Abstract
Nitric oxide synthases (NOSs) are heme-based monooxygenases that convert l-Arg to l-citrulline and nitric oxide (NO), a key signaling molecule and cytotoxic agent in mammals. Bacteria also contain NOS proteins, but the role of NO production within these organisms, where understood, differs considerably from that of mammals. For example, a NOS protein in the marine cyanobacterium Synechococcus sp. PCC 7335 (syNOS) has recently been proposed to function in nitrogen assimilation from l-Arg. syNOS retains the oxygenase (NOSox) and reductase (NOSred) domains present in mammalian NOS enzymes (mNOSs), but also contains an N-terminal globin domain (NOSg) homologous to bacterial flavohemoglobin proteins. Herein, we show that syNOS functions as a dimer and produces NO from l-Arg and NADPH in a tetrahydrobiopterin (H4B)-dependent manner at levels similar to those produced by other NOSs but does not require Ca2+-calmodulin, which regulates NOSred-mediated NOSox reduction in mNOSs. Unlike other bacterial NOSs, syNOS cannot function with tetrahydrofolate and requires high Ca2+ levels (>200 μm) for its activation. NOSg converts NO to NO3− in the presence of O2 and NADPH; however, NOSg did not protect Escherichia coli strains against nitrosative stress, even in a mutant devoid of NO-protective flavohemoglobin. We also found that syNOS does not have NOS activity in E. coli (which lacks H4B) and that the recombinant protein does not confer growth advantages on l-Arg as a nitrogen source. Our findings indicate that syNOS has both NOS and NO oxygenase activities, requires H4B, and may play a role in Ca2+-mediated signaling.
Keywords: signal transduction, cyanobacteria, cytochrome P450, nitric-oxide synthase, multifunctional enzyme, reactive nitrogen species (RNS), dioxygenase, denitrification, flavohemoglobin, nitrogen assimilation, nitrogen utilization
Introduction
Nitric oxide (NO) is a gaseous free radical involved in numerous biological processes; it is an intermediate in the denitrification pathway (1), a precursor to protein post-translational modification via S-nitrosylation (2), and is the activator of soluble guanylate cyclase in animals or H-NOX proteins in bacteria (3, 4). In mammals, NO is the product of arginine oxidation by nitric oxide synthases (NOSs)2 (5, 6). The three mammalian isoforms, endothelial (eNOS), neuronal (nNOS), and inducible (iNOS) (6–8), share a heme-containing oxygenase domain (NOSox) and a C-terminal reductase domain (NOSred). NOSred, which functions to reduce NOSox using NADPH, is composed of an FMN-binding domain and a ferredoxin-NADP+-reductase (FNR)-like domain. NOSs function as N-terminal homodimers, whereby the NOSred of one subunit reduces the NOSox of the opposite subunit (9). Electron transfer is activated by Ca2+-loaded calmodulin (CaM) (10) that binds at a conserved sequence between NOSox and NOSred, and is also facilitated by the essential cofactor tetrahydrobiopterin (H4B) that acts to supply electrons to the NOS heme for oxygen activation (11). The mammalian NOS isoforms play key roles in many biological processes, such as vasoconstriction, immune response, and neuronal plasticity (12–14) and are also involved in several pathologies, including tumorigenesis, septic shock, and cerebral ischemia (15–18).
Although NOS is ubiquitous in the animal kingdom, it is infrequently found in bacteria. The occurrence and purpose of bacterial NOS is highly species-dependent, ranging from recovery from UV damage (Deinococcus radiodurans nitric oxide synthase (drNOS)) (19) to signaling biofilm formation (Silicibacter nitric oxide synthase (siliNOS)) (20), protection from oxidative stress (Bacillus subtilis nitric oxide synthase (bsNOS)) (21), aiding pathogen virulence (Bacillus anthracis nitric oxide synthase (baNOS)) (22), and controlling oxygen-based respiration (Staphylococcus aureus nitric oxide synthase (saNOS)) (23, 24). Although their heme domain structure and catalytic mechanisms are similar to that of mammalian NOS, most bacterial NOSs lack a dedicated reductase domain, instead relying on promiscuous cellular reductases (25, 26). One NOS found in Sorangium cellulosum has an N-terminal reductase domain containing a 2Fe-2S cluster and ferredoxin-like FAD and NADPH domain, dissimilar to mammalian NOSred (27). No bacterial NOS with a covalently attached FMN/FNR reductase domain has been biochemically characterized thus far.
NOSs are also found in photosynthetic organisms. A mammalian NOS homolog was characterized from the algae Ostreococcus tauri (otNOS) (28), which is intriguing because NOS has not yet been identified in higher plants, despite NO having an undisputed role in plant signaling (29). Recently, a mammalian-like NOS with a C-terminal P-450 reductase domain was identified in the photosynthetic diazotroph Synechococcus sp. PCC 7335 (syNOS) (30). syNOS is the first prokaryotic NOS to contain a mammalian NOSred homolog; in addition, syNOS contains a somewhat unusual globin domain (NOSg) N-terminal to NOSox, as well as a 342-residue N-terminal region of unknown properties (Fig. 1 and Fig. S1). The syNOS-harboring Synechococcus strain was shown to produce NO in an l-Arg–dependent manner, and this activity was inhibited by known NOS inhibitors (30). Based on genetic experiments in Synechococcus and heterologous expression experiments in Escherichia coli, syNOS was proposed to function in nitrogen utilization from l-Arg (30). Specifically, this model asserts that syNOS first converts l-Arg to NO with NOSox and then from NO to NO3− with NOSg. Nitrate would then be assimilated back into reduced forms of nitrogen. Herein, we report the first recombinant expression, purification, and biochemical characterization of syNOS. The enzyme indeed acts as a bona fide NO synthase and also has strong NO dioxygenase (NOD) activity; however, it cannot utilize the general folate cofactor tetrahydrofolate as do other bacterial NOSs and instead requires tetrahydrobiopterin, like mammalian NOS. Although activation does not depend on CaM, it does strongly rely on Ca2+. Importantly, syNOS does not appear to aid in nitrogen utilization from l-Arg when recombinantly expressed in E. coli and also has minimal impact on NO detoxification.
Figure 1.
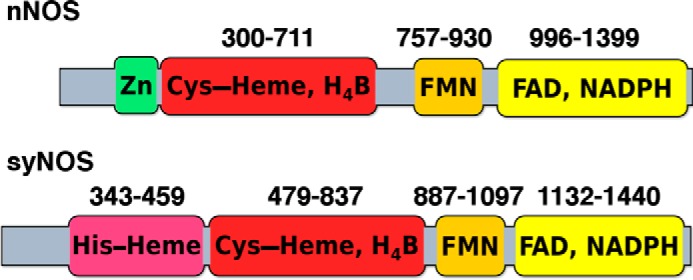
Domain map of nNOS and syNOS. The flavin and heme domains typical of nNOS are also present in syNOS; however, the zinc-ligating cysteines (Cys-327 and Cys-332 in nNOS) are absent from syNOS. Instead, syNOS also contains a globin domain not found in typical NOSs.
Results
Expression, purification, and oligomeric state of syNOS
Full-length syNOS (residues 1–1468) was co-expressed with the chaperonin GroEL/ES in E. coli BL21 DE3 cells; excess chaperonin was necessary to produce consistently well-folded and active enzyme. The yield (∼3 mg/liter of culture) and activity were very similar when the protein was expressed from two different vectors (pET28a or pCW-LIC). Affinity chromatography with ADP-Sepharose targeting the reductase domain was more effective as a first purification step than with Ni-NTA resin targeting the His6 tag. On size-exclusion chromatography (SEC), syNOS eluted in two major peaks presumably corresponding to monomer and dimer (and a minor amount of aggregate) (Fig. 2A). Nonreducing SDS-PAGE of the trailing peak produced one band corresponding to the syNOS monomer at ∼166 kDa, whereas the second leading peak produced two bands, representing the monomer and the syNOS dimer (Fig. 2A, inset). SEC coupled with multiangle light scattering (SEC-MALS) confirmed formation of a syNOS dimer, and its sensitivity to factors known to influence NOS dimerization (Fig. 2, B and C). The measured molecular mass of the first peak in the elution trace, 333 ± 4 kDa, equates to that of a syNOS dimer, and the second peak, 191 ± 6 kDa, corresponds to the monomer. A small peak was also observed at intermediate mass between those of the dimer and monomer. The sample does not appear to suffer from contamination or degradation, and thus this third peak may represent a third syNOS species in rapid oligomeric exchange. The addition of substrate or calcium did not significantly affect the population of dimer; however, H4B modestly decreased the amount of dimer.
Figure 2.
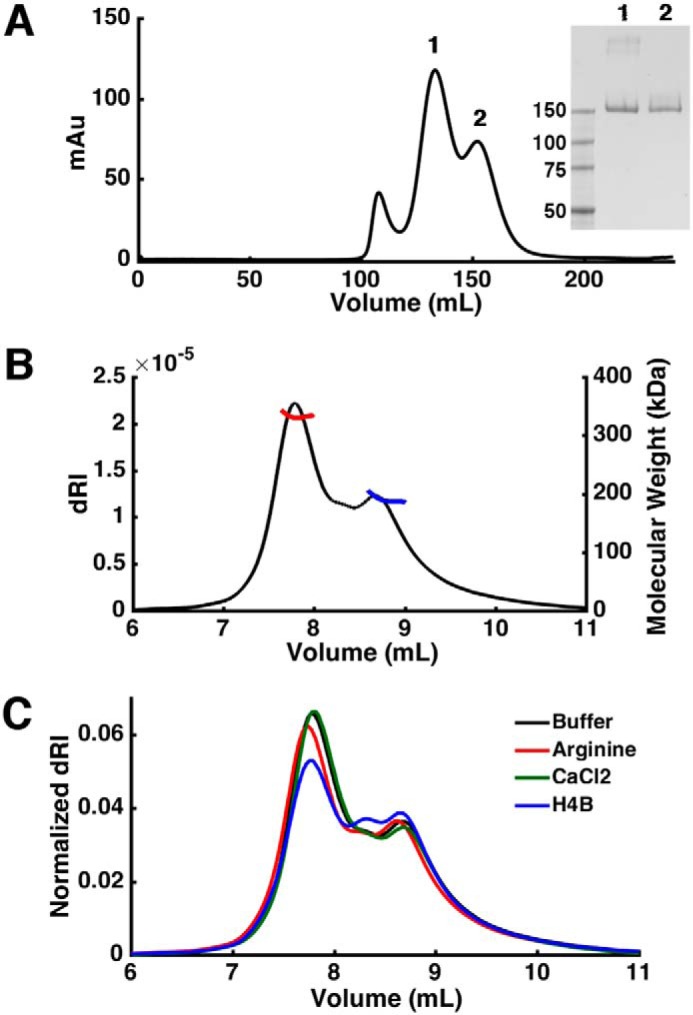
Hydrodynamic properties of syNOS. A, elution profiles of syNOS on SEC. Inset, SDS-PAGE of peaks 1 and 2. B, SEC-MALS of syNOS: dimer (red line), 333 ± 4 kDa; monomer (blue line), 191 ± 6 kDa. C, SEC-MALS of syNOS in the presence of arginine (red line), Ca2+ (green line), or H4B (blue line).
To isolate contributions from the two independent heme domains, variant proteins that removed key heme-binding ligands were also expressed and purified in the same manner as the WT enzyme. The NOSox proximal cysteine (Cys-539) was identified by alignment to NOS sequences (Fig. S1) and was substituted for alanine (C539A). The NOSg-ligating histidine was identified as His-422 with sequence alignments to globins of known structure (Fig. S2) and was also substituted for alanine (H422A) in a separate variant. Heme content of the WT and each variant, measured with the pyridine hemochrome assay, indicated that the mutations substantially reduced heme binding in the targeted domains: syNOS, 1.00 ± 0.10 μm heme/μm protein; H422A, 0.39 ± 0.08 μm heme/μm protein; and C539A, 0.72 ± 0.07 μm heme/μm protein (Table S2). A syNOS variant with both heme ligand substitutions (H422A and C539A) bound very little heme (Table S2 and Fig. S4). The sum of the heme content in the H422A and C539A variants approximately equaled that of the WT; therefore, 39% of the syNOS Soret was attributed to the NOSox heme. The concentration of NOSox-bound heme and the Soret intensity at 415 nm were used to calculate an extinction coefficient for quantifying active protein in subsequent assays.
Spectroscopic properties
Purified syNOS has a Soret band at 415 nm, which is red-shifted compared with the ferric heme absorption of typical globins (∼405 nm) and the high-spin thiol-ligated ferric heme of NOSox (∼397 nm) (Fig. 3A). The Soret for the globin heme (the C539A variant) is observed at 413 nm (Fig. 3C), similar to a flavohemoglobin from M. tuberculosis (414 nm) (31, 32), and the NOSox heme Soret at 417 nm (Fig. 3B) is more similar that of the NOS protein from S. cellulosum (416 nm) (27). Broad α-bands characteristic of ferric globin hemes are observed around 540 and 580 nm in all three proteins. A single band at ∼550 nm, expected for a NOS-type heme, is not prominent in H422A, perhaps due to remaining globin heme and protonation or dissociation of the NOSox proximal cysteine to form an inactive P420 state (33, 34). After reduction with dithionite, the Soret shifts to 425 nm, and peaks at 530 and 560 nm are observed for syNOS and C539A; this is similar to spectra of hexacoordinate neuroglobin, known to oxidize NO to nitrate (35, 36). These peaks are not observed for H422A, indicating that there is little globin heme bound in this variant. NOSs are thiolate-ligated P450-type heme proteins with a characteristic ferrous-CO Soret band at ∼450 nm (37). For WT syNOS, this species was observed as a shoulder at 444 nm corresponding to the NOSox heme and another intense absorbance at 420 nm corresponding to the ferrous-CO NOSg heme; however, the NOSox heme in the P420 state may also contribute to the intensity at 420 nm. As expected, H422A exhibits greater P450 Soret intensity at 444 nm compared with WT syNOS; albeit, there is still significant absorbance at 420 nm, most likely caused by the inactive P420 species. The ferrous-CO complex of C539A has no Soret peak at 444 nm, and only a band from NOSg is observed. These spectral features are evident in the Fe(II)CO − Fe(III) difference spectra (Fig. 3D).
Figure 3.
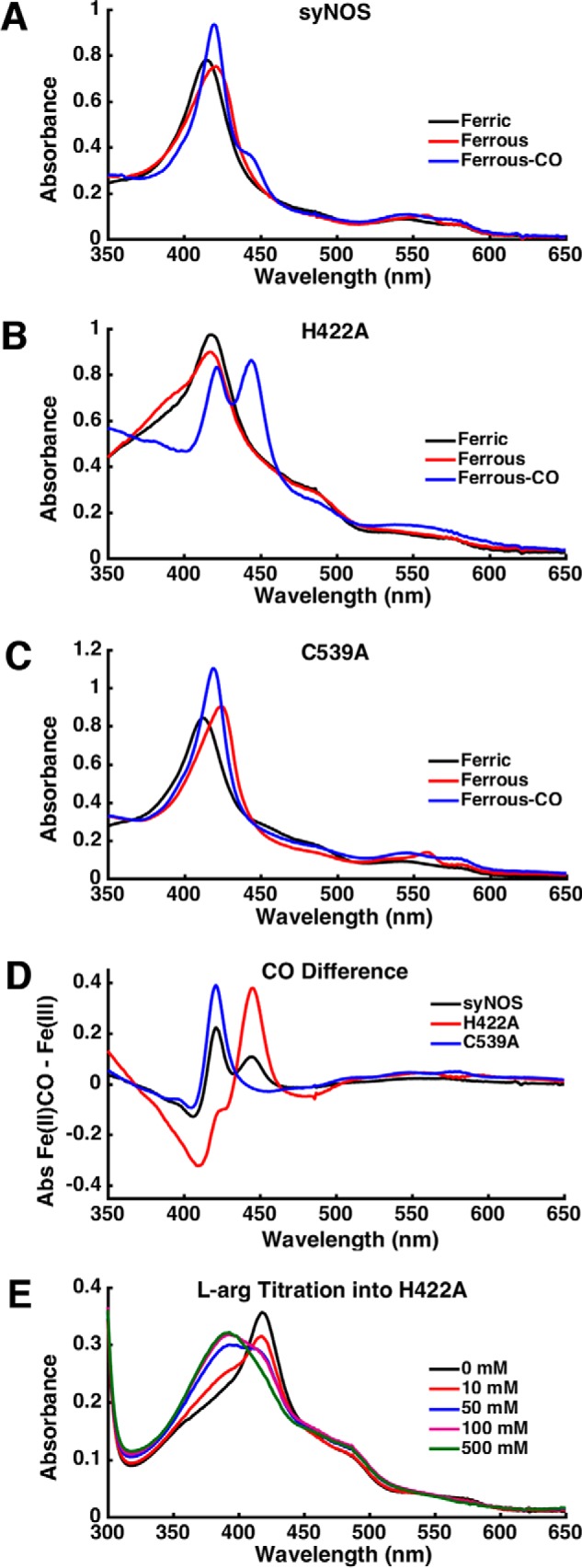
A–C, UV-visible spectra of WT syNOS (A), H422A (B), and C539A (C) in the ferric (black), ferrous (red), or ferrous-CO (blue) state. Samples were purged with argon before reduction with dithionite. D, difference spectrum of the ferrous-CO minus the ferric form of syNOS (black), H422A (red), and C539A (blue). E, arginine titration into H422A.
Mammalian and bacterial NOSs primarily contain five-coordinate low-spin hemes that exhibit a shift to high spin (Soret band at ∼390 nm) upon binding l-Arg; such a Soret shift was difficult to observe in WT syNOS. However, H422A undergoes a blue shift to ∼391 nm upon the addition of excess l-Arg (Fig. 3E). A large amount of l-Arg (500 mm) is required for complete conversion. This far exceeds the observed Michaelis constant for l-Arg (101 ± 12 μm; Fig. S3A) but may reflect the fact that, without the globin domain, the protein is destabilized (the activity drops by a factor of 8; see Table 1), and the NOS heme at least partially converts to the P420 state. Large amounts of l-Arg may stabilize the protein fold and heme center in a nonspecific manner so that a substrate-induced transition to a high-spin state can be observed. No Soret shift is observed when l-Arg is added to the NOSox heme-deficient C539A variant.
Table 1.
Specfic activity of syNOS for production of NO2− + NO3− in nmol min−1 mg−1
| Ca2+, H4B | Ca2+, H4B, CaM | H4B | Ca2+, THF | Ca2+, H4B, l-NNA | Ca2+, H4B, l-NAA | |
|---|---|---|---|---|---|---|
| syNOS | 36 ± 5, 34 ± 9a | 14.8 ± 1.0 | NDb | ND | ND | ND |
| H422A | 4.6 ± 0.8 | |||||
| C539A | 0.5 ± 0.2 | |||||
| nNOS | 1.0 ± 0.2 | 64 ± 2 |
a Citrulline production as quantified with HPLC.
b ND, no product detected.
Recombinant syNOS produces nitric oxide from l-arginine
NO production by full-length syNOS was first measured through the detection of its oxidized products, nitrate and nitrite, with the Griess assay. The specific activity of syNOS was 35.7 ± 5 nmol/min/mg (Table 1), approximately half that of the nNOS control 64.0 ± 2 nmol/min/mg, which is low compared with literature values (100–400 nmol/min/mg (37)). The syNOS C539A variant had very little measurable activity, and the activity of syNOS H422A was attenuated by about a factor of 8 compared with WT, in keeping with the results above (Table 1). The loss of NOS activity due to the globin substitution H422A likely reflects a general destabilization of the full-length protein when the globin domain is disrupted.
syNOS activity requires l-arginine, H4B, calcium, and NADPH (Table 1). Unlike analogous mammalian NOS, syNOS activity was independent of Ca2+-calmodulin (bovine), perhaps not surprisingly given that the calmodulin binding site of mNOS is not conserved in syNOS (Fig. S1) and Synechococcus does not contain an obvious homolog of calmodulin. Remarkably, syNOS is substantially activated by calcium (>10-fold); in fact furthermore, activity was reduced in the presence of calmodulin, presumably due to competition for calcium. However, the apparent Michaelis constant for Ca2+ activation is 228 ± 9 μm (Fig. S3), which may indicate that Ca2+ serves as a proxy for another physiological factor that activates the enzyme at lower concentration. syNOS cofactor utilization also differs from other bacterial NOSs in that syNOS cannot substitute tetrahydrofolate (THF) for H4B. The NOS inhibitors l-NNA and l-NAA, which mimic the substrate l-Arg, completely inhibited syNOS. This is in keeping with previous observations that l-NAME inhibits syNOS in vivo (30), as l-NAME requires hydrolysis to l-NNA (typically by cellular esterases) for inhibition of NOS (38).
l-Citrulline, the byproduct of l-Arg–based NO production, was detected as the product of the syNOS reaction with analytical HPLC (Fig. 4). After derivatization with the fluorophore ortho-phthaldialdehyde (OPA), samples were applied to a reverse-phase column, and citrulline (8.58 min) was resolved from substrate l-Arg (13.34 min). The amount of citrulline detected by HPLC was roughly equivalent to the amount of NO2− + NO3− measured by Griess, 0.95:1.
Figure 4.
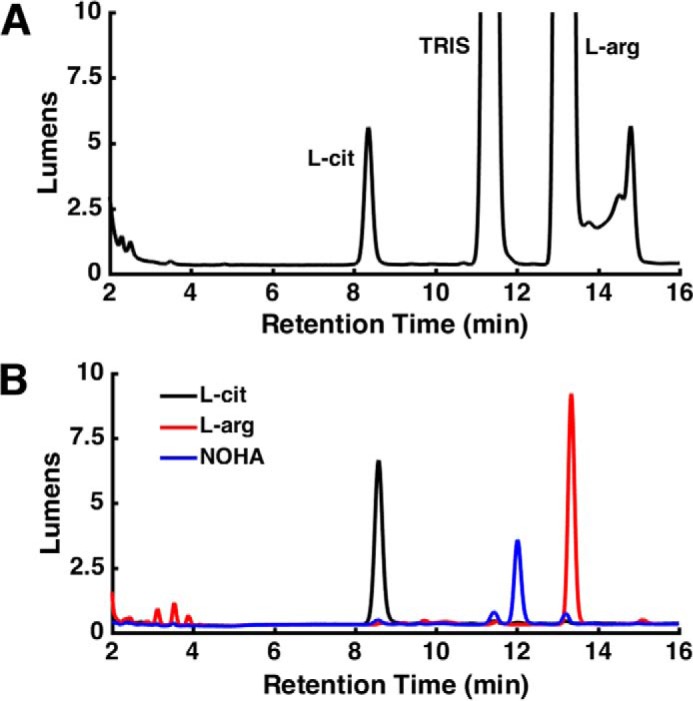
syNOS produces NOHA and citrulline. A, HPLC trace of syNOS products. B, HPLC trace of l-citrulline (black line), l-arginine (red line), and N-hydroxy-l-arginine (blue line) standards.
To directly detect NO production from syNOS, NO was chelated by the spin-trap Fe2+-MGD and detected by continuous-wave electron spin resonance (ESR) spectroscopy (Fig. 5). The NO-releasing small-molecule NOC-7 was used as a positive control. syNOS produced an NO signal identical to that of NOC-7. Moreover, the addition of the spin trap reacted with nearly all of the product NO and prevented conversion to NO2− or NO3− (Table S1).
Figure 5.
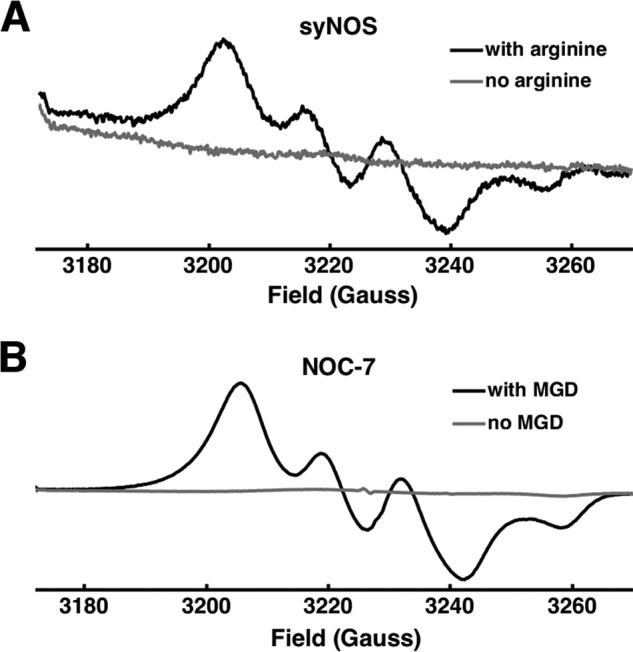
NO production by syNOS. Continuous-wave ESR of the NO complex of spin-trap Fe-MGD for syNOS with (black line) and without (gray line) L-arginine (A) and NO donor NOC-7 with (black line) and without (gray line) MGD (B).
NOSox and NOSg are both directly reduced by NOSred
In mammalian NOS, NOSox is reduced by NOSred and NADPH. To evaluate whether syNOSred can reduce syNOSox and syNOSg independent of each other, the syNOSox and syNOSg domains (residues 475–795 and residues 337–469, respectively) were cloned and expressed in isolation and then tested for their ability to accept electrons from NOSred (residues 856–1468). In the case of NOSox, the reduction experiment was carried out in the presence of CO to trap the reduced heme as a characteristic thiolate-ligated Fe(II)-CO (Soret band at 444 nm). In the presence of NADPH, NOSred produced some reduction of NOSox, as indicated by a small Soret shift to 444 nm (Fig. 6A). However, Ca2+ addition substantially increased the reduced form relative to the inactive P420 form. Thus, either Ca2+ facilitates NOSox reduction by NOSred, or Ca2+ attenuates the formation of the inactive P420 species through some means of NOSox stabilization.
Figure 6.
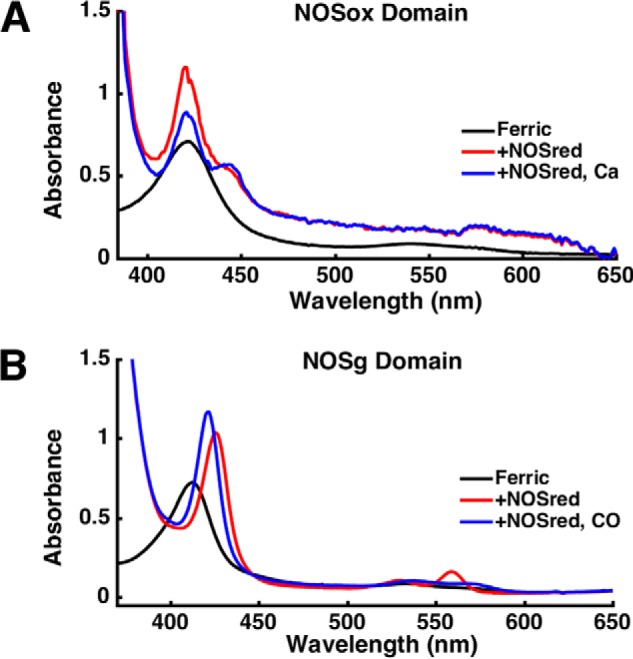
UV-visible spectra of NOSox in the presence of NOSred, NADPH, and CO with or without calcium ion (Ca2+) (A) and NOSg in the presence of NOSred and NADPH with or without CO (B). Samples with NADPH were measured under anaerobic conditions.
Likewise, syNOSred and NADPH directly reduce syNOSg, as indicated by the Soret shift to 426 nm and α-bands at 530 and 560 nm (Fig. 6B). The reductase domains of flavoHbs usually contain binding sites for FAD and NADH, but not FMN (39). Thus, either the FAD-containing FNR domain or the flavodoxin-like FMN module of syNOSred directly reduced NOSg.
syNOS globin oxidizes NO to nitrate
Upon assay of syNOS with the Griess reaction, it was found that the enzyme produces primarily nitrate with little to no nitrite formed, despite nitrite being the initial product of NO oxidation by air. Because related flavohemoglobins detoxify NO to nitrate, syNOSg may function as a NOD. Thus, we investigated the ability of syNOS to oxidize NO generated by NOC-7 (Table 2). NO was oxidized primarily to nitrate (74%) by syNOS in an NADPH-dependent manner during the time course of the experiment (NOS activity was stopped after 30 min, which equates to approximately three NOC-7 half-lives). Removal of the ligating cysteine from NOSox (C539A) did not decrease nitrate production, confirming NO dioxygenation by the globin heme. In contrast, removal of the proximal histidine from NOSg did reduce NO3− production, but not completely. The H422A variant produced more nitrate than nonenzymatic oxidation by air, suggesting that the NOSox domain also oxidizes NO to nitrate, as has been found for mNOS (40).
Table 2.
NO dioxygenation by syNOS and variants H422A and C539A
Samples (0.5 μm syNOs, 1 μm H422A, 0.5 μm C539A) were incubated for 30 min in the presence of the NO donor NOC-7 before heat-denaturing. The percentage of NO oxidized to NO2− + NO3− was measured by the Griess assay. The rate constants for NOD activity were measured with an NO electrode. The rate constants were averages of at least nine measurements, and a Q test was used to remove outliers. Student's t test indicated a significant difference between the rate constants of syNOS and H422A (p < 0.001) and between H422A and C539A (p < 0.001) but no significant difference between syNOS and C539A (p > 0.7). ND, not detected.
| Relative amount |
NO oxidation rate constant | ||
|---|---|---|---|
| NO2− | NO3− | ||
| % | % | s−1 nmol heme−1 | |
| No enzyme | 76 ± 3 | 23 ± 3 | |
| syNOS | 25.6 ± 0.6 | 74 ± 2 | 0.6 ± 0.3 |
| H422A | 44.6 ± 1.6 | 55 ± 3 | 0.10 ± 0.06 |
| C539A | 29.0 ± 1.6 | 71 ± 7 | 0.6 ± 0.4 |
| No NADPH | ND | ||
The rate constants for dioxygenation by syNOS and variants were measured with an NO-specific electrode (Table 2). NOC-7–derived NO produced a measurable current on the order of microamperes. Upon the addition of syNOS, the NO signal decayed rapidly under first-order kinetics. Consistent with results from the Griess assay, the rate constant for NO oxidation by the C539A variant (0.6 ± 0.4 s−1 nmol of heme−1) is approximately equal to that of WT syNOS (0.6 ± 0.3 s−1 nmol−1), and the rate constant of the H422A variant is far less than either (0.10 ± 0.06 s−1 nmol−1). These results not only reveal that NOSg is an efficient NOD, but confirm that NOSred directly reduces both the NOSox and NOSg heme cofactors.
Activity of syNOS in E. coli cells
It was previously reported that syNOS enabled E. coli to use l-Arg as its sole nitrogen source and that expression of syNOS increased cell density when growing on l-Arg, compared with an empty vector (EV) control (30). Under our conditions, this benefit of syNOS was not observed; instead, we found that syNOS expression conferred no significant advantage for growth on l-Arg compared with the no vector control (Fig. 7A). Given that syNOS requires H4B for NO production from l-Arg and E. coli does not make H4B (41), it is unclear how expression of syNOS would increase conversion of l-Arg to more oxidized forms of nitrogen, such as nitrate.
Figure 7.
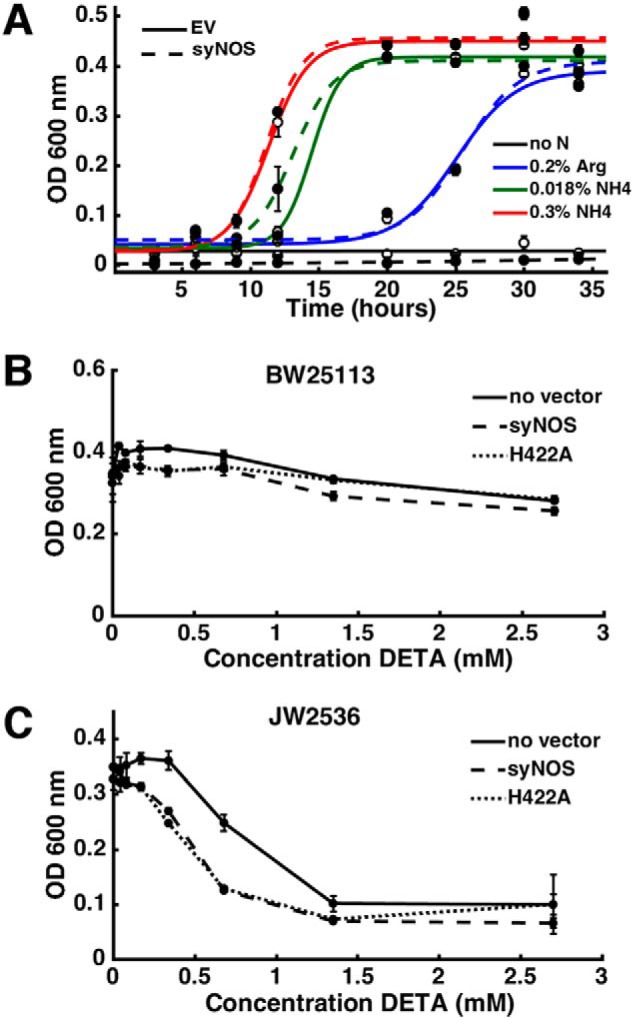
syNOS expression does not aid growth of E. coli on l-Arg or protect against nitrosative stress. A, growth of E. coli in nitrogen-limited medium. Solid lines and empty circles, EV; dashed lines and filled circles, syNOS. B, growth of E. coli (BW25113) in the presence of DETA NONOate and 26 μm IPTG. C, growth of hmp-deficient E. coli (JW2536) in the presence of DETA NONOate and 26 μm IPTG.
Although syNOS should not be active as a NOS when recombinantly expressed in E. coli, it should retain NOD activity, which may mitigate the effects of nitrosative stress. To test the ability of recombinant syNOS to protect E. coli against NO, syNOS and the NOD-deficient variant H422A were expressed from the tac promoter (pCW-LIC) in the WT E. coli strain BW25113 as well as the flavohemoglobin-deficient strain JW2536. The absence of the flavohemoglobin gene (Δhmp) renders E. coli more sensitive to NO (42), and the addition of a NOD should, in theory, allow growth at higher NO concentrations. However, this was not observed (Fig. 7, B and C). The Δhmp strain is more sensitive to nitrosative stress induced by the addition of DETA NONOate than the WT, but the added expression of syNOS actually increased sensitivity to NO. Additionally, the growth of cells containing the NOD-deficient H422A construct was indistinguishable from that of cells containing WT syNOS. Thus, syNOS cannot complement a hmp mutant of E. coli, indicating that either its NOD activity in this context is low or syNOS is detrimental in some other manner.
Discussion
Full-length syNOS proved to be a challenging protein to express recombinantly in E. coli. Although soluble protein with heme and flavin cofactors bound could be produced under several conditions, many attempts at purification produced protein with little or no synthase activity. Furthermore, active and inactive syNOS share the same spectroscopic characteristics and elute similarly on SEC. Co-expression with the chaperonin GroEL/ES was essential to consistently produce active protein. E. coli encodes GroEL/ES and several other chaperones in its genome; however, their basal level of expression was insufficient to reliably correct syNOS misfolding.
Biochemical and spectroscopic results confirm that syNOS is a genuine NOS and NOD. The NOSox ferrous-carbonmonoxy species is observed at 444 nm, as expected of a P450 type heme. In the absence of the NOSox heme, nitric oxide is not produced, and in the absence of the NOSg heme, the amount of NO dioxygenation is attenuated. syNOS shares the same substrate, products, and activating cofactors expected of a mammalian NOS; however, there are several unusual facets of syNOS enzymology.
A feature that distinguishes syNOS from animal NOSs is the absence of the calmodulin-binding sequence and auto-inhibitory loop. All three mammalian NOS isoforms require Ca2+-CaM, and in eNOS and nNOS, the calcium concentration dependence is dictated by the presence of an auto-inhibitory loop in the FMN domain (43, 44). Although syNOS does not bind mammalian calmodulin (and no protein in its genome has significant similarity to calmodulin), the addition of calcium increases NO turnover 10-fold. Moreover, NOSox reduction by NOSred is substantially enhanced by Ca2+, and thus Ca2+ alone may be playing a similar role in syNOS as Ca2+-CaM does in mNOS. No other NOS has been reported to be activated by calcium independent of calmodulin. The Swiss Institute for Bioinformatics ScanProsite tool was used to search for possible EF-hand calcium-binding motifs (Prosite accession numbers PS50222 and PS00018) in syNOS, but no such sites were identified. It is possible that calcium may have a structural role, perhaps at the dimer interface, similar to zinc in mammalian NOS (45, 46), or at interdomain contacts to facilitate electron transfer to the NOSox heme. However, the measured activation constant for calcium was high, 228 ± 9 μm, far exceeding biological concentration ranges (hundreds of nm to 10 μm (47, 48)). The mechanism of calcium binding and activation of syNOS is currently unknown; additional cofactors or proteins may be required for efficient calcium use, or Ca2+ may serve as a proxy for another factor in vivo.
Cofactor utilization also differentiates syNOS from other bacterial NOSs. All bacteria produce THF, but few produce H4B, which differs from THF in its dihydroxypropyl side chain. All bacterial NOSs characterized to date can utilize both cofactors; thus, the preference of syNOS for H4B over THF agrees with its mammalian-like NOS domain architecture. Genome analysis suggests that Synechococcus sp. PCC 7335 can produce H4B. The H4B biosynthetic pathway requires GTP cyclohydrolase I (GTPCH I), 6-pyruvoyl tetrahydropterin synthase (PTS), and sepiapterin reductase (SR) (49, 50). Both GTPCH and PTS are found in the genome of Synechococcus sp. PCC 7335 and are highly homologous to the mammalian enzymes (>50% identity); however, no gene in Synechococcus is annotated as an SR. SR belongs to the short-chain dehydrogenase/reductase (SDR) family of oxidoreductases, a large family of proteins found in all kingdoms of life (51). Synechococcus encodes many genes belonging to the SDR family. Although none share high sequence similarity (greater than 30% identity) with mammalian SR, one gene annotated as an SDR (CDS YP_002711555.1) is immediately adjacent to a gene encoding GTPCH I. Additionally, of the 15 photosynthetic prokaryotes containing gene sequences highly similar to syNOS (>60% identity), 14 also encode an SDR adjacent to a GTPCH I. Thus, it is highly likely that Synechococcus has the enzymatic machinery to produce H4B.
Mammalian NOSs cannot use THF because an N-terminal β-extension, known as the N-terminal hook, occludes the long THF p-aminobenzoyl-glutamate side chain (52). In syNOS, this region is replaced by a short linker (18 residues) to the globin domain. This raises questions concerning not only the manner of selective H4B binding, but also the manner of syNOSox dimer formation and coupling to NOSg. The NOSox motifs located at the dimer interface in other NOS, the helical lariat and helical T (52), are conserved in syNOS; however, the close proximity of NOSg suggests that it could also play a role in stabilizing the NOSox subunit, as well as the NOSox dimer. In support of a tight coupling between NOSox and NOSg, the H422A substitution in NOSg appears to also affect the stability of NOSox and/or its affinity for l-Arg.
In addition to structural implications, the NOSg domain adds a layer of complexity to syNOS chemistry and physiology. Sequence alignments assign this domain to the globin superfamily of proteins. In particular, flavoHbs catalyze reduction of nitrite to nitric oxide and reduction of nitric oxide to nitrous oxide (53) but most commonly carry out the oxidation of NO to nitrate (39, 54). syNOS catalyzes the oxidation of NO to nitrate, and this activity depends on a functional globin domain. Removal of the NOSox heme (the C539A variant) did not hinder NO oxidation; however, the NOSox domain was also capable of NO dioxygenation because nitrate production was still observed in the absence of the NOSg heme (H422A). NO dioxygenase activity by NOS enzymes has precedent; mammalian NOSs are also capable of NO dioxygenation, and chimeras composed of iNOSox and nNOSred exhibit increased NOD activity (40). By producing chimeras that coupled the fast heme reduction (kr) of nNOS with the slow NO dissociation (kd) of iNOS, as well as the addition of a V346I substitution that further slowed the NO release (55), NO dioxygenation by mNOS was substantially accelerated (40). Note that in syNOS, the equivalent position of Val-346 in the distal heme pocket contains Ile natively, typical of bacterial NOS. The NO dioxygenase activity of syNOS has two implications: 1) syNOS is the first NOS whose final product can be nitrate and not NO, and 2) the reductase domain of syNOS can reduce both the NOSox and NOSg directly. It is also worth noting that the spin trap compound intercepted nearly all NO from syNOS before it could be oxidized to NO3− by NOSg. Thus, NOD activity in syNOS is independent from NOS activity, with any NO produced by NOSox released to the solution before reaction with NOSg.
Our biochemical results confirm that syNOS oxidizes l-Arg to nitrate (30); however, we are unable to replicate the finding that syNOS allows E. coli to use l-Arg as the sole nitrogen source. The growth of E. coli transformed with empty vector is indistinguishable from that transformed with syNOS. Both strains are capable of growth on l-Arg, which is not surprising given that E. coli already contains the arginine succinyltransferase pathway to derive reduced nitrogen from arginine (56, 57). Although Synechococcus does not contain the arginine succinyltransferase pathway, it does contain alternate l-Arg salvage pathways that rely on enzymes such as arginase and deaminating l-amino acid oxidases (58, 59). Furthermore, syNOS is not expected to be active in E. coli, as the third enzyme in the H4B biosynthesis pathway, sepiapterin reductase, is absent from its genome (41), and syNOS cannot substitute H4B with THF. The proposed role of syNOS in nitrogen assimilation is somewhat questionable given the environmental conditions in which the cyanobacteria are found. Synechococcus sp. PCC 7335 is a marine organism, where the concentrations of dissolved nitrates (tens of μm (60–62)) far exceed that of arginine (tens of nm (63–65)). Additionally, this organism is capable of nitrogen fixation (66–68). It is unclear what biochemical merits result from expending reducing power (NADPH) to oxidize l-Arg to nitrate, only to then reductively assimilate nitrate back to ammonia.
Owing to the lack of H4B, syNOSox should be inactive in E. coli. However, syNOSg and syNOSred do not depend on specialized cofactors, and thus syNOS may function as an NO dioxygenase in E. coli. However, the flavohemoglobin-deficient strain (JW2536) containing syNOS is actually more susceptible to damage by NO compared with the untransformed control. If syNOS were functioning as a flavoHb we would expect the opposite. Thus, either the protein does not exhibit NOD activity in E. coli because of interfering cellular factors, or it has other additional activities that are detrimental to growth that overcome any benefit of NO oxidation to nitrate.
The dual functionality of syNOS as both a NOS and a NOD is mysterious. As syNOS is actively expressed in growing cyanobacteria (30), some regulatory mechanism or “on/off switch” may be necessary to control NO production. NOSg may participate in such a function. Globin-based regulation of NOS has precedent in animals; eNOS binds to and stabilizes α-globin at the myoendothelial junction so that α-globin can regulate NO signaling by oxidizing NO to nitrate (69). Additionally, in vitro experiments found that full-length eNOS was able to reduce α-globin to the active ferrous state at a faster rate than the methemoglobin reductase cytochrome B5 reductase (69).
In S. aureus, NOS is proposed to play a role in the transition from aerobic respiration to nitrate respiration under microaerobic conditions (23). This control is mediated by the combined action of NOS and flavoHb; at high oxygen concentrations, NOS-derived NO is detoxified by flavoHb, whereas under microaerobic conditions, flavoHb cannot bind oxygen as substrate and NO is free to inhibit cytochrome oxidase, thus inhibiting oxygen reduction and favoring nitrate respiration. Although syNOS would genetically link NOS and NOD activity for such a purpose, the NOS-containing Synechococcus species does not respire nitrate (as it lacks ccNIR and associated nrf genes). Finally, a feature of syNOS activity that may provide clues as to its biological function is its reliance on calcium, which is well-known to be a tightly regulated signaling molecule in cyanobacteria (47, 70). Interestingly, Ca2+ is used in cyanobacteria as a signal to convey changes in nitrogen utilization. Increased levels of calcium in Synechoccocus elongatus accompany acclimation to nitrogen starvation (71), and in Anabaena sp. PCC 7120, elevated calcium levels are necessary for heterocyst differentiation (72). Thus, syNOS may be poised to respond to these signals. The high Ca2+ threshold that we observe in our assays is not without note but may be a consequence of the ex vivo conditions.
In conclusion, we demonstrate that syNOS has both NOS and NOD activities. The protein represents a bacterial NOS enzyme with properties closely related to its mNOS counterparts, especially with respect to reductase coupling and cofactor utilization. However, the NOS function is not activated by Ca2+-CaM and instead appears to require only Ca2+ for activity. The enzyme's reliance on H4B calls into question any NOS activity when recombinantly expressed in E. coli, and likewise the protein is unable to aid a flavoHb null strain in tolerating nitrosative stress. The properties of syNOS in the context of Synechococcus metabolism suggest that it is unlikely to be solely involved in nitrogen utilization from arginine and may serve a more typical NOS function in signal transduction. That said, the coupling of NOS and NOD activity in a single protein indicates a genetic link between these respective activities that is beneficial to cyanobacteria.
Experimental procedures
Materials
Synechococcus PCC 7335 (ATCC 29403) was purchased from the American Type Culture Collection. E. coli strain JW2536 was purchased from Dharmacon, and BW25113 was from the Coli Genetic Stock Center at Yale University. NOC-7 and N-(dithiocarbamoyl)-N-methyl-d-glucamine (MGD) were purchased from Santa Cruz Biotechnology, Inc. Nitrate reductase was purchase from Roche Applied Science. l-NNA, l-NAA, l-citrulline, N-hydroxyarginine, and OPA were purchased from Sigma-Aldrich. H4B was purchased from Cayman Chemical. 2′,5′-ADP-Sepharose 4B and Superdex 200 resins were purchased from GE Healthcare. Ni-NTA was purchased from Thermo Scientific.
Genomic DNA extraction and cloning
Genomic DNA extraction was performed following the method of Singh et al. (73). Synechococcus (50 ml) was grown for 1 week and then harvested by centrifugation at 2,000 × g. Cells were resuspended in 400 μl of lysis buffer (4 m urea, 0.2 m Tris, pH 7.4, 20 mm NaCl, 0.2 m EDTA) supplemented with 50 μl of 20 mg/ml proteinase K. The sample was incubated for 1 h at 55 °C and mixed by gentle inversion every 15 min. One ml of the extraction buffer (3% cetyltrimethylammonium bromide, 1.4 m NaCl, 20 mm EDTA, 0.1 m Tris, pH 8.0, 1% Sarkosyl) heated to 55 °C was added to the sample. The resulting sample was subsequently incubated at 55 °C for 1 h with gentle inversion every 10 min. Once the sample cooled to room temperature, two volumes of chloroform/isoamyl alcohol (24:1) solution were added, and the sample was mixed by inversion. The sample was centrifuged for 5 min at 10,000 × g, and the upper aqueous phase was removed. Two volumes of ethanol + 0.1 volume of 3 m sodium acetate (pH 5.2) were added to the aqueous phase. This solution was incubated at −20 °C for 1 h and then centrifuged for 3 min at 10,000 × g. The pellet was washed with 500 μl of cold 70% ethanol. After evaporating the ethanol, the DNA was dissolved in 50 μl of water, and the purity was assessed by the absorbance ratio 260 nm/280 nm.
Full-length syNOS (residues 1–1468, NCBI Protein database accession number WP_006458277) was cloned from bp 486,069 to 490,475 (NCBI Nucleotide database accession number NZ_DS989905) and inserted into the following vectors: 1) pET28 (Novagen) using the restriction sites NdeI and EcoRI and 2) pCW-LIC (a gift from Cheryl Arrowsmith, Addgene plasmid 26098) using BamHI and KpnI. Point mutations were constructed by primer overlap extension PCR.
Each domain of syNOS was subcloned into expression vectors by PCR. NOSg (residues 337–469), NOSox (residues 475–795), and NOSred (residues 856–1468) were inserted into the vector pET28 using the restriction sites NdeI and XhoI.
Protein expression and purification
E. coli BL21 DE3 cells were cotransformed with syNOS (in either pCW-LIC or pET28) or its heme domain truncations, plus the chaperonins GroEL/ES in pACYCDuet. The reductase domain truncation was transformed into BL21 DE3 without GroEL/ES. Lysogeny broth Miller was inoculated with an overnight culture and incubated at 37 °C until the A600 nm reached ∼0.6. Protein expression was induced by the addition of 25 μg/ml isopropyl β-d-1-thiogalactopyranoside (IPTG), 80 μg/ml δ-aminolevulinic acid, 4 μg/ml hemin, 2 μg/ml FAD, and 2 μg/ml FMN and incubated at 17 °C overnight. Hemin and δ-aminolevulinic acid were excluded from NOSred expression, and flavins were excluded from the expression of either heme domain. Cells were harvested 18 h after induction by centrifugation at 5,000 × g and then frozen at −20 °C.
The cell pellet from a 2-liter growth was resuspended in 50 ml of lysis buffer (200 mm NaCl, 50 mm Tris, pH 7.5, 10% glycerol) with 174 μg/ml phenylmethylsulfonyl fluoride, 1.5 μg/ml pepstatin A, and 1 μg/ml leupeptin. Cells were lysed on ice by sonication using a Fisher Scientific Sonic Dismembrator 550 (amplitude of 7, pulsed 2 s on, 2 s off) for a total sonication time of 5 min. Lysate was centrifuged for 1 h at 48,000 × g at 4 °C. For the full-length constructs, the soluble portion was applied in batch to a 2′,5′-ADP-Sepharose resin pre-equilibrated with lysis buffer and then incubated at 4 °C for 2 h with gentle rocking. The resin was collected and washed with five column volumes of lysis buffer and then eluted with lysis buffer plus 5 mm NADPH. Domain truncations were lysed in the same manner, with 5 mm imidazole in the lysis buffer. After centrifugation, the soluble portion was applied to Ni-NTA, and the resin was washed with five column volumes of lysis buffer with 20 mm imidazole and then eluted with lysis buffer plus 200 mm imidazole.
The eluent was concentrated and then further purified on a Superdex 200 (full-length constructs) or 75 (truncations) 26/60 size-exclusion column by isocratic elution using gel filtration buffer (25 mm Tris, pH 7.5, 150 mm NaCl, 10% glycerol). Protein was concentrated using a 50-kDa (full-length constructs) or 10-kDa (truncations) cutoff Amicon Ultra centrifugal filter.
Multiangle light scattering
MALS was performed using a Phenomenex BioSep SEC-s3000 column (5 μm, 290 Å, 300 × 7.8 mm) with a Phenomenex SecurityGuard guard column, connected to an Agilent 1200 series HPLC with G1314D variable wavelength detector. Light scattering was detected using a Wyatt DAWN HELEOS-II, and differential refractive index was measured by a Wyatt Optilab T-rEX refractometer. The mobile phase contained 100 mm Tris, pH 7.5, 200 mm NaCl with either 5 mm arginine, 5 mm CaCl2, or 250 μm H4B with 1.5 mm DTT. A solution of BSA monomer (5 mg/ml) was used as the standard to control for peak alignment and molecular weight calculations. Data were collected for 30 min at a flow rate of 1 ml/min at 25 °C. ASTRA V software was used to analyze the molecular weight and polydispersity of each peak.
UV-visible analysis of purified syNOS
Heme content was measured using the pyridine hemochrome method. Twenty microliters of protein at 1 mg/ml were diluted to 100 μl in 20% pyridine 0.2 m NaOH. An Agilent 8453 UV-visible spectrophotometer was blanked with this solution, and then ∼0.5 mg of solid dithionite was added, and the absorbance difference at 557–573 nm was recorded (extinction coefficient 32.4 mm−1 cm−1). Protein concentration was measured using the Bradford assay (Bio-Rad protein assay dye) and the calculated extinction coefficient of the denatured protein (ϵ280 nm = 0.24053 μm−1 cm−1). The protein was denatured in 4 m urea, and then unbound cofactors were removed by concentrating in a spin column and diluting in 4 m urea, which was repeated three times. The protein concentration calculated from the absorbance at 280 nm agreed with that of the Bradford assay.
As syNOS does not appear to be fully loaded with heme, the concentration of active protein was estimated based on the amount of NOSoxy heme bound in the H422A variant. The heme concentration of a syNOS sample was measured as stated previously, and the Soret intensity at 415 nm was recorded. Thirty-nine percent of the measured heme concentration was presumed to originate from the NOSoxy heme and thus represents the concentration of active protein. This concentration and the Soret intensity were used to calculate an extinction coefficient that was used to quantify active protein in subsequent assays.
The spectra for the ferrous and ferrous-carbonmonoxy syNOS were taken after the ferric enzyme was sparged with argon and reduced with ∼0.5 mg of solid dithionite, and then carbon monoxide gas was gently bubbled into the solution. Difference spectra were constructed by subtracting the spectrum of the ferric species from that of the ferrous-carbonmonoxy species.
NOS enzymatic reaction
NOS activity was assayed following the method of Moreau et al. (74), using 250 μm H4B, 5 mm arginine, 5 mm CaCl2, 1.5 mm DTT, and 1 mm NADPH in 100 mm Tris, pH 7.5, 200 mm NaCl. Where specified, THF was used at 250 μm, and CaM was used at 10 μg/ml. The reaction proceeded for 30 min at room temperature and was stopped by rapid heating to 60–70 °C for 5 min. Samples were centrifuged at 16,000 × g for 5 min to remove any insoluble debris.
Nitrate and nitrite measurement by Griess assay
Nitrate was measured by adding 75 μl of 0.2 units/ml nitrate reductase, 1 mm NADPH, and 0.1 mm FAD to 150 μl of sample and then incubated for 2 h at 37 °C. Nitrite was quantified similarly but in the absence of nitrate reductase. NADPH was removed by zinc acetate precipitation (75, 76). To each sample, 100 μl of 0.5 m zinc acetate in 50% ethanol was added, and the samples were vortexed. Subsequently, 100 μl of 0.5 m sodium carbonate was added, followed by vortexing. After 5 min of centrifugation at 16,000 × g, 150 μl of each sample were plated in duplicate, and 25 μl of 2% sulfanilamide, 1 m HCl were added, followed by 25 μl of 0.2% naphthyl-ethylenediamine. The absorbance at 540 nm was recorded immediately afterward using a Biotek Synergy HT plate reader. A standard curve was prepared from nitrate and nitrite standards under the same conditions as the experimental samples. The standard curve was used to convert the absorbance at 540 nm to concentration.
HPLC product detection
The NOS reaction was performed as stated above, with the substitution of 50 mm HEPES (pH 7.5) for Tris. HPLC detection of derivatized citrulline was performed using the method of Davydov et al. (77) with the following modifications. Using an Agilent 1100 series HPLC equipped with an Agilent 1260 fluorescence detector, an Agilent Eclipse Plus reverse phase column (150 × 4.6 mm; equipped with a Supelguard LC-18-DB guard column) was equilibrated with 50 mm TCA (pH 4.0) and 15% acetonitrile at 1 ml/min. The derivatization agent OPA was dissolved in methanol (8 mg/ml), and then 100 μl of the OPA reagent was added to 900 μl of 100 mm sodium borate (pH 10.0) and 6 μl of β-mercaptoethanol. Then 20 μl of OPA was added to 10 μl of filtered sample. Following 3 min of incubation at room temperature, the mixture was injected onto the column. After injection, the concentration of acetonitrile was increased to 25% over 20 min, and fluorescence was detected at λex = 360 nm and λem = 455 nm. Derivatized citrulline eluted at 8.58 min, followed by NOHA at 12.01 min and l-Arg at 13.34 min. A standard curve was prepared from citrulline standards under the same conditions as the experimental samples. The standard curve was used to convert the peak area to concentration.
Fe-MGD spin trap and ESR detection
The NOS reaction was performed as stated above, with the addition of 0.7 mm Fe(II) sulfate and 2.7 mm MGD (78). After reacting for 30 min at room temperature, glycerol was added to the sample to 15% (w/v). The sample was immediately transferred to an X-band ESR tube and flash-frozen in liquid nitrogen. Continuous-wave ESR spectra were acquired using a Bruker Elexsys E500 CW ESR spectrometer with an ER4131VT variable temperature unit at 150 K and 9.4 GHz, with a modulation amplitude of 1.5 G and modulation frequency of 100 kHz.
NO detection by electrochemistry
NO oxidation was measured using an Innovated Instruments amiNO-2000 electrode and a CH Instruments Electrochemical Analyzer CH1630B potentiostat. The electrode was submerged in buffer (100 mm Tris, pH 7.5, 200 mm NaCl, 1 mm NADPH) while stirring, and a 0.85-V potential was applied for about 5 min to prime the electrode. Data collection was initiated, and the current was measured at a sampling interval of 2/s. The baseline current was recorded for about 5 min. Upon baseline stabilization, 10 μm NOC-7 was added, and the current was allowed to plateau (about 5 min). syNOS was then added to initiate NO oxidation, and data collection continued for approximately 10 min. Signal decay was fit to a monoexponential equation using Matlab, and the rate constants were extracted.
Reduction of NOSg and NOSox by NOSred
All solutions were sparged with argon and then degassed in an anaerobic COY chamber for 6 h. NOSox was added to 2 molar eq of NOSred and 1 mm NADPH, with or without 5 mm CaCl2. NOSg was added to 2 molar eq of NOSred with 1 mm NADPH. Samples were transferred to an anaerobic cuvette, and the absorbance spectra were recorded before and after sparging with carbon monoxide.
Minimal medium growth assay
The growth assay described by Lamattina and co-workers (30) was carried out with the following modifications. E. coli BL21 (DE3) pLysS cells were transformed with either pET-28a containing syNOS or the empty vector. Ten milliliters of LB were inoculated with 100 μl of an overnight culture, with 50 μg/ml kanamycin and 30 μg/ml chloramphenicol, and the culture was incubated at 37 °C until the A600 nm was ∼0.3–0.4. Protein expression was induced with 0.1 mm IPTG, 500 μm δ-aminolevulinic acid, and 1 mm arginine and incubated at 37 °C for 1.5 h. Cells were pelleted by centrifugation at 2,000 × g for 10 min and then washed three times with 5 ml of minimal medium (5.44 g of KH2PO4, 2 g of glucose, and 6 ml of salt solution dissolved in 1 liter of distilled water, pH 7.4; the salt solution contained 10 g of MgSO4·7H2O, 1.0 g of MnCl2·4H2O, 0.4 g of FeSO4·7H2O, and 0.1 g of CaCl2·2H2O dissolved in 1 liter of distilled water). The cells were resuspended in minimal medium and supplemented with 0.1 mm IPTG, 500 μm δ-aminolevulinic acid, and 1 mm arginine in an effort to maintain the conditions specified by Lamattina and co-workers (30). The culture was diluted 1:100 in minimal medium containing either 0.2% (w/v) arginine, 0.3% NH4Cl, or 0.018% NH4Cl and incubated at 37 °C. (Washing with minimal medium was strictly required; the 1:100 dilution in minimal medium contained enough nitrogen to allow for significant growth.) A600 nm was recorded using an Agilent 8453 UV-visible spectrophotometer to monitor cell density.
NO minimum inhibitory concentration
The vector pCW-LIC containing syNOS was transformed into cells of the flavohemoglobin-deficient E. coli strain JW2536 or of the WT strain BW25113. LB containing 100 μg/ml ampicillin (and 50 μg/ml kanamycin for strain JW2536) was inoculated with an overnight culture (1:200 dilution) and incubated at 37 °C until the A600 nm was ∼0.6. Cells were diluted to 106 cfu/ml (A600 nm 1 = 108 cfu/ml) in LB supplemented with antibiotics and 0, 6.25, 12.5, or 25 μg/ml IPTG. DETA NONOate was diluted in 10 mm NaOH, and 20 μl of each dilution was added to 180 μl of each solution of cells in a 96-well plate. Final concentrations of DETA NONOate were 2.7, 1.35, 0.675, 0.338, 0.169, 0.084, 0.042, and 0 mm. Microplates were wrapped with parafilm to prevent excess evaporation and incubated at 37 °C for 18 h. The A600 nm was recorded using a Biotek Synergy HT plate reader.
Author contributions
A. L. P. data curation; A. L. P. formal analysis; A. L. P. validation; A. L. P. and B. R. C. investigation; A. L. P. visualization; A. L. P. methodology; A. L. P. and B. R. C. writing-original draft; A. L. P. and B. R. C. writing-review and editing; B. R. C. conceptualization; B. R. C. resources; B. R. C. supervision; B. R. C. funding acquisition.
Supplementary Material
Acknowledgments
We thank Joanne Widom for help with cloning and Sarah Chobot Hokanson and Magali Moreau for initiating biochemical studies of syNOS.
This work was supported by National Institutes of Health Grant R35GM122535 and National Science Foundation Grant MCB1715233. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Figs. S1–S4.
- NOS
- nitric oxide synthase
- CaM
- calmodulin
- FNR
- ferredoxin-NADP+ reductase
- GTPCH I
- guanosine triphosphate cyclohydrolase I
- H4B
- (6R,1′R,2′S)-5,6,7,8-tetrahydrobiopterin
- iNOS
- inducible nitric oxide synthase
- l-Cit
- l-citrulline
- l-NAA
- NG-amino-l-arginine
- l-NAME
- NG-nitro-l-arginine methyl ester
- l-NNA
- NG-nitro-l-arginine
- mNOS
- mammalian NOS
- NOD
- nitric oxide dioxygenase
- NOHA
- Nω-hydroxy-l-arginine
- nNOS
- neuronal NOS
- NOSg
- nitric oxide synthase globin domain
- NOSox
- nitric oxide synthase oxygenase domain
- NOSred
- nitric-oxide synthase reductase domain
- OPA
- o-phthaldialdehyde
- PTS
- 6-pyruvoyl tetrahydrobiopterin synthase
- SDR
- short-chain dehydrogenase/reductase
- SR
- sepiapterin reductase
- syNOS
- Synechococcus sp. PCC 7335 nitric oxide synthase
- THF
- tetrahydrofolate
- LB
- lysogeny broth
- eNOS
- endothelial NOS
- Ni-NTA
- nickel-nitrilotriacetic acid
- SEC
- size-exclusion chromatography
- MALS
- multiangle light scattering
- flavoHb
- flavohemoglobin
- EV
- empty vector
- MGD
- N-(dithiocarbamoyl)-N-methyl-d-glucamine
- IPTG
- isopropyl β-d-thiogalactopyranoside
- ESR
- electron spin resonance
- DETA
- diethylenetriamine.
References
- 1. Zumft W. G. (1997) Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gusarov I., and Nudler E. (2018) Protein S-nitrosylation: enzymatically controlled, but intrinsically unstable, post-translational modification. Mol. Cell. 69, 351–353 10.1016/j.molcel.2018.01.022 [DOI] [PubMed] [Google Scholar]
- 3. Plate L., and Marletta M. A. (2013) Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem. Sci. 38, 566–575 10.1016/j.tibs.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denninger J. W., and Marletta M. A. (1999) Guanylate cyclase and the •NO/cGMP signaling pathway. Biochim. Biophys. Acta 1411, 334–350 10.1016/S0005-2728(99)00024-9 [DOI] [PubMed] [Google Scholar]
- 5. Knowles R. G., Palacios M., Palmer R. M., and Moncada S. (1989) Formation of nitric oxide from l-arginine in the central nervous system: a transduction mechanism for stimulation of the soluble guanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 86, 5159–5162 10.1073/pnas.86.13.5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palmer R. M., and Moncada S. (1989) A novel citrulline-forming enzyme implicated in the formation of nitric oxide by vascular endothelial cells. Biochem. Biophys. Res. Commun. 158, 348–352 10.1016/S0006-291X(89)80219-0 [DOI] [PubMed] [Google Scholar]
- 7. Bredt D. S., and Snyder S. H. (1990) Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc. Natl. Acad. Sci. U.S.A. 87, 682–685 10.1073/pnas.87.2.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yui Y., Hattori R., Kosuga K., Eizawa H., Hiki K., Ohkawa S., Ohnishi K., Terao S., and Kawai C. (1991) Calmodulin-independent nitric oxide synthase from rat polymorphonuclear neutrophils. J. Biol. Chem. 266, 3369–3371 [PubMed] [Google Scholar]
- 9. Siddhanta U., Presta A., Fan B., Wolan D., Rousseau D. L., and Stuehr D. J. (1998) Domain swapping in inducible nitric-oxide synthase. J. Biol. Chem. 273, 18950–18958 10.1074/jbc.273.30.18950 [DOI] [PubMed] [Google Scholar]
- 10. Daff S. (2003) Calmodulin-dependent regulation of mammalian nitric oxide synthase. Biochem. Soc. Trans. 31, 502–505 10.1042/bst0310502 [DOI] [PubMed] [Google Scholar]
- 11. Tejero J., and Stuehr D. (2013) Tetrahydrobiopterin in nitric oxide synthase. IUBMB Life 65, 358–365 10.1002/iub.1136 [DOI] [PubMed] [Google Scholar]
- 12. Bogdan C. (2001) Nitric oxide and the immune response. Nat. Immunol. 2, 907–916 10.1038/ni1001-907 [DOI] [PubMed] [Google Scholar]
- 13. Chachlaki K., Garthwaite J., and Prevot V. (2017) The gentle art of saying NO: how nitric oxide gets things done in the hypothalamus. Nat. Rev. Endocrinol. 13, 521–535 10.1038/nrendo.2017.69 [DOI] [PubMed] [Google Scholar]
- 14. Ignarro L. J. (2002) Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J. Physiol. Pharmacol. 53, 503–514 [PubMed] [Google Scholar]
- 15. Hobbs A. J., Higgs A., and Moncada S. (1999) Inhibition of nitric oxide synthase as a potential therapeutic target. Annu. Rev. Pharmacol. Toxicol. 39, 191–220 10.1146/annurev.pharmtox.39.1.191 [DOI] [PubMed] [Google Scholar]
- 16. Vannini F., Kashfi K., and Nath N. (2015) The dual role of iNOS in cancer. Redox Biol. 6, 334–343 10.1016/j.redox.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Connelly L., Madhani M., and Hobbs A. J. (2005) Resistance to endotoxic shock in endothelial nitric-oxide synthase (eNOS) knock-out mice: a pro-inflammatory role for eNOS-derived NO in vivo. J. Biol. Chem. 280, 10040–10046 10.1074/jbc.M411991200 [DOI] [PubMed] [Google Scholar]
- 18. Choudhari S. K., Chaudhary M., Bagde S., Gadbail A. R., and Joshi V. (2013) Nitric oxide and cancer: a review. World J. Surg. Oncol. 11, 118 10.1186/1477-7819-11-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel B. A., Moreau M., Widom J., Chen H., Yin L., Hua Y., and Crane B. R. (2009) Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proc. Natl. Acad. Sci. U.S.A. 106, 18183–18188 10.1073/pnas.0907262106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao M., Smith B. C., and Marletta M. A. (2015) Nitric oxide mediates biofilm formation and symbiosis in silicibacter. MBio 6, e00206–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gusarov I., and Nudler E. (2005) NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 13855–13860 10.1073/pnas.0504307102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shatalin K., Gusarov I., Avetissova E., Shatalina Y., McQuade L. E., Lippard S. J., and Nudler E. (2008) Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 105, 1009–1013 10.1073/pnas.0710950105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kinkel T. L., Ramos-Montañez S., Pando J. M., Tadeo D. V., Strom E. N., Libby S. J., and Fang F. C. (2016) An essential role for bacterial nitric oxide synthase in Staphylococcus aureus electron transfer and colonization. Nat. Microbiol. 2, 16224 10.1038/nmicrobiol.2016.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaudhari S. S., Kim M., Lei S., Razvi F., Alqarzaee A. A., Hutfless E. H., Powers R., Zimmerman M. C., Fey P. D., and Thomas V. C. (2017) Nitrite derived from endogenous bacterial nitric oxide synthase activity promotes aerobic respiration. MBio 8, e00887–17 10.1128/mBio.00887-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crane B. R., Sudhamsu J., and Patel B. A. (2010) Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79, 445–470 10.1146/annurev-biochem-062608-103436 [DOI] [PubMed] [Google Scholar]
- 26. Gusarov I., Starodubtseva M., Wang Z. Q., McQuade L., Lippard S. J., Stuehr D. J., and Nudler E. (2008) Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283, 13140–13147 10.1074/jbc.M710178200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agapie T., Suseno S., Woodward J. J., Stoll S., Britt R. D., and Marletta M. A. (2009) NO formation by a catalytically self-sufficient bacterial nitric oxide synthase from Sorangium cellulosum. Proc. Natl. Acad. Sci. U.S.A. 106, 16221–16226 10.1073/pnas.0908443106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foresi N., Correa-Aragunde N., Parisi G., Caló G., Salerno G., and Lamattina L. (2010) Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell. 22, 3816–3830 10.1105/tpc.109.073510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Domingos P., Prado A. M., Wong A., Gehring C., and Feijo J. A. (2015) Nitric oxide: a multitasked signaling gas in plants. Mol. Plant. 8, 506–520 10.1016/j.molp.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 30. Correa-Aragunde N., Foresi N., Del Castello F., and Lamattina L. (2018) A singular nitric oxide synthase with a globin domain found in Synechococcus PCC 7335 mobilizes N from arginine to nitrate. Sci. Rep. 8, 12505 10.1038/s41598-018-30889-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta S., Pawaria S., Lu C., Yeh S. R., and Dikshit K. L. (2011) Novel flavohemoglobins of mycobacteria. IUBMB Life 63, 337–345 10.1002/iub.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta S., Pawaria S., Lu C., Hade M. D., Singh C., Yeh S. R., and Dikshit K. L. (2012) An unconventional hexacoordinated flavohemoglobin from Mycobacterium tuberculosis. J. Biol. Chem. 287, 16435–16446 10.1074/jbc.M111.329920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sabat J., Stuehr D. J., Yeh S. R., and Rousseau D. L. (2009) Characterization of the proximal ligand in the P420 form of inducible nitric oxide synthase. J. Am. Chem. Soc. 131, 12186–12192 10.1021/ja901016a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., and Dawson J. H. (2003) Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 10.1073/pnas.0737142100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kakar S., Hoffman F. G., Storz J. F., Fabian M., and Hargrove M. S. (2010) Structure and reactivity of hexacoordinate hemoglobins. Biophys. Chem. 152, 1–14 10.1016/j.bpc.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brunori M., Giuffrè A., Nienhaus K., Nienhaus G. U., Scandurra F. M., and Vallone B. (2005) Neuroglobin, nitric oxide, and oxygen: functional pathways and conformational changes. Proc. Natl. Acad. Sci. U.S.A. 102, 8483–8488 10.1073/pnas.0408766102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stuehr D. J., and Ikeda-Saito M. (1992) Spectral characterization of brain and macrophage nitric oxide synthases: cytochrome P-450-like hemeproteins that contain a flavin semiquinone radical. J. Biol. Chem. 267, 20547–20550 [PubMed] [Google Scholar]
- 38. Pfeiffer S., Leopold E., Schmidt K., Brunner F., and Mayer B. (1996) Inhibition of nitric oxide synthesis by NG* nitro-l-arginine methyl ester (l-NAME): requirement for bioactivation to the free acid. Br. J. Pharmacol. 118, 1433–1440 10.1111/j.1476-5381.1996.tb15557.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonamore A., and Boffi A. (2008) Flavohemoglobin: structure and reactivity. IUBMB Life 60, 19–28 10.1002/iub.9 [DOI] [PubMed] [Google Scholar]
- 40. Wang Z. Q., Haque M. M., Binder K., Sharma M., Wei C. C., and Stuehr D. J. (2016) Engineering nitric oxide synthase chimeras to function as NO dioxygenases. J. Inorg. Biochem. 158, 122–130 10.1016/j.jinorgbio.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto K., Kataoka E., Miyamoto N., Furukawa K., Ohsuye K., and Yabuta M. (2003) Genetic engineering of Escherichia coli for production of tetrahydrobiopterin. Metab. Eng. 5, 246–254 10.1016/S1096-7176(03)00046-6 [DOI] [PubMed] [Google Scholar]
- 42. Membrillo-Hernández J., Coopamah M. D., Anjum M. F., Stevanin T. M., Kelly A., Hughes M. N., and Poole R. K. (1999) The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274, 748–754 10.1074/jbc.274.2.748 [DOI] [PubMed] [Google Scholar]
- 43. Salerno J. C., Harris D. E., Irizarry K., Patel B., Morales A. J., Smith S. M. E., Martasek P., Roman L. J., Masters B. S. S., Jones C. L., Weissman B. A., Lane P., Liu Q., and Gross S. S. (1997) An autoinhibitory control element defines calcium-regulated isoforms of nitric oxide synthase. J. Biol. Chem. 272, 29769–29777 10.1074/jbc.272.47.29769 [DOI] [PubMed] [Google Scholar]
- 44. Daff S., Sagami I., and Shimizu T. (1999) The 42-amino acid insert in the FMN domain of neuronal nitric-oxide synthase exerts control over Ca2+/calmodulin-dependent electron transfer. J. Biol. Chem. 274, 30589–30595 10.1074/jbc.274.43.30589 [DOI] [PubMed] [Google Scholar]
- 45. Li H., Raman C. S., Glaser C. B., Blasko E., Young T. A., Parkinson J. F., Whitlow M., and Poulos T. L. (1999) Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. J. Biol. Chem. 274, 21276–21284 10.1074/jbc.274.30.21276 [DOI] [PubMed] [Google Scholar]
- 46. Raman C. S., Li H., Martásek P., Král V., Masters B. S. S., and Poulos T. L. (1998) Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell 95, 939–950 10.1016/S0092-8674(00)81718-3 [DOI] [PubMed] [Google Scholar]
- 47. Domínguez D. C., Guragain M., and Patrauchan M. (2015) Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 57, 151–165 10.1016/j.ceca.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 48. Clapham D. (2007) Calcium signaling. Cell 131, 1047–1058 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 49. Wang H., Yang B., Hao G., Feng Y., Chen H., Feng L., Zhao J., Zhang H., Chen Y. Q., Wang L., and Chen W. (2011) Biochemical characterization of the tetrahydrobiopterin synthesis pathway in the oleaginous fungus Mortierella alpina. Microbiology 157, 3059–3070 10.1099/mic.0.051847-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thöny B., Auerbach G., and Blau N. (2000) Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347, 1–16 10.1042/bj3470001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujimoto K., Hara M., Yamada H., Sakurai M., Inaba A., Tomomura A., and Katoh S. (2001) Role of the conserved Ser-Tyr-Lys triad of the SDR family in sepiapterin reductase. Chem. Biol. Interact. 130, 825–832 [DOI] [PubMed] [Google Scholar]
- 52. Crane B. R., Arvai A. S., Ghosh D. K., Wu C., Getzoff E. D., Stuehr D. J., and Tainer J. A. (1998) Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science 279, 2121–2126 10.1126/science.279.5359.2121 [DOI] [PubMed] [Google Scholar]
- 53. Kim S. O., Orii Y., Lloyd D., Hughes M. N., and Poole R. K. (1999) Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445, 389–394 [DOI] [PubMed] [Google Scholar]
- 54. Gardner P. R., Gardner A. M., Martin L. A., and Salzman A. L. (1998) Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U.S.A. 95, 10378–10383 10.1073/pnas.95.18.10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z. Q., Wei C. C., Sharma M., Pant K., Crane B. R., and Stuehr D. J. (2004) A conserved Val to Ile switch near the heme pocket of animal and bacterial nitric-oxide synthases helps determine their distinct catalytic profiles. J. Biol. Chem. 279, 19018–19025 10.1074/jbc.M311663200 [DOI] [PubMed] [Google Scholar]
- 56. Schneider B. L., Kiupakis A. K., and Reitzer L. J. (1998) Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180, 4278–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reitzer L. (2005) Catabolism of amino acids and related compounds. EcoSal Plus 10.1128/ecosalplus.3.4.7 10.1128/ecosalplus.3.4.7 [DOI] [PubMed] [Google Scholar]
- 58. Schriek S., Rückert C., Staiger D., Pistorius E. K., and Michel K. P. (2007) Bioinformatic evaluation of l-arginine catabolic pathways in 24 cyanobacteria and transcriptional analysis of genes encoding enzymes of l-arginine catabolism in the cyanobacterium Synechocystis sp. PCC 6803. BMC Genomics 8, 437 10.1186/1471-2164-8-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gau A. E., Heindl A., Nodop A., Kahmann U., and Pistorius E. K. (2007) l-Amino acid oxidases with specificity for basic l-amino acids in cyanobacteria. Z. Naturforsch. C 62, 273–284 10.1515/znc-2007-3-419 [DOI] [PubMed] [Google Scholar]
- 60. Patey M. D., Rijkenberg M. J. A., Statham P. J., Stinchcombe M. C., Achterberg E. P., and Mowlem M. (2008) Determination of nitrate and phosphate in seawater at nanomolar concentrations. Trends Anal. Chem. 27, 169–182 10.1016/j.trac.2007.12.006 [DOI] [Google Scholar]
- 61. Watanabe M., Ohtsu J., and Otsuki A. (2000) Daily variations in nutrient concentrations of seawater at 321 m depth in Toyama Bay, Japan Sea. J. Oceanogr. 56, 553–558 10.1023/A:1011153111688 [DOI] [Google Scholar]
- 62. Hansell D. A., and Follows M. J. (2008) Nitrogen in the Atlantic Ocean. in Nitrogen in the Marine Environment, 2nd Ed. (Capone D. G., Bronk D. A., Mulholland M. R., and Carpenter E. J., eds) pp. 597–630, Academic Press, San Diego, CA [Google Scholar]
- 63. Clark M. E., Jackson G. A., and North W. J. (1972) Dissolved free amino acids in Southern California coastal waters. Limnol. Oceanogr. 17, 749–758 10.4319/lo.1972.17.5.0749 [DOI] [Google Scholar]
- 64. Kaiser K., and Benner R. (2009) Biochemical composition and size distribution of organic matter at the Pacific and Atlantic time-series stations. Mar. Chem. 113, 63–77 10.1016/j.marchem.2008.12.004 [DOI] [Google Scholar]
- 65. Hubberten U., Lara R. J., and Kattner G. (1994) Amino acid composition of seawater and dissolved humic substances in the Greenland Sea. Mar. Chem. 45, 121–128 10.1016/0304-4203(94)90096-5 [DOI] [Google Scholar]
- 66. Rippka R., and Waterbury J. B. (1977) The synthesis of nitrogenase by non-heterocystous cyanobacteria. FEMS Microbiol. Lett. 2, 83–86 10.1111/j.1574-6968.1977.tb00913.x [DOI] [Google Scholar]
- 67. Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R., Herdman M., Sivonen K., Coursin T., Laurent T., Goodwin L., et al. (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058 10.1073/pnas.1217107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rippka R., Deruelles J., Waterbury J. B., Herdman M., and Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. J. Gen. Microbiol. 111, 1–61 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- 69. Lechauve C., Butcher J. T., Freiwan A., Biwer L. A., Keith J. M., Good M. E., Ackerman H., Tillman H. S., Kiger L., Isakson B. E., and Weiss M. J. (2018) Endothelial cell α-globin and its molecular chaperone α-hemoglobin-stabilizing protein regulate arteriolar contractility. J. Clin. Invest. 128, 5073–5082 10.1172/JCI99933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walter J., Lynch F., Battchikova N., Aro E. M., and Gollan P. J. (2016) Calcium impacts carbon and nitrogen balance in the filamentous cyanobacterium Anabaena sp. PCC 7120. J. Exp. Bot. 67, 3997–4008 10.1093/jxb/erw112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leganés F., Forchhammer K., and Fernández-Piñas F. (2009) Role of calcium in acclimation of the cyanobacterium Synechococcus elongatus PCC 7942 to nitrogen starvation. Microbiology 155, 25–34 10.1099/mic.0.022251-0 [DOI] [PubMed] [Google Scholar]
- 72. Zhao Y., Shi Y., Zhao W., Huang X., Wang D., Brown N., Brand J., and Zhao J. (2005) CcbP, a calcium-binding protein from Anabaena sp. PCC 7120, provides evidence that calcium ions regulate heterocyst differentiation. Proc. Natl. Acad. Sci. U.S.A. 102, 5744–5748 10.1073/pnas.0501782102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Singh S. P., Rastogi R. P., Häder D.-P., and Sinha R. P. (2011) An improved method for genomic DNA extraction from cyanobacteria. World J. Microbiol. Biotechnol. 27, 1225–1230 10.1007/s11274-010-0571-8 [DOI] [Google Scholar]
- 74. Moreau M., Takahashi H., Sari M. A., Boucher J. L., Sagami I., Shimizu T., and Mansuy D. (2004) Importance of valine 567 in substrate recognition and oxidation by neuronal nitric oxide synthase. J. Inorg. Biochem. 98, 1200–1209 10.1016/j.jinorgbio.2004.03.015 [DOI] [PubMed] [Google Scholar]
- 75. Sohn O. S., and Fiala E. S. (2000) Analysis of nitrite/nitrate in biological fluids: denitrification of 2-nitropropane in F344 rats. Anal. Biochem. 279, 202–208 10.1006/abio.1999.4471 [DOI] [PubMed] [Google Scholar]
- 76. Medina A., and Nicholas D. J. D. (1957) Interference by reduced pyridine nucleotides in the diazotization of nitrite. Biochim. Biophys. Acta 23, 440–442 10.1016/0006-3002(57)90355-4 [DOI] [PubMed] [Google Scholar]
- 77. Davydov R., Sudhamsu J., Lees N. S., Crane B. R., and Hoffman B. M. (2009) EPR and ENDOR characterization of the reactive intermediates in the generation of NO by cryoreduced oxy-nitric oxide synthase from Geobacillus stearothermophilus. J. Am. Chem. Soc. 131, 14493–14507 10.1021/ja906133h [DOI] [PubMed] [Google Scholar]
- 78. Huisman A., Vos I., van Faassen E. E., Joles J. A., Gröne H. J., Martasek P., van Zonneveld A. J., Vanin A. F., and Rabelink T. J. (2002) Anti-inflammatory effects of tetrahydrobiopterin on early rejection in renal allografts: modulation of inducible nitric oxide synthase. FASEB J. 16, 1135–1137 10.1096/fj.01-0890fje [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


