Abstract
Purpose:
To design a deep learning algorithm that automatically delineates lung tumors seen on weekly magnetic resonance imaging (MRI) scans acquired during radiotherapy and facilitates the analysis of geometric tumor changes.
Methods:
This longitudinal imaging study comprised 9 lung cancer patients who had 6–7 weekly T2-weighted MRI scans during radiotherapy. Tumors on all scans were manually contoured as the ground truth. Meanwhile, a patient-specific adaptive convolutional neural network (A-net) was developed to simulate the workflow of adaptive radiotherapy and to utilize past weekly MRI and tumor contours to segment tumors on the current weekly MRI. To augment the training data, each voxel inside the volume of interest was expanded to a 3 × 3 cm patch as the input, whereas the classification of the corresponding patch, background or tumor, was the output. Training was updated weekly to incorporate the latest MRI scan. For comparison, a population-based neural network was implemented, trained, and validated on the leave-one-out scheme. Both algorithms were evaluated by their precision, DICE coefficient, and root mean square surface distance between the manual and computerized segmentations.
Results:
Training of A-net converged well within 2 h of computations on a computer cluster. A-net segmented the weekly MR with a precision, DICE, and root mean square surface distance of 0.81 ± 0.10, 0.82 ± 0.10, and 2.4 ± 1.4 mm, and outperformed the population-based algorithm with 0.63 ± 0.21, 0.64 ± 0.19, and 4.1 ± 3.0 mm, respectively.
Conclusion:
A-net can be feasibly integrated into the clinical workflow of a longitudinal imaging study and become a valuable tool to facilitate decision-making in adaptive radiotherapy.
Keywords: Lung tumor, Deep learning, Longitudinal study, MRI
Understanding the temporal patterns of tumor evolution during radiotherapy is critical to the design of adaptive radiotherapy (ART) such as determining the timing and frequency of reproducing a treatment plan that conforms to the newly observed target. Monitoring tumor response via weekly imaging surveillance, such as cone beam computed tomography (CBCT) during radiotherapy of locally advanced non-small cell lung cancer (LA-NSCLC), is currently a routine process in the clinic because CBCT is readily integrated into the functionality of modern linacs. Although CBCT is very helpful in clearly defining parenchymal tumors, visualizing and segmenting a mediastinal tumor are extremely challenging. Mediastinal tumors may invade the surrounding structures and attach to the pleural or mediastinal wall, therefore, having very irregular shapes and often indistinct boundaries. Magnetic resonance imaging (MRI) is increasingly utilized in the setting of radiation oncology, primarily for better soft tissue contrast. There is a dedicated MRI simulator in our clinic that is routinely used for MR-based simulation (including MR only), patient monitoring, and therapy response assessment [1]. Based on our early clinical experience, MRI does provide detailed internal tissue constitutions of tumor anatomy and can differentiate tumor from surrounding soft tissue. Therefore, we developed a non-invasive weekly MRI surveillance protocol to monitor radiotherapy of LA-NSCLC.
In a busy clinic, timely segmentation of tumors and critical organs is a highly demanding task for the radiotherapy team, who would rather spend spare time on optimizing a personalized treatment. An automatic, unbiased, accurate, and prompt process for segmentation of lung tumor is essential to enable a viable clinical longitudinal imaging protocol. Convolutional neural network (CNN) is widely applied to medical image classification, segmentation, and object detection and has been very successful in many radiotherapy sites [2–5]. There exist two major CNN types for image segmentation: semantic- and patch-based. Semantic image segmentation, such as fully convolutional CNN [6], U-net [7], V-net [8] and Dcan [9], takes a whole image (two-dimensional (2D) or three-dimensional (3D)) as an input and generates the labels of tumors as output. Consequently, the semantic-based method requires a large number of training samples to obtain a high performance. On the contrary, the patch-based CNN method is designed for a relatively small dataset. It extracts patches from all pixels/voxels of a 2D/3D image and classifies each patch into different labels [10–15]. However, because the tumor size is relatively small compared with the whole image, this method is sensitive to the selection of the volume of interest (VOI). As a newly formed longitudinal imaging study, there are limited data available for the segmentation exercise. To overcome this challenge, a patient-specific adaptive tumor segmentation method was designed by adopting a patch-based CNN model and taking full advantage of the historical information in a longitudinal study. The patch-based model was applied to augment the data using every voxel inside the VOI as an input. The historical information, i.e., segmentation results in past weekly images, was used as a prior input to define a relevant VOI for the current weekly image, then to confine and simplify the segmentation problem. The CNN model was used to learn the hidden features of tumor that are difficult to directly visualize and appreciate with the naked eye. In this paper, the development of such a novel tool of tumor segmentation is reported, specifically for a longitudinal imaging study, and its performance on a variety of LA-NSCLC patients is evaluated.
Method and materials
MRI longitudinal imaging study
Nine LA-NSCLC patients with involved mediastinal lymph nodes undergoing conventionally fractionated external beam radiotherapy to 60 Gy were enrolled in a prospective 1RB approved study to undergo weekly MRI scans. Each patient underwent a MRI at the time of their radiation therapy simulation on a 3 Tesla Philips Ingenia scanner (Philips Medical Systems, Best, Netherlands). Simulation MR1 was followed by voluntary weekly MR1 scans during treatment. The patients were scanned using a 16-element phased array anterior coil and a 44-element posterior coil. The imaging protocol included a respiratory triggered 2D axial T2-weighted (T2W) turbo spin-echo sequence (TR/TE = 3000–6000/120 ms, slice thickness = 2.5 mm and in-plane resolution of 1.1 × 0.97 mm2, flip angle = 90, number of slices = 43, number of signal averages = 2, and field of view = 300 × 222 × 150 mm3).
Throughout the course of radiotherapy, 6–7 weekly MRI scans were acquired for each patient. All weekly T2W-MR1 scans were manually registered to the simulation MRI by aligning the spine and outer lung. Tumors on all image sets were manually contoured by a radiation oncologist, serving as the ground truth. Gross tumor volume (GTV) at the point of simulation ranged from 4.8 cc to 257.2 cc, with a median of 89.1 cc. On average, tumor shrunk 59.8 ± 26.6% at the end of treatment as shown in Fig. 1.
Fig. 1.
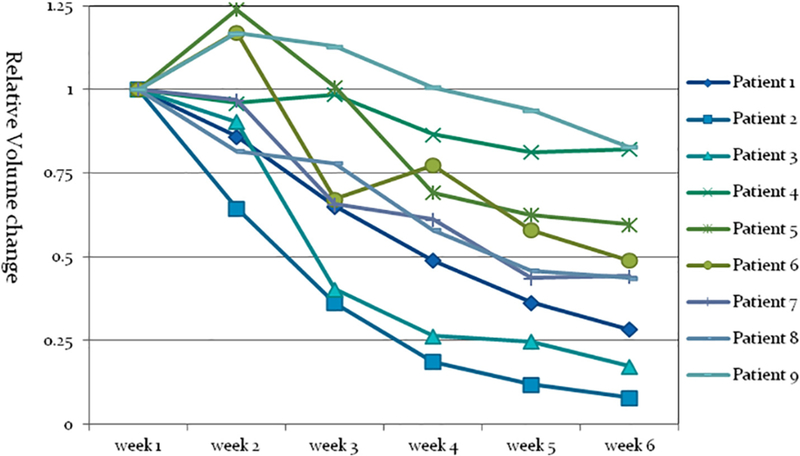
Relative lung tumor shrinkage during radiotherapy monitored via a longitudinal weekly magnetic resonance imaging study. Although volumes of lung tumors increased at the first couple of weeks among 3 patients, they all eventually decreased between 17% and 93%.
Image preprocessing
Segmentation relies heavily on the intensities of the observed images. As the first step of image preprocessing, the T2W-MRI images were standardized to remove the scanner/sequence/patient-dependent variations of image intensities so that the segmentation algorithm can be generalized among different sessions within a particular patient and across patients in the imaging study [16]. A slice based normalization method was implemented to compensate for the variations:
| (1) |
where Imean and Istd are the mean and standard deviation of the intensity within a slice, respectively.
It is critical to the segmentation algorithm to define a VOI that covers all the possible variations seen in the training and testing sessions with relatively balanced samples of both tumor and background. By taking advantage of the historical information in the longitudinal study, VOI for each individual MRI scan was derived as the union of GTVs on previous weekly scans expanded by a patient-specific and heterogeneous margin, which was one-sixth of the maximal GTV length in the lateral and anterior-posterior direction and 7.5 mm in the superior-inferior direction. This margin accounts for registration errors, respiratory uncertainties, and tumor growth among certain patients. The propagation of VOI between weeks was automatically completed after the new weekly MRI was registered to the previous weekly MRI scans.
Segmentation via CNN
A patient-specific adaptive patch-based segmentation CNN (A-net, Fig. 2) was developed to segment the tumor and avoid difficulties in handling inter-patient variations. A population-based CNN, used as a reference, was also implemented for comparison.
Fig. 2.
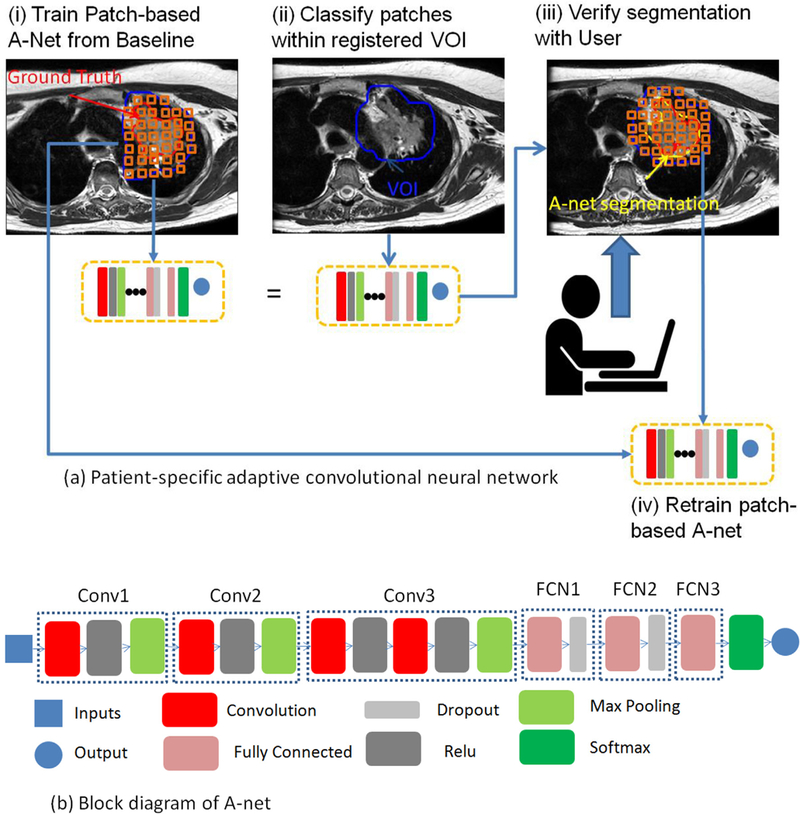
(a) Design of a patient-specific adaptive convolutional neural network for tumor segmentation. (b) Block diagram of a 6-level deep A-net, which was implemented via the MatConvNet on the MATLAB platform.
A-net
The inputs to the A-net are 2D patches extracted from the weekly MRI scans. A particular 2D patch is a 3 cm × 3 cm image centered at each voxel within the VOI. The selection of the patch size emphasizes the local rather than the global image features and strikes a balance between computation burden and performance. The patch is classified into two categories: tumor or background, according to the classification of its center. All the patches were normalized to avoid intensity variations across patches. We further expanded a 2D patch with 6 immediate slices above and below and formed a 3D patch of 3 cm × 3 cm × 3 cm. We compared the model performance and computational cost of using 2D or 3D patches as inputs of A-net to select the most suitable model parameters for this segmentation task.
Inspired by CNN architectures, such as VGG [17], Google Net [18], and Res Net [19], an architecture of A-net was designed using the MatConvNet toolbox [20] on a MATLAB platform for this longitudinal imaging study (Fig. 2b). A-net was constructed 6 levels deep, with four convolution layers, four rectifier linear units (ReLU) layers, three max pooling layers, three fully connected layers, two dropout layers, and one soft max layer. Four convolutional layers were used to down-sample the input and extract the relevant features to classify the tumor from the background. Three max pooling layers were used to refine the most representative features and avoid over-fitting. The ReLU layer, a computationally efficient, non-linear activation function, was used to learn more complex patterns. The dropout layer was used to improve the over-fitting problem by dropping out redundant units in the architecture. The major difference between A-net and other architectures were: (1) the stride for the convolutional layer and max pooling layer were configured as 2 and 1, respectively. In order to extract more global information and less local information from the relatively small input of patches, both the convolutional layer and the max pooling layer were used to downsize the input and extract representative features. The first three convolutional layers with stride 2 were used to downsize the input to nearly half to extract important global features, and the max pooling layer was used to refine the most representative local features from the global features. (2) A convolutional layer was directly followed by a ReLU layer only in the first three main layers, and a fully connected layer was directly followed by a dropout layer instead of a ReLU in the last three main layers. Potential over-fitting is reduced by simplifying the patient-specific model with linear, rather than non-linear, activation functions in the last three levels.
The direct output of A-net was a binary mask image of segmented tumors. A post-processing procedure was applied to fill the holes and remove the background noise and small isolated islands via morphological filters.
Training and testing
The allocation of data to either training or testing simulates the clinical workflow of weekly surveillance in ART. After an acquisition of a particular weekly MRI scan, the immediate clinical goal is to obtain the tumor contour. The previous weekly MRI scans, including their segmented GTVs, were allocated as training data, and the current weekly MRI became testing data, i.e., A-net was trained on data of week 1, 2... N-1, and tested on a week N (N = 2, 3, 4, 5, 6) scan. As a result, A-net was updated weekly to accommodate for new observations.
The training and testing of A-net were performed on an institutional Lilac GPU cluster with a Ge-force GTX 1080 GPU, 8 GB memory, and a batch size of 200. A loss function of SoftMax log [20] was utilized for training, which was optimized via a stochastic gradient descent algorithm with an initial learning rate of 0.001. All available patches were processed in a random order. The training process was terminated if it met the early stopping rule [21], or the maximal number of iteration (100) was reached. Time spent on the update of training was within hours and therefore did not impose any stress on the ART workflow.
Population-based CNN
The limitation of the patient-specific model is that the segmentation approved by clinicians is required up to the testing week. On the contrary, a population-based CNN model can learn useful features from existing patients, and transfer the knowledge to segment tumors for new patients without input from human experts. However, the performance of a population-based model highly depends on whether the training dataset is large enough to cover all the possible variations in future patients. Only 9 patients are available in this newly formed imaging protocol, which may compromise the applicability of the algorithm. Nevertheless, we implemented both semantic- and patch-based deep learning algorithm to segment tumor using the population data. In the patch-based approach, to obtain a fair comparison with the patient-specific model, the architecture adopted was the same as A-net. In the semantic-based approach, a U-net [7] was trained and tested. The input of U-net was a 2D slice (256 × 256 pixels) of a weekly MRI scan, and the output was a binary mask of tumor. Both population-based deep learning algorithms were validated using a leave-one-out strategy.
Evaluation
Comparing tumor segmented via CNN to that manually contoured by a radiation oncologist, we calculated the precision, sensitivity, DICE, and root mean square surface distance (RMSD) for the purpose of evaluation. In addition, we performed deformable registration to transfer tumor contours (manual segmentation) from the immediate previous weekly scan to the current weekly scan using a commercial software called MIM VISTA (Beachwood, OH) since deformable registration is an alternative clinical route for contour propagation. In the manual task of registration, we ran rigid registration first to align the outer lung and spine, followed by a deformable registration. The segmentation via deformable registration was compared to the ground truth and served as a clinical benchmark.
Results
When only the first weekly MRI was utilized to segment the second weekly MRI, the number of patches used for each individual patient ranged from 4941 to 230,296 with a median of 59,151. The training process finished within 30 minutes. As the radiotherapy treatment progressed, the number of weekly data involved in training increased. At week 6, when all previous weekly MRI were utilized, the median number of patches increased fivefold to 303,916, and the time spent on training increased to 2 h. We experimented an alternative training strategy which fine-tuned the A-net already trained on the previous week with the incremental data only to shorten the training time. However, this faster approach resulted in an averaged 0.03 loss of DICE compared to training from scratch, when the weekly tumor regression pattern oscillates such as patient #6 shown in Fig. 1, potentially due to the overfitting to the latest week in the non-linear process of training. To pursue the best performance of segmentation, we trained A-net from scratch throughout this study. After the patient-specific A-net was trained, the computational time for segmenting one weekly MRI was less than 30 s on the GPU cluster. For the population-based A-net model, the number of patches in training ranged from 1,107,841 to 1,550,969 with a median of 1,201,730. The training process completed within 8 h.
Table 1 shows the segmentation results on a weekly basis for both patient-specific and population-based algorithms. At a particular week N (N = 2, 3, 4, 5, 6), based on training with data from previous weeks, A-net can robustly segment the tumor with a precision of 0.82 ± 0.08, DICE of both 0.82 ± 0.10, and the geometric errors measured by 2.4 ± 1.4 mm. The variations of performance between weeks were caused by the uneven shrinkage of tumors between weeks as well as the much smaller tumor towards the end of treatment. Feeding A-net with 3D patches resulted in a DICE of 0.80 ± 0.12, which is statistically indistinguishable from using 2D patches. However, training time increased to 190%. This observation is similar to what is reported in a brain segmentation study [22], and justifies the use of 2D patches as a faster and convenient approach. By contrast, the population-based A-net underperformed with a lower DICE and precision of 0.64 ± 0.19 and 0.63 ± 0.21, respectively, and a significantly higher RMSD of 4.1 ± 3.0 mm. The population-based U-net with a DICE of 0.59 ± 0.28, and RMSD of 3.4 ± 2.4 mm, inferior compared to A-net, demonstrating that patch-based approach is more appropriate for this longitudinal imaging study.
Table 1.
Performance comparisons between A-net and the population-based CNN.
| 1. A-net | ||||||
|---|---|---|---|---|---|---|
| W2 | W3 | W4 | W5 | W6 | Average | |
| DICE Prec. RMSD (mm) Sen. |
0.84 ± 0.10 0.80 ± 0.15 1.9 ± 0.6 0.89 ± 0.03 |
0.82 ± 0.08 0.75 ± 0.11 2.3 ± 1.3 0.90 ± 0.06 |
0.81 ± 0.11 0.82 ± 0.08 2.9 ± 1.7 0.83 ± 0.15 |
0.81 ± 0.14 0.83 ± 0.05 2.1 ± 0.9 0.82 ± 0.18 |
0.81 ± 0.09 0.85 ± 0.08 2.5 ± 2.2 0.79 ± 0.14 |
0.82 ± 0.10 0.82 ± 0.08 2.4 ± 1.4 0.85 ± 0.13 |
| Population-based A-net | ||||||
| W2 | W3 | W4 | W5 | W6 | Average | |
| DICE Prec. RMSD (mm) Sen. |
0.70 ± 0.22 0.69 ± 0.25 2.9 ± 1.4 0.78 ± 0.09 |
0.68 ± 0.13 0.65 ± 0.15 3.7 ± 2.6 0.74 ± 0.13 |
0.64 ± 0.17 0.64 ± 0.21 4.1 ± 3.3 0.69 ± 0.14 |
0.59 ± 0.26 0.60 ± 0.26 4.8 ± 3.5 0.60 ± 0.25 |
0.58 ± 0.23 0.58 ± 0.23 5.1 ± 4.5 0.60 ± 0.25 |
0.64 ± 0.19 0.63 ± 0.21 4.1 ± 3.0 0.68 ± 0.18 |
Fig. 3 shows the segmentation results with A-net trained with all previous weekly data, only week 1 data, and the population-based model. Two representative cases: an average case (#6) and the worst case (#2) are selected as examples. The coronal view of the segmentation is shown on a weekly basis, with the segmentation, ground truth, and VOI plotted in red, yellow, and blue, respectively. A-net trained up to the current week achieved the best performance, and A-net trained using only week 1 data was comparable to the population-based algorithm, highlighting the challenge of accounting for large inter-patient variations in segmentation and the advantage of a patient-specific approach. Patient #2 is an outlier in terms of segmentation performance, which is beyond two standard deviations. The under-segmentation of patient #2 is mainly due to its characteristics, including its relatively small tumor size and the thinly spread- out contours on many slices. As indicated by Fig. 4, patient #2 is the only one whose DICE is under 0.8 when A-net is trained with only one week data. By removing patient #2 from the cohort, the performance increased significantly to 0.85 ± 0.06, 0.83 ± 0.08, 2.3 ± 1.4 mm, and 0.88 ± 0.06 on DICE, precision, RMSD, and sensitivity, respectively.
Fig. 3.
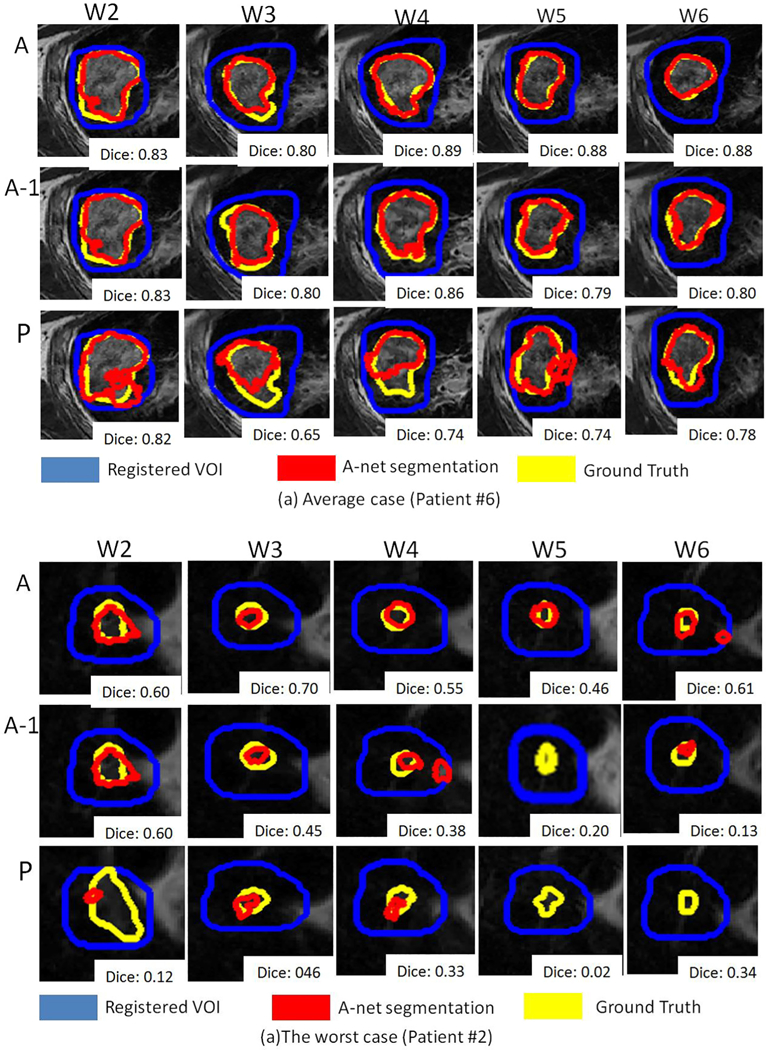
Examples of segmentation via A-net trained with data up to the current week (A), A-net trained with only the first week (A-1), and the population-based model (P).
Fig. 4.

The DICE performance versus the number of training patches with A-net trained by 1 week data.
The performance of contour propagation via deformable registration measured by DICE was 0.79 ± 0.12, as compared to 0.82 ± 0.10 obtained from A-net, with a p-value of 0.07 in a paired t-test. The underperformance was more noticeable among patients with small tumor or large volume changes along the treatment course. Manual editing by a radiation oncologist to correct the algorithm-derived (registration or A-net) contours is about 10–30 min depending on the size of the tumor.
Discussion
Quickly and reliably defining a new target of treatment is the cornerstone of ART. The segmentation process runs through the longitudinal imaging studies, goes beyond the course of radiotherapy, and extends to the follow-up quantitative imaging studies. Therefore, an adaptive model of segmentation gradually incorporating all available patient-specific data is intuitive yet potentially more effective than a population-based one-size-fits-all approach. The concern of limited patient data at the early stage of weekly surveillance can be mitigated by adopting a patch-based CNN architecture. Furthermore, at the first couple weeks of radiotherapy treatment, geometrical changes of tumor, either shrinkage or growth, are not significant, which makes the learning of A-net relatively easy compared with the end of radiotherapy treatment when shrinkage finally matures. As a result, A-net can segment lung tumors with comparable performance as reported using radiomics features [23]. A-net as an auto-segmentation algorithm has tremendous potential for fast contouring for MR-guided adaptive radiotherapy on an MR-linac. The spatial accuracy of A-net was 2.4 mm, which can potentially be used as a guidance to determine the necessary margin in generating the planning target volume for appropriate tumor coverage. Nevertheless, we are continuing our effort to recruit more patients in the longitudinal study. The newly accumulated data would help us build a more robust population-based model. We envision using the population-based model to segment the first weekly MRI scan, a missing component in the current workflow. In addition, the population-based algorithm can be used as a baseline model to obtain preliminary segmentation results, which can be thereafter fed to A-net for fine-tuning and eventually improve overall segmentation accuracy.
Even though the A-net is designed to tolerate certain registration errors, a better registration method would improve performance and especially help the definition and propagation of an accurate VOI. The current rigid registration is completed by human users. As on-going research, a patient-specific registration method is designed using a deep learning model [24,25] to make the system work on a consistent platform with minimal human intervention. Furthermore, the deep learning network can be extended to predict the spatial and temporal distributions of tumor at a later stage of treatment [ 26]. After the entire image processing of the longitudinal study is fully automated, it can be naturally integrated into the quality assurance process. One immediate application is to segment tumors on the images acquired during treatment delivery to assist motion management. After the completion of each treatment fraction, dose delivered to target and critical organs can be accurately calculated using the machine log file, the CBCT/MRI scan, and available motion traces. Dose that will be delivered during the rest of treatment can also be estimated using the predicted tumor distribution. The analysis of the accumulated/predicted dose will serve as the foundation to determine the timing of ART.
In conclusion, the patient-specific adaptive CNN algorithm can be feasibly integrated into the clinical workflow of a longitudinal imaging study. The algorithm produces useful segmentation results for evaluating the response of tumor to radiotherapy and facilitates decision-making in ART.
Acknowledgments
This research was partially supported by the MSK Cancer Center Support Grant/Core Grant (P30 CA008748).
Footnotes
Conflict of Interest
Memorial Sloan-Kettering Cancer Center has a research agreement with Varian Medical Systems.
References
- [1].Tyagi N, Fontenla S, Zelefsky M, Chong-Ton M, Ostergren K, Shah N, et al. Clinical workflow for MR-only simulation and planning in prostate. Radiat Oncol 2017;12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lau S, Lu W, et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One 2017;12: e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys 2017;44:547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qin W, Wu J, Han F, Yuan Y, Zhao W, Ibragimov B, et al. Superpixel-based and boundary-sensitive convolutional neural network for automated liver segmentation. Phys Med Biol 2018;63:095017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ibragimov B, Toesca D, Chang D, Koong A, Xing L. Combining deep learning with anatomical analysis for segmentation of the portal vein for liver SBRT planning. Phys Med Biol 2017;62:8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Long J, Shelhamer E, Darrell T, Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, p. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [7].Ronneberger O, Fischer P, Brox T. In: U-net: Convolutional networks for biomedical image segmentation. Springer; 2015. p. 234–41. [Google Scholar]
- [8].Milletari F, Navab N, Ahmadi S-A, V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 3D Vision (3DV), 2016 Fourth International Conference on, IEEE, 2016, p. 565–571. [Google Scholar]
- [9].Chen H, Qi X, Yu L, Heng P-A, Dcan: Deep contour-aware networks for accurate gland segmentation. In: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, 2016, p. 2487–2496. [Google Scholar]
- [10].Vivanti R, Ephrat A, Joskowicz L, Karaaslan O, Lev-Cohain N, Sosna J, Automatic liver tumor segmentation in follow-up ct studies using convolutional neural networks. In: Proc. Patch-Based Methods in Medical Image Processing Workshop, Vol. 2, 2015. [Google Scholar]
- [11].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH, Patch-based convolutional neural network for whole slide tissue image classification. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, p. 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- [13].Chen L, Wu Y, DSouza AM, Abidin AZ, Wismüller A, Xu C. Mri tumor segmentation with densely connected 3D CNN. Medical Imaging 2018: Image Processing. International Society for Optics and Photonics; 2018. 105741F. [Google Scholar]
- [14].Moeskops P, Wolterink JM, van der Velden BH, Gilhuijs KG, Leiner T, Viergever MA, et al. In: Isgum, deep learning for multi-task medical image segmentation in multiple modalities. Springer; 2016. p. 478–86. [Google Scholar]
- [15].Trebeschi S, van Griethuysen JJ, Lambregts DM, Lahaye MJ, Parmer C, Bakers FC, et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric mr. Sci Rep 2017;7:5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nyúl LG, Udupa JK, et al. , On standardizing the Mr Image intensity scale, image 1081. [DOI] [PubMed] [Google Scholar]
- [17].Simonyan K, Zisserman A, Very deep convolutional networks for large- scale image recognition, arXiv preprint arXiv: 1409.1556. [Google Scholar]
- [18].Szegedy C, Liu W,Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D,Vanhoucke V, Rabinovich A, et al. , Going deeper with convolutions, CVPR, 2015. [Google Scholar]
- [19].He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, p. 770–778. [Google Scholar]
- [20].Vedaldi A, Lenc K, Matconvnet: Convolutional neural networks for matlab. In: Proceedings of the 23rd ACM international conference on Multi-media, ACM, 2015, p. 689–692. [Google Scholar]
- [21].Mahsereci M, Ballers L, Lassner C, Henning P. Early stopping without a validation set. arXiv preprint arXiv: 1703.09580. [Google Scholar]
- [22].Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders I Tsgum MJ, Automatic segmentation of MR brain images with a convolutional neural network, IEEE Transactions on Medical Imaging 35, p. 1252–1261. [DOI] [PubMed] [Google Scholar]
- [23].Owens CA, Peterson CB, Tang C, Koay EJ, Yu W, Mackin DS, et al. Lung tumor segmentation methods: Impact on the uncertainty of radiomics features for non-small cell lung cancer. PloS One. 2018;13(10).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li H, Fan Y, Non-rigid image registration using fully convolutional networks with deep self-supervision, arXiv preprint arXiv: 1709.00799. [Google Scholar]
- [25].Roĥe M-M, Datar M, Heimann T, Sermesant M, Pennec X. In: Svf-net: Learning deformable image registration using shape matching. Springer; 2017. p. 266–74. [Google Scholar]
- [26].Wang C, Hu Y, Rimner A, Tyagi N, Yorker E, Mageras G, Deasy J, Zhang P. Predicting lung tumor shrinkage during radiotherapy seen in a longitudinal MR imaging study via a deep learning algorithm. AAPM 2018 annual meeting. [DOI] [PMC free article] [PubMed] [Google Scholar]


