Abstract
The gastrointestinal (GI) tract, in simplest terms, can be described as an epithelial-lined muscular tube extending along the cephalocaudal axis from the oral cavity to the anus. Although the general architecture of the GI tract organs is conserved from end to end, the presence of different epithelial tissue structures and unique epithelial cell types within each organ enables each to perform the distinct digestive functions required for efficient nutrient assimilation. Spatiotemporal regulation of signaling pathways and downstream transcription factors controls GI epithelial morphogenesis during development to confer essential regional-specific epithelial structures and functions. Here, we discuss the fundamental functions of each GI tract organ and summarize the diversity of epithelial structures present along the cephalocaudal axis of the GI tract. Next, we discuss findings, primarily from genetic mouse models, that have defined the roles of key transcription factors during epithelial morphogenesis, including p63, SOX2, SOX15, GATA4, GATA6, HNF4A, and HNF4G. Additionally, we examine how the Hedgehog, WNT, and BMP signaling pathways contribute to defining unique epithelial features along the cephalocaudal axis of the GI tract. Lastly, we examine the molecular mechanisms controlling regionalized cytodifferentiation of organ-specific epithelial cell types within GI tract, concentrating on the stomach and small intestine. The delineation of GI epithelial patterning mechanisms in mice has provided fundamental knowledge to guide the development and refinement of three-dimensional GI organotypic culture models such as those derived from directed differentiation of human pluripotent stem cells and those derived directly from human tissue samples. Continued examination of these pathways will undoubtedly provide vital insights into the mechanisms of GI development and disease and may afford new avenues for innovative tissue engineering and personalized medicine approaches to treating GI diseases.
Keywords: gastrointestinal epithelium, gastrointestinal development, regionalization, morphogenesis, cytodifferentiation, transcription factors, signaling pathways
1. Introduction
The gastrointestinal (GI) tract is an epithelial-lined muscular tube extending from the oral cavity to the anus. Organs of the Gl tract including the esophagus, stomach, small intestine, and colon function in coordination with the pancreas, liver, and gall bladder to perform the life-sustaining tasks of digestion and absorption. The GI tract also contains the body's largest reservoir of lymphoid tissue and an enteric nervous system with more neurons than the spinal cord. The general architecture of GI tract organs is conserved with all embryonic germ layers contributing to tissues of each organ (Figure 1). The innermost mucosal layer consists of an endoderm-derived epithelium. The epithelium is supported by lamina propria, a connective tissue layer, and muscularis mucosae, a thin, smooth muscle layer, both derived from mesoderm. Underlying the mucosa is the submucosa, which contains mesoderm-derived loose collagenous tissue, adipose tissue, and large blood and lymphatic vessels. Ectoderm-derived enteric ganglia also reside within the submucosa. The muscularis externa, a thick layer of mesoderm-derived muscle containing additional enteric ganglia, wraps around the mucosal and submucosal layers and generates the peristaltic forces that move contents through the gut. Generally, the muscularis externa consists of smooth muscle; however, the esophagus and anal sphincter contain skeletal muscle. Finally, the GI tract is covered by a mesoderm-derived adventitia or serosa.
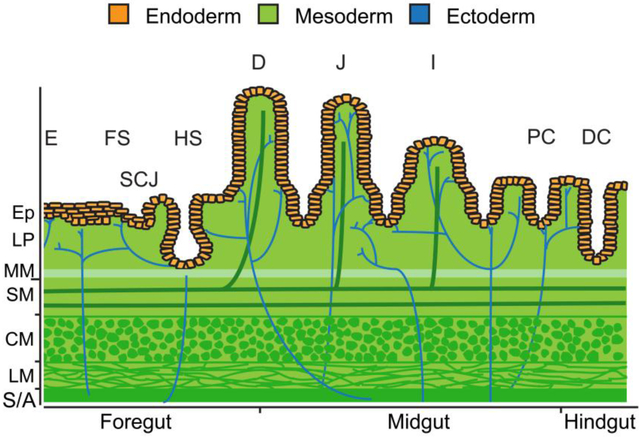
Figure 1.
All embryonic germ layers contribute to the tissue layers of GI organs. Divided into three regions, the endoderm (orange) gives rise to the innermost epithelial layer of all GI organs. The stratified squamous epithelium of the esophagus (E) and forestomach (FS) as well as the simple columnar epithelium of the hindstomach (HS) and proximal duodenum (D) of the small intestine originate from foregut endoderm. The junction between the stratified squamous epithelium and simple columnar epithelium is referred to as the squamocolumnar junction (SCJ). Midgut endoderm gives rise to the simple columnar epithelium of the distal duodenum, jejunum (J), and ileum (I) of the small intestine as well as to the simple columnar epithelium of the proximal colon (PC). The simple columnar epithelium of the distal colon (DC) arises from hindgut endoderm. Beneath the epithelium (Ep), the lamina propria (LP), muscularis mucosae (MM), submucosa (SM), vasculature, lymphatics, and muscularis externa (CM, circular muscle; LM, longitudinal muscle) layers develop from mesoderm (green). The enteric nervous system is derived from ectoderm and ganglia reside within the submucosa and the muscularis externa (blue). The gut tube is covered by a mesoderm derived adventitia (A) or serosa (S).
Although the general three-dimensional structure of GI organs is conserved, each organ, and even specific regions within an organ, perform specialized functions that together culminate in efficient nutrient assimilation. In particular, unique features of each organ’s epithelium provide the foundation for the regional-specific organ functions essential for digestion and absorption. The structure of the epithelium and the presence of organ-specific epithelial cell types drive regional-specific functions along the GI tract. The goal of this review is to explain how key transcription factors and signaling pathways contribute to regionalization of the GI epithelium. Investigations using genetically modified mouse models have been the primary source of information regarding the mechanisms through which specific transcription factors and signaling pathways orchestrate regionalization of the GI epithelium during development. More recently, studies using in vitro three-dimensional organoid cultures have also shed light on regionalizing mechanisms. The first half of this review will discuss how transcription factors and upstream signaling pathways shape the various three-dimensional structures of each GI organ’s epithelium. The second half will discuss how transcription factors and upstream signaling pathways control specific cytodifferentiation programs in the GI tract, with particular attention given to the gastric epithelium and the intestinal epithelium.
2. Generating uniquely structured epithelia along the cephalocaudal axis of the GI tract
A. The diversity of epithelial structures within the GI tract
Before discussing the molecular mechanisms underlying architectural patterning of the GI epithelium, we must first appreciate the structural differences that exist among epithelia of the GI tract and how each GI organ’s epithelial structure directly supports its unique digestive and absorptive functions. The esophagus is essentially a muscular tube that transports ingested materials from the oral cavity to the stomach. Its epithelium is stratified, with multiple cell layers providing the protection needed in the face of the frictional forces generated by the passage of undigested and partially digested materials. In rodents, the esophageal epithelium is keratinized, and it extends into the proximal stomach forming a gastric region known as the forestomach. Absent in human stomach, the rodent forestomach serves as a holding zone for food and is responsible for some degree of mechanical digestion (San Roman and Shivdasani, 2011). The simple columnar nascent epithelium of the esophagus/forestomach undergoes morphogenesis during development to give rise to a mature multi-layered epithelium that consists of a basal layer of proliferating cells and suprabasal layers of differentiated cells (Figure 1) (Zhang et al., 2016).
There is a distinct change in the epithelial architecture of the GI tract at the transition from the esophagus/forestomach to the glandular hindstomach (Figure 1). As the site of chemical and mechanical digestion, the simple columnar glandular epithelium of the stomach is ideally suited for highly efficient secretion of acid and production of digestive enzymes. The boundary between the stratified squamous epithelium of the esophagus/forestomach and the simple columnar epithelium of the glandular stomach is referred to as the squamocolumnar junction (SCJ) (Figure 1). Understanding how these different epithelial structures are generated and maintained at the SCJ is essential because metaplasia occurs in these tissues in GI diseases including Barrett’s esophagus, esophageal adenocarcinoma, and gastric adenocarcinoma.
The small intestine is the site of nutrient absorption and is also a fundamental component of the innate immune system. Essential for the function of the small intestine is the large surface area of its simple columnar epithelium, which has many folds with deep grooves, called crypts, and finger-like projections, called villi (Figure 1) (Chin et al., 2017; Noah et al., 2011; Thompson and Battle, 2014; Walton et al., 2016a). The small intestine can be divided into three functional regions: duodenum, jejunum, and ileum. The epithelial cells of the duodenum synthesize digestive enzymes, which are secreted or localized to the brush border membrane. These enzymes, together with bile produced by the liver and pancreatic digestive enzymes, complete the digestion of proteins, fats, and carbohydrates (Binder and Reuben, 2005; Jeejeebhoy, 2002). The jejunal epithelium accomplishes the bulk of nutrient absorption (Binder and Reuben, 2005; Davis and Attie, 2008; Jeejeebhoy, 2002). The ileal epithelium is responsible for absorption of vitamin B12 in addition to bile salts, which are recycled to the liver (Binder and Reuben, 2005; Davis and Attie, 2008; Jeejeebhoy, 2002). Understanding how the small intestinal epithelium is appropriately regionalized should provide fundamental insights into short bowel syndrome (SBS) and may lead to new SBS treatment options.
Similar to the small intestine, the function of the epithelium of the large intestine is absorption. Lined by a simple columnar epithelium arranged as crypts but lacking villi, these epithelial cells primarily absorb water and electrolytes (Figure 1) (Jeejeebhoy, 2002). Like the small intestine, the large intestine can also be divided into three regions: cecum, colon, and rectum. Although the epithelia of the small and large intestines share many features, the presence of villi and Paneth cells in the small intestine is one distinguishing feature. Understanding how the epithelium of these organs is specialized and regionalized is important considering that this process can be disrupted in disease. For example, Paneth cell metaplasia, or the abnormal presence of Paneth cells in the large intestine, sometimes occurs in inflammatory bowel disease and colorectal cancer (Tanaka et al., 2001; Wada et al., 2005).
B. Key transcription factors and signaling molecules that control structural patterning of GI tract epithelia
i. p63, master regulator of stratified squamous epithelial morphogenesis.
The transcription factor p63 is considered to be a master regulator of stratified squamous epithelium development (Figure 2) (Daniely et al., 2004; Koster et al., 2004; Mills et al., 1999; Romano et al., 2012; Senoo et al., 2007; Wang et al., 2011; Yang et al., 1999; Yu et al., 2005). p63 is required to establish stratified epithelia and, therefore, is typically absent from columnar epithelia. Studies of p63 knockout mouse embryos reveal that proper epithelial morphogenesis in the esophagus and forestomach requires p63, with its absence posteriorizing this domain (Wang et al., 2011). Specifically, esophageal or forestomach epithelium lacking p63 fails to stratify and instead remains columnar. Columnar-cell type cytokeratins persist in p63−/− esophagus and forestomach in place of stratified-cell type cytokeratins. Alterations in cytokeratin expression profiles likely directly contribute to changes cell shape in mutant tissues. Moreover, Hutton et al. have shown that the distinct set of cytokeratins expressed in a particular epithelial tissue structure is necessary and specific to that tissue’s structural and functional needs (Hutton et al., 1998). For example, the columnar-type cytokeratin CK18 could not fully compensate for loss of the stratified-type cytokeratin CK14 in CK14 mutant skin (Hutton et al., 1998). Beyond changes in cell shape and epithelial structure, p63-null esophageal and forestomach epithelia further display features of mucous metaplasia including the presence of intestinal-type goblet cells and the expression of intestinal markers such as Villin suggesting p63 loss as a possible mechanistic step in metaplasia observed in Barrett’s esophagus. Further emphasizing the crucial role of p63 in stratified epithelium development, ectopic p63 expression in the simple epithelium of the lung is sufficient to convert it to stratified (Romano et al., 2012).
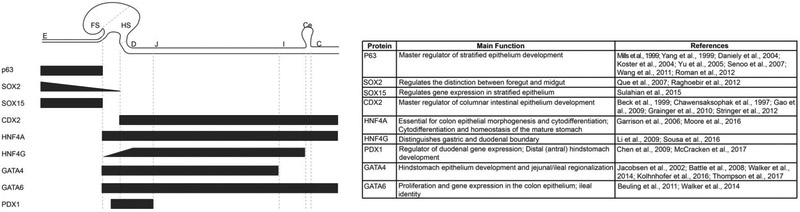
Figure 2.
Expression and functions of key transcription factors required for regionalization of the GI tract epithelium. Black bars represent transcription factor expression profiles and gradients along the cephalocaudal axis of the GI tract including the esophagus (E), forestomach (FS), hindstomach (HS), duodenum (D), jejunum (J), ileum (I), cecum (Ce) and colon (C). The table summarizes each transcription factor’s role(s) in GI tract epithelial patterning and development.
ii. SOX2 and SOX15, transcription factors with essential roles in regionalizing stratified squamous epithelium.
SOX2, a transcription factor of the Sry-like HMG box family, is abundant in the early dorsal foregut endoderm as the gut tube separates into the esophagus and trachea (Que et al., 2007; Sherwood et al., 2009). SOX2 remains highly expressed in the esophagus during epithelial stratification suggesting a role for it in this process (Que et al., 2007) (Figure 2). Because SOX2 null embryos die before gastrulation (Avilion et al., 2003), the role of SOX2 in GI development has been studied using hypomorphic mutants (Que et al., 2007). These studies provide compelling evidence that morphogenesis and differentiation of the esophageal epithelium require SOX2. While SOX2 hypomorphic mutant embryos with the most severe reduction of SOX2 (~5% normal SOX2 level) develop esophageal atresia (EA) and tracheoesophageal fistula (TEF) due to defects in tube separation, those retaining slightly higher SOX2 levels (~18% normal SOX2 level) instead have defects in stratification and development of the esophageal epithelium. In these mutants, the esophageal epithelium contains regions of abnormal columnar cells rather than stratified squamous cells. Accordingly, the number of p63+ cells is decreased. Moreover, some SOX2-deficient esophageal cells express markers of gastric and intestinal columnar epithelial cells. These studies implicate downregulation of SOX2 as a putative mechanism involved in Barrett’s esophagus and its transition to adenocarcinoma (Chen et al., 2008; Que et al., 2007). Loss of SOX2 has been associated with poorer survival in patients with esophageal adenocarcinoma (Honing et al., 2014). Conversely, up-regulation of SOX2 has been associated with squamous cell carcinoma (Bass et al., 2009; Liu et al., 2013).
SOX2 expression extends beyond the esophageal epithelium into the murine forestomach, which expresses high levels of SOX2, and into the murine/human glandular stomach, which expresses relatively low levels of SOX2 compared with the esophagus and forestomach (Figure 2) (Que et al., 2007). Like the esophagus, SOX2 deficiency in the developing forestomach posteriorizes the epithelium. Mutants contain regions of simple columnar epithelium with characteristics mirroring that of the glandular columnar stomach (Que et al., 2007). Surprising, perhaps, is the fact that glandular stomach development appears to be normal in SOX2 hypomorphic embryos (Que et al., 2007). It is possible that the level of SOX2 retained in the developing glandular stomach of mutants is sufficient to guide proper epithelium development. With the advent of in vitro stem cell-based models that mimic stomach development in a dish, the absolute requirement for SOX2 in glandular stomach development can be more readily addressed (McCracken et al., 2014; McCracken et al., 2017).
Although SOX2 is normally absent in the intestinal epithelium, studies of embryos ectopically expressing SOX2 in the intestine have been informative about SOX2 as a dominant driver of foregut epithelial development. The presence of SOX2 in the developing small intestine anteriorizes the epithelium suggesting that SOX2 has a dominant effect on driving esophageal/gastric epithelial phenotypes (Raghoebir et al., 2012). Expression of intestinal epithelial cell type markers is reduced, and expression of both gastric and forestomach/esophageal epithelial cell type markers is stimulated. Epithelial architecture, although remaining columnar, is abnormal with villus number and shape being affected. Taken together, these studies implicate SOX2 as a transcription factor with an essential role in specifying stratified squamous epithelial identity.
Recent work has shown that SOX15, a SOX family transcription factor with 75% amino acid identity to SOX2, also controls gene expression in human stratified esophageal epithelial cells by regulating the expression of a significant fraction of the esophageal stratified squamous epithelial cell transcriptome (Figure 2) (Sulahian et al., 2015; van de Wetering and Clevers, 1993). This study was performed using an in vitro deletion of SOX15 in an esophageal cell culture model, and so it will be important to assess the role of SOX15 in GI epithelial regionalization using in vivo mouse models or esophageal organoid models. Although Sox15 null mice have been reported to be grossly normal and fertile, an in-depth analysis of esophageal development in the absence of SOX15 has not been performed (Maruyama et al., 2005). Consistent with SOX15’s imputed role as a stratified epithelial cell transcription factor, SOX15 is absent in columnar metaplastic lesions of Barrett's esophagus (Sulahian et al., 2015). Surprisingly, although absent in a majority of esophageal adenocarcinomas, SOX15 remains expressed in ~20-30% of esophageal adenocarcinomas (Sulahian et al., 2015). This suggests that SOX15 can be re-activated during the progression from Barret’s esophagus to cancer. As expected, SOX15+ cancers express markers of both stratified squamous and columnar lineages, and it will be important to determine the pathophysiology of such lesions.
iii. CDX2, a master regulator of columnar intestinal epithelium development.
The homeobox-domain containing transcription factor CDX2 functions as a master regulator of columnar intestinal epithelium development (Beck et al., 1999; Chawengsaksophak et al., 1997; Gao et al., 2009; Grainger et al., 2010; Stringer et al., 2012). During mouse development, CDX2 expression is prominent in the developing hindgut by E8.5, becomes restricted to the intestine at the foregut/midgut boundary by E12.5, and is maintained in this pattern throughout adulthood (Beck et al., 1995; Beck et al., 1999; Chawengsaksophak et al., 2004). The proximal boundary of CDX2 expression is met by the distal boundary of SOX2 expression at the gastro-intestinal junction (Figure 2). Studies of SOX2 and CDX2 mutants demonstrate that establishing a balance between SOX2 and CDX2 is essential for proper regional patterning at this junction. Ectopic CDX2 expression in the mouse glandular gastric mucosa induces the expression of intestinal genes, and intestine-like goblet cells emerge (Mutoh et al., 2002; Silberg et al., 2002). More recently, Jiang et al. report that ectopic CDX2 expression in a transitional zone of cells at the SCJ induces intestinal metaplasia in vivo, mimicking the metaplasia characteristic of Barrett’s esophagus (Jiang et al., 2017). On the other hand, ectopic SOX2 expression in the intestine inhibits the ability of CDX2 to bind to its target genes resulting in a foregut-like phenotype or anteriorization of the intestine (Raghoebir et al., 2012).
Further consistent with the idea that the developing gut must have appropriately balanced SOX2-CDX2 expression, elimination of CDX2 from the early developing midgut via conditional knockout induces SOX2 expression. Accordingly, the CDX2-deficient, SOX2-expressing midgut becomes anteriorized, expressing p63, a master regulator of stratified epithelial development (Gao et al., 2009). Coincident with p63 and SOX2 expression, CDX2 mutant intestinal epithelium loses columnar characteristics and is instead flattened with stratified-like characteristics. Gene expression profiling confirms that the CDX2-deficient intestine transcriptome more closely resembles an esophageal/forestomach profile rather than an intestinal profile (Gao et al., 2009). Similarly, Cdx2+/− animals can develop intestinal polyps that contain foregut-type stratified squamous epithelium (Chawengsaksophak et al., 1997).
The role of CDX2 in later stages of intestinal epithelium development is generally conserved as loss of CDX2 from the mouse intestine beginning at ~E13.5 also anteriorizes the epithelium, albeit partially. When eliminated at this later stage, CDX2-deficient intestinal epithelium expresses SOX2, but not p63, and becomes partially transformed toward a glandular stomach-type identity rather than taking on a fully stratified esophageal/forestomach identity (Grainger et al., 2010). Similarly, inducible knockout of Cdx2 from the adult intestinal epithelium induces gastric epithelial cell marker expression and reduces intestinal epithelial cell marker expression (Stringer et al., 2012). It may be that the failure to induce p63 underlies the differences between the degree of anterior transformation observed in early versus late CDX2 mutants. CDX2 is also absolutely required for colon development as conditional deletion of CDX2 in the midgut and hindgut during early embryonic development completely abrogates colon development (Gao et al., 2009).
Taken together, CDX2 function can be juxtaposed with that of p63 and SOX2 in regionalizing the GI epithelium (Figure 2). CDX2, expressed highly in the midgut and hindgut, is essential to direct development of a columnar, intestinal epithelium whereas SOX2 and p63, both expressed highly in the foregut, are essential to direct development of a stratified esophageal/forestomach epithelium. Not surprising is the observation that CDX2 is present in columnar intestinal-type metaplasia in the esophagus and stomach (Bai et al., 2002; Phillips et al., 2003). Moreover, recent work from the Que lab suggests a mechanistic link between CDX2 and BE as CDX2 expression alone induced Barrett’s esophagus type-intestinal metaplasia within the SCJ in vivo in mice (Jiang et al., 2017). The striking difference between the effects of early loss of CDX2 in the developing small intestine versus the colon may be explained by differential impacts on the expression of key downstream effectors. For example, the foregut-associated factors SOX2, PAX9, and WNT10A are highly induced in the distal midgut/hindgut of CDX2 mutants, and the columnar factor Indian Hedgehog (discussed below) is only severely diminished in the distal midgut/hindgut domain of mutants (Gao et al., 2009).
iv. The Hepatic Nuclear Factor 4 family transcription factors in columnar epithelial development.
The Hepatic Nuclear Factor 4 (HNF4) family of transcription factors, which includes HNF4 alpha (HNF4A) and HNF4 gamma (HNF4G), has also been implicated in controlling regionalization of the GI epithelium (Figure 2). HNF4A is expressed in the developing and mature epithelium of the stomach, small intestine, and colon whereas HNF4G is expressed only in the developing and mature colon (Drewes et al., 1996; Duncan et al., 1994; Li et al., 2009; Taraviras et al., 1994; Taraviras et al., 2000). The role of HNF4A in development of the colon epithelium has been examined via conditional knockout (Garrison et al., 2006). Because this model also deletes HNF4A in the liver causing a lethal phenotype (Battle et al., 2006; Parviz et al., 2003), the colon phenotype could only be studied in embryos. Elimination of HNF4A within the embryonic colon epithelium severely disrupts colon architecture with defects in both epithelial morphogenesis and epithelial cell cytodifferentiation (Garrison et al., 2006). Several groups have examined HNF4A function in patterning the small intestine epithelium at later development stages and in adult mice, but none have reported any major effect on small intestinal epithelium development (Ahn et al., 2008; Babeu et al., 2009; Darsigny et al., 2009). It is possible that deletion of HNF4A earlier in the course of small intestinal development would illuminate a new role for HNF4A in this process. It is also possible that HNF4G, which is absent in the colon epithelium but present in the small intestinal epithelium, compensates for HNF4A function in mutants. A role for HNF4A in development of the stomach epithelium has not been reported. However, the Mills group has shown that HNF4A is necessary for cytodifferentiation and homeostasis of the adult mouse gastric epithelium (Moore et al., 2016). Although GI epithelium development has yet to be fully examined in HNF4G knockout mice, HNF4G has been implicated in regulating development of the gastric-duodenal boundary. Expression of HNF4G becomes highly upregulated in the epithelium of the duodenum compared with that of the stomach during segregation of these foregut endoderm domains suggesting a role for it in this process (Li et al., 2009). Moreover, HNF4G has been shown to be upregulated in metaplastic lesions in stomach further supporting a role for high levels of HNF4G in distinguishing the gastric and intestinal domains (Sousa et al., 2016).
v. GATA4 and GATA6 in regionalizing the columnar epithelium of the stomach and intestine.
Of the six known GATA family transcription factors, GATA4, GATA5, and GATA6 are expressed in the developing endoderm that gives rise to the epithelium of the GI tract. GATA4 and GATA6, but not GATA5, have been shown to be important regulators of GI epithelial development (Aronson et al., 2014; Battle et al., 2008; Beuling et al., 2008; Beuling et al., 2010; Beuling et al., 2011; Beuling et al., 2012; Bosse et al., 2006; Kohlnhofer et al., 2016; Thompson et al., 2017; Walker et al., 2014a; Walker et al., 2014b). While GATA6 is expressed in the epithelia of the glandular hindstomach, the entire small intestine, and the colon, GATA4 expression ceases at the jejunal/ileal boundary within the small intestinal epithelium (Figure 2) (Battle et al., 2008; Beuling et al., 2012; Bosse et al., 2006; Fang et al., 2006; Jacobsen et al., 2002). Mosaic genetic analysis studies with chimeric embryos containing Gata4 null cells suggest an important role for GATA4 in driving proper columnar glandular epithelium development in the mouse hindstomach (Jacobsen et al., 2002). In this model, Gata4 null cells rarely contribute to the glandular hindstomach epithelium, implicating GATA4 as a critical regulator of overall hindstomach development. The few Gata4 null regions emerging within the chimeric hindstomachs are anteriorized, taking on a stratified squamous type epithelial morphology. Furthermore, Gata4 null cells lack expression of gastric differentiation markers. GATA4 has been shown to cooperate with the transcriptional co-factor Friend-of-GATA (FOG1) to direct gene expression within the hindstomach epithelium (Jacobsen et al., 2005). Conditional knockouts to study GATA4's role in stomach regionalization or studies to examine GATA6's role in defining the stomach epithelium have not yet been reported. Expression of GATA4 and GATA6, however, has been reported in Barrett’s esophagus and esophageal adenocarcinoma, providing justification for further studies of these factors in regulating regionalization of the squamocolumnar junction (Duggan et al., 2016; Haveri et al., 2008; Miller et al., 2003).
Within the intestinal and colon domains, GATA factors primarily control regionalization of epithelial cytodifferentiation rather than epithelial structure, although early deletion of GATA4 within the midgut delays the onset of villus morphogenesis (Kohlnhofer et al., 2016). These cytodifferentiation phenotypes are discussed below in Section 3.
vi. Hedgehog signaling in GI epithelial regionalization.
Hedgehog (HH) proteins, secreted by epithelial cells, act on underlying mesenchymal cells, which in turn signal back to the epithelium to direct its development (van den Brink, 2007). Two HH proteins, Sonic Hedgehog (SHH) and Indian Hedgehog (IHH), are dynamically expressed in the developing endoderm and its derivative epithelia (Feng et al., 2012; Khurana and Mills, 2010; Ramalho-Santos et al., 2000; van den Brink, 2007). Early in development, SHH and IHH are broadly expressed throughout the foregut, midgut, and hindgut endoderm (Figure 3). Later, SHH is primarily expressed in the esophageal and forestomach domains, while IHH is primarily expressed in the hindstomach and intestinal domains (Figure 3). By E18.5, after GI epithelial domains have undergone significant morphogenetic remodeling, SHH expression decreases in the bulk of the stratified squamous epithelium of the esophagus, becoming limited to the distal esophageal epithelium, yet it increases in the epithelium of the hindstomach and intestines. This dynamic temporal expression pattern of HH proteins has important ramifications for regionalization of the GI epithelium (Figure 3).
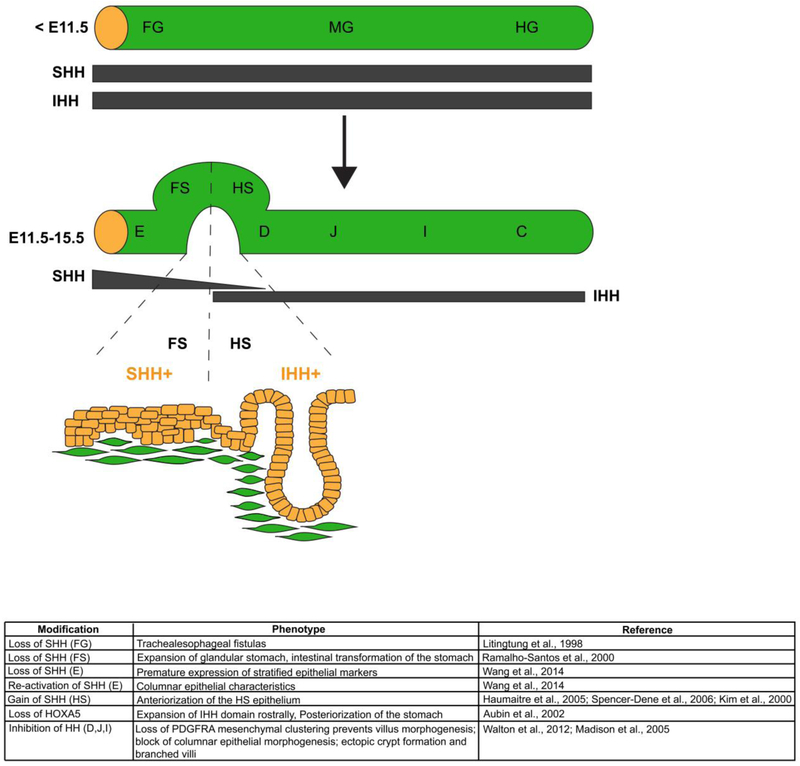
Figure 3.
A critical boundary of hedgehog signaling is required to regionalize the mouse stomach epithelium at the squamocolumnar junction. Early in development, Sonic Hedgehog (SHH) and Indian Hedgehog (IHH) are broadly expressed in all three regions of the endoderm, foregut (FG), midgut (MG) and hindgut (HG). As development progresses, SHH becomes restricted to domains that give to rise stratified squamous epithelium (esophagus, E, and forestomach, FS), and IHH becomes restricted to domains that give rise to columnar epithelium (hindstomach, HS, duodenum, D, jejunum, J, ileum, I, and colon, C). The boundary of SHH/IHH expression in the developing stomach is critical for proper SCJ development. If disrupted, the boundaries of stratified squamous epithelium and columnar epithelium shift. Loss of SHH in the forestomach domain, with concomitant induction of IHH, posteriorizes the forestomach epithelium. Conversely, loss of IHH in the hindstomach domain, with concomitant induction of SHH, anteriorizes the hindstomach domain. The table summarizes phenotypes observed in mouse mutants with HH gain and loss of function in the GI tract.
Studies of SHH gain and loss of function in the esophagus reveal an essential role for this signaling pathway in esophageal epithelial regionalization. The complete absence of SHH in the early developing foregut endoderm blocks tracheal-esophageal separation causing tracheoesophageal fistulas (Litingtung et al., 1998). A more recent examination of any remaining esophageal cells in SHH mutants shows that elimination of SHH in the esophageal domain results in premature expression of stratified epithelial cell markers (Wang et al., 2014). These data suggest that although down-regulation of SHH is detrimental during the earliest stages of esophageal development, down-regulation of SHH at later stages is key to control the onset of the stratified squamous epithelial cell differentiation program in the esophagus. Further emphasizing the necessity for SHH down-regulation in the esophageal domain to confer stratified epithelial cell development is the finding that re-activation of SHH in the esophageal epithelium drives development of columnar epithelial features, one of which is expression of the SHH-regulated transcription factor FOXA2 (Wang et al., 2014). Both SHH and FOXA2 are re-activated in the columnar metaplasia of Barrett's esophagus (Wang et al., 2014), indicating that SHH signaling is detrimental for homeostasis of the esophageal stratified squamous epithelium.
Perhaps not unexpected, HH signaling is also an essential player in regionalizing the stomach epithelium (Figure 3). Stomachs lacking SHH exhibit a greatly expanded glandular columnar stomach domain at the expense of the stratified squamous forestomach domain (Ramalho-Santos et al., 2000). Moreover, the glandular stomach of SHH mutants is also posteriorized, expressing some intestinal markers. Likewise, loss of HOXA5 from the underlying gastric mesenchyme disrupts epithelial HH signaling in the developing stomach. IHH expression expands into the forestomach domain at the expense of SHH expression in HOXA5 mutants and mutant stomachs are posteriorized, expressing the intestinal marker alkaline phosphatase even into the presumptive forestomach domain (Figure 3) (Aubin et al., 2002). On the other hand, induced expression of SHH in the hindstomach as a consequence of Hnf1b knockout, Fgf10 knockout, Fgfr2 knockout, or the expression of mutant Activin receptors compromises the IHH domain within the hindstomach and anteriorizes the epithelium (Figure 3) (Haumaitre et al., 2005; Kim et al., 2000; Spencer-Dene et al., 2006). In these cases, the mutant epithelium is thickened, and there is a loss of glandular cell types. Taken together, the balance between SHH and IHH expression is essential to pattern the stratified squamous and gastric columnar epithelia in the stomach (Figure 3).
HH signaling also regulates critical aspects of epithelial morphogenesis in the small intestine (Madison et al., 2005; Walton et al., 2012; Walton et al., 2016a). When Hedgehog signaling is blocked by expression of the pan-Hedgehog inhibitor protein Hhip, the epithelium remains immature with a pseudostratified architecture persisting (Madison et al., 2005). Furthermore, HH signals control the clustering of underlying PDGFRA+ mesenchymal cells, which is absolutely necessary for villus outgrowth, and so villus morphogenesis is abnormal in HH mutants (Walton et al., 2012). Finally, HH signaling regulates localization of the proliferative cell niche to the intestinal crypt. When blocked, proliferative cells migrate upward out of the crypt to form ectopic crypts that abnormally branch villi (Madison et al., 2005).
vii. WNT signaling in regionalizing the GI epithelium.
During foregut development, WNT signaling needs to be minimized for proper regionalization. If early mouse foregut endoderm (E7.5-8.5) is exposed to WNT signaling via expression of constitutively active B-catenin, the foregut endoderm begins to aberrantly express the intestinal master regulator CDX2, generally at the expense of foregut regulator SOX2, although rare CDX2, SOX2 co-expressing cells are present (Sherwood et al., 2011). Moreover, CDX2 expression in these cells is sufficient to induce an intestinal-like gene expression program thereby posteriorizing the foregut.
The homeodomain transcription factor BarH-like homeobox 1 (BARX1), expressed in the esophageal, forestomach, and hindstomach mesenchyme, plays a central role in inhibiting WNT signaling in the posterior foregut domain (Figure 4). Loss of mesenchymal BARX1 reduces the expression of the WNT signaling inhibitors secreted frizzled-related proteins 1 and 2 (SFRP1/2) (Kim et al., 2005; Kim et al., 2007; Woo et al., 2011). Abnormal activation of WNT signaling in the foregut endoderm of BARX1-deficient stomachs has consequences throughout the esophagus, forestomach, and hindstomach organ domains (Kim et al., 2005; Kim et al., 2007). The entire BARX1 mutant foregut is posteriorized, with anterior (esophagus/forestomach) regions containing a mixed “squamo-glandular” type epithelium with reduced levels of SOX2. Perhaps most striking is the intestinal villus-like epithelium present in BARX1 mutant hindstomachs. In this region, aberrant WNT signaling induces CDX2 expression to shift the global gene expression profile in the developing stomach toward a more posterior midgut/intestinal endoderm program. Mesenchymal expression of a related homeobox transcription factor, BAPX1, in the distal hindstomach requires BARX1, and distal gastric marker expression and pyloric constriction are lost in both Barx1 and Bapx1 null mice (Verzi et al., 2009).
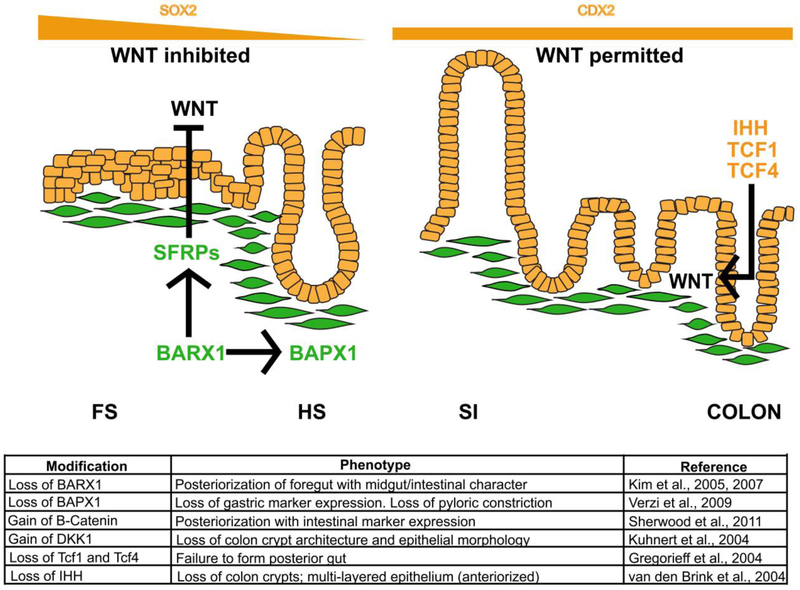
Figure 4.
WNT signaling must be inhibited in the foregut and induced in the midgut and hindgut for proper regionalization of the GI tract epithelium. In the developing stomach, WNT signaling must be inhibited for proper gastric epithelial development. The absence of WNT is critical for establishment of SOX2 expression in this domain. The mesenchymal transcription factor BARX1 is required to inhibit WNT signaling in the foregut domain. BARX1 induces expression of the WNT inhibitors known as SFRPs (secreted frizzled-related proteins). In this environment of low WNT, proper formation of the stratified squamous epithelium of the esophagus, forestomach and glandular columnar epithelium of the hindstomach occurs. In the absence of BARX1, WNT is upregulated. The esophagus and forestomach epithelia take on a “squamo-glandular” hybrid structure whereas the hindstomach epithelium becomes intestinalized. BAPX1, another gastric mesenchymal factor, also contributes to differentiation of the gastric versus intestinal domains. In posterior endoderm regions, WNT signaling is permitted, and CDX2 is induced. In the colon, loss of the WNT effectors TCF1 and TCF4 blocks posterior gut development. Additionally, cross talk between HH and WNT is required to generate the simple columnar epithelium of the colon. In the absence of IHH, WNT is perturbed, and the epithelium is anteriorized. The table summarizes phenotypes in loss and gain of function experiments involving WNT activators and repressors, as well as proteins involved in cross talk with the WNT pathway, during GI tract development and epithelial morphogenesis.
WNT signaling also plays a crucial role in colon development. Expression of the WNT inhibitor Dickkopt-1 (DKK-1) results in severely altered colon epithelial morphology, including loss of crypt architecture (Kuhnert et al., 2004). Additionally, embryos lacking the WNT effectors TCF1 and TCF4 fail to form a posterior gut (Gregorieff et al., 2004). Furthermore, there is interplay between HH and WNT signaling with HH signaling restricting WNT signaling to the crypt base. Accordingly, IHH-deficient embryonic colon lacks crypts and has a multilayered epithelium rather than a simple columnar epithelium (van den Brink et al., 2004).
viii. Bone Morphogenetic Proteins (BMPs) in regionalizing GI epithelia.
Proper temporal modulation of BMP signaling is absolutely required during esophageal and forestomach development (Figure 5A) (Que et al., 2006; Rodriguez et al., 2010). During the early period of epithelial stratification, BMP signaling must be inhibited. In fact, if present, BMP signaling represses SOX2 expression in the foregut to disrupt esophageal epithelial development (Domyan et al., 2011). To protect the esophageal epithelium from mesenchymal BMP signals, the epithelium secretes the BMP antagonists to block BMP signaling (Que et al., 2006; Rodriguez et al., 2010). Expression of a constitutively active BMP receptor in the developing esophagus and forestomach blocks epithelial stratification (Rodriguez et al., 2010). Later, active BMP signaling is required as esophageal epithelium lacking BMP receptors fails to differentiate (Rodriguez et al., 2010). Interestingly, stromal up-regulation of BMPs in the adult esophagus has been associated with columnar metaplasia in Barrett’s esophagus (Castillo et al., 2012).
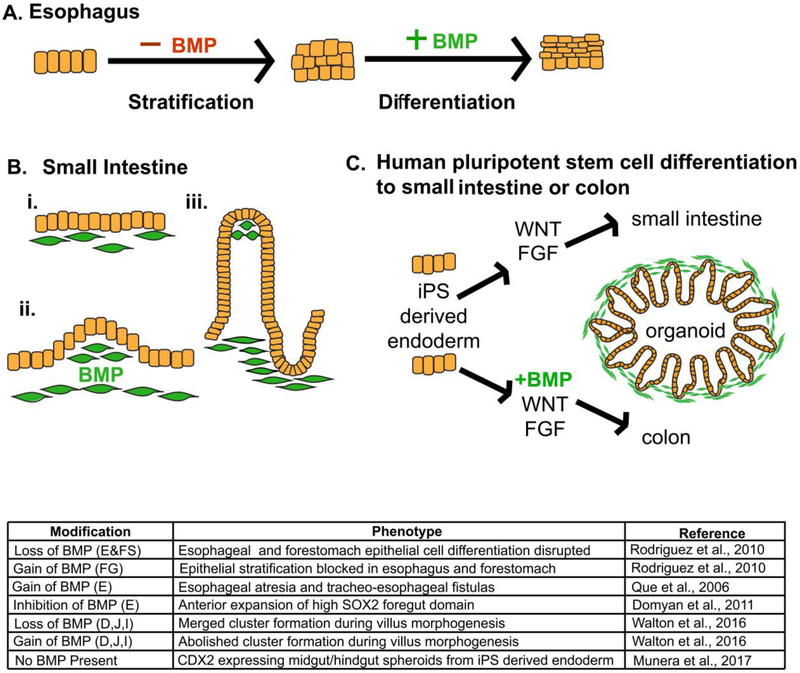
Figure 5.
Temporal modulation of BMP signaling is required for development of multiple GI epithelia. (A) Early in foregut development, BMP signaling must be inhibited to induce epithelial stratification, whereas, later in esophagus and forestomach development, BMP signaling must be active to induce differentiation of suprabasal cells within the stratified squamous epithelium of these organs. (B) BMP signaling controls villus morphogenesis in the small intestine through pattering of mesenchymal PDGFRA+ cell clusters. Mesenchymal cells beneath the developing intestinal epithelium (i) respond to BMP signaling, cluster (ii), and villus outgrowth occurs (iii). Mesenchymal clusters remain in the villus tip as the villi continue to develop (iii). (C) Differentiation of human induced pluripotent stem (iPS) cells to small intestinal organoids is driven by WNT and FGF signaling, while derivation of colonoids from iPS cells requires WNT, FGF and BMP signaling. The table summarizes the phenotypes driven by loss and gain of function studies involving key players of BMP signaling during development of GI tract organs.
In the small intestine, BMP signaling plays a key role in villus morphogenesis. However, BMP signaling is primarily focused within mesenchymal clusters, not in the epithelium (Walton et al., 2016b). While deletion of BMP receptors in the epithelium fails to disrupt villus formation, modulation of BMP signaling within the mesenchyme—either increased or decreased mesenchymal BMP—disrupts villus morphogenesis by altering the pattering of mesenchymal PDGFRA+ cell clusters (Figure 5B). BMP signaling is further required for colon development from hindgut endoderm. Recent studies using human pluripotent stem cells show that in the presence of WNT, FGF, and BMP signaling, human colonic organoids develop; in the absence of BMP, however, small intestinal organoids are the default (Figure 5C) (Múnera et al., 2017).
3. Key transcription factors and signaling molecules regulating regionalized cell differentiation gastric and intestinal epithelia
The first part of this review focused on how transcription factors and signaling pathways contribute to regionalized patterning of the three-dimensional epithelial structure. Now, we will examine another key aspect of GI epithelial regionalization, namely the mechanisms through which essential organ-specific epithelial cell types develop. We will focus on epithelial cell type regionalization in the glandular hindstomach and small intestine (Figure 6).
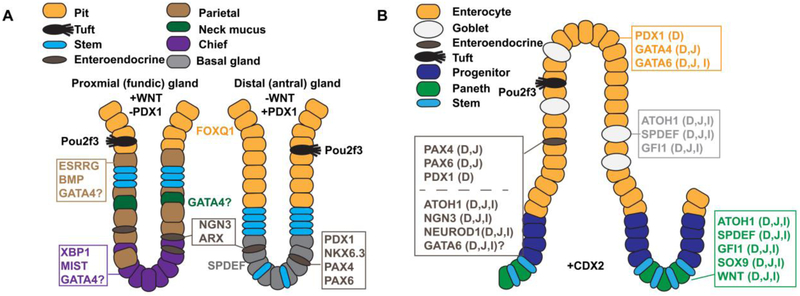
Figure 6. Key transcription factors and signaling molecules controlling cytodifferentiation in the gastric epithelium and small intestinal epithelium.
A) Illustration contrasts the cellular architecture of proximal (fundic) and distal (antral) glands of the hindstomach and shows key transcription factors and signaling molecules implicated in regulating cytodifferentiation in these regions. WNT signaling is necessary to specify the proximal (fundic) type glands and is absent in the distal (antral) region. Conversely, distal (antral) glands express the transcription factor PDX1 whereas proximal (fundic) glands lack PDX1. Note the parietal cells, chief cells, and neck mucus cells are enriched in proximal (fundic) glands compared with distal (antral) glands. B) Illustration depicts the cellular architecture of the small intestinal villus and crypt and shows key transcription factors and signaling molecules implicated in regulating intestinal cytodifferentiation. Factors are noted as D (duodenum), J (jejunum), and/or I (ileum) to describe their expression pattern. Note that enterocytes are regionalized by differential expression of PDX1 (D) and GATA4 (D, J) and that enteroendocrine cells are regionalized by differential expression of PAX4 (D,J), PAX6 (D,J), and PDX1 (D).
A. Gastric cytodifferentiation.
Specific gastric epithelial cell types synthesize acid, digestive enzymes, and hormones that enable chemical and mechanical digestion of food and regulate gastric motility (Figure 6A) (Choi et al., 2014; Kim and Shivdasani, 2016; McCracken and Wells, 2017; Willet and Mills, 2016). Two types of cells secrete mucus, neck mucus cells and surface mucus (pit/foveolar) cells. Chief cells secrete digestive enzymes including pepsinogen. Parietal cells secrete acid to control stomach pH. Enteroendocrine cells secrete hormones into the underlying lamina propria such as gastrin to promote acid secretion. Finally, as a luminal chemosensing cell type, gastric tuft cells project apical processes into the lumen and also produce secreted molecules including acetylcholine, to stimulate epithelial cell secretions, and cytokines, to mediate inflammatory responses (Gerbe and Jay, 2016; Hayakawa et al., 2017; Kaji and Kaunitz, 2017). The distribution of the specific epithelial cell types within a given gastric gland varies along the proximal to distal axis of the glandular hindstomach (Figure 6A) (Choi et al., 2014; Kim and Shivdasani, 2016; McCracken and Wells, 2017; Willet and Mills, 2016). In rodents, all differentiated cell types are present in glands of the proximal (also known as fundic) hindstomach, whereas chief cells and parietal cells are absent in the distal (also known as antral) hindstomach. Gastrin-producing enteroendocrine cells (G cells) reside exclusively in distal (antral) hindstomach glands of mice and humans. Distal (antral) glands also contain basal gland cells, which display features of both neck mucus and chief cells (Willet and Mills, 2016). Although there is an approximately 90% decrease in parietal cells along the transition zone between proximal (fundic) gastric glands and distal (antral) gastric glands in the human stomach, glands containing both gastrin-expressing G cells and acid-producing parietal cells can exist in the most distal regions of the stomach (Choi et al., 2014).
Although the mechanisms controlling epithelial cytodifferentiation in the stomach are less understood compared with the intestine, several transcription factors and signaling pathways have emerged as key players. WNT signaling plays an essential role in regionalizing epithelial cell type identity within the proximal (fundic) and distal (antral) domains of the glandular hindstomach. One key marker distinguishing these domains is the transcription factor pancreatic and duodenal homeobox 1 (PDX1), which is present in the distal (antral) glands but absent in the proximal (fundic) glands. When WNT signaling is blocked early in the developing proximal hindstomach epithelium by elimination of B-catenin using Shh-Cre, proximal gland marker expression is replaced by distal gland marker expression by E10.5, including the induction of PDX1 (McCracken et al., 2017). This suggests that WNT signaling is necessary to specify proximal versus distal gastric gland types during development. Analogous to mouse stomach, WNT signaling is also essential to promote proximal (fundic) glandular stomach development as demonstrated using human pluripotent cell-derived gastric organoids (McCracken et al., 2017). When WNT is added to the cultures during the gastric specification phase, proximal (fundic) type organoids emerge. The omission of WNT allows distal (antral) type organoids to form. Together, these data indicate that regulation of WNT signaling is essential for correct gastric gland regionalization in mice and humans.
Chief cell maturation within the proximal (fundic) hindstomach requires the transcription factor X-box binding protein 1 (XBP1) and its target, Muscle intestine and stomach expression 1 (MIST1) (Huh et al., 2010; Ramsey et al., 2007; Tian et al., 2010). Most recently, MIST1 has been identified as a transcription factor that widely controls the establishment and maintenance of a generalized secretory cell architecture in diverse cell types, including the gastric chief cell (Lo et al., 2017). Estrogen-related receptor gamma (ESRRG) null mice have reduced parietal cells in proximal (fundic) glands suggesting ESRRG-regulated production of these cells (Alaynick et al., 2010). Pit cell differentiation requires the forkhead protein FOXQ1 (Verzi et al., 2008). GATA4 may also play a role in development of gastric cell types including parietal cells, chief cells, and neck mucus-producing cells (Jacobsen et al., 2002).
Enteroendocrine cell types vary along the proximal to distal aspect of the hindstomach, and specific transcription factors regulate differentiation of region-specific types. For example, Neurogenin 3 (NGN3) and ARX are required for differentiation of distinct subsets of gastric enteroendocrine cells found in either the proximal (fundic) or distal (antral) regions (Du et al., 2012; Jenny et al., 2002; Kokubu et al., 2008; Lee et al., 2002). PDX1, NKX6.3, and PAX6 are each required specifically in the distal (antral) hindstomach for gastrin-producing G-cell differentiation, while PAX4 is necessary for serotonin and somatostatin producing enteroendocrine subtypes in this region (Choi et al., 2008; Larsson et al., 1996; Larsson et al., 1998). Tuft cells, although present throughout the hindstomach, are enriched at the squamocolumnar junction and require Pou2f3 (Gerbe et al., 2016). SPDEF is essential for differentiation of mucus cells within the distal (antral) hindstomach (Horst et al., 2010).
Finally, loss of gastric BMP signaling disrupts parietal cell development in the proximal (fundic) glands (Shinohara et al., 2010). Epithelial hyperplasia and the development of hybrid cells producing both mucins and zymogens also occurs in the absence of proper BMP signaling. Overall, BMP signaling seems to have a vital role in maintenance of the gastric epithelium because loss of BMP signaling resulting in metaplasia and dysplasia (Todisco, 2017).
B. Intestinal cytodifferentiation.
Similar to the gastric epithelium, specific types of intestinal epithelial cells work in coordination to accomplish the functions of the small intestine. The cell fate decisions that determine specification of secretory versus absorptive intestinal epithelial cells have been rigorously studied and extensively reviewed by others (Bjerknes and Cheng, 2010; Cheng and Leblond, 1974; Chin et al., 2017; De Mey and Freund, 2013; Noah et al., 2011; Qi and Chen, 2015; Simons and Clevers, 2011; Spence et al., 2011). The secretory cell lineage is composed of four subtypes: goblet, enteroendocrine, tuft, and Paneth cells. Goblet cells account for approximately 10% of the cells in the intestinal epithelium. These cells produce and secrete mucins that coat the apical surface of the epithelium providing a protective barrier against luminal pathogens (Birchenough et al., 2015; McCauley and Guasch, 2015). Enteroendocrine cells, comprising only 1% of the intestinal epithelium, secrete hormones into the lamina propria to regulate processes including cell growth and repair, glycemia, exocrine pancreatic secretion, and gut wall motility (Jenny et al., 2002). Tuft cells, comprising a rare 0.4% of the epithelium, secrete opioids to control pain, gastric emptying, intestinal secretion, and gut motility (Gerbe et al., 2012; Gerbe and Jay, 2016; Holzer, 2009; Kaji and Kaunitz, 2017). Paneth cells, which secrete antimicrobial peptides into the lumen, are the only mature cell type to migrate downward into the crypt instead of upward along the villus (Andreu et al., 2008; Ayabe et al., 2000). Finally, cells of the absorptive lineage, enterocytes, are most abundant. Enterocytes have microvilli on their apical surface, significantly expanding the luminal surface area to promote maximal efficiency in nutrient absorption.
Of these cell types, both enterocytes and enteroendocrine cells are specialized along the length of the small intestinal epithelium to have specific functions within different regions of the intestine (Figure 6B). The signaling mechanism(s) and transcription factors necessary to regionalize an enterocyte, giving it distinct functions, are not well understood. Factors differentially expressed within enterocytes in these regions likely play vital roles in determining and maintaining regional enterocyte identity. PDX is one such factor. PDX1 expression spans the SOX2/CDX2 boundary extending from the distal (antral) SOX2+ hindstomach into the CDX2+ duodenum, making PDX1 a duodenal-specific transcription factor in the small intestine (Figure 2) (Guz et al., 1995). Mice lacking PDX1 have impaired iron absorption, a specific duodenal function, as well as altered lipid absorption and lipid clearance (Chen et al., 2009). Consistent with this finding, PDX1 overexpression in the colon carcinoma-derived cell line Caco-2 causes changes in expression of genes associated with nutrient and lipid metabolism (Chen et al., 2012a). Genes directly regulated by PDX1 include the proximal enteroendocrine cell markers gastric inhibitory polypeptide (Gip) and somatostatin (Sst) as well as the duodenal expressed gene Ada (Chen et al., 2009; Dusing et al., 2001; Jepeal et al., 2005). PDX1 has also been shown to repress Sucrase isomaltase, Lactase, and Fatty acid binding protein 1 transcription, each encoding proteins primarily associated with jejunal function (Chen et al., 2012b; Heller et al., 1998; Wang et al., 2004). These data implicate PDX1 as a key regulator of duodenal enterocyte identity (Figure 6B).
Another differentially expressed transcription factor is GATA4, which is present within the epithelium of the duodenum and jejunum but absent from epithelium of the ileum (Battle et al., 2008; Bosse et al., 2006; Fang et al., 2006). Early (E11.5) conditional knockout of Gata4 in the intestine results in an epithelial cell proliferation defect via transcriptional regulation of cell cycle proteins and Frizzled class receptor 5 (Fzd5) (Kohlnhofer et al., 2016). Additionally, villus morphogenesis is delayed, and epithelial cell populations are altered. Importantly, jejunal-enriched transcripts decrease, and ileal-enriched transcripts increase. A shift away from jejunal identity and toward ileal identity also occurs when GATA4 is deleted in the intestinal epithelium later in development (E16.5) using Villin-Cre (Walker et al., 2014b). Morphologically, jejuna of E16.5 GATA4-Villin-Cre knockout intestines are predominantly normal but with altered gene expression in the enterocyte and enteroendocrine cell populations and a slight increase in goblet cell number. The changes in jejunal enterocyte gene expression observed in embryonic GATA4 mutants persist into adulthood with GATA4-deficient jejunum losing expression of 53% of the jejunal-specific gene set and gaining expression of 47% of the ileal-specific gene set (Battle et al., 2008). Importantly, the jejunal-specific set repressed includes genes encoding proteins necessary for lipid and cholesterol transport. The ileal-specific set upregulated includes genes encoding proteins involved in bile acid absorption. Consequently, these mice absorb less fat and cholesterol than wild-type mice. Inducible knockout of GATA4 from the mature epithelium results in a similar effect on gene expression (Bosse et al., 2006). Most recently, GATA4 has been shown to be both necessary and sufficient maintain jejunal and suppress ileal identity within the small intestinal epithelium (Thompson et al., 2017). Ectopic expression of GATA4 in the mouse ileum results in a loss of the ileal gene expression program and a gain in the jejunal program. Together, these findings indicate that GATA4 is essential for the establishment and maintenance of jejunal versus ileal enterocyte identities. To date, GATA4 is the only factor known to have such a principal role in regulating the jejunal-ileal boundary.
It is important to note that GATA6 is present throughout the intestinal epithelium and that the presence of GATA6 in GATA4-deficient jejunum or GATA4-expressing ileum does not compensate for loss or gain of GATA4 function in these tissues (Walker et al., 2014a; Walker et al., 2014b). Furthermore, inducible deletion of GATA6 from the adult intestinal epithelium does not alter jejunal enterocyte identity (Beuling et al., 2011). Together, these data emphasize that jejunal enterocyte regionalization is a function is unique to GATA4. When GATA6 is deleted from the ileum (where GATA4 is absent), however, ileal enterocyte gene expression changes with expression of colon epithelial cell markers Car1 and Car2 induced (Beuling et al., 2011). Loss of GATA6 from the embryonic ileal intestinal epithelium also reduces expression of ileal enterocyte markers and induces expression of the colon epithelial cell marker Car1 (Walker et al., 2014b). Simultaneous inducible deletion of both GATA4 and GATA6 from the adult intestinal epithelium results in duodenal and jejunal epithelial cells displaying a phenotype similar to the GATA6-deficient ileum (Beuling et al., 2011). A similar phenotype also occurs in embryos lacking both GATA4 and GATA6 in jejunal epithelium (Walker et al., 2014a). These data suggest that while GATA4 controls jejunal versus ileal enterocyte patterning, GATA6 refines ileal enterocyte identity within the intestinal epithelium by repressing expression of colonic genes.
Distinct subtypes of enteroendocrine cells are also differentially distributed among small intestine regions. For example, GIP-expressing and CCK-expressing enteroendocrine cells are enriched in the proximal intestine while PYY-expressing enteroendocrine cells are enriched in the distal intestine (Schonhoff et al., 2004). Several transcription factors have been found to play key roles in enteroendocrine cell development including ATOH1, NGN3, NEUROD1, PDX1, PAX4, and PAX6 (Schonhoff et al., 2004; Sheaffer and Kaestner, 2012). While ATOH1, NGN3, and NEUROD1 seem to regulate global development of enteroendocrine cells in the intestine, PDX1, PAX4, and PAX6 control enteroendocrine differentiation within the proximal intestine with enteroendocrine cells virtually eliminated from the duodenum and jejunum of mutants (Figure 6B) (Larsson et al., 1998; Schonhoff et al., 2004; Sheaffer and Kaestner, 2012). Differences in regionally-enriched enteroendocrine cell populations also occur in the small intestine of GATA4 and GATA6 conditional knockout embryos (Battle et al., 2008; Walker et al., 2014a; Walker et al., 2014b). While GATA6 is present in enteroendocrine cells and, therefore, may exert direct control over cell differentiation, GATA4 is absent from this cell population (Bosse et al., 2006; Walker et al., 2014b). Therefore, GATA4 likely regulates the enteroendocrine cell population within the intestine indirectly.
4. Concluding remarks and future perspectives
Although a handful of signaling pathways and transcription factors have been identified as key regulators of GI regional patterning, more work is needed to complete the picture. A full understanding of the mechanisms through which the GI epithelium acquires and maintains the unique gene expression patterns and tissue architectures requisite for regional-specific functions is expected to provide essential new insights into GI diseases including metaplasias, cancers, and short bowel syndrome. For example, knowledge of the normal mechanisms of esophageal and gastric epithelial development and homeostasis provides a foundation upon which new strategies to diagnose, treat, and inhibit metaplasia including Barrett's esophagus, spasmolytic polypeptideexpressing metaplasia, and intestinal metaplasia in the stomach can be elucidated. Knowledge of the patterning mechanisms that regulate duodenal, jejunal, and ileal epithelial cell identities in the small intestine provides the basis for future development of better short bowel syndrome therapies, particularly those aimed at restoring lost functions to remaining intestinal tissue. Finally, knowledge of GI epithelial patterning mechanisms has been used to guide the development of new and refinement of existing in vitro GI organ model systems including those derived from directed differentiation of pluripotent stem cells or those derived directly from human tissue samples. These models hold tremendous promise for improving our understanding human GI development itself, for bettering of our knowledge of infectious diseases, for improved pharmacology and toxicology studies, and for innovative tissue engineering and personalized medicine approaches to treating GI diseases.
Highlights.
Regionalization of the gastrointestinal (GI) epithelium is necessary for efficient nutrient assimilation.
Development of unique organ-specific epithelial structures and cell types is required for organs to perform distinct digestive functions.
Spatiotemporal regulation of signaling pathways and downstream transcription factors controls GI epithelial morphogenesis and cytodifferentiation.
This review discusses fundamental mechanisms that govern the patterning of GI epithelium along the cephalocaudal GI axis to confer regional-specific functions.
Acknowledgments
Funding: This work was supported by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (grant numbers DK087873 and DK111822, MAB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, Inoue Y, 2008. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis 14, 908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, Yu R, Jonker JW, Holt JA, Rajpal DK, Li H, Stuart J, McPherson R, Remlinger KS, Chang CY, McDonnell DP, Evans RM, Billin AN, 2010. ERRgamma regulates cardiac, gastric, and renal potassium homeostasis. Mol Endocrinol 24, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu P, Peignon G, Slomianny C, Taketo MM, Colnot S, Robine S, Lamarque D, Laurent-Puig P, Perret C, Romagnolo B, 2008. A genetic study of the role of the Wnt/beta-catenin signalling in Paneth cell differentiation. Dev Biol 324, 288–296. [DOI] [PubMed] [Google Scholar]
- Aronson BE, Rabello Aronson S, Berkhout RP, Chavoushi SF, He A, Pu WT, Verzi MP, Krasinski SD, 2014. GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27. Biochim Biophys Acta 1839, 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin J, Déry U, Lemieux M, Chailler P, Jeannotte L, 2002. Stomach regional specification requires Hoxa5-driven mesenchymal-epithelial signaling. Development 129, 4075–4087. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R, 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ, 2000. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol 1, 113–118. [DOI] [PubMed] [Google Scholar]
- Babeu JP, Darsigny M, Lussier CR, Boudreau F, 2009. Hepatocyte nuclear factor 4alpha contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am J Physiol Gastrointest Liver Physiol 297, G124–34. [DOI] [PubMed] [Google Scholar]
- Bai YQ, Yamamoto H, Akiyama Y, Tanaka H, Takizawa T, Koike M, Kenji Yagi O, Saitoh K, Takeshita K, Iwai T, Yuasa Y, 2002. Ectopic expression of homeodomain protein CDX2 in intestinal metaplasia and carcinomas of the stomach. Cancer Lett 176, 47–55. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O’Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, Ribeiro U, Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M, 2009. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 41, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Bondow BJ, Iverson MA, Adams SJ, Jandacek RJ, Tso P, Duncan SA, 2008. GATA4 is essential for jejunal function in mice. Gastroenterology 135, 1676–1686.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA, 2006. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci U S A 103, 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F, Chawengsaksophak K, Waring P, Playford RJ, Furness JB, 1999. Reprogramming of intestinal differentiation and intercalary regeneration in Cdx2 mutant mice. Proc Natl Acad Sci U S A 96, 7318–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F, Erler T, Russell A, James R, 1995. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn 204, 219–227. [DOI] [PubMed] [Google Scholar]
- Beuling E, Aronson BE, Tran LM, Stapleton KA, ter Horst EN, Vissers LA, Verzi MP, Krasinski SD, 2012. GATA6 is required for proliferation, migration, secretory cell maturation, and gene expression in the mature mouse colon. Mol Cell Biol 32, 3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Baffour-Awuah NY, Stapleton KA, Aronson BE, Noah TK, Shroyer NF, Duncan SA, Fleet JC, Krasinski SD, 2011. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology 140, 1219–1229.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Bosse T, aan de Kerk DJ, Piaseckyj CM, Fujiwara Y, Katz SG, Orkin SH, Grand RJ, Krasinski SD, 2008. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol 322, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA, Krasinski SD, 2010. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut 59, 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder HJ, Reuben A, 2005. Nutrient digestion and absorption. Medical physiology: a cellular and molecular approach. 947–974. [Google Scholar]
- Birchenough GM, Johansson ME, Gustafsson JK, Bergström JH, Hansson GC, 2015. New developments in goblet cell mucus secretion and function. Mucosal Immunol 8, 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H, 2010. Cell Lineage metastability in Gfi1-deficient mouse intestinal epithelium. Dev Biol 345, 49–63. [DOI] [PubMed] [Google Scholar]
- Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD, 2006. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol 26, 9060–9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D, Puig S, Iglesias M, Seoane A, de Bolós C, Munitiz V, Parrilla P, Comerma L, Poulsom R, Krishnadath KK, Grande L, Pera M, 2012. Activation of the BMP4 pathway and early expression of CDX2 characterize non-specialized columnar metaplasia in a human model of Barrett’s esophagus. J Gastrointest Surg 16, 227–37; discussion 237. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F, 2004. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci U S A 101, 7641–7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Köntgen F, Beck F, 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84–87. [DOI] [PubMed] [Google Scholar]
- Chen C, Fang R, Chou LC, Lowe AW, Sibley E, 2012a. PDX1 regulation of FABP1 and novel target genes in human intestinal epithelial Caco-2 cells. Biochem Biophys Res Commun 423, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Fang R, Davis C, Maravelias C, Sibley E, 2009. Pdx1 inactivation restricted to the intestinal epithelium in mice alters duodenal gene expression in enterocytes and enteroendocrine cells. Am J Physiol Gastrointest Liver Physiol 297, G1126–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Leavitt T, Sibley E, 2012b. Intestinal Pdx1 mediates nutrient metabolism gene networks and maternal expression is essential for perinatal growth in mice. Biochem Biophys Res Commun 424, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qin R, Liu B, Ma Y, Su Y, Yang CS, Glickman JN, Odze RD, Shaheen NJ, 2008. Multilayered epithelium in a rat model and human Barrett’s esophagus: similar expression patterns of transcription factors and differentiation markers. BMC Gastroenterol 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Leblond CP, 1974. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat 141, 537–561. [DOI] [PubMed] [Google Scholar]
- Chin AM, Hill DR, Aurora M, Spence JR, 2017. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol 66, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Roland JT, Barlow BJ, O’Neal R, Rich AE, Nam KT, Shi C, Goldenring JR, 2014. Cell lineage distribution atlas of the human stomach reveals heterogeneous gland populations in the gastric antrum. Gut 63, 1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MY, Romer AI, Wang Y, Wu MP, Ito S, Leiter AB, Shivdasani RA, 2008. Requirement of the tissue-restricted homeodomain transcription factor Nkx6.3 in differentiation of gastrin-producing G cells in the stomach antrum. Mol Cell Biol 28, 3208–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM, 2004. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol 287, C171–81. [DOI] [PubMed] [Google Scholar]
- Darsigny M, Babeu JP, Dupuis AA, Furth EE, Seidman EG, Lévy E, Verdu EF, Gendron FP, Boudreau F, 2009. Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One 4, e7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RA, Attie AD, 2008. Deletion of the ileal basolateral bile acid transporteridentifies the cellular sentinels that regulate the bile acid pool. Proc. Natl. Acad. Sci.USA [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey JR, Freund JN, 2013. Understanding epithelial homeostasis in the intestine: An old battlefield of ideas, recent breakthroughs and remaining controversies. Tissue Barriers 1, e24965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X, 2011. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development 138, 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes T, Senkel S, Holewa B, Ryffel GU, 1996. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol 16, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A, McCracken KW, Walp ER, Terry NA, Klein TJ, Han A, Wells JM, May CL, 2012. Arx is required for normal enteroendocrine cell development in mice and humans. Dev Biol 365, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan SP, Behan FM, Kirca M, Zaheer A, McGarrigle SA, Reynolds JV, Vaz GM, Senge MO, Kelleher D, 2016. The characterization of an intestine-like genomic signature maintained during Barrett’s-associated adenocarcinogenesis reveals an NR5A2-mediated promotion of cancer cell survival. Sci Rep 6, 32638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE, 1994. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci U S A 91, 7598–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusing MR, Florence EA, Wiginton DA, 2001. Pdx-1 is required for activation in vivo from a duodenum-specific enhancer. J Biol Chem 276, 14434–14442. [DOI] [PubMed] [Google Scholar]
- Fang R, Olds LC, Sibley E, 2006. Spatio-temporal patterns of intestine-specific transcription factor expression during postnatal mouse gut development. Gene Expr Patterns 6, 426–432. [DOI] [PubMed] [Google Scholar]
- Feng R, Xiao C, Zavros Y, 2012. The role of Sonic Hedgehog as a regulator of gastric function and differentiation. Vitam Horm 88, 473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, White P, Kaestner KH, 2009. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell 16, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA, 2006. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology 130, 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Jay P, 2016. Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol 9, 1353–1359. [DOI] [PubMed] [Google Scholar]
- Gerbe F, Legraverend C, Jay P, 2012. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci 69, 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P, 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S, Savory JG, Lohnes D, 2010. Cdx2 regulates patterning of the intestinal epithelium. Dev Biol 339, 155–165. [DOI] [PubMed] [Google Scholar]
- Gregorieff A, Grosschedl R, Clevers H, 2004. Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(−/−)/Tcf1(−/−) embryos. EMBO J 23, 1825–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G, 1995. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 121, 11–18. [DOI] [PubMed] [Google Scholar]
- Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G, Cereghini S, 2005. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci U S A 102, 1490–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveri H, Westerholm-Ormio M, Lindfors K, Mäki M, Savilahti E, Andersson LC, Heikinheimo M, 2008. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa Y, Sakitani K, Konishi M, Asfaha S, Niikura R, Tomita H, Renz BW, Tailor Y, Macchini M, Middelhoff M, Jiang Z, Tanaka T, Dubeykovskaya ZA, Kim W, Chen X, Urbanska AM, Nagar K, Westphalen CB, Quante M, Lin CS, Gershon MD, Hara A, Zhao CM, Chen D, Worthley DL, Koike K, Wang TC, 2017. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 31, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RS, Stoffers DA, Hussain MA, Miller CP, Habener JF, 1998. Misexpression of the pancreatic homeodomain protein IDX-1 by the Hoxa-4 promoter associated with agenesis of the cecum. Gastroenterology 115, 381–387. [DOI] [PubMed] [Google Scholar]
- Holzer P, 2009. Opioid receptors in the gastrointestinal tract. Regul Pept 155, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing J, Pavlov KV, Meijer C, Smit JK, Boersma-van Ek W, Karrenbeld A, Burgerhof JG, Kruyt FA, Plukker JT, 2014. Loss of CD44 and SOX2 expression is correlated with a poor prognosis in esophageal adenocarcinoma patients. Ann Surg Oncol 21 Suppl 4, S657–64. [DOI] [PubMed] [Google Scholar]
- Horst D, Gu X, Bhasin M, Yang Q, Verzi M, Lin D, Joseph M, Zhang X, Chen W, Li YP, Shivdasani RA, Libermann TA, 2010. Requirement of the epithelium-specific Ets transcription factor Spdef for mucous gland cell function in the gastric antrum. J Biol Chem 285, 35047–35055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WJ, Esen E, Geahlen JH, Bredemeyer AJ, Lee AH, Shi G, Konieczny SF, Glimcher LH, Mills JC, 2010. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 139, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E, 1998. Functional differences between keratins of stratified and simple epithelia. J Cell Biol 143, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen CM, Mannisto S, Porter-Tinge S, Genova E, Parviainen H, Heikinheimo M, Adameyko II, Tevosian SG, Wilson DB, 2005. GATA-4:FOG interactions regulate gastric epithelial development in the mouse. Dev Dyn 234, 355–362. [DOI] [PubMed] [Google Scholar]
- Jacobsen CM, Narita N, Bielinska M, Syder AJ, Gordon JI, Wilson DB, 2002. Genetic mosaic analysis reveals that GATA-4 is required for proper differentiation of mouse gastric epithelium. Dev Biol 241, 34–46. [DOI] [PubMed] [Google Scholar]
- Jeejeebhoy KN, 2002. Short bowel syndrome: a nutritional and medical approach. CMAJ 166, 1297–1302. [PMC free article] [PubMed] [Google Scholar]
- Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G, 2002. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J 21, 6338–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepeal LI, Fujitani Y, Boylan MO, Wilson CN, Wright CV, Wolfe MM, 2005. Cell-specific expression of glucose-dependent-insulinotropic polypeptide is regulated by the transcription factor PDX-1. Endocrinology 146, 383–391. [DOI] [PubMed] [Google Scholar]
- Jiang M, Li H, Zhang Y, Yang Y, Lu R, Liu K, Lin S, Lan X, Wang H, Wu H, Zhu J, Zhou Z, Xu J, Lee DK, Zhang L, Lee YC, Yuan J, Abrams JA, Wang TC, Sepulveda AR, Wu Q, Chen H, Sun X, She J, Chen X, Que J, 2017. Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature 550, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji I, Kaunitz JD, 2017. Luminal chemosensing in the gastroduodenal mucosa. Curr Opin Gastroenterol 33, 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Mills JC, 2010. The gastric mucosa development and differentiation. Prog Mol Biol Transl Sci 96, 93–115. [DOI] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA, 2005. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell 8, 611–622. [DOI] [PubMed] [Google Scholar]
- Kim BM, Miletich I, Mao J, McMahon AP, Sharpe PA, Shivdasani RA, 2007. Independent functions and mechanisms for homeobox gene Barx1 in patterning mouse stomach and spleen. Development 134, 3603–3613. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Li E, Oh SP, Schrewe H, Harmon EB, Lee JS, Melton DA, 2000. Activin receptor patterning of foregut organogenesis. Genes Dev 14, 1866–1871. [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Shivdasani RA, 2016. Stomach development, stem cells and disease. Development 143, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlnhofer BM, Thompson CA, Walker EM, Battle MA, 2016. GATA4 regulates epithelial cell proliferation to control intestinal growth and development in mice. Cell Mol Gastroenterol Hepatol 2, 189–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubu H, Ohtsuka T, Kageyama R, 2008. Mash1 is required for neuroendocrine cell development in the glandular stomach. Genes Cells 13, 41–51. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR, 2004. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang HT, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ, 2004. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A 101, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson LI, Madsen OD, Serup P, Jonsson J, Edlund H, 1996. Pancreatic-duodenal homeobox 1 -role in gastric endocrine patterning. Mech Dev 60, 175–184. [DOI] [PubMed] [Google Scholar]
- Larsson LI, St-Onge L, Hougaard DM, Sosa-Pineda B, Gruss P, 1998. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mech Dev 79, 153–159. [DOI] [PubMed] [Google Scholar]
- Lee CS, Perreault N, Brestelli JE, Kaestner KH, 2002. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev 16, 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Udager AM, Hu C, Qiao XT, Richards N, Gumucio DL, 2009. Dynamic patterning at the pylorus: formation of an epithelial intestine-stomach boundary in late fetal life. Dev Dyn 238, 3205–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C, 1998. Sonic hedgehog is essential to foregut development. Nat Genet 20, 58–61. [DOI] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, Peters JH, Rustgi A, Onaitis MW, Kiernan A, Chen X, Que J, 2013. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 12, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HG, Jin RU, Sibbel G, Liu D, Karki A, Joens MS, Madison BB, Zhang B, Blanc V, Fitzpatrick JA, Davidson NO, Konieczny SF, Mills JC, 2017. A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev 31, 154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL, 2005. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279–289. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S, 2005. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem 280, 24371–24379. [DOI] [PubMed] [Google Scholar]
- McCauley HA, Guasch G, 2015. Three cheers for the goblet cell: maintaining homeostasis in mucosal epithelia. Trends Mol Med 21, 492–503. [DOI] [PubMed] [Google Scholar]
- McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH, Wells JM, 2017. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature 541, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM, 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Wells JM, 2017. Mechanisms of embryonic stomach development. Semin Cell Dev Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Moy JR, Lin L, Schipper M, Normolle D, Brenner DE, Iannettoni MD, Orringer MB, Beer DG, 2003. Gene amplification in esophageal adenocarcinomas and Barrett’s with high-grade dysplasia. Clin Cancer Res 9, 4819–4825. [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A, 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. [DOI] [PubMed] [Google Scholar]
- Moore BD, Jin RU, Lo H, Jung M, Wang H, Battle MA, Wollheim CB, Urano F, Mills JC, 2016. Transcriptional Regulation of X-Box-binding Protein One (XBP1) by Hepatocyte Nuclear Factor 4α (HNF4A) Is Vital to Beta-cell Function. J Biol Chem 291, 6146–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, Hudson JR, Howell JC, Chatuvedi P, Spence JR, Shannon JM, Zorn AM, Helmrath MA, Wells JM, 2017. Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell 21, 51–64.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Hakamata Y, Sato K, Eda A, Yanaka I, Honda S, Osawa H, Kaneko Y, Sugano K, 2002. Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem Biophys Res Commun 294, 470–479. [DOI] [PubMed] [Google Scholar]
- Noah TK, Donahue B, Shroyer NF, 2011. Intestinal development and differentiation. Exp Cell Res 317, 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA, 2003. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34, 292–296. [DOI] [PubMed] [Google Scholar]
- Phillips RW, Frierson HF, Moskaluk CA, 2003. Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. Am J Surg Pathol 27, 1442–1447. [DOI] [PubMed] [Google Scholar]
- Qi Z, Chen YG, 2015. Regulation of intestinal stem cell fate specification. Sci China Life Sci 58, 570–578. [DOI] [PubMed] [Google Scholar]
- Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL, 2006. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation 74, 422–437. [DOI] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL, 2007. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghoebir L, Bakker ER, Mills JC, Swagemakers S, Kempen MB, Munck AB, Driegen S, Meijer D, Grosveld F, Tibboel D, Smits R, Rottier RJ, 2012. SOX2 redirects the developmental fate of the intestinal epithelium toward a premature gastric phenotype. J Mol Cell Biol 4, 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon AP, 2000. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127, 2763–2772. [DOI] [PubMed] [Google Scholar]
- Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC, 2007. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1 Development 134, 211–222. [DOI] [PubMed] [Google Scholar]
- Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J, 2010. BMP signaling in the development of the mouse esophagus and forestomach. Development 137, 4171–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, Sinha S, 2012. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139, 772–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Roman AK, Shivdasani RA, 2011. Boundaries, junctions and transitions in the gastrointestinal tract. Exp Cell Res 317, 2711–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB, 2004. Minireview: Development and differentiation of gut endocrine cells. Endocrinology 145, 2639–2644. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F, 2007. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536. [DOI] [PubMed] [Google Scholar]
- Sheaffer KL, Kaestner KH, 2012. Transcriptional networks in liver and intestinal development. Cold Spring Harb Perspect Biol 4, a008284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, Melton DA, 2009. Transcriptional dynamics of endodermal organ formation. Dev Dyn 238, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Maehr R, Mazzoni EO, Melton DA, 2011. Wnt signaling specifies and patterns intestinal endoderm. Mech Dev 128, 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Mao M, Keeley TM, El-Zaatari M, Lee HJ, Eaton KA, Samuelson LC, Merchant JL, Goldenring JR, Todisco A, 2010. Bone morphogenetic protein signaling regulates gastric epithelial cell development and proliferation in mice. Gastroenterology 139, 2050–2060.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH, 2002. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122, 689–696. [DOI] [PubMed] [Google Scholar]
- Simons BD, Clevers H, 2011. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145, 851–862. [DOI] [PubMed] [Google Scholar]
- Sousa JF, Nam KT, Petersen CP, Lee HJ, Yang HK, Kim WH, Goldenring JR, 2016. miR-30-HNF4γ and miR-194-NR2F2 regulatory networks contribute to the upregulation of metaplasia markers in the stomach. Gut 65, 914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lauf R, Shroyer NF, 2011. Vertebrate intestinal endoderm development. Dev Dyn 240, 501–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Dene B, Sala FG, Bellusci S, Gschmeissner S, Stamp G, Dickson C, 2006. Stomach development is dependent on fibroblast growth factor 10/fibroblast growth factor receptor 2b-mediated signaling. Gastroenterology 130, 1233–1244. [DOI] [PubMed] [Google Scholar]
- Stringer EJ, Duluc I, Saandi T, Davidson I, Bialecka M, Sato T, Barker N, Clevers H, Pritchard CA, Winton DJ, Wright NA, Freund JN, Deschamps J, Beck F, 2012. Cdx2 determines the fate of postnatal intestinal endoderm. Development 139, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulahian R, Chen J, Arany Z, Jadhav U, Peng S, Rustgi AK, Bass AJ, Srivastava A, Hornick JL, Shivdasani RA, 2015. SOX15 governs transcription in human stratified epithelia and a subset of esophageal adenocarcinomas. Cell Mol Gastroenterol Hepatol 1, 598–609.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Saito H, Kusumi T, Fukuda S, Shimoyama T, Sasaki Y, Suto K, Munakata A, Kudo H, 2001. Spatial distribution and histogenesis of colorectal Paneth cell metaplasia in idiopathic inflammatory bowel disease. J Gastroenterol Hepatol 16, 1353–1359. [DOI] [PubMed] [Google Scholar]
- Taraviras S, Mantamadiotis T, Dong-Si T, Mincheva A, Lichter P, Drewes T, Ryffel GU, Monaghan AP, Schütz G, 2000. Primary structure, chromosomal mapping, expression and transcriptional activity of murine hepatocyte nuclear factor 4gamma. Biochim Biophys Acta 1490, 21–32. [DOI] [PubMed] [Google Scholar]
- Taraviras S, Monaghan AP, Schütz G, Kelsey G, 1994. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev 48, 67–79. [DOI] [PubMed] [Google Scholar]
- Thompson CA, Battle MA, 2014. Basic Science of Small Intestinal Development, in: Gumucio DL, Samuelson LC, Spence JR (Eds.), Translational Gastroenterology: Organogenesis to Disease. John Wiley & Sons, Ltd, pp. 85–97. [Google Scholar]
- Thompson CA, Wojta K, Pulakanti K, Rao S, Dawson P, Battle MA, 2017. GATA4 Is Sufficient to Establish Jejunal Versus Ileal Identity in the Small Intestine. Cell Mol Gastroenterol Hepatol 3, 422–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Błazewska KM, McKenna CE, Mills JC, 2010. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol 30, 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco A, 2017. Regulation of Gastric Metaplasia, Dysplasia, and Neoplasia by Bone Morphogenetic Protein Signaling. Cell Mol Gastroenterol Hepatol 3, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Clevers H, 1993. Sox 15, a novel member of the murine Sox family of HMG box transcription factors. Nucleic Acids Res 21, 1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink GR, 2007. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev 87, 1343–1375. [DOI] [PubMed] [Google Scholar]
- van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, Peppelenbosch MP, 2004. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet 36, 277–282. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Khan AH, Ito S, Shivdasani RA, 2008. Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology 135, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, Stanfel MN, Moses KA, Kim BM, Zhang Y, Schwartz RJ, Shivdasani RA, Zimmer WE, 2009. Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development. Gastroenterology 136, 1701–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R, Yamaguchi T, Tadokoro K, 2005. Colonic Paneth cell metaplasia is preneoplastic condition of colonic cancer or not. J Carcinog 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EM, Thompson CA, Battle MA, 2014a. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol 392, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EM, Thompson CA, Kohlnhofer BM, Faber ML, Battle MA, 2014b. Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res Notes 7, 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Freddo AM, Wang S, Gumucio DL, 2016a. Generation of intestinal surface: an absorbing tale. Development 143, 2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud Å, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, Gumucio DL, 2012. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A 109, 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Whidden M, Kolterud A, Shoffner SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo AM, Schnell S, Gumucio DL, 2016b. Villification in the mouse: Bmp signals control intestinal villus patterning. Development 143, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DH, Tiwari A, Kim ME, Clemons NJ, Regmi NL, Hodges WA, Berman DM, Montgomery EA, Watkins DN, Zhang X, Zhang Q, Jie C, Spechler SJ, Souza RF, 2014. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. J Clin Invest 124, 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ouyang H, Yamamoto Y, Kumar PA, Wei TS, Dagher R, Vincent M, Lu X, Bellizzi AM, Ho KY, Crum CP, Xian W, McKeon F, 2011. Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell 145, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fang R, Olds LC, Sibley E, 2004. Transcriptional regulation of the lactase-phlorizin hydrolase promoter by PDX-1. Am J Physiol Gastrointest Liver Physiol 287, G555–61. [DOI] [PubMed] [Google Scholar]
- Willet SG, Mills JC, 2016. Stomach Organ and Cell Lineage Differentiation: from Embryogenesis to Adult Homeostasis. Cell Mol Gastroenterol Hepatol 2, 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Miletich I, Kim BM, Sharpe PT, Shivdasani RA, 2011. Barx1-mediated inhibition of Wnt signaling in the mouse thoracic foregut controls tracheoesophageal septation and epithelial differentiation. PLoS One 6, e22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F, 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718. [DOI] [PubMed] [Google Scholar]
- Yu WY, Slack JM, Tosh D, 2005. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol 284, 157–170. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiang M, Kim E, Lin S, Liu K, Lan X, Que J, 2016. Development and stem cells of the esophagus. Semin Cell Dev Biol [DOI] [PMC free article] [PubMed] [Google Scholar]