Abstract
Measles virus (MV), a paramyxovirus, is one of the most contagious human pathogens and is responsible for thousands of deaths annually. Wild-type MV evolved to counter the innate immune system by avoiding both type I interferon (IFN) induction and inhibiting IFN signaling through the JAK/STAT pathway. Flowever, virus replication is significantly inhibited in IFN-pretreated cells. Similarly, MV vaccine derived strains are also inhibited by IFN pretreatment, but vaccine strains also induce IFN. Despite the significant progress in understanding the interactions between MV and the IFN pathway, the IFN stimulated genes (ISGs) that inhibit MV replication remain largely unknown. The aim of this study is to identify specific ISGs that mediate restriction of MV. In this study, we report that Radical S-adenosyl methionine domain containing 2 (RSAD2) restricts MV infection at the stage of virus release in infected 293T cells. Furthermore, attenuated MV strains are currently being developed as a novel treatment for solid and hematological malignancies. Therefore, we tested the impact of RSAD2 expression in an oncolytic virotherapy context using a MV permissive ovarian cancer line (SR-B2). As measured in 293T cells, MV release was also impaired in SR-B2 cells transduced to express RSAD2 in vitro. Additionally, oncolytic MV therapeutic efficacy was impaired in SR-B2 cells transduced to express RSAD2 in vivo. Overall, we identify RSAD2 as a novel restriction factor for MV by inhibiting the release of virus. These results provide important information regarding the interaction between MV and the innate immune system, as well as implications for the design of oncolytic MV platforms.
Keywords: Measles, RSAD2, VIPERIN, Antiviral, Interferon, Interferon stimulated genes
1. Introduction
The negative, single-stranded Measles virus genome contains 15,894 nucleotides, consisting of 6 genes that encode 8 proteins. The viral nucleocapsid, phosphoprotein and RNA-dependent RNA-polymerase form the ribonucleoprotein (RNP), which associates with the viral RNA genome. The RNP complex is bound by the matrix protein, which facilitates transport to the cytoplasmic tails of the hemagglutinin (H) and the fusion protein (F) glycoproteins at the cell surface [1,2].
The viral glycoproteins facilitate entry through three known receptors: CD46, nectin-4 and SLAM [3–7]. Upon binding, H interacts with F to trigger a conformational change, resulting in the insertion of the fusion peptide into the target cell membrane [8]. Fusion between the viral and cellular membrane releases the RNP complex into the target cell. Spread of the virus to neighboring uninfected cells has previously been demonstrated to assemble via lipid rafts [9].
Similar to other viruses, vaccine derived MV can be detected by the innate immune system to prevent the spread of infection. MV is detected by two known pattern recognition receptors, RIG-I and MDA-5 [10]. Activation of these sensors leads to production of IFN-β and activation of the interferon system (IFN), resulting in the induction of interferon stimulated genes (ISGs). ISGs are the effector proteins in the host’s antiviral defense system that mediates restriction of virus replication. Induction of antiviral ISGs, such as MX1/2, RSAD2 and OAS proteins, have been demonstrated to restrict HIV, MV, influenza and vesicular stomatitis virus [11]. Previous studies have demonstrated that MV is sensitive to the antiviral effects of IFN; however, many of the ISGs that restrict MV remain unknown [12, 13]. While MX1 has been demonstrated to restrict MV in a cell-type specific manner, additional ISGs capable of restricting MV release have not entirely been identified [14, 15].
Vaccine derived MV strains are currently being developed as an oncolytic virus in the treatment of several tumor types, such as glioblastoma, ovarian cancer, multiple myeloma and others [16]. MV has demonstrated promising results in phase I clinical trials; however, not all patients have benefited from MV therapy. In order to maximize oncolytic MV therapeutic outcome, a further understanding of factors that impact virus replication is required. Our laboratory’s recent study identified that constitutive expression of ISGs in tumor cells can dramatically impair therapy in patients treated with oncolytic MV [17]. These results identify innate antiviral immune sensing of MV infection as a major hurdle toward optimizing oncolytic virotherapy for widespread beneficial response among patients. The aim of this study was to identify the potential ISGs that restrict MV using rationally designed assays and in vivo treatment experiments. We hypothesized that identification of ISGs that restrict MV can provide critical information to improve the design of oncolytic MV therapy, and possibly improve vaccine development.
2. Materials and Methods
2.1. Cell lines and tissue culture
Vero cells were purchased from ATCC and grown according to the recommended protocol provided. 293T cells and SR-B2 cells were cultured as previously described [18]. Primary GBM lines were harvested from patients that had primary tumor resection at Mayo Clinic and passaged as xenografts in mice [19]. All lines were grown in DMEM supplemented with 10% FBS (heat-inactivated) and penicillin-streptomycin. Stably transduced SR-B2 cells were also supplemented with 1.2 μg/ml puromycin.
2.2. LV particle generation
Lentiviral particles were produced in 293T cells, as previously described [20]. Briefly, 4 × 105 cells were plated in a 6-well dish. 16 h later, 1 μg of psg9.8-ISG, 0.4 μg gag-pol and 0.2 μg VSV-G were transfected into 293T cells using Lipofectamine 2000, according to manufacturer’s protocol. Supernatant was collected 48 h post-transfection and passed through 0.45 μm filter. Collected supernatants were used for subsequent transductions. SR-B2 and 293T cells were transduced with 400 μl of the collected supernatant. 48 h post-transduction, 1.2 μg/ml puromycin was added to the culture.
2.3. Generation of LV-NAP-Tag expression plasmid system and ISG cloning
The LV-NAP plasmid encoding the ISGs used to transduce 293T and SR-B2 was generated from the psg-9.8 (generous gift from Dr. Ikeda). First, additional cloning sites were added to the psg-9.8 plasmid. pCMV6-entry (Origene) and psg-9.8 were digested with BamHI and Notl at 37 °C. The 91 bp fragment of pCMV6-entry was inserted into the psg-9.8 vector and transformed into DH5-alpha cells (Invitrogen) and grown in LB-media supplemented with ampicillin. The fragment insertion was confirmed through sequencing.
The generation and binding domain of the monoclonal antibody (MAb) 23C8 against the neutrophil activating protein (NAP) of helicobator pylori has previously been described [21, 22]. Complementary nucleotide sequences encoding the 23C8 epitope (amino acids 97-119) were synthesized with additional BamHI and AsiSI (SgfI) restriction sites added to the 5’ and 3’ ends, respectively. The complementary strands were annealed in 1× annealing buffer (Agilent) at 55 °C for 1 h. The annealed strands were run on a 1% agarose gel and isolated using the QiaexII DNA isolation kit (Qiagen). Isolated NAP-tag DNA was treated with BamHI and AsiSI (SgfI) at 37 °C for 1 h. LV plasmid (psg9.8) was also treated with BamHI and AsiSI (SgfI) and ligated to the BamHI treated NAP-tag DNA at 14 °C overnight. The ligation reaction was then transformed in DH5-alpha cells (Invitrogen) and grown in LB media supplemented with ampicillin. The LV-NAP plasmid was confirmed through sequencing.
The selected ISGs were cloned from GBM39 cells. RNA was extracted from GBM39 cells using the RNEasy kit (Qiagen) according to manufacturer’s protocol. cDNA was generated with RT Superscript III (Invitrogen) using gene specific primers encoding AsiSI (SgfI) and MluI restriction sites at the 5’ and 3’ ends, respectively, which were used for subsequent PCR using the TA-cloning pcr2.1 kit. Plasmid DNA was transformed in DH5-alpha cells and colonies screened using a Mini-prep Kit (Qiagen). Plasmids with the inserted gene were then expanded and isolated using a Midi-prep kit (Qiagen). LV-NAP and TA-ISG were digested with AsiSI (SgfI) and MluI and run on 1% agarose gels. The corresponding bands were isolated using the Qiaexll DNA isolation kit (Qiagen) and ligated using the DNA ligation Kit (Roche) at 14°C for 1 h. Ligated DNA was transformed in DH5-alpha cells and gene sequences confirmed by sequencing.
2.4. Immunofluorescence
5 × 104 SR-B2 cells were plated in 8-well chamber slides (Thermo) and transduced with LV particles encoding the transgene of interest. The stably transduced cells (>20 days after transduction) were fixed in ice-cold methanol for 20 minutes and stored at −20°C. Cells were rinsed in PBS and blocked in 10% normal goat serum (Sigma). Cells were incubated with the MAb 27H10 [21] culture supernatant (diluted 1:5 in PBS, 2% BSA) overnight at 2-8C. The slides were rinsed three times in PBS. Cells were then incubated with the antibodies diluted in PBS, 2%BSA anti-Calnexin-Alexa Fluor 488 (Thermo MA3-027-A488) and goat-anti-Mouse IgG secondary Antibody Alexa Fluor 594 for 1 hr at room temperature. Cells were then rinsed 5 times and mounting media with DAPI (DuoLink) applied to cells prior to imaging.
2.5. Immunoblotting
Protein was isolated from samples using cell lysis buffer (Cell Signaling Technology). Protein was quantified using the BCA protein quantification assay (Pierce). Samples were run on 15% Tris-HCl Criterion gels and transferred to PVDF membranes. Membranes were blocked in 10% skim Milk/PBS for 1 hr, followed by incubation with the primary antibodies, NAP-specific MAb 23C8,8A11 (MAb recognizing MV-N generated by our laboratory, unpublished) and Tubulin (DM1A, Sigma). Membranes were washed with PBS supplemented with 0.05% Tween20 and incubated with the secondary goat-anti-mouse IgG antibody (Santa Cruz). Membranes were visualized using chemiluminescent substrate (Pierce).
2.6. Animal studies
All animal experiments were approved by the Mayo Clinic Institutional Animal Care and Use Committee. 5-week-old female SCID mice were implanted intraperitoneally with 5 × 106 SR-B2 cells. Groups of 8-9 mice were treated with 1 × 106 MV-GFP (TCID50) or inactivated virus beginning on day 10. Treatment was delivered once per week for 3 weeks. Survival was compared using the log-rank test.
2.7. Statistical analysis
Graphpad Prism (7.0) was used for statistical analysis. Student t tests were used to compare between two groups. Kaplan-Meier survival curves and log-rank tests were utilized to compare animal survival studies. P-values less than 0.05 were considered statistically significant. All tests were two-sided.
3. Results
3.1. RSAD2 inhibits infection at the step of infectious virion release
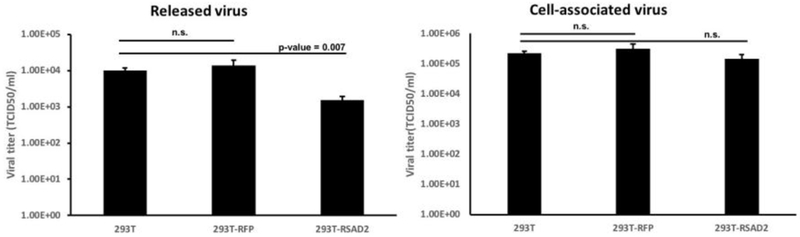
In a previous study, our laboratory identified several genes differentially expressed between Measles virus resistant and permissive cells [17]. To identify the specific ISGs that can determine permissiveness, we expressed several of these genes, including MX1, IFI44, IFI27, RSAD2 and GBP1 in 293T cells. To confirm protein expression, we generated a Tag protein that was placed on the amino-terminal of each gene. The N-terminal Tag is a 22 amino acid peptide generated from the neutrophil activating protein (NAP) of Helicobactor pylori [21,22]. The ISGs were cloned from a human primary patient-derived glioblastoma line, GBM39, and inserted into a lentivirus vector with the NAP tag (Supplemental Figure 1A). LV particles were generated and used to generate stable 293T cells (Supplemental Figure 1B). A polyclonal population of 293T cells expressing the transgenes of interest were screened for resistance to MV. Transduced 293T cells were infected with MV (MOI 0.1) and virus production measured by titration on Vero cells. Infectious virus particle release was significantly decreased in 293T cells expressing RSAD2/VIPERIN (p-value= 0.007); however, the cell associated virus remained unchanged (Figure 1).
Fig. 1. RSAD2 restricts MV release in 293T cells.
293T cells expressing RFP or RSAD2 were infected with MV-GFP (MOI=0.1). Released and cell-associated virus titers at 72-h were collected and titrated on Vero cells (n =3).
3.2. RSAD2 impairs the oncolytic MV therapeutic effect
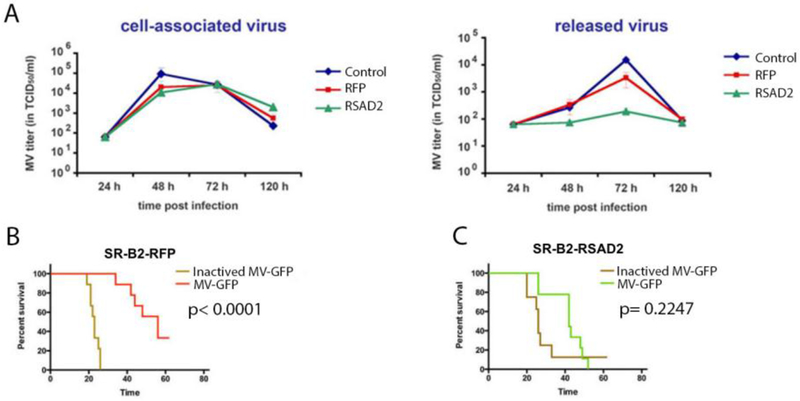
To verify these results in a tumor model, the ovarian cancer cell line SR-B2 was transduced with LV-NAP-RSAD2 or RFP control. A polyclonal population of transduced SR-B2 cells was used to determine the inhibitory effects of RSAD2. As observed in 293T cells, MV release was significantly decreased at 48 and 72 hr post-infection up to 15-fold (p-value < 0.05, experiment run in triplicate and repeated twice) (Figure 2A). A decrease in cell-associated virus was measured at 48 hr for both the RFP and RSAD2 transduced cells relative to untransduced (control) cells. This could be due to changes associated with the transduction and puromycin selection process. Importantly, however, no difference was observed in cell associated virus production between RFP and RSAD2 transduced cells.
Fig. 2.
RSAD2 restricts MV infection in OvCa SR-B2 cells in vitro and in vivo. Non-transduced SR-B2 cells (control) and SR-B2 cells transduced with RSAD2 or RFP were infected with MV-GFP (MOI 0.1). Cell-associated virus and released virus in the supernatant was collected at 24-120 h post-infection and titrated on Vero cells. Virus release in RSAD2 expressing cells was decreased 15-fold, relative to control cells (A). Mice were implanted with SR-B2 cells expressing RFP (control cells) (B) or RSAD2 expressing cells (C) and treated with 1 ×106 TCID50 MV-GFP once per week for 3 weeks and followed for survival.
To examine the impact on oncolytic MV therapy, transduced cells were implanted intraperitoneally (i.p.) into SCID mice. MV therapy significantly improved median survival of mice implanted with SR-B2 cells transduced to express RFP (p-value <0.0001); whereas median survival in mice implanted with SR-B2 cells transduced to express RSAD2 was not significantly improved (p-value = 0.2247) (Figure 2B, C).
3.3. RSAD2 Mechanism of Restriction
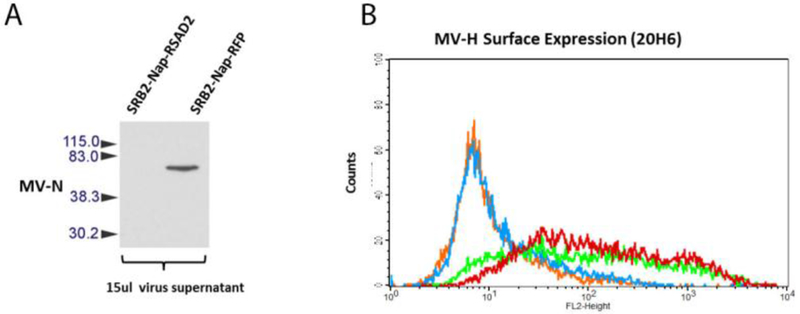
To characterize the mechanism of RSAD2 mediated restriction, we first assessed viral nucleocapsid in the supernatant of MV infected cells. As determined by western blot, the amount of virus released from cells over-expressing RSAD2 is dramatically decreased (Figure 3A). The results suggest that RSAD2 over-expression results in a reduction in the number of viral particles, as demonstrated by N protein production in the supernatant of infected cells. Previous studies have suggested that RSAD2 can inhibit glycoprotein transport to the cell surface [23]. However, 293T cells expressing RSAD2 did not impact MV hemagglutinin transport to the surface (Figure 3B).
Fig. 3.
RSAD2 does not restrict MV glycoprotein transport. SR-B2-RFP and SR-B2-RSAD2 cells were infected with MV-GFP (MOI 0.1). Release MV particles were assessed by immunoblot (A). Flow cytometry analysis of MV-H expression on the surface of 293T cells expressing MX1 (in green), RSAD2 (in red) and control non-infected 293 cells expressing MX1 (in orange) or RSAD2 (in blue) (B).
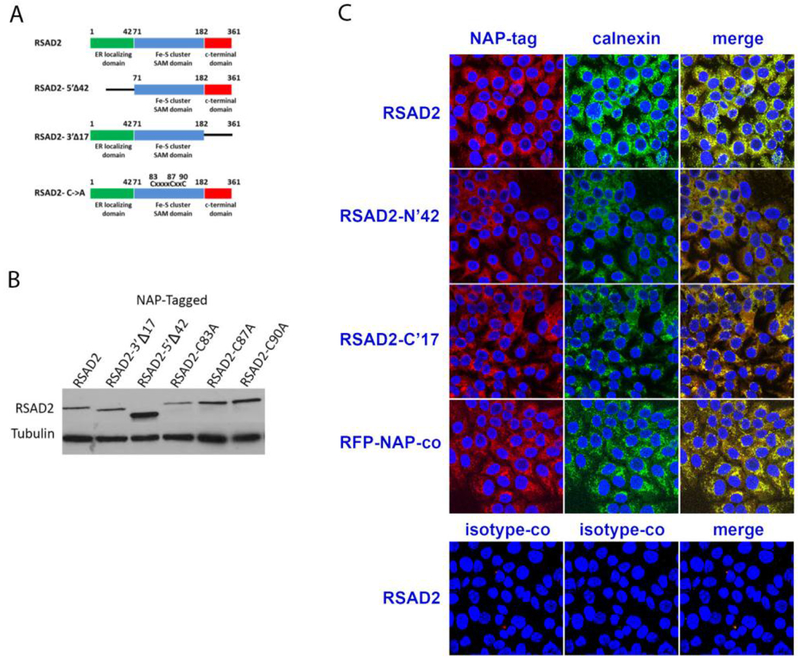
We next sought to identify the RSAD2 domains required for restricting MV replication. RSAD2 contain an amino terminal ER localizing domain (residues 1-42), a central Fe-S cluster domain (residues 71-182) and an unknown, highly conserved C-terminal domain (182-361) [24]. The domain of the N’ and C’ terminals were deleted and the Fe-S cluster domain ablated through cysteine to alanine mutations (Figure 4A). LV particles with the RSAD2 mutant constructs were generated and used to transduce SR-B2 cells (Figure 4B). Wild-type RSAD2 localized with the ER associated protein, Calnexin, as determined by immunofluorescence. Ablation of the ER localizing domain (RSAD-N’42) resulted in the mutant protein being localized to the cytoplasm and no longer co-localizing with Calnexin (Figure 4C).
Fig. 4.
Generation of RSAD2 mutants. RSAD2 mutants were generated by eliminating the N- or C-terminal domain were generated (A). Additional mutants that ablate the Fe-S cluster domain with cysteine to alanine mutations at positions 83, 87 and 90 were also generated. Protein expression of the RSAD2 mutants was confirmed by western blot (B). RSAD2 mutant proteins were analyzed for co-localization with the ER associated protein, Calnexin, by immunofluorescence (C). The SR-B2 cells were transduced with wild type (RSAD2) or truncated N and C termiunus (RSAD-N’42 or RSAD2-C’17 respectively. Cells transduced with RFP-NAP-tagged protein (RFP-NAP-co) were used as control. Isotype control antibodies were incubated with RSAD2-transduced cells (bottom row).
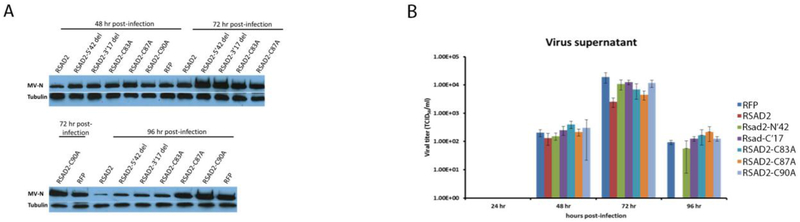
SR-B2 cells expressing the RSAD2 mutants were infected with MV (MOI 0.1) and virus production measured at 48, 72 and 96 hr post-infection. Ablation of each domain results in increased protein production, relative to cells transduced with the full-length wild-type RSAD2 construct (Figure 5A). Additionally, ablation of each domain rescued virus production, suggesting that each individual RSAD2 domain is required to mediate restriction (Figure 5B). However, mutation of Cysteine 87 to alanine still significantly restricts virus release at 72 h post-infection, similar to cells expressing wt-RSAD2 (p-value = 0.03), suggesting that this residue does not play a role in restricting MV. Cysteine 87 is an important residue of the SAM domain and plays an important role for restriction of several viruses, indicating that restriction of MV is occurring through an unknown mechanism [24].
Fig 5.
Identification of RSAD2 domains involved in restricting MV infection. SR-B2 cells expressing the RSAD2 mutants were infected with MV (MOI 0.1) and protein production measured by western blot at 48, 72 and 96 h post-infection (C). SR-B2 cells transduced with the RSAD2 mutants were infected with MV (MOI 0.1) and virus supernatant was measured by titration on Vero cells (D).
4. Discussion
The antiviral IFN defense system is the first line of defense against potential pathogens. Upon virus infection, host sensors detect viral products and induce expression of several interferon stimulated genes (ISGs) to inhibit virus replication [11]. RSAD2 is an ISG that is highly upregulated upon virus infection and type I/II IFN signaling through the JAK/STAT pathway [25]. RSAD2 has previously been demonstrated to have broad antiviral activity against several enveloped viruses, such as influenza virus, respiratory syncytial virus, hepatitis C virus, West-Nile virus, Chikungunya virus, human cytomegalovirus and HIV-1 [26–31]. Similarly, MV has been demonstrated to induce RSAD2 upon infection [17]. In addition, RSAD2 is expressed at baseline levels in tumor cells identified to have a MV infection resistant phenotype. In this study, we demonstrate that RSAD2 has antiviral activity against MV and expression can inhibit the release of MV in infected cells. These results are similar to previous observations demonstrating the restrictive properties of RSAD2 on several enveloped viruses [24]. Additionally, we demonstrate that in an oncolytic MV setting, RSAD2 expression abolishes therapeutic efficacy in vivo.
The specific mechanism of infection restriction varies among the different viruses targeted by RSAD2; however, the common inhibition effect is at the virion release step [25]. For example, during influenza infection, RSAD2 interacts with farnesyl diphosphate synthase, leading to the disruption of cholesterol biosynthesis [29]. The disruption in cholesterol synthesis was able to disrupt the formation of lipid rafts and transport of virus glycoproteins, resulting in the impaired release of influenza [29, 32]. For restriction of Dengue virus, RSAD2 co-localizes with viral protein at lipid droplets to impair virus infection [33]. Despite the extensive studies examining the antiviral properties of RSAD2 against a wide range of viruses, a unifying mechanism of restriction has not clearly been defined. Furthermore, the domains of RSAD2 required for restriction can vary across different viruses but remain undefined for several viruses [24]. Our results suggest that each domain of RSAD2 is critical to mediate restriction of MV. We recognize that we are not able to control for the level of protein expression between the different mutants (Figure 4B). However, transduction with the RSAD2 mutants results in similar or increased protein production relative to the full-length construct. Therefore, the lack of restriction mediated by the mutants is unlikely due to insufficient transgene production.
While the mechanism in which RSAD2 is able to restrict MV requires additional analysis, we are in the process of design and characterization of a recombinant MV to counter RSAD2 restriction. Several viruses are able to evade restriction of RSAD2, such as human cytomegalovirus (hCMV) [34, 35]. hCMV encodes a protein (mitochondrial inhibitor of apoptosis (MIA)) that is able to disrupt localization of RSAD2 in order to alter cellular metabolism that favors hCMV infection [35]. Recently, RSAD2 has been demonstrated to enzymatically modify ribonucleotides, resulting in replication-chain termination [36]. This does not appear to be the mechanism of restriction for MV, as demonstrated by similar levels of cell-associated virus and MV-N protein production (Supplemental Figure 1C). Additionally, while this mechanism appears to mediate restriction for a wide-range of viruses, it is not a unifying mechanism for all viruses sensitive to RSAD2 [36, 37]. Generation of a recombinant MV encoding the MIA could be a strategy to circumvent resistance in the context of oncolytic virotherapy. However, in order to fully circumvent ISG mediated restriction of MV, additional studies are required to characterize all MV restricting ISGs.
Overall, we identified RSAD2 as a novel restriction factor for MV propagation. These results are important in expanding our understanding of the MV life cycle, and possibly other Morbilliviruses. These findings could have a major impact in rational vaccine design and generation of recombinant MV platforms for oncolytic virotherapy. Recent studies have demonstrated the negative impact of the innate immune system and type I IFN triggered response on oncolytic therapy [17]. Identification of the specific ISGs that mediate restriction can provide important information to rationally design MV vectors to circumvent this restriction. As previously demonstrated, inhibition of the IFN response with a JAK1 inhibitor is able to increase virus replication >1000-fold in MV resistant GBM cells; however, systemic inhibition of JAK1 could impair the anti-tumor immune response activated upon MV mediated tumor cell lysis [17]. Therefore, identification of MV restricting ISGs is important in order to specifically target and circumvent single restricting anti-viral proteins, such as RSAD2.
Supplementary Material
Highlights.
RSAD2/VIPERIN inhibits the release of Measles virus
Expression of RSAD2 impairs the oncolytic Measles virus therapeutic effect
This study provides important information regarding the interaction between Measles virus and the interferon pathway
Acknowledgments
We wish to thank Duane Deal from Mayo Clinic, Rochester MN for the excellent assistance with confocal microscopy imaging. This work was supported by the National Institutes of Health (NIH) grants P50 CA108961, P50 CA136393, and R01 CA200507
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interests
The authors have no conflicts of interests to declare
Contributor Information
Cheyne Kurokawa, Email: kurokawa.cheyne@mayo.edu.
Ianko D. Iankov, Email: Iankov.Ianko@mayo.edu.
Evanthia Galanis, Email: Galanis.Evanthia@mayo.edu.
References
- 1.Cathomen T, et al. , A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. Embo Journal, 1998. 17(14): p. 3899–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahara M, Takeda M, and Yanagi Y, Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. Journal of virology, 2007. 81(13): p. 6827–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muhlebach MD, et al. , Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature, 2011. 480(7378): p. 530–U153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorig RE, et al. , The Human Cd46 Molecule Is a Receptor for Measles-Virus (Edmonston Strain). Cell, 1993. 75(2): p. 295–305. [DOI] [PubMed] [Google Scholar]
- 5.Tatsuo H, et al. , SLAM (CDw150) is a cellular receptor for measles virus. Nature, 2000. 406(6798): p. 893–897. [DOI] [PubMed] [Google Scholar]
- 6.Noyce RS, et al. , Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus. PLoS pathogens, 2011. 7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naniche D, et al. , Human Membrane Cofactor Protein (Cd46) Acts as a Cellular Receptor for Measles-Virus. Journal of Virology, 1993. 67(10): p. 6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navaratnarajah CK, et al. , The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nature structural & molecular biology, 2011. 18(2): p. 128–U183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manie SN, et al. , Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J Virol, 2000. 74(1): p. 305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikegame S, et al. , Both RIG-I and MDA5 RNA Helicases Contribute to the Induction of Alpha/Beta Interferon in Measles Virus-Infected Human Cells. Journal of Virology, 2010. 84(1): p. 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoggins JW and Rice CM, Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol, 2011. 1(6): p. 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naniche D, et al. , Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of Alpha/Beta interferon production. J Virol, 2000. 74(16): p. 7478–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoggins JW, et al. , Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature, 2014. 505(7485): p. 691–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider-Schaulies S, et al. , Cell type-specific MxA-mediated inhibition of measles virus transcription in human brain cells. J Virol, 1994. 68(11): p. 6910–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnorr JJ, et al. , MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J Virol, 1993. 67(8): p. 4760–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson S and Galanis E, Potential and clinical translation of oncolytic measles viruses. Expert Opin Biol Ther, 2017. 17(3): p. 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurokawa C, et al. , Constitutive Interferon Pathway Activation in Tumors as an Efficacy Determinant Following Oncolytic Virotherapy. J Natl Cancer Inst, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.lankov ID, et al. , Infected cell carriers: a new strategy for systemic delivery of oncolytic measles viruses in cancer virotherapy. Mol Ther, 2007. 15(1): p. 114–22. [DOI] [PubMed] [Google Scholar]
- 19.Giannini C, et al. , Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol, 2005. 7(2): p. 164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besnier C, Takeuchi Y, and Towers G, Restriction of lentivirus in monkeys. Proceedings of the National Academy of Sciences of the United States of America, 2002. 99(18): p. 11920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iankov ID, et al. , Development of monoclonal antibody-based immunoassays for detection of Helicobacter pylori neutrophil-activating protein. J Immunol Methods, 2012. 384(1-2): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iankov ID, Haralambieva IH, and Galanis E, Immunogenicity of attenuated measles virus engineered to express Helicobacter pylori neutrophil-activating protein. Vaccine, 2011. 29(8): p. 1710–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo JY, Yaneva R, and Cresswell P, Viperin: A Multifunctional, Interferon-Inducible Protein that Regulates Virus Replication. Cell host & microbe, 2011. 10(6): p. 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helbig KJ and Beard MR, The role of viperin in the innate antiviral response. J Mol Biol, 2014. 426(6): p. 1210–9. [DOI] [PubMed] [Google Scholar]
- 25.Seo JY, Yaneva R, and Cresswell P, Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe, 2011. 10(6): p. 534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin KC and Cresswell P, Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus (vol 98, pg 15125, 2001). Proceedings of the National Academy of Sciences of the United States of America, 2002. 99(4): p. 2461–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang D, et al. , Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. Journal of virology, 2008. 82(4): p. 1665–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang D, et al. , Identification of Five Interferon-Induced Cellular Proteins That Inhibit West Nile Virus and Dengue Virus Infections. Journal of virology, 2010. 84(16): p. 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang XY, Hinson ER, and Cresswell P, The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell host & microbe, 2007. 2(2): p. 96–105. [DOI] [PubMed] [Google Scholar]
- 30.McGillivary G, et al. , Replication of Respiratory Syncytial Virus Is Inhibited by the Host Defense Molecule Viperin. Journal of innate immunity, 2013. 5(1): p. 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasr N, et al. , HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood, 2012. 120(4): p. 778–788. [DOI] [PubMed] [Google Scholar]
- 32.Seo JY and Cresswell P, Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog, 2013. 9(8): p. e1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helbig KJ, et al. , Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl Trop Dis, 2013. 7(4): p. e2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin KC and Cresswell P, Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A, 2001. 98(26): p. 15125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo JY, et al. , Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science, 2011. 332(6033): p. 1093–7. [DOI] [PubMed] [Google Scholar]
- 36.Gizzi AS, et al. , A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature, 2018. 558(7711): p. 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proud D, et al. , Gene expression profiles during in vivo human rhinovirus infection: insights into the host response. Am J Respir Crit Care Med, 2008. 178(9): p. 962–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.