Abstract
Plant growth promoting rhizobacteria (PGPR) are potential natural alternatives to chemical fungicides in greenhouse production via inducing plant immune system against biotic stresses. In this research, 126 Streptomyces isolates were recovered from rhizosphere soils of 13 different commercial vegetable greenhouses in Iran. Streptomyces isolates were screened for in vitro Plant growth promoting (PGP) traits and ability to antagonize Fusarium oxysporum f. sp. lycopersici race 3 (FOL), the causal agent of Fusarium wilt of tomato (FWT). Six isolates with the highest antagonistic activity and at least three PGP traits were selected and compared with chemical fungicide Carbendazim® in a greenhouse experiment. All bacterial treatments mitigated FWT disease symptoms like chlorosis, stunting and wilting at the same level or better than Carbendazim®. Strains IC10 and Y28 increased shoot length and shoot fresh and dry weight compared to not inoculated control plants. Phenotypic characterization and 16S rRNA gene sequencing showed, strains IC10 and Y28 were closely related to S. enissocaesilis and S. rochei, respectively. The ability of the superior biocontrol strains to induce antioxidant enzymes activity and systemic resistance (ISR) was investigated. Increased activity of catalase (CAT) in plant treated with both strains as well as an increase in peroxidase (POX) activity in plants treated with Y28 pointed to a strain specific-induced systemic resistance (ss-ISR) in tomato against FOL. The differential induced expression of WRKY70 and ERF1 (two transcription factors involved in plant defense) and LOX and TPX by the analyzed Streptomyces strains, especially after inoculation with FOL, suggests that ss-ISR is triggered at the molecular level.
Keywords: Fusarium wilt, induced systemic resistance, Streptomyces, tomato growth promoting, siderophore production
Introduction
In recent decades, overuse of chemical fertilizers and fungicides to proliferate and protect greenhouse vegetables has been a threat to human safety. Using biological agents is the best alternative for chemical treatments (Pilkington et al., 2010). Soil-borne fungus Fusarium oxysporum Schlecht. f. sp. lycopersici (Sacc.) Snyder and Hansen (FOL) causing Fusarium wilt of tomato reduces yield in greenhouses (Fravel et al., 2003; McGovern, 2015). Invading through the vascular tissue and soil- borne feature of the pathogen make it difficult to control this disease. Besides, new races of the mentioned pathogen (e.g., race 3) have emerged that can overcome host resistance (Reis et al., 2005). The symptoms of Fusarium wilt disease caused by race 3 are yellowing the lower leaves, vascular necrosis, epinasty, defoliation, plant stunting and ultimately plant death (Sanchez-Pena et al., 2010). Biological control of this pathogen has been the subject of many studies (Ben Abdallah et al., 2016). Plant growth promoting rhizobacteria (PGPR) colonize rhizosphere or plant root and improve plant health and growth. Some of the most important plant growth promoting (PGP) activity include diazotrophic nitrogen fixation, siderophore production, solubilization of mineral phosphates and production of hormones such as indole-3-acetic acid (IAA) (Sadeghi et al., 2012; Dahal et al., 2017; Gouda et al., 2018). Plant defense strategies including physical barriers, numerous secondary metabolites and antimicrobial agents, which the effective, aggressive pathogens have to overcome them (reviewed by Bruce and Pickett, 2007). Some plant defense strategies are constitutive while others are inducible and only launch in response to a stimulating pathogen and/or beneficial microbes (Pieterse et al., 2012). Plant defense strategies keep damage of specific pests below an economic threshold; however, maintain the beneficial organisms involved in integrated pest management (IPM) (Chellemi et al., 2016). IPM considers available pest control techniques that prevent the development of pest populations and keep pesticides to levels that are economically reasonable and reduces risks to human health and the environment. Developing sustainable biocontrol measures for managing Fusarium wilt disease requires a comprehensive understanding of the molecular basis of plant–pathogen–biocontrol interactions. Hormones, PR proteins, terpenoid synthases, polyketide terpene synthases, peroxidases, lignin synthases, transcription factors, calcium signal transducers, and UDP-glucosyltransferases and ubiquitin-protein ligases are components of the plant defense-related genes (Wang et al., 2015). Generally, the plant responses to microbes are regulated through signaling pathways including salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). Jasmonic acid pathway is required for defense against necrotrophic pathogens and chewing insects, while SA pathway is involved in a wide range of plant defense responses, which ends to systemic acquired resistance (SAR) and occurs following the exposure to many biotrophic and some necrotrophic pathogens. Induced systemic resistance (ISR) is also an activated resistance process elicited by contacting with non-pathogenic microorganisms. This procedure is independent of SA and is synchronized by JA and ET (Walters and Heil, 2007). Activated induced resistance (via ISR or SAR) is a broad-spectrum and long-term resistance, which usually suppresses a disease up to 20–85% (Walters et al., 2013). Thus, inducing plants by direct interaction with rhizobacteria prior to pathogen infection, so-called priming, decreases disease severity (Beckers and Conrath, 2007). Streptomyces species, are Gram-positive filamentous bacteria, reported as PGP and biocontrol agent of Alternaria alternata (Verma et al., 2011), Rhizoctonia solani (Goudjal et al., 2014), F. oxysporum (Kim et al., 2011), Phytophthora drechsleri (Sadeghi et al., 2017), and Verticillium dahliae (Cao et al., 2016). Little is known about bio-suppression of tomato wilt by Streptomyces in greenhouse conditions.
The aims of this study were to (1) isolate and characterize Streptomyces from vegetable greenhouse soils (2) detect PGP characteristics and antagonistic activity of isolates against FOL race (3) evaluate Fusarium wilt biocontrol in tomato by superior isolates in greenhouse condition (4) establish induced systemic resistance (ISR) of tomato through inducing antioxidant enzymes and defense-related genes by inoculation of plants with biocontrol Streptomyces.
Materials and Methods
Microorganisms
Rhizosphere soil samples (500 g soil with pH = 6.2–7 and EC < 2.5 dS/m) were collected from 18 cucumber and tomato commercial greenhouses in Yazd, Isfahan and Kerman provinces of Iran in 2016. There were no symptoms of wilt and damping off diseases developing in these greenhouses. The samples were placed in plastic bags, taken to the laboratory and then air-dried for 7 days. For isolation of Streptomyces, 2 g rhizosphere soil was suspended in 100 mL of sterile saline solution (0.9% NaCl) and shaken for 30 min. Two dilutions (1:100 and 1:1000) were prepared using sterile saline solutions in a final volume of 1 mL. An aliquot of 0.1 mL of each dilution was spread on 1.8% water agar supplemented with 300 mM NaCl. The plates were incubated at 29°C for 7 days. Representative colonies were selected and streaked on plates of “International Streptomyces Project media 2” (ISP2) medium (Kampfer et al., 1991) containing 10 g/L malt extract, 4 g/L yeast extract, 4 g/L glucose, and 18 g/L agar, adjusted to pH 7.2. The plates were incubated at 29°C for 7 days. Then morphologically (color, size, and shape) distinct colonies were stored in 30% glycerol solution at −70°C (Table 1).
Table 1.
PGP properties and enzyme activity of selected isolates.
| Isolate | Host crop | Location | PGP properties |
Hydrolytic enzymes activity |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Growth on N free medium | Inorganic P solubilization | Siderophore production | IAA production | Cellulase | Protease | Chitinase | |||
| IC6 | Cucumber | Isfahan | 1 | 0.1 ± 0.1 | 0.3 ± 0.1 | 9.2 ± 0.0 | 1.0 ± 0.0 | 0.3 ± 0.0 | 1.0 ± 0.0 |
| IC10 | Cucumber | Isfahan | 1 | 0.2 ± 0.0 | 0.4 ± 0.0 | 27.1 ± 0.1 | 0.0 ± 0.0 | 1.0 ± 0.0 | 0.9 ± 0.1 |
| IC13 | Cucumber | Isfahan | 1 | 0.3 ± 0.0 | 0.4 ± 0.0 | 32.3 ± 2.6 | 1.0 ± 0.0 | 0.6 ± 0.0 | 0.9 ± 0.1 |
| IS8 | Cucumber | Isfahan | 1 | 1.9 ± 0.1 | 0.4 ± 0.0 | 21.9 ± 0.9 | 0.0 ± 0.0 | 0.4 ± 0.0 | 1.0 ± 0.0 |
| SS12 | Cucumber | Isfahan | 1 | 0.1 ± 0.0 | 0.3 ± 0.0 | 24.1 ± 1.9 | 1.0 ± 0.0 | 0.5 ± 0.0 | 0.8 ± 0.1 |
| CU122 | Cucumber | Isfahan | 1 | 0.2 ± 0.0 | 0.1 ± 0.0 | 20.5 ± 0.7 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| IC15 | Cucumber | Isfahan | 1 | 0.3 ± 0.0 | 0.3 ± 0.0 | 9.6 ± 1.2 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.9 ± 0.0 |
| SS14 | Cucumber | Isfahan | 1 | 0.0 ± 0.0 | 0.3 ± 0.0 | 24.5 ± 0.7 | 1.0 ± 0.0 | 0.5 ± 0.0 | 0.0 ± 0.0 |
| IT20 | Tomato | Isfahan | 1 | 0.2 ± 0.0 | 0.4 ± 0.0 | 24.0 ± 1.3 | 1.0 ± 0.0 | 0.5 ± 0.0 | 0.0 ± 0.0 |
| IT8 | Tomato | Isfahan | 1 | 0.0 ± 0.0 | 0.4 ± 0.0 | 25.8 ± 0.5 | 1.0 ± 0.0 | 0.2 ± 0.0 | 0.0 ± 0.0 |
| IT25 | Tomato | Isfahan | 1 | 0.2 ± 0.0 | 0.5 ± 0.0 | 25.8 ± 1.8 | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.8 ± 0.2 |
| Y7 | Cucumber | Yazd | 1 | 0.5 ± 0.1 | 0.0 ± 0.0 | 7.7 ± 0.9 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Y18 | Cucumber | Yazd | 1 | 0.1 ± 0.0 | 0.0 ± 0.0 | 12.0 ± 0.6 | 1.0 ± 0.0 | 0.9 ± 0.1 | 0.8 ± 0.2 |
| Y27 | Tomato | Yazd | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 30.8 ± 1.1 | 1.3 ± 0.4 | 0.3 ± 0.0 | 0.0 ± 0.0 |
| TO612 | Tomato | Yazd | 1 | 0.0 ± 0.0 | 0.5 ± 0.0 | 24.7 ± 0.3 | 1.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Y17 | Tomato | Yazd | 1 | 0.0 ± 0.0 | 0.4 ± 0.0 | 13.7 ± 2.8 | 1.0 ± 0.0 | 0.4 ± 0.0 | 0.6 ± 0.3 |
| Y28 | Tomato | Yazd | 1 | 0.4 ± 0.0 | 0.3 ± 0.0 | 16.8 ± 2.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.9 ± 0.0 |
| Y281 | Tomato | Yazd | 1 | 0.3 ± 0.0 | 0.3 ± 0.0 | 10.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Y16 | Tomato | Yazd | 1 | 0.0 ± 0.0 | 0.4 ± 0.1 | 11.4 ± 0.5 | 1.4 ± 0.4 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Y33 | Tomato | Yazd | 1 | 0.2 ± 0.0 | 0.0 ± 0.0 | 9.3 ± 0.2 | 1.0 ± 0.0 | 0.3 ± 0.0 | 0.8 ± 0.2 |
| Y34 | Tomato | Yazd | 1 | 0.3 ± 0.1 | 0.3 ± 0.0 | 11.8 ± 0.9 | 1.2 ± 0.2 | 0.2 ± 0.0 | 0.7 ± 0.3 |
| KH12 | Cucumber | Kerman | 1 | 0.0 ± 0.0 | 0.1 ± 0.0 | 7.5 ± 0.7 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.0 ± 0.0 |
| K40 | Cucumber | Kerman | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 27.5 ± 2.2 | 0.2 ± 0.0 | 0.4 ± 0.0 | 1.0 ± 0.0 |
| K43 | Cucumber | Kerman | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 22.5 ± 3.5 | 1.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
The means halo zone diameter/ colony diameter of three replications ± standard deviation is represented. 1, growth; 0, no growth. The data indicated for IAA production was in μg/mL.
The fungal pathogen was isolated from tomato plants displaying disease symptoms and pathogenicity test was conducted using root-dipping inoculation method on tomato seedlings. The fungus was re-isolated from the vascular tissue of a symptomatic plant. Fungal DNA extraction was carried out by the methods described by Zhang et al. (2010), Salehi et al. (2018, 2019). Polymerase chain reaction (PCR) was performed with universal pair primers (ITS4-ITS5). Race determination of fungus was done using four specific primer pairs designed by Hirano and Arie (2006) based on the sequence of the exo-polygalacturonase gene (Table 2). ITS4-ITS5 nucleotide sequence obtained by DNA amplification of fungal pathogen was deposited in NCBI GenBank with accession number MG670445. Blast analysis showed that this pathogen had 99 % similarity to F. oxysporum. The result of the race determination using four specific primer pairs showed that the fungal isolate belongs to F. oxysporum f.sp. lycopersici race 3 (Supplementary Figure S1).
Table 2.
The list of primers used in the q-RT PCR in this study.
| Gene Name | Amplicon size (bp) | Sequence |
|---|---|---|
| UniF | 670 | F-5′ATCATCTTGTGCCAACTTCAG3′ R-5′GTTTGTGATCTTTGAGTTGCCA3′ |
| Sp13 | 445 | F-5′GTCAGTCCATTGGCTCTCTC3′ R-5′TCCTTGACACCATCACAGAG3′ |
| Sp23 | 518 | F-5′CCTCTTGTCTTTGTCTCACGA3′ R-5′GCAACAGGTCGTGGGGAAAA3′ |
| Sprl | 947 | F-5′GATGGTGGAACGGTATGACC3′ R-5CCATCACACAAGAACACAGGA3′ |
| UDP-G | 122 | F-5′GATGAACGCCACCTTCTTAG3′ R-5′CTCCTTCCATAACAATCCTCAC3′ |
| WRKY70 | 131 | F-5′TGGTAAAGCATAGTGACTCAAC3′ R-5′AGAGGGAGAAGAAGGCATAA3′ |
| PAL1 | 148 | F-5′CATTGTACAGGTTGGTGAGAG3′ R-5′CATCTCTTGAGACACTCCA3′ |
| LOX E | 104 | F-5′CTTCGGATACCCTTTACCT3′ R-5′GATCTCACCCAACTTCTTTC3′ |
| TPX | 133 | F-5′AGCATTGACAACACGTACC3′ R-5′AGCACTCCCTGTCTTAACT3′ |
| PR1 | 136 | F-5′GGTAACTGGAGAGGACAA3′ R-5′GACAATCGATCACTTTATTC3′ |
| ERF1 | 126 | F-5′AGACTTGGGAGTTGAATTA3′ R-5′TACATTGCGATCTTGATTA3′ |
| TUB | 146 | F-5′CAAGAACTCGTCCTACTTTG3′ R-5′GCTCACTCACCCTTCTAA3′ |
Growth on Nitrogen-Free Medium
Screening of the free-living (non-symbiotic) diazotroph isolates was carried out according to the procedure described by Xu and Zheng (1986). Each colony was spot-inoculated on nitrogen-free agar medium incubated at 29°C for 14 days, and then growth or lack of growth was compared to ISP2 medium.
Inorganic Phosphate Solubilization
Pikovskaya’s medium (PVK) was used to measure calcium phosphate [Ca3 (PO4)2]-solubilizing activity. Sterilized PVK medium with pH 7.2 was poured into petri plates. After solidification of the medium, bacterial isolates were spot-inoculated onto the center of the plates and incubated at 29°C for 7 days. Solubilization index was evaluated according to the ratio of the clear zone diameter to colony diameter (Soltani et al., 2010).
Siderophore Production
Siderophore production was evaluated as described by Alexander and Zuberer (1991) on Chrome Azurol agar (CAS) medium. Streptomyces isolates were spot-inoculated onto the center of the plate and incubated at 29°C for 7 days. The formation of orange halo around the colonies were considered as siderophore-producing isolates. After 3 days, the ratio of the halo zone diameter to colony diameter was calculated as siderophore production.
IAA Production
To determine amounts of IAA produced by each isolate, 1 mL of bacterial culture in ISP2 broth was inoculated in Tryptic Soy Broth supplemented with 150 mg/L L-tryptophan. Approximately 1 mL of culture solution was centrifuged at 12000 rpm for 5 min, and 1mL of the supernatant was mixed with 2 mL of Salkowski’s reagent (150 mL concentrated H2SO4, 250 mL distilled water, 7.5 mL 0.5 M FeCl3.6H2O) and incubated for 20 min in darkness at room temperature (de Oliveira-Longatti et al., 2014). IAA production was qualitatively assayed as pink color development and quantified by measurement of absorbance at 530 nm using a spectrophotometer infinit M200 (Tecan, Switzerland). The calibration plot was constructed using dilutions of a standard IAA (Fluka, Switzerland) solution and the uninoculated medium with the reagent as a standard curve (0, 5, 10, 20, 50, and 100 μg/mL). The quantity of IAA in the culture was expressed as μg/mL.
Antagonistic Effect of PGPRs
Bacterial suspension of candidate PGPR isolates (Table 1) (20 μL of the 108 cfu/mL sterile saline solution) was inoculated at 29°C linearly at the two opposite sides (1 cm from the plate edge) of potato dextrose agar (PDA) plates for 48 h. Then, one fungal plug (0.5 cm diameter) was inoculated at the center of PDA plate (Kunova et al., 2016) and then incubated for 4 days. The growth inhibition percentage was calculated using the formula (a − b)/a × 100, where “a” is the fungal growth radius of a control culture (in cm) and “b” is the distance of the pathogen growth in the direction of bacteria (in cm).
Chitinase Activity
Chitinase production was determined according to the method of Hsu and Lockwood (1975). Streptomyces isolates were grown on chitin agar containing 0.4% colloidal chitin and 1.5% agar adjusted to pH 7.2. Colloidal chitin was prepared according to Berger and Reynolds (1958). Plates were incubated for 5 days at 29°C. The ability of chitinase production was shown by a clear halo around the colonies. The ratio of the clear zone diameter to colony diameter was calculated as chitinase activity.
Cellulase Activity
Carboxymethyl cellulase (CMCase) activity was determined by Mandels-Reese medium with carboxymethyl cellulose (CMC) as sole carbon source (Majidi et al., 2011). The bacteria were grown on CMC agar containing 0.4 g/L KH2PO4, 0.02 g/L CaCl2, 0.02 g/L NaCl, 0.02 g/L FeSO4 7H2O, 2.5 g/L CMC, and 15.0 g/L agar. The pH was adjusted to 7.2 with 1 M NaOH. The CMC agar plates were incubated at 29°C for 7 days to allow the secretion of cellulase. To visualize the hydrolysis zone, agar medium was flooded with an aqueous solution of Congo red (1 mg/mL) for 20 min. Congo red solution was then poured off, and the plates were further treated by flooding with 1 M NaCl for 15 min. To indicate cellulase activity, the diameter of the clear zone around each colony was measured. The ratio of the clear zone diameter to colony diameter was calculated as cellulase activity.
Protease Activity
Extracellular protease activity of Streptomyces isolates was assayed by a modification method of Mehrotra et al. (1999). Each bacterial isolate was streaked on skim milk agar containing 15 g/L skim milk powder, 0.5 g/L yeast extract and 10 g/L agar. After incubation at 29°C for 48 h, the ratio of the clear zone around bacterial colony to colony diameter was measured as protease activity.
Phenotypic and Molecular Characterization of the Selected Isolates
The potent PGP and antagonist isolates were further characterized by differential morphological traits on ISP2, ISP3, and ISP4 media (Shirling and Gottlieb, 1966), melanin formation (Suter, 1978), growth in high temperatures (37°C and 42°C), and growth on medium supplemented with 6, 10, and 12% NaCl (Kutzner, 1981).
Molecular characterization of the selected isolates was done using PCR amplification of 16S rRNA gene sequence. DNA was extracted according to the method described by Tripathi and Rawal (1998). PCR amplification was performed using the primers 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 1525R: 5′-AAAGGAGGTGATCCAGCC-3′ as described by Chun and Goodfellow (1995). Almost-complete 16S rRNA gene sequences (1400 nt) were deposited in the GenBank database under the accession numbers of MG722685 (strain IT25), MG786938 (strain TO612), MG786894 (strain Y17), MG786896 (strain Y28), MG654776 (strain IC10), and MG676358 (strain IC13). The sequences were aligned manually with corresponding sequences of available Streptomyces species deposited in the GenBank, EMBL and DDBJ databases using BLAST search tool (Altschul et al., 1997). Phylogenetic tree was constructed using the MEGA 6.0 software package (Tamura et al., 2013) based on neighbor-joining method. Bootstrap analysis was used to evaluate the stability of relationships based on 1000 resampling. Strains IC10 and Y28 were preserved and deposited in the Agricultural Biotechnology Research Institute of Iran Culture collection (ABRIICC) under accession numbers of ABRIICC 20108 and ABRIICC 20114, respectively.
Greenhouse Experiments
Tomato (Solanum lycopersicum L.) cv. Rio Grande susceptible to FOL races 2 and 3 (Barker et al., 2005), was used in greenhouse experiments. Seeds were surface sterilized with 1% sodium hypochlorite (NaOCl) and incubated in a growth chamber at 25°C for 7 days. Germinated seedlings were placed into 84-cell plug tray (50 × 30 × 5 cm deep) filled with sterilized soil and peat moss (1:2), with one seedling occupying each cell. Seedlings were watered every day with tap water and kept at 27°C and 16 h brightness/8 h darkness. After 21 days, the seedlings were transferred to pots (15 × 20 cm) filled with a sterile mixture of field soil and peat moss (2:1). For bacterial treatments, Streptomyces cell and spore were centrifuged at 8000 rpm for 15 min and then the pellet re-suspended in 10 mL sterile saline solution. Bacterial suspension was added to autoclaved sand and final cfu/g sand adjusted to 106. Seven gram of sand containing bacteria was added to the surface of each cultivated pot immediately after transferring of plant to the pot. Sterilized sand was used as a negative control. The pots were watered every 2 days. For FOL inoculation, 10 days after bacterial treats, tomato seedlings were uprooted and inoculated with conidial suspension for 30 min (root-dip method). Seedlings submerged in sterile distilled water (mock inoculation) were used for bacterial treatments and negative control. To prepare FOL suspension (pathogen inoculum), fungi was cultivated on PDA for 10 days. Conidia were harvested by scraping in sterile water (10 mL/plate) and final conidia/mL adjusted to 5 × 107 using hemocytometer. The treatments were negative control plants (mock inoculation), positive control (FOL inoculated), Carbendazim® (soil drenched with fungicide in a concentration of 1.5 g/L) and six Streptomyces treatments inoculated or non-inoculated with FOL. The plants were harvested 60 days after transferring to the pots. The growth parameters comprising of shoot length, fresh and dry weight of shoot and root were measured and recorded. Further, disease severity (DS) was assessed on a scale from 0 to 5: 1 = symptoms free = 0%; 2 = slight chlorosis, stunting, or wilting = 25%; 3 = moderate chlorosis, stunting, or wilting = 50%; 4 = severe chlorosis, stunting, or wilting = 75%; 5 = death = 100% (Marlatt et al., 1996). Percentage of control value was calculated using the formula (DC − DT)/DC × 100, where “DC” is disease index of inoculated control (FOL) and “DT” is disease index of inoculated treatment (%) (Song et al., 2004).
A greenhouse experiment was conducted separately for the next two tests. The procedure of this experiment was similar to the first one, but the experiment duration was shorter and the plants were harvested 14 days after transfer to the pots, also 1 cm of the root end of the seedlings was cut before FOL/mock inoculation. Foliar was sprayed with plant hormones (1 mM SA or 10 mM MeJA) 48 h before FOL inoculation. The treatments were negative control plants (mock inoculation), positive control (FOL inoculated), Carbendazim® (soil drenched with fungicide in a concentration of 1.5 g/L) and two Streptomyces treatments (strains IC10 and Y28) inoculated or non-inoculated with FOL. Plants leaves were harvested 2 days after FOL inoculation, frozen in liquid nitrogen and kept at −70°C for the following analysis.
Antioxidant Enzymes Activity
Frozen leaf samples (100 mg fresh weight) with 10 mg polyvinyl pyrrolydone (PVP) was transferred to 1.5 mL tube and homogenized in 1 mL Na-Pi buffer (1 mM, pH 7). The homogenate was centrifuged at 15000 × g for 15 min. All operations were performed at 4°C. The supernatant was used as a crude enzyme extract. The activities of antioxidant enzymes including peroxidase (POX: EC 1.11.1.7) and catalase (CAT: EC 1.11.3.6) were measured in a reaction containing 250 μL 0.1 M phosphate buffer (pH 7.0), 250 μL 10 mM guaiacol, 30 μL H2O2, and 40 μL crude enzyme extract (Chance and Maehly, 1955). The enzyme activity (U/mL) was measured spectrometrically (Cary 300, United States) by monitoring of the degrading H2O2 by the increase in the absorbance at 470 nm and the decrease in the absorbance at 290 nm over 3 min.
Quantitative Real-Time PCR Analysis of the Defense-Related Genes
Total RNA was extracted from shoots using RNeasy plant mini kit (QIAGEN). cDNA was synthesized using 1 μg of each RNA sample after treating with RNase-free DNase I (Invitrogen) using iScript cDNA synthesis kit (BioRad) according to manual description. Quantitative PCR was performed in a 25 μL reaction containing 1 μL of template cDNA, 0.5 μL of 10 pM of each forward and reverse specific primer (Table 2) and iQ SYBR Green Supermix kit (BioRad) on BioRad multicolor real-time PCR detection system. The PCR profile included an initial denaturation step at 95°C for 3 min, followed by 40 cycles of denaturation (95°C/30 s), primer annealing (60°C/30 s) and primer elongation (72°C/30 s), by a final elongation step (72°C/ 5 min) and recording melting curves. Results were expressed as the normalized ratio of mRNA level of target gene to internal control tubulin gene (TUB). Changes was estimated by using the Relative Expression Software Tool (REST 2009) (Pfaffl et al., 2002).
Statistical Analysis
Statistical analysis was performed using analysis of variance (ANOVA) by Microsoft Excel (Microsoft Corporation, United States) and SPSS version 16.0 (SPSS Inc. Chicago, IL, United States). All data shown are average value of three (in vitro experiments) biological replicates ± SE. The greenhouse experiments were carried out in randomized blocks design with four blocks and there were four biological replications for each treatment. The significance differences of the treatments were evaluated using multivariate generalized linear model (GLM) with Duncan multiple range test post hoc analysis at level of P < 0.05. The relationship between in vitro and in vivo data was studied using bivariate Pearson test at P ≤ 0.01.
Results
Isolation and Selection of Streptomyces Bacteria
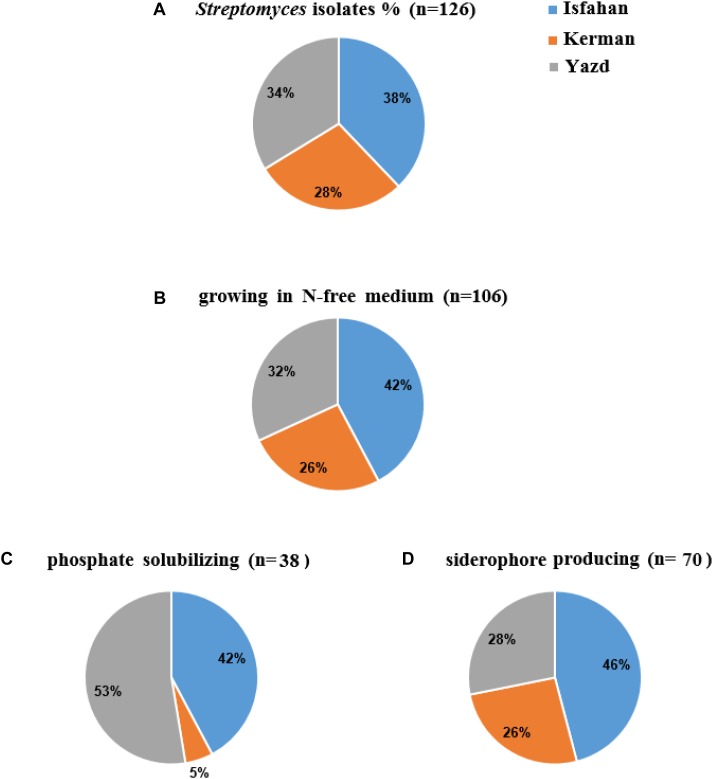
A total of 126 colonies displaying Streptomyces appearance including compact colored heaped and wrinkled, waxy, chalky, powdery or velvety colony, were isolated from tomato and cucumber rhizosphere soil collected from Isfahan, Yazd and Kerman (Figure 1A). Totally, 106 isolates (84%) were able to grow on solid nitrogen-free medium (Figure 1B) of which thirty-eight isolates were able to solubilize inorganic phosphate. Three isolates comprising of IS8, Y7, and Y28 with the ratio 1.9, 0.5, and 0.4 had the most potential to solubilize tricalcium phosphate, respectively (Figure 1C). Further, sixty-six percent of 106 isolates were able to produce siderophore (Figure 1D). The maximum orange halo due to iron chelation was recorded for isolate TO612 after 7 days of incubation. All 106 isolates produced IAA in a range of 7.0–40.9 μg/mL of which twenty percent produced IAA more than 27 μg/mL. Thirty-two isolates from 106 showed proteolytic enzyme activity. The greatest halo zone/colony diameter ratio for protease activity was 1.0 and recorded for isolate IC10 (Table 1). Forty-four isolates were found to produce cellulose. The greatest halo zone/colony diameter ratio for cellulase activity was 1.4 which recorded for Y16. Furthermore, biosynthesis of the chitinolytic enzyme was detectable for 22 isolates. Isolates IC13, K40, SS12, IC6, and Y17 were able to produce all three examined hydrolytic enzymes. Overall, 24 isolates containing at least three PGP traits and hydrolytic enzymes activity were selected for evaluation in the following experiments as shown in Table 1.
FIGURE 1.
The percent (%) of colonies isolated from Isfahan, Yazd, and Kerman provinces: colonies (A) with Streptomyces appearance (B) growing in N-free medium, (C) phosphate solubilizing, and (D) siderophore producing.
Growth Inhibition of the Selected Isolates Toward FOL
Six isolates (25% of the selected isolates) showed more than 30% inhibitory effect against FOL in dual culture test. The highest percentages of growth inhibition were 69, 49, 48, 42, 39, and 38, which were recorded for IC10, IT25, Y17, TO612, Y28, and IC13, respectively (Table 3). The biocontrol potential of these isolates (superior PGP antagonistic isolates) was evaluated in a greenhouse experiment.
Table 3.
In vitro antagonistic activity against Fusarium oxysporum f.sp. lycopersici by selected isolates.
| Isolates | Growth inhibition (%) | Isolates | Growth inhibition (%) | Isolates | Growth inhibition (%) |
|---|---|---|---|---|---|
| IC6 | 4.0 ± 0.1j∗ | IT25 | 49.1 ± 0.8b | IC34 | 1.7 ± 1.5j |
| IC10 | 68.5 ± 1.0a | Y7 | 0.6 ± 1.1j | KH12 | 30.0 ± 0.0d |
| IC13 | 38.2 ± 0.9c | Y18 | 1.3 ± 1.1j | K40 | 4.0 ± 0.0j |
| IS8 | 10.6 ± 1.1h | Y27 | 0.6 ± 1.1j | K43 | 10.0 ± 2.0h |
| SS12 | 0.6 ± 1.1j | TO612 | 41.5 ± 1.8c | ||
| CU122 | 0.0 ± 0.0k | Y17 | 47.5 ± 2.56b | ||
| IC15 | 17.9 ± 1.4g | Y28 | 38.7 ± 1.1c | ||
| SS14 | 1.0 ± 1.1j | Y281 | 2.3 ± 2.5j | ||
| IT20 | 19.3 ± 0.8f,g | Y16 | 2.6 ± 2.5j | ||
| IT8 | 9.6 ± 0.4h | Y33 | 1.6 ± 2.8j |
Values are the means (averaged from three replicates) ± SE. ∗Same letters within each column represent non-significant difference according to Duncan’s Multiple Range Test (P < 0.05).
Phenotypic and Genotypic Characterization of the Superior Isolates
Phenotypical characterization was performed using aerial hyphae and spore chains color of 10-days-old bacterial culture on ISP media. On the medium ISP2, isolates TO612 and Y17 were differentiated from IC10, IC13, IT25, and Y28 according to the color of spore chains. On the medium ISP3, isolates Y28, IT25, and IC10 were distinct from the others based on the color of aerial hyphae. Further, on the medium ISP4, isolate IC13 was differentiated from IC10 by its color of aerial hyphae. Also, these two isolates were recognizable on ISP3 according to the color of their aerial hyphae. Isolates IC10 and Y28 were well distinguished from TO612, Y17 and IC13 using ISP media and from IT25 based on melanin production. Isolates IC10 and Y28 slightly were different from each other on ISP2 medium (Supplementary Figure S2). Physiological tests showed only IC13 had potential to grow at 42°C (Table 4). All six isolates were able to grow on NaCl 6 % while only isolates Y28 and Y17 were able to grow on medium containing NaCl 10 % (Table 4). The preliminary phylogenetic analysis of the 16S rRNA gene sequences showed IC10 and Y28 were closely related to species of the genus Streptomyces. The phylogenetic tree constructed from 16S rRNA sequences showed that isolates IC10 and Y28 are two members of Streptomyces genus with more than 99.5% sequence similarity to S. enissocaesilis NRRL B-16365T and S. rochei NBRC 12908T, respectively (Figure 2).
Table 4.
Cultural characteristics of superior PGP antagonistic isolates.
| Isolate | Color of aerial hyphae – spore chains on ISP media |
Melanin production |
Growth (in/on) |
GeneBank acc. number | |||||
|---|---|---|---|---|---|---|---|---|---|
| ISP2 | ISP3 | ISP4 | Tyrosine/no tyrosine media | 42°C | NaCl 6% | NaCl 10% | NaCl 13% | ||
| IT25 | Yellow – Light purple | Colorless – Light purple | Yellow – purple | −/− | − | ++ | − | − | MG722685 |
| TO612 | Yellow – Light greenish | Yellow – Yellow | Yellow – White | +/+ | − | + | − | − | MG786938 |
| Y17 | Yellow – Greenish | Yellow – Olive | Dark yellow – White | +/+ | − | ++ | + | − | MG786894 |
| Y28 | Yellow – light purple | Colorless – Dark purple | Dark yellow – Purple | −/+ | − | ++ | + | − | MG786896 |
| IC10 | Purple –Yellow | Colorless – Purple | Yellow –Purple | −/+ | − | + | − | − | MG654776 |
| IC13 | Yellow – Light purple | Dark yellow – Purple | Yellow – Dark gray | −/− | + | + | − | − | MG676358 |
FIGURE 2.
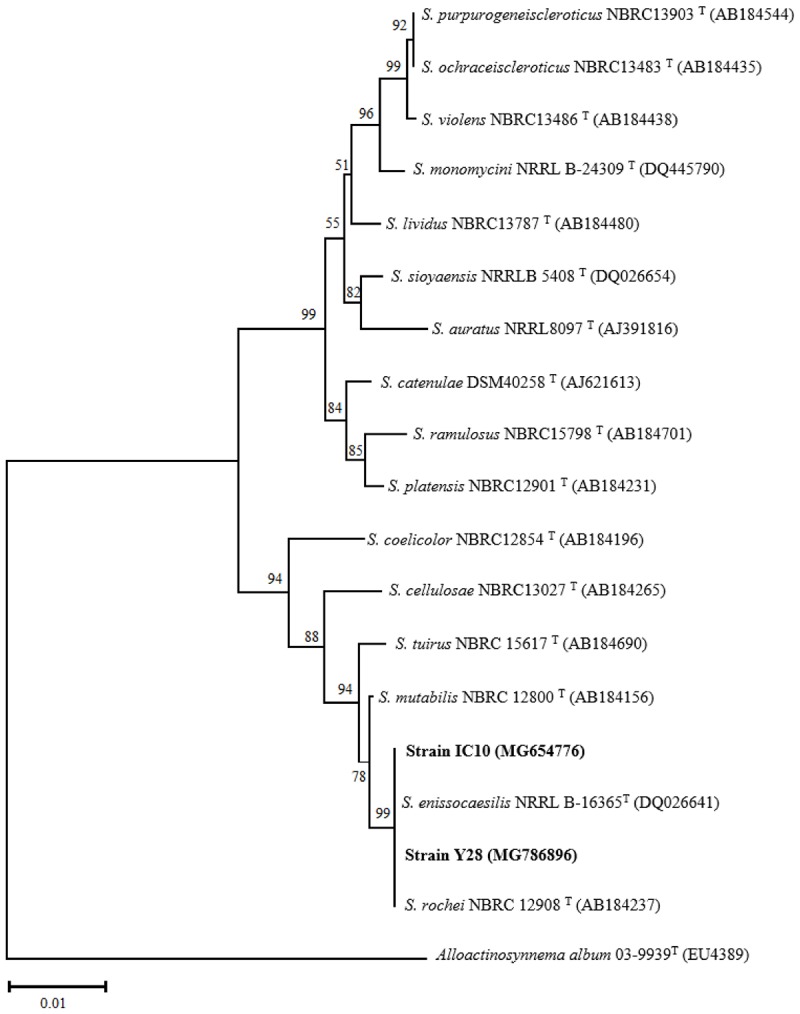
Phylogenetic tree based on almost complete 16S rRNA gene sequences of S. enissocaesilis strain IC10 and S. rochei strain Y28. Tree was calculated using neighbor-joining method illustrating the taxonomic position of the strains with related species. Accession numbers of the sequences are given in parentheses. The sequences of Alloactinosynnema album 03-9939T (EU438907) was used as outgroup. Bootstrap values are based on 1000 resampling and shown at the branching points. Bar indicates 0.01 substitutions per nucleotide position.
Tomato Growth Promotion Activity of the Superior Isolates
Strains TO612, Y28, IC10, and IC13 significantly increased shoot length compared to control plants. Strains Y28 and IC10 increased shoot length up to 20% compared to control (Figure 3 and Table 5). Strains Y17, Y28, IC10 and IC13 significantly (p < 0.05) increased fresh shoot weight while only strains IC10 and IC13 increased shoot dry weight. The increases in the shoot fresh and dry weight were in a range 29–36% and 22–37%, respectively (Table 5). Streptomyces strains did not increase root fresh and dry weight while induced root lateral branching (hairy roots) compared to control (data not shown).
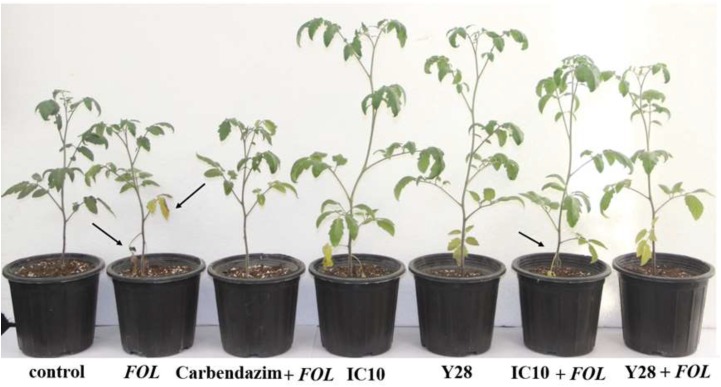
FIGURE 3.
Biocontrol of tomato wilt and stunt caused by FOL using S. enissocaesilis strain IC10 and S. rochei strain Y28 in greenhouse conditions. Data recorded 60 days after bacterial treatment. Stunting is seen in infected plants. Arrows showing wilting symptoms (chlorosis and dried leaves) in FOL inoculated plants.
Table 5.
Effect of superior PGP antagonistic isolates (IT25, TO612, Y17, Y28, IC10, and IC13) on tomato growth parameters in greenhouse conditions.
| Treatment | Shoot length (cm) | Shoot fresh weight (g)/plant | Shoot dry weight (g)/plant | Root fresh weight (g)/plant | Root dry weight (g)/plant |
|---|---|---|---|---|---|
| Negative control | 24.18 ± 0.55c∗ | 11.36 ± 0.60b | 1.79 ± 0.14b,c | 3.61 ± 0.14a | 1.72 ± 0.10a |
| IT25 | 24.87 ± 0.60c | 10.19 ± 0.35b,c | 1.39 ± 0.16c | 2.36 ± 0.35a,b | 0.90 ± 0.19d,e |
| TO612 | 29.37 ± 0.32a | 11.63 ± 0.23b | 1.87 ± 0.19b | 2.78 ± 0.17a,b | 0.81 ± 0.16d,e |
| Y17 | 26.50 ± 0.20b,c | 15.47 ± 0.30a | 2.18 ± 0.10a,b | 3.61 ± 0.19a | 1.51 ± 0.25b |
| Y28 | 29.00 ± 0.56a,b | 14.97 ± 0.49a | 2.16 ± 0.15a,b | 2.88 ± 0.12a,b | 1.23 ± 0.18c |
| IC10 | 28.90 ± 0.25a,b | 15.37 ± 0.38a | 2.31 ± 0.39a | 1.64 ± 0.32b | 0.92 ± 0.12d,e |
| IC13 | 28.62 ± 0.29a,b | 14.70 ± 0.65a | 2.45 ± 0.22a | 1.78 ± 0.13b | 0.80 ± 0.24d,e |
Data recorded 60 days after bacterial treatment. ∗Same letters within each column represent non-significant difference at 5%, according to Duncan’s tests using the GLM procedure (df = 9, F = 7.16, and p < 0.05).
Biocontrol Potential of the Superior Isolates
Biocontrol efficacy of the superior isolates against FOL causing wilt disease was evaluated in the greenhouse and compared to the chemical fungicide Carbendazim®. The high level of disease severity (4.3) was observed in positive control. In Carbendazim® treatment, disease severity was 2.1. Disease severity in the plants treated with IC10 and TO612 were 1.6 and 1.9, respectively. Strain IC10 increased control value by 12.5% compared to the chemical fungicide (Figure 3). Minimum shoot length, fresh and dry weight were related to positive control and minimum fresh and dry root weight were related to Carbendazim®. All superior isolates significantly increased shoot length, fresh and dry weight of shoot, compared to positive control (Table 6). Strains IC13 increased tomato total dry weight by 30 and 51% compared to control and Carbendazim® respectively. Strains Y28 and IC10 with the highest PGP and biocontrol activity were selected to determine their role in the induction of tomato-systemic resistance through antioxidant enzymes and defense-related genes.
Table 6.
Biocontrol of tomato wilt and stunt caused by F. oxysporum f.sp. lycopersici race 3 (FOL) using superior PGP antagonistic isolates (IT25, TO612, Y17, Y28, IC10, and IC13) in greenhouse conditions.
| Treatments | Shoot length (cm) | Shoot fresh weight (g)/plant | Shoot dry weight (g)/plant | Root fresh weight (g)/plant | Root dry weight (g)/plant | Disease severity | Control value (%) |
|---|---|---|---|---|---|---|---|
| Negative control | 23.62 ± 0.25b∗ | 11.20 ± 0.63a,b | 1.35 ± 0.35a,b | 3.31 ± 0.10a | 1.41 ± 0.33a | 1.0d | – |
| Positive control (FOL) | 14.00 ± 0.28d | 5.56 ± 0.34c | 0.78 ± 0.18c | 1.66 ± 0.33a,b | 0.37 ± 0.29c,d | 4.3a | – |
| Carbendazim®+FOL | 15.25 ± 0.22d | 13.00 ± 0.25a,b | 2.20 ± 0.16a | 0.23 ± 0.38b | 0.18 ± 0.14d | 2.1a,b,c | 71.9a,b |
| IT25+FOL | 20.20 ± 0.43c | 10.23 ± 0.27b,c | 1.77 ± 0.55a,b | 2.48 ± 0.49a | 1.00 ± 0.37a,b | 2.3a,b | 68.8b,c |
| TO612+FOL | 19.00 ± 0.20c | 12.77 ± 0.32a,b | 2.40 ± 0.38a | 2.90 ± 0.20a | 1.40 ± 0.38a | 1.9c,d | 78.1a |
| Y17+FOL | 19.50 ± 0.35c | 13.38 ± 0.26a,b | 2.09 ± 0.58a | 2.76 ± 0.37a | 0.94 ± 0.29a,b | 2.0b,c | 75.0b,c |
| Y28+FOL | 26.70 ± 0.15a | 11.88 ± 0.75a,b | 2.02 ± 0.60a | 2.94 ± 0.28a | 0.59 ± 0.33b,c | 2.0b,c | 75.0b,c |
| IC10+FOL | 26.60 ± 0.35a | 12.83 ± 0.10a,b | 2.07 ± 0.38a | 1.83 ± 0.18a,b | 0.96 ± 0.24a,b | 1.6c,d | 84.4a |
| IC13+FOL | 21.33 ± 0.39c | 15.00 ± 0.29a | 2.45 ± 0.41a | 2.54 ± 0.30a | 1.15 ± 0.31a,b | 2.1a,b,c | 71.9a,b |
Data recorded 60 days after bacterial treatment. Disease severity was assessed on a scale of 1 to 5 (1, symptoms free; 5, severe symptoms). ∗Same letters within each column represent non-significant difference at 5%, according to Duncan’s tests using the GLM procedure (df = 11, F = 3.63, and p < 0.05).
Antioxidant Enzymes Activity
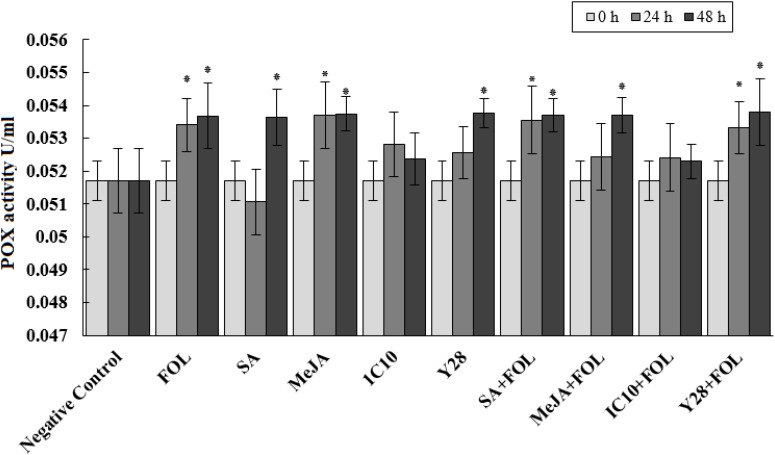
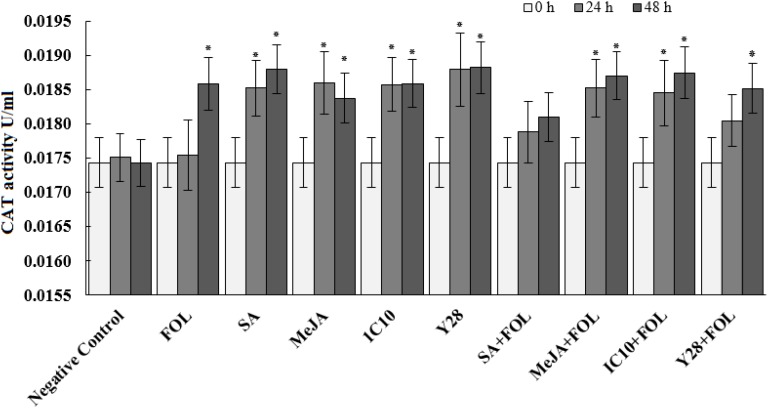
Peroxidase activity significantly increased 24 h after inoculation of plants with FOL. Further, MeJA individually increased the enzyme activity at the same time interval. Plants inoculated with FOL in SA or Y28 treatment increased POX activity after 24 h like positive control (FOL). In SA treatment, POX activity increased 48 h after exogenous application. The level of POX activity remained constant in plant treated with FOL and MeJA and also SA and Y28 inoculated with FOL. In soil treated with strain IC10 and inoculated or non-inoculated with the pathogen, POX activity did not increase during the experiment time (48 h) (Figure 4). CAT activity of tomato plants increase 48 h after FOL inoculation. Treatments including bacterial strains and plant hormones increased CAT activity after 24 h. Pathogen inoculation slightly suppressed CAT activity in the SA treatment (Figure 5).
FIGURE 4.
Effect of biological (S. enissocaesilis strain IC10 and S. rochei strain Y28) and chemical (SA and MeJA) treatments on the induction of peroxidase (POX) activity in tomato leaves non-inoculated (left) and inoculated (right) with FOL at different time intervals of inoculation (df = 29; F = 2.90; P < 0.01). Data represent the mean values ± SE of three biological replicates. Negative control: untreated and non-inoculated. In each treatment, the values marked with an asterisk are significantly (P < 0.05) different from negative control at time point 0.
FIGURE 5.
Effect of biological (S. enissocaesilis strain IC10 and S. rochei strain Y28) and chemical (SA and MeJA) treatments on the induction of catalase (CAT) activity in tomato leaves non-inoculated (left) and inoculated (right) with FOL at different time intervals of inoculation (df = 29; F = 30.73; P < 0.01). Data represent the mean values ± SE of three biological replicates. Negative control: untreated and non-inoculated. In each treatment, the values marked with an asterisk are significantly (P < 0.05) different from negative control at time point 0.
Quantitative Real-Time PCR Analysis of the Defense-Related Genes
Plant inoculation with FOL and MeJA increased UDP-glycosyltransferase (UDP) transcripts by 103- and 98-fold, respectively, compared to control. Although Streptomyces strains induced UDP transcription, they significantly suppressed gene expression in the plants inoculated with FOL (Figure 6A). Treatments including Streptomyces strains and plant hormones and not FOL significantly increased phenylalanine ammonia-lyase (PAL) transcripts compared to control. Salicylic acid and strain IC10 also retained their induction effect in plants inoculated with FOL (Figure 6B). The results revealed that plants treated with SA, IC10, Y28, SA-FOL, and IC10-FOL significantly stimulated (up-regulated) WRKY70 expression. Treatments of FOL and MeJA did not increase relative expression of WRKY70 in tomato plants (Figure 6C). Tomato peroxidase (TPX1) transcripts was significantly up-regulated in all treatments including Streptomyces strains and plant hormones. Salicylic acid and strain Y28 also induced TPX1 gene expression in plants inoculated with FOL (Figure 6D). The highest lipoxygenase (LOX) gene expression level (38- fold) was detected in Y28. There was a significant down-regulation of LOX gene in FOL and IC10 treatments. On the contrary, MeJA and Y28 stimulated LOX expression. Induced LOX expression was also observed in plants treated with bacteria and inoculated with FOL (Figure 6E). In a similar pattern, SA and Y28 significantly increased pathogenesis related protein 1 (PR1) and ethylene response factor 1 (ERF1) transcripts. Strains IC10 and Y28 also were able to stimulate the PR1 expression in the presence of FOL (Figure 6F,G).
FIGURE 6.
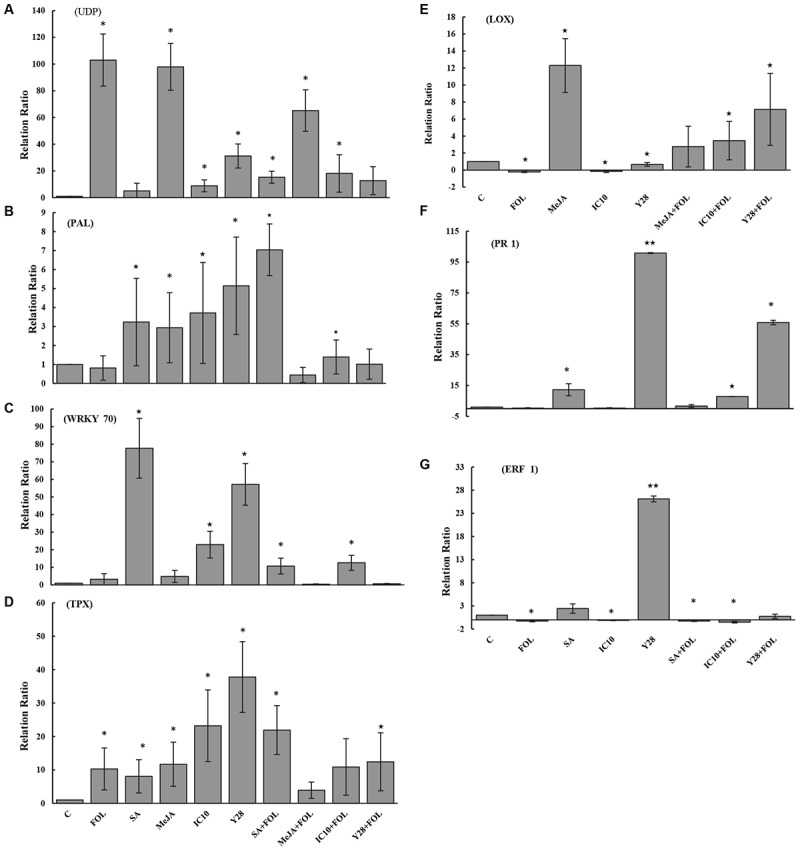
The relative level of gene expression (fold) determined by qRT-PCR of seven target genes including UDP (A), PAL (B), WRKY70 (C), TPX (D), LOX (E), PR1 (F), and ERF1 (G) versus reference control (Tubulin gene) in tomato “Riogrande” treated with chemical (MeJA and SA) or biological agents (S. enissocaesilis strain IC10 and S. rochei strain Y28) 48 h after inoculation with FOL/ distilled water. Untreated and non-inoculated plants were considered as control (C) and as reference sample. Standard error represents for three biological replicates. Positive values of fold change indicate up-regulation while negative values indicate down-regulation. The values marked with one and two asterisk are significantly different from control at P < 0.05 and P < 0.01, respectively.
Correlation Analysis
There was a significant relationship between siderophore and IAA production (r = 0.36). Besides, a correlation was found between growth inhibition and protease activity (r = 0.31). Moreover, growth inhibition of FOL in dual culture assay was significantly correlated to siderophore production (r = 0.64) (Table 7). A significant negative correlation (−0.63) was observed between growth inhibition (in vitro assay) and disease severity (in vivo assay).
Table 7.
Pearson correlation coefficients (r) between PGP traits (in vitro assays) and biocontrol activity (in vivo assay).
| Pearson correlation |
In vitro assays |
In vivo assay |
|
|---|---|---|---|
| Variables | IAA production | Fungal growth inhibition (%) | Disease severity |
| Protease activity | 0.44ns | 0.31∗ | −0.47ns |
| Siderophore production | 0.36∗ | 0.64∗∗ | 0.24ns |
| IAA production | 1 | 0.47∗ | −0.01ns |
| Fungal growth inhibition | −0.63∗ | ||
ns, non-significant; ∗P < 0.05; and ∗∗P < 0.01.
Discussion
Almost half of tomato and cucumber greenhouses in Iran (more than 7000 hectares) is located in Yazd, Isfahan, and Kerman provinces. Amount and type of organic fertilizer and soil used to prepare the greenhouse growth bed are different in these provinces. Farmers in Yazd and Kerman use sand from the bed of the nearest rivers to their greenhouse and farmers in Isfahan use soil of the greenhouse floor as a raw material to prepare culture bed (private conversation). Our results showed distribution of the bacterial isolates with the Streptomyces appearance in the three provinces is slightly different. Among the isolates that showed a PGP trait, a relatively stable share (42–46%) belonged to Isfahan. The contribution of the Streptomyces isolated from Yazd or Kerman in each PGP trait was not always constant showing Streptomyces with several PGP traits in the field soil is greater than that of rivers sand. A previous study on Streptomyces showed that bacterial diversity was more affected by the habitat than the population (Davelos et al., 2004). The restriction of some nutrients in soil may be a major contributor to the change in the microbial diversity due to natural selection.
Growth ability on a nitrogen-free medium and IAA and siderophore production were three dominant characteristics of Streptomyces isolated from vegetable rhizosphere soils. A lower percentage (30%) of the isolates had the potential for solubilizing phosphate. Besides, the location of isolation was also effective on this ratio. There was a significant positive correlation between IAA and siderophore production, which may remind a complementary role of the siderophore in plant growth promotion activity (i.e., in addition to its role in antagonistic activity). Such correlation was not observed between other traits involved in antagonist activity and IAA production.
The ability of FOL growth inhibition was very different between Streptomyces isolates showing PGP properties. Isolate CU122 with four PGP traits, was not able to inhibit FOL growth. Failure to inhibit the pathogen growth was correlated to the inability of isolate CU122 to produce chitinase and protease enzymes. On the contrary, isolate IC10 with same in vitro PGP features as well as enzymes activity inhibited FOL growth very well. The correlation of protease and chitinase activity with inhibition of FOL growth was positive and respectively, quantitative and non-quantitative. Only two exceptions did not adhere to this relationship. Isolates TO612 and KH12, which did not show enzyme activity, inhibited FOL growth in an acceptable range (equal to or greater than 30%).
This finding revealed that although chitinolytic activity of Streptomyces and digestion of the fungal cell wall is an effective mechanism of growth inhibition (Gherbawy et al., 2012), other mechanisms such as antibiosis and even an enzyme like protease (Xu et al., 2016) that is less studied are also involved in antagonism. Interestingly, there was a significant correlation between IAA and siderophore productions and FOL growth inhibition, showing relationship between PGP features and antagonistic activities in Streptomyces. IAA production is a common characteristic among antagonistic species of Streptomyces (Sreevidya et al., 2016). It seems that IAA production alone does not cause plant growth, and other traits, such as siderophore production, also has an important role in increasing plant growth. In this regard, Kunova et al. (2016) showed that the antagonistic isolates producing IAA did not increase growth of different vegetable species. These isolates were not able to produce siderophore. The significant negative correlation that was observed between FOL growth inhibition (in vitro assay) and disease severity (in vivo assay) confirmed a need for in vitro tests to select the best biocontrol strains. Recently, Wu et al. (2019) reported that a decreased percentage disease index of rice sheath blight caused by Rhizoctonia solani was associated with an increase in Streptomyces derived antifungal agent concentration. They concluded that the antifungal agent reduces sheath blight symptoms via exerting a strong antagonistic activity against R. solani both in vivo and in vitro conditions. S. rochei ACTA1551 from Greek (Kanini et al., 2013), S. miharaensis KPE62302H (Kim et al., 2012), and S. psammoticus KP1404 (Kim et al., 2011) from Korea, S. plicatus from Egypt (Abd-Allah, 2001) and S. griseorubens E44G from Saudi Arabia (Rashad et al., 2017) were reported as successful biocontrol agents to control tomato Fusarium wilt. In the previous reports, the pathogen race has not been determined and to our knowledge, this is the first report of biocontrol of Fusarium wilt caused by F. oxysporum Schlecht. f. sp. lycopersici (Sacc.) race 3 in tomato with Streptomyces strains.
Strain Y28 and IC10 were close to S. rochei and S. enissocaesilis, respectively and are grouped in a clade on the phylogenetic tree. These PGP strains were isolated from the rhizosphere soils of two greenhouses in two different locations with a distance of 300 kilometers. The high similarity in PGP, and biocontrol activity of these two strains revealed that evolutionary process can keep PGP traits and biological control activities together.
All plants treated with SA, MeJA and Y28 or inoculated with FOL increased POX activity during 48 h after FOL inoculation. The POX activity in hormone or PGPR treated plants was not affected by FOL inoculation. Despite the similar biocontrol activity, isolates IC10 and Y28 had a different effect on the induction of plant POX activity. None of the plants treated with IC10, inoculated or non-inoculated with FOL, increased POX activity. Peroxidase is considered an important pathogen-related protein (PR-protein) or defense protein involved in many physiological responses of plants to biotic stresses. Contribution to biosynthesis of lignin (Chittoor et al., 1997) and antimicrobial compounds such as phytoalexins and quinones (Mayer, 2006) are two well-known POX roles associated with ISR. Increased peroxidase activity of cucumber was reported in plants showing Fusarium wilt (Zhao et al., 2012) and also plant treated with Streptomyces as biocontrol agent (Salla et al., 2016).
Peroxidase, as well as CAT, are involved in plant antioxidant defense system and reduce the harmful effects of stresses by scavenging of ROS (Das and Roychoudhury, 2014). In this study, the effects of FOL, plant hormones and PGP strains to induce CAT activity were the same. Induction of systemic resistance through defense-related enzymes POX and CAT in tomato plants treated with salicylic acid or Pseudomonas fluorescens was reported by Nikoo et al. (2014). They showed that pathogen inoculation of plants treated with bacterial or hormonal elicitors increased both POX and CAT activity in higher levels compared to non-inoculated treated plants or plants only inoculated with pathogen. This association was not observed in our experiment.
Relative expression of several candidate genes encoding transcription factors (WRKY70 and ERF1), PR1, TPX1, UDP, LOX, and PAL was evaluated in this study. The plant-specific transcription factor WRKY70 is an important factor in Arabidopsis signaling pathways and its expression is activated by SA and repressed by JA. Ren et al. (2008) showed that overexpression of WRKY70 reduced JA responses and mutually JA treatment inhibited WRKY70 expression. On the contrary, WRKY70 is known as a positive regulator of SA-mediated defense because increases PR1 which is often studied as a marker gene for SA-dependent defense signaling (Li et al., 2004). It is reported that PGP Bacillus cereus AR156 stimulated the transcription of WRKY70 in Arabidopsis leaves. According to their study, WRKY70 modulated B. cereus-triggered ISR through activating SA signaling pathway (Wang et al., 2018). Our results are in accordance with the reported studies and showed induction of WRKY70 transcription upon treatment with SA, and not with MeJA, in FOL inoculated and non-inoculated plants. IC10 and Y28 also significantly induced transcription of WRKY70 and IC10 continued to induce gene expression after FOL inoculation. Following increased WRKY70 transcripts, expression of PR1 increased in SA and Y28 treatments. Great increase in PR1 transcript abundance by Y28 and not IC10 indicates that these two PGPRs stimulate different pathways to elicit defense priming in tomato plant.
UDP-glucose dependent hydroquinone: O-glucosyltransferase (arbutin synthase) is a member of glycosyltransferases catalyze production of arbutin in higher plants (Arend et al., 2000a). Arbutin is a phenolic glycoside that has antimicrobial and antifungal activity (Kundakovic et al., 2014). Arbutin synthase is a multifunctional enzyme converting various natural products, xenobiotics and toxins (Arend et al., 2000b). There is no report about induction of UDP in response to F. oxysporum or Streptomyces PGPRs in tomato plant. A previous report revealed that induction of members of UDP family, UGT73B3 and UGT73B5 is necessary during the hypersensitive response of Arabidopsis to plant pathogen Pseudomonas syringae (Langlois-Meurinne et al., 2005). Also treatments of Arabidopsis with SA and MeJA induced the expression of another gene family member UGT73C5 involved in mycotoxin detoxification (Poppenberger et al., 2003). Increased UDP transcript in FOL inoculated tomato can be attributed to the role of arbutin in detoxification of fungal toxin or its antimicrobial effect (Kuzniak et al., 2015). The increased expression of this gene in treatment of MeJA and not SA represents its role in ISR. Compared to FOL inoculation, a significant lower level of UDP expression was observed in plants treated with IC10 and Y28 highlighting the potential role of these PGP strains in tomato defense priming.
FOL, SA, MeJA and PGPRs induced expression of TPX1. TPX1, peroxidase encoding gene, is involved in the synthesis of lignin and suberin (Quiroga et al., 2000). Here, TPX1 expression induction by Y28 and IC10 revealed their role in ISR. TPX1 expression at the lower level was observed 48 h after FOL inoculation. Different POX activities in plants treated with PGPRs could be related to the TPX1 transcript abundance.
Phenylalanine ammonia lyase encoded by PAL is involved in the biosynthesis of salicylic acid and defense compounds including flavonoids, phenylpropanoids and lignin. Induction of PAL activity in response to various stimulants such as tissue plant pathogens and hormones was reported (Edwards et al., 1985). In this study, induced expression of PAL in bacterial and hormonal treatments was observed and there was no difference between the induction effects of bacterial strains. The induction of PAL enzymatic activity and PAL gene expression in tomato plants upon pre-treatment with Bacillus thuringiensis (Akram et al., 2013) and the mycorrhizal fungus Funneliformis mosseae (Song et al., 2015) were also reported.
Members of the ERF protein family are transcription factors involved in ET/JA or SA signaling pathways and cause moderate disease resistance responses in various plant species (Liang et al., 2008). Induced expression of ERF1 in Arabidopsis plants treated with a PGP biocontrol strain of Paraburkholderia phytofirmans in response to a model plant pathogen P. syringae pv. tomato DC3000 was reported recently (Timmermann et al., 2017). Likewise, induction of ERF1 by treatment of Y28 tomato was observed in the present study too. On the contrary, plants treated with IC10 decreased ERF1 expression. Our data supports the hypothesis that studied PGP strains regulate tomato defense responses through different molecular pathways possibly acting at transcriptional level.
LOX is a signaling molecule involved in pathogen plant resistance. Strain Y28 slightly induced LOX expression in non-pathogen inoculated plants. FOL inoculation and IC10 treatment caused a significant decreased in transcript level of LOX compared to the control plants. FOL inoculation of both bacterial treatments caused an induction of gene expression. Our finding is consistent with Mariutto et al. (2011) that showed tomato plants treated with PGP Pseudomonas putida did not induce LOX transcription before inoculation with the pathogen B. cinerea. Although transcripts of LOX for both strains were higher in FOL inoculated plants, gene expression levels in non-inoculated bacterial treatments were different significantly.
Conclusion
The superior PGP antagonistic Streptomyces strains, show biocontrol activities against Fusarium wilt of tomato caused by F. oxysporum Schlecht. f. sp. lycopersici (Sacc.) race 3. Significant positive correlation between siderophore and IAA production and siderophore accumulation and growth inhibition showed a relationship between PGP and antagonistic traits. A significant negative correlation between growth inhibition (in vitro assay) and disease severity (in vivo assay) confirmed a need for in vitro tests to select the best biocontrol strains. The correlation of PGP traits with biocontrol activity in phylogenetically close species isolated from distant habitats refers to the role of the natural selection in preserving traits that give superiority to bacteria in the rhizosphere. Here we showed biocontrol Streptomyces stimulate plant defense system through different molecular pathways at the transcriptional level.
Author Contributions
NS and AS designed and directed the research. MS gave advice during experiments. SA carried out all experiments. All authors contributed to the interpretation of the results. SA wrote the manuscript with help from NS and AS. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the Agricultural Biotechnology Research Institute of Iran (ABRII) (Grant No. 12-05-05-034-09454-950941).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01505/full#supplementary-material
Identification of Fusarium oxysporum f. sp. lycopersici (FOL) physiological race 3 by selective primers. Polymerase chain reactions carried out using unif, sp13, sp23, and sprl primer sets. 1 kb: I kb ladder; C, negative control.
Bacterial colonies, S. enissocaesilis strain IC10 and S. rochei strain Y28, on ISP2 medium 10 days after cultivation.
References
- Abd-Allah E. F. (2001). Streptomyces plicatus as a model biocontrol agent. Folia. Microbiol. 46 309–314. 10.1007/bf02815619 [DOI] [PubMed] [Google Scholar]
- Akram W., Mahboob A., Javed A. A. (2013). Bacillus thuringiensis strain 199 can induce systemic resistance in tomato against Fusarium wilt. Eur. J. Microbiol. Immunol. 3 275–280. 10.1556/EuJMI.3.2013.4.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. B., Zuberer D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fert. Soil 12 39–45. 10.1007/bf00369386 [DOI] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arend J., Warzecha H., Stockigt J. (2000a). Hydroquinone:O-glucosyltransferase from cultivated Rauvolfia cells: enrichment and partial amino acid sequences. Phytochem 53 187–193. 10.1016/s0031-9422(99)00539-7 [DOI] [PubMed] [Google Scholar]
- Arend J., Warzecha H., Hefner T., Stockigt J. (2000b). Utilizing genetically engineered bacteria to produce plant-specific glucosides. Biotechnol. Bioengin. 76 126–131. 10.1002/bit.1152 [DOI] [PubMed] [Google Scholar]
- Barker S. J., Edmonds-Tibbett T. L., Forsyth L. M., Klingler J. P., Toussaint J. P., Smith F. A., et al. (2005). Root infection of the reduced mycorrhizal colonization (rmc) mutant of tomato reveals genetic interaction between symbiosis and parasitism. Physiol. Mole. Plant Pathol. 67 277–283. 10.1016/j.pmpp.2006.03.003 [DOI] [Google Scholar]
- Beckers G. J., Conrath U. (2007). Priming for stress resistance: from the lab to the field. Curr. Opin. Plant. Biol. 10 425–431. 10.1016/j.pbi.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Ben Abdallah R. A., Mokni-Tlili S., Nefzi A., Jabnoun-Khiareddine H., Daami-Remadi M. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol. Control 97 80–88. 10.1016/j.biocontrol.2016.03.005 [DOI] [Google Scholar]
- Berger L. R., Reynolds D. M. (1958). The chitinase system of a strain of Streptomyces griseus. Biochim. Biophy. Acta. 29 522–534. 10.1016/0006-3002(58)90008-8 [DOI] [PubMed] [Google Scholar]
- Bruce T. J., Pickett J. A. (2007). Plant defence signaling induced by biotic attacks. Curr.Opin. Plant. Biol. 10 387–392. 10.1016/j.pbi.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Cao P., Liu C., Sun P., Fu X., Wang S., Wu F., et al. (2016). An endophytic Streptomyces sp. strain DHV3-2 from diseased root as a potential biocontrol agent against Verticillium dahliae and growth elicitor in tomato (Solanum lycopersicum). Anton. Leeuw. 109 1573–1582. 10.1007/s10482-016-0758-6 [DOI] [PubMed] [Google Scholar]
- Chance B., Maehly A. C. (1955). Assay of catalase and peroxidase. Methods Enzymol. 2 764–775. [Google Scholar]
- Chellemi D. O., Gamliel A., Katan J., Subbarao K. V. (2016). Development and deployment of systems-based approaches for the management of soilborne plant pathogens. Phytopathol 106 216–225. 10.1094/PHYTO-09-15-0204-RVW [DOI] [PubMed] [Google Scholar]
- Chittoor J. M., Leach J. E., White F. F. (1997). Differential Induction of a Peroxidase Gene Family During Infection of Rice by Xanthomonas oryzae pv. oryzae. Mol. Plant. Microbe Interac 10 861–871. [DOI] [PubMed] [Google Scholar]
- Chun J., Goodfellow M. (1995). A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Sys. Bacteriol 45 240–245. 10.1099/00207713-45-2-240 [DOI] [PubMed] [Google Scholar]
- Dahal B., NandaKafle G., Perkins L., Brozel V. S. (2017). Diversity of free-Living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiol. Res. 195 31–39. 10.1016/j.micres.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Das K., Roychoudhury A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2:53 10.3389/fenvs.2014.00053 [DOI] [Google Scholar]
- Davelos A. L., Xiao K., Samac D. A., Martin A. P., Kinkel L. L. (2004). Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microb. Ecol. 48 601–612. 10.1007/s00248-004-0031-9 [DOI] [PubMed] [Google Scholar]
- de Oliveira-Longatti S. M., Marra L. M., Lima Soares B., Bomfeti C. A., da Silva K., Avelar Ferreira P. A., et al. (2014). Bacteria isolated from soils of the western Amazon and from rehabilitated bauxite-mining areas have potential as plant growth promoters. World. J. Microbio.l Biotechnol. 30 1239–1250. 10.1007/s11274-013-1547-2 [DOI] [PubMed] [Google Scholar]
- Edwards K., Cramer C. L., Bolwell G. P., Dixon R. A., Schuch W., Lamb C. J. (1985). Rapid transient induction of phenylalanine ammonia-lyase mRNA in elicitor-treated bean cells. Proc. Nat. Acad. Sci. U.S.A. 82 6731–6735. 10.1073/pnas.82.20.6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fravel D. R., Olivan C., Alabouvette C. (2003). Fusarium oxysporum and its biocontrol. New Phytol. 157 493–502. 10.1046/j.1469-8137.2003.00700.x [DOI] [PubMed] [Google Scholar]
- Gherbawy Y., Elhariry H., Altalhi A., El-Deeb B., Khiralla G. (2012). Molecular screening of Streptomyces isolates for antifungal activity and family 19 chitinase enzymes. J. Microbiol. 50 459–468. 10.1007/s12275-012-2095-4 [DOI] [PubMed] [Google Scholar]
- Gouda S., Kerry G. R., Gitishree D., Paramithiotis S., Shin H.-S., Patra J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206 131–140. 10.1016/j.micres.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Goudjal Y., Toumatia O., Yekkour A., Sabaou N., Mathieu F., Zitouni A. (2014). Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res 169 59–65. 10.1016/j.micres.2013.06.014 [DOI] [PubMed] [Google Scholar]
- Hirano Y., Arie T. (2006). PCR-based differentiation of Fusarium oxysporum ff. sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. J.Gen. Plant Pathol. 72 273–283. 10.1007/s10327-006-0287-7 [DOI] [Google Scholar]
- Hsu S. C., Lockwood J. L. (1975). Powdered Chitin Agar as a Selective Medium for Enumeration of Actinomycetes in Water and Soil. Appl. Microbiol. 29 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampfer P., Kroppenstedt R. M., Dott W. (1991). A numerical classification of the genera Steptomyces and Streptoverticillium using miniaturized physiological tests. J. Gen. Microbiol. 137 1831–1891. 10.1099/00221287-137-8-1831 [DOI] [Google Scholar]
- Kanini G. S., Katsifas E. A., Savvides A. L., Karagouni A. D. (2013). Streptomyces rochei ACTA1551, an Indigenous Greek Isolate Studied as a Potential Biocontrol Agent against Fusarium oxysporum f.sp. lycopersici. BioMed. Res. Int. 387230 1–10. 10.1155/2013/387230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. D., Han J. W., Hwang I. C., Lee D., Kim B. S. (2012). Identification and biocontrol efficacy of “Streptomyces miharaensis producing filipin III against Fusarium wilt. J. Basic. Microbiol. 52 150–159. 10.1002/jobm.201100134 [DOI] [PubMed] [Google Scholar]
- Kim J. D., Han J. W., Lee S. C., Lee D., Hwang I. C., Kim B. S. (2011). Disease control effect of strevertenes produced by Streptomyces psammoticus against tomato fusarium wilt. J. Agric. Food Chem. 59 1893–1899. 10.1021/jf1038585 [DOI] [PubMed] [Google Scholar]
- Kundakovic T., Ciric A., Stanojkovic T., Sokovic M., Kovacevic N. (2014). Cytotoxicity and antimicrobial activity of Pyrus pyraster Burgsd. and Pyrus spinosa Forssk. (Rosaceae). Afr. J. Microbiol. Res. 8 511–518. [Google Scholar]
- Kunova A., Bonaldi M., Saracchi M., Pizzatti C., Chen X., Cortesi P. (2016). Selection of Streptomyces against soil borne fungal pathogens by a standardized dual culture assay and evaluation of their effects on seed germination and plant growth. BMC. Microbiol. 16:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzner H. J. (1981). “The family Streptomycetaceae,” in The Prokaryotes – A handbook on habitats, isolation and identification of bacteria, eds Starr M. P., Stolp H., Trüper H. G., Balons A., Schlegel H. G. (Berlin: Springer Verlag; ), 2028–2090. [Google Scholar]
- Kuzniak E., Wielanek M., Chwatko G., Głowacki R., Libik-Konieczny M., Piątek M., et al. (2015). Salicylic acid and cysteine contribute to arbutin-induced alleviation of angular leaf spot disease development in cucumber. J. Plant Physiol. 181 9–13. 10.1016/j.jplph.2015.03.017 [DOI] [PubMed] [Google Scholar]
- Langlois-Meurinne M., Gachon C. M., Saindrenan P. (2005). Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant. Physiol. 139 1890–1901. 10.1104/pp.105.067223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. 10.1105/tpc.016980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Lu Y., Liu H., Wang F., Xin Z., Zhang Z. (2008). A novel activator-type ERF of Thinopyrum intermedium, TiERF1, positively regulates defence responses. J. Exp. Bot. 59 3111–3120. 10.1093/jxb/ern165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majidi S., Roayaei M., Ghezelbash G. (2011). Carboxymethyl-cellulase and filter-paperase activity of new strains isolated from Persian Gulf. Microbiol. J. 1 8–16. 10.3923/mj.2011.8.16 [DOI] [Google Scholar]
- Mariutto M., Duby F., Adam A., Bureau C., Fauconnier M. L., Ongena M., et al. (2011). The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC. Plant. Biol. 11:29. 10.1186/1471-2229-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlatt M. L., Correll J. C., Kaufmann P., Cooper P. E. (1996). Two genetically distinct populations of Fusarium oxysporum f. sp. lycopersici race 3 in the United States. Plant Dis. 80 1336–1342. [Google Scholar]
- Mayer A. M. (2006). Polyphenol oxidases in plants and fungi: going places? a review. Phytochemistry 67 2318–31. 10.1016/j.phytochem.2006.08.006 [DOI] [PubMed] [Google Scholar]
- McGovern R. J. (2015). Management of tomato diseases caused by Fusarium oxysporum. Crop. Prot. 73 78–92. 10.1016/j.cropro.2015.02.021 [DOI] [Google Scholar]
- Mehrotra S., Pandey P. K., Gaur R., Darmwal N. S. (1999). The production of alkaline protease by a Bacillus species isolate. Bioresour. Technol. 67 201–203. 10.1016/s0960-8524(98)00107-2 [DOI] [Google Scholar]
- Nikoo F. S., Sahebani N., Aminian H., Mokhtarnejad L., Ghaderi R. (2014). Induction of systemic resistance and defense-related enzymes in tomato plants using Pseudomonas fluorescens CHAO and salicylic acid against root-knot nematode Meloidogyne javanica. J. Plant. Prot. Res. 4 383–389. 10.2478/jppr-2014-0057 [DOI] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids. Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell. Dev. Biol. 28 489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pilkington L. J., Messelink G., van Lenteren J. C., Le Mottee K. (2010). Protected Biological Control” – Biological pest management in the greenhouse industry. Biol. Control. 52 216–220. 10.1002/ps.5270 [DOI] [PubMed] [Google Scholar]
- Poppenberger B., Berthiller F., Lucyshyn D., Sieberer T., Schuhmacher R., Krska R., et al. (2003). Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 278 47905–47914. 10.1074/jbc.m307552200 [DOI] [PubMed] [Google Scholar]
- Quiroga M., Guerrero C., Botella M. A., Barceló A., Amaya I., Medina M. I., et al. (2000). A tomato peroxidase involved in the synthesis of lignin and suberin. Plant. Physiol. 122 1119–1128. 10.1104/pp.122.4.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashad Y. M., Al-Askar A. A., Ghoneem K. M., Saber W. I. A., Hafez E. E. (2017). Chitinolytic Streptomyces griseorubens E44G enhances the biocontrol efficacy against Fusarium wilt disease of tomato. Phytoparasit 45:227 10.1007/s12600-017-0580-3 [DOI] [Google Scholar]
- Reis A., Costa H., Boiteux L. S., Lopes C. A. (2005). First report of Fusarium oxysporum f. sp. lycopersici race 3 on tomato in Brazil. Fitopatol. Bras. 30 426–428. 10.1590/s0100-41582005000400017 [DOI] [Google Scholar]
- Ren C. M., Zhu Q., Gao B. D., Ke S. Y., Yu W. C., Xie D. X., et al. (2008). Transcription factor WRKY70 displays important but no indispensable roles in jasmonate and salicylic acid signaling. J. Integr. Plant. Biol. 50 630–637. 10.1111/j.1744-7909.2008.00653.x [DOI] [PubMed] [Google Scholar]
- Sadeghi A., Karimi E., Dahaji P. A., Javid M. G., Dalvand Y., Askari H. (2012). Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World. J. Microbiol. Biotechnol. 28 1503–1509. 10.1007/s11274-011-0952-7 [DOI] [PubMed] [Google Scholar]
- Sadeghi A., Koobaz P., Azimi H., Karimi E., Akbari A. R. (2017). Plant growth promotion and suppression of Phytophthora drechsleri damping-off in cucumber by cellulase-producing Streptomyces. BioControl 62 805–819. 10.1007/s10526-017-9838-4 [DOI] [Google Scholar]
- Salehi M., Moieni A., Safaie N. (2018). Elicitors derived from hazel (Corylus avellana L.) cell suspension culture enhance growth and paclitaxel production of Epicoccum nigrum. Sci. Rep. 8:12053. 10.1038/s41598-018-29762-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi M., Moieni A., Safaie N., Farhadi S. (2019). Elicitors derived from endophytic fungi Chaetomium globosum and Paraconiothyrium brasiliense enhance paclitaxel production in Corylus avellana cell suspension culture. Plant Cell Tissue Organ Cult. 136 161–171. 10.1007/s11240-018-1503-9 [DOI] [Google Scholar]
- Salla T. D., Astarita L. V., Santarem E. R. (2016). Defense responses in plants of Eucalyptus elicited by Streptomyces and challenged with Botrytis cinerea. Planta 243 1055–1070. 10.1007/s00425-015-2460-8 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pena P., Cauich-Pech S. O., Nunez-Farfan J., Nunez-Cebreros R. D., Hernandez-Verdugo S., Parra-Terraza S., et al. (2010). Incidence of Fusarium oxysporum f. sp. lycopersici Races in Tomato in Sinaloa, Mexico. Plant Dis. 94:1376. 10.1094/PDIS-04-10-0255 [DOI] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int J. Sys. Bacteriol. 16 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Soltani A. A., Khavazi K., Asadi-Rahmani H., Omidvari M., Abaszadeh Dahaji P., Mirhoseyni H. (2010). Plant Growth Promoting Characteristics in Some Flavobacterium spp. isolated from Soils of Iran. J. Agri. Sci. 2 1–10. [Google Scholar]
- Song W., Zhou L., Yang C., Cao X., Zhang L., Liu X. (2004). Tomato Fusarium wilt and its chemical control strategies in a hydroponic system. Crop Prot. 23 243–247. 10.1016/j.cropro.2003.08.007 [DOI] [Google Scholar]
- Song Y., Chen D., Lu K., Sun Z., Zeng R. (2015). Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant. Sci. 6:786. 10.3389/fpls.2015.00786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevidya M., Gopalakrishnan S., Kudapa H., Varshney R. K. (2016). Exploring plant growth-promotion actinomycetes from vermicompost and rhizosphere soil for yield enhancement in chickpea. Braz. J. Microbiol. 47 85–95. 10.1016/j.bjm.2015.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M. A. (1978). Isolierung von Melanin-negativen Mutanten aus Streptomyces glaucescens. dissesrtation. Zurich: ETH Zurich, 6276. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann T., Armijo G., Donoso R., Seguel A., Holuigue L., Gonzalez B. (2017). Paraburkholderia phytofirmans PsJN protects Arabidopsis thaliana against a virulent strain of Pseudomonas syringae through the Activation of induced resistance. Mol. Plant. Microbe Interac. 30 215–230. 10.1094/MPMI-09-16-0192-R [DOI] [PubMed] [Google Scholar]
- Tripathi G., Rawal S. K. (1998). A simple and efficient protocol for isolation of high molecular weight DNA from Streptomyces aureofaciens. Biotechnol. Tech. 12 629–631. [Google Scholar]
- Verma V. C., Singh S. K., Prakash S. (2011). Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol. 51:550. 10.1002/jobm.201000155 [DOI] [PubMed] [Google Scholar]
- Walters D., Heil M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant. Pathol. 71 3–17. 10.1016/j.pmpp.2007.09.008 [DOI] [Google Scholar]
- Walters D. R., Ratsep J., Havis N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64 1263–1280. 10.1093/jxb/ert026 [DOI] [PubMed] [Google Scholar]
- Wang P., Liu X., Guo J., Liu C., Fu N., Shen H. (2015). Identification and expression analysis of candidate genes associated with defense responses to Phytophthora capsici in pepper line “PI 201234”. Inter. J. Mol. Sci. 16 11417–11438. 10.3390/ijms160511417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zheng Y., Gu C., He C., Yang M., Zhang X., et al. (2018). Bacillus cereus AR156 activates defense responses to Pseudomonas syringae pv. tomato in Arabidopsis thaliana similarly to flg22. Mol. Plant. Microbe Interac. 31 311–322. 10.1094/MPMI-10-17-0240-R [DOI] [PubMed] [Google Scholar]
- Wu Z., Yang Y., Li K. (2019). Antagonistic activity of a novel antifungalmycin N2 from Streptomyces sp. N2 and its biocontrol efficacy against Rhizoctonia solani. FEMS. Microbiol. Lett. 366:fnz018. [DOI] [PubMed] [Google Scholar]
- Xu G. H., Zheng H. Y. (1986). Study Method of Soil Microbiology. Beijing: Agricultural Press. [Google Scholar]
- Xu T., Li Y., Zeng X., Yang X., Yang Y., Yuan S., et al. (2016). Isolation and evaluation of endophytic Streptomyces endus OsiSh-2 with potential application for biocontrol of rice blast disease. J. Sci. Food. Ag. 97 1149–1157. 10.1002/jsfa.7841 [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. Zhang, S. Liu X. Z., Wen H. A., Wang M. (2010). A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Let. Appl. Microbiol. 51 114–118. 10.1111/j.1472-765X.2010.02867.x [DOI] [PubMed] [Google Scholar]
- Zhao S., Du C., Tian C. (2012). Suppression of Fusarium oxysporum and induced resistance of plants involved in the biocontrol of cucumber Fusarium wilt by Streptomyces bikiniensis HD-087. World. J. Microbiol. Biotechnol. 28 2919–2927. 10.1007/s11274-012-1102-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of Fusarium oxysporum f. sp. lycopersici (FOL) physiological race 3 by selective primers. Polymerase chain reactions carried out using unif, sp13, sp23, and sprl primer sets. 1 kb: I kb ladder; C, negative control.
Bacterial colonies, S. enissocaesilis strain IC10 and S. rochei strain Y28, on ISP2 medium 10 days after cultivation.