Abstract
Polyphenism is a successful strategy adopted by organisms to adapt to environmental changes. Brown planthoppers (BPH, Nilaparvata lugens) develop two wing phenotypes, including long-winged (LW) and short-winged (SW) morphs. Though insulin receptor (InR) and juvenile hormone (JH) have been known to regulate wing polyphenism in BPH, the interaction between these regulators remains largely elusive. Here, we discovered that a conserved microRNA, miR-34, modulates a positive autoregulatory feedback loop of JH and insulin/IGF signaling (IIS) pathway to control wing polyphenism in BPH. Nlu-miR-34 is abundant in SW BPHs and suppresses NlInR1 by targeting at two binding sites in the 3’UTR of NlInR1. Overexpressing miR-34 in LW BPHs by injecting agomir-34 induces the development towards SW BPHs, whereas knocking down miR-34 in SW BPHs by injecting antagomir-34 induces more LW BPHs when another NlInR1 suppressor, NlInR2, is also suppressed simultaneously. A cis-response element of Broad Complex (Br-C) is found in the promoter region of Nlu-miR-34, suggesting that 20-hydroxyecdysone (20E) might be involved in wing polyphenism regulation. Topic application of 20E downregulates miR-34 expression but does not change wing morphs. On the other hand, JH application upregulates miR-34 expression and induces more SW BPHs. Moreover, knocking down genes in IIS pathway changes JH titers and miR-34 abundance. In all, we showed that miRNA mediates the cross talk between JH, 20E and IIS pathway by forming a positive feedback loop, uncovering a comprehensive regulation mechanism which integrates almost all known regulators controlling wing polyphenism in insects.
Author summary
Polyphenism is a fascinating phenomenon which significantly improves the ability of a species to explore various environmental resources. Brown planthopper (Nilaparvata lugens, BPH) is a notorious insect pest which causes huge damages to rice. BPH has two wing phenotypes, long-winged (LW) and short-winged (SW) morphologies. LW morphs are capable of long-distance migration, while SW morphs have high reproductive capabilities. Juvenile hormone (JH) and insulin/IGF signaling (IIS) pathway have been known to participate in regulating polyphenism in various organisms. However, how these regulators interact with each other remains largely unknown. Here, we show that a conserved microRNA, miR-34, mediates the crosstalk between JH, 20E and IIS pathway to modulate wing polyphenism in BPH. miR-34 suppresses insulin receptor-1(InR1), increases JH titer, and induces SW morphs. On the other hand, JH increases the expression of miR-34 and induces SW morphs, while 20E decreases miR-34 but does not change proportion of wing morphs. Knocking down genes in IIS pathway changes JH titer and miR-34 abundance. Therefore, miRNA, JH, 20E and IIS form an autoregulatory feedback loop to control wing polyphenism in BPH. Our work presents a comprehensive mechanism of wing polyphenism by integrating various regulators.
Introduction
The phenomenon of polyphenism is that two or more distinct phenotypes are displayed by an organism with the same genotype. This phenomenon is triggered by environmental cues such as population density, host nutrition, and temperature [1]. For example, locusts show density-dependent phenotypic plasticity and have two phases: a low-density “solitarious” phenotype and a high-density “gregarious” phenotype [2]. In beetles, horn polyphenism is a nutrition-dependent trait where some male beetles have fully-developed horns, while other males are completely hornless, depending on their nutrition status and body size [3]. Eusocial insects, including members of Hymenoptera, Blattodea (termites), often display caste differentiation, producing multiple types of offspring with different reproductive and morphological features [4]. Based on the physiological state of the mother, aphids produce winged adults in deteriorating environments and flightless morphs when environmental conditions are stable [5].
Brown planthopper (BPH, Nilaparvata lugens) is one of the most notorious planthoppers, migrating from Vietnam and the Philippines to China and Japan in summer during the rice growing season, then flying back to tropical regions during winter after rice crops have been harvested in temperate zones [6]. Wing polyphenism is a key determinant of the success of planthoppers [7]. Long-winged (LW) BPHs are capable of long-distance migration, while short-winged (SW) morphs display higher reproductive capabilities. Prior works showed that insulin/IGF-1 signaling (IIS) pathways involved in regulating wing polyphenism in BPHs [8]. Two insulin receptors (InR), NlInR1 and NlInR2, have been shown to play opposing roles in determining wing fate. NlInR2 inhibits NlInR1, switching long-winged morphs to short-winged morphs, and vice versa. These two insulin receptors modulate wing polyphenism by regulating insulin/IGF signaling (IIS) pathway [9]. Wounding causes a shift to short wing through stimulation of the forkhead transcription factor (NlFoxO) and its downstream target Nl4EBP [10]. High glucose concentration in the rice host induces long-winged females, indicating that host nutrition quality has a direct impact on wing polyphenism in female BPH. This work shows that the nutrition is a key determination factor in controlling wing polyphenism in BPH and also raises a question of sex difference in wing polyphenism [11]. Silencing the c-Jun NH2-terminal kinase (NlJNK) increased the proportion of short winged female adults, suggesting that JNK signaling involve in regulating wing polyphenism in BPH [12]. In addition, topic application of juvenile hormone (JH) in the BPH nymphs induces SW morphs [13]. Knocking down the juvenile hormone epoxide hydrolase (Nljheh) prevents the degradation of JH and induces a bias towards SW BPHs [14], providing evidence that JH might also involve in regulating wing polyphenism. However, there is a dispute over JH regulation of wing polyphenism in BPH [8, 15].
Though various regulators have been reported to involve in modulating phenotypic plasticity of wing polyphenism in BPHs, how these regulators interact with each other remains an important unsolved question. miR-34 is an important modulator involved in the cross talk between the IIS pathway and hormone in C. elegans and Drosophila. In C. elegans, miR-34 expression is regulated by IIS via a negative feedback loop between miR-34 and DAF-16 (the sole ortholog of FoxO in nematodes), providing robustness to environmental stress [16]. miR-34 is maternally inherited in Drosophila [17], and is activated by JH but is suppressed by 20E through Broad Complex (Br-C) [18]. Here, we found a microRNA (miRNA), Nlu-miR-34, modulates the crosstalk between IIS, JH and 20-hydroxyecdysone (20E) pathways to control wing polyphenism. Nlu-miR-34 suppresses NlInR1, inducing the development towards SW morph. Further, Nlu-miR-34 is activated by JH through Br-C. Moreover, silencing NlInR1 induces the expression of Nlu-miR-34, suggesting that Nlu-miR-34, IIS, and JH form an autoregulatory positive feedback loop to control wing polyphenism in BPHs.
Results
Nlu-miR-34 controls NlInR1 in BPHs
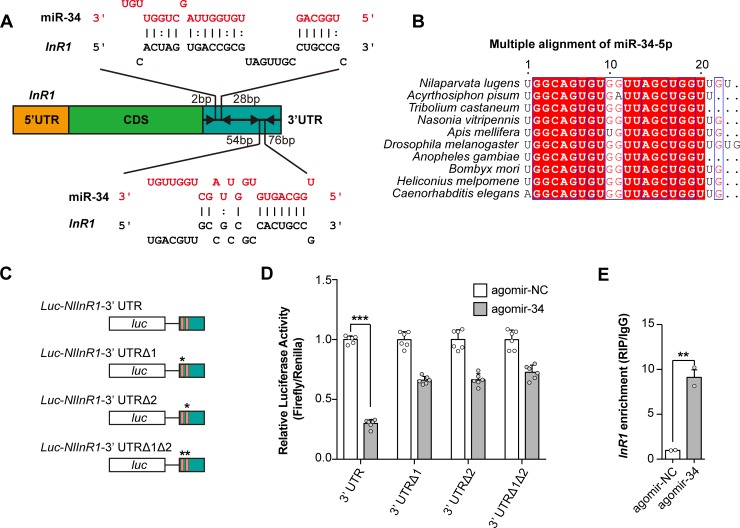
Insulin receptors in BPH (NlInRs) were reported to determine the wing polyphenism in BPH [9]. To investigate whether miRNA may play a role in regulating wing polyphenism, we carried out a bioinformatics analysis to identify miRNAs targeting at NlInR1 or NlInR2. The results showed that only Nlu-miR-34 can target NlInR1 with two putative binding sites in the 3’untranslated region (UTR) predicted by all five target prediction algorithms (Fig 1A, see methods). miR-34 is highly conserved in insects and nematodes (Fig 1B, see methods) and is predicted to have 19 target genes in BPH (S1 Table). To confirm the interaction between Nlu-miR-34 and NlInR1, we introduced the 3’UTR sequence of NlInR1 at the downstream of the firefly luciferase gene in the pMIR-REPORT vector. Constructs with or without (negative control) the 3’UTR of NlInR1 were both transfected into human embryonic kidney 293T (HEK293T) cells. The luciferase activity was significantly reduced to 30% relative to the negative control in the presence of agomir-34 (the mimics of Nlu-miR-34) (Fig 1C and 1D, Student’s t-test, p = 4.99e-12, n = 300, six replicates). Mutating any of the binding sites abolished the suppression effect of agomir-34 on the reporter activity of the NlInR1 target sites (Fig 1D), indicating that both two binding sites are essential for the interaction between Nlu-miR-34 and NlInR1. Next, we performed RNA-binding protein immunoprecipitation (RIP) assay with the antibody-Ago1 (Argonaute 1) in wing buds of BPH nymphs. The transcripts of NlInR1 was significantly enriched in the Ago1-immunoprecipitated RNAs from agomir-34-injected BPHs compared with those from the control samples (Fig 1E, 9.30-fold, Student’s t-test, p = 0.002, n = 150, two replicates), showing that Nlu-miR-34 directly interacted with NlInR1 in the wing buds in vivo. Then, we used fluorescence in situ hybridization (FISH) assay to examine whether Nlu-miR-34 co-localize with NlInR1 in wing buds. Indeed, both of Nlu-miR-34 and NlInR1 were detected in wing buds of the fourth instar BPHs (S1 Fig).
Fig 1. Nlu-miR-34 targets insulin receptor-1 (NlInR1).
(A) Two binding sites were predicted in the 3’UTR of NlInR1 by miRanda, TargetScan, microTar, PITA, and RNAhybrid. (B) Multiple alignment of miR-34-5p in insects and Caenorhabditis elegans. (C) Schematic mutation of two predicted Nlu-miR-34 binding sites within the Luc-NlInR1-3’UTR luciferase reporter. Asterisks means the mutated miRNA binding sites. (D) Dual luciferase reporter assays confirmed the interactions between Nlu-miR-34 and NlInR1 in vitro. The relative luciferase activities were significantly decreased in the presence of agomir-34. Mutating either of the binding sites abolished the repression effect of agomir-34. Data are means ± SEM, p = 4.99e-12, six replicates. (E) RNA-binding protein immunoprecipitation (RIP) confirmed the interactions between Nlu-miR-34 and NlInR1 in vivo. Transcripts of NlInR1 were significantly enriched by antibody against Ago1 in agomir-34 treated group compared with it in agomir-NC treated group. Data are means ± SEM, p = 0.002, n = 50, two replicates. **p < 0.01, ***p < 0.001 (Student’s t-test).
Nlu-miR-34 is highly expressed in SW nymphs in critical stages of wing fate determination
The critical stages of wing fate determination in BPH is the second to fourth instar of nymph [19]. However, the wing morphs cannot be discriminated at nymph stages. To examine the expression of Nlu-miR-34 in nymphs with different wing fate, we first developed two BPH strains by continually selecting SW or LW adults for crossing for more than 50 generations (SW♀ x SW♂ for SW strain, and LW♀ x LW♂ for LW strain) [14]. Almost all adults in SW strain emerge as short-winged adults while 80% of LW strain individuals become long-winged adult morphs. Because these two strains were selected from the same population, SW and LW strain should have the same genetic background. Each generation was purified in the adult stage using a previously described method [20]. We measured the expression of Nlu-miR-34 in the nymph stages of both SW and LW strain. The results show that Nlu-miR-34 was significantly highly expressed in the second and third instars nymphs in SW strain (Fig 2, Student’s t-test, p = 0.0001 for the second instar, and p = 0.002 for the third instar, four replicates), suggesting that Nlu-miR-34 might be essential to develop SW adults. However, we found that NlInR1 was also highly expressed in the second and third instar SW nymphs when using the whole body for experiments (S2 Fig).
Fig 2. Nlu-miR-34 is highly expressed in SW nymphs in critical stages of wing fate determination.
Nlu-miR-34 is highly abundant in the second and third instar SW strain nymphs, indicating that Nlu-miR-34 is expressed during critical stages of wing fate determination. The qPCR data are presented as means ± SEM, p = 0.0001 for the second instar, and p = 0.002 for the third instar, four replicates. **p < 0.01, ***p < 0.001 (Student’s t-test).
Overexpressing Nlu-miR-34 induces SW morphs by repressing NlInR1
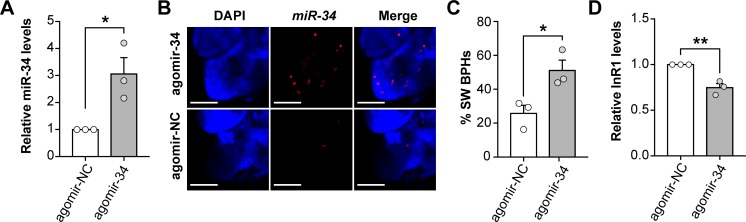
To decipher the roles of Nlu-miR-34 in controlling wing polyphenism, we increased its abundance by injecting agomir-34 into the third instar nymphs at 24 h post molt in LW strain. The abundance of Nlu-miR-34 was significantly increased by 3.06-fold compared to the negative control, which was treated with agomir-NC (a synthesized small RNA with a randomly shuffled sequence of Nlu-miR-34) (Fig 3A, Student’s t-test, p = 0.026, three replicates). Fluorescence in situ hybridization (FISH) assay showed that the abundance of Nlu-miR-34 increased in the wing bud cells at 24 h post injection (Fig 3B). Next, the agomir-34 and agomir-NC treated nymphs were kept for wing observation until emergence into adults. The results showed that overexpression of Nlu-miR-34 induced the development of SW morphs 51.08 ± 6.16% in a significantly higher proportion than those in the control group 25.78 ± 4.78% (Fig 3C, Student’s t-test, p = 0.032, n = 150, three replicates). In addition, overexpression of Nlu-miR-34 downregulated the expression of its target gene NlInR1(Fig 3D, Student’s t-test, p = 0.005, three replicates).
Fig 3. Overexpression of Nlu-miR-34 in LW strain induced the development towards SW BPHs by suppressing NlInR1.
(A) The agomir-34 was injected into early third instar nymphs in LW strain, significantly increasing the Nlu-miR-34 expression (p = 0.026). (B) Fluorescence in situ hybridization assay (FISH) showed that Nlu-miR-34 was more abundant in wing buds treated with agomir-34, relative to the negative control (agomir-NC). Scale bars:100 μm. (C) Injection of nymphs with agomir-34 induced a strong bias towards SW BPHs compared to the control (p = 0.032, n = 150). (D) The relative abundance of NlInR1 (p = 0.004) was significantly decreased after nymphs were injected with agomir-34. The qPCR and wing rate data are presented as means ± SEM, three replicates. *p < 0.05, **p < 0.01 (Student’s t-test).
Nlu-miR-34 is a supplemental suppressor on NlInR1
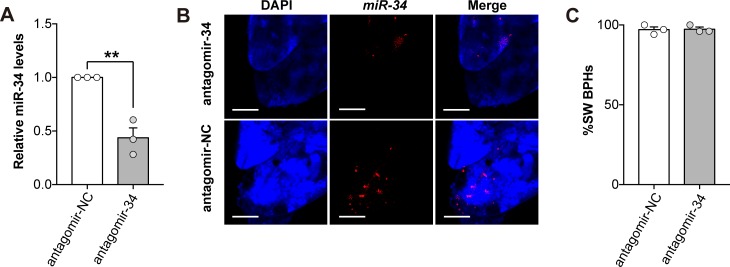
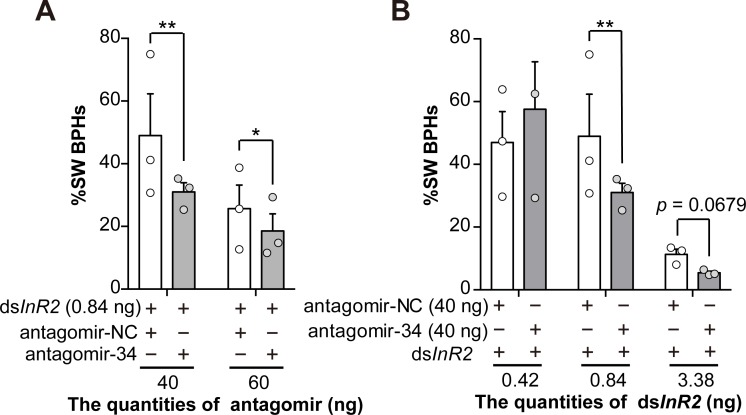
Having showed increased Nlu-miR-34 in LW strain induces SW morphs, we next inhibited Nlu-miR-34 in SW strain by injecting antagomir-34 (the inhibitor of Nlu-miR-34). The expression of Nlu-miR-34 was significantly decreased by 56.33 ± 16.16% (Fig 4A, Student’s t-test, p = 0.004, three replicates). FISH assay showed that the abundance of Nlu-miR-34 decreased in the wing bud cells at 24 h post injection (Fig 4B). However, we did not observe a decrease of SW morphs in antagomir-34 treated groups comparing with the negative control (Fig 4C, Student’s t-test, p = 0.897, n = 150, three replicates). Since NlInR2 suppresses NlInR1 by forming a heterodimer [9] and NlInR2 is highly expressed in SW strain (S3 Fig), we speculated that NlInR2 might have a more potent suppressing effect and thus impair the effect of antagomir-34. To test this hypothesis, we first knocked down NlInR2 with 0.84 ng dsNlInR2 and then inhibited Nlu-miR-34 with two different amounts of antagomir-34 (40 ng and 60 ng) in SW strain. As expected, after partially knocking down NlInR2 by injecting dsNlInR2, downregulation of Nlu-miR-34 significantly induced LW morphs (Fig 5A, Chi-square test, 40ng: χ2 = 5.8, df = 1, p = 0.003; 60ng: χ2 = 7.9, df = 1, p = 0.042; n = 150, three replicates). To test whether this suppressing effect is quantity dependent, we next inhibit NlInR2 by injecting three different amounts of dsNlInR2 (0.42 ng, 0.84 ng and 3.38 ng). The dsNlInR2-treated individuals were further injected with 40 ng antagomir-34 to inhibit Nlu-miR-34. The results showed that at a low quantity of dsNlInR2 (0.42 ng), knocking down Nlu-miR-34 with 40 ng antagomir-34 did not lead to more LW morphs (Fig 5B). However, when dsNlInR2 increased to 0.84 ng and 3.38 ng, the proportions of LW BPHs was significantly increased and SW BPHs were significantly deceased (Fig 5B, Chi-square test, 0.84 ng: χ2 = 8.4, df = 1, p = 0.004; 3.38 ng: χ2 = 3.3, df = 1, p = 0.068; n = 150, three replicates). These results suggested that both NlInR2 and Nlu-miR-34 suppress NlInR1 to control wing polyphenism. NlInR2 might have a major suppression effect whereas Nlu-miR-34 might be a “supplemental” suppressor to clear the leaky transcripts of NlInR1 in the wing bud of SW strain.
Fig 4. Inhibition of Nlu-miR-34 in SW strain by antagomir-34.
(A) The expression of Nlu-miR-34 was significantly decreased by injecting antagomir-34 (p = 0.004). (B) Fluorescence in situ hybridization assay (FISH) showed that Nlu-miR-34 was less abundant in the wing bud treated with antagomir-34, relative to the negative control (antagomir-NC). Scale bars:100 μm. (C) Inhibition of Nlu-miR-34 by antagomir-34 does not induce changes in wing morphology (n = 50). The qPCR and wing rate data are presented as means ± SEM, three replicates. **p < 0.01 (Student’s t-test).
Fig 5. Simultaneously inhibition Nlu-miR-34 and NlInR2 in SW strain induced the decrease of SW BPHs.
(A) Antagomir-34 (with two different quantities) and dsNlInR2 (0.84 ng) were injected into the third instar SW strain nymphs, showed that injection of antagomir-34 decreased the ratio of SW type BPH (Chi-square test, 40ng: χ2 = 5.8, df = 1, p = 0.003; 60ng: χ2 = 7.9, df = 1, p = 0.042). (B) The antagomir-34 (40 ng) and dsNlInR2 (with three different quantities) were simultaneously injected into the third instar SW strain nymphs. The results showed that effect of low level of dsNlInR2 (0.42 ng) with antagomir-34 is familiar with control, while higher level of dsNlInR2 (0.84 and 3.38 ng) with antagomir-34 induced a bias towards the development of LW BPHs compared to the control. Injection of antagomir-34 (40 ng) and dsNlInR2 (0.84 ng) decreased ratio of SW BPH to around 20% (Chi-square test, χ2 = 8.4, df = 1, p = 0.004). High level of dsInR2 (3.38 ng) with antagomir-34 decreased the ratio of SW BPH (Chi-square test, χ2 = 3.3, df = 1, p = 0.068). 200 insects were used for each experiment. Data are means ± SEM, three replicates. *p < 0.05, **p < 0.01.
Nlu-miR-34 is upregulated by JH through Br-C
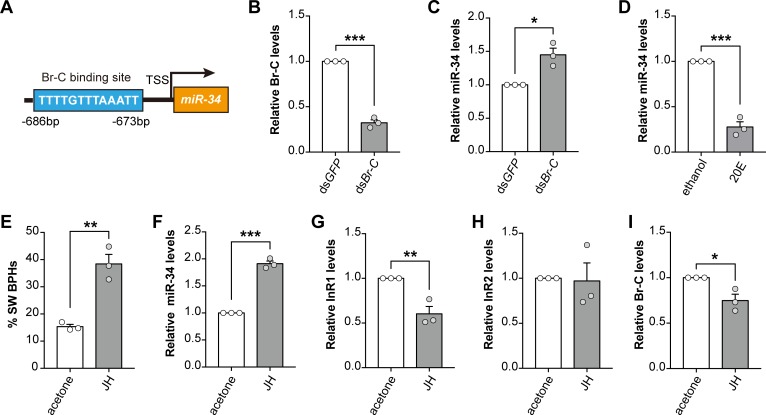
Next, to further uncover the factors regulating Nlu-miR-34 expression, we amplified the 5’UTR of Nlu-miR-34 using rapid amplification of cDNA ends (RACE). The transcription starting site (TSS) was determined by mapping the 5’UTR sequence to the BPH genome (GenBank assembly accession: GCA_000757685.1), showing that the TSS of Nlu-miR-34 is located at position 119,684 in scaffold0000007224(-). We searched for transcription binding sites (TFBS) nearby the TSS of Nlu-miR-34 (-2,000 to +100) using PROMO [21] and Match [22]. Both programs identified a potential cis-response element (CRE) of Broad Complex (Br-C), suggesting that NlBr-C might involve in regulating Nlu-miR-34 (Fig 6A). To confirm this, we knocked down NlBr-C by injecting dsNlBr-C at the third instar nymphs in SW strain (Fig 6B, Student’s t-test, p = 3.21e-5, three replicates), resulted in the increase of Nlu-miR-34 by 1.44-fold (Fig 6C, Student’s t-test, p = 0.011, three replicates). Because Br-C is an ecdysone inducible transcription factor [23], we then used 20-hydroxyecdysone (20E) to treat the third instar nymphs in SW strain. Topical application of 20E decreased Nlu-miR-34 significantly (Fig 6D, Student’s t-test, p = 0.0001, three replicates). However, similar as the effect of using antagomir-34 alone without knocking down NlInR2, 20E application did not change the proportion of wing morphs.
Fig 6. Nlu-miR-34 is upregulated by JH through Br-C.
(A) A cis-response element specific to NlBr-C Z4 activity was identified in the promoter region of Nlu-miR-34. (B) Expression of NlBr-C was successfully knocked down in LW strain after treating with dsNlBr-C (p = 3.21e-5). (C) Knockdown of NlBr-C leads to the increase of Nlu-miR-34 transcripts (p = 0.011). (D) 20E application in SW strain results in the decreased of Nlu-miR-34 (p = 0.0001, four replicates). (E) Application of JH in LW-strain induced a strong bias towards development of SW BPHs (p = 0.003, n = 100). (F) Increased expression of Nlu-miR-34 (p = 6.04e-5), (G) significantly decrease of NlInR1 (p = 0.008), (H) No effect on the expression of NlInR2. (I) JH application results in decreased abundance of NlBr-C transcripts (p = 0.022). The qPCR and wing rate data are presented as means ± SEM, three replicates. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
Since JH and 20E show antagonistic actions through Br-C [24] and JH was known to regulate wing polyphenism in BPH [13, 25], we hypothesize that JH might involve in regulating the expression of Nlu-miR-34. To test this mode, we used topical application of JH III analogue at the third instar of BPHs in LW strain. Acetone were used as the negative control. The results showed that increased JH titer led to a strong bias towards SW BPHs (increasing by 23.09 ± 6.16%) (Fig 6E, Student’s t-test, p = 0.003, n = 100, three replicates). After JH application, Nlu-miR-34 was significantly increased by 1.91-fold while NlInR1 was decreased by 39.7 ± 14.26%, but it didn’t change the NlInR2 level (Fig 6F–6H, Student’s t-test, p = 6.04e-5 and 0.008, three replicates). Moreover, NlBr-C was also decreased by 25.18 ± 12.13% at 24h post JH treatment (Fig 6I, Student’s t-test, p = 0.022, three replicates). These results showed that the upregulation of Nlu-miR-34 by JH and the downregulation of Nlu-miR-34 by 20E are likely mediated by NlBr-C in a transcriptional regulation manner.
Nlu-miR-34, JH and NlInR1 form a positive autoregulatory loop to control wing polyphenism
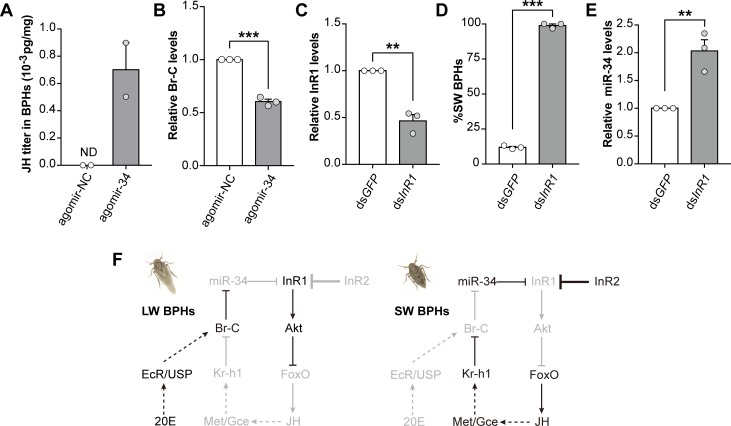
We previously showed that JH activates Nlu-miR-34. Interestingly, we found JH titer was significantly increased by the overexpression of Nlu-miR-34 (Fig 7A, n>300), suggesting that JH and Nlu-miR-34 form a positive autoregulatory loop. In addition, NlBr-C was significantly decreased by 39.60 ± 3.99% when overexpressing Nlu-miR-34 (Fig 7B, Student’s t-test, p = 6.72e-5, three replicates), suggesting the positive feedback loop between JH and Nlu-miR-34 might be mediated by Br-C.
Fig 7. Nlu-miR-34, JH, 20E and NlInR1 form a positive autoregulatory loop.
(A) JH titers were increased significantly in BPHs after treated with agomir-34 in LW strain. ND: not detected. More than 300 insects were used for each experiment. (B) Injection of agomir-34 in LW strain resulted in decreased abundance of NlBr-C transcripts (p = 6.72e-5). (C) Knockdown of NlInR1 significantly decreased the expression of NlInR1 (p = 0.001). (D) Knocking down NlInR1 in LW strain induced a strong bias towards SW BPHs (p = 2.51e-7, n = 100). (E) Higher transcriptional level of Nlu-miR-34 in the knockdown SW BPHs (p = 0.006). The qPCR and wing rate data are presented as means ± SEM, three replicates. *p < 0.05, **p < 0.01, *** p < 0.001 (Student’s t-test). (F) Schematic model of the interplay between the IIS pathway, JH, 20E and miRNA during wing determination in BPHs. In LW BPHs, low transcriptional level of Nlu-miR-34 fails to inhibit the expression of NlInR1, which activates NlAkt, inactivates NlFoxO, and reduces JH titer activates the activity of NlBr-C, which will maintain the low transcriptional level of Nlu-miR-34. In SW BPHs, high transcriptional level of Nlu-miR-34 inhibits the expression of NlInR1, fails to inactivate NlFoxO by NlAkt, stimulating to a high JH titer and repression of NlBr-C, which will maintain the high transcriptional level of Nlu-miR-34. The components that are less active or inactive are shown in grey.
To test whether NlInR1 also involve in this autoregulatory loop, we next knocked down NlInR1 by injecting dsNlInR1 in the third instar nymphs in LW strain (Fig 7C). The results showed that knocking down NlInR1 increased Nlu-miR-34 by 2.03-fold (Fig 7E, Student’s t-test, p = 0.006, three replicates) and led to a strong bias towards SW morphs (Fig 7D, Student’s t-test, p = 2.51e-7, n = 100, three replicates). Taking together, these results suggested that miR-34, JH, 20E and IIS pathway form a positive autoregulatory loop to control wing polyphenism in BPHs (Fig 7F).
Discussion
Many animals with the same genetic background exhibit polyphenism to adapt to different environment conditions [1]. It has been showed that insulin-like peptides (ILP), the IIS pathway, and JH involve in the regulation of polyphenism in various organisms [26–29]. However, it remains elusive how these regulators interact with each other. Here, we showed that Nlu-miR-34 is a mediator between the crosstalk between IIS and hormonal signals by forming a positive feedback loop. miR-34 is a conserved miRNA that plays an important regulatory role in a wide range of organisms. It seems the positive feedback loop mediated by miR-34 might be a conserved process because miR-34 is also regulated by IIS via a negative feedback loop between miR-34 and DAF-16 (the sole ortholog of FoxO in nematodes) in C. elegans [16]. We propose a regulation mode of JH -miRNA-IIS feedback loop in BPH (Fig 7F). Through this positive autoregulatory loop, minor changes in environment conditions, above a certain threshold, would be amplified significantly in vivo inducing a phenotype shift. Two pathways involved in miRNA or InR regulation are redundant or at least partially redundant. It seems that InR regulation is more sensitive to nutritive environment whereas miRNA regulation is likely to be hormonally regulated. This discovery extends our understanding of the interplay between nutrition status-based signals and hormone signals in modulating phenotypic plasticity.
Polyphenism is a phenomenon in which a single genotype can arise into two or more discrete phenotypes in response to environmental stimuli; these phenotypes do not segregate in F1 generations produced from parents with distinct phenotypes, suggesting that polyphenotypic changes are not determined by only one or two genes [30]. However, knocking down NlInR1 induces formation of 100% SW BPHs [9], suggesting that the effect of insulin or hormone regulation is normally determinative in most cases. Therefore, other regulators should participate in regulation; and these regulators should be either sensitive to environmental cues or dose dependent. Here, we showed that Nlu-miR-34 mediates the cross talk between JH and IIS, and miRNAs are normally expressed at moderate levels [31]. In contrast to other regulatory molecules, miRNAs are typically very fine-tuned, specifically binding to and regulating their targets, acting as buffers to ensure developmental robustness [32]. These features enable them to act as ideal regulators of phenotype polyphenism, responding to varied environment cues. To the best of our knowledge, we presented the first evidence that miRNAs regulate developmental plasticity by modulating the cross talk between the JH and IIS pathways. It should be noted that the experiments of overexpression of Nlu-miR-34 were repeated independently in three labs in Nanjing, Wuhan and Hangzhou, ensuring the reliability of miRNA modulation of wing polyphenism in BPH (S4 Fig).
JH was reported to regulate wing polyphenism in BPH twenty years ago by Tojo and his colleagues but without further evidence [13, 25]. A recent report showed that knocking down juvenile hormone epoxide hydrolase (Jheh) enhanced short wing formation [14]. However, Zhang and his colleagues argued that lack of direct evidence of JH regulation of wing polyphenism in BPH [8, 15]. Here we present evidence that JH participates in the regulation of wing polyphenism in BPH by upregulating Nlu-miR-34 expression. JH has been reported to regulate miRNA expression during larvae development in D. melanogaster [18]. JH can redirect the development of LW to SW/wingless morphs in BPHs [13, 25], aphids [33], and crickets [34], but the relationship between JH and wing polyphenism is inverse with that in ants [35–37]. It is interesting to notice that both winged ants and SW BPHs have higher fecundity than other morphs, inferring JH regulation of wing polyphenism varies between different insects and JH might be involved in reproduction regulation [38], which requires further investigations.
Though JH has been well understood in regulating phenotype polyphenism [1, 28], the role of 20E is less understood in this field. We showed that miR-34 is activated by JH but is suppressed by 20E in planthopper. A CRE of Br-C was found in the promoter region of Nlu-miR-34. It has been reported that downregulation of miR-34 requires Br-C in Drosophila [18]. Since we did not find any JH response element (JHRE) in the promoter region of Nlu-miR-34, JH might regulate Nlu-miR-34 in an indirectly manner. JH has been reported to show antagonistic interaction with 20E through Br-C [24], it is likely that JH may activate the expression of Nlu-miR-34 through a cross talk between JH and 20E mediated by Br-C. However, topic application of 20E did not change the proportion of wing morphs. These results open a new question whether 20E regulates wing polyphenism in BPHs, and if so, in which way the 20E may regulate wing polyphenism.
In summary, we showed, for the first time, a comprehensive regulation mechanism of wing polyphenism in BPH by integrating almost all known regulators (miRNA, IIS, JH, and 20E) into a positive autoregulatory feedback loop (Fig 7F). We also presented evidence that 20E might also involve in regulating wing polyphenism by cross talking with JH. These discoveries extend our understanding of the mechanism regulating insect polyphenism, and shed new lights on how environmental cues determine which alternative phenotypes are produced by a given genotype.
Material and methods
Insects
Wild BPH populations were obtained from rice fields in Wuhan, Hubei Province, China, and raised on a BPH-susceptible rice variety Taichuang Native 1 (TN1) in the greenhouse. SW and LW strains were selected to propagate for more than 50 generations [14]. The percentage of SW morphs in the generated SW strain was more than 80%, while LW morphs represented approximately 80% of the LW strain. Each generation was purified in the adult stage using a previously described method [18]. BPHs were raised in growth chambers at 26°C (± 2°C) under a photoperiod of 16:8h (light: dark) with the relative humidity of 75% (± 5%).
Prediction of miRNA targets
We obtained BPH miRNAs sequences that have been previously reported [39, 40]. The 3’UTR sequences of NlInR1 (KF974333) and NlInR2 (KF974334) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). We used five algorithms, including miRanda [41], TargetScan [42], microTar [43], PITA [44], RNAhybrid [45], to predict miRNAs that can target NlInR1 or NlInR2. Default parameters were used for all algorithms.
microRNAs targeting NlInR1
To investigate whether miRNA may play a role in regulating wing polyphenism, we carried out a bioinformatics analysis to identify miRNAs targeting at NlInR1 or NlInR2. The results showed that three miRNAs target on NlInR1 and four miRNAs target on NlInR2 (S2 Table). Then, we used luciferase reporter assays to confirm the interaction between miRNA and their targets, and the results confirmed the interactions between NlInRs and three miRNAs, Nlu-miR-34, Nlu-miR-989b and Nlu-miR-989c. We overexpressed these three miRNAs separately to test their effects on wing polyphenism in BPH, showing that only Nlu-miR-34 can significantly change the ratio of wing morphs (Fig 3A and S5 Fig). So, we focused on Nlu-miR-34 in further studies.
Multiple sequences alignment of miR-34 sequences
miR-34-5p sequences from several insects and nematode C. elegans were downloaded from miRBase and submitted to ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) for multiple sequence alignment and visualization.
In vitro luciferase assays
The 3’UTRs of NlInR1 and NlInR2 were amplified and introduced into the pMIR-REPORT vector (Obio, China) downstream of the firefly luciferase gene. The pRL-CMV vector (Promega, USA), which contains the Renilla luciferase gene, was used as a control luciferase reporter vector. The predicted binding site (5’-CACTAGTGACCGCGTAGTTGCCTGCCG-3’) was completely removed for mutant 1, and another binding site (5’-TGACGTTGCCGCCGCCACTGCCG-3’) was removed for generation of mutant 2. HEK293T cells (Obio, China) were cultured at 37°C in 5% CO2 in DMEM (Gibco, USA) media supplemented with 10% FBS (Hyclone) for 24 h in 96-well culture plates. Cells were transfected with pMIR-REPORT (0.2 μg), pRL-CMV (0.01μg), 0.25 μl of 100 nM miRNA mimics (RiboBio, China), and 0.25 μl Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer’s instructions. The activity of the two luciferase enzymes was measured 48 h after transfection following the manufacturer’s recommendations (Dual-Luciferase Reporter Assay System, Promega, USA) with Infinite M1000 (Tecan, Switzerland). Three replicates were performed for each experiment. Results are expressed as the means of the ratio of firefly luciferase activity/Renilla luciferase activity. Controls were set to one, and the two-tailed t-test was used for statistical analysis.
RNA-binding protein immunoprecipitation Assay (RIP)
A Magna RIP Kit (Millipore, Germany) was used to perform the RIP assay according to the manufacturer’s instructions. Approximately 50 wing buds of the fourth instar nymphs were collected and crushed using a homogenizer in ice-cold RIP lysis buffer. Magnetic beads were incubated with 5 μg of RIPAb+ Ago-1 antibody (Millipore, Germany) or normal mouse IgG (Millipore, Germany). The homogenates in the RIP lysates were centrifuged and the Ago 1-bound mRNAs in supernatants were pulled down by magnetic bead–antibody complex at 4°C overnight. The immunoprecipitated RNAs were released by digestion with protease K. Finally, the RNAs were purified by which methods and used for cDNA synthesis. The PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Japan) was used and the abundance of InR1 was quantified by qPCR.
Fluorescence in situ hybridization (FISH)
An antisense nucleic acid detection probe (5’-ACAACCAGCUAACCACACUGCCA -3’) designed to detect Nlu-miR-34 was labeled with Cy3. The probe for detecting NlInR1 (5’- GAACAGCCAGGACAGGCCGAAUCCUCCAUG-3') was labeled with FAM. The random shuffled probe (5’-UUCUCCGAACGUGUCACGUU-3’) and the probe (5’-UUGUACUACAAAAGUACUG-3’) were used as miRNA and mRNA negative controls, respectively. The wing buds were dissected from the third or fourth instar stage nymphs injected with agomir or antagomir (RiboBio, China). For fluorescence in situ hybridization, wing buds were treated for 24 h, fixed in 4% paraformaldehyde for 2 hours, and then incubated with the miRNA probes at 37°C for 24 h. The samples were washed in PBS containing Triton X-100 (5% v/v) and then incubated with the DAPI (RiboBio, China) at room temperature for 30 min. Signals were analyzed and images were recorded using a Zeiss LSM 780 confocal microscope (Carl Zeiss SAS, Germany). Figures were prepared using Zeiss LSM ZEN 2010 software (Carl Zeiss SAS, Germany).
5’UTR amplification and promoter analysis of Nlu-miR-34
Total RNA was isolated from ten fourth to fifth instar nymphs of SW BPHs using the TRIzol regent (Invitrogen, USA). The SMARTer RACE cDNA amplification kit (Clontech, USA) was used to obtain the 5’UTR of Nlu-miR-34. 5’RACE nested-PCR was performed using Ex-Taq (Takara, Japan) according to the manufacturer’s instructions. The amplified products were separated by agarose gel electrophoresis and purified using a Gel Extraction Kit (Takara, Japan). The purified DNA was ligated into the pMD19-T vector (Takara, Japan), and sent to BGI-Shenzhen for sequencing.
The transcript starting site (TSS) of miR-34 was confirmed by alignment to genomic scaffold sequences (GenBank assembly accession: GCA_000757685.1). Any unknowns “N” in the genomic sequences were confirmed by PCR validation. A 2100 bp flanking sequence (-2000 to +100) upstream of the TSS was used as the promoter region for analysis by Promoter Scan web server (http://www.cbs.dtu.dk/services/Promoter/). A putative TATA-box was identified at position -1707. To identify putative transcription factor binding sites (TFBS) in the promoter region, two algorithms, including PROMO 3.0 [18] and MATACH 1.0 [19] based on TRANSFAC database (http://gene-regulation.com/pub/databases.html), were computationally analyzed using default parameters. A Br-C Z4 biding site (TTTTGTTTAAATT) at position -699 was predicted by both programs. The Nlu-miR-34 sequence, including the intact 5’UTR, was deposited in GenBank (MG894367).
Injection of agomir-34 and antagomir-34
BPH nymphs were collected for microinjection on the first day of the third instar. 15 nl of agomir-34 (40 ng, 15 nl) or antagomir-34 (40 ng, 15 nl) were microinjected into the conjunctive of each nymph between the prothorax and mesothorax using a Nanoliter 2000 injector (WPI, USA). 150 nymphs were used for each experiment. Ten nymphs were transferred into glass tubes 24 h post injection and placed in a growth chamber. Agomir or antagomir of random shuffled sequences was injected with equivalent volume as negative control (RiboBio, China). All experiments were performed in triplicate.
RNA interference (RNAi)
In order to knock down NlInR1 and NlBr-C expression, dsRNA was synthesized using the T7 RiboMAXTM Express RNAi System (Promega, USA) following the manufacturer’s instructions. Each nymph, at the first day of the third instar, was injected with dsRNA (250 ng, 15 nl) using a Nanoliter 2000 injector (WPI). Equivalent volumes of dsGFP were injected as negative controls. 100 nymphs were used for each treatment, and all experiments were carried out in triplicate.
Co-injection of dsNlInR2 and antagomir
BPH nymphs were collected for microinjection on the first day of the third instar. The dsNlInR2, the concentration of which were divided into three groups, 0.42 ng, 0.84 ng and 3.38 ng, was injected into nymphs respectively with 40 ng antagomir-34. Either 40 ng or 60 ng antagomir-34 was injected into nymphs with 0.84 ng dsNlInR2. The volumes of mixtures were 15 nl. All of microinjections were done using a Nanoliter 2000 injector (WPI, USA). Approximately 200 nymphs were used for each experiment. Control nymphs were injected with equivalent volumes of the mixtures of dsNlInR2 and antagomir-NC. All experiments were performed in triplicate.
Topical application of JH
Juvenile hormone III (JH III, 96%) was purchased from Toronto Research Chemicals (Sigma, USA). 200 ng of JH III was dissolved in acetone and dropped 40 nl onto the mesonotum of the third instar BPH nymphs using a Nanoliter 2000 injector (WPI, USA), after anaesthetizing with carbon dioxide [14]. Equivalent volumes of acetone were used as negative controls. The rearing condition and methods of observation were same as the method of RNAi and injection of agomir-34 and antagomir-34. 100 nymphs were used for each experiment, all experiments were performed in triplicate.
Quantitative real-time PCR
Total RNA, enriched for miRNAs and mRNA, was extracted using the miRNeasy Mini kit (Qiagen, Germany) and TransZol Up Plus RNA kit (TransGen, China) from whole insect bodies (n = 30), respectively. The miScript Ⅱ RT kit (QIAGEN) and the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen) were used to prepare cDNA. Quantitative real-time PCR (qPCR) reactions were conducted with an ABI Prism 7300 (Applied Biosystems, USA) using miScript SYBR Green (Qiagen, Germany) and SYBR® Premix Ex Taq II RR820A (Takara, Japan), according to the manufacturer’s instructions. The data was analyzed using the 2-△△Ct method. Actin1 (GenBank accession No. EU179846.1) was used as the endogenous control. This gene has been studied to show it is suitable to be used as a reference gene [46]. All treatments were carried out in triplicate. Independent reactions were performed in quadruplicate for each RNA sample, and the signal intensity of the target gene is presented as the average value. The qPCR primers used in this study are listed in S3 Table.
Juvenile hormone estimation by LC-MS/MS
About 300 BPHs were collected and pooled together for analysis. Extraction of JH III was performed according to a previously described method with some modifications [47, 48]. JH III dilutions of 20, 10, 1, 0.5, 0.25, 0.125, and 0.0625 ng/ml were prepared in 70% methanol and used for calculation of the standard curve. The LC–MS/MS analyses were performed using an Agilent 6460 triple quadrupole mass spectrometer (Agilent, USA) equipped with an electrospray ionization (ESI) source, operated in the positive ion multiple-reaction monitoring (MRM) mode. Agilent Mass Hunter Workstation software was used for system operation, data acquisition, and data analysis. Chromatographic separation was carried out using a Zorbax SB-Aq column (100 mm × 2.1 mm, 3.5 μm) (Agilent, USA). JH III was separated using a binary gradient. Briefly, mobile phase A consisted of 0.1% formic acid in water and mobile phase B was comprised of 0.1% formic acid in acetonitrile. The gradient program was as follows: 0–5 min, 60%–85% of B; 5–8 min, 85%–95% of B. The flow rate of the mobile phase and the column temperature were set at 0.3 mL/min and 30°C, respectively, and the injection volume was 5 μl. The retention time of JH III was 3.82 min and the total run time was 10 min. Mass spectrometric detection was completed by use of an electrospray ionization (ESI) source in positive ion multiple-reaction monitoring (MRM) mode using the following parameters: precursor (m/z), 267; ion product (m/z), 235.2; fragmentor voltage (V), 80; collision Energy (eV), 1.
Statistical analysis
SPSS 17.0 software (IBM SPSS Inc. USA) was used for statistical analysis. The differences between treatments were compared using the Student’s t-test. The effects of antagomir on distribution of wing types in BPHs were analyzed using the Chi-square test. p < 0.05 was considered statistically significant. All results are expressed as means ± SEM, three replicates.
Supporting information
Green (NlInR1) and red (Nlu-miR-34) signals overlap to show yellow signals, suggesting Nlu-miR-34 interact directly with NlInR1 in the cells of wing buds. Control 1, Nlu-miR-34 antisense and scrambled mRNA probe; Control 2, scrambled miRNA and NlInR1 antisense probe; Control 3, scrambled miRNA and scrambled mRNA probe. Scale bars: 100 μm.
(TIF)
The data are presented as means ± SEM, three replicates. *p < 0.05 (Students’ t-test).
(TIFF)
The data are presented as means ± SEM, three replicates. *p < 0.05, **p < 0.01 (Students’ t-test).
(TIFF)
The data are presented as means ± SEM, three replicates. **p < 0.01, ***p < 0.001 (Students’ t-test).
(TIFF)
(A) Dual luciferase reporter assays confirmed the interactions between Nlu-miR-34 and NlInR1 in vitro (p = 2.35e-13). (B, C) Dual luciferase reporter assays confirmed the interactions between Nlu-miR-989b and NlInR2 (p = 1.43e-12), and the interactions between Nlu-miR-989c and NlInR2 in vitro (p = 9.48e-13). Data are means ± SEM, six replicates. (D, E) Overexpression of Nlu-miR-989b and Nlu-miR-989c in SW-strain. Wing rate data are presented as means ± SEM, three replicates. ***p < 0.001 (Student’s t-test).
(TIFF)
(DOCX)
(DOCX)
(DOCX)
Data Availability
The Nlu-miR-34 sequence, including the intact 5’UTR, was deposited in GenBank (MG894367). All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
FL was funded by the National Key Research and Development Program (2017YFD0200900, 2016YFC1200600) and the National Science Foundation of China (NSFC) (31772238). KH was funded by NSFC (31701785). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Curr Biol. 2011; 21(18): R738–749. 10.1016/j.cub.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Kang L. Molecular mechanisms of phase change in locusts. Annu Rev Entomol. 2014; 59: 225–244. 10.1146/annurev-ento-011613-162019 [DOI] [PubMed] [Google Scholar]
- 3.Emlen DJ, Corley Lavine L, Ewen-Campen B. On the origin and evolutionary diversification of beetle horns. Proc Natl Acad Sci U S A. 2007; 104 Suppl 1: 8661–8668. 10.1073/pnas.0701209104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miura T. Developmental regulation of caste-specific characters in social-insect polyphenism. Evol Dev 2005; 7: 122–129. 10.1111/j.1525-142X.2005.05014.x [DOI] [PubMed] [Google Scholar]
- 5.Brisson JA. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos Trans R Soc Lond B Biol Sci. 2010; 365: 605–616. 10.1098/rstb.2009.0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X, Chen R, Xi X, Yang L, Zhu Z, Wu J, et al. Studies on the migrations of brown planthopper Nilaparvata lugens Stal. Acta Entomologica Sinica. 1979; 22(1): 1–21. [Google Scholar]
- 7.Denno RF, Roderick GK. Populatoin biology of planthoppers. Annu Rev Entomol. 1990; 35: 489–520. [Google Scholar]
- 8.Xu HJ, Zhang CX. Insulin receptors and wing dimorphism in rice planthoppers. Philos Trans R Soc Lond B Biol Sci. 2017; 372(1713). 10.1098/rstb.2015.0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HJ, Xue J, Lu B, Zhang XC, Zhuo JC, He SF, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015; 519: 464–467. 10.1038/nature14286 [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Yao Y, Wang B, Lavine MD, Lavine LC. FOXO links wing form polyphenism and wound healing in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol. 2016; 70: 24–31. 10.1016/j.ibmb.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Xu Y, Jiang J, Lavine M, Lavine LC. Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc Natl Acad Sci U S A. 2018; 115(29): 7563–7568. 10.1073/pnas.1721473115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin X, Xu Y, Yao Y, Wang B, Lavine MD, Lavine LC. JNK signaling mediates wing form polymorphism in brown planthoppers (Nilaparvata lugens). Insect Biochem Mol Biol. 2016; 73: 55–61. 10.1016/j.ibmb.2016.04.005 [DOI] [PubMed] [Google Scholar]
- 13.Iwanaga K, Tojo S. Effects of juvenile hormone and rearing density on wing dimorphism and oöcyte development in the brown planthopper, Nilaparvata lugens. J Insect Physiol. 1986; 32(6): 585–90. 10.1016/0022-1910(86)90076-4 [DOI] [Google Scholar]
- 14.Zhao J, Zhou Y, Li X, Cai W, Hua H. Silencing of juvenile hormone epoxide hydrolase gene (Nljheh) enhances short wing formation in a macropterous strain of the brown planthopper, Nilaparvata lugens. J Insect Physiol. 2017; 102:18–26. 10.1016/j.jinsphys.2017.08.012 [DOI] [PubMed] [Google Scholar]
- 15.Zhang CX, Brisson JA, Xu HJ. Molecular mechanisms of wing polymorphism in insects. Annu Rev Entomol. 2019; 64:297–314. 10.1146/annurev-ento-011118-112448 [DOI] [PubMed] [Google Scholar]
- 16.Isik M, Blackwell TK, Berezikov E. MicroRNA mir-34 provides robustness to environmental stress response via the DAF-16 network in C. elegans. Sci Rep. 2016; 6: 36766 10.1038/srep36766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soni K, Choudhary A, Patowary A, Singh AR, Bhatia S, et al. miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013; 41(8):4470–4480. 10.1093/nar/gkt139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003; 259(1): 9–18. 10.1016/S0012-1606(03)00208-2 . [DOI] [PubMed] [Google Scholar]
- 19.Bertuso AG, Morooka S, Tojo S. Sensitive periods for wing development and precocious metamorphosis after precocene treatment of the brown planthopper, Nilaparvata lugens. J Insect Physiol. 2002; 48(2): 221–229. 10.1016/S0022-1910(01)00167-6 [DOI] [PubMed] [Google Scholar]
- 20.Morooka S, Tojo S. Maintenance and selection of strains exhibiting specific wing form and body colour under high density conditions in the brown planthopper, Nilaparvata lugens (Homoptera:Delphacidae). Appl Entomol Zool. 1992; 27: 445–454. [Google Scholar]
- 21.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003; 31(13): 3651–3653. 10.1093/nar/gkg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003; 31(13): 3576–3579. 10.1093/nar/gkg585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma H, Abbas MN, Zhang K, Hu X, Xu M, Liang H, et al. 20-Hydroxyecdysone regulates the transcription of the lysozyme via Broad-Complex Z2 gene in silkworm, Bombyx mori. Dev Comp Immunol. 2019; 94: 66–72. 10.1016/j.dci.2019.01.014 [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Li K, Gao Y, Liu X, Chen W, Ge W, et al. Antagonistic actions of juvenile hormone and 20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc Natl Acad Sci U S A. 2018; 115(1): 139–144. 10.1073/pnas.1716897115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayoade O, Morooka S, Tojo S. Enhancement of short wing formation and ovarian growth in the genetically defined macropterous strain of the brown planthopper, Nilaparvata lugens. J Insect Physiol. 1999; 45(1): 93–100. 10.1016/S0022-1910(98)00103-6 [DOI] [PubMed] [Google Scholar]
- 26.Moczek AP, Nijhout HF. Rapid evolution of a polyphenic threshold. Evol Dev. 2003; 5(3): 259–268. 10.1046/j.1525-142X.2003.03033.x [DOI] [PubMed] [Google Scholar]
- 27.Zera AJ. The endocrine regulation of wing polymorphism in insects: state of the art, recent surprises, and future directions. Integr Comp Biol. 2003; 43(5): 607–616. 10.1093/icb/43.5.607 [DOI] [PubMed] [Google Scholar]
- 28.Verma KK. Polyphenism in insects and the juvenile hormone. J Biosci. 2007; 32(2): 415–420. [DOI] [PubMed] [Google Scholar]
- 29.Corona M, Libbrecht R, Wheeler DE. Molecular mechanisms of phenotypic plasticity in social insects. Curr Opin Insect Sci. 2016; 13: 55–60. 10.1016/j.cois.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 30.Viney M, Diaz A. Phenotypic plasticity in nematodes: Evolutionary and ecological significance. Worm. 2012; 1(2):98–106. 10.4161/worm.21086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liufu Z, Zhao Y, Guo L, Miao G, Xiao J, Lyu Y, et al. Redundant and incoherent regulations of multiple phenotypes suggest microRNAs' role in stability control. Genome Res. 2017; 27(10): 1665–1673. 10.1101/gr.222505.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shomron N. MicroRNAs and developmental robustness: a new layer is revealed. PLoS Biol. 2010; 8(6): e1000397 10.1371/journal.pbio.1000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardie J. Juvenile hormone mimics the photoperiodic apterization of the alate gynopara of aphid, Aphis fabae. Nature 1980; 286: 602–604. 10.1038/286602a0 [DOI] [Google Scholar]
- 34.Zera AJ. Evolutionary genetics of juvenile hormone and ecdysteroid regulation in Gryllus: a case study in the microevolution of endocrine regulation. Comp Biochem Physiol A Mol Integr. Physiol. 2006; 144: 365–479. 10.1016/j.cbpa.2005.11.026 [DOI] [PubMed] [Google Scholar]
- 35.Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science 2002; 297(5579): 249–252. 10.1126/science.1071468 [DOI] [PubMed] [Google Scholar]
- 36.Schrempf A, Heinze J. Proximate mechanisms of male morph determination in the ant Cardiocondyla obscurior. Evol Dev. 2006; 8(3): 266–272. 10.1111/j.1525-142X.2006.00097.x [DOI] [PubMed] [Google Scholar]
- 37.Oettler J, Platschek T, Schmidt C, Rajakumar R, Favé M-J, Khila A, et al. Interruption points in the wing gene regulatory network underlying wing polyphenism evolved independently in male and female morphs in Cardiocondyla ants. J Exp Zool (Mol Dev Evol). 2018; 1–10 10.1002/jez.b.22834 [DOI] [PubMed] [Google Scholar]
- 38.Wegener J, Huang ZY, Lorenz MW, Lorenz JI, Bienefeld K. New insights into the roles of juvenile hormone and ecdysteroids in honey bee reproduction. J Insect Physiol. 2013; 59: 655–661. 10.1016/j.jinsphys.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 39.Fu X, Li T, Chen J, Dong Y, Qiu J, Kang K, et al. Functional screen for microRNAs of Nilaparvata lugens reveals that targeting of glutamine synthase by miR-4868b regulates fecundity. J Insect Physiol. 2015; 83: 22–29. 10.1016/j.jinsphys.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 40.Li T, Chen J, Fan X, Chen W, Zhang W. MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag Sci. 2017; 73(7): 1529–1537. 10.1002/ps.4492 [DOI] [PubMed] [Google Scholar]
- 41.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004; 2(11): e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thadani R, Tammi MT. MicroTar: predicting microRNA targets from RNA duplexes. BMC Bioinformatics. 2006; 7 Suppl 5: S20 10.1186/1471-2105-7-S5-S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007; 39(10): 1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- 45.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10(10): 1507–1517. 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Zhang D, Yao Q, Zhang J, Dong X, Tian H, Zhang W. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper Nilaparvata lugens. Insect Mol Biol. 2010; 19: 777–786. 10.1111/j.1365-2583.2010.01038.x [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Linse KD, Taub-Montemayor TE, Rankin MA. Comparison of radioimmunoassay and liquid chromatography tandem mass spectrometry for determination of juvenile hormone titers. Insect Biochem Mol Biol. 2007; 37(8): 799–807. 10.1016/j.ibmb.2007.05.019 [DOI] [PubMed] [Google Scholar]
- 48.Zhou J, Qi Y, Hou Y, Zhao J, Li Y, Xue X, et al. Quantitative determination of juvenile hormone III and 20-hydroxyecdysone in queen larvae and drone pupae of Apis mellifera by ultrasonic-assisted extraction and liquid chromatography with electrospray ionization tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2011; 879(25): 2533–2541. 10.1016/j.jchromb.2011.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Green (NlInR1) and red (Nlu-miR-34) signals overlap to show yellow signals, suggesting Nlu-miR-34 interact directly with NlInR1 in the cells of wing buds. Control 1, Nlu-miR-34 antisense and scrambled mRNA probe; Control 2, scrambled miRNA and NlInR1 antisense probe; Control 3, scrambled miRNA and scrambled mRNA probe. Scale bars: 100 μm.
(TIF)
The data are presented as means ± SEM, three replicates. *p < 0.05 (Students’ t-test).
(TIFF)
The data are presented as means ± SEM, three replicates. *p < 0.05, **p < 0.01 (Students’ t-test).
(TIFF)
The data are presented as means ± SEM, three replicates. **p < 0.01, ***p < 0.001 (Students’ t-test).
(TIFF)
(A) Dual luciferase reporter assays confirmed the interactions between Nlu-miR-34 and NlInR1 in vitro (p = 2.35e-13). (B, C) Dual luciferase reporter assays confirmed the interactions between Nlu-miR-989b and NlInR2 (p = 1.43e-12), and the interactions between Nlu-miR-989c and NlInR2 in vitro (p = 9.48e-13). Data are means ± SEM, six replicates. (D, E) Overexpression of Nlu-miR-989b and Nlu-miR-989c in SW-strain. Wing rate data are presented as means ± SEM, three replicates. ***p < 0.001 (Student’s t-test).
(TIFF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The Nlu-miR-34 sequence, including the intact 5’UTR, was deposited in GenBank (MG894367). All other relevant data are within the manuscript and its Supporting Information files.