Abstract
Formation of the inflammasome, a scaffolding complex that activates caspase-1, is important in numerous diseases. Pyroptotic cell death induced by anthrax lethal toxin (LT) is a model for inflammasome-mediated caspase-1 activation. We discovered 7-desacetoxy-6,7-dehydrogedunin (7DG) in a phenotypic screen as a small molecule that protects macrophages from LT-induced death. Using chemical proteomics, we identified Protein Kinase R (PKR) as the target of 7DG and show that RNAi knockdown of PKR phenocopies treatment with 7DG. Further, we show that PKR’s role in ASC assembly and caspase-1 activation induced by several different inflammasome stimuli is independent of PKR’s kinase activity, demonstrating that PKR plays a novel role in caspase-1 activation and pyroptosis that is distinct from its previously reported kinase-dependent roles in apoptosis and inflammasome formation in LPS-primed cells. Remarkably, PKR plays different roles in two distinct cell death pathways and plays a broad role in inflammasome function relevant in other diseases.
Lethal toxin (LT), elaborated by Bacillus anthracis, the causative agent of anthrax, co-opts and disrupts numerous host functions to kill host macrophages. Because LT causes cell death via an inflammasome and caspase-1 mediated mechanism1,2, it can serve as a useful tool for investigating the cell biology of the inflammasome and its role in a variety of diseases including chronic obstructive pulmonary disease, gout, type II diabetes and Alzheimer’s disease3.
B. anthracis produces two toxins, lethal toxin (LT) and edema toxin (ET). ET and LT are AB toxins that each consist of a unique enzymatic A subunit and an identical B subunit known as protective antigen (PA), which is the receptor-binding subunit that shuttles the two A subunits into the cytosol of host cells (Fig. 1a). The A subunit of ET is edema factor (EF), which is an adenylate cyclase that increases the intracellular concentration of cAMP and causes tissue edema. The corresponding A subunit of LT is lethal factor (LF), which is a zinc metalloprotease (with substrates including all the MAP kinase kinases (MEKs) except MEK54) that kills macrophages4,5. Injection of purified LT into animal models results in rapid death, demonstrating the singular importance of LT in the pathology seen with anthrax6–8. Interestingly, pyroptosis induced by LT may be a protective mechanism of the host during infection9.
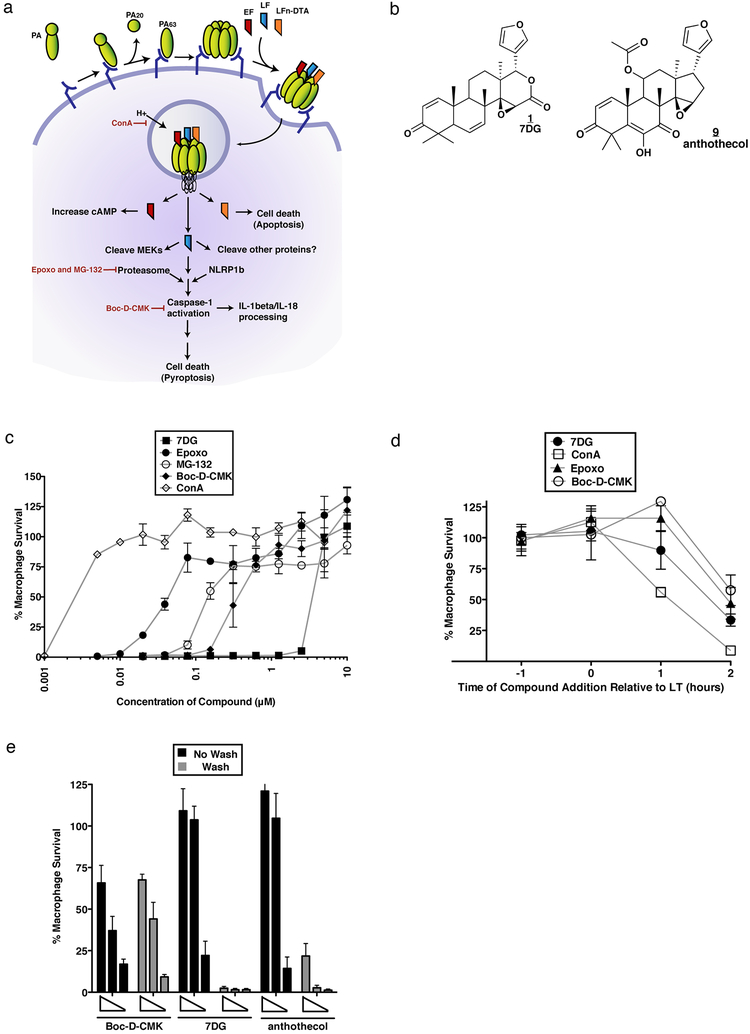
Figure 1. Chemical HTS identifies 7DG an inhibitor of LT-induced cell death.
a) Events involved in toxin entry and toxicity. PA binds to anthrax receptors on the host cell and is cleaved by a furin-like protease into PA20 and PA63, which oligomerizes and binds LF/EF/LFn-DTA proteins. Endocytosis occurs followed by trafficking to the endosome. A decrease in endosomal pH results in a conformational change in PA, forming a functional pore that allows translocation of LF/EF/LFn-DTA into the cytosol. EF causes an increase in cAMP, LFn-DTA causes apoptosis, and LF acts as a metalloprotease that requires an active proteasome complex and sensitive NLRP1b allele to activate caspase-1, resulting in cell death. ConA (concanamycin A) inhibits endosome acidification; Epoxo (epoxomicin) and MG-132 inhibit the proteasome; Boc-D-CMK inhibits caspase-1 activity (Figure adapted from Ref1). b) Structures of 7DG (1) and anthothecol (9). c) Protection of macrophages from LT-induced cell death. d) Kinetics of compounds in relation to timing of LT addition. Compounds were added 1 hour before LT addition (−1), at the same time (0), or 1 or 2 hours after LT addition (1 or 2) (7DG 20 μM, ConA 0.32 μM, Epoxo 1.25 μM, Boc-D-CMK 5 μM). e) Reversibility of compound activity. Cells were treated with decreasing concentrations of compounds, washed, and cell viability was measured after exposure to LT. Boc-D-CMK is an irreversible inhibitor. For c), d), and e) data are representative of at least 3 independent experiments, each done in triplicate. Data are shown as mean +/− StDev.
There are two main models for LT-induced cell death. In the first model, observed in macrophages from BALB/c mice, cell death proceeds through pyroptosis, a rapid (3 to 6 hours after exposure to LT), caspase-1 dependent form of cell death10,11. In this model, host factors identified for LF intoxication include NLRP110, the proteasome12 and caspase-111. NLRP1, recently shown to be a substrate of LT13, is a component of the inflammasome, a molecular platform that triggers activation of caspase-1 and subsequent processing of the proinflammatory cytokines IL-1β, IL-18, and high-mobility group box 1 (HMGB1)11,14. Upon treatment with LT, macrophages that carry a susceptible NLRP1b allele undergo pyroptosis. Chemical inhibition of either caspase-1 or proteasome activity protects susceptible NLRP1b macrophages from LT15.
In the second model, observed in C57BL/6 mouse macrophages carrying a resistant NLRP1 allele, LT-induced cell death proceeds through apoptosis, a slow process (24 to 48 hours) that requires co-stimulation of Toll-like receptors (TLR) with pathogen-associated molecular components such as lipopolysaccharide (LPS). The allelic variation seen in NLRP1 is a generalized trait and is not restricted to BALB/c and C57BL/6 mice. In this model, the primary role of LF is to cleave MEK3 and/or MEK6 to block p38 signaling16. It has been shown that inhibition of signaling through p38 synergizes with signaling through the serine-threonine Protein Kinase R (PKR), which when activated by TLR4, results in a global shutdown of protein synthesis though a PKR kinase-dependent mechanism, leading to apoptosis17. It is unclear what the relative roles of these two models of cell death are in vivo, as there are discrepancies reported between rat and mouse models18–22.
Besides the NLRP1-containing inflammasome induced by LT, there are other inflammasomes that contain different NLRP proteins (NOD-like receptor (NLR) pyrin domain–containing proteins) that are activated by different stimuli and involve the recruitment of the adaptor protein ASC (adaptor speck-like protein containing a CARD). These include the NLRP3 inflammasome induced by many stimuli (e.g. nigericin, Leu-Leu-OMe [L-leucyl-L-leucine methyl ester]23), the AIM2 (absent in melanoma 2) inflammasome stimulated by double-stranded DNA (dAdT)24, and NLRC4 (NLR family CARD domain-containing protein 4) stimulated by flagellin and often require priming with a TLR ligand such as LPS.
To further probe the mechanism by which LT induces inflammasome-mediated cell death, we performed a phenotypic chemical high-throughput screen for inhibitors that would act as chemical suppressors of the LT-induced cell death pathway. We report the identification of 7-desacetoxy-6,7-dehydrogedunin (7DG), a small molecule that protects macrophages exposed to LT by binding to and modulating signaling through Protein Kinase R (PKR). We demonstrate that this role of PKR is independent of its kinase activity and therefore distinct from the previously reported roles of PKR in cytokine processing14 and apoptosis17. We further show that 7DG disrupts a kinase-independent function of PKR that is required not only for LT-induced pyroptosis, but also for nigericin, Leu-Leu-OMe, and LT-induced inflammasome assembly, independent of LPS priming.
RESULTS
Phenotypic HTS for Inhibitors of LT-Induced Cell Death
We performed a high-throughput chemical screen to identify small molecules that protect macrophages from LT. J774A.1 BALB/c murine macrophages (J774) were pre-treated with a chemical library (final concentration ~33 μM), followed by the addition of LT. After 6 hours, viability of macrophages was determined using CellTiter-Glo (CTG). Under these conditions, ~98% of the macrophages are dead after incubation with LT (Z’ factor = 0.67). Similar screens have previously been reported25–28.
We screened 31,350 small molecules in duplicate from several commercial chemical libraries (archived at the Broad Institute). Many compounds identified in the screen are annotated as inhibitors of proteins known to be required for LT toxicity, and thus serve as positive controls within the screen (Fig. 1a). Because ConA prevents toxin cell entry (LF entry into the cytosol), we refer to it as an “upstream” inhibitor, while Epoxo, MG-132, and Boc-D-CMK act after LF has been translocated into the cytosol, and thus we refer to them as “downstream” inhibitors.
A small molecule hit of particular interest was 7DG (7-Desacetoxy-6,7-dehydrogedunin), which completely protects J774 macrophages from LT-induced death at 10 μM and has an IC50 of 5 μM (Fig. 1b, c). 7DG is able to robustly protect macrophages when added up to 1 hour after exposure to LT, but not at later time-points (Fig. 1d).
To determine if 7DG and a 7DG-related compound, anthothecol (Fig. 1b), are reversible or irreversible inhibitors, we incubated macrophages with compounds, washed several times, added fresh media with or without LT and measured viability. As a control, we used Boc-D-CMK, a known irreversible caspase-1 inhibitor, which loses no activity with washings (Fig. 1e). In contrast, 7DG and anthothecol completely lost activity, suggesting that they are reversible inhibitors.
7DG Does Not Inhibit LT Cell Entry or Protease Activity
To test whether 7DG inhibits toxin entry, we took advantage of the fact that the mechanism of EF entry is identical to LF entry, requiring PA and the same host cellular machinery. Thus, small molecules that inhibit events involved in LF entry should similarly inhibit the entry of EF, which can be detected by measuring the rise in cAMP levels when EF, an adenylate cyclase, is translocated into the cytosol (Fig. 1a). Macrophages treated with the indicated small molecule were exposed to ET and cAMP levels were measured by ELISA. While cells exposed to ET and the control upstream inhibitor ConA did not show an increase in cAMP, cells exposed to ET and DMSO control or 7DG had a significant increase in cAMP due to efficient EF entry (Supplementary Results, Supplementary Fig. 1a).
We also employed an LFn-DTA chimera, where the N-terminal domain of LF (LFn) is fused to DTA, the catalytic domain of diphtheria toxin. LFn-DTA, when combined with PA, is translocated into the cytosol in a manner identical to LF (Fig 1a). However, their respective mechanisms of killing diverge: LF cleaves its substrates leading to caspase-1 activation, while DTA ADP-ribosylates elongation factor 2, inhibiting protein synthesis. Because of the divergent downstream mechanisms of killing and the identical upstream mechanism of cell entry, inhibitors that protect macrophages against both toxins must block common upstream events. 7DG did not protect macrophages from LFn-DTA, unlike the upstream positive control ConA (Supplementary Fig. 1b). The downstream negative control Boc-D-CMK also did not protect from LFn-DTA. Thus, 7DG is a downstream inhibitor, inhibiting events after LF entry.
To determine if 7DG inhibits LF enzymatic activity in vitro, we tested its ability to inhibit LF’s proteolytic activity towards a MEK-based peptide substrate, which fluoresces upon cleavage. 7DG did not inhibit cleavage of the peptide, in contrast to galardin, a known metalloprotease inhibitor (Supplementary Fig. 1c). We also tested if 7DG inhibits LF activity in whole cells by performing immunoblot analysis for evidence of MEK3 cleavage in J774 macrophages that had been treated with LT. While ConA prevented MEK3 cleavage, 7DG did not prevent cleavage at compound concentrations that protect 100% of cells (Supplementary Fig. 1d). Thus, 7DG does not affect cell entry or translocation of LF into the cytosol, nor does it inhibit the protease activity of LF in intact cells.
One of the characterized cellular functions required for LT-induced cell death is proteasomal protease activity. To determine whether 7DG inhibits this activity, we treated J774 macrophage lysate with 7DG or a known proteasome inhibitor control (Epoxo), followed by synthetic proteasome substrates that fluoresce when cleaved by the specific protease. We found that 7DG does not directly inhibit known proteasomal activities in vitro at concentrations up to 40 μM (Supplementary Fig. 1e). To further confirm these results, we tested the ability of 7DG to inhibit the proteasome in whole cells. J774 macrophages were treated with compound and then incubated with one of three proteasome substrates that luminesce upon cleavage. Using Epoxo as a control, we found that significant inhibition of protease activity is observed before any protection of macrophages from LT is achieved (Fig. 2a and Supplementary Fig. 1f). In contrast, we found that 7DG does not inhibit proteasome activity in whole cells at concentrations affording complete protection of macrophages from LT. Thus, the mechanism of protection from LT by 7DG is not through directly inhibiting the proteasome protease activity.
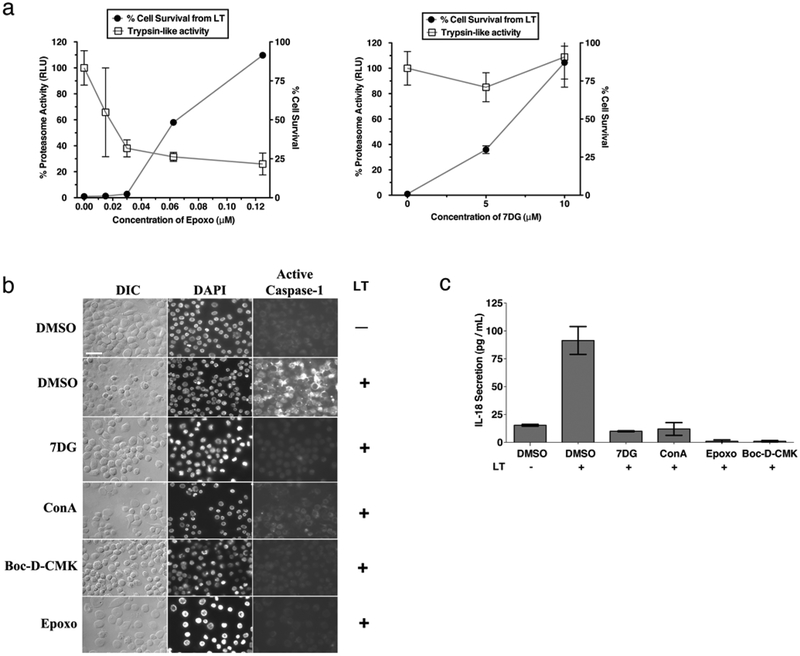
Figure 2. 7DG does not directly inhibit proteasome or caspase-1 activity, but does prevent the activation of caspase-1 by LT.
a) J774 cells were treated with different concentrations of compound abd incubated with the cell-permeable trypsin-like protease substrate Z-LRR-aminoluciferin, and luminescence was measured. A LT assay was conducted in parallel to determine the amount of protection from LT (cell survival). b) J774 macrophages were treated with compound and exposed to LT, then stained with FLICA caspase-1 reagent. Cells were then fixed, stained with DAPI, and imaged (7DG 10 μM, ConA 1 μM, Boc-D-CMK 20 μM, and Epoxo 2 μM). Scale bar represents 25 μm. c) J774 cells were incubated with compound, followed by incubation in LT. Supernatants were transferred to an IL-18 ELISA plate and the standard ELISA protocol was followed (7DG 5 μM, ConA 0.1 μM, Epoxo 2.5 μM, and Boc-D-CMK 10 μM). Data are representative of at least 3 independent experiments, each done in triplicate. Data are shown as mean +/− StDev.
7DG Inhibits LT-Induced Activation of Caspase-1
LT is known to require activation of caspase-1 to cause macrophage pyroptotic cell death, an event that is downstream of required proteasome activity. Caspase-1 exists in an inactive state in untreated cells, but when cells are exposed to LT, caspase-1 is activated and cleaves its substrates, leading to pyroptotic cell death. To test if 7DG inhibits the LT-mediated activity of caspase-1 in intact cells, we used two complementary assays. The first utilizes a cell-permeable fluorescent caspase-1 substrate (FLICA: FAM-YVAD-FMK) that covalently binds to active caspase-1 and is detected by microscopy (Fig. 2b). The second uses an IL-18 ELISA assay to measure the amount of mature IL-18 processed by active caspase-1 and secreted from cells (Fig. 2c). Using these assays, we found that 7DG, along with ConA, Epoxo, and Boc-D-CMK, all inhibit the LT-mediated activity of caspase-1. While these assays indicate that 7DG inhibits caspase-1 in the presence of LT, they do not distinguish between compounds that inhibit the activation of caspase-1 and those that directly inhibit the enzymatic activity of caspase-1. To determine if 7DG directly inhibits the enzymatic activity of caspase-1, we measured the amount of fluorescent caspase-1 substrate cleaved by recombinant active caspase-1 in vitro in the presence and absence of 7DG. Compared to the small molecule Z-YVAD-FMK, a known inhibitor of caspase-1, 7DG does not reduce the activity of caspase-1 at concentrations up to 100 μM (Supplementary Fig. 1g). Thus, 7DG inhibits steps leading up to caspase-1 activation, without directly inhibiting its enzymatic activity.
7DG Does Not Protect Cells Through HSP90 Inhibition
To try to understand the mechanism of 7DG, we tested the ability of structurally related compounds with known, annotated functions to protect cells from LT. For example, 7DG (1) is a structural analogue of gedunin (3) (Supplementary Fig. 2), which is a known HSP90 inhibitor29. Gedunin protects macrophages from LT induced death with an IC50 of 30 μM and a maximum protection of 80% at 80 μM. To determine if gedunin’s inhibition of HSP90 accounts for the observed protection, we tested whether other well-established HSP90 inhibitors protect from LT. Geldanamycin, 17-AAG, and 17-DMAG afforded no protection against LT-induced cell death (Supplementary Table 1). Radicicol did have activity, with an IC50 of 15 μM. However, this activity is unrelated to 7DG as we found that radicicol protects from LT by preventing translocation of LF (unlike 7DG) since it also inhibits EF and LFn-DTA cell entry. Furthermore, radicicol has nanomolar potency against HSP90 compared to the micromolar concentrations required to inhibit LT toxicity, suggesting that this compound is not protecting cells from LT by inhibiting HSP90. Thus, HSP90 inhibition does not protect cells from LT induced death, and is not the mechanism by which 7DG protects cells from LT.
Identification of 7DG-Interacting Proteins
To identify proteins that interact with 7DG, we sought to make a biotinylated analog of 7DG to facilitate affinity studies. Using a combination of synthesis and purchase of commercially available compounds, we tested a series of 7DG analogs in the LT macrophage survival assay to define their activity (Supplementary Fig. 2). This SAR (structure-activity relationship) analysis revealed that the lactone ring is not required for activity, since compounds 9 through 19 lack this feature and all except 12 retain activity. The epoxide is also not necessary for activity, since 4 lacks an epoxide and retains some activity. The α,β-unsaturated ketone, while important for activity since compounds 5, 7, and 12 are inactive, is not absolutely required for activity, since 6 and 14 retain some activity. The non-essentiality of the lactone, epoxide, and α,β-unsaturated ketone, all chemical functionalities with the potential to alkylate substrates, is consistent with our previous observation that 7DG does not appear to be an irreversible inhibitor.
Through SAR, we found that the 7-position is tolerant of small substitutions and that compounds lacking a lactone ring are tolerant of additions at positions 6 and 11 (refer to numbering on 9). Based on this SAR of 7DG, we synthesized a biotinylated analog of 7DG, 19, that retains potent activity, protecting cells from LT-induced death with an IC50 of 0.63 μM. Remarkably, even with the addition of the linker and biotin handle, 19 maintains cellular potency similar to 7DG. 19 acts similarly to 7DG and inhibits a downstream event related to caspase-1 activation, with no effect on LT translocation or activity (Supplementary Fig. 3b).
To identify proteins that interact with 7DG, we used our synthesized biotinylated analogue of 7DG 19 as an affinity reagent. We incubated J774 macrophages with DMSO, 7DG, 19, or 19 with 40-fold molar excess of free 7DG, lysed the cells, and incubated the lysates with streptavidin-labeled beads. After washing, proteins retained on the beads were analyzed by SDS PAGE and stained with Coomassie blue dye to detect differences in the treated samples. Proteins that were preferentially pulled down in cells treated with 19 alone compared with 19 plus excess free 7DG were cut out and analyzed by protein mass spectrometry (Supplementary Fig. 3c). We identified 11 proteins that were enriched in the pulldown by 19 and competed off by excess 7DG (Supplementary Table 2). It is noteworthy that 19, and thus 7DG, does not appear to be very promiscuous, given the fact that only a few bands are enriched and competed in the pulldown experiment, suggesting selectivity of 7DG.
PKR is Important for LT-induced Cell Death
In order to test their functional roles in mediating the effects of LT, we performed targeted knockdown of each of the 11 candidate proteins identified in the pulldown experiment using siRNA in J774 macrophages and measured survival after LT treatment. We found that knockdown of Protein Kinase R (PKR, aka: double-stranded RNA-activated protein kinase, eukaryotic translation initiation factor 2-alpha kinase 2, EIF2AK2) resulted in significant protection from LT (Fig. 3a). When we deconvoluted the pool of siRNA targeting PKR, we found that multiple siRNAs provided protection from LT (Supplementary Fig. 3f), with increasing amounts of siRNA targeting PKR resulting in increasing levels of protection from LT (Supplementary Fig. 3g). Knockdown at the protein level following siRNA treatment was confirmed by immunoblot analysis (Fig. 3b). These experiments reveal the importance of PKR in LT-induced cell death.
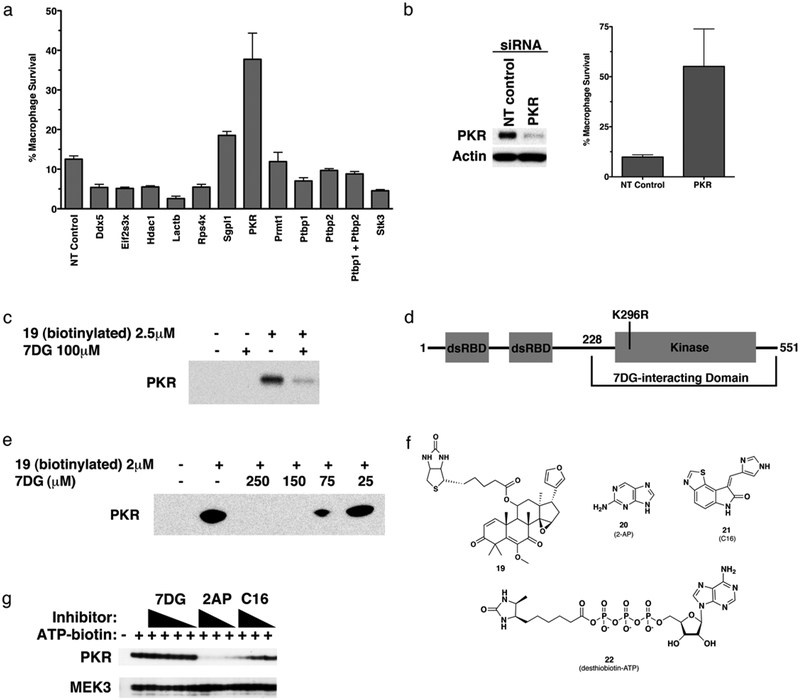
Fig 3. 7DG pulldown identifies PKR as an interacting protein important for LT-induced cell death.
a) Proteins identified from the pulldown assay with 19 were knocked down by transfection of J774 cells with siRNA targeting each identified protein and cells were tested for sensitivity to LT. NT control is a pool of siRNA oligos that do not target known mouse transcripts. b) PKR levels were determined by immunoblot after 3 days of siRNA-mediated knockdown. The amount of protection from LT afforded by the knockdown is also shown. c) J774 cells were incubated with DMSO, 7DG, 19, or 19 with 40-fold molar excess of free 7DG, lysed, and enriched with streptavidin-labeled beads. Proteins retained on the beads were analyzed by immunoblot with a specific antibody for PKR. d) Diagram of PKR, indicating two N-terminal dsRNA binding domains and a C-terminal kinase domain. The mutation K296R inactivates the kinase activity and expresses better in E. coli. The C-terminal portion found to interact with 7DG in e) is indicated. e) A recombinant C-terminal portion of PKR was expressed in E. coli, lysates were incubated with different concentrations of 7DG with or without 19 (2 μM), which was then pulled down with streptavidin beads. PKR was detected by immunoblot. f) Structures of 19, 2-AP, C16, and desthiobiotin-ATP. g) Compounds were tested for the ability to compete for binding at the ATP binding pocket of PKR using J774 cell lysate and a desthiobiotin-ATP probe and analyzed by immunoblot. Decreased intensity of PKR bands indicates an ability of the indicated compound to bind to the ATP active site of PKR and prevent binding of the desthiobiotin-ATP probe. Concentrations tested were (7DG 200, 100, 50, 25 μM; 2-AP 2000, 1000, 500 μM; C16, 10, 5, 2.5 μM). MEK3 was included as a labeling and loading control. For all assays, data are representative of at least 3 independent experiments, each done in triplicate. Data are shown as mean +/− StDev.
7DG Binds PKR Outside the ATP Catalytic Domain
To confirm that PKR is pulled down by 19, we repeated the pulldown assay and probed for the presence of PKR using immunoblot analysis. PKR antibodies detected the presence of PKR from J774 macrophage lysates treated with 19 and a much weaker band from lysates treated with 19 and 40-fold excess 7DG (Fig. 3c). These data suggest that 7DG interacts with PKR or a complex containing PKR resulting in its selective pulldown and isolation, and this interaction may account for the protective effect of 7DG on LT treated cells.
To determine if 7DG interacts with PKR directly, we expressed recombinant PKR in Escherichia coli, reasoning that other macrophage proteins required for a PKR complex are absent in E. coli. We expressed a soluble C-terminal domain of human PKR (56% identical and 70% similar to mouse PKR). The expressed protein contains a mutation (K296R) in the catalytic site of the kinase domain that has been reported to aid in expression (Fig. 3d)30. Lysates containing recombinant PKR were incubated with or without 19 in the absence or presence of increasing amounts of soluble 7DG as competitor. 19 was pulled down with streptavidin beads and PKR was detected by immunoblot analysis. Expressed PKR was pulled down with 19 and was competed off in a dose-dependent manner by 7DG (Fig. 3e), confirming that 7DG directly interacts with the C-terminal half of PKR.
We tested whether 7DG competes with ATP binding in the active site of PKR. J774 macrophage lysates were incubated with several concentrations of 7DG or either of the two known PKR kinase inhibitors 20 (2-AP) or 21 (C16)31 (Fig. 3f), reported to inhibit the kinase activity of PKR by binding to the catalytic site. These lysates were then incubated with a desthiobiotin-ATP probe (Fig. 3f), resulting in covalent labeling of kinases that are not occupied by the compound in the ATP site. Biotinylated proteins were enriched with streptavidin beads, washed, and proteins were eluted and analyzed by immunoblot. A compound that binds to the ATP active site competes with the desthiobiotin-ATP probe and results in a weaker band on the immunoblot. Because the probe labels most kinases, MEK3 was used as a labeling and loading control. While both 2-AP and C16 bound to the ATP site of PKR (and not MEK3), no concentrations of 7DG were able to compete for the desthiobiotin-ATP biotin probe binding to PKR, indicating that 7DG does not bind to the ATP catalytic pocket of PKR (Fig. 3g).
PKR Knockdown Phenocopies 7DG Exposure
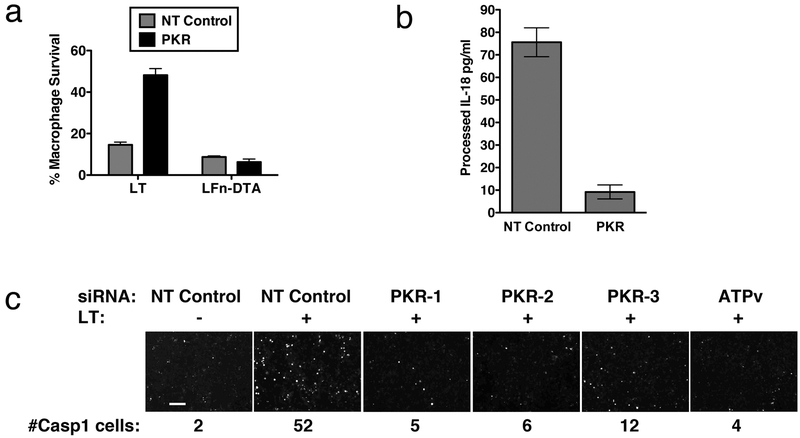
We examined whether knockdown of PKR phenocopies the effects of 7DG related to LT-induced cell death. Using the methods described above, we confirmed that PKR knockdown does not affect LT cell entry or activity, based on its lack of effect on cell survival after LFn-DTA exposure (Fig. 4a) and MEK3 cleavage in LT-treated cells (Supplementary Fig. 3i). Knockdown of PKR, like 7DG, does prevent caspase-1 activation by LT, as measured by determining IL-18 processing (Fig. 4b) and using a fluorescent caspase-1 substrate as described above (Fig. 4c). A similar siRNA dose-response in cell survival was seen with the amount of active caspase-1 (Supplementary Fig. g, h). These data confirm that PKR is downstream of LT cell entry and is involved in caspase-1 activation after LT intoxication.
Figure 4. PKR KD phenocopies treatment with 7DG.
PKR knock down in J774 cells transfected with siRNA was assayed. a) LFn-DTA entry was determined by incubating cells with LFn-DTA and PA. In parallel, cell sensitivity to LT was also tested. b) Cells were incubated with or without LT and supernatants were transferred to an IL-18 ELISA plate and the standard ELISA protocol was followed. c) After knockdown with siRNA, cells were exposed to LT and stained with FLICA caspase-1 reagent. Cells were then fixed, stained with DAPI, and imaged. Different siRNA targeting PKR are indicated, as well as an upstream control, ATPv6 siRNA. Number of caspase-1 active cells in each panel is indicated below each image. Scale bar represents 100 μm. For all assays, data are representative of at least 3 independent experiments, each done in triplicate. In graphs, data are shown as mean +/− StDev.
PKR Kinase Activity is Not Required for Pyroptosis
PKR is activated by a number of stimuli, including double-stranded RNA (dsRNA) and LPS32; and acts through a variety of effectors, including NFκB33. Upon stimulation, PKR autophosphorylates to become an active kinase32, subsequently phosphorylating its substrates. To test whether LT activates PKR kinase activity, we assessed phosphorylated PKR by immunoblot after cells were treated with LT. LT treatment did not result in PKR autophosphorylation, in contrast to treatment with a positive control that is known to cause stress-induced PKR activation34 (Supplementary Fig. 4a). Because J774 macrophages begin to die several hours after exposure to LT, it was difficult to determine late-stage PKR status in cells treated with LT alone. To allow for the analysis of PKR status at later time-points, we examined cells treated with LT and the downstream caspase-1 inhibitor Boc-D-CMK, to prevent cell death. Under these conditions, we still did not observe phosphorylated PKR by as late as 6 hours after exposure to LT, well after unprotected cells normally die. This observation suggested that while PKR is essential to intoxication, its kinase activity might be dispensable. This finding is consistent with our observation that 7DG does not compete with ATP binding in the catalytic active site of PKR.
To confirm that PKR kinase activity is not essential to LT toxicity, we tested whether the two known inhibitors of PKR kinase activity, 2-AP and C16, could protect LT treated cells from death. Neither compounds protected cells from LT at any concentration tested (Supplementary Table 1). Furthermore, neither compound enhanced nor antagonized the activity of 7DG.
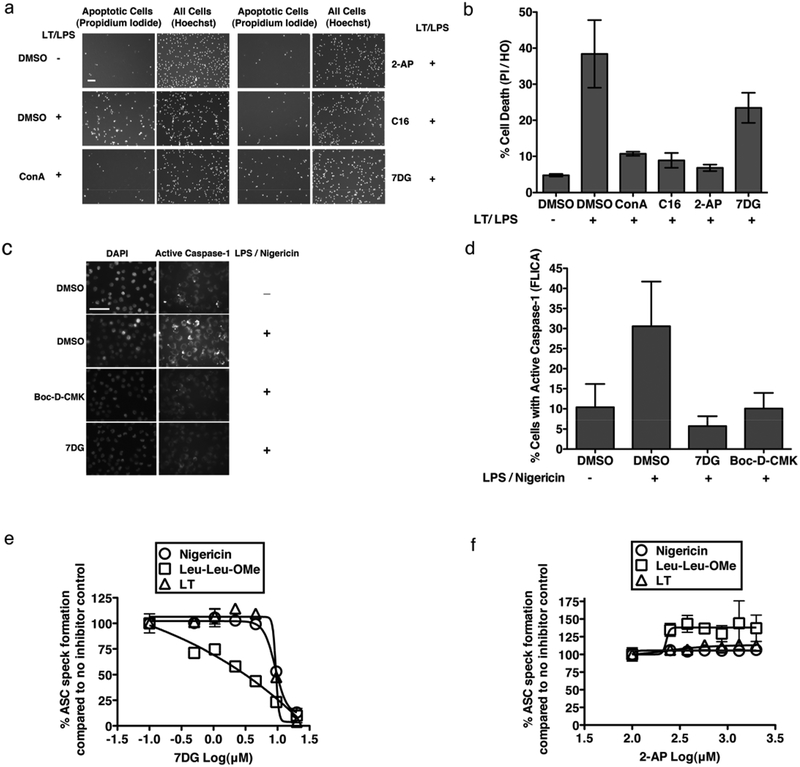
Because PKR has previously been reported to have a role in the apoptosis model of LT-mediated cell death17 (where a TLR stimulant is required and the role of LT is simply to inhibit signaling through p38), with specific implication of its kinase activity, we predicted that inhibition of the kinase activity of PKR with 2-AP and C16, but not 7DG, would protect C57BL/6 bone marrow macrophages from LPS/LT. As expected, we found that treatment with either PKR kinase inhibitor robustly protected cells from apoptosis (Fig. 5a, b), in contrast to 7DG, which did not extensively protect cells (a small, nonsignificant effect was observed, which is consistent with the previous report of a concurrent small, kinase-independent role of PKR in apoptosis17).
Figure 5. 7DG Inhibits a Kinase-Independent Function of PKR.
a) and b) C57BL/6 bone marrow derived macrophages were treated with compound, exposed to LPS and LT for 24 hours, then stained with propidium iodide and Hoechst stains and counted and graphed as % cells with propidium iodide (PI) / Hoechst stain (HO). Propidium iodide is indicative of apoptosis, while Hoechst stains all cells. p-values for one-tailed, unpaired t-tests compared to the DMSO + LT/LPS control were significant for all treatments at p=0.01, except 7DG, which was not significant at p=0.05 (ConA 0.13 μM, 2-AP 40 μM, C16 0.63 μM, 7DG 2.5 μM). Scale bar represents 75 μm. c) and d) J774 cells were treated with compound (7DG 20 μM, Boc-D-CMK 20 μM), LPS and nigericin, and incubated with FLICA caspase-1 reagent, imaged and counted. p-values for one-tailed, unpaired t-tests compared to the DMSO + LPS/nigericin control were significant for all treatments at p=0.01, and for DMSO at p=0.05. Scale bar represents 50 μm. e) and f) Immortalized mouse macrophages expressing NLRP3-FLAG and ASC-mCerulean were incubated with inhibitor and specific stimuli, fixed and visualized to determine the percentage of cells that contain ASC specks. For b, d, e, f assays, data are representative of at least 3 independent experiments, each done in triplicate. Data are shown as mean +/− StDev.
Collectively, these data (along with the desthiobiotin-ATP data) suggest that PKR plays differing roles in the pyroptotic and apoptotic cell death pathways of LT intoxication. PKR kinase activity is required in the apoptosis and the LPS-primed pyroptosis model, while it is not required in the non-primed pyroptosis model. Furthermore, these data suggest that 7DG inhibits the PKR activity required for LT-induced pyroptosis in a manner distinct from known PKR kinase inhibitors.
7DG Can Inhibit Known Interactions With PKR
Besides the well-characterized kinase activity of PKR, a kinase-independent role for PKR has been reported35–37, with PKR interacting with specific proteins to regulate their fate and function. For example, PKR plays a role in NFκB signaling by interacting with the IκK complex. This interaction results in IκB phosphorylation and proteasomal degradation35–37, with subsequent NFκB activation. Inhibition of NFκB activation has been reported to prevent LT-induced cell death38, though this was in bovine macrophages and has not been thoroughly confirmed in other model systems. To determine if 7DG might prevent NFκB activation by preventing PKR’s interaction with IκK complex, we tested whether it inhibits LPS-induced IκBβ degradation. Cells treated with 7DG or proteasome inhibitor (Epoxo) were treated with LPS to stimulate the degradation of IκBβ. While a dramatic reduction in IκBβ was observed at 30 and 40 minutes in DMSO-treated control cells, 7DG prevented this degradation, similar to Epoxo (Supplementary Fig. 4c).
These data suggest that 7DG acts by binding PKR and blocking its kinase-independent interaction with other proteins. This inhibition of NFκB activation serves as an example for how 7DG can inhibit protein-protein interactions, in this case between IkK and PKR, but likely does not account for 7DG’s ability to suppress LT-induced cell death, since inhibition of transcription and translation does not protect cells from LT (Supplementary Table 1 and Ref39). Also, we did not observe activation of NFκB-regulated TNFα when J774 cells were incubated with LT (Supplementary Fig. 4d).
PKR Interactions with Multiple Inflammasomes
Finally, we sought to determine if the interaction of PKR with other proteins might also be involved in NLRP3 assembly. While caspase-1 is activated by LT through the NLRP1 inflammasome, it can also be activated through the NLRP3 inflammasome by stimuli such as nigericin (a K+ channel toxin)40 or Leu-Leu-OMe. We exposed J774 macrophages to the NLRP3-activating combination of nigericin and LPS and measured activation of caspase-1 using the fluorescent caspase-1 dye (FLICA, mentioned above) in the presence or absence of 7DG. Cells exposed to nigericin and LPS exhibited significant caspase-1 staining detected by microscopy and 7DG and Boc-D-CMK significantly prevented this activation (Fig. 5c, d). While 7DG could be inhibiting the expression of NLRP3 through NFκB signaling, this is unlikely to be the mechanism, given the short incubation times involved (1.5 hours).
To further investigate the kinase-independent role of PKR in inflammasome activation, we took advantage of the fact that upon stimulation of cells with inflammasome activators, ASC spatially reorganizes from a diffuse cytosolic pattern into a concentrated, single dot or “speck”. Murine immortalized macrophages constitutively expressing NLRP3 (which eliminates the need for LPS priming), and fluorescently tagged ASC, were stimulated with LT (NLRP1), nigericin or Leu-Leu-OMe (NLRP3). These cells do not require cellular priming since NLRP3 is constitutively expressed in these cells41. All three stimuli result in rapid localization of ASC to form a speck that is visible by microscopy. We tested the dose-response of 7DG, 2-AP, and C16 in this assay and found that while neither 2-AP nor C16 prevented ASC assembly by all tested stimulants, 7DG prevented ASC assembly by all stimulants tested (Fig. 5e, f and Supplementary Fig. 4e – note that C16 caused partial inhibition of LT-induced ASC speck, but never reached 50% inhibition). These data demonstrate that 7DG does indeed prevent signaling through NLRP1 and NLRP3 inflammasomes, while PKR kinase inhibitors do not. Thus, PKR protein interactions, rather than PKR kinase activity, may be involved in broad mechanisms of inflammasome-mediated caspase-1 activation.
DISCUSSION
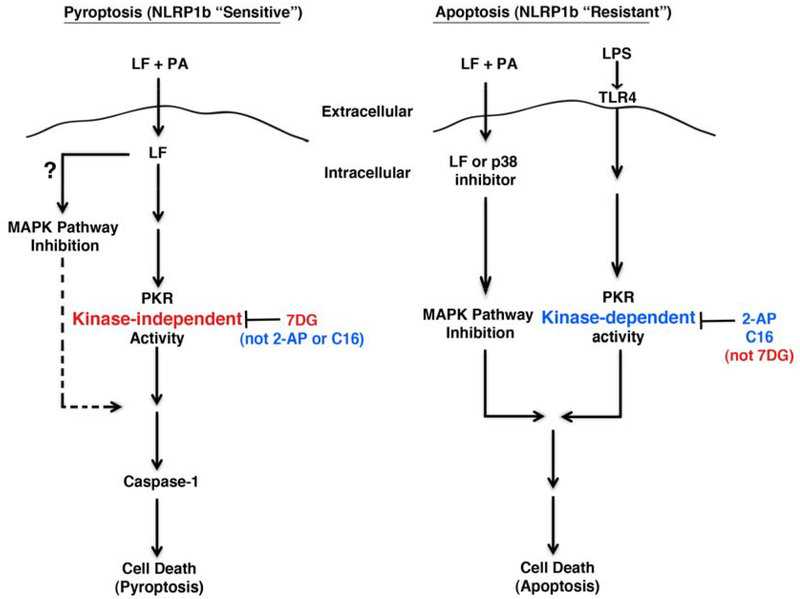
PKR has been shown to be an important global regulator of host defense mechanisms, such as mediating apoptosis in response to viral infection42. It belongs to a family of serine/threonine kinases (including PERK, HRI, and CGN2) and is known for its ability to phosphorylate eIF2a, resulting in the shutdown of protein synthesis through inhibition of translation43. PKR kinase activity is implicated in signaling in LT-induced apoptosis17 as well as inflammasome activation14. Our work confirmed a kinase role for PKR in apoptosis, similar to the reported protection from apoptosis with PKR deletion and mutation strains17. We found that the two known PKR kinase inhibitors 2-AP and C16 protect from apoptosis, while 7DG had a moderate effect. However, we did not find a kinase role for PKR in inflammasome activation or pyroptosis. Instead, neither of the known PKR inhibitors protected cells from pyroptosis or inflammasome activation, while 7DG robustly protected from both (Fig. 6). The role of PKR was confirmed to play a critical function in inflammasome activation, since knockdown of PKR resulted in significantly reduced pyroptosis.
Figure 6. Pyroptosis versus apoptosis models.
In the depicted pyroptosis model, LF enters the cell containing a LT-sensitive allele of NLRP1b and stimulates a PKR kinase-independent activity (that 7DG inhibits, but 2-AP and C16 do not), resulting in caspase-1 activation and cell death. It is unclear if LF inhibition of the MAPK pathway is required for this model. In the apoptosis model, inhibition of the MAPK pathway is required (LF can be substituted for by a p38 inhibitor) and synergizes with the PKR kinase-dependent activity stimulated by LPS/TLR4 (inhibited by 2-AP and C16, but not by 7DG), resulting in cell death.
A recent report using macrophages lacking either N- or C-terminal portions of PKR claims PKR-independent activation of caspase-1 induced by inflammasome stimuli44. It is conceivable that in this PKR deficient system, the remaining portion of PKR is sufficient for kinase-independent scaffolding functions or that one of the PKR family members compensates for the deficiency. We conclude from our study that instead of acting as a canonical kinase, PKR’s role in caspase-1 activation involves its ability to modulate the function of other proteins through protein-protein interactions, with 7DG inhibiting these interactions. Such protein-protein interactions with PKR are likely to be induced by multiple stimuli given our findings that 7DG prevented ASC speck formation in NLRP1 and NLRP3 inflammasomes.
Another recent report that PKR regulates the activation of inflammasomes through a kinase-dependent mechanism14 uses cellular experiments that rely on LPS as a priming stimulus to induce the production of NLRPs through NFκB-dependent transcriptional regulation45 and thus do not allow the differentiation between primary (e.g. LPS) and secondary (e.g. LT, nigericin, Leu-Leu-OMe) signaling pathways. Furthermore, the report does not demonstrate a requirement for PKR kinase activity in LT-induced pyroptosis, only in cytokine secretion, thus leaving the question open as to whether or not PKR kinase activity is required for pyroptosis14. It is known that PKR is involved in NFκB signaling35 and that NFκB activation results in upregulation of inflammasome components45. Thus, inhibition of PKR with known kinase inhibitors used in the report could have had their effect by inhibiting the priming event rather than the actual second signal stimuli. This possibility could explain why in our work, which does not require LPS priming, we do not find a requirement for PKR kinase activity. This highlights one of the advantages of using the murine macrophage NLRP/ASC heterologous expression system, which allows for isolated interrogation of the second signal from various stimuli.
In summary, we report the discovery of 7DG as an inhibitor of NLRP1 and NLRP3 inflammasome assembly and caspase-1 activation. We conclude that 7DG inhibits a kinase-independent role of PKR by demonstrating that 7DG binds to PKR and that this binding is outside of the ATP binding pocket, where typical PKR kinase inhibitors bind. Furthermore, we show the surprising finding that PKR is involved in both apoptosis and pyroptosis, but plays very different roles. 7DG has the potential to serve as a useful novel small molecule for studying diseases where caspase-1 activation through inflammasome stimulation results in IL-1β and IL-18 release and inflammatory damage associated with a diverse set of diseases3.
ONLINE METHODS
Cells, media, reagents
J774A.1 (J774) macrophages (ATCC) were grown in standard DMEM media supplemented with 10% FBS, glutamine, penicillin and streptomycin. PA, LF, and LFn-DTA were expressed in E. coli and purified. Reagents were purchased as follows: EF (LIST Biologicals); antibodies (Santa Cruz Biotechnologies and Cell Signaling), IL-18 ELISA kits (MBL International), cAMP ELISA kits (Assay Designs), LF substrate 3 (Calbiochem), proteasome substrates (Promega), caspase-1 fluorescent cell-permeable substrate FLICA (ImmunoChemistry Technology), compounds: Galardin, Epoxo, geldanamycin, radicicol (Biomol), 17-AAG, 17-DMAG (Calbiochem), C16 and dsRNA (EMD), 2-AP, Concanamycin A (Sigma), Boc-D-CMK (Anaspec), 8 (Analyticon), 1, 3, 4, 6, 7, 9 (Gaia), 2, 5, 10 (Spectrum), 11-19 were synthesized.
Primary LT Assay Screen
J774 macrophages were incubated overnight, compound was added for a final concentration ~33 μM and incubated for 2 hours. LT was added for a final concentration of 2 nM LF and 11nM PA and plates were incubated for 6 hours. CellTiter-Glo was added to each well and luminescence was read. Compounds were tested in duplicate, ranked by mean z-score, and analyzed separately for each day of screening. Approximately 0.5% were chosen for retesting.
ET assay
J774 macrophages were incubated overnight, compound was added for 2 hours, then 3 nM EF and 11 nM PA were added to the wells and plates were incubated for 5 hours. Standard conditions were used for the ELISA.
LFn-DTA assay
J774 macrophages were incubated overnight and compound was added and plates were incubated for 2 hours. A mix of 0.1 nM LFn-DTA and 11 nM PA was added and plates were incubated for 24 hours. Identical wells treated with compound were not given LFn-DTA/PA and were used as controls. CellTiter-Glo was added to measure cell viability.
IL-18 ELISA
J774 macrophages were seeded and incubated overnight. Cells were preincubated with compound for 2 hours, then LT (2 nM LF and 11 nM PA) was added and cells were incubated for 3 hours. Plates were spun briefly, then supernatants were transferred to an IL-18 ELISA plate and the standard ELISA protocol was followed. CellTiter-Glo was added to the cells from which the supernatant was removed to determine the amount of cell survival.
Whole cell active caspase-1 assay
J774 macrophages were treated with compound for 2 hours, then exposed to 2 hours of LT or 30 minutes with 1μg/ml LPS and 40 μM nigericin. The media was removed and replaced with FLICA caspase-1 reagent (ImmunoChemistry Technology) and incubated for 1 hour. Cells were then fixed, stained with DAPI, and imaged by fluorescence microscopy. The percentage of caspase-1 positive cells was determined using DAPI to identify all cells.
In vitro caspase-1 activity
Active, recombinant mouse caspase-1 was diluted in the provided buffer (R&D Systems) and compound was added. Fluorescent caspACE (Promega) substrate was added and kinetic reads were taken to measure the amount of cleaved substrate according to directions.
LF in vitro activity
Recombinant LF was diluted in the provided buffer and compound was added. Fluorescent substrate (LF Substrate 3, Calbiochem) was added and kinetic reads were taken to measure the amount of cleaved substrate according to directions.
MEK cleavage assay
J774 macrophages were incubated with compound for 2 hours, then LT (LF 1–2 nM and PA 11 nM) was added and cells were incubated for 2 hours. Total cell lysates were made, protein concentrations were measured and normalized, and proteins were separated on an SDS-PAGE, transferred to PVDF, and probed with indicated antibodies.
Whole cell proteasome assay
J774 macrophages were preincubated with compound for 2 hours and then mixed with cell-permeable substrates in the provided buffer (Promega). Kinetic reads of luminescence were taken every 5 minutes for 30 minutes.
Streptavidin co-precipitation
Biotinylated compound (19) was added to J774 cells that were preincubated for 60 minutes with competitor (7DG), and incubated for 1.5 hours. Cells were washed twice with PBS, lysed with modified RIPA buffer (50mM HEPES, 4mM EDTA, 150mM NaCl, 1% NP40, pH 7.5), centrifuged to clear supernatant, and then pre-washed magnetic dynabeads were added accordingly and incubated for 1 hour with motion. Beads were washed and boiled in loading buffer. Samples were run on SDS PAGE and bands of interest were excised and sequenced. Identical regions on the gel from the DMSO control lane were also sequenced.
Recombinant PKR production and pulldown
BL21(DE3) cells were transformed with a pET15b vector expressing a truncated version of PKR (delta-228 K296R) and grown on selective media. Expression was induced with 1M IPTG when cultures were at an optical density of ~1. Cells were pelleted, resuspended with modified RIPA buffer, sonicated, and centrifuged to remove debris. Lysate was incubated with the indicated compounds, allowing 1 hour preincubation with free 7DG competitor, followed by an hour incubation with 19, and finally, incubated with streptavidin beads. Beads were washed several times (twice with 0.1% Tween20 in PBS and twice with 1% NP40 in PBS) and boiled in SDS buffer. Expression of PKR was determined by immunoblot analysis using a PKR-specific antibody.
Desthiobiotin-ATP active site labeling
J774 cell lysate was incubated with compound for 1 hour at room temperature, 20μM desthiobiotin-ATP probe (ActivX ATP from Thermo Scientific) and MgCl2 were added and incubated for 10 minutes at room temperature, urea was added to stop the reaction and biotinylated proteins were enriched with streptavidin beads overnight, washed, eluted with SDS loading buffer, ran on SDS PAGE and blotted for with anti-PKR antibody or anti-MEK, using MEK as a labeling and loading control.
siRNA transfection
J774 macrophages were seeded and incubated overnight. siRNA duplex and RNAi max lipofectamine were mixed with Opti-MEM in separate tubes for 5 minutes, combined and incubated for 20 minutes, then added to cells. Media was changed after overnight incubation. Assays were conducted after 3 to 4 days of knockdown.
Apoptosis model assay
Bone marrow-derived macrophages (BMDM) were harvested from C57BL/6 mice following standard protocol. Cells were pretreated with compound for 1 hour, exposed to 100 ng/ml LPS and 2 nM LF with 11 nM PA for 24 hours, then stained with propidium iodide and Hoechst stains and counted.
ASC speck formation cellular assay
The assay utilizes immortalized mouse macrophages from NLRP3 deficient mice that have been transduced with retroviruses encoding for murine NLRP3-FLAG and human ASC-mCerulean. These cells respond to NLRP3 stimuli without priming and to NLRP1 stimuli since they are mixed C57/BL6 and 129/Sv background (sensitive to LT). Cells are incubated with inhibitor for 1 hour, then with specific stimuli for 1.5 hours (10μM nigericin) or 2 hours (1 mM Leu-Leu-OMe-HCl or LT (1 μg/ml LF + 1 μg/ml PA)). The cells are fixed with paraformaledehyde and counterstained with the nucleic acid stain DRAQ5 to assess the percentage of cells that are activated, i.e. the percentage of cells that contain ASC specks.
Statistical Analysis
Graphpad Prism software was used to generate p-values for one-tailed, unpaired t-tests to test for significance.
IκB blot
J774 macrophages were treated with compounds for 2 hours and treated with 10 ng/ml LPS for the indicated period of time. Lysates were probed by immunoblot for IkBb and actin for loading control.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank John Collier, Su Chiang, Brian Kraybill, Luke Whitesell, Susan Lindquist, Sandro Santagata, Sunita Patel-Hett, Randall King, Nevena Dimova, Deepa Patel, and Roi Avraham-Atzil for helpful discussions; Vlado Gelev for PKR constructs and for a valiant attempt to solve the structure of PKR with 7DG; Anne Clatworthy for experimental help and discussions; Adam Barker for help with production and purification of PA and LF; Yajie Wang for help with chemistry; Jenny Lee for technical assistance; Huseyin Aktas for PKR constructs; Amy Barczak for C57Bl/6 macrophages and discussions; and the Broad Institute chemical screening platform for technical help. Eugenio Fava, Philip Denner and Ana Kitanovic from the laboratory automation laboratory at the German Center for Neurodegenerative Diseases (DZNE) are thanked for the analysis of the ASC speck assays. ECH was funded by an NIH National Research Service Award fellowship F32AI084323. This work was supported in part by NIH U54 AI057159 to the New England Center of Excellence/Biodefense and Emerging Infectious Diseases (DTH).
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
ECH is currently employed by Pfizer Inc, which develops therapies for immunological diseases.
REFERENCES
- 1.Collier RJ & Young JA Anthrax toxin. Annu Rev Cell Dev Biol 19, 45–70 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Ascenzi P et al. Anthrax toxin: a tripartite lethal combination. FEBS Lett 531, 384–388 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Menu P & Vince JE The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin Exp Immunol 166, 1–15, doi: 10.1111/j.1365-2249.2011.04440.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duesbery NS et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280, 734–737 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Friedlander AM Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. J Biol Chem 261, 7123–7126 (1986). [PubMed] [Google Scholar]
- 6.Agrawal A & Pulendran B Anthrax lethal toxin: a weapon of multisystem destruction. Cell Mol Life Sci 61, 2859–2865 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Newman ZL et al. Susceptibility to anthrax lethal toxin-induced rat death is controlled by a single chromosome 10 locus that includes rNlrp1. PLoS Pathog 6, e1000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beall FA & Dalldorf FG The pathogenesis of the lethal effect of anthrax toxin in the rat. J Infect Dis 116, 377–389 (1966). [DOI] [PubMed] [Google Scholar]
- 9.Terra JK et al. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J Immunol 184, 17–20, doi: 10.4049/jimmunol.0903114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyden ED & Dietrich WF Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 38, 240–244 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Cordoba-Rodriguez R, Fang H, Lankford CS & Frucht DM Anthrax lethal toxin rapidly activates caspase-1/ICE and induces extracellular release of interleukin (IL)-1beta and IL-18. J Biol Chem 279, 20563–20566 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Tang G & Leppla SH Proteasome activity is required for anthrax lethal toxin to kill macrophages. Infect Immun 67, 3055–3060 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levinsohn JL et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog 8, e1002638, doi: 10.1371/journal.ppat.1002638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu B et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488, 670–674, doi: 10.1038/nature11290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickliffe KE, Leppla SH & Moayeri M Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell Microbiol 10, 332–343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JM, Greten FR, Li ZW & Karin M Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science 297, 2048–2051, doi: 10.1126/science.1073163 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Hsu LC et al. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 428, 341–345, doi: 10.1038/nature02405 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Hanna PC, Acosta D & Collier RJ On the role of macrophages in anthrax. Proc Natl Acad Sci U S A 90, 10198–10201 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moayeri M, Sastalla I & Leppla SH Anthrax and the inflammasome. Microbes Infect, doi: 10.1016/j.micinf.2011.12.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moayeri M, Martinez NW, Wiggins J, Young HA & Leppla SH Mouse susceptibility to anthrax lethal toxin is influenced by genetic factors in addition to those controlling macrophage sensitivity. Infect Immun 72, 4439–4447, doi: 10.1128/iai.72.8.4439-4447.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S et al. Anthrax toxin targeting of myeloid cells through the CMG2 receptor is essential for establishment of Bacillus anthracis infections in mice. Cell Host Microbe 8, 455–462, doi: 10.1016/j.chom.2010.10.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman ZL et al. Susceptibility to anthrax lethal toxin-induced rat death is controlled by a single chromosome 10 locus that includes rNlrp1. PLoS Pathog 6, e1000906, doi: 10.1371/journal.ppat.1000906 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung V et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9, 847–856, doi: 10.1038/ni.1631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornung V et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518, doi: 10.1038/nature07725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panchal RG et al. Chemical genetic screening identifies critical pathways in anthrax lethal toxin-induced pathogenesis. Chem Biol 14, 245–255, doi: 10.1016/j.chembiol.2007.01.007 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Gaddis BD, Avramova LV & Chmielewski J Inhibitors of anthrax lethal factor. Bioorg Med Chem Lett 17, 4575–4578, doi: 10.1016/j.bmcl.2007.05.089 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Newman ZL et al. Auranofin protects against anthrax lethal toxin-induced activation of the Nlrp1b inflammasome. Antimicrob Agents Chemother 55, 1028–1035, doi: 10.1128/aac.00772-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez AM et al. Amiodarone and bepridil inhibit anthrax toxin entry into host cells. Antimicrob Agents Chemother 51, 2403–2411, doi: 10.1128/aac.01184-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb J et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Thomis DC & Samuel CE Mechanism of interferon action: autoregulation of RNA-dependent P1/eIF-2 alpha protein kinase (PKR) expression in transfected mammalian cells. Proc Natl Acad Sci U S A 89, 10837–10841 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jammi NV, Whitby LR & Beal PA Small molecule inhibitors of the RNA-dependent protein kinase. Biochem Biophys Res Commun 308, 50–57 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Garcia MA et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70, 1032–1060, doi: 10.1128/mmbr.00027-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Haque J, Lacoste J, Hiscott J & Williams BR Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci U S A 91, 6288–6292 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalai M et al. The caspase-generated fragments of PKR cooperate to activate full-length PKR and inhibit translation. Cell Death Differ 14, 1050–1059, doi: 10.1038/sj.cdd.4402110 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Bonnet MC, Weil R, Dam E, Hovanessian AG & Meurs EF PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol 20, 4532–4542 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii T, Kwon H, Hiscott J, Mosialos G & Koromilas AE Activation of the I kappa B alpha kinase (IKK) complex by double-stranded RNA-binding defective and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene 20, 1900–1912, doi: 10.1038/sj.onc.1204267 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Bonnet MC, Daurat C, Ottone C & Meurs EF The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell Signal 18, 1865–1875, doi: 10.1016/j.cellsig.2006.02.010 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Liang X, Gao CF, Rutherford MS & Ji Y Activation of NF-kappaB pathway and TNF-alpha are involved in the cytotoxicity of anthrax lethal toxin in bovine BoMac macrophages. Vet Microbiol 146, 111–117, doi: 10.1016/j.vetmic.2010.04.028 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Alileche A, Squires RC, Muehlbauer SM, Lisanti MP & Brojatsch J Mitochondrial impairment is a critical event in anthrax lethal toxin-induced cytolysis of murine macrophages. Cell Cycle 5, 100–106 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Mariathasan S et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232, doi: 10.1038/nature04515 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Bauernfeind FG et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 183, 787–791, doi: 10.4049/jimmunol.0901363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaufman RJ Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci U S A 96, 11693–11695 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wek RC eIF-2 kinases: regulators of general and gene-specific translation initiation. Trends Biochem Sci 19, 491–496 (1994). [DOI] [PubMed] [Google Scholar]
- 44.He Y, Franchi L & Nunez G The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. Eur J Immunol, doi: 10.1002/eji.201243187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiao Y, Wang P, Qi J, Zhang L & Gao C TLR-induced NF-kappaB activation regulates NLRP3 expression in murine macrophages. FEBS Lett 586, 1022–1026, doi: 10.1016/j.febslet.2012.02.045 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.