SUMMARY
Integrin receptors bind collagen via metal-mediated interactions that are modulated by magnesium (Mg2+) levels in the extracellular matrix. Nuclear magnetic resonance-based relaxation experiments, isothermal titration calorimetry, and adhesion assays reveal that Mg2+ functions as both a structural anchor and dynamic switch of the α1β1 integrin I domain (α1I). Specifically, Mg2+ binding activates micro- to millisecond timescale motions of residues distal to the binding site, particularly those surrounding the salt-bridge at helix-7 and near the metal ion-dependent adhesion site. Mutagenesis of these residues impacts α1I functional activity, thereby suggesting that Mg-bound α1I dynamics are important for collagen binding and consequent allosteric rearrangement of the low-affinity closed to high-affinity open conformation. We propose a multistep recognition mechanism for α1I-Mg-collagen interactions involving both conformational selection and induced fit processes. Our findings unravel the multi-faceted role of Mg2+ in integrin-collagen recognition and assist in elucidating the molecular mechanisms by which metals regulate protein-protein interactions.
INTRODUCTION
Cells exploit transport mechanisms to promote variations in local magnesium (Mg2+) concentrations and thereby regulate protein activities (Romani, 2011). Low levels of Mg2+ cations have been linked to an onset of disorders including auto-immune diseases and inflammation (Mazur et al., 2007). Mg2+ controls a myriad of cellular processes including the functional properties of integrins (Fuhrmann et al., 2014; Zhang and Chen, 2012), which exert a principal role in anchoring cells to the extracellular matrix (ECM). The resultant cell adhesion and signaling events are associated with critical physiological processes that include cell differentiation, immune responses, wound healing, and hemostasis. While essential for vital cellular functions, integrins are normally maintained in a resting, low adhesive, bent state. Alterations in physiological conditions that perturb this inactive state are often associated with a host of diseases (Gardner, 2014). The equilibrium between low and high affinity states can be modulated by specific stimuli including extracellular levels of metal ions and collagen (Zhang and Chen, 2012). Given the pivotal role of Mg2+ in mediating integrin-collagen interactions, the molecular mechanisms by which Mg2+ modulates integrin stability/flexibility to promote ligand binding are not well established.
Integrin α1β1 is a widely expressed surface cell receptor in the immune system (Gardner, 2014) that is critical for adhesion of immune cells to collagen. Structural (Chin et al., 2013b; Nolte et al., 1999; Rich et al., 1999) and mutagenesis (Hamaia et al., 2012; Lahti et al., 2011; Shi et al., 2012; Tulla et al., 2008) studies reveal that collagen binds α1β1 integrin I domain (α1I) via coordination to Mg2+ at the metal ion-dependent adhesion site (MIDAS). The resultant complex is characterized by a shift in metal position and conformational change in α1I from the closed-unbound to open-bound state. This conformational switch induces structural changes in the α1I C-terminal helix-7 which interacts with the β-subunit, triggering a chain of allosteric events throughout α1β1 that is activated to a high-affinity upright state (Liddington, 2014). While mutations of the metal-coordinating residues abrogate collagen binding (Kamata et al., 1999), subtle changes in metal coordination at the MIDAS motif have been linked to integrin activation (Lee et al., 1995; Weinreb et al., 2012) as observed for the E317A gain of function mutant. The unique Mg2+ pentacoordinate arrangement observed in the E317A crystal structure (Lahti et al., 2011) has been proposed as a driving force for enhanced α1I:collagen affinity. Apart from mediating direct α1I:collagen interactions, structural evidence suggests that Mg2+ effects might propagate beyond the metal binding site, thereby impacting α1I stability (Gotwals et al., 1999; Nymalm et al., 2004) and conceivably protein flexibility. The latter has been proposed as a key element in recognition mechanisms regulating functional activity (Boehr et al., 2009; Kay, 2016; Kern and Zuiderweg, 2003) and our laboratory has demonstrated that intrinsic local destabilization of α1I facilitates the closed/open conformational switch induced by collagen binding (Nunes et al., 2016).
Considering the wealth of studies on metals as allosteric regulators (Arias-Moreno et al., 2011), the objective of this investigation is to illuminate the role of Mg2+ on α1I dynamic properties in addition to its well established function as a mediator of integrin-collagen recognition. Employing a multidisciplinary approach including nuclear magnetic resonance (NMR), isothermal titration calorimetry (ITC), mutagenesis, and adhesion assays, our experimental observations suggest that Mg2+ plays a dual role as both a structural anchor and activator of dynamic α1I motions within the μs to ms regime. Specifically, our data reveal that binding of Mg2+ to α1I activates allosteric μs-ms timescale motions near the MIDAS and conserved salt-bridge at helix-7, regions that are critical to the closed/open conformational switch which occurs upon collagen binding (Chin et al., 2013b). These structural fluctuations yield a minor species that may be selectively poised to interact with collagen. Our findings lead us to propose a multistep α1I:collagen recognition mechanism that is consistent with a model in which affinity regulation is achieved via a combination of conformational selection and induced fit processes. This study adds another dimension towards understanding the fundamental and intricate role of Mg2+ binding in the physiological regulation of integrin:collagen recognition events underlying cell adhesion processes.
RESULTS
Magnesium induces α1I μs-ms timescale dynamics
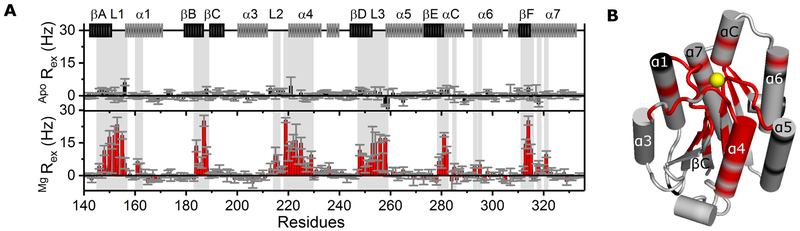
We investigated the dynamic behavior of α1I via a series of NMR experiments by monitoring a wide range of timescale motions [i.e., fast (ps-ns) to slower (hours)] in the absence of metal or at physiological (5 mM) Mg2+ concentrations (Romani, 2011). Employing in-phase Hahn echo experiments (R2HE), we evaluated α1I μs-ms dynamics and obtained relaxation exchange rates (Rex) for α1I residues in the apo- and Mg-bound forms (Figure 1A). Whereas the majority of residues in apo-α1I present Rex lower than 3 Hz and exhibit minimal or no exchange, Mg-bound α1I retains several residues with high Rex, all of which are in chemical exchange on the μs-ms timescale (Figure 1B). As confirmed by chemical shift measurements (Chin et al., 2013a), Mg2+ coordinates with residues at loops 1, 2, and 3 of the MIDAS (Figure S1A) without impacting α1I secondary structures (Figure S1B). Analysis of fast ps-ns motions via R1, R2, and NOE 15N relaxation experiments (Figure S2) reveals minimal differences between the apo and Mg-bound forms. Similarly, both species exhibit comparable dynamics for slow motions as investigated by hydrogen exchange via NMR (data not shown). These findings are consistent with mass spectrometry measurements on a rat-human chimeric α1I (Weinreb et al., 2012), which detect minimal changes in slow dynamics events between the apo and Mg-bound forms with exception of an expected decrease in flexibility at the top of α1I due to Mg2+ coordination.
Figure 1. Dynamic effect of magnesium binding on α1I.
(A) Relaxation exchange rates (Rex) obtained by R2 Hahn echo experiments for each residue of 15N-labeled α1I in the absence (top plot) and presence of 5 mM MgCl2 (bottom plot) at 20 °C. Error bars reflect propagated fitting errors. α1I secondary structure elements appear at the top with helices represented in light gray and sheets in dark gray. Shaded regions correspond to residues containing Rex values over 3 Hz. (B) The α1I crystal structure [PDB: 1PT6 (Nymalm et al., 2004)] with Mg2+ ion represented as a yellow sphere, residues in exchange colored red, and undetermined residues colored black. See also, Figure S3.
15N CPMG (Carr-Purcell-Meiboom-Gill) relaxation dispersion measurements facilitate characterization of protein motions on timescales spanning the μs-ms range (Lee, 2015; Palmer et al., 2001). In an effort to obtain kinetic, thermodynamic, and structural information on the Mg-induced exchange process occurring within the μs-ms timescale, we conducted 15N relaxation dispersion experiments in the presence of 5 mM MgCl2. The resultant dispersion profiles were analyzed quantitatively using the general Carver-Richards equation (Palmer et al., 2001) to fit each residue individually. A total of 22 residues exhibited relaxation dispersion profiles with Rex > 2 Hz and the latter match those detected via R2HE-based experiments with the exception of buried β-strand residues that are not visible in the dispersion perdeuterated samples. Residues fit individually yielded exchange rates (kex) varying from 70 to 15000 s−1 (Table S1). Assuming a single concerted chemical exchange process, one might expect nearly identical kex and populations (pA) for all residues (Farber and Mittermaier, 2015; McDonald et al., 2012). The non-uniform values observed for individual exchange parameters suggest the occurrence of multiple dynamic processes in Mg-bound α1I. Consequently, we group fit residues on the basis of comparable kex (Table 1) to correlate assignment with unique dynamic processes. Analysis of the resultant data suggest that these residues may be assigned to five specific groups, namely, three that are in fast conformational exchange (kex > 700 s−1), and two groups undergoing slower exchange (kex < 400 s−1). In subsequent sections, we demonstrate that the slow exchange process arises from Mg on/off events while the fast exchange groups reflect dynamic fluctuations intrinsic to α1I and/or induced by metal binding.
Table 1.
Group fits of 15N and 1H relaxation dispersion data for α1I residues in the presence of 5 or 100 mM MgCl2 at temperatures of 20 or 5 °C.
| kex (s−1) | χ2Red | Res. | Rex700 (Hz) | Rex600 (Hz) | R20700 (Hz) | R20600 (Hz) | α test | dωN (ppm) | [Mg] (mM) | T (°C) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 255 ± 23* | 4.89 | 15N | N153 | 12.1 ± 0.2 | 12.0 ± 0.2 | 12.0 ± 0.1 | 11.3 ± 0.2 | 0.1 ± 0.0 | 2.6 ± 0.0 | 5 | 20 |
| 15N | T222 | 6.1 ± 0.3 | 5.2 ± 0.3 | 8.7 ± 0.1 | 9.0 ± 0.0 | 1.0 ± 0.1 | 0.6 ± 0.0 | 5 | 20 | ||
| 15N | A223 | 8.0 ± 0.6 | 7.1 ± 0.6 | 9.6 ± 0.3 | 9.2 ± 0.2 | 0.8 ± 0.1 | 0.8 ± 0.1 | 5 | 20 | ||
| 15N | D253 | 10.8 ± 0.3 | 10.3 ± 0.4 | 13.2 ± 0.4 | 13.3 ± 0.2 | 0.3 ± 0.1 | 1.4 ± 0.1 | 5 | 20 | ||
| 15N | G254 | 12.4 ± 0.3 | 12.4 ± 0.3 | 14.7 ± 0.4 | 14.8 ± 0.3 | 0.0 ± 0.0 | 4.5 ± 0.3 | 5 | 20 | ||
| 15N | H257 | 8.9 ± 0.5 | 8.1 ± 0.6 | 10.1 ± 0.5 | 10.0 ± 0.2 | 0.6 ± 0.1 | 0.9 ± 0.1 | 5 | 20 | ||
| 302 ± 16 | 5.75 | 15N | I155 | 7.2 ± 0.2 | 5.8 ± 0.2 | 8.6 ± 0.1 | 8.5 ± 0.1 | 1.4 ± 0.1 | 5 | 20 | |
| 15N | G187 | 12.4 ± 0.4 | 10.7 ± 0.2 | 8.5 ± 0.1 | 8.3 ± 0.1 | 1.0 ± 0.1 | 5 | 20 | |||
| 15N | Q219 | 12.3 ± 0.3 | 10.5 ± 0.2 | 9.2 ± 0.1 | 8.9 ± 0.2 | 1.0 ± 0.1 | 5 | 20 | |||
| 15N | M221 | 5.2 ± 0.2 | 4.1 ± 0.2 | 7.5 ± 0.1 | 7.2 ± 0.1 | 1.5 ± 0.1 | 5 | 20 | |||
| 15N | G225 | 4.4 ± 0.2 | 3.4 ± 0.2 | 8.2 ± 0.1 | 8.3 ± 0.1 | 1.6 ± 0.1 | 5 | 20 | |||
| 15N | S256 | 7.6 ± 0.3 | 6.1 ± 0.3 | 10.7 ± 0.1 | 10.3 ± 0.1 | 1.4 ± 0.1 | 5 | 20 | |||
| 15N | D258 | 8.2 ± 0.3 | 6.7 ± 0.2 | 9.5 ± 0.1 | 9.3 ± 0.1 | 1.3 ± 0.1 | 5 | 20 | |||
| 15N | L282 | 5.8 ± 0.2 | 4.6 ± 0.2 | 7.5 ± 0.1 | 7.6 ± 0.1 | 1.5 ± 0.1 | 5 | 20 | |||
| 15N | Y285 | 11.5 ± 0.3 | 9.8 ± 0.2 | 11.9 ± 0.1 | 12.6 ± 0.1 | 1.0 ± 0.1 | 5 | 20 | |||
| 15N | N313 | 7.9 ± 0.2 | 6.4 ± 0.2 | 6.3 ± 0.1 | 6.3 ± 0.1 | 1.3 ± 0.1 | 5 | 20 | |||
| 15N | S315 | 4.2 ± 0.3 | 3.7 ± 0.3 | 8.3 ± 0.1 | 8.5 ± 0.1 | 1.6 ± 0.1 | 5 | 20 | |||
| 763 ± 115 | 2.71 | 15N | V314 | 15.0 ± 0.9 | 11. ± 0.6 | 15.7 ± 2.3 | 14.2 ± 1.9 | 2.0 ± 0.4 | 5 | 20 | |
| 15N | V321 | 3.4 ± 0.7 | 2.5 ± 0.5 | 13.9 ± 2.3 | 12.8 ± 1.9 | 2.0 ± 0.1 | 5 | 20 | |||
| 3520 ± 1480 | 2.47 | 15N | E317 | 2.2 ± 0.7 | 1.6 ± 0.5 | 10.5 ± 0.6 | 9.9 ± 0.5 | 1.9 ± 0.0 | 5 | 20 | |
| 15N | L318 | 18.5 ± 3.5 | 15.5 ± 2.3 | 13.9 ± 3.5 | 12.4 ± 2.4 | 1.0 ± 0.2 | 5 | 20 | |||
| 4168 ± 323 | 1.24 | 1H | H192 | 8.5 ± 0.3 | 6.4 ± 0.2 | 8.0 ± 0.2 | 9.1 ± 0.2 | 1.8 ± 0.1 | 5 | 20 | |
| 1H | E193 | 11.3 ± 0.2 | 8.6 ± 0.2 | 16.1 ± 0.2 | 17.9 ± 0.2 | 1.8 ± 0.1 | 5 | 20 | |||
| 1H | N195 | 13.3 ± 0.3 | 10.2 ± 0.2 | 9.7 ± 0.2 | 9.7 ± 0.2 | 1.6 ± 0.1 | 5 | 20 | |||
| 1H | L196 | 20.8 ± 0.4 | 16.4 ± 0.2 | 19.8 ± 0.3 | 20.3 ± 0.2 | 1.6 ± 0.1 | 5 | 20 | |||
| 2658 ± 762 | 4.70 | 15N | N153 | 4.8 ± 0.4 | 3.7 ± 0.3 | 7.0 ± 0.4 | 8.0 ± 0.3 | 1.6 ± 0.3 | 100 | 20 | |
| 15N | G254 | 6.2 ± 0.5 | 5.0 ± 0.3 | 8.6 ± 0.5 | 8.9 ± 0.4 | 1.4 ± 0.4 | 100 | 20 | |||
| 6580 ± 1731 | 1.46 | 15N | V314 | 6.1 ± 0.6 | 7.0 ± 1.3 | 3.2 ± 1.8 | 4.6 ± 1.3 | 1.8 ± 0.1 | 100 | 20 | |
| 15N | E317 | 2.7 ± 0.7 | 2.0 ± 0.5 | 7.1 ± 0.7 | 6.9 ± 0.5 | 2.0 ± 0.0 | 100 | 20 | |||
| 15N | L318 | 26.0 ± 3.9 | 20.4 ± 2.5 | 3.6 ± 4.0 | 7.2 ± 2.8 | 1.6 ± 0.3 | 100 | 20 | |||
| 15N | V321 | 2.1 ± 0.7 | 1.5 ± 0.5 | 6.1 ± 0.6 | 7.1 ± 0.5 | 2.0 ± 0.0 | 100 | 20 | |||
| 1607 ± 320 | 3.61 | 15N | W158 | 4.2 ± 0.3 | 3.3 ± 0.2 | 8.7 ± 0.2 | 9.0 ± 0.2 | 1.6 ± 0.2 | 100 | 5 | |
| 15N | V161 | 4.8 ± 0.4 | 3.8 ± 0.3 | 12.1 ± 0.2 | 12.0 ± 0.2 | 1.5 ± 0.3 | 100 | 5 | |||
| 15N | Q219 | 4.1 ± 0.3 | 3.3 ± 0.2 | 11.5 ± 0.2 | 11.8 ± 0.2 | 1.6 ± 0.2 | 100 | 5 | |||
| 15N | H257 | 2.4 ± 0.3 | 1.8 ± 0.2 | 12.7 ± 0.2 | 10.9 ± 0.2 | 1.8 ± 0.1 | 100 | 5 | |||
| 15N | N313 | 3.5 ± 0.3 | 2.7 ± 0.2 | 6.1 ± 0.2 | 6.5 ± 0.2 | 1.6 ± 0.2 | 100 | 5 | |||
| 15N | E317 | 6.7 ± 0.4 | 5.5 ± 0.2 | 12.3 ± 0.3 | 11.5 ± 0.2 | 1.3 ± 0.3 | 100 | 5 | |||
| 15N | V321 | 6.4 ± 0.4 | 5.2 ± 0.2 | 11.3 ± 0.2 | 10.0 ± 0.2 | 1.3 ± 0.3 | 100 | 5 | |||
PA = 95.2 ± 0.4 %
Mg2+ on/off exchange processes at physiological concentrations
In order to characterize specific dynamic processes that are triggered by Mg cations, we must distinguish these events from those intrinsic to apo-α1I and Mg on/off exchange processes. Our initial assessment on whether the dynamic processes correspond to Mg2+ association/dissociation employed isothermal titration calorimetry (ITC) to characterize the energetics of α1I:Mg2+ interactions (Table 2). Analysis of the ITC data via a single site binding model yields a 1:1 α1I:Mg2+ stoichiometry and dissociation constants (Kd) on the order of 0.4 mM. Low affinity binding to α1I suggests that several of the residues undergoing Rex may be involved in Mg on/off processes particularly under physiological concentrations. In an effort to resolve these exchange processes and characterize Mg2+ association/dissociation kinetics, we performed 15N ZZ-exchange experiments (Farrow et al., 1995) using a sample containing only 44.7 % Mg-bound species. Under conditions of partial saturation, four well resolved peaks are observed for the G187, G225 and G254 residues (Figure S3A), two of which are assigned to the apo and Mg-bound species, while the two extra resonances reflect slow exchange between α1I bound-unbound states during the ZZ-exchange experiment. The on- (kon) and off- (koff) rates obtained from fitting the peak intensities as a function of exchange time (Figure S3B–D)(Kloiber et al., 2011) are presented in Table S2.
Table 2.
Thermodynamic binding parameters derived from ITC profiles of the α1I-MgCl2 interaction.
| Integrin | n | Kd (μM) | Ka · 103 (M−1) | ΔG (kcal·mol−1) | ΔH (kcal·mol−1) | TΔS (kcal·mol−1) |
|---|---|---|---|---|---|---|
| α1I | 1.02 ± 0.02 | 378.6 ± 21.5 | 2.7 ± 0.2 | − 4.36 ± 0.26 | 1.92 ± 0.11 | 6.28 ± 0.37 |
| α1I / E317A | 1.00 ± 0.02 | 389.3 ± 20.4 | 2.6 ± 0.1 | − 4.34 ± 0.25 | 2.23 ± 0.13 | 6.57 ± 0.35 |
| α1I / L318A | 0.98 ± 0.04 | 393.7 ± 13.6 | 2.5 ± 0.1 | − 4.33 ± 0.25 | 1.96 ± 0.11 | 6.29 ± 0.36 |
Data correspond to average values and standard deviations determined for a minimum of three ITC experiments.
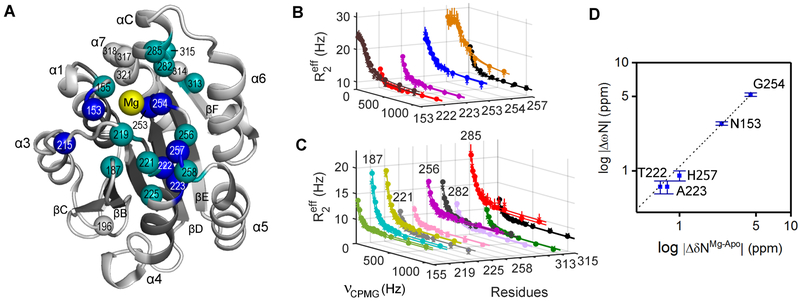
Comparison of the ZZ-exchange Mg on/off kinetics with those obtained from relaxation dispersion measurements reveals that both experiments yield similar values for residues undergoing slower exchange (kex < 400 s−1) at 5 mM Mg2+ (Table S3). At this concentration, we calculate an average kex of 247 ± 125 s−1 with 93.7 ± 3.0 % of Mg-bound species. Table 1 depicts two sets of residues with slow kinetics that participate in the Mg on/off processes. The first set includes metal binding residue D253 and its neighbors N153, T222, A223, G254, and H257 as illustrated in Figure 2A (blue residues). The second set encompasses residues I155, G187, Q219, S256 and D258 in the top loops, M221 and G225 in helix 4, L282 and Y285 in helix C, and N313 and S315 near strand-F as designated in Figure 2A (turquoise residues). These results suggest that the six residues (Figure 2B) with a kex of 255 ± 23 s−1 and eleven residues (Figure 2C) with a kex of 301 ± 16 s−1 participate in exchange processes between the apo and Mg-bound species. Our findings are further corroborated by the six residues fitting a “slow-limit” regime (α <1, Table 1) with a reasonable correlation (R2 = 0.97) observed for chemical shift differences between the major and minor species in solution (|ΔωN|) based on relaxation dispersion and chemical shift assignments deduced from analysis of the apo and Mg-bound forms (Figure 2D). Collectively, our data confirm the presence of Mg on/off events within α1I at physiological cation (1 - 20 mM) concentrations (Romani, 2011), which must be resolved from intrinsic and/or Mg-specific dynamic processes that might be involved in allosteric events.
Figure 2. Residues participating in conformational exchange between apo and Mg-bound α1I.
(A) Residues in conformational exchange at 5 mM Mg2+ are mapped into the crystal structure of Mg-bound α1I [PDB: 1PT6 (Nymalm et al., 2004)] with those undergoing “slow-limit” exchange colored blue and residues in “fast-limit” exchange colored turquoise. Fitted parameters are indicated in Table S2. (B and C) 15N CPMG relaxation dispersion curves for the “slow-limit” residues with a kex = 255 ± 23 s−1 (B) and the “fast-limit” residues a kex = 301 ± 16 s−1 (C). R215 was excluded from the group fit due to high uncertainty. Data were collected at 700 MHz (circles) and 600 MHz (crosses) with two νcpmg repeated for error estimation (vertical bars). Fitted parameters are indicated in Table 1. (D) Logarithmic |ΔωN| values of “slow-limit” residues obtained from group fits are plotted against the chemical shift difference between apo and Mg-bound species observed in the [1H-15N] NMR spectra |ΔδNMg-apo|. A linear fit of the data is characterized by R2 = 0.97 with error bars of ΔωN determined by Monte Carlo simulations.
Relaxation dispersion experiments detect a minor population sampled by Mg-bound α1I species
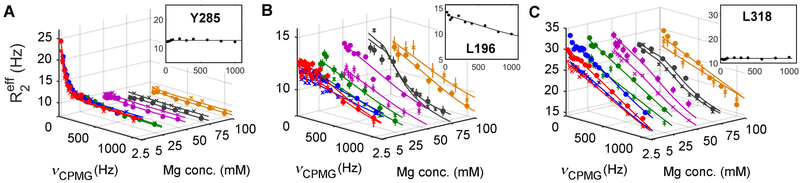
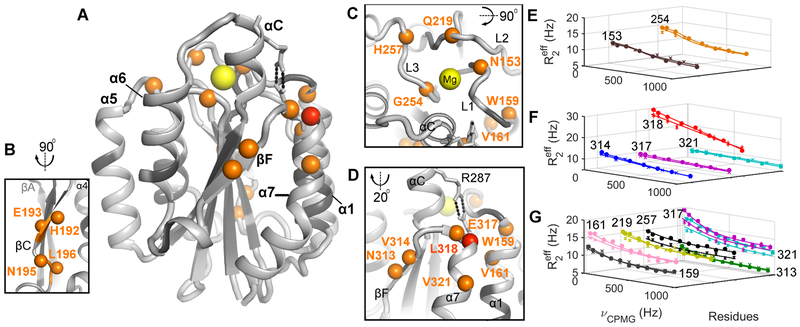
In an attempt to exclude the Mg on/off processes and thereby gain further insight regarding those residues exhibiting faster conformational exchange (i.e., kex > 700 s−1), we performed 15N relaxation dispersion experiments at varying Mg2+ concentrations. Based on dissociation constants determined via ITC analysis (Table 2), our relaxation dispersion experiments yielded α1I saturation of 0, 83.6, 91.6, 99.2, 99.5 and 99.6 % for 0, 2.5, 5, 50, 75 and 100 mM Mg2+, respectively. Assuming a two-site model, fits with reasonable precision at 0, 2.5, 5, and over 50 mM Mg2+ yielded a total of 3, 25, 22, and 7 residues with Rex > 2 Hz at 20 °C. Evaluation of dispersion curves (Figures 3 and S4) and Rex values (Figure S5) monitored as a function of Mg2+ concentration identifies three distinct relaxation dispersion profiles, namely: 1) Rex decreases with increasing Mg2+ concentration (Figure 3A) that includes all of the metal on/off residues identified previously (refer to Figure 2 and Table 1); 2) Rex is independent of Mg2+ concentration and present in the apo form (e.g., L196 in Figure 3B); and, 3) Rex is independent of Mg2+ concentration and absent in the apo form (e.g., L318 in Figure 3C). These complex Rex profiles suggest the existence of distinct exchange processes at different Mg2+ concentrations with on/off events prevailing at low concentrations and the presence of two additional exchange processes independent of Mg on/off, one of which is intrinsic to apo-α1I while the other is induced via metal coordination. General dynamic hot spots are identified in Figure 4, some of which appear in Figure 4A including the intrinsic dynamic residues (Figure 4B) in addition to the Mg-dependent slow (Figure 4C) and fast (Figure 4D) exchange processes characterized herein.
Figure 3. Relaxation dispersion profiles at different Mg2+ concentrations.
15N relaxation dispersion profiles acquired at 20 °C containing 0, 2.5, 5, 50, 75, and 100 mM Mg2+ identified three distinct profiles: (A) dispersion profiles decrease with increased Mg2+ concentration (e.g., Y285); (B) dispersion profiles independent of Mg2+ concentration (e.g., L196): and, (C) dispersion profiles independent yet requiring the presence of Mg2+ (e.g., L318). Dispersion profiles obtained in the absence of Mg2+ are depicted as insets with fitted parameters presented in Figure S6. Errors are reflected by the vertical bars and estimated by acquiring data at duplicate νcpmg values. Experiments were performed at 700 MHz (circles) and 600 MHz (crosses). See also, Figures S4 and S5.
Figure 4. Dynamic hotspots of the α1I Mg-bound species.
(A) Residues with Rex exceeding 2 Hz in the presence of 100 mM MgCl2 at 5 and/or 20 °C are mapped in orange as spheres (except the highly dynamic L318 which is depicted in red) into the α1I closed structure [PDB: 1PT6 (Nymalm et al., 2004)]. Residues undergoing μs-ms motions are located at: (B) bottom of strand-C; (C) top of α1I near the MIDAS loops (L1, L2 and L3); and, (D) surrounding the salt-bridge in strand-F, helix-7, and helix-1. (E-G) 15N CPMG relaxation dispersion profiles of the Mg-induced dynamic residues are acquired at 20 °C (E and F) and 5 °C (G). The fitted parameters are presented in Table 1 for data collected at 700 MHz (circles) and 600 MHz (crosses) with two νcpmg repeated for error estimation (vertical bars). See also, Figures S4, S5, and S6.
Amide 1H represents a more sensitive probe to characterize α1I exchange processes for residues with dispersion profiles and Rex independent of Mg2+. In addition to L196 identified by the 15N probe, 1H relaxation dispersion data reveal that the surrounding residues at strand-C (Figures 4B and S6A) are under conformational exchange with Rex > 5 Hz. The best-fit values for the global parameters at 5, 25, and 50 mM Mg2+ (Tables 1 and S4) suggest conditions of fast exchange with a kex of 4000 s-1. These data support the existence of concerted fast exchange processes intrinsic to α1I that are present in the apo (Figure S6) and Mg-bound forms involving residues located at the bottom of α1I on an opposite surface of the allosteric helix-7. To gain insight on exchange processes purely induced by Mg2+ coordination to α1I and unique to the Mg-bound species, we analyzed the 15N dispersion data obtained at 100 mM Mg2+ containing over 99.5 % of species in the Mg-bound form. Two sets of residues undergoing chemical exchange are identified (Table 1), namely: 1) N153 and G254 at the MIDAS (Figures 4C and 4E); and, 2) V314, E317, L318 and V321 at the C-terminus (Figures 4D and 4F). The former exhibits a kex of ~ 2660 s−1 that is consistent with values estimated by ZZ-exchange in 100 mM Mg2+ (Table S3), which suggests participation in the Mg on/off process. Conversely, the C-terminus yields a faster kex of 6580 s−1 that is comparable to the kinetics observed at 5 mM Mg2+ for L318 (Table S1), which retains the highest Rex of 27 Hz and thereby supports the hypothesis that such an exchange process is independent of Mg on/off rates. In an attempt to reduce the exchange rates and obtain structural information on the conformational intermediate, we acquired 15N relaxation dispersion data at lower temperatures (5 °C) in the presence of excess metal ions (100 mM Mg2+) as summarized in Table 1. Despite retaining fast exchange characteristics at low temperature, we observe improved dispersion profiles for residues H257, N313, E317 and V321 (compare Figures 4F and 4G) and identified three additional residues in fast conformational exchange that are located within helix-1 (W158 and V161) and the Q219 at MIDAS loop 2 (Figures 4C and 4D). The best group fit contains all seven residues in fast exchange at 1607 ± 320 s−1 (Figure 4G and Table 1), excluding L318 due to high data uncertainties at 5 °C. Collectively, our results suggest that the conformational fluctuations induced by Mg2+ coordination to α1I are concerted and centered around the MIDAS loops and conserved salt-bridge at helix-7 including strand-F and the top residues in helix-1 as illustrated in Figures 4A, 4C and 4D.
Minor Mg-bound α1I species adopts a unique conformation that differs from the α1I closed and open states
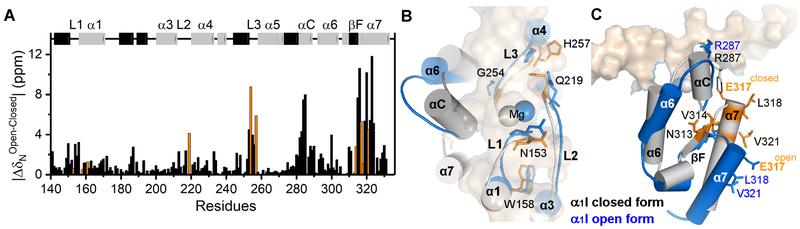
Relaxation dispersion experiments facilitate identification of two regions in α1I near the MIDAS loops and conserved salt-bridge, both of which undergo dynamic fluctuations on the μs-ms timescale. These dynamics can be related to the existence of a minor species that is in conformational exchange with the major closed species. One question that remains is whether this minor population resembles a high affinity open state observed for the α1I:collagen complex. Since the minor species is sparsely populated and these residues are within the fast exchange limit, it is not possible to obtain chemical shifts from relaxation dispersion experiments to explore this question directly (Palmer et al., 2001). In an effort to understand whether the minor species adopts a high-affinity open conformation (Chin et al., 2013b; Liddington, 2014; Siljander et al., 2004), we compared the exchanging residues of Mg-bound α1I with regions of conformational rearrangement from a closed to open state in an α1I-collagen peptide complex (Chin et al., 2013b). The chemical shift perturbation of backbone atoms within α1I caused by the binding of a collagen model peptide to Mg-bound α1I has been characterized previously (Nunes et al., 2016) and is illustrated in Figure 5. The MIDAS loops and residues in helix-C and helix-7 exhibit the largest chemical shift perturbations (ΔδN > 2 ppm, Figure 5A) as a consequence of three major α1I closed to open rearragements (Chin et al., 2013b), namely: 1) movement of the metal from loop 3 towards loop 2 at the MIDAS (Figure 5B); 2) unfolding of helix-C; and, 3) a 12 Å downward displacement of helix-7 (Figure 5C). Specifically, the Q219, G254 and H257 residues in loops 2 and 3, V314 in strand-F, and E317, L318 and V321 at the top of helix-7 experience large perturbations caused by the closed-open conformational switch (orange bars in Figure 5A). In contrast with the collagen-bound complex, helix-C in unliganded α1I at high Mg2+ concentrations does not exhibit μs-ms motions (Rex < 2 Hz), suggesting that the minor species in exchange with the major closed Mg2+ species does not adopt an α1I open activated state.
Figure 5. Structural differences between the α1I closed and open conformations.
(A) Bar plot illustrating the chemical shift differences of backbone 15N atoms between closed and open-bound forms. The α1I secondary structure elements of the closed form are presented on top with helices colored gray and sheets in black, labeling the helices, MIDAS loops (L1, L2, and L3), and C-terminus. (B, C) Overlay of the α1I closed [PDB: 1PT6, gray (Nymalm et al., 2004)] and open [1st conformer of PDB: 2M32, blue (Chin et al., 2013b)] conformations delineating structural differences in the (B) MIDAS and (C) C-terminus regions. Residues undergoing conformational exchange at 100 mM MgCl2 are highlighted in orange in the bar plot (A), and represented by orange and blue sticks in the closed and open forms, respectively (B, C). Mg2+ ions are depicted as spheres and the salt-bridge E317 residue is labeled in orange. The bound collagen peptide model is shown by a surface representation.
Probing residues in the α1I minor species population via mutagenesis and adhesion assays
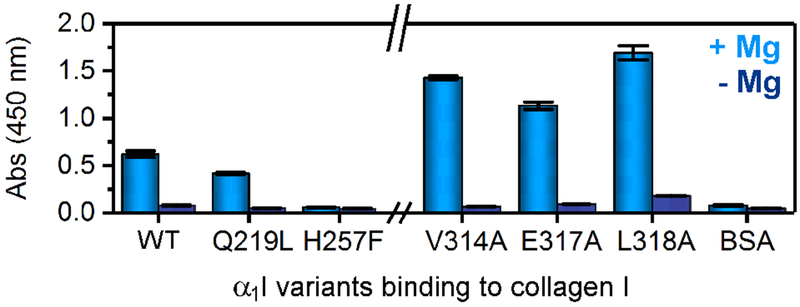
We performed single point mutations of several residues that undergo conformational exchange to explore the impact of dynamic fluctuations and employed adhesion assays to evaluate the outcomes in terms of collagen binding affinity (Figure 6). Whenever possible, we replaced the assigned dynamic residues with those of homologous αI domain integrins, thereby minimizing net changes on the α1I structure. Low adhesion in the presence of EDTA is indicative of specific metal-mediated binding for α1I variants complexed with collagen. Focusing our analysis on the MIDAS loops, Q219 is a unique residue in the α1I sequence with a Q219L mutant mimicking the homologous collagen binding α2I integrin sequence. Similarly, residue H257 is conserved amongst collagen-binding integrins and the H257F mutant mimics αMI, a leukocyte integrin with weak collagen binding properties (Hamaia and Farndale, 2014). Significantly, Q219L and H257F lead to a decrease in collagen binding with the H257F mutant completely abrogating adhesion. Our data suggest that the Q219 and H257 residues located in loops 2 and 3 of the MIDAS are important for formation of the α1I-Mg-collagen complex. In contrast, mutations of V314, E317, and L318 near the salt-bridge/helix-7 enhance collagen adhesion (Figure 6). V314 is conserved amongst collagen-binding integrins and its mutation to A illuminates the role of this hydrophobic residue on the activation process. A L318A mutation mimics the α2I sequence and exhibits comparable levels of collagen adhesion as that of E317A, a well established gain-of-function mutant characterized by absence of the conserved salt bridge and a unique transitional conformation between closed and open forms (Lahti et al., 2011; Tulla et al., 2008). ITC analysis reveals that the E317A and L318A mutations do not significantly impact Mg:α1I thermodynamic binding parameters (Table 2), effectively precluding the possibility that metal binding affinity is a determining factor in collagen adhesion.
Figure 6. Impact of mutations at α1I residues undergoing conformational exchange on collagen adhesion.
The binding of wild type α1I and selected mutants (i.e., Q219L, H257F, V314A, E317A and L318A) to collagen evaluated by ELISA assays in the presence of 5 mM MgCl2 (light color) and 5 mM EDTA (dark color) with BSA as a control of surface coating. Error bars reflect the standard deviation of triplicate measurements.
L318A gain-of-function mutation mimics structural changes of the minor species
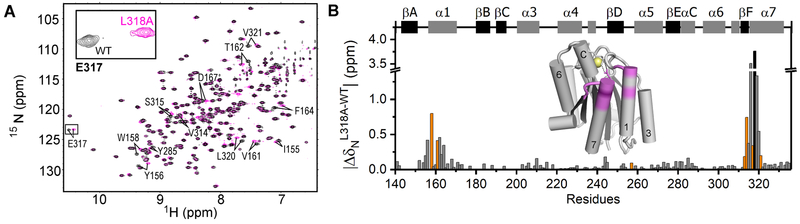
Considering the highly dynamic nature of residue L318, we have employed NMR spectroscopy to explore the impact of an L318A mutation on the α1I peptide backbone and thereby shed insight in terms of its role on structure and conformational fluctuations resulting in gain of functionality. Unlike the E317A mutant that has been crystallized in a transitional conformation with an unfolded helix-C and helix-7 positioned upward (Lahti et al., 2011; Nunes et al., 2016), similarities in the [1H-15N]-HSQC spectrum of L318A and wild type (WT) α1I (Figure 7A) suggests that the peptide backbone does not undergo major structural changes. Consequently, the L318A mutant maintains α1I in a closed conformation as characterized by the folded helix-C, helix-7 positioned upward, and a stabilizing R287-E317 salt-bridge corroborated by the E317 downfield resonance (Figure 7A inset). The L318A replacement induces large chemical shift perturbations at the mutation site on top of helix-7 (Figure 7B) and additional chemical shift changes (ΔδN > 0.1 ppm) at the top of helix-1. Comparison of these chemical shift changes with the exchanging residues in Mg-WT α1I (orange bars in Figure 7B) suggest that the structural changes elicited by an L318A activating mutation mimics those of the Mg-induced dynamics in WT α1I. These structural changes lead to an intermediate conformation that still resembles the closed state.
Figure 7. Impact of the L318A gain-of-function mutation on α1I backbone structure.
(A) Overlaid 1H-15N-HSQC spectra of wild type (WT) α1I (black) and L318A (magenta) at 800 MHz acquired at 20 °C. Residues with pronounced chemical shift differences from the WT to L318A are labeled. The inset illustrates an expanded region of E317 resonance, supporting the presence of a stabilizing salt-bridge in both α1I variants. (B) Chemical shift perturbation of the wild type α1I backbone 15N atoms (ΔδN) caused by mutation of the L318 residue (gray bars), with the mutation site represented as a black bar. Orange bars highlight residues with μs-ms dynamics in the WT α1I-Mg bound counterpart. Secondary elements of the closed α1I structure are represented at the top with β-strands and α-helices in darker and lighter color bars, respectively. At the center is a representation of the closed α1I structure [PDB: 1PT6 (Nymalm et al., 2004)], in which residues affected by the L318A mutation with ΔδN over 0.1 ppm are highlighted in pink, while the L318A mutation site is colored purple.
DISCUSSION
Magnesium triggers exchange processes in α1I yielding a minor species
The ability of metals to assume diverse coordination geometries is exploited by proteins to facilitate folding and binding processes as well as to modulate conformational structure and dynamics [as reviewed in (Jensen et al., 2007)]. Widely regarded as special allosteric effectors, metals modulate protein functional properties by altering the conformational equilibria of their targets (Arias-Moreno et al., 2011). In an effort to gain insight into the molecular mechanisms by which Mg2+ modulates integrin structural flexibility (Weinreb et al., 2012) and biological function as an adhesion molecule (Fuhrmann et al., 2014; Zhang and Chen, 2012), we have conducted a comprehensive NMR investigation on the dynamic impact of Mg2+ binding to α1I. Unlike most proteins where metal binding actually restricts motions (Capdevila et al., 2017; Jensen et al., 2007; McDonald et al., 2012), our data demonstrate that Mg2+ interactions trigger complex dynamic processes in α1I on the μs-ms scale that are likely to be associated with modulation of integrin plasticity and its ability to recognize collagen. The utility of relaxation experiments resides in their ability to characterize dynamically-controlled events on the μs-ms timescale, thereby elucidating allosteric conformational changes and detecting sparsely populated intermediate states (Boehr et al., 2009; Kay, 2016; Kern and Zuiderweg, 2003; Lee, 2015). Relaxation dispersion measurements in conjunction with chemical shift data and ZZ-exchange spectroscopy identify two distinct exchange processes in α1I induced by Mg2+ coordination at physiological concentrations as illustrated in Figure 4. These include slow exchange processes assigned to Mg2+ association/dissociation (Figure 4C) and fast exchange processes that persist at high Mg2+ concentrations (Figure 4D).
The fast exchanging residues are located at critical regions surrounding the conserved salt-bridge and share comparable exchange rates (kex ~ 6600 s−1) that are significantly faster than Mg on/off processes (kex ~ 2700 s−1) at 20 °C under Mg2+ saturating conditions (refer to Table 1). The existence of these on/off events caused by weak cation binding affinity to α1I (Kd = 0.4 mM) might function in vivo as a regulatory mechanism of α1I activity by maintaining integrins in a low adhesive state at physiological Mg2+ levels, thereby avoiding unwanted cell-matrix interactions. Conversely, residues undergoing fast exchange may be associated with an interconversion between the α1I-Mg ground state and an excited sparsely populated intermediate state. Inspection of the overall relaxation dispersion data reveals a minimum of nine residues (Figure 4D, 4F, and 4G) moving concertedly at 150 μs (kex ~ 6600 s−1) within Mg-bound α1I and six intrinsically dynamic residues moving at 250 μs (kex ~ 4000 s−1) in both the apo and holo-forms. These findings provide experimental evidence for a set of coordinated motions indicative of a sparsely populated species, herein represented as α1I-Mg*, that is in equilibrium exchange with the Mg-bound α1I ground state. While MIDAS serves as the primary collagen binding site via direct Mg2+ coordination, the salt-bridge connecting helix-C (R287) and helix-7 (E317) is a stabilizing feature of the Mg-bound α1I conformation that is disrupted in the α1I:collagen complex (Chin et al., 2013b). Moreover, structural changes in C-terminal helix-7 are critical for allosteric activation of integrins triggering structural rearrangements of the full molecule upon collagen binding (Liddington, 2014).
Allostery in α1I is facilitated by the presence of an active intermediate
Elucidation of potential Mg2+ binding effector roles on α1I allosteric activation represents a challenging task since both the metal and collagen ligands bind to similar regions within the protein. Our NMR experiments detect μs-ms dynamics induced by α1I:Mg2+ association in distal regions (over 10 Å) from the α1I metal binding site (Figure S1A) including the allosteric C-terminus. A wealth of studies suggests that the I-domain conformation plays a key role in allosterically modulating integrin-ligand affinity. Structure-based mutagenesis designed to shift the I-domain structure towards either the closed or open conformations reveal a direct link between the “opening process” and ligand affinity (Siljander et al., 2004). The conformational switch (Chin et al., 2013b; Emsley et al., 2000) has been proposed as a major triggering event in integrin allosteric activation, thereby propagating structural rearrangements to the full integrin. We observe significant relaxation dispersion for residues located in key regions of the allosteric conformational switch. Our data support the notion that Mg2+ functions as a dynamic switch by activating μs-ms timescale motions in α1I regions where collagen binding and allosteric rearrangements occur without imparting significant changes in secondary structure.
Mutagenesis is an informative tool for probing the role of dynamic protein regions on ligand binding and allosteric events (Csermely et al., 2010). We employed site directed mutagenesis to probe the relevance of each residue participating in coordinated motions. While mutations of flexible residues in the MIDAS lead to a decrease in collagen adhesion, mutations of residues surrounding the salt-bridge promote a gain-of-functionality. Enhanced collagen binding affinity exhibited by mutations of the V314, E317, and L318 residues (Figure 6) suggest that these side-chains are engaged in stabilizing contacts and thereby maintain a closed α1I conformation in the absence of collagen. Consequently, substitution of these residues unleashes the activation process that leads to ligand binding, a conformational switch, and collagen adhesion. It is worth noting that among the dynamic residues studied herein, L318 exhibits the highest motions and a L318A mutation perturbs the hydrophobic pocket underlying βF-helix-7 loop and helix-1 (Figure 7B) while remaining in a closed conformation that is comparable to the WT α1I counterpart. The regions impacted by an L318A mutation correspond to residues that exhibit transient μs-ms motions in the wild type, a finding which suggests that the minor α1I-Mg* species might correspond to an activated form of α1I differing from the ground state in the MIDAS loops and hydrophobic ratchet pocket underlying the C-terminal salt-bridge. Changes in the latter have been linked to allosteric regulation of integrin ligand affinity (Wang et al., 2017; Xiao et al., 2004). We therefore propose that α1I undergoes exchange to a dynamic intermediate that is poised for an initial collagen encounter prior to the allosteric conformational switch.
Structural insights on the sparsely populated minor species
Mutagenesis studies in conjunction with structural data reported for various members of the integrin family allow us to envision structural features of the minor species. Comparison of the residues undergoing conformational fluctuations with those impacted by the closed/open conformational switch within a α1I:Mg:collagen peptide model suggests that the α1I-Mg* species does not adopt an open form (Chin et al., 2013b; Nunes et al., 2016). Moreover, backbone relaxation dispersion experiments reveal that the minor species differs from an α1I closed state at loops 2 and 3 of MIDAS and within the salt-bridge region. Our finding that the minor species maintains a “closed” conformation sufficiently distinct from the ground state is consistent with prior studies on the I-domain in other members of the integrin superfamily (Wang et al., 2017; Xiao et al., 2004). In such cases, metal positioning and changes in the hydrophobic pockets underlying beta-F, helix-7, and helix-1 are primarily responsible for differences observed between the closed and intermediate conformations. While closed conformations presumably represent binding-incompetent states, there are notable exceptions such as the integrin-ligand interactions in an α1I:Mn:antibody complex that assumes a closed-like conformation (Karpusas et al., 2003). Significantly, the crystal structure of an unliganded E317A mutant reveals that α1I does not require positioning of helix-7 downwards to enhance its affinity for collagen (Lahti et al., 2011), thereby supporting our contention that α1I-Mg* adopts a binding-competent conformation irrespective of helix-7 orientation.
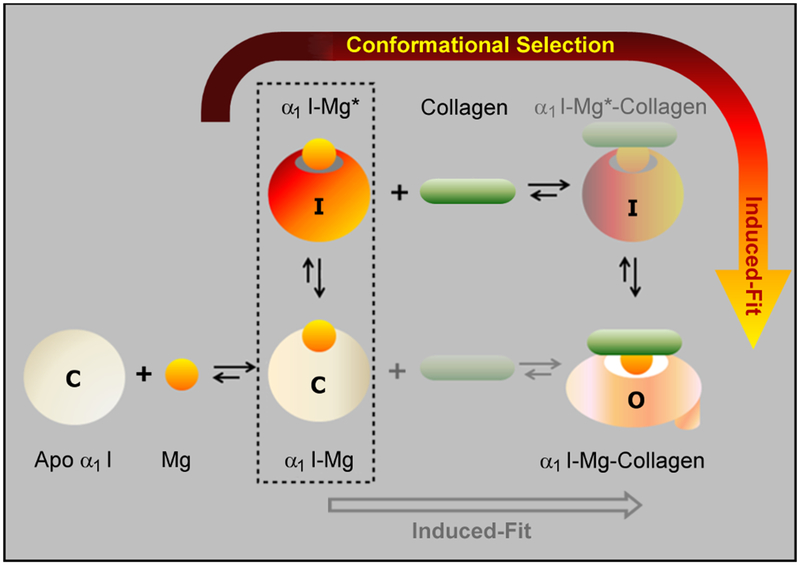
We therefore propose that the α1I-Mg* species adopts a “closed” intermediate conformation retaining the following structural features: a) folded helix-C and helix-7 positioned upward resembling the closed wild type form; b) perturbed hydrophobic pockets underlying βF-helix-7 loop and helix-1 mirroring the L318A mutant; and, c) residues within the MIDAS loops strategically positioned in an optimum collagen-binding arrangement. Our results provide compelling evidence that Mg2+ assumes an effector role in α1I-collagen recognition mechanisms by stabilizing a closed activated intermediate conformer that is prone to interact with collagen as an initial encounter in the association process leading to a final open collagen-bound form. The existence of a minor species in the unliganded ensemble of conformations resembling a binding-prone state suggests that this intermediate participates in early collagen encounter events via conformational selection. In view of these findings, we hypothesize that α1I-collagen recognition involves a multistep process in which the initial encounter with a sparsely populated minor species is followed by a binding-induced structural switch to an open conformation.
Multistep α1I:collagen recognition mechanism
Classical views on allosteric regulation encompass a wealth of studies in which conformational selection (CS) and/or induced-fit (IF) mechanisms are invoked to characterize macromolecular interactions (Boehr et al., 2009; Kay, 2016; Kern and Zuiderweg, 2003). The Mg-induced increases in α1I μs-ms timescale dynamics that we observe at the MIDAS and within proximity of the stabilizing salt bridge allow us to propose a multistep α1I:collagen recognition mechanism. In the initial phase of this multistep process represented schematically in Figure 8, α1I samples an equilibrium of dynamic species triggered by Mg2+ binding in the absence of collagen (α1I-Mg*). As a consequence of local structural adjustments at the collagen binding interface and salt bridge region, the α1I-Mg* minor species may be primed to interact with collagen via conformational selection. The inherent plasticity and less than ideal shape complementarity of α1I-Mg* with the collagen triple helical peptide leads to a subsequent conformational switch characteristic of an induced fit mechanism. At this stage, collagen binding facilitated by local destabilization of helices C and 7 (Nunes et al., 2016) induces a structural rearrangement of α1I to an open-bound state. Our proposed α1I:collagen recognition mechanism resembles extended/sequential multistep CS/IF binding models (Csermely et al., 2010; Vogt et al., 2014) that are currently available to describe macromolecular interactions. The conformational fluctuations yielding an activated α1I-Mg* state conceivably reduce the energy barrier of a closed/open allosteric switch induced by collagen binding.
Figure 8. Proposed multistep recognition mechanism of integrin α1I towards collagen.
A red/orange arrow delineates the two-step binding process: 1) a conformational selection step with collagen binding α1I-Mg*, a Mg-induced minor species in fast exchange at 6000 s−1 with the major α1I-Mg closed species (C), which adopts a high affinity state that differs from the open conformation (O); and, 2) an induced-fit step that causes major structural rearrangements and a conformational switch from α1I-Mg* species in the intermediate conformation (I) to open form α1I-Mg-collagen species (O). An alternate scenario involving a single step induced-fit process that is energetically less favorable appears in grey and is shaded for clarity. It is relevant to note that the weak interaction of Mg2+ with apo α1I (Kd ~ 0.4 mM) is characterized by on/off processes of 300 s−1 at physiological Mg2+ concentrations, which represents a limiting factor in the population of α1I-Mg* species.
Concluding Remarks
The findings presented in this study yield significant insights on the mechanisms by which Mg2+ ions control and regulate α1β1 integrin cellular adhesion. Our data suggest that Mg2+ plays a dual role in integrin-collagen interactions. In addition to functioning as a structural anchor, Mg2+ modulates integrin-collagen interactions beyond the metal binding site by activating μs-ms exchange processes, thereby yielding a minor Mg-bound species that is prone to bind collagen. Our results are consistent with a model in which the tight regulation of α1I affinity to collagen is achieved via a complex multistep recognition mechanism initiated by Mg2+ coordination to α1I. This study adds another dimension towards understanding the fundamental role of metal binding in the physiological regulation of integrin-collagen recognition events underlying cell adhesion processes.
STAR★Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jean Baum (jean.baum@rutgers.edu).
Experimental Model and Subject Detail
Microbe strains
Variants of His10-α1I proteins were overexpressed in Escherichia coli BL21(DE3) cells with Luria-Bertani (LB) broth or M9 minimal media supplemented with 15NH4Cl and D-glucose-(13C6 or 2H7-12C6 or 2H7-13C6) and containing 100 μg/mL Ampicillin in water or in 99.8% deuterium oxide (Cambridge Isotopes Laboratories).
Method Details
Protein Expression and Purification
The recombinant α1I from human integrin α1β1 used for these studies corresponds to residues 141 - 335 of the α1 subunit (NP_852478.1). Recombinant proteins were expressed in Escherichia coli BL21(DE3) cells by induction with 1 mM IPTG overnight at 20 °C (Nunes et al., 2016). Cells were harvested and lysed using a 20 % sucrose buffer solution. The His-tagged proteins were purified by Ni2+-NTA agarose (QIAGEN) affinity chromatography and buffer exchanged using PD-10 desalting columns (GE Healthcare Life) or dialysis followed by protein concentration to 0.3 - 1 mM using Amicon Ultra 3 kDa centrifugal filters (Millipore EMD). Protein concentration was determined by monitoring the absorbance at 280 nm employing an extinction coefficient of 12950 M−1 cm−1. All point mutations and truncation (containing 141 - 331 residues, NP_852478.1) were created with a standard PCR-based mutagenesis method and confirmed by DNA sequencing.
Nuclear Magnetic Resonance
Chemical Shift Perturbation
Resonance assignments were achieved acquiring TROSY versions of 3D HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH or HN(COCA)CB experiments on a Bruker 700 MHz at 25 °C on [U-13C, 15N]-labeled α1I and [U-2H, 13C, 15N]-labeled samples containing approximately 0.6 mM α1I in 50 mM phosphate buffer (NaPi, pH 6.7) with 140 mM NaCl, 20 mM BME, and 1 mM EDTA or 5 mM MgCl2. The pH of 6.7 was selected as a compromise between the minimization of solvent exchange rate with 1HN atoms and the physiological pH. Although 2H, 13C, 15N-labeled samples provided superior peak resolution, 13C, 15N labeling was crucial for identification of residues located in the core of α1I, which are inaccessible to solvent exchange. We assigned 92 % of the triple resonances for the apo and Mg-bound α1I forms which are consistent with previous studies (Chin et al., 2013a). The chemical shift perturbation (ΔδHN,N) (Jensen et al., 2007) in the TROSY spectrum caused by Mg2+ binding was calculated as follows: .
Chemical shift difference of the N backbone atoms (ΔδN) caused by the L318A mutation was obtained by acquiring [1H-15N]-TROSY-HSQC spectra on a Bruker 700 MHz of the L318A mutant and wild type α1I using [U-15N]-labeled samples containing 5 mM PIPES, 140 mM NaCl, and 50 mM MgCl2, at 20 °C. Residues with ΔδN over 0.1 ppm are mapped into the α1I structure (PDB: 1PT6) using PyMOL (Schrodinger, 2015).
Chemical shift difference of α1I backbone N amide atoms (ΔδN) between the closed and open-collagen bound conformations was obtained acquiring 15N-1H-TROSY spectra on a Bruker 700 MHz at 30 °C on a 15N-labeled α1I sample in the absence and presence of a collagen model peptide, respectively. The open conformation of α1I was prepared using 1 mM of [U-15N]-labeled α1I, comprising the 141 - 331 residues, in 5 mM PIPES buffer containing 25 mM MgCl2, 140 mM NaCl and 2 mM triple helical collagen model peptide, Ac-(GPO)4GLOGEN(GPO)4GY-NH2, containing the high affinity GLOGEN motif (Hamaia et al., 2012).
15N-R2 and R2 Hahn Echo Experiments
15N R2 relaxation rates that preserve transverse relaxation associated with chemical exchange was measured employing the Hahn echo pulse sequence (Millet et al., 2000) on a [U-15N] α1I sample in 90 % H2O / 10 % D2O, 50 mM NaPi buffer containing 140 mM NaCl, 20 mM BME, and either 5 mM MgCl2 or 1 mM EDTA (pH 6.7). Relaxation experiments were acquired at 20 °C on a Varian 800 MHz and/or Bruker 700 MHz spectrometer. The temperature was calibrated using a sample of 100 % methanol. For the α1I-Mg2+ bound sample, Rex was determined with two different concentrations of α1I (i.e., 0.6 and 0.4 mM) in order to exclude any dimerization effect on the observed Rex values. Relaxation rate constants were determined from a series of two-dimensional (2D) spectra recorded with different relaxation delays. Intensities of cross-peaks were fitted to mono-exponential or hyperbolic tangential decay functions as appropriate to yield spin relaxation rate constants. The chemical exchange (Rex) was defined for each residue (i.e., Rex = R2HE-R20), where R2HE was obtained from the in-phase Hahn echo experiment (Millet et al., 2000), and R20 is 15N-R2 with the Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence (Allerhand and Thiele, 1966). The TROSY version of the R2HE experiment was performed using eight relaxation delays ranging from 7.7 to 92.2 ms (7.7, 15.4, 23.0, 30.7, 46.1, 61.4, 76.8, and 92.2 ms), and WALTZ 16 1H decoupling was employed during the relaxation period t. The TROSY version of R2 experiment was recorded with relaxation delays of 4.8, 14.4, 24.0, 33.6, 38.4, 43.2, 52.8, 62.3, 72.0, and 81.6 ms. Two of the relaxation times were repeated for error analysis. The apparent relaxation rate constants, R2HE and R2, were calculated by fitting the ratio of the signal intensities to an exponential decay function. NMR spectra were processed using nmr-Pipe (Delaglio et al., 1995) and analyzed with Sparky (Goddard and Kneller, 2008). The mapping of dynamic residues into the α1I structure (PDB: 1PT6) was accomplished using PyMOL (Schrodinger, 2015).
Relaxation Dispersion Experiments
Experiments were recorded on [U-2H,15N]-labeled α1I samples containing 0.3 to 0.5 mM protein in 90 % H2O / 10 % D2O, employing a buffer comprised of 5 mM PIPES, 140 mM NaCl, and 1.0 mM EDTA or a standard concentration of MgCl2. TROSY-selected 15N relaxation dispersion experiments were performed using the relaxation-compensated (RC) CMPG pulse sequence (Loria et al., 1999) in the presence of 1 mM EDTA or a standard concentration of MgCl2 (i.e., 2.5, 5, 10, 50, 75 or 100 mM at 20 °C; 100 mM at 5 °C). 1H relaxation dispersion experiments were acquired using the TROSY-selected 1H CPMG pulse sequence of Arthur G. Palmer 3rd (Li et al., 2013) on samples containing 5, 25, or 50 mM MgCl2 at 20 °C. Experiments were recorded at 600 and 700 MHz frequencies (except 0 and 10 mM MgCl2 acquired at 700 MHz) with a 40 ms constant relaxation period. Approximately 13 υCPMG values and a reference experiment were used for each dispersion profile, ranging from 25 to 1000 Hz with two or three points repeated for error analysis.
Relaxation dispersion data were processed using nmr-Pipe (Delaglio et al., 1995) and extracted from peak intensities in the two-dimensional NMR spectra as a function of CPMG field strength using Sparky (Goddard and Kneller, 2008). R2eff was calculated from peak intensities with the generalized Carver-Richards equation for two-site exchange as described previously (Loria et al., 1999). The error bars for individual data points reflect propagation of the signal-to-noise ratio from duplicate measurements at one CPMG frequency. Residues were considered to exhibit significant dynamics when Rex > 2 s−1 and Rex > 5 s−1 for 15N and 1H relaxation dispersion data, respectively, and mapped into the α1I structure (PDB: 1PT6) using PyMOL (Schrodinger, 2015). The dynamic residues were fit via the GUARDD program (Kleckner and Foster, 2012) employing Monte Carlo simulations to estimate errors in the kinetic, thermodynamic, and structural parameters. Initially, the data were fitted individually for each residue with those exhibiting similar exchange rates subsequently group fit. This protocol allowed optimization of parameters Δω and R20 for the same global values of kex and pA. The quality of the fits was evaluated by measuring the ratio of χ2 values between group and individual fits (Farber and Mittermaier, 2015). Residues with large uncertainties in the measured relaxation rates were excluded from group fits.
15N ZZ-Exchange Experiments
The 15N ZZ-exchange experiments were recorded on a Bruker 700 MHz spectrometer at 20 °C using the approach developed by Tollinger’s group (Kloiber et al., 2011) combining two complementary experiments, ZZ and T1zz, both with and without resolving exchange cross peaks between unbound and Mg-bound α1I, respectively. This approach employs pulse sequences that are based on the scheme described by Farrow et al. (Farrow et al., 1995). A set of 2D spectra was recorded at ten mixing times (i.e., Tmix = 10, 20, 30, 40, 50, 60, 70, 80, 90, 100 ms) for two separate experiments. In the first experiment, 13C frequency labeling (order of indirect evolution) preceded the mixing time while in the second experiment, the 13C frequency labeling and mixing times were interchanged.
The kinetic rate constants of α1I:Mg2+ association (kon) and dissociation (koff) were determined by fitting the time dependence of auto-peak intensities obtained from both experiments applying four equations for a two site exchange system (Kloiber et al., 2011):
with
where IAA/BB (t) and IA/B (t) are the peak intensities of state A/B at different mixing times obtained by the ZZ and T1zz experiments, respectively, with IAA/BB (0) and IA/B (0) the peak intensities at zero mixing time. Parameters obtained via R software fitting (Team, 2013) include the kinetic rate constants (kAB and kBA) that describe the interconversion between states A and B, the longitudinal relaxation rates of magnetization (R1A and R1B) in sites A and B. This approach has a unique advantage in that all peaks are normalized by their respective magnitudes at the start of the mixing period. This reduces the errors associated with weak binding of Mg2+ ions to α1I and consequent inability of determining the relative quantity of unbound- and bound-Mg2+ species. Interconversion between the two states occurs even in the absence of mixing time. Thus, the longitudinal relaxation and kinetic rate constants (R1A, R1B, kAB, kBA) extracted from the four direct correlation peaks via simultaneous fitting of these four equations to the experimental data yields values not distorted by differential line-broadening effects that might be caused by exchange on the μs-ms timescales.
Using the parameters obtained from ZZ-exchange curve fitting and assuming a 1:1 model of α1I binding to Mg2+, the exchange rate (kex) and population of bound complex (fPL) was determined employing the relation kex = koff + kon⋅[L], where koff = kBA, kon⋅[L] = kAB and fPL = 1 − kBA/kex. The dissociation constants were obtained via the equation Kd = (kBA/kAB)⋅(LT − fPL⋅PT) in which LT and PT are the total concentrations of Mg2+ and α1I, respectively. Kinetic parameters at higher Mg2+ concentrations are estimated by determining the concentration of free Mg2+ in solution, L (i.e., L = LT − PL) and invoking the relation: .
Isothermal Titration Calorimetry (ITC)
Thermodynamic binding parameters for the association of Mg2+ with WT, E317A, and L318A α1I were determined via Isothermal Titration Calorimetry employing a VP-ITC (MicroCal, Northampton, MA). Protein stock solutions were depleted of metals via EDTA-column treatment and dialyzed exhaustively against a buffer comprised of 5 mM PIPES and 140 mM NaCl (pH 7.3). Protein standard solutions were filtered using a 0.22 μm pore size membrane and adjusted to a final concentration of 500 μM α1I. The titration syringe contained either a 3 or 9 mM MgCl2 standard solution prepared in the final protein dialysate. Each ITC experiment consisted of 30 consecutive 10.0 μL injections during which the reaction heats were monitored and integrated for 5.0 min. Binding isotherms were generated by recording the integrated heats normalized for Mg2+ concentration versus the metal:protein ratio. The low affinity α1I:Mg2+ complexes necessitated use of the NITPIC/SEDPHAT program suite (Brautigam et al., 2016) to facilitate unbiased baseline assignment and peak integration. A nonlinear least squares fit of the resultant profile to a single site binding model yields thermodynamic parameters for the metal:protein complex including the affinity (Ka), Gibbs free energy (ΔG), enthalpy (ΔH), entropy (ΔS), and stoichiometric ratio (n).
Adhesion assays
Adhesion of the recombinant wild type α1I and mutants Q219L, H257F, V314A, E317A and L318A to type I collagen from rat tail (BD Biosciences) was determined colorimetrically in a solid-phase assay as described previously (Nunes et al., 2016). Immulon 2HB 96-well plates (Thermo Scientific) were coated with collagen (10 μg/mL in 10 mM acetic acid) overnight at 4 °C and blocked for 1 hr with 200 μL of a 5 % BSA solution in 5 mM PIPES and 140 mM NaCl. The washing and adhesion buffers consisted of 5 mM PIPES, 140 mM NaCl and 1 mg/mL of BSA in the presence of 5 mM MgCl2 or 5 mM EDTA. Following three washings with 200 μL buffer, binding was achieved by incubating 100 μL adhesion buffer containing 10 μg/mL α1I variants with the coated collagen for 1 hr at room temperature (RT). Binding was detected by first incubating 100 μL of the mouse anti-α1I monoclonal antibody (Millipore) with a 1:2000 dilution in adhesion buffer for 1 hr at RT, followed by incubation of 100 μL for 30 min with a 1:5000 diluted goat HRP-conjugated anti-mouse IgG antibody (GenScript) in adhesion buffer at RT, incorporating washing steps between each antibody addition. Color was developed using a TMB Substrate Kit (Pierce) in accordance with the manufacturer’s instructions.
Data and Software Availability
NMR data were acquired using Topspin and VnmrJ software and pulse sequences in the Topspin and VnmrJ libraries from Bruker Biospin and Varian BioPack. Modifications to pulse sequence are mentioned and cited. Copies of the modified pulse sequence are available from Lead Contact. NMR spectra were processed using nmr-Pipe software. Data were analyzed using SPARKY for peak picking and integration. Analysis of peak intensities for determination of relaxation rates, exchange rates, populations and chemical shift differences were performed using Sparky, GUARDD, Matlab and R (sources listed in the Key Resources Table). Visualization and generation of molecular structure figures and images were created using PyMOL. ITC data were acquired on a VP-ITC manufactured by MicroCal and analyzed using the NITPIC/SEDPHAT program suite. All software are listed in the Key Resources Table and the use of each package for data analysis is described in the sub-headings of STAR Methods.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Integrin α1 antibody, clone FB12 | Millipore EMD | Cat#MAB1973; RRID: AB_2129087 |
| Goat anti-mouse IgG HRP antibody | Genescript Corporation | Cat#NC1348387; RRID: AB_1968937 |
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) competent cells | Novagen | Cat#69450 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Synthetic collagen model peptide (Ac-(GPO)4GLOGEN(GPO)4GY-NH2) | LifeTein | N/A |
| Recombinant protein: human α1I (aa 141-335, ref#NP_852478.1) | This paper | N/A |
| Recombinant protein: human α1I (aa 141-331, ref#NP_852478.1) | (Nunes et al., 2016) | N/A |
| AccuPrime™ Pfx DNA Polymerase | Fisher Scientific | Cat#12-344-024 |
| DpnI | New England Biolabs | Cat#R0176S |
| Collagen I from rat tail | BD Biosciences | Cat#354236 |
| Albumin Bovine (BSA) Fraction V 10 | VWR | Cat#97061-416 |
| Critical Commercial Assays | ||
| Pierce™ TMB Substrate Kit | ThemoFisher Scientific | Cat#34021 |
| Deposited Data | ||
| Relaxation dispersion data | Datasets in Figures 2, 3, 4, S4 and S6. | This paper |
| ZZ-exchange data | Dataset in Figure S3. | This paper |
| Oligonucleotides | ||
| Primer: L196A forward: GTGACCCATGAGTTCAACGCGAATAAGTATTCTTCCACC | Integrated DNA Technologies | N/A |
| Primer: Q219L forward: GAGAGGTGGCCGCCTAACTATGACAGCTC | Integrated DNA Technologies | N/A |
| Primer: H257F forward: GTGACAGATGGAGAGTCTTTTGACAATCATCGACTGAAG | Integrated DNA Technologies | N/A |
| Primer: E317A forward: CAATGTCTCTGATGCATTGGCTCTAGTC | Integrated DNA Technologies | N/A |
| Primer L318A forward: CAATGTCTCTGATGAAGCGGCTCTAGTCACCATTG | Integrated DNA Technologies | N/A |
| Primer V314A forward: GAAAAGCATTTCTTCAATGCGTCTGATGAATTGGCTCTAG | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Plasmid: pET-Dest42-His10-α1I | Gift from R.W. Farndale | N/A |
| Software and Algorithms | ||
| NMRPipe | (Delaglio et al., 1995) | https://www.ibbr.umd.edu/nmrpipe/index.html |
| Sparky | (Goddard and Kneller, 2008) | https://www.cgl.ucsf.edu/home/sparky/ |
| R | © The R Foundation (R Core Team, 2013) | https://www.r-project.org/ |
| GUARDD | (Kleckner and Foster, 2012) | https://research.cbc.osu.edu/foster.281/software/#GUARDD |
| PyMOL | (Schrodinger, 2015) | https://pymol.org/2/ |
| VnmrJ | Agilent | https://www.agilent.com/search/?Ntt=VnmrJ |
| TopSpin | Bruker | https://www.bruker.com/ |
| NITPIC program suite | (Brautigam et al., 2016) | http://biophysics.swmed.edu/MBR/software.html |
| SEDPHAT program suite | (Brautigam et al., 2016) | https://sedfitsedphat.nibib.nih.gov/software/default.aspx |
Quantification and Statistical Analysis
Errors within individual NMR data points reflect error-propagation of the signal-to-noise ratio from two or three points repeated during acquisition of the relaxation data. In relaxation dispersion experiments, residues with large uncertainties in the measured relaxation rates were excluded from group fits. Monte Carlo simulations were used to estimate errors associated with the parameters obtained from fits of relaxation dispersion data. The ITC data correspond to average parameters and standard deviations determined from a minimum of three separate experiments. Adhesion assays were performed in triplicate with the data corresponding to mean values ± standard deviation.
Supplementary Material
ACKNOWLEGMENTS
The authors wish to thank Seho Kim for useful discussions and technical support; Arthur G. Palmer, III for the 1H relaxation dispersion pulse sequence and helpful discussions; and, Richard W. Farndale and Samir W. Hamaia for wild type α1I cDNA and valuable discussions. This research was funded by the National Institutes of Health GM45302 to J.B.. A.M.N. was supported by an American Heart Association postdoctoral fellowship award (13POST16550007).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allerhand A, and Thiele E (1966). Analysis of Carr-Purcell spin-echo nuclear magnetic resonance experiments on multiple-spin systems. II. The effect of chemical exchange. Journal of Chemical Physics 45, 902–916. [Google Scholar]
- Arias-Moreno X, Abian O, Vega S, Sancho J, and Velazquez-Campoy A (2011). Protein-cation interactions: structural and thermodynamic aspects. Curr Protein Pept Sci 12, 325–338. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, and Wright PE (2009). The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Zhao H, Vargas C, Keller S, and Schuck P (2016). Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat Protoc 11, 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila DA, Braymer JJ, Edmonds KA, Wu H, and Giedroc DP (2017). Entropy redistribution controls allostery in a metalloregulatory protein. Proc Natl Acad Sci U S A 114, 4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin YK, Headey S, Mohanty B, Emsley J, Simpson JS, and Scanlon MJ (2013a). Assignments of human integrin alpha1I domain in the apo and Mg(2+) bound states. Biomol NMR Assign. [DOI] [PubMed] [Google Scholar]
- Chin YK, Headey SJ, Mohanty B, Patil R, McEwan PA, Swarbrick JD, Mulhern TD, Emsley J, Simpson JS, and Scanlon MJ (2013b). The Structure of Integrin alpha1I Domain in Complex with a Collagen Mimetic Peptide. J Biol Chem 288, 36796–36809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Palotai R, and Nussinov R (2010). Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci 35, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, and Liddington RC (2000). Structural basis of collagen recognition by integrin alpha2beta1. Cell 101, 47–56. [DOI] [PubMed] [Google Scholar]
- Farber PJ, and Mittermaier A (2015). Relaxation dispersion NMR spectroscopy for the study of protein allostery. Biophys Rev 7, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow NA, Zhang O, Forman-Kay JD, and Kay LE (1995). Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry 34, 868–878. [DOI] [PubMed] [Google Scholar]
- Fuhrmann A, Li J, Chien S, and Engler AJ (2014). Cation type specific cell remodeling regulates attachment strength. PLoS One 9, e102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner H (2014). Integrin alpha1beta1. Adv Exp Med Biol 819, 21–39. [DOI] [PubMed] [Google Scholar]
- Goddard TD, and Kneller DG (2008). SPARKY (San Francisco: University of California; ). [Google Scholar]
- Gotwals PJ, Chi-Rosso G, Ryan ST, Sizing I, Zafari M, Benjamin C, Singh J, Venyaminov SY, Pepinsky RB, and Koteliansky V (1999). Divalent cations stabilize the alpha 1 beta 1 integrin I domain. Biochemistry 38, 8280–8288. [DOI] [PubMed] [Google Scholar]
- Hamaia S, and Farndale RW (2014). Integrin recognition motifs in the human collagens. Adv Exp Med Biol 819, 127–142. [DOI] [PubMed] [Google Scholar]
- Hamaia SW, Pugh N, Raynal N, Nemoz B, Stone R, Gullberg D, Bihan D, and Farndale RW (2012). Mapping of potent and specific binding motifs, GLOGEN and GVOGEA, for integrin alpha1beta1 using collagen toolkits II and III. J Biol Chem 287, 26019–26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MR, Hass MA, Hansen DF, and Led JJ (2007). Investigating metal-binding in proteins by nuclear magnetic resonance. Cell Mol Life Sci 64, 1085–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Liddington RC, and Takada Y (1999). Interaction between collagen and the alpha(2) I-domain of integrin alpha(2)beta(1). Critical role of conserved residues in the metal ion-dependent adhesion site (MIDAS) region. J Biol Chem 274, 32108–32111. [DOI] [PubMed] [Google Scholar]
- Karpusas M, Ferrant J, Weinreb PH, Carmillo A, Taylor FR, and Garber EA (2003). Crystal structure of the alpha1beta1 integrin I domain in complex with an antibody Fab fragment. J Mol Biol 327, 1031–1041. [DOI] [PubMed] [Google Scholar]
- Kay LE (2016). New Views of Functionally Dynamic Proteins by Solution NMR Spectroscopy. J Mol Biol 428, 323–331. [DOI] [PubMed] [Google Scholar]
- Kern D, and Zuiderweg ER (2003). The role of dynamics in allosteric regulation. Curr Opin Struct Biol 13, 748–757. [DOI] [PubMed] [Google Scholar]
- Kleckner IR, and Foster MP (2012). GUARDD: user-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J Biomol NMR 52, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloiber K, Spitzer R, Grutsch S, Kreutz C, and Tollinger M (2011). Longitudinal exchange: an alternative strategy towards quantification of dynamics parameters in ZZ exchange spectroscopy. J Biomol NMR 51, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti M, Bligt E, Niskanen H, Parkash V, Brandt AM, Jokinen J, Patrikainen P, Kapyla J, Heino J, and Salminen TA (2011). Structure of Collagen Receptor Integrin alpha1I Domain Carrying the Activating Mutation E317A. J Biol Chem 286, 43343–43351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AL (2015). Contrasting roles of dynamics in protein allostery: NMR and structural studies of CheY and the third PDZ domain from PSD-95. Biophys Rev 7, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Bankston LA, Arnaout MA, and Liddington RC (1995). Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure 3, 1333–1340. [DOI] [PubMed] [Google Scholar]
- Li Y, Altorelli NL, Bahna F, Honig B, Shapiro L, and Palmer AG III (2013). Mechanism of E-cadherin dimerization probed by NMR relaxation dispersion. Proc Natl Acad Sci U S A 110, 16462–16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington RC (2014). Structural aspects of integrins. Adv Exp Med Biol 819, 111–126. [DOI] [PubMed] [Google Scholar]
- Loria JP, Rance M, and Palmer AG III (1999). A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. Journal of the American Chemical Society 121, 2331–2332. [Google Scholar]
- Mazur A, Maier JA, Rock E, Gueux E, Nowacki W, and Rayssiguier Y (2007). Magnesium and the inflammatory response: potential physiopathological implications. Arch Biochem Biophys 458, 48–56. [DOI] [PubMed] [Google Scholar]
- McDonald LR, Boyer JA, and Lee AL (2012). Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure 20, 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet O, Loria JP, Kroenke CD, Pons M, and Palmer AG III (2000). The Static Magnetic Field Dependence of Chemical Exchange Linebroadening Defines the NMR Chemical Shift Time Scale. Journal of the American Chemical Society 122, 2867–2877. [Google Scholar]
- Nolte M, Pepinsky RB, Venyaminov S, Koteliansky V, Gotwals PJ, and Karpusas M (1999). Crystal structure of the alpha1beta1 integrin I-domain: insights into integrin I-domain function. FEBS Lett 452, 379–385. [DOI] [PubMed] [Google Scholar]
- Nunes AM, Zhu J, Jezioro J, Minetti CA, Remeta DP, Farndale RW, Hamaia SW, and Baum J (2016). Intrinsic local destabilization of the C-terminus predisposes integrin alpha1 I domain to a conformational switch induced by collagen binding. Protein Sci 25, 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymalm Y, Puranen JS, Nyholm TK, Kapyla J, Kidron H, Pentikainen OT, Airenne TT, Heino J, Slotte JP, Johnson MS , et al. (2004). Jararhagin-derived RKKH peptides induce structural changes in alpha1I domain of human integrin alpha1beta1. J Biol Chem 279, 7962–7970. [DOI] [PubMed] [Google Scholar]
- Palmer AG III, Kroenke CD, and Loria JP (2001). Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol 339, 204–238. [DOI] [PubMed] [Google Scholar]
- Rich RL, Deivanayagam CC, Owens RT, Carson M, Hook A, Moore D, Symersky J, Yang VW, Narayana SV, and Hook M (1999). Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM. J Biol Chem 274, 24906–24913. [DOI] [PubMed] [Google Scholar]
- Romani AM (2011). Cellular magnesium homeostasis. Arch Biochem Biophys 512, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC (2015). The PyMOL Molecular Graphics System, Version 1.8.
- Shi M, Pedchenko V, Greer BH, Van Horn WD, Santoro SA, Sanders CR, Hudson BG, Eichman BF, Zent R, and Pozzi A (2012). Enhancing integrin alpha1 inserted (I) domain affinity to ligand potentiates integrin alpha1beta1-mediated down-regulation of collagen synthesis. J Biol Chem 287, 35139–35152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siljander PR, Hamaia S, Peachey AR, Slatter DA, Smethurst PA, Ouwehand WH, Knight CG, and Farndale RW (2004). Integrin activation state determines selectivity for novel recognition sites in fibrillar collagens. J Biol Chem 279, 47763–47772. [DOI] [PubMed] [Google Scholar]
- Team R.C. (2013). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Tulla M, Lahti M, Puranen JS, Brandt AM, Kapyla J, Domogatskaya A, Salminen TA, Tryggvason K, Johnson MS, and Heino J (2008). Effects of conformational activation of integrin alpha 1I and alpha 2I domains on selective recognition of laminin and collagen subtypes. Exp Cell Res 314, 1734–1743. [DOI] [PubMed] [Google Scholar]
- Vogt AD, Pozzi N, Chen Z, and Di Cera E (2014). Essential role of conformational selection in ligand binding. Biophys Chem 186, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Thinn AMM, and Zhu J (2017). A pivotal role for a conserved bulky residue at the alpha1-helix of the alphaI integrin domain in ligand binding. J Biol Chem 292, 20756–20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb PH, Li S, Gao SX, Liu T, Pepinsky RB, Caravella JA, Lee JH, and Woods VL Jr. (2012). Dynamic structural changes are observed upon collagen and metal ion binding to the integrin alpha1 I domain. J Biol Chem 287, 32897–32912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Coller BS, Wang JH, and Springer TA (2004). Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, and Chen J (2012). The regulation of integrin function by divalent cations. Cell Adh Migr 6, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR data were acquired using Topspin and VnmrJ software and pulse sequences in the Topspin and VnmrJ libraries from Bruker Biospin and Varian BioPack. Modifications to pulse sequence are mentioned and cited. Copies of the modified pulse sequence are available from Lead Contact. NMR spectra were processed using nmr-Pipe software. Data were analyzed using SPARKY for peak picking and integration. Analysis of peak intensities for determination of relaxation rates, exchange rates, populations and chemical shift differences were performed using Sparky, GUARDD, Matlab and R (sources listed in the Key Resources Table). Visualization and generation of molecular structure figures and images were created using PyMOL. ITC data were acquired on a VP-ITC manufactured by MicroCal and analyzed using the NITPIC/SEDPHAT program suite. All software are listed in the Key Resources Table and the use of each package for data analysis is described in the sub-headings of STAR Methods.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Integrin α1 antibody, clone FB12 | Millipore EMD | Cat#MAB1973; RRID: AB_2129087 |
| Goat anti-mouse IgG HRP antibody | Genescript Corporation | Cat#NC1348387; RRID: AB_1968937 |
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) competent cells | Novagen | Cat#69450 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Synthetic collagen model peptide (Ac-(GPO)4GLOGEN(GPO)4GY-NH2) | LifeTein | N/A |
| Recombinant protein: human α1I (aa 141-335, ref#NP_852478.1) | This paper | N/A |
| Recombinant protein: human α1I (aa 141-331, ref#NP_852478.1) | (Nunes et al., 2016) | N/A |
| AccuPrime™ Pfx DNA Polymerase | Fisher Scientific | Cat#12-344-024 |
| DpnI | New England Biolabs | Cat#R0176S |
| Collagen I from rat tail | BD Biosciences | Cat#354236 |
| Albumin Bovine (BSA) Fraction V 10 | VWR | Cat#97061-416 |
| Critical Commercial Assays | ||
| Pierce™ TMB Substrate Kit | ThemoFisher Scientific | Cat#34021 |
| Deposited Data | ||
| Relaxation dispersion data | Datasets in Figures 2, 3, 4, S4 and S6. | This paper |
| ZZ-exchange data | Dataset in Figure S3. | This paper |
| Oligonucleotides | ||
| Primer: L196A forward: GTGACCCATGAGTTCAACGCGAATAAGTATTCTTCCACC | Integrated DNA Technologies | N/A |
| Primer: Q219L forward: GAGAGGTGGCCGCCTAACTATGACAGCTC | Integrated DNA Technologies | N/A |
| Primer: H257F forward: GTGACAGATGGAGAGTCTTTTGACAATCATCGACTGAAG | Integrated DNA Technologies | N/A |
| Primer: E317A forward: CAATGTCTCTGATGCATTGGCTCTAGTC | Integrated DNA Technologies | N/A |
| Primer L318A forward: CAATGTCTCTGATGAAGCGGCTCTAGTCACCATTG | Integrated DNA Technologies | N/A |
| Primer V314A forward: GAAAAGCATTTCTTCAATGCGTCTGATGAATTGGCTCTAG | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Plasmid: pET-Dest42-His10-α1I | Gift from R.W. Farndale | N/A |
| Software and Algorithms | ||
| NMRPipe | (Delaglio et al., 1995) | https://www.ibbr.umd.edu/nmrpipe/index.html |
| Sparky | (Goddard and Kneller, 2008) | https://www.cgl.ucsf.edu/home/sparky/ |
| R | © The R Foundation (R Core Team, 2013) | https://www.r-project.org/ |
| GUARDD | (Kleckner and Foster, 2012) | https://research.cbc.osu.edu/foster.281/software/#GUARDD |
| PyMOL | (Schrodinger, 2015) | https://pymol.org/2/ |
| VnmrJ | Agilent | https://www.agilent.com/search/?Ntt=VnmrJ |
| TopSpin | Bruker | https://www.bruker.com/ |
| NITPIC program suite | (Brautigam et al., 2016) | http://biophysics.swmed.edu/MBR/software.html |
| SEDPHAT program suite | (Brautigam et al., 2016) | https://sedfitsedphat.nibib.nih.gov/software/default.aspx |