Abstract
Recent advances in single-particle cryo-electron microscopy (cryoEM) have resulted in determination of an increasing number of protein structures with resolved glycans. However, existing protocols for the refinement of glycoproteins at low resolution have failed to keep up with these advances. As a result, numerous deposited structures contain glycan stereochemical errors. Here, we describe a Rosetta-based approach for both cryoEM and X-ray crystallography refinement of glycoproteins that is capable of correcting conformational and configurational errors in carbohydrates. Building upon a previous Rosetta framework, we introduced additional features and score terms enabling automatic detection, setup, and refinement of glycan-containing structures. We benchmarked this approach using 12 crystal structures and showed that glycan geometries can be automatically improved while maintaining good fit to the crystallographic data. Finally, we used this method to refine carbohydrates of the human coronavirus NL63 spike glycoprotein and of an HIV envelope glycoprotein, demonstrating its usefulness for cryoEM refinement.
Keywords: glycoproteins, cryoEM, refinement, glycans
Graphical Abstract

Highlights
-
•
New method for refinement of carbohydrates with low-resolution electron density
-
•
Improved physical geometry of glycans in protein structures
-
•
Compatible with cryoEM and X-ray crystallography data
Frenz et al. have developed a new method for refinement of glycoprotein structures against low-resolution cryoEM and X-ray crystallography data. This new method is able to make significantly larger changes to the glycan geometry compared with previous methods, including the ability to changing the glycan's anomer.
Introduction
Carbohydrates are some of the most stereochemically complex biological molecules found in nature. In addition to their energetic and structural roles in living systems, their role when covalently linked to proteins is critically important in a myriad of molecular recognition processes. Viruses frequently use a glycan shield as a tool to evade the immune system, making glycoproteins the focus of intense interest for vaccinology initiatives targeting these viruses (Astronomo and Burton, 2010). Glycoproteins have historically been difficult to study structurally due to difficulties in overexpressing properly glycosylated proteins as well as their innate flexibility. Indeed, carbohydrates are frequently removed for crystallization studies (Derewenda, 2004). Despite these challenges, several thousand glycoprotein crystal structures have been determined. Recent advances in cryo-electron microscopy (cryoEM) have allowed structural studies on previously intractable glycoproteins (Walls et al., 2016a, Walls et al., 2016b; Lyumkis et al., 2013). However, the vast majority of glycoprotein structures suffer from the fact that the resolution of the electron density or electron potential maps for the carbohydrate chains is too limited to allow for accurate atomic modeling of individual glycan moieties. As a result, the implementation of prior knowledge is necessary to obtain reliable and stereochemically realistic structural models.
In 2004 a study reported that 30% of all PDB entries with covalently linked carbohydrates contain errors in nomenclature and/or chemistry (Derewenda, 2004). The potential for widespread erroneous carbohydrate structural analysis is increasing with the rise of cryoEM as a near-atomic-resolution structural technique, as exemplified by the numerous recent structures solved with unrealistic high-energy ring conformations (Agirre et al., 2015a). These observations emphasize the inadequacy and underutilization of available tools and highlight the need for the development of dedicated algorithms for glycoprotein structural refinement. To address this issue, we aimed to create a tool for the automatic detection and refinement of glycan coordinates while resolving incorrect starting glycan conformations.
We developed an approach to identify, correct, and refine glycoproteins guided by low-resolution crystallographic or cryoEM data to expedite structure determination and interpretation. The approach builds upon previous Rosetta-based structure determination tools guided by low-resolution experimental data (Frenz et al., 2017, Wang et al., 2015) and makes use of a previously developed framework for modeling of carbohydrates (Labonte et al., 2017). Compared with previous glycan refinement methods (Agirre, 2017b, Gristick et al., 2017), our approach (1) uses a physically realistic force field to ensure glycan geometry remains correct even under large conformational changes, and (2) adds the ability to change the anomer of the glycan. These protocols are publicly available with the latest Rosetta release and enable refinement of glycoprotein structures against cryoEM (Wang et al., 2016) and X-ray crystallography data, the latter taking advantage of the combined Phenix-Rosetta reciprocal-space refinement pipeline (Terwilliger et al., 2012).
Results
We developed a refinement protocol to detect and correct poorly modeled glycan configurations and to refine glycan coordinates guided by either low-resolution crystallographic data, via Phenix-Rosetta integration (DiMaio et al., 2013), or cryoEM density data. Of particular interest were the sugar rings of glycans, which may adopt a variety of different conformations with a range of energies (Figure 1 A); therefore, we designed our protocol to specifically address these ring conformations. For more on glycan ring conformations, see Agirre et al. (2017a). Several glycan refinement-specific methods have been developed, including tools for automatic detection and setup of glycan-containing structures for subsequent refinement, a score term that enables cartesian refinement of carbohydrate chains, a dedicated routine for handling sugar ring stereochemistry, and a protocol for correcting the geometry of incorrectly built glycans while optimizing the fit to density. The protocol has a large radius of convergence and is able to make substantial modifications to the input models. For example, the protocol can refine high-energy sugar ring conformers into low-energy conformers and correct erroneous anomeric configurations (see Figure 1). Runtime is about six times slower than Phenix refinement: a 300 residue protein was refined in ∼25 min compared with ∼4 min for Phenix refinement alone. The approach is fully described in STAR Methods.
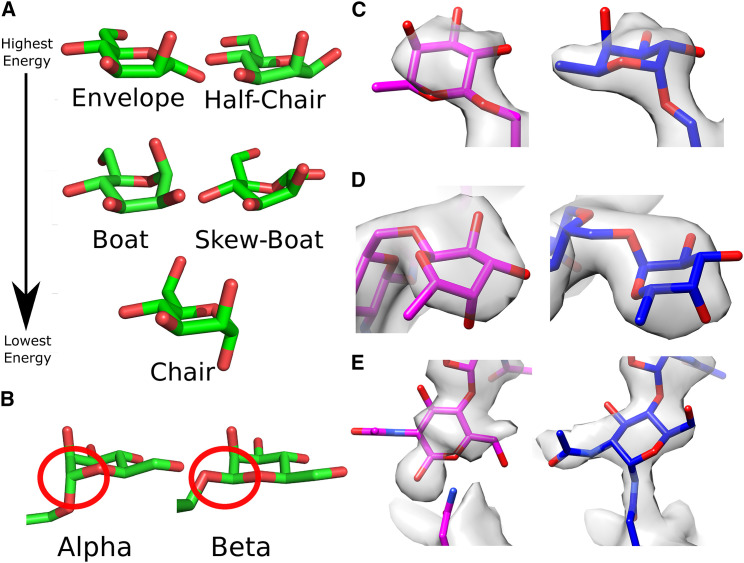
Figure 1.
Anomalies in Low-Resolution Crystal Structures from the PDB Resolved with Rosetta Glycan Refinement
(A) Mannose shown in common ring conformations that can be interconverted ranked by energy. Glycan sugar conformations should always be modeled as chairs unless there is strong evidence to the contrary.
(B) Two anomeric forms are shown, alpha with the glycosidic oxygen and the C5 carbon in trans, and beta in cis. The anomeric carbon is circled in red.
(C) Fucose 507 of PDB: 5NSC has an incorrect anomeric connection in the input (magenta), and this is resolved in the refined model (blue).
(D) Fucose 507 of PDB: 5K65 is in a high-energy ring conformation, and this is resolved in the refined model.
(E) Asp 297 of chain B in PDB: 5K65 fails to form a glycosidic bond to the N-acetyl glucosamine. This is resolved in the output model, and the connection can be seen in the density after rephasing. All density maps are shown at a threshold of 1.
To test our refinement protocol, we considered a benchmark set of 12 N-linked glycan-containing protein structures determined by X-ray crystallography at resolutions ranging between 1.9 Å and 3.5 Å and comprising a total of 133 glycan units. We identified four incorrect anomeric configurations and 23 high-energy ring conformations using the Privateer software (Agirre et al., 2015b) and detected by visual inspection that one structure was missing a glycosidic bond (Table 1 ). We compared refinement of our Rosetta-based method with that of Phenix refinement alone and, for models with high-energy ring conformations in the input, Phenix refinement with constraints generated by Privateer (Agirre, 2017b, Gristick et al., 2017). Following refinement of these 12 structures with our Phenix-Rosetta protein structure refinement pipeline, which alternates real- and reciprocal-space refinement, we were able to markedly improve the carbohydrate geometry, as assessed by Privateer. All the errors detected in the input coordinates were corrected in the output models. In particular, four incorrect anomeric carbon configurations were resolved by adjusting to the correct anomeric state and 23 high-energy ring conformations were refined into a corresponding low-energy conformation with only a slight decline in agreement to the experimental data, consistent with the idea that these glycans are being forced into poor geometry in order to over-fit the density (Figure S1). The best alternative method tested, Phenix refinement with constraints from Privateer, was able to resolve only two of the four incorrect anomers and 12 of the 23 sugars in high-energy conformations (Table 1).
Table 1.
Anomalies in Deposited Structures of Glycoprotein Conjugates
| PDB ID | # of Glycans | Reported Resolution (Å) | Original Model |
Phenix Refined |
Phenix + Privateer |
Rosetta Model |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wrong Anomer | High-Energy Ring | Wrong Anomer | High-Energy Ring | Wrong Anomer | High-Energy Ring | Wrong Anomer | High-Energy Ring | |||
| 1C1Z | 11 | 2.87 | 2 | 2 | 0 | 4 | 0 | 2 | 0 | 0 |
| 1UZG | 13 | 3.5 | 0 | 0 | 0 | 0 | NA | NA | 0 | 0 |
| 2I69 | 3 | 3.11 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5EZJ | 4 | 1.95 | 0 | 0 | 0 | 0 | NA | NA | 0 | 0 |
| 5GZ4 | 12 | 2.55 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| 5H9Y | 15 | 1.97 | 0 | 8 | 1 | 6 | 0 | 3 | 0 | 0 |
| 5K65 | 12 | 2.5 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| 5LA4 | 7 | 1.9 | 0 | 0 | 0 | 0 | NA | NA | 0 | 0 |
| 5N92 | 3 | 2.3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5NSC | 14 | 2.3 | 1 | 0 | 0 | 2 | NA | NA | 0 | 0 |
| 5VEM | 18 | 2.6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 5WBE | 21 | 2.75 | 0 | 6 | 0 | 5 | 0 | 5 | 0 | 0 |
| Total | 133 | 4 | 23 | 2 | 20 | 1 | 11 | 0 | 0 | |
This table shows the resolution of the experimental data as well as the number of incorrect anomers and high-energy ring conformations, as reported by Privateer, for each structure in our benchmark set of crystal structures before and after refinement with the three different methods, Phenix refinement alone, Phenix-Privateer (when high-energy ring conformations are present in the input), and Phenix-Rosetta glycan refinement. Cells marked NA are those for which Privateer constraints are not generated as no high-energy conformations are detected in the input.
A comparison of real-space correlations of the refined and initial models, using both 2mFo-dFc density maps as well as polder omit maps (Liebschner et al., 2017), is also shown in Figure S1. While the geometry consistently improves following Rosetta refinement, the real-space correlations show mixed results: while in some sugars, we see a better fit to the data, in other cases we see a slight worsening. This might be due to the fact our relatively more-restrained model does a worse job at explaining the density resulting from heterogeneous conformations.
To illustrate the improvements resulting from our protocol, we examined the structures of human IL-17AF (PDB: 5N92) and IgG1-Fc (PDB: 5K65). In the IL-17AF structure, fucose 507 was modeled with an incorrect beta configuration, which has been resolved to the correct alpha connection in the refined model (Figure 1C). In IgG1-Fc the fucose 507 is also problematic as it is in a high-energy boat conformation that was automatically detected and corrected in the Rosetta-refined model, which has the expected low-energy 1C4 (Figure 1D). In the IgG1-Fc structure, the most proximal N-acetyl glucosamine of the N-linked glycosylation is not bonded to residue Asn 297 and does not properly fit the density. After refinement and rephasing, the carbohydrate moiety fits with better agreement to the electron density map and its covalent linkage to Asn 297 is properly formed (Figure 1E). These residues are also shown in the polder omit map (Figure S2) (Liebschner et al., 2017).
Refinement of Glycoproteins Using CryoEM Data
In addition to the crystal structures selected for our benchmark set, this glycan refinement pipeline was applied to several viral spike glycoprotein structures recently determined using cryoEM. These include the structures of a human coronavirus (HCoV-NL63) spike glycoprotein (Walls et al., 2016b) and an HIV envelope glycoprotein determined at 3.4 Å and 3.8 Å resolution, respectively. These two proteins mediate receptor binding and fusion of the viral and host membranes at the onset of infection. Since coronavirus spike and HIV envelope glycoproteins are the targets of vaccine design initiatives, it is key to define the structure and function of their carbohydrate components.
HCoV-NL63 is a human-infecting respiratory virus decorated by a spike glycoprotein trimer covered by an extensive glycan shield that has been suggested to assist in immune evasion. We used cryoEM to image this glycoprotein and obtained a resolution at 3.4 Å. In this reconstruction, 93 N-linked glycosylation sites were visible in the map with at least two N-acetylglucosamine moieties present at most of the sites. Modeling of the protein was done using a homology model of the mouse hepatitis virus (Walls et al., 2016a) in combination with Rosetta de novo (Wang et al., 2015), RosettaES (Frenz et al., 2017), Rosetta density-guided iterative refinement (DiMaio et al., 2015), and hand-tracing using Coot (Emsley and Cowtan, 2004). The glycans were initially placed in the map at their approximate positions and the Rosetta glycan refinement protocol was subsequently used to improve the fit to density along with ensuring proper stereochemistry of the sugar rings and of the glycan bond lengths and angles. The efficiency of the protocol is illustrated by its ability to correct for inaccuracies typically observed in hand-traced models, such as the incorrect geometry of the glycosidic bond between Asn 291 and N-acetyl glucosamine 1,404 (Figure 2 A), the unrealistic length of the glycosidic bond connecting residue Asn 1,174 and the N-linked glycan chain that did not optimally fit the density (Figure 2B), and high-energy sugar conformations (Figure 2C).
Figure 2.
Correcting High-Energy Glycans in cryoEM Structures
(A) The unfavorable glycosidic bond between asparagine 241 and N-acetyl glucosamine 1,404 of NL63 (magenta) is resolved in the refined model (blue).
(B) The poor fit to the density of the glycan chain and disconnected glycosidic bond of asparagine 1,174 in the NL63 input model is resolved during refinement.
(C) The high-energy, envelope, ring conformation of mannose 1,428 of NL63, center, is resolved during refinement.
(D) NAG 1,301 of HIV does not form a proper glycosidic bond, and the glycans of the chain do not fit the density in the input (magenta). These issues are resolved in the refined model (blue).
(E) NAG 1,386 of HIV has an unfavorable glycosidic bond angle (magenta), which is resolved in the refined model (blue).
(F) MAN 1,200 of HIV fits the density poorly and the glycosidic bond angle is unfavorable (magenta). In the output model (blue) these issues are resolved.
The HIV envelope glycoprotein trimer is also decorated with an extensive glycan shield that limits access to neutralizing antibodies and thwarts the humoral immune response. Conversely, the arms race between viral evolution mechanisms and the immune systems of infected individuals has also led to the elicitation of antibodies that bind glycan-containing epitopes. To assess the efficacy of our glycan refinement protocol at lower resolutions, we analyzed a 3.8 Å resolution reconstruction of an HIV trimer in complex with an antibody antigen-binding fragment (Fab) determined by cryoEM. In this map, a total of 60 N-linked glycans were resolved and manually docked in density using Coot (Emsley and Cowtan, 2004) before refinement using Rosetta. The robustness of the refinement protocol was demonstrated in its ability to correct for the unrealistic length of the glycosidic bond connecting residue Asn 301 and the N-linked glycan chain that did not optimally fit the density (Figure 2D), the incorrect geometry of the glycosidic bond between Asn 386 and N-acetyl glucosamine 1,386 (Figure 2E), and the inadequate stereochemistry of the glycosidic bonds between Man 1,200 and Man 1,203 (Figure 2F).
Discussion
Here we describe a method for refining glycan atomic coordinates against cryoEM and X-ray crystallography data using Rosetta. Since Rosetta uses a physically realistic all-atom force field, it is well suited for modeling into near-atomic resolution density maps, which is the resolution regime achieved for most cryoEM structures. This Rosetta glycan refinement protocol expands upon previous iterations (Labonte et al., 2017) by avoiding fitting stereochemically unfavorable glycan structures into sparse experimental data and instead yielding physically realistic geometries based on prior knowledge of saccharide chemical properties. Using a benchmark set of 12 deposited crystal structures, we demonstrated our algorithm is capable of correcting models containing significant errors in glycan geometry (Table 1) above and beyond previous methods (Agirre, 2017b, Gristick et al., 2017). This is likely due to the increased radius of convergence of our refinement, as well as the ability to flip anomeric state. This functionality should prove beneficial for large-scale model validation efforts such as “PDB redo” (Joosten et al., 2011, Terwilliger et al., 2012). We further demonstrated the strength and versatility of this algorithm for improvement of glycan stereochemistry in cryoEM models determined at near-atomic resolution with two examples of glycoprotein structures recently obtained and refined using Rosetta.
The expanded Rosetta glycan framework presented here supports automated formatting of input coordinates and detection of glycan connections, as well as cartesian scoring of the most commonly found glycoprotein saccharides, including several glucose, fucose, and mannose derivatives. Future work will further expand the variety of carbohydrates handled, in order to encompass the full known spectrum of glycans found in nature. Considering the increasing number of glycoprotein structures determined at near-atomic resolution in the past few years, we anticipate this algorithm will be a valuable tool for the structural biology community. Rosetta glycan refinement is easy to use and should help novice modelers assign realistic glycan conformations while simultaneously avoiding those not supported by the experimental density.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Protein Structure | Walls et al. (2016b) | PDBID: 5SZS |
| Protein Structure | Schwarzenbacher et al. (1999) | PDBID: 1C1Z |
| Protein Structure | Modis et al. (2005) | PDBID: 1UZG |
| Protein Structure | Kanai et al. (2006) | PDBID: 2I69 |
| Protein Structure | Favuzza et al. (2017) | PDBID: 5EZJ |
| Protein Structure | Lin et al. (2017) | PDBID: 5GZ4 |
| Protein Structure | Qin et al. (2017) | PDBID: 5H9Y |
| Protein Structure | Lobner et al. (2017) | PDBID: 5K65 |
| Protein Structure | Wu et al. (2017) | PDBID: 5LA4 |
| Protein Structure | Goepfert et al. (2017) | PDBID: 5N92 |
| Protein Structure | De Nardis et al. (2017) | PDBID: 5NSC |
| Protein Structure | Gorelik et al. (2017) | PDBID: 5VEM |
| Protein Structure | Cingolani et al. (2017) | PDBID: 5WBE |
| Electron Density | Walls et al. (2016b) | EMDB: 8331 |
| Software and Algorithms | ||
| Rosetta | (Labonte et al., 2017) | N/A |
| Phenix | (Terwilliger et al., 2012) | N/A |
| Privateer | (Agirre et al., 2015b) | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Frank DiMaio (dimaio@uw.edu).
Method Details
The methods developed for this manuscript were built within the Rosetta protein structure modelling package, using a previously developed glycan-modelling framework (Labonte et al., 2017). This previous glycan-modeling framework is well suited for de novo glycan modeling where ideal glycan geometries are built and refined, but was poorly suited at refinement problems where one wishes to refine externally generated (and possibly high-energy) glycan conformers.
To develop the general-purpose refinement tools outlined in this manuscript, several problems in this framework needed to be addressed. The inability to read and write glycans in a standard format (disallowing interoperability with other software packages), limited the applicability of the approach. Furthermore, the glycan score function was unsuited to refining glycans with non-ideal bond-lengths or bond angles. In particular, the inability to energetically assess ring conformations with non-ideal bond lengths and bond angles and the inability to resolve discrepancies between the geometry and the anomeric name assignments limited the applicability of the approach. Finally, specific refinement protocols for cryoEM and crystallographic refinement needed to be developed. The remainder of this section details each of these changes.
Writing Glycans in Standard PDB Format
One of the major limitations of Rosetta’s carbohydrate framework was the inability to write the carbohydrates in standard PDB format. While the standard pdb format for glycan nomenclature has a number of limitations, including the requirement that glycans be assigned unintuitive three-letter codes in order to cover as many conformations as possible, the typical protocol for model building with glycans usually involves multiple software packages utilizing this nomenclature. To resolve this we implemented functionality to map the Rosetta glycan names back to the pdb 3 letter codes in a specialized database. This database is easily expandable to support additional glycan types following the syntax shown in Figure S3.
- These changes have been implemented Rosetta release 3.9, and are enabled by the following flags:
- •-write_glycan_pdb_codes: output glycans using standard PDB codes rather than a Rosetta-specific naming scheme
- •-output_alternate_atomids: write atom names using standard PDB nomenclature
- •-write_pdb_link_records: outputs polysaccharide connections using PDB-formatted “LINK” records.
Automatic Configuration of Glycoproteins
While the standard Rosetta carbohydrate framework is well suited for de novo glycan modeling and other problems dealing with models produced and manipulated entirely by Rosetta, it has major limitations when it comes to software compatibility. This presents significant challenges for employing Rosetta for refinement problems in concert with other software packages. These limitations include a strict syntax requirement, defined by the necessary inclusion of explicit link records for every glycan moiety, and a strict limitation on glycan ordering.
To resolve these issues, we implemented a number of improvements to Rosetta including a refactoring of the way Rosetta handles connectivity. Specifically, any standard PDB file can be read without requiring specific organization or generation of specific LINK records. We also implemented a method to automatically determine glycan connectivity using the geometry of the structure. Explicit link records will now override any automatically detected bonds, meaning a user can rely on the auto-detection code to identify all reasonable geometries while explicitly declaring connections for which the geometry is too poor for auto-detection to work properly.
The rosetta flag -auto_detect_glycan_connections is used to trigger the use of the auto-detection code, and the flag -maintain_links is used to overwrite auto-detected connections with explicit links when present.
Energy-Terms for Scoring of Glycans
The previous implementation of carbohydrate scoring only considered torsion-space refinement of glycans, in which ideal bond length and bond angle were assumed. Low-energy sugar ring conformations were sampled but were not assessed energetically and instead fixed to the sampled ideal conformation in the course of structure refinement. When attempting to develop a general refinement protocol for glycan-containing structures against cryoEM or X-ray crystallographic data, this proved to be a significant limitation. First, previous work (DiMaio et al., 2013, Conway et al., 2014) demonstrated that Cartesian-space refinement allowing for deviations in bond angle and geometry is important in both structure refinement and native structure discrimination tasks. Moreover, the restriction of fixed sugar ring conformations throughout refinement made it difficult to correct a large number of previously identified errors in glycan-containing structures.
To address this issue, several components were necessary: a) extending Rosetta’s bond geometry term to correctly model the energetics of bond geometry deviations in carbohydrates; b) a scoring term capable of assessing the energetics of different ring conformations (and deviations from those ring conformations); and c) a scoring term that ensures each sugar adopts the correct anomeric state. Combined with the protocol described in the next section, this energy function alone is sufficient to correct significant errors in manually built glycans.
To extend Rosetta’s bond geometry term for glycans ideal bond length and bond angles values were taken from phenix using elbow with am1 optimization when the default values in Rosetta were insufficient (Moriarty et al., 2009) and added to the Rosetta database for each glycan in our benchmark set. These glycans currently include alpha and beta glucose, N-acetyl glucosamine, alpha and beta mannose, and alpha and beta fucose. Other glycans are still supported (but will need to be added to the naming database to use pdb formatted outputs), however their bond lengths and angles use standard values based on atom type and should be checked carefully and ideal values updated when outliers occur. Additional constraints were added to ensure the planarity of the amide bond of N-acetyl glucosamine.
To assess the quality of carbohydrate ring conformations, we have made use of a ring conformer database to create a “rotameric” ring model in which the torsions and angles of 38 low-energy ring conformations are tabulated (French and Dowd, 1994). For a given conformation, the nearest ring “rotamer” is identified (using torsional RMSd), and harmonic constraints are generated toward that particular conformation. By adding the -ideal_sugars flag, Rosetta only considers the lowest energy ring conformation, typically the only conformation observed in glycans on a protein surface that have only minimal interaction with the protein or other chemical groups (Agirre et al., 2015a). All experiments reported in this paper make use of this flag.
Glycans adopt two anomeric states, referred to as alpha or beta, based on the position of the anomeric reference atom to the anomeric center carbon. To ensure our score function is capable of resolving incorrect anomers, chirality constraints are added around the anomeric (C1) carbon that enforce the correct anomer. For the common glycans these constraints are implemented as pseudo-torsional constraints on the O of the glycosidic bond, and the C1, C5, and C6 carbons, with an ideal torsion of -120° for the alpha form and 0° for the beta form, for glycans with non-standard naming schemes the corresponding atoms, based on the geometry, are used instead. These constraints, combined with the anomeric hydrogen flipper described in the following section, are sufficient to resolve the incorrect chirality around the anomeric position.
Protocols Enabling the Automatic Repair of Errors in Glycoproteins
To build glycoprotein conjugates into sparse cryo-electron microscopy data, we first assigned each protein residue position utilizing a combination of Rosetta de novo (Wang et al., 2015) RosettaES (Frenz et al., 2017), homology modeling with RosettaCM (Song et al., 2013), and manual model building in COOT (Emsley and Cowtan, 2004). These initial models were then run through the the Rosetta cryoEM refinement protocol (Wang et al., 2016). Next, each glycan was manually docked into the experimental density near the amino acid to which they are covalently bound. This glycosylated structure was subsequently minimized in cartesian space using a reduced weight on the repulsive score term, fa_rep = 0.05. This was followed by idealizing the anomeric hydrogen, in order to resolve any incorrect anomers, and then relaxing the system using the Rosetta “fastrelax” protocol (Tyka et al., 2011). Anomeric hydrogens were then idealized by generating a new residue free of hydrogens and then generating its ideal hydrogen positions based on the location of the non-hydrogen atoms. The anomeric hydrogen of the residue to be modified is then moved to the cartesian coordinates of its idealized counterpart.
This step is capable of changing the anomeric form of the structure to match the name provided in the input. Therefore, users are encouraged to use Privateer to detect errors in their structure before and after refinement to ensure the glycans are being refined to the correct conformation.
In order to use Rosetta glycan refinement with crystallography data, we took advantage of Rosetta’s integration with Phenix (DiMaio et al., 2013) to refine a number of previously published crystal structures containing glycans. Modifications were made to the high resolution crystal refinement protocols to account for the presence of sugars. Specifically, several steps of minimization, with ramping repulsive weights, interspersed with calls to idealize the anomeric hydrogens, as described above, were placed at the beginning of the protocol. The schedule for ramping repulsive weights in our minimization cycles was set to mimic that of the Rosetta fast relax protocol that has been shown to work well for these types of problems. The successively increasing weight of the fa_rep score term was set to 0.011, 0.1375, 0.3025, and 0.55. Calls to idealize the anomeric hydrogen position are done after minimization cycles 1 and 2 to ensure all incorrect anomers have been resolved. The two additional cycles of minimization following the second call to idealize the anomeric hydrogens ensures that all clashes with the anomeric hydrogen are resolved.
Quantification and Statistical Analysis
Anomalies in the glycoprotein structures were assessed using privateer to report mismatches between the name and conformation of the structure as well as high energy conformational states of the sugars (Table 1). Phenix was used to create polder omit maps and calculate the real space correlation to the glycans in both these maps and the 2mFo-dFc density map (Figure S1). Statistical methods of analysis were not used.
Data and Software Availability
This code is available through the Rosetta software package on any release after January 1st 2018. The package is free for academic users and information on licensing can be found at www.rosettacommons.org.
For each protocol, implementation uses “RosettaScripts” (Fleishman et al., 2011) which allows a flexible, XML syntax for describing protocols implemented in Rosetta. The XML scripts used for both protocols are included in the Rosetta distribution and will be kept up to date. The cryoEM refinement script can be found in /main/source/scripts/rosetta_scripts/cryoem/ in the Rosetta source. The script for refinement with crystallography data can be found with the other crystal refinement scripts in the public apps section of the Rosetta software package.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences (R01GM120553 to D.V., R01GM123089 to F.D., T32GM008268 to A.J.B. and A.C.W.), the National Institute of Allergy and Infectious Diseases (HHSN272201700059C to D.V., R01AI113867, U19AI117905, UM1AL100663 to W.R.S., T32AI007244 to J.A.-B.), a Pew Biomedical Scholars Award (D.V.), and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (D.V.).
Author Contributions
Conceptualization, B.F. and F.D.; Software, B.F., S.R., J.A.-B., and F.D.; Validation, B.F, A.J.B, A.C.W., D.V., and F.D. Resources, A.J.B., A.C.W., and D.V.; Investigation, B.F., A.J.B., and A.C.W.; Writing – Original Draft, B.F.; Writing – Review & Editing, B.F., S.R., A.J.B., A.C.W., J.A.-B., W.R.S., D.V., and F.D.; Supervision, W.R.S., D.V., and F.D.
Declaration of Interests
The authors declare no competing interests.
Published: October 18, 2018
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.str.2018.09.006.
Supplemental Information
References
- Agirre J. Strategies for carbohydrate model building, refinement and validation. Acta Crystallogr. D Struct. Biol. 2017;73:171–186. doi: 10.1107/S2059798316016910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre J., Davies G., Wilson K., Cowtan K. Carbohydrate anomalies in the PDB. Nat. Chem. Biol. 2015;11:303. doi: 10.1038/nchembio.1798. [DOI] [PubMed] [Google Scholar]
- Agirre J., Iglesias-Fernández J., Rovira C., Davies G.J., Wilson K.S., Cowtan K.D. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 2015;22:833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- Agirre J., Davies G.J., Wilson K.S., Cowtan K.D. Carbohydrate structure: the rocky road to automation. Curr. Opin. Struct. Biol. 2017;44:39–47. doi: 10.1016/j.sbi.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Astronomo R.D., Burton D.R. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat. Rev. Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Panella A., Perrone M.G., Vitale P., Di Mauro G., Fortuna C.G., Armen R.S., Ferorelli S., Smith W.L., Scilimati A. Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6) Eur. J. Med. Chem. 2017;138:661–668. doi: 10.1016/j.ejmech.2017.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway P., Tyka M.D., DiMaio F., Konerding D.E., Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda Z. The use of recombinant methods and molecular engineering in protein crystallization. Methods. 2004;34:354–363. doi: 10.1016/j.ymeth.2004.03.024. [DOI] [PubMed] [Google Scholar]
- DiMaio F., Echols N., Headd J.J., Terwilliger T.C., Adams P.D., Baker D. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods. 2013;10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F., Song Y., Li X., Brunner M.J., Xu C., Conticello V., Egelman E., Marlovits T., Cheng Y., Baker D. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Favuzza P., Guffart E., Tamborrini M., Scherer B., Dreyer A.M., Rufer A.C., Erny J., Hoernschemeyer J., Thoma R., Schmid G. Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody. Elife. 2017;6 doi: 10.7554/eLife.20383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman S.J., Leaver-Fay A., Corn J.E., Strauch E.-M., Khare S.D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A.D., Dowd M.K. Analysis of the ring-form tautomers of psicose with MM3(92) J. Comput. Chem. 1994;15:561–570. [Google Scholar]
- Frenz B., Walls A.C., Egelman E.H., Veesler D., DiMaio F. RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods. 2017;14:797–800. doi: 10.1038/nmeth.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert A., Lehmann S., Wirth E., Rondeau J.-M. The human IL-17A/F heterodimer: a two-faced cytokine with unique receptor recognition properties. Sci. Rep. 2017;7:8906. doi: 10.1038/s41598-017-08360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik A., Randriamihaja A., Illes K., Nagar B. A key tyrosine substitution restricts nucleotide hydrolysis by the ectoenzyme NPP5. FEBS J. 2017;284:3718–3726. doi: 10.1111/febs.14266. [DOI] [PubMed] [Google Scholar]
- Gristick H.B., Wang H., Bjorkman P.J. X-ray and EM structures of a natively glycosylated HIV-1 envelope trimer. Acta Crystallogr. D Struct. Biol. 2017;73:822–828. doi: 10.1107/S2059798317013353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten R.P., te Beek T.A.H., Krieger E., Hekkelman M.L., Hooft R.W.W., Schneider R., Sander C., Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R., Kar K., Anthony K., Gould L.H., Ledizet M., Fikrig E., Marasco W.A., Koski R.A., Modis Y. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte J.W., Adolf-Bryfogle J., Schief W.R., Gray J.J. Residue-centric modeling and design of saccharide and glycoconjugate structures. J. Comput. Chem. 2017;38:276–287. doi: 10.1002/jcc.24679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 2017;73:148–157. doi: 10.1107/S2059798316018210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.C., Wu B.S., Wu W.G. 5GZ5: crystal structure of snake venom phosphodiesterase (PDE) from Taiwan cobra (Naja atra atra) in complex with AMP. 2017. https://www.ncbi.nlm.nih.gov/Structure/pdb/5GZ5
- Lobner E., Humm A.-S., Mlynek G., Kubinger K., Kitzmüller M., Traxlmayr M.W., Djinović-Carugo K., Obinger C. Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 Å crystal structure of the complex. MAbs. 2017;9:1088–1104. doi: 10.1080/19420862.2017.1364825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D., Julien J.-P., de Val N., Cupo A., Potter C.S., Klasse P.-J., Burton D.R., Sanders R.W., Moore J.P., Carragher B. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y., Ogata S., Clements D., Harrison S.C. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nardis C., Hendriks L.J.A., Poirier E., Arvinte T., Gros P., Bakker A.B.H., de Kruif J. A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G. J. Biol. Chem. 2017;292:14706–14717. doi: 10.1074/jbc.M117.793497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z., Yang D., You X., Liu Y., Hu S., Yan Q., Yang S., Jiang Z. The recognition mechanism of triple-helical β-1,3-glucan by a β-1,3-glucanase. Chem. Commun. (Camb.) 2017;53:9368–9371. doi: 10.1039/c7cc03330c. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher R., Zeth K., Diederichs K., Gries A., Kostner G.M., Laggner P., Prassl R. Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 1999;18:6228–6239. doi: 10.1093/emboj/18.22.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., DiMaio F., Wang R.Y.-R., Kim D., Miles C., Brunette T., Thompson J., Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Dimaio F., Read R.J., Baker D., Bunkóczi G., Adams P.D., Grosse-Kunstleve R.W., Afonine P.V., Echols N. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J. Struct. Funct. Genomics. 2012;13:81–90. doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyka M.D., Keedy D.A., André I., Dimaio F., Song Y., Richardson D.C., Richardson J.S., Baker D. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.-J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y.-R., Kudryashev M., Li X., Egelman E.H., Basler M., Cheng Y., Baker D., DiMaio F. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat. Methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y.-R., Song Y., Barad B.A., Cheng Y., Fraser J.S., DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife. 2016;5 doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Jiang J., Jin Y., Kallemeijn W.W., Kuo C.-L., Artola M., Dai W., van Elk C., van Eijk M., van der Marel G.A. Activity-based probes for functional interrogation of retaining β-glucuronidases. Nat. Chem. Biol. 2017;13:867–873. doi: 10.1038/nchembio.2395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This code is available through the Rosetta software package on any release after January 1st 2018. The package is free for academic users and information on licensing can be found at www.rosettacommons.org.
For each protocol, implementation uses “RosettaScripts” (Fleishman et al., 2011) which allows a flexible, XML syntax for describing protocols implemented in Rosetta. The XML scripts used for both protocols are included in the Rosetta distribution and will be kept up to date. The cryoEM refinement script can be found in /main/source/scripts/rosetta_scripts/cryoem/ in the Rosetta source. The script for refinement with crystallography data can be found with the other crystal refinement scripts in the public apps section of the Rosetta software package.