Abstract
Graft-versus-host disease (GVHD) is the most serious complication of allogeneic hematopoietic cell transplantation. Notch signals delivered during the first 48 hours after transplantation drive proinflammatory cytokine production in conventional T cells (Tconv) and inhibit expansion of regulatory T cells (Tregs). Short-term Notch inhibition induces long-term GVHD protection. However, it remains unknown whether Notch blockade blunts GVHD through its effects on Tconv, Tregs, or both, and what early Notch-regulated molecular events occur in alloantigen-specific T cells. To address these questions, we engineered T cell grafts to achieve selective Notch blockade in Tconv vs. Tregs and evaluated their capacity to trigger GVHD in mice. Notch blockade in Tconv was essential for GVHD protection, as GVHD severity was similar in recipients of wild-type Tconv combined with Notch-deprived vs. wild-type Tregs. To identify the impact of Notch signaling on the earliest steps of T cell activation in vivo, we established a new acute GVHD model mediated by clonal alloantigen-specific 4C CD4+ Tconv. Notch-deprived 4C T cells had preserved early steps of activation, IL-2 production, proliferation, and T helper polarization. In contrast, Notch inhibition dampened IFN-γ and IL-17 production, diminished mTORC1 and ERK½ activation, and impaired transcription of a subset of Myc-regulated genes. The distinct Notch-regulated signature had minimal overlap with known Notch targets in T cell leukemia and developing T cells, highlighting the specific impact of Notch signaling in mature T cells. Our findings uncover a unique molecular program associated with pathogenic effects of Notch in T cells at the earliest stages of GVHD.
Introduction
Notch signaling is an evolutionarily conserved signaling pathway with multiple roles in immune cell development and function (1). Notch has emerged as an essential regulator of T cell alloreactivity in mouse models of graft-versus-host disease (GVHD) and allograft rejection (2–11). We previously demonstrated that genetic blockade of Notch signaling within donor CD4+ and CD8+ T cells and therapeutic neutralization of the Notch ligands Delta-like1 and Delta-like4 (Dll¼) results in long-term protection from GVHD morbidity and mortality after allogeneic hematopoietic cell transplantation (allo-HCT), while largely preserving graft-versus-leukemia (GVL) activity (2, 4, 6). We identified host fibroblastic stromal cells in secondary lymphoid organs as the critical source of Delta-like ligands that drive pathogenic Notch activity in donor T cells within 48 hours post-transplantation (10). GVHD inhibition via Notch blockade was associated with decreased IFN-γ and IL-17 production as well as expansion of pre-existing FoxP3+ regulatory T cells (Tregs). At peak expansion, Notch-deprived CD4+ and CD8+ T cells exhibited blunted Ras/MAPK signaling and upregulated several negative regulators of T cell signaling, while preserving expression of the master transcription factors T-bet and Eomesodermin (2, 4, 6). In addition, selective genetic inactivation of Notch signaling in Tregs was recently reported as sufficient to confer long-term protection from acute GVHD (9), although the existence of secondary functional changes in conventional T cells (Tconv) could not be ruled out. Therefore, further work is needed to define whether Notch signaling functions primarily to promote Tconv pathogenicity, destabilize Treg suppressive potential, or impact both populations to aggravate GVHD. Understanding the downstream molecular consequences of Notch signaling in T cells will provide new insights into its effects at the earliest stages of alloreactivity.
Studies in T cell acute lymphoblastic leukemia (T-ALL), >50% of which harbor Notch gain-of-function mutations, have provided key insights into the molecular mechanisms that operate downstream of Notch in this context (12). Chromatin immunoprecipitation and γ-secretase inhibitor washout studies revealed a range of direct transcriptional targets of Notch in T-ALL, many of which are associated with distal enhancers (13–16). However, it remains unclear if Notch regulates the same targets in mature T cells, as systematic studies have not been performed in antigen-reactive T cell subsets, which rely on a very different context-specific enhancer landscape (4, 17–19). Cleaved intracellular Notch has been proposed to function either as an amplifier of Th cell differentiation by binding to Th lineage fate master transcription factor and cytokine loci (20), or by enhancing antigen sensitivity in a B7/CD28-dependent fashion via professional hematopoietic APCs (19, 21–24). While Notch blockade failed to impact the expression of master transcription factors driving individual effector T cell lineages during GVHD (2, 4, 11), the contribution of other individual mechanisms to the role of Notch in T cell alloreactivity remains unknown (2, 4, 11).
The earliest post-transplant time window represents a critical period of alloreactive T cell priming and Notch activity that defines subsequent GVHD. Thus, we investigated the impact of Notch signaling on cellular and molecular events in alloreactive T cells during this time to gain insight into the molecular impact of Notch on alloimmunity. As Notch inhibition in mature CD4+ and CD8+ T cells exerts effects on both Tconv and Tregs, we first established the relative importance of Tconv and Tregs in mediating the protective effects of Notch inhibition in a polyclonal model of MHC-mismatched GVHD. After identifying Notch signaling in Tconv as essential for GVHD pathophysiology, we dissected alloreactive Tconv responses during the immediate post-transplant period. As the majority of donor polyclonal T cells are non-alloreactive bystander cells that do not contribute to GVHD, we established an MHC-mismatched model of acute GVHD driven by a defined clonal population of alloantigen-specific CD4+ T cells. This strategy allowed us to define the cellular and molecular effects of Notch within alloantigen-specific cells during in vivo T cell priming. Notch inhibition within Tconv preserved early T cell activation, expansion, and IL-2 production, as well as induction of tbx21, gata3, and rorc transcription. In contrast, Notch-deprived T cells had markedly decreased transcripts for multiple proinflammatory cytokines, including IFN-γ, IL-17A/F, and IL-21, showed reduced mTORC1 and MAPK activity, and exhibited features of diminished Myc function. In this relatively narrow transcriptional signature, we identified targets with broad potential impact on T cell alloimmunity that were regulated by Notch blockade. Our study highlights the molecular signature that underlies the impact of Notch signaling on the acquisition of pathogenic CD4+ T cell effector functions during GVHD.
Methods
Mice
BALB/c (H-2d, CD45.2+), BALB/c-CD45.1 (H-2d, CD45.1+), C57BL/6 (B6, H-2b, CD45.2+) and C57BL/6.Ptprca (B6-SJL, H-2b, CD45.1+) mice were bred at the University of Michigan or the University of Pennsylvania. ROSA26DNMAMLf/f mice (DNMAML; H-2b, CD45.2+) containing a Cre-inducible cassette encoding the dominant negative Mastermind-like1 (DNMAML)-GFP pan-Notch inhibitor under the ROSA26 promoter were crossed with FoxP3-internal ribosome entry site-RFP reporter mice (FoxP3-IRES-mRFP, Jackson laboratories strain 008374 (25)). Since no effect of Cre expression was observed in alloreactive T cells (data not shown), both Cd4-Cre+ or Cd4-Cre– controls were used. 4C Rag1−/− T cell receptor transgenic mice on B6 background (H-2b), reactive to I-Ad, were previously described (26). Transgenic mice expressing a GFP reporter under the control of Nur77 (Nr4a1) promoter sequences (Nur77-GFP; H-2b; Jackson laboratories strain 016617) were described before (27). All littermates (experimental and control animals) were co-housed until experimental use. Animals were 8–16 weeks old at the time of use. All protocols were approved by the University of Pennsylvania’s Office of Regulatory Affairs and the University of Michigan’s Committee on Use and Care of Animals.
Bone marrow transplantation, systemic antibody-mediated Notch inhibition, and GVHD assessment
BALB/c recipients were irradiated with 8–8.5 Gy (137Cs source) and B6-SJL recipients with 12 Gy (in two split doses separated by 3 hours). T cell-depleted bone marrow (TCD BM) was prepared as described (6). 5–10×106 TCD BM was injected alone, or with T cells in doses outlined in the appropriate figure legends. To allow sufficient cell retrieval, we transplanted 5×106 4C T cells for all experiments in which downstream T cell analyses were done prior to 48 hours post-transplant. In select experiments, T cells were labeled with CFSE or CellTracker Orange 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (CMTMR) Dye (Invitrogen). Humanized IgG1 monoclonal Abs specific for Dll1 or Dll4 (28, 29) were injected i.p. (5 mg/kg) on day 0 pre-transplant. An anti-HSV gD human IgG1 antibody was used as isotype control. The quality of each mAb batch was tested in vivo by assessing inhibition of Dll4-dependent T cell development and Dll1-dependent marginal zone B cell development (30–33). Systemic clinical GVHD was graded twice a week using an established system (34) and survival was monitored daily.
Flow cytometry
Single cell suspensions from lymphoid organs (spleen, cutaneous and mesenteric lymph nodes) were prepared using a modified version of a previously described protocol (35, 36). Briefly, organs were coarsely chopped with a scalpel and incubated at 37°C for 20’ in digestion solution containing RPMI, 2% FBS, 1.0 mg/mL collagenase IV (Invitrogen) and 40 μg/mL DNAse I (Roche), followed by 10’ of sample pipetting at 37°C to disrupt the tissue. This process was repeated up to 2 times until tissue dissolution, followed by passage through a cell strainer. Detection of cytokine production in alloreactive T cells harvested within the first 48 hours after allo-HCT was done following a 4-hour incubation with Brefeldin A (BD) only. Alloreactive T cells harvested beyond the first 48 hours post-transplant were re-stimulated for 6 hours in 96-well round bottom plates pre-coated with 2.5 μg/ml anti-CD3ε/anti-CD28 (Biolegend), with Brefeldin A added during the last 4 hours, before subsequent staining. Surface staining was performed following staining with Zombie Aqua fixable viability dye (Biolegend). Intracellular staining was done following the manufacturer’s protocol (eBioscience). Phospho-flow analysis was done as described (37). Antibodies, clones, and manufacturers are described in the Supplemental table I. Analysis was performed on an LSRFortessa (BD). Sorting was performed with FACSAria III (BD). Data were analyzed using FlowJo (TreeStar).
RNA sequencing (RNA-Seq) and bioinformatics analysis
4C T cells were sort-purified to >95% purity. Total RNA was isolated using TRIzol (Invitrogen) and RNeasy Micro kit (Qiagen). Libraries were prepared with Clontech SMARTer kit and sequencing was performed using Illumina Hi-Seq platform. Quality of the raw reads was assessed using FastQC (version 0.10.1). The first three bases from 5’ end from all reads were trimmed to remove Clontech SMARTer adapter and quality of all raw reads then assessed using FastQC (version 0.10.1). Fastq files were aligned to genome build GRCm38 using the STAR aligner (version 2.5.2b) and the Ensembl annotation (release 87) as a splicing guide. Gene counts were obtained using Subread featureCounts (version 1.5.1) using the same Ensembl annotation, counting 2nd strand alignments, not counting multi mapping alignments, and assigning reads to features with the largest overlap. Genes with less than 10 counts across all samples were discarded. Differentially expressed genes were identified using DESeq2 (version 1.14.1). All three conditions were normalized together, and results obtained by applying contrasts between any two conditions. Significant genes were filtered for an adjusted p-value < 0.01 and an absolute log2 fold change > 0.585 (1.5X). Heat maps were prepared using the regularized log2 values obtained from DESeq2 and the pheatmap function in R. GSEA was performed by first transforming the mouse identifiers into human homologs using annotation from Ensembl BioMart, and using the PreRanked function of GSEA with the log2 fold changes reported from DESeq2. RNA-Seq data (GSE126518) have been deposited in NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
Statistics
GraphPad Prism 7.03 was used for statistical analysis. Values are presented as mean ± SEM. Statistical differences were calculated using one-way ANOVA with Tukey correction for multiple comparisons, Student’s t-test, or log-rank test, as indicated for specific experiments. Only statistically significant differences with p<0.05 are highlighted.
Results
Active Notch signaling in conventional CD4+ T cells is necessary for acute GVHD development
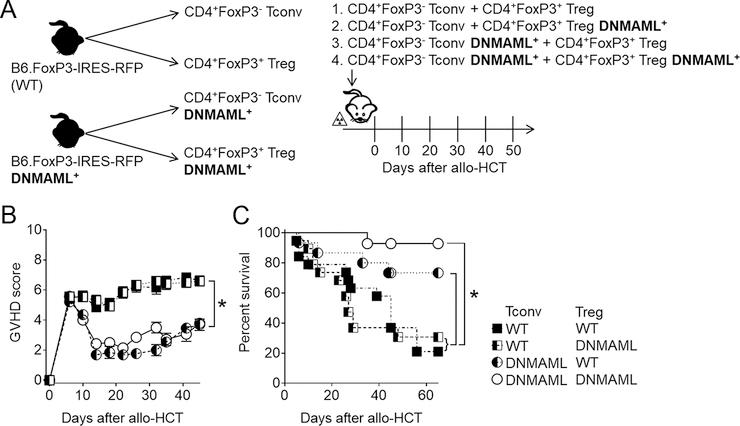
Using complementary genetic and biochemical approaches, we previously reported an essential role for Notch signaling in all mature CD4+ T cells during acute GVHD in mouse allo-HCT models (2, 4, 6). A recent study suggested that Notch inhibition specifically in Tregs was sufficient to improve acute GVHD outcomes after MHC-mismatched transplantation in mice (9). Thus, the respective importance of Notch signaling in conventional FoxP3− T cells vs. FoxP3+ Tregs during acute GVHD remains unclear. To address this question, we relied on a B6 FoxP3-IRES-mRFP reporter strain (25) and crossed it with B6-DNMAML mice (expressing GFP and lacking all canonical Notch signals in mature T cells). This strategy allowed us to engineer T cell inocula for transplantation into MHC-mismatched irradiated BALB/c mice, where Notch would be selectively inhibited in Tconv, Treg, or both (Figure 1A). We pursued this approach as we previously observed that expansion of Tregs following Notch blockade was driven by preexisting Tregs, rather than Tconv conversion to induced Tregs (4). Each inoculum was infused with T cell-depleted bone marrow (TCD BM) into lethally irradiated BALB/c recipients, followed by serial monitoring for acute GVHD severity and survival. Recipients of wild-type (WT) Tconv developed severe and lethal acute GVHD, regardless of Notch blockade in the Treg compartment (Figure 1B–C). Pan-Notch blockade via DNMAML expression in Tconv was highly protective against acute GVHD as measured by clinical scores and survival, whether or not Notch was active in co-infused Tregs (38). Thus, our data highlight an essential role for Notch signaling in CD4+ Tconv cells to mediate acute GVHD.
Figure 1. Conventional T cell-intrinsic Notch signaling is essential for GVHD induction after MHC-mismatched allogeneic bone marrow transplantation.
(A) Experimental design to assess the importance of conventional T cells (Tconv) and regulatory T cells (Tregs) in mediating the protective effects of Notch inhibition after allogeneic bone marrow transplantation. Tconv and Tregs were sort purified from CD4+-enriched cells from B6 FoxP3-IRES-mRFP or B6 FoxP3-IRES-mRFP;Tgcd4-cre;ROSADNMAML/+ (FIR-DNMAML) mice, and mixed at a ratio of 8:1 Tconv:Tregs (by analogy to the physiological ratio found in WT donors; 500,000:62,500) to generate four different donor cell groups. (B) Clinical GVHD score and (C) overall survival of lethally irradiated (8 Gy) BALB/c mice transplanted with the four experimental inocula described in (A). *p < 0.05 (Student’s t-test, A; Log-rank test, B). Data are representative of 2 experiments and at least 15 mice per group.
Establishment of an MHC-mismatched GVHD mouse model with a monoclonal population of donor alloantigen-specific CD4+ T cells
We recently reported that key Notch signals are delivered to T cells during the first 48 hours after allo-HCT (10). This narrow window provides an ideal opportunity to investigate cellular and molecular mechanisms of Notch signaling that shape pathogenic alloresponses in T cells. However, only a minor fraction of donor conventional T cells recognizes alloantigen and contributes to acute GVHD pathogenesis. Within this earliest post-transplant window, these rare cells remain difficult to track in standard models of acute GVHD. To bypass this pitfall and enable direct analyses of alloantigen-specific GVHD-causing cells, we established a new mouse model of MHC-mismatched GVHD with T cells from 4C×Rag1−/− TCR transgenic mice. 4C CD4+ T cells are a clonal population that react against I-Ad MHC class II through direct antigen recognition (26), and can be tracked easily by flow cytometry using TCR Vβ13 and/or CD90.1-directed antibodies. Use of 4C T cells on Rag1−/− background allowed us to focus specifically on Tconv, as 4C×Rag1−/− mice lack Tregs.
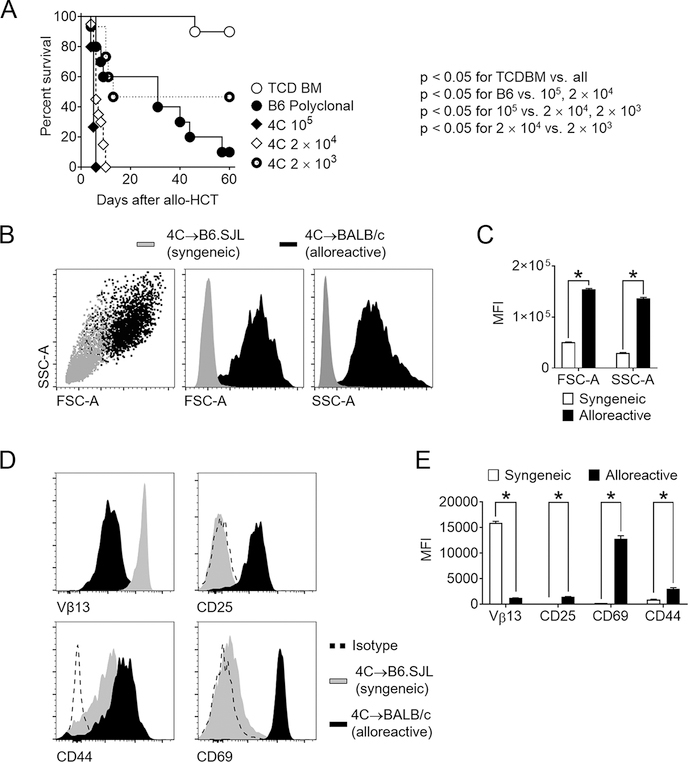
While 4C T cells have been studied in mouse models of skin transplantation (26), their capacity to cause GVHD has not been examined previously. We first studied GVHD phenotypes arising from transplantation of varying doses of 4C T cells, utilizing polyclonal WT B6 T cells as a point of comparison. Lethally irradiated BALB/c mice transplanted with WT TCD BM and 5×106 B6 splenocytes developed severe GVHD resulting in lethality of almost all recipients by day 60 after allo-HCT (Figure 2A). Transplantation of as few as 2×104 4C T cells induced universally lethal acute GVHD. Dose reduction to 2×103 4C T cells allowed partial recipient survival with evidence of acute GVHD. Thus, our new model was ideal to further investigate cellular changes in 4C T cells within the 48-hour window during which critical Notch signals are delivered (10). To document the allo-specificity of T cell activation in this model, 4C T cells were transplanted into irradiated B6.SJL (syngeneic) or BALB/c (allogeneic) hosts (Supplemental Fig. 1A). Alloreactive 4C T cells showed hallmarks of antigen-driven T cell activation, as compared to changes seen with 4C T cells undergoing homeostatic proliferation in syngeneic B6.SJL recipients. Alloreactive 4C T cells displayed greater size and granularity (Figure 2B–C). Decreased density of cell surface Vβ13+ TCR was consistent with downregulated surface TCR upon 4C T cell activation (Figure 2D–E). The allospecific T cell response was associated with significant upregulation of the early T cell activation markers CD69 and CD25, as seen both in their cell surface density (Figure 2D–E) and in the high proportion of 4C T cells expressing them (Supplemental Fig. 1B). In contrast, CD44 was induced to a larger extent in syngeneic recipients, indicating higher sensitivity of this marker to lymphopenic states independent of alloantigen encounter. Similar results were observed when 4C and BALB/c T cells were co-transferred to lethally irradiated BALB/c recipients and retrieved 36 hours after allo-HCT (Supplemental Fig. 1C–F). Thus, 4C T cells are capable of causing severe acute GVHD driven by alloantigen-specific T cell activation and effector differentiation.
Figure 2. Alloantigen-driven 4C T cells induce lethal GVHD after MHC-mismatched allogeneic bone marrow transplantation.
(A) Survival of lethally irradiated BALB/c mice transplanted with 5×106 TCD BM only, or TCD BM supplemented with 5×106 polyclonal B6 splenocytes, 105 4C T cells, 2×104 4C T cells, or 2×103 4C T cells. (B-E) Cellular hallmarks of the alloantigen-specific response in the 4C→BALB/c model of allo-HCT. Lethally irradiated recipients (B6.SJL – syngeneic; BALB/c-CD45.1 – allogeneic) received TCD BM and 5×106 4C T cells. 1.5 days after allo-HCT, secondary lymphoid organs were processed and lymphocytes isolated for analysis. (B-C) Forward scatter (FSC-A) and side scatter (SSC-A) showed dramatic increase in T cell size and granularity, consistent with acute activation. (D-E) The alloantigen-specific response led to robust 4C T cell activation. Histogram plots and cumulative data showed increased expression of the activation markers CD25, CD44, and CD69, and decrease in Vβ13 TCR chain surface density. *p < 0.05 (Log-rank test, A; Student’s t-test, C and E). Data are representative of at least 2 experiments and at least 10 mice per group.
Intrinsic Delta-like¼-mediated Notch signals in donor CD4+ T cells drive GVHD mortality
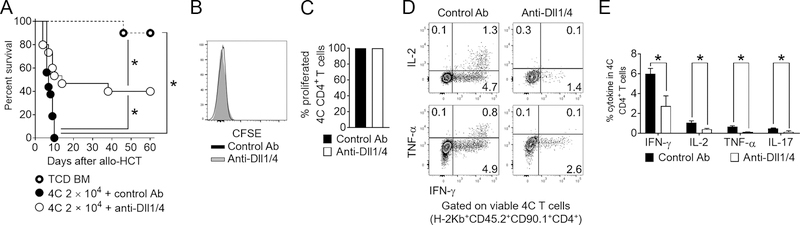
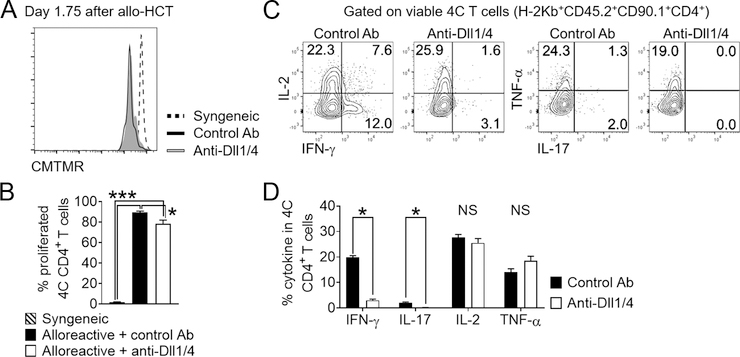
We previously reported in multiple mouse models of minor and major histocompatibility antigen-mismatched allo-HCT that Notch signaling is regulated by inputs from Dll1 and Dll4 ligands, and established Dll¼ targeting as a promising pre-clinical therapeutic strategy of GVHD prevention (6, 10, 11). In these studies, while the penetrance, severity and kinetics of disease varied, Notch inhibition and Dll¼ blockade with monoclonal antibodies resulted in profound protection from GVHD mortality/morbidity, while largely preserving GVL activity. To assess if the same Notch ligands are necessary for 4C T cell pathogenicity, lethally irradiated BALB/c recipients were treated with anti-Dll¼ or isotype control antibodies and transplanted with TCD BM plus 2×104 4C T cells (a uniformly lethal dose in the presence of Notch signaling). While 100% of isotype control recipients succumbed to GVHD within 10 days, anti-Dll¼-treated recipients showed GVHD protection, leading to long-term survival in ~40% of the recipients (Figure 3A). Thus, the degree of protection was similar to that afforded by a 10-fold reduction in T cell dose in this model (Figure 2). Similar to our results with polyclonal T cells in models of acute GVHD (6, 10), anti-Dll¼ therapy had no effect on initial 4C T cell proliferation, as shown at day 5 post-transplant (Figure 3B–C). At this time point, Dll¼ blockade inhibited the production of proinflammatory cytokines important for GVHD pathogenesis in spleen, mesenteric lymph nodes, and peripheral blood (IFN-γ, IL-2, IL-17, and TNF-α; Figure 3D–E, Figure S1G–H). Thus, Notch signals drive 4C T cell alloreactivity in vivo in a Dll¼-dependent manner.
Figure 3. Notch blockade mitigates acute GVHD driven by 4C conventional CD4+ T cells.
Lethally irradiated BALB/c mice were transplanted with 5×106 TCD BM only, or TCD BM supplemented with 2×103 4C T cells. 4C T cell recipients were treated with isotype control (blue circles) or anti-Dll1 plus anti-Dll4 (red circles) antibodies (5 mg/kg i.p. on day 0). (A) Percentage of surviving transplanted animals plotted as function of time. *p < 0.05, Log-rank test. (B-E) Lethally irradiated BALB/c mice transplanted with 4C T cells and treated with isotype control or anti-Dll¼ antibodies were sacrificed 5 days after allo-HCT and spleens retrieved for immunophenotypic analysis. (B-C) Notch blockade did not impair 4C Tconv proliferation, as assessed through dilution of CFSE. (D-E) Anti-Dll¼ treatment led to impaired cytokine production in 4C Tconv. Representative flow cytometry plots (D) and cumulative data (E) showing the percentage of donor-derived 4C T cells expressing individual intracellular cytokines after anti-CD3/CD28 restimulation. *p < 0.05, Student’s t-test.
Notch inhibition preserves early alloreactive T cell activation and TCR signaling strength, but impairs S6 and ERK½ phosphorylation
The earliest stages of CD4+ T cell priming proceed in distinct phases. Following antigen encounter, TCR signaling drives upregulation of early activation markers, nutrient sensors, and cytokine receptors followed by proliferation and cytokine production (39–42). The potential importance of Notch signaling in regulating each of these steps has been reported in other contexts (2, 6, 19, 20, 43–46). However, dominant cellular and molecular mechanisms that operate upon in vivo Notch inhibition during alloresponses remain unclear.
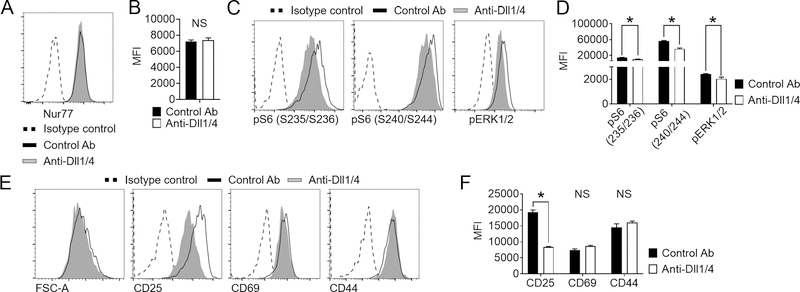
T cell fate during antigen-specific responses depends upon TCR signal strength, with stronger TCR inputs leading to enhanced effector differentiation and reactivity (47). To investigate if Notch blockade negatively regulates TCR signal strength leading to impaired effector fitness of alloreactive 4C T cells, we first evaluated intracellular expression of Nur77. Past work using Nur77(Nr4a1)-GFP allele showed that Nur77 expression is induced rapidly upon TCR activation. Unlike the early activation marker CD69, Nur77 is insensitive to inflammatory stimuli such as IL-2, TLR agonists, or type I interferons that are abundant during early allo-HCT, thus directly reflecting TCR signal strength (27, 48). Notch blockade had no impact on TCR signaling strength in 4C cells as measured through Nur77 expression at 24 hours, assessed by intracellular Nur77 staining (Figure 4A–B). Similar results were obtained upon assessing GFP expression in 4C T cells transplanted from 4C×Nur77-GFP donors (data not shown). We then assessed whether Dll¼ inhibition altered specific aspects of signal transduction during early T cell activation. Two major signaling pathways that are collectively activated by the TCR, costimulatory pathways, nutrient sensors, and cytokine receptors are the mTORC1 and the Ras/MAPK pathways. To quantify the impact of Notch blockade on these pathways, we performed phospho-flow cytometry for the mTORC1 target S6 and the MAPK targets ERK½ 24 hours post-transplantation. We co-injected both alloreactive 4C donors and syngeneic BALB/c donors into lethally irradiated BALB/c recipients and compared phosphorylation levels within the same sample, allowing us to control for non-cell autonomous effects and technical variability. Both pathways showed significantly higher activity in alloreactive 4C T cells than in co-injected syngeneic BALB/c T cells (Figure S2A–B). Dll¼ inhibition resulted in a significant decrease in both pS6(S235/S236) and pS6(S240/S244) levels, suggesting a cell-autonomous impairment in mTORC1 signaling, as well as defective Ras/MAPK activity, as seen through decreased pERK½ levels (Figure 4C–D). In contrast to the negative effect on pS6 and pERK½ levels, Notch blockade had limited impact on expression of activation markers in alloreactive 4C T cells. Aside from decreased levels of CD25 (a known direct target of Notch signaling (44, 49–51)), CD69 and CD44, as well as cell size, all hallmarks of T cell activation, were unaffected by anti-Dll¼ administration (Figure 4E–F). Collectively, these data suggested that Notch inhibition impairs select key signal transduction events during T cell priming, but not TCR signal strength nor overall T cell activation.
Figure 4. Impact of systemic Notch blockade on key cellular events in T cells during their in vivo priming by alloantigens.
(A-F) Lethally irradiated BALB/c mice were transplanted with TCD BM supplemented with 5×106 4C T cells and treated with isotype control or anti-Dll¼ antibodies on day 0. Animals were sacrificed 24 hours after allo-HCT, and secondary lymphoid organs retrieved for analysis. (A-B) Representative histogram plots (A) and cumulative data of mean fluorescence intensity (B) demonstrate that Notch blockade did not impact overall TCR signaling strength, as seen with indistinguishable Nur77 levels in 4C T cells isolated from mice treated with isotype control or anti-Dll¼ mAb. (C-D) Notch blockade impaired mTORC1 and Ras/MAPK signaling in alloreactive 4C T cells. Histogram plots (C) and cumulative data of mean fluorescence intensity (D) showing the abundance of phosphorylated S6 (S235/S236 and S240/S244 residues) and ERK½ in alloreactive 4C T cells. *p < 0.05, Student’s t-test. Data are representative of at least 3 experiments. (E-F) Notch blockade had limited impact on surface markers of alloreactive 4C T cell activation during in vivo antigen-mediated priming. Histogram plots (E) and cumulative data of mean fluorescence intensity (F) showing flow cytometric analysis of early T cell activation markers CD25 (a direct Notch target), CD69, and CD44. *p < 0.05, Student’s t-test. Data are representative of at least 3 experiments and 10 mice per group.
Notch blockade preserves early IL-2 and TNF-α production
Notch blockade did not impact early proliferation of 4C T cells, despite profoundly inhibiting production of multiple cytokines on day 5 post-transplant (Figure 3B–E), similar to our observations in polyclonal systems (2, 6, 10, 11). To better understand this finding, we assessed the effects of Notch blockade on proliferation and cytokine production during T cell priming at earlier time points. In 4C T cells retrieved 42 hours post-transplant from recipients treated with anti-Dll¼, we observed a minimal decrease in the proportion of cells that underwent initial division (Figure 5A–B). Simultaneously, Notch-blocked 4C T cells showed no impairment in the production of IL-2 or TNF-α (Figure 5C–D). In contrast, IFN-γ and IL-17A production was already markedly reduced at this early time point. These data suggest that during T cell priming, early Notch inputs are essential for inducing IFN-γ and IL-17A production. On the other hand, early Notch signals are dispensable for induction of IL-2 and TNF-α production, but necessary for sustaining IL-2 and TNF-α secretion at later time points. Thus, Notch-deprived alloreactive CD4+ T cells initially maintained the production of IL-2, although it subsequently decreased by day 5, while immediately losing the ability to produce other inflammatory cytokines with a central role in GVHD.
Figure 5. Systemic Notch blockade preserves selected early effector 4C T cell functions after allo-HCT.
B6-SJL (syngeneic) or BALB/c (allogeneic) mice were irradiated and transplanted with TCD BM supplemented with 5×106 4C T cells. BALB/c recipients were treated with isotype control or anti-Dll¼ mAb on day 0. Mice were sacrificed 42 hours after allo-HCT, and secondary lymphoid organs retrieved for analysis. (A-B) CMTMR dilution by donor 4C cells showed robust initial alloreactive T cell proliferation minimally affected by Notch blockade. (A) Representative histogram plots and (B) cumulative data with 4 animals analyzed from one of three experiments and a total of 10 animals per group. (C-D) Intracellular cytokine staining in donor 4C T cells showed no impact of Notch blockade on IL-2 and TNF-α production, but markedly decreased IFN-γ and IL-17. Representative plots (C) and cumulative data (D) from one of two experiments. * p < 0.05 (Student’s t-test, B; ANOVA, D).
Notch blockade impacts a limited number of genes regulated by the alloresponse
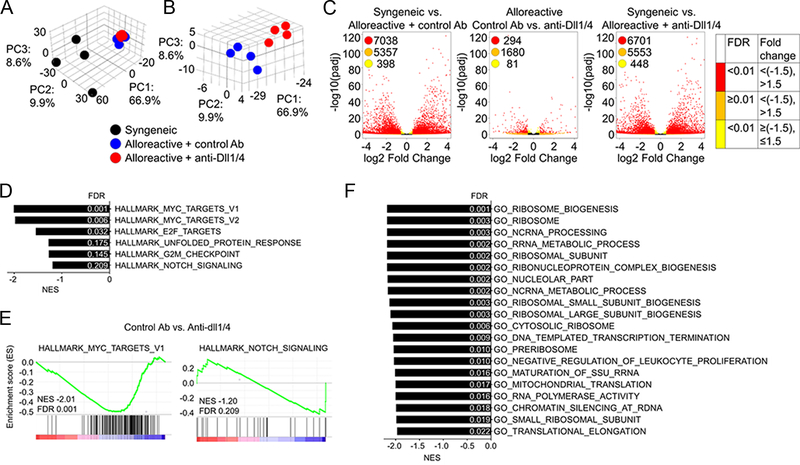
Given that all of the effects of Notch blockade are mimicked by expression of DNMAML, an inhibitor of the Notch transcriptional activation complex (2, 4), we hypothesized that transcriptional changes arising after early Notch blockade have mechanistic relevance in GVHD. To probe the effects of Notch on the 4C T cell transcriptome during T cell priming in an unbiased fashion, we performed RNA-Seq analysis. To account for non-cognate gene expression changes driven by the inflammatory milieu and the lymphopenic environment, we sort-purified 4C T cells from the following experimental groups of irradiated recipient mice: 1) 4C→B6.SJL (syngeneic control group); 2) 4C→BALB/c treated with control antibodies; 3) 4C→BALB/c treated with anti-Dll¼. 4C T cells were retrieved 36 hours after allo-HCT, followed by RNA isolation and generation of stranded amplified cDNA library before sequencing and analysis. Principal component analysis highlighted a distinct alloantigen-driven signature (Figure 6A) with limited variance caused by Dll¼ Notch ligand inhibition (Figure 6B). Consistently, we identified a robust differential gene expression between syngeneic and both alloreactive 4C T cell groups. Based on pre-defined cutoffs (gene-expression fold change [>1.5×, <−1.5×] and false discovery rate [FDR; <0.01]), Notch blockade in alloreactive T cells differentially regulated expression of 294 out of total 20,034 identified genes, whereas ~7000 genes were seen as differentially regulated between both alloreactive data sets and syngeneic 4C T cells (Figure 6C). To uncover regulation of distinct pathways by Notch blockade (thus, highlighting mechanistically relevant pathways in GVHD that are regulated by Notch), we performed Gene Set Enrichment Analysis (GSEA) for Hallmark and Gene Ontology (GO) gene sets from the Molecular Signatures Database. While myc mRNA was only mildly reduced (~20%) by anti-Dll¼ administration (Figure 8C), Notch blockade consistently downregulated expression of multiple Myc targets (Figure 6D–E). Normalized enrichment scores for the HALLMARK_NOTCH_SIGNALING gene set did not signify a major impact of Dll¼ blockade on genes previously published as regulated by Notch in other contexts (Figure 6D–E). In the GO data set, Notch blockade also broadly regulated ribosome biology associated genes (Figure 6F), suggesting a potential impact on ribosome homeostasis and translation.
Figure 6. Notch signaling has a unique but limited transcriptional footprint during priming of alloreactive T cells.
(A) 5×106 CMTMR labeled 4C T cells were transplanted into lethally irradiated BALB/c (allogeneic) or B6-SJL (syngeneic) recipient. Allogeneic recipients received anti-Dll¼ or isotype control antibody on day 0. 42 hours after transplantation 4C T cells were sort-purified for RNA isolation, library generation, and RNA-Seq. (A-B) Principal component analysis of biological samples used in this study. The percentage of variance captured by each of the three principal components demonstrated alloantigen regulation of T cell response (A) and more limited changes of the alloreactive T cell transcriptome as a result of Notch blockade (B). (C) Volcano plots displaying differentially expressed genes in 4C T cells within the three different comparison groups (syngeneic vs. alloreactive + control antibody (Ab); syngeneic vs. alloreactive + anti-Dll¼ Ab; alloreactive control vs. anti-Dll¼ Ab). Red color-coded dots represent genes with significant differential expression at pre-defined 1.5-fold change. (D-F) GSEA analysis of RNA-Seq data, showing individual gene sets and select enrichment plots from Hallmark (D-E) and gene sets from Gene Ontology (F) collections significantly regulated by Notch blockade in 4C T cells.
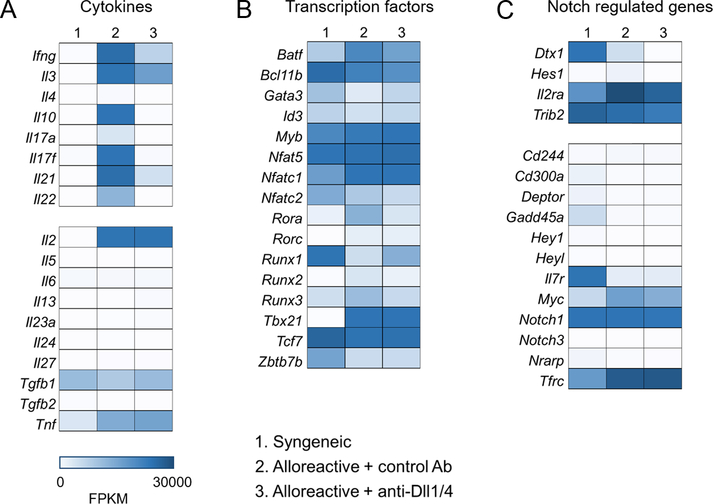
Figure 8. The transcriptional effects of Notch signaling in alloreactive T cells are distinct from those induced by oncogenic Notch.
Heatmaps showing differential expression of genes encoding cytokines (A), transcription factors important for T cell homeostasis (B), and genes regulated by Notch signaling in T-ALL (C). Notch regulated expression of many, but not all cytokines with a mechanistic role in GVHD pathogenesis (A). Notch did not impact 4C Th polarization and had no or limited impact on master transcription factors of T helper lineages (B). Notch signaling targets in conventional alloreactive T cells minimally overlapped with those observed in Notch-dependent T-ALL (C).
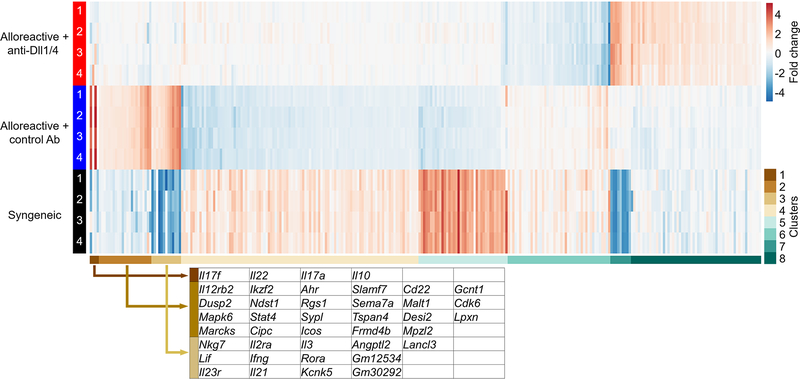
To provide further insight into genes regulated by Notch blockade and identify groups of genes with similar expression patterns, we performed k-means clustering (Figure 7, Supplemental table II). Interestingly, clustering analysis revealed grouping of genes downregulated by Notch and with emerging roles in GVHD (such as Il17 and Il10-family cytokines in cluster 1, or Ahr and Icos in cluster 2). Cluster 4 contained genes upregulated by Notch blockade, hence not direct targets, including Cblb, a negative regulator of T cell activation that we reported previously as upregulated in allogeneic T cells upon Notch blockade (4). Interestingly, this cluster also contained Bach2, another repressor of T cell function. Bach2 is particularly interesting given that its effects on regulatory and effector T cell function mimic our observations with effective Notch blockade in GVHD (52–55). Overall, k-means analyses suggest that Notch blockade synergistically regulated multiple alloantigen-induced pathogenic pathways.
Figure 7. k-means clustering analysis identifies Notch regulation of multiple GVHD-associated genes in alloreactive 4C T cells.
k-means clustering of 294 genes differentially regulated by Notch blockade in alloreactive 4C T cells during T cell priming period. Individual members of clusters 1–3 are highlighted and reveal several well-defined genes with known mechanistic relevance in GVHD (Il17f, I121, Il2ra, Ahr, Icos). Primary data from these clusters and all other clusters is presented in Supplemental Table 2.
The transcriptional landscape regulated by Notch in mature T cells differs from T-ALL
Our prior work and results presented in this study identified broad regulation of cytokine production in alloantigen-specific T cells, with no effects on IL-2 and TNF-α production within the T cell priming phase. To better characterize the impact of Notch signaling on early cytokine polarization in alloreactive 4C T cells, we extracted data on cytokine expression from our RNA-Seq dataset. Notch blockade downregulated the transcription of genes encoding cytokines with mechanistic relevance in GVHD (e.g. Il17f, Il21, and Il22). However, other cytokines with direct pathogenic importance in GVHD, including Il2, Tnfa, and Il27, remained unaffected by Dll¼ blockade (Figure 8A). Since the multifaceted role of Notch signaling in mature T cells was also ascribed to its ability to regulate multiple Th transcription factor loci (56), we analyzed the impact of Notch blockade on expression of transcription factors with known functions in T cell effector differentiation. Notch signaling did not significantly affect prototypic Th transcription factor expression (Tbx21, Gata3, Rorc; Figure 8B), indicating that cytokine production defects were not a result of skewed Th differentiation, but rather driven by alternative mechanisms. In addition, Notch signaling blockade showed limited to no impact on the expression of other transcription factors with a role in T cell homeostasis (Batf, Bcl11b, Runx1, Runx3; Figure 8B), suggesting that Notch-induced changes in alloreactive T cells were independent of Th polarization.
Notably, our dataset in the GSEA analysis did not highly correlate with published data (57–61) on Notch-regulated pathways. Moreover, prototypic Notch targets uncovered during studies of oncogenic Notch signaling in T-ALL have thus far failed to provide an obvious mechanistic explanation for decreased GVHD upon Notch inhibition in mature T cells. To investigate the regulation of individual putative Notch targets, we compared Notch targets identified in published T-ALL datasets with our RNA-Seq data. Dll¼ inhibition downregulated expression of only few well-known Notch target genes, including Dtx1, Hes, Il2ra (Cd25), and Trib2 (Figure 8C). In concordance with GSEA results, the Notch blockade signature had limited overlap with previously published data on Notch targets in T-ALL (14). Other Notch target gene transcripts, such as Tbx21, Gata3, Rorc, HeyL, Hey1, and Nrarp were either not downregulated by Dll¼ inhibition, or present below the limit of detection of RNA-Seq analysis (Figure 8B–C). Finally, Notch inhibition did not affect the abundance of Pten mRNA. Collectively, the genetic signature of active Notch in the T cell alloresponse was starkly different from that induced by oncogenic Notch. Instead, Notch signaling in alloreactive conventional T cells induced distinct molecular pathways with converging roles in pathogenic alloreactivity.
Discussion
We previously demonstrated the central role of Notch signaling in mature T cells to drive pathogenic alloresponses in multiple major and minor histocompatibility antigen-mismatched mouse models of acute and chronic GVHD (2, 4, 6, 10). However, prior to the current study, the key cellular compartment (effector vs. regulatory T cell) through which the beneficial effects of Notch inhibition was mediated was unclear. In this study, we engineered mixed T cell grafts to uncover the essential function of active Notch in conventional rather than regulatory T cells to mediate GVHD pathogenicity. Furthermore, we identified unique cellular effects and a molecular landscape associated with Notch inhibition in alloantigen-specific conventional T cells at very early stages after transplantation.
We and others have reported an increased expansion of donor-derived Tregs after allo-HCT and during GVHD protection upon genetic or biochemical Notch inhibition (2, 4, 6, 10, 11). To specifically investigate the role of Notch signaling in Tregs in this context, Charbonnier et al. used a Foxp3-Cre transgene to inactivate essential components of the Notch pathway (including Pofut1, Rbpj and Notch1) in Tregs only (9). Using this approach, they reported protection from GVHD after allogeneic transplantation of total splenocytes from mice with Notch-deficient Tregs, suggesting that Notch inhibition in Tregs was sufficient to protect from GVHD. However, this interpretation did not take into account the fact that chronic Notch inhibition in Tregs appeared to impact the composition and function of the Tconv compartment (Tconv hyporesponsiveness, decreased cytokine production, and increased prevalence of memory T cells), and that engineering of the graft would be necessary to formally restrict Notch inhibition to the Treg compartment. In our study, graft engineering offered an experimental advantage and enabled us to document directly the central pathogenic role of Notch in Tconv but not Tregs during GVHD.
In view of these findings and to better understand Notch regulation in Tconv, we developed a novel Tconv-driven MHC-mismatched model of acute GVHD, mediated by I-Ad-reactive TCR-transgenic CD4+ 4C Rag1−/− donor T cells, which lack Tregs due to the absence of I-Ad antigen during T cell development in the thymus and due to the absence of endogenous Rag1 expression. 4C T cells were potent inducers of GVHD mortality, yet remained sensitive to Notch inhibition by neutralizing antibodies against Dll¼ Notch ligands.
Thus, this model was ideal to focus our analyses on pathogenic T cells, avoid pitfalls of bulk T cell analyses at early time points, and define the cellular and molecular events underlying Notch-driven alloresponses in the absence of any potential changes in the TCR repertoire. Focusing on the previously identified early window of critical Notch activity in GVHD (10), we described relatively limited effects of Notch signaling on 4C T cells during their priming and early activation. Our analysis provided several lines of evidence that Notch inhibition preserves key aspects of T cell priming. First, Notch-deprived 4C cells displayed no defects in the upregulation of the activation markers CD69 and CD44. Second, transcription and protein synthesis of IL-2, which receives direct inputs from the TCR through the transcription factors AP-1 and NFAT, was unimpaired by systemic Dll¼ inhibition at early time points, while decreasing later. How the lack of sustained IL-2 production influences 4C T cell (or alloreactive CD4+ T cell in general) proliferation remains unclear. It is possible that in the context of strong TCR stimulation such as is seen in vivo during alloresponses, IL-2 production is dispensable for continued proliferation, particularly in the CD4+ Tconv compartment where TCR-dependent and mitogenic cytokine-independent regulation dominate (62, 63). Even if not important for Tconv proliferative responses early post-transplant, the initial burst of IL-2 in polyclonal GVHD models (10) may help explain the robust expansion of Tregs that we and others have reported upon Notch inhibition, as Treg expansion exquisitely depends on IL-2-mediated signals (64). Moreover, Dll¼ inhibition did not impact transcriptional regulation of Th skewing. While our findings conflict with a recent report that Notch enhances CD69 expression, cell size, IL-2 production, and proliferation both in vitro and in vivo via a co-stimulatory-like effect downstream of B7-CD28 signaling (19), it is possible that Notch functions as a costimulatory modulator but with different effects depending on context, antigen strength and availability, and other concomitant signals.
Both mTORC1 activity and Ras/MAPK activity were impaired in Notch-deprived alloreactive T cells during priming. mTORC1 activation is regulated by several extracellular stimuli, including amino acids, cytokine/growth factor receptors, TCR signaling, and the B7 family of costimulatory molecules (65, 66). Similarly, Ras/MAPK signaling sits downstream of TCR signaling, cytokine/growth factor receptors, and the B7/CD28 family of costimulatory molecules. Given that TCR signaling, T cell co-simulation, and cytokine/growth factor signaling simultaneously occur during T cell priming, it is unclear whether Notch inhibition regulates some or all of the aforementioned pathways to modulate mTORC1 and Ras/MAPK. Precise spatiotemporal analysis of phosphorylation signals could be helpful in dissecting contributions from each upstream signal. While Nur77-GFP levels were not altered with Notch inhibition, Nur77-GFP has been reported to mainly read out TCR-mediated Protein Kinase C (PKC) activity proportionately to TCR signal strength (48). Thus, while PKC activity is most likely intact in Notch-deprived alloreactive T cells, it is possible that Notch modulates specific arms of TCR signaling, such as Ras/MAPK activation and/or AKT-dependent mTORC1 activation. Alternatively, impairment of mTORC1 and Ras/MAPK could be due to impaired B7-mediated costimulatory signals (19) or diminished cytokine/growth factor signaling. Intact early production of IL-2, which receives direct inputs from Ca++-dependent NFAT, costimulation-dependent NF-κB and ERK½-dependent AP-1 (67), would argue that both TCR-dependent and B7-mediated ERK½ activity are initially intact.
While global Ras/MAPK and mTORC1 activity were both impaired (but not ablated) in alloreactive T cells during priming, Il2 transcription and IL-2 protein were initially preserved. These data suggest that the highly inflammatory milieu of allotransplantation conditioning, in conjunction with strong T cell receptor signaling, stimulate alternative upstream signals that are sufficient to drive IL-2 expression, at least at early time points. Given that IL-2 production by Notch-deprived alloreactive T cells was impaired at later time points after allotransplantation (Figure 3D–E), we speculate that dynamic involvement of upstream regulators of Il2 transcription, with Notch-regulated MAPK/mTORC1 signaling being dispensable early but essential later for maximal IL-2 production. Our findings should be considered in light of research on the epigenetic regulation of Il2 transcription that revealed highly dynamic involvement of proximal regulatory elements and distal enhancers through complex sequential three-dimensional rearrangements of the chromatin architecture (68).
To capture the overall impact of Notch on the molecular landscape of T cell alloreactivity, we relied on an unbiased transcriptional approach in an alloantigen-specific T cell model. Utilizing a monoclonal population of donor cells allowed us to isolate antigen-specific T cells in a precise manner, while preserving the profound impact of Notch inhibition on GVHD mortality/morbidity. Despite differences in disease kinetics and severity as compared to the polyclonal BALB/c→B6 major mismatch and B10.D2→BALB/c minor mismatch models, we observed concordant phenotypic and functional changes, as well as regulation of gene expression in both of these models. Thus, our molecular observations have broad relevance to immune pathogenesis in multiple complementary mouse GVHD models. Despite the known limitations of mouse allo-HCT models, and although more research is needed, we speculate that conserved features of Notch as an ancient signaling pathway will apply to human allogeneic transplantation as well.
Our study represents the first RNA-Seq-based transcriptional analysis of Notch effects in pathogenic T cells during GVHD. Despite its profound functional impact on alloantigen-driven responses, Notch had a surprisingly narrow effect on the transcriptional landscape, differentially regulating only 294 of ~7000 genes. Prototypic Notch targets identified in other contexts showed minimal overlap with differentially expressed genes that we identified in alloreactive T cells. In particular, alloreactive 4C cells expressed low levels of Hes1, while its putative repressed transcriptional target Pten was unaffected by Notch inhibition. Thus, the Notch-Hes1-PTEN transcriptional axis might operate in T-ALL and developing thymocytes, as reported (57, 69), but is unlikely to be essential in alloreactive T cells. Several Notch target genes, including Dtx1, Il2ra, and Trib2 were downregulated upon Dll¼ blockade in alloreactive T cells, though, aside from Il2ra, an apparent connection to GVHD pathophysiology is lacking. Consistent with our previous observations in polyclonal models of GVHD, Notch did not regulate the expression of key transcription factors that control helper CD4+ lineage fate decisions in mature T cells, including Tbx21, Rorc, and Gata3 (2, 4). On the other hand, the expression of multiple cytokines (Ifng, Il17f, Il21, Il22) and cytokine receptors (Il1r1, Il23r) with a reported role in GVHD (70–74) was downregulated early on by Dll¼ inhibition. Notch blockade also downregulated expression of Ahr and Icos (75, 76), identified as therapeutic targets in GVHD. Additional gene expression changes could be related mechanistically to the observed GVHD abrogation. A particularly interesting candidate with broad impact on adaptive immune responses and upregulated by Notch blockade in our dataset is Bach2. Beyond transcriptional regulation, our data suggest early defects in PI3K/Akt/mTOR signaling, normally responsible for BACH2 downregulation (55). BACH2 broadly regulates adaptive immune responses, stabilizing Tregs, promoting memory over effector T cell responses, and tumor immunosuppression (52–55). While transcriptional disruption is essential to confer the benefits of Notch blockade, the contribution of epigenetic mechanisms to protective benefits against GVHD cannot be ruled out and is also suggested by our identification of multiple epigenetic regulators active in adaptive immunity as impacted by Notch blockade (Dnmt3a, Mbd2, Jarid).
In summary, we have identified a CD4+ Tconv-intrinsic role for Notch signaling in alloreactivity and GVHD. Notch-deprived alloreactive CD4+ Tconv did not display overt early defects in overall antigen sensitivity, as they exhibited preserved activation marker upregulation, IL-2 production, and initial proliferation. In contrast, Notch-deprived CD4+ T cells rapidly acquired a defect in IFNγ and IL-17 production despite preserved Tbx21 and Rorc transcription, while exhibiting diminished mTORC1 and Ras/MAPK activity. Finally, our RNA-Seq data defined the Notch-regulated transcriptional landscape in mature alloreactive T cells as very distinct from that observed in T-ALL or developing T cells.
Our study emphasizes the increasingly recognized versatility of Notch signaling in the immune system that extends beyond its developmental roles. While work presented here focused on CD4+ T cells, we previously documented many parallels between Notch effects in the CD4+ and CD8+ T cell compartments in alloreactivity (4). Collectively, our data suggest that previously reported molecular mechanisms cannot account for the role of Notch in alloreactive T cells, and propose a new set of unique mechanisms through which Notch modulates T cell alloreactivity. We provide molecular cues for this functional divergence and importantly, identify not only known targets with importance in GVHD as regulated by Notch, but also others that have a potential impact on GVHD regulation but have not yet been probed in this context. Since Notch blockade affects only selected aspects of alloreactive T cell function without inducing global immunosuppression, we postulate that these targets may identify keys to successful and specific control of GVHD, as well as pathologic alloresponses beyond the field of allo-HCT.
Supplementary Material
Key points.
Notch signals in conventional CD4+ T cells are essential for GVHD pathogenesis.
Notch induces a unique molecular signature during priming of alloreactive T cells.
Acknowledgments
Supported by: National Institute of Allergy and Infectious Diseases (NIAID; R01-AI091627, to IM and R37-AI34495 to BRB), National Heart, Lung, and Blood Institute (NHLBI; R01-HL118979 and R01-HL056067, to BRB), the Leukemia and Lymphoma Society (CDP 1227–14, to IM, TRP 6462–15, to IM and BRB), Research training award from the American Society of Hematology (to VR), Career Development Award from the American Society for Blood and Marrow Transplantation (to VR); Huntsman Cancer Foundation (to VR); T32-GM007315 from the National Institute of General Medical Sciences (NIGMS) (to JC); T32-GM007863 from NIGMS (to JC and EP); and F30-AI136325 from NIAID (to EP).
Nonstandard abbreviations used
- allo-HCT
allogeneic hematopoietic cell transplantation
- BM
bone marrow
- Dll
Delta-like ligand
- DNMAML
dominant negative Mastermind-like
- GVHD
graft-versus-host disease
- GVL
graft-versus-leukemia
- T-ALL
T cell acute lymphoblastic leukemia
- TCD
T cell depleted
- Tconv
conventional CD4+FoxP3neg T cells
- Tregs
regulatory CD4+FoxP3+ T cells
- WT
wild-type
References
- 1.Radtke F, Fasnacht N, and Macdonald HR. 2010. Notch signaling in the immune system. Immunity 32: 14–27. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Sandy AR, Wang J, Radojcic V, Shan GT, Tran IT, Friedman A, Kato K, He S, Cui S, Hexner E, Frank DM, Emerson SG, Pear WS, and Maillard I. 2011. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood 117: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riella LV, Ueno T, Batal I, De Serres SA, Bassil R, Elyaman W, Yagita H, Medina-Pestana JO, Chandraker A, and Najafian N. 2011. Blockade of Notch ligand delta1 promotes allograft survival by inhibiting alloreactive Th1 cells and cytotoxic T cell generation. J Immunol 187: 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandy AR, Chung J, Toubai T, Shan GT, Tran IT, Friedman A, Blackwell TS, Reddy P, King PD, and Maillard I. 2013. T cell-specific notch inhibition blocks graft-versus-host disease by inducing a hyporesponsive program in alloreactive CD4+ and CD8+ T cells. J Immunol 190: 5818–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mochizuki K, Xie F, He S, Tong Q, Liu Y, Mochizuki I, Guo Y, Kato K, Yagita H, Mineishi S, and Zhang Y. 2013. Delta-like ligand 4 identifies a previously uncharacterized population of inflammatory dendritic cells that plays important roles in eliciting allogeneic T cell responses in mice. J Immunol 190: 3772–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, Shan GT, Radojcic V, Friedman A, Gridley T, Shelton A, Reddy P, Samuelson LC, Yan M, Siebel CW, and Maillard I. 2013. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J Clin Invest 123: 1590–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roderick JE, Gonzalez-Perez G, Kuksin CA, Dongre A, Roberts ER, Srinivasan J, Andrzejewski C Jr., Fauq AH, Golde TE, Miele L, and Minter LM. 2013. Therapeutic targeting of NOTCH signaling ameliorates immune-mediated bone marrow failure of aplastic anemia. J Exp Med 210: 1311–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood S, Feng J, Chung J, Radojcic V, Sandy-Sloat AR, Friedman A, Shelton A, Yan M, Siebel CW, Bishop DK, and Maillard I. 2015. Transient blockade of delta-like Notch ligands prevents allograft rejection mediated by cellular and humoral mechanisms in a mouse model of heart transplantation. J Immunol 194: 2899–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charbonnier LM, Wang S, Georgiev P, Sefik E, and Chatila TA. 2015. Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat Immunol 16: 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung J, Ebens CL, Perkey E, Radojcic V, Koch U, Scarpellino L, Tong A, Allen F, Wood S, Feng J, Friedman A, Granadier D, Tran IT, Chai Q, Onder L, Yan M, Reddy P, Blazar BR, Huang AY, Brennan TV, Bishop DK, Ludewig B, Siebel CW, Radtke F, Luther SA, and Maillard I. 2017. Fibroblastic niches prime T cell alloimmunity through Delta-like Notch ligands. J Clin Invest 127: 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radojcic V, Paz K, Chung J, Du J, Perkey ET, Flynn R, Ivcevic S, Zaiken M, Friedman A, Yan M, Pletneva MA, Sarantopoulos S, Siebel CW, Blazar BR, and Maillard I. 2018. Notch signaling mediated by Delta-like ligands 1 and 4 controls the pathogenesis of chronic GVHD in mice. Blood 132: 2188–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng AP, Ferrando AA, Lee W, Morris J. P. t., Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, and Aster JC. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306: 269–271. [DOI] [PubMed] [Google Scholar]
- 13.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, and Ferrando AA. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 103: 18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, Bernstein BE, Kieff E, and Aster JC. 2011. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A 108: 14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, Asp P, Hadler M, Rigo I, De Keersmaecker K, Patel J, Huynh T, Utro F, Poglio S, Samon JB, Paietta E, Racevskis J, Rowe JM, Rabadan R, Levine RL, Brown S, Pflumio F, Dominguez M, Ferrando A, and Aifantis I. 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, Blacklow SC, Liu XS, and Aster JC. 2014. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci U S A 111: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong CT, Sedy JR, Murphy KM, and Kopan R. 2008. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS One 3: e2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandy AR, Stoolman J, Malott K, Pongtornpipat P, Segal BM, and Maillard I. 2013. Notch signaling regulates T cell accumulation and function in the central nervous system during experimental autoimmune encephalomyelitis. J Immunol 191: 1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laky K, Evans S, Perez-Diez A, and Fowlkes BJ. 2015. Notch signaling regulates antigen sensitivity of naive CD4+ T cells by tuning co-stimulation. Immunity 42: 80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailis W, Yashiro-Ohtani Y, Fang TC, Hatton RD, Weaver CT, Artis D, and Pear WS. 2013. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity 39: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, Kostura L, Fauq AH, Simpson K, Such KA, Miele L, Golde TE, Miller SD, and Osborne BA. 2005. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol 6: 680–688. [PubMed] [Google Scholar]
- 22.Keerthivasan S, Suleiman R, Lawlor R, Roderick J, Bates T, Minter L, Anguita J, Juncadella I, Nickoloff BJ, Le Poole IC, Miele L, and Osborne BA. 2011. Notch signaling regulates mouse and human Th17 differentiation. J Immunol 187: 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, and Flavell RA. 2004. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117: 515–526. [DOI] [PubMed] [Google Scholar]
- 24.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, and Flavell RA. 2007. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity 27: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan YY, and Flavell RA. 2005. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A 102: 5126–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan TV, Hoang V, Garrod KR, Liu FC, Hayden T, Kim J, and Kang SM. 2008. A new T-cell receptor transgenic model of the CD4+ direct pathway: level of priming determines acute versus chronic rejection. Transplantation 85: 247–255. [DOI] [PubMed] [Google Scholar]
- 27.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, and Hogquist KA. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 208: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, and Siebel CW. 2010. Therapeutic antibody targeting of individual Notch receptors. Nature 464: 1052–1057. [DOI] [PubMed] [Google Scholar]
- 29.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, Singh M, Chien M, Tan C, Hongo JA, de Sauvage F, Plowman G, and Yan M. 2006. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 444: 1083–1087. [DOI] [PubMed] [Google Scholar]
- 30.Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, Manley NR, Duarte A, Macdonald HR, and Radtke F. 2008. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med 205: 2515–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, and Aguet M. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10: 547–558. [DOI] [PubMed] [Google Scholar]
- 32.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, and Hirai H. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18: 675–685. [DOI] [PubMed] [Google Scholar]
- 33.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, and Owen MJ. 2004. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol 5: 638–644. [DOI] [PubMed] [Google Scholar]
- 34.Cooke KR, Kobzik L, Martin TR, Brewer J, Delmonte J Jr., Crawford JM, and Ferrara JL. 1996. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood 88: 3230–3239. [PubMed] [Google Scholar]
- 35.Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, Cyster JG, and Luther SA. 2007. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol 8: 1255–1265. [DOI] [PubMed] [Google Scholar]
- 36.Fasnacht N, Huang HY, Koch U, Favre S, Auderset F, Chai Q, Onder L, Kallert S, Pinschewer DD, MacDonald HR, Tacchini-Cottier F, Ludewig B, Luther SA, and Radtke F. 2014. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med 211: 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapinski PE, Qiao Y, Chang CH, and King PD. 2011. A role for p120 RasGAP in thymocyte positive selection and survival of naive T cells. J Immunol 187: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopan R, and Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenkins MR, Tsun A, Stinchcombe JC, and Griffiths GM. 2009. The strength of T cell receptor signal controls the polarization of cytotoxic machinery to the immunological synapse. Immunity 31: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Panhuys N, Klauschen F, and Germain RN. 2014. T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity 41: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tubo NJ, and Jenkins MK. 2014. TCR signal quantity and quality in CD4(+) T cell differentiation. Trends Immunol 35: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mempel TR, Henrickson SE, and Von Andrian UH. 2004. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427: 154–159. [DOI] [PubMed] [Google Scholar]
- 43.Eagar TN, Tang Q, Wolfe M, He Y, Pear WS, and Bluestone JA. 2004. Notch 1 signaling regulates peripheral T cell activation. Immunity 20: 407–415. [DOI] [PubMed] [Google Scholar]
- 44.Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, and Pear WS. 2003. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol 171: 2896–2903. [DOI] [PubMed] [Google Scholar]
- 45.Maekawa Y, Ishifune C, Tsukumo S, Hozumi K, Yagita H, and Yasutomo K. 2015. Notch controls the survival of memory CD4+ T cells by regulating glucose uptake. Nat Med 21: 55–61. [DOI] [PubMed] [Google Scholar]
- 46.Palaga T, Miele L, Golde TE, and Osborne BA. 2003. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol 171: 3019–3024. [DOI] [PubMed] [Google Scholar]
- 47.Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, and Jameson SC. 2015. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol 16: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zikherman J, Parameswaran R, and Weiss A. 2012. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature 489: 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, and Pear WS. 2006. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med 203: 2239–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Backer RA, Helbig C, Gentek R, Kent A, Laidlaw BJ, Dominguez CX, de Souza YS, van Trierum SE, van Beek R, Rimmelzwaan GF, ten Brinke A, Willemsen AM, van Kampen AH, Kaech SM, Blander JM, van Gisbergen K, and Amsen D. 2014. A central role for Notch in effector CD8(+) T cell differentiation. Nat Immunol 15: 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathieu M, Duval F, Daudelin JF, and Labrecque N. 2015. The Notch signaling pathway controls short-lived effector CD8+ T cell differentiation but is dispensable for memory generation. J Immunol 194: 5654–5662. [DOI] [PubMed] [Google Scholar]
- 52.Tsukumo S, Unno M, Muto A, Takeuchi A, Kometani K, Kurosaki T, Igarashi K, and Saito T. 2013. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc Natl Acad Sci U S A 110: 10735–10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roychoudhuri R, Clever D, Li P, Wakabayashi Y, Quinn KM, Klebanoff CA, Ji Y, Sukumar M, Eil RL, Yu Z, Spolski R, Palmer DC, Pan JH, Patel SJ, Macallan DC, Fabozzi G, Shih HY, Kanno Y, Muto A, Zhu J, Gattinoni L, O’Shea JJ, Okkenhaug K, Igarashi K, Leonard WJ, and Restifo NP. 2016. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17: 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roychoudhuri R, Eil RL, Clever D, Klebanoff CA, Sukumar M, Grant FM, Yu Z, Mehta G, Liu H, Jin P, Ji Y, Palmer DC, Pan JH, Chichura A, Crompton JG, Patel SJ, Stroncek D, Wang E, Marincola FM, Okkenhaug K, Gattinoni L, and Restifo NP. 2016. The transcription factor BACH2 promotes tumor immunosuppression. J Clin Invest 126: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, Sciume G, Zare H, Vahedi G, Dema B, Yu Z, Liu H, Takahashi H, Rao M, Muranski P, Crompton JG, Punkosdy G, Bedognetti D, Wang E, Hoffmann V, Rivera J, Marincola FM, Nakamura A, Sartorelli V, Kanno Y, Gattinoni L, Muto A, Igarashi K, O’Shea JJ, and Restifo NP. 2013. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, and Zhao K. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zuniga-Pflucker JC, Dominguez M, and Ferrando AA. 2007. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 13: 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dohda T, Maljukova A, Liu L, Heyman M, Grander D, Brodin D, Sangfelt O, and Lendahl U. 2007. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res 313: 3141–3152. [DOI] [PubMed] [Google Scholar]
- 59.Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, Ibrahim S, Skokos D, Armstrong SA, Levine RL, Park CY, and Aifantis I. 2013. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 210: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikhailik A, Mazella J, Liang S, and Tseng L. 2009. Notch ligand-dependent gene expression in human endometrial stromal cells. Biochem Biophys Res Commun 388: 479–482. [DOI] [PubMed] [Google Scholar]
- 61.Chang WH, Ho BC, Hsiao YJ, Chen JS, Yeh CH, Chen HY, Chang GC, Su KY, and Yu SL. 2016. JAG1 Is Associated with Poor Survival through Inducing Metastasis in Lung Cancer. PLoS One 11: e0150355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Au-Yeung BB, Smith GA, Mueller JL, Heyn CS, Jaszczak RG, Weiss A, and Zikherman J. 2017. IL-2 Modulates the TCR Signaling Threshold for CD8 but Not CD4 T Cell Proliferation on a Single-Cell Level. J Immunol 198: 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Au-Yeung BB, Zikherman J, Mueller JL, Ashouri JF, Matloubian M, Cheng DA, Chen Y, Shokat KM, and Weiss A. 2014. A sharp T-cell antigen receptor signaling threshold for T-cell proliferation. Proc Natl Acad Sci U S A 111: E3679–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou T, Satake A, Corbo-Rodgers E, Schmidt AM, Farrar MA, Maltzman JS, and Kambayashi T. 2012. Cutting edge: IL-2 signals determine the degree of TCR signaling necessary to support regulatory T cell proliferation in vivo. J Immunol 189: 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chi H 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol 12: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Powell JD, Pollizzi KN, Heikamp EB, and Horton MR. 2012. Regulation of immune responses by mTOR. Annu Rev Immunol 30: 39–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HP, Imbert J, and Leonard WJ. 2006. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev 17: 349–366. [DOI] [PubMed] [Google Scholar]
- 68.Mehra P, and Wells AD. 2015. Long-Range Transcriptional Control of the Il2 Gene by an Intergenic Enhancer. Mol Cell Biol 35: 3880–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong GW, Knowles GC, Mak TW, Ferrando AA, and Zuniga-Pflucker JC. 2012. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood 120: 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jankovic D, Ganesan J, Bscheider M, Stickel N, Weber FC, Guarda G, Follo M, Pfeifer D, Tardivel A, Ludigs K, Bouazzaoui A, Kerl K, Fischer JC, Haas T, Schmitt-Graff A, Manoharan A, Muller L, Finke J, Martin SF, Gorka O, Peschel C, Ruland J, Idzko M, Duyster J, Holler E, French LE, Poeck H, Contassot E, and Zeiser R. 2013. The Nlrp3 inflammasome regulates acute graft-versus-host disease. J Exp Med 210: 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou V, Agle K, Chen X, Beres A, Komorowski R, Belle L, Taylor C, Zhu F, Haribhai D, Williams CB, Verbsky J, Blumenschein W, Sadekova S, Bowman E, Ballantyne C, Weaver C, Serody DA, Vincent B, Serody J, Cua DJ, and Drobyski WR. 2016. A colitogenic memory CD4+ T cell population mediates gastrointestinal graft-versus-host disease. J Clin Invest 126: 3541–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gartlan KH, Bommiasamy H, Paz K, Wilkinson AN, Owen M, Reichenbach DK, Banovic T, Wehner K, Buchanan F, Varelias A, Kuns RD, Chang K, Fedoriw Y, Shea T, Coghill J, Zaiken M, Plank MW, Foster PS, Clouston AD, Blazar BR, Serody JS, and Hill GR. 2018. A critical role for donor-derived IL-22 in cutaneous chronic GVHD. Am J Transplant 18: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanash AM, Kappel LW, Yim NL, Nejat RA, Goldberg GL, Smith OM, Rao UK, Dykstra L, Na IK, Holland AM, Dudakov JA, Liu C, Murphy GF, Leonard WJ, Heller G, and van den Brink MR. 2011. Abrogation of donor T-cell IL-21 signaling leads to tissue-specific modulation of immunity and separation of GVHD from GVL. Blood 118: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hippen KL, Bucher C, Schirm DK, Bearl AM, Brender T, Mink KA, Waggie KS, Peffault de Latour R, Janin A, Curtsinger JM, Dillon SR, Miller JS, Socie G, and Blazar BR. 2012. Blocking IL-21 signaling ameliorates xenogeneic GVHD induced by human lymphocytes. Blood 119: 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dant TA, Lin KL, Bruce DW, Montgomery SA, Kolupaev OV, Bommiasamy H, Bixby LM, Woosley JT, McKinnon KP, Gonzalez FJ, Blazar BR, Vincent BG, Coghill JM, and Serody JS. 2017. T-cell expression of AhR inhibits the maintenance of pTreg cells in the gastrointestinal tract in acute GVHD. Blood 130: 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor PA, Panoskaltsis-Mortari A, Freeman GJ, Sharpe AH, Noelle RJ, Rudensky AY, Mak TW, Serody JS, and Blazar BR. 2005. Targeting of inducible costimulator (ICOS) expressed on alloreactive T cells down-regulates graft-versus-host disease (GVHD) and facilitates engraftment of allogeneic bone marrow (BM). Blood 105: 3372–3380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.