Abstract
Pancreatic cancer is one of the most aggressive cancers worldwide with a high mortality rate. Prognosis remains poor even in this era of advanced medicine mainly due to early metastasis and invasion. The present study aimed to explore and validate predictors of distant metastasis and prognosis in pancreatic cancer. In our preliminary experiment, we established a novel metastatic pancreatic cancer cell line BxPC-M8 from parent BxPC-3 cells. Via whole genome sequencing, RT-qPCR, western blotting, migration and invasion assays, we initially found that BxPC-M8 shared similar biological characteristics to BxPC-3, but only differed in enhanced metastatic and invasive capabilities with a significant increase in collagen type VI α1 chain (COL6A1) expression. Knockdown of COL6A1 via small interfering RNA led to a significant decrease in migration and invasion of BxPC-M8 cells, suggesting suppressed epithelial-mesenchymal transition. Furthermore, a significant increase in COL6A1 expression was observed in cancerous tissue compared with paracancerous tissue (40.7 vs 3.7, P=0.001). Additionally, its expression was observed to be significantly associated with distant metastasis and vascular invasion at the time of surgery. Multivariate analysis revealed that COL6A1 expression (hazard ratio 1.90, 95% confidence interval 1.04-3.47, P=0.037) is an independent predictor of overall survival (OS). The median OS observed for COL6A1+ and COL6A1− patients was found to be 8±4 and 14±7 months (P=0.021), respectively. Of note, we identified that COL6A1 expression in tissue samples was associated with significantly reduced OS (P=0.001), demonstrating that COL6A1 may serve an important role in the metastatic process and could be considered as a predictor of poor outcomes in patients with pancreatic cancer. In addition, our findings suggest that COL6A1 could be an indicator of distant metastasis and a valid prognostic predictor in such patients; however, further investigation is required.
Keywords: COL6A1, pancreatic cancer, metastasis, prognostic predictor
Introduction
Pancreatic cancer (PC) has one of the highest mortality rates among all cancers worldwide with an estimated 5-year survival rate of <5% (1). In the past decade, the incidence of pancreatic cancer has been on the rise in areas such as the United States of America, China, Japan and some European countries (2,3). Prognosis remains poor due to its clinically elusive nature; patients often present with late-stage disease. Aside from surgery, other treatment options have not been reported to improve survival (4). Approximately 10% of patients are diagnosed with early stage PC, while 52% of patients often present with metastasis to surrounding tissues or organs (4). Although pancreatic cancer is the fourth leading cause of cancer-associated mortality worldwide, limited attention and funding have been placed into investigating PC; thus, determining novel biomarkers of this disease is delayed (5-7). Its genetically complex histological nature has also contributed to the slow progress in identifying new molecular targets (8,9). Various genes, such as cell-cell adhesion molecules, members of the cadherin families and integrins have been identified as modulators of metastatic progression (10). Furthermore, other molecules, including metalloproteinases and metastatic suppressor genes, have been shown to serve crucial roles in distant metastasis (11). Circulating exosomes as markers of early detection have demonstrated promising outcomes; however, they often have lower predictive efficacies and lack specificity (12,13). Therefore, further investigation into PC is required to identify novel biomarkers with higher prognostic potential. The lack of effective predictive markers for PC has been associated with poor survival rates in patients with this devastating disease (14). This suggests that identifying genes and proteins that serve key roles in the invasion and metastasis of PC is important.
Collagen type VI α1 chain (COL6A1), a gene located on chromosome 21, contributes to maintaining the integrity of various tissues (15). This gene encodes the α1 (VI) chain of type VI collagen which is a primary extracellular matrix protein (16). Its expression in tissues such as the gastrointestinal tract, brain, and pancreas is notably low (17). In tumors, however, the expression of COL6A1 transcripts and the of collagen VI proteins are markedly higher than in corresponding normal tissues (18). Overexpression of COL6A1 in these tissues has been reported to enhance motility and metastasis of lung, prostate, and cervical cancers cells, while knockdown of COL6A1 has led to the suppression of metastatic ability (18-20). In addition, COL6A1 stimulates cell motility and supports distant colonization via the expression of oncogenic transcription factors by altering tumor suppressor P53 and SMAD family member 4 (21,22). COL6A1 has been associated with invasiveness and metastatic activity in other cancers; however, its role in the metastasis and prognosis of PC remains unknown (11,18,23).
In this study, we successfully established a novel pancreatic cancer cell model, BxPC-M8, with enhanced migratory and invasive characteristics, derived from the parent cell line BxPC-3. COL6A1 was found to be significantly upregulated in BxPC-M8 cells. Furthermore, its suppressed metastatic ability became apparent after downregulating COL6A1 via RNA interference. Coupling these findings with the clinical parameters of patients, we reported an association between COL6A1 expression and shorter overall survival (OS). To the best of our knowledge, the present study is the first to demonstrate that COL6A1 could promote metastasis and portends poor prognosis in patients with PC.
Materials and methods
Patients and clinical samples
The present study was approved by The First Affiliated Hospital of Zhejiang University (FAHZJU) Ethics Committee (approval no. 2018-1033) and was conducted in accordance with the 1964 Declaration of Helsinki and its later amendments. Written informed consent was obtained from patients. The data of 65 patients with a confirmed pancreatic adenocarcinoma who underwent Whipple surgery or distal pancreatectomy were reviewed retrospectively from January 2015 to October 2017. Other pancreatic tumor types, such as neuroendocrine and acinar cell carcinomas were not included. We analyzed clinical parameters, such as age, sex, pre-surgical carbohydrate antigen 19-9 (CA199) levels, histological types, tumor size/location and stage at diagnosis according to the AJCC Cancer Staging Manual 8th Edition: Pancreas and Hepatobiliary Cancers Staging (24). Paraffin-embedded tissues samples were acquired from the tissue bank of FAHZJU. Patients were regularly followed-up by telephone or at scheduled clinic visits. Initial follow-up after surgery was conducted once a month for the first 6 months, then every 3 months for a total of 50 months. Initially, 67 patients were recruited in the study; however, as of the mismatch of clinical data for certain samples, 2 patients were omitted from analysis. Thus, only 65 patients (27 males and 38 females) with median age 62 years (range, 24-82 years) were used in the final data analysis. In addition, survival data in association with COL6A1 expression was obtained from The Cancer Genome Atlas and the Genotype-Tissue Expression databases through the Gene Expression Profiling Interactive Analysis (GEPIA) website (https://gepia.cancer-pku.cn/) (25). GEPIA data of 178 PC patients with COL6A1 expression data were used in the OS and disease-free survival (DFS) plots. Under single gene analysis, the OS and DFS analysis were set as follows: Select Gene was 'COL6A1'; group cutoff-high (%) vs. group cutoff-low (%) in respective groups were set to '40', Hazard ratio and 95% confidence interval (CI) were set to 'Yes' with 'months' as the axis unit, and 'PAAD' was selected as thecancer type in the dataset selection menu.
Cell lines and cell culture
Polycarbonate Transwell membrane inserts with 8-μm pores (6-well, Corning 3428, Corning Inc.) were used to isolate cells with high migration ability from the parent BxPC-3 cell line (China Center for Type Culture Collection). First, 24 h serum-starved BxPC-3 cells were suspended in serum-free RPMI-1640 (Meilunbio) to a final cell density of 20×104 cells/ml. Cell suspension (1 ml) was applied into the upper chamber, while the lower well beneath the polycarbonate membranes was filled with 2 ml RPMI-1640 supplemented with 10% FBS (Biological Industries) to create a chemotactic gradient to stimulate penetration of the cells. Following 24 h of incubation at 37°C, the cells on the underside of the membrane were harvested aseptically, expanded and re-seeded in RPMI-1640 containing 10% FBS. After eight rounds of harvest and expansion, the highly metastatic cells which migrated through to the underside of the membranes were harvested and labeled as BxPC-M8. BxPC-3 and BxPC-M8 cells were cultured in RPMI-1640 with 10% FBS.
Cell morphology and growth rate in vitro
BxPC-3 and BxPC-M8 cells were sub-cultured into 60-mm dishes for 48 h. Giemsa staining was carried out by a commercial kit (Nanjing Jiancheng Bioengineering Institute). Stained cells were observed and photographed with a bright field microscope (x100 magnification). To determine the cell growth rate in vitro, BxPC-3 and BxPC-M8 were plated into 6-well tissue culture plates at a density of 4×104 cells/well. Three counts for each cell line were made by Beckman Vi-CELL Automated Cell Viability Analyzer (Boston Industries, Inc.) every 24 h for 8 days. The doubling time (DT) was calculated using the equation DT= [2log/(logN-logNo)]x t, where t indicates the number of hours, while No and N represent the time at inoculation and after culture t-hours, respectively.
Wound healing assay
Cells were seeded into two 6-cm Petri dishes and incubated for 72 h in RPMI-1640 solution supplemented with 10% bovine serum at 37°C to 100% confluence. A 200-μl sterile pipette tip was used to create a scratch wound on the monolayer of cells. The detached cells were then removed by washing the supernatant with serum-free RPMI-1640 medium. Subsequently, fresh growth medium with 10% FBS was added, and cells were placed in a humidified incubator at 37°C with 5% CO2. Images were captured at 0 and 24 h. The migrating distance was recorded as the relative distance of scratch closure from time 0 and 24 h using measurements from analysis using an inverted microscope (x100 magnification) and MIPAR Image Analysis software v1 (MIPAR LLC). Three fields per view were analyzed for each cell line. Experiments were performed in triplicate.
Cell migration and invasion assays
Polycarbonate membrane Transwell (24 well, Corning 3422, Corning, Inc.) and Matrigel (BD Biosciences) were used for the migration and invasion assays. BxPC-3 and BxPC-3 M8 cells were each suspended in serum-free RPMI-1640 medium to a final cell density of 20×104 cells/ml, and 200 μl of the cell suspension was seeded to the upper chamber, while 500 μl fresh growth medium with 10% FBS was added to the lower chamber as a chemoattractant. After 24 h at 37°C with 5% CO2 in a humidified incubator, the FBS-free medium in the upper chamber was discarded, and the non-migrated or non-invasive cells were completely removed with a cotton swab. The upper chamber was then taken and placed in different wells containing 360 μl Giemsa stain A solution (Nanjing Jiancheng Bioengineering Institute) for 2 min followed by 720 μl Giemsa stain B solution (Nanjing Jiancheng Bioengineering Institute) for 6 min at room temperature. The membrane was removed and placed on respective slides (BxPC-3 and BxPC-3 M8) for microscopic observation. Cells were then counted under an inverted light microscope in three independent fields at x100 magnification. Experiments were independently performed in triplicate.
Gene sequencing and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Using TRIzol reagent (Thermo Fisher Scientific, Inc.), total RNA was extracted from BxPC-3 and BxPC-M8 cells; total RNA was sent to the Beijing Genomics Institute for gene sequencing. A 2-fold or greater difference denoted differential expression from normalized fluorescence intensity between the BxPC-3 and BxPC-M8 cells. Subsequently, total RNA was reverse transcribed into cDNA using the iScript cDNA Synthesis kit (Bio-Rad Laboratories, Inc.) according to the manufacturer's protocol. qPCR was performed on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific Inc, CA, USA) using an iTaq universal SYBR Green supermix (Bio-Rad Laboratories, Inc.). Amplification reactions required 1 μl of cDNA template, 0.3 μl each of the forward and reverse primers (10 μM), 0.2 μl of 50X ROX Reference Dye II (Takara Bio Inc.), 5 μl of 2X SYBR Premix DimerEraserin, and 3.5 μl ddH2O to a total volume of 10 μl. Sequences of the primers used are listed in Tables SI-IIII. Amplification of the transcripts involved an initial denaturation at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec, 55°C for 30 sec, and 72°C for 34 sec. Relative quantification was determined using the comparative threshold cycle method. β-actin was used as an internal control for normalization. All RT-qPCR reactions were performed in triplicate. Data were calculated using the 2−ΔΔCq method (26).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
Cells were collected and resuspended to a final concentration of 104 cells/ml in separate BxPC-M8 and BxPC-3-labeled plates respectively. Aliquots of the cell suspension were evenly distributed into 96-well plates. After incubating for 24 h at 37°C, the designated columns were treated with gemcitabine (Meilunbio) at concentrations 0, 0.2, 0.5, 1, 2, 5, 10, 20, 50 nM and incubated for 72 h at 37°C. MTT was added 4 h prior to analysusis. Formazan, the metabolite of MTT was dissolved with 150 μl dimethyl sulfoxide in each well. After 4 h of incubation at 37°C, all aliquots were discarded. The absorbance in individual wells was determined at 570 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Western blotting
Cells were seeded into a 6-cm petri dish in RPMI-1640 medium supplemented with 10% bovine serum. At ~80% confluence, cells were lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) containing protease and phosphatase inhibitors. The lysates were subjected to several freeze-thaw cycles to ensure complete lysis. The concentration of lysates was determined with the micro-bicinchoninic acid protein estimation kit (Bio-Rad Laboratories, Inc.). The protein concentrations were adjusted, and solutions were prepared under reducing conditions with 5% β-mercaptoethanol. Equal amounts (30 μg/lane) of protein were fractionated via 12% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes, blocked in 5% milk in PBS at room temperature for 1 h, and incubated with primary antibodies against COL6A1 (ab151422, Abcam), β-actin (13E5), E-cadherin (24E10), N-cadherin (D421H), vimentin (D21H3), snail (C15D3) and matrix metalloproteinase (MMP)-9 (D603H) (Cell Signaling Technology, Inc.) in a dilution of 1:1,000 overnight at 4°C. After washing with TBS containing 0.1% (v/v) Tween-20, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse (ab6728, Abcam) or rabbit (ab6721, Abcam) secondary antibodies in a dilution of 1:2,000 at room temperature for 2 h. The expression levels of COL6A1, β-actin, E-cadherin, N-cadherin, vimentin, snail and MMP-9 were analyzed. Bands were visualized using ECL kits according to the manufacturer's instructions (Abcam). Densitometric analysis was performed using Image Lab ver. 6.0.0 build 26 (Bio-Rad Laboratories, Inc.).
Tissue microarray (TMA) and immunohistochemistry (IHC) analyses
Paraffin-embedded tissue samples were sent to Shanghai Outdo Biotech Co. Ltd. For TMA construction. Immunohistochemistry was performed using an indirect immunoperoxidase method, as previously described (27). Briefly, after incubation at 60°C, slices were deparaffinized in xylene and rehydrated in alcohol. Antigen retrieval was performed in citrate buffer at 120°C for 10 min in a decloaking chamber. Endogenous peroxidase was blocked by 3% hydrogen peroxide for 10 min at room temperature. Sections (4 μm thickness) were incubated with anti-COL6A1 primary antibody (ab151422, Abcam) at 1:700 dilution overnight at 4°C. After applying horseradish peroxidase-conjugated rabbit secondary antibody (ab6721, Abcam) in a 1:2,000 dilution at room temperature for 2 h, sections were treated with a Liquid DAB+ Substrate Chromogen System (K3468; Dako; Agilent Technologies, Inc.) for the chromogenic reaction and with Mayer's hematoxylin for counterstaining according to the manufacturer's protocol. Images were captured with the NanoZoomer Digital Pathology 2.0RS and analyzed with NDP.view2 Viewing software version 2.7.25 (Hamamatsu Photonics K.K.). Three independent research pathologists semi-qualitatively categorized all samples into two groups. Similar to previously described (28) scoring method, the cytoplasmic dye color intensity of tumor cells from each sample was determined and denoted '+' for stained cytoplasm and '-' for negative staining. Samples with '−' were assigned 'COL6A1−' expression, whiles those identified as '+' were assigned 'COL6A1+'.
RNA interference
All small interfering RNAs (siRNAs) were purchased from Shanghai GenePharma Co., Ltd. Transient transfection of each siRNA for COL6A1 knockdown was conducted using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The siRNA sequences were as follows: genOFFTM st-h-COL6A1_001, 5′-GCATTGGCCTGCAGAACTT-3′; genOFFTM st-h-COL6A1_002, 5′-CGGTGCCTTTGGACTGAAA-3′; genOFFTM st-h-COL6A1_003, 5′-CCATGGACTTTATCAACGA-3′ and negative control (NC)-siRNA, 5′-UUCUCCGAACGUGUCACGUTT-3′. The cells weretransfected with st-h-COL6A1 or (NC)-siRNA for 48 h at a final concentration of 30 nM, and the efficiency of each siRNA oligo duplex was confirmed by RT-qPCR.
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics software for macOS v.25.0 (IBM Corp.) and GraphPad Prism 7.0a (GraphPad Software, Inc.). The OS was calculated as the time from the date of primary surgery to the date of reported mortality from any cause and survival was censored at the date of the last contact. Pearson's χ2 test was used in the qualitative analysis. In addition, a Student's t-test, Mann-Whitney U or Kruskal-Wallis test adjusted by the Bonferroni correction, was performed. Patient survival was analyzed via Kaplan-Meier analysis followed by a log-rank test. The Cox proportional hazard model was used for the univariate and multivariate analysis. All analysis were two-tailed; P<0.05 was considered to indicate a statistically significant difference.
Results
BxPC-M8 shares similar biological characteristics to BxPC-3 but possesses an epithelial-mesenchymal transition (EMT) morphological phenotype
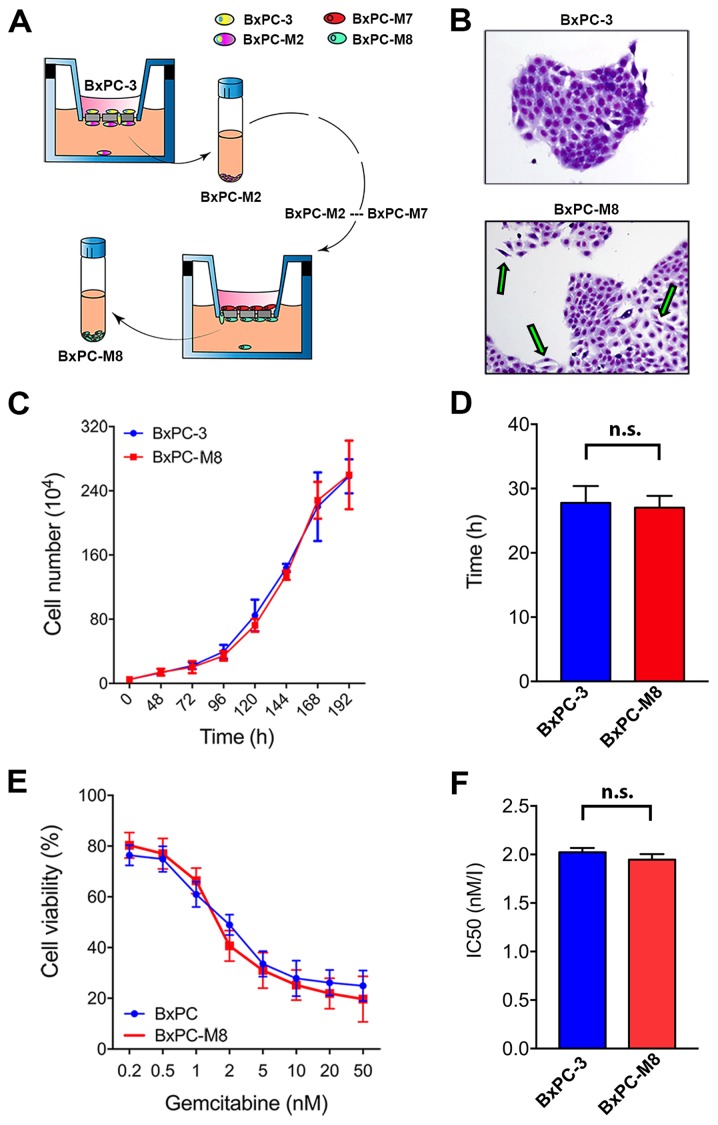
To study potential factors associated with metastasis in PC, we established a novel metastatic pancreatic cancer cell model, BxPC-M8, from BxPC-3 cells (Fig. 1A). STR profiling revealed an identity score of 1.00, validating that all allele locations of BxPC-M8 matched those of BxPC-3, indicating no cross contamination (Table SIV). As presented in Fig. 1B, BxPC-3 cells demonstrated more cohesion with clear demarcations and tight colony edges, while BxPC-M8 cells appeared spindle-shaped with a relatively reduced nucleus-to-cytoplasm ratio. These characteristics suggested that BxPC-M8 exhibited morphological features associated with EMT as described in other PC cell lines (29,30). The growth assay showed that both BxPC-3 and BXPC-M8 cells had similar proliferation rates (Fig. 1C). The DTs for the BxPC-3 and BxPC-M8 cell lines were 27.8±2.6 and 27.0±1.8 h, respectively (Fig. 1D). Additionally, sensitivity to the commonly used chemotherapeutic drug gemcitabine was comparable between BxPC-3 and BxPC-M8 with, with half-maximal inhibitory concentration values of 2.02±0.08 and 1.95±0.08 nM, respectively (Fig. 1E and F). These results indicated that BxPC-M8 has a notably similar proliferation pattern and drug sensitivity to BxPC-3.
Figure 1.
Established BxPC-M8 cell line demonstrates different morphology but a similar growth pattern compared with BxPC-3. (A) Schematic representation of the establishment of BxPC-M8 cells from the parent cell line BxPC-3. (B) Images of BxPC-3 and BxPC-M8 stained with Giemsa (magnification, x200); epithelial-mesenchymal transition phenotype with the green arrows. (C and D) Growth curve indicated a similar growth rate between the two cell lines. (E and F) Sensitivity to gemcitabine revealed no significant difference between BxPC-3 and BxPC-M8 cells as demonstrated by the IC50 value. n.s., not significant; IC50, half-maximal inhibitory concentration.
BxPC-M8 demonstrates enhanced migration and invasive abilities
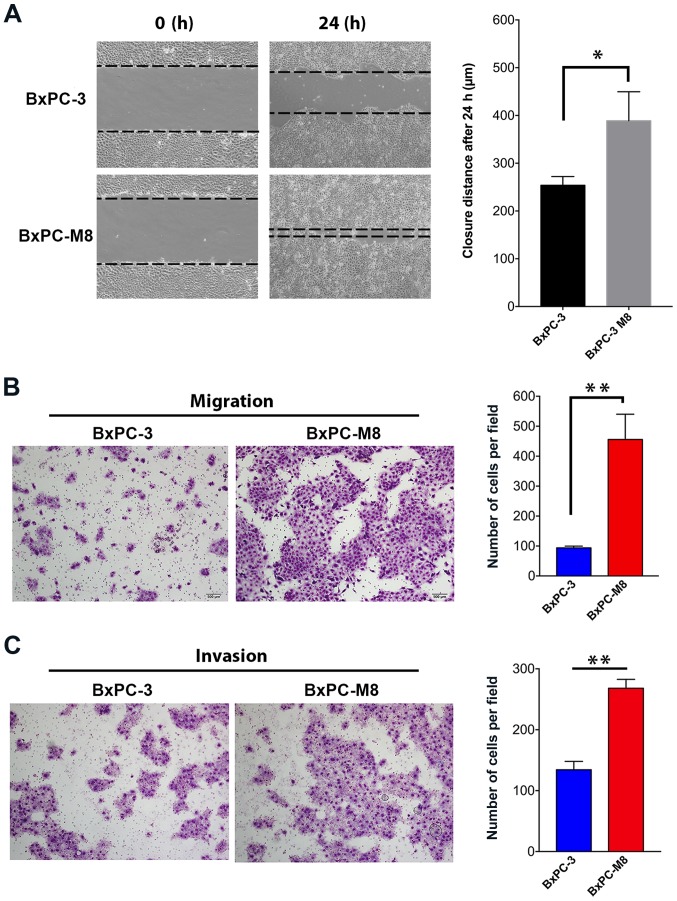
The migration abilities between the BxPC-3 and BxPC-M8 cell lines were first compared using a wound-healing assay (Fig. 2A). The size of the wound as a function of the migration ability of each cell line was determined at 0 and 24 h. Compared with the parental BxPC-3, we observed a near complete closure of the central area of confluence in BxPC-M8 cells, indicating an enhanced migration ability of BxPC-M8 cells (P=0.021; Fig. 2A). Migrated cells in the Transwell assay were quantified by microscopy (Fig. 2B). The resules revealed 93±5 versus 455±84 migrated cells per field view in BxPC-3 and BxPC-M8, respectively (P<0.01; Fig. 2B). Furthermore, similar results were also observed in the Matrigel invasion assays between BxPC-3 and BxPC-M8 cells; 135±14 invading cells were counted per field for BxPC-3, while those for BxPC-M8 were 268±14 (P<0.05; Fig. 2C). In both experiments, each field was counted in triplicate. Our findings indicated that BxPC-M8 cells exhibited significantly enhanced migration and invasive properties compared with parent BxPC-3 cells.
Figure 2.
BxPC-M8 cells possess enhanced migration and invasion abilities. (A) Wound-healing assay compared between BxPC-3 and BxPC-M8 at 0 and 24 h (*P<0.05). Inverted microscopic measurements in combination with MIPAR Image Analysis software (v1) were used to determine the distance travelled by cells. (B and C) Migratory and invasive abilities determined using Polycarbonate membrane Transwell assays without or with Matrigel, respectively (x100 magnification). All data are expressed as mean ± standard deviation of triplicate assays. Images were captured at x100 magnification; **P<0.01. All experiments were performed in triplicate.
COL6A1 is a potential metastatic gene
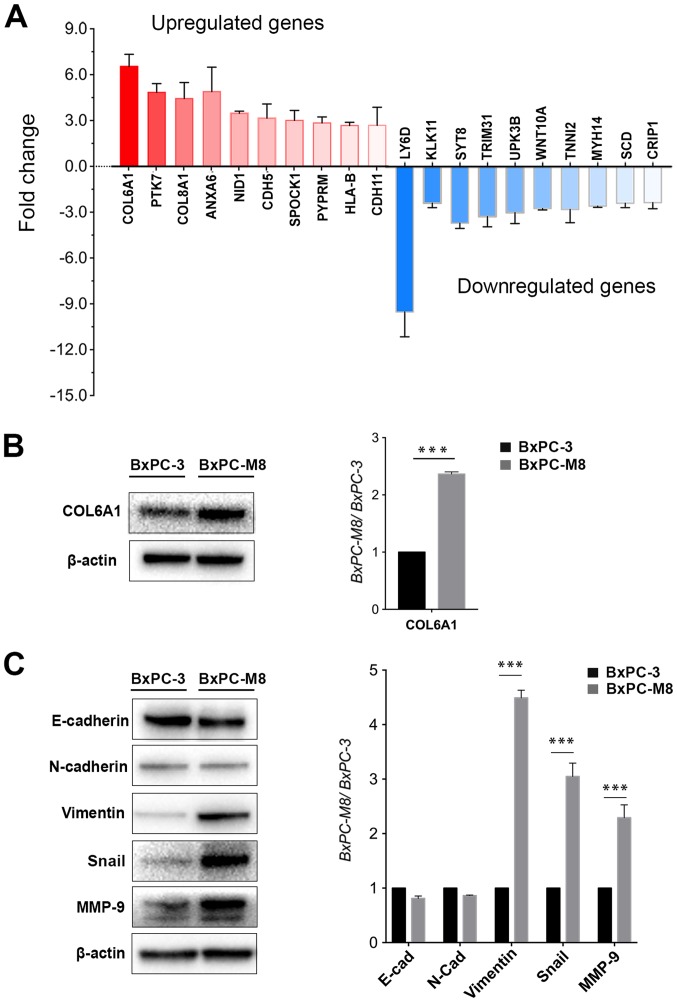
To further understand the molecular mechanisms underlying the highly metastatic activity of BxPC-M8, the differential expression of numerous genes were investigated by gene sequencing. The results revealed >1,000 genes to be differentially expressed (>1.5-fold change) between BxPC-3 and BxPC-M8 cells. A total of 20 genes with the highest difference in expression between BxPC-3 and BxPC-M8 were selected. Subsequently, RT-qPCR revealed that COL6A1, protein tyrosine kinase 7, Annexin A6, nidogen 1 and cadherin (CDH)5 were upregulated with a >3-fold increase, while osteonectin, protein tyrosine phosphatase receptor type M, major histocompatibility complex, class I, B, CDH11 and C-C motif chemokine ligand 21 had a >2-fold increase in expression. In contrast, lymphocyte antigen 6 family member D, kallikrein 11, synaptotagmin 8, and tripartite motif containing 31 were downregulated with a >3-fold difference (Fig. 3A). Western blot analysis indicated that COL6A1 was also upregulated in BxPC-M8 compared with BxPC-3 (Fig. 3B). Among the collagen type VI encoding genes COL6A1, COL6A2, COL6A3, COL6A4P1, COL6A4P2, COL6A5 and COL6A6, only COL6A1 could adequately be detected according to whole-genome sequencing of BxPC-3 and BxPC-M8 (data not shown). As established, COL6A1 only encodes the collagen VI α1 chain (31). This suggested that increased expression of COL6A1 could be linked to the enhanced migratory and invasive abilities we observed in the present study. In addition, the expression of EMT-related proteins were investigated in BxPC-3 and BxPC-M8 cells. Compared with parent BxPC-3 cells, the expression of Snail, vimentin and MMP-9 were significantly increased in BxPC-M8 (P<0.001; Fig. 3C). Our results indicated that BxPC-M8 could express a typical EMT phenotype. We proposed that COL6A1 may be associated with EMT and potentially contributes to PC metastasis.
Figure 3.
Differential expression of genes between the BxPC-3 and BxPC-M8 cell lines, and epithelial-mesenchymal transition-associated protein expression changes. (A) Expression levels of 20 selected genes were determined between the BxPC-3 and BxPC-M8 cells using reverse transcription-quantitative polymerase chain reaction. (B) Western blotting demonstrated that the expression of COL6A1 was significantly upregulated in BxPC-M8 cells compared with BxPC-3 (***P<0.001). (C) Western blotting revealed the relative expression of E-cad, N-cad vimentin, Snail and MMP-9 between BxPC-M8 and BxPC-3 cells (left panel). The results of semi-quantitative densitometric analysis were presented in the right panel (***P<0.001). COL6A1, collagen type VI α1 chain; E-cad, E-cadherin; N-cad, N-cadherin; MMP-9, matrix metalloproteinase-9.
COL6A1 silencing inhibits the migration and invasion of BxPC-M8 cells
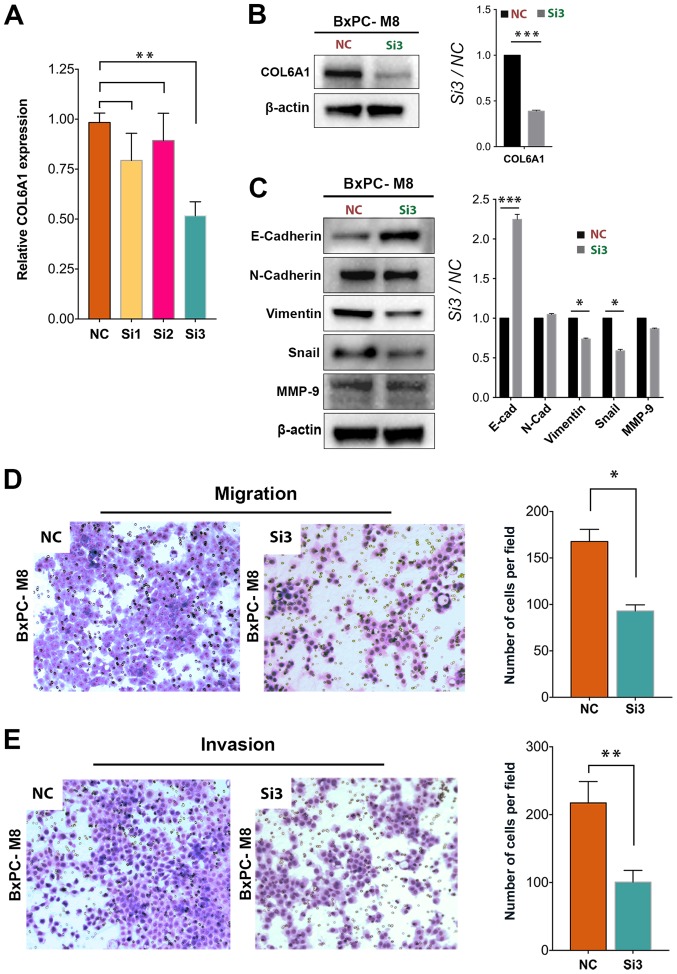
To examine whether COL6A1 plays a key role in PC metastasis, we knocked down COL6A1 expression by using three siRNA vectors targeting different regions of COL6A1 mRNA. Only the third siRNA vector (Si3 group) resulted in a signficant decrease in COL6A1 expression (1.88-fold) compared with the NC (P<0.01; Fig. 4A). Western blot analysis confirmed a signficant decrease in COL6A1 expression in the Si3 group cells compared with the NC group (Fig. 4B). As expected, expression levels of Snail and vimentin were signficantly decreased in the Si3 group compared with the NC group. The expression levels of E-cadherin were significantly increased in the Si3 group than in the NC group (P<0.001). These results suggested that COL6A1 knockdown could inhibit EMT in PC. On the contrary, no significant difference in MMP-9 expression was observed after siRNA knockdown of COL6A1, which indicates that COL6A1 expression and its pathways may exert little or no influence on MMP-9 expression in BxPC-M8 cells. Furthermore, we examined the migration ability of BxPC-M8 via a Transwell assay after silencing COL6A1. The number of migrated cells in the NC and Si3 groups were quantified under microscopy as presented in Fig. 4D. A significant decrease in cell count was seen in the Si3 group compared with in the NC group; 168±13 for the NC group vs. 93±7 cells for the Si3 cells per field (P<0.05). The invasive ability between NC and Si3 was evaluated and a quantitative assessment indicated significant reductions in cell invasion in the Si3 group compared with the NC group. Invasive cells counted per field for the NC group were 217±31, while those for Si3 group was 100±17 (P<0.01; Fig. 4E). This suggests that silencing of COL6A1 can decrease the migration and invasion of PC cells via regulation of EMT-associated proteins.
Figure 4.
Knockdown of COL6A1 significantly decreases the migration and invasion of BxPC-M8 cells. (A) Significantly decreased COL6A1 expression was observed following COL6A1 knockdown between the NC and the Si3 knockdown group, (P=0.022 by Kruskal-Wallis test; error bars indicate the standard deviation). (B and C) siRNA knockdown of COL6A1 causes significant decreases in epithelial-mesenchymal transition proteins, such as vimentin but increased the expression of E-cad in BxPC-M8 cells, as determined by western blotting (left panel). Relative semi-quantitative analysis results are also presented (right panel). (D and E) COL6A1 knockdown revealed decreases in the migration and invasion of BxPC-M8 cells as determined by Boyden Chamber and Matrigel-coated Boyden Chamber assays, respectively. Images are Giemsa stained and captured at x100 magnification. *P<0.05, **P<0.01, ***P<0.001. COL6A1, collagen type VI α1 chain; E-cad, E-cadherin; N-cad, N-cadherin; MMP-9, matrix metalloproteinase-9; NC, negative control; siRNA, small interfering RNA; Si3, third siRNA.
COL6A1 expression predicts poor prognosis in patients with PC
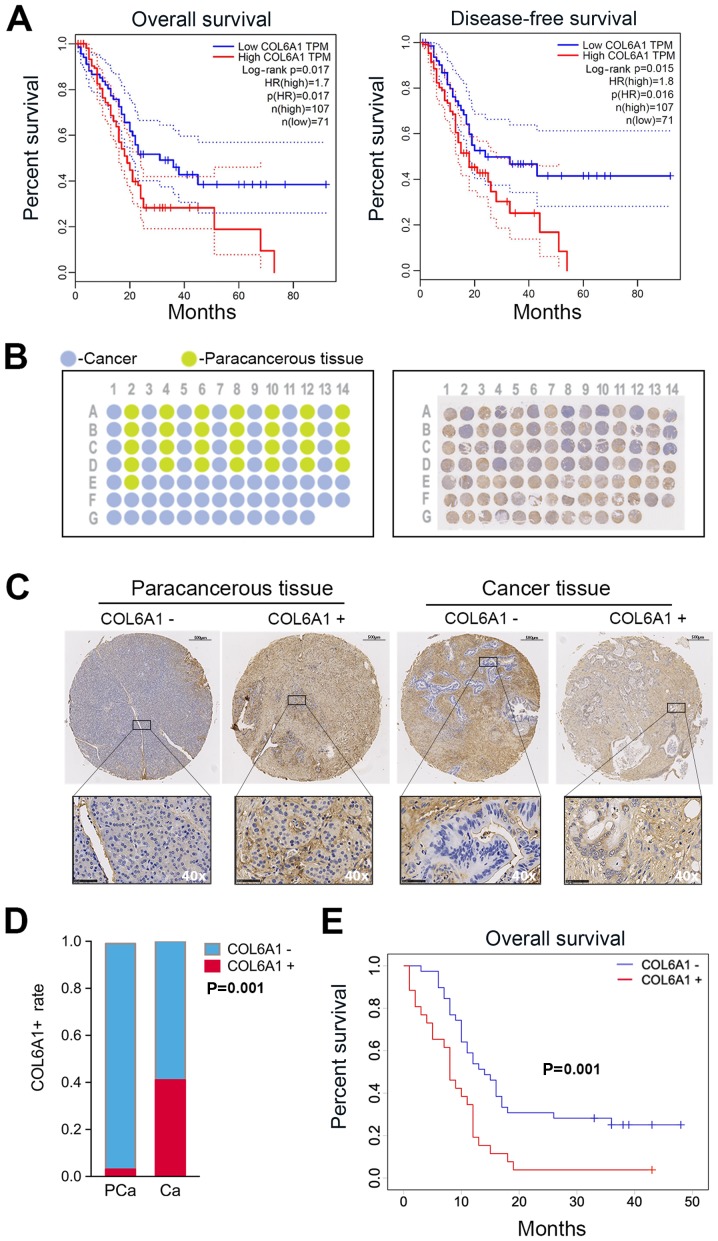
We first determined the relevance of COL6A expression in human PC by generating survival plots from the GEPIA database. The preset parameters used for the OS and DFS analysis were set as follows: Group cutoff-high (%) vs. group cutoff-low (%) in respective groups set to 40 with a 95% confidence interval (CI). The results showed a significant increase in OS in patients with low COL6A1 expression compared with high COL6A1 expression [P=0.017; hazard ratio (HR) 1.70; n=178]. DFS analysis revealed increased DFS times in the low COL6A1 expression group compared with those exhibiting high expression [P=0.016; HR 1.80; n=178] (Fig. 5A).
Figure 5.
COL6A1 expression affects the prognosis of patients with pancreatic cancer. (A) Data from the GEPIA database indicated that patients with high COL6A1 expression levels exhibited significantly decreased survival compared with those with low COL6A1 expression (P=0.017 and 0.016). (B) A representative diagram of TMA organization (left); cancer tissue and paracancerous tissue are represented as blue and green, respectively. Overview of TMA slide (right) of stained by IHC with COL6A1 antibody. (C) Representative images of immunohistochemically stained paracancerous and cancer tissues. Scale bar, 100 μm. Magnification of insert, x40. (D) COL6A1+ rate was significantly higher in cancerous tissue compared with paracancerous tissue (P=0.001). (E) Kaplan-Meier survival plot of patient specimens indicate that patients with COL6A1+ expression in tumor tissues showed significantly decreased survival compared with patients possessing COL6A1− expression (P=0.001). Data were analyzed using IBM SPSS v25. CA, cancer; COL6A1, collagen type VI α1 chain; HR, hazard ratio; PCa, paracancerous; TMA, tissue microarray.
To further determine the association between COL6A1 expression and clinical outcomes, paraffin-embedded tissue samples underwent TMA analysis. A schematic representation of the completed TMA block from patients are shown in Fig. 5B. Cancer and paracancerous tissues are represented on the schematic as blue and green circular spots respectively (Fig. 5B). IHC analysis of the tissue sections with COL6A1-antibody was performed to generate a single TMA slide with circular spots (n=96) and TMA of cancer/paracancerous tissue pairs (n=58) versus cancer only samples (n=38, Fig. 5B). Circular sections were fitted onto a single block for further analysis. Histological grade and tumor type were compared with the clinical data. Samples from 2 subjects, denoted as E01 and D01, were determined as inconsistent with clinical and histological data and were excluded in the final analysis. Representative COL6A1+ or COL6A1− cancer and paracancerous tissues have been shown in Fig. 5C. COL6A1+ versus COL6A1− rates between cancer and paracancerous sample pairs were compared. The results indicated a significant increase in COL6A1 expression among cancer samples (11/27, 40.7%) compared with paracancerous tissue pairs (1/27, 3.7%, Fig. 5D).
Of the patients confirmed for the study (n=65), the median follow-up was 11 months (range, 1-50 months). The median age in years for males and females was 62±10 (range, 24-82 years, P= 0.70), with the majority being females (n=38) 58.5% versus (n=27) 41.5%. Significant differences in COL6A1 expression were found among the the three CA199 categories (P=0.018), metastatic status (P=0.011), vascular involvement (P=0.004) and American Joint Committee on Cancer (AJCC) staging (P=0.022) (Table I). Vascular invasion was observed in 76.9% of patients with COL6A1 expression relative to patients with negative expression at the time of surgery (41.0%). In the univariate analysis, COL6A1 expression (HR 2.38, 95% CI 1.38-4.14, P=0.002), N2-category (HR 2.31, 95% CI 1.18-4.55, P=0.015), M1-category (HR 2.54, 95% CI 1.43-4.54, P=0.002), advanced AJCC stages III/IV (HR 2.80, 95% CI 1.51-5.19, P=0.001), vascular (HR 2.28, 95% CI 1.32-3.97, P=0.003) and neural invasion (HR 1.93, 95% CI 1.08-3.60, P=0.027) were associated with shorter survival, while N1/N0, M0 and earlier AJCC stages I/II were associated with relatively longer OS. Multivariate analysis also revealed that COL6A1+ (HR 1.90, 95% CI 1.04-3.47), distant metastasis (HR 2.35, 95% CI 1.24-4.47, P=0.009) and vascular invasion (HR 2.08, 95% CI 1.16-3.73, P=0.014) were independent predictors of OS and were associated with shorter survival (Table II). Kaplan-Meier survival analysis revealed that patients with COL6A1+ expression had significantly worse outcomes compared with COL6A1- patients (Fig. 5E). The median OS for the COL6A1− group was 14±7 months in contrast to 8±4 months among the COL6A1+ group. The results from survival analysis using Kaplan-Meier curves from this study were in parallel with the data of the GEPIA database. This indicates that COL6A1 expression may be associated with poor prognosis in patients with PC.
Table I.
Clinicopathological characteristics and COL6A1 expression in pancreatic tumor specimens.
| COL6A1 expression
|
|||||
|---|---|---|---|---|---|
| Clinicopathological parameters | n (%) | − [(n) %)] | + [(n) %] | P-value (χ2) | P-value (Kruskal-Wallis) |
| All cases (n) | 65 | 39 | 26 | ||
| Age (years) | |||||
| ≤55 | 14 (21.5) | 11 (28.2) | 3 (11.5) | 0.109 | |
| >55 | 51 (78.5) | 28 (71.8) | 23 (88.5) | ||
| Sex | |||||
| Male | 27 (41.5) | 19 (48.7) | 8 (30.8) | 0.150 | |
| Female | 38 (58.5) | 20 (51.3) | 18 (69.2) | ||
| Tumor location | |||||
| Head | 40 (61.5) | 25 (64.1) | 15 (57.7) | 0.603 | |
| Body/tail | 25 (38.5) | 14 (35.9) | 11 (42.3) | ||
| Tumor grade | |||||
| I | 13 (21.3) | 6 (16.2) | 7 (29.2) | 0.521 | |
| II | 37 (60.7) | 24 (64.9) | 13 (54.2) | ||
| III | 11 (18.0) | 7 (18.9) | 4 (16.6) | ||
| CA199 (U/ml) | |||||
| ≤37 | 9 (13.9) | 8 (20.5) | 1 (3.8) | 0.018a | |
| Increased <300 | 11 (16.9) | 3 (7.7) | 8 (30.9) | ||
| >300 | 45 (69.2) | 28 (71.8) | 17 (69.2) | ||
| T stage | |||||
| T1 | 4 (6.2) | 3 (7.7) | 1 (3.8) | 0.536 | |
| T2 | 36 (55.4) | 23 (59.0) | 13 (50.0) | ||
| T3 | 25 (38.4) | 13 (33.3) | 12 (46.2) | ||
| N stage | |||||
| N0/N1 | 54 (83.1) | 32 (82.1) | 22 (84.6) | 0.787 | |
| N2 | 11 (16.9) | 7 (17.9) | 4 (15.4) | ||
| M stage | |||||
| M0 | 30 (46.2) | 23 (59.0) | 7 (26.9) | 0.011a | |
| M1 | 35 (53.8) | 16 (41.0) | 19 (73.1) | ||
| AJCC staging | |||||
| IA/IB | 7 (10.8) | 7 (17.9) | 0 (0.0) | 0.022a | |
| IIA/IIB | 18 (27.7) | 13 (33.3) | 5 (19.2) | ||
| III | 4 (6.2) | 3 (7.7) | 1 (3.9) | ||
| IV | 36 (55.3) | 16 (41.1) | 20 (76.9) | ||
| Vascular invasion | |||||
| Yes | 36 (55.4) | 16 (41.0) | 20 (76.9) | 0.004a | |
| No | 29 (44.6) | 23 (59.0) | 6 (23.1) | ||
| Neural invasion | |||||
| Yes | 41 (63.1) | 25 (64.1) | 16 (64.0) | 0.834 | |
| No | 24 (36.9) | 14 (35.9) | 10 (44.0) | ||
COL6A1 relation with tumor age, sex, grade, lymph node metastasis, CA199, tumor size/location, AJCC stage and vascular invasion. In the tumor grade analysis, four tissues were removed due to unknown grade.
P<0.05. CA199, carbohydrate antigen 19-9; COL6A1, collagen type VI α1 chain.
Table II.
Cox regression analysis for overall survival.
| Univariate analysis
|
Multivariate analysis
|
|||||
|---|---|---|---|---|---|---|
| Characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age (years) | ||||||
| >55 vs ≤55 | 0.70 | 0.36-1.35 | 0.285 | |||
| Sex | ||||||
| Female vs. male | 1.55 | 0.89-2.69 | 0.121 | |||
| Tumor location | ||||||
| Body/tail vs. head | 1.09 | 0.63-1.89 | 0.758 | |||
| Tumor grade | ||||||
| II vs. I | 1.03 | 0.51-2.11 | 0.928 | |||
| III vs. I | 0.86 | 0.34-2.17 | 0.741 | |||
| COL6A1 expression | ||||||
| COL6A1+ vs. COL6A1− | 2.39 | 1.38-4.14 | 0.002a | 1.90 | 1.04-3.47 | 0.037a |
| CA199 (U/ml) | ||||||
| >37 vs ≤37 | 1.39 | 0.59-3.27 | 0.448 | |||
| T stage | ||||||
| T2 vs. T1 | 0.25 | 0.09-0.75 | 0.130 | |||
| T3 vs. T1 | 0.45 | 0.15-1.33 | 0.149 | |||
| N stage | ||||||
| N2 vs. N1/N0 | 2.32 | 1.18-4.55 | 0.015a | |||
| M stage | ||||||
| M1 vs. M0 | 2.54 | 1.43-4.54 | 0.002a | 2.35 | 1.24-4.47 | 0.009a |
| AJCC staging | ||||||
| III/IV vs. I/II | 2.80 | 1.51-5.19 | 0.001a | |||
| Vascular invasion | ||||||
| Yes vs. no | 2.28 | 1.32-3.97 | 0.003a | 2.08 | 1.16-3.73 | 0.014a |
| Neural invasion | ||||||
| Yes vs. no | 1.93 | 1.08-3.60 | 0.027a | |||
Multivariable Cox proportion hazard regression used to determine overall survival and correlation with COL6A1 expression, vascular and neural invasion, lymph node involvement tumor staging and distance metastasis.
P<0.05. CI, confidence interval; COL6A1, collagen type VI α1 chain; HR, hazard ratio.
Discussion
In this study, we explored potential metastasis-related factors in PC. By establishing the robust PC cell line BxPC-M8 from parent cells (BxPC-3), our findings may improve understanding of the metastatic and invasive characteristics between the two cell lines. Genetic profiling of the cell lines unveiled COL6A1 as a potential metastasis-associated gene. Knockdown of COL6A1 in BxPC-M8 cells led to a significant decrease in cell migration and invasive abilities confirming the metastatic role of COL6A1. Furthermore, COL6A1 expression was significantly associated with CA199 levels, distant metastasis and vascular invasion. COL6A1 was also determined to be an independent predictor of poor prognosis in patients with PC.
One of the major challenges in establishing preclinical PC cell models which can recapitulate the biological behavior in humans includes contamination and incongruity with the clinical characteristics of the disease in focus (30,32). At present, few cellular models are available for investigations into PC metastasis (33-35). BxPC-M8 demonstrated stable culture properties; its growth rate and sensitivity to conventional chemotherapeutic drugs were found to be comparable to parent BxPC-3 cells. However, BxPC-M8 differs from BxPC-3 in enhanced metastatic and invasive capabilities, making it a potential metastatic research model for PC.
Several studies have detected increased expression of COL6A1 in tumors and is associated with tumor progression (36,37). Other reports have also shown that collagen VI can promote the adhesion, penetration and invasion of tumor cells in an autocrine manner (18). In a quantitative secretomics functional analysis of metastasis-associated proteins, COL6A1 expression was reported to enhance metastatic abilities, while RNA interference-based knockdown of COL6A1 suppressed the metastatic activity of lung cancer cells (20). Furthermore, the migratory and metastatic activity of PC via runt-related transcription factor 3 (Runx3) signaling can be induced by the expression of COL6A1 (36,38). In our research, COL6A1 was found to be upregulated in BxPC-M8, and was associated with increased invasive and metastatic ability compared with parent BxPC-3 cells, suggesting a potential pro-metastatic role in PC. Additionally, an increase in EMT-associated proteins, such as Snail and vimentin, which were upregulated in BxPC-M8 compared with BxPC-3 cells, indicate a possible relation to EMT characteristics. Similar to previous reports on other cancer types (18-20), we reported that the migration and invasion of BxPC-M8 cells were significantly decreased after COL6A1 knockdown, while the expression of E-cadherin increased, but that of Snail and vimentin were decreased. The heterotrimeric structure of type VI collagen is formed by three α chains among six possible genes (COL6A1-6) (39). The present study investigated the effects of silenced COL6A1, which encodes the α1 chain of collagen type VI, and its relation to EMT protein expression. In future research, we aim to explore the potential link between EMT and other genes associated with the structure of type VI collagen, as well as its specific downstream targets.
Even though researchers have reported the correlation between high COL6A1 expression and poor clinical prognosis in patients with cervical cancer and prostate cancer (18,19), its prognostic role in PC remains unclear. To improve understanding into the relationship between COL6A1 and patient outcomes, we initially analyzed RNA sequencing data from GEPIA database (25). We observed that high COL6A1 expression was associated with shorter OS and DFS. The relationship between COL6A1 expression and clinical parameters, including the prognosis of PC patients was explored. The AJCC Cancer Staging Manual, Eighth Edition, 2017 (24) was used throughout our study for tumor staging. Clinicopathological investigations revealed that COL6A1 expression was closely associated with CA199 levels, distant metastasis, vascular invasion and AJCC stage. It has been proposed that regional lymph node metastasis has high prognostic value in relation to COL6A1 expression in cervical and prostate cancers (18,19). However, our findings did not indicate a significant association between local lymph node metastasis and COL6A1 expression. In line with studies reporting a positive relationship between COL6A1 and distant metastasis (36), we found that 73.1% of COL6A1+ patients had distant metastasis. Also, 76.9% of our patients with COL6A1 expression had vascular encroachment at the time of surgery. The median OS for COL6A1+ group was 8±4 months compared with 14±7 months in COL6A1− group. Survival analysis using Kaplan-Meier curves from this study demonstrated that expression of COL6A1 was associated with poor OS in PC patients, in accordance with the data of the GEPIA database. In addition, ~40% of patients with low COL6A1 expression survived >5 years according to GEPIA database. The follow-up in this study was conducted for up to 50 months; however, long-term follow-up is required to validate our findings with the GEPIA database.
Research regarding the metastasis of PC has been limited in recent years due to insufficient stable cell lines. The present study established a potentially useful BxPC-M8 cell model, which may provide further insight into the metastatic and prognostic molecular markers in PC. Additionally, to the best of our knowledge, our study is the first to demonstrate the clinical significance of COL6A1 in patients with PC, and highlights COL6A1 as an effective prognostic marker and a potential target for developing pharmacological therapies in the future.
Supplementary Data
Acknowledgments
We thank Dr Kenneth Goldblatt from the University Medical Center of Princeton At Plainsboro for his help in revising the final manuscript before submission. Special thanks to Dr Rongliang Tong and Dr Chuanhui Peng from the Department of Hepatobiliary and Pancreatic Surgery (The First Affiliated Hospital of Zhejiang University) for their clinical and technical input in tumor and SPSS data matching.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CA199
carbohydrate antigen 19-9
- COL6A1
collagen type VI α1 chain
- DFS
disease-free survival
- FBS
fetal bovine serum
- GEPIA
Gene Expression Profiling Interactive Analysis
- OS
overall survival
- PC
pancreatic cancer
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- siRNA
small interfering RNA
Funding
The present study was supported by The Innovative Research Groups of National Natural Science Foundation of China (grant no. 81721091); Zhejiang Provincial Natural Science Foundation (grant no. LY18H160017); Major program of National Natural Science Foundation of China (grant no. 91542205); National S&T Major Project (grant no. 2017ZX10203205); the Fundamental Research Funds for the Central Universities (grant no. 2019XZZX005-1-08) and Zhejiang International Science and Technology Cooperation Project (grant no. 2016C04003).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. Supplementary files can also be located at https://doi.org/10.6084/m9.figshare.7562813.v1
Authors' contributions
DJ and KGOA performed experiments, analyzed results and wrote the manuscript. GS performed siRNA knockdown. RC performed western blot analysis and repeated experiments. DJ and BC established the cell line. BC and LZ performed all IHC staining and TMA slide. HX and LZ and an independent pathologist performed the scoring of all TMA cores. MIAE and JL collected tissues samples and analyzed clinical data. DJ and KGOA performed statistical analysis, as well as the construction of the tables and figures. Conceptualization, methodology, and supervision of the project were performed by DJ and JW and SZ revised the final manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This research was approved by the First Affiliated Hospital of Zhejiang University Ethics Committee (approval no. 2018-1033).
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Noone AM, Cronin KA, Altekruse SF, Howlader N, Lewis DR, Petkov VI, Penberthy L. Cancer incidence and survival trends by subtype using data from the Surveillance Epidemiology and End Results (SEER) Program, 1992-2013. Cancer Epidemiol Biomarkers Prev. 2017;26:632–641. doi: 10.1158/1055-9965.EPI-16-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Yang GH, Li H, Lu XH. The changing pancreatic cancer mortality in China (1991-2000) Zhonghua Nei Ke Za Zhi. 2005;44:509–513. In Chinese. [PubMed] [Google Scholar]
- 4.Cancer.Net . Pancreatic Cancer: Statistics. American Society of Clinical Oncology ASCO; 2018. [Google Scholar]
- 5.Malvezzi M, Carioli G, Bertuccio P, Rosso T, Boffetta P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2016 with focus on leukaemias. Ann Oncol. 2016;27:725–731. doi: 10.1093/annonc/mdw022. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Jemal A, Wender RC, Gansler T, Ma J, Brawley OW. An assessment of progress in cancer control. CA Cancer J Clin. 2018;68:329–339. doi: 10.3322/caac.21460. [DOI] [PubMed] [Google Scholar]
- 7.Hidalgo M, Cascinu S, Kleeff J, Labianca R, Löhr JM, Neoptolemos J, Real FX, Van Laethem JL, Heinemann V. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology. 2015;15:8–18. doi: 10.1016/j.pan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Du YX, Liu ZW, You L, Wu WM, Zhao YP. Advances in understanding the molecular mechanism of pancreatic cancer metastasis. Hepatobiliary Pancreat Dis Int. 2016;15:361–370. doi: 10.1016/S1499-3872(15)60033-9. [DOI] [PubMed] [Google Scholar]
- 11.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao K, Liang G, Sun X, Guan L. Comparative miRNAome analysis revealed different miRNA expression profiles in bovine sera and exosomes. BMC Genomics. 2016;17:630. doi: 10.1186/s12864-016-2962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmicheal J, Hayashi C, Huang X, Liu L, Lu Y, Krasnoslobodtsev A, Lushnikov A, Kshirsagar PG, Patel A, Jain M, et al. Label-free characterization of exosome via surface enhanced Raman spectroscopy for the early detection of pancreatic cancer. Nanomedicine (Lond) 2019;16:88–96. doi: 10.1016/j.nano.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang XT, Tao HQ, Zou SC. Detection of serum tumor markers in the diagnosis and treatment of patients with pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2004;3:464–468. [PubMed] [Google Scholar]
- 15.Tanaka T, Ikari K, Furushima K, Okada A, Tanaka H, Furukawa K, Yoshida K, Ikeda T, Ikegawa S, Hunt SC, et al. Genomewide linkage and linkage disequilibrium analyses identify COL6A1, on chromosome 21, as the locus for ossification of the posterior longitudinal ligament of the spine. Am J Hum Genet. 2003;73:812–822. doi: 10.1086/378593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:357. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 17.Pontén F, Jirström K, Uhlen M. The Human Protein Atlas–a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 18.Hou T, Tong C, Kazobinka G, Zhang W, Huang X, Huang Y, Zhang Y. Expression of COL6A1 predicts prognosis in cervical cancer patients. Am J Transl Res. 2016;8:2838–2844. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu YP, Wan FN, Shen YJ, Wang HK, Zhang GM, Ye DW. Reactive stroma component COL6A1 is upregulated in castration-resistant prostate cancer and promotes tumor growth. Oncotarget. 2015;6:14488–14496. doi: 10.18632/oncotarget.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiu KH, Chang YH, Wu YS, Lee SH, Liao PC. Quantitative secretome analysis reveals that COL6A1 is a metastasis-associated protein using stacking gel-aided purification combined with iTRAQ labeling. J Proteome Res. 2011;10:1110–1125. doi: 10.1021/pr1008724. [DOI] [PubMed] [Google Scholar]
- 21.Birnbaum DJ, Adélaïde J, Mamessier E, Finetti P, Lagarde A, Monges G, Viret F, Gonçalvès A, Turrini O, Delpero JR, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50:456–465. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- 22.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: Lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–1094. doi: 10.1136/gut.2010.236026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriggi M, Giussani M, Torretta E, Capitanio D, Sandri M, Leone R, De Palma S, Vasso M, Vozzi G, Tagliabue E, et al. ECM remodeling in breast cancer with different grade: Contribution of 2D-DIGE proteomics. Proteomics. 2018;18:e1800278. doi: 10.1002/pmic.201800278. [DOI] [PubMed] [Google Scholar]
- 24.Chun YS, Pawlik TM, Vauthey JN. 8th Edition of the AJCC Cancer Staging Manual: Pancreas and Hepatobiliary Cancers. Ann Surg Oncol. 2018;25:845–847. doi: 10.1245/s10434-017-6025-x. [DOI] [PubMed] [Google Scholar]
- 25.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Escobar-Hoyos LF, Shah R, Roa-Peña L, Vanner EA, Najafian N, Banach A, Nielsen E, Al-Khalil R, Akalin A, Talmage D, et al. Keratin-17 promotes p27KIP1 nuclear export and degradation and offers potential prognostic utility. Cancer Res. 2015;75:3650–3662. doi: 10.1158/0008-5472.CAN-15-0293. [DOI] [PubMed] [Google Scholar]
- 28.Alkushi A. Validation of tissue microarray biomarker expression of breast carcinomas in Saudi women. Hematol Oncol Stem Cell Ther. 2009;2:394–398. doi: 10.1016/S1658-3876(09)50007-6. [DOI] [PubMed] [Google Scholar]
- 29.Miura K, Kimura K, Amano R, Yamazoe S, Ohira G, Murata A, Nishio K, Hasegawa T, Yashiro M, Nakata B, et al. Establishment and characterization of new cell lines of anaplastic pancreatic cancer, which is a rare malignancy: OCUP-A1 and OCUP-A2. BMC Cancer. 2016;16:268. doi: 10.1186/s12885-016-2297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres MP, Rachagani S, Souchek JJ, Mallya K, Johansson SL, Batra SK. Novel pancreatic cancer cell lines derived from genetically engineered mouse models of spontaneous pancreatic adenocarcinoma: Applications in diagnosis and therapy. PLoS One. 2013;8:e80580. doi: 10.1371/journal.pone.0080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T, Takano R, Takahara N, Tokunaka K, Saiga K, Tomura A, Sugihara H, Hayashi T, Imamura Y, Morita M. Identification of a common epitope in the sequences of COL4A1 and COL6A1 recognized by monoclonal antibody #141. J Biochem. 2019;165:85–95. doi: 10.1093/jb/mvy086. [DOI] [PubMed] [Google Scholar]
- 32.Qiu W, Su GH. Challenges and advances in mouse modeling for human pancreatic tumorigenesis and metastasis. Cancer Metastasis Rev. 2013;32:83–107. doi: 10.1007/s10555-012-9408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Y, Gao S, Hua YQ, Liu LM. Establishment of a pancreatic cancer stem cell model using the SW1990 human pancreatic cancer cell line in nude mice. Asian Pac J Cancer Prev. 2015;16:437–442. doi: 10.7314/APJCP.2015.16.2.437. [DOI] [PubMed] [Google Scholar]
- 34.Kim MJ, Kim MS, Kim SJ, An S, Park J, Park H, Lee JH, Song KB, Hwang DW, Chang S, et al. Establishment and characterization of 6 novel patient-derived primary pancreatic ductal adenocarcinoma cell lines from Korean pancreatic cancer patients. Cancer Cell Int. 2017;17:47. doi: 10.1186/s12935-017-0416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzykawski MP, Krzykawska-Serda M, Jasińska K, Marcinkiewicz J. Pan_02 murine pancreatic cancer model. Folia Med Cracov. 2015;55:15–24. [PubMed] [Google Scholar]
- 36.Jian Z, Cheng T, Zhang Z, Raulefs S, Shi K, Steiger K, Maeritz N, Kleigrewe K, Hofmann T, Benitz S, et al. Glycemic variability promotes both local invasion and metastatic colonization by pancreatic ductal adenocarcinoma. Cell Mol Gastroenterol Hepatol. 2018;6:429–449. doi: 10.1016/j.jcmgh.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita A, Sato JR, Festa F, Gomes LR, Oba-Shinjo SM, Marie SK, Ferreira CE, Sogayar MC. Identification of COL6A1 as a differentially expressed gene in human astrocytomas. Genet Mol Res. 2008;7:371–378. doi: 10.4238/vol7-2gmr432. [DOI] [PubMed] [Google Scholar]
- 38.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, Wood LD, Goggins M, Hruban RH, Chang AE, et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell. 2015;161:1345–1360. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato T, Takano R, Tokunaka K, Saiga K, Tomura A, Sugihara H, Hayashi T, Imamura Y, Morita M. Type VI collagen α1 chain polypeptide in non-triple helical form is an alternative gene product of COL6A1. J Biochem. 2018;164:173–181. doi: 10.1093/jb/mvy040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article. Supplementary files can also be located at https://doi.org/10.6084/m9.figshare.7562813.v1