Abstract
Impressive responses have been observed in patients with cancer treated with checkpoint inhibitory anti-programmed cell death-1 (PD-1) or anti-cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) antibodies through disinhibiting the immune system. However, tumors possess complex immunosuppressive tumor microenvironment to present therapeutic obstacles and the response rates to immune checkpoint inhibition remain low. One significant barrier to the efficacy of anti-PD1 treatment is the recruitment of myeloid-derived suppressor cells (MDSCs) into the tumor. MDSCs dramatically increased in peripheral blood of patients with osteosarcoma and prohibited both T-cell activation and infiltration. Here we demonstrated functional inhibition of G-MDSCs with (S)-(-)-N-[2-(3-Hydroxy-1H-indol-3-yl)-methyl]-acetamide (SNA), a specific inhibitor of PI3Kδ/γ, could prime tumor microenvironment, resultantly enhancing the therapeutic efficacy of anti-PD1 treatment in a syngeneic osteosarcoma tumor model. Combining SNA with anti-PD1 dramatically slowed osteosarcoma tumor growth and prolonged survival time of tumor-bearing mice, at least in part mediated through CD8+ T cells. Our results demonstrated that addition of SNA to anti-PD1 significantly altered infiltration and function of innate immune cells, providing the rationale for combination therapy in patients with osteosarcoma through inhibiting the function of MDSCs with a selective PI3Kδ/γ inhibitor to enhance responses to immune checkpoint blockade.
Key words: (S)-(-)-N-[2-(3-Hydroxy-1H-indol-3-yl)-methyl]-acetamide, MDSCs, Osteosarcoma, Anti-PD1
1. Introduction
Osteosarcoma (OS) is considered the most common type of primary malignant bone tumor with a high rate of mortality in adolescents and children [1], [2]. Although the survival time has significantly increased due to the advances in multimodal therapy including surgery, combination chemotherapy and external beam radiation, disease-free survival and 5-year overall survival remains 50-60%, and almost 40% of OS morality is caused by lung metastases [3], [4]. Therefore, it is definitely necessary to search for novel therapeutic agents with effective therapeutic strategy against osteosarcoma.
Immunotherapy has revolutionized treatment in a variety of cancer types by enabling infiltration of T cells into the tumor microenvironment and promoting cytotoxic signaling pathways, especially for the emergence of immune checkpoint inhibitors. However, most of cancer types are still ineffective to the immune checkpoint blockade because of tumor-mediated immunosuppression [5]. Various types of cells are involved in the immune suppression, such as tumor associated macrophages (TAMs), regulatory T cells (Treg), and myeloid derived suppressor cells (MDSCs) [6]. TAMs arise from differentiation of circulating MDSCs and monocytes, as well as from recruitment of tissue macrophages. With a spectrum of dynamic functional polarization, M1-polarized TAMs facilitate inflammation, while M2-polarized TAMs hamper adaptive immunity and facilitate invasion, metastasis, and the epithelial-to-mesenchymal transition through production of immunosuppressive cytokines, depletion of metabolic substrate, and T/NK cell inhibition [7].
MDSCs are immature monocytes and granulocytes (expressing a Gr1+CD11b+ phenotype), possessing the capacity to impede immune-mediated clearance of malignant cells through suppressing the proliferation of both CD4+ and CD8+ T cells by multiple mechanisms, including the formation of immunosuppressive reactive oxygen species via inducible nitric oxide synthase (iNOS) and arginase 1 production [8], [9]. Major subtypes of MDSCs include granulocytic MDSCs (G-MDSCs) and monocytic MDSCs (M-MDSCs). G-MDSCs have a morphology similar to that of granulocytes and M-MDSCs have a morphology similar to that of monocytes. In mice, G-MDSCs have a phenotype of CD11b+Ly6G+Ly6C−, whereas M-MDSCs have a phenotype of CD11b+Ly6G−Ly6Chigh. Although they differ in their phenotype and potency, they share a common ability to suppress effector T cell activity [10]. MDSCs are assumed to facilitate the growth and spread of tumors, and have been established to correlate with clinical metastatic burden and stage. Several approaches to target MDSCs to increase the efficacy of immunotherapy in clinic have been proposed, including eliminating MDSCs [11], blocking the recruitment of MDSCs to the tumor microenvironment [12], and targeting their immunosuppressive features [13].
Phosphatidylinositide 3-kinases (PI3Ks) are known to play important roles in hematopoietic cell biology and can activate Gr1+/CD11b+ myeloid cells [14], [15]. Class I PI3K isoforms include p110δ and p110γ. A recent study demonstrated that inhibiting tumor-infiltrating myeloid cells with p110γ-specific inhibitor sensitized tumors to anti-PD1-based therapy [16]. (S)-(-)-N-[2-(3-Hydroxy-1H-indol-3-yl)-methyl]-acetamide (SNA), a novel compound isolated from the leaves of Selaginella pulvinata [17], was found to inhibit p110δ/γ in our preliminary studies.
Therefore, it seems reasonable to investigate whether inhibiting PI3K with SNA would reverse MDSCs-mediated immunosuppression in osteosarcoma. Our results demonstrated that SNA potently inhibited the functional activity of G-MDSCs and improved responses to anti-PD1 treatment against osteosarcoma, validating the approach of targeting immunosuppressive myeloid cells with PI3K inhibitors to enhance the therapeutic efficacy of immune checkpoint inhibitors in patients with osteosarcoma.
2. Materials and Methods
2.1. Mouse tumor model and treatment
Female Balb/c mice at six-week old were purchased from Vital River Lab (Shanghai, China). Mice were maintained in a temperature controlled room with a 12/12-h light/dark schedule and had free access to water and food during the whole experimental period. All animal experimental procedures were performed in compliance with the Chinese legislation on the use and care of laboratory animals and approved by the Institutional Animal Care and Use Committee of Henan Provincial People's Hospital (protocol No.: 2017-1123).
Murine lymphoma cell line EL4 was purchased from Shanghai Cell Bank (Shanghai, China) and cultured in DMEM media (Life Technologies) supplemented with 10% FBS (Gibco), 1% streptomycin/penicillin at 37 °C, 95% air/5% CO2. Osteosarcoma cell line K7M2 was purchased from Shanghai Cell Bank (Shanghai, China) and cultured in RPMI1640 media (Life Technologies) supplemented with 10% FBS (Gibco), 1% streptomycin/penicillin at 37 °C, 95% air/5% CO2. In the s.c. tumor model, 5 × 105 K7M2 cells were subcutaneously injected into the right flank of Balb/c mice. Tumor volume was recorded by two-dimensional digital caliper measurements of the greatest longitudinal diameter (length) and the greatest transverse diameter (width), and calculated by the formula: ½ × (length × width2). When tumor volume reached 50-100 mm3, tumor-bearing mice were divided into different groups by tumor burden (5 mice per group). Mice were treated with SNA (1 mg/kg, daily I.P. injection), anti-PD1 antibody (Clone: 29F.1A12, Bio X cell, 5 mg/kg, weekly I.P. injection) or the combination, mice treated with PBS or IgG as the negative control. CD8 mAb (clone YTS 169.4) or isotype control treatments were performed via intraperitoneal injection (200 mg/injection). When the tumor volume reached 1500 mm3, all mice were sacrificed to collect blood and tumor for analysis, normal mice as the control. In the lung metastasis study, 6 × 105 cells were injected into Balb/c mice by tail vein. 14 days after cell injection, tumor-bearing mice were randomly grouped and treated with SNA (1 mg/kg, daily I.P. injection), anti-PD1 antibody (Clone: 29F.1A12, Bio X cell, 5 mg/kg, weekly I.P. injection) or the combination, mice treated with IgG as the negative control. Mice were sacrificed when they looked distressed.
2.2. Flow cytometric analysis of immune populations
Blood was collected from normal mice and tumor-bearing mice. Tumor tissues were removed from the mice after euthanasia. Single tumor cell suspension was obtained by slicing of the tumor tissues followed by digestion with tumor disassociation kit (Miltenyi Biotec, USA) at 37 °C. After digestion, the suspension was filtered through 70 µm cell strainers to get rid of debris. Mononuclear cells were enriched by percoll gradient centrifuging. Cells were stimulated with PMA/ionomyocin in the presence of Golgi stop and Monesin (eBioscience) for 4 h. Cells were washed with PBS and then stained with Live/Dead dye (Life Technologies) and fluorochrome labeled antibodies specific to cell surface markers (BioLegend). After fixation-permeabilization (Fixation/Permeabilization buffer, eBioscience), cells were stained with fluoro-conjugated antibodies specific to IFNγ and granzyme B (BioLegend) or with the isotype control antibodies. Flow cytometry analysis was performed on BD LSRFortessa and data was analyzed by FlowJo V10.
2.3. Cell viability measurement
The cytotoxicity of SNA to K7M2 cells was measured by MTT assay. Briefly, 2 × 104 cells/well were seeded to the 96-well plate and treated with SNA (50 nM-10 µM) for 48 h. After washing, 10 µL 0.5 mg/mL MTT solution was added into each well and incubated at 37 °C for 4 h. Culture media were removed and the formazan crystals were solubilized with 100 µL DMSO. The absorbance was measured at 570 nm by a microplate reader.
2.4. Proliferation assay of T-lymphocytes
T-lymphocytes were isolated from spleens of naive Balb/c mice. 1 × 106 cells/mL T-lymphocytes were stimulated by CD3/28 antibodies and then stained with 5 mM carboxyfluorescein succinimidyl ester (CFSE, Sigma). For antigen-specific experiments, T-lymphocytes were exposed to irradiated naive splenocytes (20 Gy) pulsed with OVA257-264 (SIINFEKL, 1 µg/mL; InVivoGen). T-lymphocytes were co-incubated with sorted G-MDSCs, SNA (1 µM), nor-NOHA or L-NMMA (300 µM each, Cayman Chemicals) for 4 h prior to stimulation, and quantified by flow cytometry 72 h later. Proliferation was quantified as the average number of divisions for all cells. The formula: (divisions of non-treated group - divisions of treated group)/divisions of non-treated group × 100% was used to calculate the inhibition rate of T-lymphocyte proliferation.
2.5. Killing assay of T-lymphocytes
Splenocytes of mice were cultured with SIINFEKL (2 mg/mL). Cytotoxic T-lymphocytes were exposed to SIINFEKL-pulsed (2 µg/mL) EL4 cells labeled with indium111 (10:1 E:T ratio) in the presence or absence of G-MDSCs and specific inhibitors for 4 h. Supernatants were analyzed for γ radiation counts on a WIZARD2 Automatic Gamma Counter (PerkinElmer).
2.6. Production of IFNγ from T-lymphocytes
5 × 105 T-lymphocytes from spleens of naive mice were incubated with or without G-MDSCs and specific inhibitors. Sorted lymph nodes, splenic or tumor infiltrating lymphocytes (5 × 105 cells) from tumor-bearing mice were co-incubated with irradiated (50 Gy) and IFNγ-pretreated (20 ng/mL, 24 h) K7M2 cells at a 10:1 ratio for 48 h. IFNγ levels were quantified by Mouse IFN gamma ELISA Ready-SET-Go Kit at 450 nm (eBioscience).
2.7. Immunohistochemistry
Tumor tissues were removed from the mice after euthanasia and fixed in 4% paraformaldehyde for 4 h at 4 °C. Tissues were then processed through a graded series of sucrose in PBS (10% for 1 h, 20% for 1 h and 30% for overnight) at 4°C, and embedded in optimum temperature cutting compound. Samples were frozen on dry ice and stored at -80 °C. 10 µm-thick frozen sections were cut sagittally and placed onto slides. Sections were incubated in PBS containing 5% normal goat serum, 1% BSA and 0.5% Triton X-100 for 1 h, followed by incubation with primary antibodies (anti-CD3 FITC and anti-CD8 PE) overnight at 4°C. After washing with PBS, sections were mounted with VECTASHIELD mounting medium with DAPI and visualized under the fluorescence microscope (Olympus, Shinjuku, Japan).
2.8. Quantitative real-time PCR
Total RNA was extracted with Trizol reagent and mRNA was transcribed into cDNA by adding 0.5 µg RNA with SuperScript master mix (Bio-Rad, CA, USA). Quantitative PCR was run on StepOne systems (Thermo Fisher) with SYBR green Supermix (Thermo Fisher) using comparative Ct value method. The gene expression level was normalized to the housekeeping gene Gapdh. The primer sequences are: for Nos2, forward: cctgtgttccaccaggagat; reverse: ccctggctagtgcttcagac, NCBI Reference: NM_010927.4; for Arg1, forward: ccaatggtcaggttgattcc; reverse: cccacccagtgatcttgact, GenBank: AB047402.1; for Pik3ca, forward: gagacaggatgggtcaagga; reverse: aatgcaatctgggtgactcc, NCBI Reference: NM_008839.2; for Pik3cb, forward: tgaagctagcccctgtgact; reverse: aaagggagaggaaggttgga, NCBI Reference: NM_029094.3; for Pik3cd, forward: gatgcacgtaccatcagtgg; reverse: ggtggtcttctgggaactca, NCBI Reference: NM_001164052.1; for Pik3cg, forward: ggtaccctggtgatcgagaa; reverse: agaaggcacaggtccagaga, NCBI Reference: NM_020272.2; for Gapdh, forward: acccagaagactgtggatgg; reverse: cacattgggggtaggaacac, NCBI Reference: GenBank: GU214026.1.
2.9. Western blot analysis
Protein was extracted with lysis buffer and the concentration was determined by BCA method (Bio-Rad, CA, USA). Protein with equal amounts (40 μg) was subjected to SDS-PAGE with 4-12% gel and then transferred to polyvinylidene fluoride membranes. The membranes were blocked with 1% BSA in PBS containing 0.1% Tween-20. After washing, the membranes were incubated with primary antibodies (anti-phospho-S6 (Ser240/244) rabbit mAb CST#5364, 1:1000; anti-phospho-Akt (Ser473) rabbit mAb CST#9271, 1:1000; anti-β-actin rabbit pAb ab8227, 1:3000) overnight at 4 °C, and subsequently with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG H&L HRP, ab205718, 1:5000) for 1 h at room temperature. Protein bands were detected by the ECL system (Pierce, IL, USA).
2.10. Statistical analysis
Data were analyzed with SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Kolmogorov-Smirnov test was used for normality test. Kaplan-Meier survival analysis was performed to compare differences among the groups and p value was calculated with the log-rank test. Measurement data between two groups were compared using the t-test; measurement data among multiple groups were compared using one-way analysis of variance followed by Dunnett's t-test. p < 0.05 was considered as significant difference. The experiments have been repeated to ensure reproducibility of the observations.
3. Results
3.1. MDSC subset levels increased in tumor-bearing mice and PI3K δ/γ isoforms expressed more on G-MDSCs
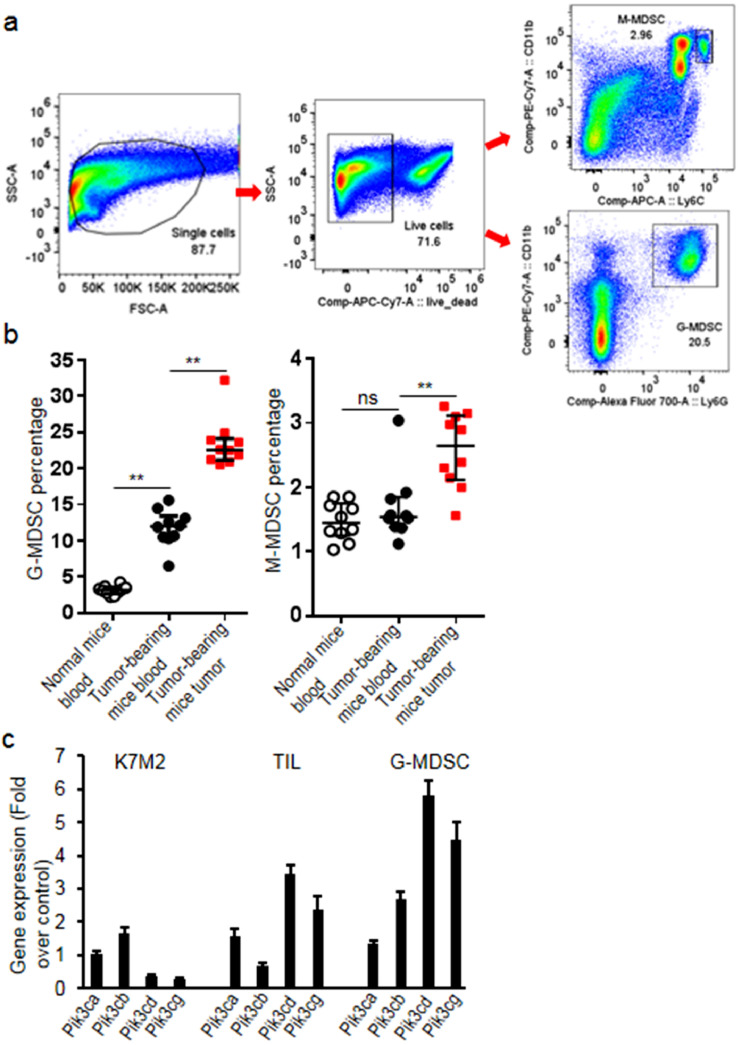
MDSC subsets were analyzed by flow staining for CD11b, Ly6G and Ly6C. Fig. 1a showed the representative flow cytometry graphics of MDSC subsets with the gating scheme. Comparing with non-tumor-bearing mice, G-MDSCs significantly increased in blood (increased 2.7 folds comparing to normal mice, p = 0.003) and tumor tissues (increased 0.99 fold comparing to the level in blood of tumor-bearing mice, p = 0.004) of tumor-bearing mice (Fig. 1b). The transcript levels of PI3K class I isoforms in K7M2 cells, sorted TILs and G-MDSCs from K7M2 tumors were measured and results demonstrated that PI3K δ/γ isoforms expressed 1 fold more on sorted G-MDSCs than TILs (p = 0.005, Fig. 1c).
Fig. 1.
G-MDSCs significantly increased in the K7M2 tumor-bearing mice. (a) Blood and single tumor lysates were stainded with anti-Ly6G AF700, anti-Ly6C APC, anti-CD11b PE-Cy7 and analyzed with flow cytometry (50000 events were acquired for blood and 100000 events were acquired for tumor cells). Representative flow cytometry graphics showing the gating strategy and MDSC subsets from normal mice and tumor-bearing mice were presented. M-MDSCs were defined as CD11b+Ly6C+ and G-MDSCs as CD11b+Ly6G+. (b) The percentage of G-MDSCs significantly increased in the tumor-bearing mice; (c) Transcript levels of PI3K subunits in K7M2 cells, sorted TILs and sorted G-MDSCs from K7M2 tumor tissues. Data were expressed as mean±SD (n = 10) for bar chart and expressed as median±interquartile (n = 10) for scatter plot. **p < 0.01. The in vivo experiments were performed independently twice. The in vitro experiments were performed independently three times.
3.2. G-MDSCs suppressed T-lymphocyte function through arginase and iNOS
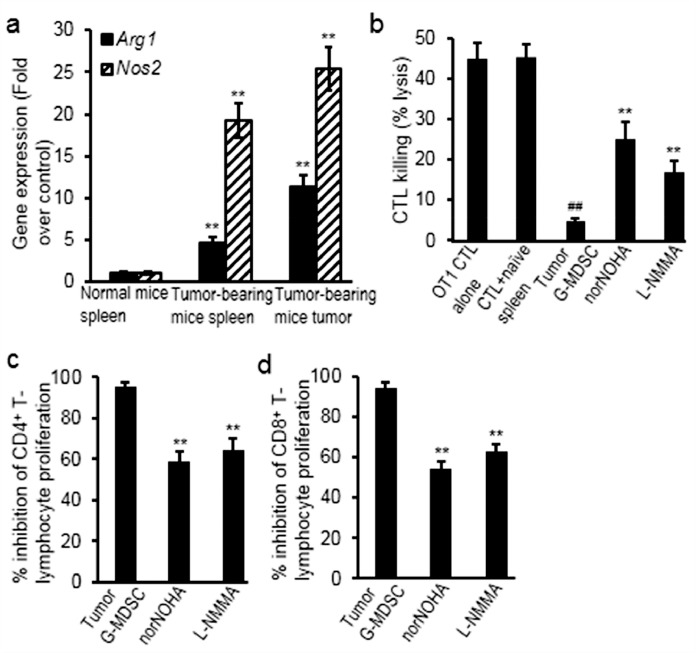
The gene expression of immunosuppressive enzymes was evaluated in splenic and tumor myeloid cells from the tumor-bearing mice, compared with levels in spleen of non-tumor-bearing mice. Results showed that expression of arginase and iNOS significantly increased (Arg1 increased 3.54 folds in spleen, p = 0.009; Arg1 increased 9.97 folds in tumor, p = 0.005; Nos2 increased 17.59 folds in spleen, p = 0.003; Nos2 increased 23.56 folds in tumor, p = 0.001) (Fig. 2a). Moreover, sorted G-MDSC cells from tumors significantly suppressed OT-1 antigen-specific CTL lysis of SIINFEKL-pulsed target cells and the suppressive effects were reversed in the presence of arginase inhibitor (norNOHA) (increased 4.2 folds, p = 0.004) and iNOS inhibitor (L-NMMA) (increased 2.4 folds, p = 0.005) (Fig. 2b). Furthermore, sorted G-MDSC cells from tumors also inhibited the proliferation of CD4+ and CD8+ T-lymphocytes, which was partially reversed by norNOHA (decreased 38% for CD4+, p = 0.005; decreased 42% for CD8+, p = 0.004) and L-NMMA (decreased 32% for CD4+, p = 0.007; decreased 33% for CD8+, p = 0.006) (Fig. 2c).
Fig. 2.
G-MDSCs suppressed the proliferation and cytolytic activities of T-lymphocytes through arginine and iNOS. (a) Gene expression of Arg1 and Nos2 significantly increased in splenic and tumor G-MDSCs from tumor-bearing mice, comparing to splenocytes from the normal mice; (b) norNOHA (arginase inhibitor) or L-NMMA (iNOS inhibitor) inhibited the suppressive effects of tumor-infiltrating G-MDSCs on the cytolytic activities of T-lymphocytes; (c) norNOHA or L-NMMA inhibited the suppressive effects of tumor-infiltrating G-MDSCs on the proliferation of T-lymphocytes. Data were expressed as mean±SD (n = 10). ##p < 0.01 vs OT1 CTL alone; **p < 0.01 vs normal mice or non-treated group. The in vitro experiments were performed independently three times.
3.3. SNA inhibited the function of G-MDSCs
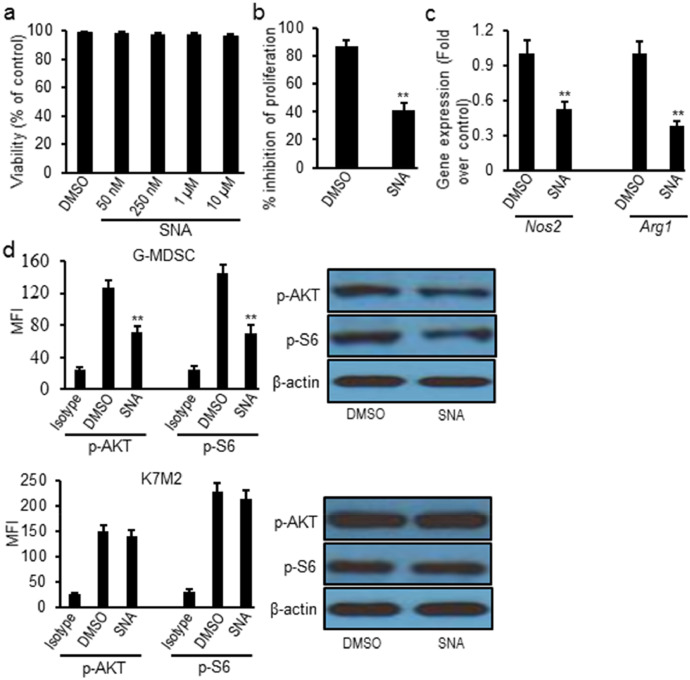
Next, we studied the effects of SNA on osteosarcoma tumor cells and found that SNA treatment didn't cause obvious cytotoxic effect on K7M2 cells comparing to the negative control, as shown by MTT assay (Fig. 3a). However, SNA treatment reversed the suppressive effects of G-MDSCs on the proliferation of T lymphocytes (decreased 53%, p = 0.006) (Fig. 3b). Further studies showed SNA treatment significantly reduced the expression of Nos2 (decreased 48%, p = 0.005) and Arg1 (decreased 62%, p = 0.003) transcript levels on G-MDSC cells from tumors (Fig. 3c). SNA treatment also decreased p-AKT (decreased 43%, p = 0.003) and p-S6 (decreased 52%, p = 0.002) expression in G-MDSCs, but not in K7M2 cells (Fig. 3d).
Fig. 3.
SNA treatment significantly reversed the suppressive capacity of tumor-infiltrating G-MDSCs ex vivo. (a) SNA treatment didn't cause cytotoxic effect on K7M2 cells measured by MTT assay; (b) SNA treatment inhibited the suppressive effects of tumor-infiltrating G-MDSCs on the proliferation of T-lymphocytes; (c) SNA treatment decreased Nos2 and Arg1 transcript levels in G-MDSCs sorted from tumor tissues; (d) SNA treatment decreased the expression of p-AKT and p-S6 on sorted tumor-infiltrating G-MDSCs, but not on K7M2 cells via intracellular flow cytometry and western blot. Data were expressed as mean±SD (n = 10). **p < 0.01 vs DMSO group. The in vitro experiments were performed independently three times.
3.4. SNA treatment primed the tumor microenvironment
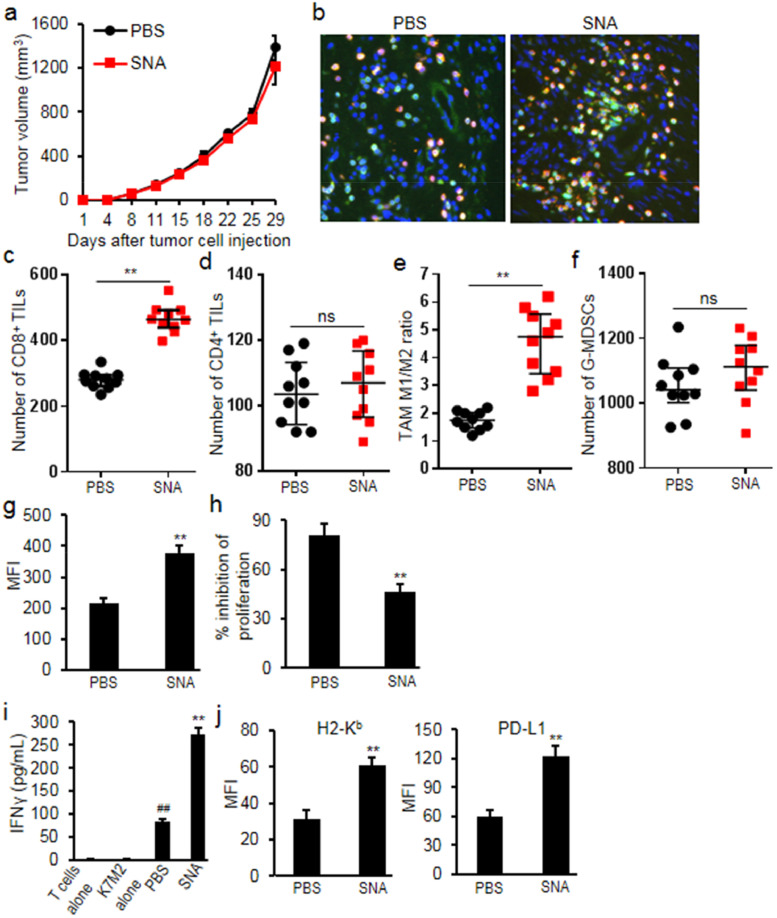
The syngeneic osteosarcoma tumor model was used to evaluate the effects of SNA treatment on tumor growth and immune response. SNA treatment at 1 mg/kg did not affect tumor growth as compared to PBS treated tumor-bearing mice (Fig. 4a). However, further analysis of tumor tissues demonstrated that SNA treatment markedly induced the CD8+ T cell infiltration (increased 0.66 fold, p = 0.009), but not the infiltration of CD4+ TILs (p = 0.635) (Fig. 4b-d). Sorted from CD11b+F4/80+ macrophages, M1 macrophages (MHCIIhigh, TAM-M1) or M2 macrophages (CD206+, TAM-M2) were analyzed in the myeloid cells, and we observed that SNA treatment shifted myeloid cells towards the TAM-M1 population (increased 1.6 folds, p = 0.005) (Fig. 4e). Although SNA treatment didn't change the accumulation of G-MDSCs into K7M2 tumors (p = 0.54) (Fig. 4f), it enhanced PD-L1 expression on G-MDSCs (increased 1.02 folds, p = 0.008) (Fig. 4g), and the ability of G-MDSCs to directly suppress the proliferation of T lymphocytes was significantly reduced (decreased 43%, p = 0.004) (Fig. 4h), which was correlated with the significant increase in the IFNγ production from T-lymphocytes when exposed to the antigen K7M2 (increased 2.29 folds, p = 0.002) (Fig. 4i). Moreover, SNA treatment significantly increased the expression of H2-Kb (MHC class I) (increased 0.95 fold, p = 0.006) and PD-L1 (increased 1.02 folds, p = 0.005) on tumor cells (Fig. 4j), suggesting that although SNA treatment did not obviously slow K7M2 tumor growth, it primed the tumor microenvironment and enhanced T-lymphocyte activation potential.
Fig. 4.
SNA treatment primed the tumor immune microenvironment in K7M2 tumor model. (a) Treatment with SNA (1 mg/kg) had minimal impact on K7M2 tumor growth; (b, c) SNA treatment increased CD8+ cell infiltration analyzed by immunohistochemistry and flow cytometry; (d) SNA treatment didn't increase CD4+ cell infiltration; (e) TAM cell population (CD11b+F4/80+) were analyzed by flow cytometry. TAM-M2 (CD206+) and TAM-M1 (MHCIIhigh) in CD11b+F4/80+ cell population were further quantified in K7M2 tumor tissues. SNA treatment increased the ratio of M1/M2; (f) SNA treatment didn't change the accumulation of tumor-infiltrating G-MDSCs; (g) SNA treatment increased the expression of PD-L1 on G-MDSC cells; (h) SNA treatment inhibited the suppressive effects of tumor-infiltrating G-MDSCs on the proliferation of T-lymphocytes; (i) SNA treatment increased the production of IFNγ. (j) SNA treatment increased the expression of H2-Kb and PD-L1 on tumor cells. Data were expressed as mean±SD (n = 10) for bar chart and expressed as median±interquartile (n = 10) for scatter plot. ##p < 0.01 vs K7M2 alone; **p < 0.01 vs PBS group. The in vivo experiments were performed independently twice. The in vitro experiments were performed independently three times.
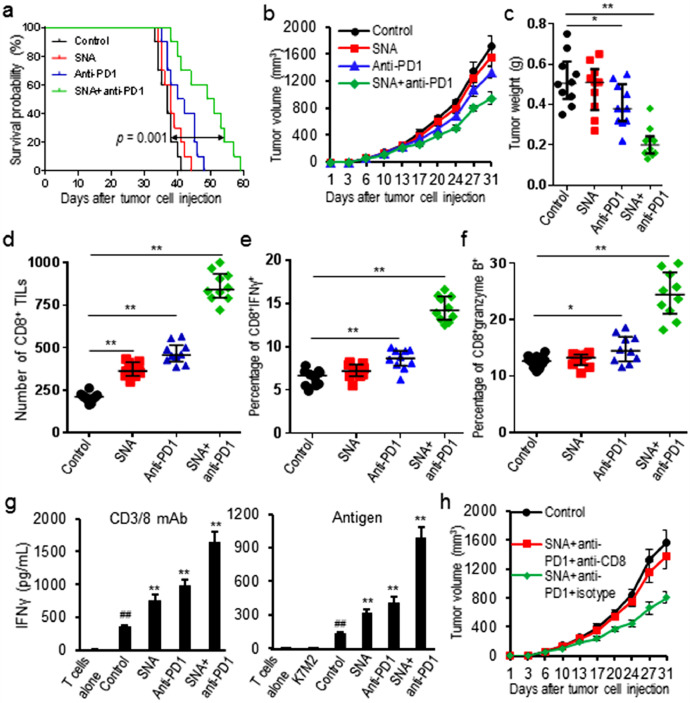
3.5. Combination treatment with SNA and anti-PD1 mAb enhanced tumor growth control and survival of K7M2 tumor-bearing mice
The observation that SNA treatment led to a strong increase of CD8+ TILs raised the question whether additional PD-1 blockade could induce long-term tumor control. Tumor-bearing mice were treated with combination of SNA and anti-PD1 mAb. Results showed combination treatment significantly increased survival time (increased median survival days from 37 days to 50 days, p = 0.001) and slowed tumor growth over either treatment alone (decreased tumor weight by 60%, p = 0.003) (Fig. 5a-c). Further analysis of tumor tissues showed that the combination treatment increased CD8+ TILs (increased 3.14 folds, p = 0.001) and IFNγ (increased 1.25 folds, p = 0.007) as well as granzyme B (increased 0.95 fold, p = 0.009) in tumor tissues (Fig. 5d-f). The combination treatment also significantly increased IFNγ production of T-lymphocytes from draining lymph nodes activated by non-specific (CD3/28 antibodies) (increased 3.68 folds, p = 0.005) or antigen-specific (IFNγ pretreated and irradiated K7M2 cells) (increased 6.44 folds, p = 0.003) stimuli, over either treatment alone (Fig. 5g). To validate whether SNA-induced CD8+ T cell infiltration is the major mechanism for synergy with PD-1 blockade, K7M2 tumor-bearing mice were treated with or without CD8+ depleting antibody. Results showed depletion of CD8+ cells nearly completely abolished the control of tumor outgrowth observed upon SNA and anti-PD1 mAb combination treatment (Fig. 5h).
Fig. 5.
Combination treatment with SNA and anti-PD1 mAb enhanced tumor control of K7M2 tumors. (a) Combination treatment increased the survival time of tumor-bearing mice; (b, c) Combination treatment slowed tumor growth and decreased tumor burden; (d-f) Combination treatment increased CD8+ TILs and IFNγ as well as granzyme B in tumor tissues; (g) Combination treatment increased the production of IFNγ from T-lymphocytes isolated from tumor-draining lymph nodes and stimulated with either CD3/28 or irradiated K7M2 cells; (h) K7M2 tumor-bearing mice were treated with SNA and anti-PD1 mAb, along with either anti-CD8 mAb or isotype control. Tumor growth curves during treatment were showed. Data were expressed as mean±SD (n = 10) for bar chart and expressed as median±interquartile (n = 10) for scatter plot. ##p < 0.01 vs T cells alone; *p < 0.05, **p < 0.01 vs control group. The in vivo experiments were performed independently twice. The in vitro experiments were performed independently three times.
4. Discussion
Osteosarcoma cells are strongly resistant to chemotherapeutics in patients with metastasis, which leads to poor clinical prognosis [18]. Combinatorial strategies that include agents with multiple mechanisms of action are potential modalities to improve efficacy and forestall the development of resistance [19]. In recent years, immune checkpoint blockade has emerged as an important and effective form of immunotherapy against human cancer [20]. However, despite the clinical potential of these inhibitors, most of patients either do not respond in the clinical trials, for example SARC028, or develop resistance [21]. To overcome these limitations, methods to improve the efficacy would be advantageous [22]. In this regard, a large number of trials are now underway combining immune checkpoint inhibitors with a wide range of therapeutic agents [23], [24]. In this study, we demonstrated that functional inhibition of G-MDSCs with SNA, an inhibitor of PI3Kδ/γ, enhanced responses to PD-1 blockade. Combination treatment induced CD8+ T-lymphocyte-dependent tumor growth delay and prolonged survival time in a syngeneic osteosarcoma tumor model.
The durable response rate following immune checkpoint inhibitors in patients with solid tumors is still small, largely because of tumor-mediated immunosuppression [5], [6]. In many solid tumors, MDSCs contribute to the tumor-associated immune suppression as key effectors in the tumor microenvironment through potently suppressing the activities of T cells and natural killer cells and have become the focus of intense study recently [25]. G-MDSCs are associated with decreased overall survival time in patients with cancer [26], [27]. Through suppressing T cell proliferation and its production of IFNγ, partially dependent on the arginase pathway [28], [29], [30], G-MDSCs have the capacity to impair the antitumor immunity, thereby promoting tumor progression and reducing the therapeutic effects of immune checkpoint blockade [31]. Recent studies demonstrated that the efficacy of immune checkpoint blockade could be dramatically enhanced through inhibiting recruitment or function of MDSCs [32], [33]. In the current study, G-MDSCs significantly increased in the peripheral blood and tumor tissues of K7M2 tumor-bearing mice, and suppressed the function of T-lymphocytes depending arginase and iNOS. SNA treatment inhibited the suppressive activity of G-MDSCs through decreasing the expression of arginase and iNOS, as well as the phosphorylation of AKT and S6 downstream of PI3K on tumor-infiltrating G-MDSCs. Moreover, SNA treatment shifted myeloid cells towards the TAM-M1 population in tumor tissues. SNA treatment also enhanced PD-L1 and H2-Kb expression on tumor cells to prime the tumor microenvironment.
PI3K signaling pathway, which is involved in the cellular growth, metabolism and survival, is known to play important roles in the regulation of iNOS and arginase expression in hematopoietic cells and can activate Gr1+/CD11b+ myeloid cells [14], [34], [35]. Previous data showed that additional blockade of PI3Kα, PI3Kδ, and PI3Kγ slightly improved tumor control and prevented resistance in melanoma [36]. Therefore, targeting PI3K to abrogate its suppressive capacity has important implications in clinic. Reports demonstrated pharmacologic inhibition of p110δ/γ led to robust inhibition of tumor-infiltrating myeloid cells in several mouse models [33], [37], [38]. Inhibition of PI3Kγ activated NF-κB to promote an immunostimulatory transcriptional program and restore cytotoxicity of CD8+ T cell, resultantly synergizing with immune checkpoint blockade therapy to increase response [38]. These studies indicated a rational approach to improve antitumor immunity of immune checkpoint blockade through functional inhibition of immunosuppressive cells with selective inhibitors of PI3K isoforms. In this study, combination treatment with SNA and anti-PD1 antibody improved anti-tumor responses over either treatment alone, at least through reversing the suppressive capacity of G-MDSCs by inhibiting PI3K.
Immunotherapies have revolutionized the treatment of cancer by priming patients’ own anti-tumor immune responses, but only a subset of patients exhibit durable responses. Our results demonstrated high infiltration of suppressive G-MDSCs in the osteosarcoma tumor tissues. SNA treatment sensitized T-cell-inflamed osteosarcoma cells to anti-PD1 treatment through inhibition of arginase and iNOS expression on G-MDSC cells. Our data provided the scientific rationale to inhibit PI3Kδ/γ to enhance the efficacy of immune checkpoint inhibitors in clinical trials against osteosarcoma.
Conflicts of interest statement
The authors declared that there is no conflict of interests in this work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2018.11.001.
Appendix. Supplementary materials
References
- 1.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown H.K., Tellez-Gabriel M., Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi: 10.1016/j.canlet.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Carrle D., Bielack S. Osteosarcoma lung metastases detection and principles of multimodal therapy. Cancer Treat. Res. 2009;152:165–184. doi: 10.1007/978-1-4419-0284-9_8. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Deng X., Yu C. Synergistic inhibitory effects of capsaicin combined with cisplatin on human osteosarcoma in culture and in xenografts. J. Exp. Clin. Cancer Res. 2018;37:251–267. doi: 10.1186/s13046-018-0922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkie K.P., Hahnfeldt P. Tumor-immune dynamics regulated in the microenvironment inform the transient nature of immune-induced tumor dormancy. Cancer Res. 2013;73:3534–3544. doi: 10.1158/0008-5472.CAN-12-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrand-Rosenberg S. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol. Immunother. 2010;59:1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulart M.R., Pluhar G.E., Ohlfest J.R. Identification of myeloid derived suppressor cells in dogs with naturally occurring cancer. PloS One. 2012;7:e33274. doi: 10.1371/journal.pone.0033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochando J.C., Chen S.H. Myeloid-derived suppressor cells in transplantation and cancer. Immunol. Res. 2012;54:275–285. doi: 10.1007/s12026-012-8335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alizadeh D., Trad M., Hanke N.T. Doxorubicin eliminates myeloid-derived suppressor cells and enhances the efficacy of adoptive T-cell transfer in breast cancer. Cancer Res. 2014;74:104–118. doi: 10.1158/0008-5472.CAN-13-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Y.M., Eissler N., Le Blanc K., Johnsen J.I., Kogner P., Kiessling R. Targeting suppressive myeloid cells potentiates checkpoint inhibitors to control spontaneous neuroblastoma. Clin. Cancer Res. 2016;22:3849–3859. doi: 10.1158/1078-0432.CCR-15-1912. [DOI] [PubMed] [Google Scholar]
- 13.Nagaraj S., Youn J.I, Weber H. Anti-inflammatory triterpenoid blocks immune suppressive function of MDSCs and improves immune response in cancer. Clin Cancer Res. 2016;16:1812–1823. doi: 10.1158/1078-0432.CCR-09-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid M.C., Avraamides C.J., Dippold H.C. Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3Kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell. 2011;19:715–727. doi: 10.1016/j.ccr.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui W., Cai Y., Zhou X. Advances in subunits of PI3K class I in cancer. Pathology. 2014;46:169–176. doi: 10.1097/PAT.0000000000000066. [DOI] [PubMed] [Google Scholar]
- 16.De Henau O., Rausch M., Winkler D. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YD., Zhang J.Z., Wang Y.H., Yang J., Li H.Y., Cai L.M. Anti-proliferative constituents from Selaginella pulvinata, Phytochem. Lett. 2016;15:26–29. [Google Scholar]
- 18.Zheng J.X., Jiang H., Xiao L.P., Wang S.S., Li X.D. miR-320 inhibits multidrug resistance of osteosarcoma cells to methotrexate by targeting XIAP. Int. J. Clin. Exp. Pathol. 2017;10:368–376. [Google Scholar]
- 19.Swart M., Verbrugge I., Beltman J.B. Combination approaches with immune checkpoint blockade in cancer therapy. Front. Oncol. 2016;6:233–248. doi: 10.3389/fonc.2016.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellman I., Coukos G., Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribas A., Hu-Lieskovan S. What does PD-L1 positive or negative mean? J. Exp. Med. 2016;213:2835–2840. doi: 10.1084/jem.20161462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner M.G., Karimi S.S., Barry-Holson K. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother. 2013;36:477–489. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott P.A., Hodi F.S., Kaufman H.L., Wigginton J.M., Wolchok J.D. Combination immunotherapy: a road map. J. Immunother. Cancer. 2017;5:16–30. doi: 10.1186/s40425-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 25.Naijar Y.G., Rayman P., Jia X. Myeloid-derived suppressor cell subset accumulation in renal cell carcinoma parenchyma is associated with intratumoral expression of IL1β, IL8, CXCL5, and Mip-1α. Clin. Cancer Res. 2017;23:2346–2355. doi: 10.1158/1078-0432.CCR-15-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arihara F., Mizukoshi E., Kitahara M. Increase in CD14+HLA-DR -/low myeloid-derived suppressor cells in hepatocellular carcinoma patients and its impact on prognosis. Cancer Immunol. Immunother. 2013;62:1421–1430. doi: 10.1007/s00262-013-1447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Mitri D., Toso A., Alimonti A. Molecular pathways: targeting tumor-infiltrating myeloid-derived suppressor cells for cancer therapy. Clin. Cancer Res. 2015;21:3108–3112. doi: 10.1158/1078-0432.CCR-14-2261. [DOI] [PubMed] [Google Scholar]
- 28.Ochoa A.C., Zea A.H., Hernandez C., Rodriguez P.C. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin. Cancer Res. 2007;13:721–726. doi: 10.1158/1078-0432.CCR-06-2197. [DOI] [PubMed] [Google Scholar]
- 29.Ko J.S., Zea A.H., Rini B.I. Sunitinib mediates reversal of myeloid-derived suppressor cell accumulation in renal cell carcinoma patients. Clin. Cancer Res. 2009;15:2148–2157. doi: 10.1158/1078-0432.CCR-08-1332. [DOI] [PubMed] [Google Scholar]
- 30.Kao J., Ko E.C., Eisenstein S., Sikora A.G., Fu S., Chen S.H. Targeting immune suppressing myeloid-derived suppressor cells in oncology. Crit. Rev. Oncol. Hematol. 2011;77:12–19. doi: 10.1016/j.critrevonc.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Highfill S.L., Cui Y., Giles A.J. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 2014;6:237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan Y., Zhang R., Peng Z., Dong D., Wei G., Wang Y.S. Inhibition of IL-18-mediated myeloid derived suppressor cell accumulation enhances anti-PD1 efficacy against osteosarcoma cancer. J. Bone Oncol. 2017;9:59–64. doi: 10.1016/j.jbo.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis R.J., Moore E.C., Clavijo P.E. Anti-PD-L1 Efficacy Can Be Enhanced by Inhibition of Myeloid-Derived Suppressor Cells with a Selective Inhibitor of PI3Kδ/γ. Cancer Res. 2017;77:2607–2619. doi: 10.1158/0008-5472.CAN-16-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin E., Haubenwallner S., Kuttke M. Macrophage PTEN regulates expression and secretion of arginase I modulating innate and adaptive immune responses. J. Immunol. 2014;193:1717–1727. doi: 10.4049/jimmunol.1302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiou W.F., Don M.J., Liao J.F., Wei B.L. Psoralidin inhibits LPS-induced iNOS expression via repressing Syk-mediated activation of PI3K-IKK-IκB signaling pathways. Eur. J. Pharmacol. 2011;650:102–109. doi: 10.1016/j.ejphar.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Deuker M.M., Marsh Durban V., Phillips W.A., McMahon M. PI30-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov. 2015;5:143–153. doi: 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali K., Soond D.R., Pineiro R. Inactivation of PI(3)K p110delta breaks regulatory T-cell-mediated immune tolerance to cancer. Nature. 2014;510:407–411. doi: 10.1038/nature13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneda M.M., Messer K.S., Ralainirina N. PI3Kgamma is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.