Abstract
Alternative splicing in tumor cells may be used as a molecular marker for the differential diagnosis of certain tumor types and assessment of prognosis. The aim of the present study was to investigate the associations among alternative splicing events, splicing factors, and the survival of patients with hepatocellular carcinoma (HCC). The alternative splicing event profiles of 371 patients with HCC were downloaded from The Cancer Genome Atlas (TCGA) SpliceSeq data, and the percent-splice-in value for each splicing event was calculated. The association between alternative splicing events and overall survival was evaluated. The most significant prognosis-related splicing events were used to build up a prognostic index (PI). A total of 3,082 survival-associated alternative splicing events were detected in HCC. The final PI based on all of the most significant candidate alternative splicing events exhibited better performance in distinguishing good or poor survival in patients compared to the PI based on a single type of splicing event. Receiver operating characteristic curves confirmed the high efficiency of the PI in predicting the survival of HCC patients, with an area under the curve of 0.914. The overexpression of 32 prognosis-related splicing factor genes could also predict poor prognosis in patients with HCC. In conclusion, the constructed computational prognostic model based on HCC-specific alternative splicing events may be used as a molecular marker for the prognosis of HCC.
Keywords: splicing, The Cancer Genome Atlas SpliceSeq, splicing factor genes, survival, hepatocellular carcinoma
Introduction
Alternative splicing, also referred to as differential splicing, refers to the process of generating various isoforms of an mRNA precursor by several approaches, i.e., by selecting diverse splice site combinations. The resulting protein products may exhibit distinguishable or antagonistic functional and structural characteristics, or they may yield various phenotypes in the same cell type due to differences in their expression levels (1,2).
The splicing isoforms of certain genes may serve as drivers for the development of tumors (3-7). Additionally, the genomes of tumor cells have more extensive alternative splicing mechanisms compared with normal cells. In most types of tumors, the expression products of genes may vary due to changes in alternative splicing, which is associated with tumor development, metastasis (8-12) and resistance to treatment (7,13-15). Additionally, alternative splicing events in tumors may act as diagnostic and prognostic molecular biomarkers (16-19), and they may also be used as targets for cancer treatment (20-24).
Alternative splicing events are regulated by splicing factors that act as oncogenes or tumor suppressor genes (25). The development of drugs targeting splicing factors is a new research focus in cancer therapy (26-32), and several studies have attempted to distinguish cancers based on the presence of abnormal RNA splicing (10,33). Furthermore, disturbances of alterative splicing are reportedly involved in the development of cancers. In addition, several survival-associated splicing events that are unique to cancer have been identified (34). RNA-binding proteins, which function as regulatory elements of splicing events, have also been associated with cancer development (35-37).
Hepatocellular carcinoma (HCC) is the fifth most common type of cancer with the third highest mortality rate worldwide. Its occurrence has been associated with viral infection, aflatoxin exposure, and other factors; in addition, a number of studies have demonstrated that alternative splicing also plays an important role in HCC (38-43). However, these studies focused on the associations between the alternative splicing events of only a few genes and HCC, whereas only few studies have analyzed alternative splicing events in HCC using large datasets. Tremblay et al (44) analyzed RNA sequencing (RNA-Seq) data from The Cancer Genome Atlas (TCGA) and found a large number of differential alternative splicing events between HCC and corresponding paracancerous tissues. Hepatitis B and C virus infections also affect alternative splicing events in HCC (44). Accordingly, alternative splicing is considered to play an important role in the occurrence of HCC. However, to the best of our knowledge, no study has yet examined the association between mRNA splicing and the prognosis of HCC based on TCGA SpliceSeq data. Therefore, by using TCGA SpliceSeq data, the present study analyzed the associations among alternative splicing events, splicing factors, and the survival of patients with HCC, with the aim of identifying splicing events that may serve as new molecular targets for the prognosis of HCC.
Materials and methods
Assortment of alternative splicing event data
The alternative splicing event profiles of HCC patients were downloaded from TCGA SpliceSeq (45), which is a resource for the investigation of transcript splicing patterns and splicing event details based on TCGA covering quantified introns or exons. Information on percent-splice-in (PSI), the ratio of normalized read counts indicating the inclusion of a transcript element over the total normalized reads for that event, was collected from the database. PSI was calculated as the ratio of reading densities of inclusions to the sum of the reading densities of inclusion and exclusion. PSI values range from 0 to 100%. Only samples with PSI values >90% were downloaded. Simultaneously, clinical data were also obtained from TCGA. A total of 7 different alternative splicing events were obtained: Exon skips (ESs), retained introns (RIs), mutually exclusive exons (MEs), alternate donor sites (ADs), alternate acceptor sites (AAs), alternate promoters (APs), and alternate terminators(ATs).
Survival analysis and production of a prognostic signature
A total of 371 HCC patients in TCGA were used to select survival-related alternative splicing events. The association between alternative splicing events and overall survival (OS) was evaluated using univariate Cox regression analysis. The most significant prognostic alternative splicing events (P<0.0001) were subjected to multivariate Cox regression analysis. The area under the curve (AUC) of a time-dependent receiver operating characteristic (ROC) curve was also calculated, which has been used widely to compare the ability of prognostic predictors. All survival analyses were conducted using the 'survival' and 'survival ROC' packages in R software. The OS rates of patients with HCC grouped by high- and low-risk alternative splicing events were plotted using Kaplan-Meier plots and the differences between groups were analyzed using log-rank tests.
UpSet plot and gene-annotation enrichment analysis
An UpSet plot, a novel visualization technique for the quantitative analysis of interactive sets, was used to analyze the intersections between the 7 types of alternative splicing. Gene-annotation enrichment analyses were used in the 'clusterProfiler' package in R to annotate and visualize biological process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of survival-associated alternative splicing genes based on the criterion of an adjusted P-value of <0.05.
Splicing factor genes
Splicing factor genes were collected from the SpliceAid 2 database (http://193.206.120.249/splicing_tissue.html). Subsequently, the expression profiles of splicing factor genes were extracted from TCGA. Cox univariate regression analysis was performed, and genes significantly associated with OS (P<0.001) were retained. An analysis of the 32-gene splicing factors was also incorporated by using the ProgGene (http://genomics.jefferson.edu/proggene/) database (46). The database helps users perform a combined analysis for a list of genes and generate a prognostic signature based on the imputed genes using Cox proportional hazard analysis. The 32 splicing factors were input and the TCGA database was selected for survival analysis. Student t-test analysis was conducted to estimate the differences of splicing factors between tumor and non-tumor samples.
Splicing correlation network construction and hub splicing factor genes
The correlation between the expression of prognosis-related splicing factor genes and the PSI values of the most significant survival-associated alternative splicing events were analyzed by Pearson's tests. The splicing events-factors axes were submitted to correlation network construction for P-values <0.05 and a Pearson's correlation coefficient >0.3. The correlation plots were generated by Cytoscape (version 3.5.1). Hub splicing factor genes were identified according to the numerical degree of each node and edge. The expression level of hub splicing factors between tumor and non-tumor tissues were calculated based on data from TCGA. The data were transformed to the form of log2(count+1) and visually displayed by OriginPro 2017 (OriginLab Corp.). Furthermore, calibration plots were generated to evaluate model calibration by using 'rms' package in R software (https://CRAN.R-project.org/package=rms).
Results
Identification of mRNA splicing event profiles in HCC
The alternative splicing event profiles of 371 HCC patients were obtained from TCGA SpliceSeq. In total, 2,6210 mRNA splicing events in 7,727 genes were collected, which comprised 8,524 ESs in 4,120 genes, 1,306 RIs in 1,879 genes, 4,359 APs in 1,775 genes, 7,736 ATs in 3,379 genes, 1,675 ADs in 1,205genes, 1,936 AAs in 1,451 genes, and 102 MEs in 101 genes. These results also indicated that one gene may have several types of mRNA splicing events, and ES was the predominant type.
Prognosis-related alternative splicing events in HCC
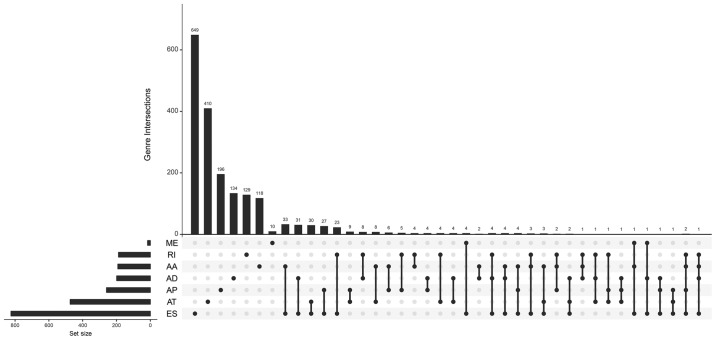
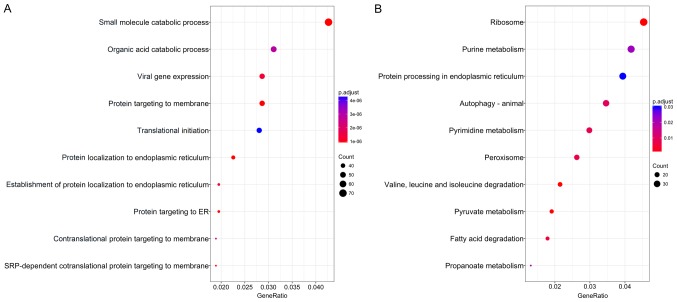
The association of these alternative splicing events with OS was studied by univariate survival analysis, and a total of 3,082 survival-associated alternative splicing events were detected in HCC (P<0.05). We observed that one gene may have two or more events that are significantly associated with survival. Thus, an intersection visualization plot was generated via UpSet (Fig. 1). The top significant survival-associated genes in HCC (P<0.0001) were piped to annotate and visualize biological process terms and KEGG pathways. The biological process terms of these genes were most enriched in small molecule catabolic process, organic acid catabolic process, and viral gene expression (Fig. 2). KEGG pathway analysis indicated that these genes were mainly enriched in ribosome, purine metabolism, protein processing in endoplasmic reticulum, and autophagy.
Figure 1.
UpSet plot of survival-related alternative splicing events. Venn diagram of the overlap between the 7 types of survival-associated alternative splicing events in HCC. HCC, hepatocellular carcinoma; AA, alternate acceptor sites; AD, alternate donor sites; AP, alternate promoters; AT, alternate terminators; ES, exon skipping; ME, mutually exclusive exons; RI, retained introns.
Figure 2.
Pathway analysis of survival-related alternative splicing genes. (A) Biological process analysis of survival-related alternative splicing genes. (B) Significantly enriched annotation from KEGG pathway analysis of survival-related alternative splicing genes. KEGG, Kyoto Encyclopedia of Genes and Genomes; ER, endoplasmic reticulum.
Prognostic indicators for HCC patients
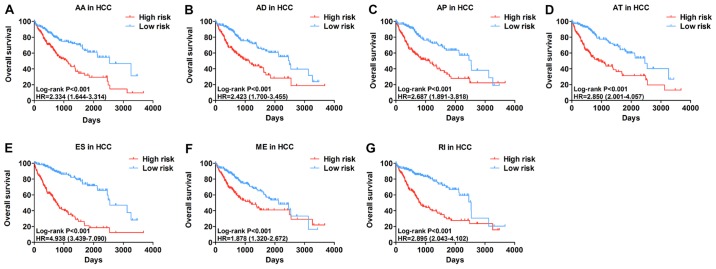
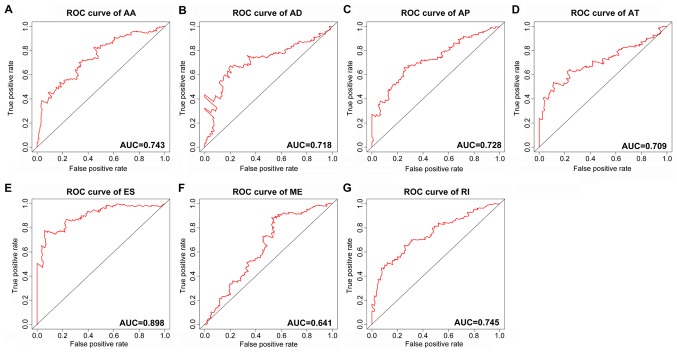
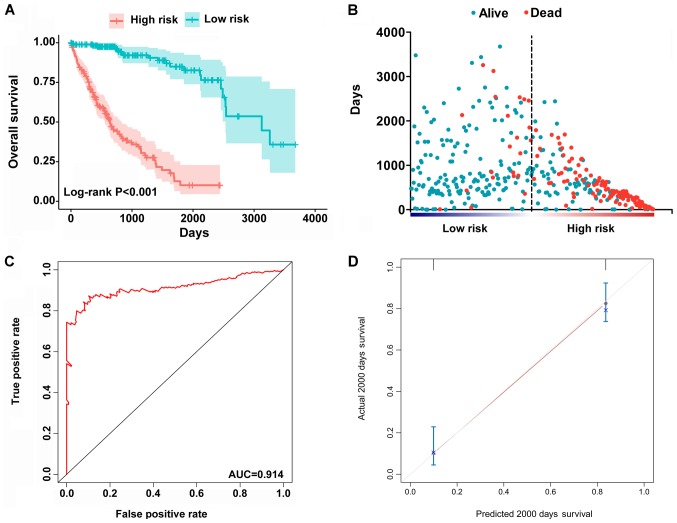
The top significant survival-associated alternative splicing events in the 7 types of alternative splicing were selected as candidate prognostic factors. Multivariate Cox regression with prognostic model construction was then applied to develop a computational model for HCC prognosis based on each individual splicing type. HCC patients were divided into subgroups with significantly different prognoses (Fig. 3) based on the 7 types of alternative splicing (Table I). These independent prognostic alternative splicing events were analyzed further to build the final prognostic computational model using a PI for HCC. Time-dependent ROC curve analyses were also applied to compare the efficiency of these prognostic models (Figs. 4 and 5). The final PI containing the most significant candidate alternative splicing events exhibited the best performance at distinguishing favorable or poor survival in patients (Fig. 6). Patients in the high-risk group had significantly worse survival compared with those in the low-risk group [hazard ratio (HR)=12.904, 95% confidence interval (CI): 7.513-22.161, P<0.001; Fig. 6A and B]. Furthermore, the PI remained an independent prognostic indicator for HCC patients in multivariate analyses after other clinicopathological characteristics were adjusted (HR=16.541, 95% CI: 7.660-35.717, P<0.001; Table II). ROC curves confirmed that the final prognostic predictor with all types of alternative splicing had the highest efficiency (Fig. 6C) with an AUC of 0.914. The specific genes involved in the final models are listed in Table III. The calibration curves exhibited good consistency between the predicted and actual survival probability (Fig. 6D).
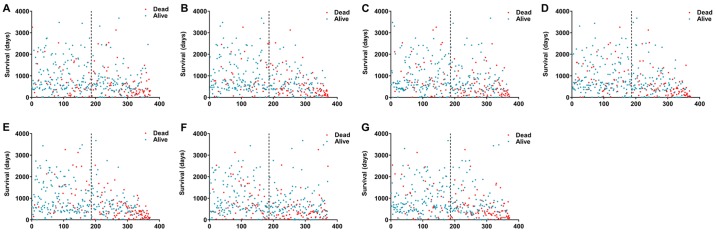
Figure 3.
Kaplan-Meier analyses indicate that HCC patients can be stratified into low- risk and high-risk groups based on the 7 types of alternative splicing. (A) AA; (B) AD; (C) AP; (D) AT; (E) ES; (F) ME; and (G) RI. HCC, hepatocellular carcinoma; AA, alternate acceptor sites; AD, alternate donor sites; AP, alternate promoters; AT, alternate terminators; ES, exon skipping; ME, mutually exclusive exons; RI, retained introns.
Table I.
Prognostic signatures based on each type of splicing event.
| Type of splicing event | Algorithm | HR (95% CI) | ROC curve |
|---|---|---|---|
| Alternate acceptor sites (AA) | 'SWI5-87732-AA' * 0.03740 + 'FDPS-8074-AA' * 0.02940 + 'COMT-61101-AA' * 0.31109 | 2.334 (1.644-3.314) | 0.743 |
| Alternate donor sites (AD) | 'VPS28-85610-AD' * 1.70e-01 + 'UBE2D3-70148-AD' * (−1.57e-02) + 'TXNDC17-38768-AD' * 9.27e-02 + 'TPT1-25803-AD' * 1.62e+01 + 'SUMF2-79789-AD' * 3.92e-02 + 'SMIM19-83740-AD' * 2.69e-02 + 'RPL26-39180-AD' * 6.06e-01 + 'NDUFB11-88904-AD' * 4.87e-02 + 'IRF3-50997-AD' * (−8.62e-03) + 'F10-26354-AD' * 2.21e-02 + 'CCDC90B-18085-AD' * 2.02e-02 + 'C1orf159-13-AD' * (−1.12e-02) + 'ADCK4-49949-AD' (-4.56e-02) | 2.423 (1.700-3.455) | 0.718 |
| Alternate promoters (AP) | 'RPL27-41172-AP' * 0.21696 + 'RCL1-85779-AP' * −0.00550 + 'NUDT6-70523-AP' * 0.01129 + 'MXI1-13080-AP' * 0.00857 + 'DUSP22-75126-AP' * 0.07876 + 'CYB5A-45803-AP' * 0.49517 + 'CHPF-57719-AP' * 0.07805 | 2.687 (1.891-3.818) | 0.728 |
| Alternate terminators (AT) | 'ZSCAN20-1679-AT' * 0.01077 + 'ZNF706-84737-AT' * 0.06539 +'WWOX-37672-AT' * −0.03181 + 'NUCB2-14523-AT' * −0.07215 +'NEIL3-71227-AT' * 0.01306 + 'MORN1-252-AT' * 0.00837 + 'FRMD4A-10810-AT' * 4.59555 + 'CRISPLD2-37867-AT' * 0.24507 + 'ACAT1-18599-AT' * 0.69847 | 2.850 (2.001-4.057) | 0.709 |
| Exon skips (ES) | 'USF2-49098-ES' * 0.03030 + 'TRAPPC6A-50409-ES' −0.03711 + 'TMEM150A-54305-ES' −0.01822 + 'TMEM120B-24894-ES' * 0.00897 + 'TCF3-46542-ES' * −0.02326 + 'SSR4-90499-ES' * 0.95791 + 'SMIM7-48191-ES' * 0.26393 + 'SHBG-39015-ES' * −0.02339 'PTRH1-87649-ES' * −0.64609 + 'PPP6R2-62825-ES' * −0.03581 + 'PPP2CB-83304-ES' * −3.17592 + 'PGAP3-40670-ES' * 0.06043 + 'PCYT2-44234-ES' * 0.12079 + 'OCEL1-48244-ES' * −0.04796 + 'SOC2-52105-ES' * 0.01667 + 'IRF3-50995-ES' * −0.03249 + 'IRF3-50990-ES' * 0.04115 + 'FKBP8-48448-ES' * −0.71033 + 'ECHDC2-3037-ES' * 0.04796 + 'CYTH1-43890-ES' * 0.01764 + 'CHRD-67959-ES' * −0.01020 + 'C16orf13-32924-ES' * 0.03301 + 'C14orf2-29536-ES' * 0.38460 + 'BAX-50838-ES' * −0.35118 + 'ARHGEF10L-862-ES' * 0.01344 + 'AFMID-94694-ES' * −0.01010 + 'ACAA1-64027-ES' * −0.71363 | 4.938 (3.439-7.090) | 0.898 |
| Mutually exclusive exons (ME) | 'SLC39A14-140283-ME' * (−0.00563) + 'H2AFY-96931-ME' * 0.02881 | 1.878 (1.320-2.672) | 0.641 |
| Retained introns (RI) | 'RPL13-38091-RI' * 0.62076 + 'ROMO1-59223-RI` * 0.07039 + 'NUDT22-16590-RI' * 0.01621 + 'NAA60-33527-RI' * (−0.00942) + 'MRPL52-26642-RI' * 0.01287 + 'MDK-15571-RI' * 0.09757 + 'MARVELD2-72372-RI' * (−2.62393) | 2.895 (2.043-4.102) | 0.745 |
HR, hazard ratio; CI, confidence interval; ROC, receiver operating characteristic.
Figure 4.
ROC curves of the PI built using the 7 types of alternative splicing in HCC. (A) AA; (B) AD; (C) AP; (D) AT; (E) ES; (F) ME; and (G) RI. ROC, receiver operating characteristic; AUC, area under the ROC curve; PI, prognostic index; HCC, hepatocellular carcinoma; AA, alternate acceptor sites; AD, alternate donor sites; AP, alternate promoters; AT, alternate terminators; ES, exon skipping; ME, mutually exclusive exons; RI, retained introns.
Figure 5.
The 7 prognostic models with single prognosis-relevant splicing events. (A) Alternate acceptor sites; (B) alternate donor sites; (C) alternate promoters; (D) alternate terminators; (E) exon skipping; (F) mutually exclusive exons; and (G) retained introns.
Figure 6.
Construction of the PI of HCC patients based on all alternative splicing events. (A) Kaplan-Meier plot indicating that patients in the high-risk group had significantly shorter OS compared with those in the low-risk group. (B) OS status and survival duration of HCC patients. (C) Predictive value of the PI for clinical outcome by ROC curve. (D) Calibration curves for predictions of OS. ROC, receiver operating characteristic; AUC, area under the ROC curve; PI, prognostic index; HCC, hepatocellular carcinoma; OS, overall survival.
Table II.
Univariate and multivariate analyses of OS in HCC patients from TCGA by Cox regression analysis.
| Univariate analysis
|
Multivariate analysis
|
|||
|---|---|---|---|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age (≥60/<60 years) | 1.212 (0.854-1.720) | 0.281 | ||
| Sex (male/female) | 0.817 (0.573-1.164) | 0.262 | ||
| Pathological stage (III-IV/I-II) | 2.446 (1.687-3.545) | <0.001 | 1.018 (0.138-7.527) | 0.986 |
| T stage (T3-T4/T1-T2) | 2.537 (1.783-3.609) | <0.001 | 1.757 (0.238-12.981) | 0.581 |
| Lymph node metastasis (yes/no) | 1.999 (0.490-8.161) | 0.334 | ||
| Distant metastasis (yes/no) | 4.033 (1.267-12.834) | 0.018 | 0.755 (0.179-3.177) | 0.701 |
| Histological grade (G3-G4/G1-G2) | 1.119 (0.780-1.604) | 0.542 | ||
| Tumor status during follow-up (tumor/tumor-free) | 2.366 (1.623-3.447) | <0.001 | 1.567 (0.979-2.508) | 0.061 |
| Vascular invasion (micro+macro/none) | 1.351 (0.892-2.047) | 0.155 | ||
| Relative family cancer history (yes/no) | 1.181 (0.818-1.704) | 0.375 | ||
| Prognostic index (high/low) | 12.904 (7.513-22.161) | <0.001 | 16.541 (7.660-35.717) | <0.001 |
OS, overall survival; HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; HR, hazard ratio; CI, confidence interval.
Table III.
Prognostic index for HCC patients based on alternative splicing events.
| Symbol | AS ID | Splice type | Coefficient | z score | P-value |
|---|---|---|---|---|---|
| ZFAND6 | 32171 | AA | −8.51E-01 | −2.83 | 0.0047 |
| SWI5 | 87732 | AA | 8.52E-01 | 2.46 | 0.01392 |
| BIRC2 | 18447 | AA | −1.28E+01 | −4.66 | 3.20E-06 |
| RPS14 | 74096 | AD | −2.55E+00 | −4.38 | 1.20E-05 |
| PGRMC2 | 70577 | AD | −3.42E+01 | −4.58 | 4.70E-06 |
| CCDC90B | 18085 | AD | 8.30E-01 | 2.57 | 0.0102 |
| APOC1 | 50361 | AD | 4.11E+00 | 3.32 | 0.00089 |
| RTN4 | 53584 | AP | −4.26E-01 | −3.29 | 0.001 |
| RPL23A | 39967 | AP | −9.34E+01 | −2.57 | 0.01003 |
| RPL23A | 39965 | AP | −3.06E+00 | −3.44 | 0.00059 |
| RCL1 | 85779 | AP | 4.21E-01 | 2.92 | 0.00347 |
| NUDT6 | 70523 | AP | 2.19E-01 | 2.16 | 0.03054 |
| NUDT6 | 70521 | AP | 6.16E-01 | 3.07 | 0.00216 |
| MXI1 | 13080 | AP | 1.53E-01 | 1.49 | 0.13594 |
| MXI1 | 13079 | AP | 4.76E-01 | 2.67 | 0.00752 |
| IAH1 | 52628 | AP | 1.41E+01 | 4.98 | 6.30E-07 |
| DUSP22 | 75126 | AP | −2.38E-01 | −1.76 | 0.07903 |
| DUSP22 | 75125 | AP | 9.50E-01 | 3.53 | 0.00041 |
| CYB5A | 45803 | AP | 1.29E+00 | 3.29 | 0.00099 |
| CHPF | 57718 | AP | −1.64E+00 | −2.21 | 0.02698 |
| CARKD | 26251 | AP | −2.49E-01 | −1.71 | 0.08786 |
| IAH1 | 52629 | AP | 4.82E-01 | 3 | 0.00273 |
| ZSCAN20 | 1679 | AT | −7.88E-01 | −3.12 | 0.00182 |
| ZNF706 | 84738 | AT | −1.42E+01 | −5.76 | 8.40E-09 |
| WWOX | 37672 | AT | −5.47E+00 | −7.13 | 9.70E-13 |
| TROAP | 21551 | AT | 3.63E-01 | 1.56 | 0.11927 |
| RAPH1 | 57075 | AT | 1.48E+00 | 1.83 | 0.0673 |
| RAPH1 | 57074 | AT | 5.54E-01 | 2.8 | 0.00504 |
| NUCB2 | 14523 | AT | −9.05E+00 | −2.22 | 0.0262 |
| FRMD4A | 10810 | AT | 5.09E+00 | 5.4 | 6.50E-08 |
| CRISPLD2 | 37867 | AT | 8.61E-01 | 3.72 | 0.0002 |
| CLSPN | 1731 | AT | −8.03E-01 | −1.74 | 0.08206 |
| BCAM | 50347 | AT | −3.64E-01 | −1.6 | 0.11049 |
| ACAT1 | 18599 | AT | 2.88E+00 | 4.6 | 4.30E-06 |
| XPC | 63523 | ES | 2.96E-01 | 1.79 | 0.0731 |
| WIPI2 | 78656 | ES | −3.47E-01 | −1.98 | 0.04758 |
| TRAPPC6A | 50409 | ES | −3.27E+00 | −4.69 | 2.70E-06 |
| TPRA1 | 66613 | ES | −4.71E+00 | −1.84 | 0.06575 |
| TMEM150A | 54305 | ES | 1.66E+00 | 2.85 | 0.00443 |
| TMEM141 | 88206 | ES | −3.62E-01 | −3.7 | 0.00022 |
| TCF3 | 46542 | ES | −1.57E+00 | −4.66 | 3.10E-06 |
| STRA13 | 44266 | ES | 2.97E+00 | 4.89 | 1.00E-06 |
| SSR4 | 90499 | ES | 2.39E+02 | 6.3 | 3.00E-10 |
| SLC7A9 | 48907 | ES | 3.61E-01 | 3.17 | 0.0015 |
| SLC25A45 | 16828 | ES | 3.71E-01 | 2.05 | 0.04004 |
| SLAIN2 | 69214 | ES | −6.78E-01 | −2.89 | 0.00385 |
| SHBG | 39015 | ES | −8.49E-01 | −7.07 | 1.50E-12 |
| PTRH1 | 87649 | ES | −6.58E+01 | −5.47 | 4.40E-08 |
| PPP2CB | 83304 | ES | −5.52E+02 | −5.72 | 1.10E-08 |
| PGAP3 | 40670 | ES | 2.10E+01 | 3.32 | 0.00089 |
| OCEL1 | 48244 | ES | −5.74E+00 | −2.97 | 0.00298 |
| MTA1 | 29647 | ES | −3.93E-01 | −2.25 | 0.02464 |
| MPPE1 | 44648 | ES | −3.09E-01 | −2.34 | 0.01921 |
| MPND | 46796 | ES | −6.87E+00 | −2.12 | 0.034 |
| LTBP3 | 16865 | ES | −4.93E-01 | −1.87 | 0.06167 |
| JOSD2 | 51205 | ES | −5.21E+00 | −2.51 | 0.0119 |
| ISOC2 | 52106 | ES | −5.98E+00 | −3.29 | 0.00099 |
| IRF3 | 50991 | ES | −7.04E+01 | −2.94 | 0.00327 |
| IRF3 | 50990 | ES | 5.08E-01 | 2.41 | 0.01599 |
| GUK1 | 10185 | ES | −2.01E+01 | −3.77 | 0.00016 |
| ECHDC2 | 3037 | ES | 6.27E-01 | 3.96 | 7.50E-05 |
| DGKZ | 15546 | ES | −8.54E-01 | −2.5 | 0.01235 |
| CSAD | 21952 | ES | 8.72E-01 | 3.91 | 9.30E-05 |
| CLASRP | 50393 | ES | −1.15E+00 | −2.99 | 0.00281 |
| CHRD | 67959 | ES | −5.44E-01 | −3.86 | 0.00011 |
| C17orf49 | 38825 | ES | 2.01E+01 | 3.52 | 0.00043 |
| C16orf13 | 32921 | ES | 2.25E+00 | 2.39 | 0.01699 |
| C16orf13 | 32919 | ES | −3.45E+00 | −3.98 | 6.80E-05 |
| BAX | 50838 | ES | −4.99E+01 | −3.86 | 0.00011 |
| ARHGEF10L | 862 | ES | 4.31E-01 | 1.83 | 0.06684 |
| APOC1 | 99361 | ES | −2.72E-01 | −3.56 | 0.00037 |
| AFMID | 94694 | ES | 2.79E-01 | 1.98 | 0.04811 |
| AFMID | 94690 | ES | 4.05E-01 | 2.62 | 0.00888 |
| AFMID | 43800 | ES | −1.21E+00 | −4.21 | 2.50E-05 |
| AFMID | 43798 | ES | 7.48E-01 | 3.04 | 0.00236 |
| AFMID | 43795 | ES | −5.04E-01 | −4.01 | 6.00E-05 |
| ACY1 | 65150 | ES | 3.56E+01 | 1.76 | 0.07779 |
| SLC39A14 | 140283 | ME | 3.82E-01 | 2.9 | 0.00372 |
| H2AFY | 96931 | ME | 2.26E+00 | 3.09 | 0.00198 |
| CYP4F3 | 48101 | ME | −3.19E-01 | −2.94 | 0.00327 |
| VPS28 | 85600 | RI | −7.11E-01 | −2.25 | 0.02443 |
| ROMO1 | 59223 | RI | 7.85E-01 | 2.64 | 0.00822 |
| NUDT22 | 16590 | RI | 2.71E+00 | 5.83 | 5.70E-09 |
| MRPL52 | 26642 | RI | 1.11E+00 | 4.03 | 5.60E-05 |
AS, alternative splicing; HCC, hepatocellular carcinoma; AA, alternate acceptor sites; AD, alternate donor sites; AP, alternate promoters; AT, alternate terminators; ES, exon skipping; ME, mutually exclusive exons; RI, retained introns.
Survival-associated splicing factor genes
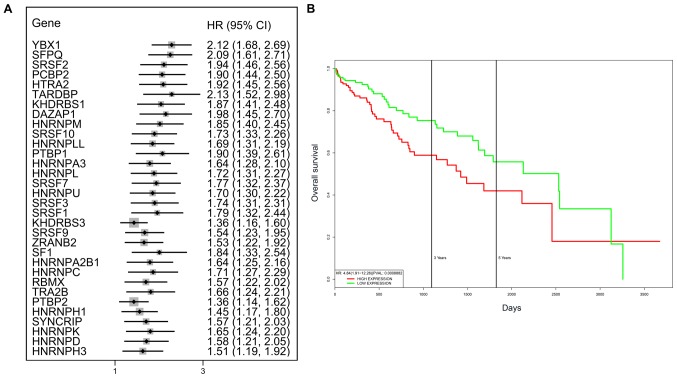
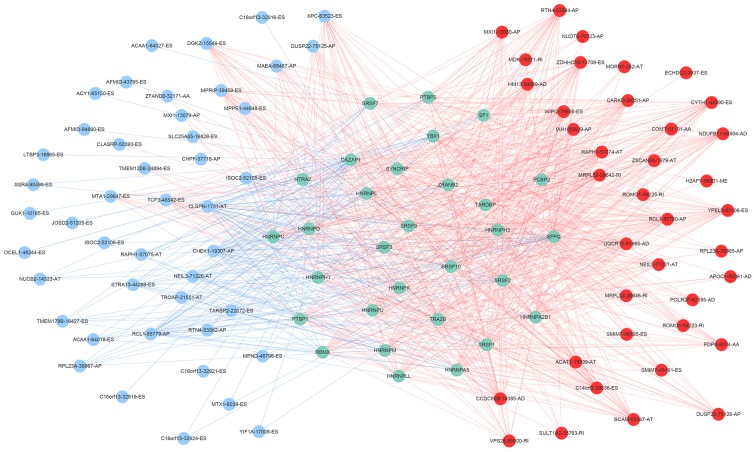
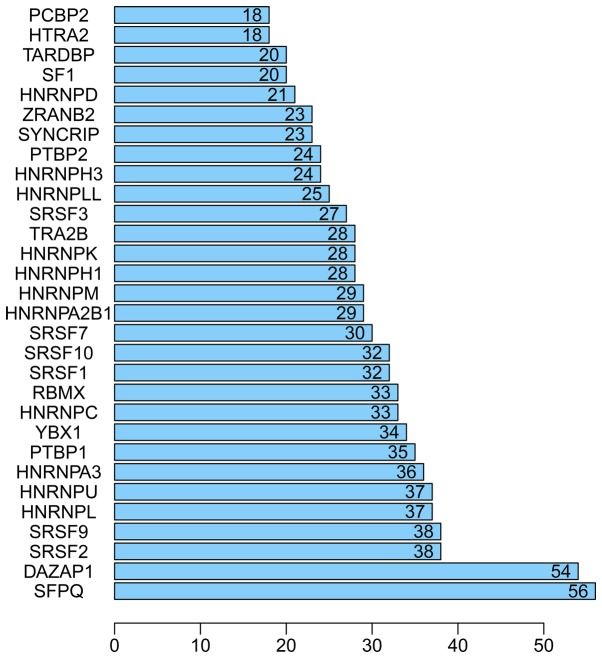
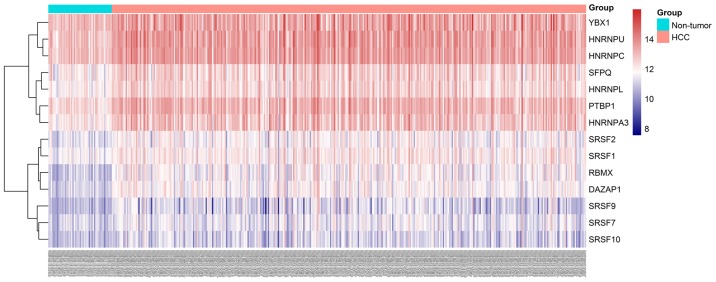
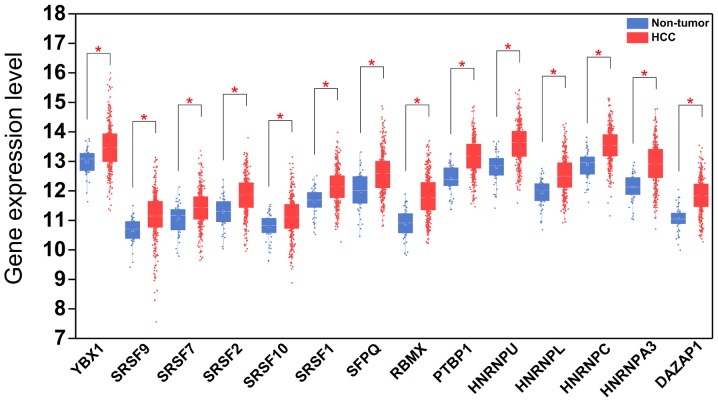
A total of 71 splicing factor genes were identified, and the associations between the expression profiles of these genes and OS were assessed based on the data obtained from TCGA, resulting in the identification of 32 prognosis-related splicing factor genes. Interestingly, overexpression of all these 32 genes may represent a risk factor for the survival of patients with HCC, as all HRs were >1 (P<0.05) (Fig. 7A). We found that the integration of the 32 SFs could also provide an effective survival risk stratification for HCC patients (Fig. 7B). HCC patients with high-risk scores for the splicing factors-based classifier had worse OS compared with those who had low-risk scores. Next, we characterized the splicing-regulatory network of the 152 (P<0.0001) most significant survival-associated alternative splicing events and 32 prognosis-related splicing factor genes in HCC (Fig. 8). Additionally, 14 core interacting genes were identified in the splicing-regulated network: SRSF7, SRSF10, SRSF1, RBMX, HNRNPC, YBX1, PTBP1, HNRNPA3, HNRNPU, HNRNPL, SRSF9, SRSF2, DAZAP1 and SFPQ (Fig. 9). Furthermore, these genes were found to not only possess prognostic significance, but to also have indispensable functions in the initiation of HCC (Figs. 10 and 11).
Figure 7.
Prognostic value of survival-associated splicing factor genes. (A) Forest plots of the HRs of survival-associated splicing factor genes in HCC. (B) The integration of the 32 splicing factor genes may achieve effective survival prediction. HR, hazard ratio; CI, confidence interval; HCC, hepatocellular carcinoma.
Figure 8.
Survival-associated splicing factors and splicing correlation network in hepatocellular carcinoma. The red circles indicate risk alternative splicing events and the blue circles indicate protective alternative splicing events. The blue circles represent splicing factor genes. The red and blue lines represent positive and negative correlations, respectively.
Figure 9.
Bar chart of the top 30 splicing factor genes in hepatocellular carcinoma.
Figure 10.
Heatmap of hub splicing factor gene expression levels in HCC and adjacent non-tumor tissues. HCC, hepatocellular carcinoma.
Figure 11.
Expression patterns of 14 hub splicing factor genes in HCC and paired non-tumor samples. Each red dot represents a distinct tumor sample and each blue dot represents a non-tumor sample. HCC, hepatocellular carcinoma. *P<0.05.
Discussion
In the present study, the PSI value and clinical outcome of patients with HCC were integrated using a novel methodology. Furthermore, prognostic signatures were developed based on splicing events to promote the development of precision treatment. To determine the underlying mechanism, we investigated the potential regulatory associations between prognosis-related splicing factors and splicing events, which provided deeper insights into the phenotype of splicing events in HCC.
With the advantage of high-throughput RNA-Seq, TCGA provides multiple sources for the investigation of whole-genome or transcriptome analyses, including the exploration of genome splicing events. Previously, Xue et al (47) and Zhu et al (48) conducted SpliceSeq analyses using TCGA (49) to generate alterative splicing profiles for non-small-cell lung cancer and ovarian cancer patients, in order to develop prognostic models with several splicing events. SpliceSeq is a Java program providing a clear view of the inclusion level of each exon and splice junction. Recently, Ryan et al (45) extended the methodology of SpliceSeq and calculated the PSI for each potential splicing event across 33 types of cancer to establish TCGA SpliceSeq database; the PSI data for HCC generated in the present study were obtained from this database. Hence, to the best of our knowledge, this is the first group to integrate PSI values from SpliceSeq with TCGA clinical prognostic parameters for the comprehensive investigation of the prognostic value of alternative splicing events in HCC. This algorithm will facilitate the discovery of novel splicing events that may be used for the prognosis of HCC and provide insights into the role of splicing events at the genome-wide level.
On the basis of the analysis of the tumor tissue data of 371 HCC cases in TCGA SpliceSeq, a large number of alternative splicing events were identified. These events do not only affect protein diversity by altering the amino acid sequences of the translated products (e.g., ES can delete a portion of the coding sequence), but they can also affect protein function by altering the yield of the translated products (e.g., AP can change the efficiency of translation). For example, the functions of the common clinical tumor suppressor genes TP53, ARID1A, PTEN and PI3K, and the proto-oncogenes MET and NOTCH1, can be changed by alternative splicing events (21). The common apoptosis-related genes Bcl-X and MLC1 can also express different products due to the occurrence of alternative splicing, showing two distinct functions of promoting apoptosis and inhibiting apoptosis (3,4,50,51). In the currently popular molecular targeting therapy for tumors, the target sequence can be lost from cancer cells due to alternative splicing, leading to drug resistance (7,13,21). In the present study, ES was the most common of the 7 alternative splicing events in HCC. Previous studies have demonstrated that the frequency of alternative splicing events is not the same in different cancers, but the frequency of ES is generally the highest (52). Although the mechanism has not yet been fully elucidated, evidence has shown that the occurrence of splicing events is not random. Chen et al (53) analyzed RNA-Seq data from 30 types of tumors and found that the frequency of alternative splicing events is markedly higher in tumors compared with that in corresponding normal tissues. These alternative splicing events are often accompanied by premature termination codons, and the probability of a premature termination codon due to alternative splicing is markedly higher in a tumor suppressor gene rather than in a proto-oncogene in cancer tissue. In the gene functional enrichment analysis, it was observed that genes with survival-associated splicing events were significant correlated with 'small molecule catabolic processes', 'organic acid processes' and 'viral gene expression'. The gene functional enrichment analysis results revealed that these genes may be involved in the metabolism of HCC. The liver is the metabolic center of the body. Hence, the onset and progression of HCC is also inseparable from metabolic abnormalities. Previously, Tremblay et al reported unique alternative splicing patterns in HBV-associated HCC, HCV-associated HCC, HBV&HCV-associated HCC, and virus-free HCC based on TCGA dataset. They also found the signatures of genes for which AS is dysregulated in different types of HCC (44). These findings also validated that the process of splicing is closely associated with viral infection status.
Accordingly, alternative splicing events may be driving the occurrence and progression of cancer. Therefore, the Cox regression method was used to analyze the association between alternative splicing events and the survival of HCC patients. The predictive power of alternative splicing on the prognosis of HCC was analyzed by considering the ROC curve. The results demonstrated that several alternative splicing events were associated with the survival of HCC patients. Thus, we attempted for the first time to use multiple alternative splicing events for the prognosis of HCC patients and obtained satisfactory results. To date, a number of studies have demonstrated that an alternative splicing event alone may be used as a biomarker for tumor diagnosis and prognosis. However, due to the heterogeneity of tumors, using a single molecule as a marker is often insufficient. Therefore, the combination of multiple molecular events for the prognosis of HCC may improve the sensitivity of diagnosis and prognosis. This follows a trend in current clinical research. Similarly, previous studies have reported several prognostic signatures based on different types of molecular events. For example, Qiao et al proposed an eight-gene signature for HCC patients' survival prediction (54). However, the AUC of their model only reached 0.77. Liao et al also developed an effective prognostic index (PI) for predicting the survival of HCC patients based on microRNAs (55). However, the AUC also only reached 0.687. Hence, our PI exhibits better discriminatory power. In the present study, the discriminatory power and model calibration were estimated to comprehensively evaluate the model accuracy. More importantly, the prognostic signature based on splicing events may provide novel insight into the monitoring of HCC patients from alternative splicing events. The splicing events included in the PI may provide promising therapeutic targets for HCC patients.
As mentioned above, alternative splicing events do not occur randomly, but they are rather regulated by multiple cellular factors, including splicing factors (56,57). Splicing factors can activate or inhibit the occurrence of splicing events by identifying regulatory sequences in exons or introns. DNA mutations, DNA methylation and aberrant histone modifications may all affect the recognition of splicing sites by splicing factors, leading to changes in the occurrence of splicing events (10,58-67). The roles of common anomalies, such as DNA mutations, in cancer can be rationally explained by selectivity. Mutations within introns and synonymous mutations in exons, which were previously believed not to affect the function of genes, have gradually been found to affect gene function by affecting splicing events (10,58,60). Splicing factors play a crucial regulatory role in the occurrence of splicing events, and mutations in their sequences or changes in their expression levels may affect splicing events (21). Changes in the expression levels of splicing factors are usually accompanied by changes in splicing events, but the pattern is not yet clear. Both the upregulation and downregulation of their expression can cause cancer or inhibit tumor growth by altering splicing events (25). Analysis of the association between splicing factors and survival in patients with HCC revealed that the expression levels of 32 splicing factors were correlated with survival time, with high expression being associated with shorter survival. It was hypothesized that the expression levels of these factors may be regulated by the similar events. For example, such an effect of the MYC oncogene has been reported (22). To elucidate the mechanism underlying the effect of splicing factors on the survival of patients with HCC, we analyzed the correlations between the expression levels of these 32 splicing factors and survival-related alternative splicing events. Surprisingly, the alternative splicing events that were positively correlated with survival were also positively correlated with the expression levels of multiple splicing factors, and most alternative splicing events that were negatively correlated with survival were also negatively correlated with the expression levels of the splicing factors. This finding indicates that the increase in the expression of multiple splicing factors can affect the survival of patients with HCC by synergistically regulating the alternative splicing events of genes.
However, there were several limitations to the present study. First, this study was based on an individual source of data from TCGA, without validation from independent cohorts, which will be carried out by our group in the future with clinical samples. Furthermore, the biological roles of the splicing events require further validation. In addition, the present study mainly focused on the splicing status of mRNAs. In fact, the splicing events of other types of RNA, including circular RNAs, also play important roles in the processes of tumors (68). Further research and effective arithmetic are required to broaden its application range (69). Finally, classic splicing factors were utilized in the present study, whereas it is possible that other RNA-binding proteins may also affect splicing events. Hence, the association between splicing events and a complete repertoire of RNA-binding proteins from established sources in the human genome should be further investigated.
Our study preliminarily demonstrated that alternative splicing events and the expression levels of splicing factors play important roles in the progression of HCC. On the basis of these results, it may be considered that splicing factors affect the occurrence and progression of HCC by regulating alternative splicing events. Some special alternative splicing events may affect the prognosis and progression of HCC by being regulated by their corresponding splicing factors. The constructed computational prognostic model based on HCC-specific alternative splicing events may be used as a molecular marker for the prognosis of HCC.
Currently, certain anticancer drugs targeting splicing factors, such as E7107 and FR901464 (29,70), have been developed, but they are not widely used in clinical practice due to their prominent side effects (21). The identified HCC-specific alternative splicing events may be used as molecular markers for the diagnosis and prognosis of HCC, and they can help physicians develop more precise targeted therapies. This approach has the potential to be widely applied in the field of HCC research.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 31760319), the Natural Science Foundation of Guangxi, China, the Guangxi Degree and Postgraduate Education Reform and Development Research Projects, China (grant no. JGY2019xxx), the Promoting Project of Basic Capacity for Young and Middle-aged University Teachers in Guangxi (grant no. 2017KY0111), the Innovation Project of Guangxi Graduate Education (grant no. YCBZ2017045), the Guangxi Medical University Training Program for Distinguished Young Scholars, the Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, and the Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-financed Scientific Research Project (grant no. Z20180979).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HW, SP and GC participated in the conception and design of the study. ZP, RH, BL, JM, XH and YD performed the statistical analysis and were involved in the preparation of the figures and tables. HW, ZP and RH reviewed the results and participated in the discussion of the data. HW, ZP, SP and GC prepared and revised the manuscript. All the authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Breitbart RE, Andreadis A, Nadal-Ginard B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 4.Bingle CD, Craig RW, Swales BM, Singleton V, Zhou P, Whyte MK. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- 5.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Zheng S, Kim H, Verhaak RGW. Silent mutations make some noise. Cell. 2014;156:1129–1131. doi: 10.1016/j.cell.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Salton M, Kasprzak WK, Voss T, Shapiro BA, Poulikakos PI, Misteli T. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat Commun. 2015;6:7103. doi: 10.1038/ncomms8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47:1242–1248. doi: 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, et al. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016;16:2605–2617. doi: 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Kornblihtt AR. Epigenetics at the base of alternative splicing changes that promote colorectal cancer. J Clin Invest. 2017;127:3281–3283. doi: 10.1172/JCI96497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015;5:1282–1295. doi: 10.1158/2159-8290.CD-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang BD, Ceniccola K, Hwang S, Andrawis R, Horvath A, Freedman JA, Olender J, Knapp S, Ching T, Garmire L, et al. Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat Commun. 2017;8:15921. doi: 10.1038/ncomms15921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebestyén E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43:1345–1356. doi: 10.1093/nar/gku1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trincado JL, Sebestyén E, Pagés A, Eyras E. The prognostic potential of alternative transcript isoforms across human tumors. Genome Med. 2016;8:85. doi: 10.1186/s13073-016-0339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen S, Wang Y, Wang C, Wu YN, Xing Y. SURVIV for survival analysis of mRNA isoform variation. Nat Commun. 2016;7:11548. doi: 10.1038/ncomms11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghigna C, De Toledo M, Bonomi S, Valacca C, Gallo S, Apicella M, Eperon I, Tazi J, Biamonti G. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: Therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol. 2010;7:495–503. doi: 10.4161/rna.7.4.12744. [DOI] [PubMed] [Google Scholar]
- 21.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabò A, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 23.Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salton M, Misteli T. Small molecule modulators of Pre-mRNA splicing in cancer therapy. Trends Mol Med. 2016;22:28–37. doi: 10.1016/j.molmed.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 27.Webb TR, Joyner AS, Potter PM. The development and application of small molecule modulators of SF3b as therapeutic agents for cancer. Drug Discov Today. 2013;18:43–49. doi: 10.1016/j.drudis.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 29.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 30.Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xargay-Torrent S, López-Guerra M, Rosich L, Montraveta A, Roldán J, Rodríguez V, Villamor N, Aymerich M, Lagisetti C, Webb TR, et al. The splicing modulator sudemycin induces a specific antitumor response and cooperates with ibrutinib in chronic lymphocytic leukemia. Oncotarget. 2015;6:22734–22749. doi: 10.18632/oncotarget.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, Wu Y, Han F, Zhang L, Yu T, et al. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut. 2019;68:118–129. doi: 10.1136/gutjnl-2017-314983. [DOI] [PubMed] [Google Scholar]
- 33.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashemikhabir S, Budak G, Janga SC. ExSurv: A web resource for prognostic analyses of exons across human cancers using clinical transcriptomes. Cancer Inform. 2016;15(Suppl 2):17–24. doi: 10.4137/CIN.S39367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neelamraju Y, Gonzalez-Perez A, Bhat-Nakshatri P, Nakshatri H, Janga SC. Mutational landscape of RNA-binding proteins in human cancers. RNA Biol. 2018;15:115–129. doi: 10.1080/15476286.2017.1391436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ, Zhou H, Yang JH, Qu LH. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018;22:286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 37.Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014;15:R14. doi: 10.1186/gb-2014-15-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo C, Cheng Y, Liu Y, Chen L, Liu L, Wei N, Xie Z, Wu W, Feng Y. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 2017;77:1168–1178. doi: 10.1158/0008-5472.CAN-16-1919. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Qian X, Peng LX, Jiang Y, Hawke DH, Zheng Y, Xia Y, Lee JH, Cote G, Wang H, et al. Corrigendum: A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18:709. doi: 10.1038/ncb3361. [DOI] [PubMed] [Google Scholar]
- 40.Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW, Chan LK, Lee JM, Man K, Cherny S, Yang WL, et al. Novel pre-mRNA splicing of intronically integrated HBV generates oncogenic chimera in hepatocellular carcinoma. J Hepatol. 2016;64:1256–1264. doi: 10.1016/j.jhep.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Luo ZL, Cheng SQ, Shi J, Zhang HL, Zhang CZ, Chen HY, Qiu BJ, Tang L, Hu CL, Wang HY, et al. A splicing variant of Merlin promotes metastasis in hepatocellular carcinoma. Nat Commun. 2015;6:8457. doi: 10.1038/ncomms9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayliss J, Lim L, Thompson AJ, Desmond P, Angus P, Locarnini S, Revill PA. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol. 2013;59:1022–1028. doi: 10.1016/j.jhep.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 44.Tremblay MP, Armero VE, Allaire A, Boudreault S, Martenon-Brodeur C, Durand M, Lapointe E, Thibault P, Tremblay-Létourneau M, Perreault JP, et al. Global profiling of alternative RNA splicing events provides insights into molecular differences between various types of hepatocellular carcinoma. BMC Genomics. 2016;17:683. doi: 10.1186/s12864-016-3029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018–D1022. doi: 10.1093/nar/gkv1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goswami CP, Nakshatri H. PROGgeneV2: Enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue L, Xie L, Song X, Song X. Identification of potential tumor-educated platelets RNA biomarkers in non-small-cell lung cancer by integrated bioinformatical analysis. J Clin Lab Anal. 2018;32:e22450. doi: 10.1002/jcla.22450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J, Chen Z, Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol. 2018;148:368–374. doi: 10.1016/j.ygyno.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 49.Ryan MC, Cleland J, Kim R, Wong WC, Weinstein JN. SpliceSeq: A resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–2387. doi: 10.1093/bioinformatics/bts452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 51.Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, He J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51. doi: 10.1016/j.canlet.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Tovar-Corona JM, Urrutia AO. Increased levels of noisy splicing in cancers, but not for oncogene-derived transcripts. Hum Mol Genet. 2011;20:4422–4429. doi: 10.1093/hmg/ddr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao GJ, Chen L, Wu JC, Li ZR. Identification of an eight-gene signature for survival prediction for patients with hepatocellular carcinoma based on integrated bioinformatics analysis. PeerJ. 2019;7:e6548. doi: 10.7717/peerj.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao X, Zhu G, Huang R, Yang C, Wang X, Huang K, Yu T, Han C, Su H, Peng T. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi: 10.2147/CMAR.S161334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: Splicing by the minor spliceosome. Wiley Interdiscip Rev RNA. 2013;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 59.Sterne-Weiler T, Sanford JR. Exon identity crisis: Disease-causing mutations that disrupt the splicing code. Genome Biol. 2014;15:201. doi: 10.1186/gb4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P, Linsenmeier M, et al. The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med. 2016;8:442–457. doi: 10.15252/emmm.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh S, Narayanan SP, Biswas K, Gupta A, Ahuja N, Yadav S, Panday RK, Samaiya A, Sharan SK, Shukla S. Intragenic DNA methylation and BORIS-mediated cancer-specific splicing contribute to the Warburg effect. Proc Natl Acad Sci USA. 2017;114:11440–11445. doi: 10.1073/pnas.1708447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23:789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest. 2017;127:3375–3391. doi: 10.1172/JCI94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding X, Liu S, Tian M, Zhang W, Zhu T, Li D, Wu J, Deng H, Jia Y, Xie W, et al. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nat Neurosci. 2017;20:690–699. doi: 10.1038/nn.4536. [DOI] [PubMed] [Google Scholar]
- 66.Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA. 2014;111:E4920–E4928. doi: 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang X, Chen W, Han Y, Shao L, Gao Y, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18:35. doi: 10.1186/s12943-019-0996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.