Abstract
Purpose of Review-
Precise and temporal expression of Runx2 and its regulatory transcriptional network is a key determinant for the intricate cellular and developmental processes in adult bone tissue formation. This review analyzes how microRNA functions to regulate this network, and how dysregulation results in bone disorders.
Recent Findings-
Similar to other biologic processes, microRNA (miRNA/miR) regulation is undeniably indispensable to bone synthesis and maintenance. There exists a miRNA–RUNX2 network where RUNX2 regulates the transcription of miRs, or is post transcriptionally regulated by a class of miRs, forming a variety of miR-RUNX2 regulatory pathways which regulate osteogenesis.
Summary-
The current review provides insights to understand transcriptional–post transcriptional regulatory network governed by Runx2 and osteogenic miRs, and is based largely from in vitro and in vivo studies. When taken together, this article discusses a new regulatory layer of bone tissue specific gene expression by RUNX2 influenced via miRNA.
Keywords: MicroRNA, Runx2, Osteoblast Differentiation, Posttranscriptional Repression, Noncoding RNA, Osteoblastogenesis
Introduction
Osteoblasts, a unique mesenchymal cell type, develop mineralized tissue bone. Rigorous gene expression programs govern the fundamental biological process of osteogenesis, which includes underlying cell fate decisions, commitment to osteogenic lineage, and differentiation and maintenance of osteogenic phenotype. Transcription of bone specific genes represents a major control point and occurs within the context of stage specific growth and differentiation of osteoblast progenitors. Precise and temporal expression of bone regulatory genes appears to be accountable for the complicated cellular processes of bone synthesis and adult bone tissue homeostasis.
The transcription factor RUNX2 encodes a nuclear transcription factor involved in osteoblastic differentiation and skeletal morphogenesis. Promoter binding transcription factors including RUNX2 belongs to an enormous class of DNA-binding proteins that play essential roles in controlling mammalian genes [1]. Runx2 binds to a bulk of osteoblast specific promoters and supports osteogenesis through transcriptional activation[2]. Inactivation of Runx2 inhibits maturation of osteoblasts and chondrocytes for both intramembranous and endochondral ossification. Haplo-insufficiency results in skeletal dysplasia characterized by hypoplastic clavicles and open fontanelles. Importantly, Runx2 post-transcriptional mRNA stability as well as RUNX2 transcriptional activity is critical for its osteogenic induction promoting bone synthesis and maintenance.
MicroRNAs (miRNAs) are 20–24 nucleotide regulatory non-coding RNAs, implicated in diverse pathophysiologic processes. Non-coding miRNAs are among the 98% of the human genome that is not transcribed into protein. A vast majority of this class of RNA molecules is significantly implicated in the regulation of gene expression [3]. Analogous to other biologic processes, microRNA regulation has been proven to regulate gene expression during bone synthesis and maintenance [4–22]. Identifying how the miRNA class of non-coding RNA regulates gene expression in bone tissue is necessary to further understand the epigenetic basis for bone development, synthesis, and maintenance. MiRNA biogenesis begins in the nucleus with RNA polymerase II transcription of miRNA genes[4–8]. The resulting transcript, primary microRNA (pri-miRNA), forms a double stranded RNA hairpin structure. Drosha/DGCR8 complex, an RNA double stranded endonuclease complex cleaves the pri-miRNA forming the double stranded precursor microRNA (pre-miRNA)[9]. Subsequently the pre-miRNA is exported to cytoplasm by RanGTP dependent Exportin 5 nuclear export cargo[10]. In the cytoplasm Dicer, a double stranded RNA endonuclease, further processes the pre-miRNA[11]. The resulting product is a short 20–24 base pair double stranded mature miRNA, which is then associated with Argonaute proteins (AGO 1, AGO2, AGO3 and AGO4)and loaded into the functional RNA Induced Silencing Complex (RISC)[12]. Prior to active RISC targeting, one strand of the mature miRNA is selected transitioning from the pre-RISC to the mature RISC complex[13]. Using the miRNA as a guide, the RISC complex binds to complementary regions on target mRNAs (3’UTR) resulting in translational repression through mechanisms including mRNA cleavage, decreased ribosomal pre and post initiation, or de-adenylation and degradation of the mRNA[13]. The cellular integration of Runx2 and miRNAs occurs at several levels: a) The regulation of miRNAs by cellular signaling that directly or indirectly control Runx2, b) regulation of miRNA expression that are transcriptionally controlled by Runx2 during osteogenic induction) direct miRNA control of Runx2 expression and translation during osteoblast differentiation. In this review, we cover our current understanding of how cellular signaling safeguards Runx2, how Runx2 promoting osteo miRs and finally how miRNA attenuates Runx2 function and stability to control the biology of osteogenesis.
Cellular signaling controls miRNAs that regulate Runx2 and osteogenesis
Bone cell differentiation is a precisely controlled process. The differentiation program of osteoblast cells is regulated by a series of osteogenic developmental signals, which commit mesenchymal stem cells (MSCs) to osteoblast cell lineage, induce growth and differentiation through stage specific induction, and ultimately attenuate Runx2 gene expression. MicroRNAs coordinate a broad spectrum of biological processes including formation and remodeling of the skeleton. Reciprocal control between Runx2 and miRNAs has emerged as an important developmental switch, controlled by osteogenic signaling pathways, for the promotion of osteoblast growth and differentiation.
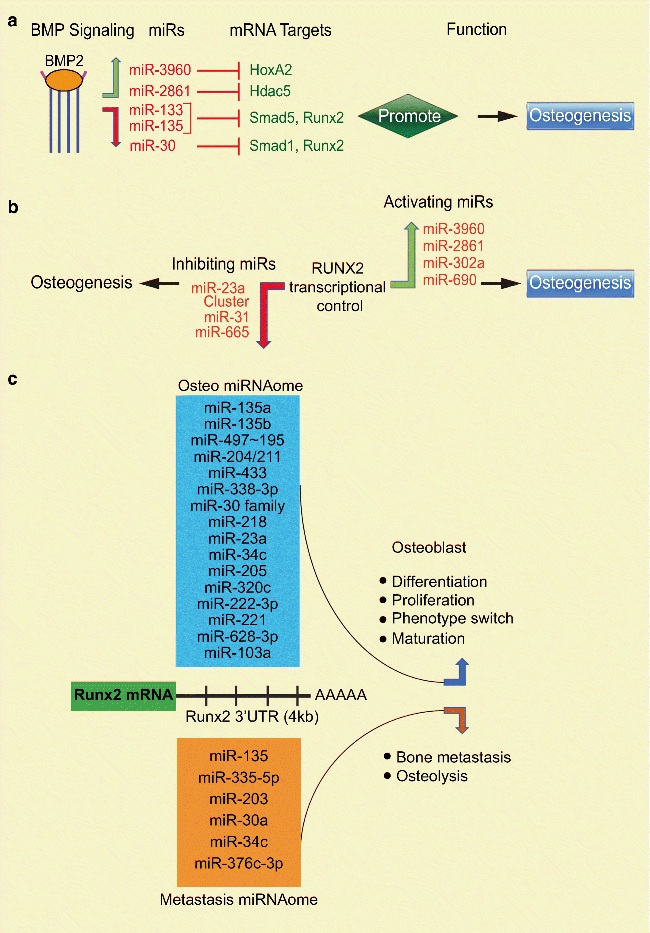
Evidence from numerous studies supports the concept that BMP signaling controls a class of miRNAs that epigenetically/post transcriptionally regulate osteoblastic differentiation[14–16]. Stromal cells (ST2) induced by BMP2 upregulate miR-3960. MiR-3960 directly represses homeobox protein A2 (Hoxa2), a transcriptional repressor of Runx2[14]. Biologically, this signaling-miR axis increases Runx2 transcripts to increase Runx2 function, ultimately enhancing osteogenesis. Analogously, BMP2 induces miR-2861 which targets histone deacetylase 5 (HDAC5) whose activity induces RUNX2 transcriptional repression, ultimately increasingRunx2 transcriptional activity to induce osteoblast differentiation (Fig. 1A)[14, 15]. Over the years it has been shown that bone formation is orchestrated by the synchronized activity of Runx2 and BMP-activated Smads. BMP2 switches premyogenic phenotype to osteogenic phenotype by down regulating miR-133 and miR-135 that suppress Runx2 and Smad5 respectively. The blockage of miR-133 and miR-135 expression and support of Runx2 function by BMP2 signaling exemplifies an epigenetic mechanism supporting osteoblast commitment and progression of differentiation (Fig. 1A)[16].
Figure 1. MiRNA Regulation of Runx2 in Skeletal Cells.
(A) Cellular signaling-miRNAs-Runx2 axis controls osteogenesis. BMP2 signaling increased the expression of miR-3960, miR-2861, which target HoxA2 and Hdac5 (inhibitors of Runx2), however, decreased the expression miR-133, miR-135 and miR-30, which directly target Runx2, Smad1 and Smad5 hence, in multiple way protectedRunx2 function that promote matrix formation and mineralization during osteoblast differentiation. (B, left) Negative regulation by Runx2 of inhibiting miRNAs, miR-23a cluster, miR-665 and miR-31 to relieve the inhibition of Runx2, Satb2, Dlx3 and Kat6a to promote osteoblast, odontoblast and dental follicular cell differentiation. (B, right) Positive regulation by Runx2 of activating miRNAs, miR-3960 and miR-2861which target inhibitors of Runx2, Hdac5, HoxA2; miR-302a targets COUP-TFII, and miR-690 targets p65 generates a feed forward circuit to induce osteogenesis. (C) miRNA-mediated inhibition of Runx2 expression. The illustration of 4 kb Runx2 3′ untranslated region (black solid line). In upper panel, a subset of Runx2-targeting miRNAs (Osteo miRNAome) express in osteoblasts, odontoblasts, bone marrow stromal cells and dental follicular cells at stages of growth and differentiation (blue box) control Runx2 mRNA stability and regulate osteogenesis and alter mesenchymal phenotypes. In lower panel, another subset of miRNAs (Metastasis miRNAome) express in breast, prostate, bone and head and neck cancer cells control Runx2 post transcriptionally to accelerate bone metastasis and osteolysis.
In BMP2 mediated bone marrow stem cell (BMSC) commitment, BMP2 enhances osteogenesis by inhibiting the miR-30 family (miRs-30 a, b, c, and d)and preserving Runx2 and Smad1function (Fig. 1A)[17]. When taken together, these findings indicate that BMP2 functionally blocks miRs that target Runx2 or activates miRs that target Runx2 repressors to ultimately support osteoblast differentiation.
RUNX2 transcriptionally regulates miRNAs for osteogenesis
RUNX2, an essential transcription factor for bone development, directly binds to its classical cis regulatory element TGTGGT (ACCACA) within proximal promoter region and controls transcription of numerous miRNAs contributing to osteogenesis. Runx2-negative regulation of miR-23a cluster (miR-23a, 27a, and 24–2) or miR-31 causes derepression of Special AT-Rich Sequence-Binding Protein 2 (SATB2) to promote differentiation of osteoblast and dental follicle cells (Fig. 1B, left) (DFCs)[18, 19]. During dentinogenesis, RUNX2 direct binding of the miR-337–miR-540–miR-665 cluster promoter is a requirement to repress miR-665 expression for the progression of odontoblast differentiation (Fig. 1B, left)[20]. Opposing to RUNX2 negative regulation (Fig. 1B, right), in BMP-2 stimulated preosteoblasts,RUNX2 directly binds to the miR-302a promoter and activates its transcription to promote osteoblast differentiation by targeting and repressing COUP-TFII, a transcription factor that represses osteoblast differentiation[21]. During stable Runx2 expression in C2C12 cells,Runx2 activated miR-690 expression by direct binding to its promoter. Furthermore, miR-690 acted as a positive regulator, repressed p65 subunit of NF-κB and promoted Runx2-induced osteogenic differentiation (Fig. 1B, right)[22]. Recent studies indicate that bone tissue-specific function of RUNX2 may operate at multiple distinct levels of gene regulatory networks and a group of miRNAs (osteo miRs) are regulated by RUNX2 in a cell- or tissue-specific fashion and may contribute to the establishment of osteoblast phenotype and differentiation[23].
MiRNA direct regulation of Runx2 mRNA
Whereas the transcriptional activity of Runx2 is requisite for both ex vivo and in vivo osteogenesis, the biological potency of miRNAs that target Runx2 is equally important to support the commitment and phenotypic maturation of osteoblasts. TargetScan bioinformatics predicted 165 miRs can potentially target Runx2. Among them 37 were highly conserved among vertebrates, 44 miRs were reasonably conserved and 84 miRs were poorly conserved (http://www.targetscan.org/cgi-bin/vert_72/view_genetable.cgi?rs=ENST00000371432.3&taxid=9606&members=&subset=1&showcnc=1&shownc=1&sortText=cs). To date, 28 miRs have been shown experimentally to inhibit Runx2posttranscriptionally through 3′ UTR binding (https://www.genecards.org/cgi-bin/carddisp.pl?gene=RUNX2)[24]. In this review, we focused on significant and promising miRNAs, which attenuate Runx2 function and stability to control the physiology of osteogenesis (Table.1).
Table 1:
miRNA regulation of Runx2 expression in osteogenesis:
| miRNA | Regulatory Function | Mode of Action | Target Gene/Pathway | References |
|---|---|---|---|---|
| MiR-10a | Promotes | Promotes ossification | ID3/Runx2 axis | [54] |
| MiR-15b | Promotes | Promotes osteoblast differentiation | Protects Runx2 protein from Smurf1 mediated degradation | [55, 56] |
| MiR-23a-27a-24–2 | Inhibits | Inhibits osteoblast differentiation and apoptosis | Runx2, Satb2, FAK | [19, 24] |
| Mir-29b | Promotes | Promotes osteogenic differentiation | Induces Runx2 expression | [57] |
| MiR-30c | Inhibits | Inhibits osteoblast differentiation | Runx2, Smad1 | [24] |
| MiR-31 | Inhibits | Inhibits osteoclast and osteoblast differentiation | Suppressed by Runx2, targets Satb2, Sp7, OSX | [58–61] |
| MiR-34c | Inhibits | Inhibits proliferation and differentiation | Runx2, SATB2, Notch Pathway | [24, 62, 63] |
| MiR-103a | Inhibits | Inhibits osteogenesis | Runx2 | [47] |
| MiR-133a | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| MiR-135a | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| MiR-137 | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| Mir-146 | Promotes | Promotes osteogenesis | Targets Smad3, enhances Runx2 | [64, 55] |
| MiR-203 | Inhibits | Inhibits heterotopic ossification | Runx2 | [65] |
| MiR-204/211 | Inhibits | Inhibits osteoblast differentiation | Runx2,Sost2 | [24] |
| MiR-205 | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| MiR-217 | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| MiR-218 | Inhibits | Inhibits osteoblast differentiation | Runx2 | [24] |
| MiR-221 | Inhibits | Inhibits osteoblast differentiation and bone formation | Runx2 | [44] |
| MiR-320 | Suppresses | Suppresses osteogenic differentiation | Runx2 | [66, 42] |
| MiR-322 | Promotes | Promotes osteogenesis in response to BMP-2 | Enhances expression of Runx2, Osx, | [67] |
| MiR-338 | Inhibits | Inhibits osteoblast differentiation | Runx2, Sfrp2, Dkk2 | [24] |
| MiR-370 | Inhibits | Inhibits osteoblast differentiation | Runx2 | [68] |
| MiR-455–3p | Promotes and suppresses | Promotes early chondrogenic differentiation and suppress maturation | Runx2 | [66, 69] |
| MiR-467g | Negatively regulates | Negatively regulates osteogenesis | Ihh/Runx2 signalling | [70] |
| MiR-628–3p | Inhibits | Inhibits osteogenesis | Runx2 | [46] |
| MiR-690 | Positively affects | Positive effect on osteogenic differentiation | Regulated by Runx2 | [22] |
| MiR-705 | Inhibits | Enhances adipogenic differentiation | Runx2 and HoxA10 | [71, 55] |
| MiR-2861 | Promotes | Promotes osteoblast differentiation by BMP-2 | Regulates Runx2 via HDAC5 | [72, 55] |
| MiR-3077–5p | Inhibits | Inhibits osteoblast differentiation | Runx2 and HoxA10 | [71] |
| MiR-3960 | Induces | Induces osteoblast differentiation | Suppresses HoxA2 which inhibits Runx2 | [14, 55] |
In MSCs, bone morphogenetic protein 9 (BMP9)-induces early osteogenesis, miR-155 expression level is increased at first and then decreased further during differentiation. Additionally, Liu et al showed that miR-155 is an inhibitor of osteogenic differentiation and this repression is mediated through direct repression of Runx2 mRNA. Thus, MSCs stemness is maintained through inhibition of Runx2 by miR-155 repression, inhibiting osteogenesis[25]. Conversely, in hMSCs, among a cohort of miRNAs, miR-155 (categorized as “Osteo miR”) was up regulated upon osteo-stimulation and gradually reduced during osteoblast differentiation. However, anti-miR-mediated knockdown of miR-155 did not show any significant changes in ALP staining. Therefore, the conflicting reports of miR-155 involvement in osteoblastogenesis warrant further study to identify its regulatory role.
In undifferentiated human mesenchymal stem cells miR-335 was identified as being up regulated by Wnt signaling and significantly down regulated later to allow distinct differentiation programs. Over expression of miR-335 inhibited hMSCs proliferation and migration, as well as their osteogenic potential. Western blot and 3’UTR reporter assays confirmed RUNX2 as a direct target of miR-335 in hMSCs. These results suggest that miR-335 down regulation is a requirement to increase Runx2 for the acquisition of the osteogenic phenotype[26]. Surprisingly, in a miR-335–5p transgenic mouse model over expressing miR-335–5p in the osteoblasts lineage, Zhang et al demonstrated higher bone mass with enhanced expression of osteogenic differentiation markers including Runx2 (already reported as a direct target). Mechanistically they showed Wnt signaling antagonist Dickkopf-1 (DKK1) was down regulated due to miR-335–5p up regulation. Taken together, this group revealed that higher expression of miR-335–5p in osteoblasts in vitro or constitutive over expression of miR-335–5p in vivo in osteoblast lineage induces osteogenic differentiation and bone formation in mice[27, 28]. These findings on miR-335from two different laboratories demonstrated a discrepancy in the miR-335 mediated regulation of Runx2-Wnt signaling function in osteogenesis. Further research and tissue specific knockdown of miR-335 are necessary to elucidate its species-specific role in osteoblast maturation.
MicroRNA-497~195 cluster, a member of the miR-15 family has been identified by Grünhagen, et al in 2014 as a regulator of osteoblast differentiation that impairs the induction of bone morphogenetic protein (BMP) responsive genes[29]. In human primary mesenchymal stromal/stem cells (MSC), miR-195 and miR-497 decrease osteogenesis. Furthermore, ALP and RUNX2 expression levels were significantly decreased in human MSC following miR-195 and miR- 497 over expression[30].
Wang et al demonstrated that miR-204 directly targets Runx2, attenuates BMP-2-induced calcification and inhibits expression of osteoblast-related genes of human aortic valve interstitial cells (VICs). Knockdown of miR-204 reinforced an osteoblast phenotype in VICs with increased alkaline phosphatase activity and osteocalcin expression. Based on these findings the authors inferred that miR-204 is a possible molecular switch to inhibit trans differentiation of human aortic VICs to osteoblast lineage[31]. In a very similar mechanism, miR-204 is repressed in vascular smooth muscle cells (VSMCs) during beta-glycerophosphate-induced calcification, whenRunx2 protein levels were elevated. Conversely, miR-204 mimics significantly decreased Runx2 protein levels and reduced osteoblastic differentiation of VSMCs. Therefore, down-regulation of miR-204 may contribute to beta-glycerophosphate-induced VSMC calcification through derepression Runx2[32]. Recently, it has been reported that in human aortic valve and primary VICs, long non-coding RNA (LncRNA) TUG1 knockdown inhibits osteoblast differentiation. TUG1 directly binds and degrades miR-204–5p. Hence, silencing of TUG1 increased miR-204–5p and subsequently inhibits Runx2 expression at the post-transcriptional level. Thus, TUG1 positively regulates the expression of Runx2, through sponging miR-204–5p, and promotes osteogenic differentiation during aortic valve calcification[33].
To study the regulation of miR-204, Zhang et al in 2012, used pluripotent C3H10T1/2 cells and overexpressed miR-204 and allowed these cells to differentiate with BMP2. MiR-204 blocks both osteogenic (e.g. Osterix and ALP) and/or chondrogenic markers (e.g. Col2A1 and Sox9) required for lineage differentiation[34]. During adipocyte differentiation of mesenchymal progenitor cells and bone marrow stromal cell (BMSC) miR-204 and its homolog miR-211 were induced, whereas Runx2 protein expression was suppressed. Forced expression of miR-204 decreased Runx2 protein while knockdown significantly elevated Runx2 protein. Additionally, this study confirmed the binding of miR-204/211 to Runx2 3’-UTR. These findings suggesting that miR-204/211 directly suppressed Runx2 to commit mesenchymal progenitor cells and BMSCs for adipogenesis[15].
At least three miR-433 binding sites have been identified on the 3’-UTR of Runx2 mRNA by computer-based prediction algorithm. ERRγ(estrogen receptor-related receptor gamma) and miR-433 expression are decreased during BMP2-induced osteogenesis of mesenchymal stem cell C3H10T1/2. Overexpression of ERRγor miR-433 inhibits osteogenic marker genes including Runx2. Inhibition of miR-433 retrieved ERRγ- mediated Runx2 suppression. Taken together these findings demonstrate that miR-433 suppressed BMP2-indcued Runx2 expression and osteoblast differentiation by decreasing the level of transcript[35].
Bioinformatics analysis of odontoblasts and bone marrow stromal cells (BMSCs) identified Runx2 as a potential target of miR-338–3p, which is increased during odontoblast differentiation. In vitro gain and loss of functional assays suggested that miR-338–3p is an attenuator of odontoblast and osteoblast differentiation through controlling Runx2 mRNA post transcriptionally[36]. Furthermore, Liu et al. showed that miR-338–3p serves as a potential modulator of osteoporosis via its effect on osteoblasts[37]. Zhang et al revealed that miR-338 expression is very cell-type and stage specific. Specifically, miR-338 is barely detectable in MC3T3-E1 osteoblasts however, several folds higher in ATDC5 chondrocytes. Notably, miR-338, which is hardly expressed in proliferating preosteoblasts, show significant increase during differentiation. Cell-type-specific and temporal expression of miR-338at various stages of differentiation with regulated Runx2 protein levels may establish a coordinated regulation during osteogenesis[24].
Studies on the miR-30 family in osteogenesis indicate that miR-30 family members (miR-30a, −30b, −30c, and −30d) mediate the inhibition of osteogenesis by targeting Runx2 and Smad[17]. For example, adipose tissue-derived stem cells can differentiate into either adipocytes or osteoblasts. During adipogenic differentiation of these cells, the expression of miR-30 family was up regulated, similarly inhibition of the miR-30 family blocked adipogenesis. Specifically, over expression of miR-30a and miR-30d among the miR-30 family members stimulated the process of adipogenesis. The inhibition of osteogenic factor RUNX2 represents a reasonable mechanism by which miR-30a and miR-30d may switch the phenotype to favor adipogenic differentiation of adipose tissue-derived stem cells[38]. Added studies revealed that miR-30a also functions as tumor suppressor in osteosarcoma by directly targeting Runx2. Inhibition of miR-30a in Saos2 cells increased proliferation, migration, and invasion; while rescue by Runx2 over expression significantly reversed the effects of either over expressing or inhibiting miR-30a. All these results suggest a critical mechanism of miR-30a in suppressing proliferation, migration, and invasion of osteosarcoma by targeting Runx2[39].
The calcification of vascular smooth muscle cells (SMCs) resembles an osteoblast-like phenotype with concomitant expression Runx2. A miR microarray and bioinformatics database independently identified miR-30b and miR-30c (miR-30b-c) as miRs that directly repress Runx2 expression in human coronary artery derived SMCs (CASMCs). BMP-2 decreases miR-30b and miR-30c expression hence increases Runx2 expression. Depletion of miR-30b-c significantly increases Runx2, intracellular calcium deposition, and mineralization. Taken together these findings indicate that the control of miRNA fate by cellular signaling and transcription factor network is important for phenotype switch required for mineralization[40].
Zhang et al. 2012 identified miR-218 among a panel of 11 Runx2-targeting miRNAs, expressed in a lineage-specific pattern in mesenchymal cell types. They found that miR-218 is highly expressed in osteoblasts and fibroblasts but very low in chondrocytes. All Runx2-targeting miRNAs (except miR-218) significantly inhibit ALP activity and osteoblast differentiation and their effects can be reversed by the corresponding anti-miRNAs. Interestingly, miR-218 reduce Runx2 expression only in chondrocyte cells[24].
Human periodontal ligament stem cells, dental pulp stem cells, gingival stem cells and human bone marrow stem cells were used to analyze miRNA expression profiles during differentiation. All cells were differentiated in osteogenic media and Runx2 expression was analyzed as a read out of osteogenesis. Analysis of 765 miRNAs demonstrated a decrease in the expression of hsa-miR-218 across all differentiated cell populations. Hsa-miR-218 directly targets and decreases RUNX2 expression in undifferentiated human dental stem cells (DSCs). Additionally, mineralized DSCs showed a decrease in hsa-miR-218 expression and increase Runx2 expression. These data reveal a central role of miR-218 for the progression and differentiation of human DSCs by modulating Runx2 expression[41].
MicroRNA-23a is constitutively expressed at high levels in both osteoblasts and chondrocytes. Zhang et al found that miR-23a is among the seven RUNX2-targeting miRNAs to directly regulate RUNX2 protein expression by targeting seed regions of the 3′ UTR of RUNX2 mRNA[24]. In a feed forward mechanism Runx2 negatively regulates the transcription of the miR-23a cluster through Runx2 binding in the cluster promoter. Moreover, in a feedback mechanism miR-23aattenuates osteoblast maturation by targeting Runx2 mRNA in the terminally differentiated osteoblasts. These findings established a vital role for the miR cluster 23a~27a~24–2 in the maintenance of the osteocyte phenotype[19].
Among numerous microRNAs that target RUNX2, miR-34c is highly expressed in osteoblast cells and controls osteoblastogenesis. Additionally, inhibition of miR-34c only increases Col2A1 levels but not alkaline phosphatase (ALP) gene expression. Opposing osteogenesis, miR-34c acts to selectively enhance expression of adipogenic markers (aP2 and PPARγ) to stimulate adipogenesis. Therefore, miR-34c can switch mesenchymal stem cells into adipogenic lineage fate by selectively blocking osteogenesis or chondrogenesis[34].
Zhang et al, in 2011 classified miR-205 as a Runx2 targeting microRNA based on SEED binding to the proximal 3’UTR region of Runx2 mRNA. Additionally, miR-205 was found to repress Runx2 expression in both osteoblast and chondrocyte cells[24].
During adipocyte differentiation of human mesenchymal stem cells (hMSCs), miR-320 family (miR-320a, 320b, 320c, 320d and 320e) were expressed significantly high. Among several biologically relevant gene targets for miR-320 family, RUNX2 was validated as a bona fide target of miR-320c. Therefore, the findings suggest that miR-320c-RUNX2 axis is a molecular switch to support the epigenetic basis for promoting adipocytes differentiation[42].
In a recent report miR-222–3p has been identified as an inhibitor of osteogenic differentiation of human mesenchymal stem cells (hBMSCs). This inhibition is mediated through direct repression of RUNX2 and Smad5. Upon over expression of miR-222–3p, hBMSCs differentiation was inhibited in-vitro. Conversely, miR-222–3p knock down promoted osteoblast specific gene expression, alkaline phosphatase activity, and mineralization. These findings suggest that miR-222–3p activity is linked toSmad5-RUNX2 signaling axis that regulates osteogenic differentiation[43].
In-vitro studies identified that miR-221 inhibits osteogenesis by directly binding the Runx2 3’UTR resulting in decreased expression. Over expression of RUNX2 significantly diminished the effect of miR-221 on osteoblast specific genes. These data suggest that miR-221 negatively regulates Runx2 expression and promotes osteoporosis in-vivo[44]. In 2016, Yeh et al studied the osteogenic potential of degenerated annulus fibrosus (DAF) cells. They identified that DAF cells possess greater osteogenic differentiation potential as compared to Normal annulus fibrosus (NAF). Interestingly, they identified that miR-221 expression is significantly higher in DAF as compared to NAF. Although this study did not identify the direct target of miR-221, it identified that forced miR-221 over expression reduces the differentiation potential of DAF cells through negatively regulating BMP2 signaling[45].
Chen et al. identified miR-628–3p from a microRNA screen from patients which had atrophic non-union fracture. They discovered that miR-628–3p was noticeably up regulated in these patients, whereas this microRNA is down regulated during osteoblast differentiation in-vitro. Furthermore, it was identified that Runx2 is a direct target of miR-628–3p, which suppressed Runx2 mRNA and protein levels through 3’UTR binding. These data indicate that miR-628–3p negatively regulates Runx2, and may contribute to osteopathologies such as atrophic non-union fracture[46].
In an investigation seeking to identify miRNA that are regulated by mechanical stimulation, Zuo et al. identified miR-103a to be down regulated during cyclic mechanical stretch (CMS)-induced osteoblast differentiation. Furthermore, miR-103a was shown to target Runx2 and negatively regulate its expression through binding its 3’UTR. In-vitro experimentation revealed miR-103a over expression to inhibit osteoblast differentiation in a CMS model; conversely miR-103a knockdown stimulated osteoblast differentiation. In-vivo, miR-103a plays an inhibitory role in bone formation during hind limb unloading in mice. Mice pretreated with a miR-103a antagonist were partly rescued from osteoporosis caused by mechanical unloading. These findings suggest that miR-103a is a mechanosensitive miRNA that targets Runx2 to inhibit osteoblast differentiation[47]. Additional studies indicate that miR-103a-3p regulates the growth and osteogenic differentiation of human derived stromal cells (hADSCs) by direct targeting of CDK6 and DICER1 partly[48].
In human osteosarcoma (OS)RUNX2 is often highly expressed. Depletion of RUNX2 inhibits growth of human OS cells. It has been reported that expression of RUNX2 is inversely linked to loss of tumor suppressor p53 in normal osteoblasts and OS cell lines. Similarly, RUNX2 protein levels decrease upon stabilization of p53. Interestingly, p53-dependent microRNA, miR-34c is significantly down regulated in OS that directly targets and represses RUNX2[49].
In normal prostate cells miR-203 repressed a cohort of pro-metastatic genes (ZEB2, Bmi and Survivin)[50], including master regulator of bone metastasis Runx2[51]. During prostate cancer bone metastasis miR-203 expression is significantly lower suggesting a fundamental anti metastatic role for this miRNA[50]. High expression of the transcription factor Runx2 is linked to breast cancer metastasis to bone. The expression profile of metastatic MDA-MB-231 revealed an undetected level of miR-203 where Runx2 is highly expressed. MDA-MB-231 cells with miR-203 over expressed results in decreased Runx2 and concomitant decreased expression of the metastasis-promoting Runx2 target genes [52].
Recently, in a cellular reconstitution assay using miR-135 Taipaleenmäki et al, showed that bone metastatic MDA-MB-231 cells had reduced abundance of Runx2 along with reduced expression of the metastasis-promoting Runx2 target genes. Additionally, orthotopic implantation of MDA-MB-231 cells delivered with miR-135, followed by an intratumoral administration of the synthetic miRNAs, reduced the tumor growth and spontaneous metastasis to bone[52]. In mesenchymal cells, miR-135 is expressed in a lineage-related pattern and during both osteogenic and chondrogenic differentiation, miR-135 strongly inhibits Runx2 protein expression[24].
Chang et al. identified that RUNX2 is commonly up regulated in the head and neck squamous cell carcinoma (HNSCC) based on micro-array analysis of patient samples. It was identified that miR-376c-3p bound to the 3’-untranslated region of RUNX2 played a pivotal role in regulating RUNX2 expression in highly metastatic HNSCC cells. Indeed, miR-376c-3p was commonly down regulated in HNSCC samples, showing an inverse correlation with RUNX2. In-vitro studies identified that restoring miR-376c-3p expression suppressed both expression of RUNX2 and metastatic capability. miR-376c-3p was shown to bind to the 3’UTR of Runx2 resulting in translational repression. Taken together, these data reveal tumor suppressive activity of miR-376c-3p in which this microRNA negatively regulates Runx2 to inhibit HNSCC metastasis[53].
Future perspective
This review addresses an emerging concept of miRNA regulation associated with Runx2 function during osteogenesis and bone metastasis (Fig. 1C). It is predicted that miRNAs regulate one third of the human genome. Yet in the bone development field, there is not a clear understanding of the role miRNAs play in cellular systems controlling osteoblast development and maintenance. One miRNA can bind hundreds of target mRNAs including Runx2, also Runx2 mRNA can have at 10s of different miRNA binding sites and can act distinctively or collectively in bone. These two basic properties predict that osteo-miRs function as powerful molecular designators that precisely modulate Runx2 functional network. Hence, detailed characterization of miRNAs that control Runx2 will provide insights for the fundamental epigenetic basis of physiological bone formation and pathologic disorders of the skeleton due to Runx2 dysregulation.
We have summarized the current understanding of miR-Runx2 regulation, yet most of these findings have been characterized using in-vitro systems. Important questions to ask next include: is the miRNA regulation of osteogenesis biologically relevant; is it establishing indispensable regulation for proper bone formation and maintenance? Do osteoblast specific signaling pathways control miRNA expression and function tailor gene expression necessary for developing bone?
Skeletal phenotypes resulting from miRNA knockdown in animal models are necessary for elucidating biological function. Science’s understanding of the fundamental mechanism of microRNA mediated repression of target genes is sound; and as communicated in this publication, there is also a firm handle on the identification of microRNA that target the master transcription factor of osteogenesis: RUNX2. Moving forward, these authors believe that there are two major focuses that will more forward our understanding of miRNA function during osteogenesis. 1) To more fully understand the role of miRNA in modulating osteoblast specific signaling systems biology. Investigators can continue to push forward this endeavor by reproducibly identifying unstudied miRNA that regulate the osteoblast specific transcription program. 2) More importantly, to elucidate the biological relevance of purported osteo-miRs through identification of skeletal phenotypes resulting from miRNA knockdown in animal models. The ultimate goal of biomedical research is to identify crux targets for therapeutic intervention in diseases. We must push the field of osteo-microRNA forward to elucidate if and how specific miRNA can be wielded clinically to intervene in diseases of skeletal development and maintenance.
Acknowledgments
We thank the members of RNA Biology and Epigenetics laboratory, School of Dentistry, UAB for assistance with critical comments, valuable suggestions, and support. We are thankful to the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (NIH/NIAMS) under Award Number 1R01AR069578 supported research for this publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
Benjamin J. Wildman, Tanner C. Godfrey, Mohammad Rehan, Yuechuan Chen, Lubana H. Afreen and Quamarul Hassan each declare that they have no conflicts of interest with the contents of this article.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References:
- 1.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 2.Komori T Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–9. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 3.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassan MQ, Tye CE, Stein GS, Lian JB. Non-coding RNAs: Epigenetic regulators of bone development and homeostasis. Bone. 2015;81:746–56. doi: 10.1016/j.bone.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Xu X, Ma Z, Huo Y, Xiao Z, Li Y et al. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA. 2011;17(8):1511–28. doi: 10.1261/rna.2732611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song MS, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J. 2017;474(10):1603–18. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284(27):17897–901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16(12):1259–66. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 14.Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011;286(14):12328–39. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105(37):13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem. 2012;287(10):7503–11. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge J, Guo S, Fu Y, Zhou P, Zhang P, Du Y et al. Dental Follicle Cells Participate in Tooth Eruption via the RUNX2-MiR-31-SATB2 Loop. J Dent Res. 2015;94(7):936–44. doi: 10.1177/0022034515578908. [DOI] [PubMed] [Google Scholar]
- 19.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24–2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107(46):19879–84. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heair HM, Kemper AG, Roy B, Lopes HB, Rashid H, Clarke JC et al. MicroRNA 665 Regulates Dentinogenesis through MicroRNA-Mediated Silencing and Epigenetic Mechanisms. Mol Cell Biol. 2015;35(18):3116–30. doi: 10.1128/MCB.00093-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang IH, Jeong BC, Hur SW, Choi H, Choi SH, Ryu JH et al. MicroRNA-302a stimulates osteoblastic differentiation by repressing COUP-TFII expression. J Cell Physiol. 2015;230(4):911–21. doi: 10.1002/jcp.24822. [DOI] [PubMed] [Google Scholar]
- 22.Yu S, Geng Q, Pan Q, Liu Z, Ding S, Xiang Q et al. MiR-690, a Runx2-targeted miRNA, regulates osteogenic differentiation of C2C12 myogenic progenitor cells by targeting NF-kappaB p65. Cell Biosci. 2016;6:10. doi: 10.1186/s13578-016-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8(4):212–27. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108(24):9863–8. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Zhong L, Yuan T, Chen S, Zhou Y, An L et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int J Mol Med. 2018;41(6):3379–93. doi: 10.3892/ijmm.2018.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tome M, Lopez-Romero P, Albo C, Sepulveda JC, Fernandez-Gutierrez B, Dopazo A et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18(6):985–95. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J et al. Effects of miR-335–5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26(8):1953–63. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Tang Y, Zhu X, Tu T, Sui L, Han Q et al. Overexpression of MiR-335–5p Promotes Bone Formation and Regeneration in Mice. J Bone Miner Res. 2017;32(12):2466–75. doi: 10.1002/jbmr.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grunhagen J, Bhushan R, Degenkolbe E, Jager M, Knaus P, Mundlos S et al. MiR-497 approximately 195 cluster microRNAs regulate osteoblast differentiation by targeting BMP signaling. J Bone Miner Res. 2015;30(5):796–808. doi: 10.1002/jbmr.2412. [DOI] [PubMed] [Google Scholar]
- 30.Almeida MI, Silva AM, Vasconcelos DM, Almeida CR, Caires H, Pinto MT et al. miR-195 in human primary mesenchymal stromal/stem cells regulates proliferation, osteogenesis and paracrine effect on angiogenesis. Oncotarget. 2016;7(1):7–22. doi: 10.18632/oncotarget.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Chen S, Deng C, Li F, Wang Y, Hu X et al. MicroRNA-204 Targets Runx2 to Attenuate BMP-2-induced Osteoblast Differentiation of Human Aortic Valve Interstitial Cells. J Cardiovasc Pharmacol. 2015;66(1):63–71. doi: 10.1097/FJC.0000000000000244. [DOI] [PubMed] [Google Scholar]
- 32.Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu X et al. MicroRNA-204 regulates vascular smooth muscle cell calcification in vitro and in vivo. Cardiovasc Res. 2012;96(2):320–9. doi: 10.1093/cvr/cvs258. [DOI] [PubMed] [Google Scholar]
- 33.Yu C, Li L, Xie F, Guo S, Liu F, Dong N et al. LncRNA TUG1 sponges miR-204–5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2018;114(1):168–79. doi: 10.1093/cvr/cvx180. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Xie RL, Gordon J, LeBlanc K, Stein JL, Lian JB et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem. 2012;287(26):21926–35. doi: 10.1074/jbc.M112.340398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fakhry M, Hamade E, Badran B, Buchet R, Magne D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J Stem Cells. 2013;5(4):136–48. doi: 10.4252/wjsc.v5.i4.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Liu H, Lin H, Yuan G, Zhang L, Chen Z. MicroRNA-338–3p promotes differentiation of mDPC6T into odontoblast-like cells by targeting Runx2. Mol Cell Biochem. 2013;377(1–2):143–9. doi: 10.1007/s11010-013-1580-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Sun Q, Wan C, Li L, Zhang L, Chen Z. MicroRNA-338–3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229(10):1494–502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- 38.Zaragosi LE, Wdziekonski B, Brigand KL, Villageois P, Mari B, Waldmann R et al. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011;12(7):R64. doi: 10.1186/gb-2011-12-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang R, Yan S, Wang J, Deng F, Guo Y, Li Y et al. MiR-30a regulates the proliferation, migration, and invasion of human osteosarcoma by targeting Runx2. Tumour Biol. 2016;37(3):3479–88. doi: 10.1007/s13277-015-4086-7. [DOI] [PubMed] [Google Scholar]
- 40.Balderman JA, Lee HY, Mahoney CE, Handy DE, White K, Annis S et al. Bone morphogenetic protein-2 decreases microRNA-30b and microRNA-30c to promote vascular smooth muscle cell calcification. J Am Heart Assoc. 2012;1(6):e003905. doi: 10.1161/JAHA.112.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gay I, Cavender A, Peto D, Sun Z, Speer A, Cao H et al. Differentiation of human dental stem cells reveals a role for microRNA-218. J Periodontal Res. 2014;49(1):110–20. doi: 10.1111/jre.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamam D, Ali D, Vishnubalaji R, Hamam R, Al-Nbaheen M, Chen L et al. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 2014;5:e1499. doi: 10.1038/cddis.2014.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan J, Guo D, Yang S, Sun H, Wu B, Zhou D. Inhibition of miR-222–3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem Biophys Res Commun. 2016;470(3):498–503. doi: 10.1016/j.bbrc.2016.01.133. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N et al. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. American journal of translational research. 2017;9(1):126–35. [PMC free article] [PubMed] [Google Scholar]
- 45.Yeh CH, Jin L, Shen F, Balian G, Li XJ. miR-221 attenuates the osteogenic differentiation of human annulus fibrosus cells. Spine J. 2016;16(7):896–904. doi: 10.1016/j.spinee.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Ji X, She F, Gao Y, Tang P. miR-628–3p regulates osteoblast differentiation by targeting RUNX2: Possible role in atrophic non-union. Int J Mol Med. 2017;39(2):279–86. doi: 10.3892/ijmm.2016.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuo B, Zhu J, Li J, Wang C, Zhao X, Cai G et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J Bone Miner Res. 2015;30(2):330–45. doi: 10.1002/jbmr.2352. [DOI] [PubMed] [Google Scholar]
- 48.Kim DS, Lee SY, Lee JH, Bae YC, Jung JS. MicroRNA-103a-3p controls proliferation and osteogenic differentiation of human adipose tissue-derived stromal cells. Exp Mol Med. 2015;47:e172. doi: 10.1038/emm.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Deen M, Taipaleenmaki H, Zhang Y, Teplyuk NM, Gupta A, Cinghu S et al. MicroRNA-34c inversely couples the biological functions of the runt-related transcription factor RUNX2 and the tumor suppressor p53 in osteosarcoma. J Biol Chem. 2013;288(29):21307–19. doi: 10.1074/jbc.M112.445890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V et al. Regulatory Role of mir-203 in Prostate Cancer Progression and Metastasis. Clin Cancer Res. 2011;17(16):5287–98. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 51.Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL et al. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25(4):589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 52.Taipaleenmaki H, Browne G, Akech J, Zustin J, van Wijnen AJ, Stein JL et al. Targeting of Runx2 by miR-135 and miR-203 Impairs Progression of Breast Cancer and Metastatic Bone Disease. Cancer Res. 2015;75(7):1433–44. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang WM, Lin YF, Su CY, Peng HY, Chang YC, Lai TC et al. Dysregulation of RUNX2/Activin-A Axis upon miR-376c Downregulation Promotes Lymph Node Metastasis in Head and Neck Squamous Cell Carcinoma. Cancer Res. 2016;76(24):7140–50. doi: 10.1158/0008-5472.CAN-16-1188. [DOI] [PubMed] [Google Scholar]
- 54.Xu C, Zhang H, Gu W, Wu H, Chen Y, Zhou W et al. The microRNA-10a/ID3/RUNX2 axis modulates the development of Ossification of Posterior Longitudinal Ligament. Scientific reports. 2018;8(1):9225. doi: 10.1038/s41598-018-27514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cellular and molecular life sciences : CMLS. 2014;71(24):4747–61. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vimalraj S, Partridge NC, Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 2014;229(9):1236–44. doi: 10.1002/jcp.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(23):15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis research & therapy. 2013;15(5):R102. doi: 10.1186/ar4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Q, Wang Z, Bi X, Zhou H, Wang Y, Gu P et al. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun. 2014;446(1):98–104. doi: 10.1016/j.bbrc.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 60.Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y et al. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem cells and development. 2013;22(16):2278–86. doi: 10.1089/scd.2012.0686. [DOI] [PubMed] [Google Scholar]
- 61.Stepicheva NA, Song JL. Function and regulation of microRNA-31 in development and disease. Molecular reproduction and development. 2016;83(8):654–74. doi: 10.1002/mrd.22678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T et al. miRNA-34c regulates Notch signaling during bone development. Human molecular genetics. 2012;21(13):2991–3000. doi: 10.1093/hmg/dds129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM et al. Dimorphic effects of Notch signaling in bone homeostasis. Nature medicine. 2008;14(3):299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheung KS, Sposito N, Stumpf PS, Wilson DI, Sanchez-Elsner T, Oreffo RO. MicroRNA-146a regulates human foetal femur derived skeletal stem cell differentiation by down-regulating SMAD2 and SMAD3. PloS one. 2014;9(6):e98063. doi: 10.1371/journal.pone.0098063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu B, Liu S, Yu B, Zhu J, Ruan H, Tang T et al. miR-203 inhibits the traumatic heterotopic ossification by targeting Runx2. Cell Death Dis. 2016;7(10):e2436. doi: 10.1038/cddis.2016.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao W, Zhang S, Wang B, Huang J, Lu WW, Chen D. Runx2 and microRNA regulation in bone and cartilage diseases. Annals of the New York Academy of Sciences. 2016;1383(1):80–7. doi: 10.1111/nyas.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gamez B, Rodriguez-Carballo E, Bartrons R, Rosa JL, Ventura F. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem. 2013;288(20):14264–75. doi: 10.1074/jbc.M112.432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Itoh T, Ando M, Tsukamasa Y, Akao Y. Expression of BMP-2 and Ets1 in BMP-2-stimulated mouse pre-osteoblast differentiation is regulated by microRNA-370. FEBS letters. 2012;586(12):1693–701. doi: 10.1016/j.febslet.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Hou C, Meng F, Zhao X, Huang G, Chen W et al. MiR-455–3p regulates early chondrogenic differentiation via inhibiting Runx2. FEBS letters. 2015;589(23):3671–8. doi: 10.1016/j.febslet.2015.09.032. [DOI] [PubMed] [Google Scholar]
- 70.Kureel J, John AA, Dixit M, Singh D. MicroRNA-467g inhibits new bone regeneration by targeting Ihh/Runx-2 signaling. The international journal of biochemistry & cell biology. 2017;85:35–43. doi: 10.1016/j.biocel.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 71.Liao L, Yang X, Su X, Hu C, Zhu X, Yang N et al. Redundant miR-3077–5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Xie H, Liu W, Hu R, Huang B, Tan YF et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. The Journal of clinical investigation. 2009;119(12):3666–77. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]