SUMMARY
Calcium signaling through calcineurin and its major transcription factor, CrzA, is integral to hyphal growth, stress response, and virulence of the pathogenic fungus Aspergillus fumigatus, the leading etiology of invasive aspergillosis. Dephosphorylation of CrzA by calcineurin activates the transcription factor, but the specific phosphorylation sites and their roles in the activation/inactivation mechanism are unknown. Mass spectroscopic analysis identified twenty phosphorylation sites, the majority of which were specific to filamentous fungi and distributed throughout the CrzA protein, with particular concentration in a serine-rich region N-terminal to the conserved DNA-binding domain (DBD). Site-directed mutagenesis of phosphorylated residues revealed that CrzA activity during calcium stimulation can only be suppressed by a high degree of phosphorylation in multiple regions of the protein. Our findings further suggest that this regulation is not solely accomplished through control of CrzA nuclear import. Additionally, we demonstrate the importance of the CrzA phosphorylation state in regulating growth, conidiation, calcium and cell-wall stress tolerance, and virulence. Finally, we identify two previously undescribed nuclear localization sequences in the DBD. These findings provide novel insight into the phosphoregulation of CrzA which may be exploited to selectively target A. fumigatus.
Keywords: Aspergillus fumigatus, Calcium, Calcineurin, CrzA, Phosphorylation, Dephosphorylation
INTRODUCTION
Calcium signaling is vital to the pathogenesis of Aspergillus fumigatus, the primary causal agent of invasive aspergillosis (IA) and a leading infectious killer in immunocompromised patients (Abad et al., 2010; Cramer et al., 2008; Denning, 1996; Soriani et al., 2008; Steinbach et al., 2006). Binding of calcium ions to calmodulin (CaM) in the cytoplasm enables interaction of the Ca2+-CaM complex with the phosphatase calcineurin, resulting in its activation. In fungi, calcineurin exerts its influence on gene expression through dephosphorylation of the calcineurin-responsive zinc finger protein CrzA/Crz1, a transcription factor (TF) analogous to mammalian NFAT, resulting in its translocation to the nucleus, where it regulates the transcription of myriad genes involved in important cellular processes (Hogan et al., 2003; Thewes, 2014). While the upstream components of this canonical pathway, including CaM and calcineurin, are strongly conserved between mammals and fungi, a high degree of variation exists among the primary TF targets of calcineurin in different species (Thewes, 2014). This implicates this juncture in the pathway as a key branching point at which the general calcium signal is translated into diverse biological responses specific to the needs of particular taxa.
In A. fumigatus, CrzA has been demonstrated to directly or indirectly influence the expression of numerous genes, including important stress response elements (de Castro et al., 2014; de Castro et al., 2017; Soriani et al., 2010 ). Deletion of CrzA results in strongly attenuated radial growth and conidiation, as well as reduced tolerance to high calcium ion concentration, alkaline pH, and cell wall and temperature stresses (Cramer et al., 2008). This deletion strain also exhibits severe attenuation of virulence in a murine model of IA (Cramer et al., 2008; Soriani et al., 2008). A. fumigatus CrzA is also critical for the caspofungin (CSP) paradoxical effect, in which filamentous growth suppression is ameliorated at higher concentrations of this cell-wall-targeting antifungal agent (Fortwendel et al., 2010). This appears to be partly due to altered regulation of cell wall composition by CrzA during CSP exposure (Ries et al., 2017). A. fumigatus CrzA displays a high degree of homology to its A. nidulans counterpart, and has also been shown to play an important role in a number of similar biological processes in this species (Hagiwara et al., 2008; Spielvogel et al., 2008).
In contrast to the highly conserved upstream components of the calcium signaling pathway, A. fumigatus CrzA shares only 15 percent amino acid sequence identity with human NFAT1 (Altschul et al., 1990). Furthermore, significant identity between CrzA and the most similar human proteins is almost entirely localized to the C-terminal DNA binding domain (DBD), containing several zinc-finger motifs common to this class of transcription factors. The N-terminal regulatory domain is highly variable between NFAT, yeast, and filamentous fungal Crz1/CrzA orthologs, and has been shown to be essential for calcineurin-mediated regulation in both mammals and fungi (Cyert, 2003; Thewes, 2014). This region contains key regulatory elements, including the calcineurin binding domain (CBD), nuclear localization signals (NLS), and nuclear export signals (NES) (Cyert, 2003; Hernández-Ortiz and Espeso, 2013). Murine NFAT was shown to be heavily phosphorylated in the regulatory domain, with complete dephosphorylation of particular regions being necessary for full nuclear translocation and activation of the TF via exposure of the NLS and masking of the NES (Okamura et al., 2000). Phosphoregulation of NFAT is proposed to occur via a large-scale conformational shift resulting from concerted phosphorylation in numerous areas rather than by modification of individual residues that serve as docking sites for regulatory proteins. NFAT activity appears to be dialed up or down along a gradient as the protein structure is brought closer to the fully active or inactive conformation. Despite a lack of homology to NFAT, the N-terminal domain of Saccharomyces cerevisiae Crz1p has also been shown to be essential for calcineurin-regulated subcellular localization (Polizotto and Cyert, 2001; Stathopoulos-Gerontides et al., 1999). An analogous regulatory mechanism also appears to be in place in the filamentous fungi (Hernández-Ortiz and Espeso, 2013, 2017 ). Truncation of the N-terminal domain of A. nidulans CrzA resulted in constitutive cytosolic localization regardless of calcium concentration or the presence of calcineurin (Hernández-Ortiz and Espeso, 2013). It was further demonstrated that the localization of CrzA in this species was controlled by dephosphorylation of the N-terminal domain by calcineurin at multiple sites, though the specific phosphorylated residues within this region were not identified. More recently, several specific sites of phosphorylation within the serine-rich region of A. nidulans CrzA were confirmed by MS analysis (Manoli and Espeso, 2019).
Both mammalian NFAT and S. cerevisiae Crz1p have been found to be targeted for phosphorylation by protein kinase A (PKA); in the former case, primary phosphorylation by PKA appears to facilitate sequential phosphorylation by glycogen synthase kinase 3-β (GSK3β; (Kafadar and Cyert, 2004; Sheridan et al., 2002; Yang et al., 2002 ). However, PKA was not found to modify the Schizosaccharomyces pombe homolog, Prz1, nor were other NFAT-targeting kinases able to suppress defects resulting from Prz1 overexpression, but the fission yeast protein was identified as being targeted by the CaM-dependent protein kinase Cmk1 (Cisneros-Barroso et al., 2014; Koike et al., 2012 ). Casein kinase I (CKI) counterparts have been implicated in phosphorylation of both S. cerevisiae and A. nidulans CrzA homologs, although in the case of A. nidulans, loss of CkiA does not affect CrzA localization, indicating an uncoupling of phosphorylation and regulation of nuclear transport in particular instances (Hernández-Ortiz and Espeso, 2013; Kafadar et al., 2003 ).
Phosphoregulation of calcineurin target TFs is complex. The data obtained from both mammalian and fungal systems indicate the presence of multiple phosphorylation sites targeted by different kinases, with both nuclear translocation and transcriptional regulation potentially being mediated by phosphorylation in partly independent manners. Detailed analysis of phosphoregulation at the residue level has only been performed for mammalian NFAT and not for any fungal CrzA orthologs. Precise understanding of these phosphoregulatory mechanisms is essential for the development of disease treatment strategies aimed at specific inhibition of fungal CrzA isoforms. In this study, we have identified novel specific phosphorylation sites of A. fumigatus CrzA that are dephosphorylated in a calcineurin-dependent and independent manner. We employed site-directed mutagenesis to examine the functional consequences of phosphorylation at these sites with regard to the growth, development, stress tolerance, and virulence of A. fumigatus. Additionally, we identified important NLS sequences in the DBD of CrzA and examined the potential for specific kinases to target CrzA using in vitro phosphorylation assays. Our data enable the formulation of a model describing a novel and complex mechanism for the phosphoregulation of CrzA, enhancing the potential for future biochemical targeting of this key contributor to fungal growth and pathogenesis.
RESULTS
The A. fumigatus CrzA N-terminal and DNA-binding domains contain putative nuclear import and export regulatory sequences
Previous studies have demonstrated the presence of particular regulatory elements within the N-terminal regions of mammalian NFAT, S. cerevisiae Crz1p, and A. nidulans CrzA (Hernández-Ortiz and Espeso, 2013; Okamura et al., 2000; Polizotto and Cyert, 2001; Stathopoulos-Gerontides et al., 1999). While in mammalian NFAT, the CBD, NLS and NES elements were functionally verified, for A. nidulans CrzA only a functional CBD was identified. A. fumigatus CrzA displays a high degree of identity (60%) to the A. nidulans protein, and the identified A. nidulans CBD sequence is conserved in A. fumigatus (amino acids 407–422, Figure 1A), suggesting that it may function similarly in both species. A key NLS identified in the DBD of S. cerevisiae Crz1p is also conserved in A. fumigatus (NLS-A, amino acids 591–593, Figure 1A). Additionally, we performed bioinformatic analysis using the cNLS Mapper program (Kosugi et al., 2009; nls-mapper.iab.keio.ac.jp) and identified an additional potential bipartite NLS sequence spanning amino acids 539–559 (NLS-B; Figure 1A) within the DBD, with a prediction score of 6.5. However, no potential NLS sequences were identified in the N-terminal regulatory domain using a cutoff score of 5.0. Finally, bioinformatic analysis using the NetNES 1.1 Server (la Cour et al., 2004; cbs.dtu.dk/services/NetNES) revealed a putative NES sequence spanning residues 52–58 (respective prediction scores of 0.695, 0.693, 0.758, 0.572, 0.614, 0.611, and 1.076 for these seven residues; Figure 1A).
Figure 1. Identified sites of phosphorylation in A. fumigatus CrzA and comparison of phosphopeptide abundance in wild-type and ΔcnaA backgrounds.
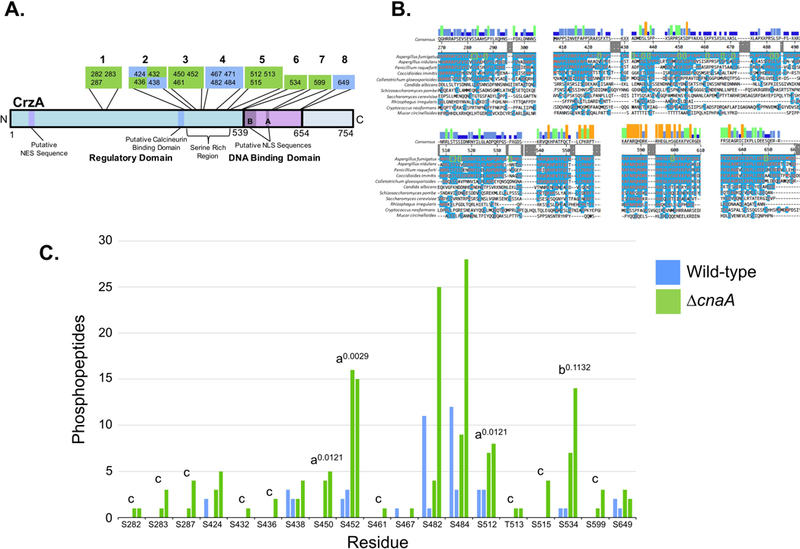
20 CrzA amino acid residues were positively identified as phosphorylated via LC-MS/MS. (A) Diagram of phosphorylation site locations in A. fumigatus CrzA. The 20 sites were grouped into 8 clusters. Sites in green boxes were phosphorylated in a calcineurin-dependent or potentially calcineurin-dependent manner while phosphorylation of sites in blue boxes was categorized as calcineurin-independent. Putative functional regions of the protein are labeled. NES = Nuclear Export Sequence, NLS = Nuclear Localization Sequence, SRR = Serine Rich Region. (B) Clustal W alignment of A. fumigatus CrzA and fungal homologues at phosphorylated regions. Blue highlighting of residues indicates identity with A. fumigatus sequence. Phosphorylated sites are indicated by green boxes. Bars above consensus sequence indicate degree of conservation (blue = less conserved, red = more conserved). (C) Number of peptides with phosphorylation at corresponding sites identified with at least 95% localization probability for each duplicate sample in the wild-type (blue) and ΔcnaA (green) genetic backgrounds. aIndicates statistically significant enrichment in ΔcnaA background compared to wild-type (student’s T-test, P < 0.05). Superscript numbers indicate specific P-values. bIndicates site with non-statistically significant enrichment in ΔcnaA background compared to wild-type still suggestive of potential calcineurin-dependent dephosphorylation. Superscript number indicates specific P-value. cIndicates sites only identified in the ΔcnaA background but for which statistically significant differences between genetic backgrounds could not be demonstrated.
A. fumigatus CrzA exhibits calcineurin-dependent and independent dephosphorylation at numerous phosphorylated residues in the N-terminal regulatory and DNA-binding domains
In order to assess the phosphorylation state of CrzA in the presence and absence of calcineurin in vivo, two A. fumigatus strains (Table S1) expressing C-terminal GFP-labeled CrzA (CrzA-GFP) fusion protein at the native crzA gene locus and from the native promoter were generated, one in the wild-type (WT) genetic background, expressing native calcineurin A catalytic subunit (CnaA), and the other in a cnaA deletion genetic background (ΔcnaA). Following 24-hour growth in liquid minimal nutrient medium at 37°C, the GFP-labeled CrzA protein from each strain was purified by GFP-Trap affinity purification and subjected to LC-MS/MS analysis to identify specific sites of phosphorylation. It may be noted that we utilized only basal growth conditions devoid of calcium stress to permit a comparison of CrzA phosphorylation between the WT and the ΔcnaA background strains. In addition we reasoned that under conditions of calcium stress A. fumigatus CrzA would exhibit dephosphorylation, preventing the detection of phosphosites (Hernández-Ortiz and Espeso, 2013). It is also clear through phenotypic comparisons of WT and CrzA-deficient A. fumigatus strains that CrzA is important for growth and development of the fungus under basal conditions and exposure to normal intracellular calcium concentrations, in addition to calcium stress (Cramer et al., 2008; Soriani et al., 2008). Thus, under the basal growth condition used in this study, one would expect varied phosphorylation states of CrzA.
A total of 20 phosphorylated sites (19 serine residues and 1 threonine residue) were identified with localization confidence scores of greater than 95% (Table 1). These 20 sites were distributed between two primary clusters in the N-terminal regulatory domain, including a cluster of 3 residues spanning positions 282–287 and a larger cluster of 15 residues spanning positions 424–534 within a highly serine-rich region (SRR), as well as two additional phosphorylated sites identified within the DBD of the protein, at positions 599 and 649 (Figure 1A and B). Clustal W alignment of CrzA homologs from 11 fungal species indicated strong conservation in the majority of these phosphorylation sites among the filamentous ascomycetes, while only a few demonstrated identity outside this taxonomic group (Figure 1B). Of the 20 phosphorylation sites, 10 sites were only identified in the ΔcnaA strain, while 9 of the remaining 10 sites were identified in both the WT and ΔcnaA genetic backgrounds (Table 1 and Figure 1C). Statistical comparisons were made between the number of individual phosphopeptides identified with a minimum of 95% localization confidence scores for each site in the WT and ΔcnaA backgrounds. For 3 of these phosphorylation sites (S450, S452, and S512), significantly greater numbers of phosphopeptides were identified in the ΔcnaA strain when compared to WT at a P value of <0.05 (Table 1 and Figure 1C), indicating a high likelihood of increased abundance of phosphorylation at these sites in the absence of calcineurin. Dephosphorylation of these residues was thus categorized as being calcineurin dependent. Phosphorylation of another site, S534, also demonstrated increased abundance in the background ΔcnaA at a value of P=0.1132, indicating a probability that this phosphorylation may also be calcineurin dependent (Figure 1C). Dephosphorylation at this site was therefore categorized as potentially calcineurin dependent for the purposes of subsequent investigation. The remaining 9 sites only identified as phosphorylated in the ΔcnaA background (S282, S283, S287, S432, S436, S461, T513, S515 and S599) were also categorized as being potentially calcineurin dependent as the data obtained do not present evidence of their phosphorylation in the CnaA-expressing strain. It should be noted that evidence of calcineurin-dependent dephosphorylation does not necessarily indicate that dephosphorylation at these residues is directly catalyzed by calcineurin, as the regulation could occur through an indirect mechanism.
Table 1.
Identified phosphopeptides for purified CrzA protein from multiple genetic backgrounds
| Phosphorylated Residue | Peptide Sequencea | Localization Probabilityb | Ambiguity Score (Ascore)c | Peptide Scored | Mascot Delta Ion Scoree | Total Peptides (WT)f | Total Peptides (ΔcnaA)f | Total Peptides (ΔpkaC1)f |
|---|---|---|---|---|---|---|---|---|
| S282 | I[pS][pS]AAPSPYLSQHE | 100% | 45.42 | 107.87 | 29.21 | 0 | 2 | 0 |
| S283 | I[pS][pS]AAPSPYLSQHE | 100% | 47.48 | 107.87 | 29.21 | 0 | 4 | 0 |
| S287 | ISSAAP[pS]PYLSQHE | 100% | 24.95 | 157.87 | 28.68 | 0 | 5 | 0 |
| S424 | IP[pS]FGPSKPASNLDSLSPPPSSTR | 100% | 94.68 | 318.75 | 73.77 | 2 | 8 | 6 |
| S432 | IPSFGP[pS]KPA[pS]NLD[pS]LSPPPSSTR | 96% | 14.24 | 103.97 | 20.98 | 0 | 1 | 0 |
| S436 | IPSFGP[pS]KPA[pS]NLD[pS]LSPPPSSTR | 99% | 24.95 | 161.18 | 33.75 | 0 | 2 | 0 |
| S438 | IPSFGPSKPASNLDSL[pS]PPPSSTR | 100% | 43.97 | 262.97 | 49.13 | 5 | 6 | 15 |
| S450 | GR[pS]K[pS]DPYAHPSTSR | 100% | 62.38 | 181.38 | 23.36 | 0 | 5 | 0 |
| S452 | GR[pS]K[pS]DPYAHPSTSR | 100% | 69.51 | 181.38 | 23.36 | 5 | 31 | 5 |
| S461 | SK[pS]DPYAHPST[pS]RLR | 97% | 20.2 | 64.31 | 15.31 | 0 | 1 | 0 |
| S467 | SS[pS]TSSSLDPLAPTTPR | 100% | 30.97 | 327.02 | 94.37 | 1 | 1 | 1 |
| S471 | SSSTSS[pS]LDPLAPTTPR | 98% | 24.95 | 191.19 | 29.27 | 0 | 0 | 1 |
| S482 | [pS]L[pS]PFDSFGR | 100% | 145.71 | 143.56 | 38.28 | 12 | 29 | 7 |
| S484 | [pS]L[pS]PFDSFGR | 100% | 122.33 | 78.55 | 19.77 | 15 | 37 | 11 |
| S512 | L[pS]TSSIDSR | 100% | 55.92 | 165.8 | 56.51 | 6 | 15 | 4 |
| T513 | RL[pS][pT]SSIDSR | 100% | 30.97 | 60.07 | 22.62 | 0 | 2 | 0 |
| S515 | RL[pS]T[pS][pS]IDSR | 100% | 20.98 | 105.24 | 31.02 | 0 | 4 | 0 |
| S534 | NYILGLADPQRPGA[pS]PNDSKR | 100% | 83.65 | 132.45 | 23.72 | 2 | 21 | 2 |
| S599 | KRHEGLH[pS]GEK | 100% | 1,000.00 | 17.15 | 13.33 | 0 | 4 | 0 |
| S649 | AGRICIKPLLDEE[pS]QERE | 100% | 1,000.00 | 222.9 | 32.63 | 3 | 5 | 5 |
Sequence of cleaved peptide containing phosphorylated residue of interest with highest localization probability. Predicted phosphorylated residues are shown as [pS/T], and residue of interest is shown in bold.
Percent probability that predicted phosphorylation site of interest is accurately identified as phosphorylated within the peptide indicated. Based on Ascore.
Statistical metric assessing probable localization accuracy for the indicated phosphorylation site. Ascore of >19 indicates >99% probability of accurate site prediction (Beausoleil et al. 2006).
Probability-based ion matching score for indicated phosphorylation site (Beausoleil et al. 2006).
Statistical metric for assigning phosphorylation to specific indicated site using Mascot proteomic analysis software (Matrix Science, Boston, MA).
Total number of peptides identified in the indicated genetic background (WT, ΔcnaA, or ΔpkaC1) via MS analysis demonstrating phosphorylation at the indicated residue with localization probabilities of >95 percent based on Ascores.
As bioinformatic analysis using the NetPhos 3.1 Server (Blom et al., 2004; cbs.dtu.dk/services/NetPhos), identified potential target motifs for protein kinase A (PKA) as well as numerous other kinases in the CrzA amino acid sequence (Table 2), we also attempted to identify specific PKA-dependent phosphorylation sites within CrzA. In a tandem experiment, we performed LC-MS/MS analysis on CrzA-GFP purified from both WT and ΔpkaC1 genetic backgrounds. However, unlike the multiple phosphorylated/dephosphorylated sites identified in the WT versus the ΔcnaA background, we found only one site (S471) that was differentially phosphorylated between the WT and ΔpkaC1 backgrounds (Table 1). Furthermore, phosphorylation at this site was only identified in the ΔpkaC1 background and not in the WT strain, which is not indicative of phosphorylation of this site by PKA but may instead suggest possible indirect regulation of this modification through targeting of other regulatory proteins. Alternatively, as only one phosphopeptide was identified with phosphorylation at S471 in any of the genetic backgrounds examined (Table 1), it may simply be that this modification is low in abundance and was only identified in the ΔpkaC1 background. While these results suggest that CrzA may not be an in vivo target of PKA despite the presence of putative PKA target motifs, we cannot exclude the possibility of PKA-dependent phosphorylation of CrzA under other conditions of growth or stress.
Table 2.
Potential protein kinases targeting A. fumigatus CrzA
| Residue | Kinase | NetPhos 3.1 Scorea |
|---|---|---|
| S287 | Cyclin-dependent kinase 5 (CDK5) | 0.544 |
| Glycogen synthase kinase 3 (GSK3) | 0.528 | |
| S424 | cGMP-dependent protein kinase (PKG) | 0.626 |
| Protein kinase C (PKC) | 0.542 | |
| GSK3 | 0.508 | |
| S436 | Cyclin-dependent kinase 1 (CDC2/CDK1) | 0.504 |
| S438 | CDK5 | 0.594 |
| p38 mitogen activated protein kinase (p38MAPK) | 0.584 | |
| CDC2 | 0.504 | |
| S450 | cAMP-dependent protein kinase (PKA) | 0.702 |
| Protein kinase B (PKB/Akt) | 0.702 | |
| PKG | 0.585 | |
| Ribosomal 26 kinase (RSK) | 0.538 | |
| CDC2 | 0.521 | |
| S452 | PKB | 0.747 |
| RSK | 0.557 | |
| CDC2 | 0.532 | |
| S461 | PKC | 0.669 |
| S467 | PKB | 0.707 |
| RSK | 0.533 | |
| S471 | CDC2 | 0.594 |
| DNA-dependent protein kinase (DNAPK) | 0.522 | |
| Calmodulin-dependent protein kinase II (CaM-II) | 0.508 | |
| S482 | PKG | 0.571 |
| S484 | CDK5 | 0.640 |
| p38MAPK | 0.516 | |
| RSK | 0.507 | |
| GSK3 | 0.500 | |
| S512 | PKA | 0.836 |
| RSK | 0.672 | |
| T513 | PKC | 0.566 |
| CDC2 | 0.503 | |
| S534 | p38MAPK | 0.554 |
| GSK3 | 0.517 | |
| S649 | DNAPK | 0.683 |
| Casein kinase II (CKII) | 0.528 |
Highest potential score = 1.000. Values over 0.650 are shown in bold.
A high degree of phosphorylation is required for inhibition of CrzA function with regard to conidiation and stress tolerance
Mass spectrometry revealed that CrzA is a highly phosphorylated protein in vivo in both the WT and the ΔcnaA genetic backgrounds, indicating a complex regulatory mechanism for this transcription factor. In order to assess the functional impact of phosphorylation at the identified residues, crzA mutant strains (Figure 1A, 2A and Table S1) were generated via site-directed mutagenesis to lock specific residues in the phosphorylated state. CrzA isoforms were expressed from the native crzA locus, driven by the native promoter, in which clusters of phosphorylated serine and threonine residues were systematically replaced with aspartic acid to mimic phosphoserine and phosphothreonine. These strains are referred to as CrzA-1 through CrzA-8, corresponding to S/T to D mutations in all of the residues in each of the eight clusters (Figure 1A), and CrzA-9 to CrzA-13 (Figure 2A), with corresponding mutations in the combinations of clusters shown. These mutant CrzA isoforms were also labeled with C-terminal GFP tags in order to facilitate investigation of subcellular localizaton. Expression of GFP-labeled CrzA protein was confirmed for combination mutant strains via Western blotting (Figure S1). Phenotypes of the 13 mutant strains were assessed with regard to radial growth and conidiation on glucose minimal medium agar (GMM; Figure 2B-D). Responses of the mutant strains to calcium and cell wall stresses were analyzed in the presence of 200mM CaCl2 or 0.5μg/mL or 4μg/mL concentrations of the cell wall-targeting antifungal agent caspofungin (CSP; Figure 2B-D). Additionally, strains CrzA-11, CrzA-12, and CrzA-13 were tested for sensitivity to the cell membrane-targeting azole antifungal compounds voriconazole and itraconazole via growth in liquid medium across several drug concentrations ranging from 0 to 0.5μg/mL. These three strains were selected for azole sensitivity testing as they represent full phosphorylation of the DBD, N-terminal regulatory domain, and both domains combined, respectively. Mutant strain phenotypes were compared to a control strain expressing a C-terminally GFP-labeled CrzA isoform with native amino acid sequence expressed from the native crzA locus and driven by the native promoter (Table S1), referred to subsequently as CrzA-C. Comparison was also made to a crzA deletion (ΔcrzA) strain (Table S1). Deletion of crzA in A. fumigatus has been shown to result in reduction in radial growth, marked attenuation of conidiation, and strongly increased sensitivity to calcium stress and CSP exposure, including attenuation of the paradoxical growth effect, in which A. fumigatus is able to grow more vigorously at higher concentrations of CSP (4μg/mL) compared to growth inhibition at lower concentrations (0.5μg/mL) (Cramer et al., 2008; Ries et al., 2017; Soriani et al., 2008). Such findings demonstrate the importance of active CrzA in maintenance of normal growth and development under basal growth conditions as well as in multiple stress responses. Mutation of the individual clusters (strains CrzA 1–8) resulted in slight radial growth defects on GMM similar to that of the ΔcrzA strain compared to the CrzA-C strain (Figure S2A), but no macroscopically evident reduction in conidiation. These mutant strains also exhibited slight but statistically significant decreases in radial growth under conditions of calcium stress compared to CrzA-C, as well as proportionally similar reductions in growth under CSP exposure, which in a few instances were also statistically significant (Figure S2A).
Figure 2. Radial growth and conidiation of CrzA phosphorylation mutants under basal and stress-inducing growth conditions and susceptibility to echinocandin and azole antifungals.
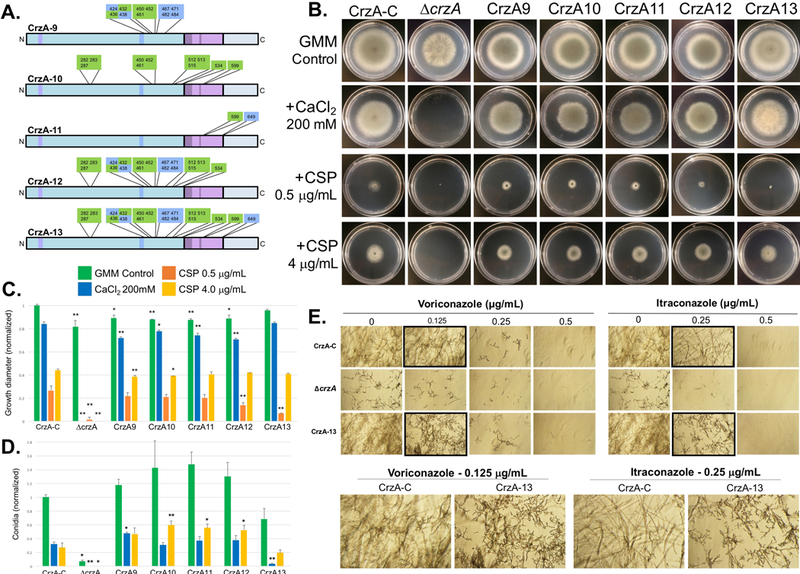
(A) Diagrams of additional mutant CrzA isoforms (CrzA-9 to CrzA-13) generated with glutamate substitutions in place of serine/threonine residues at multiple cluster combinations shown. Strain IDs are indicated for each mutant strain. Mutants CrzA-1 to CrzA-8 correspond to the 8 clusters shown in figure 1A. Color coded regions of CrzA diagram correspond to those of figure 1A. (B) Growth of selected CrzA mutant strains on solid agar media under multiple conditions. Strains CrzA9-CrzA13 as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and GMM agar containing 200mM CaCl2, 0.5μg/mL CSP, or 4μg/mL CSP, and incubated at 37°C for 4 days. (C) Quantification of radial growth on each media type (green=GMM, blue=CaCl2, orange=0.5μg/mL CSP, yellow=4μg/mL CSP). (D) Quantification of conidiation on each media type (excluding 0.5ug/mL CSP). Bars represent averages of three replicates. Error bars represent standard deviations. Asterisks indicate statistically significant differences (Dunnett’s post-test, *P < 0.05, **P < 0.01) compared to CrzA-C on the same growth medium. (E) Micrographs showing growth of fungal hyphae during exposure to voriconazole and itraconazole antifungals. Conidia of indicated strains were inoculated into liquid medium containing indicated drug concentrations and incubated at 37°C for 48 hours. Black-bordered micrographs are enlarged in lower panel to highlight concentrations at which mutant strain CrzA-13 exhibits reduced growth compared to CrzA-C. Magnification = 20X.
Combined mutation of multiple clusters (Figure 2A), including three within the SRR (strain CrzA-9), all clusters for which the dephosphorylation of all residues was calcineurin dependent or potentially calcineurin dependent (strain CrzA-10), the two phosphorylated DBD residues (CrzA-11), and the N-terminal domain clusters (strain CrzA-12), all resulted in statistically significant reduced radial growth phenotypes similar to the individual cluster mutants on GMM agar. The decrease in radial growth of the respective cluster mutants was comparable to that of the ΔcrzA strain (Figure 2B and C). However, conidiation was higher than the control strain CrzA-C for strains CrzA-9, 10, 11, and 12, an effect opposite to that observed in the ΔcrzA strain (Figure 2D), though due to variability among the replicates statistical significance could not be demonstrated. Conversely, strain CrzA-13, with 20 phosphorylation sites mutated, did not exhibit altered radial growth compared to WT, but did show decreased conidiation, although the magnitude of this attenuation was less than that observed in the ΔcrzA strain (Figure 3D) and was not demonstrated to be statistically significant. Strains CrzA-9 through CrzA-12 also showed slight but significant reductions in radial growth compared to the WT strain under calcium stress, which were roughly proportional to the decreases observed under basal conditions, while CrzA-9 and 10 also showed similar decreases in response to 4 μg/mL CSP stress (Figure 2 B and C). Conidiation was significantly increased for strains CrzA-10 through CrzA-12 at 4 μg/mL CSP exposure to approximately double the rate of the CrzA-C strain, while strain CrzA-9 showed significantly increased conidiation in the presence of 200mM CaCl2 (Figure 2D). In general, the increase in conidiation under CSP exposure was proportionally greater than that observed under basal growth conditions. Strain CrzA-13 did not show any difference in radial growth compared to CrzA-C under calcium or CSP stress, but conidiation was severely attenuated (approximately 9-fold decrease) during 200mM CaCl2 exposure and also somewhat attenuated during 4μg/mL CSP exposure, though statistical significance could not be shown in the latter case (1.37-fold average decrease; Figure 2B-D). All the crzA mutant strains showed paradoxical radial growth recovery during exposure to 4μg/mL CSP compared to 0.5μg/mL CSP, in contrast to the ΔcrzA strain which did not exhibit any macroscopically observable growth under either concentration of CSP or following exposure to 200mM CaCl2 (Figure 2B and 2C). Strain CrzA-13 also demonstrated more severe attenuation of hyphal growth compared to CrzA-C during exposure to 0.125μg/mL voriconazole and 0.25μg/mL itraconazole, though in both cases growth of this mutant strain was less attenuated than that of the ΔcrzA strain (Figure 2E). Neither strain CrzA-11 nor CrzA-12 displayed altered azole sensitivity compared to CrzA-C (Figure S2B), therefore azole sensitivity testing for strains with fewer mutated residues was not pursued.
Figure 3. Impact of CrzA phosphorylation on A. fumigatus virulence in a murine model of IA.
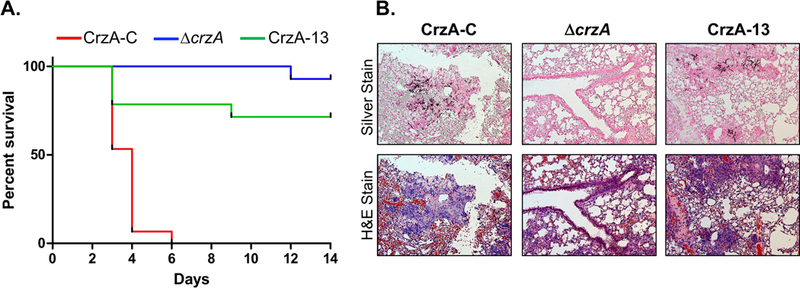
(A) Survival curve of mice inoculated with conidia of the CrzA-C control (red), ΔcrzA (blue), or mutant strain CrzA-13 (green). 15 mice immunocompromised via cyclophosphamide and triamcinolone injections were inoculated intranasally with conidia of each strain type and survival was monitored for 14 days. Survival of mice inoculated with CrzA-13 was found to be significantly higher than that of mice inoculated with CrzA-C (P < 0.0001) in a log-rank (Mantel-Cox) comparison. (B) Lung tissue histology of infected mice. Lung tissue was harvested from one additional mouse inoculated with each of the three A. fumigatus strains on day 3 post-infection and visualized microscopically following silver staining (top row) in order to assess hyphal growth (dark coloration) and H&E staining (bottom row) in order to assess tissue damage. Both hyphal invasion and tissue damage were evident in lungs infected with CrzA-C and CrzA-13 strains. Magnification = 10X.
Dynamic phosphorylation and dephosphorylation of CrzA is required for full virulence of A. fumigatus
We and others have previously independently shown that CrzA is a key factor in the virulence of A. fumigatus, as deletion of crzA results in avirulence in a murine model (Cramer et al., 2008; Soriani et al., 2008). Due to the observed phenotypic alterations following the mutation of the various phosphorylated clusters in CrzA, including reduced stress tolerance in some instances, we assessed the importance of CrzA phosphorylation/dephosphorylation in regulating pathogenesis. To accomplish this, we infected immunosuppressed mice with the CrzA-13 mutant strain, which alone of the mutants showed reduced conidiation under calcium stress and increased cell wall and azole stress sensitivity, and compared mortality to mice infected with the CrzA-C and ΔcrzA strains as positive and negative controls, respectively. Despite the relatively minor phenotypic variations from wild-type observed for the CrzA-13 strain in comparison to the ΔcrzA strain, virulence of the CrzA-13 strain was much closer to that of the ΔcrzA strain than to the CrzA-C strain (Figure 3A). While infection with the WT strain resulted in complete mortality by day 6 post-inoculation, 73.33% of mice infected with CrzA-13 survived until the end of the 14-day observation period (log-rank test, P < 0.0001). Mortality rate differences between CrzA-13 and ΔcrzA-infected mice were not statistically significant. Lung tissue was also extracted on day 3 post-infection from mice inoculated with each of the three strains and examined following methenamine silver staining and haemotoxylin and eosin (H&E) staining to observe hyphal invasion and tissue inflammation, respectively (Figure 3B). Despite the large difference observed in the virulence of the CrzA-C and CrzA-13 strains, comparable hyphal invasion and tissue damage were evident in the lungs of mice infected with both strains. Mice used in the histological analysis for both strains had been severely symptomatic, while the majority of other CrzA-13-infected mice displayed only minor symptoms, suggesting variability in disease outcome associated with this strain. By contrast, lung tissue from the ΔcrzA-infected mouse appeared healthy and did not show evidence of invasive hyphal growth, consistent with the lack of disease symptoms observed for mice in this experimental group. The histological portion of this mouse infection experiment was repeated using lung tissue extracted from three mice infected with each of the three fungal strains in order to confirm these results. Again variable degrees of invasive hyphal growth were observed in lung tissue of mice infected with CrzA-13, comparable to the CrzA-C control in some cases, while little to no hyphal growth was observed in tissue from mice infected with the ΔcrzA strain (Figure S3). The histological analysis presented for CrzA-13 may thus serve to illustrate the upper range of tissue invasion capacity for this mutant strain.
Dephosphorylation of the CrzA N-terminal regulatory domain controls calcium-induced nuclear translocation
Previous studies in yeasts and filamentous fungi have demonstrated that nuclear localization of CrzA homologs is calcineurin dependent (Hernández-Ortiz and Espeso, 2013; Stathopoulos-Gerontides et al., 1999). In response to cytoplasmic calcium, the activation of calcineurin leads to dephosphorylation of CrzA homologs and their subsequent nuclear translocation, enabling transcriptional activation of calcineurin/CrzA-dependent target genes. In order to determine how phosphorylation/dephosphorylation of CrzA influences its cytosolic to nuclear shuttling, the CrzA-C, ΔcnaA (expressing C-terminally GFP-labeled CrzA from the native crzA locus and driven by the native promoter, Table S1), and CrzA phosphorylation-locked mutant strains were examined for localization of CrzA-GFP in the presence and absence of 200mM CaCl2 using fluorescence microscopy. In the absence of CaCl2, CrzA-GFP was excluded from nuclei of both the CrzA-C and ΔcnaA control strains, as well as the CrzA mutant strains (Figure 4). Following the addition of CaCl2, strong nuclear localization of CrzA was observed in CrzA-C, but in the ΔcnaA background CrzA remained excluded from nuclei, confirming the requirement of calcineurin for nuclear localization (Figure 4 and Figure 5). Interestingly, the phosphorylation-locked CrzA mutants exhibited variable localization patterns. Among the individual cluster mutants (strains CrzA-1 through CrzA-8), none displayed a defect in calcium-stimulated nuclear localization, with nuclear accumulation in these strains closely resembling that of the CrzA-C strain (Figure S4). Among the combination cluster mutants (CrzA-9 through CrzA-13), CrzA-9, 10, and 11 also demonstrated strong calcium-induced nuclear translocation similar to CrzA-C (Figure 4). In contrast, only limited, partial CrzA nuclear accumulation was primarily observed in the CrzA-12 and 13 mutants, though in these strains CrzA was not excluded from nuclei in the presence of 200mM CaCl2 as in the case of the ΔcnaA control.
Figure 4. Calcium-stimulated nuclear translocation of mutant CrzA isoforms.
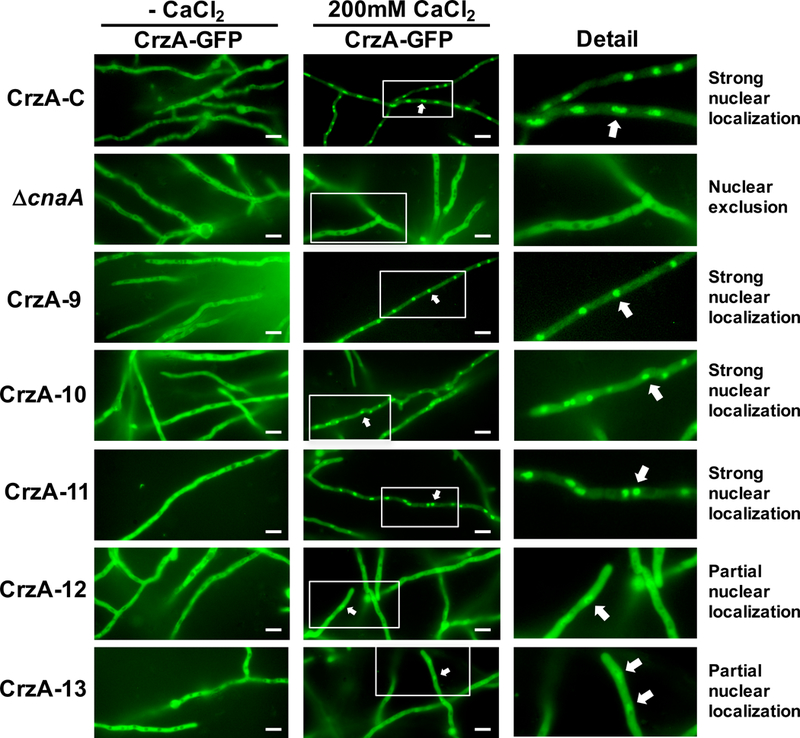
Conidia (104) of strains expressing GFP-labeled CrzA (CrzA-GFP) with native amino acid sequence (CrzA-C) or each mutant CrzA isoform CrzA-9 to CrzA-13 were inoculated into glass-bottomed Petri dishes containing liquid GMM and incubated overnight at 37°C. Hyphae were visualized microscopically prior to (CaCl -) and 15 minutes after addition of 200mM CaCl2 to the medium. A strain expressing native CrzA-GFP in a cnaA gene deletion background (ΔcnaA) was used as a negative control. Strong levels of calcium-stimulated nuclear translocation of CrzA were observed in strains CrzA-9, 10, and 11, but only partial nuclear translocation was observed in strains CrzA-12 and 13, while nuclear exclusion was observed in the ΔcnaA control. White scale bars represent 10μm distances. White arrows indicate examples of nuclear localization of CrzA-GFP where observed. CrzA-GFP = green fluorescence image. The enlarged detail images of nuclear and cytoplasmic localization of CrzA-GFP in the presence of 200mM CaCl2 are shown for clarity.
Figure 5. Quantitative comparison of calcium stimulated nuclear accumulation of CrzA in mutant strains.
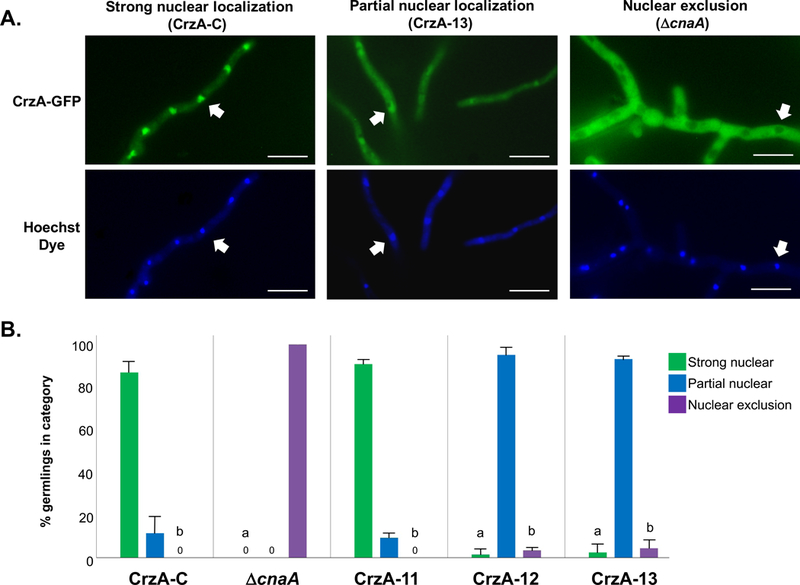
(A) Representative examples of germling hyphae in live cell cultures (9 hours at 37°C) demonstrating three categories of CrzA-GFP localization following 200mM CaCl2 exposure. Strain IDs are shown in parentheses. Upper panels show localization of CrzA-GFP, while lower panels show corresponding locations of nuclei stained with Hoechst dye. White arrows indicate locations of specific selected nuclei in corresponding micrographs. White scale bars represent 10μm. Strong nuclear localization was defined as nuclear fluorescence notably brighter than surrounding cytoplasmic fluorescence. Partial nuclear localization was defined as nuclear fluorescence equal to or only marginally brighter than surrounding cytoplasmic fluorescence. Nuclear exclusion was defined as nuclear fluorescence notably less bright than surrounding cytoplasmic fluorescence. (B) Quantification and statistical comparison of nuclear localization categories between strains. Bars represent the average number of germlings for three replicate cultures in each of the three categories for the strains examined. Green = strong nuclear localization, Blue = partial nuclear localization, Purple = nuclear exclusion. Strong nuclear localization counts were compared between CrzA-C and each of the other strains, and nuclear exclusion counts were compared between ΔcnaA and each of the other strains via Dunnett’s post-test. aIndicates statistically significant differences in strong nuclear localization (P < 0.01). bIndicates statistically significant differences in nuclear exclusion (P < 0.01)
In order to quantify these observations, triplicate cultures of strains CrzA-11, CrzA-12, and CrzA-13, as well as control strains CrzA-C and ΔcnaA were grown to germling stage and 50 germlings from each culture were scored as displaying strong nuclear localization, partial nuclear localization, or nuclear exclusion of CrzA-GFP following the addition of 200mM CaCl2 to the growth medium (Figure 5). These mutant strains were selected as CrzA-12 and CrzA-13 both express phosphomimetic aspartic acid in place of all identified phosphorylated residues in the N-terminal regulatory domain, differing only in that CrzA-13 possesses two additional aspartic acid substitutions at the two identified sites within the DBD, while CrzA-11 possesses mutations at only the two DBD sites (Figure 2A). Thus strains CrzA-11, 12 and 13 represent full phosphorylation at the DBD, N-terminal regulatory domain, and both domains combined, respectively. Statistical comparisons were made between each strain and the CrzA-C control with regard to the numbers of germlings displaying strong nuclear localization of CrzA, and between each strain and the ΔcnaA control with regard to the numbers of germlings displaying nuclear exclusion of CrzA. In the CrzA-C control, the vast majority of germlings displayed strong nuclear localization, while no strong or partial nuclear localization of CrzA was observed in the ΔcnaA strain. In CrzA-11, strong nuclear localization was similar to that of CrzA-C and nuclear exclusion was not observed. In contrast, both CrzA-12 and CrzA-13 displayed dramatically reduced levels of strong nuclear localization compared to CrzA-C, and dramatically reduced levels of nuclear exclusion compared to ΔcnaA. These results imply that dephosphorylation of the N-terminal domain is crucial for calcium-stimulated nuclear translocation of CrzA, while also suggesting that phosphorylation within the DBD is less relevant to this process. As strains CrzA-9 and 10 each also express phosphomimetic residues at three of the five N-terminal domain clusters identified yet display strong nuclear accumulation of CrzA in response to calcium (Figure 4), these results also suggest that a high degree of concerted phosphorylation distributed throughout this entire domain is required for suppression of nuclear translocation to be maintained under calcium stress conditions.
It should be noted that in similar experiments to those described but in which fungal hyphae were mounted on slides with coverslips instead of being visualized as actively growing cultures in glass-bottomed dishes, strong nuclear accumulation of CrzA was also observed in strains CrzA-12 and 13, just as in the other mutant strains and CrzA-C, though this was not observed in the ΔcnaA strain (data not shown), indicating that other environmental factors such as mechanical stress due to coverslip placement can potentially induce nuclear translocation of CrzA independently of dephosphorylation at these particular clusters. Therefore, in this study we specifically address only the role of dephosphorylation/phosphorylation of CrzA in regulating calcium-induced nuclear translocation in live-cell cultures.
Nuclear import of CrzA is governed by multiple nuclear localization signal sequences
Our results confirmed that the nuclear localization of CrzA is calcineurin dependent and that the capacity for dephosphorylation of the N-terminal domain clusters is important for nuclear localization in response to calcium stress. Previous work in A. nidulans has revealed that both the N-terminal regulatory domain and the DBD of CrzA are able to translocate to the nucleus in truncated forms of the protein lacking one region or the other, indicating that both regions likely contain NLS sequences, though neither specific NLS has been identified (Hernández-Ortiz and Espeso, 2013). Expression of the N-terminus alone allowed weak nuclear translocation that was calcium/calcineurin dependent, while expression of the DBD alone allowed strong constitutive nuclear localization independent of calcineurin. S. cerevisiae Crz1p has also been found to have NLS sequences in both of these regions (Polizotto and Cyert, 2001), but while these particular sequences were identified in yeast, only the sequence of the DBD NLS is conserved in the Aspergilli, which we termed NLS-A in A. fumigatus CrzA (Figure 1A). Expressed DBD fragments of Crz1p in S. cerevisiae lacking the sequence homologous to NLS-A did not undergo nuclear translocation in response to calcium, although when expressed alone this sequence was constitutively nuclear (Polizotto and Cyert, 2001). Bioinformatically, we identified only one potential bipartite NLS spanning 21 amino acids within the conserved DBD (NLS-B, Figure 1A). In order to determine whether either of these sequences represents a functional NLS, we generated mutant strains expressing CrzA-GFP isoforms in which either the NLS-A or NLS-B sequence alone was deleted (ΔNLS-A and ΔNLS-B, respectively; Figure 6A) or in which both putative NLS sequences were simunltaneously deleted (ΔNLS-A/B). Mutant CrzA isoforms were expressed from the native crzA locus under control of the native promoter, and expression of GFP-labeled CrzA protein in these strains was confirmed via Western blot (Figure S1). These three strains phenotypically resembled the ΔcrzA strain, with strongly reduced conidiation on solid GMM agar medium (Figure 6B). Also similarly to the ΔcrzA strain, both the ΔNLS-A and ΔNLS-B mutant strains lacked significant paradoxical growth recovery in response to higher concentrations of caspofungin (Figure S5). These phenotypic effects seem likely to be due primarily to disrupted association of the mutated CrzA DBD with target genes and consequent impairment of their transcriptional activation. Given that phenotypic effects resulting from impaired DNA binding would not be distinguishable from those resulting from impaired nuclear localization, additional phenotypic testing was not pursued for these strains.
Figure 6. Identification of sequences mediating calcium-stimulated nuclear translocation of CrzA.
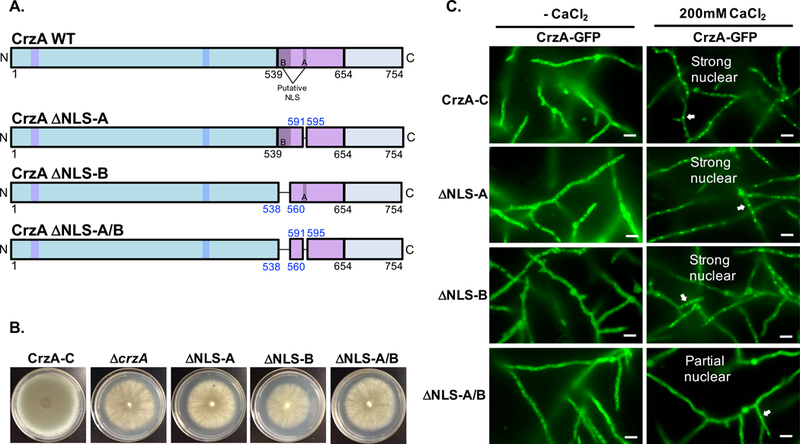
(A) Diagrams of mutant CrzA isoforms in which putative nuclear localization sequences (NLS) A, B, or both A and B are excised (ΔNLS-A, ΔNLS-B, and ΔNLS-A/B, respectively). Color coded regions on diagrams correspond to those of Figure 1A. (B) Growth phenotype of NLS mutant strains. Conidia (104) of the three mutant strains as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and incubated at 37°C for 4 days. Growth phenotypes of NLS mutants qualitatively resembled that of ΔcrzA. (C) Nuclear translocation of CrzA NLS mutant isoforms. Conidia (104) of strains expressing CrzA-GFP with native amino acid sequence (WT) or the three GFP-labeled NLS excision CrzA isoforms were inoculated into glass-bottomed Petri dishes containing liquid GMM and incubated overnight at 37°C. Hyphae were visualized microscopically prior to (CaCl -) and 15 minutes after addition of 200mM CaCl2 to the medium. Wild-type levels of calcium-stimulated nuclear translocation of CrzA were observed in strains ΔNLS-A and ΔNLS-B, but only minimal nuclear translocation was observed in strains ΔNLS-A/B. White scale bars represent 10μm distances. White arrows indicate examples of nuclear localization of CrzA-GFP where observed. CrzA-GFP = green fluorescence image.
We next determined whether nuclear translocation of CrzA is attenuated in the respective NLS mutants in the presence of calcium (Figure 6C). CrzA was excluded from nuclei in all strains prior to the addition of CaCl2, but strongly translocated to the nucleus following calcium stimulation in both the ΔNLS-A and ΔNLS-B mutant strains, as well as in the CrzA-C control strain. However, only partial nuclear accumulation of CrzA was observed in response to calcium in the ΔNLS-A/B strain. These results indicate that both sequences NLS-A and NLS-B are important for the calcium-dependent nuclear import of CrzA, but that their roles in this process may be redundant, with either NLS alone enabling efficient translocation of the protein. The lower level of observable nuclear accumulation still seen in the ΔNLS-A/B strain suggests that other additional sequences or elements may also be involved in the nuclear translocation of CrzA.
DISCUSSION
Transcription factors and their regulation are critical for the adaptation of fungi to external stresses and other environmental conditions. Here we define novel regulatory elements demonstrating the complexity of calcium signaling through the calcineurin-dependent transcription factor CrzA in the human fungal pathogen A. fumigatus. This complexity underscores the importance of finely tuned control of this critical signaling pathway, as we have now identified multiple unique levels of regulation, including the reciprocal activation/inhibition of CrzA by dephosphorylation/phosphorylation status mediated by calcineurin and multiple protein kinases and also the nuclear import of CrzA. Our results suggest these to be dependent on both phosphorylation status and on non-canonical nuclear localization sequences, as well as other factors that have yet to be characterized. In view of these findings, we propose a novel regulatory paradigm for A. fumigatus CrzA in which numerous diverse processes must work synergistically in order to modulate its activity by different degrees.
While A. fumigatus CrzA shares little identity with its yeast homolog and is only functionally analogous to mammalian NFAT proteins, previous studies suggest related mechanisms for the regulation of A. fumigatus and A. nidulans CrzA, S. cerevisiae Crz1p, and mammalian NFAT. All of these calcineurin-dependent transcription factors share a similar basic structure, including a partially conserved domain possessing DNA-binding motifs and a more variable N-terminal domain containing a highly serine-rich region and numerous regulatory elements such as calcineurin-binding motifs and nuclear localization and export signals. In the yeast and mammalian systems, previous work has indicated that phosphorylation of the N-terminal regulatory domain is sufficient to near-completely suppress activity of the relevant transcription factor in a gradual manner, with the strength of suppression increasing proportionally to the degree of phosphorylation (Okamura et al., 2000; Stathopoulos-Gerontides et al., 1999). Suppression of activity via phosphorylation was also shown to occur concomitantly with suppressed nuclear translocation of the protein in these systems.
In striking contrast, we have observed a different pattern of inhibition in the deadly fungal pathogen A. fumigatus. Instead of a gradual and proportional inhibition of CrzA activity with increasing phosphorylation, our work revealed near-normal functionality of the transcription factor regardless of phosphorylation status until a particular threshold of phosphorylation is reached, at which point a marked (though still incomplete) suppression of CrzA function is observed. Our findings suggest that phosphorylation of all the 20 identified target sites (or potentially, of specific residues within each of the clusters targeted for mutagenesis in this study) is necessary for this threshold to be reached, as only the mutant strain with phosphomimetic substitutions at each of the 20 residues presented a phenotype with significantly reduced conidiation under calcium stress and increased sensitivity to echinocandin and azole antifungal stresses. Retention of dephosphorylatable residues at any individual clusters resulted in a phenotype generally similar to that of a strain expressing wild-type CrzA. Phosphomimicry at all N-terminal domain clusters alone was insufficient to produce the more strongly suppressed phenotype. Instead, additional phosphomimicry at the two identified residues within the DNA-binding domain was also required. Likewise, phosphomimicry of these two residues on their own was insufficient to produce a CrzA suppression phenotype. This again is in contrast to findings in yeast and mammals, in which phosphorylation was detected in the N-terminal regulatory domains of the respective transcription factors and not in the DNA-binding domains, and in which N-terminal regulatory domain phosphorylation was sufficient alone to almost fully suppress activity. While our results indicate that concurrent phosphorylation at several specific regions of CrzA is required for suppression of nuclear translocation and CrzA activity during calcium stimulation, a caveat remains in that the consequences of mutation at each identified phosphorylated residue or possible combination of residues have not yet been fully explored. Additionally, it is also likely the case that other post-translational modifications not investigated in the current study (e.g. ubiquitination) may influence the subcellular localization of CrzA in conjuction with its phosphorylation status.
Another unique aspect of CrzA regulation in A. fumigatus we uncovered is that the modulation of CrzA activity via phosphorylation/dephosphorylation does not appear to occur only through control of its cytosolic/nuclear localization. In yeast and mammals, phosphoregulation of Crz1p and NFAT occurs primarily through nuclear exclusion (Okamura et al., 2000; Polizotto and Cyert, 2001; Stathopoulos-Gerontides et al., 1999), but this does not seem to be the case in A. fumigatus or possibly in other related filamentous fungi. While the CrzA-13 mutant strain expressing phosphomimetic substitutions in CrzA at all identified phosphorylation sites was deficient for nuclear translocation in response to calcium stimulation, this was also true for CrzA-12, despite the stronger phenotypic defects indicative of CrzA suppression (reduced conidiation and stress tolerance) in the CrzA-13 strain compared to the CrzA-12 strain. Suppression of nuclear translocation, therefore, does not appear to be the sole means by which phosphorylation inhibits the activity of CrzA in A. fumigatus. That phosphoregulation of CrzA activity can occur via additional mechanisms beyond the modulation of subcellular localization is further supported by the finding that a fully phosphorylated but artificially nuclear localized form of mammalian NFAT possessing an additional NLS was found to be unable to effect transcriptional activation of known target genes despite constitutive localization to the nucleus (Okamura et al., 2000).
One possible underlying mechanism might instead be the induction of a conformational shift that inhibits function through disruption of DNA binding or interaction with other transcription-associated proteins within the nucleus. A large scale conformational shift has been proposed to explain the incremental suppression of yeast Crz1p activity with accumulation of phosphorylation, though in this case the shift was hypothesized to act through the gradual obscurement of nuclear localization sequences (Cyert, 2003; Stathopoulos-Gerontides et al., 1999). The question also remains as to how the CrzA-12 strain apparently retains near-normal CrzA activity, based on phenotypic evidence, despite strongly impaired calcium-induced nuclear translocation. The fact that a low degree of nuclear accumulation was still observable in this strain in response to high calcium exposure suggests that even a small amount of active nuclear CrzA may be sufficient to maintain calcium signaling at the required functional level. As the CrzA-12 and 13 strains differ only at the two phosphorylated DBD residues, substitutions at which had no strong phenotypic effects alone (i.e. in the CrzA-11 strain), it may be the case that the phenotypic defects observed in CrzA-13 are a result of the combined effects of impaired nuclear translocation and a potential impairment of DNA binding associated with DBD phosphorylation. That the CrzA-11 strain did not show strong evidence of suppressed CrzA functionality may indicate that if the identified DBD phosphorylation does interfere with DNA binding, it is not of a sufficient degree to significantly disrupt calcium signaling on its own. Based on this speculation, a potential mechanism by which phosphorylation of the regulatory domain and DBD could act in tandem to suppress activity of the transcription factor through partial inhibition of both nuclear import and DNA-binding is illustrated in Figure 7. More research is needed to validate this model, including determination of the effect, if any, of DBD phosphorylation on DNA-binding efficiency of CrzA. If this effect is minor, as our observations suggest, a highly sensitive, quantitative assay would be required for such an experiment.
Figure 7. Hypothetical model for phosphoregulation of A. fumigatus CrzA during calcium stress.
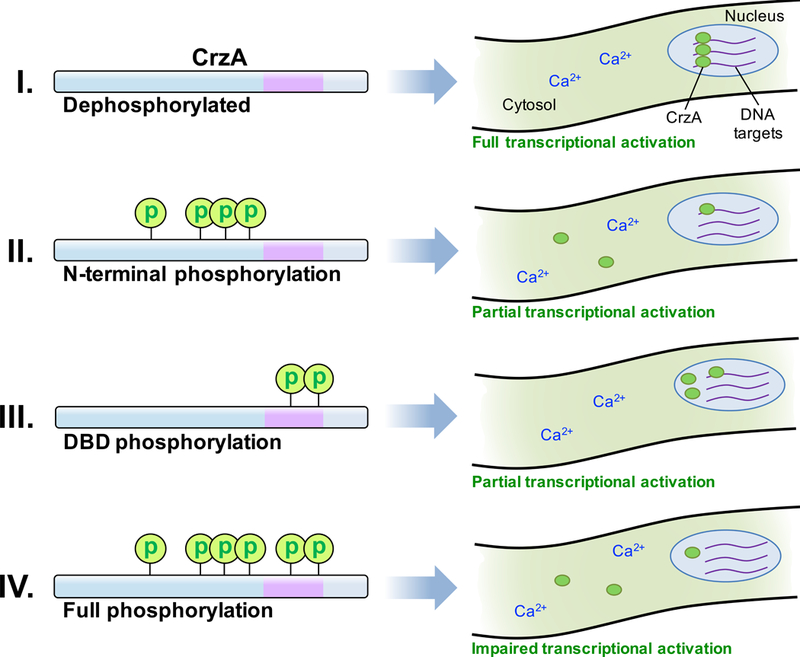
(I) When fully dephosphorylated in the presence of a high calcium ion concentration, CrzA is in the most active state, strongly localized to the nucleus and able to efficiently interact with the promoters of target genes to enable transcription following calcium stimulation. (II) When the N-terminal regulatory domain is phosphorylated, nuclear translocation is attenuated, but some protein may still enter the nucleus and bind efficiently to target genes. (III) When the DBD is phosphorylated, CrzA translocates to the nucleus, but efficiency of binding to target genes may be reduced. The high amount of nuclear protein present may enable sufficient expression of target genes. (IV) When CrzA is fully phosphorylated, both nuclear translocation and DNA-binding efficiency may be impaired, leading to insufficient activation of target gene expression and consequent phenotypic defects.
It is important to emphasize that our findings concerning the effects of CrzA phosphorylation on nuclear exclusion relate to the environmental condition of high calcium ion concentration. Under this stress condition, in which CrzA is expected to be strongly activated, our data indicate that a high degree of phosphorylation is required to block nuclear import and activity of the protein. However, our mass spectroscopic analysis indicates only partial phosphorylation of CrzA under basal environmental conditions, with only 10 of the 20 total sites demonstrated to be phosphorylated in the wild-type genetic background. Despite this, CrzA was observed to be primarily cytosolic under these same growth conditions, suggesting that partial phosphorylation is sufficient to prevent nuclear translocation to a large extent in the absence of a strong calcium stimulus, possibly due to reduced activity of other, as of yet undetermined calcium-induced factors involved in CrzA activation. In the future, it would be of interest to define the specific sites at which phosphorylation is required to maintain nuclear exclusion in the absence of excess calcium by means of reciprocal experiments in which phosphorylated residues are substituted with nonphosphorylatable alanines in order to potentially produce a constitutively nuclear localized CrzA mutant isoform. For this study, we have chosen to focus on defining the capacity for phosphorylation-mediated inhibition of CrzA under activating stress conditions, as this is likely to be of greater relevance to the inhibition of pathogenesis. It is notable, however, that phosphomimicry at sites for which dephosphorylation was not found to be regulated by calcineurin was necessary to produce phenotypic defects under both basal conditions (reduced conidiation) and stress conditions (calcium and antifungal drug sensitivity). This suggests that some degree of dynamic dephosphorylation/phosphorylation of CrzA is necessary to promote growth and conidiation under both types of environmental conditions. It would be valuable to determine whether any of the calcineurin-independent sites identified in this study are targeted by calcineurin specifically under stress conditions and/or within conidia.
The two potential NLS sequences identified (NLS-A and NLS-B) were within the DNA-binding domain of CrzA. One of these, NLS-A, is conserved in yeast Crz1p, where it was found to be sufficient to enable nuclear accumulation of a truncated C-terminal portion of the protein (Polizotto and Cyert, 2001). Our observations in A. fumigatus revealed that deletion of this NLS-A sequence alone had no effect on localization. However, deletion of the NLS-A site in conjunction with NLS-B, a predicted bipartite nuclear localization sequence, resulted in strong but still incomplete suppression of nuclear accumulation, suggesting the possible presence of other unidentified NLS sequence/s mediating this process. This is consistent with findings in both S. cerevisiae and A. nidulans, wherein the N-terminal regulatory domains were also found to be involved in nuclear trafficking of CrzA homologs (Hernández-Ortiz and Espeso, 2013; Polizotto and Cyert, 2001). Though a specific regulatory domain NLS sequence was identified in the yeast, no homologous NLS sequence has been identified in either A. nidulans or A. fumigatus, nor did our bioinformatic analysis identify any potential NLS sequences in the N-terminal domain. Moreover, no arginine/lysine clusters characteristic of NLS sequences were notable in this region suggesting that any NLS sequence located in the regulatory domain must be of a non-canonical nature. The potential presence of several NLS sequences in disparate regions of CrzA further suggests the potential for finely-tuned regulation of its activity at multiple levels. Whether these NLS sequences are involved in the phosphoregulation of subcellular CrzA localization remains to be determined. Potentially phosphorylation-independent control of nuclear translocation has been noted for specific mammalian NFAT isoforms, so it is possible that multiple localization mechanisms may exist for CrzA as well (Shen et al., 2006 ).
The phenotypic resemblance of each of the three NLS mutant strains (ΔNLS-A, ΔNLS-B, and ΔNLS-A/B) to the ΔcrzA strain indicates that all these have strongly impaired transcriptional regulatory function, highly likely to be a result of disrupted DNA binding. Despite this, only the ΔNLS-A/B strain, in which both of the two sequences are deleted, displayed altered nuclear localization, suggesting the possibility that this localization defect is not simply a side effect of impaired DNA binding capacity, but instead points to an independent role for these sequences in directing subcellular shuttling. This is in keeping with findings in S. cerevisiae, in which truncation of the entire DBD did not interfere with calcium-induced nuclear accumulation of Crz1p, indicating that the DNA-binding capacity of the yeast protein is irrelevant to its localization (Polizotto and Cyert, 2001).
The requirement of a high degree of phosphorylation at clustered residues spanning both the N-terminal regulatory and DNA-binding domains of CrzA for effective suppression of its activity suggests the need for cooperative involvement of multiple protein kinases in this process, as no single kinase is expected to target all identified sites. Indeed, our bioinformatic analysis suggested the capability of several kinases to phosphorylate CrzA (Table 2). Additionally, previous studies have implicated a number of kinases in the phosphorylation of mammalian NFAT, and yeast and filamentous fungal CrzA homologs. Despite the presence of several canonical PKA target motifs, in our LC-MS/MS analysis we did not identify any CrzA sites that were phosphorylated in a PKA-dependent manner. While it is possible that CrzA is simply not an in vivo target of PKA, these findings do not rule out the possibility that PKA may target CrzA at sites that were not efficiently covered by the LC-MS/MS analytical process. However, this seems unlikely given that the strongest predicted PKA target sites in CrzA were identified as phosphorylated in our analysis, but were not found to differ significantly in phosphorylation status between the WT and ΔpkaC1 genetic backgrounds. Alternatively, PKA may phosphorylate CrzA only under particular conditions of environmental stress where suppression of calcium signaling is desirable. Another possibility is that PKA could regulate CrzA function indirectly via phosphorylation of other regulatory proteins that interact with and control CrzA activity through other mechanisms. Further investigation is required to elucidate the complex mechanism of CrzA phosphorylation in A. fumigatus, including identification of specific kinases responsible for targeting specific sites in vivo, and particular conditions under which phosphorylation occurs.
Our novel findings indicate that the phosphoregulation of A. fumigatus CrzA is more intricate than expected. Despite our site-specific data on the phosphorylation of A. fumigatus CrzA, how precisely the localization and function of CrzA is regulated via the phosphorylation/dephosphorylation of specific residues under different environmental conditions remains to be addressed. Taking into account the large number of phosphorylated residues identified and the number of kinases predicted to phosphorylate these residues, we hypothesize that numerous safeguards must be overcome in order for the activity of this vital transcription factor to be effectively suppressed under stimulatory conditions. These safeguards include: (i) the requirement for full phosphorylation of the N-terminal regulatory domain in multiple areas in order to significantly inhibit nuclear translocation (mediated by multiple redundant NLS sequences), (ii) the concurrent phosphorylation of the DBD (potentially to interfere with the efficiency of association with target gene promoters), and (iii) additional regulatory measures yet to be discovered, as even phosphomimetic mutations at all identified phosphorylation sites did not result in complete suppression of CrzA functionality. Overall, the presence of such safeguards against inhibition of CrzA function underscores the importance of the calcium signaling pathway in the selective fitness of A. fumigatus, in keeping with the importance demonstrated here of full CrzA activity for promoting virulence of this fungus in a murine model. Although minor under normal growth conditions, the effects of phosphorylation-mediated suppression of CrzA were highly pronounced under stress conditions, including exposure to antifungal treatment and during invasive growth within the hostile environment of a mammalian host system. These findings indicate an essential role for CrzA under the conditions most relevant to disease progression and treatment, of which even minor perturbation can potentially have a major impact on outcome. Future studies should seek to better understand the specific means by which phosphorylation and other posttranslational modifications influence CrzA function with regard to the biochemical mechanisms underlying nuclear translocation and exit, DNA binding capacity, and transcriptional activation of target genes. CrzA represents a promising yet challenging target for developing new strategies to combat invasive aspergillosis, and additional research into the complex mechanisms of its regulation is essential if this potential is to be realized.
EXPERIMENTAL PROCEDURES
Protein extraction and purification
A. fumigatus recombinant strains expressing CrzA-GFP fusion protein were cultured in liquid glucose minimal medium (GMM) with 250 rpm shaking for 24 h at 37°C. Total cell lysate was obtained by homogenizing mycelia (1g wet weight) using a mortar and pestle as previously described (Juvvadi et al., 2015; Juvvadi et al., 2013; Shwab et al., 2017). Total protein in the crude extracts was quantified by the Bradford method and samples were normalized to contain 10 mg protein each. GFP-Trap® (ChromoTek, Planegg-Martinsried, Germany) affinity purifications were performed from crude extracts according to the manufacturer’s instructions as previously described (Juvvadi et al., 2015).
Phosphopeptide enrichment and LC-MS/MS analysis
Phosphopeptide enrichment and LC-MS/MS were carried out as described previously (Juvvadi et al., 2013; Juvvadi et al., 2015; Shwab et al., 2017). GFP-Trap® affinity purified protein was processed for TiO2 phosphopeptide enrichment and mass spectrometry. Proteolytic digestion was accomplished by the addition of 500 ng sequencing grade trypsin (Promega, Madison, WI) directly to the resin, with incubation at 37°C for 18 h. Peptides were subjected to phosphopeptide enrichment using a 10 μl GL Sciences TiO2 Spin Tip. The dried phosphopeptide enriched samples were resuspended and subjected to chromatographic separation on a Waters NanoAquity UPLC equipped with a 1.7 μm HSS T3 C18 75 μm I.D. x250 mm reversed-phase column. The analytical column was connected to a fused silica PicoTip emitter (New Objective, Cambridge, MA) with a 10 µm tip orifice. Phosphopeptide-enriched samples were analyzed on a QExactive Plus mass spectrometer using a data-dependent mode of acquisition. MS/MS spectra of the 10 most abundant precursor ions were acquired with a CID energy setting of 27 and a dynamic exclusion of 20 s was employed for previously fragmented precursor ions.
Qualitative identifications of selected ion chromatograms from raw LC-MS/MS data
Raw LC-MS/MS data files were processed in Mascot distiller (Matrix Science) and then submitted to independent Mascot database searches (Matrix Science) against a custom NCBI_Aspergillus database containing both forward and reverse entries of each protein as described previously (Shwab et al., 2017). Search tolerances were 5 or 10 ppm for precursor ions and 0.02 or 0.04 Da for product ions using trypsin specificity with up to two missed cleavages for phosphopeptide enriched or non-phoshopeptide enriched data, respectively. Carbamidomethylation (+57.0214 Da on C) was set as a fixed modification, whereas oxidation (+15.9949 Da on M), deamidation (+0.98 Da on N and Q) and phosphorylation (+79.98 Da on S, T, and Y) were considered variable modifications. All searched spectra were imported into Scaffold (Proteome Software) and scoring thresholds were set to achieve a protein false discovery rate of 0% using the PeptideProphet algorithm. Normalized spectral counts were used to estimate relative protein abundances for interaction studies. This was accomplished by adjusting the sum of the selected quantitative value for all proteins in the list within each MS sample to the average of sums of all MS samples present in the experiment. Only those proteins with at least two unique peptides to match were considered to be correct. Phosphorylation site localization was assessed by exporting peak lists directly from Scaffold into the online ambiguity score (Ascore) algorithm (ascore.med.harvard.edu/ascore.html).
Construction of crzA mutations in Aspergillus fumigatus
Mutant strains were generated as described previously (Shwab et al., 2017). Briefly, GFP-labeling of CrzA was accomplished by insertion of the appropriate coding region (lacking a stop codon) 5’ to, and in frame with, the egfp coding region within the pUCGH vector, followed by transformation into the akuBKU80 strain of A. fumigatus and screening via hygromycin B selection as described (Juvvadi et al., 2013). CrzA gene deletion (ΔcrzA) was accomplished via replacement of the crzA open reading frame with the A. parasiticus pyrG selectable marker in the akuBKU80 pyrG- genetic background (Table S1) followed by screening for prototrophic transformants and verification via amplification of the deletion cassette from genomic DNA and Southern blotting as described previously (Cramer et al., 2008). Site-directed mutagenesis and putative NLS deletions of crzA were accomplished by amplifying coding regions of the gene containing the desired point mutations or deleted regions via fusion PCR as described previously (Shwab et al., 2017). Expression of GFP-labeled CrzA in the cnaA and pkaC1 deletion backgrounds was accomplished by co-transformation of an akuBKU80 pyrG- auxotrophic strain with linearized plasmids for each modification, followed my hygromycin B selection of transformants. All mutant strains, as well as the crzA-egfp strain expressing GFP-tagged native CrzA protein, were verified for homologous integration of mutations and egfp coding sequence at the native crzA locus. Genomic DNAs amplified from mutant strains using a forward primer targeting a genomic DNA sequence upstream of the transformation construct and a reverse primer targeting the egfp coding region were sequenced to confirm mutations and in-frame gfp fusion. The presence of GFP-labeled mutant protein isoforms was verified for key strains by western blot as described previously (Juvvadi et al., 2013). Briefly, protein was extracted from 24-hour liquid cultures followed by separation of protein in crude extracts via SDS-PAGE, followed by transfer to a PVDF membrane and probing with anti-GFP primary antibodies and horseradish peroxidase-conjugated secondary antibodies, followed by exposure to chemiluminescent substrate and visualization on x-ray film.
Assessment of radial growth, conidiation rate, and CaCl2 and antifungal drug sensitivity
For radial growth assays, conidia (104) were point inoculated on solid agar media in petri plates and incubated for 4 days at 37°C. Media used included standard GMM (1% glucose, pH 6.5) as well as modified GMM containing 200mM CaCl2, 0.5μg/mL CSP, or 4μg/mL CSP. Conidiation was quantified by collecting conidia from each plate in equal volumes of 0.05% Tween 20 followed by dilution of collected spore stocks and counting of spores via haemocytometer. The mean radial growth and conidiation rates for each of the strains were compared statistically by Dunnett’s post test following analysis of variance (ANOVA). For voriconazole and itraconazole sensitivity determinations, CLSI M38-A2 in vitro antifungal susceptibility standards were used (Institute, 2008).
Murine invasive aspergillosis virulence assay and histopathological analysis
15 male mice (CD1, Charles River Laboratory, Raleigh, NC, USA) were immunosuppressed with cyclophosphamide (150 mg/kg, intraperitoneally, days −2 and +3) and triamcinolone acetonide (40 mg/kg, subcutaneously, days −1 and +6). 40 μl of 1×108 conidia/mL suspensions of the relevant strains were delivered intra-nasally following a brief isoflurane anesthesia induction. Survival was plotted on a Kaplan–Meier curve and analyzed using log rank pair-wise comparison (P < 0.05). To characterize in vivo disease histopathology, additional mice were infected with each of the analyzed strains as above. Mice were euthanized on day +3 after inoculation and lungs were harvested. Lung sections were stained with Gomori’s methenamine silver stain to visualize fungal hyphae and with hematoxylin and eosin stains to examine inflammation and tissue damage as previously described (Steinbach et al., 2006). Animal studies at Duke University Medical Center were in full compliance with all of the guidelines of the Duke University Medical Center Institutional Animal Care and Use Committee (IACUC) and in full compliance with the United States Animal Welfare Act (Public Law 98–198). Duke University Medical Center IACUC approved all of the vertebrate studies under the protocol number A-249–16-11. The studies were conducted in the Division of Laboratory Animal Resources (DLAR) facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Fluorescence Microscopy
Conidia (104) of CrzA-GFP-labeled strains were cultured in 35mm cover-glass-bottomed petri dishes containing 3 mL of liquid GMM and incubated for 16 h at 37°C prior to visualization. Where noted, 1 mL of 800mM CaCl2 in liquid GMM was added to the growth medium 15 minutes prior to visualization, to produce a final concentration of 200mM CaCl2. Where noted, GSK-3β Inhibitor VII kinase inhibitor (Calbiochem), was added to the growth medium to a final concentration of 50μM one hour prior to visualization. Hyphae were visualized using an Axio Observer 3 microscope (Carl Zeiss, Oberkochen, Germany) equipped with ZEN lite imaging software.
For quantitation of CrzA-GFP nuclear localization, 103 conidia of CrzA-GFP labeled strains were cultured in 35mm cover-glass-bottomed petri dishes containing 3 mL of liquid GMM and incubated for 9 h at 37°C. 1 mL of 800mM CaCl2 in liquid GMM was added to the growth medium 15 minutes prior to visualization, to produce a final concentration of 200mM CaCl2. For visualization of nuclei, Hoechst 33342 dye (Thermo Fisher Scientific, Walthon, MA) was added to the growth medium to a final concentration of 2μg/mL, and Triton X-100 (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 1% solution. 50 germlings of each strain culture were categorized as displaying strong nuclear localization of CrzA-GFP (defined as nuclear fluorescence clearly brighter than surrounding cytoplasmic fluorescence), partial nuclear localization of CrzA-GFP (defined as nuclear fluorescence equal to or only marginally brighter than surrounding cytoplasmic fluorescence), or nuclear exclusion of CrzA-GFP (defined as nuclear fluorescence less bright than surrounding cytoplasmic fluorescence). The experiment was repeated three times on three separate occasions and the average counts of each of the three replicates were calculated for each strain. Statistical comparisons to the CrzA-C and ΔcnaA strains were made via Dunnett’s post-test following ANOVA.
Supplementary Material
Figure S1. Western blot analyis of GFP-labeled CrzA mutant isoform expression. Cultures of indicated strains were grown for 24 hours in liquid GMM at 37C prior to extraction of total protein from filtered mycelial tissue frozen in liquid nitrogen and pulverized using a mortar and pestle. Total protein concentrations were normalized via Bradford assay and approximately equivalent amounts of crude extract for each strain were probed via western blot with both anti-GFP primary antibodies to detect GFP-labeled CrzA and anti-β-tubulin primary antibodies to serve as a control. Detection was accomplished by probing with horseradish peroxidase-conjugated secondary antibodies, followed by exposure to chemiluminescent substrate and visualization using x-ray film.
Figure S2. Radial growth and conidiation of single-cluster CrzA phosphorylation mutants under basal and stress-inducing growth conditions and susceptibility to echinocandin and azole antifungals. (A) Quantification of radial growth of single-cluster CrzA mutant strains on solid agar media under multiple conditions. Strains CrzA-1 through CrzA-8 as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and GMM agar containing 200mM CaCl2, 0.5μg/mL CSP, or 4μg/mL CSP, and incubated at 37°C for 4 days. Bars represent averages of three replicates, normalized against growth of CrzA-C on GMM. Green = GMM, Blue = CaCl2, Orange = 0.5μg/mL CSP, Yellow = 4μg/mL CSP). Error bars represent standard deviations. Asterisks indicate statistically significant differences (Dunnett’s post-test, *P < 0.05, **P < 0.01) compared to CrzA-C on the same growth medium. (B) Micrographs showing growth of fungal hyphae during exposure to voriconazole and itraconazole antifungals. Conidia of indicated strains were inoculated into liquid medium containing indicated drug concentrations and incubated at 37°C for 48 hours. Magnification = 20X.
Figure S3. Additional histological analysis of infected murine lung tissue. Lung tissue histology of infected mice. Mice were immunocompromised via cyclophosphamide and triamcinolone injections and three mice per A. fumigatus strain were inoculated intranasally with conidia of strain types CrzA-C, ΔcrzA, and CrzA-13. Lung tissue was harvested from all mice on day 3 post-infection and visualized microscopically following silver staining in order to assess invasive hyphal growth (dark coloration) and H&E staining in order to assess tissue damage. Both hyphal invasion and tissue damage were evident in lungs infected with CrzA-C and CrzA-13 strains, but not ΔcrzA. Magnification = 10X.
Figure S4. Calcium-stimulated nuclear translocation of single-cluster mutant CrzA isoforms. Conidia (104) of strains expressing GFP-labeled CrzA (CrzA-GFP) with native amino acid sequence (CrzA-C) or each mutant CrzA isoform CrzA-1 to CrzA-8, as well as the ΔcnaA strain, were inoculated into glass-bottomed Petri dishes containing liquid GMM and incubated overnight at 37°C. Hyphae were visualized microscopically 15 minutes after addition of 200mM CaCl2 to the medium. Strong levels of calcium-stimulated nuclear translocation of CrzA were observed in strains all strains with the exception of the ΔcnaA control, which exhibited nuclear exclusion. White scale bars represent 10μm distances.
Figure S5. Susceptibility of CrzA NLS deletion mutant strains to caspofungin. (A) Radial growth of selected CrzA mutant strains on solid agar media containing multiple caspofungin concentrations. Strains CrzA ΔNLS-A and CrzA ΔNLS-B as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and GMM agar containing 0.5μg/mL CSP, or 4μg/mL CSP, and incubated at 37°C for 3 days. (B) Quantification of radial growth on each media type (green=GMM, orange=0.5μg/mL CSP, yellow=4μg/mL CSP). Column heights represent averages of three replicates. Error bars represent standard deviations. Letters over columns indicate statistically significant differences (Dunnett’s post-test, P < 0.05) in comparison to aradial growth of CrzA-C on standard GMM without CSP, bradial growth of the same strain under comparison on standard GMM without CSP, cradial growth of the same strain under comparison on media with 0.5μg/mL CSP. A significant paradoxical growth increase from 0.5μg/mL to 4μg/mL CSP was only observed for CrzA-C.
ACKNOWLEDGEMENTS
Thanks to D. Chris Cole for the performance of azole susceptibility tests.
FUNDING
This work was funded by the Tri-Institutional Molecular Mycology and Pathogenesis Training Program (T32 AI052080) post-doctoral fellowship to E.K.S. and through an NIH/NIAID R21 award AI127551 to P.R.J. and W.J.S.
ABBREVIATIONS
- CaM
calmodulin
- CBD
calcineurin-binding domain
- CDK
cyclin-dependent kinase
- CK
casein kinase
- CSP
caspofungin
- DBD
DNA-binding domain
- GFP
green-fluorescent protein
- GMM
glucose minimal medium
- GSK
glycogen synthase kinase
- H&E
haemotoxylin and eosin
- IA
invasive aspergillosis
- JNK
Jun N-terminal kinase
- LC-MS/MS
liquid chromatography-tandem mass spectroscopy
- MAPK
mitogen-activated protein kinas;. NES nuclear export signal
- NFAT
nuclear factor of activated T-cells
- NLS
nuclear localization signal
- PKA
protein kinase A
- SRR
serine-rich region
- TF
transcription factor
- WT
wild-type
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Abad A, Fernández-Molina JV, Bikandi J, Ramirez A, Margareto J, Sendino J, Hernando FL, Pontón J, Garaizar J, and Rementeria A (2010). What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Revista Iberoamericana de Micología 27, 155–182. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, and Brunak S (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Cisneros-Barroso E, Yance-Chávez T, Kito A, Sugiura R, Gómez-Hierro A, Giménez-Zaragoza D, and Aligue R (2014. ). Negative feedback regulation of calcineurin-dependent Prz1 transcription factor by the CaMKK-CaMK1 axis in fission yeast. Nucleic Acids Research 42, 9573–9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer RAJ, Perfect BZ, Pinchai N, Park S, Perlin DS, Asfaw YG, Heitman J, Perfect JR, and Steinbach WJ (2008). Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryotic Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS (2003). Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochemical and Biophysical Research Communications 311, 1143–1150. [DOI] [PubMed] [Google Scholar]
- de Castro PA, Chen C, de Almeida RS, Freitas FZ, Bertolini MC, Morais ER, Brown NA, Ramalho LN, Hagiwara D, Mitchell TK, et al. (2014). ChIP-seq reveals a role for CrzA in the Aspergillus fumigatus high-osmolarity glycerol response (HOG) signalling pathway. Molecular Microbiology 94, 655–674. [DOI] [PubMed] [Google Scholar]
- de Castro PA, Chiaratto J, Morais ER, Dos Reis TF, Mitchell TK, Brown NA, and Goldman GH (2017. ). The putative flavin carrier family FlcA-C is important for Aspergillus fumigatus virulence. Virulence 8, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW (1996). Therapeutic outcome in invasive aspergillosis. Clinical Infectious Disease 23, 608–615. [DOI] [PubMed] [Google Scholar]
- Fortwendel JR, Juvvadi PR, Perfect BZ, Rogg LE, Perfect JR, and Steinbach WJ (2010). Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrobial Agents and Chemotherapy 54, 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Kondo A, Fujioka T, and Abe K (2008). Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Current Genetics 54, 325–338. [DOI] [PubMed] [Google Scholar]
- Hernández-Ortiz P, and Espeso EA (2013). Phospho-regulation and nucleocytoplasmic trafficking of CrzA in response to calcium and alkaline-pH stress in Aspergillus nidulans. Molecular Microbiology 89, 532–551. [DOI] [PubMed] [Google Scholar]
- Hernández-Ortiz P, and Espeso EA (2017. ). Spatiotemporal dynamics of the calcineurin target CrzA. Cellular Signalling 29, 168–180. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, and Rao A (2003). Transcriptional regulation by calcium, calcineurin, and NFAT. Genes and Development 17, 2205–2232. [DOI] [PubMed] [Google Scholar]
- Institute, C.a.L.S. (2008). Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed. CLSI document; M38–A2. [Google Scholar]
- Juvvadi PR, Belina D, Soderblom EJ, Moseley MA, and Steinbach WJ (2013). Filamentous fungal-specific septin AspE is phosphorylated in vivo and interacts with actin, tubulin and other septins in the human pathogen Aspergillus fumigatus. Biochemical and Biophysical Research Communications 431, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvvadi PR, Ma Y, Richards AD, Soderblom EJ, Moseley MA, Lamoth F, and Steinbach WJ (2015). Identification and mutational analyses of phosphorylation sites of the calcineurin-binding protein CbpA and the identification of domains required for calcineurin binding in Aspergillus fumigatus. Frontiers in Microbiology 6, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar KA, and Cyert MS (2004). Integration of stress responses: modulation of calcineurin signaling in Saccharomyces cerevisiae by protein kinase A. Eukaryotic Cell 3, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar KA, Zhu H, Snyder M, and Cyert MS (2003. ). Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes and Development 17, 2698–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike A, Kato T, Sugiura R, Ma Y, Tabata Y, Ohmoto K, Sio SO, and Kuno T (2012). Genetic screening for regulators of Prz1, a transcriptional factor acting downstream of calcineurin in fission yeast. Journal of Biology and Chemistry 287, 19294–19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, and Yanagawa H (2009). Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proceedings of the National Academy of Sciences of the USA 106, 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, and Brunak S (2004). Analysis and prediction of leucine-rich nuclear export signals. Protein Engineering, Design, and Selection 17, 527–536. [DOI] [PubMed] [Google Scholar]
- Manoli MT, and Espeso EA (2019). Modulation of calcineurin activity in Aspergillus nidulans: the roles of high magnesium concentrations and of transcriptional factor CrzA. Molecular Microbiology (E-published ahead of print) [DOI] [PubMed]
- Okamura H, Aramburu J, García-Rodríguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, and Rao A (2000). Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Molecular Cell 6, 539–550. [DOI] [PubMed] [Google Scholar]
- Polizotto RS, and Cyert MS (2001). Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. Journal of Cell Biology 154, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, de Assis LJ, Bertolini MC, Malavazi I, and Goldman GH (2017). The Aspergillus fumigatus CrzA Transcription Factor Activates Chitin Synthase Gene Expression during the Caspofungin Paradoxical Effect. MBio 8, e00705–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Liu Y, Cseresnyés Z, Hawkins A, Randall WR, and Schneider MF (2006). Activity- and Calcineurin-independent Nuclear Shuttling of NFATc1, but Not NFATc3, in Adult Skeletal Muscle Fibers. Molecular Biology of the Cell 17, 1570–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan CM, Heist EK, Beals CR, Crabtree GR, and Gardner P (2002). Protein kinase A negatively modulates the nuclear accumulation of NF-ATc1 by priming for subsequent phosphorylation by glycogen synthase kinase-3. Journal of Biology and Chemistry 277, 48664–48676. [DOI] [PubMed] [Google Scholar]
- Shwab EK, Juvvadi PR, Waitt G, Soderblom EJ, Moseley MA, Nicely NI, Asfaw YG, and Steinbach WJ (2017). A Novel Phosphoregulatory Switch Controls the Activity and Function of the Major Catalytic Subunit of Protein Kinase A in Aspergillus fumigatus. MBio 8, e02319–02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani FM, Malavazi I, da Silva Ferreira ME, Savoldi M, Von Zeska Kress MR, de Souza Goldman MH, Loss O, Bignell E, and Goldman GH (2008). Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Molecular Microbiology 67, 1274–1291. [DOI] [PubMed] [Google Scholar]
- Soriani FM, Malavazi I, Savoldi M, Espeso E, Dinamarco TM, Bernardes LA, Ferreira ME, Goldman MH, and Goldman GH (2010). Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiology 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielvogel A, Findon H, Arst HN, Araújo-Bazán L, Hernández-Ortíz P, Stahl U, Meyer V, and Espeso EA (2008). Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochemistry Journal 414, 419–429. [DOI] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A, Guo JJ, and Cyert MS (1999). Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes and Development 13, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Cramer RAJ, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, Kirkpatrick WR, Patterson TF, Benjamin DKJ, Heitman J, et al. (2006). Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryotic Cell 5, 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S (2014). Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryotic Cell 13, 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Xiong Q, Enslen H, Davis RJ, and Chow CW (2002. ). Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Molecular Cell Biology 22, 3892–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Western blot analyis of GFP-labeled CrzA mutant isoform expression. Cultures of indicated strains were grown for 24 hours in liquid GMM at 37C prior to extraction of total protein from filtered mycelial tissue frozen in liquid nitrogen and pulverized using a mortar and pestle. Total protein concentrations were normalized via Bradford assay and approximately equivalent amounts of crude extract for each strain were probed via western blot with both anti-GFP primary antibodies to detect GFP-labeled CrzA and anti-β-tubulin primary antibodies to serve as a control. Detection was accomplished by probing with horseradish peroxidase-conjugated secondary antibodies, followed by exposure to chemiluminescent substrate and visualization using x-ray film.
Figure S2. Radial growth and conidiation of single-cluster CrzA phosphorylation mutants under basal and stress-inducing growth conditions and susceptibility to echinocandin and azole antifungals. (A) Quantification of radial growth of single-cluster CrzA mutant strains on solid agar media under multiple conditions. Strains CrzA-1 through CrzA-8 as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and GMM agar containing 200mM CaCl2, 0.5μg/mL CSP, or 4μg/mL CSP, and incubated at 37°C for 4 days. Bars represent averages of three replicates, normalized against growth of CrzA-C on GMM. Green = GMM, Blue = CaCl2, Orange = 0.5μg/mL CSP, Yellow = 4μg/mL CSP). Error bars represent standard deviations. Asterisks indicate statistically significant differences (Dunnett’s post-test, *P < 0.05, **P < 0.01) compared to CrzA-C on the same growth medium. (B) Micrographs showing growth of fungal hyphae during exposure to voriconazole and itraconazole antifungals. Conidia of indicated strains were inoculated into liquid medium containing indicated drug concentrations and incubated at 37°C for 48 hours. Magnification = 20X.
Figure S3. Additional histological analysis of infected murine lung tissue. Lung tissue histology of infected mice. Mice were immunocompromised via cyclophosphamide and triamcinolone injections and three mice per A. fumigatus strain were inoculated intranasally with conidia of strain types CrzA-C, ΔcrzA, and CrzA-13. Lung tissue was harvested from all mice on day 3 post-infection and visualized microscopically following silver staining in order to assess invasive hyphal growth (dark coloration) and H&E staining in order to assess tissue damage. Both hyphal invasion and tissue damage were evident in lungs infected with CrzA-C and CrzA-13 strains, but not ΔcrzA. Magnification = 10X.
Figure S4. Calcium-stimulated nuclear translocation of single-cluster mutant CrzA isoforms. Conidia (104) of strains expressing GFP-labeled CrzA (CrzA-GFP) with native amino acid sequence (CrzA-C) or each mutant CrzA isoform CrzA-1 to CrzA-8, as well as the ΔcnaA strain, were inoculated into glass-bottomed Petri dishes containing liquid GMM and incubated overnight at 37°C. Hyphae were visualized microscopically 15 minutes after addition of 200mM CaCl2 to the medium. Strong levels of calcium-stimulated nuclear translocation of CrzA were observed in strains all strains with the exception of the ΔcnaA control, which exhibited nuclear exclusion. White scale bars represent 10μm distances.
Figure S5. Susceptibility of CrzA NLS deletion mutant strains to caspofungin. (A) Radial growth of selected CrzA mutant strains on solid agar media containing multiple caspofungin concentrations. Strains CrzA ΔNLS-A and CrzA ΔNLS-B as well as CrzA-C and ΔcrzA controls were inoculated onto standard GMM agar plates and GMM agar containing 0.5μg/mL CSP, or 4μg/mL CSP, and incubated at 37°C for 3 days. (B) Quantification of radial growth on each media type (green=GMM, orange=0.5μg/mL CSP, yellow=4μg/mL CSP). Column heights represent averages of three replicates. Error bars represent standard deviations. Letters over columns indicate statistically significant differences (Dunnett’s post-test, P < 0.05) in comparison to aradial growth of CrzA-C on standard GMM without CSP, bradial growth of the same strain under comparison on standard GMM without CSP, cradial growth of the same strain under comparison on media with 0.5μg/mL CSP. A significant paradoxical growth increase from 0.5μg/mL to 4μg/mL CSP was only observed for CrzA-C.


