Abstract
Background: Gastrointestinal stromal tumors (GISTs) represent a worldwide paradigm of target therapy. The introduction of tyrosine kinase inhibitors has deeply changed the prognosis of GIST patients, however, the majority of them acquire secondary mutations and progress. Unfortunately, besides tyrosine-kinase inhibitors, no other therapeutic options are available. Therefore, it is mandatory to identify novel molecules and/or strategies to overcome the inevitable resistance. In this context, after promising preclinical data on the novel PI3K inhibitor BYL719, the NCT01735968 trial in GIST patients who had previously failed treatment with imatinib and sunitinib started. BYL719 has attracted our attention, and we comprehensively characterized genomic and transcriptomic changes taking place during resistance.
Methods: For this purpose, we generated two in vitro GIST models of acquired resistance to BYL719 and performed an omic-based analysis by integrating RNA-sequencing, miRNA, and methylation profiles in sensitive and resistant cells.
Results: We identified novel epigenomic mechanisms of pharmacological resistance in GISTs suggesting the existence of pathways involved in drug resistance and alternatively acquired mutations. Therefore, epigenomics should be taken into account as an alternative adaptive mechanism.
Conclusion: Despite the fact that currently we do not have patients in treatment with BYL719 to verify this hypothesis, the most intriguing result is the involvement of H19 and PSTA1 in GIST resistance, which might represent druggable targets.
Keywords: gastrointestinal stromal tumors, GIST, BYL719, PI3K inhibitor, tyrosine-kinase inhibitors
Introduction
Gastrointestinal stromal tumors (GISTs) are rare mesenchymal tumors harboring KIT/PDGFRA-driver mutations in 85%–90% of cases.1 GISTs are the example par excellence of targeted therapy in solid tumors. Indeed, with the introduction of the tyrosine-kinase inhibitor (TKI) imatinib, patients’ prognosis have dramatically improved.2 Despite imatinib effectiveness, the majority of patients with advanced GISTs have persistent measurable disease and eventually develop progressive disease within 24–36 months.3 About 20 years after its approval, imatinib is still the gold standard in GIST treatment. However, to cope with the emergence of pharmacological resistance, the multi-TKIs sunitinib and regorafenib have been introduced, as second- and third-line treatment, respectively, in GIST management. Sunitinib and regorafenib share a mechanism of action with imatinib, adding clinical benefit to GIST patients after imatinib failure. Even with clinical aids, the majority of patients experience tumor progression, due to the emergence of multiple drug–resistant KIT/PDGFRA mutations.4–9 Unfortunately, all the approved treatments for GIST management are TKIs, and to date there are no additional therapeutic options. Therefore, the identification of novel druggable targets, favored by better characterization of the resistance process, may represent a key starting point to achieve a different clinical approach. In this regard, a novel PI3K inhibitor — BYL719 (Novartis) — is currently being tested in a phase IB trial in GIST patients who have previously failed imatinib and sunitinib (NCT01735968).10 Specifically, BYL719 is a selective inhibitor of the PI3K catalytic p110a subunit.11 Indeed, the PI3K pathway, which is downstream of KIT/PDGFRA receptors, is frequently activated in GIST and thought to be related to imatinib resistance.12–16 Therefore, PI3K-pathway inhibition represents an attractive target and a promising strategy to counteract imatinib resistance in GIST. In this study, we comprehensively characterized genomic and transcriptomic changes taking place during resistance and how GIST cells evolve from being drug-sensitive to drug-resistant, from an “omic” point of view. For this purpose, we generated two in vitro GIST models of acquired resistance to BYL719 and performed an omic-based analysis by integrating RNA sequencing, miRNA profiling, and methylation profiling in sensitive and resistant cells.
Methods
Cell culture and treatment
Two established human imatinib-resistant GIST cell lines, GIST48 and GIST48B, were used (Table 1). These were authenticated by KIT-sequencing and TKI-sensitivity experiments, routinely grown in adhesion, and cultured at 37°C in a 5% CO2 humidified atmosphere. To generate BYL719-resistant sublines, GIST48 and GIST48B were exposed to increasing concentrations of BYL719 (Selleck Chemicals, Houston, TX, USA), starting with a concentration of 0.05 µM and increasing gradually to 5 µM. Fresh drug was provided every 3–4 days, when the medium was replaced. After the cells had acquired the ability to grow in the presence of a specific concentration of BYL719, a proportion of them was frozen and the remaining grown at the next-highest drug level. After approximately 50 weeks, sublines of cells growing in 5 µM BYL719 were maintained continuously in culture at this dose. Upon receipt and before each change of BYL719 concentration, GIST lines were tested for mycoplasma contamination and were found to be negative.
Table 1.
GIST cell-lines a characteristics
| Origin | KIT mutations and other characteristics | Growth mediumb | |
|---|---|---|---|
| GIST48 | GIST primary/patientc | Primary, homozygous KIT exon 11 (V560D) mutation; secondary, heterozygous KIT exon 17 (D820Ad) mutation. | IMDM + 15% FBS |
| GIST48B | Subline of GIST48 | Retains primary mutation (KIT V560D) in all cells; nearly undetectable KIT transcript and protein; secondary, heterozygous KIT exon 17 (D820Ad) mutation; keeps downstream KIT signaling active |
IMDM + 15% FBS |
Notes: aKindly provided by Dr Fletcher (Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA); breagents purchased from Thermo Fisher Scientific (Waltham, MA); cestablished from a GIST patient that had progressed after initial response to imatinib therapy; dlocated in the kinase-activation loop, confers resistance to imatinib and sunitinib.
Abbreviations: IMDM, Iscove's Modified Dulbecco's Medium; FBS, Fetal Bovine Serum.
IC50: MTT-assay method
IC50 was evaluated by MTT assays using a standard protocol.17 Briefly, 104 cells/well were seeded in triplicate in a 96-well plate and incubated for 24 hours. After incubation, the medium was removed and replaced with a fresh one containing BYL719 in serial dilution and incubated for 48 hours. Subsequently, cells were washed with PBS and incubated with MTT (5 mg/mL) in PBS for 2 hours. Following MTT removal, the formazan crystals were dissolved in isopropanol and absorbance measured at 570 nm with a Tecan spectrophotometer (Spectra Model Classic, Salzburg, Austria).
DNA and RNA isolation
Genomic DNA and total RNA were extracted from parental (GIST48 and GIST48B) and BYL719-resistant (GIST48-R and GIST48B-R) cultured cells using the QIAamp DNA minikit and RNeasy minikit, respectively, following manufacturer’s instructions. Both kits were purchased from Qiagen (Hilden, Germany).
KIT and PI3KCA mutational analysis
Genomic DNA from parental and BYL719-resistant cell lines was screened for the presence of hot-spot mutations in KIT (exons 9, 11 13, 14, 17, and 18) and PI3KCA (exons 9 and 20). Selected exons were amplified by PCR using specific primers. Amplified PCR products were purified and sequenced on both strands using a Big Dye Terminator version 1.1 cycle-sequencing kit (Thermo Fisher Scientific). Sanger sequencing was performed on an ABI 310 genetic analyzer (Applied Biosystems) and analyzed using Chromas (Technelysium). Sequencing data were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to match sequences with reference sequences of KIT (NM_000222) and PI3KCA (NM_006218) genes obtained from Ensembl genome browser 94 (https://www.ensembl.org/index.html).
Western blot analysis
Whole-cell protein lysates were prepared from parental and BYL719-resistant cell-monolayers using NP40 buffer containing protease inhibitors (Halt protease- and phosphatase-inhibitor cocktail; Thermo Fisher Scientific) and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Proteins were separated on SDS-PAGE and transferred onto nitrocellulose membranes. Transfer efficiency was demonstrated by Ponceau S staining (Sigma-Aldrich). Membranes were blocked by 5% skimmed milk, followed by incubation at 4°C overnight with the presence of a primary antibody against KIT (A4502; Dako, Ely, UK), phospo-KIT (3391; Cell Signaling Technology, Leiden, Netherlands), AKT (9272; Cell Signaling Technology), phospo-AKT (9271; Cell Signaling Technology), MAPK (9102; Cell Signaling Technology), phospo-MAPK (9101; Cell Signaling Technology), mTOR (2972; Cell Signaling Technology), phospo-mTOR (2448; Cell Signaling Technology), PTEN (138G6; Cell Signaling Technology), and actin (A1978; Sigma-Aldrich). After rinsing, membranes were incubated with horseradish peroxidase–conjugated secondary antibody (Thermo Fisher Scientific) at room temperature for 2 hours. After further rinsing, immunoreactive bands were visualized by enhanced chemiluminescence (BioRad, Hercules, CA, USA) and signals captured and quantified using ChemiDoc (BioRad).
ABC transporter gene–expression analysis
Total RNA was reverse-transcribed to cDNA using a high capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer’s instructions. cDNA was loaded into a TaqMan human ABC-transporter array, which allows quantitative gene-expression analysis of human ABC-transporter genes important in drug discovery and resistance. In particular, it contains assays in triplicate for 50 human genes and 14 endogenous controls in a 348-well array. mRNA-expression levels were normalized using GAPDH and 18S as endogenous controls, and data were analyzed using the 2−ΔΔCt method. mRNAs with Ct>35 were considered unexpressed and excluded from further analysis.
RNA sequencing (RNA-seq)
Whole-transcriptome RNA libraries were prepared in accordance with the TruSeq RNA Sample Prep version 2 protocol (Illumina, San Diego, CA, USA). Poly(A)-RNA molecules from 500 ng total RNA were purified using oligo-dT magnetic beads. Subsequently, mRNA was fragmented and randomly primed for reverse transcription followed by second-strand synthesis to generate double-stranded cDNA fragments. The cDNA fragments generated went through a terminal end–repair process and ligation using paired-end sequencing adapters. The obtained products were amplified to enrich for fragments carrying adapters ligated on both ends, and to add additional sequences complementary to the oligonucleotides on the flow cell, thus creating the final cDNA library. 12pM paired-end libraries were amplified and ligated to the flow cell by bridge PC and sequenced at 2×75bp read length for RNA using Illumina sequencing-by-synthesis technology.
RNA-seq : bioinformatic analysis
After demultiplexing and FASTQ generation, through the bcltofastq function developed by Illumina, paired-end reads were trimmed using AdapterRemoval (https://github.com/MikkelSchubert/adapterremoval) with the aim of removing stretches of low-quality bases (<Q10) and Truseq/Nextera rapid-capture adapters present in the sequences. Sequences coming from RNA-seqwere mapped with TopHat/BowTie pipeline and PCR or optical duplicates were removed with the function rmdup of SAMtools.
RNA-seq: SNV calling
Single nucleotide variant (SNV) calling was performed with SAMtools and SNVMix2, which allows the identification of all point mutations and insertion/deletion variants. Variants in the dbSNP, 1000 Genomes, ExAc, and EVS databases with frequency >1% were excluded. The possible functional effects of identified variants were analyzed with three in silico tools: SIFT, PolyPhen2, and MutationTaster2.18–20
Gene-expression analysis
In order to compare gene-expression profile (GEP) between BYL719-sensitive and -resistant GIST cell lines, RNA-seq data were analyzed. After the alignment procedure, the BAM file obtained was manipulated with SAMtools to remove the optical/PCR duplicate and to sort and index it. The HTSeq count (Python HTSeq package) was adopted to count the number of reads mapped on known genes included in Ensembl 72 annotation features.
Functional annotation, GO, and pathway analysis
NetworkAnalyst (https://www.networkanalyst.ca/faces/home.xhtml) was used to identify molecular pathways and functional groupings. Gene-interaction networks, biofunction, and pathway analysis were generated using differentially expressed genes (DEGs) into known functions, pathways, and networks, primarily based on human studies. The DEGs were organized in Gene Ontology (GO) biofunction and regulatory effect networks. Significance was set at P<0.05. NetworkAnalyst uses a comprehensive high-quality protein–protein interaction database based on InnateDB. This database contains manually curated protein-interaction data from published literature, as well as experimental data from several protein–protein interaction databases, including IntAct, MINT, DIP, BIND, and BioGRID. The database currently contains 14,755 proteins and 145,955 interactions for humans.
qRT-PCR
Gene-expression levels of H19 and PSAT1 were evaluated through quantitative reverse-transcription (qRT) PCR. Briefly, RNA from both parental and BYL719-resistant cell lines was reverse-transcribed to cDNA using a high-capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacturer's instructions. qRT-PCR was performed with a Fast SYBR Green Master Mix (Applied Biosystems) using the 7900HT real-time PCR system. qRT-PCR assays for H19 and PSAT1 were performed using the primers H19-Fwd 5ʹ-ATCGGTGCCTCAGCGTTCGG-3ʹ, H19-Rev 5ʹ-CTGTCCTCGCCGTCACACCG-3ʹ, PSAT1-Fw 5ʹ-ATACAGAGAATCTTGTGCGGG-3ʹ, PSAT1-Rev 5ʹ-CATAGTCAGCACACCTTCCTG-3ʹ, GAPDH-Fwd 5ʹCGGGAAGCTTGTCATCAAT-3ʹ, and GAPDH-Rev 5ʹ-GACTCCACGACGTACTCAGC-3ʹ. All primers were obtained from Integrated DNA Technologies. Relative expression levels were evaluated by the 2–ΔΔCt method using GAPDH as a housekeeping gene.
miRNA-expression evaluation
Total RNA (300 ng) was reverse-transcribed using a TaqMan miRNA reverse-transcription kit (Thermo Fisher Scientific) using Megaplex RT primers Human Pool A and Pool B. cDNAs were load on the TaqMan arrays Human MicroRNA A and B Cards and run on a 7900HT real-time PCR system in accordance with the manufacturer's procedure. miRNA data were analyzed with SDS relative quantification software version 2.4 (Applied Biosystems) and miRNAs with Ct>35 were considered unexpressed and excluded from further analysis. miRNA-expression levels were normalized using U6 and RNU48 as endogenous controls. Normalization was carried out by subtracting the mean Ct from individual Ct values. R-Bioconductor (package Limma) was adopted to evaluate differential expression profiles between the parental and BYL719-resistant GIST cell lines.
Global methylation profile
Genomic DNA (600 ng/sample) were bisulfite-converted using EZ DNA methylation kits (Zymo Research), and DNA methylation was measured using the Illumina Infinium HD-methylation assay with Infinium Methylation Epic BeadChips according to Illumina's protocol. Raw data (Idat files) were processed in R Bioconductor (minfi27). The quality of each sample was analyzed and probe signals removed when: i) P>0.05; ii) >1% of the data set contained no data; or iii) probes contained single-nucleotide polymorphisms. None of the samples included in the study was flagged as an outlier.21 Statistical analyses were carried out using GenomeStudio, normalizing Idat values with controls provided by Illumina. The methylation score of each CpG is represented as a β-value, and differences between β-values of treated and untreated cells represent alterations in methylation level. The CpGs selected were those with absolute methylation differential value of >0.2 or <–0.2.21,22 To identify CpGs on promoter regions, we considered only UCSC reference-gene groups TSS200 or TSS1500.
Identification of validated miRNA targets
Targets of significant miRNAs were identified through specific in silico tools that allowed prediction of the most probable targets. Specifically, to limit false-positive results, we used the miRTarBase tool, which encloses more than 400,000 miRNA-target experimentally verified interactions, collected by manually surveying pertinent literature after systematic data mining of the text.
miRNA-profile and gene-expression correlation
miRNA and mRNA arrays were analyzed to highlight pairs of mRNAs and miRNAs that were discordant (up- versus downregulated and vice versa). Potential miRNA–mRNA interactions and miRNA/mRNA-expression profiles were used to construct functional interaction networks.
Methylation-profile and gene-expression correlation
To integrate methylation profiles and GEP, we considered only CpGs on promoter regions that shown absolute methylation differentials of >0.2 or <–0.2 in parental lines versus BYL719-resistant ones. Promoter regions were defined as upstream 1,500 bp and downstream 200 bp from the transcription-start site (TSS) of each gene. For each differentially methylated gene, we checked the expression level derived from RNA sequencing.
Results
MTT assay
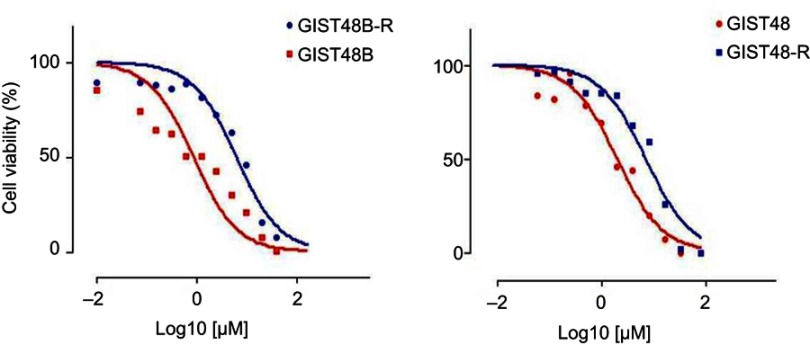
GIST48 and GIST48B cell lines were exposed to BYL719 using doses that were increased in a stepwise manner. We thus established two BYL719-resistant GIST cell lines (48-R and 48B-R), which exhibited BYL719 IC50 values that were about 15-fold higher than the parental cell lines (Figure S1).
Figure S1.
MTT assay.
Notes: Cell viability in GIST48 and GIST48B (red curves) and BYL719-resistant counterparts (blue curves).
KIT and PI3KCA mutational analysis
The involvement of KIT and PI3KCA mutations as mechanisms driving BYL719 resistance in GIST cells in vitro was investigated. Mutational analysis did not reveal additional KIT mutations in BYL719-resistant cell lines by Sanger sequencing or SNV calling. With regard to PI3KCA, we Sanger sequenced exons 9 and 20, both codifying for the catalytic p110a subunit and associated with drug resistance.23–26 All samples were wild type. PI3KCA wild-type status was also confirmed by SNV calling.
Western blot analysis
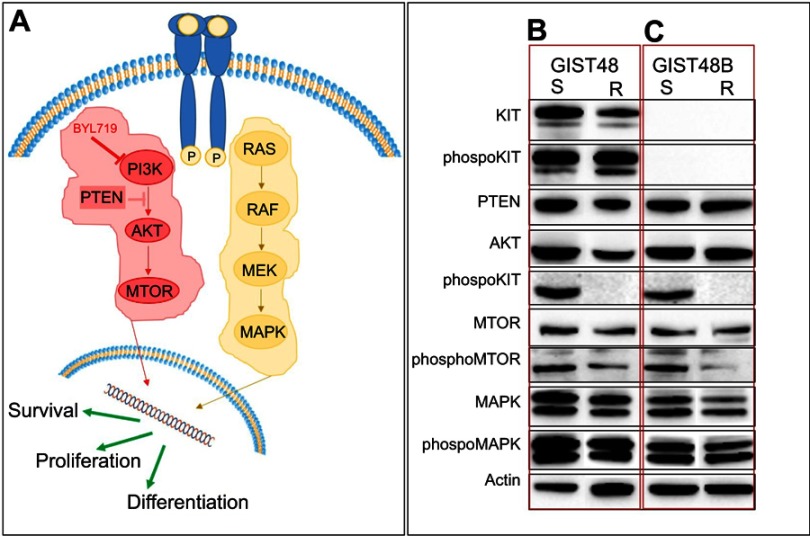
To better characterize the BYL719-resistant GIST48-R and GIST48B-R lines, we performed Western blot analysis, evaluating in particular the signaling pathway downstream of KIT (Figure 1A). In the imatinib-resistant GIST48 line (as expected, considering that the targeted PI3K is downstream of KIT), we observed comparable KIT phosphorylation between GIST48 and GIST48B-R (Figure 1B). Absence of KIT phosphorylation was observed, as expected, in both GIST48B and GIST48B-R. Indeed this GIST model had entirely lost KIT expression (ie, KIT-negative, despite activation of downstream signaling [Figure 1C]). No differences were observed in PTEN status between parental and BYL719-resistant cell lines. On the contrary, in both GIST models, we detected activated AKT in BYL719-sensitive cell lines and inactivation in the resistant ones. We also observed activation of mTOR in both parental and BYL719-resistant cell lines. Finally, we observed MAPK activation in both BYL-sensitive and -resistant GIST models. All immunoblots are shown in Figure 1, B and C.
Figure 1.
Western blot analysis.
Notes: (A) Simplified KIT-signaling pathway. Immunoblot evaluation of KIT, phospho-KIT, PTEN, AKT, phospho-AKT, mTOR, phospho-mTOR, MAPK, and phospho-MAPK in GIST48 (B) and GIST48B (C). S, BYL719-sensitive; R, BYL719-resistant.
RNA-seq: SNV calling
We identified ten novel mutations in BYL719-resistant cells compared to parental cell lines. In particular, four mutations were identified in GIST48-R cells and six in GIST48B-R (Table 2). None of the mutations was common to both the two resistant GIST cell lines. With regard to GIST48-R, one mutation was identified as benign (KIFC2) and one probably damaging (RPGR) by the three tools. One mutation (RIF1) was predicted as probably damaging by two tools. Lastly, no agreement was reached for one mutation (DPF1). Concerning GIST48B-R, one mutation (FRY) was predicted as probably damaging by the three tools, two mutations (PDGFB and SYT14) were identified as benign by two tools, and no agreement was reached for two mutations (DCHS1 and EEF2KMT). Lastly, one mutation (RARS2) was a gain of stop codon. Data are reported in Table 2. However, none of them was related to the PI3KCA pathway, had been previously identified in GIST, or associated with resistance in other tumor types; therefore, we did not consider them driver mutations for BYL719 resistance.
Table 2.
Acquired mutations in BYL719-resistant GIST cells
| GIST 48 vs GIST48-R | GIST 48B vs GIST48B-R | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Exon | AA change | Prediction* | Gene | Exon | AA change | Prediction* |
| DPF1 | 9 | p.C313G | Ba, PDc | DCHS1 | 2 | p.R168C | Ba, PDb,c |
| KIFC2 | 17 | p.R732C | Ba–c | EEF2KMT | 2 | p.H52Q | Ba,b, PDc |
| RIF1 | 19 | p.V666A | PDa,c | FRY | 5 | p.D168Y | PDa–c |
| RPGR | 6 | p.Q171R | PDa–c | PDGFB | 3 | p.R51H | Ba,c |
| RARS2 | 1 | p.C11X | Gain of stop codonb | ||||
| SYT14 | 6 | p.V359A | Bb,c | ||||
Note: *Prediction of the deleterious potential through aSIFT, bPolyPhen, and cMutationTaster2.
Abbreviations: AA, amino acid; B, benign; PD, probably damaging.
Gene expression: ABC-transporter genes and RNA-seq analysis
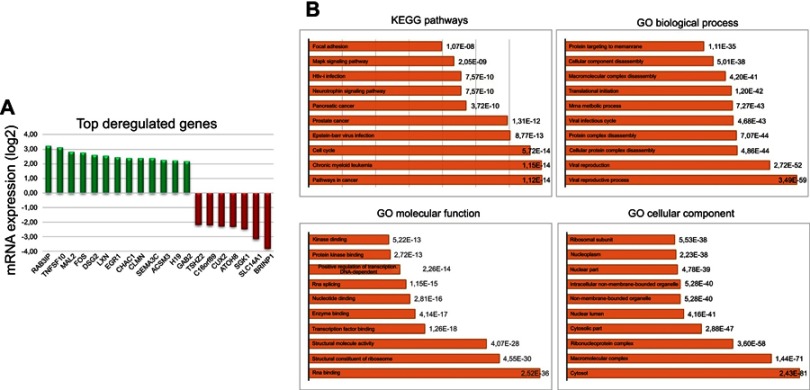
Given the well-recognized role of ABC-transporter genes in mediating drug resistance, first we analyzed a custom panel of 50 genes in BYL719-sensitive and -resistant GIST lines. None was significantly deregulated. Therefore, we performed a RNA-seq analysis, which showed 95 differentially expressed genes with P≤0.001 and a false discovery rate <0.1 in BYL719-sensitive compared to BYL719-resistant GIST lines. In particular, 48 genes were upregulated, while 47 were downregulated. The significant DEGs are reported in Table S1. The 20 top deregulated genes — 13 up- and 7 downregulated — are shown in Figure 2A. GO functional enrichment analyses of deregulated genes showed cancer, chronic myeloid leukemia, and cell cycle as the top three deregulated pathways (Kyoto Encyclopedia of Genes and Genomes pathways (KEGG), Figure 2B). In addition, we found viral reproductive process and viral reproduction to be the top biological processes involved. RNA binding, structural constituents of ribosome, and structural molecular activity were the top three molecular functions. Finally, cytosol, macromolecular complex, and ribonucleoprotein complex were the top three cellular components involved. Interestingly, among the top upregulated genes in resistant cells, there was the long non coding RNA (lncRNA) H19. RNA-seq data were confirmed by qRT-PCR, which demonstrated overexpression of H19 in GIST48-R and GIST48B-R compared with the parental BYL719-sensitive lines, with fold changes of 20.8 and 6.9, respectively.
Table S1.
List of genes differentially expressed in parental and resistant cell lines
| Upregulated genes in BYL719-resistant cell lines | Downregulated genes in BYL719-resistant cell lines | ||||||
|---|---|---|---|---|---|---|---|
| LogFC | P-value | FDR | LogFC | P-value | FDR | ||
| H19 | 2.23 | 8.61–12 | 1.08–7 | C16ORF89 | −2.21 | 0 | 0.000001 |
| RAB3IP | 3.23 | 1.19–9 | 3.93–6 | MXRA5 | −1.90 | 0 | 0.000004 |
| ANXA3 | 1.81 | 8.24–8 | 0.000148 | BRINP1 | −3.82 | 0 | 0.000121 |
| LXN | 2.57 | 2.9–7 | 0.000304 | CUX2 | −2.27 | 0 | 0.000215 |
| CHAC1 | 2.41 | 1–6 | 0.000841 | SLC4A7 | −1.57 | 0 | 0.000215 |
| TNFSF10 | 3.12 | 1.53–6 | 0.001043 | PCSK6 | −1.78 | 0 | 0.000215 |
| CLMN | 2.40 | 1.8–6 | 0.001078 | SGK1 | −2.46 | 0.000001 | 0.000535 |
| ACSM3 | 2.26 | 5.71–6 | 0.002987 | GPC2 | −1.79 | 0.000001 | 0.001010 |
| PDE5A | 1.52 | 9.67–6 | 0.004328 | FN1 | −1.54 | 0.000003 | 0.001451 |
| DSG2 | 2.60 | 1.39–5 | 0.005618 | RHBDF1 | −1.43 | 0.000007 | 0.003245 |
| ASNS | 1.30 | 1.58–5 | 0.00619 | SAMD11 | −1.38 | 0.000008 | 0.003622 |
| PSAT1 | 1.48 | 1.96–5 | 0.007033 | VCAM1 | −1.67 | 0.000017 | 0.006486 |
| SNX10 | 2.00 | 2.51–5 | 0.008495 | ID3 | −1.69 | 0.000019 | 0.006898 |
| LMO4 | 1.29 | 2.64–5 | 0.008495 | SMAD9 | −1.28 | 0.000029 | 0.008963 |
| SEMA3C | 2.39 | 3–5 | 0.009034 | SLC14A1 | −3.15 | 0.000030 | 0.009034 |
| GLRB | 2.02 | 3.22–5 | 0.009306 | TSHZ2 | −2.18 | 0.000033 | 0.009306 |
| ADD2 | 1.62 | 4.99–5 | 0.012541 | MME | −1.47 | 0.000035 | 0.009897 |
| EGR1 | 2.46 | 7.05–5 | 0.017026 | RP11-54O7.1 | −2.01 | 0.000041 | 0.010885 |
| MAL2 | 2.82 | 7.84–5 | 0.017893 | PHF2P2 | −1.39 | 0.000042 | 0.011066 |
| PRIMA1 | 2.06 | 8.88–5 | 0.018896 | TYRO3 | −1.42 | 0.000097 | 0.019908 |
| TBC1D2B | 1.47 | 9.25–5 | 0.019348 | NUAK1 | −1.45 | 0.000105 | 0.020765 |
| GAB2 | 2.19 | 0.000108 | 0.020816 | PHLDB2 | −1.64 | 0.000106 | 0.020765 |
| PROCR | 1.89 | 0.000129 | 0.023094 | DACT3 | −1.43 | 0.000104 | 0.020765 |
| ICA1 | 2.05 | 0.000132 | 0.023302 | GTF2IP3 | −2.01 | 0.000109 | 0.020816 |
| PRKCQ-AS1 | 1.56 | 0.000142 | 0.024358 | ID2 | −1.25 | 0.000117 | 0.021688 |
| DOCK9 | 1.77 | 0.000171 | 0.0276 | EFNB2 | −1.59 | 0.000137 | 0.023811 |
| LAMP3 | 1.22 | 0.000232 | 0.035097 | IQGAP2 | −1.14 | 0.000144 | 0.024358 |
| TRIB1 | 1.25 | 0.000232 | 0.035097 | RBM18 | −1.17 | 0.000170 | 0.027600 |
| TBC1D8B | 1.62 | 0.000266 | 0.038374 | SOSTDC1 | −1.84 | 0.000189 | 0.030100 |
| SFRP1 | 1.10 | 0.000358 | 0.048283 | F2R | −1.48 | 0.000225 | 0.034926 |
| TNFAIP8 | 1.29 | 0.000407 | 0.052689 | ADGRD1 | −1.54 | 0.000262 | 0.038374 |
| SAMD12 | 2.06 | 0.000424 | 0.053714 | GAREM | −1.46 | 0.000297 | 0.042399 |
| JUNB | 1.43 | 0.000444 | 0.055782 | SOX4 | −1.23 | 0.000306 | 0.043207 |
| DAAM2 | 1.21 | 0.000483 | 0.05878 | SMAD6 | −1.21 | 0.000350 | 0.047754 |
| IGSF3 | 1.13 | 0.000487 | 0.05878 | SH3BP5-AS1 | −1.66 | 0.000347 | 0.047754 |
| LGI2 | 1.90 | 0.000521 | 0.061597 | PREX2 | −1.06 | 0.000369 | 0.049242 |
| CALB2 | 1.72 | 0.000525 | 0.061597 | BCRP3 | −1.76 | 0.000420 | 0.053714 |
| PDE3B | 1.39 | 0.000552 | 0.063588 | COL16A1 | −1.15 | 0.000449 | 0.055782 |
| GPC3 | 1.71 | 0.000574 | 0.064953 | PDLIM3 | −1.45 | 0.000463 | 0.056947 |
| TNFRSF19 | 2.17 | 0.000571 | 0.064953 | ADAMTS2 | −1.65 | 0.000512 | 0.061195 |
| FOXP2 | 1.57 | 0.00061 | 0.06843 | WTIP | −1.16 | 0.000531 | 0.061674 |
| BST2 | 1.18 | 0.000619 | 0.068795 | MYO1B | −1.27 | 0.000626 | 0.068960 |
| HUNK | 1.30 | 0.000647 | 0.070631 | SORBS1 | −1.55 | 0.000703 | 0.075396 |
| FAM19A4 | 1.76 | 0.000709 | 0.075396 | PPARGC1B | −1.01 | 0.000794 | 0.081682 |
| ARRB1 | 1.27 | 0.000737 | 0.07712 | ATOH8 | −2.31 | 0.000808 | 0.082507 |
| CCDC181 | 1.55 | 0.000893 | 0.088251 | ZDHHC11 | −1.33 | 0.000832 | 0.084194 |
| GLTSCR2 | 0.98 | 0.001004 | 0.098527 | CTGF | −1.16 | 0.000869 | 0.087235 |
| FOS | 2.77 | 0.001014 | 0.098663 | ||||
Abbreviation: FDR, fold discovery rate.
Figure 2.
RNA-seq analysis.
Notes: (A) Top deregulated genes in BYL719-sensitive and -resistant GIST cell lines. (B) Go functional enrichment analysis of up- and downregulated genes.
Abbreviations: GIST, gastrointestinal stromal tumors; GO, gene ontology.
miRNA-expression profile
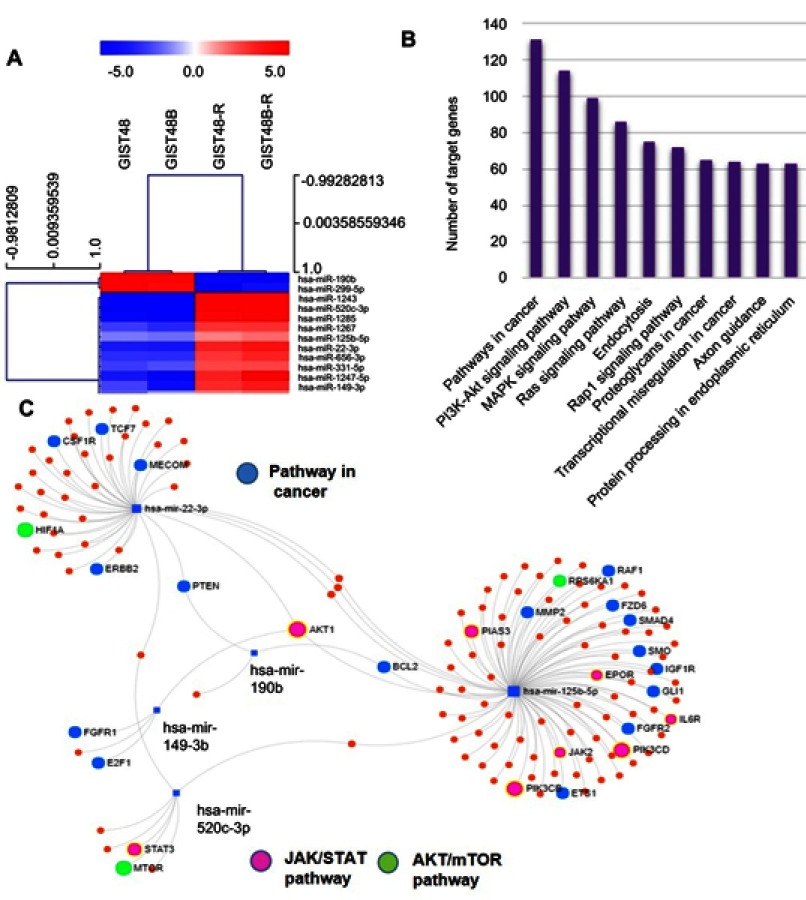
The array highlighted a total of 44 deregulated miRNAs of the 754 analyzed (P<0.05); however, after adjustment, only 13 miRNAs maintained statistical significance. In particular, two miRNAs — has-miR190b and has-miR299-5p — were upregulated and eleven miRNAs downregulated in resistant GIST cell lines compared to parental ones. All differentially expressed miRNAs are reported in Table 3. Hierarchical clustering of all samples dichotomized sensitive and resistant GIST cell lines into two distinct clusters (Figure 3A). Through miRPath 3.0, we assessed miRNA regulatory roles and identification of controlled pathways.27 Interestingly, among the pathways potentially modulated by the deregulated miRNAs (summarized in Figure 3B), the most significant and those with the greatest number of target genes were PI3K/AKT, MAPK, and RAS cascades, which are involved in BYL719's mechanism of action. Figure 3C shows the main miRNAs involved in cancer (highlighted in blue) and PI3K–AKT (in green) and JAK–STAT (in pink) pathways. In particular, among the 13 deregulated miRNAs retrieved by our data, we identified a signature of five miRNAs mainly involved in the aforementioned pathways: miR22-3p, miR125b-5p, miR149-3p, miR190b, and miR520c-3p.
Table 3.
The most significantly deregulated miRNAs
| miRNA | P-value | Adjusted P-value |
|---|---|---|
| hsa-miR1243 | 2.94–6 | 0.002232799 |
| hsa-miR520c-3p | 1.27–5 | 0.003316809 |
| hsa-miR190b | 1.31–5 | 0.003316809 |
| hsa-miR1289 | 2.72–5 | 0.005164803 |
| hsa-miR1247-5p | 0.000118 | 0.016017348 |
| hsa-miR22-3p | 0.000126 | 0.016017348 |
| hsa-miR1267 | 0.000188 | 0.020418143 |
| hsa-miR299-5p | 0.000237 | 0.022470219 |
| hsa-miR125b-5p | 0.000294 | 0.024789505 |
| hsa-miR656-3p | 0.000447 | 0.033993852 |
| hsa-miR331-5p | 0.000591 | 0.040837133 |
| hsa-miR149-3p | 0.000868 | 0.054998185 |
| hsa-miR30d-5p | 0.001126 | 0.065812471 |
Figure 3.
miRNAs profile .
Notes: (A) miRNA hierarchical clustering. miRNAs identified as differentially expressed between BYL719-sensitive and -resistant GIST cell lines were selected as markers for unsupervised hierarchical clustering. (B) GO functional enrichment analysis of up- and downregulated miRNAs. (C) Main miRNAs involved in cancer (blue), AKT–mTOR (green) and JAK signaling cascades (pink) pathways.
Global methylation profile
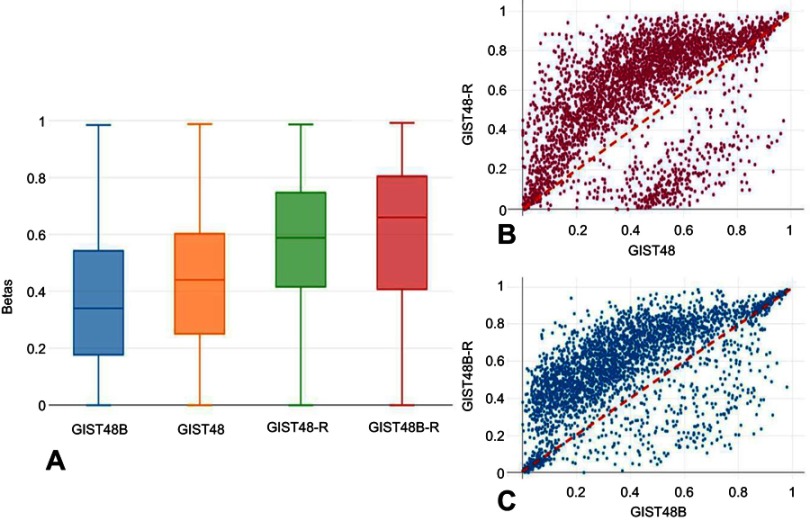
To determine whether acquired resistance involved modifications in DNA methylation, we performed genome-wide DNA-methylation profiling in BYL719-sensitive and -resistant GIST cell lines. We identified 3,305 differentially methylated CpGs. Among these, 2,817 were hypermethylated and 488 hypomethylated. Of the hypermethylated CpGs, 547 were in promoter regions of 379 genes, while 102 hypomethylated sites were in promoter regions of 70 genes. Unsupervised hierarchical cluster analysis of the demethylated genes divided the sensitive and resistant cell lines into two main clusters (Figure 4). Interestingly, as shown in Figure 5, A–C, the resistant cells showed significantly more hypermethylated CpG-island promoters compared to the sensitive counterparts.
Figure 4.

Global methylation profile.
Notes: Heat map showing DNA-methylation profiles in parental and BYL719-resistant cell lines (Δβ<–0.2 or >0.2).
Figure 5.
Global methylation profile.
Notes: (A) Box plots showing the β-values of CpGs islands at promoters only . General hypermethylation was more common in the BYL719-resistant cell lines. (B, C) Scatterplots of genome-wide DNA methylation levels (β). To highlight general hypermethylation, CpG islands with Δβ<–0.2 or >0.2 are shown.
miRNAs and mRNA network
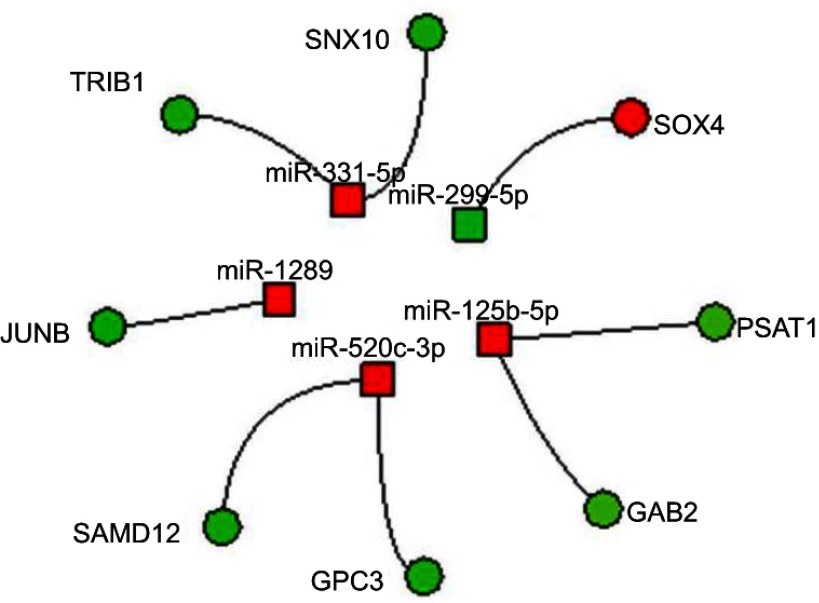
To construct miRNA–DEG networks, we downloaded the experimentally verified associations between human miRNAs and their targets from miRTarBase. This data set consists of 4,076 miRNAs and 23,054 mRNAs,28 and encloses more than 400,000 miRNA–target interactions collected by manually surveying pertinent literature after systematic data mining of the text. Among the 13 deregulated miRNAs retrieved by our data, seven (miR331-5p, miR125b-5p, miR520c-3p, miR1289, miR299-5p, miR30d-5p, and miR149-3p) had verified associations with their targets. However, taking into account the canonical inverse correlation between miRNA and target expression, we were able to identify eight mRNA–miRNA networks. In particular, we found four miRNAs downregulated and the corresponding seven targets overexpressed (three miRNAs targeted two distinct mRNAs). One miRNA was upregulated and the corresponding target downregulated. The miRNAs and their corresponding targets are presented in Figure 6.
Figure 6.
Integrated miRNA–mRNA regulatory networks.
Notes: Integration of GEP and miRNA profiles. Circles and squares represent genes and miRNAs, respectively. Red and green indicate downregulation and upregulation, respectively.
Abbreviation: GEP, gene expression profile.
DNA methylation and mRNA network
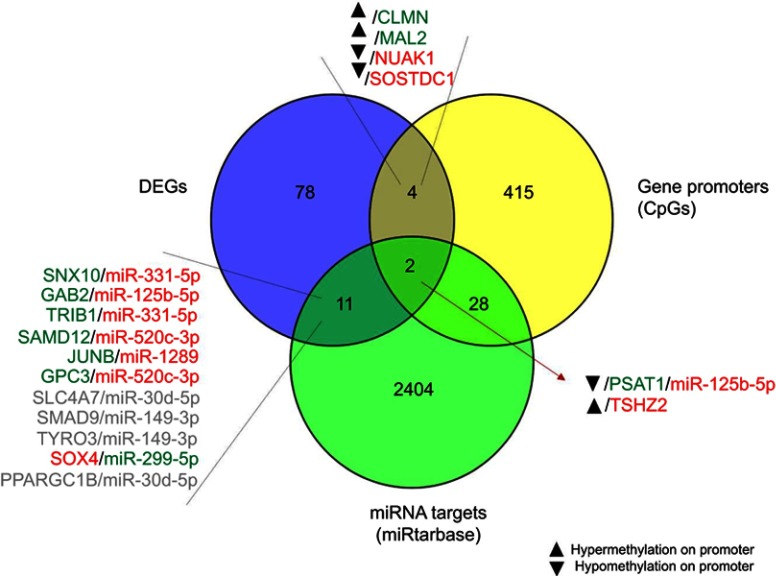
To clarify the relationship between DNA methylation and differentially expressed mRNAs, we integrated the global methylation profile with DEGs. Results are shown in Figure 7. Among the genes with the strongest promoter hypermethylation and concomitant increased expression in resistant cells were CLMN, a member of the hedgehog-interacting protein family MAL, which encodes the T-lymphocyte maturation-associated protein and functions in T-cell differentiation. On the contrary, promoter hypermethylation and concomitant TSHZ2 downregulation was observed, whereas PSAT1 showed strong promoter hypomethylation associated with increased gene expression after treatment. Overexpression of PSAT1 was confirmed by qRT-PCR, which demonstrated overexpression of PSAT1 in GIST48-R and GIST48B-R compared with the parental BYL719-sensitive lines, with fold changes of 4.1 and 5.8, respectively.
Figure 7.
Integrated miRNA–mRNA–methylation regulatory networks.
Notes: Integration of GEP, miRNA, and methylation profiles. In gray are shown genes and miRNAs with the same trend of expression, which were excluded. Red and green indicate downregulation and upregulation, respectively.
Abbreviation: GEP, gene expression profile.
Construction of the miRNA–mRNA–DNA methylation network
In order to identify a more comprehensive network, we integrated data deriving from GEP, miRNA, and methylation profiles. After intersection of all the data, the PSAT1 gene was significantly upregulated in BYL719-resistant cell lines and showed promoter hypomethylation and a potential modulation by miR125b-5p (Figure 7).
Discussion
GISTs represent a worldwide paradigm of target therapy. The introduction of imatinib to clinical practice has deeply revolutionized its management, leading GISTs from an incurable disease to a sort of chronic disease. Imatinib has determined terrific improvement in GIST prognosis; however, as often happens with TKIs, the majority of patients acquire secondary mutations and the disease progresses. Unfortunately, no therapeutic options are available for patients who have failed on imatinib and the subsequent therapeutic lines: sunitinib and regorafenib. Therefore, it is pivotal and mandatory to identify novel molecules — different from TKIs — and/or strategies to overcome the inevitable resistance. In this context, after the promising preclinical data on the novel PI3K inhibitor BYL719,10 the NCT01735968 trial in GIST patients who had previously failed treatment with imatinib and sunitinib started. BYL719 attracted our attention, and we comprehensively characterized genomic and transcriptomic changes taking place during resistance. Understanding how GIST cells evolve from being drug-sensitive to drug-resistant, from an omic point of view might accelerate the identification of novel druggable targets. For this purpose, we generated two in vitro GIST models of acquired resistance to BYL719 and performed an omic-based analysis by integrating RNA-seq, miRNA, and methylation profiling in sensitive and resistant cells. In particular, we selected GIST48 and GIST48B, sharing activation of KIT downstream signaling, including the PI3K pathway, targeted by BYL719.29 Furthermore, both cell lines are resistant to the second-line treatment sunitinib,30 making them good models, mirroring GIST patients who hypothetically will receive BYL719 treatment after imatinib and sunitinib failure.
TKI resistance is largely due to accumulation of additional kinase-domain mutations.31 Therefore, we sequenced KIT and PI3KCA hot-spot exons — in addition to RNA-seq — to evaluate the appearance of novel point mutations or insertion/deletion variants possibly able to explain BYL719 resistance. Different alterations were found in BYL719-resistant lines; however, none was shared by both models, or harbored in PI3KCA and KIT genes, or downstream effectors. In addition, none of the mutated genes has been involved in drug resistance; therefore, we did not consider them driver events for BYL719 unresponsiveness, pinpointing a potential novel mechanism of resistance in GISTs. Afterward, we analyzed a precast panel of 50 well-characterized ABC-transporter genes. Overexpression of genes involved in drug efflux is among the most common mechanisms of drug resistance.32–34 However, we did not observe any significant difference in BYL719-resistant cell lines compared to sensitive ones. This prompted us to perform a more comprehensive gene-expression analysis through the RNA-seq approach. We identified the H19 gene as one of the most significantly upregulated in BYL719-resistant cells compared to sensitive ones. H19 has been described as an oncogenic lncRNA involved in cell proliferation, metastasis, epithelial–mesenchymal transition, and poor prognosis in several cancer types, including esophageal squamous-cell carcinoma, osteosarcoma, colorectal and gastric cancer, and others.35–38
A recent study reported H19 overexpression in GIST samples compared to normal paired tissue.39 In addition, H19 upregulation showed a high correlation with ETV1 expression, which is crucial in GIST growth and survival. ETV1 cooperates with KIT through the MAPK-signaling pathway, and together they promote GIST tumorigenesis.40 Furthermore, it has been reported that H19 might promote migration and invasion in colorectal cancer by activating the RAS protein and upregulating levels of phospho-RAF, phospho-MEK and phospho-MAPK.41 Considering that RAS is upstream of both the PI3K and MAPK pathways,42 this could represent an alternative mechanism through which resistant cells bypass PI3K and maintain their tumorigenic status. This assumption was confirmed by Western blot analysis, highlighting MAPK activation in both parental and BYL719-resistant cell lines. Interestingly, we also observed a weak activation of mTOR in BYL719-resistant cell lines, despite no AKT activity. This finding agrees with the recognized complex and dynamic cross talk between PI3K/AKT and MAPK.43,44 Pathway cross talk allows a cell to achieve robust activation of key downstream targets or compensatory signaling. Particularly, the latter might be important in the context of drug resistance, allowing growth and viability of cancer cells. Indeed, combined inhibition of PI3K–Akt and MAPK pathways has shown efficacy in preclinical models,43–46 and could be of interest in the management of imatinib-resistant GISTs.
Considering the lack of acquired PI3KCA mutations, we further evaluated the hypothesis that BYL719 resistance could be mediated by epigenetic mechanisms. Studies reported that epigenetic modifications may represent alternative mechanisms to evade the pharmacological response.47–49 In this context, different trials are evaluating epigenetic therapies as drug-resistance modulators in solid tumors.47 With regard to GISTs, in recent years epigenetic treatments raised among the future perspective50,51 alternatively to TKIs and/or to bypass TKI resistance. However, the results are still at an early stage, and further investigations are essential in this novel field. Consequently, we performed deep epigenomic characterization of these two in vitro models, looking for novel mechanisms of acquired resistance and potential druggable targets in GISTs. For this purpose, we performed miRNA profiling, identifying 13 miRNAs significantly deregulated in resistant cells. Among the pathways potentially modulated by these miRNAs, the most significant and those with the greatest number of target genes were PI3K/AKT–MAPK, and RAS cascades, which are involved in BYL719's mechanism of action. Subsequently, integration of miRNA arrays and GEP revealed five miRNAs targeting eight genes (TRIB1, GAB2, SNX10, SAMD12, JUNB, GPC3, and SOX4). Among those genes, TRIB1 is a downstream effector of PI3K, and GAB252 cooperates with the PI3K–AKT pathway in promoting malignant behavior in cells.53,54
Subsequently, we integrated methylation and GEP, identifying two networks of interest involving TSHZ2 and PSAT1. The TSHZ2 gene resulted in downregulated BYL719-resistant cells compared to sensitive ones. TSHZ2, a zinc-finger homeobox nuclear protein, is supposed to be a tumor suppressor and is downregulated in breast cancer.55 We speculated that TSHZ2 might bind transcriptional regulators that control the expression of crucial genes in tumorigenesis and resistance acquisition. Moreover, PSAT1 was upregulated in BYL719-resistant cells, and concomitant hypomethylation on the PSAT1 promoter was found. However, on integration of miRNA and methylation profiles with gene-expression analysis, we pinpointed overexpression of PSAT1 associated with both promoter hypomethylation and miR125b-5p downregulation. In particular, PSAT1 overexpression in BYL719-resistant cell lines could represent an advantage for resistant cells, which have to find novel ploys to live.
Here, we showed that both methylation and miRNA modulated PSAT1 overexpression. Additional studies are needed to clarify if the epigenetic mechanisms act concurrently or in a mutual manner, with the final goal of increasing PSAT1 levels, which confer a metabolic-related growth advantage to tumor cells. PSAT1 overexpression is involved in drug resistance in different tumors, including colorectal cancer, melanoma, pancreatic cancer, and non-small-cell lung cancer.56,57 PSAT1 belongs to the serine biosynthesis pathway, which has a key role in nucleotide and amino-acid metabolism. Levels of enzymes involved in serine synthesis have been shown to increase under conditions of DNA damage and genomic instability.57,58 In addition, a recent study reported that serine supports one-carbon metabolism and proliferation of cancer cells.59 Moreover, different evidence suggests that the serine pathway is crucial in cancer metabolism, using glycolysis-derived glucose for serine production and tumor growth.60–64 In addition to these studies, which clearly ascribed to PSAT1 a role in tumorigenesis, a recent study on esophageal squamous-cell carcinoma patients linked PSAT1 overexpression to upregulation of the PI3K–AKT–GSK3β–Snail pathway.65 Therefore, targeting PSAT1 might have potential therapeutic implication in GIST patients.
In conclusion, we identified novel epigenomic mechanisms of pharmacological resistance in GISTs, suggesting the existence of pathways involved in drug resistance and alternative to acquired mutations. We are aware that prolonged exposure and high BYL719 concentrations used to generate the resistant models can be de facto a substantial source of numerous off-target effects. However, prolonged inhibition of the PI3K pathway is probably accompanied by profound transcriptomic, proteomic, and metabolomic changes involved in drug resistance. Therefore, epigenomics should be taken into account as an alternative adaptive mechanism. Currently we do not have access to GIST patients in treatment with BYL719, and thus we are not able to verify the hypothesis in vivo. However, we consider these results intriguing, particularly the involvement of H19 and PSTA1 in GIST resistance, which might represent novel druggable targets for GIST patients.
Acknowledgments
This work was supported by the Ministry of Education, University, and Research of Italy (MIUR; grant 2015Y3C5KP_002 to SA). Gloria Ravegnini is supported by an MSD Italia fellowship granted by and on behalf of MSD and L’Oréal–UNESCO for Women in Science. Giulia Sammarini is supported by Fondazione Famiglia Parmiani.
Disclosure
The authors report no conflicts of interest in this work.
Supplementary materials
References
- 1.Mei L, Smith SC, Faber AC, et al. Gastrointestinal stromal tumors: the GIST of precision medicine. Trends Cancer. 2018;4(1):74–91. doi: 10.1016/j.trecan.2017.11.006 [DOI] [PubMed] [Google Scholar]
- 2.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. nat rev cancer. 2011;11(12):865–878. doi: 10.1038/nrc3143 [DOI] [PubMed] [Google Scholar]
- 3.Serrano C, George S. Recent advances in the treatment of gastrointestinal stromal tumors. Ther Adv Med Oncol. 2014;6(3):115–127. doi: 10.1177/1758834014522491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet (London, England). 2006;368(9544):1329–1338. doi: 10.1016/S0140-6736(06)69446-4 [DOI] [PubMed] [Google Scholar]
- 5.Demetri GD, Reichardt P, Kang Y-K, et al. Efficacy and Safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381(9863):295–302. doi: 10.1016/S0140-6736(12)61857-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemming ML, Heinrich MC, Bauer S, George S. Translational insights into gastrointestinal stromal tumor and current clinical advances. Ann Oncol Off J Eur Soc Med Oncol. 2018. doi: 10.1093/annonc/mdy309 [DOI] [PubMed] [Google Scholar]
- 7.Liegl B, Kepten I, Le C, et al. Heterogeneity of Kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216(1):64–74. doi: 10.1002/path.2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo C, Ryu M-H, Jo J, Park I, Ryoo B-Y, Kang Y-K. Efficacy of imatinib in patients with platelet-derived growth factor receptor alpha–mutated gastrointestinal stromal tumors. Cancer Res Treat. 2016;48(2):546–552. doi: 10.4143/crt.2015.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassier PA, Fumagalli E, Rutkowski P, et al. Outcome of patients with platelet-derived growth factor receptor alpha-mutated gastrointestinal stromal tumors in the tyrosine kinase inhibitor era. Clin Cancer Res. 2012;18(16):4458–4464. doi: 10.1158/1078-0432.CCR-11-3025 [DOI] [PubMed] [Google Scholar]
- 10.Van Looy T, Wozniak A, Floris G, et al. Phosphoinositide 3-kinase inhibitors combined with imatinib in patient-derived xenograft models of gastrointestinal stromal tumors: rationale and efficacy. Clin Cancer Res. 2014;20(23):6071–6082. doi: 10.1158/1078-0432.CCR-14-1823 [DOI] [PubMed] [Google Scholar]
- 11.Fritsch C, Huang A, Chatenay-Rivauday C, et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13(5):1117–1129. doi: 10.1158/1535-7163.MCT-13-0865 [DOI] [PubMed] [Google Scholar]
- 12.Serrano C, Wang Y, Mariño-Enríquez A, et al. KRAS and KIT gatekeeper mutations confer polyclonal primary imatinib resistance in GI stromal tumors: relevance of concomitant phosphatidylinositol 3-kinase/AKT dysregulation. J Clin Oncol. 2015;33(22):e93–6. doi: 10.1200/JCO.2013.48.7488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer S, Duensing A, Demetri GD, Fletcher JA. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 2007;26(54):7560–7568. doi: 10.1038/sj.onc.1210558 [DOI] [PubMed] [Google Scholar]
- 14.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- 15.Patel S. Exploring novel therapeutic targets in GIST: focus on the PI3K/Akt/MTOR pathway. Curr Oncol Rep. 2013;15(4):386–395. doi: 10.1007/s11912-013-0316-6 [DOI] [PubMed] [Google Scholar]
- 16.Ríos-Moreno MJ, Jaramillo S, Díaz-Delgado M, et al. Differential activation of MAPK and PI3K/AKT/MTOR pathways and IGF1R expression in gastrointestinal stromal tumors. Anticancer Res. 2011;31(9):3019–3025. [PubMed] [Google Scholar]
- 17.Ulukaya E, Colakogullari M, Wood EJ. Interference by anti-cancer chemotherapeutic agents in the MTT-tumor chemosensitivity assay. Chemotherapy. 2004;50(1):43–50. doi: 10.1159/000077285 [DOI] [PubMed] [Google Scholar]
- 18.Kumar P, Henikoff S, Ng PC. predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 21.Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2017. doi: 10.1136/gutjnl-2016-313372 gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 cpg sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389–399. doi: 10.2217/epi.15.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omarini C, Filieri ME, Bettelli S, et al. Mutational profile of metastatic breast cancer tissue in patients treated with exemestane plus everolimus. Biomed Res Int. 2018;2018:3756981. doi: 10.1155/2018/3756981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessitore A, Bruera G, Mastroiaco V, et al. KRAS and 2 rare PI3KCA mutations coexisting in a metastatic colorectal cancer patient with aggressive and resistant disease. Hum Pathol. 2018;74:178–182. doi: 10.1016/j.humpath.2018.01.021 [DOI] [PubMed] [Google Scholar]
- 25.Lai K, Killingsworth MC, Lee CS. Gene of the Month: PIK3CA. J Clin Pathol. 2015;68(4):253–257. doi: 10.1136/jclinpath-2015-202885 [DOI] [PubMed] [Google Scholar]
- 26.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502 [DOI] [PubMed] [Google Scholar]
- 27.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-MiRPath v3.0: deciphering MicroRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou C-H, Shrestha S, Yang C-D, et al. MiRTarBase update 2018: a resource for experimentally validated MicroRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühlenberg T, Zhang Y, Wagner AJ, et al. Inhibitors of deacetylases suppress oncogenic KIT signaling, acetylate HSP90, and induce apoptosis in gastrointestinal stromal tumors. Cancer Res. 2009;69(17):6941–6950. doi: 10.1158/0008-5472.CAN-08-4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrich MC, Maki RG, Corless CL, et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26(33):5352–5359. doi: 10.1200/JCO.2007.15.7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wardelmann E, Merkelbach-Bruse S, Pauls K, et al. Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin Cancer Res. 2006;12(6):1743–1749. doi: 10.1158/1078-0432.CCR-05-1211 [DOI] [PubMed] [Google Scholar]
- 32.Beretta GL, Cassinelli G, Pennati M, Zuco V, Gatti L. Overcoming ABC transporter-mediated multidrug resistance: the dual role of tyrosine kinase inhibitors as multitargeting agents. Eur J Med Chem. 2017;142:271–289. doi: 10.1016/j.ejmech.2017.07.062 [DOI] [PubMed] [Google Scholar]
- 33.Choi YH, Yu A-M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20(5):793–807. doi: 10.2174/138161282005140214165212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klukovits A, Krajcsi P. Mechanisms and therapeutic potential of inhibiting drug efflux transporters. Expert Opin Drug Metab Toxicol. 2015;11(6):907–920. doi: 10.1517/17425255.2015.1028917 [DOI] [PubMed] [Google Scholar]
- 35.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14(1):184. doi: 10.1186/s12943-015-0458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huarte M. The emerging role of LncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging MiR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50(5):1801–1809. doi: 10.3892/ijo.2017.3941 [DOI] [PubMed] [Google Scholar]
- 38.Li H, Yu B, Li J, et al. Overexpression of LncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue L, Wang Y, Yue S, Zhang J. The expression of miRNA-221 and miRNA-222 in gliomas patients and their prognosis. Neurol Sci. 2017;38(1):67–73. doi: 10.1007/s10072-016-2710-y [DOI] [PubMed] [Google Scholar]
- 40.Chi P, Chen Y, Zhang L, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467(7317):849–853. doi: 10.1038/nature09409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Redpath R, Zhang C, Ning N. Long non‑coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol Lett. 2018;16(3):3365–3372. doi: 10.3892/ol.2018.9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolcher AW, Peng W, Calvo E. Rational approaches for combination therapy strategies targeting the MAP kinase pathway in solid tumors. Mol Cancer Ther. 2018;17(1):3–16. doi: 10.1158/1535-7163.MCT-17-0349 [DOI] [PubMed] [Google Scholar]
- 43.Aksamitiene E, Kiyatkin A, Kholodenko B.N. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans. 2012;40(5):139–146. doi: 10.1042/BST20110609 [DOI] [PubMed] [Google Scholar]
- 44.Alonso N, Diaz Nebreda A, Monczor F, et al. PI3K pathway is involved in ERK signaling cascade activation by histamine H2R agonist in HEK293T cells. Biochim Biophys Acta. 2016;1860(9):1998–2007. doi: 10.1016/j.bbagen.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 45.Serra V, Scaltriti M, Prudkin L, et al. PI3K Inhibition Results in Enhanced HER Signaling and Acquired ERK Dependency in HER2-Overexpressing Breast Cancer. Oncogene. 2011;30(22):2547–2557. doi: 10.1038/onc.2010.626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 2014;14(11):747–753. doi: 10.1038/nrc3819 [DOI] [PubMed] [Google Scholar]
- 48.Knoechel B, Roderick JE, Williamson KE, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46(4):364–370. doi: 10.1038/ng.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salgia R, Kulkarni P. The genetic/non-genetic duality of drug ‘resistance’ in cancer. Trends Cancer. 2018;4(2):110–118. doi: 10.1016/j.trecan.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin Epigenetics. 2015;7:127. doi: 10.1186/s13148-015-0157-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauer S, Hilger RA, Mühlenberg T, et al. Phase I study of panobinostat and imatinib in patients with treatment-refractory metastatic gastrointestinal stromal tumors. Br J Cancer. 2014;110(5):1155–1162. doi: 10.1038/bjc.2013.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Marco C, Laudanna C, Rinaldo N, et al. Specific Gene expression signatures induced by the multiple oncogenic alterations that occur within the PTEN/PI3K/AKT pathway in lung cancer. PLoS One. 2017;12(6):e0178865. doi: 10.1371/journal.pone.0178865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H. Gab2 regulates the migratory behaviors and e-cadherin expression via activation of the pi3k pathway in ovarian cancer cells. Oncogene. 2012;31(20):2512–2520. doi: 10.1038/onc.2011.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang WJ, Mou K, Wu XF, et al. Grb2-associated binder 2 silencing impairs growth and migration of H1975 cells via modulation of PI3K-Akt signaling. Int J Clin Exp Pathol. 2015;8(9):10575–10584. [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto M, Cid E, Bru S, Yamamoto F. Rare and frequent promoter methylation, respectively, of TSHZ2 and 3 genes that are both downregulated in expression in breast and prostate cancers. PLoS One. 2011;6(3):e17149. doi: 10.1371/journal.pone.0017149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vié N, Copois V, Bascoul-Mollevi C, et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008;7(1):14. doi: 10.1186/1476-4598-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross KC, Andrews AJ, Marion CD, Yen TJ, Bhattacharjee V. Identification of the serine biosynthesis pathway as a critical component of BRAF inhibitor resistance of melanoma, pancreatic, and non–small cell lung cancer cells. Mol Cancer Ther. 2017;16(8):1596–1609. doi: 10.1158/1535-7163.MCT-16-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markkanen E, Fischer R, Ledentcova M, Kessler BM, Dianov GL. Cells deficient in base-excision repair reveal cancer hallmarks originating from adjustments to genetic instability. Nucleic Acids Res. 2015;43(7):3667–3679. doi: 10.1093/nar/gkv222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labuschagne CF, van Den Broek NJF, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7(4):1248–1258. doi: 10.1016/j.celrep.2014.04.045 [DOI] [PubMed] [Google Scholar]
- 60.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706 [DOI] [PubMed] [Google Scholar]
- 61.Luo J. Cancer’s sweet tooth for serine. Breast Cancer Res. 2011;13(6):317. doi: 10.1186/bcr2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Marchi T, Timmermans MA, Sieuwerts AM, et al. Phosphoserine aminotransferase 1 is associated to poor outcome on tamoxifen therapy in recurrent breast cancer. Sci Rep. 2017;7(1):2099. doi: 10.1038/s41598-017-02296-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi: 10.1038/ng.890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu B, Jia Y, Cao Y, et al. Overexpression of phosphoserine aminotransferase 1 (PSAT1) predicts poor prognosis and associates with tumor progression in human esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;39(1):395–406. doi: 10.1159/000445633 [DOI] [PubMed] [Google Scholar]