Summary
Two major functions of the epigenome are to regulate gene expression and to suppress transposons. It is unclear how these functions are balanced during physiological challenges requiring tissue regeneration, where exquisite coordination of gene expression is essential. Transcriptomic analysis of seven time points following partial hepatectomy identified the epigenetic regulator, UHRF1, which is essential for DNA methylation, as dynamically expressed during liver regeneration in mice. UHRF1 deletion in hepatocytes (Uhrf1HepKO) caused genome-wide DNA hypomethylation but, surprisingly, had no measurable effect on gene or transposon expression or liver homeostasis. Partial hepatectomy of Uhrf1HepKO livers resulted in early and sustained activation of pro-regenerative genes and enhanced liver regeneration. This was attributed to redistribution of H3K27me3 from promoters to transposons, effectively silencing them and, consequently, alleviating repression of liver regeneration genes, priming them for expression in Uhrf1HepKO livers. Thus, epigenetic compensation safeguards the genome against transposon activation, indirectly affecting gene regulation.
Keywords: Tissue regeneration, epigenomics, epigenetic compensation, transposons, DNA methylation, UHRF1, H3K27me3, liver biology, partial hepatectomy
eTOC blurb
It is not clear how complex epigenetic functions are coordinated to both regulate gene expression and mitigate transposon threat. Wang et al. discovered that to compensate for loss of DNA methylation mediated transposon silencing, another repressive epigenetic mark is diverted from pro-regenerative genes, resulting in enhanced liver regeneration.
Introduction
Cell type specific epigenetic profiles dictate cell fate and the response to stimuli. Both widespread and focal changes to the epigenome have been cataloged in cells during differentiation, and much is known about how these epigenetic changes shape cell fate. For instance, extensive work on the dynamic pattern of DNA methylation during early development (Roadmap Epigenomics et al., 2015) and the epigenetic landscape of cancer (Bergman and Cedar, 2013; Mouse Genome Sequencing et al., 2002; Yue et al., 2014) has provided a framework by which complex combinations of activating and repressive epigenetic marks dictate the differences between cells undergoing dramatic changes in identity or function. An interesting insight from these many genome wide studies is that majority of the epigenetic changes that occur during physiological and pathological transitions are outside of both gene coding sequences and adjacent regulatory regions. This largely reflects the emerging understanding that distal regulatory sequences are important for gene expression and that only a small fraction of most genomes code for proteins. The vast majority of sequences labeled as intergenic regions that are littered with transposable elements (TEs), which are remnants of ancient viruses. If mobilized, TEs can restructure the genome, cause genomic instability and cell death (Chuong et al., 2016). Evolution has effectively countered the danger of TEs by leveraging repressive epigenetic marks, including DNA methylation, to ensure that young and potentially mobile TEs remain dormant. These marks serve dual roles in regulating gene expression, but little is known about how different epigenetic marks are coordinated to balance the demands on the epigenome to tightly regulate gene expression and to suppress expression of TEs. This is especially relevant given the widespread epigenetic changes that accompany developmental transitions and tissue regeneration.
DNA methylation on cytosine residues (CpGs) is the mainstay defense system against transposon activation in vertebrates (Chernyavskaya et al., 2017; Walter et al., 2016; Yoder et al., 1997). Since the bulk of CpG methylation occurs in transposons (Ziller et al., 2013) and this repressive function of DNA methylation is required in all cells, the pattern of DNA methylation is largely static across cell types. Therefore, the effect size of DNA methylation changes across developmental stages or disease states is relatively minor compared to the massive restructuring of histone modifications and variants that shape the unique epigenome of distinct cell types (Yue et al., 2014). The conclusion that TE repression is a major role of DNA methylation is also based on the finding that transposons undergo dramatic transcriptomic changes in in cells where DNA methylation is depleted (Chernyavskaya et al., 2017; Chiappinelli et al., 2015; Ohtani et al., 2018; Roulois et al., 2015; Walsh et al., 1998; Walter et al., 2016; Yoder et al., 1997). This is in contrast to the dramatic changes in gene expression that occur as a direct response to changes in other epigenetic marks (Roadmap Epigenomics et al., 2015; Soshnev et al., 2016). Thus, the epigenome appears to be partitioned between the role of DNA methylation, which remains largely static due to its accumulation mainly at TEs to suppress the damaging effects of their expression, and histone modifications and histone variants, which are more dynamic and more intimately involved in regulating gene expression. However, it is clear that in many settings, epigenetic marks can function redundantly, and this is perhaps most relevant to ensuring TE repression.
Tissue regeneration in response to injury or tissue loss is accompanied by widespread epigenetic changes (Kang et al., 2016; Sun et al., 2016), which raises the question of how the different roles of the epigenome are balanced and intersected. In some cases, when a repressive epigenetic mechanism becomes compromised, a different one can compensate, suggesting that cells employ redundant mechanisms to safe guard the genome against transposon activation (Cooper et al., 2014; Leeb et al., 2010; Sharif et al., 2016; Walter et al., 2016). However, it is not known whether cooperation between epigenetic marks when one is called in to offset for the loss of another has any impact on cell or tissue behavior.
The mammalian liver retains an amazing regenerative capacity that depends on hepatocytes both growing (hypertrophy) and re-entering the cell cycle (hyperproliferation) to restore liver mass (Michalopoulos, 2013; Miyaoka et al., 2012). This switch from quiescence to proliferation is accompanied by the altered expression of thousands of genes and dozens of transcription factors (Fukuhara et al., 2003; Kelley-Loughnane et al., 2003; Li et al., 2009; Michalopoulos, 2007; White et al., 2005), such as the E2F transcription factors to induce G1-S transition, DNA replication, and mitosis (Delgado et al., 2011; Sun et al., 2016). Since epigenetic marks regulate coordinated changes in gene expression, we hypothesize that the co-regulation of pro-regenerative genes is embedded in the epigenome. Here, we addressed the understudied question of epigenetic regulation of liver regeneration by comprehensively profiling transcriptomic changes associated with liver regeneration using the well-characterized mouse partial hepatectomy (PH) model, where surgical removal of 2/3 of liver mass causes hepatocytes to synchronously re-enter the cell cycle and proliferate to regenerate the lost liver mass within seven days (Michalopoulos, 2007; Taub, 2004). We identified a group of epigenetic regulators that display the same temporal pattern as genes that control hepatocyte proliferation. Within this group, we identified both members of the maintenance DNA methylation machinery, ubiquitin-like with phd and ring finger domains 1 (UHRF1) and DNA methyltransferase 1 (DNMT1). We tested the effects of UHRF1 loss-of-function during liver regeneration by deleting UHRF1 from hepatocytes (Uhrf1hepKO). While Uhrf1hepKO mice develop normally into viable adults, they have an augmented regenerative response following PH characterized by a premature and more robust activation of cell cycle genes, earlier onset of hepatocyte proliferation, and enhanced liver regeneration. Surprisingly, despite genome-wide DNA hypomethylation in UHRF1 deficient hepatocytes, there was no induction of TE expression. ChIP-seq analysis of repressive histone marks showed that H3K27me3 repositioned to hypomethylated transposons to suppress them. This compensatory action reduced H3K27me3 at gene promoters, priming pro-regenerative genes for activation. These findings suggest that enhancing cell cycle entry may be a secondary consequence of epigenetic compensation to protect against damage from activated transposons.
Results
We reasoned that genes that are co-expressed during liver regeneration would share a common epigenetic mechanism of regulation. To identify clusters of co-expressed genes, we analyzed the transcriptomic changes in control male mice (Uhrf1fl/fl or Uhrf1fl/+) across seven time points following PH (24, 30, 40, 48, 96 hours, and 7 and 28 days). During this time course, liver mass is restored by synchronous induction of the hepatocyte cell cycle, detected by markers of cell proliferation which peak at 48 hours after PH (Figure 1A).
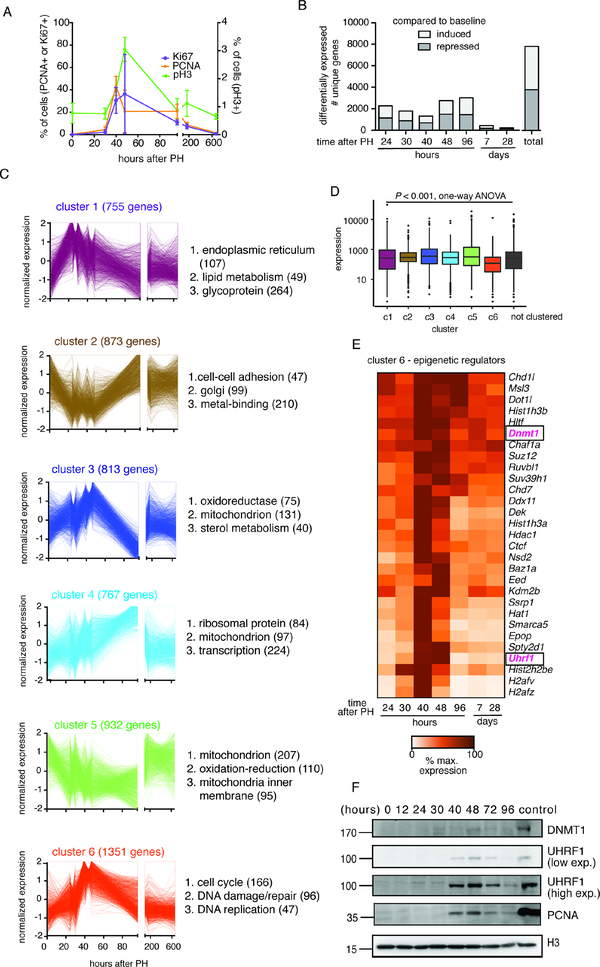
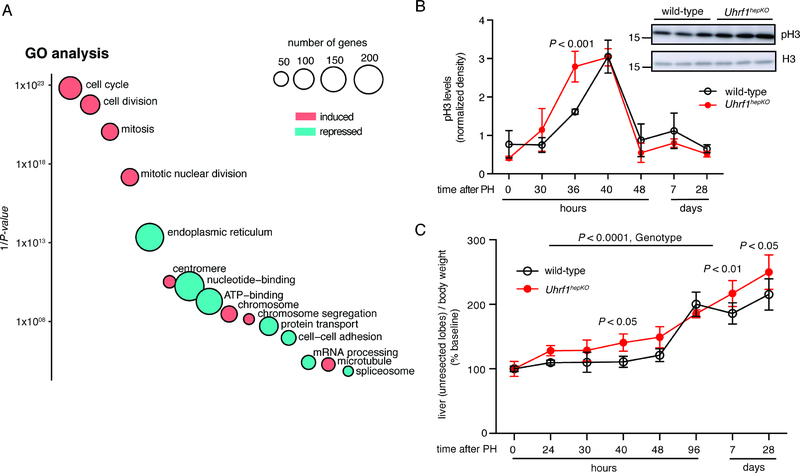
Figure 1: Comprehensive transcriptomic profiling of mouse liver regeneration identifies a group of epigenetic regulators including Uhrf1.
(A) Cell cycle markers detected by immunohistochemistry (IHC, Ki67, and PCNA) and Western blot (pH3) on control liver samples following PH. (B) All DEGs at each time point following PH were compared to quiescent livers. (C) Gene clusters of co-regulated genes identified by unsupervised clustering of significantly changed genes in regenerating control livers. The total number of gens in each cluster and those in the top 3 GO categories are noted. (D). Average normalized counts of genes from each gene cluster in quiescent livers by RNA-seq. (E). Curated list of well-established epigenetic regulators found in cluster 6. (F) UHRF1 and DNMT1 protein expression with respect to the well- established cell cycle marker PCNA during liver regeneration. Mouse ES cells which express a high level of UHRF1 were used as a positive control for blotting and histone H3 was used as a loading control. Error bars represent s.d. See also Figure S1, Table S1, S2.
We compared each time point after PH to quiescent livers (i.e. T=0) and identified 7006 unique genes that were significantly differentially expressed at any one of these time points, with some genes being differentially regulated at more than one time point (Figure 1B, Table S1). Unsupervised clustering of these differentially expressed genes (DEG) identified 6 unique clusters characterized by distinct temporal expression patterns (Figure 1C, Table S2). Gene ontology (GO) classification of the DEGs in each cluster showed that they were categorized by unique functions (Figure 1C, Figure S1). We found cluster 6 to be the most interesting because these genes peaked between 40–48 hours after PH (Figure 1C), were expressed at the lowest levels in quiescent livers compared to all other clusters of DEGs, in addition to the 1516 DEGs that did not fall into any cluster (Figure 1D), and were involved in regulating cellular proliferation (Figure 1C and S1). We reasoned that epigenetic regulatory genes contained in cluster 6 could be responsible for the regulation of this set of important proregenerative genes. We queried cluster 6 for genes categorized by GO as “regulation of gene expression, epigenetic” and found 27 out of 165 (P = 0.0645). Of these, the maintenance DNA methylation machinery genes, Uhrf1 and Dnmt1 (Figure 1E, 1F), were particularly interesting as we previously reported uhrf1 as a key regulator of cell cycle gene expression and liver development in zebrafish embryos (Jacob et al., 2015; Sadler et al., 2007). Western blot analysis showed that UHRF1 and DNMT1 proteins are not detectable in quiescent livers or in early stages of regeneration, but are markedly induced by 40 hours and return to baseline levels by 96 hours after PH (Figure 1F). Thus, both the mRNA and protein of these two important epigenetic regulators change dynamically during liver regeneration in a pattern suggestive of their role regulating this process.
To test whether Uhrf1 was involved in the gene expression clusters that characterize liver regeneration, we generated mice with Loxp sites flanking exon 6 and 10 of the Uhrf1 gene (Uhrf1fl/fl; Figure S2A). The Uhrf1fl/fl line was crossed to the Tg(Alb:Cre) line to generate hepatocyte specific deletion of these exons which creates a frameshift that generates a stop codon following amino acid 294 (Figure S2B, Table S3). We demonstrated the Uhrf1 locus is effectively deleted in genomic DNA from whole liver samples of Alb:CreTg/+;Uhrf1fl/fl mice (i.e.Uhrf1hepKO; Figure S2C–D). Consistent with the increased expression of uhrf1 in developing zebrafish livers (Jacob et al., 2015; Sadler et al., 2007) we found that Uhrf1 expression was higher in post-natal mouse livers than in adults; in Uhrf1hepKO livers, we found that the mRNA to be significantly reduced as early as post-natal day 10 (Figure 2A). In adult livers, UHRF1 protein is undetectable in quiescent livers (Figs. 1F, 2B) and peaks between 40–48 hours after PH (Figure 1F, 2B). In Uhrf1hepKO livers, both UHRF1 protein (Figure 2B and S2E) and mRNA (Figure S2F) were dramatically reduced at 48 hours after PH, demonstrating the efficacy of this knock out strategy.
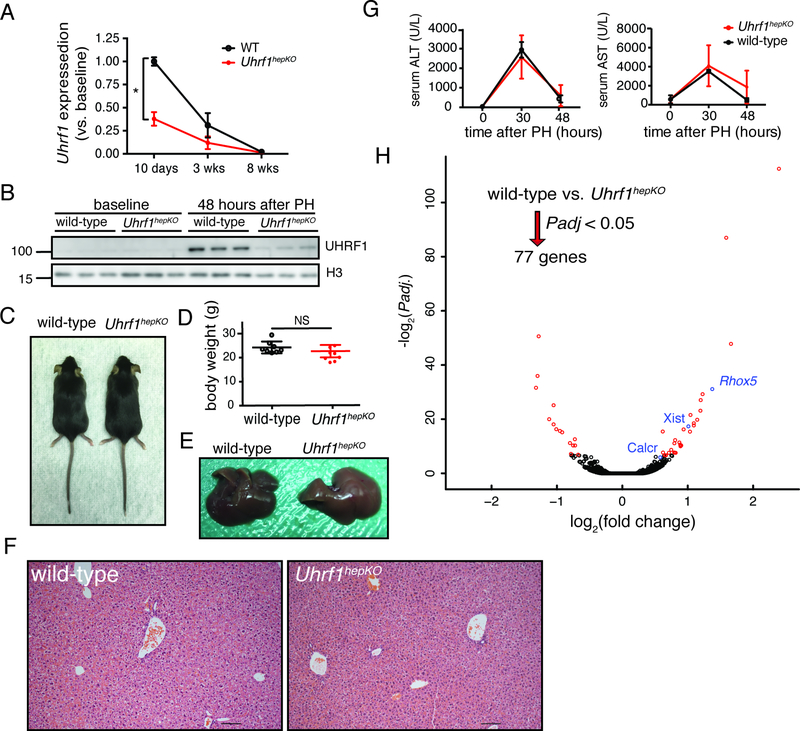
Figure 2: Uhrf1hepKO mouse livers appear normal.
(A) Normalized expression of Uhrf1 transcript at 10 days, 3 weeks, and 8 weeks in control and Uhrf1hepKO mouse livers measured by qPCR. * P < 0.0001 for the effect of genotype by two-way ANOVA. (B) Expression of UHRF1 protein in the liver of control or Uhrf1hepKO mice at 48 hours post-PH (N=3, time point of maximum UHRF1 detection in regenerating liver of control mice). (C) Representative pictures of 8 week old control and Uhrf1hepKO mice. (D) Body weight of control and Uhrf1hepKO mice at quiescence. (E) Representative pictures of dissected livers from 8 week old control and Uhrf1hepKO mice. (F) Representative hematoxylin and eosin staining of control and Uhrf1hepKO quiescent livers taken at 100X zoom. (G) Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) measurements in serum collected from control and Uhrf1hepKO mice before and at 30 and 48 hours following PH (N=3). (H) Volcano plot comparing RNA-seq data from quiescent control and Uhrf1hepKO livers. Red dots denote the significantly changed genes (Padj. < 0.05). Significantly changed imprinted genes are indicated in blue. Error bars represent s.d. See also Figure S2 and Key Resources Table for genotyping primers.
Given the dramatic phenotype in the liver of uhrf1 mutant zebrafish (Jacob et al., 2015; Sadler et al., 2007), we were surprised that Uhrf1hepKO mice appear phenotypically normal (Figure 2C). There was no difference from age-matched controls in body weight (Figure 2D), gross appearance of the liver (Figure 2E), histologically assessed hepatic architecture (Figure 2F), or the serum markers used to detect liver injury (AST and ALT), which rise and fall during regeneration following PH in both controls and Uhrf1hepKO mice (Figure 2G). Further RNA-seq analysis of quiescent livers revealed virtually no gene expression changes in Uhrf1hepKO mice compared to controls (only 30 DEG, Figure 2H and Table S4). Interestingly, some of the upregulated genes (Figure 2H labeled in blue) are imprinted, representing a category of genes that requires DNA methylation for suppression. This data reflects the efficacy of DNA hypomethylation in Uhrf1hepKO livers and indicates that loss of DNA methylation has little direct impact on gene expression in the mouse liver.
We further assessed DNA hypomethylation in Uhrf1hepKO livers using slot blot for bulk DNA methylation levels (Figure 3A) and enhanced Reduced Representation Bisulfite Sequencing (eRRBS; Figure 3B) to examine CpG methylation at base pair resolution (Garrett-Bakelman et al., 2015; Smith et al., 2009). Both approaches showed widespread loss of DNA methylation in Uhrf1hepKO livers (Figure 3A–C), with hypomethylated CpGs distributed throughout the genome (Figure 3B). Nearly all CpGs that changed their methylation status in Uhrf1hepKO livers were those fully methylated in control livers (Figure 3C), with the majority having shifted from >80% methylation in controls to <25% methylation in Uhrf1hepKO livers. However, we also detected partially methylated CpGs, which could represent a stochastic distribution of methylated CpGs in hepatocytes or could reflect methylation in other liver cell types which retain Uhrf1 in this model.
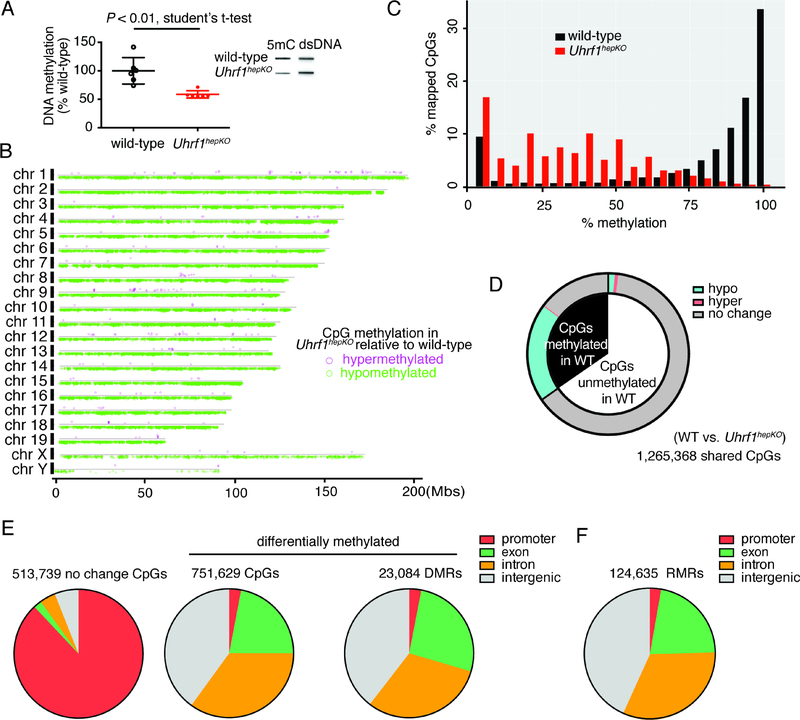
Figure 3: Uhrf1hepKO mouse livers display genome-wide loss of DNA methylation in non-promoter regions.
(A) Bulk DNA methylation levels in control and Uhrf1hepKO quiescent livers as determined by slot blot (N=6; example of one blot provided in inset). (B) Genome-wide distribution of CpGs that were hyper- (pink) or hypo-methylated (green) in Uhrf1hepKO livers compared to controls. (C) Histogram of all RRBS mapped CpGs binned by percentage methylation. (D) Distribution of CpG methylation changes in Uhrf1hepKO livers based on eRRBS analysis, comparing CpGs which were methylated >80% (black) to those that were not (white, <20%) in control livers. (E) Annotation of genomic elements based on CpGs methylation status in Uhrf1hepKO mice compared to controls. (F) Residually methylated regions (RMRs) in Uhrf1hepKO quiescent livers were distributed across the genome in a pattern similar to the DMRs.
Like many other tissues (Long et al., 2016; Long et al., 2013), the hepatic methylome is characterized by heavy methylation of intergenic regions and introns and a near complete absence of methylation in promoters (Zhang et al., 2016). The DNA methylation changes detected in Uhrf1hepKO livers reflects this pattern: there were 1 million CpGs adequately covered by eRRBS in both controls and Uhrf1hepKO samples, and the majority of these did not change their methylation status because they were unmethylated in wild-type animals. However, those CpGs that changed methylation status in Uhrf1hepKO livers, nearly all had lost DNA methylation (Figure 3B–C). Moreover, of those CpGs that were highly methylated (>80%) in controls, over half lost methylation (Figure 3D). We found that 95% of CpGs in promoters were unmethylated in control livers and remained so in Uhrf1hepKO samples (Figure 3E), reflecting the fact that promoters are protected from methylation across species (Long et al., 2016; Long et al., 2013). The CpGs that became hypomethylated mostly distributed in intergenic regions, introns, and exons (Figure 3E). A similar pattern was observed when analyzing the distribution of differentially methylated regions (DMRs) and single CpGs (Figure 3E). Interestingly, some regions of the genome remained methylated in Uhrf1hepKO livers and these residually methylated regions (RMRs) were also distributed in non-promoter regions (Figure 3F). These RMRs could emanate from other cell types in the liver that retained or could represent regions that are methylated independent of Uhrf1.
To test whether the DNA hypomethylation altered the response to mitotic signaling during regeneration, we subjected Uhrf1hepKO mice to PH surgery, where ~70% of the liver mass is removed. Unlike what we observed in quiescent livers, Uhrf1hepKO mice displayed widespread gene expression changes compared to wild-type controls following PH. Comparison of the gene expression profiles between wild-type and Uhrf1hepKO livers at each time point following PH revealed major differences at 30 and 96 hours after PH (Figure 4A, Table S4). This corresponds to the two waves of differential gene expression and cell divisions during WT liver regeneration (Figure 1A).
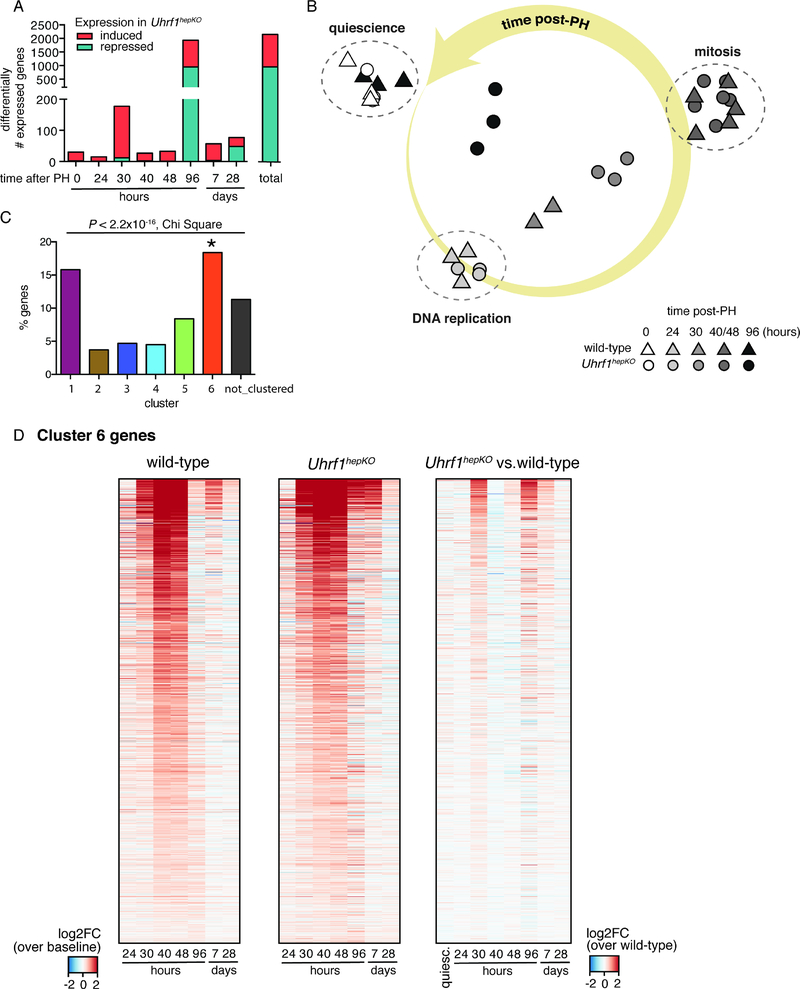
Figure 4: Hypomethylated DNA methylome leads to an earlier and more sustained activation of liver regeneration transcriptome.
(A) The number of DEGs that are induced (red) or repressed (teal) in Uhrf1hepKO mice compared to control livers collected at the same time points post-PH. (B) Principle component analysis of gene expression in quiescent and regenerating livers (24, 30, 40, 48, and 96 hours post-PH) from control and Uhrf1hepKO mice. Triangle = control. Circle = Uhrf1hepKO. Shading of data points (RNA-seq samples) is proportional to time post-PH (darker = later). Large yellow arrow indicate progression of time following surgery. Annotations were added for clarification of cell cycle events occurring in control mice at the relevant time points. (C) Percent of genes from each gene cluster identified in figure 1 that overlap with liver regeneration genes significantly induced in Uhrf1hepKO mice. (D) Heatmap of cluster 6 genes across all regeneration time points in control and Uhrf1hepKO mice. Red represents higher at each time point compared to quiescent livers (left 2 heatmaps) or higher in Uhrf1hepKO compared to control (right heatmap). Error bars represent s.d. See also Table S3.
We asked if this pattern reflected a shift in the timing of genes that are normally differentially expressed during regeneration of wild-type livers or whether it was due to the deregulation of genes that do not normally change during regeneration. Principal component analysis (PCA) of the global gene expression patterns in wild-type and Uhrf1hepKO mice during and after regeneration revealed two important findings. First, loss of UHRF1 does not change the canonical regeneration program, as most of the same genes that change their expression in wild-type mice are also differentially expressed in Uhrf1hepKO mice. Figure 4B illustrates this feature: a modified PCA graph of all the samples shows that samples from Uhrf1hepKO mice at 24 hours after PH cluster with controls at this same time point, but at 30 hours after PH, the Uhrf1hepKO samples are differentiated from controls collected at the same time point. Similarly, at 96 hours after PH, the pattern of gene expression in controls has largely returned to baseline, while in Uhrf1hepKO mice, it has not. Second, while the identity of the genes that change during regeneration is largely similar between controls and Uhrf1hepKO, the major difference in gene expression is the kinetics of when these genes are induced or repressed. The program of genes that are differentially expressed during liver regeneration in wild-type mice is shifted so that they show an earlier and more sustained activation in Uhrf1hepKO mice (Figure 4B). We next asked whether the changes in gene expression in Uhrf1hepKO livers were distributed in any of the clusters identified during wild-type liver regeneration and found that Cluster 6 was significantly enriched in this dataset (Figure 4C), displaying earlier and stronger activation in the regenerating livers of Uhrf1hepKO mice (Figure 4D).
GO and gene set enrichment analysis (GSEA) of all DEGs between controls and Uhrf1hepKO mice across time points identified cell cycle and proliferation as the most affected biological processes in Uhrf1hepKO mice (Figure 5A, Figure S3A). Specifically, the cell cycle regulatory network essential for liver regeneration in Uhrf1hepKO mice was activated in a way that would promote accelerated entry into the cell cycle following PH. Most strikingly, Uhrf1hepKO hepatocytes enter mitosis earlier than controls (Figure 5B) and have a faster recovery of liver mass following PH (Figure 5C). Importantly, at 40 hours post-PH, Uhrf1hepKO mice displayed 26% faster recovery of liver/body ratio than wild-type. This was comparable to changes reported in other models where liver regeneration is enhanced (Apte et al., 2009; Shang et al., 2016). Unlike these other models, however, enhanced liver/body ratio was still detectable in Uhrf1hepKO mice at 4 weeks after PH (Figure 5C). Thus, UHRF1 loss accelerates cell cycle entry and enhances liver regeneration.
Figure 5: Increased expression of cell cycle genes and enhanced liver regeneration in Uhrf1hepKO mice.
(A) GO DAVID analysis of all genes differentially expressed between control and Uhrf1hepKO livers post-PH. Circles represent GO categories that are most significantly enriched with y-axis being 1/P-value and size of circle proportional to number of genes in each category. Pink and cyan circles represent categories that are enriched when only genes that are overexpressed or repressed in Uhrf1hepKO livers are analyzed, respectively. (B) Densitometric quantification of western blots comparing pH3 levels between control and Uhrf1hepKO livers post-PH (N=3 mice per genotype per time point), inset showing Western blot at the 36 hr time point. (C) Liver-body weight ratio of the intact, non-resected liver lobes in control and Uhrf1hepKO mice post-PH normalized to quiescent livers of the same genotype (black = control, red = Uhrf1hepKO, N=3–5 mice per time point). P-values calculated by two-way ANOVA followed by Sidak’s multiple comparison tests. Error bars represent s.d. See also Figure S3.
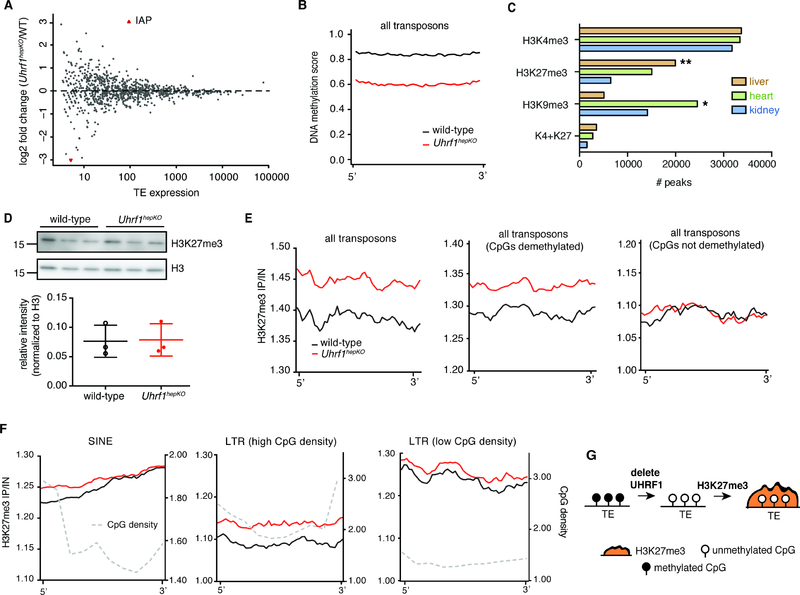
We hypothesized that DNA hypomethylation could underlie the regenerative phenotype in Uhrf1hepKO mice and investigated whether the effect of DNA methylation loss on TEs could be involved. Our previous work showed that loss of uhrf1 in zebrafish led to massive up-regulation of TEs (Chernyavskaya et al., 2017). To our surprise, RNA-seq analysis from libraries generated using RiboZero to include all RNA species only detected a single member of the IAP family of TEs - IAP-d-int (Figure S3A)- that was overexpressed in the quiescent Uhrf1hepKO liver (Figure 6A), despite a widespread loss of DNA methylation across all TEs (Figure 6B, Figure S3B). This suggests that another mechanism served to prevent TE activation when DNA methylation is stripped from them. Recent reports suggest cooperation between repressive epigenetic marks when DNA methylation is abrogated (He et al., 2019; Sharif et al., 2016; Walter et al., 2016), thus we investigated other epigenetic mechanisms that could compensate for the loss of DNA methylation on TEs. Comparison of Encyclopedia of DNA Elements (ENCODE) data for the liver, heart and kidney (Figure 6C) revealed that histone 3 lysine 27 trimethyl (H3K27me3) is the predominant repressive mark in the liver, while H3K9me3 dominates the epigenetic landscape in the heart and kidney (Yue et al., 2014). We confirmed this by our own ChIP-seq analysis of quiescent wild-type livers, where we found much higher H3K27me3 enrichment at all gene promoters compared to H3K9me3 (Figure 6D). In most cases, H3K27me3 was inversely correlated with H3K4me3, a mark associated with active genes (Figure S3C), but there were over 1000 genes that were bivalently marked with both (Figure S3D). These bivalent genes were expressed at low levels, comparable to genes marked with only H3K27me3 (Figure S3E). We asked whether any of the DEG clusters that characterize gene expression during liver regeneration were preferentially marked by H3K27me3 and H3K4me3. We found that genes in cluster 6 had the highest level of both H3K27me3 alone (Figure S3F) and bivalent marked (K4me3 + K27me3) genes (Figure S3G–H). These bivalently marked genes could be poised for activation.
Figure 6: Epigenetic compensation in Uhrf1hepKO mice protects against TE activation.
(A) Transposon expression in quiescent control and Uhrf1hepKO livers detected by RNA-seq with Ribo-Zero, red dots = FDR < 0.05. (B) Average DNA methylation enrichment profiles for control and Uhrf1hepKO mouse livers at all eRRBS mapped TEs. (C) Overlap of H3K27me3, H3K9me3, H3K4me3, and bivalent (K4+K27) peaks in mouse liver, heart, and kidney taken from the ENCODE database. (P < 2.2×10−16, by Chi-squared test with * and ** depicting the two groups that contributed the most to significance by residual calculations). (D) Liver H3K27me3 and H3K9me3 enrichment at promoters of all reference genes separated into 2 groups based on k-means clustering of enrichment scores to segregate genes that have high (top) and low (bottom) H3K27me3 occupancy surrounding the TSS. (E) Total H3K27me3 in quiescent control and Uhrf1hepKO mice as detected by Western blot with quantification by densitometry shown on the bottom (N=3 mice per group). (F) Average H3K27me3 ChlP-seq enrichment profiles for control and Uhrf1hepKO mouse livers at all eRRBS mapped TEs, those with demethylated CpGs, and those with no change in CpGs. (G) Average H3K27me3 ChIP-seq enrichment profiles for all eRRBS mapped IAP, DNA, LINE, SINE, and LTR TEs with high or low CpG density. CpG densities are also shown by grey dotted line. Since TEs are of unequal length and not always highly conserved in sequence, we created “metagenes” for each TE family that divided TEs into 40 equal bins for DNA methylation analysis and 50 equal bins for H3K27me3 enrichment analysis (default parameters) and plotted mean values for each bin. (H) Model depicting epigenetic compensation by H3K27me3 to repress transposons that lose DNA methylation. Error bars represent s.d. See also Figure S3.
We next examined H3K27me3 and H3K9me3 occupancy on transposons and found significantly higher levels of H3K27me3 on TEs in Uhrf1hepKO livers (Figure 6F), while no changes were seen with H3K9me3 (Figure S3I). Strikingly, H3K27me3 was only increased at hypomethylated TEs in Uhrf1hepKO mice but not at TEs in RMDs (Figure 6F). Second, H3K27me3 in Uhrf1hepKO livers was largely redistributed to those hypomethylated TEs with high CpG density, such as in the 5’ region of short interspersed nuclear elements (SINEs) and across CpG dense long-terminal repeats (LTRs), but not on TEs with low CpG density (Figure 6G). Notably, H3K27me3 was not as enriched on lAPs compared to other TEs (Figure 6G), possibly explaining the derepression of lAP-d-int (Figure 6A, Figure S3A). This was not due to a change in global H3K27me3 levels in Uhrf1hepKO livers (Figure 6G), suggesting that H3K27me3 is redistributed to CpG rich, hypomethylated TEs as a mechanism to compensate for their lack of repression by DNA methylation (Figure 6H).
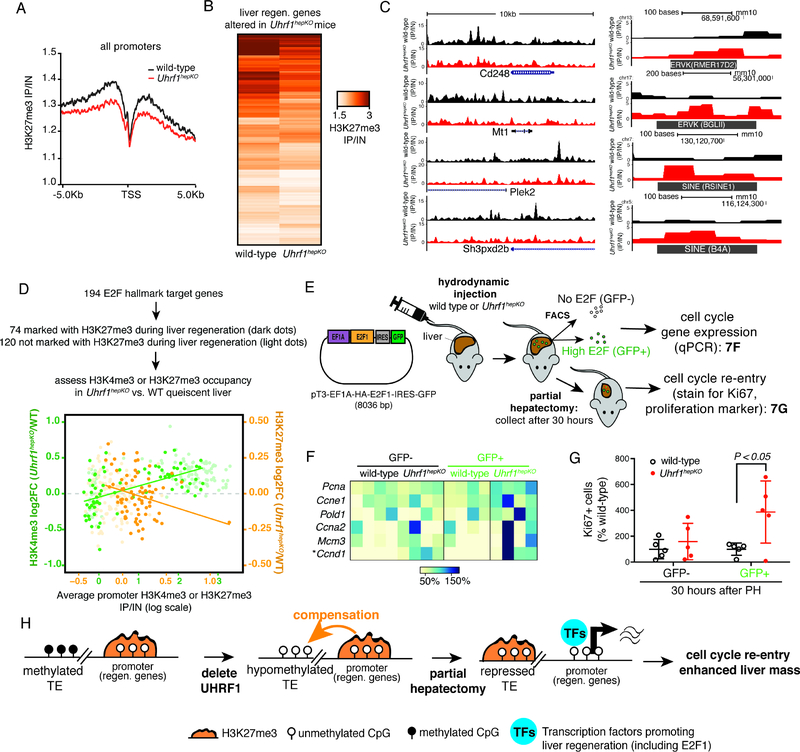
The increase in H3K27me3 occupancy of hypomethylated TEs in Uhrf1hepKO livers was accompanied by a reduction of H3K27me3 at gene promoters (Figure 7A), and was most prominent for those promoters which had the highest level of H3K27me3 promoter occupancy in controls, such as the cluster 6 genes that were marked by H3K27me3, either alone or bivalently with H3K4me3 (Figure S4A–D). Of these, we identified 201 genes occupied by H3K27me3 at promoters in wild-type mice which showed a pattern of being differentially expressed between wild-type and Uhrf1hepKO at any time point during liver regeneration (Figure 7B). Uhrf1 deletion resulted in lower occupancy of H3K27me3 in the promoters of many of these genes (Figure 7B–C, Table S5), in contrast to TEs, which gained this same mark (Figure 6F, 7C). Our results suggest that UHRF1 loss results in redistribution of this repressive mark from the promoters of genes that are required for regeneration. Similar analyses for H3K9me3 revealed no global changes at gene promoters between wild-type and Uhrf1hepKO livers (Figure S4A).
Figure 7: Reprogramming of the epigenome via the loss of UHRF1 primes cell cycle genes for expression.
(A) Average H3K27me3 ChIP-seq enrichment plots for all gene promoters in control and Uhrf1hepKO livers. (B) Comparison of H3K27me3 enrichment scores for 201 genes that are marked with H3K27me3 and are differentially expressed between control and Uhrf1hepKO livers during regeneration. (C) UCSC genome browser screenshots of 5 genes from (B) that lost H3K27me3 in the promoter region and 5 TEs that gained H3K27me3. (D) Control versus Uhrf1hepKO liver H3K27me3 (orange) and H3K4me3 (green) enrichment at promoters of 74 H3K27me3-regulated (darker dots) and 120 not H3K27me3-regulated (lighter dots) E2F target genes. Lines represent line-of-best-fit based on 74 H3K27me3-regulated E2F target genes. (E) Cartoon describing the hydrodynamic injection of a plasmid encoding E2F1-IRES:GFP into control and Uhrf1hepKO mice. This was followed by FACS sorting quiescent livers for GFP positive and negative cells, and qPCR for E2F1 target genes or PH and then immunofloruescene staining of the cell cycle marker, Ki67. (F) Expression of E2F1 target genes in GFP+ hepatocytes from hydrodynamic injected control and Uhrf1hepKO mice measured by qPCR, normalized to maximum expression as 100% (N=4 mice per genotype, P < 0.0001 for the effect of genotype by two-way ANOVA, “*” P < 0.001 for subsequent Sidak’s comparisons between wild-type and Uhrf1hepKO livers sorted for GFP- (negative for E2F1 expression) and GFP+ (E2F1 overexpressing) hepatocytes. (G) Percent of GFP+ cells that are also Ki67+ in the liver of mice that had hydrodynamic tail vein injection and collected 30 hours after PH (N=3). P-value calculated by two-way ANOVA followed by Sidak’s multiple comparison tests. (H) Model showing that epigenetic compensation by H3K27me3 to repress hypomethylated transposons in Uhrf1hepKO mice enhances liver regeneration. Error bars represent s.d. See also Figure S4, Table S4.
We hypothesized that the altered epigenetic environment in cells lacking UHRF1 primes genes that promote liver regeneration for expression when the relevant transcription factor is present. E2F1 is one of the best characterized transcription factor responsible for activating cell cycle gene expression during cell proliferation (van den Heuvel and Dyson, 2008). We compared levels of H3K27me3 and H3K4me3 enrichment of the GSEA dataset representing 194 E2F target genes (HALLMARK_E2F_TARGETS) and then carried out ChIPseq of H3K27me3 in wild-type livers at 30, 40 and 96 hours after PH to identify those E2F1 target genes that dynamically changed this mark (Figure S4E). Interestingly, in Uhrf1hepKO quiescent livers, many of these genes had reduced H3K27me3 and elevated H3K4me3 occupancy (Figure 7D), suggesting that Uhrf1 deletion results in a more permissive chromatin environment for E2F target genes relevant to liver regeneration. To test this, we performed hydrodynamic tail vein injection (Zhang et al., 1999) to deliver a plasmid encoding human E2F1 co-transcribed with an IRES-GFP into control and Uhrf1hepKO mice. GFP positive cells which were effectively transduced with this plasmid were assessed for E2F target gene expression and the cell proliferation marker, Ki67 (Figure 7E). We found that following HDT injection, E2F1 target genes were expressed at higher levels in Uhrf1hepKO quiescent livers compared to controls (Figure 7F); however, it was not enough to trigger cell division as detected using Ki67 staining (not shown). We therefore provided additional mitogenic stimulus by carrying out PH on these injected mice and found that there was a significant increase in Ki67 staining in E2F1-GFP transduced hepatocytes in Uhrf1hepKO livers compared to GFP- cells or to GFP+ cells in livers of control mice (Figure 7G). This demonstrates that E2F1 action is facilitated by Uhrf1 deficiency, and suggests that the permissive epigenetic landscape on E2F1 target genes primes them for induction to promote regeneration.
Discussion
Most regenerating tissues in mammals rely on stem cells, but the liver represents an exception to this paradigm. Under most circumstances, differentiated hepatocytes proliferate to restore liver mass following resection or injury. The switch from quiescence to regenerating hepatocyte does not evoke gene expression changes associated with “stem-ness”, but instead relies on having a facile program enabling a rapid switch to a pro-regenerative gene expression program (Otu et al., 2007; Yanger et al., 2014). The hypothesis that epigenetics facilitates these changes is supported by previous studies focused on the SWI/SNF family of chromatin remodelers and their interactions with liver master transcription factors such as CEBPα and CEBPβ (Iakova et al., 2003; Jakobsen et al., 2013; Jin et al., 2010; Orellana et al., 2010; Sun et al., 2016; Wang et al., 2008a, b). Here, we took an unbiased approach by a detailed time-course study of transcriptomic changes during liver regeneration. This uncovered UHRF1, a gene we have previously shown in zebrafish to be involved in DNA methylation, transposon suppression, and liver development (Feng et al., 2010; Jacob et al., 2015; Kent et al., 2016; Sadler et al., 2007), which we report here is critical for regulating the epigenome and liver regeneration by mediating the cross talk between multiple epigenetic marks.
We found that UHRF1 loss in the liver caused DNA hypomethylation, as predicted by work in many other systems (Bostick et al., 2007; Feng et al., 2010; Ohno et al., 2013; Sharif et al., 2007). However, to our surprise, this did not lead to massive TE activation or activate an immune response to mitigate the unleashing of these endogenous parasites as we found in zebrafish (Chernyavskaya et al., 2017). Instead, we uncovered an additional layer of protection against TE expression encoded in the repressive histone code (Figure 7H): when DNA methylation is stripped from TEs, H3K27me3 is redistributed from the promoters of genes required for liver regeneration to hypomethylated TEs. This is consistent with findings from cultured cells, where H3K27me3 compensates to repress a subset of TEs during extended culture with DNA hypomethylation (Cooper et al., 2014; Ohtani et al., 2018; Walter et al., 2016) and work that points to a preferential occupancy of non-methylated CpG islands by H3K27me3 (Blackledge et al., 2014; Farcas et al., 2012; Hagarman et al., 2013; Jermann et al., 2014; Li et al., 2017; Mendenhall et al., 2010). In the case where tens of thousands of CpG-rich TEs are unmasked by DNA hypomethylation - either by manipulating UHRF1 (Chernyavskaya et al., 2017; Mudbhary et al., 2014; Ramesh et al., 2016; Sharif et al., 2016) or other mechanisms leading to DNA hypomethylation such as those found in cancer or autoimmune disease (Chiappinelli et al., 2015; Roulois et al., 2015; Volkman and Stetson, 2014) - H3K27me3 redistribution could be accounted for simply by competition for the overwhelming number of potential binding sites compared to the relatively few promoters in the genome. The resulting reduction of H3K27me3 at promoters of cell proliferation genes poised for activation in the quiescent liver creates a favorable epigenetic environment for subsequent activation during liver regeneration and enhanced liver regeneration kinetics (Figure 7H). The recent report that E2F recruits EZH2 to repetitive regions to silence TE expression (Ishak et al., 2016) raises the interesting but untested question of whether E2F1 in Uhrf1 deficient cells may have a role in repositioning H3K27me3 to repetitive sequences. Future studies with E2F1 transduced cells will enable us to address this.
The finding that the livers of Uhrf1hepKO mice contain a globally hypomethylated genome yet appear healthy is interesting and unexpected. This is different from observations made in embryonic stem cells and some other adult tissues where the lack of UHRF1 led to genomic instability and cell death (Maenohara et al., 2017; Obata et al., 2014; Ramesh et al., 2016; Xiang et al., 2017; Yamashita et al., 2018; Zhao et al., 2017). We observed Uhrf1hepKO mice for up to 18 months and detect no phenotypic anomalies or spontaneous tumor formation (SW, unpublished). This could be attributed to tissue specific differences where the adult liver is known to more tolerable towards genomic anomalies and represents one of the few tissues where majority of cells exist in the polyploid state (Duncan et al., 2010). In addition, Uhrf1hepKO mice are markedly different from the dramatic liver injury reported for mice with Dnmt1 deficient hepatocytes (Kaji et al., 2016). Since DNA methylation of TEs was not fully explored and the status of other epigenetic marks is not known in the Dnmt1 deficient model, direct comparison to our study is difficult. In particular, we report genomic regions that remain fully methylated in the absence of Uhrf1 (Figure 3F). These RMRs may represent regions of active de novo methylation by Dnmt3a/b or Uhrf1-independent methylation activities of Dnmt1, and the latter could also serve as explanation for the dramatic phenotypic differences between Uhrf1 and Dnmt1 deficient mice. It is also possible these represent residual methylation from other liver cell types that are not Uhrf1 deficient.
Our observation of epigenetic compensation in vivo with a significant impact on liver regeneration extends previous observations from cultured cells and may be therapeutically relevant, as drugs that modify the DNA methylome are clinically available. Future studies should test whether these drugs can induce epigenetic compensation in vivo as means to augment regenerative capacity in animal models of liver disease and liver failure (Campbell and Tummino, 2014). It is becoming increasingly clear that no direct correlation exists between promoter DNA méthylation and gene expression. Further exploration of this phenomenon of epigenetic compensation may better link DNA methylation changes to gene expression changes in both experimental and clinical settings.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kirsten Sadler Edepli (kirsten.edepli@nyu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice maintenance and experiments were approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (IUCUC). Temperature, humidity, and light:dark cycles were controlled and mice were fed food and water ad libitum.
Since Uhrf1 knockout mice are embryonic lethal (Muto et al., 2002), we deleted Uhrf1 specifically in hepatocytes by generating a floxed Uhrf1 allele (Uhrf1fl/fl). The flox sites were engineered to flank exons 6–10 so that when the loxP sites are recombined, it creates a stop codon in exon 6, deleting the nuclear localization signal and all the domains required for DNA methylation (FIgure S2). Mice with hepatocyte-specific deletion of the Uhrf1 gene (Uhrf1fl/fl;Alb-Cre; referred to as Uhrf1HepKO mice) were generated by crossing mice homozygous for floxed Uhrf1 (Uhrf1fl/fl) with mice expressing the Cre recombinase under the liver-specific albumin promoter backcrossed onto the Uhrf1fl/fl background (Alb-cre Tg/+; Uhrf1 fl/+).
Unless otherwise specified in the results section, male mice on a congenic C57Bl/6 background were used between 8 to 12 weeks of age. Partial hepatectomy (PH) surgery was carried out following the protocols described in Mitchell et al. to remove 70% of liver mass (Mitchell and Willenbring, 2008). Briefly, surgery was performed on pairs of Uhrf1HepKO males with littermates WT controls (mice with one or two copies of the floxed Uhrf1 targeting vector but no Cre transgene). All mice were anesthetized before surgery and then their left and median lobes removed. All surgery were performed between 8am-12pm to control for circadian effects. At 24, 30, 40, 48, and 96 hours, 7 days, and 4 weeks following surgery, different mice were sacrificed and their livers were collected, flash frozen in liquid nitrogen, and stored at −80°C for subsequent analysis. Liver samples were also fixed in 10% formalin and then paraffin- or OCT - embedded for immunohistochemistry or immunofluorescence, respectively.
Hydrodynamic injections experiments were carried out on pairs of Uhrf1HepKO males with littermates WT controls (mice with one or two copies of the floxed Uhrf1 targeting vector but no Cre transgene) at 6–8 weeks old, with some of these mice subjected to partial hepatectomy at 72 hours post injection.
For eRRBS studies, we used 6 to 8 week old male mice on a mixed undefined background that include C57BL/6J which we previously reported to have essentially identical DNA methylation pattern as 6 to 8 week old C57BL/6J males (Zhang et al., 2016).
METHOD DETAILS
Genotyping
Mouse tail clips were sent to Transnetyx Inc. for genotyping using primers listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-H3K4me3 | Abcam | Cat#ab1012 |
| Mouse monoclonal anti-H3K27me3 | Active Motif | Cat#61017 |
| Rabbit polyclonal anti-H3K9me3 | Active Motif | Cat#39161 |
| Mouse monoclonal anti-5MeC | Eurogentec | Cat#BI-MECY-100 |
| Mouse monoclonal anti-dsDNA | Abcam | Cat#ab27156 |
| Rabbit polyclonal anti-UHRF1 | Bonapace lab | PMID:12058012 |
| Rabbit monoclonal anti-DNMT1 | Cell Signaling | Cat#D63A6 |
| Mouse monoclonal anti-PCNA | Sigma | Cat#p8825 |
| Rabbit polyclonal PCNA | Santa Cruz | Cat#SC7907 |
| Rabbit polyclonal anti-pH3 | Santa Cruz | Cat#sc-8656-R |
| Rabbit polyclonal anti-H3 | Sigma | Cat#H0164 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#ab15580 |
| Chicken polyclonal anti-GFP | Abcam | Cat#ab13970 |
| Bacterial and Virus Strains | ||
| pCMVHA-E2F1 | Addgene | plasmid # 24225 |
| pT3-EF1a-NRAS-IRES-GFP | Dr. Scott Lowe | N/A |
| CMV-SB13 | Dr. Scott Lowe | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 10% neutral buffered formalin | Sigma | Cat#HT501128 |
| Collagenase B | Roche | Cat#11088831001 |
| Micrococcal nuclease | Fisher Scientific | Cat#70196Y 15ku |
| Magna ChIP Protein A+G Magnetic beads | Millipore | Cat#16–663 |
| Ampure XP beads | Beckman Coulter | Cat#A63880 |
| Propidium iodide | Sigma | Cat#P4864 |
| Critical Commercial Assays | ||
| Zymo ZR-Duet™ DNA/RNA MiniPrep Kit | Zymo | Cat#D7003 |
| Quantabio qScript cDNA SuperMix | Quantabio | Cat#95048–025 |
| SYBR green | ThermoFisher | Cat#4309155 |
| TruSeq RNA Library Prep Kit v2 | Illumina | Cat#RS-122–2001 |
| In-Fusion® HD Cloning Plus | Takarabio | Cat# 638910 |
| Deposited Data | ||
| RNAseq | This paper | GSE125006 |
| ChIPseq | This paper | GSE125006 |
| ERRBS | This paper | GSE125006 |
| Experimental Models: Organisms/Strains | ||
| Uhrf1fl/fl mice | This paper | N/A |
| Tg(Alb:Cre) mice | The Jackson Laboratory | stock #016832 |
| Oligonucleotides | ||
| GENOTYPING: Uhrf1FloxedAllele F CCAGACAACAAGAACAGAGACACTT | This paper | N/A |
| GENOTYPING: Uhrf1FloxedAllele R GGAACTTCGGAATT CGAT AT CAAG CT | This paper | N/A |
| GENOTYPING: Uhrf1WTAllele F CCAGACAACAAGAACAGAGACACTT | This paper | N/A |
| GENOTYPING: Uhrf1WTAllele R CT CAGAG CAATTTT CCTTAT AAAAT CAAGACTT ATT | This paper | N/A |
| GENOTYPING: CreTransgene F TTAATCCATATTGGCAGAACGAAAACG | This paper | N/A |
| GENOTYPING: CreTransgene R CAGGCTAAGTGCCTTCTCTACA | This paper | N/A |
| QPCR: Uhrf1_Forward CTAGCAGCTGGAAGGAACCC | This paper | N/A |
| QPCR: Uhrf1_Reverse GCCGATGTACTCTCTCACGG | This paper | N/A |
| QPCR: Cre_Forward CGACCAGGTTCGTTCACTCA | This paper | N/A |
| QPCR: Cre_Reverse CAGCGTTTTCGTTCTGCCAA | This paper | N/A |
| Software and Algorithms | ||
| FastQC | Barbraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| Trimmomatic | Bolger et al. 2014 | https://github.com/timflutre/trimmomatic |
| MFuzz | Kumar & Futschik, 2007 | http://mfuzz.sysbiolab.eu |
| MethylKit | Akalin 2012 | https://bioconductororg/packages/release/bioc/html/methylKit.html |
| BWA-MEM | Li and Durban 2010 | https://github.com/lh3/bwa |
| PICARD tools | Broad Institute | http://broadinstitute.github.io/picard/ |
| Deeptools2 | Richter et al., 2016; Afgan et al., 2016 | http://deeptools.ie-freiburg.mpg.de/ |
| GO DAVID | Huang et al. 2008 | DAVID 6.8 |
| GSEA | Subramanian et al. 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| GeneOverlap | Li 2013 | https://www.bioconductor.org/packages/release/bioc/html/GeneOverlap.html |
| Genomation | Akalin et al. 2015 | https://bioconductororg/packages/release/bioc/html/genomation.html |
| Bioquant | BIOQUANT | https://bioquant.com/ |
| Prism 6 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
Animal procedures
Partial Hepatectomy
Partial hepatectomy were carried out as described by Mitchell et al. (Mitchell and Willenbring, 2008). Mice were anaesthetized using isoflurane. A small incision was made in the abdomen to open the peritoneal cavity. Two cuts were made to free the liver from the falciform ligament and the membrane that links the caudate and the left lateral lobe. 4–0 silk thread (Ethicon, SA10) is placed at the base of the left lobe and a knot is tied before the left lobe is resected. 4–0 silk thread is then placed and tied around the median lobe right above the gall bladder, followed by resection to remove 2/3 of the median lobe. Finally the peritoneal was closed using 5–0 suture (Ethicon, JV389) and the skin was closed using wound clips (BrainTree Scientific, EZC APL). For measuring the weight of the regenerating liver following surgery, only the intact, non-resected lobes were measured to improve accuracy and consistency between different animals.
Isolation of Primary Hepatocytes
Mouse primary hepatocytes were isolated using a two-step collagenase liver perfusion method. Mice were anaesthetized using Ketamine(100 mg/kg, ) / Xylazine (10 mg/kg, ). Following opening of the abdomen, perfusion solutions warmed to 37°C were pumped through a catheter placed into the portal vein and drained out the inferior vena cava. Mice were perfused with 25 mL of Liver Perfusion Medium (Invitrogen, 17701038) followed by 25 mL of Hepatocyte Wash Medium (Invitrogen, 17704024) supplemented with 0.05% collagenase B (Roche, 11088831001). The liver was then excised and its capsules were torn to release hepatocytes. The isolated hepatocytes were then filtered through 100μM cell strainers (BD Biosciences, 352360), washed 3 times with Hepatocyte Wash Medium by centrifuging at 50 × g and then finally resuspended in 1%BSA PBS with 0.1% propidium iodide (Sigma, P4864) and sorted using FACS (BD Biosciences, FACSAria). For FACS, hepatocytes were gated by size using FSC/SSC and live/dead using propidium iodide staining followed by GFP fluorescence into GFP+ versus GFP- live hepatocytes directly into TRIzol (ThermoFisher, 15596026).
Hydrodynamic Injection
To generate the pT3-EF1a-E2f1-IRES-GFP vector, the pT3-EF1a-NRAS-IRES-GFP plasmid digested with XhoI and EcoRIrestriction enzymes was used as a donor vector. The “E2f1” sequence was PCR-amplified from the pCMVHA-E2F1 vector (Addgene plasmid # 24225) and the cloning of the fragments into the donor vector was performed by In-Fusion cloning (Takara Bio, 638910). The CMV-SB13 and pT3-EF1a-NRAS-IRES-GFP were kindly provided by Dr. Scott Lowe (MSKCC, New York). The pCMVHA-E2F1 vector was a gift from Kristian Helin (Addgene plasmid # 24225, (Lukas et al., 1996)). All constructs were verified by nucleotide sequencing and vector integrity was confirmed by restriction enzyme digestion. The vectors will be made available through Addgene.
A sterile 0.9% NaCl solution/plasmid mix was prepared containing DNA. We injected 30 μg of pT3-EF1a-E2f1-IRES-GFP and a 4:1 ratio of transposon to SB13 transposase-encoding plasmid dissolved in 2 ml of 0.9% NaCl solution. Mice were injected with the 0.9% NaCl solution/plasmid mix into the lateral tail vein with a total volume corresponding to 10% of body weight in 5–7 seconds. Vectors for hydrodynamic delivery were produced using the QIAGEN plasmid PlusMega kit (QIAGEN, 12981). Equivalent DNA concentration between different batches of DNA was confirmed to ensure reproducibility among experiments.
RNA and DNA extraction
RNA were isolated from FACS sorted hepatocytes using TRIzol (Invitrogen, 15596026). RNA and genomic DNA were isolated from liver tissues stored at −80°C by first homogenizing using a dounce homogenizer in DNA/RNA shield (provided by Zymo ZR-Duet™kit), incubated with protease K (provided by kit) for 30 min at 55°C, then column extracted using the Zymo ZR-Duet™ DNA/RNA MiniPrep kit following the manufacture’s instructions for silicon column-based RNA and gDNA extraction.
For eRRBS, total livers were isolated from mice between 6–8 weeks old and frozen at −80°C for DNA isolation. Genomic DNA was extracted from liver samples using Qiagen DNeasy kit according to the manufacturer’s instruction.
Quantitative reverse-transcription PCR (qPCR)
RNA samples were reverse-transcribed into cDNA using Quantabio qScript cDNA SuperMix (Quantabio, 95048–025). QPCR was performed using SYBR green (ThermoFisher, 4309155) on a LightCycler 480 (Roche, 05015243001) with primers listed in Key Resources Table. Samples ran in triplicates and relative expression was quantified by calculating ΔΔCt values against Gapdh, the endogenous control.
Gene expression profiling
RNA-seq libraries were prepared with PolyA capture unless otherwise stated (i.e. with Ribo-Zero for TE analysis). RNA-seq libraries were prepared according to the Illumina TruSeq RNA sample preparation version 2 protocol. RNA from whole liver was used to generate libraries, which were analyzed on an Agilent 2100 Bioanalyzer. Sequencing quality was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and the reads were quality trimmed using Trimmomatic (Bolger et al., 2014) to remove low Q scores, adapter contamination and systematic sequencing errors. 100 base pair single-end reads that passed quality control were aligned to reference genome GRCm38.p4. Overall alignment rates were above 95%. After alignment, read counts were generated using HTseq count and the counts were analyzed using the DESeq2 (Love et al., 2014) for differential gene expression. Quality control of results from the differential gene expression analysis was carried out by looking at alignment rates, coverage, and clustering of replicates. Three biological replicates were adopted for all experiments and replicates out of clustering were removed for analysis. For multiple testing we applied Benjamin-Hochberg correction in DEseq2 and treated adjusted p-values less 0.05 as statistically significant. Significant DEG compared to baseline were clustered to reveal the expression pattern across different time points with a unsupervised learning method which is implement by the R package ‘Mfuzz’ (Kumar and E Futschik, 2007).
DNA methylation profiling
ERRBS was performed on whole liver gDNA following published protocols (Garrett-Bakelman et al., 2015; Smith et al., 2009). Bisulfite-converted DNA sequencing libraries were generated for each of the studied samples, which measure both 5hmC and 5mC in methylated fraction and 5fC, 5caC, and C in unmethylated fraction. In brief, 50 ng of high quality genomic DNA in 50 μl of DNase-free water was used as starting material. The whole library preparation includes enzyme digestion of genomic DNA which enriched CpG rich regions, phenol:chloroform clean up, end-repair, A-tailing, adapter ligation, size selection, bisulfite conversion, enrichment PCR, and quality control. These amplified libraries were sequenced on the Hiseq2000 platform for 50 cycles single end read runs at the Epigenomics Core facility in the Department of Medicine, Weill Cornell Medical College (New York, NY, USA). Image capture, analysis, and base calling were performed using Illumina’s CASAVA 1.8. Differentially methylated bases were detected with logistic regression and SLIM method which is implemented in an R package ‘methylKit’ (Akalin et al., 2012). CpGs with methylation level below 0.1 were treated as unmethylated. Differentially methylated CpGs are defined as having methylation differences larger than 0.25 and q-value < 0.05. Since transposons are of unequal length and not always highly conserved in sequence, we created “metagenes” for each transposon family that divided transposons into 40 equal bins for analysis and plotted mean values for each bin.
ChIP-sequencing
For isolation of nuclei from flash-frozen liver tissue, 100–200 mg of frozen tissue were cut into ~ 10 mg chunks on ice and then homogenized in ice-cold buffer I (0.32 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris, pH 7.5, 1:1,000 protease inhibitor cocktail (Roche, 11697498001)) using a Dounce homogenizer until nuclei is released into solution and no visible tissue chunks remain (5–6 slow plunges). Subsequent micrococcal digest, chromatin immunoprecipitation, and library preparation were carried out exactly as described (Alonso et al., 2018). Antibodies used are anti-H3K4me3 (Abcam, ab1012), anti-H3K27me3 (Active Motif, 61017), and anti-H3K9me3 (Active Motif, 39161). 75 base pair single-end or 100 base pair paired-end reads that passed quality trimming were then aligned against the mouse reference genome (GRCm38.p4) using BWA-MEM (H. Li and Durbin, 2010). The resulting BAM alignments were then processed through PICARD tools (to clean, sort, and deduplicate (PCR and Optical duplicates)) and exported to the Galaxy/Deeptools2 (version 2.5.0.0, (Richter et al., 2016)) public server at http://deeptools.ie-freiburg.mpg.de/ (Afgan et al., 2016). bamCompare 2.5.1 in Deeptools2 was used to generated the ratio of each histone IP to input control with default parameters. Histone enrichment plots and heatmaps were generated by using Deeptools2 computeMatrix with default parameters followed by plotEnrichment or plotHeatmap with K means clustering into 2 groups. Details of the ChIP-seq experiments are shown here:
| SAMPLE NAME | MAPPED READS | ALIGNMENT RATE | LIBRARY PREP (Single-End/Paired-End) |
|---|---|---|---|
| Input (round 1) | 126,305,490 | 99.56% | SE |
| baseline WT H3K27me3 | 82,830,166 | 99.58% | SE |
| baseline Uhrf1hepKO H3K27me3 | 72,476,182 | 99.60% | SE |
| baseline WT H3K4me3 | 47,584,177 | 99.28% | SE |
| Input (round 2) | 189,953,193 | 94.73% | PE |
| baseline WT H3K9me3 (round 2) | 124,010,776 | 90.53% | PE |
| baseline Uhrf1hepKO H3K9me3 (round 2) | 126,087,162 | 89.49% | PE |
| baseline WT H3K4me3 (round 2) | 60,208,280 | 90.77% | PE |
| baseline Uhrf1hepKO H3K4me3 (round 2) | 65,070,982 | 94.12% | PE |
| Input (round 3) | 203,303,611 | 99.96% | PE |
| Regenerating WT H3K27me3 – 0hr (round 3) | 82,659,374 | 93.87% | PE |
| Regenerating WT H3K27me3 – 30hr (round 3) | 67,682,622 | 99.97% | PE |
| Regenerating WT H3K27me3 – 40hr (round 3) | 63,809,278 | 99.96% | PE |
| Regenerating WT H3K27me3 – 96hr (round 3) | 77,672,450 | 99.96% | PE |
Since transposons are of unequal length and not always highly conserved in sequence, we created “metagenes” for each transposon family that divided transposons into 50 equal bins for H3K27me3 enrichment analysis (default parameters in Deeptools2) and plotted mean values for each bin.
Slot blot
1–3 ng of gDNA were denatured in 0.4 M NaOH/10 mM EDTA, neutralized with 2 mM ammonium acetate and loaded in duplicate onto a nitrocellulose membrane using a slot blot apparatus. Membranes were baked at 80°C, blocked with 5% milk, incubated overnight in either anti-5MeC (Eurogentec, BI-MECY-100; 1:2000) or anti-dsDNA as control (Abcam, ab27156; 1:8000), washed in TBST (37 mM NaCl, 20 mM Tris pH 7.5, 0.1% Tween 20), and probed with anti-mouse HRP secondary antibody (Promega, W4021; 1:5000) for 1 h at room temperature followed by development in ECL (ThermoFisher, 32106). Total 5MeC and dsDNA was averaged between technical duplicates and the 5MeC:dsDNA ratio was calculated for at least three livers at each time point and averaged.
Immunoblotting
Livers were collected, snap frozen in liquid nitrogen, and stored at −80°C until processing. Using a Dounce homogenizer, 50 mg of liver was homogenized in 2.5 ml of homogenization buffer (5 mM MgCl2, 50 mM Tris-HCl pH 7.6, 50 mM NaCl, 1 mM EDTA, 5% v/v glycerol, 0.1% w/w Triton X-100, and 0.1% v/v β-mercaptoethanol). Contents were kept on ice at all times. The liver homogenate was centrifuged at 1100 × g for 10 min, pelleted nuclei were resuspended in 500 μL of sonication buffer (150 mM NaCl, 150 mM Tris-HCl, pH 7.4, 1 × protease inhibitors (Roche, 11697498001), and 1 × phosphatase inhibitors (Sigma, 4906845001)). Samples were kept on ice at all times. Cells were lysed via sonication using a probe sonicator (ThermoFisher Scientific Inc) at 25% amplitude with 4 × 5 s bursts on ice. Protein concentrations in cell lysates were determined using the Bradford Protein Assay (Bio-Rad, 5000006). Samples were mixed with SDS-PAGE loading buffer, boiled at 95°C for 10 min, and loaded onto 10% SDS-PAGE gels, electrophoresed, transferred onto nitrocellulose membranes, blocked with 5% w/v powdered milk in TBST buffer (20 mM Tris-HCl, 150 mM NaCl, 0.1% v/v Tween 20, pH 8.0) for 1 hr at room temperature, and incubated with primary antibodies diluted in blocking buffer overnight at 4°C. The next day blots were washed for 3 × 5 min with TBST buffer, incubated with secondary antibodies diluted in blocking buffer, washed for 3 × 10 min, incubated for 2 min with ECL prime, and imaged using Bio-Rad ChemiDoc. Immunoblot bands are quantified by densitometry using GelAnalyzer (http://www.gelanalyzer.com). Antibodies used are anti-UHRF1 (Bonapace lab, 1:1000), anti-DNMT1 (Santa Cruz D63A6, 1:1000), anti-PCNA (Sigma p8825, 1:1000), anti-pH3 (Santa Cruz sc-8656-R, 1: 1000), and anti-H3 (Sigma H0164, 1: 10000) used as loading control.
Histology, immunohistochemistry, and immunofluorescence
Liver tissues were fixed in 10% neutral buffered formalin (Sigma HT501128) overnight and then stored in 70% ethanol until paraffin embedding, sectioning, and hematoxylin and eosin staining at the Mount Sinai Biorepository and Pathology Core (https://icahn.mssm.edu/research/portal/resources/deans-cores/biorepository-and-pathology). Immunohistochemical or immunofluorescence staining for PCNA (Santa Cruz SC7907; 1:500), Ki67 (Abcam ab15580; 1:2500), and GFP (Abcam ab13970; 1:200) were also performed on formalin-fixed, paraffin-embedded (FFPE) liver sections. FFPE sections were deparaffinized with xylene, steamed in 10 mM sodium citrate pH8 for 30 min for antigen retrieval, blocked in 3% BSA in PBS for 10 minutes, then incubated in primary antibody overnight. Next day, the primary antibody was washed off with 3 × PBS washes. From here, immunohistochemical staining (for PCNA, and Ki67) proceed with incubation in HRP conjugated anti-mouse/rabbit secondary antibody (ThermoFisher; 1:200), 3 washes in PBS, incubation with DAB reagent (DAKO K4063), counterstain with hematoxylin (Sigma, HHS16), dH2O washes followed by a final 5 minute incubation in PBS before mounting coverslip in Permount (Fisher Scientific, SP15–500). For immunofluorescence staining (for Ki67 and GFP), sections were incubated with anti-rabbit/chicken secondary antibody coupled with Alexa Fluor Dyes (ThermoFisher; 1:200), washed 3 times in PBS, then mounted with coverslip in Fluoromount-G with DAPI (ThermoFisher, 00–4959-52). Images were taken on a fluorescence microscope (Zeiss Observer 7).
Liver tissues were also frozen in OCT (ThermoFisher, 23–730-571) directly by placing on dry ice and stored at −80°C. Immunofluorescence staining for UHRF1 (Bonapace lab, 1:500) was performed on direct OCT-embedded liver sections. Frozen sections were incubated in methanol at −20°C for 20 min followed by 3 washes in PBS. From this step onwards, frozen sections were blocked and incubated in antibodies the same as described above for immunofluorescence from FFPE sections.
ALT/AST assays
ALT and AST measurements were performed in the clinical core facilities at Mount Sinai using standard clinically established protocols (https://icahn.mssm.edu/about/departments/pathology/diagnostic).
QUANTIFICATION AND STATISTICAL ANALYSIS
For quantification of immunohistochemistry and immunofluorescence, samples were de-identified and quantified in a blind fashion by experimenters. Morphometry was used to quantify the number of nuclei with Bioquant image analysis software, which allowed for standardized quantification criteria for all samples from each experiment (Bioquant Image Analysis Corporation, Nashville, TN, USA).
All statistical tests, error bars, and N numbers are reported in the corresponding figure legends with P-values indicated in the figure when significant. Researchers were blinded to genotype during the acquisition of data. Sample sizes were not pre-determined, but were chosen based on previous publications. To address randomness, any available (mutant or control) mice were included in the study and no mice were excluded. One-way, two-tailed analysis of variance tests (ANOVA), followed by post-hoc tests were computed in Prism 6 (GraphPad). Chi-squared test and other more sophisticated statistical analysis were performed in R.
DATA AND SOFTWARE AVAILABILITY
GO DAVID analysis were carried out using DAVID 6.8 using default parameters for function annotation (Huang et al., 2008). GSEA (gene set enrichment analysis) was carried out using default parameters (Subramanian et al., 2005). The R package GeneOverlap was used to overlap gene sets and calculate significance of overlap (S. Li, 2013). The R package Genomation was used to annotate CpGs and H3K27me3 peaks into different genomic elements (Akalin et al., 2015).
Supplementary Material
Table S1. Related to Figure 1. 7006 unique genes that were differentially expressed compared to baseline at any time point during wild-type mouse liver regeneration.
Table S2. Related to Figure 1. 6 unique clusters of wild-type regeneration genes that display distinct temporal expression patterns as determined by unsupervised clustering.
Table S3. Related to Figure 4. DEG between wild-type and Uhrf1hepKO regenerating livers at each time point following partial hepatectomy.
Table S4. Related to Figure 7. All 201 genes differentially expressed between wild-type and Uhrf1hepKO regenerating livers and occupied by H3K27me3 at promoters. 74 HALLMAKR_E2F_TARGET genes that are potentially regulated by H3K27me3 during liver regeneration. 120 HALLMAKR_E2F_TARGET genes that are potentially not regulated by H3K27me3 during liver regeneration
Highlights.
Liver regeneration is characterized by distinct patterns of gene expression.
Uhrf1 loss in hepatocytes enhances liver regeneration despite DNA hypomethylation.
H3K27me3 accumulates on hypomethylated transposons in Uhrf1 deficient livers.
Promoters of pro-regenerative genes lose H3K27me3, facilitating their activation.
Acknowledgments
The authors are grateful to Bhavani Madakashira and Marina Ruiz De Galarreta for technical help, to Nizar Drou in the NYUAD bioinformatics core and the NYUAD genomics facility for essential support with sample processing and genomics data, and to Scott Friedman for expert insight and support.
Funding: NIH/NIDDK (5R01DK080789 to KCS and CU, 2R01CA154683 to EB, R37CA230636 to AL) and NYUAD Investigator Funding (to KCS).
Footnotes
Competing interests: Authors declare no competing interests
Data and materials availability: All datasets generated in this study have been uploaded into the GEO database under accession number GSE125006 and will be released to the public upon publication. All code used for analysis is available on github https://github.com/zcmit/Liver-Regeneration.
ADDITIONAL RESOURCES
RNA-seq, ChIP-seq, ATAC-seq, and eRRBS raw data have been deposited at the Gene Expression Omnibus under the ID GSE125006.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Franke V, Vlahovicek K, Mason CE, and Schubeler D (2015). genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 31, 1127–1129. [DOI] [PubMed] [Google Scholar]
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, and Mason CE (2012). MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Bernstein E, and Hasson D (2018). Histone native chromatin immunoprecipitation. in histone variants: methods and protocols, Orsi GA, and Almouzni G, eds. (Springer; New York: ), pp. 77–104. [DOI] [PubMed] [Google Scholar]
- Apte U, Gkretsi V, Bowen William C, Mars Wendy M, Luo JH, Donthamsetty S, Orr A, Monga Satdarshan PS, Wu C, and Michalopoulos George K (2009). Enhanced liver regeneration following changes induced by hepatocyteD specific genetic ablation of integrin□linked kinase. Hepatology 50, 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Y, and Cedar H (2013). DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol 20, 274. [DOI] [PubMed] [Google Scholar]
- Blackledge Neil P., Farcas Anca M., Kondo T, King Hamish W., McGouran Joanna F., Hanssen Lars L.P., Ito S, Cooper S, Kondo K, Koseki Y, et al. (2014). Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Usadel B, and Lohse M (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, and Jacobsen SE (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. [DOI] [PubMed] [Google Scholar]
- Campbell RM, and Tummino PJ (2014). Cancer epigenetics drug discovery and development: the challenge of hitting the mark. J Clin Invest. 124, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyavskaya Y, Mudbhary R, Zhang C, Tokarz D, Jacob V, Gopinath S, Sun X, Wang S, Magnani E, Madakashira BP, et al. (2017). Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development 144, 2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli Katherine B., Strissel Pamela L., Desrichard A, Li H, Henke C, Akman B, Hein A, Rote Neal S., Cope Leslie M., Snyder A, et al. (2015). Inhibiting DnA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, and Feschotte C (2016). Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 18, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge Neil P., De Marco V, Elderkin S, Koseki H, Klose R, et al. (2014). Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I, Fresnedo O, Iglesias A, Rueda Y, Syn W-K, Zubiaga AM, and Ochoa B (2011). A role for transcription factor E2F2 in hepatocyte proliferation and timely liver regeneration. Am J Physiol. 301, G20–G31. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, and Grompe M (2010). The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. (2012). KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 1, e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 107, 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara Y, Hirasawa A, Li X-K, Kawasaki M, Fujino M, Funeshima N, Katsuma S, Shiojima S, Yamada M, Okuyama T, et al. (2003). Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 38, 784–792. [DOI] [PubMed] [Google Scholar]
- Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, Mason CE, Figueroa ME, and Melnick AM (2015). Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. JoVE, e52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagarman JA, Motley MP, Kristjansdottir K, and Soloway PD (2013). Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLOS ONE 8, e53880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Fu X, Zhang M, He F, Li W, Abdul MM, Zhou J, Sun L, Chang C, Li Y, et al. (2019). Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat Commun. 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4, 44. [DOI] [PubMed] [Google Scholar]
- Iakova P, Awad SS, and Timchenko NA (2003). Aging reduces proliferative capacities of liver by switching pathways of C/EBPa growth arrest. Cell 113, 495–506. [DOI] [PubMed] [Google Scholar]
- Ishak CA, Marshall AE, Passos DT, White CR, Kim SJ, Cecchini MJ, Ferwati S, MacDonald WA, Howlett CJ, Welch ID, et al. (2016). An RB-EZH2 complex mediates silencing of repetitive DNA sequences. Mol Cell. 64, 1074–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob V, Chernyavskaya Y, Chen X, Tan PS, Kent B, Hoshida Y, and Sadler KC (2015). DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development 142, 510–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, and Porse BT (2013). Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 23, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermann P, Hoerner L, Burger L, and Schubeler D (2014). Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci USA. 111, E3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang G-L, Iakova P, Shi X, Haefliger S, Finegold M, and Timchenko NA (2010). Epigenetic changes play critical role in age-associated dysfunctions of the liver. Aging Cell 9, 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Factor VM, Andersen JB, Durkin ME, Tomokuni A, Marquardt JU, Matter MS, Hoang T, Conner EA, and Thorgeirsson SS (2016). DNMT1 is a Required Genomic Regulator for Murine Liver Histogenesis and Regeneration. Hepatology 64 582–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, and Poss KD (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, and Bezerra JA (2003). Independent and overlapping transcriptional activation during liver development and regeneration in mice. Hepatology 35, 525–534. [DOI] [PubMed] [Google Scholar]
- Kent B, Magnani E, Walsh MJ, and Sadler KC (2016). UHRF1 regulation of Dnmt1 is required for pre-gastrula zebrafish development. Dev Biol. 412, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, and E Futschik M (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, and Wutz A (2010). Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 24, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. (2017). Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Campbell JS, Mitchell C, McMahan RS, Yu X, Riehle KJ, Bumgarner RE, and Fausto N (2009). Relationships between deficits in tissue mass and transcriptional programs after partial hepatectomy in mice. Am J Pathol. 175, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S (2013). GeneOverlap: Test and visualize gene overlaps In GeneOverlap: Test and visualize gene overlaps (github), pp. R package [Google Scholar]
- Long HK, King HW, Patient RK, Odom DT, and Klose RJ (2016). Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 44, 6693–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grutzner F, Odom DT, Patient R, Ponting CP, et al. (2013). Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2, e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Petersen BO, Holm K, Bartek J, and Helin K (1996). Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 16, 1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenohara S, Unoki M, Toh H, Ohishi H, Sharif J, Koseki H, and Sasaki H (2017). Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 13, e1007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, and Bernstein BE (2010). GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK (2007). Liver regeneration. J Cell Physiol. 213, 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK (2013). Principles of liver regeneration and growth homeostasis. Compr Physiol. 3, 485–513. [DOI] [PubMed] [Google Scholar]
- Mitchell C, and Willenbring H (2008). A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 3, 1167–1170. [DOI] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, and Miyajima A (2012). Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 22, 1166–1175. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing C, Chinwalla AT, Cook LL, Delehaunty KD, Fewell GA, Fulton LA, Fulton RS, Graves TA, Hillier LW, Mardis ER, et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520. [DOI] [PubMed] [Google Scholar]
- Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson RT, et al. (2014). UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, and Tatsumi K (2002). Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and dNa replication blocks. J Biol Chem. 277, 34549–34555. [DOI] [PubMed] [Google Scholar]
- Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi M, et al. (2014). The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nature Immunology 15, 571. [DOI] [PubMed] [Google Scholar]
- Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, and Seki Y (2013). A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 140, 2892–2903. [DOI] [PubMed] [Google Scholar]
- Ohtani H, Liu M, Zhou W, Liang G, and Jones PA (2018). Switching roles for DNA and histone methylation depend on evolutionary ages of human endogenous retroviruses. Genome Res. 28, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana D, Liu X, Wang G-L, Jin J, Iakova P, and Timchenko NA (2010). Calmodulin controls liver proliferation via interactions with C/EBPβ-LAP and C/EBPβ-LIP. J Biol Chem. 285, 23444–23456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, and Karp SJ (2007). Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem. 282, 11197–11204. [DOI] [PubMed] [Google Scholar]
- Ramesh V, Bayam E, Cernilogar FM, Bonapace IM, Schulze M, Riemenschneider MJ, Schotta G, and Gotz M (2016). Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev. 30, 2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Ryan DP, Kilpert F, Ramírez F, Heyne S, Manke T, Bhardwaj V, Grüning B, and Dündar F (2016). DeepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen Shu Y., Han H, Liang G, Jones Peter A., Pugh Trevor J., et al. (2015). DNA-Demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Krahn KN, Gaur NA, and Ukomadu C (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci USA. 104, 1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang N, Arteaga M, Chitsike L, Wang F, Viswakarma N, Breslin P, and Qiu W (2016). FAK deletion accelerates liver regeneration after two-thirds partial hepatectomy. Sci Rep. 6, 34316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Endo Takaho A., Nakayama M, Karimi Mohammad M., Shimada M, Katsuyama K, Goyal P, Brind’Amour J, Sun M-A, Sun Z, et al. (2016). Activation of endogenous retroviruses in Dnmt1−/− ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell 19, 81–94. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Gu H, Bock C, Gnirke A, and Meissner A (2009). High-throughput bisulfite sequencing in mammalian genomes. Methods 48, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev Alexey A., Josefowicz Steven Z., and Allis CD (2016). Greater than the sum of parts: complexity of the dynamic epigenome. Mol Cell 62, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Chuang J-C, Kanchwala M, Wu L, Celen C, Li L, Liang H, Zhang S, Maples T, Nguyen Liem H., et al. (2016). Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R (2004). Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 5, 836–847. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, and Dyson NJ (2008). Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 9, 713. [DOI] [PubMed] [Google Scholar]
- Volkman HE, and Stetson DB (2014). The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. 15, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, and Bestor TH (1998). Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 20, 116–117. [DOI] [PubMed] [Google Scholar]
- Walter M, Teissandier A, Perez-Palacios R, and Bourc’his D (2016). An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife 5, e11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko L, Medrano EE, and Timchenko NA (2008a). HDAC1 cooperates with C/EBPa in the inhibition of liver proliferation in old mice. J Biol Chem. 283, 26169–26178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-L, Salisbury E, Shi X, Timchenko L, Medrano EE, and Timchenko NA (2008b). HDAC1 promotes liver proliferation in young mice via interactions with C/EBPp. J Biol Chem. 283, 26179–26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P, Brestelli JE, Kaestner KH, and Greenbaum LE (2005). Identification of transcriptional networks during liver regeneration. J Biol Chem. 280, 3715–3722. [DOI] [PubMed] [Google Scholar]
- Xiang H, Yuan L, Gao X, Alexander PB, Lopez O, Lau C, Ding Y, Chong M, Sun T, Chen R, et al. (2017). UHRF1 is required for basal stem cell proliferation in response to airway injury. Cell Discov. 3, 17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Inoue K, Saeki N, Ideta-Otsuka M, Yanagihara Y, Sawada Y, Sakakibara I, Lee J, Ichikawa K, Kamei Y, et al. (2018). Uhrf1 is indispensable for normal limb growth by regulating chondrocyte differentiation through specific gene expression. Development 145. [DOI] [PubMed] [Google Scholar]
- Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, Pikarsky E, and Stanger BZ (2014). Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, and Bestor TH (1997). Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340. [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hoshida Y, and Sadler KC (2016). Comparative epigenomic profiling of the DNA methylome in mouse and zebrafish uncovers high interspecies divergence. Front Genet 7, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Budker V, and Wolff JA (1999). High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 10, 1735–1737. [DOI] [PubMed] [Google Scholar]
- Zhao J, Chen X, Song G, Zhang J, Liu H, and Liu X (2017). Uhrf1 controls the selfrenewal versus differentiation of hematopoietic stem cells by epigenetically regulating the cell- division modes. Proc Natl Acad Sci USA. 114, E142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. (2013). Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Related to Figure 1. 7006 unique genes that were differentially expressed compared to baseline at any time point during wild-type mouse liver regeneration.
Table S2. Related to Figure 1. 6 unique clusters of wild-type regeneration genes that display distinct temporal expression patterns as determined by unsupervised clustering.
Table S3. Related to Figure 4. DEG between wild-type and Uhrf1hepKO regenerating livers at each time point following partial hepatectomy.
Table S4. Related to Figure 7. All 201 genes differentially expressed between wild-type and Uhrf1hepKO regenerating livers and occupied by H3K27me3 at promoters. 74 HALLMAKR_E2F_TARGET genes that are potentially regulated by H3K27me3 during liver regeneration. 120 HALLMAKR_E2F_TARGET genes that are potentially not regulated by H3K27me3 during liver regeneration
Data Availability Statement
GO DAVID analysis were carried out using DAVID 6.8 using default parameters for function annotation (Huang et al., 2008). GSEA (gene set enrichment analysis) was carried out using default parameters (Subramanian et al., 2005). The R package GeneOverlap was used to overlap gene sets and calculate significance of overlap (S. Li, 2013). The R package Genomation was used to annotate CpGs and H3K27me3 peaks into different genomic elements (Akalin et al., 2015).