Abstract
R-loops are prevalent three-stranded non-B DNA structures composed of an RNA:DNA hybrid and a single-strand of DNA. R-loops are implicated in various basic nuclear processes such as class-switch recombination, transcription termination and chromatin patterning. Perturbations in R-loop metabolism have been linked to genomic instability and implicated in human disorders, including cancer. As a consequence, the accurate mapping of these structures has been of increasing interest over recent years. Here, we describe two related immunoprecipitation-based methods to map R-loop structures: basic DRIP-seq (DNA:RNA ImmunoPrecipitation followed by sequencing), an easy, robust, but resolution-limited technique, as well as DRIPc-seq (DNA:RNA Immunoprecipitation followed by cDNA conversion and sequencing), a high-resolution and strand-specific iteration of the method that permits accurate R-loop mapping genome-wide. Briefly, after gentle DNA extraction and restriction digestion with a cocktail of enzymes, R-loop structures are immunoprecipitated with the anti-RNA:DNA hybrid S9.6 antibody. Compared to DRIP-seq in which the immunoprecipitated DNA is directly sequenced, DRIPc-seq permits the recovery of the RNA moiety of R-loops and these RNA strands are subjected to strand-specific RNA-seq analysis. Accurately mapping R-loop distribution in various cell lines and under varied conditions is essential to understanding the formation, roles, and dynamic resolution of these important structures.
Keywords: R-loops, DNA:RNA hybrids, S9.6, DRIP, DRIPc, library, mapping, strand-specific RNA-sequencing
EDITORIAL SUMMARY:
R-loops are DNA:RNA hybrid structures found throughout the genome and relevant to both normal and disease states. DRIPc-seq, which is based on immunoprecipitation with the S9.6 antibody recognizing DNA:RNA hybrids, permits genome-wide mapping of R-loops.
TWEET:
DRIPc-seq accurately maps genome-wide distribution of R-loops following immunoprecipitation with an anti-DNA:RNA hybrid antibody.
COVER TEASER:
Genome-wide mapping of R-loops with DRIPc-seq
Introduction
R-loops are three-stranded nucleic acid structures composed of an RNA:DNA hybrid and a single-stranded DNA loop. These structures form primarily during transcription upon hybridization of the nascent transcript to the template DNA strand 1. Historically, R-loops were discovered as an important intermediate involved in the initiation of DNA replication at the replication origins of bacteriophage, plasmid, and mitochondrial genomes 2-8. Later on, R-loops were shown to form at repeated class switch sequences during the process of immunoglobulin class switch recombination 9-12. It was then recognized that R-loop structures can be extremely long, reaching up to kilobase length 12, and that they form over specific regions owing to the thermodynamic favorability of RNA:DNA versus DNA:DNA base-pairing for specific sequences 13,14. Besides their proposed physiological roles in DNA replication initiation and class switch recombination in B cells, R-loop structures have long been considered to be rare, resulting from accidental entanglements of RNA with DNA during transcription in the nucleus. However, the development of DNA:RNA Immunoprecipitation (DRIP) coupled to high-throughput DNA sequencing (DRIP-seq) allowed the first genome-wide mapping of R-loops in human cells, which showed that those structures are far more prevalent than expected 15. In addition, R-loop-favorable sequence characteristics such as GC skew were shown to be a conserved hallmark of numerous R-loop loci in vertebrate genomes 16. Concurrently, aberrant R-loop formation has been implicated in various processes linked to genomic instability such as hyper recombination, transcription-replication collisions, replication and transcriptional stress 17-20. Not surprisingly, R-loops have also been linked to several human diseases, including fragile X syndrome, ataxia with ocular apraxia type 2 and some cancers 21. As a consequence, R-loop biology during the last decade has been of increasing interest and made the accurate mapping of R-loop an exciting and important challenge to better understand the distribution and function of these structures.
DNA:RNA immunoprecipitation (DRIP) relies on the S9.6 monoclonal antibody which recognizes DNA:RNA hybrids with sub-nanomolar affinity 22. DRIP-seq permits robust genome-wide profiling of R-loop formation 15. While useful, this technique suffers from limited resolution and does not provide information on strand-specificity. To overcome these limitations, we developed a near base-pair resolution and strand-specific method to accurately map R-loops in any cell population called DRIPc-seq 23 (DNA:RNA immunoprecipitation followed by cDNA conversion coupled to high throughput sequencing) (see Figure 1 for a general outline of the method). This method confirmed that R-loops are prevalent and dynamic structures that can occupy collectively up to 3% of mammalian genomes. R-loops are formed co-transcriptionally over conserved genic regions with a predilection for promoter and terminal regions which represent hotspots of R-loop formation. DRIPc-seq maps have allowed the identification of specific in vivo epigenomic signatures of R-loop formation. Compared to position-matched, expression-matched loci devoid of DRIPc-seq signal, R-loop forming promoters are enriched specifically for RNA polymerase pausing, open chromatin (low nucleosome occupancy), and for co-transcriptionally deposited chromatin marks H3K4me1 and H3K36me3 23. Likewise, R-loop forming gene termini show substantially higher RNA polymerase stalling and higher transcription termination efficiencies, suggesting that terminal R-loop formation associates with efficient transcription termination 23,24. Overall, this evidence supports a role for R-loop structures in the regulation of gene expression 1,19,20,23.
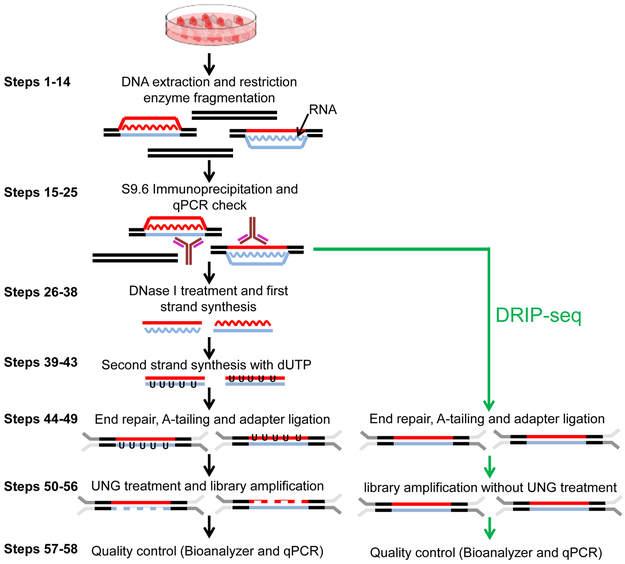
Figure 1∣.
Overview of the DRIPc-seq protocol. After gentle DNA extraction, the DNA is fragmented with restriction enzymes and RNA:DNA hybrids are immunoprecipitated with the S9.6 antibody. The RNA strand of the hybrid is released by DNase I treatment and subjected to reverse transcription with dUTP. Uracil-N-glycosylase (UNG) treatment creates abasic sites in that strand (gapped lines) which ensures that libraries are only built from one strand. The cDNA is then indexed, amplified and sequenced after meeting quality controls. The green arrows represents the DRIP-seq protocol, a simpler but lower resolution alternative method to DRIPc-seq.
Applications of the method
The protocol presented here can easily be applied to perform DRIP-seq instead of the higher resolution, strand-specific, DRIPc-seq. DRIP-seq is technically less demanding, requires less starting material, and is less time-consuming, but still provides useful and robust information on R-loop distribution in a genome. We typically recommend that users first successfully perform DRIP-seq before attempting the DRIPc-seq protocol. The major difference between both methods pertains to the construction of high-throughput sequencing libraries. An alternate library construction step is provided for DRIP-seq. Other DRIP protocols have been published recently and could be useful references for users 25,26.
R-loop mapping by DRIP-seq and DRIPc-seq enables users to measure the steady-state distribution of R-loop structures in any genome of interest. More importantly, these methods allow understanding global changes in R-loop distribution or dynamics caused by genetic perturbations (gene knockouts or knockdowns) or by chemical treatments (drugs, hormones). Published examples include the response of human breast cancer cells to transcription induction by estrogen 27 or the consequences of silencing DNA topoisomerase 1 in human HEK293 cells 28. These techniques can be applied to any cell type for which sufficient starting material can be obtained. R-loop profiles have been successfully generated using DRIP-seq and or DRIPc-seq in murine cells 23 and in Schizzosaccharomyces pombe 29. DRIP methods, when followed by quantitative PCR (qPCR) instead of high-throughput sequencing, are useful to estimate the quantity of R-loops at specific loci in a cell population. This is best expressed as the fraction of the input DNA that was immunoprecipitated (% input), with no further normalization, as is commonly used in chromatin immunoprecipitation (ChIP) assays (see Box 1). In all cases, and in particular when changes in R-loop patterns under perturbed conditions are of interest, it is essential to profile gene expression globally in parallel to performing DRIP analysis. Since R-loops form co-transcriptionally, true changes in R-loop formation or resolution must be disentangled from simple changes caused by transcriptional effects.
Box1: qPCR analysis ● TIMINC 2 hours.
Procedure
A. The first qPCR analysis will verify that the DRIP procedure was effective (step 25). This is a critical validation test.
From the 50 μL of immunoprepitated DNA (step 24), take 2 μL and add 8 μL of water. As an input, use any one of the 5 tubes you saved in step 15.
For each PCR reaction, prepare the following mix:
Plan on testing three R-loop positive loci (for human cells RPL13A, TFPT, CALMS) and 2 R-loop negative loci (for human cells EGR1neg, SNRPNneg) – see Table 1.
Run the following program on a Real-Time PCR machine:
| Cycle number |
Duration | Temperature |
| 1 | 30 sec | 95°C |
| 2-39 | 10 sec | 95°C |
| 30 sec | 60°C | |
| 40 | Melting curve | |
▲CRITICAL STEP: The SsoAdvanced SYBR Green mix works with a two-step PCR cycle where the extension and annealing steps are combined but depending on the master mix used, you might have to change the program to a more classic three-step PCR cycle.
qPCR results are analyzed two ways. First, the raw efficiency of the DRIP can be expressed as a percentage of input DNA precipitated. Second, the specificity of the DRIP can be measured as a ratio of the recovery efficiency for a positive locus over a negative locus. A successful DRIP will show both high efficiency and high specificity.
To calculate the percentage of input, apply the following formula for each locus:
% input = 100*2^(Ct Input(corrected) - Ct DRIPedDNA), where Ct input(corrected) = (Ct Input - log2(10)) (we subtract log2(10) because the input represents 1/10th of the IPed DNA, see step 15)
To calculate fold enrichment, use one of the negative loci as a reference and apply the following formula:
Fold-enrichment = [2^(Ct Input (Positive Locus (corrected)) - Ct DRIPedDNA (Positive Locus)] / [2^(Ct (Input Negative Locus (corrected)) - Ct DRIPedDNA (Negative Locus)].
As shown in the figure, R-loop positive loci are typically recovered with an efficiency ranging from 1 to 15% of input depending on the locus (left), whereas negative loci typically show values lower than 0.1% 23. This recovery is an estimate of the percentage of cells carrying an R-loop structure at the time of lysis for a given locus. Thus the dynamic range of the method exceeds two orders of magnitude. Fold-enrichments for positive loci range from 20 to over 300-fold (right).
B. Another validation is performed after second strand synthesis (step 41). At this step, you will only be able to check the fold enrichment.
From the 42.5 μL in step 41, take 2 μL and add 8 μL of water. Then, proceed as in part A and calculate the fold enrichment. Use the same input as the one you used for step 25.
▲CRITICAL STEP The DNase I treatment will degrade most of the background DNA. As a result, negative loci become mostly undetectable (Ct values for negative loci should be higher than 36 cycles, if detectable at all). A lower Ct value could mean that the DNase I treatment was incomplete; this could in turn result in a higher background after sequencing.
C. A final qPCR validation is performed after the library amplification (step 56). From the 13 μL in step 56, take 1 μL and add 9 μL of water. Proceed as in part A and calculate the fold enrichment. Use the same input as the one you used for step 25.

Limitations of the method
In the DRIP-based methods described here, restriction enzyme cocktails are chosen to fragment the genome because they offer the gentlest fractionation, preserving fragile R-loops and ensuring the highest possible yields (10% of input is routinely recovered over strong positive loci). In the DRIP-seq method, where restriction fragments are directly sequenced after immunoprecipitation, this leads to lower resolution and as noted 30 can bias the signal towards larger fragments. This can be remedied in part by combining data produced with different restriction cocktails 31. In the context of DRIPc-seq, this issue is of little consequence since the DNA is entirely degraded and only the RNA moiety of R-loops is recovered and converted into strand-specific DNA sequencing libraries. A thorough exploration of parameters relevant to DRIP has been published 30.
DRIP-based approaches are heavily reliant on the S9.6 monoclonal antibody. This antibody shows sub-nanomolar affinity for RNA:DNA hybrids 22 and little to no affinity for dsDNA (F.C. unpublished observations), highlighting a clear strength of the approach. Reports that the antibody possesses binding preferences for short specific sequence motifs 32 are only a concern if the desired targets of the analysis are short R-loops (<50 bp). In agreement with evidence that R-loops are large structures 10,12,15 composed of a diverse multitude of potential epitopes, we find that the sequences captured in DRIPc-seq do not show any measurable bias for or against any of the more or less preferred binding motifs, respectively (Supplementary Figure 1).
S9.6 however, possesses significant residual affinity for double-stranded RNA (dsRNA), particularly AU-rich dsRNA 22. This binding can in some instances be problematic, especially when sequencing libraries are constructed from material derived originally from RNA, as in DRIPc-seq 29. Fortunately, this issue is easy to diagnose and to remedy. The telltale sign that dsRNA was immunoprecipitated by S9.6 and made into sequencing libraries is that the resulting signal is not stranded (i.e. signal maps to both strands) and is not responsive to pre-treatment by Ribonuclease H, an enzyme that exclusively degrades RNA in the context of RNA:DNA hybrids in vitro 33. This was observed in recent R-loop profiling studies in fission yeast 29. To remedy this problem, extracted nucleic acids were treated with RNase III, which specifically cleaves dsRNA, prior to DRIP. As expected, the signal became stranded and co-directional with transcription and was also sensitive to RNase H treatment in vivo and in vitro 29.
In mammalian systems, this issue has never been encountered and the DRIPc-seq signal is overwhelmingly strand-specific and so exquisitely sensitive to RNase H pre-treatment that sequencing libraries could not be built from samples treated with RNase H prior to immunoprecipitation 23. In DRIP-seq, where sequencing libraries are built from material derived originally from DNA, this issue matters far less as any immunoprecipitated RNA species will not be sequenced. DRIP-seq signal, like DRIPc-seq signal is highly sensitive to RNase H pre-treatment and results from both procedures agree well with each other 15,23,31 (Figure 2). It has been suggested that RNase III pre-treatment should be performed prior to DRIP to alleviate any concern about dsRNA interfering with the immunoprecipitation or the resulting sequencing signal 34. This is likely a useful suggestion but caution is advised as we observed that some RNase III preparations can display non-negligible activity towards RNA:DNA hybrids depending on buffers (John Smolka and F.C., unpublished observations). Thus, the specificity of the RNase III preparations must be thoroughly checked first using oligonucleotide substrates. Likewise, it was suggested that pre-treatment with the broad spectrum RNase A enzyme was advisable to allay any concern that RNA could interfere with the efficiency of DRIP 35. We and others have observed that RNase A treatment can lead to profound loss of signal depending on incubation time, concentration, and buffer conditions 30 which isn’t surprising given that RNase A possesses significant RNase H activity36. RNase A treatment therefore has to be carefully calibrated and is not-well suited for a standard quality control. Regardless, we note that the possibility that single-stranded RNA species interfere with DRIP efficiency was not found to be of concern in this protocol as judged from highly reproducible DRIP yields over hundreds of DRIP experiments. In agreement, comparison of DRIP efficiency by qPCR with and without RNase A treatment under controlled conditions revealed no significant differences, and DRIPc-seq profiles after RNase A treatment were very similar to those obtained without treatment23.
Figure 2∣.
Mapping DNA:RNA hybrids after DRIPc-seq. A. A typical DRIPc-seq outcome for human NTERA-2 cells is shown over a representative screenshot spanning a 200 kb region. Reads were processed as per the pipeline in Box 2 and mapped to the human genome (hg19 reference). Each track represents DRIPc-seq signal broken down per strand with red corresponding to + strand genes and blue corresponding to − strand genes. Two independent replicates are shown. DRIP-seq data over the same region is displayed in green. DRIP-seq data after RNase H pre-treatment is shown below (teal). B. (Left) XY correlation plot between two DRIPc-seq replicates showing strong reproducibility (r, Pearson correlation). (Right) XY correlation plot between DRIP-seq pre-treated or not with RNase H. RNase H pre-treatment abrogates the signal genome-wide.
A related, but distinct potential limitation concerns the possibility that free portions of the RNA strand involved in R-loop formation may be immunoprecipitated along with R-loops, thereby producing false positive “trailing” peaks in DRIPc-seq. This possibility may again be mitigated by the use of ribonuclease treatment specific towards ssRNA such as RNase T1. Orthogonal approaches such as non-denaturing bisulfite probing 12, that query R-loops through their single-stranded looped out DNA can also be useful to address this possibility.
DRIP-based methods described here require a relatively large number of cells to ensure the recovery of sufficient material for library preparation. DRIP-seq can be performed with a starting material of 12 to 15 μg of DNA, whereas DRIPc-seq requires 30 to 40 μg of DNA, corresponding to a minimum of 5 million cells. Thus, a limitation of the method is that it is not compatible with samples with low cell counts. In addition, the method only provides a population-average snapshot of R-loop formation; the positions and lengths of individual R-loop structures cannot be deduced from the data. The method is also limited to measuring RNA:DNA hybrids of sufficient length, typically above ~70 bp, as the library construction steps impose size constraints. The protocol presented here therefore doesn’t lend itself to the study of short R-loops, which, on a positive note, ensures that short RNA:DNA hybrids such as Okazaki fragments do not contribute to DRIPc-seq signal.
Finally, the method requires the extraction of nucleic acids and the fractionation of the DNA by enzymatic digestion. DRIP-based methods are therefore not suited to query in vivo R-loop formation and it is formally possible that some R-loops, particularly short, unstable structures, may fall apart during the DNA extraction and fragmentation process. Thus, R-loop formation could be under-estimated. The opposite concern, namely that R-loops may form de novo during the process of DNA extraction is highly unlikely. First, making artificial R-loops from purified plasmid DNA and RNA requires heating the DNA to its denaturing point in 60-70% formamide37,38. Any deviation from the optimal temperature saw rates of R-loop formation plummet. Thus, the energy barrier to R-loop formation from DNA and RNA outside of the immediate vicinity of the transcription bubble is extremely high. In agreement, incubation of complementary RNA with supercoiled plasmid DNA does not result in any R-loops (39 and F.C., unpublished observations). This agrees with clear and consistent evidence that in vitro, R-loops form in cis during transcription 40. In a highly complex mixture of genomic DNA and RNA, promoting such RNA strand invasion would in addition require a homology search process and energy to melt the duplex DNA over hundreds of base-pairs. The likelihood that such a process could explain the highly robust and reproducible signals seen in DRIP-based approaches is extremely low.
Advantages of DRIPc-seq and comparison to other methods
DRIPc-seq offers reproducible, high-resolution, strand-specific R-loop maps on a genome-wide scale. The signal is exquisitely sensitive to RNase H pre-treatment which establishes that the material sequenced derives from RNA:DNA hybrid containing species, presumably R-loops. The DRIP procedure at the heart of the method is highly reproducible and offers a greater than 100-fold dynamic range in human cells between positive and negative loci 23 (Box 1). Other DRIP-based methods aimed at high-resolution, strand-specific mapping have been described, including S1-DRIP in yeast 41 and ssDRIP-seq in Arabidopsis thaliana 42. The relative performance of these three methods has not been systematically compared.
Two additional methods aimed at providing genome-wide, high-resolution R-loop maps have recently been published. bisDRIP-seq 43, exploits the fact that the single-stranded DNA on the looped out DNA strand of an R-loop is sensitive to non-denaturing bisulfite treatment 12. Following bisulfite treatment during lysis, a DRIP step was performed to enrich for R-loops, followed by library construction and high-throughput sequencing, scoring for strand-specific patterns of C to T conversions 43. This method, like other DRIP-based approaches, relies on successful immunoprecipitation of R-loops using the S9.6 antibody. In addition, and unlike other methods, it requires the ssDNA strand of an R-loop to survive the bisulfite treatment and immunoprecipitation steps intact so it can be amplified and sequenced. Any nick introduced in the displaced strand during the bisDRIP-seq procedure will render that strand un-amplifiable and therefore undetectable. DRIPc-seq, by contrast, targets the RNA moiety of the RNA:DNA hybrid component of R-loops and evidence shows that RNA: DNA hybrids derived from R-loops can be robustly recovered even after complete loss of the displaced strand 41. A preliminary comparison of bisDRIP-seq and DRIPc-seq revealed some agreement between both methods in calling promoter R-loops over highly active, GC-skewed, CpG island promoters 43. However, results from bisDRIP-seq were interpreted to suggest that R-loops at promoters were constrained to the first exon of genes, which was not observed in DRIPc-seq. Instead, promoter R-loops in DRIPc-seq most often peak downstream of the first exon, where GC skew is maximal 16, and extend for hundreds of base-pairs 23. More importantly, bisDRIP-seq could seldom identify thousands of gene body and terminal R-loop peaks that featured prominently among the highest R-loop hotspots detected by DRIPc-seq. The reasons for such discrepancies are not clear yet but point to differential sensitivities of the methods. Further work will be needed to understand the source of these discrepancies.
A second recent method termed R-ChIP-seq 44 used a ChIP-seq approach to map the genomic binding sites of a catalytically inactive RNase H1 protein (dRNASEH1) expressed in human cells. The underlying assumption behind the method is that dRNase H1 will be able to bind sites wherever RNA:DNA hybrids, including R-loops, are formed. R-ChIP-seq therefore uses RNase H1 binding as a proxy for R-loop locations in the genome, in contrast to S9.6-based approaches which directly query R-loops through DNA:RNA hybrid immunoprecipitation. R-ChIP-seq, which like DRIPc-seq is high-resolution and strand-specific, showed that dRNASEH1 predominantly bound to promoter regions, overlapping with the site of RNA Polymerase II promoter-proximal pause sites 44. A mechanistic connection between transcriptional pausing and dRNASEH1 recruitment was further revealed. R-ChIP-seq did not identify thousands of R-loop hotspots over gene bodies and terminal regions that are consistently highlighted by DRIPc-seq. One possibility to account for the discrepancies is that R-ChIP-seq possesses lower sensitivity than DRIP-based methods. Indeed, DRIP permits 10-fold higher recovery yields over positive loci compared to R-ChIP 23,44. Alternatively, it is possible that dRNASEH1 is not free to bind to all R-loops or hybrids owing to as-yet-unknown accessibility or targeting mechanisms. Further work will be required to fully address these discrepancies and to flesh out the biology of the RNase H1 protein. We note that contrary to DRIP-based approaches which can be used for many cell types including primary cells and hard to transfect cells, R-ChIP-seq requires the creation of stable cell lines expressing the dRNaseH1 protein. Likewise, because R-ChIP-seq uses dRNASEH1 to capture RNA:DNA hybrids, the sensitivity of R-ChIP-seq signal to endogenous or exogenous RNase H addition cannot be evaluated. However, in contrast to DRIP-based approaches which require DNA extraction, RChIP-seq permits the capture of dRNASEH1 targets in cells upon rapid protein-DNA crosslinking.
Experimental design
The DRIPc-seq protocol is composed of six main parts and takes roughly five days to complete. It starts with cell samples and ends with the sequencing libraries. A flowchart of the protocol is displayed on Figure 1.
Cell harvest and lysis (steps 1-4).
This protocol begins with growing cells to a confluency of 75 to 80%. As R-loops are mostly co-transcriptional structures, it is important that your cells are not too confluent since some cells show contact growth inhibition. We recommend starting with 7 to 8 million cells and to plan for a minimal of two biological replicates with proper controls (RNase H-treated) to achieve meaningful statistical analysis. The protocol can be performed on any cells, tissues, or organoids provided they can be dissociated to single cells prior to DNA extraction.
DNA extraction and fragmentation (steps 5-14).
After overnight lysis, the DNA is extracted through a classic salt/ethanol precipitation. The use of phase lock gel tubes during this step allows us to clean up the DNA faster and to avoid additional centrifugation and manipulation steps. One of the key goals of early steps is to preserve otherwise fragile R-loop structures. It is well appreciated that mechanical shearing can lead to fragmentation of the looped-out ssDNA strand 41. Likewise, it is possible that the RNA strand spontaneously dissociates. Once genomic DNA is rehydrated in TE buffer, fragmentation is performed in the gentlest possible way using a cocktail of restriction enzymes. The bias introduced by the non-random enzymatic fragmentation is not a concern for DRIPc-seq as the DNA moiety of R-loops will ultimately be digested by DNase I to release the RNA strand that will be sequenced after reverse-transcription. During restriction fragmentation, we highly recommend adding RNase H to one of your tubes to generate a specificity control. The ability to effectively remove the epitope for the antibody to be used in immunoprecipitation is virtually unique to DRIP and provides a powerful safeguard against non-specific binding. RNase H pre-treatment should lead to complete loss of S9.6 signal as measured post immunoprecipitation by qPCR during quality controls steps (Steps 57-58).
S9.6 immunoprecipitation (Step 15-24).
After fragmentation, RNA:DNA hybrids including R-loops are immunoprecipitated by S9.6 and the complexes are bound to agarose beads (magnetic beads could be used at this step instead of agarose beads without significant changes). The immunoprecipitated material is then eluted, cleaned-up, and resuspended in RNase-free water to prevent RNase contamination in the following parts of the protocol. We highly recommend performing a qPCR validation step at this step to check the efficiency of the immunoprecipitation. It is important that test loci fall within the indicated range of recovery and fold enrichment (Box 1) to ensure the construction of good DRIPc-seq libraries. A low recovery will inevitably lead to poor libraries and poor signal-to-noise ratios post-sequencing.
DNase I treatment and RNA reverse transcription (Step 26-43).
The immunoprecipitated material is next incubated with DNase I to digest away the DNA portion of R-loops and to remove any potential non-specific DNA that may have carried through the DRIP. This step dramatically reduces the background thereby increasing significantly the sensitivity of DRIPc-seq compared to DRIP. After precipitation, the RNA is subjected to first strand synthesis and clean-up using AMPure beads. Since the quantity of RNA at this step is very low, we prefer using AMPure beads rather than standard clean-up columns due to the higher recovery achieved from AMPure beads. The second strand synthesis is next performed in the presence of dUTP instead of dTTP to allow the construction of a strand-specific library. After cleaning the cDNA with AMPure beads, we recommend performing another qPCR validation to evaluate the fold enrichments after second strand synthesis, before starting library construction. As a final step, the cDNAs are sonicated. This is to ensure that R-loops, which are heterogeneous in length and range from 100 bp to 1-2 kilobases, are fragmented to an average length of 200-300 bp. This step is particularly useful if case single-end 100 bp sequencing is performed. The sonication could be adapted to obtain longer fragments in the event a different type of sequencing such as paired end sequencing is desired.
Strand specific library construction (Step 44-56).
The sequencing library is built after the sonication of the cDNA following end repair, dATP-tailing and adaptor ligation. The library is indexed during the adaptor ligation step. Prior to the last PCR amplification, the uracil-containing DNA strand of the library is eliminated with Uracil N-Glycosylase thereby providing strand specificity information regarding the RNA engaged in R-loop formation during sequencing. The library is amplified from 10 to 15 cycles depending on starting material. A more accurate number of cycles can be determined by qPCR, if necessary. Finally the library is cleaned up with AMPure beads using two different ratios to select fragments ranging from 200 to 500 bp.
Quality control (Step 57-58).
The last quality control check on an Agilent BioAnalyzer is essential to ensure good sequencing library quality. A good library should have a concentration over 1 ng/μl and a distribution of sizes between 200 and 500 bp over a single broad peak. A final qPCR validation will allow you to confirm fold-enrichments between positive and negative loci, which in successful cases can reach over a thousand fold for a hotspot like RPL13A. If the sample meets the quality control, the library will be sequenced on a HiSeq Illumina sequencer.
DRIP-seq and DRIPc-seq in non-human cells.
The protocol can in theory be adapted to any cells and organisms. So far, most work has focused on a variety of human and murine cells. The protocol used to perform R-loop mapping in mouse cells strictly follows the protocol described here, with the only difference in the sonication during step 43 (see the associated CRITICAL STEP) and in the primer sets used to validate the immunoprecipitation in step 25. The sequence of primers used for mouse DRIPc-seq can be found in Table 1.
Table 1.
Primers used for qPCR validation (human and mouse). All sequences are listed in the 5’ to 3’ direction. Primers are resuspended in TE at a 100 μM concentration and then further diluted in molecular biology grade water at a working concentration of 10 μM.
| Species | Type | Name | Sequence |
|---|---|---|---|
| human | positive control locus | RPL13A F | AGGTGCCTTGCTCACAGAGT |
| human | positive control locus | RPL13A R | GGTTGCATTGCCCTCATTAC |
| human | positive control locus | TFPT F | TCTGGGAGTCCAAGCAGACT |
| human | positive control locus | TFPT R | AAGGAGCCACTGAAGGGTTT |
| human | positive control locus | CALM3F | GAGGAATTGTGGCGTTGACT |
| human | positive control locus | CALM3R | AGAGTGGCCAAATGAGCAGT |
| human | negative control locus | EGR1neg F | GAACGTTCAGCCTCGTTCTC |
| human | negative control locus | EGR1neg R | GGAAGGTGGAAGGAAACACA |
| human | negative control locus | SNRPNneg F | GCCAAATGAGTGAGGATGGT |
| human | negative control locus | SNRPNneg R | TCCTCTCTGCCTGACTCCAT |
| mouse | positive control locus | ACTIN F | TGCTCCCCGGGCTGTATT |
| mouse | positive control locus | ACTIN R | ACATAGGAGTCCTTCTGACCCATT |
| mouse | positive control locus | EIF5A F | CCACTTACATCTGGCTGGAC |
| mouse | positive control locus | EIF5A R | CCTTGGGTCTCACTCATCC |
| mouse | negative control locus | Chr1neg F | TTCCAACAAAGCAGCAAATG |
| mouse | negative control locus | Chr1neg R | GGGTCACCAGACCTGTTTTT |
Materials
Biological Materials
REAGENTS
Cell culture medium (DMEM Gibco, cat. No. 11960-044; 10% FBS Foundation B Gemini, cat. No. 900-208 (50 ml for 500 ml of DMEM); 1X L-Glutamine 200 mM Gibco, cat. No. 25030-081; 1X Pen/Strep Gibco, cat. No. 15140-122) ▲CRITICAL: The medium may need to be adapted to the cell line of interest.
0.05% (wt/vol) Trypsin-EDTA 1X (Gibco, cat. No. 25300-054)
DPBS 1X (Gibco, cat. No. 14190-144)
S9.6 Antibody at a concentration of 1 mg/mL (Kerafast, cat. No. ENH001 or Millipore/Sigma, cat. No. MABE1095) CRITICAL: The S9.6 antibody can also be purified directly from the monoclonal hybridoma (ATCC® HB-8730™) according to standard protocols. Initial comparisons between commercial aliquots and purified stocks did not reveal significant differences. For all subsequent experiments, a purified stock of S9.6 obtained from Antibodies Inc. (Davis, CA) was used.
Tris base (1 M, Sigma-Aldrich, cat. No. EC 201-064-4)
EDTA pH 8 (0.5 M, Sigma-Aldrich, cat. No. E4884-500g)
Sodium Acetate (NaOAc) 3 M pH 5.2 (Sigma-Aldrich, cat. No. S7899-100ML)
SDS 20% (Sigma-Aldrich, cat. No. 05030-500ML-F)
Proteinase K 20 mg/mL (Roche, cat. No. 03115828001)
Spermidine 0.1 M (Sigma-Aldrich, cat. No. 05292-1ML-F)
BSA 100X (New England BioLabs, cat. No. B9001)
Phenol/Chloroform Isoamyl alcohol 25:24:1 (Affymetrix, cat. No. 75831-400ML)
Sodium Chloride (NaCl) (5 M, MilliporeSigma, cat. No. SX0420-5)
Agarose A/G beads (ThermoFisher Scientific, cat. No. 20421)
Ribonuclease A (RNase A) (10 mg/mL, Sigma-Aldrich, cat. No. 4875)
Ribonuclease H (RNase H) (NEB, cat. No. M0297S) CRITICAL: commercial RNase H preparations are sometimes too dilute to completely digest genomic R-loops and their activity tends to decrease with time. Human RNase H2 can be purified with relative ease 45 and the corresponding preparations typically possess higher specific activities. Here, we use purified RNase H2 stock (0.7 mg/ml) but no difference is observed when using fresh commercial RNase H.
DEPC-treated water (ThermoFisher Scientific, cat. No. AM9915G)
DNase I and DNase I 10X buffer (New England BioLabs, cat. No. M0303S)
RNAZap (ThermoFisher Scientific, cat. No. AM9780)
iScript reverse transcription supermix (BioRad, cat. No. 1708840)
SsoAdvanced Universal SYBR Green supermix (BioRad, cat. No. 1725272)
Agencourt AMPure XP beads (Beckman Coulter, cat. No. A63881) ▲CRITICAL: AM Pure XP beads are stored at 4°C but must be brought to room temperature before use.
DNA polymerase I (New England BioLabs, cat. No. M0209)
dNTP solution mix 10 mM (New England BioLabs, cat. No. N0447S)
dNTP solution set 100 mM (New England BioLabs, cat. No. N0446S)
dATP solution 100 mM (New England BioLabs, cat. No. N0440S)
dUTP solution 100 mM (ThermoFisher Scientific, cat. No. R0133)
ATP solution 10 mM (New England BioLabs, cat. No. P0756S)
Nuclease free water (New England BioLabs, cat. No. B1500S)
E. coli DNA ligase (New England BioLabs, cat. No. M0205S)
ß-Nicotinamide adenine dinucleotide sodium salt (16 mM, Sigma-Aldrich, cat. No. N0632)
Glycogen (ThermoFisher Scientific, cat. No. R0561)
NEBNext End repair module (New England BioLabs, cat. No. E6050)
Klenow fragment (3’ to 5’ exo-) (New England BioLabs, cat. No. M0212S)
Quick Ligation Kit (New England BioLabs, cat. No. M2200S)
AmpErase Uracil N-glycosylase (ThermoFisher Scientific, cat. No. N8080096)
Phusion Flash High-Fidelity PCR master mix (ThermoFisher Scientific, cat. No. F548S)
Ethanol absolute 200 proof (VWR, cat. No. V1001)
Triton X-100 (ThermoFisher Scientific, cat. No. BP151-500)
Potassium Chloride (ThermoFisher Scientific, cat. No. P217-500)
Magnesium Chloride Hexahydrate (ThermoFisher Scientific, cat. No. M35-500)
Sodium Phospate Dibasic Anhydrous (ThermoFisher Scientific, cat. No. S374-500)
Sodium Phosphate Monobasic (ThermoFisher Scientific, cat. No. S397-500)
Restriction enzymes (New England BioLabs): BsrGI (cat. No. R0575S), EcoRI (cat. No. R0101S), HindIII (cat. No. R0104S), SspI (cat. No. R0132S), XbaI (cat. No. R0145S) ▲CRITICAL: This restriction enzyme cocktail was designed to digest the human genome in fragments of ~3-5 kb on average. Other cocktails have been used to increase the resolution of DRIP-seq 31. In principle any enzyme cocktail can be used provided it cuts the genome in fragments of 2-5 kb or less. Make sure that you adjust buffer and enzyme concentrations if you change one or several restriction enzymes. You also might have to design new primers for PCR amplification to check DRIP efficiency as a restriction sites could fall between the primers presented here.
Index adapters (Illumina). The adapters used for the libraries are the TruSeq Single indexes. For details about the sequence, see https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/experiment-design/illumina-adapter-sequences-1000000002694-07.pdf
-
PCR primers for library amplification (Illumina). The libraries are amplified using the PCR primer 1.0 P5
(5’ AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA 3’) and the PCR primer 2.0 P7 (5’ CAAGCAGAAGACGGCATACGAGAT 3’)
Oligonucleotides (Invitrogen, see Table 1 for sequences).
EQUIPMENT
15 mL tube High density Maxtract phase lock gel (Qiagen, cat. No. 129065)
2 mL tube phase lock gel light (VWR, cat. No. 10847-800)
Forma Series II 3110 water-jacketed CO2 incubator (ThermoFisher Scientific, cat. No. 3110). The incubator is set up for 37°C and 5% of CO2.
Barnstead Nanopure (ThermoFisher Scientific, cat. No. D11971)
Fisherbrand Mini-tube rotator (Fisher Scientific, cat. No. 05-450-127)
UVP HL-2000 HybriLinker oven (Fisher Scientific, cat. No. UVP95003101)
Fisherbrand Isotemp Digital Dry bath (Fisher Scientific cat. No. 88-860-022)
Magnetic rack Dynamag-2 magnet (ThermoFisher Scientific, cat. No. 12321D)
SimpliNano Spectrophotometer (Genesee Scientific cat. No. 85-544)
C1000 Touch Thermal cycler (BioRad, cat. No. 1851148)
CFX96 Touch Real-Time PCR Detection System (BioRad, cat. No. 1855195)
Bioruptor Plus sonication (Diagenode, cat. No. B01020001)
Bioanalyzer 2100 (Agilent, cat. No. G2939BA)
Sorvall ST16 Centrifuge (Fisher Scientific, cat. No. 75-410-885)
Eppendorf 5424R Centrifuge (Genesee Scientific, cat. No. 86-370A)
BioRad mini centrifuge (BioRad, cat. No. 1660603)
REAGENT SETUP
TE buffer. TE buffer is 10 mM Tris-HCl pH 8 and 1 mM EDTA. It can be stored at room temperature for up to a month.
1M Na-Phosphate pH 7 is made by mixing 39 mL of 2 M monobasic sodium phosphate, 61 mL of 2 M dibasic sodium phosphate and 100 mL of nanopure water. CRITICAL: These solutions tend to crystalize - warm up if needed to help resolubilize. The 1M Na-Phosphate pH 7 solution can be stored indefinitely at room temperature.
10x DRIP Binding buffer is made by mixing 100 mM NaPO4 pH 7.0, 1.4 M NaCl and 0.5% (vol/vol) Triton X-100. CRITICAL: Filter the solution with a 0.22 μm filter. The 1X binding buffer consists of the 10x binding buffer diluted 10 times in TE buffer. These solutions can be stored at room temperature for up to a month.
DRIP Elution buffer is made of 50 mM Tris pH 8.0, 10 mM EDTA pH 8.0 and 0.5% SDS (vol/vol). It can be stored at room temperature for up to a year.
RNase-free reagents: from step 24 to 31, you will need RNase-free 1 M Tris-HCl pH 8, 0.5 M EDTA and 3 M NaOAc. To ensure the quality of these reagents, dilute the respective powder in DEPC-treated water and make sure that the glassware is adequately clean. Autoclave your reagents once made. Always handle these reagents with gloves and pipette using filter tips.
5X Second Strand Synthesis buffer is made of 200 mM Tris-HCl pH 7, 22 mM MgCl2 and 425 mM KCl as a 5x stock. It can be stored at room temperature for up to a year.
dNTP mix. Make a 10 mM solution stock by mixing 10 μL each of 100 mM stocks for dATP, dCTP, dGTP, and dUTP with 60 μL of nuclease free water. CRITICAL This dNTP mix is only used during the second strand synthesis step (step 39). For all other steps, a pre-made dNTP solution is used (NEB, cat. No. N0446S)
80% (vol/vol) ethanol is prepared by diluting absolute ethanol (4 parts) with molecular biology grade water (1 part). CRITICAL: Always prepare fresh the day of DNA purification with AMPure beads.
Ice cold 100% and 80% ethanol Cool down absolute ethanol or 80% ethanol (vol/vol) in a freezer the day before using. ! CAUTION Ethanol is flammable and needs to be stored in a flammable proof freezer.
PROCEDURE
CRITICAL: All centrifugation steps are performed at room temperature (18-23°C) unless otherwise specified.
Cell harvest and lysis ● TIMING 14-15 hours
1∣ Culture the NTERA-2 cells to 75-80% confluency in a 10 cm dish. Remove the culture medium and wash the cells once with warm DBPS 1x. Aspirate the DPBS and add 1.5 mL of 0.05% Trypsin-EDTA 1X for 2 minutes at 37°C to dissociate the cells from the dish. Add 5 mL of warm complete media to stop the reaction and, after pipetting well, transfer the content in a 15 mL tube and pellet the cells gently at 1,000 rpm for 3 minutes.
▲CRITICAL STEP For NTERA-2 cells, a 75% confluent dish corresponds roughly to 5 million cells. For peace of mind, we advise to start with 7 to 8 million cells. Count cells and pool multiple dishes as needed when working with other cell types.
2∣ Wash the cells by resuspending in 5 mL of 1X DPBS. Pellet the cells gently at 1,000 rpm for 3 minutes.
3∣ Resuspend the cells in 1.6 mL of TE buffer. Add 50 μL of SDS 20% (wt/vol) and 5 μL of Proteinase K 20 mg/mL. Invert gently the tube 5-6 times until solution become viscous.
▲CRITICAL STEP: Make sure to fully resuspend the cells, avoiding any clumps, before adding SDS and proteinase K. An incomplete resuspension will lower the extraction efficiency.
▲CRITICAL STEP: After adding SDS, do not try to pipet the solution, only mix by gently inverting.
4∣ Incubate at 37°C overnight (12-14h).
DNA extraction and digest ● TIMING 24 hours
5∣ Spin a 15 mL high density Maxtract phase lock gel tube for 1 minute at 1,500 g to pellet the gel.
6∣ Pour your DNA lysate directly into the phase lock gel tube and add 1 volume (1.6 mL) of Phenol/Chloroform Isoamyl alcohol 25:24:1. Invert gently 4-5 times and spin down at 1,500 g for 5 minutes.
▲CRITICAL STEP: After this step, the supernatant should be clear. If not, spin 5 more minutes.
7∣ Add 1/10 volume 3 M NaOAc pH 5.2 (160 μL) and 2.5 volumes 100 % Ethanol (4 mL) to a new 15 mL tube. Pour in the DNA (top aqueous phase) from phase lock gel tube and mix the DNA and ethanol gently by inverting the tube until DNA fully precipitates (visible by eye as a white precipitate). Spool the DNA out of the mixture with cut tips and transfer to a clean 2 mL tube.
▲CRITICAL STEP: It is essential that the DNA is fully precipitated; any unprecipitated DNA will result in poor digestion efficiency. This step can take up to 10 minutes depending on the amount of starting material. Fully precipitated DNA is white; no translucent portions should be visible.
▲CRITICAL STEP: Do not centrifuge to pellet the DNA, it will result in higher loads of RNA contamination, which could in part titrate the S9.6 antibody.
▲CRITICAL STEP: As most of the tips are now graduated, we advise to cut a 1000 μL tip at the 250 μL line for spooling the DNA.
8∣ Wash the DNA by adding 1.5 mL of ethanol 80%, invert gently the tube 2 or 3 times and let it stand for 10 minutes. Carefully discard the supernatant by avoiding pipetting the DNA. Repeat this step twice. Carefully remove as much ethanol as possible by pipetting after last wash. CRITICAL: Do not centrifuge between the washes.
9∣ Allow the DNA to air dry completely while inverting the tube (may take up to an hour according to the amount of DNA) and add 125 μL of TE directly on the DNA pellet. Keep on ice for an hour. Gently resuspend DNA by pipetting two or three times with a 200 μL cut tip. Leave on ice at least another hour before starting the restriction enzyme digest.
▲CRITICAL STEP: Do not attempt to resuspend the genomic DNA by over-pipetting or vortexing. Genomic DNA is kept viscous at that stage to ensure maximal preservation of RNA:DNA hybrids. This implies that precise quantification of genomic DNA going into the digest is not possible at that stage. Instead, DNA will be quantified after digestion and cleanup (step 14). CRITICAL: If the DNA “clump” appears too large (i.e. larger than 100 μL in volume), use a cut tip to split the clump in two pieces that can be then digested separately in step 10.
10∣ Digest 50 to 100 μL of extracted genomic DNA using a cocktail of restriction enzymes according to supplier’s instructions. Incubate overnight in a 37°C room.
? TROUBLESHOOTING
| Component | Amount (μL) | Final concentration |
| DNA (step 9) | 50-100 | |
| NEB Buffer 2 | 15 | 1X |
| BSA | 1.5 | 1X |
| Spermidine | 1.5 | 0.5X |
| Restriction Enzymes (NEB; Reagents) | 30U each | |
| Water | Up to 150 μL |
▲CRITICAL STEP: Make sure to adjust restriction enzyme volumes to add the correct number of units. CRITICAL: Spermidine should be added last at the proper concentration; a higher concentration can make DNA precipitate. CRITICAL: the incubation is done in a 37°C enclosed chamber or room instead of a water bath to avoid condensation in the lid and to improve the digest.
▲CRITICAL STEP: After overnight incubation, the viscosity of the mixture should have disappeared if the digest is complete; you should be able to pipet the DNA with ease. We encourage users to run an aliquot on an agarose gel (0.8%, wt/vol) to verify that the digest is complete (Supplementary Figure 2). Incompletely digested DNA will lead to lower resolution in the DRIP-seq procedure.
11∣ Spin a 2 mL “phase lock gel light” tube for 1 minute at 16,000 g to pellet the gel. Gently pipet the DNA from the previous step into phase lock gel tube. Add 100 μL of water and one volume (250 μL) of Phenol/Chloroform Isoamyl alcohol 25:24:1. Invert gently the tube 4-6 times and spin down at 16,000 g for 10 minutes at room temperature.
▲CRITICAL STEP: 100 μL of water is added to facilitate the pipetting of the aqueous phase during the following step.
12∣ In a clean 1.5 mL tube, mix 1.5 μL of glycogen, 1/10 volume of 3M NaOAc pH 5.2 (25 μL) and 2.5 volumes of 100 % Ethanol (625 μL). Gently pipet in the DNA (top aqueous phase) from the phase lock gel tube and mix by inverting 4-6 times. Incubate at least an hour at −20°C to increase precipitation yields. CRITICAL: Make sure to not carry over any of the phase-lock gel; this will result in poor precipitation efficiency.
13∣ Spin at 16,000 g for 35 minutes at 4°C. Discard the supernatant and add 200 μL of room temperature 80% ethanol. Spin 10 minutes at 16,000 g at 4°C and discard supernatant.
14∣ Air dry the pellet for 15 to 25 minutes depending on DNA concentration and resuspend in 50 μL of TE buffer. Leave the tube on ice for 30 minutes to an hour and then gently resuspend. Measure the concentration (OD260) of your DNA on a SimpliNano spectrophotometer or equivalent. Typically, you should expect a concentration around 1 μg/μL.
? TROUBLESHOOTING
▲CRITICAL STEP: To resuspend DNA, do not vortex the tube or over-pipet. You can add 50 μL more of TE buffer or leave on ice longer to help resuspension.
▲CRITICAL STEP: An essential specificity control is provided by pre-treating the nucleic acids with RNase H to remove any RNA:DNA hybrids from the mixture. For this, treat 10 μg of digested DNA with 4 μL of NEB RNase H for 4-6 hours at 37°C. Then, proceed to the S9.6 immunoprecipitation step below.
■ PAUSE POINT: DNA can be stored at −80°C for a month.
S9.6 DNA:RNA ImmunoPrecipitation (DRIP) ● TIMING 20 hours
15∣ In five separate 1.5 mL tubes, dilute in each tube 8 μg of digested DNA in 500 μL of TE buffer per tube. Save 1/10 volume for each tube (50 μL) to use as input in later qPCR (steps 25, 42 and 57); store at −20°C.
▲CRITICAL STEP: We recommend carrying out five immunoprecipitations in parallel so as to ensure the recovery of sufficient amounts of material. Reducing the amount of recovered material might cause you to over-amplify the library during sequencing library preparation. Performing five parallel immunoprecipitations was found to be superior to simply performing one immunoprecipitation with five times the amount of material and antibody.
▲CRITICAL STEP: If your goal is to perform DRIP-seq, perform only one immunoprecipitation with 8 μg of digested DNA instead of five immunoprecipitations in parallel.
▲CRITICAL STEP: For the RNase H control, use the RNase H-pretreated DNA, and dilute 8 μg as described above. Then proceed to DRIP (Step 16) and follow the exact same procedures for all tubes. This specificity control is critical to ensure the immunoprecipitation is specific and will be required for any publication.
16∣ Add 52 μL of 10X binding buffer and 20 μL of S9.6 antibody to the 450 μL of diluted DNA.
17∣ Incubate 14 to 17 hours at 4°C while gently inverting on a mini-tube rotator (about 10 rpm).
18∣ For each tube in step 15, wash 100 μL of the agarose bead slurry with 700 μL of 1X binding buffer by inverting the tubes on a mini-tube rotator for 10 minutes at room temperature. Spin down the beads one minute at 1,100 g and discard the supernatant. Repeat this step once. CRITICAL: Make sure not to lose beads by pipetting during washes.
19∣ Add the DNA from step 17 to the 100 μL of washed beads from step 18 and incubate 2 hours at 4°C while gently inverting on a mini-tube rotator (about 10 rpm).
20∣ Spin down the beads for one minute at room temperature at 1,100 g and discard the supernatant.
21∣ Add 750 μL of 1X binding buffer and invert the tubes on a mini-tube rotator for 15 minutes at room temperature. Spin down the beads for one minute at 1,100 g and discard the supernatant. Repeat once. CRITICAL: Make sure not to lose beads by pipetting during the washes.
22∣ Add to the beads 300 μL of DRIP elution buffer and 7 μL of proteinase K (20 mg/mL). Seal your tubes with parafilm to avoid any leaking and incubate with rotation at 55°C for 45 minutes in a temperature-controlled rotating oven (UVP HL-2000 HybriLinker or equivalent).
23∣ After incubation, spin down the beads one minute at 1,100 g. Meanwhile, spin five 2 mL “phase lock gel light” tubes for 1 minute at 16,000 g to pellet the gel. Transfer the supernatant of each tube to the 2 mL phase lock tube, add one volume (300 μL) of Phenol/Chloroform Isoamyl alcohol 25:24:1. Invert gently 4-5 times and spin down at 16,000 g for 10 minutes at room temperature. Recover DNA (aqueous phase) and precipitate DNA as in steps 12 and 13.
24∣ Air dry pellets for 10 to 15 minutes and add 10 μL of RNase-free TE buffer in each tube. Leave tubes on ice for 15 to 30 minutes and gently resuspend. Combine the 5 tubes in one (50 μL).
▲CRITICAL STEP: For DRIP-seq, resuspend the DNA from step 24 in 50 μL.
▲CRITICAL STEP: For the RNase H control, resuspend the DNA from step 24 in 50 μL.
▲CRITICAL STEP: See the setup reagent section to prepare RNase-free buffers.
■ PAUSE POINT The immunoprecipitated DNA can be stored at −80°C for a week.
25∣ Check DRIP efficiency by qPCR. Before proceeding, it is imperative to ensure that the DRIP procedure worked. For this, use two negative and three positive loci and measure their immunoprecipitation as a fraction of input DNA. Relative enrichments are calculated using the Pfaffl method 46 (see Box 1A). The RNase H-treated sample should lead to very low yields (>90% reduction in DRIP efficiency).
? TROUBLESHOOTING
▲CRITICAL STEP: We highly encourage making sure strong, reproducible, DRIP yields can be routinely obtained before proceeding to the next step. In our experience, it can take 3-6 trials before achieving consistency in performing the DNA extraction and DRIP. You should be able to reproduce the yields indicated in Box 1 for primers targeting positive control regions.
▲CRITICAL STEP: For DRIP-seq or the RNase H control, use 2 μL per qPCR reaction (3 positive loci and 2 negative loci, see Box 1) to check the DRIP efficiency. Add 0.5 μL of water to bring the remaining volume to 40.5 μL and proceed directly to step 43.
DNase treatment and RNA reverse transcription ● TIMING 7 hours
▲CRITICAL STEP: From this step until step 32, take every precaution to work with RNase-free buffers and reagents as the DNase treatment will release the RNA moiety of R-loops. This represents a very minute quantity of RNA compared to other RNA-based protocols such as RNA-seq. Every reagent you use in the following steps must be RNase-free and everything that enters your working space needs to be sprayed with RNAZap. Make sure to use all supplementary proper personal protective equipment such as mask to ensure the most RNase-free environment possible.
26∣ In a 1.5mL tube, mix:
| Component | Amount (μL) |
Final concentration |
| DNA (from step 24) | 48 | |
| 10X DNase Buffer | 10 | 1X |
| DNase I | 3 | 6U |
| Water | 39 | |
| Total | 100 |
Incubate at 37°C for 45 minutes.
27∣ Add 1 μL of 0.5M EDTA pH 8, mix and incubate at 75°C for 15 minutes to heat inactivate the DNase I enzyme.
28∣ Add 1 μL of glycogen, 10 μL of 3M NaOAc pH 5.2 and 240 μL of ice cold 100% Ethanol. Mix by inverting gently 4-6 times. Incubate at −20°C for an hour.
29∣ Spin 35 minutes at 16,000 g at 4°C. Discard the supernatant, add 200 μL of ice cold 80% ethanol. Spin 10 minutes at 16,000 g at 4°C. Discard supernatant.
30∣ Air dry the pellet for about 15 minutes and add 16 μL of 10 mM Tris-HCl pH 8. Leave on ice for 15 minutes and resuspend the pellet by pipetting gently.
31∣ Transfer the 16μL in a PCR tube and add 4μL of IScript reverse transcription supermix. Run the following program provided by the manufacturer in a thermal cycler (Bio-Rad C1000 Touch or equivalent).
| Duration | Temperature |
| 5 min | 25°C |
| 30 min | 42°C |
| 5 min | 85°C |
| Hold | 4°C |
32∣ Clean up the reaction with AMPure XP beads. AMPure beads allow for efficient size selection purification depending on the sample to bead volume ratios used 47. In this protocol here, we will use three different ratios: 1.6X to recover DNA fragments of all sizes, 1X to recover DNA fragments over 200 bp and 0.65X to eliminate fragments over 500 bp.
In this step, we simply clean our reaction using a 1.6X ratio. Transfer the 20 μL reaction from Step 31 in a 1.5 mL tube, add 80 μL of RNase-free water and 160 μL of AMPure beads. CRITICAL: Bring AMPure beads to room temperature before use.
33∣ Mix by pipetting 5-6 times and let stand at room temperature for 15 minutes. CRITICAL: Briefly spin the tube with a bench mini centrifuge to bring down drops on the side of the tube.
34∣ Place the tube on a magnetic rack until the solution is clear (5 minutes). Discard supernatant.
35∣ Freshly prepare 1 mL of 80% ethanol. While the tube is on the magnetic rack, add 200 μL of freshly made 80% ethanol. Incubate at room temperature for 30 seconds and discard supernatant. Repeat this step once (2 washes total).
36∣ Air dry the beads for 5 to 10 minutes (or until the beads take a light brown color) and add 40 μL of 10 mM Tris-HCl pH 8. CRITICAL: Do not overdry the beads.
37∣ Mix by vortexing or pipetting well. Spin briefly, incubate 5 minutes at room temperature and put the tube back on the magnetic rack.
38∣ When the solution is clear, transfer the 40 μL containing your eluted DNA in a new PCR tube and proceed directly to the next step.
39∣ In the PCR tube, mix:
| Component | Amount (μL) | Final concentration |
| DNA | 40 | |
| 5X second strand Buffer | 20 | 1X |
| 10 mM dNTP | 5 | 0.5 mM |
| 16 mM NAD | 1 | 160 μM |
| Water | 32.2 | |
| Total | 98.2 |
Mix well and incubate on ice for 5 minutes. CRITICAL: In your dNTP mix, don’t forget to replace dTTP by dUTP to be able to build a strand-specific library.
40∣ Add 0.3 μL of RNase H (1.6 U), 0.5 μL of E. coli DNA ligase (5U) and 1 μL of DNA polymerase I (10 U). Mix well and incubate at 16°C for 30 minutes. CRITICAL: Longer treatments will decrease the quality of the cDNA, most likely due to the low amount of material and the exonuclease activity of the DNA polymerase I. CRITICAL: After the 30 minutes incubation, do not leave the tube standing either in the PCR machine or on your bench, proceed directly to the next step.
41∣ Clean up your reaction with AMPure XP beads (1.6X). Transfer your 100 μL to a new 1.5 mL tube and add 160 μL of AMPure beads. Follow steps 33 to 38 with an elution volume of 42.5 μL (in 10 mM Tris-HCl pH 8).
42∣ As in step 25, before proceeding to the next step, check the R-loop enrichments with qPCR on two negative and three positive loci using the Pfaffl method (see Box 1B) 46 The samples can be kept on ice until the qPCR is performed. In case of overnight storage, we advise to keep the DNA at −80°C.
? TROUBLESHOOTING
43∣ Transfer the 40.5 μL of your cDNA from Step 41 in a 0.5 mL tube and proceed to sonication using a Diagenode Bioruptor NGS. Perform 12 cycles of 15 sec ON / 60 sec OFF HIGH in 4°C water bath. Spin the tube after 6 cycles to ensure homogeneous sonication.
CRITICAL: If you are using mouse DNA, perform 15 cycles, spin after 5 and 10 cycles.
CRITICAL: At this step, the sonication efficiency is hardly possible to check due to the low amount of material. The only way to visualize fragment sizes is to run an Agilent High sensitivity DNA 1000 kit following the provider’s instructions. You should obtain a size distribution ranging from 100 to 500 bp. The high number of sonication cycles ensures that the DNA is brought down to a size compatible with Illumina library construction and checking sonication efficiency of fragment size distribution is not required.
Strand-specific library construction ● TIMING 6 hours
44∣ The first step of the library construction is to end-repair your sonicated DNA. In a 1.5 mL tube, mix:
| Component | Amount (μL) | Final concentration |
| Sonicated DNA from step 43 | 40.5 | - |
| NEB 10X end repair module Buffer | 5 | 1X |
| ATP 10 mM | 2 | 0.4 mM |
| NEBNext End repair module enzyme | 2.5 | - |
| Total | 50 |
Mix well and incubate at room temperature for 30 minutes. CRITICAL: ATP is included in the NEB 10X end repair module buffer, but as it tends to degrade over repeated freeze-thaw cycles, we add fresh ATP in our mix.
45∣ Clean up the reaction using AMPure beads (1.6X). Add 80 μL of the beads to the 50 μL reaction from Step 44 and follow step 33 to 38 with an elution volume of 34 μL.
46∣ Perform the A-tailing step by mixing in a 1.5mL tube:
| Component | Amount (μL) | Final concentration |
| DNA from step 45 | 34 | - |
| 10X NEB Buffer 2 | 5 | 1X |
| 1mM dATP | 10 | 200 μM |
| NEB Klenow exo- | 1 | 5 U |
| Total | 50 |
Mix well and incubate 30 minutes at 37°C.
47∣ Clean up the reaction using AMPure beads (1.6X). Add 80 μL on the beads to the 50 μL and follow step 33 to 38 with an elution volume of 12 μL.
48∣ To ligate index adapters, prepare the following mix in a 1.5mL tube:
| Component | Amount (μL) | Final concentration |
| DNA from step 47 | 12 | - |
| NEB 2X quick ligation buffer | 15 | 1X |
| Illumina index adapters (60 μM) | 1 | 2 μM |
| NEB quick ligase | 2 | |
| Total | 30 |
Mix well and incubate 20 minutes at room temperature.
49∣ Clean up the reaction using AMPure beads (1X). To your reaction, add 70 μL of 10 mM Tris-HCI pH 8 and 100 μL of the beads and follow step 32 to 38 with an elution volume of 20 μL.
50∣ To the 20 μL of eluted DNA, add 1.5 μL of AmpErase Uracil N-glycosylase (1.5U). Incubate 20 minutes at 37°C. CRITICAL: The activity of the AmpErase Uracil N-glycosylase decreases over time so make sure to use enzyme prior to the supplier’s expiration date. CRITICAL: The AmpErase Uracil N-glycosylase does not need any specific buffer to work, so you can add it directly to the 10 mM Tris-HCl pH 8 buffer and proceed directly to the PCR amplification step without any clean up.
▲CRITICAL STEP: Only half of the eluted DNA is used for PCR amplification in step 51 to keep a backup in the event the library is under- or over-amplified. Store at −80°C.
▲CRITICAL STEP: If you are performing DRIP-seq, skip this step and go directly to step 51
51∣ PCR amplification of the library. Prepare the following mix:
| Component | Amount (μL) | Final concentration |
| DNA from step 50 | 10 | - |
| PCR primer 1.0 P5 | 1 | 0.3 μM |
| PCR primer 2.0 P7 | 1 | 0.3 μM |
| Phusion master mix | 15 | 1X |
| Water | 3 | - |
| Total | 30 |
52∣ Mix well and run the following program in a thermal cycler (Bio-Rad C1000 Touch or equivalent).
| Cycle number |
Duration | Temperature |
| 1 | 30 sec | 98°C |
| 2-15 | 10 sec | 98°C |
| 30 sec | 60°C | |
| 30sec | 72°C | |
| 16 | 5 min | 72°C |
| 17 | Hold | 12°C |
▲CRITICAL STEP: Try to avoid over-amplifying your library. 15-17 cycles should be the maximum number of cycles to avoid over-amplification and biases.
53∣ Proceed to a two-step clean-up of your library using AMPure beads. The first step (0.65X) will remove fragments over 500 bp (step 54) and the second step will remove fragments under 200 bp (step 56).
54∣ For the first step, in a 1.5 mL tube, add 70 μL of 10 mM Tris-HCl pH 8 to your reaction from step 52 and 65 μL of the beads. Mix well and incubate at room temperature for 15 minutes.
55∣ Place the tube on a magnetic rack until the solution is clear (5 minutes). Transfer the supernatant (165 μL) in a new 1.5mL tube and discard the beads.
56∣ For the second step, add 100 μL of 10 mM Tris-HCI pH 8 and 135 μL of AMPure beads (400 μL total). Follow step 32 to 38 with a washing volume of 400 μL of freshly made 80% ethanol and an elution volume of 13 μL.
Quality control ● TIMING 3 hours
57∣ As in step 25, check R-loop enrichments with qPCR on two negative and three positive loci using the Pfaffl method (see Box 1C)46
58∣ Check the size distribution of your library from step 56 using an Agilent High sensitivity DNA 1000 kit following the instructions provided with the kit.
? TROUBLESHOOTING
▲CRITICAL STEP Make sure that your library fits the common requirements for Illumina sequencing: fragments must be distributed between 200 and 500 bp and the library must possess a concentration over 1 ng/μl. In case of adapter or primer contamination, proceed to an additional AMPure clean-up using a 1X ratio.
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2:
Troubleshooting table
| Step | Problem | Possible reasons | Possible solutions |
|---|---|---|---|
| 10 | DNA is undigested or still viscous | The precipitation was not complete (step 7) | Make sure that the DNA is fully precipitated (white and not translucent at all). |
| The DNA to enzyme ratio was too high. | Reduce DNA in half. You can also add 15U of each enzyme & leave at 37°C for 2 hours, pipet gently a few times to resuspend DNA, then add the other 15U of enzyme and leave overnight | ||
| The DNA was overdried | Make sure to not overdry the DNA at step 9 | ||
| 14 | Not enough DNA | Most likely insufficient starting material. | Start from beginning with more cells. |
| 25 | No enrichment in known R-loop forming regions | Reasons can be multiple | Using the same DNA, retry the immunoprecipitation steps. If the outcome is the same, start over with a new DNA extraction from the beginning. Verify your S9.6 batch using oligonucleotides in gel shift assays 32. Practice DRIP on in vitro transcribed substrates 15 |
| 42 | Decrease in enrichment between step 25 and this step | The second strand synthesis in step 40 was too long | Reduce incubation time to 20 or 25 minutes. You can also reduce the amount of DNA polymerase I. |
| 58 | The sequencing library is not good (over-amplified [characterized by spiky Bioanalyzer profile] or under amplified [concentration < 200 pg/ μL]) | The number of amplification cycles was not correct. | Using half of the unamplified library from step 51, adjust the number of cycles. Avoid over-amplification to prevent PCR biases. |
● TIMING - DRIPc-seq
Step 1-4: Cell harvest and lysis: 30 min and overnight
Step 5-14: DNA extraction and fragmentation: 6h, overnight and 3 hrs
Step 15-24: S9.6 immunoprecipitation: 30 minutes, overnight and 7 hrs
Step 25: qPCR check: 2hrs
Step 26-43: DNase I treatment and RNA reverse transcription: 7 hrs
Step 44-56: Strand specific library construction: 6 hrs
Step 57-58: Quality control: 3 hrs
Box 1: qPCR analysis: 2 hrs
Box 2: Data analysis: 1 day
● TIMING - DRIP-seq
Step 1-4: Cell harvest and lysis: 30 min and overnight
Step 5-14: DNA extraction and fragmentation: 6h, overnight and 3 hrs
Step 15-24: S9.6 immunoprecipitation: 30 minutes, overnight and 7 hrs
Step 25: qPCR check: 2hrs
Step 44-56: Non strand specific library construction: 6 hrs
Step 57-58: Quality control: 3 hrs
Box 1: qPCR analysis: 2 hrs
Box 2: Data analysis: 1 day
Anticipated results:
Post sequencing, extracting R-loop mapping information can be performed in a manner similar to most ChIP mapping procedures using standard computational pipelines (Box 2). After adapter trimming, removal of PCR duplicates and mapping to a reference genome, the mapping results can be uploaded to a genome browser. A typical expected output of DRIPc-seq and DRIP-seq is shown in Figure 2A. This screenshot represents a 200 kb region with R-loops mapping to the positive and negative strands indicated in red and blue, respectively for two independent replicates. A corresponding output for DRIP-seq is shown below in green along with a control RNase H-treated sample (teal) also obtained after DRIP-seq. The strong reduction of peaks in the RNase H track confirms that the technique captures signals derived from RNA:DNA hybrids (Figure 2A and 2B).
Box 2: Data analysis ● TIMING 1day.
After the size and quality of the libraries has been checked on a Bioanalyzer and enrichments have been validated, we recommend Single Read 100 sequencing. Since the expected library size is between 300 and 400 bp, SR150 or PE150 can also be considered. Several samples can be pooled in one sequencing lane as long as 35 to 45 million reads can be generated per sample. The number of samples pooled will depend on the sequencer used.
Here we describe the data analysis pipeline for single-end sequencing (for paired-end, the pipeline will need to be adapted slightly according to the standard usage statements of each tool).
Data analysis is directly performed on .fastq or .fastq.gz files and can be performed using freely available bioinformatic tools. Before starting, make sure you have access to fastq-mcf53, bowtie254, samtools55 and bedtools56 and to a UNIX compatible environment with a minimal 16 Gb RAM. In the following pipeline, the name of your sample is stipulated between “” for each step.
1. From the sequencing data (.fastq or .fastq.gz files), the first step is to trim adapter sequences from reads. To do this step, run the following command line using fastq-mcf.
fastq-mcf adaptor.fa “sample.fastq” -o “sample_filtered.fastq”
In this command line, adaptor.fa needs to contain the sequences of the adapters you used when building libraries (see below for example).
>Illumina_Single_End_Apapter_1
ACACTCTTTCCCTACACGACGCTGTTCCATCT
>Illumina_Single_End_Apapter_2
CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT
2. In the following step, reads will be aligned to the reference genome using bowtie 2. The reference_genome contains all necessary chromosome annotations from your genome of reference (human, mouse, yeast…). The output of this command line will be transferred into samtools for duplicate removal. The reads will then be sorted and finally the read duplicates will be removed.
bowtie2 –x reference_genome -U “sample_filtered.fastq” ∣ samtools view -bS -t “reference_genome_fasta_index.fa.fai” - > “sample_filtered_aligned.bam” samtools sort “sample_filtered_aligned.bam” -o “sample_filtered_aligned_sorted.bam” samtools rmdup -s “sample_filtered_aligned_sorted.bam” “sample_filtered_aligned_sorted_rmdup”
3. The final step will be to create a file that can be uploaded on a genome browser to visualize your sequencing data. The duplicate-removed bam file and bam index file can be directly uploaded onto an ftp or https storage (e.g. Amazon S3) for visualization. To create bam index file using samtools run the following:
samtools index “sample_filtered_aligned_sorted_rmdup.bam” “sample_filtered_aligned_sorted_rmdup.bam.bai”
4. Since bam files are usually large, we can also create bigWig files which usually are hundred times smaller in size. For stranded samples (as in DRIPc-seq), reads will be assorted into positive and negative strands using samtools before being turned into bedGraph using bedtools genomecov. The option “-scale N” is to normalize samples to each other based on number of mapped reads in each sample (e.g. to normalize to 50 million reads, we use N = 5e7/mapped_reads).
samtools view “sample_filtered_aligned_sorted_rmdup.bam” ∣ awk ‘$2 ~ /^(16∣83∣163)$/57‘ ∣ samtools view –bS –t “reference_genome.fa.fai” − ∣ bedtools genomecov –ibam - -split -scale N -bg -g “reference_genome.fa.fai” > “sample_filtered_aligned_sorted_rmdup_pos.bedGraph” gzip “sample_filtered_aligned_sorted_rmdup_pos.bedGraph” samtools view “sample_filtered_aligned_sorted_rmdup.bam” ∣ awk ‘$2 ~ /^(0∣99∣147)$/57‘ ∣ samtools view –bS –t “reference_genome.fa.fai” − ∣ bedtools genomecov –ibam - -split -scale N -bg -g “reference_genome.fa.fai” > “sample_filtered_aligned_sorted_rmdup_neg.bedGraph” gzip “sample_filtered_aligned_sorted_rmdup_neg.bedGraph”
5. For DRIP-seq, which is not strand-specific, users only need to run bedtools genomecov on the duplicate-removed bam file generated in step 2.
bedtools genomecov –ibam “sample_filtered_aligned_sorted_rmdup.bam” -split -scale N -bg -g “reference_genome.fa.fai” > “sample_filtered_aligned_sorted_rmdup.bedGraph” gzip “sample_filtered_aligned_sorted_rmdup.bedGraph”
6. Finally, files will be changed from bedGraph into bigWig using wigToBigWig.
wigToBigWig –clip “sample_filtered_aligned_sorted_rmdup_pos.bedGraph.gz” “reference_genome.fa.fai” “sample_filtered_aligned_sorted_rmdup_pos.bigWig” wigToBigWig –clip “sample_filtered_aligned_sorted_rmdup_neg.bedGraph.gz” “reference_genome.fa.fai” “sample_filtered_aligned_sorted_rmdup_neg.bigWig” wigToBigWig –clip “sample_filtered_aligned_sorted_rmdup.bedGraph.gz” “reference_genome.fa.fai” “sample_filtered_aligned_sorted_rmdup.bigWig”
7. The output files can be then uploaded to the genome browser. The graphical output is presented in Figure 2A.
DRIPc-seq signal often appears as a mixture of sharp and broad peaks depending on the loci that were analyzed. Adequate peak calling can be performed using by standard packages such as MACS 48, which preferentially identify short peaks with high signal to noise ratios or with Sicer49, which uses spatial clustering to identify broad peaks. Alternatively, we have developed a Hidden Markov Model-based peak calling algorithm that can detect both types of peaks and is well-adapted to DRIP-seq and DRIPc-seq analysis 23. This algorithm is available online from GitHub (https://github.com/chedinlab/DRIPc/tree/master/peak_calling/DRIPc).
Experimental approaches aimed at identifying changes in R-loop distributions between a set of different conditions will require the identification of loci exhibiting statistically significant differential R-loop formation. This analysis can be performed using standard packages like DEseq 50. In order to be able to confidently and directly ascribe a change in R-loop presence to a particular condition (absence of a protein of interest for instance), it is imperative that users conduct side-by-side measurement of potential transcription variation between conditions. Indeed, since R-loops are primarily formed co-transcriptionally, any change to the transcriptional landscape will likely result in a change at the R-loop level. Depending on the conditions considered (long term genetic perturbations versus short drug treatments), approaches such as RNA-seq or methods aimed at measuring nascent transcription (GRO-seq51 or 4SU-seq52) should be considered.
Supplementary Material
Supplementary Figure 1. Sequences captured by DRIPc-seq show no correlation with S9.6 intrinsic binding preferences. 6-mers found to be poorly or tightly bound by S9.6 were curated from Konig et al., (2017) and grouped as low and high binding. We evaluated each 6-mer frequency in the R-loop forming sequence space identified by DRIPc-seq (Sanz et al., 2016), resulting in observed frequencies. As a comparison, we retrieved non-R-loop forming genic regions derived from loci that were matched for expression, length and location and measured 6-mer frequencies over this control set. For each R-loop peak, 25 random, matched peaks were extracted and the average frequency determined for each 6-mer. This resulted in expected frequencies.
A. The graph shows the log2 fold ratio of observed (R-loop forming) over expected (matched non-R-loop forming) frequencies for each 6-mer. Some 6-mers are clearly more or less represented than others in DRIPc-seq data compared to expectations from control non-R-loop loci. This could reflect the intrinsic sequence preference of R-loop formation and/or the intrinsic preference of S9.6 antibody. If the latter is true, we expected S9.6-highly bound epitopes (red) to be over-represented and S9.6-poorly bound epitopes (blue) to be underrepresented. This was not observed, however. Instead, S9.6 tightly or poorly bound 6-mers were equally likely to be under- or over-represented. This suggests that DRIPc-seq data does not suffer from systematic biases caused by S9.6 sequence preference.
B. To account for what could be driving the over- or under-representation of certain 6-mers, we simply calculated the GA content of the motifs. As shown below, depleted motifs tend to be GA-poor (CT-rich), while enriched motifs tend to be GA-rich irrespective of whether they are tightly or poorly bound by S9.6 (the dashed grey line represents 50% GA content). Given that GA-rich regions are favorable for R-loop formation, the observed trends are most likely to reflect the intrinsic sequence biases underlying R-loop formation, not S9.6 binding. Similar results were observed when 8-mers were considered.
Supplementary Figure 2: Genomic DNA digestion profiles. DNA digestion profiles after Step 10 were visualized after agarose gel electrophoresis through a 0.8% agarose gel run in 1x TAE buffer. DNA was extracted from human NTERA-2 cells and digested with restriction enzyme cocktail indicated in Step 10. Lanes 1 and 2 show an example of incomplete digestion, as evidenced by the high molecular weight bands above 20 kilobases. Lanes 3 and 4 show an example of fully digested DNA as judged from the disappearance of the top band. The leftmost lane (M) corresponds to a 1kb plus GeneRuler ladder from ThermoFisher.
Acknowledgements
We thank Dr. Stella R. Hartono, John Smolka and Maika Malig for constructive comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM120607).
Footnotes
Competing Interests
The authors declare no competing interests as defined by Nature Research, or other interests that might be perceived to influence the interpretation of the article.
REFERENCES
- 1.Santos-Pereira JM & Aguilera A R loops: new modulators of genome dynamics and function. Nat Rev Genet 16, 583–97 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kreuzer KN & Brister JR Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J 7, 358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carles-Kinch K & Kreuzer KN RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J Mol Biol 266, 915–26 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Masukata H & Tomizawa J A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62, 331–8 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Itoh T & Tomizawa J Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77, 2450–4 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akman G et al. Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc Natl Acad Sci U S A 113, E4276–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DY & Clayton DA Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem 273, 30614–21 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Xu B & Clayton DA A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cell Biol 15, 580–9. (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels GA & Lieber MR RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res 23, 5006–11. (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang FT, Yu K, Hsieh CL & Lieber MR Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A 103, 5030–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reaban ME & Griffin JA Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348, 342–4. (1990). [DOI] [PubMed] [Google Scholar]
- 12.Yu K, Chedin F, Hsieh CL, Wilson TE & Lieber MR R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4, 442–51 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Ratmeyer L, Vinayak R, Zhong YY, Zon G & Wilson WD Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry 33, 5298–304 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Roberts RW & Crothers DM Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258, 1463–6. (1992). [DOI] [PubMed] [Google Scholar]
- 15.Ginno PA, Lott PL, Christensen HC, Korf I & Chedin F R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45, 814–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartono SR, Korf IF & Chedin F GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Res 43, 9729–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilera A & Garcia-Muse T R loops: from transcription byproducts to threats to genome stability. Mol Cell 46, 115–24 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Sollier J & Cimprich KA Breaking bad: R-loops and genome integrity. Trends Cell Biol 25, 514–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costantino L & Koshland D The Yin and Yang of R-loop biology. Curr Opin Cell Biol 34, 39–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skourti-Stathaki K & Proudfoot NJ A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28, 1384–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard P & Manley JL R Loops and Links to Human Disease. J Mol Biol 429, 3168–3180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips DD et al. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26, 376–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanz LA et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63, 167–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skourti-Stathaki K, Proudfoot NJ & Gromak N Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42, 794–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Hage A & Tollervey D Immunoprecipitation of RNA:DNA Hybrids from Budding Yeast. Methods Mol Biol 1703, 109–129 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Rubio M, Barroso SI & Aguilera A Detection of DNA-RNA Hybrids In Vivo. Methods Mol Biol 1672, 347–361 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Stork CT et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzo SG et al. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol 19, 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartono SR et al. The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J Mol Biol 430, 272–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halasz L et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res 27, 1063–1073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginno PA, Lim YW, Lott PL, Korf I & Chedin F GC skew at the 5' and 3' ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23, 1590–600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konig F, Schubert T & Langst G The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS One 12, e0178875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerritelli SM & Crouch RJ Ribonuclease H: the enzymes in eukaryotes. Febs J 276, 1494–505 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanoosthuyse V Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Noncoding RNA 4(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K & Lieber MR Complexities due to single-stranded RNA during antibody detection of genomic rna:dna hybrids. BMC Res Notes 8, 127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F et al. Current Protocols in Molecular Biology Page 3.13.1 Suppl. 8 (1995). [Google Scholar]
- 37.Thomas M, White RL & Davis RW Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A 73, 2294–8 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White RL & Hogness DS R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell 10, 177–92 (1977). [DOI] [PubMed] [Google Scholar]
- 39.Drolet M, Bi X & Liu LF Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J Biol Chem 269, 2068–74 (1994). [PubMed] [Google Scholar]
- 40.Duquette ML, Handa P, Vincent JA, Taylor AF & Maizels N Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18, 1618–29 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahba L, Costantino L, Tan FJ, Zimmer A & Koshland D S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30, 1327–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants 3, 704–714 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Dumelie JG & Jaffrey SR Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell 68, 745–757 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loomis EW, Sanz LA, Chedin F & Hagerman PJ Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet 10, e1004294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, 2004–07 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lis JT & Schleif R Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res 2, 383–9 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu S, Grullon S, Ge K & Peng W Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol 1150, 97–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S & Huber W Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Core LJ, Waterfall JJ & Lis JT Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabani M et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29, 436–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aronesty E Command-line tools for processing biological sequencing data. (2011). [Google Scholar]
- 54.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding F et al. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome 16, 424–31 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Sequences captured by DRIPc-seq show no correlation with S9.6 intrinsic binding preferences. 6-mers found to be poorly or tightly bound by S9.6 were curated from Konig et al., (2017) and grouped as low and high binding. We evaluated each 6-mer frequency in the R-loop forming sequence space identified by DRIPc-seq (Sanz et al., 2016), resulting in observed frequencies. As a comparison, we retrieved non-R-loop forming genic regions derived from loci that were matched for expression, length and location and measured 6-mer frequencies over this control set. For each R-loop peak, 25 random, matched peaks were extracted and the average frequency determined for each 6-mer. This resulted in expected frequencies.
A. The graph shows the log2 fold ratio of observed (R-loop forming) over expected (matched non-R-loop forming) frequencies for each 6-mer. Some 6-mers are clearly more or less represented than others in DRIPc-seq data compared to expectations from control non-R-loop loci. This could reflect the intrinsic sequence preference of R-loop formation and/or the intrinsic preference of S9.6 antibody. If the latter is true, we expected S9.6-highly bound epitopes (red) to be over-represented and S9.6-poorly bound epitopes (blue) to be underrepresented. This was not observed, however. Instead, S9.6 tightly or poorly bound 6-mers were equally likely to be under- or over-represented. This suggests that DRIPc-seq data does not suffer from systematic biases caused by S9.6 sequence preference.
B. To account for what could be driving the over- or under-representation of certain 6-mers, we simply calculated the GA content of the motifs. As shown below, depleted motifs tend to be GA-poor (CT-rich), while enriched motifs tend to be GA-rich irrespective of whether they are tightly or poorly bound by S9.6 (the dashed grey line represents 50% GA content). Given that GA-rich regions are favorable for R-loop formation, the observed trends are most likely to reflect the intrinsic sequence biases underlying R-loop formation, not S9.6 binding. Similar results were observed when 8-mers were considered.
Supplementary Figure 2: Genomic DNA digestion profiles. DNA digestion profiles after Step 10 were visualized after agarose gel electrophoresis through a 0.8% agarose gel run in 1x TAE buffer. DNA was extracted from human NTERA-2 cells and digested with restriction enzyme cocktail indicated in Step 10. Lanes 1 and 2 show an example of incomplete digestion, as evidenced by the high molecular weight bands above 20 kilobases. Lanes 3 and 4 show an example of fully digested DNA as judged from the disappearance of the top band. The leftmost lane (M) corresponds to a 1kb plus GeneRuler ladder from ThermoFisher.