Genome integrity is fundamental to viability and health and can be impacted by metabolic alterations that affect chromatin composition. Saatchi and Kirchmaier present evidence that loss of fumarase, an ortholog of the tumor suppressor...
Keywords: fumarate, DNA replication stress, histone methylation, HTZ1, JHD2
Abstract
Fumarase is a well-characterized TCA cycle enzyme that catalyzes the reversible conversion of fumarate to malate. In mammals, fumarase acts as a tumor suppressor, and loss-of-function mutations in the FH gene in hereditary leiomyomatosis and renal cell cancer result in the accumulation of intracellular fumarate—an inhibitor of α-ketoglutarate-dependent dioxygenases. Fumarase promotes DNA repair by nonhomologous end joining in mammalian cells through interaction with the histone variant H2A.Z, and inhibition of KDM2B, a H3 K36-specific histone demethylase. Here, we report that Saccharomyces cerevisiae fumarase, Fum1p, acts as a response factor during DNA replication stress, and fumarate enhances survival of yeast lacking Htz1p (H2A.Z in mammals). We observed that exposure to DNA replication stress led to upregulation as well as nuclear enrichment of Fum1p, and raising levels of fumarate in cells via deletion of FUM1 or addition of exogenous fumarate suppressed the sensitivity to DNA replication stress of htz1Δ mutants. This suppression was independent of modulating nucleotide pool levels. Rather, our results are consistent with fumarate conferring resistance to DNA replication stress in htz1Δ mutants by inhibiting the H3 K4-specific histone demethylase Jhd2p, and increasing H3 K4 methylation. Although the timing of checkpoint activation and deactivation remained largely unaffected by fumarate, sensors and mediators of the DNA replication checkpoint were required for fumarate-dependent resistance to replication stress in the htz1Δ mutants. Together, our findings imply metabolic enzymes and metabolites aid in processing replicative intermediates by affecting chromatin modification states, thereby promoting genome integrity.
ALL organisms have developed mechanisms to detect, signal and repair damaged DNA to ensure accurate and complete duplication, and inheritance of their genome. Genomic instability is a major driver of tumorigenesis, and multiple factors contribute to genome instability, including failure to properly repair damaged DNA caused by endogenous sources like errors during DNA replication or exogenous agents including ultraviolet (UV) light or chemicals. Perturbed replication contributes to early genomic instability in cancers (Bartkova et al. 2005; Gorgoulis et al. 2005), and replication stress can promote tumorigenesis in mice (Bilousova et al. 2005). In the past few years, metabolic enzymes and metabolites including fumarate hydratase (also called fumarase) and fumarate, succinate dehydrogenase (SDH) and succinate, as well as isocitrate dehydrogenase (IDH) and R-2-hydroxyglutarate (R-2-HG) have emerged as modulators of DNA damage responses in bacteria, yeast, and mammals (Yogev et al. 2010; Jiang et al. 2015; Singer et al. 2017; Sulkowski et al. 2017, 2018; Leshets et al. 2018).
Fumarase is a well-characterized TCA cycle enzyme found in the mitochondria that catalyzes the reversible reaction of converting fumarate to malate (Woods et al. 1988). Fumarase is also present in the cytosol in organisms ranging from yeast to humans (Tolley and Craig 1975; Akiba et al. 1984; Yogev et al. 2011). In yeast, both cytosolic and mitochondrial fumarase are encoded by a single gene, FUM1 (Wu and Tzagoloff 1987). Yeast Fum1p contains an N-terminal sequence that is processed in the mitochondrial matrix (Stein et al. 1994; Sass et al. 2001). Rapid folding of mature Fum1p inhibits its import into mitochondria, and a subset of processed fumarase is released back into the cytosol by retrograde movement (Knox et al. 1998; Karniely and Pines 2005).
Fumarase also acts as a tumor suppressor, and defects in the gene encoding fumarase (FH) in humans are commonly found in hereditary leiomyomatosis and renal cell cancer (HLRCC) (Launonen et al. 2001; Kiuru et al. 2002; Tomlinson et al. 2002; Lehtonen et al. 2004; Menko et al. 2014) as well as in glioblastomas, neuroblastomas, and other cancers (Khalil 2007; Fieuw et al. 2012). Recent studies have provided a link between fumarase plus the metabolite fumarate and genome integrity, revealing a previously underappreciated way in which such metabolic defects have the potential to contribute to tumorigenesis. Among the first evidence for the role of fumarase in maintaining genome integrity in eukaryotes emerged from studies in the budding yeast Saccharomyces cerevisiae that showed fumarase promotes growth upon exposure to various types of DNA damage, and that fumarate, but not malate, suppresses double-stranded DNA break (DSB) sensitivity of cells lacking the cytosolic form of fumarase (Yogev et al. 2010). Recently, yeast fumarase has been shown to promote homologous recombination through interaction with, and stabilization of, Sae2p, which is associated with the MRX complex at DSBs during DNA end resection (Leshets et al. 2018).
Chromatin modifications, including histone methylation, also play a central role in regulation of DNA damage responses for various types of DNA damage in organisms ranging from yeast to humans (House et al. 2014; Hauer and Gasser 2017), and recent studies have begun to uncover links between metabolic enzymes plus metabolites, including fumarase plus fumarate, and chromatin during DNA damage responses. Fumarate can modulate histone methylation by acting as a competitive inhibitor of α-ketoglutarate (α-KG)-dependent dioxygenases, including JmjC-domain-containing histone demethylases (Xiao et al. 2012; Jiang et al. 2015). During DSB repair by nonhomologous end joining (NHEJ) in humans, fumarase is recruited to chromatin at the site of DSBs through DNA-PK-dependent phosphorylation as well as interaction with the histone variant H2A.Z (Jiang et al. 2015). H2A.Z transiently becomes associates with DSBs (Kalocsay et al. 2009; Xu et al. 2012) through the actions of the H2A.Z/Htz1p-specific chromatin remodeling complexes SWR1C and INO80C (Krogan et al. 2003; Mizuguchi et al. 2004; Papamichos-Chronakis et al. 2006, 2011; van Attikum et al. 2007; Lademann et al. 2017). Mammalian H2A.Z and the budding yeast ortholog Htz1p can promote DNA repair by NHEJ as well as homologous recombination (Papamichos-Chronakis et al. 2011; Xu et al. 2012). In human cells, fumarase is proposed to produce high local concentrations of fumarate at sites of DNA damage as the activity of fumarase can be detected in chromatin fractions after exposure to irradiation, IR, and fumarate (but not malate) improves repair by NHEJ through inhibition of the H3 K36-specific lysine demethylase KDM2B (Jiang et al. 2015). Moreover, nuclear localization of human fumarase or depletion of KDM2B promotes cell survival after exposure to IR (Jiang et al. 2015).
Deletion of FUM1 in yeast, and loss of the catalytic activity of fumarase in human cells, or loss of function mutations in fumarase in HLRCC tumors, cause accumulation of fumarate to high cellular levels (several hundred-fold increase in yeast, millimolar levels in humans) (Arikawa et al. 1999; Pollard et al. 2005; Lin et al. 2011; Sulkowski et al. 2018). Also, elevated levels of fumarate or succinate [another competitive inhibitor of α-KG-dependent dioxygenases (Xiao et al. 2012; Laukka et al. 2016)] correlate with elevated levels of DSBs in patient-derived HLRCC and SDH-related hereditary paraganglioma and pheochromocytoma, SDH PGL/PCC (Sulkowski et al. 2018). However, how such changes in metabolite availability affect DNA repair and other cellular functions is poorly understood.
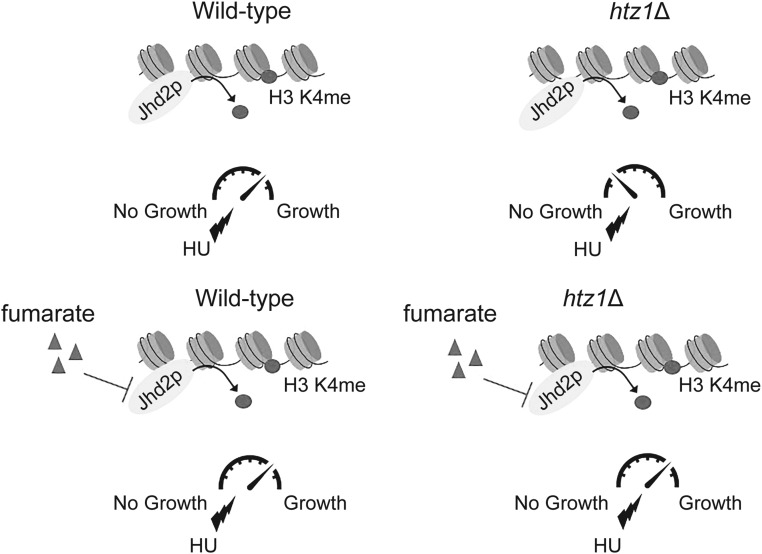
Here, we explored the relationship between yeast Fum1p, the metabolite fumarate, and Htz1p during DNA replication stress. We demonstrate that yeast fumarase was induced, and enriched in the nuclei upon treatment with hydroxyurea (HU), and observed synthetic genetic interaction upon exposure to HU in cells lacking FUM1 and HTZ1. We further demonstrate that exogenous fumarate suppressed the DNA replication stress sensitivity of htz1Δ mutants in a manner independent of modulating nucleotide pools, but dependent on components required for activation of the intra-S phase checkpoint, also known as the S Phase checkpoint. In the presence of fumarate, intra-S phase checkpoint activation and adaptation (as measured by phosphorylation status of Rad53p) remained largely intact. Consistent with fumarate promoting histone methylation to confer resistance to DNA replication stress, deletion of the JmjC domain-containing Jhd2p, a H3 K4 demethylase (Liang et al. 2007; Seward et al. 2007; Tu et al. 2007), was sufficient to confer resistance to HU in htz1Δ mutants, and this suppression required H3 K4 methylation. Moreover, loss of FUM1, or addition of exogenous fumarate, inhibited Jhd2p-dependent demethylation of H3 K4 in vivo. Together, our findings highlight a fumarate-sensitive role for Jhd2p and histone methylation in responses to DNA replication stress as well as link Htz1p to proper processing of replicative intermediates through the DNA replication checkpoint during intra-S phase checkpoint activation.
Materials and Methods
Yeast strains and plasmid construction
Yeast strains and plasmids used in this study are listed in Supplemental Material, Table S1 and Table S2, respectively. Oligonucleotides used to generate yeast strains or plasmids are listed in Table S3. Yeast strains containing deletions of open reading frames were generated by standard PCR-based gene disruption strategies (Guthrie and Fink 1991). Strains expressing histone mutants were made by plasmid shuffling (Adams et al. 1998).
Growth assay of sensitivity to DNA damaging agents
Cells were grown overnight in rich (YPD) medium, diluted to 104 cells/µl, and 3 µl of 10-fold serial dilutions were spotted onto YPD containing 2×PBS (274 mM NaCl, 16 mM Na2HPO4, 4 mM KH2PO4, 5.4 mM KCl) with or without noted amounts of HU or camptothecin, or were exposed to indicated doses of UV in the presence or absence of monoethyl fumarate (Cat. no. 128422; Sigma). Images were taken after 2–3 days of growth at 30°. The ethyl ester modification on fumarate facilitates cell permeability (MacKenzie et al. 2007). This ethyl group can then be removed by cytosolic esterases, releasing the metabolite fumarate.
Preparation of yeast nuclear extracts
Nuclear extracts were prepared from 200 ml cultures grown in YPD to an OD600 of 0.6 and treated with or without 200 mM HU for 3 hr as described by Miller et al. (2008). Briefly, cells were harvested, washed with ice-cold water and resuspended in 3 ml of spheroplasting buffer (1 mM sorbitol, 50 mM potassium phosphate pH 6.5, 14 mM β-mercaptoethanol). Next, cells were pelleted, resuspended in 3 ml of spheroplasting buffer containing 5 mg/ml of lyticase (Cat. no. L4025; Sigma), and then incubated at 30° until spheroplasted. Sepheroplasted cells were pelleted at 5000 × g for 5 min at 4°, and then washed in 3 ml of spheroplasting buffer. Cells were pelleted, resuspended in 5 ml of lysis buffer (18% Ficoll 400, 20 mM potassium phosphate pH 6.8, 1 mM MgCl2, 0.5 mM EDTA, 1 mM PMSF, 1 μg/ml Leupeptin/Pepstatin mix), lysed with 20 strokes using a Dounce homogenizer, and separated by centrifugation at 3000 × g for 10 min to remove cell debris. The nuclei were pelleted at 50,000 × g for 30 min at 4° using a SW-41 rotor. Nuclei were resuspended in 200 μl of NP buffer (0.34 mM sucrose, 20 mM KCl, 5 mM MgCl2, 1 mM PMSF, 1.0 μg\ml Leupeptin/Pepstatin mix) for storage at 4°.
Preparation of whole cell extracts
Three milliliters of yeast cultures grown overnight in YPD to an OD600 of 0.8 were harvested, flash frozen on dry ice, and stored at −80°. Cell pellets were resuspended in 250 μl of 2.0 M NaOH containing 8% β-mercaptoethanol, incubated on ice for 5 min, and pelleted by centrifugation at 14,500 × g at 4° for 2 min. Cell pellets were resuspended in 250 μl of high salt extraction buffer (40 mM HEPES NaOH pH 7.5, 350 mM NaCl, 0.1% Tween 20, 10% glycerol), and repelleted by centrifugation at 14,500 × g at 4° for 2 min. Pellets were resuspended in 2×SDS sample buffer (200 mM Tris-HCl pH 6.8, 20% SDS, 20% glycerol, 0.08% bromophenol blue, 10% β-mercaptoethanol), prior to loading onto 7 or 12% SDS-PAGE gels for immunoblotting (see below).
Immunoblotting
Analyses of Fum1-GFP localization:
Whole cell extracts or nuclear fractions were prepared from logarithmically growing cultures (as described above) from indicated genotypes and treated with or without 200 mM HU for 3 hr, and separated by electrophoresis on 12% SDS-PAGE gels. Following electrophoresis, proteins were transferred to PVDF membrane that had been presoaked in methanol for 5 min followed by soaking in transfer buffer (25 mM Tris and 1.44% glycine pH 8.3, 20% methanol, 0.02% SDS). Next, membranes were incubated with 5% milk in PBS-T (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, 0.1%Tween 20) for 1 hr at room temperature followed by incubation with anti-GFP antibody (ab290, 1:5000 in 2% milk in PBS-T; Abcam) overnight at 4°. Membranes were washed three times in PBS-T for 10 min each. ECL anti-rabbit Horseradish-peroxidase-linked IgG (NA934, 1:10,000; Amersham) was used as secondary antibody. Membranes were washed again as above and visualized by adding 1 ml Luminata Crescendo Western HRP Substrate (Cat. no. WBLUR0500; Millipore) on the membrane for 1 min followed by imaging using the ChemiDoc XRS+. Blots were then quantified using Image Lab software. Membranes were stripped by 0.2 M NaOH at room temperature and reprobed with anti-Proliferating Cell Nuclear Antigen (1:10,000) antibody (Daganzo et al. 2003; Franco et al. 2005) and ECL anti-rabbit Horseradish-peroxidase-linked IgG (NA934, 1:10,000; Amersham) as secondary antibody. Fold enrichment of Fum1p was calculated relative to PCNA as shown in Figure 1 legend.
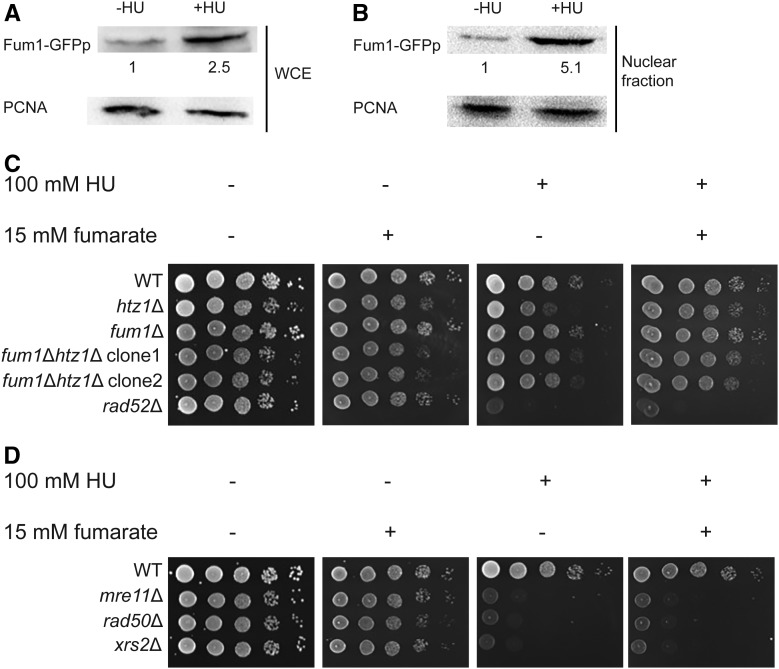
Figure 1.
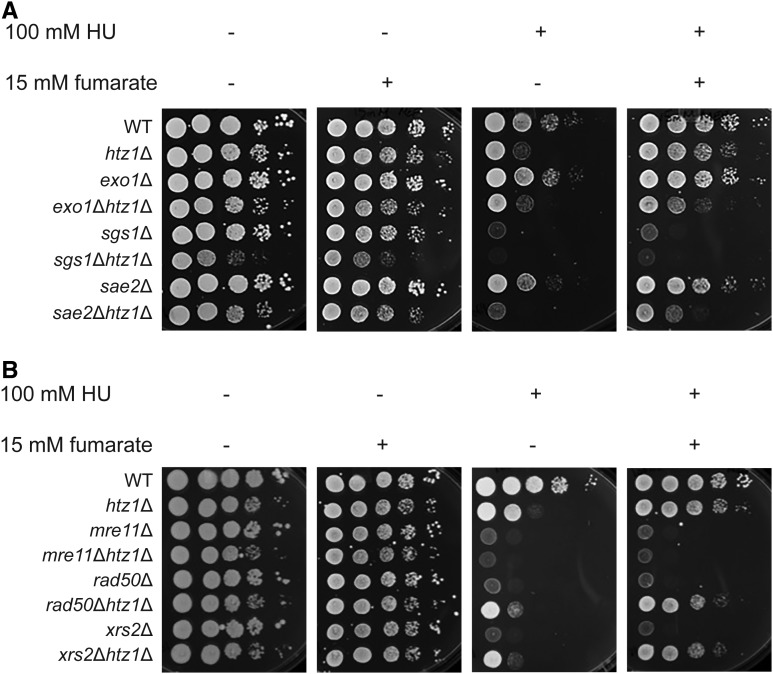
Fumarate can complement sensitivity of htz1Δ mutants to DNA replication stress. (A and B) Expression of Fum1p is induced, and Fum1p becomes enriched in the nuclear fraction upon exposure to hydroxyurea, HU. Yeast expressing Fum1-GFPp were incubated in the absence or presence of 200 mM HU at 30° for 3 hr. Whole cell extracts (A), or nuclear fractions (B) were analyzed by immunoblotting using anti-GFP, and anti-PCNA antibodies. A representative immunoblot and fold enrichment of Fum1p from two independent experiments is shown. Levels of Fum1-GFPp were normalized to levels of PCNA (loading control), then expressed relative to signal that was observed in the absence of HU, which was set to 1. . (C) Genetic interaction between fum1Δ and htz1Δ mutants. (D) The effect of exogenous fumarate on DNA replication stress in mre11Δ, rad50Δ and xrs2Δ mutants. (C and D) Cells with genotypes as indicated were grown overnight in rich (YPD) medium, then 3 μl of 10-fold serial dilutions were spotted onto YPD medium containing the indicated concentrations of fumarate and/or HU, and incubated at 30° for 2 days prior to imaging.
Analyses of H3 methylation levels:
Logarithmically growing cultures (1 × 107 cells) were harvested before and after a 6-hr treatment with 5 mM monoethyl fumarate. Whole cell extracts were prepared as described above, and separated on 12% SDS-PAGE gels. Transfer and blocking steps were performed as described above, and membranes were incubated with anti-H3K4me3 antibody (39159, 1:5000; Active Motif). Membranes were washed, incubated with ECL anti-rabbit secondary antibody, developed and imaged as described above. Membranes were stripped and reprobed using anti-H3 antibody (ab1791, 1:5000; Abcam), H3 K4me2 antibody (07-030, 1:5000; EMD Millipore), H3 K36me3 (ab9050, 1:5000; Abcam, or 9763S, 1:1000; Cell Signaling Technology), or H3 K79me3 (ab195500, 1:5000; Abcam) for 1 hr at room temperature or overnight at 4°. ECL anti-rabbit horseradish-peroxidase-linked IgG (NA934, 1:10,000; Amersham) were used as secondary antibody, and blots were washed, developed, and imaged as above.
Analyses of FLAG-Jhd2p levels:
Indicated strains carrying plasmids for expression of FLAG-Jhd2p (Mersman et al. 2009) were grown in selective media (lacking leucine), and 1 × 107 cells from logarithmically growing cultures were harvested. Whole cell extracts were prepared as described previously and loaded on 10% SDS-PAGE gels. Transfer and blocking steps were done as above and membranes were incubated with anti-FLAG antibody (ab1162, 1:5000; Abcam) for 1 hr at room temperature. Membranes were washed, incubated with ECL anti-rabbit horseradish-peroxidase-linked IgG (NA934, 1:10,000; Amersham), developed and imaged as above. Next, membranes were stripped and reprobed using anti-PGK1 antibody (A-6457, 1:5000; Molecular Probes) for 1 hr at room temperature, washed, incubated with ECL anti-mouse horseradish-peroxidase-linked IgG (NA931, 1:10,000; Amersham) as secondary antibody, and developed and imaged as described earlier.
Analyses of Rad53p phosphorylation:
Logarithmically growing cells were treated with noted amounts of DNA damaging agents with or without 5 mM monoethyl fumarate in YPD containing 2× PBS. Aliquots of 3 × 107 cells were collected before treatment, plus 30 min, 1 hr and then every 2 hr after treatment for 8 hr, and whole cells extracts were prepared as described above, then loaded onto 7% SDS-PAGE gels. Proteins were transferred to PVDF membranes as described above. Membranes were blocked in 5% milk in PBS-T for 1 hr at room temperature, then incubated with anti-Rad53p antibodies (ab104232, 1:2000; Abcam) overnight at 4°. Membranes were washed three times in PBS-T for 10 min each, followed by incubation with ECL anti-rabbit Horseradish-peroxidase-linked IgG (NA934, 1:10,000; Amersham). Blots were developed and imaged as described above.
Analysis of JHD2 mRNA expression by qRT-PCR
Indicated strains carrying an empty vector or a plasmid for overexpression of FLAG-JHD2 were grown in selective medium (Complete Supplement Medium lacking leucine). Logarithmically growing cultures (5 × 107 cells) were harvested as described by Schmitt et al. (1990). Total RNA (1 μg) was incubated with 1 U DNaseI (M6101; Promega) for 1 hr at 37°, and DNase I was inactivated by addition of stop solution (20 mM EGTA, pH 8.0) and incubation at 65° for 10 min. DNase I-treated RNA was used for cDNA synthesis using random hexamer primers, and 200 unit M-MLV reverse transcriptase (28025013; Thermofisher). cDNAs were diluted 1:100 and used to analyze transcript levels by qPCR using primers listed in Table S3 and SYBR Green PCR master mix (A25741; Fisher Scientific) following manufacturer’s instructions. Quantification was analyzed by the comparative CT method. Average and SD of three independent experiments were reported.
Analysis of cell cycle by flow cytometry
Logarithmically growing cells with the indicated genotypes were grown at 30° in rich medium (YPD) to OD600 ∼0.4. Cells were arrested in G1 by addition of α-factor at final concentration of 10 μg/ml for 3 hr. Cells were washed three times with YPD, resuspended in YPD containing 2× PBS and 100 μg/ml protease, and treated with 100 or 200 mM HU, with or without 5 mM monoethyl fumarate. One ml aliquots of cells were collected prior to HU treatment, and again every 20 min after release from G1 for 4 hr. Cells were pelleted by centrifugation, resuspended in 70% ethanol, and incubated at room temperature for 1 hr before storing overnight at 4°. Cells were then washed twice in FACS buffer (200 mM Tris-HCl pH 7.5, 20 mM EDTA), and resuspended in 100 μl of FACS buffer containing 0.1% RNase, then incubated for 2 hr at 37°. Cells were washed with 1× PBS, and incubated in 100 μl of propidium iodide solution (0.05 mg/ml propidium iodide in 1× PBS) overnight in the dark at 4°. Prior to analysis, 400 μl of 1×PBS was added to each sample. Samples were briefly sonicated (Branson Sonifier 450; VWR Scientific) and analyzed by Beckman Coulter Cytoflex S, and FlowJo software (version 7.6.5).
Statistical analysis
H3 K4 me3 and H3 K4me2 levels were normalized to H3 levels, and JHD2 trancript levels were normalized to ACT1. Assays were conducted in at least triplicate, and statistical analyses for immunoblots and qRT-PCR were conducted using the Wilcoxon Rank Sum test with MSTAT v6.3 (https://mcardle.wisc.edu/mstat/).
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the findings of the article are present within the article, figures and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8109437.
Results
Loss of FUM1 suppresses sensitivity to replication stress in htz1 mutants
Fumarase has previously been implicated in DSB repair (Yogev et al. 2010; Jiang et al. 2015; Leshets et al. 2018; Sulkowski et al. 2018). To assess the impact of fumarase on responses to DNA replication stress, we first analyzed expression and cellular localization of Fum1p in S. cerevisiae upon exposure to HU. Logarithmically growing yeast expressing Fum1p C-terminally tagged with GFP were treated with HU for 3 hr, and expression and nuclear localization of Fum1p were monitored by quantitative protein blots. As shown in Figure 1A, after treatment with HU, Fum1p levels in whole cell extracts increased more than twofold, and Fum1p became enriched in the nuclear fraction by more than fivefold (Figure 1B). This further enrichment of Fum1p in the nuclear fraction implied DNA replication stress had triggered localization of Fum1p to the nucleus (see also Yogev et al. 2010).
In humans, fumarase is recruited to chromatin during NHEJ-mediated repair of DSBs through interaction with the histone variant H2A.Z, and depletion of H2A.Z reduces enrichment of fumarase at sites of DSB (Jiang et al. 2015). In yeast, Htz1p promotes genome stability and chromosome segregation (Krogan et al. 2004; Kalocsay et al. 2009). Similarly, the chromatin remodeling complexes SWR1C and INO80C, which regulate the deposition and eviction of Htz1p, also contribute to genome integrity (Krogan et al. 2003; Mizuguchi et al. 2004; Papamichos-Chronakis et al. 2006, 2011; van Attikum et al. 2007; Lademann et al. 2017).
To explore the relationship between Htz1p and yeast fumarase during DNA replication stress, we examined genetic interactions in cells lacking HTZ1 and/or FUM1 in growth assays of 10-fold serial dilutions onto rich medium lacking or containing HU. Thus, differences in growth by one spot, after normalizing to no treatment controls, reflect phenotypic differences on the order of around one magnitude in this assay. In the presence of HU, htz1Δ, but not fum1Δ, mutants exhibited growth defects relative to wild-type yeast and relative to the absence of HU (Figure 1C first vs. third panel), and this growth defect of the htz1Δ mutants was suppressed by complementation with exogenous expression of HTZ1 (Figure S1 top row, first vs. third and fifth panels). This sensitivity of htz1Δ mutants in the presence of HU was suppressed in fum1Δ htz1Δ mutants (Figure 1C first vs. third panel), and restored by the exogenous expression of FUM1 in fum1Δ htz1Δ mutants (Figure S1 bottom row, first vs. third and fifth panels), implying that loss of FUM1 had partially bypassed a requirement for HTZ1 during replication stress.
Exogenous fumarate suppresses the DNA replication stress sensitivity of htz1Δ mutants
As deletion of FUM1 causes accumulation of fumarate in the cell (Pollard et al. 2005; Lin et al. 2011), the above observation (Figure 1C) raised the possibility that elevated levels of fumarate caused by deletion of FUM1 had conferred resistance to HU in the htz1Δ fum1Δ mutants. To test this possibility, we compared growth of htz1Δ mutants to wild-type yeast in the presence and absence of HU, and with or without adding exogenous fumarate to the growth medium at concentrations comparable to the levels found in HLRCC tumors (Pollard et al. 2005). As shown in Figure 1C (third vs. fourth panel), the addition of exogenous fumarate largely suppressed the sensitivity of htz1Δ mutants to HU as well as further enhanced growth of htz1Δ fum1Δ mutants in HU. Similar to htz1Δ mutants, strains lacking SWR1 exhibited growth defects relative to wild-type on rich (YPD) medium as well as medium containing HU, and the sensitivity of swr1Δ mutants to HU was also partially suppressed by the addition of exogenous fumarate (Figure S2). Exogenous fumarate alone did not adversely affect the number of colonies in the absence of HU in these experiments; however, colony sizes of all strains tested decreased in the presence of exogenous fumarate (e.g., Figure 1). The cause of this decrease in colony size is unknown.
To assess further the effect of exogenous fumarate on sensitivity to DNA replication stress, we repeated our growth assays using cells lacking RAD52 or components of the MRX complex. Rad52p promotes loading of the nucleoprotein filament recombinase Rad51p onto ssDNA and strand exchange at DSBs, and is essential for homology-dependent DNA repair (Game and Mortimer 1974; New et al. 1998; Shinohara and Ogawa 1998; Pâques and Haber 1999; Symington 2002). In Schizosaccharomyces pombe, Rad52 is also required for recombination-independent restart of replication from terminally arrested forks in which nascent DNA is protected by Rad51 from excessive ssDNA formation by the exonucleases Exo1 or Mre11, and for properly merging a converging fork with a terminally arrested fork (Hashimoto et al. 2010; Lambert et al. 2010; Schlacher et al. 2011, 2012; Iraqui et al. 2012; Higgs et al. 2015; Ait Saada et al. 2017). In contrast to what had been observed for htz1Δ mutants, addition of exogenous fumarate did not suppress the sensitivity to HU in cells lacking RAD52 (Figure 1C). Genes encoding the MRX complex are also members of the RAD52 epistasis group. The MRX complex, consisting of Mre11p, Rad50p, and Xrs2p, acts as a major DSB sensor, but also functions to stabilize components of the replication machinery at stalled forks (Lisby et al. 2004; Tittel-Elmer et al. 2009). Like rad52Δ mutants, mre11Δ, rad50Δ, and xrs2Δ mutants exhibited severe growth defects in the presence of HU, and addition of exogenous fumarate did not suppress these defects (Figure 1D and Figure S3). These results indicated that fumarate complemented sensitivity to DNA replication stress created by the absence of the histone variant, but not Rad52p or the MRX complex.
Suppression of the DNA replication stress sensitivity of htz1Δ mutants by fumarate is not due to modulation of nucleotide pools
Exposure to HU results in stalled replication forks via inhibition of ribonucleotide reductase, which leads to depletion of nucleotide pools (Krakoff et al. 1968). This depletion is thought to result in the creation of stretches of ssDNA at stalled forks that become coated with RPA, which is required to recruit Mec1p-Ddc2p and promote activation of the kinase Mec1p, and activation of the intra-S phase checkpoint (Zou and Elledge 2003). Therefore, we tested the possibility that fumarate had promoted growth of htz1Δ mutants upon exposure to HU by modulating the nucleotide pools. We first tested whether increasing the dNTP pool by deletion of SML1, which encodes an inhibitor of ribonucleotide reductase (Zhao et al. 1998; Chabes et al. 1999), could promote growth of htz1Δ mutants in the presence of HU by comparing wild-type, sml1Δ, htz1Δ, and sml1Δ htz1Δ mutants in growth assays. Instead, we observed a negative synthetic genetic interaction in the absence of SML1 and HTZ1 as the sml1Δ htz1Δ mutants exhibited a severe growth defect in rich medium (YPD) compared to the sml1Δ or htz1Δ single mutants, whereas the growth of sml1Δ htz1Δ mutants in the presence of HU was comparable to that of htz1Δ mutants (Figure 2A first vs. third panel). Addition of exogenous fumarate suppressed the sensitivity to DNA replication stress of both htz1Δ and sml1Δ htz1Δ mutants (Figure 2A third vs. fourth panel), implying that fumarate suppressed the sensitivity to HU through a mechanism independent of elevating nucleotide pools.
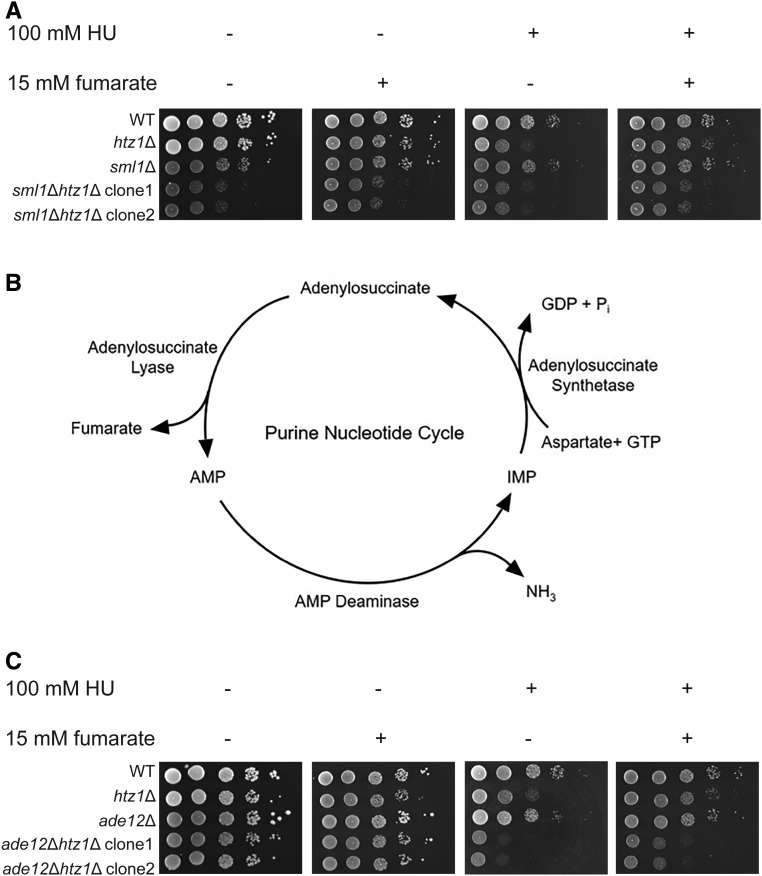
Figure 2.
Fumarate-mediated suppression of sensitivity to DNA replication stress of htz1Δ mutants is independent of modulation of nucleotide pools. (A) Fumarate suppresses the sensitivity to DNA replication stress of htz1Δ mutants in the absence of an inhibitor of ribonucleotide reductase. (B) Fumarate is a product in the purine nucleotide synthesis cycle. In the purine nucleotide cycle, aspartate becomes converted to fumarate in a two-stage reaction, which is facilitated by hydrolysis of GTP. This two-stage reaction involves generation of adenylosuccinate from inosine monophosphate (IMP) and aspartate, which is then converted to fumarate and adenosine monophosphate (AMP). This reaction is followed by deamination of AMP to IMP by AMP deaminase. (C) Fumarate suppresses the sensitivity to DNA replication stress of htz1Δ mutants in the absence of adenylosuccinate synthase (ADE12). Strains with genotypes as indicated in (A and C) were analyzed in serial dilution growth assays as described in Figure 1.
Fumarate is also a product in the purine nucleotide cycle, and can function as a weak inhibitor of adenylosuccinate lyase, which converts adenylosuccinate into adenosine monophosphate (AMP) plus fumarate in the purine nucleotide cycle (Barnes and Bishop 1975) (Figure 2B). This raised the possibility that regulation of AMP production by inhibition of adenylosuccinate lyase activity and/or accumulation of adenylosuccinate upon addition of exogenous fumarate had suppressed the sensitivity of htz1Δ mutants to HU. Therefore, we tested whether decreased production of adenylosuccinate and disruption of the purine nucleotide cycle by deletion of the gene encoding adenylosuccinate synthase (ADE12) could block fumarate-dependent suppression of the sensitivity of htz1Δ mutants to HU. As shown in Figure 2C, ade12Δ mutants did not exhibit growth defects compared to wild type in presence of HU, whereas ade12Δ htz1Δ mutants were hypersensitive to HU relative to wild-type yeast or htz1Δ mutants, indicating a negative synthetic genetic interaction during DNA replication stress. However, addition of exogenous fumarate to the medium partially suppressed the sensitivity of ade12Δ htz1Δ mutants to HU, indicating that adenylosuccinate and the integrity of the purine nucleotide cycle were dispensable for fumarate-mediated suppression in htz1Δ mutants. Taken together, these results implied that fumarate suppressed the sensitivity to DNA replication stress of htz1Δ mutants independently of modulating nucleotide levels.
Loss of JHD2 phenocopies fumarate-dependent suppression of DNA replication stress sensitivity in htz1Δ mutants
Fumarate is a competitive inhibitor of α-KG dependent dioxygenases, including JmjC domain-containing protein demethylases (Xiao et al. 2012; Yang et al. 2013; Jiang et al. 2015). Histone H3 K4, H3 K36, and H3 K79 methylation play important roles in maintaining genome stability including during DNA replication, DNA damage responses and repair, as well as in activation of DNA damage checkpoints (Wysocki et al. 2005; Lazzaro et al. 2008; Faucher and Wellinger 2010; Rizzardi et al. 2012; Jha and Strahl 2014; Pai et al. 2014). Therefore, we hypothesized that fumarate had suppressed the sensitivity to DNA replication stress in htz1Δ mutants in the above experiments by modulating the levels of histone lysine methylation through inhibition of one or more JmjC domain-containing histone demethylases. We reasoned that if inhibition of a JmjC histone demethylase by fumarate promoted growth upon DNA replication stress in the htz1Δ mutants, then deletion of that histone demethylase might act similarly. Therefore, we tested the sensitivity of mutants lacking individual JmjC histone demethylases to HU in the presence or absence of HTZ1, including cells lacking JHD1 (removes H3 K36me2 and me1; Tsukada et al. 2006; Fang et al. 2007; Tu et al. 2007), JHD2 (removes H3 K4me3 and me2; Ingvarsdottir et al. 2007; Liang et al. 2007; Tu et al. 2007), RPH1 (removes H3 K36me3 and me2; Kim and Buratowski 2007; Tu et al. 2007), ECM5 (unknown target) or GIS1 (predicted to remove H3 K36 methylation; Tu et al. 2007; Kwon and Ahn 2011; Sein et al. 2015) (Figure 3A).
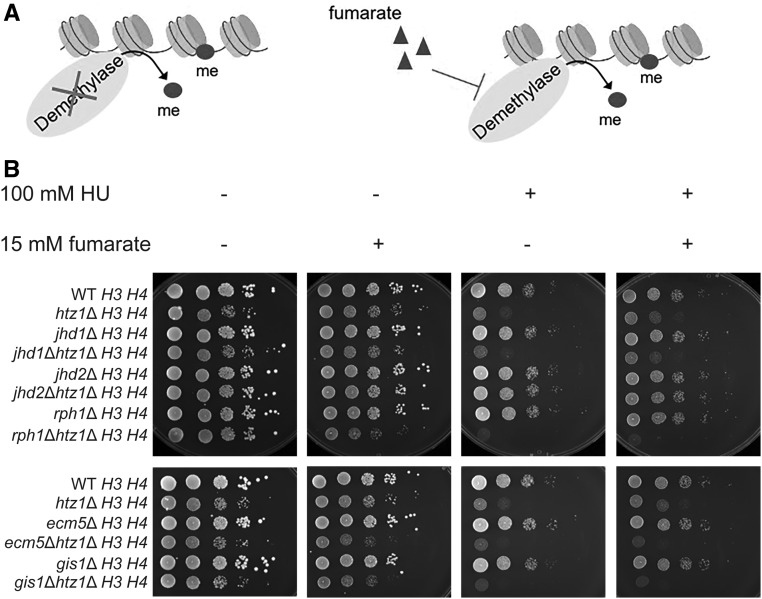
Figure 3.
Loss of JmjC domain-containing histone demethylase Jhd2p suppresses the sensitivity to DNA replication stress of htz1Δ mutants. (A) Enhancing histone methylation by deletion of histone demethylase(s) or enzyme inhibition by fumarate. (B) Genetic interaction analyses between htz1Δ mutants and histone demethylase mutants. Strains with indicated genotypes were analyzed in serial dilution growth assays as described in Figure 1.
When grown on rich medium (YPD), smaller colonies were observed in htz1Δ, jhd1Δ htz1Δ, rph1Δ htz1Δ, ecm5Δ htz1Δ, and gis1Δ htz1Δ mutants compared to wild-type, whereas the colony sizes of jhd2Δ htz1Δ mutants were similar to wild-type, implying that deletion of JHD2 complemented growth defects caused by loss of HTZ1 (Figure 3B first panels). In contrast to htz1Δ mutants, deletion of single histone demethylases did not result in sensitivity to HU compared to wild-type (Figure 3B first vs. third panels). jhd1Δ htz1Δ mutants were as sensitive to HU as htz1Δ mutants, whereas rph1Δ htz1Δ, ecm5Δ htz1Δ, and gis1Δ htz1Δ mutants were slightly more sensitive to HU compared to htz1Δ mutants. In contrast, jhd2Δ htz1Δ mutants showed no sensitivity to HU compared to wild type, and deletion of JHD2 relieved the sensitivity of htz1Δ mutants to DNA replication stress (Figure 3B first vs. third panels). Addition of exogenous fumarate had no further effect on sensitivity of jhd2Δ htz1Δ mutants to DNA replication stress. In contrast, deletion of RPH1, ECM5, or GIS1 in strains lacking HTZ1 resulted in sensitivity to exogenous fumarate alone (Figure 3B first vs. second panels). This sensitivity precluded our ability to determine the impact of loss of RPH1, ECM5 or GIS1 on fumarate-dependent suppression of sensitivity to HU in the htz1Δ mutants under the conditions tested. Overall, the results of our analyses indicate that deletion of JHD2 was sufficient to alleviate the sensitivity to replication stress of htz1Δ mutants, and implied that inhibition of the histone demethylase Jhd2p by fumarate may have conferred resistance to DNA replication stress in the htz1Δ mutants by promoting histone H3 K4 methylation.
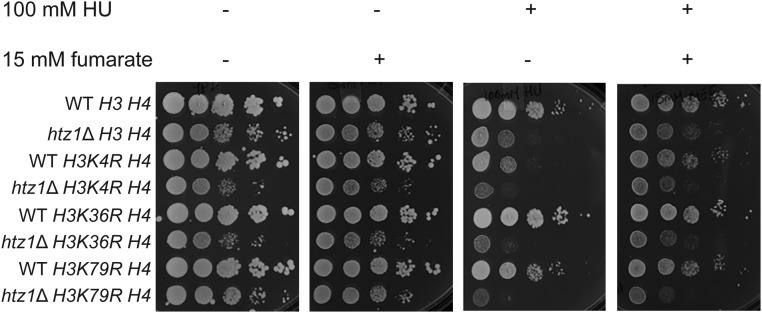
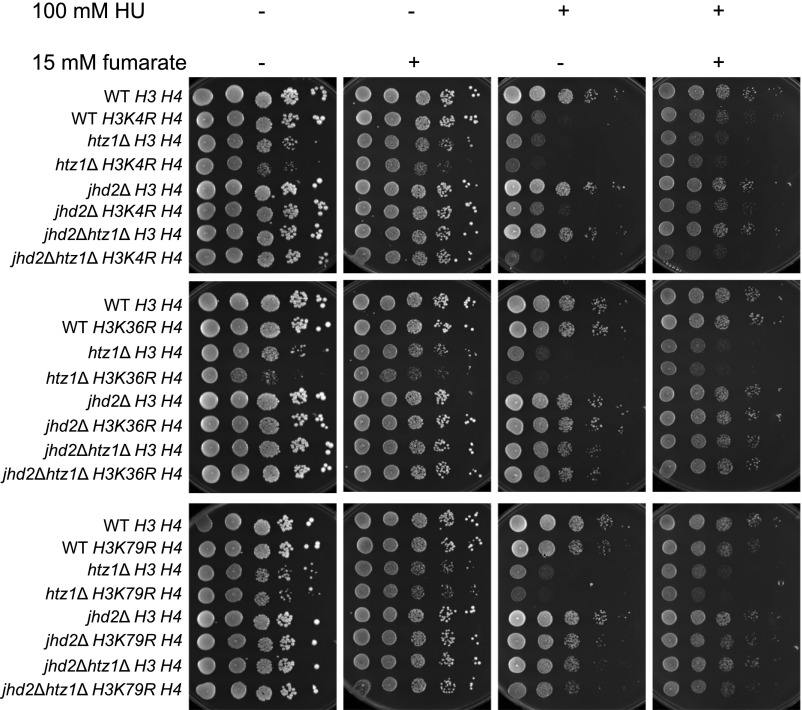
To explore this possibility, we compared sensitivity to HU in wild type or htz1Δ mutants lacking chromosomal copies of genes encoding histones H3/H4 and expressing wild-type H3/H4 or H3 mutants in which lysine methylation sites had been mutated to arginine plus wild-type H4 from a plasmid. Yeast lacking H3 K4 methylation showed growth defects in the presence of HU, consistent with previously reported sensitivity of set1Δ or H3 K4R mutants to HU (Faucher and Wellinger 2010). In contrast, yeast lacking H3 K36 or H3 K79 methylation did not (Figure 4, first vs. third panel), consistent with previously reported lack of sensitivity of set2Δ mutants (Biswas et al. 2008; Jha and Strahl 2014), and dot1Δ or H3 K79R mutants to HU (Rossodivita et al. 2014; Stulemeijer et al. 2015). Moreover, we observed synthetic growth defects between htz1Δ and H3 K4R or H3 K36R mutants when grown on rich medium (YPD) relative to single mutants or wild type (Figure 4, first panel), consistent with previous reports of synthetic growth defects between set1Δ and htz1Δ mutants (Venkatasubrahmanyam et al. 2007), or set2Δ and swr1Δ mutants (Fuchs et al. 2012). These growth defects were exacerbated in the presence of HU (Figure 4, first vs. third panel). In contrast, no growth defects were observed in htz1Δ H3 K79R relative to htz1Δ mutants in rich medium, but htz1Δ K79R mutants were hypersensitive to DNA replication stress relative to either single mutant (Figure 4, first vs. third panel). Addition of fumarate suppressed the sensitivity to DNA replication stress of wild-type strains expressing H3 K4R as well as htz1Δ strains expressing H3 K4R, H3 K36R or H3 K79R mutants, but to varying degrees (Figure 4). Together, these results implied that fumarate could suppress sensitivity to replication stress caused by defects in multiple methylation events, and that methylation of an individual residue was not solely required for this suppression. We have been unable to generate and test triple mutants lacking HTZ1 with histone H3 K4R,K36R or H3 K4R,K79R as these mutant combinations appear to be lethal. Together, these data were consistent with fumarate-dependent suppression functioning through multiple pathways involving different histone methylation sites, or nonhistone protein methylation (see Discussion).
Figure 4.
Fumarate-dependent suppression of sensitivity to DNA replication stress of strains expressing H3 mutants with lysine to arginine mutations at H3 K4, K36, or K79. Strains with genotypes as indicated were analyzed in serial dilution growth assays as described in Figure 1.
Suppression of DNA replication stress sensitivity of htz1Δ mutants by deletion of JHD2 requires H3 K4 methylation
In addition to methylated H3 K4 being enriched in transcriptionally active regions, both methylated H3 K4 and Set1p, the sole H3 K4-specific methyltransferase in yeast, become enriched at DSB sites, and mutants lacking SET1 show growth defects in the presence of HU as well as genotoxic agents that induce DSBs (Faucher and Wellinger 2010). To test whether suppression of sensitivity to HU in jhd2Δ htz1Δ mutants observed in Figure 3 required methylated H3 K4, synthetic interaction analyses were conducted using wild-type yeast, plus htz1Δ, jhd2Δ, and jhd2Δ htz1Δ mutants expressing wild-type H3/H4 or H3 mutants in which individual lysine methylation sites had been mutated to arginine plus H4 from a plasmid. These analyses, shown in Figure 5, indicated that the observed increase in colony size in jhd2Δ htz1Δ mutants compared to htz1Δ mutants when grown on rich medium (YPD) did not require H3 K4 (Figure 5, top row, first panel), H3 K36 (Figure 5, middle row, first panel) or H3 K79 (Figure 5, bottom row, first panel) methylation, implying a histone methylation-independent role for Jhd2p in promoting growth exists. However, unlike jhd2Δ htz1Δ mutants expressing wild-type histones, jhd2Δ htz1Δ mutants expressing H3 K4R were as sensitive to HU as htz1Δ mutants expressing H3 K4R (Figure 5, top row, third panel compared to first). In contrast, jhd2Δ htz1Δ mutants expressing H3 K36R (Figure 5, middle row, third panel compared to first) or H3 K79R mutants (Figure 5, bottom row, third panel compared to first) were not sensitive to HU, similar to jhd2Δ htz1Δ mutants expressing wild-type H3/H4. Together, these findings were consistent with jhd2Δ-dependent suppression of growth sensitivity in htz1Δ mutants upon DNA replication stress requiring H3 K4, but not H3 K36 or H3 K79 methylation. Consistent with our results in Figure 3 and Figure 4, addition of exogenous fumarate partially suppressed the sensitivity of jhd2Δ htz1Δ H3 K4R mutants to HU (Figure 5), implying that fumarate could confer resistance to DNA replication stress by multiple mechanisms, one of which was H3 K4 methylation-independent.
Figure 5.
Impact of histone methylation, loss of JHD2 and exogenous fumarate on sensitivity to DNA replication stress of htz1Δ mutants. Strains with indicated genotypes were analyzed in serial dilution growth assays as described in Figure 1.
Fumarate is a modulator of Jhd2p activity and H3 K4me3 levels
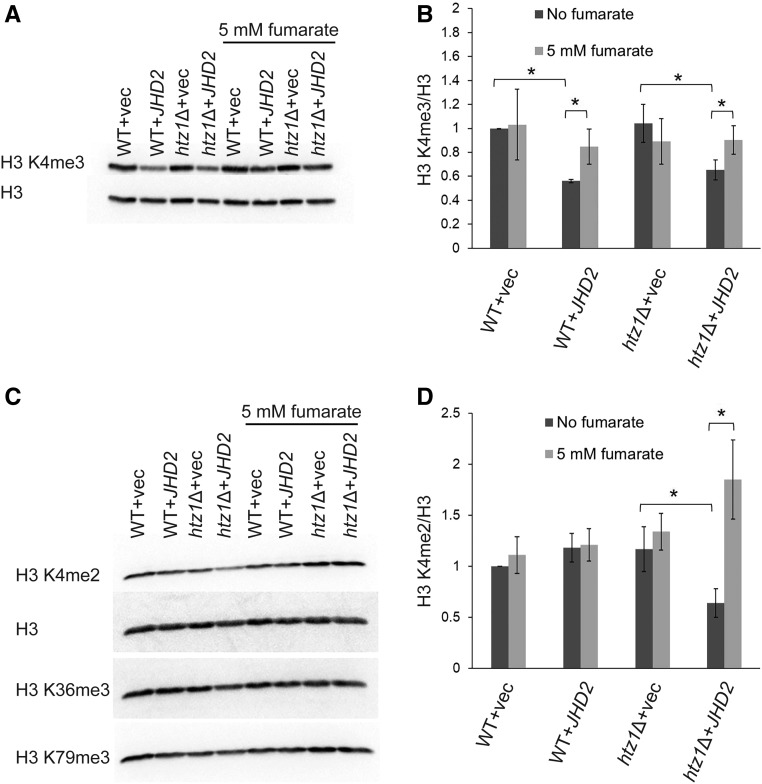
Collectively, the above findings led us to predict that elevated H3 K4 methylation levels, via either deletion of JHD2 or exposure to exogenous fumarate, could suppress replication stress sensitivity in htz1Δ mutants. To test whether exogenous fumarate could inhibit Jhd2p activity, we analyzed the effect of exogenous fumarate on H3 K4 methylation in wild-type or htz1Δ strains overexpressing FLAG-Jhd2p as this background facilitates detection of changes in methylation states of H3 K4 because endogenously expressed Jhd2p is subject to efficient proteosomal degradation (Mersman et al. 2009). Logarithmically growing cultures from strains harboring an empty vector or a plasmid for overexpression of FLAG-Jhd2p were harvested before and after exposure to fumarate for 6 hr, and whole cell extracts were used to analyze global levels of H3 K4me3 or H3 K4me2 relative to H3 in immunoblots. As shown in Figure 6, overexpression of FLAG-Jhd2p results in reduced levels of H3 K4me3 in both wild type (see also Mersman et al. (2009)) and htz1Δ mutants (P = 0.029 and 0.050, respectively) (Figure 6, A and B) as well as H3 K4me2 (Figure 6, C and D) in htz1Δ mutants (P = 0.029). This difference in effect on H3 K4me2 levels in wild-type cells vs. htz1Δ mutants may have been due to enhanced overexpression of JHD2 mRNA from the heterologous PYK1 promoter in htz1Δ mutants (Figure S4A), which led to enhanced overexpression of FLAG-Jhd2p (Figure S4, B and C). In contrast, expression of JHD2 mRNA from its endogenous promoter was similar in wild-type cells and htz1Δ mutants containing vector alone (Figure S4A).
Figure 6.
Fumarate modulates levels of JDH2-dependent H3 K4 methylation. Wild-type yeast and htz1Δ mutants carrying an empty vector or a plasmid for overexpression of FLAG-Jhd2p were grown logarithmically in selective medium with or without 5 mM fumarate. (A and C) Whole cell extracts of strains with indicated genotypes were analyzed in at least three independent experiments by immunoblotting against H3 K4me3 (A), or H3 K4me2, H3K36me3, H3 K79me3 (C) and H3 (loading control). (B and D) Levels of H3 K4me3 (B) or H3 K4me2 (D) were normalized to H3, and expressed relative to that observed in wild-type with vector (vec), which was set to 1 (Avg. ± STD, n = 3; representative independent experiments shown in (A and C). The level of H3 K4me3 or H3 K4me2 relative to H3 was calculated as . The statistical analysis was performed using Wilcoxon Rank Sum test and P-value ≤ 0.05 is shown by *.
After treatment with fumarate, the global levels of H3 K4me3 in wild-type yeast or htz1Δ mutants overexpressing FLAG-Jhd2p, as well as H3 K4me2 in htz1Δ mutants overexpressing FLAG-Jhd2p were significantly increased relative to in the absence of fumarate (P = 0.029, 0.05 and 0.029, respectively; see also Figure S5, A and B), implying that fumarate had inhibited Jhd2p in vivo. A similar significant increase in H3 K4me2 levels was observed in fum1Δ htz1Δ relative to htz1Δ mutants overexpressing FLAG-Jhd2p (Figure S5; P = 0.029). In contrast to H3 K4 methylation, global levels of H3 K36me3 or H3 K79me3 were not affected by exposure to fumarate or loss of FUM1 in these experiments (Figure 6, C and D, or Figure S5A, respectively). Together, these results were consistent with previous reports of elevation of H3 K4 methylation levels in mammalian cells upon treatment with fumarate or siRNA targeting fumarase (Xiao et al. 2012).
The impact of fumarate on cell cycle progression and checkpoint activation upon DNA replication stress
Like wild-type yeast, htz1Δ and swr1Δ mutants do not accumulate spontaneous DSBs as measured by Pulsed-Field Gel Electrophoreses (Morillo-Huesca et al. 2010), and, upon release into HU, htz1Δ and swr1Δ mutants exhibit wild-type replication bubbles and forks, with no evidence of accumulation of stalled, broken, or recessed forks at, or near, early origins by two-dimensional (2D) gel analyses, indicating initiation and fork progression per se in these mutants is relatively normal (Dhillon et al. 2006; Srivatsan et al. 2018). Also, like in wild type, late origins fail to fire in HU in htz1Δ mutants, consistent with late origin firing being negatively regulated by Rad53p via proper intra-S phase checkpoint activation in HU in the absence of Htz1p (Dhillon et al. 2006). However, although the early and late origin replication program is conserved (Dhillon et al. 2006), the timing of origin firing is delayed in htz1Δ mutants (Dhillon et al. 2006), and loss of HTZ1 or SWR1 delays completion of replication relative to wild type (Dhillon et al. 2006; Srivatsan et al. 2018). In addition, although htz1Δ mutants exhibit a decreased rate of progression through S phase, the efficiency of checkpoint-dependent cell cycle arrest in early S phase in HU (Dhillon et al. 2006), and recovery from exposure to HU in htz1Δ mutants remains similar to wild type (Srivatsan et al. 2018). To test whether fumarate affected cell cycle kinetics in htz1Δ mutants, we analyzed cell cycle progression of wild-type yeast and htz1Δ mutants in the presence or absence of HU and/or fumarate. To do so, we first synchronized the cells in G1 with α-factor before releasing into rich medium with or without HU or fumarate. Cells were harvested before, and at 20-min intervals after release, and their DNA content was analyzed by flow cytometry. As shown in Figure S6, addition of HU slowed progression of both wild-type yeast and htz1Δ mutants through S phase, as expected. In the absence of HU, fumarate did not dramatically affect the cell cycle profile of wild-type yeast or the htz1Δ mutants during the first S phase. These results implied fumarate did not function by promoting entry into Start in the htz1Δ mutants via eliminating a delay in the induction of G1 cyclins (Dhillon et al. 2006). In the presence of HU, addition of fumarate also did not dramatically affect the cell cycle profile in wild-type yeast or htz1Δ mutants during the first cell cycle.
Reports of defects in phosphorylation of Rad53p in htz1Δ mutants in response to DNA damage (Dhillon et al. 2006; Kalocsay et al. 2009) prompted us to assess the impact of fumarate on the DNA damage checkpoint activation and deactivation as well. We analyzed checkpoint responses by monitoring the phosphorylation status of Rad53p in wild-type yeast and htz1Δ mutants grown in rich medium following exposure to HU in the presence or absence of exogenous fumarate (Figure S7). Cultures were first synchronized in G1 by addition of α-factor, then released into HU-containing growth medium containing or lacking fumarate. Cells were then collected before and every 30 min to 2 hr after addition of HU for a total of 8 hr. The phosphorylation status of Rad53p was then analyzed from each timepoint by immunoblotting using Rad53p-specific antibodies and whole cell extracts. As shown in Figure S7, phosphorylated Rad53p was detected within 1 hr of treatment with HU in wild-type yeast and htz1Δ mutants in the presence or absence of fumarate, indicating that exogenous fumarate had little or no impact on activation of Rad53p. In htz1Δ mutants, phosphorylated Rad53p diminished after 6 hr of treatment with HU, whereas phosphorylated Rad53p was still detectable in wild-type yeast, indicating that htz1Δ mutants had defects in maintaining checkpoint activation, and had adapted to DNA replication stress earlier than had wild-type. Addition of fumarate did not dramatically affect this early checkpoint deactivation. Taken together, checkpoint activation and deactivation in wild-type yeast or htz1Δ mutants were largely unaffected upon addition of exogenous fumarate under these conditions.
Suppression of sensitivity to DNA replication stress of htz1Δ mutants by fumarate requires intra-S phase checkpoint sensors and mediators
The intra-S phase checkpoint consists of two branches: the DNA damage checkpoint (DDC) and the DNA replication checkpoint (DRC). In both DDC and DRC, induced phosphorylation of Rad53p is dependent on signaling events that act upstream of the sensor Mec1p kinase, which is recruited to ssDNA coated with RPA via Mec1p’s interacting partner Ddc2p (Rouse and Jackson 2002; Zou and Elledge 2003), although DDC tends to be a slower, sustained response, whereas DRC is rapid, but transient (Pardo et al. 2016). In DDC, Mec1p uses the adaptor Rad9p to transduce the signal to Rad53p in response to DNA damage (Weinert and Hartwell 1988; Gilbert et al. 2001; Sweeney et al. 2005) [see Pardo et al. (2016) for review]. In contrast, Mrc1p travels with the DNA replication fork (Katou et al. 2003; Lou et al. 2008; Komata et al. 2009), and acts as a “threshold-driven” sensor during DRC. Upon phosphorylation by Mec1p, Mrc1p mediates a signal to Rad53p that is strong enough to activate the intra-S phase checkpoint only when numerous forks are impeded (Alcasabas et al. 2001; Tanaka and Russell 2001, 2004; Duncker et al. 2002; Shimada et al. 2002; Osborn and Elledge 2003; Tercero et al. 2003; Smolka et al. 2006; Xu et al. 2006; Chen and Zhou 2009) [see Pardo et al. (2016) for review]. DDC and DRC appear to regulate downstream targets somewhat differently during intra-S phase checkpoint activation. For example, MRC1 is required to inhibit late origin firing in HU and MMS, whereas RAD9 is not (Alcasabas et al. 2001; Bacal et al. 2018).
Multiple factors act as DNA damage sensors for both the DDC and DRC branches of the intra-S phase checkpoint signaling pathway to activate Mec1p, including the Ddc1p–Mec3p–Rad17p complex, and Rad24p. Ddc1p–Mec3p–Rad17p is analogous to the 9-1-1 complex in mammals. Rad24p is the large subunit of an alternative RF-C complex that loads Ddc1p–Mec3p–Rad17p onto DNA at the 5′ junction between RPA-bound ssDNA and dsDNA (Majka and Burgers 2003; Zou et al. 2003; Furuya et al. 2004), such as those present at Okazaki fragments. Loss of these factors results in defects in or loss of phosphorylation of Rad53p upon DNA replication stress (Paciotti et al. 1998; Shimomura et al. 1998; Kondo et al. 1999; Alcasabas et al. 2001; Gilbert et al. 2001).
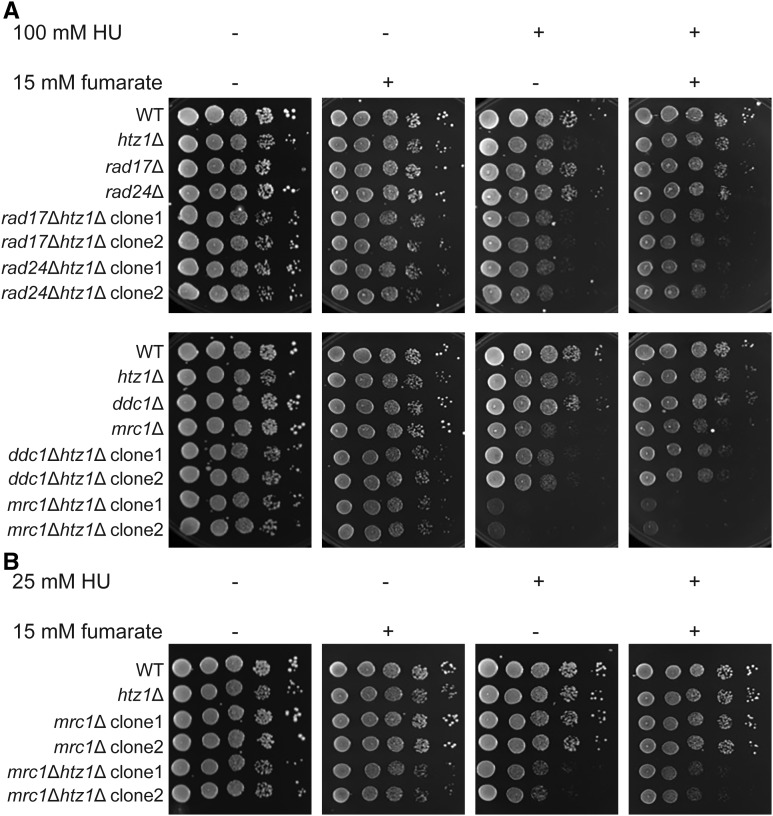
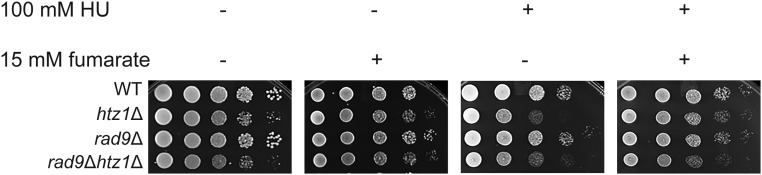
To test whether the fumarate-dependent resistance to HU in htz1Δ mutants required sensors of the checkpoint signaling pathway, we examined sensitivity to replication stress in strains in which components of the intra-S phase checkpoint had been deleted. We found that deletion of RAD17, RAD24, or DDC1 from wild-type did not result in sensitivity to HU under the conditions tested, and their deletion in htz1Δ mutants did not increase the sensitivity of htz1Δ mutants to HU (Figure 7A, first vs. third panel). However, deletion of RAD17, RAD24, or DDC1 prevented fumarate-dependent resistance to HU in the htz1Δ mutants (Figure 7A), indicating the 9-1-1 complex was required for fumarate to confer resistance to DNA replication stress. Similarly, cells lacking the DDC mediator Rad9p (the ortholog of mammalian 53BP1) did not exhibit sensitivity to HU under the conditions tested, and inactivation of DDC by loss of RAD9 instead partially restored growth of htz1Δ mutants in HU. Addition of exogenous fumarate did not enhance this effect (Figure 8).
Figure 7.
Fumarate-dependent suppression of sensitivity to DNA replication stress of htz1Δ mutants requires components of the intra-S phase checkpoint. (A) The 9-1-1 complex and the 9-1-1 loader Rad24p are required for fumarate to suppress the sensitivity to DNA replication stress of htz1Δ mutants. (A and B) htz1Δ mutants require the DRC mediator Mrc1p during DNA replication stress. Strains with genotypes as indicated were analyzed as described in Figure 1.
Figure 8.
htz1Δ mutants do not require the DDC mediator Rad9p during DNA replication stress. Strains with genotypes as indicated were analyzed as described in Figure 1.
We also assessed the impact of loss of HTZ1 and/or exposure to fumarate on DRC. Deletion of the DRC mediator MRC1 (Figure 7A, bottom row, first vs. third panel) caused a growth defect in medium containing HU (see also Alcasabas et al. 2001; Tanaka and Russell 2001; Osborn and Elledge 2003), and mrc1Δ htz1Δ mutants showed enhanced sensitivity to HU as compared to wild-type yeast or htz1Δ mutants [see also Srivatsan et al. (2018) for mrc1Δ swr1Δ mutants], implying that the presence of Htz1p was critical to limit abnormal replication intermediates when Mrc1p was not present to stabilize the fork. We therefore repeated these growth assays using a lower concentration of HU to assess the effect of fumarate on this sensitivity (Figure 7B); however, addition of fumarate to the growth medium could not suppress the sensitivity of mrc1Δ htz1Δ mutants to HU. Together, these results implied that the resistance to replication stress conferred to htz1Δ mutants by exposure to fumarate required several intact intra-S phase checkpoint sensors and mediators, and were consistent with fumarate having promoted a different step in the intra-S phase checkpoint signaling pathway that had been compromised by loss of HTZ1.
We next examined the relationship between SGS1 and HTZ1 plus fumarate (Figure 9A). Sgs1p, a 3′–5′ RecQ helicase and yeast ortholog of the Bloom Syndrome protein BLM, is a stable component of the replication fork, where it interacts with RPA and Dna2p (Cejka et al. 2010; Hegnauer et al. 2012). Upon intra-S phase checkpoint activation with HU, Sgs1p binds Rad53p, stabilizes DNA pol α and DNA pol ε association with the stalled forks, and may contribute to fork stability by reversing recessed forks and preventing inappropriate recombination by resolving strand exchange (Cobb et al. 2003; Versini et al. 2003; Bjergbaek et al. 2005; Bernstein et al. 2009, 2010; Hegnauer et al. 2012). When phosphorylated by Mec1p, Sgs1p functions in the DRC pathway with Mrc1p to activate Rad53p, although Sgs1p’s helicase activity per se is not required for this phosphorylation event (Bjergbaek et al. 2005; Hegnauer et al. 2012). In the absence of replication stress, sgs1Δ mutants grew with similar efficiency as wild type, whereas sgs1Δ htz1Δ mutants exhibited a growth defect relative to either single mutant in the absence of replication stress (Figure 9A, first panel). Growth defects of sgs1Δ and sgs1Δ htz1Δ mutants in HU could not be suppressed by the addition of fumarate (Figure 9A, third vs. fourth panel), implying that Htz1p was critical for ensuring survival during replication stress in the absence of Sgs1p, like the other DRC component Mrc1p (Figure 7). However, fumarate could not bypass this role of Htz1p.
Figure 9.
Impact of loss of HTZ1 and exogenous fumarate on sensitivity to DNA replication stress of mutants with defects in DRC and processing and restart of aberrant replication forks. (A) Fumarate suppresses the sensitivity to DNA replication stress in cells lacking EXO1, or SAE2 in the presence or absence of HTZ1. (B) Loss of HTZ1 confers resistance to DNA replication stress of cells lacking subunits of the MRX complex, and fumarate enhances this effect. Strains with genotypes as indicated were analyzed as described in Figure 1.
While Sgs1p is epistatic to Mrc1p in activation of Rad53p, Sgs1p also binds Rad51p (Wu et al. 2001) and functions in a pathway parallel to Mrc1p during replication fork recovery to stabilize association of DNA polymerase ε at forks (Bjergbaek et al. 2005). As Sgs1p-dependent stabilization of DNA polymerase ε at forks requires the helicase activity of Sgs1p as well as Rad51p, this pathway has been proposed to be involved in resolving reversed forks and promoting recombination-dependent restart of forks, prompting us to examine the relationship between Htz1p and factors involved in processing forks for replication restart.
Suppression of sensitivity to DNA replication stress of htz1Δ mutants by fumarate and end resection
During replication stress, MRN-Ctp1, the S. pombe ortholog of MRX-Sae2p, facilitates replication restart in a DSB-independent pathway by limiting uncontrolled resection by the exonuclease Exo1 at terminally arrested forks (Teixeira-Silva et al. 2017). In the absence of DSBs at stalled forks, short-range resection by MRN-Ctp1 creates ssDNA gaps that enable loading of RPA, Rad52, and Rad51, as well as replication restart (Lambert et al. 2010; Tsang et al. 2014; Nguyen et al. 2015; Teixeira-Silva et al. 2017). To test whether the fumarate-dependent resistance to HU in the htz1Δ mutants could be related to fork resection and restart, we revisited sensitivity to replication stress in strains in which components of the MRX complex had been deleted (Figure 9B). In the absence of replication stress, rad50Δ, xrs2Δ, and mre11Δ mutants grew with similar efficiency as wild-type, and rad50Δ htz1Δ, xrs2Δ htz1Δ, and mre11Δ htz1Δ mutants grew with similar efficiency as htz1Δ mutants (Figure 9B, first panel), consistent with previous reports that loss of HTZ1 does not inherently result in accumulation of broken forks (Dhillon et al. 2006; Srivatsan et al. 2018). Consistent with a critical role of the MRX complex in processing replicative intermediates arising during replication stress, cells lacking RAD50, XRS2 or MRE11 were hypersensitive to HU relative to wild-type or htz1Δ mutants. In contrast, deletion of HTZ1 in rad50Δ or xrs2Δ, but not mre11Δ mutants partially suppressed their sensitivity to HU (Figure 9B, first vs. third panel), and addition of exogenous fumarate further suppressed this sensitivity, but only in the absence of HTZ1 (Figure 9B). The significance of this difference in phenotypes between the subunits of the MRX complex is not understood. However, upon γ-irradiation, Mre11p can form weak nuclear foci in the absence of XRS2, and is nuclear in cells lacking RAD50 (Lisby et al. 2004), plus a Rad50-independent function of Mre11 in repair of DSBs has previously been documented in Archaea (Kish and DiRuggiero 2008). Regardless, these results implied chromatin composition impacted the fate of arrested forks in the absence of a functional MRX complex, and, therefore, viability.
Sae2p and the exonuclease Exo1p promote survival during replication stress by counteracting the formation of aberrant branched structures at stalled forks associated with DSB formation and fork collapse (Colosio et al. 2016). Sae2p stimulates Mre11p and promotes end resection with the MRX complex during DSB repair (Lengsfeld et al. 2007; Mimitou and Symington 2008; Cannavo and Cejka 2014), and facilitates release of the MRX complex from DNA ends to promote repair (Puddu et al. 2015). Sae2p promotes processing of structures mimicking replicative intermediates in vitro, and appears to have functions distinct from Mre11p in counteracting reversed fork cleavage (Colosio et al. 2016; Ghodke and Muniyappa 2016). Exo1p can resect nascent strands in reversed forks, thereby limiting their formation into structures that could lead to DSBs (Colosio et al. 2016). In S. pombe, long-range resection at terminally arrested forks requires Exo1 [but not Rqh1 (Sgs1p)] and is Ctp1 (Sae2p)-dependent (Teixeira-Silva et al. 2017). When examining the relationship between SAE2, EXO1, and HTZ1 plus fumarate, we found sae2Δ and exo1Δ mutants grew with similar efficiency as wild-type, and sae2Δ htz1Δ plus exo1Δ htz1Δ mutants grew with similar efficiency as htz1Δ mutants on rich medium (Figure 9A, first panel). sae2Δ mutants were mildly sensitive to HU, and sae2Δ htz1Δ mutants were more sensitive than either single mutant to replication stress (Figure 9A, first vs. third panel). However, like in htz1Δ mutants, fumarate fully suppressed the hypersensitivity of sae2Δ mutants to replication stress, and partially suppressed the hypersensitivity of sae2Δ htz1Δ mutants (Figure 9A, third vs. fourth panel), consistent with exposure to fumarate leading to bypass of a defect caused by the absence of either Sae2p or Htz1p. Unlike sae2Δ mutants, cells lacking EXO1 [in which resection of a regressed, terminally arrested fork would be expected to be limited as in Schizosaccharomyces pombe (Teixeira-Silva et al. 2017)] grew similar to wild-type on HU, indicating Exo1p was not required to process/restart stalled forks under the conditions tested [Figure 9A, see also Doerfler and Schmidt (2014)]. exo1Δ htz1Δ and htz1Δ mutants were similarly sensitive to replication stress, implying that limiting resection by Exo1p and Htz1p function may fall in the same pathway at replication forks (see Adkins et al. 2013) for exo1Δ swr1Δ interactions with UV and zeocin). This defect in exo1Δ htz1Δ mutants was partially suppressed by exogenous fumarate (Figure 9A third vs. fourth panel). Together, these results were consistent with exposure to fumarate having enabled bypass of defects related to processing stalled forks.
Suppression of sensitivity to DNA replication stress of htz1Δ mutants by fumarate is independent of displacement of Ku from replicative intermediates
YKU70 encodes a component of the Ku complex, which is best known for its ability to bind DSB ends and promote NHEJ (Boulton and Jackson 1996). Ku70p also has a NHEJ-independent function during replication stress, in which Ku70p binds reversed forks to regulate end resection by limiting homology-directed repair (Foster et al. 2011; Teixeira-Silva et al. 2017). At terminally arrested forks in S. pombe, MRN-Ctp1, is proposed to displace Ku via short-range resection. This short-range resection is Ctp1-dependent and Exo1-independent, and Exo1 is not required for replication restart at stalled forks lacking DSBs (Teixeira-Silva et al. 2017). However, in the absence of Ku70, Rad50, and Ctp1 are no longer required to promote initial resection of a stalled fork lacking a DSB. Instead, stalled forks now can be resected by Exo1, but HR-mediated fork restart becomes delayed. Consistent with conservation of this process, loss of YKU70 in budding yeast suppresses MMS sensitivity and mildly suppresses HU sensitivity of mre11 nuclease dead and sae2Δ mutants (Foster et al. 2011).
To test the relationship between YKU70, HTZ1, and fumarate during replication stress, we conducted analogous growth assays in htz1Δ strains containing or lacking YKU70. However, the sensitivity to replication stress of htz1Δ mutants did not require YKU70, and fumarate could suppress the sensitivity to HU of htz1Δ mutants similarly in the presence and absence of YKU70 (Figure S8). These results implied fumarate did not suppress sensitivity to replication stress in the htz1Δ mutants by promoting removal of Ku from ends of reversed forks, but rather suppressed a Ku70p-independent defect in the htz1Δ mutants.
Varying effects of fumarate and loss of FUM1 on sensitivity to UV and camptothecin in htz1Δ mutants
Exposure to UV can create lesions, including pyrimidine dimers and (6–4) photoproducts, throughout the cell cycle. These lesions are repaired by multiple pathways, depending on their location and their presence during different points in the cell cycle, including Nucleotide Excision Repair (NER), through either transcription-coupled NER or Global Genome NER pathways (Waters et al. 2015). However, if UV lesions are present in S phase, they can result in arrest of replication forks and uncoupling of the replicative helicase from DNA polymerase (Byun et al. 2005), in contrast to HU, in which the fork remains intact and travels slowly (Sogo et al. 2002). Such UV lesions can be repaired by multiple postreplicative repair strategies that are error prone, involving specialized translesion synthesis DNA polymerases, or error free, involving template switching, fork regression, and gap-filling (Boiteux et al. 2013). To test the impact of loss of HTZ1 and exposure to fumarate on sensitivity to UV, we compared growth of htz1Δ mutants to wild-type yeast in untreated cells vs. after exposing cells to UV, and with or without adding exogenous fumarate. As shown in Figure S9A (top row, first vs. third panel), htz1Δ mutants were sensitive to exposure to UV (see also Deng et al. 2005). This UV sensitivity could be suppressed by expression of HTZ1 exogenously (Figure S9A top row, first vs. third panels), but not via deletion of FUM1 (Figure S9A top row, first vs. third panels) or the addition of exogenous fumarate (Figure S9A top row, third vs. fourth panels). Loss of FUM1 also did not alter sensitivity to UV relative to wild-type (Figure S9A row, first vs. third panels), nor did exposure to exogenous fumarate (Figure S9A, third vs. fourth panels).
We next evaluated the impact of fumarate and FUM1 on sensitivity to camptothecin. Camptothecin is a topoisomerase I (TopI) inhibitor that causes ssDNA nicks by stabilizing TopI cleavage complexes and preventing them from religating DNA. This then results in replication-dependent formation of DSBs on the leading strand when forks collide with the TopI cleavage complexes (Covey et al. 1989; Hsiang et al. 1989; Strumberg et al. 2000). Thus, camptothecin activates the RAD9-dependent (Lancelot et al. 2007) DDC branch of the intra-S phase checkpoint, in contrast to the DRC-related pathway implicated in relation to HU discussed above. As shown in Figure S9B (top row, panels one and three) htz1Δ mutants were hypersensitive to camptothecin (see also Deng et al. 2005), and this sensitivity was suppressed by the addition of exogenously expressed HTZ1. In contrast to what had been observed for HU, this sensitivity was increased by loss of FUM1. Cells lacking both FUM1 and HTZ1 were hypersensitive to camptothecin relative to either single mutant or wild type, but this sensitivity could be partially suppressed by the addition of exogenous fumarate (Figure S9B first vs. third and fourth rows). Together, these results imply that FUM1 and fumarate influence repair of multiple forms of DNA damage, but not all repair pathways (see Discussion).
Discussion
Collectively, our findings are consistent with a role for metabolism in maintaining genome integrity. Here, we demonstrated that yeast fumarase, and fumarate, the product of catalysis by fumarase, act as intra-S phase checkpoint response factors (summarized in Table 1). Consistent with fumarate promoting replication fork integrity, we found that an increase in cellular levels of fumarate by deletion of FUM1, or addition of exogenous fumarate relieved the sensitivity to replication stress of yeast lacking HTZ1 (Figure 1 and Figure S1), or SWR1 (Figure S2). Evidence from our genetic studies are consistent with fumarate conferring resistance to HU by modulation of histone methylation levels primarily through inhibition of the JmjC domain-containing histone demethylase Jhd2p (Figure 3) rather than via modulation of nucleotide pools (Figure 2), cell cycle progression (Figure S6), or checkpoint activation (Figure S7). We have shown that deletion of JHD2 suppressed the sensitivity of htz1Δ mutants to replication stress (Figure 3 and Figure 5), and exogenous fumarate, or loss of FUM1, could modulate H3 K4 methylation levels in vivo (Figure 6 and Figure S5). These results are consistent with elevated histone H3 K4 methylation conferring resistance to replication stress in htz1Δ mutants. Our findings revealed fumarate could also promote resistance to replication stress through a second pathway in htz1Δ mutants that was H3 K4 methylation- or JHD2-independent (Figure 4 and Figure 5), implying that multiple mechanisms exist by which fumarate promoted growth during replication stress. Synthetic interaction analyses with intra-S phase checkpoint factors were consistent with the sensitivity to replication stress of htz1Δ mutants, and suppression of this sensitivity by fumarate, being primarily associated with defects in one or more events involved in processing and restart of intact, stalled forks, rather than recognition of damage or activation of the intra-S phase checkpoint (Figure 7, Figure 8, Figure 9, Figure S3, Figure S7, Figure S8, and Table 1).
Table 1. Summary of genetic interactions between htz1Δ and DNA replication stress response components or histone demethylases.
| Function | Mutant tested | Genetic interaction with htz1Δ on rich medium | Genetic interaction with htz1Δ on HU | Suppression of HU sensitivity by fumarate in wild-type background | Suppression of HU sensitivity by fumarate in htz1Δ background |
|---|---|---|---|---|---|
| Sensor of DNA replication stress | rad17Δ | Nonea | None | N/Db | Noc |
| ddc1Δ | None | None | N/D | No | |
| rad24Δ | None | None | N/D | No | |
| Mediator of checkpoint signaling | mrc1Δ | None | −d | N/D | No |
| rad9Δ | None | +d | N/D | N/D | |
| DNA processing factor | exo1Δ | None | None | N/D | Yesc |
| sgs1Δ | — | N/D | No | No | |
| sae2Δ | None | — | Yes | Yes | |
| yku70Δ | None | None | N/D | Yes | |
| mre11Δ | None | — | No | No | |
| rad50Δ | None | + | No | Yes | |
| xrs2Δ | None | + | No | Yes | |
| rad52Δ | Not testede | Not tested | No | Not tested | |
| JmjC domain enzyme | jhd1Δ | None | None | N/D | Yes |
| jhd2Δ | + | + | N/D | N/D | |
| rph1Δ | NONEf | — | N/D | No | |
| gis1Δ | NONE | None | N/D | No | |
| ecm5Δ | NONE | None | N/D | No | |
| Other | fum1Δ | None | + | N/D | Yes |
| swr1Δ | Not tested | Not tested | Yes | Not tested | |
| sml1Δ | — | + | N/D | Yes | |
| ade12Δ | None | — | N/D | Yes |
No genetic interaction observed.
Not detectable under conditions of assay.
No detectable suppression (No), or suppression (Yes) of HU sensitivity by fumarate.
Negative (−), or positive (+) genetic interaction.
Mutant was not tested.
No genetic interaction on rich medium, negative genetic interaction on rich medium containing fumarate.
In humans, fumarase has been described as a tumor suppressor where its loss has been associated with stabilization of HIF1-α under normoxic conditions through inhibition of α-KG-dependent prolyl hydroxylases (Tomlinson et al. 2002; Isaacs et al. 2005; Koivunen et al. 2007; Gaude and Frezza 2014; Laurenti and Tennant 2016). However, a growing body of evidence points toward an additional integral role of this tumor suppressor in responses to DNA damage. In the past few years, a direct link between fumarase deficiency and genome instability has emerged from studies in yeast as well as in mammalian cells. Yeast expressing Fum1p exclusively in mitochondria are sensitive to ionizing radiation, HU, and DSBs created by expression of the HO endonuclease (Yogev et al. 2010; Leshets et al. 2018), and exhibit dramatically reduced stability of Sae2p, leading to defects in resection at HO-mediated DSBs (Leshets et al. 2018). Fum1p binds Sae2p in vitro and in vivo (Leshets et al. 2018); however, whether the catalytic activity of Fum1p, in addition to binding, is required to stabilize Sae2p has yet to be tested. In this study, we have demonstrated that Fum1p acted as an intra-S phase checkpoint response factor that became induced and enriched in the nucleus upon exposure to stress during DNA replication [Figure 1, see also Yogev et al. (2010)], and that fumarate could suppress sensitivity to DNA replication stress in yeast lacking SAE2 (Figure 9A), collectively implying Fum1p promotes genome integrity both through stabilizing Sae2p, and through the production of fumarate to modulate chromatin modification states. Interestingly, human fumarase also becomes enriched in chromatin extracts after exposure to ionizing radiation, and is recruited to DSBs created by the restriction endonuclease I-SceI (Jiang et al. 2015). Upon induction of a DSB in human cells, chromatin association of fumarase is facilitated by its interaction with H2A.Z, but whether CtIP (Sae2p) is also required is unknown. During DSB repair by NHEJ, chromatin-associated fumarase promotes association of the NHEJ factor Ku70 by production of fumarate and inhibition of KDM2B, a histone demethylase that targets H3 K36 methylation (Jiang et al. 2015). In contrast, during the intra-S phase checkpoint response in budding yeast, sensitivity to replication stress in htz1Δ mutants was independent of YKU70, and fumarate promoted resistance to replication stress in htz1Δ mutants through a YKU70-independent pathway (Figure S8) that involved inhibition of Jhd2p, a H3 K4-specific demethylase (Figure 3, Figure 4, Figure 5, and Figure 6). As fumarate could also suppress the sensitivity to DNA replication stress of htz1Δ sae2Δ mutants, which contained FUM1 but lacked these anticipated partners for targeting Fum1p to sites of damage (Figure 9A), our findings are consistent with a model (Figure 10) in which a critical role of fumarase during the DNA replication stress response is to modify chromatin composition by generating fumarate to inhibit Jhd2p, and potentially other dioxygenases (Figure 3, Figure 4, Figure 5, and Figure 6).
Figure 10.
Model: Fumarate promotes cell survival in htz1Δ mutants upon DNA replication stress by inhibition of the JmjC domain-containing histone demethylase Jhd2p.
Additional investigation will be required to decipher the mechanism(s) by which modulation of H3 K4 methylation by fumarate contributes to resistance to HU, but our data are consistent with a role for this modification in facilitating processing and restart of stalled forks (Figure 9). While replication fork reversal during replication stress protects genome stability by facilitating replication restart, reversed forks resemble one end of a DSB, and are susceptible to excessive resection of nascent strands, resulting in genome instability (Thangavel et al. 2015; Zellweger et al. 2015; Giannattasio and Branzei 2017; Quinet et al. 2017; Menin et al. 2018). Thus, mechanisms induced during intra-S phase checkpoint activation also serve to limit resection during replication restart (Sogo et al. 2002; Rossi et al. 2015). Precedence exists for a role of H3 K4 methylation in intra-S phase checkpoint response and replication restart. In budding yeast, cells lacking SET1 and expressing wild-type H3 or H3 K4R are similarly sensitive to replication stress, and set1Δ mutants exhibit a defect in recovery from exposure to HU as well as defects in recruiting YKu80p to DSBs (Faucher and Wellinger 2010). In mammals, SETD1A localizes to replication forks, and SETD1A-dependent H3 K4 methylation at forks stalled with HU protects such compromised forks from excessive Dna2-dependent resection via promoting histone mobilization by the chaperone FANCD2, and by negatively regulating the remodeler CHD4. This, in turn, promotes recruitment of RAD51, or RAD51 filament stability, at stalled or arrested forks in HU or MMS (Higgs et al. 2018). Additionally, SETD1A may play a role in the mammalian transcriptional response during DDR, but the impact of this effect is unclear (Arndt et al. 2018; Higgs et al. 2018; Hoshii et al. 2018). Other examples for a role of H3 K4 methylation in fork restart during the intra-S phase checkpoint response can be found with the mammalian H3 K4-specific methyltransferases MLL2/3, which also function at stalled forks by enhancing recruitment of MRE11 and influencing fork processing in the absence of BRCA2 (Ray Chaudhuri et al. 2016; Higgs et al. 2018), as well as in the H3 K4- and K36-specific methyltransferase Metnase, which participates in restarting stalled forks after arrest in HU (De Haro et al. 2010). The relationship between these methyltransferases, H2A.Z, and fumarate at stalled forks in mammals awaits investigation.
Prior to the identification of H2A.Z as a binding partner of human fumarase (Jiang et al. 2015), Htz1p/H2A.Z had been identified as a participant in responses to DNA damage in yeast and human cells (Mizuguchi et al. 2004; Dhillon et al. 2006; Kalocsay et al. 2009; Xu et al. 2012; Adkins et al. 2013), and yeast HTZ1 had been found to exhibit synthetic genetic interactions with various DNA damage response factors including MEC1, MRC1, RAD53, and EXO1 (Figure 7 and Figure 9, and see Pan et al. 2006; Collins et al. 2007; Adkins et al. 2013). During DSB repair, this histone variant is transiently incorporated around DSBs (Kalocsay et al. 2009), and Htz1p as well as the chromatin remodeling complexes SWR1C and INO80C, which regulate deposition of Htz1p onto chromatin, are involved in NHEJ, homologous recombination, and fork stability (Kobor et al. 2004; Papamichos-Chronakis et al. 2006, 2011; van Attikum et al. 2007; Papamichos-Chronakis and Peterson 2008; Xu et al. 2012; Adkins et al. 2013). Like in htz1Δ mutants, fumarate suppresses the sensitivity to replication stress of swr1Δ mutants (Figure S2), which are defective in incorporation of Htz1p into nucleosomes (Krogan et al. 2003; Kobor et al. 2004; Mizuguchi et al. 2004).
The chromatin remodeler INO80C is a target of the DRC; INO80C binds and is phosphorylated by Rad53p (Morrison et al. 2007; Chen et al. 2010; Poli et al. 2016), and participates in removal of Htz1p adjacent to DSBs (van Attikum et al. 2007; Lademann et al. 2017), which promotes Rad51p presynaptic filament formation for homologous recombination (Lademann et al. 2017). Together with Mec1p and the PAF1 complex, INO80C has also been implicated in replication fork progression and restart during collisions between the replication and transcription machinery through a mechanism that involves eviction of the initiating form (phosphorylated on Ser 5) of RNA PolI from DNA during replication stress in HU (Poli et al. 2016). Consistent with this observation, during recovery from HU, DSBs in mec1 mutants are more likely to occur within genes induced by replication stress (Hoffman et al. 2015). How the substrate of INO80C—nucleosomes containing Htz1p, such as those found at transcriptional start sites—affect the efficiency of resolving such collisions is unknown. Interestingly, the PAF1 complex is also essential for monubiquitination of H2B at promoters by Bre1p-Rad6p (Wood et al. 2003). Bre1p-Rad6p, in turn, is required for H3 K4 methylation by COMPASS/Set1p (Dover et al. 2002; Sun and Allis 2002; Krogan et al. 2003; Ng et al. 2003; Wood et al. 2003) and for association of COMPASS with RNA Pol II (Krogan et al. 2003). htz1Δ H3 K4R mutants exhibit synthetic growth defects (Figure 4 and Venkatasubrahmanyam et al. 2007), and set1Δ and H3 K4R mutants exhibit growth defects in HU (Figure 4 and Figure 5 and Faucher and Wellinger 2010). In addition, loss of FUM1 or exogenous fumarate promoted H3 K4 methylation (Figure 6 and Figure S5), and fumarate suppressed sensitivity of htz1Δ mutants to HU through a JHD2-dependent pathway (Figure 3). Thus, it is tempting to speculate that, by promoting H3 K4 methylation, fumarate could facilitate processing of such replication-transcription machinery collisions occurring in the htz1Δ mutants during replication stress.
In addition to having growth defects upon exposure to HU, set1Δ mutants display an increased rate of plasmid loss relative to wild-type that could be suppressed by multiple copies of an origin of replication on a plasmid in mini-chromosome maintenance assays (Faucher and Wellinger 2010; Rizzardi et al. 2012). Additionally, H3 K4me2 and me3 are enriched at origins of replication and H3 K4 methylation promotes efficient origin function (Rizzardi et al. 2012). As htz1Δ mutants show delays in origin firing (Dhillon et al. 2006), these observations collectively support a model in which high levels of H3 K4 methylation (by deletion of JHD2 or inhibition of Jhd2p by elevated levels of cellular fumarate) could promote firing from inefficient or late origins to complement htz1Δ-dependent delays in replication during replication stress. Such a mechanism could involve instilling characteristics normally associated with early origins to late origins to advance timing of firing, or bypass of the Mrc1p-activated Rad53p inhibitory signal during DDR that normally blocks late origin firing in HU. However, we did not observe dramatic fumarate-dependent effects on cell cycle progression during replication stress in htz1Δ mutants (Figure S6), and this scenario would require late origin regulation in yeast to be somewhat different than in mammalian cells, which require SETD1A to prevent late origin firing after exposure to MMS during the DRC response (Higgs et al. 2018).
Thus, we instead favor a model in which fumarate confers resistance to replication stress primarily by bypassing a defect in replication fork processing and restart caused by the absence of HTZ1 through upregulating histone methylation, in part, by inhibition of Jhd2p (Figure 10). However, a second possible mechanism by which growth defects of htz1Δ mutants during replication stress may be suppressed by fumarate is by complementing defects in spindle assembly. In addition to inducing DNA replication stress, HU can cause defects in spindle formation (Liu et al. 2008). Moreover, Htz1p localizes to pericentric chromatin, and htz1Δ mutants show defects in spindle assembly and sensitivity to benomyl (Krogan et al. 2004; Keogh et al. 2006), but, like wild-type cells, arrest in HU with short mitotic spindles, a single nucleus, and large buds (Dhillon and Kamakaka 2000).
Future studies will also be required to reveal whether fumarase and fumarate can act in histone methylation independent pathway(s) to modulate cellular responses to DNA damage and replication stress. For example, fumarate has the potential to regulate protein function (e.g., Blatnik et al. 2008) through reacting with cysteine residues to create a post-translational modification known as succination (Alderson et al. 2006). In fact, high levels of protein succination have been found in tumors with fumarase deficiency from patients with HLRCC (Bardella et al. 2011), but potential regulatory roles of protein succination on cysteine residues during responses to DNA damage largely await characterization.
Collectively, the results of this study and others (Yogev et al. 2010; Leshets et al. 2018) indicate that cellular responses to DNA replication stress are sensitive to fumarate, and imply that metabolism is intimately linked to the intra-S phase checkpoint response. Consistent with our findings, other studies have reported that addition of exogenous antagonists of α-KG including fumarate, succinate, or the oncometabolite R-2-HG as well as expression of tumor derived FH, SDH, or IDH mutants cause a global increase of histone methylation levels (Xu et al. 2011; Xiao et al. 2012). Moreover, fumarate, succinate and 2-HG can modulate numerous cellular functions ranging from gene expression and silencing to DNA damage responses (Xu et al. 2011; Xiao et al. 2012; Jiang et al. 2015; Janke et al. 2017; Sulkowski et al. 2017). Thus, changes in metabolite availability through normal signal relay pathways, exogenous sources or genetic metabolic defects have the potential to impact genome integrity upon DNA replication stress by regulation of histone methylation, supporting a model in which metabolites broadly play crucial roles as chemical messengers in signaling pathways triggering cellular responses to various types of stress—the examples illustrated herein being DNA replication stress and DNA damage.
Acknowledgments
We thank Jill Hutchcroft and Jordan Page for technical assistance, and Scott Briggs, Paul Kaufman, and Mark Parthun for plasmids. We are also thankful to Joe Ogas and Scott Briggs for their input into this manuscript. This work was supported by the National Science Foundation (NSF) (A.L.K.), Purdue University Office of Vice Provost for Research (A.L.K.), Purdue University Department of Biochemistry, and the Cancer Prevention Internship Program [National Cancer Institute (NCI) R25CA128770]. This research was also supported by the National Cancer Institute (NCI P30 CA23168) for data acquired in the Purdue University Center for Cancer Research (PCCR) DNA Sequencing Facility and Flow Cytometry facility as well as Bird Stair Research Fellowships (F.S.).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8109437.
Communicating editor: D. Bishop
Literature Cited
- Adams A., Gottschling D. E., Kaiser C., Stearns T., 1998. Methods in Yeast Genetics : A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- Adkins N. L., Niu H., Sung P., Peterson C. L., 2013. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20: 836–842. 10.1038/nsmb.2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait Saada A., Teixeira-Silva A., Iraqui I., Costes A., Hardy J., et al. , 2017. Unprotected replication forks are converted into mitotic sister chromatid bridges. Mol. Cell 66: 398–410.e4. 10.1016/j.molcel.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Akiba T., Hiraga K., Tuboi S., 1984. Intracellular distribution of fumarase in various animals. J. Biochem. 96: 189–195. 10.1093/oxfordjournals.jbchem.a134812 [DOI] [PubMed] [Google Scholar]
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J. H., et al. , 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. 10.1038/ncb1101-958 [DOI] [PubMed] [Google Scholar]
- Alderson N. L., Wang Y., Blatnik M., Frizzell N., Walla M. D., et al. , 2006. S-(2-Succinyl)cysteine: a novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 450: 1–8. 10.1016/j.abb.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Arikawa Y., Kuroyanagi T., Shimosaka M., Muratsubaki H., Enomoto K., et al. , 1999. Effect of gene disruptions of the TCA cycle on production of succinic acid in Saccharomyces cerevisiae. J. Biosci. Bioeng. 87: 28–36. 10.1016/S1389-1723(99)80004-8 [DOI] [PubMed] [Google Scholar]
- Arndt K., Kranz A., Fohgrub J., Jolly A., Bledau A. S., et al. , 2018. SETD1A protects HSCs from activation-induced functional decline in vivo. Blood 131: 1311–1324. 10.1182/blood-2017-09-806844 [DOI] [PubMed] [Google Scholar]
- Bacal J., Moriel‐Carretero M., Pardo B., Barthe A., Sharma S., et al. , 2018. Mrc1 and Rad9 cooperate to regulate initiation and elongation of DNA replication in response to DNA damage. EMBO J. 37: pii: e99319. 10.15252/embj.201899319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardella C., El-Bahrawy M., Frizzell N., Adam J., Ternette N., et al. , 2011. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 225: 4–11. 10.1002/path.2932 [DOI] [PubMed] [Google Scholar]
- Barnes L. B., Bishop S. H., 1975. Adenylosuccinate lyase from human erythrocytes. Int. J. Biochem. 6: 497–503. 10.1016/0020-711X(75)90030-0 [DOI] [Google Scholar]
- Bartkova J., Hořejší Z., Koed K., Krämer A., Tort F., et al. , 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870. 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- Bernstein K. A., Shor E., Sunjevaric I., Fumasoni M., Burgess R. C., et al. , 2009. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 28: 915–925. 10.1038/emboj.2009.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K. A., Gangloff S., Rothstein R., 2010. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 44: 393–417. 10.1146/annurev-genet-102209-163602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilousova G., Marusyk A., Porter C. C., Cardiff R. D., DeGregori J., 2005. Impaired DNA replication within progenitor cell pools promotes leukemogenesis. PLoS Biol. 3: e401 10.1371/journal.pbio.0030401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Xin H., Dutta-Biswas R., Yu Y., et al. , 2008. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178: 649–659. 10.1534/genetics.107.084202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb J. A., Tsai-Pflugfelder M., Gasser S. M., 2005. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24: 405–417. 10.1038/sj.emboj.7600511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatnik M., Thorpe S. R., Baynes J. W., 2008. Succination of proteins by fumarate: mechanism of inactivation of glyceraldehyde-3-phosphate dehydrogenase in diabetes. Ann. N. Y. Acad. Sci. 1126: 272–275. 10.1196/annals.1433.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Jinks-Robertson S., Hartwell L. H., Crouse G. F., 2013. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193: 1025–1064. 10.1534/genetics.112.145219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double‐strand break repair and serves as a barrier to error‐prone DNA repair pathways. EMBO J. 15: 5093–5103. 10.1002/j.1460-2075.1996.tb00890.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun T. S., Pacek M., Yee M., Walter J. C., Cimprich K. A., 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19: 1040–1052. 10.1101/gad.1301205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Cejka P., 2014. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514: 122–125. 10.1038/nature13771 [DOI] [PubMed] [Google Scholar]
- Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., et al. , 2010. DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 467: 112–116. 10.1038/nature09355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A., Domkin V., Thelander L., 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274: 36679–36683. 10.1074/jbc.274.51.36679 [DOI] [PubMed] [Google Scholar]
- Chen S., Albuquerque C. P., Liang J., Suhandynata R. T., Zhou H., 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285: 12803–12812. 10.1074/jbc.M110.106989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-H., Zhou H., 2009. Reconstitution of Rad53 activation by Mec1 through adaptor protein Mrc1. J. Biol. Chem. 284: 18593–18604. 10.1074/jbc.M109.018242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M., 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22: 4325–4336. 10.1093/emboj/cdg391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. 10.1038/nature05649 [DOI] [PubMed] [Google Scholar]
- Colosio A., Frattini C., Pellicanò G., Villa-Hernández S., Bermejo R., 2016. Nucleolytic processing of aberrant replication intermediates by an Exo1-Dna2-Sae2 axis counteracts fork collapse-driven chromosome instability. Nucleic Acids Res. 44: 10676–10690. 10.1093/nar/gkw858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey J. M., Jaxel C., Kohn K. W., Pommier Y., 1989. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 49: 5016–5022. [PubMed] [Google Scholar]
- Daganzo S. M., Erzberger J. P., Lam W. M., Skordalakes E., Zhang R., et al. , 2003. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13: 2148–2158. 10.1016/j.cub.2003.11.027 [DOI] [PubMed] [Google Scholar]
- De Haro L. P., Wray J., Williamson E. A., Durant S. T., Corwin L., et al. , 2010. Metnase promotes restart and repair of stalled and collapsed replication forks. Nucleic Acids Res. 38: 5681–5691. 10.1093/nar/gkq339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Brown J. A., You D., Brown J. M., 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170: 591–600. 10.1534/genetics.104.028795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Kamakaka R. T., 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6: 769–780. 10.1016/S1097-2765(00)00076-9 [DOI] [PubMed] [Google Scholar]
- Dhillon N., Oki M., Szyjka S. J., Aparicio O. M., Kamakaka R. T., 2006. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 26: 489–501. 10.1128/MCB.26.2.489-501.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler L., Schmidt K. H., 2014. Exo1 phosphorylation status controls the hydroxyurea sensitivity of cells lacking the Pol32 subunit of DNA polymerases delta and zeta. DNA Repair (Amst.) 24: 26–36. 10.1016/j.dnarep.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., et al. , 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277: 28368–28371. 10.1074/jbc.C200348200 [DOI] [PubMed] [Google Scholar]
- Duncker B. P., Shimada K., Tsai-Pflugfelder M., Pasero P., Gasser S. M., 2002. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA 99: 16087–16092. 10.1073/pnas.252093999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Hogan G. J., Liang G., Lieb J. D., Zhang Y., 2007. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Mol. Cell. Biol. 27: 5055–5065. 10.1128/MCB.00127-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher D., Wellinger R. J., 2010. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 6: e1001082 10.1371/journal.pgen.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieuw A., Kumps C., Schramm A., Pattyn F., Menten B., et al. , 2012. Identification of a novel recurrent 1q42.2–1qter deletion in high risk MYCN single copy 11q deleted neuroblastomas. Int. J. Cancer 130: 2599–2606. 10.1002/ijc.26317 [DOI] [PubMed] [Google Scholar]
- Foster S. S., Balestrini A., Petrini J. H. J., 2011. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol. Cell. Biol. 31: 4379–4389. 10.1128/MCB.05854-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A. A., Lam W. M., Burgers P. M., Kaufman P. D., 2005. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 19: 1365–1375. 10.1101/gad.1305005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S. M., Kizer K. O., Braberg H., Krogan N. J., Strahl B. D., 2012. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J. Biol. Chem. 287: 3249–3256. 10.1074/jbc.M111.273953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Poitelea M., Guo L., Caspari T., Carr A. M., 2004. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 18: 1154–1164. 10.1101/gad.291104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K., 1974. A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. Mol. Mech. Mutagen. 24: 281–292. 10.1016/0027-5107(74)90176-6 [DOI] [PubMed] [Google Scholar]
- Gaude E., Frezza C., 2014. Defects in mitochondrial metabolism and cancer. Cancer Metab. 2: 10 10.1186/2049-3002-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke I., Muniyappa K., 2016. Genetic and biochemical evidences reveal novel insights into the mechanism underlying Saccharomyces cerevisiae Sae2-mediated abrogation of DNA replication stress. J. Biosci. 41: 615–641. 10.1007/s12038-016-9642-9 [DOI] [PubMed] [Google Scholar]
- Giannattasio M., Branzei D., 2017. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cell. Mol. Life Sci. 74: 2361–2380. 10.1007/s00018-017-2474-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C. S., Green C. M., Lowndes N. F., 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8: 129–136. 10.1016/S1097-2765(01)00267-2 [DOI] [PubMed] [Google Scholar]
- Gorgoulis V. G., Vassiliou L.-V. F., Karakaidos P., Zacharatos P., Kotsinas A., et al. , 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913. 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego. [Google Scholar]
- Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V., 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17: 1305–1311. 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer M. H., Gasser S. M., 2017. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 31: 2204–2221. 10.1101/gad.307702.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegnauer A. M., Hustedt N., Shimada K., Pike B. L., Vogel M., et al. , 2012. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 31: 3768–3783. 10.1038/emboj.2012.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs M. R., Reynolds J. J., Winczura A., Blackford A. N., Borel V., et al. , 2015. BOD1L is required to suppress Deleterious resection of stressed replication forks. Mol. Cell 59: 462–477. 10.1016/j.molcel.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Higgs M. R., Sato K., Reynolds J. J., Begum S., Bayley R., et al. , 2018. Histone methylation by SETD1A protects nascent DNA through the nucleosome chaperone activity of FANCD2. Mol. Cell 71: 25–41.e6. 10.1016/j.molcel.2018.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E. A., McCulley A., Haarer B., Arnak R., Feng W., 2015. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 25: 402–412. 10.1101/gr.180497.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshii T., Cifani P., Feng Z., Huang C.-H., Koche R., et al. , 2018. A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell 172: 1007–1021.e17. 10.1016/j.cell.2018.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- House N. C. M., Koch M. R., Freudenreich C. H., 2014. Chromatin modifications and DNA repair: beyond double-strand breaks. Front. Genet. 5: 296 10.3389/fgene.2014.00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y. H., Lihou M. G., Liu L. F., 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49: 5077–5082. [PubMed] [Google Scholar]
- Ingvarsdottir K., Edwards C., Lee M. G., Lee J. S., Schultz D. C., et al. , 2007. Histone H3 K4 demethylation during activation and attenuation of GAL1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 7856–7864. 10.1128/MCB.00801-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui I., Chekkal Y., Jmari N., Pietrobon V., Fréon K., et al. , 2012. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 8: e1002976 10.1371/journal.pgen.1002976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs J. S., Jung Y. J., Mole D. R., Lee S., Torres-Cabala C., et al. , 2005. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153. 10.1016/j.ccr.2005.06.017 [DOI] [PubMed] [Google Scholar]
- Janke R., Iavarone A. T., Rine J., 2017. Oncometabolite D-2-Hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. eLife 6: pii: e22451. 10.7554/eLife.22451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha D. K., Strahl B. D., 2014. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 5: 3965 [corrigenda: Nat. Commun. 6: 6015 (2015)]. 10.1038/ncomms4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Qian X., Shen J., Wang Y., Li X., et al. , 2015. Local generation of fumarate promotes DNA repair through inhibition of histone H3 demethylation. Nat. Cell Biol. 17: 1158–1168 [corrigenda: Nat. Cell Biol. 20: 1226 (2018)]. 10.1038/ncb3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N. J., Jentsch S., 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 33: 335–343. 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Karniely S., Pines O., 2005. Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 6: 420–425. 10.1038/sj.embor.7400394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. 10.1038/nature01900 [DOI] [PubMed] [Google Scholar]
- Keogh M.-C., Kim J.-A., Downey M., Fillingham J., Chowdhury D., et al. , 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501. 10.1038/nature04384 [DOI] [PubMed] [Google Scholar]
- Khalil A. A., 2007. Biomarker discovery: a proteomic approach for brain cancer profiling. Cancer Sci. 98: 201–213. 10.1111/j.1349-7006.2007.00374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Buratowski S., 2007. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J. Biol. Chem. 282: 20827–20835. 10.1074/jbc.M703034200 [DOI] [PubMed] [Google Scholar]
- Kish A., DiRuggiero J., 2008. Rad50 is not essential for the Mre11-dependent repair of DNA double-strand breaks in halobacterium sp. strain NRC-1. J. Bacteriol. 190: 5210–5216. 10.1128/JB.00292-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M., Lehtonen R., Arola J., Salovaara R., Järvinen H., et al. , 2002. Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. 62: 4554–4557. [PubMed] [Google Scholar]
- Knox C., Sass E., Neupert W., Pines O., 1998. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J. Biol. Chem. 273: 25587–25593. 10.1074/jbc.273.40.25587 [DOI] [PubMed] [Google Scholar]
- Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., et al. , 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131 10.1371/journal.pbio.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P., Hirsilä M., Remes A. M., Hassinen I. E., Kivirikko K. I., et al. , 2007. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 282: 4524–4532. 10.1074/jbc.M610415200 [DOI] [PubMed] [Google Scholar]
- Komata M., Bando M., Araki H., Shirahige K., 2009. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol. Cell. Biol. 29: 5008–5019. 10.1128/MCB.01934-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Matsumoto K., Sugimoto K., 1999. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol. 19: 1136–1143. 10.1128/MCB.19.2.1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakoff I. H., Brown N. C., Reichard P., 1968. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 28: 1559–1565. [PubMed] [Google Scholar]
- Krogan N. J., Keogh M.-C., Datta N., Sawa C., Ryan O. W., et al. , 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12: 1565–1576. 10.1016/S1097-2765(03)00497-0 [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Baetz K., Keogh M.-C., Datta N., Sawa C., et al. , 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101: 13513–13518 [corrigenda: Proc. Natl. Acad. Sci. USA 103: 6410 (2006)]. 10.1073/pnas.0405753101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D.-W., Ahn S. H., 2011. Role of yeast JmjC-domain containing histone demethylases in actively transcribed regions. Biochem. Biophys. Res. Commun. 410: 614–619. 10.1016/j.bbrc.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Lademann C. A., Renkawitz J., Pfander B., Jentsch S., 2017. The INO80 complex removes H2A.Z to promote presynaptic filament formation during homologous recombination. Cell Rep. 19: 1294–1303. 10.1016/j.celrep.2017.04.051 [DOI] [PubMed] [Google Scholar]
- Lambert S., Mizuno K., Blaisonneau J., Martineau S., Chanet R., et al. , 2010. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 39: 346–359. 10.1016/j.molcel.2010.07.015 [DOI] [PubMed] [Google Scholar]
- Lancelot N., Charier G., Couprie J., Duband-Goulet I., Alpha-Bazin B., et al. , 2007. The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA. Nucleic Acids Res. 35: 5898–5912. 10.1093/nar/gkm607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukka T., Mariani C. J., Ihantola T., Cao J. Z., Hokkanen J., et al. , 2016. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 291: 4256–4265. 10.1074/jbc.M115.688762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launonen V., Vierimaa O., Kiuru M., Isola J., Roth S., et al. , 2001. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc. Natl. Acad. Sci. USA 98: 3387–3392. 10.1073/pnas.051633798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti G., Tennant D. A., 2016. Isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH), fumarate hydratase (FH): three players for one phenotype in cancer? Biochem. Soc. Trans. 44: 1111–1116. 10.1042/BST20160099 [DOI] [PubMed] [Google Scholar]
- Lazzaro F., Sapountzi V., Granata M., Pellicioli A., Vaze M., et al. , 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 27: 1502–1512. 10.1038/emboj.2008.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen R., Kiuru M., Vanharanta S., Sjöberg J., Aaltonen L.-M., et al. , 2004. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am. J. Pathol. 164: 17–22. 10.1016/S0002-9440(10)63091-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T., 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28: 638–651. 10.1016/j.molcel.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshets M., Ramamurthy D., Lisby M., Lehming N., Pines O., 2018. Fumarase is involved in DNA double-strand break resection through a functional interaction with Sae2. Curr. Genet. 64: 697–712. https://www.ncbi.nlm.nih.gov/pubmed/29204698 [DOI] [PubMed] [Google Scholar]
- Liang G., Klose R. J., Gardner K. E., Zhang Y., 2007. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 14: 243–245 [corrigenda: Nat. Struct. Mol. Biol. 14: 351 (2007)]. 10.1038/nsmb1204 [DOI] [PubMed] [Google Scholar]
- Lin A.-P., Anderson S. L., Minard K. I., McAlister-Henn L., 2011. Effects of excess succinate and retrograde control of metabolite accumulation in yeast tricarboxylic cycle mutants. J. Biol. Chem. 286: 33737–33746. 10.1074/jbc.M111.266890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R., 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Liu H., Liang F., Jin F., Wang Y., 2008. The coordination of centromere replication, spindle formation, and kinetochore–microtubule interaction in budding yeast. PLoS Genet. 4: e1000262 10.1371/journal.pgen.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., Komata M., Katou Y., Guan Z., Reis C. C., et al. , 2008. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32: 106–117. 10.1016/j.molcel.2008.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie E. D., Selak M. A., Tennant D. A., Payne L. J., Crosby S., et al. , 2007. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol. Cell. Biol. 27: 3282–3289. 10.1128/MCB.01927-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J., Burgers P. M. J., 2003. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 100: 2249–2254. 10.1073/pnas.0437148100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menin L., Ursich S., Trovesi C., Zellweger R., Lopes M., et al. , 2018. Tel1/ATM prevents degradation of replication forks that reverse after topoisomerase poisoning. EMBO Rep. 19: e45535 10.15252/embr.201745535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menko F. H., Maher E. R., Schmidt L. S., Middelton L. A., Aittomäki K., et al. , 2014. Hereditary leiomyomatosis and renal cell cancer (HLRCC): renal cancer risk, surveillance and treatment. Fam. Cancer 13: 637–644. 10.1007/s10689-014-9735-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman D. P., Du H.-N., Fingerman I. M., South P. F., Briggs S. D., 2009. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 23: 951–962. 10.1101/gad.1769209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A., Yang B., Foster T., Kirchmaier A. L., 2008. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics 179: 793–809. 10.1534/genetics.107.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Symington L. S., 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.-H., Sen S., et al. , 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- Morillo-Huesca M., Clemente-Ruiz M., Andújar E., Prado F., Antonellis K., 2010. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One 5: e12143 10.1371/journal.pone.0012143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. J., Kim J.-A., Person M. D., Highland J., Xiao J., et al. , 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130: 499–511. 10.1016/j.cell.2007.06.010 [DOI] [PubMed] [Google Scholar]
- New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C., 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. 10.1038/34950 [DOI] [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K., 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709–719. 10.1016/S1097-2765(03)00092-3 [DOI] [PubMed] [Google Scholar]
- Nguyen M. O., Jalan M., Morrow C. A., Osman F., Whitby M. C., 2015. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. eLife 4: e04539 10.7554/eLife.04539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17: 1755–1767. 10.1101/gad.1098303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V., Lucchini G., Plevani P., Longhese M. P., DeMaggio A., et al. , 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 17: 4199–4209. 10.1093/emboj/17.14.4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C.-C., Deegan R. S., Subramanian L., Gal C., Sarkar S., et al. , 2014. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 5: 4091 10.1038/ncomms5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., et al. , 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [corrigenda: Cell 124: 1069–1081 (2013)]. 10.1016/j.cell.2005.12.036 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Peterson C. L., 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 15: 338–345. 10.1038/nsmb.1413 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Krebs J. E., Peterson C. L., 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20: 2437–2449. 10.1101/gad.1440206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L., 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Crabbé L., Pasero P., 2016. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 17: fow101 10.1093/femsyr/fow101 [DOI] [PubMed] [Google Scholar]
- Poli J., Gerhold C.-B., Tosi A., Hustedt N., Seeber A., et al. , 2016. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30: 337–354. 10.1101/gad.273813.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P. J., Brière J. J., Alam N. A., Barwell J., Barclay E., et al. , 2005. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 14: 2231–2239. 10.1093/hmg/ddi227 [DOI] [PubMed] [Google Scholar]
- Puddu F., Oelschlaegel T., Guerini I., Geisler N. J., Niu H., et al. , 2015. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 34: 1509–1522. 10.15252/embj.201590973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A., Lemaçon D., Vindigni A., 2017. Replication fork reversal: players and guardians. Mol. Cell 68: 830–833. 10.1016/j.molcel.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A. A., et al. , 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535: 382–387 [corrigenda: Nature 539: 456 (2016)]. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi L. F., Dorn E. S., Strahl B. D., Cook J. G., 2012. DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae. Genetics 192: 371–384. 10.1534/genetics.112.142349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S. E., Ajazi A., Carotenuto W., Foiani M., Giannattasio M., 2015. Rad53-mediated regulation of Rrm3 and Pif1 DNA helicases contributes to prevention of aberrant fork transitions under replication stress. Cell Rep. 13: 80–92. 10.1016/j.celrep.2015.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossodivita A. A., Boudoures A. L., Mecoli J. P., Steenkiste E. M., Karl A. L., et al. , 2014. Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae. Nucleic Acids Res. 42: 6286–6299. 10.1093/nar/gku242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J., Jackson S. P., 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9: 857–869. 10.1016/S1097-2765(02)00507-5 [DOI] [PubMed] [Google Scholar]
- Sass E., Blachinsky E., Karniely S., Pines O., 2001. Mitochondrial and cytosolic isoforms of yeast fumarase are derivatives of a single translation product and have identical amino termini. J. Biol. Chem. 276: 46111–46117. 10.1074/jbc.M106061200 [DOI] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., et al. , 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., Jasin M., 2012. A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22: 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L., 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18: 3091–3092. 10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sein H., Värv S., Kristjuhan A., 2015. Distribution and maintenance of histone H3 lysine 36 trimethylation in transcribed locus. PLoS One 10: e0120200 10.1371/journal.pone.0120200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seward D. J., Cubberley G., Kim S., Schonewald M., Zhang L., et al. , 2007. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 14: 240–242. 10.1038/nsmb1200 [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S. M., 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16: 3236–3252. 10.1101/gad.239802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura T., Ando S., Matsumoto K., Sugimoto K., 1998. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol. Cell. Biol. 18: 5485–5491. 10.1128/MCB.18.9.5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa T., 1998. Stimulation by Rad52 of yeast Rad51- mediated recombination. Nature 391: 404–407. 10.1038/34943 [DOI] [PubMed] [Google Scholar]
- Singer E., Silas Y. B., Ben-Yehuda S., Pines O., 2017. Bacterial fumarase and L-malic acid are evolutionary ancient components of the DNA damage response. eLife 6: pii: e30927. 10.7554/eLife.30927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M. B., Chen S., Maddox P. S., Enserink J. M., Albuquerque C. P., et al. , 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175: 743–753. 10.1083/jcb.200605081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Lopes M., Foiani M., 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Srivatsan A., Li B.-Z., Szakal B., Branzei D., Putnam C. D., et al. , 2018. The Swr1 chromatin-remodeling complex prevents genome instability induced by replication fork progression defects. Nat. Commun. 9: 3680 10.1038/s41467-018-06131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein I., Peleg Y., Even-Ram S., Pines O., 1994. The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 4770–4778. 10.1128/MCB.14.7.4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumberg D., Pilon A. A., Smith M., Hickey R., Malkas L., et al. , 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20: 3977–3987. 10.1128/MCB.20.11.3977-3987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulemeijer I. J. E., De Vos D., van Harten K., Joshi O. K., Blomberg O., et al. , 2015. Dot1 histone methyltransferases share a distributive mechanism but have highly diverged catalytic properties. Sci. Rep. 5: 9824 10.1038/srep09824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski P. L., Corso C. D., Robinson N. D., Scanlon S. E., Purshouse K. R., et al. , 2017. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 9: pii: eaal2463. 10.1126/scitranslmed.aal2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski P. L., Sundaram R. K., Oeck S., Corso C. D., Liu Y., et al. , 2018. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 50: 1086–1092. 10.1038/s41588-018-0170-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.-W., Allis C. D., 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. 10.1038/nature00883 [DOI] [PubMed] [Google Scholar]
- Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., et al. , 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15: 1364–1375. 10.1016/j.cub.2005.06.063 [DOI] [PubMed] [Google Scholar]
- Symington L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. 10.1128/MMBR.66.4.630-670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Russell P., 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3: 966–972. 10.1038/ncb1101-966 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Russell P., 2004. Cds1 phosphorylation by Rad3-Rad26 kinase is mediated by forkhead-associated domain interaction with Mrc1. J. Biol. Chem. 279: 32079–32086. 10.1074/jbc.M404834200 [DOI] [PubMed] [Google Scholar]
- Teixeira-Silva A., Ait Saada A., Hardy J., Iraqui I., Nocente M. C., et al. , 2017. The end-joining factor Ku acts in the end-resection of double strand break-free arrested replication forks. Nat. Commun. 8: 1982 10.1038/s41467-017-02144-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Longhese M. P., Diffley J. F. X., 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11: 1323–1336. 10.1016/S1097-2765(03)00169-2 [DOI] [PubMed] [Google Scholar]
- Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., et al. , 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 208: 545–562. 10.1083/jcb.201406100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M., Alabert C., Pasero P., Cobb J. A., 2009. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 28: 1142–1156. 10.1038/emboj.2009.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley E., Craig I., 1975. Presence of two forms of fumarase (fumarate hydratase E.C. 4.2.1.2) in mammalian cells: immunological characterization and genetic analysis in somatic cell hybrids. Confirmation of the assignment of a gene necessary for the enzyme expression to human chromosome 1. Biochem. Genet. 13: 867–883. 10.1007/BF00484417 [DOI] [PubMed] [Google Scholar]
- Tomlinson I. P. M., Alam N. A., Rowan A. J., Barclay E., Jaeger E. E., et al. , 2002. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30: 406–410. 10.1038/ng849 [DOI] [PubMed] [Google Scholar]
- Tsang E., Miyabe I., Iraqui I., Zheng J., Lambert S. A. E., et al. , 2014. The extent of error-prone replication restart by homologous recombination is controlled by Exo1 and checkpoint proteins. J. Cell Sci. 127: 2983–2994. 10.1242/jcs.152678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., et al. , 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816. 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- Tu S., Bulloch E. M. M., Yang L., Ren C., Huang W.-C., et al. , 2007. Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 282: 14262–14271. 10.1074/jbc.M609900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Gasser S. M., Bird A., Yu D., et al. , 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26: 4113–4125. 10.1038/sj.emboj.7601835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H. Y., Madhani H. D., 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A. Z. Proc. Natl. Acad. Sci. U. S. A. 104: 16609–16614. 10.1073/pnas.0700914104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versini G., Comet I., Wu M., Hoopes L., Schwob E., et al. , 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22: 1939–1949. 10.1093/emboj/cdg180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters R., van Eijk P., Reed S., 2015. Histone modification and chromatin remodeling during NER. DNA Repair (Amst.) 36: 105–113. 10.1016/j.dnarep.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. 10.1126/science.3291120 [DOI] [PubMed] [Google Scholar]
- Wood A., Schneider J., Dover J., Johnston M., Shilatifard A., 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278: 34739–34742. 10.1074/jbc.C300269200 [DOI] [PubMed] [Google Scholar]
- Woods S. A., Schwartzbach S. D., Guest J. R., 1988. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 954: 14–26. 10.1016/0167-4838(88)90050-7 [DOI] [PubMed] [Google Scholar]
- Wu L., Davies S. L., Levitt N. C., Hickson I. D., 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276: 19375–19381. 10.1074/jbc.M009471200 [DOI] [PubMed] [Google Scholar]
- Wu M., Tzagoloff A., 1987. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J. Biol. Chem. 262: 12275–12282. [PubMed] [Google Scholar]
- Wysocki R., Javaheri A., Allard S., Sha F., Côté J., et al. , 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25: 8430–8443. 10.1128/MCB.25.19.8430-8443.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Yang H., Xu W., Ma S., Lin H., et al. , 2012. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26: 1326–1338 [corrigenda: Genes Dev. 29: 887 (2015)]. 10.1101/gad.191056.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Yang H., Liu Y., Yang Y., Wang P., et al. , 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Davenport M., Kelly T. J., 2006. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev. 20: 990–1003. 10.1101/gad.1406706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ayrapetov M. K., Xu C., Gursoy-Yuzugullu O., Hu Y., et al. , 2012. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48: 723–733. 10.1016/j.molcel.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Soga T., Pollard P. J., 2013. Oncometabolites: linking altered metabolism with cancer. Clin. Investig. (Lond.) 123: 4–10. 10.1172/JCI67228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Yogev O., Singer E., Shaulian E., Goldberg M., et al. , 2010. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8: e1000328 10.1371/journal.pbio.1000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Naamati A., Pines O., 2011. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278: 4230–4242. 10.1111/j.1742-4658.2011.08359.x [DOI] [PubMed] [Google Scholar]
- Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J. A., et al. , 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208: 563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Muller E. G., Rothstein R., 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329–340. 10.1016/S1097-2765(00)80277-4 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
- Zou L., Liu D., Elledge S. J., 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 100: 13827–13832. 10.1073/pnas.2336100100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the findings of the article are present within the article, figures and tables. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8109437.