Abstract
Background
Acetyl xylan esterase plays an important role in the complete enzymatic hydrolysis of lignocellulosic materials. It hydrolyzes the ester linkages of acetic acid in xylan and supports and enhances the activity of xylanase. This study was conducted to identify and overexpress the acetyl xylan esterase (AXE) gene revealed by the genomic sequencing of the marine bacterium Ochrovirga pacifica.
Results
The AXE gene has an 864-bp open reading frame that encodes 287 aa and consists of an AXE domain from aa 60 to 274. Gene was cloned to pET-16b vector and expressed the recombinant AXE (rAXE) in Escherichia coli BL21 (DE3). The predicted molecular mass was 31.75 kDa. The maximum specific activity (40.08 U/mg) was recorded at the optimal temperature and pH which were 50 °C and pH 8.0, respectively. The thermal stability assay showed that AXE maintains its residual activity almost constantly throughout and after incubation at 45 °C for 120 min. The synergism of AXE with xylanase on beechwood xylan, increased the relative activity 1.41-fold.
Conclusion
Resulted higher relative activity of rAXE with commercially available xylanase on beechwood xylan showed its potential for the use of rAXE in industrial purposes as a de-esterification enzyme to hydrolyze xylan and hemicellulose-like complex substrates.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1169-y) contains supplementary material, which is available to authorized users.
Keywords: Acetyl xylan esterase, Marine bacteria, Ochrovirga pacifica, Synergism, Beech wood xylan
Introduction
Hemicellulose is the second most abundant polysaccharide type in land plant cell walls and it consisted of about 25–35% of forest and agricultural residues [1, 2]. Hemicellulose differs from cellulose in its heterogeneous chemical composition associated with β-1,4-xylan, which has high polymerization ability and is highly branched [3]. Hemicellulose consists of a linear backbone of β-1,4-linked xyloses and short-chain branches of O-acetyl, α-l-arabinofuranosyl, and α-d-glucuronyl residues [4]. Enzymatic hydrolysis of xylan is catalyzed by endoxylanase and various side chain-cleaving enzymes, such as β-xylosidase, α-glucuronidase, α-arabinofuranosidase, and acetyl xylan esterase [5]. Hardwood xylans (e.g., beechwood and birchwood xylan) are highly acetylated, and acetyl xylan esterase plays a major role in making them partially soluble in water [6] by turning them to short chains through enzymatic degradation. Determining different degradation strategies for these complex and economically important substances is important for their industrial exploitation.
The first use of xylanases [7] obtained from microbes by the pulp and paper industry garnered much interest; this was followed by many studies in this domain over the past few decades [8–10]. Enzymes derived from bacteria [11, 12], including actinomycetes [13, 14] as well as yeast [15, 16], have been used in these studies and are recently being applied at industrial scales [17]. Acetyl xylan esterase facilitates the action of endoxylanases by increasing the approachability to the xylan backbone by cleaving the ester bonds of acetyl groups [6]. Therefore, the synergistic action of acetyl xylan esterase and endoxylanases increases the efficient hydrolysis of xylan [18, 19]. The marine bacterium Ochrovirga pacifica belongs to the family Flavobacteriaceae and was identified from a seaweed sample during our previous study [20]. During genome sequence analysis of O. pacifica, an acetyl xylan esterase (AXE) gene was found. This study was conducted to characterize this AXE gene and perform cloning, expression, and biochemical characterization of the expressed recombinant enzyme. Finally, the synergistic effect of recombinant AXE was tested with commercially available xylanase on beechwood xylan as the substrate.
Methods
Bacterial strains, culture conditions, plasmid, and reagents
Escherichia coli DH5α and BL21 (DE3) strains were used as the cloning and expression hosts, respectively. Both strains were grown in Luria–Bertani broth (LB broth) at 37 °C with agitation at 180 rpm. The pET-16b (Novagen, Madison, USA) vector was used for enzyme expression and purification. Buffers and enzymes used for the polymerase chain reaction (PCR) and DNA manipulation were purchased from Takara (Takara Bio Inc., Shiga, Japan). PCR products were purified using an AccuPrep® Gel Purification Kit (Bioneer, Daejeon, South Korea), and cloned plasmids were extracted using an AccuPrep® Plasmid MiniPrep DNA Extraction Kit (Bioneer). Substrates and other reagents used for the enzyme assay, including p-nitrophenyl acetate (p-NPA), p-nitrophenol (p-NP), d-xylose, and dinitrosalicylic acid, were purchased from Sigma-Aldrich (St Louis, MO, USA). The synergistic effect was verified with commercially available endo-1,4-β-xylanase derived from Aspergillus niger (Megazyme Int., Wicklow, Ireland) using beechwood xylan (Tokyo Chemical Industry Co. Ltd., Tokyo, Japan) as the substrate.
Identification and molecular characterization of AXE
The marine bacterium O. pacifica was isolated from a seaweed sample collected from Chuuk State, Federated States of Micronesia [20], and its genome was sequenced [21]. The AXE gene was identified, and conserved domains were predicted using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD; http://www.ncbi.nlm.nih.gov/cdd/). The Signal IP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) [22] was used to predict the N-terminal signal peptide of the AXE amino acid sequence, and the EMBOSS Pairwise Sequence Alignment Tool (https://www.ebi.ac.uk/Tools/psa/) [23] was used to calculate the identity, similarity, and gap percentages of the AXE amino acid sequence against the closest neighbor proteins identified by the NCBI BLAST program (https://blast.ncbi.nlm.nih.gov) as well as several acetyl xylan esterases characterized by other works [24]. The isoelectric point and molecular weight were determined using the protein isoelectric point calculator (http://isoelectric.org/calculate.php) [25] and DNA Dynamo (Blue Tractor Software, North Wales, UK), respectively. The nucleotide and amino acid sequence of AXE were submitted to Genbank under accession number MH937751.
Cloning of the AXE gene
PCR was performed to amplify the targeted AXE gene from the genomic DNA of O. pacifica without the predicted N-terminal signal sequence. Forward (GAGAGACATATGCAAAAAGAAGTAAAGTTGGCC) and reverse (GAGAGAGGATCCTTATTCTACTTTGCTTATAGGAAC) primers were designed to bind the pET-16b cloning site using the pET-16b sequence. The PCR mixture consisted of 1 μL of genomic DNA template (200 ng/μL), 35.5 μL sterile deionized water, 5 μL 10× Ex Taq buffer (20 mM Mg2+), forward and reverse primers (20 pmol each), 4 μL dNTPs (2.5 mM), and Ex Taq DNA polymerase (3 U). PCR amplification conditions were as follows: initial denaturation at 94 °C for 5 min; 30 cycles of denaturation at 94 °C for 30 s, annealing at 48 °C for 30 s, extension at 72 °C for 1 min 20 s; and a final extension at 72 °C for 5 min. PCR products were purified using an AccuPrep Gel Purification Kit. The pET-16b vector and purified PCR products were digested with NdeI and BamHI restriction enzymes (Takara Bio Inc.) according to the manufacturer’s instructions. Digested PCR product was ligated into the digested pET-16b vector using T4 DNA ligase according to the manufacturer’s protocol. Recombinant plasmid was transformed into E. coli DH5α cells by heat shock [26]. Clones of recombinant plasmid were purified using the plasmid extraction kit following the manufacturer’s instructions; it was then transformed to the expression host E. coli BL21 (DE3) by heat shock [26]. The nucleotide sequence of the constructed recombinant plasmid was confirmed by sequencing (Macrogen, Korea).
Protein expression and purification
The clone harboring pET16b-AXE was incubated overnight at 37 °C in 4 mL LB broth supplemented with 100 µg/mL ampicillin (LB-amp); 200 mL LB-amp broth was inoculated with total volume of the overnight culture and incubated at 37 °C with agitation until the optical density at 600 nm reached ~ 0.6. Then, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a 1 mM final concentration. The culture was incubated for another 24 h at 20 °C under conditions designed to induce expression of the recombinant acetyl xylan esterase (rAXE). Cells were harvested by centrifugation at 8000×g for 10 min at 4 °C. Preparation of cell lysate and purification of histidine-tagged rAXE were performed according to the user protocol for the His·Bind® Resin Chromatography Kit (Novagen). Purified protein was quantified by Bradford reagent (Sigma, USA) with bovine serum albumin (BSA) as a standard. The molecular mass and purity were evaluated using 12% Sodium dodecyl sulfate poly-acrylamide gel electrophoresis (SDS-PAGE). A pre-stained protein marker (Lonza ProSieve™, Rockland, USA) was used as the reference.
Enzyme assay
p-NPA was used as the substrate for the rAXE activity assay. The reaction was performed on a microplate at 30 °C in a 200 µL total reaction volume containing 195 µL phosphate buffer (10 mM, pH 6.7), 5 µL purified rAXE enzyme, and 100 µL substrate (0.3423 mM). p-NP released within 10 min was measured by monitoring the absorbance at 405 nm (BioTek Instruments, Winooski, USA). p-NP was used as the standard. One unit of enzyme activity was defined as the amount of rAXE required to release 1 µmol p-NP in 10 min under standard conditions. Each and every assay was performed with a blank contained inactivated enzyme which was run under same pH and temperature conditions.
Biochemical characterization of rAXE
The optimum temperature and pH were determined using p-NPA as the substrate at different temperatures (25–55 °C) and pHs (phosphate citrate buffer for pH 2.0–7.0 and glycine–NaOH buffer for pH 8.0–10.0 at 50 °C) under standard conditions.
Effect of temperature and pH on rAXE stability
To analyze the temperature stability of rAXE, 5 µL purified enzyme was pre-incubated in 195 µL phosphate buffer (pH 6.7) at 45 °C, 50 °C, and 55 °C over various time durations (0–120 min at 20 min intervals); this was followed by cooling on ice for 5 min. Finally, residual activity was measured under standard assay conditions. To measure the effect of pH on rAXE stability, the enzyme was incubated in a series of buffers (phosphate–citrate buffer for pH 2–7.0 and glycine–NaOH buffer for pH 8–10) at 4 °C for 12 h, and residual activity was tested under standard assay conditions. rAXE without pretreatment was used as the control for both tests.
Effect of metal ions and salt on rAXE
The effect of different metal ions on rAXE activity was evaluated using 1 mM and 5 mM solutions of Ca2+, Mg2+, Fe2+, Mn2+, Cu2+, Zn2+ and finally ethylenediaminetetraacetic acid (EDTA). The metal ions were incorporated into the buffer solution, and the enzyme activity assay was performed under standard conditions. The relative enzyme activity was calculated using the activity of the untreated sample as 100%. The effect of salt on rAXE was analyzed under standard assay conditions with different NaCl concentrations (0.05–0.5 M). The effect of NaCl on rAXE stability was analyzed by incubating the enzyme (5 µL) in different concentrations of NaCl (0.05–0.5 M) solutions at 4 °C for 12 h and assaying the residual activity of the enzyme in each solution.
Synergistic effect of rAXE
The synergistic effect of rAXE on the activity of a commercially available endo-1,4-β-xylanase was assayed by measuring the xylose released using a modified 3,5-dinitrosalicylic acid (DNS) method [27] with d-xylose as the standard. The reaction mixture was prepared in an Eppendorf tube containing 1% beechwood xylan in phosphate buffer (pH 8.0), 5 U xylanase, and 1.7 U AXE. Incubation was carried out at 50 °C, and activity was checked at 30 min intervals for 2 h. The amount of enzyme required to produce 1 μmol of reduced sugar per minute was defined as one unit of enzyme activity.
Results
Identification and molecular characterization of rAXE
The AXE gene has an 864-bp open reading frame that encodes 287 aa the acetyl esterase domain can be found from aa 60 to 274. The predicted molecular mass and isoelectric point were 31.75 kDa and 8.35, respectively. The N-terminal region of the protein contains a signal peptide consisting of 21 aa (Fig. 1). Conserved domain analysis [28] revealed that the enzyme belongs to the alpha/beta hydrolase family and has a conserved acetyl esterase/lipase (COG0657) belonging to the acetyl esterase superfamily. Other nonspecific hits, such as alpha/beta hydrolase fold (pfam07859), dienelactone hydrolase (COG0412), predicted esterase (COG0400), and carboxylesterase type B (COG2272), have also been associated with the sequence. AXE showed the highest sequence identity (55.9%) with an alpha/beta hydrolase of Wenyingzhuangia fucanilytica (GenBank Accession No. WP_068826614.1); additional closest matching proteins were 1,4-beta-xylanase (54.9%) of W. fucanilytica (ANW96467.1), alpha/beta hydrolase (51.5%) of Muricauda antarctica (WP_072877402.1), and alpha/beta hydrolase (51.2%) of Reichenbachiella versicolor (WP_109831252.1). These sequences were also compared to amino acid sequences of four characterized acetyl xylan esterases (Table 1).
Fig. 1.
Nucleotide and deduced amino acid sequence of AXE. The N-terminal signal sequence is underlined; the acetyl esterase domain (aa 60 to 274) is highlighted; asterisk indicates a stop codon
Table 1.
Identity and similarity comparisons of the AXE amino acid sequence with its uncharacterized closest neighbor enzymes identified by NCBI BLAST, as well as characterized acetyl xylan esterases from Bacillus pumilus [29], Flavobacterium johnsoniae UW101 [30], Butyrivibrio proteoclasticus B316 [31], Ruminococcus flavefaciens 17 [32], and Streptomyces lividans 1326 [33]
| Organism | Accession no. | Identity (%) | Similarity (%) | Gap (%) | Remarks |
|---|---|---|---|---|---|
| W. fucanilytica | WP_068826614.1 | 55.9 | 70.9 | 6.0 | Uncharacterized |
| W. fucanilytica | ANW96467.1 | 54.9 | 68.7 | 10.4 | Uncharacterized |
| M. antarctica | WP_072877402.1 | 51.5 | 69.4 | 5.4 | Uncharacterized |
| R. versicolor | WP_109831252.1 | 51.2 | 66.1 | 5.4 | Uncharacterized |
| B. pumilus | CAB76451.2 | 14.9 | 27.7 | 49.8 | Characterized |
| F. johnsoniae UW101 | ABQ06890.1 | 14.1 | 24.01 | 43.05 | Characterized |
| B. proteoclasticus B316 | ADL35669.1 | 10.1 | 19.4 | 63.3 | Characterized |
| R. flavefaciens 17 | CAB55348.1 | 8.3 | 13.3 | 71.5 | Characterized |
| S. lividans 1326 | AAC06115.2 | 7.3 | 12.7 | 70.7 | Characterized |
Expression and purification of rAXE
The AXE gene was expressed in pET-16b vector with an N-terminal 10-histidine tag supplied with the vector to facilitate the affinity purification of the target protein. Figure 2 shows the high expression level and purity of the recombinant protein, which gave a strong band on SDS-PAGE. The protein was purified by one-step affinity purification using a His·Bind® Resin Chromatography Kit (Novagen). The molecular mass of the protein estimated by SDS-PAGE was ~ 32.0 kDa, roughly equal to the predicted molecular mass (32.1 kDa).
Fig. 2.
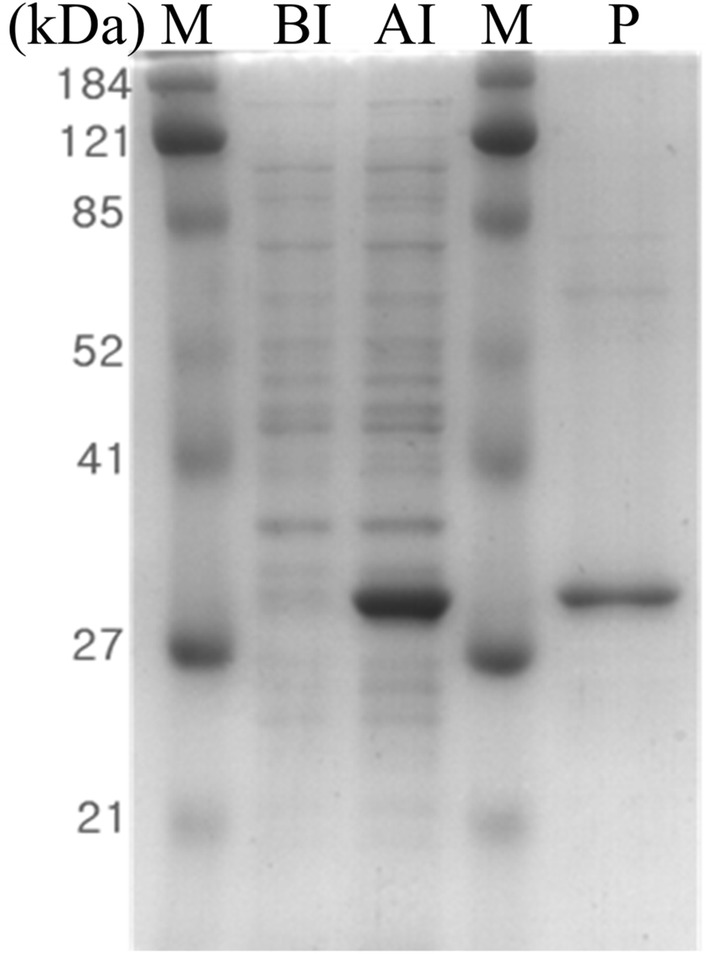
SDS-PAGE analysis of rAXE. M molecular weight marker, BI whole cell lysate before induction, AI whole cell lysate after induction (incubated at 20 °C for 16 h with 180 rpm agitation), P purified rAXE using His·Bind® Resin Chromatography Kit
Biochemical characterization of rAXE
The optimal temperature for rAXE was 50 °C; its activity decreased around 20% at temperatures > 50 °C (Fig. 3a). The thermal stability assay showed that rAXE maintains its residual activity almost constantly throughout and after incubation at 45 °C for 120 min (Fig. 3b). Incubation at 55 °C for 20 min reduced its activity by more than 50%. rAXE showed maximum activity at pH 8.0 (assayed at 50 °C) and exhibited less than 25% relative activity at pHs < 6.0 (Fig. 3c). Activity was drastically decreased above pH 8.0, and 23% relative activity was observed at pH 10.0. After 12 h incubation at 4 °C and pH 3.0–5.0, there was 0% remaining enzyme activity. Residual activities at pHs 7.0, 8.0, and 9.0 were > 40%; stability dramatically decreased at pHs > 9.0 (Fig. 3d). The maximum specific activity of rAXE towards p-NPA was recorded at pH 8.0 (assayed at 50 °C) as 40.08 U/mg.
Fig. 3.
Effects of pH and temperature on rAXE activity. a Effect of temperature on relative enzyme activity (relative activity was calculated using activity at 50 °C as 100%). b Thermal stability assay (relative activity was calculated using activity of untreated enzyme as 100%). c Effect of pH on activity at 50 °C (relative activity was calculated using activity at pH 8.0 as 100%). d pH stability assay (relative activity was calculated using activity of enzyme treated with pH 8.0 buffer as 100%). Data are shown as mean ± standard deviation (sd), n = 3
Effect of metal ions and salt on rAXE activity
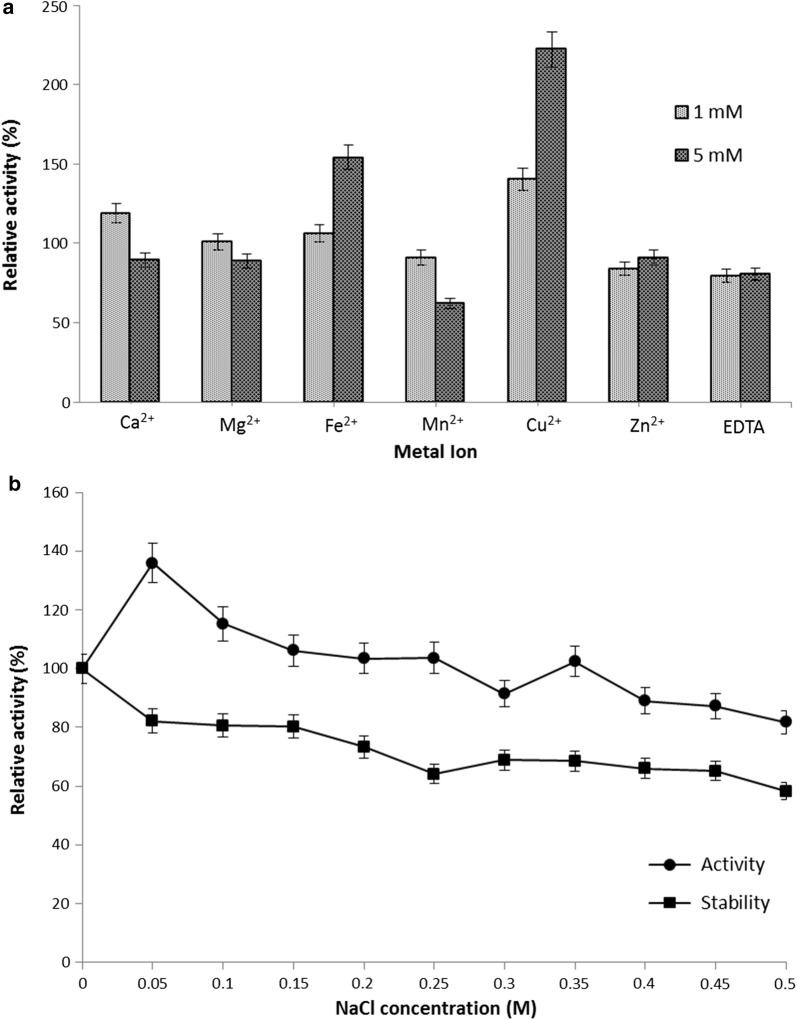
The effect of metal ions on rAXE activity was determined using the activity of the untreated enzyme as the control (100%). Relative activity was examined at two concentrations (1 mM and 5 mM) of each metal ion. Reaction mixtures containing 1 mM Ca2+, 1 or 5 mM Cu2+, and 5 mM Fe2+ showed strong stimulatory effects on enzyme activity; 5 mM Ca2+, 5 mM Mg2+, both 1 mM and 5 mM Mn2+ or Zn2+ and EDTA showed inhibitory effects on enzyme activity (Fig. 4a). The effect of NaCl on rAXE activity and stability was examined; as shown in Fig. 4b, enzymatic activity was strongly stimulated at 0.05 M NaCl, whereas activity gradually decreased at higher concentrations (Fig. 4b). However, relative activity higher than 100% was recorded at NaCl concentrations between 0.05 and 0.25 M. Incubation in NaCl for 12 h had no positive effect on enzyme activity, and the residual activity for all concentrations was < 100% relative activity.
Fig. 4.
a Effect of various metal ions on the relative activity of rAXE. The enzyme reaction was performed with final concentrations of 1 mM and 5 mM for each metal ion. The activity in the absence of a metal ion was taken as the control (100%). b Effect of NaCl on the relative activity and stability of rAXE. The activity at 0 M NaCl in the reaction mixture was taken as the control (100%). Data are presented as mean ± standard deviation (sd), n = 3
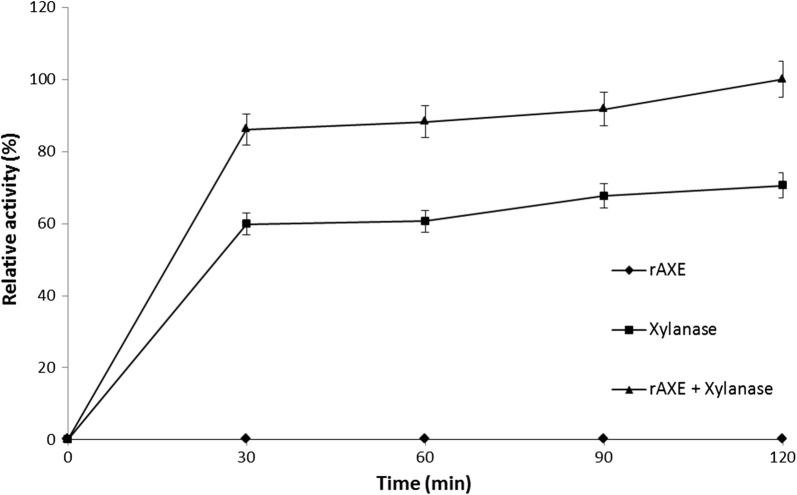
Synergistic effect of rAXE
The synergistic effect of rAXE on the activity of a commercially available xylanase was assayed (Fig. 5). The reaction mixture prepared with rAXE only (1.7 U) showed no relative activity; xylanase only (5 U) showed ~ 70% relative activity. The reaction mixture containing both xylanase and rAXE showed 29.47% higher relative activity compared to xylanase alone (a 1.41-fold increase).
Fig. 5.
Synergism of AXE with a commercially available xylanase on beechwood xylan as the substrate. The reaction mixtures were prepared in Eppendorf tubes containing 1% beechwood xylan in phosphate buffer (pH 8.0) and incubated over the time at 50 °C. Combination of rAXE and xylanase was included 1.7 U of rAXE and 5 U of commercially available endo-1,4-β-xylanase derived from A. niger. Only rAXE and xylanase contained reaction mixtures were included 1.7 U and 5 U respectively. Relative activity was determined with the activity of rAXE + xylanase at 120 min as 100%. Data are given as means ± SD, n = 3
Discussion
This study was conducted to characterize the acetyl xylan esterase gene from the marine bacterium O. pacifica, isolated from a seaweed sample [20]. This is the first report of such an enzyme from the genus Ochrovirga (family: Flavobacteriaceae), and further biochemical characterization of the expressed enzyme within an E. coli expression system was performed. Genomic DNA analysis of O. pacifica revealed the presence of the AXE gene, and NCBI conserved domain analysis [34] revealed the acetyl esterase domain within the amino acid sequence (aa 60–274). The acetyl esterase domain belongs to the alpha/beta hydrolase superfamily. This superfamily consists of lipases, peroxidases, proteases, epoxide hydrolases, and dehalogenases, which are functionally diversified for hydrolyzing different substrates [35]. Several other nonspecific hits, such as alpha/beta hydrolase protein fold, were also associated with the amino acid sequence, which may have developed evolutionarily to hydrolyze substrates with different chemical and physicochemical properties. The highest sequence identity (55.9%) matched the uncharacterized alpha/beta hydrolase of W. fucanilytica (GenBank Accession No. WP_068826614.1), demonstrating the uniqueness of the AXE gene of O. pacifica. A phylogenetic tree of the AXE amino acid sequence along with the closest matches identified by BLAST and five characterized acetyl xylan esterases reported previously showed that AXE from O. pacifica is positioned in a separate clade (Additional file 1: Fig. S1). The family Flavobacteriaceae is well known for the degradation of macromolecules, such as complex carbohydrates [36]. Most species of the family grow on algal thalli and have the ability to degrade algal cells [37]. Razeq et al. [30] have characterized and reported an acetyl xylan esterase encoded in the Flavobacterium johnsoniae genome (FjoAcXE), which is a member of same family. However, the phylogenetic relationship between AXE and FjoAcXE is not close (see Additional file 1).
The presence of the N-terminal signal sequence suggests that the AXE enzyme may be secreted for extracellular hydrolysis processes; expression for the experiment was done without the signal sequence. The optimum temperature of rAXE was 50 °C, and it maintained more than 80% of its residual activity after incubation for 2 h at 50 °C. In this experiment optimum temperature was examined within 25 °C to 55 °C range. Reason was substrate converted to products rapidly in higher temperatures. Previously characterized acetyl xylan esterases from different microorganisms [19, 38] showed optimal temperatures < 50 °C and lower thermal stability compared to rAXE; this may be advantageous in industrial applications of rAXE. The optimal pH of rAXE at the optimal temperature was pH 8.0. Most fungal and rumen bacterial-derived acetyl xylan esterases show neutral optimal pHs [39, 40], whereas bacteria living in harsh conditions and marine environments show alkaline pHs of 8.0–9.5 [29, 41, 42]. Characteristics of the enzymes and proteins derived from the microbes are highly depended on their ecological conditions [43–45]. Naturally they use their enzymes to degrade the substrates in environment and play a major role in nutrient cycling [46]. At present there is an increasing attention on the marine microbial enzymes because of their good performances in hard conditions like high or low temperatures, pressure, pH and high salt concentrations [47, 48].
The effect of different metal ions on the activity of rAXE was evaluated. Strong stimulation was observed with the presence of 1 mM of Ca2+, 1 and 5 mM Cu2+, and 5 mM Fe2+ in the reaction mixture. Further studies of metal ion interactions with rAXE are recommended. According to Taylor et al. [49], some acetyl xylan esterases from microorganisms show special metal ion preferences that enhance their catalytic activities. Furthermore, 5 mM Ca2+, 5 mM Mg2+, and both 1 and 5 mM Mn2+ and Zn2+ showed inhibitory effects on enzyme activity; the inhibitory effect of Zn2+ on esterase has been reported by several authors [50, 51]. The effects of salt on rAXE activity and stability were tested and 0.05 M NaCl strongly stimulated rAXE activity. Most esterases derived from microorganisms are more salt tolerant than rAXE; enhanced activity has been demonstrated within 0.0–4.0 M NaCl [52, 53]. According to Steiner and Lindskog [54], the stimulatory activity of salt occurs due to the increased chemical potential of p-NPA in aqueous solutions rather than participation in the enzyme-related catalytic steps.
The synergistic effect of xylanase and rAXE on beechwood xylan was investigated. The cooperative effect of rAXE and xylanase on xylan initiates with the de-esterification of acetyl substitutes on side chains, making them more accessible to xylanase for hydrolysis of the glycosidic bonds [2]. Shorter polysaccharide fragments resulted from the increase in the substrate preference of rAXE attributable to xylanase activity [6]. Furthermore, decreased polymerization caused by xylanase may increase the reaction rate by decreasing the viscosity of the reaction mixture, allowing more substrate-rAXE interactions [18]. In our study, the mixture of rAXE and xylanase exhibited a 1.41-fold increase in relative activity on beechwood xylan compared to xylanase alone. Therefore, rAXE has good potential as an accessory enzyme for hydrolyzing xylan and hemicellulose.
Conclusion
The present study identified and biochemically characterized rAXE from the marine bacterium O. pacifica, which was isolated from a seaweed sample. This is the first report of such an enzyme from the genus Ochrovirga. The synergistic activity of rAXE with commercially available xylanase showed higher relative activity on beechwood xylan. Therefore, there is strong potential for the use of rAXE in industrial purposes as a de-esterification enzyme to hydrolyze xylan and hemicellulose-like complex substrates.
Additional file
Additional file 1. Phylogenetic analysis of O. pacifica AXE along with the uncharacterized closest amino acid sequences identified by NCBI BLAST and several characterized acetyl xylan esterases. The neighbor-joining tree was constructed using the bootstrap method with 1000 replications.
Acknowledgements
Not applicable.
Abbreviations
- AXE
acetyl xylan esterase
- rAXE
recombinant acetyl xylan esterase
- p-NPA
p-nitrophenyl acetate
- p-NP
p-nitrophenol
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- BSA
bovine serum albumin
- DNS
3,5-dinitrosalicylic acid
- EDTA
ethylenediaminetetraacetic acid
- FjoAcXE
acetyl xylan esterase encoded in the Flavobacterium johnsoniae genome
Authors’ contributions
SAH and YKK performed the majority of the laboratory works. YL, EJ and TYE contributed to the interpretation of the results. YHK and SDM also performed the laboratory experiments. DHK and MDZ helped to revise the manuscript. SAH wrote the manuscript and overall work was supervised by CO. All authors read and approved the final manuscript.
Funding
This work was supported by the grant from Korea Institute of Ocean Science & Technology (KIOST) (Research grant—PE99722).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deutschmann R, Dekker RF. From plant biomass to bio-based chemicals: latest developments in xylan research. Biotechnol Adv. 2012;30:1627–1640. doi: 10.1016/j.biotechadv.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Tong X, Lange L, Grell MN, Busk PK. Hydrolysis of wheat arabinoxylan by two acetyl xylan esterases from Chaetomium thermophilum. Appl Biochem Biotechnol. 2015;175:1139–1152. doi: 10.1007/s12010-014-1348-6. [DOI] [PubMed] [Google Scholar]
- 3.Selvarajan E, Veena R. Recent advances and future perspectives of thermostable xylanase. Biomed Pharmacol J. 2017;10:261–279. doi: 10.13005/bpj/1106. [DOI] [Google Scholar]
- 4.Walia A, Guleria S, Mehta P, Chauhan A, Parkash J. Microbial xylanases and their industrial application in pulp and paper biobleaching: a review. 3 Biotech. 2017;7:11. doi: 10.1007/s13205-016-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juturu V, Wu JC. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv. 2012;30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Beg Q, Kapoor M, Mahajan L, Hoondal G. Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol. 2001;56:326–338. doi: 10.1007/s002530100704. [DOI] [PubMed] [Google Scholar]
- 7.Viikari L, Ranua M, Kantelinen A, Sundquist J, Linko M. Bleaching with enzymes. In: Proceedings of 3rd international conference on biotechnology in the pulp and paper industry, STFI, Stockholm. 1986. p. 67–69.
- 8.Nair SG, Sindhu R, Shashidhar S. Enzymatic bleaching of kraft pulp by xylanase from Aspergillus sydowii SBS 45. Indian J Microbiol. 2010;50:332–338. doi: 10.1007/s12088-010-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sridevi A, Ramanjaneyulu G, Devi PS. Biobleaching of paper pulp with xylanase produced by Trichoderma asperellum. 3 Biotech. 2017;7:266. doi: 10.1007/s13205-017-0898-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangwar AK, Prakash NT, Prakash R. Applicability of microbial xylanases in paper pulp bleaching: a review. BioResources. 2014;9:3733–3754. doi: 10.15376/biores.9.2.3733-3754. [DOI] [Google Scholar]
- 11.McDermid KP, Forsberg C, MacKenzie C. Purification and properties of an acetylxylan esterase from Fibrobacter succinogenes S85. Appl Environ Microbiol. 1990;56:3805–3810. doi: 10.1128/aem.56.12.3805-3810.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shareck F, Biely P, Morosoli R, Kluepfel D. Analysis of DNA flanking the xlnB locus of Streptomyces lividans reveals genes encoding acetyl xylan esterase and the RNA component of ribonuclease P. Gene. 1995;153:105–109. doi: 10.1016/0378-1119(94)00763-I. [DOI] [PubMed] [Google Scholar]
- 13.Halgasová N, Kutejová E, Timko J. Purification and some characteristics of the acetylxylan esterase from Schizophyllum commune. Biochem J. 1994;298:751–755. doi: 10.1042/bj2980751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linden J, Samara M, Decker S, Johnson E, Boyer M, Pecs M, Adney W, Himmel M. Purification and characterization of an acetyl esterase from Aspergillus niger. Appl Biochem Biotechnol. 1994;45:383–393. doi: 10.1007/BF02941813. [DOI] [PubMed] [Google Scholar]
- 15.Morais CG, Cadete RM, Uetanabaro APT, Rosa LH, Lachance M-A, Rosa CA. d-xylose-fermenting and xylanase-producing yeast species from rotting wood of two Atlantic Rainforest habitats in Brazil. Fungal Genet Biol. 2013;60:19–28. doi: 10.1016/j.fgb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Adsul MG, Bastawde KB, Gokhale DV. Biochemical characterization of two xylanases from yeast Pseudozyma hubeiensis producing only xylooligosaccharides. Bioresour Technol. 2009;100:6488–6495. doi: 10.1016/j.biortech.2009.07.064. [DOI] [PubMed] [Google Scholar]
- 17.Viikari L, Kantelinen A, Sundquist J, Linko M. Xylanases in bleaching: from an idea to the industry. FEMS Microbiol Rev. 1994;13:335–350. doi: 10.1111/j.1574-6976.1994.tb00053.x. [DOI] [Google Scholar]
- 18.Biely P, MacKenzie CR, Puls J, Schneider H. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Nat Biotechnol. 1986;4:731. doi: 10.1038/nbt0886-731. [DOI] [Google Scholar]
- 19.Blum DL, Li X-L, Chen H, Ljungdahl LG. Characterization of an acetyl xylan esterase from the anaerobic fungus Orpinomyces sp. strain PC-2. Appl Environ Microbiol. 1999;65:3990–3995. doi: 10.1128/aem.65.9.3990-3995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon YK, Kim JH, Kim JJ, Yang SH, Ye BR, Heo SJ, Hyun JH, Qian ZJ, Park HS, Kang DH, Oh C. Ochrovirga pacifica gen. nov., sp. nov., a novel agar-lytic marine bacterium of the family Flavobacteriaceae isolated from a seaweed. Curr Microbiol. 2014;69:445–450. doi: 10.1007/s00284-014-0598-4. [DOI] [PubMed] [Google Scholar]
- 21.Oh C, Kwon Y-K, Heo S-J, De Zoysa M, Affan A, Lee Y, Lee J, Choi Y-U, Park H-S, Jung K-H. Complete genome sequence of strain S85, a novel member of the family Flavobacteriaceae. J Bacteriol. 2011;193:6107. doi: 10.1128/JB.05993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozlowski LP. IPC—isoelectric point calculator. Biol Direct. 2016;11:55. doi: 10.1186/s13062-016-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. 2007;6:253. doi: 10.3791/253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 28.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2006;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degrassi G, Kojic M, Ljubijankic G, Venturi V. The acetyl xylan esterase of Bacillus pumilus belongs to a family of esterases with broad substrate specificity. Microbiol. 2000;146:1585–1591. doi: 10.1099/00221287-146-7-1585. [DOI] [PubMed] [Google Scholar]
- 30.Razeq FM, Jurak E, Stogios PJ, Yan R, Tenkanen M, Kabel MA, Wang W, Master ER. A novel acetyl xylan esterase enabling complete deacetylation of substituted xylans. Biotechnol Biofuels. 2018;11:74. doi: 10.1186/s13068-018-1074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly WJ, Leahy SC, Altermann E, Yeoman CJ, Dunne JC, Kong Z, Pacheco DM, Li D, Noel SJ, Moon CD. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS ONE. 2010;56:56. doi: 10.1371/journal.pone.0011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aurilia V, Martin JC, McCrae SI, Scott KP, Rincon MT, Flint HJ. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology. 2000;146:1391–1397. doi: 10.1099/00221287-146-6-1391. [DOI] [PubMed] [Google Scholar]
- 33.Shareck F, Roy C, Makoto Y, Morosoli R, Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991;107:75–82. doi: 10.1016/0378-1119(91)90299-Q. [DOI] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nardini M, Dijkstra BW. α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 36.Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, Huettel B, Stüber K, Reinhardt R, Harder J. The genome of the algae-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for the degradation of algal polysaccharides. Appl Environ Microbiol. 2013;79:6813–6822. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol. 2012;14:2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 38.Kam DK, Jun H-S, Ha JK, Inglis GD, Forsberg CW. Characteristics of adjacent family 6 acetylxylan esterases from Fibrobacter succinogenes and the interaction with the Xyn10E xylanase in hydrolysis of acetylated xylan. Can J Microbiol. 2005;51:821–832. doi: 10.1139/w05-074. [DOI] [PubMed] [Google Scholar]
- 39.Chung H-J, Park S-M, Kim H-R, Yang M-S, Kim D-H. Cloning the gene encoding acetyl xylan esterase from Aspergillus ficuum and its expression in Pichia pastoris. Enzyme Microb Technol. 2002;31:384–391. doi: 10.1016/S0141-0229(02)00122-9. [DOI] [Google Scholar]
- 40.Pouvreau L, Jonathan M, Kabel M, Hinz S, Gruppen H, Schols H. Characterization and mode of action of two acetyl xylan esterases from Chrysosporium lucknowense C1 active towards acetylated xylans. Enzyme Microb Technol. 2011;49:312–320. doi: 10.1016/j.enzmictec.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 41.De Santi C, Willassen NP, Williamson A. Biochemical characterization of a family 15 carbohydrate esterase from a bacterial marine arctic metagenome. PLoS ONE. 2016 doi: 10.1371/journal.pone.0159345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cieśliński H, Białkowska AM, Długołęcka A, Daroch M, Tkaczuk KL, Kalinowska H, Kur J, Turkiewicz M. A cold-adapted esterase from psychrotrophic Pseudoalteromas sp. strain 643A. Arch Microbiol. 2007;188:27–36. doi: 10.1007/s00203-007-0220-2. [DOI] [PubMed] [Google Scholar]
- 43.Tchigvintsev A, Reva O, Polaina J, Bargiela R, Canet A, Valero F, Eguizabal ER, Yakunin AF, Ferrer M. Biochemical diversity of carboxyl esterases and lipases from Lake Arreo (Spain): a metagenomic approach. Appl Environ Microbiol. 2013;79:3553–3562. doi: 10.1128/AEM.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, Yuan M, Wei L, Zhang Y, Lin Y, Zhang L, Liu Z. Characterization of a novel cold-adapted phosphinothricin N-acetyltransferase from the marine bacterium Rhodococcus sp. strain YM12. J Mol Catal B Enzym. 2014;104:23–28. doi: 10.1016/j.molcatb.2014.03.001. [DOI] [Google Scholar]
- 45.Beier S, Jones CM, Mohit V, Hallin S, Bertilsson S. Global phylogeography of chitinase genes in aquatic metagenomes. Appl Environ Microbiol. 2011;77:1101–1106. doi: 10.1128/AEM.01481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2004;437:349. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 47.Demirjian DC, Morís-Varas F, Cassidy CS. Enzymes from extremophiles. Curr Opin Chem Biol. 2001;5:144–151. doi: 10.1016/S1367-5931(00)00183-6. [DOI] [PubMed] [Google Scholar]
- 48.Trincone A. Marine biocatalysts: enzymatic features and applications. Mar Drugs. 2011;9:478–499. doi: 10.3390/md9040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor EJ, Gloster TM, Turkenburg JP, Vincent F, Brzozowski AM, Dupont C, Shareck F, Centeno MS, Prates JA, Puchart V. Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J Biol Chem. 2006;281:10968–10975. doi: 10.1074/jbc.M513066200. [DOI] [PubMed] [Google Scholar]
- 50.Fu J, Leiros H-KS, de Pascale D, Johnson KA, Blencke H-M, Landfald B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl Microbiol Biotechnol. 2013;97:3965–3978. doi: 10.1007/s00253-012-4276-9. [DOI] [PubMed] [Google Scholar]
- 51.Ko K-C, Rim S-O, Han Y, Shin BS, Kim G-J, Choi JH, Song JJ. Identification and characterization of a novel cold-adapted esterase from a metagenomic library of mountain soil. J Ind Microbiol Biotechnol. 2012;39:681–689. doi: 10.1007/s10295-011-1080-y. [DOI] [PubMed] [Google Scholar]
- 52.Esteban-Torres M, Santamaría L, de las Rivas B, Muñoz R. Characterisation of a cold-active and salt-tolerant esterase from Lactobacillus plantarum with potential application during cheese ripening. Int Dairy J. 2014;39:312–315. doi: 10.1016/j.idairyj.2014.08.004. [DOI] [Google Scholar]
- 53.Jeon JH, Lee HS, Kim JT, Kim S-J, Choi SH, Kang SG, Lee J-H. Identification of a new subfamily of salt-tolerant esterases from a metagenomic library of tidal flat sediment. Appl Microbiol Biotechnol. 2012;93:623–631. doi: 10.1007/s00253-011-3433-x. [DOI] [PubMed] [Google Scholar]
- 54.Steiner H, Lindskog S. Effects of high concentrations of salt on the esterase activity of human carbonic anhydrase. FEBS Lett. 1972;24:85–88. doi: 10.1016/0014-5793(72)80832-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Phylogenetic analysis of O. pacifica AXE along with the uncharacterized closest amino acid sequences identified by NCBI BLAST and several characterized acetyl xylan esterases. The neighbor-joining tree was constructed using the bootstrap method with 1000 replications.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Additional file.