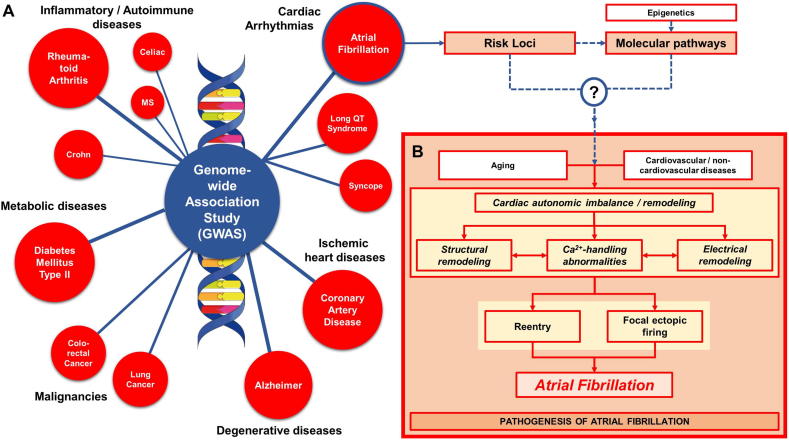
Genome-wide association studies (GWAS) have identified potential pathogenic variants associated with several complex traits/phenotypes in multi-ethnic populations (Fig. 1A), which has helped redesign therapeutic strategies for those particular diseases [1]. For example, the association of genetic variants in PCSK9, identified in classical genetic studies and GWAS, with LDL cholesterol levels and coronary artery disease has led to new therapeutic options [1]. Similarly, in type-2 diabetes mellitus, GWAS have been employed to identify novel risk loci [2], while in rheumatoid arthritis, GWAS have detected the genetic overlap with other immunologic diseases, such as human primary immunodeficiency and hematological cancer, suggesting the potential benefit of repurposing drugs targeting those diseases [3]. GWAS have also been applied to study several electrocardiographic traits, including PR, RR and QT intervals, as well as complex arrhythmias, such as atrial fibrillation (AF) and sudden cardiac death [4].
Fig. 1.
A) Examples of complex traits/phenotypes investigated using GWAS. B) The pathogenesis of atrial fibrillation (AF). Genetic predisposition, advancing age, and acquired factors including cardiovascular and non-cardiovascular diseases promote AF through a variety of mechanisms, notably structural, electrical, autonomic and Ca2+-handling remodeling, leading to the generation of AF-maintaining reentrant waves and/or focal ectopic firing.
AF is a progressive arrhythmia with multifactorial etiologies and pathogenesis, and a significant impact on morbidity and mortality [5]. Since rhythm control with catheter ablation or cardiac surgery can be performed in only a limited number of AF patients [6], antiarrhythmic drugs [7] along with anticoagulation for stroke prevention [8] remain the cornerstone of AF management. Several pathological mechanisms, including electrical remodeling, structural remodeling, autonomic imbalance and calcium-handling abnormalities, contribute to the initiation, maintenance and progression of AF (Fig. 1B) [5]. AF shows a significant heritability, highlighting a genetic predisposition, particularly in the case of lone AF (i.e., AF in the apparent absence of overt cardiovascular disease or atrial pathology). To date, GWAS have identified more than 100 susceptibility loci associated with AF and pathway analyses have revealed the association with cardiac developmental, structural and electrophysiological pathways [9,10], although the magnitude of their contribution and the precise mechanistic role remain largely unknown [11].
In the current issue of the Journal, Ebana et al. [12] employed meta-analysis gene-set enrichment of variant associations (MAGENTA), a computational tool that tests for enrichment of genetic associations in specific biological processes or gene sets, to identify the biological pathways linked to AF-associated loci previously identified in a GWAS of a Japanese population with or without lone AF. Two novel biological pathways were found to be associated with AF: the mammalian target of rapamycin (mTOR), a serine/threonine kinase involved in the regulation of cellular growth and metabolism in response to a wide range of environmental cues, and the CCCTC-binding factor (CTCF) pathway, a transcriptional repressor. Further microarray analyses using human atrial samples and gene-set enrichment analysis (GSEA) supported a role for mTOR, revealing down-regulation of mTOR signaling in patients with AF, while the CTCF pathway was randomly distributed with low enrichment score [12]. These data suggest a potential role for mTOR signaling in AF, in line with previous animal studies showing that cardiac-specific deletion of LKB1, a negative regulator of mTOR, increased mTOR levels and AF susceptibility in mice [13].
Although the MAGENTA and GSEA analyses provide independent evidence for a role of mTOR in two different patient cohorts and methodologies, it is noteworthy that the mTOR pathway was not identified in other, much larger GWAS [9,10]. Conversely, pathways involving genes commonly associated with AF in other GWAS (notably PITX2 and TBX5) could not be identified in the analysis by Ebana et al. [12]. These findings suggest that differences in patient characteristics (including race/ethnicity and type of AF), the origin of the tissue samples, as well as the employed methodologies (e.g., GWAS and GSEA software) might significantly affect the identified biological/molecular pathways. For example, the fact that the mTOR pathway was found in a study of a Japanese population, but has never been reported before in larger multi-ethnic studies suggests a potential sensitivity of pathway analyses to the race/ethnic diversity of the sample populations. Likewise, the failure to detect pathways involving PITX2 and TBX5 could indicate that their contribution to lone AF may be more subtle than in patients with paroxysmal or persistent AF, making the GWAS underpowered to detect a significant association. In this respect, it is interesting that the current study [12] identified a role for mTOR in both the GWAS of lone-AF patients and in atrial samples of patients undergoing open heart surgery with valvular AF.
Although the extensive employment of the GWAS technology has generated multiple novel hypotheses of AF pathophysiology, the mechanistic role of most risk loci for AF identified using GWAS remains unknown. The AF risk locus at 4q25, the strongtest signal in all AF-related GWAS typically attributed to PITX2, represents a prime example, with modest associations between genetic variants, PITX2 levels and outcomes, as well as multiple proposed mechanisms [14]. Compared to other applications, the use of GWAS-based pathway analysis in AF might be particularly challenging due to the large number of potential mechanisms that can promote AF. Moreover, the genetic predisposition to AF interacts with numerous acquired factors such as age, life-style (including exercise [15]) and other comorbidities [14]. These mechanisms are distinct for different types of AF (e.g., paroxysmal vs. persistent AF) and operate over different time courses [14]; information which is not typically taken into account in GWAS populations. Another challenge for the application of GWAS in AF is the likely contribution of functional alterations of the genome (i.e., epigenetics), including DNA methylation, histone modifications and microRNA regulation. For example, in recent years, several microRNAs have been identified to regulate AF-associated electrical, structural and Ca2+-handling remodeling [16]. Nevertheless, it should be noted that almost all of the risk loci and pathways associated with AF that have been identified using GWAS to date are not considered as targets of traditional antiarrhythmic drugs, which might partially explain the limited efficacy of currently-available anti-AF therapies [7].
Taken together, these findings highlight the complexity of AF and the importance of understanding the contribution of its genetic predisposition for better management of the disease. The application of novel pathway analyses, including MAGENTA and Bayesian network analysis [12,17], in large, well-phenotyped cohorts with specific AF subtypes together with hypothesis-driven experimental studies may facilitate a better mechanistic understanding of AF risk loci. Ultimately, this may contribute to improved therapeutic options, although this remains a distant goal at present.
Conflict of interest
None (all authors).
Acknowledgments
Acknowledgments
The authors’ work is supported by the Netherlands Organization for Scientific Research (ZonMW Veni 91616057 to J.H.), the National Institutes of Health (R01-HL131517 and R01-HL136389 to D.D.), the German Research Foundation (DFG, Do 769/4-1 to D.D.).
References
- 1.Lieb W., Vasan R.S. Scientific contributions of population-based studies to cardiovascular epidemiology in the GWAS era. Front. Cardiovasc. Med. 2018;5:57. doi: 10.3389/fcvm.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imamura M., Takahashi A., Yamauchi T., Hara K., Yasuda K., Grarup N., Zhao W., Wang X., Huerta-Chagoya A., Hu C., Moon S., Long J., Kwak S.H., Rasheed A., Saxena R., Ma R.C., Okada Y., Iwata M., Hosoe J., Shojima N., Iwasaki M., Fujita H., Suzuki K., Danesh J., Jorgensen T., Jorgensen M.E., Witte D.R., Brandslund I., Christensen C., Hansen T., Mercader J.M., Flannick J., Moreno-Macias H., Burtt N.P., Zhang R., Kim Y.J., Zheng W., Singh J.R., Tam C.H., Hirose H., Maegawa H., Ito C., Kaku K., Watada H., Tanaka Y., Tobe K., Kawamori R., Kubo M., Cho Y.S., Chan J.C., Sanghera D., Frossard P., Park K.S., Shu X.O., Kim B.J., Florez J.C., Tusie-Luna T., Jia W., Tai E.S., Pedersen O., Saleheen D., Maeda S., Kadowaki T. Genome-wide association studies in the Japanese population identify seven novel loci for type 2 diabetes. Nat. Commun. 2016;7 doi: 10.1038/ncomms10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R.R., Manoharan A., Ortmann W., Bhangale T., Denny J.C., Carroll R.J., Eyler A.E., Greenberg J.D., Kremer J.M., Pappas D.A., Jiang L., Yin J., Ye L., Su D.F., Yang J., Xie G., Keystone E., Westra H.J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E.A., Diogo D., Cui J., Liao K., Guo M.H., Myouzen K., Kawaguchi T., Coenen M.J., van Riel P.L., van de Laar M.A., Guchelaar H.J., Huizinga T.W., Dieude P., Mariette X., Bridges S.L., Jr., Zhernakova A., Toes R.E., Tak P.P., Miceli-Richard C., Bang S.Y., Lee H.S., Martin J., Gonzalez-Gay M.A., Rodriguez-Rodriguez L., Rantapaa-Dahlqvist S., Arlestig L., Choi H.K., Kamatani Y., Galan P., Lathrop M., R. consortium, G. consortium, Eyre S., Bowes J., Barton A., de Vries N., Moreland L.W., Criswell L.A., Karlson E.W., Taniguchi A., Yamada R., Kubo M., Liu J.S., Bae S.C., Worthington J., Padyukov L., Klareskog L., Gregersen P.K., Raychaudhuri S., Stranger B.E., De Jager P.L., Franke L., Visscher P.M., Brown M.A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T.W., Siminovitch K.A., Momohara S., Matsuda F., Yamamoto K., Plenge R.M. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husser D., Buttner P., Stubner D., Ueberham L., Platonov P.G., Dinov B., Arya A., Hindricks G., Bollmann A. PR interval associated genes, atrial remodeling and rhythm outcome of catheter ablation of atrial fibrillation-a gene-based analysis of GWAS data. Front. Genet. 2017;8:224. doi: 10.3389/fgene.2017.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heijman J., Voigt N., Nattel S., Dobrev D. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ. Res. 2014;114(9):1483–1499. doi: 10.1161/CIRCRESAHA.114.302226. [DOI] [PubMed] [Google Scholar]
- 6.Engelsgaard C.S., Pedersen K.B., Riber L.P., Pallesen P.A., Brandes A. The long-term efficacy of concomitant maze IV surgery in patients with atrial fibrillation. Int. J. Cardiol. Heart Vasc. 2018;19:20–26. doi: 10.1016/j.ijcha.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dan G.A., Dobrev D. Antiarrhythmic drugs for atrial fibrillation: imminent impulses are emerging. Int. J. Cardiol. Heart Vasc. 2018;21:11–15. doi: 10.1016/j.ijcha.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang T.Y., Liao J.N., Chao T.F., Vicera J.J., Lin C.Y., Tuan T.C., Lin Y.J., Chang S.L., Lo L.W., Hu Y.F., Chung F.P., Chen S.A. Oral anticoagulant use for stroke prevention in atrial fibrillation patients with difficult scenarios. Int. J. Cardiol. Heart Vasc. 2018;20:56–62. doi: 10.1016/j.ijcha.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roselli C., Chaffin M.D., Weng L.C., Aeschbacher S., Ahlberg G., Albert C.M., Almgren P., Alonso A., Anderson C.D., Aragam K.G., Arking D.E., Barnard J., Bartz T.M., Benjamin E.J., Bihlmeyer N.A., Bis J.C., Bloom H.L., Boerwinkle E., Bottinger E.B., Brody J.A., Calkins H., Campbell A., Cappola T.P., Carlquist J., Chasman D.I., Chen L.Y., Chen Y.I., Choi E.K., Choi S.H., Christophersen I.E., Chung M.K., Cole J.W., Conen D., Cook J., Crijns H.J., Cutler M.J., Damrauer S.M., Daniels B.R., Darbar D., Delgado G., Denny J.C., Dichgans M., Dorr M., Dudink E.A., Dudley S.C., Esa N., Esko T., Eskola M., Fatkin D., Felix S.B., Ford I., Franco O.H., Geelhoed B., Grewal R.P., Gudnason V., Guo X., Gupta N., Gustafsson S., Gutmann R., Hamsten A., Harris T.B., Hayward C., Heckbert S.R., Hernesniemi J., Hocking L.J., Hofman A., Horimoto A., Huang J., Huang P.L., Huffman J., Ingelsson E., Ipek E.G., Ito K., Jimenez-Conde J., Johnson R., Jukema J.W., Kaab S., Kahonen M., Kamatani Y., Kane J.P., Kastrati A., Kathiresan S., Katschnig-Winter P., Kavousi M., Kessler T., Kietselaer B.L., Kirchhof P., Kleber M.E., Knight S., Krieger J.E., Kubo M., Launer L.J., Laurikka J., Lehtimaki T., Leineweber K., Lemaitre R.N., Li M., Lim H.E., Lin H.J., Lin H., Lind L., Lindgren C.M., Lokki M.L., London B., Loos R.J.F., Low S.K., Lu Y., Lyytikainen L.P., Macfarlane P.W., Magnusson P.K., Mahajan A., Malik R., Mansur A.J., Marcus G.M., Margolin L., Margulies K.B., Marz W., McManus D.D., Melander O., Mohanty S., Montgomery J.A., Morley M.P., Morris A.P., Muller-Nurasyid M., Natale A., Nazarian S., Neumann B., Newton-Cheh C., Niemeijer M.N., Nikus K., Nilsson P., Noordam R., Oellers H., Olesen M.S., Orho-Melander M., Padmanabhan S., Pak H.N., Pare G., Pedersen N.L., Pera J., Pereira A., Porteous D., Psaty B.M., Pulit S.L., Pullinger C.R., Rader D.J., Refsgaard L., Ribases M., Ridker P.M., Rienstra M., Risch L., Roden D.M., Rosand J., Rosenberg M.A., Rost N., Rotter J.I., Saba S., Sandhu R.K., Schnabel R.B., Schramm K., Schunkert H., Schurman C., Scott S.A., Seppala I., Shaffer C., Shah S., Shalaby A.A., Shim J., Shoemaker M.B., Siland J.E., Sinisalo J., Sinner M.F., Slowik A., Smith A.V., Smith B.H., Smith J.G., Smith J.D., Smith N.L., Soliman E.Z., Sotoodehnia N., Stricker B.H., Sun A., Sun H., Svendsen J.H., Tanaka T., Tanriverdi K., Taylor K.D., Teder-Laving M., Teumer A., Theriault S., Trompet S., Tucker N.R., Tveit A., Uitterlinden A.G., Van Der Harst P., Van Gelder I.C., Van Wagoner D.R., Verweij N., Vlachopoulou E., Volker U., Wang B., Weeke P.E., Weijs B., Weiss R., Weiss S., Wells Q.S., Wiggins K.L., Wong J.A., Woo D., Worrall B.B., Yang P.S., Yao J., Yoneda Z.T., Zeller T., Zeng L., Lubitz S.A., Lunetta K.L., Ellinor P.T. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018;50(9):1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G., Surakka I., Mathis M.R., Yamazaki M., Crawford R.D., Gabrielsen M.E., Skogholt A.H., Holmen O.L., Lin M., Wolford B.N., Dey R., Dalen H., Sulem P., Chung J.H., Backman J.D., Arnar D.O., Thorsteinsdottir U., Baras A., O'Dushlaine C., Holst A.G., Wen X., Hornsby W., Dewey F.E., Boehnke M., Kheterpal S., Mukherjee B., Lee S., Kang H.M., Holm H., Kitzman J., Shavit J.A., Jalife J., Brummett C.M., Teslovich T.M., Carey D.J., Gudbjartsson D.F., Stefansson K., Abecasis G.R., Hveem K., Willer C.J. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018;50(9):1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng L.C., Choi S.H., Klarin D., Smith J.G., Loh P.R., Chaffin M., Roselli C., Hulme O.L., Lunetta K.L., Dupuis J., Benjamin E.J., Newton-Cheh C., Kathiresan S., Ellinor P.T., Lubitz S.A. Heritability of atrial fibrillation. Circ. Cardiovasc. Genet. 2017;10(6) doi: 10.1161/CIRCGENETICS.117.001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebana Y., Sun Y., Yang X., Watanabe T., Makita S., Ozaki K., Tanaka T., Arai H., Furukawa T. Pathway analysis with genome-wide association study data detected the association of atrial fibrillation with the mTOR signaling pathway. Int. J. Cardiol. Heart Vasc. 2019;24 doi: 10.1016/j.ijcha.2019.100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda Y., Sato K., Pimentel D.R., Sam F., Shaw R.J., Dyck J.R., Walsh K. Cardiac-specific deletion of LKB1 leads to hypertrophy and dysfunction. J. Biol. Chem. 2009;284(51):35839–35849. doi: 10.1074/jbc.M109.057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heijman J., Guichard J.B., Dobrev D., Nattel S. Translational challenges in atrial fibrillation. Circ. Res. 2018;122(5):752–773. doi: 10.1161/CIRCRESAHA.117.311081. [DOI] [PubMed] [Google Scholar]
- 15.Ayinde H., Schweizer M.L., Crabb V., Ayinde A., Abugroun A., Hopson J. Age modifies the risk of atrial fibrillation among athletes: a systematic literature review and meta-analysis. Int. J. Cardiol. Heart Vasc. 2018;18:25–29. doi: 10.1016/j.ijcha.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N. Editorial commentary: the emerging role of epigenetics in atrial fibrillation. Trends Cardiovasc. Med. 2016;26(4):319–320. doi: 10.1016/j.tcm.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Heijman J., Dobrev D. Bayesian network analyses in atrial fibrillation - a path to better therapies? Int. J. Cardiol. Heart Vasc. 2019;22:210–211. doi: 10.1016/j.ijcha.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]