Abstract
Ribosome tagging has become a very useful in vivo approach for analyzing gene expression and mRNA translation in specific cell types that are difficult and time consuming to isolate by conventional methods. The approach is based on selectively expressing an HA-tagged ribosomal protein in a target cell type and then using antibodies against HA to purify the polysomes and associated mRNAs from the target cell. The original approach makes use of a mouse line (RiboTag) harboring a modified allele of Rpl22 (Rpl22-HA) that is induced by the action of Cre recombinase. The Rpl22-HA gene can also be introduced into the animal by stereotaxic injections of an AAV-DIO-Rpl22-HA that is then activated in Cre-expressing cells. Both methods for tagging ribosomes facilitate the immunoprecipitation of ribosome bound mRNAs and their analysis by qRT-PCR or RNA-Seq. This protocol will discuss the technical procedures and describe important considerations relevant to the analysis of the data.
Introduction:
The analysis of gene expression provides a description of a cell’s potential functions and its response to extracellular signals. However, when researchers attempt to analyze gene expression in vivo they are confronted with the problem that all tissues contain a diverse array of different cell types with unique functions. Efforts to isolate a specific cell type for analysis based on sorting techniques can be successful but these additional manipulations are both time consuming and have been shown to alter transcription (Haimon et al., 2018). The brain is a particularly challenging tissue in which to quantitate gene expression and translation. A typical mouse brain is made up of around 75 million neurons, 23 million glial cells, 7 million endothelial cells and several million other miscellaneous cell types (Williams, 2000). Neurons are extremely diverse in terms of their expression of various neurotransmitters, neuropeptides, and receptors and typically these neurons are anatomically intermingled making them very difficult to separate for molecular analysis. In situ analysis of mRNA expression has been very useful in identifying the overall anatomical distribution of neurons expressing specific genes and the Allen Brain Institute has assembled a comprehensive database for both the mouse and human brain which is very useful. However, anatomical localization cannot define specific neuronal cell types. Characterization of the mRNA expression phenotype of specific neurons has progressed with the development of ribosome-tagging approaches, cell sorting based on promoter specific expression of fluorescent marker proteins, and single-cell RNA-Seq. Each approach has advantages and disadvantages. In this protocol we describe a ribosome-tagging approach (RiboTag) which our lab developed (Sanz et al., 2009) and discuss the application of this approach to the analysis of polyribosome associated mRNA transcripts in specific populations of neurons. The RiboTag approach takes advantage of the hundreds of specific promoter-driven Cre recombinase transgenics currently available (Jackson Laboratories has more than 480 lines) and others are being added at a rapid pace.
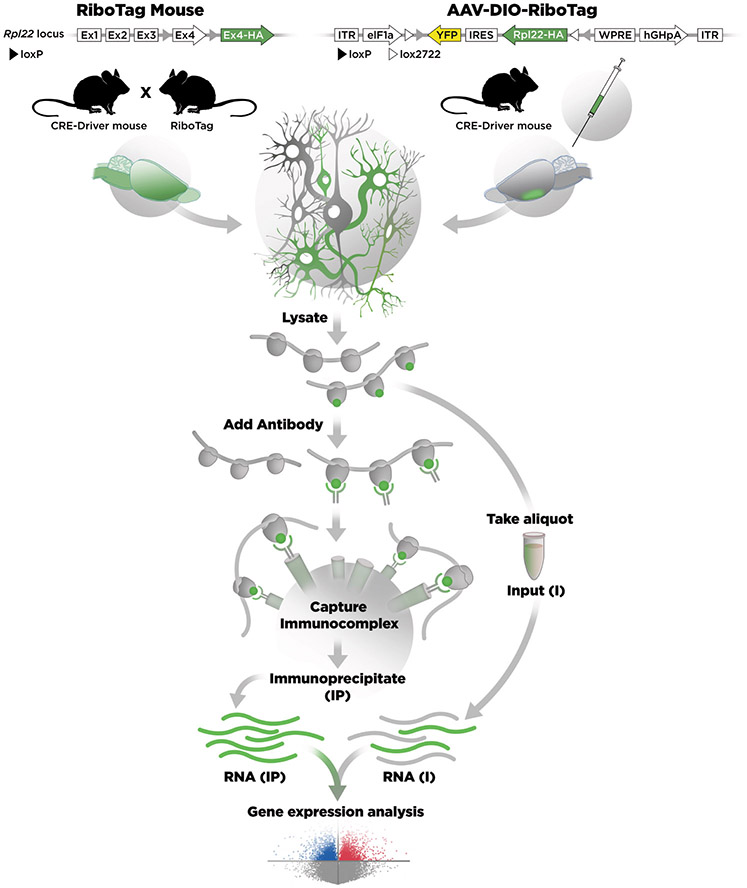
There are two basic approaches involving the use of RiboTag as diagrammed in Figure 1 and each has advantages and disadvantages. In the first approach we use a RiboTag mouse line in which the 60S ribosomal gene, Rpl22, was modified so that in the presence of Cre recombinase the last exon of Rpl22 will be replaced with an HA-tagged exon effectively labeling the cell’s polyribosomes so that they can be isolated by immunoprecipitation. These RiboTag mice are crossed to a Cre recombinase driver mouse line and the double heterozygotes are used for analysis. Immunoprecipitation of polyribosomes from neuronal tissue purifies only the polysomes that are tagged in the Cre-expressing cells. The mRNA from these polysomes is isolated and the transcripts analyzed by qRT-PCR or RNA-Seq. The second approach utilizes a Rpl22-HA construct in a flex or DIO virus (AAV) flanked by two different sets of loxP sites (Sanz et al., 2015). When the AAV-DIO-RiboTag is injected into a specific anatomical region of the brain of an adult mouse the Rpl22-HA is only expressed in the neurons that are actively expressing Cre recombinase and also infected by the AAV. After waiting several weeks for the Rpl22-HA to be expressed and incorporated into polyribosomes, tissue samples are isolated and used to immunoprecipitate HA-tagged polyribosomes followed by qRT-PCR or RNA-Seq. The viral approach requires accurate stereotaxic injections of virus and is more labor intensive but has the advantage that only adult neurons that still express the Cre transgenic will be targeted. The direct cross of RiboTag mice with Cre driver mice is easier to do but has the potential disadvantage that the Rpl22-HA could continue to be expressed in neurons that have differentiated from a progenitor population that expressed Cre recombinase at an earlier stage during development. One can take advantage of this difference by comparing results from the same brain region using the RiboTag mouse and the RiboTag virus using the same Cre driver transgenic. We have used this approach to demonstrate that embryonic Pomc-expressing progenitor neurons differentiate into both Agrp and Kiss1 expressing neurons in the adult arcuate nucleus of the hypothalamus (Sanz et al., 2015).
Figure 1. Cartoon comparing the RiboTag mouse and AAV-DIO-RiboTag approaches.
On the left side, the Rpl22 locus was modified by homologous recombination to insert LoxP sites on either end of the last exon (Ex4) of the Rpl22 gene and a duplicated Ex4 with an HA tag was inserted downstream. The CRE-Driver mouse removes the WT Ex4 sequence in specific cells and the transcript now splices in Ex4-HA (shown in green). Polyribosomes from cells expressing the Ex4-HA are recovered from a lysate by immunoprecipitation (IP) with an antibody directed against HA and magnetic beads coated with an artificial Protein A/G peptide. RNA is isolated from the IP polyribosomes and RNA is also isolated from the lysate. Gene expression analysis is done by RNA-Seq. On the right side the DIO-RiboTag viral vector is shown schematically. In the presence of Cre-recombinase the Rpl22-HA-IRES-YFP cassette is flipped and locked in the ON configuration so that the eIF1a promoter now drives expression of Rpl22-HA in specific cells. RNA is isolated from the IP and compared with the input.
Basic Protocol 1 outlined below describes our methods in detail using the RiboTag mouse crossed with a specific Cre-recombinase expressing transgenic mouse to isolate polyribosomes by immunoprecipitation from brain homogenates and then further isolate RNA from both the input (original homogenate) and the immunoprecipitated (IP) polysomes. Alternative Protocol 1 describes the viral approach using stereotaxic injection of AAV-DIO-RiboTag into adult Cre-recombinase expressing mouse brain to avoid developmental activation of RiboTag expression. In Support Protocol 1 we describe the breeding and genotyping of RiboTag mice and in Support Protocol 2 we describe the analysis of captured RNA using qRT-PCR. Basic Protocol 2 discusses the use of RiboTag mice crossed with a specific Cre transgenic line to identify the anatomical localization of RiboTag expression by immunohistochemistry. In Basic Protocol 3 we describe the current approach our lab is using to produce DNA libraries for RNA-Seq. Basic Protocol 4 describes the bioinformatic tools that we are using to analyze the RNA-Seq data. In the Commentary we will provide a discussion of these methods and identify some of the most critical parameters and potential trouble spots.
BASIC PROTOCOL 1
The RiboTag Assay
This approach has been described and updated in several publications from our laboratory (Quintana et al., 2012; Sanz et al., 2013; Sanz et al., 2015; Sanz et al., 2009) and is shown schematically in Figure 1. We have now introduced modifications that improve the efficiency of the immunocapture and therefore increase the RNA yields obtained in the immunoprecipitates. These modifications include using an indirect method for the immunocapture of HA-tagged polysomes (instead of the direct immunoprecipitation procedure described in the original publication) and using Protein A/G beads with a higher binding capacity for IgGs.
Materials:
Mice:
RiboTag mouse line (JAX stock No. 011029)
Cre-driver mouse line of choice
Equipment:
Large sharp surgical scissors (FST cat No. 14001–14)
Small extra fine scissors (FST cat No. 14084–08)
Spatula (FST cat No. 10091–12)
Harris Uni-Core (Electron Microscopy Sciences cat No. 69036)
Razor blades (Electron Microscopy Sciences cat No. 71960)
Mouse brain matrix (Electron Microscopy Sciences cat No. 69022)
Glass Dounce homogenizers (2ml or 7ml) (Sigma cat. No. 8938 or 9063)
Refrigerated centrifuge (Eppendorf; cat. No. 5417R) with rotor for 1.5 ml microcentrifuge tubes.
Microcentrifuge tubes (Axygen; MCT-175-C)
10 ul ART Barrier pipette tips (Thermo Fisher Sci.; cat. No. 2140-HR)
20 ul ART Barrier pipette tips (Thermo Fisher Sci.; cat. No. 2149-HR)
200 ul ART Barrier pipette tips (Thermo Fisher Sci.; cat. No. 2069-HR)
1000 ul ART Barrier pipette tips (Thermo Fisher Sci.; cat. No. 2179-HR)
Microtube magnetic stand (Thermo Fisher Sci.; cat. No. 12320D)
Microtube rotator (Thermo Fisher Sci.; cat No. 88881002)
Nanodop One (Thermo Fisher Sci.; cat. No. 13–400-518)
Fluorescence microplate reader (for Quant-iT Ribogreen RNA assays)
2100 Bioanalyzer Instrument (Agilent; cat. No. G2939BA)
Materials:
Sterile water for irrigation (Baxter, cat. No. 2F7114)
1M DTT, 10X 0.5ml ampules (Sigma; cat. No. 646563–10X 0.5ml). Aliquot open ampules and store at −80ºC
Protease inhibitor cocktail (Sigma, cat. No. P8340–5ml). Aliquot and store at −20C
RNAsin (40U/ul) (Promega, cat. No. N2115)
b-mercaptoethanol (Bio-Rad; cat. No.1610710
Pierce protein A/G magnetic beads (Thermo Fisher Sci.; cat. No. 88803)
Covance Anti-HA.11 Epitope Tag Antibody (BioLegend; cat. No. MMS-101R). Aliquot and store at −20ºC
RNeasy Mini Kit (Qiagen; cat. No. 74104) or RNeasy Micro kit (Qiagen; cat. No. 74004)
Quant-iT RiboGreen RNA Assay kit (Thermo Fisher Sci.; cat. No. R11490)
RNA 6000 Nano kit or RNA 6000 Pico kit (Agilent; cat. No. G2939BA, 5067–1511 or 5067–1513)
Ethanol (200 proof)
Protocol steps:
Harvest tissue
Euthanize mice using IACUC approved procedures such as carbon dioxide inhalation
Decapitate mice using large sharp surgical scissors
Make a midline incision in the skin and expose the skull
Using small scissors and a spatula remove the skull
Lift the brain out of the skull and use a spatula to section the optic and other cranial nerves
Place the brain in a cold block or brain matrix for further dissection. Sampling tools such as Harris Uni-cores can be used to punch the area of interest.
Lysate preparation
-
7.
Clean Dounce tissue grinder and dissection tools and rinse thoroughly with RNase-free water (sterile water for irrigation can be used at this step). Air dry and place Dounce homogenizer on ice.
-
8.
Prepare the homogenization buffer as described in the Reagents and Solutions section and keep on ice.
-
9.Dissect the tissue containing the cell type of interest. Specific areas or whole tissue can be used in this approach.
- A brain matrix and sampling tools such as brain punches or Harris Uni-Cores can be used to precisely dissect the area of interest. Dissected tissue or punches of the area of interest can either be flash-frozen in liquid nitrogen right after dissection and stored at −80ºC for months or processed fresh.
-
10.Place the tissue in the ice-cold Dounce homogenizer vessel containing a defined volume of pre-chilled homogenization buffer and homogenize using 40 strokes of the loose-fitting pestle (type A pestle), followed by 40 more strokes with the tight-fitting pestle (type B pestle).
- We typically use a 2–5% w/v ratio (for areas with a limited number of cells from the cell type of interest a pool of punches of the same area from different animals can be used to increase RNA yields). For small amounts of tissue (10–50mg tissue) we use the 2 ml Dounce and for 50–300mg we use the 7 ml Dounce.
-
11.
After homogenization, transfer lysate to a microtube on ice.
-
12.
Clear the lysate at 10,000 x g for 10 min at 4ºC using a refrigerated centrifuge.
-
13.
Transfer the post-mitochondrial supernatant to a pre-chilled microcentrifuge tube on ice. Discard resulting pellet.
-
14.
Take a small aliquot of the cleared lysate (40–80 ul) and store it at −80ºC for subsequent Input analysis.
Immunoprecipitation
-
15.
Add 4 ul of the anti-Hemagglutinin (HA) antibody (Covance Anti-HA.11 Epitope Tag Antibody) to 800 ul of the remaining cleared lysate and incubate for 4 hours at 4ºC in a microtube rotator with gentle mixing (end-over-end).
-
16.Prepare the magnetic protein A/G beads:
- Thoroughly resuspend the beads by repeated inversion or gentle vortexing.
- Place a microtube magnetic stand on ice and transfer 200 ul of the resuspended beads to a clean microcentrifuge tube.
- Place the tube into the magnetic stand on ice to collect the beads and remove and discard the storage buffer.
- Add 400 ul of homogenization buffer and rotate end-over-end for 5 min to wash the beads.
- Collect the beads with the magnetic stand, discard buffer.
-
17.
Add the cleared lysate with the antibody to the beads. Incubate samples overnight at 4ºC in a tube rotator with gentle end-over-end mixing.
-
18.Next day, prepare the high salt buffer as described in the Reagents and Solutions section. Keep on ice.
- Place samples into the magnetic stand on ice and remove supernatant (supernatant from the immunoprecipitation can be discarded or saved at −80ºC for subsequent analysis).
- Leave tubes in the magnet for at least one minute to collect all the beads against the side of the tube.
-
19.
With the tubes on the stand, add 800 ul of high salt buffer (Reagents and Solutions) to remove non-specific binding from the immunoprecipitate (IP). Transfer tubes to the rotator and wash the IP with gentle end-over-end mixing for 5 min at 4ºC.
-
20.
Place samples into the magnetic stand on ice and remove supernatant
-
21.
Repeat high salt washes two more times with the same volume of high salt buffer for 5 min each, for a total of three washes.
-
22.Resuspend beads in 800 ul of high salt buffer, mix on rotator for 5 min and transfer beads and buffer to a clean tube.
- This step helps remove any RNA or polyribosomes that might be sticking to the original tube.
-
23.
Place tubes on the magnetic stand for at least 1 min, carefully remove all high salt buffer
-
24.
Add 350 ul of RLT buffer from the Qiagen’s RNeasy extraction kit (mini or micro) supplemented with β-mercaptoethanol (provided with the kit).
-
25.Remove tubes from stand and vortex samples for 30 seconds at RT.
- At this step samples can be stored at −80ºC for subsequent RNA extraction or processed through the RNA extraction procedure.
RNA extraction
-
26.
Place tubes containing IPs in RLT buffer (from Qiagen kit) into the magnetic stand for 1 min at RT to allow beads to be collected against the side of the tube
-
27.
Transfer the clear RLT buffer containing RNA to a new microcentrifuge tube.
-
28.
Proceed with RNA isolation with DNAse digestion following Qiagen’s RNeasy extraction kit directions.
-
29.To extract RNA from the Inputs (Step 6), bring input sample to 350 ul with RLT buffer supplemented with β-mercaptoethanol, vortex extensively and proceed to RNA extraction as described for IPs.
- RNA extraction using column-based kits should be performed at RT. However, eluted RNA should be kept on ice at all times. For long term storage, RNA is safely kept at −80ºC for years. Whenever working with isolated RNA, always wear gloves to minimize RNase contamination, decontaminate working surfaces and use clean nuclease-free tubes and barrier pipette tips.
-
30.Quantitate RNA using Quant-iT RiboGreen RNA Reagent (Thermo Fisher) or a Nanodrop-One (Thermo Fischer Sci) following the manufacturer’s protocols.
- If very low levels of RNA are expected (less than 2 ng/ul) the samples can be accurately quantitated by doing qRT-PCR using a house keeping gene such as Actb and comparing the results to known standard RNA solutions prepared with input RNA from the same tissue as detailed in Support Protocol 2.
-
31.Analyze RNA integrity on an Agilent 2100 Bioanalyzer system with the associated RNA 6000 nano and pico kits (chose nano chips for samples 5–500 ng/ul, for samples with RNA concentrations < 5 ng/ul (down to 50 pg/ul) the pico chips should be used). Follow manufacturer’s directions.
- Since most of the RNA isolated in Inputs and IPs is rRNA, the quality of mRNA is indirectly estimated by assessing the integrity of rRNA. A measure of RNA quality, the RNA integrity number (RIN), is also calculated by the software using an algorithm that assesses the electrophoretic traces of each sample. A RIN value of 7–10 indicates high quality RNA but RNA-Seq libraries can still be made and successfully analyzed starting with partially degraded RNA.
ALTERNATE PROTOCOL 1
Messenger RNA profiling using the RiboTag viral vector
To overcome the limitations associated with developmental activation of Cre recombinase transgenes and allow cell type-specific mRNA profiling of adult cells using Cre-Driver lines, we have developed a Cre-dependent RiboTag viral vector based on AAV1 (AAV1-DIO-RiboTag) (Sanz et al., 2015), where RiboTag expression is dependent on the activity of the promoter driving Cre recombinase expression in adult mice. This viral vector was created by inserting the cDNA of the mouse ribosomal protein Rpl22 gene fused to three hemagglutinin (HA) sequences (Rpl22 · 3HA), into a double-floxed inverse open reading frame (DIO) AAV construct. For easy visualization of transduced cells, this construct also contains an IRES sequence (IRES2) followed by the YFP coding sequence as shown in Figure 1.
Materials:
Mice:
Cre-driver mouse line of choice.
Reagents:
RiboTag viral vector (AAV1-DIO-RPL22HA-IRES-YFP). Available upon request from authors
Equipment:
Stereotaxic apparatus with microsyringe pump (e.g. from World Precision Instruments)
Injection syringe (e.g. 5 ul Hamilton syringe)
Surgical tools
Protocol steps:
Define stereotaxic coordinates by locating the injection target in a mouse brain atlas such as the Paxinos and Franklin’s “the Mouse Brain in Stereotaxic Coordinates” (Paxinos & Franklin, 2013) or the Allen institute for Brain Science’s “Adult Mouse Brain Reference Atlas” (Dong)
- Inject 0.2 to 0.5 ul of a RiboTag viral vector preparation (7 × 1011 viral genomes/ml) at the target site using the defined coordinates.
- Follow a stereotaxic injection protocol such as (Gore, Soden, & Zweifel, 2013). When targeting bilateral structures, inject each side. Injection volumes can be adjusted to the targeted area, or multiple injections performed to cover larger areas. Injecting different mice using the same coordinates and pooling the targeted areas after dissection helps correct for variability resulting from stereotaxic viral vector injections.
Allow at least 2 weeks for the viral vector to express Rpl22-HA. For RiboTag assays, good results are obtained when sacrificing animals at least 3 weeks after injections to allow for robust incorporation of Rpl22-HA into polyribosomes.
Perform RiboTag immunoprecipitations as described in Basic Protocol 1
All surgical procedures should be performed in accordance with protocols approved by your Institutional Animal Care Use Committee (IACUC).
SUPPORT PROTOCOL 1 Breeding and Genotyping of RiboTag Mice
Breeding of RiboTag Mice:
We maintain a live colony of RiboTag Breeders on a C57BL/6J background by mating homozygotes since they have no adverse phenotypes. Experimental mice for RiboTag assays are usually obtained by breeding male Cre recombinase-expressing mice to female homozygous RiboTag mice (Rpl22HA/HA), Therefore, experimental mice are always heterozygous for the RiboTag allele (Rpl22HA/+).
PCR Genotyping:
To genotype the RiboTag mice, a PCR was developed that includes the loxP site 5’ to the wild-type exon 4 of Rpl22; the wild-type PCR product is 242 bp, while the mutant product is about 290 bp. This PCR also distinguishes homozygous RiboTag mice, as the 5’ oligo corresponding to the intron between exons 3 and 4 of Rpl22 is not present in the repeated segment of intron preceding the C-terminal Rpl22HA exon 4. Resulting offspring from RiboTag mice bred to Cre recombinase-expressing mouse lines (experimental mice) are genotyped for the presence of the Cre recombinase gene and the RiboTag allele using the PCR oligonucleotides and reaction conditions described below
Materials:
NucleoSpin Tissue DNA extraction kit (Macherey-Nagel cat No. 740952)
DreamTaq DNA polymerase (Thermo Fisher Sci. cat No. EP0702 and B71)
dNTPs 10 mM each (Thermo Fisher Sci. cat No. R0192)
(dilute to 2.5 mM for each dNTP)
Alternative:
Phire Tissue Direct PCR Master Mix kit (Thermo Fisher Sci. cat No. F170
Oligonucleotides for RiboTag genotyping:
RiboTag loxP For: 5’-GGGAGGCTTGCTGGATATG-3’
RiboTag loxP Rev: 5’-TTTCCAGACACAGGCTAAGTACAC-3’
Oligonucleotides For Cre recombinase genotyping:
Cre cDNA For: 5’-GCATTACCGGTCGATGCAACGAGTG-3’
Cre cDNA Rev: 5’-GAACGCTAGAGCCTGTTTTGCACGTTC-3’
Acrylamide-bis solution 40% (37.5:1) (Merck cat No. 1.00638.1000)
Tris-Borate-EDTA (TBE) 10x (Fisher Scientific cat No. BP1333)
Ammonium Persulfate (APS; Sigma cat No. A3678).
(Prepare a 10% solution. Store at 20ºC.)
TEMED (Merck cat No. 1.10732.0100)
SYBR Safe DNA gel stain (Thermo Fisher Sci. S33102)
100 bp DNA ladder (Thermo Fisher Sci. cat No. SM0241)
Equipment:
Thermal cycler (Thermo Fisher Sci. cat No. A24811)
Mini protean vertical gel electrophoresis system (Bio-Rad cat No. 1658003)
Power supply (Bio-Rad cat No. 1645050)
UV Gel imaging system
Strip tubes and caps for PCR
Protocol steps:
- Extract DNA from mouse tail snips (2–5 mm) using a silica membrane column-based kit such as the NucleoSpin Tissue following manufacturer’s directions.
- We have also successfully used direct PCR kits that do not require prior DNA extraction such as the Phire Tissue Direct PCR Master Mix kit for RiboTag and Cre recombinase genotyping. DNA can also be extracted from tail samples by conventional methods using SDS/Protease K digestion, phenol/chloroform extraction, and ethanol precipitation.
Prepare the DreamTaq PCR Master mix as follows: (per reaction) 13 µl of water, 2 µl of 10x DreamTaq green buffer, 2 µl of 10mM dNTPs, 1 µl of the forward oligo (10uM), 1 µl of the reverse oligo (10uM), 0.1 µl of DreamTaq polymerase. Mix gently and briefly centrifuge.
Add 19 µl of the master mix to each well.
Add 1 µl of tail DNA (~2 ng) to each reaction.
Mix and briefly centrifuge.
Place strips in the thermal cycler and run the appropriate program for each reaction. Thermal profile for RiboTag reactions is: 95°C for 2 min followed by 30 cycles of 95°C for 20 s, 57°C for 20 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. Thermal profile for Cre reactions is 95°C for 2 min followed by 30 cycles of 95°C for 10 s, 63°C for 10 s, and 72°C for 40 s, with a final extension at 72°C for 5 min.
Prepare a 10% acrylamide gel by mixing (for two gels) 14 ml of water, 5 ml of 40% acrylamide-bis, 1ml of TBE 10x, 30 µl of TEMED and 150 µl of 10% APS. Allow gels to polymerize for 10–15 min.
Place gels into a vertical gel electrophoresis system, add buffer (0.5X TBE), and load DNA ladder and PCR reactions.
Run the electrophoresis at 175v (constant voltage) for 45 min.
Stain gels with SYBR safe and analyze PCR products under UV light.
Detecting “Ectopic” Cre Recombination in tail DNA
Mouse lines driving Cre recombinase expression under cell type specific promoters should typically restrict RiboTag activation to the cell type of interest. However, transient expression of Cre recombinase during development is often observed in different Cre driver lines and can lead to recombination of the RiboTag allele in unexpected cell types. If this “ectopic” expression takes place in the fertilized egg or during early development, it will be detected during the RiboTag genotyping PCR and a “recombination” band will show up at ~350 bp. If recombination occurs in the fertilized egg the loxP band will be replaced by the “recombination” band. If Cre recombinase is expressed during early development, recombination of the RiboTag allele will only take place in a portion of the cells and both the loxP and the recombination band will show up during PCR analysis. If genotyping heterozygous animals (Rpl22HA/+), the wild-type band will always be present in the PCR assay independently of the potential recombination events that may take place on the floxed allele.
SUPPORT PROTOCOL 2 qRT-PCR analysis
The success of the RiboTag procedure can be assessed by using qRT-PCR to evaluate whether specific mRNAs have been enriched during the isolation. Transcripts from the gene that is driving the Cre-recombinase in the specific cell types should be enriched in the IP RNA sample compared to the input RNA. We also recommend monitoring mRNAs that are not expected to be expressed in the targeted cells to verify that they are de-enriched in the IP. Quantitative RT-PCR may substitute for a more expensive and time-consuming analysis by RNA-Seq when only a small subset of transcripts need to be assessed.
Materials:
Equipment:
Real-Time PCR machine
Reagents:
We use the one step Brilliant II or III SYBR green kits from Agilent or RNA-Ct one step kits from Thermo Fisher Sci.
To confirm the specific isolation of transcripts from the cell type of study, we analyze RNA extracted from Inputs and IPs by single-step qRT-PCR. The enriched presence of known cell type-specific transcripts in the IPs compared to inputs indicates that the RiboTag isolation was successful.
Test samples for expression of transcripts specific to cell types in the dissected area other than the one of interest such as a glial marker when using a neuron-specific Cre transgenic. These are expected to appear depleted in the IPs when compared to inputs.
Perform qRT-PCR analysis for a housekeeping gene such as Actb. Normalizing all data to that standard can help correct for errors in RNA quantitation or differences in RNA quality. Adding equal amounts of RNA to the qRT-PCR reaction for all samples results in little variation in the CTs obtained for the housekeeping gene (less than 1 CT of difference is expected among all samples), which confirms RNA integrity in all samples and suitability of the housekeeping gene for data normalization.
BASIC PROTOCOL 2: RiboTag mice as a reporter mouse line for direct visualization of Cre recombination
Materials:
Mice:
RiboTag mouse line (JAX stock No. 011029)
Cre-driver mouse line of choice
Reagents:
Normal Donkey Serum (Jackson ImmunoResearch cat. No. 017–000-121)
Zymed’s rabbit anti-HA antibody (Thermo Fisher Sci., cat. No. 71–5500
Covance’s Alexa Fluor 488-conjugated mouse anti-HA antibody (BioLegend,cat. No. A488–101L)
Alexa Fluor 488 donkey anti-rabbit antibody (Thermo Fisher Sci., cat. No. A21206)
SuperFrost plus slides (Thermo Fisher Sci. cat No. J1800AMNT)
Coverslips 24 × 60 mm #1 (Menzel-Glaser, available through VWR cat No. 631–1339)
Fluoromount G with DAPI (Electron Microscopy Sciences, cat. No. 17984–24)
Equipment:
Netwells (Corning CLS3477)
Nutator mixer (e.g. Boekel or Clay Adams)
Paint brush
Protocol steps:
Tissue preparation
- Transcardially perfuse mice with ice-cold phosphate-buffered saline (PBS) pH 7.4 containing 4% paraformaldehyde (PFA).
- Perfusion with fixative solution reduces background resulting from endogenous mouse IgG detection when using anti-mouse secondary antibodies. If no primary antibodies raised in mouse are used for staining, brains can by directly fixed after removal by immersion in fixative for 12–24 h.
Remove brains and postfix overnight at 4ºC in PBS pH 7.4 containing 4% PFA.
Wash brains twice in ice-cold PBS pH 7.4
Immerse brains in an ice-cold solution of 30% sucrose in PBS pH 7.4. Specimens will float. Keep at 4ºC until they sink (about 24 h)
Remove brains from the cryoprotective solution and pat dry to remove excess solution
Freeze in powdered dry ice for 5 min and store at −80 ºC until sectioning.
Cryo-section brains in 30 um-slices using a freezing microtome. Collect sections from the area of interest for free-floating staining in PBS pH 7.4 with 0.02% sodium azide as preservative. Store sections at 4º.
Imunofluorescence
-
8.
Use a paint brush to transfer sections to wells containing Netwells inserts filled with PBS pH 7.4. Rinse sections once with PBS to remove preservative.
-
9.
Block sections in PBS with 0.2% Triton X-100 and 10% normal donkey serum (NDS) for 1 h at room temperature with agitation.
-
10.Incubate overnight at 4ºC with a primary antibody solution containing an anti-HA antibody such as the Zymed’s rabbit anti-HA (#71–5500 Thermo Fisher Sci; use at 1:1000), or with a fluorophore-conjugated anti-HA antibody such as the Covance’s Alexa Fluor 488-conjugated mouse anti-HA antibody (# A488–101L BioLegend, use at 1:500), in PBS pH 7.4 with 1% NDS and 0.2% Triton X-100.
- Fluorophore-conjugated antibodies allow for a direct visualization of the signal without secondary antibody detection. Using fluorophore-conjugated secondary antibodies will however provide an amplified signal.
-
11.
Wash sections three times for 5 minutes in PBS pH 7.4 with 0.2% Triton X-100. If a fluorophore-conjugated anti-HA antibody is used proceed to step 13.
-
12.
Incubate with secondary antibody solution containing an anti-rabbit antibody conjugated to the fluorophore of choice (e.g., Alexa Fluor 488; Invitrogen; 1:500 in PBS pH 7.4 with 1% NDS and 0.2% Triton X-100) for 1 h at room temperature.
-
13.
Wash sections three times for 5 minutes in PBS pH 7.4 with 0.2% Triton X-100
-
14.
Mount stained sections onto slides for imaging using Fluoromount G with DAPI.
-
15.
Seal the edge with nail polish and allow slides to dry for 1–2 hours before visualizing. Protect from light.
-
16.
Slides can be stored at 4ºC protected from light. For best results, slides should be analyzed within 1–2 weeks using the microscope of choice. Staining using anti-HA antibodies in Cre-positive RiboTag mice results in a strong perinuclear signal in the cell type of interest.
-
17.Image slides on a fluorescent microscope (standard or confocal) to visualize the Alexa Fluor 488 and DAPI.
- DAPI will bind DNA and light up the nuclei in all cells and Alexa fluorescence will only be present in the cells that are expressing RiboTag. Expect to see most of the RiboTag labeling in the cytoplasm where polyribosomes are abundant and in the nucleolar structures in the nucleus where ribosomes are being assembled.
BASIC PROTOCOL 3: RNA-Seq Library Preparation
There are many library preparation kits on the market but Trio RNASeq (NuGEN) has worked well in our hands. A detailed protocol is available on NuGEN’s website (NuGEN). Briefly, 500 pg – 50 ng of total RNA in a volume of 10 ul is used as starting material for samples. We recommend using similar quantities of RNA for all samples that are going to be compared with each by RNA-Seq. We have good results starting with ~2 ng of total RNA.
Materials
Trio RNA-Seq, mouse rRNA, part number 0507-X; (X = 8, 32, or 96), (NuGEN)
AMPure XP, reference number A63880, (Beckman Coulter)
NEBNext Library Quant Kit for Illumina, catalog number E7630X (X = S, or L), (NEB)
NextSeq 500/550 High Output Kit v2.5 (75 Cycles), catalog number 20024906, (Illumina)
200 mM NaOH
200 mM Tris, pH 7.0
0.2 mL PCR tubes (flat top optically clear)
1.5 mL microtubes
0.2 mL tube magnetic rack
Equipment:
Thermal cycler (PCR machine)
Quantitative thermal cycler
NextSeq Sequencer (Illumina)
Convert isolated RNA from RiboTag into cDNA libraries
- Perform DNase treatment using the Trio RNASeq kit.
- Although DNA digestion is performed on the RNA purification column, the first step of the Trio RNASeq kit is to perform DNase treatment. It is extremely important to eliminate any residual DNA and the second DNase treatment does not result in any loss of sample.
- Synthesize cDNA from the total RNA using reverse transcriptase primed with both oligo dT and random hexamers and then use DNA polymerase to produce a double stranded cDNA molecule
- These primers include an RNA sequence at the 5’ end that is not complementary and does not hybridize to the sample RNA. Then a second strand is made with a DNA polymerase (Dafforn et al., 2004; Kurn et al., 2005).
- Purify double stranded cDNA using silicon coated magnetic beads (AMPure XP, Beckman Coulter).
- These beads are used several times throughout the protocol adding various amounts of a salt containing bead binding solution. By varying the salt concentration in the reaction, one is selecting for different fragment sizes, with more salt attracting almost all fragment sizes and less preferentially selecting for larger fragments (Boom et al., 1990; Melzak, Sherwood, Turner, & Haynes, 1996).
- Amplify the double-stranded cDNA using single primer isothermal amplification (SPIA).
- RNase H is used to digest the RNA portion of the RNA / DNA heteroduplex at one end of the double stranded cDNA. A 5’ RNA 3’ DNA primer and a strong strand displacing DNA polymerase are used to make a single stranded cDNA that is the compliment to the original RNA. The process is continually repeated all at one temperature making many such copies (Dafforn et al., 2004; Kurn et al., 2005).
- Fragment and repair ends of the cDNA
- The kit accomplishes this using an enzymatic process. After fragmentation adapters are ligated to either side of the cDNA with the 3’ side receiving a unique nucleotide barcode that will be used to identify samples after sequencing.
- PCR Amplification of cDNA products.
- The kit uses two PCR amplifications with a targeted step in between designed to deplete any cDNAs that would have been made from ribosomal RNA. Oligo probes are designed to bind to rRNA derived cDNA, extend that product, then restriction enzymes target these cleaving the adaptors off these products essentially eliminating them from the library.
Quantify cDNA libraries
Even starting at the low end of the kit’s range 500 pg of total RNA, one ends up with micrograms of cDNA. This needs to be greatly diluted before sequencing. To get the best results from the sequencer the cDNA libraries must be loaded on at a precise quantity. As well, each sample that will be multiplexed together should be loaded in equal quantities. The most accurate way to quantify the cDNA libraries is using qPCR against the adaptors on the cDNA fragments. Again, there are many kits on the market that can accomplish this but NEBNext has worked well in our hands.
Four standard samples come with the kit ranging from 0.01 pM to 10 pM. In order to fall within these one needs to make serial dilutions of the libraries and use a range of those samples to quantify. In our hands 1:100,000 and 1:1,000,000 are the two dilutions that will fall between the given standards. Our libraries are most often between 0.1 and 1 µM.
A detailed user manual is available on NEB’s website (NEB, 2017).
Denature and dilute libraries
The day of the sequencing run, libraries need to be denatured and diluted. A detailed protocol is available on Illumina’s website (Illumina, 2018a).
Make a 1 nM dilution of each sample to be multiplexed together using a portion of the libraries made earlier. Add an equal amount of each 1 nM sample to a master sample.
Add 20 uL of the master sample to 20 uL of 0.2 M NaOH. Mix by pipetting half of the volume 10 to 20 times. Incubate for 5 minutes.
Add 20 uL 0.2 M Tris pH 7 to neutralize the NaOH. Mix well by pipetting.
Add 940 uL of prechilled HT buffer provided with the NextSeq reagents to make a 20 pM dilution. Vortex briefly then spin the contents down. Keep the 20 pM sample on ice until you are ready to make the final dilution.
Add 117 uL of the 20 pM sample to 1183 uL of prechilled HT buffer for a final dilution of 1.8 pM in 1.3 mL. Vortex briefly then spin the contents down. Keep this on ice until you are ready to load this on the sequencer.
Sequencing
Thaw reagents cartridge at 4 degrees overnight. Take the reagents cartridge and flow cell out of the refrigerator 30 minutes to an hour before your sequencing run. Ensure all reagent have thawed before proceeding.
From the NextSeq500’s software interface select Sequence.
Following the instructions on screen, load the flow cell.
Empty the spent reagents container into the appropriate waste container, then reload. Load buffer cartridge.
Pierce the foil covering reservoir 10 on the reagent cartridge with a pipette tip. Then load the full volume of the 1.8 pM sample into reservoir 10. Load the reagent cartridge onto the NextSeq500.
- Specify the run parameters on the setup screen. Assign a name to Run Name, enter 75 into Read 1 and 8 into Index 1.
- Single end reads may be sufficient for gene expression quantification and differential gene expression analysis. Running paired end reads will increase alignment confidence for specific isoforms and for novel transcript discovery. In this case you would enter 75 into “Read 2”. NextSeq high output cartridges are rated for 400 million single end or 200 million paired end reads.
- Select Next, the system will run through a number of checks. Once it has finished with these select Start.
- A detailed user guide including instructions for loading reagents and sample onto the NextSeq sequencer is available on Illumina’s website (Illumina, 2018b).
- Once reagents and sample have been loaded on the NextSeq and the sequencing run has begun, the multiplexed sample is washed over the flow cell where cDNA molecules with adaptors hybridize to an oligo lawn with complementary sequences. A DNA polymerase makes a compliment to the original cDNA molecule primed from the oligo attached to the flow cell, the double stranded cDNA is denatured, and the original cDNA molecule is washed away. The attached compliment binds a second oligo attached to the flow cell, and is copied by a DNA polymerase in a process called bridge amplification. The process is repeated many times over until a cluster of cDNA molecules grows that will be large enough for the sequencer to read. Fluorescently labeled dNTPs are washed over the flow cell where they incorporate into the growing DNA strand primed off the template attached to the flow cell. This process is used to identify the sequence of both the DNA library fragments made from the original samples and the nucleotide barcode ligated to that DNA that identifies which sample it comes from (Illumina, 2016).
BASIC PROTOCOL 4: RNA-Seq Bioinformatics
The analysis of RNA-Seq data can be done using a variety of open source or proprietary programs. If you are using a Core Facility to do RNA-Seq they are also likely to be able to analyze the data for you or at least assist you in the basic steps to obtain the aligned datasets. We build the libraries and run the RNA-Seq ourselves and use the methods described below to analyze the data.
Software
Software for Linux OS:
Bcl2fastq2 (Illumina, 2017a)
Software for Linux OS or Mac OS:
FASTX-Toolkit (Hannon_Lab, 2014)
TopHat2 (Trapnell et al., 2016)
Bowtie2 (Langmead, Kim, Charles, Wilks, & Antonescu, 2018)
Cufflinks (Trapnell et al., 2014)
Software for Linux OS, Mac OS, or Windows OS:
RStudio (RStudio, 2018)
tidyr (Wickham & Henry, 2018)
dplyr (Wickham, François, Henry, & Müller, 2018)
ggplot2 (Wickham, Chang, et al., 2018)
Convert BCL files to FASTQ files
Illumina sequencers generate per cycle base call (BCL) files. However, most informatic programs take FASTQ or FASTA files as input. Bcl2fastq2 software is able to covert the files, as well as, demultiplex samples. A detailed user guide is available on Illumina’s website including system requirements for the software (Illumina, 2017b).
-
Create a sample sheet with sample names, numbers, and corresponding nucleotide barcodes. This is saved as a .csv in the following format:
[Data],,
Sample_Name,Sample_ID,Index
Sample1,1,CGCTACAT
Sample2,2,AATCCAGC
Sample3,3,CGTCTAAC
Etc.
-
Run the blc2fastq2 in Terminal (denoted with $ hereafter) with the following options:
$ bcl2fastq --minimum-trimmed-read-length 0 --mask-short-adapter-reads 0 --no-lane-splitting -o /output_folder -R run_folder --sample-sheet run_folder/SampleSheet.csv
Explanation:
blc2fastq – calls the program.
minimum-trimmed-read-length – specifies a length to not trim if trimming adapters will result in a read of less than this length. In these cases, the adapter portion will be masked by replacing base calls with Ns. By specifying 0 we are telling the program to trim adapters no matter the remaining length of the read. Default is 35.
mask-short-adapter-reads – specifies a length of a read that if it falls below will be masked by Ns. By specifying 0 we are instructing the program not to do this. Default is 22 (must be set equal or lower than minimum-trimmed-read-length).
no-lane-splitting – this option instructs the program to include all reads for each given sample in one file. Without this option the NextSeq will give four files for each sample, one for each of lanes 1–4. If the researcher is planning to upload FASTQs to Illumina’s basespace.com they should not call this option as basespace will only accept sample files that do have lanes split.
-o – specifies the output folder.
-R – specifies the run folder or where the BCL files are located.
--sample-sheet – points to where the sample sheet is located.
Trim and filter FASTQ files
-
3.
Trim the first 5 base calls and filter out low quality reads. FASTX-Toolkit tools can accomplish this with the following command:
$ for i in sample1 sample2 sample3 etc; do zcat $i_S1_R1_001.fastq.gz | fastx_trimmer -f 6 -v -Q33 | fastq_quality_filter -q 25 -p 80 -v -Q33 -z -o $i.fastq.gz; done
Explanation:
Commonly known as a “for loop”.
for i in sample1 sample2 sample3 etc – is a list of your samples to iterate over.
do X – could be any command. This command will then iterate over your list.
done – tells the command to stop.
$i – is the placeholder where the list of samples will be substituted in.
zcat – looks at a compressed file.
| – called a pipe, it sends information from first command to the next command.
fastx_trimmer -f 6 – this option says trim the first 5 base calls and keep everything from 6 onward.
fastq_quality_filter -q 25 -p 80 – this option says only keep reads with a quality score of 25 or higher on at least 80% of the base calls.
-z – is the option to compress output file.
-o – is the name of file or directory name/name of file.
Align FASTQ files to a reference genome
We use TopHat2 (Kim et al., 2013; Trapnell, Pachter, & Salzberg, 2009) for alignment against GRCm38/mm10 UCSC. Useful links to reference genomes with prebuilt bowtie indexes and GTF transcript annotation files can be found here: http://ccb.jhu.edu/software/tophat/igenomes.shtml. A detailed protocol for TopHat has been published elsewhere (Trapnell et al., 2012).
-
4.
Use the following command for alignments:
$ for i in sample1 sample2 sample3 etc; do tophat2 -p 8 -G genes.gtf -o output_folder/$i Bowtie2Index/genome folder_with_fastqs/$i.fastq.gz; done
explanation:
tophat2 – is a program that performs alignment.
-p 8 – specifies the number of CPU processors to use. Check your system information and set this at or below the total number of logical cores on the computer running the software.
-G – points to directory and .gtf file.
-o – points to output directory and /$i – is the placeholder for sample.
Sequence/Bowtie2Index/genome – points to directory with index files.
folder_with_fastqs/$i.fastq.gz – points to directory and fastq files. This is the actual sequencing data.
Quantify and compute differential gene expression
We use cuffdiff of cufflinks (Roberts, Trapnell, Donaghey, Rinn, & Pachter, 2011; Trapnell et al., 2013; Trapnell et al., 2010) to compute differential gene expression. A detailed protocol has been published elsewhere (Trapnell et al., 2012).
-
5.
Use the following command to calculate differential expression:
$ cuffdiff -p 8 -no-update-check -o output_folder -b Bowtie2Index/genome.fa -u genes.gtf -L condition1,condition2 \./tophat_folder/condition1_S1/accepted_hits.bam,./tophat_folder/condition1_S2/accepted_hits.bam,tophat_folder/condition1_S3/accepted_hits.bam,etc \./tophat_folder/condition2_S1/accepted_hits.bam,./tophat_folder/condition2_S2/accepted_hits.bam,tophat_folder/condition2_S3/accepted_hits.bam,etc
explanation:
cuffdiff – is a program to test for differential gene expression.
-p 8 – this option specifies how many CPU processors to use.
-no-update-check – use this option as the software is no longer being updated.
-o – points to folder you want your results saved in.
-L – name of your two conditions e.g. control, treatment or input, IP.
-b – this option corrects for RNA-seq read coverage bias (Roberts et al., 2011).
-u – this option performs an initial estimate to improve accuracy when weighting reads mapping to multiple locations.- We run cuffdiff first on inputs vs IPs within conditions and then IP vs IP across conditions. In some cases, one may only be asking what is unique about these cells within this tissue. In which case one would only compare input to IP. It is important to note that genes that are expressed widely in the total population of neurons will not show up as differentially expressed even if they are expressed in the targeted neuron population. The difference between IP and input will only be significant when a transcript is preferentially expressed in the HA-tagged neuron population.
Visualize results
Use R within RStudio to visualize your results utilizing tidyr, dplyr, and ggplot2 packages (Wickham, 2016).
-
6.
If necessary, install packages.
> install.packages(“tidyr”)
> install.packages(“dplyr”)
> install.packages(“ggplot2”)
Load packages.
> load(tidyr)
> load(dplyr)
> load(ggplot2)
-
7.
Import output files “gene_exp.diff” and “genes.read_group_tracking” from cuffdiff and save them as an object. The names of the objects are arbitrary; however, good object names should be short but descriptive. Below the first named object “cdstats_aipvn” describes cuffdiff statistical output for condition A, IP vs input. The example below assumes the three cuffdiff output folders are saved in the working directory the R-project is started in.
> cdstats_aipvn <- read.delim(“cuffdiff_A_IP_vs_N/gene_exp.diff”)
> cdstats_bipvn <- read.delim(“cuffdiff_B_IP_vs_N/gene_exp.diff”)
> cdstats_avb <- read.delim(“cuffdiff_A_vs_B/gene_exp.diff”)
> cdfpkms <- read.delim(“cuffdiff_A_vs_B/genes.read_group_tracking”)
-
8.
Below are three examples of creating a list of genes expressed at certain thresholds in the IP relative to the input in condition A.
> A_2fold <- filter(cdstats_aipvn, q_value < 0.05, log2.fold_change. > 1, value_2 > 5) %>% select(“test_id”)
> A_1fold <- filter(cdstats_aipvn, log2.fold_change. > 0, value_2 > 5) %>% select(“test_id”)
> A_half_fold <- anti_join(dr1stats, filter(dr1stats, log2.fold_change. < −1, q_value < 0.05), by = “test_id”) %>% filter(value_2 > 5) %>% select(“test_id”)
-
9.
Filter condition B the same way, then join the 2 lists to create a list of genes that could be important in either condition A or condition B.
> AB_2fold <- full_join(A_2fold, B_2fold)
-
10.
Now the new list of genes that could be important in either condition can be used to filter for genes that one would want to compare across conditions.
> AIP_vs_BIP <- inner_join(AB_2fold, cdstats_avb, by = “test_id”)- Cuffdiff includes a column in the output file gene_exp.diff labeled “significant” that can be useful while graphing data assigning different colors to the binomial values (“yes” or “no”). However, we find it useful to append additional columns that can specify other criteria, for example, both statistically significant and meaningfully different.
-
11.
In the example below, we are using 2-fold again.
> temp1 <- filter(AIP_vs_BIP, abs(log2.fold_change.) > 1, q_value < 0.05) %>% mutate(DE = “yes”)
> temp2 <- anti_join(AIP_vs_BIP, temp1, by = “test_id”) %>% mutate(DE = “no”)
> AIP_vs_BIP2 <- full_join(temp1, temp2)
-
12.
A common graph made with RNA-Seq data is a volcano plot. This plots p-value vs fold change, usually both have been log transformed. One can now use the newly created “DE” column to highlight genes that are both 2-fold different and statistically different.
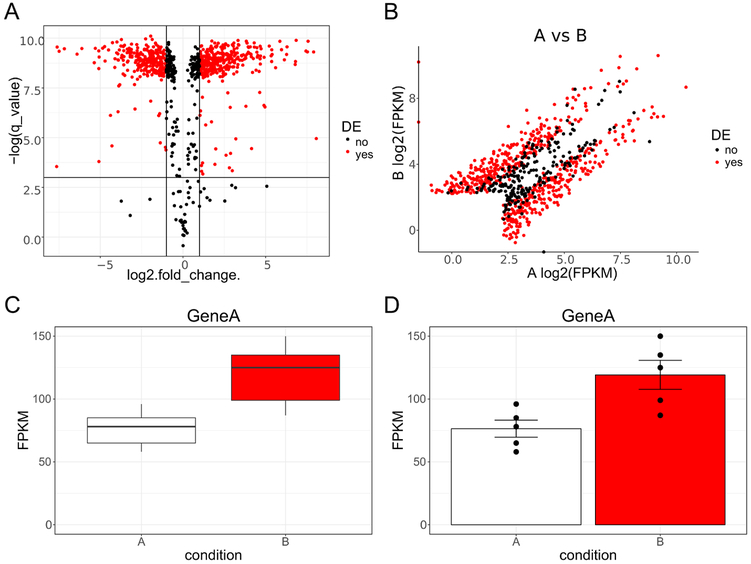
> volcplot <- ggplot2(AIP_vs_BIP2, aes(log2.fold_change., -log(q_value), color = DE) + geom_point(alpha = 0.5)- By saving the plot as an object (“volcplot”) it is easy to call up the object and modify various aspects experimenting with different options until one is satisfied with the plot’s appearance. Below we have modified the default colors of the points to black and red, drawn a horizontal line where the corrected p-value is 0.05, drawn lines indicating where the fold change is 2-fold, and selected a different theme (Figure 2A). We like the way theme_classic and theme_bw look but this is purely a matter of preference.
-
> volcplot + scale_color_manual(values=c(“black”, “red”)) + geom_hline(yintercept = -log(0.05)) + geom_vline(xintercept = c(−1,1)) + theme_bw()
Another common plot one could make is a scatter plot, plotting log transformed values for a list of genes with condition A on one axis and condition B on the other.
> scatplot <- ggplot(AIP_vs_BIP2, aes(log2(value_1), log2(value_2), color = DE)) +
geom_point(alpha = 0.3) +
scale_color_manual(values = c(“black”, “red”)) +
theme_classic()
One can relabel the axis and give the plot a title if they wish (Figure 2B).
> scatplot + labs(x = “A log2(FPKM)”, y = “B log2(FPKM)”, title = “A vs B”) +
theme(plot.title = element_text(hjust = 0.5)) +
theme(text = element_text(size = 20))
Finally, one may wish to plot individual genes of interest:
Figure 2. Example plots of hypothetical RNA Seq data.
A, Volcano plot of data with log base 2 transformed fold change (condition A vs condition B) plotted on the x axis and negative natural log transformed corrected p-value (q-value) plotted on the y axis. The solid horizontal line denotes where q-value is equal to 0.05. The two solid vertical lines denote where fold change is equal to 2 (or −2). Data points are colored red if they are both 2-fold different and have a q-value less than 0.05; otherwise they are black. B, Scatter plot of data with log2 transformed fragments per kilobase million (FPKM) plotted on the x axis for condition A and the y axis for condition B. As in A data points are colored red if they are both 2-fold different and have a q-value less than 0.05; otherwise they are black. C, Box plots for Gene A for condition A and B. FPKM are plotted on the y axis with the two conditions represented side by side on the x axis. D, Bar graphs denoting mean plus or minus standard error of the mean (SEM) for Gene A for condition A and B. FPKM are plotted on the y axis with the two conditions represented side by side on the x axis. Individual data points are superimposed on top of the bars for greater data reporting transparency. DE, differentially expressed.
Boxplot
> box_geneA <- filter(cdfpkm, tracking_id == “GeneA”) %>%
ggplot(aes(condition, FPKM, fill = condition)) +
geom_boxplot() +
theme_bw() +
theme(legend.position=“none”) +
scale_fill_manual(values = c(“white”, “red”)) +
ylim(0,NA)
Add a title using labs, center justify using hjust, and change the font size with element text (Figure 2C).
> box_geneA + labs(title = “GeneA”) +
theme(plot.title = element_text(hjust = 0.5)) +
theme(text = element_text(size = 20))
Bar chart
> bar_geneA <- filter(cdfpkm, tracking_id == “GeneA”) %>%
ggplot(aes(condition,FPKM)) +
stat_summary(fun.data=“mean_se”, geom=“bar”, color=“black”, aes(fill=condition)) +
stat_summary(fun.data=“mean_se”, geom=“errorbar”, width=0.3, color=“black”) +
theme_bw() +
theme(legend.position=“none”) +
scale_fill_manual(values = c(“white”, “red”)) +
labs(title = “GeneA”) +
theme(plot.title = element_text(hjust = 0.5)) +
theme(text = element_text(size = 20))
Add in individual data points for greater data reporting transparency (Figure 2D).
> bar_geneA + geom_point(size = 3)
REAGENTS AND SOLUTIONS
Tris base (Sigma; cat. No. T1503).
Prepare a 1.5M pH 7.4 stock solution (pH with HCL) and store at 4ºC
KCl (Sigma; cat. No. P9541).
Prepare a 1M stock solution and store at 4ºC
MgCl2 (Sigma; cat. No. 63068).
Prepare a 1M stock solution and store at 4ºC
Nonidet P-40 Substitute, 10% (w/v) solution (Roche, cat. No. 11 332 473 001)
Cycloheximide (Sigma, cat. No. C7698).
Prepare a 5mg/ml stock solution and store at −20ºC.
Heparin (Sigma; cat. No. H3393).
Prepare a 100 mg/ml stock solution and store at 4ºC.
Phosphate buffered saline (PBS), tablets (Millipore, cat. No. 524650–1EA).
Prepare at 1x with water.
Paraformaldehyde (Sigma cat. No. P6148).
Prepare a 4% solution in PBS 1x. Adjust pH to 7.4. Prepare fresh and store at 4ºC.
Sucrose (Sigma cat. No. 84097).
Prepare a 30% solution in PBS 1x pH 7.4. Store at 4ºC.
Sodium Azide (Sigma, cat. No. S2002).
Prepare a 0.02% solution in PBS pH 7.4
Triton X-100 (Millipore, cat. No. 648466).
Prepare a 0.2% solution in PBS pH 7.4.
Homogenization buffer:
50 mM Tris pH7.5
100 mM KCl
12 mM MgCl2
1% Nonidet P-40
1 mM DTT
200 U/mL Promega RNasin
1 mg/mL heparin
100 ug/mL cycloheximide
-
protease inhibitor mixture (1x)
Optional: The addition of 1% sodium deoxycholate (DOC) to the homogenization buffer reduces some of the non-specific background. We have shown that DOC does not significantly reduce the purification of specific polysomes and suggest testing it in your experiments.
Homogenization buffer is prepared by adding defined volumes of stock solutions for Tris (1.5M pH 7.4), KCl (1M) and MgCl2 (1M) to nuclease free water to achieve the final concentrations described above in the desired volume. Right before homogenization, buffer is supplemented with DTT, protease inhibitors, RNasin, heparin and cycloheximide.
High salt buffer:
50 mM Tris pH 7.5
300 mM KCl
12 mM MgCl2
1% Nonidet P-40
0.5 mM DTT
-
100 ug/mL cycloheximide
Prepare using Tris, KCl and MgCl2 stock solutions. Add DTT and Cycloheximide right before using.
COMMENTARY
Background Information
It has been a little over 10 years since the first development of cell-type specific ribosome tagging strategies that permitted the analysis of a cell’s translatome (the mRNAs engaged on polyribosomes) (Heiman et al., 2008). The original technique utilized BAC transgenic mice that were engineered to express a GFP-tagged 60S ribosomal subunit protein (Rpl10a) behind a cell type specific promoter. Antibodies directed against GFP were used to immunoprecipitate the tagged polyribosomes from which mRNA could then be isolated and analyzed by qRT-PCR, microarray, or RNA-Seq. This approach required the generation of novel BAC transgenic mice specific for each cell type and did not take advantage of the many tissue specific Cre recombinase mouse lines that were already developed.
We developed an alternative approach described in this RiboTag protocol in which an HA-tagged ribosomal protein (Rpl22) is conditionally expressed from the mouse genome by the action of Cre recombinase (Sanz et al., 2009). This approach takes advantage of the constantly expanding diversity of cell type specific Cre recombinase expressing mouse lines. Both approaches suffer from the same limitation: the investigator must have already identified a gene promoter that is specific for the cell type of interest. Although it is unlikely that subtypes of neurons can be uniquely defined by the expression of a single Cre recombinase transgene, the ribosome tagging strategies have facilitated analysis of the mRNAs engaged with polyribosomes in many neuronal cell types and can also be used to study the regulation of specific genes in vivo.
One alternative approach that has rapidly been adopted by neuroscientists is single cell transcript analysis by direct RNA-Seq. A single anatomical structure such as the arcuate nucleus of the hypothalamus is dissociated into single cells, the transcripts from each cell are uniquely barcoded, and the RNA is analyzed by RNA-Seq. Clustering analysis is used to identify unique cell types including neurons and glial cells, and then the neurons can be further clustered into discreet sub-types based on their transcript phenotype. This unbiased approach is very useful to explore the full range of neurons in a specific anatomical location and will rapidly leading to transcriptome phenotypes for all of the neurons and glial cells in a mouse brain. One limitation of this approach is that relatively non-abundant transcripts escape detection due to the large number of different cell types being sequenced en masse. Another limitation is that the techniques that must be used to obtain single cells, especially starting with the brain, can cause alterations in gene expression (e.g. induction of immediate early genes) (Moffitt et al., 2018; van den Brink et al., 2017) and may also lose transcripts that are sequestered in neuronal processes where they are also likely to be under translational control. The ribosome capture approaches like RiboTag will only isolate mRNAs that have ribosomes bound to them and are expected to be actively translated into protein.
Critical Parameters and Troubleshooting
1. Cell type Specificity:
The specificity of the RiboTag analysis depends on the specificity of the Cre recombinase transgenic mouse line and every effort should be made to verify that the Cre transgenic drives expression of Rpl22-HA in the appropriate cell type and does not give rise to ectopic expression in other tissues that would interfere with the analysis. It is important to check that recombination of the RiboTag allele is not occurring during early development giving rise to mice with variable expression in many other cell types. This can be done by analyzing expression of Rpl22-HA by immunohistochemistry and comparing it to a marker for the cells of interest as described in Basic Protocol 2. We also routinely screen for RiboTag recombination in tail DNA during genotyping as described in the Support Protocol 1. This will help identify mice that have undergone recombination of the RiboTag allele during early development and they should be removed from the cohort to be studied as adults.
2. Background and Controls:
When we analyze the RNA from a RiboTag experiment we find that the immunoprecipitated polysomes still contain mRNAs from cells that are not expressing Cre recombinase and Rpl22-HA (e.g. glial cells). These transcripts are depleted in the IP (IP/input ratios <0.3) but since some are abundantly expressed, they will still appear at significant levels in the IP. As described in Basic Protocol 4 we advise analyzing both the input and IP RNA and then considering whether to filter out background gene transcripts that are depleted. In some of our experiments we are interested in gene transcripts which are enriched in the cell type being studied and help define a unique phenotype for those neurons. In addition to the background of depleted mRNAs seen in the IP we have observed that the IP may also contain enriched transcripts from genes that are clearly not expressed in the cell type of interest. These false positives can confound the data and we suggest doing a control IP experiment starting with equivalent tissue from a mouse that does not express RiboTag. Transcripts that enrich in the absence of Rpl22-HA expression are artifacts of the technique and should be eliminated from the specific transcripts isolated when Rpl22-HA is expressed. As described in the protocol we have recently been able to reduce the capture of these artifactually enriched genes by including an ionic detergent, sodium deoxycholate, in the initial homogenization buffer.
3. Anticipated Results and Statistical Analysis:
Each tissue homogenate yields two RNA-Seq libraries, an input and an IP. We typically run 8–16 RNA-Seq libraries on a NextSeq 500/550 flow cell and expect to have 25–50 million aligned reads for each library. Statistical analysis of this data requires at least 3 replicates making these experiments quite expensive in terms of reagents. If one is trying to phenotype the cells the first analysis would be to determine all of the expressed genes comparing IP to input and then to rank those transcripts by the ratio of IP to input. Transcripts that come up as enriched in a control IP from a mouse that does not express RiboTag should be removed from this transcript phenotype. If the animals have been treated with pharmacologic agents or exposed to physiological challenges, transcripts that change between samples can be identified and statistically analyzed as described in Basic Protocol 4. It is very helpful to correlate the specific transcripts that are enriched in the Cre-expressing cells with the in situ hybridization results from the Allen Mouse Brain Atlas. Specificity can also be confirmed using RNAscope (Advanced Cell Diagnostics) to colocalize transcripts in the anatomical region of interest. One can also confirm results by qRT-PCR starting with either the original RNA samples or qPCR with the DNA libraries.
The RiboTag approach is ideally suited to studying the dynamic regulation of genes in a specific neuronal population. Transcripts can be induced within minutes after the addition of hormones and drugs or changes in nutritional or environmental conditions. When an mRNA is specifically induced in the RiboTag targeted cell but constitutively expressed in neighboring cells, the level of transcripts in the IP will increase and the enrichment (IP/input) will also increase. If a gene like Fos is induced in all of the cells in the tissue being analyzed, then you will see an increase in IP transcripts for Fos but no change in enrichment.
Translational control:
RiboTag techniques will only isolate mRNAs that have ribosomes bound and this implies that these mRNAs are being actively translated. However, it is important to consider that RiboTag is not going to be very sensitive to how many ribosomes are engaged on a particular mRNA. For example, if an mRNA is under translational control and only has 2–3 ribosomes in the repressed state and 12 or more ribosomes when induced, the efficiency of immunoprecipitation may not change enough to be a quantitative indicator of translational activity. Ribosomes may also remain attached to mRNAs even when elongation and active synthesis of protein is repressed.
4. Time Considerations:
One of the advantages of the RiboTag procedure is that once the animals are obtained either by mating RiboTag mice with a specific Cre driver transgenic or stereotaxically injecting AAV-RiboTag virus and waiting for RiboTag expression, the immunoprecipitation of specific RNA can be completed in a day. Production of DNA libraries for RNA-Seq and DNA sequencing can be completed in a few more days. The most time-consuming step is the analysis of results and this depends on what questions are being asked and the number of different tissues and conditions being studied.
ACKNOWLEDGEMENT
Supported by NIH grants GM032875 and MH086386 (GSM), F32DK116432 (JCB) Marie Sklodowska-Curie Individual Fellowship (H2020-MSCA-IF-2014–658352)(ES), and Ramón y Cajal fellowship (RyC-2012–11873) European Research Council (Starting grant Neuromito, ERC-2014-StG-638106 and Proof of Concept ERC-2018-PoC-825569) Mineco Proyectos I+D de Excelencia (SAF2014–57981P; SAF2017–88108-R) Agaur (2017SGR-323)(AQ)
LITERATURE CITED
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, & van der Noordaa J (1990). Rapid and simple method for purification of nucleic acids. Journal of clinical microbiology, 28(3), 495–503. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1691208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, . . . Kurn N (2004). Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. BioTechniques, 37(5), 854–857. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/15560142. doi: 10.2144/04375PF01 [DOI] [PubMed] [Google Scholar]
- Dong HW (2019). Allen Mouse Brain Atlas. Retrieved from http://mouse.brain-map.org/static/atlas [Google Scholar]
- Gentleman R, Ihaka R, Bates D, Chambers J, Dalgaard P, Hornik K, . . . Murdoch D (2018). R (Version 3.5.1) [language and environment for statistical computing]: The R Foundation. Retrieved from https://www.r-project.org/ [Google Scholar]
- Gore BB, Soden ME, & Zweifel LS (2013). Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Curr Protoc Neurosci, 65, 4 35 31–20. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/25429312. doi: 10.1002/0471142301.ns0435s65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimon Z, Volaski A, Orthgiess J, Boura-Halfon S, Varol D, Shemer A, . . . Jung S (2018). Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat Immunol, 19(6), 636–644. doi: 10.1038/s41590-018-0110-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon_Lab GJ (2014). FASTX-Toolkit (Version 0.0.14): Hannon Lab. Retrieved from http://hannonlab.cshl.edu/fastx_toolkit/download.html [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, . . . Heintz N (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell, 135(4), 738–748. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/19013281. doi: 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina. (2016). Introduction to sequencing by synthesis. Retrieved from https://www.youtube.com/watch?v=fCd6B5HRaZ8 [Google Scholar]
- Illumina. (2017a). bcl2fastq2 (Version 2.2): Illumina. Retrieved from https://support.illumina.com/downloads/bcl2fastq-conversion-software-v2-20.html [Google Scholar]
- Illumina. (2017b). bcl2fastq Conversion v2.19. 2. Retrieved from https://support.illumina.com/content/dam/illumina-support/documents/documentation/software_documentation/bcl2fastq/bcl2fastq2_guide_15051736_v2.pdf [Google Scholar]
- Illumina. (2018a). Denature and dilute libraries guide. NextSeq system. 6 Retrieved from https://support.illumina.com/content/dam/illumina-support/documents/documentation/system_documentation/nextseq/nextseq-denature-dilute-libraries-guide-15048776-06.pdf [Google Scholar]
- Illumina. (2018b). NextSeq 500: System guide. 4. Retrieved from https://support.illumina.com/content/dam/illumina-support/documents/documentation/system_documentation/nextseq/nextseq-500-system-guide-15046563-04.pdf [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, & Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology, 14(4), R36 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/23618408. doi: 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, & Wang S (2005). Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clinical chemistry, 51(10), 1973–1981. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/16123149. doi: 10.1373/clinchem.2005.053694 [DOI] [PubMed] [Google Scholar]
- Langmead B, Kim D, Charles R, Wilks C, & Antonescu V (2018). Bowtie2 (Version 2.3.4.3) [Sequencer aligner]. Retrieved from http://bowtie-bio.sourceforge.net/bowtie2/index.shtml [Google Scholar]
- Melzak KA, Sherwood CS, Turner RFB, & Haynes CA (1996). Driving forces for DNA adsorption to silica in perchlorate solutions. Journal of Colloid and Interface Science, 181(2), 635–644. Retrieved from <Go to ISI>://A1996UZ53600031. doi:DOI 10.1006/jcis.1996.0421 [DOI] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, . . . Zhuang X (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science, 362(6416). Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/30385464 http://science.sciencemag.org/content/sci/362/6416/eaau5324.full.pdf. doi: 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEB. (2017). NEBNext: Library quant kit for illumina. 1.2. Retrieved from https://www.neb.com/-/media/catalog/datacards-or-manuals/manuale7630.pdf
- NuGEN. (2017). User guide: Trio RNA-seq. 2. Retrieved from https://www.nugen.com/sites/default/files/M01440v2_User_Guide%3A_Trio_RNA-Seq_4270.pdf
- Paxinos G, & Franklin KBJ (2013). The Mouse Brain in Stereotaxic Coordinates. San Diego, CA: Academic Press. [Google Scholar]
- Quintana A, Sanz E, Wang W, Storey GP, Guler AD, Wanat MJ, . . . Palmiter RD (2012). Lack of GPR88 enhances medium spiny neuron activity and alters motor- and cue-dependent behaviors. Nat Neurosci, 15(11), 1547–1555. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/23064379. doi: 10.1038/nn.3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, & Pachter L (2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome biology, 12(3), R22 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/21410973. doi: 10.1186/gb-2011-12-3-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio. (2018). RStudio (Version 1.1.463) [integrated development environment (IDE)]: RStudio. Retrieved from https://www.rstudio.com/products/rstudio/download/#download [Google Scholar]
- Sanz E, Evanoff R, Quintana A, Evans E, Miller JA, Ko C, . . . McKnight GS (2013). RiboTag analysis of actively translated mRNAs in Sertoli and Leydig cells in vivo. PLoS One, 8(6), e66179 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/23776628. doi: 10.1371/journal.pone.0066179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Quintana A, Deem JD, Steiner RA, Palmiter RD, & McKnight GS (2015). Fertility-regulating Kiss1 neurons arise from hypothalamic POMC-expressing progenitors. J Neurosci, 35(14), 5549–5556. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/25855171. doi: 10.1523/JNEUROSCI.3614-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, & Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A, 106(33), 13939–13944. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/19666516. doi: 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson D, Sauvageau M, Goff L, Rinn JL, & Pachter L (2014). Cufflinks (Version 2.2.1) [Transcriptome assembly and differential expression analysis for RNA-Seq]. Retrieved from http://cole-trapnell-lab.github.io/cufflinks/install/ [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, & Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology, 31(1), 46–53. Retrieved from <Go to ISI>://000313563600021. doi: 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Kim D, Pertea G, Pimentel H, Kelley R, Pachter L, & Salzberg S (2016). TopHat 2 (Version 2.1.1) [Splice junction mapper]. Retrieved from http://ccb.jhu.edu/software/tophat/index.shtml [Google Scholar]
- Trapnell C, Pachter L, & Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25(9), 1105–1111. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/19289445. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, . . . Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols, 7(3), 562–578. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/22383036. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, . . . Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology, 28(5), 511–515. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/20436464. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Sage F, Vertesy A, Spanjaard B, Peterson-Maduro J, Baron CS, . . . van Oudenaarden A (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods, 14(10), 935–936. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/28960196. doi: 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. New York,NY: Springer-Verlag New York. [Google Scholar]
- Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, . . . Yutani H (2018). ggplot2 (Version 3.1.0). Retrieved from https://ggplot2.tidyverse.org/ [Google Scholar]
- Wickham H, François R, Henry L, & Müller K (2018). dplyr (Version 0.7.8): RStudio. Retrieved from https://dplyr.tidyverse.org/ [Google Scholar]
- Wickham H, & Henry L (2018). tidyr (Version 0.8.2.9000): RStudio. Retrieved from https://tidyr.tidyverse.org/
- Williams RW (2000). Mapping genes that modulate mouse brain development: a quantitative genetic approach (Goffinet AF & Rakic P Eds.). New York: Springer. [DOI] [PubMed] [Google Scholar]