Abstract
Protein synthesis is crucial for maintaining synaptic plasticity and synaptic signalling. Here we have attempted to understand the role of RNA binding proteins, Fragile X Mental Retardation Protein (FMRP) and Moloney Leukemia Virus 10 (MOV10) protein in N-Methyl-D-Aspartate Receptor (NMDAR) mediated translation regulation. We show that FMRP is required for translation downstream of NMDAR stimulation and MOV10 is the key specificity factor in this process. In rat cortical synaptoneurosomes, MOV10 in association with FMRP and Argonaute 2 (AGO2) forms the inhibitory complex on a subset of NMDAR responsive mRNAs. On NMDAR stimulation, MOV10 dissociates from AGO2 and promotes the translation of its target mRNAs. FMRP is required to form MOV10-AGO2 inhibitory complex and to promote translation of MOV10 associated mRNAs. Phosphorylation of FMRP appears to be the potential switch for NMDAR mediated translation and in the absence of FMRP, the distinct translation response to NMDAR stimulation is lost. Thus, FMRP and MOV10 have an important regulatory role in NMDAR mediated translation at the synapse.
Electronic supplementary material
The online version of this article (10.1186/s13041-019-0473-0) contains supplementary material, which is available to authorized users.
Keywords: FMRP/MOV10/NMDAR mediated translation
Introduction
In mature neurons, protein synthesis in the dendrites and spines outweigh that of the cell body due to their sheer volume [1]. Protein synthesis at dendrites and spines is regulated by the activation of many different neurotransmitter receptors such as glutamate, dopamine, and serotonin [2–4] also termed as activity mediated protein synthesis. Thus, it is important to decipher the specificity of translational response to a given neurotransmitter receptor stimulation. This task has gained significance since the dysregulation of protein synthesis is thought to be a common cause for multiple neurodevelopmental disorders [5]. Glutamate is the major excitatory neurotransmitter in the mammalian brain and NMDAR and the group I metabotropic Glutamate Receptor (mGluR) are two of its primary receptors that mediate synaptic plasticity. Both NMDAR and mGluR regulate protein synthesis, group I mGluR leading to global translation activation and NMDAR to translation inhibition shown through metabolic labelling of proteins [6–9]. At transcriptome level, both group I mGluR and NMDAR stimulation leads to translation activation of specific subset of mRNAs. Group I mGluR stimulation leads to translation of mRNAs such as Fragile X mental retardation 1 (Fmr1), postsynaptic density 95 (Psd-95), activity regulated cytoskeleton associated protein (Arc) [10–12] and NMDAR stimulation leading to translation of β-actin, Glutamate receptor ionic epsilon 1 (Grin2a), Fmr1, Calcium/calmodulin dependent kinase II alpha (camk2a) and Arc mRNAs [9, 13–17]. Group I mGluR mediated translation activation is well studied, however, the mechanistic insight of NMDAR mediated translation is poorly explored [18, 19]. In the current study, we tried to elucidate NMDAR mediated control over the translation machinery by determining the factors involved in it.
MicroRNAs and microRNA induced silencing complex (miRISC) are thought to play an important role in regulating activity mediated protein synthesis. MicroRNA-AGO2 mediated translation inhibition can be reversed by dissociation of miRISC from the mRNA and promoting its translation [11, 20, 21]. This reversibility of miRISC is of particular interest in the context of synaptic plasticity as it can inhibit translation until an appropriate stimulus relieves the inhibition. Under these conditions, microRNAs provide the sequence specificity while several RNA binding proteins (RBP) which are not part of miRISC core complex will act as a molecular switch through their dynamic interaction with AGO2. FMRP is one such RBP which has a significant role in synaptic protein synthesis. Previously, it was shown that FMRP along with AGO2 regulate translation in response to the group I mGluR stimulation at the synapse [11]. While FMRP is also reported to regulate translation through multiple mechanisms [22, 23], its role in reversibility of miRISC mediated inhibition is likely to be of relevance for synaptic translation. The loss of FMRP and the subsequent synaptic dysfunction is the hallmark of Fragile X Syndrome (FXS) [24]. Interestingly, FMRP is reported to interact with a large number of mRNAs [25] and thus potentially regulates translation beyond mGluR signalling. Another RBP known to regulate translation downstream of synaptic signalling is MOV10 and is also known to interact with both FMRP and AGO2 [19, 26]. Since both NMDAR and group I mGluR mediated plasticity involve protein synthesis, it is also essential to study the role of FMRP and MOV10 in NMDAR mediated protein synthesis at the synapse.
In the current study, we show that the dynamic interaction between AGO2-MOV10-FMRP determines the translation response to NMDAR stimulation. This study highlights the involvement of FMRP and its phosphorylation status in NMDAR mediated signalling and provides a molecular mechanism to explain the specificity of translation on NMDAR stimulation.
Results
MOV10 dissociates from AGO2 and moves to polysomes on NMDAR stimulation
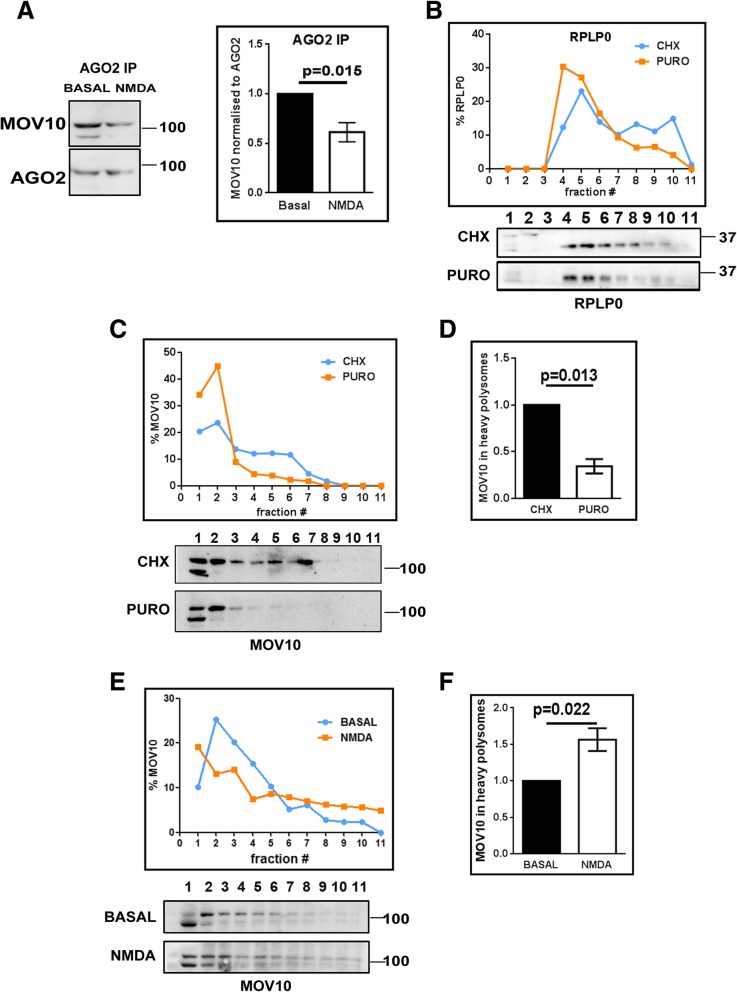
In order to understand the mechanism of NMDAR mediated translation, we chose to investigate the role of MOV10 because of its implication in previous studies [19, 27]. MOV10 is an RNA helicase and is also shown to regulate the translation of its target mRNAs [19, 27, 28] Though MOV10 is proposed to play a role in NMDAR mediated translation in these studies, the molecular mechanism was not clear. In order to characterize its regulatory role, we looked at the association of MOV10 with miRISC protein AGO2 and with polysomes. We used post-natal day 30 (P30) rat cortical synaptoneurosomes for this study. Synaptoneurosomes were characterised by electron microscopy for the presence of postsynaptic density (PSD) and synaptic vesicles (SV) and for the enrichment of PSD-95 protein (Additional file 1: Figures S1A and S1B). The synaptoneurosome preparation used here is based on rapid filtration method. This method results in a relatively crude prep of synaptoneurosomes which are intact and are responsive to neurotransmitter stimulation [6, 11]. This preparation is suitable for our work since the focus is to study stimulation mediated changes in translation. We show that MOV10 co-precipitates with AGO2 from cortical synaptoneurosomes preparation (Additional file 1: Figure S1C). Further, we did AGO2 immunoprecipitation (IP) from synaptoneurosome lysate after NMDAR stimulation and quantified MOV10 association with AGO2 through western blot analysis (densitometric MOV10 values were normalized to that of AGO2). On NMDAR stimulation, there was a significant decrease in the association of MOV10 with AGO2 compared to basal (Fig. 1a) while there was no change in the levels of MOV10 (input) on NMDAR stimulation in synaptoneurosomes (Additional file 1: Figure S1E). These results hold true when we reversed the IP. For this, we did MOV10 IP, and looked for AGO2 in the pellet on NMDAR stimulation (Additional file 1: Figure S1D). We observed a decrease in the association of MOV10 and AGO2 on NMDAR stimulation, confirming that MOV10 dissociates from the inhibitory complex (AGO2) on NMDAR stimulation.
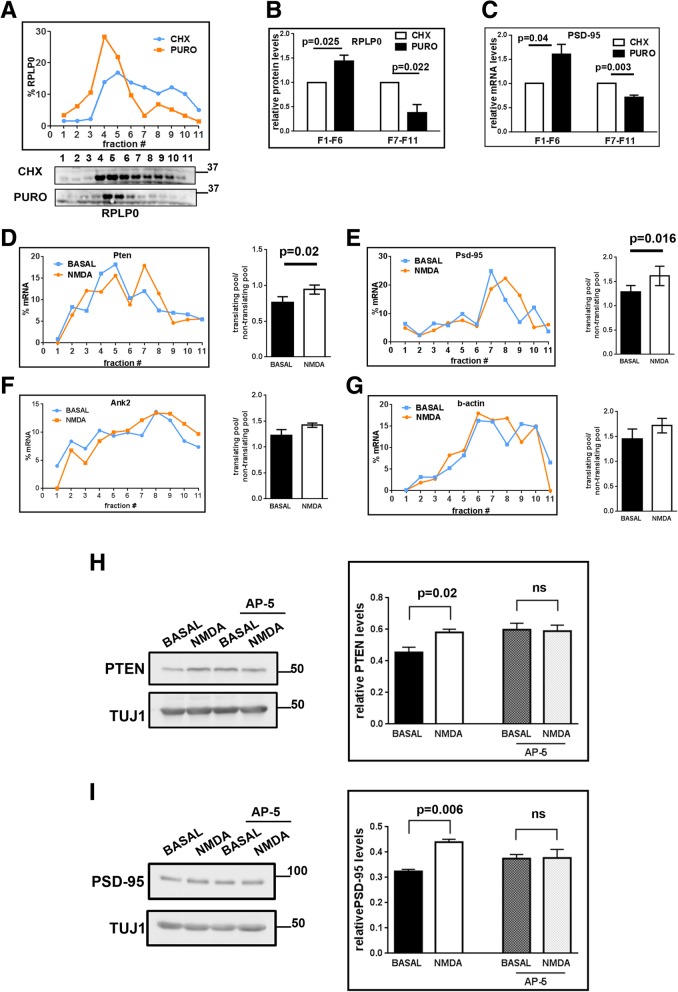
Fig. 1.
Effect of NMDAR stimulation on MOV10 interaction with AGO2 and distribution in polysomes. a Immunoblots for MOV10 and AGO2 after AGO2-immunoprecipitation on NMDAR stimulation. Quantitative analysis of MOV10 association with AGO2 following AGO2 immunoprecipitation on NMDAR stimulation in rat cortical synaptoneurosomes (n = 5, paired Student’s t-test, ±SEM). Values are normalized to basal levels. b Distribution of RPLP0 on linear sucrose gradient from rat cortical synaptoneurosomes after cycloheximide or puromycin treatment based on immunoblots shown below. c Distribution of MOV10 on linear sucrose gradient from rat cortical synaptoneurosomes after cycloheximide or puromycin treatment based on immunoblots shown below (representative of three experiments, also see Additional file 1: Figure S2A). d Quantification of MOV10 in heavy polysomes (fraction 7–11) for cycloheximide or puromycin treatment (n = 3, paired Student’s t-test, ±SEM). Values are normalised to cycloheximide levels. e Distribution of MOV10 on linear sucrose gradient from rat cortical synaptoneurosomes after NMDAR stimulation based on immunoblots shown below (representative of five experiments, also see Additional file 1: Figure S2B). f Quantification of MOV10 in heavy polysomes (fraction 7–11) for NMDAR stimulation as compared to basal condition (n = 5, paired Student’s t-test, ±SEM). Values are normalised to basal levels
To understand the role of MOV10 on synaptic translation, we looked at the association of MOV10 protein with polysomes in synaptoneurosomes. In synaptoneurosomes, on puromycin (PURO) treatment, actively translating polysomes shift to lighter fractions compared to cycloheximide (CHX) treatment, as shown by ribosomal protein lateral stalk subunit P0 (RPLP0) (Fig. 1b). MOV10 was present in polysomal fractions but puromycin treatment led to significant reduction of MOV10 from heavy polysomes and a shift to lighter fractions (Fig. 1c, d and Additional file 1: Figure S2A) indicating that MOV10 is associated with actively translating polysomes. MOV10 distribution in polysomes was further validated using a sucrose step gradient method [29] in Neuro 2a cells (Additional file 1: Figures S2C-S2E). Thus, we found that MOV10 associates with AGO2 as well as with puromycin sensitive polysomes. Interestingly, in synaptoneurosomes, the percentage of MOV10 in translating polysomes (puromycin-sensitive) was significantly increased on NMDAR stimulation compared to basal condition (Fig. 1e, f, Additional file 1: Figures S2B and S2F). These results show that on NMDAR stimulation MOV10 dissociates from inhibitory protein AGO2 and moves into translating polysomes.
FMRP is required for the translation response downstream of NMDAR stimulation
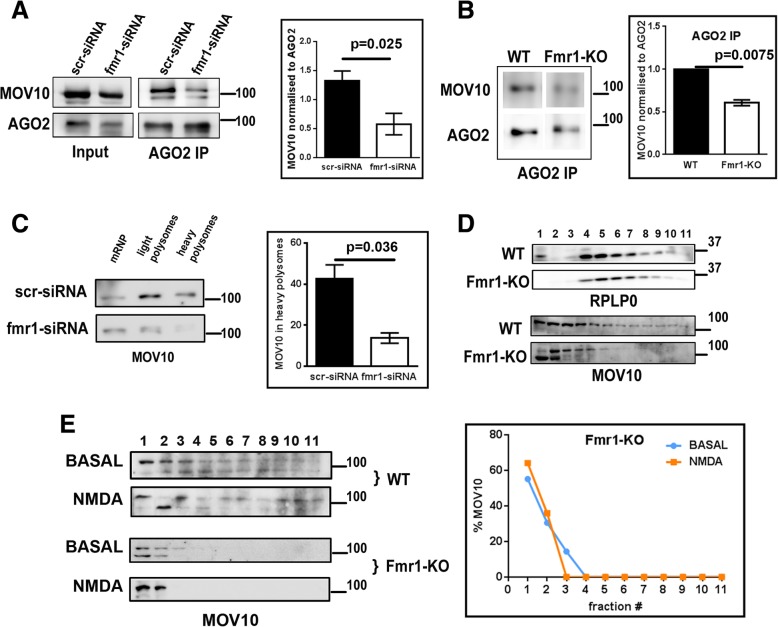
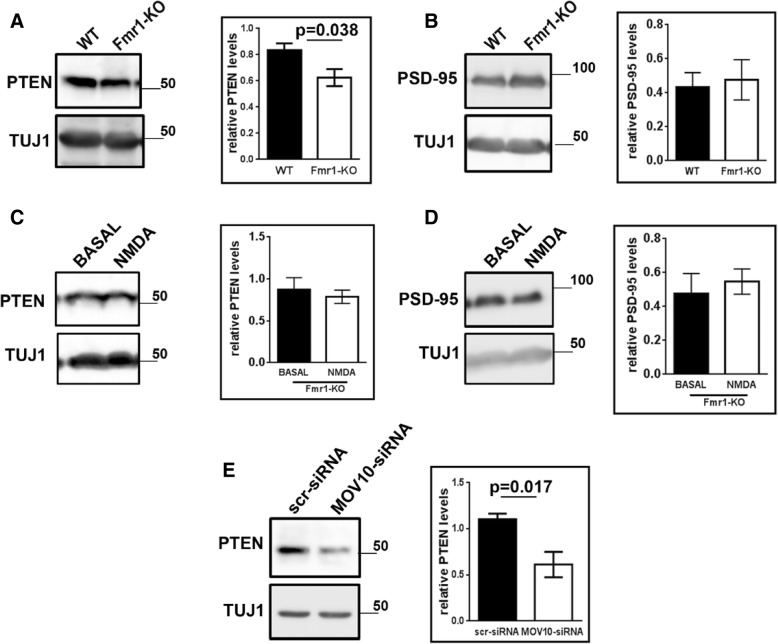
It is previously reported that MOV10 interacts with FMRP and AGO2 independently [26, 30]. Here we investigated the role of FMRP in AGO2-MOV10 interaction. Interestingly, when FMRP was knocked down in Neuro-2a cells by specific small interfering RNA (siRNA) against Fmr1 mRNA (Additional file 1: Figure S3A), MOV10 co-precipitation with AGO2 was significantly reduced (Fig. 2a) compared to the scramble siRNA treated cells. AGO2 levels did not show significant change on Fmr1 knockdown (Additional file 1: Figure S3B). To study the role of FMRP in the MOV10-AGO2 interaction in neurons, we used Fmr1-Knockout (Fmr1-KO) rat (Additional file 1: Figure S3C) synaptoneurosomes for AGO2 immunoprecipitation and polysome profiling assays. AGO2-MOV10 interaction was reduced in Fmr1-KO synaptoneurosomes compared to wild-type (WT) corroborating our Neuro 2a results (Fig. 2b). These results show that the absence of FMRP reduces the interaction of MOV10 with AGO2 both in Neuro 2a cells and in cortical synaptoneurosomes.
Fig. 2.
MOV10 interaction with AGO2 and its distribution in polysomes requires FMRP. a Immunoblots for MOV10 and AGO2 for input and after AGO2-immunoprecipitation on fmr1 knockdown in Neuro2a cells. Quantitative analysis of MOV10 association with AGO2 following AGO2 immunoprecipitation from Neuro 2a cells transfected with scramble siRNA (scr-siRNA) or fmr1 siRNA (fmr1-siRNA) (n = 4, unpaired Student’s t-test, ±SEM). b Immunoblots for MOV10 and AGO2 after AGO2-immunoprecipitation from wild type (WT) or Fmr1-KO synaptoneurosomes (WT and Fmr1-KO samples were run on separate immunoblots). Quantitative analysis of MOV10 association with AGO2 following AGO2 immunoprecipitation from WT or Fmr1-KO synaptoneurosomes (n = 3, unpaired Student’s t-test, ±SEM). c Immunoblots for MOV10 after sucrose step gradient on fmr1 knockdown in Neuro2a cells. Quantification of MOV10 in 30% fraction (heavy polysomes) separated on a sucrose step gradient from Neuro 2a cells transfected with scramble siRNA (scr-siRNA) or fmr1 siRNA (fmr1-siRNA). Values normalised to samples from scrambled siRNA transfected cells (n = 3, unpaired Student’s t-test, ±SEM). d Immunoblots of ribosomal protein RPLP0 and MOV10 on a linear sucrose gradient from wild-type (WT) or Fmr1 KO rat cortical synaptoneurosomes. e Immunoblots of MOV10 on a linear sucrose gradient from wild-type (WT) and Fmr1 KO rat cortical synaptoneurosomes on NMDAR stimulation. Distribution of MOV10 on linear sucrose gradient from Fmr1-KO rat synaptoneurosomes on NMDAR stimulation based on adjacent immunoblots
FMRP knockdown (Fmr1-siRNA) in Neuro 2a cells resulted in a significantly reduced association of MOV10 with polysomes (Fig. 2c). In the Fmr1-KO synaptoneurosomes, we could detect MOV10 only in the lighter fractions (fractions 1–5) of the linear sucrose gradient and absent in the polysomes (Fig. 2d) while there was no change in the distribution of ribosomes (based on RPLP0 western blot) (Fig. 2d). Earlier we showed that when cortical synaptoneurosomes were stimulated with NMDA, there was a significant increase in the percentage of MOV10 in the heavy polysomes (Fig. 1e and f). This shift of MOV10 to polysomes on NMDAR stimulation was absent in the Fmr1-KO synaptoneurosomes (Fig. 2e). Further, we also studied the role of AGO2 in the distribution of MOV10 in polysomes (Additional file 1: Figure S3D). In the absence of AGO2, the presence of MOV10 in polysomes was not affected (Additional file 1: Figures S3E). These results confirm that FMRP is not only required for the association of MOV10 with AGO2 and translating polysomes at basal state but also for the shift of MOV10 from AGO2 to polysomes in response to NMDAR stimulation.
Translation of specific mRNAs is affected by the absence of MOV10 and FMRP
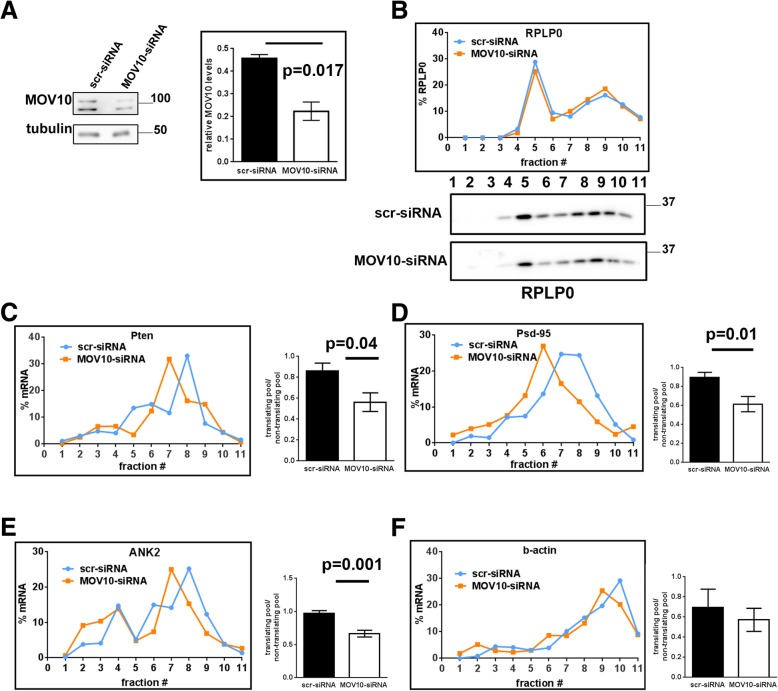
MOV10 is reported to interact with a large number of mRNAs [28, 31]. Since MOV10 is specifically dissociated from AGO2 on NMDAR stimulation and is shifted to the polysomal fraction (Fig. 1a, e), we investigated whether MOV10 has any effect on the translation of certain mRNAs. For this, we knocked down MOV10 (using siRNA) in primary neurons (Fig. 3a), and looked at the distribution of mRNAs in polysomes as compared to scrambled siRNA. Ribosomal protein RPLP0 did not show any change on MOV10 knockdown (Fig. 3b). Polysomal fractions were determined by the sensitivity to puromycin (fractions 8–11 were puromycin-sensitive in the case of primary neurons (Additional file 1: Figures S4A and S4B)). We chose mRNAs that were targets of MOV10 and/or FMRP from earlier reports [25, 26]. We saw a decrease in translation for the mRNAs phosphatase tensin homolog (Pten), Psd-95 and ankyrin 2 (Ank2) but no change for β-actin mRNA implying a role for MOV10 for these candidates (Fig. 3c-f and average line graphs in Additional file 1: Figures S4D-S4G). These candidates are also enriched in the pellet of MOV10 immunoprecipitation (Additional file 1: Figure S4C) as compared to that of Immunoglobulin G (IgG). These results indicate that MOV10 plays a role in translation activation of a specific set of mRNAs.
Fig. 3.
Translation of specific mRNAs is affected on MOV10 knockdown. a Immunoblots showing MOV10 knockdown from primary neuronal cultures transfected with scrambled siRNA (scr-siRNA) or MOV10 siRNA (MOV10-siRNA). Quantification of MOV10 in from primary neurons transfected with scramble siRNA (scr-siRNA) or MOV10 siRNA (MOV10-siRNA). Values normalised to samples from scrambled siRNA transfected cells (n = 3, unpaired Student’s t-test, ±SEM). b Distribution of RPLP0 on a linear sucrose gradient from primary neurons transfected with scrambled or MOV10 siRNA based on immunoblots shown below. c-f Distribution of selected mRNAs on linear sucrose gradient from primary neurons transfected with scrambled siRNA (scr-siRNA) or MOV10 siRNA (MOV10-siRNA) followed by quantification of mRNAs in polysomes (bar graphs, n = 4–6, unpaired Student’s t-test, ±SEM) for the mRNAs Pten (c), Psd-95 (d), Ank2 (e) and b-actin (f). Also see Additional file 1: Figure S4D-S4G
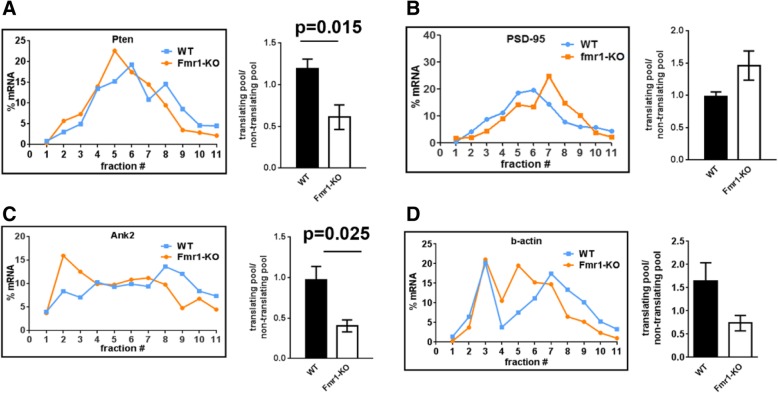
To study the role of FMRP in this context, we analysed the translation of MOV10 target mRNAs in Fmr1-KO synaptoneurosomes. Our selected MOV10 target mRNAs are also previously reported to be FMRP targets [25] and we further validated their association with FMRP (Additional file 1: Figure S5A) by immunoprecipitation. Interestingly, in the absence of FMRP, except for Psd-95 mRNA, both Pten and Ank2 mRNAs showed significant a reduction in translation as per their distribution in polysomal fractions (Fig. 4a and c, with average line graphs in Additional file 1: Figure S5B-S5E). The translation of Psd-95 mRNA increased in the absence of FMRP as previously shown [11] but this was not statistically significant (Fig. 4b). β-actin mRNA also showed a trend of decrease on Fmr1-KO polysomes which was not statistically significant (Fig. 4d). These results indicate that the combination of FMRP and MOV10 have both distinct and convergent roles in the translation of mRNAs.
Fig. 4.
Translation of MOV10 target mRNAs is affected in Fmr1-KO synaptoneurosomes. a-d Distribution of selected (MOV10 targets) mRNAs on linear sucrose gradient from WT and Fmr1 KO rat synaptoneurosomes followed by quantification of mRNAs in polysomes (bar graph, n = 3–5, unpaired Student’s t-test ±SEM) for the mRNAs Pten (a), Psd-95 (b), Ank2 (c) and b-actin (d). Also see Additional file 1: Figure S5B-S5E
NMDAR stimulation leads to translation of FMRP-MOV10 target mRNAs
On NMDAR stimulation, MOV10 moves into polysomes and absence of which leads to translation inhibition of specific mRNAs. To check whether these mRNAs undergo translation on NMDAR stimulation, we did polysome profiling from synaptoneurosomes after NMDAR stimulation. Translating polysomal fractions were determined by the sensitivity to puromycin (fractions 7–11 were puromycin-sensitive in the case of synaptoneurosomes (Fig. 5a) which is reflected by the decreased ribosomal protein RPLP0 from fraction 7–11 (Fig. 5a and b) and the corresponding increase in the early fractions). Similarly, the distribution of Psd-95 mRNA was also shifted out of fractions 7–11 in puromycin treated samples compared to cycloheximide-treated samples (Fig. 5c). Hence we considered the mRNAs present in fractions 7–11 as actively translating pool for further quantification, as also discussed previously [6, 32]. Next, we did polysome profiling from synaptoneurosomes after NMDAR stimulation. The profile for ribosomal protein RPLP0 did not significantly change for both basal and NMDAR stimulation conditions (Additional file 1: Figure S6A and S6B).
Fig. 5.
NMDAR stimulation leads to translational upregulation of MOV10-FMRP target mRNAs. a Ribosome distribution on linear sucrose gradient on cycloheximide and puromycin treatment based on RPLP0 immunoblot (below) from rat cortical synaptoneurosomes. b, c Quantitative distribution of RPLP0 protein or Psd-95 mRNA on linear sucrose gradient cycloheximide and puromycin treatment normalized to cycloheximide levels (n = 3, unpaired Student’s t test, ±SEM). d-g Distribution of mRNAs on linear sucrose gradient from synaptoneurosomes at basal state and on NMDAR stimulation followed by quantification of mRNAs in polysomes (bar graphs, n = 3, paired Student’s t-test, ±SEM) for the mRNAs; Pten (d), Psd-95 (e), Ank2 (f) and b-actin (g). Also see Additional file 1: Figure S6C-S6F. h Immunoblot showing PTEN protein after NMDAR stimulation with or without AP-5. Quantification of PTEN levels on NMDAR stimulation normalized to Tuj1 (n = 6, one way ANOVA, Tukey’s multiple comparison test, ±SEM). i Immunoblot showing PSD-95 protein after NMDAR stimulation with or without AP-5. Quantification of PSD-95 levels on NMDAR stimulation normalized to Tuj1 (n = 4, one way ANOVA, Tukey’s multiple comparison test, ±SEM)
Among the four candidates that we tested in this assay, we found that Pten and Psd-95 mRNAs showed an increase in translating fractions on NMDAR stimulation and Ank2 and β-actin did not show any change (Fig. 5d-g, with average line graphs in Additional file 1: Figure S6C-S6F). Thus, NMDAR leads to translation activation of Pten and Psd-95 mRNAs which is mediated through both MOV10 and FMRP.
Among the mRNA candidates that we tested, we found Pten mRNA to be consistently significant for all the assays. To further validate this process, we looked at PTEN protein levels through western blotting (whole blots in Additional file 1: Figure S7A-S7B to show antibody specificity). Interestingly, we observed a significant increase in protein levels for both PTEN and PSD-95 on NMDAR stimulation in synaptoneurosomes as compared to basal conditions (Fig. 5h and i). This increase was lost when the stimulation was done in the presence of specific NMDAR inhibitor 2-Amino-5-Phosphonopentanoic acid (AP-5), confirming the role of NMDAR in translation upregulation of these mRNAs (Fig. 5h and i). We also observed a decrease in PTEN protein levels from Fmr1-KO synaptoneurosomes as compared to that in WT (Fig. 6a). PSD-95 protein levels showed an increase but were not significant from Fmr1-KO synaptoneurosomes (Fig. 6b) similar to the polysome profiling data (Fig. 4b). We also looked at the translation response to NMDAR stimulation in FMR1-KO synaptoneurosomes. We saw no change in both PTEN and PSD-95 protein levels on NMDAR stimulation in Fmr1-KO synaptoneurosomes (Fig. 6c and d), implying that FMRP is required for NMDAR mediated protein synthesis of Pten and Psd-95 mRNAs. In primary neurons, on knockdown of MOV10, we observed a decrease in PTEN protein levels, as compared to scrambled siRNA levels (Fig. 6e). Overall, these results confirm the translation activation of a specific subset of mRNAs on NMDAR stimulation mediated through MOV10 and FMRP.
Fig. 6.
Pten and Psd-95 mRNAs are translationally regulated by FMRP-MOV10 on NMDAR stimulation. a Immunoblots showing PTEN protein from WT and Fmr1-KO synaptoneurosomes. Quantification of PTEN levels from WT and Fmr1-KO synaptoneurosomes normalized to Tuj1 (n = 5, unpaired Student’s t test, ±SEM). b Immunoblots showing PSD-95 protein from WT and Fmr1-KO synaptoneurosomes. Quantification of PSD-95 levels from WT and Fmr1-KO synaptoneurosomes normalized to Tuj1 (n = 3, unpaired Student’s t test, ±SEM). c Immunoblost showing PTEN protein after NMDAR stimulation from Fmr1-KO synaptoneurosomes. Quantification of PTEN levels on NMDAR stimulation normalized to Tuj1 (n = 3, paired Student’s t test, ±SEM). d Immunoblots showing PSD-95 protein after NMDAR stimulation from Fmr1-KO synaptoneurosomes. Quantification of PSD-95 levels on NMDAR stimulation normalized to Tuj1 (n = 3, paired Student’s t test, ±SEM). e Immunoblot showing PTEN protein from neurons transfected with scramble or MOV10 siRNA. Quantification of PTEN levels on MOV10 knockdown normalized to Tuj1 (n = 4, unpaired Student’s t test, ±SEM)
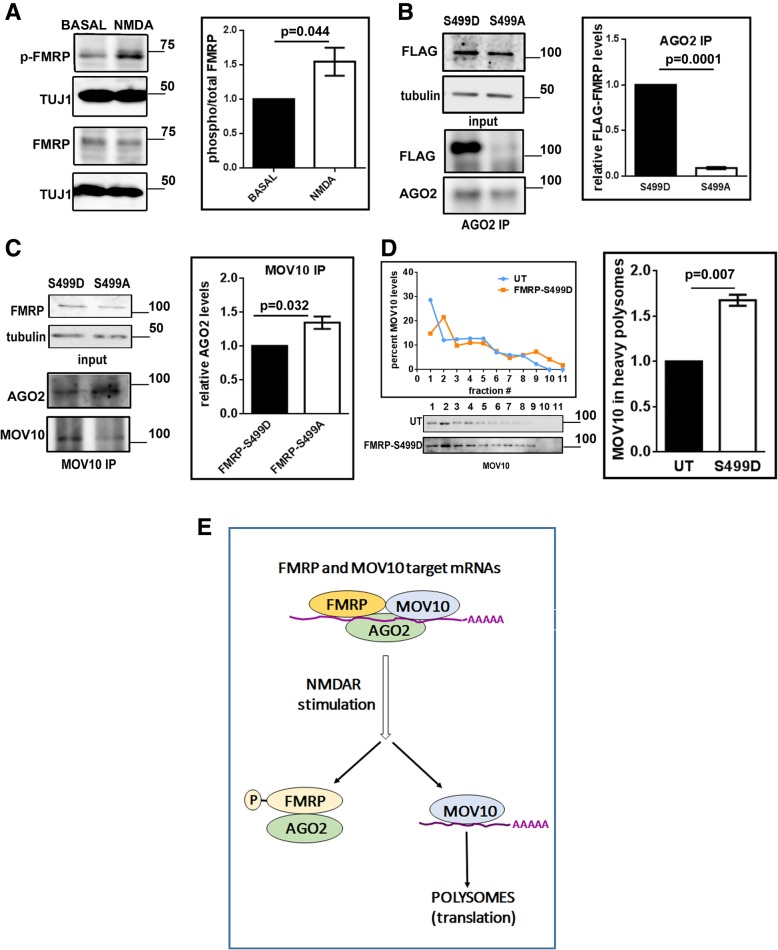
Dephosphorylated FMRP forms the inhibitory complex with MOV10-AGO2 and phosphorylation of FMRP dissociates this complex
Previously, it has been shown that the FMRP gets dephosphorylated downstream of mGluR signaling [11, 33]. In this study, we wanted to investigate the role of the phosphorylation state of FMRP in NMDAR mediated translation. For this, we quantitated the change in phosphorylation status of FMRP on NMDAR stimulation from cortical synaptoneurosomes. For this we used the antibody which specifically recognises the phosphorylated form of FMRP at S499 (Fig. 7a and Additional file 1: Figure S8A). On NMDAR stimulation there was a significant increase in phosphorylation of FMRP (Fig. 7a) with no change in total FMRP levels (Additional file 1: Figure S8B). To study the effect of this result on MOV10-FMRP-AGO2 interaction, we overexpressed phospho-mimetic or dephospho-mimetic form of FMRP (FMRP-S499D and FMRP-S499A respectively), in Neuro 2a cells. In this condition, as previously reported [11] AGO2 interacts more with phospho-mimetic form of FMRP (FMRP-S499D) compared to dephospho-mimetic form (FMRP-S499A) (Fig. 7b and Additional file 1: Figure S8C2). We did MOV10 immunoprecipitation in the overexpression background and quantitated the amount of AGO2 co-precipitated with MOV10. Here we observed increased AGO2 co-precipitation with MOV10 from the cells overexpressing FMRP-S499A (dephosphorylated FMRP mimetic) compared to the cells overexpressing FMRP-S499D (phosphorylated FMRP mimetic) (Fig. 7c and Additional file 1: Figure S8C1). Thus, phosphorylated FMRP appears to promote the dissolution of AGO2-MOV10 inhibitory complex which is in contrast to the previous result that phosphorylated FMRP promotes FMRP-AGO2 inhibitory complex [11]. Polysome profiling from Neuro 2a cells overexpressing FMRP-S499D led to an increase in the percentage of MOV10 in polysomes (Fig. 7d and Additional file 1: Figure S8D) as compared to un-transfected cells indicating that overexpression of FMRP-S499D shifts MOV10 to translating polysomes. On the other hand overexpression of FMRP-S499A led to a decrease of MOV10 in polysomes (Additional file 1: Figure S8E). Overexpression of FMRP-S499D or FMRP-S499A did not affect the overall polysome profile based on RPLP0 western blot (Additional file 1: Figure S8E) but has a significant impact on the distribution of MOV10 in polysomes. Thus, these results indicate that phosphorylation of FMRP is likely the switch downstream of NMDAR that shifts MOV10 to polysomes and promotes translation of its target mRNAs.
Fig. 7.
Phosphorylation of FMRP is the switch for NMDAR mediated translation. a Immunoblots for phospho FMRP (at S499) and total FMRP from synaptoneurosomes after NMDAR stimulation. Quantification of the ratio of phospho-FMRP to total-FMRP normalized to tuj1 for NMDAR stimulation (n = 6, paired Student’s t test, ±SEM).b Immunoblots for FLAG showing overexpression of FMRP-S499D, FMRP-S499A and tubulin from Neuro 2a cells. Bottom panel: Immunoblots for FLAG-FMRP and AGO2 after AGO2 immunoprecipitation from Neuro 2a cells transfected with FMRP-S499D or FMRP-S499A. Quantitative analysis of AGO2 association with FLAG-FMRP following AGO2 immunoprecipitation normalised to levels in the FMRP-S499D overexpressing cells. (n = 3, unpaired Student’s t-test, ±SEM). c Immunoblots for FMRP showing overexpression of FMRP-S499D, FMRP-S499A and tubulin from Neuro 2a cells. Bottom panel: Immunoblots for AGO2 and MOV10 after MOV10 immunoprecipitation from Neuro 2a cells transfected with FMRP-S499D or FMRP-S499A. Quantitative analysis of AGO2 association with MOV10 following MOV10 immunoprecipitation normalised to levels in the FMRP-S499D overexpressing cells. (n = 3, unpaired Student’s t-test, ±SEM). d Distribution of MOV10 separated on a linear sucrose gradient from Neuro 2a cells untransfected (UT) or transfected with FMRP-S499D based on immunoblots shown below followed by quantification of MOV10 in polysomes (n = 3, unpaired Student’s t-test, ±SEM). e Model illustrating the role of MOV10 in response to NMDAR stimulation which is mediated by FMRP and its phosphorylation status
Discussion
Protein synthesis is known to play an important role downstream of both NMDAR and mGluR stimulation [34]. But currently, there is no clear understanding regarding the distinct translational response downstream of these pathways. Though mGluR stimulation is correlated with global translation activation and NMDAR stimulation with translation inhibition, there are many contrasting reports when it comes to individual transcripts [9, 12, 35].
Since there is an overlap of many signalling components between NMDAR and mGluR [34], we hypothesized that similar to mGluR mediated translation regulation, specificity of NMDAR mediated translation is regulated at the messenger ribonuclearprotein (mRNP)-mRNA level [11]. RNA binding proteins such as FMRP, Human antigen R (HuR), Staufen2, and MOV10 play a crucial role in regulating translation of target mRNAs in a reversible manner. Staufen2 (Stau 2) is required for the transport and translation of microtubule associated protein 1b (Map1b) mRNA downstream of mGluR activation [36] whereas interaction of HuR with cationic amino acid transporter (CAT-1) mRNA was shown to relieve it from miRISC mediated inhibition in response to stress [20]. MOV10 is the mammalian homolog of Drosophila Armitage protein which is shown co-localize with AGO2 in HEK cells and is a component of miRISC [30]. An earlier study in hippocampal neurons has linked MOV10 to NMDAR mediated translation activation [19]. MOV10 is known to bind to mRNAs [31] and regulate the translation of CamK2a, lysophospholipase 1 (lypla1) mRNAs via their 3′ untranslated region (3’UTRs) [19]. In the above studies, the RNA binding proteins are shown to influence microRNA mediated inhibition and the translation of specific mRNA in response to particular signalling cues. In this regard, MOV10 was an ideal candidate for NMDAR mediated translation regulation as its role has been established previously [19, 27]. In order to characterise its regulatory role, we looked at MOV10 association with miRISC protein AGO2 and polysomes in synaptoneurosomes.
We found that MOV10 associates with both miRISC (AGO2) and polysomes and thus is involved in both translation inhibition and activation. On NMDAR signaling, the association of MOV10 with AGO2 decreases and it concomitantly increases in translating polysomes. Thus, MOV10 promotes the active translation of its bound mRNAs on NMDAR stimulation. In agreement with this, MOV10 knockdown showed a decrease in translation of its selected target mRNAs shown by polysome profiling. These results indicate that MOV10 not only acts an inhibitory RBP as shown previously [19, 27] but also has a role in translation activation of mRNAs downstream of NMDAR stimulation.
We also show that another RBP, FMRP, has a crucial role in MOV10 mediated translation regulation downstream of NMDAR activation. The role of FMRP as a translational regulator is well established in response to mGluR stimulation [24], but very little is known about the role of FMRP in the context of NMDAR stimulation. In this study, we show that there is an active translation of a specific subset of mRNAs downstream of NMDAR stimulation and FMRP along with MOV10 is critical for this regulation. Our data shows that FMRP is essential for the formation of MOV10-AGO2 inhibitory complex and for the shift of MOV10 (along with its target mRNAs) to translating polysomes on NMDAR stimulation. Studying the phosphorylation status of FMRP seems to provide the key molecular insight in understanding synaptic translation. In contrast to mGluR stimulation as shown previously [11], we found that on NMDAR stimulation there is an increase in the phosphorylation of FMRP at S499. Overexpression of phospho-mimetic of FMRP (FMRP-S499D) increases the FMRP-AGO2 complex but leads to the shift of MOV10 from AGO2 to translating polysomes. In contrast, dephospho-mimetic FMRP (FMRP-S499A) leads to an increase in MOV10-FMRP-AGO2 complex and decreased the MOV10 in translating polysomes. Based on these data, we propose a model (Fig. 7e) that indicates a possible mechanism for NMDAR mediated translation activation through FMRP and MOV10. These results indicate that NMDAR mediated FMRP phosphorylation has an effect on MOV10 mediated translation of mRNAs. Phosphorylation of FMRP is the switch downstream of NMDAR that leads to MOV10 moving into polysomes promoting translation of its target mRNAs.
We were also able to show that translation of Pten mRNA is upregulated on NMDAR activation and is regulated by FMRP and MOV10. PTEN is a known inhibitor of Protein Kinase B (Akt/PKB) pathway and pten mutations have been linked with autism spectrum disorders (ASD) [37]. We have used polysome profiling and immunoprecipitation along with MOV10 knockdown and FMRP knockout systems to show their role in NMDAR mediated translation but there is a scope to further test the roles of these RBPs in NMDAR signalling. Thus, in summary, this work draws attention to the importance of studying the role of FMRP beyond mGluR stimulation and particularly in NMDAR mediated signalling which will have a clear bearing on the molecular pathology of Fragile X Syndrome (FXS) and autism spectrum disorders (ASD).
Materials and methods
Cell line and primary neuronal culture: Primary neuronal cultures were prepared from cerebral cortices of embryonic day 18 (E18) rats (Sprague-Dawley) according to the established protocol [38]. 2-3 × 106 dissociated cells were plated on poly-L-lysine (0.2 mg/ml in borate buffer, pH 8.5) coated 10 cm culture dishes. Neurons were attached to the substrate in minimal essential medium with FBS (10%) for 3 h, then later grown in defined Neurobasal Medium (Invitrogen) with GlutaMAX™ supplement (Gibco™) and B-27 supplements (Invitrogen). Neurons were cultured for 14d at 37 °C in a 5% CO2 environment. For knockdown studies in neurons, NeuroMag (OZ Biosciences) was used as the transfection reagent. Silencer select siRNAs from Ambion against MOV10 transcript were transfected on days in vitro (DIV) 12 and the neurons were lysed on DIV 14.
Neuro2a cells were maintained in DMEM (Gibco®) with 10% FBS (Sigma) and GlutaMAX™ supplement (Gibco™). For knockdown studies, Silencer Select siRNA from Ambion were used. siRNA transfections were done using Lipofectamine® 2000 transfection reagent. For overexpression studies, phosphomutants of FMRP, FMRP-S499D and FMRP-S499A plasmid constructs [11] were transfected using Lipofectamine® 2000 and cells were lysed 24 h after the transfection.
Immunoprecipitation
Immunoprecipitation was done using anti-EiF2C2 (Abnova H00027161-MO1), anti-FMRP (Sigma-F4055), anti-MOV10 (Abcam-ab80613), Mouse IgG (Abcam-ab37355) and protein G Dyna beads (Invitrogen). Samples were processed for either western blotting or quantitative (real-time) PCR following immunoprecipitation as described previously [11]. The above antibodies including RPLP0 (Abcam-ab101279), α-tubulin (Sigma T9026), FLAG M2 (Sigma Millipore F3165) and β-III tubulin (Tuj1, Sigma T8578), phospho FMRP-S499 (Abcam-ab183319) were used for immunoblotting.
Sucrose step gradient
800 μl of 20% sucrose solution was overlaid on 800 μl of 30% sucrose solution. 400 μl cell lysate was added and the centrifugation was carried out at 40,200 rpm for 2 h in SW 50.1 rotor (Beckman Coulter) [29]. Fractions were then collected and analyzed by qPCR and western blotting. All sucrose solutions were made in gradient buffer (20 mM Tris-Cl pH 7.4, 100 mM KCl, 5 mM MgCl2 0.1 mg/ml cycloheximide, protease inhibitor and RNase inhibitor). The lysis buffer consisted of the gradient buffer with 1% Nonidet P-40 (NP40). For puromycin treatment, 1 mM puromycin was added to Neuro 2a cells or synaptoneurosomes and incubated for 2 h or 30 min respectively at 37 °C before lysis.
Synaptoneurosome preparation
Cortical synaptoneurosomes were prepared by differential filtration method [6] from Sprague Dawley (SD) WT or fmr1 KO [39] rats. For stimulation, synaptoneurosome solution was pre-warmed at 37 °C for 5 min and then stimulated with N- Methyl-D-Aspartate (NMDA, Sigma 20 μM) for 5 min at 37 °C with mock stimulation considered as the basal condition.
For PTEN and PSD-95 protein detection, post NMDAR stimulation, the synaptoneurosomes were pelleted, the buffer was replaced with fresh synaptoneurosomes buffer and the synaptoneurosomes were incubated at 37 °C for additional 20 min. Synaptoneurosomes were then lysed and denatured by SDS-denaturing buffer. For AP-5 treatment, the synaptoneurosomes were pre-incubated with AP-5 (100 μM) for 10 min at 37 °C, stimulated with NMDA for 5 min at 37 °C. Post NMDAR stimulation, the synaptoneurosomes were further incubated at 37 °C for 20 min in fresh synaptoneurosome buffer and then lysed and denatured by SDS-denaturing buffer. Anti-PTEN (CST 9552S) and anti-PSD-95 (Abcam 76,115) antibodies were used for western blotting.
Electron microscopy
Electron microscopy was done from synaptoneurosomes as described earlier [40]. Synaptoneurosomes were fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1M sodium cacodylate. Fixed samples after washes were then embedded in epoxy resin at 60C for 48h. Blocks were sectioned and imaged using TEM (FEI-Technai biotwin T12) at 100kV.
Polysome profiling
Polysome assay was done from synaptoneurosome samples after stimulation [6]. In brief, synaptoneurosome/cell lysate was separated on a 15–45% linear sucrose gradient in the presence of cycloheximide or puromycin. 1.0 mL fractions were collected and used for further analysis through western blot and qPCR.
Quantitative analysis for polysome profiling: qPCR data was analysed by the absolute quantification method using a standard curve as mentioned previously [41]. Absolute copy numbers for a particular mRNA were obtained from each of the 11 fractions. These copy numbers were then represented as percentage distribution across the 11 fractions.
For synaptoneurosomes
Fractions 7 to 11 were considered as translating pool based on sensitivity to puromycin (Fig. 5a).
Translating pool/non-translating pool = sum of the percentage of mRNA from fraction 7 to fraction 11 ÷ sum of the percentage of mRNA from fraction 1 to fraction 6.
For primary neurons (Additional file 1: Figure S3A)
Translating pool/non-translating pool = sum of the percentage of mRNA from fraction 8 to fraction11 ÷ sum of the percentage of mRNA from fraction 1 to fraction 7.
Quantitative PCR primers
For 18S ribosomal RNA (rRNA) quantification, cDNA samples were diluted one thousand times and then used for qPCR.
List of Primers
| Transcript | Forward sequence (5’→3′) | Reverse Sequence (5’→3′) |
|---|---|---|
| Psd-95 | ATGGCAGGTTGCAGATTGGA | GGTTGTGATGTCTGGGGGAG |
| 18S rRNA | GGTGACGGGGAATCAGGGTTCGAT | TTTTCGTCACTACCTCCCCGGGTC |
| Pten | AGGACCAGAGATAAAAAGGGAGT | CCTTTAGCTGGCAGACCACA |
| Ankyrin2 | ACCCTGCCAATTTATGCCAAG | GTTTCTGTCGACTCTGTCTCA |
| β-actin | GGCTCCTAGCACCATGAAGAT | AAACGCAGCTCAGTAACAGTC |
Statistical analysis
Group comparisons were made using one way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Statistical significance was calculated using paired/unpaired Student’s t-test for biochemical experiments as mentioned. Data are presented as mean ± Standard error of Mean (SEM). p values less than 0.05 were considered statistically significant.
Additional file
Figure S1. Effect of NMDAR stimulation on MOV10 interaction with AGO2 (related to Fig. 1). Figure S2. Sucrose step gradient to isolate mRNPs, light and heavy polysomes (related to Fig. 1). Figure S3. FMRP regulates translation of NMDAR target mRNAs through MOV10 (related to Fig. 2). Figure S4. Puromycin sensitive fractions in primary neurons and validation of MOV10 targets by RNA-IP and (related to Fig. 3). Figure S5. Validation of FMRP targets by RNA-IP (related to Fig. 4). Figure S6. NMDAR stimulation leads to no change in the RPL0 distribution (related to Fig. 5). Figure S7. Whole blots for PTEN and PSD-95 to show antibody specificity (related to Fig. 6). Figure S8. Phosphorylation of FMRP is the switch for NMDAR mediated translation (Related to Fig. 7). (DOCX 3830 kb)
Acknowledgements
We would like to thank the Shanta Wadhwani Foundation for supporting travel to present this work internationally. We are grateful to the sequencing facility, electron microscopy facility and animal facility of NCBS/inStem for the various technical help.
Funding
The work has been funded by Dept. of Biotechnology (BT/PR8723/AGR/36/776/2013), India, and NeuroStem grant (BT/IN/Denmark/07/RSM/2015–2016) to RSM and Centre for Brain Development and Repair (CBDR), Dept. of Biotechnology (BT/MB-CNDS/2013) to SC. Animal work in the NCBS/inStem Animal Care and Resource Center was partially supported by the National Mouse Research Resource (NaMoR) grant# BT/PR5981/MED/31/181/2012;2013-2016 & 102/IFD/SAN/5003/2017-2018 from the Indian Department of Biotechnology.
Availability of data and materials
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- AGO2
Argonaute 2
- Ank2
Ankyrin 2
- ANOVA
Analysis Of Variance
- AP-5
2-Amino-5-Phosphonopentanoic acid
- Arc
Activity regulated cytoskeleton associated protein
- ASD
Autism Spectrum Disorder
- Camk2a
Calcium/calmodulin dependent kinase II alpha
- CHX
cycloheximide
- DIV
days in vitro
- E18
Embryonic day 18
- Fmr1
fragile X mental retardation 1
- FMRP
Fragile X Mental Retardation Protein
- FXS
Fragile X Syndrome
- Grin2a
Glutamate receptor ionic epsilon 1
- HuR
Human antigen R
- IgG
Immunoglobulin G
- IP
Immunoprecipitation
- KO
Knockout
- lypla1
lysophospholipase 1
- mGluR
metabotropic Glutamate Receptor
- miRISC
miRNA Induced Silencing Complex
- MOV10
Moloney Leukemia Virus 10
- mRNP
messenger ribonucleprotein
- NMDAR
N-Methly-D-Aspartate Receptor
- NP40
Nonidet P-40
- P30
postnatal day 30
- PKB
Protein Kinase B
- PSD
Postsynaptic density
- Psd-95
postsynaptic density 95
- Pten
phosphatase tensin homolog
- PURO
Puromycin
- qPCR
quantitative PCR
- RBP
RNA Binding Protein
- RPLP0
Ribosomal Protein Lateral stalk subunit P0
- rRNA
ribosomal RNA
- SD
Sprague Dawley
- siRNA
small interfering RNA
- Stau2
Staufen 2
- SV
Synaptic Vesicles
- UTR
Untranslated region
- WT
Wild-Type
Authors’ contribution
PMK and RSM designed the experiments. PMK, SR., and NN carried out biochemical experiments. SC provided resources and suggestions. PMK and RSM wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval
All the work was done with due approval from the Institutional Animal Ethics committee (IAEC) and the Institutional Biosafety Committee (IBSC), InStem, Bangalore, India.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Preeti Madhav Kute, Email: preetimk@instem.res.in.
Sarayu Ramakrishna, Email: sarayur@instem.res.in.
Nagammal Neelagandan, Email: nagammaln10@gmail.com.
Sumantra Chattarji, Email: shona@ncbs.res.in.
Ravi. S. Muddashetty, Email: ravism@instem.res.in
References
- 1.Hanus C, Schuman EM. Proteostasis in complex dendrites. Nat Rev Neurosci. 2013;14:638–648. doi: 10.1038/nrn3546. [DOI] [PubMed] [Google Scholar]
- 2.Eskin A., Yeung S. J., Klass M. R. Requirement for protein synthesis in the regulation of a circadian rhythm by serotonin. Proceedings of the National Academy of Sciences. 1984;81(23):7637–7641. doi: 10.1073/pnas.81.23.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin P, Nastiuk KL, Daniel N, Girault JA, Czernik AJ, Glowinski J, Nairn AC, Prémont J. Glutamate-dependent phosphorylation of elongation factor-2 and inhibition of protein synthesis in neurons. J Neurosci. 1997;17:3445–3454. doi: 10.1523/JNEUROSCI.17-10-03445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith W. Bryan, Starck Shelley R., Roberts Richard W., Schuman Erin M. Dopaminergic Stimulation of Local Protein Synthesis Enhances Surface Expression of GluR1 and Synaptic Transmission in Hippocampal Neurons. Neuron. 2005;45(5):765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luchelli L., Thomas M. G., Boccaccio G. L. Synaptic control of mRNA translation by reversible assembly of XRN1 bodies. Journal of Cell Science. 2015;128(8):1542–1554. doi: 10.1242/jcs.163295. [DOI] [PubMed] [Google Scholar]
- 8.Weiler IJ, Greenough WT. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc Natl Acad Sci U S A. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheetz A. J., Nairn Angus C., Constantine-Paton Martha. NMDA receptor-mediated control of protein synthesis at developing synapses. Nature Neuroscience. 2000;3(3):211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 10.Weiler IJ, S a I, Klintsova a Y, Spencer CM, Brazelton a D, Miyashiro K, T a C, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buxbaum A. R., Wu B., Singer R. H. Single -Actin mRNA Detection in Neurons Reveals a Mechanism for Regulating Its Translatability. Science. 2014;343(6169):419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloomer Wendy A. C., VanDongen Hendrika M. A., VanDongen Antonius M. J. Arc/Arg3.1 Translation Is Controlled by ConvergentN-Methyl-D-aspartate and Gs-coupled Receptor Signaling Pathways. Journal of Biological Chemistry. 2007;283(1):582–592. doi: 10.1074/jbc.M702451200. [DOI] [PubMed] [Google Scholar]
- 15.Sutton Michael A., Taylor Anne M., Ito Hiroshi T., Pham Anh, Schuman Erin M. Postsynaptic Decoding of Neural Activity: eEF2 as a Biochemical Sensor Coupling Miniature Synaptic Transmission to Local Protein Synthesis. Neuron. 2007;55(4):648–661. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Todd PK, Malter JS, Mack KJ. Whisker stimulation-dependent translation of FMRP in the barrel cortex requires activation of type I metabotropic glutamate receptors. Mol Brain Res. 2003;110:267–278. doi: 10.1016/s0169-328x(02)00657-5. [DOI] [PubMed] [Google Scholar]
- 17.Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chmielewska Joanna J., Kuzniewska Bozena, Milek Jacek, Urbanska Katarzyna, Dziembowska Magdalena. Neuroligin 1, 2, and 3 Regulation at the Synapse: FMRP-Dependent Translation and Activity-Induced Proteolytic Cleavage. Molecular Neurobiology. 2018;56(4):2741–2759. doi: 10.1007/s12035-018-1243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Schratt GM, Tuebing F, E a N, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 22.Zalfa Francesca, Giorgi Marcello, Primerano Beatrice, Moro Annamaria, Di Penta Alessandra, Reis Surya, Oostra Ben, Bagni Claudia. The Fragile X Syndrome Protein FMRP Associates with BC1 RNA and Regulates the Translation of Specific mRNAs at Synapses. Cell. 2003;112(3):317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 23.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S. MOV10 and FMRP regulate AGO2 association with MicroRNA recognition elements. Cell Rep. 2014;9:1729–1742. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell. 2014;54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Khatter H, Myasnikov AG, Mastio L, Billas IML, Birck C, Stella S, Klaholz BP. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res. 2014;42:1–11. doi: 10.1093/nar/gkt1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 31.Sievers Cem, Schlumpf Tommy, Sawarkar Ritwick, Comoglio Federico, Paro Renato. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Research. 2012;40(20):e160–e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzniewska B, Chojnacka M, Milek J, Dziembowska M. Preparation of polysomal fractions from mouse brain synaptoneurosomes and analysis of polysomal-bound mRNAs. J Neurosci Methods. 2018;293:226–233. doi: 10.1016/j.jneumeth.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci. 2012;32:2582–7. doi: 10.1523/JNEUROSCI.5057-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santini E., Klann E. Reciprocal signaling between translational control pathways and synaptic proteins in autism spectrum disorders. Science Signaling. 2014;7(349):re10–re10. doi: 10.1126/scisignal.2005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27:13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebeau G, Miller LC, Tartas M, McAdam R, Laplante I, Badeaux F, DesGroseillers L, Sossin WS, Lacaille JC. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn Mem. 2011;18:314–326. doi: 10.1101/lm.2100611. [DOI] [PubMed] [Google Scholar]
- 37.Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal Arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaech Stefanie, Banker Gary. Culturing hippocampal neurons. Nature Protocols. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 39.Till SM, Asiminas A, Jackson AD, Katsanevaki D, Barnes SA, Osterweil EK, Bear MF, Chattarji S, Wood ER, Wyllie DJA, et al. Conserved hippocampal cellular pathophysiology but distinct behavioural deficits in a new rat model of FXS. Hum Mol Genet. 2015;24:5977–5984. doi: 10.1093/hmg/ddv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villasana LE, Klann E, Tejada-Simon MV. Rapid isolation of synaptoneurosomes and postsynaptic densities from adult mouse hippocampus. J Neurosci Methods. 2006;158:30–36. doi: 10.1016/j.jneumeth.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, D a R, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of NMDAR stimulation on MOV10 interaction with AGO2 (related to Fig. 1). Figure S2. Sucrose step gradient to isolate mRNPs, light and heavy polysomes (related to Fig. 1). Figure S3. FMRP regulates translation of NMDAR target mRNAs through MOV10 (related to Fig. 2). Figure S4. Puromycin sensitive fractions in primary neurons and validation of MOV10 targets by RNA-IP and (related to Fig. 3). Figure S5. Validation of FMRP targets by RNA-IP (related to Fig. 4). Figure S6. NMDAR stimulation leads to no change in the RPL0 distribution (related to Fig. 5). Figure S7. Whole blots for PTEN and PSD-95 to show antibody specificity (related to Fig. 6). Figure S8. Phosphorylation of FMRP is the switch for NMDAR mediated translation (Related to Fig. 7). (DOCX 3830 kb)
Data Availability Statement
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.