Abstract
Hereditary hemochromatosis (HH) is a genetic disorder that causes excess absorption of iron and can lead to a variety of complications including liver cirrhosis, arthritis, abnormal skin pigmentation, cardiomyopathy, hypogonadism, and diabetes. Hemojuvelin (HJV) is the causative gene of a rare subtype of HH worldwide. This study aims to systematically review the genotypic and phenotypic spectra of HJV-HH in multiple ethnicities, and to explore the genotype–phenotype correlations. A comprehensive search of PubMed database was conducted. Data were extracted from 57 peer-reviewed original articles including 132 cases with HJV-HH of multiple ethnicities, involving 117 biallelic cases and 15 heterozygotes. Among the biallelic cases, male and female probands of Caucasian ancestry were equally affected, whereas males were more often affected among East Asians (P=1.72×10-2). Hepatic iron deposition and hypogonadism were the most frequently reported complications. Hypogonadism and arthropathy were more prevalent in Caucasians than in East Asians (P=9.30×10-3, 1.69×10-2). Among the recurrent mutations, G320V (45 unrelated cases) and L101P (7 unrelated cases) were detected most frequently and restricted to Caucasians. [Q6H; C321*] was predominant in Chinese patients (6 unrelated cases). I281T (Chinese and Greek), A310G (Brazilian and African American), and R385* (Italian and North African) were reported across different ethnicities. In genotype–phenotype correlation analyses, 91.30% of homozygotes with exon 2-3 mutations developed early-onset HH compared to 66.00% of those with exon 4 mutations (P=2.40×10-2). Hypogonadism occurred more frequently in homozygotes with missense mutations (72.55%) than in those with nonsense mutations (35.71%; P=2.43×10-2). Liver biopsy was accepted by more probands with frame-shift or missense mutations (85.71% and 60.78%, respectively) than by those with nonsense mutations (28.57%; P=2.37×10-2, 3.93×10-2). The present review suggests that patients’ ethnicity, geographical region, and genetic predisposition should be considered in the diagnosis, prognosis and management of HJV-HH.
Electronic supplementary material
The online version of this article (10.1186/s13023-019-1097-2) contains supplementary material, which is available to authorized users.
Keywords: Iron overload, Hereditary hemochromatosis, Juvenile hemochromatosis, Hemojuvelin
Background
Hereditary hemochromatosis (HH) is a genetic disorder characterized by disturbed regulation of iron, which leads to systemic iron overload and a wide spectrum of serious complications including cardiomyopathy, liver cirrhosis, arthritis, and skin pigmentation, as well as endocrine disorders such as diabetes, hypogonadotropic hypogonadism, hypothyroidism, etc. [1, 2]. To date, five genes, HFE, HAMP, HJV, TFR2, and SLC40A1, have been identified as causative genes for various HH subtypes [2, 3].
The hemojuvelin gene (HJV) maps to chromosome 1q21.1, and its coding sequence consists of three exons (exon 2, 3 and 4). HJV encodes a co-receptor of bone morphogenetic proteins (BMPs) that regulates the circulating level of hepcidin, the principal hormone in the mediation of iron homeostasis [4]. Pathogenic mutations in the HJV gene cause hemochromatosis in an autosomal recessive hereditary pattern. Mutation of this gene was first identified in Caucasian families with primary hemochromatosis in 2004 [5]. A previous study based on public databases indicated that the homozygous or compound heterozygous status of HJV pathogenic mutations is estimated to cause iron overload in approximately 1 in 5–6 million people worldwide [3]. Biallelic HJV mutations were estimated to cause up to 90% of the juvenile hemochromatosis (JH, also known as type 2 HH), which is the most severe HH form with an onset before 30 years of age [4]. If not appropriately treated, JH can lead to death from hemochromatosis-related cardiomyopathy [6]. In addition to the autosomal recessive mode of inheritance, studies in recent years also reported that HJV heterozygotes can develop middle age-onset hemochromatosis [7–12]. The phenotypic spectrum and the outcomes in HJV-HH cases are of special interest for clinicians, as this information could better guide the diagnosis, prognosis and management of the patients.
During the past 15 years, more than 70 genetic variants related to HJV-HH were identified in patients with iron-overload, including non-synonymous, non-sense, frame-shift and in-frame mutations in the coding region of exons 2–4, as well as mutations in the 5′ untranslated region (UTR) and intron. About two-thirds of the mutations were identified in a single proband or family, and recurrent mutations also were detected. Most of the recurrent mutations were restricted to the given race and origin of the family and were diverse and distinct among geographic regions around the world. For example, G320V, the most commonly reported mutation in HJV, has been frequently reported in Caucasians (mainly in the North European population), but has never been detected in East Asians [13, 14]. C321*, the most frequently reported mutation in Chinese patients, has not been found in patients of any other race [13]. Given such a wide spectrum of mutations, diagnosis of HJV-HH can be challenging. Therefore, the phenotypic variations with respect to ethnicity and genotype need to be investigated in the pursuit for more personalized screening and management strategies for HJV-HH.
In the present review, we complied cases of HJV-HH published in peer-reviewed journals worldwide in order to summarize the genotypic and phenotypic spectra of HJV-HH for multiple ethnicities, as well as the outcomes, for the purpose of improving the current understanding of the disease. This review also provides a comparison of phenotypes between Caucasian and East Asian patients, along with a comparison between early-onset and late-onset HJV-HH. All HJV mutations described to date were reviewed in terms of ethnic and geographic association. Genotype–phenotype correlations were explored. As HJV-HH is a rare hereditary disease in both Caucasians and non-Caucasians, this review provides valuable information for identifying HJV-related iron overload both before and after genetic testing.
Materials and methods
Literature search strategy
To identify all published HH cases that were genetically confirmed to be HJV pathogenic mutation cases (OMIM *608374), we conducted an extensive literature review, which included a comprehensive search of the National Center for Biotechnology Information PubMed database (http://www.ncbi.nlm.nih.gov/pubmed) for articles published through March 20, 2019. The search strategy consisted of multiple queries combining “hemojuvelin” or “HJV” or “HFE2” or “Hemochromatosis, type 2” or “juvenile hemochromatosis” without a language restriction. A list of the search terms employed in the database searches is presented in Additional file 1. A manual search of references from articles including relevant review articles, systematic reviews and meta-analyses was performed. The titles and abstracts of the 546 generated articles were carefully screened to remove the obviously ineligible studies.
Two investigators independently assessed the eligibility of the remaining studies by reviewing their full text. Only peer-reviewed publications providing both genotype and phenotype data related to HJV hemochromatosis were included in this review. Finally, 57 articles were included for further investigation. A list of the excluded articles with the associated reasons for exclusion is provided in Additional file 2.
Duplicate exclusion
To minimize the bias through reporting of the same case from multiple publications, we used the mutation information (both cDNA and protein change), sex, age at onset, serum ferritin (SF) and transferrin saturation (TS) measurements to identify duplicated cases. When a case was included in more than one article, we included only the publication with more detailed case information in the analysis, and all reference was assigned to the individual. Seven cases were found to be reported in more than one publication.
Eligibility assessment
Among the 167 cases with primary iron overload reported by the 57 eligible publications, six cases that also had genetically diagnosed alpha-thalassemia, beta-thalassemia or congenital dyserythropoietic anemia II were excluded [7, 15–18], and four cases with unavailable sex information were excluded [5]. An additional 25 cases with compound heterozygous or homozygous status of pathogenic mutations in other HH genes (e.g., C282Y, H63D, S65C of HFE gene) were removed [8, 9, 19–28]. Thus, 132 eligible iron overload cases were included for data extraction.
Data extraction
Using a standardized data collection form, the following data from the eligible cases regarding the genetic, phenotypic, demographic and clinical outcomes of individual patients were manually extracted and compiled in a database: genotype of HJV and other HH-related genes, sex, age at diagnosis, age at presentation, SF, TS, heart disease, skin hyperpigmentation (including freckled skin), arthropathy, hypogonadism, diabetes or glucose intolerance, osteopathy, thyroid abnormality, abnormality on liver function test, liver iron deposition, liver fibrosis or cirrhosis, therapies and outcomes. Early-onset was defined by an age at presentation of 30 years or younger; all other ages were considered late-onset. If the age at presentation was not provided, the age at diagnosis was used for classification as early-onset or late-onset. The pathogenicity of the reported HJV variants was assessed based on the available phenotypic data and the opinions of the study authors. All data were extracted and verified by two investigators.
Annotation and prediction of the pathogenicity of variants
The variants were mapped to the reference genome (GRCh37/hg19 and GRCh38/hg38) and annotated in the following public databases: 1000 genome (http://www.internationalgenome.org/), Exome Sequencing Project (ESP; Exome Variant Server, NHLBI GO ESP, Seattle, WA; http://evs.gs.washington.edu/EVS/), and Exome Aggregation Consortium (ExAC; Cambridge, MA; http://exac.broadinstitute.org/) data sets. Two computational algorithms, PROVEAN v1.1.3 (http://provean.jcvi.org/index.php) and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), were applied to predict the deleterious effects of the missense mutations.
Nomenclature
The variants were named according to the GenBank HJV reference sequences NM_213653.3 and NP_998818.1 (http://www.ncbi.nlm.nih.gov), following the guideline provided by the Human Genome Variation Society (http://www.hgvs.org/).
Statistical analysis
Statistical analyses were performed using SAS (version 9.2; SAS Institute, Cary, NC, USA). Differences among continuous data were analyzed using factorial analysis of variance (ANOVA) or Student’s t-test. Non-parametric tests were utilized when the normality assumption was not met. Differences among categorical variables were analyzed using chi-squared test of independence or Fisher's exact test.
Results
Literature search and inclusion of cases
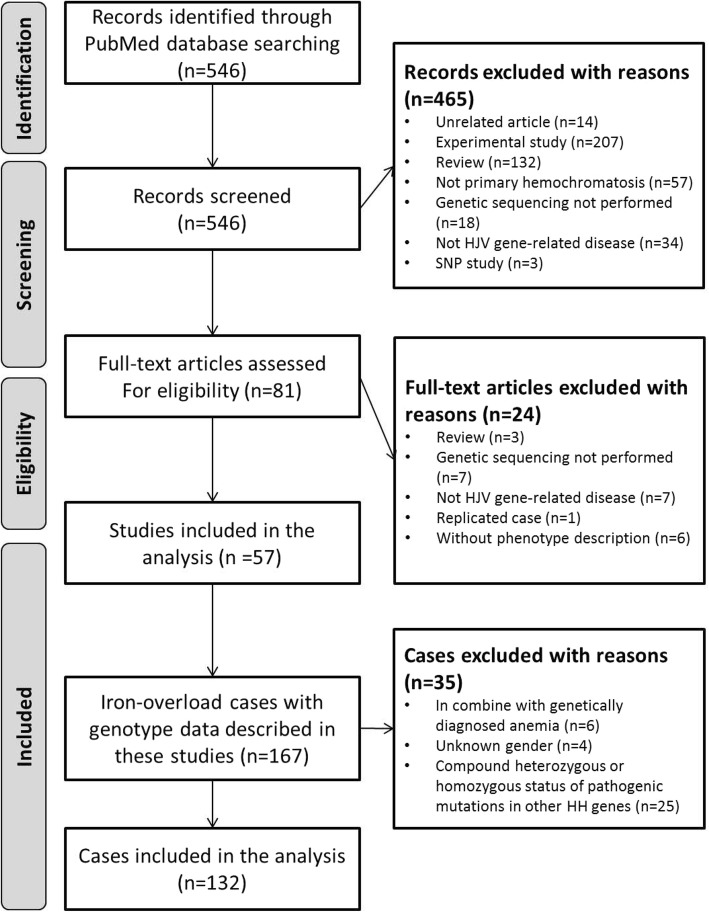
The process used to search the literature and identify eligible studies is described in the flow chart in Fig. 1. The search strategy initially identified 546 unique articles. Based on screening of titles and abstracts, 81 were kept for further evaluation. On examination of the full text, 57 articles met the inclusion criteria, and a total of 132 cases were eligible for data extraction. These cases included 117 cases of biallelic mutation and 15 cases with heterozygous mutation. The clinical characteristics of all 132 HJV-variant cases are presented in Table 1, and the extracted information for each patient was collected in Additional files 3 and 4.
Fig. 1.
Flow chart of literature search and study selection
Table 1.
Clinical characteristics of the included HJV-HH cases
| HJV-HH Cases with biallelic mutations | HJV-HH Cases with a single mutation | P | |||||
|---|---|---|---|---|---|---|---|
| All | Probands | Non-probands | All | Probands | Non-probands | ||
| N | 117 | 97 | 20 | 15 | 9 | 6 | |
| Male, n (%) | 61 (52.14) | 52 (53.61) | 9 (45.00) | 10 (66.67) | 5 (55.56) | 5 (83.33) | 9.11 × 10-1C |
|
Ethnicities Caucasian/East Asian/African/ND, (%) |
98/18/1/0 (83.76/15.38/8.55/0.00) | 83/13/1/0 (85.57/13.40/1.03/0.00) | 15/5/0/0 (75.00/25.00/0.00/0.00) | 3/9/0/3 (20.00/60.00/0.00/20.00) | 2/4/0/3 (22.22/44.44/0.00/33.33) | 1/5/0/0 (16.67/83.33/0.00/0.00) | 6.71 × 10 -3 F a |
| Homozygotes, n (%) | 89 (76.07) | 73 (75.26) | 16 (80.00) | — | — | — | — |
| Age at diagnosis (year) | 25.00 (20.00, 32.00) | 26.00 (21.00, 32.00) | 21.00 (16.00, 28.50) | 53.00 (36.00, 65.00) | 58.00 (36.00, 62.00) | 48.00 (46.00, 70.00) | 8.02 × 10 -4 W |
| Age at presentation (year) | 21.50 (18.00, 28.00) | 22.50 (19.50, 27.50) | 16.50 (13.00, 32.00) | — | — | — | — |
| Disease onset before 30 years, n (%) | 88 (75.21) | 72 (74.23) | 16 (80.00) | 4 (26.67) | 3 (33.33) | 1 (16.67) | 1.75 × 10 -2 F |
| Serum parameters at presentation | |||||||
| Serum ferritin (ng/ml) | 3541.80 (2270.00, 5293.00) | 3700.00 (2329.00, 5520.00) | 1971.50 (1117.50, 4314.00) | 444.00 (356.00, 1402.00) | 784.50 (388.50, 1512.50) | 382.50 (266.00, 554.00) | 2.70 × 10 -5 W |
| Transferrin saturation (%) | 94.00 (90.00, 100.00) | 95.00 (89.00, 100.00) | 92.00 (90.00, 97.00) | 49.00 (40.00, 68.00) | 64.00 (47.00, 83.00) | 40.00 (34.00, 46.00) | 1.03 × 10 -4 W |
| Complications | |||||||
| Cardiomyopathy, n (%) | 40 (34.19) | 36 (37.11) | 4 (20.00) | 1 (6.67) | 1 (11.11) | 0 (0.00) | 1.57 × 10-1F |
| Skin hyperpigmentation, n (%) | 48 (41.03) | 40 (41.24) | 8 (40.00) | 2 (13.33) | 1 (11.11) | 1 (16.67) | 1.49 × 10-1F |
| Arthropathy, n (%) | 32 (27.35) | 27 (27.84) | 5 (25.00) | 1 (6.67) | 1 (11.11) | 0 (0.00) | 4.40 × 10-1F |
| Endocrine abnormality | |||||||
| Hypogonadism, n (%) | 70 (59.83) | 63 (64.95) | 7 (35.00) | 2 (13.33) | 1 (11.11) | 1 (16.67) | 2.95 × 10 -3 F |
| Glucose intolerance, n (%) | 36 (30.77) | 33 (34.02) | 3 (15.00) | 2 (13.33) | 1 (11.11) | 1 (16.67) | 2.66 × 10-1F |
| Osteopathy, n (%) | 11 (9.40) | 7 (7.22) | 4 (20.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1.00 × 10-1F |
| Thyroid abnormality, n (%) | 9 (7.69) | 7 (7.22) | 2 (10.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1.00 × 10-1F |
| Liver disease | |||||||
| Abnormal liver function test, n (%) | 39 (33.33) | 35 (36.08) | 4 (20.00) | 4 (26.67) | 3 (33.33) | 1 (16.67) | 1.00 × 10-1F |
| Liver iron deposition, n (%) | 74 (63.25) | 67 (69.07) | 7 (35.00) | 4 (26.67) | 4 (44.44) | 0 (0.00) | 1.53 × 10-1F |
| Liver fibrosis, n (%) | 43 (36.75) | 39 (40.21) | 4 (20.00) | 1 (6.67) | 1 (11.11) | 0 (0.00) | 1.49 × 10-1F |
| Liver cirrhosis, n (%) | 28 (23.93) | 26 (26.80) | 2 (10.00) | 1 (6.67) | 1 (11.11) | 0 (0.00) | 4.43 × 10-1F |
| Liver biopsy, n (%) | 62 (52.99) | 56 (57.73) | 6 (30.00) | 3 (20.00) | 3 (33.33) | 0 (0.00) | 1.81 × 10-1F |
|
Therapy Phlebotomy/Chelating agent/Phlebotomy & Chelating agent/ND, n (%) |
60/3/7/47 (51.28/2.56/5.98/40.17) | 46/3/7/41 (47.42/3.09/7.22/42.27) | 14/0/0/6 (70.00/0.00/0.00/30.00) | 3/0/0/12 (20.00/0.00/0.00/80.00) | 2/0/0/7 (22.22/0.00/0.00/77.78) | 1/0/0/5 (16.67/0.00/0.00/83.33) | — |
Abbreviations: HJV-HH HJV-related hereditary hemochromatosis, ND not described
Data are shown as n (%) or median (interquartile range). P values were calculated to assess the differences between probands with biallelic mutations and probands with a single mutation using chi-square test, Fisher’s exact test or Wilcoxon test as appropriate. C, on chi-square test; F, on Fisher’s exact test; W, on Wilcoxon test. a, compared the proportions of Caucasians and East Asians. P values <0.05 are denoted in bold and underlined
Phenotypic spectrum of the biallelic HJV mutation cases
As shown in Table 1, 97 of the 117 cases with biallelic mutations were the probands of their family with comparable proportions of males (53.61%) and females (46.39%). Of the probands, 83 were Caucasians, 13 were East Asians, and one was African. Seventy-three of the probands had homozygous mutation, and 24 had compound heterozygous mutation. Presentation of iron overload or its related traits occurred at age 30 years or earlier in 72 probands, whereas 25 cases were late-onset. Notably, hepatic iron deposition and hypogonadism were the most frequently reported complications. Moreover, more than one-third of cases had complications including liver fibrosis, skin hyperpigmentation, abnormal liver function test results, cardiomyopathy, and impaired glucose regulation. Arthropathy, liver cirrhosis, osteopathy, and thyroid abnormality were also reported.
We further compared the phenotypes of the early-onset and late-onset probands (Table 2). The early-onset probands showed a significantly higher prevalence of hypogonadism (75.00%) than the late-onset probands (36.00%; P=4.30×10-4). Liver iron deposition was more frequently detected in late-onset cases (96.00%) and was significantly higher in these cases than in the early-onset ones (59.72%; P=7.22×10-4). A greater proportion of late-onset cases developed glucose intolerance (including diabetes; 48.00%) compared with that among early-onset cases (29.17%), but the difference was not statistically significant (P=8.68×10-2).
Table 2.
Comparisons of the clinical characteristics of early-onset versus late-onset HJV-HH cases with biallelic mutations
| Probands | Non-probands | |||||
|---|---|---|---|---|---|---|
| Onset age ≤30 years | Onset age >30 years | P | Onset age ≤30 years | Onset age >30 years | P | |
| N | 72 | 25 | 16 | 4 | ||
| Male, n (%) | 37 (51.39) | 15 (60.00) | 4.57 × 10-1C | 8 (50.00) | 1 (25.00) | 5.91 × 10-1F |
| Homozygotes, n (%) | 54 (75.00) | 19 (76.00) | 9.21 × 10-1C | 12 (75.00) | 4 (100.00) | 5.38 × 10-1F |
| Age at diagnosis (year) | 24.00 (19.50, 27.50) | 39.00 (32.00, 49.00) | 1.53 × 10 -12 W | 19.00 (16.00, 22.50) | 45.50 (36.00, 53.00) | 2.86 × 10 -3 W |
| Age at presentation (year) | 21.00 (18.00, 25.00) | 33.50 (32.00, 39.00) | 7.10 × 10 -7 W | 15.00 (12.00, 17.00) | 34.00 (32.00, 49.00) | 2.18 × 10 -2 W |
| Serum parameters at presentation | ||||||
| Serum ferritin (ng/ml) | 3520.90 (2291.50, 5782.50) | 3987.00 (2680.00, 4959.00) | 6.16 × 10-1W | 1971.50 (1117.50, 3498.50) | 2753.50 (869.50, 6981.50) | 9.52 × 10-1W |
| Transferrin saturation (%) | 95.50 (89.00, 100.00) | 93.50 (89.00, 99.50) | 4.73 × 10-1W | 92.00 (90.00, 98.00) | 92.00 (75.00, 94.90) | 5.46 × 10-1W |
| Complications | ||||||
| Cardiomyopathy, n (%) | 29 (40.28) | 7 (28.00) | 2.74 × 10-1C | 3 (18.75) | 1 (25.00) | 1.00 × 10-0F |
| Skin hyperpigmentation, n (%) | 29 (40.28) | 11 (44.00) | 7.45 × 10-1C | 6 (37.50) | 2 (50.00) | 1.00 × 10-0F |
| Arthropathy, n (%) | 18 (25.00) | 9 (36.00) | 2.90 × 10-1C | 4 (25.00) | 1 (25.00) | 1.00 × 10-0F |
| Endocrine abnormality | ||||||
| Hypogonadism, n (%) | 54 (75.00) | 9 (36.00) | 4.30 × 10 -4 C | 6 (37.50) | 1 (25.00) | 1.00 × 10-0F |
| Glucose intolerance, n (%) | 21 (29.17) | 12 (48.00) | 8.68 × 10-2C | 2 (12.50) | 1 (25.00) | 5.09 × 10-1F |
| Osteopathy, n (%) | 6 (8.33) | 1 (4.00) | 6.73 × 10-1F | 4 (25.00) | 0 (0.00) | 5.38 × 10-1F |
| Thyroid abnormality, n (%) | 4 (5.56) | 3 (12.00) | 3.69 × 10-1F | 3 (18.75) | 1 (25.00) | 1.00 × 10-0F |
| Liver disease | ||||||
| Abnormal liver function test, n (%) | 23 (31.94) | 12 (48.00) | 1.50 × 10-1C | 3 (18.75) | 1 (25.00) | 1.00 × 10-0F |
| Liver iron deposition, n (%) | 43 (59.72) | 24 (96.00) | 7.22 × 10 -4 C | 6 (37.50) | 1 (25.00) | 1.00 × 10-0F |
| Liver fibrosis, n (%) | 31 (43.06) | 8 (32.00) | 3.31 × 10-1C | 3 (18.75) | 1 (25.00) | 1.00 × 10-0F |
| Liver cirrhosis, n (%) | 20 (27.78) | 6 (24.00) | 7.13 × 10-1C | 1 (6.25) | 1 (25.00) | 3.68 × 10-1F |
| Liver biopsy, n (%) | 41 (56.94) | 15 (60.00) | 7.90 × 10-1C | 5 (31.25) | 1 (25.00) | 1.00 × 10-0F |
|
Therapy Phlebotomy/Chelating agent/Phlebotomy & Chelating agent/ND, n (%) |
28/2/6/36 (38.89/2.78/8.33/50.00) | 18/1/1/5 (72.00/4.00/4.00/20.00) | — | 11/0/0/5 (68.75/0.00/0.00/31.25) | 3/0/0/1 (75.00/0.00/0.00/25.00) | — |
Abbreviations: HJV-HH HJV-related hereditary hemochromatosis, ND not described
Data are shown as n (%) or median (interquartile range). P values were calculated to assess the differences between cases with onset age ≤30 years and cases with onset age >30 years among probands or non-probands using chi-square test, Fisher’s exact test or Wilcoxon test as appropriate. C, on chi-square test; F, on Fisher’s exact test; W, on Wilcoxon test. P values <0.05 are denoted in bold and underlined
Comparisons of phenotypes between Caucasian and East Asian probands with biallelic HJV mutation
In Caucasian probands with biallelic HJV mutation, similar proportions of male (48.19%) and female (51.81%) were observed, whereas the proportion of male cases (84.62%) was significantly greater among East Asians (P=1.72×10-2). The age at diagnosis of iron overload and the age at the first presentation of symptoms, as well as the pretreatment levels of SF and TS, were comparable between the populations. Notably, hypogonadism was diagnosed in 71.08% of the Caucasian probands, but in only 33.33% of the East Asians patients (P=9.30×10-3). Also, 32.53% of the Caucasian probands, but none of the East Asian probands, complained of arthralgia or were diagnosed with arthropathy such as arthritis (P=1.69×10-2). In addition, East Asian probands showed higher prevalence rates of complications including liver iron deposition, abnormal liver function test results, glucose intolerance, and cardiomyopathy than did Caucasian probands, whereas osteopathy and thyroid involvement were only reported in Caucasian probands. However, the prevalence rates for these complications were not statistically different between the populations (Table 3).
Table 3.
Comparisons of the clinical characteristics of Caucasian versus East Asian HJV-HH cases with biallelic mutations
| Probands | Non-probands | |||||
|---|---|---|---|---|---|---|
| Caucasians | East Asians | P | Caucasians | East Asians | P | |
| N | 83 | 13 | 15 | 5 | ||
| Male, n (%) | 40 (48.19) | 11 (84.62) | 1.72 × 10 -2 F | 6 (40.00) | 3 (60.00) | 6.17 × 10-1F |
| Homozygotes, n (%) | 64 (77.11) | 8 (61.54) | 3.00 × 10-1F | 12 (80.00) | 4 (80.00) | 1.00 × 10-1F |
| Age at diagnosis (year) | 26.00 (21.00, 32.00) | 26.00 ( 19.00, 37.00) | 6.72 × 10-1W | 20.00 ( 16.00, 24.00) | 27.00 ( 22.00, 51.00) | 8.03 × 10-2W |
| Age at presentation (year) | 22.50 (20.00, 27.00) | 21.50 ( 14.00, 48.00) | 8.73 × 10-1W | — | — | — |
| Disease onset before 30 years, n (%) | 63 (75.90) | 8 (61.54) | 3.13 × 10-1F | 13 (86.67) | 3 (60.00) | 2.49 × 10-1F |
| Serum parameters at presentation | ||||||
| Serum ferritin (ng/ml) | 3553.00 (2329.00, 5293.00) | 4974.00 (2337.00,6678.00) | 2.21 × 10-1W | 1648.00 (1117.50, 3498.50) | 3389.00 (1505.00,5273.50) | 6.71 × 10-1W |
| Transferrin saturation (%) | 95.00 (88.00, 100.00) | 94.40 ( 92.05, 97.55) | 9.80 × 10-1W | 92.00 (80.00, 98.00) | 93.70 ( 91.00, 95.60) | 8.01 × 10-1W |
| Complications | ||||||
| Cardiomyopathy, n (%) | 29 (34.94) | 7 (53.85) | 2.26 × 10-1F | 3 (20.00) | 1 (20.00) | 1.00 × 10-1F |
| Skin hyperpigmentation, n (%) | 35 (42.17) | 5 (38.46) | 1.00 × 10-0F | 6 (40.00) | 2 (40.00) | 1.00 × 10-1F |
| Arthropathy, n (%) | 27 (32.53) | 0 (0.00) | 1.69 × 10 -2 F | 5 (33.33) | 0 (0.00) | 2.66 × 10-1F |
| Endocrine abnormality | ||||||
| Hypogonadism, n (%) | 59 (71.08) | 4 (30.77) | 9.30 × 10 -3 F | 6 (40.00) | 1 (20.00) | 6.13 × 10-1F |
| Glucose intolerance, n (%) | 26 (31.33) | 7 (53.85) | 1.27 × 10-1F | 1 (6.67) | 2 (40.00) | 1.40 × 10-1F |
| Osteopathy, n (%) | 7 (8.43) | 0 (0.00) | 5.88 × 10-1F | 4 (26.67) | 0 (0.00) | 5.30 × 10-1F |
| Thyroid abnormality, n (%) | 7 (8.43) | 0 (0.00) | 5.88 × 10-1F | 2 (13.33) | 0 (0.00) | 1.00 × 10-1F |
| Liver disease | ||||||
| Abnormal liver function test, n (%) | 28 (33.73) | 7 (53.85) | 2.17 × 10-1F | 2 (13.33) | 2 (40.00) | 2.49 × 10-1F |
| Liver iron deposition, n (%) | 55 (66.27) | 11 (84.62) | 3.34 × 10-1F | 5 (33.33) | 2 (40.00) | 1.00 × 10-1F |
| Liver fibrosis, n (%) | 36 (43.37) | 3 (23.08) | 2.29 × 10-1F | 4 (26.67) | 0 (0.00) | 5.30 × 10-1F |
| Liver cirrhosis, n (%) | 22 (26.51) | 4 (30.77) | 7.45 × 10-1F | 1 (6.67) | 1 (20.00) | 4.47 × 10-1F |
| Liver biopsy, n (%) | 48 (57.83) | 8 (61.54) | 1.00 × 10-1F | 4 (26.67) | 2 (40.00) | 6.13 × 10-1F |
|
Therapy Phlebotomy/Chelating agent/Phlebotomy & Chelating agent/ND, n (%) |
41/2/6/34 (49.40/2.41/7.23/40.96) | 5/0/1/7 (38.46/0.00/7.69/53.85) | — | 12/0/0/3 (80.00/0.00/0.00/20.00) | 2/0/0/3 (40.00/0.00/0.00/60.00) | — |
Abbreviations: HJV-HH HJV-related hereditary hemochromatosis, ND not described
Data are shown as n (%) or median (interquartile range). P values were calculated to assess the differences between Caucasians and East Asians using Fisher’s exact test or Wilcoxon test as appropriate. F, on Fisher’s exact test; W, on Wilcoxon test. P values <0.05 are denoted in bold and underlined
Phenotypic spectrum of the monoallelic HJV mutation cases
As shown in Table 1, 15 monoallelic HJV mutation-related iron-overload cases were reported, including nine probands (5 men and 4 women) and six of their relatives. Two of the probands were Caucasian, four were East Asian, and three originated from Brazil with no race information provided. Early-onset occurred in only three of the probands. Signs of organ damage other than hepatic iron deposition and abnormal liver function were seldom reported in these patients (Table 1).
The ethnicity distributions were significantly different between the monoallelic mutation probands and biallelic mutation probands (P=6.71×10-3). As expected, the SF and TS levels at presentation for the monoallelic mutation probands were significantly lower than those for biallelic mutation probands (P=2.70×10-5, 1.03×10-4). The age at diagnosis among the monoallelic mutation probands was later (P=8.02×10-4), and more individuals with late-onset were identified (P=1.75×10-2). Iron overload-related complications were less prevalent in monoallelic mutation cases than in biallelic mutation cases, and this was especially true for hypogonadism (P=2.95×10-3; Table 1).
Mutation profiles in different ethnicities
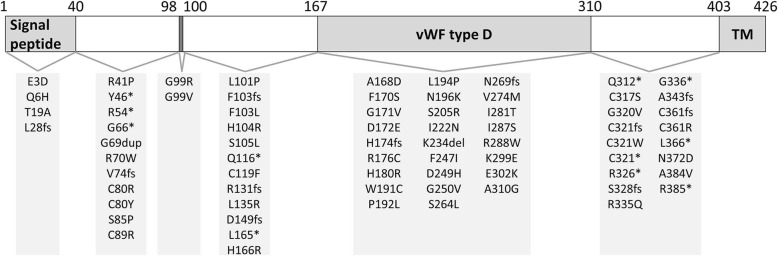
In total, 72 variants in the coding sequence as well as 1 mutation in the splicing site, 2 mutations in the non-coding region of 5′ UTR, and 1 case of whole gene deletion were reported in HJV patients as related to hemochromatosis before March 20, 2019 in publications found in the PubMed database. The locations of the mutations in the HJV gene are shown in Fig. 2.
Fig. 2.
Structure of the HJV protein and mutations in HJV. Abbreviations: TM, transmembrane domain; vWF type D: von Willebrand factor type D domain
The 72 coding sequence variants included 11 frame-shift mutations, 11 nonsense mutations, 48 missense mutations, 1 deletion, and 1 insertion. Respectively, 4, 37 and 31 mutations were located in exons 2, 3, and 4. Except for A310G, the variants in the coding sequence were either absent in the databases or reported with a very low minor allele frequency.
A detailed list of the information and family origin of each variant is provided in Table 4. Twenty-six of the reported mutations were observed in more than one family. Three mutations were reported across different ethnicities, including I281T (in Chinese and Greek), A310G (in Brazilian and African American), and R385* (in Italian and North African). G320V, L101P, D149fs, E302K, G336*, G99R, and R176C were the most frequently recurrent variations in Caucasians, and each mutation was detected in at least three unrelated probands. In East Asians, HJV mutations were reported only in families of Chinese or Japanese origin. [Q6H; C321*] in cis was the most frequently observed mutation in Chinese families with hemochromatosis. E3D was identified in five families of Chinese origin. Both Q6H and E3D were predicted as benign by bioinformatics tools. I281T was reported in two families from China. D249H and Q312* were the most frequently reported mutations in Japanese cases, and Q312* was also observed in the Chinese population. R385* and A310G were the commonest variations in the African population, and both were also reported in Caucasians from Brazil or Italy. However, very few cases of African ancestry that underwent genetic diagnosis have been published.
Table 4.
Information for HJV mutations
| Nucleotide change | Amino acid change | Coordinates (GRCh37/hg19) |
RSID | Allele frequency reported | Bioinformatics prediction of missense variant | Number of reported probands | Family of origin | Ethnicity reported | Reference | Included and excluded cases in the present study ref | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1000G | ESP | ExAC | GnomAD | PolyPhen-2 (HumDiv) | PolyPhen-2 (HumVar) | Provean | |||||||||
| c.9G>C |
p.Glu3Asp (p.E3D) |
chr1:145414790 | rs12025510 | 3.00×10-3 | NA | 1.25×10-3 | 1.19×10-3 | Benign | Benign | Neutral | 5 | China | East Asian | 9 | Included: Heterozygote: 5 [9] |
| c.18G>C |
p.Gln6His (p.Q6H) |
chr1:145414799 | rs376970642 | NA | 7.70×10-5 | 2.47×10-5 | 2.00×10-5 | Benign | Benign | Neutral | 6 | China | East Asian | 9,10,11,29 | Included: Homozygote: 5 [9, 10, 29]; Heterozygote: 1 [11] |
| c.55A>G |
p.Thr19Ala (p.T19A) |
chr1:145414836 | rs781981862 | NA | NA | 8.24×10-6 | 8.16×10-6 | Benign | Benign | Neutral | 1 | China | East Asian | 15 | Excluded: Heterozygote: 1 [15] |
| c.81delG |
p.Leu28SerfsTer24 (p.L28fs) |
chr1:145414862 | NA | NA | NA | NA | NA | — | — | — | 1 | England/Ireland | Caucasian | 30 | Included: Homozygote: 1 [30] |
| c.122G>C |
p.Arg41Pro (p.R41P) |
chr1:145415303 | rs781785800 | NA | NA | 8.30×10-6 | 4.08×10-6 | Probably damaging | Probably damaging | Deleterious | 1 | France | Caucasian | 26 | Included: Compound heterozygote: 1 [26] |
| c.138C>A |
p.Tyr46Ter (p.Y46*) |
chr1:145415319 | NA | NA | NA | NA | NA | — | — | — | 1 | China | East Asian | 31 | Included: Homozygote: 1 [31] |
| c.160A>T |
p.Arg54Ter (p.R54*) |
chr1:145415341 | rs121434375 | NA | NA | NA | NA | — | — | — | 1 | African American | African American | 17 | Excluded: Heterozygote: 1 [17] |
| c.196G>T |
p.Gly66Ter (p.G66*) |
chr1:145415377 | rs1469129426 | NA | NA | NA | NA | — | — | — | 2 | Czech, Romania: | Caucasian | 32,33 | Included: Homozygote: 2 [32, 33] |
| c.204_205insGGA |
p.Gly69dup (p.G69dup) |
chr1:145415386_145415387insGGA | NA | NA | NA | NA | NA | — | — | — | 1 | United States (African American) | African American | 34 | Excluded: Heterozygote: 1 [34] |
| c.208C>T |
p.Arg70Trp (p.R70W) |
chr1:145415389 | rs377513689 | NA | 1.54×10-4 | 5.06×10-5 | 3.31×10-5 | Probably damaging | Possibly damaging | Neutral | 1 | France | Caucasian | 35 | Excluded: Heterozygote: 1 [35] |
| c.220delG |
p.Val74TrpfsTer40 (p.V74fs) |
chr1:145415401 | NA | NA | NA | NA | NA | — | — | — | 1 | England | Caucasian | 18 | Included: Compound heterozygote: 1 [18] |
| c.238T>C |
p.Cys80Arg (p.C80R) |
chr1:145415419 | rs28940586 | NA | NA | 8.50×10-6 | 4.16×10-6 | Probably damaging | Probably damaging | Deleterious | 2 | Southeast United States, Australia | Caucasian | 28,36 | Included: Compound heterozygote: 2 [28, 36] |
| c.239G>A |
p.Cys80Try (p.C80Y) |
chr1: 145415420 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Bangladesh/United Kingdom | Caucasian | 37 | Included: Compound heterozygote: 1 [37] |
| c.253T>C |
p.Ser85Pro (p.S85P) |
chr1:145415434 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 18 | Included: Homozygote: 1 [18] |
| c.265T>C |
p.Cys89Arg (p.C89R) |
chr1:145415446 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Western Iran | Caucasian | 38 | Included: Homozygote: 1 [38] |
| c.295G>A |
p.Gly99Arg (p.G99R) |
chr1:145415476 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 3 | Pakistan, Albania | Caucasian | 18,37 | Included: Homozygote: 2; Compound heterozygote: 1 [18, 37] |
| c.296G>T |
p.Gly99Val (p.G99V) |
chr1:145415477 | rs1451187897 | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Greece | Caucasian | 5 | Included: Homozygote: 1 [5] |
| c.302T>C |
p.Leu101Pro (p.L101P) |
chr1: 145415483 | rs74315327 | NA | NA | 8.65×10-6 | 8.41×10-6 | Probably damaging | Probably damaging | Deleterious | 7 | Southeast United, Albania, France | Caucasian | 18,26,27,36 |
Included: Compound heterozygote: 5 [18, 26, 36] Excluded: Heterozygote: 2 [27] |
| c.306delC |
p.Phe103SerfsTer11 (p.F103fs) |
chr1:145415487 | NA | NA | NA | NA | NA | — | — | — | 1 | Italy | Caucasian | 7 | Included: Heterozygote: 1 [7] |
| c.309C>G |
p.Phe103Leu (p.F103L) |
chr1:145415490 | NA | NA | NA | NA | NA | Possibly damaging | Benign | Deleterious | 1 | China | East Asian | 9 | Included: Homozygote: 1 [9] |
| c.311A>G |
p.His104Arg (p.H104R) |
chr1:145415492 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | China | East Asian | 9 | Included: Compound heterozygote: 1 [9] |
| c.314C>T |
p.Ser105Leu (p.S105L) |
chr1:145415495 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | France | Caucasian | 27 | Excluded: Heterozygote: 1 [27] |
| c.346C>T |
p.Gln116Ter (p.Q116*) |
chr1:145415527 | NA | NA | NA | NA | NA | — | — | — | 1 | Ireland | Caucasian | 39 | Included: Compound heterozygote: 1 [39] |
| c.356G>T |
p.Cys119Phe (p.C119F) |
chr1:145415537 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Germany | Caucasian | 40 | Included: Homozygote: 1 [40] |
| c.391_403del |
p.Arg131PhefsTer111 (p.R131fs) |
chr1:145415572_145415584del | rs1486905702 | NA | NA | NA | NA | — | — | — | 1 | Italy | Caucasian | 18 | Included: Homozygote: 1 [18] |
| c.404T>G |
p.Leu135Arg (p.L135R) |
chr1:145415585 | rs782182681 | NA | NA | 2.75×10-5 | 1.29×10-5 | Benign | Benign | Neutral | 1 | Spanish | Caucasian | 25 | Excluded: Heterozygote: 1 [25] |
| c.445delG |
p.Asp149ThrfsTer97 (p.D149fs) |
chr1:145415626 | NA | NA | NA | NA | NA | — | — | — | 4 | Italy | Caucasian | 8,18 |
Included: Homozygote: 2 [18]; Heterozygote: 1 [8] Excluded: Homozygote: 1 [18] |
| c.494T>A |
p.Leu165Ter (p.L165*) |
chr1:145415675 | rs782431871. | NA | NA | 8.36×10-6 | 8.32×10-6 | — | — | — | 2 | Netherland | Caucasian | 16,21 | Included: Compound heterozygote: 2 [16, 21] |
| c.497A>G |
p.His166Arg (p.H166R) |
Chr1:145415678 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Arab | Caucasian | 41 | Included: Homozygote: 1 [41] |
| c.503C>A |
p.Ala168Asp (p.A168D) |
chr1:145415684 | rs782125244 | NA | NA | NA | NA | Probably damaging | Probably damaging | Neutral | 1 | Australia/England | Caucasian | 18 | Included: Homozygote: 1 [18] |
| c.509T>C |
p.Phe170Ser (p.F170S) |
chr1:145415690 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 2 | Italy | Caucasian | 18 | Included: Homozygote: 2 [18] |
| c.512G>T |
p.Gly171Val (p.G171V) |
chr1:145415693 | rs782077224 | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | NA | Not mentioned | Not mentioned | 42 | NA [42] |
| c.516C>G |
p.Asp172Glu (p.D172E) |
chr1:145415697 | rs782708481 | NA | NA | 8.35×10-6 | 4.16×10-6 | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 18 | Included: Compound heterozygote: 1 [18] |
| c.515_516insC |
p.His174ProfsTer23 (p.H174fs) |
chr1:145415696_145415697insC | NA | NA | NA | NA | NA | — | — | — | 1 | Japan | East Asian | 43,44 | Included: Homozygote: 1 [43, 44] |
| c.526C>T |
p.Arg176Cys (p.R176C) |
chr1:145415707 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 3 | France | Caucasian | 19,22,26,45 |
Included: Homozygote: 1 [45] ; Compound heterozygote: 1 [26] |
| c.539A>G |
p.His180Arg (p.H180R) |
chr1:145415720 | rs1395419937 | NA | NA | NA | 2.07×10-5 | Benign | Benign | Neutral | 1 | France | Caucasian | 26 | Included: Compound heterozygote: 1 [26] |
| c.573G>T |
p.Trp191Cys (p.W191C) |
chr1:145415754 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 18 | Included: Homozygote: 1 [18] |
| c.575C>T |
p.Pro192Leu (p.P192L) |
chr1:145415756 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Pakistan | Caucasian | 46 | Included: Homozygote: 1 [46] |
| c.581T>C |
p.Leu194Pro (p.L194P) |
chr1:145415762 | rs782682271 | NA | NA | 8.34×10-6 | 8.26×10-6 | Probably damaging | Probably damaging | Deleterious | 1 | Pakistan | Caucasian | 46 | Included: Homozygote: 1 [46] |
| c.588T>G |
p.Asn196Lys (p.N196K) |
chr1:145415769 | rs1020058448 | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 8 | Excluded: Heterozygote: 1 [8] |
| c.615C>G |
p.Ser205Arg (p.S205R) |
chr1:145415796 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Neutral | 1 | Italy | Caucasian | 18 | Included: Compound heterozygote: 1 [18] |
| c.665T>A |
p.Ile222Asn (p.I222N) |
chr1:145416320 | rs74315325 | NA | 7.70×10-5 | 8.24×10-6 | 2.44×10-5 | Probably damaging | Probably damaging | Deleterious | 2 | Southeast United States, Canada | Caucasian | 5,36 |
Included: Compound heterozygote: 1 [36] Excluded: Compound heterozygote: 1 [5] |
| c.700_702delAAG |
p.Lys234del (p.K234del) |
chr1:145416355_145416357delAAG | NA | NA | NA | NA | NA | — | — | — | 1 | Europe | Caucasian | 47 | Included: Homozygote: 1 [47] |
| c.739T>A |
p.Phe247Ile (p.F247I) |
chr1:145416394 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Turkey | Caucasian | 16 | Included: Homozygote: 1 [16] |
| c.745G>C |
p.Asp249His (p.D249H) |
chr1:145416400 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 2 | Japan | East Asian | 43,48-50 | Included: Homozygote: 2 [43, 48–50] |
| c.749G>T |
p.Gly250Val (p.G250V) |
chr1:145416404 | rs863224819 | NA | NA | NA | 3.23×10-5 | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 18 | Included: Compound heterozygote: 1 [18] |
| c.791C>T |
p.Ser264Leu (p.S264L) |
chr1:145416446 | rs782576713 | NA | NA | 1.65×10-5 | 2.03×10-5 | Possibly damaging | Benign | Neutral | 1 | Spanish | Caucasian | 23 | Excluded: Heterozygote: 1 [23] |
| c.806dupA |
p.Asn269LysfsTer43 (p.N269fs) |
chr1:145416461dupA | NA | NA | NA | NA | NA | — | — | — | 1 | England | Caucasian | 18 | Included: Compound heterozygote: 1 [18] |
| c.820G>A |
p.Val274Met (p.V274M) |
chr1:145416475 | rs187777957 | 5.99×10-4 | NA | 9.88×10-5 | 8.53×10-5 | Probably damaging | Probably damaging | Deleterious | 1 | China | East Asian | 9 | Included: Compound heterozygote: 1 [9] |
| c.842T>C |
p.Ile281Thr (p.I281T) |
chr1:145416497 | rs74315326 | NA | NA | NA | 8.12×10-6 | Probably damaging | Probably damaging | Deleterious | 3 | Greece, China | Caucasian, East Asian | 5,9,10 | Included: Homozygote: 1 [5]; Compound heterozygote: 2 [9, 10] |
| c.860T>G |
p.Ile287Ser (p.I287S) |
chr1:145416515 | NA | NA | NA | NA | NA | Possibly damaging | Possibly damaging | Deleterious | 1 | China | East Asian | 31 | Included: Homozygote: 1 [31] |
| c.862C>T |
p.Arg288Trp (p.R288W) |
chr1:145416517 | rs782493762 | NA | NA | 1.65×10-5 | 1.62×10-5 | Probably damaging | Probably damaging | Deleterious | 2 | France | Caucasian | 18,26,51 | Included: Homozygote: 2 [18, 26, 51] |
| c.895A>G |
p.Lys299Glu (p.K299E) |
chr1:145416550 | NA | NA | NA | NA | NA | Benign | Benign | Neutral | 1 | France | Caucasian | 26 | Included: Compound heterozygote: 1 [26] |
| c.904G>A |
p.Glu302Lys (p.E302K) |
chr1:145416559 | rs143496559 | 3.99×10-4 | 3.08×10-4 | 2.64×10-4 | 2.72×10-4 | Probably damaging | Possibly damaging | Deleterious | 4 | Brazil, France | Caucasian | 12,27 |
Included: Heterozygote: 2 [12] Excluded: Heterozygote: 2 [27] |
| c.929C>G |
p.Ala310Gly (p.A310G) |
chr1:145416584 | rs7540883 | 2.66×10-2 | 2.52×10-2 | 6.97×10-3 | 5.43×10-3 | Probably damaging | Possibly damaging | Neutral | 22 | Brazil, United States (African American) | Caucasian, African American | 12,34 |
Included: Heterozygote: 1 [12] Excluded: Heterozygote: 21 [34] |
| c.934C>T |
p.Gln312Ter (p.Q312*) |
chr1:145416589 | NA | NA | NA | NA | NA | — | — | — | 3 | Japan, China | East Asian | 9,48,49,52 |
Included: Homozygote: 1 [49, 52]; Compound heterozygote: 1 [48] Excluded: Homozygote: 1 [9] |
| c.950G>C |
p.Cys317Ser (p.C317S) |
chr1:145416605 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | Italy | Caucasian | 53 | Included: Homozygote: 1 [53] |
| c.959G>T |
p.Gly320Val (p.G320V) |
chr1:145416614 | rs74315323 | NA | 3.08×10-4 | 1.73×10-4 | 1.58×10-4 | Probably damaging | Probably damaging | Deleterious | 45 | Bangladesh/United Kingdom, Ireland, Netherland, Southeast United States, United States, Slovakia, Kosovo, Germany, Denmark, Belgium, Canada, Hungary, Australia, Croatia, Brazil, Romania, Greece, Serbia, Italy, France, India | Caucasian | 5,12,16,18-20,22,24,26-28,34,36,37,39,40,46,54-65 |
Included: Homozygote: 30 [5, 12, 16, 18–20, 22, 24, 26–28, 34, 36, 37, 39, 40, 46, 54–65]; Compound heterozygote: 7 [16, 34, 36, 37, 39, 40] Excluded: Compound heterozygote: 4 [5, 22, 24, 26]; Homozygote: 3 [5]; Heterozygote: 1 [27] |
| c.960dupG |
p.Cys321ValfsTer21 (p.C321fs) |
chr1:145416615_145416616insG | NA | NA | NA | NA | NA | — | — | — | 2 | Italy | Caucasian | 7,18 |
Included: Compound heterozygote: 1 [18] Excluded: Heterozygote: 1 [7] |
| c.963C>G |
p.Cys321Trp (p.C321W) |
chr1:145416618 | rs121434374 | NA | NA | 1.65×10-5 | 1.22×10-5 | Probably damaging | Probably damaging | Deleterious | 1 | United States | Caucasian | 34 | Included: Compound heterozygote: 1 [34] |
| c.962_963delGCinsAA |
p.Cys321Ter (p.C321*) |
chr1:145416617_145416618delinsAA | NA | NA | NA | 8.24×10-6 | NA | — | — | — | 6 | China | East Asian | 9-11,29 | Included: Homozygote: 5 [9, 10, 29]; Heterozygote: 1 [11] |
| c.976C>T |
p.Arg326Ter (p.R326*) |
chr1:145416631 | rs74315324 | NA | 7.70×10-5 | 1.65×10-5 | 1.22×10-5 | — | — | — | 2 | Australia, Greece | Caucasian | 5,28 |
Included: Compound heterozygote: 1 [28] Excluded: Compound heterozygote: 1 [5] |
| c.982_985delTCTC |
p.Ser328AspfsTer10 (p.S328fs) |
chr1: 145416637_145416640delTCTC | rs786205063 | NA | NA | NA | NA | — | — | — | 1 | Slovakia | Caucasian | 40 | Included: Compound heterozygote: 1 [40] |
| c.1004G>A |
p.Arg335Gln (p.R335Q) |
chr1:145416659 | rs377109351 | NA | 1.54×10-4 | 7.41×10-5 | 5.69×10-5 | Possibly damaging | Benign | Neutral | 2 | France | Caucasian | 27 | Excluded: Heterozygote: 2 [27] |
| c.1006G>T |
p.Gly336Ter (p.G336*) |
chr1:145416661 | NA | NA | NA | NA | NA | — | — | — | 4 | India | Caucasian | 66 | Included: Homozygote: 4 [66] |
| c.1026delT |
p.Ala343ProfsTer24 (p.A343fs) |
chr1:145416681delT | NA | NA | NA | NA | NA | — | — | — | 1 | Sri Lanka | Caucasian | 37 | Included: Homozygote: 1 [37] |
| c.1080delC |
p.Cys361ValfsTer6 (p.C361fs) |
chr1:145416735delC | NA | NA | NA | NA | NA | — | — | — | 1 | Greece | Caucasian | 5 | Included: Homozygote: 1 [5] |
| c.1081T>C |
p.Cys361Arg (p.C361R) |
chr1:145416736 | NA | NA | NA | NA | NA | Probably damaging | Probably damaging | Deleterious | 1 | France | Caucasian | 26 | Excluded: Compound heterozygote: 1 [26] |
| c.1097T>A |
p.Leu366Ter (p.L366*) |
chr1:145416752 | rs1205047449 | NA | NA | NA | NA | — | — | — | 1 | Canada | Caucasian | 20 | Included: Homozygote: 1 [20] |
| c.1114A>G |
p.Asn372Asp (p.N372D) |
chr1:145416769 | rs782156457 | NA | NA | 8.24×10-6 | 1.22×10-5 | Possibly damaging | Benign | Deleterious | 1 | France | Caucasian | 27 | Excluded: Heterozygote: 1 [27] |
| c.1151C>T |
p.Ala384Val (p.A384V) |
chr1:145416806 | NA | NA | NA | NA | NA | Possibly damaging | Possibly damaging | Neutral | 1 | France | Caucasian | 26 | Included: Compound heterozygote: 1 [26] |
| c.1153C>T |
p.Arg385Ter (p.R385*) |
chr1:145416808 | rs782803011 | NA | NA | 8.24×10-6 | 2.03×10-5 | — | — | — | 3 | Italy, North Africa | Caucasian, North African | 18,26 | Included: Homozygote: 3 [18, 26] |
| c.-89-4dupT | — | chr1:145414649 | NA | NA | NA | NA | NA | — | — | — | 1 | France | Caucasian | 19 | Excluded: Heterozygote: 1 [19] |
| c.-36G>A | — | chr1:145414746 | NA | NA | NA | NA | NA | — | — | — | 2 | India | Caucasian | 66 | Included: Compound heterozygote: 2 [66] |
| c.-1624G>A | — | chr1:145413158 | NA | NA | NA | NA | NA | — | — | — | 2 | India | Caucasian | 66 | Included: Compound heterozygote: 2 [66] |
| Gene deletion | — | — | — | NA | NA | NA | NA | — | — | — | 1 | Canada | Caucasian | 20 | Included: Heterozygote: 1 [20] |
Abbreviations: 1000G 1000 genome project, ESP exome sequencing project, ExAC Exome Aggregation Consortium, GnomAD Genome Aggregation Database, HJV-HH HJV-related hereditary hemochromatosis, NA not available
Genotype–phenotype correlation in HJV-variant homozygotes
No significant differences in clinical features were observed between cases with compound heterozygous mutation versus homozygous mutation. Next, 73 probands with homozygous mutation status were included in an exploration of genotype–phenotype correlation. Among them, 22 and 50 of the probands had pathogenic mutations in exon 3 and exon 4, respectively, and only one had exon 2 mutations (Table 5). Moreover, there were 51 cases with missense mutations, 14 cases with nonsense mutations, 7 cases with frame-shift mutations, and 1 case caused by deletion (Table 6).
Table 5.
Correlations of mutation locations with phenotypes in HJV-HH homozygous cases
| Exon 2-3 | Exon 4 | P | |
|---|---|---|---|
| N | 23 | 50 | |
| Male, n (%) | 14 (60.87%) | 29 (58.00%) | 8.17 × 10-1C |
|
Ethnicities Caucasian/East Asian/African, n (%) |
20/3/0 (86.96/13.04/0.00) | 44/5/1 (88.00/10.00/2.00) | 7.04 × 10-1Fa |
| Age at diagnosis (year) | 23.00 (20.00, 26.00) | 28.00 (24.00, 37.00) | 6.96 × 10 -3 W |
| Age at presentation (year) | 22.50 (20.00, 25.50) | 25.00 (20.00, 32.00) | 1.71 × 10-1W |
| Disease onset before 30 years, n (%) | 21 (91.30%) | 33 (66.00%) | 2.40 × 10 -2 C |
| Serum parameters at presentation | |||
| Serum ferritin (ng/ml) | 3065.00 (2000.00, 4485.00) | 4018.50 (2500.00, 6008.50) | 1.23 × 10-1W |
| Transferrin saturation (%) | 95.00 (90.00, 100.00) | 96.00 (91.00, 100.00) | 5.62 × 10-1W |
| Complications | |||
| Cardiomyopathy, n (%) | 7 (30.43%) | 21 (42.00%) | 3.45 × 10-1C |
| Skin hyperpigmentation, n (%) | 5 (21.74%) | 25 (50.00%) | 3.92 × 10 -2 F |
| Arthropathy, n (%) | 4 (17.39%) | 14 (28.00%) | 3.94 × 10-1F |
| Endocrine abnormality | |||
| Hypogonadism, n (%) | 18 (78.26%) | 31 (62.00%) | 1.70 × 10-1C |
| Glucose intolerance, n (%) | 6 (26.09%) | 16 (32.00%) | 6.09 × 10-1C |
| Osteopathy, n (%) | 1 (4.35%) | 4 (8.00%) | 1.00 × 10-1F |
| Thyroid abnormality, n (%) | 0 (0.00%) | 3 (6.00%) | 5.47 × 10-1F |
| Liver disease | |||
| Abnormal liver function test, n (%) | 8 (34.78%) | 19 (38.00%) | 7.91 × 10-1C |
| Liver iron deposition, n (%) | 15 (65.22%) | 35 (70.00%) | 6.83 × 10-1C |
| Liver fibrosis, n (%) | 12 (52.17%) | 20 (40.00%) | 3.30 × 10-1C |
| Liver cirrhosis, n (%) | 7 (30.43%) | 13 (26.00%)) | 6.93 × 10-1C |
| Liver biopsy, n (%) | 17 (73.91%) | 25 (50.00%) | 5.48 × 10-2C |
|
Therapy Phlebotomy/Chelating agent/Phlebotomy & Chelating agent/ND, n (%) |
6/0/2/15 (26.09/0.00/8.70/65.22) | 28/3/4/15 (56.00/6.00/8.00/30.00) | — |
Abbreviations: HJV-HH HJV-related hereditary hemochromatosis, ND not described
Data are shown as n (%) or median (interquartile range). P values were calculated to assess the intergroup differences between homozygotes with exon 2-3 mutation and homozygotes with exon 4 mutation using chi-square test, Fisher’s exact test, or Wilcoxon test as appropriate. C, on chi-square test; F, on Fisher’s exact test; W, on Wilcoxon test. a, compared the proportions of Caucasians and East Asians. P values <0.05 are denoted in bold and underlined
Table 6.
Correlations of mutation types with phenotypes in HJV-HH homozygous cases
| Frameshift | Nonsense | Missense | Deletion | P frameshift-nonsense | P frameshift -missense | P nonsense-missense | |
|---|---|---|---|---|---|---|---|
| N | 7 | 14 | 51 | 1 | |||
| Male, n (%) | 5 (71.43%) | 11 (78.57%) | 27 (52.94%) | 0 (0.00%) | 1.00 × 10-0F | 4.42 × 10-1F | 1.27 × 10-1F |
|
Ethnicities Caucasian/East Asian/African, n (%) |
6/1/0 (85.71/14.29/0.00) | 9/4/1 (64.29/28.57/7.14) | 48/3/0 (94.12/5.88/0.00) | 1/0/0 (100.00/0.00/0.00) | 6.13 × 10-1Fa | 4.11 × 10-1Fa | 2.68 × 10 -2 F a |
| Age at diagnosis (year) | 21.00 (17.00, 25.00) | 25.00 (19.00, 45.00) | 28.00 (23.00, 32.00) | 20.00 | Pframeshift-nonsense-missense = 1.81 × 10-1K | ||
| Age at presentation (year) | 21.00 (17.00, 25.00) | 24.00 (19.00, 25.00) | 25.00 (21.00, 31.00) | — | Pframeshift-nonsense-missense = 2.34 × 10-1K | ||
| Disease onset before 30 years, n (%) | 7 (100.00%) | 9 (64.29%) | 37 (72.55%) | 1 (100.00%) | 1.24 × 10-1F | 1.78 × 10-1F | 5.29 × 10-1F |
| Serum parameters at presentation | |||||||
| Serum ferritin (ng/ml) | 4840.00 (2800.00, 5999.00) | 2736.50 (1700.00, 7000.00) | 3728.0 (2500.00, 5574.50) | 1955.00 | Pframeshift-nonsense-missense = 6.93 × 10-1K | ||
| Transferrin saturation (%) | 96.50 (94.00, 100.00) | 94.00 (89.50, 97.30) | 96.00 (90.00, 100.00) | — | Pframeshift-nonsense-missense = 3.73 × 10-1K | ||
| Complications | |||||||
| Cardiomyopathy, n (%) | 3 (42.86%) | 6 (42.86%) | 18 (35.29%) | 1 (100.00%) | 1.00 × 10-0F | 6.96 × 10-1F | 6.04 × 10-1C |
| Skin hyperpigmentation, n (%) | 2 (28.57%) | 7 (50.00%) | 20 (39.22%) | 1 (100.00%) | 6.42 × 10-1F | 6.98 × 10-1F | 4.68 × 10-1C |
| Arthropathy, n (%) | 0 (0.00%) | 2 (14.29%) | 15 (29.41%) | 1 (100.00%) | 5.33 × 10-1F | 1.73 × 10-1F | 3.23 × 10-1F |
| Endocrine abnormality | |||||||
| Hypogonadism, n (%) | 6 (85.71%) | 5 (35.71%) | 37 (72.55%) | 1 (100.00%) | 6.35 × 10-2F | 6.64 × 10-1F | 2.43 × 10 -2 F |
| Glucose intolerance, n (%) | 3 (42.86%) | 7 (50.00%) | 12 (23.53%) | 0 (0.00%) | 1.00 × 10-0F | 3.60 × 10-1F | 5.37 × 10-2C |
| Osteopathy, n (%) | 0 (0.00%) | 0 (0.00%) | 4 (7.84%) | 1 (100.00%) | — | 1.00 × 10-0F | 1.00 × 10-0F |
| Thyroid abnormality, n (%) | 0 (0.00%) | 1 (7.14%) | 2 (3.92%) | 0 (0.00%) | 1.00 × 10-0F | 1.00 × 10-0F | 5.23 × 10-1F |
| Liver disease | |||||||
| Abnormal liver function test, n (%) | 3 (42.86%) | 4 (28.57%) | 19 (37.25%) | 1 (100.00%) | 6.38 × 10-1F | 1.00 × 10-0F | 7.54 × 10-1F |
| Liver iron deposition, n (%) | 5 (71.43%) | 11 (78.57%) | 33 (64.71%) | 1 (100.00%) | 1.00 × 10-0F | 1.00 × 10-0F | 5.20 × 10-1F |
| Liver fibrosis, n (%) | 5 (71.43%) | 3 (21.43%) | 24 (47.06%) | 0 (0.00%) | 5.55 × 10-2F | 4.23 × 10-1F | 1.27 × 10-1F |
| Liver cirrhosis, n (%) | 2 (28.57%) | 3 (21.43%) | 14 (27.45%) | 1 (100.00%) | 1.00 × 10-0F | 1.00 × 10-0F | 7.45 × 10-1F |
| Liver biopsy, n (%) | 6 (85.71%) | 4 (28.57%) | 31 (60.78%) | 1 (100.00%) | 2.37 × 10 -2 F | 4.03 × 10-1F | 3.93 × 10 -2 F |
|
Therapy Phlebotomy/Chelating agent/Phlebotomy & Chelating agent/ND, n (%) |
3/0/1/3 (42.86/0.00/7.14/42.86) | 9/1/0/4 (64.29/7.14/0.`00/28.57) | 21/2/5/23 (41.18/3.92/9.80/45.10) | 1/0/0/0 (100.00/0.00/0.00/0.00) | — | — | — |
Abbreviations: HJV-HH HJV-related hereditary hemochromatosis, ND not described
Data are shown as n (%) or median (interquartile range). P values were calculated to assess the intergroup differences among homozygotes with frame-shift mutation, nonsense mutation, and missense mutation using chi-square test, Fisher’s exact test, or Kruskal-Wallis test as appropriate. C, on chi-square test; F, on Fisher’s exact test; K, on Kruskal-Wallis test. a, compared the proportions of Caucasians and East Asians. P values <0.05 are denoted in bold and underlined
Compared to those with exon 4 mutation, homozygotes with mutations in exons 2 and 3 displayed a significantly earlier age at diagnosis (median [interquartile range, IQR]: 23.00 [20.00, 26.00] vs. 28.00 [24.00, 37.00], P=6.96×10-3). Twenty-one (91.30%) cases with exon 2-3 variants were early-onset, whereas 66.00% of those with exon 4 variants were early-onset (P=2.40×10-2). The SF and TS levels were comparable, as well as the prevalence rates of most of the complications, except that the homozygotes with exon 4 variants showed a higher prevalence of skin hyperpigmentation or freckles than did those with exon 2-3 variants (P=3.92×10-2; Table 5).
On comparisons among mutation types, no significant differences were detected in age at diagnosis, age at presentation, SF, or TS. Notably, a greater proportion of probands with missense mutations presented with hypogonadism (72.55%) compared with that among those with nonsense mutations (35.71%; P=2.43×10-2). Liver biopsy was accepted by more probands with frame-shift or missense mutations accepted liver biopsy (85.71%, 60.78%, respectively) than by those with nonsense mutations (28.57%, P=2.37×10-2, 3.93×10-2; Table 6).
Therapies and outcomes among biallelic HJV mutation cases
As shown in Additional file 3, therapy information was provided for 70 cases. Among them, 60 cases were treated with phlebotomy, 3 with chelating agents, and 7 with phlebotomy in combination with chelating agents.
The outcome data were provided for 40 cases. Among them, five cases who had previously diagnosed with cardiomyopathy expired after diagnosis of HH-HJV (three of cardiac failure and two of sepsis). The patients in all other cases experienced varying degrees of improvement after therapy administration, and all cases reported complete or partial iron depletion.
Fifteen cases reported improvement after treatment, and in 12 of these cases, liver function was restored to normal. Seven cases that presented with cardiomyopathy showed significant improvement in cardiac function after therapeutic phlebotomy in combine with or without iron chelation agent administration, of which six achieved completely normalized or nearly normalized heart function. One achieved complete recovery of hypogonadism after treatment with phlebotomy and deferasirox. Two cases achieved complete recovery by iron depletion, one of which had been previously treated with insulin and was able to discontinue insulin therapy. In addition, two cases experienced improvement in bone density after phlebotomy.
Discussion
HH is one of the most common genetic disorders in the Northern European population, affecting approximately 1 in 200 people [67]. In European populations, HFE homozygous and compound heterozygous mutations account for 60–95% of iron overload cases [68]. Due to the high frequency of the HFE C282Y mutation, a simple genetic test can confirm the diagnosis in most of these patients. However, in other parts of the world, HFE mutation is less common. For example, the majority of Asian HH cases are due to non-HFE mutation [13, 37]. A previous study based on data from the publicly available next-generation sequencing (NGS) databases demonstrated that all recessive inherited non-HFE forms of HH were predicted to be extremely rare around the world [3]. This rarity makes the diagnosis and management of HJV-HH a great challenge, and indeed, this diagnosis was frequently delayed in the affected individuals. This review systematically characterized the reported genotypic and phenotypic spectra of HJV-HH in multiple ethnicities by analyzing the data for 132 eligible HJV-variant cases. To our knowledge, this is the first study to investigate the phenotype–genotype correlation of HJV-HH.
The HJV gene was first identified in 2004 in a group of JH patients from Greece, Canada, and France [5]. HJV protein is a membrane protein that is highly expressed in the liver, skeletal muscle and heart and plays a role in iron absorption and release from cells in addition to having anti-inflammatory properties [4]. As a member of the repulsive guidance molecule (RGM) protein family, the longest isoform of the HJV protein consists of several functional motifs with an N-terminal signal peptide (amino acids [aa] 1–40), a conserved RGD motif (aa 98–100), a partial von Willebrand type D (vWD) domain (aa 167–310), and a C-terminal glycosylphosphatidylinositol (GPI) anchor domain (aa 403–426) [4]. It is highly conserved between species, except for the signal peptide and GPI anchor domain [5]. To date, 75 HJV variants of multiple mutation types have been identified in HJV-related iron-overload cases, including missense, nonsense, frame-shift, and in-frame mutations in exons 2, 3 and 4, as well as insertion in the splicing site and substitutions in the 5′ UTR. Most of the mutations are localized in evolutionally conserved residues [5]. Interestingly, multiple alterations at the positions of cysteine 80 [28, 36, 37], glycine 99 [5, 18, 37], phenylalanine 103 [7, 9], cysteine 321 [7, 9–11, 18, 29, 34], and cysteine 361 [5, 26] have been identified, which further strengthens the evidence that these regions have essential biological functions. In addition to the deleterious variants, loss-of-heterozygosity (LOH) resulting from gene deletion and uniparental disomy also has been detected in HJV-HH cases [20, 32], which emphasizes the involvement of novel mechanisms in the pathogenesis of HH. Thus, the multiplex ligation-dependent probe amplification method for detecting LOH should be considered in combination with sequencing of targeted gene regions when appropriate.
By associating the variants with the family origins, we observed that approximately two-thirds of the variations have been reported only in single cases or families, which was in accordance with the sporadic nature of the disease. Notably, one-third of them were recurrent mutations, most of which were detected only in the geographic origin of the respective family. Strong ethnic differences were identified in the genotype profile. G320V [5, 12, 16, 18–20, 22, 26–28, 34, 36, 37, 39, 40, 46, 54–65] and L101P [18, 26, 27, 36], the most commonly reported substitutions, were restricted to Caucasian ancestry. Among those reported in Caucasians more than three times, D149fs, R176C, and G336* were observed in Italian, French, or Indian cases only [8, 18, 19, 22, 26, 45, 66], suggesting that cases originating from different nations had their own recurrent mutations. [Q6H; C321*] in cis was the predominant genotype in Chinese HJV-HH [9–11, 29]. C321* was predicted to produce a truncated HJV protein that fails to present on the cell surface [69], whereas Q6H, which is localized in the signal peptide region, caused no or only slight alteration in HJV localization and was identified in cases with C321* only. Similarly, the other frequently detected variant in the signal peptide region, E3D, was only identified in those with SUGP2 R639Q mutation, a deleterious mutation that affects the BMP/SMAD pathway [9]. Both Q6H and E3D were predicted to be benign and were suggested as indicators of causative variants of HH [9]. D249H and Q312* were the most common mutations in Japanese cases [43, 48–50, 52]. Q312* was also detected in Chinese cases [9], suggesting its potential as a hyper mutant spot shared by the East Asian populations. Moreover, only three variations were reported across different races, including I281T [5, 9, 10], A310G [12, 34], and R385* [18, 26]. To some extent, these findings imply the existence of important hyper mutant regions across different ethnicities.
The clinical spectrum of HJV-related HH varied with respect to ethnicity. First, our results confirmed that males and females were equally affected in Caucasian probands with biallelic mutation [14]. However, interestingly, males accounted for 11 of the 13 probands with biallelic mutation of East Asian ancestry, indicating the first time that East Asian males are more vulnerable than females. The phenomenon could be explained by iron loss through menstruation after puberty, which is believed to result in a less severe phenotype in females. In Caucasians, this natural advantage among females could be partially diminished because of the relatively high consumption of red meat in the Western diet, which might interact with the risk of hemochromatosis. However, considering the limited sample size of East Asians in this review, whether the findings can reflect the true distribution among East Asians requires further investigation.
Second, ethnic discrepancy in the clinical complications of HJV-HH was identified. Hypogonadism and arthropathy were more prevalent in Caucasian probands than in East Asian probands, despite the comparable age at diagnosis and SF and TS levels. In contrast, the prevalence rates of glucose intolerance and cardiac and hepatic complications appeared relatively higher in East Asians, but the differences were not statistically significant. The underlying mechanisms remain unclear. However, we suspect that ethnic differences in the susceptibility of target tissues might contribute to the potential differences. Considering diabetes as an example, β-cell function is considered more vulnerable in East Asians than in Caucasians [70]. In this study, in addition to a higher rate of diabetes, we found that insulin was used to treat diabetes only in East Asian cases [10, 43, 49], whereas oral anti-diabetic drugs were used in the two Caucasian cases with diabetes [16, 21, 34]. In addition, better awareness of hemochromatosis among Caucasians than among other ethnicities and populations with different cultural backgrounds could confound the above results.
Biallelic HJV mutations are known as the major cause of JH, but not all of the HJV biallelic mutation cases developed JH. In this review, over three-fourths of the probands were early-onset cases. Hypogonadism was more prevalent in early-onset HJV-HH, whereas liver iron deposition occurred with a higher prevalence in the late-onset cases, suggesting phenotypic variation and potential differences in pathogenesis between the two groups. In the genotype–phenotype correlation study, although we were unable to explore and compare the phenotypes of single variants due to the sporadic nature and complexity of disease, we were able to identify that more than 90% of the homozygotes with mutation in exons 2-3 developed hemochromatosis before age 30, compared with only 66% among homozygotes with exon 4 mutation, indicating that the genetic defects in exons 2-3 may have a greater deleterious impact to HJV function. Although the underlying biological mechanisms have yet to be elucidated, our findings emphasize the existence of genotype–phenotype correlation.
Experimental studies indicated that loss of HJV membrane export is central to the pathogenesis of iron overload [69, 71]. Currently reported nonsense mutations, such as Q312*, C321*, R326*, G336*, L366*, and R385*, were suspected to induce a truncated HJV protein lacking the GPI anchor motif, resulting in defective targeting of HJV to the plasma membrane. For example, the R326* mutant is mainly retained in the endoplasmic reticulum in vitro [69]. Frame-shift mutations that alter the reading frame of the protein are generally considered deleterious, because this leads to completely altered translation. The missense mutations can have diverse effects in the pathogenesis of iron overload. In vivo studies demonstrated that some, including G320V, W191C, and F170S [69], as well as mutant at TMPRSS6 cleavage site arginine 176 and 288, can cause a significant reduction of membrane-anchored HJV, which then impairs the activation of the signaling pathway involved in hepcidin up-regulation. Moreover, autoproteolysis does not occur for the arginine 176 and 288 mutant, resulting in the loss of its ability as a BMP co-receptor in the regulation of hepcidin [71]. Researchers also hypothesized that G99V and C119F cause a defect in the binding to BMP or to co-receptors, which generates an inadequate activation signal for hepcidin production [69]. L101P, located near the RGD region, is speculated to result in an alteration of RGD function [5]. Ideally, stratifying the mutations by their biological function in the regulation of hepcidin may be a better way to investigate the phenotype–genotype correlation, because of the tremendous differences in the pathogenic mechanisms. However, in the present study, we found a tendency toward a relatively earliest onset age for the frame-shift mutation homozygotes compared to that of the nonsense or missense mutation homozygotes. Interestingly, the nonsense mutation homozygotes showed the lowest prevalence of hypogonadism, as well as the lowest rate of accepting liver biopsy, compared to the missense or frameshift groups, indicating the nonsense mutations could be less harmful to the target tissues. Overall, further comparisons of biological mechanisms among the different types of mutations are needed.
Therapeutic phlebotomy and/or chelating agents were commonly used for treating hemochromatosis. Previous studies suggested that it may take longer for excess iron to removed and target levels of iron parameters to be reached following phlebotomy in JH. If not appropriately treated in the early stages of symptomatic presentation, cardiac failure becomes a leading cause of death in these patients [6]. In this review, seven HJV-HH cases showed significant improvement in cardiac function after therapy administration, most of which even achieved normalized or nearly normalize heart function [6], suggesting that the currently available therapies, if adequately applied, can provide a great chance of recovery from cardiomyopathy in HJV-HH. Notably, while some patients died from complications of cardiomyopathy [5, 33, 36, 56], two cases with cardiomyopathy died of sepsis [18, 49, 51]. This emphasizes the need for heightened vigilance not only for cardiac dysfunction but also for sepsis among HJV-HH patients, especially those who have already presented myocardial involvement.
The results further suggested that the reversal of endocrine dysfunction is a greater challenge compared to the improvement of hepatic function in HJV-HH. In hemochromatosis, hypogonadism is mainly secondary to the selective deposition of iron in the gonadotropin-producing cells of the pituitary gland, leading to impaired hormone secretion [1]. It was previously reported that gonadotropic cell dysfunction induced by the excessive iron overload of HH could be reversed if appropriate treatment is initiated early enough [72]. Sex hormone replacement therapy offers rapid correction of sexual problems and is commonly used in these patients, but iron depletion is the key to normalization of this disorder. In this review, hypogonadism was resolved in one of the juvenile-onset cases by applying phlebotomy in combination with deferasirox [12, 60]. Previous cohort studies suggested a favorable effect could be achieved for glucose control in hemochromatosis cases [10, 12, 16, 21, 43, 49, 60]. Diabetes in hemochromatosis involves two major mechanisms, beta-cell damage and insulin resistance, due to hepatic damage. In our study, two HJV-HH cases achieved diabetes recovery with iron depletion [10, 12, 60]. One had abnormal liver function test results but did not have cirrhosis, while the other had hepatic cirrhosis and was previously treated with insulin. After therapeutic phlebotomy, both patients achieved euglycemia and improved liver function. Osteoporosis in hemochromatosis might attributed to iron overload and its-related hypogonadism, liver failure, or parathyroid defects [1]. In the present review, most cases with osteoporosis or osteopenia also had hypogonadism, while half the cases also had liver cirrhosis. Two cases achieved partial improvement in bone density after phlebotomy [16]. Based on these results, although complete resolution of endocrine disorders was possible in HJV-HH, it was seldom achieved. Efficient strategies to prevent, early diagnose and reverse endocrine dysfunction in HJV-HH are urgently needed. However, the relationships of age, sex, genotype, and clinical features with disease outcomes are difficult to further clarify due to the complexities of the clinical manifestations and the limitation of a review study design; these relationships are of clinical significance though and need to be investigated in the future.
Finally, although the classic mode of HJV-HH inheritance follows an autosomal recessive pattern, the heterozygous status of HJV was recently reported to cause iron-overload phenotypes or even middle-age onset hemochromatosis in some cases [7–12], supporting the additive effect of the HJV gene in the determination of phenotype. A similar hereditary pattern was previously observed in HFE-HH. Studies indicated that individuals who were heterozygous for HFE C282Y or H63D showed elevated SF and TS levels, but did not develop complications of iron overload [73, 74]. In this review, six cases with one HJV mutation developed complications [7, 9, 11]. Ten HJV heterozygous cases also had heterozygous mutations in another HH-related gene, which implies a digenic inheritance mode [8, 9, 12]. In addition, the possibility of the existence of an unknown causal gene cannot be excluded. More heterozygous probands and a higher prevalence of complications were reported in East Asians, suggesting that East Asians might be more vulnerable to the monoallelic gene deficiency than Caucasians. Thus, these findings emphasize the importance of screening and the management of asymptomatic heterozygotes in HJV-HH families to avoid unwanted outcomes.
In conclusion, the current study provides a systematic review of the genotypic and phenotypic spectra of HJV-HH worldwide. Although the sporadic nature of HJV-HH determined that sequencing of the targeted genes related to HH will be the best approach to pursue a definitive diagnosis for quite a long time, knowledge of the cross-ethnicity and ethnicity-specific mutations will be beneficial for the interpretation of sequencing data as well as the establishment of ethnicity-specific approaches to genetic testing. Heterozygous relatives of HJV-HH patients should also be accessed for iron-overload disease. Given the large genetic and phenotypic diversity of this disease, the accumulation of data from future clinical cases is of great significance to provide better guidance for the diagnosis, prognosis and management of HJV-related disease, which should take into consideration patients’ ethnicity, geographical region, and genetic predisposition.
Additional files
Search terms used for database searches. (DOCX 18 kb)
Included and excluded articles. (DOCX 161 kb)
Clinical findings for cases with biallelic mutations. (DOCX 96 kb)
Clinical findings for cases with monoallelic mutation. (DOCX 56 kb)
Acknowledgements
The study was supported by grant from the Ministry of Science and Technology of the People’s Republic of China (2016YFC0901204).
Abbreviations
- 1000G
1000 genome project
- ESP
Exome sequencing project
- ExAC
Exome aggregation consortium
- GnomAD
Genome aggregation database
- HGVS
Human genome variation society
- HJV-HH
HJV-related hereditary hemochromatosis
- NA
Not available
- ND
Not described
- TM
Transmembrane domain
- vWF Type D
von Willebrand factor type D domain
Authors’ contributions
XC, WY, and XK developed the study design. LX and XK preformed the literature search. HZ and LS extracted the data. XK, XX, LX, HZ, and LS analyzed and interpreted the data. XK and XC drafted the manuscript. All of the authors critically revised the manuscript and contributed to the discussion. XK, XC and WY completed the final version of manuscript. All authors read and approved the final manuscript, as well as the submission of this work. XC is the guarantor of this work. As the corresponding author, XC have had full access to the data in the study and final responsibility for the decision to submit for publication.
Funding
None.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaomu Kong, Email: yazikate@163.com.
Lingding Xie, Email: hatagirl1979@163.com.
Haiqing Zhu, Email: zhu_haiqing@126.com.
Lulu Song, Email: qy2067@163.com.
Xiaoyan Xing, Email: xingxy221@126.com.
Wenying Yang, Email: ywying_1010@163.com.
Xiaoping Chen, Phone: +86-10-84205254, Email: chenxp1995@163.com.
References
- 1.Pelusi C, Gasparini DI, Bianchi N, Pasquali R. Endocrine dysfunction in hereditary hemochromatosis. J Endocrinol Invest. 2016;39:837–847. doi: 10.1007/s40618-016-0451-7. [DOI] [PubMed] [Google Scholar]
- 2.Bardou-Jacquet E, Ben Ali Z, Beaumont-Epinette M, Loreal O, Jouanolle A, Brissot P. Non-HFE hemochromatosis: Pathophysiological and diagnostic aspects. Clin Res Hepatol Gastroenterol. 2014;38:143–154. doi: 10.1016/j.clinre.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DF, Subramaniam VN. The global prevalence of HFE and non-HFE hemochromatosis estimated from analysis of next-generation sequencing data. Genet Med. 2016;18:618–626. doi: 10.1038/gim.2015.140. [DOI] [PubMed] [Google Scholar]
- 4.Malyszko J. Hemojuvelin: The Hepcidin Story Continues. Kidney Blood Press Res. 2009;32:71–76. doi: 10.1159/000208988. [DOI] [PubMed] [Google Scholar]
- 5.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 6.De Gobbi M, Roetto A, Piperno A, Mariani R, Alberti F, Papanikolaou G, et al. Natural history of juvenile haemochromatosis. Br J Haematol. 2002;117:973–979. doi: 10.1046/j.1365-2141.2002.03509.x. [DOI] [PubMed] [Google Scholar]
- 7.Pelusi S, Rametta R, Della CC, Congia R, Dongiovanni P, Pulixi EA, et al. Juvenile hemochromatosis associated with heterozygosity for novel hemojuvelin mutations and with unknown cofactors. Ann Hepatol. 2014;13:568–571. doi: 10.1016/S1665-2681(19)31259-1. [DOI] [PubMed] [Google Scholar]
- 8.Biasiotto G, Roetto A, Daraio F, Polotti A, Gerardi GM, Girelli D, et al. Identification of new mutations of hepcidin and hemojuvelin in patients with HFE C282Y allele. Blood Cells Mol Dis. 2004;33:338–343. doi: 10.1016/j.bcmd.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Lv T, Zhang W, Xu A, Li Y, Zhou D, Zhang B, et al. Non-HFE mutations in haemochromatosis in China: combination of heterozygous mutations involving HJV signal peptide variants. J Med Genet. 2018;55:650–660. doi: 10.1136/jmedgenet-2018-105348. [DOI] [PubMed] [Google Scholar]
- 10.Huang FW, Rubio-Aliaga I, Kushner JP, Andrews NC, Fleming MD. Identification of a novel mutation (C321X) in HJV. Blood. 2004;104:2176–2177. doi: 10.1182/blood-2004-01-0400. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Xue J, Chen B, Wang Q, Shi M, Xie X, et al. Two middle-age-onset hemochromatosis patients with heterozygous mutations in the hemojuvelin gene in a Chinese family. Int J Hematol. 2014;99:487–492. doi: 10.1007/s12185-014-1547-5. [DOI] [PubMed] [Google Scholar]
- 12.Santos PC, Cancado RD, Pereira AC, Schettert IT, Soares RA, Pagliusi RA, et al. Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients. Blood Cells Mol Dis. 2011;46:302–307. doi: 10.1016/j.bcmd.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 13.McDonald CJ, Wallace DF, Crawford DHG, Subramaniam VN. Iron storage disease in Asia-Pacific populations: The importance of non-HFE mutations. J Gastroenterol Hepatol. 2013;28:1087–1094. doi: 10.1111/jgh.12222. [DOI] [PubMed] [Google Scholar]
- 14.Sandhu K, Flintoff K, Chatfield MD, Dixon JL, Ramm LE, Ramm GA, et al. Phenotypic analysis of hemochromatosis subtypes reveals variations in severity of iron overload and clinical disease. Blood. 2018;132:101–110. doi: 10.1182/blood-2018-02-830562. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Niu S, Dong A, Cai H, Anderson GJ, Han B, et al. A Chinese family carrying novel mutations in SEC23B and HFE2, the genes responsible for congenital dyserythropoietic anaemia II (CDA II) and primary iron overload, respectively. Br J Haematol. 2012;158:143–145. doi: 10.1111/j.1365-2141.2012.09102.x. [DOI] [PubMed] [Google Scholar]
- 16.Smit SL, Peters T, Gisbertz I, Moolenaar W, Hendriks Y, Vincent HH, et al. Variable workup calls for guideline development for type 2A hereditary haemochromatosis. Neth J Med. 2018;76:365–373. [PubMed] [Google Scholar]
- 17.Murugan RC, Lee PL, Kalavar MR, Barton JC. Early age-of-onset iron overload and homozygosity for the novel hemojuvelin mutation HJV R54X (exon 3; c.160A-->T) in an African American male of West Indies descent. Clin Genet. 2008;74:88–92. doi: 10.1111/j.1399-0004.2008.01017.x. [DOI] [PubMed] [Google Scholar]
- 18.Lanzara C, Roetto A, Daraio F, Rivard S, Ficarella R, Simard H, et al. Spectrum of hemojuvelin gene mutations in 1q-linked juvenile hemochromatosis. Blood. 2004;103:4317–4321. doi: 10.1182/blood-2004-01-0192. [DOI] [PubMed] [Google Scholar]
- 19.Cunat S, Giansily-Blaizot M, Bismuth M, Blanc F, Dereure O, Larrey D, et al. Global sequencing approach for characterizing the molecular background of hereditary iron disorders. Clin Chem. 2007;53:2060–2069. doi: 10.1373/clinchem.2007.090605. [DOI] [PubMed] [Google Scholar]
- 20.Lanktree MB, Sadikovic B, Waye JS, Levstik A, Lanktree BB, Yudin J, et al. Clinical evaluation of a hemochromatosis next-generation sequencing gene panel. Eur J Haematol. 2017;98:228–234. doi: 10.1111/ejh.12820. [DOI] [PubMed] [Google Scholar]
- 21.van Dijk BA, Kemna EH, Tjalsma H, Klaver SM, Wiegerinck ET, Goossens JP, et al. Effect of the new HJV-L165X mutation on penetrance of HFE. Blood. 2007;109:5525–5526. doi: 10.1182/blood-2006-11-058560. [DOI] [PubMed] [Google Scholar]
- 22.Aguilar-Martinez P, Lok CY, Cunat S, Cadet E, Robson K, Rochette J. Juvenile hemochromatosis caused by a novel combination of hemojuvelin G320V/R176C mutations in a 5-year old girl. Haematologica. 2007;92:421–422. doi: 10.3324/haematol.10701. [DOI] [PubMed] [Google Scholar]
- 23.Altes A, Bach V, Ruiz A, Esteve A, Felez J, Remacha AF, et al. Mutations in HAMP and HJV genes and their impact on expression of clinical hemochromatosis in a cohort of 100 Spanish patients homozygous for the C282Y mutation of HFE gene. Ann Hematol. 2009;88:951–955. doi: 10.1007/s00277-009-0705-y. [DOI] [PubMed] [Google Scholar]
- 24.Brakensiek K, Fegbeutel C, Malzer M, Struber M, Kreipe H, Stuhrmann M. Juvenile hemochromatosis due to homozygosity for the G320V mutation in the HJV gene with fatal outcome. Clin Genet. 2009;76:493–495. doi: 10.1111/j.1399-0004.2009.01261.x. [DOI] [PubMed] [Google Scholar]
- 25.de Diego C, Opazo S, Sanchez-Castano A, Martinez-Castro P. New HJV mutation in a patient with hyperferritinemia and H63D homozygosity for the HFE gene. Int J Hematol. 2007;86:379–380. doi: 10.1532/IJH97.E0748. [DOI] [PubMed] [Google Scholar]
- 26.Hamdi-Roze H, Ben AZ, Ropert M, Detivaud L, Aggoune S, Simon D, et al. Variable expressivity of HJV related hemochromatosis: “Juvenile” hemochromatosis? Blood Cells Mol Dis. 2018;74:30–33. doi: 10.1016/j.bcmd.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Le Gac G, Scotet V, Ka C, Gourlaouen I, Bryckaert L, Jacolot S, et al. The recently identified type 2A juvenile haemochromatosis gene (HJV), a second candidate modifier of the C282Y homozygous phenotype. Hum Mol Genet. 2004;13:1913–1918. doi: 10.1093/hmg/ddh206. [DOI] [PubMed] [Google Scholar]
- 28.Wallace DF, Dixon JL, Ramm GA, Anderson GJ, Powell LW, Subramaniam N. Hemojuvelin (HJV)-associated hemochromatosis: analysis of HJV and HFE mutations and iron overload in three families. Haematologica. 2005;90:254–255. [PubMed] [Google Scholar]
- 29.Yuanfeng L, Hongxing Z, Haitao Z, Xiaobo P, Lili B, Fuchu H, et al. Mutation analysis of the pathogenic gene in a Chinese family with hereditary hemochromatosis. Yi Chuan. 2014;36:1152–1158. doi: 10.3724/SP.J.1005.2014.1152. [DOI] [PubMed] [Google Scholar]
- 30.Lee P, Promrat K, Mallette C, Flynn M, Beutler E. A juvenile hemochromatosis patient homozygous for a novel deletion of cDNA nucleotide 81 of hemojuvelin. Acta Haematol. 2006;115:123–127. doi: 10.1159/000089479. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Du Y, Liu G, Guo S, Hou B, Jiang X, et al. Identification of novel mutations in HFE, HFE2, TfR2, and SLC40A1 genes in Chinese patients affected by hereditary hemochromatosis. Int J Hematol. 2017;105:521–525. doi: 10.1007/s12185-016-2150-8. [DOI] [PubMed] [Google Scholar]
- 32.Neroldova M, Frankova S, Stranecky V, Honsova E, Luksan O, Benes M, et al. Hereditary haemochromatosis caused by homozygous HJV mutation evolved through paternal disomy. Clin Genet. 2015;87:96–98. doi: 10.1111/cge.12346. [DOI] [PubMed] [Google Scholar]
- 33.Janosi A, Andrikovics H, Vas K, Bors A, Hubay M, Sapi Z, et al. Homozygosity for a novel nonsense mutation (G66X) of the HJV gene causes severe juvenile hemochromatosis with fatal cardiomyopathy. Blood. 2005;105:432. doi: 10.1182/blood-2004-09-3508. [DOI] [PubMed] [Google Scholar]
- 34.Lee PL, Barton JC, Brandhagen D, Beutler E. Hemojuvelin (HJV) mutations in persons of European, African-American and Asian ancestry with adult onset haemochromatosis. Br J Haematol. 2004;127:224–229. doi: 10.1111/j.1365-2141.2004.05165.x. [DOI] [PubMed] [Google Scholar]
- 35.Le Lan C, Mosser A, Ropert M, Detivaud L, Loustaud-Ratti V, Vital-Durand D, et al. Sex and acquired cofactors determine phenotypes of ferroportin disease. Gastroenterology. 2011;140:1199–1207. doi: 10.1053/j.gastro.2010.12.049. [DOI] [PubMed] [Google Scholar]
- 36.Lee PL, Beutler E, Rao SV, Barton JC. Genetic abnormalities and juvenile hemochromatosis: mutations of the HJV gene encoding hemojuvelin. Blood. 2004;103:4669–4671. doi: 10.1182/blood-2004-01-0072. [DOI] [PubMed] [Google Scholar]
- 37.Lok CY, Merryweather-Clarke AT, Viprakasit V, Chinthammitr Y, Srichairatanakool S, Limwongse C, et al. Iron overload in the Asian community. Blood. 2009;114:20–25. doi: 10.1182/blood-2009-01-199109. [DOI] [PubMed] [Google Scholar]
- 38.Malekzadeh MM, Radmard AR, Nouroozi A, Akbari MR, Amini M, Navabakhsh B, et al. Juvenile Hemochromatosis, Genetic Study and Long-term Follow up after Therapy. Middle East J Dig Dis. 2014;6:87–92. [PMC free article] [PubMed] [Google Scholar]
- 39.Daraio F, Ryan E, Gleeson F, Roetto A, Crowe J, Camaschella C. Juvenile hemochromatosis due to G320V/Q116X compound heterozygosity of hemojuvelin in an Irish patient. Blood Cells Mol Dis. 2005;35:174–176. doi: 10.1016/j.bcmd.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Gehrke SG, Pietrangelo A, Kascak M, Braner A, Eisold M, Kulaksiz H, et al. HJV gene mutations in European patients with juvenile hemochromatosis. Clin Genet. 2005;67:425–428. doi: 10.1111/j.1399-0004.2005.00413.x. [DOI] [PubMed] [Google Scholar]
- 41.Ramzan K, Imtiaz F, Al-Ashgar HI, AlSayed M, Sulaiman RA. Juvenile hemochromatosis and hepatocellular carcinoma in a patient with a novel mutation in the HJV gene. Eur J Med Genet. 2017;60:308–311. doi: 10.1016/j.ejmg.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Klomp C, Abbes AP, Engel H. Gene symbol: HFE2a (HJV). Disease: juvenile hemochromatosis. Hum Genet. 2005;118:545–546. [PubMed] [Google Scholar]
- 43.Hattori A, Miyajima H, Tomosugi N, Tatsumi Y, Hayashi H, Wakusawa S. Clinicopathological study of Japanese patients with genetic iron overload syndromes. Pathol Int. 2012;62:612–618. doi: 10.1111/j.1440-1827.2012.02848.x. [DOI] [PubMed] [Google Scholar]
- 44.Maeda T, Nakamaki T, Saito B, Nakashima H, Ariizumi H, Yanagisawa K, et al. Hemojuvelin hemochromatosis receiving iron chelation therapy with deferasirox: improvement of liver disease activity, cardiac and hematological function. Eur J Haematol. 2011;87:467–469. doi: 10.1111/j.1600-0609.2011.01693.x. [DOI] [PubMed] [Google Scholar]
- 45.Ka C, Le Gac G, Letocart E, Gourlaouen I, Martin B, Ferec C. Phenotypic and functional data confirm causality of the recently identified hemojuvelin p.r176c missense mutation. Haematologica. 2007;92:1262–1263. doi: 10.3324/haematol.11247. [DOI] [PubMed] [Google Scholar]
- 46.Burri E, Decker M, Eriksson U, Buser P, Hunziker L. 28-year old patient with successfully treated dilatative cardiomyopathy. Internist (Berl) 2008;49:349–352. doi: 10.1007/s00108-007-2007-6. [DOI] [PubMed] [Google Scholar]
- 47.Lee PL, Beutler E. Regulation of hepcidin and iron-overload disease. Annu Rev Pathol. 2009;4:489–515. doi: 10.1146/annurev.pathol.4.110807.092205. [DOI] [PubMed] [Google Scholar]
- 48.Ikuta K, Hatayama M, Addo L, Toki Y, Sasaki K, Tatsumi Y, et al. Iron overload patients with unknown etiology from national survey in Japan. Int J Hematol. 2017;105:353–360. doi: 10.1007/s12185-016-2141-9. [DOI] [PubMed] [Google Scholar]
- 49.Koyama C, Hayashi H, Wakusawa S, Ueno T, Yano M, Katano Y, et al. Three patients with middle-age-onset hemochromatosis caused by novel mutations in the hemojuvelin gene. J Hepatol. 2005;43:740–742. doi: 10.1016/j.jhep.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Kaneko Y, Miyajima H, Piperno A, Tomosugi N, Hayashi H, Morotomi N, et al. Measurement of serum hepcidin-25 levels as a potential test for diagnosing hemochromatosis and related disorders. J Gastroenterol. 2010;45:1163–1171. doi: 10.1007/s00535-010-0259-8. [DOI] [PubMed] [Google Scholar]
- 51.Filali M, Le Jeunne C, Durand E, Grinda JM, Roetto A, Daraio F, et al. Juvenile hemochromatosis HJV-related revealed by cardiogenic shock. Blood Cells Mol Dis. 2004;33:120–124. doi: 10.1016/j.bcmd.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Nagayoshi Y, Nakayama M, Suzuki S, Hokamaki J, Shimomura H, Tsujita K, et al. A Q312X mutation in the hemojuvelin gene is associated with cardiomyopathy due to juvenile haemochromatosis. Eur J Heart Fail. 2008;10:1001–1006. doi: 10.1016/j.ejheart.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Ravasi G, Pelucchi S, Mariani R, Silvestri L, Camaschella C, Piperno A. A severe hemojuvelin mutation leading to late onset of HFE2-hemochromatosis. Dig Liver Dis. 2018;50:859–862. doi: 10.1016/j.dld.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 54.Eisold M, Gehrke S, Stremmel W, Gugler R. A young diabetic with small-nodule liver cirrhosis, high transferrin saturation and negative HFE test. Dtsch Med Wochenschr. 2005;130:1494–1496. doi: 10.1055/s-2005-870845. [DOI] [PubMed] [Google Scholar]
- 55.Berg LB, Milman NT, Friis-Hansen L, Jensen PD, Frund T. Juvenile haemochromatosis caused by a homozygous Gly320Val mutation in the haemojuvelin gene. Ugeskr Laeger. 2013;175:1113–1114. [PubMed] [Google Scholar]
- 56.Cherfane C, Lee P, Guerin L, Brown K. A late presentation of a fatal disease: juvenile hemochromatosis. Case Rep Med. 2013;2013:875093. doi: 10.1155/2013/875093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pauwels R, Vandecasteele E, Devos D, Pauwels W, De Pauw M. An unexpected cause of liver cirrhosis and cardiomyopathy in a young man. Acta Clin Belg. 2018;73:393–397. doi: 10.1080/17843286.2017.1409474. [DOI] [PubMed] [Google Scholar]
- 58.Farrell CP, Parker CJ, Phillips JD. Exome sequencing for molecular characterization of non-HFE hereditary hemochromatosis. Blood Cells Mol Dis. 2015;55:101–103. doi: 10.1016/j.bcmd.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varkonyi J, Lueff S, Szucs N, Pozsonyi Z, Toth A, Karadi I, et al. Hemochromatosis and hemojuvelin G320V homozygosity in a Hungarian woman. Acta Haematol. 2010;123:191–193. doi: 10.1159/000297214. [DOI] [PubMed] [Google Scholar]
- 60.Santos PC, Cancado RD, Pereira AC, Chiattone CS, Krieger JE, Guerra-Shinohara EM. HJV hemochromatosis, iron overload, and hypogonadism in a Brazilian man: treatment with phlebotomy and deferasirox. Acta Haematol. 2010;124:204–205. doi: 10.1159/000321493. [DOI] [PubMed] [Google Scholar]
- 61.Militaru MS, Popp RA, Trifa AP. Homozygous G320V mutation in the HJV gene causing juvenile hereditary haemochromatosis type A. A case report. J Gastrointestin Liver Dis. 2010;19:191–193. [PubMed] [Google Scholar]
- 62.Cooray SD, Heerasing NM, Selkrig LA, Subramaniam VN, Hamblin PS, McDonald CJ, et al. Reversal of end-stage heart failure in juvenile hemochromatosis with iron chelation therapy: a case report. J Med Case Rep. 2018;12:18. doi: 10.1186/s13256-017-1526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Angelopoulos NG, Goula AK, Papanikolaou G, Tolis G. Osteoporosis in HFE2 juvenile hemochromatosis. A case report and review of the literature. Osteoporos Int. 2006;17:150–155. doi: 10.1007/s00198-005-1920-6. [DOI] [PubMed] [Google Scholar]
- 64.Angelopoulos NG, Goula A, Dimitriou E, Tolis G. Reversibility of hypogonadotropic hypogonadism in a patient with the juvenile form of hemochromatosis. Fertil Steril. 2005;84:1744. doi: 10.1016/j.fertnstert.2005.05.070. [DOI] [PubMed] [Google Scholar]
- 65.Angelopoulos N, Papanikolaou G, Noutsou M, Rombopoulos G, Goula A, Tolis G. Glucose metabolism, insulin secretion and insulin sensitivity in juvenile hemochromatosis. A case report and review of the literature. Exp Clin Endocrinol Diabetes. 2007;115:192–197. doi: 10.1055/s-2007-970426. [DOI] [PubMed] [Google Scholar]
- 66.Dhillon BK, Chopra G, Jamwal M, Chandak GR, Duseja A, Malhotra P, et al. Adult onset hereditary hemochromatosis is associated with a novel recurrent Hemojuvelin (HJV) gene mutation in north Indians. Blood Cells Mol Dis. 2018;73:14–21. doi: 10.1016/j.bcmd.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 67.Milman N, Pedersen P, Ovesen L, Melsen G, Fenger K. Frequency of the C282Y and H63D mutations of the hemochromatosis gene (HFE) in 2501 ethnic Danes. Ann Hematol. 2004;83(10):654–657. doi: 10.1007/s00277-004-0874-7. [DOI] [PubMed] [Google Scholar]
- 68.Bacon BR, Powell LW, Adams PC, Kresina TF, Hoofnagle JH. Molecular medicine and hemochromatosis: at the crossroads. Gastroenterology. 1999;116:193–207. doi: 10.1016/S0016-5085(99)70244-1. [DOI] [PubMed] [Google Scholar]
- 69.Silvestri L, Pagani A, Fazi C, Gerardi G, Levi S, Arosio P, et al. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007;109:4503–4510. doi: 10.1182/blood-2006-08-041004. [DOI] [PubMed] [Google Scholar]
- 70.Chan JC, Malik V, Jia W, Kadowaki T, Yajnik CS, Yoon KH, et al. Diabetes in Asia: epidemiology, risk factors, and pathophysiology. JAMA. 2009;301:2129–2140. doi: 10.1001/jama.2009.726. [DOI] [PubMed] [Google Scholar]
- 71.Rausa M, Ghitti M, Pagani A, Nai A, Campanella A, Musco G, et al. Identification of TMPRSS6 cleavage sites of hemojuvelin. J Cell Mol Med. 2015;19:879–888. doi: 10.1111/jcmm.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly TM, Edwards CQ, Meikle AW, Kushner JP. Hypogonadism in hemochromatosis: reversal with iron depletion. Ann Intern Med. 1984;101:629–632. doi: 10.7326/0003-4819-101-5-629. [DOI] [PubMed] [Google Scholar]
- 73.Evangelista AS, Nakhle MC, de Araujo TF, Abrantes-Lemos CP, Deguti MM, Carrilho FJ, et al. HFE genotyping in patients with elevated serum iron indices and liver diseases. Biomed Res Int. 2015;2015:164671. doi: 10.1155/2015/164671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olynyk JK, Cullen DJ, Aquilia S, Rossi E, Summerville L, Powell LW. A population-based study of the clinical expression of the hemochromatosis gene. N Engl J Med. 1999;341:718–724. doi: 10.1056/NEJM199909023411002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search terms used for database searches. (DOCX 18 kb)
Included and excluded articles. (DOCX 161 kb)
Clinical findings for cases with biallelic mutations. (DOCX 96 kb)
Clinical findings for cases with monoallelic mutation. (DOCX 56 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.