Abstract
Utilization of microalgae has been hampered by limited tools for creating loss-of-function mutants. Furthermore, modified strains for deployment into the field must be free of antibiotic resistance genes and face fewer regulatory hurdles if they are transgene free. The oleaginous microalga, Nannochloropsis oceanica CCMP1779, is an emerging model for microalgal lipid metabolism. We present a one-vector episomal CRISPR/Cas9 system for N. oceanica that enables the generation of marker-free mutant lines. The CEN/ARS6 region from Saccharomyces cerevisiae was included in the vector to facilitate its maintenance as circular extrachromosal DNA. The vector utilizes a bidirectional promoter to produce both Cas9 and a ribozyme flanked sgRNA. This system efficiently generates targeted mutations, and allows the loss of episomal DNA after the removal of selection pressure, resulting in marker-free non-transgenic engineered lines. To test this system, we disrupted the nitrate reductase gene (NR) and subsequently removed the CRISPR episome to generate non-transgenic marker-free nitrate reductase knockout lines (NR-KO).
Keywords: CRISPR/Cas9, Nannochloropsis, episome, marker-free, gene targeting, algal genetic engineering
Microalgae are some of the most productive biomass sources on Earth and can generate many high-value bioproducts such as omega-3 fatty acids, carotenoids, and unusual polysaccharides (1). One genus of microalgae that has distinguished itself for productivity and genetic engineering potential is Nannochloropsis (2–4). Genetic engineering of microalgae has been hampered by a limited ability to disrupt genes and by the challenges of generating transformed algae that meet biocontainment requirements for open pond production (5). Recently, the use of endogenous elements, marker-free strategies, and DNA free genome editing have allowed the generation of modified organisms that lack antibiotic selection markers (6–8) or even any heterologous DNA (9, 10). Such organisms satisfy the criteria for non-transgenic organisms, making deployment of modified strains into open systems feasible.
Gene targeting in microalgae has been performed by homologous recombination (11), TALENs (12–14), and CRISPR/Cas9 (10, 15–20). Gene disruption by these methods in microalgae has usually required the integration of a transgenic selection marker into the genome. A CRISPR/Cas9 system for N. oceanica IMET had low efficiency (~1–4%), possibly due to low targeting efficiency of the single guide RNA (sgRNA) produced from a protein-coding gene promoter (18). Another CRISPR/Cas9 system for N. gaditana utilized introduction of synthesized sgRNAs and required two selection markers for targeted disruption (17). Efficient CRISPR/Cas9 based mutagenesis requires a sgRNA without modified ends or extraneous sequences for efficient interaction with the Cas9 nuclease and DNA target. Therefore, sgRNAs flanked by the hammerhead (HH) and hepatitis delta virus (HDV) self-cleaving ribozymes are used to generate precise 5’ and 3’ ends (21, 22), and enable the production of sgRNAs under the control of protein-coding gene promoters (RNA polymerase II class).
Episomes (extrachromosomal nuclear DNA that is usually circular) occur naturally in some red algae (23), and diatoms are capable of maintaining synthetic circular episomes containing a low GC (guanine cytosine) centromere-like region (24–26). In diatoms, an extrachromosomal episome with a centromere and autonomous replication sequence fusion developed from Saccharomyces cerevisiae (CEN/ARS) is replicated and segregated into daughter cells (24, 25). Furthermore, this episomal system is able to produce proteins that can be localized throughout the cell and results in more uniform transgene expression levels between independent transformants compared to integrated vectors, possibly by avoiding integration site-specific effects (25). While circular episomes are an effective expression platform in diatoms, without selection pressure, they are lost over several generations providing a way to “cure” cells of them (25–27).
We developed a CRISPR/Cas9 system that can be expressed from an episome in the heterokont microalgae Nannochloropsis oceanica CCMP1779. The subsequent removal of the episome after the mutation was produced generates marker-free non-transgenic gene disruption mutants that can be modified repeatedly.
Results/Discussion
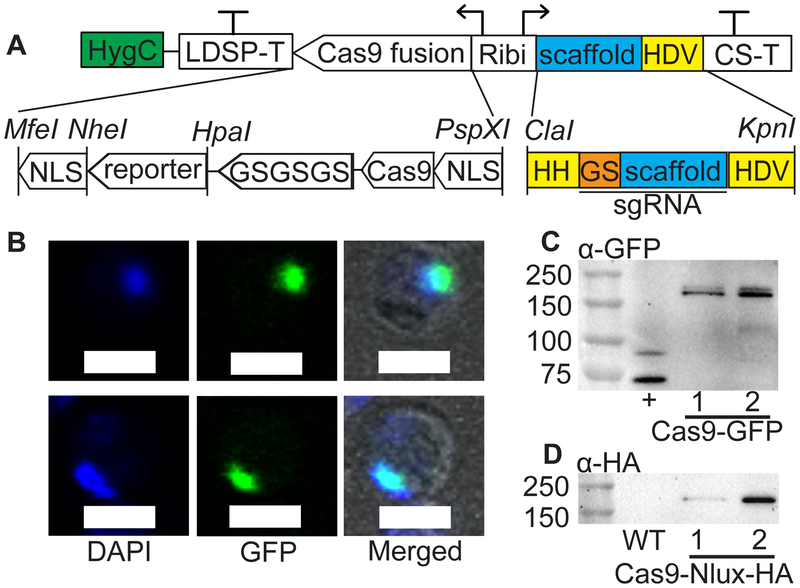
The linearized pNOC-CRISPR-GFP vector, containing a Cas9-GFP expression cassette and a Hygromycin B resistance marker (HygR) (3), generated Hygromycin B-resistant transformants when introduced into N. oceanica (Figure 1A). To establish a highly efficient CRISPR/Cas9 gene editing system in N. oceanica we confirmed the nuclear localization of a Cas9-GFP nuclease with C’ and N’ terminal SV40 nuclear localization signals (NLS) by confocal microscopy. Wild-type lines did not possess any GFP signal, while an untargeted GFP expressing line had GFP signal throughout the cytosol with partial overlap with the DAPI stain (Figure S1A–B). The Cas9-GFP signal was punctate and co-localized with a DNA stain (4’, 6-Diamidino-2-Phenylindole, DAPI), demonstrating Cas9-GFP nuclear localization in N. oceanica (Figure 1B, Figure S1C). To facilitate Cas9 detection in vivo, we then generated a Cas9 nuclease fused to the ultra-bright NanoLuciferase reporter protein with an HA tag (Nlux-HA) in the vector pNOC-CRISPR. Nlux has high luminescence activity (28) and accordingly, N. oceanica transformants can be screened for the presence of the Cas9-Nlux-HA fusion in a 96-well plate using a small number of cells (2). Immunoblotting confirmed the production of Cas9-Nlux-HA (186.2 kDa) and Cas9-GFP proteins (191.8 kDa) (Figure 1C–D) in lines transformed with the integrating empty vector constructs (iEV) pNOC-CRISPR and pNOC-CRISPR-GFP respectively. These constructs lack a sgRNA sequence for a specific genomic locus.
Figure 1.
A one-vector CRISPR system for gene disruption in N. oceanica. (A) The pNOC-CRISPR vector series includes a Hygromycin B resistance cassette (HygC, green). A bidirectional promoter (Ribi) drives the transcription of the Cas9-reporter fusion and gRNA scaffold (scaffold, blue), along with the LDSP and CS terminators (LDSP-T, CS-T) respectively. Cas9 is fused to either the GFP or Nlux (with HA tag) reporters by a 3x glycine-serine linker (GSGSGS) and contains SV40 nuclear localization signals on the N’ and C’ termini (NLS). The gRNA scaffold and the 3’ self-cleaving HDV ribozyme is integrated into the vector. The 5’ hammerhead ribozyme (HH) specific for each guide sequence (GS, orange) is fused to the gRNA scaffold (scaffold) to form a sgRNA. Ribozymes are highlighted in yellow. Unique restriction sites are shown with an upwards line and the name in italics. (B) Confocal analysis of DAPI nuclear staining, Cas9-GFP signal, and merged brightfield, DAPI and GFP signal in N. oceanica cells. Scale bar of 2 μM. (C) Immunoblotting with an α-GFP antibody detected the Cas9-GFP produced in N. oceanica transformed with pNOC-CRISPR-GFP. A N. oceanica line producing a delta-5 fatty acid desaturase (~75 kDa) fused with CFP was used as a GFP positive control (+). (D) Immunoblotting with an α-HA antibody detected the appropriately sized Cas9-Nlux-HA in N. oceanica transformed with pNOC-CRISPR. Wild-type N. oceanica was included as a negative control (WT). For (C) and (D) numbers on the left of immunoblots indicate size markers (KDa).
The pNOC-CRISPR vector contains the Cas9-Nlux-HA and a gRNA scaffold with a 3’ fusion to the HDV ribozyme (21, 22) under the control of the recently characterized endogenous ribosomal subunit bidirectional promoter (Ribi, located between NannoCCMP1779_9669 and NannoCCMP1779_9668) (2). To facilitate cleavage precisely at the 5’ end of as gRNA sequence, a hammerhead ribozyme (HH) specific to the guide sequence is introduced during sgRNA generation (Figure S2). Strategies for cloning sgRNAs are described in the Methods section.
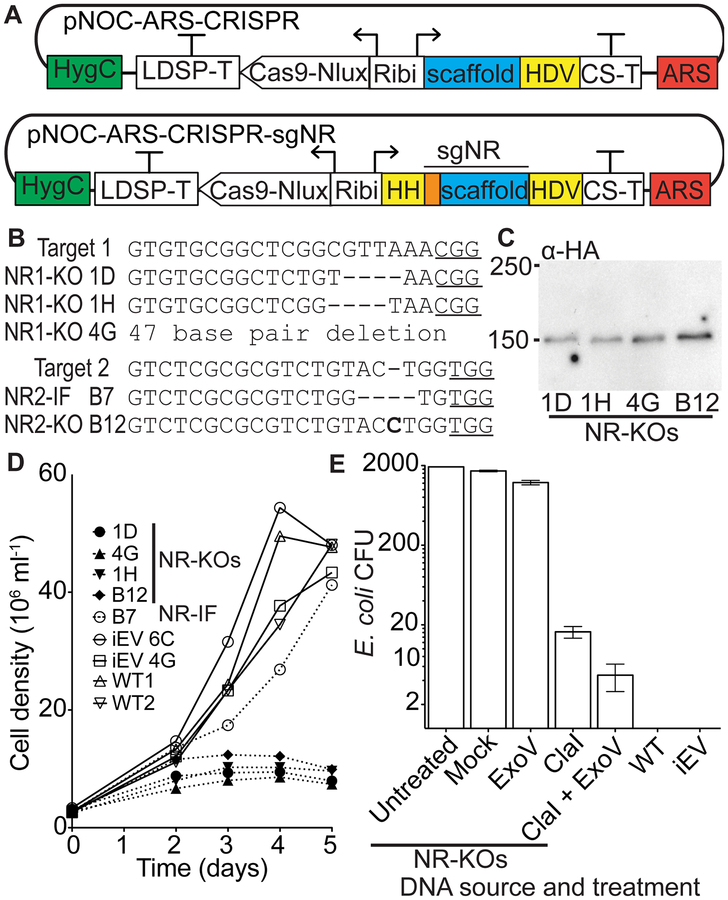
We next designed an episome-based CRISPR/Cas9 system, by incorporating the CEN/ARS region from S. cerevisiae into the pNOC-CRISPR-Nlux vector, generating the vector pNOC-ARS-CRISPR (Figure 2A). To test this system, we targeted the nitrate reductase gene (NR) for the known phenotype of NR disruption lines (NR-KO) (Figure 2A) (6, 29, 30). Nannochloropsis NR-KO lines can grow on ammonium (NH4) but are not able to sustain growth when provided with only nitrate (NO3) as a nitrogen source (11, 18). Two sgRNAs flanked by self-cleaving ribozymes (sgNR1 and sgNR2) were used to independently target two sites at the 5’ end of the NR gene. We introduced the pNOC-ARS-CRISPR-sgNR1 and pNOC-ARS-CRISPR-sgNR2 episomes into N. oceanica and generated the NR1-KO and NR2-KO lines respectively. Introduction of the circular episomal CRISPR plasmids into N. oceanica resulted in Hygromycin B-resistant lines, which were screened for Cas9-Nlux-HA signal (Figure S3A–B).
Figure 2.
Development of an episomal CRISPR system. (A) The S. cerevisiae CEN/ARS6 region (ARS, red) was included in the pNOC-ARS-CRISPR construct for episomal maintenance. Guide sequences(orange) for nitrate reductase targets, with a 5’hammerhead ribozyme (HH),were fused to the gRNA scaffold to form the NR sgRNAs (sgNR). Ribozymes are highlighted in yellow. The sgNR1 and sgNR2were added to pNOC-ARS-CRISPR to form pNOC-ARS-CRISPR-sgNR1 and pNOC-ARS-CRISPR-sgNR2, respectively. N. oceanica is transformed with circular episomal CRISPR constructs. (B) Mutations in the two target sites in the NR genomic locus (Target 1 and Target 2) of N. oceanica transformed with the respective pNOC-ARS-CRISPR-sgNR construct. Mutant lines are identified by 96-well plate location (Figure S3). Deleted nucleotides are represented with dashes and inserted nucleotides are shown in bold. Protospacer adjacent motifs (PAM sites) are underlined. (C) Immunoblot using an α-HA antibody of N. oceanicaNR1-KO and NR2-KO lines producing Cas9-Nlux-HA from the CRISPR episome. Numbers on the left indicate size markers (KDa). (D) Growth curves after transfer from NH4 to NO3 containing medium of NR-KO frame-shifted lines (1D, 1H, 4G, B12) and NR2-IF B7 in-frame line, empty vector integrated CRISPR control lines (iEV), and wildtype (WT). (E) Episome rescue by E. coli transformation using equal quantities of DNA isolated from episomal (NR-KO) lines, integrated empty-vector (iEV), and wild-type (WT) N. oceanica lines. Values are the average colonies generated ± SE (NR-KOs n = 3 independent lines, WT n = 2 biological replicates, and iEV n = 2 independent lines). Equal quantities of DNA from NR-KO lines after treatment were used for E. coli transformation, and the resulting colonies counted (n = 3 independent lines). Exonuclease V (ExoV), ClaI endonuclease (ClaI), and ClaI endonuclease with Exonuclease V (ClaI+ExoV).
Lines displaying luminescence were examined for mutations in the NR gene by PCR amplification of the genomic locus and Sanger sequencing (Figure S3). We found cumulatively that 47% of lines with Nlux signal had frame-shift mutations and 13% had in-frame mutations, while 13% had no mutation and 27% resulted in low-quality sequencing outcomes (Figure S3C). The two sgRNAs had different efficiencies with sgNR1 (in NR1-KO lines) having a high mutation efficiency with 9/10 screened colonies containing a mutation and no wild-type sequences recovered, while sgNR2 (in NR2-KO lines) had a lower mutation efficiency with 9/20 screened lines possessing a mutation and 4/20 lines lacking a mutation (Figure S3C). The low-quality sequencing reactions often had good quality chromatographs up to the target site and then became unreadable. This observation suggests that a heterogeneous population of mutants was recovered from single colonies, possibly due to mutagenesis occurring after plating. It is likely that pure genotypic lines could be obtained from these populations by streaking to single colonies.
We selected N. oceanica mutants with small deletions (1D, 1H), a larger (47 bp) deletion (4G), and a small insertion (B12) as frame-shifted null mutants (NR-KO lines), and the B7 line with a 3 bp deletion as an in-frame mutant (NR-IF) for further analyses (Figure 2B, Figure 3D–E). Immunoblotting detected the Cas9-Nlux-HA protein in all the lines indicating that proteins can be produced from an episomal DNA in N. oceanica (Figure 2C). To test for the loss of NR activity, cell growth was analyzed in liquid cultures after transfer from NH4 to NO3 containing medium. The frame-shifted NR-KO mutants could not grow, while the wild-type (WT) lines, the iEV lines, and the in-frame mutant B7 grew in NO3-containing liquid medium (Figure 2D). This indicates frame-shifts introduced into the coding sequence of targeted genes ablate the function of the resulting protein, while in-frame mutations may still produce functional proteins.
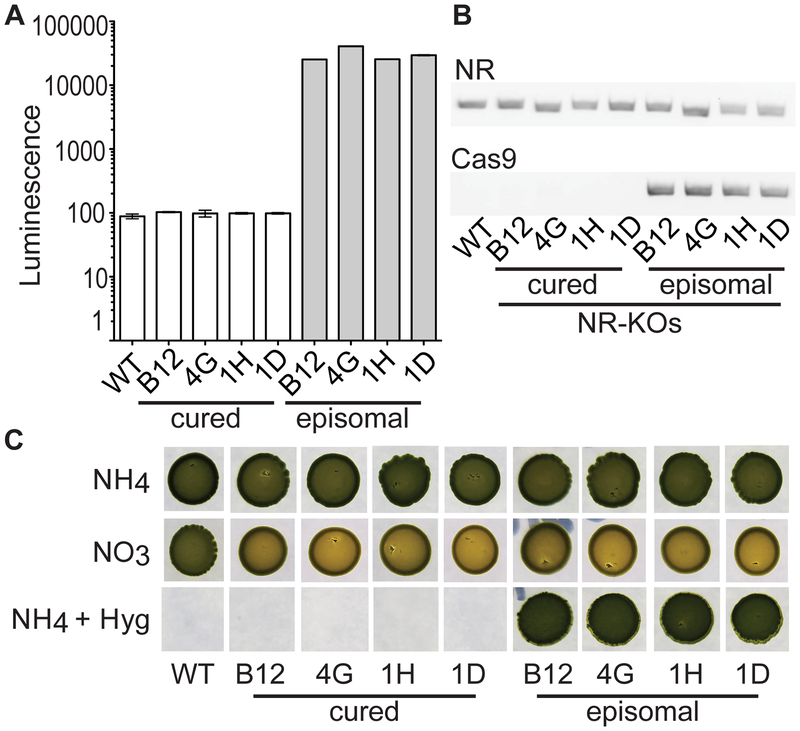
Figure 3.
Generation of marker-free non-transgenic mutants by episomal removal (curing). (A) Luminescence from equal number of cells of wild-type (WT), and NR-KO lines either containing the episome or cured of the episome. (B) PCR for detection of a positive control NR genomic locus and the Cas9 regions on the episome conducted on the same DNA extract obtained from WT and, NR-KO episomal and cured lines. (C) Plating of an equal number of cells of WT, NR-KO and NR-KO cured lines on NH4, NO3, and NH4 with Hygromycin B on F/2 solid medium after 1 month of growth.
To determine if the episomes were maintained as circular DNA, we conducted an episome rescue experiment by transforming Escherichia coli with DNA isolated from N. oceanica episome-carrying lines (NR-KOs). WT and iEV lines were used as negative controls. E. coli transformants were only obtained from untreated DNA isolated from NR-KO lines, but not from untreated DNA of iEV lines, or the WT strain (Figure 2E). To further confirm that the plasmids rescued from N. oceanica NR-KO lines were circular, an equal amount of DNA from the NR-KO lines was subjected to endonuclease ClaI restriction digest and/or treated with Exonuclease V, which acts on free DNA ends(25). Due to the ClaI site in the pNOC-ARS-CRISPR plasmid, ClaI treatment of DNA isolated from NR-KO lines strongly decreased the number of E. coli transformants (Figure 2E). Treatment with Exonuclease V resulted in a small reduction of E. coli transfomants compared to mock treatments, while a combined endonuclease and exonuclease treatment resulted in very few colonies being recovered (Figure 2E). Restriction fragment analysis of plasmids recovered from the episome rescue indicated that the episomes were faithfully maintained in N. oceanica (Figure S4). Restriction enzyme recognition sites by the CEN/ARS region (AgeI), in the Cas9 and HygR genes (EcoRI), and backbone (NotI), indicated that the episome had no apparent insertions or deletions (Figure S4A–B). Sequencing of the recovered episomes further confirmed their authenticity (Figure S4C–D).
We performed southern blots to further characterize the CRISPR episomal lines (Figure S5). As a control, we used iEV lines containing the genomic integrated pNOC-CRISPR vector. DNA was isolated from NR-KO, iEV and WT cells and was then digested with SacI and hybridized with either a HygR or an AmpR gene probe (Figure S5A). In all the NR-KO lines tested, both probes generated one fragment of ~13,000 bp that matched the episome CRISPR vector size (Figure S5B). However, the iEV lines generated different size fragments when using the HygR probe and only one line had a detectable fragment when the AmpR probe was used (Figure S5B). The AmpR probe hybridizes close to the AseI cut site used for linearizing the pNOC-CRISPR vector before transformation to generate the iEV lines. Therefore, the absence of a band in AmpR probed iEV DNA indicates that the AmpR was partially lost during integration or that the fragment is too small to be detected in the current experiment. None of the WT lines had a detectable signal in these assays. This experiment further confirmed the presence of a circular DNA molecule in the CRISPR episomal lines.
After generating the NR-KO lines we sought to remove the episome to form marker-less non-transgenic mutants. Nlux signal is a convenient measure for Cas9 presence in the transformed lines and to monitor for the loss of the episomes. Four NR-KO lines were grown for 10 days without selection in liquid medium and plated on solid medium to generate independent colonies. The colonies were screened for Nlux luminescence, followed by a PCR test for the presence of the episome and a control genomic locus (Figure S6A). After growth in non-selective medium, the NR-KO lines had various levels of Nlux signal and 25–75% of the lines had low luminescence signal (Figure S6B). The rate of successful inheritance of an episome during each cell division (segregation efficiency) is ascertained by removing selection for a known number of cell divisions and determining the percentage of episome carrying individuals (25, 31). In order to determine the number of generations that occurred during the episome curing procedure, the doubling time (1.023 days) of N. oceanica grown in NH4 media was measured (Figure S7). After ~30 generations, the segregation efficiency of the pNOC-CRISPR constructs was 96–99% based on the percentage of recovered colonies that maintained luminescence signal (Figure S6B).
To confirm that these cured low Nlux luminescent lines had lost the episome, we conducted PCR for the Cas9 gene contained in the episome, and the NR gene as a genomic positive control (Figure 3A–B). PCR detected the NR gene in all lines, while the Cas9 coding sequence was only present in the episomal lines but not in the cured lines (Figure 3B). Furthermore, growth tests were used to demonstrate the different phenotypes of the generated lines. First, the cured lines that had lost the episome were unable to grow on Hygromycin B-containing medium, while the episome-carrying parental lines were Hygromycin B resistant (Figure 3C). Second, growth on solid medium containing either NO3 or NH4 revealed a chlorotic phenotype of the episome-carrying and cured NR-KO lines only when grown on NO3 (Figure 3C) (11).
We have developed a system to produce marker-less non-transgenic gene knockout strains that not only theoretically allows for the generation of lines with multiple modifications but also facilitates the transfer of strains developed in the lab to the field. Our CRISPR/Cas9 components have been optimized for efficient detection, with a Cas9-Nlux-HA reporter fusion that has high signal to noise ratio and can be screened in a 96-well plate format soon after transformant isolation. Furthermore, the Cas9-Nlux-HA and sgRNA are coregulated by a bidirectional promoter, thus linking Cas9 production to sgRNA expression (Figure 1A) (2). The pNOC-CRISPR vector series contains unique restriction sites between the elements making it an ideal platform for further development of CRISPR/Cas9 as a system for transcriptional reprogramming and other DNA targeting techniques.
The use of self-cleaving ribozymes facilitates the flexible expression sgRNAs. The ribozyme flanked sgRNAs can be expressed from different types of promoters, in our case a bidirectional promoter, but also possibly from conditional promoters. The differing targeting efficiencies of the sgRNAs tested in this study demonstrate that sgRNA design plays an important role in Cas9 gene disruption rates (Figure S3C), and indicates that the development of sgRNA design tools (32, 33) for this genus is a route for further optimization of CRISPR/Cas9 in Nannochloropsis. Our final episomal vector, pNOC-ARS-CRISPR-v2, contains a multi-cloning site with a pair of BspQI type IIS restriction sites for scarless insertion of a guide sequence, for one-step cloning (Figure S8A–B). Self-cleaving ribozyme sequences for expression of the sgRNA could be used for the production of multiple sgRNAs from a single transcript (22). Production of multiple sgRNAs can be used to target multiple genes for simultaneous disruption, used in pairs with nickase Cas9 variants for enhanced specificity, or for deletion of a region between two targeted sites on a chromosome.
Transgenic expression in algae is most often performed by integrating a DNA construct into the genome. However, this can result in site-specific effects, or the disruption of genes at the insertion site. Therefore, episomes are an emerging expression platform in microalgae that avoids these issues. In diatoms, effective centromere sequences have the simple requirements of a GC content <30%, a length of >500 bp, with long stretches of A-T sequences (24–26). These centromere-like regions interact with the centromeric histone protein (CENH3) likely facilitating segregation during cell division (24). The segregation efficiency of the pNOC-CRISPR system was 96–99% (Figure S6B) which is comparable to the 97% segregation efficiency reported for episomal constructs developed for diatoms (25). However, the molecular processes for replication of synthetic episomes in algae are yet to be determined. The discovery of the relatively simple sequence requirements for centromeres in diatoms and the observation that the yeast CEN/ARS region is effective in diatoms and Nannochloropsis species suggest that heterokont algae have the potential to maintain newly acquired DNA, facilitating gene acquisition.
While first-generation synthetic episomes can robustly express transgenes, they are gradually lost from the population when selection pressure is not applied. These characteristics are ideal for transient CRISPR/Cas9 gene disruption, making it easy to screen transformants for mutations in the target site, followed by curing of the episome (8, 9). The cured mutants, thus contain a scar in the target site but lack an antibiotic resistance gene or any other transgenic elements, and can be used for subsequent modifications and do not need further biocontainment strategies. This technique will aid in the development of chassis strains for biotechnology, and makes marker-free genomic integration or endogenous locus tagging of expression constructs foreseeable.
To make the generated non-transgenic NR-KO mutant available for wide use, the NR1-KO 4G-5A strain is deposited with NCMA (Table S1). To make the high-efficiency, CRISPR/Cas9 dual component pNOC-CRISPR vectors widely available, the plasmids developed in this study are deposited with Addgene (Table S1).
Methods
Strains and growth conditions
N. oceanica CCMP1779 was used in all experiments. Cells were grown in F/2 medium under constant 100 μmol m−2 s−1 light at 22° C, on a shaker set at 120 rpm. F/2 medium with 2.5 mM KNO3 or 2.5 mM NaNH4 was used. Hygromycin B (Sigma-Aldrich) was utilized at a concentration of 100 μg/ml. Cell concentrations were measured with a Z2 Coulter Counter (Beckman Coulter) with a range of 1.8–3.6 μM. To determine whether growth could be sustained on nitrate, N. oceanica strains were inoculated to 5×106 cells/ml in F/2 with 2.5 mM KNO3 and cell density measured every 24 hours. For assessment of antibiotic resistance, growth after plating 30,000 cells on solid medium was monitored for 1 month.
CRISPR plasmid construction
The details of the construction of the plasmids produced in this study is contained in Data S1.
N. oceanica transformation
Integrating vectors were digested with AseI and concentrated by ethanol precipitation. Transformations were conducted as previously described (2), with 3 μg of DNA and 30 μg of blocking DNA (Ultrapure salmon sperm DNA - Invitrogen). Transformants were plated using the top agar method on 100 μg/ml Hygromycin B and grown under 100 μmol m−2 s−1 for 3 weeks. Individual lines were transferred to 500 μl of F/2 with 100 μg/ml Hygromycin B (GoldBio) in a 96 deep well plate (Evergreen Biotech). Cultures were then maintained on solid plates.
NanoLuciferase luminescence assays
For Nlux activity screening, 100 μl of cell culture was transferred to a luminescence plate, and 100 μl of F/2 containing Nanoglo substrate (Promega) at a 10,000X dilution. For normalized luminescence measurements, 1.5 million cells per well were transferred to the 96-well luminometer plate, the volume was adjusted to 100 μl with F/2 medium, and 100 μl of F/2 containing Nanoglo substrate (Promega) at a 10,000X dilution was added. Luminescence was measured after 180 sec delay with a 0.3 sec exposure using a Centro XS3 LB960 (Berthold).
N. oceanica colony PCR
A 1X Q5 buffered solution (NEB) with 1 μl of N. oceanica culture per 10 μl of required sample was boiled (100° C) for 10 min. A 10 μl PCR mastermix was added to the 10 μl of sample and PCR conducted according to the manufacturer’s suggestions (NEB). The primers NR F+ and NR R- were used to amplify the NR gene (Table S2).
Episomal DNA isolation from N. oceanica
For DNA isolation 10 ml of culture at mid-log phase (~ 3×107 cells ml−1) was collected and frozen. The protocol of Karas et al. (25) was utilized with some modifications. Briefly, for each reaction 235 μl Qiagen P1 solution was combined with 5 μl lysozyme (25 mg/ml) (Sigma-Aldrich), 2.5 μl macerozyme (100 mg/ml)(Yakult Pharmaceuticals), 2.5 μl zymolyase (10 mg/ml) (Zymo Research), and 5 μl cellulase (100mg/ml)(Yakult Pharmaceuticals) and used to resuspend the cell pellets. The resuspended cell culture was incubated at 37° C for 30 min. After addition of the P2 solution, and S3 solution (Qiagen), the cell debris was pelleted at 14000 × G for 10 min. Supernatant was aspirated and combined with an equal volume of isopropanol, mixed and centrifuged at 14,000 × G at 4° C for 10 min. After decanting the pellet was washed with 750 μl 70% ethanol, centrifuged, decanted, and dried. The isolated DNA was resuspended in Tris-EDTA buffer (TE) and re-isolated by phenol:chloroform extraction. The final DNA was resuspended in 40 μl TE, and the concentration determined using a NanoDrop Lite (Thermo Scientific).
Episome rescue
Equal amounts of DNA isolated from NR-KO, iEV, and WT lines were used for E. coli transformations. For enzyme treatment of the DNA extracted from NR-KO lines, 2 μg of DNA was treated with 10 units of Exonuclease V (NEB) and/or 10 units of ClaI (NEB) in a 20 μl reaction for 1 hour at 37° C. Reactions were heat inactivated at 75° C for 30 min. An equal amount of DNA (500 ng) from enzyme treated samples, mock treated, and untreated samples were used to transform E. coli (DH5α high-efficiency efficiency, NEB). Resulting colonies were counted and random colonies selected for plasmid isolation. Resulting plasmids and control pNOC-ARS-CRISPR-sgNR2 DNA, were digested with AgeI, NotI, and EcorI (NEB) according to manufacturer’s instructions (NEB), and separated on a 0.9% agarose gel.
Episome curing
Episome containing NR-KO lines were grown in liquid 2.5 mM NaNH4 F/2 without Hygromycin B for 10 days. Cultures were diluted to 3,000 cells per ml, and 3,000 cells were plated on NaNH4 F/2. Single colonies developed over 3 weeks, and 48 colonies were isolated and grown in NaNH4 F/2. After 1 week, the Nlux luminescence was measured and low Nlux signal lines were further screened by colony PCR. Primers used were NR F+, NR R-, and epi Cas9 F+and epi Cas9 R- (Table S2).
Segregation efficiency
A growth curve was conducted using an inoculum of ~5×106 cells/ml in F/2 with 2.5 mM NaNH4 and cell density was measured every 24 hours. The doubling time was determined using a best-fit exponential curve and the number of generations deduced by dividing the duration of the experiment by the doubling time. The segregation efficiency was determined according to Iwanaga et al. (25, 31) and using the percentage of cells that maintained luminescence in the curing screen.
Immunoblotting
Immunoblotting was conducted as described previously (2). Protein extract (50 μg) was separated on 8–10% SDS-polyacrylamide gels. After transfer to PVDF membranes Cas9-GFP was detected with α-GFP (Abcam ab5450) 1:1,000 in TBST with 5% BSA followed by donkey α-goat-HRP (Santa Cruz sc-2020) 1:10,000 in TBST with 5% milk, and Cas9-Nlux-HA with α-HA-HRP (Roche 3F10) 1:1,000 in TBST with 5% milk. Detection was conducted by chemilumiscence with Femto substrate (ThermoFisher). After detection, total protein was stained with Direct Blue 71 (34).
Confocal microscopy
Confocal microscopy was performed using an inverted Olympus FluoView1000 confocal laser scanning microscope (Olympus Corporation, USA). Cells grown in F/2 medium were fixed in 70% ethanol overnight at 4°C. Cell pellets were collected and stained with 4’, 6-Diamidino-2-Phenylindole (DAPI, 358/461 - Life Technologies) with a final concentration of 0.2 μg/μl in PBS for 2 hours at 4°C. Cells were washed with PBS and observed using a 100x UPlanSApo oil objective (N.A. 1.4). DAPI fluorochromes were excited using a 405 nm blue diode laser, and the emission signals were filtered using the BA430–470 band pass filter. Cas9-GFP was excited using an argon 488 nm laser, and the emission signals from GFP were filtered using a BA500–530 band pass filter. Post-imaging analyses were performed using Olympus FluoView1000 software.
Southern-blot analysis
We used a procedure described previously (3). Briefly, N. oceanica DNA was isolated by CTAB and 20 μg was digested with SacI and was separated on an agarose gel (0.9%, 70 Volts overnight), followed by blotting to a Hybond Nylon membrane overnight (GE Health Care). Subsequent hybridization and detection was performed with a DIG labeling and detection kit according to the manufacturer’s instructions (Roche Applied Sciences). Two probes were amplified by PCR, one for the HygR gene (with the primers HygR probe F+ and HygR probe R-) and one for the AmpR gene (AmpR probe F+ and AmpR probe R-), and were used for hybridization in PefectHyb Plus hybridization buffer (Sigma-Aldrich).
Supplementary Material
Data S1 contains the description of vector construction and the sequences of the vectors generated in this study (.txt)
Figure S1 showing confocal imaging of wildtype, untargeted GFP expressing, andCas9-GFP expressing N. oceanica cells with the nucleus stained by DAPI; Figure S2 showing cloning strategies for the generation of a ribozyme flanked sgRNA; Figure S3 showing identification of NR knockout mutants by CRISPR/Cas9; Figure S4 showing further verification of rescued episomes; Figure S5 showing southern blot analysis of the episomal and integrated empty-vector CRISPR mutants; Figure S6 showing the selection of cured mutant lines; Figure S7 showing a growth curve in ammonium containing medium; Figure S8 showing a one-vector CRISPR system for scarless cloning of targets
Table S1 showing the list of constructs generated in this study; Table S2 showing the primers used in the study.
Acknowledgements:
We thank Bill Henry for his analysis of the N. oceanica U6 region and Kathryn Meeks for the gift of the hCas9 plasmid. This work was supported by a National Science Foundation grant (IOS-1354721) to EF. T.T. was partly supported by the Plant Biotechnology for Health and Sustainability Training Program at MSU (NIH T32 GM110523). In addition, parts of this work were supported by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (DE-FG02–91ER20021) and MSU-AgBioResearch.
Abbreviations:
- sgRNA
single-guide RNA
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats, single guide RNA
- KO
knockout
- HDV
hepatitis delta virus ribozyme
- HH
hammerhead ribozyme
- CEN/ARS
Saccharomyces cerevisiae centromere and autonomous replication sequence fusion
- NR
nitrate reductase
- HR
homologous recombination
- GFP
Green Fluorescent Protein
- Nlux
NanoLuciferase
- PCR
polymerase chain reaction
- DAPI - 4’
6-Diamidino-2-Phenylindole
- CLSM
confocal laser scanning microscopy
- NLS
nuclear localization signal
Footnotes
The authors report no financial conflicts of interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website
References
- 1.Chew KW, Yap JY, Show PL, Suan NH, Juan JC, Ling TC, Lee D-J, and Chang J-S (2017) Microalgae biorefinery: High value products perspectives, Bioresource Technology 229, 53–62 DOI: 10.1016/j.biortech.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 2.Poliner E, Pulman JA, Zienkiewicz K, Childs K, Benning C, and Farré EM (2017) A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production, Plant Biotechnology Journal DOI: 10.1111/pbi.12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vieler A, Wu G, Tsai C-H, Bullard B, Cornish AJ, Harvey C, Reca I-B, Thornburg C, Achawanantakun R, Buehl CJ, Campbell MS, Cavalier D, Childs KL, Clark TJ, Deshpande R, Erickson E, Armenia Ferguson A, Handee W, Kong Q, Li X, Liu B, Lundback S, Peng C, Roston RL, Sanjaya, Simpson JP, TerBush A, Warakanont J, Zäuner S, Farre EM, Hegg EL, Jiang N, Kuo M-H, Lu Y, Niyogi KK, Ohlrogge J, Osteryoung KW, Shachar-Hill Y, Sears BB, Sun Y, Takahashi H, Yandell M, Shiu S-H, and Benning C (2012) Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779, PLoS Genet. 8, e1003064DOI: 10.1371/journal.pgen.1003064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, and Tredici MR (2009) Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor, Biotechnology and Bioengineering 102, 100–112 DOI: 10.1002/bit.22033 [DOI] [PubMed] [Google Scholar]
- 5.Kumar S (2015) GM Algae for Biofuel Production: Biosafety and Risk Assessment, Vol. 9 DOI: [Google Scholar]
- 6.Kindle KL, Schnell RA, Fernández E, and Lefebvre PA (1989) Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase, J. Cell Biol 109, 2589–2601 DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheah YE, Albers SC, and Peebles CAM (2013) A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803, Biotechnology Progress 29, 23–30 DOI: 10.1002/btpr.1661 [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Tong Y, Pan J, Yang Y, Liu Q, Tan X, Zhao S, Qin L, and Chen X (2016) A redesigned CRISPR/Cas9 system for marker-free genome editing in Plasmodium falciparum, Parasites & Vectors 9 DOI: 10.1186/s13071-016-1487-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Gao F, and Wu S (2016) An episomal CRISPR/Cas9 system to derive vector-free gene modified mammalian cells, Protein & Cell 7, 689–691 DOI: 10.1007/s13238-016-0299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim J-S, Jin E, and Bae S (2016) DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins, Scientific Reports 6, 30620DOI: 10.1038/srep30620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilian O, Benemann CSE, Niyogi KK, and Vick B (2011) High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp, Proceedings of the National Academy of Sciences 108, 21265–21269 DOI: 10.1073/pnas.1105861108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, Voytas DF, Cavarec L, and Duchateau P (2014) Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology, Nature Communications 5 DOI: 10.1038/ncomms4831 [DOI] [PubMed] [Google Scholar]
- 13.Diner RE, Schwenck SM, McCrow JP, Zheng H, and Allen AE (2016) Genetic Manipulation of Competition for Nitrate between Heterotrophic Bacteria and Diatoms, Frontiers in Microbiology 7 DOI: 10.3389/fmicb.2016.00880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weyman PD, Beeri K, Lefebvre SC, Rivera J, McCarthy JK, Heuberger AL, Peers G, Allen AE, and Dupont CL (2015) Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis, Plant biotechnology journal 13, 460–470 DOI: 10.1111/pbi.12254 [DOI] [PubMed] [Google Scholar]
- 15.Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, and Weeks DP (2014) Successful Transient Expression of Cas9 and Single Guide RNA Genes in Chlamydomonas reinhardtii, Eukaryotic Cell 13, 1465–1469 DOI: 10.1128/ec.00213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin S-E, Lim J-M, Koh HG, Kim EK, Kang NK, Jeon S, Kwon S, Shin W-S, Lee B, Hwangbo K, Kim J, Ye SH, Yun J-Y, Seo H, Oh H-M, Kim K-J, Kim J-S, Jeong W-J, Chang YK, and Jeong B. r. (2016) CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii, Scientific Reports 6, 27810DOI: 10.1038/srep27810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ajjawi I, Verruto J, Aqui M, Soriaga LB, Coppersmith J, Kwok K, Peach L, Orchard E, Kalb R, Xu W, Carlson TJ, Francis K, Konigsfeld K, Bartalis J, Schultz A, Lambert W, Schwartz AS, Brown R, and Moellering ER (2017) Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator, Nature Biotechnology 35, 647–652 DOI: 10.1038/nbt.3865 [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Lu Y, Xin Y, Wei L, Huang S, and Xu J (2016) Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9, Plant J 88, 1071–1081 DOI: 10.1111/tpj.13307 [DOI] [PubMed] [Google Scholar]
- 19.Hopes A, Nekrasov V, Kamoun S, and Mock T (2016) Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana, Plant Methods 12 DOI: 10.1186/s13007-016-0148-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nymark M, Sharma AK, Sparstad T, Bones AM, and Winge P (2016) A CRISPR/Cas9 system adapted for gene editing in marine algae, Scientific Reports 6, 24951DOI: 10.1038/srep24951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W-W, and Matlashewski G (2015) CRISPR-Cas9-Mediated Genome Editing in Leishmania donovani, mBio 6, e00861–00815 DOI: 10.1128/mBio.00861-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weninger A, Hatzl A-M, Schmid C, Vogl T, and Glieder A (2016) Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris, Journal of Biotechnology 235, 139–149 DOI: 10.1016/j.jbiotec.2016.03.027 [DOI] [PubMed] [Google Scholar]
- 23.Moon DA, and Goff LJ (1997) Molecular characterization of two large DNA plasmids in the red alga Porphyra pulchra, Curr. Genet 32, 132–138 DOI: 10.1007/s002940050257 [DOI] [PubMed] [Google Scholar]
- 24.Diner RE, Noddings CM, Lian NC, Kang AK, McQuaid JB, Jablanovic J, Espinoza JL, Nguyen NA, Anzelmatti MA, Jansson J, Bielinski VA, Karas BJ, Dupont CL, Allen AE, and Weyman PD (2017) Diatom centromeres suggest a mechanism for nuclear DNA acquisition, Proceedings of the National Academy of Sciences 114, E6015–E6024 DOI: 10.1073/pnas.1700764114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karas BJ, Diner RE, Lefebvre SC, McQuaid J, Phillips APR, Noddings CM, Brunson JK, Valas RE, Deerinck TJ, Jablanovic J, Gillard JTF, Beeri K, Ellisman MH, Glass JI, Hutchison Iii CA, Smith HO, Venter JC, Allen AE, Dupont CL, and Weyman PD (2015) Designer diatom episomes delivered by bacterial conjugation, Nature Communications 6, 6925DOI: 10.1038/ncomms7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diner RE, Bielinski VA, Dupont CL, Allen AE, and Weyman PD (2016) Refinement of the Diatom Episome Maintenance Sequence and Improvement of Conjugation-Based DNA Delivery Methods, Frontiers in bioengineering and biotechnology 4, 65DOI: 10.3389/fbioe.2016.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slattery SS, Diamond A, Wang H, Therrien JA, Lant JT, Jazey T, Lee K, Klassen Z, Desgagne-Penix I, Karas BJ, and Edgell DR (2018) An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum, ACS Synth Biol DOI: 10.1021/acssynbio.7b00191 [DOI] [PubMed] [Google Scholar]
- 28.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, and Wood KV (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate, ACS chemical biology 7, 1848–1857 DOI: 10.1021/cb3002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy JK, Smith SR, McCrow JP, Tan M, Zheng H, Beeri K, Roth R, Lichtle C, Goodenough U, Bowler CP, Dupont CL, and Allen AE (2017) Nitrate Reductase Knockout Uncouples Nitrate Transport from Nitrate Assimilation and Drives Repartitioning of Carbon Flux in a Model Pennate Diatom, The Plant Cell 29, 2047–2070 DOI: 10.1105/tpc.16.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernández E, Schnell R, Ranum LP, Hussey SC, Silflow CD, and Lefebvre PA (1989) Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii, Proceedings of the National Academy of Sciences of the United States of America 86, 6449–6453 DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwanaga S, Kato T, Kaneko I, and Yuda M (2012) Centromere plasmid: a new genetic tool for the study of Plasmodium falciparum, PLoS ONE 7, e33326DOI: 10.1371/journal.pone.0033326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, and Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases, Nat Biotechnol 31, 827–832 DOI: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing, Nucleic Acids Res 42, W401–407 DOI: 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong H-Y, Yoo G-S, and Choi J-K (2002) Detection of Proteins on Blots Using Direct Blue 71, 0, 387–392 DOI: 10.1385/1-59259-169-8:387 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 contains the description of vector construction and the sequences of the vectors generated in this study (.txt)
Figure S1 showing confocal imaging of wildtype, untargeted GFP expressing, andCas9-GFP expressing N. oceanica cells with the nucleus stained by DAPI; Figure S2 showing cloning strategies for the generation of a ribozyme flanked sgRNA; Figure S3 showing identification of NR knockout mutants by CRISPR/Cas9; Figure S4 showing further verification of rescued episomes; Figure S5 showing southern blot analysis of the episomal and integrated empty-vector CRISPR mutants; Figure S6 showing the selection of cured mutant lines; Figure S7 showing a growth curve in ammonium containing medium; Figure S8 showing a one-vector CRISPR system for scarless cloning of targets
Table S1 showing the list of constructs generated in this study; Table S2 showing the primers used in the study.