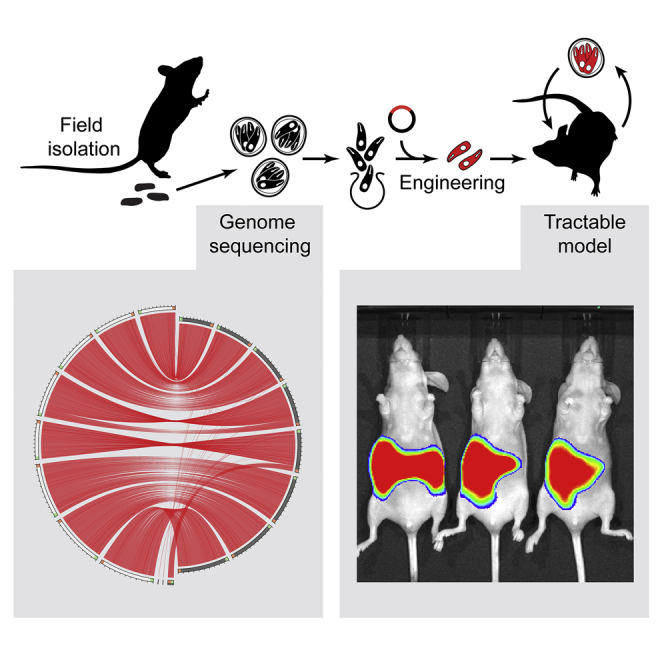
Summary
Cryptosporidium is a leading cause of diarrheal disease and an important contributor to early childhood mortality, malnutrition, and growth faltering. Older children in high endemicity regions appear resistant to infection, while previously unexposed adults remain susceptible. Experimental studies in humans and animals support the development of disease resistance, but we do not understand the mechanisms that underlie protective immunity to Cryptosporidium. Here, we derive an in vivo model of Cryptosporidium infection in immunocompetent C57BL/6 mice by isolating parasites from naturally infected wild mice. Similar to human cryptosporidiosis, this infection causes intestinal pathology, and interferon-γ controls early infection while T cells are critical for clearance. Importantly, mice that controlled a live infection were resistant to secondary challenge and vaccination with attenuated parasites provided protection equal to live infection. Both parasite and host are genetically tractable and this in vivo model will facilitate mechanistic investigation and rational vaccine design.
Keywords: cryptosporidiosis, Cryptosporidium, parasite, host-pathogen, immunity, intestine, Apicomplexa, diarrhea
Graphical Abstract
Highlights
-
•
We isolated and sequenced Cryptosporidium tyzzeri, a natural mouse pathogen
-
•
C. tyzzeri can be genetically manipulated using CRISPR-driven homologous repair
-
•
C. tyzzeri models human cryptosporidiosis with T cell- and IFN-γ-dependent resolution
-
•
Mice develop protective immunity following both live infection and vaccination
Cryptosporidiosis is an important diarrheal disease that lacks effective treatment and vaccination. Sateriale et al. isolate Cryptosporidium tyzzeri from naturally infected wild mice and derive a model of infection in immunocompetent mice in which both parasite and host are genetically tractable. The mechanisms of protective immunity are now open to analysis.
Introduction
Cryptosporidiosis, initially recognized as an opportunistic infection associated with advanced HIV-AIDS, is now known as a leading cause of diarrheal disease in immunocompetent individuals around the world (Checkley et al., 2015, Striepen, 2013). Young children are particularly vulnerable to infection with the apicomplexan parasite Cryptosporidium. In the Global Enteric Multicenter Study, the largest study of its kind to assess the etiology of early childhood diarrheal disease in sub-Saharan Africa and southeast Asia, Cryptosporidium was strongly associated with moderate-to-severe diarrhea and death across all study sites (Kotloff et al., 2013). Children that survive these life-threatening Cryptosporidium infections remain prone to malnutrition and stunted growth, with measurable decreases in height-for-age scores (Checkley et al., 1998, Mølbak et al., 1997, Mondal et al., 2009, Platts-Mills et al., 2015). A recent meta-analysis that accounted for growth faltering estimated over 12 million disability adjusted life years attributable to cryptosporidiosis in 2016 alone (Khalil et al., 2018). This massive impact on public health is exacerbated by the lack of tools to manage the disease; the current treatment for Cryptosporidium is of limited efficacy and new more potent medicines are still under development (Huston et al., 2015, Manjunatha et al., 2017) and no vaccine is available to prevent the infection.
Cryptosporidiosis in humans is predominantly caused by Cryptosporidium hominis and Cryptosporidium parvum. Epidemiology in children, as well as studies in human volunteers and in different animal species, have shown the development of immunity and suggest that vaccination may prevent the disease (Mead, 2010, Okhuysen et al., 1998), but the lack of animal models has limited vaccine development. Gnotobiotic piglets are susceptible to C. hominis and calves to C. parvum, but large animal models require specialized facilities (Tzipori, 1998, Widmer et al., 2000). Experiments in immunocompetent mice have been performed with Cryptosporidium muris; however, this species differs significantly from those infecting humans: phylogenetically, biochemically, and in the anatomical site of infection (stomach versus the small intestine). Mice are naturally resistant to C. hominis and C. parvum, but can be rendered susceptible to the latter by chemical immune suppression (Petry et al., 1995) or genetic immune insufficiency (Griffiths et al., 1998, Mead et al., 1991). These models have been critical to our understanding of innate mechanisms of resistance to Cryptosporidium and affirmed the important role of T cells in parasite control (Borad and Ward, 2010, Mead, 2014). However, because these studies are performed in neonates, immunocompromised or naturally resistant mice, we know little about the mechanisms required for long-lived protection.
Robust rodent models with natural progression that replicate human disease have proven transformative for many fields of infection biology (Martina et al., 2003, Ploss et al., 2009). In order to develop such a model for cryptosporidiosis, we isolated a species of Cryptosporidium that naturally colonizes the mouse small intestine. This parasite, Cryptosporidium tyzzeri, genetically resembles those that cause human cryptosporidiosis and causes similar disease pathology. We then use CRISPR-driven genome engineering to create stable transgenic C. tyzzeri that express luminescent and fluorescent reporters. With these transgenic parasites we demonstrate acquired resistance to infection in mice and importantly such resistance can be elicited through vaccination with an attenuated parasite. Moving forward this fully tractable mouse model of cryptosporidiosis will permit rigorous investigation of potential vaccination strategies and allow the dissection of the complex host-pathogen interactions of Cryptosporidium infection.
Results
Isolation of Cryptosporidium tyzzeri and De Novo Genome Assembly
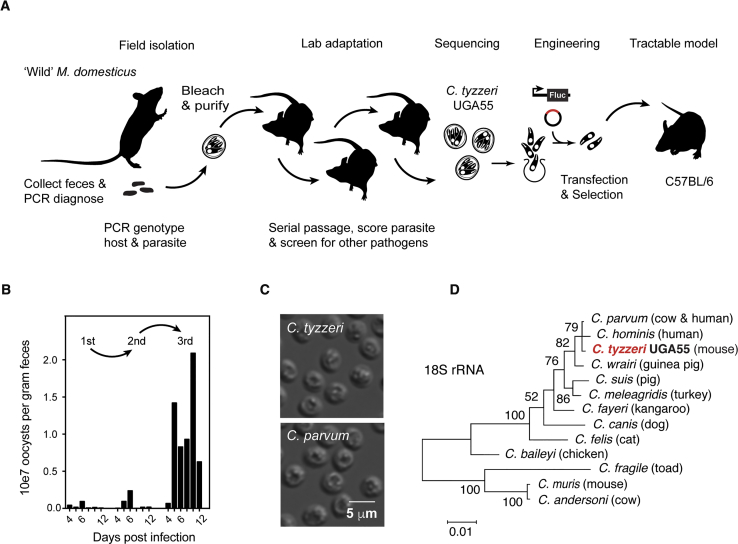
Mouse fecal pellets were collected from farms in Athens, Georgia in the United States; PCR analyses showed the presence of DNA from mice of the species Mus musculus domesticus (Figures S1A and S1B), and for 30% of the samples, Cryptosporidium DNA. Oocysts were purified from feces, bleach treated, and then used to infect adult C57BL/6 mice, housed in quarantine, by gavage (Figure 1A). Cryptosporidium burden was monitored by measuring fecal parasite shedding by qPCR (Figure 1B) and mice were subjected to a panel of diagnostic tests and veterinary pathology to rule out co-infection with other pathogens (Table S1). Parasites were passaged three times in this fashion in isolation, with oocyst yield increasing in each passage and Cryptosporidium remained the sole pathogen detected.
Figure 1.
A Natural Mouse Model for Cryptosporidiosis
(A) Schematic overview of isolation and characterization of Cryptosporidium tyzzeri.
(B) Fecal shedding of C. tyzzeri in quarantined mice. Oocysts were purified from field-collected mouse feces and used to infect C57BL/6 mice, followed by serial passage. Fecal parasite burden is measured by qPCR.
(C) Differential interference contrast microscopy of C. tyzzeri and C. parvum oocysts. Note that C. tyzzeri oocysts isolated in this study are of similar shape and size as C. parvum.
(D) Maximum likelihood phylogenetic tree of the 18s rRNA locus of different Cryptosporidium species (see Figure S1C for a phylogenetic tree constructed with full genome alignments).
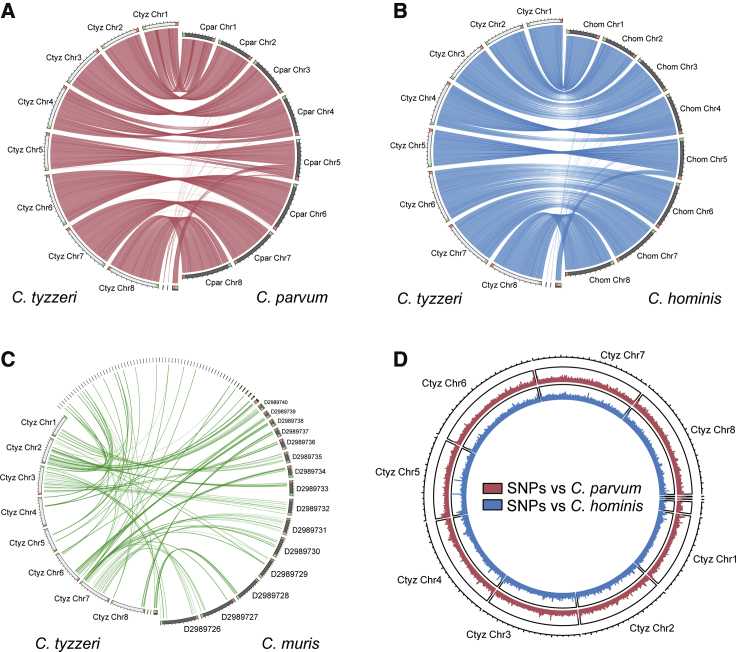
Morphologically, oocysts from the current isolated strain, designated UGA55, are similar to those of C. parvum ([4.84 + 0.16] × [4.13 + 0.14] μm, Figure 1C) and distinct from the larger and more oval shaped C. muris oocysts. Genetically, sequencing of the 18S ribosomal RNA locus revealed a match to C. tyzzeri, a natural parasite of house mice (Ren et al., 2012, Kváč et al., 2013). The genome of C. tyzzeri isolate UGA55 was sequenced, assembled de novo, and manually annotated. This is the first assembled genome sequence for C. tyzzeri, which was made publicly available through GenBank and the Cryptosporidium Genomic Resource, CryptoDB (Heiges et al., 2006). Phylogenetic analyses place C. tyzzeri UGA55 in close proximity to the most prevalent human pathogens, C. parvum and C. hominis. This holds true regardless of whether this analysis is focused on 18S rRNA (Figure 1D) or expanded to the whole genome by using 2764 single-copy orthologous genes shared among the seven Cryptosporidium species for which full genome sequences are available (Figure S1C). When compared to the genomes of C. parvum and C. hominis, the genome of C. tyzzeri is remarkably similar in size, structure, and sequence. The genome of C. tyzzeri is distinct from C. muris as illustrated by the pronounced difference in the ability to map the genome sequences to each other in synteny plots when using a local alignment e-value cut-off of 1e-20 (Figure 2). There are 3977 predicted protein encoding genes in a highly AT-rich 9.02-Mb genome, with 95%–96% sequence identity at the nucleotide level to C. hominis and C. parvum (Figure 2; see Table 1 for a more thorough comparison). We identified 181,187 single-nucleotide polymorphisms relative to C. parvum and 223,396 for C. hominis, which are most abundant in telomeric regions that carry genes for highly variable secretory proteins (Abrahamsen et al., 2004, Xu et al., 2004). Note that in Figure 2D, SNP abundance peaks between C. tyzzeri and C parvum correlate with those between C. tyzzeri and C. hominis, and they also correlate with such peaks between C. parvum and C. hominis (data not shown).
Figure 2.
The Genome of Cryptosporidium tyzzeri Is Very Similar to the Genomes of C. parvum and C. hominis
(A–C) Synteny maps comparing the assembled genomic contigs of C. tyzzeri UGA55 and C. parvum UKP6 (A), C. hominis 30976 (B), and C. muris RN66 (C). Each line connects a genomic segment with a local alignment e-value cut-off of 1e−20. This correlates to >90% overall nucleotide identity.
(D) Single-nucleotide polymorphism (SNP) density map comparing C. tyzzeri with C. parvum (red) and hominis (blue) with a 1 kb rolling window.
Table 1.
Comparative Analysis of Cryptosporidium Genomes
| C. tyzzeri | C. hominis | C. parvum | C. muris | |
|---|---|---|---|---|
| Genome size (Mb) | 9,015,713 | 9,059,225 | 9,102,324 | 9,245,251 |
| Number of scaffolds | 11 | 53 | 8 | 84 |
| GC content (%) | 30.25 | 30.13 | 30.22 | 28.47 |
| Percent coding (%) | 77.96 | 81.86 | 81.09 | 75.23 |
| Gene density (genes per 100 Kb) | 44.13 | 43.59 | 43.31 | 42.55 |
| Total predicted protein encoding genes | 3,977 | 3,949 | 3,941 | 3,934 |
| Genes with introns (%) | 15.31 | 14.84 | 14.59 | 20.36 |
| Average gene length (bp) | 1,736 | 1,852 | 1,834 | 1,747 |
| Predicted proteins with: | ||||
| signal peptide | 698 | 678 | 716 | 603 |
| transmembrane domain | 865 | 851 | 870 | 836 |
| functional domain (interpro) | 2,315 | 2,317 | 2,311 | 2,428 |
| EC number | 918 | 915 | 908 | 945 |
| In comparison to C. tyzzeri: | ||||
| % genome aligned with >75% identity | - | 99.2 | 99 | 7.2 |
| Total SNPs within aligned region | - | 223,396 | 181,187 | 18,054 |
Pathology of Cryptosporidium tyzzeri Infection in Mice Closely Resembles Human Disease
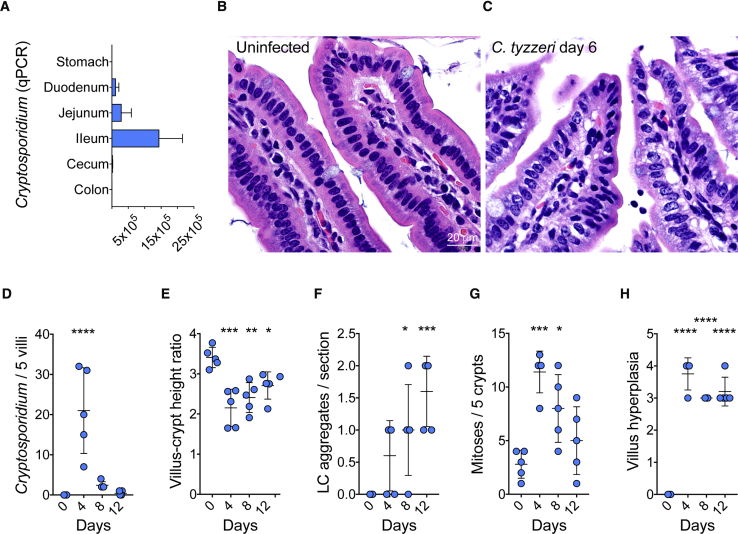
In C57BL/6 mice infected with C. tyzzeri parasites detected by qPCR were found throughout the small intestine with the highest burden in the ileum (Figure 3A) and they were absent from the stomach and the colon. Histological analysis of the small intestine during a time course of infection revealed pronounced pathological changes. Infected tissue showed a marked loss of the glycocalyx of the epithelial cells, effacement and blunting of villi, and a significant influx of leukocytes (see STAR Methods for the detailed scoring rubric that was developed). Within the crypts of the small intestine there was a significant increase in mitoses upon infection with C. tyzzeri, with crypt hyperplasia and increased crypt branching (Figures 3B–3H). Similar pathology has been reported from infected humans (Lumadue et al., 1998) and the associated histological and physiological changes have been linked to malabsorption and environmental enteropathy in children as a cause of growth stunting (Checkley et al., 1998, Khalil et al., 2018).
Figure 3.
Cryptosporidium tyzzeri Infects the Small Intestine and Causes Pathology
(A) The intestinal tract from infected mice was resected and 1-cm2 segments were tested for the presence of parasite DNA by qPCR. n = 7 mice from 2 separate experiments with tissue harvested at day 10 post infection with 10,000 oocysts.
(B and C) Hematoxylin and eosin stained sections of the distal small intestine of mice. Note tall columnar villus epithelium and intact apical microvilli border with associated glycocalyx in uninfected (B) mice, in contrast to severe villus attenuation, loss of the apical glycocalyx and effacement of microvilli in infected (C) animals.
(D–H) Mice were infected with 50,000 C. tyzzeri oocysts, killed at 0, 4, 8, and 12 days post infection and histologically scored for pathological changes over the course of infection. In the small intestine of infected mice, parasite burden peaked on day 4 post infection (D). Infection resulted in a marked reduction in villus:crypt height ratio (villus blunting) (E), lymphocyte aggregation (F), epithelial dysplasia with increased mitoses (G), and villus hyperplasia (H). n = 5 mice per group, with each dot representing the score for a single mouse, data are mean ± standard deviation (SD). Significance is determined using one-way ANOVA with Dunnett multiple comparison test (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, and ∗∗∗∗p < 0.00005).
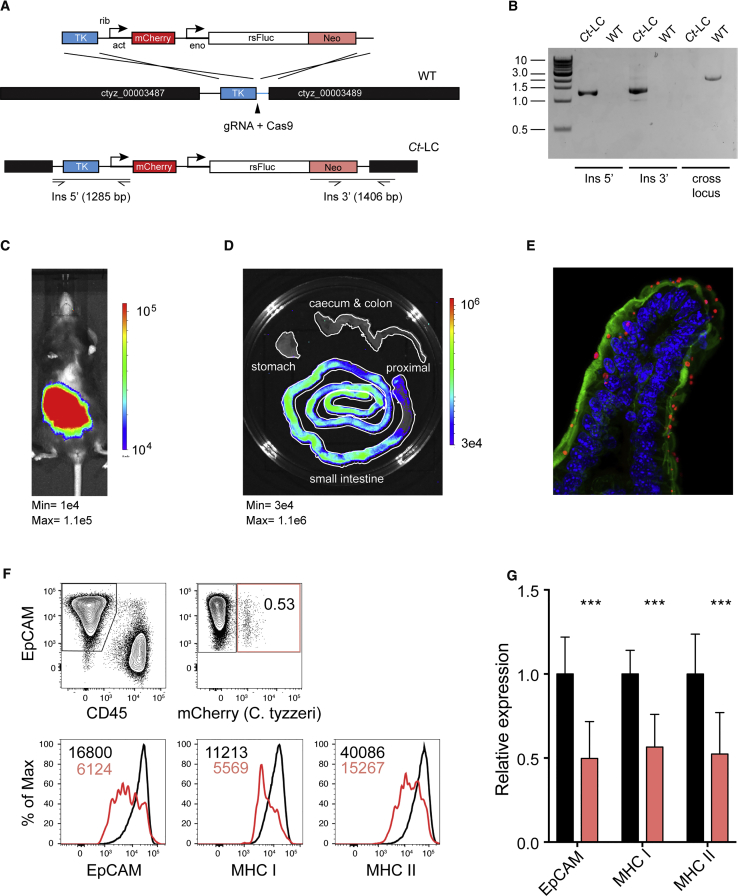
C. tyzzeri Is Genetically Tractable Using a CRISPR/Cas9 Approach
To make C. tyzzeri tractable for immunological studies we genetically engineered reporter parasites that can be detected in vivo to allow for precise longitudinal measurements of parasite burden. We harnessed a CRISPR/Cas9-driven homologous recombination strategy (Vinayak et al., 2015) to introduce a dual reporter and drug selection cassette into the thymidine kinase locus (Figure 4A). Transgenic parasites were then selected and propagated in vivo using paromomycin and PCR mapping demonstrated successful integration of the cassette into the C. tyzzeri genome at the desired locus (Figure 4B). Infection with these transgenic C. tyzzeri expressing red-shifted luciferase (strain Ct-LC) is readily detected by whole animal imaging upon injection of luciferin (Figure 4C) and can be seen in the small intestine (Figure 4D) but not the stomach or colon. The strain also expresses the fluorescent protein reporter mCherry, allowing for parasite visualization within the infected tissue and cells (Figure 4E). It is also possible to dissociate sections of the small intestine into individual cells (see STAR Methods for detail) and to exploit the fluorescence of these transgenic parasites to specifically identify infected epithelial cells by flow cytometry (Figure 4F). Previous in vitro studies suggested that Cryptosporidium might interfere with IFNγ induced expression of the major histocompatibility complex (MHC) (Choudhry et al., 2009), but infection of mice led to global upregulation of epithelial cell MHC class I and II expression (data not shown). Taking advantage of the ability to distinguish infected and uninfected cells, we compared MHC expression of these populations. Uninfected (cherry negative) epithelial cells from the small intestine expressed high levels of MHC class I and II (Figure 4G). However, infected epithelial cells showed reduced MHC class I and II surface expression; these data provide in vivo evidence that Cryptosporidium may modulate or suppress aspects of immune detection within its natural host.
Figure 4.
Genetic Manipulation of Cryptosporidium tyzzeri
(A) Map of the thymidine kinase (TK) locus targeted in C. tyzzeri by insertion of a construct that includes the selection marker Neomycin phosphotransferase (Neo), redshifted firefly luciferase (rsFLuc), and the fluorescent protein reporter mCherry. The solid black arrow indicates the position of the Cas9 induced double-strand break.
(B) PCR analysis using genomic DNA from wild-type (WT) and transgenic (Ct-LC) parasites. Note that insertion of the multi-gene cassette into the locus precludes amplification across the locus because of the excessive size of the amplicon. Therefore, amplification is only apparent in WT.
(C–E) The resulting transgenic strain, Ct-LC, can be used to locate and measure parasite burden in (C) the whole animal, (D) the intestinal tract, and (E) in infected tissue (immunofluorescence of small intestine with red-mCherry, green-actin, blue-nuclei).
(F and G) Small intestines of infected mice were disaggregated into single cell suspension and analyzed by flow cytometry. EpCAM and CD45 distinguish enterocytes and leukocytes (F). Infected enterocytes are identified by mCherry and comparison of uninfected (black) and infected (red) cells harvested from the same animal show downregulation of EpCAM and MHC I and II (G). Data are mean ± SD and significance is determined using a Student’s t test comparing infected and uninfected epithelial cells (∗∗∗p < 0.001).
Resolution of C. tyzzeri Infection Is Dependent on Interferon Gamma and T Cells
Next, we explored the role of host immune function in colonization and clearance in normal and immunocompromised mice. When immunocompetent, adult C57BL/6 mice were challenged with C. parvum, the infection was undetectable by in vivo imaging and fecal parasite shedding (Figures 5A and 5B). Note that this was a viable and virulent inoculum that readily produced a patent infection in mice that lacked the gene for IFN-γ (Figure 5C). In contrast, challenge of C57BL/6 mice with the same dose of C. tyzzeri resulted in robust infection by either measurement (Figures 5A–5C). C57BL/6 mice controlled C. tyzzeri infection within 2–3 weeks, after which parasites were no longer detectable by whole animal imaging or fecal qPCR (Figures 5D, S2A, and S2B). We note that in healthy human adults, cryptosporidiosis is a self-limiting infection and disease that resolves with similar kinetics to those observed here for C. tyzzeri in mice (Jokipii and Jokipii, 1986).
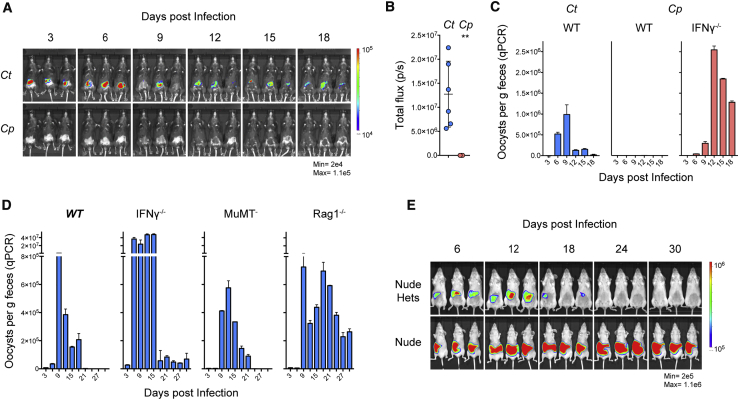
Figure 5.
Interferon γ and T Cells Govern Susceptibility and Resolution of Murine Cryptosporidiosis
(A–C) Immunocompetent C57BL/6 mice infected with 10,000 oocysts of C. tyzzeri (Ct) or C. parvum (Cp). Infection was monitored by whole animal imaging following parasite luciferase (A, quantified over entire infection in B) or by qPCR detecting parasite shed in the feces (C). Note that Cp produces robust infection in Ifnγ−⁄−. For A and B, n = 9 mice total over 2 experiments, images shown are representative, and data represent mean ± SD with significance determined using a two-tailed t test (∗p < 0.05). For C, n = 12 mice total, 4 per group, with qPCR performed on pooled fecal material with two technical replicates.
(D) C57BL/6 wild-type mice (WT) and strains lacking IFNγ (Ifnγ−⁄−), mature B cells (μMt−), or mature B and T cells (Rag1−⁄−) were infected with 10,000 C. tyzzeri oocysts and infection was monitored by qPCR from collected feces. n = 16 mice total, 4 per group, with qPCR performed on pooled fecal material with two technical replicates, and data represent mean ± SD.
(E) Homozygous (Nude) and heterozygous (Nude Hets) Foxn1nu mice were infected with 10,000 transgenic C. tyzzeri oocysts and monitored for one month. n = 6 mice total; quantification of luminescence and fecal parasite shedding by qPCR in Figure S3. Mice lacking T cells (Rag1−⁄− and Nude) fail to clear the infection.
While C. tyzzeri produces robust infection in wild-type mice, this infection is significantly exacerbated in Ifnγ−⁄− mice. During the first two weeks of infection, parasite burden in Ifnγ−⁄− mice infected with C. tyzzeri was approximately 100-fold higher than that of wild-type mice (Figure 5D). C. tyzzeri infection of Ifnγ−⁄− mice produces enhanced tissue burden and pathology observed by histological analysis of intestinal sections (Figures S2C–S2F) and Ifnγ−⁄− mice infected with more than 10,000 oocysts succumbed to the infection. While at lower doses there was eventual control of parasite numbers in Ifnγ−⁄− mice, we noted a failure to fully eradicate this infection. Rag1−⁄− mice, which lack functional B and T cells, show a similar early parasite burden to wild-type controls, yet fail to control C. tyzzeri and remain chronically infected. We monitored infection of Rag1−⁄− mice for 6 months and did not observe resolution over this time (Figure S3A). To assess the individual contribution of B and T cells, μMt− mice that lack mature B cells and homozygous Foxn1nu mice that lack thymus derived T cells were infected. The ability of μMt− mice to control C. tyzzeri was comparable to wild-type mice, whereas homozygous Foxn1nu mice remained chronically infected (Figures 5E, S3B, and S3C). Together this data demonstrates that T cells are critical for control in this mouse model, mirroring what is seen in humans. Indeed, cryptosporidiosis is a common opportunistic infection in HIV-AIDS patients, as well as immunosuppressed organ transplant recipients, causing severe and chronic disease in those with low numbers of CD4 T cells (Manabe et al., 1998, Flanigan et al., 1992, Schmidt et al., 2001). Overall, this comparison highlights the IFNγ dependent component of host specificity in Cryptosporidium and illustrates the importance of IFNγ for early and late resistance to C. tyzzeri. In addition, similar to what has been observed in HIV-AIDS patients (Manabe et al., 1998), T cells and not B cells have a critical role for clearance.
Primary Infection and Vaccination Induce Resistance to Subsequent Challenge
Children in endemic areas acquire resistance to cryptosporidiosis and while a single infection typically does not produce sterile immunity, it can prevent or lessen disease (Checkley et al., 2015, Okhuysen et al., 1998, Korpe et al., 2018). To test if this resistance can be modeled in mice, we assessed whether primary infection protects from secondary challenge (see schematic outline in Figure 6A). C57BL/6 mice were inoculated with live or heat-killed C. tyzzeri oocysts and infection was monitored by qPCR and by day-15 mice had cleared the infection (Figure S4A). Note that the parasites used in this primary infection were not marked by a genetic reporter. After 4 weeks, mice were challenged with transgenic C. tyzzeri expressing red-shifted luciferase and this secondary challenge was monitored by imaging (Figure S4B). In these experiments, mice that received heat-killed C. tyzzeri oocysts were not protected and had a parasite burden comparable to naive mice. However, mice that experienced a live C. tyzzeri infection demonstrated significantly lower infection upon secondary challenge (Figures 6B and 6C shows a summary of multiple experiments, p < 0.0001). For some intracellular pathogens, resistance to secondary challenge is dependent on concomitant immunity, which is lost with clearance of the initial infection (Belkaid et al., 2002). To assess whether a cryptic infection contributed to the resistance here, mice were treated for 2 weeks with paromomycin to eradicate potential persisting parasites (see Figure 6A in brackets). Even after this drug treatment, previously infected mice were resistant to secondary challenge (Figures 6D, S4C, and S4D).
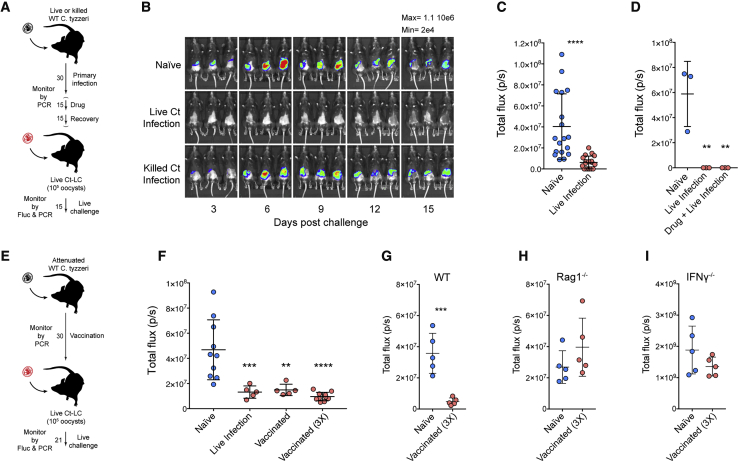
Figure 6.
Infection Results in Protective Immunity That Can Be Induced with an Attenuated Vaccine
(A) Schematic outline of challenge experiments for B, C, and D.
(B) C57BL/6 mice were subjected to primary infection as indicated, followed by a challenge with 100,000 oocysts of transgenic C. tyzzeri. n = 9 total mice, 3 per group.
(C) Summary of parasite burden (total flux over the two-week imaging period) comparing naive and previously infected mice. n = 36 total mice, 18 per group, over 5 independent experiments. Data represent mean ± SD with significance determined using a Students t test (∗∗∗∗p < 0.00005).
(D) Parasite burden of challenge with and without intervening paromomycin treatment. n = 9 total mice, 3 per group, and data represent mean ± SD with significance determined using one-way ANOVA with Dunnett multiple comparison test (∗∗p < 0.005). Individual parasite burden by imaging and fecal parasite shedding by qPCR in Figure S4.
(E) Schematic outline of vaccination experiments for F–I.
(F) Mice were infected or vaccinated as indicated and then challenged with 100,000 transgenic C. tyzzeri oocysts. Total parasite burden from whole animal imaging is shown. Individual parasite burden by imaging and fecal parasite shedding by qPCR in Figure S5. Note that live infection and vaccination results in comparable protection. n = 30 mice, with each dot representing the total parasite burden for a single mouse. Data are mean ± SD and significance is determined using one-way ANOVA with Dunnett multiple comparisons test (∗∗p < 0.005, ∗∗∗p < 0.0005, and ∗∗∗∗p < 0.00005).
(G–I) Mice were vaccinated with oocysts attenuated by 300 gray of gamma irradiation then challenged with 1000,000 transgenic C. tyzzeri oocysts. Total parasite burden from whole animal imaging is shown. Note the large difference in parasite tissue burden of mice deficient IFNγ (I) compared to both WT C57BL/6 mice (G) and mice that lack mature B and T cells (H). n = 10 mice for each strain, with each dot representing the total parasite burden for a single mouse. Data are mean ± SD and significance is determined using Student’s t test comparing naive and vaccinated mice within each strain (∗∗∗p < 0.0005).
A vaccine to prevent or limit the severity of cryptosporidiosis could significantly reduce child mortality and impact stunting and its many detrimental consequences (Khalil et al., 2018). Previous studies in mice have used Salmonella vectors to deliver Cryptosporidium antigens with limited efficacy (Bartelt et al., 2016) and perhaps the use of irradiated oocysts in calves has proven the most effective strategy (Jenkins et al., 2004). Indeed, parasites attenuated through irradiation, natural mutation, or genetic engineering show promising activity for related parasites including Toxoplasma and Plasmodium (Fox and Bzik, 2002, Mueller et al., 2005). Therefore, to assess whether C. tyzzeri would provide a system to establish a live-attenuated vaccine, wild-type C. tyzzeri oocysts were subjected to increasing doses of gamma irradiation and used to infect C57BL/6 mice. Irradiation at 200gray decreased total parasite shedding in mice to 11% of non-irradiated controls, and at doses of 300gray and above parasite shedding was not detected (Figure S5A). Mice that received parasites attenuated with irradiation at 300gray demonstrated significant protection when rechallenged with transgenic Ct-LC parasites (Figures 6E, 6F, and S5B–S5F). Single or multiple exposures to this attenuated parasite vaccine led to protection equal to that observed in mice that experienced a live infection. To begin to define the requirements for vaccine induced immunity we conducted vaccination studies in T cell or IFNγ-deficient mouse strains lacking the key players we identified for resolution of the infection. In contrast to wild-type C57BL/6, Rag1−⁄− and Ifnγ−⁄− mice showed no protection from challenge following multiple vaccine exposures (Figures 6G and 6H). These results underscore the central role of IFNγ and T cells in resistance to Cryptosporidium (Figure 6I) and they show that this natural model provides an experimental system to rigorously define the host and parasite factors required for a vaccine to generate the pathogen-specific T cells necessary to protect from cryptosporidiosis.
Discussion
Cryptosporidium is an important pathogen with a global public health impact, but the numerous technical difficulties to study Cryptosporidium have limited progress (Striepen, 2013). However, recent advances in genetic manipulation (Vinayak et al., 2015), cryo-preservation of viable parasites (Jaskiewicz et al., 2018), and production of infectious oocyst within intestinal organoid cultures (Heo et al., 2018) have transformed the ability to manipulate Cryptosporidium. In the infective oocyst stage, the parasite is sheltered by a chemically and environmentally resilient wall that renders water chlorination ineffective in protecting drinking and recreation water, in fact we routinely use bleach in the laboratory when purifying oocysts to kill bacteria and viruses with no detriment to the parasite. Outbreaks, even in countries with advanced water treatment efforts, are common and can occur at massive scale (>460,000 cases in the 1993 Milwaukee outbreak; Mac Kenzie et al., 1994). In the United States, for example, despite the efforts of the Centers of Disease Control and others the incidence of cryptosporidiosis has been steady if not rising in recent years (Painter et al., 2015). Studies in India found that providing bottled water alone did little to curb cryptosporidiosis in children in a high endemicity setting (Sarkar et al., 2013). While improved water sanitation is of obvious benefit to pediatric health, additional tools are needed to protect children from cryptosporidiosis and others have highlighted the potential impact of a vaccine (Khalil et al., 2018). The model of cryptosporidiosis reported here replicates many features of human infection in a system where parasite and host are genetically tractable and provides the opportunity to develop a rational approach to test different vaccination strategies.
Cryptosporidiosis has epidemiological and clinical overlap with rotavirus infection, as both are common infections of young children (Kotloff et al., 2013). Several rotavirus vaccines are now available, including formulations optimized for populations outside of the United States and Europe. As an example, India has developed and now produces its own vaccine (Bhandari et al., 2014). These vaccines are safe and effective, and their use is recommended by the WHO (2013). A vaccine for cryptosporidiosis could have a similar life-saving impact for children in low and medium income countries. Rotavirus vaccines do not afford complete protection (Bar-Zeev et al., 2015, Bhandari et al., 2014), but nonetheless carry significant benefit. Similarly, natural infection with rotavirus does not result in sterile immunity yet provides protection from subsequent severe disease (Velázquez et al., 1996). This is remarkably similar to cryptosporidiosis; children can experience multiple infections and human volunteers can be infected in a secondary challenge, however, older children and adults with prior exposure show enhanced protection from disease (Chappell et al., 1999, Korpe et al., 2018, Kotloff et al., 2013). Infection with C. tyzzeri accurately matches the level of immunity observed in children, not sterile and yet significantly protective, providing a realistic test for vaccine candidates in a natural host-parasite relationship.
C. tyzzeri produces self-limiting infections restricted to the small intestine, and while mice do not develop watery diarrhea in response to this (or most other) infections, the associated pathology closely resembles what is seen in humans. Villus blunting, along with loss of the apical glycocalyx and effacement of microvilli can be readily detected in response to infection. There is also significant increase in the turnover of enterocytes manifesting as crypt hyperplasia. This could reflect an anaplerotic response simply replenishing cells lost because of infection. Alternatively, this could also be part of an orchestrated physiological response to limit the parasite through an accelerated villus elevator leading to enhanced shedding. While such physiological mechanisms are now a well-established as response to intestinal nematodes (Cliffe et al., 2005), examples for a role in the control of intracellular pathogens (Rauch et al., 2017) are still rare.
In the intestine the immune system faces important challenges that it has to balance. Microbes are extremely abundant as part of the commensal flora and they and their products must not trigger inflammation to avoid autoimmunity and allergy. At the same time, the intestine is a major entry point for infections and the immune system has to mount a vigorous response in an overall tolerogenic environment. This challenge may be reflected in the lack of ready sterile immunity to intracellular intestinal pathogens, but ultimately this is overcome. This natural mouse model of an infection that is truly limited to the intestinal epithelium offers a unique opportunity to mechanistically understand how immunity can overcome this challenge and may allow us to discovery ways to manipulate the process to better prevent disease.
The mouse model of cryptosporidiosis presented here allows for the study of immunity and pathogenesis in mice with mature and fully functional immune systems. Mouse strains with specific immune deficiencies can now be infected and compared against appropriate wild-type control. Two key players have already emerged from this early study: T cells and IFNγ. T cells are required to clear a primary infection as well as for the protective effect of the vaccine. IFNγ was previously implicated in innate control of C. parvum in mice and humans (Griffiths et al., 1998, White et al., 2000) and we similarly observe an important role early in C. tyzzeri infection. We further note that mice lacking IFNγ fail to fully clear C. tyzzeri infection and show no protection when vaccinated. IFNγ thus plays a central and complex role in both innate and adaptive immunity to Cryptosporidium, but the mechanism by which IFNγ acts to restrict Cryptosporidium within an infected enterocyte is unknown (Pollok et al., 2001). Another poorly understood aspect of Cryptosporidium immunity is which types of parasite antigen would be appropriate targets for protective immunity. Cryptosporidium occupies a peculiar, intracellular yet extracellular, position at the very periphery of the epithelium, and parasites are shed into the intestinal lumen and it is unclear what cellular processes are involved in antigen sampling and presentation required for T cell priming. The enterocyte itself could have an important role in this process, as they do express MHC class I and II, but their ability to prime CD4 and CD8 T cells in the context of an enteric pathogen has not been tested. In addition, there are specialized sub-sets of dendritic cells in the gut that have been shown to be able to sample microbial antigens from the lumen (Niess et al., 2005, Rescigno et al., 2001). It is now possible to assess their contribution to role in the development of Cryptosporidium specific T cell responses during infection or vaccination.
Recently, there has been a vigorous and concerted effort to develop new and more effective Cryptosporidium treatments (Baragaña et al., 2019, Buckner et al., 2019, Lee et al., 2019, Love et al., 2017, Manjunatha et al., 2017). One barrier to drug development is the high cost of in vivo drug testing; immunocompromised mice, gnotobiotic piglets, and calves are all suitable yet expensive models to test drug efficacy. Cryptosporidium tyzzeri infects a wide range of inbred and outbred mice (data not shown) and thus will lower the bar to measuring efficacy of compounds. Furthermore, malnutrition, a common complication in cryptosporidiosis infection, treatment, and recovery, can be effectively modeled using the natural mouse model (Costa et al., 2011). We note that while there is a significant change in fecal consistency during infection, mice do not develop watery diarrhea during C. tyzzeri infection. Because of this limitation, calves remain the model of cryptosporidiosis to evaluate the impact of diarrhea on pharmacokinetics.
C. tyzzeri is, phylogenetically, very closely related to the parasites that cause human infection, C. hominis and C. parvum. In fact, our de novo assembly for C. tyzzeri indicated that over 99% of its genome aligns with the genome sequences of C. hominis and parvum with >75% nucleotide identity. This high degree of conservation suggests that much of the parasite biology and metabolism are shared, which will facilitate in vivo analysis when investigating the function of specific genes and pathways. Despite the high level of identity and synteny, there are differences and they are most apparent in polymorphic regions that encode families of secretory proteins of still largely unknown function (Abrahamsen et al., 2004, Templeton et al., 2004, Xu et al., 2004). These differences may hold answers to the question of what determines host specificity. Mice are not a natural host for C. parvum, yet can be infected when immunosuppressed, most pronounced when IFNγ is removed. This suggests that, at least in part, the difference between C. tyzzeri and C. parvum may be explained by the ability of the parasite to appropriately interact with a host’s innate immune response. Proteins or protein families that distinguish parasite species thus may be strong candidates for factors that act to modulate or overcome immunity and their diversification is likely driven by a coevolution of host-parasite interaction that favors specialization. The ability to genetically engineer both C. parvum and C. tyzzeri offers an opportunity to experimentally dissect the molecular basis of their interaction with their murine host.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-I-A/I-E | BioLegend | Clone M5/114.15.2; RRID: AB_2565976 |
| anti-EpCAM | BioLegend | Clone G8.8; RRID: AB_1134105 |
| anti-H-2Db | BioLegend | Clone KH95; RRID: AB_313510 |
| anti-CD45.2 | eBioscience | Clone 104; RRID: AB_2534956 |
| anti-H-2Kb | BD Biosciences | Clone AF6-88.5; RRID: AB_394927 |
| Alexa Fluor 488 Phalloidin | Thermofisher | Cat# A12379; RRID: AB_2315147 |
| Rat monoclonal anti-mCherry | Thermofisher | Clone 16D7; RRID: AB_2536611 |
| Goat-anti-Rat polyclonal Alexa Fluor 594 | Thermofisher | Cat# A-11007; RRID: AB_10561522 |
| Critical Commercial Assays | ||
| Quick-DNA Fecal/Soil Microbe Kit | Zymo Research | Cat# D6010 |
| anti-Cryptosporidium magnetic beads | Thermofisher | Cat# 73011 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69504 |
| Deposited Data | ||
| Raw data from WGS of Cryptosporidium tyzzeri | NCBI | SRR5683558 |
| Assembled and annotated genome | EuPathDB | UGA55 |
| Experimental Models: Organisms/Strains | ||
| Cryptosporidium tyzzeri | This paper | UGA55 strain |
| Cryptosporidium parvum | Bunchgrass Farms, ID | IOWA strain |
| C57BL/6 mice | Jackson Laboratory | Strain 000664 |
| Ifng-/- mice | Jackson Laboratory | Strain 002287 |
| Rag1-/- mice | Jackson Laboratory | Strain 002216 |
| μMt- mice | Jackson Laboratory | Strain 002288 |
| athymic mice | Jackson Laboratory | Strain 002019 |
| Oligonucleotides | ||
| Primers for cloning, see Table S2 | This paper | N/A |
| 18 s probe and primer for parasite quantification, see Table S2 | Jothikumar et al., 2008 | N/A |
| Recombinant DNA | ||
| Cas9 expression vector with Cryptosporidium tyzzeri thymidine kinase locus guide | This paper and Vinayak et al., 2015 | N/A |
| Cryptosporidium mCherry/RE9 expression cassette vector | This paper | N/A |
| Software and Algorithms | ||
| MEGA software package | Kumar et al., 2016 | https://www.megasoftware.net/ |
| Genome Analysis Toolkit v3.8.0 | McKenna et al., 2010 | https://software.broadinstitute.org/gatk/ |
| Picard tools v2.16.0 | https://broadinstitute.github.io/picard/ | https://broadinstitute.github.io/picard/ |
| Burrows-Wheeler Aligner | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| Bedtools v2.26 | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| Samtools v1.6 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| R with circos package | Gu et al., 2014 | https://www.r-project.org/ |
| PRISM | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v10 software | TreeStar | https://www.flowjo.com/ |
| Other | ||
| 137-Cs irradiator | J.L Shepard | Mark1-30 |
| LSRFortessa | BD Biosciences | N/A |
| IVIS | Caliper Life Sciences | Lumina II |
| ViiA 7 Real-Time System | Thermofisher | N/A |
Contact for Reagent and Resource Sharing
For access to reagents or parasite strains used in this study please contact the Lead Contact, Boris Striepen (striepen@upenn.edu).
Experimental Model and Subject Details
Mouse Models of Infection
C57BL/6 (stock no:000664), Ifng-/- (stock no:002287), Rag1-/- (stock no:002216), μMt- (stock no:002288), and athymic mice (stock no:002019) were purchased from Jackson Laboratory. Unless otherwise noted, mice used in this study were females ranging from 4 – 8 weeks, however, we note that we did not measure a difference in infection burden between male and female mice in both C57BL/6 and Ifng-/- mice. All mice were gender and age matched within individual experiments. All protocols for animal care were approved by the Institutional Animal Care and Use Committee of the University of Georgia (protocol A2016 01-028-Y1-A4) and the Institutional Animal Care and Use Committee of the University of Pennsylvania (protocol #806292).
Parasite Strains
Cryptosporidium tyzzeri wild-type and transgenic strains are propagated within infected Ifng-/- mice (stock no:002287). Oocysts are then purified from fecal collections using sucrose flotation followed by a cesium chloride gradient (see Method Details). Cryptosporidium parvum oocysts used in this study are Iowa strain, purchased from Bunchgrass Farms (Dreary, ID).
Method Details
Isolation of Parasites
Mouse fecal pellets were collected and stored in water overnight. Collected pellets were then assayed by qPCR for the presence of the Cryptosporidium using a 18S probe and primer set (see Table S2) from fecal isolated with Quick-DNA Fecal/Soil Microbe Kit (Zymo Research, Irvine, CA, USA). If positive for the presence of Cryptosporidium, sample DNA was subjected to further genotyping (gp60, SSU, and COWP1) to determine the species. During this time fecal pellets were stored in 2.5% potassium dichromate at 4°C. To inoculate mice, fecal samples were first washed 3× with cold water to remove potassium dichromate, then filtered through a 200um strainer. Strained fecal material was treated with a 1:4 dilution of household bleach for 5min on ice, washed 3× with cold tap water, and used to gavage C57BL/6 mice housed in quarantine. Parasite shedding was monitored by qPCR, and infected mice underwent a full screen by a veterinary pathologist to test for concurrent infections (see Table S1). This procedure was repeated twice to transfer the infection to successive cages, with an additional purification step utilizing anti-Cryptosporidium magnetic beads (Thermofisher, Waltham, MA). Again, mice underwent a full screen by a veterinary pathologist following each infection.
18S Phylogenetic Analyses
18S rRNA from UGA55 was amplified using specific primers (see Table S2) and phylogenetic tree was created with the MEGA software package (Kumar et al., 2016). Alignment was performed using MUSCLE (Edgar, 2004) under default settings, and phylogenetic reconstruction was performed using Maximum Likelihood, Neighbor-Joining, and Maximum Parsimony to ensure consensus. Maximum Likelihood is displayed in the manuscript using 500 bootstrap replications. Sequences used for alignment are as follows: Cryptosporidium fragilis GenBank: EU162753.1, Cryptosporidium andersoni GenBank: AY954885.1, Cryptosporidium muris GenBank: L19069.1, Cryptosporidium felis GenBank: AF108862.1, Cryptosporidium canis GenBank: AF112576.1, Cryptosporidium meleagridis GenBank: AF112574.1, Cryptosporidium wrairi GenBank: AF115378.1, Cryptosporidium baileyi GenBank: L19068.1, Cryptosporidium suis GenBank: AF115377.1, Cryptosporidium fayari GenBank: AF108860.1, Cryptosporidium hominis CryptoDB: Chro.rrn016, and Cryptosporidium parvum CryptoDB: cgd7_7.
Genome Sequencing
107 C. tyzzeri oocysts were incubated at 37°C for 1 hr in 0.8% sodium taurocholate in PBS to induce excystation. DNA was then purified using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), and a DNA library was prepared with a Nextera XT Prep Kit (Illumina, San Diego, CA, USA). Reads were generated using 150-bp pared-end sequencing on a MiSeq platform (Illumina, San Diego, CA, USA) at the Georgia Genomics and Bioinformatics Core.
Genome Assembly and Annotation
All reads obtained by the Illumina Miseq sequencing platform had their adapters and low-quality sequences removed using (Bolger et al., 2014). The threshold used was a minimum read length of 100 bases and a phred-scale quality score > 30. The genome was assembled de novo with Velvet assembler v1.2.10 using a k-mer value of 87 and a minimum contig length of 800 bases. Scaffolding was performed using two lines of evidence, firstly using the paired-end information of 300 insert size followed by a guided scaffolding using the close related C. parvum genome PacBio long reads to order these resulted scaffolds from the previous step using SSPACE-long reads v.1.1 (Boetzer and Pirovano, 2014). The final scaffolds were submitted to GapFiller v.1.10 (Boetzer and Pirovano, 2012) with 10 interactions to extend end fill the gaps using the Illumina sequence, and each base was corrected by using ICORN v.0.97 (Otto et al., 2010).
The genome was annotated using three different gene prediction tools: (i) an ab initio prediction using GeneMark-ES (Lomsadze et al., 2005); (ii) evidence-trained predictions by SNAP/Maker (Cantarel et al., 2008, Johnson et al., 2008) and (iii) Augustus (Stanke and Morgenstern, 2005). The training-based step for these predictions was made by using evidence from publicly available data from several Cryptosporidium species and a manual curation of all genes in the context of existing molecular evidence (Baptista et al., unpublished data).
SNP Analyses and SNP/Synteny Map Generation
Cryptosporidium Illumina reads (Sequence Read Archive accession numbers: SRR5683558 C.tyzzeri UGA55; SRR1557959 C.hominis 30976; SRR6147945 C. parvum UKP6; and SRR001350 C. muris RN66) were mapped against the C. tyzzeri UGA55 genome sequence generated in this project using BWA v. 0.7.15 (Li and Durbin, 2009) with the mem mode. The resulting alignment was then processed to remove PCR duplicates (which can introduce bias in variant calling) and also increases the quality and reliability of the analysis using Picard tools v2.16.0 (http://broadinstitute.github.io/picard/). Variants were called using the Genome Analysis Toolkit v3.8.0 (GATK) Haplotypecaller (McKenna et al., 2010) with a ploidy of one. Variants with low mapping quality (<25), allele frequency lower than 70%, or less than 10x coverage were discarded. The variant call files (vcf) were then processed using samtools v1.6 (Li et al., 2009) and bedtools v.2.26 (Quinlan and Hall, 2010). SNPs were then grouped into 1 kb windows and plotted in R using the circlize package (Gu et al., 2014). For synteny analysis, genome sequences were aligned using blastn with an e-value of 1e-20, and then the alignments were plotted using the Rcircos package for R (Krzywinski et al., 2009).
Generation of Transgenic Parasites
107 C. tyzzeri oocysts were first incubated at 37°C for 1 hr in 0.8% sodium taurocholate to induce excystation. Excysted sporozoites were then transfected using an Amaxa 4D electroporater (Lonza, Basel, Switzerland) with parasites suspended in SF buffer and using the program EH100. 50 μg of each plasmid were used for transfection, one encoding Cas9/gRNA, and another encoding a selection cassette surrounded by 1 kb of homologous DNA for insertion into the genome (Pawlowic et al., 2017, Vinayak et al., 2015). Transfected parasites were surgically implanted (Vinayak et al., 2015) into the small intestine of anesthetized Ifng-/- mice. Paromomycin, was given to the mice in their drinking water (16 mg/mL) ad libitum to select for parasites rendered resistant through expression of neomycin phosphotransferase. Parasite shedding was monitored by fecal qPCR and feces were collected daily during peak shedding.
To purify transgenic parasites from collected feces we used sucrose flotation followed by a cesium chloride gradient (Pawlowic et al., 2017). Collected mouse feces was homogenized in tap water using a LabGen 125 homogenizer (Cole-Parmer, Vernon Hills, IL, USA) and filtered through a 250-μm mesh filter. This resulting filtrate was mixed 1:1 with a saturated sucrose solution (specific gravity 1.33) and centrifuged at 1,000 g for 5 min. Supernatant was collected, resuspended in 0.85% saline solution and overlaid onto CsCl solution (specific gravity 1.15), and centrifuged at 16,000 g for 3 min. Purified oocysts were collected from the saline-CsCl interface and resuspended in cold PBS.
Measuring Parasite Shedding by Fecal qPCR
Parasite DNA was isolated from 100 mg of collected fecal material using a Quick-DNA Fecal/Soil Microbe Kit (Zymo Research, Irvine, CA, USA). Parasite shedding was measured using a Cryptosporidium specific probe and primer set (Jothikumar et al., 2008) (see Table S2). SsoAdvanced Universal Probes Supermix (BioRad, Hercules, CA, USA) was used with a ViiA 7 Real-Time System (Thermofisher, Waltham, MA, USA) to perform qPCR. To quantify parasites samples were run with a standard curve of DNA from fecal samples spiked with a known concentration of oocysts.
Measuring Parasite Burden in Tissue by In Vivo Imaging
Mice were subcutaneously injected with D-luciferin (Gold Biotechnology, St Louis, MO, USA) at 125 mg/kg, then anesthetized in an induction chamber using 2.5% isoflurane. After five minutes mice were moved into an IVIS Lumina II instrument (Caliper Life Sciences, Waltham, MA, USA) and luminescence was measured using 5 min exposure, medium binning, and 1/16 F-stop.
Measuring Parasite Burden in Tissue by qPCR
Mice were killed according to our approved animal use protocol and sections were dissected from the small intestine and colon, 1 cm in length, and thoroughly washed in PBS. From the stomach and the caecum, 1 cm x 1 cm square sections were cut. DNA was purified from each section using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and parasite burden was measured using a Cryptosporidium specific probe and primer set (see Table S2). SsoAdvanced Universal Probes Supermix (BioRad, Hercules, CA, USA) was used with a ViiA 7 Real-Time System (Thermofisher, Waltham, MA, USA) to perform qPCR.
Flow Cytometry
Single-cell suspensions were prepared from intestinal sections by shaking diced tissue at 37°C for 25 minutes in Hank’s Balanced Salt Solution with 5 mM EDTA and 1 mM DTT. Cell pellets were then passed through 70 μm and 40 μm filters. Cells were surface stained using the following fluorochrome-conjugated Abs: anti-I-A/I-E (M5/114.15.2), anti-EpCAM (G8.8), and anti-H-2Db (KH95) from BioLegend, anti-CD45.2 (104) from eBioscience, and anti-H-2Kb (AF6-88.5) from BD Biosciences. Data were collected on a LSRFortessa (BD Biosciences) and were analyzed with FlowJo v10 software (TreeStar).
Histology of Intestinal Tissue
Mice were killed and small intestine was dissected. This tissue was flushed with 10% neutral buffered formalin (Sigma, St Louis, MO, USA), then ‘swiss-rolled’ and fixed overnight. Fixed samples were paraffin-embedded, sectioned, and stained with haematoxylin and eosin. Slides were evaluated by a board-certified veterinary pathologist in a blinded fashion for quantitative measurements of number of parasites, villus/crypt architectural features and inflammatory infiltrates, and semi-quantitative scores for villus epithelium lesions (see Figure S6). Intestinal segments with highest number of parasites comprising distal jejunum to proximal ileum, which were consistent for all mice in the study, were selected for detailed histologic evaluation. To avoid inconsistent intra-segmental measurements due to differing orientations of villi and crypts within the section, five consecutive and complete villus-crypt structures, meaning villus and crypt lumens within the same plane of section, were selected for scoring. C. tyzzeri organisms within the apical regions of villus epithelium within each of the five villi were totaled. Measurements obtained for villus height, villus width taken at the mid-point of the villus height, crypt depth and crypt width taken at the mid-depth of the crypt were averaged, and villus:crypt height ratios were calculated. Numbers of villus and crypt goblet cells, crypt mitotic figures and lamina propria inflammatory infiltrates (neutrophils, eosinophils, plasma cells) for 5 villus/crypt structures were tallied and averaged. Gastro-Intestinal Lymphoid Tissue (GALT) foci and non-follicular lymphocyte aggregates for each section of distal jejunum/ileum segments were also enumerated. Semi-quantitative scores for the following parameters were generated as follows: villus epithelial separation/sloughing (0 = none; 1 = cell separation from villus tip of 1-2 out of 5 villi evaluated; 2 = cell separation from villus tip of 3-5 out of 5 villi evaluated; 3 = denuded villus tips; 4 = denuded villus tips with fibrin and/or inflammatory exudate); villus epithelial dysplasia (0 = none; cells well-aligned in a monolayer; 1 = cytoplasmic basophilia with high nuclear:cytoplamic ratio; 2 = mild, multifocal epithelial disorganization; 3 = regional moderate epithelial malalignment, 4 = marked regional malalignment with cell piling); villus epithelial attenuation (0 = none, epithelium are uniformly tall and columnar-shaped; 1 = multifocal cuboidal-shaped epithelium; 2 = regional-multiregional cuboidal shaped-epithelium; 3 = flattened villus epithelium); and degree of crypt branching (0 = none; 1 = 1-2 branches observed for 1–2 out of 5 crypts evaluated; 2 = 1-2 branches observed for 3-5 out of 5 crypts evaluated; 3 = greater than 2 branches within 1 or more crypts out of 5 crypts evaluated.
Immunofluorescence of Intestinal Tissue
Mice were killed and the distal 1/3 of the small intestine was dissected. This tissue was flushed with 10% neutral buffered formalin (Sigma, St Louis, MO, USA), then ‘swiss-rolled’ and fixed overnight. Samples were then placed 30% sucrose in phosphate buffered saline (PBS) for cryoprotection, and mounted with OCT compound (Tissue-Tek, Sakura Finetek, Japan). Sections were permeabilized and blocked with 10% BSA in PBS with 0.1% triton-x (Sigma, St Louis, MO, USA) for 1 hr, then incubated with primary and secondary antibody in 1% BSA in PBS with 0.1% triton-x. Primary label used: 1:50 Alexa Fluor 647 Phalloidin (Thermofisher, Waltham, MA), 1:1000 rat monoclonal anti-mCherry, 1-50 rabbit polyclonal anti-CD3 (Abcam, Cambridge, UK). Secondary antibodies used: Goat-anti-Rat polyclonal Alexa Fluor 594 (Thermofisher, Waltham, MA) and Goat-anti-Rabbit polyclonal Alexa Fluor 488 (Thermofisher, Waltham, MA). Nuclei of parasite and host were stained using Hoeschst 33342 (Thermofisher, Waltham, MA).
Gamma Irradiation of Parasites
Oocysts were exposed to gamma irradiation using a J.L Shepard Mark 1-30 self-shielded irradiator with a 137Cs source and were kept cold throughout within a chilled tube rack.
Quantification and Statistical Analysis
GraphPad PRISM was used for all statistical analyses. When measuring the difference between two populations, we used a standard T-test. For data sets with 3 or more experimental groups we used a one-way ANOVA with Dunnett’s multiple comparison’s test. No statistical tests were used to predetermine sample size and no animals were excluded from results. Replicates were as follows: Figure 3A, N = 7 from two separate experiments; Figure 3D–3H, N = 20 with 5 mice per experimental group; Figures 5A and 5B, N = 9 mice from two separate experiments; Figures 4F and 4G, for EpCAM measurements n = 16 mice from 6 experiments, for MHCI measurements n = 8 mice from 3 experiments, for MHCII measurements n = 12 mice from 5 experiments; Figure 5C, N = 12 with 4 mice per experimental group; Figure 5D, N = 16 with 4 mice per experimental group; Figure 5E, N = 6 with 3 mice per experimental group; Figure 6A, N = 9 with 3 mice per experimental group; Figure 6C, N = 36 with 18 mice per experimental group over 5 separate experiments; Figure 6D, N = 9 with 3 mice per experimental group; Figure 6F, N = 30 mice over two separate experiments; Figure 6G, N = 10 with 5 mice per experimental group; Figure 6H, N = 10 with 5 mice per experimental group; Figure 6I, N = 10 with 5 mice per experimental group.
Date and Software Availability
Raw sequencing data is available through NCBI (SRR5683558) and a fully assembled and annotated genome of Cryptosporidium tyzzeri is available through the Eukaryotic Pathogen Database (https://eupathdb.org/eupathdb/). Please refer to the Key Resources Table for links to the software used for genome assembly, annotation, and analysis.
Acknowledgments
We thank Dr. Lorraine Fuller for help and advice on oocyst collection and isolation and Dr. Cameron Koch for access to and help with gamma irradiation. This work was funded in part by grants from the National Institutes of Health (R01AI112427) and the Bill & Melinda Gates Foundation to B.S. (OPP1161001) and to J.C.K. (OPP1151701). A.S. was supported by a fellowship (F32AI124518) and a pathway to independence award (K99AI137442), and J.A.G. was supported by a fellowship (T32A1055400) from the National Institutes of Health.
Author Contributions
J.S. and A.S. collected field samples, isolated oocysts, and conducted initial passage and typing. A.S. developed transgenic model, sequenced genome, and conducted mouse studies into susceptibility, resolution, immunity, and vaccination with support from G.T.H., C.F.B., and E.M.K. R.B. and J.C.K. assembled and annotated the genome sequence and performed comparative analyses, J.B.E. conducted histopathological analysis and J.A.G. and C.A.H. flow cytometry. A.S., J.S., and B.S. conceived of the study and A.S. and B.S. wrote the manuscript with contributions from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: June 20, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2019.05.006.
Supporting Citations
The following references appear in the Supplemental Information: Emms and Kelly, 2015, Guindon et al., 2010, Stamatakis, 2014.
Supplemental Information
References
- WHO Rotavirus vaccines WHO position paper: January 2013-recommendations. Vaccine. 2013;31:6170–6171. doi: 10.1016/j.vaccine.2013.05.037. [DOI] [PubMed] [Google Scholar]; WHO, (2013). Rotavirus vaccines WHO position paper: January 2013-recommendations. Vaccine 31, 6170-6171. [DOI] [PubMed]
- Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A., Deng M., Liu C., Widmer G., Tzipori S. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]; Abrahamsen, M.S., Templeton, T.J., Enomoto, S., Abrahante, J.E., Zhu, G., Lancto, C.A., Deng, M., Liu, C., Widmer, G., Tzipori, S., et al. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441-445. [DOI] [PubMed]
- Baragaña B., Forte B., Choi R., Nakazawa Hewitt S., Bueren-Calabuig J.A., Pisco J.P., Peet C., Dranow D.M., Robinson D.A., Jansen C. Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis. Proc. Natl. Acad. Sci. USA. 2019;116:7015–7020. doi: 10.1073/pnas.1814685116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baragaña, B., Forte, B., Choi, R., Nakazawa Hewitt, S., Bueren-Calabuig, J.A., Pisco, J.P., Peet, C., Dranow, D.M., Robinson, D.A., Jansen, C., et al. (2019). Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis. Proc. Natl. Acad. Sci. USA 116, 7015-7020. [DOI] [PMC free article] [PubMed]
- Bartelt L.A., Bolick D.T., Kolling G.L., Roche J.K., Zaenker E.I., Lara A.M., Noronha F.J., Cowardin C.A., Moore J.H., Turner J.R. Cryptosporidium priming is more effective than vaccine for protection against cryptosporidiosis in a murine protein malnutrition model. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004820. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bartelt, L.A., Bolick, D.T., Kolling, G.L., Roche, J.K., Zaenker, E.I., Lara, A.M., Noronha, F.J., Cowardin, C.A., Moore, J.H., Turner, J.R., et al. (2016). Cryptosporidium priming is more effective than vaccine for protection against cryptosporidiosis in a murine protein malnutrition model. PLoS Negl. Trop. Dis. 10. [DOI] [PMC free article] [PubMed]
- Bar-Zeev N., Kapanda L., Tate J.E., Jere K.C., Iturriza-Gomara M., Nakagomi O., Mwansambo C., Costello A., Parashar U.D., Heyderman R.S. Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic roll-out: an observational and case-control study. Lancet Infect. Dis. 2015;15:422–428. doi: 10.1016/S1473-3099(14)71060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bar-Zeev, N., Kapanda, L., Tate, J.E., Jere, K.C., Iturriza-Gomara, M., Nakagomi, O., Mwansambo, C., Costello, A., Parashar, U.D., Heyderman, R.S., et al. (2015). Effectiveness of a monovalent rotavirus vaccine in infants in Malawi after programmatic roll-out: an observational and case-control study. Lancet Infect. Dis.. 15, 422-428. [DOI] [PMC free article] [PubMed]
- Belkaid Y., Piccirillo C.A., Mendez S., Shevach E.M., Sacks D.L. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]; Belkaid, Y., Piccirillo, C.A., Mendez, S., Shevach, E.M., and Sacks, D.L. (2002). CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature 420, 502-507. [DOI] [PubMed]
- Bhandari N., Rongsen-Chandola T., Bavdekar A., John J., Antony K., Taneja S., Goyal N., Kawade A., Kang G., Rathore S.S. Efficacy of a monovalent human-bovine (116E) rotavirus vaccine in Indian infants: a randomised, double-blind, placebo-controlled trial. Lancet. 2014;383:2136–2143. doi: 10.1016/S0140-6736(13)62630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bhandari, N., Rongsen-Chandola, T., Bavdekar, A., John, J., Antony, K., Taneja, S., Goyal, N., Kawade, A., Kang, G., Rathore, S.S., et al. (2014). Efficacy of a monovalent human-bovine (116E) rotavirus vaccine in Indian infants: a randomised, double-blind, placebo-controlled trial. Lancet 383, 2136-2143. [DOI] [PMC free article] [PubMed]
- Boetzer M., Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13, R56. [DOI] [PMC free article] [PubMed]
- Boetzer M., Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boetzer, M., and Pirovano, W. (2014). SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15, 211. [DOI] [PMC free article] [PubMed]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bolger, A.M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. [DOI] [PMC free article] [PubMed]
- Borad A., Ward H. Human immune responses in cryptosporidiosis. Future Microbiol. 2010;5:507–519. doi: 10.2217/fmb.09.128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Borad, A., and Ward, H. (2010). Human immune responses in cryptosporidiosis. Future Microbiol. 5, 507-519. [DOI] [PMC free article] [PubMed]
- Buckner F.S., Ranade R.M., Gillespie J.R., Shibata S., Hulverson M.A., Zhang Z., Huang W., Choi R., Verlinde C.L.M.J., Hol W.G.J. Optimization of methionyl tRNA-synthetase inhibitors for treatment of cryptosporidium infection. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02061-18. [DOI] [PMC free article] [PubMed] [Google Scholar]; Buckner, F.S., Ranade, R.M., Gillespie, J.R., Shibata, S., Hulverson, M.A., Zhang, Z., Huang, W., Choi, R., Verlinde, C.L.M.J., Hol, W.G.J., et al. (2019). Optimization of methionyl tRNA-synthetase inhibitors for treatment of cryptosporidium infection. Antimicrob. Agents Chemother. 63. [DOI] [PMC free article] [PubMed]
- Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cantarel, B.L., Korf, I., Robb, S.M., Parra, G., Ross, E., Moore, B., Holt, C., Sanchez Alvarado, A., and Yandell, M. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188-196. [DOI] [PMC free article] [PubMed]
- Chappell C.L., Okhuysen P.C., Sterling C.R., Wang C., Jakubowski W., Dupont H.L. Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am. J. Trop. Med. Hyg. 1999;60:157–164. doi: 10.4269/ajtmh.1999.60.157. [DOI] [PubMed] [Google Scholar]; Chappell, C.L., Okhuysen, P.C., Sterling, C.R., Wang, C., Jakubowski, W., and Dupont, H.L. (1999). Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am. J. Trop. Med. Hyg.. 60, 157-164. [DOI] [PubMed]
- Checkley W., Epstein L.D., Gilman R.H., Black R.E., Cabrera L., Sterling C.R. Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am. J. Epidemiol. 1998;148:497–506. doi: 10.1093/oxfordjournals.aje.a009675. [DOI] [PubMed] [Google Scholar]; Checkley, W., Epstein, L.D., Gilman, R.H., Black, R.E., Cabrera, L., and Sterling, C.R. (1998). Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am. J. Epidemiol.. 148, 497-506. [DOI] [PubMed]
- Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Checkley, W., White, A.C., Jr., Jaganath, D., Arrowood, M.J., Chalmers, R.M., Chen, X.M., Fayer, R., Griffiths, J.K., Guerrant, R.L., Hedstrom, L., et al. (2015). A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis.. 15, 85-94. [DOI] [PMC free article] [PubMed]
- Choudhry N., Korbel D.S., Edwards L.A., Bajaj-Elliott M., McDonald V. Dysregulation of interferon-gamma-mediated signalling pathway in intestinal epithelial cells by Cryptosporidium parvum infection. Cell. Microbiol. 2009;11:1354–1364. doi: 10.1111/j.1462-5822.2009.01336.x. [DOI] [PubMed] [Google Scholar]; Choudhry, N., Korbel, D.S., Edwards, L.A., Bajaj-Elliott, M., and McDonald, V. (2009). Dysregulation of interferon-gamma-mediated signalling pathway in intestinal epithelial cells by Cryptosporidium parvum infection. Cell. Microbiol. 11, 1354-1364. [DOI] [PubMed]
- Cliffe L.J., Humphreys N.E., Lane T.E., Potten C.S., Booth C., Grencis R.K. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]; Cliffe, L.J., Humphreys, N.E., Lane, T.E., Potten, C.S., Booth, C., and Grencis, R.K. (2005). Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science 308, 1463-1465. [DOI] [PubMed]
- Costa L.B., JohnBull E.A., Reeves J.T., Sevilleja J.E., Freire R.S., Hoffman P.S., Lima A.A.M., Oriá R.B., Roche J.K., Guerrant R.L. Cryptosporidium-malnutrition interactions: mucosal disruption, cytokines, and TLR signaling in a weaned murine model. J. Parasitol. 2011;97:1113–1120. doi: 10.1645/GE-2848.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; Costa, L.B., JohnBull, E.A., Reeves, J.T., Sevilleja, J.E., Freire, R.S., Hoffman, P.S., Lima, A.A.M., Oria, R.B., Roche, J.K., Guerrant, R.L., et al. (2011). Cryptosporidium-malnutrition interactions: mucosal disruption, cytokines, and TLR signaling in a weaned murine model. J. Parasitol.. 97, 1113-1120. [DOI] [PMC free article] [PubMed]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Edgar, R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792-1797. [DOI] [PMC free article] [PubMed]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Emms, D.M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157. [DOI] [PMC free article] [PubMed]
- Flanigan T., Whalen C., Turner J., Soave R., Toerner J., Havlir D., Kotler D. Cryptosporidium infection and CD4 counts. Ann. Intern. Med. 1992;116:840–842. doi: 10.7326/0003-4819-116-10-840. [DOI] [PubMed] [Google Scholar]; Flanigan, T., Whalen, C., Turner, J., Soave, R., Toerner, J., Havlir, D., and Kotler, D. (1992). Cryptosporidium infection and CD4 counts. Ann. Intern. Med.. 116, 840-842. [DOI] [PubMed]
- Fox B.A., Bzik D.J. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature. 2002;415:926–929. doi: 10.1038/415926a. [DOI] [PubMed] [Google Scholar]; Fox, B.A., and Bzik, D.J. (2002). De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415, 926-929. [DOI] [PubMed]
- Griffiths J.K., Theodos C., Paris M., Tzipori S. The gamma interferon gene knockout mouse: a highly sensitive model for evaluation of therapeutic agents against Cryptosporidium parvum. J. Clin. Microbiol. 1998;36:2503–2508. doi: 10.1128/jcm.36.9.2503-2508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]; Griffiths, J.K., Theodos, C., Paris, M., and Tzipori, S. (1998). The gamma interferon gene knockout mouse: a highly sensitive model for evaluation of therapeutic agents against Cryptosporidium parvum. J. Clin. Microbiol. 36, 2503-2508. [DOI] [PMC free article] [PubMed]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B. Circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]; Gu, Z., Gu, L., Eils, R., Schlesner, M., and Brors, B. (2014). Circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811-2812. [DOI] [PubMed]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]; Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307-321. [DOI] [PubMed]
- Heiges M., Wang H., Robinson E., Aurrecoechea C., Gao X., Kaluskar N., Rhodes P., Wang S., He C.Z., Su Y. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heiges, M., Wang, H., Robinson, E., Aurrecoechea, C., Gao, X., Kaluskar, N., Rhodes, P., Wang, S., He, C.Z., Su, Y., et al. (2006). CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 34, D419-D422. [DOI] [PMC free article] [PubMed]
- Heo I., Dutta D., Schaefer D.A., Iakobachvili N., Artegiani B., Sachs N., Boonekamp K.E., Bowden G., Hendrickx A.P.A., Willems R.J.L. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Heo, I., Dutta, D., Schaefer, D.A., Iakobachvili, N., Artegiani, B., Sachs, N., Boonekamp, K.E., Bowden, G., Hendrickx, A.P.A., Willems, R.J.L., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814-823. [DOI] [PMC free article] [PubMed]
- Huston C.D., Spangenberg T., Burrows J., Willis P., Wells T.N.C., van Voorhis W. A proposed target product profile and developmental cascade for new cryptosporidiosis treatments. PLoS Negl. Trop. Dis. 2015;9:e0003987. doi: 10.1371/journal.pntd.0003987. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huston, C.D., Spangenberg, T., Burrows, J., Willis, P., Wells, T.N.C., and van Voorhis, W. (2015). A proposed target product profile and developmental cascade for new cryptosporidiosis treatments. PLoS Negl. Trop. Dis. 9, e0003987. [DOI] [PMC free article] [PubMed]
- Jaskiewicz J.J., Sandlin R.D., Swei A.A., Widmer G., Toner M., Tzipori S. Cryopreservation of infectious Cryptosporidium parvum oocysts. Nat. Commun. 2018;9:2883. doi: 10.1038/s41467-018-05240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jaskiewicz, J.J., Sandlin, R.D., Swei, A.A., Widmer, G., Toner, M., and Tzipori, S. (2018). Cryopreservation of infectious Cryptosporidium parvum oocysts. Nat. Commun.. 9, 2883. [DOI] [PMC free article] [PubMed]
- Jenkins M., Higgins J., Kniel K., Trout J., Fayer R. Protection of calves against cryptosporiosis by oral inoculation with gamma-irradiated Cryptosporidium parvum oocysts. J. Parasitol. 2004;90:1178–1180. doi: 10.1645/GE-3333RN. [DOI] [PubMed] [Google Scholar]; Jenkins, M., Higgins, J., Kniel, K., Trout, J., and Fayer, R. (2004). Protection of calves against cryptosporiosis by oral inoculation with gamma-irradiated Cryptosporidium parvum oocysts. J. Parasitol.. 90, 1178-1180. [DOI] [PubMed]
- Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I.W. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johnson, A.D., Handsaker, R.E., Pulit, S.L., Nizzari, M.M., O'Donnell, C.J., and de Bakker, P.I.W. (2008). SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938-2939. [DOI] [PMC free article] [PubMed]
- Jokipii L., Jokipii A.M. Timing of symptoms and oocyst excretion in human cryptosporidiosis. N. Engl. J. Med. 1986;315:1643–1647. doi: 10.1056/NEJM198612253152604. [DOI] [PubMed] [Google Scholar]; Jokipii, L., and Jokipii, A.M. (1986). Timing of symptoms and oocyst excretion in human cryptosporidiosis. N. Engl. J. Med.. 315, 1643-1647. [DOI] [PubMed]
- Jothikumar N., da Silva A.J., Moura I., Qvarnstrom Y., Hill V.R. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol. 2008;57:1099–1105. doi: 10.1099/jmm.0.2008/001461-0. [DOI] [PubMed] [Google Scholar]; Jothikumar, N., da Silva, A.J., Moura, I., Qvarnstrom, Y., and Hill, V.R. (2008). Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol. 57, 1099-1105. [DOI] [PubMed]
- Khalil I.A., Troeger C., Rao P.C., Blacker B.F., Brown A., Brewer T.G., Colombara D.V., De Hostos E.L., Engmann C., Guerrant R.L. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health. 2018;6:e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khalil, I.A., Troeger, C., Rao, P.C., Blacker, B.F., Brown, A., Brewer, T.G., Colombara, D.V., De Hostos, E.L., Engmann, C., Guerrant, R.L., et al. (2018). Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health 6, e758-e768. [DOI] [PMC free article] [PubMed]
- Korpe P.S., Gilchrist C., Burkey C., Taniuchi M., Ahmed E., Madan V., Castillo R., Ahmed S., Arju T., Alam M. Case-control study of cryptosporidium transmission in Bangladeshi households. Clin. Infect. Dis. 2018 doi: 10.1093/cid/ciy593. [DOI] [PMC free article] [PubMed] [Google Scholar]; Korpe, P.S., Gilchrist, C., Burkey, C., Taniuchi, M., Ahmed, E., Madan, V., Castillo, R., Ahmed, S., Arju, T., Alam, M., et al. (2018). Case-control study of cryptosporidium transmission in Bangladeshi households. Clin. Infect. Dis.. [DOI] [PMC free article] [PubMed]
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]; Kotloff, K.L., Nataro, J.P., Blackwelder, W.C., Nasrin, D., Farag, T.H., Panchalingam, S., Wu, Y., Sow, S.O., Sur, D., Breiman, R.F., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209-222. [DOI] [PubMed]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., Jones, S.J., and Marra, M.A. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639-1645. [DOI] [PMC free article] [PubMed]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis 7.0 version for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar, S., Stecher, G., and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis 7.0 version for bigger datasets. Mol. Biol. Evol. 33, 1870-1874. [DOI] [PMC free article] [PubMed]
- Kváč M., McEvoy J., Loudová M., Stenger B., Sak B., Květoňová D., Ditrich O., Rašková V., Moriarty E., Rost M. Coevolution of Cryptosporidium tyzzeri and the house mouse (Mus musculus) Int. J. Parasitol. 2013;43:805–817. doi: 10.1016/j.ijpara.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kvač, M., McEvoy, J., Loudova, M., Stenger, B., Sak, B., Květoňova, D., Ditrich, O., Raškova, V., Moriarty, E., Rost, M., et al. (2013). Coevolution of Cryptosporidium tyzzeri and the house mouse (Mus musculus). Int. J. Parasitol.. 43, 805-817. [DOI] [PMC free article] [PubMed]
- Lee S., Ginese M., Girouard D., Beamer G., Huston C.D., Osbourn D., Griggs D.W., Tzipori S. Piperazine-Derivative MMV665917: an effective drug in the diarrheic piglet model of Cryptosporidium hominis. J. Infect. Dis. 2019 doi: 10.1093/infdis/jiz105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee, S., Ginese, M., Girouard, D., Beamer, G., Huston, C.D., Osbourn, D., Griggs, D.W., and Tzipori, S. (2019). Piperazine-Derivative MMV665917: an effective drug in the diarrheic piglet model of Cryptosporidium hominis. J. Infect. Dis.. [DOI] [PMC free article] [PubMed]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760. [DOI] [PMC free article] [PubMed]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., and Durbin, R., and Genome Project Data Processing. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078-2079. [DOI] [PMC free article] [PubMed]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, Y.O., and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33, 6494-6506. [DOI] [PMC free article] [PubMed]
- Love M.S., Beasley F.C., Jumani R.S., Wright T.M., Chatterjee A.K., Huston C.D., Schultz P.G., McNamara C.W. A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl. Trop. Dis. 2017;11:e0005373. doi: 10.1371/journal.pntd.0005373. [DOI] [PMC free article] [PubMed] [Google Scholar]; Love, M.S., Beasley, F.C., Jumani, R.S., Wright, T.M., Chatterjee, A.K., Huston, C.D., Schultz, P.G., and McNamara, C.W. (2017). A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl. Trop. Dis. 11, e0005373. [DOI] [PMC free article] [PubMed]
- Lumadue J.A., Manabe Y.C., Moore R.D., Belitsos P.C., Sears C.L., Clark D.P. A clinicopathologic analysis of AIDS-related cryptosporidiosis. AIDS. 1998;12:2459–2466. doi: 10.1097/00002030-199818000-00015. [DOI] [PubMed] [Google Scholar]; Lumadue, J.A., Manabe, Y.C., Moore, R.D., Belitsos, P.C., Sears, C.L., and Clark, D.P. (1998). A clinicopathologic analysis of AIDS-related cryptosporidiosis. AIDS 12, 2459-2466. [DOI] [PubMed]
- Mac Kenzie W.R., Hoxie N.J., Proctor M.E., Gradus M.S., Blair K.A., Peterson D.E., Kazmierczak J.J., Addiss D.G., Fox K.R., Rose J.B. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]; Mac Kenzie, W.R., Hoxie, N.J., Proctor, M.E., Gradus, M.S., Blair, K.A., Peterson, D.E., Kazmierczak, J.J., Addiss, D.G., Fox, K.R., and Rose, J.B. (1994). A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med.. 331, 161-167. [DOI] [PubMed]
- Manabe Y.C., Clark D.P., Moore R.D., Lumadue J.A., Dahlman H.R., Belitsos P.C., Chaisson R.E., Sears C.L. Cryptosporidiosis in patients with AIDS: correlates of disease and survival. Clin. Infect. Dis. 1998;27:536–542. doi: 10.1086/514701. [DOI] [PubMed] [Google Scholar]; Manabe, Y.C., Clark, D.P., Moore, R.D., Lumadue, J.A., Dahlman, H.R., Belitsos, P.C., Chaisson, R.E., and Sears, C.L. (1998). Cryptosporidiosis in patients with AIDS: correlates of disease and survival. Clin. Infect. Dis.. 27, 536-542. [DOI] [PubMed]
- Manjunatha U.H., Vinayak S., Zambriski J.A., Chao A.T., Sy T., Noble C.G., Bonamy G.M.C., Kondreddi R.R., Zou B., Gedeck P. A Cryptosporidium PI(4) K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017;546:376–380. doi: 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]; Manjunatha, U.H., Vinayak, S., Zambriski, J.A., Chao, A.T., Sy, T., Noble, C.G., Bonamy, G.M.C., Kondreddi, R.R., Zou, B., Gedeck, P., et al. (2017). A Cryptosporidium PI(4) K inhibitor is a drug candidate for cryptosporidiosis. Nature 546, 376-380. [DOI] [PMC free article] [PubMed]
- Martina B.E.E., Haagmans B.L., Kuiken T., Fouchier R.A.M., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S.M., Lim W., Osterhaus A.D.M.E. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]; Martina, B.E.E., Haagmans, B.L., Kuiken, T., Fouchier, R.A.M., Rimmelzwaan, G.F., Van Amerongen, G., Peiris, J.S.M., Lim, W., and Osterhaus, A.D.M.E. (2003). Virology: SARS virus infection of cats and ferrets. Nature 425, 915. [DOI] [PMC free article] [PubMed]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]; McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297-1303. [DOI] [PMC free article] [PubMed]
- Mead J.R. Challenges and prospects for a Cryptosporidium vaccine. Future Microbiol. 2010;5:335–337. doi: 10.2217/fmb.09.115. [DOI] [PubMed] [Google Scholar]; Mead, J.R. (2010). Challenges and prospects for a Cryptosporidium vaccine. Future Microbiol. 5, 335-337. [DOI] [PubMed]
- Mead J.R. Prospects for immunotherapy and vaccines against Cryptosporidium. Hum. Vaccin. Immunother. 2014;10:1505–1513. doi: 10.4161/hv.28485. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mead, J.R. (2014). Prospects for immunotherapy and vaccines against Cryptosporidium. Hum. Vaccin. Immunother.. 10, 1505-1513. [DOI] [PMC free article] [PubMed]
- Mead J.R., Arrowood M.J., Sidwell R.W., Healey M.C. Chronic Cryptosporidium parvum infections in congenitally immunodeficient SCID and nude mice. J. Infect. Dis. 1991;163:1297–1304. doi: 10.1093/infdis/163.6.1297. [DOI] [PubMed] [Google Scholar]; Mead, J.R., Arrowood, M.J., Sidwell, R.W., and Healey, M.C. (1991). Chronic Cryptosporidium parvum infections in congenitally immunodeficient SCID and nude mice. J. Infect. Dis.. 163, 1297-1304. [DOI] [PubMed]
- Mølbak K., Andersen M., Aaby P., Højlyng N., Jakobsen M., Sodemann M., da Silva A.P. Cryptosporidium infection in infancy as a cause of malnutrition: a community study from Guinea-Bissau, west Africa. Am. J. Clin. Nutr. 1997;65:149–152. doi: 10.1093/ajcn/65.1.149. [DOI] [PubMed] [Google Scholar]; Molbak, K., Andersen, M., Aaby, P., Hojlyng, N., Jakobsen, M., Sodemann, M., and da Silva, A.P. (1997). Cryptosporidium infection in infancy as a cause of malnutrition: a community study from Guinea-Bissau, west Africa. Am. J. Clin. Nutr.. 65, 149-152. [DOI] [PubMed]
- Mondal D., Haque R., Sack R.B., Kirkpatrick B.D., Petri W.A., Jr. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 2009;80:824–826. [PMC free article] [PubMed] [Google Scholar]; Mondal D., Haque R., Sack R.B., Kirkpatrick B.D., and Petri W.A. Jr. Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh, 2009, Am. J. Trop. Med. Hyg., 80, 824-826. [PMC free article] [PubMed]
- Mueller A.K., Labaied M., Kappe S.H.I., Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]; Mueller, A.K., Labaied, M., Kappe, S.H.I., and Matuschewski, K. (2005). Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433, 164-167. [DOI] [PubMed]
- Niess J.H., Brand S., Gu X., Landsman L., Jung S., McCormick B.A., Vyas J.M., Boes M., Ploegh H.L., Fox J.G. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]; Niess, J.H., Brand, S., Gu, X., Landsman, L., Jung, S., McCormick, B.A., Vyas, J.M., Boes, M., Ploegh, H.L., Fox, J.G. (2005). CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254-258. [DOI] [PubMed]
- Okhuysen P.C., Chappell C.L., Sterling C.R., Jakubowski W., DuPont H.L. Susceptibility and serologic response of healthy adults to reinfection with Cryptosporidium parvum. Infect. Immun. 1998;66:441–443. doi: 10.1128/iai.66.2.441-443.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]; Okhuysen, P.C., Chappell, C.L., Sterling, C.R., Jakubowski, W., and DuPont, H.L. (1998). Susceptibility and serologic response of healthy adults to reinfection with Cryptosporidium parvum. Infect. Immun.. 66, 441-443. [DOI] [PMC free article] [PubMed]
- Otto T.D., Sanders M., Berriman M., Newbold C. Iterative correction of reference nucleotides (iCORN) using second generation sequencing technology. Bioinformatics. 2010;26:1704–1707. doi: 10.1093/bioinformatics/btq269. [DOI] [PMC free article] [PubMed] [Google Scholar]; Otto, T.D., Sanders, M., Berriman, M., and Newbold, C. (2010). Iterative correction of reference nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26, 1704-1707. [DOI] [PMC free article] [PubMed]
- Painter J.E., Hlavsa M.C., Collier S.A., Xiao L., Yoder J.S., Centers for Disease, C., and Prevention Cryptosporidiosis surveillance – United States, 2011–2012. MMWR Suppl. 2015;64:1–14. [PubMed] [Google Scholar]; Painter, J.E., Hlavsa, M.C., Collier, S.A., Xiao, L., Yoder, J.S., and Centers for Disease, C., and Prevention. (2015). Cryptosporidiosis surveillance - United States, 2011-2012. MMWR Suppl. 64, 1-14. [PubMed]
- Pawlowic M.C., Vinayak S., Sateriale A., Brooks C.F., Striepen B. Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol. 2017;46:20B 22 21–20B 22 32. doi: 10.1002/cpmc.33. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pawlowic, M.C., Vinayak, S., Sateriale, A., Brooks, C.F., and Striepen, B. (2017). Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol. 46, 20B 22 21-20B 22 32. [DOI] [PMC free article] [PubMed]
- Petry F., Robinson H.A., McDonald V. Murine infection model for maintenance and amplification of Cryptosporidium parvum oocysts. J. Clin. Microbiol. 1995;33:1922–1924. doi: 10.1128/jcm.33.7.1922-1924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petry, F., Robinson, H.A., and McDonald, V. (1995). Murine infection model for maintenance and amplification of Cryptosporidium parvum oocysts. J. Clin. Microbiol. 33, 1922-1924. [DOI] [PMC free article] [PubMed]
- Platts-Mills J.A., Babji S., Bodhidatta L., Gratz J., Haque R., Havt A., McCormick B.J.J., McGrath M., Olortegui M.P., Samie A. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED) Lancet Glob. Health. 2015;3:E564–E575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Platts-Mills, J.A., Babji, S., Bodhidatta, L., Gratz, J., Haque, R., Havt, A., McCormick, B.J.J., McGrath, M., Olortegui, M.P., Samie, A., et al. (2015). Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. Health 3, E564-E575. [DOI] [PMC free article] [PubMed]
- Ploss A., Evans M.J., Gaysinskaya V.A., Panis M., You H., de Jong Y.P., Rice C.M. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ploss, A., Evans, M.J., Gaysinskaya, V.A., Panis, M., You, H., de Jong, Y.P., and Rice, C.M. (2009). Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457, 882-886. [DOI] [PMC free article] [PubMed]
- Pollok R.C.G., Farthing M.J.G., Bajaj-Elliott M., Sanderson I.R., McDonald V. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology. 2001;120:99–107. doi: 10.1053/gast.2001.20907. [DOI] [PubMed] [Google Scholar]; Pollok, R.C.G., Farthing, M.J.G., Bajaj-Elliott, M., Sanderson, I.R., and McDonald, V. (2001). Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology 120, 99-107. [DOI] [PubMed]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Quinlan, A.R., and Hall, I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. [DOI] [PMC free article] [PubMed]
- Rauch I., Deets K.A., Ji D.X., von Moltke J., Tenthorey J.L., Lee A.Y., Philip N.H., Ayres J.S., Brodsky I.E., Gronert K. NAIP-NLRC4 Inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity. 2017;46:649–659. doi: 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rauch, I., Deets, K.A., Ji, D.X., von Moltke, J., Tenthorey, J.L., Lee, A.Y., Philip, N.H., Ayres, J.S., Brodsky, I.E., Gronert, K., et al. (2017). NAIP-NLRC4 Inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46, 649-659. [DOI] [PMC free article] [PubMed]
- Ren X., Zhao J., Zhang L., Ning C., Jian F., Wang R., Lv C., Wang Q., Arrowood M.J., Xiao L. Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus) Exp. Parasitol. 2012;130:274–281. doi: 10.1016/j.exppara.2011.07.012. [DOI] [PubMed] [Google Scholar]; Ren, X., Zhao, J., Zhang, L., Ning, C., Jian, F., Wang, R., Lv, C., Wang, Q., Arrowood, M.J., and Xiao, L. (2012). Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus). Exp. Parasitol.. 130, 274-281. [DOI] [PubMed]
- Rescigno M., Urbano M., Valzasina B., Francolini M., Rotta G., Bonasio R., Granucci F., Kraehenbuhl J.P., Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]; Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., Granucci, F., Kraehenbuhl, J.P., and Ricciardi-Castagnoli, P. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol.. 2, 361-367. [DOI] [PubMed]
- Sarkar R., Ajjampur S.S.R., Prabakaran A.D., Geetha J.C., Sowmyanarayanan T.V., Kane A., Duara J., Muliyil J., Balraj V., Naumova E.N. Cryptosporidiosis among children in an endemic semiurban community in southern India: does a protected drinking water source decrease infection? Clin. Infect. Dis. 2013;57:398–406. doi: 10.1093/cid/cit288. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sarkar, R., Ajjampur, S.S.R., Prabakaran, A.D., Geetha, J.C., Sowmyanarayanan, T.V., Kane, A., Duara, J., Muliyil, J., Balraj, V., Naumova, E.N., et al. (2013). Cryptosporidiosis among children in an endemic semiurban community in southern India: does a protected drinking water source decrease infection? Clin. Infect. Dis.. 57, 398-406. [DOI] [PMC free article] [PubMed]
- Schmidt W., Wahnschaffe U., Schäfer M., Zippel T., Arvand M., Meyerhans A., Riecken E.O., Ullrich R. Rapid increase of mucosal CD4 T cells followed by clearance of intestinal cryptosporidiosis in an AIDS patient receiving highly active antiretroviral therapy. Gastroenterology. 2001;120:984–987. doi: 10.1053/gast.2001.22557. [DOI] [PubMed] [Google Scholar]; Schmidt, W., Wahnschaffe, U., Schafer, M., Zippel, T., Arvand, M., Meyerhans, A., Riecken, E.O., and Ullrich, R. (2001). Rapid increase of mucosal CD4 T cells followed by clearance of intestinal cryptosporidiosis in an AIDS patient receiving highly active antiretroviral therapy. Gastroenterology 120, 984-987. [DOI] [PubMed]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-1313. [DOI] [PMC free article] [PubMed]
- Stanke M., Morgenstern B. Augustus: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki458. W465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stanke, M., and Morgenstern, B. (2005). Augustus: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465-467. [DOI] [PMC free article] [PubMed]
- Striepen B. Parasitic infections: time to tackle cryptosporidiosis. Nature. 2013;503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]; Striepen, B. (2013). Parasitic infections: time to tackle cryptosporidiosis. Nature 503, 189-191. [DOI] [PubMed]
- Templeton T.J., Iyer L.M., Anantharaman V., Enomoto S., Abrahante J.E., Subramanian G.M., Hoffman S.L., Abrahamsen M.S., Aravind L. Comparative analysis of Apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]; Templeton, T.J., Iyer, L.M., Anantharaman, V., Enomoto, S., Abrahante, J.E., Subramanian, G.M., Hoffman, S.L., Abrahamsen, M.S., and Aravind, L. (2004). Comparative analysis of Apicomplexa and genomic diversity in eukaryotes. Genome Res. 14, 1686-1695. [DOI] [PMC free article] [PubMed]
- Tzipori S. Cryptosporidiosis: laboratory investigations and chemotherapy. Adv. Parasitol. 1998;40:187–221. doi: 10.1016/s0065-308x(08)60121-9. [DOI] [PubMed] [Google Scholar]; Tzipori, S. (1998). Cryptosporidiosis: laboratory investigations and chemotherapy. Adv. Parasitol.. 40, 187-221. [DOI] [PubMed]
- Velázquez F.R., Matson D.O., Calva J.J., Guerrero M.L., Morrow A.L., Carter-Campbell S., Glass R.I., Estes M.K., Pickering L.K., Ruiz-Palacios G.M. Rotavirus infection in infants as protection against subsequent infections. N. Engl. J. Med. 1996;335:1022–1028. doi: 10.1056/NEJM199610033351404. [DOI] [PubMed] [Google Scholar]; Velazquez, F.R., Matson, D.O., Calva, J.J., Guerrero, M.L., Morrow, A.L., Carter-Campbell, S., Glass, R.I., Estes, M.K., Pickering, L.K., and Ruiz-Palacios, G.M. (1996). Rotavirus infection in infants as protection against subsequent infections. N. Engl. J. Med.. 335, 1022-1028. [DOI] [PubMed]
- Vinayak S., Pawlowic M.C., Sateriale A., Brooks C.F., Studstill C.J., Bar-Peled Y., Cipriano M.J., Striepen B. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vinayak, S., Pawlowic, M.C., Sateriale, A., Brooks, C.F., Studstill, C.J., Bar-Peled, Y., Cipriano, M.J., and Striepen, B. (2015). Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477-480. [DOI] [PMC free article] [PubMed]
- White A.C., Robinson P., Okhuysen P.C., Lewis D.E., Shahab I., Lahoti S., DuPont H.L., Chappell C.L. Interferon-gamma expression in jejunal biopsies in experimental human cryptosporidiosis correlates with prior sensitization and control of oocyst excretion. J. Infect. Dis. 2000;181:701–709. doi: 10.1086/315261. [DOI] [PubMed] [Google Scholar]; White, A.C., Robinson, P., Okhuysen, P.C., Lewis, D.E., Shahab, I., Lahoti, S., DuPont, H.L., and Chappell, C.L. (2000). Interferon-gamma expression in jejunal biopsies in experimental human cryptosporidiosis correlates with prior sensitization and control of oocyst excretion. J. Infect. Dis.. 181, 701-709. [DOI] [PubMed]
- Widmer G., Akiyoshi D., Buckholt M.A., Feng X., Rich S.M., Deary K.M., Bowman C.A., Xu P., Wang Y., Wang X. Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol. Biochem. Parasitol. 2000;108:187–197. doi: 10.1016/s0166-6851(00)00211-5. [DOI] [PubMed] [Google Scholar]; Widmer, G., Akiyoshi, D., Buckholt, M.A., Feng, X., Rich, S.M., Deary, K.M., Bowman, C.A., Xu, P., Wang, Y., Wang, X., et al. (2000). Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol. Biochem. Parasitol.. 108, 187-197. [DOI] [PubMed]
- Xu P., Widmer G., Wang Y., Ozaki L.S., Alves J.M., Serrano M.G., Puiu D., Manque P., Akiyoshi D., Mackey A.J. The genome of Cryptosporidium hominis. Nature. 2004;431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]; Xu, P., Widmer, G., Wang, Y., Ozaki, L.S., Alves, J.M., Serrano, M.G., Puiu, D., Manque, P., Akiyoshi, D., Mackey, A.J., et al. (2004). The genome of Cryptosporidium hominis. Nature 431, 1107-1112. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.