Abstract
Single-cell gene expression analyses of mammalian tissues have uncovered profound stage-specific molecular regulatory phenomena that have changed the understanding of unique cell types and signaling pathways critical for lineage determination, morphogenesis, and growth. We discuss here the case for a Pediatric Cell Atlas as part of the Human Cell Atlas consortium to provide single-cell profiles and spatial characterization of gene expression across human tissues and organs. Such data will complement adult and developmentally focused HCA projects to provide a rich cytogenomic framework for understanding not only pediatric health and disease but also environmental and genetic impacts across the human lifespan.
Introduction
In recent years, there have been dramatic advances in technologies to profile molecules in single cells. Efforts to profile single cells were first introduced nearly three decades ago, pioneered in the 1990s by groups headed by James Eberwine (Van Gelder et al., 1990, Eberwine et al., 1992) and Norman Iscove (Brady et al., 1990). In the past few years, the field has been transformed by a series of advances that combine next-generation sequencing and massively parallel processing of single cells, first with single-cell RNA sequencing (RNA-seq) (Macosko et al., 2015, Klein et al., 2015, Gierahn et al., 2017), chromatin organization (Buenrostro et al., 2015), and sequence variation (Yuan et al., 2017) as well as in combination for multimodal readouts (Stuart and Satija, 2019). Other experimental technologies under development in proteomics (Specht, and Slavov, 2018), chromosomal conformation (Lando et al., 2018), dynamic cell imaging (Fermie et al., 2018), and lineage tracing (Woodworth et al., 2017) present great promise for studying transient processes in single cells that complement longstanding histological characterization methods. These technologies can provide views into cellular and tissue physiology and pathology that would be only apparent at single-cell resolution (Figure 1), with exceptional potential for producing transformative insights across fields such as developmental biology, genetics, disease pathology, and evolutionary biology (Baslan and Hicks, 2017, Marioni and Arendt, 2017, Behjati et al., 2018).
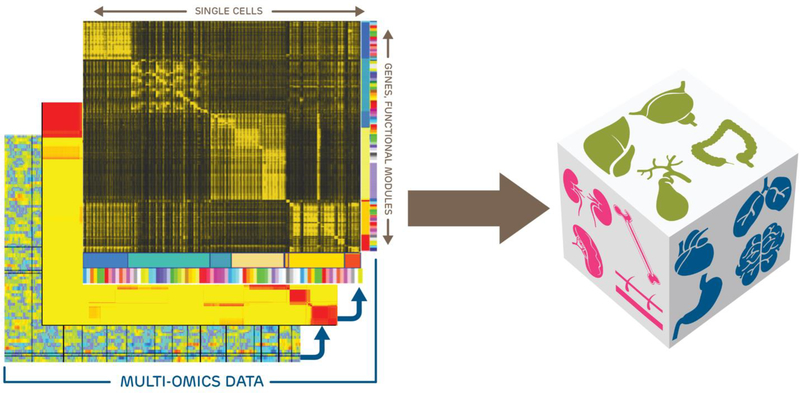
Figure 1. Compiling a Pediatric Single-Cell Atlas.
A pediatric single-cell atlas can consist of multi-omics data from hundreds to many thousands of cells isolated from multiple tissues from normally developing and disease-affected individuals. Single cells can be grouped into cell types that have unique molecular profiles representing primary programs for that cell type as well as sub-state-specific additional programming. The utility of a single-cell atlas is the possibility to map molecular signatures driving developmental, physiological, and pathological processes. Thus, single cell-based signatures can reveal the roles and responses of multiple cell lineages that dictate a given organ’s and/or tissue’s collective biology.
Given these advances, whole-organism tissue maps at the single-cell level are now feasible (Cao et al., 2017, Sebé-Pedrós et al., 2018, Plass et al., 2018, Tabula Muris Consortium et al., 2018). A case for the creation of a comprehensive human cell atlas, including the scientific history, technologies, challenges, and promise for a project of that scale has been recently well described (Regev et al., 2017, Regev et al., 2018). The construction of the Human Cell Atlas (HCA), which focuses on single-cell profiles and spatial characterization of all adult, pediatric, and human developmental tissues, systems, and organs, is now underway, and is organized under a global “coalition of the willing” where researchers will generate data under different funding sources to be deposited into a central data coordination platform (Rozenblatt-Rosen et al., 2017, Regev et al., 2018). Across the world, multiple initiatives will contribute to the creation of a human cell atlas, as well as to applications in specific disease areas. For example, the National Institutes of Health (NIH) supports programs such as the Human Biomolecular Atlas Program (HuBMAP), the Human Tumor Atlas Network (HTAN), and the BRAIN Initiative Cell Census Network programs. However, while the Pediatric Cell Atlas (PCA) is a cornerstone of the full HCA (Regev et al., 2018), with few exceptions (INSERM, 2018, MRC, 2018, LungMap, 2019) to date most initiatives do not focus on normal pediatric tissues.
The Case for a Pediatric Cell Atlas
Support for research on the health of children still proportionally lags behind that for adults, including in funding from the NIH (Gitterman et al., 2018a, Gitterman et al., 2018b). The inevitable scientific advances driven by the HCA are expected to profoundly influence translational and precision medicine research (Shalek and Benson, 2017). Likewise, developmental atlases will offer new insight into the unique molecular and cellular processes operating during embryonic and fetal stages (Behjati et al., 2018). However, without a systematic inclusion of children in the current atlassing endeavors, advancements in pediatric precision medicine and therapeutic development will continue to fall behind. To also secure these breakthrough discoveries for children, we propose a longitudinal pediatric component within the HCA consortium, a PCA. The plan for a PCA was originally outlined in the Human Cell Atlas White Paper (Regev et al., 2018) to represent a distributed and interdisciplinary research effort into studying the unique biology of children in the context of child health and human development (Figure 2). We expect the data generated from healthy tissues for a PCA would help to directly address many important questions in biology and medicine, some of which we discuss below.
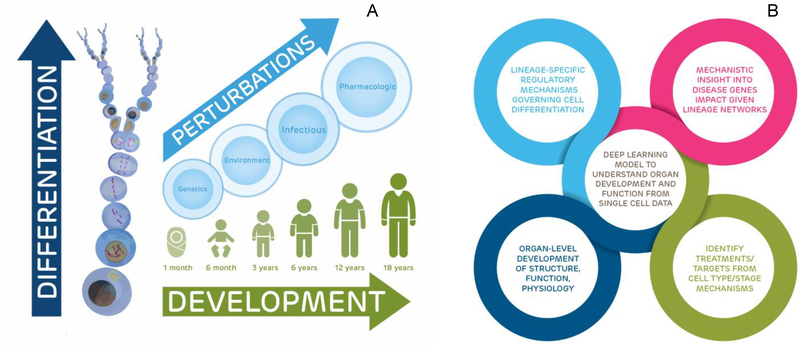
Figure 2. Potential Applications for a PCA.
The PCA has the potential to map and illuminate the cellular basis of normal and abnormal development, cell- and organ-level differentiation, and compensatory and causal processes of disease.
(A) Healthy children are frequently in a global state of growth activation compared to adults through the effects of growth factors, leading to profound impacts on gene expression and cell and tissue interactions, especially in the context of perturbations due to genetics, acquired somatic mutations, environment, infectious disease, and pharmacologics.
(B) All of the outputs of a pediatric single-cell atlas are interrelated to provide a holistic outlook on how cells and tissues interact, differentiate, and function with each other in times of normal versus disease states
How do cell-specific developmental programs vary over the human lifespan?
Embryonic, fetal, juvenile, adolescent, and adult tissues have unique classes of gene expression and developmental programs (Ranzoni and Cvejic, 2018). This is well demonstrated in the Functional Annotation of the Mammalian Genome (FANTOM5) collection, which has utilized CAGE (Cap Analysis of Gene Expression) sequencing from all major organs, primary cell types, and 30 time courses of cellular differentiation (Lizio et al., 2015). It is believed that disruptions to developmental programs operating during fetal and postnatal growth may strongly influence health later in life, especially in metabolic, respiratory, and cardiovascular systems (Barker, 2004, Hanson and Gluckman, 2014, Visentin et al., 2014), but it is not yet clear how these effects carry forward in tissues from early development to adulthood. Epigenetic patterns differ between stem-like and differentiated cell types, but it is unclear how lineage-specific and somatic stem cells are altered during postnatal maturation, aging, and as a function of environmental exposures (Meissner et al., 2008). Furthermore, despite technological advancements in mapping epigenetic landscapes, the epigenetic factors that drive tissue maturation and aging remain largely undefined (Todhunter et al., 2018).
Other open questions include how stage-specific differences in healthy tissues vary by internal and external factors, such as growth factor signaling, or how nutritional and environmental factors may impact regulatory mechanisms. In Figure 3, we show how a stage-specific emphasis on single-cell healthy tissue atlas data can be used to derive novel modules of differentially expressed genes, revealing new insights into pathways and networks of fundamental importance that distinguish between prenatal, postnatal, and adult-stage differentiated neurons. As further shown in Figure 4, even a simple comparative re-analysis of late fetal-versus-adult neurons using single-cell transcriptome data (Darmanis et al., 2015) reveals completely unique gene signatures that are deeply enriched for biological processes and networks whose dysfunction leads to human nervous system developmental disorders (full data access at https://toppcell.cchmc.org/). We present these results to show the power of this approach for any developing system.
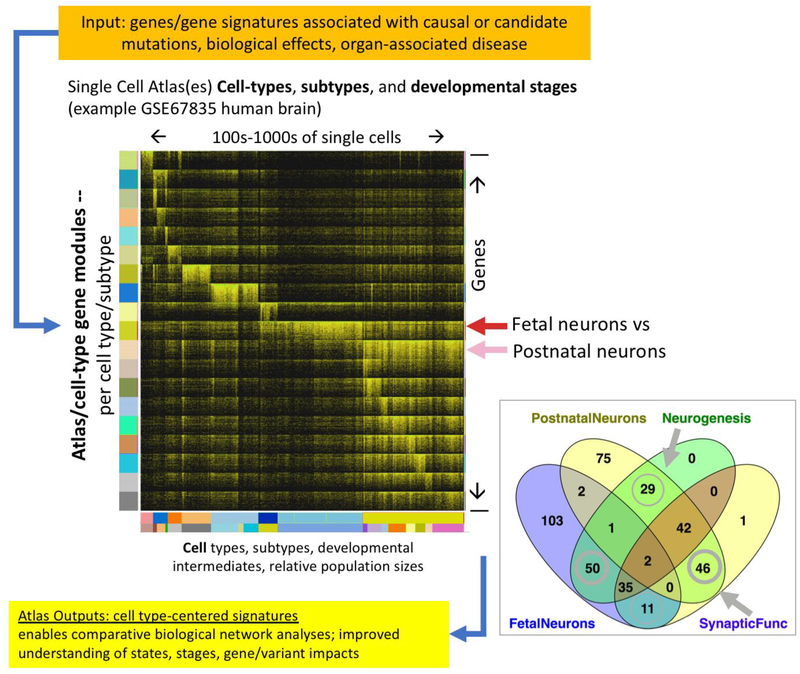
Figure 3. Example of Data Reuse When Datasets Are Analyzed from the Perspective of Building the PCA.
Reanalysis of the Human Brain Single-Cell Survey Study (Darmanis et al., 2015; NIH GEO GSE84465) yields a series of gene expression modules that exhibit the greatest differential expression between cell classes, subclasses, and stages. The heatmap shows the top 200 differentially expressed genes per each cell type, subtype, and stage (log2(TPM+1)) and highlights the the major signatures of fetal and postnatal neurons while contrasting the lack of representation of mature differentiated neuron subtypes (cells on the right side of heatmap; signature modules on the lower half of heatmap) in fetal neurons (middle portion of the heatmap). Very few of the top stage-specific neuronal genes overlap (fetal neurons versus postnatal neurons) despite enrichment of similar functions with completely different genes comprising those categories. Moreover, there are also subtle, albeit fundamental, shifts in the biological functions of the developmental stage gene modules. An interactive view of this data can be seen at http://toppcell.cchmc.org/.
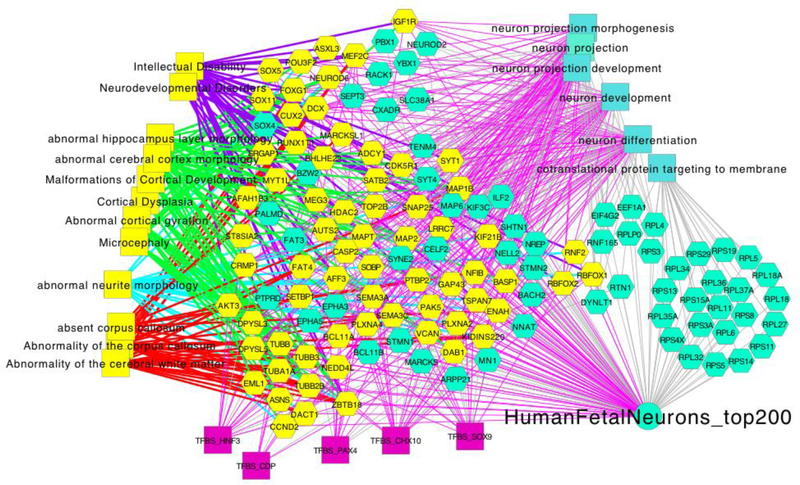
Figure 4. Enrichment Analysis of the Major Signature Overexpressed in Fetal Neurons versus Those from Postnatal and Adult Human Brain.
Modular analysis of data shown in Figure 3 yields functional associations (rectangles) shared by the top 200 contrasting genes (hexagons) and their links to Gene Ontology, mouse gene knockout phenotype, or human OMIM gene-associated phenotype terms (phenotype-associated genes [yellow hexagons] which are connected by separately colored edges per phenotype group. This example highlights the necessity of profiling fetal and pediatric cells and genes, which have similar functions and processes compared to their adult counterparts but impact development, function, and physiology at different stages of development through different gene and regulatory programs. It also indicates that critical genetic associations can only be fully appreciated in the context of fetal stage neurons rather than their mature counterparts. Network analysis carried out using the “top 200 fetal quiescent neuron” gene-expression signature shown in Figure 3 as analyzed using the http://toppcluster.cchmc.org/ multiple-annotations biological network analysis functions to generate XGMML output that was then clustered in Cytoscape (Shannon et al., 2003).
What cell populations are present in and distinguish pediatric tissues?
For the large majority of healthy pediatric tissues, there exists little or no understanding of how cellular processes affect the course of development and maturation, or how pediatric cell populations contrast with those from adults. Signaling, transcriptional, and epigenetic factors are believed to act differently in children’s tissues leading to a global state of growth-guiding development (Stevens et al., 2013), while most adult tissues are believed to be quiescent with respect to growth, and replication serves to maintain established tissue architecture (Clevers and Watt, 2018). As a consequence, the cell types and their molecular states contributing to tissue growth in children might well differ from those during homeostatic cell renewal that serves to maintain a tissue in adult life, though specific concepts of “cell type” and “cell state” still require rigorous scientific definition (Trapnell, 2015) and ontological classification, such as that found in the Cell Ontology (Meehan et al., 2011, Osumi-Sutherland, 2017). Significant differences in cell populations between pediatric and adult tissues have been observed, for instance, in bone marrow (Choumerianou et al., 2010), but it is unclear if the observed differences are a result of specific developmental cell states, unique pediatric or adult cell types, or differences in tissue distribution or proportion of cell populations, all of which may vary in tissues by age, sex, genetics, or developmental stage. Cellular heterogeneity in pediatric tissue cell populations may also contribute to the regulation of growth while maintaining organ function. Regulatory control may be due to changes in inter-cellular variability of gene expression for key pathways or due to the action of rare cell subpopulations that have not yet been discovered (Hasegawa et al., 2015), both of which would be impossible to resolve with ensemble data generated from bulk tissue.
How is pediatric physiology distinguished in health and disease?
The mantra that “children are not just small adults” is evident from critical differences in pediatric pharmacology and physiology as seen by responses to therapeutic interventions and by treatment outcomes. These differences are not well understood at the tissue level (Fernandez et al., 2011). Age-dependent responses to anesthesia and medications (Batchelor and Marriott, 2015, Andropoulos, 2018), environmental exposures (Wright, 2017), traumatic injuries (Luerssen et al., 1988, Resch et al., 2019), or surgical outcomes (Imura et al., 2001) have been observed, but the reasons for these differences are still unclear. Childhood diseases also have age-dependent symptoms, prognoses, and outcomes (Wheeler et al., 2011) even when stratified for key clinical variables, possibly due to tissue developmental composition at the time of dysfunction (Mann et al., 2010). Since many childhood-onset diseases become lifelong, non-communicable chronic diseases, it is key to improve our understanding of disease inception in early childhood. Undetected disorders in neonates, such as sudden infant death syndrome, are restricted to a relatively small postnatal period during which organs are undergoing rapid maturation, and these disorders are likely impacted by intrauterine factors (Athanasakis et al., 2011). There is also little known about the effects of genomic variants, environmental effectors, or their interactions on individual populations of cells within tissues and how these effects might vary by age to alter normal development, cellular function, or therapeutic efficacies in children.
How is pediatric physiology distinguished in health and disease?
The mantra that “children are not just small adults” is evident from critical differences in pediatric pharmacology and physiology as seen by responses to therapeutic interventions and by treatment outcomes. These differences are not well understood at the tissue level (Fernandez et al., 2011). Age-dependent responses to anesthesia and medications (Batchelor and Marriott, 2015, Andropoulos, 2018), environmental exposures (Wright, 2017), traumatic injuries (Luerssen et al., 1988, Resch et al., 2019), or surgical outcomes (Imura et al., 2001) have been observed, but the reasons for these differences are still unclear. Childhood diseases also have age-dependent symptoms, prognoses, and outcomes (Wheeler et al., 2011) even when stratified for key clinical variables, possibly due to tissue developmental composition at the time of dysfunction (Mann et al., 2010). Since many childhood-onset diseases become lifelong, non-communicable chronic diseases, it is key to improve our understanding of disease inception in early childhood. Undetected disorders in neonates, such as sudden infant death syndrome, are restricted to a relatively small postnatal period during which organs are undergoing rapid maturation, and these disorders are likely impacted by intrauterine factors (Athanasakis et al., 2011). There is also little known about the effects of genomic variants, environmental effectors, or their interactions on individual populations of cells within tissues and how these effects might vary by age to alter normal development, cellular function, or therapeutic efficacies in children.
Applications in Research and Medicine
The PCA component of the HCA would generate fundamental contributions to our understanding of pediatric physiology in health and disease and to the development of precise therapeutic interventions for children. Below are some of the many potential applications to research and medicine.
Provide age-matched single-cell profiles of non-diseased tissues as reference maps
Single-cell surveys of bioenergetics, growth, and functional programs from typical fetal and pediatric tissues will contribute to a greater understanding of complex diseases arising from conditions such as congenital birth defects, developmental delays, inborn errors of metabolism, or pediatric cancer. As some pediatric diseases differ in presentation and outcome by age, an atlas of cells in healthy pediatric tissues organized by developmental age would thus be an important and broadly useful data resource. The PCA would promote an “age and stage” approach to sample ascertainment by supporting the creation of indexes for coordinated tissue banks and study populations among participating groups. Data from the PCA would be useful for comparative analyses of normal tissue data versus that from disease and dysregulated states in pediatric tissues, including helping to identify suitable normal controls (Zeng et al., 2019) as well as for concurrent studies on matched adult and developmental tissues within the HCA’s Biological Networks. There is increasing excitement about the use of stem-cell-derived “organoids” as in vitro models of human organ development and disease (Clevers, 2016). However, these cells and tissues represent early, usually fetal, stages of development and will require reference datasets based on single-cell RNA sequencing of normal developing tissues to calibrate cellular fidelity.
Map developmental trajectories of pediatric cells and tissues
The PCA would make important contributions toward our understanding of human growth and development across the human lifespan. The PCA could support generation and analyses of a virtual “time course” of multi-omics data to provide insights into the specifics of pediatric cell regulatory networks (Packer and Trapnell, 2018) across different developmental stages. For example, predicted cellular trajectories in organs such as kidney during human fetal development suggest highly consistent developmental programs in age-matched samples (Wang et al., 2018). Models of cellular processes and their tissue locality would greatly enhance our understanding of changing cellular composition during normal and perturbed development. For instance, how pathways and effectors are regulated within and across different children’s tissues to promote healthy growth and development could be extended to study these processes across the lifespan. Understanding these processes can help inform many aspects of human biology and medicine, including wound healing, tissue regeneration, and capacity to respond to physiological challenge. Comparisons of pediatric, adolescent, and adult single-cell data may also provide insight into how cell types transition from “growing” to “adult homeostatic” states.
Contribute insights in public health
Many chronic diseases that affect specific tissues, such as diabetes, asthma, and neuropsychiatric disorders, often first manifest in childhood or adolescence. Environmental exposures during development, the so-called exposome, may have long-term effects on children’s and adult’s health and tissue function at the cellular level (Balshaw et al., 2017, Vineis et al., 2017), especially during specific developmental windows (Dietert et al., 2000). Neuroimmunologic cell and tissue responses to lower socioeconomic status, stress, inflammation, and air pollution may be linked to observed health disparities (Olvera Alvarez et al., 2018). Thus, targeted studies of single cells along with genetic, demographic, socio-economic, and exposome data may reveal biomarkers and therapeutic opportunities to improve health and outcomes. Nutrition during childhood can impact adult tissue development and function in clinically relevant ways (Rytter et al., 2014) and may have long-term implications for public health (Eriksson et al., 2001, van Abeelen et al., 2012, Lelijveld et al., 2016). A lifespan epidemiological approach to studying human health (Ben-Shlomo and Kuh, 2002) would benefit from the PCA’s contributions to the span of normal tissue growth and cellular function on a stage-by-stage basis.
Increase precision in pediatric drug discovery and toxicology
Children’s responses to medications can differ from those of adults because of differences in drug metabolism and differences in cellular responses to drugs (Stephenson, 2005). These differences can cause substantial morbidity, as was the case for children with SHH-driven medulloblastoma who received vismodegib and subsequently suffered from irreversible growth plate fusions (Robinson et al., 2017). The majority of pediatric patients who require pharmaceutical treatment receive medications that are either not approved or incompletely labeled for pediatric use (Ward et al., 2018). The PCA would be particularly suited to help address these disparities. Partnerships with the pharmaceutical and biotech industries could help accelerate pediatric drug development by providing an index of available pediatric tissues and specialist researchers across participating institutions. The PCA would also provide a pediatric tissue catalog reporting on cell composition and genomic profiles organized by cell type and developmental age. Single-cell data itself could increase the precision of physiologically based pharmacokinetic modeling for pediatric systems by deeply profiling tissue metabolism and unique pathways that underlie developmental programs in key pediatric tissues such as liver, kidney, and gut. This may yield better predictions of adverse effects and provide insight into key differences in responses in pediatric clinical trials and effectiveness studies compared to adults (Yellepeddi et al., 2019).
Establish linked platforms for a global pediatric research network
As a biological network within the HCA, the PCA will bring together scientists across the pediatric research community to enable, innovate, and accelerate research in single-cell-based pediatric health and disease, which would otherwise be infeasible for individual laboratories or institutions to accomplish. Data shared among global partners through the Data Coordination Platform (DCP) for the HCA will allow for novel and flexible context-specific exploratory re-analyses that can yield entirely new insights on molecular patterns and mechanisms responsible for system-specific and multi-system growth and development, as shown in Figures 3 and 4. Within the HCA, the PCA will connect with complimentary organ-, tissue-, and system-specific biological networks to relate pediatric and adult data. Thus, partnerships across an international network of participating children’s hospitals, pediatric-focused research centers, biorepositories, foundations, and consortia would be able to accelerate pediatric research by helping to connect patients and families to research efforts, source rare tissues, integrate and disseminate novel platforms and approaches, develop and support common data harmonization standards, and widely share data to empower collaborative and diverse research teams to address pressing basic and translational science problems. An existing collaborative endeavor among three of the largest children’s hospitals in the country, The Genomics Research and Innovation Network (GRIN), has already developed collaborative instruments and environments for pediatric genetic and clinical data exchange and can be useful in designing similar instruments for the PCA (GRIN, 2019). Membership in the PCA community is open to anyone willing to commit to values of the HCA: transparency and open data sharing; support of community, diversity, equity, and inclusion; commitment to patient privacy; technology development for preparing and analyzing pediatric samples; and dedication to computational excellence (Regev et al., 2018). Information on the effort is open to the community at the PCA component of the HCA web site at http://www.humancellatlas.org/PCA and members can register at https://www.humancellatlas.org/joinHCA.
Expected Challenges And Timelines
Organization and Scope
Developing the PCA toward understanding human tissue growth and development will require significant resources for profiling and analyzing millions of cells from potentially thousands of tissue samples from pediatric donors, with a focus on longitudinal age. To ensure success, the broad scale of the endeavor outlined here would require contributions from diverse teams spanning clinical and basic research across multiple institutions, which together enlist recruitment of samples spanning a range of ages in many pediatric tissues. Legal, ethical, and resource challenges would be encountered during establishment of a PCA biological network that synergizes through collaboration, freely and fairly shares data and best practices, and strives for compliance reciprocity. An effective PCA, as part of the HCA, would create new opportunities for tissue-specific sub-projects to share and maximize resources, including expertise, workflows, and tools, while encouraging appropriate academic attributions and output.
Representing PCA data in the Human Cell Atlas Data Coordination Platform
The HCA Data Coordination Platform supports ingestion of rich and extensible metadata, cloud-scalable and multi-platform mirrored data stores, cloud-native data processing pipelines generating standardized and harmonized data, and user-friendly data access for users. PCA data require specific metadata and an emphasis on establishing data access patterns that accommodate data that is not openly consented. Pediatric research may also emphasize experimental models that may motivate or prioritize additional analysis elements under access control, such as raw RNA-seq data and DNA-sequencing data for probands and families.
Ethics
The PCA must consider the ethics of obtaining samples from healthy children who are not able to provide informed consent themselves and releasing pediatric tissue data under informed consent while simultaneously safeguarding patient identity. The unique challenges of the PCA will be addressed by the HCA Ethics Working Group to address frameworks on ethical management and sharing of pediatric data and release recommendations. Professional pediatric and genetics organizations have recently offered input into ethical issues of genetic testing and research in children (Botkin, 2016), but until recently, there have been relatively few formal opinions on pediatric open-data sharing. Recently, an interdisciplinary Paediatric Task Team as part of the Global Alliance for Genomics and Health (GA4GH; https://www.ga4gh.org/), has developed the Key Implications for Data Sharing (KIDS) framework for pediatric genomics data sharing (Rahimzadeh et al., 2018), which will be part of the developing ethical framework.
Since children grow to be adults and may wish to withdraw their samples at a later date, the PCA must also consider developing procedures for secure sample extraction and removal while retaining strict privacy of pediatric samples for those studies opting to restrict open-sample access. Privacy control will be of particular importance to donor parents, who are often very concerned about open-data sharing of their children’s data and its future consequences (Burstein et al., 2014). Additional protection of patient privacy through genetic de-identification could be accomplished by variant removal from raw transcriptional data while keeping original data available in a controlled-access repository with a Data Access Committee (such as dbGAP or similar). The HCA Ethics Working Group will continue to work with clinicians, researchers, and organizations to support ethical best practices while ensuring data will have the greatest impact on science while respecting patient privacy.
Technology
Single-cell profiling technologies are undergoing rapid evolution. Both dissociation-based and in situ spatial assays are gaining throughput. The molecular types potentially interrogated by these assays include DNA, RNA, protein, metabolite, chromatin spatial conformation, and epigenetic modifications (Stuart and Satija, 2019). Moreover, to acquire a more complete picture of gene activities in single cells, measurements of gene expression should ultimately go beyond changes in overall levels and also interrogate changes in gene isoforms arising from pervasive alternative processing and modifications of RNA (Park et al., 2018). Functional states of cells are also determined by their constellation of protein abundances and by the myriad post-translational modifications that regulate their associations and activities. Single-cell proteome analysis is at an earlier phase of development, mainly limited by losses associated with material handling and chromatographic separations which adversely impact the sensitivity and hence depth of analysis. Advances in these areas along with continual rapid development of evermore sensitive mass spectrometry methods show some promise for this field (Stoeckius et al., 2017, DeLaney et al., 2018, Budnik et al., 2018, Couvillion et al., 2019). Continued improvement across all platforms has allowed for simultaneous “multi-omic” sampling from single cells (Kelsey et al., 2017, Macaulay et al., 2017, Hu et al., 2018b, Clark et al., 2018, Stuart and Satija, 2019). The average costs of an experiment, although significant at this moment, are expected to decrease over time as technologies are further developed and may also be decreased through multiplexing strategies (Gehring et al., 2018, Regev et al., 2018, Stoeckius et al., 2018).
Other rapidly evolving areas of research in the single-cell community include meta-scale approaches to information management, for which the pediatric community must provide input as necessary to their development. For instance, without attention to age- and stage-specific pediatric anatomy, useful spatial atlassing of cells to their tissue and physical locations may be impeded. Another important consideration for PCA projects, where individual genetics and environmental exposures are important variables, is ensuring the use of methods and strategies to maintain the value of community investments by minimizing non-informative experimental replication and maximizing cross-compatibility of data through validation and calibration of new and enhanced technologies for both primary and secondary data generation. Calibration standards between studies and platforms and technology groups will allow for compatibility and linkage between older standards and evolving technological platforms. As part of the HCA, the PCA will reuse other HCA protocols for pediatric tissues as appropriate and follow the guidelines provided by the HCA Standards and Technology Working Group.
Sample ascertainment, biopsy, and management
Collaborative biological PCA networks across various institutions will need to define strategies for tissue procurement, preservation, and single-cell sample preparation while maintaining assay fidelity and throughput. Obtaining pediatric samples is non-trivial but regularly accomplished in studies and clinical trials. Approaches to obtaining pediatric samples have been described across many studies, utilizing tissues from surgical biopsy, transplantation byproducts, and parental donations following accidents (Bandyopadhyay et al., 2018). Methods for easily identifying available pediatric tissues from PCA collaborators with attention to inclusion of demographic diversity, such as sex and ethnicity, will be a critical component of the PCA’s biobanking efforts.
While protocols for single-cell analysis suggest processing either newly acquired or cryopreserved tissues, there have been advances in processing single-cell DNA from formalin-fixed paraffin-embedded tissues to study whole-genome copy number variation in cancer (Martelotto et al., 2017), as well as recovery of transcriptomic data from cryopreserved tissues (Konnikova et al., 2018), both of which open up opportunities for similar analyses utilizing samples from pediatric tissue repositories. The PCA could assist projects by linking them to other well-established biobanks such as those founded by the Children’s Oncology Group (https://www.childrensoncologygroup.org/), the Children’s Brain Tumor Tissue Consortium (https://cbttc.org/), and Alex’s Lemonade Stand (http://www.cccells.org/).
Computation and Information Management
Like other HCA projects and biological networks, the computational challenges of the PCA are a critical consideration. New, improved protocols for tissue preparation, cell isolation, and computational analysis contribute to the growing size and complexity of single-cell data sets (Svensson et al., 2018), an example of which is the HCA’s recent release of transcriptome data for approximately 530,000 cells from umbilical cord blood and bone marrow (https://preview.data.humancellatlas.org/). To standardize our approach and allow for comparison with adult components of the HCA, the PCA will leverage the HCA’s DCP, designed to harmonize deposited data and provide data and portals for access, analysis tools, and visualization. A key analytical need for the PCA, analyses of longitudinal datasets, would be driven by PCA researcher needs. Computational methods with the ability to cluster and visualize cellular heterogeneity across millions of cells have been recently introduced (Cho et al., 2018, Wolf et al., 2018, Zhang and Taylor, 2018), which could be adapted for longitudinal data, for instance using machine learning approaches (Hu and Greene, 2018, van Dijk et al., 2018, Lin et al., 2017, Amodio et al., 2017, Schiebinger et al., 2017, Schiebinger et al., 2019) to help map cell lineages and trajectories particular to pediatric development.
Data Standardization, Harmonization, and Reuse
In-depth anatomical knowledge of how individual cells relate to tissues, organs, and organismal development has been encoded using the Uberon anatomy and stage ontologies, which relate anatomical entities at all spatio-temporal scales to developmental stages. A PCA tissue map that supports “age and stage” requires novel approaches that can combine spatiotemporal classification of cellular attributes in a “bottom-up” fashion with existing “top-down” anatomical and developmental knowledge. Exploratory data analysis of highly complex PCA data will be critical for generating novel hypotheses on growth and development in pediatric tissues. Traditional models for cellular classification during development need to work harmoniously with omics-based single-cell classification strategies in order to maximize utility across the broadest set of scientific use cases aimed at understanding development and pediatric disease progression (Aevermann et al., 2018, Osumi-Sutherland, 2017). Despite the rich resources provided by the Gene Ontology and Cell Ontology to record cellular and subcellular gene functions, much work remains in defining and linking across new and existing ontological terms and frameworks on normal and abnormal development, gene function, cell type, and cell state (Gene Ontology Consortium, 2017). The PCA will work with the HCA’s metadata team to ensure that the HCA metadata schema in the DCP reflects the richness of pediatric-specific metadata.
Funding and Resources
The opportunity to contrast children’s tissues at a single-cell level across ages would provide a unique and unprecedented window into pediatric diseases and their treatment. The PCA represents a project with challenges in tissue procurement and single-cell preparation but offers significant rewards in the potential contributions to our understanding of children’s health and disease. As a diverse and interdisciplinary effort, the PCA would benefit from programs such as the NIH Common Fund or the recently formed trans-NIH Pediatric Research Consortium (N-PeRC), which was founded to coordinate pediatric research programs across NIH’s 27 institutes and centers. It is expected that some fundraising efforts will often be specific to participating institutions or groups and not PCA-wide, as has been the case across the HCA.
Pilot Organ Systems
There exist a substantial number of tissues and organs poised for exploration by a coordinated PCA, each with significant opportunity for producing new insights along with translational and clinical promise. While the PCA pilot studies should focus on normal (non-diseased) tissue, this list is not intended to be exhaustive or directive for all possible tissue, organ, and disease-oriented atlases.
Brain and nervous system
Recent studies of CNS development are richly implicative of the roles of genomic maturation — transcriptomic and epigenetic — of neuron, astrocyte, oligodendrocyte, microglial, and endothelial lineages. Tissue-level processes such as synaptogenesis, myelination, and pruning start during gestation and continue through to adulthood (Emery and Lu, 2015, Sharma et al., 2016). Cellular models of neuronal development used for spatiotemporal reasoning and classification will require reconciliation with molecular characterization (Osumi-Sutherland et al., 2012). Moreover, given that known genetic and non-genetic risk factors only explain a fraction of overall disease risk (Manolio et al., 2009), genomic variability between individual neurons of the brain has long been suspected to contribute to the diversification of neuronal complexity and possibly the burden of neurological disease (Muotri and Gage, 2006, D’Gama and Walsh, 2018). Molecular characterization of cell types in the developing human brain has recently begun and has already led to the identification of novel cell types, novel cell-type-specific markers, and insight into gene networks enriched in specific cells (Pollen et al., 2015, Thomsen et al., 2016, La Manno et al., 2016, Nowakowski et al., 2017). As of now, there have been few deep molecular characterizations of cell types in the pediatric brain as compared to adult, including in critical neurological and brain anatomical locations (Lake et al., 2018), such that hypotheses linking children’s neurological health and disease to cell types and cell states remain limited by our lack of foundational knowledge.
Single-cell DNA sequencing of the human cortex has been used to follow a neuron’s developmental lineage, through tracing mutational patterns (Lodato et al., 2015) or using pseudotime methods (Zhong et al., 2018). These methods could help to reveal developmental lineages and anatomic complexities in the brain with respect to organogenesis, maturation, and the attainment of a fully functional central nervous system.
For the field of neurology, mapping neuronal lineage and assessing the variability within the genetic and epigenetic landscape will be relevant for progress in common neurological disorders in children such as epilepsy, autism, and developmental delay. For example, studies into somatic mosaicism — which is increasingly recognized as a source for focal epilepsies and brain malformations — led to the discovery of the mTOR pathway as a crucial mechanism for epilepsies (Lee et al., 2012, Mirzaa et al., 2016, Alcantara et al., 2017, Lim et al., 2017a). In addition, somatic brain mutations unrelated to the mTOR pathway have also recently been discovered in lesional and non-lesional epilepsy, illustrating the potential power of a PCA for characterizing previously opaque brain disorders through sequencing (Winawer et al., 2018). In patients with autism, postzygotic mutations, only affecting a subset of cells, are increasingly recognized as a disease mechanism (Lim et al., 2017b). In addition, the cellular architecture of human neurons may have unique features, such as human-specific interneuron subtypes (DeFelipe et al., 2006, Boldog et al., 2018), suggesting that elucidating the genetic and epigenetic variability in the human brain through a PCA may discover previously unrecognized cellular patterns that are unique to the developing human brain.
Cell signaling in the neural stem cells, such as the Notch pathway, can be detected in pediatric brain tumors (Lasky and Wu, 2005) and single-cell RNA sequencing of developing human cortex has revealed a diversity of neural stem and progenitor cells with potential roles in neurodevelopmental diseases (Nowakowski et al., 2017). In the field of neuro-oncology, pediatric CNS cancers arise in distinct neuroanatomical locations, different but predictable ages of onset, and are frequently associated with lineage-specific developmental transitions. Profiles of normal brain tissue and of neural stem cells by age as provided by the PCA is necessary in order to develop hypotheses as to cell types and processes giving rise to CNS tumors and the associated risks involved (Gage and Temple, 2013).
Gut
The normal human gut manages fluid, electrolyte, and nutrient absorption needed for survival while protecting us from intestinal microbes. Cells performing these tasks include epithelial absorptive cells, mucus-producing cells, stem cells, transit amplifying cells, Paneth cells, enteroendocrine cells, endothelial cells, fibroblasts, smooth muscle cells, intestinal pacemaker cells, enteric neurons, enteric glia, and a wide array of immune system cells in the bowel wall (Smillie et al., 2018, Haber et al., 2017). Some of these cell types can be further subdivided. For example, there are at least 10 types of enteroendocrine cells that sense luminal contents and mechanical distension and then secrete a wide array of hormones and cytokines that impact bowel epithelial function, motility, mucosal immunology, pancreatic endocrine and exocrine function, gallbladder contractility, and appetite (Worthington et al., 2018, Psichas et al., 2015). The enteric nervous system contains about as many neurons as the spinal cord, and there are dozens of neuronal and glial cell types that control motility, regulate blood flow, and impact immune and epithelial cell function (Furness, 2012). Intestinal absorption, patterns of motility, bowel movement frequency, and susceptibility to intestinal injury all change as children grow in response to microbes, nutrients, and genetic programming, so PCA data for the bowel will illuminate many aspects of human health. For example, late fetal bowel is particularly susceptible to serious inflammation called necrotizing enterocolitis (NEC), a problem that does not occur in term infants, older children, or adults (Niño et al., 2016). NEC susceptibility is presumed to reflect age-dependent changes in intestinal barrier function, bowel motility, and immune system function that may be reflected in changes in cell populations as well as single-cell gene expression patterns at various ages. In the first 5 years of a child’s life, the bowel changes dynamically with the length of the small and large intestine nearly doubling during this interval (Struijs et al., 2009). Remarkably, individual cells must maintain regional identity in this dynamically growing organ because patterns of bowel motility and epithelial function differ markedly along the bowel length. Intestinal microbiota changes dramatically in the first 3 years of life, in part because of diet changes but also because of changes in immune and epithelial cell function. In contrast, intestinal microbiota is relatively stable during adulthood (Yatsunenko et al., 2012). These differences are important because the composition of the gut microbiome has been shown to significantly impact health. Supporting the hypothesis that critical changes occur during late fetal and postnatal stages, a recent single-cell study on human fetal intestine revealed over 30 cell types and demonstrated important differences between fetal and adult intestine (Gao et al., 2018). Defining what is normal in varied bowel regions at distinct pediatric ages is essential, especially as we consider disease processes unique to the pediatric age group including congenital anomalies, metabolic and mitochondrial diseases, and single-gene defects that cause bowel dysfunction.
As another example where PCA data for the bowel will be valuable, chronic disruption in the gut’s cellular function and composition can lead to very early onset and pediatric inflammatory bowel disease (IBD). IBD affects over 80,000 American children. It manifests before 18 years of age in 25% of the ∼2 million affected Americans (Loftus, 2003), and the disease is increasing most rapidly in very young children less than 6 years of age (Benchimol et al., 2017). The specifics of the tissue environment and the genetic origins of IBD vary between individuals, with many identified genetic variants that result in disease development by altering the intestinal barrier or immune cell function causing hyperimmunity, autoimmunity, or immunodeficiency (Peloquin et al., 2016, Jostins et al., 2012). The phenotype of pediatric IBD can be different, (Billiet, and Vermeire, 2015) and more severe (Rosen et al., 2015), as compared to older patients. There can also be differences in patterns of bowel motility (Chumpitazi, and Nurko, 2008). The immune system may play a more prominent role in pediatric IBD, particularly in children with the disease, as the immune system develops in the first 3 years of life (Simon et al., 2015). The Gut Cell Atlas program supported by the Helmsley Charitable Trust will focus on such efforts (Helmsley Trust, 2018).
Collectively, these observations suggest a critical need to acquire pediatric bowel data (e.g., pooled and single-cell RNA sequencing, metabolomic data, proteomic data, and information about cellular organization) for a wide array of intestinal cell types, at varied ages, and in many bowel regions. The PCA can provide these critical data about cell populations using mucosal biopsies and surgically resected tissue from healthy (e.g., organ donor) and diseased children (e.g., inflammatory bowel disease and Hirschsprung disease resection specimens). Comparisons between PCA and adult HCA data will provide valuable insight as we develop new approaches to rebuild and repair damaged gut epithelium and establish innovative treatment strategies for intestinal diseases. PCA data will also facilitate generation of new organs from stem cells to replace or repair injured bowel.
Heart
The heart undergoes a period of postnatal remodeling involving significant gene splicing and expression changes that are distinct from those in adults (Xu et al., 2005). In children, congenital heart disease (CHD) makes up nearly one-third of all major congenital anomalies, with an occurrence of 8 out of 1,000 live births (van der Linde et al., 2011), and although significant progress has been made in understanding genetic contributions to CHD (Jin et al., 2017), it is not yet well understood how abnormal tissue development leads to CHD. The vital function of the heart has been known for hundreds of years, but the molecular and cellular underpinnings of postnatal heart maturation and function remain poorly understood. Hearts of different pediatric stages exhibit significant anatomical, genetic, and functional heterogeneity. Cell-type composition and metabolic states in the heart undergo significant changes and remodeling for functional maturation during the whole pediatric period. Single-nucleus RNA-seq of nearly 20,000 nuclei has been used to study different postnatal developmental stages of the mouse heart, which has revealed major and rare cardiac cell types as well as significant anatomical and functional heterogeneity among all cell types in the postnatal developing heart (Hu et al., 2018a, DeLaughter et al., 2016). Moreover, when applied to a mouse model of pediatric mitochondrial cardiomyopathy, profound cell-type-specific modifications of the cardiac transcriptome were revealed at single-cell resolution, including changes of subtype composition, maturation states, and functional remodeling of each cell type (Hu et al., 2018a). Similar experimental approaches can also be applied to studying the pediatric heart. A PCA of the heart can fill these knowledge gaps and bring novel insights into many pediatric heart diseases that affect tens of thousands of children each year.
Hematopoietic and immune systems
Immune homeostasis has been shown to shift dramatically as children lose maternally transferred antibodies, acquire new infections and antigenic exposures, and experience changes in microbiota over time (Putignani et al., 2014, Kollmann et al., 2017, Olin et al., 2018). The hematopoietic and immune system undergo remarkable changes in the first years of a child’s life, with profound impact on health throughout the rest of the lifespan. Children are born with a relatively naive T/B cell compartment, hypofunctional innate immunity (Collins et al., 2018), and fetal red blood cells. Intrinsic developmental gene expression programs and extrinsic environmental influences over the following years shape the developing hematopoietic system (Schatorjé et al., 2011, Belkaid and Hand, 2014). Perturbed development of immune competence and tolerance has been linked to allergies, atopic disease, autoimmune disease, cancer, and metabolic diseases (Renz et al., 2017), accounting for a large burden of human morbidity and health spending. Vaccination, our primary effort to modify immunity throughout the world, is almost exclusively performed in the young child and in early adolescence (Levy et al., 2013). The neonatal, pediatric and adult immune systems respond very differently to infection, trauma, and other inflammatory insults (Maddux and Douglas, 2015, Beura et al., 2016); therefore, it is critical to define and understand the shifting pediatric hematopoietic system separately from that of adults.
Recently, single-cell transcriptomic studies of the adult hematopoietic and immune systems (reviewed in Papalexi and Satija, 2018) have provided new insights into the immune system, including leukocyte developmental and differentiation hierarchies in the bone marrow (Velten et al., 2017), the effect of myelodysplastic clones on normal hematopoiesis, and the specificity and functional response to infection and antigenic challenges (Stubbington et al., 2017, Villani et al., 2017). A recent unbiased transcriptomic survey of over 100,000 hematopoietic bone marrow cells from eight healthy adults revealed significant variation in the relative proportion of distinct cell lineage between donors as well as lineage-dependent gene expression with donor age (Hay et al., 2018). Another study on bone marrow included a sample from a single child who had a remarkably different distribution of transcriptionally defined stem and progenitor populations compared to the adult samples, making a strong case for the necessity of a pediatric-specific immune cell atlas (Zhao et al., 2017). The HCA preview data include cord blood samples from multiple individuals, showing substantial variation (https://preview.data.humancellatlas.org/).
Understanding normal pediatric hematopoietic and immune development would thus have critical impact on child and adult health. Pediatric leukemia is the most common pediatric cancer and can have an age-dependent prognosis, perhaps linked to the immune system’s development in ways that are not yet understood. While the field of hematopoiesis has learned from the study of model organisms such as mice, many of the developmental dependencies discussed above are specific to humans and cannot be modeled or understood without looking at the human pediatric hematopoietic and immune system as it matures. The pediatric immune atlas will sample from primary (bone marrow, thymus) and secondary (lymphatic fluid, tonsil, peripheral blood, and in rare cases, spleen) lymphoid tissues, as well as immune cells obtained from tissues such as lung, liver, intestine, and skin, across a wide range of ethnic backgrounds and ages including newborns and preterm infants.
Kidney
The kidneys receive 20% of the heart’s blood output and in adults generate approximately 180 liters of glomerular filtrate per day. In mammals, the normal kidney forms from a lineage-specific set of progenitor cells that finish differentiating soon after birth (Lindström et al., 2018). There is considerable individual variation in the number of nephrons per kidney, with important medical consequences (Bertram et al., 2011). The basis for this variation is not understood. Children can suffer a number of kidney diseases including structural malformations, glomerulonephritis, and focal segmental glomerulosclerosis (FSGS) (Harambat et al., 2012). FSGS, the most common glomerular disease in pediatric patients, is poorly understood, progressive, challenging to treat, and the risk of recurrence in transplanted kidneys can be as high as 30%–40% (Kiffel et al., 2011). Comparison of the cellular composition of primary and recurrent FSGS kidneys with normal age-matched kidney tissue may help contrast the cell-level pathophysiological processes operating in FSGS. The PCA would contribute to our understanding of the normal and abnormal development of the human kidney by providing samples of cell types and population distributions from normal fetal and early neonatal tissues. The PCA would also work in a synergistic and complementary fashion with the Kidney Precision Medicine Initiative (https://kpmp.org/), which aims to provision deep classification of adult kidney disease patients and their cellular attributes in a kidney tissue atlas.
Liver
The liver is the largest solid organ in the body, critical for metabolic function. For children with end-stage liver disease, transplantation is often the only option. 33% of pediatric liver transplantations in North America from 1995–2002 were performed on children less than 1 year of age (McDiarmid et al., 2004). Biliary atresia (BA), a progressive fibrosing cholangiopathy presenting in infancy, is the leading indication for liver transplantation in children (Asai et al., 2015, Verkade et al., 2016). Recent studies have shown that damage to bile ducts in BA begins at or before birth (Harpavat et al., 2011). A detailed map of gene expression in infant bile ducts, which continue to develop in the weeks and months postnatally, could shed light on the pathogenesis of BA and other developmental disorders of bile ducts presenting during childhood. Targeted interventions to prevent the development and progression of end-stage liver disease in children will require a detailed understanding of the cellular components of the normal pediatric liver. Recently, a map of the adult human liver, as identified by single-cell RNA sequencing revealed distinct liver resident macrophages in the human liver, with inflammatory and non-inflammatory properties (MacParland et al., 2018). The rationale for developing a pediatric-specific liver map lies in the fact that there are differences in metabolic functions in the pediatric and adult liver although the mechanisms are unclear (Blanco et al., 2000). Drug metabolism differs substantially in infants and children as compared with adults (Stephenson, 2005); a map of gene expression in hepatocytes in the infant liver would provide detailed information about the timeline for hepatocyte maturation during early development. Moreover, there are specific liver diseases that are found in adults, for example, non-alcoholic fatty liver disease (NAFLD), that can have a unique histological presentation in children (Schwimmer et al., 2005), possibly suggesting distinct underlying dysfunctions are promoting disease in children. A baseline map of the healthy pediatric liver will help in the identification of pathological cellular processes driving the development and progression of pediatric liver diseases leading to liver failure.
Lung
The human lung is a wonderfully complicated organ required for the transition to air breathing at birth and for ventilation throughout our lifetime. Moreover, the lung constitutes a primary interface with the environment, generating innate and adaptive immune responses to danger signals. Lung function is inherently linked to its remarkable tubular-alveolar structure of the airways and the lung parenchyma and dependent upon the contributions and interactions among a great diversity of cells and the matrix scaffolds that support them. During lung morphogenesis and until maturity, cells proliferate, migrate, differentiate, and interact to form and maintain lung architecture (Hogan et al., 2014). A diversity of mesenchymal, epithelial, endothelial, vascular, neuronal, and hematopoietic immune cells are present in precise numbers and stereotyped locations, interacting to maintain tissue homeostasis and regeneration after injury. The lung is vulnerable to environmental challenges, being directly exposed to inhaled gases, particles, toxins, and microbial pathogens that underlie the pathogenesis of acute and common chronic lung disorders affecting children. Many of these disorders have life-long health consequences. Respiratory Distress Syndrome and Bronchopulmonary Dysplasia are relatively common disorders affecting newborns and preterm infants (Reuter et al., 2014). Chronic disorders of mucociliary clearance and host defense, for example, cystic fibrosis and primary ciliary dyskinesia, are associated with chronic inflammation, infection, and tissue remodeling with lifelong consequences. Reduced lung growth during pediatric development, such as observed in children with asthma, may even predispose to chronic obstructive pulmonary disease (COPD) later in life (McGeachie et al., 2016). Asthma and COPD are chronic inflammatory lung diseases that arise from the interaction of a genetic predisposition to develop the disease with specific environmental factors that trigger disease inception. While asthma typically starts in the first few years of life (Holt and Sly, 2012), most asthma studies, including the first single-cell analyses (Vieira Braga et al., 2019), study the chronic phase of the disease, thereby potentially missing crucial causative mechanism operating in children developing the disease. Also for COPD, where expression of the disease is generally only observed after the age of 40 (Rabe and Watz, 2017), understanding the cellular mechanisms governing lung growth during pediatric development is of critical importance to be able to design novel interventions aimed at lung repair.
The roles of multiple cell types and their interactions in normal and diseased lung can be explored by single-cell analytic approaches. For example, single-cell RNA profiling of human epithelial cells has provided insights into the pathogenesis of pulmonary fibrosis, cystic fibrosis, and asthma (Xu et al., 2016, Montoro et al., 2018, Reyfman et al., 2018, Vieira Braga et al., 2019). Atypical non-lineage restricted epithelial cells were identified that contribute to tissue remodeling in idiopathic pulmonary fibrosis. Likewise, a new epithelial cell type, the ionocyte, which expresses CFTR, was identified by single-cell sequencing, supporting its role in the pathogenesis of cystic fibrosis (Montoro et al., 2018, Plasschaert et al., 2018).
One notable reference atlas project, funded by the National Heart, Lung, and Blood Institute (NHLBI), LungMAP (LungMap, 2019) has provided detailed molecular portraits of distal lung cell class and subclasses of the mouse and is beginning to cover proximal bronchial and human samples (https://www.lungmap.net/about/lungmap-team/nhlbi). Data from the efforts thus far includes more than 40,000 single cells from developing and postnatal stages as well as bulk cell profiles of mRNA, miR, proteins, lipids, metabolites, and epigenomic profiling (Ardini-Poleske et al., 2017, Du et al., 2017). An initial analysis of neonatal and pediatric human lung samples has also begun (Bandyopadhyay et al., 2018), with key initial goals of providing profiles of sorted cell types, via mRNA, protein, lipid, metabolite, and epigenomic measures and technologies representing key goals. Micro- and macro-2D and micro- and macro-3D imaging modalities are also part of the overall effort with goals that include developing integrative models of cell and tissue morphogenesis, signaling networks, and multi-scale physiology, to understand lung formation, maturation, function, injury, and repair (Xi et al., 2017, Zacharias et al., 2018). Substantial differences of cell-type-specific signatures between embryonic, fetal, and postnatal cell types occur that demonstrate a high degree of lineage specialization reflective of different transcription factor drivers and important developmental pathways that are not shown in corresponding cell types of postnatal lung (Treutlein et al., 2014, Ding et al., 2018). The ability to accurately analyze large numbers of cells by single-nuclear RNA sequencing, chromatin accessibility, and high-resolution imaging will enable the use of human tissues, including our archival tissues from children with rare lung diseases, to identify the cells, genes, and gene networks involved in the pathogenesis of acute and chronic lung diseases.
Placenta
The placenta is a unique temporary organ, which acts as lungs, liver, gut, kidneys, and endocrine glands for the fetus, supplying it with nutrients and oxygen, and harmonizes the cross-talk between the fetal and maternal immune system (Burton, and Fowden, 2015). The structure of the maternal-fetal interface features a complex relationship between fetal cells and maternal tissue. Specifically, fetal trophoblasts invade into the maternal decidua, orchestrated by maternal immune cells, stromal cells, and glandular epithelial cells. While it is essential for intrauterine fetal growth and development, it also plays a major role in perinatal and pediatric health outcomes. Dysfunction at the maternal-fetal interface can have significant health implications to mothers and offspring, such as increased risk of cardiovascular disease in offspring after maternal hypertension in pregnancy (McDonald et al., 2008, Seely et al., 2015), or increased risk of type 2 diabetes in the offspring during late adult life (Kajantie et al., 2017). One such dysfunction is preeclampsia, a syndrome manifested by a sudden increase in blood pressure and accompanied by proteinuria or in association with thrombocytopenia, impaired liver function, development of renal insufficiency, pulmonary edema, or new-onset cerebral or visual disturbances (American College of Obstetricians and Gynecologists et al., 2013). It affects 7%–10% of pregnancies worldwide and is the leading cause of maternal and fetal morbidity and mortality. The cause of preeclampsia, effective treatment, and reliable onset prediction are still unknown; however, it is evident that the placenta plays a major role in the development of this disease (Huppertz, 2008). Furthermore, disruption of the fine balanced cross-talk between immune cells and trophoblast early in pregnancy might cause abnormal placentation leading to faulty placental perfusion and disturbed fetal nutrition (Hiby et al., 2010, Przybyl et al., 2016). The effect of preeclampsia on a child requires more attention in order to minimize risks in adulthood. For example, Reveret et al. reported an immediate impact of preeclampsia on the children’s blood pressure, measured within days of birth, necessitating long-term cardiovascular follow-up and targeted preventive strategies for affected offspring (Reveret et al., 2015). The statement that child health starts from the placenta would not be an exaggeration, but the details of that dependency are not well understood.
Studies featuring placental tissues at single-cell resolution have been recently published in mouse (Nelson et al., 2016) and human (Pavličev et al., 2017). They revealed the cell communication network together with functionally distinct cell types of the maternal-fetal interface at term; used transcriptomic information derived from single cells for the interpretation of cell-free plasma RNA, thus opening new possibilities for non-invasive diagnostics (Tsang et al., 2017); and described human trophoblast differentiation. These results advanced our understanding of human placentation as well as the stark differences between rodent and human (Liu et al., 2018). Reproductive success is dependent on the maternal-fetal interface in the first weeks of pregnancy (Smith, 2010). Recently, a comprehensive single-cell map of first-trimester maternal-fetal interface has revealed novel subsets of maternal and fetal cells and their crucial tolerogenic role in this environment (Vento-Tormo et al., 2018). The PCA will focus on expanding this knowledge in healthy tissues to help inform later work targeting diseases of pregnancy, which account for the large proportion of child health complications. Since the placenta undergoes major molecular, structural, and functional rearrangements during three trimesters, the PCA would work with the greater HCA community to perform a fine-grain map of all stages of placenta development. One of the major implications of the placenta PCA subproject would be improved cell-type profiles and interpretation of their interactions, which are important not only for future placenta single-cell studies but also for re-annotating accumulated bulk RNA-seq data, introducing an added value to the whole project.
Skeletal Muscle
At over a third of the human body weight, skeletal muscle is not just the single largest tissue in the human body, but it also contributes the most to metabolic and physical health by regulating energy usage, storage, production, and regulation of body temperature to name just a few of its functions (Zurlo et al., 1990, Rowland et al., 2015). While skeletal muscle is home to a variety of cell types, its minimum functional unit is the multinucleated syncytial cell called the myofiber. This mesodermal cell lineage is formed by the fusion of mono-nucleated precursors called myoblasts, derived primarily from satellite cells. The myogenic process begins early during human growth by a developmental program that leads to the addition of nuclei to the growing muscle fiber, and it is followed by the growth of the muscle fibers to its optimized size based on the extent of use (or disuse) of the individual muscle. While the satellite cells remain quiescent during adult life and much of the muscle activity maintains status quo, the muscle maintains the full capacity to grow and regenerate in response to stress, trauma, or disease that damages the myofibers (Lepper et al., 2011, Sambasivan et al., 2011). Thus, the cellular composition and the expressome of skeletal muscle changes from early development and growth to a homeostatic state during adulthood while, at all times, maintaining its ability to efficiently respond to changes that may impinge upon it. The knowledge of these skeletal muscle abilities has come from the cellular, molecular, and genetic analysis of cultured satellite cells and a variety of model organisms including C. elegans, D. melanogaster, D. rerio, and M. musculus. Such analyses have identified a wide array of myogenic regulatory factors and cell types that contribute to the process of developmental or regenerative myogenesis. Single-cell RNA analyses have been performed on adult satellite cell and non-myogenic cells isolated from mice and humans (Cho and Doles, 2017, Tabula Muris Consortium et al., 2018, Giordani et al., 2018, Heuvel et al., 2018). While offering some insights into the molecular complexity and diversity, it does not include the multi-nuclear myofibers responsible for the majority of muscle function. In contrast, single-nucleus sequencing has only been performed on human myotubes (Zeng et al., 2016), which supports the use of single nuclear extraction, sequencing, and analysis in skeletal muscle tissue. Furthermore, with myofibers being the minimal functional entity of muscle use, disuse, damage, or regeneration, there is a pressing need to understand the genetic diversity of the myofibers. This diversity likely exists between the nuclei of different types of the myofibers (e.g., slow- and fast-twitch muscle fibers) and also perhaps among the multiple nuclei that populate individual myofibers, as reported by cultured cell analysis (Zeng et al., 2016). Little attention has also been focused on analyzing the developing muscle, which is of greater relevance to the pediatric population. In addition, the puzzling facts that muscles at different anatomical locations are differentially affected in inherited muscular dystrophies may be explained by intrinsic differences in the muscle cells. The PCA effort is required to develop an atlas of such single-cell profiles for mono-nucleated cells in human muscle as well as single-nucleus RNA profiles for human myofibers during development. Such information will be indispensable for understanding how the molecular program of developing muscle allows for the balancing act required to maintain the terminally differentiated status of the muscle while having the capacity to regenerate fully. Disruptions in this balancing act have been identified in congenital and degenerative skeletal muscle disorders. Analyses of normal healthy developing muscle tissue will provide the required benchmark for this process and support the ongoing efforts in developing treatments that restore the balance lost in diseased muscle due in genetic and other diseases.
Skin
As one of the most accessible human organs, skin provides the perfect system to obtain tissue to understand the changes and specialization of single cells and how they vary over age from prematurity through adolescence, from multiple sites on the body, over multiple ethnic backgrounds, and between sexes. Skin plays six primary functions: protection, absorption, excretion, secretion, regulation, and sensation (Proksch et al., 2008). The outermost layer of the skin, the epidermis, is composed of multiple cell types including keratinocytes, melanocytes, Langerhans cells, mast cells, and Merkel cells (Matsui, and Amagai, 2015), and below that, the dermis is made of fibroblasts which secrete the connective tissue matrix and is a home for nerve and endothelial cells. The composition of these layers changes significantly from infancy to adulthood in terms of cell distribution, connective density, caliber and distribution of hair follicles, and number and location of apocrine and sebaceous glands as well as in inherent elasticity and ability to remodel. Children with rare disorders like ichthyosis and epidermolysis bullosa have provided insight into the careful interplay of cells, lipids, collagen, and proteins needed to provide the barrier function and strength of the skin (Matsui, and Amagai, 2015).
The human skin has been profiled in the Human Protein Atlas where tissue sections for histology and tissue cDNAs for microarrays were used to explore proteins expressed in the skin, follicular units, sweat glands, and sebaceous glands (Uhlén et al., 2015). Through this project, over 400 genes have been shown to be highly enriched in the skin compared to other organs. In-depth analysis of the elevated genes using antibody-based protein profiling allowed creation of a portrait of in-situ protein expression for the different layers of the epidermis and in the dermis (Edqvist et al., 2015). Despite these advances, proteomic and expression analysis of normal skin tissue from multiple sites on the human body from infancy through adolescence has not been performed in a systematic way and is critical for understanding developmental changes in pigmentation, skin regeneration, hair cycle, and connective tissue structure. A pediatric skin atlas would be able to provide normal skin references for studies on pathological skin conditions related to disrupted growth and development, including congenital melanocytic lesions, Spitz nevi, epidermal nevi, vascular tumors, vascular malformations, and lipomas and also would be able to inform research on tissue-engineered skin substitutes for wounds and impaired barrier conditions.
Pediatric Cancer
Childhood cancers are the leading cause of disease-related mortality, where leukemia and brain tumors lead the list as the deadliest pediatric cancers in absolute numbers (NCI, 2018). Childhood is one of the two peaks of leukemia incidence, and a number of developmental hematopoietic abnormalities first manifest in childhood. Children’s and adult tumors are genomically and genetically distinct (Ma et al., 2018, Liu et al., 2016) and children tumors have a strong developmental component. The age at which a child develops leukemia or neuroblastoma is one of the strongest predictors of outcome (Riehm et al., 1987, Mastrangelo et al., 1986, Smith et al., 1996). While several frequent leukemia-imitating genomic alterations are common between adults and children, specific classes of variation, such as splicing factor mutations in acute myeloid leukemia, are restricted to adults (Farrar et al., 2016). A recent study using single cells comparing normal renal tissue and renal tumors from both children and adults has shown that adult kidney cancers appear to be derived from a subtype of proximal convoluted tubular cells, while pediatric Wilms tumors likely arise from aberrant fetal cells (Young et al., 2018). Single-cell studies of pediatric H3K27M diffuse gliomas have identified predominant oligodendrocyte precursor cell sub-populations, from which stem-like cycling cells propagate the disease (Filbin et al., 2018). A subset of metastatic neuroblastoma often spontaneously regresses without treatment in infants under 12 months of age, but not in older children, highlighting the developmental influence on survival in patients with this cancer (Nickerson et al., 2000).
Although not in the direct scope of profiling normal tissues, the PCA can partner with researchers driving single-cell pediatric tumor atlases to help source normal tissues for comparison. These comparisons would help to produce high-resolution insights into oncogenic mechanisms at the sub-clonal level. Any pediatric tumor atlas would benefit from the PCA’s normal single-cell profiles and can compare these to the most common cancers of childhood including, but not limited to, leukemia, neuroblastoma, high-grade glioma, medulloblastoma, neurofibromatosis, and ependymoma. The comprehensive analysis of very high-risk pediatric lymphoblastic leukemia, neuroblastoma, and high-grade glioma is currently part of the NIH’s Cancer Moonshot Human Tumor Atlas Network (HTAN). A PCA would provide a much-needed framework of normal development that will be needed to understand how aberrant development leads to pediatric cancer.
Summary
Pediatric tissues are different from adult tissues in several important ways, most notably because they are still undergoing development. Here, we highlight several unique opportunities and advantages in the PCA for the medical and research communities. Systematic analysis of pediatric tissues and organ systems at different developmental stages would improve our understanding of cell-type-specific responses to clinical situations, even those where collecting single-cell data on temporal and perturbational responses is infeasible. The PCA would provide integrated observational atlas data and develop algorithms to mine these data in the context of both early and advanced developmental dynamic processes. Deep single-cell-based profiling of pediatric tissues of both dissociated cells and cells within tissues, the goal of the PCA as part of the overall HCA effort, would provide the framework to define and characterize cell types, cell states, and their interactions that, together, lead to normal childhood development and are subverted by childhood disease states and processes.
Acknowledgements
D.M.T.: NIH 5P01HD070454–09, 5U2CHL138346–02; B.J.A.: NIH P30 DK078392, U01 HL122642, HL122638, U19 MH104172, MH194173; K.T.: NIH U2C CA233285 and U01 CA226197; C.S.G.: Alex’s Lemonade Stand Foundation, grant 2018–182718 from the Chan-Zuckerberg Initiative DAF, Gordon and Betty Moore Foundation grant GBMF 4552, and NIH R01 HG010067; K.E.H.: NIH K01DK100485 and NIH R03DK114463; S.E.H.: NIH 5K12HD043245 and K08 AI135091; L.P.: DoD PRMRP award W81XWH-16–1-0400 and NIH DK111495; A.H.B.: NIH R01AR044345 and MDA602235 from the Muscular Dystrophy Association (USA); N.S., B.J.A.: the Cincinnati Children’s Research Foundation; L.X.G.: NIH K01ES025434, P20 COBRE GM103457, R01 LM012373, R01 HD084633; D.M.T., Y.Z., M.S.K., B.D.: the Department of Biomedical and Health Informatics and the CHOP Research Institute; S.W.K.: NIH R01MH107205; A.R. is an Investigator of the Howard Hughes Medical Institute.
Declaration of Interests
A.R. is an SAB member of ThermoFisher Scientific and Syros Pharmaceuticals and a founder and equity holder of Celsius Therapeutics. S.W.K. receives sponsored research support from Pfizer Inc.
References
- Aevermann BD, Novotny M, Bakken T, Miller JA, Diehl AD, Osumi-Sutherland D, Lasken RS, Lein ES, and Scheuermann RH (2018). Cell type discovery using single-cell transcriptomics: implications for ontological representation. Hum Mol Genet 27, R40–R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara D, Timms AE, Gripp K, Baker L, Park K, Collins S, Cheng C, Stewart F, Mehta SG, Saggar A, et al. (2017). Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain 140, 2610–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American College of Obstetricians and Gynecologists, Task Force on Hypertension in Pregnancy (2013). Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy; pp. 1122–1131. [DOI] [PubMed] [Google Scholar]
- Amodio M, Srinivasan K, van Dijk D, Mohsen H, Yim K, Muhle R, Moon KR, Kaech S, Sowell R, Montgomery R, et al. (2017). Exploring Single-Cell Data with Multitasking Deep Neural Networks. bioRxiv 237065. [Google Scholar]
- Andropoulos DB (2018). Effect of Anesthesia on the Developing Brain: Infant and Fetus. Fetal Diagn Ther 43, 1–11. [DOI] [PubMed] [Google Scholar]
- Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, Hagood JS, Kaminski N, Mariani TJ, Potter SS, et al. (2017). LungMAP: The Molecular Atlas of Lung Development Program. American Journal of Physiology-Lung Cellular and Molecular Physiology 313, L733–L740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai A, Miethke A, and Bezerra JA (2015). Pathogenesis of biliary atresia: defining biology to understand clinical phenotypes. Nature Reviews Gastroenterology & Hepatology 12, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasakis E, Karavasiliadou S, and Styliadis I (2011). The factors contributing to the risk of sudden infant death syndrome. Hippokratia 15, 127–131. [PMC free article] [PubMed] [Google Scholar]
- Balshaw DM, Collman GW, Gray KA, and Thompson CL (2017). The Children’s Health Exposure Analysis Resource: enabling research into the environmental influences on children’s health outcomes. Curr Opin Pediatr 29, 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay G, Huyck HL, Misra RS, Bhattacharya S, Wang Q, Mereness J, Lillis JA, Myers JR, Ashton J, Bushnell T, et al. (2018). Dissociation, Cellular Isolation and Initial Molecular Characterization of Neonatal and Pediatric Human Lung Tissues. American Journal of Physiology-Lung Cellular and Molecular Physiology 126, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP (2004). The Developmental Origins of Adult Disease. Journal of the American College of Nutrition 23, 588S–595S. [DOI] [PubMed] [Google Scholar]
- Baslan T, and Hicks J (2017). Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat. Rev. Cancer 17, 557–569. [DOI] [PubMed] [Google Scholar]
- Batchelor HK, and Marriott JF (2015). Paediatric pharmacokinetics: key considerations. British Journal of Clinical Pharmacology 79, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Lindsay S, Teichmann SA, and Haniffa M (2018). Mapping human development at single-cell resolution. Development 145, dev152561. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the Microbiota in Immunity and Inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shlomo Y (2002). A life course approach to chronic disease epidemiology: conceptual models, empirical challenges and interdisciplinary perspectives. International Journal of Epidemiology 31, 285–293. [PubMed] [Google Scholar]
- Benchimol EI, Bernstein CN, Bitton A, Carroll MW, Singh H, Otley AR, Vutcovici M, El-Matary W, Nguyen GC, Griffiths AM, et al. (2017). Trends in Epidemiology of Pediatric Inflammatory Bowel Disease in Canada: Distributed Network Analysis of Multiple Population-Based Provincial Health Administrative Databases. The American Journal of Gastroenterology 112, 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram JF, Douglas-Denton RN, Diouf B, Hughson MD, and Hoy WE (2011). Human nephron number: implications for health and disease. Pediatr. Nephrol. 26, 1529–1533. [DOI] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiet T, and Vermeire S (2014). Differences between adults and children: genetics and beyond. Expert Review of Gastroenterology & Hepatology 9, 191–196. [DOI] [PubMed] [Google Scholar]
- Blanco JG, Harrison PL, Evans WE, and Relling MV (2000). Human Cytochrome P450 Maximal Activities In Pediatric versus Adult Liver. Drug Metab Dispos 28, 379. [PubMed] [Google Scholar]
- Boldog E, Bakken TE, Hodge RD, Novotny M, Aevermann BD, Baka J, Bordé S, Close JL, Diez-Fuertes F, Ding S-L, et al. (2018). Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci 21, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botkin JR (2016). Ethical issues in pediatric genetic testing and screening. Curr Opin Pediatr 28, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G, Barbara M, and Iscove N (1990). Representative in vitro cDNA amplification from individual hemopoietic cells and colonies. Methods Mol Cell Biol 2, 17–25. [Google Scholar]
- Budnik B, Levy E, Harmange G, and Slavov N (2018). SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein MD, Robinson JO, Hilsenbeck SG, McGuire AL, and Lau CC (2014). Pediatric data sharing in genomic research: attitudes and preferences of parents. Pediatrics 133, 690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton GJ, and Fowden AL (2015). The placenta: a multifaceted, transient organ. Philos Trans R Soc Lond B Biol Sci 370, 20140066–20140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS and Doles JD (2017). Single cell transcriptome analysis of muscle satellite cells reveals widespread transcriptional heterogeneity. Gene 636, 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Berger B, and Peng J (2018). Generalizable and Scalable Visualization of Single-Cell Data Using Neural Networks. Cell Systems 7, 185–191.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choumerianou DM, Martimianaki G, Stiakaki E, Kalmanti L, Kalmanti M, and Dimitriou H (2010). Comparative study of stemness characteristics of mesenchymal cells from bone marrow of children and adults. Cytotherapy 12, 881–887. [DOI] [PubMed] [Google Scholar]
- Chumpitazi B, and Nurko S (2008). Pediatric Gastrointestinal Motility Disorders: Challenges and a Clinical Update. Gastroenterology & Hepatology 4, 140–148. [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Argelaguet R, Kapourani C-A, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. (2018). scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Comms 9, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2016). Modeling Development and Disease with Organoids. Cell 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Clevers H, and Watt FM (2018). Defining Adult Stem Cells by Function, not by Phenotype. Annual Review of Biochemistry 87, 1015–1027. [DOI] [PubMed] [Google Scholar]
- Collins A, Weitkamp J-H, and Wynn JL (2018). Why are preterm newborns at increased risk of infection? Arch. Dis. Child. Fetal Neonatal Ed 103, F391–F394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion SP, Zhu Y, Nagy G, Adkins JN, Ansong C, Renslow RS, Piehowski PD, Ibrahim YM, Kelly RT, and Metz TO (2018). New mass spectrometry technologies contributing towards comprehensive and high throughput omics analyses of single cells. Analyst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Gama AM, and Walsh CA (2018). Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci 21, 1504–1514. [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J, Ballesteros-Yanez I, Inda MC, and Munoz A (2006). Double-bouquet cells in the monkey and human cerebral cortex with special reference to areas 17 and 18. Prog Brain Res 154, 15–32. [DOI] [PubMed] [Google Scholar]
- DeLaney K, Sauer CS, Vu NQ, and Li L (2018). Recent Advances and New Perspectives in Capillary Electrophoresis-Mass Spectrometry for Single Cell “Omics”. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. (2016). Single-Cell Resolution of Temporal Gene Expression during Heart Development. Developmental Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietert RR, Etzel RA, Chen D, Halonen M, Holladay SD, Jarabek AM, Landreth K, Peden DB, Pinkerton K, Smialowicz RJ, et al. (2000). Workshop to identify critical windows of exposure for children’s health: immune and respiratory systems work group summary. Environmental Health Perspectives 108, 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Aronow BJ, Kaminski N, Kitzmiller J, Whitsett JA, and Bar-Joseph Z (2018). Reconstructing differentiation networks and their regulation from time series single-cell expression data. Genome Research 28, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, Pryhuber GS, Mariani TJ, Bhattacharya S, Guo M, et al. (2017). Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, and Coleman P (1992). Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist P-HD, Fagerberg L, Hallström BM, Danielsson A, Edlund K, Uhlén M, and Pontén F (2014). Expression of Human Skin-Specific Genes Defined by Transcriptomics and Antibody-Based Profiling. J Histochem Cytochem. 63, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, and Lu QR (2015). Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb Perspect Biol 7, a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JG, Forsén T, Tuomilehto J, Osmond C, and Barker DJ (2001). Early growth and coronary heart disease in later life: longitudinal study. Bmj 322, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, Alonzo TA, Guidry Auvil JM, Davidsen TM, Gesuwan P, et al. (2016). Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res 76, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fermie J, Liv N, Brink, ten C, van Donselaar EG, Müller WH, Schieber NL, Schwab Y, Gerritsen HC, and Klumperman J (2018). Single organelle dynamics linked to 3D structure by correlative live-cell imaging and 3D electron microscopy. Traffic 19, 354–369. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Perez R, Hernandez A, Tejada P, Arteta M, and Ramos JT (2011). Factors and Mechanisms for Pharmacokinetic Differences between Pediatric Population and Adults. Pharmaceutics 3, 53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, et al. (2018). Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB (2012). The enteric nervous system and neurogastroenterology. Nature Reviews Gastroenterology & Hepatology 9, 286–294. [DOI] [PubMed] [Google Scholar]
- Gage FH, and Temple S (2013). Neural Stem Cells: Generating and Regenerating the Brain. Neuron 80, 588–601. [DOI] [PubMed] [Google Scholar]
- Gao S, Yan L, Wang R, Li J, Yong J, Zhou X, Wei Y, Wu X, Wang X, Fan X, et al. (2018). Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat Cell Biol 20, 721–734. [DOI] [PubMed] [Google Scholar]
- Gehring J, Park JH, Chen S, Thomson M, and Pachter L (2018). Highly Multiplexed Single-Cell RNA-seq for Defining Cell Population and Transcriptional Spaces. bioRxiv 315333. [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, and Shalek AK (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Meth 14, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L, He GJ, Negroni E, Sakai H, Law JY, Siu MM, Wan R, Tajbakhsh S, Cheung TH, and Le Grand F (2018). High-dimensional single-cell cartography reveals novel skeletal muscle resident cell populations. bioRxiv 304683. [DOI] [PubMed] [Google Scholar]
- Gitterman DP, Langford WS, and Hay WW Jr (2018). The Fragile State of the National Institutes of Health Pediatric Research Portfolio, 1992–2015. JAMA Pediatr 172, 287–293. [DOI] [PubMed] [Google Scholar]
- Gitterman DP, Langford WS, and Hay WW (2018). The uncertain fate of the National Institutes of Health (NIH) pediatric research portfolio. Pediatr. Res. 84, 328–332. [DOI] [PubMed] [Google Scholar]
- GRIN. (2019). The Genomics Research and Innovation Network. https://www.grinnetwork.org/
- Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, and Gluckman PD (2014). Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol. Rev 94, 1027–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harambat J, van Stralen KJ, Kim JJ, and Tizard EJ (2012). Epidemiology of chronic kidney disease in children. Pediatr. Nephrol 27, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpavat S, Finegold MJ, and Karpen SJ (2011). Patients with biliary atresia have elevated direct/conjugated bilirubin levels shortly after birth. Pediatrics 128, e1428–e1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Taylor D, Ovchinnikov DA, Wolvetang EJ, de Torrenté L, and Mar JC (2015). Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development. PLoS Genet 11, e1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay SB, Ferchen K, Chetal K, Grimes HL, and Salomonis N (2018). The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol 68, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- helmsleytrust.org (2018). Human Gut Cell Atlas Funding Opportunity by the Helmsley Trust. https://helmsleytrust.org/rfa/gut-cell-atlas
- Heuvel AVD, Mahfouz A, Kloet SL, Balog J, Engelen BGMV, Tawil R, Tapscott SJ, and Maarel SMV (2018). Single-cell RNA-sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Hum. Mol. Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiby Susan E., Apps Richard, Sharkey Andrew M., Farrell Lydia E., Gardner Lucy, Mulder Arend, Claas Frans H., et al. (2010). “Maternal Activating KIRs Protect against Human Reproductive Failure Mediated by Fetal HLA-C2.” The Journal of Clinical Investigation 120 (11): 4102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, Niklason L, Calle E, Le A, Randell SH, et al. (2014). Repair and Regeneration of the Respiratory System: Complexity, Plasticity, and Mechanisms of Lung Stem Cell Function. Cell Stem Cell 15, 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt PG, and Sly PD (2012). Viral infections and atopy in asthma pathogenesis: new rationales for asthma prevention and treatment. Nat Med 18, 726–735. [DOI] [PubMed] [Google Scholar]
- Hu P, Liu J, Zhao J, Wilkins BJ, Lupino K, Wu H, Pei L. (2018a). Single-nucleus transcriptomic survey of cell diversity and functional remodeling in the postnatal developing hearts. Genes & development 32, 1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, and Greene CS (2018) Parameter tuning is a key part of dimensionality reduction via deep variational autoencoders for single cell RNA transcriptomics. bioRxiv. http://biorxiv.org/lookup/doi/10.1101/385534 [PMC free article] [PubMed] [Google Scholar]
- Hu Y, An Q, Sheu K, Trejo B, Fan S, and Guo Y (2018b). Single Cell Multi-Omics Technology: Methodology and Application. Front Cell Dev Biol 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Huang K, An Q, Du G, Hu G, Xue J, Zhu X, Wang C-Y, Xue Z, and Fan G (2016). Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biology 17, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz B (2008). Placental Origins of Preeclampsia: Challenging the Current Hypothesis. Hypertension 51, 970–975. [DOI] [PubMed] [Google Scholar]
- Imura H, Caputo M, Parry A, Pawade A, Angelini GD, and Suleiman MS (2001). Age-dependent and hypoxia-related differences in myocardial protection during pediatric open heart surgery. Circulation 103, 1551–1556. [DOI] [PubMed] [Google Scholar]
- INSERM (2018). Programme transversal Human Development Cell Atlas 2018 : ouverture de l’appel à projets. https://www.inserm.fr/actualites-et-evenements/actualites/programme-transversal-human-development-cell-atlas-2018-ouverture-appel-projets
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. (2017). Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature Genetics 49, 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajantie E, Osmond C, and Eriksson JG (2017). Gestational hypertension is associated with increased risk of type 2 diabetes in adult offspring: the Helsinki Birth Cohort Study. American Journal of Obstetrics and Gynecology 216, 281.e1–281.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G, Stegle O, and Reik W (2017). Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75. [DOI] [PubMed] [Google Scholar]
- Kiffel J, Rahimzada Y, and Trachtman H (2011). Focal Segmental Glomerulosclerosis and Chronic Kidney Disease in Pediatric Patients. Advances in Chronic Kidney Disease 18, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann TR, Kampmann B, Mazmanian SK, Marchant A, and Levy O (2017). Protecting the Newborn and Young Infant from Infectious Diseases: Lessons from Immune Ontogeny. Immunity 46, 350–363. [DOI] [PubMed] [Google Scholar]
- Konnikova L, Boschetti G, Rahman A, Mitsialis V, Lord J, Richmond C, Tomov VT, Gordon W, Jelinsky S, Canavan J, et al. (2018). High-dimensional immune phenotyping and transcriptional analyses reveal robust recovery of viable human immune and epithelial cells from frozen gastrointestinal tissue. Mucosal Immunol 11, 1684–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, et al. (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Stevens TJ, Basu S, and Laue ED (2018). Calculation of 3D genome structures for comparison of chromosome conformation capture experiments with microscopy: An evaluation of single-cell Hi-C protocols. Nucleus 9, 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JL, and Wu H (2005). Notch signaling, brain development, and human disease. Pediatr. Res 57, 104R–109R. [DOI] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, et al. (2012). De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nature Genetics 44, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelijveld N, Seal A, Wells JC, Kirkby J, Opondo C, Chimwezi E, Bunn J, Bandsma R, Heyderman RS, Nyirenda MJ, et al. (2016). Chronic disease outcomes after severe acute malnutrition in Malawian children (ChroSAM): a cohort study. The Lancet Global Health 4, e654–e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, and Fan CM (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Goriely S, and Kollmann TR (2013). Immune response to vaccine adjuvants during the first year of life. Vaccine 31, 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D’Gama AM, Kim SN, Hill RS, Goldberg AP, et al. (2017a). Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci 20, 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JS, Gopalappa R, Kim SH, Ramakrishna S, Lee M, Kim W-I, Kim J, Park SM, Lee J, Oh J-H, et al. (2017b). Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. The American Journal of Human Genetics 100, 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Jain S, Kim H, and Bar-Joseph Z (2017). Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic Acids Res 45, e156–e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, Ransick A, Parvez RK, Thornton ME, Basking L, et al. (2018). Conserved and Divergent Features of Mesenchymal Progenitor Cell Types within the Cortical Nephrogenic Niche of the Human and Mouse Kidney. Journal of the American Society of Nephrology 29, 806–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fan X, Wang R, Lu X, Dang Y-L, Wang H, Lin H-Y, Zhu C, Ge H, Cross JC, et al. (2018). Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28, 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-F, Wang B-Y, Zhang W-N, Huang J-Y, Li B-S, Zhang M, Jiang L, Li J-F, Wang M-J, Dai Y-J, et al. (2016). Genomic Profiling of Adult and Pediatric B-cell Acute Lymphoblastic Leukemia. EBioMedicine 8 IS -, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. (2015). Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biology 16, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Woodworth MB, Lee S, Evrony GD, Mehta BK, Karger A, Lee S, Chittenden TW, D’Gama AM, Cai X, et al. (2015). Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus EV Jr (2003). Mortality in inflammatory bowel disease: peril and promise. Gastroenterology 125, 1881–1883. [DOI] [PubMed] [Google Scholar]
- Luerssen TG, Klauber MR, and Marshall LF (1988). Outcome from head injury related to patient’s age. A longitudinal prospective study of adult and pediatric head injury. J Neurosurg 68, 409–416. [DOI] [PubMed] [Google Scholar]
- lungmap.org (2019). The LungMAP project website.
- Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, et al. (2018). Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Ponting CP, and Voet T (2017). Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends in Genetics 33, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, et al. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Comms 9, 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddux AB, and Douglas IS (2015). Is the developmentally immature immune response in paediatric sepsis a recapitulation of immune tolerance? Immunology 145, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann G, Attarbaschi A, Schrappe M, De Lorenzo P, Peters C, Hann I, De Rossi G, Felice M, Lausen B, LeBlanc T, et al. (2010). Improved outcome with hematopoietic stem cell transplantation in a poor prognostic subgroup of infants with mixed-lineage-leukemia (MLL)-rearranged acute lymphoblastic leukemia: results from the Interfant-99 Study. Blood 116, 2644–2650. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, and Arendt D (2017). How Single-Cell Genomics Is Changing Evolutionary and Developmental Biology. Annu. Rev. Cell Dev. Biol 33, 537–553. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Baslan T, Kendall J, Geyer FC, Burke KA, Spraggon L, Piscuoglio S, Chadalavada K, Nanjangud G, Ng CKY, et al. (2017). Whole-genome single-cell copy number profiling from formalin-fixed paraffin-embedded samples. Nat Med 23, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo R, Poplack D, Bleyer A, Riccardi R, Sather H, and D’Angio G (1986). Report and recommendations of the rome workshop concerning poor-prognosis acute lymphoblastic leukemia in children: Biologic bases for staging, stratification, and treatment. Medical and Pediatric Oncology 14, 191–194. [DOI] [PubMed] [Google Scholar]
- Matsui T, and Amagai M (2015). Dissecting the formation, structure and barrier function of the stratum corneum. Int. Immunol 27, 269–280. [DOI] [PubMed] [Google Scholar]
- McDiarmid SV, Anand R, Lindblad AS, SPLIT Research Group (2004). Studies of Pediatric Liver Transplantation: 2002 update. An overview of demographics, indications, timing, and immunosuppressive practices in pediatric liver transplantation in the United States and Canada. Pediatr Transplant 8, 284–294. [DOI] [PubMed] [Google Scholar]
- McDonald SD, Malinowski A, Zhou Q, Yusuf S, and Devereaux PJ (2008). Cardiovascular sequelae of preeclampsia/eclampsia: A systematic review and meta-analyses. American Heart Journal 156, 918–930. [DOI] [PubMed] [Google Scholar]
- McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, Wise RA, Szefler SJ, Sharma S, Kho AT, et al. (2016). Patterns of Growth and Decline in Lung Function in Persistent Childhood Asthma. N Engl J Med 374, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, and Diehl AD (2011). Logical Development of the Cell Ontology. BMC Bioinformatics 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. (2008). Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Campbell CD, Solovieff N, Goold C, Jansen LA, Menon S, Timms AE, Conti V, Biag JD, Adams C, et al. (2016). Association of MTOR Mutations With Developmental Brain Disorders, Including Megalencephaly, Focal Cortical Dysplasia, and Pigmentary Mosaicism. JAMA Neurol 73, 836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRC (2018) Funding boost for initiative mapping entire human body. https://mrc.ukri.org/news/browse/funding-boost-for-initiative-mapping-entire-human-body/
- Muotri AR, and Gage FH (2006). Generation of neuronal variability and complexity. Nature 441, 1087–1093. [DOI] [PubMed] [Google Scholar]
- National Cancer Institute, DCCPS, Surveillance Research Program (2018). Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) Research Data (1973–2015).
- Nelson AC, Mould AW, Bikoff EK, and Robertson EJ (2016). Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal–foetal interface during pregnancy. Nat Comms 7, 11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson HJ, Matthay KK, Seeger RC, Brodeur GM, Shimada H, Perez C, Atkinson JB, Selch M, Gerbing RB, Stram DO, et al. (2000). Favorable biology and outcome of stage IV-S neuroblastoma with supportive care or minimal therapy: a Children’s Cancer Group study. Jco 18, 477–486. [DOI] [PubMed] [Google Scholar]
- Niño DF, Sodhi CP, and Hackam DJ (2016). Necrotizing enterocolitis: new insights into pathogenesis and mechanisms. Nature Reviews Gastroenterology & Hepatology 13, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, Gustafsson A, Bernhardsson AK, Zhang C, Bohlin K, et al. (2018). Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera Alvarez HA, Kubzansky LD, Campen MJ, and Slavich GM (2018). Early life stress, air pollution, inflammation, and disease: An integrative review and immunologic model of social-environmental adversity and lifespan health. Neuroscience & Biobehavioral Reviews 92, 226–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Sutherland D (2017). Cell ontology in an age of data-driven cell classification. BMC Bioinformatics 18, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi-Sutherland D, Reeve S, Mungall CJ, Neuhaus F, Ruttenberg A, Jefferis GSXE, and Armstrong JD (2012). A strategy for building neuroanatomy ontologies. Bioinformatics 28, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Packer J, and Trapnell C (2018). Single-Cell Multi-omics: An Engine for New Quantitative Models of Gene Regulation. Trends in Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexi E, and Satija R (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol 18, 35–45. [DOI] [PubMed] [Google Scholar]
- Park E, Pan Z, Zhang Z, Lin L, and Xing Y (2018). The Expanding Landscape of Alternative Splicing Variation in Human Populations. The American Journal of Human Genetics 102, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavličev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, and Jones H (2017). Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Research 27, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin JM, Goel G, Villablanca EJ, and Xavier RJ (2016). Mechanisms of Pediatric Inflammatory Bowel Disease. Annual Review of Immunology 34, 31–64. [DOI] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, and Rajewsky N (2018). Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360. [DOI] [PubMed] [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proksch E, Brandner JM, and Jensen J-M (2008). The skin: an indispensable barrier. Exp. Dermatol. 17, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Przybyl L, Haase N, Golic M, Rugor J, Solano ME, Arck PC, Gauster M, Huppertz B, Emontzpohl C, Stoppe C, et al. (2016). CD74-Downregulation of Placental Macrophage-Trophoblastic Interactions in Preeclampsia. Circ. Res 119, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psichas A, Reimann F, and Gribble FM (2015). Gut chemosensing mechanisms. J. Clin. Invest 125, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putignani L, Del Chierico F, Petrucca A, Vernocchi P, and Dallapiccola B (2014). The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood. Pediatr. Res 76, 2–10. [DOI] [PubMed] [Google Scholar]
- Rabe KF, and Watz H (2017). Chronic obstructive pulmonary disease. The Lancet 389, 1931–1940. [DOI] [PubMed] [Google Scholar]
- Rahimzadeh V, Schickhardt C, Knoppers BM, Sénécal K, Vears DF, Fernandez CV, Pfister S, Plon S, Terry S, Williams J, et al. (2018). Key Implications of Data Sharing in Pediatric Genomics. JAMA Pediatr 172, 476–481. [DOI] [PubMed] [Google Scholar]
- Ranzoni AM, and Cvejic A (2018). Single-cell biology: resolving biological complexity, one cell at a time. Development 145, dev163972. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, et al. (2017). Science Forum: The Human Cell Atlas. Elife 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann S, Rozenblatt-Rosen O, Stubbington M, Ardlie K, Amit I, Arlotta P, Bader G, Benoist C, Biton M, et al. (2018). The Human Cell Atlas White Paper. arXiv q-bio.TO, arXiv:1810.05192. [Google Scholar]
- Renz H, Holt PG, Inouye M, Logan AC, Prescott SL, and Sly PD (2017). An exposome perspective: Early-life events and immune development in a changing world. J. Allergy Clin. Immunol 140, 24–40. [DOI] [PubMed] [Google Scholar]
- Resch C, Anderson VA, Beauchamp MH, Crossley L, Hearps SJC, van Heugten CM, Hurks PPM, Ryan NP, and Catroppa C (2019). Age-dependent differences in the impact of paediatric traumatic brain injury on executive functions: A prospective study using susceptibility-weighted imaging. Neuropsychologia 124, 236–245. [DOI] [PubMed] [Google Scholar]
- Reuter S, Moser C, and Baack M (2014). Respiratory distress in the newborn. Pediatr Rev 35, 417–28–quiz429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveret M, Boivin A, Guigonnis V, Audibert F, and Nuyt AM (2015). Preeclampsia: effect on newborn blood pressure in the 3 days following preterm birth: a cohort study. J Hum Hypertens 29, 115–121. [DOI] [PubMed] [Google Scholar]
- Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen C-I, Ren Z, et al. (2018). Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med rccm.201712–2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehm H, Feickert HJ, Schrappe M, Henze G, and Schellong G (1987). Therapy Results in Five ALL-BFM Studies Since 1970: Implications of Risk Factors for Prognosis In Leukemias Acute, Büchner T, Schellong G, Hiddemann W, Urbanitz D, and Ritter J, eds. (Berlin, Heidelberg: Springer Berlin Heidelberg; ), pp. 139–146. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Kaste SC, Chemaitilly W, Bowers DC, Laughton S, Smith A, Gottardo NG, Partap S, Bendel A, Wright KD, et al. (2017). Irreversible growth plate fusions in children with medulloblastoma treated with a targeted hedgehog pathway inhibitor. Oncotarget 8, 69295–69302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MJ, Dhawan A, and Saeed SA (2015). Inflammatory Bowel Disease in Children and Adolescents. JAMA Pediatrics 169, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LA, Bal NC, and Periasamy M (2015). The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol. Rev. Camb. Philos. Soc 90, 1279–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Stubbington MJT, Regev A, and Teichmann SA (2017). The Human Cell Atlas: from vision to reality. Nature 550, 451–453. [DOI] [PubMed] [Google Scholar]
- Rytter MJH, Kolte L, Briend A, Friis H, and Christensen VB (2014). The Immune System in Children with Malnutrition—A Systematic Review. PLoS ONE 9, e105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van WL, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, and Galy A (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. [DOI] [PubMed] [Google Scholar]
- Schatorjé EJH, Gemen EFA, Driessen GJA, Leuvenink J, van Hout RWNM, van der Burg M, and de Vries E (2011). Age-matched Reference Values for B-lymphocyte Subpopulations and CVID Classifications in Children. Scandinavian Journal of Immunology 74, 502–510. [DOI] [PubMed] [Google Scholar]
- Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Liu S, Lin S, Berube P, Lee L, et al. (2017). Reconstruction of developmental landscapes by optimal-transport analysis of single-cell gene expression sheds light on cellular reprogramming. bioRxiv 191056. [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. (2019). Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming. Cell (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer JB, Behling C, Newbury R, Deutsch R, Nievergelt C, Schork NJ, and Lavine JE (2005). Histopathology of pediatric nonalcoholic fatty liver disease. Hepatology 42, 641–649. [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhé M-P, Renno J, Loe-Mie Y, Lifshitz A, Mukamel Z, Schmutz S, et al. (2018). Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell 173, 1520–1534.e1520. [DOI] [PubMed] [Google Scholar]
- Seely EW, Tsigas E, and Rich-Edwards JW (2015). Preeclampsia and future cardiovascular disease in women: How good are the data and how can we manage our patients? Pregnancy as a Window to Future Health 39, 276–283. [DOI] [PubMed] [Google Scholar]
- Shalek AK, and Benson M (2017). Single-cell analyses to tailor treatments. Science Translational Medicine 9, eaan4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Klein SS, Barboza L, Lohdi N, and Toth M (2016). Principles Governing DNA Methylation during Neuronal Lineage and Subtype Specification. J. Neurosci 36, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AK, Hollander GA, and McMichael A (2015). Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci 282, 20143085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2018). Rewiring of the cellular and inter-cellular landscape of the human colon during ulcerative colitis. bioRxiv 455451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GCS (2010). First-trimester determination of complications of late pregnancy. Jama 303, 561–562. [DOI] [PubMed] [Google Scholar]
- Smith M, Arthur D, Camitta B, Carroll AJ, Crist W, Gaynon P, Gelber R, Heerema N, Korn EL, Link M, et al. (1996). Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. Journal of Clinical Oncology 14, 18–24. [DOI] [PubMed] [Google Scholar]
- Specht H, and Slavov N (2018). Transformative Opportunities for Single-Cell Proteomics. J. Proteome Res 17, 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson T (2005). How children’s responses to drugs differ from adults. British Journal of Clinical Pharmacology 59, 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A, Hanson D, Whatmore A, Destenaves B, Chatelain P, and Clayton P (2013). Human growth is associated with distinct patterns of gene expression in evolutionarily conserved networks. BMC Genomics 14, 547–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, and Smibert P (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat Meth 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, Smibert P, and Satija R (2018). Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struijs M-C, Diamond IR, de Silva N, and Wales PW (2009). Establishing norms for intestinal length in children. J. Pediatr. Surg 44, 933–938. [DOI] [PubMed] [Google Scholar]
- Stuart T, and Satija R (2019). Integrative single-cell analysis. Nat Rev Genet. [DOI] [PubMed] [Google Scholar]
- Stubbington MJT, Rozenblatt-Rosen O, Regev A, and Teichmann SA (2017). Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V, Vento-Tormo R, and Teichmann SA (2018). Exponential scaling of single-cell RNA-seq in the past decade. Nature Protocols 13, 599–604. [DOI] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, et al. (2016). Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Meth 13, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhunter ME, Sayaman RW, Miyano M, and LaBarge MA (2018). Tissue aging: the integration of collective and variant responses of cells to entropic forces over time. Curr Opin Cell Biol 54, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C (2015). Defining cell types and states with single-cell genomics. Genome Res 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, and Quake SR (2014). Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni Y-B, To KF, Cheng YKY, Chiu RWK, et al. (2017). Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci U S A 114, E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419–1260419. [DOI] [PubMed] [Google Scholar]
- van Abeelen AFM, Elias SG, Bossuyt PMM, Grobbee DE, van der Schouw YT, Roseboom TJ, and Uiterwaal CSPM (2012). Famine exposure in the young and the risk of type 2 diabetes in adulthood. Diabetes 61, 2255–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde D, Konings EEM, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJM, and Roos-Hesselink JW (2011). Birth Prevalence of Congenital Heart Disease Worldwide. Journal of the American College of Cardiology 58, 2241–2247. [DOI] [PubMed] [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, Zastrow, von ME, Yool A, Dement WC, Barchas JD, and Eberwine JH (1990). Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, et al. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 19, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA (2018). Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade HJ, Bezerra JA, Davenport M, Schreiber RA, Mieli-Vergani G, Hulscher JB, Sokol RJ, Kelly DA, Ure B, Whitington PF, et al. (2016). Biliary atresia and other cholestatic childhood diseases: Advances and future challenges. J. Hepatol 65, 631–642. [DOI] [PubMed] [Google Scholar]
- Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polański K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. (2019). A cellular census of healthy lung and asthmatic airway wall identifies novel cell states in health and disease. bioRxiv 527408. [DOI] [PubMed] [Google Scholar]
- Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Chadeau-Hyam M, Gmuender H, Gulliver J, Herceg Z, Kleinjans J, Kogevinas M, Kyrtopoulos S, Nieuwenhuijsen M, Phillips DH, et al. (2017). The exposome in practice: Design of the EXPOsOMICS project. Special Issue: Human Biomonitoring 2016 220, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin S, Grumolato F, Nardelli GB, Di Camillo B, Grisan E, and Cosmi E (2014). Early origins of adult disease: Low birth weight and vascular remodeling. Atherosclerosis 237, 391–399. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen Y, Yong J, Cui Y, Wang R, Wen L, Qiao J, and Tang F (2018). Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing. Cell Rep 24, 3554–3567.e3. [DOI] [PubMed] [Google Scholar]
- Ward RM, Benjamin DK Jr., Davis JM, Gorman RL, Kauffman R, Kearns GL, Murphy MD, and Sherwin CMT (2018). The Need for Pediatric Drug Development. The Journal of Pediatrics 192, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DS, Wong HR, and Zingarelli B (2011). Pediatric Sepsis - Part I: “Children are not small adults!.” Open Inflamm J 4, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winawer MR, Griffen NG, Samanamud J, Baugh EH, Rathakrishnan D, Ramalingam S, Zagzag D, Schevon CA, Dugan P, Hegde M, et al. (2018). Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann Neurol 83, 1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biology 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Girskis KM, and Walsh CA (2017). Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat Rev Genet 18, 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington JJ, Reimann F, and Gribble FM (2018). Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol 11, 3–20. [DOI] [PubMed] [Google Scholar]
- Wright RO (2017). Environment, susceptibility windows, development and child health. Current Opinion in Pediatrics 29, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee D-K, Gotts JE, et al. (2017). Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol 19, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang D, Ding J-H, Wang W, Chu P-H, Dalton ND, Wang H-Y, Bermingham JR, Ye Z, Liu F, et al. (2005). ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120, 59–72. [DOI] [PubMed] [Google Scholar]
- Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl A-KT, Funari VA, Gokey JJ, et al. (2016). Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellepeddi V, Rower J, Liu X, Kumar S, Rashid J, and Sherwin CMT (2019). State-of-the-Art Review on Physiologically Based Pharmacokinetic Modeling in Pediatric Drug Development. Clin Pharmacokinet 58, 1–13. [DOI] [PubMed] [Google Scholar]
- Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, Collord G, Botting RA, Popescu D-M, Loudon KW, et al. (2018). Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G-C, Cai L, Elowitz M, Enver T, Fan G, Guo G, Irizarry R, Kharchenko P, Kim J, Orkin S, et al. (2017). Challenges and emerging directions in single-cell analysis. Genome Biology 18, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Jiang S, Kong X, El-Ali N, Ball AR Jr., Ma CI, Hashimoto N, Yokomori K, and Mortazavi A (2016). Single-nucleus RNA-seq of differentiating human myoblasts reveals the extent of fate heterogeneity. Nucleic Acids Res. 44, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WZD, Glicksberg BS, Li Y, and Chen B (2019). Selecting precise reference normal tissue samples for cancer research using a deep learning approach. BMC Med Genomics 12, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Taylor DM (2018). Scedar: a scalable Python package for single-cell RNA-seq exploratory data analysis. bioRxiv 375196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Gao S, Wu Z, Kajigaya S, Feng X, Liu Q, Townsley DM, Cooper J, Chen J, Keyvanfar K, et al. (2017). Single-cell RNA-seq reveals a distinct transcriptome signature of aneuploid hematopoietic cells. Blood 130, 2762–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
- Zurlo F, Larson K, Bogardus C, and Ravussin E (1990). Skeletal muscle metabolism is a major determinant of resting energy expenditure. J. Clin. Invest 86, 1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]