Abstract
Background
Little is known about epigenetic regulation of intracranial aneurysms (IAs). Circular non-coding RNAs (circRNAs) play crucial roles in cardiovascular diseases, but they have received scant research attention regarding their relationship with IAs. This study aimed to explore new pathological mechanisms of IA through circRNA expression profiles and to provide novel therapeutic strategies.
Material/Methods
The comprehensive circRNA and mRNA expression profiles were detected by RNA-Seq in human IA walls and superficial temporal arteries (STAs). The RNA-Seq findings were validated by qRT-PCR. GO and KEGG analyses indicated the functions of these circRNAs. A competing endogenous RNA (ceRNA) network was constructed to reveal the circRNA-miRNA-mRNA relationship. Two newly discovered circRNAs were further detected in peripheral blood of IA patients and healthy people to clarify their expression patterns in the periphery.
Results
Many differentially expressed circRNAs are closely involved in immune/inflammatory response and cell adhesion/adherens junction. The novel circRNAs (hsa_circ_0072309 and hsa_circ_0008433) regulate DDR2 and MMP2, respectively, which are associated with SMC dysfunction and vascular injury through ceRNA. Moreover, we found differential expression of these 2 circRNAs in the peripheral blood of IA patients, and the expression pattern of hsa_circ_0072309 had central and peripheral consistency.
Conclusions
To the best of our knowledge, this is the first study to perform circRNA sequencing analysis of IAs. hsa_circ_0072309 and hsa_circ_0008433 are novel and pivotal circRNAs related to IAs. This study provides new insights into therapeutic targets and biomarkers for IA patients.
MeSH Keywords: High-Throughput Nucleotide Sequencing, Intracranial Aneurysm, RNA Polymerase III
Background
Intracranial aneurysm (IA) is a local pathological expansion of the cerebral arteries. Its prevalence is approximately 3% of the population worldwide, with a mean age of 50 years [1]. Rupture of IAs is an acute stroke and critical condition called spontaneous subarachnoid hemorrhage (SAH), which has a mortality rate of nearly 50% [2]. There has been much recent research exploring the molecular mechanisms of IAs, but the genetic pathology of IA remains unclear.
CircRNA is a novel type of endogenous non-coding RNA without a 5′ cap and 3′ poly (A) end. It is widely present in human cells, regulating transcriptional or posttranscriptional gene expression [3]. They are widely expressed in the central nervous system (CNS) and enrich biological functions, such as the function of “miRNA sponge”, which subsequently affects mRNA expression and interacts with proteins through CeRNA [4]. Multiple studies have revealed that the ceRNA mechanism is involved in many crucial pathological processes, including inflammation, smooth muscle cell (SMC) phenotype conversion, and extracellular matrix (ECM), which play important roles in vascular diseases [5–7].
Using next-generation high-throughput sequencing technology, this study is the first to define the circRNA expression profiles of IAs. Compared with microarrays, sequencing methodology provides more comprehensive and reliable data [8], which will improve our knowledge of the epigenetic mechanisms of IAs. We found that some novel circRNA molecules are closely involved in the pathogenesis of IAs and show a certain consistency both in central and peripheral. These findings were further verified by the subsequent peripheral blood PCR experiments in the IA population with enlarged sample sizes. In summary, these results may provide novel insights into the molecular mechanisms of IAs, shed light on new treatments, and promote exploration of circRNAs as biomarkers.
Material and Methods
Patient selection and specimen collection for RNA sequencing
A total of 4 human IA walls (2 were ruptured IAs and the other 2 were unruptured IAs) and 4 normal human superficial temporal arteries (STAs) were harvested from patients in the Department of Neurosurgery, the Second Affiliated Hospital of Fujian Medical University between December 2017 and June 2018. Their respective peripheral blood samples were collected at the same time. All selected patients were confirmed to have IAs by head computed tomography angiography (CTA) or digital subtraction angiography (DSA) and they consented to microsurgical clipping. We excluded those who had cerebrovascular diseases other than IA, systemic malignant tumor, or severe complications. In case of ruptured IAs, surgeries were performed within the acute phase (24 h) after initial subarachnoid hemorrhage [9]. The aneurysmal domes were obtained during the operation without endangering patients. As controls, we collected STA tissues from patients with craniocerebral trauma if the tissues had been injured during craniotomy [10–12]. All control patients underwent head CTA to exclude IAs before surgery. The included patients’ baseline information and clinical features of IAs are shown in Table 1. Tissue samples were immediately stored in liquid nitrogen (<1 min after resection) and transferred to a −80°C freezer. All included volunteers signed informed consent and the study was approved by the Ethics Committee of the Second Affiliated Hospital of Fujian Medical University.
Table 1.
Baseline data of patients for sequencing and the clinical features of IAs.
| Items | IAs (n=4) | STAs (n=4) | T/χ2 | P Value |
|---|---|---|---|---|
| Mean age | 57.50±3.000 | 56.75±2.754 | −0.368 | 0.725 |
| Sex | 0 | 1.000* | ||
| Male | 1 (25%) | 1 (25%) | ||
| Female | 3 (75%) | 3 (75%) | ||
| SBP (mmHg) | 162.75±31.01 | 153.25±29.07 | −0.447 | 0.671 |
| DBP (mmHg) | 83.50±14.39 | 87.75±14.39 | 0.418 | 0.691 |
| TG (mmol/L) | 0.62±0.31 | 1.55±0.94 | 1.873 | 0.110 |
| LDL (mmol/L) | 1.73±0.58 | 2.83±1.00 | 1.903 | 0.106 |
| HDL (mmol/L) | 1.45±0.69 | 1.66±0.44 | 0.509 | 0.629 |
| Hypertension | 1.000 * | |||
| Yes | 3 (75%) | 2 (50%) | ||
| No | 1 (25%) | 2 (50%) | ||
| Smoking | 1.000* | |||
| Yes | 1 (25%) | 2 (50%) | ||
| No | 3 (75%) | 2 (50%) | ||
| IA sample ID | Location | Ruptured/Unruptured | Type | Size (mm) |
| IA 1 | AcoA | Ruptured | Saccular | 7 |
| IA 2 | MCA | Ruptured | Irregular | 11 |
| IA 3 | PcoA | Unruptured | Saccular | 15 |
| IA 4 | MCA | Unruptured | Saccular | 9.5 |
AcoA – anterior communicating artery aneurysm; PcoA – posterior communicating artery aneurysm; MCA – middle cerebral artery aneurysm; SBP – systolic blood pressure; DBP – diastolic blood pressure; TG – triglyceride; HDL – high-density lipoprotein; LDL – low-density lipoprotein; Size – Maximum Diameter;
Fisher test.
Study population for exploration
Peripheral blood samples were collected from 30 pairs of matched IA patients (24 were ruptured IAs and 6 were unruptured IAs) and healthy people between August 2018 and November 2018 at our institute. The inclusion criteria for the IA group were the same as those for RNA-Seq. The control group was selected from people who were attending a regular health check-up, and all of them underwent head CTA/DSA to exclude IA. To ensure the quality of samples, we excluded those with factors that could affect the state of peripheral blood, including pregnancy, chemotherapy, and fever (≥37°C). Neutrophils were further isolated and extracted from the blood (<1 h after collection) and stored in TRIzol at −80°C.
RNA isolation and purification
Total RNA was extracted from each tissue and peripheral blood sample using TRIzol reagent (Takala, USA) according to the kit’s instructions. The quality of the RNA samples was assessed using a NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and OD260/OD280 ratios between 1.8 and 2.1 were accepted. Agarose gel electrophoresis and the Nanodrop spectrophotometer were used to check the quantity of RNA before sequencing and qRT-PCR.
CircRNA and mRNA sequencing
The Ribo-Zero™ Removal Kit (Epicentre, USA) was used to ribo-consume the total RNA. The rRNA elimination was assessed using an Agilent bioanalyzer (RNA 6000 Pico chip). The ribo-consumed RNA was treated with RNaseR (20 U/L, 1 h at 37°C) and further purified with RNA Cleanup magnetic beads (Geneaid, USA). Next, circRNA and mRNA libraries were constructed according to the manufacturer’s instructions (New England BioLabs, NEBNext Ultra Directional RNA Library Prep Kit). We used the Library Quantification Kit (Kapa Biosystems, USA) to identify and quantify sequencing using the Illumina HiSeq4000 system. Finally, circRNA sequences were predicted using CircRNA identifier (CIRI) software.
Verification with qRT-PCR
Reverse transcription of quantified RNA was performed using SuperScript® III Reverse Transcriptase (RT) according to the manufacturer’s instructions. The expression levels of circRNAs were verified using SYBR Green assays (ArrayStar). β-actin was used as an internal control and the experiment was independently repeated 3 times. The 2−ΔΔCT method was applied to calculate the relative expression of each circRNA.
Gene ontology and KEGG pathway analysis
Gene ontology enrichment analysis (http://www.geneongoloty.org/) was conducted to explore potential biological functions of the target genes. KEGG pathway analysis (http://www.genome.jp/kegg/) revealed the signaling pathways of these genes at the molecular level. Genes with corrected P<0.05 were considered enriched.
CircRNA-miRNA-mRNA network construction
miRNA-binding sites of circRNA were predicted in both TargetScan and miRanda. Information on miRNA-mRNA regulatory relationships was identified using the miRTarBase database. Cytoscape was used to draw the graph of circRNA-miRNA-mRNA interaction networks.
Statistical analysis
We compared the expression of circRNAs between the IA group and the control group by Mann-Whitney U test. Categorical variables were assessed by the chi-square (χ2) test or two-tailed t test for normally distributed data, and the Mann-Whitney U test was used for skewed data. Pearson coefficient analysis was used to assess the CeRNA network construction. A p value <0.05 (two-tailed) was considered significant. All statistical analyses were performed using SPSS 24.0 software.
Results
CircRNA and mRNA expression profiles
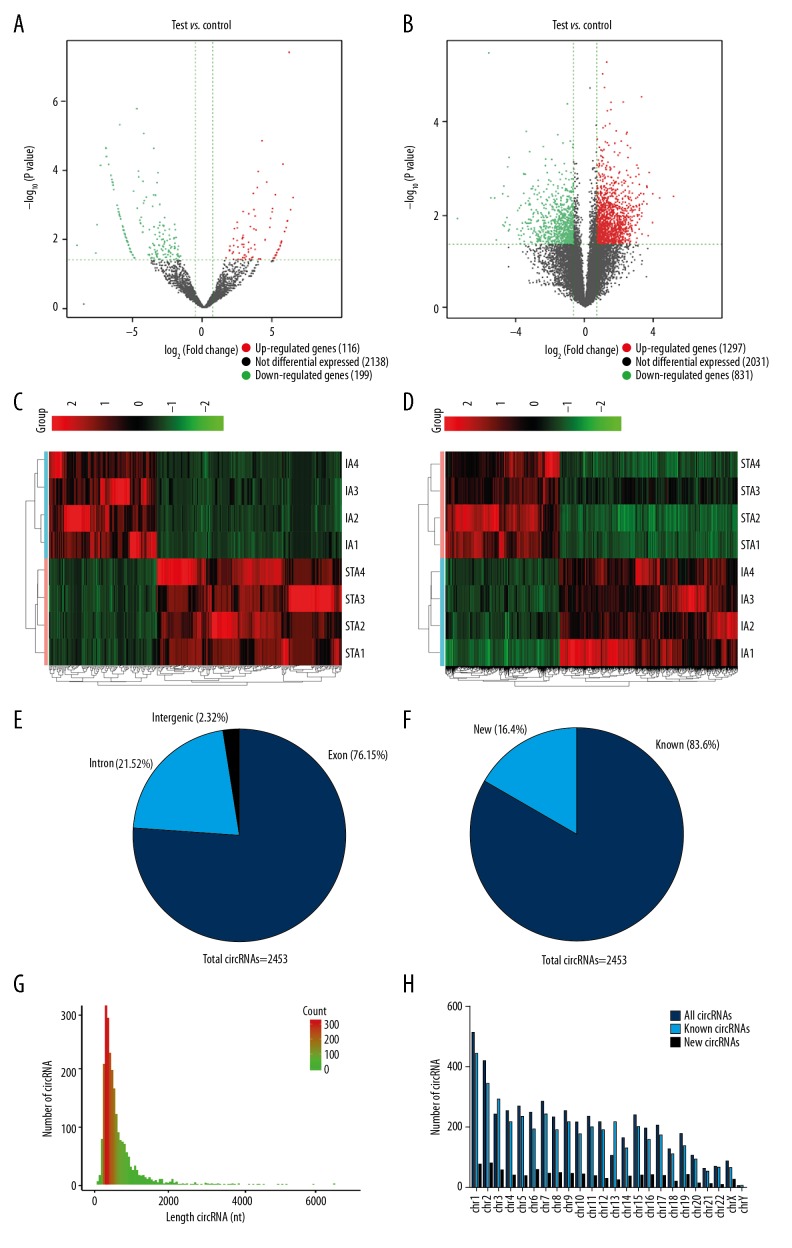
Compared with the STA group, 315 circRNAs and 4159 mRNAs were found to be significantly differentially expressed in the IAs group (FC>1.5, P<0.05). Among them, 116 circRNAs were upregulated and 199 were downregulated, while 1297 mRNAs were upregulated and 831 were downregulated (Figure 1A, 1B). Heatmaps assessed and compared the expression changes of circRNAs and mRNAs between the 2 groups (Figure 1C, 1D). We found that 76.15% of circRNAs are derived from exons (Figure 1E), 83.6% of circRNAs have been identified in circBase, and 16.4% are novel (Figure 1F). Most circRNAs had a predicted spliced length of <1000 nt (Figure 1G). These dysfunctional circRNAs were widely distributed among all chromosomes, including sex chromosomes (Figure 1H). The top 5 most significantly differentially expressed circRNAs and mRNAs are listed in Table 2.
Figure 1.
The circRNA and mRNA expression profiles compared between IAs and STAs. (A, B) Volcano plots show significantly differentially expressed circRNAs (A) and mRNAs (B) between IAs and STAs (FC>1.5, P<0.05). The red and green dots represent upregulated and downregulated circRNAs and mRNAs. (C, D) Heatmaps classified the differential expressed circRNAs and mRNAs between IAs and STAs. (E) The percentage of differentially expressed circRNAs arising from genomic locus. (F) The length distribution of differentially expressed circRNAs. (G) Proportion of previously known and newly discovered circRNAs detected by RNA-Seq. (H) Chromosomal location of circRNAs.
Table 2.
The top 5 up- and downregulated circRNAs in the 2 groups.
| CircRNA ID | Regulation | Log2 FC | P | Horm | CircBase ID | Host gene | Length |
|---|---|---|---|---|---|---|---|
| chr17: 7480128-7480270: + | Up | 5.9628048 | <0.001 | chr17 | SNORD10 | 142 | |
| chr14: 23371395-23371591: − | Up | 5.7986801 | 0.002 | chr14 | RBM23 | 196 | |
| chr1: 66378927-66384518: + | Up | 5.7025993 | <0.001 | chr1 | hsa_circ_0008433 | PDE4B | 351 |
| chr17: 80992910-81006661: − | Up | 5.5964359 | 0.004 | chr17 | B3GNTL1 | 282 | |
| chr19: 18285849-18286507: + | Up | 5.5674489 | 0.004 | chr19 | hsa_circ_0005571 | IFI30 | 351 |
| chr11: 92085261-92088570: + | Down | −8.5141271 | 0.020 | chr11 | hsa_circ_0000348 | FAT3 | 3309 |
| chr2: 179542851-179542935: − | Down | −7.2466032 | 0.032 | chr2 | TTN | 84 | |
| chr12: 56094682-56094938: − | Down | −7.1576707 | <0.001 | chr12 | hsa_circ_0026782 | ITGA7 | 256 |
| chr2: 179515969-179516047: − | Down | −6.9305180 | <0.001 | chr2 | TTN | 78 | |
| chr5: 38523520-38530768: − | Down | −6.5743179 | <0.001 | Chr5 | hsa_circ_0072309 | LIFR | 580 |
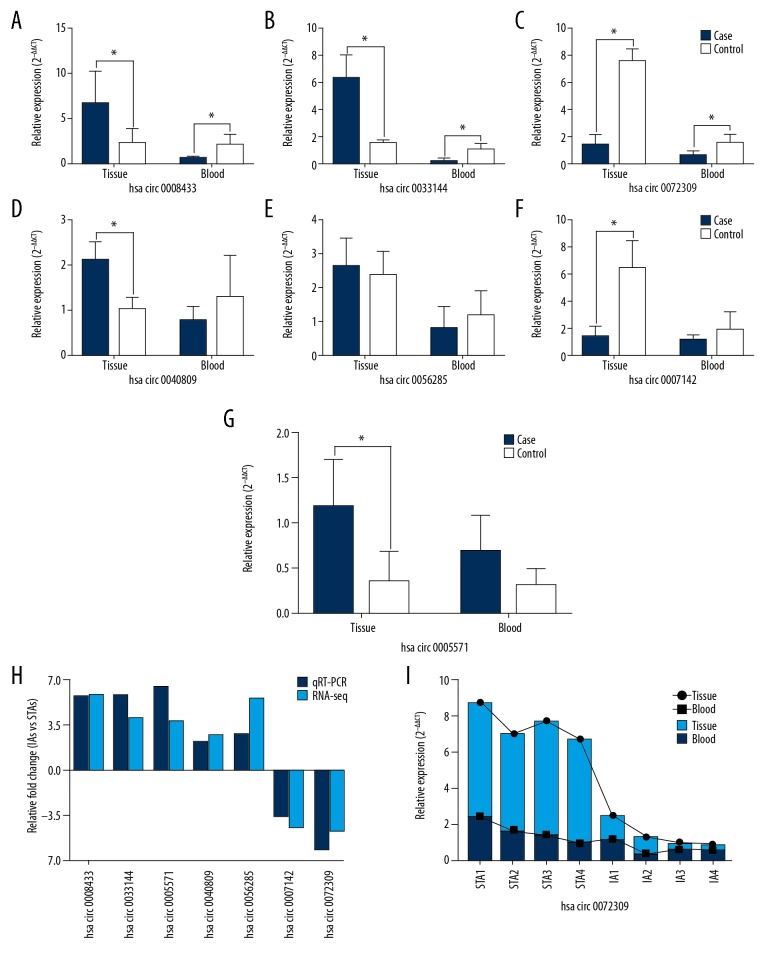
Verification of candidate circRNAs by qRT-PCR
Seven differentially expressed circRNAs were selected to confirm the sequencing data by qRT-PCR in the same tissue samples used for the sequencing, including 5 upregulated (hsa_circ_0008433, hsa_circ_0033144, hsa_circ_0005571, hsa_circ_0040809, hsa_circ_0056285) and 2 downregulated (hsa_circ_0072309, hsa_circ_0007142) circRNAs. We also explored the expression of these candidate circRNAs in the peripheral blood of sequencing patients (Figure 2A–2G). The results showed that all selected circRNAs expression was consistent with the RNA-seq results, which confirmed the accuracy of sequencing (Figure 2H). Hsa_circ_0072309 showed the same expression patterns in IA tissues and peripheral blood (P<0.05) (Figure 2G), while the expression patterns of hsa_circ_0008433 and hsa_circ_0033144 in blood were different from those in tissues (P<0.05) (Figure 2A, 2B). Interestingly, the expression trend of hsa_circ_0072309 in each sample showed some similarity (Figure 2I).
Figure 2.
PCR validation of circRNAs in tissue and peripheral blood. (A–G) The PCR results of the 7 selected circRNAs in IA tissues and peripheral blood. (H) Relative fold changes of circRNAs by RNA-seq and qRT-PCR. (I) The expression trends of hsa_circ_0072309 in each IA and STA sample from tissues and peripheral blood. * P<0.05
GO and pathway analysis
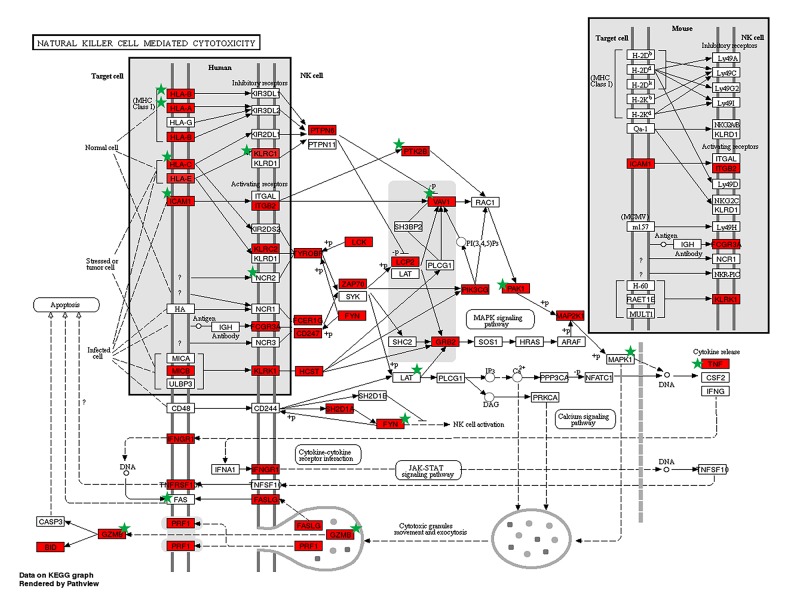
GO analysis indicated that most of the up- and downregulated functional terms were related to immune/inflammatory response and cell adhesion/adherence junction (Table 3). The top 5 upregulated biological processes were also involved in defense response to virus and T cell/TNF receptor binding, while the top 5 downregulated mainly included extracellular matrix, cytoskeleton, and actin filament binding. KEGG analysis consistently revealed that most upregulated target genes were enriched in inflammatory pathway, including classical ones such as chemokine and NF-κB. Among them, the most relevant pathways were NK cell-mediated cytotoxicity (Figure 3). As shown in the figure, many genes involved in this pathway [13] were significantly differentially expressed in our RNA-Seq and targeted by circRNAs. For downregulated genes, the most affected pathways were cell adhesion/adherence junction. In addition, it was also involved in regulation of lipolysis and vascular smooth muscle contraction, which is related to arterial wall strength and integrity (Table 4).
Table 3.
Top 5 GO terms of up-and downregulated target genes in IAs and STAs.
| GO term | Gene count | Fold enrichment | P value* |
|---|---|---|---|
| Upregulated target genes | |||
| GO: 0006954: Inflammatory response | 75 | 3.4 | 1.4E-21 |
| GO: 0051607: Defense response to virus | 52 | 5.1 | 1.1E-23 |
| GO: 0060337: Type I interferon | 31 | 8.4 | 6.9E-22 |
| GO: 0005164: TNF receptor binding | 14 | 7.7 | 5.2E-10 |
| GO: 0042608: T cell receptor binding | 6 | 15.2 | 2.0E-07 |
| Downregulated target genes | |||
| GO: 0007155: Cell adhesion | 44 | 2.4 | 6.7E-08 |
| GO: 0031012: Extracellular matrix | 42 | 3.5 | 1.0E-12 |
| GO: 0005856: Cytoskeleton | 41 | 2.8 | 2.8E-09 |
| GO: 0003735: Ribosomal structure | 23 | 3.3 | 3.0E-07 |
| GO: 0051015: Actin filament binding | 22 | 3.1 | 1.9E-06 |
IAs vs. STA.
Figure 3.
Upregulated target genes in the NK cell-mediated cytotoxicity pathway. Target genes are marked with a green star. Image produced using DAVID.
Table 4.
Top 10 pathways of up-and downregulated target genes in IAs and STAs.
| KEGG term | Gene count | Fold enrichment | P value* |
|---|---|---|---|
| Upregulated target genes | |||
| hsa04650: NK cell-mediated cytotoxicity | 33 | 10.1 | 6.6E-11 |
| hsa04660: T cell receptor | 28 | 9.7 | 1.9E-10 |
| hsa04060: Cytokine-cytokine receptor interaction | 48 | 9.0 | 9.8E-10 |
| hsa04621: NOD-like receptor | 35 | 8.5 | 2.9E-09 |
| hsa04217: Necroptosis | 34 | 8.3 | 4.7E-09 |
| hsa05340: Primary immunodeficiency | 37 | 7.1 | 7.5E-08 |
| hsa05168: Chemokine | 35 | 7.6 | 2.3E-08 |
| hsa05167: NF-kappa B signaling pathway | 34 | 8.2 | 5.6E-09 |
| hsa04380: TNF signaling pathway | 26 | 7.0 | 9.0E-08 |
| hsa05340: Cell adhesion molecules | 14 | 7.3 | 4.0E-08 |
| Downregulated target genes | |||
| hsa03010: Ribosome | 21 | 6.0 | 9.2E-07 |
| hsa04520: Adherens junction | 12 | 4.5 | 2.9E-05 |
| hsa04923: Regulation of lipolysis in adipocytes | 10 | 4.2 | 5.4E-05 |
| hsa04360: Axon guidance | 19 | 4.0 | 8.9E-05 |
| hsa05012: Parkinson’s disease | 16 | 3.6 | <0.001 |
| hsa04360: Rap1 signaling pathway | 19 | 4.0 | 8.9E-05 |
| hsa05012: Vascular smooth muscle contraction | 16 | 3.6 | <0.001 |
| hsa05412: Oxidative phosphorylation | 11 | 3.8 | <0.001 |
| hsa05410: cGMP-PKG signaling pathway | 11 | 3.2 | <0.001 |
| hsa00280: Focal adhesion | 8 | 3.1 | <0.001 |
IAs vs. STAs.
Prediction of circRNA-miRNA-mRNA network
TargetScan and miRanda were utilized to predict miRNAs targeted by the differentially expressed circRNAs. The first 5 miRNAs combined with the 7 selected circRNAs were predicted using Tot Energy detection, in which lower Tot Energy is associated with a closer combination between circRNAs and miRNAs (Table 5). The miRNA-mRNA regulatory relationships were further identified using miRTarBase. The mRNAs were found to be significantly differentially expressed in RNA-Seq, and those predicted by the bioinformatics analyses were further selected for ceRNA construction. In this network, hsa_circ_0008433 can regulate inflammatory gene MMP2 by sponging hsa-miR-181c-5p and hsa-miR-181b-5p. Hsa_circ_0072309 can act as a ceRNA to affect SMC regulators DDR2 and SAT2 through hsa-miR-519e-5p and hsa-miR-516b-5p, respectively (Figure 4).
Table 5.
Predicted miRNAs for the 7 up- or downregulated circRNAs in IAs.
| circRNA ID | miRNA1 | miRNA2 | miRNA3 | miRNA4 | miRNA5 |
|---|---|---|---|---|---|
| hsa_circ_0008433 | hsa-miR-181c-5p | hsa-miR-499b-5p | hsa-miR-34c-5p | hsa-miR-499a | hsa-miR-181b-5p |
| hsa_circ_0033144 | hsa-miR-588 | hsa-miR-193a-3p | hsa-miR-361-3p | hsa-miR-125a-3p | hsa-miR-516a-3p |
| hsa_circ_0005571 | hsa-miR-371a-5p | hsa-miR-483-3p | hsa-miR-532-3p | hsa-miR-33b-5p | hsa-miR-216a-3p |
| hsa_circ_0040809 | hsa-miR-519d-5p | hsa-miR-525-5p | hsa-miR-520a-5p | hsa-miR-515-5p | hsa-miR-519e-5p |
| hsa_circ_0056285 | hsa-miR-708-5p | hsa-miR-28-5p | hsa-miR-539-3p | hsa-miR-485-3p | hsa-miR-328-3p |
| hsa_circ_0072309 | hsa-miR-519e-5p | hsa-miR-515-5p | hsa-miR-516b-5p | hsa-miR-603 | hsa-miR-584-3p |
| hsa_circ_0007142 | hsa-miR-128-3p | hsa-miR-216a-3p | hsa-miR-576-3p | hsa-miR-376a-3p | hsa-miR-376b-3p |
Figure 4.
Simplified sub-network containing high-score interactions of circRNA-miRNA-mRNA. The brown, red, and blue points represent circRNAs, miRNAs, and protein-coding RNAs, respectively.
Expression of candidate circRNAs in peripheral blood of IA population
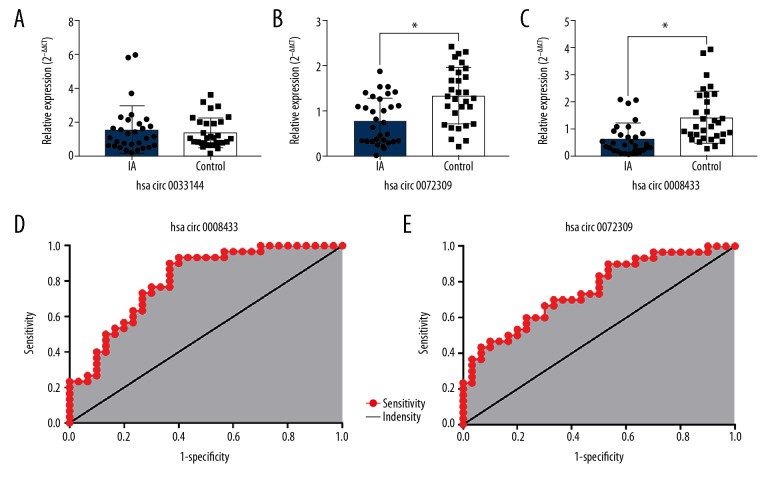
Based on the findings of sequencing, we selected 3 circRNAs (hsa_circ_0072309, hsa_circ_0008433, and hsa_circ_0033144) with significant difference in expression (P<0.05) in tissues and blood for further verification. In 30 pairs of IA patients and healthy people, the expression level of hsa_circ_0033144 in peripheral blood was not significantly different between the 2 groups (Figure 5A). Hsa_circ_0072309 and hsa_circ_0008433 still showed significant differences in expression between the 2 groups. The expression pattern of hsa_circ_0072309 in blood was consistent with that in tissues, while the expression trend of hsa_circ_0008433 in blood differed from that in tissues. This was consistent with the small samples test results (Figure 5B, 5C). Areas under the ROC curve of hsa_circ_0008433 and hsa_circ_0072309 were 0.8022 (95% CI: 0.6913~0.9132; P<0.001) and 0.7544 (95% CI: 0.6337~0.8752; P<0.001), respectively (Figure 5D, 5E).
Figure 5.
qRT-PCR validation of specific circRNAs expression in peripheral blood of IA patients and healthy people. (A–C) The expression of hsa_circ_0033144, hsa_circ_0072309, and hsa_circ_0008433 in peripheral blood in IA vs. control groups. * P<0.05. IA group (n=30) vs. control group (n=30). (D, E) The ROC curve of hsa_circ_0008433 and hsa_circ_0072309.
Discussion
Recent research has shown that circRNAs are involved in various neurological diseases [14]. In this study, we are the first to present expression profiles of circRNAs in IA, and revealed that most dysfunctional circRNAs are mainly involved in immunity/inflammation and cell adhesion/adherens junction processes that are known to be critical for the pathogenesis of IA [15,16].
Inflammatory response and degeneration of vascular media layer are the pathological basis for the development of IAs [17]. CircRNAs have been verified to exert significant roles in inflammation, including regulating inflammation genes, recruiting macrophages, and adjusting crucial signal pathways [18]. Our results suggest that several inflammation-related biological processes or pathways are potentially regulated by circRNAs. The most affected pathway was NK/T cell-mediated cytotoxicity. Infiltration of inflammatory cells is the primary pathological change in early stages of IAs [19]. Related studies also confirmed that NK cells intensively infiltrate in abdominal aortic aneurysm (AAA) and damage the vessel by producing cytotoxicity to arterial SMCs [20]. However, inflammatory cells release cytokines and promote the overexpression of MMPs, which degrades the extracellular matrix (ECM) and weakens vessels, and MMP2 and MMP9 are closely associated with formation of aneurysms [21,22]. According to our ceRNA network analysis, hsa_circ_0008433 targeted hsa-miR-181c-5p to regular MMP2, which has been found to be highly expressed in IA walls, and these result in arterial elastic fiber destruction and pathological remodeling of vessels [23].
The necrosis and apoptosis of SMCs contribute to the formation, progression, and rupture of IAs [24]. In our study, the necroptosis pathway enriched 34 genes targeted by several circRNAs. Among them, hsa_circ_0072309 regulates discoid domain receptor 2 (DDR2) and systemic amino acid transporter 2 (SAT2) through competitively sponging hsa-miR-519e-5p and hsa-miR-516b-5p, respectively. DDR2 is a critical factor in SMC-mediated regulation of collagen turnover in atherosclerosis, which is the pathological basis of IAs [25]. SAT2 regulates vascular SMCs growth and collagen synthesis by mediating l-valine transport and promotes pathological remodeling of vessels, which is one of the molecular mechanisms involved in vascular diseases [26]. Moreover, studies have shown that hsa_circ_0072309 is directly involved in the regulation of cell proliferation and migration by sponging miR-492 [27]. These findings elucidate the regulation of SMCs by circRNA in IAs.
The downregulated genes targeted by circRNAs in our research were most enriched in cell adhesion/adherens junction biological pathways, which are closely related to vascular strength and integrity. Numerous cell adhesion molecules, such as VCAM-1 and ICAM-1, participate in aneurysm production through recruiting NK/T cells and basophils [28]. Adherence junctions (AJs) are critical in maintaining the morphology and function of endothelial cells (ECs). Early hemodynamic changes result in abnormal expression of VE-cadherin/α-catenin mediated by p120-catenin, which promotes the development of IAs [29]. We found that lymphoid enhancer factor 1 (LEF1) and transforming growth factor beta induced TGFBI, which was targeted by several circRNAs in the ceRNA network. Binding sites of LEF1 act as promoter regions of fibronectin and α5/β1 integrin, which affects adherence junctions and is related to the completeness of cerebral vascular and blood-brain barriers. TGFBI acts on arterial ECM through cell adhesion to affect vascular stability [30]. These are the critical mechanisms for IAs and subarachnoid hemorrhage [31,32].
Finally, circRNA has superior structural stability and is released into extracellular spaces through exosomes, which can be detected in saliva, milk, and plasma [33]. These characteristics make it a good candidate for use as a disease biomarker. In this study, we verified the sequencing results in multiple samples (tissue and blood) and revealed the expression patterns of several critical IA-related circRNAs in center and periphery. Hsa_circ_0008433 and hsa_circ_0072309 were significantly differentially expressed in tissues and peripheral blood. The expression pattern of hsa_circ_0072309 in blood was highly consistent with that in tissues, which revealed its expression characteristics of central and peripheral homogeneity, and this could be very helpful for the noninvasive diagnosis of IA in the future. It remains unclear why hsa_circ_0008433 is expressed in blood as opposed to tissues, and this may be related to the heterogeneous expression of circRNA and the chemotaxis of central inflammation [34]. Further functional experiments are needed to confirm this speculation. Nevertheless, we still believe that these 2 circRNAs have broad research prospects and importance, and might provide crucial clues for the study of IA diagnostic and therapeutic markers.
However, due to the particularity of human IA samples and difficulty in collecting, the tissue sample sizes were relatively small. For this reason, we specifically collected tissue samples from the commonly preferred position of IA for RNA-Seq and included ruptured and unruptured patients to maximize objectivity and representativeness of the experiment. In addition, we also referred to other sequencing experiments of human and animal tissues that were the same or smaller than this sample size [35,36]. STAs as control materials are yet another limitation, although there are certain differences between STAs and intracranial arteries. The method of using STAs for similar experiments has been established previously, and is still the best choice [37]. Moreover, research is currently based on bioinformatics, and further in vitro and in vivo functional experiments are needed.
Conclusions
To the best of our knowledge, this is the first study to identify the differential expression profiles of circRNAs in IAs and reveal their expression patterns in center and periphery. Hsa_circ_0072309 and hsa_circ_0008433 are 2 novel and pivotal IA-related circRNAs that might be valuable for therapy and noninvasive diagnosis of IA in the future.
Footnotes
Source of support: Departmental sources
References
- 1.Vlak MH, Algra A, Brandenburg R, et al. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: A systematic review and meta-analysis. Lancet Neurol. 2011;10(7):626–36. doi: 10.1016/S1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwkamp DJ, Setz LE, Algra A, et al. Changes in case fatality of aneurismal subarachnoid haemorrhage over time, according to age, sex, and region: A meta-analysis. Lancet Neurol. 2009;8:635–42. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen I, Chen CY. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–79. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floris G, Zhang L, Follesa P. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54:5156–65. doi: 10.1007/s12035-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Cui L, Yuan J, et al. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem Biophys Res Commun. 2017;494:126–32. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 6.Hosaka K, Hoh BL. Inflammation and cerebral aneurysms. Transl Stroke Res. 2014;5:190–98. doi: 10.1007/s12975-013-0313-y. [DOI] [PubMed] [Google Scholar]
- 7.Starke RM, Chalouhi N, Ding D, et al. Vascular smooth muscle cells in cerebral aneurysm pathogenesis. Transl Stroke Res. 2014;5:338–46. doi: 10.1007/s12975-013-0290-1. [DOI] [PubMed] [Google Scholar]
- 8.‘t Hoen PA, Ariyurek Y, Thygesen HH, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakaoka H, Tajima A, Yoneyama T, et al. Gene expression profiling reveals distinct molecular signatures associated with the rupture of intracranial aneurysm. Stroke. 2014;45:2239–45. doi: 10.1161/STROKEAHA.114.005851. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Yue H, Hao Y, et al. Expression profile of long noncoding RNAs in human cerebral aneurysms: A microarray analysis. J Neurosurg. 2017;127:1055–62. doi: 10.3171/2016.9.JNS16839. [DOI] [PubMed] [Google Scholar]
- 11.Shi C, Awad IA, Jafari N, et al. Genomics of human intracranial aneurysm wall. Stroke. 2009;40(4):1252–61. doi: 10.1161/STROKEAHA.108.532036. [DOI] [PubMed] [Google Scholar]
- 12.Pera J, Korostynski M, Krzyszkowski T, et al. Gene expression profiles in human ruptured and unruptured intracranial aneurysms: What is the role of inflammation? Stroke. 2010;41(2):224–31. doi: 10.1161/STROKEAHA.109.562009. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: Novel regulators of neuronal development. Front Mol Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawlowska E, Szczepanska J, Wisniewski K, et al. NF-κB-mediated inflammation in the pathogenesis of intracranial aneurysm and subarachnoid hemorrhage. Does autophagy play a role? Int J Mol Sci. 2018;19(4):1245. doi: 10.3390/ijms19041245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Wang W, Zhang L, et al. Identification of a long non-coding RNA-associated competing endogenous RNA network in intracranial aneurysm. World Neurosurg. 2016;97:684–92. doi: 10.1016/j.wneu.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Chalouhi N, Hoh BL, Hasan D. Review of cerebral aneurysm formation, growth, and rupture. Stroke. 2013;44:3613–22. doi: 10.1161/STROKEAHA.113.002390. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Wang J, Wang S, et al. DNA methylation regulates gene expression in intracranial aneurysms. World Neurosurg. 2017;105:28–36. doi: 10.1016/j.wneu.2017.04.064. [DOI] [PubMed] [Google Scholar]
- 19.Liu YF, Zhang Y, Dai D, et al. Expression of NF-κB, MCP-1 and MMP-9 in a cerebral aneurysm rabbit model. Can J Neurol Sci. 2014;41:200–5. doi: 10.1017/s0317167100016589. [DOI] [PubMed] [Google Scholar]
- 20.Jagadesham VP, Scott DJA, Carding SR. Abdominal aortic aneurysms: An autoimmune disease? Trends Mol Med. 2008;14:522–29. doi: 10.1016/j.molmed.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Turjman AS, Turjman F, Edelman ER. Role of fluid dynamics and inflammation in intracranial aneurysm formation. Circulation. 2014;129:373–82. doi: 10.1161/CIRCULATIONAHA.113.001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Srivastava SK, Chaudhuri TK, et al. Multifaceted role of matrix metalloproteinases (MMPs) Front Mol Biosci. 2015;2:19. doi: 10.3389/fmolb.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L, Zhu YQ, Li MH, et al. Geometric, hemodynamic, and pathological study of a distal internal carotid artery aneurysm model in dogs. Stroke. 2013;44:2926–29. doi: 10.1161/STROKEAHA.113.002290. [DOI] [PubMed] [Google Scholar]
- 24.Starke RM, Chalouhi N, Ding D, et al. Vascular smooth muscle cells in cerebral aneurysm pathogenesis. Transl Stroke Res. 2014;5:338–46. doi: 10.1007/s12975-013-0290-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Yang J, Yang J, et al. Circular RNAs: Novel rising stars in cardiovascular disease research. Int J Cardiol. 2016;202:726–27. doi: 10.1016/j.ijcard.2015.10.051. [DOI] [PubMed] [Google Scholar]
- 26.Ensenat D, Hassan S, Reyna SV. Transforming growth factor-beta 1 stimulates vascular smooth muscle cell L-proline transport by inducing system A amino acid transporter 2 (SAT2) gene expression. Biochem J. 2001;360:507–12. doi: 10.1042/0264-6021:3600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–41. doi: 10.2147/CMAR.S186857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan J, Li X, Zhong L, et al. MCP-1, ICAM-1 and VCAM-1 are present in early aneurysmal dilatation in experimental rats. Folia Histochem Cytobiol. 2010;48:455–61. doi: 10.2478/v10042-010-0042-y. [DOI] [PubMed] [Google Scholar]
- 29.Zhao JL, Xiao ZP, Yu NZ, et al. Knockdown of P120 catenin aggravates endothelial injury under an impinging flow by inducing breakdown of adherens junctions. Mol Med Rep. 2019;19:541–48. doi: 10.3892/mmr.2018.9657. [DOI] [PubMed] [Google Scholar]
- 30.Ruigrok YM, Rinkel GJ, van’t Slot R, et al. Evidence in favor of the contribution of genes involved in the maintenance of the extracellular matrix of the arterial wall to the development of intracranial aneurysms. Hum Mol Genet. 2006;15:3361–68. doi: 10.1093/hmg/ddl412. [DOI] [PubMed] [Google Scholar]
- 31.Chang CC, Lee PS, Chou Y, et al. Mediating effects of aryl-hydrocarbon receptor and RhoA in altering brain vascular integrity: the therapeutic potential of statins. Am J Pathol. 2012;181:211–21. doi: 10.1016/j.ajpath.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 32.Wisniewska MB, Misztal K, Michowski W, et al. LEF1/beta-catenin complex regulates transcription of the Cav3.1 calcium channel gene (Cacna1g) in thalamic neurons of the adult brain. J Neurosci. 2010;30:4957–69. doi: 10.1523/JNEUROSCI.1425-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–84. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenner AJ, Stevens B, Woodruff TM. New tricks for an ancient system: Physiological and pathological roles of complement in the CNS. Mol Immunol. 2018;102:3–13. doi: 10.1016/j.molimm.2018.06.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J, Chen GJ, Jing YW, et al. LncRNA expression profile of human thoracic aortic dissection by high-throughput sequencing. Cell Physiol Biochem. 2018;46:1027–41. doi: 10.1159/000488834. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesha SH, Dudics S, Song Y, et al. The miRNA expression profile of experimental autoimmune encephalomyelitis reveals novel potential disease biomarkers. Int J Mol Sci. 2018;19(12) doi: 10.3390/ijms19123990. pii: E3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassam AB, Horowitz M, Chang YF, et al. Altered arterial homeostasis and cerebral aneurysms: A molecular epidemiology study. Neurosurgery. 2004;56(5):1450–60. doi: 10.1227/01.neu.0000125005.67850.f8. [DOI] [PubMed] [Google Scholar]