Abstract
Anencephaly is a fatal human neural tube defect (NTD) in which the anterior neural tube remains open. Zebrafish embryos with reduced Nodal signaling display an open anterior neural tube phenotype that is analogous to anencephaly. Previous work from our laboratory suggests that Nodal signaling acts through induction of the head mesendoderm and mesoderm. Head mesendoderm/mesoderm then, through an unknown mechanism, promotes formation of the polarized epithelium which is capable of undergoing the movements required for closure. We compared the transcriptome of embryos treated with a Nodal signaling inhibitor at sphere stage, which causes NTDs, to those treated at 30% epiboly, which does not cause NTDs. This screen identified over 3,000 transcripts with potential roles in anterior neurulation. Expression of several genes encoding components of tight and adherens junctions was significantly reduced, supporting the model that Nodal signaling regulates formation of the neuroepithelium. mRNAs involved in Wnt, FGF, and BMP signaling were also differentially expressed, suggesting these pathways might regulate anterior neurulation. In support of this, we found that pharmacological inhibition of FGF-receptor function causes an open anterior NTD as well as loss of mesodermal derivatives. This suggests that Nodal and FGF both promote anterior neurulation through induction of head mesoderm.
Keywords: Zebrafish, RNA-sequencing, neural tube defects, neurulation, Nodal
Introduction
Neural tube defects (NTDs) are the second most common birth defect in humans, ranging between 1.2-124 per 10,000 births, depending upon geographic location (Zaganjor et al., 2016). The most common NTDs are anencephaly and spina bifida. Anencephaly occurs when the anterior neural tube fails to close and is characterized by partial or complete absence of the cranial vault and cerebral hemisphere (Detrait et al., 2005). Spina bifida occurs when there is defective closure of the neural tube in the spinal column (Detrait et al., 2005).
In many cases, human NTDs have been linked to underlying genetic factors. Studies in model organisms such as mice, chicks, zebrafish, and frogs have been used to identify potential genes that may be linked to the human defects. For instance, in mice, there are over 250 different models with NTDs and over 200 genes that are known to cause NTDs upon misexpression (Copp et al., 2013; Greene and Copp, 2014; Harris and Juriloff, 2010). These genes are being tested for relationships to human NTDs, but in most cases, this link has not yet been found.
Mechanisms of neurulation vary between anterior and posterior regions of the embryo. Primary neurulation forms the anterior portion of the neural tube down to the sacral region and secondary neurulation forms the neural tube posterior to this, including the neural tube of the tail (Smith and Schoenwolf, 1997). Only disruptions in primary neurulation result in NTDs (Copp et al., 2003). Basic morphological events of primary neurulation are similar across species, beginning with formation of the neural plate from columnarization of ectodermal cells. Next, the lateral edges of the neural plate thicken and the neural plate goes through convergent extension and other movements, which assist with bending, extension, and closure to form the neural tube (Colas and Schoenwolf, 2001; Lowery and Sive, 2004).
Primary neurulation varies slightly at different axial levels of the same embryo. For instance, in mice, spinal neurulation at the cervical/hindbrain boundary occurs via bending only at the medial hinge point (MHP) at what will become the ventral-most part of the neural tube. In the region of the lower spine, neurulation occurs via bending the MHP as well as two dorsal lateral hinge points (Shum and Copp, 1996; Ybot-Gonzalez et al., 2007). These variations are also reflected by genetics. In mice, knockout of some genes results in anencephaly, while loss of others result in spina bifida or even craniorachischisis, where primary neurulation fails along the whole length of the embryo (Copp et al., 2003). Therefore, study of the genetics involved at each axial level is necessary to completely understand the mechanisms of neurulation.
Previous work in our laboratory found that zebrafish with decreased Nodal signaling display a phenotype analogous to that of humans with anencephaly (Aquilina-Beck et al., 2007; Ciruna et al., 2006). This phenotype is characterized by an open anterior neural tube, a lack of anterior mesendodermal/mesodermal tissue, decreased membrane localization of the cell adhesion protein N-cadherin in the mesoderm, and disrupted organization of the cells within the neural tube. Our initial characterization of Nodal deficient mutants enabled us to propose a three step model for zebrafish anterior neurulation. In the first step, Nodal signals induce the head mesendoderm/mesoderm during early- to mid-blastula stages. Second, the mesendoderm/mesoderm interacts with the overlying neuroectoderm. This interaction probably begins between shield and tailbud stages (6-10 hpf), as neuroepithelium morphology is already abnormal at neural plate stage (10 hpf) in Nodal mutants (Aquilina-Beck et al., 2007). Third, this mesendodermal/mesodermal signal support formation of a polarized neuroepithelium. This neuroepithelium is then competent to undergo all of the cellular and tissue level changes needed for closure of the anterior neural tube. Consistent with this model, an open neural tube phenotype is present only when Nodal signaling is inhibited before late-blastula stages, the same time Nodal is inducing mesendodermal/mesodermal tissues and before the formation of neuroepithelium (Araya et al., 2014; Gonsar et al., 2016).
In this study, RNA-sequencing was used to identify genetic factors involved in zebrafish neurulation. This screen identified over 3,000 transcripts that were differentially expressed between embryos that would have a closed versus open neural tube. We found genes potentially involved in (1) mesendoderm/mesoderm induction (2) signaling from the mesendoderm/mesoderm to the neuroectoderm, and (3) adhesion between neural tube cells. These transcripts fell into several subgroups including genes involved in several developmentally important pathways (Nodal, BMP, Wnt (canonical, PCP, and Ca++), and FGF as well as genes encoding components of protein complexes involved in neuroepithelium formation (adherens junctions, tight junctions). Previous studies have found that Nodal and FGF act synergistically in complex feedback loops to induce and maintain mesodermal tissues (Mathieu et al., 2004). We found that pharmacologic inhibition of FGF receptor (FGFR) function resulted in open anterior neural tubes. FGF signaling was required for neurulation up to early gastrula stages (6.0 hpf), during the time when the mesodermal germ layer is being induced. Further, presence of NTD in FGF signaling deficient embryos correlated with decreased notochord, a mesoderm-derived tissue. Thus, we propose that FGF signaling acts with Nodal to induce the mesodermal tissues required for anterior neurulation.
Results
Identification of over 3,000 genes potentially involved in anterior neurulation
This study compared the transcriptome of developing zebrafish embryos that would have a closed neural tube to the transcriptome of those that would have an open neural tube. To do this, we used the drug SB505124. SB505124 blocks function of Nodal and Activin receptors, the Activin receptor-Like Kinases (ALKs) 4, 5, and 7, by binding to their ATP binding site (DaCosta Byfield et al., 2004; Hagos et al., 2007) at different stages of development. Our previous studies found that embryos treated with SB505124 starting at developmental stages approximately 40 minutes apart resulted in different neural tube phenotypes (Gonsar et al., 2016). Treatment with SB505124 beginning at sphere stage (mid-blastula, 4.0 hpf) resulted in an open neural tube, as indicated by an elongated or divided pineal anlage (Figure 1). In contrast, treatment beginning at the 30% epiboly stage) (late-blastula, 4.7 hpf) resulted in a closed anterior neural tube, as indicated by a normal, oval pineal organ at the dorsal midline of the (Figure 1). Hereafter, embryos treated at sphere will be referred to as Open Neural Tube (ONT) and those treated at 30% epiboly will be referred to Closed Neural Tube (CNT), even though they were typically assayed at stages before anterior neurulation was completed.
Figure 1: ONT and CNT cause open and closed neural tube phenotypes, respectively.
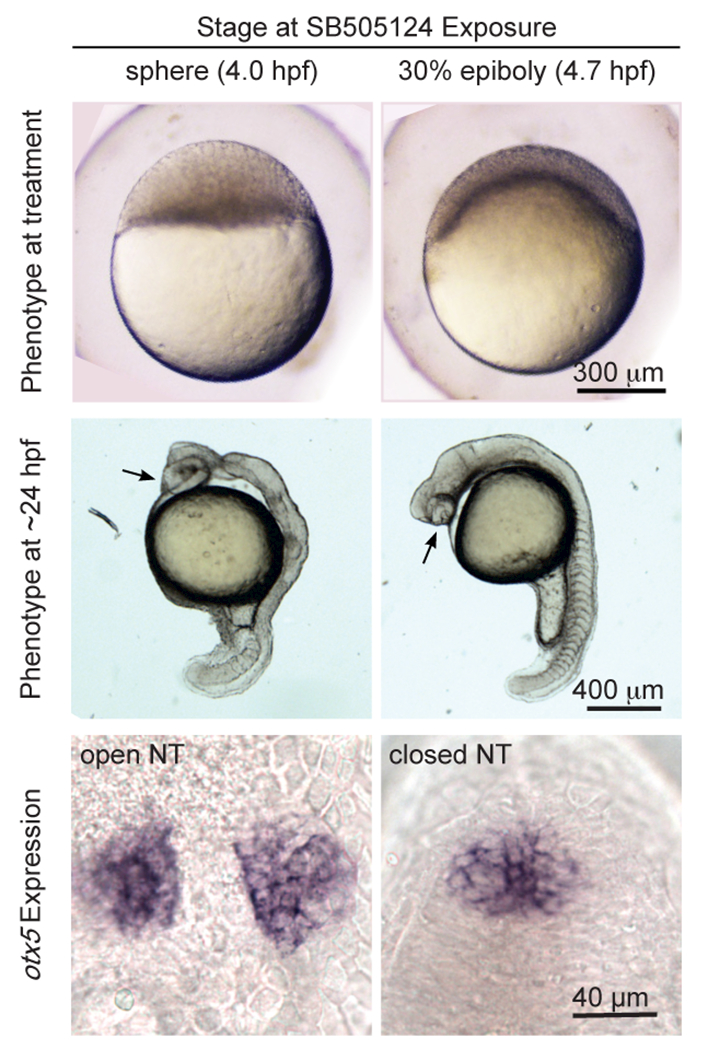
Treatment with SB505124 at sphere stage always results in an open neural tube while treatment at 30% epiboly stage always results in a closed neural tube. Top row: Lateral views, animal pole to top. Middle row: Lateral views, anterior towards top. Bottom row: Dorsal view, anterior towards top.
Although the differences in neural tube phenotype were the focus of this study, there were many other phenotypic differences between embryos treated at sphere and 30% epiboly. For instance, embryos treated at the earlier stage had a greater loss of mesodermal tissue, as indicated by the shorter tail and fewer number of somites (Figure 1). Our previous studies found that embryos with a greater loss of mesendoderm/mesoderm had an increased chance of having an open anterior neural tube (Gonsar et al., 2016). In case changes in gene expression within mesendoderm/mesoderm or other tissues are important for anterior neurulation, our RNAseq experiments used whole embryos.
To identify transcripts potentially involved in different steps of neurulation, ONT and CNT embryos were snap frozen at three stages important for neurulation: shield stage (6.0 hpf), when neuroectoderm induction initiates, tailbud stage (10.0 hpf), when the neural plate starts to fold, and 7 somite stage (around 12.0 hpf), when the closed neural rod is formed in the forebrain (Grinblat et al., 1998; Kimmel et al., 1995). Thus, there were a total of six experimental conditions (ONT frozen at 6, 10, and 12 hpf and CNT frozen at 6, 10, and 12 hpf). Three independent biological replicates for each experimental condition were sequenced, and the average transcript number for each condition was used as the expression level. Differentially expressed mRNAs were analyzed by comparing transcripts between ONT and CNT embryos at each of the three stages. These mRNAs were then identified by mapping the raw reads to NCBI’s RefSeq Danio rerio mRNA sequences. Only mRNAs that differed in expression by 50% or more between the ONT and CNT samples and had at least 100 total counts in one collection point with a false discovery rate of < 0.05 were considered significantly different between ONT and CNT samples.
After all criteria were applied, a total of 3,187 transcripts were significantly differentially expressed between the CNT and ONT embryos (Table 1). Expression differences were viewed as ratios of counts in CNT embryos divided by counts in ONT embryos. Thus, CNT/ONT ratios of > 1 indicated higher expression in CNT embryos while ratios of < 1 indicated higher expression in ONT embryos. For six genes, differential expression results from RNA-sequencing were compared to those produced using relative, reverse transcriptase polymerase chain reaction (RT-PCR). The CNT/ONT expression ratio matched between RNAseq and RT-PCR in all cases, with the exception of fgf8a at shield stage (Supplemental Table 1).
Table 1:
Over 3,000 transcripts were significantly differentially expressed between embryos that would have closed and open neural tubes.
| Shield (6.0 hpf) | Tailbud (10.0 hpf) | 7 somites (12.0 hpf) | All 3 stages | |
|---|---|---|---|---|
| Total significantly differentially expressed | 2,553 | 423 | 634 | 3,186 |
| Higher expression CNT versus ONT embryos | 1,427 | 288 | 290 | 1,792 |
Genes with significant differential expression had to have at least 100 average counts in at least one treatment group, had to differ in expression by at least 50%, and have a false discovery rate of <0.05.
Nodal signals act as morphogens, and the response of target cells depends upon both the dose of Nodal they receive and the length of exposure to the Nodal signal (Gritsman et al., 2000; Hagos and Dougan, 2007; Robertson, 2014; Rogers and Schier, 2011). Since ONT embryos had greater inhibition of Nodal signaling than CNT embryos, our screen should have identified several Nodal downstream genes. Consistent with our screen functioning as designed, several Nodal target genes were differentially expressed (Table 2). For instance, Cyclops and Squint, two of the three zebrafish Nodal ligands, promote the expression of the transcription factor genes mixl1 and sox32. Mixl1 and Sox32 then promote expression of sox17, which leads to endoderm formation (Alexander and Stainier, 1999). As expected for genes positively regulated by Nodal signaling, all three of these genes had CNT/ONT ratios of > 1 (Table 2). Since SB505124 blocks ALK receptor function, we did not expect to see differential expression of ALK transcripts. Consistent with this, none of the ALKs involved in Nodal signaling were differentially expressed (data not shown). However, acvr1l mRNA, which encodes a component of the ALK receptor complex, was differentially expressed, suggesting there may be some transcriptional regulation of receptor function.
Table 2:
Genes known to be affected by Nodal were differentially expressed in the RNA-sequencing screen.
| Differential Expression Ratio CNT / ONT | ||||
|---|---|---|---|---|
| Gene | Shield (6.0 hpf) | Tailbud (10.0 hpf) | 7 Somites (12.0 hpf) | References |
| sox17 | 8.63 | - | - | (Alexander and Stainier, 1999; David and Rosa, 2001) |
| sox32 | - | - | 1.90 | (Dickmeis et al., 2001; Kikuchi et al., 2001; Sakaguchi et al., 2001) |
| mixl1 | 3.23 | - | - | (Alexander and Stainier, 1999; Kikuchi et al., 2001) |
| goosecoid | 6.12 | 2.04 | - | (Agius et al., 1999; Thisse et al., 1994) |
| noggin1 | 5.09 | 6.12 | 1.58 | (Ragland and Raible, 2004; Szeto and Kimelman, 2006) |
| follistatin a | - | 36.57 | 2.94 | (Ragland and Raible, 2004; Szeto and Kimelman, 2006) |
| pitx2 | - | - | 2.27 | (Liang et al., 2000; Yan et al., 1999) |
Our data also contained genes known to cause NTDs in mice and humans, suggesting these genes might have a conserved role in neurulation (Table 3) (Copp and Greene, 2010; Copp et al., 2013). For instance, of four genes with loss-of-function mutations associated with human anencephaly, three were differentially expressed in our screen (Lemay et al., 2015).
Table 3:
Genes associated with NTDs in other vertebrates were differentially expressed in developing zebrafish.
| Zebrafish Gene | Zebrafish | Mouse | Humans | Citations |
|---|---|---|---|---|
| Direction of CNT/ONT ratio | ||||
| plld | >1 | >1 | - | (Luo et al., 2005) |
| vcla | >1 | >1 | - | (Xu et al., 1998) |
| shroom3 | <1 | >1 | >1 | (Hildebrand and Soriano, 1999; Lemay et al., 2015) |
| brd2a | >1 | >1 | - | (Shang et al., 2009) |
| nf1b | >1 | >1 | - | (Lakkis et al., 1999) |
| casp9 | >1 | >1 | - | (Kuida et al., 1998) |
| cited2 | >1 | >1 | - | (Bamforth et al., 2001; Yin et al., 2002) |
| crebbpa | <1 | >1 | - | (Tanaka et al., 2000) |
| gli2a | <1 | >1 | - | (Pan et al., 2009) |
| prkacab | >1 | >1 | - | (Huang et al., 2002) |
| ptch1 | <1 | >1 | - | (Goodrich et al., 1997; Milenkovic et al., 1999) |
| cyp26a1 | <1 | >1 | - | (Niederreither et al., 2002) |
| grhl3 | >1 | - | >1 | (Lemay et al., 2015) |
| ptprsa | >1 | - | >1 | (Lemay et al., 2015) |
Identification of candidate signaling pathways
One of the main goals for this study was to identify potential signals that could act downstream of Nodal to induce or maintain mesendoderm/mesoderm or to mediate the interaction between the mesendoderm/mesoderm and the neuroectoderm. Since these signals would be involved in promoting neural tube closure, we expected their associated genes would be decreased in ONT embryos compared to CNT embryos. Genes mapping to four pathways fit these criteria: the Nodal (Table 2, Figure 2), Wnt (Canonical, PCP, and Ca++) (Figure 3), BMP (Figure 2), and FGF (Figure 4) signaling pathways.
Figure 2: Differential expression of transcripts encoding components of the BMP and Nodal signaling pathways.
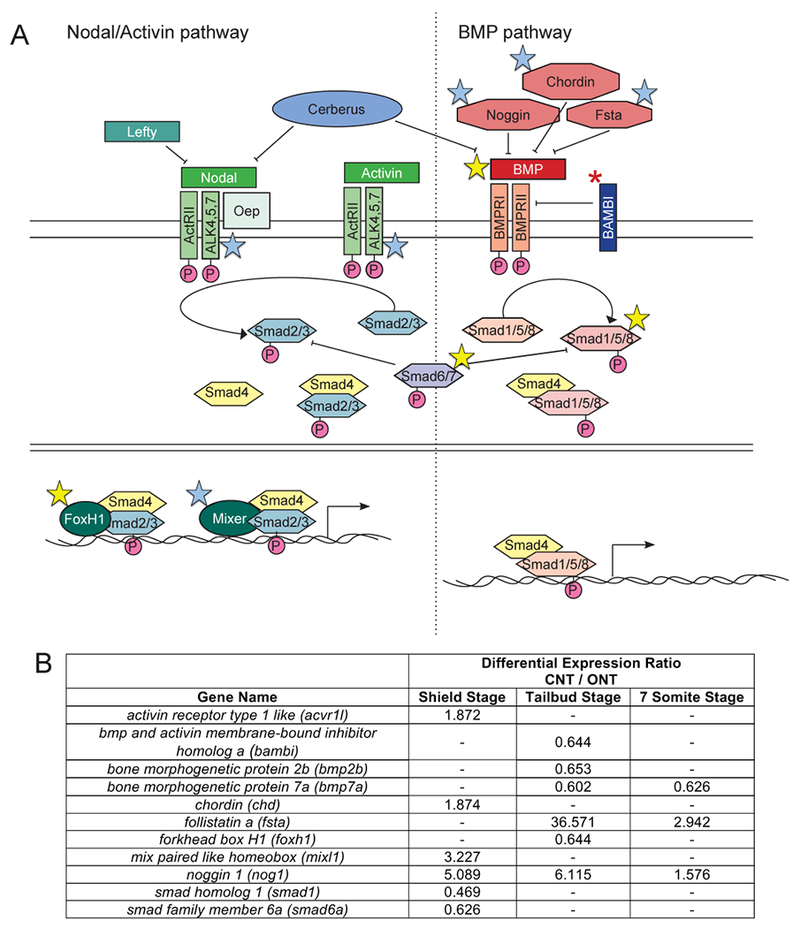
A) Danio rerio Nodal/Activin and BMP signaling pathways. Blue stars indicate the corresponding gene had higher expression in CNT embryos. Yellow stars indicate the corresponding gene had lower expression in CNT embryos. B) Table of differentially expressed genes in these pathways and the corresponding expression ratios from the RNA-sequencing data analysis.
Figure 3: Differential expression of transcripts encoding portions of the Wnt signaling pathway.
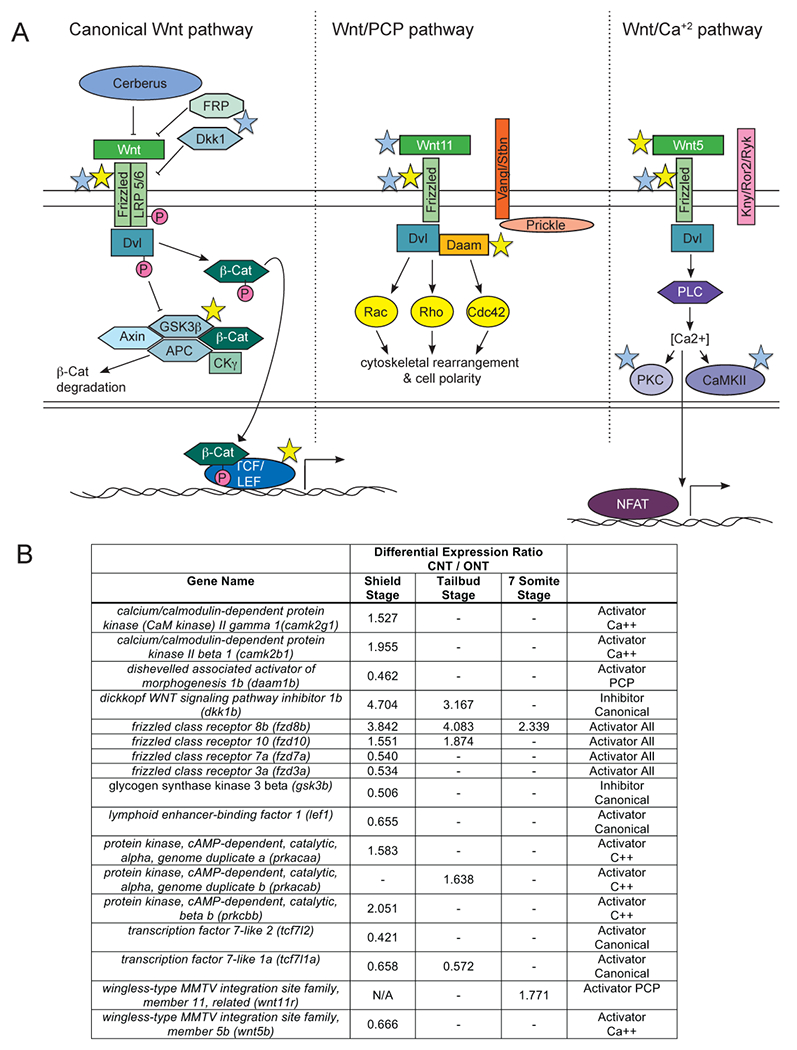
A) Danio rerio Wnt signaling pathway. Colored stars used as in Figure 2. B) Table of differentially expressed genes in the Wnt pathway and corresponding expression ratios from the RNA-sequencing data analysis.
Figure 4: Differential expression of transcripts encoding portions of the FGF signaling pathway.
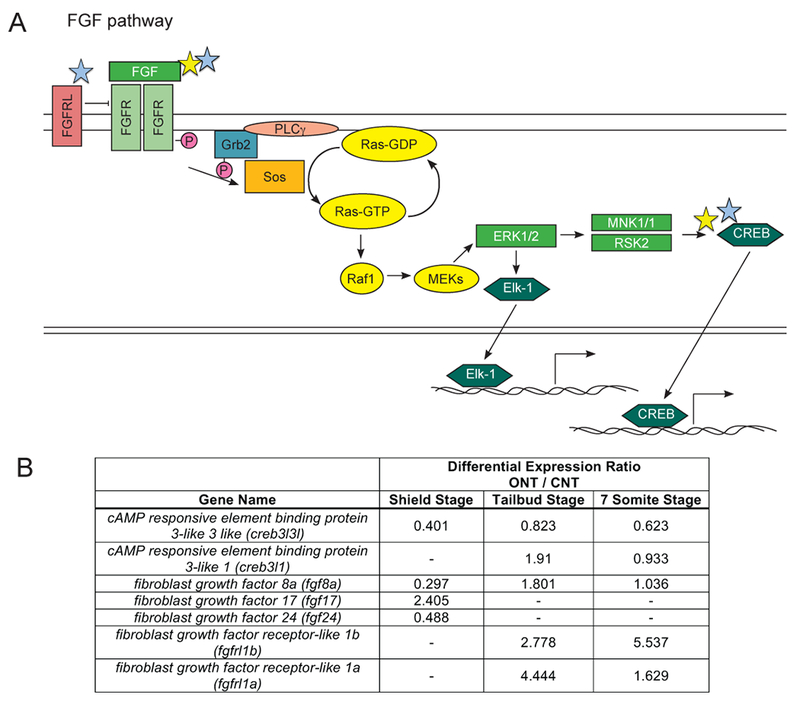
A) Danio rerio FGF signaling pathway. Colored stars used as in Figure 2. B) Table of differentially expressed genes represented in the FGF pathway and corresponding expression ratios from the RNA-sequencing data analysis.
BMP signaling works in a gradient to specify the dorsoventral mesodermal tissues, with high BMP signaling causing ventral fates and low causing dorsal fates (De Robertis, 2009). Differential expression of genes in the BMP pathway suggests signaling is increased in embryos with open neural tubes (Figure 2). Several genes encoding negative regulators of the BMP pathway had CNT/ONT > 1 (chd, fsta, nog1), while several genes encoding positive regulators had a CNT/ONT < 1 (bmp7a, bmp2a, smad1, smad6a).
The Wnt family of secreted signals has several distinct downstream pathways, all of which have roles during development. Differential expression of genes common to all three Wnt signaling pathways did not suggest a clear role for Wnt signaling in anterior neurulation (Figure 3). For instance, zebrafish have 14 genes encoding Frizzled (Fzd) proteins, the receptor component common to all of the Wnt pathways. Four genes encoding some fzd genes were higher in embryos with closed neural tubes at shield stage, while others were lower (Figure 3).
Canonical Wnt signaling is involved in many events during development, including dorsal-ventral axis induction, organ development, tail formation, and neural patterning (Komiya and Habas, 2008). The RNAseq data suggest inhibition of canonical Wnt signaling could be important for anterior neurulation. Genes encoding positive regulators (bambia) or downstream effectors (lef1, tcf7l2, tcf7l1a) had CNT/ONT ratios < 1, demonstrating lower expression when the neural tube is closed. In contrast, dkk1b, which encodes a secreted inhibitor of the canonical Wnt pathway, had a CNT/ONT ratio > 3 at shield and tailbud stages. Further, dkk1b is expressed in the dorsal, anterior mesendoderm at these stages, placing it in the right place and time to be involved in regulating anterior neurulation in addition to its known role in head development (Komiya and Habas, 2008). Consistent with our screen, overexpression of the zebrafish Nodal signal Squint caused expanded dkk1b expression, while loss of Nodal signaling caused reduced expression (Hashimoto et al., 2000).
The Wnt/PCP pathway is essential for neurulation through its role in polarizing the cells of the neuroepithelium (Cai and Shi, 2014; Ciruna et al., 2006; Tawk et al., 2007; Wallingford and Harland, 2002). The Wnt/Ca++ pathway shares many components with the PCP pathway, including being activated by the same subset of Wnt ligands (Wnt4-7, 11) (Li et al., 2005). Our screen did not give a clear prediction for these two related pathways, as some positive components were expressed more highly in ONT and others in CNT embryos (Figure 3).
Inhibition of FGF signaling caused defects in the anterior neural tube
Of the candidate pathways, we chose to further test the potential role of FGF signaling in anterior neurulation. The RNAseq data in part supported higher FGF signaling in embryos with closed neural tubes, as fgf17 was expressed more highly in CNT embryos at shield stage and fgf8a was expressed more highly at tailbud and 7 somite stages (Figure 4). However, other aspects of the RNAseq data support an association between decreased FGF signaling and anterior neurulation. For example, FGFR-like proteins lack the intracellular kinase domain and are therefore thought to inhibit FGF signaling. At tailbud and 7 somite stages, two fgf receptor-like genes, fgfrl1b, fgfrl1a, had CNT/ONT ratios > 1 (Figure 4).
To test the function of FGF signaling in neurulation, we used the small molecule SU5402 to inhibit the tyrosine kinase activity of FGFR (Londin et al., 2005; Mathieu et al., 2004; Mohammadi et al., 1997; Raible and Brand, 2001; Rentzsch et al., 2004; Sun et al., 1999). Initiation of treatment occurred at mid blastula to mid gastrula stages and continued until the embryos were fixed at 24 hpf.
There was significant variation in effectiveness among different batches of SU5402, however all batches caused similar phenotypes, which we organized into four classes based on their phenotype at 24 hpf (Supplemental Table 2, Table 4, Figure 5). Class I embryos had a slightly truncated tail, normal somites, and no apoptosis. Class II embryos had a more severely truncated, immotile tail, absent or misshapen somites, small eyes, and apoptosis in the head and tail. Class III embryos had very small tail, no or misshapen somites, malformed eyes, and apoptosis in the head and tail. Class IV embryos were composed of a head and a tail with very little tissue in the trunk of the embryo. There was a strong relationship between severity of phenotype and stage when treatment was initiated (Table 4, Figure 5). Class I embryos were found only in the group treated at the latest developmental time point (70% epiboly, 8.0 hpf) (Table 4, Figure 5). The majority of embryos with inhibitor treatment starting between dome (4.3 hpf) and shield (6.0 hpf) stages fell into class II (Table 4, Figure 5). Embryos with treatment starting at the earliest time point, sphere (4.0 hpf), fell into classes III and IV.
Table 4.
Embryos with later initiation of the FGF inhibitor treatment have less severe phenotypes.
| Stage at Onset of SU5402 Exposure | WT | Class I | Class II | Class III | Class IV | Total |
|---|---|---|---|---|---|---|
| DMSO Control | 298 | 0 | 0 | 0 | 0 | 298 |
| Sphere | 0 | 0 | 0 | 23 | 36 | 59 |
| Dome | 0 | 0 | 51 | 26 | 0 | 77 |
| 30% Epiboly | 0 | 0 | 270 | 6 | 0 | 276 |
| 50% Epiboly | 0 | 0 | 276 | 0 | 0 | 276 |
| Shield | 0 | 0 | 267 | 0 | 0 | 267 |
| 70% Epiboly | 0 | 249 | 0 | 0 | 0 | 249 |
Figure 5. FGF signaling at or before the onset of gastrulation is required for anterior neural tube closure.
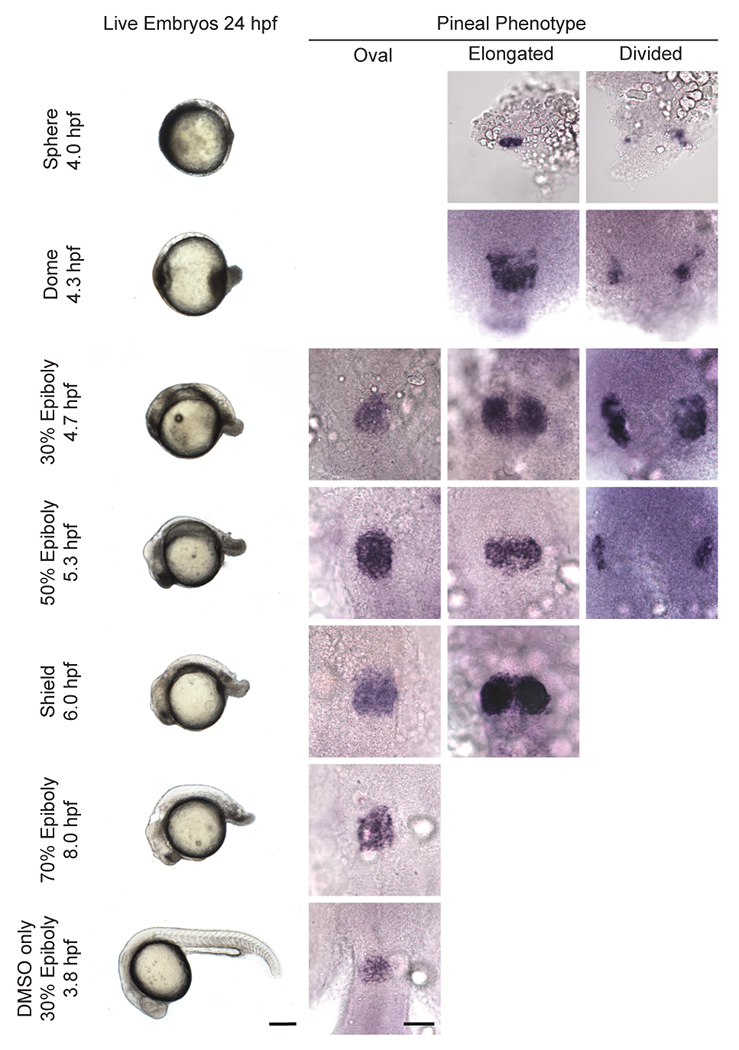
Embryos were treated with SU5402 starting at time points indicated and then analyzed for anterior neural tube phenotype by pineal morphology. First column: lateral views of whole embryos with anterior to the left and dorsal to the top. Scale bar: 250 μm. Second-fourth columns: dorsal view with anterior to the top. Scale bar: 50 μm. Refer to Table 5 for quantitative data.
Consistent with an essential role for FGF signaling in neurulation, inhibition of FGF with SU5402 signaling caused pineal phenotypes similar to those in Nodal signaling deficient embryos (Table 5, Figure 5). Treatment starting at mid blastula stages (sphere, 4.0 hpf and dome, 4.3 hpf) caused an open elongated or divided pineal phenotype, suggesting an open anterior neural tube. As the initiation of treatment moved later in development, the percentage of embryos with NTD decreased, from 49% of embryos whose treatment started at 30% epiboly (4.7) to only 8% of embryos whose treatment started at shield stage (6.0 hpf). A two-tailed Fisher’s exact test indicated an association between SU5402 treatment initiation and neural tube phenotype as well as a significant sample size with P = 2.2 × 10−16.
Table 5.
Initiating inhibition of FGF signaling at or before the onset of gastrulation causes open neural tubes.
| Closed NT | Open NT | |||
|---|---|---|---|---|
| Stage at Onset of SU5402 Exposure | Oval Pineal | Elongated Pineal | Divided Pineal | Total |
| DMSO Control | 298 (100%) | 0 (0%) | 0 (0%) | 298 |
| Sphere (4.0 hpf) | 0 (0%) | 4 (36%) | 7 (67%) | 11* |
| Dome (4.3 hpf) | 0 (0%) | 17 (28%) | 43 (72%) | 60 |
| 30% Epiboly (4.7 hpf) | 119 (51%) | 74 (32%) | 39 (17%) | 232 |
| 50% Epiboly (5.3 hpf) | 184 (67%) | 28 (10%) | 63 (23%) | 275 |
| Shield (6.0 hpf) | 242 (92%) | 22 (8%) | 0 (0%) | 264 |
| 70% Epiboly (8.0 hpf) | 243 (100%) | 0 (0%) | 0 (0%) | 243 |
A total of 59 embryos were analyzed for pineal phenotype but only 11 embryos had detectable otx5 expression in the pineal.
FGF deficient embryos have a correlation with notochord presence and neural tube closure but not with hatching glands
Previous research found a correlation between several anterior mesendodermal/mesodermal tissues and neural tube closure in Nodal deficient embryos. FGF signaling acts coordinately with Nodal signaling to induce and maintain mesodermal tissue (Mathieu et al., 2004), suggesting lack of mesodermal tissue could be the cause of the NTD in FGF-signaling deficient embryos. As has been previously reported, expression of one-eyed pinhead (oep), which encodes an essential component of the Nodal receptor complex, was reduced in a concentration dependent manner in SU5402 treated embryos (Figure 6) (Mathieu et al., 2004). In contrast, the level of expression of the nodal related 2 gene (ndr2), which encodes one of the zebrafish Nodal ligands, was not affected, although the shape of the ndr2-expressing tissue was altered at the highest inhibitor concentrations.
Figure 6: Expression of the oep gene is decreased in embryos with deficient FGF signaling.
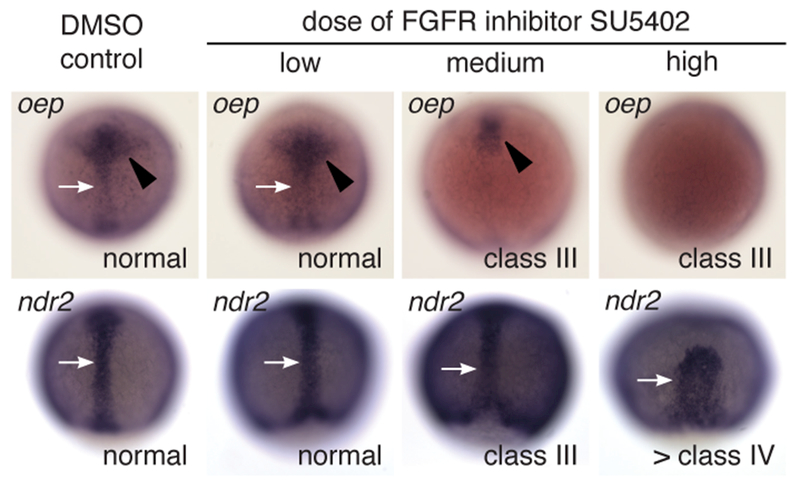
Embryos were treated with SU5402 starting at dome stage and then fixed at 75% epiboly and assayed for expression of the Nodal receptor complex gene oep and the Nodal ligand gene ndr2. oep expression has a dose dependent decrease in the midline tissue (white arrows) and prechordal plate/anterior neuroectoderm (black arrowheads). In contrast, expression levels of ndr2 remain high, although the shape of the midline tissue is disrupted at the highest concentration. Scale bar: 100 μM.
FGF and Nodal signaling deficient embryos with anterior NTD had similar morphological phenotypes, suggesting they could have similar loss of mesodermal tissues (compare Figure 5 with Figure 1). However, one difference is that Nodal signaling deficient embryos lack endoderm and mesendoderm in addition to mesoderm (Mathieu et al., 2004). This results in some prominent morphological differences, such as cyclopia in Nodal deficient embryos and two eyes upon loss of FGF signaling (Figures 1 and 5). To determine if the loss of mesodermal tissue was related to the neural tube defect, we tested for a correlation between failure in anterior neural tube closure and loss of mesoderm-derived notochord in FGFR inhibitor treated embryos. The phenotypes of inhibitor treated embryos ranged from no notochord to almost normal notochord (Table 6, Figure 7). Consistent with the loss of mesoderm being a cause of anterior NTD in embryos with decreased FGF signaling, inhibitor treated embryos had a strong correlation between notochord presence and neural tube closure (two-tailed Fisher’s exact test, P = 2.2 × 10−16). As expected, all inhibitor treated embryos had normal mesendoderm-derived hatching glands (Supplemental Figure 1), and there was no correlation between hatching gland presence and a closed neural tube (two-tailed Fisher’s exact test, P = 1).
Table 6.
Notochord tissue increases with later initiation of FGF signaling inhibitor treatment.
| Stage at Onset of SU5402 Exposure | Notochord phenotype | ||||
|---|---|---|---|---|---|
| None | Some | Most | Full | Total | |
| DMSO Control | |||||
| Closed NT | 0 | 0 | 0 | 108 | 108 |
| Open NT | 0 | 0 | 0 | 0 | 0 |
| 30% Epiboly | |||||
| Closed NT | 2 | 17 | 36 | 0 | 55 |
| Open NT | 2 | 28 | 3 | 0 | 33 |
| 50% Epiboly | |||||
| Closed NT | 0 | 32 | 59 | 0 | 91 |
| Open NT | 0 | 4 | 2 | 0 | 6 |
| Shield | |||||
| Closed NT | 0 | 2 | 79 | 64 | 145 |
| Open NT | 0 | 1 | 1 | 0 | 2 |
| 70% Epiboly | |||||
| Closed NT | 0 | 0 | 0 | 130 | 130 |
| Open NT | 0 | 0 | 0 | 0 | 0 |
Figure 7. Correlation between notochord presence and neural tube closure in FGF signaling deficient embryos.
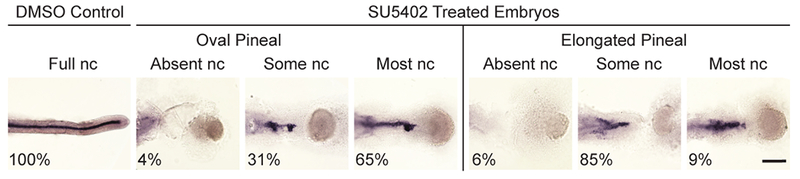
Embryos were assayed with the notochord marker col2a1 to determine the amount of notochord (nc) present and the pineal marker otx5 to report the neural tube phenotype. Representative images of embryos treated at 30% epiboly with DMSO or the FGFR inhibitor SU5402. Pineal phenotype is indicated but not shown. Dorsal views of deyolked embryo trunks with anterior to the left. Scale bar: 150 μm. Refer to Table 6 for quantitative data.
Loss of posterior neuroectoderm in embryos FGF signaling deficient embryos
Despite the similarities in their pineal and mesoderm phenotypes, FGF and Nodal signaling deficient embryos had many differences in their neuroectoderm. Notably, the anterior-posterior patterning of Nodal deficient embryos was largely normal, while FGF deficient embryos had a concentration-dependent, progressive loss of posterior tissue (Figure 8). For instance, ndr1;ndr2 double mutants, which had a severe loss of Nodal signaling, had an overall morphology similar to Class III SU5402-treated embryos (Figure 8). However, patterning in their hindbrains were different. In the ndr1;ndr2 mutants, rhombomeres 1-5 were present and in the correct anterior-posterior orientation. In the Class III SU5402-treated embryos, only one rhombomere, most likely the most anterior, was evident (Figure 8). In contrast, the morphology of the brain in Nodal signaling deficient fish appeared to be more disrupted. For instance, a left and right side of the neural tube could be distinguished in the FGFR inhibitor Class III embryos, but in not the ndr1;ndr2 mutants (Figure 8).
Figure 8: Comparison of embryos treated with a variety of FGFR inhibitors reveals a common phenotype of loss of posterior structures.
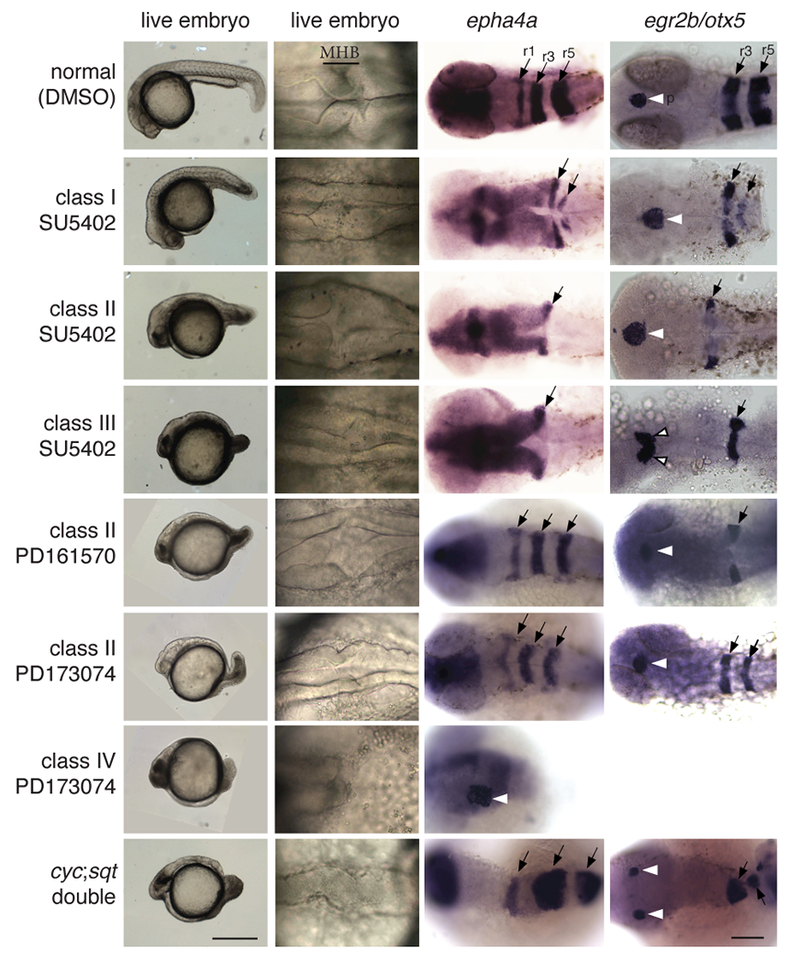
WT embryos were treated with the indicated FGFR inhibitor or vehicle alone (DMSO) starting at dome stage and continuing until fixation at ~30 hpf. ndr1;ndr2 mutants were produced by natural spawnings. Embryos treated with PD161570 and PD173074 treatment have phenotypes that similar to those caused by SU5402, including loss of hindbrain rhombomerers (r’s, black arrows) that increases in severity with increasing phenotypic class & inhibitor concentration. In contrast, ndr1;ndr2 mutants have no evidence of loss of hindbrain tissues even though they have a similar shortening of the anterior-posterior axis as the FGFR inhibitor treated embryos and more disrupted morphology (compare region of the midbrain/hindbrain boundary (MHB)). Pineal precursors are indicated by white arrowheads. First column, lateral views. Second-fourth columns, dorsal views. Scale bars: First column 0.5 mm, remaining columns 0.1 mm. Refer to Table 8 and Supplemental Table 2 for quantitative data.
Other FGFR inhibitors cause phenotypes similar to SU5402
To verify the results using SU5402, we treated developing embryos with two other inhibitors of FGFRs, PD161570 and PD173074 (Batley et al., 1998; Mohammadi et al., 1998; Rodriguez et al., 2012; Shifley et al., 2012). These inhibitors cause similar phenotypes to those produced from SU5402 treatment, including similar overall morphology, posterior truncation of the neuroectoderm, and elongated and divided pineal organs (Figure 8, Table 7)
Table 7.
Treatment with FGFR inhibitors PD161570 and PD173074 at dome stage caused open neural tubes in the forebrain.
| Phenotypic Class of Embryos | Pineal Phenotype | |||||
|---|---|---|---|---|---|---|
| Oval | Elongated | Divided | Undetectable | Total | ||
| DMSO control | Normal | 33 | 0 | 0 | 0 | 33 |
| PD161570 | ||||||
| 20 μM | Normal | 0 | 0 | 0 | 0 | 0 |
| 50 μM | Normal & I | 14 | 0 | 0 | 0 | 14 |
| 100 μM | I & II | 9 | 0 | 0 | 0 | 9 |
| 200 μM | III & IV | 2 | 3 | 1 | 1 | 7 |
| 300 μM | 1 | 1 | 0 | 0 | 2 | |
| 400 & 600 μM | IV & >IV | 0 | 0 | 1 | 3 | 4 |
| PD173074 | ||||||
| 20 μM | Normal | 6 | 0 | 0 | 0 | 6 |
| 50 μM | Normal & I | 8 | 0 | 0 | 0 | 8 |
| 100 μM | I & II & III | 19 | 1 | 0 | 0 | 20 |
| 200 μM | III & IV | 3 | 2 | 1 | 2 | 8 |
| 250-400 μM | IV | 0 | 4 | 3 | 1 | 8 |
Differential expression of genes involved in cell adhesion
Cell-cell adhesion is responsible for organizing the developing neural tube into a polarized epithelium and ultimately for the movements that close the neural tube. Formation of epithelia is a progressive process. The first step in the formation of many epithelia is interactions among Nectin proteins on opposing cells. Nectin interactions initiate the formation of Cadherin based adherens junctions, which in turn seed the formation of adjacent tight junctions. Our screen identified many adherens and tight junction genes with known and unknown roles in neurulation (Figures 9 and 10).
Figure 9: Differential expression of transcripts encoding portions of Adherens Junctions.
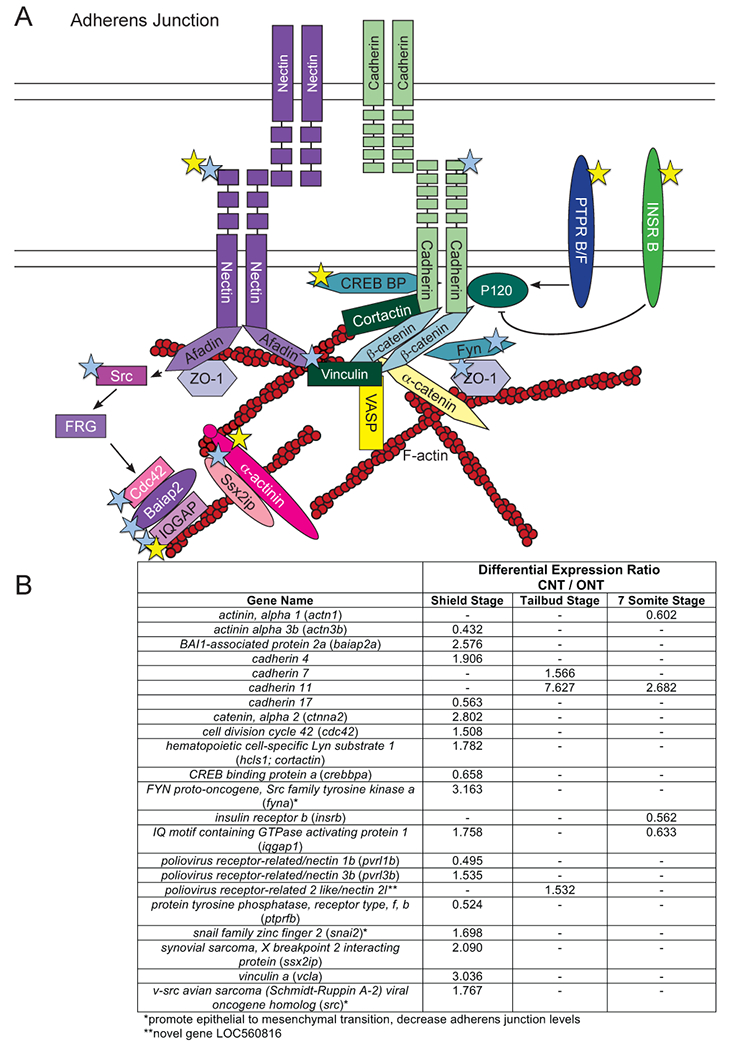
A) Schematic of an adherens junction. Colored stars used as in Figure 2. B) Table of differentially expressed adherens junction genes and corresponding expression ratios from the RNA-sequencing data analysis.
Figure 10: Differential expression of transcripts encoding portions of the Tight Junction pathway.
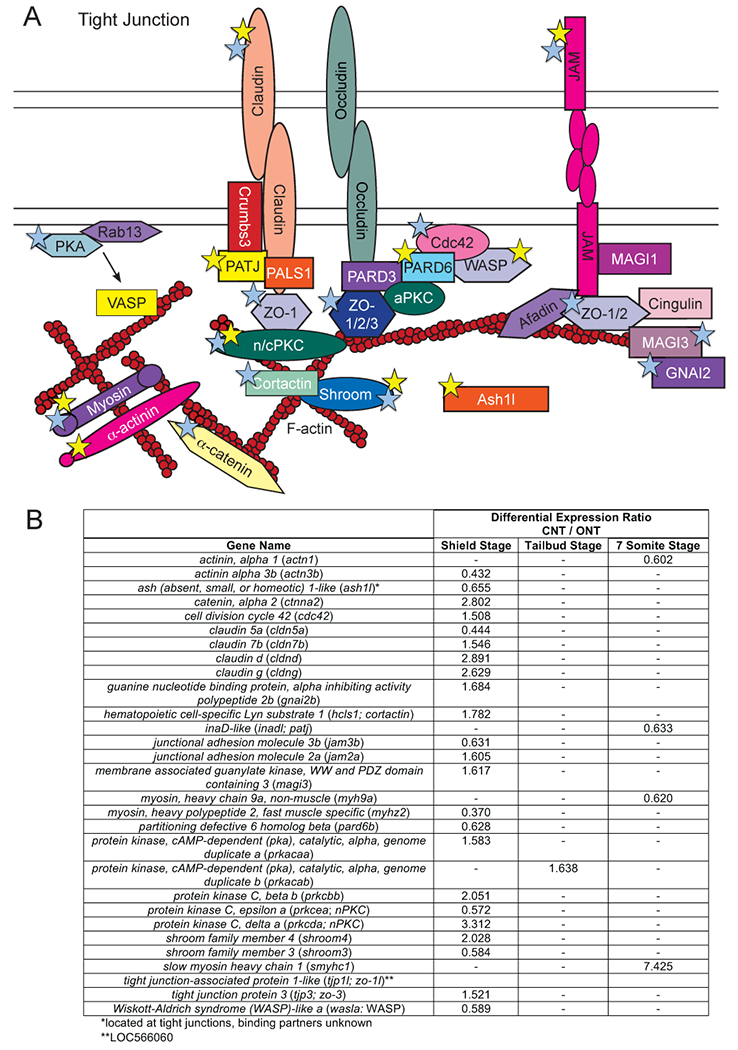
A) Schematic of a tight junction. Colored stars used as in Figure 2. B) Table of differentially expressed tight junction genes and corresponding expression ratios from the RNA-sequencing data analysis.
Two Nectin genes (nectin1b, 3b) were differentially expressed at shield stage, although in different directions (Figure 9). Nectin proteins also act as juxtracrine signals (Huang and Lui, 2016). Several genes encoding proteins that act downstream of Nectin in juxtracrine signaling (src, cdc42, baiap2, iqgap) had a CNT/ONT > 1 at shield stage, suggesting that Nectin signaling could have a positive role in anterior neurulation.
Two cadherin genes (cdh7 and cdh11) were expressed at lower levels at tailbud stage and one (cdh11) at the seven somite stage in ONT embryos (Figure 9). However, there are no reports of these genes being expressed in the developing neural tube. In contrast, cdh2/n-cad mRNA, which encodes the central component of neural adhesion junctions, was not differentially expressed. The majority of differentially expressed genes involved in tight junction formation varied only at shield stage, with no genes being differentially expressed at more than one time point (Figure 10). This suggests that transcription regulation could be very dynamic at shield stages, but plays a lesser role at later stages.
Discussion
Comparison of the transcriptome between embryos with and without neurulation defects identified over 3,000 candidate genes that could act downstream of Nodal signaling in zebrafish anterior neurulation. In particular, there were several sets of differentially expressed genes that mapped to the same signaling pathway, suggesting these pathways could regulate neurulation. For instance, the pattern of differential expression suggests FGF signaling could have a positive role in anterior neurulation, while Wnt signaling could have a negative or inhibitory role. In support of this, treatment of embryos with a FGF signaling inhibitor caused open neural tube defects. Although previous studies suggest Nodal deficient mutants have defects in cell adhesion within the neuroepithelium, genes involved in making adherens and tight junctions were differentially expressed only at the onset of gastrulation, when the neuroepithelium was being induced. This suggests that any defects in formation of a polarized epithelium are due to post-transcriptional mechanisms. Finally, this screen identified many genes that have not yet been linked to neurulation, thus providing many new candidate genes that could be involved in human NTDs.
Advantages of RNAseq approach
RNA-sequencing has been proven to be a very useful approach for identifying candidates involved in neurulation downstream of Nodal. Since few embryos are needed for sequencing, this technique is very accessible. 30-60 embryos per sample in our study, and around 100 embryos in other recent studies contained over 1 μg of high quality mRNA (Harvey et al., 2013; Vesterlund et al., 2011). Also, the cost of RNA-sequencing has decreased over the last few years, making sequencing of multiple samples more cost effective. Therefore, RNA-sequencing has become a much more accessible technique and will be very useful for a variety of future forward genetic studies. Importantly, since Nodal signaling is involved in so many developmental events, we compared embryos that differed by only 20 minutes in the timing of their Nodal inhibitor treatment. These closely related treatments resulted in embryos that had many of the same phenotypes (cyclopic eyes, reduced mesoderm, etc.), but differed in having open and closed anterior neural tubes and in the severity of some of the other phenotypes, such as the extent of mesendoderm/mesoderm loss. Although the function of most of the genes identified remains to be tested, the fact that our first functional tests found a new signaling pathway (FGF) suggests that this approach enriched for genes involved in anterior neurulation.
FGF signaling is required for anterior neurulation in zebrafish
Our study suggests FGF signaling is required for anterior neurulation. Treating embryos with the FGFR inhibitor SU5402 at or before early gastrulation stages (6.0 hpf) resulted in open neural tubes. Other aspects of SU5402 phenotypes closely match those found by others using SU5402 in zebrafish (Londin et al., 2005; Mathieu et al., 2004; Rentzsch et al., 2004), those produced by two other FGFR inhibitors in our study, those found in zebrafish mutants with a severe loss of FGF signaling (Roy and Sagerstrom, 2004), and those found in medaka embryos that lack both maternal and zygotic FGFR1 mRNA (Shimada et al., 2008). Thus, it is likely that the open anterior neural tube phenotype is caused by inhibition of the FGF signaling pathway.
SU5402 was originally found as a specific inhibitor of FGFR1, but has since been shown to act on a well-conserved region that is present in all four types of FGFR (FGFR1-4) (Johnson and Williams, 1993; Mohammadi et al., 1997). The five zebrafish FGFR all fall into one of these receptor types, so all FGF signaling is likely affected in the inhibitor treated embryos.
In support for a role for FGF signaling in neurulation, FGFR-1 KO mice die before neurulation occurs. However, chimeric mice with a low level of FGFR1−/− cells have a number of neural tube defects including duplication of the spinal cord neural tube and spina bifida. No defects were found in the anterior neural tube (Deng et al., 1997). However, mice with a large percentage of cells lacking FGFR1 died, and so it is possible that this masked a role for FGF signaling in anterior neurulation.
The penetrance of neural tube defects was high only when relatively high concentrations of SU5402 were used (data not shown). The simplest possibility is that a low level of FGF signaling is sufficient for anterior neurulation. FGF signaling has a well-established role posteriorizing tissues during vertebrate development. Consistent with this, posterior tissues were more sensitive to FGF signaling defects than anterior tissues [This study and (Ota et al., 2009)]. Thus, a very high level of FGFR inhibition may be required to affect neurulation in the forebrain. Another possibility is that a FGFR relatively insensitive to SU5402 is involved in anterior neurulation. Zebrafish FGFR1a, FGFR1b, and FGFR4 all have one amino acid difference from the SU5402 binding site in human FGFR1 (Johnson and Williams, 1993; Mohammadi et al., 1997). Although these changes are all conservative, they could affect inhibitor binding.
Potential roles for FGF signaling in anterior neurulation
Research in a number of vertebrate systems, including zebrafish, indicates that FGF and Nodal signaling work coordinately to promote mesoderm development (Kiecker et al., 2016). Since anterior mesendoderm/mesoderm are required for anterior neurulation, the most likely cause of the NTD in the FGF signaling inhibitor treated embryos is the loss of mesoderm. Consistent with this, most embryos with open neural tubes had only a small amount of notochord present while embryos a closed neural tube had most of their notochord tissue or a full notochord. Statistical analysis found a significant correlation between notochord presence and a closed anterior neural tube. The expression patterns of fgf and fgfr genes are consistent with FGF signaling acting through the development of mesoderm. Several genes encoding FGF ligands are expressed in the anterior mesendoderm/mesoderm during the period when anterior neurulation is occurring, including fgf8a and fgf17, which were differentially expressed in our screen. fgf8a is expressed in the cephalic paraxial mesoderm, a tissue involved in promoting anterior neural tube closure, during gastrulation (Gonsar et al., 2016; Thisse and Thisse, 2004). fgf17 is expressed in the presumptive dorsal mesoderm at sphere stage (Reifers et al., 1998; Shimizu et al., 2006; Thisse and Thisse, 2004; Warga et al., 2013).
Using a similar approach, the requirement for Nodal signaling in anterior neurulation was mapped to up to late blastula stages (4.3 hpf) (Gonsar et al., 2016). In this study, inhibition of FGFR activity starting before the onset of gastrulation (6.0 hpf) caused anterior NTD. This suggests that the requirement for FGF signaling occurs in part after the requirement for Nodal signaling.
Work in zebrafish and other vertebrates suggests that FGF and Nodal signaling act in complex feedback loops. For instance, in zebrafish, the genes fgf3, fgf8, and fgf17b can be induced by ectopic activation of Nodal signaling and expression is lost in the anterior regions of embryos that lack Nodal signaling, suggesting FGF signaling is in part downstream of Nodal (Cao et al., 2004; Mathieu et al., 2004). Our previous studies suggested that both mesoderm and mesendoderm were involved in anterior neurulation (Gonsar et al., 2016). However, loss of FGF signaling affects only mesoderm, suggesting that a severe loss of mesoderm is sufficient to cause an open neural tube.
Although there is a great deal of evidence suggesting FGF signaling is required for anterior neurulation through its role in mesoderm development, we cannot rule out other roles for FGF signaling. The two fgf genes that were differentially expressed in our screen are not exclusively transcribed in the mesendoderm/mesoderm. fgf8a is expressed in the forebrain neural rod at ~5-9 somite stages as well as later in the neural tube while fgf17 is expressed in the presumptive telencephalon from 1-13 somite stages in zebrafish (Guo et al., 1999; Heisenberg et al., 1999; Jovelin et al., 2010; Lun and Brand, 1998; Reifers et al., 1998; Thisse and Thisse, 2004). The binding affinity of FGF17 for different FGFRs has not yet been tested. However, modular scanning in zebrafish demonstrated that FGF8 binds FGFR1 and FGFR4 in vivo with affinities appropriate with them being FGF8 receptors (Ries et al., 2009). Although not differentially expressed, fgfr1a and fgfr1b are both are expressed in all or almost all cells during cleavage stages up to early gastrulation. After this, expression becomes more restricted, but persists in the anterior neural plate during gastrulation (Ota et al., 2010; Ota et al., 2009; Rohner et al., 2009; Thisse et al., 2001). These data are consistent with fgf8a, fgf17, fgfr1a, and fgfr1b being part of the pathway that mediates mesendoderm/mesoderm to neuroectoderm communication or acting within the neuroectoderm to promote anterior neurulation.
Potential roles for other cellular interactions
Previous studies indicate the developing neural tube is disorganized in Nodal signaling deficient embryos with an open neural tube (Aquilina-Beck et al., 2007; Araya et al., 2014). The organization of the neural tube is mediated by adherens and tight junctions, which form in a step wise fashion. Our RNAseq data suggest transcriptional regulation plays a role in the regulation of adherens and tight junction genes only at the onset of gastrulation (shield stage). This is consistent with earlier studies on Nodal-deficient embryos. For instance, N-cadherin protein was decreased at the membrane of Nodal signaling-deficient embryos at tailbud and early somite stages, but mRNA levels were normal (Aquilina-Beck et al., 2007). Similarly, ZO1 (TJP1a), which is associated with both adherens and tight junctions, was expressed at normal levels in embryos treated with SB505124 at similar stages to our study (Araya et al., 2014).
There are several post-transcriptional regulatory processes that could be affected upon loss of Nodal signaling. These include incorrect apical-basal polarity of the neuroepithelium, negative regulation of adherens junctions by the canonical Wnt signaling pathway, and failure of newly generated neural cells to intercalate into the neuroepithelium. Our RNAseq data gives insight into only one of these. We found that expression of dkk1b, which encodes a Wnt signaling inhibitor, was lower in embryos with open neural tubes. This suggests that Wnt signaling could be increased when the neural tube is open. As Wnt signaling acts in competes with adherens junctions for the use of β-catenin (Nelson and Nusse, 2004), increased Wnt signaling could disrupt the integrity or number of adherens junctions and cause disorganization of the anterior neural tube.
Methods
All methods using animals were approved by the University of Minnesota IACUC.
Animal Care
Zebrafish (Danio rerio) were maintained in a fish facility with circulating water at 28.5 oC and a 14:10 light:dark cycle using standard methods (Westerfield, 2000). For this project, we used the wild-type strain Zebrafish Danio rerio (ZDR, Aquatica Tropical, Plant City, FL) and Tg (flh:EGFP)C161 fish (Gamse et al., 2003). Embryos were obtained by natural spawning of 2-10 adult fish. Embryonic stages were defined by morphology (Kimmel et al., 1995).
Pronase Treatment
Embryos used in the RNAseq experiments were pronase treated to remove their chorions before 2-(5-benzo [1,3] dioxol-5-yl-2-terbutyl-3H-imidazol-4-yl)-6-methylpyridine hydrochloride hydrate (SB505124) inhibitor treatment or before snap freezing in liquid nitrogen. A concentrated stock of previously frozen pronase (15 mg/mL, Roche) was activated by incubation at 37 °C for at least 30 minutes. To make a working solution, the pronase stock was diluted to a concentration of 2 mg/mL in embryo media (5 mM NaCl, 0.17 mM KCl, 0.33 mM MgSO4, 1 × 10−5% methylene blue in fish water) to make a total of 17 ml. Water overlying embryos in a 2% agarose-coated, 100 mm × 20 mm Petri dish was replaced the pronase solution. Embryos were incubated at 28.5 °C. After 4 minutes, embryos were swirled around the Petri dish using a plastic transfer pipet. The embryos were placed back into the 28.5 °C incubator and swirled every 30 seconds until approximately 75% of the embryos were out of their chorions.
The pronase reaction was stopped by pipetting the embryos into a 2% agarose-coated fish tank containing ~1 L embryo media. Embryos were transferred using a plastic pipet to 100 mm × 20 mm 2% agarose-lined Petri dishes containing at least 20 ml embryo media. Embryos with were gently agitated with forceps until the chorion disassociated. Intact embryos were maintained at 28.5 °C until they were at the appropriate stage for SB505124 treatment or snap freezing. Fire polished glass Sigma cote (Sigma-Aldrich) coated pipets were used to transfer dechorionated embryos into 2.0 mL Eppendorf tubes for snap freezing.
SB505124 Treatment
For RNA-sequencing experiments, SB505124 from Sigma-Aldrich was used at a concentration of 100 μM in 12 mL embryo media in a 15 mm × 60 mm Petri dish (DaCosta Byfield et al., 2004; Hagos et al., 2007). For RT-PCR and whole mount in situ hybridization, SB505124 from Tocris was used at a concentration of 20 μM, which caused a similar phenotype to 100 μM of the Sigma drug, in a total of 30 mL embryo media in a 100 mm × 20 mm Petri dish. For both SB505124 stocks, Dimethyl Sulfoxide (DMSO) was used to make the 10 mM stock solution of SB505124. SB505124 treatment/solution (12 mL for Sigma-Aldrich and 30 mL for Tocris, with a final concentration of 1% DMSO) was added to pooled groups of ZDR embryos at either sphere (4.0 hpf) or 30% epiboly (4.7 hpf) stages and left on until the embryos were snap frozen in liquid nitrogen or fixed in 4% paraformaldehyde. Control embryos were treated with 1% DMSO in embryo media and raised in parallel to inhibitor treated embryos. In addition, at least 20 embryos from each treatment group were left in SB505124 until 24 hpf and scored for pineal phenotype by whole mount in situ hybridization (WISH) to ensure that the embryos had the expected neural tube phenotype. Phenotypes at 24 hpf between both SB50124 stocks were similar at the concentrations used.
RNA Preparation
Whole embryos were treated with SB505124 starting at sphere or 30% epiboly stages until they were frozen with liquid nitrogen at the shield, tailbud, or 7 somite stage. Three samples for each treatment group at each stage were collected for a total of 18 samples for sequencing. 30-60 embryos were frozen per biological replicate, with a greater number of embryos in samples with earlier developmental stages or treated at sphere stage. Samples were stored at −80 °C until RNA was isolated using the Qiagen RNeasy Mini Kit. Samples were quantified using a NanoDrop Spectrophotometer to ensure at least 1μg of RNA was present and were sent on dry ice to the University of Minnesota Biomedical Genomics Center (UMGC, St. Paul, MN) for sequencing with an Illumina HiSeq 2500.
RNA-Sequencing
A fluorimetric RiboGreen assay was used to requantify the isolated RNA once received by the UMGC. For each sample, an RNA integrity number (RIN) was generated by capillary electrophoresis via the Agilent BioAnalyzer 2100. Only samples that had a RIN of 8 or more and a mass of greater than 1 μg were considered high quality and made into Illumina sequencing libraries through Illumina’s Truseq RNA Preparation v2 kit (RS-122-2001). Briefly, to make these libraries, 1 μg of RNA went through two rounds of purification for polyA containing mRNA using oligo-dT attached magnetic beads. mRNA was fragmented and then primed with random hexamers. RNA fragments were reverse transcribed into first strand cDNA, RNA template was removed, and a replacement strand was synthesized to make ds cDNA. AMPure XP beads were used to separate the ds cDNA from the reaction mix. cDNA ends were blunt ended and repaired. A single adenylate was added to the 3’ ends and multiple RNA adaptor indexes were ligated. DNA fragments with adapter indexes were PCR amplified. All libraries were validated, normalized, pooled, and size selected to approximately 200 bp using Caliper’s XT instrument before sequencing.
Libraries were clustered on the HiSeq 2500 at 10 pM through hybridization to a paired end flow cell. Next, the flow cell was sequenced using Illumina’s Rapid Run SBS chemistry. When read 1 was completed, a 7 base pair index read was performed. In order to produce the template for paired end read 2, library fragments were resynthesized in the reverse direction and sequenced from the opposite end of the read 1 fragment.
For each cycle of sequencing, a base call (.bcl) file was generated by Illumina Real Time Analysis (RTA) software. These base call files and run folders were exported to Minnesota Supercomputing Institute (MSI) servers. Primary analysis and de-multiplexing were performed using Illumina’s CASAVA software 1.8.2, which resulted in de-multiplexed FASTQ files. FASTQ files were released to our account for bioinformatic analysis.
Bioinformatics and Statistical Analysis
Reads from each sample were mapped to NCBI’s RefSeq database of mRNA for Danio rerio using NCBI’s megablast. After genes were identified, they were quantified resulting in counts. Counts for each gene were upper quartile normalized to obtain accurate expression levels. Normalized counts were fitted to a negative binomial distribution using DESeq v1.6.1 (Anders and Huber, 2012).
Differences between the counts for each gene in CNT and ONT embryos were determined using an analysis of deviance in DESeq (command: nbinomGLMTest) which generated a P-value (Anders and Huber, 2010; Anders and Huber, 2012). P-values were independently filtered to restrict for genes with a 50% or greater change in expression and at least 100 or more mean reads in either the open or closed neural tube sample at the time point being analyzed (shield, tailbud, or 7 somite stage). The Benjamini-Hochberg method was used to control the false discovery rate to 0.05 (Benjamini and Hochberg, 1995). Only mRNAs meeting all of these criteria were further analyzed.
Lists of differentially expressed transcripts were put into DAVID for analysis to get tissue expression as well as functional groups and pathways represented (Huang da et al., 2009a, 2009b). The Kyoto encyclopedia of genes and genomes (KEGG) pathways were accessed through our DAVID analysis to visualize pathways represented and then through the KEGG pathways website to visualize specific genes in our data in pathways of interest (Ogata et al., 1999).
Relative Reverse Transcription PCR
RNA-sequencing results were confirmed by relative reverse transcription polymerase chain reaction (RT-PCR). Transcripts chosen for RT-PCR were of specific interest to us as potential candidates in our model and covered genes found in each of the three time points. Each time point had a transcript with a RNAseq CNT/ONT expression ratio > 1 and another with a ratio < 1. We determined average expression of the transcripts relative to our two reference genes over three replicates with a 95% confidence interval. RNA was isolated from SB505124 treated embryos as previously described. Isolated RNA was reverse transcribed into cDNA through Qiagen’s Omniscript Reverse Transcription Kit. Briefly, 1-1.5 μg of RNA was incubated with dNTPs, random nonamers, and RNase inhibitor for 60 minutes at 37 °C. Resulting cDNA was diluted 1:10 and added to a mixture containing 2× Rotor-Green SYBR Green PCR Master Mix and primers (fgf8a forward 5’-CGACGTTTGTGCCAAGCTTAT-3’, fgf8a reverse 5’-TCTGCAGAGCCGTGTAGTTG-3’; hatn10 forward 5’-TGAAGACAGCAGAAGTCAATG-3’, hatn10 reverse 5’-CAGTAAACATGTCAGGCTAAATAA-3’, loopern4 forward 5’-TGAGCTGAAACTTTACAGACACAT-3’, loopern4 reverse 5’-AGACTTTGGTGTCTCCAGAATG-3’, dkk1b forward 5’-AAGAGTTCGTGTCCATCGCC-3’, dkk1b reverse 5’-GGCCCTCTTTTAGGACAGGC-3’, szl forward 5’-GATGCGTTTACCCAACCTGC-3’, szl reverse 5”-GCAGCTCTCCTTTACTGCCA-3’, pcdh8 forward 5’-GGAGGAGCTCAGAAACCTGG-3’, pcdh8 reverse 5’-GTGCGAGTGGCTGTAGAGTG-3’, cdh11 forward 5’-TAAGGACGAAATGGCCCACC-3’, cdh11 reverse 5’-GGTCATTCTGCTGAAGCCCT-3’, pcdh10b forward 5’-AACCTCAATGCTTCCACGCT-3’, pcdh10b reverse 5’-TGTCCACTCGGAAGGAGCTA-3’, tbx1 forward 5’-CAAGCTCAACGCCGAACAAA-3’, tbx1 reverse 5’-TCTGCCATTGGGTCCATTCC-3’). All tubes were put in the RotorGene3000 with an inactivation step of 5 minutes at 95 °C followed by 40 2-step cycles of denaturation for 5 seconds at 95 °C and annealing/extension for 15 seconds at 60 °C and lastly a melting curve analysis from 60 to 95 °C. Results were analyzed using delta/delta CT and the three independent replicates were normalized to the expressed repeat elements hatn10 and loopern4 (Vanhauwaert et al., 2014).
WISH
Embryos were assayed for expression of orthodenticle homobox 5 (otx5) (Gamse et al., 2002), collagen type 2 alpha1a (col2a1) (Yan et al., 1995), cathepsin L 1b (ctsl1b) (Vogel and Gerster, 1997), ndr2 (Sampath et al., 1998), oep (Zhang et al., 1998), eph receptor A4a (epha4a) (Xu, 1994), and early growth response 2b (egr2b) (Oxtoby and Jowett, 1993) mRNA using an established protocol (Thisse and Thisse, 2014). Briefly, whole embryos were incubated with digoxygenin (DIG)-labeled antisense RNA probe in hybridization mix (50% formamide) at 70 °C overnight. DIG was detected by an anti-DIG antibody covalently linked to Alkaline Phosphatase (AP). Cells expressing mRNA of interest were visualized after the addition of the AP substrates 4-nitro blue tetrazonium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) which is converted into a purple product by the removal of phosphate (Thisse and Thisse, 2014).
FGFR Inhibitor Treatments
The FGFR inhibitors SU5402, PD161570, and PD173074 (Tocris) were dissolved in DMSO to make a concentrated stock solution. The stock solution was diluted with embryo media and DMSO to obtain a working solution of SU5402 (6.75-675 μM-Supplemental Table 2), PD161570 or PD173074 (100 μM to produce Class II embryos and 200-300 μM to produce Class IV embryos) and either 1 or 2% DMSO. Embryos were maintained in embryo media at 28.5 °C until treatment. Once desired treatment stage was reached, embryo media was removed and replaced with the appropriate working solution. Control embryos were treated and raised in embryo media with 1% or 2% DMSO. Embryos were raised at 28.5 °C and then analyzed by light microscopy and WISH.
Statistical Analysis
Embryos were treated with SU5402 at time points ranging from mid blastula to mid gastrula stages and scored for an open or closed anterior neural tube were analyzed with a 2 × 6 contingency table. The cells in the first dimension corresponded to the neural tube phenotype and in the second dimension to the stage at initiation of SU5402 treatment. A two-tailed Fisher’s exact test was used to determine association between neural tube closure and timing of SU5402 treatment. Sample size was considered to be adequate when a significant association (P < 0.05) was observed.
The relationship between FGF signaling deficiency effects on mesendodermal/mesodermal tissue presence were tested by assaying SU5402-treated embryos for presence of hatching gland and notochord using WISH. Individual tissue presence, notochord or hatching gland, was scored as follows: 0 equaled no tissue present, 1 equaled some tissue present, 2 equaled most tissue present, and 3 equaled full tissue present. Separate 2 × 4 contingency tables with the first dimension corresponding to neural tube closure and the second to tissue presence scores were analyzed by a two-tailed Fisher’s exact test to determine if there was a significant association between tissue presence and neural tube closure. All Fisher’s exact tests were performed using R version 3.1.2 software.
Imaging
Embryos were imaged using a Nikon Eclipse 80i microscope with a SPOT Insight Fire Wire camera. Live embryos were mounted in methylcellulose on glass depression slides. Fixed embryos were mounted in 100% glycerol on glass coverslips. Images were processed using Adobe InDesign CS6 and image clarity adjustments were made using Adobe Photoshop CS6 (Adobe Systems Inc.).
Supplementary Material
Supplemental Figure 1. Embryos treated with SU5402 have normal hatching gland tissue. SU5402 treated embryos were assayed for cstl1b expression to determine amount of hatching gland present and the pineal marker otx5 to determine whether the forebrain neural tube was open or closed. All SU5402 treatments resulted in normal hatching glands (n= 88, 30% epiboly; n= 97, 50% epiboly, n=147, shield, n= 130, 70% epiboly, and n= 108, DMSO control) even when the anterior neural tube was open. Frontal views with anterior to the top. Scale bar: 100 μm.
Acknowledgements
The authors would like to thank Jenna Ruzich, Michael Schoenenberger, Abbygail Coyle, and Jake Billings for technical assistance. The University of Minnesota Genomics Core, especially Aaron Becker, helped with the RNA-sequencing.
Funding
This work was sponsored in part by National Institutes of Health [R15 HD068176-01A1 & 2R15HD068176-02 to JOL] and by a University of Minnesota Grant in-Aid of Research, Artistry, and Scholarship (JOL).
Footnotes
Data Availability: RNA sequencing results are available on the GEO site https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=azqnqagexzajhqf&acc=GSE92440.
The authors have no competing interests.
References Cited
- Agius PE, Piccolo S, De Robertis EM. 1999. [The head inducer Cerberus in a multivalent extracellular inhibitor]. J Soc Biol 193: 347–354. [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. 1999. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol 9: 1147–1157. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2012. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL). [Google Scholar]
- Aquilina-Beck A, Ilagan K, Liu Q, Liang JO. 2007. Nodal signaling is required for closure of the anterior neural tube in zebrafish. BMC Dev Biol 7: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C, Tawk M, Girdler GC, Costa M, Carmona-Fontaine C, Clarke JD. 2014. Mesoderm is required for coordinated cell movements within zebrafish neural plate in vivo. Neural Dev 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC, Bhattacharya S. 2001. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29: 469–474. [DOI] [PubMed] [Google Scholar]
- Batley BL, Doherty AM, Hamby JM, Lu GH, Keller P, Dahring TK, Hwang O, Crickard K, Panek RL. 1998. Inhibition of FGF-1 receptor tyrosine kinase activity by PD 161570, a new protein-tyrosine kinase inhibitor. Life Sci 62: 143–150. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57. [Google Scholar]
- Cai C, Shi O. 2014. Genetic evidence in planar cell polarity signaling pathway in human neural tube defects. Front Med 8: 68–78. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zhao J, Sun Z, Zhao Z, Postlethwait J, Meng A. 2004. fgf17b, a novel member of Fgf family, helps patterning zebrafish embryos. Dev Biol 271: 130–143. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. 2006. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas JF, Schoenwolf GC. 2001. Towards a cellular and molecular understanding of neurulation. Dev Dyn 221: 117–145. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. 2010. Genetics and development of neural tube defects. J Pathol 220: 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene NDE, Murdoch JN. 2003. The genetic basis of mammalian neurulation. Nature Reviews Genetics 4: 784–793. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Stanier P, Greene ND. 2013. Neural tube defects: recent advances, unsolved questions, and controversies. Lancet Neurol 12: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCosta Byfield S, Major C, Laping NJ, Roberts AB. 2004. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol 65: 744–752. [DOI] [PubMed] [Google Scholar]
- David NB, Rosa FM. 2001. Cell autonomous commitment to an endodermal fate and behaviour by activation of Nodal signalling. Development 128: 3937–3947. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. 2009. Spemann’s organizer and the self-regulation of embryonic fields. Mech Dev 126: 925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Bedford M, Li C, Xu X, Yang X, Dunmore J, Leder P. 1997. Fibroblast growth factor receptor-1 (FGFR-1) is essential for normal neural tube and limb development. Dev Biol 185: 42–54. [DOI] [PubMed] [Google Scholar]
- Detrait ER, George TM, Etchevers HC, Gilbert JR, Vekemans M, Speer MC. 2005. Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol Teratol 27: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmeis T, Mourrain P, Saint-Etienne L, Fischer N, Aanstad P, Clark M, Strahle U, Rosa F. 2001. A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev 15: 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamse JT, Shen YC, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JO. 2002. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat Genet 30: 117–121. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. 2003. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development 130: 1059–1068. [DOI] [PubMed] [Google Scholar]
- Gonsar N, Coughlin A, Clay-Wright JA, Borg BR, Kindt LM, Liang JO. 2016. Temporal and spatial requirements for Nodal-induced anterior mesendoderm and mesoderm in anterior neurulation. Genesis 54: 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109–1113. [DOI] [PubMed] [Google Scholar]
- Greene ND, Copp AJ. 2014. Neural tube defects. Annu Rev Neurosci 37: 221–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinblat Y, Gamse J, Patel M, Sive H. 1998. Determination of the zebrafish forebrain: induction and patterning. Development 125: 4403–4416. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Talbot WS, Schier AF. 2000. Nodal signaling patterns the organizer. Development 127: 921–932. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. 1999. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron 24: 555–566. [DOI] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST. 2007. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagos EG, Fan X, Dougan ST. 2007. The role of maternal Activin-like signals in zebrafish embryos. Dev Biol 309: 245–258. [DOI] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. 2010. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res A Clin Mol Teratol 88: 653–669. [DOI] [PubMed] [Google Scholar]
- Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, Smith JC. 2013. Identification of the zebrafish maternal and paternal transcriptomes. Development 140: 2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Itoh M, Yamanaka Y, Yamashita S, Shimizu T, Solnica-Krezel L, Hibi M, Hirano T. 2000. Zebrafish Dkk1 functions in forebrain specification and axial mesendoderm formation. Dev Biol 217: 138–152. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Brennan C, Wilson SW. 1999. Zebrafish aussicht mutant embryos exhibit widespread overexpression of ace (fgf8) and coincident defects in CNS development. Development 126: 2129–2140. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99: 485–497. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang K, Lui WY. 2016. Nectins and nectin-like molecules (Necls): Recent findings and their role and regulation in spermatogenesis. Semin Cell Dev Biol [DOI] [PubMed] [Google Scholar]
- Huang Y, Roelink H, McKnight GS. 2002. Protein kinase A deficiency causes axially localized neural tube defects in mice. J Biol Chem 277: 19889–19896. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Williams LT. 1993. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res 60: 1–41. [DOI] [PubMed] [Google Scholar]
- Jovelin R, Yan YL, He X, Catchen J, Amores A, Canestro C, Yokoi H, Postlethwait JH. 2010. Evolution of developmental regulation in the vertebrate FgfD subfamily. J Exp Zool B Mol Dev Evol 314: 33–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C, Bates T, Bell E. 2016. Molecular specification of germ layers in vertebrate embryos. Cell Mol Life Sci 73: 923–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, Yelon D, Thisse B, Stainier DY. 2001. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev 15: 1493–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of Embryonic Development of the Zebrafish. Developmental Dynamics 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Komiya Y, Habas R. 2008. Wnt signal transduction pathways. Organogenesis 4: 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. 1998. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94: 325–337. [DOI] [PubMed] [Google Scholar]
- Lakkis MM, Golden JA, O’Shea KS, Epstein JA. 1999. Neurofibromin deficiency in mice causes exencephaly and is a modifier for Splotch neural tube defects. Dev Biol 212: 80–92. [DOI] [PubMed] [Google Scholar]
- Lemay P, Guyot MC, Tremblay E, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, De Marco P, Merello E, Massicotte C, Desilets V, Michaud JL, Rouleau GA, Capra V, Kibar Z. 2015. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J Med Genet 52: 493–497. [DOI] [PubMed] [Google Scholar]
- Li F, Chong ZZ, Maiese K. 2005. Vital elements of the Wnt-Frizzled signaling pathway in the nervous system. Curr Neurovasc Res 2: 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. 2000. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development 127: 5101–5112. [DOI] [PubMed] [Google Scholar]
- Londin ER, Niemiec J, Sirotkin HI. 2005. Chordin, FGF signaling, and mesodermal factors cooperate in zebrafish neural induction. Dev Biol 279: 1–19. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. 2004. Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mech Dev 121: 1189–1197. [DOI] [PubMed] [Google Scholar]
- Lun K, Brand M. 1998. A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 125: 3049–3062. [DOI] [PubMed] [Google Scholar]
- Luo H, Liu X, Wang F, Huang Q, Shen S, Wang L, Xu G, Sun X, Kong H, Gu M, Chen S, Chen Z, Wang Z. 2005. Disruption of palladin results in neural tube closure defects in mice. Mol Cell Neurosci 29: 507–515. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Griffin K, Herbomel P, Dickmeis T, Strahle U, Kimelman D, Rosa FM, Peyrieras N. 2004. Nodal and Fgf pathways interact through a positive regulatory loop and synergize to maintain mesodermal cell populations. Development 131: 629–641. [DOI] [PubMed] [Google Scholar]
- Milenkovic L, Goodrich LV, Higgins KM, Scott MP. 1999. Mouse patched1 controls body size determination and limb patterning. Development 126: 4431–4440. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, Green D, Schlessinger J, Hubbard SR. 1998. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J 17: 5896–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. 1997. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276: 955–960. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303: 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. 2002. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet 31: 84–88. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S, Tonou-Fujimori N, Nakayama Y, Ito Y, Kawamura A, Yamasu K. 2010. FGF receptor gene expression and its regulation by FGF signaling during early zebrafish development. Genesis 48: 707–716. [DOI] [PubMed] [Google Scholar]
- Ota S, Tonou-Fujimori N, Yamasu K. 2009. The roles of the FGF signal in zebrafish embryos analyzed using constitutive activation and dominant-negative suppression of different FGF receptors. Mech Dev 126: 1–17. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. 1993. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res 21: 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Wang C, Wang B. 2009. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol 326: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JW, Raible DW. 2004. Signals derived from the underlying mesoderm are dispensable for zebrafish neural crest induction. Dev Biol 276: 16–30. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. 2001. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev 107: 105–117. [DOI] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. 1998. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125: 2381–2395. [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Bakkers J, Kramer C, Hammerschmidt M. 2004. Fgf signaling induces posterior neuroectoderm independently of Bmp signaling inhibition. Dev Dyn 231: 750–757. [DOI] [PubMed] [Google Scholar]
- Ries J, Yu SR, Burkhardt M, Brand M, Schwille P. 2009. Modular scanning FCS quantifies receptor-ligand interactions in living multicellular organisms. Nat Methods 6: 643–645. [DOI] [PubMed] [Google Scholar]
- Robertson EJ. 2014. Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin Cell Dev Biol 32: 73–79. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Allegrucci C, Alberio R. 2012. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS One 7: e49079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, Schier AF. 2011. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol 27: 377–407. [DOI] [PubMed] [Google Scholar]
- Rohner N, Bercsenyi M, Orban L, Kolanczyk ME, Linke D, Brand M, Nusslein-Volhard C, Harris MP. 2009. Duplication of fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr Biol 19: 1642–1647. [DOI] [PubMed] [Google Scholar]
- Roy NM, Sagerstrom CG. 2004. An early Fgf signal required for gene expression in the zebrafish hindbrain primordium. Brain Res Dev Brain Res 148: 27–42. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kuroiwa A, Takeda H. 2001. A novel sox gene, 226D7, acts downstream of Nodal signaling to specify endoderm precursors in zebrafish. Mech Dev 107: 25–38. [DOI] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. 1998. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature 395: 185–189. [DOI] [PubMed] [Google Scholar]
- Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. 2009. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn 238: 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifley ET, Kenny AP, Rankin SA, Zorn AM. 2012. Prolonged FGF signaling is necessary for lung and liver induction in Xenopus. BMC Dev Biol 12: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Yabusaki M, Niwa H, Yokoi H, Hatta K, Kobayashi D, Takeda H. 2008. Maternal-zygotic medaka mutants for fgfr1 reveal its essential role in the migration of the axial mesoderm but not the lateral mesoderm. Development 135: 281–290. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Hibi M. 2006. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development 133: 4709–4719. [DOI] [PubMed] [Google Scholar]
- Shum AS, Copp AJ. 1996. Regional differences in morphogenesis of the neuroepithelium suggest multiple mechanisms of spinal neurulation in the mouse. Anat Embryol (Berl) 194: 65–73. [DOI] [PubMed] [Google Scholar]
- Smith JL, Schoenwolf GC. 1997. Neurulation: coming to closure. Trends Neurosci 20: 510–517. [DOI] [PubMed] [Google Scholar]
- Sun L, Tran N, Liang C, Tang F, Rice A, Schreck R, Waltz K, Shawver LK, McMahon G, Tang C. 1999. Design, synthesis, and evaluations of substituted 3-[(3- or 4-carboxyethylpyrrol-2-yl)methylidenyl]indolin-2-ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. Journal of medicinal chemistry 42: 5120–5130. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Kimelman D. 2006. The regulation of mesodermal progenitor cell commitment to somitogenesis subdivides the zebrafish body musculature into distinct domains. Genes Dev 20: 1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. 2000. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev 95: 133–145. [DOI] [PubMed] [Google Scholar]
- Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD. 2007. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature 446: 797–800. [DOI] [PubMed] [Google Scholar]
- Thisse B, Pfumio S, Furthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. 2001. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission. [Google Scholar]
- Thisse B, Thisse C. 2004. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. [Google Scholar]
- Thisse B, Thisse C. 2014. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol Biol 1211: 53–67. [DOI] [PubMed] [Google Scholar]
- Vanhauwaert S, Van Peer G, Rihani A, Janssens E, Rondou P, Lefever S, De Paepe A, Coucke PJ, Speleman F, Vandesompele J, Willaert A. 2014. Expressed repeat elements improve RT-qPCR normalization across a wide range of zebrafish gene expression studies. PLoS One 9: e109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterlund L, Jiao H, Unneberg P, Hovatta O, Kere J. 2011. The zebrafish transcriptome during early development. BMC Dev Biol 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel AM, Gerster T. 1997. Expression of a zebrafish cathepsin L gene in anterior mesendoderm and hatching gland. Deve Genes Evol 206: 477–479. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Harland RM. 2002. Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development 129: 5815–5825. [DOI] [PubMed] [Google Scholar]
- Warga RM, Mueller RL, Ho RK, Kane DA. 2013. Zebrafish Tbx16 regulates intermediate mesoderm cell fate by attenuating Fgf activity. Dev Biol 383: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M 2000. The zebrafish book.
- Xu W, Baribault H, Adamson ED. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development 125: 327–337. [DOI] [PubMed] [Google Scholar]
- Yan YL, Hatta K, Riggleman B, Postlethwait JH. 1995. Expression of a type II collagen gene in the zebrafish embryonic axis. Dev Dyn 203: 363–376. [DOI] [PubMed] [Google Scholar]
- Yan YT, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF, Shen MM. 1999. Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev 13: 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybot-Gonzalez P, Gaston-Massuet C, Girdler G, Klingensmith J, Arkell R, Greene ND, Copp AJ. 2007. Neural plate morphogenesis during mouse neurulation is regulated by antagonism of Bmp signalling. Development 134: 3203–3211. [DOI] [PubMed] [Google Scholar]
- Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, Yuan S, Prabhakar NR, Herrup K, Conlon RA, Hoit BD, Watanabe M, Yang YC. 2002. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A 99: 10488–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaganjor I, Sekkarie A, Tsang BL, Williams J, Razzaghi H, Mulinare J, Sniezek JE, Cannon MJ, Rosenthal J. 2016. Describing the Prevalence of Neural Tube Defects Worldwide: A Systematic Literature Review. PLoS One 11: e0151586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. 1998. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell 92: 241–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Embryos treated with SU5402 have normal hatching gland tissue. SU5402 treated embryos were assayed for cstl1b expression to determine amount of hatching gland present and the pineal marker otx5 to determine whether the forebrain neural tube was open or closed. All SU5402 treatments resulted in normal hatching glands (n= 88, 30% epiboly; n= 97, 50% epiboly, n=147, shield, n= 130, 70% epiboly, and n= 108, DMSO control) even when the anterior neural tube was open. Frontal views with anterior to the top. Scale bar: 100 μm.


