
Table 3. Results of chi-squared tests for differences in the frequencies of sex-biased vs. -unbiased genes, male- vs. female-biased genes, and candidate sex-determining genes (CG) vs. other genes (not CG) between the pseudoautosomal region (PAR), the sex-linked region (SL), linkage group 1 (LG1), and autosomes (Au).
| Sex-biased vs. unbiased | Female-biased vs. male-biased | Candidate gene vs. other gene | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Unbiased | Sex-biased | Total | Female biased | Male biased | Total | Not CG | CG | Total | |
| PAR | 1168 | 41 | 1,209 | 22 | 19 | 41 | 1,177 | 32 | 1,209 |
| 96.6% | 3.4% | 100% | 53.7% | 46.3% | 100% | 97.4% | 2.6% | 100% | |
| SL | 536 | 32 | 568 | 16 | 16 | 32 | 546 | 22 | 568 |
| 94.4% | 5.6% | 100% | 50% | 50% | 100% | 96.1% | 3.9% | 100% | |
| Total | 1704 | 73 | 1777 | 38 | 35 | 73 | 1,723 | 54 | 1,777 |
| 95.9% | 4.1% | 100% | 52.1% | 47.9% | 100% | 97% | 3% | 100% | |
| Test | χ2 = 4.381 df = 1 ϕ = 0.053 P = 0.036* | χ2 = 0.006 df = 1 ϕ = 0.036 P = 0.941 | χ2 = 1.578 df = 1 ϕ = 0.033 P = 0.209 | ||||||
| Au | 8056 | 243 | 8,299 | 90 | 153 | 243 | 8094 | 207 | 8,301 |
| 97.1% | 2.9% | 100% | 37% | 63% | 100% | 97.5% | 2.5% | 100% | |
| LG1 | 1704 | 73 | 1,777 | 38 | 35 | 73 | 1,723 | 54 | 1,777 |
| 95.9% | 4.1% | 100% | 52.1% | 47.9% | 100% | 97% | 3% | 100% | |
| Total | 9760 | 316 | 10,076 | 128 | 188 | 316 | 9817 | 261 | 10,078 |
| 96.9% | 3.1% | 100% | 40.5% | 59.5% | 100% | 97.4% | 2.6% | 100% | |
| Test | χ2 = 6.326 df = 1 ϕ = 0.026 P = 0.012* | Χ2 = 4.649 df = 1 ϕ = 0.129 P = 0.031* | χ2 = 1.515 df = 1 ϕ = 0.013 P = 0.218 | ||||||
| Au | 8056 | 243 | 8,299 | 90 | 153 | 243 | 8094 | 207 | 8,301 |
| 97.1% | 2.9% | 100% | 37% | 63% | 100% | 97.5% | 2.5% | 100% | |
| SL | 536 | 32 | 568 | 16 | 16 | 32 | 546 | 22 | 568 |
| 94.4% | 5.6% | 100% | 50% | 50% | 100% | 96.1% | 3.9% | 100% | |
| Total | 8,592 | 275 | 8,867 | 106 | 169 | 275 | 8640 | 229 | 8,869 |
| 96.9% | 3.1% | 100% | 38.5% | 61.5% | 100% | 97.4% | 2.6% | 100% | |
| Test | χ2 = 12.066 df = 1 ϕ = 0.038 P < 0.001* | χ2 = 1.496 df = 1 ϕ = 0.085 P = 0.221 | χ2 = 3.493 df = 1 ϕ = 0.021 P = 0.062 | ||||||
Significant difference.
Sex-linked sequence analysis of individuals sampled across the species range
To confirm the lack of recombination in the sex-linked region, we estimated FST and sex-biased heterozygosity for genome capture data from 20 males and 20 females sampled from natural populations across the species’ range (Table S2), based on 6557 transcripts assigned to the linkage map. The sex-linked region showed clear differentiation between males and females using both metrics. FST between males and females was elevated in the sex-linked region relative to the PAR and the autosomes (Wilcoxon, P = 4.79 × 10−5, P = 1.099 × 10−7, respectively; Figure 7A), although the FST values were very small. Similarly, males were more heterozygous than females in the sex-linked region relative to the PAR and the autosomes (Wilcoxon, P < 2.2 × 10−16 in both; Figure 7C). A sliding window analyses of FST and sex-biased heterozygosity identified a region of elevated FST at ∼65 cM of LG1 in the female recombination map (Figure 7B)—a value exceeding that for any autosomal region. We also identified a region between 50 and 65 cM on LG1 (Figure 7D) with higher heterozygosity in males than females (Figure 7E). Finally, both FST and male-biased heterozygosity were greatest in the sex-linked region of LG1 (Table S12).
Figure 7.
Summary of the effect of sex linkage on SNP metrics from genome capture data across the species range. (A) FST for sex linkage category, (B) distribution of FST across the female recombination map for LG1, (C) ratio of male/female heterozygosity for sex linkage category, (D) ratio of male/female heterozygosity across the female recombination map of LG1, (E) observed heterozygosity in males (blue) and females (red) across LG1. Asterisks in (A and C) indicate significant Wilcoxon tests (P < 0.001), the sex-linked region is indicated by vertical lines in (B, D, and E).
Discussion
A genome assembly, annotation, and genetic map for dioecious M. annua
Our study provides a draft assembly and annotation of diploid dioecious M. annua (2n = 16). Cytogenetic analysis has confirmed the haploid number of n = 8 chromosomes for diploid M. annua (Durand 1963), with ∼10,000 ORFs assigned to the corresponding eight linkage groups by genetic mapping. Similar to other plants, ∼two-thirds of the M. annua genome comprises repetitive sequences, mostly gypsy LTR, copia LTR, and L1 LINE retrotransposons (Chan et al. 2010; Sato et al. 2011; J. Wang et al. 2012; Rahman et al. 2013). The number of genes in M. annua (34,105) is similar to that of diploid species such as Arabidopsis thaliana (>27,000 genes) and R. communis (>31,000 genes) and is thus compatible with having a long recent history under diploidy rather than polyploidy; for comparison, the gene content of diploid Gossypium is >40,000 genes, pointing to a polyploid history (J. Wang et al. 2012). The draft annotated genome of M. annua will be useful for understanding potential further links between sex determination and sexual dimorphism, as well as the consequences of genome duplication and hybridization (Obbard et al. 2006) for sexual-system transitions in polyploid lineage of the species complex (Pannell et al. 2008).
The size of the nonrecombining Y-linked region of M. annua
Patterns of SNP segregation in small families from 568 ORFs suggests that a substantial length (one-third) of the M. annua Y chromosome has ceased recombining with the X. Lack of recombination on the sex-linked region is further supported by the existence of a Y-specific nonfunctional gene duplication, which could be amplified in populations ranging from Israel to Britain, as well as additional population-genetic data from males and females sampled from across the diploid species’ range. The sex-linked region had slightly, but significantly, higher FST, a higher male/female SNP ratio, and higher heterozygote frequency in males than females, indicating a history of low recombination. Previous mapping of bacterial artificial chromosomes (BACs) found male-specific PCR products distributed over a region of between 52 and 66.82 cM of the sex-linked region, corresponding to 4.86% of the genome, and implying a physical length of between 14.5 and 19 Mb for the nonrecombining genome (Veltsos et al. 2018). The large nonrecombining region of the M. annua Y chromosome contrasts with that of other plants with homomorphic sex chromosomes, which typically have <1% of their Y not recombining, e.g., V. vinifera (Fechter et al. 2012; Picq et al. 2014), F. chiloensis (Tennessen et al. 2016), and Populus species (Paolucci et al. 2010; Geraldes et al. 2015).
Limited gene loss, pseudogenization, and mild purifying selection on the M. annua Y
We found only limited evidence for Y-chromosome degeneration within the nonrecombining region of the M. annua Y chromosome, despite its substantial length. Very few genes have been lost or have become nonfunctional, and only one of the 528 X-linked genes corresponding to the nonrecombining region of the Y (0.2%) did not have a Y-linked homolog. Our analysis may have overlooked genes that have been lost from the Y, because their detection from RNAseq data relies on the presence of polymorphisms on the X copy (see Blavet et al. (2015)) and requires their detectable expression in our sampled tissues. Nevertheless, the absence of evidence of substantial gene loss is consistent with other signatures of only mild Y-chromosome degeneration in M. annua, and is likely reliable.
There was only slightly lower relative expression from the Y compared to the X allele. Allele-specific expression was inferred on the basis of mapping to the reference transcriptome, and we expect only limited mapping bias against expressed Y-specific genes. The fact that different levels of Y/X expression depended on sex-bias (it was reduced for female-biased genes) further suggests that the result is probably not an artifact of the analysis. We found some evidence for relaxed purifying selection on Y alleles compared to X alleles, as expected in genomic regions experiencing limited recombination (Charlesworth and Charlesworth 2000). Specifically, pairwise comparisons of X and Y alleles (after excluding the few with inframe stop codons) showed higher dN/dS than pairwise comparisons of M. annua orthologs with M. huetii or R. communis. Moreover, our tree-based analysis of common orthologs in all species indicated that 28% of the identified Y-linked alleles have experienced an accelerated rate of molecular evolution in the M. annua lineage since its divergence from its sister species M. huetii, which is also dioecious. However, we found no evidence for differential codon usage bias between the Y-linked and other sequences. Codon usage bias is expected to be lower for nonrecombining regions of the genome experiencing weaker purifying selection (Hill and Robertson 1966), and there appears to have been a shift toward less preferred codon usage in R. hastatulus Y-linked genes, increasing in severity with time since the putative cessation of recombination between X and Y chromosomes (Hough et al. 2014). Although our analysis involves multiple statistical tests of significance, so that some results might represent Type 1 error, taken together they point to a very recent cessation of recombination for much of the Y chromosome. In contrast, up to 28% of R. hastatulus Y-linked genes have been lost (Hough et al. 2014) and ∼50% of S. latifolia Y-linked genes are dysfunctional (Papadopulos et al. 2015; Krasovec et al. 2018).
Sex-biased gene expression in M. annua
We found substantial differences in gene expression between males and females of M. annua, consistent with the observation of sexual dimorphism for a number of morphological, life-history, and defense traits in M. annua (Hesse and Pannell 2011; Labouche and Pannell 2016). There were at least four times more male-biased than female-biased genes; this is probably conservative because we did not use a minimum log2FC threshold in our analysis, which would have further increased the proportion of male-biased genes. The observed slight decrease in expression of Y- compared with X-linked alleles suggests that dosage compensation could have begun evolving. However, any such dosage compensation would likely be at a very early stage, not least because YY males (which lack an X chromosome) are viable (Kuhn 1939; Li et al. 2019). Interestingly, sex linkage sometimes influenced male- and female-biased genes differently, with male-biased genes having a higher nucleotide diversity (π) than female-biased genes when sex linked, again perhaps reflecting a history of relaxed purifying selection. Male- and female-biased genes also differed in pairwise dN/dS relative to M. huetii, which was higher for female-biased genes, contrary to expectations under a hypothesis of faster male evolution (Wu and Davis 1993).
Evidence for evolutionary strata on the M. annua Y?
We were unable to find any direct evidence for evolutionary strata with different levels of divergence on the M. annua Y chromosome, but phylogenetic comparisons point to the likely existence of at least two such strata on the M. annua Y. Dioecy evolved in a common ancestor to M. annua and M. huetii (Krähenbühl et al. 2002; Obbard et al. 2006), and both species share the same Y chromosome and sex-determining region, as revealed by crosses (Russell and Pannell 2015) and the possession of common male-specific PCR markers (Veltsos et al. 2018). The phylogeny of sex-linked genes within the shared sex-determining region should have the topology [(M. annua-Y, M. huetii), M. annua-X]. In contrast, our PAML analysis clearly indicates that the phylogeny of most genes in the nonrecombining region of the M. annua Y have the topology [(M. annua-Y, M. annua-X), M. huetii], indicating divergence between the X and Y copies of these genes in M. annua more recently than the species split between M. annua and M. huetii. These results suggest that M. annua and M. huetii share an old (and possibly small) nonrecombining stratum that includes the sex-determining locus, and that a larger stratum has been added to it in the M. annua lineage. Another possible interpretation is gene conversion between X and Y copies within M. annua, as has been reported for humans and some plants (Rautenberg et al. 2008; Wu et al. 2010; Trombetta et al. 2014). This would lead to an underestimate of the size of the ancestral stratum and may have erased evidence of a longer common shared history between the nonrecombining regions of M. annua and M. huetii.
Although most Y-linked genes that we sampled in M. annua point to an X/Y divergence that postdates the species’ split from M. huetii, we found seven genes with a phylogeny consistent with the hypothesized old stratum shared with M. huetii. If these genes are indeed within an older stratum, we might expect them to colocalize in the same part of the nonrecombining region. However, we found that they were quite scattered on the female recombination map. We have no explanation for this pattern, but we note that it would be consistent with a break of synteny between the M. annua Y and X chromosomes (and thus potentially also between the M. huetii and M. annua Y chromosomes) in the region corresponding to the nonrecombining region.
Patterns of divergence in terms of tree-based dS and dN between the X and Y chromosomes of M. annua and their corresponding orthologs in M. huetii and R. communis were largely consistent with the expected phylogenetic relationships between these species (Krähenbühl et al. 2002; Obbard et al. 2006). The increased dS for the Y branch is consistent with a higher mutation rate in male meiosis. Nevertheless, the dS branch length for R. communis was lower than expected for a simple molecular clock across the phylogeny. The recent transition to an annual life cycle in the clade that includes M. annua and M. huetii may be partially responsible for the larger number of mutations that have accumulated along their branches compared to the perennial R. communis (Smith and Donoghue 2008).
Age of the nonrecombining Y-linked region of M. annua
The age of the M. annua nonrecombining region can be estimated on the basis of inferred mutation rates in plants. Ossowski et al. (2010) estimated the mutation rate for A. thaliana to be between 7 × 10−9 and 2.2 × 10−8 per nucleotide per generation. More recently, Krasovec et al. (2018) estimated the mutation rate for S. latifolia to be 7.31 × 10−9. The similarity of these estimates suggests that they might be broadly applicable. Accepting a mutation rate for plants of ∼7.5 × 10−9 and using the synonymous site divergence between X- and Y-linked sequences of M. annua of 0.011 (Table 2), we infer that recombination between most of the X- and Y-linked genes in M. annua may have ceased as recently as 1.5 million generations ago [more recently if we adopt the upper end of the mutation-rate range estimated by Ossowski et al. (2010)]. Given that M. annua is an annual plant, the putative second nonrecombining stratum of the Y chromosome of diploid M. annua could be less than 1 million years old. Krasovec et al. (2018) used their mutation-rate estimate to infer an age of 11 million years for the oldest sex-linked stratum of S. latifolia, i.e., an order of magnitude older than the putatively expanded sex-linked region of M. annua. The few estimates for the time since recombination suppression in other plants with homomorphic sex chromosomes, and those with smaller nonrecombining regions, range from 15 to 31.4 MYA (reviewed in Charlesworth 2016). M. annua thus appears to have particularly young sex chromosomes.
Concluding remarks
We conclude by speculating on a possible role for sexually antagonistic selection in favoring suppressed recombination on the M. annua Y. The canonical model for sex-chromosome evolution supposes that suppressed recombination originally evolves in response to selection to bring sexually antagonistic loci into linkage with the sex-determining locus, and that the Y chromosome begins to degenerate after recombination has ceased (Rice 1987). Given this model, we might expect to see signatures of sexual antagonism in the sex-linked region before the onset of substantial Y-chromosome degeneration. While we do not observe obvious signatures of sexually antagonistic selection, some of the observed patterns in the sex-linked region are difficult to interpret as the outcome of degeneration. For example, while the Y-linked alleles showed slightly lower expression than X-linked alleles, this was not true for male-biased genes. One possible explanation is that the female-biased Y-linked alleles in males might have been driven to lower expression by sexually antagonistic selection. If female-biased expression is beneficial in females (Parisi et al. 2003; Connallon and Clark 2010), the observed enrichment of female-biased genes on the X would be consistent with sexually antagonistic selection because X-linked alleles occur twice as often in females than in males. Similarly, the magnitude of sex-biased gene expression (absolute log2FC) was higher in the sex-linked region, as might be expected if levels of gene expression were the result of an escalation of antagonistic gene expression between the sexes. The possibility that sexually antagonistic selection played a role in the evolution of suppressed recombination might be investigated further by seeking sex-linked QTL for sexually antagonistic traits (Delph et al. 2010), or functional analysis of candidate genes. It would also be valuable to compare expression levels of sex-linked genes with those in a related species with ancestral levels of gene expression, such as orthologs in M. huetii that still lie outside the nonrecombining region of the Y.
Acknowledgments
We thank John Russell for setting up the crosses, Jos Käfer for help with the Galaxy workflow for the SEX-DETector analysis, Jerome Goudet for statistical advice, and Guillaume Cossard, Wen-Juan Ma, Deborah Charlesworth, Stephen Wright, and anonymous reviewers for valuable comments on the manuscript. K.E.R. was supported by grants 31003A_163384 and 31003A_141052 to J.R.P. from the Swiss National Science Foundation, and by the University of Lausanne. P.V. was supported by Sinergia grant 26073998 from the Swiss National Science Foundation to J.R.P., Nicolas Perrin and Mark Kirkpatrick. J.R.P. and D.A.F. acknowledge support from the Natural Environment Research Council (NERC) and Biotechnology & Biological Sciences Research Council (BBSRC), United Kingdom, which funded the early stages of this project. The computations were performed at the server at the department of Plant Sciences, University of Oxford, the Vital-IT Center for high-performance computing of the SIB (Swiss Institute of Bioinformatics; http://www.vital-it.ch), and the Texas Advanced Computing Center. A.M. and G.A.B.M. acknowledge support from Agence nationale de la Recherche (ANR) (grant number ANR-14-CE19-0021-01). V.H., R.H., and B.V. were supported by the Czech Science Foundation (grant 18-06147S). M.A.T. was supported by the National Institutes of Health (NIH) grant R01GM116853 (to Mark Kirkpatrick) and the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement number 715257). Author contributions: conceived the project: J.R.P. and D.A.F.; wet labo-ratory work: D.A.F. and S.C.G.-M.; cytogenetics work: V.H., R.H., and P.V.; genome assembly and annotation: K.E.R.; gene expression analysis: K.E.R., O.E., and P.V.; segregation analysis and mapping: P.V., A.M., P.R., and G.A.B.M.; pop-ulation genetic and divergence analysis: K.E.R. and P.V.; analysis of natural populations: S.C.G.-M. and M.A.T.: wrote the paper: J.R.P., K.E.R., and P.V.; supervised the research: G.A.B.M., D.A.F., and J.R.P; commented and contributed to the final manuscript: all authors.
Footnotes
Supplemental material available at: https://osf.io/a9wjb/.
Communicating editor: S. Wright
Literature Cited
- Akagi T., Henry I. M., Tao R., Comai L., 2014. A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. 10.1126/science.1257225 [DOI] [PubMed] [Google Scholar]
- Albritton S. E., Kranz A. L., Rao P., Kramer M., Dieterich C., et al. , 2014. Sex-biased gene expression and evolution of the x chromosome in nematodes. Genetics 197: 865–883. 10.1534/genetics.114.163311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J., 2010. topGO: enrichment analysis for gene ontology. R package version 2: https://bioconductor.org/packages/release/bioc/html/topGO.html
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Au K. F., Underwood J. G., Lee L., Wong W. H., 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7: e46679 10.1371/journal.pone.0046679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. A., Meadows L. A., Wang J., Dow J. A., Russell S., 2007. Variable sexually dimorphic gene expression in laboratory strains of Drosophila melanogaster. BMC Genomics 8: 454 10.1186/1471-2164-8-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102. 10.1016/j.tree.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Bergero R., Forrest A., Kamau E., Charlesworth D., 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175: 1945–1954. 10.1534/genetics.106.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Gardner J., Bader B., Yong L., Charlesworth D., 2019. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc. Natl. Acad. Sci. USA 116: 6924–6931. 10.1073/pnas.1818486116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blavet N., Blavet H., Muyle A., Käfer J., Cegan R., et al. , 2015. Identifying new sex-linked genes through BAC sequencing in the dioecious plant Silene latifolia. BMC Genomics 16: 546 10.1186/s12864-015-1698-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527 (erratum: Nat. Biotechnol. 34: 888). 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Chan A. P., Crabtree J., Zhao Q., Lorenzi H., Orvis J., et al. , 2010. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 28: 951–956. 10.1038/nbt.1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1991. The evolution of sex chromosomes. Science 251: 1030–1033. 10.1126/science.1998119 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997. 10.1086/283342 [DOI] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. 10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2015. Plant contributions to our understanding of sex chromosome evolution. New Phytol. 208: 52–65. 10.1111/nph.13497 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., 2016. Plant sex chromosomes. Annu. Rev. Plant Biol. 67: 397–420. 10.1146/annurev-arplant-043015-111911 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G. A., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. 10.1038/sj.hdy.6800697 [DOI] [PubMed] [Google Scholar]
- Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., et al. , 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64: 3417–3442. 10.1111/j.1558-5646.2010.01136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dag O., Asar O., Ilk O., 2014. A methodology to implement Box-Cox transformation when no covariate is available. Commun. Stat. Simul. Comput. 43: 1740–1759. 10.1080/03610918.2012.744042 [DOI] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson T. E., Geber M. A., 1999. Sexual dimorphism in physiology and morphology, pp. 175–215 in Gender and Sexual Dimorphism in Flowering Plants, edited by Geber M. A., Dawson T. E., Delph L. F. Springer, Heidelberg, Germany: 10.1007/978-3-662-03908-3_7 [DOI] [Google Scholar]
- Delph D. F., 1999. Sexual dimorphism in life history, pp. 149–173 in Gender and Sexual Dimorphism in Flowering Plants, edited by Geber M. A., Dawson T. E., Delph L. F. Springer, Heidelberg, Germany: 10.1007/978-3-662-03908-3_6 [DOI] [Google Scholar]
- Delph L. F., Arntz A. M., Scotti-Saintagne C., Scotti I., 2010. The genomic architecture of sexual dimorphism in the dioecious plant Silene latifolia. Evolution 64: 2873–2886. [DOI] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B., 1963. Le complèxe Mercurialis annua L. s.l.: Une étude biosystématique. Annales des Sciences Naturelles, Botanique, Paris 12: 579–736. [Google Scholar]
- Durand B., Durand R., 1991. Sex determination and reproductive organ differentiation in Mercurialis. Plant Sci. 80: 49–65. 10.1016/0168-9452(91)90272-A [DOI] [Google Scholar]
- Durand B., Louis J. P., Hamdi S., Cabre E., Yu L. X., et al. , 1987. Major regulator genes, phytohormone levels and specific gene-expression for reproductive organogenesis in Mercurialis annua L. (2n=16). J. Cell. Biochem. 1987: 18–20. [Google Scholar]
- Eckhart V. M., 1999. Sexual dimorphism in flowers and inflorescences, pp. 123–148 in Gender and Sexual Dimorphism in Flowering Plants, edited by Geber M. A., Dawson T. E., Delph L. F. Springer, Heidelberg, Germany: 10.1007/978-3-662-03908-3_5 [DOI] [Google Scholar]
- English A. C., Richards S., Han Y., Wang M., Vee V., et al. , 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7: e47768 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter I., Hausmann L., Daum M., Sörensen T. R., Viehöver P., et al. , 2012. Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol. Genet. Genomics 287: 247–259. 10.1007/s00438-012-0674-z [DOI] [PubMed] [Google Scholar]
- Geraldes A., Hefer C. A., Capron A., Kolosova N., Martinez-Nuñez F., et al. , 2015. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol. Ecol. 24: 3243–3256. 10.1111/mec.13126 [DOI] [PubMed] [Google Scholar]
- Gibson J. R., Chippindale A. K., Rice A. M., 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. Biol. Sci. 269: 499–505. 10.1098/rspb.2001.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Martínez S. C., Ridout K., Pannell J. R., 2017. Range expansion compromises adaptive evolution in an outcrossing plant. Curr. Biol. 27: 2544–2551.e4. 10.1016/j.cub.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R. S., 2007 Improved pairwise alignment of genomic DNA. Ph.D. Thesis, The Pennsylvania State University, State College, PA. [Google Scholar]
- Hesse E., Pannell J., 2011. Sexual dimorphism in a dioecious population of the wind-pollinated herb Mercurialis annua: the interactive effects of resource availability and competition. Ann. Bot. 107: 1039–1045. 10.1093/aob/mcr046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. 10.1017/S0016672300010156 [DOI] [PubMed] [Google Scholar]
- Hobza R., Cegan R., Jesionek W., Kejnovsky E., Vyskot B., et al. , 2017. Impact of repetitive elements on the Y chromosome formation in plants. Genes (Basel) 8: E302 10.3390/genes8110302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz S., Jiménez H., 1967. Cruzamientos interespecíficos e intergenéricos en Caricaceas y sus implicaciones fitotécnicas. Agron. Trop. 17: 323–343. [Google Scholar]
- Hough J., Hollister J. D., Wang W., Barret S. C. H., Wright S. I., 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc. Natl. Acad. Sci. USA 111: 7713–7718. 10.1073/pnas.1319227111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M., Kikuchi T., Sanders M., Newbold C., Berriman M., et al. , 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14: R47 10.1186/gb-2013-14-5-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama Y., Ishii K., Aonuma W., Ikeda T., Kawamoto H., et al. , 2016. A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 6: 18917 10.1038/srep18917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., et al. , 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadka D. K., Nejidat A., Tal M., Golan-Goldhirsh A., 2002. DNA markers for sex: molecular evidence for gender dimorphism in dioecious Mercurialis annua L. Mol. Breed. 9: 251–257. 10.1023/A:1020361424758 [DOI] [Google Scholar]
- Khadka D. K., Nejidat A., Tal M., Golan-Goldhirsh A., 2005. Molecular characterization of a gender-linked DNA marker and a related gene in Mercurialis annua L. Planta 222: 1063–1070. 10.1007/s00425-005-0046-6 [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Chen K., Wylie T., Larson D. E., McLellan M. D., et al. , 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähenbühl M., Yuan Y. M., Küpfer P., 2002. Chromosome and breeding system evolution of the genus Mercurialis (Euphorbiaceae): implications of ITS molecular phylogeny. Plant Syst. Evol. 234: 155–169. 10.1007/s00606-002-0208-y [DOI] [Google Scholar]
- Krasovec M., Chester M., Ridout K., Filatov D. A., 2018. The mutation rate and the age of the sex chromosomes in Silene latifolia. Curr. Biol. 28: 1832–1838.e4. 10.1016/j.cub.2018.04.069 [DOI] [PubMed] [Google Scholar]
- Kuhn E., 1939. Selbstbestäubungen subdiöcischer Blütenpflanzen, ein neuer Beweis für die genetische Theorie der Geschlechtsbestimmung. Planta 30: 457–470. 10.1007/BF01917065 [DOI] [Google Scholar]
- Labouche A. M., Pannell J. R., 2016. A test of the size-constraint hypothesis for a limit to sexual dimorphism in plants. Oecologia 181: 873–884. 10.1007/s00442-016-3616-3 [DOI] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1999. Four evolutionary strata on the human X chromosome. Science 286: 964–967 (erratum: Science 286: 2273). 10.1126/science.286.5441.964 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder E. H., Cano J. M., Leinonen T., O’Hara R. B., Nikinmaa M., et al. , 2010. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol. Biol. Evol. 27: 1495–1503. 10.1093/molbev/msq031 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Veltsos P., Cossard G., Gerchen J., Pannell J.R., 2019 YY males of the dioecious plant Mercurialis annua are fully viable but produce largely infertile pollen. New Phytol. (in press). 10.1101/658708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Moore P. H., Ma H., Ackerman C. M., Raglba M., et al. , 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. 10.1038/nature02228 [DOI] [PubMed] [Google Scholar]
- Loeptien H., 1979. Identification of the sex chromosome pair in asparagus (Asparagus officinalis L.). Zeitschrift fur Pflanzenzüchtung 82: 162–173. [Google Scholar]
- Lüdecke, D., 2017 sjPlot: data visualization for statistics in social science. R package version 2.4.0. Accessed November 2018. Available at: https://CRAN.R-project.org/package=sjPlot.
- Luo R., Liu B., Xie Y., Li Z., Huang W., et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18 [corrigenda: Gigascience 4: 30 (2015)]. 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J. E., 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29: 677–683. 10.1016/j.tig.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Mank J. E., Ellegren H., 2009. Are sex-biased genes more dispensable. Biol. Lett. 5: 409–412. 10.1098/rsbl.2008.0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., Malone J. H., Clark A. G., 2012. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 22: 1255–1265. 10.1101/gr.132100.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty J. N., Nayak S., Jha S., Joshi R. K., 2017. Transcriptome profiling of the floral buds and discovery of genes related to sex-differentiation in the dioecious cucurbit Coccinia grandis (L.). Voigt. Gene 626: 395–406. 10.1016/j.gene.2017.05.058 [DOI] [PubMed] [Google Scholar]
- Montgomery S. H., Mank J. E., 2016. Inferring regulatory change from gene expression: the confounding effects of tissue scaling. Mol. Ecol. 25: 5114–5128. 10.1111/mec.13824 [DOI] [PubMed] [Google Scholar]
- Morgulis A., Gertz E. M., Schäffer A. A., Agarwala R., 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J. Comput. Biol. 13: 1028–1040. 10.1089/cmb.2006.13.1028 [DOI] [PubMed] [Google Scholar]
- Muyle A., Käfer J., Zemp N., Mousset S., Picard F., et al. , 2016. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biol. Evol. 8: 2530–2543. 10.1093/gbe/evw172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K., Ellegren H., 2008. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics 180: 1131–1136. 10.1534/genetics.108.090324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z., 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Harris S. A., Buggs R. J. A., Pannell J., 2006. Hybridization, polyploidy, and the evolution of sexual systems in Mercurialis (Euphorbiaceae). Evolution 60: 1801–1815. 10.1111/j.0014-3820.2006.tb00524.x [DOI] [PubMed] [Google Scholar]
- Ono T., 1939. Polyploidy and sex determination in Melandrium. I. Colcicine-induced polyploids of Melandrium album. Botanical Magazine 53: 549–556. 10.15281/jplantres1887.53.549 [DOI] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. 10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell J., Dorken M. E., Pujol B., Berjano R., 2008. Gender variation and transitions between sexual systems in Mercurialis annua (Euphorbiaceae). Int. J. Plant Sci. 169: 129–139. 10.1086/523360 [DOI] [Google Scholar]
- Paolucci I., Gaudet M., Jorge V., Beritognolo I., Terzoli S., et al. , 2010. Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genetics &. Genomes 6: 863–875. [Google Scholar]
- Papadopulos A. S., Chester M., Ridout K., Filatov D. A., 2015. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc. Natl. Acad. Sci. USA 112: 13021–13026. 10.1073/pnas.1508454112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., et al. , 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700. 10.1126/science.1079190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J., Ellegren H., 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 14: 83–87. 10.1038/nrg3376 [DOI] [PubMed] [Google Scholar]
- Picq S., Santoni S., Lacombe T., Latreille M., Weber A., et al. , 2014. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol. 14: 229 10.1186/s12870-014-0229-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucholt P., Rönnberg-Wästljung A. C., Berlin S., 2015. Single locus sex determination and female heterogamety in the basket willow (Salix viminalis l.). Heredity 114: 575–583. 10.1038/hdy.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucholt P., Wright A. E., Liu Conze L., Mank J. E., Berlin S., 2017. Recent sex chromosome divergence despite ancient dioecy in the willow Salix viminalis. Mol. Biol. Evol. 34: 1991–2001. 10.1093/molbev/msx144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J., Kubat Z., Kejnovsky E., Jesionek W., Cizkova J., et al. , 2018. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19: 153 10.1186/s12864-018-4547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Bergero R., Forrest A., Kaiser V. B., Charlesworth D., 2010. Nucleotide diversity in Silene latifolia autosomal and sex-linked genes. Proc. Biol. Sci. 277: 3283–3290. 10.1098/rspb.2010.0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt H. J., Pühler A., Broer I., 1993. Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 6: 699–706. 10.1094/MPMI-6-699 [DOI] [Google Scholar]
- Rahman A. Y., Usharraj A. O., Misra B. B., Thottathil G. P., Jayasekaran K., et al. , 2013. Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics 14: 75 10.1186/1471-2164-14-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 33: 3726–3732. 10.1093/bioinformatics/btx494 [DOI] [PubMed] [Google Scholar]
- Rautenberg A., Filatov D., Svennblad B., Heidari N., Oxelman B., 2008. Conflicting phylogenetic signals in the SlX1/Y1 gene in Silene. BMC Evol. Biol. 8: 299 10.1186/1471-2148-8-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2007 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing. Vienna. ISBN 3–900051–07–0, Available at: http://www.R-project.org.
- Renner S. S., 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101: 1588–1596. 10.3732/ajb.1400196 [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. 10.1111/j.1558-5646.1987.tb05864.x [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. R. W., Pannell J., 2015. Sex determination in dioecious Mercurialis annua and its close diploid and polyploid relatives. Heredity 114: 262–271. 10.1038/hdy.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson B. J., Wang L., Tiffin P., Wu Z., Olson M. S., 2018. Sex-biased gene expression in flowers, but not leaves, reveals secondary sexual dimorphism in Populus balsamifera. New Phytol. 221: 527–539. [DOI] [PubMed] [Google Scholar]
- Sato S., Hirakawa H., Isobe S., Fukai E., Watanabe A., et al. , 2011. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 18: 65–76. 10.1093/dnares/dsq030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R., 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E., Künstner A., Fraser B. A., Zipprich G., Kottler V. A., et al. , 2014. Transcriptome assemblies for studying sex-biased gene expression in the guppy, Poecilia reticulata. BMC Genomics 15: 400 10.1186/1471-2164-15-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smarda P., Bureš P., Smerda J., Horová L., 2012. Measurements of genomic GC content in plant genomes with flow cytometry: a test for reliability. New Phytol. 193: 513–521. 10.1111/j.1469-8137.2011.03942.x [DOI] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 2013 RepeatMasker Open-4.0. Accessed February 2019. Available at: http://www.repeatmasker.org.
- Smith B., 1955. Sex chromosomes and natural polyploidy in dioecious Rumex. J. Hered. 46: 226–232. 10.1093/oxfordjournals.jhered.a106563 [DOI] [Google Scholar]
- Smith S. A., Donoghue M. J., 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322: 86–89. 10.1126/science.1163197 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber A., Jamsari A., Kinney M. S., Pires J. C., Jung C., 2007. Genetic and physical maps around the sex-determining M-locus of the dioecious plant Asparagus. Mol. Genet. Genomics 278: 221–234. 10.1007/s00438-007-0235-z [DOI] [PubMed] [Google Scholar]
- Tennessen J. A., Govindarajulu R., Liston A., Ashman T. L., 2016. Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex‐determining region. New Phytol. 211: 1412–1423. 10.1111/nph.13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toups M., Rodrigues N., Perrin N., Kirkpatrick M., 2019. A reciprocal translocation radically reshapes sex‐linked inheritance in the common frog. Mol. Ecol. 28: 1877–1889. 10.1111/mec.14990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta B., Sellitto D., Scozzari R., Cruciani F., 2014. Inter- and intraspecies phylogenetic analyses reveal ex-tensive X-Y gene conversion in the evolution of gametologous sequences of human sex chromosomes. Mol. Biol. Evol. 31: 2108–2123. 10.1093/molbev/msu155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkogeorga G., Cahais V., Galtier N., 2012. The population genomics of a fast evolver: high levels of diversity, functional constraint, and molecular adaptation in the tunicate Ciona intestinalis. Genome Biol. Evol. 4: 740–749. 10.1093/gbe/evs054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P., Cossard G., Beaudoing E., Beydon G., Savova Bianchi D., et al. , 2018. Size and content of the sex-determining region of the Y chromosome in dioecious Mercurialis annua, a plant with homomorphic sex chromosomes. Genes (Basel) 9: E277 10.3390/genes9060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurture G. W., Sedlazeck F. J., Nattestad M., Underwood C. J., Fang H., et al. , 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33: 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Na J. K., Yu Q., Gschwend A. R., Han J., et al. , 2012. Sequencing papaya X and Y-h chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl. Acad. Sci. USA 109: 13710–13715. 10.1073/pnas.1207833109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wang Z., Li F., Ye W., Wang J., et al. , 2012. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44: 1098–1103. 10.1038/ng.2371 [DOI] [PubMed] [Google Scholar]
- Westergaard M., 1958. The mechanism of sex determination in dioecious splants. Adv. Genet. 9: 217–281. 10.1016/S0065-2660(08)60163-7 [DOI] [PubMed] [Google Scholar]
- Wright A. E., Darolti I., Bloch N. I., Oostra V., Sandkam B., et al. , 2017. Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nat. Commun. 8: 14251 10.1038/ncomms14251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Davis A. W., 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. Am. Nat. 142: 187–212. 10.1086/285534 [DOI] [PubMed] [Google Scholar]
- Wu M., Moore R. C., 2015. The evolutionary tempo of sex chromosome degradation in Carica papaya. J. Mol. Evol. 80: 265–277. 10.1007/s00239-015-9680-1 [DOI] [PubMed] [Google Scholar]
- Wu T. D., Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Wu X., Wang J., Na J. K., Yu Q., Moore R. C., et al. , 2010. The origin of the non-recombining region of sex chromosomes in Carica and Vasconcellea. Plant J. 63: 801–810. 10.1111/j.1365-313X.2010.04284.x [DOI] [PubMed] [Google Scholar]
- Xue W., Li J. T., Zhu Y. P., Hou G. Y., Kong X. F., et al. , 2013. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics 14: 604 10.1186/1471-2164-14-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Oda Y., Haseda A., Fujito S., Mikami T., et al. , 2014. Molecular evidence that the genes for dioecism and monoecism in Spinacia oleracea L. are located at different loci in a chromosomal region. Heredity 112: 317–324. 10.1038/hdy.2013.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zemp N., Tavares R., Muyle A., Charlesworth D., Marais G. A., et al. , 2016. Evolution of sex-biased gene expression in a dioecious plant. Nat. Plants 2: 16168 10.1038/nplants.2016.168 [DOI] [PubMed] [Google Scholar]
- Li X., Veltsos P., Cossard G., Gerchen J., Pannell J. R., 2019. YY males of the dioecious plant Mercurialis annua are fully viable but produce largely infertile pollen. New Phytol. (in press). .org/10.1101/658708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw DNA and RNA sequence data generated in this study have been submitted to NCBI under accession SRP098613 BioProject ID PRJNA369310. Scripts, supplemental files, and intermediate files are available at https://osf.io/a9wjb/. File S1 contains the genetic map data and other information on each mapped transcript. File S2 contains the Sanger sequence of clones containing the X and Y variants, where there is a premature stop codon on the Y, and the script used to make the phylogenetic tree. File S3 summarizes the GO results for sex-biased genes. File S4 contains transcriptome annotation information. File S5 contains the assembled transcriptome and X and Y haplotype predictions from SEX-DETector. File S6 contains the genome and predicted genes on it. File S7 is the M. annua repeat library.