Abstract
Whether conventional dendritic cells (cDC) acquire subset identity under direction of Wnt family glycoproteins is unknown. We demonstrate that Wnt4, a beta-catenin independent Wnt ligand, is produced by both hematopoietic and non-hematopoietic cells and is both necessary and sufficient for pre-cDC1/cDC1 maintenance. Whereas BM cDC precursors undergo phosphoJNK/c-Jun activation upon Wnt4 treatment, loss of cDC Wnt4 in CD11cCreWnt4flox/flox mice impaired differentiation of CD24+, Clec9A+, CD103+ cDC1 compared to CD11cCre controls. Conversely, sc-RNAseq analysis of BM revealed a 2-fold increase in cDC2 gene signature genes and flow cytometry demonstrated increased numbers of SIRP-α+ cDC2 amid lack of Wnt4. Increased cDC2 numbers due to CD11c-restricted Wnt4 deficiency increased interleukin 5 production, group 2 innate lymphoid cell (ILC2) expansion, and host resistance to the hookworm parasite Nippostrongylus brasiliensis. Collectively, these data uncover a novel and unexpected role for Wnt4 in cDC subset differentiation and Type 2 immunity.
Introduction
Tissue-resident cDC are a heterogeneous population that can be broadly classified into cDC1 and cDC2 subsets that arise from a common DC progenitor (CDP) in the bone marrow (1). FLT3-ligand drives conventional DC1 (cDC1) development under control of interferon regulatory factor 8 (IRF8) autoactivation stabilized by Batf3-Jun heterodimers to form a trimolecular complex (2–5). CD24, Clec9A, and CD103 together mark the cDC1 subset that can cross-present antigen and produce IL-12p70 for Th1 and CD8+ T cell driven immune responses. On the other hand, CD172a+ cDC2 require IRF-4 for development and this subset promotes Th2 and Th17 responses in a variety of contexts (1). Although the transcriptional machinery that regulates cDC-lineage commitment has received much attention (1, 6), it is less well understood whether secreted proteins direct cDC1 vs. cDC2 specification and/or expansion during development from pre-cDC precursors.
The mammalian Wingless/Integrated (Wnt) family of secreted glycoproteins are regenerative factors that serve important roles in development, cancer, and tissue repair through regulation of cell proliferation and cell fate specification (7). Wnt signaling drives transcription through either a canonical β-catenin-dependent or non-canonical calcium dependent planar cell polarity (PCP) pathway, the latter of which involves c-Jun N-terminal kinase (JNK) (8). JNK functions to activate the AP-1 family transcription factors such as c-Jun (9, 10). There has been some investigation of Wnt proteins in DC biology, but the data are somewhat controversial. While there is evidence that CD11c (Itgax) promoter-driven deletion of β-catenin deletion increases Th1/Th17 responses and increases disease severity in the context of colitis (11), others have reported that constitutive activation of β-catenin in CD11c+ cells also enhances Th1 responses to regulate the outcome of infection with Toxoplasma gondii (12). Exposure to Wnts alters DC cytokine production (13, 14) and tumor cells secrete Wnt5a to promote tolerogenic DC activity (15). However, non-canonical Wnt proteins such as Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7, and Wnt11 have received little attention in DC biology. Curiously, Wnt4 overexpression in hematopoietic stem cells (HSC) increased the frequency of FLT3+ cells (16) and increased progenitor frequency through Rac1 and JNK-dependent mechanisms (17).
Herein, we use a loss of function approach to explore the hypothesis that Wnt4 regulated distinct aspects of cDC development, tissue homeostasis, and infection. Our data show that CD11c-driven Wnt4 deficiency disrupts the pre-cDC 1 vs. pre-cDC2 ratio in BM and reduces the numbers of intestinal cDC1 under steady state conditions. We attribute the imbalance of cDC1/cDC2 ratio to impairment of the JNK/c-Jun activation pathway that promotes cDC1 development from nascent progenitors. Indeed, Wnt4 deficiency basally increases cDC2, but decreases in cDC1 within the BM and small intestine. This perturbation accompanies rapid development of Type 2 immunity in response to the hookworm parasite Nippostrongylus brasiliensis. Accelerated Type 2 immunity marked by enhanced ILC2, TH2, and increased IL-5 production together demonstrates a previously unrecognized and biologically important role for Wnt4 in regulating cDC development.
Methods
Mice
CD11cCre(B6.Cg-Tg(Itgax-cre)1–1Reiz/J) and Wnt4flox/flox (B6;129S-Wnt4tm1.1Bhr/BhrEiJ) mice were purchased from JAX, bred to homozygosity and compared to age-/gender- matched CD11cCre and Wnt4flox/flox mice (controls) housed under SPF conditions at San Francisco General Hospital or University of Pennsylvania. All procedures were IACUC-approved (UCSF #AN109782–01) and (UPenn #805911).
Single-cell RNA Sequencing
Bone marrow cells were enriched with CD11c MicroBeads (Miltenyi Biotec, Germany). Single-cell RNA sequencing was performed using 10× Chrome platform with 10× Single Cell 3’ v1 chemistry (10× Genomics, Pleasanton, CA). The feature-barcode matrices from Cell Ranger (Pipeline) were read into R (v3.4.4) and analyzed with Seurat 3.0 (18). Seurat objects were created keeping only cells with at least 200 features and requiring features be found in at least 3 cells. Control and CD11cCre-Wnt4flox/flox Seurat objects were merged and the features were log-normalized with a scale factor of 10,000. Variable features were identified using a mean variability plot as the selection method with default parameters for the mean cutoff and the dispersion cutoff. The data was scaled and PCA was performed with default options. Neighbors were determined using 15 dimensions and 15 clusters were identified using a resolution of 0.55. The number of cells for Control and CD11cCre-Wnt4flox/flox were nearly identical (3,265 and 3,390, respectively) and so the fold-change between strains were calculated by dividing the number of cells in each cluster for the CD11cCre-Wnt4flox/flox by the numbers of cells in that cluster from the control. A t-SNE calculation was performed using 15 dimensions for the input features and Control and CD11cCre-Wnt4flox/flox samples were plotted separately.
Using the fold-change calculations for each cluster, the t-SNE results were replotted with a custom color palette denoting the degree of fold-change with warm colors designating high representation of cells in the CD11cCre-Wnt4flox/flox and cool colors designating low representation of cells in CD11cCre-Wnt4flox/flox. Expression of Itgax in each cell was plotted using “FeaturePlot”. Differentially expressed genes for each cluster was calculated using “FindAllMarkers” and the top genes for each cluster were submitted to BioGPS to examine their expression in a multitude of different cell-types using the Mouse MOE430 Gene Atlas dataset. The cell-type most closely representing each cluster’s pattern of gene expression was used to determine cluster identity. The same methods were used to process the control data without merging to the CD11cCre-Wnt4flox/flox data. After identifying cell-type identities for each cluster in the control, the cell-types in the control were used as a reference to classify cell-types in CD11cCre-Wnt4flox/flox. This was accomplished by using the commands “FindTransferAnchors”, “TransferData”, and “AddMetaData” in Seurat 3.0. Separate t-SNE calculations and plots were then made for each sample.
Flow cytometry and PrimeFlow
Flow cytometry were performed on LSRII or LSRFortessa (BD Biosciences, San Jose, CA) and analyzed in FlowJo 9.9.6 (FlowJo LLC, Ashland, OR). PrimeFlow™ kit was purchased from Affymetrics (Santa Clara, CA); mouse Actb, Wnt4 and Wnt16 ViewRNA® probes were generated by Affymetrics. Antibodies: CD45 (30-F11), CD115 (AFS98), CD117 (2B8), CD135 (A2F10), CD11c (N418), MHCII (M5/114.15.2), Siglec-H (551), CD172a (P84), Ly6C (HK1.4), CD24 (M1/69), CD64 (X54–5/7.1), F4/80 (BM8), B220 (RA3–6B2), CD3 (145–2C11), Ter-119 (TER-119), Ly6G (1A8), CD19 (1D3), NK1.1 (PK136), XCR1 (ZET), CD103 (2E7), CD8a (53–6.7), CD11b (M1/70), Thy1.2 (30-H12), ICOS (15F9), CD127 (A7R34), ST2 (D1H9), GATA3 (16E10A23), CD4 (GK1.5). Gr-1 (RB6–8C5), CD5 (53–7.3), CD26 (H194–112), Clec9A (42D2). Phospho-cJun (Ser73, clone D47G9) was from Cell Signaling Technology (Danvers, MA). The live/dead fixable Aqua dye was from Invitrogen (Carlsbad, CA).
Microscopy
Small intestines “Swiss rolls” were prepared, cut into 7 μm thick sections and fixed in cold 4% PFA. The sections were then incubated and stained with the following primary Antibodies: Hamster Anti-mouse CD11c (N418, BioLegend), Mouse Anti-mouse CD45.2 FITC (104, BioLegend)-FITC conjugated, Rat Anti-mouse Wnt4 (MAB4751, R&D Systems), followed by incubation with the Alexa Fluor and Cyanine dye series of secondary antibodies (Alexa Fluor® 488 AffiniPure F(ab’)₂ Fragment Donkey Anti-Mouse IgG (H+L), Allophycocyanin (APC) AffiniPure F(ab’)₂ Fragment Donkey Anti-Rat IgG (H+L), Cy™3 AffiniPure Goat Anti-Armenian Hamster IgG (H+L) supplied by Jackson ImmunoResearch. Nuclei were stained with DAPI and pseudo-colored. Detection of fluorescence was observed under Cy2, Cy3, and Cy5 filters on a Leica Inverted Microscope DMi8 S Platform and a Leica DM6000B microscope with an automated stage coupled with a Leica DFC350FX camera. Exposure time and fluorescence intensities were normalized to appropriate isotype control images. Each fluorescent channel was photographed separately, then merged and overlaid them atop the corresponding images.
Cell isolation and culture
The protocol to culture iCD103 BMDC was modified from (19). In brief, lineage positive cells were depleted from total BM cells using lineage depletion kit (Miltenyi Biotec). Enriched progenitors were plated at 0.5– 1 ×105/ml in RPMI CM containing 100 ng/ml Flt3L +10 ng/ml GM-CSF (both from Peprotech, Rocky Hill, NJ) for 12 days. Non-adherent cells were then collected for experiments. To grow Flt3L-induced BMDC, 3– 5×105 cells were plated in IMDM +10% FBS+ 1% Pen/Strep containing 100 ng/ml Flt3L for 7 days. Treatment groups also received 100 ng/ml recombinant mWnt4 or 100 ng/ml R-Spondin1 (R&D Systems, Minneapolis, MN) or both.
Splenic DC was isolated as previously described (20). Lungs were digested in DMEM+ 0.15 mg/ml Liberase TL+ 14 ug/ml DNase I+ 0.4 mg/ml Dispase for 1h at 37C on a shaker. For small intestine cells, approximately 10 cm of jejunum was collected and opened longitudinally then washed in HBSS+ 5 mM EDTA+ 2.5 mM DTT for 3×10 min at 37C on a shaker followed by digestion in DMEM+ 2% FBS+ 0.05 mg/ml Liberase TM+ 20 μg/ml DNase I for 50 min. Mesenteric lymph node cells were re-stimulated with plate-bound α-CD3 and α-CD28 (both from Biolegend) for 72h.
Phospho-JNK ELISA
BM cells enriched with Histopaque-1083 gradient were rested in IMDM without FBS for 2h before treating with vehicle (PBS), R-Spondin1 (100 ng/ml), Wnt4 (100 ng/ml), or the combination of both. Cell lysates were collected at various timepoints following the manufacturer’s protocol. Phospho-JNK ELISA kit was used (R&D Systems).
T cell differentiation and cytokine ELISA
BM progenitors were cultured for 7d in RPMI complete media with 20 ng/ml mGM-CSF (PeproTech). Spleen and lymph node cells from OT-II mice were enriched with a naïve CD4 T cell isolation kit (Miltenyi Biotec). DC were pulsed with OVA (50 ug/well) for 8h, washed, then co-cultured with T cells (DC: T= 1:10) for 3d. Cells were then re-stimulated with α-CD3 for 72h. Mouse IL-5 ELISA kit was from eBioscience.
Real-time qPCR
0.5 cm of duodenum or 2 × 106 BM cells was used for RNA extraction using NucleoSpin RNA Plus kit (Macherey-Negel, Dueren, Germany), and cDNA was generated with Maxima H Minus RTase (Thermo Fisher). Primers: Gapdh FW 5’-AGGTCGGTGTGAACGGATTTG-3’, RV 5’-TGTAGACCATGTAGTTGAGGT- 3’; Il5 FW 5’-CTCTGTTGACAAGCAATGAGA- 3’, RV 5’- TCTTCAGTATGTCTAGCCCCT- 3’; Ccl2 FW 5”-TAAAAACCTGGATCGGAACCAAA- 3”, RV 5”-GCATTAGCTTCAGATTTACGG GT - 3’.
Nippostrongylus brasiliensis (N.b.) infection
Parasites were cultured and passaged as previously described (21), washed in PBS+ 1% Pen/Strep and 650–700 L3 larvae per mouse were injected s.q. Fecal pellets were collected to determine egg counts. To assess worm counts, small intestines were opened longitudinally and incubated in PBS for 2h at 37C.
Statistics
Statistical analyses were performed using Prism7 (GraphPad, La Jolla, CA).
Results
Expression of Wnt4 in BM progenitors and tissue DC
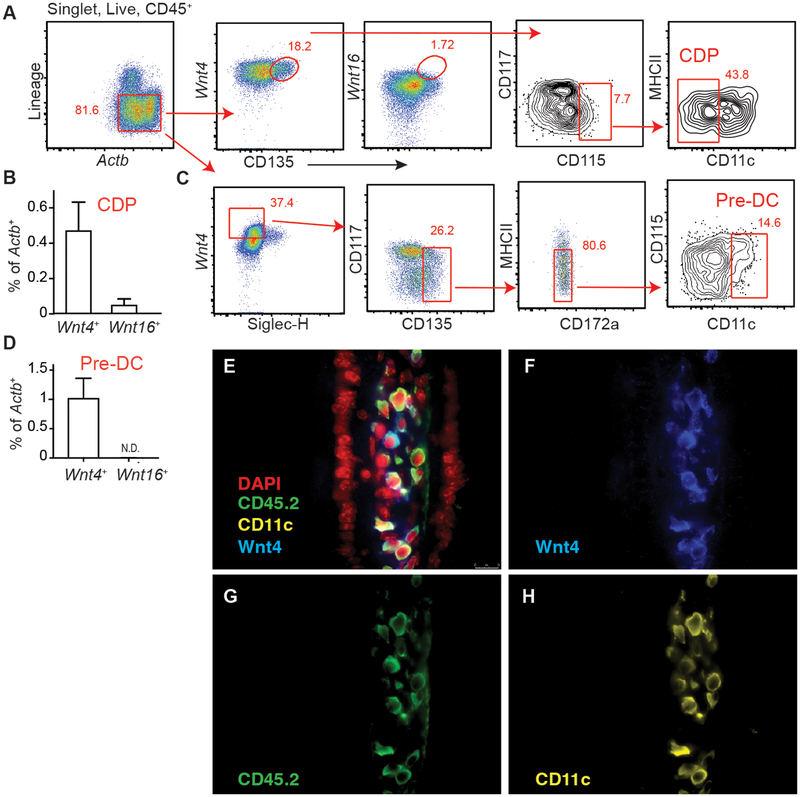
Evidence that Wnt4 can drive Flt3 expression in BM precursors (16) prompted us to speculate its involvement in DC development. Using a flow-based PrimeFlow™ approach, we found Wnt4 expression in both CDP and pre-cDC (Fig. 1A–D, Supplement Fig. 1A–B). Although previously shown to be induced by Wnt4 (22), Wnt16 was not detected in either progenitor population (Fig. 1BD and 1D). To test whether Wnt4 protein was expressed in CD11c positive cells and/or in non-hematopoietic cells in peripheral tissues, fluorescence immunostaining was done using Wnt-4 specific pAb co-stained with mAb specific for CD45 and CD11c on small intestines of wild-type mice. Data showed that Wnt4 positive cells were both CD45− as well as CD45+CD11c+, indicating DC-extrinsic and DC-intrinsic sources of Wnt4 exist (Fig. 1E–H, Supplement Fig.1C–E).
FIGURE 1.
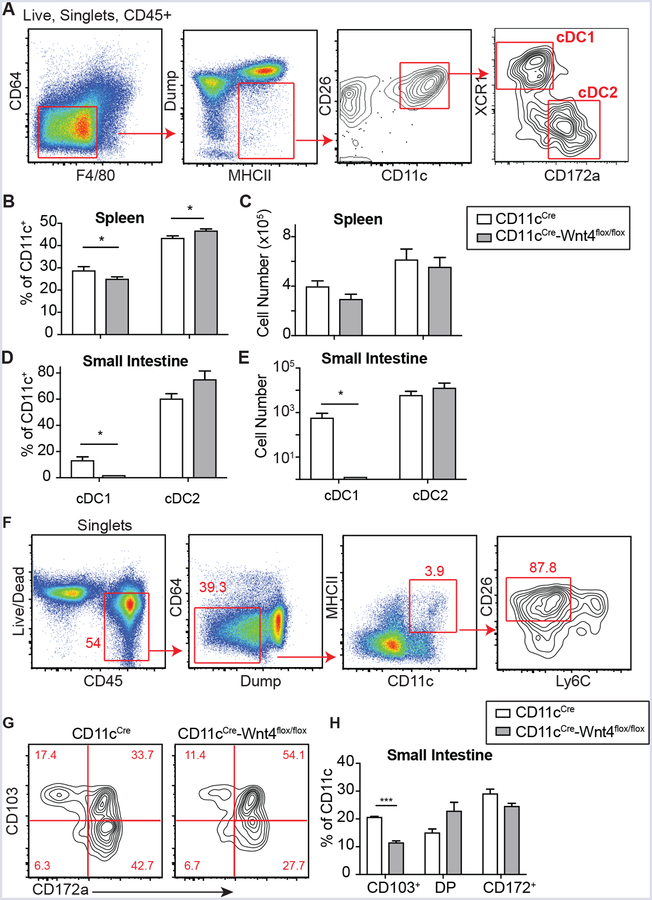
Wnt4 is expressed in BM DC progenitors and mature tissue cDC. (A) Gating and (B) Quantification of Wnt4+/Wnt16+ CDP in naïve CD11cCre BM. (C) Gating to identify Wnt4+/Wnt16+ pre-cDC in naïve CD11cCre BM. (D) Expression of Wnt4 and Wnt16 in pre-cDC as gated in “C”. (E-H) Expression of Wnt4, CD45.2, and CD11c in small intestines of naïve CD11cCremice. Images show merged (E) and single staining for Wnt4 (F), CD45.2 (G) and CD11c (H), respectively. All images were at 40×. Lineage markers: B220, CD3, CD5, CD11b, CD19, NK1.1, Ly6G, and Ter-119. Bar graphs show Mean ± SEM. N=3–5/group. Representative results from 3 independent experiments.
Wnt4 controls the balance of pre-cDC1 vs. pre-cDC2
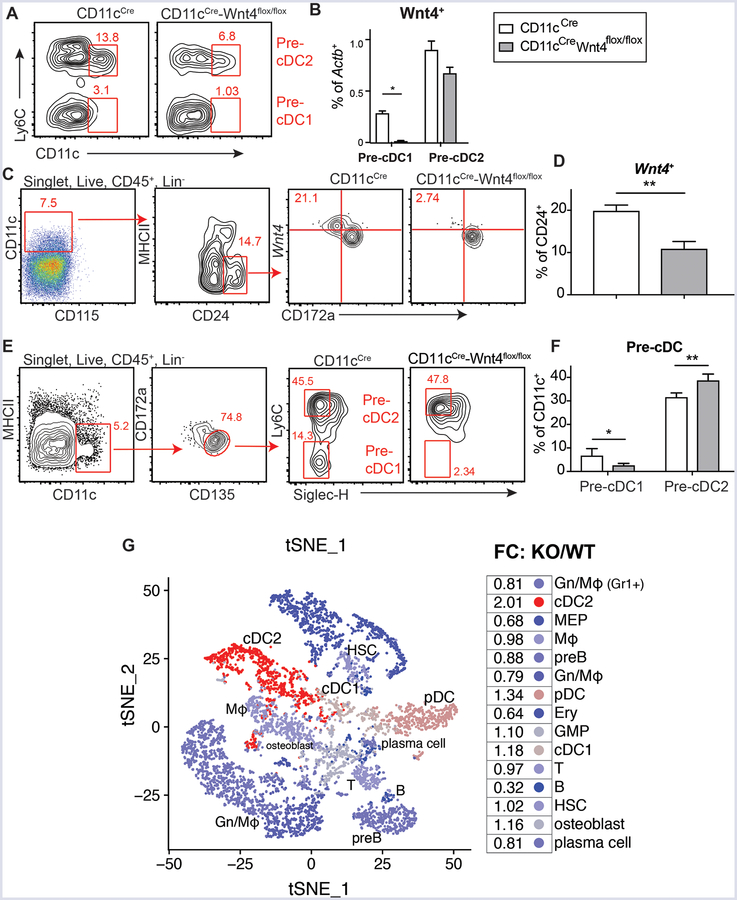
CD11cCreWnt4flox/flox mice were generated to understand whether cell-intrinsic Wnt4 served any role in cDC development and/or function. Comparison of Wnt4 levels in pre-cDC- defined by SiglecH−CD135+CD117medCD172alowMHCIIlowCD11c+ using Ly6C−/low to define pre-cDC1 and Ly6C+ to define pre-cDC2 (5), revealed significant reduction in pre-cDC1 within CD11cCre-Wnt4flox/flox compared to CD11cCre controls (Fig. 2A–B, Supplemental Fig. 1B). The loss of Wnt4 was most evident in the CD45+Lin−CD11c+CD115−MHCIIlowCD24+ population (Fig. 2C–D, Supplemental Fig. 2A). CD24 is mainly expressed on pre-cDC1-primed, but not pre-cDC2-primed cells within DC progenitors (5), hence CD11cCre-Wnt4flox/flox BM had a lower percentage of pre-cDC1, but a relatively intact pre-cDC2 population (Fig. 2E–F, Supplemental Fig. 2B). As an alternative approach to test whether Wnt4 shaped the pre-cDC1vs. pre-cDC2 ratio early in ontogeny, single-cell RNA-Seq analysis was performed. Data based on ~4,000 CD11c+ enriched BM cells using magnetic beads revealed that loss of Wnt4 resulted in a 2-fold enrichment of cDC2 transcriptome bearing cells at the steady-state compared to controls (Fig. 2G, Supplemental Fig. 2C–E). Taken together, the lack of Wnt4 expression in CD11c+ DC progenitors was associated with a decrease in the BM pre-cDC1/pre-cDC2 ratio.
FIGURE 2.
CD11c-dependent Wnt4 deficiency affects cDC progenitors in BM. (A) Representative flow plots showing Wnt4+ pre-cDC1 and pre-cDC2 populations in CD11cCre vs CD11cCre-Wnt4flox/flox BM. Cells were pre-gated on CD45+Lin−Actb+Siglec-H−Wnt4+CD135+CD117medCD172a−MHCIIlowCD115− as described in Figure 1C. (B) Quantification of Wnt4+ pre-cDC1 and pre-cDC2 in CD11cCre vs CD11cCre-Wnt4flox/flox BM as gated in “A”. (C and D) (C) Gating and (D) frequency of Wnt4+ cell within CD11c+CD115−CD24+ population. (E) Gating and representative flow plots and (F) quantification of pre-cDC1 and pre-cDC2 cells in CD11cCre vs CD11cCre-Wnt4flox/flox BM. Lineage markers: B220, CD3, CD5, CD11b, CD19, NK1.1, Ly6G, and Ter-119. Bar graphs show mean ± SEM with * p< 0.05 and ** p<0.01 by Students’ t-test. Representative results from 3–4 independent experiments. (G) Visualization of single-cell transcriptome from bone marrow cells after CD11c MACS enrichment. Projection of the clusters identified by modularity optimization of the shared nearest neighbors onto a t-SNE plot of the merged CD11cCre and CD11cCre-Wnt4flox/flox datasets. The fold-change of each cluster for KO (CD11cCre-Wnt4flox/flox) compared to WT (CD11cCre) was calculated and projected onto the merged t-SNE plot.
Wnt4 is critical for cDC1 development in vitro
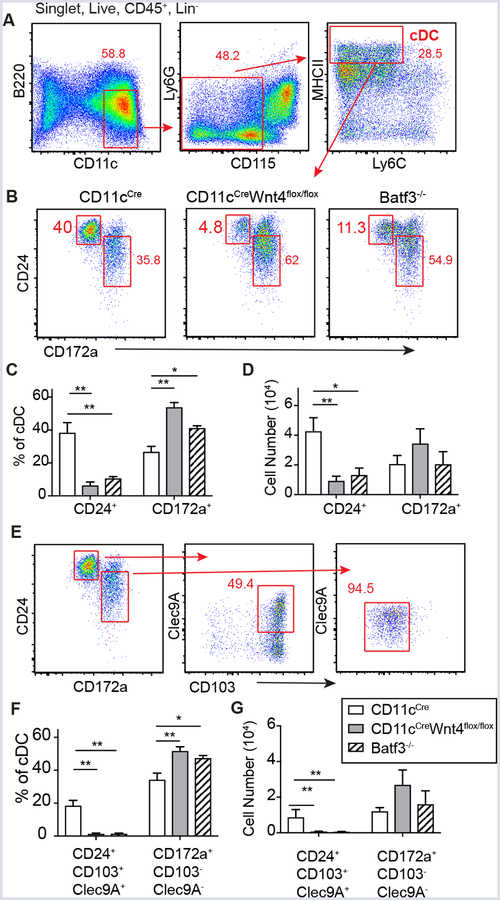
To test whether CD11cCre-Wnt4flox/flox progenitors could normally develop into the cDC1 subset, we employed a culture system reported to favor CD103+ cDC1 differentiation in an IRF8- and Batf3-dependent manner (19). Strikingly, data show that mature cDC (CD11c+B220−CD115−Ly6G−Ly6C−MHCIIhigh) in CD11cCre-Wnt4flox/flox cultures (Fig. 3A, Supplement Fig. 2F) were devoid of CD24+ (cDC1) DC similar to Batf3 (Fig. 3B–D). Upon further stratifying CD24+ cells for CD103 and Clec9A we found that most cells were also CD103+ and ~50% also expressed Clec9A, whereas CD172a+ cells (marker for cDC2) were mostly CD103− with low levels of Clec9A expression (Figure 3E–G). This indicated that CD11cCre-Wnt4flox/flox BM progenitors had defective ability to development into cDC1.
FIGURE 3.
Defective generation of cDC1 from CD11cCreWnt4flox/flox BM progenitors cultured with Flt3L plus GM-CSF. (A) Gating strategy to identify cDC from BM progenitors cultured for 12 days in RPMI CM+ 100 ng/ml Flt3L+ 10 ng/ml GM-CSF. (B) Representative flow plots showing CD24+ (cDC1) and CD172a+ (cDC2) in control vs CD11cCreWnt4flox/flox vs. Batf3−/− after gating as in “A”. (C and D) Quantification of cDC populations showing (C) percentage and (D) cell number of each cDC subset as gated in “B”. (E) Expression of CD103 and Clec9A within each cDC population. (F) Frequency and (G) Cell number of cDC subsets as shown in “E”. Representative results from 2 experiments. Graphs show mean ± SEM n=5/group with * p< 0.05 and ** p<0.01 by ANOVA.
Wnt4 promotes cJun and JNK activation to promote cDC1 commitment
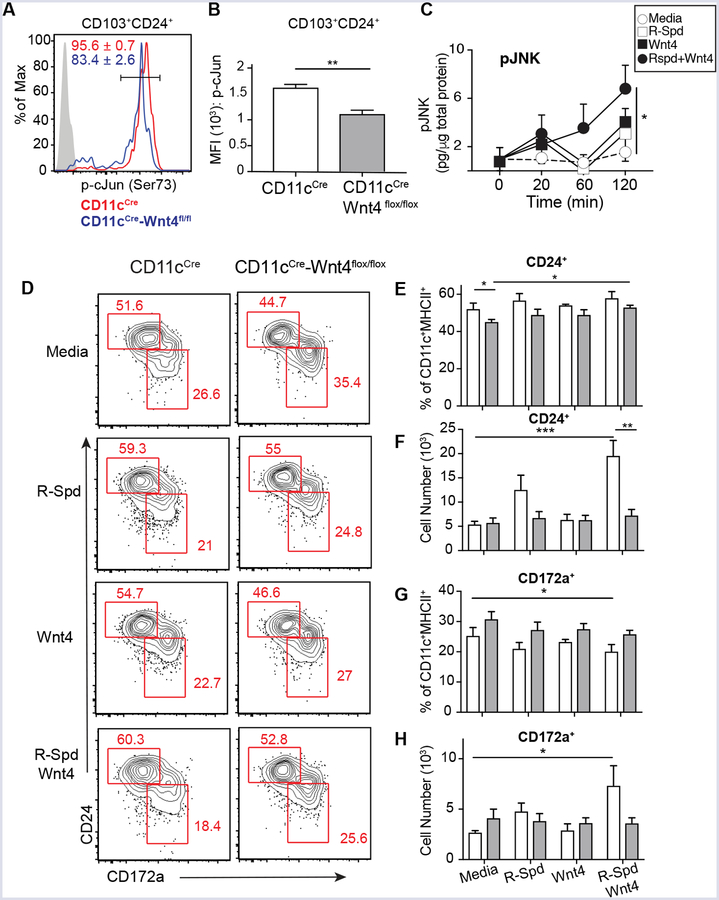
Wnt4 utilizes the Planer cell polarity (PCP) signaling to activate JNK and Jun in hematopoietic stem cells (17). Therefore, we asked whether the JNK/Jun axis was defective in CD11cCre-Wnt4flox/flox DC. Indeed, CD103+CD24+ BMDC derived from CD11cCre-Wnt4flox/flox had fewer phospho-cJun+ cells and lower phospho-cJun MFI levels in comparison to controls (Fig. 4A–B). Congruent with this finding, experiments involving treatment of BM cultures with either rWnt4 or rWnt4 plus R-spondin showed rapid induction of phospho-JNK (Fig. 4C). Therefore, we next asked whether Wnt4 was sufficient to promote cDC1 differentiation from BM cDC progenitors. Culture of total BM from either CD11cCre or CD11cCre-Wnt4flox/flox strains with Flt3L in addition to R-Spondin1 (R-Spd), Wnt4, or a combination of the two resulted in both CD24+ (cDC1) and CD172a+ (cDC2) subsets that expanded within 7 days (Fig. 4D) (23). We noted however, that the combined addition of Wnt4/R-Spd treatment led to a preferential 3-fold increase in CD24+ cells in controls, but only partially expanded CD24+ cells in CD11cCre-Wnt4flox/flox cultures (Fig. 4E–H). Although no significant differences in cDC percentage were found between CD11cCre and CD11cCre-Wnt4flox/flox strains, the CD172a+ population was clearly more prevalent in CD11cCre-Wnt4flox/flox cultures following Wnt4/R-Spd treatment (Fig. 4E–H) and moderately in the Flt3/GMCSF culture system (Fig. 3). Thus, exogenous Wnt4 promotes a signaling cascade that favors cDC1 development, perhaps through autocrine Wnt4 production.
FIGURE 4.
Wnt4 regulates activation of cJun and JNK and Flt3L-induced BMDC differentiation. (A) Representative histogram showing levels of phosphorylated cJun in CD103+CD24+ cDC in Flt3L+GM-CSF culture as gated in Figure 3E. Numbers show frequencies (Mean ± SEM from 5 biological replicates) of p-cJunhigh cells. (B) Median fluorescence intensity (MFI) of p-cJun in CD103+CD24+ cDC from Flt3L+GM-CSF culture. (C) Quantification of pJNK at indicated time points from BM cells treated with IMDM complete media alone or media containing R-Spd, Wnt4, or R-Spd+ Wnt4. (D) Representative flow plots from BM culture of for 7 days in IMDM complete media + 100 ng/ml Flt3L supplemented with R-Spd1 (100 ng/ml), Wnt4 (100 ng/ml), R-Spd1+ Wnt4(100 ng/ml each), or vehicle. Cells were pre-gated on CD45+F4/80−B220−CD11c+MHCIIhigh population. (E-H) (E and G) Frequencies and (F and G) cell number of CD24+ (E, F) and CD172a+ (G, H) from conditions in “D”. Graphs show Mean ± SEM from N=3–5 biological replicates per condition with * p< 0.05, ** p< 0.01 and *** p< 0.005 as determined by t-test or two-way ANOVA with Tukey correction for multiple comparison. Representative results from 3 independent experiments.
CD11c-dependent Wnt4 expression is critical for cDC homeostasis and regulates helminth burden after Nippostrongylus brasiliensis infection
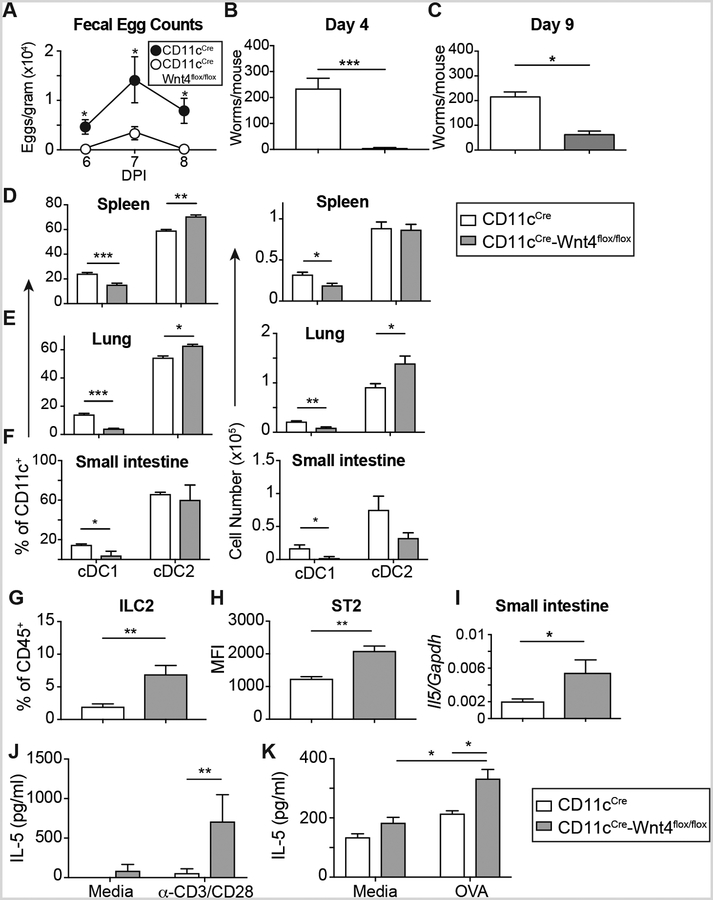
CD11c-restricted Wnt4 deficiency reduced cDC1 percentages in both spleen and small intestine with relatively modest increases in splenic cDC2 (Fig. 5A–E, Supplemental Fig. 3A). There was a selective reduction in number and percentage of intestinal CD103+ DC in CD11cCre-Wnt4flox/flox mice, consistent with expression of CD103 on cDC1 (24) (Fig. 5F–H, Supplemental Fig. 3B). Because cDC1-derived IL-12 inhibits host protective Type 2 immune responses, we tested whether Wnt4 deficiency would alter host protection against the hookworm N. brasiliensis (N.b.). In this model, inoculation of wild-type mice with infectious stage larvae (L3) results in spontaneous clearance of adult parasites and termination of egg production between 9–12 days (25). Data in Figure 6A show CD11cCre-Wnt4flox/flox mice had significantly lower fecal egg counts than CD11cCre controls between d6 through d8 post-infection. CD11cCre-Wnt4flox/flox mice also eliminated their intestinal worms significantly faster than controls at both early (d4) and late (d9) stages of infection (Fig. 6B–C).
FIGURE 5.
CD11c-dependent Wnt4 disrupts tissue cDC homeostasis. (A) Gating strategy to identify cDCs. (B-E) Frequencies (B, D) and cell number (C, E) of cDC populations in spleen (B, C) and small intestine (SI) (D,E). (F- H) (F) Gating strategy, (G) representative flow plots and (H) Quantification of small intestine cDC populations from CD11cCre vs CD11cCre-Wnt4flox/flox based on expression of CD172a and CD103 (DP= CD103 and CD172a double positive). Graphs show Mean ± SEM from N=3–4 biological replicates per condition with * p< 0.05 and *** p< 0.005 as determined by t-test or two-way. Representative results from 3 independent experiments.
FIGURE 6.
CD11cCre-Wnt4flox/flox animals mount stronger Th2 immune responses and show accelerated worm clearance after N.b. infection. (A) Fecal egg counts on d6–8 from mice infected with 700 L3 N.b. larvae. (B, C) Adult worm counts from CD11cCre vs CD11cCre-Wnt4flox/flox mice on (B) d4 and (C) d9 after N.b. infection. (D-F) Percentage (left) and number (right) of cDC1 and cDC2 in spleen (D), lung (E), and small intestines (F) at d4 post-N.b. infection. (G) Percentage of lung ILC2 and (H) Median fluorescence intensity (MFI) of ST2 expression on lung ILC2 from CD11cCre vs. CD11cCre-Wnt4flox/flox mice at d4 of N.b. infection. (I) Transcript levels of Il5 mRNA in small intestine from CD11cCre vs. CD11cCre-Wnt4flox/flox mince at d9 post-N.b. infection. (J) IL-5 levels from d9 N.b.-infected mesenteric lymph node cells after 72h re-stimulation (N=3–5/group). (K) IL-5 levels of OT-II T cells stimulated with OVA-pulsed CD11cCre or CD11cCre-Wnt4flox DC (N=3–4/group). Representative results from 3 independent experiments. Bar graphs show mean ± SEM of n=5/group with * p< 0.05, ** p< 0.01, and *** p< 0.005 as determined by Students’ t-test or two-way ANOVA with Tukey correction for multiple comparison.
To determine whether infection reversed the basal defects observed in naïve CD11cCre-Wnt4flox/flox mice, we probed tissues for cDC1 vs. cDC2 at day 4 post-N.b. infection. cDC1 percentage and number were both significantly reduced in CD11cCre-Wnt4flox/flox mice as compared to CD11cCre controls, accompanied by an increased percentage and number of cDC2 in lung and spleen (Fig. 6D–F). Although Th2 cell and Th2 cytokine levels were comparable between strains at the steady state (Supplemental Fig. 4E–I), after infection, both ILC2 percentages and ILC2 ST2 expression levels were significantly increased in CD11cCre-Wnt4flox/flox compared to controls (Fig. 6G–H, Supplemental Fig. 4A–D). Higher levels of ST2, the IL-33 receptor, implied increased sensitivity to the alarmin cytokine IL-33 (26). Moreover, intestinal Il5 levels and T cell mitogen (α-CD3)-stimulated IL-5 release from mesenteric lymph nodes of infected mice were both higher in CD11cCre-Wnt4flox/flox compared to controls (Fig. 6I–J).
Lastly, OVA-pulsed BMDC from CD11cCre vs. CD11cCre-Wnt4flox/flox mice were used to test whether DC lacking Wnt4 intrinsically promoted naïve CD4+OTII cells to become Th2 cells. Data show increased IL-5 from CD4+OTII cultured with Wnt4 deficient DC as compared to CD11cCre counterparts (Fig. 6K). Collectively, this indicates that lack of Wnt4 in DC accelerates innate and adaptive Type 2 immunity.
Discussion
These data indicate that Wnt4 serves a role as both an intrinsic and extrinsic regulator of cDC identity. Loss of Wnt 4 dysregulated the balance between the CD24+, Clec9a+, CD103+ cDC1 population vs. CD172a+ cDC2 in favor of the latter under steady-state conditions in BM and peripheral organs. Consistent with the notion that the heterogeneous cDC2 population promotes both Th2 and Th17 responses, our data show that CD11cCreWnt4flox/flox mice were highly resistant to the hookworm parasite N. brasiliensis with augmented ILC2 responses, TH2 cell development, and IL-5 production. To our knowledge, this is the first demonstration that a non-canonical Wnt ligand influences cDC fate. More work needs to be done to better understand how Wnt4 integrates into cDC developmental regulation by transcription factors such as c-Jun, IRF-4, IRF8, and Batf3.
BATF3, an ATF family transcriptional factor, is crucial for the development of cDC1 after the pre-DC stage (1). Lack of Batf3 affects both lymphocyte and DC function, most notably impairing cDC1 development in both lymphoid and non-lymphoid tissues (27, 28). To promote commitment of pre-cDC to the cDC1 subset, BATF3 forms a complex with two other transcription factors, JUN that partners with IRF8 to stabilize the early auto-activation of IRF8 during cDC1 development (2). In the absence of BATF3, the initial activation of IRF8 is not sustained and pre-cDC1 can reverse developmental path to become pre-cDC2 (2). Stabilization of this complex via JUN is a potential mechanism for how Wnt4 promotes balance between cDC1 and cDC2. Our experiments of DC differentiation from BM using Flt3L plus GM-CSF, which favors cDC1 differentiation, revealed that CD11cCre-Wnt4flox/flox mice exhibited an equivalent phenotype as Batf3−/− cultures in that both showed ablated cDC1 differentiation compared to controls. Intriguingly, although we found lower frequencies of pre-cDC1 in CD11cCre-Wnt4flox/flox BM, the expression levels of IRF8 in these progenitors was comparable to controls while splenic cDC1 from CD11cCre-Wnt4flox/flox showed decreased IRF8 level (data not shown). This suggests that Wnt4 is critical for both pre-cDC and tissue cDC, but perhaps only the later involves the regulation on cJun/IRF8/Batf3 complex and expression of IRF8. We also found that Wnt4 is expressed as early as in the CDP stage, however, because CD11c only turns on expression after the CDP stage, we cannot interrogate the role of Wnt4 in earlier progenitors such as CDP and macrophage-DC precursor (MDP) within the current model.
Wnt4 can activates non-canonical and potentially canonical signaling pathways depending on the context (29, 30); however in HSC, the progenitor expansion driven by Wnt4 is independent of β-catenin (16) and utilizes the PCP pathway to activate JNK2 and induce expression of cJun and cFos (17). It was demonstrated cross multiple models that Wnt4-induced PCP signaling is mediated by Frizzled 6 (Fzd6) receptor (8) and that knocking down Fzd6 blocked Wnt4-dependent HSC expansion (17). Intriguingly, Fzd6 is highly expressed in BM cells pre-committed to become cDC1 (5). Fzd6 itself is critical for the survival and self-renewal of BM progenitor cells, as Fzd6−/− BM cells showed increased apoptosis and defective engraftment (31). Whether Wnt4 signals through Fzd6 in either BM pre-cDC or mature cDC and whether Wnt4 signaling intersects with BATF3 for cDC1 development remain open questions. Although Wnt16 can also bind to Fzd6 (32, 33), we did not find Wnt16 expression in DC during our analysis. Future studies are required to determine whether other non-canonical Wnts (Wnt5, Wnt7, Wnt11 etc.) are also expressed in DC and whether they have a similar effect as Wnt4 on DC development.
We demonstrate that the combination of Wnt4 plus R-Spondin1 promoted maximum DC expansion in Flt3L cultures (Fig. 4D–H) and that treatment of Wnt4 and R-Spondin1 also induced the highest JNK activation in BM cells (Fig. 4C). There are four R-Spondins in both human and mouse (34). Specifically, R-Spondin1 binds to Lgr5, which associates with Fzd/LRP Wnt receptor complex, and inhibits Dickkopf (DKK)-1-dependent internalization of LPR5/6 (35, 36). Therefore, despite not strongly activating Wnt signaling cascade by itself (36, 37), R-Spondin1 greatly enhances Wnt signals. Previous work indicated that R-spondin1 enhanced Wnt4-induced signaling (38, 39) and our data implies that R-spondin1 augments Wnt4 mediated non-canonical signaling in DC precursors.
We show that dysregulation between cDC1/cDC2 numbers due to Wnt4 deficiency augmented Type 2 immunity. These data are consistent with studies showing that loss of IRF8-dependent cDC1 lead to a strong Th2 response following infection with Trichuris muris (T.m.), whereas the lack of IRF4-dependent cDC2 delayed the clearance of T.m. (40). Evert et al demonstrated that the BATF3-dependent cDC1 is a critical source of IL-12, the absence of which also increased resistance to helminth infection (41). IL-12 antagonizes Th2 responses (42) and exogenous IL-12 delays immunity against N. brasiliensis (43). Some suggest Type 2 immunity is deliberate, as TSLP (44–46) and IL-25 (47, 48) and IL-33 can each initiate Th2 immune responses. Alternatively, Type 2 immune responses can occur as a default mechanism as it is known that exposure to IL-4, early activation of GATA3, low strength of TCR signaling and absence of Th1 cytokines such as IFN-γ and IL-12 are all mechanisms that can promote Th2 response (49–51). We found that even with the defect in cDC1, naïve CD11cCre-Wnt4flox/flox mice did not have more Th2 or ILC2 nor higher Th2 cytokine production in comparison to controls, which was similar to the phenotype in Batf3−/− (41). Further investigations will seek to understand whether DC-intrinsic Wnt4 also regulates production of Th2-polarizing cytokines. Although mice lacking cDC1 succumb to sub-lethal Toxoplasma gondii (T.g.) infection due to lack of early IL-12 production (52), CD11cCre-Wnt4flox/flox mice only had a very moderate defect in clearing T.g. infection (data not shown). It is therefore likely that provision of IL-12 by the remaining cDC1 or other DC subsets in CD11cCre-Wnt4flox/flox mice was sufficient to induce host protection against Toxoplasma.
In summary, we demonstrate Wnt4 as a critical regulator of cDC1 vs cDC2 development in mice. It would be interesting to know whether Wnt4 has a similar role in human DC biology where expansion of autologous DC for tumor immunotherapy is a topic of considerable interest. Thus, it is possible that manipulation of Wnt4 activity in DC precursors could be used to shape CD8+ T cell responses for immunity against tumors, viruses, or microbes.
Supplementary Material
Key Points.
Wnt4 is a noncanonical Wnt protein that is expressed in stroma and cDC
Wnt4 is both necessary and sufficient for maintenance of cDC1
Lack of DC-derived Wnt4 accelerates Type 2 immunity
Acknowledgments
This work is supported by grants: UO1AI125940, AI095289, GM083204, and Burroughs Wellcome Fund.
Reference
- 1.Murphy TL, Grajales-Reyes GE, Wu X, Tussiwand R, Briseño CG, Iwata A, Kretzer NM, Durai V, and Murphy KM. 2016. Transcriptional control of dendritic cell development. Annu. Rev. Immunol 34: 93–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, Kretzer NM, Briseño CG, Durai V, Bagadia P, Haldar M, Schönheit J, Rosenbauer F, Murphy TL, and Murphy KM. 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol 16: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tailor P, Tamura T, Morse HC, and Ozato K. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 111: 1942–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, Nussenzweig M, and Merad M. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med 206: 3115–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HRB, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, Renia L, Poidinger M, Naik S, Newell EW, Robson P, and Ginhoux F. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol 16: 718–728. [DOI] [PubMed] [Google Scholar]
- 6.Mildner A, and Jung S. 2014. Development and function of dendritic cell subsets. Immunity 40: 642–656. [DOI] [PubMed] [Google Scholar]
- 7.Bejsovec A 2018. Wingless Signaling: A Genetic Journey from Morphogenesis to Metastasis. Genetics 208: 1311–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corda G, and Sala A. 2017. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis 6: e364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, and Woodgett JR. 1991. Phosphorylation of c-jun mediated by MAP kinases. Nature 353: 670–674. [DOI] [PubMed] [Google Scholar]
- 10.Ip YT, and Davis RJ. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)--from inflammation to development. Curr. Opin. Cell Biol 10: 205–219. [DOI] [PubMed] [Google Scholar]
- 11.Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang Y-C, and Pulendran B. 2010. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science (80-.). 329: 849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen SB, Smith NL, McDougal C, Pepper M, Shah S, Yap GS, Acha-Orbea H, Jiang A, Clausen BE, Rudd BD, and Denkers EY. 2015. Beta-catenin signaling drives differentiation and proinflammatory function of IRF8-dependent dendritic cells. J. Immunol 194: 210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oderup C, LaJevic M, and Butcher EC. 2013. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J. Immunol 190: 6126–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valencia J, Hernández-López C, Martínez VG, Hidalgo L, Zapata AG, Vicente Á, Varas A, and Sacedón R. 2011. Wnt5a skews dendritic cell differentiation to an unconventional phenotype with tolerogenic features. J. Immunol 187: 4129–4139. [DOI] [PubMed] [Google Scholar]
- 15.Zhao F, Xiao C, Evans KS, Theivanthiran T, DeVito N, Holtzhausen A, Liu J, Liu X, Boczkowski D, Nair S, Locasale JW, and Hanks BA. 2018. Paracrine Wnt5a-β-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 48: 147–160.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis I, Heinonen KM, Chagraoui J, Vainio S, Sauvageau G, and Perreault C. 2008. The signaling protein Wnt4 enhances thymopoiesis and expands multipotent hematopoietic progenitors through beta-catenin-independent signaling. Immunity 29: 57–67. [DOI] [PubMed] [Google Scholar]
- 17.Heinonen KM, Vanegas JR, Lew D, Krosl J, and Perreault C. 2011. Wnt4 enhances murine hematopoietic progenitor cell expansion through a planar cell polarity-like pathway. PLoS One 6: e19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, Berod L, Stüve P, Puttur F, Merad M, and Sparwasser T. 2014. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood 124: 3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stagg AJ, Burke F, Hill S, and Knight SC. 2001. Isolation of mouse spleen dendritic cells. Methods Mo.l Med. 64: 9–22. [DOI] [PubMed] [Google Scholar]
- 21.Hung L-Y, Oniskey TK, Sen D, Krummel MF, Vaughan AE, Cohen NA, and Herbert DR. 2018. Trefoil Factor 2 Promotes Type 2 Immunity and Lung Repair through Intrinsic Roles in Hematopoietic and Nonhematopoietic Cells. Am. J. Pathol 188: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YC, Clark RJ, Pelegri F, and Alexander CM. 2009. Wnt4 is not sufficient to induce lobuloalveolar mammary development. BMC Dev. Biol 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durai V, Bagadia P, Briseño CG, Theisen DJ, Iwata A, Davidson JT, Gargaro M, Fremont DH, Murphy TL, and Murphy KM. 2018. Altered compensatory cytokine signaling underlies the discrepancy between Flt3−/− and Flt3l−/− mice. J. Exp. Med 215: 1417–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steimle A, and Frick J-S. 2016. Molecular mechanisms of induction of tolerant and tolerogenic intestinal dendritic cells in mice. J. Immunol. Res 2016: 1958650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkelman FD, Shea-Donohue T, Goldhill J, Sullivan CA, Morris SC, Madden KB, Gause WC, and Urban JF. 1997. Cytokine regulation of host defense against parasitic gastrointestinal nematodes: lessons from studies with rodent models. Annu. Rev. Immunol 15: 505–533. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, and Kastelein RA. 2005. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23: 479–490. [DOI] [PubMed] [Google Scholar]
- 27.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung S-SJ, Murphy TL, Hildner K, and Murphy KM. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med 207: 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, and Murphy KM. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science (80-.). 322: 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardi H, Gay S, Fedon Y, Vernus B, Bonnieu A, and Bacou F. 2011. Wnt4 activates the canonical β-catenin pathway and regulates negatively myostatin: functional implication in myogenesis. Am. J. Physiol. Cell Physiol 300: C1122–38. [DOI] [PubMed] [Google Scholar]
- 30.Tanigawa S, Wang H, Yang Y, Sharma N, Tarasova N, Ajima R, Yamaguchi TP, Rodriguez LG, and Perantoni AO. 2011. Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev. Biol 352: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abidin BM, Owusu Kwarteng E, and Heinonen KM. 2015. Frizzled-6 Regulates Hematopoietic Stem/Progenitor Cell Survival and Self-Renewal. J. Immunol 195: 2168–2176. [DOI] [PubMed] [Google Scholar]
- 32.Kilander MBC, Dahlström J, and Schulte G. 2014. Assessment of Frizzled 6 membrane mobility by FRAP supports G protein coupling and reveals WNT-Frizzled selectivity. Cell Signal. 26: 1943–1949. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Zhu D, Chen F, Qian M, Wei H, Chen W, and Xu J. 2016. SFRP2 augments WNT16B signaling to promote therapeutic resistance in the damaged tumor microenvironment. Oncogene 35: 4321–4334. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Kim K-A, Wagle M, Tran K, Zhan X, Dixon MA, Liu S, Gros D, Korver W, Yonkovich S, Tomasevic N, Binnerts M, and Abo A. 2008. R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell 19: 2588–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RBM, Mohri Y, Nishimori K, Mohammed S, Heck AJR, and Clevers H. 2011. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476: 293–297. [DOI] [PubMed] [Google Scholar]
- 36.Binnerts ME, Kim K-A, Bright JM, Patel SM, Tran K, Zhou M, Leung JM, Liu Y, Lomas WE, Dixon M, Hazell SA, Wagle M, Nie W-S, Tomasevic N, Williams J, Zhan X, Levy MD, Funk WD, and Abo A. 2007. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl. Acad. Sci. USA 104: 14700–14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glinka A, Dolde C, Kirsch N, Huang Y-L, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat C-M, and Niehrs C. 2011. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12: 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomizuka K, Horikoshi K, Kitada R, Sugawara Y, Iba Y, Kojima A, Yoshitome A, Yamawaki K, Amagai M, Inoue A, Oshima T, and Kakitani M. 2008. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet 17: 1278–1291. [DOI] [PubMed] [Google Scholar]
- 39.Chassot A-A, Bradford ST, Auguste A, Gregoire EP, Pailhoux E, de Rooij DG, Schedl A, and Chaboissier M-C. 2012. WNT4 and RSPO1 together are required for cell proliferation in the early mouse gonad. Development 139: 4461–4472. [DOI] [PubMed] [Google Scholar]
- 40.Demiri M, Müller-Luda K, Agace WW, and Svensson-Frej M. 2017. Distinct DC subsets regulate adaptive Th1 and 2 responses during Trichuris muris infection. Parasite Immunol 39. [DOI] [PubMed] [Google Scholar]
- 41.Everts B, Tussiwand R, Dreesen L, Fairfax KC, Huang SC-C, Smith AM, O’Neill CM, Lam WY, Edelson BT, Urban JF, Murphy KM, and Pearce EJ. 2016. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med 213: 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vignali DAA, and Kuchroo VK. 2012. IL-12 family cytokines: immunological playmakers. Nat. Immunol 13: 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finkelman FD, Madden KB, Cheever AW, Katona IM, Morris SC, Gately MK, Hubbard BR, Gause WC, and Urban JF. 1994. Effects of interleukin 12 on immune responses and host protection in mice infected with intestinal nematode parasites. J. Exp. Med 179: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor BC, Zaph C, Troy AE, Du Y, Guild KJ, Comeau MR, and Artis D. 2009. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J. Exp. Med 206: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou B, Comeau MR, De Smedt T, Liggitt HD, Dahl ME, Lewis DB, Gyarmati D, Aye T, Campbell DJ, and Ziegler SF. 2005. Thymic stromal lymphopoietin as a key initiator of allergic airway inflammation in mice. Nat. Immunol 6: 1047–1053. [DOI] [PubMed] [Google Scholar]
- 46.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, Gilliet M, Ho S, Antonenko S, Lauerma A, Smith K, Gorman D, Zurawski S, Abrams J, Menon S, McClanahan T, de Waal-Malefyt Rd R, Bazan F, Kastelein RA, and Liu Y-J. 2002. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat. Immunol 3: 673–680. [DOI] [PubMed] [Google Scholar]
- 47.Fort MM, Cheung J, Yen D, Li J, Zurawski SM, Lo S, Menon S, Clifford T, Hunte B, Lesley R, Muchamuel T, Hurst SD, Zurawski G, Leach MW, Gorman DM, and Rennick DM. 2001. IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity 15: 985–995. [DOI] [PubMed] [Google Scholar]
- 48.Angkasekwinai P, Park H, Wang Y-H, Wang Y-H, Chang SH, Corry DB, Liu Y-J, Zhu Z, and Dong C. 2007. Interleukin 25 promotes the initiation of proallergic type 2 responses. J. Exp. Med 204: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamane H, Zhu J, and Paul WE. 2005. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J. Exp. Med 202: 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noben-Trauth N, Hu-Li J, and Paul WE. 2000. Conventional, naive CD4+ T cells provide an initial source of IL-4 during Th2 differentiation. J. Immunol 165: 3620–3625. [DOI] [PubMed] [Google Scholar]
- 51.Ouyang W, Löhning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, and Murphy KM. 2000. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12: 27–37. [DOI] [PubMed] [Google Scholar]
- 52.Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, Sher A, Ploegh HL, Murphy TL, Sibley LD, and Murphy KM. 2011. CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35: 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.