Abstract
Objectives
The objectives of this study were to confirm the prevalence of feline immunodeficiency virus (FIV) infection in domestic cats in the region north of Ceará, Brazil, and to determine the factors associated with infection and the major circulating subtypes of the virus in this area.
Methods
Samples from 148 cats were collected and tested using anti-FIV antibody screening, with confirmation of positive results by PCR. Univariate analysis was performed considering the epidemiological characteristics and FIV status. Sequencing and phylogenetic analysis of the gag and pol genes were performed to confirm the FIV subtype.
Results
Nine cats (6.1%) tested positive for FIV – one female (0.7%) and eight males (5.4%). Male cats were significantly more likely to be infected (P <0.05). Phylogenetic analysis of gag and pol gene sequences indicated that the FIV isolates circulating in the study area belonged to subtype B.
Conclusions and relevance
In this study, we demonstrated a low prevalence for FIV in the northwest of Ceará, north-eastern Brazil. Male sex is a significant risk factor for FIV infection and the best predictive factor for FIV status. All isolates examined in this study clustered within subtype B, which is the predominant subtype in Brazil. This is the first report of genetic characterization of FIV in the state of Ceará, Brazil.
Keywords: Retrovirus, feline immunodeficiency virus, subtype B, domestic cat, Ceará, north-eastern Brazil
Introduction
Feline immunodeficiency virus (FIV) is a member of the genus Lentivirus of the Retroviridae family.1 FIV is an important viral pathogen that infects domestic and wild cats and can cause a slow, progressive degeneration of the immune system, which eventually leads to a disease comparable to AIDS in humans.2,3 The major characteristic of FIV infection is the CD4+ T-cell loss and reduced CD4+/CD8+ ratios that precede the development of immunodeficiency in a proportion of infected cats. FIV has been studied widely as both an important veterinary pathogen and an animal model for HIV/AIDS.1,4 The outcome of infection depends on the balance between the viral destruction of the immune system and the ability of the remaining immune system to eliminate the virus.
The virus persists in the cat host through active replication and is able to avoid clearance by the immune system. Active replication is an important component of the rapid evolutionary potential of FIV, a potential that manifests itself in the evolution of immune escape variants, drug-resistant variants and variants with the ability to use cell surface receptors in a different way.5,6 Lentiviruses are complex retroviruses containing accessory genes that encode regulatory proteins, in addition to three large open reading frames, gag, pol and env. Isolates can be grouped into at least five distinct clades, according to variations; the diversity in the env gene sequence (range of average diversity of subtypes 15–30%) can also influence their pathogenicity. These are designated A–E and recombinant strains,7–9 a number that can be expected to increase as further studies reveal additional diversity.10,11 Subsequently, studies have shown that it is possible to perform genetic classification for FIV based on the nucleotide sequence of gag, which is useful as a simple method for FIV subtyping.12–14 Although less frequently used, pol can be used to confirm the genetic classification of FIV,15 and, recently, inter- and intragenic recombination of FIV subtypes were detected when gag, pol and env were studied.16
FIV infection occurs in domestic cats worldwide, with prevalence rates varying from 1–44% in different regions.2,17,18 Outdoor, adult, male, sick cats are at increased risk of infection, because transmission occurs following bites from infected animals.19 Although FIV was first recognized in 1993 in Brazil,20 there are few data describing the prevalence, ecology, clinical aspects or genetic analyses of FIV in the country. It is important to emphasize that Brazil is a country with great territorial extension. Here, the reported prevalence estimates range from 2–37.5%. Preliminary studies suggested that FIV infection is widespread in the domestic cat population of Brazil, predominantly with subtype B, with a single report of subtype A circulation in cats in Maranhão, north-eastern Brazil,5,21–24 but little is known about the prevalence and phylogeny of FIV in most states in the northeast region of Brazil.5,25 Finally, a better characterization of FIV strains circulating within Brazil is required to augment our understanding of the significance of FIV in felids in this country.
The aim of this study was to confirm the prevalence of FIV infection in domestic cats in the state of Ceará, Brazil, as well as to determine the factors associated with infection and the major circulating subtypes of the virus in the study area.
Materials and methods
Ethical Aproval
The present study was submitted to, and approved by, the Ethics Committee of Animal Experimentation and Animal Welfare of Federal University of Minas Gerais (CEUA/UFMG - 315/2014).
Animals and samples
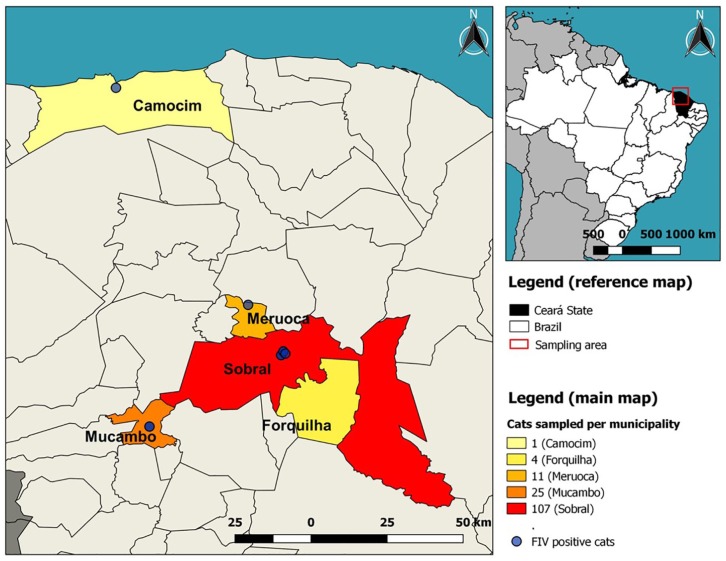
This study was carried out in north region of Ceará, north-eastern Brazil (Figure 1). Samples from 148 domestic shorthair cats were collected from 2014–2015. The cats in this study consisted of client-owned cats recruited through participating veterinary clinics in Sobral, Ceará. From this total, 63 cats came from four animal shelters and 12 from the zoonosis control center of Sobral. To evaluate factors associated with FIV infection, such as age, sex, sterilization and outdoor access, a questionnaire was completed for each cat and all cats underwent a general physical and clinical examination. Blood samples were obtained by jugular venipuncture using 25 G needles and 2 ml vacuum tubes with and without anticoagulant EDTA, following handling and supervision protocols, and with the owner’s consent. All blood samples without anticoagulant were centrifuged for 10 mins at 1200 × g to obtain serum. DNA extraction was performed using DNeasy Blood and Tissue Kit following the manufacturer’s instructions from blood samples in anticoagulant (Qiagen). Serum and DNA were stored at −80°C until diagnostic testing was performed.
Figure 1.
The study area in north-west Ceará, north-eastern Brazil. Colors indicate the number of cats sampled per municipality and feline immunodeficiency virus-positive cats are highlighted by the blue circles
Diagnostic tests and molecular characterization
The presence of FIV infection in cats was determined using the SNAP FIV/FeLV Combo Test (IDEXX Laboratories). Seropositive samples were tested using a PCR-based confirmatory test that amplified gag.13 Gag and pol were used for phylogenetic analysis.
A semi-nested PCR using a forward primer targeting the primer biding site of the FIV genome and primers described by Rosati et al,26 without restriction sites and transmembrane epitope sequences, were used to amplify the gag region, which encodes the entire FIV capsid (p24). The PCR reaction was performed in a total volume of 25 µl, containing 1 × Go Taq Mastermix (Promega), 10 pmol of each primer (FIVpbs and FIVP24rv) and 2.5 µl of extracted DNA. Amplification conditions were 94°C for 3 mins, 40 cycles at 94°C for 15 s, 52°C for 30 s and 72°C for 1 min and 30 s, followed by a final extension at 72°C for 10 mins. The semi-nested PCR was performed in a total of 25 µl, containing 1 × Go Taq Mastermix (Promega), 10 pmol of each primer (FIVP24fw and FIVP24rv) and 1 µl of PCR product. Amplification conditions were 94°C for 3 mins, 40 cycles of 94°C for 15 s, 50°C for 30 s and 72°C for 45 s, followed by a final extension at 72°C for 10 mins. A partial pol sequence was amplified using outer primers P1F/P2R and inner primers P2F/P1R, as described by Troyer et al.27 Ultrapure water was used as a negative control and DNA from a naturally infected cat was used as a positive control (samples provided by Professor Margaret J. Hosie’s laboratory).
The p24 or pol gene amplicons (669 or 577 bp, respectively) were purified from agarose gels using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare), according the manufacturer’s recommendations. Bidirectional sequencing was performed using an BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI 3500 sequencer (Applied Biosystems). The quality of the sequences was determined using Phred Electropherogram Quality Analysis software (available at http://asparagin.cenargen.embrapa.br/phph),28 and chromatograms were checked manually and edited with MEGA program version 5.2.2.29 Final sequence was obtained with Cap-Contig application of the Bioedit Sequence Alignment Editor program version 7.2.5.30
The FIV p24 and pol nucleotide sequences obtained in this study were aligned with FIV sequences retrieved from GenBank, using the CLUSTAL/W method implemented in MEGA program version 5.2.2.29 Phylogenetic trees for FIV nucleotide sequences were constructed with MEGA 5 program, using the maximum likelihood method and Hasegawa–Kishino–Yano evolutive model with 1000 bootstrap replicates and gamma distribution. Values for nucleotide identity were calculated using the Bioedit Sequence Alignment Editor program version 7.2.5.30 The FIV nucleotide sequences of this data set have been deposited in the GenBank data base under accession numbers included in Figures 2 and 3.
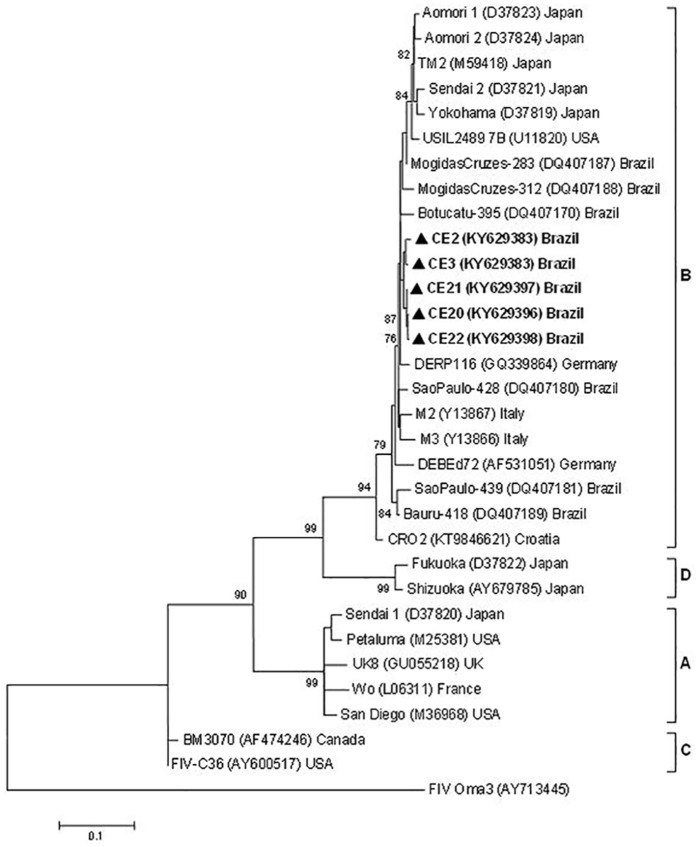
Figure 2.
Maximum likelihood tree based on 459 nucleotide alignment of 32 feline immunodeficiency virus (FIV) gag sequences. The phylogenetic tree for FIV nucleotide sequences was constructed with the MEGA 5 program, using the maximum likelihood method and the Hasegawa–Kishino–Yano evolutive model with 1000 bootstrap replicates and gamma distribution. FIV Oma was used as the outgroup, and the triangle indicates FIV sequences from Ceará. Bootstrap values >70 are shown at the nodes. Bar represents the number of substitutions per site
Figure 3.
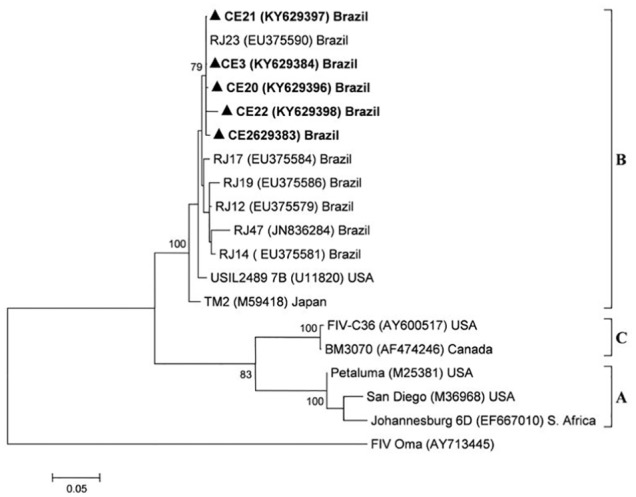
Phylogenetic tree based on 504 nucleotide alignment of 19 feline immunodeficiency virus (FIV) pol sequences. The tree was constructed with the MEGA 5 program, using the maximum likelihood method and the Hasegawa–Kishino–Yano evolutive model with 1000 bootstrap replicates and gamma distribution. FIV Oma was used as the outgroup, and the triangle indicates FIV sequences from Ceará. Bootstrap values >70 are shown at the nodes. Bar represents substitutions number per site
Statistical analysis
A questionnaire was created using EpiInfo software to gather and analyze data that could help to describe epidemiologically the FIV presence (age categories [young, adult and senior; sex; outdoor/street access; neutering status; presence or absence of clinical signs [weight loss, lymphadenopathy, gingivitis, stomatitis, diarrhea, vomiting, fever, anorexia, nasal discharge, coughing, sneezing, ocular discharge, conjunctivitis, otitis, behavioral changes, alopecia, pruritus and changes in the urinary system]; deworming; rabies vaccine; panleukopenia, rhinotracheitis and calicivirus triple vaccine). The association of all variables with the presence of FIV was assessed with the χ2 test and odds ratio (OR). All analyses were performed according a minimal significance level of 5% (P <0.05). The results were interpreted with the help of graphics and tables elaborated in EPI INFO version 3.5.2.
Results
Of 148 cats analyzed, nine (6.1%) tested positive for FIV infection (eight males and one female) (Figure 4). Univariate analysis of the associations between the epidemiological and animal characteristics and positive tests for FIV infection revealed that male cats were significantly more likely to be infected (χ2 = 7.86; P = 0.005; OR = 11.5). Phylogenetic analysis of gag and pol sequences from five samples indicated that FIV isolates circulating in the study area belonged to subtype B, with 98.6–100% and 98.4–100% of nucleotide identity, respectively. FIV gag sequences showed close phylogenetic relationships with other sequences from Brazil (95.8–98.4% nucleotide identity) and Germany (98% nucleotide identity) (Figure 2). FIV pol sequences showed close phylogenetic relationship to isolates from Brazil, the USA and Japan (100–98.4% and 98.4–97% nucleotide identity, respectively) (Figure 3).
Figure 4.
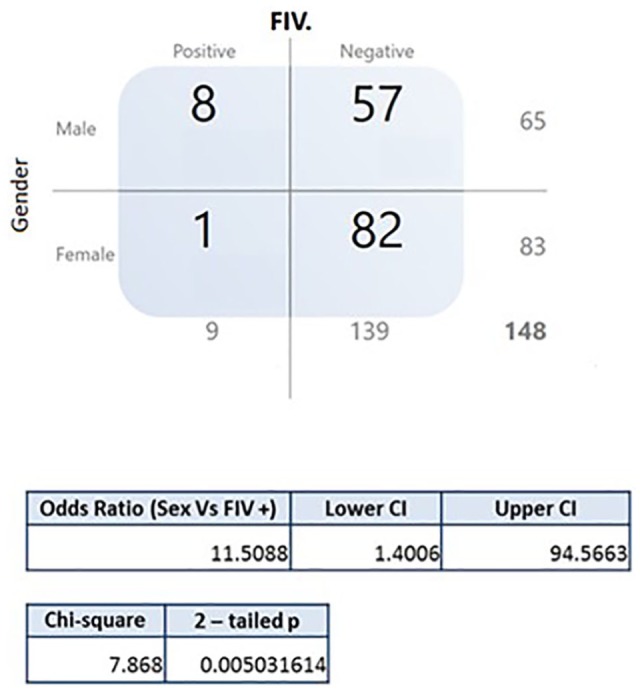
Association of the epidemiological and animal characteristics and positivity for feline immunodeficiency virus (FIV). Positive animals and the variable of sex (P <0.05). CI = confidence interval
Discussion
FIV infection is a serious issue in feline medicine, typically involving chronic immune dysfunction and opportunistic infections. FIV occurs worldwide, with prevalence estimates of 1–14% in cats with no clinical signs and up to 44% in sick cats.2 The overall FIV prevalence determined for the domestic cat population described here (6.1%) is lower than most previous reports for naturally infected cats in Brazil.23,31–33 However, the prevalence observed in the present study is similar to studies performed in Minas Gerais and Bahia.21,25 Nevertheless, it is not possible to compare the prevalence reported in these studies because of the differences in the cat populations studied and the different diagnostic methods used in each study. Studies addressing the prevalence and epidemiology of FIV have been mainly limited to southern and south-eastern Brazil.5,23,31,33 No studies have examined the prevalence and epidemiology of FIV within feline populations in Ceará, north-eastern Brazil. It is important to emphasize that we expected to find a higher prevalence of FIV infection, given that these cats may have frequent interaction with other cats, owing the lifestyle of domestic cats in the study area, where most cats are not neutered and have outdoor access. More studies of naturally infected cats are needed to determine whether a lower prevalence could, indeed, be a characteristic of the state of Ceará.
Factors such as age, sex, behavior, lifestyle, type of habitat and health status have been shown to be associated with risk of FIV infection.2,18 As the number of FIV-positive cats identified was small, no significant associations between disease and risk factors were observed, with the exception of sex (OR 11.5, χ2= 7.86, P = 0.005; Figure 4), although many risk factors occurred in the majority of the infected population, such as not being castrated (n = 6/9) and having access to the streets (n = 6/9). FIV-infected cats were in relatively good health, with no specific clinical abnormalities, compared with the uninfected cats. Thus, the positive cats were considered to be in the asymptomatic phase of infection. Again, further studies examining a larger number of animals from Ceará are needed to better characterize the risks of FIV infection.
As a complementary study, nested PCR was performed on all positive samples. All seropositive cats tested positive by both PCR methods used, amplifying gag and pol. All PCR products were sequenced, although only five samples gave rise to sequence data that could be analyzed phylogenetically. As independent FIV isolates were identified and characterized, it became evident that genomic heterogeneity existed among different isolates.7,9 Based on the diversity of the V3–V5 region of env, FIV is currently classified into five subtypes, A–E.7–9 Within a subtype, there can exist sub-subtypes, which are distinct lineages that are very closely related to one subtype.22 Preliminary studies have suggested that subtype B is predominant in the domestic cat population of Brazil, but confirmation of the circulating subtype(s) is essential owing the extension of the country and lack of data covering the whole territory.
Results from this first phylogenetic analysis of FIV sequences from the state of Ceará, Brazil, revealed that they clustered within subtype B and were closely related phylogenetically with Germany isolates (gag sequences; Figure 2) and isolates from the USA and Japan (pol sequences; Figure 3).
Conclusions
Previously, there have been no reports of the genetic characterization of FIV in the state of Ceará, Brazil. Here, we demonstrate a low prevalence of FIV infection in the state of Ceará, north-eastern Brazil. Male sex is a significant risk factor for FIV infection and the best predictive factor for FIV status. All isolates examined in this study clustered within subtype B, which is the predominant subtype in Brazil. It would be interesting to test a larger number of the cat population in the coming years to better understand the epidemiology and circulation of FIV strains in Ceará, Brazil.
Acknowledgments
We would like to thank all members of Hospital Veterinário de Pequenos Animais do Centro Universitário INTA - UNINTA - for their full support during this study. Thanks are also due to all members of Projeto José de Melo for their delightful cooperation and support.
Footnotes
Accepted: 13 May 2019
Conflict of interest: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was supported by the CNPq (Brazil’s National Research Council), [401617 2012-2].
ORCID iD: Poliana Marisa Miranda Menezes  https://orcid.org/0000-0003-2538-5800
https://orcid.org/0000-0003-2538-5800
References
- 1. Pedersen NC, Ho EW, Brown ML, et al. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 1987; 235: 790–793. [DOI] [PubMed] [Google Scholar]
- 2. Hosie MJ, Addie D, Belák s, et al. Feline immunodeficiency. ABCD guidelines on prevention and management. J Feline Med Surg 2009; 11: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O’Brien SJ, Troyer JL, Brown MA, et al. Emerging viruses in the Felidae: shifting paradigms. Viruses 2012; 4: 236–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamamoto JK, Pu R, Sato E, et al. Feline immunodeficiency virus pathogenesis and development of a dual-subtype feline-immunodeficiency-virus vaccine. AIDS 2007; 21: 547–563. [DOI] [PubMed] [Google Scholar]
- 5. Martins AN, Medeiros SO, Simonetti JP, et al. Phylogenetic and genetic analysis of feline immunodeficiency virus gag, pol, and env genes from domestic cats undergoing nucleoside reverse transcriptase inhibitor treatment or treatment-naive cats in Rio de Janeiro, Brazil. J Virol 2008; 82: 7863–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Willett BJ, Kraase M, Logan N, et al. Modulation of the virus-receptor interaction by mutations in the V5 loop of feline immunodeficiency virus (FIV) following in vivo escape from neutralising antibody. Retrovirology 2010; 7: 38 DOI: 10.1186/1742-4690-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kakinuma S, Motokawa K, Hohdatsu T, et al. Nucleotide sequence of feline immunodeficiency virus: classification of Japanese isolates into two subtypes which are distinct from non-Japanese subtypes. J Virol 1995; 69: 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pecoraro MR, Tomonaga K, Miyazawa T, et al. Genetic diversity of Argentine isolates of feline immunodeficiency virus. J Gen Virol 1996; 77: 2031–2035. [DOI] [PubMed] [Google Scholar]
- 9. Sodora DL, Shpaer EG, Kitchell BE, et al. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J Virol 1994; 68: 2230–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayward JJ, Taylor J, Rodrigo AG. Phylogenetic analysis of feline immunodeficiency virus in feral and companion domestic cats of New Zealand. J Virol 2007; 81: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weaver EA, Collisson EW, Slater M, et al. Phylogenetic analyses of Texas isolates indicate an evolving subtype of the clade B feline immunodeficiency viruses. J Virol 2004; 78: 2158–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duarte A, Tavares L. Phylogenetic analysis of Portuguese feline immunodeficiency virus sequences reveals high genetic diversity. Vet Microbiol 2006; 114: 25–33. [DOI] [PubMed] [Google Scholar]
- 13. Hohdatsu T, Motokawa K, Usami M, et al. Genetic subtyping and epidemiological study of feline immunodeficiency virus by nested polymerase chain reaction-restriction fragment length polymorphism analysis of the gag gene. J Virol Methods 1998; 70: 107–111. [DOI] [PubMed] [Google Scholar]
- 14. Steinrigl A, Klein D. Phylogenetic analysis of feline immunodeficiency virus in Central Europe: a prerequisite for vaccination and molecular diagnostics. J Gen Virol 2003; 84: 1301–1307. [DOI] [PubMed] [Google Scholar]
- 15. Carpenter MA, Brown EW, MacDonald DW, et al. Phylogeographic patterns of feline immunodeficiency virus genetic diversity in the domestic cat. Virology 1998; 251: 234–243. [DOI] [PubMed] [Google Scholar]
- 16. Hayward JJ, Rodrigo AG. Recombination in feline immunodeficiency virus from feral and companion domestic cats. Virol J 2008; 5: 76 DOI: 10.1186/1743-422X-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bandecchi P, Dell’Omodarme M, Magi M, et al. Feline leukaemia virus (FeLV) and feline immunodeficiency virus infections in cats in the Pisa district of Tuscany, and attempts to control FeLV infection in a colony of domestic cats by vaccination. Vet Rec 2006; 158: 555–557. [DOI] [PubMed] [Google Scholar]
- 18. Levy J, Crawford C, Hartmann K, et al. 2008 American Association of Feline Practitioners’ feline retrovirus management guidelines. J Feline Med Surg 2008; 10: 300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hartmann K. Feline immunodeficiency virus infection: an overview. Vet J 1998; 155: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagiwara MK, Reche Junior A, Lucas SRR, et al. Feline immunodeficiency virus infection in cats from São Paulo, Brazil. Brazi J Vet Res Anim Sci 1993; 30: 217–220. [Google Scholar]
- 21. Caxito FA, Coelho FM, Oliveira ME, et al. Feline immunodeficiency virus subtype B in domestic cats in Minas Gerais, Brazil. Vet Res Commun 2006; 30: 953–956. [DOI] [PubMed] [Google Scholar]
- 22. Lara VM, Taniwaki as, Araújo JP., Jr. Phylogenetic characterization of feline immunodeficiency virus (FIV) isolates from the state of São Paulo. Pesq Vet Bras 2007; 27: 467–470. [Google Scholar]
- 23. Reche A, Jr, Hagiwara MK, Lucas SRR. Clinical study of acquired immunodeficiency syndrome in domestic cats in São Paulo. Brazil J Vet Res Anim Sci 1997; 34: 152–155. [Google Scholar]
- 24. Martins N dos S, Rodrigues AP de S, da Luz LA, et al. Feline immudeficiency virus subtypes B and A in cats from São Luis, Maranhão, Brazil. Arch Virol 2018; 163: 549–554. [DOI] [PubMed] [Google Scholar]
- 25. Lacerda LC, Silva AN, Freitas JS, et al. Feline immuno‑deficiency virus and feline leukemia virus: Frequency and associated factors in cats in northeastern Brazil. Genet Mol Res 2017; 16: 1–8. [DOI] [PubMed] [Google Scholar]
- 26. Rosati S, Profiti M, Lorenzetti R, et al. Development of recombinant capsid antigen/transmembrane epitope fusion proteins for serological diagnosis of animal lentivirus infections. J Virol Methods 2004; 121: 73–78. [DOI] [PubMed] [Google Scholar]
- 27. Troyer JL, Pecon-slattery J, Roelke ME, et al. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol 2005; 79: 8282–8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Togawa RC, Brigido MM. PHPH: web based tool for simple electropherogram quality analysis. International Conference on Bioinformatics and Computational Biology; 2003. May 14–16; Ribeirão Preto, Brazil. [Google Scholar]
- 29. Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95–98. [Google Scholar]
- 31. Lara VM, Taniwaki SA, Júnior JPA. Occurrence of feline immunodeficiency virus infection in cats. Ciência Rural 2008; 38: 2245–2249. [Google Scholar]
- 32. Martins N, dos S, Rodrigues APS, Gonçalves SA, et al. Occurrence of feline immunodeficiency virus (FIV) and feline leukemia (FeLV) in São Luís-MA. Am J Anim Vet Sci 2015; 10: 187–192. [Google Scholar]
- 33. Silva FS, Castro CC, Finger P, et al. Ocorrência do subtipo B do vírus da imunodeficiência felina em gatos domésticos da região sul do estado do Rio Grande do Sul, Brasil. Arq Bras Med Vet Zootec 2014; 66: 1–6. DOI: 10.1590/S0102-09352014000100001. [Google Scholar]