Abstract
Despite recent advances in asthma management with anti-IL-5 therapies, many patients with eosinophilic asthma remain poorly controlled. IL-3 shares a common beta subunit receptor with both IL-5 and GM-CSF, but through alpha subunit-specific properties, uniquely influences eosinophil biology and may serve as a potential therapeutic target. We aimed to globally characterize the transcriptomic profiles of GM-CSF, IL-3 and IL-5 stimulation on human circulating eosinophils and identify differences in gene expression using advanced statistical modeling. Human eosinophils were isolated from the peripheral blood of healthy volunteers and stimulated with either GM-CSF, IL-3 or IL-5 for 48 hours. RNA was then extracted and bulk sequencing performed. DESeq analysis identified differentially expressed genes and weighted gene co-expression network analysis independently defined modules of genes that are highly co-expressed. GM-CSF, IL-3 and IL-5 commonly upregulated 252 genes and downregulated 553 genes, producing a pro-inflammatory and survival phenotype that was predominantly mediated through TWEAK signaling. IL-3 stimulation yielded the most numbers of differentially expressed genes that were also highly co-expressed (n = 119). These genes were enriched in pathways involving JAK/STAT signaling. GM-CSF and IL-5 stimulation demonstrated redundancy in eosinophil gene expression. In conclusion, IL-3 produces a distinct eosinophil gene expression program among the beta-chain receptor cytokines. IL-3 upregulated genes may provide a foundation for research into therapeutics for patients with eosinophilic asthma who do not respond to anti-IL-5 therapies.
Keywords: Human, Eosinophils, Cytokines, IL-3, Asthma
Introduction
Asthma is a chronic disease of the airway associated with significant morbidity. In the United States alone, costs related to asthma have amounted to more than eighty billion dollars annually [1]. While a significant proportion of asthmatics maintain control via inhaled corticosteroid therapy, disease heterogeneity has caused many individuals to remain poorly controlled with either high symptom scores and/or frequent exacerbations. The recent approval of monoclonal antibodies against IL-5 has improved control of severe asthmatics with eosinophilia, demonstrating lower exacerbation rates, reduced oral glucocorticoid use, improved lung function, and improved asthma control scores in randomized control trials [2-3]. Nevertheless, not all patients with eosinophilic asthma respond to anti-IL-5 therapies clinically, and populations of functional eosinophils have been shown to remain in the airway despite anti-IL-5 treatment [4-5]. It has therefore been suggested that alternative mechanisms remain relevant to the pathogenesis of eosinophilic asthma in certain individuals and that further investigation is necessary to optimize precision treatment strategies.
One area of investigative focus has involved the cytokines IL-3 and GM-CSF. Similar to IL-5, these two cytokines are produced as part of the Th2 inflammatory response and are crucial to eosinophil development and function [6-7]. Despite all three cytokines sharing a common beta (β)-chain receptor subunit, each differentially affects eosinophil biology as the result of divergent downstream intracellular signaling events through alpha (α)-chain subunit-specific properties [6,8]. Of these cytokines, IL-3 has been shown to most strongly and differentially affect eosinophil function, especially over prolonged stimulation periods [6]. This, together with the observation that eosinophils recruited to the airway following allergen challenge have increased surface levels of the α-chain subunit specific to IL-3 but reduced levels of the α-chain subunit specific to IL-5, has made IL-3 an attractive potential target in asthma therapeutics [5,9]. While knowledge of eosinophil biology as it relates to IL-3 stimulation has been rapidly growing, no study to date has utilized large scale gene expression profiling to define the IL-3 signature in eosinophils. In the present work, we utilize whole transcriptome bulk RNA sequencing methods and advanced statistical modeling to demonstrate a unique transcriptional profile of human blood eosinophils stimulated by IL-3 ex vivo after being harvested from normal subjects. Since IL-3 has been previously described to prolong intracellular signaling in eosinophils [6], we specifically report results following cytokine stimulation for 48 hours.
Materials and Methods
Subject characteristics
Venous blood specimens were obtained after informed consent under a protocol approved by the Institutional Review Board at the University of California San Diego. Whole blood (160 ml) was drawn from 13 healthy volunteers without known allergic disease (7 for gene expression studies and 6 for flow cytometry experiments).
Eosinophil isolation and stimulation
Eosinophils were purified from the peripheral blood of normal donors by negative selection (StemCell Technologies, Vancouver, Canada) as previously described [10]. Briefly, red blood cells (RBCs) were depleted by hetastarch incubation followed by gravity separation. Granulocytes were isolated by centrifugation of RBC-depleted blood over a Ficoll gradient. Eosinophils were then isolated from the granulocyte fraction by incubation with a cocktail of negative selection antibodies followed by passage over a magnetized column. Eosinophil purity was routinely > 99% by Hema 3 staining (Fisher Scientific, Medford, MA), and eosinophil viability was routinely > 99% by trypan blue exclusion after cell isolation. Purified eosinophils from each donor were split equally into unstimulated, GM-CSF-stimulated, IL-3-stimulated, and IL-5-stimulated groups (1-2×106 eosinophils per group). Human recombinant IL-3, IL-5 (R&D Systems, Minneapolis, MN) and GM-CSF (BioLegend, San Diego, CA) were used for stimulation, all for 48 hours at a concentration of 10 ng/ml in RPMI-1640 (supplemented with 10% FBS and 1% penicillin/streptomycin) at 37 ˚C. Cell viability remained > 90% for all conditions after 48 hours of stimulation, as assessed by propidium iodide staining. For gene expression experiments, eosinophils were resuspended in a phenol/guanidine-based lysis reagent (QIAzol, Qiagen), either immediately after purification for unstimulated cells or after 48 hours of stimulation, and stored at −80 ˚C prior to RNA extraction. For flow cytometry experiments, unstimulated eosinophils were stored at 4 ˚C prior to staining, and stimulated cells were stained after 48 hours. Unstimulated eosinophils remained > 98% viable after storage at 4 ˚C for 48 hours.
RNA extraction
Isolated and stimulated eosinophils from 7 donors were stored in QIAzol lysis reagent as previously described. Total RNA was isolated using miRNeasy micro kit (Qiagen) loaded on an automated platform (Qiacube, Qiagen). Samples were quantified as described previously [11-12] and quality of RNA assessed by Fragment Analyzer (Advance Analytical). All samples had an RNA integrity number (RIN) > 8.0 and passed our quality and quantity control steps as described previously [12-13].
mRNA sequencing library preparation
Purified total RNA (≈5 ng) was amplified following the Smart-seq2 protocol [13-14]. Briefly, mRNA was captured using poly-dT oligos and directly reverse-transcribed into full-length cDNA using the described template-switching oligo [13-14]. cDNA was amplified by PCR for 14-15 cycles and purified using AMPure XP magnetic bead (0.9:1 (vol:vol) ratio, Beckman Coulter). From this step, for each sample, 1 ng of cDNA was used to prepare a standard NextEra XT sequencing library (NextEra XT DNA library prep kit and index kits; Illumina). Barcoded Illumina sequencing libraries (Nextera; Illumina) were generated utilizing an automated platform (Biomek FXP, Beckman Coulter). Both whole-transcriptome amplification and sequencing library preparations were performed in a 96-well format to reduce assay-to-assay variability. Quality control steps were included to determine total RNA quality and quantity, the optimal number of PCR preamplification cycles, and fragment library size [13]. Samples that failed quality controls were eliminated from downstream steps. Libraries that passed strict quality controls were pooled at equimolar concentration, loaded and sequenced on the Illumina Sequencing platform, HiSeq2500 (Illumina). Libraries were sequenced to obtain more than 8 million 50-bp single-end reads (HiSeq Rapid Run Cluster and SBS Kit V2; Illumina) mapping uniquely to mRNA reference, generating a total of ~252 million mapped reads (median of ~8 million filtered mapped reads per sample). Approximately 40% of the total reads were duplicates. Given the inability to separate artifactual read duplicates from true biological duplicates in RNA-sequencing (unlike in DNA sequencing where the initial number of copies is known), we did not filter out duplicate reads during analysis. This approach is consistent with prior studies that show many duplicate reads reflect biological reality and that removal of such duplicates can worsen the power and false discovery rate for differential gene expression [15-16]. The sequences presented in this article have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE128027.
RNA sequencing analysis
RNA-seq data was mapped against the hg38 reference using TopHat [17] (v1.4.1., --librarytype fr-secondstrand -C) and the RefSeq gene annotation downloaded from the UCSC Genome Browser site. The read coverage per gene was computed using HTSeq-count (-m union -s yes -texon -i gene_id,http://www-huber.embl.de/users/anders/HTSeq). To identify genes differentially expressed between GM-CSF, IL-3, and IL-5-stimulated, as well as unstimulated eosinophils, negative binomial tests for pairwise comparisons employing the Bioconductor package DESeq2 (v1.16.1) were performed [18]. Genes were considered differentially expressed between any pairwise comparison when DESeq2 analysis resulted in a Benjamini-Hochberg-adjusted p-value < 0.01 (1% false discovery rate) and ∣Log2 (fold change)∣ ≥ 1. Moreover, any gene identified as “commonly upregulated” or “commonly downregulated” passed this threshold in all individual pairwise comparisons. Pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen) [19].
Weighted gene co-expression network analysis (WGCNA)
Weighted correlation network analysis using the R package WGCNA (version 1.61) was performed on the transcripts per million data (TPM) matrix [20]. Genes whose expression values were less than 10 TPM in all samples were removed as established in the third phase of the MAQC project (MAQC-III), also called Sequencing Quality Control (SEQC) [21]. A total of 11,889 well-expressed genes were used for generating the co-expression network. A β = 10 was selected following the scale-free topology criterion [22]. Gene modules were generated using blockwiseModules function (parameters: checkMissingData = TRUE, power = 10, TOMType = “unsigned”, minModuleSize = 30, maxBlockSize = 11889, mergeCutHeight = 0.25). A total of 24 different modules were generated, including a ‘grey’ module for non-co-expressed genes which was excluded from further analysis. Since each module by definition is comprised of highly correlated genes, their combined expression may be usefully summarized by eigengene profiles, effectively the first principal component of a given module. A small number of eigengene profiles may therefore summarize the principle patterns within the cellular transcriptome with minimal loss of information. This dimensionality-reduction approach facilitated calculation of a Spearman correlation for modules and each clinical trait (Unstimulated, Stimulated, IL-3, IL-5, GM-CSF) used in the analysis.
To visualize co-expression networks, the function exportNetworkToCytoscape at weighted = true, threshold = 0.05 was used. A soft thresholding power was chosen based on the criterion of approximate scale-free topology. Networks were generated in Gephi (version 0.9.2) using the Fruchterman Reingold layout algorithm followed by Noverlap to eliminate individual node overlap [23]. The size of each gene node was scaled according to the ‘average degree’ as calculated in Gephi.
Flow cytometry
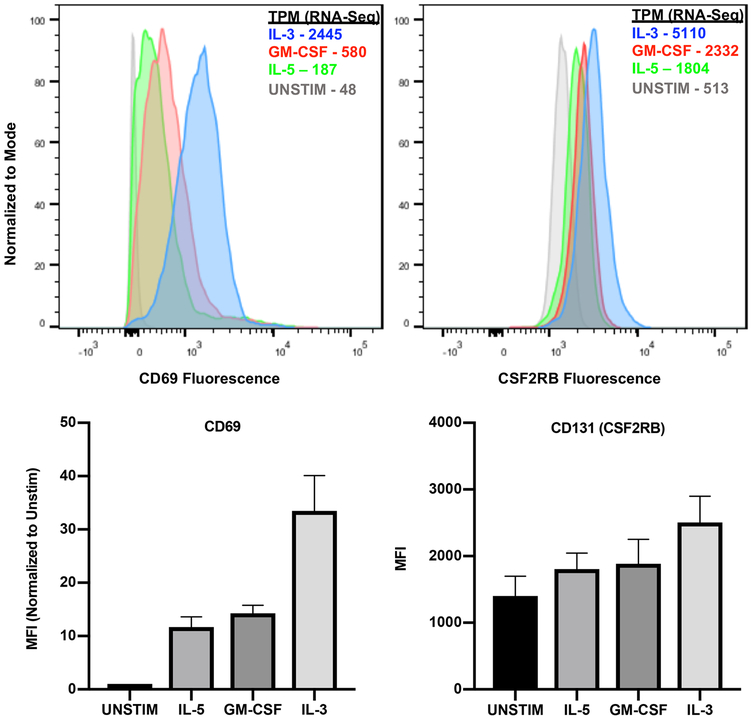
CD69 and CD131 (CSF2RB) were selected as proteins of interest based on the RNA-sequencing analysis presented in this manuscript and their expression was measured by flow cytometry. Peripheral eosinophils from 6 donors were isolated as described above and stained using specific fluorescently conjugated antibodies: mouse anti-human CD69 coupled to APC-Cy7 (clone FN50, BioLegend) and mouse anti-human CD131 coupled with PE (clone 1C1, BioLegend). Providing 3 biologic replicates, stimulated eosinophils from 3 donors were surface stained with anti-CD69. Similarly, stimulated eosinophils from 3 separate donors were permeabilized using 100% methanol and stained with anti-CD131 (CSF2RB). Flow cytometry data were obtained with a BD Biosciences LSRII, gated to eosinophils using forward and side scatter characteristics in this already highly purified population, and analyzed with FlowJo 10.5.0 Tree Star, Ashland, OR).
Results
β-chain receptor cytokines share common eosinophil activation signals
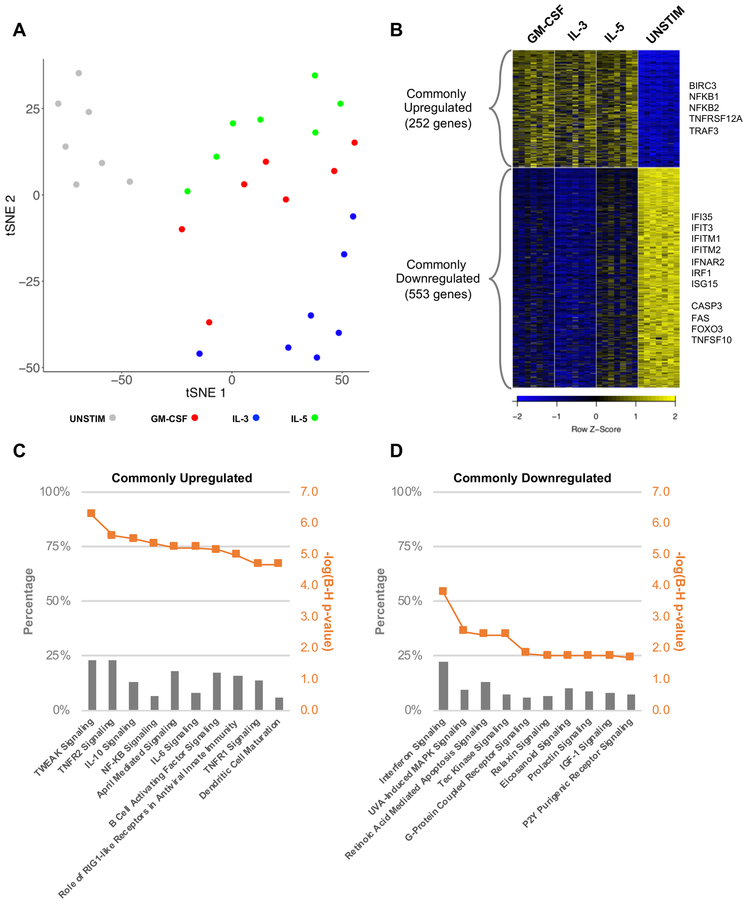
We first aimed to categorize the transcriptomic profiles distinguishing eosinophils stimulated with β-chain receptor cytokines from those left unstimulated ex vivo. Principle component analysis of the 2,000 most variable genes showed clear separation of unstimulated eosinophils from the GM-CSF, IL-3 and IL-5-stimulated groups (Figure 1A). DESeq2 analysis, which assesses for differences in average gene expression across groups, identified differentially expressed genes based on pairwise comparisons among stimulated and unstimulated conditions (Benjamini-Hochberg-adjusted p-value < 0.01). To identify a gene expression profile specific to the stimulated condition (“β-chain receptor cytokine specific genes”), we identified genes for which all pairwise comparisons between the stimulated and unstimulated groups met statistical significance with a Benjamini-Hochberg-adjusted p-value < 0.01 and ∣Log2 (fold change)∣ ≥ 1. Since DESeq2 does not account for absolute transcript levels and genes exhibiting low expression under stimulated conditions are less likely to have a meaningful biologic effect, we eliminated any gene with less than 10 TPM in a stimulated group. This analysis revealed 252 genes commonly upregulated and 553 genes commonly downregulated by β-chain receptor cytokines (Figure 1B, Supplemental Table I).
Figure 1. β-chain receptor cytokines share common activation signals on eosinophils.
A. tSNE plot of the top 2000 differentially expressed genes (5 principle components; 9 perplexity) B. Heatmap depicting the 252 commonly upregulated and 553 commonly downregulated genes in response to β-chain receptor cytokine stimulation. C,D. Top 10 pathways represented by the commonly upregulated and downregulated genes. The number of genes identified by differential expression analysis in relation to the total number of genes known for any given pathway are represented by percentages.
Pathway analysis of the commonly upregulated genes revealed multiple pathways utilizing NF-κB signaling, notable for promoting cell survival and proliferation as well as immune mediated inflammation [24]. Of these, the TWEAK (TNFSF12) signaling pathway was most significant (Benjamini-Hochberg-adjusted p-value = 5.49E-07) (Figure 1C). TWEAK is a cytokine belonging to the TNF superfamily and can either be weakly pro-apoptotic by signaling through Death Receptor 3 (DR3/TNFRSF25) or pro-survival and inflammatory by signaling through Fibroblast Growth Factor-Inducible 14 (Fn14/TNFRSF12A) [25]. We discovered upregulation of TNFRSF12A transcripts as well as its downstream signaling mediators (TRAF3) and effectors (NFKB1/NFKB2) [26]. BIRC3, encoding an inhibitor of apoptosis along the TWEAK pathway, was additionally upregulated. Finally, genes commonly upregulated in response to cytokine stimuli were overrepresented in several pathways independent of NF-κB signaling, with the top five pathways including the unfolded protein response (CD82; CEBPG; INSIG1; NFE2L2; SREBF2), phagosome maturation (CTSC; CTSD; CTSL; RAB5A; TUBA1B; TUBA1C; TUBB4B), tyrosine degradation (FAH; HPD), granulocyte adhesion and diapedesis (CCL1; CCL22; CCL24; CXCL1; ICAM1; IL1A; IL1B), and 14-3-3 mediated signaling (ELK1; RRAS; TUBA1B; TUBA1C; TUBB4B; YWHAG) (Supplemental Table II).
Pathway analysis of the commonly downregulated genes showed an overrepresentation of genes involved in interferon signaling (Figure 1D). We further discovered downregulation of several pro-apoptotic genes including those encoding for the TNF family ligands TNFSF10 and FAS, CASP3 and the PARP family cleavage products, and the transcription factor FOXO3 (Supplemental Table II). Collectively, β-chain receptor cytokines stimulate eosinophils to promote a pro-inflammatory and pro-survival phenotype.
IL-3 stimulation yields a unique eosinophil gene expression signature
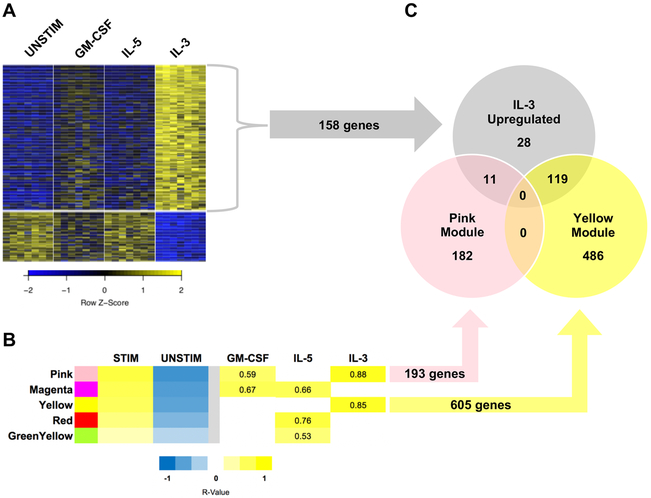
Similarly focusing on genes that maintained significance in differential expression across all pairwise comparisons, we next aimed to categorize cytokine-specific signals. Supported by IL-3 separation in the principle component analysis (Figure 1A), IL-3 stimulation resulted in the most unique gene expression profile with 158 upregulated genes and 36 downregulated genes relative to all other conditions including unstimulated (Figure 2A, Supplemental Table I). IL-5 analysis revealed one unique downregulated gene (PMAIP1) but no upregulated genes. GM-CSF did not produce any unique differentially expressed genes in either direction.
Figure 2. IL-3 stimulation yields a unique eosinophil gene expression signature.
A. Heatmap depicting the 158 upregulated and 36 downregulated genes in response to IL-3 stimulation. B. Select WGCNA modules reaching significance (p < 0.01). Numerical cells indicate the Spearman correlation coefficient of the stimulus group to the module. C. Genes similarly identified by DESeq2 and WGCNA analysis.
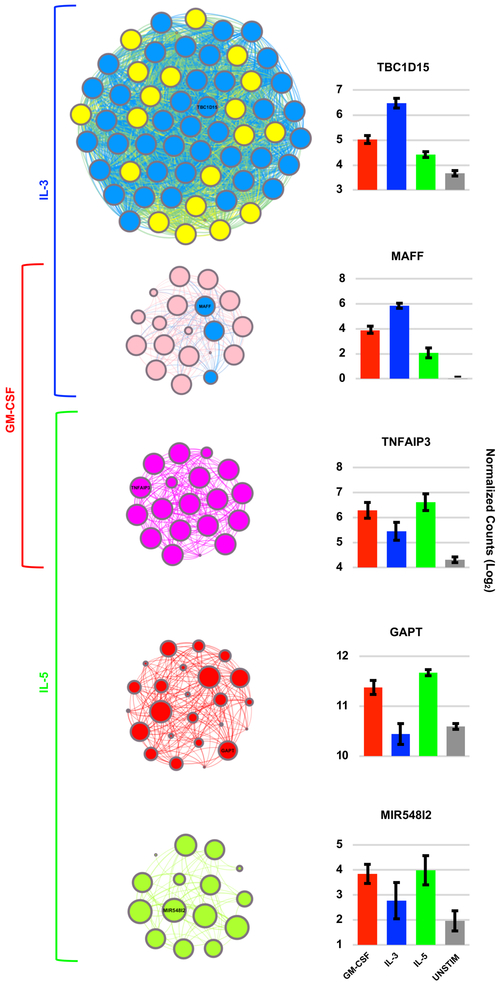
Weighted gene co-expression network analysis (WGCNA), which assigns genes into modules based on similar patterns of change in expression across samples, yielded further support for an IL-3-specific gene expression signal. Of the 24 modules identified by WGCNA, five modules were highly correlated with β-chain receptor cytokine stimulus groups (Figure 2B, r > 0.5, p < 0.01). The two strongest correlations occurred in relation to IL-3 and are represented by the pink and yellow modules (Figure 2B). These two modules contained 82% of the IL-3 upregulated genes identified by DESeq2 analysis, with most of such genes belonging to the yellow module (Figure 2C). Interestingly, the hub genes of the yellow module were predominantly genes deemed IL-3 specific by DESeq2 analysis (Figure 3, Supplemental Table III, blue nodes/font). Hub genes are the genes that are most tightly co-expressed with other genes within a given module, and therefore, are thought to be key regulators of their corresponding module’s biology. The green-yellow and red modules, correlating most strongly with IL-5, and the magenta module, correlating strongly with both GM-CSF and IL-5, also revealed module-specific hub genes but without an overlapping group-specific signal per DESeq2 analysis (Figure 3).
Figure 3. Hub genes of the yellow WGCNA module include a large number of differentially expressed IL-3 specific genes.
Gene connectivity plots of the five significant WGCNA modules along with the mean gene expression for the most representative (“hub”) gene of each module. Top 10% of genes from each module are shown. Large nodes indicate “hub genes” of increased connectivity. Blue nodes represent genes that were group-specific based on DESeq2 analysis. For example, in the yellow module nodes that are depicted in blue rather than yellow represent IL-3 specific genes based on DESeq2 analysis. In this particular module, there were a large number of IL-3 specific differentially expressed genes amongst the WGCNA hub genes. Error bars indicate standard error of the mean.
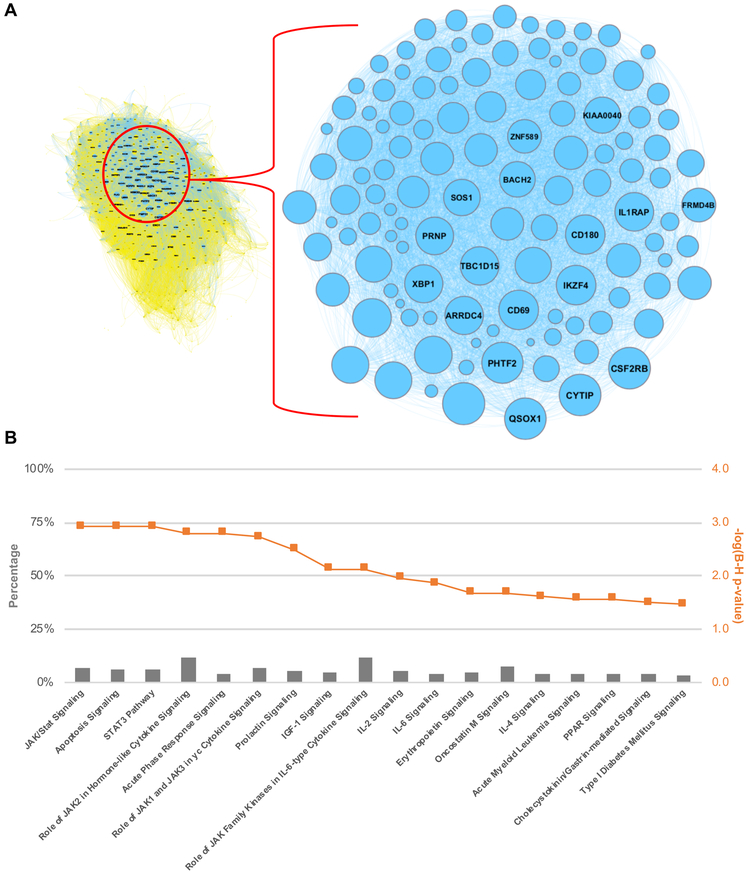
To define the biology of IL-3 stimulation on eosinophils, we focused on the 119 genes that are both differentially upregulated (DESeq2) and highly co-expressed (yellow module) (Figure 2C, Figure 4A). The top hub genes in the yellow module, defined by a module membership r ≥ 0.95, included 17 genes that were also IL-3 specific by differential expression (Figure 4A, Table I). Included in this cohort were transcripts encoding the common β-chain receptor subunit for GM-CSF, IL-3, and IL-5 (CSF2RB), surface activation markers CD69 and CD180, two transcriptional regulators extensively linked to the biology of regulatory T cells (IKZF4, BACH2), a sulfhydryl oxidase responsible for post-translational disulfide bond formation (QSOX1), a regulator of lysosomal dynamics (TBC1D15), and a transcription factor required for early eosinophil differentiation (XBP1) among other protein coding genes with either known or undefined functions [27-33]. Fluorescence-activating cell sorting (FACS) analysis corroborated our sequencing data at the protein level for two of these genes - CD69 and CD131 (CSF2RB), and mean fluorescence intensity (MFI) for both markers met statistical significance by one-way ANOVA (p < 0.05) (Figure 5). Given that CD69 is a surface marker of leukocyte activation, FACS was performed for surface expression of this protein, while whole cell protein expression was interrogated for CD131 to ensure that any intracellular stores were assayed.
Figure 4. IL-3 stimulation yields 119 differentially expressed and highly co-expressed genes.
A. Genes both differentially upregulated (DESeq2 analysis) and highly co-expressed (yellow module) following IL-3 stimulation (n=119). B. Pathway enrichment analysis of these 119 genes.
Table I. IL-3 specific hub genes.
Top hub genes of the yellow module as defined by a correlation coefficient ≥ 0.95. All 17 genes were also IL-3 specific based on DESeq2 analysis.
| Gene Symbol | Gene Significance (IL-3) | Module Membership | Normalized Mean Counts | |||||
|---|---|---|---|---|---|---|---|---|
| GS (r) | P-Value | MM (r) | P-Value | GM-CSF | IL-3 | IL-5 | UNSTIM | |
| TBC1D15 | 0.82 | 1.06E-07 | 0.98 | 5.06E-19 | 33 | 93 | 22 | 13 |
| CD180 | 0.81 | 1.65E-07 | 0.97 | 4.33E-18 | 48 | 287 | 6 | 2 |
| CD69 | 0.82 | 8.02E-08 | 0.97 | 5.24E-18 | 580 | 2445 | 187 | 48 |
| IL1RAP | 0.77 | 1.75E-06 | 0.97 | 1.23E-17 | 54 | 169 | 35 | 26 |
| IKZF4 | 0.85 | 1.30E-08 | 0.97 | 8.24E-17 | 17 | 69 | 7 | 1 |
| QSOX1 | 0.90 | 9.91E-11 | 0.96 | 2.89E-16 | 55 | 137 | 42 | 14 |
| ARRDC4 | 0.82 | 1.09E-07 | 0.96 | 3.87E-16 | 144 | 546 | 51 | 55 |
| PHTF2 | 0.91 | 3.48E-11 | 0.96 | 6.65E-16 | 70 | 159 | 56 | 17 |
| SOS1 | 0.84 | 2.10E-08 | 0.96 | 8.23E-16 | 30 | 89 | 25 | 10 |
| KIAA0040 | 0.79 | 7.47E-07 | 0.95 | 3.74E-15 | 34 | 120 | 23 | 12 |
| BACH2 | 0.79 | 6.24E-07 | 0.95 | 3.79E-15 | 2 | 14 | 2 | 1 |
| FRMD4B | 0.73 | 9.13E-06 | 0.95 | 8.55E-15 | 15 | 69 | 11 | 3 |
| CSF2RB | 0.86 | 5.37E-09 | 0.95 | 1.90E-14 | 2332 | 5110 | 1804 | 513 |
| PRNP | 0.86 | 6.73E-09 | 0.95 | 2.19E-14 | 135 | 598 | 104 | 38 |
| XBP1 | 0.86 | 3.20E-09 | 0.95 | 2.90E-14 | 241 | 572 | 151 | 96 |
| ZNF589 | 0.79 | 5.07E-07 | 0.95 | 3.52E-14 | 9 | 33 | 6 | 6 |
| CYTIP | 0.87 | 1.25E-09 | 0.95 | 3.64E-14 | 756 | 1554 | 567 | 248 |
Figure 5. Protein levels of CD69 and CD131 in human eosinophils.
Fluorescence-activated cell sorting (FACS) of surface CD69 and total cell CD131 (CSF2RB) with mean fluorescence intensity (MFI) for each cytokine-stimulated condition. Both met statistical significance by one-way ANOVA (p < 0.05). Corresponding sequencing data for each gene is included in transcripts per million (TPM).
Subsequent pathway analysis of all 119 genes connected to IL-3 by both DESeq2 and WGCNA revealed an overrepresentation of genes involved in JAK/STAT signaling as well as of genes linked to apoptosis mechanisms (Figure 4B, Supplemental Table IV). The later finding contrasts with the collective pro-survival signal we observed in cytokine stimulated eosinophils as a whole. Three non-coding RNAs (AC090152.1, AC078846.1, LINC01943) were part of this 119-gene group, and therefore, not recognized by pathway analysis.
Discussion
This study is the first of our knowledge to use two advanced, independent statistical models to analyze whole transcriptome RNA sequencing and categorize the downstream effects of β-chain receptor cytokine signaling. Using an unbiased pathway analysis approach, we discovered an overrepresentation of upregulated pro-survival genes common to all three β-chain receptor cytokines. Genes involved in TWEAK signaling pathway were most enriched. While TWEAK/Fn14 interactions have been connected to several inflammatory diseases including asthma, direct evidence supporting its role in eosinophils is lacking [34-35]. A recent study however linked single nucleotide polymorphisms in BIRC3, the inhibitor of apoptosis associated with the TWEAK pathway, with reduced asthma susceptibility and reduced loads of circulating eosinophils [36]. Furthermore, BIRC3 has been identified as a potential pathogenic gene in childhood asthma based on a molecular interaction network study [37].
Downregulated genes common to all three β-chain receptor cytokines were similarly enriched in several pathways, with the most significant pathway related to interferon signaling. INFα has been demonstrated to inhibit airway eosinophilia and hyperresponsiveness as well as inhibit eosinophil mediator release [38-39]. This finding, together with our observed downregulation of several pro-apoptotic genes, supports a role for inhibition of apoptosis as one method by which β-chain receptor cytokines promote eosinophil’s role in asthma pathogenesis.
Most striking in our analysis was the gene expression profile produced by IL-3. Consistent with prior work, we found that IL-3 stimulation most uniquely influences eosinophil gene expression relative to the other β-chain receptor cytokines following prolonged stimulation. Using two independent techniques, we identified a cohort of 119 genes separating IL-3 based both on transcript expression levels and co-expression networks. The top genes in this cohort included a range of genes with previously described roles in eosinophils, with defined roles in other cell lines but with undescribed function in eosinophils, or with undescribed functions altogether. For instance, XBP1 has been demonstrated as essential for early eosinophil development but less is known about its role in eosinophils that have fully matured in the blood or airway [33]. QSOX1 has gained interest in cancer research as a disulfide bond catalyst with an atypical localization to the Golgi apparatus and extracellular space [30-31]. The importance of such an enzyme in eosinophils may relate to eosinophil disulfide-bond containing secretory proteins (e.g. eosinophil peroxidase and major basic protein) with cytotoxic and immunomodulatory roles in asthma [40-42]. Two transcription factors – IKZF4 and BACH2 – have been connected to regulatory T cell stability, with the later only indirectly linked to eosinophils in the current literature [27-29, 43]. The interaction between CD69 and its ligands (myosin light chains 9 and 12 - the former also a member of our IL-3-defining gene cohort) has been shown to be crucial in allergic airway inflammation and the recruitment of activated T cells to sites of inflammation [44]. Lastly, upregulation of the common β-chain receptor subunit (CSF2RB) may provide a contributing mechanism by which IL-3 uniquely prolongs intracellular signaling in eosinophils [6, 45].
We acknowledge that these 119 genes identified by cytokine stimulation of peripheral blood eosinophils in non-asthmatic subjects may not reflect the true biology of airway eosinophils in asthma patients. In different tissue compartments, eosinophils exist in subsets, each displaying distinct phenotypes and gene expression patterns [46]. While the role of IL-3 stimulation of eosinophils in the airway remains an area of active investigation, there is reason to believe its role is important, particularly in patients who do not respond to anti-IL-5 therapy. Not only does IL-3 increase in the airway of asthmatic subjects following allergen challenge, eosinophils also display increased levels of surface IL-3 receptor following mepolizumab therapy [5,47]. Esnault et al previously published results of microarray analyses on human BAL and sputum cells following allergen challenge to the lung. Additionally, they assessed for gene downregulation following mepolizumab therapy. Among others, they discovered upregulation in genes that are also members of our IL-3 defining cohort. CD69, RHOH, and ST6GAL1 were upregulated in both BAL and sputum eosinophils subjected to allergen challenge, IL2RA, NDFIP2, PIM2, SLC2A1, SOCS1 and SVIP were upregulated in the BAL samples, and CYTIP, IL1RAP, MAST4, OSBPL3, OSM and SOCS2 were upregulated in the sputum samples. Of these genes, CD69, NDFIP2 and SOCS1 were the only to decrease following mepolizumab treatment [48].
We believe the value of our study lies in its genome-wide and unbiased approach. Such methodology allowed us to identify protein-coding genes, some previously associated with asthma, and make novel connections with either IL-3 or the eosinophil cell type. Furthermore, we linked the expression of a few non-coding RNAs to β-chain receptor cytokine stimuli and foresee potential implications as their gene regulatory and epigenetic roles become further defined. These coding and non-coding genes may provide the foundation for future investigation on therapeutic targets in eosinophilic asthma, and with the recognized increase in IL-3-Rα expression in airway eosinophils, may address the current shortcomings of anti-IL-5 therapy. Limitations of our study include the snapshot picture that does not account for the presence of multiple cytokines simultaneously, at varying concentrations over time. Our study of peripheral blood eosinophils from healthy subjects may additionally not account for the effects of the local tissue environment or of the host. Finally, we are limited by the lack of post-transcript analysis, as prior work has described IL-3 dependent translational modifications that would be missed in the present work [49].
In summary, these results highlight the redundant cytokine signaling mechanisms involved in eosinophil development and function but support a unique role for IL-3. The identification of several genes strongly correlated with IL-3 may provide the foundation for future therapeutic advancements in eosinophilic asthma.
Supplementary Material
Key Points.
Beta-chain receptor cytokines produce a pro-inflammatory and survival phenotype
IL-3 stimulation yields a distinct eosinophil gene expression program
IL-3-upregualted “hub genes” may have implications in future asthma therapeutics
Acknowledgements
We thank the eosinophil donors for their contributions to this project.
Grant Support: This work was supported by National Institutes of Health Grants NIH K08HL116429 (P.A.) and NIH S10OD016262 (G.S.).
Abbreviations:
- TPM
transcripts per million
- WGCNA
weighted gene co-expression network analysis
Footnotes
Conflict of Interest: The authors whose names are listed certify that they have no financial or non-financial interests in the subject matter or materials discussed in this manuscript.
References
- 1.Nurmagambetov T, Kuwahara R, and Garbe P. 2018. The economic burden of asthma in the United States, 2008-2013. Ann. Am. Thorac. Soc 15: 348–356. [DOI] [PubMed] [Google Scholar]
- 2.Bel EH, Wenzel SE, Thompson PJ, Prazma CM, Keene ON, Yancey SW, Ortega HG, and Pavord ID; SIRIUS Investigators. 2014. Oral glucocorticoid-sparing effect of mepolizumab in eosinophilic asthma. N. Engl. J. Med 371: 1189–1197. [DOI] [PubMed] [Google Scholar]
- 3.Ortega HG, Liu MC, Pavord ID, Brusselle GG, FitzGerald JM, Chetta A, Humbert M, Katz LE, Keene ON, Yancey SW, and Chanez P; MENSA Investigators. 2014. Mepolizumab treatment in patients with severe eosinophilic asthma. N. Engl. J. Med 371: 1198–1207. [DOI] [PubMed] [Google Scholar]
- 4.Flood-Page PT, Menzies-Gow AN, Kay AB, and Robinson DS. 2003. Eosinophil’s role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am. J. Respir. Crit. Care Med 167: 199–204. [DOI] [PubMed] [Google Scholar]
- 5.Kelly EA, Esnault S, Liu LY, Evans MD, Johansson MW, Mathur S, Mosher DF, Denlinger LC, and Jarjour NN. 2017. Mepolizumab attenuates airway eosinophil numbers, but not their functional phenotype, in asthma. Am. J. Respir. Crit. Care Med 196: 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esnault S, and Kelly EA. 2016. Essential mechanisms of differential activation of eosinophils by IL-3 compared to GM-CSF and IL-5. Crit. Rev. Immunol 36: 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asquith KL, Ramshaw HS, Hansbro PM, Beagley KW, Lopez AF, and Foster PS. 2008. The IL-3/IL-5/GM-CSF common receptor plays a pivotal role in the regulation of Th2 immunity and allergic airway inflammation. J. Immunol 180: 1199–1206. [DOI] [PubMed] [Google Scholar]
- 8.Geijsen N, Uings IJ, Pals C, Armstrong J, McKinnon M, Raaijmakers JA, Lammers JW, Koenderman L, and Coffer PJ. 2001. Cytokine-specific transcriptional regulation through an IL-5Ralpha interacting protein. Science. 293: 1136–1138. [DOI] [PubMed] [Google Scholar]
- 9.Liu LY, Sedgwick JB, Bates ME, Vrtis RF, Gern JE, Kita H, Jarjour NN, Busse WW, and Kelly EA. 2002. Decreased expression of membrane IL-5 receptor alpha on human eosinophils: I. Loss of membrane IL-5 receptor alpha on airway eosinophils and increased soluble IL-5 receptor alpha in the airway after allergen challenge. J. Immunol 169: 6452–6458. [DOI] [PubMed] [Google Scholar]
- 10.Akuthota P, Capron K, and Weller PF. 2014. Eosinophil purification from peripheral blood. Methods Mol. Biol 1178: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seumois G, Vijayanand P, Eisley CJ, Omran N, Kalinke L, North M, Ganesan AP, Simpson LJ, Hunkapiller N, Moltzahn F, Woodruff PG, Fahy JV, Erle DJ, Djukanovic R, Blelloch R, and Ansel KM. 2012. An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol 1: 70–89. [PMC free article] [PubMed] [Google Scholar]
- 12.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, Vedanayagam M, Ganesan AP, Chawla A, Djukanović R, Ansel KM, Peters B, Rao A, and Vijayanand P. 2014. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol 15: 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosales SL, Liang S, Engel I, Schmiedel BJ, Kronenberg M, Vijayanand P, and Seumois G. 2018. A sensitive and integrated approach to profile messenger RNA from samples with low cell numbers. Methods Mol. Biol 1799: 275–301. [DOI] [PubMed] [Google Scholar]
- 14.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9: 171–181. [DOI] [PubMed] [Google Scholar]
- 15.Parekh S, Ziegenhain C, Vieth B, Enard W, and Hellmann I. 2016. The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep 6: 25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Wu PH, Beane T, Zamore PD, and Weng Z. 2018. Elimination of PCR duplicates in RNA-seq and small RNA-seq using unique molecular identifiers. BMC Genomics. 19: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer A, Green J, Pollard J Jr., and Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langfelder P, and Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Z, ŁP., Li S, Thierry-Mieg J, Thierry-Mieg D, Shi W, Wang C, Schroth GP, Setterquist RA, Thompson JF, Jones WD, Xiao W, Xu W, Jensen RV, Kelly R, Xu J, Conesa A, Furlanello C, Gao H, Hong H, Jafari N, Letovsky S, Liao Y, Lu F, Oakeley EJ, Peng Z, Praul CA, Santoyo-Lopez J, Scherer A, Shi T, Smyth GK, Staedtler F, Sykacek P, Tan XX, Thompson EA, Vandesompele J, Wang MD, Wang J, Wolfinger RD, Zavadil J, Auerbach SS, Bao W, Binder H, Blomquist T, Brilliant MH, Bushel PR, Cai W, Catalano JG, Chang CW, Chen T, Chen G, Chen R, Chierici M, Chu TM, Clevert DA, Deng Y, Derti A, Devanarayan V, Dong Z, Dopazo J, Du T, Fang H, Fang Y, Fasold M, Fernandez A, Fischer M, Furió-Tari P, Fuscoe JC, Caimet F, Gaj S, Gandara J, Gao H, Ge W, Gondo Y, Gong B, Gong M, Gong Z, Green B, Guo C, Guo L, Guo LW, Hadfield J, Hellemans J, Hochreiter S, Jia M, Jian M, Johnson CD, Kay S, Kleinjans J, Lababidi S, Levy S, Li QZ, Li L, Li L, Li P, Li Y, Li H, Li J, Li S, Lin SM, López FJ, Lu X, Luo H, Ma X, Meehan J, Megherbi DB, Mei N, Mu B, Ning B, Pandey A, Pérez-Florido J, Perkins RG, Peters R, Phan JH, Pirooznia M, Qian F, Qing T, Rainbow L, Rocca-Serra P, Sambourg L, Sansone SA, Schwartz S, Shah R, Shen J, Smith TM, Stegle O, Stralis-Pavese N, Stupka E, Suzuki Y, Szkotnicki LT, Tinning M, Tu B, van Delft J, Vela-Boza A, Venturini E, Walker SJ, Wan L, Wang W, Wang J, Wang J, Wieben ED, Willey JC, Wu PY, Xuan J, Yang Y, Ye Z, Yin Y, Yu Y, Yuan YC, Zhang J, Zhang KK, Zhang W, Zhang W, Zhang Y, Zhao C, Zheng Y, Zhou Y, Zumbo P, Tong W, Kreil DP, Mason CE, and Shi L. 2014. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 32: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, and Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl. Genet. Mol. Biol 4: Article17. [DOI] [PubMed] [Google Scholar]
- 23.Bastian M, Heymann S, and Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media. [Google Scholar]
- 24.Gupta SC, Sundaram C, Reuter S, and Aggarwal BB. 2010. Inhibiting NF-ΚB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta 1799: 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkly LC, Michaelson JS, Hahm K, Jakubowski A, and Zheng TS. 2007. TWEAKing tissue remodeling by a multifunctional cytokine: role of TWEAK/Fn14 pathway in health and disease. Cytokine. 40: 1–16. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee M, Raju R, Radhakrishnan A, Nanjappa V, Muthusamy B, Singh K, Kuppusamy D, Lingala BT, Pan A, Mathur PP, Harsha HC, Prasad TS, Atkins GJ, Pandey A, and Chatterjee A. 2012. A bioinformatics resource for TWEAK-Fn14 signaling pathway. J. Signal Transduct. 2012: 376470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, Drake CG, Liu JO, Ostrowski MC, and Pardoll DM. 2009. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 325: 1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, Sciumè G, Zare H, Vahedi G, Dema B, Yu Z, Liu H, Takahashi H, Rao M, Muranski P, Crompton JG, Punkosdy G, Bedognetti D, Wang E, Hoffmann V, Rivera J, Marincola FM, Nakamura A, Sartorelli V, Kanno Y, Gattinoni L, Muto A, Igarashi K, O’Shea JJ, and Restifo NP. 2013. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 498: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Lao Y, Teng XL, Li S, Zhou Y, Wang F, Guo X, Deng S, Chang Y, Wu X, Liu Z, Chen L, Lu LM, Cheng J, Li B, Su B, Jiang J, Li HB, Huang C, Yi J, and Zou Q. 2018. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat. Commun 9: 3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alon A, Grossman I, Gat Y, Kodali VK, DiMaio F, Mehlman T, Haran G, Baker D, Thorpe C, and Fass D. 2012. The dynamic disulfide relay of quiescin sulphydryl oxidase. Nature. 488: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilani T, Alon A, Grossman I, Horowitz B, Kartvelishvily E, Cohen SR, and Fass D. 2013. A secreted disulfide catalyst controls extracellular matrix composition and function. Science. 341: 74–76. [DOI] [PubMed] [Google Scholar]
- 32.Wong YC, Ysselstein D, and Krainc D. 2018. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 554: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bettigole SE, Lis R, Adoro S, Lee AH, Spencer LA, Weller PF, and Glimcher LH. 2015. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat. Immunol 16: 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidler D, Wu P, Herro R, Claus M, Wolf D, Kawakami Y, Kawakami T, Burkly L, and Croft M. 2017. TWEAK mediates inflammation in experimental atopic dermatitis and psoriasis. Nat. Commun 8: 15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu C, Zhang L, Liu Z, Li C, and Bai Y. 2018. TWEAK/Fn14 interaction induces proliferation and migration in human airway smooth muscle cells via activating the NF-ΚB pathway. J. Cell Biochem 119: 3528–3536. [DOI] [PubMed] [Google Scholar]
- 36.Roscioli E, Hamon R, Ruffin RE, Grant J, Hodge S, Zalewski P, and Lester S. 2017. BIRC3 single nucleotide polymorphism associate with asthma susceptibility and the abundance of eosinophils and neutrophils. J. Asthma 54: 116–124. [DOI] [PubMed] [Google Scholar]
- 37.Gao XM 2016. A network approach predicts NFKBIA and BIRC3 as pathogenic genes in childhood asthma. Genet. Mol. Res 15. [DOI] [PubMed] [Google Scholar]
- 38.Kikkawa Y, Sugiyama K, Obara K, Hirata H, Fukushima Y, Toda M, and Fukuda T. 2012. Interferon-alpha inhibits airway eosinophilia and hyperresponsiveness in an animal asthma model. Asia Pac. Allergy 2: 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aldebert D, Lamkhioued B, Desaint C, Gounni AS, Goldman M, Capron A, Prin L, and Capron M. 1996. Eosinophils express a functional receptor for interferon alpha: inhibitory role of interferon alpha on the release of mediators. Blood. 87: 2354–2360. [PubMed] [Google Scholar]
- 40.Wagner LA, Ohnuki LE, Parsawar K, Gleich GJ, and Nelson CC. 2007. Human eosinophil major basic protein 2: location of disulfide bonds and free sulfhydryl groups. Protein J. 26: 13–18. [DOI] [PubMed] [Google Scholar]
- 41.Thomsen AR, Sottrup-Jensen L, Gleich GJ, and Oxvig C. 2000. The status of half-cystine residues and locations on N-glycosylated asparagine residues in human eosinophil peroxidase. Arch. Biochem. Biophys 379: 147–152. [DOI] [PubMed] [Google Scholar]
- 42.McBrien CN, and Menzies-Gow A. 2017. The biology of eosinophils and their role in asthma. Front. Med 4: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato Y, Kato H, Ebina-Shibuya R, Itoh-Nakadai A, Okuyama R, and Igarashi K. 2017. Bach2 controls homeostasis of eosinophils by restricting the type-2 helper function of T cells. Tohoku J. Exp. Med 241: 175–182. [DOI] [PubMed] [Google Scholar]
- 44.Hayashizaki K, Kimura MY, Tokoyoda K, Hosokawa H, Shinoda K, Hirahara K, Ichikawa T, Onodera A, Hanazawa A, Iwamura C, Kakuta J, Muramoto K, Motohashi S, Tumes DJ, Iinuma T, Yamamoto H, Ikehara Y, Okamoto Y, and Nakayama T. 2016. Myosin light chains 9 and 12 are functional ligands for CD69 that regulate airway inflammation. Sci. Immunol 1 :1–10. [DOI] [PubMed] [Google Scholar]
- 45.Esnault S, Hebert AS, Jarjour NN, Coon JJ, and Mosher DF. 2018. Proteomic and phospho-proteomic changes induced by prolonged activation of human eosinophils with IL-3. J. Proteome Res 17: 2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, Janss T, Starkl P, Ramery E, Henket M, Schleich FN, Radermecker M, Thielemans K, Gillet L, Thiry M, Belvisi MG, Louis R, Desmet C, Marichal T, and Bureau F. 2016. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Invest 126: 3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson MW, Kelly EA, Busse WW, Jarjour NN, and Mosher DF. 2008. Up-regulation and activation of eosinophil integrins in blood and airway after segmental lung antigen challenge. J. Immunol 180: 7622–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esnault S, Kelly EA, Schwantes EA, Liu LY, DeLain LP, Hauer JA, Bochkov YA, Denlinger LC, Malter JS, Mathur SK, and Jarjour NN. 2013. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One. 8: e67560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esnault S, Kelly EA, Johansson MW, Liu LY, Han S-H, Akhtar M, Sandbo N, Mosher DF, Denlinger LC, Mathur SK, Malter JS, and Jarjour NN. 2014. Semaphorin 7A is expressed on airway eosinophils and upregulated by IL-5 family cytokines. Clin. Immunol 150: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.