Abstract
Promoter is a fundamental DNA element located around the transcription start site (TSS) and could regulate gene transcription. Promoter recognition is of great significance in determining transcription units, studying gene structure, analyzing gene regulation mechanisms, and annotating gene functional information. Many models have already been proposed to predict promoters. However, the performances of these methods still need to be improved. In this work, we combined pseudo k-tuple nucleotide composition (PseKNC) with position-correlation scoring function (PCSF) to formulate promoter sequences of Homo sapiens (H. sapiens), Drosophila melanogaster (D. melanogaster), Caenorhabditis elegans (C. elegans), Bacillus subtilis (B. subtilis), and Escherichia coli (E. coli). Minimum Redundancy Maximum Relevance (mRMR) algorithm and increment feature selection strategy were then adopted to find out optimal feature subsets. Support vector machine (SVM) was used to distinguish between promoters and non-promoters. In the 10-fold cross-validation test, accuracies of 93.3%, 93.9%, 95.7%, 95.2%, and 93.1% were obtained for H. sapiens, D. melanogaster, C. elegans, B. subtilis, and E. coli, with the areas under receiver operating curves (AUCs) of 0.974, 0.975, 0.981, 0.988, and 0.976, respectively. Comparative results demonstrated that our method outperforms existing methods for identifying promoters. An online web server was established that can be freely accessed (http://lin-group.cn/server/iProEP/).
Keywords: promoter, pseudo k-tuple nucleotide composition, position-correlation scoring function, feature selection, web server
Introduction
In a genome, promoters are important regions of DNA that locate near the transcription start sites (TSSs) of genes.1 They are essentially nucleotide sequences of approximately extending dozens to hundreds base pairs upstream and downstream of the TSS. They always serve as regulatory elements for the assembly of transcription machinery, especially combining with RNA polymerase2 for promoting accurate initiation of transcription. Additionally, evidence has proved that promoters play crucial roles in the regulation of gene expression, such as alternative splicing, stability of transcripts, mRNA localization, and translation.3 The identification of promoters in a gene is an important part of the recognition of a gene’s complete structure. Hence, the mapping of promoters to genome is usually the first step in unraveling the mechanisms of gene transcriptional and expressional regulation. Therefore, research on promoter prediction is full of significance and deserves to be pushed forward.
DNA elements in promoters are different between eukaryotes and prokaryotes. In eukaryotes, most protein-coding genes and some nuclear small RNAs have binding sites for RNA polymerase II. The core region of RNA polymerase II-dependent promoters usually contains several regulatory units: the TATA element, which is located 25 bp upstream of the TSS; the initiator; and the downstream promoter element (DPE), usually located 30 bp downstream of the TSS.4 In prokaryotes, most genes are regulated by the σ70 promoter, which contains three basic elements: the Pribnow Box with the consensus sequence 5′-TATAAT-3′ located 10 bp upstream of the TSS, the −35 region with the consensus sequence 5′-TTGACA-3′ located 35 bp upstream of the TSS, and the initiator adjacent to the TSS.5, 6 Distinct gene-regulatory mechanisms and sequence compositions among species promote us to use different methods to identify promoters in their genomes.7, 8
With the development of high-throughput sequencing technology, increasing genomes need to be annotated. It is costly, laborious, and time consuming to use experimental methods to characterize promoters, however, which promotes the development of the computational methods in promoter identification. There have been many attempts to predict promoters in different species. Some models were based on the principle of sequence similarity, and others converted the original sequences into numeric sequences and then adopted machine learning approaches to perform recognition. The latter extracted features according to various promoter properties, such as CpG content,9 free energy,10 consensus sequence,11 and global descriptor,10 and built the prediction programs based on machine learning approaches, such as Fisher’s linear discriminant,10 decision tree,12 support vector machine (SVM),13 Hidden Markov Model,11 neural network,14 pattern-based nearest neighbor search approach,15 and so on. Recently, deep learning has been used to grasp complex promoter sequence characteristics16, 17 and related bioinformatics identification problems.18, 19, 20, 21, 22 Although existing algorithms have exhibited encouraging performance, most of those predictors focused on only one species, and there is still space for prediction performance improvement.
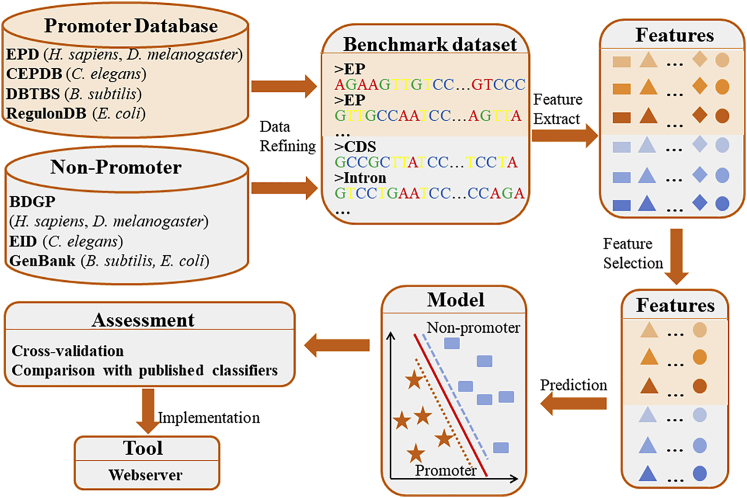
In this study, according to the steps shown in Figure 1, we developed an effective and powerful computational promoter prediction program for eukaryote and prokaryote species. We firstly collected promoter and non-promoter sequences in five species to construct the reliable benchmark datasets. The features extracted from the primary sequences were filtered according to the ability of distinguishing promoters from non-promoters by using feature selection technique. Subsequently, the optimal features were inputed into the SVM to train, test, and build models. Finally, based on the proposed model, we established a user-friendly web server iProEP, which can be freely accessed at http://lin-group.cn/server/iProEP/.
Figure 1.
A Flowchart to Outline the Promoter Prediction Program Construction
Results
Optimization of Three PseKNC-Related Parameters
As indicated in the PseKNC section (see Materials and Methods), three parameters, k, λ, and ω, must be determined when using PseKNC to formulate promoters and non-promoters. In the PseKNC, the k and λ describe the short-range and long-range sequence-order effect, respectively, and ω is the weight factor to adjust the ratio of the two effects. In this work, the optimal values of the three parameters for five species can be obtained by searching the following scopes:
| (Equation 1) |
For each species, the performances of 1,500 (5 × 30 × 10) different combinations of three parameters were examined to obtain their optimal combination that could produce best accuracy. Thus, we constructed 1,500 SVM classifiers based on 5-fold cross-validation for each species. The optimal combinations of the three parameters for five species were reported in Table 1.
Table 1.
The Optimal Values of Three PseKNC Parameters for Five Species
| Kingdom | Species | k | λ | ω | ACC (%) |
|---|---|---|---|---|---|
| Eukaryotes | H. sapiens | 4 | 24 | 0.1 | 90.9 |
| D. melanogaster | 5 | 9 | 0.1 | 89.5 | |
| C. elegans | 4 | 22 | 0.1 | 81.4 | |
| Prokaryotes | B. subtilis | 4 | 12 | 0.2 | 83.8 |
| E. coli | 4 | 12 | 0.1 | 80.7 |
The Ultimate Five Promoter Classifiers
By combining PseKNC with position-correlation scoring function (PCSF), promoter and non-promoter samples can be formulated by (4k + 6λ + n) dimension features. In the 4k + 6λ dimension PseKNC features, 4k reflects the DNA short-range correlation information, and 6λ describes the long-range correlation information. The position information is characterized by n dimension PCSF (see Materials and Methods). When incorporating these features into a prediction model, redundant information or noise might influence the performance of the model. Therefore, Minimum Redundancy Maximum Relevance (mRMR) combined with the increment feature selection (IFS) process was adopted to eliminate these unrelated features for improving the accuracy and robustness of promoter recognition models.
Ultimately, by constructing a great number of SVM-based models and comparing these models’ performance using 5-fold cross-validation, the optimal feature subsets for five species were screened out and shown in Table 2. It is obvious that the accuracies were indeed improved after removing noise features. It was also noted that the feature dimensions for C. elegans, B. subtilis, and E. coli were dramatically decreased after feature selection. However, only 13 and 204 features were excluded for H. sapiens and D. melanogaster. The reason for these phenomena may be that promoter sequences of H. sapiens and D. melanogaster are much more complex than those of the other three species.
Table 2.
The Feature Numbers and Accuracies for Five Species before and after mRMR Feature Selection
| Kingdom | Species | Original Features |
Optimal Features |
||
|---|---|---|---|---|---|
| Feature Number | ACC (%) | Feature Number | ACC (%) | ||
| Eukaryotes | H. sapiens | 423 | 93.4 | 410 | 93.5 |
| D. melanogaster | 1,097 | 93.3 | 893 | 93.8 | |
| C. elegans | 405 | 94.4 | 65 | 95.6 | |
| Prokaryotes | B. subtilis | 345 | 94.0 | 55 | 95.5 |
| E. coli | 345 | 92.1 | 44 | 93.2 | |
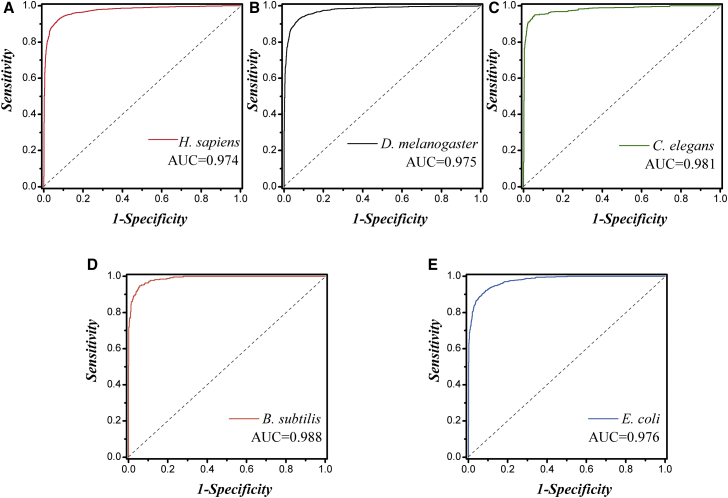
After determining the optimal feature subsets, for convenience in subsequent comparisons, the 10-fold cross-validation was applied to seek the best SVM-related parameters (c and γ) and to evaluate those models. For H. sapiens, D. melanogaster, C. elegans, B. subtilis, and E. coli, the optimal values of c and γ are 2 and 2−3, 2 and 2−2, 25 and 2−1, 25 and 2−7, and 2−1 and 2−1, respectively. The detailed results were listed in Table 3. In addition, receiver operating characteristics (ROC) curves were also plotted in Figure 2 to visually show the prediction capability of our model on discrimination between promoters and non-promoters.
Table 3.
The Results for Five Species by Using 10-Fold Cross-Validation
| Kingdom | Species | ACC (%) | Sn (%) | Sp (%) | AUC |
|---|---|---|---|---|---|
| Eukaryotes | H. sapiens | 93.3 | 92.3 | 92.7 | 0.974 |
| D. melanogaster | 93.9 | 92.6 | 92.6 | 0.975 | |
| C. elegans | 95.7 | 95.0 | 94.4 | 0.981 | |
| Prokaryotes | B. subtilis | 95.2 | 94.8 | 94.3 | 0.988 |
| E. coli | 93.1 | 92.2 | 91.2 | 0.976 |
Figure 2.
Evaluating the iProEP by Using ROC Curve
ROC curves for promoter prediction in (A) H. sapiens, (B) D. melanogaster, (C) C. elegans, (D) B. subtilis, and (E) E. coli.
Comparison with Existing Promoter Classifiers
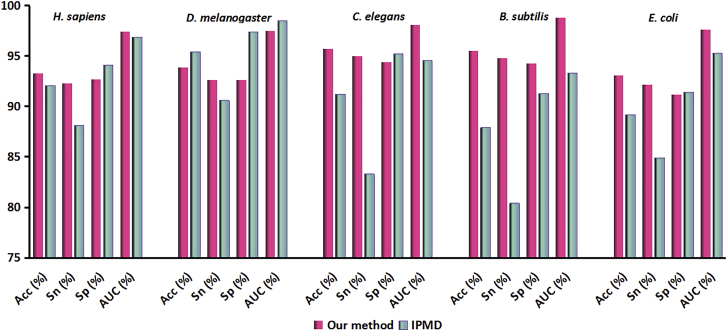
Comparison with other existing methods is an important strategy to highlight the merits of proposed models. Currently, several computational methods have been developed for eukaryote and prokaryote promoter prediction.17, 23 To provide a fair comparison of the same data, only a method called IPMD24 was used to make comparisons, because the same benchmark datasets and same cross-validation rule were used in both works. Furthermore, comparison in the paper24 has demonstrated that IPMD is superior to other existing predictors, such as NNPP2.2, McPromoter. The IPMD is a hybrid method that combined PCSF and increment of diversity (ID) with the modified Mahalanobis Discriminant. Figure 3 recorded the results obtained by our proposed method and IPMD. The results show that our model is superior to the IPMD model, especially for C. elegans, B. subtilis, and E. coli.
Figure 3.
The Comparison between Our Proposed Method with IPMD Classifiers in 10-Fold Cross-Validation
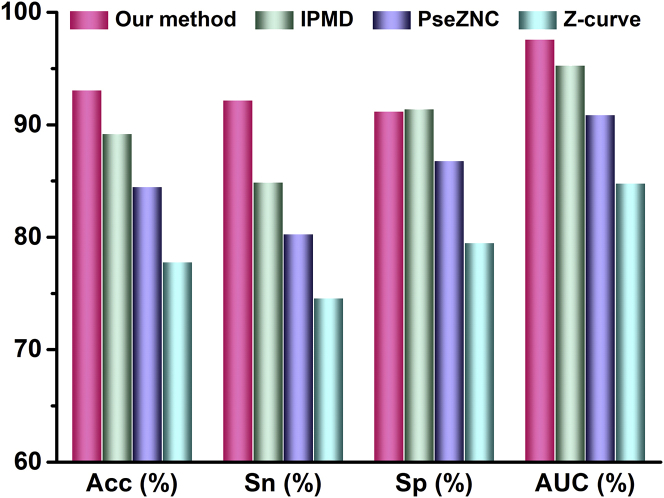
Moreover, multi-window Z-curve25 and PseZNC26 have been proposed as feature extraction approaches for σ70 promoter prediction in E. coli. Based on the same E. coli data, multi-window Z-curve was re-evaluated in Lin et al.26 Its overall accuracy is only 77.81% with the area under receiver operating curve (AUC) of 0.8480, which is lower than those of our proposed method. PseZNC is a feature extraction technique that combines multi-window Z-curve with PseKNC. The accuracy of the PseZNC-based method is also lower than our method. Detailed comparison was exhibited in Figure 4. Z-curve theory has been successfully applied in prokaryotic gene prediction because of the characteristics of period-3 in codon. However, promoter sequence cannot code amino acids and dose not obey the codon rule. This is why the two Z-curve-based methods cannot produce better results on promoter prediction.
Figure 4.
The Prediction Results of Four Methods on the Same E. Coli σ70 Promoter Data
Recently, two predictors called iPromoter-2L27 and MULTiPly28 were also designed for E. coli promoter prediction. We could make a raw comparison because the benchmark data in these studies were all derived from RegulonDB. Both predictors could provide multi-layer prediction for recognizing promoters and their subtypes. The former was based on multi-window-based PseKNC and Random Forest, which produced the accuracy (ACC), sensitivity (Sn), and specificity (Sp) of 81.68%, 79.20%, and 84.16%, respectively. The latter obtained the related three indexes of 86.92%, 87.27%, and 86.57% by a SVM-based model. It was found that our proposed model yielded ACC, Sn, and Sp of 93.1%, 92.2%, and 91.2%, respectively (Table 3), which are superior to the two predictors.
Cross-Species Evaluation
Cross-species evaluation on eukaryote and prokaryote was performed to assess the generalization ability of the proposed method. It should be noted that because of the different sequence structure, composition, and regulatory mechanism between eukaryote and prokaryote, the following experiments were performed. We first evaluated the H. sapiens-based model on D. melanogaster and C. elegans data. Results (Table 4) showed that the accuracies are only 77.10% and 66.63% for the two test datasets. Subsequently, we investigated the prediction performances of the D. melanogaster-based model on H. sapiens and C. elegans data. Only 68.41% of H. sapiens sequences and 65.68% of C. elegans sequences can be correctly identified. Finally, we performed similar examinations and obtained similar results on the models from C. elegans, B. subtilis, and E. coli. The unsatisfactory results are mainly due to the species-specificity property of promoter sequences.
Table 4.
The Results for Cross-Species Examination
| Kingdom | Model Training | Model Test | ACC (%) |
|---|---|---|---|
| Eukaryotes | H. sapiens | D. melanogaster | 77.19 |
| C. elegans | 66.63 | ||
| D. melanogaster | H. sapiens | 68.41 | |
| C. elegans | 65.68 | ||
| C. elegans | H. sapiens | 66.57 | |
| D. melanogaster | 69.58 | ||
| Prokaryotes | B. subtilis | E. coli | 75.95 |
| E. coli | B. subtilis | 80.92 |
Web Server and Tutorial
A user-friendly and publicly accessible web server could provide convenience for researchers.29, 30, 31 Thus, based on our proposed method, we established a powerful web server called iProEP, by which researchers can identify promoters by uploading DNA sequences. A step-by-step guide on how to use the web server is given as follows:
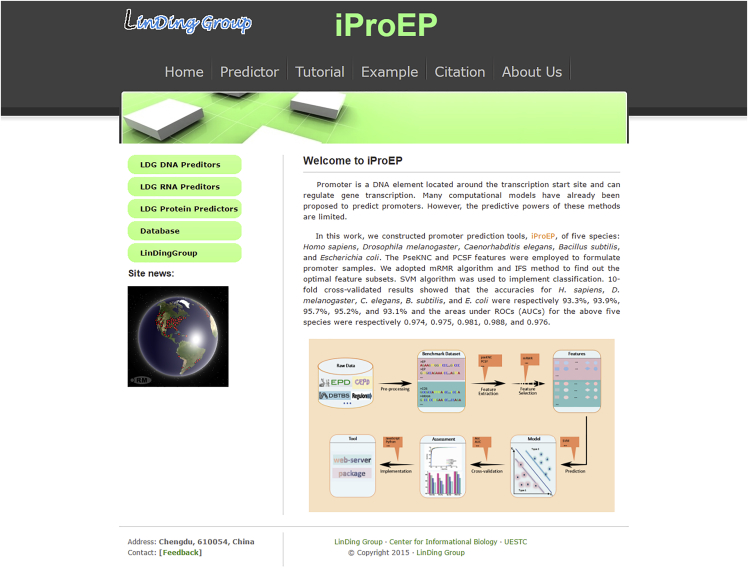
Step 1. Click on the web address http://lin-group.cn/server/iProEP/ and the user will see the brief summary about iProEP (Figure 5).
Step 2. Click on the “Predictor” on the navigation bar, then choose a suitable species and input the query DNA sequences into the input box for prediction. It should be noted that the sequences must be FASTA format with the length of >300 bp for eukaryote and >81 bp for prokaryote. Click on the “example” button below the input box to see the sample sequence in the FASTA format.
Step 3. Click on the “submit” button to obtain the predicted result. If the sequence is longer than 300 or 81 bp, the predictor will scan the sequence using the 300- or 81-bp window with the step of 1 bp for eukaryote or prokaryote, respectively. The result for each subsequence will be displayed on the result page.
Figure 5.
The Homepage of the iProEP Web Server
Available at http://lin-group.cn/server/iProEP/.
Discussion
Computationally identifying promoters has attracted scholars’ attention for many years, and many encouraging results were obtained. However, it is still a challenging topic in bioinformatics.17 In this work, we proposed a new feature extraction technique that combines PseKNC with PCSF for improving prediction ACC. A series of examinations demonstrated that our proposed method can distinguish promoter from non-promoter sequences with good performance. Thus, we established a predictor iProEP for providing convenience to scholars.
In the future work, many more promoters derived from other species will be collected for species-specific promoter prediction.17, 32 Moreover, although the combination of PseKNC and PCSF worked well in this study, new feature extraction techniques should be developed to further improve the performance of promoter prediction. Finally, with accumulation of more and more data and the development of a deep learning technique in many biological problems,17, 21, 33, 34, 35 it is suitable to identify promoters by using a deep learning technique.
Materials and Methods
Benchmark Dataset
A key step for constructing a powerful and robust prediction model is to construct an objective and strict benchmark dataset. In this work, we established five benchmark datasets including promoter and non-promoter sequences for five species (Table 5).
Table 5.
The Detail Information of the Training Datasets for Five Species
| Kingdom | Species | Promoter | Non-promoter |
Location | |
|---|---|---|---|---|---|
| CDS | Non-CDSa | ||||
| Eukaryotes (300 bp) | H. sapiens | 1,787 | 1,800 | 1,800 | [−249, +50] |
| D. melanogaster | 1,886 | 1,799 | 2,859 | [−249, +50] | |
| Prokaryotes (81 bp) | C. elegans | 598 | 600 | 600 | [−249, +50] |
| B. subtilis | 270 | 300 | 300 | [−60, +20] | |
| E. coli | 741 | 700 | 700 | [−60, +20] | |
CDS, coding sequences.
Intron for eukaryotes and convergent intergenetic region for prokaryotes.
Eukaryotic Promoter Database (EPD)36 is a high-quality and non-redundant promoter resource and can be freely accessed at https://epd.epfl.ch//EPD_database.php. The 1,787 H. sapiens and 1,886 D. melanogaster Pol II promoter sequences were obtained from the EPD database. The 598 C. elegans promoter sequences were extracted from CEPDB (C. elegans promoter database; http://rulai.cshl.edu/cgi-bin/CEPDB/home.cgi). Each eukaryotic promoter is 300 bp long from 249 bp upstream to 50 bp downstream regions of TSS (TSS is regarded as 0-th site).
For prokaryote, 270 B. subtilis σ43 promoters were collected from DBTBS37 (http://dbtbs.hgc.jp), and 741 E. coli K-12 σ70 promoter sequences were gained from RegulonDB38 (http://regulondb.ccg.unam.mx/). All prokaryotic promoters have 81 nt with the region from −60 to +20 flanking TSS (TSS is regarded as the 0-th site).
The negative datasets were taken from the five species genome sequences. We randomly extracted 1,800 coding sequences and 1,800 introns from human DNA sequences from http://www.fruitfly.org/sequence/human-datasets.html 39 to generate the non-promoter dataset for H. sapiens. For D. melanogaster, a negative dataset including 2,859 coding sequences and 1,799 introns was downloaded from the website (http://www.fruitfly.org/sequence/drosophila-datasets.html).40 The negative sample of C. elegans contains 600 coding sequences, and 600 introns were randomly extracted from Exon-Intron Database (EID).41 For prokaryotes, all negative samples were randomly taken from the well-known database GenBank.42 The number of non-promoter sequences for B. subtilis and E. coli are 600 (including 300 coding sequences and 300 convergent intergenic sequences) and 1,400 (including 700 coding sequences and 700 convergent intergenic sequences), respectively.
To get rid of the influence of noise data, we eliminated the sequences that contain other IUPAC code letters, such as “N,” “S,” and “W,” from both positive and negative datasets. In order to ensure that the format of negative sequences can match the promoters, the lengths of eukaryotic and prokaryotic non-promoter sequences are also 300 and 81 bp, respectively. The details of the benchmark datasets were listed in Table 5.
It is well known that sequence similarity could influence the evaluation on the proposed mode.43 We investigated the sequence similarity of the five species promoters by using CD-HIT. After setting the cutoff of sequence identity to 0.8 to exclude high similar promoters, we found that 98.0%, 99.3%, 95.0%, 98.5%, and 96.0% promoters for H. sapiens, D. melanogaster, C. elegans, B. subtilis, and E. coli remained, suggesting that the original datasets are objective enough to construct prediction models. Moreover, for the purpose of providing an objective comparison with the previous promoter prediction method IPMD, the same benchmarking datasets as used by IPMD are also provided. All data used in this study can be freely downloaded from http://lin-group.cn/server/iProEP/pages/download.php.
Pseudo k-Tuple Nucleotide Composition (PseKNC)
In general, the input of almost all the existing machine learning classification methods, such as SVM,44, 45, 46 Random Forest,47 and Artificial Neural Network,48, 49, 50 must be a numeric value rather than a string sequence. Thus, each sample must be transferred into a fixed length of the feature vector.
A simple and common strategy to transform a DNA sample into a vector is to use its k-tuple nucleotide composition, which can be formulated by a vector D with 4k elements according to the following formula:
| (Equation 2) |
where the symbol T means the transposition of a vector, and is the normalized frequency of the i-th k-tuple nucleotide component occurring in the DNA sequence.
In order to take both local and global sequence-order information of a DNA sequence into consideration, PseKNC51, 52 was proposed and has been widely utilized to represent DNA or RNA sequences.53, 54 Its basic principle is to combine the correlation of physiochemical properties of oligonucleotides and k-mer composition to formulate DNA sequences. There are two kinds of PseKNCs: type I and type II PseKNC. The former is also called the parallel correlation type, which mixes different physicochemical properties together to represent a nucleotide sequence with a vector containing 4k + Λ components. The latter is named the series correlation type, which describes a nucleotide sequence by a vector containing 4k + λΛ factors. Comparing with the type I PseKNC, which has been widely and successfully applied in various bioinformatics fields,8, 55 few works focused on the application of type II PseKNC.54, 56 Considering the merit of type II PseKNC that different correlation information was separated independently, this work employed the type II PseKNC to transform sample sequences into vectors given as below:
| (Equation 3) |
where
| (Equation 4) |
has the same meaning as in Equation 2; λ is an integer number less than L − k, which reflects the correlation tiers or correlation rank along a DNA sequence; ω is a weight factor used to balance the effect of global correlation information and local property; and τj represents the m-tier correlation factor, which describes the sequence-order correlation between all the m-tier contiguous k-tuple nucleotides along a DNA sequence. Here τj can be calculated by
| (Equation 5) |
where
| (Equation 6) |
where is a numerical value of the ξ-th physicochemical property for the dinucleotide at position i, is the corresponding value for the dinucleotide at position i + m, and Λ is the number of physicochemical properties. In this study, six DNA local structural properties of the 16 DNA dinucleotides were utilized in this work; the concrete values of three local translational properties (slide, shift, rise) and three local angular properties (roll, tilt, twist) were taken from Goñi et al.’s57 work. It should be noted that the original values of six DNA local structural properties should be subjected to a standard version by Equation 7 and then can be used in Equation 6 to calculate PseKNC:
| (Equation 7) |
where is the original value of the ξ-th DNA local structural property for the dinucleotide at position i, the symbol means taking the average of the quantity therein for the 16 different combinations of A, G, C, T for , and SD represents the corresponding SD. The standard version of these physicochemical property values can be also found in many other DNA-related studies.55 The superiority of the final standard 16 values converted by Equation 7 is that they will have a zero mean value over the 16 different dinucleotides and will not be changed if going through the same conversion procedure again.58
PCSF
By aligning promoter sequences for every species, we can construct a position-correlation scoring matrix (PCSM).24, 59 Each row in the PCSM consisted of factor pxi, which is the probability of k-mer x at the i-th site of promoter samples. pxi can be calculated by the following formula:
| (Equation 8) |
where nxi is the actual count of x appearing at the i-th site, and bxi is the corresponding pseudocount. Ni indicates the sum of real counts of all k-mers at the i-th site (namely, positive sample number), and Bi is the corresponding sum of the pseudocount. If the sample size is not large enough, some k-mers will not be present when k increases. Hence, the pseudocount could improve estimation of the probability pxi for k-mer x at the i-th site. Bi and bxi can be given by
| (Equation 9) |
in which p0 is the background frequency of k-mer, which is equal to 1/4k. With the increasing sample number Ni, the influence of pseudocounts will weaken, because of the slow increase of .
Some conservation sites of trimers for five species have been screened out by a great number of complex conservation analyses and ACC evaluations in Lin and Li.24 Based on these sites and PCSM, the PCSF feature of positive and negative samples for five species can be expressed as
| (Equation 10) |
where n is the number of selected conservation sites, and each element is defined as
| (Equation 11) |
In this equation, p0 is the background probability of each trimer (p0 = 1/43), and pxi can be obtained on the basis of PCSM.
mRMR
Commonly, picking out of the most useful features from the high-dimension data is a requisite step to exclude noise, improve prediction ACC and efficiency, avoid model overfitting, as well as build a robust model. In the present work, with the increase of two variables in Equation 4, k and λ, the dimension of PseKNC features will raise sharply, which may result in the curse of dimensionality. Therefore, it is absolutely necessary to find out the optimal features that could produce a robust model with highest ACC. mRMR is a popular feature selection technique that could calculate a score for each feature for measuring the importance of the feature.60, 61 It used a series of intuitive measures of relevance and redundancy to find a very compact subset from candidate features and has been widely used in data mining of biological processes.62, 63, 64, 65 For discrete features, two selection criteria, Mutual Information Difference criterion (MID) and Mutual Information Quotient criterion (MIQ), can be used to calculate the score of a feature. In the study, we chose the score from MIQ.
After scoring the PseKNC and PCSF features by mRMR, the IFS strategy with 5-fold cross-validation was applied to obtain the best feature subset that could produce the maximum prediction ACC. During the IFS procedure, the ranked features were added in the training set one by one according to mRMR rank; IFS strategy evaluates the performance of the top k-ranked features. The 5-fold cross-validation was to seek the best penalty coefficient c and width parameter γ for SVM models when obtaining the best feature subset.54, 56
SVM
SVM is a widely employed machine learning algorithm based on statistical learning theory66 and has been extended in bioinformatics fields.67, 68, 69, 70, 71, 72, 73 The core idea of SVM is to seek out a classification hyperplane that can maximize the margin of the feature space. LibSVM is a popular softpackage for executing SVM74 and can be freely downloaded from https://www.csie.ntu.edu.tw/∼cjlin/libsvm/. This study used LibSVM with radial basis function (RBF) to perform classification. We employed the grid search method with cross-validation to seek the best penalty coefficient c and width parameter γ. The searching space is as follows:
| (Equation 12) |
Performance Evaluation Metrics
In order to assess the quality of a predictor and compare different prediction tools, the following three indexes,75 namely, the overall ACC, Sn, and Sp, were used and formulated as
| (Equation 13) |
| (Equation 14) |
| (Equation 15) |
where TP (true positive) and TN (true negative) present the numbers of correctly identified promoters and non-promoters, respectively, and FP (false positive) and FN (false negative) denote the number of non-promoters incorrectly classified as promoters and the number of promoters incorrectly classified as non-promoters.
ROC analysis was used to measure the performance of the model with the varying of decision thresholds.76
Author Contributions
H.D., W.C., and H.L. conceived and designed the study. H.-Y.L., Z.-Y.Z., Z.-D.S., and W.S. conducted the experiments. H.-Y.L. and Z.-Y.Z. implemented the algorithms. H.-Y.L., Z.-Y.Z., and Z.-D.S. established the web server. H.-Y.L., Z.-Y.Z., W.C., and H.L. performed the analysis and wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work has been supported by the National Natural Scientific Foundation of China (grants 61772119 and 31771471), the Natural Science Foundation for Distinguished Young Scholar of Hebei Province (no. C2017209244), and the Science Strength Promotion Programme of UESTC.
Contributor Information
Wei Chen, Email: chenweiimu@gmail.com.
Hao Lin, Email: hlin@uestc.edu.cn.
References
- 1.Haberle V., Lenhard B. Promoter architectures and developmental gene regulation. Semin. Cell Dev. Biol. 2016;57:11–23. doi: 10.1016/j.semcdb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Thomas M.C., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 3.Slobodin B., Agami R. Transcription initiation determines its end. Mol. Cell. 2015;57:205–206. doi: 10.1016/j.molcel.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen A.G., Baldi P., Chauvin Y., Brunak S. The biology of eukaryotic promoter prediction—a review. Comput. Chem. 1999;23:191–207. doi: 10.1016/s0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- 5.Hawley D.K., McClure W.R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He W., Jia C., Duan Y., Zou Q. 70ProPred: a predictor for discovering sigma70 promoters based on combining multiple features. BMC Syst. Biol. 2018;12(Suppl 4):44. doi: 10.1186/s12918-018-0570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Z.Y., Lai H.Y., Yang H., Zhang C.J., Yang H., Wei H.H., Chen X.X., Zhao Y.W., Su Z.D., Li W.C. Pro54DB: a database for experimentally verified sigma-54 promoters. Bioinformatics. 2017;33:467–469. doi: 10.1093/bioinformatics/btw630. [DOI] [PubMed] [Google Scholar]
- 8.Lin H., Deng E.Z., Ding H., Chen W., Chou K.C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res. 2014;42:12961–12972. doi: 10.1093/nar/gku1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abeel T., Saeys Y., Bonnet E., Rouzé P., Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18:310–323. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J.Y., Zhou Y., Yu Z.G., Anh V., Zhou L.Q. Human Pol II promoter recognition based on primary sequences and free energy of dinucleotides. BMC Bioinformatics. 2008;9:113. doi: 10.1186/1471-2105-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davuluri R.V., Grosse I., Zhang M.Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- 13.Anwar F., Baker S.M., Jabid T., Mehedi Hasan M., Shoyaib M., Khan H., Walshe R. Pol II promoter prediction using characteristic 4-mer motifs: a machine learning approach. BMC Bioinformatics. 2008;9:414. doi: 10.1186/1471-2105-9-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burden S., Lin Y.X., Zhang R. Improving promoter prediction for the NNPP2.2 algorithm: a case study using Escherichia coli DNA sequences. Bioinformatics. 2005;21:601–607. doi: 10.1093/bioinformatics/bti047. [DOI] [PubMed] [Google Scholar]
- 15.Gan Y., Guan J., Zhou S. A pattern-based nearest neighbor search approach for promoter prediction using DNA structural profiles. Bioinformatics. 2009;25:2006–2012. doi: 10.1093/bioinformatics/btp359. [DOI] [PubMed] [Google Scholar]
- 16.Xu W., Zhang L., Lu Y. SD-MSAEs: Promoter recognition in human genome based on deep feature extraction. J. Biomed. Inform. 2016;61:55–62. doi: 10.1016/j.jbi.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Umarov R.K., Solovyev V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE. 2017;12:e0171410. doi: 10.1371/journal.pone.0171410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou Q., Xing P., Wei L., Liu B. Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA. 2019;25:205–218. doi: 10.1261/rna.069112.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L., Su R., Wang B., Li X., Zou Q., Gao X. Integration of Deep Feature Representations and Handcrafted Features to Improve the Prediction of N6-Methyladenosine Sites. Neurocomputing. 2019;324:3–9. [Google Scholar]
- 20.Su R., Liu X., Wei L., Zou Q. Deep-Resp-Forest: A deep forest model to predict anti-cancer drug response. Methods. 2019 doi: 10.1016/j.ymeth.2019.02.009. Published online February 14, 2019. [DOI] [PubMed] [Google Scholar]
- 21.Peng L., Peng M.M., Liao B., Huang G.H., Li W.B., Xie D.F. The Advances and Challenges of Deep Learning Application in Biological Big Data Processing. Curr. Bioinform. 2018;13:352–359. [Google Scholar]
- 22.Long H.X., Wang M., Fu H.Y. Deep Convolutional Neural Networks for Predicting Hydroxyproline in Proteins. Curr. Bioinform. 2017;12:233–238. [Google Scholar]
- 23.Singh S., Kaur S., Goel N. A Review of Computational Intelligence Methods for Eukaryotic Promoter Prediction. Nucleosides Nucleotides Nucleic Acids. 2015;34:449–462. doi: 10.1080/15257770.2015.1013126. [DOI] [PubMed] [Google Scholar]
- 24.Lin H., Li Q.Z. Eukaryotic and prokaryotic promoter prediction using hybrid approach. Theory Biosci. 2011;130:91–100. doi: 10.1007/s12064-010-0114-8. [DOI] [PubMed] [Google Scholar]
- 25.Song K. Recognition of prokaryotic promoters based on a novel variable-window Z-curve method. Nucleic Acids Res. 2012;40:963–971. doi: 10.1093/nar/gkr795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin H., Liang Z.Y., Tang H., Chen W. IEEE/ACM Trans. Comput. Biol. Bioinform; 2017. Identifying Sigma70 promoters with novel pseudo nucleotide composition. Published online February 8, 2017. [DOI] [PubMed] [Google Scholar]
- 27.Liu B., Yang F., Huang D.S., Chou K.C. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 2018;34:33–40. doi: 10.1093/bioinformatics/btx579. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M., Li F., Marquez-Lago T.T., Leier A., Fan C., Kwoh C.K., Chou K.C., Song J., Jia C. MULTiPly: a novel multi-layer predictor for discovering general and specific types of promoters. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz016. 2019, btz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B., Han L., Liu X., Wu J., Ma Q. IEEE/ACM Trans. Comput. Biol. Bioinform; 2018. Computational prediction of sigma-54 promoters in bacterial genomes by integrating motif finding and machine learning strategies. Published online March 15, 2018. [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Chen X., McDermaid A., Ma Q. DMINDA 2.0: integrated and systematic views of regulatory DNA motif identification and analyses. Bioinformatics. 2017;33:2586–2588. doi: 10.1093/bioinformatics/btx223. [DOI] [PubMed] [Google Scholar]
- 31.Ma Q., Zhang H., Mao X., Zhou C., Liu B., Chen X., Xu Y. DMINDA: an integrated web server for DNA motif identification and analyses. Nucleic Acids Res. 2014;42:W12–W19. doi: 10.1093/nar/gku315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahmuradov I.A., Umarov R.K., Solovyev V.V. TSSPlant: a new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 2017;45:e65. doi: 10.1093/nar/gkw1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Zhao Y., Liao X., Shi W., Li K., Zou Q., Peng S. Deep learning in omics: a survey and guideline. Brief. Funct. Genomics. 2018;18:41–57. doi: 10.1093/bfgp/ely030. [DOI] [PubMed] [Google Scholar]
- 34.Yu L., Sun X., Tian S.W., Shi X.Y., Yan Y.L. Drug and Nondrug Classification Based on Deep Learning with Various Feature Selection Strategies. Curr. Bioinform. 2018;13:253–259. [Google Scholar]
- 35.Wei L., Ding Y., Su R., Tang J., Zou Q. Prediction of Human Protein Subcellular Localization Using Deep Learning. J. Parallel Distrib. Comput. 2018;117:212–217. [Google Scholar]
- 36.Dreos R., Ambrosini G., Cavin Périer R., Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013;41:D157–D164. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sierro N., Makita Y., de Hoon M., Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36:D93–D96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44(D1):D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohler U., Liao G.C., Niemann H., Rubin G.M. Computational analysis of core promoters in the drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. RESEARCH0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepelev V., Fedorov A. Advances in the Exon-Intron Database (EID) Brief. Bioinform. 2006;7:178–185. doi: 10.1093/bib/bbl003. [DOI] [PubMed] [Google Scholar]
- 42.Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2015;43:D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou Q., Lin G., Jiang X., Liu X., Zeng X. Sequence Clustering in Bioinformatics: An Empirical Study. Brief. Bioinform. 2019 doi: 10.1093/bib/bby090. 2019, bby090. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X.J., Feng C.Q., Lai H.Y., Chen W., Lin H. Predicting Protein Structural Classes for Low-Similarity Sequences by Evaluating Different Features. Knowl. Base. Syst. 2019;163:787–793. [Google Scholar]
- 45.Yang H., Lv H., Ding H., Chen W., Lin H. iRNA-2OM: A Sequence-Based Predictor for Identifying 2′-O-Methylation Sites in Homo sapiens. J. Comput. Biol. 2018;25:1266–1277. doi: 10.1089/cmb.2018.0004. [DOI] [PubMed] [Google Scholar]
- 46.Li D., Ju Y., Zou Q. Protein Folds Prediction with Hierarchical Structured SVM. Curr. Proteomics. 2016;13:79–85. [Google Scholar]
- 47.Kandaswamy K.K., Chou K.C., Martinetz T., Möller S., Suganthan P.N., Sridharan S., Pugalenthi G. AFP-Pred: A random forest approach for predicting antifreeze proteins from sequence-derived properties. J. Theor. Biol. 2011;270:56–62. doi: 10.1016/j.jtbi.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 48.Cao R., Freitas C., Chan L., Sun M., Jiang H., Chen Z. ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network. Molecules. 2017;22:e1732. doi: 10.3390/molecules22101732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao R., Bhattacharya D., Hou J., Cheng J. DeepQA: improving the estimation of single protein model quality with deep belief networks. BMC Bioinformatics. 2016;17:495. doi: 10.1186/s12859-016-1405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang L., Zhang J., Xuan P., Zou Q. BP Neural Network Could Help Improve Pre-miRNA Identification in Various Species. BioMed Res. Int. 2016;2016:9565689. doi: 10.1155/2016/9565689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen W., Lei T.Y., Jin D.C., Lin H., Chou K.C. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014;456:53–60. doi: 10.1016/j.ab.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Chen W., Zhang X., Brooker J., Lin H., Zhang L., Chou K.C. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics. 2015;31:119–120. doi: 10.1093/bioinformatics/btu602. [DOI] [PubMed] [Google Scholar]
- 53.Yu C.Y., Li X.X., Yang H., Li Y.H., Xue W.W., Chen Y.Z., Tao L., Zhu F. Assessing the Performances of Protein Function Prediction Algorithms from the Perspectives of Identification Accuracy and False Discovery Rate. Int. J. Mol. Sci. 2018;19:183. doi: 10.3390/ijms19010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dao F.Y., Lv H., Wang F., Feng C.Q., Ding H., Chen W., Lin H. Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics. 2019;35:2075–2083. doi: 10.1093/bioinformatics/bty943. [DOI] [PubMed] [Google Scholar]
- 55.Chen W., Feng P.M., Lin H., Chou K.C. iSS-PseDNC: identifying splicing sites using pseudo dinucleotide composition. BioMed Res. Int. 2014;2014:623149. doi: 10.1155/2014/623149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng C.Q., Zhang Z.Y., Zhu X.J., Lin Y., Chen W., Tang H., Lin H. Iterm-Pseknc: A Sequence-Based Tool for Predicting Bacterial Transcriptional Terminators. Bioinformatics. 2019;35:1469–1477. doi: 10.1093/bioinformatics/bty827. [DOI] [PubMed] [Google Scholar]
- 57.Goñi J.R., Pérez A., Torrents D., Orozco M. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 2007;8:R263. doi: 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chou K.C., Shen H.B. Recent progress in protein subcellular location prediction. Anal. Biochem. 2007;370:1–16. doi: 10.1016/j.ab.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Li Q.Z., Lin H. The recognition and prediction of sigma70 promoters in Escherichia coli K-12. J. Theor. Biol. 2006;242:135–141. doi: 10.1016/j.jtbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 61.Zou Q., Zeng J., Cao L., Ji R. A Novel Features Ranking Metric with Application to Scalable Visual and Bioinformatics Data Classification. Neurocomputing. 2016;173:346–354. [Google Scholar]
- 62.Kabir M., Ahmad S., Iqbal M., Hayat M. iNR-2L: A two-level sequence-based predictor developed via Chou’s 5-steps rule and general PseAAC for identifying nuclear receptors and their families. Genomics. 2019 doi: 10.1016/j.ygeno.2019.02.006. Published online February 16, 2019. 10.1016/j.ygeno.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Yuan F., Lu L., Zhang Y., Wang S., Cai Y.D. Data mining of the cancer-related lncRNAs GO terms and KEGG pathways by using mRMR method. Math. Biosci. 2018;304:1–8. doi: 10.1016/j.mbs.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Li B.Q., Hu L.L., Chen L., Feng K.Y., Cai Y.D., Chou K.C. Prediction of protein domain with mRMR feature selection and analysis. PLoS ONE. 2012;7:e39308. doi: 10.1371/journal.pone.0039308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S.P., Zhang Q., Lu J., Cai Y.D. Analysis and Prediction of Nitrated Tyrosine Sites with the Mrmr Method and Support Vector Machine Algorithm. Curr. Bioinform. 2018;13:3–13. [Google Scholar]
- 66.Cortes C., Vapnik V. Support-Vector Networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- 67.Manavalan B., Shin T.H., Lee G. PVP-SVM: Sequence-Based Prediction of Phage Virion Proteins Using a Support Vector Machine. Front. Microbiol. 2018;9:476. doi: 10.3389/fmicb.2018.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W., Lv H., Nie F., Lin H. i6mA-Pred: Identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz015. 2019, btz015. [DOI] [PubMed] [Google Scholar]
- 69.Tang H., Zhao Y.W., Zou P., Zhang C.M., Chen R., Huang P., Lin H. HBPred: a tool to identify growth hormone-binding proteins. Int. J. Biol. Sci. 2018;14:957–964. doi: 10.7150/ijbs.24174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song J., Wang Y., Li F., Akutsu T., Rawlings N.D., Webb G.I., Chou K.C. Iprot-Sub: A Comprehensive Package for Accurately Mapping and Predicting Protease-Specific Substrates and Cleavage Sites. Brief. Bioinform. 2019;20:638–658. doi: 10.1093/bib/bby028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manavalan B., Shin T.H., Lee G. DHSpred: support-vector-machine-based human DNase I hypersensitive sites prediction using the optimal features selected by random forest. Oncotarget. 2017;9:1944–1956. doi: 10.18632/oncotarget.23099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W., Yang H., Feng P., Ding H., Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33:3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 73.Cao R., Wang Z., Wang Y., Cheng J. SMOQ: a tool for predicting the absolute residue-specific quality of a single protein model with support vector machines. BMC Bioinformatics. 2014;15:120. doi: 10.1186/1471-2105-15-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang C.C., Lin C.J. Libsvm: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011;2:27. [Google Scholar]
- 75.Lv H., Zhang Z.M., Li S.H., Tan J.X., Chen W., Lin H. Evaluation of different computational methods on 5-methylcytosine sites identification. Brief. Bioinform. 2019:bbz048. doi: 10.1093/bib/bbz048. [DOI] [PubMed] [Google Scholar]
- 76.Metz C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]