Abstract
MicroRNAs (miRNAs) are known to modulate gene expression, but their activity at the tissue-specific level remains largely uncharacterized. To study their contribution to tissue-specific gene expression, we developed novel tools to profile putative miRNA targets in the Caenorhabditis elegans intestine and body muscle. We validated many previously described interactions and identified ∼3500 novel targets. Many of the candidate miRNA targets curated are known to modulate the functions of their respective tissues. Within our data sets we observed a disparity in the use of miRNA-based gene regulation between the intestine and body muscle. The intestine contained significantly more putative miRNA targets than the body muscle highlighting its transcriptional complexity. We detected an unexpected enrichment of RNA-binding proteins targeted by miRNA in both tissues, with a notable abundance of RNA splicing factors. We developed in vivo genetic tools to validate and further study three RNA splicing factors identified as putative miRNA targets in our study (asd-2, hrp-2, and smu-2), and show that these factors indeed contain functional miRNA regulatory elements in their 3′UTRs that are able to repress their expression in the intestine. In addition, the alternative splicing pattern of their respective downstream targets (unc-60, unc-52, lin-10, and ret-1) is dysregulated when the miRNA pathway is disrupted. A reannotation of the transcriptome data in C. elegans strains that are deficient in the miRNA pathway from past studies supports and expands on our results. This study highlights an unexpected role for miRNAs in modulating tissue-specific gene isoforms, where post-transcriptional regulation of RNA splicing factors associates with tissue-specific alternative splicing.
Keywords: miRNA, alternative splicing, hrp-2, asd-2, smu-2, transcriptome, ALG-1, intestine, body muscle, Caenorhabditis elegans
MULTICELLULAR organisms have evolved complex forms of gene regulation achieved at different stages throughout development, and equally executed at pre-, co-, and post-transcriptional levels. Alternative splicing, which leads to the production of different protein isoforms using single mRNA precursors, fine tunes these regulatory networks and contributes to the acquisition of tissue identity and function. In humans, >95% of genes undergo alternative splicing (Pan et al. 2008; Wang et al. 2008), and this mechanism is required to ensure that each tissue possesses the correct gene expression pattern needed to thrive (Baralle and Giudice 2017). Many aberrant alternative splicing events are linked to diseases (Scotti and Swanson 2016; Montes et al. 2019).
While several tissue-specific splicing factors are known to directly promote RNA splicing, most of the alternative splicing events are achieved through differential expression of particular classes of RNA-binding proteins (RBPs), which in turn bind specific cis-acting elements located within exon/intron junctions in a combinatorial manner, promoting or inhibiting splicing. Serine Arginine (SR) proteins recognize exon-splicing enhancers (ESEs) and are important in promoting constitutive and alternative pre-mRNA splicing, while heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large class of nuclear RBPs that bind exon-splicing silencers (ESSs) and usually promote exon retention (Matlin et al. 2005). The relative expression levels of members from these two classes of splicing factors vary between tissues, and this imbalance is believed to promote the outcome of tissue-specific alternative splicing events (Caceres et al. 1994; Zhu et al. 2001).
Tissue identity is also achieved through post-transcriptional gene regulation events, mostly occurring through 3′ untranslated regions (3′UTRs), which are portions of genes located between the STOP codon and the poly(A) tail of mature eukaryotic mRNAs. 3′UTRs have been recently subjected to intense study as they were found to be targeted by a variety of factors, which recognize small regulatory elements in these regions and are able to modulate the dosage of gene output at the post-transcriptional level (Matoulkova et al. 2012; Oikonomou et al. 2014; Mayr 2017). While these regulatory mechanisms are still poorly characterized, and the majority of functional elements remain unknown, disorders in the 3′ end processing of mRNAs have been found to play key roles in the loss of tissue identity and the establishment of major diseases, including neurodegenerative diseases, diabetes, and cancer (Conne et al. 2000; Mayr and Bartel 2009; Delay et al. 2011; Rehfeld et al. 2013).
3′UTRs are frequently targeted by a class of repressive molecules named microRNAs (miRNAs). miRNAs are short noncoding RNAs, ∼22 nt in length, that are incorporated into a large protein complex named the microRNA-induced silencing complex (miRISC), where they guide the interaction between the miRISC and the target mRNA by base pairing, primarily within the 3′UTR (Bartel 2009). The final outcome of miRNA targeting can be context-dependent; however, mRNAs targeted by the miRISC are typically held in translational repression prior to degradation of the transcript (Ambros and Ruvkun 2018; Bartel 2018). Initial studies showed that although mismatches between miRNAs and their targets are common, many interactions make use of perfect complementarity at a small conserved heptametrical motif located at position 2–7 at the 5′ end of the miRNA (seed region) (Ambros and Ruvkun 2018; Bartel 2018). Later findings showed that while important, the seed region may also contain one or more mismatches while pairing with its target mRNA, and that this element alone is not a sufficient predictor of miRNA targeting (Ha et al. 1996; Reinhart et al. 2000; Didiano and Hobert 2006; Grimson et al. 2007). Compensatory base pairing at the 3ʹ end of the miRNA (nucleotides 10–13) can also play a role in target recognition (Shin et al. 2010; Chi et al. 2012) and has been implicated in conferring target specificity to miRNAs that share the same seed regions (Broughton et al. 2016; Wolter et al. 2017).
miRNAs and their 3′UTR targets are frequently conserved and play a variety of roles in modulating fundamental biological processes across metazoans. Bioinformatic algorithms, such as miRanda (Betel et al. 2008), TargetScan (Lewis et al. 2005), and PicTar (Lall et al. 2006), use evolutionary conservation and thermodynamic principles to identify miRNA target sites, and are the preferred tools for miRNA target identification. Based on these algorithms it was initially predicted that each miRNA controls hundreds of gene products (Chen and Rajewsky 2007). Recent high-throughput wet bench approaches have validated and expanded on these initial predictions and provide further evidence that miRNAs can indeed target hundreds of genes, and regulate molecular pathways throughout development and in diseases (Selbach et al. 2008; Helwak et al. 2013; Wolter et al. 2014, 2017; Brown et al. 2017).
In the past few years, several groups produced tissue-specific miRNA localization data in mouse, rat, and human tissues (Eisenberg et al. 2007; Landgraf et al. 2007) and in cancer (Jima et al. 2010). A previous low-throughput study has identified hundreds of Caenorhabditis elegans intestine- and muscle-specific miRNAs and their targets, which are mostly involved in the immune response to pathogens (Kudlow et al. 2012). This study used a microarray-based approach, which unfortunately does not provide enough depth to fully understand miRNA function in a tissue-specific manner. In addition, this study identified only a subset of miRNA targets, which rely on the scaffolding proteins AIN-1 and AIN-2, later found to be only present at specific developmental stages (Kudlow et al. 2012; Jannot et al. 2016).
In C. elegans there are three known Argonaute proteins that execute the miRNA pathway, which are named alg-1, alg-2, and alg-5. A recent transcriptome analysis in strains deficient in each of these members shows a remarkable difference in function, where alg-1 and alg-2 are mostly expressed in somatic tissues and are functionally redundant, and alg-5 is expressed exclusively in the gonads, interacts with only a subset of miRNAs, and is required for optimal fertility (Brown et al. 2017). A more recent study used a novel methylation-dependent sequencing approach (mime-Seq) and identified high-quality tissue-specific miRNAs in the intestine and body muscle tissues (Alberti et al. 2018).
Taken together, these studies unequivocally show that there are indeed distinct functional miRNA populations in tissues, which are in turn capable of reshaping transcriptomes and contributing to the acquisition and maintenance of cell identity. Since most miRNAs’ targets are only predicted, it is still unclear how these events are initiated and maintained.
Our group has pioneered the use of the round worm C. elegans to systematically study tissue-specific gene expression (Blazie et al. 2015, 2017). In a previous study, we optimized a method to isolate and sequence high-quality tissue-specific mRNA from worms, and published several integrative analyses of gene expression in most of the C. elegans somatic tissues, including the intestine and body muscle (Blazie et al. 2015, 2017). In these studies, we found an abundance of several tissue-specific RNA splicing factors, which could explain tissue-specific alternative splicing events. For example, we detected the RNA splicing factors asd-2 and sup-12, previously shown to regulate splicing patterns of the unc-60 gene in the C. elegans body muscle (Ohno et al. 2012), and hrp-2, a hnRNP known to induce alternative splicing isoforms of the three widely expressed genes unc-52, lin-10, and ret-1 (Kabat et al. 2009; Heintz et al. 2017). The human orthologs of hrp-2, hnRNPQ and hnRNPR, have been shown to act in a dosage-dependent manner to regulate the alternative splicing of the widely expressed gene PKM, demonstrating the importance of regulating the dosage of hnRNPs (Chen and Cheng 2012). Studies performed using human cell lines have revealed that miRNA-based regulation of splicing factor dosage can drive tissue development (Makeyev et al. 2007).
To better understand the tissue-specific contribution of miRNA-based regulation to gene dosage, RBP’s functions, and tissue identity, we performed RNA immunoprecipitation of the C. elegans Argonaute ortholog ALG-1, isolated and sequenced the tissue-specific targets of miRNAs in C. elegans, and used them to identify putative miRNA targets from two of its largest and most well-characterized tissues, the intestine and body muscle.
As expected, we found that the number of ALG-1 interacting genes in each tissue correlates with its transcriptome size. However, there is a greater proportion of the transcriptome in the intestine that interacts with ALG-1 when compared to the body muscle, suggesting that the degree of regulation by miRNA in tissues is heterogeneous. In addition, many identified targets possess RNA-binding domains and include several mRNA splicing factors such as hnRNPs and SR proteins. We also detected and validated several tissue-specific regulatory networks involved in tissue-specific alternative splicing of genes. The analysis of splice junctions in transcriptomes from miRNA-deficient strains from past studies supports and expands these observations, suggesting a potential role for miRNA in regulating mRNA biogenesis in addition to mRNA turnover.
Materials and Methods
Preparation of MosSCI vectors and GFP::ALG-1 strains
The strains used for the ALG-1 pull-down were prepared using a modified version of the previously published polyA-pull construct (Blazie et al. 2015, 2017). We produced a second-position Entry Gateway vector containing the genomic sequence of alg-1 tagged at its N-terminus with the GFP fluorochrome. Briefly, we designed primers flanking the coding sequence of alg-1 and performed a polymerase chain reaction (PCR) amplification to clone the alg-1 locus from genomic DNA extracted from N2 wild type (wt) worms (primer 1 and 2 in Supplemental Material, Table S2). The resulting PCR product was analyzed on a 1% agarose gel, which displayed a unique expected band at ∼3500 nucleotides. This band was then isolated using the QIAquick Gel Extraction Kit (cat. 28704; QIAGEN, Valencia, CA) according to the manufacturer’s protocol. Upon recovery, we digested the purified PCR product with the restriction enzymes SacI and BamHI and then cloned it into the modified polyA-pull construct (Blazie et al. 2015, 2017), replacing the gene pab-1. The ligation reaction was performed using the NEB Quick Ligation Kit (cat. MS2200S) according to the manufacturer’s protocol. We used the QuikChange II Site-Directed Mutagenesis Kit (cat. 200523; Agilent, Santa Clara, CA) to remove the unnecessary C-terminal 3xFLAG tag from the polyA-pull vector (primers 3 and 4 in Table S2). We then cloned the previously described endogenous alg-1 promoter (Vasquez-Rifo et al. 2012) by designing primers to add Gateway BP cloning elements, and then performed PCR using N2 wt genomic DNA as a template (primers 5 and 6 in Table S2). Using the resulting PCR product, we performed a Gateway BP cloning reaction into the pDONR P4P1R vector (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. To assemble the final injection clones, we performed several Gateway LR Clonase II plus reactions (cat. 12538-013; Invitrogen) using the destination vector CFJ150 (Frøkjær-Jensen et al. 2012), the tissue-specific or endogenous promoters (alg-1 for endogenous, ges-1 for the intestine, and myo-3 for the body muscle), the gfp-tagged alg-1 coding sequence, and the unc-54 3′UTR as previously published (Blazie et al. 2017).
Microinjections and screening of transgenic C. elegans strains
To prepare single-copy integrated transgenic strains we used the C. elegans strain EG6699 [ttTi5605 II; unc-119(ed3) III; oxEx1578] (Frøkjær-Jensen et al. 2012), which is designed for MosI-mediated single-copy integration (MosSCI) insertion, using standard injection techniques. These strains were synchronized by bleaching (Porta-de-la-Riva et al. 2012), then grown at 20° for 3 days to produce young adult (YA) worms. YA worms were then picked and subjected to microinjection using a plasmid mix containing: pCFJ601 (50 ng/μl), pMA122 (10 ng/μl), pGH8 (10 ng/μl), pCFJ90 (2.5 ng/μl), pCFJ104 (5 ng/μl), and the transgene (22.5 ng/μl) (Frøkjaer-Jensen et al. 2008). Three injected worms were isolated and individually placed into single small nematode growth media (NGM) plates (cat 8609-0160; USA Scientific) seeded with OP50-1 and were allowed to grow and produce progeny until the worms had exhausted their food supply. The plates were then screened for progenies that exhibited wild-type movement and proper GFP expression, and single worms exhibiting both markers were picked and placed onto separate plates to lay eggs overnight. To select for single-copy integrated worms, an additional screen was performed to select for worms that lost the mCherry fluorochrome expression (extrachromosomal injection markers).
Genotyping of transgenic C. elegans strains
Single adult worms were isolated and allowed to lay eggs overnight and then genotyped for single-copy integration of the transgene by single worm PCR as previously described (Broughton et al. 2016) (primers 7–9 in Table S2). Progeny from worms that contained the single-copy integrations were propagated and used for this study. A complete list of worm strains produced in this study is shown in Table S3.
Validating expression of the transgenic construct
To validate the expression of our transgenic construct and to evaluate our ability to immunoprecipitate GFP-tagged ALG-1, we performed an immunoprecipitation (as described below) followed by a western blot. For the western blot we used a primary anti-GFP antibody (NB600-303; Novus) (1:2000) and a fluorescent secondary antibody (LI-COR, 925–32211) (1:5000), followed by imaging using the ODYSSEY CLX system (LI-COR Biosciences, NE) (Figure S1).
In vivo validation of GFP::ALG-1 functionality by layoff experiment
To validate the in vivo functionality of our transgenic GFP-tagged ALG-1, we used a genetic approach. It was previously shown that the knockout alg-1 strain RF54 [alg-1(gk214) X] led to a decrease in fertility (Bukhari et al. 2012). We rescued this decrease in fertility in the alg-1 knockout strain RF54 [alg-1(gk214) X] by crossing it into our strain MMA17 (Table S3), which expresses our green fluorescent protein (GFP)-tagged transgenic ALG-1, driven by the endogenous alg-1 promoter. The resulting strain MMA20 [alg-1(gk214)X; alg-1p::gfp::alg-1::unc-54 II] only expresses our cloned alg-1 gene tagged with the GFP fluorochrome. We validated the genotype of MMA20 using single worm PCRs as previously described (Broughton et al. 2016) (primers 10 and 11 in Table S2 and Figure S2). The layoff experiment was used to evaluate the ability of our transgenic GFP-tagged ALG-1 construct to rescue the loss in fertility seen in the alg-1 knockout strain (RF54). The layoff experiment was performed by first synchronizing N2 (wt), RF54, and MMA20 strains to arrested L1 larvae, through bleaching followed by starvation overnight in M9 solution. We then plated the L1 arrested worms on NGM plates seeded with OP50-1 and allowed the worms to develop to the adult stage for 48 hr after which single worms were isolated onto OP50-1 seeded plates. The adult worms were left to lay eggs overnight (16 hr) after which the adult worms were removed. The eggs were allowed to hatch and develop for 24 hr, and the number of larvae in each plate was counted.
Sample preparation and cross-linking
A 0.5-ml sample of mixed stage C. elegans of each strain was grown on five large 20-cm plates (cat 8609-0215; USA Scientific) and harvested by centrifugation at 400 rcf for 3 min. The pellets were initially washed in 15 ml dH2O water and spun down at 400 rcf for 3 min and then resuspended in 10 ml of M9+0.1%Tween20 and then cross-linked three times on ice, with energy setting: 3000 × 100 μJ/cm2 (3 kJ/m2) (Stratalinker 2400; Stratagene, La Jolla, CA) (Moore et al. 2014). After the cross-linking, each C. elegans strain was recovered by centrifugation at 400 rcf for 3 min, and resuspended in two volumes (1 ml) of lysis buffer [150 mM NaCl, 25 mM HEPES(NaOH) pH 7.4, 0.2 mM DTT, 10% glycerol, 25 units/ml of RNasin Ribonuclease Inhibitor (cat N2611; Promega, Madison, WI)], 1% Triton X-100, and one tablet of protease inhibitor for every 10 ml of lysis buffer (Roche cOmplete ULTRA Tablets, cat 5892791001; Sigma, St . Louis, MO). The lysed samples were subjected to sonication using the following settings: amplitude 40%; 5× with 10-sec pulses; 50-sec rest between pulses (Q55 Sonicator; Qsonica). After the sonication, the cell lysate was cleared of debris by centrifugation at 21,000 rcf at 4° for 15 min, and the supernatants were then transferred to new tubes.
GFP-TRAP bead preparation and immunoprecipitation
Twenty-five microliters of GFP-TRAP beads (gtma-10; Chromotek) (total binding capacity 7.5 μg) per immunoprecipitation were resuspended by gently vortexing for 30 sec, and washed three times with 500 μl of cold Dilution/Wash buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA). The beads were then resuspended in 100 μl/per Immunoprecipitation of Dilution/Wash buffer. The 100 μl of resuspended beads was then incubated with 0.5 ml of lysate for 1 hr on the rotisserie at 4°. At the completion of the incubation step, the beads were collected using magnets. The unbound lysate was saved for PAGE analysis. The beads containing the immunoprecipitated alg-1 associated to the target mRNAs were then washed three times in 200 μl of Dilution/Wash buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA), and then the RNA/protein complex was eluted using 200 μl of Trizol (cat 15596026; Invitrogen) and incubated for 10 min at room temperature.
Trizol/driectzol RNA purification
The RNA purification was performed using the RNA MiniPrep kit (cat ZR2070; Zymo Research) as per the manufacturer’s protocol. All centrifugation steps were performed at 21,000 × g for 30 sec. We added an equal volume of ethanol (95–100%) to each sample in Trizol and mixed thoroughly by vortexing (5 sec, level 10). The samples were then centrifuged, recovered using a magnet, and the supernatant was transferred into a Zymo-Spin IIC Column in a Collection Tube and centrifuged. The columns were then transferred into a new collection tube and the flow-through was discarded. Four hundred microliters of RNA wash buffer was added into each column and centrifuged. In a separate RNase-free tube, we added 5 μl of DNase I (6 unit/µl) and 75 μl of DNA Digestion Buffer, mixed and incubated at room temperature (20–30°) for 15 min. Then 400 µl of DirectZol RNA PreWash (cat ZR2070; Zymo Research) was added to each sample and centrifuged twice. The flow-through was discarded in each step. RNA wash buffer (700 µl) was then added to each column, and it was centrifuged for 2 min to ensure complete removal of the wash buffer. The columns were then transferred into RNase-free tubes, and the RNAs were eluted with 30 μl of DNase/RNase-Free Water added directly to the column matrix and centrifuge.
Complementary DNA (cDNA) library preparation and sequencing
Each cDNA library was prepared using a minimum of 500 pg of immunoprecipitated RNA from each tissue. The total RNA was reverse transcribed using the IntegenX’s (Pleasanton, CA) automated Apollo 324 robotic preparation system using previously optimized conditions (Blazie et al. 2015). The cDNA synthesis was performed using a SPIA (Single Primer Isothermal Amplification) kit (IntegenX and NuGEN, San Carlos, CA) (Kurn et al. 2005). The cDNA was then sheared to ∼300-bp fragments using the Covaris S220 system (Covaris, Woburn, MA). We used the Agilent 4200 TapeStation instrument (Agilent) to quantify the abundance of cDNAs and calculate the appropriate amount of cDNA necessary for library construction. Tissue-specific barcodes were then added to each cDNA library, and the finalized samples were pooled and sequenced using the HiSeq platform (Illumina, San Diego, CA) with a 1 × 75 bp HiSeq run.
Data analysis
We obtained ∼15 M unique reads per sample (∼130 M reads total). The software Bowtie 2 (Langmead et al. 2009) run with default parameters was used to perform the alignments to the C. elegans genome WS250. We used custom Perl scripts and Cufflinks (Trapnell et al. 2010) algorithm to study the differential gene expression between our samples. A summary of the results is shown in Figure S3. Mapped reads were further converted into a bam format and sorted using SAMtools software run with generic parameters (Li et al. 2009) and used to calculate Fragments Per Kilobase Million (FPKM) values, as an estimate of the abundance of each gene per sample. We used an FPKM ≥1 on the median from each replicate as a threshold for identifying positive hits. This stringent approach discarded ∼50–75% of mapped reads for each sample (Figure S3B). The quality of our finalized list of target genes was tested using a principal component analysis vs. our N2 wt negative control (Figure S3C).
Validation of sequencing results by qPCR
We prepared cDNA from tissue-specific, ALG-1 immunoprecipitated, RNA using the SPIA kit as previously described (IntegenX and NuGEN) (Kurn et al. 2005). We performed 10 µl qPCRs in triplicate, using 0.5 µl of cDNA per reaction, 250 nM of gene-specific primers, and a SYBR Green qPCR master mix (cat RT-SN2X-03T; Eurogentec). The primers used (Table S2 37–40) were designed using the software Primer-BLAST, targeting 100- to 120-bp product sizes and default parameters. The mean cycle-threshold (CT) between the three replicates for each gene, and SD, were used in Figure S3D.
Molecular cloning and assembly of the expression constructs
The promoters of candidate genes were extracted from genomic DNA using genomic PCR and cloned into Gateway-compatible entry vectors (Invitrogen). We designed Gateway-compatible primers (primers 12–19 in Table S2) targeting 2000 bp upstream of a given transcription start site, or up to the closest gene. Using these DNA primers, we performed PCRs on C. elegans genomic DNA, amplified these regions, and analyzed the PCR products by gel electrophoresis. Successful DNA amplicons were then recombined into the Gateway entry vector pDONR P4P1R using Gateway BP Clonase reactions (Invitrogen). The reporter construct pAPAreg has been previously described in Blazie et al. (2017). The coding region of this construct was prepared by joining the coding sequence of the mCherry fluorochrome to the SL2 splicing element found between the gpd-2 and gpd-3 genes, and to the coding sequence of the GFP gene. The entire cassette was then PCR amplified with Gateway-compatible primers and cloned into pDONR P221 by Gateway BP Clonase reactions (Invitrogen).
The 3ʹUTRs of the genes in this study were cloned by anchoring the Gateway-compatible primers at the translation STOP codon of each gene to the longest annotated 3UTR. We have included 50 bp downstream of the annotated PAS site to include 3ʹ end processing elements (primers 20–27 in Table S2). The PCR products were analyzed using gel electrophoresis analysis and used to perform Gateway BP Clonase reactions (cat. 11789020; Invitrogen) into pDONR P2RP3 as per the manufacturer’s protocol. The unc-54 3ʹUTR used in this study was previously described in Blazie et al. (2017). The constructs injected to produce transgenic strains were assembled by performing Gateway LR reactions (Invitrogen). We cloned each promoter, reporter, and 3ʹUTR construct per the manufacturer's protocol into the MosSCI compatible destination vector CFJ150. We then microinjected each reporter construct (100 ng/μl) with CFJ601 (100 ng/μl) into MosSCI-compatible C. elegans strains using standard microinjection techniques (Evans and Thomas 2006).
Fluorescent imaging and analysis of nematodes
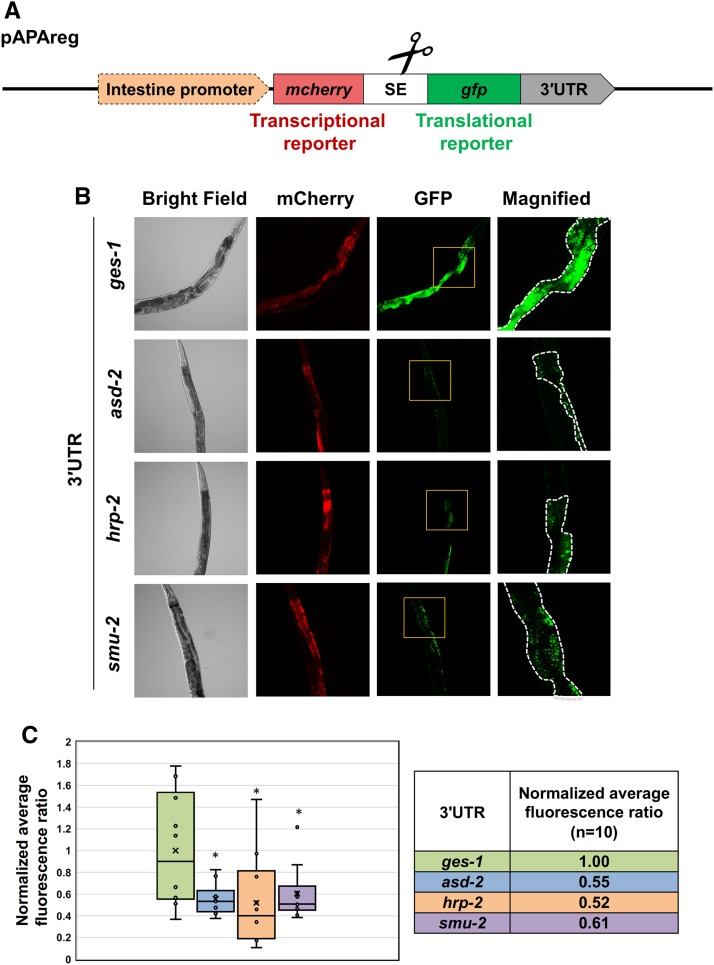
Confocal images shown in Figure 4 were acquired in the Biodesign Imaging Core, Division of the Arizona State University Bioimaging Facility. We made use of the previously described APAreg construct, which uses a single promoter to drive the transcription of a polycistronic pre-mRNA where the coding sequence of the mCherry fluorochrome is separated from the coding sequence of GFP by a SL2 trans-splicing element (SE) (Blazie et al. 2017). The test 3′UTR is cloned downstream of the GFP gene. Since the mCherry transcript is trans-spliced, it reports transcription activation. The GFP gene instead reports translational activity, since its expression is dictated by the downstream tested 3′UTR. If a given miRNA targets the test 3′UTR, the GFP intensity decreases when compared with an untargeted 3′UTR (ges-1). By comparing the ratio of the mCherry (indicating transcription) to the GFP (indicating translation) fluorochromes, we are able to define the occurrence of post-transcriptional silencing triggered by the tested 3′UTR (Figure 4A) (Blazie et al. 2017).
Figure 4.
asd-2, hrp-2, and smu-2 3′UTRs regulate post-transcriptional gene expression in the intestine. (A) Diagram of the construct used in these experiments (pAPAreg). An intestine-specific promoter drives the expression of a bicistronic dual fluorochrome vector in the intestine. The mCherry fluorochrome reports transcription activity of the construct, while the GFP reports post-transcriptional activity through the test 3′UTR cloned downstream of the GFP reporter sequence. If the test 3′UTR is targeted by repressive regulatory factors such as miRNAs, the GFP fluorochrome lowers in its expression. SE, trans-splicing element extracted from the intergenic region located between the genes gpd-2 and gpd-3. (B) Representative images of C. elegans strains generated with pAPAreg constructs expressing one of the following 3′UTRs: ges-1, asd-2, hrp-2, or smu-2 downstream of the GFP fluorochrome. Yellow boxes indicate magnified regions. White dotted lines highlight the intestine tissue. (C) The whisker plots show the quantified and normalized mean fluorescence ratio between the GFP and the mCherry fluorochromes. The mean fluorescence ratio is calculated from 10 worms per strain. The error bars indicate the SEM. Student’s t-test *P < 0.05. We observed ∼40% reduction in normalized GFP intensity modulated by asd-2, hrp-2, and smu-2 3′UTRs.
Transgenic strains were grown at room temperature on NGM plates seeded with OP50-1. The mixed stage worms were washed twice with M9 and resuspended in 1 mM of levamisole before imaging using a Nikon C1 Ti-E microscope with 488- and 561-nm lasers, 0.75 numerical aperture, and 90-μM pinhole microscope with a 40× magnification objective lens. We acquired 10 images for each transgenic strain (total 40 images) using the same microscope settings. The fluorescence of GFP and mCherry fluorochromes from the acquired images were individually quantified using the integrated density (ID) function of the ImageJ software (Schneider et al. 2012). Fluorescence ratios were then calculated for each worm (n = 10, total 40 images) by dividing the ID for GFP by the ID for mCherry. The finalized result for each strain is the averaged fluorescence ratio calculated across all 10 imaged worms. We performed a two-tailed Student’s t-test to compare the mean fluorescence ratios for each strain with a P-value cut off <0.05 to establish the presence of post-transcriptional gene regulation.
Bioinformatic analysis of tissue-specific miRNA targeting biases
The tissue-specific miRNA studies were performed in two steps. First, we utilized custom-made Perl scripts to scan across the longest 3ʹUTR of each C. elegans protein-coding gene (WS250) in our data sets, searching for perfect sequence complementarity to the seed regions of all C. elegans miRNAs present in the miRBase database (release 21) (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Kozomara and Griffiths-Jones 2011, 2014). This result was then used to calculate the percentage of seed presence in the intestine and body muscle data sets. To calculate the percentage of predicted targets, we extracted both predicted target genes, and their target miRNA name from the miRanda database (Betel et al. 2008) and compared the results with our study. A complete list of miRNA predictions for each tissue profiled is shown in Table S1.
Comparison with other data sets
We extracted the WormBase IDs of genes in the intestine and body muscle transcriptomes previously published by our group (Blazie et al. 2017), and most abundant miRNA targets (transcript names) identified by Kudlow et al. (2012) in these tissues. We then translated the transcript names from Kudlow et al. (2012) into WormBase IDs using custom Perl scripts, and compared how many genes in each of these groups overlap with our ALG-1 pull-downs. The results are shown in Figure S4. For the analysis shown in Figure S6 we extracted the names of the miRNAs previously identified by Alberti et al. (2018) in the C. elegans intestine and body muscle tissues (Alberti et al. 2018). We then used custom Perl scripts to search for the presence of the seed regions of these miRNAs in the 3′UTRs of the genes identified in this study (Figure S6).
Reannotation of alg-1 and alg-2 knockout transcriptome data sets and splice junction identification
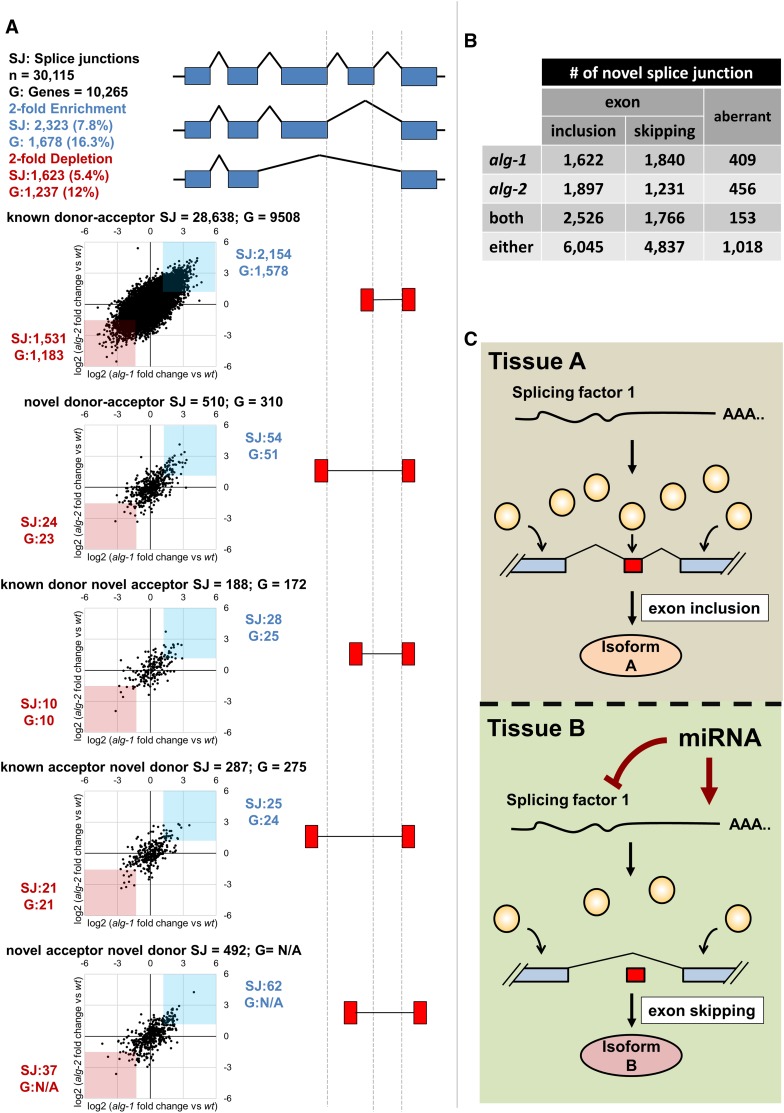
We downloaded from the GEO database the following transcriptome data sets published by Brown et al. (2017): Project number GSE98935, Wild type Rep 1–3 (GSM2628055, GSM2628056, GSM2628057); alg-1(gk214) Rep 1–3 (GSM2628061, GSM2628062, GSM2628063); alg-2(ok304) Rep 1–3 (GSM2628064, GSM2628065, GSM2628066). We used in-house Perl scripts to prepare the reads for mapping and then these reads as input to the TopHat algorithm (Trapnell et al. 2012) to map splice junctions in all nine data sets independently. The TopHat algorithm mapped between 30 and 56 M reads to splice junctions in each sample. wt_rep1; 43,721,355 mapped reads (64% of total input reads), wt_rep2; 44,440,441 (64%), wt_rep3; 37,248,408(62.7%), alg-1_rep1; 30,808,645 (62.3%), alg-1_rep2; 35,914,514 (63.2%), alg-1_rep3; 43,721,355(63.9%), alg-2_rep1; 54,471,761(63.2%), alg-2_rep2; 56,000,173 (66.8%), alg-2_rep3; 46,638,369 (63.9%). We then combined the mapped reads obtained in the three replicates for each strain and used the open source software regtools (Griffith Lab, McDonnell Genome Institute) to annotate these splice junctions using the following command “regtools junctions annotate junctions.bed WS250.fa WS250.gtf.” The software produced ∼41.8K splice junctions supported by at least 10 reads for the combined N2 wt data set, ∼42.3K splice junctions for the alg-1 data set, and 46.3K for the alg-2 data set. We analyzed the three resulting cumulative data sets normalized by dividing each score by the total number of mapped reads within each sample. This approach produced 36.7K high-quality splice junctions for the combined N2 wt data set, ∼37K for the alg-1 combined data set, and ∼38K for the combined alg-2 data set. The analysis in Figure 6A was performed using splice junctions that are present in all three data sets (30,115 total). To calculate the fold-change for each splice junction, we divided the normalized scores of each splice junction in the alg-1 and alg-2 combined data sets by the corresponding scores in the wild-type combined data set. The fold-change of each splice junction was then plotted on a log2 scale as shown in Figure 6.
Figure 6.
(A) Genome-wide changes in splice junction usage in C. elegans strains deficient in the miRNA pathway. Analysis of splice junction (SJ) usage in miRNA-deficient strains [alg-1(gk214) or alg-2(ok304)] reannotated from Brown et al. (2017). The graphs illustrate the changes in splice junction abundance for different types of splicing events (red boxes). The x-axis represents the fold-change of the normalized number of reads for each splice junction, comparing the alg-1(gk214) strain to wt, while the y-axis represents the fold change obtained when comparing alg-2(ok304) to wt. Splice junctions with >2-fold enrichment in both strains are highlighted in blue, while splice junctions with 2-fold depletion in both strains are highlighted in red. The number of genes (G) with enriched or depleted splice junctions and the total number of splice junctions mapped (SJ) are indicated next to the graphs. There is a 2-fold change in ∼13% of all the splicing events mapped in these knockout strains, affecting 3301 genes, when compared to the N2 wt control. (B) Table summarizing the number of novel splicing events detected in alg-1 and alg-2 data sets from Brown et al. (2017). These splicing events are not observed in the N2 wt control and indicate an increase in novel and aberrant splicing events in miRNA-deficient strains. (C) A proposed role for miRNAs in the modulation of tissue-specific alternative splicing. The abundance of RNA splicing factors (yellow circles) dictates the splicing events in a given tissue A. The presence of an miRNA in tissue B may lower the dosage of these splicing factors resulting in tissue-specific alternative splicing.
RNAi experiments
The RNAi experiments shown in Figure 5 and Figure S7 were performed as follows. N2 worms were synchronized by bleaching and starving overnight in M9 buffer until they reached the L1 stage and then transferred to agar plates containing OP50-1 bacteria, HT115 bacteria with hrp-2 RNAi or asd-2 RNAi (Kamath and Ahringer 2003). We used par-2 RNAi as a positive control for the experiments, which results in 100% embryonic lethality. To perform the layoff experiments, individual synchronized young adult worms were left overnight (16 hr) to lay eggs. Hatched larvae were counted 24 hr later. Total RNA was extracted from N2 worms treated with either hrp-2 or asd-2 RNAi at the adult stage in triplicate.
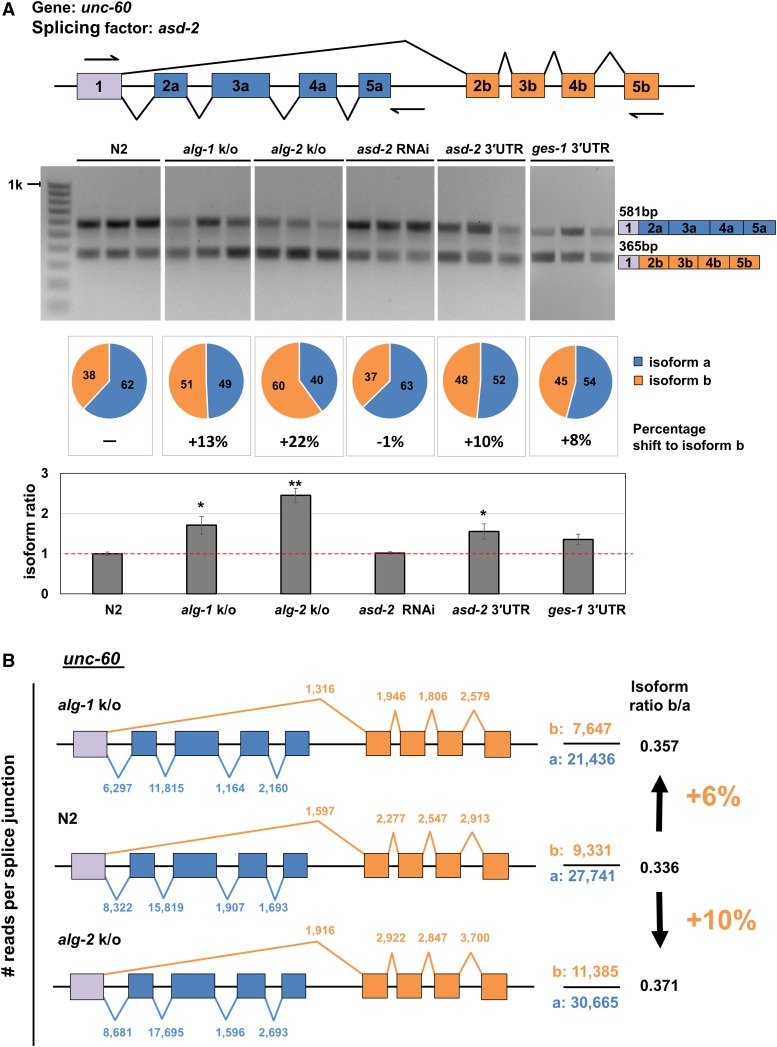
Figure 5.
The splicing pattern of unc-60 is modulated by miRNA activity in the intestine tissue. (A) Top panel: Schematic of the genomic locus of unc-60. This gene is expressed as a longer unc-60a isoform (blue boxes) and a shorter unc-60b isoform (orange boxes). Arrows mark the binding sites of the primers used to detect the two isoforms. Middle panel: RT-PCR performed from total RNA extracted from biological replicates in triplicate and visualized in a 1% agarose gel. (1) N2: wt worms. (2) alg-1 k/o: RF54[alg-1(gk214) X]. (3) alg-2 k/o: WM53[alg-2(ok304) II]. (4) asd-2 RNAi: N2 worms subjected to asd-2 RNAi. (5) overexpression of asd-2 3′UTR in the intestine. (6) Overexpression of the unspecific ges-1 3′UTR in the intestine. The pie charts below each gel show a quantification of each of the occurrence of the two isoforms. The percentage below the pie chart is the change in unc-60 isoform b abundance when compared to N2 (wt). The bar chart shows the change in isoform ratio between strains. The y-axis shows the abundance ratio (shorter isoform/longer isoform) of the two alternatively spliced isoforms examined. Exon skipping increases in alg-1 and alg-2 k/o strains, in asd-2 3′UTR overexpression strains and is not observed overexpressing the unspecific ges-1 3′UTR. Error bars indicate SEM. Student’s t-test *P < 0.05, **P < 0.01. (B) A comparison of the splice junction usage in unc-60 as observed in the reannotated in transcriptome data for alg-1 and alg-2 knockout strains (Brown et al. 2017). The numbers above each splice junction indicate the number of reads mapped. The total reads for each isoform are indicated to the right of the gene model. The isoform ratios (b/a) are calculated by dividing the total number of reads mapped for each isoform. There is a ∼6–10% increase in the expression of the shorter unc-60b isoform in the miRNA-deficient strains. Blue: reads corresponding to unc-60a; orange: reads corresponding to unc-60b.
RNA extraction for detection of intestine-specific splicing variants
We extracted total RNA using the Direct-zol RNA MiniPrep Plus kit (cat ZR2070; Zymo Research) from (1) N2 wt worms, (2) RF54 [alg-1(gk214) X] strain, (3) WM53 [alg-2(ok304) II] strain, (4) N2 strain subjected to RNAi as previously described (Ahringer 2006) for asd-2 and hrp-2, and (5) transgenic worms overexpressing the asd-2 3′UTR or the hrp-2 3′UTR under the control of an intestinal promoter (ges-1p::pAPAreg::3′UTR). Each strain was synchronized by growing in M9 media to the L1 stage then transferred to plates containing HT115. We extracted RNA 48 hr later from adult worms in triplicate for each condition.
cDNA preparation, image acquisition, and splicing isoform analysis
At the completion of the RNA extractions, the cDNA was synthesized from each sample using SuperScript III RT (cat 18080093; Life Technologies) according to the manufacturer’s protocol. Briefly, 200 ng of each RNA sample was incubated with 1 μl of 50 mM poly dT anchor and 1 μl of 10 mM dNTP mix and brought to a total volume of 14 μl with nuclease-free H2O and incubated for 5 min at 60° then iced for 1 min. Four microliters of 5× first strand buffer, 1 μl of 0.1 M DTT, and 1 μl (200 units) of SuperScript III reverse transcriptase were added to each sample and incubated at 50° for 60 min then heat inactivated at 70° for 15 min. Two hundred nanograms of cDNA from each sample was used in PCRs consisting of 34 cycles using HiFi Taq Polymerase (cat 11304011; Invitrogen) according to the manufacturer’s protocols. Primers used to test alternative splicing of unc-60, unc-52, lin-10, and ret-1 were designed to flank the alternatively spliced exons and were adapted from previous studies (Kabat et al. 2009; Ohno et al. 2012; Heintz et al. 2017) (Table S2 primers 28–36). We then acquired images of the PCR amplicons (5 μl) separated by agarose gel electrophoresis and assigned the alternatively spliced isoforms using the ImageJ software package (Schneider et al. 2012). We used the ID function of ImageJ by defining equally sized regions of interest around each band in the images and compared the ID values by normalizing the smaller bands to the larger bands. The resulting isoform ratios are displayed in Figure S8. Each strain was quantified in triplicate and subjected to a two-tailed Student’s t-test. Statistical significance was assigned for P-values <0.05.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures and supplemental figures, and tables and supplemental tables. Raw reads were submitted to the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/), under the SRP Study Accession # PRJNA517741. The results of our analyses are available in Excel format in Tables S1 and S4, and in our 3′UTR-centric website www.UTRome.org. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8095457.
Results
A method for the identification of putative, tissue-specific miRNA targets
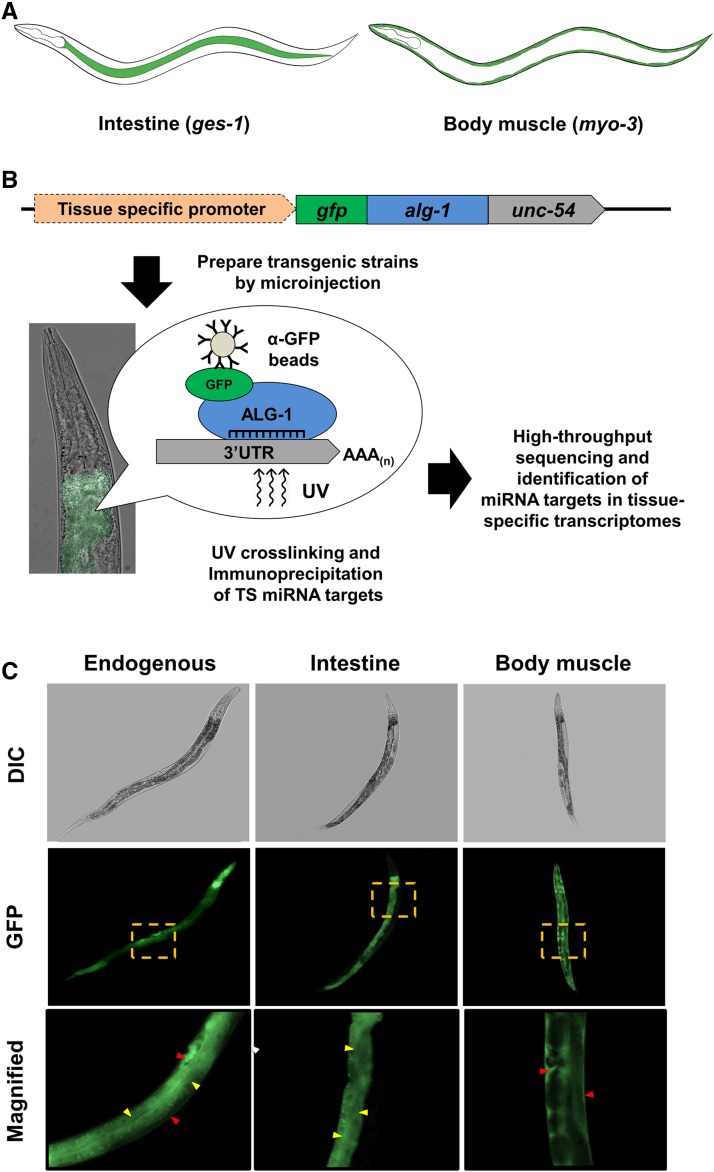
To study the contribution of miRNA activity in producing and maintaining tissue identity, we performed RNA immunoprecipitations of miRNA target genes in two of the largest, morphologically different, and most well-characterized tissues in C. elegans: the intestine (McGhee 2007) and body muscle (Gieseler et al. 2017) (Figure 1A). We took advantage of the ability of the Argonaute protein to bind miRNA target genes and cloned alg-1, one of the worm orthologs of the human Argonaute 2 protein, downstream of the GFP. The expression of this construct was then driven by the endogenous promoter (alg-1p) or restricted to the intestine (ges-1p) or body muscle (myo-3p) using tissue-specific promoters (Figure 1B).
Figure 1.
Identification of putative miRNA targets by tissue-specific immunoprecipitation and sequencing. (A) The anatomical location of the two somatic tissues used in this study. (B) Workflow for the identification of tissue-specific miRNA targets. We cloned the C. elegans Argonaute 2 ortholog alg-1 and fused with the GFP fluorochrome and the unspecific unc-54 3′UTR. The expression of this cassette was driven in the intestine and body muscle by using tissue-specific promoters. These constructs were microinjected into MosSCI-compatible C. elegans strains to produce single-copy integrated transgenic animals. These strains were then subjected to UV cross-linking and lysed by sonication. The resulting lysate was subjected to RNA immunoprecipitations with α-GFP antibodies. The resultant tissue-specific miRNA target transcripts were purified; the cDNA libraries were prepared and sequenced using Illumina HiSeq. (C) Representative images of C. elegans single-copy integrated strains showing the expression of GFP tagged alg-1 in endogenous (alg-1p), intestine (ges-1p), and body muscle (myo-3p) tissues. Yellow box indicates magnified regions, yellow arrows mark intestine cells, and red arrows mark body muscle cells.
We produced transgenic strains for each construct (Figure 1C) using single-copy integration technology (MosSCI) (Frøkjær-Jensen et al. 2012, 2014). Single-copy integration produces uniform expression of a selected construct at endogenous level, minimizing the expression mosaics induced by repetitive extrachromosomal arrays. The strains were validated for integration using genomic PCRs and Western blots (Figure S1).
We then examined the functionality of our cloned alg-1 in rescue experiments using the alg-1−/− strain RF54(gk214). This strain has a decrease in fertility caused by the loss of functional alg-1 (Bukhari et al. 2012), which was fully rescued by our cloned alg-1 construct in a layoff experiment (Figure S2), suggesting that our cloned alg-1 is functional and able to fully mimic endogenous alg-1.
We then used our strains to perform tissue-specific RNA immunoprecipitations. Each tissue-specific ALG-1 IP and control IPs were performed in duplicate using biological replicates (total six sequencing runs). We obtained ∼25 M reads on average for each tissue, of which ∼80% were successfully mapped to the C. elegans genome (WS250) (Figure S3). To maximize our success, we used a stringent approach to determine gene presence (Materials and Methods) and used a FPKM ≥1 on the median from each replicate as a threshold for identifying positive hits. This led us to use for our analysis only the top 25–50% of genes mapped in each data set (Figure S3, B and C) (Blazie et al. 2015, 2017).
Our analysis resulted in 3681 different protein-coding genes specifically targeted by the miRISC using the endogenous alg-1 promoter or in the intestine or body muscle. The complete list of genes detected in this study is shown in Table S1.
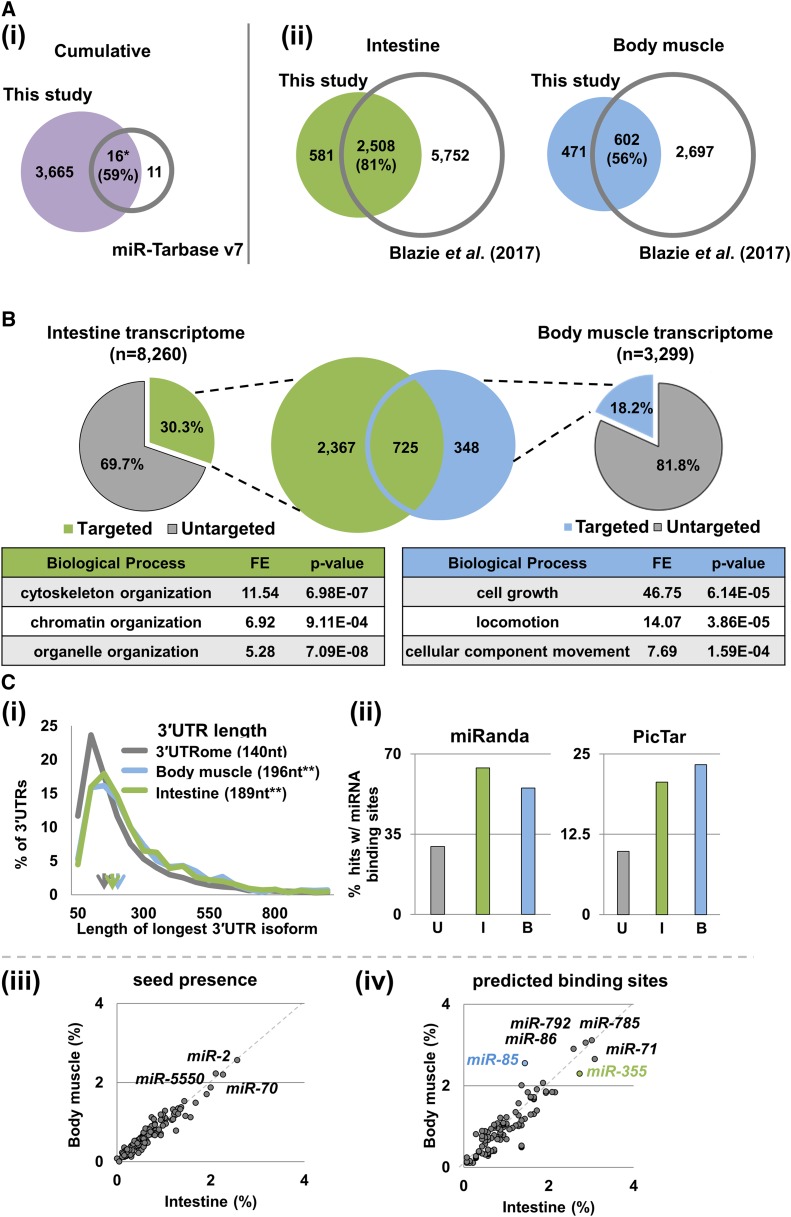
There are only 27 validated C. elegans miRNA–target interactions with strong evidence reported in the miRNA target repository miR-TarBase v7, and our study confirmed 16 of these interactions (59%), which is a 3-fold enrichment when compared to a random data set of similar size (P < 0.05, chi-squared test) (Figure 2Ai). When compared to genes present in the C. elegans intestine and body muscle transcriptomes (Blazie et al. 2017), 81% of the intestine and 56% of the body muscle targets identified in this study match with their respective tissues (Figure 2Aii). A comparison between our hits and a previously published ALG-1 IP data set in all tissues also supports our results (Figure S4) (Zisoulis et al. 2010).
Figure 2.
A comparative analysis of ALG-1 immunoprecipitations to identify putative miRNA targets in the intestine and body muscle tissues. (A) Comparison of putative miRNA targets identified in this study to other previously published data sets. The numbers indicate protein-coding genes targeted by miRNA in each tissue. (i) Venn diagram showing the comparison of the genes identified in this study to miR-TarBase v7, a compendium of all experimentally validated miRNA targets. Fifty-nine percent of the genes in this database match those identified in this study. (ii) Venn diagrams showing the comparison of the genes identified in this study to previously published tissue-specific intestine and body muscle transcriptomes. The majority of the targets in both data sets were previously assigned to each tissue. Green – intestine (ges-1p); blue – body muscle (myo-3p). (B) Pie charts showing the proportion of putative miRNA targets detected in this study compared to tissue-specific transcriptomes previously characterized by Blazie et al. (2017). The Venn diagram in the center shows the number of protein-coding genes identified in this study as putative miRNA targets between the intestine and body muscle tissues. The tables show a Gene Ontology analysis for pathway enrichment using the top 100 genes from each data set used in this study. Green – intestine (ges-1p); blue – body muscle (myo-3p). (C) (i) The length of 3′UTRs from protein-coding genes as from the C. elegans 3′UTRome v1 (Mangone et al. 2010) compared to the intestine and body muscle targets identified in this study. The arrows indicate the median 3′UTR length. Genes targeted in the intestine and body muscle have longer 3′UTRs on average than those published in the C. elegans 3′UTRome v1. ANOVA Single factor test **P < 0.01. (ii) Proportion of 3′UTRs with predicted miRNA binding sites (miRanda and PicTar) (Lall et al. 2006; Betel et al. 2008) in the 3′UTRome (gray), and in 3′UTRs of genes identified in this study. Green – intestine; blue – body muscle. The genes identified in this study in the intestine and body muscle tissues are enriched for predicted miRNA binding sites. (iii and iv) Analysis of miRNA target sites identified in this study. The two axes show the proportion of 3ʹUTRs with perfect seeds or with predicted target sites (miRanda) (Betel et al. 2008), normalized to the total number of genes targeted in each tissue for each miRNA. miRNAs that target >2% of the genes are listed. The blue mark denotes miR-85, a body muscle specific miRNA. The green mark denotes miR-355, an intestine-specific miRNA.
To further validate the quality of our hits, we used GFP-based approaches to confirm the tissue localization of a few tissue-specific genes identified in our study, and found with the exception of one, their observed localization matched the expected tissue (Figure S5). A qPCR analysis of trap-4 and grd-5, two hits selectively detected in the two tissues, is in agreement with our findings (Figure S3D). In addition, to further test the quality of our data, we compared our results with the intestine and body muscle-specific miRNA localization data from past studies (Alberti et al. 2018) (Figure S6). We found that >70% of the genes identified in our study possess predicted binding sites in their 3′UTRs for miRNAs detected in each tissue, suggesting strong correlation between our results and Alberti et al. (2018) (Figure S6).
ALG-1 targets in the intestine regulate key metabolic enzymes
The C. elegans intestine is composed of 20 cells that begin differentiation early in embryogenesis and derive from a single blastomere at the eight-cell stage (McGhee 2007). As the primary role of the intestine is to facilitate the digestion and absorption of nutrients, many highly expressed genes in this tissue are digestive enzymes, ion transport channels, and regulators of vesicle transport (McGhee 2007).
In our intestinal ALG-1 pull-down we identified 3089 protein-coding genes as putative miRNA targets, 2367 of which were uniquely isolated from this tissue when compared to the body muscle (Figure 2B). As expected, and consistent with the function of the intestine, we find a number of enzymes involved with glucose metabolism, such as enol-1 an enolase, ipgm-1 a phosphoglycerate mutase, and three out of four glyceraldehyde-3-phosphate dehydrogenases (gpd-1, gpd-2, and gpd-4). The human ortholog of the C. elegans gene enol-1, eno1 has been previously identified as a target of miR-22 in the context of human gastric cancer (Qian et al. 2017). In addition, some of our top hits are the fatty acid desaturase enzymes fat-1, fat-2, fat-4, and fat-6, which are all involved with fatty acid metabolism, suggesting that these metabolic pathways are subjected to a high degree of regulation in the intestine. All these genes contain seed elements in their 3′UTRs (Table S1). Additionally, we found five out of six vitellogenin genes (vit-1, vit-2, vit-3, vit-5, and vit-6), with vit-2 and vit-6 being the most abundant (Table S1). vit-2 was shown to be targeted by ALG-1 in a previous study (Kudlow et al. 2012), and both possess miRanda (Betel et al. 2008, 2010) and/or PicTar (Lall et al. 2006) predicted binding sites (Table S1). These vitellogenin genes produce yolk proteins and are energy carrier molecules synthesized in the intestine. These yolk proteins are then transported to the gonads and into the oocytes to act as an energy source for the developing embryos (DePina et al. 2011). Accordingly, we also find a number of RAB family proteins that are responsible for intracellular vesicular transport (rab-1, rab-6.1, rab-7, rab-8, rab-21, rab-35, and rab-39).
Several transcription factors were also identified as putative miRNA targets in the intestine. skn-1 is a bZip transcription factor that is initially required for the specification of cell identity in early embryogenesis and then later plays a role in modulating insulin response in the intestine of adult worms (Blackwell et al. 2015). This gene has already been found to be targeted by miRNA in many past studies (Zisoulis et al. 2010; Kudlow et al. 2012) and contains many predicted miRNA binding sites and seed regions from both miRanda (Betel et al. 2008, 2010) and PicTar (Lall et al. 2006) prediction software (Table S1). A second transcription factor pha-4 is expressed in the intestine, where it has an effect on dietary restriction-mediated longevity (Smith-Vikos et al. 2014). pha-4 is a validated target of let-7 in the intestine (Grosshans et al. 2005), and along with skn-1, is also targeted by miR-228 (Smith-Vikos et al. 2014). Additionally, pha-4 is targeted by miR-71 (Smith-Vikos et al. 2014).
We also find as a putative target of miRNA, the gene die-1, which is associated with the attachment of the intestine to the pharynx and the rectum (Heid et al. 2001), and the chromatin remodeling factor lss-4 (let seven suppressor), which is able to prevent the lethal phenotype induced by knocking out the miRNA let-7 (Grosshans et al. 2005). These two genes were also validated by others as miRNA targets (Grosshans et al. 2005).
The role of miRNAs in regulating the innate immune response through the intestine and these genes has been reported in multiple studies (Ding et al. 2008; Kudlow et al. 2012; Sun et al. 2016). Consistent with those findings, the genes atf-7, pmk-1, and sek-1 were all identified as putative targets of miRNAs in the intestine. These three genes act together to produce a transcriptional innate immune response where the transcription factor atf-7 is activated through phosphorylation by kinases pmk-1 and sek-1.
Muscle ALG-1 targets modulate locomotion and cellular architecture
C. elegans possess 95 striated body wall muscle cells, which are essential for locomotion (Gieseler et al. 2017). Its sarcomeres are composed of thick filaments containing myosin associated with an M-line and thin filaments containing actin associated with the dense body. The pulling of actin filaments by myosin heads generates force that produces locomotion (Moerman and Williams 2006).
Our ALG-1 pull-down identified 1047 protein-coding genes as putative miRNA targets in the body muscle tissue (Table S1). Within this group, 348 genes were not present in our intestine data set and are specifically restricted to the body muscle tissue (Table S1). Our top hits include genes involved in locomotion and general DNA maintenance (grd-5, gcc-1, gop-2, etc.), and several with unknown function. Consistent with muscle functions, we detected mup-2 (muscle contractile protein troponin T), myo-3 (myosin heavy chain), dlc-1 (dynein light chain 1), and F22B5.10 (a poorly characterized gene involved in striated muscle myosin thick filament assembly). mup-2, myo-3, and dlc-1 were all found to be associated with ALG-1 in previous studies (Zisoulis et al. 2010; Kudlow et al. 2012). Consistent with muscle function, a GO term analysis of this data set highlights an enrichment of genes involved in locomotion (Table S1), suggesting a potential role for miRNAs in this biological process.
We also identified numerous actin gene isoforms (act-1, act-2, act-3, and act-4), which are required for maintenance of cellular architecture within the body wall muscle, and the Rho GTPase rho-1, which is required for regulation of actin filament-based processes including embryonic polarity, cell migration, cell shape changes, and muscle contraction (Table S1). Small GTPase are a gene class heavily targeted by miRNAs (Enright et al. 2003; Liu et al. 2012). The human ortholog of rho-1 is a known target for miR-31, miR-133, miR-155, and miR-185 (Liu et al. 2012).
Importantly, we also found several muscle-specific transcription factors including mxl-3, a basic helix–loop–helix transcription factor and K08D12.3, an ortholog of the human gene ZNF9. These genes are known to regulate proper muscle formation and cell growth. mxl-3 is targeted by miR-34 in the context of stress response (Chen et al. 2015). Both genes have been detected in past ALG-1 immunoprecipitation studies (Zisoulis et al. 2010).
Our top hit in this tissue is the zinc finger CCCH-type antiviral gene pos-1, a maternally inherited gene necessary for proper fate specification of germ cells, intestine, pharynx, and hypodermis (Farley et al. 2008). pos-1 contains several predicted miRNA binding sites in its 3′UTR (Table S1) and based on our GFP reporter validation study is strongly expressed in the body muscle (Figure S5). We also found the KH domain-containing protein gld-1, the homolog of the human gene QKI, which is targeted by miR-214 (Wu et al. 2017), miR-200c, and miR-375 (Pillman et al. 2018).
miRNA targeting is more extensive in the intestine than it is in the body muscle
By comparing the percentage of the putative tissue-specific miRNA targets identified in our study to the previously published intestine and body muscle transcriptomes (Blazie et al. 2015, 2017), we found that the hits in the intestine are almost twice the number of hits we obtained in the body muscle tissue (30.3% vs. 18.2%) (Figure 2B). The length of the 3′UTRs of genes identified as putative miRNA targets in the intestine and the body muscle tissues are similar when comparing the two tissues, but are on average longer and have more predicted miRNA binding sites than the overall C. elegans transcriptome (Figure 2Ci). Our results indicate that despite similarity in average 3′UTR length in tissues, the extent of miRNA-based regulatory networks is not similar across tissues. In this specific case, we found that the intestine utilizes miRNA-based gene regulation to a greater extent when compared to the body muscle.
Putative miRNA targets in the intestine and body muscle are enriched for miR-355 and miR-85 binding sites
A bioinformatic analysis of the longest 3′UTR isoforms of the targeted genes showed there was no specific requirement for the seed regions in either tissue (Figure 2Ciii). However, the use of predictive software showed that in addition to others, there is an intestine-specific bias for miR-355 targets (Figure 2Civ, green mark). This miRNA is involved in insulin signaling and innate immunity (Zhi et al. 2017), which in C. elegans are both mediated through the intestine.
In contrast, we observed an enrichment of targets for the poorly characterized miR-85 in the body muscle data set (Figure 2Civ, blue mark). These two miRNAs are uniquely expressed in the respective tissues (Martinez et al. 2008).
While miR-85 and miR-355 were the most abundant and tissue-restricted miRNAs identified in this study, several other miRNAs, including miR-71, miR-86, miR-785, and miR-792 were also found highly expressed but less spatially restricted.
Putative intestine and body muscle miRNAs target RBPs
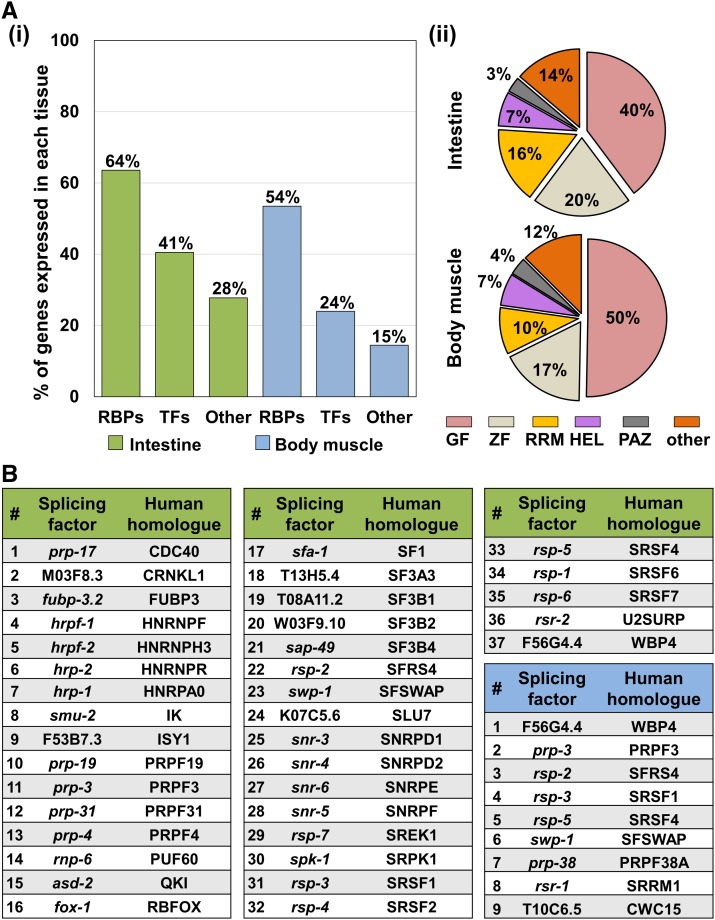
Upon further analysis, we observed an unexpected enrichment of genes containing RNA-binding domains in both data sets (Table S1). RBPs are known to play an important role in producing tissue-specific gene regulation by controlling gene expression at both the co- and post-transcriptional levels (Tamburino et al. 2013), and out of the ∼887 RBPs defined in C. elegans (Tamburino et al. 2013), we identified almost half as targets of miRNAs across both tissues (45%).
We found that out of the 599 known RBPs present in the intestine transcriptome (Blazie et al. 2015, 2017), 380 (64%) were present in our intestine data set as putative targets of miRNAs (Figure 3A). This is a notable enrichment when compared to transcription factors and non-RBP genes found in these tissues by Blazie et al. (2017), of which only a fraction were identified in our study as putative miRNA targets (Figure 3Ai). A similar trend is also present in the body muscle, with 170 (55%) of RBPs identified as putative miRNA targets (Figure 3Aii). Importantly, the largest pool of targeted RBPs in both tissues was composed of general factors (GFs), such as translation factors, tRNA interacting proteins, ribosomal proteins, and ribonucleases (Figure 3Aii). To a lesser extent we also detected other RBPs containing zinc-fingers (ZF), RNA-recognition motifs (RRM), Helicases (HEL) and PIWI/PAZ PIWI, and Argonaute-containing proteins (PAZ), suggesting extensive miRNA regulatory networks are in place in these tissues.
Figure 3.
An enrichment of RBPs and RNA splicing factors targeted by miRNAs in the intestine and body muscle tissues. (A) (i) Proportion of tissue-specific RBPs (Blazie et al. 2015, 2017) in each tissue that were identified as putative targets of miRNAs in this study. There is an enrichment of RBPs targeted in the intestine (green 63.6%) and the body muscle (blue 53.5%) tissues. “TFs” represents genes annotated as transcription factors while “Other” represents protein-coding genes that are not RBPs. (ii) Subtypes of RBPs targeted by miRNAs in the intestine and body muscle tissues. GF, general Factors, including translation factors, tRNA proteins, ribosomal proteins and ribonucleases; ZF, zinc finger; RRM, RNA recognition motif; HEL, RNA Helicase; PAZ, PIWI PAZ, PIWI, Argonautes. The majority of the targeted RBPs are general and zinc finger-containing factors. (B) RNA splicing factors identified as putative miRNA targets in the intestine (green) and body muscle (blue) tissues. More than half of the RNA splicing factors examined are targeted by miRNAs in the intestine as compared to the body muscle tissue.
miRNAs target RNA splicing factors
A further analysis of the RNA-binding proteins targeted in each tissue revealed that one of the most abundant classes of RBPs detected in our ALG-1 pull-down in intestine and body muscle data sets was RNA splicing factors (Figure 3B). The C. elegans transcriptome contains at least 78 known RNA splicing factors involved in both constitutive and alternative splicing (Tamburino et al. 2013). Sixty-four RNA splicing factors (82%) have been previously assigned by our group in the intestine (Blazie et al. 2015, 2017) and presumably are responsible for tissue-specific RNA splicing; 31 RNA splicing factors (40%) were also previously assigned by our group to the body muscle tissue (Blazie et al. 2015, 2017).
Our tissue-specific ALG-1 pull-down identified 37 RNA splicing factors as putative miRNA targets in the intestine (∼47%) (Figure 3B), and 34 of these were also previously identified by our group as being expressed in this tissue (Blazie et al. 2015, 2017). In contrast, we have detected only nine RNA splicing factors in our body muscle tissue ALG-1 pull-down, five of which were previously assigned by our group in the body muscle transcriptome (Blazie et al. 2015, 2017) (Figure 3B).
The difference in RNA splicing factors targeted by miRNA in these two tissues is significant as the intestine contains three orders of magnitude more putative miRNA-targeted RNA splicing factors than the body muscle. Of note, many different subtypes of RNA splicing factors identified in this study have human homologs, such as well-known snRNPs, hnRNPs, and SR proteins (Figure 3B).
Expression of the RNA splicing factors asd-2, hrp-2, and smu-2 is modulated through their 3′UTRs
To validate that RNA splicing factors found in our ALG-1 pull-down IPs are targeted by miRNAs in the intestine, we used the pAPAreg dual fluorochrome vector we developed in a past study (Blazie et al. 2017) (Figure 4A). We selected three representative RNA splicing factors identified in our study in the intestine (asd-2, hrp-2, and smu-2) (Table 1) and prepared transgenic strains to validate their expression and regulation (Figure 4B). We used the ges-1 3ʹUTR as a negative control for miRNA targeting, as it is strongly transcribed and translated in the intestine, with no predicted miRNA binding sites (PicTar), and poorly conserved seed regions (TargetScan), suggesting minimal post-transcriptional gene regulation (Egan et al. 1995; Marshall and McGhee 2001). ges-1 was not significantly abundant in our intestine ALG-1 pull-down (Table S1). The presence of the ges-1 3ʹUTR in the pAPAreg vector led to the expression of both mCherry and GFP fluorochromes, indicating robust transcription and translation of the construct as expected (Figure 4B).
Table 1. Summary of expression pattern, miRNA targets, and predicted miRNA binding sites for asd-2, hrp-2, and smu-2.
| Gene | Transcriptome | miRNAome | miRNA predictions | Known targets | |||
|---|---|---|---|---|---|---|---|
| Body muscle | Intestine | Body muscle | Intestine | miRanda | TargetScan | ||
| asd-2 | Y | Y | — | Y | miR-86, miR-255, miR-259, miR-785 | miR-1/796, miR-46/47 | let-2, unc-60 |
| hrp-2 | Y | Y | — | Y | miR-58, miR-62, miR-80, miR-81, miR-82, miR-83, miR-84, miR-85, miR-86, miR-90, miR-232, miR-244, miR-259, miR-357, miR-785 | miR-1018, miR-1821, miR-4809 | ret-1, unc-52, lin-10 |
| smu-2 | — | Y | — | Y | — | miR-60-3p, miR-234, miR-230, miR-392, miR-789, miR-792, miR-1020, miR-1818, miR-1828 | unc-52, unc-73 |
We then cloned asd-2, hrp-2, and smu-2 3ʹUTRs downstream of the GFP fluorochrome in our pAPAreg vector, prepared transgenic worms expressing these constructs, and studied the fluctuation of the expression level of the GFP fluorochrome in these transgenic strains. All three 3ʹUTRs were able to significantly lower GFP expression when compared to the control strain with the ges-1 3ʹUTR, with ∼40% repression, while the mCherry signal was similar in all strains (Figure 4B). These results suggest that these three RBPs contain regulatory binding sites within their 3ʹUTRs potentially able to repress their expression.
MiRNAs target intestine RNA splicing factors promoting tissue-specific alternative splicing
We then tested changes to tissue-specific alternative splicing in the intestine caused by the STAR protein family member asd-2, which regulates the alternative splicing pattern of the gene unc-60. unc-60 is expressed as two alternatively spliced isoforms in a tissue-specific manner (Ohno et al. 2012) (Figure 5A); unc-60a is expressed predominantly in the body muscle while unc-60b is expressed in many other tissues including the intestine (Ohno et al. 2012).
We first tested the unc-60 RNA isoform ratio in wt N2 worms. We extracted total RNA from N2 worms in triplicate and performed RT-PCR experiments using primers flanking the two unc-60 isoforms (Figure 5A). As expected, we found that the unc-60a longer isoform was more abundantly expressed in wt worms (62%) (Figure 5A).
We then investigated if the miRNA pathway has a role in regulating these splicing events, by testing changes in unc-60 isoform abundance in the alg-1 and alg-2 knockout strains RF54 [alg-1(gk214) X] and WM53[alg-2(ok304) II]. These strains are deficient in miRNA-based gene regulation. We found that loss of these miRNA effectors leads to a 10–20% shift in the expression of the two unc-60 isoforms (Figure 5A), indicating the importance of the miRNA pathway in regulating alternative splicing of this gene.
We then used a genetic approach to test the alternative splicing of this gene in the context of miRNA regulation. We reasoned that if ALG-1 targets the asd-2 3′UTR in the intestine lowering the expression of asd-2, which in turn causes unc-60 alternative splicing pattern, we should be able to interfere with this mechanism by overexpressing the asd-2 3′UTR in this tissue and in turn test the role of the miRNA pathway in this process.
As expected, the overexpression of the asd-2 3ʹUTR in the intestine led to changes in the unc-60 alternative splicing pattern, indicating that post-transcriptional regulation of asd-2 through its 3ʹUTR is important for the alternative splicing pattern of unc-60 in the intestine (Figure 5A). Conversely, asd-2 RNAi and overexpressing an unspecific 3′UTR (ges-1) did not induce changes in unc-60 alternative splicing pattern (Figure 5A). We validated the efficiency of our RNAi experiments by performing a layoff experiment, which indicated strong RNAi activity (Figure S7). Similar results were observed by testing a second splicing factor (hrp-2) known to direct alternative splicing of the genes ret-1, lin-10, and unc-52 (Kabat et al. 2009; Heintz et al. 2017) (Figure S8).
Loss of miRNA function leads to dispersed changes in splice junction usage
Since our data support a role for the miRNA pathway in modulating mRNA biogenesis, we were interested in testing the extent of these effects at the transcriptome level. We decided to download and map splicing junctions in genes from alg-1 and alg-2 knockout strains previously published by Brown et al. (2017). These worm strains are viable but are severely impaired. We reasoned that if the miRNA pathway contributes at some level to mRNA biogenesis, we should be able to see widespread changes in the usage of splice junctions in these data sets. To test this hypothesis, we downloaded the alg-1 and alg-2 data sets (three replicates for each strain plus wt N2 control) and extracted splice junction information. We first tested if the effects we observed in unc-60 with our biochemical and genetic approaches (Figure 5A) could also be detected in these data sets. In the case of unc-60, there is a 6–10% change in splice junction usage between isoforms consistently in all reannotated replicates, in both alg-1 and alg-2 knockout strains (Figure 5B). This result is in line with our analysis in Figure 5A. A similar and more striking aberrant splice junction usage is observed in the case of lin-10 and unc-52, and with a less pronounced effect in ret-1 (Figure S9). These results are also in agreement with our study illustrated in Figure S8, since an increase of splice isoform change in lin-10, unc-52, and ret-1 was also present in these two Argonaute-deficient strains (Figure S9).
We then expanded this analysis to all splicing junctions we were able to map using these transcriptomes. From a total of 30,115 high-quality known splice junctions present in all three data sets (Table S4), we identified ∼3946 of them in ∼2915 protein-coding genes that were affected by >2-fold change in usage in both alg-1 and alg-2 knockout data sets (∼13.2% of the total mapped splice junctions) (Figure 6A). In addition, we detect several cases of exon inclusion, skipping, and aberrant splicing events that occur exclusively in the alg-1 and/or alg-2 mutant strains (Figure 6B).
Discussion
In this manuscript we have developed tools and techniques to identify putative tissue-specific miRNA targets and applied them to uniquely define the genes targeted by miRNAs in the C. elegans intestine and body muscle tissues. We validated previous findings and mapped ∼3000 of novel tissue-specific interactions (Figure 2 and Table S1).
To perform these experiments, we have prepared worm strains expressing ALG-1 fused to GFP and expressed this cassette in the intestine and body muscle using tissue-specific promoters. We validated the ALG-1 expression (Figure S1) and the viability of our ALG-1 construct in in vivo studies using a layoff experiment (Figure S2). While not strictly a brood size assay where progenies are scored throughout the lifespan of the worms, the layoff experiment provides a simpler measure of the functionality of our alg-1 construct. While its results may be generally affected by egg retention, developmental delays, and slow ovulation rates, we did not observe these issues in our rescue strain.
We have then performed ALG-1 immunoprecipitations in duplicate, separated the miRNA complex from their targets, and sequenced the resultant RNA using Illumina sequencing (Figure 1 and Figure S3). To confirm our results, we validated a few selected hits with expression localization studies in both tissues (Figure S5).
Our ALG-1 pull-down results are mostly in agreement with previous studies on a large group of previously identified hits also present in our data sets (Figure 2A and Figure S4). Of note, Zisoulis et al. (2010) have performed their ALG-1 immunoprecipitations using cell extracts from intact and stage-specific (L4) worms, in contrast with this study, which used mixed-stages and tissue-specific conditions. The biological and technical differences in the methodology may account for some of the discrepancies observed when comparing these two studies.
The genes identified in this study overall match with the intestine transcriptome previously published by our group (81%) (Blazie et al. 2015, 2017). Of note, only 56% of genes identified as putative miRNA targets in the body muscle match the body muscle transcriptome (Figure 2B). Perhaps, the remaining targets are genes strongly down-regulated by miRNAs in this tissue, leading to rapid deadenylation and mRNA degradation that make them undetectable using our PAB-1-based pull-down approach. Given the fact that the body muscle transcriptome is significantly smaller than the intestine transcriptome, it may also be subjected to less regulation through miRNA. However, if we normalize the number of genes expressed in each tissue and study the proportion of the transcriptome targeted by miRNA, we still find significantly more regulation in the intestine (Figure 2A), suggesting that this tissue may indeed employ miRNA-based gene regulation to a greater extent.
We found a disparity in the proportion of each tissue-specific transcriptome targeted by miRNAs, with a notably larger proportion of genes targeted in the intestine. The majority of the targeted genes in our intestine pull-down IP are unique to the intestine and share only a handful of genes with our body muscle data set (725 genes, 23% of the total intestine data set) (Figure 2B). Conversely, very few genes are unique in the body muscle pull-down IP. The small pool of shared genes includes housekeeping genes that are most likely regulated similarly in both tissues. Of note, this minimal overlap between our tissue-specific data sets indicates that our ALG-1 pull-down is indeed tissue-specific with marginal cross-contamination.
Intriguingly, when we look at the miRNA population predicted to target the genes in our data sets as from miRanda (Figure 2Ciii and iv), we found an enrichment of known tissue-specific miRNA targets (Betel et al. 2010), which is in agreement with miRNA localization data sets (Alberti et al. 2018) (Figure S6). This in turn indicates that there is a tissue-specific miRNA targeting bias in C. elegans, with unique tissue-specific miRNAs, targeting unique populations of genes.
Approximately 40% of the identified hits were not predicted by miRanda to possess miRNA target sites (Figure 2Cii). Our group and others have shown in the past that prediction algorithms, while useful as a first step in characterizing miRNA targets, possess high levels of false positive and negative hits (Wolter et al. 2014), which in turn may have introduced noise in our analysis. Importantly, ∼70% of genes with predicted miRanda sites contain seed regions for miRNAs previously assigned by Alberti et al. (2018) in these tissues (Figure S6), suggesting that the majority of the genes identified in this study are indeed putative miRNA targets.
Our experimental approach was designed for tissue-specific mRNA target identification and unfortunately did not provide miRNA data. We assign tissue-specific targets to miRNAs relying primarily on prediction software and correlation to past-published data sets. These comparative approaches required conversion between genomic releases and data consolidation across different developmental stages and conditions, which may have added unwanted variability to our comparative analysis.
One of the most surprising findings of this study is that many targets obtained with our tissue-specific ALG-1 pull-down are RBPs. Sixty-four percent of the intestinal RBPs were found in our intestine ALG-1 pull-down, while 54% of the muscle RBPs were in our muscle ALG-1 pull-down. This result was unexpected given the small number of RBPs previously identified in the C. elegans genome (n = 887) (Tamburino et al. 2013), which amounted to only 4% of the total C. elegans protein-coding genes. However, previous studies have hinted at a strong regulatory network between miRNAs and RBPs, as the 3′UTRs of RBPs were found to contain on average more predicted miRNA binding sites than other gene classes (Tamburino et al. 2013).
Some of these RBPs in our top hits are well-characterized factors involved in C. elegans fertilization and early embryogenesis but are not well documented in somatic tissues. For example, within our top 100 hits we obtained the genes pos-1 and mex-5 in the body muscle, and gld-1 and oma-2 in the intestine. We were surprised by these results, but at least in the case of pos-1, which is our top hit in our body muscle data set, we validated its presence in the body muscle (Figure S5), suggesting a potential role for this and other RBPs outside the gonads.
RNA-binding-domain-containing proteins are involved in many biological processes, and their role is not limited to RNA biogenesis (Tamburino et al. 2013). RBPs can bind single- or double-strand RNAs and associate with proteins forming ribonucleoprotein complexes (RNPs). Longevity, fat metabolism, and development are all processes controlled by RNPs (Lee and Schedl 2006; Masuda et al. 2009; Aryal et al. 2017), and in the context of miRNA regulation, the ability of miRNAs to control RBPs abundance and function allow for an increased control of fundamental cellular core processes. Two hundred thirty-four RBPs are uniquely detected as putative miRNA targets in the intestine, while 147 RBPs are shared between both data sets.
Within this intestinal data set we mapped a surprising number of RBPs involved in RNA splicing (Figure 3B). We performed a literature search for known RNA splicing factors in C. elegans; out of the 72 total proteins identified, 37 of them were detected at a different level of strength in our intestine ALG-1 pull-down. In contrast, we do not observe this level of complexity in the body muscle, with only nine RNA splicing factors identified in this data set (Figure 3B).
asd-2 and smu-2 are well-known RNA splicing factors that induce exon retention in a dosage-dependent manner (Spartz et al. 2004; Ohno et al. 2012), while hrp-2 abundance leads to exon skipping (Kabat et al. 2009). Here we show that all three RNA splicing factors possess regulatory targets within their 3′UTRs that amount to ∼40% silencing activity in the intestine (Figure 4). Although we do not know which miRNAs target the asd-2 and hrp-2 3′UTRs, in Figure 5A and Figure S8 we show that alg-1 and alg-2 influence splice junction usage, perhaps through modulating the expression levels of these genes. In addition, the depletion of miRNAs targeting these RNA splicing factors using a sponge approach led to defects in the alternative splicing pattern of downstream genes regulated by asd-2 and hrp-2. Interestingly, the miRNAs predicted to target most splicing factors were not found highly expressed in this study. miR-85 and miR-355, the most abundant and tissue-restricted miRNAs identified, are only predicted to target <10% of all the RBPs found. This perhaps suggests that the abundance of miRNAs involved in RNA alternative splicing may be tightly regulated in tissues to make sure splicing events are properly executed.
Our genome-wide splice junction mapping effort in miRNA-deficient strains shows similar trends of aberrant splicing of unc-60, unc-52, lin-10, and ret-1 (Figure 5B and Figure S9), and displays an overall disruption of splicing events (∼13.2% of all splice junctions mapped) (Figure 6, A and B). Most of these defects are in known donor–acceptor splicing events, perhaps because RNA surveillance mechanisms may hide more severe disruptions.
Intriguingly, a GO term analysis on these aberrantly spliced genes highlights an enrichment in genes involved in development and morphogenesis (Figure S10), suggesting that perhaps, in addition to the well-known role in modulating gene abundance, the miRNA pathway may also sustain their activity, by allowing the proper splicing of these developmental genes. More experiments need to be performed to study the full extent of these effects.
Unfortunately, our in vivo approach does not reach the resolution needed to conclusively pinpoint the extent of the miRNA pathway in this process. To perform in vivo experiments, we used total RNA extracted from transgenic worms, and studied change in exon abundance occurring in a single tissue within a whole animal, which prevented us from reaching the same resolution obtainable with in vitro splicing experiments and mini-genes. In addition, the effects we observe are ameliorated by the presence of at least one functional Argonaute protein, which can compensate for the loss of the other. Knockout of the entire miRNA pathway is lethal in C. elegans, and while aberrant splicing may play a role in producing this phenotype, these activities are challenging to detect in vivo.
Taken together, our results support a role for miRNAs in regulating alternative splicing in the intestine, where their presence in a tissue-specific manner may lead to alteration of the dosage balance of RNA splicing factor, leading to tissue-specific alternative splicing (Figure 6C). MiRNAs typically alter gene expression dosage, rather than induce complete loss of protein function (Wolter et al. 2017; Bartel 2018). On the other hand, many RNA splicing factors involved with constitutive and alternative splicing are ubiquitously expressed (Shin and Manley 2004) but are somehow able to induce tissue-specific alternative splicing in a dosage dependent manner. In this context, it is feasible that miRNAs may alter the dosage of RNA splicing factors, leading to tissue-specific alternative splicing (Figure 6C).
We have uploaded our intestine and body muscle miRNA target data sets into the 3′UTRome database (www.UTRome.org), which is the publicly available resource for the C. elegans community interested in 3′UTR biology (Mangone et al. 2008, 2010). To provide a more comprehensive overview, we have also manually curated and included results from several available data sets including PicTar (Lall et al. 2006) and TargetScan (Lewis et al. 2005) miRNA target predictions, experimentally validated ALG-1 interaction (Zisoulis et al. 2010; Kudlow et al. 2012), and tissue-specific gene expression and expanded 3′UTR isoform annotation data (Jan et al. 2011; Blazie et al. 2015, 2017).
Acknowledgments
We thank Stephen Blazie for the cloning of the alg-1 coding sequence used as the backbone for the development of the vectors used in this manuscript. We thank Honor Glenn and Jordan Yaron for their advice and assistance when using the confocal microscopes and image acquisition. We thank Cherie Alissa Lynch and Alan Rawls for advice and assistance in performing the qPCR experiments. We also thank Heather Hrach for insights and review of the manuscript. We thank Gabrielle Richardson for maintaining the C. elegans strains used in this manuscript. This work was supported by the National Institutes of Health grant number 1R01GM118796. The authors declare that they have no competing interests.
Author contributions: MM and KK designed the experiments. KK executed the experiments. ALS executed a portion of the experiments. HSS assisted with the experiments and the imaging of the C. elegans transgenic lines and performed the analysis in Figure S4. MM and KK performed the bioinformatics analysis and uploaded the results to the UTRome.org database. MM and KK led the analysis and interpretation of the data, assembled the figures, and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8095457.
Communicating editor: V. Reinke
Literature Cited
- Ahringer, J., 2006 Reverse genetics, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.47.1, http://www.wormbook.org./10.1895/wormbook.1.47.1 [Google Scholar]
- Alberti C., Manzenreither R. A., Sowemimo I., Burkard T. R., Wang J., et al. , 2018. Cell-type specific sequencing of microRNAs from complex animal tissues. Nat. Methods 15: 283–289. 10.1038/nmeth.4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Ruvkun G., 2018. Recent molecular genetic explorations of Caenorhabditis elegans MicroRNAs. Genetics 209: 651–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal B., Singh A. K., Rotllan N., Price N., Fernandez-Hernando C., 2017. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 28: 273–280. 10.1097/MOL.0000000000000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle F. E., Giudice J., 2017. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18: 437–451. 10.1038/nrm.2017.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2018. Metazoan microRNAs. Cell 173: 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Wilson M., Gabow A., Marks D. S., Sander C., 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res. 36: D149–D153. 10.1093/nar/gkm995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C., Leslie C., 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11: R90 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., Isik M., 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88: 290–301. 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie S. M., Babb C., Wilky H., Rawls A., Park J. G., et al. , 2015. Comparative RNA-Seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 13: 4 10.1186/s12915-015-0116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazie S. M., Geissel H. C., Wilky H., Joshi R., Newbern J., et al. , 2017. Alternative polyadenylation directs tissue-specific miRNA targeting in Caenorhabditis elegans somatic tissues. Genetics 206: 757–774. 10.1534/genetics.116.196774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Lovci M. T., Huang J. L., Yeo G. W., Pasquinelli A. E., 2016. Pairing beyond the seed supports microRNA targeting specificity. Mol. Cell 64: 320–333. 10.1016/j.molcel.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. C., Svendsen J. M., Tucci R. M., Montgomery B. E., Montgomery T. A., 2017. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 45: 9093–9107. 10.1093/nar/gkx536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari S. I., Vasquez-Rifo A., Gagne D., Paquet E. R., Zetka M., et al. , 2012. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res. 22: 1034–1045. 10.1038/cr.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J. F., Stamm S., Helfman D. M., Krainer A. R., 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709. 10.1126/science.8085156 [DOI] [PubMed] [Google Scholar]
- Chen H. C., Cheng S. C., 2012. Functional roles of protein splicing factors. Biosci. Rep. 32: 345–359. 10.1042/BSR20120007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Rajewsky N., 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 8: 93–103. 10.1038/nrg1990 [DOI] [PubMed] [Google Scholar]
- Chen X., Chen S., Xiu Y. L., Sun K. X., Zong Z. H., et al. , 2015. RhoC is a major target of microRNA-93–5P in epithelial ovarian carcinoma tumorigenesis and progression. Mol. Cancer 14: 31 10.1186/s12943-015-0304-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S. W., Hannon G. J., Darnell R. B., 2012. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 19: 321–327. 10.1038/nsmb.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conne B., Stutz A., Vassalli J. D., 2000. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat. Med. 6: 637–641. 10.1038/76211 [DOI] [PubMed] [Google Scholar]
- Delay C., Calon F., Mathews P., Hebert S. S., 2011. Alzheimer-specific variants in the 3′UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 6: 70 10.1186/1750-1326-6-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina A. S., Iser W. B., Park S. S., Maudsley S., Wilson M. A., et al. , 2011. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 11: 11 10.1186/1472-6793-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O., 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13: 849–851. 10.1038/nsmb1138 [DOI] [PubMed] [Google Scholar]
- Ding X. C., Slack F. J., Grosshans H., 2008. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 7: 3083–3090. 10.4161/cc.7.19.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C. R., Chung M. A., Allen F. L., Heschl M. F., Van Buskirk C. L., et al. , 1995. A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Dev. Biol. 170: 397–419. 10.1006/dbio.1995.1225 [DOI] [PubMed] [Google Scholar]
- Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., et al. , 2007. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 104: 17016–17021 [corrigenda: Proc. Natl. Acad. Sci. USA 105: 399 (2008)]. 10.1073/pnas.0708115104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., John B., Gaul U., Tuschl T., Sander C., et al. , 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. C., and C. Thomas, 2006 Transformation and microinjection, WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.108.1, http://www.wormbook.org./10.1895/wormbook.1.108.1 [Google Scholar]
- Farley B. M., Pagano J. M., Ryder S. P., 2008. RNA target specificity of the embryonic cell fate determinant POS-1. RNA 14: 2685–2697. 10.1261/rna.1256708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K., Qadota H., Benian G. M., 2017. Development, structure, and maintenance of C. elegans body wall muscle. WormBook 2017: 1–59. 10.1895/wormbook.1.81.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., 2004. The microRNA Registry. Nucleic Acids Res. 32: D109–D111. 10.1093/nar/gkh023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J., 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34: D140–D144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J., 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158. 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., et al. , 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27: 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H., Johnson T., Reinert K. L., Gerstein M., Slack F. J., 2005. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell 8: 321–330. 10.1016/j.devcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Ha I., Wightman B., Ruvkun G., 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 10: 3041–3050. 10.1101/gad.10.23.3041 [DOI] [PubMed] [Google Scholar]
- Heid P. J., Raich W. B., Smith R., Mohler W. A., Simokat K., et al. , 2001. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev. Biol. 236: 165–180. 10.1006/dbio.2001.0315 [DOI] [PubMed] [Google Scholar]
- Heintz C., Doktor T. K., Lanjuin A., Escoubas C., Zhang Y., et al. , 2017. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature 541: 102–106 (erratum: Nature 547: 476). 10.1038/nature20789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A., Kudla G., Dudnakova T., Tollervey D., 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C. H., Friedman R. C., Ruby J. G., Bartel D. P., 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. 10.1038/nature09616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannot G., Michaud P., Quévillon Huberdeau M., Morel-Berryman L., Brackbill J. A., et al. , 2016. GW182-free microRNA silencing complex controls post-transcriptional gene expression during Caenorhabditis elegans embryogenesis. PLoS Genet. 12: e1006484 (erratum: PLoS Genet. 12: e1006534). 10.1371/journal.pgen.1006484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jima D. D., Zhang J., Jacobs C., Richards K. L., Dunphy C. H., et al. , 2010. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood 116: e118–e127. 10.1182/blood-2010-05-285403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat J. L., Barberan-Soler S., Zahler A. M., 2009. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J. Biol. Chem. 284: 28490–28497. 10.1074/jbc.M109.023101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow B. A., Zhang L., Han M., 2012. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol. Cell 46: 530–541. 10.1016/j.molcel.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurn N., Chen P., Heath J. D., Kopf-Sill A., Stephens K. M., et al. , 2005. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 51: 1973–1981. 10.1373/clinchem.2005.053694 [DOI] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y. L., et al. , 2006. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 16: 460–471. 10.1016/j.cub.2006.01.050 [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., et al. , 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. H., and T. Schedl, 2006 RNA-binding proteins. WormBook, ed. The C. elegans Research Community WormBook, 18: 1–13. 10.1895/wormbook.1.79.1 10.1895/wormbook.1.79.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P., 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Bi F., Zhou X., Zheng Y., 2012. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol. 22: 365–373. 10.1016/j.tcb.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T., 2007. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27: 435–448. 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Macmenamin P., Zegar C., Piano F., Gunsalus K. C., 2008. UTRome.org: a platform for 3′UTR biology in C. elegans. Nucleic Acids Res. 36: D57–D62. 10.1093/nar/gkm946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. 10.1126/science.1191244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S. D., McGhee J. D., 2001. Coordination of ges-1 expression between the Caenorhabditis pharynx and intestine. Dev. Biol. 239: 350–363. 10.1006/dbio.2001.0442 [DOI] [PubMed] [Google Scholar]
- Martinez N. J., Ow M. C., Reece-Hoyes J. S., Barrasa M. I., Ambros V. R., et al. , 2008. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 18: 2005–2015. 10.1101/gr.083055.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Marasa B., Martindale J. L., Halushka M. K., Gorospe M., 2009. Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging (Albany N.Y.) 1: 681–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F., Smith C. W., 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6: 386–398. 10.1038/nrm1645 [DOI] [PubMed] [Google Scholar]
- Matoulkova E., Michalova E., Vojtesek B., Hrstka R., 2012. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 9: 563–576. 10.4161/rna.20231 [DOI] [PubMed] [Google Scholar]
- Mayr C., 2017. Regulation by 3′-untranslated regions. Annu. Rev. Genet. 51: 171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Mayr C., Bartel D. P., 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee, J. D., 2007 The C. elegans intestine. WormBook, ed. The C. elegans Research Community WormBook, 27: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman, D. G., and B. D. Williams, 2006 Sarcomere assembly in C. elegans muscle. WormBook, ed. The C. elegans Research Community WormBook, 16: 1–16. 10.1895/wormbook.1.81.1 10.1895/wormbook.1.81.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes M., Sanford B. L., Comiskey D. F., Chandler D. S., 2019. RNA splicing and disease: animal Models to therapies. Trends Genet. 35: 68–87. 10.1016/j.tig.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Zhang C., Gantman E. C., Mele A., Darnell J. C., et al. , 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 9: 263–293. 10.1038/nprot.2014.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G., Ono K., Togo M., Watanabe Y., Ono S., et al. , 2012. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet. 8: e1002991 10.1371/journal.pgen.1002991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou P., Goodarzi H., Tavazoie S., 2014. Systematic identification of regulatory elements in conserved 3′ UTRs of human transcripts. Cell Rep. 7: 281–292. 10.1016/j.celrep.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J., 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415 [corrigenda: Nat. Genet. 41: 762 (2009)]. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Pillman K. A., Phillips C. A., Roslan S., Toubia J., Dredge B. K., et al. , 2018. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 37(13) 10.15252/embj.201899016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-de-la-Riva M., Fontrodona L., Villanueva A., Ceron J., 2012. Basic Caenorhabditis elegans methods: synchronization and observation. J. Vis. Exp. (64): 4019 https://www.ncbi.nlm.nih.gov/pubmed/22710399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Xu W., Xu J., Shi Q., Li J., et al. , 2017. Enolase 1 stimulates glycolysis to promote chemoresistance in gastric cancer. Oncotarget 8: 47691–47708. 10.18632/oncotarget.17868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeld A., Plass M., Krogh A., Friis-Hansen L., 2013. Alterations in polyadenylation and its implications for endocrine disease. Front. Endocrinol. (Lausanne) 4: 53 10.3389/fendo.2013.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J., Slack F. J., Basson M., Pasquinelli A. E., Bettinger J. C., et al. , 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M. M., Swanson M. S., 2016. RNA mis-splicing in disease. Nat. Rev. Genet. 17: 19–32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., et al. , 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Shin C., Manley J. L., 2004. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 5: 727–738. 10.1038/nrm1467 [DOI] [PubMed] [Google Scholar]
- Shin C., Nam J. W., Farh K. K., Chiang H. R., Shkumatava A., et al. , 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell 38: 789–802. 10.1016/j.molcel.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Vikos T., de Lencastre A., Inukai S., Shlomchik M., Holtrup B., et al. , 2014. MicroRNAs mediate dietary-restriction-induced longevity through PHA-4/FOXA and SKN-1/Nrf transcription factors. Curr. Biol. 24: 2238–2246. 10.1016/j.cub.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A. K., Herman R. K., Shaw J. E., 2004. SMU-2 and SMU-1, Caenorhabditis elegans homologs of mammalian spliceosome-associated proteins RED and fSAP57, work together to affect splice site choice. Mol. Cell. Biol. 24: 6811–6823. 10.1128/MCB.24.15.6811-6823.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhi L., Shakoor S., Liao K., Wang D., 2016. microRNAs involved in the control of innate immunity in Candida infected Caenorhabditis elegans. Sci. Rep. 6: 36036 10.1038/srep36036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburino A. M., Ryder S. P., Walhout A. J., 2013. A compendium of Caenorhabditis elegans RNA binding proteins predicts extensive regulation at multiple levels. G3 (Bethesda) 3: 297–304. 10.1534/g3.112.004390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [corrigenda: Nat. Protoc. 9: 2513 (2014)]. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Rifo A., Jannot G., Armisen J., Labouesse M., Bukhari S. I., et al. , 2012. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One 7: e33750 10.1371/journal.pone.0033750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter J. M., Kotagama K., Pierre-Bez A. C., Firago M., Mangone M., 2014. 3′LIFE: a functional assay to detect miRNA targets in high-throughput. Nucleic Acids Res. 42: e132 10.1093/nar/gku626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter J. M., Le H. H., Linse A., Godlove V. A., Nguyen T. D., et al. , 2017. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res. 27: 53–63. 10.1101/gr.209361.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li Z., Yang M., Dai B., Hu F., et al. , 2017. MicroRNA-214 regulates smooth muscle cell differentiation from stem cells by targeting RNA-binding protein QKI. Oncotarget 8: 19866–19878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi L., Yu Y., Jiang Z., Wang D., 2017. mir-355 functions as an important link between p38 MAPK signaling and insulin signaling in the regulation of innate immunity. Sci. Rep. 7: 14560 10.1038/s41598-017-15271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A. R., 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8: 1351–1361. 10.1016/S1097-2765(01)00409-9 [DOI] [PubMed] [Google Scholar]
- Zisoulis D. G., Lovci M. T., Wilbert M. L., Hutt K. R., Liang T. Y., et al. , 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17: 173–179. 10.1038/nsmb.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures and supplemental figures, and tables and supplemental tables. Raw reads were submitted to the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/), under the SRP Study Accession # PRJNA517741. The results of our analyses are available in Excel format in Tables S1 and S4, and in our 3′UTR-centric website www.UTRome.org. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8095457.