Abstract
Gut lymphocytes and the microbiota establish a reciprocal relationship that impacts the host immune response. Class I-restricted T cell-associated molecule (CRTAM) is a cell adhesion molecule expressed by intraepithelial T cells and is required for their retention in the gut. Here we show that CRTAM expression affects gut microbiota composition under homeostatic conditions. Moreover, Crtam−/− mice infected with the intestinal pathogen Salmonella exhibit reduced Th17 responses, lower levels of inflammation, and reduced Salmonella burden, which is accompanied by expansion of other microbial taxa. Thus, CRTAM enhances susceptibility to Salmonella, likely by promoting the inflammatory response that promotes the pathogen’s growth. We also found that the gut microbiota from wild-type mice, but not from Crtam−/− mice, induces CRTAM expression and Th17 responses in ex-germ-free mice during Salmonella infection. Our study demonstrates a reciprocal relationship between CRTAM expression and the gut microbiota, which ultimately impacts the host response to enteric pathogens.
Introduction
Resident T cells in the gut are key orchestrators of mucosal immunity. Located between epithelial cells (intraepithelial lymphocytes) and in the lamina propria, these T cells are comprised of several subpopulations, including CD4+ T cells, CD8+ T cells, γδ T cells, natural killer T cells (NKT), and a unique population of CD4+CD8+ T cells (1, 2). A consequence of CD4+ T cell depletion in gut-associated lymphoid tissue (GALT) is a disruption of the gut epithelial barrier, a scenario common among human immunodeficiency virus (HIV)-infected patients (3). As a result, HIV patients exhibit an increased susceptibility to bacteremia caused by intestinal pathogens such as Salmonella and Campylobacter (4–6), indicating that CD4+ T cells are required in order to develop an optimal immune response against enteric pathogens.
Th17 cells are an important subset of CD4+ T cells residing in the gut. These cells constitute a distinct lineage from Th1 and Th2 cells, and are characterized by the production of interleukin (IL)-17A, IL-17F, and IL-22, collectively known as Th17 cytokines (7). Th17 cytokines are expressed in the intestinal mucosa in response to enteric pathogens such as Salmonella and Citrobacter rodentium (8–10), and orchestrate a host response that strengthens the mucosal barrier and protects from systemic dissemination of these pathogens. In the mouse model of inflammatory diarrhea, mice deficient in the IL-17A receptor exhibit higher levels of Salmonella translocation from the gut to systemic sites such as the spleen. Moreover, Th17 deficiency results in a reduction of neutrophil recruitment to the intestinal mucosa during infection (10), which may explain the increased systemic dissemination of Salmonella in the absence of IL-17 signaling. For Citrobacter, the highly homologous IL-17A and IL-17F (8), and IL-17C (11) are important to reduce the pathogen’s colonization of the colon. This reduction in colonization is mediated by IL-17A/F-induced β-defensins (8), and by IL-17C/IL-22-induced proinflammatory cytokines, chemokines, and antimicrobial proteins including calprotectin, lipocalin-2, RegIIIβ, and RegIIIγ (11). Together, these studies underscore the importance of Th17 cells in host defense against gut pathogens.
Homing and residency of T cells to the gut require the expression of specialized chemokines, chemokine receptors, and adhesion molecules (reviewed in (12)). For instance, entry of naïve and effector T cells into the intestinal mucosa is mediated by integrin α4β7, which binds to mucosal vascular addressin cell adhesion molecule-1 (MAdCAM-1) expressed in the lamina propria venules (13). Integrin αEβ7 is involved in the retention of effector and memory lymphocytes to the gut epithelium through its interaction with E-cadherin (14). In addition to the interactions described above, it has been shown that the interaction between Class I-restricted T cell-associated molecule (CRTAM) in T cells and Cell Adhesion Molecule 1 (CADM1; also known as NECL2 and TSLC1) contributes to the residency and maintenance of T cell populations in the gut mucosa (15).
Located on the cell surface, CRTAM is an Immunoglobulin superfamily member that was originally identified in activated CD8+ T cells and NKT cells (hence its name, Class-I restricted) (16). Further reports described CRTAM on NK cells (17). More recently, CRTAM has been described on intraepithelial CD4+CD8α/α+ and CD4+ T cells, as well as on CD8+ T cells of the intestinal mucosa (15). CADM1, the only CRTAM ligand described to date (17), is a cell-surface molecule of the nectin and NECL families that is expressed on CD8α dendritic cells (DCs), CD103+ DCs, epithelial cells, neurons, and certain tumor cells (18–21). CRTAM–CADM1 interactions strengthen NK cell and CD8+ T cell effector functions (17, 19, 22, 23). Moreover, it has been proposed that CRTAM is essential for the establishment of CD4+ T cell polarization after TCR engagement. During this process, CD4+ T cell proliferation is blocked, and these cells begin to produce effector cytokines, including IFN-γ, IL-17A, and IL-22 (24). Given CRTAM’s role in mediating T cell retention to the gut and in the production of effector cytokines, two prior studies investigated the role of CRTAM during intestinal infection. In this context, CRTAM was found to confer protection against the parasite Toxoplasma gondii (15), whereas its role during infection with Citrobacter rodentium is less clear, as one study showed CRTAM-mediated protection (24), but a second study did not confirm these results (15).
The intestinal mucosa is the body’s largest immunological organ and is constantly exposed to antigens from food, pathogens, and commensal microbes. Interactions between commensal microbes (collectively known as the microbiota) and the intestinal mucosa play a fundamental role in the induction, education and function of the immune system (25). The intestinal mucosa, in turn, has evolved to tolerate the microbiota and keep a homeostatic relationship. To maintain this delicate balance, the gut microbiota and the intestinal immune system engage in bidirectional interactions, whereby reciprocal signals between the gut immune system and the gut microbiota are exchanged and mutually shape the immune system and the composition of the microbiota. Specific members of the gut microbiota shape different aspects of innate and adaptive immunity in the gut, including the development and function of particular regulatory and effector T-cell lineages. For example, segmented filamentous bacteria (SFB), commensals of the phylum Firmicutes, are required for T helper (Th)17 differentiation, which provides increased resistance to infection with the gut pathogen Citrobacter rodentium (26).
Because the interaction between gut T cells and the microbiota plays an important role in maintaining gut homeostasis, we hypothesized that CRTAM expression modulates the balance between the gut microbiota and mucosal immunity. Here, we show that CRTAM shapes the gut microbiota under homeostatic conditions. As gut microbes and intestinal T cells contribute to host defense against intestinal pathogens, we tested whether CRTAM plays a role during Salmonella infection. We found that Crtam−/− mice exhibit reduced Th17 responses, lower levels of inflammation, decreased intestinal colonization by Salmonella, and an outgrowth of other microbial taxa. Moreover, germ-free mice colonized with fecal microbiota collected from Crtam−/− mice exhibited lower expression of CRTAM on T cells, and a reduced Th17 response, upon Salmonella infection. Collectively, our data reveal an interplay between CRTAM expression and the gut microbiota, which ultimately impacts the mucosal immune system and the host response to enteric pathogens.
Materials and Methods
Mouse experiments
Mice heterozygous for the Crtamtm1(KOMP)Wtsi allele (referred in the text as Crtam−/−) were obtained from the Knockout Mouse Project (KOMP) repository (targeting project ID CSD67690) at the University of California, Davis. We obtained these founder mice and performed the following experiments on their WT and Crtam−/− littermate progeny. Male and female mice were orally gavaged with streptomycin 24 hours before oral gavage with 109 colony-forming units (CFUs) of Salmonella enterica serovar Typhimurium IR715, as previously described (10, 27). Fecal samples were collected at 24, 48, 72, and 96 hours post-infection. The cecal content was collected at 96 hours post-infection. Fecal samples and cecal content were serially diluted and plated on appropriate antibiotic-containing LB agar plates to determine bacterial counts (CFUs). Spleen, mesenteric lymph nodes (MLN), Peyer’s patches and terminal ileum were collected, weighed, homogenized, serially diluted, and plated on appropriate antibiotic-containing LB agar plates to determine CFUs. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees at the University of California, Irvine and at the University of California, San Diego.
Cell extraction from the gut and cell analysis
Terminal ileum, cecum, and colon were collected and kept in IMDM medium supplemented with 10% FBS and 1% antibiotic/antimycotic solution. Next, Peyer’s patches were removed, cut open longitudinally, and washed with HBSS supplemented with 15 mM HEPES and 1% antibiotic/antimycotic. Then, the tissue was shaken in 10 ml of HBSS/ 15 mM HEPES/ 5 mM EDTA/ 10% FBS solution at 37 °C in a water bath for 15 min. Supernatant was removed and kept on ice. Remaining tissue was cut into small pieces, then digested in a 10 ml mixture of collagenase (Type VII, 1 mg/ml), Liberase (20 μg/ml), and DNAse (0.25 mg/ml) in IMDM medium for 15 min in a shaking water bath at 37 °C. Afterwards, both fractions were strained through a 70 μm cell strainer, pooled, and then the extracted cells were stained for analysis by flow cytometry. When indicated, terminal ileum, cecum and colon were processed separately, and the IEL and LPL fractions were kept separated, and then the extracted cells were stained for analysis by flow cytometry. Briefly, cells were blocked with a CD16/32 antibody (eBioscience), stained with the viability dye eFluor780 (eBioscience), then extracellularly stained using the following monoclonal antibodies: CD45 (clone 30-F11), CD3 (clone 17A2), CD4 (clone RM4–5), CD8α (clone 53–6.7) – each from BioLegend; TCR γδ (clone eBio-GL3) and CD25 (clone PC61.5) – each from eBioscience. After surface staining, cells were fixed and permeabilized according to the manufacturer’s instructions (“Fix and Perm” kit, eBioscience), then stained intracellularly with anti-IL17 antibody (clone TC11–18H10.1), anti-IFNγ antibody (clone XMG1.2), and anti-IL-10 antibody (clone JES5–16-E3) from eBioscience. Cells were analyzed on an LSRII flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo software (TreeStar, Ashland, OR).
RNA extraction and qPCR
Total RNA was extracted from mouse cecal tissue using Tri-Reagent (Molecular Research Center). Reverse transcription of 1 μg of total RNA was performed using the Transcriptor First Strand cDNA Synthesis kit (Roche). Quantitative real-time PCR (qRT-PCR) for the expression of Actb, Cxcl1, Il1b, Il6, Il17, Il22, Il23, Lcn2, and Tgfb was performed using the LightCycler 480 SYBR Green Master Mix on the LightCycler 480 II (Roche). Primer pair sequences are shown in Supplementary Table 1. Gene expression was normalized to Actb (β-actin). Fold changes in gene expression were relative to uninfected controls and calculated using the ΔΔ-Ct method.
Histopathology
Cecal tissue samples were fixed in formalin, processed according to standard procedures for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin and eosin. The pathology score of cecal and liver samples was determined by blinded examinations of cecal and liver sections from a board-certified pathologist using previously published methods (27). Each cecal section was evaluated for the presence of neutrophils, mononuclear infiltrate, submucosal edema, surface erosions, inflammatory exudates, and cryptitis. Inflammatory changes were scored from 0 to 4, according to the following scale: 0=none; 1=low; 2=moderate; 3=high; 4=extreme. The inflammation score was calculated by adding up all of the scores obtained for each parameter and interpreted as follows: 0–2=within normal limit; 3–5=mild; 6–8=moderate; >8=severe.
Analysis of the Microbiota
Fecal samples were collected before infection and 96 hours post-infection from littermate co-housed mice. The samples were snap-frozen in liquid nitrogen, and then DNA was later extracted using the QIAamp DNA Stool Kit (Qiagen) according to manufacturer’s instructions with modifications as previously described (28). 16S rDNA (V4 region) was then amplified by PCR with primers 515F and 806R (modified by addition of barcodes for multiplexing), then sequenced on an Illumina MiSeq system (UC Davis HMSB Facility). Sequences were processed and analyzed by employing the SILVA rRNA gene database v123 (29) and the QIIME pipeline v1.9.1 (30) with default settings, except as noted. In brief, paired-end sequences were joined, quality-filtered, reverse-complemented and chimera-filtered (usearch61 option; reference file 97_otus_16S.fasta from SILVA v123). Then, operational taxonomic units (OTUs) were picked de novo at 97% similarity, and taxonomy was assigned with the RDP classifier (“pick_otus” options: enable_rev_strand_match True, otu_picking_method usearch61; “align_seqs” option: template_fp core_alignment_SILVA123.fasta; “filter_alignment” options: allowed_gap_frac 0.80, entropy_threshold 0.10, suppress_lane_mask_filter True; “assign_taxonomy” options: assignment_method rdp, confidence 0.8, rdp_max_memory 24000, reference_seqs_fp 97_otus_16S.fasta, id_to_taxonomy_fp consensus_taxonomy_7_levels.txt). Samples were rarefied to 10,000 reads. Alpha (Shannon index) and beta (unweighted and weighted UniFrac) diversity were assessed via QIIME. Prism 7 software (GraphPad) was used for statistical analyses (Mann–Whitney U test).
Fecal transplant and infection in germ-free mice
Fresh fecal pellets were obtained from WT and Crtam−/− littermate co-housed mice, and placed in an Eppendorf tube containing sterile PBS supplemented with 10% glycerol. Fecal pellets were shaken for 20 minutes, then the fecal suspension was strained through a 70 μm cell strainer to eliminate the fiber present in the fecal pellet. Aliquots of the fecal suspension were then snap-frozen in liquid nitrogen and stored at −80 °C until the day of the transplant. For the fecal transplant, 10-week-old Swiss Webster germ-free mice were used. Male and female germ-free mice were inoculated orally with 100 μL of the fecal suspension at day 0, day 2, and day 4, then the microbiota was allowed to engraft for 6 days. Mice were then orally gavaged with streptomycin 24 hours before oral gavage with Salmonella enterica serovar Typhimurium (1×109 CFU per mouse). At 96 hours post-infection, tissues were collected and cell analysis was performed as described above.
Statistical analysis
Statistical significance was determined using unpaired, two-tailed Student’s t test on log-transformed data, or Mann-Whitney U test. Differences were considered statistically significant if the P value was <0.05.
Results
CRTAM expression impacts the gut microbiota in homeostatic conditions.
The gut microbiota plays a critical role in gut homeostasis, which in turn influences a wide variety of host responses, including the immune response. Of particular note, members of the gut microbiota play a central role in regulating T cell responses in the gut (26, 31, 32). For instance, the microbiota regulates expression of the homing receptor GPR15, which contributes to the retention of T cells in specific regions of the intestinal mucosa (33). Previous studies have shown that CRTAM contributes to the retention of T cells in the gut (15). To determine whether CRTAM-dependent retention of T cells in the gut has an impact on the gut microbiota composition, we compared Crtam−/− mice with wild-type co-housed littermates.
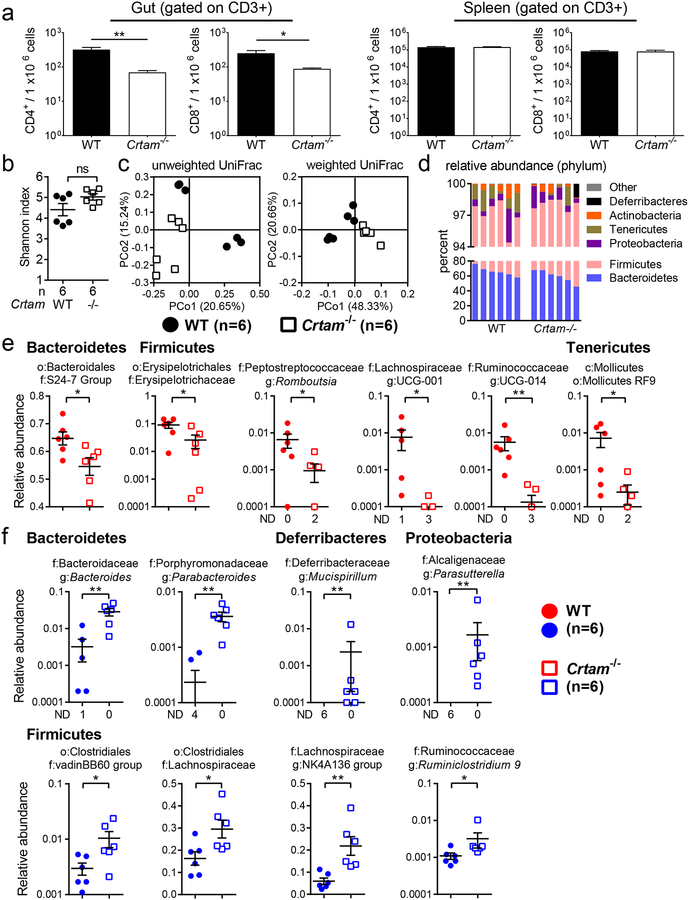
First, we collected the small intestine, large intestine, and spleen from WT and Crtam−/− mice to assess T cell populations. We observed a significant reduction in the frequency of CD4+ and CD8+ T cells in the small and large intestine of Crtam−/− mice (Figure 1a, left panel). As expected, we observed similar frequencies of CD4+ and CD8+ T cells from the spleens of Crtam−/− mice (Figure 1a, right panel). These findings indicated that CRTAM is important to retain T cells in the small and large intestine, but not in the spleen, which may be due to the microbiota activating T cells in the gut to express CRTAM.
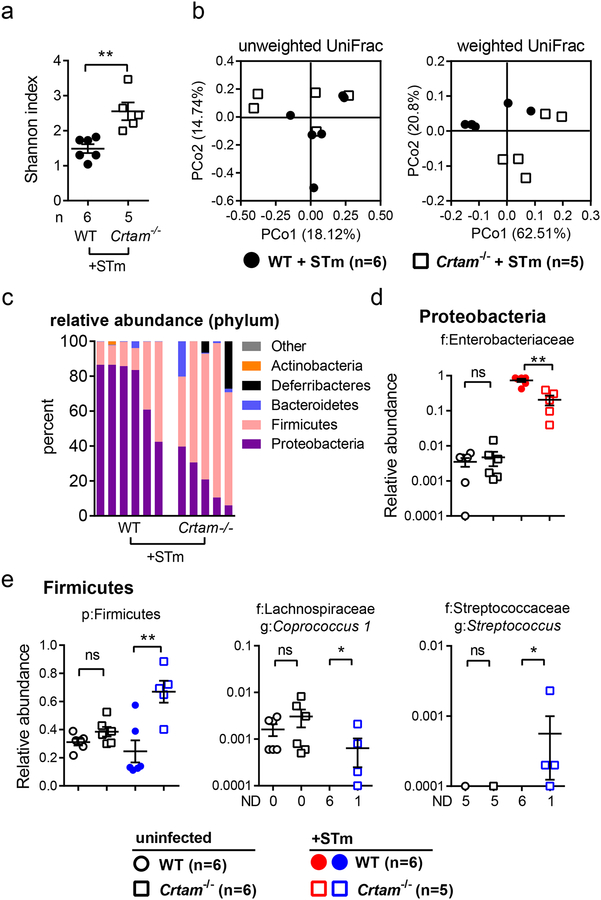
Figure 1. Composition of the gut microbiota is modulated by CRTAM.
Composition of the gut microbiota is modulated by CRTAM. (a) Frequency of T cells per million live cells analyzed for CD4+ (CD3+CD4+) and CD8+ (CD3+CD8+) T cells in the gut (left panel) or in the spleen (right panel) of WT (black bar) or Crtam−/− (white bar) mice (n=3 per group). Data displayed are representative of two independent experiments. (b) Alpha diversity (Shannon index) in the gut microbiota of WT (black circles) or Crtam−/− (white squares) mice. (c) PCoA (principal coordinate analysis) plot based on the unweighted (left panel) or weighted (right panel) UniFrac metric. (d) Bar chart of phylum relative abundance within each sample. (e, f) Relative abundance (as a fraction of 1) of eubacterial taxa that exhibited a statistically significant change in Crtam−/− mice relative to WT mice; either (e) diminished relative abundance or (f) increased relative abundance (ND, not detected); n per group is indicated in the figure. For (d-f), only taxa present at >0.1% relative abundance in at least one sample were considered. (a, b, e, f) Bars represent the mean ± standard error; a significant change (t-test in (a), Mann-Whitney-U in b, e, f) relative to WT control is indicated by * (P ≤ 0.05) or ** (P ≤ 0.01).
Because T cells shape the gut microbiota through different mechanisms, we hypothesized that the reduced frequency of T cells in the gut of Crtam−/− mice could change the composition of the intestinal microbiota. To test this hypothesis, we collected fecal samples from WT and Crtam−/− littermate co-housed mice to perform total DNA extraction and 16S sequencing. The intrasample Eubacterial diversity was slightly increased in Crtam−/− mice (Figure 1b), and the intersample community composition and structure were different in the absence of CRTAM (Figure 1c). In general, composition of the gut microbiota reflected what is usually observed in homeostatic conditions and in the absence of gut inflammation, with a predominance of the phyla Bacteroidetes and Firmicutes (Figure 1d). Nevertheless, we observed a significant relative increase of phylum Firmicutes (p < 0.0087) and relative reduction of Tenericutes (p < 0.039) in Crtam−/− mice. Moreover, deeper taxonomic analysis revealed significantly altered families and genera in the Crtam−/− mouse. Families with significantly reduced relative abundance in the absence of CRTAM (Figure 1e) included the S24–7 Group (Candidatus Homeothermaceae (34)) and Erysipelotrichaceae. Significantly reduced genera included Romboutsia (family Peptostreptococcaceae) and UCG-014 (family Ruminococcaceae) (Figure 1e). We also observed a significantly increased relative abundance of families and genera in Crtam−/− mice, including the families Lachnospiraceae and vadin BB60 Group, as well as the genera Bacteroides, Parabacteroides, Mucispirillum, Parasutterella, Ruminiclostridium 9, and the Lachnospiraceae NK4A136 group (Figure 1f). The analysis of the gut microbiota in WT and Crtam−/− littermate co-housed mice indicated that the absence of CRTAM altered the gut microbial taxonomic composition, a finding that has potential implications on colonization resistance to pathogens.
CRTAM enhances susceptibility to Salmonella infection.
Members of the gut microbiota provide colonization resistance to enteric pathogens through mechanisms that include immune modulation, barrier maintenance, nutrient utilization, and direct growth inhibition (35, 36). In turn, enteric pathogens have evolved mechanisms to outcompete the gut microbiota to cause disease. One of the best-studied examples of competition between an intestinal pathogen and the microbiota is during infection with Salmonella enterica. In homeostatic conditions, the gut microbiota provides effective colonization resistance to Salmonella. However, the inflammatory response triggered by Salmonella in the gut enables the pathogen to outcompete the gut microbiota by a variety of mechanisms (28, 37–40). For instance, intestinal inflammation provides Salmonella with novel electron acceptors and nutrients to thrive in the inflamed gut (40, 41).
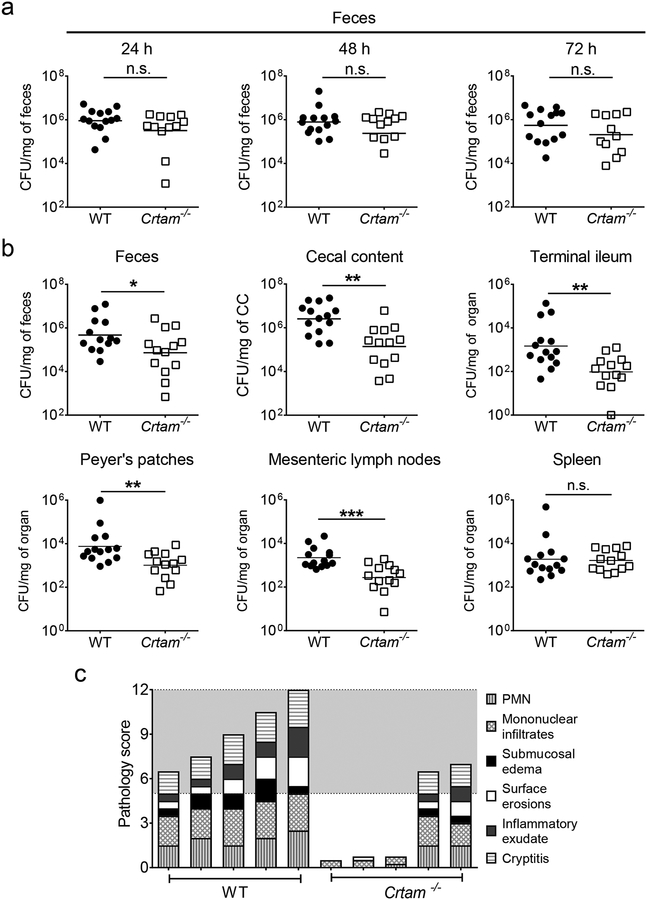
Because the absence of CRTAM results in reduced levels of gut-resident T cells and changes the gut microbiota composition, we tested whether Crtam−/− mice exhibited altered susceptibility to Salmonella infection. Here, we infected WT and Crtam−/− littermate mice with Salmonella enterica serovar Typhimurium by using a well-established colitis model (10, 27, 42). Although we did not observe significant differences in the Salmonella fecal burden at 24, 48, and 72 hours post-infection (Figure 2a), we recovered significantly lower Salmonella numbers from the feces and cecal content of Crtam−/− mice at 96 hours post-infection (Figure 2b). Moreover, the Salmonella burden in the terminal ileum, mesenteric lymph nodes, and Peyer’s patches was also lower in Crtam−/− mice (Figure 2b). By contrast, there was no difference in the number of Salmonella recovered from the spleen of WT and Crtam−/− mice (Figure 2b), which is consistent with our finding of Crtam−/− mice exhibiting a lower frequency of T cells in the gut, but not in the spleen, relative to WT mice (Figure 1a).
Figure 2. Colonization of Salmonella in WT and Crtam−/− mice.
WT mice (black circles) and Crtam−/− mice (white squares) were treated with streptomycin, then 24h later were infected with wild-type Salmonella enterica serovar Typhimurium (STm). Colony-forming units (CFU) in (a) feces at the indicated time points, (b) feces, cecal content (CC), terminal ileum, Peyer’s patches, mesenteric lymph nodes, and spleen were determined at 96h post-infection. (a, b) Data shown comprise four independent experiments (WT n=14, Crtam−/− n=13). Bars represent the geometric mean ± standard error. n.s. = not significant. A significant change (Mann-Whitney-U) relative to WT control is indicated by * (P ≤ 0.05), ** (P ≤ 0.01), or *** (P ≤ 0.001). (c) Overall pathology score of cecum collected from STm-infected WT or Crtam−/− mice (WT n=5, Crtam−/− n=5). PMN = polymorphonuclear leukocytes. The grey region indicates moderate to severe inflammation.
Whereas the 96-hour timepoint corresponds to the peak of inflammation during Salmonella infection in WT mice (10, 27), the cecum of Crtam−/− mice at 96 hours showed lower levels of inflammation (Figure 2c). Taken together, our results are consistent with previous findings that intestinal inflammation boosts Salmonella colonization (38, 40, 43), and that gut T cells are important to amplify the immune response during Salmonella infection (44). Furthermore, our data demonstrates that Crtam−/− mice are more resistant to Salmonella colonization.
CRTAM impacts the Th17 response during Salmonella infection.
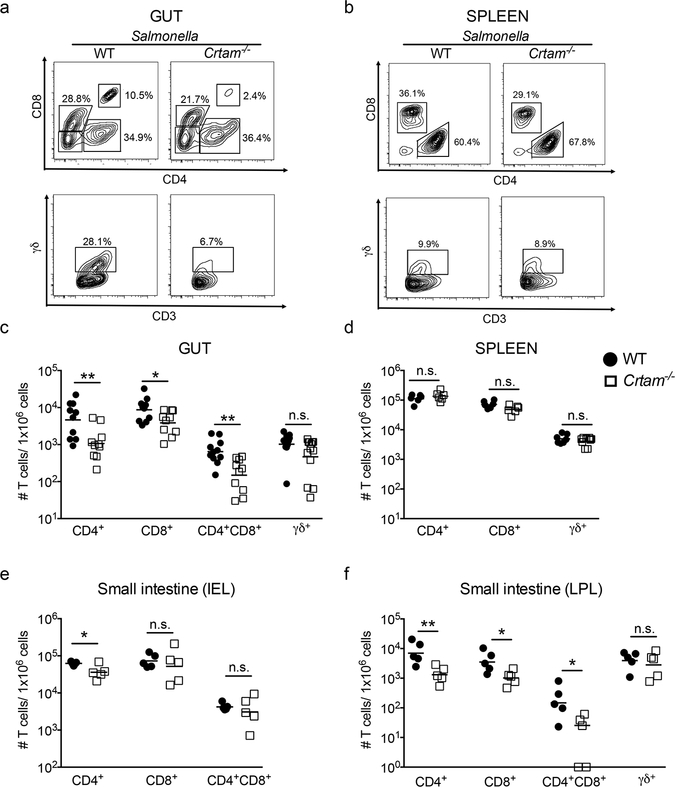
As we discussed earlier, the CRTAM-CADM1 interaction has a major impact on the residency and maintenance of T cells, including Th17 cells, in the gut mucosa. Previous studies have shown that the microbiota shapes Th17 responses in the gut (26), and we have shown that Th17 responses play a central role during Salmonella infection (10). In light of these findings, we analyzed the subpopulations of T cells in the gut and in the spleen of WT and Crtam−/− mice 96 hours after Salmonella infection. We observed that the percentage of some T cell subpopulations (CD4+CD8α/α+ and TCRγδ+) is reduced in the gut of Crtam−/− mice during Salmonella infection (Figure 3a). By contrast, there was no difference in the percentage of TCRγδ+ T cells in the spleen of WT and Crtam−/− mice infected with Salmonella (Figure 3b). When we calculated the frequencies of T cell subpopulations (CD4+, CD8+, CD4+CD8α/α+ and TCRγδ+), we found a significant reduction of the CD4+, CD8+, CD4+CD8α/α+ T cell subsets, and a trend towards a reduction of the TCRγδ+ subset, in the gut of Crtam−/− mice (Figure 3c). No differences among T cell subsets were observed in the spleen (Figure 3d). As Salmonella exploits T cell-mediated inflammatory responses, these results track with our findings of a reduced Salmonella burden in the gut and no reduction in the spleen (Figures 2a and 2b).
Figure 3. T cells in the gut of WT and Crtam−/− mice during Salmonella infection.
T cell subsets (gated on live, CD3+ cells) from the (a) gut or (b) spleen were analyzed by flow cytometry. (a, b) Representative contour plots of CD4+, CD8+ and CD4+CD8+ cells (upper panel) and TCRγδ+ cells (lower panel) obtained from S. Typhimurium-infected WT or Crtam−/− mice are shown. (c, d) Frequency of each T cell subpopulation per million live cells obtained from the (c) gut or (d) spleen of S. Typhimurium-infected WT (black circles) or Crtam−/− (white squares) mice (n=7–9 mice per group). Data shown comprise two independent experiments. (e, f) Cells from small intestine were fractionated into (e) intraepithelial lymphocytes (IEL) and (f) lamina propria lymphocytes (LPL). T cell subsets were analyzed by flow cytometry and frequency of each T cell subpopulation per million live cells was determined. Data represent the mean ± standard error. n.s. = not significant. A significant difference (t-test) is indicated by * (P ≤ 0.05) or ** (P ≤ 0.01).
To determine whether the reduction in T cell subpopulations in Crtam−/− mice could be ascribed to a specific intestinal region and/or cell fraction, we analyzed the same T cell subsets from the small intestine, cecum, and colon compartments after separation of intraepithelial lymphocytes (IEL) and lamina propria lymphocytes (LPL). We found that, in the small intestine of infected Crtam−/− mice, CD4+ T cells were significantly reduced in both the IEL and LPL fractions, and that the CD8+ and CD4+CD8α/α+ T cell subsets were reduced in the LPL fraction (Figures 3e and 3f). In the infected cecum, we observed a significant reduction in CD4+ and CD8+ T cells in the IEL fraction, whereas no significant changes were observed in the cecal LPL fraction or in the colon IEL and LPL fractions (Supplementary figure 1a-d). Collectively, these data demonstrate that, during Salmonella infection, mice lacking CRTAM exhibit lower frequencies of T cells in the gut, in particular in the small intestine LPL (CD4+, CD8+, CD4+CD8α/α+) and in the cecum IEL (CD4+, CD8+).
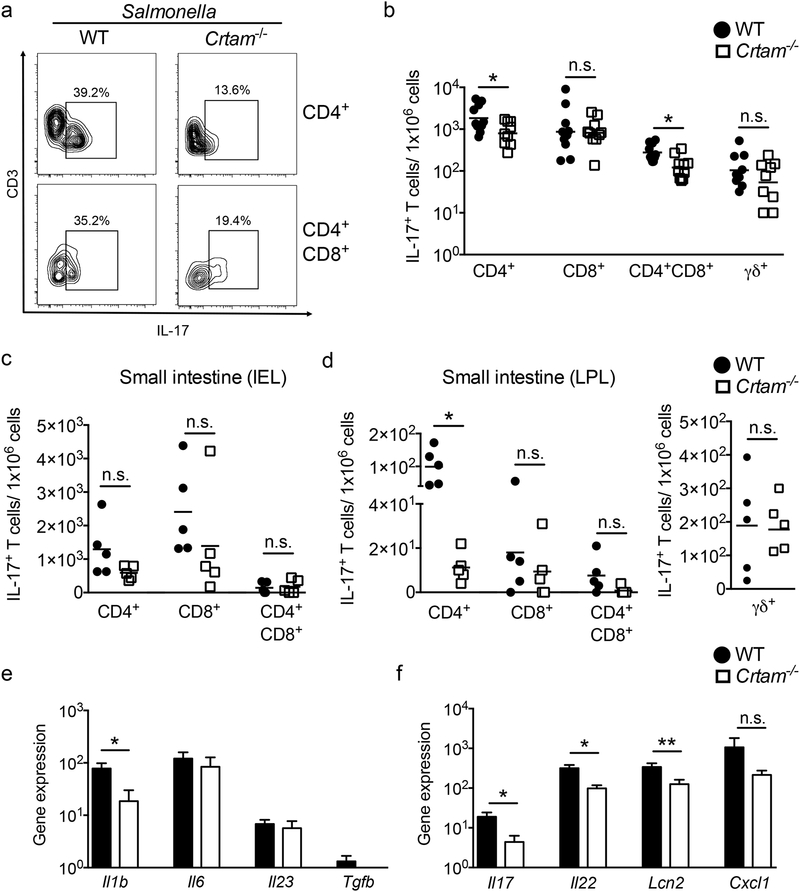
Next, we analyzed the IL-17-producing T cells in the gut of WT and Crtam−/− mice 96 hours after Salmonella infection. Crtam−/− mice exhibited a significantly lower percentage and frequency of IL-17A-producing CD4+ and CD4+CD8α/α+ T cells, but not of IL-17-producing CD8+ or TCRγδ+ T cells (Figures 4a and 4b). As we observed a more profound reduction of T cells in the small intestine of Crtam−/− mice during Salmonella infection, we determined the frequency of IL-17-producing T cells in this compartment. While we found a general reduction of all IL-17-producing T cell subsets in the small intestine, only IL-17-producing CD4+ T cells from the LPL fraction were significantly reduced (Figures 4c and 4d).
Figure 4. IL-17 production during Salmonella infection in WT and Crtam−/− mice.
Immune cells from the gut of infected mice were isolated, then stimulated with PMA and ionomycin in the presence of brefeldin-A. After 6 hours of stimulation, cells were stained for intracellular IL-17, then analyzed by flow cytometry. (a) Representative contour plots of CD3+CD4+IL-17+ and CD3+CD4+CD8+IL-17+ cells obtained from the gut of S. Typhimurium-infected WT (n=12) or Crtam−/− (n=11) mice are shown. (b) Frequency of IL-17-producing cells per million live cells was calculated for each T cell subset. Data shown comprise three independent experiments. (c, d) Cells from the small intestine were fractionated into (c) intraepithelial lymphocytes (IEL) and (d) lamina propria lymphocytes (LPL), then treated as mentioned above. Frequency of IL-17-producing cells per million live cells was calculated for each indicated T cell subset. (b, c, d) Each black circle (WT) or white square (Crtam−/−) represents a mouse, and bars represent the average of each group. (e) Relative expression levels (qPCR) of Il1b, Il6, Il23, and Tgfb in the cecum of WT (black bars, n=8) or Crtam−/− (white bars, n=11) mice at 96h post-S. Typhimurium infection, compared to uninfected controls. (f) Relative expression levels (qPCR) of Il17, Il22, Lcn2, and Cxcl1 in the cecum of WT (black bars, n=10) or Crtam−/− (white bars, n=8) mice at 96h post-S. Typhimurium infection, compared to uninfected controls. Expression of Actb was used as a housekeeping control. Data represent the mean ± standard error. Data shown comprise three independent experiments. n.s. = not significant. A significant difference (t-test) is indicated by * (P ≤ 0.05) or ** (P ≤ 0.01).
In addition to IL-17-producing T cells, we determined whether CRTAM also influences the abundance of Th1 and T regulatory (Treg) cells during Salmonella infection. Although we found a small but insignificant increase in the frequency of IFNγ-producing T cells in Crtam−/− mice (Supplementary Figure 1e), we did not observe differences in the frequency of Tregs (CD4+, CD25+, FoxP3+ T cells) or IL-10-producing Tregs (Supplementary Figure 1f, g). Overall, our analysis of cells from the entire gut, and from the cecum, colon, and small intestine individually, suggest that during Salmonella infection CRTAM globally impacts the levels of particular T cell subsets (CD4+, CD8+, CD4+CD8α/α+), and of IL-17-producing T cells (Figure 3, Figure 4a–d, and Supplementary figure 1). Moreover, our findings indicate that these alterations are more profound in the lamina propria of the small intestine.
A possible explanation for the lower frequency of IL-17-producing T cells in Crtam−/− mice is a reduced expression of Th17-polarizing cytokines (IL-1β, IL-6, IL-23, and TGFβ (Reviewed in (45)). Although we did not detect differences in the expression of these cytokine genes under homeostatic conditions (data not shown), we found that expression of Il1b was significantly reduced in the absence of CRTAM during infection (Figure 4e). We also evaluated the expression of the Th17 cytokine genes Il17 and Il22, as well as of two genes whose expression is upregulated in response to these cytokines: the CXC chemokine CXCL-1 (Cxcl1), and the antimicrobial protein lipocalin-2 (Lcn2) (46). In agreement with our data pointing to diminished Th17 responses in Crtam−/− mice, each of these genes was highly upregulated in the cecum during Salmonella infection, but was lower in the cecum of infected Crtam−/− mice (Figure 4f).
Collectively, these data indicate that CRTAM favors the retention of T cells in the gut during Salmonella infection, resulting in an overall increased frequency of gut T cells, and in particular of IL-17-producing T cells. The increased gut T cell responses exacerbate intestinal inflammation, creating an environment that enhances Salmonella colonization of the gut (38–40, 43). As we recovered similar Salmonella numbers in the spleen, our data suggest that Crtam−/− mice exhibit sufficient inflammation to limit Salmonella dissemination. However, due to the lower frequency of T cells in the gut, Crtam−/− mice have a significantly reduced inflammatory response. Because Salmonella exploits inflammation to thrive in the intestine, Crtam−/− mice are thus less permissive to maximal gut colonization by Salmonella. As such, we hypothesized that Salmonella’s ability to outcompete the microbiota would be impaired in Crtam−/− mice.
CRTAM-mediated responses favor the expansion of Proteobacteria in the gut during infection.
During Salmonella infection, intestinal inflammation results in a significant reduction of microbial diversity, dominated by an expansion of Proteobacteria, the phylum to which Salmonella belongs (28, 37, 39, 40). We next tested whether the composition of the gut microbiota differed during Salmonella infection of WT and Crtam−/− littermate co-housed mice. Consistent with earlier findings, we observed low bacterial diversity in Salmonella-infected WT mice (Figure 5a). By contrast, bacterial diversity was significantly increased in Crtam−/− mice upon Salmonella infection (Figure 5a), and the intersample community structure was different in mice lacking CRTAM (Figure 5b, weighted UniFrac).
Figure 5. Gut microbiota composition in WT and Crtam−/− mice following Salmonella infection.
WT and Crtam−/− mice (from the microbiota analysis in Figure 1) were treated with streptomycin, then infected 24h later with wild-type S. Typhimurium (STm). Fecal samples were collected 96h post-infection for DNA sequence analysis. (a) Alpha-diversity (Shannon index) of the gut microbiota from WT (black circles) or Crtam−/− (white squares) mice. (b) PCoA (principal coordinate analysis) plot based on the unweighted (left panel) or weighted (right panel) UniFrac metric. (c) Bar chart of phylum relative abundance within each sample. (d, e) Relative abundance (as a fraction of 1) of eubacterial taxa that exhibited a statistically significant change in Crtam−/− mice relative to WT mice; either (d) diminished relative abundance or (e) increased relative abundance (ND, not detected); n per group is indicated in the figure. For (c-e), only taxa present at >0.1% relative abundance in at least one sample were considered. (a, d, e) A significant (Mann-Whitney-U) change relative to WT control is indicated by * (P ≤ 0.05) or ** (P ≤ 0.01).
At the phylum level, we observed the expected clear predominance of Proteobacteria in infected WT mice (Figure 5c). However, the relative abundance of Proteobacteria was significantly reduced in infected Crtam−/− mice, and a concomitant relative expansion of phylum Firmicutes was detected (Figure 5c). Further analysis showed that the relative abundance of the family Enterobacteriaceae was higher in infected WT mice when compared with infected Crtam−/− mice (Figure 5d), which is consistent with the higher Salmonella burden observed by colony-forming unit assessment of the feces and cecal content from infected WT mice (Figure 2b).
Although the relative abundance of phylum Firmicutes as a whole was significantly elevated among Salmonella-infected Crtam−/− mice, the Firmicutes sub-taxa varied from sample to sample. Nevertheless, members of family Lachnospiraceae (genus Coprococcus) and of family Streptococcaceae (genus Streptococcus) were significantly elevated in Crtam−/− mice upon Salmonella infection (Figure 5e). Collectively, these results suggest that CRTAM plays an important role in the interplay between the host and the gut microbiota, which in turn shapes the host response and microbial composition during Salmonella infection.
The microbiota shapes T cell responses and CRTAM expression during infection.
The gut microbiota sets the immunological tone of the intestine through a number of feedback loops that exist between members of the microbiota and the mucosal immune system (47). To determine whether the observed differences in the gut microbiota of WT and Crtam−/− mice influence gut T cell responses, we transplanted germ-free mice (GF) with a suspension prepared from the feces of either WT or Crtam−/− littermate mice. Mice that received the fecal transplant (ex-germ-free mice) were then infected with Salmonella or mock, according to the experimental design shown in Figure 6a.
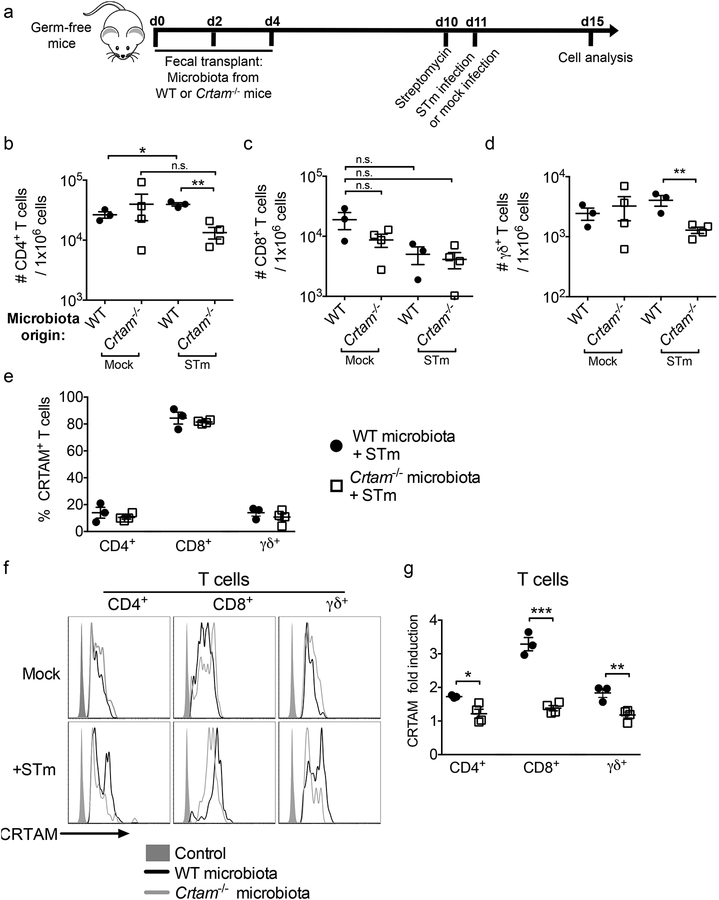
Figure 6. The gut microbiota influences gut T cell abundance in ex-germ-free mice.
Germ-free mice were transplanted with the gut microbiota from either WT (black circles) or Crtam−/− (white squares) mice by oral gavage of a fecal suspension, and then infected with wild-type S. Typhimurium according to the scheme presented in (a). (b-d) Flow cytometry analysis was performed to determine the absolute number of T cell subpopulations in the gut of the aforementioned microbiota-transplanted mice that were either mock-infected or infected. Frequency of (b) CD4+ T cells, (c) CD8+ T cells, or (d) TCRγδ+ T cells per million live cells are shown. (e) Percentage of CRTAM+ cells among either CD4+, CD8+ or TCRγδ+ T cells in the gut of ex-germ-free mice that had been transplanted with microbiota from WT mice or Crtam−/− mice, then infected with S. Typhimurium for 96h. (f) Representative histograms showing expression of CRTAM on the surface of either CD4+, CD8+ or TCRγδ+ T cells from ex-germ-free mice transplanted with microbiota from WT (black histogram) or Crtam−/− (grey histogram) mice, which were mock-infected (upper panel) or infected with S. Typhimurium (lower panel). (g) Fold induction of CRTAM expression (as a ratio of mean fluorescence intensity) on T cell subpopulations from the gut of mice in (f) (infected transplanted mice divided by mock-infected transplanted mice; WT microbiota = black circles, Crtam−/− microbiota = white squares). Data shown are representative of two independent experiments. Each circle or square represents a mouse, and bars represent the average (n=3–4 per group). n.s. = not significant. (b-d, g) A significant difference (t-test) is indicated by * (P ≤ 0.05), ** (P ≤ 0.01), or *** (P ≤ 0.001).
We did not detect significant differences in the frequency of T cells within the different cell subpopulations isolated from the gut of mock-infected, ex-germ-free mice transplanted with the fecal microbiota from either WT or Crtam−/− mice (Figure 6b–d). However, when the ex-germ-free mice were infected with Salmonella, we observed significant differences between the fecal transplant recipients. At 96 h post-infection, we observed a significant increase in the frequency of CD4+ T cells in the gut of ex-germ-free mice transplanted with the gut microbiota from WT mice, but not in ex-germ-free mice transplanted with the gut microbiota from Crtam−/− mice (Figure 6b). In the same mice, we did not observe significant differences in CD8+ T cells (Figure 6c), but there was a significant difference in TCRγδ+ T cells (Figure 6d).
Because the frequency of CD4+ T cells was reduced in the gut of Salmonella-infected mice that had been transplanted with the Crtam−/− mouse microbiota, and because CRTAM contributes to T cell adhesion to the gut mucosa, we next analyzed whether the gut microbiota influences the expression of CRTAM on the surface of T cells. To this end, the gut microbiota did not influence the percentage of T cells expressing CRTAM in ex-germ-free mice (Figure 6e). Moreover, during mock infection, the fecal transplant alone did not influence the magnitude of CRTAM expression in any of the T cell subpopulations analyzed (Figure 6f, upper panel). However, during Salmonella infection, we observed the appearance of CD4+ T cells, CD8+ T cells, and TCRγδ+ T cells that express high levels of CRTAM, but only in the mice transplanted with WT mouse microbiota (Figure 6f, bottom panel). When we calculated the fold induction of CRTAM expression, we observed that it was significantly higher in T cells of mice that were transplanted with the WT mouse microbiota (Figure 6g). Together, these data suggest that the gut microbiota influences CRTAM expression levels on T cells during Salmonella infection. Therefore, our study provides evidence of crosstalk between the gut microbiota and cells expressing CRTAM, wherein the presence of CRTAM influences the gut microbiota composition, and the gut microbiota impacts the expression of CRTAM.
Because the gut microbiota influences the expression of CRTAM, we next evaluated IL-17 production during Salmonella infection of ex-germ-free mice transplanted with WT or Crtam−/− mouse gut microbiota. When we analyzed IL-17 expression by different T cell subpopulations, we observed a reduction in the percentage of CD4+ T cells that produce IL-17 (Figure 7a). We also observed a significant difference in the frequency of IL-17-producing CD4+ T cells (Figure 7b), but not of IL-17-producing CD8+ or TCRγδ+ T cells (Figure 7c and 7d). As we discussed earlier, Salmonella exploits IL-17-dependent inflammation to thrive in the gut, and we recovered fewer Salmonella CFUs in Crtam−/− mice, where IL-17 production was lower (Figure 2). We thus hypothesized that the lower levels of IL-17-producing CD4+ T cells in ex-germ-free mice transplanted with Crtam−/− mouse gut microbiota would lead to a reduction in Salmonella colonization.
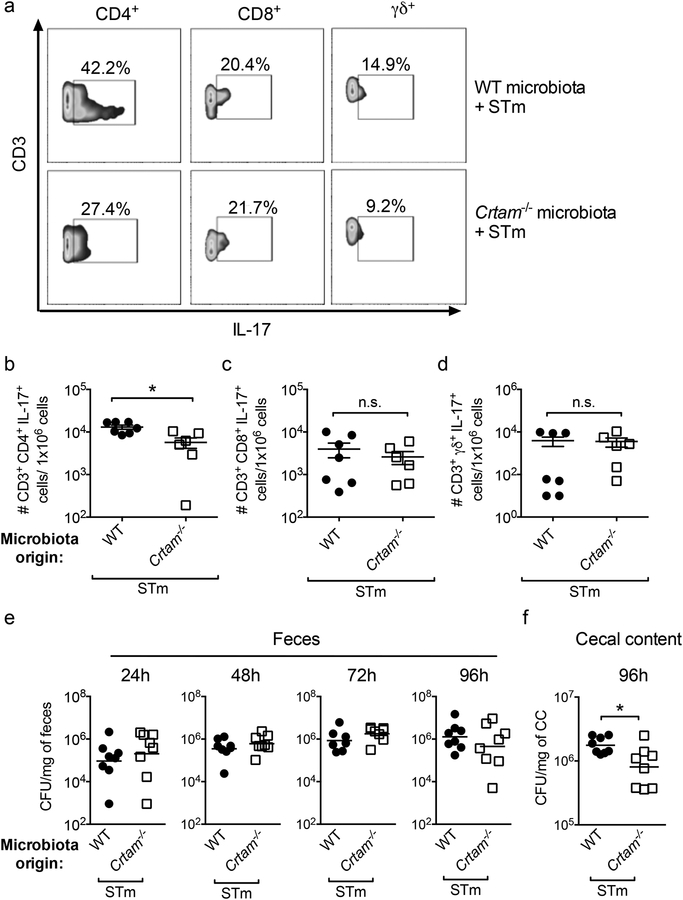
Figure 7. The gut microbiota of Crtam−/− mice influences IL-17 production in ex-germ-free mice.
Germ-free mice were transplanted with the gut microbiota from either WT (black circles) or Crtam−/− (white squares) mice by oral inoculation of a fecal suspension, then infected with wild-type S. Typhimurium (STm). IL-17-producing T cells were then analyzed by flow cytometry. (a) Representative contour plots of IL17-producing CD4+, CD8+ or TCRγδ+ T cells from S. Typhimurium-infected (96h post-infection), ex-germ-free mice previously transplanted with WT (upper panel) or Crtam−/− (lower panel) microbiota. (b-d) Frequency per million live cells of the indicated IL-17-producing T cell subpopulations was determined. (e, f) Colony-forming units (CFU) in (e) feces at the indicated time points, and (f) cecal content at 96h post-infection, were determined. Data shown comprise two independent experiments. Each circle or square represents a mouse, and bars represent the average of each group (n=6–7 per group). n.s. = not significant. A significant difference (t-test for IL-17-producing cells or Mann-Whitney-U for bacterial CFU) is indicated by * (P ≤ 0.05).
Comparable to what we observed during Salmonella infection of WT and Crtam−/− mice (Figure 2a), we found no difference in fecal Salmonella numbers at 24, 48 and 72 h post-infection of ex germ-free mice transplanted with WT or Crtam−/− gut microbiota (Figure 7e). In line with our hypothesis, Salmonella colonization was significantly reduced in the cecal content of ex germ-free mice transplanted with Crtam−/− microbiota (Figure 7f). All in all, this finding is in agreement with our results showing that absent (in Crtam−/− mice) or reduced (in ex-germ-free mice transplanted with Crtam−/− mouse microbiota) expression of CRTAM results in a reduced frequency of gut T cells and in a diminished Th17 response during Salmonella infection.
Discussion
The gut microbiota influences a wide variety of host responses, including the immune response. In the gut, members of the microbiota have been found to regulate a variety of T cell responses (26, 31, 32); for instance, the gut microbiota regulates expression of the homing receptor GPR15, which contributes to the recruitment of T regulatory cells in specific regions of the intestinal mucosa (33). As a previous study showed that CRTAM contributes to the retention of T cells in the gut (15), we sought to determine whether CRTAM impacts the composition of the gut microbiota. Although we did not observe differences in the expression of antimicrobial peptides (lipocalin-2, RegIIIγ, calprotectin) between WT and Crtam−/− mice in homeostatic conditions (data not shown), we did detect changes in the gut microbiota of Crtam−/− mice. In the absence of CRTAM, there was a relative increase of phylum Firmicutes with a concomitant reduction of Tenericutes. In a deeper taxonomic analysis, we found an increased relative abundance of genus Bacteroides (phylum Bacteroidetes) in Crtam−/− mice. It is known that some species of Bacteroides modulate the immune response in the gut; for example, polysaccharide A (PSA), a capsular polysaccharide from Bacteroides fragilis, has been shown to promote immunological tolerance (48). Additionally, lipopolysaccharide (LPS) from Bacteroides dorei was shown to inhibit the immunostimulatory activity of E. coli LPS (49). The genus Parabacteroides, a member of the family Porphyromonadaceae (phylum Bacteroidetes), was also increased in Crtam−/− mice relative to WT mice. Members of this latter family have been shown to provide protection against Salmonella-induced colitis (50). Taken together, we hypothesized that changes in the gut microbiota of Crtam−/− mice could influence colonization resistance to pathogens.
Colonization resistance comprises the set of mechanisms by which the microbiota prevents pathogen colonization (reviewed in (36)). These mechanisms involve direct interactions between the microbiota and the pathogen, as well as microbiota-dependent stimulation and development of the mucosal immune system (36). Nevertheless, pathogens have evolved mechanisms to subvert colonization resistance and cause disease. On this front, the intestinal pathogen Salmonella employs specific virulence factors to trigger inflammation in the intestinal mucosa. This inflammatory environment alters the intestinal lumen and favors growth of Salmonella over its competitors by generating new nutrients for the pathogen and by creating a generally inhospitable environment for competing commensal microbes (38–41, 43). Thus, intestinal inflammation confers a competitive advantage to Salmonella over the gut microbiota.
Increased gut Th17 responses exacerbate intestinal inflammation, creating an environment that enhances Salmonella colonization (38–40, 43). Here, we show that CRTAM is required to induce a robust Th17 response during infection and to promote intestinal colonization by Salmonella. The changes observed in the gut microbiota composition of Crtam−/− mice, together with the decreased Th17 responses, likely contribute to the Crtam−/− mouse’s increased colonization resistance to Salmonella. Of note, both WT and Crtam−/− mice exhibited similar levels of SFB in the gut, both in homeostatic conditions and during Salmonella infection (data not shown). Due to the lower frequencies of T cells in the gut, Crtam−/− mice exhibit a significantly reduced inflammatory response (Figure 4), which hampers Salmonella’s ability to outcompete the microbiota (Figure 5), and results in reduced gut colonization by the pathogen (Figure 2b). Nevertheless, our data demonstrate that Crtam−/− mice still exhibit sufficient inflammation to limit Salmonella dissemination, as we observed comparable Salmonella colonization of the spleen in WT and Crtam−/− mice (Figure 2b).
Analysis of the gut microbiota in WT mice during infection revealed a bloom of Proteobacteria, the phylum to which Salmonella belongs. By contrast, the relative abundance of phylum Firmicutes predominated in Crtam−/− mice. These results are in line with previous studies showing that Salmonella requires an inflammatory environment to outcompete gut anaerobes, including Firmicutes, and thus establish maximal colonization of the gut (39, 40, 42). In our study, we found that the relative abundance of Coprococcus and Streptococcus was increased in infected Crtam−/− mice (Figure 5e). These members of the gut microbiota have been implicated in mediating colonization resistance and in the regulation of immune responses. For example, an increase in the relative abundance of Coprococcus is associated with resistance to Campylobacter and Salmonella infection in humans (51), and Coprococcus also plays a role in colonization resistance against Clostridium difficile infection (52). In addition, Coprococcus is an active producer of short-chain fatty acids (SCFAs) (53), metabolites that are known to have anti-inflammatory effects (54) and to provide colonization resistance to Salmonella by maintaining a hypoxic epithelium (55). For genus Streptococcus, some members isolated from the human small intestine have been shown to have immunomodulatory properties, including the ability to downregulate NF-кB in human intestinal cells (56). Therefore, the higher relative abundance of Coprococcus and Streptococcus in Crtam−/− mice likely helps the host to control Salmonella infection by enhancing colonization resistance. These findings strongly suggest that CRTAM plays an important role in the interplay between the host and the microbiota, which in turn shapes the host response and microbial composition during Salmonella infection.
After transplantation of the gut microbiota into germ-free mice, and then infection with Salmonella, we observed a reduction in the frequencies of T cells (in particular, CD4+ T cells) in mice transplanted with the Crtam−/− mouse gut microbiota in comparison to mice transplanted with the WT mouse gut microbiota (Figure 6). Moreover, we found that ex-germ-free mice transplanted with the gut microbiota from WT mice exhibited significantly more CRTAM on the T cell surface, which was accompanied by increased production of IL-17 upon infection and by higher Salmonella colonization (Figures 6 and 7). By contrast, these responses were not observed in ex-germ-free mice transplanted with the Crtam−/− microbiota. Our data thus indicate that the gut microbiota modulates CRTAM expression, and consequently IL-17 production. These results are consistent with a prior study showing that naïve Crtam−/− CD4+ T cells, when activated and cultured in Th17 differentiation media, produce less IL-17 than WT CD4+ T cells (24). Another finding that could be linked to differences in the gut microbiome is the reduced expression of the Il1b gene in Crtam−/− mice during infection (Figure 4). IL-1β is a cytokine required for Th17 cell differentiation (57), and IL-1β production is influenced by the gut microbiome (58). Taken together, these results suggest a reciprocal modulation between the gut microbiota, CRTAM expression, and the host response to Salmonella infection in the gut.
The mechanism by which CRTAM is induced in vivo remains to be elucidated. Prior analysis of the Crtam promoter has shown that the transcription factor AP-1, an important regulator of inflammatory responses, positively regulates Crtam expression (59). As microbiota-dependent AP-1 activity is required for the expression of genes including Il1b, Il6, and Reg3g (60), it is possible that the gut microbiota regulates Crtam expression in gut T cells by a similar mechanism. In accordance with this idea, during infection, we observed a microbiota-dependent induction of CRTAM expression, which was enhanced in ex-germ-free mice transplanted with microbiota from WT mice, but not from Crtam−/− mice (Figure 6). Future studies are needed to identify the underlying mechanisms, including the specific microbe(s) that modulate CRTAM expression in gut T cells.
Our study is consistent with prior studies showing that CRTAM is a key regulator of the host response against gut infections. While CRTAM is protective against Toxoplasma gondii (15), CRTAM’s role is controversial during Citrobacter rodentium infection. One report showed that CRTAM protects the host against this intestinal pathogen (24), while another showed that CRTAM does not have a role (15). In contrast to these studies, we show that CRTAM confers higher susceptibility to Salmonella gut infection. Because expression of CRTAM shapes the gut microbiota and influences T cell responses, the outcome observed in each of these infections is different, as it also depends on the pathogen’s susceptibility to CRTAM-dependent responses. In this sense, we have shown that CRTAM is required for the induction of a robust Th17 response in the gut during Salmonella infection. The enhanced Th17 response, in turn, exacerbates gut inflammation that enables Salmonella to outcompete the gut microbiota and to grow to high levels.
Collectively, our study demonstrates a previously unknown reciprocal interplay between CRTAM and the gut microbiota, which ultimately impacts colonization resistance to Salmonella.
Supplementary Material
Key points:
The presence of CRTAM impacts the gut microbiota during homeostasis and infection
CRTAM-mediated responses enhance Salmonella gut infection
Transplanted gut microbiota modulates CRTAM expression in ex-germ-free mice
Acknowledgements
We would like to thank Matthew Rolston at the UC Davis School of Medicine Host-Microbe Systems Biology Core for processing samples for sequencing, and Dean Nguyen for his help performing some experiments.
Funding
Work in the Raffatellu lab was supported by Public Health Service Grants AI114625, AI121928, AI126277, and AI126465, by the Chiba University-UCSD Center for Mucosal Immunology, Allergy, and Vaccines, and by the UCSD Department of Pediatrics. M.R. also holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. APL was partly supported by a UC MEXUS-CONACYT award, and by a Mucosal Immunology Studies Team (MIST) Scholar Award in Mucosal Immunity.
Footnotes
Competing interests
The authors declare no conflict of interest.
References
- 1.Agace WW 2008. T-cell recruitment to the intestinal mucosa. Trends Immunol 29: 514–522. [DOI] [PubMed] [Google Scholar]
- 2.Rocha B 2007. The extrathymic T-cell differentiation in the murine gut. Immunol Rev 215: 166–177. [DOI] [PubMed] [Google Scholar]
- 3.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, and Markowitz M. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med 200: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaser JB, Morton-Kute L, Berger SR, Weber J, Siegal FP, Lopez C, Robbins W, and Landesman SH. 1985. Recurrent Salmonella typhimurium bacteremia associated with the acquired immunodeficiency syndrome. Ann Intern Med 102: 189–193. [DOI] [PubMed] [Google Scholar]
- 5.Gordon MA 2008. Salmonella infections in immunocompromised adults. J Infect 56: 413–422. [DOI] [PubMed] [Google Scholar]
- 6.Tee W, and Mijch A. 1998. Campylobacter jejuni bacteremia in human immunodeficiency virus (HIV)-infected and non-HIV-infected patients: comparison of clinical features and review. Clin Infect Dis 26: 91–96. [DOI] [PubMed] [Google Scholar]
- 7.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, and de Waal Malefyt R. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8: 950–957. [DOI] [PubMed] [Google Scholar]
- 8.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, Sudo K, Nakae S, Sasakawa C, and Iwakura Y. 2009. Differential roles of interleukin-17A and −17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30: 108–119. [DOI] [PubMed] [Google Scholar]
- 9.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, and Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14: 282–289. [DOI] [PubMed] [Google Scholar]
- 10.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, and Bäumler AJ. 2008. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med 14: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, and Qian Y. 2011. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat Immunol 12: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 12.Zimmerman NP, Vongsa RA, Wendt MK, and Dwinell MB. 2008. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis 14: 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner N, Lohler J, Kunkel EJ, Ley K, Leung E, Krissansen G, Rajewsky K, and Muller W. 1996. Critical role for beta7 integrins in formation of the gut-associated lymphoid tissue. Nature 382: 366–370. [DOI] [PubMed] [Google Scholar]
- 14.Schon MP, Arya A, Murphy EA, Adams CM, Strauch UG, Agace WW, Marsal J, Donohue JP, Her H, Beier DR, Olson S, Lefrancois L, Brenner MB, Grusby MJ, and Parker CM. 1999. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J Immunol 162: 6641–6649. [PubMed] [Google Scholar]
- 15.Cortez VS, Cervantes-Barragan L, Song C, Gilfillan S, McDonald KG, Tussiwand R, Edelson BT, Murakami Y, Murphy KM, Newberry RD, Sibley LD, and Colonna M. 2014. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J Exp Med 211: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy J, Vicari AP, Saylor V, Zurawski SM, Copeland NG, Gilbert DJ, Jenkins NA, and Zlotnik A. 2000. A molecular analysis of NKT cells: identification of a class-I restricted T cell-associated molecule (CRTAM). J Leukoc Biol 67: 725–734. [DOI] [PubMed] [Google Scholar]
- 17.Arase N, Takeuchi A, Unno M, Hirano S, Yokosuka T, Arase H, and Saito T. 2005. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int Immunol 17: 1227–1237. [DOI] [PubMed] [Google Scholar]
- 18.Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, Keller AM, Joffre O, Zelenay S, Nye E, Le Moine A, Faure F, Donckier V, Sancho D, Cerundolo V, Bonnet D, and Reis e Sousa C. 2010. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med 207: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galibert L, Diemer GS, Liu Z, Johnson RS, Smith JL, Walzer T, Comeau MR, Rauch CT, Wolfson MF, Sorensen RA, Van der Vuurst de Vries AR, Branstetter DG, Koelling RM, Scholler J, Fanslow WC, Baum PR, Derry JM, and Yan W. 2005. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J Biol Chem 280: 21955–21964. [DOI] [PubMed] [Google Scholar]
- 20.Sakisaka T, and Takai Y. 2004. Biology and pathology of nectins and nectin-like molecules. Curr Opin Cell Biol 16: 513–521. [DOI] [PubMed] [Google Scholar]
- 21.Mizutani K, Kawano S, Minami A, Waseda M, Ikeda W, and Takai Y. 2011. Interaction of nectin-like molecule 2 with integrin alpha6beta4 and inhibition of disassembly of integrin alpha6beta4 from hemidesmosomes. J Biol Chem 286: 36667–36676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boles KS, Barchet W, Diacovo T, Cella M, and Colonna M. 2005. The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood 106: 779–786. [DOI] [PubMed] [Google Scholar]
- 23.Murakami Y 2005. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci 96: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh JH, Sidhu SS, and Chan AC. 2008. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell 132: 846–859. [DOI] [PubMed] [Google Scholar]
- 25.Belkaid Y, and Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157: 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, and Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, and Bäumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, and Raffatellu M. 2014. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40: 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, and Glockner FO. 2013. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42: D643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, and Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazmanian SK, Liu CH, Tzianabos AO, and Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118. [DOI] [PubMed] [Google Scholar]
- 32.Round JL, and Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 107: 12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M, Sarpel U, Rifkin DB, Xu R, and Littman DR. 2013. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340: 1456–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CG, Palfreyman RW, Nielsen LK, Cooper MA, Morrison M, Hansbro PM, and Hugenholtz P. 2016. Genomic characterization of the uncultured Bacteroidales family S24–7 inhabiting the guts of homeothermic animals. Microbiome 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenney PT, and Pamer EG. 2015. From Hype to Hope: The Gut Microbiota in Enteric Infectious Disease. Cell 163: 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sassone-Corsi M, and Raffatellu M. 2015. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J Immunol 194: 4081–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, and Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, von Mering C, Robinson MD, Stecher B, and Hardt WD. 2013. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 14: 641–651. [DOI] [PubMed] [Google Scholar]
- 39.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, and Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, and Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, and Baumler AJ. 2016. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534: 697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, and Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, and Bäumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. MBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, and Bäumler AJ. 2008. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun 76: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanov II, Zhou L, and Littman DR. 2007. Transcriptional regulation of Th17 cell differentiation. Semin Immunol 19: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Lopez A, Behnsen J, Nuccio SP, and Raffatellu M. 2016. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol 16: 135–148. [DOI] [PubMed] [Google Scholar]
- 47.Thaiss CA, Zmora N, Levy M, and Elinav E. 2016. The microbiome and innate immunity. Nature 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 48.Mazmanian SK, Round JL, and Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453: 620–625. [DOI] [PubMed] [Google Scholar]
- 49.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, Group DS, and Xavier RJ. 2016. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165: 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, and Finlay BB. 2011. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6: e20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kampmann C, Dicksved J, Engstrand L, and Rautelin H. 2016. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin Microbiol Infect 22: 61 e61–68. [DOI] [PubMed] [Google Scholar]
- 52.Perez-Cobas AE, Artacho A, Ott SJ, Moya A, Gosalbes MJ, and Latorre A. 2014. Structural and functional changes in the gut microbiota associated to Clostridium difficile infection. Front Microbiol 5: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pryde SE, Duncan SH, Hold GL, Stewart CS, and Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217: 133–139. [DOI] [PubMed] [Google Scholar]
- 54.Roy CC, Kien CL, Bouthillier L, and Levy E. 2006. Short-chain fatty acids: ready for prime time? Nutr Clin Pract 21: 351–366. [DOI] [PubMed] [Google Scholar]
- 55.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, and Bäumler AJ. 2016. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 19: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, Renault P, Blottiere HM, Daniel C, and Delorme C. 2014. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol 80: 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, Ma L, Watowich SS, Jetten AM, Tian Q, and Dong C. 2009. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30: 576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seo SU, Kamada N, Munoz-Planillo R, Kim YG, Kim D, Koizumi Y, Hasegawa M, Himpsl SD, Browne HP, Lawley TD, Mobley HL, Inohara N, and Nuñez G. 2015. Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity 42: 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valle-Rios R, Patino-Lopez G, Medina-Contreras O, Canche-Pool E, Recillas-Targa F, Lopez-Bayghen E, Zlotnik A, and Ortiz-Navarrete V. 2009. Characterization of CRTAM gene promoter: AP-1 transcription factor control its expression in human T CD8 lymphocytes. Mol Immunol 46: 3379–3387. [DOI] [PubMed] [Google Scholar]
- 60.Mukherji A, Kobiita A, Ye T, and Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153: 812–827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.