Abstract
Constitutive activation of the epidermal growth factor receptor (EGFR) because of somatic mutations of the EGFR gene is commonly observed in tumors of non–small cell lung cancer (NSCLC) patients. Consequently, tyrosine kinase inhibitors (TKI) targeting the EGFR are among the most effective therapies for patients with sensitizing EGFR mutations. Clinical responses to the EGFR-targeting TKIs are evaluated through 2-[18F]fluoro-2-deoxy-glucose (18FDG)-PET uptake, which is decreased in patients responding favorably to therapy and is positively correlated with survival. Recent studies have reported that EGFR signaling drives glucose metabolism in NSCLC cells; however, the precise downstream effectors required for this EGFR-driven metabolic effect are largely unknown. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3) is an essential glycolytic regulator that is consistently overexpressed in lung cancer. Here, we found that PFKFB3 is an essential target of EGFR signaling and that PFKFB3 activation is required for glycolysis stimulation upon EGFR activation. We demonstrate that exposing NSCLC cells harboring either WT or mutated EGFR to EGF rapidly increases PFKFB3 phosphorylation, expression, and activity and that PFKFB3 inhibition markedly reduces the EGF-mediated increase in glycolysis. Furthermore, we found that prolonged NSCLC cell exposure to the TKI erlotinib drives PFKFB3 expression and that chemical PFKFB3 inhibition synergizes with erlotinib in increasing erlotinib's anti-proliferative activity in NSCLC cells. We conclude that PFKFB3 has a key role in mediating glucose metabolism and survival of NSCLC cells in response to EGFR signaling. These results support the potential clinical utility of using PFKFB3 inhibitors in combination with EGFR-TKIs to manage NSCLC.
Keywords: lung cancer; glycolysis; metabolism; epidermal growth factor (EGF); epidermal growth factor receptor (EGFR); 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase; erlotinib; PFKFB3; tumorigenesis; tyrosine kinase inhibitor; small molecule antagonist
Introduction
Non–small cell lung cancer (NSCLC)3 accounts for 80% of all lung cancers and is the leading cause of cancer-related death in men and in women (1). Aberrant activation of the epidermal growth factor receptor (EGFR) plays a central role in NSCLC tumorigenesis; therefore, EGFR is regarded a critical target in the clinical management of NSCLCs. EGFR mutations occur in 10–20% of NSCLCs, where the two most common alterations are deletions in exon 19 (delE746-A750) and L858R substitution in exon 21 (2). These mutations favor an “active” kinase conformation that promotes ligand-independent transactivation of the EGFR and are closely associated with favorable clinical responses to EGFR tyrosine kinase inhibitors (TKIs) (3, 4). Although the use of EGFR-TKIs like erlotinib and gefitinib have improved progression-free survival in patients with EGFR-driven NSCLC, they have failed to show an overall survival benefit largely because of primary or secondary resistance to this therapy (5, 6).
Recent studies have shown that activation of the EGFR increases glucose consumption and metabolism in NSCLC cells (7). Clinically, alterations in glucose metabolism measured by 2-[18F]fluoro-2-deoxy-glucose (18FDG)-PET uptake are observed within days of initiating EGFR-TKI therapy in NSCLC patients (8). These changes in tumor metabolism precede those in tumor size, thus 18FDG-PET is routinely used to predict responses to EGFR-TKIs in xenografts and lung cancer patients (9–12). Taken together, these data indicate that glucose metabolism is an early target of EGFR-TKI and suggest that suppression of glucose metabolism may be essential to achieve therapeutic responses to EGFR-TKIs. Although the stimulation of glucose metabolism driven by EGFR signaling has recently been found to involve increased GLUT3 expression (7), relatively little is known about the requirement of other downstream glycolytic regulators to stimulate glucose metabolism in response to EGFR activation in NSCLC.
The 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases (PFKFB) family of enzymes are essential glycolytic activators whose function is to catalyze the synthesis and degradation of fructose-2,6-bisphosphate (F26BP) (13). F26BP is an allosteric activator of 6-phosphofructo-1-kinase (PFK-1), a rate-limiting enzyme and essential control point in the glycolytic pathway (14, 15). The PFKFB3 family member is of particular interest because it is consistently overexpressed in transformed tissues, has the highest kinase to bisphosphatase ratio, and is required for tumorigenic growth (16, 17). Although it has been known that EGF stimulation leads to high levels of glycolysis in cells (18) and PFKFB3 expression is up-regulated in lung tumors relative to normal tissue (19), a direct link between this major glycolytic regulator and EGFR has not been established.
Here, we identify EGFR as a novel regulator of PFKFB3 and demonstrate that EGFR activation rapidly increases PFKFB3 phosphorylation and expression. Additionally, we show that PFKFB3 is required for the increase of glycolysis in response to EGF and for the survival of NSCLC cells. Importantly, we demonstrate that persistent expression of PFKFB3 in response to MAPK-pathway activation confers resistance to erlotinib in NSCLC cells expressing mutated EGFR (mutEGFR). Inhibition of PFKFB3 using a small molecule antagonist developed by our group (PFK158) increases sensitivity to EGFR-TKIs in NSCLC cells with either WT or mutEGFR genotype. Our results indicate that targeting PFKFB3 in combination with EGFR-TKIs may have a clinical role in the management of NSCLC.
Results
EGFR activation promotes PFKFB3 phosphorylation and expression in NSCLCs
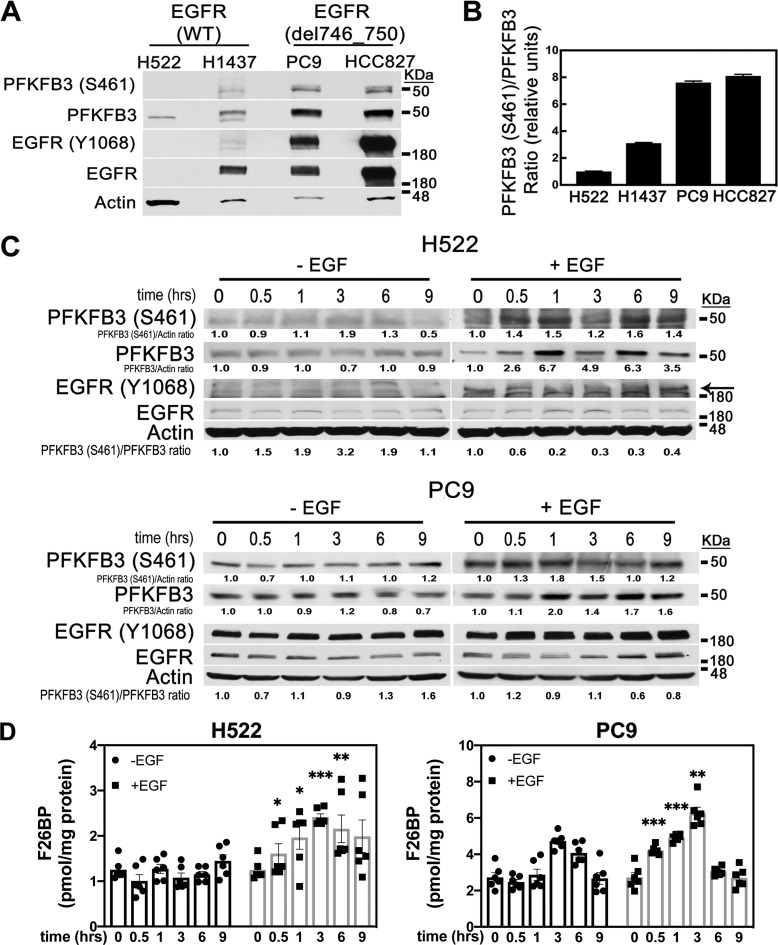
Given that EGFR signaling regulates glucose metabolism in NSCLC cells (20), we postulated that PFKFB3, a key glycolytic driver, might be regulated by EGFR. Initially, we evaluated PFKFB3 phosphorylation and expression levels in a panel of NSCLC cell lines expressing either WT EGFR (H522, H1437) or EGFR harboring the delE746-A750 activating deletion (PC9, HCC827). Immunoblot analysis of serum-starved NSCLCs revealed that the cell lines expressing mutEGFR had significantly higher levels of PFKFB3 relative to the NSCLC cells expressing WT-EGFR (Fig. 1A). Increased phosphorylation of Y1068 EGFR confirmed constitutive activation of EGFR in both cell lines expressing mutEGFR. Notably, elevated PFKFB3 expression was accompanied with a marked increase in phosphorylation at S461 (Fig. 1, A and B) and correlated with elevated levels of EGFR phosphorylation in mutEGFR cell lines. Importantly, H1437 cells with increased copy number of WT-EGFR failed to sustain elevated PFKFB3 levels in the absence of EGF, indicating that activated EGFR is required to induce PFKFB3 expression and phosphorylation.
Figure 1.
Regulation of PFKFB3 by EGFR activation in WT and mutEGFR cell lines. A, NSCLCs were serum starved for 24 h and analyzed for PFKFB3 and EGFR expression and phosphorylation by Western blotting. B, quantitative densitometry analysis is shown as the ratio of S461 PFKFB3 to PFKFB3 protein normalized to H522 cells ± S.E. from three independent experiments (n = 3). C, H522 and PC9 cells were serum starved for 24 h followed by stimulation with 50 ng/ml EGF for 0–9 h. Protein levels were evaluated by Western blotting at the indicated times post-EGF stimulation. Arrow indicates Y1068 EGFR (top band). PFKFB3 (S461)/actin; PFKFB3/actin, and PFKFB3(S461)/PFKFB3 ratios were quantified and normalized to time zero for each experimental condition. D, F26BP was measured using an enzyme-coupled reaction dependent on NADH oxidation. Error bars, mean ± S.E. of three independent experiments (n = 6). p values are shown as follows: *, <0.05; **, <0.01; and ***, <0.001.
Ligand-stimulated EGFR increases PFKFB3 phosphorylation, expression, and activity in NSCLCs
Activation of WT-EGFR requires ligand-dependent dimerization of the receptor resulting in the phosphorylation of the tyrosine residues within its cytoplasmic tail (21). To investigate the effects of EGF on PFKFB3 phosphorylation and expression, we stimulated H522 (WT-EGFR) and PC9 (mutEGFR) cells with EGF and monitored PFKFB3 phosphorylation and expression status over a period of 9 h. EGF exposure resulted in a significant increase in PFKFB3 S461 phosphorylation after 30 min of EGF stimulation in both H522 and PC9 cells. This increase in PFKFB3 phosphorylation was dynamic as shown by the decrease in PFKFB3 S461 levels 3 h post EGF treatment followed by an increase at the 6-h time point in H522 cells. We also observed an up-regulation of PFKFB3 protein levels in both H522 and PC9 cells upon EGF treatment with a maximum increase noted 1 h post stimulation (Fig. 1C). Notably, although EGF stimulation increases PFKFB3 phosphorylation, the ratio of phospho-PFKFB3 to PFKFB3 either remained relatively the same (PC9 cells) or decreased (H522 cells) given the increase in PFKFB3 levels. Thus, we conclude that EGF stimulation increases PFKFB3 expression leading to the observed increase in PFKFB3 phosphorylation. Next, we measured the effects of EGF stimulation on the steady-state concentration of the PFKFB3 enzyme product F26BP. In the absence of EGF stimulation, we observed a 2-fold increase in basal F26BP levels in PC9 cells as compared with H522 cells (Fig. 1D). These results are consistent with elevated basal phosphorylation and PFKFB3 levels in nonstimulated PC9 cells. Stimulation with EGF resulted in a rapid increase in the intracellular concentration of F26BP in both H522 and PC9 cells. Given that both PFKFB3 and phospho-PFKFB3 have enzymatic activity, we believe that the increase in F26BP production upon EGF stimulation is predominantly driven by the increase in PFKFB3 expression (22). Taken together, these results confirm that activation of EGFR by EGF drives PFKFB3 phosphorylation, expression, and activity.
Active EGFR is required to increase PFKFB3 phosphorylation and protein levels in NSCLCs
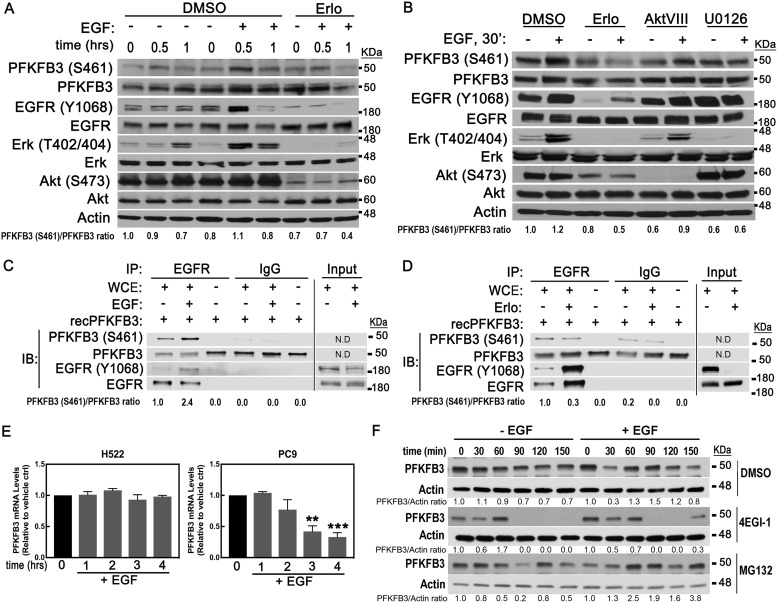
To validate that activation of EGFR is required to induce PFKFB3 phosphorylation and protein expression, we stimulated PC9 cells with EGF in the presence of the EGFR inhibitor erlotinib. Treatment with erlotinib effectively blocked EGFR activation and downstream signaling as shown by the lack of phosphorylation of EGFR, ERK, and Akt. Consequently, we observed a block of the EGF-driven increase in PFKFB3 phosphorylation (Fig. 2A). Knowing that EGFR activation drives multiple signaling pathways, we sought to evaluate whether PI3K/Akt and/or RAS/MAPK were required for the EGF-driven increase in PFKFB3 phosphorylation and expression. Treatment of PC9 cells with either erlotinib or U0126 (MEK inhibitor) prior to stimulation with EGF abrogated the increase in PFKFB3 phosphorylation in response to EGF (Fig. 2B). Akt inhibition caused a significant drop in basal PFKFB3 phosphorylation but failed to block EGF stimulation of PFKFB3 phosphorylation, indicating that Akt does not contribute to the EGF-driven phosphorylation of PFKFB3. Importantly, we noted that U0126 increased basal phosphorylation levels of EGFR and completely blocked further activation of EGFR in response to EGF (Fig. 2B). Taken together, these data indicate that EGF activates EGFR, which in turn rapidly increases PFKFB3 protein and phosphorylation levels.
Figure 2.
Activated EGFR regulates PFKFB3 phosphorylation and expression. A, PC9 cells were serum starved for 24 h, treated with either vehicle (DMSO) or an EGFR inhibitor (erlotinib; 5 μm) 30 min prior to stimulation with EGF and harvested at the indicated times for Western blot analysis. B, PC9 cells were treated with either vehicle (DMSO), EGFR inhibitor (erlotinib; 5 μm), Akt inhibitor (AktVIII; 10 μm), or MEK inhibitor (U0126; 10 μm) 30 min prior to EGF stimulation. Samples were collected 30 min post EGF stimulation and immunoblotted with the indicated antibodies. PFKFB3(S461)/PFKFB3 ratios were quantified and normalized to nontreated samples (time zero). C and D, PC9 cells were treated with either EGF for 2 min or 0.5 μm erlotinib for 1 h prior to harvesting. Phosphorylation of recombinant PFKFB3 by EGFR was assessed by incubating immunoprecipitated EGFR with recombinant PFKFB3 in a kinase reaction mixture and allowed to incubate for 1 h. Reactions were stopped with 2× Laemmli buffer followed by elution at 60 °C for 30 min and immunoblotting. N.D., not determined. E, PFKFB3 mRNA levels were evaluated by real time PCR in H522 and PC9 cells treated with EGF for the indicated times. Error bars, mean ± S.E. of three independent experiments (n = 9). **, p < 0.01; ***, p < 0.001; compared with untreated control. F, PC9 cells were serum starved for 24 h, stimulated with EGF and, 15 min later, cells were treated with either DMSO, a translation inhibitor (4EGI-1; 50 μm), or a proteasome inhibitor (MG132; 10 μm). Lysates were collected 0–150 min post treatment and subjected to Western blot analysis. PFKFB3/actin ratios were quantified and normalized to nontreated samples.
Next, we assessed whether activated EGFR can phosphorylate PFKFB3 using an in vitro kinase assay. EGFR was immunoprecipitated from PC9 cells treated with either EGF or erlotinib and incubated with recombinant PFKFB3. We found that incubation of recombinant PFKFB3 with immunoprecipitated EGFR resulted in the phosphorylation of recombinant PFKFB3. Notably, stimulation of EGFR with EGF for as short as 2 min (to avoid recruitment of downstream kinase effectors) resulted in increased phosphorylation of recombinant PFKFB3 (Fig. 2C). In contrast, incubation with EGFR exposed to erlotinib resulted in decreased PFKFB3 phosphorylation despite the increase in immunoprecipitated EGFR levels (Fig. 2D). Based on previous studies we speculate that this increase in immunoprecipitated EGFR in the erlotinib-treated sample reflects a change in antigen-antibody binding because of a conformational change in EGFR protein (23–25). Altogether, these results suggest that PFKFB3 is a substrate of EGFR for phosphorylation at S461.
To elucidate the mechanism by which EGFR activation increases PFKFB3 protein levels, we next examined changes in PFKFB3 mRNA levels in EGF-stimulated H522 and PC9 cells. Interestingly, EGF stimulation had no effect on PFKFB3 mRNA levels in H522 cells and resulted in a marked decrease in PFKFB3 mRNA levels in PC9 cells (Fig. 2E). The lack of correlation between PFKFB3 mRNA and protein levels in response to EGF suggests a posttranscriptional regulation of PFKFB3 by EGF. To test whether EGF stimulation results in increased protein translation or decreased degradation, serum-starved PC9 cells were treated with either a translation inhibitor (4EGI-1) or a proteasome inhibitor (MG132) 15 min after stimulation with EGF. We found that treatment with the translation inhibitor led to a near complete loss of PFKFB3 protein, indicating that de novo protein synthesis is required to maintain both basal and EGF-driven PFKFB3 levels because of constitutive degradation of PFKFB3 (Fig. 2F). Culture of PC9 cells in the presence of EGF followed by proteasomal inhibition resulted in PFKFB3 accumulation suggesting an increase in PFKFB3 protein synthesis driven by EGF. Taken together, these results show that PFKFB3 has a short half-life time and that transactivation of EGFR by EGF increases PFKFB3 by a translation dependent mechanism.
PFKFB3 is required for EGFR-mediated up-regulation of glycolysis
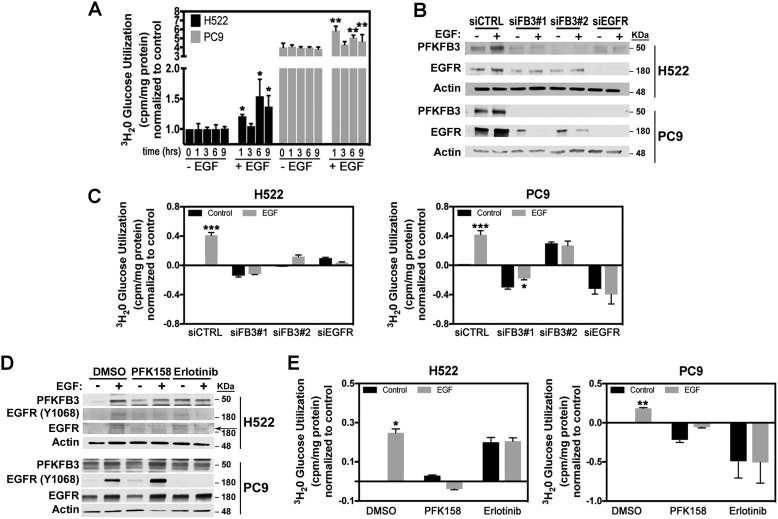
Previous observations showed that EGFR regulates glucose metabolism in lung adenocarcinoma cells (7, 26). Given that PFKFB3 protein and its product F26BP exert significant control of the glycolytic flux to lactate in vivo and in vitro, we postulated that PFKFB3 might regulate the EGFR-driven increase in glucose metabolism. To determine whether increased PFKFB3 expression and activation promote EGFR-driven glucose metabolism we first measured glycolytic flux rate in H522 and PC9 cells in the absence or presence of EGF. We found that mutEGFR PC9 cells displayed a 3-fold increase in basal glycolysis compared with the H522 WT-EGFR cells in line with their increased PFKFB3 expression and kinase activity (Fig. 3A). Additionally, EGF stimulation resulted in a marked increase in glycolysis after 1 h of exposure in both H522 and PC9 cells. Consistent with the fluctuations in PFKFB3 phosphorylation and expression observed in response to EGF (Fig. 1C), we noted a drop in glycolytic flux after 3 h of EGF exposure followed by a second increase in glycolytic flux after 6 to 9 h of treatment.
Figure 3.
Increased glycolysis in response to EGF requires PFKFB3. A, H522 and PC9 cells were serum starved for 24 h followed by stimulation with EGF for 0–9 h. Glycolysis was measured by the release of 3H2O by enolase. Data were normalized to nonstimulated H522 cells. Error bars, mean ± S.E. of three independent experiments (n = 12). p values were calculated against the vehicle-treated sample for the corresponding time point. B, H522 and PC9 cells were transfected with either a control siRNA (siCTRL), two independent PFKFB3 siRNA (siFB3#1 and siFB3#2) or a pool of three siRNAs against EGFR (siEGFR) 48 h prior to EGF stimulation. Shown are PFKFB3 and EGFR protein expression by Western blot analysis after either 9 h (H522) or 6 h (PC9) of EGF stimulation. C, cells were treated as described in B and glycolysis was measured by the release of 3H2O by enolase. Results were normalized to non–EGF-stimulated cells transfected with siCTRL. Error bars, mean ± S.E. of three independent experiments (n = 12). p values were calculated against the vehicle-treated sample for each transfection condition. D, H522 and PC9 cells were treated with either PFK158 (5 μm) or erlotinib (5 μm) 1 h prior to stimulation with EGF. Shown are PFKFB3 and EGFR protein expression by Western blot analysis after 1 h of EGF stimulation. Arrow indicates EGFR. E, cells were treated as described in D and glycolysis was measured 1 h post EGF treatment and data were normalized to nonstimulated cells treated with DMSO. Error bars, H522, mean ± S.E. of three independent experiments (n = 12); PC9, mean ± S.E. of two independent experiments (n = 8). p values were calculated against the non–EGF-stimulated sample for each treatment. p values are shown as follows: *, <0.05; **, <0.01; and ***, <0.001.
To evaluate the requirement of PFKFB3 in EGFR-mediated glucose metabolism, we used two PFKFB3-specific siRNAs and examined glycolysis in H522 and PC9 cells stimulated with EGF. Additionally, to ensure that the EGF-driven metabolic effect is dependent solely on EGFR and not on other ERBB family members, we suppressed EGFR by using a pool of EGFR-specific siRNAs. Initially, we confirmed selective suppression of PFKFB3 or EGFR relative to negative control siRNA in transfected H522 and PC9 cells (Fig. 3B). Moreover, silencing of EGFR resulted in a marked decrease in PFKFB3 protein levels in both H522 and PC9 cells, confirming that basal and EGF-induced PFKFB3 expression are both maintained by EGFR-dependent pathways in NSCLCs (Fig. 3B). Interestingly, we found that transfection with both PFKFB3-specific siRNAs decreased EGFR levels. Silencing of PFKFB3 or EGFR completely abrogated the EGF-mediated increase in glycolysis in H522 and PC9 cells (Fig. 3C).
Our group previously characterized a small molecule inhibitor of PFKFB3 (PFK158) that suppresses glucose metabolism (27, 28). We predicted that acute inhibition of PFKFB3 would block the increase in glycolysis driven by EGFR activation. We then examined the effect of PFK158 on PFKFB3, EGFR (Y1068), and EGFR protein expression in the absence and presence of EGF and observed relatively no effect on the EGF-induced expression of PFKFB3, EGFR (Y1068) and EGFR (Fig. 3D). We measured glycolytic flux in H522 and PC9 cells pretreated for 1 h with either PFK158 or erlotinib and stimulated with EGF (1 h). Addition of either PFK158 or erlotinib dramatically attenuated glycolysis levels in PC9 cells, confirming higher dependence of mutEGFR cells on glucose metabolism. Under EGF stimulation, we observed an increase in the glycolytic rate in H522 and PC9 vehicle-treated cells which was attenuated by the acute treatment with either PFK158 or erlotinib (Fig. 3E). Surprisingly, treatment of H522 cells with erlotinib resulted in a marked increase in basal glycolysis but effectively blocked the effect of EGF. Taken together, these results demonstrate that PFKFB3 is required for the stimulation of glucose metabolism driven by the activation of EGFR. Collectively, these results demonstrate that EGF stimulation and activating mutations of EGFR increase glucose metabolism by regulating PFKFB3 expression and activity. Furthermore, our findings suggest that PFKFB3 expression may be essential for tumorigenesis in EGFR-driven NSCLC.
Activation of the MAPK pathway sustains PFKFB3 expression in response to erlotinib treatment
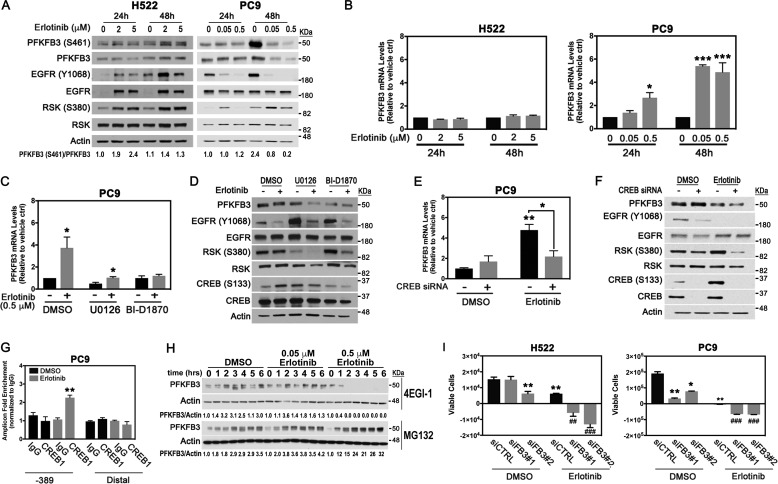
Based on our observations that EGFR silencing dramatically attenuated PFKFB3 expression in H522 and PC9 cells (Fig. 3B), we next examined the relative expression of PFKFB3 in cells treated with erlotinib for 24 to 48 h. We found that prolonged exposure of H522 cells to erlotinib caused a dose-dependent increase in PFKFB3 phosphorylation at 24 and 48 h, followed by an increase in expression after 48 h of treatment (Fig. 4A). Notably, the increase in PFKFB3 phosphorylation and expression was coincident with increased EGFR phosphorylation and expression. These observations are in agreement with previously published data showing that treatment with erlotinib increases affinity of EGFR to EGF, which in turn, stimulates receptor activation (23, 29). We speculate that increased PFKFB3 phosphorylation and expression in response to erlotinib in WT-EGFR cells is a result of increased EGFR activity driven by erlotinib. Next, we examined PFKFB3 expression in PC9 cells treated with erlotinib and found that erlotinib exposure blocked PFKFB3 phosphorylation and decreased PFKFB3 expression after 24 and 48 h of exposure (Fig. 4A). Notably, we have consistently observed an increase in EGFR Y1068 in H522 and PC9 cells after 48 h in culture in the absence of treatment. These results agree with previous studies indicating that EGFR expression and phosphorylation increases with higher cell density (30–32). Accordingly, we believe that the increase in EGFR Y1068 levels and the corresponding increase in phosphorylated PFKFB3 at the 48-hour time point is because of an increase in cell number in vehicle-treated cells. Importantly, although prolonged erlotinib treatment suppressed EGFR phosphorylation it promoted MAPK pathway reactivation in both cell lines (note increased RSK phosphorylation, Fig. 4A). Feedback activation of MAPK in response to TKIs has been previously demonstrated to contribute to an incomplete response to erlotinib (33, 34).
Figure 4.
PFKFB3 expression modulates response to erlotinib in NSCLC cells. A, H522 and PC9 cells were treated with the indicated concentrations of erlotinib for 24 to 48 h. Whole cell lysates were evaluated by Western blotting with the indicated antibodies. PFKFB3(S461)/PFKFB3 ratios were quantified and normalized to vehicle-treated samples at 24 h. B, PFKFB3 mRNA levels were evaluated by real-time PCR. Error bars, mean ± S.E. of three independent experiments, normalized to vehicle-treated samples (n = 9). C, PC9 cells were treated with either a MEK inhibitor (U0126; 20 μm) or an RSK inhibitor (BI-D1870; 20 μm) in the presence of 0.5 μm erlotinib for 48 h. PFKFB3 mRNA levels were evaluated by real time PCR and normalized to DMSO untreated control. Error bars, mean ± S.E. of three independent experiments (n = 9). D, PC9 cells were treated as described in C and protein levels were evaluated by Western blotting. E, PC9 cells were transfected with either a control siRNA or a pool of CREB1-specific siRNAs and treated with DMSO or erlotinib for 48 h. PFKFB3 mRNA levels were evaluated by real-time PCR and normalized to vehicle-treated control siRNA. Error bars, mean ± S.E. of three independent experiments (n = 9). F, PC9 cells were treated as described in E and protein levels were evaluated by Western blotting. G, ChIP assay experiments were performed with PC9 cells treated with 0.5 μm erlotinib for 24 h using antibodies to CREB1 or control mouse IgG and primers flanking −389 and distal regions (lacking CRE site) of the PFKFB3 promoter. Real-time PCR data were normalized to control IgG signal and shown as -fold enrichment. Error bars, mean ± S.E. of three independent experiments (n = 9). H, PC9 cells were treated with indicated concentrations of erlotinib for 24 h and exposed to a translation inhibitor (4EGI-1; 50 μm) or a proteasome inhibitor (MG132; 50 μm) for 0 to 6 h. Whole cell lysates were collected and immunoblotted with the indicated antibodies. PFKFB3/actin ratios were quantified and normalized to the corresponding time zero. I, H522 and PC9 cells were transfected with PFKFB3 siRNAs (siFB3#1, siFB3#2) followed by treatment with either vehicle or erlotinib (H522, 2 μm; PC9, 0.5 μm) for 48 h. Cell survival was evaluated by trypan blue exclusion. Shown are changes in absolute amounts of viable cells between 24 and 48 h post treatment. Error bars, mean ± S.E. of three independent experiments (n = 12). p values are shown as follows: *, <0.05; **, <0.01, and ***, p < 0.001 compared with vehicle treatment. ##, p <0.01, ###, p< 0.001, compared with indicated erlotinib treatment.
To investigate the mechanism regulating PFKFB3 expression in response to EGFR inhibition, we measured PFKFB3 mRNA expression levels in H522 and PC9 cells treated with erlotinib for 24 and 48 h. We found that prolonged exposure to erlotinib significantly up-regulated PFKFB3 mRNA expression in PC9 cells, whereas in H522 cells, PFKFB3 mRNA levels remained relatively unchanged (Fig. 4B). To evaluate the contribution of activated MAPK signaling to increased PFKFB3 mRNA expression, we examined the effect of inhibiting the MAPK pathway in combination with erlotinib in PC9 cells for 48 h. Inhibition of either MEK (U0126) or RSK (BI-D1870) drastically blocked the increase in PFKFB3 mRNA in response to erlotinib, indicating that MAPK signaling promotes PFKFB3 mRNA transcription upon erlotinib treatment (Fig. 4C). Furthermore, blockage of MEK or RSK activity in PC9 cells resulted in further loss of PFKFB3 as shown by the marked decrease in PFKFB3 levels in cells treated with U0126 or BI-D1870 and exposed to erlotinib (Fig. 4D).
The cAMP response element-binding (CREB) protein is one of the several transcription factors stimulated by the ERK-RSK axis of the MAPK pathway (35, 36). In our studies, erlotinib treatment of PC9 cells resulted in a sharp increase in CREB1 phosphorylation which was fully blocked by inhibiting MEK or RSK activation (Fig. 4D). Importantly, a pool of siRNAs against CREB1 completely inhibited the increase in mRNA (Fig. 4E) and led to reduced PFKFB3 protein levels (Fig. 4F) in response to erlotinib treatment. We speculated that heightened PFKFB3 mRNA expression in erlotinib-treated PC9 cells might be because of increased recruitment of CREB1 to the PFKFB3 promoter. We analyzed the PFKFB3 5′-promoter sequence using the TRANSFAC software (Biobase) (37) and identified several putative CREB1-binding sites located at the −389, −1890, and −2188 from the PFKFB3 transcription start site. We performed ChIP assay using a CREB1-specific antibody and appropriate primers sets for amplification of the sequences for each binding site, a distal region was used as a negative control. Our results demonstrate a 2-fold enrichment in total CREB1 binding to −389 region of PFKFB3 promoter after 24 h of erlotinib treatment (Fig. 4G). These results indicate that reactivation of the MAPK pathway in response to prolonged erlotinib exposure increases activation and recruitment of CREB1 to the promoter of the PFKFB3 gene resulting in increased PFKFB3 gene transcription.
Although prolonged treatment with erlotinib significantly increased PFKFB3 mRNA levels in PC9 cells, the protein levels remained lower than vehicle-treated samples. To investigate whether the discrepancy in between mRNA expression (Fig. 4B) and protein levels (Fig. 4, A and D) was because of increased PFKFB3 protein degradation, we treated cells with erlotinib for 24 h and evaluated PFKFB3 expression in the presence of a proteasome inhibitor (MG132) or a translation inhibitor (4EGI-1). Proteasomal inhibition for up to 6 h caused a time- and dose-dependent accumulation of PFKFB3 protein in PC9 cells treated with erlotinib, whereas inhibition of translation resulted in PFKFB3 loss after 1 h in cells treated with 0.5 μm erlotinib (Fig. 4H). These results indicate that erlotinib increases proteasomal degradation of PFKFB3 protein and that continuous translation of PFKFB3 is required to maintain PFKFB3 levels. All together, these results suggest that in mutEGFR cells erlotinib-activated MAPK signaling acts as a compensatory mechanism that promotes PFKFB3 transcription and ensures continuous translation of PFKFB3 thus minimizing its enhanced proteasomal degradation.
PFKFB3 supports NSCLC survival in response to erlotinib treatment
Given that prolonged exposure to erlotinib can sustain PFKFB3 expression in WT and mutEGFR cell lines by independent mechanisms, we next assessed the combined effect of PFKFB3 silencing with erlotinib on the survival of NSCLC cells. Transfection of H522 and PC9 cells with two PFKFB3-specific siRNAs in combination with erlotinib resulted in significant cytotoxicity on cell proliferation in both cell lines, whereas erlotinib alone showed a cytostatic effect (Fig. 4I). Our results suggest that PFKFB3 inhibition enhances erlotinib-driven toxicity.
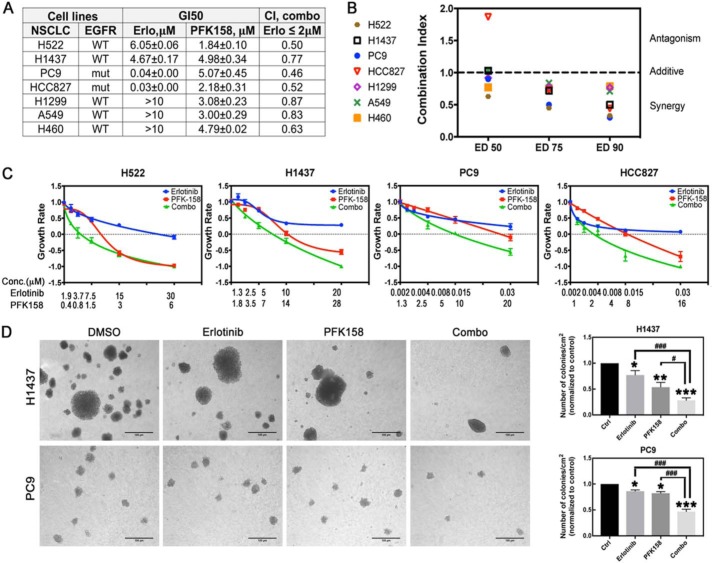
PFKFB3 inhibition synergistically increased the anti-proliferative effect of erlotinib in WT and mutEGFR cell lines
Our observations suggested that targeting PFKFB3 in combination with erlotinib may be a viable therapeutic approach for NSCLC cells to enhance sensitivity to erlotinib. To evaluate the efficacy of combining a PFKFB3 inhibitor with erlotinib, we treated WT-EGFR (H522, H1437, H1299, H460, A549) and mutEGFR (PC9, HCC827) cell lines alone or in combination and evaluated cell viability in trypan exclusion experiments 48 h post treatment. As expected, cell lines harboring an EGFR mutation were highly sensitive to erlotinib treatment, whereas WT-EGFR cell lines showed no effective growth inhibition (GI) at clinically relevant concentrations (2 μm) (Fig. 5A). Importantly, PFKFB3 inhibition effectively suppressed cell growth in all cell lines, suggesting high reliance of cells on PFKFB3 function (Fig. 5A). The effect of dual treatment was evaluated using the Chou-Talalay method (38). We observed synergy between erlotinib and PFK158 in all the NSCLC cell lines evaluated (combination indexes ≤1) when using clinically relevant concentrations of both drugs (Fig. 5A). Moreover, synergism was maintained at high effective dose levels (ED75, ED90) suggesting its therapeutic utility for cancer treatment (Fig. 5B) (38). To distinguish cytostatic from cytotoxic drug effects, we calculated growth rate values as described in Ref. 39, and found that erlotinib partially inhibited cell growth in WT-EGFR cell lines and exhibited a cytostatic effect in cells expressing mutEGFR at the drug concentrations tested (growth rate (GR) values ≥0) (Fig. 5C). In contrast, treatment with PFK158 as a single agent resulted in a cytotoxic effect (GR values <0) in all the cell lines except PC9 cells. Dual treatment significantly increased cytotoxicity in all the cell lines (Fig. 5C), suggesting the clinical potency of the drug combination. Next, we evaluated the efficacy of combined EGFR and PFKFB3 inhibition on tumor growth in vitro using an anchorage-independent growth assay in WT-EGFR (H1437, given that H522 cells were unable to grow in soft agar) and mutEGFR (PC9) cell lines. We found that the drug combination significantly inhibited colony formation compared with individual treatment (Fig. 5D). These observations suggest that PFKFB3 plays a role in mediating resistance to EGFR inhibition and validate the utility of dual inhibition of PFKFB3 and EGFR activity in tumor cells with various EGFR mutation status.
Figure 5.
PFK158 synergizes with erlotinib in WT and mutEGFR cell lines. A, WT-EGFR (H522, H1437, H1299, H460, A549) and mutEGFR (PC9, HCC827) cells were exposed to either erlotinib, PFK158, or the combination for 48 h. Cell viability was evaluated by trypan blue exclusion and used to determine the GI50 (drug concentration that causes 50% growth inhibition) relative to the vehicle control cells. Combination index for drug combination with erlotinib ≤2 μm was calculated using the CalcuSyn program based on the Chou-Talalay method. Mean ± S.D. of three independent experiments (n = 6). B, combination index at various effective doses (ED) for the constant drug ratio for different NSCLCs lines was calculated. Data shown are from a representative experiment. C, NSCLC cells were treated with PFK158 and erlotinib as single drugs or in combination at the indicated concentrations for 48 h. Cell viability was evaluated by trypan blue exclusion experiments. Data are plotted as growth rate values as indicated in methods, where GR >0 indicates inhibited growth, GR = 0, cytostatic effect and GR <0, cytotoxic effect. Error bars, mean ± S.E. of three independent experiments (n = 6). D, H1437 cells were seeded in soft-agar plates and exposed to 0.45 μm erlotinib and 0.62 μm PFK158 alone or in combination for 15 days. PC9 cells were seeded in soft-agar plates and exposed to 3.12 nm erlotinib and 0.31 μm PFK158 alone or in combination for 10 days. Left panel, representative images of formed colonies are shown (20× magnification). Scale bar, 100 μm. Right panel, numbers of colonies/cm2 normalized to vehicle-treated cells. Error bars, mean ± S.E. of three independent experiments (n = 18). p values are shown as follows: *, <0.05; **, <0.01; and ***, <0.001 (compared with vehicle treatment). #, p < 0.05; ###, p < 0.001, compared with indicated drug treatment.
Discussion
In this study, we demonstrate that EGFR activation leads to the up-regulation of PFKFB3 expression and activity and that PFKFB3 inhibition significantly improves the efficacy of erlotinib in NSCLC cells. Adding to the understanding that activation of EGFR stimulates glycolytic flux and regulates the expression of key glycolytic enzymes, including hexokinase and pyruvate kinase (20, 40), we provide evidence that PFKFB3 is stimulated by EGFR signaling and is required for the EGFR-driven glycolysis increase in NSCLC cells. Alteration in glucose metabolism is a common feature of the metabolic reprogramming exhibited by tumor cells to support aberrant proliferation. Increased glucose utilization relies largely on the up-regulation of PFKFB3 activity, a powerful regulator of the entire glycolytic pathway. From a therapeutic perspective, our findings imply that targeting PFKFB3 in combination with EGFR inhibition may be essential to positively impact the clinical efficacy and outcomes of EGFR-TKIs.
Targeted therapies against EGFR have been approved as a standard first-line therapy in NSCLC patients with sensitive EGFR mutations (41). Although initially designed to recognize WT kinase active site of EGFR, the first generation of TKIs, such as erlotinib and gefitinib, showed clinical benefits in patients with NSCLCs bearing mutated EGFR (42). Although the presence of mutations in the EGFR gene is associated with a favorable response to EGFR-TKIs, many patients do not respond to treatment because of intrinsic resistance. Importantly, almost all patients that have initial responses to EGFR-TKIs eventually develop resistance largely because of acquired mutations in exons 19 and 20 (T790M, most commonly) (43, 44). Patients treated with a third-generation drug developed to target secondary T790M mutations, osimertinib, develop resistance after 1 to 2 years of treatment (45). Beyond the intrinsic and genetically acquired resistance to EGFR-TKIs, bypass activation of downstream pathways such as MAPK and PI3K have been shown to result in adaptive resistance. Our results showed that MAPK reactivation upon prolonged erlotinib exposure increases PFKFB3 transcription thereby attenuating the erlotinib-induced PFKFB3 degradation in mutEGFR cells. Elevated PFKFB3 mRNA levels in response to erlotinib were maintained by increased recruitment of CREB1 to the PFKFB3 promoter. The CREB1 transcription factor provides exclusive mRNA transcription and further translation under stress conditions and its increased activity is associated with negative prognosis in NSCLCs (46). These findings are in agreement with recently published data showing that prolonged treatment of EGFR-addicted cancer cells with TKIs displayed a metabolic shift toward increased glycolysis and lactate production (47) and that glucose inhibition can overcome acquired resistance to EGFR TKIs (40). Based on our results we speculate that persistent expression of PFKFB3 because of MAPK reactivation allows cancer cells to survive the effects of EGFR-TKIs and ultimately contribute to its resistance. Importantly, we discovered that chronic exposure of erlotinib in WT-EGFR cells up-regulates PFKFB3 expression likely because of increased EGFR signaling. These results raise the question of whether the lack of response of patients with WT-EGFR to TKIs is in part because of increased PFKFB3 and glycolysis. Although more intensive investigation is required to address this question, we have demonstrated that inhibiting PFKFB3 significantly increases cellular responses to erlotinib and reduces survival of WT-EGFR NSCLC cells.
Although high expression of PFKFB3 is observed in a multitude of cancers and is associated with increased tumor proliferation and survival, the molecular mechanisms regulating PFKFB3 expression and activity are not completely understood. At the transcriptional level, PFKFB3 expression is regulated by HIF-1α, the progesterone and estrogen receptors (28, 48, 49). Moreover, loss of phosphatase and tensin homolog is known to reduce degradation of PFKFB3 whereas activation of protein kinase B (Akt) results in phosphorylation of PFKFB3 at S461 leading to its activation (50, 51). Here, we describe that EGF stimulation of both WT and mutEGFR regulate PFKFB3 expression via its posttranslational regulation. Importantly, we also show that the steady-state concentration of the PFKFB3 product F26BP correlates with PFKFB3 phosphorylation and expression in response to EGF. Although the role of EGF in regulating glucose metabolism has been described previously, the downstream targets required to promote glycolysis in response to EGF remain largely unknown. Here we identify PFKFB3 as a molecular effector of EGF-driven signaling. Furthermore, we provide evidence that enhanced glycolysis in response to EGF depends on PFKFB3 activity, as determined using a small molecule inhibitor of PFKFB3. Interestingly, although mutEGFR cells display higher basal levels of PFKFB3 phosphorylation and expression compared with WT-EGFR cells, stimulation with EGF further increases PFKFB3 expression and activity. These results are in concert with previous studies showing that whereas mutEGFR have decreased dependence on ligand stimulation for activation, the presence of EGF further increases receptor activation (52).
In our experiments we observed that EGFR silencing dramatically decreases PFKFB3 expression. Surprisingly, PFKFB3 knockdown in serum-starved cells resulted in a marked reduction of EGFR expression in PC9 cells and a modest effect on EGFR expression in H522 cells (Fig. 3B). EGFR expression at the cell membrane is regulated by the presence of ligand and by the levels of endocytosis and trafficking of the receptor. In response to stimulation or drug treatment, internalized receptor could proceed to recycling or lysosomal degradation (53). Recently, it has been shown that glucose co-transporter SGLT1 stabilizes EGFR at the membrane in glucose-deprived cells, although enhanced glycolytic flux driven by mutEGFR increases receptor stability and is critical for NSCLC survival (40, 54). Our finding suggests that PFKFB3 might play some role in regulating EGFR expression or recycling; however, this speculation requires further investigation.
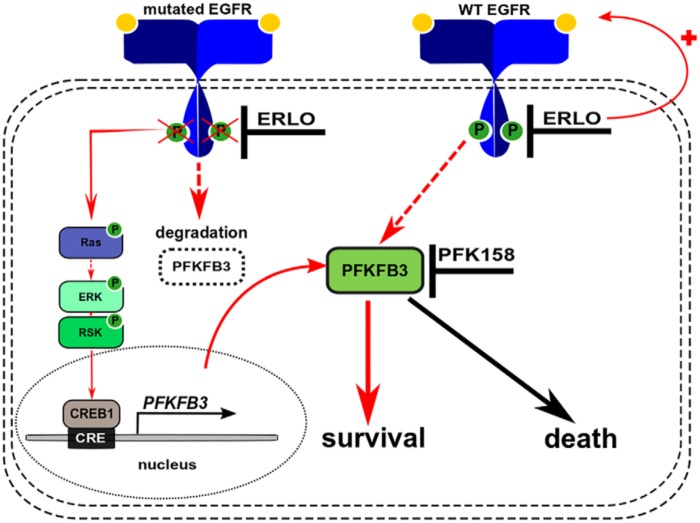
PFKFB3 can be used as a prognostic marker of lung adenocarcinomas and PFKFB3 inhibition with a small-molecule inhibitor reduces tumor growth in vivo (55–57). Here, we evaluated the efficacy of PFK158, the first-in-class small molecule inhibitor of PFKFB3, which has already shown some efficacy in a phase I clinical trial in patients with advanced solid tumors (27, 58, 59). Our in vitro experiments confirmed that PFK158 decreases cell survival of NSCLC cells (WT and mutEGFR). In dual treatment experiments, we confirmed the synergistic effect and improved cytotoxicity of drug combinations in cell lines with different EGFR mutation status. Although the mechanism by which PFKFB3 inhibition potentiates erlotinib activity in NSCLC cells is still under investigation, the results presented here support the superiority of the combined therapy and will be tested further in vivo. A model describing the role of PFKFB3 in modulating cellular responses to erlotinib is shown in Fig. 6. Synergy in treatment of NSCLCs cell lines with EGFR and PFKFB3 inhibitors opens the discussion of the putative role of PFKFB3 in regulating overall drug response to EGFR-TKIs, development of initial compensation, and its link to the secondary resistance to TKIs that requires further investigation.
Figure 6.
Model describing the putative mechanism of synergetic interaction between PFK158 and erlotinib in WT and mutEGFR NSCLC cell lines. In mutEGFR cells, chronic inhibition of EGFR signaling by erlotinib promotes reactivation of the MAPK pathway thereby stimulating PFKFB3 transcription via CREB1 binding to the PFKFB3 promoter. Enhanced PFKFB3 transcription compensates for the loss of protein because of targeted degradation by the proteasome and promotes cell survival. In WT-EGFR cells, prolonged erlotinib exposure stimulates EGFR signaling which, in turn, up-regulates PFKFB3 expression by an unknown mechanism and promotes cell survival. PFKFB3 inhibition with PFK158 blocks the prosurvival function of PFKFB3 resulting in a marked increase in cytotoxicity when used in combination with erlotinib. Solid arrows indicate established direct regulations of downstream targets. Dashed arrows indicate indirect activation where intermediate steps are involved but are not specified in this schematic.
In summary, our studies indicate that PFKFB3 is an essential regulatory effector of the EGFR pathway that serves to promote the survival of NSCLC cells. Consequently, our findings may have identified a novel and potentially efficacious strategy for the treatment of NSCLC lung cancer that uses an inhibitor of PFKFB3 in combination with EGFR-TKIs.
Experimental procedures
Reagents
EGF (E9644) was purchased from Sigma-Aldrich. Erlotinib (S1023, S7786) and BI-D1870 (S2843) were obtained from Selleck Chemicals; U0126 (v1121) was purchased from Promega. 4EGI-1 (15362) and MG132 (10012628) were obtained from Cayman Chemical Co. PFK158 (HY-12203) was purchased from MedChem Express.
Cell culture
H522, PC9, H1437, HCC827, A549, H460, and H1299 were purchased from the American Type Culture Collection (ATCC). H522, PC9, HCC827, H460, and H1299 cells were cultured in RPMI medium (Invitrogen) supplemented with 10% FBS (Clontech) and 50 μg/ml gentamicin (Life Technologies). H1437, A549 were cultured in DMEM (Invitrogen) supplemented with 10% FBS and gentamicin. Cells were incubated at 37 °C with 5% CO2. Cells were serum-starved in RPMI medium for 24 h prior to stimulation with 50 ng/ml EGF or 10 mm acetic acid as vehicle control.
siRNA transfection
PC9 and H522 cells were seeded in 6-well plates at a density of 6 × 104 and 3.5 × 105 cells/well, respectively, in 2 ml of complete medium 24 h prior to transfection. Transfections were performed using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instructions and allowed to incubate for the indicated times. For EGF stimulation experiments, the medium was changed to serum-free 24 h after transfection and allowed to incubate for 24 h prior to stimulation. The following siRNAs were used: PFKFB3 siRNA1 (HSS107860, Invitrogen), PFKFB3 siRNA2 (HSS107862, Invitrogen), EGFR siRNA (sc-29301, Santa Cruz Biotechnology), CREB1 siRNAs (sc-29281, Santa Cruz Biotechnology); control siRNAs that have no homology to any sequence in the human genome were used as the controls (12935-112, Invitrogen; sc-37007, sc-44231, Santa Cruz Biotechnology).
Antibodies and Western blotting
Whole cell lysates were harvested using RIPA buffer (Thermo Fisher) supplemented with protease inhibitors. Protein concentration was determined using the BCA protein assay kit (Thermo Fisher) following the manufacturer's instructions. Proteins were separated on 10% or 4–20% Mini PROTEAN TGX gels under reducing conditions and transferred to Immun-Blot PVDF Membranes (Bio-Rad). The membranes were blocked with 5% BSA or 5% nonfat milk in TBS-T (0.1% Tween 20) and immunoblotted with the indicated antibodies. HRP-conjugated goat anti-rabbit (Invitrogen) and anti-mouse IgG (Sigma) were used as secondary antibodies. Amersham Biosciences ECL Prime Western blotting detection reagent (GE Healthcare) was used to detect immunoreactive bands. The membranes were visualized on autoradiography film BX (MidSci). PFKFB3 antibody was obtained from Proteintech. Antibodies to detect Y1068 EGFR, EGFR, T402/T404 ERK, ERK, S473 Akt, Akt, S380 RSK, RSK were purchased from Cell Signaling Technology. S133 CREB1 and CREB1 antibodies were obtained from Santa Cruz Biotechnology. Antibody against S461 PFKFB3 was generated by Proteintech. We confirmed the specificity of the antibody by using an in vitro kinase assay where recombinant PFKFB3 was incubated with active recombinant Akt1 in the presence or absence of Lambda Protein Phosphatase and by using A549 cell lysates from PFKFB3 siRNA transfected cells in the presence or absence of Lambda Protein Phosphatase. Quantitative densitometry was performed with ImageJ (National Institutes of Health); signal density was normalized to the corresponding β-actin loading control.
RNA extraction and real-time RT-qPCR
Total RNA was extracted using the RNeasy kit (Qiagen) following the manufacturer's instructions. Reverse transcription was performed using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). Quantitative RT-PCR was performed with the ABI PRISM 7500 using TaqMan primers and probes for PFKFB3 and control gene β-actin (Thermo Fisher). Samples were analyzed using the comparative CT method, where -fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCt. Data are presented as mean ± S.E. of three experiments with three technical replicates and statistical significance was assessed by the two-sample t test (independent variable).
Chromatin immunoprecipitation (ChIP) assay
ChIP experiments were performed using the SimpleChIP Chromatin IP Kit (Cell Signaling Technology). Briefly, 3 × 106 PC9 cells/IP were treated with vehicle or 0.5 μm erlotinib for 24 h. Chromatin was crosslinked by incubating cells with 1% formaldehyde for 10 min. Immunoprecipitation of the lysed extracts was performed using anti-CREB1 ChIP grade antibody or normal mouse IgG (Santa Cruz Biotechnology) as a control. Oligonucleotides used for RT-qPCR flanking PFKFB3 gene regions: (−373 to −355), GCCAGCCCGGACTCTTTA (forward), GCTCGTCCGCTCGGAAA (reverse); (−1924 to −1719), GCCTCTCAAGGTTCCCAGTC (forward), CAGGTAAGGACATGGGCCAG (reverse); (−2489 to −2281), CTAGACTCGGAACGCGTTATT (forward), CTTCTTCCACACTCCCTACAAG (reverse); (−10343 to −10153, distal region), TGGGTTCCAGTTTCTGTTCTC (forward), GATCTGGGCTGCTCTATGATTT (reverse); primers were purchased from Integrated DNA Technologies. -Fold enrichment was calculated using the comparative CT method. Data are presented as mean ± S.E. of three experiments with three technical replicates and statistical significance was assessed by the two-sample t test (independent variable).
In vitro PFKFB3 phosphorylation with immunoprecipitated EGFR
106 PC9 cells were cultured in either complete medium or serum-starved for 24 h (for the EGF-stimulated experiments). Cells in complete medium were exposed to 0.5 μm erlotinib for 1 h; serum-starved cells were stimulated with 50 ng/ml EGF for 2 min prior to harvesting. Whole cell lysates were collected using RIPA buffer, sonicated and standardized for protein content across all samples. EGFR was immunoprecipitated from lysates using anti-EGFR antibody with overnight rotation at 4 °C. A normal rabbit IgG antibody was used as a negative control. Prewashed Protein G Sepharose (Life Technologies) was then added to the IP samples for an additional 3 h at 4 °C and subsequently washed twice with kinase buffer: 25 mm Tris, pH 7.5, 2 mm DTT, 10 mm Na3VO4, 10 mm MgCl2. Immunoprecipitates were resuspended in a kinase reaction mixture containing 10 μm ATP and 1.4 μg recombinant PFKFB3 and incubated at room temperature for 1 h. Reactions were stopped by the addition of 2× Laemmli buffer followed by incubation at 60 °C for 30 min on a shaker. Samples were subjected to SDS-PAGE and Western blot analysis using a VeriBlot IP Detection Reagent (HRP) (Abcam).
F26BP assay
Cells stimulated with either EGF or vehicle for the indicated times were harvested and snap frozen in liquid nitrogen. Pellets were lysed in NaOH/Tris acetate by heating at 80 °C for 5 min. Extracts were neutralized to pH 7.2 by adding ice-cold 1 m acetic acid in the presence of 20 mm HEPES. Intracellular F26BP levels were measured using a method described previously (15).
Glycolysis assay
H522 or PC9 cells growing in 6-well plates were incubated in 500 μl of medium containing 1 μCi of 5-[3H]glucose for 60 min in 5% CO2 at 37 °C. Glycolysis assay was performed as described in Ref. 28. Data are presented as mean ± S.E. of independent experiments with technical and biological duplicates.
Cell viability assay
Cells were seeded in complete medium in 24-well plates prior to the addition of the indicated treatments. Cell viability was evaluated by trypan blue exclusion assay at the time of treatment (0 h) and 24–48 h later. Cell numbers were corrected according to the dilution factor. To evaluate the drug effect on cell survival, GR values were calculated using the equation: GR(d) = 2∧(log2(xd/x0)/log2(xctrl/x0)) − 1, where xd and xctrl = amounts of viable cells after treatment with drug (d) or vehicle (ctrl); x0 = cell amount at the time of treatment. Data are presented as mean ± S.E. of three independent experiments with biological and technical duplicates.
Anchorage-independent growth assay
Cell growth in soft agar was evaluated as described previously (60). Cells were harvested with trypsin/EDTA, resuspended in 0.3% noble agar (BD Biosciences) in complete medium containing drug treatments and seeded on top of the feeder layer, containing drug treatments. A total of 5 × 103 cells were suspended in the complete medium containing 0.3% agar and appropriate drugs. Media (including drug treatments) were replenished once in 5–6 days until colonies became visible. Colonies were allowed to grow for 10–15 days. Colonies were counted in three random 1 cm2 areas per plate under 20× magnification. Cells were imaged using a Nikon Eclipse TE200-S microscope, images were taken with ACT-1 software. Data were plotted as % decrease in colonies number compared with vehicle-treated cells. All data are expressed as the mean ± S.E. of three independent experiments with biological duplicates.
Statistics
Results are reported as the mean ± S.E. Statistical analysis was performed by the two-tailed Student's t test (independent variable) using GraphPad Prism, version 7.0 (GraphPad). p values <0.05 were considered to be statistically significant.
Author contributions
N. L., J. C., and Y. I.-F. conceptualization; N. L. and Y. I.-F. formal analysis; N. L. investigation; N. L. writing-original draft; S. T., J. C., and Y. I.-F. writing-review and editing; J. C. and Y. I.-F. resources; J. C. supervision; J. C. and Y. I.-F. funding acquisition.
Acknowledgment
We thank Dr. Brian Clem for assisting with the interpretation of our data and manuscript preparation.
This work was supported by Kentucky Lung Cancer Research Program Grant (to Y. I.-F.) and James Graham Brown Foundation Grant (to J. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- NSCLC
- non–small cell lung cancer
- CREB
- cAMP-response element-binding protein
- ED
- effective doses
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- ERK
- extracellular signal-regulated kinase
- F26BP
- fructose-2,6-bisphosphate
- 18FDG-PET
- PET with 2-[18F]fluoro-2-deoxy-glucose
- GI50
- concentration giving half-maximal growth inhibition
- GR
- growth rate
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- mutEGFR
- mutated EGFR
- PFKFB
- 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
- RSK
- 90 kDa ribosomal S6 kinase
- TKI
- tyrosine kinase inhibitor.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2017) Cancer statistics, 2017. CA-Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2. Lynch T. J., Bell D. W., Sordella R., Gurubhagavatula S., Okimoto R. A., Brannigan B. W., Harris P. L., Haserlat S. M., Supko J. G., Haluska F. G., Louis D. N., Christiani D. C., Settleman J., and Haber D. A. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 3. Paez J. G., Jänne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., Naoki K., Sasaki H., Fujii Y., Eck M. J., Sellers W. R., Johnson B. E., and Meyerson M. (2004) EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 4. Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., and Varmus H. (2004) EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 101, 13306–13311 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dempke W. C. (2015) Targeted therapy for NSCLC—a double-edged sword? Anticancer Res. 35, 2503–2512 [PubMed] [Google Scholar]
- 6. Scagliotti G. V., Bironzo P., and Vansteenkiste J. F. (2015) Addressing the unmet need in lung cancer: The potential of immuno-oncology. Cancer Treat. Rev. 41, 465–475 10.1016/j.ctrv.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 7. Makinoshima H., Takita M., Matsumoto S., Yagishita A., Owada S., Esumi H., and Tsuchihara K. (2014) Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J. Biol. Chem. 289, 20813–20823 10.1074/jbc.M114.575464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hachemi M., Couturier O., Vervueren L., Fosse P., Lacœuille F., Urban T., and Hureaux J. (2014) [18F]FDG positron emission tomography within two weeks of starting erlotinib therapy can predict response in non-small cell lung cancer patients. PloS One 9, e87629 10.1371/journal.pone.0087629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su H., Bodenstein C., Dumont R. A., Seimbille Y., Dubinett S., Phelps M. E., Herschman H., Czernin J., and Weber W. (2006) Monitoring tumor glucose utilization by positron emission tomography for the prediction of treatment response to epidermal growth factor receptor kinase inhibitors. Clin. Cancer Res. 12, 5659–5667 10.1158/1078-0432.CCR-06-0368 [DOI] [PubMed] [Google Scholar]
- 10. Zander T., Scheffler M., Nogova L., Kobe C., Engel-Riedel W., Hellmich M., Papachristou I., Toepelt K., Draube A., Heukamp L., Buettner R., Ko Y. D., Ullrich R. T., Smit E., Boellaard R., et al. (2011) Early prediction of nonprogression in advanced non–small-cell lung cancer treated with erlotinib by using [18F]fluorodeoxyglucose and [18F]fluorothymidine positron emission tomography. J. Clin. Oncol 29, 1701–1708 10.1200/JCO.2010.32.4939 [DOI] [PubMed] [Google Scholar]
- 11. Tiseo M., Ippolito M., Scarlattei M., Spadaro P., Cosentino S., Latteri F., Ruffini L., Bartolotti M., Bortesi B., Fumarola C., Caffarra C., Cavazzoni A., Alfieri R. R., Petronini P. G., Bordonaro R., Bruzzi P., Ardizzoni A., and Soto Parra H. J. (2014) Predictive and prognostic value of early response assessment using 18FDG-PET in advanced non-small cell lung cancer patients treated with erlotinib. Cancer Chemother. Pharmacol. 73, 299–307 10.1007/s00280-013-2356-x [DOI] [PubMed] [Google Scholar]
- 12. Sunaga N., Oriuchi N., Kaira K., Yanagitani N., Tomizawa Y., Hisada T., Ishizuka T., Endo K., and Mori M. (2008) Usefulness of FDG-PET for early prediction of the response to gefitinib in non-small cell lung cancer. Lung Cancer 59, 203–210 10.1016/j.lungcan.2007.08.012 [DOI] [PubMed] [Google Scholar]
- 13. Rider M. H., Bertrand L., Vertommen D., Michels P. A., Rousseau G. G., and Hue L. (2004) 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 381, 561–579 10.1042/BJ20040752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Schaftingen E., Hue L., and Hers H. G. (1980) Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem. J. 192, 897–901 10.1042/bj1920897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Schaftingen E., Hue L., and Hers H. G. (1980) Control of the fructose-6-phosphate/fructose 1,6-bisphosphate cycle in isolated hepatocytes by glucose and glucagon. Role of a low-molecular-weight stimulator of phosphofructokinase. Biochem. J. 192, 887–895 10.1042/bj1920887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atsumi T., Chesney J., Metz C., Leng L., Donnelly S., Makita Z., Mitchell R., and Bucala R. (2002) High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 62, 5881–5887 [PubMed] [Google Scholar]
- 17. Vivanco I. (2014) Targeting molecular addictions in cancer. Br. J. Cancer 111, 2033–2038 10.1038/bjc.2014.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diamond I., Legg A., Schneider J. A., and Rozengurt E. (1978) Glycolysis in quiescent cultures of 3T3 cells. Stimulation by serum, epidermal growth factor, and insulin in intact cells and persistence of the stimulation after cell homogenization. J. Biol. Chem. 253, 866–871 [PubMed] [Google Scholar]
- 19. Minchenko O. H., Ogura T., Opentanova I. L., Minchenko D. O., Ochiai A., Caro J., Komisarenko S. V., and Esumi H. (2005) 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene family overexpression in human lung tumor. Ukr. Biokhim. Zh. 77, 46–50 [PubMed] [Google Scholar]
- 20. De Rosa V., Iommelli F., Monti M., Fonti R., Votta G., Stoppelli M. P., and Del Vecchio S. (2015) Reversal of Warburg effect and reactivation of oxidative phosphorylation by differential inhibition of EGFR signaling pathways in non-small cell lung cancer. Clin. Cancer Res. 21, 5110–5120 10.1158/1078-0432.CCR-15-0375 [DOI] [PubMed] [Google Scholar]
- 21. Lu H. S., Chai J. J., Li M., Huang B. R., He C. H., and Bi R. C. (2001) Crystal structure of human epidermal growth factor and its dimerization. J. Biol. Chem. 276, 34913–34917 10.1074/jbc.M102874200 [DOI] [PubMed] [Google Scholar]
- 22. Bando H., Atsumi T., Nishio T., Niwa H., Mishima S., Shimizu C., Yoshioka N., Bucala R., and Koike T. (2005) Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin. Cancer Res. 11, 5784–5792 10.1158/1078-0432.CCR-05-0149 [DOI] [PubMed] [Google Scholar]
- 23. Park J. H., Liu Y., Lemmon M. A., and Radhakrishnan R. (2012) Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem. J. 448, 417–423 10.1042/BJ20121513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu C., Mi L. Z., Schürpf T., Walz T., and Springer T. A. (2012) Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J. Biol. Chem. 287, 38244–38253 10.1074/jbc.M112.414391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valley C. C., Arndt-Jovin D. J., Karedla N., Steinkamp M. P., Chizhik A. I., Hlavacek W. S., Wilson B. S., Lidke K. A., and Lidke D. S. (2015) Enhanced dimerization drives ligand-independent activity of mutant epidermal growth factor receptor in lung cancer. Mol. Biol. Cell 26, 4087–4099 10.1091/mbc.E15-05-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Momcilovic M., Bailey S. T., Lee J. T., Fishbein M. C., Magyar C., Braas D., Graeber T., Jackson N. J., Czernin J., Emberley E., Gross M., Janes J., Mackinnon A., Pan A., Rodriguez M., et al. (2017) Targeted inhibition of EGFR and glutaminase induces metabolic crisis in EGFR mutant lung cancer. Cell Rep. 18, 601–610 10.1016/j.celrep.2016.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clem B. F., O'Neal J., Tapolsky G., Clem A. L., Imbert-Fernandez Y., Kerr D. A. 2nd, Klarer A. C., Redman R., Miller D. M., Trent J. O., Telang S., and Chesney J. (2013) Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol. Cancer Ther. 12, 1461–1470 10.1158/1535-7163.MCT-13-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imbert-Fernandez Y., Clem B. F., O'Neal J., Kerr D. A., Spaulding R., Lanceta L., Clem A. L., Telang S., and Chesney J. (2014) Estradiol stimulates glucose metabolism via 6-phosphofructo-2-kinase (PFKFB3). J. Biol. Chem. 289, 9440–9448 10.1074/jbc.M113.529990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macdonald-Obermann J. L., and Pike L. J. (2018) Allosteric regulation of epidermal growth factor (EGF) receptor ligand binding by tyrosine kinase inhibitors. J. Biol. Chem. 293, 13401–13414 10.1074/jbc.RA118.004139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ray P., Tan Y. S., Somnay V., Mehta R., Sitto M., Ahsan A., Nyati S., Naughton J. P., Bridges A., Zhao L., Rehemtulla A., Lawrence T. S., Ray D., and Nyati M. K. (2016) Differential protein stability of EGFR mutants determines responsiveness to tyrosine kinase inhibitors. Oncotarget 7, 68597–68613 10.18632/oncotarget.11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang L., Ji X., Yang X., and Zhang X. (2017) Cell type- and density-dependent effect of 1 T static magnetic field on cell proliferation. Oncotarget 8, 13126–13141 10.18632/oncotarget.14480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ranayhossaini D. J., Lu J., Mabus J., Gervais A., Lingham R. B., and Fursov N. (2014) EGF potentiation of VEGF production is cell density dependent in H292 EGFR wild type NSCLC cell line. Int. J. Mol. Sci. 15, 17686–17704 10.3390/ijms151017686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ercan D., Xu C., Yanagita M., Monast C. S., Pratilas C. A., Montero J., Butaney M., Shimamura T., Sholl L., Ivanova E. V., Tadi M., Rogers A., Repellin C., Capelletti M., Maertens O., et al. (2012) Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2, 934–947 10.1158/2159-8290.CD-12-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma P., Fu Y., Chen M., Jing Y., Wu J., Li K., Shen Y., Gao J. X., Wang M., Zhao X., and Zhuang G. (2016) Adaptive and acquired resistance to EGFR inhibitors converge on the MAPK pathway. Theranostics 6, 1232–1243 10.7150/thno.14409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Cesare D., Jacquot S., Hanauer A., and Sassone-Corsi P. (1998) Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc. Natl. Acad. Sci. U.S.A. 95, 12202–12207 10.1073/pnas.95.21.12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xing J., Ginty D. D., and Greenberg M. E. (1996) Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959–963 10.1126/science.273.5277.959 [DOI] [PubMed] [Google Scholar]
- 37. Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., and Wingender E. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chou T. C. (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 58, 621–681 10.1124/pr.58.3.10 [DOI] [PubMed] [Google Scholar]
- 39. Hafner M., Niepel M., and Sorger P. K. (2017) Alternative drug sensitivity metrics improve preclinical cancer pharmacogenomics. Nat. Biotechnol. 35, 500–502 10.1038/nbt.3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J. H., Nam B., Choi Y. J., Kim S. Y., Lee J. E., Sung K. J., Kim W. S., Choi C. M., Chang E. J., Koh J. S., Song J. S., Yoon S., Lee J. C., Rho J. K., and Son J. (2018) Enhanced glycolysis supports cell survival in EGFR-mutant lung adenocarcinoma by inhibiting autophagy-mediated EGFR degradation. Cancer Res. 78, 4482–4496 10.1158/0008-5472.CAN-18-0117 [DOI] [PubMed] [Google Scholar]
- 41. Nan X., Xie C., Yu X., and Liu J. (2017) EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 8, 75712–75726 10.18632/oncotarget.20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Soler R., Piperdi B., Haigentz M., and Ling Y.-H. (2004) Determinants of sensitivity to the EGFR TK inhibitor erlotinib (E) in a panel of NSCLC cell lines. J. Clin. Oncol. 22, 7026 10.1200/jco.2004.22.90140.7026 [DOI] [Google Scholar]
- 43. Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., Kris M. G., and Varmus H. (2005) Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu S.-G., and Shih J.-Y. (2018) Management of acquired resistance to EGFR TKI–targeted therapy in advanced non-small cell lung cancer. Mol. Cancer 17, 38 10.1186/s12943-018-0777-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oxnard G. R., Hu Y., Mileham K. F., Husain H., Costa D. B., Tracy P., Feeney N., Sholl L. M., Dahlberg S. E., Redig A. J., Kwiatkowski D. J., Rabin M. S., Paweletz C. P., Thress K. S., and Jänne P. A. (2018) Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 4, 1527–1534 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seo H. S., Liu D. D., Bekele B. N., Kim M. K., Pisters K., Lippman S. M., Wistuba I. I., and Koo J. S. (2008) Cyclic AMP response element-binding protein overexpression: A feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 68, 6065–6073 10.1158/0008-5472.CAN-07-5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Apicella M., Giannoni E., Fiore S., Ferrari K. J., Fernández-Pérez D., Isella C., Granchi C., Minutolo F., Sottile A., Comoglio P. M., Medico E., Pietrantonio F., Volante M., Pasini D., Chiarugi P., Giordano S., and Corso S. (2018) Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 28, 848–865.e6 10.1016/j.cmet.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 48. Minchenko A., Leshchinsky I., Opentanova I., Sang N., Srinivas V., Armstead V., and Caro J. (2002) Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 277, 6183–6187 10.1074/jbc.M110978200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Novellasdemunt L., Obach M., Millán-Ariño L., Manzano A., Ventura F., Rosa J. L., Jordan A., Navarro-Sabate A., and Bartrons R. (2012) Progestins activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) in breast cancer cells. Biochem. J. 442, 345–356 10.1042/BJ20111418 [DOI] [PubMed] [Google Scholar]
- 50. Cordero-Espinoza L., and Hagen T. (2013) Increased concentrations of fructose 2,6-bisphosphate contribute to the Warburg effect in phosphatase and tensin homolog (PTEN)-deficient cells. J. Biol. Chem. 288, 36020–36028 10.1074/jbc.M113.510289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manes N. P., and El-Maghrabi M. R. (2005) The kinase activity of human brain 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase is regulated via inhibition by phosphoenolpyruvate. Arch. Biochem. Biophys. 438, 125–136 10.1016/j.abb.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 52. Red Brewer M., Yun C. H., Lai D., Lemmon M. A., Eck M. J., and Pao W. (2013) Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 110, E3595–E3604 10.1073/pnas.1220050110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tan X., Lambert P. F., Rapraeger A. C., and Anderson R. A. (2016) Stress-induced EGFR trafficking: Mechanisms, functions, and therapeutic implications. Trends Cell Biol. 26, 352–366 10.1016/j.tcb.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cossu-Rocca P., Muroni M. R., Sanges F., Sotgiu G., Asunis A., Tanca L., Onnis D., Pira G., Manca A., Dore S., Uras M. G., Ena S., and De Miglio M. R. (2016) EGFR kinase-dependent and kinase-independent roles in clear cell renal cell carcinoma. Am. J. Cancer Res. 6, 71–83 [PMC free article] [PubMed] [Google Scholar]
- 55. Li X., Liu J., Qian L., Ke H., Yao C., Tian W., Liu Y., and Zhang J. (2018) Expression of PFKFB3 and Ki67 in lung adenocarcinomas and targeting PFKFB3 as a therapeutic strategy. Mol. Cell Biochem. 445, 123–134 10.1007/s11010-017-3258-8 [DOI] [PubMed] [Google Scholar]
- 56. Clem B., Telang S., Clem A., Yalcin A., Meier J., Simmons A., Rasku M. A., Arumugam S., Dean W. L., Eaton J., Lane A., Trent J. O., and Chesney J. (2008) Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol. Cancer Ther. 7, 110–120 10.1158/1535-7163.MCT-07-0482 [DOI] [PubMed] [Google Scholar]
- 57. Klarer A. C., O'Neal J., Imbert-Fernandez Y., Clem A., Ellis S. R., Clark J., Clem B., Chesney J., and Telang S. (2014) Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2, 2 10.1186/2049-3002-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Telang S., O'Neal J., Tapolsky G., Clem B., Kerr A., Imbert-Fernandez Y., and Chesney J. (2014) Discovery of a PFKFB3 inhibitor for phase I trial testing that synergizes with the B-Raf inhibitor vemurafenib. Cancer Metab. 2, (Suppl. 1), P14 10.1186/2049-3002-2-S1-P14 [DOI] [Google Scholar]
- 59. Redman R., Pohlmann P., Kurman M., Tapolsky G. H., and Chesney J. (2015) Abstract CT206: PFK-158, first-in-man and first-in-class inhibitor of PFKFB3/ glycolysis: A phase I, dose escalation, multi-center study in patients with advanced solid malignancies. Cancer Res. 75, CT206–CT206 [Google Scholar]
- 60. Telang S., Yalcin A., Clem A. L., Bucala R., Lane A. N., Eaton J. W., and Chesney J. (2006) Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene 25, 7225–7234 10.1038/sj.onc.1209709 [DOI] [PubMed] [Google Scholar]