Abstract
Macrophages respond to chemical/metabolic and physical stimuli, but their effects cannot be readily decoupled in vivo during pro-inflammatory activation. Here, we show that preventing macrophage spreading by spatial confinement, as imposed by micropatterning, microporous substrates or cell crowding, suppresses late lipopolysaccharide (LPS)-activated transcriptional programs (biomarkers IL-6, CXCL9, IL-1β, and iNOS) by mechanomodulating chromatin compaction and epigenetic alterations (HDAC3 levels and H3K36-dimethylation). Mechanistically, confinement reduces actin polymerization, thereby lowers the LPS-stimulated nuclear translocation of MRTF-A. This lowers the activity of the MRTF-A-SRF complex and subsequently downregulates the inflammatory response, as confirmed by chromatin immunoprecipitation coupled with quantitative PCR and RNA sequencing analysis. Confinement thus downregulates pro-inflammatory cytokine secretion and, well before any activation processes, the phagocytic potential of macrophages. Contrarily, early events, including activation of the LPS receptor TLR4, and downstream NF-κB and IRF3 signalling and hence the expression of early LPS-responsive genes were marginally affected by confinement. These findings have broad implications in the context of mechanobiology, inflammation and immunology, as well as in tissue engineering and regenerative medicine.
During several pathological conditions, macrophages are activated into a spectrum of states, from pro-inflammatory (M1) to pro-healing (M2) phenotypes1, with gradually altered transcriptional profiles and functional outputs2–6. Soluble biochemical and metabolic factors have been seen as the major drivers and determinants of the resulting states7. Moreover, tissue-resident macrophages also encounter major physical stimuli and spatial constraints, from extracellular matrix stiffness, architecture and from neighbouring cells8–12. The same is true for macrophages that infiltrate tissue/implant interfaces after surgery to orchestrate the healing process. However, surprisingly little is known about whether and how the spatial confinement of macrophages might be associated with their transcriptional control, functional outputs and hence activation. For other cell types, physical factors have been shown to have a direct bearing on cellular physiology and differentiation13–17, but how these mechanical aspects influence macrophage activation and their functional outputs is still largely unclear and has found attention only recently18–22. This is due to the inherently heterogeneous microenvironments that macrophages experience in their native tissue niches, for example in terms of soluble biochemical factors, the chemical composition of the extracellular matrix, the physical parameters of their surroundings and, finally, cell-cell contacts. Furthermore, common immunology-based assays that average over millions of cells are incapable of clarifying how spatial confinement in tissues or in animal models affects the inflammatory activation of macrophages. Only recently, macrophage elongation has been observed in some disease models like atherosclerosis23,24, and a synergetic effect between macrophage elongation and interleukin IL-4/IL-13 induced signalling during M2 activation has been observed in vitro20. In contrast, how macrophage shape and spatial confinement impact the M1 activation process and downstream functional outputs remains unexplored.
Pro-inflammatory M1 activation is a multi-step process starting with the activation of Toll-like receptors (TLRs), followed by further downstream signalling cascades1,4, to enable macrophages to fight infections efficiently. Since much new biology has been learned from the study of cells on micro- and nano-engineered substrates, we have focused here on 2D adhesive islands and on microporous substrates, because an engineering approach ensures that the micro-environmental niche is the same for all cells analysed. Both approaches allow us to tightly regulate spatial and biochemical factors to quantitatively control the size and shape of single macrophages, thereby broadening the findings towards material science applications. We asked whether any of the sequential steps during M1 activation are affected by spatial confinement, and found that the M1 activation process has at least two temporal phases that are differentially regulated by spatial confinement. The early phase within the first 3 h after lipopolysaccharide (LPS) activation is not significantly impacted, including TLR4 activation and the activation of transcription factors like p65 and interferon regulatory transcription factor 3 (IRF3). In contrast, a few hours after LPS activation, the upregulation of various late-responsive genes is reduced for spatially confined macrophages, which we show is due to alterations in chromatin compaction, histone deacetylase 3 (HDAC3) levels and histone modifications. Analysis of the underpinning mechanism by chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) and RnA sequencing (RNA-seq) revealed that reduced actin polymerization in spatially confined macrophages resulted in lower activity of the myocardin-related transcription factor A-serum response factor (MRTF-A-SRF) complex. Lastly, and to test their capability to stage an inflammatory response to fight infections, we investigated the functional implications of spatial confinement on a macrophage’s phagocytic potential and cytokine secretion. Taken together, this study provides the first comprehensive evidence that macrophage confinement has critical implications for the activation and function of pro-inflammatory macrophages.
Confinement downsizes late pro-inflammatory gene expression
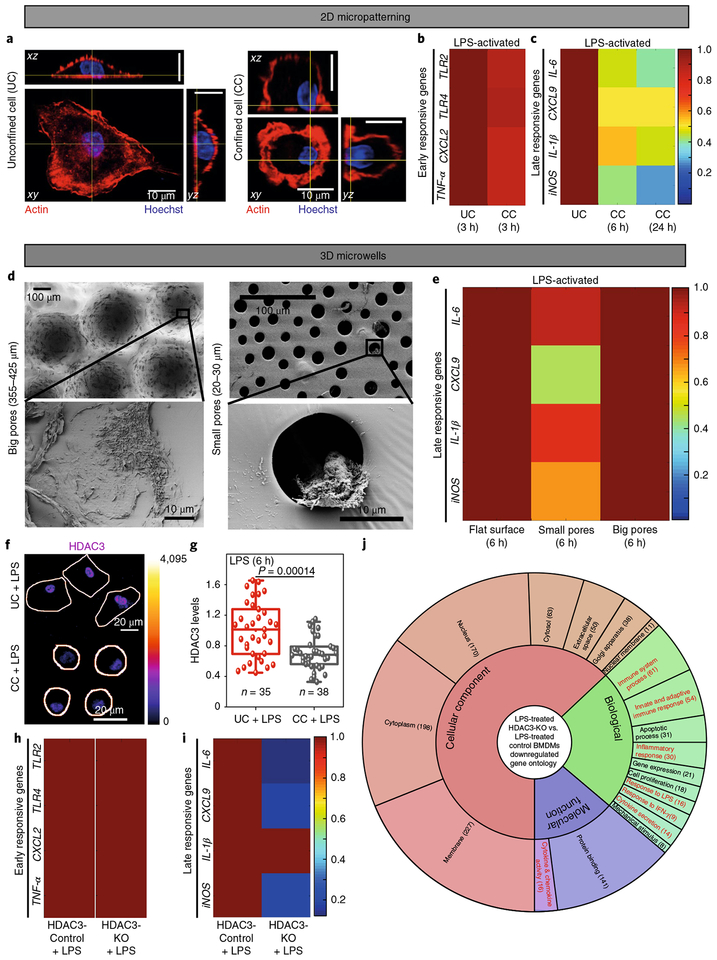
M1 activation is marked by the biphasic regulation of several gene clusters (Fig. 1), whereby the first cluster of genes responds to LPS treatment within the first few hours, while others respond only later (4–24 h)4–6. To elucidate the effect of confinement on these gene clusters, single bone-marrow-derived macrophages (BMDMs) were confined on circular fibronectin-coated substrates with an adhesive area of ~200 μm2 (Supplementary Fig. 1a–d), which restricts cell spreading. Unconfined cells (UCs) were more spread than confined cells (CCs) (Fig. 1a). Following LPS treatment, no difference was found in the expression of the early-responsive genes, including TLR2, TLR4, CXCL2 and TNF-α, between UCs and CCs (Fig. 1b and Supplementary Fig. 1e). Interestingly, expression of the late-responsive genes IL-6, CXCL9, IL-1β and iNOS, activated fully only after 4–6 h of LPS treatment6, were all significantly lower in LPS-treated CCs than in the UCs (Fig. 1c and Supplementary Fig. 1e). This effect remained well pronounced even after 24 h of LPS treatment (Fig. 1c and Supplementary Fig. 1e). Similarly, in microwells (Fig. 1d and Supplementary Figs. 2 and 3), which mimic the structure of porous implant materials, downregulation of CXCL9 and iNOS was observed in microwells with small pore sizes (20–30 μm) as compared to larger ones (355–425 μm) (Fig. 1e). This suggests that spatial confinement of macrophages directly controls the differential expression of gene clusters during progressive M1 activation and that sufficient spreading of macrophages is required for an efficient pro-inflammatory activation.
Fig. 1 |. Confinement controls the biphasic regulation of M1 transcriptional programs via HDAC3.
a, Representative orthogonal views of UC and CC BMDMs stained for F-actin and nuclei. Scale bars, 10 μm. b,c, Colour-coded arrays show the expression levels of early (b) and late (c) responsive gene clusters in UCs and CCs treated with LPS, as normalized to LPS-treated UCs. d, Representative electron microscopy images of BMDMs cultured in microwells with big (left) and small (right) pores, respectively, as further described in Supplementary Figs. 2 and 2. e, Colour-coded arrays show the expression levels of late-responsive genes in LPS-stimulated BMDMs cultured on flat polydimethylsiloxane (PDMS) surfaces, small and big hemispherical pores, normalized to the LPS-treated BMDMs cultured on a flat PDMS surface. f, Colour-coded representative images of UCs and CCs treated with LPS for 6 h and stained for HDAC3. Colour bar indicates pixel intensity values. Cell edges are marked in white. Scale bar, 20 μm. g, Box plots show HDAC3 levels in UCs and CCs treated with LPS for 6 h, normalized to LPS-treated UCs (the boxes show 25th and 75th percentiles, the middle horizontal line shows the median, small open squares show the mean, and whiskers indicate s.d.). n, number of cells analysed per condition. h,i, Colour-coded arrays show the expression levels of early (h) and late (i) responsive genes in LPS-treated HDAC3-Control and HDAC3-KO (knockout) BMDMs (reanalysed from previously published microarray data32, GEO accession no. GSE33162). Levels are normalized to the LPS-treated HDAC3-Control BMDMs. j, Gene ontology analysis of all downregulated genes in HDAC3-KO versus HDAC3-Control BMDMs, both treated with LPS, on flat surfaces. The number of genes in each category is shown in parentheses (Supplementary Data 1). P values were accessed with two-tailed Student’s t-test. All experiments were independently repeated three times with similar results.
Confinement tunes M1 specific epigenetic modifications
Because cell and nuclear morphologies are tightly coupled for other cell types25,26, we confirmed that LPS-treated UCs demonstrated higher nuclear projection areas, lower nuclear heights and slightly higher nuclear volumes (Supplementary Fig. 4a–e) compared to CCs, but found that the 2D nuclear shape (circularity) remained unchanged (Supplementary Fig. 4f). We next measured apparent chromatin spatial density, a measure of average chromatin compaction27, and found it to be much higher in LPS-treated CCs than in UCs (Supplementary Fig. 5b), suggesting a significantly higher chromatin compaction in CCs. Because H3K36-dimethylation (H3K36me2) is a marker for chromatin compaction and downregulates LPS-induced target genes28, the effect of confinement on H3K36me2 levels was probed. LPS-treated CCs had significantly higher levels of H3K36me2 than UCs (Supplementary Fig. 5a,b). Furthermore, ChIP-qPCR experiments showed higher enrichment of H3K36me2 at the IL-6 and iNOS gene promoter regions (Supplementary Fig. 5c) in Control BMDMs as compared to LPS-treated BMDMs, showing its role in gene repression. This was also found to be confinement-dependent, since, for the same genes, LPS-treated CCs showed higher fold-enrichment than UCs (Supplementary Fig. 5d). Confinement thus upregulates chromatin compaction and increases the levels of H3K36 dimethylation; however, CXCL9 and IL-1β expression was not found to be dependent on H3K36me2, which suggests the involvement of other possible epigenetic modifications and mechanisms.
Confinement limits HDAC3-regulated late M1 activation
In development and disease, changes in chromatin compaction and gene expression have been attributed to the altered activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs)29. Higher activity of Class-I HDACs30,31, mainly HDAC332,33, has been shown during pro-inflammatory gene expression. Although, in fibroblasts, biophysical regulation of gene expression by HDAC3 has recently been shown26, the function of HDAC3 in the differential regulation of gene clusters during M1 activation, and also its regulation by confinement, has not been explored. To probe this, macrophages were stimulated with LPS for different periods of time (1–9 h) and stained for HDAC3 (Supplementary Fig. 6a). Total levels of HDAC3 were found to increase following LPS treatment (Supplementary Fig. 6b), which correlated well with the timescale of late-responsive gene expression. Furthermore, LPS-treated UCs showed significantly higher HDAC3 levels than CCs (Fig. 1f,g), confirming that confinement downregulates the LPS-induced increase of HDAC3 expression, as seen for UCs. A similarly strong confinement-dependent regulation was not observed for HDAC1 (Supplementary Fig. 6a,c–e).
To check whether HDAC3 activity is indeed required for the biphasic expression of gene clusters, published microarray data for HDAC3-KO macrophages32 were reanalysed, confirming that the LPS-induced expression of early-responsive genes was largely independent of HDAC3 (ref.32 and Fig. 1h). In contrast, the late-responsive genes, which were found here to be sensitive to cell confinement (Fig. 1i), were severely downregulated in these HDAC3-KO macrophages (ref.32 and Fig. 1j), as further confirmed here by specifically inhibiting HDCA3 activity with RGFP966 (Supplementary Fig. 7a). Confinement-induced lower expression of late-responsive genes could be rescued in RAW 264.7 macrophages following transfection with a HDAC3-overexpressing plasmid (Supplementary Fig. 7b). Microarray analysis further showed that the expression levels of several inflammatory genes and late-responsive gene were downsized in LPS-induced HDAC3-KO BMDMs, with minimal effect on early-responsive genes (Supplementary Fig. 7c,d). Gene ontology analysis also confirmed that all the downregulated genes in LPS-induced HDAC3-KO BMDMs largely belong to the immune and LPS response system and inflammatory genes (Fig. 1j).
Morphological changes during M1 activation are reversible
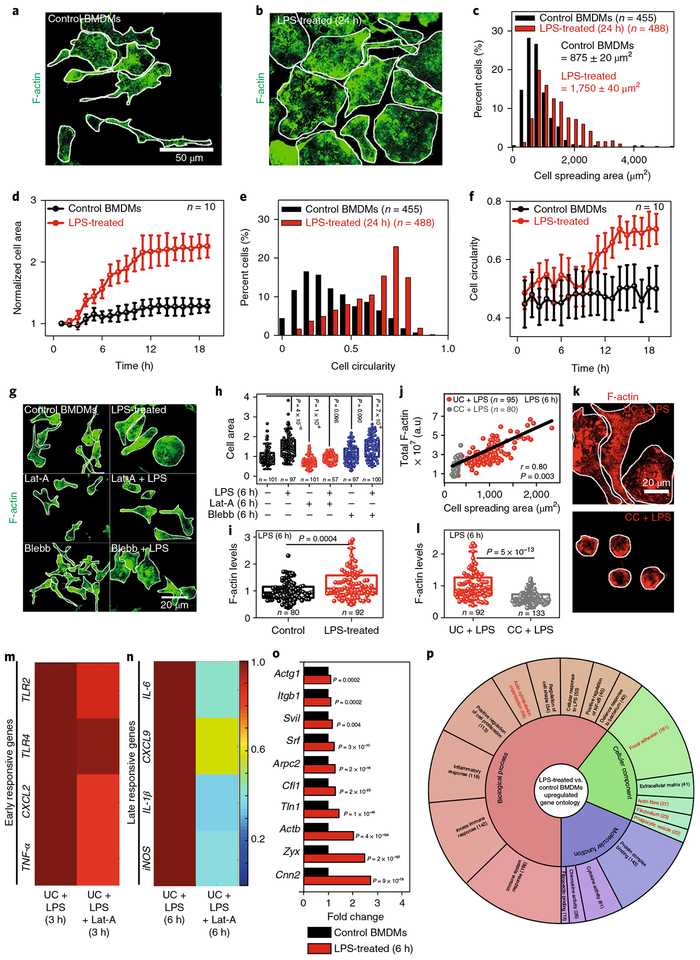
To gain a mechanistic understanding, we next checked whether macrophage M1 activation is also associated with morphological changes and whether these changes correlate with the observed timescales. After LPS stimulation for 24 h BMDMs increased their cell (Fig. 2a–c) and nuclear (Supplementary Fig. 8a,b) areas. BMDM activation showed two distinct phases: the cell area remained mostly unchanged up to 3 h and then increased significantly (4 h onwards) to more than twofold within 12–14 h of stimulation (Fig. 2d and Supplementary Videos 1 (Control BMDMs) and 2 (LPS-treated BMDMs)). The 2D cell shape also changed dramatically, from elongated to more circular (Fig. 2e), but was significant only after 12–14 h of activation (Fig. 2f). Unlike the cell spreading area, the cell shape transition was not a prerequisite for efficient transcription of late-responsive genes, as verified on micropatterned adhesive stripes (Supplementary Fig. 9a–c). Physiologically important, M1 activation was found to be reversed following LPS removal, both at the transcriptional and morphological levels (Supplementary Figs. 8a–c and 10a–e).
Fig. 2 |. Confinement downregulates LPS-induced actin polymerization, which tames down the expression of late-responsive genes.
a,b, Representative images of Control and LPS-treated BMDMs stained for F-actin. Scale bar, 50 μm. C–f, Normalized distributions and live dynamic changes of cell spreading areas and cell circularity in Control and LPS-treated BMDMs. Data are shown as mean±s.e. Cell circularity is defined as 4π(area) / (perimeter)2. g, Representative images of BMDMs treated with different drugs in the presence or absence of LPS (6 h). Scale bar, 20μm. h, Cell spreading area as normalized to untreated Control BMDMs. i, Box plots showing levels of F-actin in Control and LPS-treated BMDMs, as normalized to untreated Control BMDMs. j, Cell spreading area versus total F-actin content in LPS-treated UC and CC BMDMs. r, Pearson’s correlation coefficient. k,l, Representative images and the corresponding quantification of F-actin in LPS-treated UCs and CCs as normalized to LPS-treated UCs. Scale bar, 20μm. m,n, Colour-coded arrays showing the expression levels of early (m) and late (n) responsive genes in LPS-treated and Lat-A + LPS-treated BMDMs, as normalized to LPS-treated BMDMs. o, Expression levels of actin-related genes in LPS-treated versus Control BMDMs. p, Gene ontology analysis of all the upregulated genes in LPS-treated versus Control BMDMs. The number of genes in each category is shown in parentheses (see also Supplementary Data 2). In all the box plots, boxes show 25th and 75th percentiles, the middle horizontal line shows the median, small open squares show the mean, and whiskers indicate s.d. n, number of cells analysed per condition. P values were obtained with the two-sided Student’s t-test. Cell edges are marked in white. All the experiments were independently repeated three times.
We then confirmed using RAW 264.7 macrophages that LPS-induced morphological and transcriptional changes were not specific to BMDMs (Supplementary Fig. 11a–g). Furthermore, contrary to M1 activation, M2 activation of BMDMs with IL-4/IL-13 has no measurable influence on cell and nuclear size (Supplementary Fig. 12a–d), even though a significant cell shape transition towards highly elongated was observed (Supplementary Fig. 12e,f and Supplementary Video 3), the functional consequences of which have recently been probed20.
Early LPS signalling not impacted by confinement
Because LPS activates the transmembrane TLR434, followed by downstream activation of p65, a subunit of NF-κB5, and it also upregulates TLR4 expression35, we next asked which of the critical signalling events might be influenced by macrophage confinement. Nuclear translocation of p65 preceded the changes in cell spreading area, peaked at 30 min, remained high even after 3–6 h of LPS treatment, and was independent of cell confinement (Supplementary Fig. 13a–e). Similarly, nuclear levels of phospho-interferon regulatory factor 3 (pIRF3), another crucial signalling event during M1 activation36, were independent of cell confinement (Supplementary Fig. 13f,g). Together, these results suggest that cell confinement regulates gene expression by signalling events further downstream to TLR4 activation and p65 nuclear translocation.
Confinement downsizes actin-regulated late M1 transcription
Increases in cell and nuclear size induced by LPS were found to be dependent on actin polymerization and independent of myosin activity, as confirmed by pharmacological inhibitors (Fig. 2g,h and Supplementary Fig. 8d). As the role of actin polymerization in M1 activation is poorly understood, we first demonstrated that LPS-stimulated BMDMs have significantly higher levels of F-actin (Fig. 2i and Supplementary Fig. 14c). A strong correlation was observed between cell spreading areas and the total F-actin levels (Fig. 2j), whereby cell confinement reduced the total levels of F-actin (Fig. 2k,l and Supplementary Figs. 11h–j and 14a,b–d). In contrast, levels of phospho-myosin (p-MLC) remained unchanged following LPS treatment, with no impact resulting from cell confinement (Supplementary Fig. 15a–d). As this clearly suggests a possible and previously unexplored role of polymerized actin during M1 activation, actin polymerization was inhibited by latrunculin-A (Lat-A) and the expression levels of early and late-responsive genes were quantified in M1 macrophages. Interestingly, late-responsive genes were found to be significantly downregulated in Lat-A + LPS-treated cells as compared to only LPS-treated cells, but not early-responsive genes (Fig. 2m,n). This confirms that downregulation of late-responsive genes by cell confinement is due to reduced levels of polymerized actin. Again, inhibition of myosin II activity by Blebbistatin (Blebb) had no influence on gene expression profiles (Supplementary Fig. 15e).
Along with higher F-actin levels, RNA-seq analysis further revealed upregulation of genes involved in actin polymerization (Arpc2), F-actin repair (Zyx), cytoskeletal dynamics (Cfl1, Cnn2) and integrin binding (Tln1) (Fig. 2o) along with the transcription factor SRF following M1 activation. Gene ontology analysis of all the upregulated genes confirms actin-cytoskeleton organization and actin fibres as the major drivers of the mechanotransduction response towards confinement (Fig. 2p).
Confinement reduces MRTF-A-dependent M1 transcription
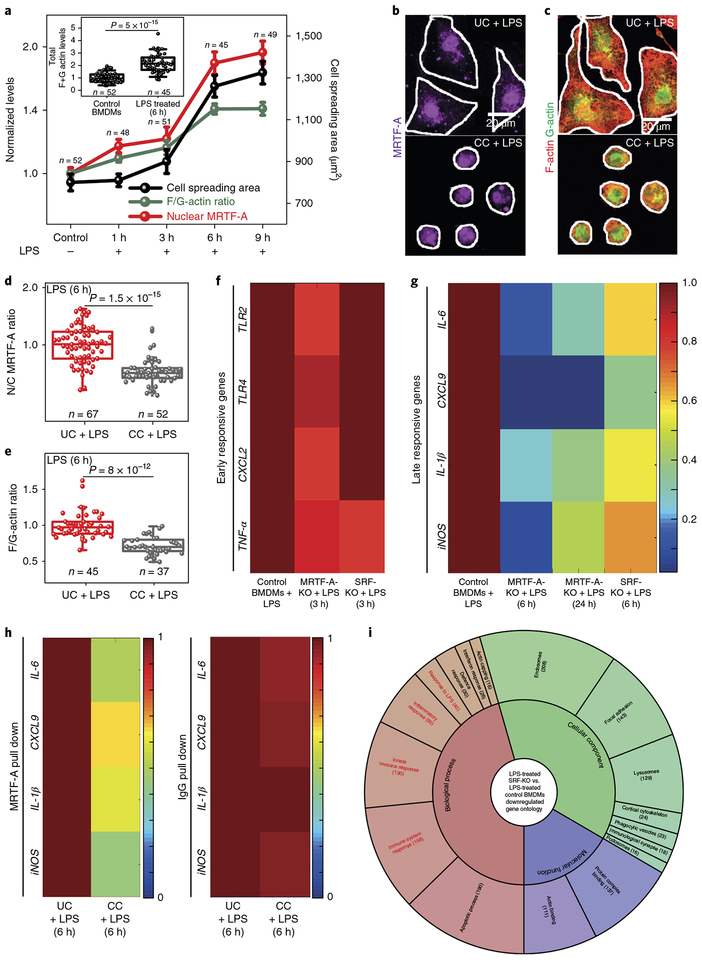
Myocardin-related transcription factor-A (MRTF-A) is an actin-dependent transcription co-factor37,38 that forms a complex with cytoplasmic G-actin. Following actin polymerization, MRTF-A is released and translocates into the nucleus, where it complexes with SRF and drives the expression of targets genes39. Because recent reports suggest that MRTF-A mediates pro-inflammatory transcription40,41 and SRF regulates cytoskeletal gene expression in macrophages42, we next probed whether confinement-induced effects are mediated via the MRTF-A-SRF pathway. Following LPS stimulation, nuclear translocation of MRTF-A was found to be a slow process, unlike early transcription factors (Supplementary Fig. 13), and ramped up significantly only after 3–4 h of activation (Fig. 3a and Supplementary Fig. 16). Its nuclear translocation correlated with spreading-associated increases in the F/G-actin ratios and total actin levels (F + G) (Fig. 3a and Supplementary Fig. 16). Importantly, nuclear translocation of MRTF-A for CCs is downsized (Fig. 3b,d and Supplementary Videos 4 and 5), as is the F/G-actin ratio (Fig. 3c,e and Supplementary Videos 4 and 5). However, all the single cell data taken together do not suggest the existence of a critical cell spreading area threshold for the F/G-actin ratio (Supplementary Fig. 17).
Fig. 3 |. Confinement reduces MRTF-A nuclear translocation, which downregulates late M1 transcriptional programs.
a, Time course changes in cell spreading areas, nuclear levels of MRTF-A and F/G-actin ratios in BMDMs treated with LPS for different periods of time. Inset, Total levels of actin (F + G) in Control and LPS-treated (6 h) BMDMs, as normalized to the Control BMDMs. b,c, Representative images of UC and CC BMDMs treated with LPS for 6 h and stained for MRTF-A (b) and G-actin and F-actin (c). Cell edges are marked in white. Scale bars, 20 μm. See also Supplementary Videos 4 and 5. d,e, Box plots showing nuclear to cytoplasmic (N/C) ratios of MRTF-A (d) and of F/G-actin in (e) UCs and CCs treated with LPS for 6 h, normalized to LPS-treated UCs. In all box plots, the box shows 25th and 75th percentiles, middle horizontal line shows the median, small open squares show the mean, and whiskers indicate s.d. f,g, Colour-coded arrays showing expression levels of early (f) and late (g) responsive genes in Control, MRTF-A-KO and SRF-KO BMDMs treated with LPS, normalized to LPS-treated Control. h, Colour-coded arrays showing ChIP-qPCR analysis of MRTF-A at the promoter region of late-responsive genes in LPS-treated UCs and CCs as normalized to LPS-treated UCs. IgG served as a ChIP control. i, Gene ontology analysis of all downregulated genes in LPS-treated SRF-KO versus LPS-treated Control BMDMs. The number of genes in each category is shown in parentheses (see also Supplementary Data 3). n, number of cells analysed per condition. P values were obtained with the two-tailed Student’s t-test. All experiments were independently repeated twice with similar results.
To check whether gene expression regulation by cell confinement is indeed mediated by MRTF-A-SRF activity, BMDMs were isolated from MRTF-A KO (MRTF-A-KO) and SRF KO (SRF-KO) mice. KO BMDMs showed smaller spreading areas and lower nuclear projection areas compared to the Control BMDMs, before and after LPS treatment, along with a diminished tendency of cell shape changes following LPS treatment (Supplementary Figs. 18a–e and 19a–e). This was accompanied by lower total F-actin levels (Supplementary Figs. 17, 18f,g and 19f,g) and also a lower expression of actin-related genes (Supplementary Fig. 19h). Furthermore, in MRTF-A-KO macrophages, early signalling events (Supplementary Fig. 20a–e) and expression of early-responsive genes (Fig. 3f) remained unperturbed, while all the late-responsive genes were significantly downregulated (Fig. 3g). The confinement-induced downsizing of late-responsive genes could be rescued in RAW 264.7 macrophages following transfection with a constitutively active MRTF-A overexpressing plasmid43 (Supplementary Fig. 11k). ChIP-qPCR analysis on BMDMs confirmed MRTF-A binding at the promoter region of late-responsive genes, which was downsized by macrophage confinement in comparison to the higher fold-enrichment in LPS-treated UCs (Fig. 3h).
The late-responsive genes were also severely downregulated in LPS-stimulated SRF-KO cells, with minimal effect on early-responsive genes (Fig. 3f,g). RNA-seq analysis revealed that the expression levels of a large number of pro-inflammatory genes, which get upregulated following LPS induction, were downsized in LPS-treated SRF-KO cells (Supplementary Fig. 21a,b). Furthermore, more than 300 late-responsive genes showed lower expression in SRF-KO cells (Supplementary Fig. 21c). The majority of downregulated genes in LPS-treated SRF-KO cells fall into the category of inflammatory genes and actin-dependent processes (Fig. 3i). Taken together, these data show for the first time not only a crucial role of the MRTF-A-SRF pathway in the regulation of pro-inflammatory gene expression, but also that MRTF-A-SRF-complex-regulated gene activity is downsized by cell confinement.
Confinement by cell crowding downsizes M1 activation
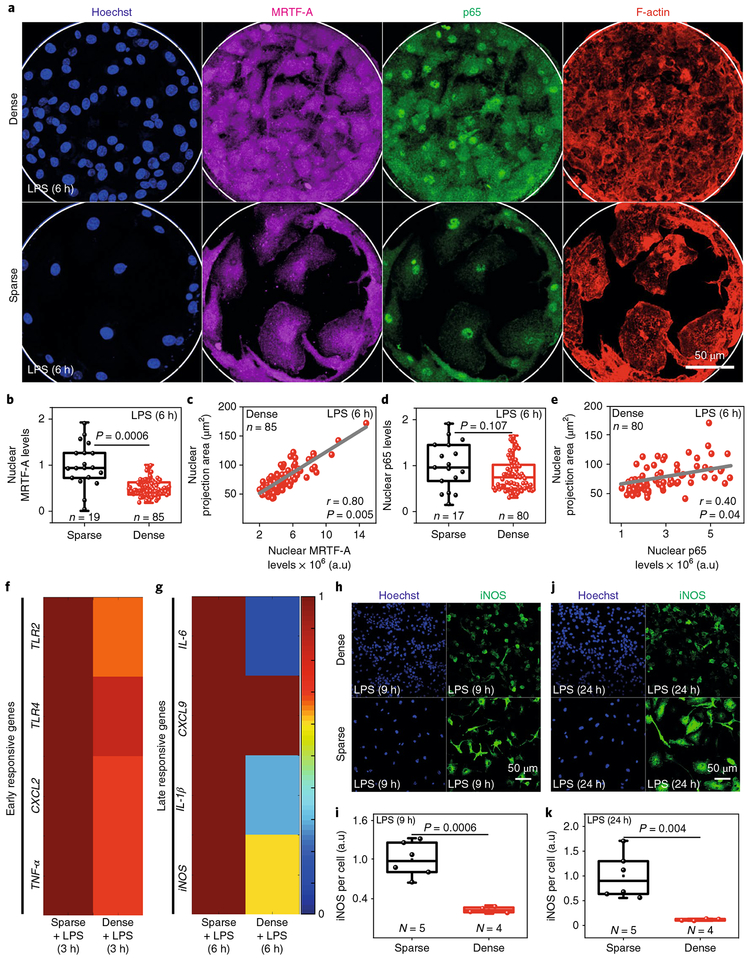
Macrophages infiltrate dense tissues during homeostasis, inflammation and disease progression, where, along with other factors, cell crowding can also limit their spreading. To mimic cell crowding, BMDMs were cultured on large microfabricated circular patterns of ~200 μm diameter with varying cell densities (Fig. 4a). Interestingly, nuclear MRTF-A levels were significantly higher in sparse and hence more spread-out cells as compared to densely packed cells (Fig. 4a,b). Even within densely packed cells, a strong correlation was observed between nuclear areas and nuclear MRTF-A levels (Fig. 4c). Contrary to this, nuclear translocation of p65 largely remained similar (Fig. 4d,e). Since changes in cell and nuclear areas were highly correlated (Supplementary Fig. 8e), nuclear projection areas were used as a measure of cell spreading. Further, the expression of early-responsive genes remained the same whereas expression levels of late-responsive genes were significantly down-regulated in crowded condition (Fig. 4f,g), along with the levels of iNOS per cell (Fig. 4h–k). Cell crowding thus recapitulates confinement-induced downregulation of M1 activation. Importantly, iNOS production is one of the most prominent biomarkers of pro-inflammatory macrophage activation, yet our findings illustrate that iNOS expression is significantly downregulated by confinement, independent of the method of confinement.
Fig. 4 |. Cell crowding recapitulates confinement-induced downregulation of M1 activation.
a, Representative images of BMDMs cultured on microfabricated islands of ~200 μm diameter with different cell densities, treated with LPS and stained for nuclei, p65, MRTF-A and F-actin. Scale bar, 50 μm. b, Box plots show levels of nuclear MRTF-A in sparsely and densely seeded BMDMs treated with LPS for 6 h, as normalized to LPS-treated sparsely seeded cells. c, Correlation plot between nuclear projection areas and nuclear levels of MRTF-A in densely seeded macrophages treated with LPS for 6 h. r, Pearson’s correlation coefficient. d, Box plots showing levels of nuclear p65 in sparsely and densely seeded macrophages treated with LPS for 6 h, as normalized to LPS-treated sparse cells. e, Correlation plot between nuclear projection areas and nuclear levels of p65 in densely seeded macrophages treated with LPS for 6 h. f,g, Colour-coded arrays show the expression levels of early (f) and late (g) responsive genes in LPS-treated densely and sparsely seeded macrophages. Levels are normalized to the LPS-treated sparsely seeded cells. h,j, Representative images of densely and sparsely seeded BMDMs treated with LPS for 9 h (h) and 24 h (j) and stained for nuclei and iNOS. Scale bars, 50μm. i,k, Box plots showing iNOS levels per cell in sparsely and densely seeded macrophages, as normalized to LPS-treated sparsely seeded cells. n, number of cells analysed per condition. N, number of fields of view analysed per condition. In all box plots, boxes show 25th and 75th percentiles, middle horizontal line shows median, small open squares show the mean, and whiskers indicate s.d. P values were obtained with the two-tailed Student’s t-test. All experiments were independently repeated three times with similar results.
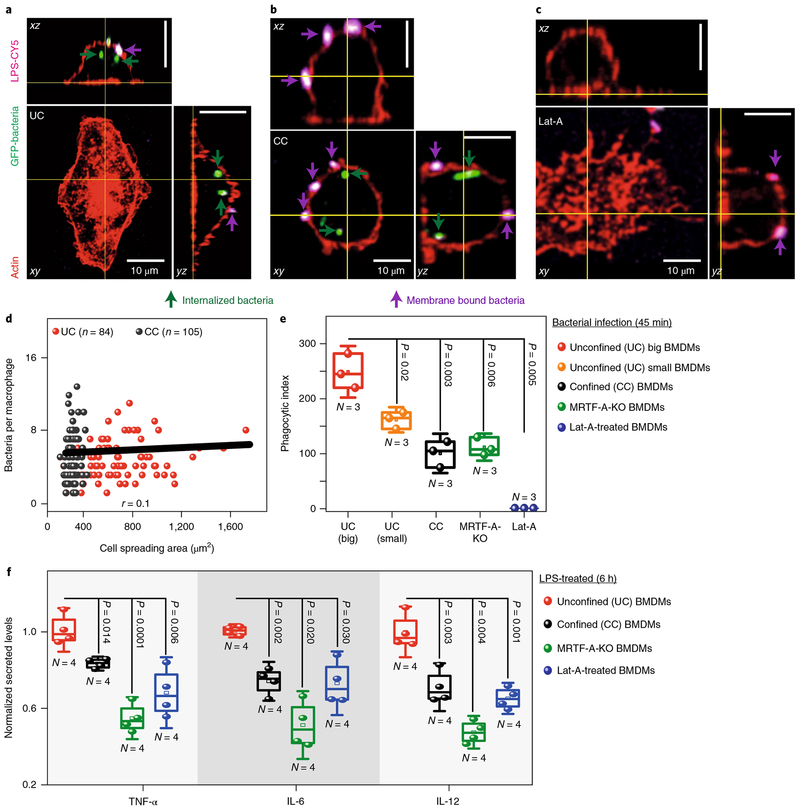
Confinement reduces bacterial uptake and cytokine secretion
Two central functions of pro-inflammatory macrophages include their ability to clear foreign bodies and to secrete pro-inflammatory cytokines. To check whether macrophage confinement affects their phagocytic potential, UCs and CCs were infected with green fluorescent protein (GFP)-expressing E. coli and were stained with an anti-LPS antibody to differentiate between membrane-bound and internalized bacteria (Fig. 5a,b). The phagocytosis assay was performed after only 45 min of bacterial exposure, which was not sufficient to observe activation differences in UCs versus CCs. Within this early phase, a weak correlation between cell spreading area and bacteria per macrophage suggests that cell confinement (Fig. 5d and Supplementary Fig. 22a) has little impact on the ability of macrophages to bind bacteria. However, the phagocytic potential, as measured by quantifying the phagocytic index, was largely reduced for CCs (Fig. 5e and Supplementary Fig. 22b) and was dependent on MRTF-A and actin polymerization (Fig. 5c–e). Similar inhibitory effects of SRF-KO on phagocytosis have been shown recently42.
Fig. 5 |. Macrophage confinement downregulates bacterial phagocytosis and secreted cytokine levels.
a–c, Representative images of UC (a), CC (b) and latrunculin-A (Lat-A) (c) treated BMDMs infected with GFP-tagged E. coli for 45 min and stained with anti-LPS antibody and phalloidin, without permeabilization of the plasma membrane, to distinguish between membrane-bound adherent (violet) and internalized bacteria (green). Scale bars, 10 μm. d, Cell spreading area versus total bacteria per macrophage, in UCs and CCs, where the plot shows only a weak correlation. e, Box plots showing early-phase differences in the phagocytic index in UCs big, UCs small (cell area less than 500 μm2), CCs, MRTF-A-KO and Lat-A-treated BMDMs. Phagocytic index was calculated as follows: Phagocytic index = ((total number of engulfed bacteria/total number of counted macrophages) × (number of macrophages containing engulfed bacteria/total number of counted macrophages)) × 100. Data are pooled from N = 3 independent experiments. f, Box plots showing the difference in secreted cytokine levels between UCs, CCs, MRTF-A-KO and Lat-A-treated BMDMs, all treated with LPS for 6 h. Levels are normalized to the LPS-treated UCs. N, number of biological replicates. In all box plots, boxes show 25th and 75th percentiles, middle horizontal line shows the median, small open squares show the mean, and whiskers indicate s.d. P values were obtained with the two-tailed Student’s t-test. All experiments were independently repeated three times with similar results.
Because cytokine secretion is downstream of epigenetic transcriptional regulation, levels of known M1 specific cytokines were quantified between LPS-treated UCs and CCs by enzyme-linked immunosorbent assay (ELISA). To confirm that confinement downregulates the pro-inflammatory response of macrophages, the levels of secreted TNF-α, IL-6 and IL-12 were significantly lower in LPS-stimulated CCs as compared to UCs (Fig. 5f) and were also found to be dependent on MRTF-A and actin polymerization (Fig. 5f). Unlike the protein product of other early-responsive genes such as TLR4 (Supplementary Fig. 13h–l), TNF-α might be post-translationally regulated by cell confinement as it has been suggested that actin polymerization regulates the expression of a few LPS-induced genes post-translationally44,45. This illustrates, for the first time, a direct functional coupling between macrophage confinement and the pro-inflammatory response.
The detailed mechanisms that tune tissue-specific macrophage polarization and their functional plasticity remain elusive, even though tissue-specific differences are well documented. Since gradual transitioning from the pro-inflammatory to the pro-healing phenotype plays a pivotal role in tissue repair and metabolic homeostasis, the physiological significance of our findings is far-reaching. We discovered here that the LPS-induced late-responsive genes are significantly downregulated by spatially confining macrophages (Fig. 6), including the expression of the most frequently used biomarkers of M1 macrophages such as iNOS, which has been implicated in several critical functions, including microbial killing, immune regulation46,47, secretion of reactive oxygen species and differentiation21. Recognizing that the pro-inflammatory response of macrophages can be significantly suppressed by limiting their spreading, for example by tuning adhesive chemistries or topographical features of implants as illustrated here on microporous substrates, or by cell crowding, also opens new perspectives for the design of immunosuppressive or immunomodulating biomaterials. As for tissue-specific niches, it was well established that the physicochemical nature of a biomaterial can regulate the release of cytokines by innate immune cells, specifically macrophages48,49, yet the underpinning mechanisms remain poorly understood. The mechanisms described here might thus also contribute to the improved healing of topographically structured and porous materials. We thus show that spatial constraints can bring about modularity and down tune the magnitude of pro-inflammatory gene expression programs and hence the functional outputs by which macrophages stage their pro-inflammatory response (Fig. 6). While we have illustrated this here by using the most well-known activator of TLR4, namely LPS, a cohort of other ligands can activate TLR4 too. This includes a variety of endogenous proteins such as low-density lipoprotein and β-defensins50, known as drivers of atherosclerosis and to trigger the release of histamines and prostaglandin D2, respectively, which further widens the implications of our findings.
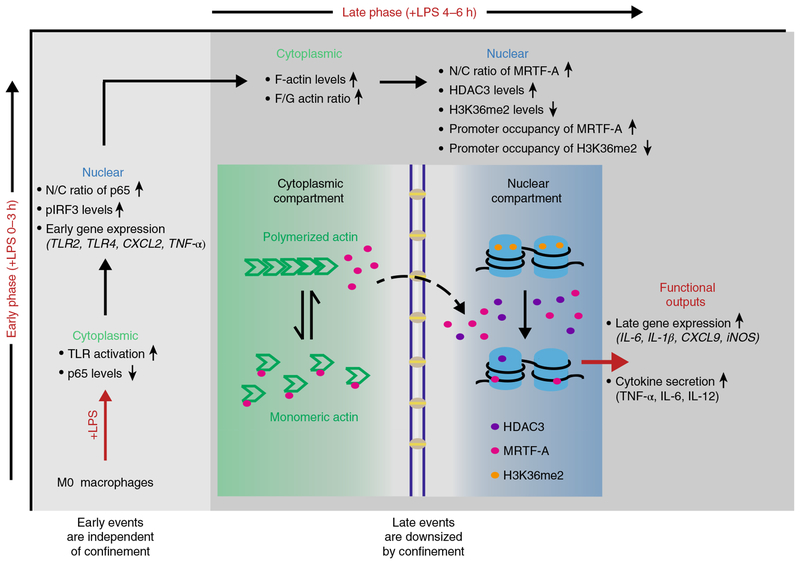
Fig. 6 |. Schematic overview of how the spatial confinement of pro-inflammatory macrophages downregulates their late, but not early transcriptional programs, and thus their pro-inflammatory functional outputs.
LPS stimulation upregulates pro-inflammatory early and late-responsive gene clusters, as indicated, if macrophages are free to spread. As macrophage spreading increases significantly only a few hours after LPS stimulation, spatial confinement of M1 macrophages does not affect TLR4 activation nor nuclear factor (NF)-κB nuclear translocation, both of which precede LPS-induced macrophage spreading. As time progresses, restricting cell spreading significantly downregulates the LPS-stimulated activity of late transcription regulators such as that of HDAC3 and of the MRTF-A–SRF complex, and consequently cytokine expression. This includes the expression of frequently used biomarkers to identify M1 macrophages, including iNOS. Spatial confinement thus tames the capability of M1 macrophages to stage an inflammatory response as required to effectively fight infections.
Methods
Micropatterning.
To prepare PDMS stamps, PDMS elastomer (SYLGARD 184 Dow Corning) was used in a 1:10 ratio of curative to precursor, according to the manufacturer’s protocol, and moulded in silicon wafers microfabricated with the desired geometries. Micropatterned PDMS stamps were treated with air-plasma under vacuum in a plasma cleaner (Model PDC-002 Harrick Scientific); 100 μgml−1 fibronectin solution (F0895 Sigma) was allowed to adsorb onto the surface of each PDMS stamp under sterile conditions for 10 min. The PDMS stamp was allowed to dry for further 10 min before depositing it onto the surface of hydrophobic cell culture dishes (ibidi) for 15 min to allow the fibronectin-coated micropatterns to be transferred. The surface was then treated with 1% Pluronic F-127 (P2443 Sigma) for 30 min to passivate non-fibronectin-coated regions.
Bone marrow isolation and primary macrophage differentiation.
Femurs were isolated from postmortem healthy male mice (10–14 weeks old C57BL/6) and bone marrow was flushed with PBS. Bone marrow was further passed through a 7 μm cell strainer to obtain a single-cell suspension. Bone marrow was centrifuged and suspended in BMDM medium. Two million cells were seeded in 60 mm nontreated plastic dishes for 7 days in 5% CO2 at 37 °C. Equal volumes of medium were again added on day 4. BMDMs were used on day 7 for further experiments. BMDM culture medium contained DMEM, 10% FBS, glutamine, non-essential amino acids, sodium pyruvate, β-mercaptoethanol, penicillin-streptomycin and L929 medium. L929 medium was prepared by culturing 2 million L929 cells in 200 ml medium in a 300 cm2 flask for 8 days without changing the medium. The medium used for L929 cell culture contained RPMI, 10% FBS, glutamine, nonessential amino acids, sodium pyruvate, HEPES, and β-mercaptoethanol. L929 cells were cultured and maintained in 5% CO2 at 37 °C.
Before seeding the macrophages on the micropatterned dishes, excessive Pluronic F-127 was washed off. Dishes were washed three times with sterile 1× PBS and were incubated under ultraviolet light for 30 min. Cells (30,000) were seeded in these dishes, containing 2 ml of culture medium, for 60 min with gentle shaking after every 15 min. Non-adherent cells were then removed, and the remaining cells were further incubated in the cell culture medium. More than 90% of cells on the microfabricated islands were thus found to be attached to an island as single cells (Supplementary Fig. 1d).
Raw 264.7 cell culture.
Raw 264.7 cells were cultured in DMEM supplemented with 10% FBS and penicillin-streptomycin. Cells were cultured at 37 °C under 5% CO2.
Fabrication of microporous PDMS substrate.
Ultraviolet-curable polymeric glue (Norland optical glue) was spin-coated to create an evenly thick layer of glue. Poly(methyl methacrylate) (PMMA) microspheres (PMPMS-1.2 Cospehric) with two different size ranges (20–30 μm and 355–425 μm) were dispersed on top of the glue layer. The plates were placed under ultraviolet light (360 nm, LED) for 30 s to polymerize the glue and fix the microsphere to the surface. PDMS (curative-to-precursor ratio of 1:10, Sylgard 184) was poured on top of the microsphere-coated dishes and cured for 60 min at 80 °C. PDMS was then peeled off the dishes, plasma-treated in air for 30 s (Harrick Plasma) to create a hydrophilic surface for subsequent fibronectin coating, and then ultraviolet-sterilized for 30 min before seeding the cells (Supplementary Fig. 2). Flat PDMS, without any microspheres, was used as a control.
Macrophage activation.
All studies were performed with primary BMDMs, unless stated. Unlike the frequently used immortalized cell lines, BMDMs more closely mimic macrophage behaviour in vivo. Pro-inflammatory M1 activation was achieved by stimulating macrophages with LPS (25 ng ml−1) (L3129 Sigma). The pro-regenerative M2 phenotype was achieved by inducing BMDMs with IL-4/IL-13 (20 ng ml−1) (404-ML-010 & 413-ML-005 R&D Systems) for 24 h.
Drug treatment.
Actin depolymerization studies were performed with 100 nM Lat-A (L5163 Sigma). BMDMs were treated with Lat-A for 60 min before the addition of LPS, and Lat-A was kept in the medium along with LPS during M1 activation. Myosin II motor activity was inhibited with 50 μM blebbistatin (B0560 Sigma). BMDMs were treated with blebbistatin for 60 min before the addition of LPS, and the blebbistatin was kept in the medium along with LPS during M1 activation. For HDAC3 inhibition studies, BMDMs were treated with 20 μM RGFP966 (S7229 Selleckchem) for 24 h before LPS addition, and RGFP966 was kept in the medium along with LPS during M1 activation.
Transfection.
RAW 264.7 cells were transfected with the Nucleofector Kit for Mouse Macrophages (VAPA 1009 Lonza), according to the manufacturer’s instructions. In brief, 1 million cells were transfected with 10 μg of the desired plasmid, HDAC351 or CA-MRTF-A52, using reagents provided in the kit, for 18 h before seeding on micropatterned substrates.
Immunostaining.
Cells were rinsed twice with 1× PBS, followed by fixation using 4% paraformaldehyde (Sigma) in PHEM buffer (500 ml of 2× PHEM buffer contains 18.14 g PIPES, 6.5 g HEPES, 3.8 g EGTA, 0.99 g MgSO4; pH 7 adjusted with KOH) for 20 min. Cells were washed and permeabilized with 0.3% Triton X-100 (Sigma) in 1× PBS for 10 min. After washing twice with 1× PBS, cells were treated with 2% BSA (blocking solution) for 60 min before incubating with primary antibody (diluted in 2% BSA) at 4 °C overnight. Primary antibodies against iNOS (1:100; Cell Signaling), HDAC3 (1:300; sc-136290 SantaCruz), phosphorylated-myosin light chain (p-MLC) (1:75; #3671S Cell Signaling), TLR4 (1:200; Abcam), TLR2 (1:50; sc-21760 Santa Cruz), p65 (1:300; sc-8008 Santa Cruz), MRTF-A (1:100; ab49311 Abcam), pIRF3 (1:50; SAB4504031 Sigma) and H3K36 dimethylation (1:500; ab9049 Abcam) were used for staining. Cells were then washed with blocking solution and incubated with the corresponding secondary antibodies, diluted in blocking solution, along with the nuclear stain Hoechst-33342 (1:1,000; 62249 Life Technologies) for 45 min. Filamentous actin (F-actin) was labelled using phalloidin Alexa-Fluor 488 or 568 (1:200; Life Technologies) and globular actin (G-actin) was labelled using DNase-I, Alexa-Fluor 488 conjugate (1:500; D12371 Life Technologies). All antibodies used in this study have previously been extensively used and validated for immunofluorescence studies (for example, MRTF-A51,53,54 p-MLC51,55,56, HDAC351,57,58, HDAC159 and p6560,61).
Cytokine secretion.
Cell culture medium was collected from unconfined, confined, MRTF-A-KO and Lat-A-treated BMDMs after 6 h of LPS stimulation. To remove cell debris, cell culture medium was centrifuged and the supernatant was stored at −80 °C till further processing. All ELISA kits were purchased from Peprotech (TNF-α (900-TM54), IL-6 (900-TM50), IL-12 (900-TM97) and CXCL2 (900-M152)). The experiment was performed according to the manufacturer’s protocol. All data fitting and quantifications were done using OriginPro8.1 and MATLAB2016b (MathWorks), respectively.
Bacterial phagocytic assay.
Bacterial strains MG1655 were employed here as E. coli wild type. Constitutive GFP expression was achieved by transforming with PrpsM-GFP, as previously described62. GFP-transfected E. coli strain was grown in LB medium and diluted in antibiotic-free BMDM culture medium before experiments after overnight growth to an optical density of 0.1. UC, CC, MRTF-A-KO and Lat-A-treated BMDMs were infected with bacteria to macrophage ratios of 10:1 for 45 min. Non-adherent bacteria were then removed by washing three times with ice-cold 1× PBS (pH 7.4) supplemented with 5 mM EDTA. BMDMs were then fixed with 4% paraformaldehyde for 20 min. After fixation, BMDMs were washed twice with 1× PBS and incubated with 2% BSA for 60 min. Without permeabilization of the plasma membrane, BMDMs were incubated with anti-LPS primary antibody (1:100; ab137967 abcam) at 4 °C overnight. BMDMs were then permeabilized with 0.3% Triton X-100 (Sigma) in 1× PBS for 10 min after washing twice with 1× PBS and incubated with Cy5 conjugated secondary antibody along with phalloidin Alexa-Fluor 568 (1:200; Life Technologies) for 45 min. For bacterial uptake assays, Z-stack images were captured using a ×63 objective, ×4 magnification and a z-step of 0.30 |am. The number of phagocytic bacteria and membrane-bound bacteria were then counted per cell.
Confocal imaging and analysis.
Images of single cells were taken using a Leica SP5 confocal microscope. To estimate cell spreading areas, 2D images were taken using a ×40 (0.7 NA) air objective. For nuclear volume analyses, images were captured using ×100 objective, ×4 magnification and a z-step of 0.30 μm, as shown in Supplementary Fig. 8f, which also shows the threshold nuclear images generated using MATLAB2016b (MathWorks), used to quantify nuclear volume. Similarly, Supplementary Fig. 14a,b shows detailed slice-by-slice (z-step of 0.7 μm) images of BMDMs stained with phalloidin (to stain F-actin) for quantifying total F-actin levels in LPS-treated UC and CC BMDMs. Imaging conditions were kept the same during all experiments.
To estimate protein expression levels (nuclear and cytoplasmic), cells were stained with the desired antibodies and 3D confocal images were captured rather than projected images. Fluorescence images were captured on a confocal Leica SP8 microscope using a ×63 (1.25 NA) oil objective with a z-step of 0.7 μm and a confocal pinhole of 1 a.u. Supplementary Fig. 14e shows detailed slice-by-slice (z-step of 0.7 μm) images of LPS-treated UCs and CCs stained with MRTF-A and Hoechst-33342. Also shown are the segmented nuclear images, generated using MATLAB2016b (MathWorks), for proper demarcation of nuclear volumes. Using these segmented nuclear images, MRTF-A nuclear protein levels were then defined as the sum of the total pixel intensity of MRTF-A in this nuclear volume (hence nuclear MRTF-A), while the remaining signal from the cell was taken as the cytoplasmic level of the protein (hence cytoplasmic MRTF-A). Total levels of selected proteins in a cell were thus defined as the sum of the nuclear and cytoplasmic fractions of the proteins. Nuclear levels, nuclear-to-cytoplasmic levels and total expression levels of HDAC1, HDAC3, p65 and of other proteins were quantified using a similar approach. Imaging conditions were kept the same during all experiments. All of the quantifications per cell were carried out using MATLAB2016b (MathWorks) or ImageJ (NIH).
SEM sample preparation.
The samples were fixed in 2.5% glutaraldehyde (G7651 Sigma) in 1× PBS pH 7.4 for 30 min at room temperature. After three rinses in 1× PBS followed by 0.1 M cacodylate buffer pH 7.4 (Electron Microscopy Sciences cat. no. 11652) for 5 min each, the samples were post-fixed in 1% osmium tetroxide (19152 Electron Microscopy Sciences) in 0.1 M cacodylate buffer pH 7.4 for 1 h at room temperature. Subsequently, the samples were washed three times with distilled water for 10 min each. Dehydration was carried out in a series of 50%, 70%, 80%, 90%, twice 100% and twice 100% ethanol dried over a molecular sieve, with each step lasting 10 min. Critical point drying was performed in a Tousimis 931 device. The samples were sputter-coated with 10 nm platinum/palladium 80/20 using a CCU-010 high-vacuum coating unit by Safematic. Images were taken in a Zeiss Leo 1530 scanning electron microscope.
RNA isolation and real-time qPCR analysis.
RNA was isolated with a Qiagen RNA Isolation Kit according to the manufacturer’s protocol. cDNA was prepared using iScript Advanced cDNA Synthesis Kit (172–5038 BioRad). Real-time qPCR experiments were performed using SsoFast EvaGreen Supermix (172–5203 BioRad) in a CFX96 Real-Time PCR Detection System (Bio-Rad). The primer and their corresponding sequences used for real-time qPCR analysis are given in Supplementary Table 1, with 18S rRNA as a control.
ChIP.
BMDMs were crosslinked and chromatin immunoprecipitated using a True MicroChip kit (Diagenode) and antibodies specific for H3K36me2 (ab9049 Abcam) and MRTF-A (ab49311 Abcam), according to the manufacturer’s instructions. In brief, 60,000 BMDMs were crosslinked with formaldehyde (1% final concentration), and the reaction was stopped with the addition of glycine to 125 mM final concentration. BMDMs were lysed and chromatin sheared using a Bioruptor instrument (Diagenode) with the following sonication conditions: five cycles of 30 s on/30 s off at high power. Samples were vortexed before and after performing five sonication cycles, followed by a short centrifugation at 4 °C. Ten percent of sheared chromatin was reserved as the input fraction, and the remaining was immunoprecipitated by incubating with 3 μg of desired antibodies. For each immunoprecipitation series, a negative control using an antibody against IgG was run with sheared chromatin obtained from the same number of cells. After 18 h incubation at 4 °C, chromatin was immunoprecipitated using magnetic beads, washed, and released by incubating at 65 °C for 4 h. DNA was purified using MicroChip columns (Diagenode), according to the manufacturer’s instructions. The performance of the immunoprecipitation reaction was assessed by qPCR. Primer sequences are listed in Supplementary Table 2.
RNA sequencing.
RNA was isolated with the Qiagen RNA Isolation Kit according to the manufacturer’s protocol. For library preparation, the quantity and quality of the isolated RNA were determined with a Qubit (1.0) Fluorometer (Life Technologies) and a Tapestation 4200 (Agilent). The TruSeq Stranded HT mRNA Sample Prep Kit (Illumina) was used in subsequent steps following the manufacturer’s protocol. Briefly, total RNA samples (1 μg) were ribosome-depleted and then reverse-transcribed into double-stranded cDNA with actinomycin added during the first-strand synthesis. The cDNA samples were fragmented, end-repaired and polyadenylated before ligation of TruSeq adapters. The adapters contain the index for dual multiplexing. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR. The quality and quantity of the enriched libraries were validated using a Qubit (1.0) fluorometer and the Tapestation 4200 (Agilent). The product is a smear with an average fragment size of ~360 bp. The libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5 with 0.1% Tween 20. For cluster generation and sequencing, a Hiseq 4000 SR Cluster Kit (Illumina) was used using 8 pM of pooled normalized libraries on the cBOT V2. Sequencing was performed on the Illumina HiSeq with single-end 125 bp using the HiSeq 3000/4000 SBS Kit (Illumina).
Reads quality was checked with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and sequencing adapters were trimmed using Trimmomatic63. Reads at least 20 bases long, and an overall average Phred quality score greater than 10 were aligned to the reference genome and transcriptome (FASTA and GTF files, respectively, downloaded from Ensemble, genome build GRCm38) with STAR v2.5.164, with default settings for single-end reads. The distribution of reads across genomic features was quantified using the R package Genomic Ranges from Bioconductor Version 3.065. Differentially expressed genes were identified using the R package edgeR from Bioconductor Version 3.066.
Functional analysis of genes.
The functional analysis of differentially regulated genes was performed using DAVID software67. Based on gene ontology analysis, genes were organized and were summarized using KRONA plots.
Statistical analysis.
The cell number assayed for quantification is given in individual figures. Comparisons were performed by means of a two-tailed Student t-test.
Data availability
Microarray data for HDAC3-KO BMDMs were obtained from ref.32 and the GEO accession no. is GSE33162. RNA-seq data for Control untreated BMDMs, LPS-treated BMDMs and SRF-KO-LPS-treated BMDMs have been deposited in GEO. The accession codes are GSM3373977-GSM3373979, GSM3373980-GSM3373982 and GSM3373983-GSM3373985 for Control untreated BMDMs, LPS-treated BMDMs and SRF-KO-LPS-treated BMDMs, respectively. The remaining data supporting the findings of this study are available within the Article and its Supplementary Information files and from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements
The authors thank A. Oxenius and the Mice Facility both at ETH Zurich for donating postmortem animals for bone marrow isolation, S. Farmer (Boston University) for providing bone marrow from MRTF-A-KO mice and for helpful discussions, S. Halene (Animal Modeling Core, Yale Cooperative Center of Excellence in Hematology (YCCEH) NIDDK grant no. U54DK106857, Yale University) for providing bone marrow from SRF-KO mice, and K. Sobue and T. Mayanagi (Iwaka Medical University) for providing CA-MRTF-A plasmid. The authors thank G. V. Shivashankar (MBI, NUS, Singapore) for providing HDAC3-overexpressing plasmid, L. Philipp and U. Kutay (ETH Zurich) and M. O. Hottiger (University of Zurich) for helpful discussions on ChIP experiments. In the Vogel group, I. A. Vizcarra is acknowledged for initial help with BMDM culture, J.-Y. Shiu and V. Hosseini for their advice and the manufacturing of 3D-microwells, C. Spencer for technical support, I. Gerber for acquiring the electron microscopy images, L. A. Branco and I. Schoen for critical comments on the manuscript and finally the FIRST and ScopeM facilities at ETH Zurich for access to microfabrication and confocal microscopy. This work was supported by ETH Zurich and by the Swiss National Science Foundation Grant (grant no. CR32I3_156931 to V.V.) and also in part by the Swiss NCCR Molecular Systems Engineering.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41563-018-0190-6
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41563-018-0190-6.
Publisher's Disclaimer: Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mosser DM & Edwards JP Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass CK & Natoli G Molecular control of activation and priming in macrophages. Nat. Immunol 17, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards JP, Zhang X, Frauwirth KA & Mosser DM Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol 80, 1298–1307 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence T & Natoli G Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol 11, 750–761 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R & Horng T Transcriptional control of the inflammatory response. Nat. Rev. Immunol 9, 692–703 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Raza S et al. Analysis of the transcriptional networks underpinning the activation of murine macrophages by inflammatory mediators. J. Leukoc. Biol 96, 167–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez FO & Gordon S The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammermann T & Germain RN The multiple faces of leukocyte interstitial migration. Semin. Immunopathol 36, 227–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makino A et al. Mechanotransduction in leukocyte activation: a review. Biorheology 44, 221–249 (2007). [PubMed] [Google Scholar]
- 10.Gruber E, Heyward C, Cameron J & Leifer C Toll-like receptor signaling in macrophages is regulated by extracellular substrate stiffness and Rho-associated coiled-coil kinase (ROCK1/2). Int. Immunol 30, 267–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I & Le Cabec V Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J. Immunol 184, 1049–1061 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Mennens SFB, van den Dries K & Cambi A Role for mechanotransduction in macrophage and dendritic cell immunobiology. Results Probl. Cell Differ 62, 209–242 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Chen CS, Mrksich M, Huang S, Whitesides GM & Ingber DE Geometric control of cell life and death. Science 276, 1425–1428 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Downing TL et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater 12, 1154–1162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engler AJ, Sen S, Sweeney HL & Discher DE Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fu J et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 7, 733–736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel V & Sheetz M Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol 7, 265–275 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Huse M Mechanical forces in the immune system. Nat. Rev. Immunol 17, 679–690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McWhorter FY, Davis CT & Liu WF Physical and mechanical regulation of macrophage phenotype and function. Cell. Mol. Life Sci 72, 1303–1316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McWhorter FY, Wang T, Nguyen P, Chung T & Liu WF Modulation of macrophage phenotype by cell shape. Proc. Natl Acad. Sci. USA 110, 17253–17258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata K et al. Effect of the release from mechanical stress on osteoclastogenesis in RAW264.7 cells. Int. J. Mol. Med 28, 73–79 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sussman EM, Halpin MC, Muster J, Moon RT & Ratner BD Porous implants modulate healing and induce shifts in local macrophage polarization in the foreign body reaction. Ann. Biomed. Eng 42, 1508–1516 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Chinetti-Gbaguidi G et al. Human atherosclerotic plaque alternative macrophages display low cholesterol handling but high phagocytosis because of distinct activities of the PPARy and LXRa pathways. Circ. Res 108, 985–995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoger JL et al. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis 225, 461–468 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Ihalainen TO et al. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat. Mater 14, 1252–1261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain N, Iyer KV, Kumar A & Shivashankar GV Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl Acad. Sci. USA 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascetti G, Carrara S & Vergani L Relationship between chromatin compactness and dye uptake for in situ chromatin stained with DAPI. Cytometry 44, 113–119 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Xu G et al. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) production. J. Biol. Chem 290, 5414–5423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haberland M, Montgomery RL & Olson EN The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet 10, 32–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das Gupta K, Shakespear MR, Iyer A, Fairlie DP & Sweet MJ Histone deacetylases in monocyte/macrophage development, activation and metabolism: refining HDAC targets for inflammatory and infectious diseases. Clin. Transl. Immunol 5, e62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shakespear MR, Halili MA, Irvine KM, Fairlie DP & Sweet MJ Histone deacetylases as regulators of inflammation and immunity. Trends Immunol 32, 335–343 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Chen X et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl Acad. Sci. USA 109, E2865–E2874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeksema MA et al. Targeting macrophage histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol. Med 6, 1124–1132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow JC, Young DW, Golenbock DT, Christ WJ & Gusovsky F Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem 274, 10689–10692 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Dai Q & Pruett SB Ethanol suppresses LPS-induced Toll-like receptor 4 clustering, reorganization of the actin cytoskeleton, and associated TNF-α production. Alcohol. Clin. Exp. Res 30, 1436–1444 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Reimer T, Brcic M, Schweizer M & Jungi TW Poly(I:C) and LPS induce distinct IRF3 and NF-κB signaling during type-I IFN and TNF responses in human macrophages. J. Leukoc. Biol 83, 1249–1257 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Connelly JT et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol 12, 711–718 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Olson EN & Nordheim A Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol 11, 353–365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selvaraj A & Prywes R Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol 5, 13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L et al. MRTF-A mediates LPS-induced pro-inflammatory transcription by interacting with the COMPASS complex. J. Cell Sci 127, 4645–4657 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Xie L MKL1/2 and ELK4 co-regulate distinct serum response factor (SRF) transcription programs in macrophages. BMC Genomics 15, 301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan AL et al. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol. Cell. Biol 31, 861–875 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishi T, Mayanagi T, Iwabuchi S, Akasaka T & Sobue K Myocardin-related transcription factor A (MRTF-A) activity-dependent cell adhesion is correlated to focal adhesion kinase (FAK) activity. Oncotarget 7, 72113–72130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han J, Brown T & Beutler B Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J. Exp. Med 171, 465–475 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruys V, Marinx O, Shaw G, Deschamps J & Huez G Translational blockade imposed by cytokine-derived UA-rich sequences. Science 245, 852–855 (1989). [DOI] [PubMed] [Google Scholar]
- 46.MacMicking JD et al. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81, 641–650 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Gabay C Interleukin-6 and chronic inflammation. Arthritis Res. Ther 8(Suppl. 2), S3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartneck M, Heffels KH, Bovi M, Groll J & Zwadlo-Klarwasser G The role of substrate morphology for the cytokine release profile of immature human primary macrophages. Mater. Sci. Eng. C 33, 5109–5114 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Wynn TA & Vannella KM Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brubaker SW, Bonham KS, Zanoni I & Kagan JC Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain N, Iyer KV, Kumar A & Shivashankar GV Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl Acad. Sci. USA 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kishi T, Mayanagi T, Iwabuchi S, Akasaka T & Sobue K Myocardin-related transcription factor A (MRTF-A) activity-dependent cell adhesion is correlated to focal adhesion kinase (FAK) activity. Oncotarget 7, 72113–72130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang F et al. Myocardin-related transcription factor A mediates OxLDL-induced endothelial injury. Circ. Res 108, 797–807 (2011). [DOI] [PubMed] [Google Scholar]
- 54.O’Sullivan NC, Pickering M, Di Giacomo D, Loscher JS & Murphy KJ Mkl transcription cofactors regulate structural plasticity in hippocampal neurons. Cereb. Cortex 20, 1915–1925 (2010). [DOI] [PubMed] [Google Scholar]
- 55.John GR et al. Interleukin-1β induces a reactive astroglial phenotype via deactivation of the Rho GTPase-Rock axis. J. Neurosci 24, 2837–2845 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamijo K et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol. Biol. Cell 17, 43–55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bardai FH & D’Mello SR Selective toxicity by HDAC3 in neurons: regulation by Akt and GSK3β. J. Neurosci 31, 1746–1751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de la Vega L et al. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 46, 472–483 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Montgomery RL et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21, 1790–1802 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clement M et al. CD31 is a key coinhibitory receptor in the development of immunogenic dendritic cells. Proc. Natl Acad. Sci. USA 111, E1101–E1110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yim N et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun 7, 12277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Avalos Vizcarra I et al. How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics. Sci. Rep 6, 18109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lawrence M et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data for HDAC3-KO BMDMs were obtained from ref.32 and the GEO accession no. is GSE33162. RNA-seq data for Control untreated BMDMs, LPS-treated BMDMs and SRF-KO-LPS-treated BMDMs have been deposited in GEO. The accession codes are GSM3373977-GSM3373979, GSM3373980-GSM3373982 and GSM3373983-GSM3373985 for Control untreated BMDMs, LPS-treated BMDMs and SRF-KO-LPS-treated BMDMs, respectively. The remaining data supporting the findings of this study are available within the Article and its Supplementary Information files and from the corresponding authors upon reasonable request.