Abstract
Dissolution dynamic nuclear polarization (d-DNP) is a versatile method to enhance nuclear magnetic resonance (NMR) spectroscopy. It boosts signal intensities by four to five orders of magnitude thereby providing the potential to improve and enable a plethora of applications ranging from the real-time monitoring of chemical or biological processes to metabolomics and in-cell investigations. This perspectives article highlights possible avenues for developments and applications of d-DNP in biochemical and physicochemical studies. It outlines how chemists, biologists and physicists with various fields of interest can transform and employ d-DNP as a powerful characterization method for their research.
Introduction
Dissolution dynamic nuclear polarization (d-DNP) is a technique that aims at overcoming experimental limitations of solution state nuclear magnetic resonance (NMR) spectroscopy imposed by its intrinsically low sensitivity. Weak signal intensities in NMR are a major bottleneck that often impede rapid detection schemes by necessitating signal averaging or high (e.g., non-physiological) concentrations of target molecules. d-DNP1 mitigates these constraints by providing hyperpolarized nuclear spin states, which feature dramatic signal intensity boosts by several orders of magnitude. It is probably the most versatile method among other established hyperpolarization techniques, such as para-hydrogen induced polarization (PHIP),2 signal amplification by reversible exchange (SABRE)3 or spin exchange or metastability exchange optical pumping (SEOP or MEOP)4. Several reviews describing these methods are already available5.
d-DNP is based on the general concepts of DNP6 making use of the high polarization of electron spins by transferring it to nuclear spins through microwave irradiation at low temperatures. The original idea behind d-DNP1 consists in a fast dissolution of the sample after DNP, which preserves most of the hyperpolarization. This approach enables a plethora of solution-state applications in analytical NMR or magnetic resonance imaging (MRI). Here again several reviews are available in the literature.7,8,9,10
d-DNP provides the potential to boost signal intensities by three to four orders of magnitude thereby opening possibilities to access materials and properties that cannot be studied by conventional NMR.11,12 Hence, potential applications in various fields of research ranging from physical chemistry over nuclear physics to biochemistry are imaginable.
While one of the currently most prominent applications of d-DNP relies on the use of hyperpolarized biomolecular imaging tracers for in-vivo metabolic MRI13,14, this article targets the in-vitro analytical, physiochemical and biochemical applications and perspectives of d-DNP thereby trying to raise the attention of a multidisciplinary community. After a brief introduction, current uses in biochemical, chemical and physical contexts will be outlined and perspectives will be developed for future applications, such as real-time chemical reaction monitoring, investigation of biological substrates, metabolomics and nuclear spin physics of solids. Subsequently, based on the current state-of-the-art a perspective on future instrumentation and methods improvements will be developed in view of improving dDNP to make it more efficient, general, or transportable.
The d-DNP methodology - merits and pitfalls
d-DNP goes back to an invention by Ardenkjaer-Larsen, Golman and co-workers in 2003,1 who combined DNP of frozen liquids at low-temperatures (see 15and references therein for a detailed discussion of DNP mechanisms) with a subsequent dissolution and rapid transition to ambient temperatures to increase signal intensities in liquid state NMR spectroscopy. The combination of temperature jumps with DNP can spectacularly increase nuclear spin polarizations PI (vide infra for a more detailed definition), yet such an experiment also requires peculiar instrumentation and workflows. Currently, a prototypical d-DNP experiment for hyperpolarized NMR consists of three principal steps:
-
i)
Dynamic nuclear polarization of a sample at a low temperature TDNP ≤ 4.2 K within a dedicated DNP apparatus that maintains a static magnetic field
-
ii)
rapid heating and dissolution of the sample to the liquid state prior to fast transfer to an NMR spectrometer operating at typical magnetic fields or dedicated low-field devices;
-
iii)
NMR detection at close to ambient temperature TNMR > 273 K.
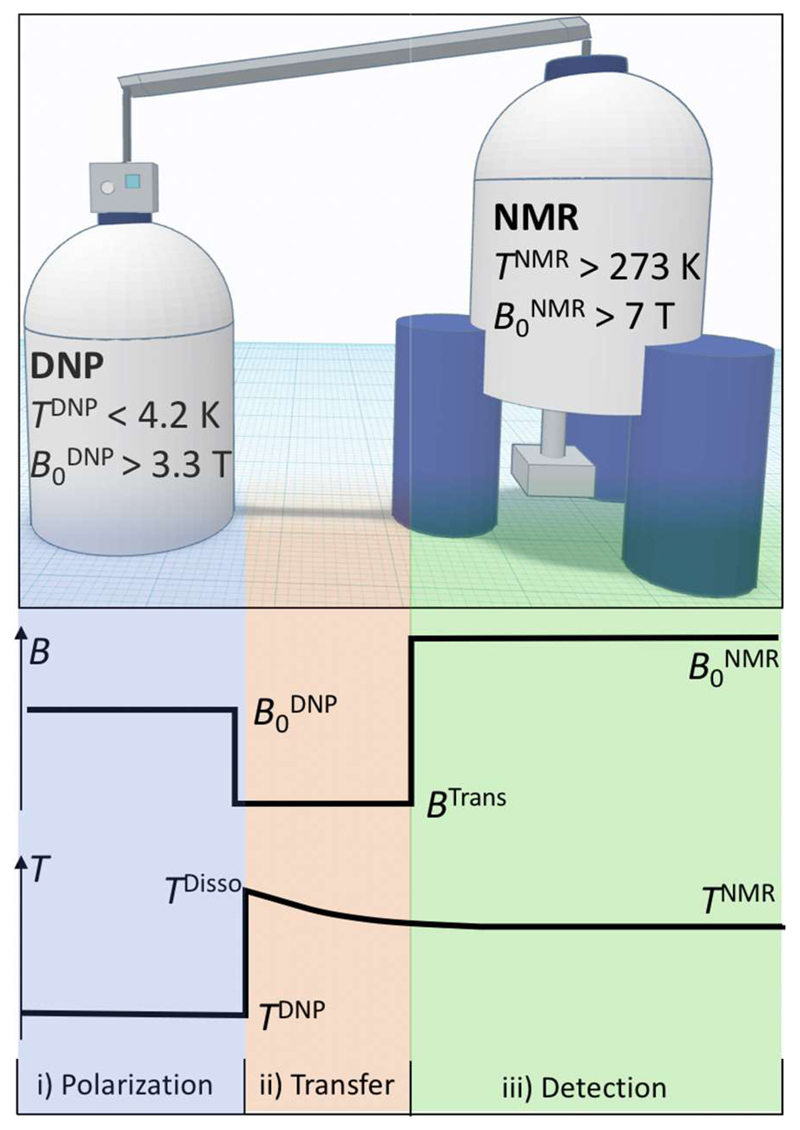
The basic setup of such an experiment is sketched in Fig. 1, which depicts a DNP apparatus connected to a conventional NMR spectrometer via a dissolution and transfer system.
Figure 1.
Principle setup of a d-DNP experiment. The DNP apparatus (left) typically operates at comparable or slightly lower magnetic fields than the NMR spectrometer used for detection (right). Qualitative magnetic field, B, and temperature, T, profiles as a function of time are sketched below together with the association of the different phases (i, ii and iii) of a d-DNP experiment with the different parts of the setup.
To give the reader an overview over the requirements of a d-DNP experiment, a brief description of steps i) , ii) and iii) is provided in the following:
i) The experiment starts with the preparation of a sample that contains paramagnetic molecules that are often coined polarization agents (PAs). Stable radicals such as nitroxides or tri-aryl methyl compounds often serve as PAs. After freezing of the sample, partial saturation of the electron paramagnetic resonance (EPR) line of a PA by slightly off-resonant microwave irradiation will entail an increased nuclear spin polarization P due to a transfer of polarization from the unpaired electrons of the PAs to the nuclei in their vicinity (the ratio of gyromagnetic ratios is γ(e-)/γ(1H) ≈ 668). This process normally takes place in a dedicated DNP apparatus, as it requires probes with microwave irradiation and often NMR capabilities (see for example references 1,16,17). A more or less homogeneous distribution of PAs in the DNP samples usually leads to optimal hyperpolarization. Therefore, crystallization, which leads to uncontrolled local PA-enriched and -depleted phases needs to be avoided, if necessary enforced by addition of vitrification agents such as glycerol.18
The sections “Sample Throughput” and “Transportable Hyperpolarization” provide a closer look at current developments concerning instrumentation and state-of-the-art PAs. For the moment it is sufficient to consider that microwave irradiation induces DNP by means of various mechanisms, most prominent, the cross effect and thermal mixing.19,20 The effectiveness of the different mechanisms depends on various factors such as the EPR line-shape, the concentration and the electron T1e of the PA. In d-DNP applications, one typically operates at temperatures close to 1 K, where mixtures of several mechanisms may be operative. A detailed description of these processes and their relation to d-DNP is beyond the scope of this perspective article. The reader is referred to the seminal work by Abragam and Goldman6 as well as to monographs21, articles15,22–26 and reviews.5,27 Yet, for the following perspectives the reader should keep in mind that DNP of samples that contain millimolar quantities of PAs entails a transfer of polarization from a PA to nuclear spins in its vicinity causing a boost in NMR signal intensity by increasing the nuclear polarization PI. The resulting spin state is then denoted as “hyperpolarized”.
ii) After the hyperpolarization procedure is complete, the sample is rapidly dissolved and diluted in hot heavy water and transferred to a conventional NMR spectrometer. The influence of the temperature jump during a d-DNP experiment can directly be deduced from eq. 1:
| (1) |
with εDNP the polarization enhancement brought about by DNP, and ℏ and kB the Planck and Boltzmann constants, respectively, and γ the gyromagnetic ratio of the nuclear spin I. The lower the temperature TDNP at which the DNP process takes place, the higher the thermal equilibrium polarization. The large signal enhancement in a d-DNP experiment can therefore be traced back to the multiplicative effects of DNP and the temperature jump.
The speed of the dissolution and transfer process is critical due to two major factors. Firstly, large fractions of hyperpolarization can be lost during the dissolution/heating process, if it proceeds to slowly. The target substance may pass through a regime in which nuclear spin relaxation is very effective (e.g., through slow tumbling of the molecule during the dissolution and exacerbate paramagnetic relaxation) causing a rapid decay of the hyperpolarization. Secondly, the sample needs to be rapidly transferred from the DNP apparatus to the detection NMR device during a transfer time ttrans through the magnetic field Btrans. The latter might be uncontrolled or too low, which may cause enhanced losses of hyperpolarization via enhanced paramagnetic relaxation28 or scalar relaxation of the first and second kind.29,30 Several recent developments try to mitigate this bottleneck of the d-DNP technology. For example, the use of magnetic tunnels31 that maintain a stable magnetic field Btrans during the transfer or high-pressure transfer systems in which the sample is propelled within only 0.5 to 2 s (depending on the setup) from the DNP apparatus to the NMR spectrometer.32,33
Additionally, it has recently been proposed that the dissolution process can take place in the detection NMR spectrometer after the cold solid sample has been transferred and not as usual within the DNP apparatus before the transfer.34 Several developments and perspectives in this regard will be discussed in more detail below in section “Solid transfer, liquid transfer”.
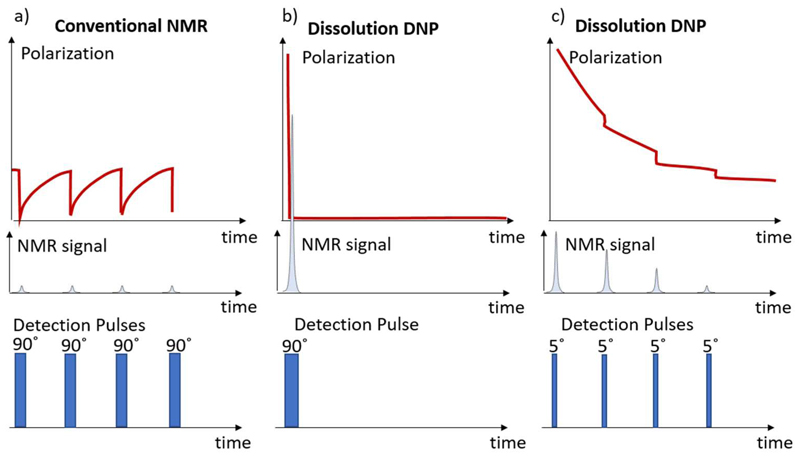
iii) After the sample has been dissolved and transferred, NMR detection proceeds. The hyperpolarized spin states can be exploited in NMR experiments if their duration does not exceed the lifetime of the hyperpolarization. A single pulse experiment with a 90°detection pulse will yield maximum sensitivity in a single scan (see Fig. 2), but it is also very common to detect the hyperpolarized magnetization with a train of small angle detection pulses, especially when the experiment aims at monitoring kinetic or dynamic processes.
Figure 2.
Comparison of a conventional NMR experiment with a d-DNP experiment. a) Traditionally, NMR detects the nuclear spin polarization in thermal equilibrium by means of a 90° detection pulse that reads the entire polarization. Subsequently, one waits for rebuild-up of the nuclear spin polarization to repeat the experiment. Typical, recovery delays are between a few hundreds of milliseconds to minutes. In contrast, in d-DNP the entire experiment is executed during a single period (b, c). One may use a single read-out pulse, which yields a single very intense spectrum. Yet, typically for 1D NMR by dDNP, the experiment starts with such an intense polarization that allows for small angle detection such that only small shares of the polarization are used to read the signal (or Ernst angles in cases of continuously replenished intermetdaites). The detection can be repeated rapidly, e.g. every 100 ms, before the hyperpolarization has decayed to thermal equilibrium polarization. This strategy is useful for monitoring dynamic/kinetic processes on a T1 timescale, but if sensitivity enhancement is the main goal, a single 90° can likewise be applied to obtain a maximum signal in a single detection.
In contrast to conventional NMR, where the experiment may last indefinitely within spectrometer stability and schedule constraints, signal detection in a d-DNP experiment needs to be completed within a short time interval after the sample has been transferred (see Fig. 1). The hyperpolarization has a limited lifetime as it inevitably decays towards thermal equilibrium with the time constant of longitudinal nuclear relaxation T1.
The limited lifetime of hyperpolarization and the irreversible nature of d-DNP imposes limitations in the application of conventional 2D and 3D experiments, which typically take longer then 2 min. Several approaches have been developed to resolve this dilemma. They will be discussed in more detail in the section “High-resolution d-DNP vs. high-resolution NMR” as well as “d-DNP and ultrafast NMR detection”. While, on the one hand, a d-DNP experiment is time-constrained, it is, on the other hand, boosted by signal enhancements εd-DNP of three to four orders of magnitude. εd-DNP is here understood as the ratio between nuclear spin polarization Pd-DNP of a hyperpolarized sample and corresponding polarization in thermal equilibrium PTE. This ratio can reach over 1 000 in proton NMR and over 10 000 in NMR of heteronuclei.11,12 Hence, novel detection strategies for higher-dimensional or time-resolved NMR become conceivable.
Small angle detection pulses of typically 1 - 30° are sufficient to observe an intense signal although only a small share of the nuclear polarization is used for detection. The readout can therefore be repeated rapidly, e.g. every 100 ms, before the polarization has vanished. In contrast, conventional NMR typically employs longer waiting times between every detection together with a readout of the entire polarization by 90°pulses. These relations are visualized in Fig. 2.
Applications of d-DNP for chemists, biologists and physicists
With a general outline of the d-DNP method at hand, we will highlight in following selected state-of-the-art examples and extrapolate these with respect to potential avenues to future d-DNP applications. The selection of examples in this article is of course of limited extent and does not claim to cover all important recent developments in the field.
High-resolution d-DNP vs. high-resolution NMR – Applications to structural biology of proteins
The established solution to overcome the intrinsically low sensitivity in conventional NMR is signal averaging. To this end, a spectrum is several times sequentially acquired such that the result features an improved signal-to-noise ratio (SNR). Obviously, the higher the desired SNR, the longer is the necessary acquisition time. d-DNP is – to a large extent - not limited anymore by signal intensities such that the need for signal accumulation becomes outdated and novel possibilities emerge.
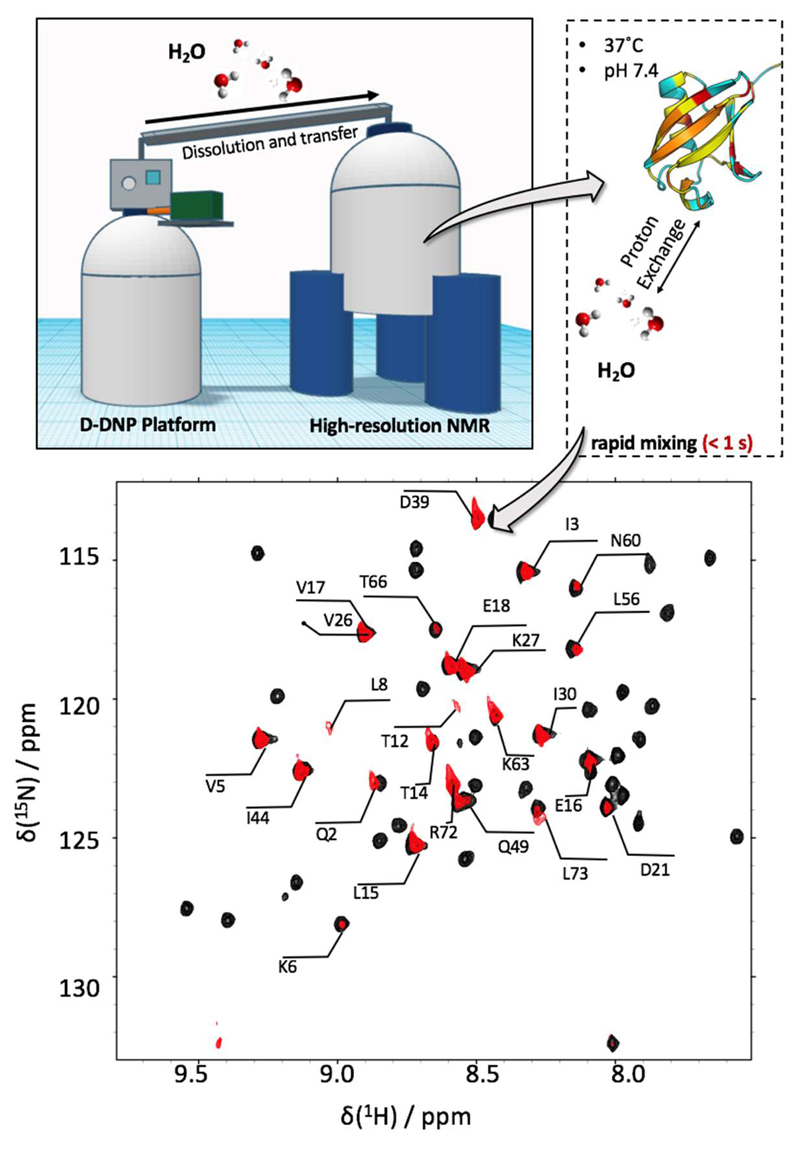
Due to the necessity for rapid detection, often only one-dimensional spectra are recorded by means of d-DNP. However, recently it has been shown that high-resolution multidimensional NMR of proteins by d-DNP is also possible by using hyperpolarized solvents as vectors to transport hyperpolarization from the low-temperature conditions within a DNP apparatus to a protein solution and finally into a protein (see Fig. 3 for a sketch of the approach).33,35–37,38,39 Indeed, by dissolving a well-folded or intrinsically disordered protein (IDP) in a buffer that contains labile hyperpolarized protons, the hyperpolarization is introduced into the protein by chemical exchange between labile protein side-chain or backbone protons and the solvent. With this technique, multidimensional NMR spectra with increased intensity can be measured due to two key points: 1) The approach is applicable to any water-soluble protein, as only the buffer must undergo the d-DNP procedure. This renders this approach broadly applicable to a plethora of substrates. The major limitation is imposed by the proton exchange rate between solvent and the protein’s backbone amide protons, which needs to ranging between ca. 0.5 s-1 and 100 s-1 to provide substantial signal enhancements.35,37 2) The hyperpolarization of the protons in a protein buffer decays with a time constant T1 that is much longer than the one of protons attached to the protein, which is of the order of 1 s. Typically, partially deuterated buffers are employed to prolong T1 times and hyperpolarization lifetimes of ca. 2 min. can be achieved with this approach. Hence, rapid multidimensional, high-resolution detection schemes become feasible.
Figure 3.
Experimental strategy for hyperpolarized multi-dimensional NMR of proteins. Top: To couple D-DNP with high-resolution protein NMR applications, hyperpolarized aqueous buffers are produced and subsequently mixed with a protein solution. Rapid exchange of hyperpolarized protons between the water in the buffer and the target proteins introduced hyperpolarization into the protein. As proton exchange is quite efficient at pH 7.4 and 37°C, NMR under near-physiological is feasible by this approach. Bottom: 1H-15N HMQC of ubiquitin detected through hyperpolarized 96% deuterated PBS buffer (red) superimposed to a conventional HSQC in 10% deuterated PBS (black). Evidently, not all residues can be detected by hyperpolarization-based detection as the signal intensity depends on the efficiency of the exchange of hyperpolarization between the solvent and the protein. The resulting spectra are, hence, sparse and reminiscent of selective labeling techniques based on selectively isotope enriched amino acids.
It was shown that 1H-15N correlation spectroscopy (HMQC) of >200 amino acid long IDPs is possible at residue resolution. Very recently, the approach has been extended to three-dimensional NMR (HNCO) and carbon-direct detection in 13C-15N correlation experiments (HN-CON).40 Enhancement factors ε in such experiments vary between ca. 10 to 500 with respect to signal intensities in thermal equilibrium ceteris paribus. The signal enhancements translate into a squared reduction in experimental time.
The result of a hyperpolarized multidimensional protein NMR experiment differs from a conventional NMR experiment. As the hyperpolarization of the protein is introduced through exchange between hyperpolarized solvent protons and labile protein protons (or through exchange-relayed nuclear Overhauser effects)37, the signal enhancement is – like the exchange rate – residue-dependent. For example, in 1H-15N correlation spectra only such proton-nitrogen pairs are detected that exchange hyperpolarization efficiently with the solvent during the recovery delay between each detection. Hence, only a subset of signals is detected in comparison to the conventional counterpart of a 1H-15N correlation spectrum. The “hyperpolarization-selective” detection can be compared with selective labeling approaches, such as targeted 13C or 15N-enrichment of selected amino acids.41,42 Such techniques render spectra sparse and, hence, allow for residue-specific analysis of larger proteins, for which uniform labeling would lead to strong signal overlap. Similar applications are concievable for large proteins, where signal broadening due to relaxation would be mitigated. The hyperpolarization exchange approach to high-resolution NMR might turn out to be very powerful supplement to established techniques, which suffer from overlapping signals.
Reducing concentrations, NMR optimized conditions
Hyperpolarized protein NMR is not likely to be superior to conventional NMR, instead it is of a complementary nature. For example, it can be employed to support NMR under physiological conditions (pH 7.4, 37°C), where conventional techniques may suffer from too low signal intensities. Such penalties can be compensated by hyperpolarization.
Another potential of hyperpolarized protein NMR lies in the possible reduction of substrate concentrations. Typically, NMR of proteins requires that substrate concentrations in the micromolar range (>50 µM). However, physiological concentrations are often much lower, even nanomolar and such variations can lead to significant shifts in the conformational ensemble of a protein. For example, the transcription factor MAX exists predominantly as a well-folded homodimer at higher, NMR-typical concentrations, but at physiological concentrations it mostly adopts an intrinsically disordered conformation.43 Hyperpolarized protein NMR can enable a reduction of the necessary substrate quantities to physiological concentrations to mitigate such biases. Possibly even in-cell NMR at physiological protein concentrations can be envisaged for future developments of d-DNP rendering this method even more versatile and a possible tool for characterization of low-concentrated and low-yield substrates.
Applications of hyperpolarized NMR to protein interactions
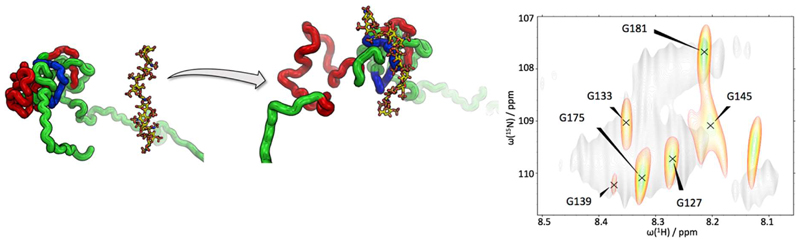
As the nature of hyperpolarized protein NMR is somewhat differing from established applications, novel possibilities to investigate protein interactions become conceivable.38,44,45 Among other excellent examples, this has recently been demonstrated at hand of the binding of the extracellular matrix protein osteopontin (OPN) to its ligand heparin. OPN is an intrinsically disordered protein (IDP) that features a disordered yet compacted core region. While the core’s increased level of compression shields it from the solvent, other regions of the protein are strongly exposed to the solvent. Binding of heparin to OPN leads to dissolution of the core and consequently changed solvent exposure of the core residues.46 As a consequence, hyperpolarization-selective detection leads to an observation of residues that are localized in the core region only in the heparin-bound state. This relation is visualized in Fig. 4.
Figure 4.
Sketch of the IDP OPN. In the ligand free form, OPN populates a compacted core (red and blue patches). When heparin binds to the positively charged part of the core (blue), the negatively charged part of the core (red) becomes more solvent exposed, which leads to the detection of glycine situated within the red marked stretch by means of hyperpolarization-boosted 1H-15N HMQC (right). In the absence of heparin, these glycines could not be detected.
Glycine residues located between aa 100 and 180, which take part in the formation of OPN’s core region cannot be detected by means of hyperpolarized solvents in the heparin-free form. However, in the presence of heparin, the proton exchange between the core residues and the solvent is accelerated and the glycines become observable.36
Hence, the set of residues that pass the “hyperpolarization-selective” filter varies due to conformational changes induced by the binding event. This feature provides an accurate indication for a protein interaction and can reveal adaptions of structural dynamics due to population shifts in conformational spaces of IDPs. Such a method might develop into a complement to conventional approaches, such as chemical shift perturbations in ligand titration experiments. Due to the rapid nature of the experiment, which typically takes no longer than 2 min. it also appears to be especially well-suited for metastable targets or transient intermediates allowing for residue-resolved interaction studies in short periods of time.
Potential of d-DNP for metabolomics
Metabolomics is a growing application field that could benefit in the near future from the sensitivity boost offered by d-DNP. Metabolomics –which aims at identifying and quantifying as many metabolites as possible in biological samples– currently relies on mass spectrometry (MS) and NMR as analytical tools. However, the metabolomics field is largely dominated by MS due to the much higher sensitivity of this technique. NMR is still popular thanks to its ease-of-use and to its higher reproducibility and identification capabilities, but the MS/NMR complementarity in metabolomics would certainly be reinforced by more sensitive NMR methods.
Recent studies highlighted the potential of d-DNP for the analysis of biological samples such as those commonly encountered in metabolomics. This requires to successfully hyperpolarize complex mixtures and to detect the multiple corresponding signals with a sufficient analytical performance to monitor biological variations between samples from different groups. A first approach consists in incubating the targeted biological samples with 13C-labeled substrates, followed by d-DNP on the resulting samples.47 This approach was applied to determine the concentration of several metabolites in cell extracts. Since it is based on isotope labeling, this method aims at targeting specific metabolic pathways. An untargeted strategy has also been suggested, which consists in hyperpolarizing samples at natural 13C abundance by applying cross-polarization (CP) from 1H to 13C spins. This approach has the double advantage that it does not require 13C labeling and that a high and reproducible 13C polarization of metabolites can in principle be reached within a few tens of minutes (versus a few hours for the direct 13C hyperpolarization of labeled samples). Dumez et al. demonstrated the potential of this approach on plant and cancer cell extracts at natural abundance, both with 1D and ultrafast (UF) 2D NMR detection.48
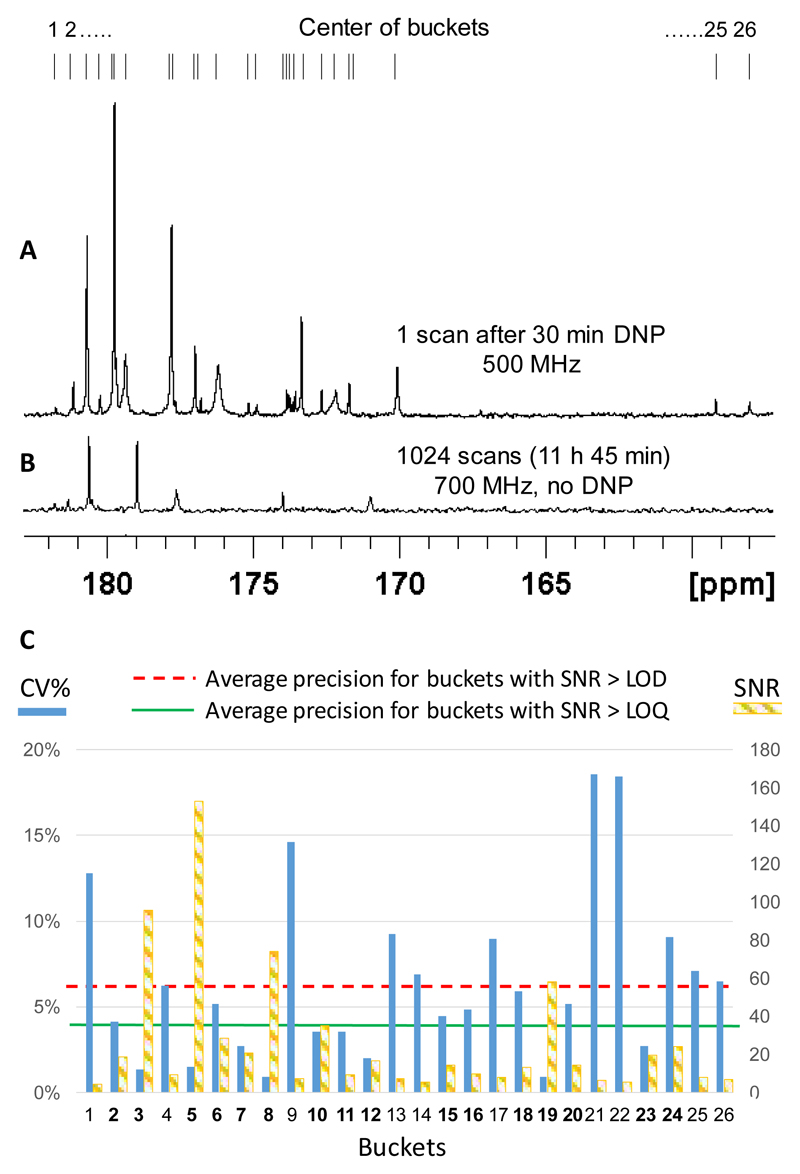
While these recent papers open interesting perspectives, several challenges must be addressed before d-DNP can be applied to real metabolomic studies. A notable characteristic of the approaches mentioned above is that only quaternary carbons could be detected thanks to their longer relaxation times. Faster injection systems that could enable the detection of broadband 13C spectra of hyperpolarized metabolite mixtures will certainly enhance the applicability of the method. Another key requirement for metabolomics is the precision of the method in terms of repeatability and reproducibility. The signal variation between successive experiments must be significantly smaller than the targeted biological variations (generally of at least 20%). An encouraging result has recently been obtained by Bornet et al., who demonstrated the very good repeatability (3.6% in average for signals above the limit of quantification) of d-DNP assisted by CP on tomato extracts (Fig. 5).49 Note that the trueness of d-DNP is not an issue since only relative concentration variations between samples are sought. In particular, the T1-weighting of metabolite signals is unavoidable but is not an issue if the T1 variations between samples within a metabolomics study are negligible and if the experiment is reproducible. Still, absolute metabolite concentrations can in principle be measured by applying adequate correction factors determined with refernce samples.36
Figure 5.
Potential of CP-assisted D-DNP for metabolomics. (a) Quaternary region of a 13C NMR spectrum of a green tomato fruit pericarp extract. (b) Thermal spectrum obtained without hyperpolarization on an identical extract. (c) Single-scan 13C NMR spectrum of a 20 mg extract. (c) Repeatability and sensitivity of D-DNP 13C NMR evaluated on 8 successive experiments performed on identical tomato extracts. The average precision, expressed in terms of repeatability, is 6.4% for all buckets containing signals above the limit of detection (LOD), and 3.6% when only the 16 buckets with signals above the limit of quantification (LOQ) are taken into account. The dashed orange bars correspond to the average SNR of the corresponding buckets, showing that the repeatability is proportional to the inverse of the SNR. The buckets are numbered from 1 to 26, indicated by lines on top of the spectra. Those in bold indicate buckets containing signals above the LOQ. Reproduced with permission from Reference 49.
While these studies pave the way towards hyperpolarized NMR metabolomics, future work will need to explore if d-DNP can bring more than conventional 1H metabolomics, both in terms of sample classification and biological interpretation. Such exploration will require embedding d-DNP in a meticulous analytical workflow including careful sample preparation, hyperpolarized NMR and data analysis.
Interaction monitoring – Applications to General Chemistry and Biochemistry
The potential of real-time monitoring has given rise to some remarkable studies in the recent past. For example, tracking of fast folding-upon-binding processes of proteins, of in-cell metabolic conversion,50 enzyme kinetics51, pH jumps52 or biomineralization processes.53
The general principle in all these applications is to hyperpolarize one interaction partner and to mix it with another interaction partner upon dissolution and transfer to the detection spectrometer followed by real-time NMR detection by either low-dimensional or higher-dimensional (ultrafast-detected) techniques (cf. Fig. 6) to follow the event in time. Among many excellent examples, one very interesting demonstration by Hilty and co-workers54 is based on monitoring signal intensities of live anionic chain ends in the polymerization of polystyrene. The d-DNP approach allowed to deduce details about reaction mechanisms and kinetic parameters with high precision. Another example by Abergel and co-workers is based on enzymatic conversion of hyperpolarized metabolites that are mixed upon dissolution with a protein cocktail that mimics parts of the physiological metabolic network.51,55 The conversion of the metabolite can then be monitored by following the build-up of NMR signals of the reaction products. Such, parts of the pentose phosphate pathway (PPP) have been studied by using hyperpolarized glucose (GLU) as tracer molecule that is mixed with a mixture of hexokinase (HA) and adenosine-triphosphate (ATP). By analyzing the decay of the educt GLU signal and the signal build-up of the product glucose-6-phosphate (G6P) with elaborate kinetic models, unprecedented insights (e.g. that phosphorylation of both α and β glucose anomers proceeds with comparable efficiencies) into kinetics and mechanism behind the PPP could be revealed.
Figure 6.
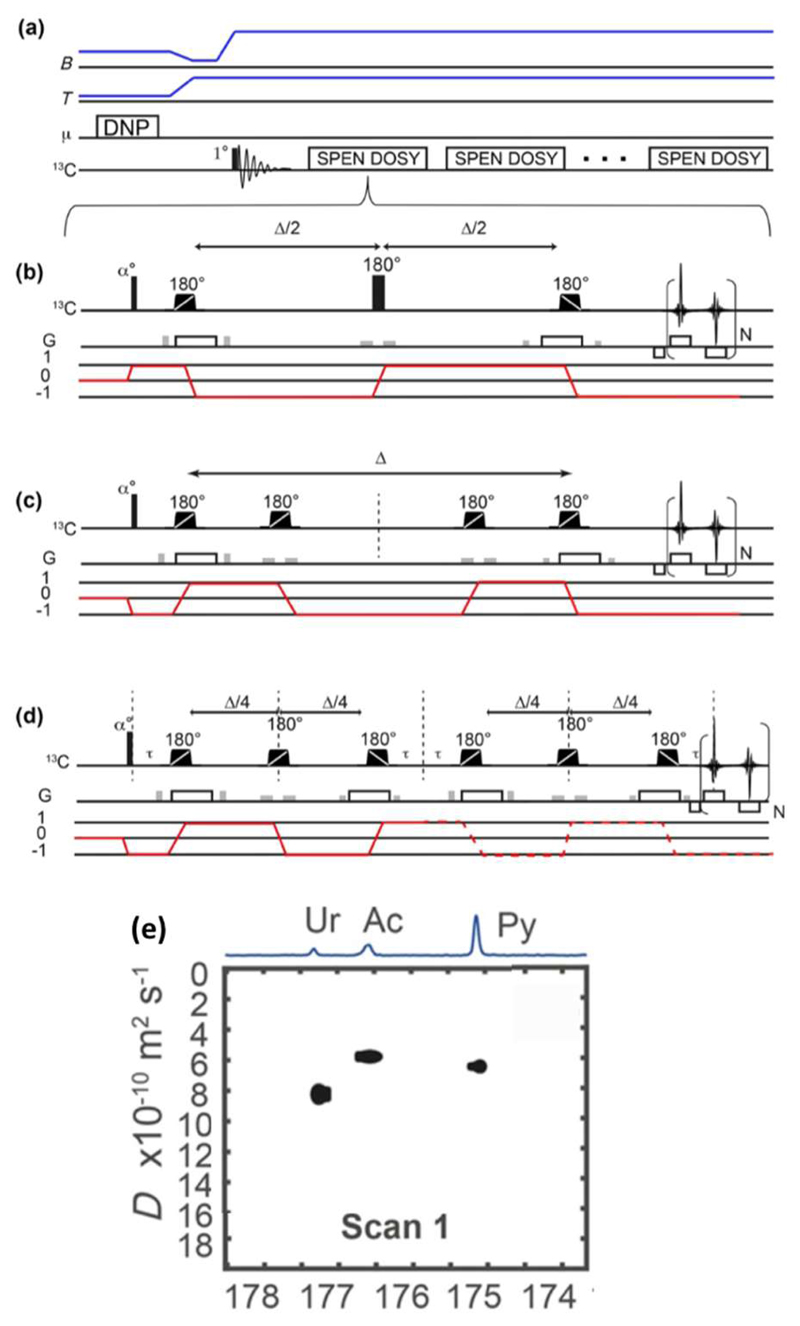
Description of the d-DNP SPEN DOSY experiment. a) Timing of the polarization, dissolution and acquisition steps, with the evolution of the temperature and magnetic field. b) Spin-echo SPEN DOSY pulse sequence. c) Double spin-echo SPEN DOSY pulse sequence. d) Double diffusion encoding SPEN DOSY pulse sequence. Crusher gradients are shown in grey. The selected coherence transfer pathway is shown in red. Reproduced with permission from reference 60. e) DOSY spectrum for the double-diffusion-encoding SPEN DOSY pulse sequence, obtained in a single D-DNP experiment.
d-DNP is a tool with potentially universal scope for reaction monitoring on milliseconds to seconds time scales and it might possibly develop into a versatile technique to monitor and study the early onset of a plethora of chemical processes.
Perspective on instrumentation and methods developments
Like NMR spectroscopy in general, d-DNP devices and techniques are undergoing continuous developments. In the following, we will give an account of the current state-of-the-art and possible future avenues as well as consequences for possible applications of d-DNP.
d-DNP and ultrafast NMR detection
The fast decaying and, more importantly, non-renewable nature of DNP-enhanced polarization, once in solution, makes it challenging, and in many cases, impossible, to collect multidimensional NMR spectra with classic time-incremented experiments. An important exception is the HMQC experiment, which is compatible with multi-scan acquisition when implemented with small-tip angle excitation pulses.56
A general solution to the problem of collecting 2D spectra from hyperpolarized substrates is to use single-scan, so called “ultrafast” NMR methods, based on the principle of spatial encoding. This was demonstrated early on by Frydman and co-workers with the acquisition of HSQC spectra.57 Further demonstrations include the acquisition of 13C-1H spectra from hyperpolarized natural products or complex metabolic mixtures.58,48 Spatial encoding is also the basis for ultrafast Laplace NMR (UFLNMR), where diffusion and/or relaxation information is encoded spatially.57 UFLNMR was used by Telkki and co-workers to obtain chemical information in heterogeneous media from hyperpolarized compounds, or to speed up the acquisition of water diffusivity and relaxation data in low-field NMR using single-sided magnets.57 Diffusion data can also be obtained in a single scan with chemical shift resolution from hyperpolarized substrates. This requires the use of convection-compensated diffusion encoding schemes. (Fig. 6).57 Overall, single-scan 2D NMR offers many promising options to increase the information content of DNP-enhanced experiments, although these have been little exploited so far. One limitation of the approach is that many single-scan 2D methods are not compatible with the acquisition of multiple consecutive spectra, while most applications of d-DNP concern time-evolving samples. While the sensitivity penalty associated with the gradient-based acquisition of UF2DNMR may also be a concern, it is counter-balanced in d-DNP by the single vs multi scan nature of the experiment.59
Several strategies have also been reported to obtain effectively multidimensional information from a small number of scans, in a way that is compatible with new renewable hyperpolarized signals. For example, Hilty and co-workers have used off resonance 13C decoupling to scale the apparent 13C-1H coupling in 1H 1D spectra and provide access to the chemical shift of the coupled carbon for each proton.61 They also showed how the selective inversion of a signal can result in a new type of correlation between, e.g. reactant and product, for systems undergoing a chemical reaction.62 This strategy was used to help in the mechanistic analysis of metal catalyzed polymerization reactions.63
Sample throughput
A main shortcoming of the d-DNP state-of-the-art is its low sample throughput. Hyperpolarization buildup times are often on the order of hours such that only few experiments are possible per day. To overcome this bottleneck, an instrumentation workaround has recently been developed that allows one to hyperpolarize several samples at the same time, but to dissolve them sequentially such that high experimental repetition rates and multi-shot experiments become possible.64,65 Another effective way of enhancing sample throughput consists in the use of 1H-13C cross polarization together with a rapid and efficient hyperpolarization of 1H spins with broad lines PAs such as nitroxides.66,67 By further increasing the polarizing magnetic field68, implementing suitable frequency modulation of the microwave irradiation69 and performing microwave gating during CP70, record 13C polarizations > 60% have been achieved in less than 10 minutes. It is most likely that these advances will be further improved in the near future by better radiofrequency coil designs. (CP experiments are currently performed with B1 < 20 kHz whereas > 50 kHz values would be desirable.) Another route to improve the throughput would be the use of more efficient PAs that would polarize 1H-nuclei faster and higher, possibly even at higher temperatures. For the moment, the best PAs for proton DNP are simplest forms of nitroxide radicals (such as TEMPO, amino-TEMPO, or TEMPOL) but one can hope that well-chosen or newly designed PAs may surpass the existing ones (see for example the current effort to improve PAs in MAS-DNP).71–73 These developments may pave the way towards applications of d-DNP with enhanced sample throughput such as compound screening applications in combinatorial research.
Solid transfer, liquid transfer – Applications to spin physics
Typically for d-DNP, samples are dissolved within the DNP apparatus and subsequently transferred in the liquid state to the detection NMR spectrometer. However recently, it has been demonstrated that a transfer of some solid samples is likewise feasible if the transfer is very rapid, i.e. on the order of 100 ms, and the magnetic field Btrans does not drop below a critical values where nuclear thermal mixing occurs.34 While, d-DNP of solutes in liquid state samples is currently further developed and applicable to various NMR active nuclei, the solid-transfer development opens new avenues for applications of d-DNP, not only with respect to NMR of hyperpolarized liquids and solids,74 but also with respect to hyperpolarized NMR of dissolved gases. Fig. 7 displays a sketch of the setup developed to transfer a DNP hyperpolarized solid sample to the NMR spectrometer.
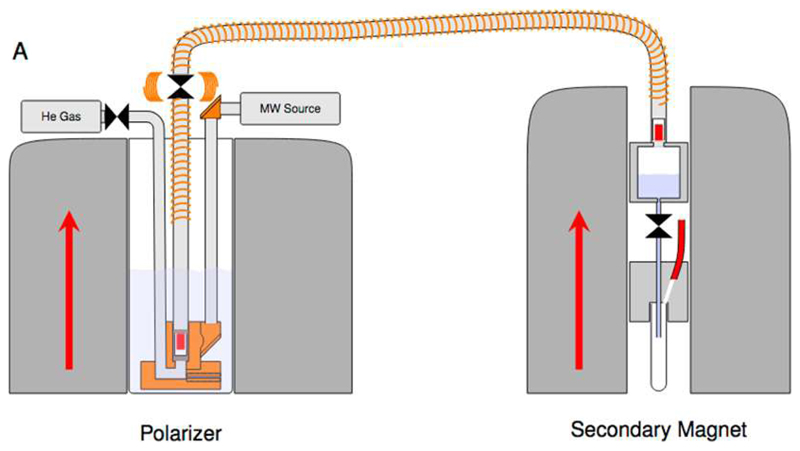
Figure 7.
Setup for transfer of a DNP hyperpolarized solid sample to an NMR spectrometer as proposed by Meier and co-workers. After DNP (left) the solid sample is pushed by pressurized helium through a tube surrounded by a solenoid that maintains a non-zero magnetic field during the transfer within ca. 100 ms to a detection spectrometer (right), where it is dissolved prior to injection into an NMR tube. Reproduced with permission from reference 34.
The transfer of solids is a promising method to study the effect of temperature jumps on spin dynamics, as has been shown in a recent example where the para-to-ortho conversion of proton nuclear spins was studied in hyperpolarized water molecules enclosed in C60 fullerene cages (H2O@C60). Such conversions have been subject to many speculations due to the omnipresence of ortho- and para-water,75 but have so far been cumbersome to access due to fast proton exchange in water. Yet, Meier and co-wokers showed that hyperpolarized H2O@C60 facilitated the access to these states and to study their properties, albeit somewhat artificial conditions.76 After rapid transfer of solid H2O@C60 from cold temperatures close to 1 K to an NMR spectrometer operating at ambient temperatures TNMR, a conversion of para water to ortho water can be observed at hand of the intensity of the water NMR signal. Similar principles can be applied to a plethora of other substrates to study the effect of rapid temperature changes on nuclear polarizations and dynamics. Especially in highly symmetric systems interesting results can be anticipated.77,78
Another potential provided by the transfer-then-dissolution approach lies in the study of dissolved gases. The traditional dissolution-then-transfer strategy often suffers from imminent degassing of the solution. Typically, pressurized helium is used to rapidly push the dissolved sample in the liquid state through a capillary towards the detection NMR spectrometer. This process has undergone intense optimization in recent years.32 However, the applied pressure leads to a removal of the gas from the solution. This problem could be overcome by first transferring a hyperpolarized frozen gas pellet to the detection NMR spectrometer and then dissolving it in a suitable liquid in-situ within the spectrometer. Such, NMR of important substances such as methane, ethylene or 15N2, might become possible.79 This might render dDNP more versatile and applicable to a broader range of scientific problems.
Transportable hyperpolarized substrates
The concept of solid-state transfer of the DNP sample is also currently opening new avenues for remote transport of hyperpolarized samples, which will ultimately make the presence of a DNP machine at the point of the NMR experiment unnecessary. Three proof of concept papers that were recently published indeed demonstrated that transport of hyperpolarized substances could be possible over tens of hours timescales. This was done either by brute force polarization (with PA-free sample formulation)80, by 1H-13C CP-DNP (on sub-micron heterogenous sample formulations)81, or by thermal annihilation of photo-induced radicals (using UV excitation)82. The key in these approaches always is the absence of paramagnetic relaxation during the transport step. It is likely that in the next couple of years, these methods will develop further and enable remote hyperpolarized applications (such as real-time metabolic imaging, metabolomics, drug discovery, etc.). The prospect of hyperpolarization in dedicated DNP centers may raise the question of a scaling-up of DNP sample quantities and throughput to much higher extents than today, which may lead to a radical transformation of the DNP instrumentation towards high volume cryostats, high power microwave sources, lower cryogenic fluid consumption, etc.
Outlook
This article tried to outline selected possible applications of d-DNP that might be of interest for a broader community covering physical, chemical and biological research. Many potentials emerged up to now due to the progress d-DNP has undergone since its invention in 2003, and its future developments seem promising in view of the current state-of-the-art. However, d-DNP remains a complicated method, which is often cumbersome to use, and expert operators are often necessary as many factors need to be considered ranging from low-temperature nuclear physics, over hydrodynamics to rapid NMR detection. A possible avenue for a more widespread application of d-DNP could be hyperpolarization user facilities, where external users can apply and be supported by local expert staff. Examples in this direction are (among others) the d-DNP facilities at EPFL and CRMN Lyon48,83,84, at the ENS Paris,19,60,85 and at the MagLab in Talahassee where numerous biologically, physically and chemically oriented projects of external users of d-DNP have been successfully executed with the help of local operators. If such direction were to be pursued, developments that NMR spectroscopy has undergone in the last 40 years, from an expert method to almost completely automated facilities could be envisaged for future developments of d-DNP. Alternatively, the prospects of transportable hyperpolarization might also definitely eliminate the need for an on-site d-DNP access and simply transform hyperpolarization into a consumable (such as PET tracers).
Acknowledgments
The authors acknowledge support of the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC Grant Agreements n°714519/HP4all and n°801936/HYPROTIN). Support from the French National Research Agency (Grant ANR-17-ERC2-0011) is also acknowledged. Support from the Corsaire metabolomics core facility is also acknowledged.
References
- (1).Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in Signal-to-Noise Ratio of > 10,000 Times in Liquid-State NMR. Proc Natl Acad Sci. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Buljubasich L, Franzoni MB, Münnemann K. Parahydrogen Induced Polarization by Homogeneous Catalysis: Theory and Applications. In: Kuhn LT, editor. Hyperpolarization Methods in NMR Spectroscopy. Vol. 338. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. pp. 33–74. [DOI] [PubMed] [Google Scholar]
- (3).Rayner PJ, Duckett SB. Signal Amplification by Reversible Exchange (SABRE): From Discovery to Diagnosis. Angew Chem Int Ed. 2018;57(23):6742–6753. doi: 10.1002/anie.201710406. [DOI] [PubMed] [Google Scholar]
- (4).Barskiy DA, Coffey AM, Nikolaou P, Mikhaylov DM, Goodson BM, Branca RT, Lu GJ, Shapiro MG, Telkki VV, Zhivonitko VV, et al. NMR Hyperpolarization Techniques of Gases. Chem Eur J. 2017;23(4):725–751. doi: 10.1002/chem.201603884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kovtunov KV, Pokochueva EV, Salnikov OG, Cousin SF, Kurzbach D, Vuichoud B, Jannin S, Chekmenev EY, Goodson BM, Bar-skiy DA, et al. Hyperpolarized NMR Spectroscopy: D -DNP, PHIP, and SABRE Techniques. Chem - Asian J. 2018;13(15):1857–1871. doi: 10.1002/asia.201800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Abragam A, Goldman Maurice. Principles of Dynamic Nuclear-Polarization. 1978;41(3):395–467. [Google Scholar]
- (7).Bornet A, Jannin S. Optimizing Dissolution Dynamic Nuclear Polarization. J Magn Reson. 2016;264:13–21. doi: 10.1016/j.jmr.2015.12.007. [DOI] [PubMed] [Google Scholar]
- (8).Ardenkjaer-Larsen JH. On the Present and Future of Dissolution-DNP. J Magn Reson. 2016;264:3–12. doi: 10.1016/j.jmr.2016.01.015. [DOI] [PubMed] [Google Scholar]
- (9).Jähnig F, Kwiatkowski G, Ernst M. Conceptual and Instrumental Progress in Dissolution DNP. J Magn Reson. 2016;264:22–29. doi: 10.1016/j.jmr.2015.12.024. [DOI] [PubMed] [Google Scholar]
- (10).Keshari KR, Wilson DM. Chemistry and Biochemistry of 13 C Hyperpolarized Magnetic Resonance Using Dynamic Nuclear Polarization. Chem Soc Rev. 2014;43(5):1627–1659. doi: 10.1039/C3CS60124B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lumata L, Jindal AK, Merritt ME, Malloy CR, Sherry AD, Kovacs Z. DNP by Thermal Mixing under Optimized Conditions Yields >60 000 Fold Enhancement of 89 Y NMR Signal. J Am Chem Soc. 2011;133(22):8673–8680. doi: 10.1021/ja201880y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Vuichoud B, Milani J, Chappuis Q, Bornet A, Bodenhausen G, Jannin S. Measuring Absolute Spin Polarization in Dissolution-DNP by Spin PolarimetrY Magnetic Resonance (SPY-MR) J Magn Reson. 2015;260:127–135. doi: 10.1016/j.jmr.2015.09.006. [DOI] [PubMed] [Google Scholar]
- (13).Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-13C]Pyruvate. Sci Transl Med. 2013;5(198):198ra108–198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wilson DM, Kurhanewicz J. Hyperpolarized 13C MR for Molecular Imaging of Prostate Cancer. J Nucl Med. 2014;55(10):1567–1572. doi: 10.2967/jnumed.114.141705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jannin S, Comment A, van der Klink JJ. Dynamic Nuclear Polarization by Thermal Mixing Under Partial Saturation. Appl Magn Reson. 2012;43(1-2):59–68. doi: 10.1007/s00723-012-0363-4. [DOI] [Google Scholar]
- (16).Baudin M, Vuichoud B, Bornet A, Bodenhausen G, Jannin S. A Cryogen-Consumption-Free System for Dynamic Nuclear Polarization at 9.4 T. J Magn Reson. 2018;294:115–121. doi: 10.1016/j.jmr.2018.07.001. [DOI] [PubMed] [Google Scholar]
- (17).Ardenkjaer-Larsen JH, Bowen S, Petersen JR, Rybalko O, Vinding MS, Ullisch M, Nielsen NC. Cryogen-Free Dissolution Dynamic Nuclear Polarization Polarizer Operating at 3.35 T, 6.70 T and 10.1 T. Magn Reson Med. 2019;81(3):2184–2194. doi: 10.1002/mrm.27537. [DOI] [PubMed] [Google Scholar]
- (18).Weber EMM, Sicoli G, Vezin H, Frébourg G, Abergel D, Bodenhausen G, Kurzbach D. Sample Ripening through Nanophase Separation Influences the Performance of Dynamic Nuclear Polarization. Angew Chem Int Ed. 2018;57(18):5171–5175. doi: 10.1002/anie.201800493. [DOI] [PubMed] [Google Scholar]
- (19).Guarin D, Marhabaie S, Rosso A, Abergel D, Bodenhausen G, Ivanov KL, Kurzbach D. Characterizing Thermal Mixing Dynamic Nuclear Polarization via Cross-Talk between Spin Reservoirs. J Phys Chem Lett. 2017;8(22):5531–5536. doi: 10.1021/acs.jpc-lett.7b02233. [DOI] [PubMed] [Google Scholar]
- (20).Shimon D, Hovav Y, Feintuch A, Goldfarb D, Vega S. Dynamic Nuclear Polarization in the Solid State: A Transition between the Cross Effect and the Solid Effect. Phys Chem Chem Phys. 2012;14(16):5729. doi: 10.1039/c2cp23915a. [DOI] [PubMed] [Google Scholar]
- (21).Wenckebach T. Essentials of Dynamic Nuclear Polarization. Spindrift Publications; 2016. [Google Scholar]
- (22).Hovav Y, Feintuch A, Vega S. Theoretical Aspects of Dynamic Nuclear Polarization in the Solid State – Spin Temperature and Thermal Mixing. Phys Chem Chem Phys. 2013;15(1):188–203. doi: 10.1039/C2CP42897K. [DOI] [PubMed] [Google Scholar]
- (23).Serra SC, Rosso A, Tedoldi F. Electron and Nuclear Spin Dynamics in the Thermal Mixing Model of Dynamic Nuclear Polarization. Phys Chem Chem Phys. 2012;14(38):13299. doi: 10.1039/c2cp41947e. [DOI] [PubMed] [Google Scholar]
- (24).Wenckebach WT. Spectral Diffusion and Dynamic Nuclear Polarization: Beyond the High Temperature Approximation. J Magn Reson. 2017;284:104–114. doi: 10.1016/j.jmr.2017.10.001. [DOI] [PubMed] [Google Scholar]
- (25).Lumata L, Kovacs Z, Sherry AD, Malloy C, Hill S, van Tol J, Yu L, Song L, Merritt ME. Electron Spin Resonance Studies of Trityl OX063 at a Concentration Optimal for DNP. Phys Chem Chem Phys. 2013;15(24):9800. doi: 10.1039/c3cp50186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Weber EMM, Vezin H, Kempf JG, Bodenhausen G, Abergél D, Kurzbach D. Anisotropic Longitudinal Electronic Relaxation Affects DNP at Cryogenic Temperatures. Phys Chem Chem Phys. 2017;19(24):16087–16094. doi: 10.1039/C7CP03242K. [DOI] [PubMed] [Google Scholar]
- (27).Zhang G, Hilty C. Applications of Dissolution Dynamic Nuclear Polarization in Chemistry and Biochemistry. Magn Reson Chem. 2018;56(7):566–582. doi: 10.1002/mrc.4735. [DOI] [PubMed] [Google Scholar]
- (28).Miéville P, Jannin S, Bodenhausen G. Relaxometry of Insensitive Nuclei: Optimizing Dissolution Dynamic Nuclear Polarization. J Magn Reson. 2011;210(1):137–140. doi: 10.1016/j.jmr.2011.02.006. [DOI] [PubMed] [Google Scholar]
- (29).Chiavazza E, Kubala E, Gringeri CV, Düwel S, Durst M, Schulte RF, Menzel MI. Earth’s Magnetic Field Enabled Scalar Coupling Relaxation of 13C Nuclei Bound to Fast-Relaxing Quadrupolar 14N in Amide Groups. J Magn Reson. 2013;227:35–38. doi: 10.1016/j.jmr.2012.11.016. [DOI] [PubMed] [Google Scholar]
- (30).Elliott SJ, Bengs C, Brown LJ, Hill-Cousins JT, O’Leary DJ, Pileio G, Levitt MH. Nuclear Singlet Relaxation by Scalar Relaxation of the Second Kind in the Slow-Fluctuation Regime. J Chem Phys. 2019;150(6):064315. doi: 10.1063/1.5074199. [DOI] [PubMed] [Google Scholar]
- (31).Milani J, Vuichoud B, Bornet A, Miéville P, Mottier R, Jannin S, Bodenhausen G. A Magnetic Tunnel to Shelter Hyperpolarized Fluids. Rev Sci Instrum. 2015;86(2):024101. doi: 10.1063/1.4908196. [DOI] [PubMed] [Google Scholar]
- (32).Bowen S, Hilty C. Rapid Sample Injection for Hyperpolarized NMR Spectroscopy. Phys Chem Chem Phys. 2010;12(22):5766. doi: 10.1039/c002316g. [DOI] [PubMed] [Google Scholar]
- (33).Olsen G, Markhasin E, Szekely O, Bretschneider C, Frydman L. Optimizing Water Hyperpolarization and Dissolution for Sensitivity-Enhanced 2D Biomolecular NMR. J Magn Re son. 2016;264:49–58. doi: 10.1016/j.jmr.2016.01.005. [DOI] [PubMed] [Google Scholar]
- (34).Kouřil K, Kouřilová H, Levitt MH, Meier B. Dissolution-Dynamic Nuclear Polarization with Rapid Transfer of a Polarized Solid. ArXiv180700223 Phys. 2018 doi: 10.1038/s41467-019-09726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Szekely O, Olsen GL, Felli IC, Frydman L. High-Resolution 2D NMR of Disordered Proteins Enhanced by Hyperpolarized Water. Anal Chem. 2018;90(10):6169–6177. doi: 10.1021/acs.anal-chem.8b00585. [DOI] [PubMed] [Google Scholar]
- (36).Kurzbach D, Canet E, Flamm AG, Jhajharia A, Weber EMM, Konrat R, Bodenhausen G. Investigation of Intrinsically Disordered Proteins through Exchange with Hyperpolarized Water. Angew Chem Int Ed. 2017;56(1):389–392. doi: 10.1002/anie.201608903. [DOI] [PubMed] [Google Scholar]
- (37).Kadeřávek P, Ferrage F, Bodenhausen G, Kurzbach D. High-Resolution NMR of Folded Proteins in Hyperpolarized Physiological Solvents. Chem Eur J. 2018;24(5):13418–13423. doi: 10.1002/chem.201802885. [DOI] [PubMed] [Google Scholar]
- (38).Chappuis Q, Milani J, Vuichoud B, Bornet A, Gossert AD, Bodenhausen G, Jannin S. Hyperpolarized Water to Study Protein–Ligand Interactions. J Phys Chem Lett. 2015;6(9):1674–1678. doi: 10.1021/acs.jpc-lett.5b00403. [DOI] [PubMed] [Google Scholar]
- (39).Vuichoud B, Bornet A, de Nanteuil F, Milani J, Canet E, Ji X, Miéville P, Weber E, Kurzbach D, Flamm A, et al. Filterable Agents for Hyperpolarization of Water, Metabolites, and Proteins. Chem - Eur J. 2016;22(41):14696–14700. doi: 10.1002/chem.201602506. [DOI] [PubMed] [Google Scholar]
- (40).Olsen GL. Hyperpolarized 3D NMR for Biomolecular Applications. 2018.
- (41).Lichtenecker RJ, Coudevylle N, Konrat R, Schmid W. Selective Isotope Labelling of Leucine Residues by Using α-Ketoacid Precursor Compounds. ChemBioChem. 2013;14(7):818–821. doi: 10.1002/cbic.201200737. [DOI] [PubMed] [Google Scholar]
- (42).Tugarinov V, Kanelis V, Kay LE. Isotope Labeling Strategies for the Study of High-Molecular-Weight Proteins by Solution NMR Spectroscopy. Nat Protoc. 2006;1(2):749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- (43).Kizilsavas G, Ledolter K, Kurzbach D. Hydrophobic Collapse of the Intrinsically Disordered Transcription Factor Myc Associated Factor X. Biochemistry. 2017;56(40):5365–5372. doi: 10.1021/acs.bio-chem.7b00679. [DOI] [PubMed] [Google Scholar]
- (44).Ragavan M, Iconaru LI, Park CG, Kriwacki RW, Hilty C. Real-Time Analysis of Folding upon Binding of a Disordered Protein by Using Dissolution DNP NMR Spectroscopy. Angew Chem Int Ed. 2017;56(25):7070–7073. doi: 10.1002/anie.201700464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Karlsson M, Ardenkjær-Larsen JH, Lerche MH. Hyperpolarized 133 Cs Is a Sensitive Probe for Real-Time Monitoring of Biophysical Environments. Chem Commun. 2017;53(49):6625–6628. doi: 10.1039/C7CC02943H. [DOI] [PubMed] [Google Scholar]
- (46).Kurzbach D, Schwarz TC, Platzer G, Höfler S, Hinderberger D, Konrat R. Compensatory Adaptations of Structural Dynamics in an Intrinsically Disordered Protein Complex. Angew Chem Int Ed Engl. 2014;53(15):3840–3843. doi: 10.1002/anie.201308389. [DOI] [PubMed] [Google Scholar]
- (47).Lerche MH, Yigit D, Frahm AB, Ardenkjær-Larsen JH, Malinowski RM, Jensen PR. Stable Isotope-Resolved Analysis with Quantitative Dissolution Dynamic Nuclear Polarization. Anal Chem. 2018;90(1):674–678. doi: 10.1021/acs.analchem.7b02779. [DOI] [PubMed] [Google Scholar]
- (48).Dumez JN, Milani J, Vuichoud B, Bornet A, Lalande-Martin J, Tea I, Yon M, Maucourt M, Deborde C, Moing A, et al. Hyperpolarized NMR of Plant and Cancer Cell Extracts at Natural Abundance. The Analyst. 2015;140(17):5860–5863. doi: 10.1039/C5AN01203A. [DOI] [PubMed] [Google Scholar]
- (49).Bornet A, Maucourt M, Deborde C, Jacob D, Milani J, Vuichoud B, Ji X, Dumez JN, Moing A, Bodenhausen G, et al. Highly Repeatable Dissolution Dynamic Nuclear Polarization for Heteronuclear NMR Metabolomics. Anal Chem. 2016;88(12):6179–6183. doi: 10.1021/acs.anal-chem.6b01094. [DOI] [PubMed] [Google Scholar]
- (50).Harris T, Eliyahu G, Frydman L, Degani H. Kinetics of Hyperpolarized 13C1-Pyruvate Transport and Metabolism in Living Human Breast Cancer Cells. Proc Natl Acad Sci. 2009;106(43):18131–18136. doi: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sadet A, Weber EMM, Jhajharia A, Kurzbach D, Bodenhausen G, Miclet E, Abergel D. Rates of Chemical Reactions Embedded in a Metabolic Network by Dissolution Dynamic Nuclear Polarisation NMR. Chem Eur J. 2018;24(21):5456–5461. doi: 10.1002/chem.201705520. [DOI] [PubMed] [Google Scholar]
- (52).Nardi-Schreiber A, Gamliel A, Harris T, Sapir G, Sosna J, Gomori JM, Katz-Brull R. Biochemical Phosphates Observed Using Hyperpolarized 31P in Physiological Aqueous Solutions. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-00364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Weber EMM, Loh E, Kress T, Bodenhausen G, Abergel D, Sewsurn S, Azaïs T, Kurzbach D. Real-Time Monitoring of Calcium Phosphate Pre-Nucleation Clusters for-Mation by Dissolution Dynamic Nuclear Polarization. 2019 [Google Scholar]
- (54).Lee Y, Heo GS, Zeng H, Wooley KL, Hilty C. Detection of Living Anionic Species in Polymerization Reactions Using Hyperpolarized NMR. J Am Chem Soc. 2013;135(12):4636–4639. doi: 10.1021/ja4001008. [DOI] [PubMed] [Google Scholar]
- (55).Miclet E, Abergel D, Bornet A, Milani J, Jannin S, Bodenhausen G. Toward Quantitative Measurements of Enzyme Kinetics by Dissolution Dynamic Nuclear Polarization. J Phys Chem Lett. 2014;5(19):3290–3295. doi: 10.1021/jz501411d. [DOI] [PubMed] [Google Scholar]
- (56).Zeng H, Bowen S, Hilty C. Sequentially Acquired Two-Dimensional NMR Spectra from Hyperpolarized Sample. J Magn Reson. 2009;199(2):159–165. doi: 10.1016/j.jmr.2009.04.011. [DOI] [PubMed] [Google Scholar]
- (57).Frydman L, Blazina D. Ultrafast Two-Dimensional Nuclear Magnetic Resonance Spectroscopy of Hyperpolarized Solutions. Nat Phys. 2007;3(6):415–419. doi: 10.1038/nphys597. [DOI] [Google Scholar]
- (58).Giraudeau P, Shrot Y, Frydman L. Multiple Ultrafast, Broadband 2D NMR Spectra of Hyperpolarized Natural Products. J Am Chem Soc. 2009;131(39):13902–13903. doi: 10.1021/ja905096f. [DOI] [PubMed] [Google Scholar]
- (59).Mishkovsky M, Frydman L. Progress in Hyperpolarized Ultrafast 2D NMR Spectroscopy. ChemPhysChem. 2008;9(16):2340–2348. doi: 10.1002/cphc.200800461. [DOI] [PubMed] [Google Scholar]
- (60).Guduff L, Kurzbach D, van Heijenoort C, Abergel D, Dumez JN. Single-Scan 13 C Diffusion-Ordered NMR Spectroscopy of DNP-Hyperpolarised Substrates. Chem - Eur J. 2017;23(66):16722–16727. doi: 10.1002/chem.201703300. [DOI] [PubMed] [Google Scholar]
- (61).Bowen S, Zeng H, Hilty C. Chemical Shift Correlations from Hyperpolarized NMR by Off-Resonance Decoupling. Anal Chem. 2008;80(15):5794–5798. doi: 10.1021/ac8004567. [DOI] [PubMed] [Google Scholar]
- (62).Bowen S, Hilty C. Temporal Chemical Shift Correlations in Reactions Studied by Hyperpolarized Nuclear Magnetic Resonance. Anal Chem. 2009;81(11):4543–4547. doi: 10.1021/ac900456q. [DOI] [PubMed] [Google Scholar]
- (63).Chen C-H, Shih W-C, Hilty C. In Situ Determination of Tacticity, Deactivation, and Kinetics in [Rac-(C2H4 (1-Indenyl)2)ZrMe][B(C6F5)4] and [Cp2ZrMe][B(C6F5)4]-Catalyzed Polymerization of 1-Hexene Using 13 C Hyperpolarized NMR. J Am Chem Soc. 2015;137(21):6965–6971. doi: 10.1021/jacs.5b04479. [DOI] [PubMed] [Google Scholar]
- (64).Krajewski M, Wespi P, Busch J, Wissmann L, Kwiatkowski G, Steinhauser J, Batel M, Ernst M, Kozerke S. A Multisample Dissolution Dynamic Nuclear Polarization System for Serial Injections in Small Animals: Multisample Dissolution DNP Polarizer for Small Animals. Magn Reson Med. 2017;77(2):904–910. doi: 10.1002/mrm.26147. [DOI] [PubMed] [Google Scholar]
- (65).Hu S, Larson PEZ, VanCriekinge M, Leach AM, Park I, Leon C, Zhou J, Shin PJ, Reed G, Keselman P, et al. Rapid Sequential Injections of Hyperpolarized [1-13C]Pyruvate in Vivo Using a Sub-Kelvin, Multi-Sample DNP Polarizer. Magn Reson Imaging. 2013;31(4):490–496. doi: 10.1016/j.mri.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Batel M, Krajewski M, Däpp A, Hunkeler A, Meier BH, Kozerke S, Ernst M. Dissolution Dynamic Nuclear Polarization Efficiency Enhanced by Hartmann–Hahn Cross Polarization. Chem Phys Lett. 2012;554:72–76. doi: 10.1016/j.cplett.2012.10.018. [DOI] [Google Scholar]
- (67).Bornet A, Melzi R, Jannin S, Bodenhausen G. Cross Polarization for Dissolution Dynamic Nuclear Polarization Experiments at Readily Accessible Temperatures 1.2 < T < 4.2 K. Appl Magn Reson. 2012;43(1–2):107–117. doi: 10.1007/s00723-012-0358-1. [DOI] [Google Scholar]
- (68).Vuichoud B, Milani J, Bornet A, Melzi R, Jannin S, Bodenhausen G. Hyperpolarization of Deuterated Metabolites via Remote Cross-Polarization and Dissolution Dynamic Nuclear Polarization. J Phys Chem B. 2014;118(5):1411–1415. doi: 10.1021/jp4118776. [DOI] [PubMed] [Google Scholar]
- (69).Bornet A, Milani J, Vuichoud B, Perez Linde AJ, Bodenhausen G, Jannin S. Microwave Frequency Modulation to Enhance Dissolution Dynamic Nuclear Polarization. Chem Phys Lett. 2014;602:63–67. doi: 10.1016/j.cplett.2014.04.013. [DOI] [Google Scholar]
- (70).Bornet A, Pinon A, Jhajharia A, Baudin M, Ji X, Emsley L, Bodenhausen G, Ardenkjaer-Larsen JH, Jannin S. Microwave-Gated Dynamic Nuclear Polarization. Phys Chem Chem Phys. 2016;18(44):30530–30535. doi: 10.1039/C6CP05587G. [DOI] [PubMed] [Google Scholar]
- (71).Lesage A, Lelli M, Gajan D, Caporini MA, Vitzthum V, Miéville P, Alauzun J, Roussey A, Thieuleux C, Mehdi A, et al. Surface Enhanced NMR Spectroscopy by Dynamic Nuclear Polarization. J Am Chem Soc. 2010;132(44):15459–15461. doi: 10.1021/ja104771z. [DOI] [PubMed] [Google Scholar]
- (72).Rosay M, Tometich L, Pawsey S, Bader R, Schauwecker R, Blank M, Borchard PM, Cauffman SR, Felch KL, Weber RT, et al. Solid-State Dynamic Nuclear Polarization at 263 GHz: Spectrometer Design and Experimental Results. Phys Chem Chem Phys. 2010;12(22):5850. doi: 10.1039/c003685b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Shimon D, Feintuch A, Goldfarb D, Vega S. Static 1 H Dynamic Nuclear Polarization with the Biradical TOTAPOL: A Transition between the Solid Effect and the Cross Effect. Phys Chem Chem Phys. 2014;16(14):6687–6699. doi: 10.1039/C3CP55504F. [DOI] [PubMed] [Google Scholar]
- (74).Eills J, Alonso-Valdesueiro J, Salazar Marcano DE, Ferreira da Silva J, Alom S, Rees GJ, Hanna JV, Carravetta M, Levitt MH. Preservation of Nuclear Spin Order by Precipitation. Chem Phys Chem. 2018;19(1):40–44. doi: 10.1002/cphc.201701189. [DOI] [PubMed] [Google Scholar]
- (75).Mammoli D, Salvi N, Milani J, Buratto R, Bornet A, Sehgal AA, Canet E, Pelupessy P, Carnevale D, Jannin S, et al. Challenges in Preparing, Preserving and Detecting Para-Water in Bulk: Overcoming Proton Exchange and Other Hurdles. Phys Chem Chem Phys. 2015;17(40):26819–26827. doi: 10.1039/C5CP03350K. [DOI] [PubMed] [Google Scholar]
- (76).Meier B, Kouřil K, Bengs C, Kouřilová H, Barker TC, Elliott SJ, Alom S, Whitby RJ, Levitt MH. Spin-Isomer Conversion of Water at Room Temperature and Quantum-Rotor-Induced Nuclear Polarization in the Water-Endofullerene H 2 O @ C 60. Phys Rev Lett. 2018;120(26) doi: 10.1103/PhysRevLett.120.266001. [DOI] [PubMed] [Google Scholar]
- (77).Dumez J-N, Håkansson P, Mamone S, Meier B, Stevanato G, Hill-Cousins JT, Roy SS, Brown RCD, Pileio G, Levitt MH. Theory of Long-Lived Nuclear Spin States in Methyl Groups and Quantum-Rotor Induced Polarisation. J Chem Phys. 2015;142(4):044506. doi: 10.1063/1.4906273. [DOI] [PubMed] [Google Scholar]
- (78).Jhajharia A, Weber EMM, Kempf JG, Abergel D, Bodenhausen G, Kurzbach D. Communication: Dissolution DNP Reveals a Long-Lived Deuterium Spin State Imbalance in Methyl Groups. J Chem Phys. 2017;146(4):041101. doi: 10.1063/1.4974358. [DOI] [PubMed] [Google Scholar]
- (79).Vuichoud B, Canet E, Milani J, Bornet A, Baudouin D, Veyre L, Gajan D, Emsley L, Lesage A, Copéret C, et al. Hyperpolarization of Frozen Hydrocarbon Gases by Dynamic Nuclear Polarization at 1.2 K. J Phys Chem Lett. 2016;7(16):3235–3239. doi: 10.1021/acs.jpclett.6b01345. [DOI] [PubMed] [Google Scholar]
- (80).Hirsch ML, Smith BA, Mattingly M, Goloshevsky AG, Rosay M, Kempf JG. Transport and Imaging of Brute-Force 13 C Hyperpolarization. J Magn Reson. 2015;261:87–94. doi: 10.1016/j.jmr.2015.09.017. [DOI] [PubMed] [Google Scholar]
- (81).Ji X, Bornet A, Vuichoud B, Milani J, Gajan D, Rossini AJ, Emsley L, Bodenhausen G, Jannin S. Transportable Hyperpolarized Metabolites. Nat Commun. 2017;8:13975. doi: 10.1038/ncomms13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Capozzi A, Cheng T, Boero G, Roussel C, Comment A. Thermal Annihilation of Photo-Induced Radicals Following Dynamic Nuclear Polarization to Produce Transportable Frozen Hyperpolarized 13C-Substrates. Nat Commun. 2017;8:15757. doi: 10.1038/ncomms15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gajan D, Bornet A, Vuichoud B, Milani J, Melzi R, van Kalkeren HA, Veyre L, Thieuleux C, Conley MP, Gruning WR, et al. Hybrid Polarizing Solids for Pure Hyperpolarized Liquids through Dissolution Dynamic Nuclear Polarization. Proc Natl Acad Sci. 2014;111(41):14693–14697. doi: 10.1073/pnas.1407730111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Jannin S, Helm L, Bodenhausen G. Kinetics of Yttrium–Ligand Complexation Monitored Using Hyperpolarized 89 Y as a Model for Gadolinium in Contrast Agents. J Am Chem Soc. 2010;132(14):5006–5007. doi: 10.1021/ja1013954. [DOI] [PubMed] [Google Scholar]
- (85).Ivanov KL, Kress T, Baudin M, Guarin D, Abergel D, Bodenhausen G, Kurzbach D. Relaxation of Long-Lived Modes in NMR of Deuterated Methyl Groups. J Chem Phys. 2018;149(5):054202. doi: 10.1063/1.5031177. [DOI] [PubMed] [Google Scholar]