Abstract
We have reported child anxiety sensitivity (CASI) predicts chronic post-surgical pain (CPSP). Here, we evaluated DNA methylation profiles to understand gene-environmental interactions underlying CPSP and CASI, in order to identify shared, enriched, genomic pathways. In 73 prospectively recruited adolescents undergoing spine fusion, preoperative CASI, and pain data over 12 months post-surgery were collected. DNA from peripheral blood of evaluable subjects with (n=16) and without CPSP (n=40) were analyzed using MethylationEPIC arrays. We identified 637 and 2,445 differentially methylated positions (DMPs) associated with CPSP and CASI respectively (p≤0.05). Ingenuity pathway analysis of 39 genes with DMPs for both CPSP and CASI revealed enrichment of several canonical pathways, including GABA receptor (p=0.00016 (CPSP); 0.0008 (CASI)) and Dopamine-DARPP32 Feedback in cAMP (p=0.004 (CPSP) and 0.00003 (CASI)) Signaling. Gene–gene interaction network enrichment analysis revealed participation of pathways in cell signaling, molecular transport, metabolism and neurological diseases (p-value <10–8). Bioinformatic approaches to identify histone marks and transcription factor (TF) binding events underlying DMPs, showed their location in active regulatory regions in pain pathway relevant brain cells. Using Enrichr/Pinet enrichment and Library of Integrated Network-Based Cellular Signatures (LINCS) knockdown signatures, we identified TFs regulating genes with DMPs in association with CPSP and CASI. In conclusion, we identified epigenetically enriched pathways associated with CPSP and anxiety sensitivity in children undergoing surgery. Our findings support GABA hypofunction and Dopamine-DARPP32 pathway’s roles in emotion/reward and pain. This pilot study provides new epigenetic insights into the pathophysiology of CPSP, and a basis for future studies in biomarker development and targetable interventions.
Perspective: Differential DNA methylation in regulatory genomic regions enriching shared neural pathways were associated with chronic post-surgical pain and anxiety sensitivity in adolescents undergoing spine surgery. Our findings support GABA hypofunction and Dopamine-DARPP32 pathway’s roles in emotion/reward contributing to behavioral maintenance of pain 10–12 months after surgery.
Keywords: Bioinformatics, epigenetics, anxiety, chronic postsurgical pain, DNA methylation, functional genomics
Introduction
Chronic postsurgical pain (CPSP) is often defined as pain that lasts beyond 3–6 months post-surgery, in the absence of other preexisting problems or postoperative complications.[46; 77] In children, the median prevalence of CPSP is 20%,[57] however the incidence ranges from 11–54% after spine fusion,[9; 40; 66] a painful surgery that adolescents undergo. CPSP involves multiple peripheral and central signaling and modulatory pathways regulated by genes.[32] While chronic pain conditions have a heritable risk of 45%,[78] and genetic factors explain some of the individual differences in pain perception,[1; 53] a genetic basis for CPSP has been elusive,[35] attributed partly to lack of replicability[38] and inconsistent findings[61] in genetic association studies,[4; 74] and lack of consideration of gene-environmental interactions. In addition, especially in children, caregiving environment and psychological factors like anxiety, prime children’s pain responses influence his or her response to further surgical stress.[15; 30] Twin studies have shown that environmental factors are involved in the inter-personal differences in pain sensitivity.[1] Since epigenetic mechanisms such as DNA methylation (addition of a methyl group to the 5’ position of a cytosine - guanine residue (CpG dinucleotide))[70] are known to mediate the influence of environmental factors on genetic expression,[60] and have been known to influence pain processing and the transition of acute to chronic pain, [6] it leads us to hypothesize that elucidating gene-environmental influences through epigenetics will explain critical gaps in predisposition and mechanisms involved in CPSP.[13; 17]
We recently identified psychological and perioperative factors associated with CPSP in adolescents undergoing spine fusion surgery. Among psychosocial factors studied, we reported that child anxiety sensitivity index (CASI) was significantly associated with CPSP in the spine cohort. µ-In addition, we identified opioid receptor gene (OPRM1) DNA methylation markers as predictors of acute and chronic postsurgical pain. However, effect sizes of single CpG sites are small, and sometimes identify associations that cannot be replicated. Hence, in this pilot study, we compare DNA methylation profiles and use a global bioinformatics-based approach to identify pathways, gene-gene interactions, histone marks, and protein-DNA binding events enriched in DNA methylation differences associated with CPSP and CASI. This approach integrates epigenetic-level data with biologic processes, pathways, and networks, and overcomes pitfalls of hypothesis-driven candidate marker association studies, which overlook unknown possible causal variants.[80] Such approaches have been used previously to study epigenetics of chronic pain conditions (such as fibromyalgia)[11] and psychological conditions (such as panic disorder),[65] but not CPSP or anxiety. We will test the hypothesis that biological processes enriched in gene sets with differentially DNA methylated positions (DMPs) will be shared by CPSP and anxiety, thus suggesting new avenues for preventing and treating CPSP.
Methods
An observational prospective cohort study was conducted in 73 adolescents with idiopathic scoliosis undergoing posterior spine fusion. The surgical, anesthetic and pain plans were standardized (see Supplement). The studies are registered with ClinicalTrials.gov (Identifier: NCT01839461, NCT01731873), as part of a larger pharmacogenomics study. The study was approved by the institutional review board. Written informed consent was obtained from parents and assent was obtained from children before enrollment.
Participants:
Healthy non-obese children aged 10–18 years were recruited for the study if they fulfilled following criteria: American Society of Anesthesiologists (ASA) physical status less than or equal to two (mild systemic disease), a diagnosis of idiopathic scoliosis and/or kyphosis, scheduled to undergo elective spinal fusion. We excluded females who were pregnant or breastfeeding, diagnosis of chronic pain or opioid use in the past six months, had hepatic or renal disease or developmental delays.
Data Collection
Prior to surgery, following data were collected: demographics (sex, age, race), weight, pain scores (numerical rating scale/0–10 NRS)[72] and home medications. We assessed anxiety in both child and a parent using the 0–10 visual analog scale (VAS), a simple scale validated for this use.[5] Questionnaires to assess pain catastrophizing and anxiety sensitivity were administered (Table 1). Surgical and anesthetic data collected included propofol and remifentanil doses, duration of surgery, and number of vertebral levels fused. On postoperative days (POD) one and two, we recorded pain scores (every four hours), and doses of morphine equivalents and diazepam administered. After hospital discharge, research coordinators administered questionnaires over phone in a standard fashion, at 10–12 months after surgery (Table 1) to obtain pain measures.
Table 1:
Data collection schema
| Data variables | Pre-operative | Intra-operative | Over 48 hours after surgery | 10–12 months after surgery |
|---|---|---|---|---|
| Demographics VAS Anxiety scores (parent and child) Pain score (child) | x | |||
| Surgical duration Vertebral levels fused Propofol dose Remifentanil dose | x | |||
| Pain assessment Opioid consumption Diazepam use Analgesic adjuncts | x | x | x | |
| Child Questionnaires CASI | x | |||
| Parent Questionnaires PCS-P | x |
time calculated from end of surgery
Abbreviations: VAS: Visual analog scale; CASI = Childhood Anxiety Sensitivity Index; PCS-P = Pain Catastrophizing Scale (Parent version)
Outcomes
Outcomes evaluated were a) CPSP, defined as NRS>3/10 at 10–12 months post-surgery.[47; 77] NRS cut-offs of 3/10 were used because they depict moderate/severe pain, are associated with functional disability, and have been described as a predictor for persistence of pain.[24] b) Child Anxiety Sensitivity Index (CASI), an 18-item self-report validated measure of symptoms of anxiety in children and adolescents (range 18–54), was chosen as the anxiety measure, because CASI has previously been shown to be strongly correlated with state and trait anxiety.[58] It measures how anxiety-related symptoms are interpreted as being physically, psychologically or socially harmful. [67] The CASI has been shown to have good test-retest reliability, and has been validated with high internal consistency in clinical and nonclinical pediatric samples (aged 8– 15.8 years).[67] Our own studies have shown that the odds of pain persistence at 1 year after spine surgery was 1.24 times higher per unit increase in CASI score (95% CI 1.09–1.42, p=0.002),[10] and is supported by studies in other pediatric cohorts.[54] Higher anxiety sensitivity is associated with fear of pain, avoidance behavior and maladaptive coping styles, leading to increased pain persistence and disability.
Measurement of DNA methylation
Blood samples were collected before surgery in EDTA. Genomic DNA was isolated and frozen at −20 °C. To study DNA methylation, 500ng of genomic DNA (measured by Thermo Scientific NanoDrop spectrophotometer), with quality controls maintaining a purity of 260/280 ratio from 1.6 – 2.0, was extracted, and treated with bisulfite using Zymo EZ DNA Methylation Gold kit (Zymo Research, Orange, CA, USA), according to manufacturer’s instructions. Bisulfite-converted samples were hybridized in the Human Infinium MethylationEPIC BeadChip microarrays (©Illumina Inc., San Diego, CA). This array provides unparalleled coverage of CpG islands, genes, and enhancers.
Data analysis
The clinical characteristics and demographics of the cohort were described using mean (with standard deviation), median (IQR) and frequency (percentage) depending on data distribution. Prior to the DNA methylation analysis, the quality of the methylation arrays was assessed using sample-independent and dependent internal control probes included on the array for staining, extension, hybridization, specificity and bisulfite conversion. The number of probes with detection P value ≤0.05 was examined for each sample. Only samples that passed the quality control with >95% probes detected were included in the analysis. If CpG sites were not detected in all samples at p=0.01 level, or if they were located on the X and Y chromosomes, they were excluded. Of the 73 samples, one was excluded from analysis as it did not pass all quality control steps. The remaining samples all had more than 99% of the probes detected. The signal intensities were background-adjusted using out-of-band probes (noob) and normalized using subset-quantile within array normalization (swan in R 3.4.4’minfi_1.22.1’). [2] Beta values, calculated as , and M values, the logit transformation of the beta values,[19] were used. Surrogate variable analysis (SVA) was used to control batch effect and unknown confounders such as cell composition. The method “irw” in SVA_3.24.4 was used[42].[41] Age and race were included in the full model when generating the SVs. Models on beta and M values with different clinical measures generated different numbers of SVs, ranging from 6 to 7. To test whether the surrogate variables are associated with the outcome (CPSP and CASI), we performed Pearson and Spearman correlation.
For each of the CpG sites, the association of DNAm with CPSP and CASI was tested with linear regression. The linear models were adjusted to include age, sex, race and significant surrogate variables. CpG sites whose DNAm (both beta and M values) were associated with CPSP or CASI at p≤0.05 level were selected for further evaluation. The selected DMPs should also have differences ≥ 0.05 in beta between CPSP yes and no groups. (Figure 1) As impact of non-genetic covariates were previously found on CPSP and CASI,[9] to ensure the robustness of the association identified from the above analyses in which the DNAm was used as the dependent variable, we conducted logistic and linear regression for CPSP and CASI, respectively, in which CPSP and CASI were used as dependent variables and beta value as primary independent variable. Models were adjusted to include non-genetic co-variables. Significant non-genetic co-variables were identified by univariate analysis for CPSP (factors tested: age, sex, race, morphine dose in mg/kg POD1 and 2, preoperative anxiety score (VAS) for child and parent, duration of surgery, vertebral levels fused, PCS-P and CASI) and CASI (factors tested: age, sex, race, PCS-P, diazepam doses and parent anxiety score), and selection of co-variables associated at p < 0.10. Analyses were performed using Statistical Analysis System (SAS), version 9.4 (SAS Institute Inc., Cary, NC) and R 3.4.4. Only CpG sites showing significant association with beta values in these models (p<0.05) were extracted from Methylation EPIC array annotation files and imported into Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA) for pathway mapping, gene network detection, and upstream regulator identification. Gene networks mapped by overlapping genes (with p-score>8; p-score = −log10(p-value)) and hence, p-values < 10−8 were identified and constructed in IPA.
Figure 1:
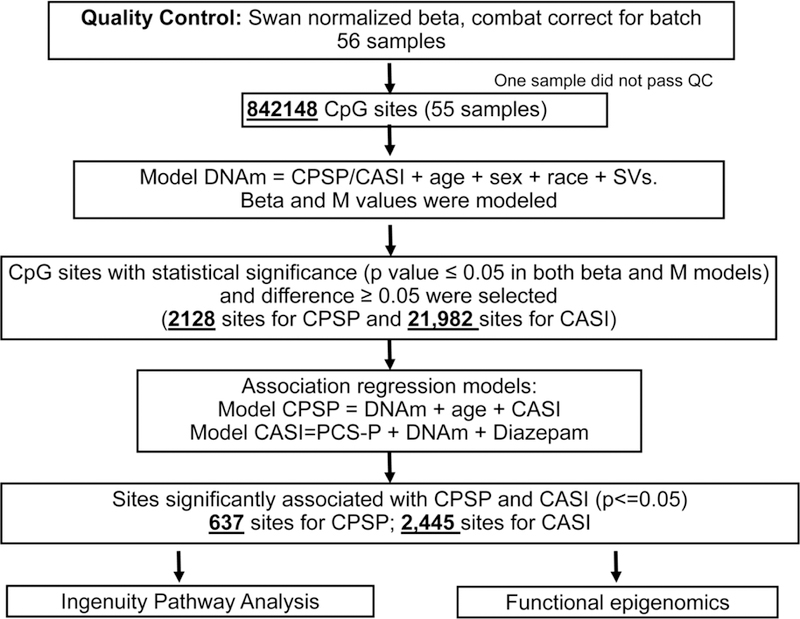
Workflow for the statistical analysis of MethylationEPIC array data for chronic postsurgical pain (CPSP) and anxiety sensitivity (CASI) outcomes. One sample did not pass quality control. DNA methylation beta and M values were modeled and CpG sites with methylation satisfying certain criteria were included in logistic (for CPSP) and linear (CASI) regression models adjusted for other covariates. Methylation at sites significantly associated with outcomes were then included in the pathway and functional analyses.
To identify potential regulatory mechanisms altered by CpG methylation differences, we evaluated CpG sites that were significant in the previous step (p<0.05) against a control set of CpG sites (p>0.4) using a compiled large collection of functional genomics datasets from various sources, including ENCODE[12], Roadmap Epigenomics[3], Cistrome[43], and ReMap-ChIP[26]. In total, this database contains 4,045 datasets performed in 1,069 different cell types and conditions. 1,544 datasets monitor binding interactions of proteins such as transcription factors with the human genome using ChIP-seq; 1,213 measure the presence of histone marks using ChIP-seq; 277 measure open chromatin through DNase-seq; 55 measure expression quantitative loci (eQTLs); and 558 predict “ActiveChromatin” states using combinations of histone marks[20]. Overall, 240 experiments were performed in cell lines and cell types related to brain regions relevant to pain/anxiety outcomes.
We next used the RELI algorithm to estimate the statistical enrichment of histone marks and protein binding events at the genomic loci displaying altered DNA methylation.[28] As input, the method took a set of genomic loci (in this case, regions with differential methylation marks). The coordinates of each locus were padded by 100 bases in either direction (to account for experimental resolution). The resulting loci were then systematically intersected with the ChIP-seq and epigenetic data set libraries described above, and the number of input regions overlapping each dataset by at least one base was counted. Next, a P-value describing the significance of this overlap was estimated using a simulation-based procedure. To this end, the control set of CpG sites that do not change (p>0.4) was used as a negative, background set. A distribution of expected overlap values was then created from 2,000 iterations of randomly sampling from the negative set, each time choosing a set of negative examples that match the input set in terms of the total number of genomic loci and the length of each locus. The distribution of the expected overlap values from the randomized data resembles a normal distribution and can thus be used to generate a Z-score and corresponding P-value estimating the significance of the observed number of input regions that overlap each data set. Collectively, this procedure controlled for the count and sizes of the input loci, and the count and sizes of each individual dataset in the library. The final output of the method is a p-value based ranking of all the functional genomics datasets, in terms of their overlap with the input set.
With the goal of further elucidating pathways and potential regulatory mechanisms underlying the observed epigenetic changes, we performed enrichment analysis using a comprehensive, curated library of transcription factor targets that combines results from ENCODE and literature-based CHEA ChIP-seq experiments, available through Enrichr (http://amp.pharm.mssm.edu/Enrichr/). Next, we used the Library of Integrated Network-based Cellular Signatures (LINCS) of genetic perturbations (gene knockdowns of the 39 genes common to both outcomes with DNA methylation changes) and connectivity analysis, with the focus on kinase signaling pathways, available through Pinet (http://pinet-server.org) and Enrichr.[39] One of the goals of LINCS library is to enable analysis of connectivity between genetic [36] and chemical perturbations by measuring correlations between their transcriptional echo (correlation between landmark gene expression vectors). Here, we use signatures of genetic knock-downs of gene encoding protein kinases, which consist of genes whose mRNA expression is downregulated in response to the loss of function for each kinase.
Results
The mean age of participants was 14.4 years (SD 1.6); they were mostly white (82%) and female (84%). Demographics and description of variables evaluated, are presented in Table 2. Median preoperative pain score was 0.0 (IQR 0.0–1.0) and mean (SD) for AUC on POD1 and 2 was 202.6 (84.3). As expected, there was a significant difference in NRS pain scores at 10–12 months between the non-CPSP (0.0 (0.0–1.0)) and CPSP (5.0 (4.0–6.0)) groups (p<0.001). Of 73 subjects recruited, follow-up for CPSP outcomes was successful for 56 subjects. Incidence of CPSP in this cohort was 15/56 (29%).
Table 2:
Demographics and other variables considered in univariate analysis for outcomes
| All | Chronic Post-surgical pain (CPSP)* | CASI (N=56) | ||||
|---|---|---|---|---|---|---|
| N=73 | No (N=40) | Yes (N=15) | p value | dCorrelation coefficient | p value | |
| aAge (years) | 14.4 ± 1.6 | 14.3 ± 1.8 | 15.2 ± 1.3 | 0.07 | 0.01 | 0.96 |
| bSex (Male) | 9 (16%) | 8 (20%) | 1 (7%) | 0.42 | 0.06 | 0.68 |
| bRace (White) | 45 (82%) | 33 (83%) | 12 (80%) | 1.00 | 0.04 | 0.76 |
| cWeight (Kg) | 53.7 (50.4–58.0) | 53.5 (50.4–59.8) | 53.9 (51.0–58.0) | 0.84 | −0.19 | 0.25 |
| cPreoperative pain score | 0.0 (0.0–0.1) | 0.0 (0.0–0.0) | 1.0 (0.0–2.0) | 0.006 | - | - |
| cVAS Anxiety (Child) | 4.4 (3.0–6.9) | 4.4 (3.0–6.8) | 3.3 (3.0–8.5) | 0.75 | 0.26 | 0.14 |
| cVAS Anxiety (Parent) | 6.7 (4.7–8.1) | 5.4 (4.4–8.0) | 7.6 (5.0–8.8) | 0.37 | 0.25 | 0.15 |
| cNumber of vertebral levels fused | 12.0 (10.0–13.0) | 12.0 (11.0–13.0) | 12.0 (10.0–12.0) | 0.40 | −0.01 | 0.95 |
| aSurgical duration (hours) | 4.2 ± 1.1 | 4.3 ± 1.0 | 3.9 ± 1.4 | 0.38 | −0.12 | 0.47 |
| aPain AUC POD1&2 | 201.8 ± 85.8 | 181.0 ± 76.6 | 261.1 ± 85.4 | 0.002 | - | - |
| cMorphine dose POD1&2 mg/kg | 1.2 (0.9–1.7) | 1.2 (1.0–1.8) | 1.7 (0.9–2.9) | 0.13 | 0.15 | 0.27 |
| cCASI | 28.4 (24.0–32.5) | 27.0 (24.0–31.0) | 29.8 (26.0–34.3) | 0.09 | - | - |
| cPain scores at 6–12 months | 1.0 (0.0–4.0) | 0.0 (0.0–1.0) | 5.0 (4.0–6.0) | <0.001 | 0.15 | 0.38 |
| aPCS-P | 21.7 ± 11.6 | 20.4 ± 12.5 | 24.1 ± 9.7 | 0.48 | 0.40 | 0.015 |
| cDiazepam use mg/kg | 0.1 (0.1–0.2) | 0.1 (0.1–0.2) | 0.2 (0.1–0.2) | 0.12 | 0.26 | 0.09 |
Note:
data exhibited normal distribution; shown as mean ± SD and compared using t tests for PP.
shown as frequency (proportion) and compared using Fisher’s exact tests for PP.
data did not exhibit a normal distribution; shown as median (IQR) and compared using Wilcoxon rank sum tests for PP.
Spearman correlation coefficient.
Data from 55 subjects who had CPSP outcomes and evaluable Methylation EPIC array data are presented here
Abbreviations: VAS: Visual Analog Scale; POD: Postoperative Day; CASI: Childhood anxiety sensitivity index; PCS-P: Pain catastrophizing scale -parents; AUC: Area under curve; POD: postoperative day
Non-genetic covariates for DNAm-outcome analyses
At a significance threshold of p<0.1, univariate analyses identified age and CASI as significant determinants of CPSP (p= 0.070 and 0.090 respectively) (Table 2); and PCS-P (p=0.015) and diazepam dose (p=0.090) for CASI. Preoperative pain, AUC and pain at 10–12 months were all significantly higher in the CPSP group compared to the non-CPSP group (p=0.006, 0.002 and <0.001 respectively). Since preoperative pain, AUC and CPSP are correlated pain outcomes, with possible overlap of DNA methylation associations, we did not include them as co-variables in the multivariate DNAm model for CPSP.
DMPs associated with CPSP/CASI
Based on SVA analyses, no surrogate variables were associated with CPSP or CASI at nominal level (p<0.05). Based on the workflow for analyses of DMPs associated with CPSP/CASI, adjusted for non-genetic covariates (Figure 1), we identified 637 DMPs for CPSP and 2,445 DMPs for CASI. The distribution of differentially methylated regions in association with CPSP and CASI (p-value<0.05 and delta-beta>5) with regard to genomic location, are presented in Table 3. The entire list of CpG sites (DMPs) including location, effect size and p-values, selected for IPA analysis for CPSP and CASI shared pathway analyses, will be provided as supplementary viewing files.
Table 3:
Distribution of differentially methylated regions associated with chronic post-surgical pain (CPSP) and anxiety sensitivity (CASI) based on genomic location
| Genomic location | CPSP – differentially methylated sites | CASI – differentially methylated sites | ||
|---|---|---|---|---|
| Number | % | Number | % | |
| TSS1500 | 211 | 11.01 | 103 | 11.77 |
| TSS200 | 93 | 4.85 | 35 | 4.00 |
| 5’UTR | 40 | 2.09 | 64 | 7.31 |
| 1st exon | 17 | 0.89 | 2 | 0.23 |
| Gene body | 729 | 38.03 | 318 | 36.34 |
| 3’UTR | 3 | 0.16 | 22 | 2.51 |
| N_Shelf | 31 | 1.62 | 14 | 1.60 |
| N_Shore | 57 | 2.97 | 18 | 2.06 |
| S_Shelf | 24 | 1.25 | 12 | 1.37 |
| S_Shore | 42 | 2.19 | 19 | 2.17 |
| CpG island | 117 | 6.10 | 29 | 3.31 |
| Open sea | 553 | 28.85 | 239 | 27.31 |
CpG: 5’-C-phosphate-G-3’: cytosine and guanine separated by only 1 phosphate.
Pathway enrichment and gene network analyses
Annotation information on the DMPs was used for the analysis; in total, 310 genes (CPSP) and 1526 genes (CASI) were annotated to the DMPs. The genes associated with DMPs were over-represented in several canonical pathways (p-value<0.05) for CPSP and CASI. Pathways for CPSP included GABA receptor signaling, Protein kinase C signaling, dopamine receptor and cAMP mediated signaling among others. Pathways for CASI included beta-adrenergic, CDK5, protein kinase A, dopamine receptor and G-protein coupled receptor signaling among others.
Shared, enriched genomic pathways – CPSP and CASI
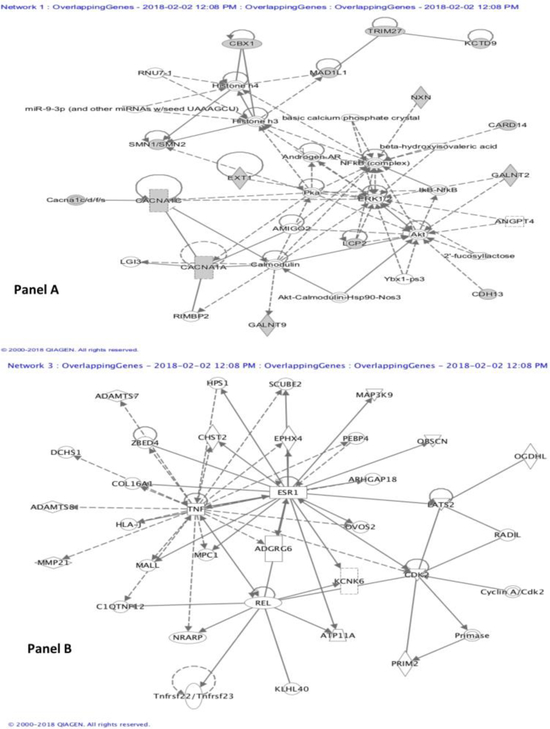
At gene level, 39 genes had DMPs associated with both outcomes, and they enriched 14 pathways (Table 4), among which the key ones were GABA receptor signaling and Dopamine-DARPP32 Feedback in cAMP Signaling. Gene network analyses of genes with DMPs common to CASI and CPSP revealed three networks associated with these phenotypes (p-value<10−8). Of the three, two play a role in cell signaling, molecular transport, cancer, vitamin and mineral metabolism (p-scores of 33 and 19), while the third is involved in neurological disease and connective tissue function (p-score of 12). Two representative networks are illustrated in Fig 2.
Table 4:
Shared overlapping pathways identified by gene overlap for CpG sites associated with both chronic post surgical pain and child anxiety sensitivity index.
| Shared Ingenuity Canonical Pathways for CPSP and CASI* | P-value CPSP | Ratio | Molecules | P-value CASI | Ratio |
|---|---|---|---|---|---|
| GABA Receptor Signaling | 0.0001 6 | 0.07 4 | ABAT,ADCY5,CACNA1H,CACNA1C,GABBR1, KCNH2, CACNA1A | 0.0008 | 0.15 8 |
| Dopamine-DARPP32 Feedback in Camp Signaling | 0.004 | 0.04 3 | PPP1R1B, ADCY5, PLCG2, CAMKK1, CACNA1C, DRD4, CACNA1A | 0.00003 | 0.15 2 |
| Cellular Effects of Sildenafil (Viagra) | 0.005 | 0.04 6 | ADCY5, PLCG2, CACNA1C, NPPA, KCNH2, CACNA1A | 0.0002 | 0.15 3 |
| GPCR-Mediated Nutrient Sensing in Enteroendocrine Cells | 0.012 | 0.04 5 | ADCY5,PLCG2,CACNA1H,CACNA1C,CACNA1A | 0.0002 | 0.16 1 |
| Calcium Signaling | 0.013 | 0.03 4 | CAMKK1, TRDN, CACNA1H, CACNA1C, CACNA1A, ATP2B2, CAMK2B | 0.0026 | 0.11 7 |
| nNOS Signaling in Skeletal Muscle Cells | 0.013 | 0.07 3 | CACNA1H, CACNA1C, CACNA1A | 0.0008 | 0.22 0 |
| Dopamine Receptor Signaling | 0.015 | 0.05 2 | PPP1R1B, ADCY5, DRD4, SLC18A2 | 0.00007 | 0.19 5 |
| Synaptic Long Term Depression | 0.020 | 0.03 5 | PLBD1,PLCG2,PLA2G4C,CACNA1H,CACNA1C, CACNA1A | 0.02570 | 0.10 3 |
| cAMP-mediated signaling | 0.022 | 0.03 1 | PDE9A, ADCY5, VIPR2, PTH1R, GABBR1, DRD4, CAMK2B | 0.00000 2 |
0.15 0 |
| Corticotropin Releasing Hormone Signaling | 0.028 | 0.03 6 | ADCY5,PLCG2,CACNA1H,CACNA1C,CACNA1A | 0.0064 | 0.12 2 |
| Netrin Signaling | 0.045 | 0.04 6 | CACNA1H, CACNA1C, CACNA1A | 0.0006 | 0.18 5 |
| CREB Signaling in Neurons | 0.046 | 0.02 8 | ADCY5,PLCG2,CACNA1H,CACNA1C,CACNA1A, CAMK2B | 0.0008 | 0.12 3 |
Two pathways not included in the table above include Gustatory pathway and sperm motility which are not relevant to the outcomes being studied
Figure 2:
Gene-gene interaction networks. Genes associated with the differentially methylated sites were uploaded to Ingenuity pathway analysis. Based on p-value cutoffs of 10−8, three networks were identified. Two of them were similar in function with several overlapping molecules. Hence, two of the different networks are presented here. The network in panel A is associated with Cell Signaling, Molecular Transport, Vitamin and Mineral Metabolism. It had a p-score of 33 and 14 focus molecules (including CACNA1A, CACNA1C, Calmodulin, ERK1/2, Histone h3, Histone h4, IkB-NfkB, NFkB (complex), miR-9–3p). The network in Panel B is associated with Neurological Disease, Organismal Injury and Abnormalities, Connective Tissue Development and Function; with a p-score of 12, and 6 focus molecules (including ESR1, KCNK6, PRIM2, TNF).
Functional genomics analyses
DMPs associated with CPSP are located in active regulatory regions with open chromatin marked by H3K27ac, H3K4me1 and H3K4me3 in brain cells from the hippocampus, frontal lobe, temporal lobe, anterior cingulate cortex, etc. (Table 5). Also depicted in Table 5 are the significant (p<0.05, after correction for multiple testing) protein (e.g., transcription factor) binding events identified to overlap significantly at the CpG sites, significant for CASI. Of note, many involve the RNA polymerase subunit POLR2A, suggesting that many differential methylation events might result in altered gene expression. The results of the Enrichr experiments are shown in Figure 3, where for each transcription factor, e.g., REST, its (here ENCODE mapped) targets among genes identified in our study are indicated by red squares (and include in this case RIMS2, CDH13, SPTBN4 etc.). Note that statistical significance of the enrichment is indicated by red vertical bars associated with each transcription factor. The results of the LINCS analysis are summarized in Table 6.
Table 5:
Overlap between CpG sites associated with chronic postsurgical pain (CPSP) and childhood anxiety sensitivity index (CASI) with functional genomics datasets in cells derived from brain tissue
| CpG sites associated with Chronic postsurgical pain (Histone markers) | ||||
|---|---|---|---|---|
| Dataset | Cell type | Epigenetic mark | Ratio | Corrected P-value |
| Roadmap Epigenomics (Histone narrow) | Brain (Hippocampus, Middle) | H3K27me3 | 0.226 | 1.37E-14 |
| Roadmap Epigenomics (Histone narrow) | Brain (Mid Frontal Lobe) | H3K27me3 | 0.190 | 3.56E-13 |
| Roadmap Epigenomics (Histone narrow) | Fetal (Brain, Male) | H3K27me3 | 0.187 | 1.96E-09 |
| Roadmap Epigenomics (Histone narrow) | Brain (Inferior Temporal Lobe) | H3K27me3 | 0.154 | 1.11E-07 |
| Roadmap Epigenomics (Histone narrow) | Brain (Cingulate Gyrus) | H3K27me3 | 0.146 | 5.68E-07 |
| Roadmap Epigenomics (Histone narrow) | Fetal (Brain, Male) | H3K4me1 | 0.349 | 6.03E-07 |
| Roadmap Epigenomics (Histone narrow) | Fetal (Brain, Female) | H3K27me3 | 0.185 | 8.03E-06 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Germinal Matrix) | Bivalent enhancer | 0.046 | 9.36E-05 |
| Roadmap Epigenomics (Histone narrow) | Brain (Substantia Nigra) | H3K27me3 | 0.119 | 0.00014 |
| Roadmap Epigenomics (Histone narrow) | Brain (Angular Gyrus) | H3K27me3 | 0.127 | 0.0015 |
| Roadmap Epigenomics (Active Chromatin) | Fetal (Brain, Male) | Bivalent enhancer | 0.096 | 0.0055 |
| Roadmap Epigenomics (Active Chromatin) | Fetal (Brain, Male) | Bivalent TSS | 0.019 | 0.0109 |
| eQTLs (GTEx V6) | Brain (Anterior cingulate cortex BA24) | eQTL | 0.025 | 0.015 |
| CpG sites associated with Childhood anxiety Sensitivity Index (Histone markers) | ||||
| Roadmap Epigenomics (Histone narrow) | Brain (Cingulate Gyrus) | H3K4me3 | 0.321 | 6.76E-10 |
| Roadmap Epigenomics (Histone narrow) | Brain (Mid Frontal Lobe) | H3K4me3 | 0.321 | 8.98E-09 |
| Roadmap Epigenomics (Dnase narrow) | H1 Derived Neuronal Progenitor Cells | Dnase | 0.381 | 1.37E-08 |
| Roadmap Epigenomics (Histone narrow) | Brain (Inferior Temporal Lobe) | H3K4me3 | 0.321 | 2.26E-07 |
| Roadmap E pigenomics (Histone narrow) | H9_Derived_Neuron_Cultured_Cells | H2A.Z | 0.251 | 4.26E-07 |
| Roadmap Epigenomics (Histone narrow) | H9 Derived Neuronal Progenitor Cells | H2A.Z | 0.334 | 5.73E-07 |
| Roadmap Epigenomics (Histone narrow) | Brain (Hippocampus Middle) | H3K4me3 | 0.333 | 7.51E-07 |
| Roadmap Epigenomics (Histone narrow) | Brain (Anterior Caudate) | H3K4me3 | 0.330 | 1.09E-06 |
| Roadmap Epigenomics (Histone narrow) | Brain (Angular Gyrus) | H3K4me3 | 0.299 | 1.49E-06 |
| Roadmap Epigenomics (Histone narrow) | Brain (Substantia Nigra) | H3K4me3 | 0.294 | 2.00E-06 |
| Roadmap Epigenomics (Histone narrow) | H9_Derived_Neuron_Cultured_Cells | H3K4me3 | 0.256 | 1.11E-05 |
| Roadmap Epigenomics (Histone narrow) | H9 Derived Neuronal Progenitor Cells | H3K4me3 | 0.250 | 3.48E-05 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Hippocampus Middle) | 2_TssAFlnk | 0.103 | 0.0001 |
| Roadmap Epigenomics (Histone narrow) | H1 Derived Neuronal Progenitor Cells | H3K4me2 | 0.283 | 0.0002 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Anterior Caudate) | 1_TssA | 0.232 | 0.0006 |
| DNaseI Duke | Cerebellum | Dnase | 0.314 | 0.0007 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Cingulate Gyrus) | ActiveChromatin | 0.381 | 0.0007 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Inferior Temporal Lobe) | ActiveChromatin | 0.383 | 0.0009 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Inferior Temporal Lobe) | 2_TssAFlnk | 0.091 | 0.0010 |
| Roadmap Epigenomics (Active Chromatin) | H9_Derived_Neuron | ActiveChromat in | 0.356 | 0.0014 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Substantia Nigra) | 1_TssA | 0.211 | 0.0019 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Anterior Caudate) | ActiveChromatin | 0.391 | 0.0022 |
| Roadmap Epigenomics (Active Chromatin) | H1_Derived_Neuronal_Progenitor | ActiveChromatin | 0.317 | 0.0024 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Angular Gyrus) | 1_TssA | 0.220 | 0.0028 |
| Roadmap Epigenomics (Histone narrow) | Brain (Substantia Nigra) | H3K9ac | 0.264 | 0.0052 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Hippocampus Middle) | 1_TssA | 0.201 | 0.0078 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Substantia Nigra) | ActiveChromatin | 0.363 | 0.0085 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Cingulate Gyrus) | 2_TssAFlnk | 0.088 | 0.0112 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Cingulate Gyrus) | 1_TssA | 0.203 | 0.0116 |
| Roadmap Epigenomics (Histone narrow) | Brain (Anterior Caudate) | H3K9ac | 0.302 | 0.0227 |
| DNaseI Duke | Frontal cortex | Dnase | 0.386 | 0.0232 |
| Roadmap Epigenomics (Histone narrow) | Brain (Mid Frontal Lobe) | H3K9ac | 0.267 | 0.0246 |
| Roadmap Epigenomics (Histone narrow) | Brain (Inferior Temporal Lobe) | H3K9ac | 0.310 | 0.0279 |
| Roadmap Epigenomics (Histone narrow) | Brain (Cingulate Gyrus) | H3K9ac | 0.294 | 0.0314 |
| Roadmap Epigenomics (Active Chromatin) | H9_Derived_Neuronal_Progenitor | ActiveChromatin | 0.340 | 0.0329 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Hippocampus Middle) | ActiveChromatin | 0.402 | 0.0395 |
| Roadmap Epigenomics (Active Chromatin) | Brain (Angular Gyrus) | 2_TssAFlnk | 0.069 | 0.0478 |
| CpG sites associated with Childhood anxiety Sensitivity Index (Transcription factors) | ||||
| Dataset | Cell type | Protein | Ratio | Corrected P-value |
| ENCODE | HepG2+forskolin | POLR2A | 0.175 | 0.00032 |
| ENCODE | MCF10A-Er-Src+4OHTAM_1uM_36hr | POLR2A | 0.169 | 0.00053 |
| ENCODE | MCF10A-Er-Src+EtOH_0.01pct | POLR2A | 0.171 | 0.00058 |
| Cistrome | HEK293 | BRD2 | 0.209 | 0.0012 |
| ENCODE | MCF-7 | POLR2A | 0.155 | 0.0013 |
| ENCODE | MCF-7+serum_stimulated_media | POLR2A | 0.172 | 0.0017 |
| Cistrome | CUTLL1 | ETS1 | 0.169 | 0.0028 |
| ENCODE | ProgFib | POLR2A | 0.160 | 0.0044 |
| ENCODE | HCT-116 | POLR2A | 0.190 | 0.0047 |
| ENCODE | HeLa-S3 | POLR2A | 0.186 | 0.0068 |
| ENCODE | Gliobla | POLR2A | 0.165 | 0.0078 |
| ENCODE | A549+DEX_100nM | POLR2A | 0.191 | 0.0078 |
| ENCODE | A549+EtOH_0.02pct | POLR2A | 0.191 | 0.0082 |
| ENCODE | ECC-1+DMSO_0.02pct | POLR2A | 0.174 | 0.0093 |
| ENCODE | MCF-7+serum_starved_media | POLR2A | 0.167 | 0.0093 |
| ENCODE | NB4 | POLR2A | 0.156 | 0.011 |
| ENCODE | H1-hESC | POLR2A | 0.173 | 0.013 |
| ReMap | hek293 | BRD3 | 0.077 | 0.014 |
| ENCODE | A549 | POLR2A | 0.169 | 0.014 |
| ENCODE | H1-hESC | RBBP5 | 0.112 | 0.014 |
| ENCODE | HUVEC | POLR2A | 0.198 | 0.016 |
| Misc (GEO) | LoVo | ASCL2 | 0.124 | 0.020 |
| ENCODE | SK-N-MC | POLR2A | 0.137 | 0.020 |
| ENCODE | GM19099 | POLR2A | 0.170 | 0.023 |
| ENCODE | GM15510 | POLR2A | 0.165 | 0.025 |
| ENCODE | MCF-7+serum_starved_media | CTCF | 0.113 | 0.026 |
| ENCODE | GM12878 | POLR2A | 0.188 | 0.034 |
| Misc (GEO) | LoVo | GMEB2 | 0.166 | 0.035 |
| Pazar | CD4+ | HMGN1 | 0.198 | 0.039 |
| Misc (GEO) | LoVo | MEF2C | 0.039 | 0.039 |
| Misc (GEO) | LoVo | RAD21 | 0.261 | 0.044 |
‘Ratio’ indicates the fraction of CPSP or CASI differentially methylated CpGs whose genomic coordinates intersect the indicated dataset. P-value is based on the significance of the ratio, and is adjusted for multiple testing, based on simulations (see Methods). No significant transcription factors in brain cells were identified for CpG sites associated with CPSP
Figure 3:
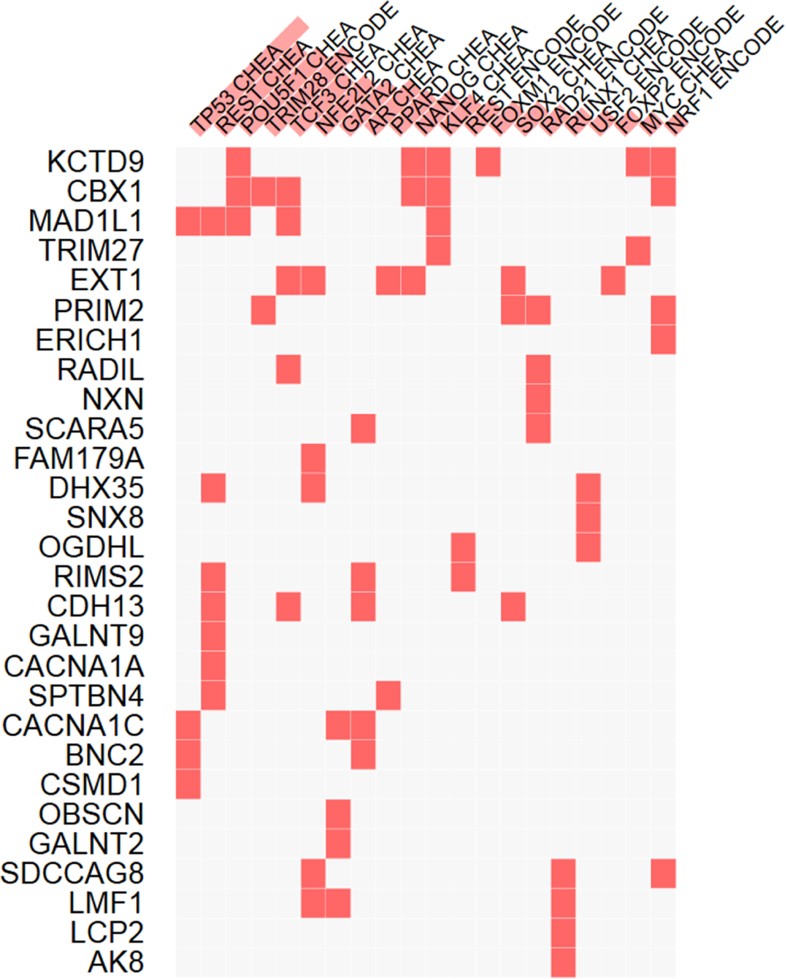
The results of enrichment analysis using Enrichr are shown here, where for each transcription factor, e.g., REST, its (here ENCODE mapped) targets among genes identified in our study are indicated by red squares (and include in this case RIMS2, CDH13, SPTBN4 etc.). Note that statistical significance of the enrichment is indicated by red vertical bars associated with each transcription factor.
Table 6:
Signatures of gene knockdowns for protein kinases and potential downstream targets among genes with DNA methylation associated with Chronic postsurgical pain and Childhood anxiety sensitivity index
| Pathway | P Value | Gene List |
| ABL1_knockdown_96h_HEPG2 | 0.0003 | TNXB, NXN, OGDHL, TRIM27, ADAMTS8, |
| DAPK3_knockdown_96h_HA1E | 0.0003 | SPTBN4, GALNT2, NXN, OGDHL, ADAMTS8, |
| RARB_knockdown_96h_HA1E | 0.003 | EXT1, PRIM2, ZNF814, LMF1, |
| GPR31_knockdown_96h_PC3 | 0.003 | RIMS2, ZNF814, CACNA1C, ADAMTS8, |
| GABRG1_knockdown_96h_HCC515 | 0.003 | BNC2, ERICH1, ZNF814, CARD14, |
| RIPK2_knockdown_96h_HCC515 | 0.003 | EXT1, OBSCN, TNXB, TRIM27, |
| RIOK3_knockdown_96h_HA1E | 0.003 | SPTBN4, OBSCN, TNXB, ADAMTS8, |
| GABBR1_knockdown_96h_HA1E | 0.003 | OBSCN, NXN, ZNF814, TRIM27, |
| GPRC5C_knockdown_96h_PC3 | 0.003 | RIMS2, PRIM2, SPTBN4, CACNA1C, |
| GPR151_knockdown_96h_A375 | 0.003 | EXT1, SPTBN4, ERICH1, CARD14, |
| ADRA2A_knockdown_96h_A375 | 0.003 | ADRA2A_knockdown_96h_A375 |
| CHRM3_knockdown_96h_PC3 | 0.003 | SPTBN4, OBSCN, ERICH1, CACNA1C, |
| FLT3_knockdown_96h_HA1E | 0.003 | PRIM2, SPTBN4, NXN, ADAMTS8, |
| CSNK1G2_knockdown_96h_HA1E | 0.003 | PRIM2, NXN, OGDHL, ADAMTS8, |
| TNIK_knockdown_96h_HEPG2 | 0.003 | SPTBN4, GALNT2, NXN, OGDHL, |
LINCS L1000 Kinase Perturbations show genes identified in this study as undergoing epigenetic regulation (right column) being significantly downregulated by kinase knock-down signatures in the left column, thus representing potential downstream targets of the respective kinase signaling cascade. Note that for each kinase, the corresponding cell line is also indicated, e.g., GABRG1 knock-down in HCC515 cells in the 5th row
Discussion
We have previously shown that psychological variables (CASI),[9] clinical variables and OPRM1 promoter DNA methylation[10] are associated with CPSP. In this study, we evaluated DNA methylation profiles for association with CPSP and CASI (anxiety sensitivity) in children undergoing spine surgery and followed up on our findings with an integrative computational analysis to identify common, targetable pathways enriched by the genes with differentially methylated CpG sites associated with these outcomes.
Epigenetic research into acute to chronic pain transitions[6] is still in its infancy. To our knowledge, there is only a handful of clinical epigenetic studies in postsurgical patients. DNA methylation of the Secreted Protein, Acidic, Rich in Cysteine (SPARC) promoter was shown to play a role in chronic low back pain related to degenerated intervertebral discs. [69] CpG methylation within the Tumor Necrosis Factor (TNF) gene promoter has also been identified as a mechanism by which TNF alters the risk for mild persistent breast pain in subjects with breast cancer undergoing surgery.[68] We previously reported on two CpG sites in an active regulatory region of the OPRM1 gene that binds multiple transcription factors, to be predictive of CPSP in another subset of the spine surgery cohort.[10] Hence, although the study sample size is small, we believe this study provides novel insight and evidence for the role of epigenetics in CPSP. Moreover, similar analyses have been reported with smaller sample sizes (n=47; case + controls) in prior epigenetic-chronic pain studies.[11] Another epigenome based pathway analyses used whole blood DNA for DNAm in a large cohort of adults, with chronic widespread musculoskeletal pain.[45] They found that 6% of variance for the pain phenotype was explained by epigenetic factors, and showed enrichment for neurological pathways, including synaptic long-term depression, axonal guidance signaling, CREB, neuropathic pain signaling and melatonin signaling.[45] While some of the pathways are similar to what we have identified for CPSP, the differences may be reflective of differences in the nature of pain and cohorts evaluated.
We will focus our discussion on shared enriched pathways common to CPSP and CASI. Of great interest is that the top canonical pathways enriched by genes with DMPs common to both these outcomes were the GABA receptor signaling and Dopamine-DARPP32 pathways. This is aligned with previous literature citing hypofunction of GABAergic inhibitory tone in the dorsal horn of the spinal cord as a key factor in central neuropathic pain after spinal cord injury. [18] Mechanisms proposed for GABAergic hypofunction include decreased number of GABA receptors (through apoptosis),[62] downregulation of GABA synthesizing enzyme (GAD), [48] and decreased GABA concentrations.[27] Several studies, both in vitro and in vivo, support the role of DNA methyltransferases in the regulation of GABAergic gene expression in brain regions relevant for pain and anxiety (cortex, striatum and hippocampus).[33] DNA epigenetic modifications of GABAergic interneurons in the basolateral amygdala were shown to be involved in the anxiety-like phenotypes in prenatal stress mice, and importantly, this was shown to be reversible with a demethylating agent, 5-Aza deoxycytidine.[79] Further, our functional genomics analysis show that many of the CpG sites identified are located in regions of the brain marked by lysine 27 tri-methylation (H3K27me3), which is known to negatively regulate gene expression. Our study thus provides new evidence for DNA methylation as a mechanism for possibly reduced function of the GABA receptor pathway genes and its role in CPSP and anxiety pathogenesis.
Our findings are also aligned with postulated roles for the DARPP-32 dopamine pathway (Supplementary figure 1) in the actions of drugs of abuse,[49; 76] inflammatory states,[75] and psychiatric conditions like schizophrenia and bipolar disorder.[31] DARPP-32 is a substrate of cAMP-dependent protein kinase (PKA) highly concentrated in dopamine-innervated brain areas, which functions as a PKA-regulated inhibitors of protein phosphatase-1 (PP1). The identification of epigenetic enrichment of this pathway is exciting as animal studies suggest a role for this phosphoprotein as intracellular detector of convergent dopamine-1 receptor and N-methyl-D-aspartate (NMDA) receptor activation,[7] which are target receptors for pain medications (opioids) and antipsychotic medications (for example haloperidol), and may suggest therapeutic interventions for CPSP, based on epigenetic profile.[23] Cyclin-dependent kinase5 (Cdk5) inhibitor (roscovitine) has been shown to decrease DARPP-32 phosphorylation,[75] and present exciting opportunities for future research, as its intrathecal use was found to decrease formalin-induced nociceptive response in rats[75] and remifentanil-induced hyperalgesia.[44] Since dopamine is involved in reward-mechanisms[64] and motivation to engage in pain self-management behaviors is an important predictor of adaptation/coping with acute pain, anxiety induced avoidance or lack of motivation[50] is the plausible mechanism by which dopamine signaling might be a player in development of CPSP in the presence of anxiety.
Genes with common DMPs associated with CPSP and CASI were also over-represented in nitric oxide signaling (NOS), which deserves mention. Nitric oxide is formed by N-methyl-d-aspartate (NMDA)-receptor activation. It has been shown to be an analgesic [55] and algesic [29] mediator at spinal, supraspinal and systemic sites in experimental animals. Pu et. al. postulated a dual control mechanism, wherein, excitatory NMDA is counteracted by inhibitory μ-opioid receptor signaling to modulate cyclic GMP/nitric oxide release, [56] thus influencing neuronal plasticity. Moreover, NOS also plays a role in morphine dependence and tolerance, which has been shown to be prevented using NOS inhibitors. [59]
There is evidence from prior studies for the role of pathways we have identified to be enriched by genes with DMPs associated with CASI. Bioinformatics analysis of microRNAs with differential expression in association with anxiety disorder used gene ontology and KEGG pathway analysis to predict target genes and functions. Epigenetically enriched pathways were elucidated which involved those related to neuronal brain functions, similar to our findings (GnRH signaling pathway).[21] Protein Kinase A signaling pathway, closely related to the DARPP-32 pathway described above, is involved in neuronal plasticity in the amygdala, is responsible for amplification of anxiety behaviors in response to stressful stimuli. Several clinical studies have shown that alterations in PKA are associated with anxiety, depression, and other psychiatric disorders,[37] which supports our findings.
The limitations of this study are the use of blood samples for DNA methylation measurement. While measurement in the primary target tissue (brain) would be ideal, these are inaccessible in clinical human studies. Some evidence for use of blood samples as a correlate for brain samples comes from a previous study compared methylation profiles derived from 12 tissues and found high correlation of DNAm between somatic tissues.[22] Davies et. al. concluded that peripheral tissues may be useful in studies of complex neurobiological phenotypes,[14] based on their findings that inter-individual variation in DNA methylation was reflected across brain and blood. However, the translational relevance of findings in easily available tissue like blood cannot be overemphasized. In addition, we have used bioinformatics approaches to provide evidence for functional relevance of our findings in the brain. ChIP assay findings show that the DMPs identified are located in active chromatin areas in pain pathway relevant brain cells, which is a strong indicator of DNAm in these CpG sites possibly affecting gene expression and function in neural tissue. Moreover, Enrichr and Pinet analyses revealed that the genes with DMPs (common to CPSP and CASI) are regulated by several TFs previously associated with neuronal phenotypes. These TFs include REST, TRIM28, POU5F1, NFE2L2, GATA2 and NANOG (Figure 3). NANOG is also associated with POU5F1, KLF4 etc. Further evidence for the GABA pathway involvement in CPSP and CASI comes from the analysis of overlaps with LINCS knockdowns. This reveals several GABA receptor subunits (including GPRC5C), among other potential positive upstream regulators of genes with shared DMPs (Table 6). GPRC5C has been previously shown to be an activator of NANOG,[25] which seems to be consistent with NANOG’s (and related TFs) targets being among (in this case predicted to be) positively regulated genes. Also, LINCS knockdowns of NANOG are strongly positively correlated (in multiple cell lines) with SMAD1/2/3, POLR2A (and other units of PolIIa), EP300 and other putative TFs targeting the overlap genes, which adds supporting evidence for them working together (not shown in the figures). The results of pathways analyses are limited due to being highly dependent on the input list of CpG loci – hence, validation/replication of the sites will be needed for translational impact. Nevertheless, the findings of this pilot study provide a significant basis for future research in predictive biomarkers and development of DNA methylation integrated prediction models for CPSP. In addition, it demonstrates that epigenetic markers could be used to identify downregulated and upregulated pathways which can then be manipulated using drugs, another step towards individualized medicine. Cross sectional studies are potentially affected by reverse causation bias and genetic variation confounders[51] and do not provide causation inferences. Hence, future longitudinal studies will be necessary to identify epigenetic changes over time resulting from surgical insult and pain/opioid exposures.
In conclusion, our findings provide a better understanding for the shared role of epigenetic regulation of CPSP and anxiety. While future studies are required in larger prospective cohorts with longitudinal evaluation of DNAm with replication of our findings, these pilot results are promising, and open new avenues of epigenetics-based pain research, since DNA methylation mechanisms can be modified by factors like diet, exercise, stress and meditation.[34; 73] Our findings provide a basis for biopsychosocial profiles involved in CPSP and suggest consideration of behavioral and other pathway-targeted strategies, based on the individual’s methylation profile.[52] There is promise from animal models for epigenetic modification to prevent the progression to chronic postsurgical pain,[15; 16] and use of demethylating drugs in other diseases,[63; 71] for such therapies to be useful for the treatment of chronic pain. Recent advent of targeted epigenetic modification[8] also provides hope for decreasing nonspecific effects and poor delivery of epigenetic modulating to target cells and tissues, a major impediment to the development and clinical application of such analgesics.
Supplementary Material
Highlights.
First pilot epigenome wide study in pediatric chronic postsurgical pain (CPSP).
DNA methylation in shared pathways associated with CPSP and anxiety sensitivity.
Differential DNA methylation enrich GABA and Dopamine-DARPP32 pathways.
GABA hypofunction and dopamine risk-reward pathways may be involved in CPSP.
Functional bioinformatics supports results and transcription factor targets proposed.
Acknowledgement:
We would like to acknowledge Ashley Ulm and Veda Yadagiri (Pyrosequencing core, CCHMC), Diane Kissell for their role in analyzing the DNA extraction and pyrosequencing, under supervision by Hong Ji (Director, Pyrosequencing Core) and Kejian Zhang (Director of Molecular Genetics Lab, Cincinnati Children’s Hospital). We would also like to acknowledge Kayla Stallworth and Hope Esslinger, CCRC IV, previous research coordinators for the Department of Anesthesia, Cincinnati Children’s Hospital, for their help with patient recruitment in the earlier stages of the study.
Disclosure The project was supported by the 5K23HD082782 through the EUNICE KENNEDY SHRIVER NATIONAL INSTITUTE OF CHILD HEALTH & HUMAN DEVELOPMENT, National Institutes of Health (PI: Chidambaran), Center for Pediatric Genomics and Shared Facility Discovery Award from Cincinnati Children’s Hospital Medical Center (PI: Chidambaran). MTW was supported by NIH R21 HG008186, NIH R01 NS099068–01A1, Cincinnati Children’s Hospital “Center for Pediatric Genomics” pilot study award, and a Cincinnati Children’s Hospital Research Fund “Endowed Scholar” award. HJ was supported by NIH/NIAID R21AI119236, ALA/AAAAI Respiratory Diseases Research Award 515708, and Center for Pediatric Genomics and Shared Facility Discovery Award from Cincinnati Children’s Hospital Medical Center (PI: Chidambaran). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None of the authors have any conflicts of interest to disclose.
References
- [1].Angst MS, Phillips NG, Drover DR, Tingle M, Ray A, Swan GE, Lazzeroni LC, Clark JD. Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain;153:1397–1409 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics;30:1363–1369 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol;28:1045–1048 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Branford R, Droney J, Ross JR. Opioid genetics: the key to personalized pain control? Clin Genet;82:301–310 2012 [DOI] [PubMed] [Google Scholar]
- [5].Bringuier S, Dadure C, Raux O, Dubois A, Picot MC, Capdevila X. The perioperative validity of the visual analog anxiety scale in children: a discriminant and useful instrument in routine clinical practice to optimize postoperative pain management. Anesth Analg;109:737–744 2009 [DOI] [PubMed] [Google Scholar]
- [6].Buchheit T, Van de Ven T, Shaw A. Epigenetics and the transition from acute to chronic pain. Pain medicine;13:1474–1490 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Buesa I, Ortiz V, Aguilera L, Torre F, Zimmermann M, Azkue JJ. Disinhibition of spinal responses to primary afferent input by antagonism at GABA receptors in urethane-anaesthetised rats is dependent on NMDA and metabotropic glutamate receptors. Neuropharmacology;50:585–594 2006 [DOI] [PubMed] [Google Scholar]
- [8].Chen H, Kazemier HG, de Groote ML, Ruiters MHJ, Xu G-L, Rots MG. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic acids research;42:1563–1574 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chidambaran V, Ding L, Moore DL, Spruance K, Cudilo EM, Pilipenko V, Hossain M, Sturm P, Kashikar-Zuck S, Martin LJ, Sadhasivam S. Predicting the pain continuum after adolescent idiopathic scoliosis surgery: A prospective cohort study. European journal of pain;21:1252–1265 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chidambaran V, Zhang X, Martin LJ, Ding L, Weirauch MT, Geisler K, Stubbeman BL, Sadhasivam S, Ji H. DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmgenomics Pers Med;10:157–168 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ciampi de Andrade D, Maschietto M, Galhardoni R, Gouveia G, Chile T, Victorino Krepischi AC, Dale CS, Brunoni AR, Parravano DC, Cueva Moscoso AS, Raicher I, Kaziyama HHS, Teixeira MJ, Brentani HP. Epigenetics insights into chronic pain: DNA hypomethylation in fibromyalgia-a controlled pilot-study. Pain;158:1473–1480 2017 [DOI] [PubMed] [Google Scholar]
- [12].Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature;489:57–74 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Crow M, Denk F, McMahon SB. Genes and epigenetic processes as prospective pain targets. Genome medicine;5:12 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol;13:R43 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Denk F, McMahon SB. Chronic pain: emerging evidence for the involvement of epigenetics. Neuron;73:435–444 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Denk F, McMahon SB, Tracey I. Pain vulnerability: a neurobiological perspective. Nature neuroscience;17:192–200 2014 [DOI] [PubMed] [Google Scholar]
- [17].Doehring A, Geisslinger G, Lotsch J. Epigenetics in pain and analgesia: an imminent research field. European journal of pain;15:11–16 2011 [DOI] [PubMed] [Google Scholar]
- [18].Drew GM, Siddall PJ, Duggan AW. Mechanical allodynia following contusion injury of the rat spinal cord is associated with loss of GABAergic inhibition in the dorsal horn. Pain;109:379–388 2004 [DOI] [PubMed] [Google Scholar]
- [19].Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics;11:587 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature;473:43–49 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fan H, Niu W, He M, Kong L, Zhong A, Zhang Q, Yan Y, Zhang L. [Bioinformatics analysis of differently expressed microRNAs in anxiety disorder]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi;32:641–646 2015 [DOI] [PubMed] [Google Scholar]
- [22].Fan S, Zhang X. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Commun;383:421–425 2009 [DOI] [PubMed] [Google Scholar]
- [23].Fernandez E, Schiappa R, Girault JA, Le Novere N. DARPP-32 is a robust integrator of dopamine and glutamate signals. PLoS Comput Biol;2:e176 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gerbershagen HJ, Rothaug J, Kalkman CJ, Meissner W. Determination of moderate-to-severe postoperative pain on the numeric rating scale: a cut-off point analysis applying four different methods. British journal of anaesthesia;107:619–626 2011 [DOI] [PubMed] [Google Scholar]
- [25].Gonzales KA, Liang H, Lim YS, Chan YS, Yeo JC, Tan CP, Gao B, Le B, Tan ZY, Low KY, Liou YC, Bard F, Ng HH. Deterministic Restriction on Pluripotent State Dissolution by Cell-Cycle Pathways. Cell;162:564–579 2015 [DOI] [PubMed] [Google Scholar]
- [26].Griffon A, Barbier Q, Dalino J, van Helden J, Spicuglia S, Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic acids research;43:e27 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gwak YS, Hulsebosch CE. GABA and Central Neuropathic Pain following Spinal Cord Injury. Neuropharmacology;60:799–808 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, Magnusen AF, Lynch A, Chetal K, Yukawa M, Barski A, Salomonis N, Kaufman KM, Kottyan LC, Weirauch MT. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nature genetics; 2018 [DOI] [PMC free article] [PubMed]
- [29].Hervera A, Negrete R, Leanez S, Martin-Campos JM, Pol O. Peripheral effects of morphine and expression of mu-opioid receptors in the dorsal root ganglia during neuropathic pain: nitric oxide signaling. Molecular pain;7:25 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hettema JM, Prescott CA, Myers JM, Neale MC, Kendler KS. The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry;62:182–189 2005 [DOI] [PubMed] [Google Scholar]
- [31].Ishikawa M, Mizukami K, Iwakiri M, Asada T. Immunohistochemical and immunoblot analysis of Dopamine and cyclic AMP-regulated phosphoprotein, relative molecular mass 32,000 (DARPP-32) in the prefrontal cortex of subjects with schizophrenia and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry;31:1177–1181 2007 [DOI] [PubMed] [Google Scholar]
- [32].James SK. Chronic postsurgical pain: is there a possible genetic link? British Journal of Pain 1;11 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kadriu B, Guidotti A, Chen Y, Grayson DR. DNA methyltransferases1 (DNMT1) and 3a (DNMT3a) colocalize with GAD67-positive neurons in the GAD67-GFP mouse brain. J Comp Neurol;520:1951–1964 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kanherkar RR, Stair SE, Bhatia-Dey N, Mills PJ, Chopra D, Csoka AB. Epigenetic Mechanisms of Integrative Medicine. Evidence-Based Complementary and Alternative Medicine;2017:19 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Katz J One man’s risk factor is another man’s outcome: difference in risk factor profiles for chronic postsurgical pain maintenance vs transition. Pain;153:505–506 2012 [DOI] [PubMed] [Google Scholar]
- [36].Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, Kuleshov MV, Ma’ayan A, Stathias V, Terryn R, Cooper D, Forlin M, Koleti A, Vidovic D, Chung C, Schurer SC, Vasiliauskas J, Pilarczyk M, Shamsaei B, Fazel M, Ren Y, Niu W, Clark NA, White S, Mahi N, Zhang L, Kouril M, Reichard JF, Sivaganesan S, Medvedovic M, Meller J, Koch RJ, Birtwistle MR, Iyengar R, Sobie EA, Azeloglu EU, Kaye J, Osterloh J, Haston K, Kalra J, Finkbiener S, Li J, Milani P, Adam M, Escalante-Chong R, Sachs K, Lenail A, Ramamoorthy D, Fraenkel E, Daigle G, Hussain U, Coye A, Rothstein J, Sareen D, Ornelas L, Banuelos M, Mandefro B, Ho R, Svendsen CN, Lim RG, Stocksdale J, Casale MS, Thompson TG, Wu J, Thompson LM, Dardov V, Venkatraman V, Matlock A, Van Eyk JE, Jaffe JD, Papanastasiou M, Subramanian A, Golub TR, Erickson SD, Fallahi-Sichani M, Hafner M, Gray NS, Lin JR, Mills CE, Muhlich JL, Niepel M, Shamu CE, Williams EH, Wrobel D, Sorger PK, Heiser LM, Gray JW, Korkola JE, Mills GB, LaBarge M, Feiler HS, Dane MA, Bucher E, Nederlof M, Sudar D, Gross S, Kilburn DF, Smith R, Devlin K, Margolis R, Derr L, Lee A, Pillai A. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst;6:13–24 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Keil MF, Briassoulis G, Stratakis CA, Wu TJ. Protein Kinase A and Anxiety-Related Behaviors: A Mini-Review. Front Endocrinol (Lausanne);7:83 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim H, Clark D, Dionne RA. Genetic contributions to clinical pain and analgesia: avoiding pitfalls in genetic research. J Pain;10:663–693 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Koleti A, Terryn R, Stathias V, Chung C, Cooper DJ, Turner JP, Vidovic D, Forlin M, Kelley TT, D’Urso A, Allen BK, Torre D, Jagodnik KM, Wang L, Jenkins SL, Mader C, Niu W, Fazel M, Mahi N, Pilarczyk M, Clark N, Shamsaei B, Meller J, Vasiliauskas J, Reichard J, Medvedovic M, Ma’ayan A, Pillai A, Schurer SC. Data Portal for the Library of Integrated Network-based Cellular Signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic acids research;46:D558–D566 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Landman Z, Oswald T, Sanders J, Diab M. Prevalence and predictors of pain in surgical treatment of adolescent idiopathic scoliosis. Spine (Phila Pa 1976);36:825–829 2011 [DOI] [PubMed] [Google Scholar]
- [41].Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics;28:882–883 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Leek JTJW, Parker HS, Fertig EJ, Jaffe AE, Storey JD, Zhang Y, Torres LC. sva: Surrogate Variable Analysis. R package version 3.24.4, 2017.
- [43].Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol;12:R83 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu X, Liu Y, Zhang J, Zhang W, Sun YE, Gu X, Ma Z. Intrathecal administration of roscovitine prevents remifentanil-induced postoperative hyperalgesia and decreases the phosphorylation of N-methyl-D-aspartate receptor and metabotropic glutamate receptor 5 in spinal cord. Brain Res Bull;106:9–16 2014 [DOI] [PubMed] [Google Scholar]
- [45].Livshits G, Malkin I, Freidin MB, Xia Y, Gao F, Wang J, Spector TD, MacGregor A, Bell JT, Williams FMK. Genome-wide methylation analysis of a large population sample shows neurological pathways involvement in chronic widespread musculoskeletal pain. Pain;158:1053–1062 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Macrae WA. Chronic post-surgical pain: 10 years on. British journal of anaesthesia;101:77–86 2008 [DOI] [PubMed] [Google Scholar]
- [47].Macrae WA, Davies HTO. Chronic postsurgical pain Seattle: IASP Press, 1999. [Google Scholar]
- [48].Meisner JG, Marsh AD, Marsh DR. Loss of GABAergic interneurons in laminae I-III of the spinal cord dorsal horn contributes to reduced GABAergic tone and neuropathic pain after spinal cord injury. J Neurotrauma;27:729–737 2010 [DOI] [PubMed] [Google Scholar]
- [49].Nairn AC, Svenningsson P, Nishi A, Fisone G, Girault JA, Greengard P. The role of DARPP-32 in the actions of drugs of abuse. Neuropharmacology;47 Suppl 1:14–23 2004 [DOI] [PubMed] [Google Scholar]
- [50].Navratilova E, Xie JY, Okun A, Qu C, Eyde N, Ci S, Ossipov MH, King T, Fields HL, Porreca F. Pain relief produces negative reinforcement through activation of mesolimbic reward–valuation circuitry. Proceedings of the National Academy of Sciences of the United States of America;109:20709–20713 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ng JW, Barrett LM, Wong A, Kuh D, Smith GD, Relton CL. The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities. Genome Biol;13:246 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Niederberger E, Resch E, Parnham MJ, Geisslinger G. Drugging the pain epigenome. Nature reviews Neurology;13:434–447 2017 [DOI] [PubMed] [Google Scholar]
- [53].Norbury TA, MacGregor AJ, Urwin J, Spector TD, McMahon SB. Heritability of responses to painful stimuli in women: a classical twin study. Brain;130:3041–3049 2007 [DOI] [PubMed] [Google Scholar]
- [54].Page MG, Stinson J, Campbell F, Isaac L, Katz J. Identification of pain-related psychological risk factors for the development and maintenance of pediatric chronic postsurgical pain. J Pain Res;6:167–180 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pataki I, Telegdy G. Further evidence that nitric oxide modifies acute and chronic morphine actions in mice. Eur J Pharmacol;357:157–162 1998 [DOI] [PubMed] [Google Scholar]
- [56].Pu S, Horvath TL, Diano S, Naftolin F, Kalra PS, Kalra SP. Evidence showing that beta-endorphin regulates cyclic guanosine 3’,5’-monophosphate (cGMP) efflux: anatomical and functional support for an interaction between opiates and nitric oxide. Endocrinology;138:1537–1543 1997 [DOI] [PubMed] [Google Scholar]
- [57].Rabbitts JA, Fisher E, Rosenbloom BN, Palermo TM. Prevalence and Predictors of Chronic Postsurgical Pain in Children: A Systematic Review and Meta-Analysis. The journal of pain : official journal of the American Pain Society;18:605–614 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rabian B, Embry L, MacIntyre D. Behavioral validation of the Childhood Anxiety Sensitivity Index in children. J Clin Child Psychol;28:105–112 1999 [DOI] [PubMed] [Google Scholar]
- [59].Rahmati B, Beik A. Prevention of morphine dependence and tolerance by Nepeta menthoides was accompanied by attenuation of Nitric oxide overproduction in male mice. J Ethnopharmacol;199:39–51 2017 [DOI] [PubMed] [Google Scholar]
- [60].Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, de Nijs L, Houtepen LC, Eijssen L, Jaffe AE, Kenis G, Viechtbauer W, van den Hove D, Schraut KG, Lesch KP, Kleinman JE, Hyde TM, Weinberger DR, Schalkwyk L, Lunnon K, Mill J, Cohen H, Yehuda R, Baker DG, Maihofer AX, Nievergelt CM, Geuze E, Boks MPM. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Molecular Psychiatry;23:1145 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sadhasivam S, Chidambaran V. Pharmacogenomics of opioids and perioperative pain management. Pharmacogenomics;13:1719–1740 2012 [DOI] [PubMed] [Google Scholar]
- [62].Sah R, Galeffi F, Ahrens R, Jordan G, Schwartz-Bloom RD. Modulation of the GABA(A)-gated chloride channel by reactive oxygen species. J Neurochem;80:383–391 2002 [DOI] [PubMed] [Google Scholar]
- [63].Saunthararajah Y, Lavelle D, DeSimone J. DNA hypo-methylating agents and sickle cell disease. Br J Haematol;126:629–636 2004 [DOI] [PubMed] [Google Scholar]
- [64].Scott DJ, Stohler CS, Egnatuk CM, Wang H, Koeppe RA, Zubieta JK. Placebo and nocebo effects are defined by opposite opioid and dopaminergic responses. Arch Gen Psychiatry;65:220–231 2008 [DOI] [PubMed] [Google Scholar]
- [65].Shimada-Sugimoto M, Otowa T, Miyagawa T, Umekage T, Kawamura Y, Bundo M, Iwamoto K, Tochigi M, Kasai K, Kaiya H, Tanii H, Okazaki Y, Tokunaga K, Sasaki T. Epigenome-wide association study of DNA methylation in panic disorder. Clinical Epigenetics;9:6 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sieberg CB, Simons LE, Edelstein MR, DeAngelis MR, Pielech M, Sethna N, Hresko MT. Pain prevalence and trajectories following pediatric spinal fusion surgery. The journal of pain : official journal of the American Pain Society;14:1694–1702 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Silverman WK, Goedhart AW, Barrett P, Turner C. The facets of anxiety sensitivity represented in the childhood anxiety sensitivity index: confirmatory analyses of factor models from past studies. J Abnorm Psychol;112:364–374 2003 [DOI] [PubMed] [Google Scholar]
- [68].Stephens KE, Levine JD, Aouizerat BE, Paul SM, Abrams G, Conley YP, Miaskowski C. Associations between genetic and epigenetic variations in cytokine genes and mild persistent breast pain in women following breast cancer surgery. Cytokine;99:203–213 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tajerian M, Alvarado S, Millecamps M, Dashwood T, Anderson KM, Haglund L, Ouellet J, Szyf M, Stone LS. DNA methylation of SPARC and chronic low back pain. Molecular pain;7:65 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol;41:401–407 2006 [DOI] [PubMed] [Google Scholar]
- [71].Viet CT, Dang D, Ye Y, Ono K, Campbell RR, Schmidt BL. Demethylating drugs as novel analgesics for cancer pain. Clinical cancer research : an official journal of the American Association for Cancer Research;20:4882–4893 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].von Baeyer CL. Numerical rating scale for self-report of pain intensity in children and adolescents: recent progress and further questions. Eur J Pain;13:1005–1007 2009 [DOI] [PubMed] [Google Scholar]
- [73].Vucetic Z, Kimmel J, Reyes TM. Chronic high-fat diet drives postnatal epigenetic regulation of mu-opioid receptor in the brain. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology;36:1199–1206 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain;146:270–275 2009 [DOI] [PubMed] [Google Scholar]
- [75].Wang CH, Chou WY, Hung KS, Jawan B, Lu CN, Liu JK, Hung YP, Lee TH. Intrathecal administration of roscovitine inhibits Cdk5 activity and attenuates formalin-induced nociceptive response in rats. Acta Pharmacol Sin;26:46–50 2005 [DOI] [PubMed] [Google Scholar]
- [76].Wang WW, Cao R, Rao ZR, Chen LW. Differential expression of NMDA and AMPA receptor subunits in DARPP-32-containing neurons of the cerebral cortex, hippocampus and neostriatum of rats. Brain Res;998:174–183 2004 [DOI] [PubMed] [Google Scholar]
- [77].Werner MU, Kongsgaard UE. I. Defining persistent post-surgical pain: is an update required? British journal of anaesthesia;113:1–4 2014 [DOI] [PubMed] [Google Scholar]
- [78].Young EE, Lariviere WR, Belfer I. Genetic basis of pain variability: recent advances. J Med Genet;49:1–9 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhu C, Liang M, Li Y, Feng X, Hong J, Zhou R. Involvement of Epigenetic Modifications of GABAergic Interneurons in Basolateral Amygdala in Anxiety-like Phenotype of Prenatally Stressed Mice. Int J Neuropsychopharmacol; 2018 [DOI] [PMC free article] [PubMed]
- [80].Zorina-Lichtenwalter K, Meloto CB, Khoury S, Diatchenko L. Genetic predictors of human chronic pain conditions. Neuroscience;338:36–62 2016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.