Abstract
The role of mitochondria in cancer continues to be debated, and whether exploitation of mitochondrial functions is a general hallmark of malignancy or a tumor- or context-specific response is still unknown. Using a variety of cancer cell lines and several technical approaches, including siRNA-mediated gene silencing, ChIP assays, global metabolomics and focused metabolite analyses, bioenergetics, and cell viability assays, we show that two oncogenic Myc proteins, c-Myc and N-Myc, transcriptionally control the expression of the mitochondrial chaperone TNFR-associated protein-1 (TRAP1) in cancer. In turn, this Myc-mediated regulation preserved the folding and function of mitochondrial oxidative phosphorylation (OXPHOS) complex II and IV subunits, dampened reactive oxygen species production, and enabled oxidative bioenergetics in tumor cells. Of note, we found that genetic or pharmacological targeting of this pathway shuts off tumor cell motility and invasion, kills Myc-expressing cells in a TRAP1-dependent manner, and suppresses primary and metastatic tumor growth in vivo. We conclude that exploitation of mitochondrial functions is a general trait of tumorigenesis and that this reliance of cancer cells on mitochondrial OXPHOS pathways could offer an actionable therapeutic target in the clinic.
Keywords: metabolism, mitochondria, Myc (c-Myc), metastasis, invasion, oxidative phosphorylation, TRAP1
Introduction
A hallmark of cancer is the reprogramming of cellular metabolism (1). Much work has been devoted to the preferential utilization of glycolysis by tumor cells, even when oxygen is present (2), and its impact on cancer progression (3). However, we now know that mitochondria remain functional in most malignancies and that oxidative bioenergetics fuel important tumor traits (4), including metastasis (5, 6). Although both metabolic states are likely to coexist during tumor growth (7), a direct, mechanistic link between oncogenic signaling and mitochondrial functions has not been determined, and the question of how general is the exploitation of mitochondria in cancer (4) has remained unanswered.
In this context, Myc proteins comprise a family of ubiquitous, transforming oncogenes (8) that are amplified, deregulated, or translocated in most human cancers (9). Oncogenic Myc drives a plethora of transcriptional and nontranscriptional responses that promote tumor growth and proliferation (10), linked to worse disease outcome in the clinic (11). Although the Myc target gene(s) in tumor progression has not been completely elucidated, and controversy still exists about the requirements of Myc-directed gene expression (12), this pathway connects to multiple aspects of tumor metabolism (13), including mitochondrial functions (14, 15).
In this study, we investigate a role of oncogenic Myc in exploiting mitochondria for cancer progression.
Results and discussion
Myc regulation of mitochondrial metabolism
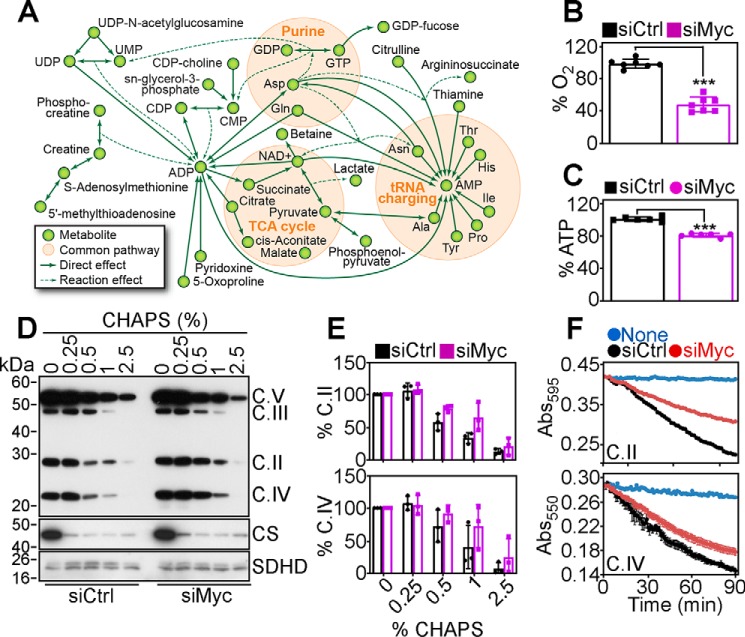
Oncogenic Myc drives aggressive and metastatic prostate cancer (16). Therefore, we began this study by conducting a global metabolomics screening (∼5,000 metabolites, first metabolomic screen) in prostate adenocarcinoma PC3 cells transfected with nontargeted siRNA or Myc-directed siRNA. Loss of Myc in these conditions down-regulated several metabolic clusters (clusters 9 and 4) (Fig. S1A). Bioinformatics analysis of this dataset (Fig. S1B) demonstrated that Myc loss reduced the levels of purine (AMP ↓1.9-fold; ADP ↓1.7-fold; GDP ↓1.9-fold; GTP ↓1.3-fold) and pyrimidine (UDP ↓2.6-fold; UTP ↓1.6-fold) bases, TCA2 cycle intermediates (citrate ↓1.7-fold, malate ↓1.4-fold; cis-aconitic acid ↓1.7-fold; succinate ↓1.3-fold), and catabolites (urea ↓1.5-fold; creatinine ↓1.3-fold) (Fig. 1A, Fig. S1B). A second focused metabolomics screen of ∼300 metabolites gave similar results (Fig. S1B), as Myc silencing in PC3 cells reduced the levels of oxidative phosphorylation and TCA cycle biochemicals, along with metabolites involved in purine and pyrimidine biosynthetic pathways and amino acid levels (Fig. 1A and Fig. S1B). Conversely, Myc knockdown did not affect glutamine levels in PC3 cells (13), suggesting tumor-specific differences in metabolic reprogramming. Consistent with these data, Myc-silenced PC3 cells exhibited decreased oxygen consumption rates (OCR) (Fig. S1C) and overall reduced oxygen consumption (Fig. 1B). Together with lower glucose utilization (Fig. S1D), decreased lactate generation (Fig. S1E), and reduced extracellular acidification rates (ECAR) (Fig. S1F), these cells showed diminished ATP production, compared with control transfectants (Fig. 1C).
Figure 1.
Myc regulation of mitochondrial bioenergetics and protein folding. A, networks of associations between metabolites significantly affected by siRNA silencing of Myc in PC3 cells. The metabolites shown are concordantly modulated in a global metabolomics screen (∼5,000 metabolites) and a focused metabolite analysis (∼300 metabolites). B and C, PC3 cells transfected with control nontargeting siRNA (siCtrl) or Myc-directed siRNA (siMyc) were analyzed for total oxygen consumption (B) or ATP production (C). Mean ± S.D. (n = 3–7). ***, p < 0.0001. D, PC3 cells were transfected as in B, and detergent (CHAPS)-insoluble proteins were analyzed. The position of the oxidative phosphorylation complex (C) subunits is indicated. Citrate synthase (CS) and oxidative phosphorylation complex II subunit, SDHD, were used as controls. E, conditions are as in C, and protein bands corresponding to complex II subunit, SDHB, or complex IV subunit, CoxII, were quantified by densitometry (n = 2). F, PC3 cells transfected as in B were analyzed for citrate synthase–normalized mitochondrial oxidative phosphorylation complex II (top) or complex IV (bottom) activity. Representative tracings out of three independent experiments are shown.
Control of mitochondrial protein folding by Myc
Next, we looked at the mechanism(s) of Myc regulation of oxidative phosphorylation. We found that Myc silencing in PC3 cells caused the accumulation of detergent-insoluble subunits, i.e. misfolded oxidative phosphorylation complex subunits, including succinate dehydrogenase B (SDHB, complex II) and cytochrome c oxidase II (CoxII, complex IV) (Fig. 1, D and E). Other oxidative phosphorylation subunits, ubiquinol-cytochrome c reductase core protein 2 (UQCRC2, complex III) or ATP synthase F1 subunit (ATP5A, complex V), were unaffected (Fig. S2A). Consistent with these results, Myc knockdown resulted in reduced mitochondrial complex II and complex IV activity, compared with control transfectants (Fig. 1F; Fig. S2B). Instead, complex I activity was unchanged in control or Myc-silenced cells (Fig. S2, C and D). In line with the defects of oxidative bioenergetics, Myc loss was accompanied by increased production of mitochondrial ROS (Fig. S2E), hyperoxidation of peroxiredoxin-3 (Prx3) (Fig. S2F), a marker of oxidative stress, and phosphorylation of the energy sensor, AMPK (Thr-172), indicative of nutrient deprivation (Fig. S2F).
Mitochondrial chaperone TRAP1 is a novel Myc target gene
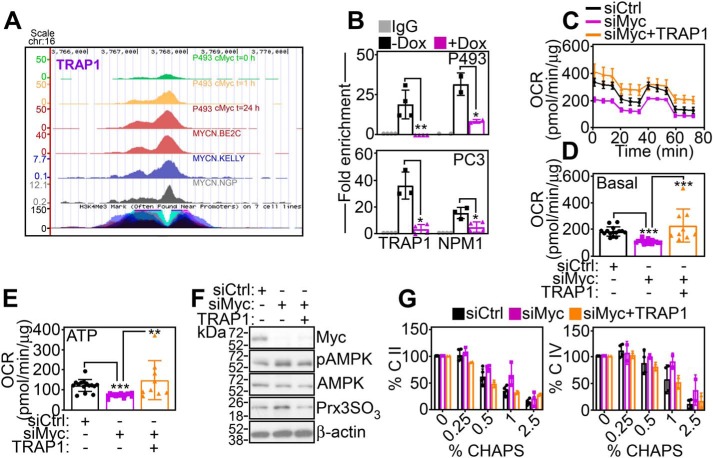
Previous studies have shown that SDHB protein folding requires the activity of the mitochondrial Hsp90-like chaperone, TNFR-associated protein-1 (TRAP1) (17). Analysis of ChIP-Seq tracks demonstrated time-dependent accumulation of Myc at the TRAP1 promoter in Burkitt's lymphoma P493 cells as well as neuroblastoma BE2C, Kelly, and NGP cell lines (Fig. 2A). Consistent with this, Myc bound to the promoter of TRAP1, as well as NPM1, a known Myc target gene, by ChIP in P493 or PC3 cells, in a reaction abolished by siRNA knockdown of Myc (Fig. 2B). As a result, Myc silencing reduced the expression of TRAP1 mRNA (Fig. S3A) and protein (Fig. S3B) in various tumor cell types. Bioinformatics analysis of four independent cancer cell line data bases demonstrated that high levels of Myc strongly correlated (p = 2.6 × 10−20–6.1 × 10−117) with TRAP1 expression (Fig. S3C), reinforcing the generality of this response across genetically disparate tumors.
Figure 2.
Identification of TRAP1 as a Myc target gene. A, ChIP-Seq tracks of Myc accumulation at the TRAP1 promoter in P493 at three time points after removal of Dox or neuroblastoma BE2C, Kelly, or NGP cell lines. B, ChIP of Myc accumulation at TRAP1 or NPM1 promoter in P493 cells containing a Dox-regulated inducible Myc transgene (Dox-off system) in the presence (+) or absence (−) of Dox (top panel) or PC3 cells transfected with siCtrl or siMyc (bottom panel). IgG, nonbinding IgG. Mean ± S.D. (n = 3). *, p = 0.04–0.01; **, p = 0.005. C–E, PC3 cells transfected with siCtrl or siMyc were reconstituted with TRAP1 cDNA and analyzed for OCR using an Agilent Seahorse XFe96 analyzer (C, representative tracings) and basal respiration (D) or total ATP production (E) was quantified. Mean ± S.D., **, p = 0.005; ***, p < 0.0001. F, reconstitution conditions are as in C, and transfected PC3 cells were analyzed by Western blotting. p, phosphorylated. G, reconstitution conditions are as in C, and isolated mitochondrial extracts were analyzed for accumulation of detergent (CHAPS)-insoluble SDHB (C. II, left panel) or CoxII (C. IV, right panel) by Western blotting followed by densitometric quantification of protein bands. Mean ± S.D.
We next asked whether the Myc homolog, N-Myc, also regulated TRAP1 in the model of neuroblastoma (Fig. 2A). Conditional silencing of N-Myc in neuroblastoma SHEP21 cells reduced TRAP1 mRNA (Fig. S4A) and protein expression (Fig. S4B). Conversely, 4OHT-regulated induction of N-Myc in SHEP or SKNAS cells increased TRAP1 mRNA (Fig. S4C) and protein levels (Fig. S4B). Sequence analysis of a human TRAP1 locus revealed the presence of putative c-Myc- and N-Myc–binding sites within ∼1.4 kb of the first ATG (Fig. S4D). Transfection of this TRAP1 promoter region upstream of a luciferase reporter gene (TRAP1-Luc) produced luciferase activity in PC3 or P493 cells, in a response abolished by siRNA knockdown of Myc (Fig. S4E). In addition, mutagenesis of a putative Myc-binding site at position −60 abolished TRAP1-Luc luciferase activity in control and Myc-knockdown PC3 cells (Fig. S4F).
Next, we asked whether TRAP1 was required for mitochondrial bioenergetics in Myc-expressing tumors. We found that re-expression of non-siRNA–inhibitable TRAP1 in Myc knockdown cells rescued the defect of OCR (Fig. 2C) and corrected basal respiration in these settings (Fig. 2D). Consistent with improved bioenergetics, reconstitution with TRAP1 restored ATP production in Myc knockdown cells (Fig. 2E) and reversed the expression of markers of cellular starvation (AMPK phosphorylation) and oxidative stress (Prx3 hyperoxidation) (Fig. 2F). As a prerequisite of these responses, re-expression of TRAP1 in Myc knockdown PC3 cells corrected the folding of SDHB (complex II) and CoxII (complex IV) subunits (Fig. 2G; Fig. S5A), restoring complex II (Fig. S5, B and C) and complex IV (Fig. S5, D and E) activity, quantitatively comparable with control transfectants.
Myc–TRAP1 signaling fuels tumor chemotaxis and invasion
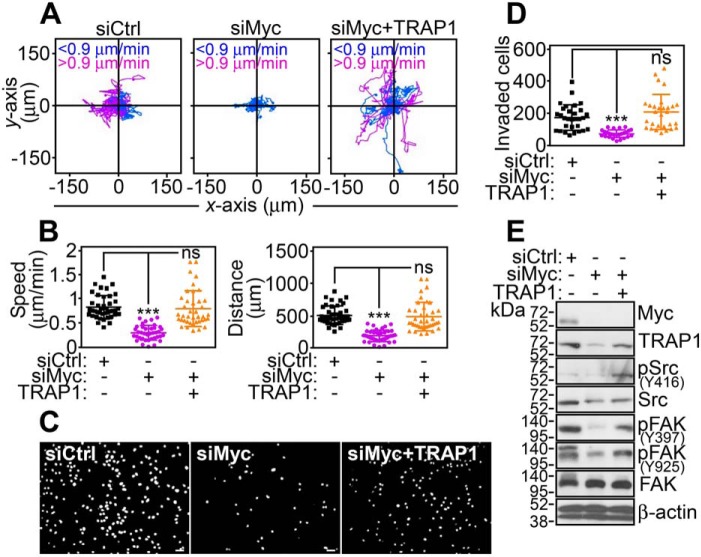
TRAP1-directed bioenergetics has been implicated in tumor traits (18), including tumor cell motility (19), and this possibility was next investigated. We found that Myc silencing with two independent siRNA sequences (Fig. S6A) or pooled siRNA (Fig. 3A) potently inhibited tumor cell motility (Fig. S6A; Fig. 3A). Quantitative analysis of time-lapse video microscopy demonstrated that Myc loss suppressed the speed of cell movements and shortened the total distance traveled by individual cells (Fig. 3B; Fig. S6B). Consistent with these data, Myc knockdown suppressed tumor cell invasion across Matrigel-coated inserts (Fig. 3, C and D; Fig. S6, D and E). In reconstitution experiments, re-expression of TRAP1 in Myc-silenced cells was sufficient to correct the defect in tumor cell motility (Fig. 3A; Fig. S6A), restore the speed of cell movements and the total distance traveled by individual cells (Fig. 3B; Fig. S6B), and increase tumor cell invasion across Matrigel to levels of control transfectants (Fig. 3, C and D; Fig. S6, C and D). Biochemically, PC3 cells reconstituted with TRAP1 also exhibited phosphorylation of cell motility kinases, Src and focal adhesion kinase (FAK), which was suppressed by Myc knockdown (Fig. 3E).
Figure 3.
Myc–TRAP1 regulation of tumor cell motility. A, PC3 cells transfected with siCtrl or siMyc were reconstituted with TRAP1 cDNA and analyzed for 2D cell motility by time-lapse video microscopy. Each tracing corresponds to the movements of an individual cell (n = 19–20). The cutoff velocities for slow (<0.9 μm/min)- or fast (>0.9 μm/min)-moving cells are indicated. The siCtrl panel is the same as siCtrl in Fig. S6A. B, conditions are as in A, and PC3 cells were analyzed for speed of cell movements (left panel) or total distance traveled by individual cells (right panel). Each symbol corresponds to an individual determination (n = 38–41). ***, p < 0.0001; ns, not significant. C and D, conditions are as in A, and transfected PC3 cells were analyzed for invasion across Matrigel-coated inserts (C, representative images of 4′,6-diamidino-2-phenylindole–stained nuclei of invaded cells) and quantified (D). Each symbol corresponds to an individual determination (siCtrl, n = 30; siMyc, n = 26; siMyc + TRAP1, n = 29). The siCtrl panel is the same as siCtrl in Fig. S6C. Scale bars, 50 μm. Mean ± S.D. ***, p < 0.0001; ns, not significant. E, conditions are as in A, and transfected PC3 cells were analyzed by Western blotting.
Pharmacologic targeting of Myc–TRAP1 for cancer therapy
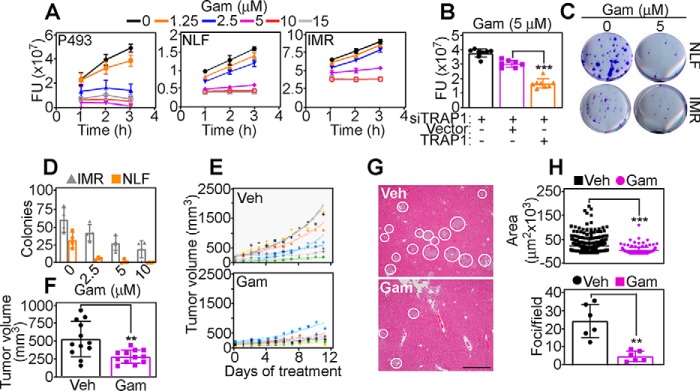
Analysis of neuroblastoma patient cohorts revealed that TRAP1 expression was strongly associated with N-Myc levels (p = 1 × 10−63–2 × 10−96) (Fig. S7A) and correlated with shortened overall survival (Fig. S7B), reinforcing the clinical relevance of this pathway. To test whether this could provide an actionable therapeutic target, we next treated P493 or neuroblastoma NLF or IMR cells with a mitochondria-targeted small molecule TRAP1 antagonist, Gamitrinib (18). In these experiments, Gamitrinib treatment caused concentration-dependent dissipation of mitochondrial inner membrane potential (Fig. S7C) and complete loss of tumor cell viability (Fig. 4A). This response was specific because siRNA silencing of TRAP1 attenuated Gamitrinib-mediated cell killing, whereas reconstitution with TRAP1 strongly increased cell death in these settings (Fig. 4B). Consistent with these data, Gamitrinib suppressed neuroblastoma NLF or IMR colony formation (Fig. 4, C and D) and inhibited P493 xenograft tumor growth in immunocompromised mice (Fig. 4, E and F). When analyzed in a model of disseminated disease, systemic administration of Gamitrinib suppressed liver metastases of PC3 cells (Fig. 4G), reducing both the surface area and number of metastatic foci (Fig. 4H), compared with vehicle-treated animals.
Figure 4.
Myc–TRAP1 signaling promotes primary and metastatic tumor growth. A, P493, NLF, or IMR cells were incubated with increasing concentrations of Gamitrinib (Gam) and analyzed for cell viability after the indicated time intervals. FU, fluorescence units. Mean ± S.D. (n = 3). B, IMR cells were transfected with TRAP1-directed siRNA, reconstituted with vector or TRAP1, and analyzed for cell viability in the presence of Gamitrinib. Mean ± S.D. (n = 8). ***, p < 0.0001. C and D, NLF or IMR cells were treated with the indicated concentrations of Gamitrinib and analyzed for colony formation (B, representative images) and quantified (C). Mean ± S.D. (n = 3). E, P493 cells were engrafted s.c. in athymic nude mice, and randomized animal groups were treated with vehicle (Veh) or Gamitrinib (10 mg/kg/daily/i.p.) with quantification of tumor growth at the indicated time intervals. Trend lines correspond to an individual tumor. F, conditions are as in E, and tumor growth in the indicated animal groups was quantified after 12 days of treatment. Mean ± S.E. (n = 12). **, p = 0.003. G, immunocompromised mice injected with PC3 cells into the spleen were treated with vehicle or Gamitrinib (10 mg/kg/daily/i.p.), and metastatic foci in the liver were analyzed by H&E staining after 11 days. White circles, metastatic foci. Representative images are shown. Scale bar, 500 μm. H, conditions are as in G, and the surface area (top panel) and number (bottom panel) of liver metastases in each treatment group were quantified (top, n = 156–344; bottom, n = 4–7). Mean ± S.D.**, p = 0.002; ***, p < 0.0001.
In sum, the identification of TRAP1 as a direct transcriptional target of a ubiquitous oncogene, i.e. Myc (11), demonstrates that mitochondrial reprogramming is a universal trait of cancer, required for primary and metastatic tumor growth. This is consistent with an expanding role of mitochondria as tumor drivers (4), and mechanisms of intra-organelle protein folding, oxidative bioenergetics, and ROS buffering (18) as key requirements of cancer progression. The regulation by Myc explains why TRAP1 is prominently overexpressed in genetically disparate malignancies, compared with normal tissues (20), and validates its role as a tumor driver. Data from genetically engineered mouse models support this conclusion, as transgenic expression of TRAP1 accelerated prostatic tumorigenesis (21), whereas homozygous deletion of TRAP1 delayed age-associated pathologies, including cancer (22). On the other hand, the Myc–TRAP1 axis of mitochondrial reprogramming is druggable, and selective disruption of intra-organelle protein folding with Gamitrinib (18) delivered potent tumor cell killing and anticancer activity in preclinical models. This therapeutic vulnerability may be exploited for personalized therapy of common, hard-to-treat Myc-driven cancers (16).
Experimental procedures
Cells and cell culture
Prostate adenocarcinoma PC3 or DU145 cells were obtained from the American Type Culture Collection and maintained in culture according to the supplier's specifications. A human Burkitt's lymphoma P493-6 cell line was described previously (23). Clones of P493 cells containing a doxycycline (Dox)-regulated Myc transgene induced after Dox removal (Dox-off system) were as described previously (24). Neuroblastoma SHEP21N, SHEP21-NMycER (25), and SKNAS-N-MycER (26) cells containing a conditionally-regulated N-Myc transgene were as described. In these cells, treatment with 50 ng/ml Dox for 48 h suppresses N-Myc expression, whereas addition of 4-hydroxytamoxifen (4OHT, 0.5 μg/ml) results in strong N-Myc induction.
Antibodies and reagents
Antibodies to Myc and TRAP1 were purchased from Abcam and BD Biosciences, respectively. Antibodies to AMPK, pAMPK (Thr-172), Src, pSrc (Tyr-416), FAK, pFAK (Tyr-925 or Tyr-397), and N-Myc were from Cell Signaling Technology. An antibody to β-actin was from Sigma. Antibodies to Prx3 (Santa Cruz Biotechnology) or 2-Cys PrxSO3 (Thermo Fisher Scientific Inc.) were used. Dox, β-estradiol, and 4OHT were purchased from Sigma. The complete chemical synthesis, HPLC profile, and MS of mitochondria-targeted small molecule Hsp90/TRAP1 antagonist Gamitrinib (GA miochondrial matrx inhibitors) has been reported previously (19).
Plasmid and siRNA transfection
Gene knockdown experiments by small interfering RNA (siRNA) were as described previously (19). The following siRNA sequences were used: control; ON-TARGETplus nontargeting siRNA pool (D-001810); human Myc-directed siRNA (L-020010); human Myc siRNA #1 (J-003282-23); human Myc siRNA #2 (J-003282-24); and human N-Myc directed siRNA (L-003913-01).
Chromatin immunoprecipitation (ChIP) assay
P493 cells treated with β-estradiol plus Dox for 48 h or PC3 cells transfected with control nontargeting siRNA or Myc-directed siRNA for 72 h were used for ChIP experiments as described previously (27)
TRAP1 promoter analysis
Genomic DNA sequences were downloaded from the University of California Santa Cruz Genome browser (http://genome.ucsc.edu)3 (37). Putative Myc- or N-Myc–binding sites were identified using Factorbook motif. A 1,397-bp region of the human TRAP1 gene (−1330 to +66, +1 corresponds to the transcription start site) containing two putative Myc-binding sites (5′-Ca/gCGTG-3′) was amplified and cloned in pGL4.15-promoterless vector upstream from a luciferase reporter gene. For analysis of promoter activity, PC3 or P493 cells transfected with control nontargeting siRNA or Myc-directed siRNA were reconstituted with 2 μg of TRAP1-luciferase reporter plasmid (TRAP1-Luc), and luciferase activity was quantified following the manufacturer's protocol (Promega).
Global metabolomics screening
PC3 cells (2 × 106) were transfected with control siRNA or Myc-directed siRNA followed by incubation with 800 μl of methanol for 30 s. The cell extract was further incubated with 550 μl of Milli-Q water containing internal standards (H3304-1002, Human Metabolome Technologies, Inc., Tsuruoka, Japan) for 30 s and centrifuged at 2,300 × g for 5 min at 4 °C. Proteins were removed by centrifugation of an 800-μl upper aqueous layer through a Millipore 5-kDa cutoff filter at 9,100 × g for 120 min at 4 °C, and the filtrate was centrifugally concentrated and suspended in 50 μl of Milli-Q water for capillary electrophoresis-mass spectrometry analysis. Global metabolome measurements were carried out at Human Metabolome Technology Inc. (Tsuruoka, Japan) (28). Hierarchical cluster analysis and principal component analysis were performed using proprietary PeakStat and SampleStat software, respectively. Detected metabolites were plotted on metabolic pathway maps using VANTED (Visualization and Analysis of Networks containing Experimental Data) software (29).
Focused metabolite analysis
LC-MS analysis was performed on a Thermo Fisher Scientific Q Exactive HF-X mass spectrometer equipped with a HESI II probe and coupled to a Thermo Fisher Scientific Vanquish Horizon UHPLC system. Polar metabolites were extracted using 80% methanol and separated at 0.2 ml/min by HILIC chromatography at 45 °C on a ZIC-pHILIC 2.1 inner diameter × 150-mm column using 20 mm ammonium carbonate, 0.1% ammonium hydroxide, pH 9.2, and acetonitrile with a gradient of 0 min, 85% B; 2 min, 85% B; 17 min, 20% B; 17.1 min, 85% B; and 26 min, 85% B. Relevant MS parameters were as follows: sheath gas, 40; auxiliary gas, 10; sweep gas, 1; auxiliary gas heater temperature, 350 °C; spray voltage, 3.5 kV for the positive mode and 3.2 kV for the negative mode; capillary temperature, 325 °C; and funnel RF level at 40. A sample pool (quality control) was generated by combining an equal volume of each sample and analyzed using a full MS scan at the start, middle, and end of the run sequence. For full MS analyses, data were acquired with polarity switching at: scan range 65 to 975 m/z; 120,000 resolution; automated gain control (AGC) target of 1E6; and maximum injection time (IT) of 100 ms. Data-dependent MS/MS was performed without polarity switching; a full MS scan was acquired as described above, followed by MS/MS of the 10 most abundant ions at 15,000 resolution, AGC target of 5E4, maximum IT of 50 ms, isolation width of 1.0 m/z, and stepped collision energy of 20, 40, and 60. Metabolite identification and quantitation were performed using Compound Discoverer 3.0. Metabolites were identified from a mass list of 206 verified compounds (high confidence identifications) as well as by searching the MS/MS data against the mzCloud database and accepting tentative identifications with a minimum score of 50.
Mitochondrial protein folding
Protein lysates were prepared in RIPA buffer, and equal amounts of protein lysates were separated by SDS-gel electrophoresis as described previously (30). Changes in mitochondrial protein folding were assessed at increasing detergent (CHAPS) concentrations (0–2.5%) as described previously (17).
Mitochondrial bioenergetics
PC3 cells were analyzed for changes in oxidative phosphorylation complex activity using Abcam reagents (complex I, catalog no. ab109721; complex II, catalog no. ab109908; complex III, catalog no. ab109909; complex IV, catalog no. ab109905), as described previously (17). Complex V activity was quantified using Cayman reagent catalog no. 701000. OCR, ECAR, and ATP production were quantified using an Agilent Seahorse XFe96 analyzer (Agilent Technologies, Wilmington, DE). Changes in mitochondrial ROS production (MitoSox Red) and inner membrane potential (tetramethylrhodamine, ethyl ester) were assessed as described previously (31). Assays for glucose (eEnzyme CAG005) and lactate (Abcam Ab65331) production and oxygen consumption (ENZO Life Sciences ENZ-51045-1) were used.
Cell viability assay and colony formation
Changes in cell viability in control or Gamitrinib (0–15 μm)-treated P493, NLF, or IMR cultures were quantified by a fluorogenic Alamar Blue dye, as described previously (32). NLF and IMR neuroblastoma cells were treated with vehicle or Gamitrinib (0–10 μm) and plated at 200 cells per condition, and macroscopically-visible colonies stained with 0.5% w/v crystal violet/methanol were manually counted after 14 days.
Tumor cell motility and invasion
PC3 cells transfected with control nontargeting siRNA or Myc-directed siRNA were analyzed for 2D motility in 4-well Ph+ chambers (Ibidi) by time-lapse video microscopy over 10 h, as described previously (30). Tracking data were exported into the Chemotaxis and Migration Tool version 2.0 (Ibidi) for graphing and calculation of mean and standard deviation of speed accumulated distance of movement. Tumor cell invasion was determined using growth factor reduced Matrigel-coated 8 μm PET Transwell chambers (Corning), as described previously (30).
Bioinformatics analysis
Differential expressions for metabolomics data were tested using Student's t test, and p values were corrected for multiple testing using Benjamini-Hochberg method. Metabolites that passed a false discovery rate <5% cutoff were considered significant. Overlap between the two metabolomics experiments was performed using KEGG and HMDB identifications. All known interactions between overlapped metabolites were derived from the Ingenuity Knowledge Base. CHIP-seq data for c-Myc in P493 Burkitt's lymphoma cells at three independent time points after Dox addition were derived from NCBI GEO database set GSE36354 (24). CHIP-seq analysis of N-Myc distribution in neuroblastoma BE2C, Kelly, and NGP cells was from GEO database set GSE36354 (33). Data were aligned using bowtie (34) against hg19 genome version and analyzed using the HOMER (35) algorithm to generate bigwig files used for visualizing CHIP-seq tracks in the UCSC browser.
Animal studies
Studies involving mice were carried out in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC) of The Wistar Institute (protocol nos. 112625 and 112610). P493 cells (2 × 107) were injected s.c. in athymic nude mice. Once tumors reached 100 mm3 in volume, animals were randomized into two groups to receive 20% Cremophor (vehicle) or Gamitrinib (10 mg/kg) daily i.p. for 10 days. For a liver metastasis model, PC3 cells (1 × 106) were injected into the spleen of SCID/beige mice, and randomized animal groups were treated with vehicle or Gamitrinib for 11 days before histological quantification of liver metastases, as described previously (36).
Statistical analysis
Data are expressed as means ± S.D. of multiple independent experiments or replicates of representative experiments out of a minimum of two or three independent determinations. Two-tailed Student's t test was used for two-group comparative analyses. Statistical analyses were performed using the GraphPad software package (Prism 6.0). A p value of <0.05 was considered as statistically significant.
Author contributions
E. A., B. J. A., C. V. D., and D. C. A. conceptualization; E. A. data curation; E. A., A. V. K., H.-Y. T., L. R. L., and D. I. G. formal analysis; E. A., J. H. S., J. C. G., and D. W. S. investigation; E. A., J. H. S., and J. C. G. methodology; E. A. and D. C. A. writing-review and editing; B. J. A. and C. V. D. resources; A. V. K. and S. R. K. software; L. R. L. validation; D. I. G. and D. W. S. funding acquisition; D. C. A. supervision; D. C. A. writing-original draft; D. C. A. project administration.
Supplementary Material
Acknowledgments
We thank James Hayden and Frederick Keeney at the Wistar Institute Imaging Core Facility. The support for Shared Resources was provided by Cancer Center Support Grant P30 CA010815, and National Institutes of Health Grants S10 OD023586 and S10 OD023658 to The Wistar Institute.
This work was supported by National Institutes of Health Grants P01 CA140043 (to D. C. A., D. W. S., L. R. L., and D. I. G.), R35 CA220446 (to D. C. A.), R50 CA221838 (to H.-Y. T.), R00 CA204593 (to B. J. A), and R50 CA211199 (to A. V. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Figs. S1–S7.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TCA
- trichloroacetic acid
- ROS
- reactive oxygen species
- OCR
- oxygen consumption rate
- Prx3
- peroxiredoxin-3
- SDHB
- succinate dehydrogenase B
- ECAR
- extracellular acidification rate
- Dox
- doxycycline
- 4OHT
- 4-hydroxytamoxifen
- Cox
- cytochrome c oxidase
- AMPK
- AMP-activated protein kinase
- FAK
- focal adhesion kinase
- AGC
- automated gain control
- IT
- injection time.
References
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Potter M., Newport E., and Morten K. J. (2016) The Warburg effect: 80 years on. Biochem. Soc. Trans. 44, 1499–1505 10.1042/BST20160094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gatenby R. A., and Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 4. Vyas S., Zaganjor E., and Haigis M. C. (2016) Mitochondria and cancer. Cell 166, 555–566 10.1016/j.cell.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caino M. C., Ghosh J. C., Chae Y. C., Vaira V., Rivadeneira D. B., Faversani A., Rampini P., Kossenkov A. V., Aird K. M., Zhang R., Webster M. R., Weeraratna A. T., Bosari S., Languino L. R., and Altieri D. C. (2015) PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc. Natl. Acad. Sci. U.S.A. 112, 8638–8643 10.1073/pnas.1500722112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. LeBleu V. S., O'Connell J. T., Gonzalez Herrera K. N., Wikman H., Pantel K., Haigis M. C., de Carvalho F. M., Damascena A., Domingos Chinen L. T., Rocha R. M., Asara J. M., and Kalluri R. (2014) PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003, 1–15 10.1038/ncb3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehuédé C., Dupuy F., Rabinovitch R., Jones R. G., and Siegel P. M. (2016) Metabolic plasticity as a determinant of tumor growth and metastasis. Cancer Res. 76, 5201–5208 10.1158/0008-5472.CAN-16-0266 [DOI] [PubMed] [Google Scholar]
- 8. Dang C. V. (2012) MYC on the path to cancer. Cell 149, 22–35 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Ng P. K., Jeong K. J., Cao S., Wang Z., Gao J., et al. (2018) Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 10.1016/j.cell.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabò A., Kress T. R., Pelizzola M., de Pretis S., Gorski M. M., Tesi A., Morelli M. J., Bora P., Doni M., Verrecchia A., Tonelli C., Fagà G., Bianchi V., Ronchi A., Low D., et al. (2014) Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walz S., Lorenzin F., Morton J., Wiese K. E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M., Roels F., Wüstefeld T., Fischer M., Teichmann M., Zender L., et al. (2014) Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 10.1038/nature13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D. R., Tessarollo L., Casellas R., Zhao K., and Levens D. (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., and Dang C. V. (2015) MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 10.1158/2159-8290.CD-15-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee K. M., Giltnane J. M., Balko J. M., Schwarz L. J., Guerrero-Zotano A. L., Hutchinson K. E., Nixon M. J., Estrada M. V., Sánchez V., Sanders M. E., Lee T., Gómez H., Lluch A., Pérez-Fidalgo J. A., Wolf M. M., et al. (2017) MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 26, 633–647.e7 10.1016/j.cmet.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li F., Wang Y., Zeller K. I., Potter J. J., Wonsey D. R., O'Donnell K. A., Kim J. W., Yustein J. T., Lee L. A., and Dang C. V. (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25, 6225–6234 10.1128/MCB.25.14.6225-6234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez-Bravo V., Pippa R., Song W. M., Carceles-Cordon M., Dominguez-Andres A., Fujiwara N., Woo J., Koh A. P., Ertel A., Lokareddy R. K., Cuesta-Dominguez A., Kim R. S., Rodriguez-Fernandez I., Li P., Gordon R., et al. (2018) Nuclear pores promote lethal prostate cancer by increasing POM121-driven E2F1, MYC, and AR nuclear import. Cell 174, 1200–1215.e20 10.1016/j.cell.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chae Y. C., Angelin A., Lisanti S., Kossenkov A. V., Speicher K. D., Wang H., Powers J. F., Tischler A. S., Pacak K., Fliedner S., Michalek R. D., Karoly E. D., Wallace D. C., Languino L. R., Speicher D. W., and Altieri D. C. (2013) Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat. Commun. 4, 2139 10.1038/ncomms3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altieri D. C. (2013) Hsp90 regulation of mitochondrial protein folding: from organelle integrity to cellular homeostasis. Cell. Mol. Life Sci. 70, 2463–2472 10.1007/s00018-012-1177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caino M. C., Chae Y. C., Vaira V., Ferrero S., Nosotti M., Martin N. M., Weeraratna A., O'Connell M., Jernigan D., Fatatis A., Languino L. R., Bosari S., and Altieri D. C. (2013) Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J. Clin. Invest. 123, 2907–2920 10.1172/JCI67841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang B. H., Plescia J., Dohi T., Rosa J., Doxsey S. J., and Altieri D. C. (2007) Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 131, 257–270 10.1016/j.cell.2007.08.028 [DOI] [PubMed] [Google Scholar]
- 21. Lisanti S., Garlick D. S., Bryant K. G., Tavecchio M., Mills G. B., Lu Y., Kossenkov A. V., Showe L. C., Languino L. R., and Altieri D. C. (2016) Transgenic expression of the mitochondrial chaperone TNFR-associated protein 1 (TRAP1) accelerates prostate cancer development. J. Biol. Chem. 291, 25247–25254 10.1074/jbc.M116.745950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lisanti S., Tavecchio M., Chae Y. C., Liu Q., Brice A. K., Thakur M. L., Languino L. R., and Altieri D. C. (2014) Deletion of the mitochondrial chaperone TRAP-1 uncovers global reprogramming of metabolic networks. Cell Rep. 8, 671–677 10.1016/j.celrep.2014.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuhmacher M., Kohlhuber F., Hölzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G. W., Laux G., Polack A., Weidle U. H., and Eick D. (2001) The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29, 397–406 10.1093/nar/29.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin C. Y., Lovén J., Rahl P. B., Paranal R. M., Burge C. B., Bradner J. E., Lee T. I., and Young R. A. (2012) Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ushmorov A., Hogarty M. D., Liu X., Knauss H., Debatin K. M., and Beltinger C. (2008) N-myc augments death and attenuates protective effects of Bcl-2 in trophically stressed neuroblastoma cells. Oncogene 27, 3424–3434 10.1038/sj.onc.1211017 [DOI] [PubMed] [Google Scholar]
- 26. Valentijn L. J., Koppen A., van Asperen R., Root H. A., Haneveld F., and Versteeg R. (2005) Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 65, 3136–3145 10.1158/0008-5472.CAN-04-2469 [DOI] [PubMed] [Google Scholar]
- 27. Altman B. J., Hsieh A. L., Sengupta A., Krishnanaiah S. Y., Stine Z. E., Walton Z. E., Gouw A. M., Venkataraman A., Li B., Goraksha-Hicks P., Diskin S. J., Bellovin D. I., Simon M. C., Rathmell J. C., Lazar M. A., et al. (2015) MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 10.1016/j.cmet.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sugimoto M., Wong D. T., Hirayama A., Soga T., and Tomita M. (2010) Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95 10.1007/s11306-009-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Junker B. H., Klukas C., and Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7, 109 10.1186/1471-2105-7-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo J. H., Agarwal E., Bryant K. G., Caino M. C., Kim E. T., Kossenkov A. V., Tang H. Y., Languino L. R., Gabrilovich D. I., Cohen A. R., Speicher D. W., and Altieri D. C. (2018) Syntaphilin ubiquitination regulates mitochondrial dynamics and tumor cell movements. Cancer Res. 78, 4215–4228 10.1158/0008-5472.CAN-18-0595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo J. H., Rivadeneira D. B., Caino M. C., Chae Y. C., Speicher D. W., Tang H. Y., Vaira V., Bosari S., Palleschi A., Rampini P., Kossenkov A. V., Languino L. R., and Altieri D. C. (2016) The mitochondrial unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. PLoS Biol. 14, e1002507 10.1371/journal.pbio.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bryant K. G., Chae Y. C., Martinez R. L., Gordon J. C., Elokely K. M., Kossenkov A. V., Grant S., Childers W. E., Abou-Gharbia M., and Altieri D. C. (2017) A mitochondrial-targeted purine-based HSP90 antagonist for leukemia therapy. Oncotarget 8, 112184–112198 10.18632/oncotarget.23097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeid R., Lawlor M. A., Poon E., Reyes J. M., Fulciniti M., Lopez M. A., Scott T. G., Nabet B., Erb M. A., Winter G. E., Jacobson Z., Polaski D. R., Karlin K. L., Hirsch R. A., Munshi N. P., et al. (2018) Enhancer invasion shapes MYC N-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 10.1038/s41588-018-0044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map format and SAM tools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caino M. C., Seo J. H., Aguinaldo A., Wait E., Bryant K. G., Kossenkov A. V., Hayden J. E., Vaira V., Morotti A., Ferrero S., Bosari S., Gabrilovich D. I., Languino L. R., Cohen A. R., and Altieri D. C. (2016) A neuronal network of mitochondrial dynamics regulates metastasis. Nat. Commun. 7, 13730 10.1038/ncomms13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wong N., and Wang X. (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–D152 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.