Summary
Bacteria in soils encounter redox-active compounds, such as phenazines, that can generate oxidative stress, but the mechanisms by which different species tolerate these compounds are not fully understood. Here, we identify two transcription factors, ActR and SoxR, that play contrasting yet complementary roles in the tolerance of the soil bacterium Agrobacterium tumefaciens to phenazines. We show that ActR promotes phenazine tolerance by proactively driving expression of a more energy-efficient terminal oxidase at the expense of a less-efficient alternative, which may affect the rate at which phenazines abstract electrons from the electron transport chain and thereby generate reactive oxygen species. SoxR, on the other hand, responds to phenazines by inducing expression of several efflux pumps and redox-related genes, including one of three copies of superoxide dismutase and five novel members of its regulon that could not be computationally predicted. Notably, loss of ActR is far more detrimental than loss of SoxR at low concentrations of phenazines, and also increases dependence on the otherwise functionally redundant SoxR-regulated superoxide dismutase. Our results thus raise the intriguing possibility that the composition of an organism’s electron transport chain may be the driving factor in determining sensitivity or tolerance to redox-active compounds.
Keywords: phenazines, oxidative stress, gene expression regulation, Agrobacterium tumefaciens, transcription factors, oxidation-reduction
Abbreviated summary:
In this study, we show that tolerance of redox-active phenazines (Phz) in A. tumefaciens is modulated by two transcription factors with contrasting yet complementary modes of action. ActR promotes phenazine tolerance by proactively upregulating energy-efficient cytochrome o oxidase (Cyo) at the expense of less-efficient cytochrome d oxidase (Cyd), and is important even at low concentrations of phenazines. Conversely, SoxR upregulates several proteins in response to phenazines, but only becomes essential at high phenazine concentrations.
Introduction
Soil-dwelling bacteria commonly secrete redox-active secondary metabolites, including a variety of phenazine derivatives (Turner and Messenger, 1986; Mavrodi et al., 2010). Such metabolites can reversibly accept electrons from and donate electrons to a variety of substrates in a process known as redox cycling. This property enables them to benefit their producers by promoting iron acquisition (Hernandez et al., 2004; Wang et al., 2011), anaerobic survival (Wang et al., 2010; Glasser et al., 2014), and biofilm development (Ramos et al., 2010). At the same time, because of their ability to abstract electrons from redox enzymes (such as those in the respiratory electron transport chain (ETC)), reduced quinones, and other cellular reductants, and subsequently to transfer those electrons to oxygen, these metabolites can also suppress competing microbes by generating reactive oxygen species (ROS) or interfering with respiration (Hassan and Fridovich, 1980; Baron and Rowe, 1981; Baron et al., 1989; Hassett et al., 1992). Phenazine producers can promote crop productivity in a phenazine-dependent manner, in part by suppressing fungal plant pathogens (Audenaert et al., 2002; Chin-A-Woeng et al., 2003; Khare and Arora, 2011), and phenazine biosynthesis has been observed on plant roots colonized by these bacteria (Chin-A-Woeng et al., 1998; Séveno et al., 2001; LeTourneau et al., 2018). Phenazines have also been shown to accumulate in dryland agricultural soils and the rhizosphere of cereal crops (Mavrodi et al., 2012). These observations imply that tolerance of phenazines may be an important fitness determinant in certain soils and plant-associated environments. Yet aside from their producers, little is known about whether and how other soil bacteria tolerate these compounds.
Previous studies on phenazine toxicity and tolerance in bacteria have largely focused on the effects of the phenazine pyocyanin (PYO) on either generic model organisms like Escherichia coli or opportunistic human pathogens like Staphylococcus aureus. The latter frequently co-infects chronic wounds and cystic fibrosis patients along with the PYO producer Pseudomonas aeruginosa (Noto et al., 2017), which is itself an opportunistic human pathogen found in soils and aquatic environments (Green et al., 1974; Römling et al., 1994; Alonso et al., 1999). PYO is the most toxic among the phenazines commonly secreted by P. aeruginosa, presumably because of its high reactivity with oxygen (Meirelles and Newman, 2018). A comparative study of E. coli and P. aeruginosa suggested that antioxidant defenses and slower redox cycling of PYO in P. aeruginosa contribute to its resistance to PYO, whereas PYO-sensitive E. coli experiences higher levels of ROS due to faster redox cycling (Hassett et al., 1992). In addition, a study based on co-evolution with P. aeruginosa revealed that mutations in an efflux pump regulator, a porin, and a flavodoxin NADP+ reductase enzyme can modulate PYO tolerance in E. coli (Khare and Tavazoie, 2015). S. aureus, on the other hand, can escape PYO toxicity to some extent by adopting a non-respiring phenotype, as well as by expressing genes involved in quinone detoxification (Noto et al., 2017), while certain non-pathogenic staphylococci are inherently resistant to PYO due to expression of a PYO-insensitive variant of cytochrome bd oxidase (Voggu et al., 2006). However, whether these mechanisms are at work in other bacteria that naturally tolerate phenazines is unknown, making it difficult to predict how these molecules may shape bacterial communities in soil.
To address this question, we set out to identify genes that are necessary for phenazine tolerance in Agrobacterium tumefaciens, a common gram-negative soil bacterium and plant pathogen. We chose A. tumefaciens because it is relatively tolerant of phenazines, compared to several other genetically tractable bacterial species (An et al., 2006; Costa et al., 2015). We employed PYO as our primary model compound because it is the most toxic among the phenazines that have been used to enhance crop productivity (Audenaert et al., 2002; Chin-A-Woeng et al., 2003; Khare and Arora, 2011; Meirelles and Newman, 2018). Using a forward genetic screen, we found two transcriptional regulators that both play crucial roles in tolerance of PYO, but at different concentrations and through contrasting yet complementary mechanisms. Our results point toward possible broader themes underlying tolerance of phenazines and other redox-active secondary metabolites, such as the importance of controlling their interactions with the respiratory ETC, which could have important consequences for understanding and predicting ecological impacts of these widely-produced molecules.
Results
Transposon mutagenesis reveals genes necessary for PYO tolerance
To identify genes necessary for wild-type (WT) levels of PYO tolerance, we performed random transposon insertion mutagenesis in A. tumefaciens NT1 using a mariner-based transposon that confers kanamycin resistance (Chiang and Rubin, 2002). Kanamycin-resistant mutants were screened for PYO sensitivity using a colorimetric 96-well plate assay. As WT or PYO-tolerant mutants grow to a high cell density in static liquid cultures containing 100 µM PYO, they consume the oxygen in the medium and concurrently reduce PYO from its blue oxidized form to its colorless reduced form. Conversely, PYO-sensitive mutants are unable to grow to a high enough density to fully effect the color change from blue to clear within 48 hrs (Fig. 1A). Putative PYO-sensitive mutants were verified by comparing their growth in liquid cultures to WT across a range of PYO concentrations, from 0 to 200 µM. From roughly 5000 screened mutants, 12 (~0.2%) proved to be disproportionately sensitive to PYO; that is, while a few of these mutants exhibited growth defects even without PYO, the ratio of their growth (i.e. optical density after 24 hrs) with PYO to their growth without PYO was much lower than it was for WT (Fig. 1B).
Fig. 1.
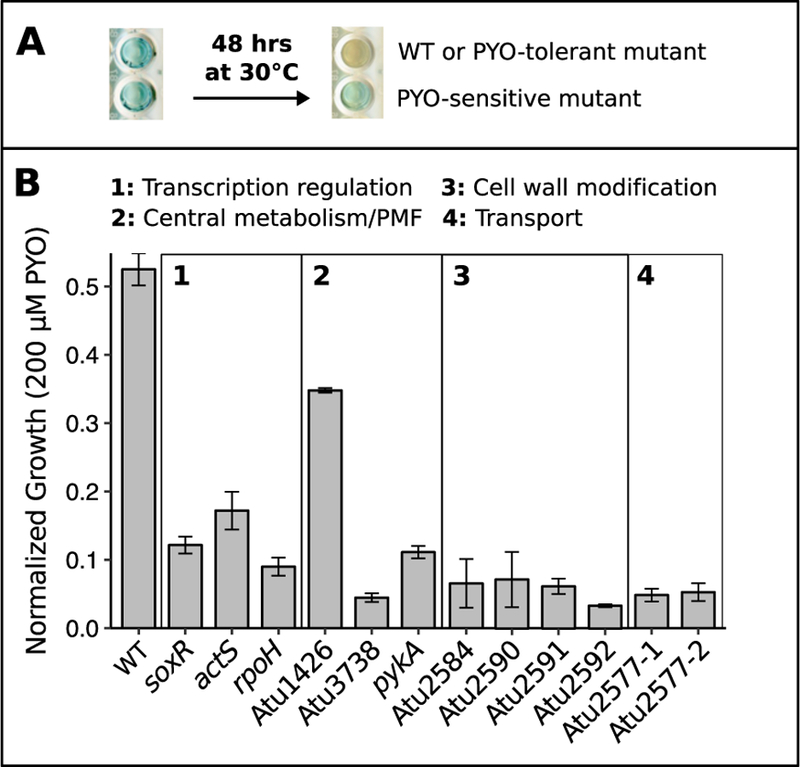
Transposon mutagenesis reveals genes necessary for tolerance of PYO.
A) An example 96-well plate demonstrating the colorimetric strategy used to identify PYO-sensitive transposon mutants. As WT or PYO-tolerant mutants grow to a high density over 48 hrs in static cultures, cellular reductants transform PYO from its blue oxidized form to its colorless reduced form. Wells containing PYO-sensitive mutants that cannot grow to a high density remain blue. B) Growth of WT and the 12 PYO-sensitive transposon mutants after 24 hrs in the presence of 200 µM PYO. Cultures were either in or approaching stationary phase at this time point, and growth was normalized to growth in parallel cultures without PYO. Differences between WT and mutants were generally greater at 200 µM PYO than at lower concentrations. Mutants are named by the gene containing the transposon insertion and categorized by predicted function (see Table S1 for further details). Error bars represent standard deviations of biological replicates (n = 3).
The locus of the transposon insertion for each of the 12 verified PYO-sensitive mutants was determined using arbitrary PCR. Three of the insertions were in genes encoding transcriptional regulators, three were in genes putatively related to carbon metabolism or maintenance of protonmotive force, four were likely related to cell wall modification, and the remaining two were in a single gene, Atu2577, encoding an ABC transporter (Table S1). This list of genes important for PYO tolerance is not comprehensive, as in A. tumefaciens NT1, approximately 24,000 transposon insertion mutants would need to be screened to achieve 99% confidence of having disrupted every gene in the genome. Nevertheless, we were particularly interested in following up on the transcriptional regulators identified in our screen, as the genes they regulate could reveal broader insights about mechanisms that are important for PYO tolerance. Specifically, we focused on the mutants with insertions in actS, the sensor in the actSR two-component sensor-response system, and soxR, which has previously been implicated in the oxidative stress response of A. tumefaciens (Eiamphungporn et al., 2006) and as a sensor of redox-active antibiotics in P. aeruginosa and Streptomyces coelicolor (Dietrich et al., 2006; Dietrich et al., 2008; Singh et al., 2013). We did not pursue the third transcriptional regulator, rpoH, as the transposon mutant exhibited a growth defect without PYO (Fig. S1A).
∆actR and ∆soxR mutants are differentially sensitive to diverse redox-active small molecules
To characterize the roles of actSR and soxR in protecting against PYO, we first generated in-frame deletions of actS, actR, and soxR via allelic replacement (Morton and Fuqua, 2012a). Both ∆actS and ∆actR were similarly sensitive to PYO (Fig. S1B); thus, we used ∆actR for all further experiments as ActR is the transcriptional regulator in this two-component system. Intriguingly, ∆actR and ∆soxR displayed strikingly different profiles of sensitivity to PYO. ∆actR is significantly more sensitive to PYO than WT at concentrations as low as 10 µM; conversely, ∆soxR is no more sensitive to PYO than WT up to at least 100 µM, and in fact grows better than WT at up to 50 µM PYO, yet its growth is severely inhibited by 200 µM PYO (Fig. 2A). Complementation by inducing expression of actR or soxR in ∆actR or ∆soxR, respectively, from a plasmid rescued PYO tolerance in each mutant (Fig. S1C).
Fig. 2.
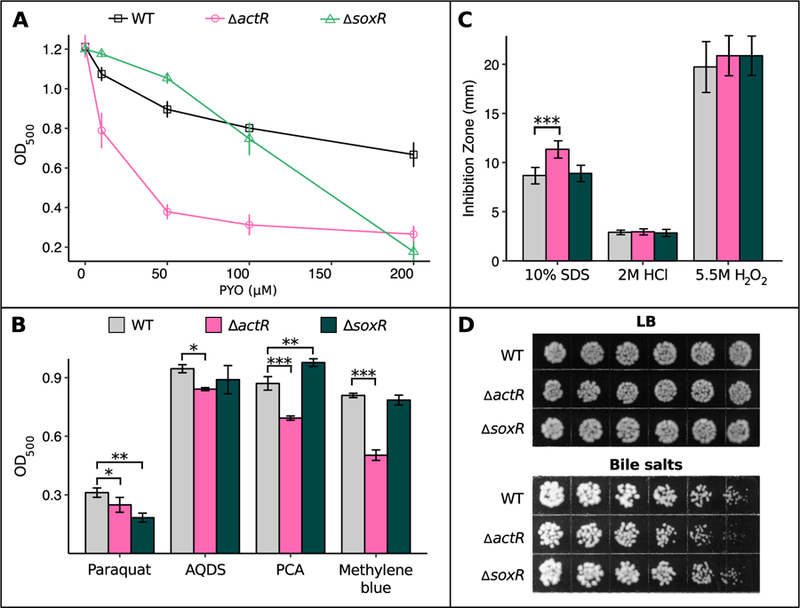
∆actR and ∆soxR mutants exhibit differential sensitivity to redox-active small molecules.
A) Growth of WT, ∆actR, and ∆soxR after 24 hrs in the presence of different concentrations of PYO, measured by optical density at 500 nm (n = 3). B) Growth of WT, ∆actR, and ∆soxR after 24 hrs in the presence of 20 mM paraquat, 10 mM AQDS, 500 µM PCA, or 200 µM methylene blue (n = 3). For each molecule, the chosen concentration was the lowest tested dose at which growth of either ∆actR or ∆soxR was statistically significantly different from WT. C) Diameter of growth inhibition zone around a disk infused with 10% SDS, 2 M HCl, or 5.5 M H2O2 (n ≥ 6). The measurements represent the diameter of the zone of clearing minus the diameter of the disk itself. D) Growth of WT, ∆actR, and ∆soxR on agar plates containing either plain LB or a concentration gradient (low-high, left to right) of bile salts (up to 2%). Images are representative of eight biological replicates. In A and B, cultures were in stationary phase at the reported time point. In B and C, * p < 0.05, ** p < 0.01, *** p < 0.001 (in B, linear regression with dummy variable coding using WT as the reference group; in C, Kruskal-Wallis test followed by pairwise Wilcox rank sum test with the Benjamini-Hochberg procedure for controlling the false discovery rate). Error bars in A-C represent standard deviations of biological replicates.
Extending beyond PYO, we also investigated the relative sensitivity of ∆actR and ∆soxR to diverse redox-active molecules spanning a range of standard reduction potentials (Eo’ vs. that of the normal hydrogen electrode) from −446 mV (paraquat) to +11 mV (methylene blue) (Fig. S2) (Wang et al., 2010). For each molecule, we tested increasing concentrations until we found a dose at which the growth of either ∆actR or ∆soxR was statistically significantly different from WT. Similar to the results with PYO (Eo’ −40 mV), ∆actR was more sensitive than either WT or ∆soxR to anthraquinone-2,6-disulfonate (AQDS, Eo’ −184 mV), phenazine-1-carboxylic acid (PCA, Eo’ −114 mV), and methylene blue, whereas ∆soxR was no more sensitive than WT to these molecules at the tested concentrations (Fig. 2B). However, ∆soxR was more sensitive than ∆actR to paraquat. Finally, to determine whether actR and soxR specifically affect sensitivity to redox-active molecules, we tested whether these mutants were more sensitive than WT to a variety of other stresses, including detergents (SDS or bile salts), low pH (HCl), hydrogen peroxide, the membrane stressor EDTA, and osmotic stressors (myo-inositol or NaCl). Neither ∆actR nor ∆soxR was noticeably more sensitive than WT to any of these stresses (Fig. 2C and Fig. S3), with the exception that ∆actR was more sensitive to detergents (Fig. 2C, D).
SoxR regulates multiple previously overlooked genes
To understand why ActR and SoxR differentially affect sensitivity to phenazines, we began to characterize the genetic mechanisms by which SoxR contributes to PYO tolerance. It is known from studies in other bacteria that redox-cycling drugs activate SoxR by directly oxidizing its [2Fe-2S] cluster (Dietrich and Kiley, 2011; Gu and Imlay, 2011; Singh et al., 2013), and that oxidized SoxR binds to a conserved “SoxR box” sequence in the promoter regions of target genes (Dietrich et al., 2008). Using this sequence, SoxR in A. tumefaciens has variously been predicted to regulate up to four putative major facilitator superfamily transporters (Atu0942, Atu2361, Atu4895, and Atu5152) along with an operon containing a superoxide dismutase (Atu4583, a.k.a. sodBII), ferredoxin (Atu4582), and a putative flavin reductase (Atu4581) (Eiamphungporn et al., 2006; Dietrich et al., 2008; Novichkov et al., 2013). However, of these computationally predicted members of the SoxR regulon, SoxR-dependent expression has previously only been confirmed for Atu5152 (Eiamphungporn et al., 2006) and sodBII (Saenkham et al., 2007). SoxR has also been shown to autoregulate by binding to a SoxR box in its own promoter (Eiamphungporn et al., 2006). To validate the other computationally predicted members of the SoxR regulon, we performed quantitative reverse-transcription PCR (qRT-PCR) on WT and ∆soxR cultures treated with either 100 µM PYO or a solvent control for 20 minutes. We expected true SoxR-regulated genes to be highly expressed only in PYO-treated WT. This was indeed the case for all but one of the genes computationally predicted to be SoxR-regulated based on the presence of a SoxR box-containing promoter (Fig. 3A, non-bolded genes). Atu4895 alone was not upregulated by PYO, suggesting that it is not in fact regulated by SoxR (Fig. S4A).
Fig. 3.
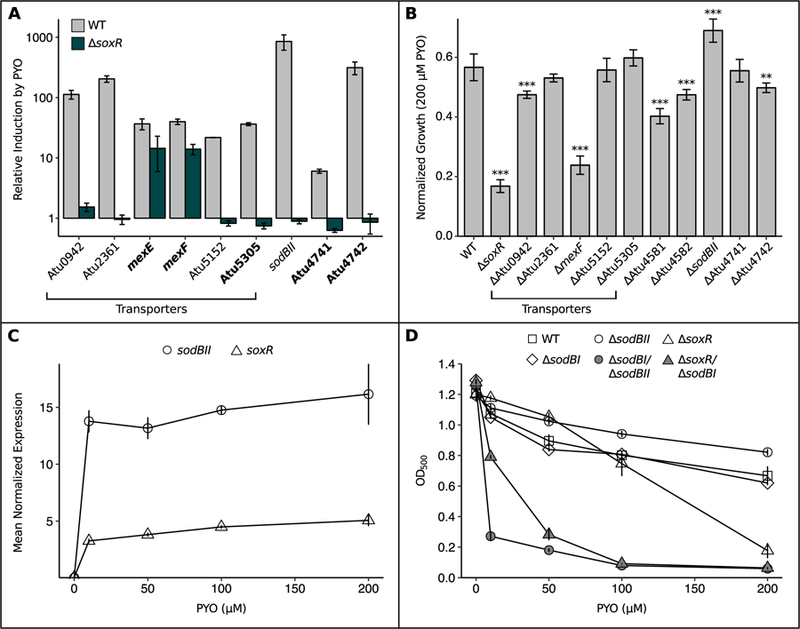
SoxR protects A. tumefaciens against PYO by upregulating functionally redundant superoxide dismutase, transporters and redox-related genes.
A) qRT-PCR validation of putative members of the SoxR regulon, showing that induction of these genes upon a 20 min exposure to 100 µM PYO was partially or fully abrogated in the ∆SoxR mutant (n = 3). Bolded genes lack a SoxR box and thus could not be identified as SoxR-regulated by computational approaches in earlier studies. qRT-PCR validation was not performed for Atu4581 and Atu4582, as these genes are predicted to be co-transcribed with sodBII (Mao et al., 2009). Expression levels were normalized to the housekeeping gene rpoD, and induction was calculated as the expression in the presence of PYO divided by the expression without PYO. B) Growth of single knockout mutants for members of the SoxR regulon after 24 hrs in the presence of 200 µM PYO, normalized to growth of parallel cultures without PYO. ** p < 0.01, *** p < 0.001 (n = 3, linear regression with dummy variable coding using WT as the reference group). C) Normalized expression levels of sodBII and soxR in WT after 20 min exposure to different concentrations of PYO (n = 3). Expression levels were determined by qRT-PCR and normalized to rpoD. D) Growth of WT, ∆sodBI, ∆sodBII, ∆soxR, and the ∆sodBI/∆sodBII and ∆soxR/∆sodBI mutants after 24 hrs in the presence of different concentrations of PYO (n ≥ 3). Error bars in all panels represent standard deviations of biological replicates. In B and D, cultures were in stationary phase at the reported time point.
The discrepancy between the presence of a SoxR box upstream of Atu4895 and its lack of response to PYO led us to ask whether there might conversely be “cryptic” members of the SoxR regulon that lack a SoxR box but exhibit a SoxR-dependent response to PYO. Given that all of the qRT-PCR-validated, SoxR box-containing members of the SoxR regulon exhibited large fold changes in WT upon PYO treatment (Fig. 3A, non-bolded genes), we expected that any cryptic member(s) of the SoxR regulon would exhibit a similarly strong response to PYO. We therefore performed an RNA-seq experiment to identify genes that are highly induced by PYO in WT. Based on the fold changes of soxR itself and the already-validated members of the SoxR regulon, we defined candidate cryptic members of the SoxR regulon as those genes with a log2 fold change > 4 in PYO-treated cultures relative to the solvent-treated control (Table S2). We then tested the resulting seven candidates for a SoxR-dependent transcriptional response by performing qRT-PCR under the four conditions described above (WT + PYO, WT – PYO, ∆soxR + PYO, and ∆soxR – PYO). Remarkably, we found that the PYO-mediated induction of three of the seven candidates (Atu4741, Atu4742, and Atu5305) was completely abolished in the absence of SoxR, while the PYO-mediated induction of two others (Atu2482 and Atu2483, annotated as mexE and mexF respectively) appeared to partially depend on SoxR (Fig 3A, bolded genes). Using the motif scanning program FIMO (Grant et al., 2011), we confirmed the lack of a typical SoxR box upstream of these genes. Thus, we have expanded the known set of genes with SoxR-dependent regulation in A. tumefaciens to include five genes that could not have been found with previously applied computational methods.
Among the five identified cryptic members of the SoxR regulon, Atu4741 is particularly interesting as it is a σ54-dependent Fis family transcriptional regulator, and hence could potentially be involved in regulating the other cryptic members. Although Atu4741 is annotated as acoR due to its similarity to the acoR gene in Bacillus subtilis, A. tumefaciens does not possess annotated homologs of the acetoin catabolism genes that are regulated by AcoR in B. subtilis, suggesting that Atu4741 plays a different regulatory role in A. tumefaciens. Atu4742, located just upstream of Atu4741, is predicted to encode a small (108 amino acid) hypothetical protein containing tetratricopeptide repeats, which often mediate protein-protein interactions (Zeytuni and Zarivach, 2012); this, along with its proximity to Atu4741, suggests that the two may interact. Atu5305 is annotated as a permease belonging to the major facilitator family of transporters, and thus could be involved in transporting PYO out of the cell. Finally, mexE and mexF encode homologs of the cytoplasmic membrane protein and membrane fusion protein components, respectively, of the MexEF-OprN efflux pump originally discovered in P. aeruginosa. In the latter, MexEF-OprN has been shown to transport chloramphenicol, fluoroquinolones, and a precursor to a quorum sensing signal (Köhler et al., 1997; Lamarche and Déziel, 2011; Llanes et al., 2011); it is also known to be induced by nitrosative (Fetar et al., 2011) or disulfide stress (Fargier et al., 2012). To our knowledge, SoxR-dependent expression of mexEF has not previously been reported in other organisms; rather, in P. aeruginosa, mexEF is regulated at least in part by MexT, a LysR family transcriptional activator (Köhler et al., 1999).
Functional redundancy rationalizes the ∆soxR phenotype
Altogether, the SoxR regulon in A. tumefaciens contains five putative efflux pumps (Atu0942, Atu2361, mexEF, Atu5152, and Atu5305), a putative flavin reductase (Atu4581), ferredoxin (Atu4582), superoxide dismutase (sodBII), an uncharacterized transcriptional regulator (Atu4741), and a hypothetical protein (Atu4742). To investigate the contributions of each of these genes to PYO tolerance, we generated in-frame deletions and challenged the resulting mutants with 200 µM PYO. Only five mutants were significantly more sensitive to 200 µM PYO than WT, and no single mutant was as sensitive as ∆soxR itself, suggesting considerable functional redundancy among the SoxR regulon (Fig. 3B). Surprisingly, ∆sodBII was actually less sensitive to PYO than WT. This was counterintuitive, given that superoxide has been proposed to be a toxic byproduct of PYO treatment (Hassan and Fridovich, 1980; Hassett et al., 1992; Rada and Leto, 2013; Managò et al., 2015).
We wondered whether functional redundancy might explain both the surprising phenotype of ∆sodBII and the fact that ∆soxR only becomes sensitive to PYO at a relatively high dose. First, we confirmed via qRT-PCR that SoxR strongly induces expression of its regulon even at 10 µM PYO, using soxR itself and sodBII as representative examples (Fig. 3C). Thus, the tolerance of ∆soxR to low levels of PYO is in spite of considerable transcriptional differences between WT and ∆soxR under these conditions, suggesting that loss of SoxR may be compensated by functionally redundant genes outside of its regulon. Indeed, besides SoxR-regulated SodBII, A. tumefaciens possesses two other superoxide dismutases, SodBI and SodBIII (Saenkham et al., 2007). SodBI is constitutively highly expressed at all stages of growth, whereas SodBIII is primarily expressed during stationary phase (Saenkham et al., 2007). We therefore asked whether loss of SodBI would sensitize ∆sodBII or ∆soxR to lower concentrations of PYO. Indeed, while ∆sodBI itself was not sensitive to PYO, ∆sodBII/∆sodBI and ∆soxR/∆sodBI were significantly more sensitive than WT not only to high doses of PYO, but also to low doses, unlike the ∆sodBII and ∆soxR single mutants (Fig. 3D). Thus, the functions of SodBI and SodBII appear to be largely redundant at the tested concentrations of PYO, and this functional redundancy is a key factor in the tolerance of ∆soxR to low concentrations of PYO.
Loss of ActR causes constitutive dysregulation of the cytochrome o and d ubiquinol oxidases
Having rationalized why SoxR only becomes essential at high concentrations of PYO, we next sought to understand why the ∆actR mutant is hypersensitive to PYO. ActR homologs (known as ArcA, PrrA, RegA, or RoxR) are present in diverse proteobacteria and have been well-studied in several species (Elsen et al., 2004; Gralnick et al., 2005; Wong et al., 2007; Fernández-Piñar et al., 2008; Gao et al., 2008; Park et al., 2013; Lunak and Noel, 2015). In general, they regulate elements of the respiratory ETC in response to changes in cellular redox balance (for example, during transitions from aerobiosis to anaerobiosis), although the specific elements and directionality of regulation vary across species (Tseng et al., 1996; Swem and Bauer, 2002; Elsen et al., 2004; Fernández-Piñar et al., 2008; Gao et al., 2008). In Sinorhizobium medicae, which belongs to the same taxonomic order as A. tumefaciens, ActR regulates a glutathione S-transferase, cytochrome oxidase components, and an assimilatory nitrate reductase (Fenner et al., 2004), while in A. tumefaciens, ActR is known to co-regulate expression of nitrite reductase and pseudoazurin, an electron donor to the denitrification pathway, during anaerobic growth (Baek et al., 2008). However, to our knowledge, the ActR regulon has not been comprehensively defined in A. tumefaciens. Thus, to investigate how ActR contributes to PYO tolerance during aerobic growth, we performed RNA-seq to compare the transcriptomes of WT and ∆actR both with and without 100 µM PYO treatment. Among the genes that were up- or downregulated by PYO in each strain (comparing WT + PYO vs. WT – PYO, or ∆actR + PYO vs. ∆actR - PYO), there was a strong correlation between the fold changes in WT and ∆actR; in fact, there were no genes that were strongly up- or downregulated by PYO in WT but not in ∆actR (Fig. 4A). This suggested that ActR does not regulate a specific transcriptional response to PYO. However, we noted that among the genes that were up- or downregulated by PYO, the magnitude of the fold changes was often greater in ∆actR than in WT (such genes lie above the y = x line in Fig. 4A), possibly indicating that ∆actR was experiencing more stress than WT.
Fig. 4.
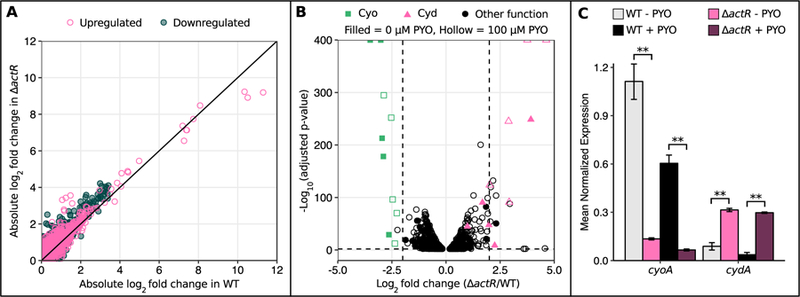
Expression of cytochrome o oxidase and cytochrome d oxidase is dysregulated in ∆actR.
A) Plot of the absolute values of log2 fold changes in gene expression upon 100 µM PYO treatment in WT against absolute values of log2 fold changes in gene expression upon 100 µM PYO treatment in ∆actR. Only genes that were statistically significantly differentially expressed (adjusted p-value < 0.01) upon PYO treatment in at least one strain are plotted. Each point represents a single gene. The black line is y = x. B) Volcano plots of RNA-seq data comparing gene expression levels in ∆actR to WT, with either 0 µM PYO or 100 µM PYO. The vertical dashed lines mark a log2 fold change of −2 or 2, and the horizontal dashed line marks an adjusted p-value of 0.01. Only genes with statistical significance below the adjusted p-value cutoff are plotted. C) qRT-PCR data confirming the expression patterns of the cyo and cyd operons in ∆actR vs. WT. The + PYO condition represents treatment with 100 µM PYO. Only cyoA and cydA are shown for brevity, as they are co-transcribed with other members of these operons. Expression levels were normalized to the housekeeping gene rpoD. ** p < 0.01 (Welch’s t-test followed by the Benjamini-Hochberg procedure for controlling the false discovery rate; n = 3). Error bars represent standard deviations of biological replicates.
We then compared the transcriptomic profile of ∆actR to WT either with or without PYO (i.e. ∆actR + PYO vs. WT + PYO, or ∆actR - PYO vs. WT - PYO). Interestingly, several genes related to oxidative stress were upregulated in ∆actR relative to WT only upon PYO treatment, including catalase, ferredoxin I, a putative DNA oxidation protective protein, and genes involved in amino acid metabolism (Table S3). Similar to our above observation, this could imply that PYO exerts greater toxicity in ∆actR. We also found that two sets of genes are strongly dysregulated in ∆actR regardless of PYO treatment: the cyo operon that encodes cytochrome o ubiquinol oxidase (Cyo) is downregulated > 4 fold, while the cyd operon that encodes cytochrome d ubiquinol oxidase (Cyd) is upregulated > 2 fold, with the latter being exaggerated upon PYO treatment (Fig. 4B). These expression patterns were confirmed with qRT-PCR (Fig. 4C). Other elements of the ETC or central metabolism that were upregulated in ∆actR regardless of PYO treatment included succinate dehydrogenase, ubiquinol-cytochrome c reductase, and cbb3 cytochrome c oxidase, along with several genes involved in sugar transport/metabolism, amino acid metabolism, fatty acid metabolism, carbon oxidation or the tricarboxylic acid (TCA) cycle (Table S4). On the other hand, caa3 cytochrome c oxidase, pseudoazurin, and biosynthetic genes for cytochrome c and heme were downregulated in ∆actR (Table S5). While the fold changes for these genes were not as dramatic as for cyo and cyd, the overall pattern suggests that ActR is broadly involved in regulating flux through central metabolic pathways in A. tumefaciens, as it is in E. coli, Salmonella enterica, Pseudomonas putida, and other species (Fernández-Piñar et al., 2008; Evans et al., 2011; Morales et al., 2013; Park et al., 2013).
Downregulation of cytochrome o ubiquinol oxidase sensitizes cells to PYO
Given that the fold changes were most striking for the terminal ubiquinol oxidases, we next asked whether the dysregulation of these genes in ∆actR is causally related to PYO sensitivity. Indeed, a mutant lacking the cyo operon grew similarly to ∆actR during exponential phase across all tested concentrations of PYO (Fig. 5A, left), although it achieved a higher optical density in stationary phase (Fig. 5A, right), possibly due to compensatory upregulation of the cyd operon to levels considerably higher than that in ∆actR itself (Fig. S4B). Interestingly, the ∆actR/∆cyo double mutant is more sensitive to PYO than either ∆actR or ∆cyo alone (Fig. 5A). This result likely arises from a combination of two factors: 1) total loss of cyo is presumably more severe than the downregulation imposed by loss of ActR; and 2) without ActR, expression of genes to compensate for total loss of cyo may be impaired. Conversely, overexpression of the cyo operon in the ∆actR background partially rescued tolerance to PYO, with a greater effect at the lower doses of PYO (Fig. 5B). On the other hand, increased overexpression of the cyd operon from an inducible plasmid-borne promoter in ∆actR did not further sensitize ∆actR to PYO, and in fact conferred a mild benefit at low doses of PYO (Fig. 5B). We therefore hypothesized that cyd may play a compensatory role upon abnormal downregulation of cyo. To further test this hypothesis, we attempted to determine whether a ∆actR/∆cyd double mutant would be more sensitive to PYO than ∆actR alone, but were unable to perform this experiment due to the severe growth defect of this double mutant in liquid culture even without PYO (data not shown). Nevertheless, the cyd overexpression results suggest that the natural upregulation of cyd in ∆actR does not augment its PYO sensitivity, but rather is a compensatory response or else a passive side effect of losing actR (ActR might normally repress cyd during aerobic growth). Overall, ActR-mediated regulation of cyo appears to be important for PYO tolerance, although the fact that overexpressing cyo in ∆actR only partially rescued PYO tolerance suggests that other ActR-regulated genes likely also contribute to the ∆actR phenotype.
Fig. 5.
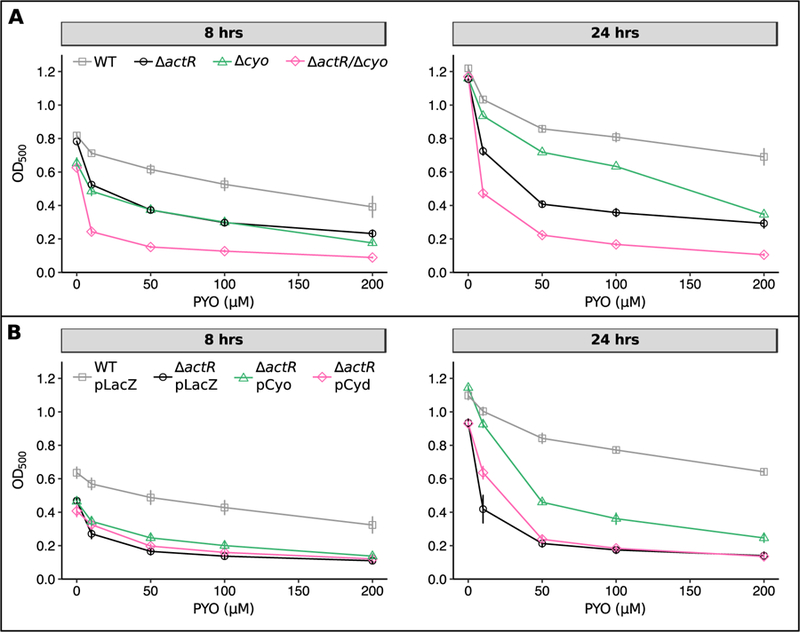
Loss of cytochrome o oxidase, but not overexpression of cytochrome d oxidase, increases sensitivity to PYO.
A) Growth of WT, ∆actR, ∆cyo, and ∆actR/∆cyo after 8 hrs (left) or 24 hrs (right) in the presence of different concentrations of PYO. 8 hrs corresponds to late exponential phase while 24 hrs corresponds to stationary phase. B) Growth of WT pLacZ (vector control for overexpression constructs), ∆actR pLacZ, ∆actR pCyo (overexpression construct for the cyo operon), and ∆actR pCyd (overexpression construct for the cyd operon). Overexpression was induced by adding 1 mM IPTG at the start of the experiment. Growth of ∆actR pCyo is statistically significantly higher than growth of ∆actR pLacZ at both time points and across all concentrations of PYO, except for 0 µM and 200 µM PYO after 8 hrs (p < 0.05, Welch’s t-test followed by the Benjamini-Hochberg procedure for controlling the false discovery rate). All data points are plotted with error bars representing standard deviations of biological replicates (n = 3).
In considering candidates for other genes besides cyo that might contribute to the ∆actR phenotype, we were particularly intrigued by the upregulation of multiple dehydrogenases involved in carbon oxidation and the TCA cycle (Table S4). These enzymes oxidize various substrates while concomitantly reducing NAD+ to NADH. Thus, upregulation of these genes could lead to a higher NADH/NAD+ ratio in the cell. However, upon measuring the NADH/NAD+ ratios in exponentially growing cultures of WT and ∆actR, we found only a slight trend towards a higher ratio in ∆actR, which was not statistically significant (Fig. S5A). We also asked whether ∆actR exhibits increased redox cycling of PYO independently of the action of the terminal oxidases in the ETC. Specifically, we measured the rate of PYO-dependent oxygen consumption by WT and ∆actR in the presence of cyanide, which inhibits the terminal oxidases. This rate has previously been correlated both with the rate of redox cycling of PYO and with PYO toxicity (Hassett et al., 1992), as the apparent oxygen consumption under this condition results from reduced PYO donating electrons to oxygen and thereby generating ROS. Interestingly, we found that WT and ∆actR cultures treated with PYO and cyanide consumed oxygen at nearly identical rates (Fig. S5B), implying similar rates of PYO redox cycling in the two strains. However, this result does not exclude the possibility that the terminal oxidases themselves might influence the rate of PYO redox cycling.
Cellular ATP levels correlate with Cyo expression but not necessarily with PYO sensitivity
To rationalize the link between Cyo levels and phenazine tolerance, we first asked whether loss of ActR alters the rate of aerobic respiration, as Cyd possesses a much higher affinity for oxygen than Cyo (Matsushita et al., 1984; D’mello et al., 1996; Borisov et al., 2011). We measured oxygen consumption rates in exponential-phase cultures of WT and ∆actR using a Clark-type electrode. These measurements revealed that WT and ∆actR actually consume oxygen at similar rates, both with and without PYO treatment (Fig. 6A). However, besides differing in their affinities for oxygen, Cyo and Cyd have different coupling ratios for proton translocation (2 H+/e- vs. 1 H+/e-, respectively) (Calhoun et al., 1993) (Fig. 6B). Hence, given similar rates of oxygen consumption, ∆actR should be less efficient at generating protonmotive force (PMF) than WT. This would be expected to impair PMF-dependent processes, such as ATP synthesis via FoF1-ATP synthase. Indeed, we found that both ∆actR and ∆cyo possess less ATP than WT during exponential growth, supporting the hypothesis that the electron transport chain (ETC) is less efficient without Cyo (Fig. 6C). This difference could potentially explain why ∆actR is also more sensitive to detergents: tolerance of SDS and bile salts typically involves energy-intensive processes like efflux and DNA repair (Nickerson and Aspedon, 1992; Begley et al., 2005). Interestingly, the ATP pools of all three strains decreased upon treatment with PYO, but this difference was only statistically significant in ∆actR. This could suggest that PYO decouples oxidative phosphorylation more effectively in ∆actR, and/or that ∆actR spends more ATP on defenses against PYO toxicity.
Fig. 6.
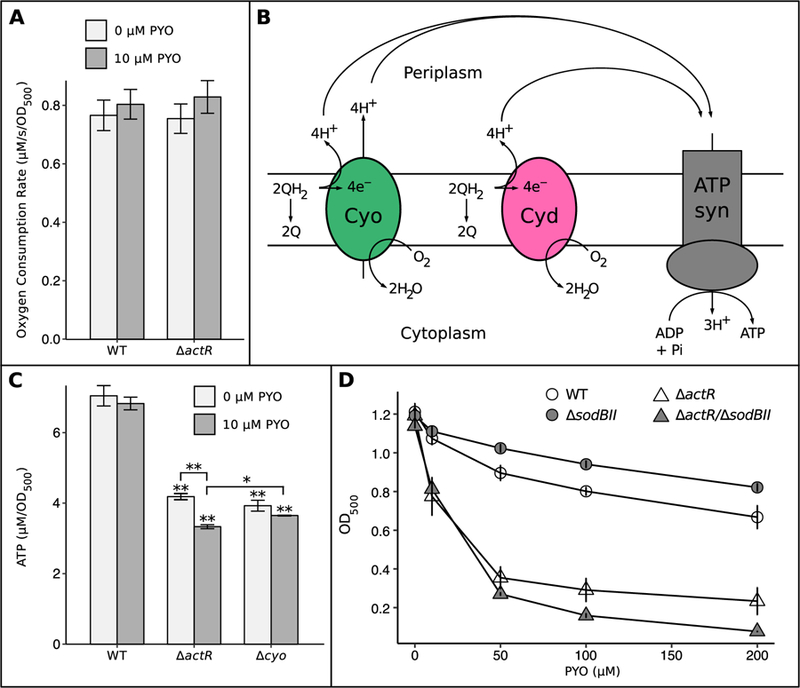
Loss of ActR decreases cellular ATP levels and increases dependence on SoxR-regulated SodBII.
A) Oxygen consumption rates of exponential-phase WT and ∆actR with and without 10 µM PYO (n = 3). B) Simplified cartoon showing the relationship between Cyo, Cyd, and ATP synthase, as well as the different coupling constants of Cyo and Cyd (2H+/e- vs. 1H+/e-, respectively). QH2 = ubiquinol (reduced), Q = ubiquinone (oxidized). C) Bulk ATP levels in exponential phase cultures of WT, ∆actR, and ∆cyoABCD, with and without 10 µM PYO. * p < 0.05, * p < 0.01 (Welch’s t-test followed by the Benjamini-Hochberg procedure for controlling the false discovery rate; n = 3). D) Growth of WT, ∆sodBII, ∆actR, and ∆actR/∆sodBII after 24 hrs in the presence of different concentrations of PYO, showing the increased dependency of ∆actR on SodBII (n ≥ 3). Cultures were in stationary phase at this time point. Data for WT and ∆actR are from Fig. 2A. Error bars in A, B, and D represent standard deviations of biological replicates.
To test whether the rate of oxidative phosphorylation modulates PYO sensitivity, we treated WT and ∆actR with DCCD (N,N’-dicyclohexylcarbodiimide), a classical inhibitor of FoF1-ATP synthase. Both strains compensated for inhibited ATP synthesis by growing more slowly in the presence of DCCD (Fig. S6A), and consequently the bulk ATP levels decreased only slightly (Fig. S6B). Nevertheless, we were intrigued to note that while the DCCD treatment increased sensitivity to PYO in ∆actR, DCCD actually improved tolerance to 10 µM PYO in WT (Fig. S6C). Thus, the link between lower rates of ATP synthesis, smaller ATP pools, and increased PYO sensitivity seems to be specific to the ∆actR genetic background, although it is also possible that ATP only becomes a limiting factor in PYO tolerance below a certain level.
Loss of ActR increases dependence on SodBII-mediated protection against PYO
Taken together, the above evidence suggested that PYO might exert toxic effects more readily in ∆actR than in WT, rather than ∆actR simply being deficient in responding to PYO-induced toxicity. This would be consistent with the fact that i) PYO treatment affected the ATP pool of ∆actR more severely, ii) inhibiting ATP synthesis decreased PYO tolerance in ∆actR but not in WT, and iii) several genes related to oxidative stress were more strongly transcriptionally induced by PYO in ∆actR than in WT. To test this hypothesis directly, we first attempted to measure whether PYO generates ROS more rapidly in ∆actR compared to WT, as superoxide and hydroxyl radical have previously been identified as toxic byproducts of PYO redox cycling (Hassett et al., 1992; Noto et al., 2017). However, we found that technical limitations prevented reliable quantification of intracellular ROS in suspensions of PYO-treated A. tumefaciens (see Experimental Procedures and Fig. S7). We therefore turned instead to a genetic approach to infer the relative toxicity of PYO in ∆actR compared to WT. Specifically, we asked whether the SoxR-regulated, functionally redundant superoxide dismutase SodBII is more important for damage control in ∆actR compared to WT, which could imply greater production of superoxide by PYO in ∆actR. Indeed, while loss of SodBII actually increased growth at all tested concentrations of PYO in the WT genetic background, loss of SodBII further sensitized ∆actR to PYO at concentrations above 10 µM (Fig. 6D). The trend was similar, though weaker, when comparing the effects of losing SodBII in WT versus ∆cyo: the growth advantage conferred by loss of SodBII was smaller for ∆cyo than for WT at PYO concentrations above 10 µM, suggesting increased dependency on SodBII in ∆cyo, albeit to a lesser extent than in ∆actR (Fig. S1D). Thus, the downregulation of cyo in ∆actR may contribute to its increased dependence on SodBII, though other genes are likely also involved. These findings are particularly notable given the evidence that ∆actR and ∆cyo conserve energy less efficiently than WT. Induction of SodBII, along with the rest of the SoxR regulon, may incur significant energy costs associated with protein synthesis, especially considering that SodBII is the second-most highly expressed non-ribosomal gene in the entire A. tumefaciens transcriptome under PYO treatment (Table S6). Given the functional redundancy of SodBII, this cost could be gratuitous at low concentrations of PYO, which would explain why loss of either SoxR or SodBII benefits WT under those conditions. If PYO were similarly toxic in WT, ∆actR, and ∆cyo, we would have expected the comparatively energy-limited ∆actR and ∆cyo mutants to benefit even more than WT from the energy savings associated with avoiding gratuitous induction of SodBII. For this not to be the case suggests that PYO may behave differently in WT compared to ∆actR and ∆cyo, likely with increased superoxide-related toxicity in the latter two.
Discussion
Here we have identified two transcription factors in A. tumefaciens that contribute to tolerance of PYO and other redox-active molecules in markedly different fashions: SoxR responds to oxidative stressors by upregulating expression of transporters and redox-related genes, whereas ActR exerts its protective effect at least in part by proactively regulating a key component of the aerobic respiratory ETC. While SoxR has previously been established as an oxidative stress response regulator in A. tumefaciens, we have expanded its known regulon to include genes that were hidden to computational approaches—a result that should encourage caution when predicting the SoxR regulons of other bacteria. In addition, the SoxR regulon we have identified in A. tumefaciens exhibits considerable functional overlap with genes that also modulate PYO tolerance in E. coli (Hassan and Fridovich, 1980; Khare and Tavazoie, 2015), suggesting that these functional classes—efflux pumps, superoxide dismutase, and electron transfer proteins such as flavodoxin and ferredoxin—are likely to be important for PYO tolerance in diverse bacteria. Yet the finding that ActR affects phenazine tolerance is perhaps more notable, for two reasons. First, ∆actR is susceptible to a wide range of redox-active molecules at lower concentrations than ∆soxR, bringing to mind the old adage, “An ounce of prevention is worth a pound of cure.” This is in contrast to the historical emphasis on studying inducible, rather than intrinsic, bacterial defenses against oxidative stress (Farr and Kogoma, 1991; Scandalios, 2002; Imlay, 2008; Chiang and Schellhorn, 2012; Imlay, 2013). Second, the correlation between the ∆actR phenotype and dysregulation of the ETC recalls prior studies in which phenazines appeared to directly interfere with the flow of electrons through the ETC, suggesting that phenazine toxicity and ETC composition may be intimately linked (Hassan and Fridovich, 1980; Baron and Rowe, 1981; Baron et al., 1989; Voggu et al., 2006; Biswas et al., 2009).
ActR belongs to a family of transcription factors including ArcA, PrrA, RegA, and RoxR, which are known for their ability to respond to the redox state of the cell, and to drive or repress the expression of different genes accordingly (Elsen et al., 2004; Fenner et al., 2004). Thus, in A. tumefaciens, it is possible that ActR not only activates cyo and represses cyd during aerobic growth, but also represses cyo and/or activates cyd under microoxic or reducing conditions. If true, this could help explain why our ∆cyo mutant more closely resembles ∆actR during exponential growth than during stationary phase: all of our RNA-seq and qPCR experiments were performed during exponential growth, when cyo appears to be the dominant terminal oxidase in WT, but during stationary phase, it is possible that ActR normally promotes expression of cyd at the expense of cyo as oxygen tensions drop due to high cell density. This would be beneficial as cyd possesses a much higher affinity for oxygen than cyo, and hence is generally the preferred terminal oxidase under oxygen-limited conditions (Borisov et al., 2011). Under this model, A. tumefaciens would require ActR in order to efficiently couple oxygen respiration to oxidative phosphorylation regardless of growth phase, as without ActR, it would neither be able to fully activate cyo under the well-oxygenated conditions that prevail during exponential growth, nor would it be able to fully activate cyd under the microoxic conditions that may arise during stationary phase. In both scenarios, failure to optimize the ETC could result in energy limitation and an excess of cellular reductants, which in turn could decrease the cell’s capacity to respond to oxidative damage while simultaneously promoting toxic redox cycling of PYO.
Notably, the actR homolog arcA in E. coli was originally named dye because mutations in this gene increase sensitivity to redox-active dyes such as toluidine blue O and methylene blue (Buxton et al., 1983; Alvarez et al., 2010). An investigation into the molecular basis of this phenotype found that overexpression of ROS-scavenging enzymes could not restore tolerance to these dyes, but that ∆arcA displayed normal tolerance to these dyes during respiration-independent growth (Alvarez et al., 2010). The authors consequently inferred that the dominant mode of toxicity of these dyes in ∆arcA is not ROS production, but rather the diversion of electrons from the ETC to the redox cycling of the dye, resulting in uncoupling of oxidative phosphorylation. By contrast, superoxide production appears to be an important mechanism of toxicity for PYO, given that superoxide dismutase is required for tolerance to PYO in A. tumefaciens, and that loss of ActR increases dependence on the SoxR-regulated superoxide dismutase; however, redox cycling of PYO may also uncouple oxidative phosphorylation. In addition, although ArcA in E. coli is not known to regulate the terminal oxidases during aerobic growth (Tseng et al., 1996; Park et al., 2013), ectopic overexpression of cyd in the E. coli ∆arcA mutant restores tolerance to redox-active dyes, while ectopic overexpression of cyo does not (Alvarez et al., 2010). This is again in contrast to our finding that overexpression of cyo in the A. tumefaciens ∆actR mutant restores tolerance to PYO to a greater extent than overexpression of cyd. Separately, Cyo itself has also previously been linked to oxidative stress tolerance in E. coli (Lindqvist et al., 2000), with at least one study suggesting that loss of Cyo potentiates oxidant toxicity by increasing basal endogenous ROS production in E. coli (Brynildsen et al., 2013). However, our study appears to be the first to explicitly link ActR-mediated regulation of cyo to intrinsic tolerance of redox-active molecules.
To our knowledge, our findings are also the first to suggest that the preemptive protection conferred by ActR may be even more important than inducible SoxR-regulated mechanisms when challenged by certain redox-active molecules; these differing mechanisms of tolerance have previously been studied separately rather than comparatively in other organisms. Nevertheless, a preemptive role for ActR homologs in oxidative stress tolerance has also been found in Haemophilus influenzae (Wong et al., 2007) and Shewanella oneidensis (Wan et al., 2015). In H. influenzae, the ActR homolog ArcA confers protection against hydrogen peroxide during low to high oxygen transitions, partially through preemptive upregulation of a putative iron storage protein (Wong et al., 2007), while in S. oneidensis, ArcA may promote hydrogen peroxide resistance by regulating cell envelope permeability (Wan et al., 2015). In A. tumefaciens, on the other hand, no iron-related genes are downregulated in ∆actR to the extent seen by Wong et al. in H. influenzae, although we have not ruled out the possibility that the few mildly downregulated iron-related genes we found (negative fold change < 2.5) might contribute to the ∆actR phenotype. We have also found no evidence that ActR in A. tumefaciens regulates homologs of the putative cell envelope-related genes identified in S. oneidensis by Wan et al. Interestingly, unlike in S. oneidensis, our ∆actR mutant was no more sensitive than WT to hydrogen peroxide in a disk diffusion assay (Fig. 2C); instead, the putative link between ActR and oxidative stress tolerance in A. tumefaciens seems to be specific to redox-active molecules.
Although we found that ActR does not regulate a response to PYO in A. tumefaciens, the ActR homolog ArcA does appear to regulate a transcriptional response to hydrogen peroxide in other organisms such as E. coli and S. enterica (Loui et al., 2009; Morales et al., 2013). In hydrogen peroxide-treated E. coli, ArcA represses flagellin (fliC) and stimulates expression of two genes involved in amino acid transport and metabolism, gltI and oppA (Loui et al., 2009). In S. enterica, ArcA responds to hydrogen peroxide treatment by downregulating several PEP-PTS and ABC transporters, while upregulating genes involved in glutathione and glycerolipid metabolism and nucleotide transport (Morales et al., 2013). Consequently, hydrogen peroxide treatment oxidizes the glutathione pool to a greater extent in ∆arcA compared to the WT strain (Morales et al., 2013). The same study also indicated that loss of ArcA leads to an approximately ten-fold increase in the NADH/NAD+ ratio during aerobic growth, with concomitantly increased ROS production (Morales et al., 2013). However, loss of ActR does not appear to have this effect in A. tumefaciens, implying that a different mechanism underpins the PYO sensitivity of our ∆actR mutant. Our RNA-seq data set also provides no indication that ActR regulates glutathione reductases in A. tumefaciens, unlike in S. enterica or even the more closely related S. medicae (Fenner et al., 2004; Morales et al., 2013).
The genetic evidence linking ActR and Cyo to phenazine tolerance is particularly interesting in light of other evidence suggesting that phenazines directly interact with and accept electrons from the respiratory ETC (Hassan and Fridovich, 1980; Baron and Rowe, 1981; Baron et al., 1989; Voggu et al., 2006; Biswas et al., 2009). Perhaps most relevant are the observations that specific sequence variants in a subunit of cytochrome bd oxidase confer PYO tolerance in staphylococci (Voggu et al., 2006), and that cbb3-type terminal oxidases are necessary and sufficient for normal phenazine reduction in P. aeruginosa (Jo et al., 2017). While technical limitations prevented us from quantitatively comparing intracellular ROS formation in WT and ∆actR, multiple observations from our ∆actR mutant—such as the fact that SodBII-mediated protection is more important in the ∆actR background—hint that the composition of the ETC, and in particular the pool of terminal oxidases, may influence how readily phenazines can “steal” electrons and thereby exert toxic effects. Importantly, as alluded to above, the toxic effects of stealing electrons from the ETC may not be limited to the generation of ROS, as redox cycling of phenazines may subvert the coupling of electron transfer to proton pumping and hence inhibit oxidative phosphorylation. This possibility is suggested by the fact that the ATP pool in ∆actR shrinks upon PYO treatment. At the same time, loss of Cyo might also increase basal ROS production to levels that do not affect growth under non-stressful conditions, but that nevertheless compromise the cell’s ability to cope with exogenous oxidative stress, as has been observed in E. coli (Brynildsen et al., 2013). To further add insult to injury, the effects of any increase in ROS production—whether basal or PYO-mediated—would be compounded by the less-efficient coupling of the ETC to oxidative phosphorylation in ∆actR, as many oxidative damage repair systems require ATP (Selby and Sancar, 1995; Galletto et al., 2006; Heinze et al., 2009; Dahl et al., 2015), not to mention the energy that is presumably required to synthesize the proteins in the SoxR regulon.
Overall, we propose a model in which ActR-mediated regulation of the ETC in A. tumefaciens helps to preemptively mitigate PYO toxicity, likely by minimizing the ability of PYO to decouple oxidative phosphorylation and generate ROS, while also promoting efficient energy conservation to power energy-dependent defense mechanisms such as the activation of the SoxR regulon (Fig. 7). Notably, loss of ActR has also been linked to downregulation of cyo in Rhizobium etli, a fellow member of the Rhizobiaceae (Lunak and Noel, 2015). Moreover, in the much more distantly related betaproteobacterium Burkholderia cenocepacia, a transposon insertion in an ActR homolog (BCAL0499) dramatically increased sensitivity to PYO and PCA (Bernier et al., 2018), although this gene was not recognized as an ActR homolog in that study, nor was it further characterized; instead, we recognized this connection after performing a BLAST-P search with the BCAL0499 sequence against the A. tumefaciens genome. Together, these observations suggest that the role we have found for ActR in A. tumefaciens may extend to diverse organisms. Our results also raise the intriguing possibility that the composition of an organism’s ETC may be a key determinant of its sensitivity to phenazines and other redox-active compounds. Future comparative studies will shed light on the usefulness of this principle for understanding and predicting ecological impacts of this important class of bacterial secondary metabolites.
Fig. 7.
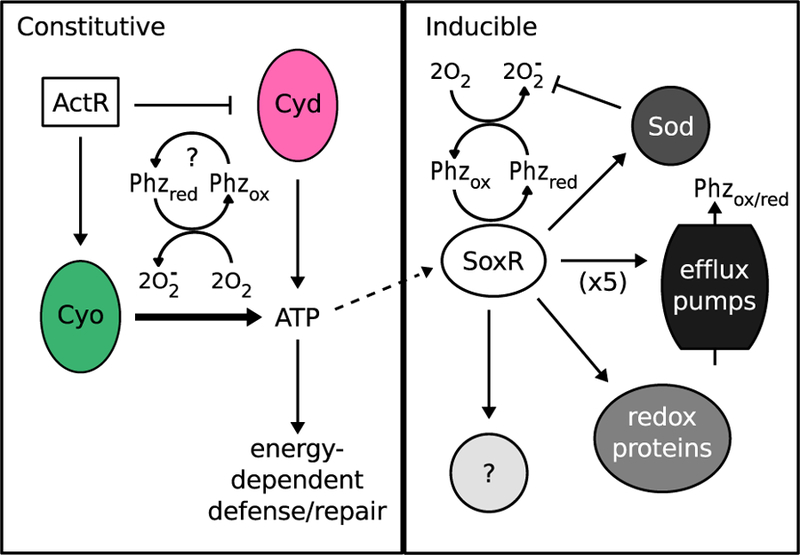
Proposed model for regulation of phenazine tolerance in A. tumefaciens.
Phenazine tolerance is regulated in both a constitutive (ActR-mediated) and inducible (SoxR-mediated) manner. ActR promotes phenazine tolerance in part by upregulating expression of cytochrome o oxidase (Cyo) at the expense of cytochrome d oxidase (Cyd) during aerobic growth. Cyo is more efficient at powering ATP synthesis and thereby supports energy-dependent defense and repair mechanisms. Levels of Cyo and Cyd may also affect how readily phenazines “steal” electrons from the electron transport chain and generate toxic superoxide radicals. Phenazines can directly oxidize and thereby activate SoxR. Activation of the SoxR regulon is likely energy dependent due to massive upregulation of several proteins, including superoxide dismutase (Sod), at least five efflux pumps, redox-related proteins such as ferredoxin, and other proteins that are as yet uncharacterized. The SoxR-regulated Sod becomes more important in the absence of ActR. Phzox = oxidized phenazines, Phzred = reduced phenazines.
Experimental procedures
Strains and media
All experiments with A. tumefaciens were performed with strain NT1, a derivative of strain C58 that lacks the pathogenicity-conferring Ti plasmid (Watson et al., 1975). A. tumefaciens was routinely grown at 30°C, with shaking at 250 rpm for liquid cultures. Unless otherwise stated, the medium for liquid cultures was Luria-Bertani (LB) Miller broth (Difco) buffered with 50 mM MOPS (pH 7), while solid plates were made with LB containing 1.5% agar. Kanamycin was added where appropriate at 50 µg/mL to make LB-Kan. LB was chosen as it is a commonly used rich medium and because we found that A. tumefaciens is highly sensitive to PYO in minimal glucose media such as AT medium (Morton and Fuqua, 2012b), possibly due to the harmful effects of superoxide on amino acid biosynthetic enzymes (Carlioz and Touati, 1986). The donor strain for transposon mutagenesis was E. coli β2155 pSC189, which was grown at 37°C in LB containing 300 µM diaminopimelic acid (DAP) and 100 µg/mL carbenicillin. Molecular cloning was carried out in E. coli DH10b using restriction enzyme-based standard protocols, except for pCyo, which was constructed by Gibson assembly (Gibson et al., 2009). Plasmids (except for pSC189) were transferred to A. tumefaciens by electroporation. All restriction enzymes and Gibson assembly reagents were purchased from New England Biolabs (NEB). PYO was synthesized and purified as previously described (Cheluvappa, 2014), and PCA was purchased from Princeton Biomolecular Research, Inc. For all experiments involving PYO, a 50x stock solution was made in 20 mM HCl, and the latter was used as a solvent control (2% of final culture volume) in 0 µM PYO conditions. Paraquat and AQDS were directly dissolved in MOPS-buffered LB at the desired final concentration. Stock solutions (50x) of methylene blue and PCA were made in water, with the latter dissolved by adding an equimolar amount of NaOH.
Transposon mutagenesis screen and identification of transposon insertion loci
To generate random transposon insertion mutants, we conjugated A. tumefaciens with the donor strain for pSC189 as follows. First, A. tumefaciens and E. coli β2155 pSC189 were grown overnight to early stationary phase, then a 1 mL aliquot of E. coli was centrifuged and washed twice with LB to avoid carryover of carbenicillin and DAP. A 1 mL aliquot of A. tumefaciens was then added to the same microcentrifuge tube and centrifuged to generate a mixed cell pellet. The pellet was resuspended in 20 µL of LB, spotted directly on LB agar, incubated for 24 hr at 30°C, and resuspended in 1 mL of LB by scraping and pipetting. This procedure was repeated approximately 25 times to generate independent libraries of transposon mutants. The libraries were diluted by a factor of 2 × 10−4 and 100 µL was spread on LB-Kan plates. After two days at 30°C, colonies were picked directly into 96-well microtiter plates containing 100 µL of LB-Kan in each well. The 96-well plates were wrapped in plastic film and incubated at 30°C for 24 hrs without shaking, in plastic containers containing damp paper towels for humidity. Subsequently, using a 48-pronged replicator flame-sterilized with ethanol, a small aliquot (1–2 µL) of each culture was transferred to a fresh 96-well plate containing 200 µL aliquots of LB-Kan + 100 µM PYO and incubated as above. After two days, the plates were assessed for the color change from blue to clear. Mutants in wells that remained blue were recovered from the original 96-well plates (that were grown without PYO) and stored at −80°C in 15% glycerol until further analysis.
The transposon insertion location in each mutant of interest was determined using arbitrary PCR. Genomic DNA was extracted from overnight cultures of the mutants using the QIAamp DNA Mini Kit (Qiagen) and PCR-amplified in 50 µL reactions with the primers Mar4 and SS9arb1 (Table S7). 5 µL of the PCR product was treated with ExoSAP-IT (Applied Biosystems) and 1 µL of cleaned-up product was re-amplified using the nested primers Mar4–2 and arb3 (Table S7). Following gel electrophoresis in 1% agarose, 1–2 distinct bands per mutant were gel extracted with the QIAquick Gel Extraction Kit (Qiagen) and submitted for Sanger sequencing (Laragen). The resulting sequences were aligned with BLAST against the A. tumefaciens C58 genome on the MicroScope platform (Vallenet et al., 2017).
In-frame deletion, complementation, and overexpression experiments
In-frame deletion mutants were generated via allelic replacement according to a previously published protocol (Morton and Fuqua, 2012a), using the plasmid pNPTS138 (M. R. K. Alley, unpublished), except that plating steps were done on LB agar containing 5% sucrose and/or 50 µg/mL kanamycin as appropriate. Complementation and overexpression experiments were performed by cloning the coding sequence(s) of the relevant gene(s) into the plasmid pSRKKm in place of lacZα, such that expression of the gene(s) of interest would be driven by the lacIq-lac promoter-operator complex (Khan et al., 2008). Expression from these plasmids in A. tumefaciens was induced by adding 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) to the culture medium at the beginning of the experiment. All plasmids used in this study are described in Table S8.
Bacterial growth-based assays
To assess sensitivity to PYO or other redox-active molecules in liquid cultures, stationary-phase overnight cultures of A. tumefaciens were diluted to an OD500 of 1 in fresh medium and inoculated 1:10 (initial OD500 = 0.1) into 200 µL aliquots of medium containing different concentrations of the redox-active molecules in 96-well flat-bottom tissue culture plates. Each plate was then transferred to a small humidity cassette (Tecan) containing 4 mL of water and incubated in a Spark 10M plate reader (Tecan) with shaking at 30°C for 24 hr, monitoring OD500 as a proxy for growth. OD500 measurements were corrected by subtracting the OD500 of “blank” wells containing the respective treatment without cells. Each strain was assessed with at least three biological replicates (derived from independent overnight cultures). Throughout this study, differences in growth after 24 hrs approximately correspond to differences in maximum achievable cell density, as cultures were either in or approaching stationary phase at this time point.
Sensitivity to acid, hydrogen peroxide, or SDS was assessed with a disk diffusion assay. Stationary-phase overnight cultures were diluted to an OD500 of 1, mixed 1:50 with 42°C pre-warmed LB soft agar (0.8% agar), and evenly pipetted in 5 mL aliquots onto solid LB agar plates. Disks of Whatman #1 filter paper (7 mm diameter) were soaked in 2 M HCl, 5.5 M H2O2, or 10% SDS and placed in the center of the plates. Diameters of growth inhibition zones were measured after two days of incubation at room temperature.
Sensitivity to bile salts, EDTA, and osmotic stresses was assessed using a gradient plate assay. To prepare the gradient plates, 25 mL of molten LB agar containing 2% bile salts (HiMedia Laboratories), 1 mM EDTA, 0.4 M NaCl (total concentration including the salt already present in LB), or 1 M myo-inositol was poured into square grid plates (Fisherbrand) tilted by propping against their lids. Once this layer had set, the plates were topped with 30 mL of plain LB agar and left at room temperature to equilibrate for at least 24 hrs. LB-only agar plates were prepared in the same manner as a control. Exponential-phase cultures were diluted to an OD500 of 0.5, then diluted again 10−4, and 5 µL aliquots of the suspension were spotted on the gradient plates. The plates were incubated at 30°C for 2–3 days before imaging with an Epson Perfection V550 Photo flatbed scanner. For both the disk diffusion and gradient plate experiments, each strain was tested in duplicate at least three independent times.
RNA extraction, RNA-seq, and qRT-PCR
To obtain RNA for RNA-seq or qRT-PCR, overnight cultures (duplicate for RNA-seq, triplicate for qRT-PCR) were diluted to an OD500 of 0.1 and grown to exponential phase (OD500 ~0.5–0.7), then treated with PYO or 20 mM HCl (solvent control) for 20 min before collecting 1 mL aliquots. Cell pellets obtained by centrifuging the aliquots were flash-frozen in liquid nitrogen and stored at −80°C until RNA extraction. RNA was extracted from the pellets using the RNeasy Mini Kit (Qiagen) with a modified protocol. Briefly, the cell pellets were resuspended in 200 µL of TE buffer (30 mM Tris-HCl, 1 mM EDTA, pH 8) containing 15 mg/mL lysozyme, plus 15 µL of 20 mg/mL proteinase K (Qiagen). After 10 min at 37°C with occasional mixing, 700 µL of Buffer RLT containing 1% β-mercaptoethanol was added, vortexing to mix, followed by 500 µL of absolute ethanol. The manufacturer’s protocol for bacteria was then followed starting at step 7 (loading the lysate onto the spin column), including the optional on-column digestion with DNase I (Qiagen). After eluting the RNA in RNase-free water, residual genomic DNA was removed with the TURBO DNA-free Kit (Invitrogen), following the manufacturer’s protocol.
For RNA-seq, rRNA was depleted by treatment with the Ribo-Zero rRNA Removal Kit for Bacteria (Illumina). The remaining RNA was then fragmented to an average size of 200 bp by incubation at 94°C for exactly 60 s with the RNA Fragmentation Buffer from the NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB). Fragmentation was halted by placing the samples on ice and adding the RNA Fragmentation Stop Buffer. The concentrations and average sizes of the fragmented RNA samples were determined using the Agilent RNA 6000 Pico Kit on an Agilent 2100 Bioanalyzer. Subsequent library preparation steps were carried out with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB), following the manufacturer’s protocol for rRNA-depleted FFPE RNA. Sequencing was performed at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology to a depth of 10–15 million reads on an Illumina HiSeq2500 and processed using the Illumina HiSeq control software (HCS version 2.0). Low-quality bases were removed using Trimmomatic (LEADING:27 TRAILING:27 SLIDINGWINDOW:4:20 MINLEN:35) (Bolger et al., 2014). Alignment to the A. tumefaciens C58 genome and calculation of the number of reads per gene was performed with Rockhopper 2 (Tjaden, 2015), with 0.04 allowed mismatches and a minimum seed length of 0.33. Differential gene expression analysis was performed using DESeq2 (Love et al., 2014). Sequence data were submitted to the NCBI Sequence Read Archive under the accession number PRJNA521160.
For qRT-PCR, cDNA was synthesized from TURBO DNase-treated RNA using the iScript cDNA Synthesis Kit (Bio-Rad). The PCR reactions were set up in a 20 µL volume with iTaq Universal SYBR Green Supermix (Bio-Rad) (10 µL of the supermix, 4 µL of a 1:10 dilution of cDNA [~10 ng], 0.4 µL each of 10 µM forward and reverse primers, and 5.2 µL of water). Primers for qRT-PCR were designed using Primer-BLAST (Ye et al., 2012) and are listed in Table S7. Reactions were run using a Fast 7500 Real-Time PCR System machine (Applied Biosystems). A standard curve for each primer pair was generated using known concentrations of A. tumefaciens genomic DNA, enabling calculation of the relative concentrations of cDNA for each gene of interest. The expression levels (relative concentrations) for each gene were normalized to the expression of the housekeeping gene rpoD.
Oxygen consumption rate measurements
To measure oxygen consumption rates, each overnight culture was diluted to an OD500 of 0.1 in four 5 mL cultures (total 20 mL) and grown to exponential phase (OD500 ~0.5–0.7). Meanwhile, 19 mL of MOPS-buffered LB per sample was aliquoted into autoclaved scintillation vials and pre-equilibrated by stirring with a magnetic stir bar at 500 rpm for at least 1 hr. Once the cultures had reached exponential phase, all 20 mL of each sample was centrifuged in a 50 mL conical tube. The cell pellet was resuspended in 1 mL of MOPS-buffered LB and adjusted to an OD500 of 5, then 1 mL of the suspension was added to 19 mL of pre-equilibrated medium with continued stirring at 500 rpm. The concentration of oxygen in the medium was recorded over time at 1 s intervals using an OX-500 microsensor (Unisense), which was submerged to a fixed depth below the surface. After 7 min, the cultures were spiked with 10 µM PYO and the oxygen concentration was recorded for another 7 min. The measurements were calibrated by using a solution of 0.1 M sodium ascorbate + 0.1 M NaOH as the zero-oxygen reference, and MOPS-buffered LB that had equilibrated with stirring at 500 rpm for several hours as the saturated reference. The microsensor response was converted to dissolved oxygen concentration using the salinity-temperature-solubility tables provided by the manufacturer. Oxygen consumption rates were calculated using an approach similar to a previously published method (Riedel et al., 2013), except that we treated only the first 2 min of each 7 min recording period as an equilibration period due to the rapid response of the microsensor, and that we fitted separate linear regressions for each 10 µM interval of oxygen concentration.
To measure PYO-mediated oxygen consumption rates in the presence of cyanide, experiments were performed as above except that 5 mM potassium cyanide (KCN) was added to the cell suspensions after the first 7 min (to ensure that cells were respiring normally prior to cyanide addition). Upon addition of cyanide, oxygen consumption halted within 10 s, and oxygen concentrations steadily rose over time due to leakage of oxygen back into the system. After 5 min of cyanide treatment, 10 µM PYO was added to the cell suspensions. As this led to no visible change in the rate of oxygen leakage/consumption, after another 5 min more PYO was added to a final concentration of 100 µM. Plating for CFUs before and after the experiments confirmed that 5 mM KCN did not kill the cells (data not shown).
NADH and NAD+ extraction and measurement
To determine the NADH/NAD+ ratios of WT and ∆actR, overnight cultures were diluted to an OD500 of 0.1 and grown to exponential phase (5 hrs) in 5 mL MOPS-buffered LB. NADH and NAD+ were then extracted and quantified according to a previously published protocol (Kern et al., 2014). To monitor the progress of the enzyme-mediated colorimetric reaction, absorbance at 570 nm was tracked in a Spark 10M plate reader (Tecan) over a period of 20 min, with the minimum possible time between readings (~45 s).
ATP measurements and ATP synthesis inhibition
To measure ATP levels, overnight cultures were diluted to an OD500 of 0.1 and grown to exponential phase (5 hrs) in 5 mL MOPS-buffered LB with or without 10 µM PYO. The ATP measurements were then performed using BacTiter-Glo Reagent (Promega) in opaque white 96-well plates, according to the manufacturer’s instructions. Luminescence readings were taken at 30°C using a Spark 10M plate reader (Tecan). ATP concentrations were calculated using a standard curve generated with known quantities of pure ATP dissolved in water, and were normalized to the OD500 of each culture. To test whether inhibiting ATP synthesis can increase sensitivity to PYO, the experiment was performed in the same manner except that the cultures were treated with 0 or 100 µM N,N-dicyclohexylcarbodiimide (DCCD), in combination with 0 or 10 µM PYO. These experiments were performed in triplicate.
Measurement of ROS production
We first measured ROS production by using electron paramagnetic resonance (EPR) spectroscopy in conjunction with the spin trap 5-tert-butoxycarbonyl-5-methyl-1-pyrroline-N-oxide (BMPO), which reacts with superoxide and hydroxyl radical to form relatively stable radical adducts (BMPO/•OOH and BMPO/•OH, respectively) (Zhao et al., 2001). However, we found that this approach could only detect extracellular ROS in suspensions of A. tumefaciens (Fig. S7A), consistent with a previous study that used a similar spin trap to detect superoxide formation in PYO-treated E. coli (Hassett et al., 1992). To perform these measurements, overnight cultures were first grown in AT minimal medium modified to contain 50 mM phosphate buffer and no added iron (Morton and Fuqua, 2012b). The cultures reached mid-exponential phase after approximately 20–24 hrs, at which point the cells were pelleted by centrifugation and washed twice with Chelex 100 (Bio-Rad)-treated 0.1 M phosphate buffer (pH 7.4) containing 25 µM diethylenetriaminepentaacetic acid (DTPA). The cells were then resuspended in the buffer to an OD500 of 1. For each sample, 176 µL of cell suspension was combined with 20 µL of 250 mM BMPO (dissolved in the phosphate buffer), 2 µL of 50% glucose (Chelex 100-treated), and 2 µL of 10 mM PYO, and mixed by vortexing for 5 s every minute for three minutes. The sample was then loaded into a flat quartz EPR spectroscopy cuvette and the spectrum was recorded on a Bruker EMX X-band CW-EPR spectrometer starting at 10 min 30 s after the addition of PYO. The instrument settings were as follows: microwave power, 5.08 mW; microwave frequency, 9.836 GHz; modulation frequency, 100 kHz; modulation amplitude, 1 G; time constant, 81.92 msec; receiver gain, 5.02 × 104; sweep time, 83.89 s. Spectra shown in Fig. S6A are averages of 6 consecutive scans from single samples that are representative of at least three biological replicates, except for the cell-free and exogenous SOD control samples, for which only the initial spectrum was recorded.
To measure intracellular superoxide formation, we considered using dihydroethidium (DHE), a cell-permeable dye that reacts with superoxide to form the fluorescent derivative 2-hydroxyethidium, although it can also be nonspecifically oxidized to similarly fluorescent ethidium (Zielonka et al., 2007). Unfortunately, we found that addition of PYO to DHE in cell-free phosphate buffer was sufficient to increase fluorescence, suggesting that PYO can abiotically oxidize DHE and that DHE oxidation would therefore not be a reliable reporter for intracellular superoxide (Fig. S7B). To test DHE for oxidation upon exposure to PYO, 1.5 µL of 5 mM DHE (Invitrogen) was combined in a microcentrifuge tube with 5 µL of 1 mM PYO or 5 µL of 20 mM HCl (solvent control) in 493.5 µL of Chelex 100-treated 0.1 M phosphate buffer (pH 7.4) containing 100 µM DTPA. The samples were incubated at 30°C with shaking at 250 rpm for 1 hr. Subsequently, 200 µL of each sample was transferred to a black plastic flat transparent-bottomed 96-well plate and fluorescence (excitation at 510 nm, emission at 580 nm) was recorded in a Spark 10M plate reader (Tecan). All steps were performed in the dark or under dim lighting to minimize photooxidation of DHE.
Statistical analysis
All statistical analyses were performed using R (R Core Team, 2018). In all cases involving multiple pairwise comparisons within an experiment, the Benjamini-Hochberg procedure was used to control the false discovery rate. For Figs. 2B and 3B, statistical significance was calculated by generalized linear regression with dummy variable coding, using WT as the reference group within each treatment. The function ‘ncvTest’ from the package ‘car’ (Fox and Weisberg, 2011) was used to verify the homoscedasticity of the residuals, while the Shapiro-Wilk normality test was used to verify the normality of the residuals. For Fig. 2C, the Shapiro-Wilk test revealed non-normality of the residuals following linear regression, so instead the Kruskal-Wallis test was performed within each treatment, followed by pairwise comparisons using the Wilcox rank sum test. For Fig. 4C, Welch’s t-test was used to make pairwise comparisons between WT and ∆actR within each condition (i.e. + PYO or – PYO). For Fig. 6C, Welch’s t-test was used to make pairwise comparisons between strains within each condition (i.e. + PYO or – PYO), as well as within strains but between conditions. For Fig. S6B, ncvTest revealed heteroscedasticity of the residuals following linear regression, so instead samples were grouped by strain and PYO treatment and Welch’s t-test was then used to make pairwise comparisons between 0 µM and 100 µM DCCD treatments within each group. For Fig. S6C, the assumptions of linear regression were met, so linear regression was performed with a custom contrast matrix to specify comparisons between 0 µM and 100 µM DCCD treatments within each strain.
Supplementary Material
Acknowledgements
We thank all members of the Newman lab for helpful advice, discussions, and feedback on the manuscript, and Clay Fuqua for generously providing pNPTS138. We also thank the Marine Biological Laboratory Microbial Diversity Course, Class of 2017, for assistance with screening transposon mutants. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE‐1745301. This work was also supported by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology (Caltech), the Caltech Electron Paramagnetic Resonance Spectroscopy Facility, and grants to DKN from the ARO (W911NF-17-1-0024) and NIH 341 (1R01AI127850-01A1).
Footnotes
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Alonso A, Rojo F, and Martínez JL (1999) Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ Microbiol 1: 421–430. [DOI] [PubMed] [Google Scholar]
- Alvarez AF, Malpica R, Contreras M, Escamilla E, and Georgellis D (2010) Cytochrome d but not cytochrome o rescues the toluidine blue growth sensitivity of arc mutants of Escherichia coli. J Bacteriol 192: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Danhorn T, Fuqua C, and Parsek MR (2006) Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci 103: 3828–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K, Pattery T, Cornelis P, and Höfte M (2002) Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol Plant-Microbe Interact 15: 1147–1156. [DOI] [PubMed] [Google Scholar]
- Baek SH, Hartsock A, and Shapleigh JP (2008) Agrobacterium tumefaciens C58 uses ActR and FnrN to control nirK and nor expression. J Bacteriol 190: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron SS, and Rowe JJ (1981) Antibiotic action of pyocyanin. Antimicrob Agents Chemother 20: 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron SS, Terranova G, and Rowe JJ (1989) Molecular mechanism of the antimicrobial action of pyocyanin. Curr Microbiol 18: 223–230. [Google Scholar]
- Begley M, Gahan CGM, and Hill C (2005) The interaction between bacteria and bile. FEMS Microbiol Rev 29: 625–651. [DOI] [PubMed] [Google Scholar]
- Bernier SP, Son S, and Surette MG (2018) The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200: e00156–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas L, Biswas R, Schlag M, Bertram R, and Götz F (2009) Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol 75: 6910–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J, and Verkhovsky MI (2011) The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta - Bioenerg 1807: 1398–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, and Collins JJ (2013) Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol 31: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RS, Drury LS, and Curtis CAM (1983) Dye sensitivity correlated with envelope protein changes in dye (sfrA) mutants of Escherichia coli K12 defective in the expression of the sex factor F. J Gen Microbiol 129: 3363–3370. [DOI] [PubMed] [Google Scholar]
- Calhoun MW, Oden KL, Gennis RB, Mattos M.J.T. de, and Neijssel OM (1993) Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J Bacteriol 175: 3020–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, and Touati D (1986) Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J 5: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheluvappa R (2014) Standardized chemical synthesis of Pseudomonas aeruginosa pyocyanin. MethodsX 1: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, and Rubin EJ (2002) Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296: 179–185. [DOI] [PubMed] [Google Scholar]
- Chiang SM, and Schellhorn HE (2012) Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys 525: 161–169 10.1016/j.abb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng TFC, Bloemberg GV, and Lugtenberg BJJ (2003) Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol 157: 503–523. [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng TFC, Bloemberg GV, Bij AJ van der Drift, van der KMGM, Schripsema J, Kroon B, et al. (1998) Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact 11: 1069–1077. [Google Scholar]
- Costa KC, Bergkessel M, Saunders S, Korlach J, and Newman DK (2015) Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. MBio 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’mello R, Hill S, and Poole RK (1996) The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142: 755–763. [DOI] [PubMed] [Google Scholar]
- Dahl J-U, Gray MJ, and Jakob U (2015) Protein quality control under oxidative stress conditions. J Mol Biol 427: 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LEP, and Kiley PJ (2011) A shared mechanism of SoxR activation by redox-cycling compounds. Mol Microbiol 79: 1119–1122. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Price-Whelan A, Petersen A, Whiteley M, and Newman DK (2006) The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol 61: 1308–1321. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Teal TK, Price-Whelan A, and Newman DK (2008) Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science (80- ) 321: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, Charoenlap N, Vattanaviboon P, and Mongkolsuk S (2006) Agrobacterium tumefaciens soxR is involved in superoxide stress protection and also directly regulates superoxide-inducible expression of itself and a target gene. J Bacteriol 188: 8669–8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen S, Swem LR, Swem DL, and Bauer CE (2004) RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev 68: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, and Hassan HM (2011) Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol 11: 58 http://www.biomedcentral.com/1471-2180/11/58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier E, Aogáin M, Mooij MJ, Woods DF, Morrissey JP, and Dobson ADW (2012) MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J Bacteriol 194: 3502–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr SB, and Kogoma T (1991) Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev 55: 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner BJ, Tiwari RP, Reeve WG, Dilworth MJ, and Glenn AR (2004) Sinorhizobium medicae genes whose regulation involves the ActS and/or ActR signal transduction proteins. FEMS Microbiol Lett 236: 21–31. [DOI] [PubMed] [Google Scholar]
- Fernández-Piñar R, Ramos JL, Rodríguez-Herva J, and Espinosa-Urgel M (2008) A two-component regulatory system integrates redox state and population density sensing in Pseudomonas putida. J Bacteriol 190: 7666–7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetar H, Gilmour C, Klinoski R, Daigle DM, Dean CR, and Poole K (2011) mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob Agents Chemother 55: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, and Weisberg S (2011) An R companion to applied regression 2nd ed., Sage, Thousand Oaks, CA. [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, and Kowalczykowski SC (2006) Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443: 875–878. [DOI] [PubMed] [Google Scholar]
- Gao H, Wang X, Yang ZK, Palzkill T, and Zhou J (2008) Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, and Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Glasser NR, Kern SE, and Newman DK (2014) Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol 92: 399–412 http://doi.wiley.com/10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick JA, Brown CT, and Newman DK (2005) Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol 56: 1347–1357. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SK, Schroth MN, Cho JJ, Kominos SK, and Vitanza-Jack VB (1974) Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl Microbiol 28: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, and Imlay JA (2011) The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol 79: 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HM, and Fridovich I (1980) Mechanism of the antibiotic action of pyocyanine. J Bacteriol 141: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Charniga LL, Bean K, Ohman DE, and Scohen M (1992) Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun 60: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze RJ, Giron-Monzon L, Solovyova A, Elliot SL, Geisler S, Cupples CG, et al. (2009) Physical and functional interactions between Escherichia coli MutL and the Vsr repair endonuclease. Nucleic Acids Res 37: 4453–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez ME, Kappler A, and Newman DK (2004) Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl Environ Microbiol 70: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77: 755–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Cortez KL, Cornell WC, Price-Whelan A, and Dietrich LEP (2017) An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6: e30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Price-Whelan A, and Newman DK (2014) Extraction and measurement of NAD(P)+ and NAD(P)H. In Pseudomonas Methods and Protocols Filloux A, and Ramos J-L (eds). Springer New York, New York, NY: pp. 311–323. [Google Scholar]
- Khan SR, Gaines J, Roop RM, and Farrand SK (2008) Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74: 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare A, and Tavazoie S (2015) Multifactorial competition and resistance in a two-species bacterial system. PLoS Genet 11: e1005715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare E, and Arora NK (2011) Dual activity of pyocyanin from Pseudomonas aeruginosa — antibiotic against phytopathogen and signal molecule for biofilm development by rhizobia. Can J Microbiol 57: 708–713. [DOI] [PubMed] [Google Scholar]
- Köhler T, Epp SF, Curty LK, and Pechère J-C (1999) Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181: 6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty LK, and Pechère J-C (1997) Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol 23: 345–354. [DOI] [PubMed] [Google Scholar]
- Lamarche MG, and Déziel E (2011) MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6: e24310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeTourneau MK, Marshall MJ, Cliff JB, Bonsall RF, Dohnalkova AC, Mavrodi DV, et al. (2018) Phenazine-1-carboxylic acid and soil moisture influence biofilm development and turnover of rhizobacterial biomass on wheat root surfaces. Environ Microbiol 20: 2178–2194. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Membrillo-Hernández J, Poole RK, and Cook GM (2000) Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek 78: 23–31. [DOI] [PubMed] [Google Scholar]
- Llanes C, Kohler T, Patry I, Dehecq B, Delden C. Van, and Plesiat P (2011) Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother 55: 5676–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loui C, Chang AC, and Lu S (2009) Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunak ZR, and Noel KD (2015) Quinol oxidase encoded by cyoABCD in Rhizobium etli CFN42 is regulated by ActSR and is crucial for growth at low pH or low iron conditions. Microbiology 161: 1806–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Managò A, Becker KA, Carpinteiro A, Wilker B, Soddemann M, Seitz AP, et al. (2015) Pseudomonas aeruginosa pyocyanin induces neutrophil death via mitochondrial reactive oxygen species and mitochondrial acid sphingomyelinase. Antioxid Redox Signal 22: 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F, Dam P, Chou J, Olman V, and Xu Y (2009) DOOR: A database for prokaryotic operons. Nucleic Acids Res 37: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Patel L, and Kaback HR (1984) Cytochrome o type oxidase from Escherichia coli. Characterization of the enzyme and mechanism of electrochemical proton gradient generation. Biochemistry 23: 4703–4714. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak YS, Paulitz TC, et al. (2012) Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl Environ Microbiol 78: 804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, et al. (2010) Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol 76: 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles LA, and Newman DK (2018) Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Mol Microbiol 110: 995–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales EH, Collao B, Desai PT, Calderón IL, Gil F, Luraschi R, et al. (2013) Probing the ArcA regulon under aerobic/ROS conditions in Salmonella enterica serovar Typhimurium. BMC Genomics 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, and Fuqua C (2012a) Genetic manipulation of Agrobacterium. Curr Protoc Microbiol 25: 3D.2.1–3D.2.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, and Fuqua C (2012b) Laboratory maintenance of Agrobacterium. Curr Protoc Microbiol 24: 3D.1.1–3D.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson KW, and Aspedon A (1992) Detergent-shock response in enteric bacteria. Mol Microbiol 6: 957–961. [DOI] [PubMed] [Google Scholar]
- Noto MJ, Burns WJ, Beavers WN, and Skaar EP (2017) Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J Bacteriol 199: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. (2013) RegPrecise 3.0 - A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14: 745 BMC Genomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DM, Akhtar S, Ansari AZ, Landick R, and Kiley PJ (2013) The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9: e1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018) R: A language and environment for statistical computing .
- Rada B, and Leto TL (2013) Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol 21: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I, Dietrich LEP, Price-Whelan A, and Newman DK (2010) Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol 161: 187–191 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel TE, Berelson WM, Nealson KH, and Finkel SE (2013) Oxygen consumption rates of bacteria under nutrient-limited conditions. Appl Environ Microbiol 79: 4921–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Wingender J, Müller H, and Tümmler B (1994) A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol 60: 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenkham P, Eiamphungporn W, Farrand SK, Vattanaviboon P, and Mongkolsuk S (2007) Multiple superoxide dismutases in Agrobacterium tumefaciens: functional analysis, gene regulation, and influence on tumorigenesis. J Bacteriol 189: 8807–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios J (2002) Oxidative stress responses - what have genome-scale studies taught us? Genome Biol 3: reviews1019.1–1019.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, and Sancar A (1995) Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem 270: 4882–4889. [DOI] [PubMed] [Google Scholar]
- Séveno NA, Morgan JAW, and Wellington EMH (2001) Growth of Pseudomonas aureofaciens PGS12 and the dynamics of HHL and phenazine production in liquid culture, on nutrient agar, and on plant roots. Microb Ecol 41: 314–324. [DOI] [PubMed] [Google Scholar]
- Singh AK, Shin J-H, Lee K-L, Imlay JA, and Roe J-H (2013) Comparative study of SoxR activation by redox-active compounds. Mol Microbiol 90: 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem DL, and Bauer CE (2002) Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J Bacteriol 184: 2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B (2015) De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-P, Albrecht J, and Gunsalus RP (1996) Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol 178: 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM, and Messenger AJM (1986) Occurence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol 27: 211–275. [DOI] [PubMed] [Google Scholar]
- Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, et al. (2017) MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 45: D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, and Götz F (2006) Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188: 8079–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Mao Y, Dong Y, Ju L, Wu G, and Gao H (2015) Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis. Sci Rep 1–14. [DOI] [PMC free article] [PubMed]
- Wang Y, Kern SE, and Newman DK (2010) Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol 192: 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, and Newman DK (2011) Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193: 3606–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B, Currier TC, Gordon MP, Chilton M-D, and Nester EW (1975) Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol 123: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SMS, Alugupalli KR, Ram S, and Akerley BJ (2007) The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol 64: 1375–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, and Madden TL (2012) Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, and Zarivach R (2012) Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20: 397–405 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Zhao H, Joseph J, Zhang H, Karoui H, and Kalyanaraman B (2001) Synthesis and biochemical applications of a solid cyclic nitrone spin trap: a relatively superior trap for detecting superoxide anions and glutathiyl radicals. Free Radic Biol Med 31: 599–606. [DOI] [PubMed] [Google Scholar]
- Zielonka J, Vasquez-Vivar J, and Kalyanaraman B (2007) Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat Protoc 3: 8–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


