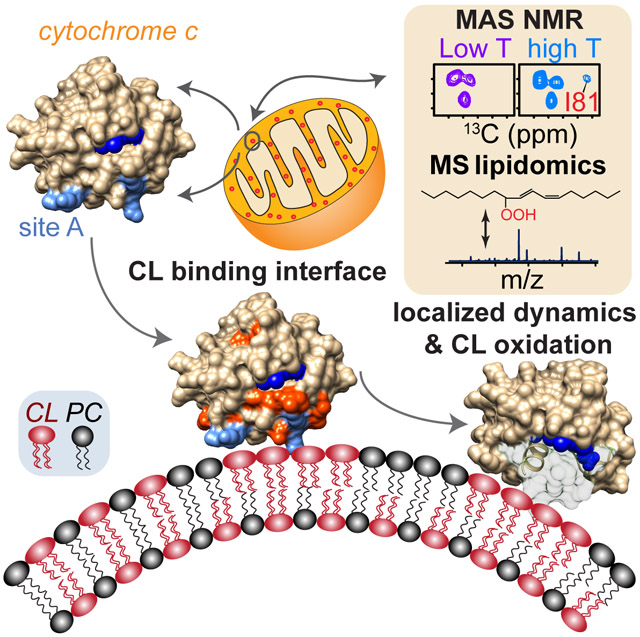
Summary
The peroxidation of cardiolipins by reactive oxygen species (ROS), which is regulated and enhanced by cytochrome c (cyt c), is a critical signaling event in mitochondrial apoptosis. We probe the molecular underpinnings of this mitochondrial death signal through structural and functional studies of horse heart cyt c binding to mixed-lipid membranes containing cardiolipin with mono- and polyunsaturated acyl chains. Lipidomics reveal the selective oxidation of polyunsaturated fatty acid (PUFA) cardiolipin, while multidimensional solid state NMR (ssNMR) probes the structure and dynamics of the membrane and the peripherally bound protein. The hydrophilic milieu at the membrane interface stabilizes a native-like fold, but also leads to localized flexibility at the membrane-interacting protein face. PUFA CL acts as both a preferred substrate and a dynamic regulator by impacting the dynamics of the cyt c N70-I85 omega loop, which covers the heme cavity.
Keywords: Cytochrome c, cardiolipin, mitochondrial protein, apoptosis, membrane protein, PUFA, protein structure and dynamics, membrane oxidation, lipidomics, solid state NMR
Graphical Abstract
eTOC Blurb
Li et al. show how cardiolipin acts as both a substrate and a dynamic regulator of cytochrome c’s pro-apoptotic lipid peroxidase functionality. Combining solid-state NMR and mass spectrometry lipidomics, they find that localized motion of the 70-85 Ω loop regulates heme cavity access in the peroxidase-active peripherally membrane-bound protein.
Apoptosis is a chain of regulated intracellular events that lead to the degradation of unhealthy or unwanted cells (Taylor et al., 2008). It is central in many neurodegenerative disorders, including Alzheimer’s, Parkinson’s and Huntington’s diseases (Mattson, 2000; Taylor et al., 2008). Resistance of cells to apoptosis is a characteristic of tumorigenic cells in several cancer types (Fulda, 2010). A better understanding of apoptotic mechanisms can provide new perspectives for disease intervention. Mitochondria are key regulators of apoptosis, with the release of cytochrome c (cyt c) from mitochondria marking the point of no return (Kagan et al., 2005; Ow et al., 2008; Taylor et al., 2008). Whilst protein-based factors play important roles in cyt c release, one essential participant is the mitochondria-specific lipid cardiolipin (CL) (Kagan et al., 2009). In healthy mitochondria CL is predominantly in the inner leaflet of the mitochondrial inner membrane (MIM) (de Kroon et al., 1997). Under apoptotic conditions, CL comes to increasingly face cyt c in the inter-membrane space (IMS) (Kagan et al., 2005), enabling tight binding by cyt c, which in turn activates the protein’s pro-apoptotic lipid peroxidase function (Abe et al., 2011; Belikova et al., 2006). Cyt c-mediated lipid oxidation selectively affects polyunsaturated fatty acid-containing (PUFA) CL, yielding oxygenated products that are key signaling components for apoptosis (Kagan et al., 2005; Maguire et al., 2017). CL and its oxidation products similarly act as messengers in mitophagy and immune responses (Maguire et al., 2017). Thus, better understanding of CL signaling has widespread applications, including the design of drugs that modulate apoptosis (Atkinson et al., 2011; Jiang et al., 2014).
The structural changes in cyt c caused by binding to CL-containing membrane that facilitate its on-demand lipid peroxidase activity, remain elusive. As delineated in a recent comprehensive review (Schweitzer-Stenner, 2018), one challenge relates to the fact that the CL-cyt c interaction is modulated strongly by experimental context. There is also a lack of techniques to probe membrane-bound cyt c with atomic resolution. In solution NMR studies, the membrane-bound protein tumbles too slowly to be observable, even though changes in the spectra of soluble protein reveal aspects of the initial membrane interactions (Brown and Wuthrich, 1977; Kobayashi et al., 2016; Mohammadyani et al., 2018). As such, current insight into membrane-bound cyt c is derived from diverse complementary lower-resolution techniques (Schweitzer-Stenner, 2018). These suggest that cyt c can interact with CL via multiple binding sites (Alvarez-Paggi et al., 2017; Schweitzer-Stenner, 2018). Most prominent is the so-called “site A”, featuring lysines K72, K73, K86, and K87, which would facilitate an electrostatic interaction with the negatively charged CL (Rytomaa and Kinnunen, 1994; Sinibaldi et al., 2017). Other studies de-emphasize the role of particular binding sites, reporting partial if not complete unfolding upon membrane binding, with a notable dependence on experimental conditions and especially the lipid-to-protein (L/P) ratio (Belikova et al., 2006; Hong et al., 2012 ; Muenzner et al., 2013; Oellerich et al., 2002). This also leads to the coexistence of compact and partially unfolded cyt c states on the membrane (Hanske et al., 2012; Hong et al., 2012; Malyshka and Schweitzer-Stenner, 2017; Pandiscia and Schweitzer-Stenner, 2015a, b).
Here, we set out to probe in some detail the interaction of cyt c with mitochondrial mimetic membranes under a particular set of conditions. Following up on our prior work (Mandal et al., 2015), we examine membranes with an elevated amount of protein-accessible CL, leading to a substantial increase in peroxidase activity, to recapitulate the lipid peroxidase activity seen in pro-apoptotic mitochondria. Mass spectrometry lipidomics reveal the selective lipid oxidation catalyzed by this CL/cyt c complex. 2D and 3D magic angle spinning (MAS) solid-state NMR (ssNMR) experiments reveal a native-like fold and identify sites perturbed by membrane binding. Variable temperature studies finetune our spectral quality and point to a key role for localized dynamics. Our ssNMR structure-function studies elucidate how interactions with CL-rich nanodomains modulate cyt c structure and dynamics to enable its CL peroxidase activity.
Results
Selective CL Peroxidation by Membrane-bound Cyt c.
The peroxidase activity of horse heart cyt c (Figure 1a) in presence of mixed-lipid large unilamellar vesicles (LUVs) has a strong CL dependence (Figure 1b-c). LUVs containing 20 mol-% TOCL trigger a boost of peroxidase function (Figure 1b), compared to the unbound wild-type cyt c. The latter lacks most of the background activity seen in a previously studied double mutant (Mandal et al., 2015; Rumbley et al., 2002). Remarkably, the activity increases strongly with 33% and 50% CL-containing LUVs (Figure 1c). This dramatic activity increase is not explained merely by an equivalent increase in binding (Figure 2a), implying a structural difference behind the pronounced peroxidase activation.
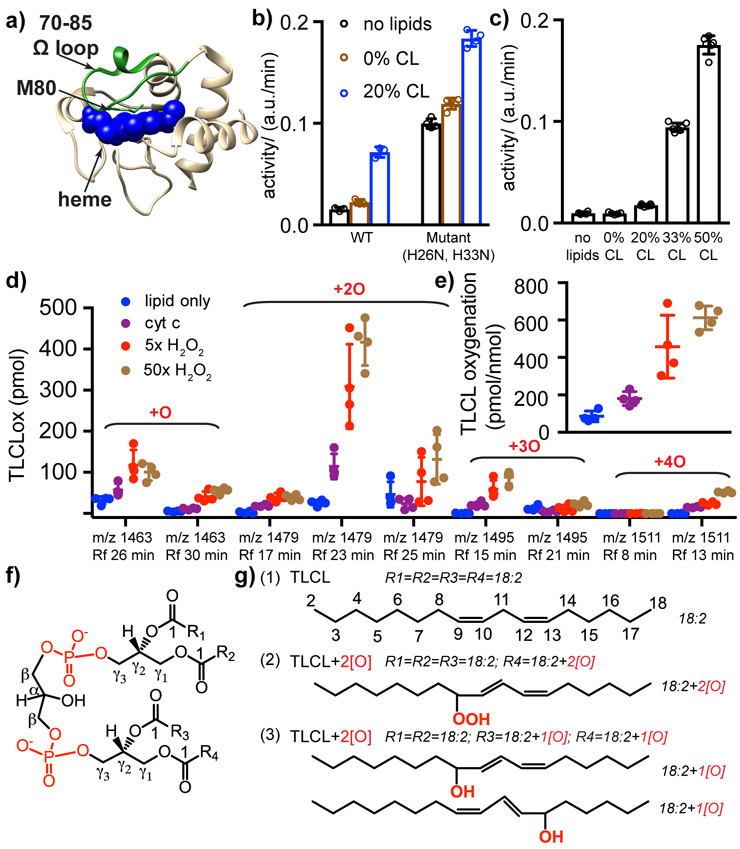
Figure 1. Lipid peroxidase activity of pro-apoptotic cyt c.
(a) X-ray crystal structure of horse heart cyt c (Bushnell et al., 1990), showing the heme (blue spheres), M80 (green sticks), and the 70-85 Ω loop (green highlight). (b) Peroxidase activities of wild-type (WT) and a double mutant (H26N, H33N) cyt c, measured with a fluorescence-based amplex red assay at room temperature. Activities were measured for 0.5 μM cyt c, either without lipids or with TOCL/DOPC LUVs featuring the indicated mol-% of CL at an effective CL/cyt c molar ratio of 4. “Effective CL” accounts for the CL on the outer leaflet of the LUVs. (c) WT cyt c (0.5 μM) peroxidase activity in presence of TOCL/DOPC LUVs with 0, 20, 33, and 50 mol-% CL (effective CL/cyt c ratio = 0, 4, 6.6, 10). Activity values in (b-c) are internally normalized and not comparable to each other. (d) Oxygenated products of TLCL detected by mass spectrometry lipidomics, revealing variants with one to four oxygens added. The amount of product is shown for each mass-to-charge ratio (m/z) and retention time (RT). Control sample lacks cyt c or H2O2, while P/L = 1:25 for other samples, with H2O2 at 0, 5, and 50 times the cyt c concentration of 1 μM. (e) Quantification of TLCL oxidation by mass spectrometry lipidomics, showing the molar ratio of the sum of oxygenated products to the starting TLCL. (f) CL structure with carbon nomenclature, showing four acyl chains (R1, R2, R3, and R4) connected to the diphosphatidylglycerol through two glycerols. (g) (1) Four identical acyl chains [18:2] of intact TLCL with carbons numbered. (2) Mono-hydroperoxy at C9 of the acyl chain; one predominant oxygenated product of TLCL (m/z 1479, RT 23 min). (3) Oxidative modification of two acyl chains with the addition of one hydroxyl group on each acyl chain at position C9 and C13, respectively (m/z 1479, RT 17 min). All replicates together with their mean and s.d. are plotted in (b)-(e). See also Figure S1 and S2.
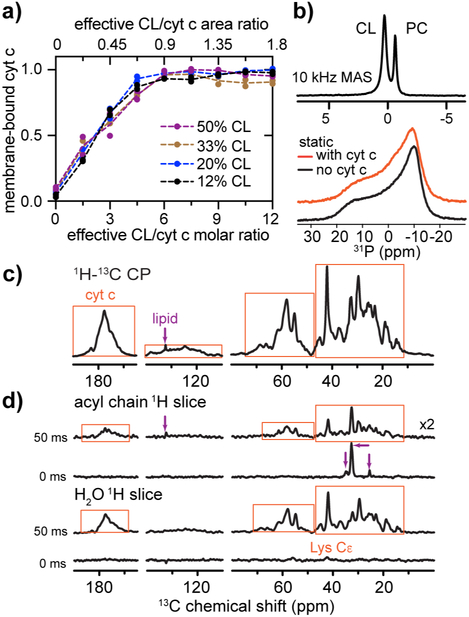
Figure 2. Interaction of cyt c with lipid vesicles of different CL compositions.
(a) Binding of cyt c to TOCL/DOPC vesicles with 12%, 20%, 33%, and 50 mol-% TOCL. The membrane-bound fraction of cyt c is plotted as a function of the effective CL/cyt c molar ratio (bottom axis). The corresponding CL/cyt c area ratio is shown on the top. (b) 31P ssNMR spectra of 50 mol-% CL-containing liposome (TOCL and DOPC) with bound cyt c acquired at 10 kHz MAS (top) and under static condition (bottom; orange). The molar L/P ratio is 25:1. The static spectrum of isolated liposomes is shown in black. MAS and static spectra were acquired at a magnetic field of 15.6 T and temperature of 275 K, and 14.1 T and 282 K, respectively. (c) 1D 1H-13C cross polarization (CP) MAS NMR spectrum of U-13C,15N cyt c bound to TLCL/DOPC (1:1) LUVs (L/P = 25:1), at 14.1 T, MAS 8 kHz, and temperature 265 K. Protein and lipid peaks are marked with orange boxes and purple arrows, respectively. (d) SSNMR shows the 13C labeled protein (orange boxes) interacts with the membrane and is surrounded by mobile water. 1D ssNMR spectral slices showing 13C sites in proximity to mobile acyl chain and water solvent protons, respectively, in absence and presence of 50 ms 1H-1H polarization exchange (from 2D 1H-13C spectrum in Figure S3d). 2D spectra were acquired at 8 kHz MAS, 14.1 T, and 265 K at which the bulk water solvent was frozen while lipid-proximal water stayed mobile.
When plotting the proportion of membrane-bound cyt c for different CL/PC compositions against the number of surface-accessible CL per cyt c protein, the binding curves become remarkably self-consistent (Figure 2a). At low CL/cyt c ratios (0 to 4.5) the fraction of bound cyt c increases in proportion to the availability of CL. The increase plateaus until high CL:cyt c ratios (7.5 to 12) allow near-quantitative binding. Thus, binding saturates when approximately six CL lipids are available per protein. Considering the area-per-lipid for CL (Pan et al., 2015), and the size of native cyt c (Bushnell et al., 1990), we calculated an approximate area ratio that reflects the collective CL surface relative to the cyt c radial surface area (Figure 2a; upper axis). Notably, the area of a nanocluster of approximately six CL (~ 780 Å2) is equivalent to 0.9 x the projection area of natively folded cyt c.
Noting that the abovementioned fluorescence assays rely on a soluble substrate quite distinct from the natural lipid targets, we also tested whether our in vitro conditions recapitulate the lipid peroxidation seen in pro-apoptotic mitochondria (Crimi and Esposti, 2011; Kagan et al., 2005). We note that cyt c’s enzymatic activity in context of lipid oxidation is mechanistically different from its processing of non-CL substrates. When CL gets oxidized, its hydroperoxy-group (CL-OOH) becomes a highly effective source of oxidizing equivalents for further PUFA oxidation. In this process, CL-OOH-driven oxidation occurs at ~1,000 higher rates than that with H2O2 (Belikova et al., 2009). Moreover, initiation of cyt c/CL peroxidase activity is associated with oxidative modification of the protein and its self-activation, which further enhances CL oxidation activity (Barayeu et al., 2019). Thus, we used mass spectroscopy-based lipidomics (Maguire et al., 2017; Tyurin et al., 2009) to detect lipid-specific peroxidation products in our samples (Figure 1d-e). In presence of cyt c, lipid oxidation occurs with a preference for the PUFA tetralinoleoyl-CL (TLCL; C18:2). Excess H2O2 (5x and 50x cyt c) leads to increased and faster oxidation. After 1 hr with 50x H2O2, the extent of oxidation is 612 pmol/nmol lipid, far exceeding the residual oxidation activity seen in absence of cyt c (Figure 1e), which is partially due to pre-existing lipid hydroperoxides in bovine heart CL (Barayeu et al., 2019). In-depth analysis by LC-ESI-MS/MS methods (Figure 1d and Figure S1) reveals multiple oxidized TLCL species containing one to four oxygens at m/z 1463.9599; 1479.9543; 1495.9491 and 1511.9436 as the major species. Quantitatively, TLCL oxygenation products with m/z 1479, containing a hydroperoxy-group, were predominant. The ranking order of abundance for the oxidized CL products is 1479 >> 1463 > 1495 > 1511. The oxidation reaction does not alter the overall chemical architecture of CLs (Figure 1f). In LC-MS the oxygenated CL products have shorter LC retention times (RT) due to the decreased hydrophobicity (Figure S1). Oxygenated TLCLox containing two oxygens with m/z 1479 were recorded at RT 17, 23, and 25 min. Oxidation of one acyl chain of CL yielded a hydroperoxy-group (18:2/18:2/18:2/18:2+2[O]) (m/z 1479, RT 23 min) (Figure 1g). The oxidative modification of two acyl chains of CL (18:2/18:2/18:2+1[O] /18:2+1[O]) (m/z 1479, RT 17 min) was also observed. A more detailed analysis of the products is in Figures S1 and S2. Overall, we observe that our in vitro conditions recapitulate the intra-membrane PUFA CL peroxidase activity of cyt c.
The Lipid Peroxidase-Active Cyt c is Located at the Periphery of the Target Membrane.
Next we use ssNMR to probe the structure and dynamics of the CL/cyt c complex. Dynamics-based spectral editing (DYSE) ssNMR (Matlahov and van der Wel, 2018) can distinguish the signals from labeled protein, unlabeled lipids and solvent. Figure 2c shows the 1D 13C MAS NMR spectrum of uniformly 13C,15N labeled (U-13C,15N) cyt c bound to LUVs containing 1:1 (molar) of TLCL (C18:2 PUFA chains) and DOPC. Peaks from protein and lipids are seen, with protein signals dominating, as marked. At the same conditions, a 2D MAS NMR measurement of signal exchange between labeled cyt c and its environment probed the protein-lipid and protein-solvent interactions (Yao et al., 2016). This approach selects 1H signals from highly mobile water and acyl chains based on long transverse (T2) relaxation times. These “mobile” 1H signals are transferred to the protein via 1H-1H spin diffusion, and finally detected via 1H-13C cross-polarization (CP). Protein-lipid correlations are absent when no 1H-1H mixing is applied (Figure 2d). Some exchange between Lys and mobile water occurs due to the side chain solvent exposure and their rapid chemical exchange (Sivanandam et al., 2011). When 1H-1H polarization exchange is allowed for 50 ms, strong correlations between protein and both lipids and membrane-proximal water are observed. The detected lipid peaks show no significant chemical shift changes for either PC or CL lipids upon cyt c binding (Figure S3). We also applied 31P static NMR which is very sensitive to the presence of non-bilayer lipid phases (van der Wel et al., 2000). The 31P static spectrum (Figure 2b) is characteristic of a fluid lipid bilayer, with no indication of highly curved cubic or hexagonal phases. In summary, there is no indication of a significant disruption of the lipid structure by cyt c, which is bound at the membrane-water interface.
Structural Changes in the Activated Protein by Multidimensional MAS NMR.
We next use ssNMR to examine the conformation and dynamics of the cyt c bound to CL-containing LUVs near the binding saturation that we noted above (Figure 2a). At an effective CL to cyt c ratio of ~6.3, we bound fully 13C- and 15N-labeled (U-13C,15N) cyt c to TOCL/DOPC (1:1) LUVs (L/P = 25:1). This L/P ratio is slightly lower than our prior work (Mandal et al., 2015) to maximize the protein signal, while we set out to avoid low L/P ratios that overload the membrane and may lead to membrane disruption and protein aggregation (Oellerich et al., 2004). The fluidity of hydrated LUVs leads to sample dynamics that impact the effectiveness of many standard ssNMR experiments, causing CP ssNMR spectra of membrane-associated cyt c to feature weak signals at ambient conditions (Matlahov and van der Wel, 2018). Also solution-NMR-like J-mediated INEPT experiments (Figure S3c) that detect the flexible components of the sample (Matlahov and van der Wel, 2018) show little protein signal. This argues against the presence of isotropically flexible protein segments. Consistent with this finding, freezing of the sample yields remarkably high-quality NMR spectra (Figure 3b) without the overly broad peak shapes that are expected when one freezes unfolded polypeptides (Su and Hong, 2011). Dispersed and narrow off-diagonal cross peaks suggest a high level of structural homogeneity. Partial resonance assignments for 52 out of 104 residues across the sequence (Table S1) were obtained with 2D and 3D ssNMR. Some of the assignments were supported by comparison to solution NMR shifts of unbound protein (O'Brien et al., 2015) (Figure S4a), verified by backbone connections in 3D ssNMR datasets (e.g. Figure S4b).
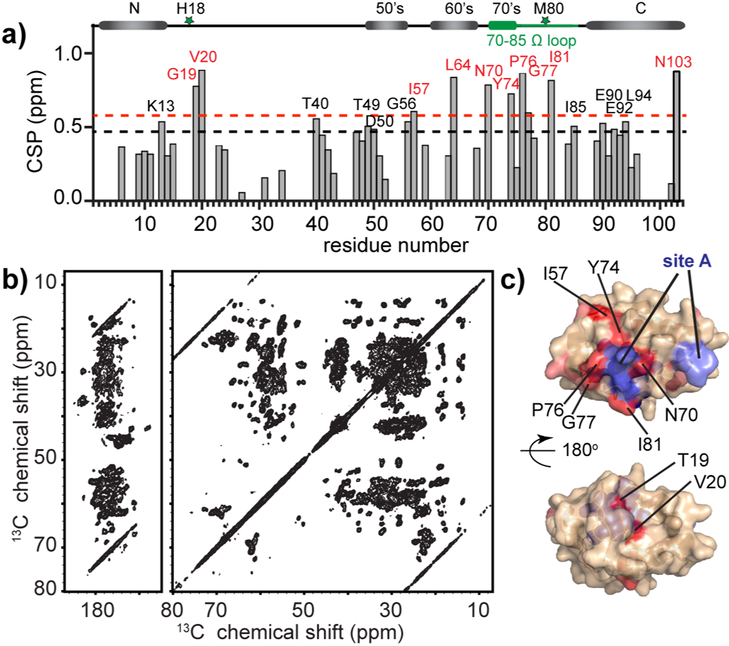
Figure 3. SSNMR of cyt c bound to TOCL/DOPC LUVs.
(a) Residue-specific NMR chemical shift perturbations (CSP) of cyt c upon binding to the membrane compared to cyt c chemical shifts in solution. Strong (>1.3x average) and medium (>1.1x average) CSP values shown in red and black. The secondary structure of native cyt c (Bushnell et al., 1990) is plotted at top, with heme coordination residues H18 and M80 marked as green stars, and the 70-85 Ω loop in green. (b) 2D 13C-13C DARR MAS ssNMR spectrum acquired at 252 K, MAS frequency of 10 kHz, and 17.6 T, probing the sedimented LUV-bound protein (1:25 P/L ratio). (c) Strong and medium chemical shift perturbation sites mapped on the cyt c structure in red and salmon, with strong perturbations marked with black labels. The previously proposed “site A” is highlighted in slate blue and labeled in blue. See also Figure S4 and Table S1.
A comparison to the NMR shifts of soluble cyt c reveals chemical shift perturbations (CSP) induced by membrane binding (Figure 3a). Residues with notable CSPs are shown on the cyt c structure in Figure 3c (salmon and red). Strong perturbations occur in the 70-85 Ω loop with N70, Y74, P76, G77 and I81 as the dominant perturbation sites. Neighboring the strongly perturbed I81, the signal of M80, is either completely missing or greatly shifted compared to the soluble protein (Figure S4a). Residue G56 and I57, spatially close to the 70-85 Ω loop, also show CSPs. Elsewhere, several residues of the 40-54 Ω loop and the C-terminal helix show moderate perturbations. Strong CSPs affect T19-V20 following the heme coordinating H18. Collectively, most of the strong perturbation sites reside on one face of cyt c (Figure 3c), clustered around the abovementioned “site A” (shown slate blue).
Dynamics Changes Affect Specific Regions of Membrane-bound Cyt c.
Although we thus far discuss the ssNMR in terms of changes in structure, previous studies noted dynamic changes upon membrane binding and peroxidase activation (Hanske et al., 2012; Pinheiro et al., 1997). To gain more insight into the protein motional properties under the current experimental conditions, we performed temperature-dependent ssNMR. 1D 1H NMR tracks the freezing of bulk and lipid-associated water (Figure S5d), revealing a progressive freezing of sample water (intensity losses of the narrow signal of mobile water), with membrane-associated water resisting full freezing down to 240 K (Mandal et al., 2015; Mandal and van der Wel, 2016). Only upon cooling to 264 K and below, we observe the dramatic appearance of strong 15N CP and 13C-15N double CP protein signals (Figure S5a-b).
For a more in-depth look, we measured 2D spectra at select temperatures. 2D NCA spectra yield a 13Cα-15N intra-residue (backbone) peak for each residue (Figure 4a). As the temperature lowers, we see a broadening of a defined subset of residues (red labels), around “site A” and the 70-85 Ω loop (Figure 4b), while others appear unaffected. This signal broadening is characteristic of a “freezing out” of localized motion. Corresponding variable-temperature 13C-13C DARR spectra, featuring signals from both backbone and side chain carbons, permit a similar analysis for the protein sidechains (Figure 4c). Clusters of residues near site A, (Y74, P76, I81, I85 and T89) show analogous CSPs. We interpret these line broadening events to arise from local dynamics in the affected loop regions (Figure 4b and 4d).
Figure 4. Residues in 70-85 Ω loop and close to “site A” are affected by local dynamics detected by 2D MAS NMR.
(a) 2D NCA spectra of LUV-bound cyt c (50/50 mol-% TOCL/DOPC; L/P 25:1) acquired at 240 K (mauve), 252 K (green), and 264 K (blue) temperatures. Temperature-dependent changes in peak intensity or position are labeled. (b) Cyt c structure with the affected residues highlighted in cyan. Site A (in slate blue) and heme (in blue) are noted. (c) Spectral regions from 2D 13C-13C DARR spectra at the same temperatures, showing residues with changing sidechain dynamics. Signals from specific residues labeled in red (Y74 in (1), I81 and I85 in (2) and P76 in (3)) are affected, while others (e.g. F10 in (1), I95 and I75 in (2), and I9 and V11 in (3)) are not. The latter signals are consistent at the different temperatures. (d) Location of residues from (c) highlighted in the cyt c structure in magenta. The spectra were acquired at 15.6 T magnetic field and 10 kHz MAS. See also Figure S5.
Dynamics of Cyt c Bound to Liposomes with Different Acyl Chain Flexibilities.
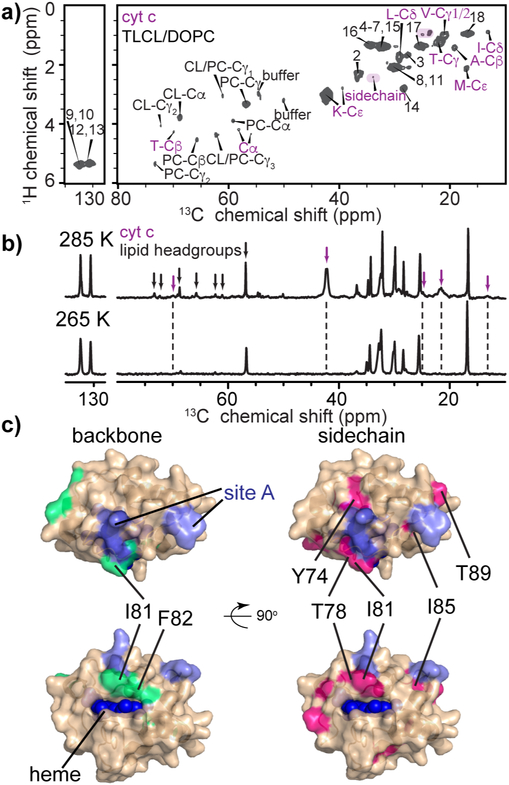
We speculated that such dynamics would be sensitive to changes in the membrane fluidity. We probed this using samples with a PUFA CL (TLCL), which displays increased flexibility and a lower melting temperature compared to TOCL. In mobility-selective INEPT ssNMR we see an increased, though still limited, number of protein signals. Alongside the dominant side chain carbons (residues V, K, E, A, M, and I), weak backbone Cα signals are seen, including one assigned to T (Figures 5a and S5h). We acquired 2D NCA and DARR CP-based ssNMR spectra at 265 K, after again acquiring a series of variable temperature 1D spectra (Figure S5e-g). Seemingly, spectral quality is optimized when we eliminate collective sample motions (e.g. membrane undulation) by freezing most bulk solvent, while retaining a layer of fluid hydration water around the protein of interest. In comparison to the TOCL/DOPC LUVs, spectral changes in both NCA and DARR spectra are seen for the I81 and F82 backbones and side chains of residues Y74, T78, I81, and I89 (Figure 5c), visualized in Figure S6. Remarkably, the spectral changes associated with dynamically different membranes are very similar to those seen as a function of temperature for (1:1) TOCL/DOPC membranes in Figure 4, further supporting their origins being a dynamic flexibility in the protein that is coupled to the dynamic membrane interfacial milieu.
Figure 5. PUFA CL and its impact on cyt c dynamics.
(a) INEPT-based ssNMR 2D spectrum of U-13C,15N-labeled cyt c bound to TLCL/DOPC (1:1) vesicles, P/L ratio 1:25, with protein peaks highlighted in purple labels. The spectrum was acquired at 14.1 T, 285 K, and MAS rate of 10 kHz. (b) INEPT-based 1D 13C spectra of TLCL/DOPC (1:1) vesicles with U-13C,15N-labeled cyt c bound, acquired at 285 K and 265 K at 14.1 T and 10 kHz. Cyt c and lipid headgroup signals are marked in the spectra with purple and black arrows, respectively, with unmarked peaks being due to lipid acyl chains. The protein and lipid headgroup signals are missing in the spectrum at 265 K, whereas lipid acyl chain signals are retained. (c) Lipid-dependent perturbations in cyt c backbone (green) and sidechain (pink) comparing binding to TLCL/DOPC (1:1) LUVs, to TOCL/DOPC membrane binding. Site A and heme are marked. Refer also to Figure S5 and S6.
Discussion
CL Binding and Peroxidation
We used lipid vesicles with varying CL content to probe the binding and pro-apoptotic lipid peroxidase activity of cyt c. Binding showed a characteristic dependence on CL/cyt c ratio, with saturation at about 6 CL per cyt c (Figure 2a), which we interpret as selective binding to CL nanoclusters. A similar stoichiometry was found by a similar approach (Muenzner et al., 2013) and other experimental methods (Belikova et al., 2006; Borisenko et al., 2008; Mohammadyani et al., 2018). However, other computational and experimental studies report a range of values, suggesting a dependence on experimental conditions and measurement techniques (Schweitzer-Stenner, 2018). While an electrostatically bound peripheral protein should occupy more space than its physical footprint (Mbamala et al., 2005; Mohammadyani et al., 2018), cyt c binds membranes at levels that match and exceed full occupancy (Oellerich et al., 2004), which ultimately causes membrane disruption and protein aggregation.
Cyt c peroxidase activity dramatically increased for LUVs containing elevated CL amounts as high as 50 mol-% (Figure 1c). Since 20- and 50 mol-% CL-containing LUVs bind 80% and 95% of added cyt c, increased binding does not explain the almost order of magnitude increase in activity (Figure 1c). This observation suggests that such high-CL conditions trigger a hyper-activated state of cyt c that is not accessible for more limiting amounts of CL. Importantly, by mass spectrometry lipidomics, we detected lipid peroxidation that leads to the selective modification of TLCL and yielded specific TLCL oxygenation products (Figure 1d-g). Similar CL oxidation levels (of 130-140 pmol/nmol) were detected in mitochondria isolated from mouse liver and when mouse embryonic cells were exposed to apoptotic stimulus (Kagan et al., 2005). Thus, our in vitro experimental conditions recapitulate features of the early stages of mitochondrial apoptosis.
Our peroxidase-active CL/cyt c complexes gave ssNMR correlation spectra with remarkably good spectral quality (Figure 3b and Figure S4), enabling previously-unavailable residue-specific assignments for 50% of the protein. The N- and C-terminal α-helices as well as the 60s’ α-helix are well preserved, while residue-specific CSPs (relative to unbound protein) are less than 1.0 ppm indicating the absence of large-scale structural rearrangements. Perturbation of M80 (and neighboring residues) indicates a disruption of heme ligation upon CL binding. From the ssNMR the fate of M80 is not clear, but prior studies have shown it to be displaced (Alvarez-Paggi et al., 2017; Schweitzer-Stenner, 2018). The strongest CSPs affect residues clustering on specific faces of the protein (Figure 3c), most notably, a patch near the “site A” membrane interaction site. We observed lesser perturbations near Lys patch K53/55, consistent with the idea that it can extend the primary “site A” binding site (Elmer-Dixon and Bowler, 2017). We saw by ssNMR (Figure 2d) that the protein is in direct contact with both the (fluid) membrane and a membrane-proximal hydration water layer, indicative of peripheral membrane binding. This places the protein in a hydrophilic environment that presumably plays a key role in the preservation of the native fold. Thus, our data support the idea that “site A” is the primary site for surface-mediated electrostatic binding to the negatively charged CL headgroups.
It is important to note that the cyt c - CL interaction is sensitive to the conditions such as salt, pH, L/P ratios, and the membrane model used (Alvarez-Paggi et al., 2017; Schweitzer-Stenner, 2018). Alternative binding modes prevail under other conditions. For example, the “site L” (opposite “site A”) has been implicated under acidic conditions (Kawai et al., 2005), but plays a lesser role at pH 7 (Mohammadyani et al., 2018). Consistent with the latter, our measurements at neutral pH show little evidence of perturbations in the assigned residues G23, G24, K27, and N31 near site L. Caution should be applied when extrapolating our findings to other sample conditions.
Cyt c is Dynamic but not Unfolded under the Examined Conditions.
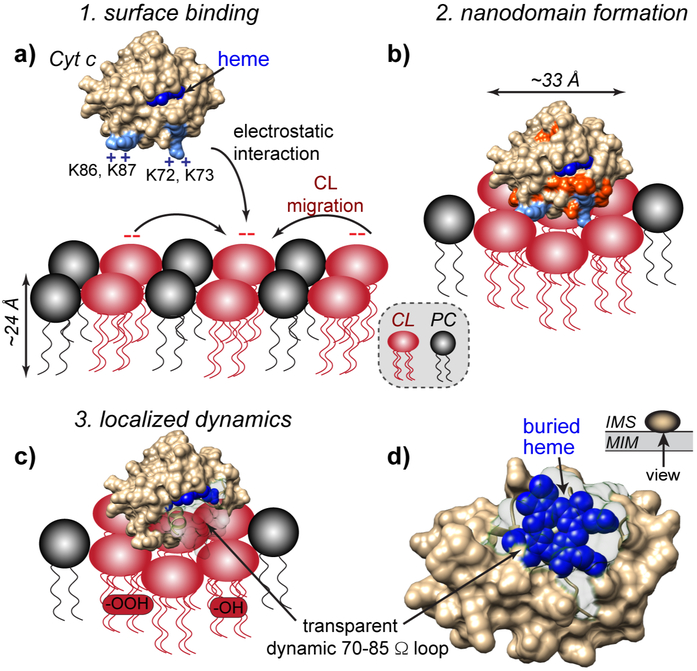
We also used ssNMR to probe the dynamic properties of the CL/cyt c complexes in our samples. We observed site-selective peak broadening, indicative of a freezing-out of dynamics of loop residues, as we modulated the membrane interface mobility by varying the temperature or changing lipid fluidity (Figures 4-5). Specifically, we repeatedly noted dynamic changes in residues of the 70-85 Ω loop. This finding is noteworthy since this region is close to the “site A” residues responsible for binding to the CL headgroups, and also acts as a “lid” over the heme crevice (Figure 6d). Thus, our data point to local dynamic changes at the membrane interface near the “site A” membrane-binding site.
Figure 6. Schematic model of pro-apoptotic interplay of cyt c and CL in the mitochondrial membrane.
(a) At physiological pH, cyt c binds to negatively charged CL headgroups in the outer leaflet of the mitochondrial inner membrane (MIM) through electrostatic interactions with the so-called “site A” (K72, K73, K86, and K87). Strong association with the membrane involves a cluster of ~6 CL molecules, forming CL-enriched nanodomains. (b) The interaction causes structural and dynamic perturbations of the membrane-facing side of cyt c, surrounding the “site A” lysines (shown orange-red). (c-d) In presence of increased ROS (characteristic of dysfunctional mitochondria), pro-apoptotic lipid peroxidation is catalyzed by the PUFA-CL/cyt c nanocomplex. Although positioned on the surface, cyt-c’s binding to the CL lipids induces dynamics in the 70-85 Ω loop, thereby making the heme cavity more open and accessible. Selective access to the heme cavity is facilitated by the CL nanocluster formation along with the remarkable innate flexibility of the PUFA acyl chains, which are also highly peroxidation-prone.
Protein unfolding is known to induce high peroxidase activity in cyt c, and has been reported to occur upon membrane binding. The impact of membrane binding is highly dependent on sample conditions (Schweitzer-Stenner, 2018). Several prior studies have highlighted the importance of the effective CL/cyt c ratio. Time-resolved FRET measurements showed a compact conformation is dominant at low CL/cyt c ratios, but conformational exchange between compact and extended states at a CL/cyt c ratio of 375 (Hanske et al., 2012; Hong et al., 2012). At a lower CL/cyt c ratio near 10, cyt c was shown to undergo heme proximal structural changes, while higher CL/cyt c ratios lead to partially unfolded conformations (Elmer-Dixon and Bowler, 2017). High and low L/P ratios induce different binding modes with other phospholipids (Oellerich et al., 2004). We here focused on a CL/cyt c ratio of 6.3:1, with sufficient CL to achieve saturated binding by cyt c (Figure 2) and substantial lipid peroxidation activity (Figure 1). This L/P ratio maximizes the signal/noise ratio (SNR) for the LUV-bound protein’s ssNMR signals and facilitated thereby the in-depth examination of the bound protein. The resulting observation that the protein is mostly folded under these conditions is consistent with prior studies at similar CL/cyt c ratios (Elmer-Dixon and Bowler, 2017; Hanske et al., 2012; Hong et al., 2012; Malyshka and Schweitzer-Stenner, 2017; Pandiscia and Schweitzer-Stenner, 2015a). These high-occupancy conditions may apply spatial or motional constraints on the protein that reduce its propensity to unfold, as previously discussed in context of cyt c encapsulated within reverse micelles (O'Brien et al., 2015). One important aspect of our findings is that we found the largely folded, superficially bound state of cyt c to be nonetheless an active CL peroxidase in our lipidomics assays.
Similar to our prior studies at 4:1 CL/cyt c ratio (Mandal et al., 2015), the current ssNMR data make it clear that the protein is mostly folded. DYSE ssNMR spectra based on through-bond polarization transfers (i.e. using the INEPT scheme) are effective at detecting dynamically disordered proteins or protein segments in membrane-associated and aggregated proteins (Boatz et al., 2017; Lin et al., 2017; Matlahov and van der Wel, 2018). They yield high-quality 1D and 2D spectra of fluid lipid membranes (Figure 5a and S3a-c), even when they are surrounded by partially frozen aqueous solvent (Figure S5c, S5f) (Mandal et al., 2015; Mandal and van der Wel, 2016). However, a limited number of dynamic protein residues are observed in the membrane-bound cyt c, in contrast with membrane-bound proteins featuring dynamic and disordered segments (Gopinath and Veglia, 2018; Xu et al., 2010) and the peripherally bound myelin basic protein (Ahmed et al., 2010). The few flexible residues include several hydrophobic ones, which could be rendered flexible due to a hydrophobic interaction with the highly fluid PUFA. Note that tight interactions with fluid lipids gives (flexible) polypeptides relaxation properties similar to the fluid bilayer itself (Banigan et al., 2015).
Notably, higher CL content (50%) induces substantial increase of cyt c peroxidase activity compared to 20% CL (Figure 1c). Previous ssNMR of cyt c bound to liposomes containing 20% TOCL and 80% DOPC had pointed to a mostly folded state for cyt c, although the spectral quality limited our ability for a close inspection of the binding sites and local dynamics. Within the limits of the spectral quality, and the different protein variants and other sample conditions, the similarity of the ssNMR spectra (and observed chemical shifts) suggest that there is no dramatic change in protein structure, however. As such, we propose that the primary reason for the increased activity is related to increased dynamics of the protein when associated with the more CL-rich membranes, and in particular the most fluid PUFA CL-containing vesicles. As discussed in the Results section, it is worth reiterating that CL peroxidation by cyt c is mechanistically distinct from soluble substrates, as it is facilitated and enhanced by the CL-derived products.
The ssNMR peaks detected for a dynamic structural ensemble reflect a time-averaged signal that is weighted by the relative occupancy of the underlying conformational states. Proteins that occupy multiple structural (or dynamic) states that are not in rapid exchange, yield multiple signals for the structurally distinct residues, with signal intensities that scale with relative populations. Therefore, the currently available ssNMR data allow for the presence of as-yet undetected minor populations (<10 %) that are nonetheless functionally relevant. This may include the previously noted coexistence of compact and extended forms of the protein, as discussed above. Additional ssNMR studies are needed to fully dissect the complex structure-function relationship and the dynamic ensemble of the bound cyt c.
Interplay of CL Binding and Peroxidation under Pro-apoptotic Conditions.
Thus, we recapitulated here the binding of cyt c to CL-containing membrane at limiting CL concentrations and show that the cyt c/CL complex acquires a high level of CL-specific peroxidase activity (Figure 1). This activated protein state reflects a near-native fold bound to the membrane surface, though with a notable induced flexibility in specific loop residues. We now propose a model of how this interaction of cyt c with CL may relate to this folded CL/cyt c complex’s proapoptotic peroxidase function, as shown in Figure 6. Whilst mitochondrial CL are normally not free to engage with cyt c in the IMS, proapoptotic conditions place increasing amounts of free CL accessible to the IMS in the inner and outer mitochondrial membranes. Then, a newfound CL-cyt c interaction increasingly outcompetes the electrostatic interaction between cyt c and complex III and IV as the CL amount increases (Figure 6a). The complete binding of cyt c to the membrane involves multiple CL molecules, suggesting CL nanodomain formation and rationalizing a strong dependence on the local CL concentration and the availability of free CL. At pH 7.4, binding site A is the primary interacting site based on the ssNMR perturbations (Figure 6b). The membrane interaction appears to render the heme-binding 70-85 Ω loop dynamic (Figure 6c), with notable implications for the CL-specific peroxidase activity of the complex. As shown in Figure 6c-d, this loop region not only faces the membrane interface but also masks the heme within cyt c. Flexibility of this loop therefore increases access of the peroxidase active site to nearby lipid molecules. A supplementary video illustrating the relative locations of the heme and the 70-85 Ω loop is available online. With cyt c positioned atop negatively charged CL nanodomains, the CL concentration is locally enriched crowding out other lipids (Heimburg and Marsh, 1995; Mbamala et al., 2005). This provides one rationale for the observed selectivity of lipid peroxidation for CL. We also noted the preference for the PUFA over monounsaturated CL. On the one hand, PUFA are inherently more reactive towards oxidation (Abe et al., 2011; Tyurina et al., 2006). However, we also propose a possible role for their increased flexibility, as we observed the latter to directly influence the dynamics of the lipid-bound protein. Moreover, acyl chain dynamics are instrumental for achieving interface-proximal interactions between the oxidizable double bonds and the active site of a surface-bound protein (Bachar et al., 2004; Mandal et al., 2015). Thus, membrane fluidity can play multiple crucial roles in modulating and enhancing proapoptotic processes in mitochondrial membranes.
In summary our model emphasizes that cyt c does not necessarily have to abandon its native fold in order to switch its role from an electron shuttle to a lipid peroxidase generating pro-apoptotic signals. Our findings highlight the importance of localized dynamics and membrane fluidity in this orchestrated functional switch, with mitochondrial CL playing a crucial role as both substrate and a dynamic regulator.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Patrick C.A. van der Wel (p.c.a.van.der.wel@rug.nl). The datasets generated and analyzed associated with this study are available from the corresponding author upon reasonable request.
METHOD DETAILS
Materials
See also the Key Resource Table below. Common chemicals were purchased from Fisher Scientific (Pittsburgh, PA) or Sigma-Aldrich (St. Louis, MO). Horse heart cytochrome c (without isotopic labeling) was purchased from Sigma-Aldrich (catalog number C7752). Dioleoyl phosphatidylcholine (1,2-dioleoyl-sn-glycero-3-phosphocholine, DOPC; C18:1), 1-Palmitoyl(D31)-2-oleoyl-sn-glycero-3-phosphocholine, (16:0D31-18:1 PC), tetramyristoyl-cardiolipin (1',3'-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt), TMCL), monounsaturated tetraoleoyl cardiolipin (1',3'-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt), TOCL; C18:1), and bovine heart cardiolipin (BHCL; natural mix with mostly C18:2) were obtained from Avanti Polar Lipids (Alabaster, Alabama). Tetralinoleoyl-cardiolipin (1',3'-bis[1,2-dilinoleoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt) TLCL (C18:2) was obtained as a custom synthesis from Avanti Polar Lipids. Control samples of oxygenated TLCLs (containing 1-4 oxygen) were biosynthesized from TLCL in the reaction catalyzed by soybean lipoxidase (LOX, Sigma-Aldrich) in 50 mM HEPES buffer containing 100 μM DTPA (for transition metals chelation) and saturated with oxygen (before addition of LOX) at pH 7.4. Oxygenated TLCL molecular species were purified by preparative reverse phase HPLC.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E.coli | EMD Millipore | Cat#713974 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ammonium chloride (15N, 99%) | Cambridge Isotope Laboratories, Inc. | Cat#NLM-467–10 |
| D-Glucose (U-13C6, 99%) | Cambridge Isotope Laboratories, Inc. | Cat#CLM-1396–10 |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine | Avanti | Cat#850375C |
| 1-Palmitoyl(D31)-2-oleoyl-sn-glycero-3-phosphocholine | Avanti | Cat#860399 |
| 1',3'-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt) | Avanti | Cat#750332 |
| 1',3'-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt) | Avanti | Cat#710335 |
| Bovine heart cardiolipin | Avanti | Cat#840012 |
| 1',3'-bis[1,2-dilinoleoyl-sn-glycero-3-phospho]-sn-glycerol (sodium salt) | Avanti | Custom synthesis |
| HEPES sodium salt | Sigma-Aldrich | Cat#H3784 |
| Diethylenetriaminepentaacetic acid | Sigma-Aldrich | Cat#D6518 |
| Potassium hexa-cyanoferrate (III) | Sigma-Aldrich | Cat#60299 |
| Amplex red | Life Technologies | Cat#A12222 |
| 5-aminolevulinic acid hydrochloride | Sigma-Aldrich | Cat#A7793 |
| Horse heart cytochrome c | Sigma-Aldrich | Cat#C7752 |
| Soybean lipoxidase | Sigma-Aldrich | Cat#L7395 |
| Deposited Data | ||
| NMR chemical shifts of horse heart cytochrome c bound to 1:1 (molar) TOCL/DOPC vesicles | BMRB | Entry 27556 |
| Recombinant DNA | ||
| pETDuet-1 DNA | EMD Millipore | Cat#71146 |
| Software and Algorithms | ||
| TopSpin v3.5 | Bruker Biospin | https://www.bruker.com/products/mr/nmr/nmr-software/nmr-software/topspin/overview.html |
| Sparky 3 | Goddard et al., UCSF | https://www.cgl.ucsf.edu/home/sparky/ |
| CcpNmr v2 | Stevens et al., 2011 | https://www.ccpn.ac.uk/ |
| NMRPipe | Delaglio et al., 1995 | https://www.ibbr.umd.edu/nmrpipe/install.html |
| CSI 3.0 | Hafsa et al., 2015 | http://csi3.wishartlab.com |
| PyMol | Schrodinger LLC | https://pymol.org/2/ |
| UCSF Chimera v1.11.2 | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| Compound Discoverer | ThermoFisher | https://www.thermofisher.com/us/en/home/products-and-services/promotions/industrial/compound-discoverer.html |
| Xcalibur | ThermoFisher | https://www.thermofisher.com/order/catalog/product/OPTON-30487 |
Mixed Lipid Vesicle Preparation
Mixtures of CL and PC phospholipids were used to prepare mitochondrial mimetic liposomes as previously described (Mandal et al., 2015). TOCL and DOPC in chloroform were mixed in a 1:1 molar ratio and dried under N2 gas for 15 – 20 min and placed under vacuum in a desiccator overnight to remove residual solvent. The dried lipid films were resuspended in HEPES buffer (20 mM HEPES, pH 7.4) to form large unilamellar vesicles (LUVs) by vortexing. Afterwards, the lipid suspension was quickly frozen in a cold bath made from dry ice and ethanol, and thawed completely by transferring to a hot water bath about 52 °C. The freeze-and-thaw cycle was repeated 10 times. The final TOCL/DOPC (1:1) liposome stock was prepared by extrusion through a 200 nm NanoSizer MINI extruder unit (T&T Scientific Corp, Knoxville, TN) for 11 times. Other liposomes used throughout our studies were prepared consistently by the same protocol.
Fluorescence-based Peroxidase Activity Assays
The cyt c peroxidation activities induced by CL-containing vesicles were characterized using amplex red (AR) assays as previously reported by us and others (Abe et al., 2011; Kapralov et al., 2011; Mandal et al., 2015). First, lipid vesicles containing TOCL and DOPC were prepared with four different levels of TOCL content, including 0 mol-%, 20 mol-%, 33 mol-%, and 50 mol-%. in HEPES buffer (20 mM HEPES, 100 μM DTPA, pH 7.4). The same buffer was used to prepare all reagent stock solutions. The substrate 50 μM AR was first added to a cuvette, after which liposomes and cyt c stock solutions were added to reach final concentrations of 20 μM and 0.5 μM, respectively, and mixed well. The cyt c peroxidase activity measurement was initiated by adding freshly prepared H2O2 stock solution to a final concentration of 50 μM. Fluorescence intensity during the initial reaction period of 5 min was recorded with the excitation wavelength fixed at 535 nm and emission detected at 585 nm. Background reactions were recorded prior to the addition of H2O2. To determine the lipid-bound cyt c peroxidase activity this background reaction rate was subtracted. The fluorescence data were first normalized within each experimental series according to the highest fluorescence reached and then slopes determined by linear fitting were used to evaluate the peroxidase activity. The comparative measurements of the peroxidase activities of WT and double mutant (H26N, H33N) cyt c were performed under different lipid conditions including the absence of lipids, the presence of 0% and 20% TOCL-containing DOPC/TOCL liposomes. Three to six replicates were performed for each condition, which are plotted in the column scatter plot together with the mean value and standard deviation.
Mass Spectrometry Lipidomics
To characterize the in vitro peroxidation of CL catalyzed by CL-induced cyt c peroxidase, we deployed an assay to quantify lipid oxidation and characterize CL oxygenated products using mass spectroscopy lipidomics. Liposomes containing BHCL, TOCL, and DOPC at 0.33:0.67:1 molar ratio were prepared, and the lipid concentration was kept at 25 μM in the assay. HEPES buffer (20 mM HEPES, 100 μM DTPA, pH 7.4) was used to prepare stock solutions. Cyt c stock was added to reach a lipid/cyt c molar ratio of 25:1, consistent with the ssNMR samples. Freshly prepared H2O2 stock was added to reach final concentrations of 5 μM (5x H2O2 sample) and 50 μM (50x H2O2 sample) to initiate the oxidation, and was added every 15 min during 1 hour of incubation. Upon quenching the reaction, lipids from liposomes including both reactants and products were separated from the protein following Folch procedure (Folch et al., 1957) under nitrogen atmosphere. CL and oxygenated CL species were analyzed by LC-ESI-MS/MS as previously described (Kim et al., 2011; Tyurina et al., 2014) with small modifications. LC-ESI-MS/MS in negative mode was performed using a Dionex UltimateTM 3000 RSLCnano system coupled online Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) using a reverse phase C18 column (Luna (2) 3 μm, 100 Å, 150 × 1 mm, Phenomenex, Torrance, CA). The column was eluted at a flow rate 0.060 mL/min using gradient of solvent system consisting of mobile phase A (acetonitrile/water/ triethylamine, 45:5:2.5 v/v) and B (2-propanol/ water/ trimethylamine, 45:5:2.5 v/v). Both mobile phases contained 5 mM acetic acid and 0.01% formic acid. The resolution was set up at 140000 that corresponds to 5 ppm in m/z measurement error. M/Z values for CLs and their oxidation species are presented to 4 decimal points. TMCL and 1-palmitoyl(D31)-2-oleoyl-sn-glycero-3-phosphocholine were used as internal MS standards. Additionally, oxygenated molecular species of CL products were identified by MS/MS analysis using an Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific, Waltham, MA). Data analysis were performed using Xcalibur (ThermoFisher Scientific) and Compound Discoverer (ThermoFisher Scientific).
Measurement of Lipid Vesicle Binding
Liposomes consisting of TOCL and DOPC with TOCL compositions of 12%, 20%, 33%, and 50% were prepared following the protocol described above. Different ratios of liposomes and cyt c (kept at 6 μM) were incubated in sample buffer (20 mM HEPES, 100 μM DTPA, pH 7.4) at room temperature for 15 min in 230 μL polypropylene ultracentrifuge tubes. The unbound cyt c was separated from liposome-bound cyt c by ultracentrifugation at 435,000 g for 2 hours at 4 °C in a Optima TLX ultracentrifuge with a TLA-100 rotor (Beckman Coulter, Indianapolis, IN). Immediately after the ultracentrifugation, the supernatant containing unbound cyt c was carefully removed and saved for UV-vis spectrophotometer (Beckman Coulter) measurement. Cyt c solution without liposome or centrifugation was prepared in parallel as a reference. UV-vis absorbance at 409 nm was recorded for samples (S) and references (S0), and the ratio of membrane-bound cyt c was calculated as 1-S/S0 and plotted as a function of the “effective” CL/cyt c molar ratio. The effective CL/cyt c ratio represents the molar ratio of CL on the outer leaflet of the LUVs. Experiments were performed in duplicate with individual measurements plotted in Figure 2a. We also report the corresponding area ratio comparing the surface area of the CL to the projection of cyt c onto the membrane. The area ratio calculations approximate each folded cyt c to be a sphere with a radius of 16.5 Å, yielding a projection area of 854 Å2. We used a TOCL cross-sectional area of 129.8 Å2 based on prior molecular dynamics simulations (Pan et al., 2015).
Production of 13C, 15N Enriched Protein
The expression and purification of uniformly 13C, 15N enriched horse heart cyt c were carried out similar to the procedures previously reported for the folding-optimized mutant cyt c (H26N, H33N) (Mandal et al., 2015). A pETDuet plasmid (EMD Millipore, Billerica, MA) encoding both wild type horse heart cyt c and the heme lyase was used to express cyt c with covalently bound heme in E. coli (Pollock et al., 1998; Rumbley et al., 2002). To produce isotopically labeled cyt c, cells were first grown at 37 °C in minimal media containing 0.1% (w/v) 15NH4Cl and 0.4% (w/v) 13C D-glucose (Cambridge Isotope Laboratories, Inc. Tewksbury, MA) and 0.5 mM FeCl3, and 580 μM unlabeled 5-aminolevulinic acid hydrochloride. When an optical density of ~1.1 was reached, protein overexpression was induced with 1 mM IPTG accompanied with the addition of more 15NH4Cl and 13C D-glucose (0.1 and 0.2 g/L, respectively). The temperature was turned down to 18 °C and the cell cultures continued to grow at 18 °C for ~16 h before pelleting down by centrifugation and resuspending in 25 mM sodium phosphate buffer (pH 6.5) with 0.02% sodium azide. The cells were lysed by homogenizer and the cell debris was removed by centrifugation. Cyt c purification was carried out using a cation exchange HiTrap SP HP column (GE Healthcare, Chicago, IL) with phosphate buffer (25 mM sodium phosphate, 10% (v/v) glycerol, 0.02% sodium azide, pH 6.5) and a 0 – 1.0 M NaCl salt gradient. The eluate containing cyt c was further purified through a gel filtration Superdex 200 16/600 column (GE Healthcare, Chicago, IL) using phosphate buffer (25 mM sodium phosphate, 150 mM NaCl, 5% glycerol, 0.02% sodium azide, pH 6.5). Fully oxidized cyt c was obtained by incubation with 5-fold molar amount of potassium hexacyanoferrate (III) (Sigma-Aldrich). The final 13C, 15N cyt c stock in HEPES buffer (20 mM HEPES, pH 7.4) was obtained by the removal of excess K3Fe(CN)6 through buffer exchange in 10 kDa Amicon Ultra-15 centrifugal filters (MilliporeSigma, Burlington, MA).
Preparation of Samples for ssNMR
Natural abundance (i.e., not isotopically labeled) cyt c bound to TOCL/DOPC (1:1) liposomes, U-13C,15N-labeled cyt c bound to TOCL/DOPC (1:1) liposomes, and U-13C,15N-labeled cyt c bound to TLCL/DOPC (1:1) liposomes for ssNMR studies were prepared as follows. Cyt c stock solution was gently added to CL-containing liposomes, prepared as described above, to reach a molar ratio of 1:25 where the effective CL/cyt c ratio was ~6.3. HEPES buffer (20 mM HEPES pH 7.4) was used to prepare both cyt c and different liposome stocks. The resulting suspension was incubated and vortexed for ~15 min at room temperature. The cyt c/CL-containing liposomes complexes were transferred to 3.2 mm thin-wall MAS ssNMR rotors using a home-build rotor packing tool (Mandal et al., 2017). The packing tool combines sample pelleting and simultaneous transfer into the rotor through one step of ultracentrifugation. A typical centrifugation force of 175,000 g was applied in a Beckman Coulter Optima L-100 XP ultracentrifuge with a SW-32 Ti rotor (Beckman Coulter, Indianapolis, IN), with a total centrifugation time of 3-4 h at 4 °C.
Solid-state NMR Measurements
1D, 2D, and 3D ssNMR measurements are described briefly below, with more detailed experimental parameters in Table S2. MAS and static 31P ssNMR (Figure 2b) was employed to test the integrity of bilayer structure of TOCL/DOPC (1:1) membrane upon cyt c binding. 31P single pulse excitation spectra were acquired at 275 K, MAS rate of 10 kHz, and magnetic field of 15.6 T, using a 31P 90° pulse of 7 μs. 31P static spectra of TOCL/DOPC (1:1) liposomes and cyt c/ TOCL/DOPC (1:1) liposomes complexes were acquired at 282 K and 14.1 T, using a 31P 90° pulse of 9.7 μs and high power (85 kHz) CW 1H decoupling. 1H-13C INEPT spectra of TOCL/DOPC (1:1) liposomes in the presence and absence of unlabeled cyt c were acquired with τ1 = 1.6 ms or 1.8 ms and τ2 = 0.8 ms. 2D proton-diffused 1H-13C cross polarization heteronuclear correlation spectra (HH-CPHETCOR, Figure S3d) were acquired on cyt c/TLCL/DOPC (1:1) complexes at 265 K, 14.1 T, and MAS rate of 8 kHz. A pulse sequence was employed in which the initial 1H 90° pulse was followed by a 1H transverse evolution period, followed by a longitudinal 1H-1H spin diffusion mixing time of 0 or 50 ms, and finally a 1H-13C CP transfer step. No homonuclear decoupling was applied during the 1H evolution time, which effectively suppresses signals from protons lacking the fast dynamics required for slow T2 relaxation. 1D 13C (Figure 2d) slices corresponding to the lipid acyl chain CH2 protons (1.4 ppm) and the mobile water protons (5.2 ppm) were extracted using Bruker Topspin software. Chemical shift assignments of membrane-bound cyt c were performed on cyt c/TOCL/DOPC (1:1) complexes by combining 2D 13C-13C DARR (Figure 3b), 2D NCA (Figure 4a), 3D NCACX, and 3D NCOCX experiments (Figure S4b) acquired at 252 K, 15.6 T, and 10 kHz. For variable temperature analysis, additional 2D DARR and NCA spectra at temperatures of 240 K and 264 K were acquired. A systematic characterization of the temperature-dependence of structural and dynamics changes of the water, lipid, and protein was carried out by applying a set of 1D MAS NMR measurements to the cyt c/TOCL/DOPC (1:1) complexes shown in Figure S5a-d (at temperatures ranging from 282 K to 240 K). Similar 1D measurements were performed on cyt c bound to TLCL/DOPC (1:1) complexes from 285 K to 265 K (Figure S5e-g). The MAS NMR measurements involved 1H single pulse, 1H-13C INEPT, 13C-15N SPECIFIC CP, and 15N CP experiments. The highly mobile residues of cyt c associated with the membrane were detected in 2D 13C-13C INEPT-TOBSY (Total Through-Bond Correlation Spectroscopy) experiments, using U-13C,15N-cyt c/TLCL/DOPC (1:1) complexes (Figure S5h). 2D 1H-13C INEPT-based HETCOR spectra of U-13C,15N-cyt c bound to both TLCL/DOPC (1:1) and TOCL/DOPC (1:1) liposomes were acquired (Figure 5a and S3b). Reported temperatures reflect the estimated sample temperature based on calibration measurements using an external KBr test sample (Mandal and van der Wel, 2016; Thurber and Tycko, 2009), unless indicated otherwise (Table S2). Due to variations in spinning friction and RF heating between experiments the actual sample temperature may vary a few (i.e. ± 5) degrees.
NMR spectra were processed by NMRPipe software (Delaglio et al., 1995). Apodizations with exponential line broadening or sine-bell function were applied in 1D and 2D processing. 1D spectra were processed with 10 Hz exponential line broadening, except that 30 Hz line broadening was applied in 1D 31P spectra. 3D and 2D spectra were processed involving apodization with 90° or 60° sine-bell function, linear prediction of the indirect dimension(s) with once the original data size, zero filling, and Fourier transformation. The chemical shifts were referenced to dilute aqueous 2,2-dimethylsilapentane-5-sulfonic acid (DSS) and liquid ammonia, using adamantane 13C chemical shifts as an external reference, as previously described (Lin et al., 2017; Mandal et al., 2015). NMR data analysis were performed in the Sparky (T.D. Goddard and D. G. Kneller, Sparky 3, UCSF) and CcpNmr software suites (Stevens et al., 2011). Secondary structure analysis based on the NMR resonance frequencies was performed with the online CSI server (Hafsa et al., 2015). The chemical shift perturbations (CSPs) for membrane-bound cyt c were determined as the r.m.s.d. between membrane-bound cyt c chemical shifts and cyt c solution NMR shifts (BMRB entry ID 25640) (O'Brien et al., 2015). The average CSP multiplied by 1.1 and 1.3 have been used as cutoffs to determine medium and strong perturbations, separately. Molecular graphics of cyt c shown in Figure 1 and 6 were generated with UCSF Chimera (Pettersen et al., 2004), and those in Figure 3, 4, and 5 with PyMol (The PyMOL Molecular Graphics System).
QUATIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean values with S.D., with the individual replicates also plotted on the graphs in Figure 1b-d and Figure 2a. Additional statistical details can be found in the figure legends and Method Details, above.
DATA AND SOFTWARE AVAIBILITY
The NMR chemical shifts of horse heart cytochrome c bound to 1:1 (molar) TOCL/DOPC vesicles, listed in Supplementary Table S1, have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) as entry 27556.
Supplementary Material
Supplementary video. A video illustration of how 70-85 Ω loop shields the heme cavity of the natively folded protein, such that loop dynamics can facilitate heme exposure. Related to Figure 6. The video is generated using the UCSF Chimera program (Pettersen et al., 2004).
Highlights.
Cytochrome c is a cardiolipin-selective lipid peroxidase in intrinsic apoptosis.
Lipidomics probe oxygenation products of cardiolipin peroxidation in vitro.
Solid-state NMR shows folded, but dynamically perturbed, protein on lipid surface.
PUFA lipid-coupled dynamics of 70-85 Ω loop regulate access to heme cavity.
Acknowledgements
We thank Mike Delk for advice and support with the NMR measurements. The authors acknowledge funding support from the National Institutes of Health R01 GM113908 to P.C.A. v.d. Wel, P01 HL114453 and U19AI068021 to V.E. Kagan, and NIH instrument grant S10 OD012213-01 for the 750 MHz NMR spectrometer.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe M, Niibayashi R, Koubori S, Moriyama I, and Miyoshi H (2011). Molecular mechanisms for the induction of peroxidase activity of the cytochrome c-cardiolipin complex. Biochemistry 50, 8383–8391. [DOI] [PubMed] [Google Scholar]
- Ahmed MAM, Bamm VV, Harauz G, and Ladizhansky V (2010). Solid-State NMR Spectroscopy of Membrane-Associated Myelin Basic Protein—Conformation and Dynamics of an Immunodominant Epitope. Biophys. J 99, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Paggi D, Hannibal L, Castro MA, Oviedo-Rouco S, Demicheli V, Tortora V, Tomasina F, Radi R, and Murgida DH (2017). Multifunctional cytochrome c: Learning new tricks from an old dog. Chem. Rev 117, 13382–13460. [DOI] [PubMed] [Google Scholar]
- Atkinson J, Kapralov AA, Yanamala N, Tyurina YY, Amoscato AA, Pearce L, Peterson J, Huang Z, Jiang J, Samhan-Arias AK, et al. (2011). A mitochondria-targeted inhibitor of cytochrome c peroxidase mitigates radiation-induced death. Nat. Commun 2, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachar M, Brunelle P, Tieleman DP, and Rauk A (2004). Molecular Dynamics Simulation of a Polyunsaturated Lipid Bilayer Susceptible to Lipid Peroxidation. J. Phys. Chem. B 108, 7170–7179. [Google Scholar]
- Banigan JR, Gayen A, and Traaseth NJ (2015). Correlating lipid bilayer fluidity with sensitivity and resolution of polytopic membrane protein spectra by solid-state NMR spectroscopy. Biochim. Biophys. Acta - Biomembranes 1848, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barayeu U, Lange M, Méndez L, Arnhold J, Shadyro OI, Fedorova M, and Flemmig J (2019). Cytochrome c auto-catalyzed carbonylation in the presence of hydrogen peroxide and cardiolipins J. Biol. Chem, in press, DOI: 10.1074/jbc.RA1118.004110 (online publication date Dec. 12, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikova NA, Tyurina YY, Borisenko G, Tyurin V, Samhan Arias AK, Yanamala N, Furtmuller PG, Klein-Seetharaman J, Obinger C, and Kagan VE (2009). Heterolytic reduction of fatty acid hydroperoxides by cytochrome c/cardiolipin complexes: antioxidant function in mitochondria. J. Am. Chem. Soc 131, 11288–11289. [DOI] [PubMed] [Google Scholar]
- Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, and Kagan VE (2006). Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry 45, 4998–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatz JC, Whitley MJ, Li M, Gronenborn AM, and van der Wel PCA (2017). Cataract-associated P23T γD-crystallin retains a native-like fold in amorphous-looking aggregates formed at physiological pH. Nat. Commun 8, 15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko GG, Kapralov AA, Tyurin VA, Maeda A, Stoyanovsky DA, and Kagan VE (2008). Molecular design of new inhibitors of peroxidase activity of cytochrome c/cardiolipin complexes: fluorescent oxadiazole-derivatized cardiolipin. Biochemistry 47, 13699–13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LR, and Wuthrich K (1977). NMR and ESR studies of the interactions of cytochrome c with mixed cardiolipin-phosphatidylcholine vesicles. Biochim. Biophys. Acta 468, 389–410. [DOI] [PubMed] [Google Scholar]
- Bushnell GW, Louie GV, and Brayer GD (1990). High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol 214, 585–595. [DOI] [PubMed] [Google Scholar]
- Crimi M, and Esposti MD (2011). Apoptosis-induced changes in mitochondrial lipids. Biochim. Biophys. Acta 1813, 551–557. [DOI] [PubMed] [Google Scholar]
- de Kroon AI, Dolis D, Mayer A, Lill R, and de Kruijff B (1997). Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim. Biophys. Acta 1325, 108–116. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Elmer-Dixon MM, and Bowler BE (2017). Site A-Mediated Partial Unfolding of Cytochrome c on Cardiolipin Vesicles Is Species-Dependent and Does Not Require Lys72. Biochemistry 56, 4830–4839. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 226, 497–509. [PubMed] [Google Scholar]
- Fulda S (2010). Exploiting mitochondrial apoptosis for the treatment of cancer. Mitochondrion 10, 598–603. [DOI] [PubMed] [Google Scholar]
- Gopinath T, and Veglia G (2018). Probing membrane protein ground and conformationally excited states using dipolar- and J-coupling mediated MAS solid state NMR experiments. Methods 148, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafsa NE, Arndt D, and Wishart DS (2015). CSI 3.0: a web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res. 43, W370–W377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, and Pletneva EV (2012). Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. U S A 109, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg T, and Marsh D (1995). Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys. J 68, 536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Muenzner J, Grimm SK, and Pletneva EV (2012). Origin of the conformational heterogeneity of cardiolipin-bound cytochrome C. J. Am. Chem. Soc 134, 18713–18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Bakan A, Kapralov AA, Silva KI, Huang Z, Amoscato AA, Peterson J, Garapati VK, Saxena S, Bayir H, et al. (2014). Designing inhibitors of cytochrome c/cardiolipin peroxidase complexes: mitochondria-targeted imidazole-substituted fatty acids. Free Radic. Biol. Med 71, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang JF, Stoyanovsky DA, Wipf P, Kochanek PM, et al. (2009). Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radi. Biol. Med 46, 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang JF, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, et al. (2005). Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol 1, 223–232. [DOI] [PubMed] [Google Scholar]
- Kapralov AA, Yanamala N, Tyurina YY, Castro L, Samhan-Arias A, Vladimirov YA, Maeda A, Weitz AA, Peterson J, Mylnikov D, et al. (2011). Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim. Biophys. Acta - Biomembranes 1808, 2147–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai C, Prado FM, Nunes GL, Di Mascio P, Carmona-Ribeiro AM, and Nantes IL (2005). pH-Dependent interaction of cytochrome c with mitochondrial mimetic membranes: the role of an array of positively charged amino acids. J. Biol. Chem 280, 34709–34717. [DOI] [PubMed] [Google Scholar]
- Kim J, Minkler PE, Salomon RG, Anderson VE, and Hoppel CL (2011). Cardiolipin: characterization of distinct oxidized molecular species. J. Lipid. Res 52, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Nagao S, and Hirota S (2016). Characterization of the cytochromec membrane-binding site using cardiolipin-containing bicelles with NMR. Angew. Chem. Intl. Edit 55, 14019–14022. [DOI] [PubMed] [Google Scholar]
- Lin H-K, Boatz JC, Krabbendam IE, Kodali R, Hou Z, Wetzel R, Dolga AM, Poirier MA, and van der Wel PCA (2017). Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nat. Commun 8, 15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire JJ, Tyurina YY, Mohammadyani D, Kapralov AA, Anthonymuthu TS, Qu F, Amoscato AA, Sparvero LJ, Tyurin VA, Planas-Iglesias J, et al. (2017). Known unknowns of cardiolipin signaling: The best is yet to come. Biochim. Biophys. Acta 1862, 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshka D, and Schweitzer-Stenner R (2017). Ferrocyanide-Mediated Photoreduction of Ferricytochrome C Utilized to Selectively Probe Non-native Conformations Induced by Binding to Cardiolipin-Containing Liposomes. Chemistry 23, 1151–1156. [DOI] [PubMed] [Google Scholar]
- Mandal A, Boatz JC, Wheeler TB, and van der Wel PCA (2017). On the use of ultracentrifugal devices for routine sample preparation in biomolecular magic-angle-spinning NMR. J. Biomol. NMR 67, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A, Hoop CL, DeLucia M, Kodali R, Kagan VE, Ahn J, and van der Wel PC (2015). Structural changes and proapoptotic peroxidase activity of cardiolipin-bound mitochondrial cytochrome c. Biophys. J 109, 1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A, and van der Wel PCA (2016). MAS (1)H NMR probes freezing point depression of water and liquid-gel phase transitions in liposomes. Biophys. J 111, 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlahov I, and van der Wel PCA (2018). Hidden motions and motion-induced invisibility: dynamics-based spectral editing in solid-state NMR. Methods 148, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP (2000). Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Bio 1, 120–129. [DOI] [PubMed] [Google Scholar]
- Mbamala EC, Ben-Shaul A, and May S (2005). Domain Formation Induced by the Adsorption of Charged Proteins on Mixed Lipid Membranes. Biophys. J 88, 1702–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadyani D, Yanamala N, Samhan-Arias AK, Kapralov AA, Stepanov G, Nuar N, Planas-Iglesias J, Sanghera N, Kagan VE, and Klein-Seetharaman J (2018). Structural characterization of cardiolipin-driven activation of cytochrome c into a peroxidase and membrane perturbation. Biochim. Biophys. Acta 1860, 1057–1068. [DOI] [PubMed] [Google Scholar]
- Muenzner J, Toffey JR, Hong YN, and Pletneva EV (2013). Becoming a peroxidase: Cardiolipin-induced unfolding of cytochrome c. J. Phys. Chem. B 117, 12878–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien ES, Nucci NV, Fuglestad B, Tommos C, and Wand AJ (2015). Defining the Apoptotic Trigger: The interaction of cytochrome c and cardiolipin. J. Biol. Chem 290, 30879–30887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellerich S, Lecomte S, Paternostre M, Heimburg T, and Hildebrandt P (2004). Peripheral and integral binding of cytochrome c to phospholipids vesicles. J. Phys. Chem. B 108, 3871–3878. [Google Scholar]
- Oellerich S, Wackerbarth H, and Hildebrandt P (2002). Spectroscopic characterization of nonnative conformational states of cytochrome c. J. Phys. Chem. B 106, 6566–6580. [Google Scholar]
- Ow YP, Green DR, Hao Z, and Mak TW (2008). Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol 9, 532–542. [DOI] [PubMed] [Google Scholar]
- Pan JJ, Cheng XL, Sharp M, Ho CS, Khadka N, and Katsaras J (2015). Structural and mechanical properties of cardiolipin lipid bilayers determined using neutron spin echo, small angle neutron and X-ray scattering, and molecular dynamics simulations. Soft Matter 11, 130–138. [DOI] [PubMed] [Google Scholar]
- Pandiscia LA, and Schweitzer-Stenner R (2015a). Coexistence of native-like and non-native cytochrome c on anionic liposomes with different cardiolipin content. J. Phys. Chem. B 119, 12846–12859. [DOI] [PubMed] [Google Scholar]
- Pandiscia LA, and Schweitzer-Stenner R (2015b). Coexistence of native-like and non-native partially unfolded ferricytochrome c on the surface of cardiolipin-containing liposomes. J. Phys. Chem. B 119, 1334–1349. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pinheiro TJ, Elöve GA, Watts A, and Roder H (1997). Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry 36, 13122–13132. [DOI] [PubMed] [Google Scholar]
- Pollock WB, Rosell FI, Twitchett MB, Dumont ME, and Mauk AG (1998). Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry 37, 6124–6131. [DOI] [PubMed] [Google Scholar]
- Rumbley JN, Hoang L, and Englander SW (2002). Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants. Biochemistry 41, 13894–13901. [DOI] [PubMed] [Google Scholar]
- Rytomaa M, and Kinnunen PK (1994). Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J. Biol. Chem 269, 1770–1774. [PubMed] [Google Scholar]
- Schweitzer-Stenner R (2018). Relating the multi-functionality of cytochrome c to membrane binding and structural conversion. Biophys. Rev 10, 1151–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi F, Milazzo L, Howes BD, Piro MC, Fiorucci L, Polticelli F, Ascenzi P, Coletta M, Smulevich G, and Santucci R (2017). The key role played by charge in the interaction of cytochrome c with cardiolipin. J. Biol. Inorg. Chem 22, 19–29. [DOI] [PubMed] [Google Scholar]
- Sivanandam VN, Jayaraman M, Hoop CL, Kodali R, Wetzel R, and van der Wel PCA (2011). The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc 133, 4558–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Fogh RH, Boucher W, Higman VA, Eisenmenger F, Bardiaux B, Rossum B-J, Oschkinat H, and Laue ED (2011). A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR 51, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, and Hong M (2011). Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes. J. Phys. Chem. B 115, 10758–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, and Martin SJ (2008). Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Bio 9, 231–241. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, V.r.p., Schrödinger, LLC. [Google Scholar]
- Thurber KR, and Tycko R (2009). Measurement of sample temperatures under magic-angle spinning from the chemical shift and spin-lattice relaxation rate of 79Br in KBr powder. J. Magn. Reson 196, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurin VA, Tyurina YY, Jung MY, Tungekar MA, Wasserloos KJ, Bayir H, Greenberger JS, Kochanek PM, Shvedova AA, Pitt B, and Kagan VE (2009). Mass-spectrometric analysis of hydroperoxy- and hydroxy-derivatives of cardiolipin and phosphatidylserine in cells and tissues induced by pro-apoptotic and pro-inflammatory stimuli. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 877, 2863–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina YY, Kini V, Tyurin VA, Vlasova II, Jiang J, Kapralov AA, Belikova NA, Yalowich JC, Kurnikov IV, and Kagan VE (2006). Mechanisms of cardiolipin oxidation by cytochrome c: relevance to pro- and antiapoptotic functions of etoposide. Mol. Pharmacol 70, 706–717. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang J, Anthonymuthu TS, Kapralova VI, Vikulina AS, Jung MY, Epperly MW, et al. (2014). A mitochondrial pathway for biosynthesis of lipid mediators. Nat. Chem 6, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel PCA, Pott T, Morein S, Greathouse DV, Koeppe II RE, and Killian JA (2000). Tryptophan-anchored transmembrane peptides promote formation of nonlamellar phases in phosphatidylethanolamine model membranes in a mismatch-dependent manner. Biochemistry 39, 3124–3133. [DOI] [PubMed] [Google Scholar]
- Xu J, Soong R, Im S-C, Waskell L, and Ramamoorthy A (2010). INEPT-Based Separated-Local-Field NMR Spectroscopy: A Unique Approach To Elucidate Side-Chain Dynamics of Membrane-Associated Proteins. J. Am. Chem. Soc 132, 9944–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Lee M, Liao SY, and Hong M (2016). Solid-State Nuclear Magnetic Resonance Investigation of the Structural Topology and Lipid Interactions of a Viral Fusion Protein Chimera Containing the Fusion Peptide and Transmembrane Domain. Biochemistry 55, 6787–6800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary video. A video illustration of how 70-85 Ω loop shields the heme cavity of the natively folded protein, such that loop dynamics can facilitate heme exposure. Related to Figure 6. The video is generated using the UCSF Chimera program (Pettersen et al., 2004).